Abstract
We have previously reported the transcriptomic and lipidomic profile of the first-generation, hygromycin-resistant (HygR) version of the BCGΔBCG1419c vaccine candidate, under biofilm conditions. We recently constructed and characterized the efficacy, safety, whole genome sequence, and proteomic profile of a second-generation version of BCGΔBCG1419c, a strain lacking the BCG1419c gene and devoid of antibiotic markers. Here, we compared the antibiotic-less BCGΔBCG1419c with BCG. We assessed their colonial and ultrastructural morphology, biofilm, c-di-GMP production in vitro, as well as their transcriptomic and lipidomic profiles, including their capacity to activate macrophages via Mincle and Myd88. Our results show that BCGΔBCG1419c colonial and ultrastructural morphology, c-di-GMP, and biofilm production differed from parental BCG, whereas we found no significant changes in its lipidomic profile either in biofilm or planktonic growth conditions. Transcriptomic profiling suggests changes in BCGΔBCG1419c cell wall and showed reduced transcription of some members of the DosR, MtrA, and ArgR regulons. Finally, induction of TNF-α, IL-6 or G-CSF by bone-marrow derived macrophages infected with either BCGΔBCG1419c or BCG required Mincle and Myd88. Our results confirm that some differences already found to occur in HygR BCGΔBCG1419c compared with BCG are maintained in the antibiotic-less version of this vaccine candidate except changes in production of PDIM. Comparison with previous characterizations conducted by OMICs show that some differences observed in BCGΔBCG1419c compared with BCG are maintained whereas others are dependent on the growth condition employed to culture them.
Keywords: BCG, Tuberculosis, RNASeq, Transcriptomic, c-di-GMP, Lipids, Cytokines
Subject terms: Bacterial genetics, Transcriptomics
Introduction
Tuberculosis (TB) is a transmissible, predominantly respiratory disease where cases and related mortality increased worldwide to an estimated 10.6 million cases with around 1.3 million associated deaths in 20221. The only vaccine currently in use to prevent disseminated and miliary TB during childhood, Mycobacterium bovis Bacille Calmette-Guèrin (BCG) has diverse shortcomings2 that have led to development of several novel vaccine candidates, which are at different stages of preclinical and clinical characterization3 (https://newtbvaccines.org/tb-vaccine-pipeline/preclinical-stage/). Among the strategies intended to replace or improve BCG, there are novel, live, attenuated, mycobacteria-based vaccine (LAV) candidates, which aim to increase safety, immunogenicity, and efficacy of current BCG4.
Among said novel LAVs is BCGΔBCG1419c, for which we have developed and characterized two different versions, where the BCG1419c gene has been: (1) partially removed and replaced by a hygromycin resistance gene (first generation version5) or (2) completely deleted with no resistance marker incorporated (second generation version6). BCG1419c is predicted to encode for a c-di-GMP phosphodiesterase, therefore leading us to hypothesize that BCGΔBCG1419c might produce more c-di-GMP than its parental BCG, as we showed to occur in a growth-phase dependent-manner in planktonic cultures7. Of note, determination of c-di-GMP levels was reported by indirect measurement based on the activity of a reporter gene in said work.
We showed that the first-generation version of BCGΔBCG1419c increased in vitro biofilm production, while it lacked production of phthiocerol dimycocerosates (PDIM) and produced longer phenol glycolipid (PGL) species when cultured as biofilms in Sauton medium with no detergent5. Transcriptional profiling of this same version of BCGΔBCG1419c also in biofilm cultures showed decreased expression of genes involved in mycolic acids (MAs) metabolism and antigenic chaperones8 compared with BCG. These non-clinical characterizations were conducted with biofilm cultures because of overarching hypothesis is that in vitro biofilms produced by mycobacteria resemble yet not fully explored aspects of TB pathogenesis, which should be considered as an alternate approach to produce a vaccine candidate against this disease9. In fact, biofilm-like structures were recently observed in TB lesions from mice, guinea pigs and humans10, therefore strengthening the validity of our further development of BCGΔBCG1419c.
Based on this background, here we evaluated colonial morphology, biofilm production and lipidomic profile of planktonic and biofilm cultures of the second-generation version of BCGΔBCG1419c and its parental BCG Pasteur ATCC 35734 strain. The lipidomic analyses included evaluation of PDIM production, as we previously found no transcriptional changes in in vitro produced biofilm cultures8, therefore complicating an explanation for the absence of this compound in the hygromycin-marked BCGΔBCG14195.
On the other hand, considering that most preclinical assays of novel LAV are conducted with bacteria cultured in planktonic conditions, employing shaken cultures in Middlebrook 7H9 medium with ADC/OADC supplement and detergent, in this work we decided to characterize the transcriptional profile of BCGΔBCG1419c and BCG when cultured in conditions that our group has recently employed to characterize the genome11, safety, immunogenicity, efficacy, and proteome of the antibiotic-less, second-generation version of BCGΔBCG1419c7. We went further in this work to directly assess c-di-GMP production in vitro. Finally, as we previously found transcriptional changes in the expression of genes involved in mycolic acids synthesis in biofilm cultures8, which can be incorporated into trehalose dimycolate (TDM, also known as cord factor) signaling via Mincle12 and Myd8813, we decided to compare the capacity of these strains to induce TNF-α, IL-6 and G-CSF in primary, bone-marrow derived murine macrophages obtained from wild type mice and knock-outs (KO) in Mincle or Myd88.
Our results confirm that some microbiological and transcriptomic differences already found to occur in the first-generation version of BCGΔBCG1419c compared with BCG are maintained in the antibiotic-less version of this vaccine candidate, except for changes in production of PDIM. This work also added additional information about transcriptional adaptation in BCG to the absence of BCG1419c, further suggesting some of these changes occur regardless of the cells being within mature biofilms or as planktonic cultures whereas other changes are likely dependent on the growth conditions the bacterial cells are encountered. Finally, we observed that for full induction of TNF-α, IL-6 or G-CSF by bone-marrow derived macrophages, both BCGΔBCG1419c and BCG required Mincle and Myd88.
Results
Microbiological characterization and increased c-di-GMP content in planktonic cultures compared with BCG of the antibiotic-less version of BCG∆BCG1419c
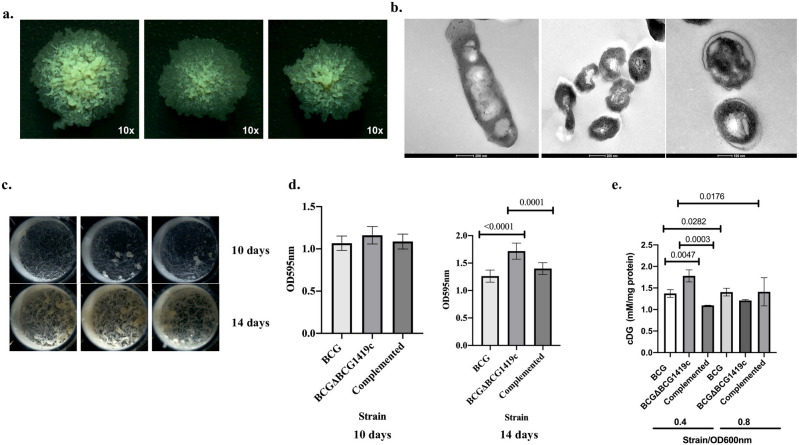
As we have mentioned before, we constructed a novel, antibiotic-less version of the BCGΔBCG1419c vaccine candidate based on the fact that attenuation without the presence of antibiotic-resistance markers is required to fulfill the Geneva consensus criteria for novel TB vaccine candidates14. In an attempt to determine which phenotypes already reported for the hygromycin-marked version of BCGΔBCG1419c are maintained in the antibiotic-less version, we decided to evaluate colony morphology and biofilm production in vitro. We observed that the BCG Pasteur ATCC 35734 strain showed a greater elevation starting from the center and decreasing its density when reaching the edge. The edges of this strain were irregular and the shape of the colonies of this strain were rounder compared to the mutant and complemented strain (Fig. 1a, left panel) As for the BCG∆BCG1419c strain, colonies were flatter and thinner, their highest elevation was in the center with a wide edge, which was also irregular. The shape of this colony tended to be ovoid (Fig. 1a, middle panel). As for the complemented strain, it was smaller in size than the previous two strains, its center presented greater elevation that was not maintained most of the colony as opposed to BCG Pasteur WT strain, therefore restoring partially colonial morphology (Fig. 1a, right panel). An additional characterization we conducted this time was evaluating bacterial ultrastructure by Transmission Electron Microscopy (TEM). Here, we found that the morphology of BCGΔBCG1419c showed anomalies in comparison with the parent BCG Pasteur strain. BCGΔBCG1419c showed a smaller size and irregular shape with constriction and concavities of the cell wall, which was widened and electron-lucid in some bacteria, while in others the cell wall was thinner than that of the parent BCG strain (Fig. 1b).
Figure 1.
Phenotypic changes in colonial and ultrastructural morphology, biofilm and c-di-GMP production in the presence or absence of BCG1419c. (a) Isolated, single colonies obtained after 3 weeks of incubation at 37 °C on 7H10 OADC agar plates. (b) TEM of BCG and BCGΔBCG1419c planktonic cells cultured in 7H9 OADC Tween 80 and harvested at OD600nm 0.8. Representative electron microscopy micrographs of parental BCG and mutant BCGΔBCG1419c. (Left panel) typical morphology of the BCG strain. (Middle panel) abnormal morphology of BCGΔBCG1419c showing smaller bacilli with irregular shape, some cells show constrictions or cavities on the bacterial surface (arrows). (Right panel) high power magnification of mutant bacilli showing widened electron-lucid cell wall (arrows). (c) Surface pellicles formed in Sauton media with no detergent at 37 °C, 5% CO2, for 10 (top panel) or 14 days (bottom panel) in tissue culture flasks with vented caps. (d) Biofilm quantification of the different BCG strains at 10 days or 14 days of culture in Sauton media, in 48-well plates. For colonies, images at ×10 are shown. All experiments were performed three different times, with duplicates (c,d), and one representative image is shown in all instances; error bars represent standard deviations of the. One-Way ANOVA followed by Dunnett’s multiple comparison test was used to assess significance of changes among BCG strains. Statistically significant p actual values are shown on top of the bars depicting the means. (e) c-di-GMP content (mM) was normalized to mg of protein per sample and was determined by HPLC for in vitro cultures of BCG, BCGΔBCG1419c, and its complemented strain, at OD600 nm 0.4 and 0.8 (triplicate cultures). Data are shown as means with bars indicating standard deviation (SD). OD600nm refers to the optical density at 600 nm at which samples were harvested. Statistically significant differences are indicated by the p values shown.
We also compared the amount of biofilm produced at days 10 and 14 of incubation. Even though no major morphological differences were found (Fig. 1c), crystal violet staining allowed us to confirm that BCG∆BCG1419c produced more biofilm than parental BCG but only at day 14 post-incubation (Fig. 1d, p < 0.0001 for BCG∆BCG1419c vs BCG, and 0.0001 for BCG∆BCG1419c vs complemented strain, respectively).
On the other hand, because of the deletion of BCG1419c, we expected BCG∆BCG1419c to modify its content of the second messenger c-di-GMP. We indirectly confirmed this by means of the activity of a reporter gene7. To directly evaluate whether this nucleotide was affected by the presence or absence of BCG1419c, we quantitated this molecule by HPLC. As can be seen in Fig. 1e, we found that, in comparison to parental BCG, BCG∆BCG1419c had an increased amount of c-di-GMP at OD600nm 0.4 (p = 0.0047), an increase that was also observed when compared to its complemented derivative (p = 0.0003), suggesting a reversion to a wild-type like phenotype. However, this was partial as BCG had higher c-di-GMP amount compared with the complemented strain (p = 0.0282). Of note, the increased amount of c-di-GMP observed at OD600nm 0.4 in BCG∆BCG1419c showed a significant decrease when this strain reached OD600nm 0.8 (p = 0.0176), therefore showing that BCG∆BCG1419c indeed produced more c-di-GMP than BCG but only early in its growth in planktonic conditions.
