Abstract
We report attempts to analyze interactions between components of the pullulanase (Pul) secreton (type II secretion machinery) from Klebsiella oxytoca encoded by a multiple-copy-number plasmid in Escherichia coli. Three of the 15 Pul proteins (B, H, and N) were found to be dispensable for pullulanase secretion. The following evidence leads us to propose that PulE, PulL, and PulM form a subcomplex with which PulC and PulG interact. The integral cytoplasmic membrane protein PulL prevented proteolysis and/or aggregation of PulE and mediated its association with the cytoplasmic membrane. The cytoplasmic, N-terminal domain of PulL interacted directly with PulE, and both PulC and PulM were required to prevent proteolysis of PulL. PulM and PulL could be cross-linked as a heterodimer whose formation in a strain producing the secreton required PulG. However, PulL and PulM produced alone could also be cross-linked in a 52-kDa complex, indicating that the secreton exerts subtle effects on the interaction between PulE and PulL. Antibodies against PulM coimmunoprecipitated PulL, PulC, and PulE from detergent-solubilized cell extracts, confirming the existence of a complex containing these four proteins. Overproduction of PulG, which blocks secretion, drastically reduced the cellular levels of PulC, PulE, PulL, and PulM as well as PulD (secretin), which probably interacts with PulC. The Pul secreton components E, F, G, I, J, K, L, and M could all be replaced by the corresponding components of the Out secretons of Erwinia chrysanthemi and Erwinia carotovora, showing that they do not play a role in secretory protein recognition and secretion specificity.
Gram-negative bacteria use several different pathways for protein secretion. In the two-step general secretory pathway, proteins are first translocated across the cytoplasmic membrane via the Sec machinery. Selected secretory proteins are subsequently transported across the outer membrane by one of several branch pathways (27). In Klebsiella oxytoca, pullulanase secretion by the main terminal branch of the general secretory pathway requires a putative multiprotein, envelope-associated complex, the Pul secreton (also called type II secretion machinery), encoded by 15 pul genes (the 13-gene pulC to -O operon, pulB, and pulS) that flank the pullulanase structural gene pulA (30). The organization of secreton genes is conserved in different gram-negative bacteria (27), most of which use the same secreton to secrete several secretory proteins lacking sequence-conserved segments that could correspond to common secretion signals. The identity of the secreton component that recognizes secretion signals (40) in secretory proteins has not been firmly established (8, 41).
Here, we report attempts to study interactions between components of the Pul secreton. Previously, we demonstrated multimerization of the type IV pilin-like (pseudopilin) product of the pulG gene (28, 32), processing of pre-PulG by the pulO gene product (prepilin peptidase) (29), intimate association of PulD with PulS (11, 20), and a possible interaction between PulC and PulD (secretin) (26). Studies reported here center around interactions involving PulE protein, which is associated with the cytoplasmic membrane (25) but does not have hydrophobic transmembrane segments. The membrane association of PulE homologues has already been studied in other systems. For example, the PulE homologue EpsE of Vibrio cholerae is anchored to the cytoplasmic membrane via the PulL homologue EpsL (36). Analysis of a chimera between the PulE homologue ExeE from Aeromonas hydrophila and EpsE led Sandkvist et al. to conclude that membrane association occurred via the amino-terminal part of EpsE (36). Similar results were observed in Pseudomonas aeruginosa, in which XcpR (XcpE) associates with the cytoplasmic membrane via XcpY (XcpL) (1). Although the XcpR (XcpE) component of Pseudomonas putida could not substitute for the corresponding P. aeruginosa protein, secretion could be promoted by a chimera composed of the N-terminal region of the E protein from P. aeruginosa and the C-terminal region from the E protein of P. putida (5). Further studies of both Xcp and Eps secretons revealed that protein L is unstable in the absence of the M protein (EpsM in Vibrio cholerae and XcpZ in P. aeruginosa) (18, 37). Furthermore, coimmunoprecipitation revealed that EpsL interacts directly with EpsM (37). Recent two-hybrid and protease accessibility studies of secreton components from Erwinia chrysanthemi also indicated that the E, L, and M components interact (34).
The reconstitution and amplification of the complete Pul secreton in Escherichia coli (7) provide a unique opportunity to examine interactions between secreton components in a readily accessible system. We began by creating nonpolar mutations in each of the pul genes on a plasmid carrying the complete Pul secreton gene cluster. The effects of the mutations on the stability, multimerization (as determined by chemical cross-linking), and localization of PulE were also examined. These studies pointed to a specific role for PulL in stabilizing and anchoring PulE to the cytoplasmic membrane. Therefore, we examined the role of different domains of PulE and PulL in the interaction between the two proteins. The mutants were also used in complementation experiments with the out secreton genes from E. chrysanthemi and Erwinia carotovora (17) to identify components that were specific to the Pul secreton.
MATERIALS AND METHODS
Bacterial strains, plasmids, culture conditions, and pullulanase assay.
E. coli K-12 strains are listed in Table 1, and plasmids are listed in Table 2. Strains HS2019, PAP7460, and PAP7500B (Table 1) were employed in many of the experiments using antibodies raised against MalE hybrid proteins. These strains do not produce MalE protein (which reacts with these antibodies) and carry a malG mutation that allows MalE-independent uptake of maltose (44) in order to activate MalT, the positive activator of pulAp and pulCp (7). Details of the construction of other clones and mutants not previously reported are given below. Plasmids carrying cloned out genes under lacZ control used here (Table 3) were described by Lindeberg et al. (17).
TABLE 1.
E. coli K-12 strains
| Strain | Relevant characteristics | Reference or source |
|---|---|---|
| MC4100 | Δ(lac-argF)U169 araD139 relA1 respL150 | Lab collection |
| PAP105 | Δ(lac-pro) pAPIP501 [F′ (lacIqΔlacZM15 pro+ Tn10)] (Tcr) | Lab collection |
| PAP7232 | MC4100 malP::(pulS pulA-B pulC to -O) F′ (lacIqpro+ Tn10) (Tcr) | 29 |
| PAP3175 | PAP7232::ΔpulE3 | 22 |
| PAP3389 | PAP7232::ΔpulF | This study |
| PAP3390 | PAP7232::ΔpulL | This study |
| PAP7452 | PAP7232 pulB::kan1 | 26 |
| HS2019 | MC4100 ΔmalE444 malG501 | H. Shuman |
| PAP7460 | HS2019 pAPIP501 [F′ (lacIqΔlacZM15 pro+ Tn10)] (Tcr) | This study |
| PAP7500B | PAP7460 malP::(pulS pulA pulB::kan1 pulC to -O) | This study |
TABLE 2.
