Abstract
Clubroot caused by the protist Plasmodiophora brassicae is a major disease affecting cultivated Brassicaceae. Using a combination of quantitative trait locus (QTL) fine mapping, CRISPR-Cas9 validation, and extensive analyses of DNA sequence and methylation patterns, we revealed that the two adjacent neighboring NLR (nucleotide-binding and leucine-rich repeat) genes AT5G47260 and AT5G47280 cooperate in controlling broad-spectrum quantitative partial resistance to the root pathogen P. brassicae in Arabidopsis and that they are epigenetically regulated. The variation in DNA methylation is not associated with any nucleotide variation or any transposable element presence/absence variants and is stably inherited. Variations in DNA methylation at the Pb-At5.2 QTL are widespread across Arabidopsis accessions and correlate negatively with variations in expression of the two genes. Our study demonstrates that natural, stable, and transgenerationally inherited epigenetic variations can play an important role in shaping resistance to plant pathogens by modulating the expression of immune receptors.
Key words: methylation, clubroot, Plasmodiophora brassicae, AT5G47260, AT5G47280, ADR1-L3, ddm1
This study reports a natural, stable, transgenerationally inherited epimutation in Arabidopsis that affects two adjacent genes encoding immune receptors, AT5G47260 and AT5G47280, which control broad-spectrum partial resistance to the root pathogen Plasmodiophora brassicae. This methylation variation is widespread across Arabidopsis ecotypes and negatively correlated with variations in expression of both genes.
Introduction
Intraspecific diversity in plant immune interactions is associated with a high level of sequence variation at hundreds of NLRs (nucleotide-binding and leucine-rich repeats), one of the largest and most rapidly evolving plant gene families (Meyers et al., 2003; Yue et al., 2012; Shao et al., 2016). On the basis of their N-terminal domains, NLRs have been classified into four subclasses: Toll/interleukin-1 receptor type (TIR-NLR or TNL), coil-coiled type (CC-NLR or CNL), RPW8-type CC-NLR (CCRPW8 NLR or RNL), and G10-type CC-NLR (CCG10 NLR) (Contreras et al., 2023). Many NLR proteins are involved in recognition of a small range of effector proteins secreted by specific strains of plant pathogens, potentially triggering the induction of strong plant defense responses that can rapidly stop pathogen invasion (Maekawa et al., 2011; Jones et al., 2016). The catalog of NLR genes expressed in a given plant genotype thus globally shapes the range of isolate-specific total resistances (incompatible interactions). However, this general rule has a few exceptions, including the existence of non-NLR-driven resistances (Thomas, 1998; Xiao et al., 2001; Larkan et al., 2013) and broad-spectrum NLR-driven resistances (Ernst et al., 2002; Qu et al., 2006).
Effectors may be recognized in different ways: (1) NLRs can monitor the effect of pathogen effectors on their cellular targets; (2) pathogen effectors can be recognized by their direct interaction with one canonical NLR domain; or, alternatively, (3) effectors can be recognized by one non-canonical NLR domain, called an integrated decoy (ID), which mimics a protein domain of the effector target (Kourelis and van der Hoorn, 2018). Effector-activated CNLs then assemble into pentameric oligomers called resistosomes, driving a rapid intracellular inward Ca2+ flux that triggers downstream cellular defense responses (Förderer et al., 2022). Activated TNLs drive similar Ca2+-mediated defense responses by an indirect pathway: assembled into tetrameric oligomers, their TIR domain mediates the biosynthesis of small signaling molecules, leading to downstream assembly of pentameric CCRPW8 NLR-based resistosomes that mediate Ca2+-mediated defenses (Essuman et al., 2022). CCRPW8 NLRs thus play a central role in the integration of hub-connected TNL-based non-self-recognition processes and have therefore been called “helper NLRs” (Wu et al., 2017).
In contrast to R-gene-driven resistance, quantitative resistance is polygenic, i.e., it involves allelic variation at several quantitative trait loci (QTLs), which collectively contribute to post-invasive partial resistance in compatible plant–pathogen interactions. The nature of the few resistance QTLs cloned to date supports the premise that quantitative resistance genes (QRGs) are functionally more diverse than R genes (Nelson et al., 2017; Pilet-Nayel et al., 2017; Delplace et al., 2022). Among these QRGs, however, there are still genes encoding NLRs (Hayashi et al., 2010; Fukuoka et al., 2014; Xu et al., 2014; Debieu et al., 2016) and other receptors (Diener and Ausubel, 2005; Hurni et al., 2015) or co-receptors (Huard-Chauveau et al., 2013). Thus, variation in NLR genes (or other non-self-recognition loci) also appears to contribute to variations in basal resistance levels during compatible interactions.
To trigger effective resistance, cellular levels of NLR proteins must reach minimum thresholds. However, high levels of NLRs can also lead to autoimmunity drawbacks, including spontaneous hypersensitive response and retarded plant growth (Li et al., 2015; Lai and Eulgem, 2018). NLR abundance is thus tightly controlled by multiple mechanisms at the transcriptional, post-transcriptional (i.e., alternative splicing), and post-translational levels (i.e., ubiquitin-dependent proteolytic regulation) (Zhang and Gassmann, 2007; Li et al., 2015; Lai and Eulgem, 2018). NLR regulation also involves a multitude of epigenetic-related cellular processes, including redundant networks of small RNAs (sRNAs) (Shivaprasad et al., 2012; Fei et al., 2013; Deng et al., 2018; Huang et al., 2019), histone modifications (Palma et al., 2010; Xia et al., 2013; Zou et al., 2014; Ramirez-Prado et al., 2018), histone-mark-dependent alternative splicing (Tsuchiya and Eulgem, 2013), and regulation of chromatin structure and DNA methylation (Li et al., 2010; Deleris et al., 2016). There is increasing evidence that epigenetic processes can play a role in the transitory imprinting of some plant biotic stress responses, at least for a few generations (Molinier et al., 2006; Slaughter et al., 2011; Luna et al., 2012; López Sánchez et al., 2016, 2021; Morán-Diez et al., 2021). It is, however, not yet clear to what extent stable transgenerational inheritance of epigenetically regulated gene expression contributes to the natural intraspecific diversity of plant–pathogen interactions.
The few available examples of transgenerational epigenetically controlled traits are found mainly in plant species, where the association between natural or induced differentially methylated regions (DMRs) and phenotypic traits was shown to be stably or (more often) metastably inherited across generations (Quadrana and Colot, 2016; Furci et al., 2019; Liégard et al., 2019). Such regions, designated epialleles, can have an effect on relevant agronomic traits: compatibility, accumulation of vitamin E, and fruit ripening in tomato; starch metabolism, disease resistance, and sex determination in melon; and fruit productivity in oil palm (Manning et al., 2006; Martin et al., 2009; Durand et al., 2012; Silveira et al., 2013; Quadrana et al., 2014; Ong-Abdullah et al., 2015; He et al., 2018; Bhat et al., 2020).
Plant DNA methylation can occur at cytosines in the three sequence contexts CG, CHG, and CHH (Henderson and Jacobsen, 2007) (where H can be A, C, or T), and its effect varies depending on the targeted genomic features (i.e., transposable elements [TEs], gene promoters, or gene bodies). DNA methylation patterns result from the dynamic combination of de novo methylation, maintenance methylation, and demethylation. De novo DNA methylation is catalyzed by the canonical and non-canonical RNA-directed DNA methylation (RdDM) pathways, which are both guided by small interfering RNAs (Cuerda-Gil and Slotkin, 2016; Zhang et al., 2018). Maintenance of DNA methylation relies mainly on RNA-independent pathways and requires the activity of DDM1, MET1, and VIM proteins at CG sites and of DDM1, KYP, CMT2/3, and the histone mark HK9me2 at CHG and CHH sites (Law and Jacobsen, 2010; Matzke and Mosher, 2014). Previous studies have noted that natural DMRs are over-represented on genes of the NLR disease-resistance gene family (Kawakatsu et al., 2016). However, it remains unclear whether natural epigenetic variation in NLR genes can influence the outcome of interactions between plants and pathogens.
Here, we report the identification of two adjacent NLR genes controlled by a naturally occurring stable epigenetic variation underlying a QTL involved in partial resistance to clubroot in Arabidopsis. Clubroot is a root gall disease caused by the telluric biotrophic pathogen Plasmodiophora brassicae (Rhizaria) and affects all Brassicaceae crops such as oilseed rape, kale, and turnip. The infection process involves a primary infection in root hairs that lasts only a few days. Secondary plasmodia then develop in root cortical cells, causing hyperplasia and hypertrophy that ultimately impair plant water and nutrient uptake. The reference accessions Columbia-0 (Col-0) and Burren-0 (Bur-0) are fully susceptible and partially resistant to P. brassicae isolate eH, respectively (Alix et al., 2007; Jubault et al., 2008b) (Supplemental Figure 1). Four main QTLs, which act additively, determine this difference. Here, we combined fine mapping of the QTL Pb-At5.2, which had the strongest effect on resistance, with CRISPR-Cas9 validation to identify two adjacent NLR genes, AT5G47260 and AT5G47280, both involved in the control of clubroot partial resistance. Expression levels of the two genes vary between the susceptible and resistant parents and are linked to the DNA methylation status of a small region that includes these two genes and a neighboring TE sequence. The methylation status of the two resistance genes is stable over generations and is not associated with any structural variation in the intervening transposon. Epiallelic variation at this locus is frequent among natural Arabidopsis accessions, and the low methylated state was correlated with the expression levels of the two NLR genes and with increased quantitative resistance to P. brassicae among 126 accessions. We showed that the RNA-independent pathway involving DDM1, MET1, VIM, and CMT2/3 maintains the hypermethylated epiallele in the clubroot-susceptible Col-0 accession. Overall, our findings demonstrate that quantitative resistance to a major root disease of Brassicaceae is associated, in Arabidopsis, with stable inheritance of a natural epigenetic variation involved in controlling the constitutive expression of an NLR gene pair.
