Abstract
Salmonella enterica encodes a type III secretion system within a pathogenicity island located at centisome 63 that is essential for virulence. All type III secretion systems require the function of a family of low-molecular-weight proteins that aid the secretion process by acting as partitioning factors and/or secretion pilots. One such protein is SicA, which is encoded immediately upstream of the type III secreted proteins SipB and SipC. We found that the absence of SicA results in the degradation of both SipB and SipC. Interestingly, in the absence of SipC, SipB was not only stable but also secreted at wild-type levels in a sicA mutant background, indicating that SicA is not required for SipB secretion. We also found that SicA is capable of binding both SipB and SipC. These results are consistent with a SicA role as a partitioning factor for SipB and SipC, thereby preventing their premature association and degradation. We also found that introduction of a sicA null mutation results in the lack of expression of SopE, another type III-secreted protein. Such an effect was shown to be transcriptional. Introduction of a loss-of-function sipC mutation into the sicA mutant background rescued sopE expression. These results indicate that the effect of sicA on sopE expression is indirect and most likely exerted through a regulatory factor(s) partitioned by SicA from SipC. These studies therefore describe a surprisingly complex function for the Salmonella enterica type III secretion-associated chaperone SicA.
Salmonella spp., as well as other pathogenic gram-negative bacteria, have evolved a specialized protein secretion system, termed type III, that mediates the delivery of bacterial proteins into the host cell (11, 17). These bacterial proteins either antagonize or stimulate host-cell responses for the pathogen's benefit. Salmonella enterica encodes at least two of these systems, one at centisome 63 and the other at centisome 31 (10). The centisome 63 type III secretion system is required for the interaction of Salmonella with the intestinal epithelium, while that at centisome 31 appears to be essential for the establishment of systemic infection.
Type III secretion systems are very complex and require the function of more than 20 proteins (11, 17). A subset of these proteins form a supramolecular structure resembling a needle (needle complex) that spans the bacterial envelope (22). One feature of type III secretion systems is the requirement of a unique family of cytoplasmic proteins that share a number of structural features: (i) low molecular weight, (ii) low isoelectric point, and (iii) predominantly α-helical secondary structure (33). Although the actual function of this protein family is poorly understood and the subject of some controversy, it is clear that they act as chaperone-like molecules and are required for the stability and/or the secretion of their cognate proteins. Unlike conventional chaperone molecules, however, the type III secretion-associated chaperones do not have the capacity to hydrolyze ATP and exert their activity over a discrete number (most often one) of cognate proteins. Thus, in these systems, the absence of a given chaperone results in either the premature degradation of the cognate protein(s) and/or the abolition of its secretion (3, 6, 8, 25, 26, 31, 32, 34). For many members of this protein family, the physical interaction with their cognate proteins as well as their binding sites have been established (8, 26, 31, 32, 34).
One member of this type III secretion-associated chaperone family is the S. enterica serovar Typhimurium SicA protein (21). SicA has primary amino acid sequence similarity with IpgC from Shigella spp. (1) and SycD (LcrH) from Yersinia spp. (2), which are known to exert chaperone-like functions in related type III secretion systems. Although SicA has been shown to be required for Salmonella entry into host cells (21), nothing is known about its function. Its sequence similarity with type III secretion-associated chaperones coupled to the fact that it is encoded immediately adjacent to the type III secreted proteins SipA, SipB, SipC, and SipD suggest that SicA may exert its function by serving as a chaperone for any or all of these type III secreted proteins.
In this paper, we describe a complex function for SicA. We show that SicA functions to partition and stabilize the SipB and SipC type III secreted proteins. In addition, we show that SicA plays an indirect role in the expression of SopE, which is also a target of the type III secretion system. We postulate that SicA exerts this latter function by partitioning a factor(s) required for sopE expression.
MATERIALS AND METHODS
Basic media and growth conditions for bacterial strains.
Bacterial strains were grown in L-broth or on L-agar plates (23). Inducing medium refers to L-broth with the final NaCl concentration increased to 0.3 M from 0.09 M. Incubation was at 37°C, with rotation at 30 rpm. Antibiotics, when appropriate for selection, were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; kanamycin, 50 μg/ml; streptomycin, 100 μg/ml; and tetracycline, 10 μg/ml.
Preparation of culture supernatant proteins and whole-cell extracts from S. enterica serovar Typhimurium by TCA precipitation.
Bacterial overnight cultures were grown in L-broth with appropriate antibiotics and subcultured at a dilution of 1:17 in 10 ml of inducing medium. At an optical density of 0.8 at 600 nm, bacteria were harvested in an SS34 rotor in a Sorval superspeed centrifuge at 9,200 rpm for 30 min at 4°C. Supernatants were subjected to an additional spin in a fresh tube, and 8.5 ml was subsequently filtered through a 0.45-μm-pore-size low protein binding syringe filter (Gelman Sciences, Ann Arbor, Mich.). An aliquot of filtrate was plated on L-agar to verify the absence of bacterial cells from the preparation, and then 100% trichloroacetic acid (TCA) was added to the filtrate to a final concentration of 12% and incubated on ice for either 2 h or overnight. Supernatant proteins were precipitated at 11,500 rpm at 4°C for 30 min. The resultant protein film was washed and resuspended in 26 μl of phosphate-buffered saline (PBS) (pH 7.0) containing 77 mM Tris-HCl, pH 8.0.