Plasmids carrying cloned pul genes
| Plasmid | Vector/origin/resistance | Cloned genes and/or mutations | Reference and/or source of cloned fragment |
|---|---|---|---|
| pCHAP231 | pBR322/ColE1/Ap | pBR322::(pulS pulA-B pulC to -O) | 7 |
| pCHAP1219 | pBR322/ColE1/Ap | pCHAP231 pulS::Tn5 | 6 |
| pCHAP1201 | pBR322/ColE1/Ap | pCHAP231 pulB::kan1 | This study; 26 |
| pCHAP1218 | pBR322/ColE1/Ap | pCHAP231 pulA::Tn5 | 7 |
| pCHAP1229 | pBR322/ColE1/Ap | pCHAP231 pulC::kan2 | 26 |
| pCHAP1226 | pBR322/ColE1/Ap | pCHAP231 pulB::kan1 ΔpulD | 26 |
| pCHAP1230 | pBR322/ColE1/Ap | pCHAP231 pulB::kan1 ΔpulE3 | This study |
| pCHAP1227 | pBR322/ColE1/Ap | pCHAP231 pulB::kan1 ΔpulF | This study |
| pCHAP1216 | pBR322/ColE1/Ap | pCHAP231 pulB::kan1 ΔpulG1 | This study |
| pCHAP1324 | pBR322/ColE1/Ap | pCHAP231 pulH::kan2 | This study |
| pCHAP1357 | pBR322/ColE1/Ap | pCHAP231 pulI::kan2 | This study |
| pCHAP1323 | pBR322/ColE1/Ap | pCHAP231 pulJ::kan2 | This study |
| pCHAP1325 | pBR322/ColE1/Ap | pCHAP231 pulK::kan2 | This study |
| pCHAP1217 | pBR322/ColE1/Ap | pCHAP231 pulB::kan1 ΔpulL | This study |
| pCHAP1359 | pBR322/ColE1/Ap | pCHAP231 pulM::kan2 | This study |
| pCHAP1350 | pBR322/ColE1/Ap | pCHAP231 pulN::kan2 | This study |
| pCHAP1225 | pBR322/ColE1/Ap | pCHAP231 pulO::Mud | 7 |
| pCHAP580 | pSU18/p15/Cm | lacZp-pulS | This study; 6 |
| pCHAP1228 | pSU18/p15/Cm | lacZp-pulA | 14 |
| pCHAP5001 | pSU18/p15/Cm | lacZp-pulC | 26 |
| pCHAP3635 | pSU18/p15/Cm | lacZp-pulD | 8 |
| pCHAP2273 | pSU18/p15/Cm | lacZp-pulE | This study; 25 |
| pCHAP1206 | pSU18/p15/Cm | lacZp-pulF | This study |
| pCHAP1205 | pSU18/p15/Cm | lacZp-pulG | This study; 29 |
| pCHAP1321 | pSU18/p15/Cm | lacZp-pulH | This study |
| pCHAP1352 | pSU18/p15/Cm | lacZp-pulI | This study |
| pCHAP1329 | pSU18/p15/Cm | lacZp-pulJ | This study |
| pCHAP1271 | pSU18/p15/Cm | lacZp-pulK | This study |
| pCHAP2245 | pSU18/p15/Cm | lacZp-pulL | This study |
| pCHAP1354 | pSU18/p15/Cm | lacZp-pulM | This study |
| pCHAP1356 | pSU18/p15/Cm | lacZp-pulN | This study |
| pCHAP1224 | pSU18/p15/Cm | lacZp-pulO | This study |
| pCHAP2014 | pBGS18/ColE1/Km | lacZp-pulE pulF′ | 22 |
| pCHAP2045 | pBGS18/ColE1/Km | lacZp-pulE pulF pulG′ | 22 |
| pCHAP2230 | pBGS18/ColE1/Km | lacZp-pulL | This study |
| pCHAP2235 | pHSG575/pSC101/Cm | lacZp-pulL | This study |
| pCHAP5107 | p733/ColE1/Ap | tetp-lexA-pulE′ | This study |
| pCHAP5112 | pSU18/p15/Cm | lacZp-pulL′ | This study |
| pCHAP125 | pHSG576/pSC101/Cm | lacZp-′pulK-pulL-pulM-pulN-pulO | 33 |
TABLE 3.
Complementation of single-locus pul mutations by corresponding pul genes and by out genes from E. chrysanthemi and E. carotovoraa
| Mutation | Secretion (%) | Complementation (% secretion) after incubation with (+) or without (−) IPTG
|
|||||
|---|---|---|---|---|---|---|---|
|
pul gene
|
E. ch out gene
|
E. ca out gene
|
|||||
| − | + | − | + | − | + | ||
| pulS | <10 | >80 | >80 | >80* | >80* | NT | NT |
| pulB | >80 | NT | NT | NT | NT | NT | NT |
| pulC | <10 | >80 | >80 | <10 | <10 | <10 | <10 |
| pulD | <10 | >80 | >80 | <10 | <10 | <10 | <10 |
| pulE | <10 | >80 | >80 | 30 | >80 | >80 | 50 |
| pulF | <10 | >80 | <10 | 40 | <10 | >80 | 50 |
| pulG | <10 | >80 | >80 | <10 | >80 | <10 | >80 |
| pulH | >80 | NT | NT | NT | NT | NT | NT |
| pulI | <10 | >80 | >80 | >80 | >80 | >80 | >80 |
| pulJ | <10 | >80 | >80 | >80 | >80 | >80 | >80 |
| pulK | <10 | >80 | >80 | <10 | >80 | <10 | >80 |
| pulL | <10 | >80 | >80 | >80 | 60 | >80 | 70 |
| pulM | 20 | >80 | >80 | >80 | >80 | 70 | 60 |
| pulN | >80 | NT | NT | NA | NA | NT | NT |
| pulO | <10 | >80 | >80 | NT | NT | NT | NT |
Assays were performed with derivatives of strain PAP105. The pul mutations were present on pCHAP231 derivatives (Table 2). Strains carrying pCHAP231 secreted >80% of the pullulanase produced. All data are averages from three or more independent assays. All complementing plasmids were derivatives of pSU vectors carrying cloned pul genes (Table 2) or cloned out genes (17). NT, not tested; NA, not applicable; E. ch, E. chrysanthemi; E. ca, E. carotovora; *, see reference 11.
Bacteria were grown at 30°C in Luria-Bertani medium buffered at pH 7.2 with 10 mM phosphate or with 10% M63 medium (19) and supplemented, where appropriate, with 0.4% maltose to induce expression of pulA and the pulC to -O operon or 1 mM isopropyl thiogalactoside (IPTG) to induce expression of genes under lacZ promoter control. Expression of the lexA-pulE′ fusion was induced with 200 μg of anhydrotetracycline (Acros Organics) per ml. Antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 25 μg/ml; spectinomycin, 50 μg/ml; and tetracycline, 16 μg/ml.
Pullulanase secretion was measured as the proportion of the enzyme accessible to its substrate (pullulan) in whole cells compared to that in cells lysed with 0.5% octylpolyoxyethylene (22).
Construction of pulL and pulF gene deletions in the chromosomal pul locus.
In order to create an in-frame deletion in pulL, a 2.0-kb EcoRI-MscI fragment of pCHAP125 bearing the full-length pulL flanked by the 3′ end of pulK and the 5′ end of pulM (33) was cloned into pBGS18 (42). The resulting plasmid (pCHAP2230) was then cleaved at the two EcoRV sites in pulL and religated with an 8-bp SacII adapter, generating a 384-bp deletion between codons 201 and 355 of the 398-codon pulL gene (pCHAP2236). The entire cloned fragment in pCHAP2236 was then subcloned into M13mp11 and used to create a homologous exchange (3) with the chromosomal pul region of PAP7232 (29). The resulting strain, PAP3390, was shown to have a single, internally deleted copy of pulL by Southern hybridization.
To create an in-frame deletion in pulF, two site-directed mutagenesis reactions (16) were performed with pCHAP2014 and pCHAP2045 (25), generating BamHI sites at codons 195 and 275 of pulF (401 codons). After BamHI and EcoRI cleavage of both plasmids, a recombinant plasmid carrying a 237-bp pulF deletion (pCHAP2238) was reconstituted by ligating the purified DNA fragments. The mutated pulF allele was introduced in place of wild-type pulF in PAP7232 as described above to give strain PAP3389 (ΔpulF), which was shown by Southern blotting to carry a single, internally deleted copy of pulF.
Construction of pCHAP231 derivatives carrying mutations in specific pul genes.
The strategy used to transfer the chromosomal deletions in pulE (25), pulF, pulG (29), and pulL onto pCHAP231 (pBR322 with the full set of pul genes [6]) was the same as that described previously for pulD carried by pCHAP1226 (26), resulting in plasmids pCHAP1230, pCHAP1227, pCHAP1216, and pCHAP1217, respectively (Table 2).
All other mutations in pCHAP231 were created by a strategy similar to that used previously to create the pulC mutation carried by pCHAP1229 (26). DNA fragments carrying the complete pulH, pulI, pulJ, pulK, pulM, or pulN gene together with approximately 1 kb of flanking DNA were amplified by PCR and cloned into pUC18Cm (S. Lory). The resulting plasmids were linearized using restriction enzymes that cleaved only within the gene of interest as follows: pulH, MluI site at codon 93 of the 160-codon gene; pulI, NheI site at codon 47 of the 121-codon gene; pulJ, MscI site at codon 18 of the 198-codon gene; pulK, BglII site at codon 93 of the 326-codon gene; pulM, BstEII site at codon 138 of the 161-codon gene; pulN, SacII site at codon 125 of the 252-codon gene. The linearized DNA was then ligated with compatible SnaBI adapters. The kan2 cassette was inserted at these sites as previously described (26). (The kan2 cassette was positioned so that the ATG codon immediately after the kanamycin resistance gene is fused to the reading frame of the gene into which it is inserted.) The resulting plasmids were then linearized and electroporated into recD or recB sbcA strains carrying pCHAP231, with selection for the homologous exchange reaction that replaced the wild-type gene on pCHAP231 by the mutated copy as described previously (26). The presence of the desired mutation on the resulting plasmids (Table 2) was verified by PCR using primers hybridizing to sites in pCHAP231 and in the kan2 cassette.
Construction of pulE and pulL gene fusions.
