Significance
The G protein-coupled receptors (GPCRs) form a prominent group of cell receptors, deeply involved in the regulation of key physiological mechanisms in all major organ systems. Among them, class C GPCRs are notable for their large extracellular domains and constitutive dimerization. Metabotropic glutamate receptor 2(mGlu2), a member of class C GPCRs, is of particular interest for its potential role in treating psychiatric conditions like schizophrenia and depression. Our comprehensive simulations using an electrostatic coarse-grained model have yielded the free-energy landscapes for several key events during the activation process of mGlu2, contributing insightful knowledge to the field of class C GPCR activation mechanisms.
Keywords: mGlu2, activation mechanism, agonist binding, free-energy landscapes, mutational effects
Abstract
Homomeric dimerization of metabotropic glutamate receptors (mGlus) is essential for the modulation of their functions and represents a promising avenue for the development of novel therapeutic approaches to address central nervous system diseases. Yet, the scarcity of detailed molecular and energetic data on mGlu2 impedes our in-depth comprehension of their activation process. Here, we employ computational simulation methods to elucidate the activation process and key events associated with the mGlu2, including a detailed analysis of its conformational transitions, the binding of agonists, Gi protein coupling, and the guanosine diphosphate (GDP) release. Our results demonstrate that the activation of mGlu2 is a stepwise process and several energy barriers need to be overcome. Moreover, we also identify the rate-determining step of the mGlu2’s transition from the agonist-bound state to its active state. From the perspective of free-energy analysis, we find that the conformational dynamics of mGlu2’s subunit follow coupled rather than discrete, independent actions. Asymmetric dimerization is critical for receptor activation. Our calculation results are consistent with the observation of cross-linking and fluorescent-labeled blot experiments, thus illustrating the reliability of our calculations. Besides, we also identify potential key residues in the Gi protein binding position on mGlu2, mGlu2 dimer’s TM6–TM6 interface, and Gi α5 helix by the change of energy barriers after mutation. The implications of our findings could lead to a more comprehensive grasp of class C G protein-coupled receptor activation.
G protein-coupled receptors (GPCRs), the largest family of cell surface receptors in eukaryotes, are indispensable in numerous physiological activities (1). Their pivotal role in a vast array of physiological processes makes them prime targets for pharmaceutical innovation (2, 3). According to their sequence homology, mammalian GPCRs are sorted into four classes: class A, B, C, and F. Among all GPCRs, class C GPCRs exhibit a unique structure, operate in dimeric forms, and consist of several structural domains. Compared with the other family members in GPCRs, class C GPCRs are distinguished by their relatively larger extracellular domains (ECD) which contain the binding sites for ligands. As representatives of the class C GPCRs, Metabotropic glutamate receptors (mGlus) are essential for the regulation of synaptic transmission and neuronal excitability, with dimerization being an obligatory process for their efficacy (4). Further evidence suggests the clinical potential of mGlus in treating central nervous system diseases, which has led to their recognition as significant pharmacological targets for a variety of neurological and psychiatric disorders, ranging from depression to schizophrenia and addiction (5–10). Therefore, it is necessary and crucial for us to understand their working mechanisms.
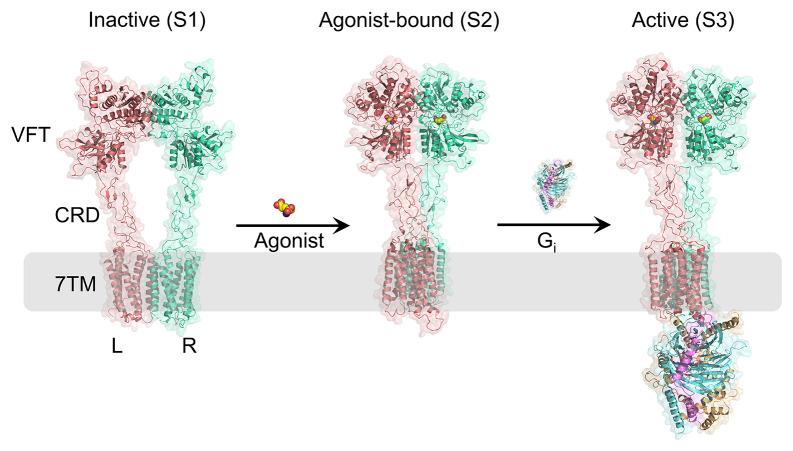
The mGlus are composed of two subunits, each of which encompasses a variety of multiple functional domains (Fig. 1). The large ECD has a Venus flytrap domain (VFT), which is responsible for the binding sites for intrinsic ligands, along with a cysteine-rich domain (CRD), that acts as a bridge between the VFT and the seven-helical transmembrane domain (7TM) (11). Previous crystallographic studies on VFT in isolation have shown that the binding of agonist induces significant conformational alterations within the dimeric structure (12, 13). In the absence of agonists, the mGlus remain in an inactive state (S1 in Fig. 1) that involves an open conformation for VFTs and two proximal 7TMs form interaction with each other. Once the agonist binds to the VFTs, it initiates a notable compaction of the receptor, prompting the VFTs and CRDs into proximity with their equivalent components in the other subunits. The closure of VFT is imperative for the receptor’s activation. Specifically, the closure of one VFT is adequate to initiate signaling, whereas the closure of both VFTs is required achieve full activation (14). These conformational changes result in the rearrangement of the 7TMs, resulting in the rotation of both 7TMs and the formation of a dimer interface along the TM6 (11, 12, 15). Furthermore, the coupling of Gi protein is facilitated by the 7TMs through an additional movement, resulting in the formation of an asymmetric interface between the dimer. The binding site for G-proteins is localized to only one of the 7TMs, suggesting an asymmetric transmission of signals in mGlus. This marks the completion of mGlu2;s transition from its inactive to its active state (S3 in Fig. 1). With the aid of cryogenic electron microscopy (cryo-EM) and X-ray diffraction (XRD) techniques, the structural profiles of the mGlus in different states have been obtained (11, 13, 16–18). Moreover, advancements in fluorescence resonance energy transfer (FRET) have provided a clearer picture of mGlus’ conformational dynamics, indicating that these receptors are highly dynamic proteins that sample multiple conformation states (19–22). However, molecular and energetic details of the mGlus activation are still unknown. These constraints impede a comprehensive comprehension of the signal transduction and regulatory processes of mGlus and also pose challenges to the advancement of future therapeutic agents specifically aimed at the homodimer mGlus.
Fig. 1.
