ABSTRACT
During embryonic development, lymphatic endothelial cell (LEC) precursors are distinguished from blood endothelial cells by the expression of Prospero-related homeobox 1 (Prox1), which is essential for lymphatic vasculature formation in mouse and zebrafish. Prox1 expression initiation precedes LEC sprouting and migration, serving as the marker of specified LECs. Despite its crucial role in lymphatic development, Prox1 upstream regulation in LECs remains to be uncovered. SOX18 and COUP-TFII are thought to regulate Prox1 in mice by binding its promoter region. However, the specific regulation of Prox1 expression in LECs remains to be studied in detail. Here, we used evolutionary conservation and chromatin accessibility to identify enhancers located in the proximity of zebrafish prox1a active in developing LECs. We confirmed the functional role of the identified sequences through CRISPR/Cas9 mutagenesis of a lymphatic valve enhancer. The deletion of this region results in impaired valve morphology and function. Overall, our results reveal an intricate control of prox1a expression through a collection of enhancers. Ray-finned fish-specific distal enhancers drive pan-lymphatic expression, whereas vertebrate-conserved proximal enhancers refine expression in functionally distinct subsets of lymphatic endothelium.
Keywords: Prox1, Enhancers, Evolutionary conservation, Gene regulation, Lymphatic endothelial cell, Transcription factor, Zebrafish
Summary: Although Prox1 is a key factor in lymphatic endothelial cell development, little is known about its cis-regulation. Here, we characterise multiple prox1a enhancers active in subsets of zebrafish lymphatic vasculature.
INTRODUCTION
Transcription factor (TF) regulation plays a pivotal role in cell specification and the acquisition of tissue identity during development (Spitz and Furlong, 2012). The TF PROX1 is involved in lymphatic vasculature development. In mouse, PROX1 is expressed by the lymphatic progenitors and is the first marker of specified lymphatic endothelial cells (LECs) (Wigle and Oliver, 1999). Prox1 mutants are characterised by the loss of all lymphatic structures (Wigle and Oliver, 1999). However, during embryonic development, prox1 is expressed in multiple tissues, such as the central nervous system, liver, retina and skeletal muscles (Glasgow and Tomarev, 1998; Pistocchi et al., 2008), implying the necessity for mechanisms restricting its expression in a tissue-specific manner. Although TFs regulating Prox1 expression in the lymphatics have been described in mammals (François et al., 2008; Srinivasan et al., 2010), the upstream cis-regulation of Prox1 remains largely unexplored, leaving a comprehensive understanding of the regulatory logic of the gene incomplete.
Enhancers are cis-regulatory elements that can be located proximally or distally to a gene locus. Enhancers can function in a modular fashion, with tissue-specific enhancers activated independently (Long et al., 2016). Enhancer regulation is particularly interesting in a developmental context, as many ‘developmental toolbox’ genes are differentially regulated through enhancer activity. Important enhancer elements active in lymphatic and blood vascular development have been identified in genes such as gata2a (Shin et al., 2019), flt1 (Bussmann et al., 2010), notch1b (Chiang et al., 2017), fli1 (Villefranc et al., 2007) and etsrp (Veldman and Lin, 2012). However, to date only one lymphatic-specific Prox1 enhancer (Kazenwadel et al., 2023), with activity enriched in the lymphatic valves, has been described. Therefore, the number, contextual nature and identity of the enhancers driving Prox1 expression in lymphatics remains to be investigated.