The 5′ end of pulE, encoding the first 177 amino acids of the protein, was amplified by PCR using pCHAP231 as template. The resulting DNA fragment was flanked by BamHI and XhoI sites at the 5′ and 3′ ends, respectively, and was subcloned into p733 carrying the full-length lexA gene (15) to give plasmid pCHAP5107. Expression of the resulting in-frame lexA-pulE′ fusion was induced with anhydrotetracycline; the stability of the hybrid protein was confirmed by immunodetection using LexA antibodies (Santa Cruz Biotechnology).
The 5′ end of pulL, encoding the cytoplasmic domain of the protein (amino acids 1 to 247), was amplified by PCR using a 5′ oligonucleotide that introduced a DNA sequence encoding a hexahistidine tag. The amplified fragment was flanked by EcoRI and HindIII sites at its 5′ and 3′ ends, respectively. The PCR product was subsequently subcloned into pSU18, giving pCHAP5112.
Cloning of pul genes for complementation assays.
pul genes that were not previously cloned separately were amplified by PCR using pCHAP231 as a template and cloned into pSU18 using restriction sites introduced by the primers. The 5′ primer was engineered with an EcoRI site in the same reading frame as the EcoRI site in the vector so that the cloned gene was fused directly to the lacZ open reading frame. A second set of plasmids in which a 60- to 80-bp segment of the upstream pul gene was cloned in frame with the lacZ reading frame of the vector gave identical results.
Antibodies.
DNA corresponding to the predicted periplasmic domain of PulL was excised from pCHAP231, and DNA corresponding to the predicted periplasmic domain of PulM was amplified by PCR. Both fragments were cloned into pMAL-p2 (New England Biolabs) to generate malE-′pulL and malE-′pulM gene fusions coding for periplasmic hybrids. DNA coding for the entire sequence of PulE was PCR amplified and cloned into pMAL-c2 to create a malE-pulE gene fusion coding for a cytoplasmic hybrid protein. All three hybrid proteins were purified from total soluble protein extracts of IPTG-induced cells by amylose chromatography (New England Biolabs) and injected into rabbits to obtain antibodies.
Gel electrophoresis and immunoblotting.
After solubilization in sample buffer containing 2% sodium dodecyl sulfate (SDS) and heating at 100°C, proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred onto nitrocellulose. Ponceau red staining allowed visualization of porins (outer membrane markers) in membrane fractions. Pul proteins and DjlA were detected by incubation with specific primary antisera diluted to 1:2,000 to 1:10,000, followed by secondary horseradish peroxidase-linked anti-rabbit immunoglobulin G (IgG) (Amersham-Pharmacia; 1:10,000) and enhanced chemiluminescence (ECL kit; Amersham-Pharmacia). The LexA-PulE′ hybrid was detected with anti-LexA primary antibodies (Santa Cruz Biotechnology) diluted 1:2,000 and horseradish peroxidase-linked anti-goat IgG diluted 1:10,000 (Santa Cruz Biotechnology). The His-tagged truncated PulL protein was detected using monoclonal (His)6 antibodies (Clontech; 1:1,000) and horseradish peroxidase-linked anti-mouse IgG (1:10,000; Amersham-Pharmacia). Before the membrane was reprobed, bound antibodies were stripped off as described by the manufacturer (Amersham-Pharmacia).
Immunoprecipitation.
The MalE-PulM antibodies were affinity purified using the MalE-PulM hybrid protein covalently linked to CNBr-activated Sepharose 4B (Amersham-Pharmacia). Protein A Dynabeads (Dynal) were washed three times with phosphate-buffered saline containing 1% bovine serum albumin and incubated overnight with the affinity-purified anti-PulM antibodies. Cross-linking of antibodies to the beads was performed according to the instructions of the manufacturer using dimethyl pimelimidate (Sigma). Whole-cell extracts of E. coli K-12 harboring pCHAP231 were generated as described previously (37). Two hundred microliters of bacterial extract was incubated with different volumes of antibody-loaded beads at 4°C for 2 h on a rotating device. After being washed three times with TBST buffer (10 mM Tris-Cl, 150 mM NaCl, 0.05% Tween 20, pH 8.0), the beads were resuspended in SDS sample buffer and heated for 5 min at 100°C. Proteins in the loaded material and in the nonadsorbed and immunoadsorbed fractions were separated on a 12% polyacrylamide gel in SDS. After they were blotted onto a nitrocellulose membrane, different Pul proteins were visualized using specific polyclonal antibodies.
Cross-linking and proteolysis.
Four cross-linkers [ethylene glycol bis(succimidylsuccinate) (EGS; spacer length, 16.1 Å; Pierce), dithiobis(succimidyl proprionate) (DSP; 12.0 Å; Sigma), disuccimidyl tartrate (6.4 Å; Pierce), and formaldehyde] were used essentially as described previously (26, 28). In some cases, one of each pair of samples prepared in parallel was treated with phenol to dissociate PulD multimers prior to analysis by SDS-PAGE and immunoblotting (9, 26). Nitrocellulose membranes onto which the proteins had been blotted were probed successively with antibodies recognizing different Pul secreton components. All DSP-induced cross-links described here could be reversed by treatment with 10 mM dithiothreitol.
To analyze the stability and protease accessibility of the Pul secreton components, bacteria that were washed and resuspended in 25 mM Tris (pH 7.5) to an optical density at 600 nm of 1.0 were disrupted by sonication and then incubated at 37°C with or without 100 μg of proteinase K per ml for 30 min. Samples were then chilled on ice, and proteins were precipitated with 12.5% trichloroacetic acid. Precipitated proteins were examined by SDS-PAGE and immunoblotting.
Cellular fractionation and flotation sucrose gradients.
Two hundred milliliters of E. coli cultures was grown to an optical density at 600 nm of 1.0 to 1.2 before being lysed with or without prior conversion to spheroplasts (4) by passage through a French press at 5 × 107 or 1.2 × 108 Pa, respectively. DNase, RNase, and Pefabloc (Interchim) were added to a final concentration of 0.1 mg/ml. Cell debris was removed by low-speed centrifugation. The clarified lysate was then fractionated by ultracentrifugation at 180,000 × g for 45 min at 4°C, resulting in separation of soluble material (cytoplasmic and periplasmic contents) from particulate material (aggregated and membrane fractions).
For flotation gradients, the particulate fraction was first washed with 25 mM HEPES (pH 7.4) and then resuspended in the same buffer adjusted at 60% (wt/wt) in sucrose. Membranes were then loaded at the bottom of Beckman SW55 centrifuge tubes and successively overlaid with 56 to 32% (wt/vol) sucrose solutions (steps of 3%). The gradients were centrifuged at 220,000 × g for 36 h at 10°C. Fractions of 250 μl were collected from the top of the tubes.
RESULTS
PulB, PulH, and PulN are nonessential.
Nonpolar mutations in each of the 15 pul genes (pulA, pulB, pulC to pulO, and pulS) carried by pCHAP231 (Table 2) were tested for effects on pullulanase secretion in E. coli K-12 (Table 3). Three of the mutations, pulB, pulH, and pulN, had no effect on pullulanase secretion. The absence of a requirement for pulB was observed previously in studies of Tn5 insertions and deletions in pCHAP231 and other multiple-copy-number plasmids (7, 14). To determine whether pulB is required for secretion when the secreton genes are integrated in the E. coli K-12 chromosome, we constructed strain PAP7452 (26), a derivative of PAP7232 [malP::(pulS pulAB pulC to -O)] (7, 28) in which pulB was disrupted by a kanamycin resistance cassette (Table 1). Pullulanase secretion levels in PAP7452 and PAP7232 were indistinguishable (data not shown). Thus, pulB is not required for pullulanase secretion, at least in E. coli.
Previous studies indicated that both pulH and pulN were required for pullulanase secretion (33, 35). However, the transposon insertion mutations analyzed previously might have had a polar effect on the expression of downstream genes. pulN homologues are frequently absent from secreton gene clusters in other bacteria (27). Furthermore, a mutation in the E. chrysanthemi pulH homologue outH did not abolish pectate lyase secretion promoted by the reconstituted Out secreton on a plasmid in E. coli, although the chromosomal outH mutation in Erwinia did block secretion (17).
All other mutations except pulA (which abolished pullulanase production) and pulM (see below) abolished pullulanase secretion (<10% of wild-type levels). The pulM mutation reduced secretion only to 20%. The failure of polyclonal PulM-specific antiserum to detect a truncated form of the protein in extracts of the pulM mutant (data not shown) suggested that the PulM was not synthesized by the mutant, although the antibodies might not recognize epitopes present in the remaining, N-terminal part of the truncated protein. In all cases, pullulanase secretion was fully restored by complementation by a cloned copy of the wild-type gene in trans on a compatible plasmid (Table 3), indicating that the mutations are nonpolar.