Different conformational states of mGlu2. Models of the inactive state (S1, PDB ID:7EPA), agonist-bound state (S2, PDB ID:7EPB), and active state (S3, PDB ID:7E9G) are shown. The L and R subunits of mGlu2 are shown in pink and green cartoon representations, respectively. The agonist is shown in the space-filling model. The Gi protein is represented by three different colors corresponding to the three subunits of the Gi protein: light orange for the alpha subunit, blue for the beta subunit, and purple for the gamma subunit.
The rapid evolution of cryo-EM and XRD techniques has brought about the successful structural characterization of full-length mGlus (18, 23–27). This has made it possible to explore the molecular details of the working mechanism of mGlu2 by computational modeling. However, obtaining reliable free-energy landscapes from all-atom simulations still remains challenging due to the large scale of the membrane protein and the sophisticated nature of its conformational movements. Typically, the coarse-grained (CG) models are employed to describe these large biological systems. The CG model we utilized in our research was the outcome of the collaborative efforts of Warshel and his coworkers (28–32). It has proven to be a well-behaved and powerful tool for large biophysical systems. This CG model is designed to characterize protein stabilities and conformational energies, which also enhances the model’s electrostatic properties (31), and has been refined specially for the analysis of membrane protein (32). Furthermore, the CG model has been extensively employed to investigate the molecular mechanism of biological functions in a broad array of complex systems, like ATPase (33–35), GPCRs (36–38), Hv1 channel (39), and SARS-Cov2 spike protein (40–43). Its extensive application has affirmed it as a reliable tool to depict the energy landscapes during the conformational transitions of proteins. We will apply it to investigate the activation of mGlu2.
In this study, we used computational tools to simulate the activation process of the mGlu2. First, we built the models of the endpoint structures and simulated the transition process to obtain the intermediate structures using the targeted molecular dynamics (TMD) method. Then, we obtained a comprehensive view of the free-energy landscape for mGlu2’s activation, along with a clear depiction of the energy barriers. Our CG models of the mGlu2 effectively emulated the free-energy profiles for the binding of agonists to the mGlu2 and also predicted a possible minimum energy pathway for the binding of agonists to the VFTs in mGlu2. Furthermore, we investigated the mechanisms of Gi protein binding and the GDP release event. We obtained the free-energy landscapes, determined the minimum energy pathway and energy barrier for Gi protein binding to the receptor, and also identified a low-barrier pathway for GDP release. Ultimately, we deciphered the energetic basis for the experimental mutational effects and predicted new potential mutational sites that affect the activation process and the stability of the mGlu2–Gi complex. Overall, our work systematically describes the activation process of mGlu2, explains its activation mechanism from a free-energy perspective and provides additional dynamic and kinetic information. Furthermore, our results and conclusions may open a new avenue to inspire and aid the design and development of drugs targeting mGlu2.
Results
Mechanisms of the Conversion between the Inactive and the Agonist-Bound State.
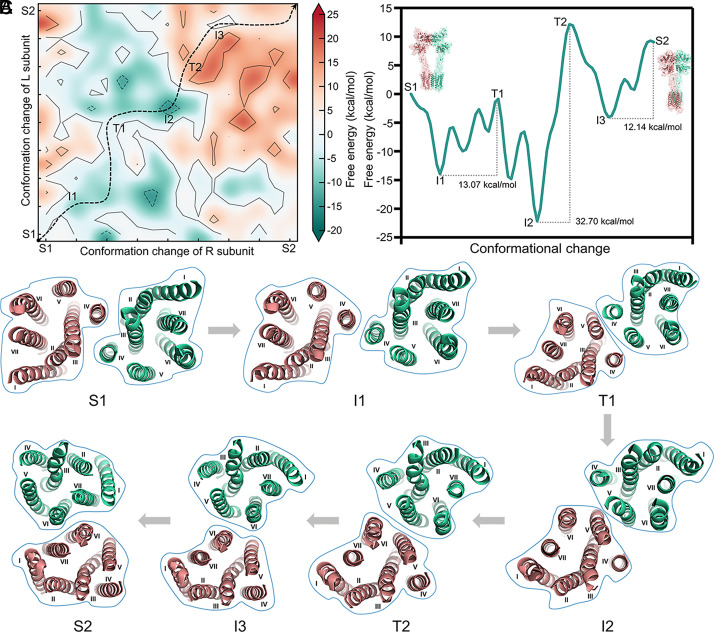
Previous studies (15) have suggested that mGlus undergo significant conformational changes when they transition from the inactive (S1) state to the agonist-bound (S2) state. However, the structural changes and energy basis of the conversion between these two states remain unknown. Here, we first constructed the conformational changes of mGlu2 from an inactive state (PDB ID: 7EPA) to an agonist-bound (PDB ID: 7EPB) state (23) using TMD method without incorporating any effects of ligands. Subsequently, we calculated the CG free energy for each conformation, which allowed us to trace the conformational change trajectory. We then picked 17 conformations at equal intervals for each subunit of mGlu2, which included 15 intermediates and 2 endpoints. By integrating the conformations of the two constituent subunits, we developed a two-dimensional conformational map of the conformational changes of the L and R subunit of mGlu2. Then, we calculated the conformational free energy at every point on this map using CG models (computational details are mentioned in SI Appendix, section S1). The CG free-energy landscape representing the transition from state S1 to S2 is depicted in Fig. 2A, where the black dashed line represents the possible minimal energy path.
Fig. 2.
The energetic/structural coupling of the conformational changes for the conversion between inactive (S1) and agonist-bound (S2) state. (A) The landscape of free energy associated with the conformational changes of each subunit in mGlu2. The black dashed line indicates the possible minimal energy path. The energy barrier for the transition from I1 to T1, I2 to T2, and I3 to T3 are 13.07 kcal/mol, 32.70 kcal/mol, and 12.14 kcal/mol, respectively. (B) Free-energy profiles along the minimal energy path in panel A. (C) Multiple dimerization structures of the mGlu2 transmembrane domain (the extracellular view) from the S1 to S2 state.