BCG∆BCG1419c does not show alterations in cell envelope lipid components compared with BCG Pasteur ATCC 35734
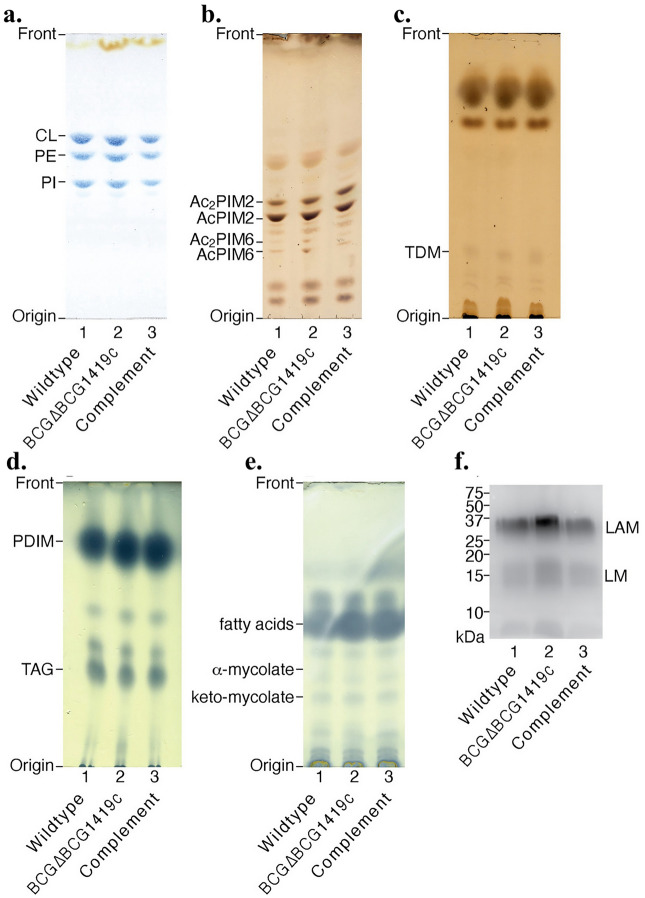
The first generation, hygromycin-resistant version of BCGΔBCG1419c lacked PDIM production in biofilm cultures5, although we did not find transcriptional changes between that mutant and its parental BCG strain that could explain the lack of this compound8. The mycobacterial plasma membrane is composed of major glycerophospholipids such as cardiolipin (CL), phosphatidylethanolamine (PE), and phosphatidylinositol (PI), therefore we characterized the production of these lipids in the second-generation BCGΔBCG1419c and its parental BCG Pasteur ATCC 35734 strain. For this, we extracted cellular lipids and analyzed them by high-performance thin layer chromatography. There were no obvious differences in the major plasma membrane phospholipids between the wildtype and BCG∆BCG1419c (Fig. 2a). Major glycolipid species such as AcPIM2, Ac2PIM2, AcPIM6, and Ac2PIM6 were also detected at comparable levels between the wildtype and the mutant (Fig. 2b). The outer membrane of mycobacteria, known as mycomembrane, is composed of trehalose dimycolates (TDM) (Fig. 2c), phthiocerol dimycocerosates (PDIM), and triacylglycerols (TAG) (Fig. 2d) among other lipids. These outer membrane lipids were also detected in the mutant at comparable levels to the wildtype. Total contents of fatty acids and mycolic acids were analyzed as methyl ester derivatives (α- and keto-mycolic acids) and were also not significantly different between the wildtype and the mutant (Fig. 2e). Mannose-based lipoglycans such as lipomannan (LM) and lipoarabinomannan (LAM) are uniquely found in Mycobacterium species. We observed a slightly increased amount of LAM and LM in BCGΔBCG1419c compared with the wild type and complemented strains, although we did not determine whether this was quantitatively significant or not (Fig. 2f). While we cannot exclude the possibility that subtle structural features of cell envelope lipids are altered in the mutant, our analysis did not detect any differences in major cell envelope lipids in BCG∆BCG1419c compared with BCG Pasteur ATCC 35734 in biofilm cultures.
Figure 2.
Analysis of cell envelope lipids. The lipids of the different BCG strains sampled from planktonic cultures in 7H9 OADC 0.05% Tween 80 at OD600nm 0.8 were extracted and analyzed by HPTLC as described in “Methods”. (a) Phospholipids. (b) PIMs. (c) TDM. (d) PDIMs and TAGs. (e) FAMEs and MAMEs. (f) LM/LAM. Analyses was performed with triplicate samples and a representative image is shown for each lipid class.
We next compared the production of these lipids when BCG∆BCG1419c and BCG Pasteur ATCC 35734 were cultured as planktonic cells in 7H9 OADC Tween 80 or mature (2 weeks-old) biofilm cultures in Sauton media with no detergent. Overall, we did not detect changes in cardiolipin (CL), phosphatidylethanolamine (PE), phosphatidylinositol (PI, supplementary Fig. 1a), Ac2PIM2, and AcPIM2, while Ac2PIM6 and AcPIM6 seemed to differ in their content in planktonic versus biofilm cells but in a strain-independent manner (supplementary Fig. 1b). A slight variation could be observed in TDM present in BCG∆BCG1419c compared to parental BCG in both planktonic and biofilm cultures (supplementary Fig. 1c) although we cannot claim this to be significant as more precise analyses would need to be conducted. All the original TLC plates comparing planktonic versus biofilm cells (lipids) and SDS-PAGE (15% gel) visualized by Pro-Q Emerald 488 Glycoprotein Gel and Blot Stain Kit (Thermo Fisher) (for LM/LAM) are shown in Supplementary Fig. 1.
Transcriptional profiling of BCG∆BCG1419c and BCG during planktonic growth conditions
We previously reported the transcriptional profile differences occurring between the first generation, hygromycin resistant version of BCG∆BCG1419c and its parental strain, BCG Pasteur 1173P2 when grown as biofilms in vitro8. Considering that we produced a second-generation, antibiotic-less version of BCG∆BCG1419c, now in BCG Pasteur ATCC 35734, for further vaccine development15, coupled with the fact that most preclinical studies of mycobacteria-based vaccine safety and/or efficacy are conducted with bacteria grown in planktonic conditions using Middlebrook 7H9 media with 10% ADC/OADC and 0.05% Tween 80, here we decided to investigate the transcriptional response of antibiotic-less BCG∆BCG1419c and its parental strain BCG Pasteur ATCC 35734 grown in planktonic conditions and harvesting cells for RNA isolation at the same stage (OD600nm 0.8) recently reported in an efficacy study against M. tuberculosis HN8787.
The BCG Pasteur 1173P2 genome, used as a reference, has 4109 protein and RNA-encoding genes. Here, we were able to detect gene transcription from all BCG genes (Supplementary Table 1). The most significant functional clusters were defined by DAVID16 indicating significant enrichments for three different functions, being downregulation of transmembrane (72 genes), arginine biosynthesis (7 genes), and cell wall organization (12 genes, Benjamini–Hochberg adjusted p values 0.00026, 0.0015, and 0.036, respectively) in BCG∆BCG1419c compared with its parental strain BCG Pasteur ATCC 35734 (Supplementary Table 1).
Regarding differential gene expression [considered as significant (when both Log2-fold change ≥ 0.585 or ≤ − 0.585 plus p < 0.05) or not] overall, we found relatively few differences and mostly gene downregulation (36 gene upregulated, 123 genes downregulated, Table 1). Of note, among the 123 downregulated genes, we observed that 13 genes belong to the MtrA regulon, and 4 genes belongs to the ArgR regulon (Table 1). Among the genes that were significantly upregulated in BCGΔBCG1419c compared with BCG WT (Log2 Fold-change ≥ 0.585 plus p < 0.05) we found rrs (16S rRNA), rpsN1 (30S ribosomal protein), rrl (23S rRNA), 5S rRNA, rpmC (50S ribosomal protein), glyU (tRNA Gly), serT (tRNA Ser), groES, mbtH, BCG_3249c (a possible anti-sigma factor RshA regulates the activity of the mycobacterial stress response sigma factor SigH), BCG_1418c (hypothetical protein) and BCG_1420 (LuxR family transcriptional regulator, with an adenylate/guanylate cyclase domain, an ATPase domain, and a helix-turn-helix DNA-binding domain) (Table 1).
Table 1.
Differentially expressed genes with significant changes found between BCGΔBCG1419c and BCG in planktonic cultures.