Effects of pul mutations on Pul protein stability.
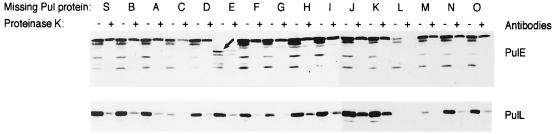
We next examined the possibility that some Pul proteins might be destabilized by the absence of another protein with which they would normally interact. The only reproducible effect observed by immunoblotting of total protein extracts from the mutant strains was a modest reduction in the level of PulE in the pulL mutant (data not shown). The instability of PulE in the absence of PulL was amplified by sonicating the bacteria and incubating them with or without proteinase K (Fig. 1). However, no other pul mutation affected the stability of PulE. The pulE mutation did not exert a reciprocal effect on the level of PulL (Fig. 1), possibly because a truncated PulE protein is produced (25). The absence of PulM also destabilized PulL (Fig. 1), indicating a possible interaction between these two proteins. Finally, PulL was also destabilized in the pulC mutant (Fig. 1), a novel finding that suggests a previously unsuspected link between PulC and PulL. The levels of PulC and PulM were unaffected by the pulL mutation (data not shown).
FIG. 1.
Immunodetection of PulE and PulL in sonicated, untreated, or proteinase K-treated cells of strain HS2019 carrying pCHAP231 with mutations in specific pul genes. The sample buffer contained a 10 mM concentration of the reducing agent dithiothreitol. The double band of PulE is due to incomplete reduction of the intramolecular disulfide bond that forms when the cells are disrupted by sonication (24). The arrow indicates the internally truncated form of PulE produced in the pulE mutant.
Overproduction of PulG destabilizes several Pul proteins.
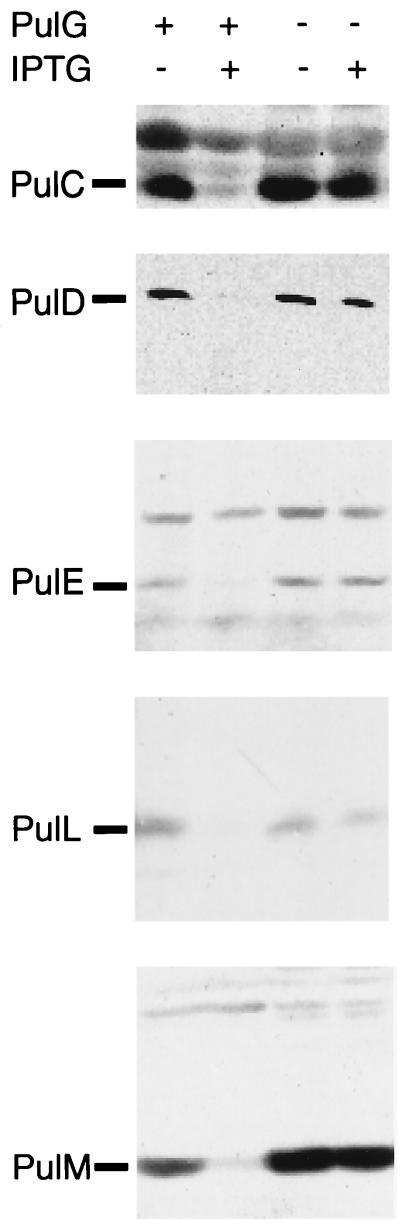
Overproduction of certain secreton components has been reported to diminish secretion, possibly because stoichiometric imbalance disrupts interactions between secreton components and causes loss of secreton function (2, 29). Since such an approach could indicate interactions within the secreton, we examined the effects of increased expression of individual pul genes in strain PAP7500B on pullulanase secretion and the levels of other Pul proteins. Overexpression of pul genes S, C, D, E, H, I, J, K, M, and O had no effect on pullulanase secretion. However, PulG overproduction blocked secretion, as observed previously (29), and reduced PulC, PulD, PulE, PulL, and PulM to low or barely detectable levels (Fig. 2). The level of coregulated pullulanase produced by the cells (as measured by specific enzymatic activity) was unaffected by high-level production of PulG (data not shown), indicating that expression of the pul gene cluster is unaffected. Thus, overproduction of PulG seems to destabilize Pul proteins C, D, E, L, and M.
FIG. 2.
Effect of increased expression of pulG carried by pCHAP1205 on the levels of PulC, PulD, PulE, PulL, and PulM proteins in strain PAP7500B grown in maltose to induce expression of the chromosomal pul genes. Paired samples from the same number of bacteria from noninduced and IPTG-induced cultures are shown. Control cells (PulG −) carried the vector pSU18. Protein extracts were blotted onto nitrocellulose and detected with antibodies against the indicated Pul proteins. In some panels, bands that are recognized nonspecifically by the antibodies serve as internal controls.
Increased expression of pul genes F and L also reduced secretion to <30% (pulL) or abolished it completely (pulF). In none of these cases did we observe a dramatic effect on the levels of other Pul proteins similar to that caused by PulG overproduction (data not shown). The inhibitory effects of increased pulL expression could not be suppressed by increasing pulE expression (data not shown), which is in contrast to results reported recently in similar studies with the E and L components of the Eps, Xcp, and Out secretons (1, 34, 36, 37). The negative interference phenotype caused by increased pulL expression could be due to interactions involving the cytoplasmic N-terminal domain of PulL, because high-level production of this domain (in strains carrying pCHAP5112) also caused a 90% drop in pullulanase secretion. Once again, however, the negative interference effect was not overcome by simultaneously increasing PulE levels.
Cross-linking and immunoprecipitation to probe interactions involving PulC, PulE, PulG, PulL, and PulM.
The data presented so far suggest possible interactions involving Pul secreton components C, E, G, L, and M. In an attempt to obtain more direct evidence for such interactions, we studied the effects of the different pul gene mutations on the formation of multimers that could be stabilized by cross-linking. The mutants were treated with four different amine-specific cross-linkers with different spacer lengths (EGS, DSP, disuccimidyl tartrate, and formaldehyde) under optimized conditions, and proteins were blotted onto nitrocellulose sheets that were probed with antibodies to specific Pul proteins.
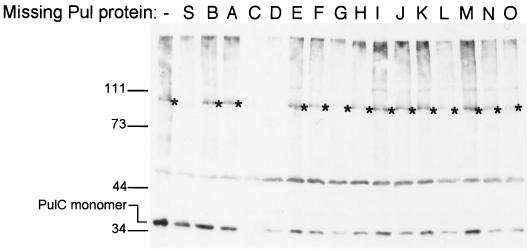
PulC formed the previously observed 110-kDa complex (26) after treatment with all four cross-linkers except when PulS or PulD was absent (Fig. 3; note that variations in the level of the 110-kDa band detected in other strains were not reproducible). Since PulS is required for PulD assembly into the outer membrane (9), these results indicate that formation of the 110-kDa complex requires PulC and correctly assembled PulD and does not involve any other Pul protein. As reported previously (26), we conclude that the 110-kDa band is either a PulC trimer or, less likely, a PulC-PulD heterodimer.
FIG. 3.
Formation of the 110-kDa multimer containing PulC after cross-linking with DSP requires PulS and PulD but not any of the other components of the Pul secreton. The bacteria were maltose-induced HS2019 carrying pCHAP231 derivatives lacking different pul genes (Table 2). The position of the ca.-110-kDa cross-linked PulC multimer is indicated by an asterisk. Differences in yields of PulC monomer and multimer in different strains were not reproducible. Size markers (kilodaltons) and PulC monomers are indicated on the left.