There are three major barriers in this path. By analyzing different terms of free energy, we found that hydrophobic energy contributes the most to these three major barriers. (SI Appendix, Table S1). In the S1 state, the VFT domains display an open conformation, distant CRDs, and an asymmetric 7TM dimer with a TM3-TM4 interface (S1 in Fig. 2C). Note that the I1 state is more stable compared to the experimental structure (S1 state) on the energy curve (Fig. 2B). This could be a result of the experimental structure that is stabilized by its selective negative allosteric modulator, a factor not included in our modeled structure. Researchers always use its selective negative allosteric modulator to stabilize the inactive state of mGlu2 (18, 23, 27). The energy barrier between I1 and T1 state is 13.07 kcal/mol, which may contribute to the spatial hindrance of hydrophobic interaction between the TM4–TM4 interface (I1 and T1 in Fig. 2C and SI Appendix, Fig. S2A). Our calculation data, along with findings from existing studies (23) on mGlu2 confirm that the dimerization mediated by TM4 contributes to the stabilization of the receptor’s inactive conformation. The highest barrier (32.70 kcal/mol) occurs between the I2 to T2. The dimer of 7TMs changes from an asymmetric interface involving TM4, TM5, and TM6 to a TM5–TM6 interface (I2 and T2 in Fig. 2C). These results suggest that the highest barrier is contributed to the lock of the dimer interface involving TM4, TM5, and TM6 interface (SI Appendix, Fig. S2B) which impairs receptor activation (44). Another barrier, 12.14 kcal/mol, occurs between the I3 to the S2 state. The 7TM dimer interface changes from TM5–TM6 to TM6–TM6 (I3 and S2 in Fig. 2C). Overall, our results demonstrate that the activation of mGlu2 proceeds in a stepwise manner, and several energetic barriers need to be overcome to achieve the conversion from the S1 to the S2 state.
Exploring the Binding Path of Agonists to the VFTs in mGlu2.
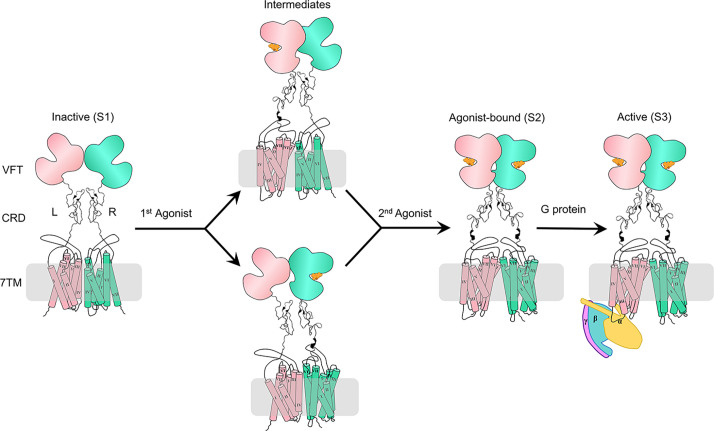
Once the agonist binds to the VFTs, the mGlu2 dimer undergoes significant conformational change. This interaction prompts the VFTs and CRDs to come into proximity with their counterparts in the other subunit, which in turn results in a rotation of both 7TMs and a transition of the dimer interface from the asymmetric TM3–TM4 to the TM6–TM6. Previous studies (45) have demonstrated that there is an intermediate state during mGlu2 activation, characterized by one VFT domain in a “closed” conformation while the other remains an “open” conformation. In this state, the interface of VFTs maintains a relaxed form and the receptor is in an inactive state (Fig. 3). Only when both VFTs are bound with the agonist, the receptor can maximally activate the mGlu2 dimer (14). However, how agonists bind to mGlu2 and the effects of agonists binding on energy barriers are still unclear.
Fig. 3.
Schematic representation of the mGlu2 activation process. Initially, the mGlu2 is inactive, characterized by open VFTs, separated CRDs, and an asymmetric 7TM dimers interface which is composed of TM3 and TM4. The binding of an agonist to one VFT results in an intermediate state where the 7TMs remain inactive. Full activation is achieved when agonist binds to both VFTs, leading to the compaction of the mGlu2 dimer and the VFTs, bringing the VFTs and CRDs proximity to their counterparts, and generating a dimer interface along TM6. Finally, the Gi protein couples to the receptor and the 7TMs undergo a further movement.
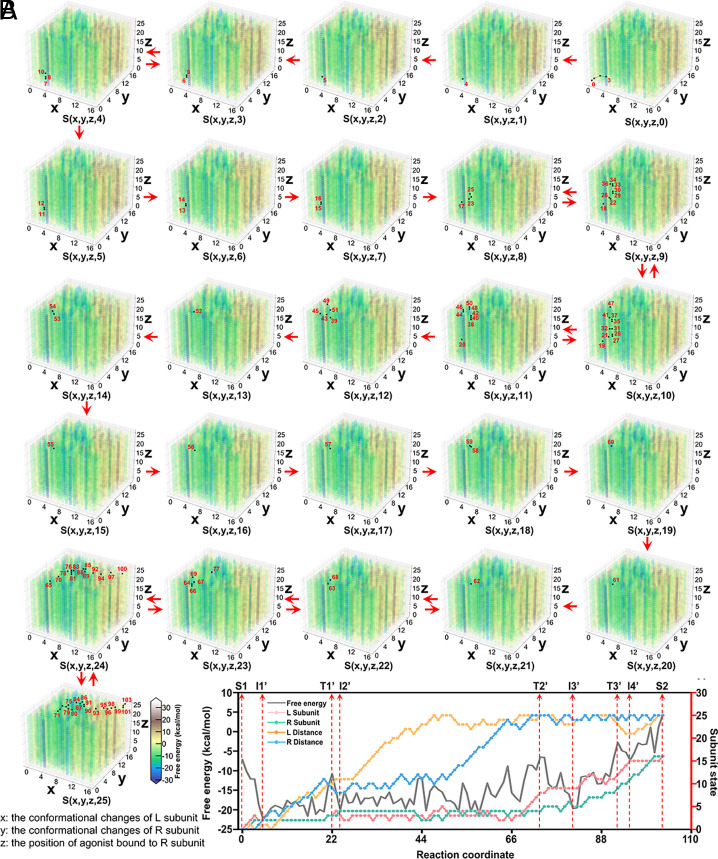
Based on the conformations in the 2D free-energy landscape (Fig. 2A), we docked two agonists to both subunits and constructed the possible conformation combinations of the agonist positions and the conformations of mGlu2 homodimer (details are mentioned in SI Appendix, section S1.1). A total of 195,364 conformations were constructed and each conformation is depicted as S(x, y, z, i) where x, y represent the conformational change of the L and R subunit of mGlu2 dimer, z, i indicate the agonist position for the R and L subunit. The complete free-energy landscape for mGlu2 conformations is shown in Fig. 4A, where the agonist’s binding free energy has been added to the folding energy for each conformation. Next, a Monte Carlo (MC) pathway searching procedure was performed to identify the minimum-energy pathway from S(0, 0, 0, 0) where the mGlu2 is in the S1 state to S(16, 16, 25, 25) where the receptor is in the S2 state. More details about the procedure are available in SI Appendix, section S1.4.