Western immunoblot analysis.
Samples were separated by discontinuous sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Schleicher and Schuell, Keene, N.H.) per standard methods. Immobilized proteins were probed with specific monoclonal antibodies and visualized by enhanced chemiluminescence (Pierce Chemical Co., Rockford, Ill.).
Coimmunoprecipitation.
Bacterial overnight cultures were subcultured 1:20 in 20 ml of inducing medium with antibiotics and grown to an optical density of 0.8 at 600 nm. Bacterial cells were pelleted and washed with an equal volume of Hanks buffered saline solution (HBSS; Gibco-BRL, Gaithersburg, Md.). The final pellet was resuspended in 1.5 ml of HBSS and maintained on ice during sonication for the minimum amount of time required to observe cleared lysate (40- to 50-s pulse) with an HSU200LF probe in a W380 Ultrasonic Processor (Heat Systems Ultrasonics, Inc., Farmingdale, N.Y.). The cycle time was continuous with a 50% duty cycle. Unbroken cells were removed by two 10-min centrifugations at maximum speed in a microcentrifuge. A 0.5-ml volume of cleared lysate was transferred to a fresh Eppendorf tube and brought to a final volume of 0.9 ml with HBSS. To this was added 70 μl of a 50% slurry of Protein A Sepharose (CL-4B; Pharmacia Biotech AB, Uppsala, Sweden) that had been washed four times with a 10× bed volume of HBSS. Polyclonal antibody directed against SipB and/or SipC was added with the slurry simultaneously at a dilution of 1:100. Immunoprecipitation was carried out at 4°C with end-over-end rotation for 1 h. The Protein A Sepharose was gently pelleted for 30 s at 5,000 rpm in a microcentrifuge, and 0.1 ml of supernatant was reserved in a final concentration of 1× SDS-PAGE protein loading buffer on ice for later analysis. Sepharose beads were then washed four consecutive times with 1 ml of HBSS by inverting three times quickly by hand and gently pelleting as above. Washed beads were resuspended in 65 μl of 50 mM Tris (pH 8)–0.4% SDS loading buffer and boiled. Samples were resolved in SDS–10 or 15% polyacrylamide gels at a 30% acrylamide-to-0.8% bisacrylamide ratio.
Metabolic labeling and immunoprecipitation.
Metabolic labeling and pulse-chase experiments were carried out as described before (8). Immunoprecipitations were carried out as described elsewhere (8) by using polyclonal antibodies directed against SipB and SipC.
C2,3O and β-galactosidase assays.
Bacterial overnight cultures were subcultured at a 1:20 dilution in 20 ml of inducing medium and grown to an optical density of 0.8 at 600 nm. Bacterial cells were pelleted, washed with 5 ml of cold 20 mM potassium phosphate buffer (pH 7.2), and repelleted. Pellets were resuspended in 1.5 ml of cold APB (10% acetone, 100 mM potassium phosphate buffer [pH 7.5]) and sonicated on ice for 1 min to disrupt cells. Extracts were centrifuged at maximum speed in a microcentrifuge for 10 min at 4°C to remove cellular debris. Total protein concentration was determined with the Pierce BCA Protein Assay Reagent, and known concentrations of bovine serum albumin were used as standards as dictated by the manufacturer (Pierce Chemical Co.). C2,3O activity was determined by following the increase in absorbance at 375 nm at room temperature due to accumulation of 2-hydroxymuconic semi-aldehyde in 3-ml polypropylene reaction cuvettes. Briefly, 2.5 ml of 100 μM potassium phosphate buffer (pH 8.0), 0.45 ml of APB, 50 μl of extract, and 10 μl of 100 mM catechol were mixed, normalized against a blank containing all of the above ingredients excluding extract, and immediately read at an optical density of 375 nm. Extract concentration was adjusted to obtain a reaction rate at which product formation increased the optical density by no more than 0.005 per s. One mUnit corresponds to the formation at room temperature of 1 nmol of 2-hydroxymuconic semialdehyde per min per mg of protein. The molar absorption coefficient was 42,000. Calculations were performed with the following formula: mUnit = 7.1 × 104 × (VBCA/VC2,3O) × (A375/T) × (DC2,3O/Y) × (1/DBCA), where VBCA is the volume of extract used to determine total protein concentration, VC2,3O is the volume of extract used in the C2,3O assay, A375 is the absorbance at 375 nm at end of time T, T is the time required to reach A375, Y is the amount (in micrograms) of protein in VBCA, as calculated from the linear quadratic equation of the protein standard curve, and D is the dilution factor (i.e., Vfinal/Vsample). The levels of β-galactosidase were measured as described previously (35).
Recombinant DNA techniques and plasmid and strain constructions.