Results
Fine mapping of the Pb-At5.2 locus responsible for clubroot resistance
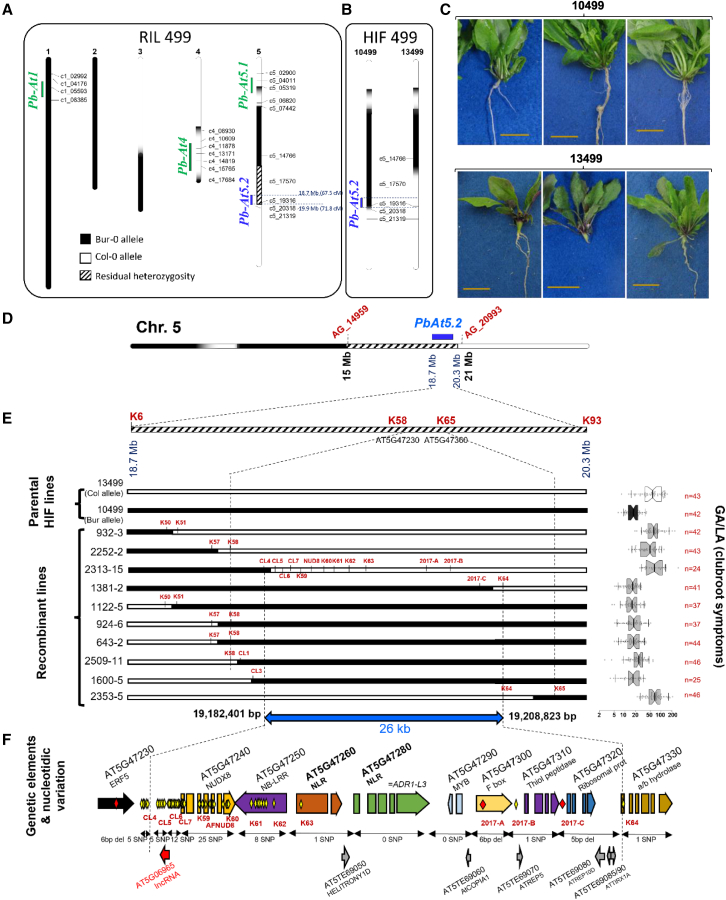
In previous work, we used a population of F7 recombinant inbred lines (RILs) between the partially resistant accession Bur-0 and the susceptible Col-0, to map a QTL (Pb-At5.2) on chromosome 5 between 67.5 and 71.8 cM that explained a significant proportion (R2 = 20%) of the resistance (Figure 1A) (Alix et al., 2007; Jubault et al., 2008b). In TAIR10, this interval (between AT5G46260 and AT5G47690) contains 158 annotated sequences, including protein-coding genes, TE genes, pre-tRNAs, and small nuclear RNAs. The effect and confidence interval of this QTL were also confirmed previously in the heterogeneous inbred family (HIF) lines 10499 and 13499 (Lemarié et al., 2015). Both lines were derived from the RIL 499, which harbors the homozygous Bur-0 (resistance) allele on the QTLs Pb-At1 and Pb-At5.1, the homozygous Col-0 (susceptibility) allele on the QTL Pb-At4, and residual heterozygosity in the Pb-At5.2 region. The lines 10499 and 13499 inherited the homozygous Bur-0 (resistance) allele and the Col-0 (susceptibility) allele, respectively, at the QTL Pb-At5.2 (Figure 1B and 1C; Supplemental Text 1).
Figure 1.
Fine mapping of Pb-At5.2.
(A) Genetic map and residual heterozygosity in the recombinant inbred line (RIL) 499 derived from Bur-0 and Col-0 (Alix et al., 2007), and genetic and physical positions of clubroot resistance QTLs (Jubault et al., 2008b). Black, Bur-0 allele; white, Col-0 allele; hatched, heterozygous (Col-0/Bur-0).
(B) Allele configuration at Pb-At5.2 in the two derived HIF lines 10499 and 13499.
(C) Photos showing that Pb-At5.2 conferred partial resistance to the eH isolate in the HIF 499 genetic background. HIF 10499 and 13499 harbored Bur-0 and Col-0 alleles, respectively, at Pb-At5.2. Observations were made 21 days post inoculation.
(D) First round of fine mapping: allelic structure in the F1 lines derived from reciprocal crosses between 10499 and 13499. A total of 554 F3 lines with recombination in the confidence interval were screened using 10 SNP markers between AG_14959 and AG_20993. High-density genotyping (94 SNPs from K1 to K94) in 88 recombinant F3 lines and clubroot phenotyping of their bulked segregating F4 progenies led to the identification of a new interval between markers K58 (AT5G47230) and K65 (AT5G47360).
(E) Second round of fine mapping: recombination positions in homozygous individuals obtained from selected recombinant lines. For each line, the GA/LA index (disease symptoms, log scale) is indicated on the right panel. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range (IQR) from the 25th and 75th percentiles; outliers are represented by dots; data points are plotted as open circles. The number of individual plants analyzed for each genotype is indicated (n). The notches are defined as +/− 1.58 × IQR/sqrt(n) and represent the 95% confidence interval for each median. GA/LA values statistically different from those of 13499 are indicated by asterisks (t-test, ∗∗∗p < 0.001). Genetic markers are indicated for each recombination position. Markers between CL5 and 2017-C were used in every line but are shown only for 2313-15.
(F) New 26-kb interval of Pb-At5.2 between markers CL4 (excluded) and K64 (excluded), containing eight annotated ORFs, six transposons, and one long non-coding RNA. Yellow and red diamonds indicate SNPs and nucleotide deletions, respectively.
The initial aim of the present work was to fine map Pb-At5.2, starting with reciprocal crosses between HIF lines 10499 and 13499. Clubroot symptoms in individuals of the F1 progeny were as severe as those in the susceptible parental line HIF 13499, suggesting that the Bur-0 resistance allele was recessive (Supplemental Figure 2). The boundaries of the Pb-At5.2 resistance locus were further refined through several rounds of genotyping and clubroot phenotyping (generations F3–F5 downstream of crosses 10499/13499) (details are given in Figure 1D–1F, Supplemental Figure 3, Supplemental Text 1, Supplemental Data 1, and Supplemental Text 3). This enabled us to narrow down the confidence interval to 26 kb between the markers CLG4 (19 182 401 bp, in the promoter region of AT5G47240) and K64 (19 208 823 bp, in AT5G47330), with the genetic markers being defined using the available de novo genome assembly of Bur-0 (Schneeberger et al., 2011). Comparison of the genetic sequences of Bur-0 and Col-0 in this 26-kb region revealed the absence of any structural variation and a low frequency of single-nucleotide polymorphisms (SNPs) (Figure 1F; details in Supplemental Text 3). This region contained eight annotated open reading frames (ORFs), including three NLR-encoding genes (AT5G47250, AT5G47260, and AT5G47280), six annotated TE sequences, and one long non-coding RNA gene (Figure 1F). The two F5 homozygous progeny lines 1381-2 and 2313-15, harboring the closest recombination events from both sides of the 26-kb interval (see Figure 1E), also showed partial resistance to a series of additional P. brassicae isolates (pathotypes 1, 4, and 7 following the classification of Some et al., 1996) and P1(+), which is representative of the new virulent strains that are emerging in Europe following breaking of clubroot resistance in oilseed rape varieties derived from the cultivar “Mendel” (Zamani-Noor et al., 2022). This result highlighted the broad spectrum of resistance conferred by the Bur-0 allele of Pb-At5.2 (Supplemental Figure 4).
RNA-seq analysis revealed a constitutive expression polymorphism of two NLR genes in the 26-kb QTL confidence interval
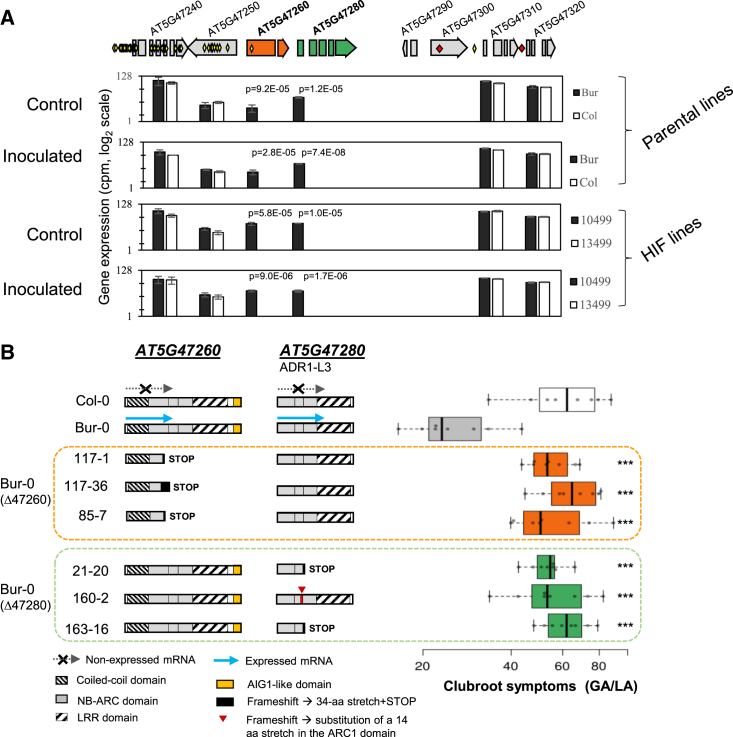
RNA-sequencing (RNA-seq) analysis was performed on Bur-0 and Col-0 accessions and on recombinant HIF lines 10499 and 13499. Pathogen-induced gene expression patterns differed markedly between genotypes harboring alleles Pb-At5.2BUR and Pb-At5.2COL (Supplemental Figure 5 and Supplemental Data 2). This regulation was consistent with our previously published studies, i.e., suggesting a role of camalexin biosynthesis and salicylic acid–mediated responses in Pb-At5.2BUR-mediated resistance and a role of jasmonic acid–driven induction of ARGAH2 in Pb-At5.2COL-mediated basal resistance (for details see Supplemental Text 2). We then focused on the eight ORFs in Pb-At5.2. Genes AT5G47290 and AT5G47300 showed no expression, and genes AT5G47240, AT5G47250, AT5G47310, and AT5G47320 showed similar expression levels in all four accessions (Figure 2A). In the 26-kb interval, only two genes, AT5G47260 and AT5G47280, both encoding proteins from the non-TIR-NLR gene family, were differentially expressed between resistant and susceptible accessions: these two genes were constitutively expressed in Bur-0 and 10499 roots (i.e., with the Bur-0 allele), but their expression was undetectable in Col-0 and 13499 (i.e., with the Col-0 allele) (Figure 2A). AT5G47280 encodes ADR1-L3, an NBS-LRR protein related to the small family of ADR1-type RNLs (although the encoded protein lacks the N-ter RPW8 domain). AT5G47260, encodes a CC-NBS-LRR-X protein with an ID-like C-ter extension domain (Figure 2B) homologous to members of the IAN family of AIG1 (= AvrRpt2-Induced Gene1, AT1G33960)-related proteins (Martin et al., 2023). These two adjacent genes are separated by a helitron, AT5TE69050 (Figures 1F and 3A). AT5G47280 contained no SNPs, and AT5G47260 contained only one non-synonymous SNP (Supplemental Figures 7 and 8, Supplemental Text 3). There was also no sequence variation in the helitron AT5TE69050 located between the two genes.
Figure 2.
Identification and CRISPR-Cas9 validation of two NLR-encoding genes controlling clubroot resistance at QTL Pb-At5.2.
(A) Sequence variations and expression levels of genes in the Pb-At5.2 region. Gene expression values are from RNA-seq analyses conducted under inoculated and control conditions at 14 days post inoculation (log2 normalized CPM) with parental lines Col-0/Bur-0 and HIF lines 13499/10499 (the last two were derived from RIL 499 and are homozygous Pb-At5.2Col/Col and Pb-At5.2Bur/Bur, respectively) (Supplemental Data 2). False discovery rate–adjusted p values are shown if less than 0.05. Yellow and red diamonds indicate SNP and INDEL variations, respectively.