In zebrafish, the most studied lymphatic beds are the trunk and facial lymphatics (Eng et al., 2019; Hogan et al., 2009; Küchler et al., 2006; Okuda et al., 2012; Yaniv et al., 2006) which require prox1 for correct development (Grimm et al., 2023). As a result of the teleost-specific genome duplication event, two zebrafish co-orthologues of mammalian Prox1 have been described in the literature, called respectively prox1a and prox3 (previously referred to as prox1b) (Giacco et al., 2010). prox1a is an early marker of lymphatic identity, being expressed by lymphatic progenitors in the posterior cardinal vein (PCV) at 32 h post-fertilization (hpf), before the onset of sprouting (Dunworth et al., 2014; Koltowska et al., 2015a). Its expression marks all described lymphatic beds for the duration of larval development. Mutants for prox1a and prox3 show a reduction in the number of LECs (Grimm et al., 2023), confirming the importance of these factors for LEC development in zebrafish. In particular, it is prox1a that has a prominent developmental role in zebrafish lymphangiogenesis (Grimm et al., 2023; Koltowska et al., 2015a). Despite this, little is known about prox1a regulation. Although the TFs Coup-TFII (Nr2f2) and Sox18 do not seem to regulate prox1a (van Impel et al., 2014), a role of Vegfc signalling upstream of prox1a in the specification of LECs has been reported (Koltowska et al., 2015a). However, the cis-regulatory elements of prox1a and their evolution are still to be described.
In this study, we aimed to characterise the enhancers regulating the expression of prox1a in the developing lymphatics. We identified several elements driving expression in different subsets of the lymphatic endothelium. These include both conserved elements across vertebrates and elements specific to actinopterygians. Our results suggest that prox1a is tightly regulated in a spatially patterned manner by a cohort of enhancers acting in concert.
RESULTS
The prox1a locus is enriched in evolutionarily conserved non-coding regions
To identify enhancer elements in the zebrafish prox1a locus, we analysed DNA conservation, as conserved non-coding elements (CNE) close to a gene can indicate the presence of enhancers. We aligned the region of the PROX1/prox1a locus of seven Osteichthyes species against that of the zebrafish using mVISTA (Fig. 1A; Table S1). The species were selected to have a balanced spread along the phylogeny. Local microsynteny, which is the conservation of the identity and position of the features surrounding the gene of interest, was verified by comparing the identities of neighbouring loci (Fig. S1A). Microsynteny indicates that no major genomic rearrangements, such as insertions, deletions or inversions of whole regions, have occurred around the considered locus, and therefore supports the presence of evolutionarily conserved enhancers. In all species analysed, an SMYD2 homolog was located downstream of prox1a. Similarly, RPS6KC1 was located upstream of the locus in all cases except zebrafish. Consequently, we defined the regions of interest for the conservation analysis as the sequences between the two loci adjacent to PROX1/prox1a, encompassing the long intergenic region upstream of prox1a (300 kbp). We recovered 25 conserved non-coding sequences, located upstream, downstream and in the intronic regions of PROX1/prox1a (Fig. 1A). We also investigated conservation in the other described PROX1 co-orthologue, prox3. Interestingly, prox3 presents very low levels of conservation outside of exon sequences, with only two Teleost-conserved elements identified in the analysis and none conserved with other vertebrates in intergenic regions (Fig. S1B).
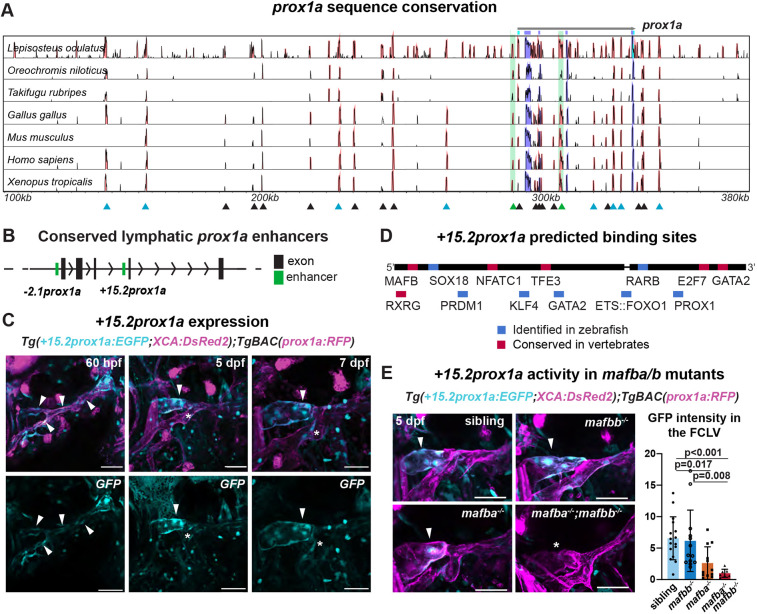
Fig. 1.