Recombinant DNA techniques were performed according to standard procedures (24). Bacterial strains are listed in Table 1. All strains were made electrocompetent as described elsewhere (28) and transformed with DNA via Gene Pulser (Bio-Rad Laboratories, Richmond, Calif.). S. enterica serovar Typhimurium sipD mutants were constructed as follows. A Pseudomonas putida Tol plasmid gene, xylE, that encodes catechol-2,3-dioxygenase (C2,3O) (18) and which lacks a promoter and transcriptional terminator was excised with BglII from pSB383 (19) and inserted into the compatible BamHI site of sipD in pSB412 (20), yielding plasmid pSB1484. A 4.1-kb HindIII fragment was moved from pSB1484 into the same sites of pBSL86 to yield pSB1487. This was digested with SmaI, and a 4.1-kb fragment was moved into the same site of pSB890, a suicide vector derived from pSB377 that can be counterselected for with sucrose (19), to yield pSB1488. This plasmid was maintained in, or conjugated into, SB300 (wild type) or SB221 (sicA) with the E. coli strain SM10 λpir. Transconjugants that were streptomycin resistant, sucrose resistant, tetracycline sensitive, and positive when tested for C2,3O activity were confirmed for allelic exchange by Southern blot analysis (data not shown) and designated SB320 and SB267, respectively.
TABLE 1.
S. enterica serovar Typhimurium strains used in this study
| Strain | Relevant characteristic | Reference or source |
|---|---|---|
| SB136 | invA::aphT | 12 |
| SB169 | sipB::aphT | 21 |
| SB220 | sipC::aphT | 21 |
| SB221 | sicA::aphT | 21 |
| SB227 | sipC::xylE | 21 |
| SB265 | Δ(sicAsipB)::aphT | This study |
| SB266 | sicA::aphT, sipC::xylE | This study |
| SB267 | sicA::aphT, sipD::xylE | This study |
| SB268 | sicA::aphT, ΔsipA | This study |
| SB269 | sicA::aphT, sptP::xylE | This study |
| SB300 | Mouse-passaged SL1344 | 15 |
| SB848 | ΔsipA | 36 |
| SB550 | sptP::xylE | 8 |
| SB319 | sicA::M45 | This study |
| SB320 | sipD::xylE | This study |
| SB876 | sopE::lacZ | Hardt and Galán, (unpublished data) |
| SB879 | sopE::lacZ sicA::aphT | This study |
| SB1235 | sopE::lacZ sicA::aphT sipC::xylE | This study |
| SB804 | sitB::lacZ | 35 |
| SB1234 | sitB::lacZ sicA::aphT | This study |
To construct the S. enterica serovar Typhimurium strain SB265, in which sicA and sipB are deleted and replaced with a cassette that confers kanamycin resistance, the following strategy was employed. An NsiI-EcoRI fragment was deleted from pSB810 (21), and the remaining vector was made blunt with T4 polymerase. An aphT cassette lacking the transcriptional terminator for this gene but containing its promoter (12) was subcloned from pSB80 as a HincII fragment and inserted into filled-in pSB810, resulting in pSB701. A 2.7-kb XbaI-KpnI fragment of pSB701 was blunted as above and moved into the SmaI site of the R6K-derived suicide vector pSB890 (14) to yield pSB1489. This vector was conjugated into SB300 (wild type) via SM10 λpir. Transconjugants that were streptomycin resistant, kanamycin resistant, sucrose resistant, and tetracycline sensitive were confirmed for allelic exchange by Southern blot analysis (data not shown).
P22HTint-mediated transduction (30) was used to construct SB266, an S. enterica serovar Typhimurium strain that carries null mutations in both sicA and sipC, and the allelic exchange was confirmed by Southern hybridization analysis.
SB319, which expresses a chromosomally encoded M45-epitope-tagged SicA, was constructed by allelic replacement with the R6K-derived suicide vector pSB890 (13). M45 epitope tagging of SicA and SipD was accomplished with the tagging vector pSB616 (5). The M45 epitope is derived from the adenovirus E4-6/7 protein and is recognized by the M45.7 monoclonal antibody (29).
RESULTS
sicA affects the intracellular and extracellular levels of SipB and SipC.
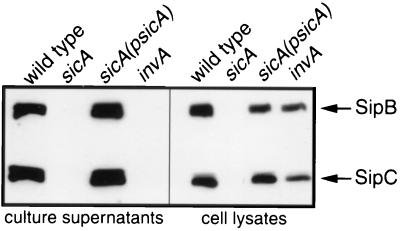
We have previously shown that mutations in sicA significantly impair the ability of S. enterica serovar Typhimurium to invade cultured Henle-407 cells (21). sicA is encoded within the centisome 63 pathogenicity island immediately upstream of sipB and sipC, two genes required for Salmonella entry into host cells. Because of the proximity of these genes to one another, their shared invasion phenotype, and the sequence similarity between SicA and known chaperones of type III secretion systems, the effect of a sicA null mutation on the levels of Sip proteins was examined. SipB and SipC appeared to be absent in culture supernatants and whole-cell lysates of the sicA mutant strain, as examined by Western immunoblot analysis with monoclonal antibodies directed against SipB and SipC (Fig. 1). The presence of SipB and SipC in the culture supernatants and whole-cell lysates of the sicA mutant was restored by the introduction of a sicA-complementing plasmid indicating that the observed effect of the sicA mutation is not due to polarity on downstream genes (Fig. 1). An invA mutant that is defective for type III secretion was included as a control (Fig. 1). These results indicate that SicA influences the levels of SipB and SipC.
FIG. 1.
