Abstract
Extracellular matrix (ECM) stiffening is a typical characteristic of cartilage aging, which is a quintessential feature of knee osteoarthritis (KOA). However, little is known about how ECM stiffening affects chondrocytes and other molecules downstream. This study mimicked the physiological and pathological stiffness of human cartilage using polydimethylsiloxane (PDMS) substrates. It demonstrated that epigenetic Parkin regulation by histone deacetylase 3 (HDAC3) represents a new mechanosensitive mechanism by which the stiffness matrix affected chondrocyte physiology. We found that ECM stiffening accelerated cultured chondrocyte senescence in vitro, while the stiffness ECM downregulated HDAC3, prompting Parkin acetylation to activate excessive mitophagy and accelerating chondrocyte senescence and osteoarthritis (OA) in mice. Contrarily, intra-articular injection with an HDAC3-expressing adeno-associated virus restored the young phenotype of the aged chondrocytes stimulated by ECM stiffening and alleviated OA in mice. The findings indicated that changes in the mechanical ECM properties initiated pathogenic mechanotransduction signals, promoted the Parkin acetylation and hyperactivated mitophagy, and damaged chondrocyte health. These results may provide new insights into chondrocyte regulation by the mechanical properties of ECM, suggesting that the modification of the physical ECM properties may be a potential OA treatment strategy.
Subject terms: Bone quality and biomechanics, Bone
Introduction
Osteoarthritis (OA) is the most common age-related and post-traumatic degenerative joint disease.1 There are more than 300 million patients with knee osteoarthritis (KOA) and is projected to become the disease with the highest disability rate globally by 2030.2,3 Although most of the etiological factors are still under investigation,4 advanced age and mechanical overloading recognized as the two most significant risk factors for OA development.5 All cells in the human body are subject to mechanical influences.6 This is particularly true of articular cartilage, given that its primary role is to transmit force to the underlying bone and decrease friction in the joint.7,8 Appropriate mechanical loading is essential for the health of articular cartilage, while mechanical overloading can result in degenerative lesions of articular cartilage, which can lead to the occurrence and development of OA.9,10
However, the specific mechanism behind the role of mechanical overloading in cartilage degeneration remains unclear. This is partially due to several factors. (1) Gaps in our understanding of whether the currently studied mechanical models of OA can summarize human disease. So far, most mechanical studies have used compression, tension, and shear force models for chondrocytes, while there is limited exploration on the stress of chondrocytes in patients with OA at daily resting state11,12; (2) The knowledge regarding the molecular mechanisms driving disease development is incomplete, particularly for KOA. KOA is characterized by interference between cells and the extracellular matrix (ECM) where they are located, leading to anabolic and catabolic imbalances. No clear inciting event is evident in most cases.
The primary constituents of cartilage are chondrocytes and their ECM. They are crucial for maintaining cartilage homeostasis and are frequently compromised in the malfunctioning cartilage associated with OA.13,14 Previous studies have demonstrated that the upregulated advanced glycation end-products (AGEs) in joint aging and increased lysyl oxidase (LOX) in cartilage injuries both accelerated matrix stiffening in vitro, due to the matrix-crosslinking and loss of glycosaminoglycans (GAGs).15,16 In fact, it has been demonstrated that matrix stiffness can directly regulate the behavior and phenotype of cells in vitro.17,18 Since chondrocytes represent the only cell type in articular cartilage, they can cause cartilage damage when they exhibit metabolic dysfunction.19 OA cartilage lesions display higher senescent chondrocyte levels than young, healthy cartilage, suggesting a strong correlation between chondrocyte senescence and OA severity.20 Interestingly, the removal of senescent cells from mouse joints not only prevents disease progression but also maintains tissue architecture.21 However, the role of ECM stiffness in the regulation of chondrocyte senescence and osteoarthritis progression is unclear. In addition, specific molecular targets directly related to ECM stiffness in osteoarthritis chondrocytes were not revealed. Therefore, the studies are needed to determine the chondrocyte phenotype under stimulation of ECM stiffening and the molecular targets for maintaining cartilage homeostasis.
In this study, we sought to clarify the biomechanics of ECM and the role of chondrocyte mechanotransduction in the initiation and progression of OA. Given the immediacy of ECM-cell contact, it is hypothesized that ECM stiffening is one of the earliest events of OA initiation, leading to mechanical chondrocyte regulation. Therefore, the ECM stiffness changes in a clear time range after injury, and its chondrocyte phenotype regulation is investigated. We found that ECM stiffening stimulated senescence in chondrocyte and in articular cartilage, and identified that histone deacetylase 3 (HDAC3) is a key factor in the regulation of chondrocyte senescence phenotype by matrix stiffness. Matrix stiffening downregulates HDAC3 to activate phosphatase and tensin homolog-induced kinase 1 (PINK1) and Parkin (PINK1/Parkin) mediated mitophagy, thereby stimulating chondrocyte senescence and accelerating the initiation and progression of osteoarthritis. Targeting HDAC3 or PINK1/Parkin signaling may represent a novel therapeutic approach for OA treatment.
Result
ECM stiffening induces chondrocyte senescence in OA patients and model mice
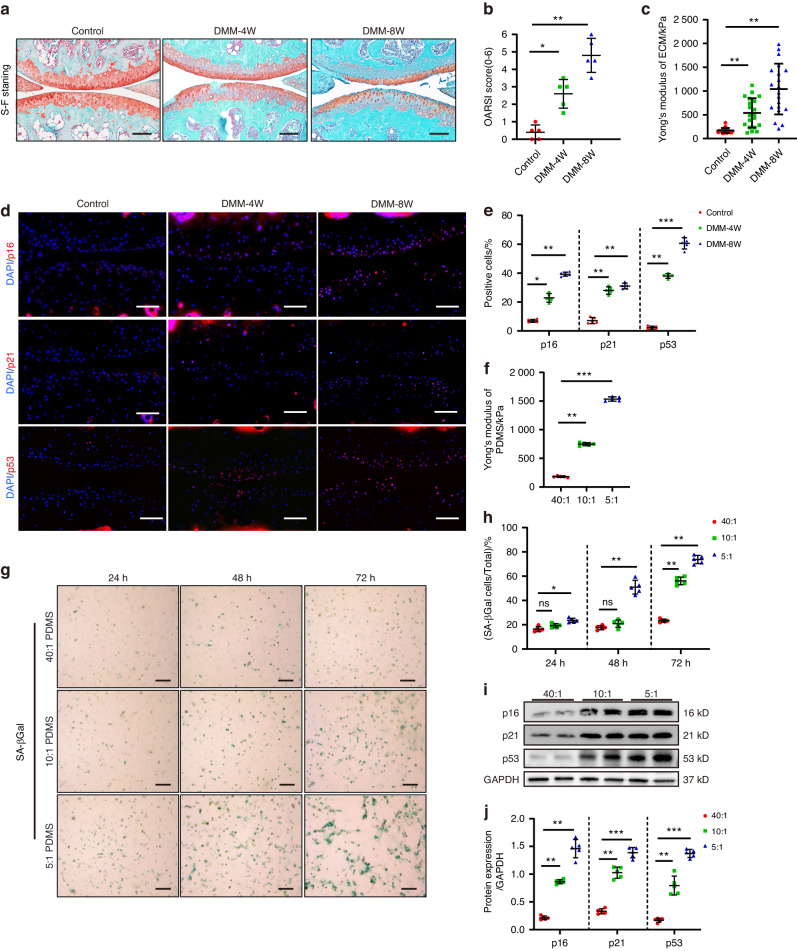
To examine the mechanical properties of cartilage ECM during OA in mice, this study used atomic force microscopy (AFM) to measure the ECM stiffness (Young’s modulus) of mouse medial tibial cartilage in a mechanical load inducted OA model (destabilization of the medial meniscus, DMM), since it reflected the most common area of human KOA.22 The results showed that the OA model mice displayed obvious surface fibrosis and structural cartilage defects at 4 weeks (4 W) and 8 weeks (8 W) (Fig. 1a, b), with cartilage stiffness values of 537.9 kPa and 1 040.5 kPa, respectively, which were about three- and sixfold higher than in normal cartilage (Fig. 1c). Consistent with these results, the ECM stiffness also increased in human OA cartilage samples (Fig. S1A). Therefore, increased ECM stiffness was positively correlated with the degree of cartilage damage. In addition, the number of stained articular chondrocytes for p16INK4a, p21, and p53 increased markedly in the DMM groups (Fig. 1d, e), which was characteristic of senescent cells.23,24 This suggests an association between ECM stiffness and the cartilage senescent phenotype.
Fig. 1.
ECM stiffening induced chondrocyte senescence in vitro and in mice. a Representative images of safranin O/fast green staining of controls and mice at 4 and 8 weeks after DMM surgery. Scale bars: 100 µm. b Quantitative analysis of the OARSI scale (n = 5). c Measurement of matrix stiffness (Young’s modulus) for controls and mice at 4 and 8 weeks after DMM surgery using atomic force microscopy (AFM) (n = 20). d Representative images of immunofluorescence of p16INK4a, p21, p53 in chondrocytes of controls and mice at 4 and 8 weeks after DMM surgery. Scale bars: 50 µm. e Quantification of p16 INK4a, p21, p53-positive chondrocytes as a proportion of the total chondrocytes of control and DMM mice (n = 5). Data are shown as mean ± SD. f Measurement of matrix stiffness (Young’s modulus) for 40:1, 10:1 and 5:1 PDMS using atomic force microscopy (AFM) (n = 5). g, h Representative images and quantification of SA-βGal staining in primary chondrocytes cultivated on PDMS substrates of different stiffness (40:1,10:1, 5:1) for 24, 48, and 72 h (n = 5). Scale bar: 50 µm. i, j Western Blotting analysis of p16INK4a, p21 and p53 expression in mouse primary chondrocytes cultivated on 40:1,10:1, 5:1PDMS for 48 h (n = 5). *P < 0.05, **P < 0.01, ***P < 0.001. ns not significant; Con control, DMM destablization of the medial meniscus, DAPI 4’,6-diamidino-2-phenylindole, OARSI Osteoarthritis Research Society International, 40:1, 10:1, 5:1, 40:1, 10:1, 5:1 polydimethylsiloxane (PDMS) substrates
To further evaluate the direct effect of different degrees of ECM stiffening on the chondrocyte phenotype, primary mouse chondrocytes were implanted on polydimethylsiloxane (PDMS) substrates at 40:1(≈180 kPa), 10:1(≈750 kPa), 5:1(≈1 515 kPa) since they overlapped with the physiological and pathological ECM stiffness range of mouse and human knee cartilage in this study (Fig. 1f). Compared with the chondrocytes cultivated on soft substrates, those grown on stiffness substrates exhibited senescence, phenotypes and slow cell proliferation, while propagation almost ceased entirely 48 h after inoculation. The chondrocytes cultivated on 5:1 PDMS substrates exhibited decreased activity 72 h after inoculation (Fig. S1B). The number of senescence-associated β-galactosidase (SA-βGal) -stained cells, a classic indicator of senescence, increased in a time-dependent manner after culturing on stiffness substrates (Fig. 1g, h). In addition, stiffness substrates increased the p16INK4a, p21 and p53 protein expression in the primary mouse chondrocytes (Fig. 1i, j), which was consistent with the findings regarding the proteins extracted from the articular cartilage of OA patients (Fig. S1C, D). Furthermore, the chondrocyte senescence phenotype regulation by matrix stiffness was consistent with the degree of cartilage component degradation, Col2a1 expression and upregulated MMP13 expression in the primary chondrocytes inoculated onto a stiffness matrix (Fig. S1E, F). These results demonstrated that ECM stiffening accelerated chondrocyte senescence in vitro and in articular cartilage, suggesting a potential mechanism in OA pathogenesis and development.