| Upregulated in BCGΔBCG1419c vs BCG WT fold-change ≥ 0.585 plus p < 0.05 | ||||||
|---|---|---|---|---|---|---|
| GENE_NAME | GENE_ID | Mtb ortholog | Product | LOG2FOLD | AVG_PVALUE | Regulon |
| rrs | rrs | 16S rRNA | 3.4982 | 0.000104738 | ||
| EBG00001157317 | EBG00001157317 | SSU_rRNA_archaea | 2.9089 | 0.000387952 | ||
| EBG00001157343 | EBG00001157343 | PK-G12rRNA | 2.2264 | 0.034778283 | ||
| secE2 | BCG_0417 | Rv0379 | Possible protein transport protein secE2 with Calcium dodecin domain | 1.6343 | 0.012587538 | |
| EBG00001157360 | EBG00001157360 | 5_8S_rRNA | 1.5695 | 0.005169291 | ||
| BCG_3831c | BCG_3831c | Probable remnant of a transposase | 1.4757 | 0.049151559 | ||
| BCG_3079c | BCG_3079c | Rv3054c | Conserved hypothetical protein with NAD(P)H-dependent FMN reductase domain | 1.3741 | 0.000116064 | |
| EBG00001157348 | EBG00001157348 | LSU_rRNA_bacteria | 1.2775 | 0.005406273 | ||
| rpsN1 | BCG_0767 | Rv0717 | Probable 30S ribosomal protein S14 rpsN1 | 1.2582 | 0.015651912 | |
| BCG_2010c | BCG_2010c | Rv1993c | Conserved hypothetical protein | 1.1703 | 0.013705724 | |
| rrl | rrl | 23S rRNA | 1.1078 | 0.016674466 | ||
| groES | BCG_3488c | Rv3418c | 10 kDa chaperonin groES | 1.098 | 0.020911838 | |
| EBG00001157315 | EBG00001157315 | 5S_rRNA | 1.05 | 0.031607557 | ||
| BCG_2922c | BCG_2922c | Rv2901c | Conserved hypothetical protein | 1.0287 | 0.037531455 | |
| mbtH | BCG_2391c | Rv2377c | Putative conserved protein mbtH | 1.004 | 0.011346227 | |
| BCG_0026c | BCG_0026c | Rv3920c | Hypothetical protein similar to jag protein | 0.9649 | 0.02941768 | |
| phoY1_1 | BCG_3330c | Rv3301c | Probable phosphate-transport system transcriptional regulatory protein phoU homolog 1 phoY1 | 0.9622 | 0.007044978 | |
| PE_PGRS43b | BCG_2509c | Rv2490c | PE-PGRS family protein [second part] | 0.9184 | 0.047561816 | |
| rpmC | BCG_0759 | Rv0709 | Probable 50S ribosomal protein L29 rpmC | 0.9129 | 0.041923764 | |
| glyU | glyU | tRNA-Gly | 0.9079 | 0.000861898 | ||
| phoY1_2 | BCG_3366c | Rv3301c | Probable phosphate-transport system transcriptional regulatory protein phoU homolog 1 phoY1 | 0.8864 | 0.010088291 | |
| BCG_2220c | BCG_2220c | Rv2204c | Conserved hypothetical protein with iron-sulfur cluster assembly accessory protein domain | 0.8801 | 0.04788665 | |
| BCG_0840 | BCG_0840 | Rv0787A | Conserved hypothetical protein with phosphoribosylformylglycinamidine (FGAM) synthase domain | 0.8727 | 0.029136225 | |
| BCG_1418c | BCG_1418c | Rv1356c | Hypothetical protein | 0.8671 | 0.000299023 | |
| serT | serT | tRNA-Ser | 0.8631 | 0.000637902 | ||
| BCG_2827 | BCG_2827 | Rv2809 | Hypothetical protein | 0.8464 | 0.041819341 | |
| PE_PGRS22 | BCG_1151 | Rv1091 | PE-PGRS family protein | 0.8331 | 0.028777497 | |
| PE_PGRS45b | BCG_2641c | Rv2615c | PE-PGRS family protein | 0.7918 | 0.012061469 | |
| TB7.3_1 | BCG_3248c | Rv3221c | Biotinylated protein TB7.3 contains acetyl-CoA carboxylase biotin carboxyl carrier protein subunit domain | 0.791 | 0.013039128 | |
| TB7.3_2 | BCG_3341c | Rv3221c | Biotinylated protein TB7.3 contains acetyl-CoA carboxylase biotin carboxyl carrier protein subunit domain | 0.7188 | 0.019029613 | |
| BCG_2053 | BCG_2053 | Rv2034 | Probable ArsR-type repressor protein | 0.6551 | 0.018687862 | |
| BCG_1420 | BCG_1420 | Rv1358 | Probable transcriptional regulatory protein | 0.6283 | 0.000717219 | |
| BCG_3348c | BCG_3348c | Rv3225c | Possible transferase, GCN5-related N-acetyltransferase, phosphorylase domain | 0.6262 | 0.003293885 | |
| BCG_3227c | BCG_3227c | Rv3202c | Possible ATP-dependent DNA helicase | 0.5937 | 0.000595553 | |
| BCG_3249c | BCG_3249c | Rv3221A | Possible anti-sigma factor RshA regulates the activity of the mycobacterial stress response sigma factor SigH | 0.5886 | 0.042051519 | |
| gid_1 | BCG_3977c | Probable glucose-inhibited division protein B gid | 0.5873 | 0.421759769 | ||
| Downregulated BCGΔBCG1419c vs BCG WT fold-change ≤ − 0.585 plus p < 0.05 | ||||||
|---|---|---|---|---|---|---|
| GENE_NAME | GENE_ID | Mtb ortholog | Product | LOG2FOLD | AVG_PVALUE | Regulon |
| BCG_3715c | BCG_3715c | Rv3657c | Possible conserved alanine rich membrane protein | − 1.1444 | 0.023526035 | |
| BCG_3154 | BCG_3154 | Rv3131 | Conserved hypothetical protein. Putative nitroreductase, member of the DosR-regulon | − 1.0879 | 0.000785114 | |
| cobLa | BCG_2092c | Probable precorrin-6y methyltransferase CobLa [first part] | − 1.0652 | 0.047535781 | ||
| BCG_0295 | BCG_0295 | Rv0257 | Conserved hypothetical protein | − 0.9771 | 0.027386067 | |
| BCG_0788 | BCG_0788 | Rv0738 | Conserved hypothetical protein with DNA damage-inducible protein DinB domain | − 0.9704 | 0.009118011 | |
| BCG_1419c | BCG_1419c | Rv1357c | Conserved hypothetical protein with cyclic diguanylate phosphodiesterase domain | − 0.9519 | 0.009820681 | |
| pirG | BCG_3872 | Rv3810 | Exported repetitive protein precursor pirG/erp | − 0.9464 | 0.000708503 | MtrA regulon |
| lipF | BCG_3551c | Rv3487c | Probable esterase/lipase lipF | − 0.9198 | 0.000144981 | |
| BCG_1496c | BCG_1496c | Rv1435c | Probable conserved Proline, Glycine, Valine-rich secreted protein | − 0.9068 | 0.006275716 | MtrA regulon |
| BCG_1462 | BCG_1462 | Rv1401 | Possible membrane protein | − 0.8878 | 0.002947653 | |
| BCG_3695 | BCG_3695 | Putative transposase | − 0.8857 | 0.041641661 | ||
| murG | BCG_2170c | Rv2153c | UPD-N-acetylglucosamine-N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol-N-acetylglucosamine transferase MurG | − 0.8737 | 0.009443607 | |
| BCG_2763 | BCG_2763 | Rv2747 | Possible transferase, Probable L-glutamate alpha-N-acetyltranferase ArgA (alpha-N-acetylglutamate synthase) | − 0.8719 | 0.005494031 | |
| BCG_0711c | BCG_0711c | Rv0662c | Conserved hypothetical protein Possible antitoxin VapB7 | − 0.865 | 0.031849363 | |
| lpqZ | BCG_1304 | Rv1244 | Putative lipoprotein lpqZ | − 0.8632 | 0.004945189 | |
| serA2 | BCG_0778c | Rv0728c | Possible D-3-phosphoglycerate dehydrogenase serA2 | − 0.8559 | 0.011718358 | |
| BCG_3153c | BCG_3153c | Rv3130c | Conserved hypothetical protein Triacylglycerol synthase (diacylglycerol acyltransferase) Tgs1 | − 0.8421 | 0.034329997 | |
| nirD | BCG_0291 | Rv0253 | Probable nitrite reductase [NAD(P)H] small subunit nirD | − 0.8362 | 0.018275726 | |
| BCG_0518 | BCG_0518 | Rv0477 | Possible conserved secreted protein | − 0.8313 | 0.005587559 | |
| lipR | BCG_3109 | Rv3084 | Probable acetyl-hydrolase/esterase lipR | − 0.828 | 0.010400861 | |
| argB | BCG_1693 | Rv1654 | Probable Acetylglutamate kinase argB | − 0.8205 | 0.003955123 | ArgR regulon |
| BCG_0699 | BCG_0699 | Rv0650 | Possible sugar kinase with ROK family protein domain | − 0.8087 | 0.027255813 | |
| BCG_1762 | BCG_1762 | Rv1723 | Probable hydrolase with CubicO group peptidase, beta-lactamase class C family domain | − 0.8087 | 0.009155488 | |
| BCG_3465 | BCG_3465 | Rv3395A | Probable membrane protein | − 0.7883 | 0.035013964 | |
| BCG_0008c | BCG_0008c | Rv0008c | Possible membrane protein with cell wall synthesis protein CwsA domain | − 0.7879 | 0.037588749 | |
| BCG_1219c | BCG_1219c | Rv1158c | Conserved hypothetical ala-, pro-rich protein | − 0.7862 | 0.005061423 | MtrA regulon |
| BCG_0702c | BCG_0702c | Rv0653c | Possible transcriptional regulatory protein (probably tetR-family) | − 0.7705 | 0.024929043 | |
| BCG_0501 | BCG_0501 | Rv0461 | Probable transmembrane protein | − 0.7693 | 0.005563857 | |
| BCG_2782 | BCG_2782 | Rv2765 | Probable alanine rich hydrolase with dienelactone hydrolase family protein domain | − 0.7689 | 0.026772117 | |
| speE | BCG_2625 | Rv2601 | Putative spermidine synthase speE | − 0.7643 | 0.008314323 | |
| BCG_0673 | BCG_0673 | Rv0627 | Conserved hypothetical protein Possible toxin VapC5 | − 0.7584 | 0.010403475 | |
| BCG_3664c | BCG_3664c | Rv3600c | Conserved hypothetical protein with type III pantothenate kinase domain | − 0.7558 | 0.009655975 | |
| yrbE1A | BCG_0204 | Rv0167 | Conserved hypothetical integral membrane protein yrbE1A | − 0.7506 | 0.004801835 | |
| adhE1 | BCG_0198c | Rv0162c | Putative zinc-type alcohol dehydrogenase (e subunit) adhE | − 0.7478 | 0.024960044 | |
| BCG_1844 | BCG_1844 | Rv1810 | Conserved hypothetical protein | − 0.7437 | 0.011342536 | |
| purT | BCG_0426 | Rv0389 | Probable phosphoribosylglycinamide formyltransferase 2 purT | − 0.7374 | 0.002402391 | |
| argF | BCG_1695 | Rv1656 | Probable Ornithine carbamoyltransferase, anabolic ArgF | − 0.7363 | 0.010148525 | ArgR regulon |
| BCG_2741c | BCG_2741c | Rv2728c | Conserved hypothetical alanine rich protein with class III extradiol ring-cleavage dioxygenase family protein domain | − 0.7323 | 0.016000596 | |
| BCG_0284 | BCG_0284 | Rv0246 | Probable conserved integral membrane protein | − 0.72 | 0.027263273 | |
| cobD | BCG_2253c | Rv2236c | Probable conserved membrane protein CobD | − 0.7136 | 0.033002992 | |
| deoD | BCG_3372 | Rv3307 | Probable purine nucleoside phosphorylase deoD | − 0.7087 | 0.006876059 | |
| pks16 | BCG_1070 | Rv1013 | Putative polyketide synthase pks16 | − 0.7077 | 0.003716129 | |
| BCG_1059c | BCG_1059c | Rv1002c | Conserved membrane protein with dolichyl-phosphate-mannose–protein mannosyltransferase domain | − 0.707 | 0.005819792 | |
| xerC | BCG_2915c | Rv2894c | Probable integrase/recombinase xerC | − 0.7054 | 0.009092191 | MtrA regulon |
| BCG_0891 | BCG_0891 | Rv0839 | Conserved hypothetical protein with class I SAM-dependent methyltransferase domain | − 0.7016 | 0.001141476 | |
| BCG_1349c | BCG_1349c | Rv1290c | Conserved hypothetical protein | − 0.7016 | 0.005788882 | |
| celA1 | BCG_0093 | Rv0062 | Possible cellulase celA1 (ENDOGLUCANASE) | − 0.7013 | 0.011925226 | |