(B) Effect of AT5G47260 or AT5G47280 knockout on GA/LA index (disease symptoms) in the Bur-0 background. Cas9-mediated mutations were obtained in the Bur-0 genetic background. For each targeted gene, three independent lines harboring independent homozygous mutations were used. Lines 117-1 and 21-20 no longer have the CRISPR-Cas9 cassette. For each line, the mean clubroot symptom score (GA/LA, log scale) was obtained by modeling raw data of eight biological replicates (with 10–12 individual plants per replicate). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the IQR from the 25th and 75th percentiles; data points are plotted as open circles. GA/LA values of edited lines that are statistically different from the Bur-0 GA/LA value are indicated by asterisks (Dunnett’s test) as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
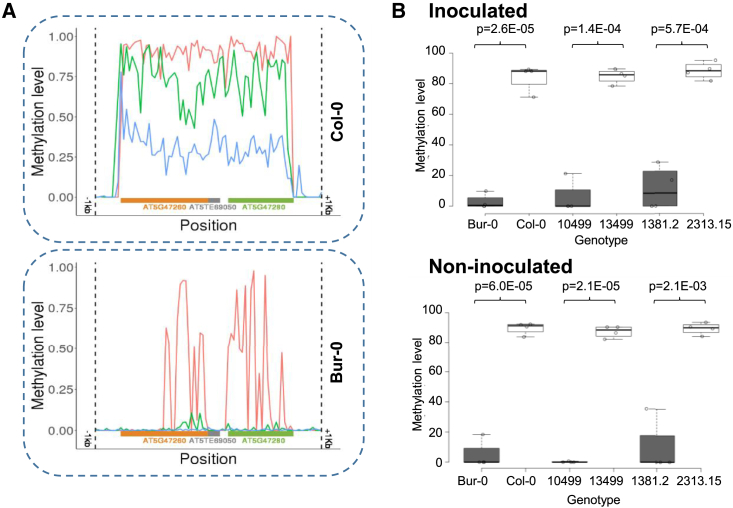
Figure 3.
Methylation of the region surrounding the two NLR-encoding genes that control clubroot resistance at QTL Pb-At5.2.
(A) Methylation profiles in the AT5G47260 and AT5G47280 region of Col-0 and Bur-0 accessions inferred from bisulfite data previously reported in Kawakatsu et al. (2016). Average methylation level was calculated within non-overlapping 100-bp windows starting 1 kb before the transcription start site (TSS) of AT5G47260 and stopping 1 kb after the trancription site end (TSE) site of AT5G47280. Red: methylation in the CG context. Green: methylation in the CHG context. Blue: methylation in the CHH context.
(B) Methylation profiles on AT5G47260 obtained by CHOP–qPCR in inoculated and non-inoculated roots of Bur-0/Col-0, 10499/13499, and the homozygous recombinant lines 2313-15 (Pb-At5.2Col/Col) and 1381-2 (Pb-At5.2Bur/Bur). The last two genotypes harbor the narrowest recombination events from either side of Pb-At5.2 (between markers CLG4 and K64, details in Figure 1). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the IQR from the 25th and 75th percentiles; outliers are represented by dots; data points are plotted as open circles. n = 4 bulks of six plants, and p values are shown (two-sided t-test).
CRIPSR-Cas9 validation of the role of NLR genes AT5G47260 and AT5G47280 in clubroot resistance
Given the contrasting expression levels of AT5G47260 and AT5G47280 in Bur-0 and Col-0, we next addressed their functional significance in clubroot resistance by generating knockout lines via CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 (CRISPR-associated protein 9) technology. We targeted the regions encoding the NB-ARC domain of both genes with two single guide RNAs in both resistant Bur-0 and HIF 10499 (which contains the Bur-0 allele) accessions. The CRISPR-Cas9-generated mutations in AT5G47260 and AT5G47280 gave rise to premature stop codons in most mutant lines and to substitution of a stretch of 14 amino acids in the ARC1 domain in line 160-2 (Figure 2B, Supplemental Figures 6‒8, and Supplemental Text 4). For the following experiments, we used three mutants from separate transformation events for each gene and each background (Bur-0 or HIF 10499). For each gene, one mutant without a T-DNA insertion was obtained in the Bur-0 background. The AT5G47260 and AT5G47280 CRISPR-knockout mutants were then evaluated for clubroot resistance in a complete randomized design. For both genes, clubroot symptoms were significantly higher in all lines edited in the Bur-0 genetic background than in the wild-type resistant Bur-0 accession and were as high as those of the susceptible accession Col-0 (Figure 2B), demonstrating the involvement of both AT5G47260 and AT5G47280 in clubroot resistance. Similar results were obtained for CRISPR-edited lines in the 10499 HIF genetic background (Supplemental Figure 9).
Expression polymorphism of both NLR genes is associated with stably inherited methylation variation
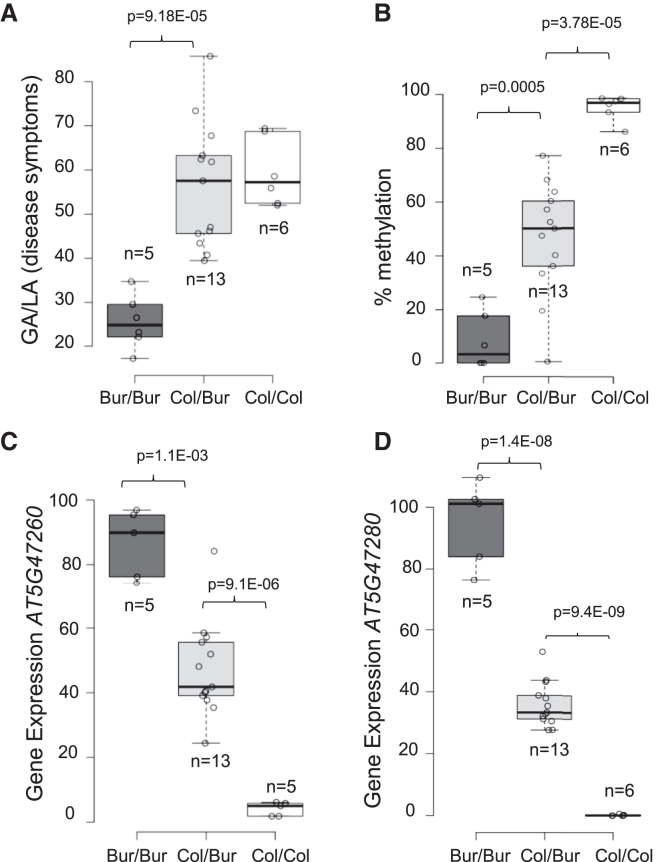
To understand why the two NLR genes AT5G47260 and AT5G47280 were differentially expressed in Bur-0 and Col-0, despite the absence of any sequence variation in their putative promoter regions, we analyzed the DNA methylation level of these regions in the two accessions using publicly available methylome data (Kawakatsu et al., 2016). The genomic interval between 19 188 411 and 19 196 559, which includes the two genes AT5G47260 and AT5G47280 and the intervening transposon AT5TE69050, was hypermethylated in Col-0 and hypomethylated in Bur-0 (Figure 3A). These contrasting methylation states were confirmed experimentally using DNA extracted from infected and non-infected roots of Col-0 and Bur-0 plants (Figure 3B) and CHOP–qPCR. The differences in DNA methylation were also found between the progeny HIF lines 10499 and 13499 and in the pair of HIF-derived homozygous near-isogenic lines 1381-2 and 2313-15 (Figure 3B), indicating that they are stably inherited independently of any DNA sequence polymorphism outside the locus. Moreover, the “Col-like” hypermethylation of AT5G47260 and AT5G47280 was systematically associated with low expression of the two NLR genes and a lower level of partial resistance to P. brassicae infection. To further investigate the inheritance of this epiallelic variation and its penetrance on gene expression and clubroot resistance, we investigated two groups of 100 individual plants, corresponding to the progenies derived from selfing the heterozygous 2509 and 1381 lines (harboring heterozygosity at the locus). Evaluation of plant disease for each individual plant in the two progenies revealed a 3:1 Mendelian segregation of the partial resistance phenotype. Clubroot symptoms in individuals with only one Bur-0 resistance allele were the same as in individuals with two susceptible Col-0 alleles (Figure 4A). In clubroot-inoculated roots of each individual plant from the 2509 progeny, the methylation state of the Pb-At5.2 region was monitored by CHOP–qPCR on AT5G47260. The SNP allele status at Pb-At5.2 was also investigated for each individual plant (for details of markers see Supplemental Data 1). Heterozygous Bur/Col individuals displayed intermediate parental methylation and expression values (Figure 4B–4D), providing a molecular explanation for the recessivity of the Bur-0 resistance allele. Together, these results suggested a link between partial resistance to P. brassicae and stably inherited epiallelic variation at Pb-At5.2, which controls the expression of two NLR genes.
Figure 4.
Intermediate methylation and transcript levels of candidate genes in heterozygous plants are associated with full clubroot susceptibility.
Eighty-three individual plants from the segregating progeny of recombinant line 2509 (heterozygous in Chr.5 region between genetic markers K58 and K93) were sampled at 21 days post inoculation. Leaves from each individual plant were used for genotyping (PCR marker CL_N8), which defined n pools of >3 plants of each zygosity profile: Bur/Bur (n = 5), Col/Bur (n = 13), and Col/Col (n = 6) (black, gray, and white boxes, respectively). Each plant pool was evaluated for (A) clubroot resistance (GA/LA), (B) percent methylation at the locus, and (C and D) candidate gene expression (AT5G47260 and AT5G47280). Gene expression was quantified by RT–qPCR, and data were normalized over mean-Cp from the pools Bur/Bur following Pfaffl’s method with two reference genes (Pfaffl, 2001). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the IQR from the 25th and 75th percentiles; outliers are represented by dots; data points are plotted as open circles.
The Bur-like hypomethylated epiallele is well represented among Arabidopsis accessions and contributes to reduction in clubroot symptoms
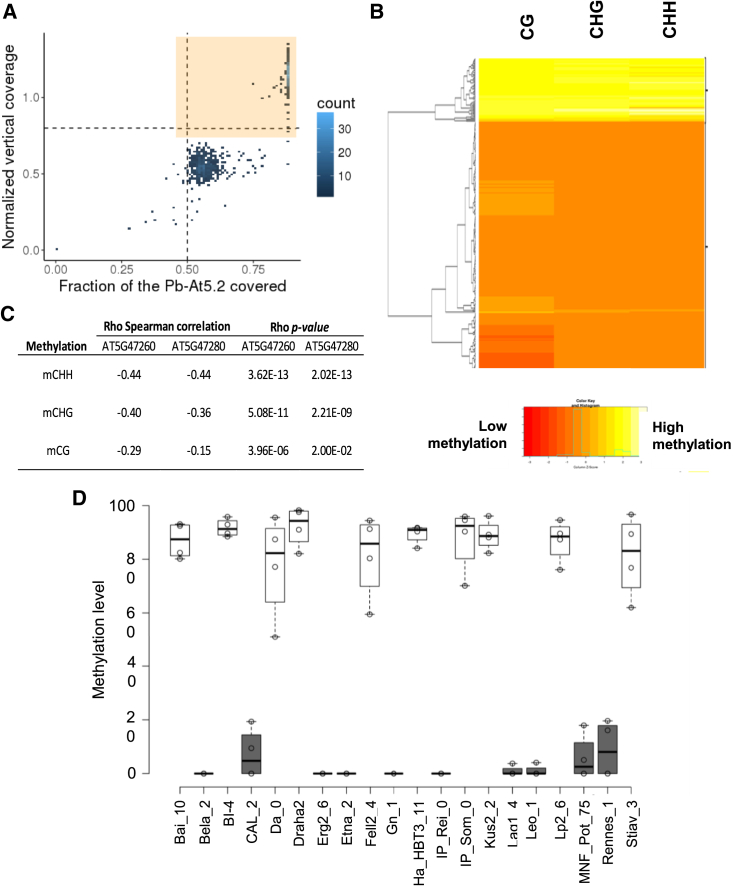
To assess the relative contribution of changes in DNA sequence and DNA methylation at Pb-At5.2 to clubroot resistance, we investigated natural allelic and epiallelic diversity across Arabidopsis accessions. We took advantage of recently published Illumina short genome sequence reads obtained from 1135 Arabidopsis accessions (1001 Genomes Consortium, 2016) to document the species-wide molecular diversity of the Pb-At5.2 genomic region. On the basis of quantitative horizontal and vertical coverage of short reads aligned to the Col-0 reference genome sequences, we identified two discrete groups of accessions. One group, containing 401 accessions, was characterized by high vertical and horizontal coverage (>0.75) and included the reference accession Col-0 as well as the partially clubroot-resistant Bur-0 (Figure 5A; for a detailed list of genotypes see Supplemental Data 3, sheet 1). The remaining 734 accessions contained diverse structural rearrangements, principally long deletions that translate into poor horizontal and vertical coverage compared with the reference Col-0 genome. Closer examination of coverage plots for the 401 Col-0/Bur-0-like accessions revealed a uniform haplotype structure that was present at high frequency at the species level (minor allele frequency [MAF] ∼0.37). Nonetheless, the haplotype frequency varied among geographic groups, ranging from 52.7% in Spain to 17.7% in Asia (Supplemental Figure 10). We then analyzed DNA methylation levels in 287 accessions from among the 401 accessions that contained the Col-0/Bur-0-like Pb-At5.2 and for which bisulfite data were publicly available (Kawakatsu et al., 2016). From these data, we could distinguish a group of 228 accessions, including Bur-0, that showed hypomethylation of Pb-At5.2 and another group of 59 accessions, including Col-0, that displayed hypermethylation (Figure 5B). The prevalence of accessions with the Col-like (epi)haplotype varied considerably with geographic origin, ranging from 1.8% in Spain to 16.8% in central Europe (Supplemental Data 3 and Supplemental Figure 10). Consistent with a causal role for DNA methylation in the transcriptional regulation of AT5G47260 and AT5G47280, reanalysis of publicly available RNA-seq data revealed a pronounced negative correlation between methylation level and AT5G47260 and AT5G47280 expression (Figure 5C). These results were further validated in infected roots from 20 natural accessions (Figure 5D).