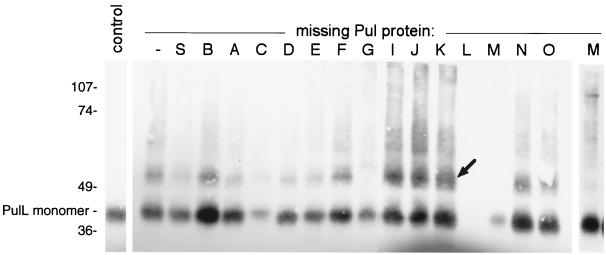
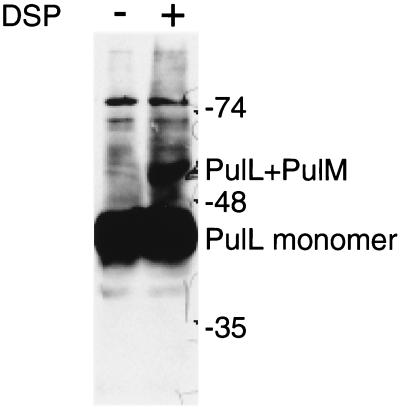
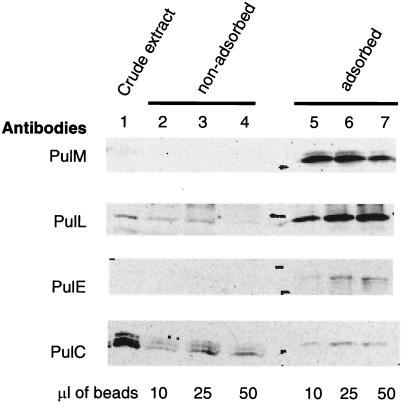
A 52-kDa cross-linked product that reacted with PulL antibodies was observed after treatment with EGS (Fig. 4) and DSP but not with the other cross-linkers (data not shown). Formation of this band did not occur in mutants lacking PulG or PulM and was increased in mutants devoid of PulI, PulJ, and PulK (Fig. 4). The size of this band is close to that expected for PulL-PulG or PulL-PulM heterodimers. Both PulG and PulM produced cross-linked bands (possibly homomultimers) of ca. 52 kDa even in the absence of PulL, making it impossible to determine whether the 52-kDa band detected by the PulL antibodies band contained one or another of these proteins. However, three lines of evidence suggest that it is a heterodimer of PulL and PulM. First, the size of the band was unaffected by a mutation in pulO, which abolishes processing of PulG and thereby causes it to migrate more slowly during SDS-PAGE (reference 29 and data not shown). Second, a 52-kDa band was also observed after DSP (Fig. 5) or EGS (data not shown) cross-linking of cells carrying pCHAP2230 (pulL) plus pCHAP1354 (pulM). Third, purified antibodies coupled to magnetic beads could immunoprecipitate not only PulM but also PulL and, to a lesser extent, PulC and PulE from detergent (Triton X-100)-solubilized extracts of E. coli K-12(pCHAP231) (Fig. 6). We could not apply this assay to other Pul proteins because they were either insoluble in the detergent used or bound nonspecifically to the magnetic beads.
FIG. 4.
Formation of a 52-kDa band that reacts with PulL antibodies after treatment of cells producing secreton components with EGS. The bacteria were maltose-induced HS2019 carrying pCHAP231 derivatives lacking different pul genes (Table 2). The control sample was HS2019(pCHAP231) that was not treated with EGS. The lane at the extreme right (labeled M) shows a prolonged chemiluminescence exposure of the sample from the strain lacking PulM. Proteins were separated by SDS-PAGE on an 8% acrylamide gel. Size markers (kilodaltons) and the position of the PulL monomer are indicated on the left, and the position of the 52-kDa band is indicated by an arrow. The higher yields of cross-linked product in strains lacking PulI, PulJ, or PulK were reproducible.
FIG. 5.
Formation of a cross-linked multimer of ca. 52 kDa in IPTG-induced cells of PAP7500B carrying pCHAP2230 (pulL) and pCHAP1354 (pulM), as detected by SDS-PAGE and immunoblotting with PulL antibodies. The cross-linker was DSP. The positions of the PulL monomer, the presumed PulL-PulM heterodimer, and molecular size markers (in kilodaltons) are indicated at the right.
FIG. 6.
Coimmunoprecipitation of Pul proteins L, E, and C together with PulM by purified antibodies directed against PulM. Lane 1 shows the relative abundance of the proteins in the crude extracts. Lanes 2 to 4 show the unadsorbed proteins after incubation with different volumes of magnetic beads coated with the antibodies. Lanes 5 to 7 show the proteins adsorbed onto the beads. Note that the PulC antiserum detected three bands, only one of which (the upper band) corresponds to PulC and was immunoprecipitated.
In no case did we find any evidence for the presence of cross-linked PulL or PulE homomultimers. However, small amounts of what appeared to be PulM homodimers were observed after cross-linking of E. coli K-12(pCHAP231) with DSP and EGS. Formation of these homodimers was also observed in cells expressing only PulM, where they could be detected even in samples that had not been treated with cross-linkers (data not shown).
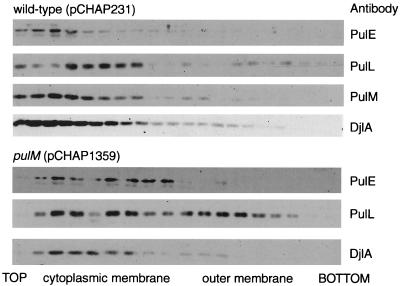
Factors required for cytoplasmic membrane association of PulE.
The locations of PulE, PulL, and PulM were determined by floating membranes through centrifuged sucrose gradients. PulE, PulL, and PulM were all found exclusively in the membrane (pellet) fraction after ultracentrifugation of French press-disrupted E. coli producing the full set of Pul proteins (pCHAP231) and were recovered almost exclusively from the cytoplasmic membrane fraction near the top of the sucrose gradients (Fig. 7). The deletion in pulM (pCHAP1359) reduced the amounts of PulE and especially PulL recovered after fractionation, but they remained cytoplasmic membrane associated (Fig. 7). Deletion of pulF (pCHAP1227) had no effect whatsoever on the location of PulE, PulL, or PulM (data not shown), while deletion of pulL (pCHAP1217) resulted in failure to detect PulE in membranes after fractionation (see above) without affecting the localization of PulM (data not shown).
FIG. 7.
Localization of PulE, PulL, and PulM proteins in HS2019(pCHAP231) (wild type) and effect of ΔpulM mutation introduced into pCHAP231 (to give pCHAP1359) on the localization of PulE and PulL. Membranes were separated by flotation through centrifuged sucrose gradients, and proteins were detected by immunoblotting. Antibodies against the cytoplasmic membrane protein DjlA were used as a control. The outer membrane fraction was identified by the presence of porins and OmpA protein. The PulE and PulL immunoblots in the ΔpulM strain were developed for longer than were the immunoblots of membranes from the wild-type strain to compensate for the presence of lower amounts of these proteins.
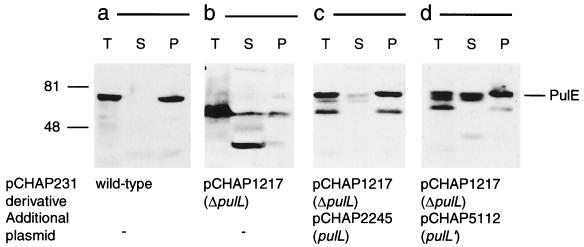
The instability of PulE provoked by the pulL deletion (pCHAP1217) (Fig. 8b) was corrected by introducing pCHAP2245 (pulL) (Fig. 8c). PulE was also stabilized in E. coli(pCHAP1217) by pCHAP5112, which encodes only the N-terminal, cytoplasmic region of PulL (PulL′ [Fig. 8d]), but pullulanase secretion was not restored (data not shown). In this strain, approximately 50% of PulE was in the soluble fraction (Fig. 8d). This is the first time that we have observed a soluble form of PulE that might be amenable to biochemical analysis.
FIG. 8.
Protection of PulE by PulL and its sedimentation in total membrane fraction. French press lysates from spheroplasts of E. coli strain PAP105 carrying the complete pul gene cluster in pCHAP231 (a), pCHAP231 ΔpulL (pCHAP1217) (b), pCHAP231 ΔpulL (pCHAP1217) and pCHAP2245 in which pulL is under IPTG-inducible lacZp control (c), or pCHAP231 ΔpulL (pCHAP1217) and pCHAP5112 in which the 5′ end (247 codons) of pulL is under IPTG-inducible lacZp control (d) were fractionated by ultracentrifugation. PulE was detected by immunoblotting. Molecular size markers (kilodaltons) are indicated on the left. T, whole bacteria; S, soluble fraction; P, total membrane (pellet) fraction.
To investigate whether the PulE in the pellet fraction was membrane associated or aggregated, it was subjected to flotation through sucrose gradients. In control membranes from strains carrying both pCHAP1217 (pCHAP231 ΔpulL) and pCHAP2245 (pulL), PulE was found mainly in the cytoplasmic membrane (Fig. 9a), as expected, whereas some PulL was found in intermediate-density and outer membrane fractions as well as in the cytoplasmic membrane (Fig. 9b). In contrast, in cells carrying both pCHAP1217 and pCHAP5112 (pulL′), the carboxy-terminally truncated form of PulL that was recovered in the pellet (membrane fraction) remained at the bottom of the gradients, indicating that it is not membrane associated (Fig. 9d). Nevertheless, PulE was membrane associated in these cells, forming a broad peak that spanned from the cytoplasmic membrane through the outer membrane to the soluble-aggregated fraction at the bottom of the gradient (Fig. 9c). Thus, PulE is stable and associates to at least some extent with the cytoplasmic membrane even when PulL itself is unable to associate with this membrane.