The conserved +15.2prox1a enhancer drives expression in a subset of the facial lymphatics. (A) Conservation analysis of the 380 kbp region surrounding the zebrafish prox1a locus compared with seven vertebrate species. Blue peaks, exons; red peaks, conserved non-coding DNA; black arrowheads, conserved peaks; blue arrowheads, tested peaks; green arrowheads, identified −2.1prox1a and +15.2prox1a lymphatic enhancers. In the 5′ the first 100 kbp of the alignment contain no conservation peak and has been omitted from the graph. (B) prox1a locus showing the position of the identified conserved lymphatic enhancers. Green boxes, −2.1prox1a and +15.2prox1a enhancers; black boxes, exons. (C) Confocal projections of facial lymphatics labelled with Tg(+15.2prox1a:EGFP; XCA:DsRed2)uu7kk (cyan) and Tg(prox1a:RFP)nim5 (magenta) at 60 hpf, 5 dpf and 7 dpf. Arrowheads show expression in the facial LECs (60 hpf) and FCLV (5 and 7 dpf). Asterisks show expression in facial lymphatic endothelium. (D) Predicted endothelial TF binding sites in +15.2prox1a. Blue, binding sites identified in zebrafish (P<1e-04); red, conserved binding sites within vertebrates. (E) Quantification of +15.2prox1a activity in mafba/b mutants. Left: confocal projections of facial lymphatics labelled with Tg(+15.2prox1a:EGFP; XCA:DsRed2)uu7kk (cyan) and Tg(prox1a:RFP)nim5 (magenta) at 5 dpf showing enhancer activity (arrowhead) or lack thereof (asterisk) in the FCLV. Right: quantification of signal intensity in the FCLV normalised to the ganglia in 5 dpf embryos. Sibling (n=16) versus mafbb−/− (n=13), mafba−/− (n=12) and mafba−/−;mafbb−/− (n=7). Mean±s.d. Sibling versus mafbb−/−, not significant (ns) (P>0.999); sibling versus mafba−/−, P=0.017; sibling versus mafba−/−;mafbb−/−, P<0.001; mafbb−/− versus mafba−/−, ns (P=0.146); mafbb−/− versus mafba−/−;mafbb−/−, P=0.008; mafba−/− versus mafba−/−;mafbb−/−, ns (P>0.999) (Kruskal–Wallis test with Dunn's multiple comparison test). Four technical replicates, biological replicates correspond to the number of data points per condition. Scale bars: 50 μm.
We then tested selected prox1a CNEs for regulatory activity. Histone modifications have been linked to non-coding elements such as promoters and enhancers. Specifically, H3K4me1 marks primed and active enhancers (Heintzman et al., 2007) and H3K27ac indicates active enhancers (Bonn et al., 2012; Creyghton et al., 2010). Currently, no LEC-specific zebrafish databases for H3K4me1 and H3K27ac are available. Whole-body databases were instead used to identified ten of the selected prox1a CNEs as primed enhancers (Aday et al., 2011; Bogdanović et al., 2012) (Fig. 1A; Fig. S1C). These were subsequently cloned into the Zebrafish Enhancer Detection (ZED) vector (Bessa et al., 2009) and tested in vivo in F1 (Table S2). Two of the tested sequences drove GFP expression in the subsets of lymphatic endothelium. The conservation of the identified elements was further confirmed using PhyloP and the Multiz alignment to vertebrate tracks in the UCSC Genome browser (Fig. S1D). Of the two elements, +15.2prox1a [located 15.2 kbp downstream of the transcription start site (TSS)] was enriched in the facial collecting lymphatic vessel (FCLV) (Fig. 1B,C; Fig. S1E; Table 1; Table S2), and −2.1prox1a (located 2.1 kbp upstream of the TSS) was enriched in the lymphatic valve (Figs 1B, 2A-C; Fig. S2A; Table 1; Table S2).