Figure 5.
Natural epigenetic variation at Pb-At5.2 affects expression of AT5G47260/AT5G47280 in Arabidopsis accessions.
(A) Screening for 1001 Genomes Arabidopsis accessions that display a Col/Bur-like genomic structure at Pb-At5.2 (chr5: 19 185 600–19 200 600). x axis: horizontal coverage region covered by at least one read. y axis: vertical coverage in read percentage. The 401 accessions framed in the northeast intercardinal region delimited by dotted lines have a vertical read coverage >0.8 and a horizontal DNA-seq >0.5 (DNA-seq data from Alonso-Blanco et al. (2016)).
(B) Clustering of a series of accessions harboring Col/Bur-like genomic structure at Pb-At5.2 by their level of methylation on AT5G47260 and AT5G47280. Bisulfite data were obtained from the 1001 Genomes project (Supplemental Data 3, sheet 2). Average methylation level was calculated beginning 1 kb before the TSS of AT5G47260 and stopping 1 kb after the TSE site of AT5G47280 for each context.
(C) Spearman correlation between methylation and gene expression of AT5G47260 and AT5G47280 in a subset of 253 Arabidopsis accessions for which expression data were available (RNA-seq data from Kawakatsu et al., 2016). The correlation between gene expression and methylation level is given for all three DNA methylation contexts in the interval from 1 kb before the TSS of AT5G47260 to 1 kb after the TSE site of AT5G47280.
(D) Confirmation of methylation profiles at AT5G47260 in inoculated roots from 20 ecotypes. Methylation level was determined using CHOP–qPCR. Black and white bars indicate genotypes with Bur-like and Col-like methylation patterns, respectively. Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the IQR from the 25th and 75th percentiles; outliers are represented by dots; data points are plotted as open circles. n = 4 bulks of six plants.
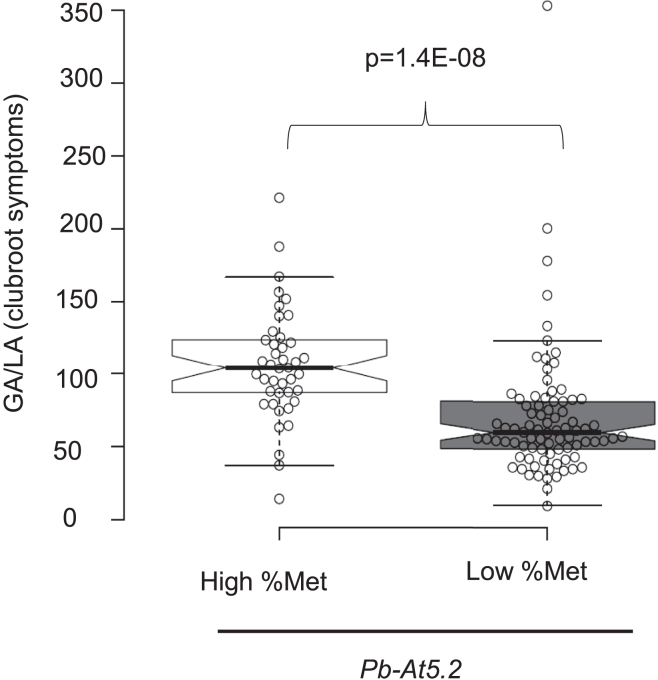
Both Col-like and Bur-like epialleles were significantly represented among the natural accessions, offering interesting genetic material with which to determine the actual contributions of DNA sequence and DNA methylation to the control of clubroot partial resistance. One hundred and twenty-six accessions were selected for their methylation levels at the AT5G47260–AT5G47280 region in data from Kawakatsu et al. (2016), including 42 accessions with the Col-like epiallele and 85 accessions with the Bur-like epiallele, and then assessed for their resistance to P. brassicae isolate eH. Whereas no DNA sequence polymorphisms in Pb-At5.2 showed an association with clubroot resistance (Supplemental Data 4), the low DNA methylation state of the AT5G47260/AT5G47280 locus was significantly associated with enhanced resistance levels (Figure 6). Together, these results corroborate and extend the conclusions obtained by fine mapping of Pb-At5.2 and provide strong evidence that natural epiallelic variations contribute to the quantitative differences in clubroot resistance observed among Arabidopsis accessions.
Figure 6.
Variation in clubroot symptoms among Arabidopsis accessions is linked to epivariation at Pb-At5.2.
Effect of Pb-At5.2 epiallele variation on clubroot susceptibility, evaluated in 126 Arabidopsis accessions with a similar Bur/Col-like genomic structure at the locus. Each open circle represents one accession. In total, 42 accessions had a Col-like epiallele (high percentage of methylation, High % Met), and 84 had a Bur-like epiallele (low percentage of methylation, Low % Met). For each accession, the mean GA/LA was obtained by modeling raw data of two biological replicates with two blocks (six individual plants in each block). Center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the IQR from the 25th and 75th percentiles; outliers are represented by dots; data points are plotted as open circles. The notches are defined as +/− 1.58 × IQR/sqrt(n) and represent the 95% confidence interval for each median. The p value (Wilcoxon test) is indicated.
Pb-At5.2 epivariation is independent of cis-genetic variations
At the Pb-At5.2 locus, the transposon AT5TE69050 was present in both parental genotypes, with no sequence variation that might have been the primary cause of variation in DNA methylation on the two adjacent genes. Analysis of 34 out of the 287 accessions with the Col-0/Bur-0-like haplotype did not reveal the presence/absence of TE insertion variants within the 26-kb Pb-At5.2 region (Quadrana et al., 2016; Stuart et al., 2016), with the exception of a helitron insertion in the accession NFA-10. Moreover, 18 and 16 of these accessions displayed hypermethylated and hypomethylated epialleles, respectively, indicating that variation in DNA methylation is not associated with TE presence/absence variants. In addition, the cis-nucleotide polymorphism located within the coding sequence of AT5G47260 and detected in Bur-0 was absent in at least five other accessions sharing the hypomethylated epiallele (Supplemental Figure 11), indicating that the hypomethylated state of Pb-At5.2 is not correlated with any specific DNA sequence polymorphism at the locus.
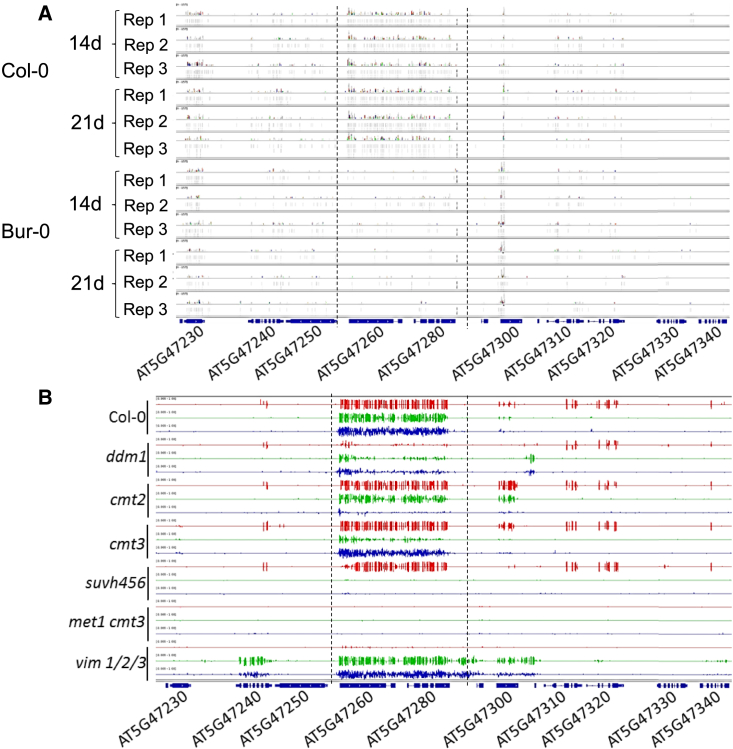
The hypermethylated epigenetic variant is maintained by the RNA-independent pathway
Analysis of sRNAs identified in Col-0 (Stroud et al., 2013) revealed that the AT5G47260/AT5G47280 region is targeted mostly by 24-nt sRNAs, which prompted us to generate sRNA profiles from non-inoculated roots of Col-0 and Bur-0 and from roots inoculated with P. brassicae isolate eH 14 and 21 days after inoculation. Consistent with the previously observed pattern of DNA methylation, we found high levels of sRNAs only in Col-0 (Figure 7A). To further explore the mechanisms involved in the maintenance of methylation at this locus, we made use of publicly available methylomes of Col-0 mutant plants defective in one or several DNA methylation pathways (Stroud et al., 2013). Despite the high levels of sRNAs detected over the AT5G47260/AT5G47280 region, mutations affecting the RdDM pathway did not influence its DNA methylation level (Supplemental Figure 12). Conversely, DNA methylation was largely lost in mutants defective in sRNA-independent maintenance of DNA methylation, i.e., ddm1, cmt2/3, met1, and suvh456 (Figure 7B). These results raise questions about the role of sRNAs targeted to the Col-like hypermethylated region whereas methylation maintenance depends solely on the RNA-independent pathway.
Figure 7.
Epigenetic variation at Pb-At5.2 correlates with the abundance of locus-targeted sRNA but is maintained by the RNA-independent methylation pathway.
(A) Mapping of sRNA-seq reads. Reads were obtained from roots of Col-0 and Bur-0 accessions at two time points 14 and 21 days after sowing. For each condition, three bulks (numbered from Rep 1 to Rep 3) of six plants were used.