FIG. 9.
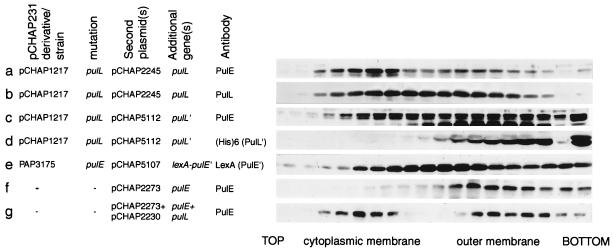
Association of PulE and PulL and a LexA-PulE′ hybrid protein with the cytoplasmic membrane in strains with different combinations of secreton components. Membranes prepared by disrupting whole bacteria or spheroplasts in a French press were separated by flotation through density sucrose gradients. (a and b) E. coli PAP105 carrying pCHAP1217 bearing the full set of pul genes except pulL and with pulL provided in trans on pCHAP2245; (c and d) like gradients a and b except that the in trans plasmid was pCHAP5112 coding for truncated PulL (PulL′); (e) PAP3175 with the complete pul gene cluster in the chromosome but with the ΔpulE mutation and bearing pCHAP5107 (lexA-pulE′); (f) E. coli PAP105 expressing pulE alone (pCHAP2273); (g) E. coli PAP105 expressing pulE (pCHAP2273) and pulL (pCHAP2230) from compatible plasmids (note that, for gradients f and g, secreton components other than PulE and PulL are absent). Proteins were detected by immunoblotting with antibodies specific for PulE, PulL, LexA (for LexA-PulE′ hybrid), or (His)6 (for PulL′). Only those parts of the immunoblots displaying PulE, LexA-PulE′, PulL, or PulL′ are shown.
Previous studies suggested that the amino-terminal region of PulE and its homologue EpsE might be involved in their association with the cytoplasmic membrane (25, 36). Therefore, we examined the location of the amino-terminal region of PulE (amino acids 1 to 177) by expressing a lexA-pulE′ fusion in a strain carrying the full set of pul genes with the ΔpulE3 deletion in the chromosome [PAP3175(pCHAP5107)]. The hybrid protein was recovered in both cytoplasmic membrane and intermediate fractions (Fig. 9e), indicating that it is able to associate with the membrane (presumably via PulL). Some of the LexA-PulE′ formed aggregates that appeared in higher-density fractions.
To determine whether PulL alone is sufficient for the association of PulE with the cytoplasmic membrane, we examined the location of these proteins in the absence of all other secretion components. For these experiments, we used pCHAP2273 coding for wild-type PulE lacking the 38-amino-acid N-terminal extension resulting from the fusion of lacZ and pulE coding regions in previously used plasmids (25). As reported previously (25), PulE aggregated when produced alone (Fig. 9f), but when pulE and pulL were coexpressed [PAP105(pCHAP2273, pCHAP2230)], PulE was evenly distributed between the cytoplasmic membrane and high-density (aggregated) fractions (Fig. 9g). PulL alone was found in the cytoplasmic membrane (data not shown; note that PulL synthesized alone is apparently stable in the absence of PulM). Thus, PulL prevents PulE aggregation and colocalizes with PulE in the cytoplasmic membrane. This result confirms the direct interaction between PulE and PulL.
Erwinia out genes can complement pul mutations.
The data presented above suggest the existence of a tight interaction between PulE and PulL that is modulated by PulM and possibly by PulC and PulG. Heterologous complementation can provide valuable information on secreton specificity and interactions between different secreton components (5, 17, 36). Here, we extended previous studies (8, 11, 26) by analyzing the ability of E. chrysanthemi and E. carotovora out genes to substitute for their pul homologues. The results (Table 3) show that all pul genes except pulC and pulD but including pulE, pulG, pulL, and pulM could be replaced by the corresponding out genes, although full complementation was sometimes obtained only at high levels of (IPTG-induced) expression.
DISCUSSION
We report several different approaches (protease protection, cross-linking, immunoprecipitation, and subcellular fractionation) to probe protein-protein interactions involving components of the pullulanase secretion complex, the secreton. The main objective was to study the pivotal PulE protein and its interactions with the cytoplasmic membrane. Before discussing the broader implications and specific questions raised by our work, we will summarize the data and indicate the limitations of the different techniques.
The protease protection data provide indirect evidence of protein-protein interactions. PulE is destabilized in the absence of PulL, and PulL is destabilized in the absence of PulC and PulM. Therefore, one can conclude that these four proteins probably form a complex, but one cannot determine which proteins interact directly, nor can one exclude the involvement of other proteins in the complex. No other new interactions were revealed by these assays.
Cross-linking data provide more direct information on protein-protein interactions. However, the fact that the absence of one protein from the secreton changes the ability of a particular cross-linker to stabilize multimers formed by another does not necessarily indicate that the two proteins interact directly, although it is probably safe to assume that they are in the same multiprotein complex. For the four proteins mentioned above, PulM formed homodimers, even in the absence of all other secreton components (data not shown), and PulL and PulM probably form a heterodimer (Fig. 4 and 5). PulG seems to influence the formation of this heterodimer when all other secreton components are present (see below). The cross-linking data do not provide any evidence of PulE or PulL homodimers or of an interaction between PulE and PulL. Formation of the previously described 110-kDa cross-linked product detected by antibodies against PulC is shown here to be unaffected by mutations in any of the other pul genes except D and S. Therefore, we can extend the previous interpretation that the 110-kDa product could represent a PulC homotrimer by adding that its detection requires the presence of PulD that is correctly localized and stabilized by the previously established interaction with PulS. Formation of this 110-kDa complex is independent of any interaction between PulC and PulE, PulL, or PulM.
The immunoprecipitation data show unambiguously that PulC, PulE, and PulL interact (directly or indirectly) with PulM but do not exclude the possibility that other proteins are present in the complex. The fractionation data, which confirm the intimate association between PulE and PulL, reveal several different states of PulE, depending on which other secreton components are present. PulE alone forms aggregates that float up the sucrose gradient to a position near that of the outer membrane fragments. When the rest of the secreton is present, aggregation is prevented and PulE associates with the cytoplasmic membrane. Removal of PulL from the secreton causes degradation of PulE. Finally, PulL alone can anchor PulE to the cytoplasmic membrane. The N-terminal, cytoplasmic domain of PulL alone did not promote association of PulE with the cytoplasmic membrane, but it could do so if all other secreton components (except full-length PulL) were present. This observation might indicate that, although PulE can associate with the membrane via PulL, it can also do so through another component of the secreton, provided it is protected from proteolysis and aggregation by interacting transitorily with the cytoplasmic domain of PulL. Finally, the fractionation data show that association of PulE with the cytoplasmic membrane involves the N-terminal region of the protein.
These data allow us to build up a first picture of the interactions between some of the Pul secreton components and lead us to propose that Pul proteins E, L, and M form a core subcomplex. Recent studies in several other laboratories have begun to reveal similar interactions between secreton components E, L, and M (1, 18, 34, 36, 37, 43). Taken together, the data indicate protein L-mediated cytoplasmic membrane association of protein E via the N-terminal regions of the two proteins and indicate that interaction between the L and M proteins probably occurs through their periplasmic domains (34). In addition, dimerization of XcpR (XcpE) via its amino-terminal domain was also reported (45), OutE and OutL both form homodimers in a two-hybrid system (34), PulE forms disulfide-bonded dimers when cells are disrupted (24, 25), and PulM forms homodimers (data not shown). Thus, the E-L-M subcomplex probably contains two copies of each of its components.