| BCG_1518c | BCG_1518c | Rv1457c | Probable unidentified antibiotic-transport integral membrane ABC transporter | − 0.7011 | 0.017966363 | |
| BCG_0352 | BCG_0352 | Rv0312 | Conserved hypothetical proline and threonine rich protein with Hsp70 family protein domain | − 0.7007 | 0.006180045 | MtrA regulon |
| BCG_0517 | BCG_0517 | Rv0476 | Possible conserved transmembrane protein | − 0.7003 | 0.008443131 | |
| argJ | BCG_1692 | Rv1653 | Probable Glutamate n-acetyltransferase argJ | − 0.6962 | 0.011937422 | ArgR regulon |
| BCG_1061c | BCG_1061c | Rv1004c | Probable membrane protein | − 0.6961 | 0.028415351 | |
| prrA | BCG_0955c | Rv0903c | Two component response transcriptional regulatory protein prrA | − 0.6901 | 0.002751943 | |
| BCG_0934 | BCG_0934 | Rv0882 | Probable transmembrane protein | − 0.6794 | 0.038109383 | |
| BCG_2713 | BCG_2713 | Rv2700 | Possible secreted alanine rich protein with LytR cell envelope-related transcriptional attenuator domain | − 0.6777 | 0.015900774 | |
| BCG_1310 | BCG_1310 | Rv1250 | Probable drug-transport integral membrane protein | − 0.6758 | 0.013796646 | |
| BCG_3054 | BCG_3054 | Rv3031 | Conserved hypothetical protein with glycoside hydrolase family 57 protein domain | − 0.6735 | 0.012038058 | |
| add | BCG_3379c | Rv3313c | Probable adenosine deaminase add | − 0.6725 | 0.022782705 | |
| panC | BCG_3666c | Rv3602c | Probable pantoate–beta-alanine ligase panC | − 0.6717 | 0.008371359 | |
| BCG_1539 | BCG_1539 | Rv1477 | Hypothetical invasion protein (ripA) | − 0.6698 | 0.00493817 | MtrA regulon |
| ephC | BCG_1185 | Rv1124 | Probable epoxide hydrolase ephC | − 0.6668 | 0.024392971 | |
| BCG_0265 | BCG_0265 | Rv0228 | Probable integral membrane acyltransferase | − 0.6665 | 0.031158919 | |
| BCG_1619c | BCG_1619c | Rv1566c | Possible inv protein | − 0.6661 | 0.007192759 | |
| BCG_1001c | BCG_1001c | Probable mycolyl transferase | − 0.6654 | 0.045990406 | ||
| ugpBa | BCG_2853c | Rv2833c | Probable Sn-glycerol-3-phosphate-binding lipoprotein ugpB [first part] | − 0.6645 | 0.035959294 | |
| BCG_1624 | BCG_1624 | Rv1571 | Conserved hypothetical protein with 2′-5′RNA ligase family protein domain | − 0.663 | 0.035162251 | |
| sugI | BCG_3401 | Rv3331 | Probable sugar-transport integral membrane protein sugI | − 0.6629 | 0.018548577 | |
| BCG_3854 | BCG_3854 | Rv3792 | Probable conserved transmembrane protein Arabinofuranosyltransferase AftA | − 0.6629 | 0.007871683 | |
| BCG_3676c | BCG_3676c | Rv3612c | Conserved hypothetical protein | − 0.6601 | 0.003454172 | |
| dedA | BCG_2664 | Rv2637 | Possible transmembrane protein dedA | − 0.6587 | 0.039418929 | |
| BCG_3491c | BCG_3491c | Rv3421c | Conserved hypothetical protein with tRNA (adenosine(37)-N6)-threonylcarbamoyltransferase complex dimerization subunit type 1 TsaB domain | − 0.6525 | 0.031652896 | |
| BCG_1863 | BCG_1863 | Rv1828 | Conserved hypothetical protein with MerR family transcriptional regulator domain | − 0.6512 | 0.007243893 | |
| BCG_0842c | BCG_0842c | Rv0789c | Hypothetical protein | − 0.65 | 0.042091095 | |
| PPE9 | BCG_0425c | PPE family protein | − 0.6477 | 0.024967853 | ||
| cut4 | BCG_3518 | Rv3452 | Probable cutinase precursor cut4 | − 0.6474 | 0.02589246 | |
| BCG_1218c | BCG_1218c | Rv1157c | Conserved hypothetical ala-, pro-rich protein | − 0.6467 | 0.006357262 | MtrA regulon |
| BCG_3691 | BCG_3691 | Rv3633 | Conserved hypothetical protein with Ectoine hydroxylase-related dioxygenase, phytanoyl-CoA dioxygenase domain | − 0.6456 | 0.004480936 | |
| cobS | BCG_2224 | Rv2208 | Probable cobalamin 5'-phosphate synthase CobS | − 0.6448 | 0.020180231 | |
| BCG_3851 | BCG_3851 | Rv3789 | Conserved hypothetical protein with GtrA domain-containing protein | − 0.6412 | 0.02717627 | |
| echA16 | BCG_2851 | Rv2831 | Probable enoyl-CoA hydratase echa16 | − 0.6407 | 0.040951984 | |
| BCG_3745c | BCG_3745c | Rv3686c | Conserved hypothetical protein | − 0.6383 | 0.023684945 | |
| BCG_0241c | BCG_0241c | Rv0204c | Probable conserved transmembrane protein | − 0.6362 | 0.010034117 | |
| BCG_3015 | BCG_3015 | Rv2994 | Probable conserved integral membrane protein with MFS transporter domain | − 0.6361 | 0.016319392 | |
| leuB | BCG_3016c | Rv2995c | 3-isopropylmalate dehydrogenase leuB | − 0.636 | 0.02535765 | |
| BCG_1657 | BCG_1657 | Rv1619 | Conserved membrane protein | − 0.6359 | 0.029269187 | |
| argD | BCG_1694 | Rv1655 | Probable Acetylornithine aminotransferase argD | − 0.635 | 0.010948399 | ArgR regulon |
| fadD33 | BCG_1407 | Rv1345 | Possible polyketide synthase fadD33 | − 0.6338 | 0.007716787 | |
| BCG_1724c | BCG_1724c | Rv1686c | Probable conserved integral membrane protein ABC transporter | − 0.6334 | 0.03708873 | |
| BCG_2589 | BCG_2589 | Rv2566 | Long conserved hypothetical protein [second part] | − 0.6334 | 0.033570675 | |
| BCG_0388 | BCG_0388 | Rv0349 | Hypothetical protein | − 0.6315 | 0.025410477 | |
| BCG_1540 | BCG_1540 | Rv1478 | Hypothetical invasion protein (ripB) | − 0.6313 | 0.004572077 | MtrA regulon |
| BCG_0330 | BCG_0330 | Rv0290 | Probable conserved transmembrane protein 3 ESX conserved component EccD3. ESX-3 type VII secretion system protein | − 0.6302 | 0.015606343 | |
| BCG_1711c | BCG_1711c | Rv1673c | Conserved hypothetical protein with Transglutaminase-like enzyme, putative cysteine protease domain | − 0.6289 | 0.043524332 | |
| yrbE1B | BCG_0205 | Rv0168 | Conserved hypothetical integral membrane protein yrbE1B | − 0.6288 | 0.016757565 | |
| narL | BCG_0896c | Rv0844c | Possible nitrate/nitrite response transcriptional regulatory protein narL | − 0.627 | 0.010270002 | |
| BCG_0055 | BCG_0055 | Putative secreted protein P60-related protein [second part] | − 0.6268 | 0.026680451 | ||
| BCG_2812c | BCG_2812c | Rv2794c | Conserved hypothetical protein phosphopantetheinyl transferase PptT | − 0.6226 | 0.015274191 | |
| cydD | BCG_1659c | Rv1621c | Probable 'component linked with the assembly of cytochrome' transport transmembrane ATP-binding protein ABC transporter cydD | − 0.6197 | 0.026880206 | |
| BCG_3754 | BCG_3754 | Rv3695 | Possible conserved membrane protein RDD family protein | − 0.6188 | 0.020087176 | |
| BCG_2071c | BCG_2071c | Rv2052c | Conserved hypothetical protein | − 0.6154 | 0.018909791 | |
| BCG_2196 | BCG_2196 | Rv2181 | Probable conserved integral membrane protein Alpha(1→2)mannosyltransferase/manT | − 0.6154 | 0.007679435 | |
| BCG_2692 | BCG_2692 | Rv2679 | Probable enoyl-CoA hydratase echA15 | − 0.6145 | 0.025973179 | |
| pra | BCG_1136 | Rv1078 | Probable Proline-rich antigen homolog pra | − 0.6119 | 0.014551011 | |
| mmsB | BCG_0802c | Rv0751c | Probable 3-hydroxyisobutyrate dehydrogenase mmsB | − 0.6115 | 0.008050415 | |
| mtc28 | BCG_0071c | Rv0040c | Secreted proline rich protein MTC28 (PROLINE RICH 28 KDA ANTIGEN) | − 0.6097 | 0.005413412 | MtrA regulon |
| BCG_3443 | BCG_3443 | Rv3371 | Conserved hypothetical protein possible triacylglycerol synthase (diacylglycerol acyltransferase) | − 0.6089 | 0.006884392 | |
| arsB2 | BCG_3643 | Rv3578 | Possible arsenical pump integral membrane protein arsB2 | − 0.6069 | 0.029200206 | |
| mutA | BCG_1555 | Rv1492 | Probable methylmalonyl-CoA mutase small subunit mutA | − 0.6052 | 0.012106302 | |
| menE | BCG_0586c | Rv0542c | Possible o-succinylbenzoic acid–CoA ligase menE | − 0.6049 | 0.01997206 | |
| BCG_3879 | BCG_3879 | Rv3817 | Putative phosphotransferase | − 0.6039 | 0.027736949 | |
| BCG_1587 | BCG_1587 | Rv1535 | Hypothetical protein | − 0.6026 | 0.024513436 | |
| BCG_1802c | BCG_1802c | Rv1761c | Hypothetical exported protein | − 0.5977 | 0.028422702 | |
| BCG_2479 | BCG_2479 | Rv2459 | Probable conserved integral membrane transport protein MFS transporter | − 0.5973 | 0.007794219 | |
| BCG_3777 | BCG_3777 | Rv3717 | Conserved hypothetical protein with N-acetylmuramoyl-L-alanine amidase domain | − 0.5972 | 0.016730339 | MtrA regulon |
| BCG_1618c | BCG_1618c | Rv1565c | Conserved hypothetical membrane protein with acyltransferase and SGNH domains | − 0.5945 | 0.013232892 | MtrA regulon |
| BCG_0263c | BCG_0263c | Rv0226c | Probable conserved transmembrane protein | − 0.5931 | 0.009766941 | |
| plcD | BCG_1794c | Rv2351c | Probable phospholipase C 4 plcD | − 0.5912 | 0.038388997 | MtrA regulon |
| rfe | BCG_1362 | Rv1302 | Putative undecapaprenyl-phosphate alpha-n-acetylglucosaminyltransferase rfe/wecA | − 0.5904 | 0.032022205 | |
| BCG_0697 | BCG_0697 | Rv0648 | Alpha-mannosidase | − 0.588 | 0.007643403 | |
| BCG_1575 | BCG_1575 | Rv1523 | Probable methyltransferase | − 0.5879 | 0.027733217 | |
| BCG_3514 | BCG_3514 | Rv3448 | Probable conserved integral membrane protein | − 0.5864 | 0.044909115 | |
| BCG_3772 | BCG_3772 | Rv3712 | Possible ligase | − 0.5864 | 0.019375521 | |
| sapM | BCG_3375 | Rv3310 | Possible acid phosphatase | − 0.5853 | 0.010551491 | MtrA regulon |
Genes highlighted in bold correspond to those matching potential pseudogenes/interrupted genes.
Genes that were significantly downregulated in BCGΔBCG1419c compared with BCG WT (Log2 Fold-change ≤ − 0.585 plus p < 0.05) included diverse functions, among others: BCG_3154 (Rv3131, putative nitroreductase, member of the DosR-regulon), BCG_1419c (conserved hypothetical protein with cyclic diguanylate phosphodiesterase domain), lipF (Probable esterase/lipase), murG (UPD-N-acetylglucosamine-N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol-N-acetylglucosamine transferase), argA (alpha-N-acetylglutamate synthase), serA2 (d-3-phosphoglycerate dehydrogenase), and BCG_3153c (Rv3130c, tgs1, triacylglycerol synthase, member of the DosR-regulon) (Table 1).