(B) Methylation state at the Pb-At5.2 locus in knockout lines (Col-0 genomic background) defective for the RdDM or non-RdDM pathway (Stroud et al., 2013).
Red: methylation in CG context. Green: methylation in CHG context. Blue: methylation in CHH context.
Discussion
To date, only a very small number of resistance QTLs have been characterized at the molecular level (Delplace et al., 2022). Detection and fine mapping of resistance QTLs is typically challenging not only because of the difficulties associated with measuring small variations in partial resistance in large numbers of individual progeny but also because resistance QTLs can be sensitive to environmental changes (Aoun et al., 2017; Laperche et al., 2017; Aigu et al., 2018). However, technical issues may have been only part of the problem. Recent developments in the field of epigenetics suggest that some inherited resistance factors may not be detected by classic genetic approaches that are based solely on DNA sequence variation. In the present work, a genome-wide association study (GWAS) failed to identify any nucleotide variation in the 26-kb interval of Pb-At5.2 associated with clubroot response. By contrast, clubroot resistance was clearly related to epigenetic variation at two NLR genes in this interval. This work thus reveals for the first time an epigenetically driven expression polymorphism that makes a substantial contribution to the natural diversity of plant immune response.
Many examples of epialleles are metastable, i.e., they can be reversed by stochastic or unidentified factors (Weigel and Colot, 2012). Stability over multiple generations is a primary concern from both evolutionary and breeding perspectives. The epiallele described here seems to be extremely stable, as it was robustly detected in all our previous QTL investigations in Arabidopsis. This included two independent segregating progenies derived from Bur-0 and Col-0 (Jubault et al., 2008a) and additional studies with the HIF lines 10499/13499 (Lemarié et al., 2015; Gravot et al., 2016). The high level of methylation and absence of AT5G47260 and AT5G47280 expression observed in Col-0 were also found in a set of publicly available data obtained in different laboratories from diverse plant tissues and conditions (Winter et al., 2007; Stroud et al., 2013; Klepikova et al., 2016; 1001 Genomes Consortium, 2016; Kawakatsu et al., 2016). It was also confirmed by our own data generated from inoculated roots, non-inoculated roots, and leaf samples. Finally, this methylation pattern was also robustly found in multiple replicates of individual plants. Thus, Pb-At5.2 can be classified as a stable epiallele without reservation.
There has only been one report of a plant disease resistance caused by an inherited methylation variant that affects expression of a resistance-related gene (Nishimura et al., 2017). In that study, a stable expression polymorphism (between Ler-0 and Ag-0 accessions) on the TIR-only encoding gene RBA1 (AT1G47370) affected effector-triggered immunity responses to the Pseudomonas syringae effector hopBA1. This expression polymorphism was linked to the nearby presence/absence of a TE sequence in the promoter region of the gene and to MET1-dependent DNA methylation variation. However, because DNA methylation was reversed when the TE sequence was segregated away (Supplemental Figure 2 in Nishimura et al., 2017), this DNA methylation variation is not “epigenetic,” as it is an obligate consequence of sequence variation (i.e., presence/absence of the TE sequence). In the present study, we showed that DNA methylation variation in the region between AT5G47260 and AT5G47280, including the TE sequence AT5TE69050, is not linked to any nucleotide/structural variation at the locus or elsewhere in the genome. Thus, Pb-At5.2COL and Pb-At5.2BUR can be considered “pure epialleles” as defined by Richards (2006).
From available genomic and epigenomic data from the 1001 Genomes Project, it can be extrapolated that the Bur-like clubroot resistance epiallele is present in about half of the accessions from the “Relict,” “Spain,” and “Italy/Balkans/Caucasus” groups and 39% of the accessions from the “North Sweden” group (Supplemental Data 3). By contrast, the Bur-like epiallele is likely (taking into account missing methylation data) present at about 10% of the “Germany” group. On the other hand, the clubroot susceptibility of the Col-like epiallele was absent from the accessions in the “Relict” and “North Sweden” groups but reached at least 16.8% in the “Central Europe” group. This geographic structure suggests that both epialleles can confer fitness gains, depending on the environmental context. However, it does not appear to be obviously related to the incidence of clubroot in Brassica culture (usually low in the warm southern European regions). Keeping in mind that NLRs can detect unrelated effectors from distinct microbial species (Narusaka et al., 2009) and echoing previous work (Karasov et al., 2014), we hypothesize that maintenance of this epivariation in natural populations may reflect additional roles played by Pb-At5.2 against other plant pathogens (besides the control of clubroot infection).
AT5G47280 has been annotated as ADR1-Like 3 on the basis of its phylogenetic relationship with the small family of helper CCRPW8-NLRs, including ADR1, ADR1-L1, and ADR1-L2 (Saile et al., 2020, 2021). However, the absence of RPW8 in ADR1-L3 and the absence of ADR1-L3 expression in Col-0 have raised questions about its actual role in plant immunity. AT5G47260 and AT5G47280 belong to a small heterogeneous cluster of three non-TIR-NLRs, which also includes AT5G47250. This small cluster is located on chromosome 5, not far from the largest NLR hotspot in the Arabidopsis genome (Meyers et al., 2003). None of these three genes has previously been shown to participate in plant–pathogen interactions. Here, we showed that expression of both AT5G47260 and AT5G47280 is necessary for partial resistance to P. brassicae. There are a few examples of tandem NLR genes encoding pairs of proteins that function as heterodimers (Cesari et al., 2014; Williams et al., 2014; Saucet et al., 2015). Similarly, the proteins encoded by these two jointly epigenetically regulated genes may function together in the control of cell defense responses during clubroot infection. Although the underlying molecular mechanisms are unknown, the canonical example of the TIR-NLR heterodimer RRS1/RPS4 corresponds to a recessive resistance locus, similar to Pb-At5.2.
In Arabidopsis, DNA methylation is widely distributed in both the promoters and the bodies of most NB-LRR-encoding genes (Kong et al., 2018, 2020), predominantly in the CG sequence context. This suggests that plant genomes contain multiple functional resistance genes whose possible roles in biotic interactions are locked by epigenetic processes. This hypothesis is also supported by our previous study, in which we demonstrated that ddm1-triggered hypomethylation at different genomic loci resulted in the unlocking of genetic factors that ultimately exert significant control over clubroot symptom development (Liégard et al., 2019). It would now be interesting to carry out a careful genome-wide analysis of methylation profiles of all NLR genes among Arabidopsis accessions, which would take into account the structural diversity of all these individual genes (supported by additional targeted resequencing of NLR loci). The intraspecific diversity in methylation patterns of NLR and RLK/RLP genes in plants, their heritability, and their consequences for plant biotic interactions also deserve further attention in future studies.
Methods
Plant materials and growth conditions
The HIF lines 10499 and 13499 and their parental accessions Col-0 (186AV) and Bur-0 (172AV) were provided by Versailles Arabidopsis Stock Center (http://publiclines.versailles.inrae.fr/). Arabidopsis thaliana accessions were all purchased from the Nottingham Stock Center. Individuals in the panel of 126 accessions were selected according to their methylation levels at the region of interest (Kawakatsu et al., 2016). All accessions and in-house-generated recombinant lines used in this study are listed in Supplemental Data 1 and Supplemental Data 3. Seed germination was synchronized by placing seeds on wet blotting paper in Petri dishes for 2 days at 4°C. Seeds were sown individually in pots (4-cm diameter) containing a sterilized mixture of two-thirds compost and one-third vermiculite. Growth chamber conditions of 16-h light (110 μmol m−2 s−1) at 20°C and 8-h darkness at 18°C were used to grow plants. The 126 Arabidopsis accessions and HIF lines were challenged with P. brassicae in two biological replicates in a completely randomized block design (with two blocks per replicate, each block consisting of six plants per genotype). The Arabidopsis accessions Col-0 and Bur-0 and the HIFs 10499, 13499, 1381-2, and 2313-15 used in RNA-seq and sRNA-seq approaches were assessed when infected with P. brassicae or in the uninfected condition in three randomized blocks. The CRISPR-Cas9 edited lines and corresponding wild-type lines were challenged with P. brassicae in eight replicates in a completely randomized block design (each replicate consisting of 10–12 plants per genotype). Almost all clubroot tests were performed with the eH isolate of P. brassicae described by Fähling et al. (2003), which belongs to the most virulent pathotype, P1. The resistance spectrum of Pb-At5.2 was also assessed using the additional isolates Pb137-522, Ms6, K92-16, and P1(+). For every isolate used in this study, the pathotype was validated in every experiment using the differential host set according to Some et al. (1996), also including two genotypes of Brassica oleracea ssp. acephala C10 and CB151. One milliliter of resting spore suspension (107 spores ml−1) prepared according to Manzanares-Dauleux et al. (2000) was used for pathogen inoculation 10 days after germination (stage 1.04; Boyes et al., 2001). This inoculum was applied to the crown of each seedling.
Phenotyping
HIF lines and Arabidopsis accessions were phenotyped 3 weeks after inoculation (21 days post inoculation) for their susceptibility to P. brassicae. Plants were thoroughly rinsed with water and photographed. Infected roots were removed and frozen in liquid nitrogen. Clubroot symptoms were evaluated by image analysis using the gall area/leaf area (GA/LA) index calculated according to Gravot et al. (2011).
Fine mapping of the locus responsible for clubroot resistance
Fine mapping of Pb-At5.2 was performed starting from crosses between HIF lines 10499 and 13499, followed by successive rounds of genotyping and clubroot phenotyping in subsequent plant generations (full details are given in Supplemental Text 1).
RNA isolation, mRNA sequencing, and differential gene expression analysis
Total RNA was extracted from frozen and lyophilized roots (collected 14 days after inoculation) using the mirVana miRNA Isolation Kit (Invitrogen) according to the manufacturer’s instructions. After extraction, the RNA samples were quantified using NanoDrop ND-1000 technology, and their quality was assessed using the RNA 6000 assay kit (Agilent). Samples with an RNA integrity number (RIN) greater or equal to 7 were used for sequencing. cDNA-sequencing (cDNA-seq) library construction and sequencing were performed by the NGS platform at the Marie Curie Institute of Paris. Each library was sequenced in paired-end mode using the Illumina HiSeq 2500 platform. Reads were aligned to the TAIR10 genome annotation and assembly of Col-0 A. thaliana concatenated with the P. brassicae genome using STAR software version 2.5.3.a (Dobin et al., 2013). Alignment conditions were selected according to the Arabidopsis genome. A maximum of five multiple read alignments was accepted, and no more than three mismatches were allowed for each alignment. The resulting BAM files were used to determine read counts using the counts function in featureCounts software (version 1.4.6) and the TAIR10 gff file of Arabidopsis concatenated with the gff file of P. brassicae. Differentially expressed genes were determined using the edgeR package (Robinson et al., 2010) in R software version 3.3.0 (R Core Team, 2013). Raw counts obtained as described previously were used as input data for edgeR. After CPM (counts per million) values were determined, only genes with at least one CPM in three samples were retained. Expression signals were normalized using the TMM method (trimmed mean of M values) with the CalcNormFactors function in edgeR. Finally, differentially expressed genes were identified using the decideTests function of edgeR with one minimum fold change between −1.5 and 1.5.