There is also increasing evidence that the putative E-L-M subcomplex interacts with other secreton components. For example, we show for the first time that PulC might interact with the PulELM subcomplex, as indicated by the fact that its absence destabilizes PulL and that it can be immunoprecipitated by antibodies against PulM. It will be recalled that PulC also interacts (directly or indirectly) with PulD (Fig. 3 and reference 26). PulG protein also seems to interact with the ELM complex. Kagami et al. reported a conditional mutation in the P. aeruginosa secreton gene encoding the pseudopilin XcpT (XcpG) that was suppressed by a second mutation in xcpR (xcpE) (13). Here, we extend this observation by reporting that PulG is required to form the cross-linked PulL-PulM heterodimer when the rest of the secreton is present. Interestingly, the effect of inactivating pulG (failure to form the PulL-M heterodimer) was the opposite of that observed when three other pseudopilin genes (pulI, pulJ, and pulK) were inactivated (consistently increased levels of this heterodimer) (Fig. 4). Presumably, the pseudopilins interact with the PulELM complex but in ways that differentially affect the contact between proteins L and M. Surprisingly, PulL and PulM can be cross-linked to form a heterodimer in the complete absence of pseudopilins and all other secreton components (Fig. 5), which suggests that the pseudopilins affect this heterodimer only when the rest of the secreton is present. One possible interpretation of these results is that the PulELM complex is involved in the assembly of pseudopilins into a putative supramolecular structure that we have tentatively called the pseudopilus (28, 29, 32). The interaction between PulL and PulM or between PulM monomers might occur when only these two proteins are present or when the pseudopilins are being assembled. However, the absence of PulG might prevent pseudopilus assembly, causing a change in PulL-PulM interactions such that cross-linking can no longer stabilize them. The absence of the other pseudopilins might provoke the opposite effect, causing increased assembly of PulG into pseudopili and increasing the interactions between PulL and PulM.
It is interesting that overproduction of PulG reduced the cellular levels of at least five Pul proteins (C, D, E, L, and M), causing a total block in pullulanase secretion (29). Previously, we reported that the secretion defect resulting from overproduction of PulG could not be suppressed by increasing the expression of other, individual pul genes (29), implying that no particular Pul protein was titrated by high levels of PulG. The fact that increasing the level of PulG causes a decrease in the levels of several secreton components provides an explanation for this observation.
The Pul secreton cannot secrete the cellulase CelZ, which is a substrate for the E. chrysanthemi Out secreton (39). However, the corresponding out genes from either E. chrysanthemi or E. carotovora can complement mutations in all but two of the pul genes (C and D). These results have three important implications. First, the only secreton components that can be responsible for substrate specificity are C and D, as suggested previously by Lindeberg et al. (17). Second, the proposed PulELM subcomplex can form even when one of the partners is replaced by the homologous Out protein. Third, Out pseudopilins can substitute for their Pul counterparts and be assembled in functional form by the PulELM subcomplex. It is well established that type IV pilins from one species of bacterium can be assembled into pili by the type IV pilus assembly machinery of other species (38). It will be interesting to determine whether homologous components of the type IV pilus machinery and the secreton are interchangeable, as is the case for prepilin peptidase (31), and whether the secreton can be used to assemble type IV pili.
Secreton components E and L are not interchangeable between the closely related species V. cholerae and A. hydrophila (36), despite the fact that they can secrete proteins normally secreted by the other (10, 12, 21). This situation is different from that described here for the Pul and Out secretons, presumably reflecting the independent evolution of substrate-specific determinants (proteins C and D [17]) and other, structural or mechanical components of the secreton (E and L). From this, we predict that secreton components C and D might be interchangeable between V. cholerae and A. hydrophila and that the incompatibility between the E and L proteins of Vibrio and Aeromonas probably represents divergent coevolution of these two components in the different species.
The data do not provide any evidence for a link between PulF and the PulELM complex, which is surprising in view of the fact that genes encoding PulF homologues are invariably adjacent to a pulE homologue (27). The role of PulF in pullulanase secretion remains to be clarified, but it is the most likely candidate for the protein that anchors PulE to the cytoplasmic membrane when PulL is synthesized without its periplasmic domain and its transmembrane anchor.
On the basis of the data presented here and elsewhere, we can begin to construct a conceptual model of the secreton. We propose that the ELM subcomplex in the cytoplasmic membrane is involved in the export of pseudopilins and/or their assembly into a pilus-like structure. This process could be driven by ATP hydrolysis via PulE. The pilus-like structure could act as a scaffold for the assembly of other secreton components or could play an active role in secretion, for example, by providing a conduit linking the periplasm to the outside of the cell. The fact that two lines of evidence show a link between the cytoplasmic membrane ELM subcomplex and the C protein is particularly intriguing because it has already been proposed that this protein could form a bridge between the two membranes (2, 26). Furthermore, PulC probably interacts with PulD (secretin) (26), the protein that probably forms the channel via which pullulanase crosses the outer membrane (20). Thus, we propose that PulC could control opening and closing of the secretin channel via specific recognition of translocation substrates (in this case, pullulanase) and transduction of energy from the proton gradient across the cytoplasmic membrane (23). This model for secreton function can now be tested by purification and analysis of its component parts and by isolation of the complete machinery.
ACKNOWLEDGMENTS
We thank Jean-Michel Betton, David Clarke, Alan Collmer, Michaël Kornacker, Stephen Lory, Benedict Michel, Noreen Murray, and Howard Shuman for strains, plasmids, or antibodies. We are grateful to Nathalie Nadeau for technical assistance and to all members of the secretion laboratory for their interest and encouragement.
This work was supported by the European Union (Training and Mobility in Research grant number FMRX-CT96-0004) and by the French Research Ministry (Programme fondamental en Microbiologie et Maladies infectieuses et parasitaires). F.E. is supported by a grant from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Ball G, Chapon-Hervé V, Bleves S, Michel G, Bally M. Assembly of XcpR in the cytoplasmic membrane is required for extracellular protein secretion in Pseudomonas aeruginosa. J Bacteriol. 1999;181:382–388. doi: 10.1128/jb.181.2.382-388.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bleves S, Gérard-Vincent M, Lazdunski A, Filloux A. Structure-function analysis of XcpP, a component involved in general secretory pathway-dependent protein secretion in Pseudomonas aeruginosa. J Bacteriol. 1999;181:4012–4019. doi: 10.1128/jb.181.13.4012-4019.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blum P, Holzschu D, Kwan H S, Riggs D, Artz S. Gene replacement and retrieval with recombinant M13mp bacteriophages. J Bacteriol. 1989;171:538–546. doi: 10.1128/jb.171.1.538-546.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brockman R W, Heppel L A. On the localization of alkaline phosphatase and cyclic phosphodiesterase in Escherichia coli. Biochemistry. 1968;7:2554–2562. doi: 10.1021/bi00847a016. [DOI] [PubMed] [Google Scholar]
- 5.de Groot A, Krijger J-J, Filloux A, Tommassen J. Characterization of type II protein secretion (xcp) genes in the plant growth-stimulating Pseudomonas aeruginosa, strain WCS358. Mol Gen Genet. 1996;250:491–504. doi: 10.1007/BF02174038. [DOI] [PubMed] [Google Scholar]
- 6.d'Enfert C, Pugsley A P. Klebsiella pneumoniae pulS gene encodes an outer membrane lipoprotein required for pullulanase secretion. J Bacteriol. 1989;171:3673–3679. doi: 10.1128/jb.171.7.3673-3679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.d'Enfert C, Ryter A, Pugsley A P. Cloning and expression in Escherichia coli of the Klebsiella pneumoniae genes for production, surface localization and secretion of the lipoprotein pullulanase. EMBO J. 1987;6:3531–3538. doi: 10.1002/j.1460-2075.1987.tb02679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guilvout I, Hardie K R, Sauvonnet N, Pugsley A P. Genetic dissection of the outer membrane secretin PulD: are there distinct domains for multimerization and secretion specificity? J Bacteriol. 1999;181:7212–7220. doi: 10.1128/jb.181.23.7212-7220.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hardie K R, Lory S, Pugsley A P. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 1996;15:978–988. [PMC free article] [PubMed] [Google Scholar]