Reduced chondrocyte HDAC3 is associated with ECM stiffening in the articular cartilage of OA patients and aged mice
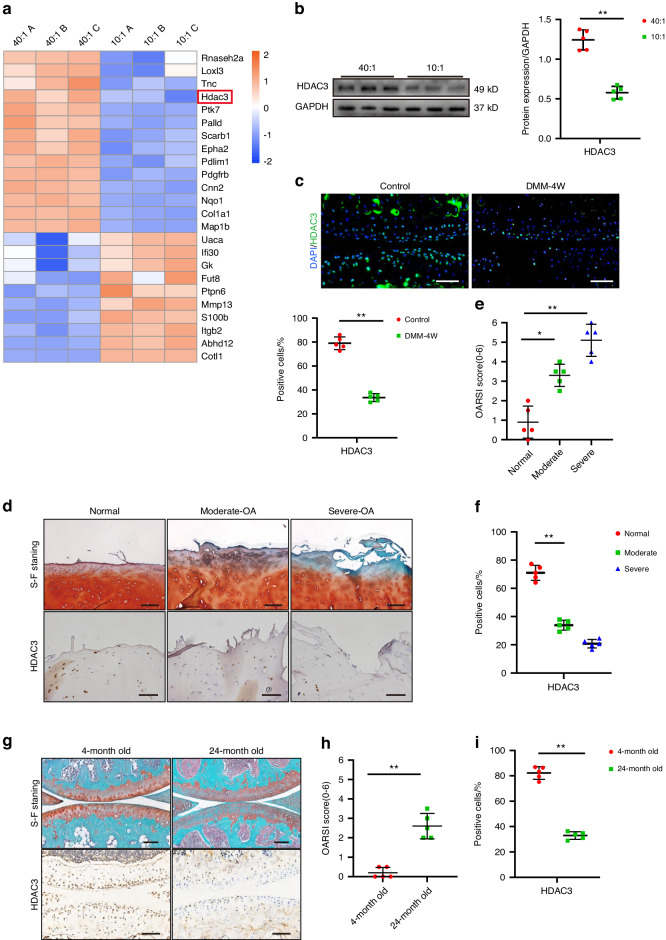
The ECM stiffening mechanism behind chondrocyte senescence stimulation was subsequently investigated. Quantitative proteomic analysis was performed to map the protein profiles of the primary mouse chondrocytes exposed to physiological and pathological stiffness substrate stimulation. Of the 24 differentially expressed proteins regulated by mechanical stimuli, the significant HDAC3 downregulation stood out (Fig. 2a). HDAC3 belongs to the histone deacetylase family and is essential for proper endochondral ossification during development and aging-related bone marrow obesity.25 However, its role in the chondrocyte senescence stimulated by ECM stiffening remains unclear. Western blotting confirmed significantly decreased HDAC3 levels in the chondrocytes stimulated by ECM stiffening (Fig. 2b). Consistent with this result, lower HDAC3 mRNA and protein levels were evident in primary human chondrocytes cultivated on stiffness substrates (Fig. S2A–C). In addition, immunofluorescence (IF) staining showed substantially decreased HDAC3 levels in the cartilage of OA mice suffering from ECM stiffening (Fig. 2c).
Fig. 2.
Chondrocyte HDAC3 is reduced by ECM stiffening and is decreased in the articular cartilage of OA patients. a Heatmap of mechanosensitive proteins differentially present in mouse chondrocytes under physiological and pathological stiffness stimuli identified by quantitative proteomics. b Western Blotting analysis of HDAC3 expression in mouse primary chondrocytes cultivated on Physiologic and Pathologic stiffness of PDMS substrates (n = 5). c Representative images and quantification of immunofluorescence of HDAC3 in chondrocytes of controls and mice at 4 weeks after DMM surgery. Scale bars: 50 µm. d Representative images of safranin O/fast green and IHC staining of HDAC3 in articular cartilage from normal and OA patients in moderately damaged and severely damaged. Scale bars: 50 µm. e Quantification of the OARSI scale based on staining results in (d) (n = 5). f Quantification of HDAC3-positive chondrocytes based on staining results in (d) (n = 5). g Representative images of safranin O/fast green and IHC staining of HDAC3 in chondrocytes of mice aged 4 and 24 months. Scale bars: 100 µm. h Quantification of the OARSI scale based on staining results in (g) (n = 5). i Quantification of HDAC3-positive chondrocytes based on staining results in (g) (n = 5). *P < 0.05, **P < 0.01. ns not signicicant. OA osteoarthritis; DAPI 4’,6-diamidino-2- phenylindole; OARSI Osteoarthritis Research Society International. 40:1, 10:1, 40:1, and 10:1 polydimethylsiloxane (PDMS) substrates
To determine whether the chondrocyte response to ECM stiffening stimuli involved decreased or increased HDAC3 protein production, primary mouse chondrocytes were treated with cycloheximide (CHX) (50 µm) to block new protein synthesis or MG132 (10 µm) to inhibit proteolysis during culturing on ECM stiffening substrates for 12 h. The HDAC3 expression level decreased significantly after treatment with CHX alone, while no significant difference was evident in the HDAC3 protein level after ECM stiffening stimulation, regardless of whether CHX treatment was employed (Fig. S2D, E). In addition, although proteolysis inhibition by MG132 increased the HDAC3 protein level in the control cells, it could not prevent the decrease in the HDAC3 protein level stimulated by ECM stiffening (Fig. S2F, G). These results indicated that the HDAC3 loss caused by ECM sclerosis was mainly due to reduced HDAC3 protein synthesis.
In addition, the reduced HDAC3 expression in OA patients was associated with cartilage damage, which was confirmed by immunohistochemical (IHC) staining and western blotting (Fig. 2d–f and Fig. S2H, I). Furthermore, lower HDAC3 expression was observed in aged mice. Compared with 6-month-old mice, the number of HDAC3-positive cells decreased significantly in the articular cartilage of 24-month-old mice (Fig. 2g–i). Taken together, these results suggest that HDAC3 may play a role in the association between ECM stiffening and chondrocyte senescence during OA development.
ECM stiffening activates PINK1/Parkin-mediated mitophagy, chondrocyte senescence, and joint degeneration in mice
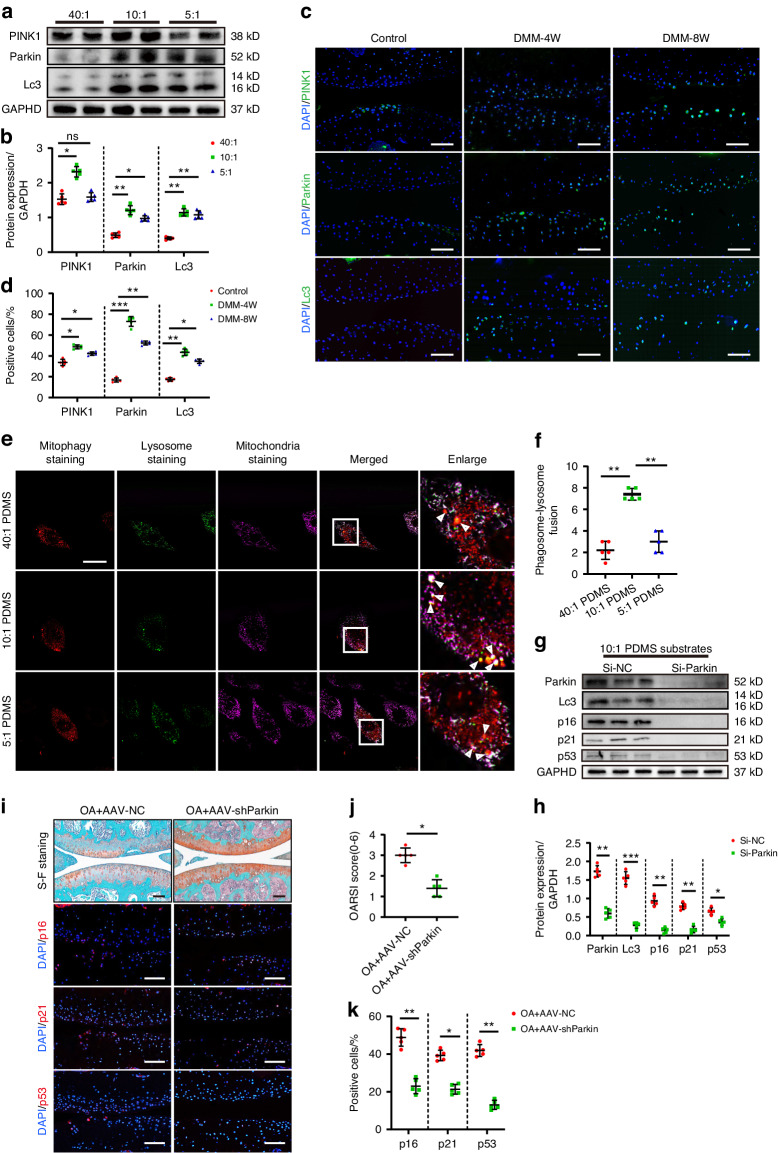
Further proteomic analysis showed that the mitochondrial compositional differences were highly correlated with those of ECM, actin fiber bundles, and stress fibers (Fig. S3A). Although studies have shown that excessive mitophagy contributes to many pathological conditions,26–28 the regulatory role and mechanism of ECM stiffening in mitophagy have not yet been clarified. Western blotting showed a significantly higher mitophagy level in the chondrocytes cultivated on pathological stiffness substrates than those on physiological stiffness substrates (Fig. 3a, b). However, the level of mitophagy was not completely consistent with the substrate stiffness. Compared with the moderate degeneration group, the chondrocyte mitophagy level was slightly reduced in the severe degeneration group, which was consistent with the observation of the chondrocytes in DMM modeled OA mice and OA patients (Fig. 3c, d and Fig. S3B, C). The mitochondrial proteins were directly extracted from the chondrocytes for validation, yielding the same results (Fig. S3D, E). The lysosomes and mitochondria were localized in the chondrocytes to further determine the mitophagy variation at different pathological stiffness stimuli. The damaged mitochondria fused with the lysosomes during the induction of mitophagy, releasing bright fluorescence. Consistent with previous results, the chondrocytes in the moderate degeneration group contained the most mitophagy vesicles (Fig. 3e, f). In addition, the moderate degeneration group exhibited the most significant decline in mitochondrial membrane potential, marking the early onset of mitochondrial damage (Fig. S3F, G). It is speculated that the excessive number of damaged mitochondria during high-stiffness stimulation leads to an absolute shortage of intracellular mitochondria, decreasing mitophagy activation. Electron microscopy confirmed the hypothesis. The number of damaged mitochondria in the severe degeneration group was significantly higher than in the moderate degeneration group, while the number of lysosome-encapsulated mitochondria decreased (Fig. S3H, I). In addition, the effect of ECM stiffening on mitochondrial homeostasis was tested. The results showed that ECM stiffness reduced the chondrocyte ability to generate mitochondria, as evidenced by a decrease in the MFN1 and MFN2 mitochondrial fusion markers in response to higher substrate stiffness, as well as the upregulation of the DRP1 and FIS1 mitochondrial division markers (Fig. S4A, B). Consistent with expectations, the level of mitochondrial fission in chondrocytes from the severe degeneration group was slightly reduced relative to the moderate degeneration group, which remains likely to be attributable to increased mitochondrial dysfunction in response to high stiffness stimulation and an absolute deficit in the number of mitochondria.