There was also downregulation of genes belonging to the MtrA regulon: pirG (exported repetitive protein precursor), BCG_1496c (probable conserved Proline, Glycine, Valine-rich secreted protein), BCG_1219c (conserved hypothetical ala-, pro-rich protein), xerC (probable integrase/recombinase), BCG_0352 (conserved hypothetical proline and threonine rich protein with Hsp70 family protein domain), BCG_1539 (hypothetical invasion protein, ripA), BCG_1218c (conserved hypothetical ala-, pro-rich protein), BCG_1540 (hypothetical invasion protein, ripB), mtc28 (secreted proline rich protein, proline rich 28 KDa antigen), BCG_3777 (conserved hypothetical protein with N-acetylmuramoyl-l-alanine amidase domain), BCG_1618c (conserved hypothetical membrane protein with acyltransferase and SGNH domains), plcD (probable phospholipase C 4), and sapM (possible acid phosphatase) (Table 1).
Finally, we also observed downregulation of genes belonging to the ArgR regulon: argB (probable acetylglutamate kinase), argF (probable ornithine carbamoyltransferase), argJ (probable glutamate N-acetyltransferase), argD (probable acetylornithine aminotransferase) (Table 1).
BCG∆BCG1419c does not show changes in macrophage activation via Mincle nor Myd88 compared with BCG as both mycobacteria require these molecules for full induction of TNF-α, IL-6 and G-CSF
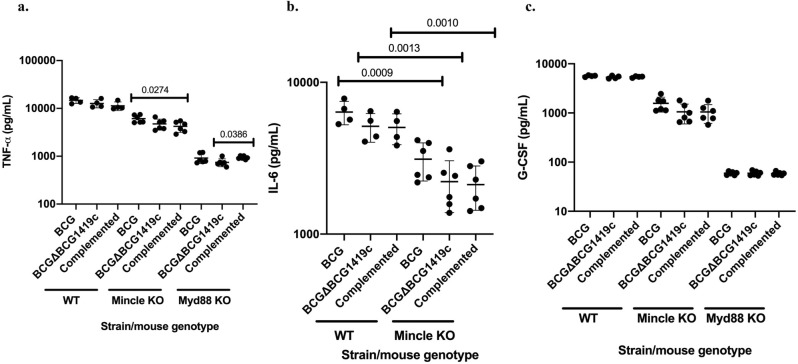
Mycolic acids can be incorporated into trehalose dimycolate (TDM, cording factor), a highly immunostimulatory mycobacterial glycolipid that can signal via Mincle12 and Myd8813. As we previously found transcriptional changes in the expression of genes involved in mycolic acids synthesis in biofilm cultures of the first-generation version of BCGΔBCG1419c8, and even though we did not observe any significant change in the amount of TDM produced between BCG and BCGΔBCG1419c (Fig. 2c), we decided to compare the capacity of BCG, BCGΔBCG1419c and its complemented strain, to induce TNF-α, IL-6 and G-CSF in primary, bone-marrow derived murine macrophages obtained from wild type mice and knock-outs (KO) in Mincle or Myd88, because we cannot rule out that subtle structural features of TDM (i.e. cyclopropanation) could be altered in the mutant. We observed that production of TNF-α, IL-6, and G-CSF was reduced in KO- compared with WT macrophages, regardless of what strain was used to infect them (Fig. 3 and Table 2). The effect of lacking Mincle was statistically significant (two–fivefold decrease) in reducing secretion of TNF-α, IL-6, and G-CSF (Fig. 3 and Table 2; table shows comparison between WT and KO macrophages to avoid crowding Fig. 3a and c). The lack of Myd88 led to significant and even more pronounced decrease in secretion of cytokines (four to sixfold decrease for TNF-α and almost or above 90-fold for G-CSF, not detectable levels for IL-6) compared with WT macrophages (Fig. 3 and Table 2; table shows comparison between WT and KO macrophages to avoid crowding Fig. 3a and c). Together, these results show that macrophage activation via Mincle or Myd88 does not differ between BCG and BCGΔBCG149c as both strains required these receptors to fully activate macrophages to produce the proinflammatory cytokines determined here.
Figure 3.
Cytokine secretion by wild type, Mincle- and Myd88-KO murine macrophages in response to infection with BCG, BCGΔBCG1419c and its complemented strain. BCG strains were obtained from planktonic cultures in 7H9 OADC 0.05% Tween 80 at OD600nm 0.8 and used to infect primary bone-marrow derived macrophages from mice as detailed in “Methods”. (a) TNF-α. (b) IL-6. (c) G-CSF. Macrophages were used for stimulation in duplicate or triplicate wells, giving an average cytokine values from 4 to 6 wells in each experiment. Bars indicate the mean cytokine values with individual values indicated as dots; error bars represent standard deviations of the mean. One-Way ANOVA followed by Dunnett’s multiple comparison test or Kruskal Wallis followed by Dunn’s multiple comparison test was used to assess significance of changes among BCG strains, depending on data distribution. An unpaired t test, Welch’s t test, or Mann–Whitney test was used to compare each cytokine produced by WT and each KO macrophage, depending on data distribution. Statistically significant p actual values are shown on top of the bars depicting the means.
Table 2.
Δ. Comparison of cytokines secretion by wild type, Mincle- or Myd88 KO primary macrophages in response to infection with BCG or BCGΔBCG1419c.
| TNF-α (pg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|
| WT | Mincle KO | Myd88 KO | ||||||
| BCG | BCGΔBCG1419c | Complemented | BCG | BCGΔBCG1419c | Complemented | BCG | BCGΔBCG1419c | Complemented |
| 14,685.25 ± 2022.29 | 12,694.92 ± 2463.62 | 11,330.98 ± 2321.5 | 6101.66 ± 1108.66 | 4737.03 ± 1213.91 | 4206.09 ± 1057.24 | 917.235 ± 216.03 | 745.483 ± 138.99 | 937.91 ± 69.9 |
| p < 0.0001 (WT vs Mincle KO) | p = 0.0001 (WT vs Mincle KO) | p = 0.0095 (WT vs Mincle KO) | p = 0.0095 (WT vs Myd88 KO) | p = 0.0023 (WT vs Myd88 KO) | p = 0.0095 (WT vs Myd88 KO) | |||
| Values shown are the means with standard deviations | p = 0.0022 (Mincle KO vs Myd88KO) | p = 0.0004 (Mincle KO vs Myd88KO) | p = 0.00046 (Mincle KO vs Myd88KO) | |||||
| Mean fold-change vs WT | 2.4 | 2.7 | 2.7 | 6.6 | 6.3 | 4.5 | ||
| IL-6 (pg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|
| WT | Mincle KO | Myd88 KO | ||||||
| BCG | BCGΔBCG1419c | Complemented | BCG | BCGΔBCG1419c | Complemented | BCG | BCGΔBCG1419c | Complemented |
| 6370 ± 1122.04 | 5128 ± 1099.71 | 5050.15 ± 1158.33 | 3114.88 ± 878.28 | 2214.77 ± 828.76 | 2120.979 ± 687.01 | Not determined (ND, below detection limits) | ND | ND |
| p < 0.0001 (WT vs Mincle KO) | p = 0.0001 (WT vs Mincle KO) | p = 0.0095 (WT vs Mincle KO) | ND | ND | ND | |||
| Values shown are the means with standard deviations | ||||||||
| Mean fold-change vs WT | 2 | 2.3 | 2.4 | |||||
| G-CSF (pg/mL) | ||||||||
|---|---|---|---|---|---|---|---|---|
| WT | Mincle KO | Myd88 KO | ||||||
| BCG | BCGΔBCG1419c | Complemented | BCG | BCGΔBCG1419c | Complemented | BCG | BCGΔBCG1419c | Complemented |
| 5639.35 ± 206.47 | 5339.43 ± 349.16 | 5412.86 ± 184.42 | 1570.77 ± 525.94 | 1055.72 ± 458.25 | 1048.38 ± 434.76 | 59.35 ± 4.53 | 59.38 ± 6.26 | 58.39 ± 4.53 |
| p < 0.0001 (WT vs Mincle KO) | p < 0.0001 (WT vs Mincle KO) | p < 0.0001 (WT vs Mincle KO) | p < 0.0001 (WT vs Myd88 KO) | p < 0.0001 (WT vs Myd88 KO) | p < 0.0001 (WT vs Myd88 KO) | |||
| Values shown are the means with standard deviations | p = 0.0009 (Mincle KO vs Myd88 KO) | p = 0.0031 (Mincle KO vs Myd88 KO) | p = 0.0026 (Mincle KO vs Myd88 KO) | |||||
| Mean fold-change vs WT | 3.6 | 5.1 | 5.2 | 95 | 89.9 | 92.7 | ||
Discussion
Mycobacterium bovis Bacille Calmette-Guèrin (BCG) remains, after over one century now, as the only vaccine in use to fight the global burden imposed by TB. Given that BCG has been unable to reduce pulmonary TB, which accounts for roughly 80% cases of this disease, a number of vaccine candidates intended to replace or improve BCG have been developed, including live, attenuated, mycobacteria-based vaccine (LAV) candidates, which aim to increase safety, immunogenicity, and efficacy of current BCG4.
One of these LAV candidates is BCGΔBCG1419c, developed following the overarching hypothesis that in vitro biofilms produced by mycobacteria resemble yet not fully explored aspects of TB pathogenesis9. This is an approach to produce a vaccine candidate against TB entirely different from most other candidates currently in the pipeline17 (https://newtbvaccines.org/tb-vaccine-pipeline/preclinical-stage/). Of note, very recent evidence showing biofilm-like structures observed in TB lesions from mice, guinea pigs and humans10 further suggest it might be worthwhile to continue developing and conducting more in-depth characterization of BCGΔBCG1419c.
Considering the differences between first- and second-generation versions of BCGΔBCG1419c (a BCG Pasteur 1173P2 background that maintained a hygromycin resistance-HygR-gene compared with a BCG Pasteur ATCC 35734 background with no antibiotic resistance gene, respectively), we wanted to continue the non-clinical characterization of antibiotic-less BCGΔBCG1419c by determining colonial morphology and biofilm production, two phenotypes that were altered in the HygR version of BCGΔBCG1419c compared with its parental BCG Pasteur 1173P25.
Here, we found that antibiotic-less BCGΔBCG1419c maintained changes in colonial morphology and biofilm production that were restored to a full (biofilm) or partial (colony morphology) extent upon reintroduction of a single copy of the homologous gene, Rv1357c, under its native regulatory region, integrated into the genome of the deletion mutant (Fig. 1a,c,d); this confirms that these phenotypes are associated with the presence or absence of the c-di-GMP phosphodiesterase-encoding BCG1419c gene. On the other hand, by directly measuring c-di-GMP by HPLC, we confirmed that BCGΔBCG1419c produced more of this second messenger in planktonic cultures at early logarithmic growth phase (Fig. 1e), a result in agreement with our prior indirect determination of c-di-GMP levels in BCGΔBCG1419c and BCG Pasteur ATCC 357347. The alterations in c-di-GMP levels might seem small in terms of the experimental value determined, yet it may represent a great scale when it comes to physiological changes. We must also bear in mind that BCGΔBCG1419c still maintains a copy of BCG1416c in its genome11. This is relevant because the Mtb homologue to BCG1416c is Rv1354c, whose enzyme has PDE activity in vitro18, which may contribute to a not so high increase in c-di-GMP content. Furthermore, it was shown that the recombinant product of Rv2837c, hydrolyzes c-di-AMP efficiently and also possesses PDE activity for c-di-GMP19, therefore potentially contributing to the relatively minor changes observed in our works. Taken together, along with the fact that we are not overexpressing any diguanylate cyclase-encoding gene in our work, these evidences may explain why the tightly controlled (and low level) of c-di-GMP detected in our direct HPLC measurements.