Effect of a loss-of-function mutation in sicA on the intracellular and extracellular levels of SipB and SipC. Whole-cell lysate and culture supernatant proteins from wild-type S. enterica serovar Typhimurium, its isogenic sicA mutant strain SB221, or the same mutant carrying a sicA-complementing plasmid pSB814 (psicA) were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and probed with a mixture of SipB and SipC monoclonal antibodies.
SicA affects SipB and SipC protein stability.
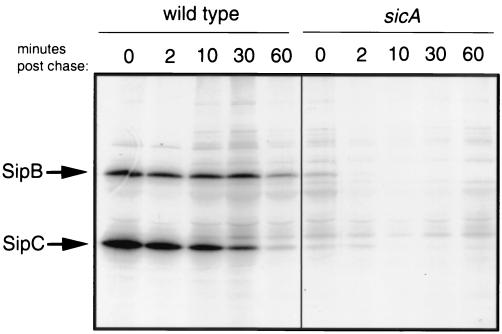
To investigate whether SicA affects the levels of SipB and SipC by influencing their stability, a pulse-chase experiment was undertaken. Cultures of wild-type and sicA mutant strains were subjected to metabolic labeling with [35S]methionine and [35S]cysteine for 2 min and subsequently chased with cold amino acids. The levels of radiolabeled SipB and SipC over time were then determined by immunoprecipitation and SDS-PAGE analysis. As shown in Fig. 2, in the sicA mutant strain the levels of SipB and SipC were drastically reduced even at the earliest sampling time and these proteins were virtually undetectable after a 10-min chase. In contrast, in the wild type, the levels of SipB and SipC change slowly over time, and these proteins could still be detected after a 60-min chase (Fig. 2). These results indicate that SicA affects the stability of SipB and SipC.
FIG. 2.
Absence of SicA results in decreased SipB and SipC stability. Wild-type S. enterica serovar Typhimurium and the isogenic sicA mutant strain SB221 were pulse-labeled with [35S]methionine for 2 min and chased with cold methionine for 60 min. At the indicated time points, samples were removed, immunoprecipitated with anti-SipB and anti-SipC polyclonal antibodies, and run on an SDS-PAGE gel. Labeled proteins were visualized by fluorography.
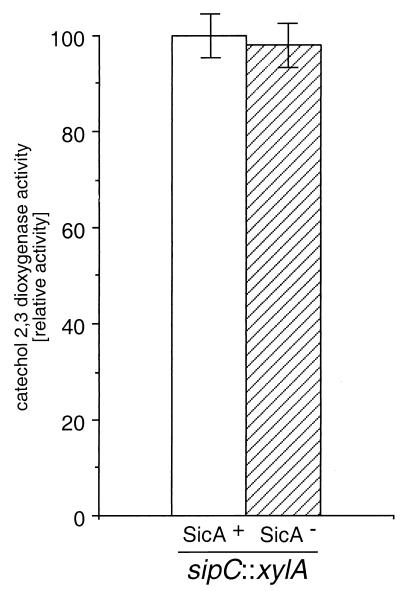
To address the possibility that the absence of SipB and SipC in the sicA mutant strain could be due to transcriptional effects of the sicA mutation (see below) on the sip operon, we measured the transcription of sipC both in the wild type as well as in a sicA::aphT mutant background. As shown in Fig. 3, the levels of sipC::xylE transcription were equivalent in both the wild-type and the sicA::aphT mutant strains, supporting a role for SicA on the stability of SipC and SipB. Nevertheless, these results do not address a potential additional role of SicA on the expression of the sip operon, since in this experimental setup, the expression of the sip genes is under the control of the aphT promoter driving the expression of downstream genes (see Materials and Methods).
FIG. 3.
Effect of a loss-of-function mutation in sicA on the expression of sipC. The levels of C2,3D in the S. enterica serovar Typhimurium strain SB227, which carries an sipC::xylE gene fusion, and in its derivative strain SB266, which carries a loss-of-function mutation in sicA, were measured as described in Materials and Methods. Activity is expressed as percentage of wild-type activity, which was considered 100.
SicA does not affect the stability of InvJ, SipA, and SipD.
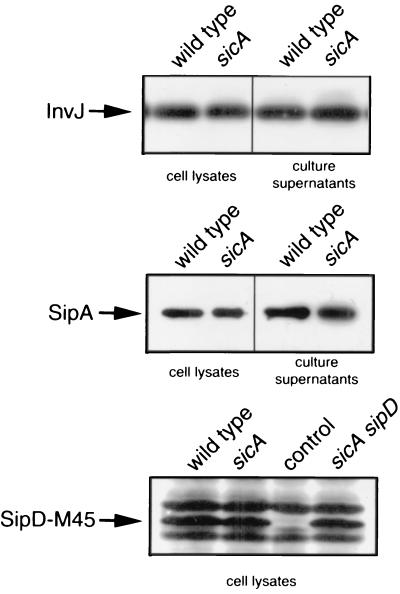
We tested the possibility that sicA may exert a more global effect on the stability of other type III-secreted proteins. We examined the levels of several type III-secreted proteins in an S. enterica serovar Typhimurium sicA mutant strain by Western immunoblot analysis with monoclonal antibodies directed to InvJ or the M-45 epitope present in chromosomally encoded tagged versions of SipA and SipD. As shown in Fig. 4, the absence of sicA did not affect the stability of either of these proteins. These results indicate that SicA influences the stability of only a subset of type III secreted proteins.