CRISPR-Cas9 constructs and plant transformation
Two guide sequences were designed for each targeted gene (i.e., AT5G47260 and AT5G47280) using CRISPOR software (Concordet and Haeussler, 2018), taking care to select sequences with very high specificity scores (Supplemental Data 5). Guide sequences were ordered as oligonucleotides (IDT) and cloned downstream of the Arabidopsis U6-26 promoter and upstream of an enhanced single guide RNA scaffold as reported previously (Chauvin et al., 2021) to produce individual guide modules. Assembly of guide modules for single genes was performed using PCR amplification with specific primers followed by classical restriction/ligation cloning (Supplemental Data 5, sheet 1). Guide assemblies were then cloned by a Gateway LR reaction (Thermo Fisher Scientific) into the pDe-Cas9 backbone (Fauser et al., 2014) harboring an nptII resistance cassette (Chauvin et al., 2021), resulting in two binary plasmids for CRISPR-mediated targeting of AT5G47260 (pDe-Cas9_T79-80) and AT5G47280 (pDe-Cas9_T81-82) (Supplemental Data 5, sheet 1). All constructs were checked by Sanger sequencing. The resulting plasmids were then transferred into Agrobacterium tumefaciens C58/GV3101pMP90 by heat shock and used to transform Arabidopsis plants via the floral dip method (Clough and Bent, 1998). Transgenic plants were screened on solid plates with half-strength Murashige–Skoog medium containing 50 mg l−1 kanamycin (Yeasen, cat. no. 60206ES10). A first screening was performed on the T1 generation using PCR and sequencing to identify plants with mutations in AT5G47260 or AT5G47280 (primer pairs are listed in Supplemental Data 5, sheet 2). A second round of screening enabled the identification of T2 plants homozygous for the mutations and free from CRISPR-Cas9 cassette T-DNA. T3 seedlings were used to evaluate P. brassicae resistance.
GWAS analyses
A conventional GWAS on GA/LA data from 126 accessions was performed with easyGWAS (Grimm et al., 2017) (https://easygwas.ethz.ch/). Association analysis was performed with EMMAX (Kang et al., 2010) using 1 806 554 SNPs with an MAF > 0.05, after correction for population structure by including the first three principal components in the additive model.
Small RNA isolation, sequencing, clustering, and differential presence determination
sRNA was extracted from frozen and lyophilized roots (collected 14 days after inoculation) using the mirVana miRNA Isolation Kit (Invitrogen) according to the manufacturer’s instructions. After extraction, the sRNAs were quantified using a NanoDrop ND-1000 and quality controlled using the Small RNA assay kit (Agilent). Samples with an RIN greater than or equal to 7 were used for sequencing. Construction and sequencing of cDNA-seq libraries were performed on the NGS platform of the Marie Curie Institute of Paris. For each sample, single-ended (50 bp) sequencing was performed using the Illumina HiSeq 2500 platform. Reads were aligned to the TAIR10 genome annotation and assembly of Col-0 A. thaliana concatenated with the P. brassicae genome using STAR version 2.5.3.a (Dobin et al., 2013), then counted and clustered using ShortStack software (Axtell, 2013). The presence of differentially expressed sRNAs was determined using edgeR (Robinson et al., 2010) in R version 3.3.0 (R Core Team, 2013). Raw counts obtained as described previously were used as input data to edgeR. After CPMs were determined, only genes with at least one CPM in three samples were retained. Expression signals were normalized using the TMM method with the CalcNormFactors function in edgeR. Finally, differentially expressed sRNAs were identified using the decideTests function in edgeR with one minimum fold change between −1.5 and 1.5.
RNA isolation and RT–qPCR analysis
Total RNA was extracted from lyophilized roots of accessions and HIF lines 21 days after infection using the TRIzol extraction protocol. Samples with residual traces of DNA were treated with DNAse (Promega ref. M6 10A). Before reverse transcription of RNA to cDNA with SuperScript II (Invitrogen), RNA quality was verified by agarose gel electrophoresis. RT–qPCR was performed in a LightCycler 480 thermocycler (Roche) with cDNA obtained as described above. Gene expression was normalized using as references two Arabidopsis genes defined as stable during infection using RNA-seq data (AT1G54610, AT5G38470) following Pfaffl’s method (Pfaffl, 2001). Primer sets were designed for each gene and are listed in Supplemental Table 1.
CHOP–PCR and qPCR assays
Gene methylation profiles were investigated using the enzyme McrBC (M0272L, BioLabs) (Zhang et al., 2014). Forty nanograms of DNA was incubated with 0.5 μl of BSA (20 mg/ml), 0.5 μl of guanosine triphosphate (20 mM), 5 μl of NE Buffer (10×), and 0.2 μl of McrBC (10 000 U/ml). For CHOP–PCR and qPCR, 2 ng of digested and undigested DNA was used. For CHOP–PCR, the temperature conditions were adjusted according to the primer design, and 35 amplification cycles were used. To determine the methylation state of the targeted region, each sample was digested or not (control) with McrBC before amplification. For CHOP–qPCR, the temperature conditions were adjusted according to the primer design, and 30 amplification cycles were used. Methylation levels of the target region were calculated as the percentage of molecules lost through McrBC digestion as described in Silveira et al. (2013) with the formula: (1 − (2 − (Ct digested sample − Ct undigested sample))) × 100. The percentages of DNA methylation for AT5G13440 and AT5G47400 were calculated in all CHOP qPCRs as controls. AT5G13440 and AT5G47400 were selected from 1001 Genomes data as hypomethylated and hypermethylated, respectively, in most Arabidopsis accessions, and their expression did not vary during clubroot infection. The primer sets designed for each gene are listed in Supplemental Table 1.
Published data
The DNA-seq data, RNA-seq data, variant sequences, and bisulfite data for the natural accessions studied here were obtained from previous studies (1001 Genomes Consortium, 2016; Kawakatsu et al., 2016) archived at the NCBI with SRA number SRP056687 and the NCBI Gene Expression Omnibus references GEO: GSE43857 and GSE80744. The bisulfite data and sRNA data for Arabidopsis mutants studied here were obtained from a previous report (Stroud et al., 2013) and are archived at the NCBI under accession GEO: GSE39901.
Statistical analysis
Data were statistically analyzed using the R program (R Core Team, 2013).
Data and code availability
The data supporting the findings of this study are available within the paper and its supplemental information files. All unique materials used are readily available from the authors.
Funding
We acknowledge the Versailles Arabidopsis Stock Center for providing HIF lines, the biological resource center BrACySol for furnishing Brassica accessions, the Gentyane Platform for their contribution in genotyping the fine-mapping population, and the Institute Marie Curie for sequencing mRNA and sRNA. Cyril Falentin is acknowledged for his help in the design of KASPAR markers. IGEPP colleagues are acknowledged for their technical support for clubroot phenotyping and sampling.
Author contributions
B.L., A.G., M.J.M.-D., and M.J. designed and conducted the experiments. C.L. and A.G. carried out the fine mapping. C.L., J.L., J.B., T.B., B.L., A.G., and M.J. performed the phenotyping and sampling. B.L., A.G., J.B., J.L., and M.J. carried out epigenetic and gene expression studies. F.V. designed and performed the CRISPR-Cas9 constructs, and C.L. and T.B. carried out the transformation, selection, and handling of edited plants. B.L., Y.A., L.Q., and V.C. conducted bioinformatics analyses. A.G., B.L., L.Q., F.V., V.C., M.J.M.-D., and M.J. participated in drafting and revision of the manuscript.
Acknowledgments
No conflict of interest is declared.
Published: January 23, 2024
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Supplemental information
References
- Alonso-Blanco C., Andrade J., Becker C., Bemm F., Bergelson J., Borgwardt K.M., Cao J., Chae E., Dezwaan T.M., Ding W., et al. 1001 Genomes Consortium 1,135 Genomes Reveal the Global Pattern of Polymorphism in Arabidopsis thaliana. Cell. 2016;166:481–491. doi: 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigu Y., Laperche A., Mendes J., Lariagon C., Guichard S., Gravot A., Manzanares-Dauleux M.J. Nitrogen supply exerts a major/minor switch between two QTLs controlling Plasmodiophora brassicae spore content in rapeseed. Plant Pathol. 2018;67:1574–1581. [Google Scholar]
- Alix K., Lariagon C., Delourme R., Manzanares-Dauleux M.J. Exploiting natural genetic diversity and mutant resources of Arabidopsis thaliana to study the A. thaliana–Plasmodiophora brassicae interaction. Plant Breed. 2007;126:218–221. [Google Scholar]
- Aoun N., Tauleigne L., Lonjon F., Deslandes L., Vailleau F., Roux F., Berthomé R. Quantitative Disease Resistance under Elevated Temperature: Genetic Basis of New Resistance Mechanisms to Ralstonia solanacearum. Front. Plant Sci. 2017;8 doi: 10.3389/fpls.2017.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell M.J. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA. 2013;19:740–751. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat R.S., Rockey J., Shirasawa K., Tilak I.S., Brijesh Patil M.P., Reddy Lachagari V.B. DNA methylation and expression analyses reveal epialleles for the foliar disease resistance genes in peanut (Arachis hypogaea L.) BMC Res. Notes. 2020;13:20. doi: 10.1186/s13104-020-4883-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes D.C., Zayed A.M., Ascenzi R., McCaskill A.J., Hoffman N.E., Davis K.R., Görlach J. Growth stage–based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell. 2001;13:1499–1510. doi: 10.1105/TPC.010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesari S., Bernoux M., Moncuquet P., Kroj T., Dodds P.N. A novel conserved mechanism for plant NLR protein pairs: the “integrated decoy” hypothesis. Front. Plant Sci. 2014;5:606. doi: 10.3389/fpls.2014.00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvin L., Sevestre F., Lukan T., Nogué F., Gallois J.-L., Chauvin J.-E., Veillet F. Gene Editing in Potato Using CRISPR-Cas9 Technology. Methods Mol. Biol. 2021;2354:331–351. doi: 10.1007/978-1-0716-1609-3_16. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Concordet J.-P., Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras M.P., Pai H., Tumtas Y., Duggan C., Yuen E.L.H., Cruces A.V., Kourelis J., Ahn H.-K., Lee K.-T., Wu C.-H., et al. Sensor NLR immune proteins activate oligomerization of their NRC helpers in response to plant pathogens. EMBO J. 2023;42:e111519. doi: 10.15252/embj.2022111519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuerda-Gil D., Slotkin R.K. Non-canonical RNA-directed DNA methylation. Nat. Plants. 2016;2:16163–16168. doi: 10.1038/nplants.2016.163. [DOI] [PubMed] [Google Scholar]