Table 1.
prox1a enhancers drive regionalised GFP expression in lymphatic endothelium
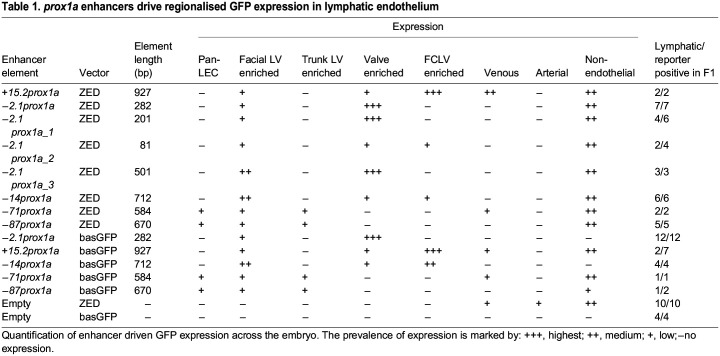
Fig. 2.
The conserved −2.1prox1a enhancer drives expression in the lymphatic valve. (A) Confocal projections of facial lymphatics labelled with Tg(−2.1prox1a:EGFP; XCA:DsRed2)uu3kk (cyan) and Tg(prox1a:RFP)nim5 (magenta) at 3, 5 and 7 dpf. Arrowheads show expression in the developing lymphatic valve. Asterisks show expression in the facial lymphatic endothelium. (B) Confocal projections of the facial lymphatics labelled with Tg(−2.1prox1a:basEGFP;ACry:GFP)uu10kk (cyan) and Tg(prox1a:RFP)nim5 (magenta) at 5 dpf. Left: expression in the lymphatic valve (arrowhead) and facial lymphatic endothelium (asterisk). Right: magnification of the boxed area in the left panel. (C) Confocal projection of the whole embryo at 5 dpf labelled with Tg(−2.1prox1a:basEGFP;ACry:GFP)uu10kk (cyan) and Tg(prox1a:RFP)nim5 (magenta). Arrowhead shows expression in the lymphatic valve. Asterisks show additional expression in the skin. (D) Predicted endothelial TF binding sites in −2.1prox1a. Blue, binding sites identified in zebrafish (P<1e-04); red, conserved binding sites within vertebrates; yellow, binding sites conserved in vertebrates but absent in zebrafish. (E) Representative images and quantification of Nfatc1 binding-dependent −2.1prox1a activity. Left: confocal projections of lymphatic valve labelled with WT, Nfatc1 binding site-mutated or scrambled −2.1prox1a:basEGFP;ACry:GFP constructs. Arrowheads show signal in the valve. Asterisk shows missing signal in the valve. Right: quantification of signal intensity in the valve cells expressing GFP in 5 dpf injected embryos. Mean±s.d. WT (n=12) versus Nfatc1 binding site-mutated (n=12) injected embryos at 5 dpf; P=0.002 (two-tailed Mann–Whitney test). Four technical replicates, biological replicates correspond to the number of data points per condition. Scale bars: 50μm (A,B,E); 500 μm (C).