HygR BCGΔBCG1419c lacked PDIM5 in biofilm cultures, yet transcriptional profiling of this same growth condition did not show significant changes in transcription of genes involved in the synthesis or export of this complex lipid, a phenotype that was not restored upon reintegration of Rv1357c into the mutant5. Perhaps not surprisingly, we found that antibiotic-less BCGΔBCG1419c was not defective in producing PDIM (Fig. 2d). This confirms that PDIM production is not dependent on the presence or absence of BCG1419c, at least under in vitro growth conditions, and potentially that the lack of PDIM in the hygromycin-resistant mutant was a product of an unexpected mutation.
Additional lipidomic analyses showed that, overall, this mutant had no major significant differences compared with BCG Pasteur ATCC 35734 in the amount of cardiolipin, phosphatidyl ethanol amine, phosphatidyl inositol (Fig. 2a), phosphatidyl inositol mannosides (Fig. 2b), TDM (Fig. 2c) and mycolic acids (Fig. 2e) produced in biofilm cultures, the only possible exception being a slightly increased amount of LAM, although we did not determine whether this was quantitatively significant or not (Fig. 2f).
On the other hand, when we compared the lipidomic profiles of biofilm and planktonic cultures of BCGΔBCG1419c and its parental BCG Pasteur ATCC 35734 strain, we observed two possible differences: (1) an apparent change in the production of Ac2PIM6 and AcPIM6, although this was in a strain-independent manner, and (2) in TDM in a growth condition-independent manner. In both instances, we would need to formally evaluate the significance of these possible changes, or lack of it (Supplementary Fig. 1). Together, these results show that there were no differences in major cell envelope lipids in BCG∆BCG1419c compared with BCG Pasteur ATCC 35734 and suggest that the lack of PDIM in HygR BCGΔBCG1419c could have been the consequence of an unanticipated spontaneous mutation that did not affect gene expression but that affected production and/or export of this complex lipid. Said hypothetical mutation could have occurred either during the mutagenesis performed to replace BCG1419c, or at any other moment after in vitro passages, as it is known that this can select for spontaneous mutations leading to PDIM loss20.
Given that most preclinical tests of novel LAV are conducted with bacteria cultured in planktonic conditions, employing shaken cultures in Middlebrook 7H9 medium with ADC/OADC supplement and detergent, in this work we decided to characterize the transcriptional profile of BCGΔBCG1419c and BCG when cultured in conditions that our group has recently employed to characterize the genome, safety, immunogenicity, efficacy, and proteome of the antibiotic-less, second-generation version of BCGΔBCG1419c6,7,11,15. RNASeq comparison of these strains found a few upregulated genes with most changes corresponding with downregulation of transcription in BCGΔBCG1419c compared with BCG Pasteur ATCC 35734. The main feature observed for upregulation was that BCGΔBCG1419c increased transcription of genes involved in ribosomal and/or protein synthesis, including rrs (16S rRNA), rpsN1 (30S ribosomal protein), rrl (23S rRNA), 5S rRNA, rpmC (50S ribosomal protein), glyU (tRNA Gly), and serT (tRNA Ser) (Table 1).
Most changes observed in BCGΔBCG1419c were reduced transcription, including that of genes belonging to the MtrA regulon (Table 1). Among the known functions of the MtrA regulon, we recently reported that it modulates cell division and intrinsic tolerance and drug resistance21. Within this regulon, pirG (exported repetitive protein precursor, erp22) has been described as required for virulence, whereas BCG_1539 (hypothetical invasion protein, ripA23) is required for peptidoglycan cleavage and virulence, BCG_1540 (hypothetical invasion protein, ripB24) is also required for peptidoglycan remodeling. On the other hand, mtc28 (secreted proline rich protein, proline rich 28 KDa antigen25) is an antigenic secreted protein, and sapM (possible acid phosphatase26) hydrolyzes PI3P, contributing to inhibition of phagosome-late endosome fusion (Table 1).
Moreover, we observed that transcription of several genes involved in cell wall synthesis (Table 1) were downregulated in BCGΔBCG1419c compared with BCG, among them: murG (BCG_2170, UPD-N-acetylglucosamine-N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol-N-acetylglucosamine transferase), cswA (BCG_0008c, possible membrane protein with cell wall synthesis protein CwsA domain27), rfe/wecA (BCG_1362, putative undecapaprenyl-phosphate alpha-n-acetylglucosaminyltransferase28), BCG_3777 (conserved hypothetical protein with N-acetylmuramoyl-L-alanine amidase domain29), aftA (BCG_3854, probable conserved transmembrane protein arabinofuranosyltransferase30), and BCG_3443 (conserved hypothetical protein possible triacylglycerol synthase (diacylglycerol acyltransferase) domain31), which are involved in synthesis of the mycolyl-arabinogalactan-peptidoglycan complex. Further to this, manT (BCG_2196, probable conserved integral membrane protein alpha(1→2) mannosyltransferase32), and BCG_1059 (conserved membrane protein with dolichyl-phosphate-mannose–protein mannosyltransferase domain33) have been described to be required for mannose-capping of lipoarabinomannan, and protein-O-mannosylation (required for virulence), respectively.
Taken together, transcriptional data supports the notion of BCGΔBCG1419c having an altered cell wall compared with BCG, which may affect its integrity, and could explain why planktonic BCGΔBCG1419c showed constriction and concavities of the cell wall in some bacteria, while in others their cell wall was thinner than that of the parental BCG strain (Fig. 1b). On the other hand, when we compared the transcriptomic data generated in this study against proteomic data under the same experimental conditions, we found that argF was significantly downregulated in transcriptome (Table 1), whereas the protein was significantly upregulated in proteome comparisons between BCGΔBCG1419c and BCG7; BCG_2220c was significantly upregulated in transcriptome (Table 1), whereas the protein was significantly downregulated in proteome comparison7, and BCG_0075c was significantly downregulated both at the transcript (Table 1) and protein levels7. Other than this, we noticed that there was a poor correlation between transcriptomic and proteomic differences found between BCGΔBCG1419c and BCG under planktonic growth conditions (Middlebrook 7H9 media with 0.2% glycerol, 10% OADC and 0.05% Tween 80). This has already been suggested to be the case due to differences in half lives and post transcription machinery utilization34.
In planktonic cells, we also observed significant downregulation in BCGΔBCG1419c compared with BCG of genes belonging to different two-component systems, which are involved in response to multiple environmental cues and that are relevant for bacterial adaption. Among them, we found BCG_3154 (Rv3131, putative nitroreductase) and BCG_3153c (Rv3130c, tgs1, triacylglycerol synthase), members of the DosR regulon, as well as reduced transcription of narL, and prrA (Table 1). Why do we think this is relevant? On the one hand, DosR and NarL have been shown to interact in vivo and co-regulate gene expression during aerobic nitrate metabolism in M. tuberculosis35. DosR is known to be induced upon hypoxia and response to NO36 while prrA also affected M. tuberculosis response to NO and hypoxia37. We also observed downregulation of genes belonging to the ArgR regulon (Table 1). Of note, in BCG, c-di-GMP was suggested to be required for adaptation to hypoxia in an ArgR-dependent manner, which induced arginine and nitrite metabolism gene clusters38; moreover, it is known that high c-di-GMP levels induce the expression of the DosR operon in Mycobacterium smegmatis39 to respond to oxidative stress. Together, we can hypothesize that the reduced transcription of BCG_3154, BCG_3153c, and the ArgR regulon, observed at DO600nm 0.8 in BCGΔBCG1419c compared with BCG (Table 1) might be the consequence of reduced c-di-GMP content at this stage (Fig. 1d). Whether this affects the capability of BCGΔBCG1419c to adapt to and survive under hypoxia, remains to be determined. This hypothesis coupled to the fact that PrrA and MtrA (which included several genes downregulated in BCGΔBCG1419c compared with BCG) are both targets of protein phosphorylation mediated by PknK40, this potentially links the activity of their target genes.
We previously found downregulation in the expression of genes involved in mycolic acids synthesis (groEL1, fas, kasA, kasB, acpM, fabD) in biofilm cultures of HygR BCGΔBCG1419c compared with BCG8. From these genes, fas (Log2 − 0.5303, p = 0.038396) and fabD (Log2 − 0.4719, p = 0.044258) were also downregulated in planktonic cultures of BCGΔBCG1419c compared with BCG, while the remaining genes were not significantly affected (Supplementary Table 1). Downregulation of transcription of these two genes, coupled with the observed downregulation of some members of the DosR- and ArgR-regulons observed in antibiotic-less BCGΔBCG1419c compared with BCG further strengthen the notion that regardless of growth condition (biofilm or planktonic cultures), our vaccine candidate has a distinct profile to that of parental BCG. The fact that we found more differentially expressed genes in this work compared to our previous report using mature biofilm cultures8 may be the consequence of a greater transcriptional activity occurring in log-phase planktonic cultures used here, or because of a higher heterogeneity that may occur in biofilm cultures, which may reduce consistent transcriptional measurement in the bulk population.
Mycolic acids be incorporated into trehalose dimycolate (TDM, cord factor), which activates macrophages via Mincle and Myd8813. We found that macrophage activation via Mincle or Myd88 did not differ between BCG and BCGΔBCG149c, with both strains requiring these receptors to fully activate macrophages to produce TNF-α, IL-6, and G-CSF, with a more pronounced decrease in macrophages lacking Myd88 (Fig. 3 and Table 2). These findings are in agreement with the known roles of these receptors in response to mycobacterial antigens, especially TDM13,41, and given the absence of any significant difference in the induction of TNF-α, IL-6, and G-CSF by BCGΔBCG149c compared with BCG, this should rule out the hypothesis of potential modifications in TDM produced by BCGΔBCG149c, at least to the extent capable of inducing differential secretion of the cytokines determined here.
Taken together, our results show that BCGΔBCG1419c has a different colonial, ultrastructural, biofilm, c-di-GMP production, and transcriptomic profile (in planktonic cultures) compared with BCG, possibly explaining structural differences in their cell wall as well as suggesting potential differences that might be relevant for in vivo conditions, which deserve further explorations in future works. On the other hand, we demonstrated that there was no major change in the lipidomic profile of BCGΔBCG1419c compared with BCG, strengthening the notion that the lack of PDIM in HygR BCGΔBCG1419c was the consequence of an unanticipated mutation. We also showed that induction of TNF-α, IL-6 or G-CSF by macrophages infected with BCGΔBCG1419c and BCG require Mincle and Myd88.
Methods
BCG culture
Mycobacterium bovis Pasteur BCG (BCG) ATCC 35734 strain and its isogenic derivative, BCGΔBCG1419c, which lacks a cyclic di-GMP phosphodiesterase encoded by the BCG1419c gene were grown in 7H9 liquid medium supplemented with OADC, 0.1% Tween 80 for 15 days at 37 °C and 5% CO2. Shaking cultures were then started at 100 rpm starting from an initial OD of 0.05 in supplemented 7H9 medium until reaching an OD of ~ 0.8. Starting from this culture, the cultures for each of the trials were prepared.
Colonial morphology
A bacterial suspension of each of the BCG Pasteur and BCGΔBCG1419c strains was prepared at OD600nm 0.03, then two serial dilutions 1:100 were made, inoculated onto 7H10 agar plates supplemented with OADC, and 0.5% glycerol and they were incubated for 21 days at 37 °C in the presence of 5% CO2. After that time, to observe colonial morphology, colonies of BCG WT, BCG∆BCG1419c and complemented strains observed and recorded using a stereoscope with incident light and background illumination with a magnification of 10×.