- Debieu M., Huard-Chauveau C., Genissel A., Roux F., Roby D. Quantitative disease resistance to the bacterial pathogen Xanthomonas campestris involves an Arabidopsis immune receptor pair and a gene of unknown function. Mol. Plant Pathol. 2016;17:510–520. doi: 10.1111/mpp.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deleris A., Halter T., Navarro L. DNA Methylation and Demethylation in Plant Immunity. Annu. Rev. Phytopathol. 2016;54:579–603. doi: 10.1146/annurev-phyto-080615-100308. [DOI] [PubMed] [Google Scholar]
- Delplace F., Huard-Chauveau C., Berthomé R., Roby D. Network organization of the plant immune system: from pathogen perception to robust defense induction. Plant J. 2022;109:447–470. doi: 10.1111/tpj.15462. [DOI] [PubMed] [Google Scholar]
- Deng Y., Liu M., Li X., Li F. microRNA-mediated R gene regulation: molecular scabbards for double-edged swords. Sci. China Life Sci. 2018;61:138–147. doi: 10.1007/s11427-017-9237-4. [DOI] [PubMed] [Google Scholar]
- Diener A.C., Ausubel F.M. RESISTANCE TO FUSARIUM OXYSPORUM 1, a Dominant Arabidopsis Disease-Resistance Gene, Is Not Race Specific. Genetics. 2005;171:305–321. doi: 10.1534/genetics.105.042218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand S., Bouché N., Perez Strand E., Loudet O., Camilleri C. Rapid Establishment of Genetic Incompatibility through Natural Epigenetic Variation. Curr. Biol. 2012;22:326–331. doi: 10.1016/j.cub.2011.12.054. [DOI] [PubMed] [Google Scholar]
- Ernst K., Kumar A., Kriseleit D., Kloos D.-U., Phillips M.S., Ganal M.W. The broad-spectrum potato cyst nematode resistance gene (Hero) from tomato is the only member of a large gene family of NBS-LRR genes with an unusual amino acid repeat in the LRR region. Plant J. 2002;31:127–136. doi: 10.1046/j.1365-313x.2002.01341.x. [DOI] [PubMed] [Google Scholar]
- Essuman K., Milbrandt J., Dangl J.L., Nishimura M.T. Shared TIR enzymatic functions regulate cell death and immunity across the tree of life. Science. 2022;377 doi: 10.1126/science.abo0001. [DOI] [PubMed] [Google Scholar]
- Fähling M., Graf H., Siemens J. Pathotype Separation of Plasmodiophora brassicae by the Host Plant. J. Phytopathol. 2003;151:425–430. [Google Scholar]
- Fauser F., Schiml S., Puchta H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014;79:348–359. doi: 10.1111/tpj.12554. [DOI] [PubMed] [Google Scholar]
- Fei Q., Xia R., Meyers B.C. Phased, Secondary, Small Interfering RNAs in Posttranscriptional Regulatory Networks. Plant Cell. 2013;25:2400–2415. doi: 10.1105/tpc.113.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förderer A., Yu D., Li E., Chai J. Resistosomes at the interface of pathogens and plants. Curr. Opin. Plant Biol. 2022;67 doi: 10.1016/j.pbi.2022.102212. [DOI] [PubMed] [Google Scholar]
- Fukuoka S., Yamamoto S.-I., Mizobuchi R., Yamanouchi U., Ono K., Kitazawa N., Yasuda N., Fujita Y., Thi Thanh Nguyen T., Koizumi S., et al. Multiple functional polymorphisms in a single disease resistance gene in rice enhance durable resistance to blast. Sci. Rep. 2014;4:4550. [Google Scholar]
- Furci L., Jain R., Stassen J., Berkowitz O., Whelan J., Roquis D., Baillet V., Colot V., Johannes F., Ton J. Identification and characterisation of hypomethylated DNA loci controlling quantitative resistance in Arabidopsis. Elife. 2019;8 doi: 10.7554/eLife.40655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravot A., Grillet L., Wagner G., Jubault M., Lariagon C., Baron C., Deleu C., Delourme R., Bouchereau A., Manzanares-Dauleux M.J. Genetic and physiological analysis of the relationship between partial resistance to clubroot and tolerance to trehalose in Arabidopsis thaliana. New Phytol. 2011;191:1083–1094. doi: 10.1111/j.1469-8137.2011.03751.x. [DOI] [PubMed] [Google Scholar]
- Gravot A., Richard G., Lime T., Lemarié S., Jubault M., Lariagon C., Lemoine J., Vicente J., Robert-Seilaniantz A., Holdsworth M.J., et al. Hypoxia response in Arabidopsis roots infected by Plasmodiophora brassicae supports the development of clubroot. BMC Plant Biol. 2016;16:251. doi: 10.1186/s12870-016-0941-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D.G., Roqueiro D., Salomé P.A., Kleeberger S., Greshake B., Zhu W., Liu C., Lippert C., Stegle O., Schölkopf B., et al. easyGWAS: A Cloud-Based Platform for Comparing the Results of Genome-Wide Association Studies. Plant Cell. 2017;29:5–19. doi: 10.1105/tpc.16.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi N., Inoue H., Kato T., Funao T., Shirota M., Shimizu T., Kanamori H., Yamane H., Hayano-Saito Y., Matsumoto T., et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010;64:498–510. doi: 10.1111/j.1365-313X.2010.04348.x. [DOI] [PubMed] [Google Scholar]
- He L., Wu W., Zinta G., Yang L., Wang D., Liu R., Zhang H., Zheng Z., Huang H., Zhang Q., et al. A naturally occurring epiallele associates with leaf senescence and local climate adaptation in Arabidopsis accessions. Nat. Commun. 2018;9:460. doi: 10.1038/s41467-018-02839-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Jacobsen S.E. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Huang C.-Y., Wang H., Hu P., Hamby R., Jin H. Small RNAs – Big Players in Plant-Microbe Interactions. Cell Host Microbe. 2019;26:173–182. doi: 10.1016/j.chom.2019.07.021. [DOI] [PubMed] [Google Scholar]
- Huard-Chauveau C., Perchepied L., Debieu M., Rivas S., Kroj T., Kars I., Bergelson J., Roux F., Roby D. An atypical kinase under balancing selection confers broad-spectrum disease resistance in Arabidopsis. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurni S., Scheuermann D., Krattinger S.G., Kessel B., Wicker T., Herren G., Fitze M.N., Breen J., Presterl T., Ouzunova M., et al. The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci. USA. 2015;112:8780–8785. doi: 10.1073/pnas.1502522112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.D.G., Vance R.E., Dangl J.L. Intracellular innate immune surveillance devices in plants and animals. Science. 2016;354 doi: 10.1126/science.aaf6395. [DOI] [PubMed] [Google Scholar]
- Jubault M., Hamon C., Gravot A., Lariagon C., Delourme R., Bouchereau A., Manzanares-Dauleux M.J. Differential Regulation of Root Arginine Catabolism and Polyamine Metabolism in Clubroot-Susceptible and Partially Resistant Arabidopsis Genotypes. Plant Physiol. 2008;146:2008–2019. doi: 10.1104/pp.108.117432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubault M., Lariagon C., Simon M., Delourme R., Manzanares-Dauleux M.J. Identification of quantitative trait loci controlling partial clubroot resistance in new mapping populations of Arabidopsis thaliana. Theor. Appl. Genet. 2008;117:191–202. doi: 10.1007/s00122-008-0765-8. [DOI] [PubMed] [Google Scholar]
- Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.-Y., Freimer N.B., Sabatti C., Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov T.L., Kniskern J.M., Gao L., DeYoung B.J., Ding J., Dubiella U., Lastra R.O., Nallu S., Roux F., Innes R.W., et al. The long-term maintenance of a resistance polymorphism through diffuse interactions. Nature. 2014;512:436–440. doi: 10.1038/nature13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T., Huang S.-S.C., Jupe F., Sasaki E., Schmitz R.J., Urich M.A., Castanon R., Nery J.R., Barragan C., He Y., et al. Epigenomic Diversity in a Global Collection of Arabidopsis thaliana Accessions. Cell. 2016;166:492–505. doi: 10.1016/j.cell.2016.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepikova A.V., Kasianov A.S., Gerasimov E.S., Logacheva M.D., Penin A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016;88:1058–1070. doi: 10.1111/tpj.13312. [DOI] [PubMed] [Google Scholar]
- Kong W., Li B., Wang Q., Wang B., Duan X., Ding L., Lu Y., Liu L.-W., La H. Analysis of the DNA methylation patterns and transcriptional regulation of the NB-LRR-encoding gene family in Arabidopsis thaliana. Plant Mol. Biol. 2018;96:563–575. doi: 10.1007/s11103-018-0715-z. [DOI] [PubMed] [Google Scholar]
- Kong W., Xia X., Wang Q., Liu L.-W., Zhang S., Ding L., Liu A., La H. Impact of DNA Demethylases on the DNA Methylation and Transcription of Arabidopsis NLR Genes. Front. Genet. 2020;11:460. doi: 10.3389/fgene.2020.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourelis J., van der Hoorn R.A.L. Defended to the Nines: 25 Years of Resistance Gene Cloning Identifies Nine Mechanisms for R Protein Function. Plant Cell. 2018;30:285–299. doi: 10.1105/tpc.17.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y., Eulgem T. Transcript-level expression control of plant NLR genes. Mol. Plant Pathol. 2018;19:1267–1281. doi: 10.1111/mpp.12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laperche A., Aigu Y., Jubault M., Ollier M., Guichard S., Glory P., Strelkov S.E., Gravot A., Manzanares-Dauleux M.J. Clubroot resistance QTL are modulated by nitrogen input in Brassica napus. Theor. Appl. Genet. 2017;130:669–684. doi: 10.1007/s00122-016-2842-8. [DOI] [PubMed] [Google Scholar]
- Larkan N.J., Lydiate D.J., Parkin I.A.P., Nelson M.N., Epp D.J., Cowling W.A., Rimmer S.R., Borhan M.H. The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 2013;197:595–605. doi: 10.1111/nph.12043. [DOI] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemarié S., Robert-Seilaniantz A., Lariagon C., Lemoine J., Marnet N., Jubault M., Manzanares-Dauleux M.J., Gravot A. Both the Jasmonic Acid and the Salicylic Acid Pathways Contribute to Resistance to the Biotrophic Clubroot Agent Plasmodiophora brassicae in Arabidopsis. Plant Cell Physiol. 2015;56:2158–2168. doi: 10.1093/pcp/pcv127. [DOI] [PubMed] [Google Scholar]
- Li Y., Tessaro M.J., Li X., Zhang Y. Regulation of the Expression of Plant Resistance Gene SNC1 by a Protein with a Conserved BAT2 Domain. Plant Physiol. 2010;153:1425–1434. doi: 10.1104/pp.110.156240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Kapos P., Zhang Y. NLRs in plants. Curr. Opin. Immunol. 2015;32:114–121. doi: 10.1016/j.coi.2015.01.014. [DOI] [PubMed] [Google Scholar]
- Liégard B., Baillet V., Etcheverry M., Joseph E., Lariagon C., Lemoine J., Evrard A., Colot V., Gravot A., Manzanares-Dauleux M.J., Jubault M. Quantitative resistance to clubroot infection mediated by transgenerational epigenetic variation in Arabidopsis. New Phytol. 2019;222:468–479. doi: 10.1111/nph.15579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López Sánchez A., Stassen J.H.M., Furci L., Smith L.M., Ton J. The role of DNA (de)methylation in immune responsiveness of Arabidopsis. Plant J. 2016;88:361–374. doi: 10.1111/tpj.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López Sánchez A., Pascual-Pardo D., Furci L., Roberts M.R., Ton J. Costs and Benefits of Transgenerational Induced Resistance in Arabidopsis. Front. Plant Sci. 2021;12 doi: 10.3389/fpls.2021.644999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E., Bruce T.J.A., Roberts M.R., Flors V., Ton J. Next-Generation Systemic Acquired Resistance. Plant Physiol. 2012;158:844–853. doi: 10.1104/pp.111.187468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa T., Kufer T.A., Schulze-Lefert P. NLR functions in plant and animal immune systems: so far and yet so close. Nat. Immunol. 2011;12:817–826. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- Manning K., Tör M., Poole M., Hong Y., Thompson A.J., King G.J., Giovannoni J.J., Seymour G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006;38:948–952. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- Manzanares-Dauleux M.J., Divaret I., Baron F., Thomas G. Evaluation of French Brassica oleracea landraces for resistance to Plasmodiophora brassicae. Euphytica. 2000;113:211–218. [Google Scholar]