Fig. 4.
The complete free-energy landscape of mGlu2 during the process of agonist binding. (A) Coupled free-energy landscape of the distance between the pockets in VFT and the agonist and the conformational changes of each subunit of mGlu2 from state S1 to S2. Totally 195,364 (17*17*26*26) conformations of mGlu2 construct the complete free-energy landscape which is displayed by 26 cubic boxes. For each cubic box, the X and Y axes indicate the conformational changes of L and R subunits of the mGlu2 dimer which vary from 0 to 16, while the Z axis reflects the distance from the pocket in VFT of the R subunits to the agonist, ranging from 0 to 25. The distance between the agonist and the pocket in VFT of the L subunits is a fixed value, which only varies in different cubic boxes. The conformations of mGlu2 sampled along the path are marked with black dots and numbered sequentially with numbers in each cubic box. Movement between black dots in the adjacent boxes is shown as red arrows. (B) The energetic and conformational changes along the optimal conformational pathway of agonist binding to VFT in mGlu2 from S1 to S2 process (gray lines). L and R distances here indicate the distance between the ligand and the pocket in VFT of the L subunit and R subunit, respectively. The states of the L subunit, R subunit, L distance, and R distance of the minimum free-energy path are represented by pink, green lines, orange, and blue lines, respectively.
The conformations of mGlu2 sampled along the minimum-energy path are marked with black dots in Fig. 4A, which contains a 103-step flip between S(0, 0, 0, 0) and S(16, 16, 25, 25) (Movie S1). The minimum energy pathway and the state of each subunit were identified and shown in Fig. 4B. Initially, the agonists are far away from the receptor and do not induce significant conformational changes of mGlu2. The conformations of mGlu2 change slightly and then remain stable. Subsequently, as the agonist gradually approaches the binding pocket in the L subunit of the mGlu2 dimer, the conformations of the L subunit start to fluctuate. Once the agonist binds to the L subunit, the conformation of the L and R subunit both undergo small changes. This is consistent with that the activation of mGlu2 may involve a conformation where only one subunit’s VFT domain is in active state (45). Later, when the second agonist binds to the other subunit, the conformations of both subunits change significantly. These two subunits cooperate with each other under the influence of agonists. Finally, the receptor finishes the transition from the S1 to the S2 state. In the minimum energy pathway, the highest barrier occurs between the I3’ and T3’, 16.88 kcal/mol. Compared with the above situation without agonists (32.70 kcal/mol), the energy barrier is significantly reduced. These results emphasize the importance of dual agonist binding from an energetic point of view. The conformational alternations of these two subunits establish a cooperative relationship rather than occurring simultaneously. In addition, we also consider other two situations as scenario 1 and scenario 2 shown in SI Appendix, Fig. S3: only one agonist binds to the L and R subunit respectively. Similar results also appeared in these two situations: the emergence of agonists caused a large decrease in the energy barrier (SI Appendix, Figs. S4 and S5).
Mechanism of Gi Protein Coupling in mGlu2.
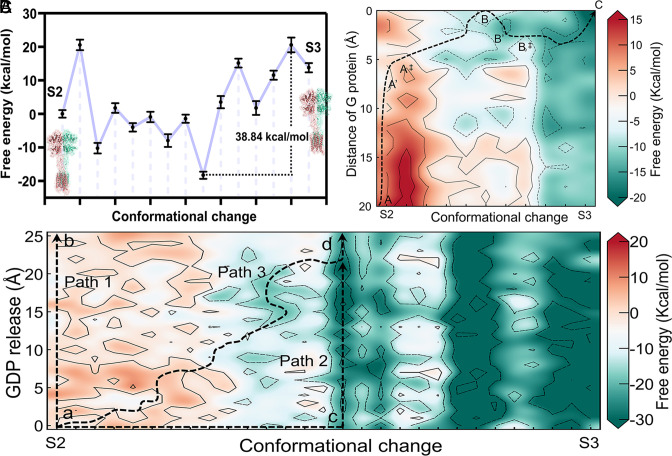
It has previously been proposed that during the activation of mGlu2, the coupling of G protein is facilitated by only one subunit of the dimer (46). However, the molecular basis for the asymmetric signaling of mGlu2 has not yet been elucidated. In this study, we first built CG models of mGlu2 to explore the conformational changes from the S2 to S3 state. Based on Cryo-EM structures (PDB ID: 7EPB, 7E9G), we built the structural models to represent these two distinct conformations of the mGlu2 homodimer. Using the TMD method, we then produced a sequence of intermediate structures that bridge the gap between these two states. Following this, we constructed the free-energy landscape that corresponds to the series of conformations. As shown in Fig. 5A, the transition of mGlu2 from the S2 to the S3 state is hindered by an energy barrier (38.84 kcal/mol) in the absence of Gi protein. This is also the highest barrier throughout the activation process from the S1 state to the S3 state. Although the receptor undergoes minor conformational changes between the S2 and S3 states, it is the rate-determining step during the entire activation process. This aligns well with the previous observation that Liauw et al. made using single-molecule FRET (45) who found the same rate-determining step.
Fig. 5.
The free-energy landscape of the coupling of the Gi protein and GDP release event. (A) The CG free-energy profiles for the transition from the agonist-bound (S2) to the active (S3) state. The data are expressed as means ± SD. (B) The free-energy landscape of the conformational changes between S2 and S3 with the distance to the G protein. The possible minimal energy path is indicated by the black dashed line. (C) The coupled free-energy landscape of the conformational changes between S2 and S3 and the GDP release. Three possible pathways are indicated by black dashed lines.
Furthermore, to understand the energetic mechanism of Gi coupling with the receptor, we constructed a set of structural models depicting the approach of the Gi protein and conformational variations in mGlu2. Details are mentioned in SI Appendix, section S1.1. As shown in Fig. 5B, the X-axis represents the conformational changes of mGlu2 while the Y-axis describes the distance between the centers of the pulled and initial Gi protein. The black dashed line depicts the minimum energy pathway that was identified. In the beginning, the conformation of mGlu2 does not change. As the Gi protein approaches, the conformation of mGlu2 starts to change. A barrier occurs when the conformation of mGlu2 begins to change (7.99 kcal/mol between states A’ and A‡ in the presence of Gi protein and agonists, SI Appendix, Fig. S6). Obviously, the presence of Gi protein promotes the activation of mGlu2, which is consistent with the observation of mGlu2 constitutive activity test experiments (18). Besides, we found that the Gi protein couples to the mGlu2 twice in the minimum energy pathway. When the Gi protein first binds to the receptor (point B in Fig. 5B), it then moves away from the receptor. This may be due to the spatial hindrance between the 7TM of the receptor and the Gi protein, which causes the Gi protein to move away temporarily. Subsequently, the conformations of the mGlu2 continue to change, such as the 7TM of the mGlu2 opens a “cleft” to facilitate the binding of the receptor (18). Finally, the receptor converts to the S3 state and couples to the Gi protein (point C in Fig. 5B).