FIG. 4.
Effect of loss-of-function mutation in sicA on levels of type III secreted proteins InvJ, SipA, and SipD. Whole-cell lysate and culture supernatant proteins from wild-type S. enterica serovar Typhimurium and isogenic sicA mutant derivatives were probed for the presence of the different proteins, as indicated by Western immunoblotting with antibodies directed to the proteins or their epitope tags as appropriate (see Materials and Methods). Lane control, sample from an otherwise identical S. enterica serovar Typhimurium strain that does not express the M45-SipD epitope-tagged protein.
SicA interacts with SipB and SipC.
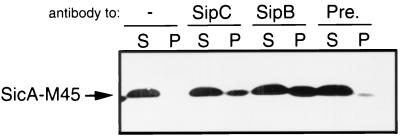
We examined whether SicA could interact with SipB and SipC as might be predicted if SicA behaves in a manner analogous to those of its homologous proteins IpgC of Shigella spp. (25) or SycD of Yersinia spp. (26). The interaction between SicA and its cognate substrates SipB and SipC was examined by coimmunoprecipitation analysis. Equal amounts of whole-cell lysates of an S. enterica serovar Typhimurium strain expressing a functional epitope-tagged SicA were immunoprecipitated with polyclonal antibodies directed to either SipB or SipC. The resulting immunoprecipitates were analyzed by Western blotting using an antibody directed to the M45 epitope tag present in SicA. As shown in Fig. 5, epitope-tagged SicA was observed in the immunoprecipitates of the samples treated with polyclonal antibodies directed to SipC and SipB but not in immunoprecipitates of the samples treated with the preimmune sera. These results indicate that SicA is capable of interacting with both SipB and SipC, and it is consistent with its postulated role as a chaperone or partitioning factor for these secreted proteins.
FIG. 5.
Interaction of SicA with SipB and SipC. Whole-cell lysates of the S. enterica serovar Typhimurium strain SB319 which expresses a functional, chromosomally encoded, M45-epitope-tagged form of SicA were immunoprecipitated with antibodies directed to SipB, SipC, or a preimmune serum (Pre), as described in Materials and Methods. Proteins that bound to the beads (P) or that remained in the supernatant (S) were probed by Western immunoblotting for the presence of M45-SicA with a monoclonal antibody directed to the M45 epitope.
SipB is stable and secreted in a sicA sipC double-mutant strain.
It has been postulated that members of the type III secretion-associated family of chaperones exert their function at least in part by acting as partitioning factors that prevent the premature cytoplasmic association between substrates of the type III secretion machinery. It is believed that this premature association leads to the degradation of the interacting proteins. If SicA prevents the cytoplasmic association of SipB with SipC, thereby preventing their premature degradation, it follows that absence of one of the cognate interacting proteins may lead to the stability of the other in the absence of SicA.
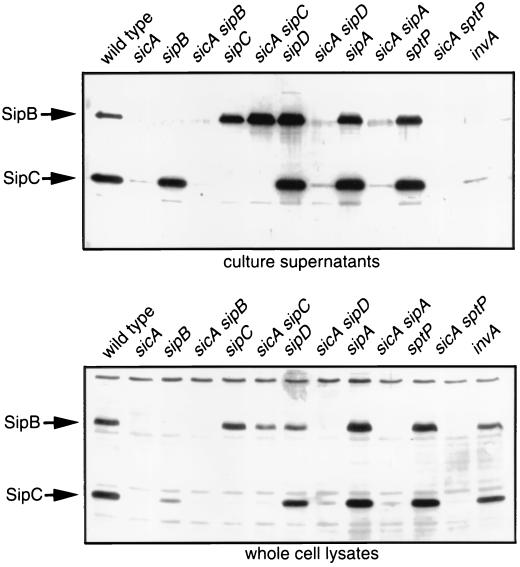
We tested this hypothesis by analyzing the stability of SipB and SipC in a sicA mutant background and in the absence of either SipB or SipC, as appropriate. As shown in Fig. 6, SipB was readily detectable in both culture supernatants and whole-cell lysates of a sicA sipC double-mutant strain. Introduction of a plasmid encoding SipC into this strain resulted in the degradation of SipB, as this protein could not be detected in culture supernatants or whole-cell lysates of the complemented strain (data not shown). These results indicate that SicA prevents the cytoplasmic association between SipB and SipC and that in the absence of such association, SipB is not only stable but also secreted at wild-type levels. These results also indicate that SicA is not essential for SipB secretion in the absence of SipC.
FIG. 6.
SipB is stable and secreted in a sicA sipC double-mutant strain. Culture supernatants and whole-cell lysates of wild-type S. enterica serovar Typhimurium and several isogenic mutant strains were probed for the presence of SipB and SipC by Western immunoblotting with monoclonal antibodies directed to these proteins.