- Martin A., Troadec C., Boualem A., Rajab M., Fernandez R., Morin H., Pitrat M., Dogimont C., Bendahmane A. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:1135–1138. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- Martin E.C., Ion C.F., Ifrimescu F., Spiridon L., Bakker J., Goverse A., Petrescu A.-J. NLRscape: an atlas of plant NLR proteins. Nucleic Acids Res. 2023;51:D1470–D1482. doi: 10.1093/nar/gkac1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M.A., Mosher R.A. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014;15:394–408. doi: 10.1038/nrg3683. [DOI] [PubMed] [Google Scholar]
- Meyers B.C., Kozik A., Griego A., Kuang H., Michelmore R.W. Genome-Wide Analysis of NBS-LRR–Encoding Genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molinier J., Ries G., Zipfel C., Hohn B. Transgeneration memory of stress in plants. Nature. 2006;442:1046–1049. doi: 10.1038/nature05022. [DOI] [PubMed] [Google Scholar]
- Morán-Diez M.E., Martínez de Alba Á.E., Rubio M.B., Hermosa R., Monte E. Trichoderma and the Plant Heritable Priming Responses. J. Fungi. 2021;7:318. doi: 10.3390/jof7040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narusaka M., Shirasu K., Noutoshi Y., Kubo Y., Shiraishi T., Iwabuchi M., Narusaka Y. RRS1 and RPS4 provide a dual Resistance-gene system against fungal and bacterial pathogens. Plant J. 2009;60:218–226. doi: 10.1111/j.1365-313X.2009.03949.x. [DOI] [PubMed] [Google Scholar]
- Nelson R., Wiesner-Hanks T., Wisser R., Balint-Kurti P. Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 2017;19:21–33. doi: 10.1038/nrg.2017.82. [DOI] [PubMed] [Google Scholar]
- Nishimura M.T., Anderson R.G., Cherkis K.A., Law T.F., Liu Q.L., Machius M., Nimchuk Z.L., Yang L., Chung E.-H., El Kasmi F., et al. TIR-only protein RBA1 recognizes a pathogen effector to regulate cell death in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2017;114:E2053–E2062. doi: 10.1073/pnas.1620973114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong-Abdullah M., Ordway J.M., Jiang N., Ooi S.-E., Kok S.-Y., Sarpan N., Azimi N., Hashim A.T., Ishak Z., Rosli S.K., et al. Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature. 2015;525:533–537. doi: 10.1038/nature15365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma K., Thorgrimsen S., Malinovsky F.G., Fiil B.K., Nielsen H.B., Brodersen P., Hofius D., Petersen M., Mundy J. Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilet-Nayel M.-L., Moury B., Caffier V., Montarry J., Kerlan M.-C., Fournet S., Durel C.-E., Delourme R. Quantitative resistance to plant pathogens in pyramiding strategies for durable crop protection. Front. Plant Sci. 2017;8:1838. doi: 10.3389/fpls.2017.01838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu S., Liu G., Zhou B., Bellizzi M., Zeng L., Dai L., Han B., Wang G.-L. The Broad-Spectrum Blast Resistance Gene Pi9 Encodes a Nucleotide-Binding Site–Leucine-Rich Repeat Protein and Is a Member of a Multigene Family in Rice. Genetics. 2006;172:1901–1914. doi: 10.1534/genetics.105.044891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L., Colot V. Plant transgenerational epigenetics. Annu. Rev. Genet. 2016;50:467–491. doi: 10.1146/annurev-genet-120215-035254. [DOI] [PubMed] [Google Scholar]
- Quadrana L., Almeida J., Asís R., Duffy T., Dominguez P.G., Bermúdez L., Conti G., Corrêa da Silva J.V., Peralta I.E., Colot V., et al. Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat. Commun. 2014;5:3027. doi: 10.1038/ncomms5027. [DOI] [PubMed] [Google Scholar]
- Quadrana L., Bortolini Silveira A., Mayhew G.F., LeBlanc C., Martienssen R.A., Jeddeloh J.A., Colot V. The Arabidopsis thaliana mobilome and its impact at the species level. Elife. 2016;5 doi: 10.7554/eLife.15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Prado J.S., Piquerez S.J.M., Bendahmane A., Hirt H., Raynaud C., Benhamed M. Modify the Histone to Win the Battle: Chromatin Dynamics in Plant–Pathogen Interactions. Front. Plant Sci. 2018;9 doi: 10.3389/fpls.2018.00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E.J. Inherited epigenetic variation--revisiting soft inheritance. Nat. Rev. Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saile S.C., Jacob P., Castel B., Jubic L.M., Salas-Gonzáles I., Bäcker M., Jones J.D.G., Dangl J.L., El Kasmi F. Two unequally redundant “helper” immune receptor families mediate Arabidopsis thaliana intracellular “sensor” immune receptor functions. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saile S.C., Ackermann F.M., Sunil S., Keicher J., Bayless A., Bonardi V., Wan L., Doumane M., Stöbbe E., Jaillais Y., et al. Arabidopsis ADR1 helper NLR immune receptors localize and function at the plasma membrane in a phospholipid dependent manner. New Phytol. 2021;232:2440–2456. doi: 10.1111/nph.17788. [DOI] [PubMed] [Google Scholar]
- Saucet S.B., Ma Y., Sarris P.F., Furzer O.J., Sohn K.H., Jones J.D.G. Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat. Commun. 2015;6:6338. doi: 10.1038/ncomms7338. [DOI] [PubMed] [Google Scholar]
- Schneeberger K., Ossowski S., Ott F., Klein J.D., Wang X., Lanz C., Smith L.M., Cao J., Fitz J., Warthmann N., et al. Reference-guided assembly of four diverse Arabidopsis thaliana genomes. Proc. Natl. Acad. Sci. USA. 2011;108:10249–10254. doi: 10.1073/pnas.1107739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z.-Q., Xue J.-Y., Wu P., Zhang Y.-M., Wu Y., Hang Y.-Y., Wang B., Chen J.-Q. Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol. 2016;170:2095–2109. doi: 10.1104/pp.15.01487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad P.V., Chen H.-M., Patel K., Bond D.M., Santos B.A., Baulcombe D.C. A microRNA superfamily regulates nucleotide binding site–leucine-rich repeats and other mRNAs. Plant Cell. 2012;24:859–874. doi: 10.1105/tpc.111.095380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveira A.B., Trontin C., Cortijo S., Barau J., Del Bem L.E.V., Loudet O., Colot V., Vincentz M. Extensive Natural Epigenetic Variation at a De Novo Originated Gene. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter A., Daniel X., Flors V., Luna E., Hohn B., Mauch-Mani B. Descendants of primed Arabidopsis plants exhibit resistance to biotic stress. Plant Physiol. 2011;158:835–843. doi: 10.1104/pp.111.191593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Some A., Manzanares M.J., Laurens F., Baron F., Thomas G., Rouxel F. Variation for virulence on Brassica napus L. amongst Plasmodiophora brassicae collections from France and derived single-spore isolates. Plant Pathol. 1996;45:432–439. [Google Scholar]
- Stroud H., Greenberg M.V.C., Feng S., Bernatavichute Y.V., Jacobsen S.E. Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell. 2013;152:352–364. doi: 10.1016/j.cell.2012.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Eichten S.R., Cahn J., Karpievitch Y.V., Borevitz J.O., Lister R. Population scale mapping of transposable element diversity reveals links to gene regulation and epigenomic variation. Elife. 2016;5 doi: 10.7554/eLife.20777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2013. R: A Language and Environment for Statistical Computing Advance Access Published 2013. [Google Scholar]
- Thomas C.M., Dixon M.S., Parniske M., Golstein C., Jones J.D. Genetic and molecular analysis of tomato Cf genes for resistance to Cladosporium fulvum. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1998;353:1413–1424. doi: 10.1098/rstb.1998.0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T., Eulgem T. An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc. Natl. Acad. Sci. USA. 2013;110:E3535–E3543. doi: 10.1073/pnas.1312545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Colot V. Epialleles in plant evolution. Genome Biol. 2012;13:249. doi: 10.1186/gb-2012-13-10-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.J., Sohn K.H., Wan L., Bernoux M., Sarris P.F., Segonzac C., Ve T., Ma Y., Saucet S.B., Ericsson D.J., et al. Structural basis for assembly and function of a heterodimeric plant immune receptor. Science. 2014;344:299–303. doi: 10.1126/science.1247357. [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-H., Abd-El-Haliem A., Bozkurt T.O., Belhaj K., Terauchi R., Vossen J.H., Kamoun S. NLR network mediates immunity to diverse plant pathogens. Proc. Natl. Acad. Sci. USA. 2017;114:8113–8118. doi: 10.1073/pnas.1702041114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Cheng Y.T., Huang S., Win J., Soards A., Jinn T.-L., Jones J.D.G., Kamoun S., Chen S., Zhang Y., Li X. Regulation of Transcription of Nucleotide-Binding Leucine-Rich Repeat-Encoding Genes SNC1 and RPP4 via H3K4 Trimethylation. Plant Physiol. 2013;162:1694–1705. doi: 10.1104/pp.113.214551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Ellwood S., Calis O., Patrick E., Li T., Coleman M., Turner J.G. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science. 2001;291:118–120. doi: 10.1126/science.291.5501.118. [DOI] [PubMed] [Google Scholar]
- Xu X., Hayashi N., Wang C.-T., Fukuoka S., Kawasaki S., Takatsuji H., Jiang C.-J. Rice blast resistance gene Pikahei-1 (t), a member of a resistance gene cluster on chromosome 4, encodes a nucleotide-binding site and leucine-rich repeat protein. Mol. Breed. 2014;34:691–700. [Google Scholar]
- Yue J.-X., Meyers B.C., Chen J.-Q., Tian D., Yang S. Tracing the origin and evolutionary history of plant nucleotide-binding site–leucine-rich repeat (NBS-LRR) genes. New Phytol. 2012;193:1049–1063. doi: 10.1111/j.1469-8137.2011.04006.x. [DOI] [PubMed] [Google Scholar]
- Zamani-Noor N., Wallenhammar A.-C., Kaczmarek J., Patar U.R., Zouhar M., Manasova M., Jędryczka M. Pathotype Characterization of Plasmodiophora brassicae, the Cause of Clubroot in Central Europe and Sweden (2016–2020) Pathogens. 2022;11:1440. doi: 10.3390/pathogens11121440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-C., Gassmann W. Alternative splicing and mRNA levels of the disease resistance gene RPS4 are induced during defense responses. Plant Physiol. 2007;145:1577–1587. doi: 10.1104/pp.107.108720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Tang K., Wang B., Duan C.-G., Lang Z., Zhu J.-K. Protocol: a beginner’s guide to the analysis of RNA-directed DNA methylation in plants. Plant Methods. 2014;10:18. doi: 10.1186/1746-4811-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Lang Z., Zhu J.-K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018;19:489–506. doi: 10.1038/s41580-018-0016-z. [DOI] [PubMed] [Google Scholar]
- Zou B., Yang D.-L., Shi Z., Dong H., Hua J. Monoubiquitination of Histone 2B at the disease resistance gene locus regulates its expression and impacts immune responses in Arabidopsis. Plant Physiol. 2014;165:309–318. doi: 10.1104/pp.113.227801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the paper and its supplemental information files. All unique materials used are readily available from the authors.