+15.2prox1a drives expression in the FCLV
+15.2prox1a is a 927 bp element located in intron 3 of prox1a (Fig. 1B; Fig. S1C). The element is composed of two separate conservation peaks divided by 17 bp (Fig. 1D). At 5 and 7 days post-fertilization (dpf) the element is able to drive GFP expression in the FCLV, with faint additional expression in the facial lymphatics (Fig. 1C; Fig. S1E; Table 1). We also observed the reporter expression at 60 hpf in the developing facial lymphatic, but not in the ventral aorta lymphangioblast (VA-L) (Fig. 1C; Fig. S1E,F). In addition, +15.2prox1a is active in the venous endothelium in the primary head sinus (PHS) (Fig. S1G) and in the PCV (Fig. S1H). In a whole-body image, additional expression can be detected in the gallbladder and pancreas (Fig. S1I). As this element is highly conserved across vertebrates, we used a P-value cut-off of 1e-02 for our MEME Suite (Bailey and Elkan, 1994) and TOMTOM (Gupta et al., 2007) to determine the motifs and putative transcription factor binding sites present. We identified conserved binding sites for multiple known lymphatic regulators, such as Gata2, Nfatc1, Mafb, Sox18, Prox1 (Arnold et al., 2022; Geng et al., 2016; Kazenwadel et al., 2015; Koltowska et al., 2015b; Shin et al., 2019) and the retinoic acid receptors Rarb and Rxrg (Bowles et al., 2014; Marino et al., 2011). We also found putative sites for TFs which have been implied in regulation of Prox1 expression or are related to endothelial cell (EC) identity such as Tfe3, Ets::Foxo1, Prdm1 and Klf4 (Dieterich et al., 2015; Niimi et al., 2019; Park et al., 2014; Pham et al., 2007; Tai-Nagara et al., 2020; De Val et al., 2008; Yoshimatsu et al., 2011) (Fig. 1D; Fig. S1J,K; Tables S4,S5). In order to test the functional importance of the predicted binding sites, we took advantage of the mafbauq4bh and mafbbub47bh mutant lines available in our lab (Arnold et al., 2022; Koltowska et al., 2015b). Quantification of the activity of +15.2prox1a in the mutant lines revealed a 60% reduction in enhancer activity in mafba mutants and 85% in double mutants (Fig. 1E). This confirms the functional relevance of the MAFB predicted binding site for +15.2prox1a activity, and suggests this enhancer is an important driver of prox1a contributing to the regulatory logic necessary for its correct spatial expression in developing lymphatics.
−2.1prox1a drives expression in the lymphatic valve across developmental stages
−2.1prox1a is a conserved sequence of 282 bp located upstream of the TSS (Fig. 1B; Fig. S1C). We recently reported (Kazenwadel et al., 2023) that this element is active in the developing tissue of the lymphatic valve at 5 dpf, and weakly in the rest of the facial lymphatics (Fig. 2A; Fig. S2A; Table 1). However, the functional characterisation of this enhancer and the timing of its expression in zebrafish are yet to be determined. Here, we uncovered that at 3 dpf the −2.1prox1a element is first active in the site of the future valve formation, preceding the gata2a onset of expression (Quillien et al., 2017), and its activity is ongoing until valve maturation at 7 dpf (Fig. 2A; Fig. S2A).
As the empty ZED vector induces low level of GFP expression in various tissues (Table 1; Fig. S2B) and it uses a gata2a minimal promoter (Bessa et al., 2009), we wanted to confirm the specificity of −2.1prox1a-driven expression. We re-cloned −2.1prox1a in a different plasmid, using an e1b TATA minimal promoter (Villefranc et al., 2007) which does not drive GFP expression without enhancer sequences (Fig. S2C; Table 1). We obtained the same enriched expression in the lymphatic valve and sparse expression in facial lymphatics (Fig. 2B,C; Table 1). We additionally investigated the enhancer activity in the other vascular valves present during embryonic development. Two lymphovenous valves (LVVs) have been described connecting the facial lymphatics to the veins (Meng et al., 2023; Shin et al., 2019). We detected weak activity of −2.1prox1a in the anterior LVV connecting the FCLV to the PHS, but none in the posterior LVV (Fig. S2D). No activity was detected in association with the developing atrio-ventricular and bulbo-ventricular cardiac valves (Fig. S2E). Whole-body imaging at 5 dpf revealed additional expression in a population of cells in the abdomen and extremely weak expression in few muscle cells (Fig. 2C), confirming the predominant activity of this enhancer in the lymphatic valve.