We also examined the stability of SipC in a sicA sipB double-mutant background. In contrast to SipB, we could not detect SipC in either culture supernatants or whole-cell lysates of this double-mutant strain (Fig. 6). The absence of SipC from this mutant strain cannot be explained by a transcriptional effect of the mutation, as expression of SipC in this mutant background is driven by the aphT promoter present in the cassette located immediately upstream of sipC (see Materials and Methods and Fig. 3). Upon reintroduction of sicA into this mutant strain, SipC was detectable in both whole-cell lysates and supernatant fractions (data not shown). These results indicate that SicA is most likely serving to protect SipB from an undesirable interaction with SipC, but that its role in stabilizing SipC may entail (i) partitioning SipC from an as-yet-unidentified interacting partner/s, (ii) capping of a degradation signal in SipC, or (iii) preventing SipC from misfolding, which would lead to degradation.
We compared the production of SipB and SipC in sicA strains carrying null mutations in other type III secreted proteins to ascertain if any other mutant combination could affect the production of these proteins in a manner similar to that of sicA sipC double mutants. As shown in Fig. 6, SipB and SipC were not detected in a sicA sptP, sicA sipA, or sicA sipD double-mutant combinations, suggesting that SicA may not partition SipC from these secreted proteins. Alternatively, more than one effector protein may need to be absent in order to reveal with this strategy the potential SicA-mediated partitioning of SipC from other effector proteins.
sopE expression is dependent on SicA.
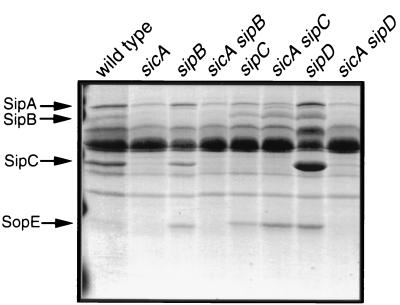
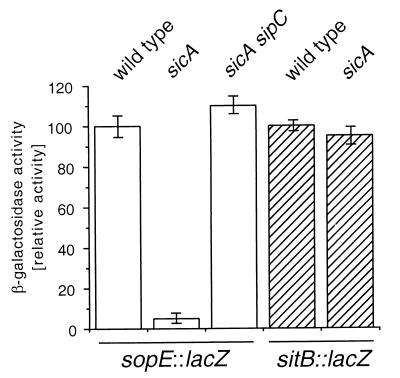
Examination of the profile of total secreted proteins present in the culture supernatants of the sicA mutant strain revealed the absence of proteins other than SipB and SipC when compared to the wild type (Fig. 7). In particular, a protein with a molecular mass of 25 kDa, which corresponds to SopE, was conspicuously absent from culture supernatants of the sicA mutant. This observation suggested the possibility that SicA may also act as a chaperone for SopE and other substrates of the type III secretion system. To begin to investigate this possibility, we examined the levels of sopE transcription in a sicA mutant background by using reporter gene fusions. Surprisingly, the transcription of sopE was completely abrogated in a sicA mutant background, indicating a requirement of sicA for sopE transcription and most likely not for its stability (Fig. 8). In contrast, the transcription of the sit operon, which encodes an iron transport system and is also contained within SPI-1, was not affected, ruling out a global effect of the absence of SicA on gene expression. Introduction of a sipC null mutation in the sicA mutant strain restored the production of SopE to wild-type levels (Fig. 7 and 8). In contrast, introduction of a sipB or sipD null mutation into the sicA mutant strains did not (Fig. 7). These results argue that SicA does not exert a direct effect on the transcription of sopE. Rather, these results are consistent with SicA influencing the stability of another factor which may partition with SipC and may be directly responsible for activating the transcription of sopE and other effector proteins. Such a factor cannot be any of the Sip proteins since SopE is produced at wild-type levels in strains carrying null mutations in sipB, sipC, sipD, or sipA (data not shown). Furthermore, the effect of such a factor must be restricted to only a subset of the type III secreted proteins since a mutation in sicA did not affect the expression of other type III secreted proteins, such as InvJ (Fig. 3).
FIG. 7.
Culture supernatant protein profiles of wild-type and isogenic mutant strains of S. enterica serovar Typhimurium. Culture supernatants proteins from the different strains of S. enterica serovar Typhimurium were separated by SDS-PAGE and visualized by Coomassie blue staining, as described in Materials and Methods.
FIG. 8.
Effect of a loss-of-function mutation in sicA on the expression of sopE. The levels of β-galactosidase in the S. enterica serovar Typhimurium strain SB876, which carries a sopE::lacZ gene fusion, and its isogenic derivative strains SB879 and SB1235, which carry loss-of-function mutations in sicA or in sicA and sipC, respectively, were measured as described in Materials and Methods. Activity is expressed as percentage of wild-type activity, which was considered 100. As negative controls, levels of β-galactosidase were measured in the S. enterica serovar Typhimurium strain SB804, which carries a sitB::lacZ gene fusion (wild type), and its isogenic derivative strains of SB1234, which carry loss-of-function mutations in sicA.
DISCUSSION
Secretion of proteins via the type III secretion systems requires the function of a family of low-molecular-weight accessory proteins that are believed to work as chaperones or partitioning factors (33). Although the function of these chaperones have been studied extensively, particularly in Yersinia spp., their mechanism of action remains poorly understood. We have shown here that SicA exerts functions consistent with its putative role as a type III secretion-associated chaperone. Our results are consistent with SicA playing a role as a partitioning factor that prevents the premature association between SipB and SipC: (i) the absence of SicA resulted in the degradation of both SipB and SipC, and (ii) SicA was shown to bind to both these proteins. In this context, the function of SicA is consistent with the role postulated for the related protein IpgC from Shigella spp. IpgC has been shown to be required for the stability of the secreted invasins IpaB and IpaC by preventing their premature association within the bacterial cytoplasm (25).