Fig. 3.
ECM stiffening activates PINK1/Parkin mediated mitophagy which its inhibition delays chondrocyte senescence and joint degeneration in mice. a, b Western Blotting analysis of PINK1, Parkin, and Lc3 expression in mouse primary chondrocytes cultivated on 40:1, 10:1, and 5:1 PDMS substrates (n = 5). c Representative images of immunofluorescence of PINK1, Parkin, Lc3 in chondrocytes of controls and mice at the end of 4 weeks and 8 weeks after DMM surgery. Scale bars: 50 µm. d Quantification of p16INK4a, p21, p53-positive chondrocytes based on staining results in (c) (n = 5). e Representative images of mitochondria staining, lysosome staining and mitophagy staining in mouse primary chondrocytes cultivated on 40:1, 10:1, and 5:1 PDMS substrates. Scale bars: 5 µm. f Quantification of phagosome-lysosome fusion in chondrocytes based on staining results in (e) (n = 5). g, h Western Blotting analysis of Parkin, Lc3, p16INK4a, p21, and p53 in mouse primary chondrocytes which transfected si-NC or si-Parkin cultivated on 10:1 PDMS substrates (n = 5). i Representative images of safranin O/fast green and immunofluorescence staining of p16INK4a, p21, and p53 in the cartilage of mice intra-articularly injected with AAV-NC or AAV-shParkin after DMM surgery. Scale bars: 50 µm. j Quantification of the OARSI scale based on staining results in (i) (n = 5). k Quantification of p16 INK4a, p21-positive chondrocytes based on staining results in (i) (n = 5). *P < 0.05, **P < 0.01, ***P < 0.001. ns not significant; Con control, DMM destablization of the medial meniscus, DAPI 4’,6-diamidino-2-phenylindole, OARSI Osteoarthritis Research Society International, 40:1, 10:1, 5:1, 40:1, 10:1, 5:1 polydimethylsiloxane (PDMS) substrates
Subsequently, the relationship between ECM stiffening-induced mitophagy and chondrocyte aging, as well as OA progression, was investigated. Considering the substantial decline in the number of mitochondria cultivated on high-stiffness substrates, it may not be possible to salvage mitochondrial loss via in vitro and in vivo regulation. Next, this work focuses on alleviating mitochondrial homeostasis disorders in the moderate degeneration group. Parkin recruitment and activation are necessary for Parkin-dependent mitophagy. After silencing Parkin expression with small-interfering RNA (siRNA), the p16INK4a, p21, and p53 protein expression of the chondrocytes cultivated on pathological stiffness substrates decreased significantly (Fig. 3g, h). Treatment with cyclosporin A (10 μmol/L, Macklin), an inhibitor of mitochondrial autophagy, similarly attenuates the senescent phenotype of chondrocytes (Fig. S4C, D). Moreover, Parkin interference reduced the positive SA-Gal staining rate of the chondrocytes cultivated on pathological stiffness matrix substrates (Fig. S4E, F).
In vivo, Adeno-associated virus interfering Parkin (AAV-shParkin) and comparable amounts of an AAV-negative control were injected intraarticularly once a week from 3 days after DMM surgery. As expected, AAV-shParkin effectively alleviated OA development in mice by reducing cartilage damage and proteoglycan loss (Fig. 3i, j). Importantly, AAV-shParkin also reduced the number of p16INK4a, p21 and p53 positive chondrocytes in the articular cartilage (Fig. 3i, k). Together, these data suggest that inhibiting PINK1/Parkin-dependent mitophagy can alleviate the chondrocyte senescence and OA progression caused by ECM stiffening.
HDAC3 loss activates PINK1/Parkin signaling to promote chondrocyte senescence and OA progression
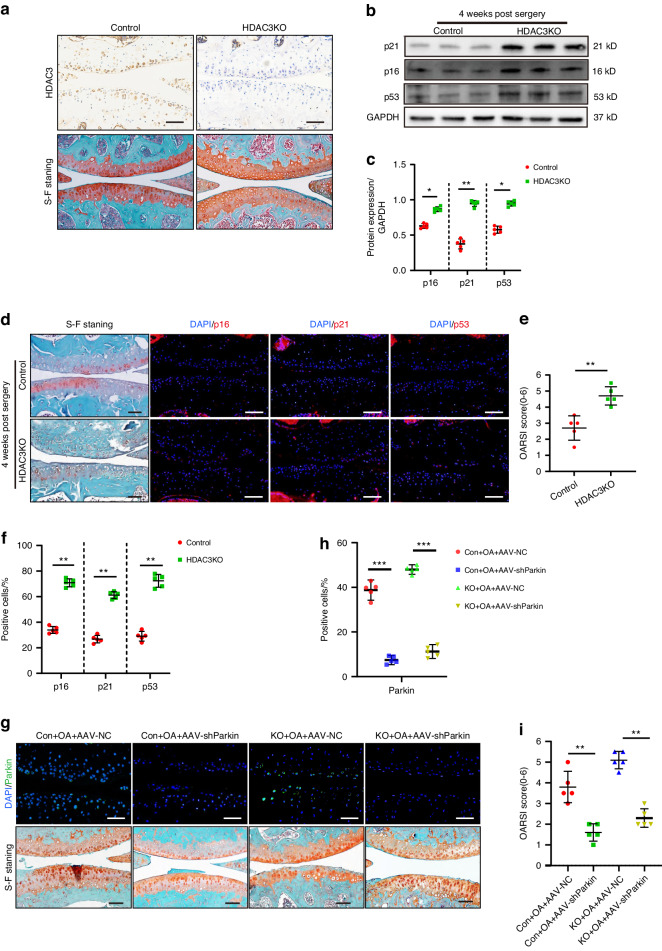
To determine the relationship between chondrocyte HDAC3 and mitophagy as well as OA progression, knockout mice (HDAC3KO) with a conditionally deleted HDAC3 gene in the chondrocytes were produced by crossing HDAC3flox/flox mice with Col2a1-cre mice, The genotypes were determined via PCR (Fig. S5A–C). The body lengths of 8-week-old HDAC3KO mice were marginally shorter than those of the control group, and their body weight was noticeably lower, suggesting that their growth of was slower (Fig. S5D–F). In addition, IHC staining further confirmed HDAC3 ablation in the articular chondrocytes of the HDAC3KO mice (Fig. 4a).
Fig. 4.
Loss of HDAC3 activates PINK1/Parkin signaling to promote chondrocyte senescence and OA progression. a Representative images of IHC and safranin O/fast green staining of HDAC3 in articular cartilage of HDAC3KO and Control mice at 3 months old. Scale bar: 50 µm. b, c Western Blotting analysis of p16INK4a, p21, p53 in the cartilage of HDAC3KO and Control mice after DMM surgery (n = 5). d Representative images of safranin O/fast green and immunofluorescence staining of p16INK4a, p21, and p53 in HDAC3KO and Control cartilage of mice after DMM surgery. Scale bars: 50 µm. e Quantification of the OARSI scale based on staining results in (d) (n = 5). f Quantification of p16INK4a, p21, and p53-positive chondrocytes based on staining results in (d) (n = 5). g Representative images of safranin O/fast green and immunofluorescence staining of Parkin in HDAC3KO and Control cartilage of mice which intra-articularly injected with AAV-NC or AAV-shParkin after DMM surgery. Scale bar: 50 µm. h Quantification of Parkin-positive chondrocytes based on staining results in (g) (n = 5). i Quantification of the OARSI scale based on staining results in (g) (n = 5). *P < 0.05, **P < 0.01. DMM destablization of the medial meniscus, DAPI 4’,6-diamidino-2-phenylindole, OARSI Osteoarthritis Research Society International, KO knockout, AAV-shParkin adenovirus expressing small hairpin Parkin, AAV-NC negative control
When cultured on physiological stiffness substrates, chondrocytes from HDAC3KO mice and controls showed no appreciable changes in senescence phenotype (Fig. S6A, B). However, when cultured on pathological stiffness substrates, the primary chondrocytes with HDAC3 gene deletion enhanced the senescence phenotype. It is shown that the protein upregulation of p16INKA4, p21, p53 and enhancement of SA-β-galactosidase staining (Fig. 4b, c and Fig. S6C, D). The data suggest that HDAC3 deficiency alone is not sufficient to induce chondrocyte senescence in the absence of mechanical stimuli. Silencing the Parkin expression in the HDAC3KO chondrocytes alleviated the aging phenotype resulting from HDAC3 deletion (Fig. S6E, F). This suggests that HDAC3 gene deletion promotes chondrocyte senescence by activating PINK1/Parkin-dependent mitophagy. HDAC3 gene deletion also encouraged chondrocyte senescence and OA progression in vivo. Compared with the control group, HDAC3 deletion accelerated the occurrence of experimental OA, showing significant cartilage erosion, as well as proteoglycan and cell density loss in the articular cartilage, which was further verified using the scale analysis of the International Osteoarthritis Research Institute (OARSI) (Fig. 4d, e). Moreover, the number of positive p16INK4a, p21, and p53 cells in the joint chondrocytes of the HDAC3KO mice was significantly higher than that of their control group offspring (Fig. 4d, f). Consistent with previous results, Parkin inhibition alleviated articular cartilage erosion in the HDAC3KO DMM model mice (Fig. 5g–i). In summary, these results indicate that the absence of HDAC3 in the chondrocytes accelerates the cell and cartilage aging caused by ECM stiffening.