Coupling between the Receptor Conformational Change and the GDP Release.
Through the facilitation of nucleotide release from the Gi protein heterotrimer, the mGlu2 receptor enables the internalization of extracellular signals within the cell, yet the precise workings of this event are not clearly defined. To explore where and when the nucleotide is released from the Gi protein, we constructed the free-energy profiles for GDP release by introducing a GDP molecule and a Mg2+ ion at various distances from the nucleotide-binding pocket to the bulk. In addition, we generated a specific intermediate structure for each combination of system conformation and the nucleotide to map the free-energy landscape. We initiated our analysis with extensive MD relaxation, followed by PDLD/S-LRA/β binding energy calculations to calculate the free energies. More details are shown in SI Appendix, sections S1.1 and S1.3. Fig. 5C illustrates the free-energy landscape that integrates the conformational changes and the dissociation of GDP. The X-axis represents the conformational change of the complex, and the Y-axis reflects the GDP/Mg2+ distance from the nucleotide-binding site.
As depicted in the free-energy landscape (Fig. 5C), three possible pathways are represented by black dashed lines. For path 1 (from point a to b), the release of GDP requires crossing a large energy barrier (22.17 kcal/mol) before any conformational change happens. This observation is consistent with the experimental finding that receptors first couple with G protein and then catalyze nucleotide release by favoring an internal structural rearrangement of the Ras domain (47). Once an active receptor binds to an inactive, GDP-bound G protein, it will dramatically accelerate GDP release (48, 49). Similar phenomena also appears in our calculations. As shown in path 2, an energy barrier (19.80 kcal/mol) occurs after the releasing pathway is opened (from point c to d). However, before that happens, the complex first needs to undergo the conformational changes (from point a to c, the highest energy is 17.88 kcal/mol). According to the energy landscape, path 3 (from point a to d) is the most likely pathway for GDP release, with a relatively small energy barrier of 16.88 kcal/mol. The GDP release occurs during the transition period from the S2 to S3 state. The coupling of the G-protein to mGlu2 induces the opening of G-protein to release GDP and initiate signaling.
Taken together, the above findings offer molecular insights into the whole activation mechanisms of mGlu2. First, agonist binding effectively brings the conformational changes of mGlu2. The conformations of both subunits change in a coupled manner rather than separately and independently of each other. Moreover, an intermediate state that only one VFT binds the agonist could exist during the activation process. The closure of the VFTs results in the close proximity of the CDRs and a change in the dimerization mode of the 7TMs. This arrangement enables their mutual interaction and promotes the formation of the asymmetric TM6-TM6 dimer interface. Finally, the G protein gradually approaches the receptor in a way that would enable its nucleotide exchange, thus transmitting intracellular signaling. The rate-determining step occurs between the agonist-bound state and the active state. Besides, the presence of agonist and G-protein induces the conformational changes of mGlu2 and promotes receptor activation.
Predicting Mutational Effects of Key Residues of mGlu2–Gi Complex.
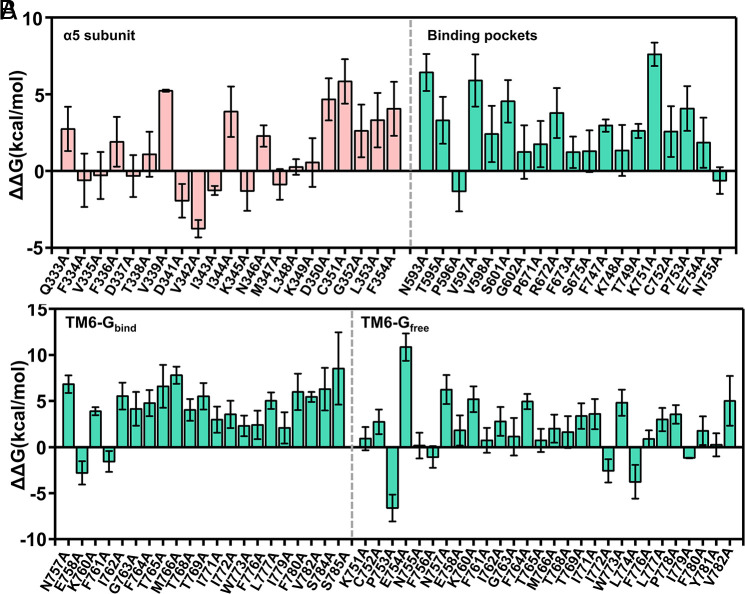
Unlike GPCRs belonging to families A and B, the α5 subunit in G protein is stabilized by a pocket formed between TM3, intracellular loops 2 and 3 (ICL2 and ICL3), and the C terminus of mGlu2 (binding pocket in SI Appendix, Fig. S8), rather than being inserted in a cavity at the 7TM bundle. The movement of one TM6 in the TM6–TM6 interface enables a conformational change in the intracellular region of the receptor to facilitate G protein recognition (25). In this work, we tried to investigate whether other residues in the Gi protein binding position of mGlu2, mGlu2 dimer’s TM6-TM6 interface, and Gi α5 helix (SI Appendix, Fig. S8) could affect the coupling of Gi protein and destabilization of the complex structure by mutating different residues to alanine. Fig. 6 depicts the relative difference in free energy of changing the residues at the positions of the α5 subunit, binding pocket, and the TM6–TM6 interface. These free energies are calculated using the PDLD/S-LRA/β method. More details are described in SI Appendix, section S1.3. Taking the coupling of Gi protein and the GDP release into account, we calculated the relative change of free-energy barrier (), in which the free-energy barrier () is defined in SI Appendix, Fig. S7. This is the difference in the free-energy barrier between the mutated mGlu2-Gi complex and the wild-type mGlu2-Gi complex. A positive difference in the free-energy barrier suggests that the residue (X) plays a role in stabilizing the formation of the complex, and the introduction of an alanine mutation at that position would be detrimental to the complex stability, leading to a higher energy barrier and inhibiting the activation of mGlu2 and vice versa.
Fig. 6.