We found that in a sipC mutant background strain, the absence of SicA did not lead to the degradation of SipB. These results indicate that SicA may prevent the premature interaction of SipB with SipC, which may lead to their targeting for degradation. In contrast, in a sicA sipB double-mutant background, SipC was still targeted for degradation. These results suggest that SicA may partition SipC from other bacterial effector proteins or may prevent the self-association of SipC, either event perhaps resulting in the degradation of this protein. These results also suggest a more central role for SipC in the organization of the complex of secreted proteins destined to be translocated into host cells. Indeed, both SipB and SipC have been shown to be essential for the translocation of effector proteins into the host cell (4, 9).
The phenotype of the sicA sipC double mutant resembles that of the sycD lcrV mutant of Yersinia (26). In Yersinia, the absence of SycD leads to the degradation of the type III secretion translocases YopB and YopD. However, in the absence of lcrV, SycD is no longer required for the stability of YopB and YopD. Thus, the parallel between these phenotypes in Yersinia and Salmonella, coupled to the amino acid sequence similarity between SicA and SycD and their association with proteins involved in the translocation of effectors into the host cell, suggests a common mechanism for this subset of the type III secretion-associated family of chaperones.
Our results do not support a role for SicA in the actual secretion process since in the sicA sipC double-mutant strain, SipB was secreted at wild-type levels. However, these results do not rule out a role for SicA in establishing the timing of secretion of the Sip proteins. Indeed, the biology of the invasion-associated type III secretion system of Salmonella indicates that it is very likely that secretion of different proteins must be temporally regulated since at least some of these effector proteins must act in a temporally coordinated fashion (7). It is possible that the type III secretion-associated chaperones may play a role in this aspect of the function of these systems.
In examining the total protein profile of culture supernatants of a sicA mutant strain, we observed the absence of several type III secreted proteins other than SipB or SipC. This observation prompted us to examine the potential role of SicA in stabilizing these effector proteins similar to its role in stabilizing SipB and SipC. Surprisingly, we found that the absence of the secreted proteins in the sicA mutant strain was the result of the abrogation of their expression rather than their degradation. However, the absence of SicA did not affect the expression of invJ, which also encodes a type III secreted protein. These results therefore indicate that SicA influences the expression of only a subset of type III secreted proteins. Introduction of a sipC loss-of-function mutation into the sicA mutant strain restored the expression of SopE. These results imply that the transcriptional effect of SicA is most likely indirect and that the expression of sopE must be controlled by a factor(s) partitioned from SipC by SicA. The absence of SicA, therefore, would result in the degradation of such a putative regulatory protein. Experiments are underway to identify such a regulatory factor. Transcriptional regulatory activities have previously been postulated for SycD, a SicA-related chaperone in Yersinia spp. (2). However, such regulatory function is thought to be indirect through its function in the type III secretion system (26). Further studies will be required to establish whether SycD and SicA exert their regulatory function in a similar manner.
In summary, our results indicate that SicA exerts a chaperone or partitioning function that prevents the premature association between SipC and SipB and perhaps other yet-unidentified proteins, including one or more that may control the expression of other type III secreted proteins. These results also suggest that SicA exerts a more complex activity than that of other type III secretion-associated chaperones, such as SicP, which seem to act on a single substrate and do not seem to affect the transcription of other type III secreted proteins (8). Thus, as previously postulated for Yersinia, it appears that in Salmonella there are also two types of type III secretion-associated chaperones: one class which exerts their function on a single effector protein represented by SicP (8) and presumably pipC/sigE (16, 27), and the other class represented by SicA, which is multivalent and exerts its function over proteins involved in translocation of effectors into host cells.
ACKNOWLEDGMENTS
We thank members of the Galán laboratory for critical review of the manuscript.
This work was supported by Public Health Service Grant AI30492 from the National Institutes of Health.