Fig. 5.
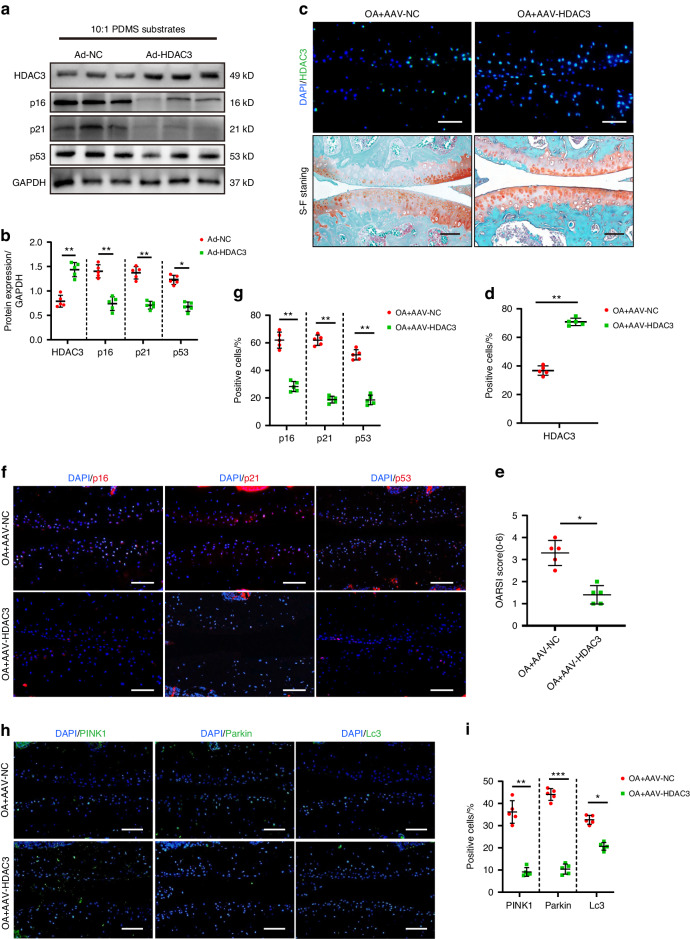
HDAC3 Overexpression delays chondrocyte aging and OA progression. a, b Western Blotting analysis of HDAC3, p16INK4a, p21, and p53 expression in the primary chondrocytes of mice which transfected adenovirus containing HDAC3 (Ad-HDAC3) or Ad-NC cultivated on 10:1 PDMS substrates (n = 5). c Representative images of safranin O/fast green and immunofluorescence staining of HDAC3 in cartilage of mice intra-articularly injected with AAV-NC or AAV-HDAC3 after DMM surgery. Scale bars: 50 µm. d Quantification of HDAC3-positive chondrocytes based on staining results in (c) (n = 5). e Quantification of the OARSI scale based on staining results in (c) (n = 5). f Representative images of immunofluorescence staining of p16INK4a, p21, and p53 in cartilage of mice intra-articularly injected with AAV-NC or AAV-HDAC3 after DMM surgery. Scale bars: 50 µm. g Quantification of p16INK4a, p21, and p53-positive chondrocytes based on staining results in (f) (n = 5). h Representative images of immunofluorescence staining of PINK1, Parkin, and Lc3 in cartilage of mice intra-articularly injected with AAV-NC or AAV-HDAC3 after DMM surgery. Scale bars: 50 µm. i Quantification of PINK1, Parkin and Lc3-positive chondrocytes based on staining results in (h) (n = 5). *P < 0.05, **P < 0.01, ***P < 0.001. OA osteoarthritis, DAPI 4’,6-diamidino-2-phenylindole, OARSI Osteoarthritis Research Society International, AAV-HDAC3 adenovirus expressing small hairpin HDAC3, AAV-NC negative control
HDAC3 overexpression delays chondrocyte aging and OA progression
The primary mouse chondrocytes cultured on pathological stiffness substrates were treated with and without HDAC3 containing adenovirus (Ad-HDAC3) for 24 h. The p16INK4a, p21, and p53 levels decreased significantly in the Ad-HDAC3-treated chondrocytes, while the number of positively SA-β-Galactosidase-stained chondrocytes decreased (Fig. 5a, b and Fig. S6G, H), Indicating that HDAC3 addition alleviated the chondrocyte aging caused by ECM stiffening. Next, starting 3 days after DMM surgery, AAV-HDAC3 and an equal amount of an AAV-negative control were injected weekly into the joints of the mice. IF showed that AAV-HDAC3 intra-articular injection mainly affected the articular cartilage. Additionally, significantly higher HDAC3 expression was evident in the deep-layer chondrocytes of the AAV-HDAC3-treated mice, confirming that AAV successfully transuded HDAC3 overexpression (Fig. 5c, d). The results showed that AAV-HDAC3 effectively alleviated chondrocyte aging and OA development in mice by decreasing the number of positive p16INK4a, p21, and p53 cells (Fig. 5f, g). Moreover, the hypertrophic chondrocyte differentiation, cartilage destruction, and proteoglycan loss were reduced in the tibial cartilage of the AAV-HDAC3-treated mice (Fig. 5c, e). As expected, HDAC3 overexpression significantly reduced PINK1/Parkin-dependent mitophagy in the mouse chondrocytes (Fig. 5h, i). In summary, these findings suggest that HDAC3 delays ECM stiffening-induced chondrocyte senescence and OA progression.
HDAC3 affects mitophagy via epigenetic Parkin regulation
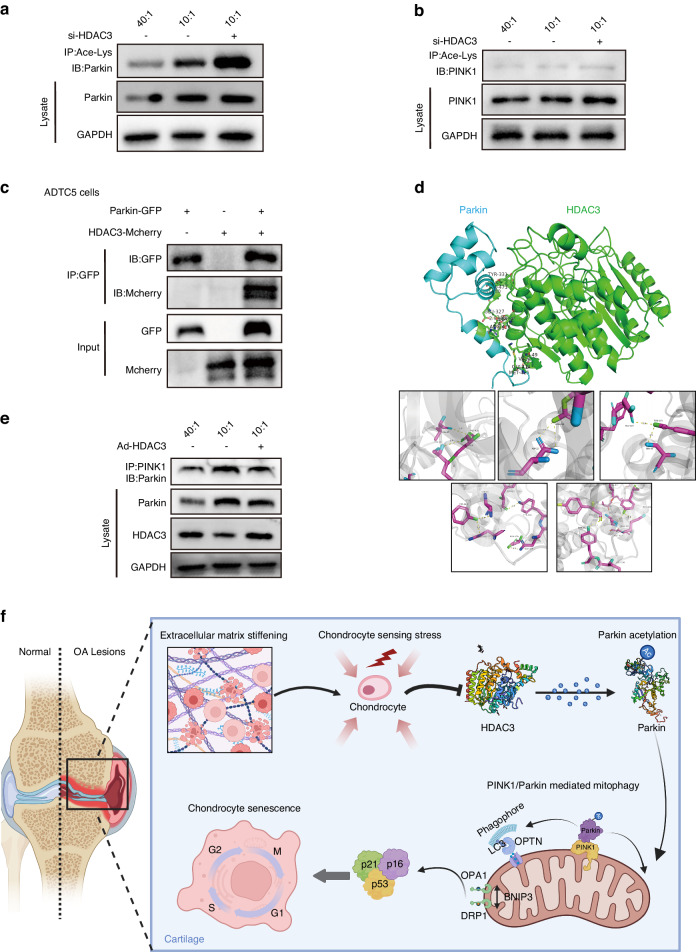
Next, this study explored the HDAC3 regulation of chondrocyte mitophagy. HDAC3 is mainly considered an epigenomic regulator of deacetylate histones, which in turn controls the transcription of the various genes required for development and physiology.29 Recent studies have shown that HDAC3 can drive non-histone deacetylation outside the nucleus.30 It is well known that protein acetylation is mainly responsible for transcriptional gene regulation. First, the transcriptional regulation of the mitophagy-related genes by HDAC3 was investigated. When stimulated by pathological ECM stiffening, the Prkn chondrocyte difference was more obvious in the HDAC3KO mice than in the control group (Fig. S6I). Subsequently, the Parkin acetylation level was investigated via immunoprecipitation. The HDAC3 loss caused by ECM stiffening activated Parkin acetylation, which increased further after HDAC3 knockdown (Fig. 6a), showing that HDAC3 significantly affected Parkin acetylation regulation. However, as a protein equally critical for mitochondrial autophagy initiation, no significant differences were evident in the acetylation level of PINK1 affected by sclerosis or HDAC3 knockdown (Fig. 6b). In addition, the interaction between the HDAC3 and Parkin in ATDC5 cells was determined (Fig. 6c). To help identify which HDAC3 and Parkin sites participated in this binding event, a 3D HDAC3-Parkin complex structure was modeled using the ZDOCK protein-docking algorithm (Fig. 6d). Protein docking results confirmed that there were multiple interaction sites on the interaction surface of HDAC3 and Parkin, including seven hydrogen bonds generated by groups such as lys49 and gly414, and the binding energy between HDAC3 and Parkin protein was −17.9 kcal/mol, indicating that they had strong affinity activity.
Fig. 6.