Ultrastructural analysis by transmission electron microscopy
Bacilli were cultured in 7H9 OADC Tween 80 medium until OD600 ≈ 1.0–1.3, harvested and fixed by immersion in a solution of 4% glutaraldehyde dissolved in cacodylate buffer for 4 h, followed by second fixation with osmium tetroxide fumes. Then, the bacterial suspension was centrifuged to form a pellet that was dehydrated with graded ethyl alcohol and embedded in Spur resin (London Resin Company, Aldermaston, UK). Sections or 70 nm to 90 nm width were obtained and placed on copper grids, contrasted with uranium salts and Reynold’s lead citrate (Electron Microscopy Sciences, Hatfield, PA, USA), and examined with an FEI Tecnai G2 Spirit Transmission Electron Microscope (Hillsboro, OR, USA).
Biofilm culture
The BCG Pasteur and BCG∆BCG1419c strains were grown in liquid Sauton media without detergent (l-Asparagine 4 g, citric acid 2 g, KH2PO4 0.5 g, MgSO4 0.5 g, ferric ammonium citrate 0.05 g, glycerol 60 mL, 1% ZnSO4, 1 mL per liter, pH adjusted to 7 with 5 N NaOH). Before this, a 20 mL pre-inoculum was prepared in Middlebrook 7H9 medium supplemented with OADC and 0.05% Tween 80 at an initial OD600nm of 0.03, cultured statically at 37 °C and 5% CO2, until reaching an optical density between 0.8 and 1.0. Then, cells were washed. The pellet was resuspended in 10 mL of Sauton without detergent and cultures were prepared in Sauton liquid medium without detergent at an OD600nm of 0.05, and 750 µL of total volume were placed in each well. The plates were incubated at 37 °C in the presence of 5% CO2 for 10 and 14 days.
Biofilm quantification by crystal violet staining
After 10 days and 14 days of incubation, the liquid medium was removed, and all surface film and biofilm adhered to the wells remained. Immediately, 1 mL of methanol was added to each well to fix the biofilm for 5 min, the solvent was removed, and it was left to dry for 24 h at room temperature. Immediately, 1 mL of 1% crystal violet was added to each well, allowing the dye to incorporate into the biofilm for 5 min. The dye was removed, and 4 washes were carried out with distilled water to remove the residual dye. The biofilm-bound dye was extracted by adding 1 mL of 30% acetic acid to each well and allowed to incubate for 24 h. The sample from each well was diluted 1:5 in distilled water for cultures in Sauton medium and read with a UV–Vis spectrophotometer at an absorbance of 595 nm using water as a blank and the 30% acetic acid solution in the appropriate proportions according to the dilution used.
c-di-GMP quantification by HPLC
After reaching the desired OD600nm (0.4 and 0.8), cells were washed thrice with 7H9, followed by cell lysis using the bead beating method, spin the lysed cells at 12,000 rpm for 30ʹ and recover the supernatant. The supernatant is injected into a C18 column. The mobile phase was acetonitrile (30% acetonitrile: 70% Water), and the detection wavelength was 254 nm, which is specific for cDG and not any other nucleotide. The half-life of other nucleotides differs from cDG. So, we can be sure that the molecule getting detected is cDG and not anything else. To quantify the absolute c-di-GMP concentration, we use the area under the curve (AUC) obtained for the sample by using the equation obtained from the standard curve of known concentrations. Values were normalized to the total protein concentration of each sample analyzed.
RNA isolation
Cells in RNA later were shipped from CIATEJ to Institute for Systems Biology (ISB), at room temperature, At ISB, samples were transferred to a tube containing Lysing Matrix B (MP Biomedicals, Santa Ana, CA) and vigorously shaken at max speed for 30 s in a FastPrep 120 homogenizer (MP Biomedicals) three times. Tubes were centrifuged for 1 min (max speed), then 66 μL of 3 M sodium acetate pH 5.2 added and mixed well. Acid phenol (pH 4.2) was added at 726 μL and tubes were inverted to mix well (~ 60 times). Samples were incubated at 65 °C for 5 min, inverting tubes to mix samples every 30 s. Then, centrifuged at 14,000 rpm for 5 min and upper aqueous phase was transferred to a new tube. 3 M sodium acetate (pH 5.2) was added at 1/10th volume along with 3× volumes of 100% ethanol. Sample was mixed well and incubated at − 20 °C for 1 h or overnight. Following incubation, samples were centrifuged at 14,000 rpm for 30 min at 4 °C, ethanol was discarded and 500 μL of 70% ethanol was added. Samples were centrifuged again at 14,000 rpm for 10 min at 4 °C, supernatant discarded, and any residual ethanol removed using pipet. Pellet was allowed to air dry, resuspended in 30–40 μL of RNase free water and quantified by Nanodrop (Thermo Scientific). This was followed by in solution genomic DNA digestion using RQ1 Dnase (Promega) following manufacturer’s recommendation. RNA quality was analyzed in a 2100 Bioanalyzer system (Agilent Technologies). Total RNA samples were depleted of ribosomal RNA using the Ribo-Zero Bacteria rRNA Removal Kit (Illumina, San Diego, CA). Samples were prepared with TrueSeq Stranded mRNA HT library preparation kit (Illumina, San Diego, CA) and sequenced on the NextSeq 2000 instrument with NextSeq P2 (200 cycles) kit.
RNA-seq analysis
Raw FASTQ read data were processed using the R package DuffyNGS as described previously42. Briefly, raw reads were filtered for rRNA transcripts and aligned against the M. bovis BCG str. Pasteur (1173P2) genome with Bowtie243, using the command line option “very-sensitive.” BAM files recorded both uniquely mapped and multiply mapped reads to each of the forward and reverse strands of the genome(s) at single-nucleotide resolution. Gene transcript abundance was then measured by summing total reads landing inside annotated gene boundaries, expressed as both RPKM and raw read counts. Two stringencies of gene abundance were provided using all aligned reads and by just counting uniquely aligned reads. The raw and processed RNA-seq data generated for this study are available in the Gene Expression Omnibus under accession number GSE251760.
Differentially expressed genes
A panel of 5 DE tools was used to identify gene expression changes between the mutant Δ1419 and wild type. The tools included (i) RoundRobin (in-house); (ii) RankProduct44; (iii) significance analysis of microarrays (SAM)45; (iv) EdgeR46; and (v) DESeq247. Each DE tool was called with appropriate default parameters and operated on the same set of transcription results, using RPKM abundance units for RoundRobin, RankProduct, and SAM and raw read count abundance units for DESeq2 and EdgeR. All 5 DE results were then synthesized, by combining gene DE rank positions across all 5 DE tools. Specifically, a gene’s rank position in all 5 results was averaged, using a generalized mean to the 1/2 power, to yield the gene’s final net rank position. Each DE tool’s explicit measurements of differential expression (fold change) and significance (p-value) were similarly combined via appropriate averaging (arithmetic and geometric mean, respectively). Genes with P-value (averaged P-value across the tools) below 0.05 were considered differentially expressed. We performed BLAST48 to identify homologs in M. tuberculosis H37Rv and functional term clusters were defined by DAVID16 for either up or down-regulated significantly expressed genes. Conserved domains were searched for proteins with Conserved Hypothetical Functions at https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi with default parameters.
Lipid extraction and analyses
Planktonic and biofilm cultures produced at CIATEJ were lyophilized and shipped at room temperature to University of Massachusetts at Amherst (UMA). There, to harvest phospholipids and glycolipids, 10 volumes (v/w) of chloroform/methanol (2:1, v/v) were added to lyophilized cells. The cell suspension was briefly vortexed, sonicated, and incubated at room temperature for at least one-hour. The suspension was spun down at 16,900×g on a microfuge for 1 min to harvest the supernatant. We repeated the extraction with10 volumes of chloroform/methanol (2:1, v/v) and 10 volumes of chloroform/methanol/water (1:2:0.8, v/v/v) against the same pellet. The combined lipid extract was further purified by n-butanol/water phase partitioning as previously49. The final lipid extract was analyzed by high-performance thin layer chromatography (HPTLC) (silica gel 60, EMD Merck) using the solvent systems as follows: phospholipids were developed on an HPTLC plate in a solvent containing chloroform/methanol/13 M ammonia/1 M ammonium acetate/water (180:140:9:9:23, v/v/v/v/v) and detected by molybdenum blue staining. PIMs were developed on an HPTLC plate in a solvent containing chloroform/methanol/13 M ammonia/1 M ammonium acetate/water (180:140:9:9:23, v/v/v/v/v) and detected by orcinol staining. TDM was developed on an HPTLC plate in a solvent containing chloroform/methanol/water (90:10:1, v/v/v) and detected by orcinol staining.
The delipidated cell pellet after the lipid extraction described above was subjected to LM/LAM extraction using hot phenol and analyzed by SDS-PAGE (15% gel) and visualized by Pro-Q Emerald 488 Glycoprotein Gel and Blot Stain Kit (Thermo Fisher) as described previously50.
For the analysis of PDIMs, we suspended 100 mg of lyophilized cells in 1 ml of methanol/0.3% (w/v) aqueous sodium chloride (10:1) and 0.5 ml of petroleum ether. The cell suspension was vortexed for 15 min to emulsify the two layers, and the top petroleum ether phase was harvested after brief centrifugation. The extraction was repeated with another 0.5 ml of petroleum ether against the same methanol/water phase. The combined petroleum ether phase was dried, resuspended in 100 µl of petroleum ether, and 10 µl was spotted on an HPTLC plate. HPTLC plate was developed in petroleum ether/diethyl ether (9:1) once, and PDIMs plus TAGs were visualized by phosphomolybdic acid staining.
For mycolic acid methyl ester (MAME) and fatty acid methyl ester (FAME) analysis, 50 mg of lyophilized cells were incubated overnight at 100 °C in 2 ml of 15% tetra butyl ammonium hydroxide. The alkaline-hydrolyzed materials were mixed with 2 ml water, 1 ml dichloromethane, and 250 µl iodomethane to produce methyl esters of fatty acids and mycolic acids. The mixture was incubated at room temperature for 30 min with mixing. The upper aqueous layer was discarded, and the lower organic layer was washed with 3 ml of 1 M HCl and 3 ml of water. The organic layer was dried and resuspended in 500 µl of toluene/acetonitrile (2:3). The MAMEs and FAMEs were separated by HPTLC with petroleum ether/acetone (95:5) once and visualized by phosphomolybdic acid staining.
Macrophage obtention and determination of cytokines
In each experiment, bone-marrow derived macrophages (BMM) were differentiated from 2 different mice for each genotype (C57BL/6, Mincle-KO (MNA), and Myd88-KO mice in complete DMEM containing 10% L-cell conditioned media as a source of M-CSF for 7 days. BMM were harvested using Accutase (Sigma-Merck), washed and plated in cDMEM without M-CSF or antibiotics in 96 flat-bottom well plates at 2 × 105 cells per well. After overnight incubation, BCG strains were added at MOI 5. 24 h later, the supernatants were harvested and analyzed by ELISA (DuoSet, R&D Systems) for TNF-α, IL-6 and G-CSF according to the manufacturer’s instructions. BMM were plated for stimulation in duplicate or triplicate wells, giving us average cytokine values from 4 to 6 wells in each experiment.
Supplementary Information
Acknowledgements
M.A.F.V. is grateful to CIATEJ for internal funding to complete this project. M.J.A.S. received a Ph.D. fellowship from CONACYT (Number 745841). Support by BayLAT to initiate the cooperation between M.A.F.V. and R.L. and technical help by Barbara Bodendorfer is acknowledged.
Author contributions
Conceptualization M.A.F.V.; Methodology M.A.F.V., M.J.A.S., E.P., N.B., Y.M., I.S. D.K.S., R.Y., R.L., and J.C.L.C.; Investigation M.A.F.V., M.J.A.S., E.P., Y.M., I.S., R.Y., D.M.E., J.C.L.C. and R.H.P.; Formal analysis M.A.F.V., M.J.A.S., E.P., N.B., Y.M., D.K.S., R.L., and R.H.P.; Writing M.A.F.V., E.P., and R.L.; Resources M.A.F.V., N.B., R.L., and R.H.P.