REFERENCES
- 1.Baudry B, Kaczorek M, Sansonetti P J. Nucleotide sequence of the invasion plasmid antigen B and C genes (ipaB and ipaC) of Shigella flexneri. Microb Pathog. 1988;4:345–357. doi: 10.1016/0882-4010(88)90062-9. [DOI] [PubMed] [Google Scholar]
- 2.Bergman T, Hakansson S, Forsberg A, Norlander L, Macellaro A, Baeckman A, Boelin I, Wolf-Watz H. Analysis of the V antigen lcrGVH-yopBD Operon of Yersinia pseudotuberculosis: evidence for a regulatory role of LcrH and LcrV. J Bacteriol. 1991;173:1607–1616. doi: 10.1128/jb.173.5.1607-1616.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng L W, Anderson D M, Schneewind O. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol Microbiol. 1997;24:757–765. doi: 10.1046/j.1365-2958.1997.3831750.x. [DOI] [PubMed] [Google Scholar]
- 4.Collazo C, Galán J E. The invasion-associated type III system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 5.Collazo C, Galán J E. Requirement of exported proteins for secretion through the invasion-associated Type III system in Salmonella typhimurium. Infect Immun. 1996;64:3524–3531. doi: 10.1128/iai.64.9.3524-3531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frithz-Lindsten E, Rosqvist R, Johansson L. The chaperone-like protein YerA of Yersinia pseudotuberculosis stabilizes YopE in the cytoplasm but is dispensible for targeting to the secretion loci. Mol Microbiol. 1995;16:635–647. doi: 10.1111/j.1365-2958.1995.tb02426.x. [DOI] [PubMed] [Google Scholar]
- 7.Fu Y, Galan J E. A salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature. 1999;401:293–297. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]
- 8.Fu Y, Galán J E. Identification of a specific chaperone for SptP, a substrate of the centisome 63 type III secretion system of Salmonella typhimurium. J Bacteriol. 1998;180:3393–3399. doi: 10.1128/jb.180.13.3393-3399.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu Y, Galán J E. The Salmonella spp. protein tyrosine phosphatase SptP is translocated into host cells and disrupts the host-cell cytoskeleton. Mol Microbiol. 1998;27:359–368. doi: 10.1046/j.1365-2958.1998.00684.x. [DOI] [PubMed] [Google Scholar]
- 10.Galán J E. Interaction of Salmonella with host cells through the centisome 63 type III secretion system. Curr Opin Microbiol. 1999;2:46–50. doi: 10.1016/s1369-5274(99)80008-3. [DOI] [PubMed] [Google Scholar]
- 11.Galán J E, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:322–328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 12.Galán J E, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella typhimurium invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;17:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardt W-D, Galán J E. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc Natl Acad Sci USA. 1997;94:9887–9892. doi: 10.1073/pnas.94.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardt W-D, Urlaub H, Galán J E. A target of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc Natl Acad Sci USA. 1998;95:2574–2579. doi: 10.1073/pnas.95.5.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoiseth S K, Stocker B A. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 16.Hong K H, Miller V L. Identification of a novel Salmonella invasion locus homologous to Shigella ipgDE. J Bacteriol. 1998;180:1793–1802. doi: 10.1128/jb.180.7.1793-1802.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingram C, Brawner M, Youngman P, Westpheling J. xylE functions as an efficient reporter gene in Streptomyces spp.: use for the study of galP1, a catabolite-controlled promoter. J Bacteriol. 1989;171:6617–6624. doi: 10.1128/jb.171.12.6617-6624.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaniga K, Bossio J C, Galán J E. The Salmonella typhimurium invasion genes invF and invG encode homologues to the PulD and AraC family of proteins. Mol Microbiol. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- 20.Kaniga K, Trollinger D, Galán J E. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaniga K, Tucker S C, Trollinger D, Galán J E. Homologues of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J Bacteriol. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, Galán J E, Aizawa S-I. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 23.Lennox E S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 24.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Ménard R, Sansonetti P J, Parsot C, Vasselon T. The IpaB and IpaC invasins of Shigella flexneri associate in the extracellular medium and are partitioned in the cytoplasm by a specific chaperon. Cell. 1994;79:515–529. doi: 10.1016/0092-8674(94)90260-7. [DOI] [PubMed] [Google Scholar]
- 26.Neyt C, Cornelis G R. Role of SycD, the chaperone of the Yersinia yop translocators YopB and YopD. Mol Microbiol. 1999;31:143–156. doi: 10.1046/j.1365-2958.1999.01154.x. [DOI] [PubMed] [Google Scholar]
- 27.Norris F A, Wilson M P, Wallis T S, Galyov E E, Majerus P W. SopB, a protein required for virulence of Salmonella dublin, is an inositol phosphate phosphatase. Proc Natl Acad Sci USA. 1998;95:14057–14059. doi: 10.1073/pnas.95.24.14057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Callaghan D, Charbit A. High efficiency transformation of Salmonella typhimurium and Salmonella typhi by electroporation. Mol Gen Genet. 1990;223:157–160. doi: 10.1007/BF00315809. [DOI] [PubMed] [Google Scholar]
- 29.Obert S, O'Connor R J, Schmid S, Hearing P. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol Cell Biol. 1994;14:1333–1346. doi: 10.1128/mcb.14.2.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:74–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- 31.Wattiau P, Bernier B, Deslée P, Michiels T, Cornelis G R. Individual chaperones required for Yop secretion by Yersinia. Proc Natl Acad Sci USA. 1994;91:10493–10497. doi: 10.1073/pnas.91.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wattiau P, Cornelis G R. SycE, a chaperone-like protein of Yersinia enterocolitica involved in the secretion of YopE. Mol Microbiol. 1993;8:123–131. doi: 10.1111/j.1365-2958.1993.tb01209.x. [DOI] [PubMed] [Google Scholar]
- 33.Wattiau P, Woestyn S, Cornelis G R. Customized secretion chaperones in pathogenic bacteria. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 34.Woestyn S, Sory M P, Boland A, Lequenne O, Cornelis G R. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol Microbiol. 1996;20:1261–1271. doi: 10.1111/j.1365-2958.1996.tb02645.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhou D, Hardt W-D, Galán J E. Salmonella typhimurium encodes an iron transport system within the centisome 63 pathogenicity island. Infect Immun. 1999;67:1974–1981. doi: 10.1128/iai.67.4.1974-1981.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou D, Mooseker M, Galán J E. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science. 1999;283:2092–2095. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]