Evaluation of mutational effects of the residues of (A) the α5 subunit, binding pockets, and (B) TM6-Gbind and TM6-Gfree region. The effects were calculated using . indicates the mutation impedes Gi protein activation, and indicates the mutation facilitates Gi protein activation. The data are expressed as means ± SEM. Residues on the Gi protein are shown in pink and residues on mGlu2 are shown in green. TM6-Gbind represents the TM6 in the subunit that binds to the Gi protein while TM6-Gfree represents the TM6 in the subunit that does not bind to the Gi protein.
Previous investigations (18, 25) of the α5 subunit in mediating the G protein activation have suggested that residues I344, L348, L353, and F354 form a hydrophobic interaction core. ICL3 of mGlu2 is interacting with this hydrophobic core, which plays a significant role in mediating mGlu2-Gi recognition. The same hydrophobic interaction consisting of residues I344, L348, L353, and F354 in the Gi protein was also observed in mGlu4–Gi complex (25). Besides, ICL2 establishes a polar interaction network with Gα subunit: R665 and R670 on receptor ICL 2 form salt bridges with Gα residues D193 and D350 (25). Disruption of these interactions destabilizes the mGlu2–Gi complex, and this is consistent with our calculations: the values of mutants I344A, D350A, C351A, L353A, and F354A are all larger than 3 kcal/mol, which indicates that these mutations increase the energy barrier. For most of the positions in the binding pockets, the stability of the complex decreases by alanine mutations (positive ). Although some residues display negative values of , the magnitude of the stabilizing effect at those locations is quite marginal. Moreover, the specific single-point mutations of hydrophobic residue I762, F764, M766, I771, W773, F776, L777, F780, and V782 on the interface of TM6-TM6 exhibit an unfavorable trend in the activation of Gi protein with a positive (Fig. 6B). Before mutations, those residues on the interface of TM6–TM6 make extensive hydrophobic contacts with each other, which maintain the receptor’s active conformation that is critical for the coupling of Gi protein. In other words, the single mutation disrupts the hydrophobic interaction between the TM6–TM6 interface which contributes to the increase of the energy barrier.
Many previous mutagenesis studies have identified some key residues for stabilizing the active conformation of mGlu2 and the Gi protein recognition. Seven et al. (18) mutated hydrophobic residues F756 and F661 in the binding pocket to alanine. Notably, mutation of these two residues significantly impaired G-protein activation. They also found that the triple-alanine mutation at positions 712 to 714 of mGlu2 ECL2 residues (E712A/R713A/R714A) decreases glutamate-stimulated G-protein activity which indicates that this loop is essential in the mediating the structural changes that occur between the VFTs and the 7TMs. In addition, the mutation C770A in both subunits of mGlu2 can increase G protein activation (45) since this mutation breaks the disulfide bond between the TM6–TM6 interface. We calculated the of these mutations which have been previously confirmed by experiments (18, 23, 25, 27, 45) as control (SI Appendix, Fig. S9). Encouragingly, our calculations are consistent with the experiments, which verifies the reliability of our method.
Discussion
Herein, we have explored the activation of mGlu2 from several aspects. First, we constructed the free-energy profile to illustrate the conformational changes and determined the rate-determining step of the entire activation process. Second, in order to investigate how the agonist binds to the mGlu2, a free-energy landscape including 195,364 models of dimer conformations was constructed. The CG strategy was applied for the conformational free energy while the binding free energy of agonists was calculated by the PDLD/s-LRA/β method. Moreover, we could identify the most likely minimum-energy pathway of agonist binding by using the accelerated MC algorithm. From the minimum-energy pathway, we found that the conformations of both subunits change in a cooperative manner and several dimerization modes of mGlu2 exist during the activation process. These findings suggest asymmetric dimerization plays a vital role in controlling receptor activation.
Third, we suggested the molecular basis for the specific coupling of the mGlu2 and the Gi protein and the release of GDP. The coupling of the Gi protein promotes the activation of mGlu2, a major drop in the energy barrier (from 38.84 kcal/mol in the absence of Gi protein to 7.99 kcal/mol in the presence of Gi protein). This result is consistent with experimental observation of mGlu2 constitutive activity. Our findings provide molecular basis and insights into the mechanism of the asymmetric signal transduction of mGlus, the binding of Gi protein, as well as the activation process of class C GPCRs. Furthermore, we also identified a specific pathway for the GDP release, which encounters a modest energy barrier of 16.88 kcal/mol. This pathway involves a simultaneous conformational transition between S2 and S3 states of mGlu2. Finally, mutational effects were assessed through the shifts in the energy barriers in this specific pathway. Some other key residues were identified by the alanine mutations at the Gi protein binding position in mGlu2, mGlu2 dimer’s TM6-TM6 interface, and Gi α5 helix.
Our results could provide information and guide the direction for future mutagenesis studies (such as double and triple mutations) of mGlus and possibly other class C GPCRs. Overall, our work fully explains the conformational changes in mGlu2 as it evolves from an inactive state to an active state and has brought to light the sophisticated patterns of signal transduction within the homodimers of mGlu2. The mGlu2 plays a crucial role in numerous neurodevelopment and psychiatric disorders. However, no drugs targeting this receptor family have been approved for market due to the limited studies of its activation mechanism. The current work enhances the understanding of class C GPCRs and paves the way for further in-depth investigation into the energetics of mGlu2’s signal transduction process. In addition, the identification of several unique dimerization interfaces opens up the potential for targeting new allosteric binding pockets that favor different states. These insights may serve to hasten the process of drug development targeting the mGlu2 receptor.
Materials and Methods
Model Preparation.
In this study, three major states were generated through homology modeling with the aid of Modeller (50, 51). The relevant PDB structures are 7EPA (inactive state), 7EPB (agonist-bound state), and 7E9G (active state) (23, 25). Then, TMD was employed to construct a range of intermediate structures that connect these end states (52). The detailed modeling protocol is described in SI Appendix, sections S1.1 and S1.2. For these structures, we incorporated membrane particles and utilized the implicit solvent models. Moreover, in order to eliminate steric clashes, we conducted an additional relaxation simulation for each newly generated structure before calculating the CG energies. The PDLD/s-LRA/β method (53–56) was employed to evaluate the binding and solvation energy. Details are mentioned in SI Appendix, section S1.3.
Calculation of the CG Free Energy.
Our CG model and force field have been continuously developed, with a focus on the solvation model of ionizable residues, emphasizing the pivotal role of protein electrostatic effects. The total energy construct of our CG model is encapsulated by the following equation:
| [1] |
The terms in Eq. 1 represent the side chain van der Waals energy, main chain solvation energy, main chain hydrogen bond energy, side chain electrostatic energy, side chain polar energy, side chain hydrophobic energy, main chain/side chain electrostatic energy, and main chain/side chain van der Waals energy, respectively. The constants , , and are scaling coefficients and they have values of 0.10, 0.25, and 0.15, respectively, in this work (31, 38).