HDAC3 affects mitophagy through epigenetic regulation of Parkin. a Acetylation of endogenous Parkin in chondrocytes transfected si-NC or si-HDAC3 cultivated on 40:1 and 10:1 PDMS substrates. Parkin acetylation was analyzed by immunoprecipitation with an anti-acetyl-lys antibody followed by western blotting for Parkin. b Acetylation of endogenous PINK1 in chondrocytes transfected si-NC or si-HDAC3 cultivated on 40:1 and 10:1 PDMS substrates. PINK1 acetylation was analyzed by immunoprecipitation with an anti-acetyl-lys antibody followed by western blotting for PINK1. c Mcherry and GFP were immunoprecipitated from ATDC5 cells after transfection with both plasmids containing HDAC3-Mcherry and Parkin-GFP. The presence of Mcherry and GFP in the immunoprecipitates was evaluated by immunoblotting. d Top: optimized binding modes with the lowest binding energy generated by ZDock, and key residues for interaction between mouse HDAC3 and Parkin. Down: magnified view of boxed area. e Cell lysates of chondrocytes which transfected adenovirus containing HDAC3 (Ad-HDAC3) or Ad-NC cultivated on 40:1 and 10:1 PDMS substrates were subjected to immunoprecipitation with anti-PINK1 antibody followed by immunoblotting to detect Parkin. f Schematic diagram representing molecular pathways in which ECM stiffening induces OA development through HDAC3. IB immunoblotting, IP immunoprecipitate
As an E3 ubiquitin ligase, Parkin mediates mitophagy downstream of PINK1.31 The effect of acetylation modification on the interaction between Parkin and its upstream PINK1 protein was investigated. As expected, culturing on a stiffness substrate increased the level of interacting Parkin in the immunoprecipitated PINK1 protein complex, while HDAC3 overexpression decreased the interaction between Parkin and PINK1 (Fig. 6e). The above results indicate that HDAC3 regulated Parkin acetylation is involved in the induction of mitophagy.
Discussion
The mechanical properties of ECM regulate a variety of biological cell phenotypes. HDAC3 was found to be a critical component in the relationship between ECM mechanical properties and chondrocyte senescence. ECM stiffening during the OA process stimulated chondrocyte senescence in vitro and in mouse articular cartilage. Furthermore, ECM stiffening stimuli downregulated HDAC3, which increased Parkin acetylation and activated Parkin-dependent mitophagy, promoting chondrocyte senescence and cartilage degradation. Therefore, HDAC3 supplementation and Parkin-dependent mitophagy inhibition are potential targets for OA treatment (Fig. 6f).
This study showed that OA cartilage exhibited a stiffer cartilage matrix than normal cartilage, while the cartilage matrix stiffness increased with the OA severity. Changes in the biomechanical properties of the cartilage matrix can be attributed to changes in the cartilage composition and structure. Cartilage, mainly composed of collagen fibers, proteoglycans, and water, supports weight in joints. The collagen fibers are exposed during OA development, while water and proteoglycan loss produce cross-linked collagen fibers that confer tensile strength to tissues stiffer than intact normal cartilage. To biosimulate the effect of matrix stiffness on chondrocytes, a matrix stiffness model was established based on PDMS to examine the interaction between chondrocytes and ECM stiffness. Various studies have examined matrix stiffness in cell culture systems.32,33 In cancer, matrix stiffness can promote tumor growth by directly stimulating or inducing exosome secretion.17 In addition, previous research showed that matrix stiffness guided stem cell lineage specification and proliferation.34 The present study found that pathological matrix stiffness stimulation accelerated chondrocyte and cartilage aging in mice, indicating interaction between the biomechanical and biological environments during OA.
HDAC3 is widely regarded as an epigenetic cell regulator. It cooperates with the silencing mediator of retinoic acid and thyroid hormone receptor and nuclear receptor corepressor to promote histone deacetylation and targets a variety of pathways, such as gene transcription, cell development, cell cycle, and tumorigenesis. In addition, evidence indicates HDAC3 mechanosensitivity.29 Previous studies showed that abnormal matrix stiffness in liver cirrhosis changed HDAC3 expression and the organization of its cytoskeleton.35 Another study indicated that HDAC3 was essential during shear-induced stem cell differentiation.36 However, no studies are available on the role of HDAC3 in chondrocyte senescence regulation and OA progression. In this study, mass spectrometric proteomic analysis showed that HDAC3 differed significantly in proteins regulated by mechanical stimulation. Subsequent experiments also verified that HDAC3 was significantly downregulated by matrix sclerosis stimulation in chondrocytes in vivo and in vitro, while its deletion caused chondrocyte senescence and OA deterioration. In addition, HDAC3 deletion enhanced the chondrocyte senescence phenotype in the articular cartilage of DMM OA mice, while in vivo HDAC3 overexpression alleviated chondrocyte senescence and OA progression. However, no significant changes were evident in the chondrocyte senescence phenotype in the HDAC3KO mice and controls, suggesting that HDAC3 deficiency alone was insufficient for inducing chondrocyte senescence in the absence of mechanical stimuli.
The investigation into the role of downstream HDAC3 signaling in regulating chondrocyte senescence and OA development via ECM stiffening was noteworthy since it revealed a strong correlation between the compositional variations of the ECM, actin fiber bundles, and stress fibers and those of the mitochondria. Mitochondria represent the “power source” of cells, producing cellular energy in the form of ATP and participating in various important cellular processes.37 Since mitochondrial dysfunction is associated with cellular damage and a variety of diseases, it is essential to maintain mitochondrial homeostasis.38,39 Mitophagy is a unique form of autophagy that regulates the turnover of dysfunctional mitochondria and is a key mechanism for maintaining mitochondrial homeostasis.40 Considerable evidence indicates an association between mitophagy and OA progression.41,42 However, some controversy exists about the role of mitophagy in OA progression. Studies have shown that Parkin overexpression reduces mitochondrial ROS and chondrocyte apoptosis by scavenging dysfunctional mitochondria.43 In addition, metformin can also activate the sirt3-PINK1-prkn signaling pathway to counteract the oxidative stress caused by IL1B, as well as the anabolic and catabolic imbalance in chondrocytes.44 However, other studies have shown that PINK1-mediated mitophagy leads to mitochondrial fragmentation and cell death in human chondrocytes and rats after MIA treatment, while PINK1 knockout mice with MIA-induced OA exhibit less cartilage damage and pain behavior compared with the control group.45 The dual effect of mitophagy on cell survival and function in the above results may be attributed to the mitophagy level in pathological conditions, model intervention differences, and of disease progression stages. This study showed that the HDAC3 loss stimulated by ECM stiffening significantly activated Parkin-dependent mitophagy and accelerated cellular senescence. When stimulated by severe pathological ECM stiffening, the level of Parkin-dependent mitophagy decreased slightly compared to the moderate pathological group. It is reasonable to assume that mitophagy is essential for basal mitochondrial turnover and maintenance in physiological conditions and may also play a primary role in initiating various pathological stresses. Mitochondrial clearance activation by excessive stiffness stimulation resulted in a complete lack of mitochondria in the chondrocytes, reducing the relative level of mitophagy. In addition, Parkin interference effectively delayed the chondrocyte senescence phenotype and OA development in the HDAC3KO mice, suggesting that Parkin-dependent mitophagy was required for HDAC3 to affect cartilage aging.
The importance of HDAC3 in chromatin dynamics and gene expression regulation is well recognized.46,47 At the mechanistic level, HDAC3 regulated mitophagy by affecting Parkin acetylation modification. Previous studies have reported the important role of Parkin acetylation in Parkin-dependent mitophagy and tumor suppression.31 Consistent with this, in this study, HDAC3 inhibition upregulated the lysine acetylation modification of Parkin and activated Parkin-dependent mitochondria. These results suggested that the Parkin acetylation regulation by HDAC3 played a vital role in ECM stiffening-induced chondrocyte senescence and cartilage degeneration.
Although the data in this study suggest that HDAC3-Parkin signaling is important for chondrocyte senescence and cartilage degeneration, the regulation of mitophagy at an appropriate level requires further investigation. For genetic diseases associated with mitophagy defects, the mutation or deletion of mitophagy-related genes easily causes complex multisystem lesions and neurodegeneration.48,49 In addition, although previous studies have shown that part of HDAC3 is initially located in the cytoplasm, how the remainder approaches mitochondria remain unclear. Therefore, the activity trajectory and function of the HDAC3 that regulates Parkin acetylation require further exploration. Although the practicality and safety of applying HDAC3-Parkin signaling for OA treatment requires further verification in future research, the results of this study reveal a causal relationship between ECM stiffening and cartilage aging while determining that HDAC3 deletion and Parkin-dependent mitophagy activation are important for ECM stiffening to regulate cartilage aging. Therefore, targeting HDAC3-Parkin signaling may be a novel therapeutic approach for OA treatment. Since the effect of matrix stiffness on OA is still under investigation, this study raises many new hypotheses and expectations. Considering the mechanical sensitivity of chondrocytes to ECM stiffness, modulating the physicochemical properties of ECM to restrict cartilage degeneration using osteochondral scaffolds, which have been extensively investigated to date, is also a feasible topic for future research.50–52 Furthermore, determining whether HDAC3-dependent ECM stiffening participates in the OA process in any other way or whether HDAC3-Parkin signaling modulation alleviates cartilage ECM stiffness during OA will be investigated in continued research.
Materials and methods
Human samples
Human osteoarthritis cartilage was obtained from patients undergoing total knee arthroplasty (n = 8). Normal control cartilage was obtained from traffic accident patients with no history of arthritis (n = 5). Cartilages were excised from the tibial plateau and femoral condyles during total knee replacement surgery. Cartilage samples were fixed in 4% paraformaldehyde, decalcified in 15% EDTA and paraffin-embedded for further histological analysis.
All human samples were obtained from the Third Affiliated Hospital of Southern Medical University, Guangzhou, China. All patients provided informed consent to use their clinical information for scientific research. This study was approved by the Ethics Committee of the Third Affiliated Hospital of Southern Medical University (2024-ER-001).
Animals
The col2a1-cre mice were purchased from Cyagen Biosciences, Jiangsu, China. The primers used are shown below. Forward: CTCTTCGCTATTATTCACCCTCAGCTT; Reverse: CTTGCGAACCTCATCACTCGTTG. Unique product lengths of 431 bp were generated. HDAC3flox/flox mice were purchased from GemPharmatech, Jiangsu, China. Both mice were of C57BL/6J background. To generate chondrocyte specific HDAC3 null mice, col2a1cre mice were bred with HDAC3flox/flox, and the HDAC3 chondrocyte-specific null mice were named HDAC3KO mice. The offspring mice carrying HDAC3flox/flox without CRE were used as the control group. Routine genotyping of tail DNA was performed according to the instructions of GemPharmatech and Cyagen Biosciences. The primers used are shown below. Forward: GTTAATCCGTGGGAGGATATTTTCT; Reverse: CCACTCAAACAAGCATACAGAGAAACA. Unique product lengths of 373 bp were generated.