Competing interests
M.A.F.V., M.J.A.S., and R.H.P. are co-inventors on a patent on BCGΔBCG1419c held by the Centro de Investigación y Asistencia en Tecnología y diseño del Estado de Jalisco (CIATEJ), A.C. and Instituto Nacional de Ciencias Médicas y Nutrición “Salvador Zubirán”, which could be construed as a potential conflict of interest. M.A.F.V. is an editorial board member (Microbiology) at Scientific Reports. All other authors do not have any conflict of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-61815-8.
References
- 1.Organization, W. H. Global tuberculosis report 2023. (2023).
- 2.Flores-Valdez MA. After 100 years of BCG immunization against tuberculosis, what is new and still outstanding for this vaccine? Vaccines. 2021 doi: 10.3390/vaccines10010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasiliu A, et al. Tuberculosis prevention: Current strategies and future directions. Clin. Microbiol. Infect. 2023 doi: 10.1016/j.cmi.2023.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores-Valdez MA, Kupz A, Subbian S. Recent developments in mycobacteria-based live attenuated vaccine candidates for tuberculosis. Biomedicines. 2022 doi: 10.3390/biomedicines10112749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flores-Valdez MA, et al. The cyclic Di-GMP phosphodiesterase gene Rv1357c/BCG1419c affects BCG pellicle production and in vivo maintenance. IUBMB Life. 2015;67:129–138. doi: 10.1002/iub.1353. [DOI] [PubMed] [Google Scholar]
- 6.Velazquez-Fernandez JB, et al. Proteomic characterization of a second-generation version of the BCGDeltaBCG1419c vaccine candidate by means of electrospray-ionization quadrupole time-of-flight mass spectrometry. Pathog. Dis. 2021 doi: 10.1093/femspd/ftaa070. [DOI] [PubMed] [Google Scholar]
- 7.Aceves-Sanchez MJ, et al. BCG∆BCG1419c and BCG differ in induction of autophagy, c-di-GMP content, proteome, and progression of lung pathology in Mycobacterium tuberculosis HN878-infected male BALB/c mice. Vaccine. 2023;41:3824–3835. doi: 10.1016/j.vaccine.2023.04.065. [DOI] [PubMed] [Google Scholar]
- 8.Flores-Valdez MA, et al. The BCGDeltaBCG1419c vaccine candidate reduces lung pathology, IL-6, TNF-alpha, and IL-10 during chronic TB infection. Front. Microbiol. 2018;9:1281. doi: 10.3389/fmicb.2018.01281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flores-Valdez MA. Vaccines directed against microorganisms or their products present during biofilm lifestyle: Can we make a translation as a broad biological model to tuberculosis? Front. Microbiol. 2016;7:14. doi: 10.3389/fmicb.2016.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakraborty P, Bajeli S, Kaushal D, Radotra BD, Kumar A. Biofilm formation in the lung contributes to virulence and drug tolerance of Mycobacterium tuberculosis. Nat. Commun. 2021;12:1606. doi: 10.1038/s41467-021-21748-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Auria G, et al. Genome sequences of BCG Pasteur ATCC 35734 and its derivative, the vaccine candidate BCGDeltaBCG1419c. BMC Genomics. 2023;24:69. doi: 10.1186/s12864-023-09169-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen M, et al. Macrophage phosphoproteome analysis reveals MINCLE-dependent and -independent mycobacterial cord factor signaling. Mol. Cell Proteomics. 2019;18:669–685. doi: 10.1074/mcp.RA118.000929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoenen H, et al. Differential control of Mincle-dependent cord factor recognition and macrophage responses by the transcription factors C/EBPbeta and HIF1alpha. J. Immunol. 2014;193:3664–3675. doi: 10.4049/jimmunol.1301593. [DOI] [PubMed] [Google Scholar]
- 14.Walker KB, et al. The second Geneva Consensus: Recommendations for novel live TB vaccines. Vaccine. 2010;28:2259–2270. doi: 10.1016/j.vaccine.2009.12.083. [DOI] [PubMed] [Google Scholar]
- 15.Aceves-Sanchez MJ, et al. Vaccination with BCGDeltaBCG1419c protects against pulmonary and extrapulmonary TB and is safer than BCG. Sci. Rep. 2021;11:12417. doi: 10.1038/s41598-021-91993-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Cobelens F, et al. Accelerating research and development of new vaccines against tuberculosis: A global roadmap. Lancet Infect. Dis. 2022;22:e108–e120. doi: 10.1016/S1473-3099(21)00810-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta K, Kumar P, Chatterji D. Identification, activity and disulfide connectivity of C-di-GMP regulating proteins in Mycobacterium tuberculosis. PLoS ONE. 2010;5:e15072. doi: 10.1371/journal.pone.0015072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dey RJ, et al. Inhibition of innate immune cytosolic surveillance by an M. tuberculosis phosphodiesterase. Nat. Chem. Biol. 2017;13:210–217. doi: 10.1038/nchembio.2254. [DOI] [PubMed] [Google Scholar]
- 20.Domenech P, Reed MB. Rapid and spontaneous loss of phthiocerol dimycocerosate (PDIM) from Mycobacterium tuberculosis grown in vitro: Implications for virulence studies. Microbiology. 2009;155:3532–3543. doi: 10.1099/mic.0.029199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peterson EJR, et al. MtrA modulates Mycobacterium tuberculosis cell division in host microenvironments to mediate intrinsic resistance and drug tolerance. Cell Rep. 2023;42:112875. doi: 10.1016/j.celrep.2023.112875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Mendonca-Lima L, et al. The allele encoding the mycobacterial Erp protein affects lung disease in mice. Cell Microbiol. 2003;5:65–73. doi: 10.1046/j.1462-5822.2003.00237.x. [DOI] [PubMed] [Google Scholar]
- 23.Healy C, Gouzy A, Ehrt S. Peptidoglycan hydrolases RipA and Ami1 are critical for replication and persistence of Mycobacterium tuberculosis in the host. mBio. 2020 doi: 10.1128/mBio.03315-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Both D, Schneider G, Schnell R. Peptidoglycan remodeling in Mycobacterium tuberculosis: Comparison of structures and catalytic activities of RipA and RipB. J. Mol. Biol. 2011;413:247–260. doi: 10.1016/j.jmb.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 25.Manca C, Lyashchenko K, Colangeli R, Gennaro ML. MTC28, a novel 28-kilodalton proline-rich secreted antigen specific for the Mycobacterium tuberculosis complex. Infect. Immun. 1997;65:4951–4957. doi: 10.1128/iai.65.12.4951-4957.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vergne I, et al. Mechanism of phagolysosome biogenesis block by viable Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2005;102:4033–4038. doi: 10.1073/pnas.0409716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plocinski P, et al. Mycobacterium tuberculosis CwsA interacts with CrgA and Wag31, and the CrgA-CwsA complex is involved in peptidoglycan synthesis and cell shape determination. J. Bacteriol. 2012;194:6398–6409. doi: 10.1128/JB.01005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishizaki Y, et al. Inhibition of the first step in synthesis of the mycobacterial cell wall core, catalyzed by the GlcNAc-1-phosphate transferase WecA, by the novel caprazamycin derivative CPZEN-45. J. Biol. Chem. 2013;288:30309–30319. doi: 10.1074/jbc.M113.492173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prigozhin DM, Mavrici D, Huizar JP, Vansell HJ, Alber T. Structural and biochemical analyses of Mycobacterium tuberculosis N-acetylmuramyl-l-alanine amidase Rv3717 point to a role in peptidoglycan fragment recycling. J. Biol. Chem. 2013;288:31549–31555. doi: 10.1074/jbc.M113.510792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alderwick LJ, Seidel M, Sahm H, Besra GS, Eggeling L. Identification of a novel arabinofuranosyltransferase (AftA) involved in cell wall arabinan biosynthesis in Mycobacterium tuberculosis. J. Biol. Chem. 2006;281:15653–15661. doi: 10.1074/jbc.M600045200. [DOI] [PubMed] [Google Scholar]
- 31.Rastogi S, et al. The diacylglycerol acyltransferase Rv3371 of Mycobacterium tuberculosis is required for growth arrest and involved in stress-induced cell wall alterations. Tuberculosis. 2017;104:8–19. doi: 10.1016/j.tube.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 32.Kaur D, et al. Lipoarabinomannan of Mycobacterium: Mannose capping by a multifunctional terminal mannosyltransferase. Proc. Natl. Acad. Sci. USA. 2008;105:17973–17977. doi: 10.1073/pnas.0807761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu CF, et al. Bacterial protein-O-mannosylating enzyme is crucial for virulence of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA. 2013;110:6560–6565. doi: 10.1073/pnas.1219704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haider S, Pal R. Integrated analysis of transcriptomic and proteomic data. Curr. Genomics. 2013;14:91–110. doi: 10.2174/1389202911314020003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malhotra V, Agrawal R, Duncan TR, Saini DK, Clark-Curtiss JE. Mycobacterium tuberculosis response regulators, DevR and NarL, interact in vivo and co-regulate gene expression during aerobic nitrate metabolism. J. Biol. Chem. 2015;290:8294–8309. doi: 10.1074/jbc.M114.591800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voskuil MI, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giacalone D, Yap RE, Ecker AMV, Tan S. PrrA modulates Mycobacterium tuberculosis response to multiple environmental cues and is critically regulated by serine/threonine protein kinases. PLoS Genet. 2022;18:e1010331. doi: 10.1371/journal.pgen.1010331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Hu L, Zhang H, He ZG. Cyclic di-GMP triggers the hypoxic adaptation of Mycobacterium bovis through a metabolic switching regulator ArgR. Environ. Microbiol. 2022;24:4382–4400. doi: 10.1111/1462-2920.15987. [DOI] [PubMed] [Google Scholar]
- 39.Hu Q, et al. Cyclic di-GMP co-activates the two-component transcriptional regulator DevR in Mycobacterium smegmatis in response to oxidative stress. J. Biol. Chem. 2019;294:12729–12742. doi: 10.1074/jbc.RA119.008252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malhotra V, et al. Mycobacterium tuberculosis PknK substrate profiling reveals essential transcription terminator protein Rho and two-component response regulators PrrA and MtrA as novel targets for phosphorylation. Microbiol. Spectr. 2022;10:e0135421. doi: 10.1128/spectrum.01354-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geisel RE, Sakamoto K, Russell DG, Rhoades ER. In vivo activity of released cell wall lipids of Mycobacterium bovis bacillus Calmette-Guerin is due principally to trehalose mycolates. J. Immunol. 2005;174:5007–5015. doi: 10.4049/jimmunol.174.8.5007. [DOI] [PubMed] [Google Scholar]
- 42.Vignali M, et al. NSR-seq transcriptional profiling enables identification of a gene signature of Plasmodium falciparum parasites infecting children. J. Clin. Investig. 2011;121:1119–1129. doi: 10.1172/JCI43457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 45.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robinson MD, Smyth GK. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics. 2008;9:321–332. doi: 10.1093/biostatistics/kxm030. [DOI] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen NH, Chen ML, Chak V, Balu-Iyer SV. Biophysical characterization of tolerogenic lipid-based nanoparticles containing phosphatidylcholine and lysophosphatidylserine. J. Pharm. Sci. 2022;111:2072–2082. doi: 10.1016/j.xphs.2022.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rahlwes KC, et al. The cell envelope-associated phospholipid-binding protein LmeA is required for mannan polymerization in mycobacteria. J. Biol. Chem. 2017;292:17407–17417. doi: 10.1074/jbc.M117.804377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.