For the purpose of evaluating the CG energy, we initially converted the all-atom structures into CG models, simplifying the side chains of the residues into a single united atom. Subsequently, we carried out an additional relaxation simulation and employed MC Proton Transfer (MCPT) algorithm to determine the charges for the ionizable groups within the protein (32). MCPT was applied to model the proton transfers between the ionizable residues, continuing the simulation until the electrostatic free energy of the protein in its folded state achieved converged. Moreover, the proton acceptance probability was evaluated by the standard Metropolis criterion. Before the determination of the CG energies, extensive molecular dynamics simulations were conducted to relax each structure. The folding free energies were then calculated using the last fifteen conformations of the relaxation trajectory. The mean values of the last fifteen conformational energies were taken for further analysis. Additionally, the SDs of these fifteen free energies were used to represent the error bars. The free energy of each model was calculated based on the obtained charge distribution. The Molaris-XG package (28, 53) was employed to carry out all calculations.
Pathway Search of Agonist Binding to mGlu2 Based on the Free-Energy Landscape.
To study how the agonist binds to the mGlu2, a total of 195,364 mGlu2 conformations were constructed and the free-energy landscapes are shown in Fig. 4. Here, we executed MC sampling (57) on the free-energy landscape according to the Metropolis criteria. The acceptance condition of a possible MC move is
| [2] |
However, some conformational barriers are too high to obtain convergent sampling in a reasonable simulation time. Thus, we accelerated the sampling by using accelerated molecular dynamics simulation method (58–60). See SI Appendix, section S1.4 for details.
Supplementary Material
Appendix 01 (PDF)
Movement of the minimum-energy pathway in the complete free energy landscape of mGlu2 in the agonist-binding process.
Acknowledgments
This research was supported by the National Natural Science Foundation of Youth Fund Project (grant number: 22103066); the National Natural Science Foundation of China (grant number: 61931024); and the 2021 Basic Research General Project of Shenzhen, China (grant number: 20210316202830001). A.W. is supported by the NIH R35 GM122472 and the NSF Grant MCB 1707167.
Author contributions
X.Z., K.A., T.H., A.W., and C.B. designed research; X.Z. performed research; X.Z. and D.S. contributed new reagents/analytic tools; X.Z. analyzed data; and X.Z., M.L., K.A., T.H., A.W., and C.B. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: D.T., The University of Texas at Austin; and I.V., University of California Davis.
Contributor Information
Tingjun Hou, Email: tingjunhou@zju.edu.cn.
Arieh Warshel, Email: warshel@usc.edu.
Chen Bai, Email: baichen@cuhk.edu.cn.
Data, Materials, and Software Availability
The input files of simulations are publicly available at https://github.com/Xiaohong2991/mGlu2-iunput-files/tree/main (61). All other data are included in the manuscript and/or SI Appendix.
Supporting Information
References
- 1.Thal D. M., Glukhova A., Sexton P. M., Christopoulos A., Structural insights into G-protein-coupled receptor allostery. Nature 559, 45–53 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Dorsam R. T., Gutkind J. S., G-protein-coupled receptors and cancer. Nat. Rev. Cancer 7, 79–94 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Lagerström M. C., Schiöth H. B., Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat. Rev. Drug Discovery 7, 339–357 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Niswender C. M., Conn P. J., Metabotropic glutamate receptors: Physiology, pharmacology, and disease. Annu. Rev. Pharmacol. Toxicol. 50, 295–322 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaki S., mGlu2/3 receptor antagonists as novel antidepressants. Trends Pharmacol. Sci. 38, 569–580 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Dogra S., Conn P. J., Metabotropic glutamate receptors as emerging targets for the treatment of schizophrenia. Mol. Pharmacol. 101, 275–285 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwan C., et al. , Combined 5-HT2A and mGlu2 modulation for the treatment of dyskinesia and psychosis in Parkinson’s disease. Neuropharmacology 186, 108465 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Litim N., Morissette M., Di Paolo T., Metabotropic glutamate receptors as therapeutic targets in Parkinson’s disease: An update from the last 5 years of research. Neuropharmacology 115, 166–179 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Moussawi K., Kalivas P. W., Group II metabotropic glutamate receptors (mGlu2/3) in drug addiction. Eur. J. Pharmacol. 639, 115–122 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiang Z., et al. , Input-specific regulation of glutamatergic synaptic transmission in the medial prefrontal cortex by mGlu2/mGlu4 receptor heterodimers. Sci. Signaling 14, eabd2319 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koehl A., et al. , Structural insights into the activation of metabotropic glutamate receptors. Nature 566, 79–84 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunishima N., et al. , Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 407, 971–977 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Muto T., Tsuchiya D., Morikawa K., Jingami H., Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc. Natl. Acad. Sci. U.S.A. 104, 3759–3764 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kniazeff J., et al. , Closed state of both binding domains of homodimeric mGlu receptors is required for full activity. Nat. Struct. Mol. Biol. 11, 706–713 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Hlavackova V., et al. , Sequential inter-and intrasubunit rearrangements during activation of dimeric metabotropic glutamate receptor 1. Sci. Signaling 5, ra59 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Doré A. S., et al. , Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature 511, 557–562 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Wu H., et al. , Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344, 58–64 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seven A. B., et al. , G-protein activation by a metabotropic glutamate receptor. Nature 595, 450–454 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doumazane E., et al. , Illuminating the activation mechanisms and allosteric properties of metabotropic glutamate receptors. Proc. Natl. Acad. Sci. U.S.A. 110, E1416–E1425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vafabakhsh R., Levitz J., Isacoff E. Y., Conformational dynamics of a class C G-protein-coupled receptor. Nature 524, 497–501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Habrian C. H., et al. , Conformational pathway provides unique sensitivity to a synaptic mGluR. Nat. Commun. 10, 5572 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Habrian C., Latorraca N., Fu Z., Isacoff E. Y., Homo-and hetero-dimeric subunit interactions set affinity and efficacy in metabotropic glutamate receptors. Nat. Commun. 14, 8288 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du J., et al. , Structures of human mGlu2 and mGlu7 homo- and heterodimers. Nature 594, 589–593 (2021). [DOI] [PubMed] [Google Scholar]