- 10.Hardie K R, Schulze A, Parker M W, Buckley J T. Vibrio spp. secrete proaerolysin as a folded dimer without the need for disulphide bond formation. Mol Microbiol. 1995;17:1035–1044. doi: 10.1111/j.1365-2958.1995.mmi_17061035.x. [DOI] [PubMed] [Google Scholar]
- 11.Hardie K R, Seydel A, Guilvout I, Pugsley A P. The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol Microbiol. 1996;22:967–976. doi: 10.1046/j.1365-2958.1996.01539.x. [DOI] [PubMed] [Google Scholar]
- 12.Hirst T R, Leece R. The phenomenon of toxin secretion by vibrios and aeromonads. Experientia. 1991;47:429–431. [PubMed] [Google Scholar]
- 13.Kagami Y, Ratliff M, Surber M, Martinez A, Nunn D. Type II protein secretion by Pseudomonas aeruginosa: genetic suppression of a conditional mutation in the pilin-like component XcpT by the cytoplasmic component XcpR. Mol Microbiol. 1998;27:221–233. doi: 10.1046/j.1365-2958.1998.00679.x. [DOI] [PubMed] [Google Scholar]
- 14.Kornacker M G, Pugsley A P. Molecular characterization of pulA and its product, pullulanase, a secreted enzyme of Klebsiella pneumoniae UNF5023. Mol Microbiol. 1989;4:73–85. doi: 10.1111/j.1365-2958.1990.tb02016.x. [DOI] [PubMed] [Google Scholar]
- 15.Kornacker M G, Remsburg B, Menzel R. Gene activation by the AraC protein can be inhibited by DNA looping between AraC and a LexA repressor that interacts with AraC: possible applications as a two-hybrid system. Mol Microbiol. 1998;30:615–624. doi: 10.1046/j.1365-2958.1998.01096.x. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 17.Lindeberg M, Salmond G P C, Collmer A. Complementation of deletion mutations in a cloned functional cluster of Erwinia chrysanthemi out genes with Erwinia carotovora out homologs reveals OutC and OutD as candidate gatekeepers of species-specific secretion of proteins via the type II pathway. Mol Microbiol. 1996;20:175–190. doi: 10.1111/j.1365-2958.1996.tb02499.x. [DOI] [PubMed] [Google Scholar]
- 18.Michel G, Bleves S, Ball G, Lazdunski A, Filloux A. Mutual stabilization of the XcpZ and XcpY components of the secretory apparatus in Pseudomonas aeruginosa. Microbiology. 1998;144:3379–3386. doi: 10.1099/00221287-144-12-3379. [DOI] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 20.Nouwen N, Ranson N, Saibil H, Wolpensinger B, Engel A, Ghazi A, Pugsley A P. Secretin PulD: association with pilot protein PulS, structure and ion-conducting channel formation. Proc Natl Acad Sci USA. 1999;96:8173–8177. doi: 10.1073/pnas.96.14.8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overbye Michel, L, Sandkvist M, Bagdasarian M. Specificity of the protein secretory apparatus: secretion of the heat-labile enterotoxin B subunit pentamers by different species of Gram-negative bacteria. Gene. 1995;152:41–45. doi: 10.1016/0378-1119(94)00691-k. [DOI] [PubMed] [Google Scholar]
- 22.Possot O, d'Enfert C, Reyss I, Pugsley A P. Pullulanase secretion in Escherichia coli K12 requires a cytoplasmic protein and a putative polytopic cytoplasmic membrane protein. Mol Microbiol. 1992;6:95–105. doi: 10.1111/j.1365-2958.1992.tb00841.x. [DOI] [PubMed] [Google Scholar]
- 23.Possot O, Letellier L, Pugsley A P. Energy requirement for pullulanase secretion by the main terminal branch of the general secretory pathway. Mol Microbiol. 1997;24:457–464. doi: 10.1046/j.1365-2958.1997.3451726.x. [DOI] [PubMed] [Google Scholar]
- 24.Possot O, Pugsley A. The conserved tetracysteine motif in the general secretory pathway component PulE is required for efficient pullulanase secretion. Gene. 1997;192:45–50. doi: 10.1016/s0378-1119(97)00009-7. [DOI] [PubMed] [Google Scholar]
- 25.Possot O, Pugsley A P. Molecular characterization of PulE, a protein required for pullulanase secretion. Mol Microbiol. 1994;12:287–299. doi: 10.1111/j.1365-2958.1994.tb01017.x. [DOI] [PubMed] [Google Scholar]
- 26.Possot O M, Gérard M, Pugsley A P. Membrane association and multimerization of secreton component PulC. J Bacteriol. 1999;181:4004–4011. doi: 10.1128/jb.181.13.4004-4011.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pugsley A P. Multimers of the precursor of a type IV pilin-like component of the general secretory pathway are unrelated to pili. Mol Microbiol. 1996;20:1235–1245. doi: 10.1111/j.1365-2958.1996.tb02643.x. [DOI] [PubMed] [Google Scholar]
- 29.Pugsley A P. Processing and methylation of PulG, a pilin-like component of the general secretory pathway of Klebsiella oxytoca. Mol Microbiol. 1993;9:295–308. doi: 10.1111/j.1365-2958.1993.tb01691.x. [DOI] [PubMed] [Google Scholar]
- 30.Pugsley A P, d'Enfert C, Reyss I, Kornacker M G. Genetics of extracellular protein secretion by Gram-negative bacteria. Annu Rev Genet. 1990;24:67–90. doi: 10.1146/annurev.ge.24.120190.000435. [DOI] [PubMed] [Google Scholar]
- 31.Pugsley A P, Dupuy B. An enzyme with type IV prepilin peptidase activity is required to process a component of the general extracellular protein secretion pathway of Klebsiella oxytoca. Mol Microbiol. 1992;6:751–760. doi: 10.1111/j.1365-2958.1992.tb01525.x. [DOI] [PubMed] [Google Scholar]
- 32.Pugsley A P, Possot O. The general secretory pathway of Klebsiella oxytoca: no evidence for relocalization or assembly of pilin-like PulG protein into a multiprotein complex. Mol Microbiol. 1993;10:665–674. doi: 10.1111/j.1365-2958.1993.tb00938.x. [DOI] [PubMed] [Google Scholar]
- 33.Pugsley A P, Reyss I. Five genes at the 3′ end of the Klebsiella pneumoniae pulC operon are required for pullulanase secretion. Mol Microbiol. 1990;4:365–379. doi: 10.1111/j.1365-2958.1990.tb00604.x. [DOI] [PubMed] [Google Scholar]
- 34.Py B, Loiseau L, Barras F. Assembly of the type II secretion machinery of Erwinia chrysanthemi: direct interaction and associated conformational change between OutE, the putative ATP-binding component and the membrane protein OutL. J Mol Biol. 1999;289:659–670. doi: 10.1006/jmbi.1999.2803. [DOI] [PubMed] [Google Scholar]
- 35.Reyss I, Pugsley A P. Five additional genes in the pulC-O operon of the Gram-negative bacterium Klebsiella oxytoca UNF5023 that are required for pullulanase secretion. Mol Gen Genet. 1990;222:176–184. doi: 10.1007/BF00633815. [DOI] [PubMed] [Google Scholar]
- 36.Sandkvist M, Bagdasarian M, Howard S P, DiRita V J. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 1995;14:1664–1673. doi: 10.1002/j.1460-2075.1995.tb07155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandkvist M, Hough L P, Bagdasarian M M, Bagdasarian M. Direct interaction of the EpsL and EpsM proteins of the general secretion apparatus in Vibrio cholerae. J Bacteriol. 1999;181:3129–3135. doi: 10.1128/jb.181.10.3129-3135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauvonnet N, Gounon P, Pugsley A P. PpdD type IV pilin of Escherichia coli K-12 can be assembled into pili in Pseudomonas aeruginosa. J Bacteriol. 2000;182:848–854. doi: 10.1128/jb.182.3.848-854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sauvonnet N, Poquet I, Pugsley A P. Extracellular secretion of pullulanase is unaffected by minor sequence changes but is usually prevented by adding reporter proteins to its N- or C-terminal end. J Bacteriol. 1995;177:5238–5246. doi: 10.1128/jb.177.18.5238-5246.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sauvonnet N, Pugsley A P. Identification of two regions of Klebsiella oxytoca pullulanase that together are capable of promoting β-lactamase secretion by the general secretory pathway. Mol Microbiol. 1996;22:1–7. doi: 10.1111/j.1365-2958.1996.tb02650.x. [DOI] [PubMed] [Google Scholar]
- 41.Shevchik V E, Robert-Badouy J, Condemine G. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 1997;16:3007–3016. doi: 10.1093/emboj/16.11.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spratt B G, Hedge P J, te Heesen S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 43.Thomas J D, Reeves P J, Salmond G P C. The general secretion pathway of Erwinia carotovora subsp. carotovora: analysis of the membrane topology of OutC and OutF. Microbiology. 1997;143:713–720. doi: 10.1099/00221287-143-3-713. [DOI] [PubMed] [Google Scholar]
- 44.Treptow N A, Shuman H A. Genetic evidence for substrate and periplasmic-binding-protein recognition by the MalF and MalG proteins, cytoplasmic membrane components of the Escherichia coli maltose transport system. J Bacteriol. 1985;163:654–660. doi: 10.1128/jb.163.2.654-660.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turner L R, Olson J W, Lory S. The XcpR protein of Pseudomonas aeruginosa dimerizes via its N-terminus. Mol Microbiol. 1997;26:877–887. doi: 10.1046/j.1365-2958.1997.6201986.x. [DOI] [PubMed] [Google Scholar]