−2.1prox1a presents predicted binding sites for a variety of known lymphatic factors. At the 5′ it contains binding sites for Nfatc1 and Fox, previously reported to be conserved between zebrafish and mammals (Kazenwadel et al., 2023). In the mouse enhancer, a vertebrate-conserved Gata2 site (Fig. 2D; Tables S4,S5) has been confirmed in LECs using ChIP-seq that is functionally necessary for lymphatic vessel formation (Kazenwadel et al., 2023). However, this site is not present in zebrafish (Fig. 2D). The enhancer also includes binding sites for genes involved in vascular development such as forkhead box transcription factors (Niimi et al., 2017; Scallan et al., 2021), TCFs (Cha et al., 2016; Nicenboim et al., 2015), SNAI2 (Hultgren et al., 2020), retinoic acid receptors (Bowles et al., 2014; Marino et al., 2011) and ETS factors (Pham et al., 2007; De Val et al., 2008; Yoshimatsu et al., 2011) (Fig. 2D; Fig. S2F,G; Tables S4,S5). In order to test the functional relevance of the predicted TFs, we focused on the conserved Nfatc1 binding site. We injected embryos with the e1b TATA minimal promoter vector (Villefranc et al., 2007) containing either the wild-type (WT) −2.1prox1a sequence or one lacking the Nfatc1 site. We compared the level of activity in the valve at 5 dpf, and observed a 40% reduction of intensity in the mutated enhancer compared with the WT (Fig. 2E). No activity was observed in the scrambled-sequence controls (n=36). This suggests that Nfatc1 is indeed needed for the full activity of −2.1prox1a, but that even in its absence other factors can drive partial expression in the valve. Overall, these results show that despite some divergence, part of the upstream regulation of this enhancer is conserved between zebrafish and mammals.
Accessible regions of open chromatin at the prox1a locus drive expression in the lymphatics
Using an evolutionary conservation approach, we have identified enhancers that are active in a subset of lymphatic endothelium, but no pan-lymphatic drivers. As non-conserved enhancers also exist, we complemented our conservation analysis using a chromatin accessibility approach, which can indicate the presence of an active enhancer in a tissue. To identify additional prox1a lymphatic enhancers we used a previously published single nuclei ATAC-seq (snATAC-seq) database of zebrafish ECs (N=3155) at 4 dpf, which included arterial ECs, venous ECs and LECs (Grimm et al., 2023). From control/WT sorted ECs, we selected all ECs and endocardium, and then subsetted and re-clustered the data (Fig. S3A). Cluster identity was determined using the gene accessibility score of marker genes of lymphatic and blood endothelium (Fig. S3A). Gene Ontology (GO) terms analysis revealed an enrichment for terms connected with lymphangiogenesis in the LEC-accessible regions, and enrichment for angiogenesis terms in the less accessible regions (Fig. S3B).
We focused on the DNA region surrounding the prox1a locus and identified six regions of open chromatin in LECs (Fig. 3A). We established stable transgenic lines for three of them: −87prox1a, −71prox1a and −14prox1a (Table S2). All drove expression in the lymphatic endothelium (Table 1). Noticeably, −2.1prox1a was one of the elements retrieved by this analysis. We cloned the corresponding chromatin accessibility peak, named −2.1prox1a_3, which contained an additional 219 base pairs (bp) compared with −2.1prox1a. −2.1prox1a_3 was able to induce expression in the lymphatic valve and residual expression in the rest of the facial lymphatic vessels (Fig. S3C,D). This broader expression in the facial LECs is probably due to additional putative TF binding sites present in the sequence which regulate zebrafish lymphangiogenesis, such as Mafb (Arnold et al., 2022; Koltowska et al., 2015b). The +15.2prox1a enhancer does not appear to be accessible in either LECs or BECs at 4 dpf, which could be due to the dynamic activity of this element.