- 24.Hoogenboom B. W., Asymmetry is central to excitatory glutamate receptor activation. Nat. Struct. Mol. Biol. 28, 629–630 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Lin S., et al. , Structures of Gi-bound metabotropic glutamate receptors mGlu2 and mGlu4. Nature 594, 583–588 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Zhang J., et al. , Structural insights into the activation initiation of full-length mGlu1. Protein Cell 12, 662–667 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X., et al. , Structural insights into dimerization and activation of the mGlu2-mGlu3 and mGlu2-mGlu4 heterodimers. Cell Res. 33, 762–774 (2023), 10.1038/s41422-023-00830-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamerlin S. C., Vicatos S., Dryga A., Warshel A., Coarse-grained (multiscale) simulations in studies of biophysical and chemical systems. Annu. Rev. Phys. Chem. 62, 41–64 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Lee M., Kolev V., Warshel A., Validating a coarse-grained voltage activation model by comparing its performance to the results of Monte Carlo simulations. J. Phys. Chem. B 121, 11284–11291 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messer B. M., et al. , Multiscale simulations of protein landscapes: Using coarse-grained models as reference potentials to full explicit models. Proteins 78, 1212–1227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vicatos S., Rychkova A., Mukherjee S., Warshel A., An effective Coarse-grained model for biological simulations: Recent refinements and validations. Proteins 82, 1168–1185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vorobyov I., Kim I., Chu Z. T., Warshel A., Refining the treatment of membrane proteins by coarse-grained models. Proteins 84, 92–117 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai C., Asadi M., Warshel A., The catalytic dwell in ATPases is not crucial for movement against applied torque. Nat. Chem. 12, 1187–1192 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Bai C., Warshel A., Revisiting the protomotive vectorial motion of F0-ATPase. Proc. Natl. Acad. Sci. U.S.A. 116, 19484–19489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukherjee S., Warshel A., Dissecting the role of the γ-subunit in the rotary–chemical coupling and torque generation of F1-ATPase. Proc. Natl. Acad. Sci. U.S.A. 112, 2746–2751 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alhadeff R., Warshel A., A free-energy landscape for the glucagon-like peptide 1 receptor GLP1R. Proteins 88, 127–134 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An K., Zhu X., Bai C., The nature of functional features of different classes of G-protein-coupled receptors. Biology 11, 1839 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai C., et al. , Exploring the activation process of the β2AR-Gs complex. J. Am. Chem. Soc. 143, 11044–11051 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee M., Bai C., Feliks M., Alhadeff R., Warshel A., On the control of the proton current in the voltage-gated proton channel Hv1. Proc. Natl. Acad. Sci. U.S.A. 115, 10321–10326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.An K., et al. , A systematic study on the binding affinity of SARS-CoV-2 spike protein to antibodies. AIMS Microbiol. 8, 595 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai C., et al. , Predicting mutational effects on receptor binding of the spike protein of SARS-CoV-2 variants. J. Am. Chem. Soc. 143, 17646–17654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bai C., Warshel A., Critical differences between the binding features of the spike proteins of SARS-CoV-2 and SARS-CoV. J. Phys. Chem. B 124, 5907–5912 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu X., An K., Yan J., Xu P., Bai C., In silico optimization of SARS-CoV-2 spike specific nanobodies. Front. Biosci. (Landmark Ed) 28, 67 (2023). [DOI] [PubMed] [Google Scholar]
- 44.Xue L., et al. , Major ligand-induced rearrangement of the heptahelical domain interface in a GPCR dimer. Nat. Chem. Biol. 11, 134–140 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Liauw B. W., Afsari H. S., Vafabakhsh R., Conformational rearrangement during activation of a metabotropic glutamate receptor. Nat. Chem. Biol. 17, 291–297 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hlavackova V., et al. , Evidence for a single heptahelical domain being turned on upon activation of a dimeric GPCR. EMBO J. 24, 499–509 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dror R. O., et al. , Structural basis for nucleotide exchange in heterotrimeric G proteins. Science 348, 1361–1365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oldham W. M., Hamm H. E., Heterotrimeric G protein activation by G-protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 9, 60–71 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Sprang S. R., G protein mechanisms: Insights from structural analysis. Annu. Rev. Biochem. 66, 639–678 (1997). [DOI] [PubMed] [Google Scholar]
- 50.Martí-Renom M. A., et al. , Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29, 291–325 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Webb B., Sali A., Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinformatics 54, 5.6.1–5.6.37 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlitter J., Engels M., Krüger P., Targeted molecular dynamics: A new approach for searching pathways of conformational transitions. J. Mol. Graphics 12, 84–89 (1994). [DOI] [PubMed] [Google Scholar]
- 53.Lee F. S., Chu Z. T., Warshel A., Microscopic and semimicroscopic calculations of electrostatic energies in proteins by the POLARIS and ENZYMIX programs. J. Comput. Chem. 14, 161–185 (1993). [Google Scholar]
- 54.Muegge I., Tao H., Warshel A., A fast estimate of electrostatic group contributions to the free energy of protein-inhibitor binding. Protein Eng. 10, 1363–1372 (1997). [DOI] [PubMed] [Google Scholar]
- 55.Schutz C. N., Warshel A., What are the dielectric “constants” of proteins and how to validate electrostatic models? Proteins 44, 400–417 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Singh N., Warshel A., Absolute binding free energy calculations: On the accuracy of computational scoring of protein–ligand interactions. Proteins 78, 1705–1723 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuczera G., Parent E., Monte Carlo assessment of parameter uncertainty in conceptual catchment models: The Metropolis algorithm. J. Hydrol. 211, 69–85 (1998). [Google Scholar]
- 58.de Oliveira C. A. F., Hamelberg D., McCammon J. A., Coupling accelerated molecular dynamics methods with thermodynamic integration simulations. J. Chem. Theory Comput. 4, 1516–1525 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gedeon P. C., Thomas J. R., Madura J. D., Accelerated molecular dynamics and protein conformational change: A theoretical and practical guide using a membrane embedded model neurotransmitter transporter. Methods Mol. Biol. 1215, 253–287 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamelberg D., Mongan J., McCammon J. A., Accelerated molecular dynamics: A promising and efficient simulation method for biomolecules. J. Chem. Phys. 120, 11919–11929 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Zhu X., Bai C., mGlu2-iunput-files. GitHub. https://github.com/Xiaohong2991/mGlu2-iunput-files/tree/main. Accessed 29 April 2024.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Movement of the minimum-energy pathway in the complete free energy landscape of mGlu2 in the agonist-binding process.
Data Availability Statement
The input files of simulations are publicly available at https://github.com/Xiaohong2991/mGlu2-iunput-files/tree/main (61). All other data are included in the manuscript and/or SI Appendix.