For mice OA model, all C57BL/6J mice were purchased from the Laboratory Animal Center of Southern Medical University (Guangzhou, China). Twelve-week-old C57BL/6J mice and their littermate control mice underwent medial meniscus destabilization (DMM) surgery to induce osteoarthritis. To reduce the number of animals used, only male mice were used for experiments in this study. 20 mice were randomly assigned to experimental groups. The operative area was shaved, and the mice were selected to be fixed in the supine position with the left hind limb flexed at 90°, and the operative field was fully disinfected. The skin was cut to expose the patellar ligament, and the joint capsule was dissected along the inner edge of the patellar ligament with sharp knife scissors to bluntly clean out the intra-articular adipose tissue, and the medial meniscus was attached to the tibial plateau through the medial meniscus tibial collateral ligament (MMTL). After dissecting the MMTL and flushing the wound, the joint cavity was sutured, the skin wound was closed, and topical amoxicillin was applied to prevent wound infection. In the sham-operated group, only the joint capsule was dissected and then sutured layer by layer. The mice were also observed for anesthesia awakening and wound healing.
Mice were executed at 4 and 8 weeks postoperatively, and knee joint specimens were collected. The knee joints were fixed in 40 g/L paraformaldehyde for 24–48 h, decalcified in 100 g/L EDTA for 1 week, embedded in paraffin wax, sectioned in coronal position at 4 μm intervals, and stained with saffron O/solid green. The samples were scored using the Osteoarthritis Research Society International (OARSI) grading/staging system.
All animals were provided with standard feed and kept in pathogen-free cages with constant temperature and humidity. The circadian rhythm was maintained at 12 h. All animal experiments were approved by the animal protection and use Committee of the Ruiye model animal (Guangzhou, China) Biotechnology Co. (Guangzhou, China) and carried out according to the guidelines and regulations of the Committee (no. RYEth-20231008321).
Intra-articular delivery of HDAC3 and shParkin adeno-associated virus in experimental OA
Adeno-associated virus containing HDAC3 (AAV-HDAC3) (GENECHEM, Shanghai, China) and shParkin (AAV-shParkin) (GENECHEM, Shanghai, China) was administered to C57BL/6J mice with DMM-induced OA by intra-articular injection performed at 3-, 14-, and 21-day post DMM surgery. Specifically, a longitudinal skin incision was made to visualize the patellar ligament and the patella; then we injected 1 × 1010 AAV particles in a 10 µL volume into the knee joint cavity by inserting a small needle into the area underneath the patella of the leg. The control groups were all treated with negative control (AAV-NC) for the same periods. Mice were euthanized at 28 days post-surgery for histological analyses. The knee joints were fixed in 40 g/L paraformaldehyde for 24–48 h, decalcified in 100 g/L EDTA for 1 week, embedded in paraffin wax, and sectioned in coronal position at 4 μm intervals for further histological analysis.
Histological analyses
Tissue samples were fixed in 4% paraformaldehyde buffered with phosphate-buffered saline (pH 7.4) for 24 h at 4 °C. Specimens were decalcified with 10% EDTA (pH 7.4) for 2 weeks at 4 °C, embedded in paraffin and 4-µm thick sagittal sections were cut. Safranin O/Fast Green staining was performed according to standard protocols. Safranin O/Fast Green staining slides were used to evaluate cartilage degeneration by OARSI scoring system. Each section was assessed by two blinded, independent graders and the mean score was used for statistical analysis.
For OARSI scoring system, we recommend these 0–6 subjective scoring system to apply to all four quadrants of the joint: medial femoral condyle, medial tibial plateau, lateral femoral condyle, lateral tibial plateau. A score of 0 represents normal cartilage, 0.5 = loss of proteoglycan with an intact surface, 1 = superficial fibrillation without loss of cartilage, 2 = vertical clefts and loss of surface lamina, 3 = vertical clefts/erosion to the calcified layer lesion for 1%–25% of the quadrant width, 4 = lesion reaches the calcified cartilage for 25%–50% of the quadrant width, 5 = lesion reaches the calcified cartilage for 50%–75% of the quadrant width, 6 = lesion reaches the calcified cartilage for >75% of the quadrant width. The OA severity is expressed as a maximal score.
Immunohistochemistry and IF
Specimens were prepared as described previously. Following deparaffinization and rehydration, the sections were soaked in citrate buffer (10 mmol/L citric acid, pH 6.0) for 16–18 h at 60 °C to unmask the antigen for IHC and IF analyses. For IHC analysis, we added 3% hydrogen peroxide for 15 min. The sections were blocked with 1% sheep serum for 1 h at 37 °C and incubated with the primary antibodies (in 1% bovine serum albumin and 0.1% Triton X-100) overnight at 4 °C. For IHC staining, sections were stained with horseradish peroxidase-conjugated secondary antibodies (Proteintech, Hubei, China) and Horseradish Peroxidase Color Development Kit was used to observe the chromogen, with hematoxylin for counterstaining. For IF, sections were stained with Alexa 488 or Alexa 594 dye-labeled secondary antibodies (Thermo Fisher Scientific, MA, USA). Nuclei were labeled with 4, 6-diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific, MA, USA) and images were obtained using a FluoView FV1000 confocal microscope (Olympus, Tokyo, Japan). Sections were randomly coded and scored by two blinded observers for three sections per joint.
Cell cultures
Primary articular chondrocytes were isolated from 3-day-old C57BL/6J mice knee joint cartilage according to a standard protocol using collagenase II. Primary chondrocytes were cultured in DMEM-F12 with 15% FBS. The medium was changed every day. For most experiments, primary cells were transferred to serum-free DMEM for 24 h before being exposed to stimulation.
Polydimethylsiloxane (PDMS) substrate preparation
PDMS substrates with different stiffness were prepared from the commercially available Sylgard 184 silicone elastomer kit (Dow Corning, MI, USA) by mixing base and curing agent in varying ratios (base polymer-to-cross-linker ratios, w/w). The pre-polymer mixtures were mixed thoroughly for 5 min, degassed, and poured into culture dishes for spreading. PDMS substrates were cured at 60 °C for 2 h, and the thickness of PDMS substrate was 1 mm. They were then immersed in ethanol for 3 h for sterilization, and washed with 1xPBS three times. We coated PDMS substrates with fibronectin (Millipore, MA, USA) for cell culture according to the manufacturer’s protocol.
Mechanical characterization
For each joint, two to three sections were chosen from the central, load-bearing region of the tibial cartilage that is in direct contact with condylar cartilage during joint loading (sections 10–30 cut from the medial to lateral ends, following trimming of the first 300 μm thickness at the medial end). The removed tissue was used for frozen sections, on each section, at a distance of about 20–40 μm from the tibial surface. The middle/deep region of the uncalcified cartilage layer of identified one or two regions of interest (ROIs). Within each ROI, AFM (Bruker Co., MA, USA) nanomechanical mapping was performed in a 40 × 40 grid (1 600 indents) using polystyrene microspherical tips up to ≈100 nN maximum indentation force at 10 μm/s effective indentation depth rate. For each indentation, the effective indentation modulus, Eind, was calculated by fitting the entire loading portion of the indentation force-depth (F–D) curve to the finite thickness-corrected Hertz model. All data analyses were performed using NanoScope Analysis2.0 (Bruker Co., MA, USA).
Senescence associated β-galactosidase (SA-β-Gal) assay
Cytochemical staining for SA-β-Gal was performed using an SA-β-Gal staining kit (Beyotime, Shanghai, China). Following the cell treatment, cytochemical staining for SA-β-Gal was performed at pH 6 according to the manufacturer’s protocol, and the positive cells in four randomly selected fields per treatment were counted (n = 5).
CCK-8 assay for cell viability
The assessment of cell viability was performed using the Cell Counting Kit-8 (Beyotime, Shanghai, China) reagent to evaluate cellular activity. chondrocytes were cultivated on 40:1, 10:1, and 5:1 PDMS substrates. At specified time points, the CCK-8 reagent was added to the culture medium. The cell-CCK-8 mixture was incubated at 37 °C in a cell culture for 1 h. Using a microplate reader (Thermo Fisher Scientific, MA, USA), measure the absorbance of each well at a wavelength of 450 nm.
Transmission electron microscope
Obtain chondrocytes cultivated on 40:1, 10:1, and 5:1 PDMS substrates. After washing with PBS, chondrocytes were scraped and harvested with 4% glutaraldehyde and centrifuged at 4 °C. The precipitated chondrocytes were continued to fix with 4% glutaraldehyde for another 2 h at room temperature and stored at 4 °C. Electron photomicrographs of the chondrocytes were taken by Scientific Compass Technology Co., Zhejiang, China.
Western blotting analysis
Tissues and cells were lysed using lysis buffer (62.5 mmol/L Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, 50 mmol/L dithiothreitol, 0.01% bromophenol blue) at 96 °C for 10 min. The samples were separated by SDS-PAGE for 70 min and the proteins were subsequently transferred to membranes (Bio-Rad Laboratories, CA, USA) by the wet transfer method. Each membrane was incubated with primary antibodies overnight at 4 °C on a shaker. Following incubation with specific secondary antibodies, we detected the proteins with an enhanced chemiluminescence kit (Proteintech, Hubei, China). The following primary antibodies were used: p16INK4a (1:1 000, #18769, Cell Signaling), p21 (1:1 000, ab188224, Abcam), p53 (1:1 000 10442-1-AP, Proteintech), HDAC3 (1:1 000, 10255-1-AP, Proteintech), GAPDH (1:5 000, 10494-1-AP, Proteintech), PINK1 (1:1 000 for Western blot and 1:50 for Immunoprecipitation, #6946, Cell Signaling), Parkin (1:1 000, 14060-1-AP, Proteintech), Lc3 (1:1 000, ab48394, Abcam), MMP13 (1:1 000, 18165-1-AP, Proteintech), COL2A1 (1:1 000, #40772, SAB), Acetylated-lysine (1:100 for Immunoprecipitation, #9441, Cell Signaling) and GFP (1:200 for Immunoprecipitation, 66002-1-Ig, Proteintech).
Real-time qPCR
Total RNA was isolated from tissue and cell pellets with TRIzol Reagent (Takara Biotechnology, Tokyo, Japan) and reverse transcribed with reverse transcription reagent (Takara Biotechnology, Tokyo, Japan) according to the manufacturer’s protocol. Complementary DNA was used for real-time PCR with SYBR Premix Taq (Vazyme Biotech, Jiangsu, China) in a Light Cycler (Roche Molecular Biochemicals, Basel, Switzerland). Relative quantification of gene expression was performed with the comparative threshold method. Changes in mRNA expression levels were calculated after normalization to values for the GAPDH calibrator gene. The primers used are shown below. Pink1(Forward: CGGTCGCACACTGTTCCTCGT; Reverse: CCCTCCAGCAACTGCAAGGTCA), Prkn (Forward: TTCCGAATCACCTGACGGTT; Reverse: ATGACTTCTCCTCCGTGGT), Map1lc3a (Forward: CCCCAGTAAGATCCCGGTGA; Reverse: ATGATCTTGACCAACTCGCTCA), Ulk1 (Forward: AAACATCCTGCTGTCCAACCC; Reverse: GCCGCCATCATGTTGCTCT), Hspd1 (Forward: GTGTGAATTCCAAGATGCCTA, Reverse: TTATGACCAATGGCTTCCGAT), Sqstm1 (Forward: F: ACCCATCTACAGAGGCTGATCCC; Reverse: CCAGCCGCCTTCATCCGAGA).
siRNA transfection
Small interfering RNA (siRNA) oligonucleotides against mouse, Parkin and nontargeting scrambled control siRNA were purchased from GenePharma, Shanghai, China. Lipofectamine 3000 (Thermo Fisher Scientific, MA, USA) was used to transfect 100 nm siRNA-HDAC3 and siRNA-Parkin in opti-MEM (Thermo Fisher Scientific, MA, USA) following the manufacturer’s protocol. After 24 h of transfection, the cell culture media were replaced with fresh DMEM and incubated for a further for 48 h. The oligonucleotide sequences of Parkin siRNA were, forward: 5′GGA AGG AGC UUC CGA AUC ATT 3′; Reverse: 5′- UGA UUC GGA AGC UCC UUC CTT -3′; chondrocytes were transduced with siRNA when the cells were 30%–50% confluent.
Plasmid constructs and transfection
Mcherry-HDAC3 was prepared by cloning the corresponding HDAC3 DNA fragment into the mcherry-pcs2 vector using NheI and xhol restriction sites. EGFP-PRKN was purchased from MIAOLING BIOLOHY, Hubei, China. According to the manufacturer’s instructions, indicated plasmids were transfected using Lipofectamine 3000 (Thermo Fisher Scientific, MA, USA).
Immunoprecipitation
For immunoprecipitation assays, transduced and treated cells were lysed with RIPA lysis buffer supplemented with a completely EDTA-free protease inhibitor mixture (Beyotime, Shanghai, China) for 30 min on ice. Lysates were removed by centrifugation at 13 000 r/min for 15 min and the supernatant was collected. The protein-containing supernatant was incubated with primary antibody or 1 μg of normal mouse IgG (Proteintech, Hubei, China) as a control for 1 h at 4 °C with rotation. The protein lysates were subsequently incubated with 30 μL of pre-washed protein A-agarose beads (Beyotime, Shanghai, China) for 3 h at 4 °C with gentle rotation. The immunoprecipitates were analyzed by immunoblotting.
Mitochondrial membrane potential assay (JC-1)
We use JC-1(Beyotime, Shanghai, China) probe to detect the mitochondrial membrane potential. Chondrocytes inoculated in physiological and pathological hardness were resuspended in 0.5 mL of cell culture medium, 0.5 mL of JC-1 staining working solution was added and mixed by inverting several times. Incubate for 20 min at 37 °C in a cell culture incubator. After incubation centrifuge the cells at 600 g 4 °C for 3–4 minutes to precipitate the cells. Next, the cells were washed twice with JC-1 staining buffer: 1 mL of JC-1 staining buffer was added to resuspend the cells, centrifuged at 600 g 4 °C for 3–4 min to precipitate the cells, and the supernatant was discarded. Add 1 mL of JC-1 staining buffer to resuspend the cells, centrifuge at 600 g for 3–4 min at 4 °C, precipitate the cells and discard the supernatant. Finally, after resuspension with appropriate amount of JC-1 staining buffer, the red-green fluorescence ratio was detected by flow cytometry (BD, NJ, USA).
LC/MS-MS mass spectrometry-based proteomics
The sample was sonicated three times on ice using a high-intensity ultrasonic processor (Scientz, Zhejiang, China) in lysis buffer (8 mol/L urea, 1% protease inhibitor cocktail). The remaining debris was removed by centrifugation at 12 000 g at 4 °C for 10 min. Finally, the supernatant was collected and the protein concentration was determined with BCA kit according to the manufacturer’s instructions.
For digestion, the protein solution was reduced with 5 mmol/L dithiothreitol for 30 min at 56 °C and alkylated with 11 mmol/L iodoacetamide for 15 min at room temperature in darkness. The protein sample was then diluted by adding 100 mmol/L TEAB to urea concentration less than 2 mol/L. Finally, trypsin was added at 1:50 trypsin-to-protein mass ratio for the first digestion overnight and 1:100 trypsin-to-protein mass ratio for a second 4 h-digestion. Finally, the peptides were desalted by C18 SPE column.
The tryptic peptides were dissolved in solvent A (0.1% formic acid, 2% acetonitrile/in water), directly loaded onto a homemade reversed-phase analytical column (25-cm length, 75/100 μm i.d.). Peptides were separated with a gradient from 6% to 24% solvent B (0.1% formic acid in acetonitrile) over 70 min, 24%–35% in 14 min and climbing to 80% in 3 min then holding at 80% for the last 3 min, all at a constant flow rate of 450 nL/min on a nanoElute UHPLC system (Bruker Co., MA, USA).
The peptides were subjected to capillary source followed by the timsTOF Pro (Bruker Co., MA, USA) mass spectrometry. The electrospray voltage applied was 1.60 kV. Precursors and fragments were analyzed at the TOF detector, with a MS/MS scan range from 100 to 1 700 m/z. The timsTOF Pro was operated in parallel accumulation serial fragmentation (PASEF) mode. Precursors with charge states 0–5 were selected for fragmentation, and 10 PASEF-MS/MS scans were acquired per cycle. The dynamic exclusion was set to 30 s.
The resulting MS/MS data were processed using MaxQuant search engine (v.1.6.15.0). Tandem mass spectra were searched against the human SwissProt database (20422 entries) concatenated with reverse decoy database. Trypsin/P was specified as cleavage enzyme allowing up to two missing cleavages. The mass tolerance for precursor ions was set as 20 × 10−6 in the first search and 5 × 10–6 in the main search, and the mass tolerance for fragment ions was set as 0.02 Da. Carbamidomethyl on Cys was specified as a fixed modification, and acetylation on protein N-terminal and oxidation on Met were specified as variable modifications. FDR was adjusted to <1%.
Molecular docking
HDAC3–Parkin complex predictions were performed by Novopro, Guangdong, China. Briefly, 3D modeling of HDAC3 and Parkin was conducted in I-TASSER software. HDAC3 and Parkin docking was performed in BIOVIA Discovery Studio Visualizer software using ZDOCK algorithms. To gain more successive protein complex prediction, we reranked poses with the ZRANK scoring program: poses with a high density, high ZDOCK score and low ZRANK score were selected. The geometry of the selected docking solution was optimized using an energy minimization protocol and the Biovia Smart Minimizer algorithm. For the selected minimized solution, binding interface residues were identified and the types of interaction (for example, hydrogen bonds and electrostatic and hydrophobic interactions) were determined.
Statistical analysis
All experiments were performed at least three times. All statistical analyses were performed using SPSS Statistics for Windows, Version 28.0 (IBM Corp., NY, USA), and graphs were generated with GraphPad Prism 8.0. Except where indicated, data are displayed as means, with uncertainty expressed as 95% confidence intervals (mean ± 95% CI). For unpaired experiments, two-tailed Student t test, linear regression analysis, or two-way ANOVA was performed. For paired experiments, two-tailed paired t test or linear mixed effect models were utilized. We checked the features of the regression model by comparing the residuals vs. fitted values (i.e., the residuals had to be normally distributed around zero) and independence between observations. No correction was applied for multiple comparison because outcomes were determined a priori and were highly correlated. No statistical analyses included confounders (e.g., body mass in each animal) due to the small sample size. We conducted a complete case analysis in the case of missing data. In all experiments, P values < 0.05 were considered statistically significant. Throughout this text, “n” represents the number of independent observations of knees or cells from different animals. Specific data representation details and statistical procedures are also indicated in the figure legends.
Supplementary information
Acknowledgements
This research was supported by the National Key R&D Program of China (2022YFB4600600), National Natural Science Foundation of China (32271181, 82301785) and Guangdong Basic and Applied Basic Research Foundation (2020B1515120001), Sichuan Province Central Guided Local Science and Technology Development Special Program(2023ZYD0072).
Author contributions
Wenhua Huang and Huan Liu conceived the ideas for experimental designs and wrote the manuscript. Bowen Fu, Jianlin Shen, and Ze Zhang conducted the majority of the experiments and analyzed data and helped with manuscript preparation. Zengping Liu and Nian Sun collected human tissue samples. Canjun Zeng and Xuenong Zou supervised the project and conceived the experiments. All authors approved the final version of the manuscript.
Data availability
Data are available in a public, open-access repository. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE53 partner repository with the dataset identifier PXD046993. All data generated or analyzed during this study are included in this submitted article. and its additional files.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Bowen Fu, Jianlin Shen.
Contributor Information
Huan Liu, Email: 20016040@163.com.
Wenhua Huang, Email: huangwenhua2009@139.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41413-024-00333-9.
References
- 1.Martel-Pelletier, J. et al. Osteoarthritis. Nat. Rev. Dis. Prim.2, 16072 (2016). [DOI] [PubMed]
- 2.Sharma L. Osteoarthritis of the knee. N. Engl. J. Med. 2021;384:51–59. doi: 10.1056/NEJMcp1903768. [DOI] [PubMed] [Google Scholar]
- 3.Glyn-Jones S, et al. Osteoarthritis. Lancet. 2015;386:376–387. doi: 10.1016/S0140-6736(14)60802-3. [DOI] [PubMed] [Google Scholar]
- 4.Zhang H, et al. Maintaining hypoxia environment of subchondral bone alleviates osteoarthritis progression. Sci. Adv. 2023;9:eabo7868. doi: 10.1126/sciadv.abo7868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loeser RF, Collins JA, Diekman BO. Ageing and the pathogenesis of osteoarthritis. Nat. Rev. Rheumatol. 2016;12:412–420. doi: 10.1038/nrrheum.2016.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Belly H, Paluch EK, Chalut KJ. Interplay between mechanics and signalling in regulating cell fate. Nat. Rev. Mol. Cell Biol. 2022;23:465–480. doi: 10.1038/s41580-022-00472-z. [DOI] [PubMed] [Google Scholar]
- 7.Iijima H, et al. Age-related matrix stiffening epigenetically regulates α-Klotho expression and compromises chondrocyte integrity. Nat. Commun. 2023;14:18. doi: 10.1038/s41467-022-35359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armiento AR, Alini M, Stoddart MJ. Articular fibrocartilage—why does hyaline cartilage fail to repair? Adv. Drug Deliv. Rev. 2019;146:289–305. doi: 10.1016/j.addr.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 9.Jiang W, et al. Mechanisms linking mitochondrial mechanotransduction and chondrocyte biology in the pathogenesis of osteoarthritis. Ageing Res. Rev. 2021;67:101315. doi: 10.1016/j.arr.2021.101315. [DOI] [PubMed] [Google Scholar]
- 10.Richard D, et al. Evolutionary selection and constraint on human knee chondrocyte regulation impacts osteoarthritis risk. Cell. 2020;181:362–381.e328. doi: 10.1016/j.cell.2020.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, et al. Mechanical overloading promotes chondrocyte senescence and osteoarthritis development through downregulating FBXW7. Ann. Rheum. Dis. 2022;81:676–686. doi: 10.1136/annrheumdis-2021-221513. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, et al. Mechanical overloading induces GPX4-regulated chondrocyte ferroptosis in osteoarthritis via Piezo1 channel-facilitated calcium influx. J. Adv. Res. 2022;41:63–75. doi: 10.1016/j.jare.2022.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han S. Osteoarthritis year in review 2022: biology. Osteoarthr. Cartil. 2022;30:1575–1582. doi: 10.1016/j.joca.2022.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Peng Z, et al. The regulation of cartilage extracellular matrix homeostasis in joint cartilage degeneration and regeneration. Biomaterials. 2021;268:120555. doi: 10.1016/j.biomaterials.2020.120555. [DOI] [PubMed] [Google Scholar]
- 15.Kim JH, et al. Matrix cross-linking-mediated mechanotransduction promotes posttraumatic osteoarthritis. Proc. Natl. Acad. Sci. USA. 2015;112:9424–9429. doi: 10.1073/pnas.1505700112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stolz M, et al. Early detection of aging cartilage and osteoarthritis in mice and patient samples using atomic force microscopy. Nat. Nanotechnol. 2009;4:186–192. doi: 10.1038/nnano.2008.410. [DOI] [PubMed] [Google Scholar]
- 17.Wu B, et al. Stiff matrix induces exosome secretion to promote tumour growth. Nat. Cell Biol. 2023;25:415–424. doi: 10.1038/s41556-023-01092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patwardhan S, Mahadik P, Shetty O, Sen S. ECM stiffness-tuned exosomes drive breast cancer motility through thrombospondin-1. Biomaterials. 2021;279:121185. doi: 10.1016/j.biomaterials.2021.121185. [DOI] [PubMed] [Google Scholar]
- 19.Xu M, et al. Transplanted senescent cells induce an osteoarthritis-like condition in mice. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2017;72:780–785. doi: 10.1093/gerona/glw154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie J, et al. Cellular senescence in knee osteoarthritis: molecular mechanisms and therapeutic implications. Ageing Res. Rev. 2021;70:101413. doi: 10.1016/j.arr.2021.101413. [DOI] [PubMed] [Google Scholar]
- 21.Jeon OH, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat. Med. 2017;23:775–781. doi: 10.1038/nm.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi D, et al. Pre-radiographic osteoarthritic changes are highly prevalent in the medial patella and medial posterior femur in older persons: Framingham OA study. Osteoarthr. Cartil. 2014;22:76–83. doi: 10.1016/j.joca.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engeland K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022;29:946–960. doi: 10.1038/s41418-022-00988-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng F, et al. MicroRNA-193b-3p regulates chondrogenesis and chondrocyte metabolism by targeting HDAC3. Theranostics. 2018;8:2862–2883. doi: 10.7150/thno.23547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esteban-Martínez L, Boya P. BNIP3L/NIX-dependent mitophagy regulates cell differentiation via metabolic reprogramming. Autophagy. 2018;14:915–917. doi: 10.1080/15548627.2017.1332567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peña-Blanco A, et al. Drp1 modulates mitochondrial stress responses to mitotic arrest. Cell Death Differ. 2020;27:2620–2634. doi: 10.1038/s41418-020-0527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaltouki A, Hsieh CH, Kim MJ, Wang X. Alpha-synuclein delays mitophagy and targeting Miro rescues neuron loss in Parkinson’s models. Acta Neuropathol. 2018;136:607–620. doi: 10.1007/s00401-018-1873-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emmett MJ, Lazar MA. Integrative regulation of physiology by histone deacetylase 3. Nat. Rev. Mol. Cell Biol. 2019;20:102–115. doi: 10.1038/s41580-018-0076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narita T, Weinert BT, Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019;20:156–174. doi: 10.1038/s41580-018-0081-3. [DOI] [PubMed] [Google Scholar]
- 31.Sun X, et al. Histone deacetylase inhibitors inhibit cervical cancer growth through Parkin acetylation-mediated mitophagy. Acta Pharm. Sin. B. 2022;12:838–852. doi: 10.1016/j.apsb.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi B, Xu Q, Liu W. An overview of substrate stiffness guided cellular response and its applications in tissue regeneration. Bioact. Mater. 2022;15:82–102. doi: 10.1016/j.bioactmat.2021.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Janmey PA, Fletcher DA, Reinhart-King CA. Stiffness sensing by cells. Physiol. Rev. 2020;100:695–724. doi: 10.1152/physrev.00013.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vining KH, Mooney DJ. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. cell Biol. 2017;18:728–742. doi: 10.1038/nrm.2017.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyu C, et al. Advanced glycation end-products as mediators of the aberrant crosslinking of extracellular matrix in scarred liver tissue. Nat. Biomed. Eng. 2023;7:1437–1454. doi: 10.1038/s41551-023-01019-z. [DOI] [PubMed] [Google Scholar]
- 36.Zeng L, et al. HDAC3 is crucial in shear- and VEGF-induced stem cell differentiation toward endothelial cells. J. Cell Biol. 2006;174:1059–1069. doi: 10.1083/jcb.200605113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallace DC. A mitochondrial bioenergetic etiology of disease. J. Clin. Investig. 2013;123:1405–1412. doi: 10.1172/JCI61398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Forbes JM, Thorburn DR. Mitochondrial dysfunction in diabetic kidney disease. Nat. Rev. Nephrol. 2018;14:291–312. doi: 10.1038/nrneph.2018.9. [DOI] [PubMed] [Google Scholar]
- 39.Sorrentino V, Menzies KJ, Auwerx J. Repairing mitochondrial dysfunction in disease. Annu. Rev. Pharmacol. Toxicol. 2018;58:353–389. doi: 10.1146/annurev-pharmtox-010716-104908. [DOI] [PubMed] [Google Scholar]
- 40.Palikaras K, Lionaki E, Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat. Cell Biol. 2018;20:1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 41.Liu L, et al. The physiological metabolite α-ketoglutarate ameliorates osteoarthritis by regulating mitophagy and oxidative stress. Redox Biol. 2023;62:102663. doi: 10.1016/j.redox.2023.102663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin Z, et al. Curcumin exerts chondroprotective effects against osteoarthritis by promoting AMPK/PINK1/Parkin-mediated mitophagy. Biomed. Pharmacother. 2022;151:113092. doi: 10.1016/j.biopha.2022.113092. [DOI] [PubMed] [Google Scholar]
- 43.Ansari MY, Khan NM, Ahmad I, Haqqi TM. Parkin clearance of dysfunctional mitochondria regulates ROS levels and increases survival of human chondrocytes. Osteoarthr. Cartil. 2018;26:1087–1097. doi: 10.1016/j.joca.2017.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, et al. Protective effects of metformin against osteoarthritis through upregulation of SIRT3-mediated PINK1/Parkin-dependent mitophagy in primary chondrocytes. Biosci. Trends. 2019;12:605–612. doi: 10.5582/bst.2018.01263. [DOI] [PubMed] [Google Scholar]
- 45.Shin, H. J. et al. Pink1-mediated chondrocytic mitophagy contributes to cartilage degeneration in osteoarthritis. J. Clin. Med.8, 1849 (2019). [DOI] [PMC free article] [PubMed]
- 46.Kuang Z, et al. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Science. 2019;365:1428–1434. doi: 10.1126/science.aaw3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, et al. SETD5-coordinated chromatin reprogramming regulates adaptive resistance to targeted pancreatic cancer therapy. Cancer Cell. 2020;37:834–849.e813. doi: 10.1016/j.ccell.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrucelli L, et al. Parkin protects against the toxicity associated with mutant alpha-synuclein: proteasome dysfunction selectively affects catecholaminergic neurons. Neuron. 2002;36:1007–1019. doi: 10.1016/S0896-6273(02)01125-X. [DOI] [PubMed] [Google Scholar]
- 49.Wang R, et al. PINK1, Keap1, and Rtnl1 regulate selective clearance of endoplasmic reticulum during development. Cell. 2023;186:4172–4188.e4118. doi: 10.1016/j.cell.2023.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tamaddon M, et al. Osteochondral scaffolds for early treatment of cartilage defects in osteoarthritic joints: from bench to clinic. Biomater. Transl. 2020;1:3–17. doi: 10.3877/cma.j.issn.2096-112X.2020.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Donate R, et al. Translation through collaboration: practice applied in BAMOS project in in vivo testing of innovative osteochondral scaffolds. Biomater. Transl. 2022;3:102–104. doi: 10.12336/biomatertransl.2022.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Chen Y, Wei Y. Osteoarthritis animal models for biomaterial-assisted osteochondral regeneration. Biomater. Transl. 2022;3:264–279. doi: 10.12336/biomatertransl.2022.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perez-Riverol Y, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–d552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in a public, open-access repository. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE53 partner repository with the dataset identifier PXD046993. All data generated or analyzed during this study are included in this submitted article. and its additional files.