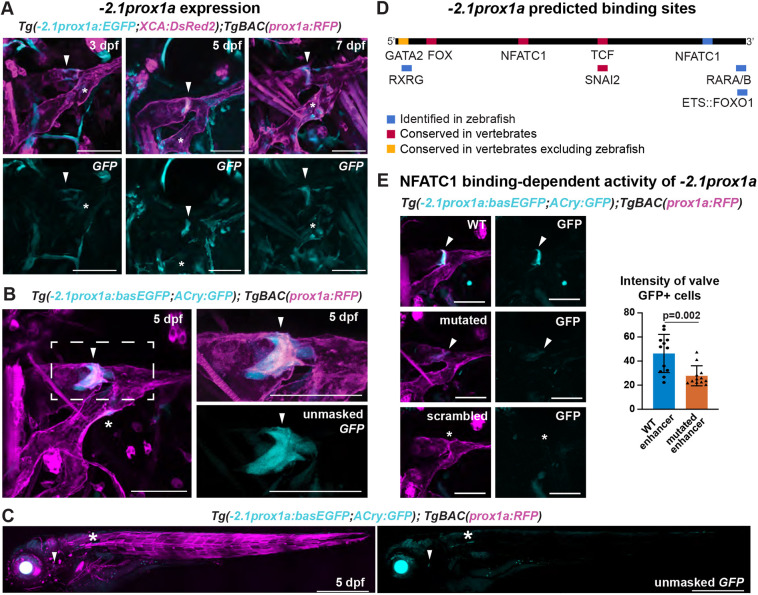
Fig. 3.
snATAC-seq identifies four lymphatic prox1a enhancers. (A) Chromatin state surrounding the prox1a locus in lymphatic endothelial cells (LECs), venous endothelial cells (VECs) and arterial endothelial cells (AECs) at 4 dpf, showing the region between 32,815,787−32,975,788 base pairs of chromosome (chr) 17. Orange, tested enhancers; purple, identified accessible chromatin sequences in LECs. (B) Confocal projections of the facial and trunk lymphatics labelled with Tg(−87prox1a:EGFP; XCA:DsRed2)uom122 (cyan) and Tg(prox1a:RFP)nim5 (magenta) at 5 dpf. Arrowheads show expression in the face and trunk lymphatics. (C) Confocal projections of the facial and trunk lymphatics labelled with Tg(−71prox1a:EGFP; XCA:DsRed2)uom121 (cyan) and Tg(prox1a:RFP)nim5 (magenta) at 5 dpf. Arrowheads show expression in the face and trunk lymphatics. Asterisks show expression in PCV. (D) Confocal projections of the facial and trunk lymphatics labelled with Tg(−14prox1a:EGFP; XCA:DsRed2)uom120 (cyan) and Tg(prox1a:RFP)nim5 (magenta) at 5 dpf. Arrowheads show expression in the facial lymphatics. (E) Predicted endothelial TF binding sites in −87prox1a. (P<1e-04), −71prox1a (P<1e-04) and −14prox1a (P<1e-04). Blue, binding sites identified in zebrafish; red, conserved binding sites within Actinopterygii. Scale bars: 50 μm.
We further investigated whether the enhancers identified by snATAC-seq present sequence conservation within Actinopterygii. Microsynteny was tested in nine ray-finned fish species (Fig. S4A) confirming that the loss of rps6kc1 upstream of prox1a we observed in zebrafish is a Otocephala-specific rearrangement and is not present in other Actinopterygii (Fig. S1A,C). The sequence conservation analysis revealed that −71prox1a is conserved across Actinopterygii, and so is −14prox1a although with a weaker signal. In contrast, −87prox1a could not be identified with sequence conservation in any of the considered acanthopterygian species (Oreochromis niloticus, Acanthochromis polycanthus, Takifugu rubipes and Amphiprion percula) (Fig. S4B). However, the presence of this enhancer in Lepisosteus oculatus suggests it was present in the ancestor of Actinopterygii, and the sequence has subsequently diverged or was lost in the acanthopterygian lineage. The reduced level of conservation of −87prox1a and −14prox1a also explains why only −71prox1a is marked as conserved in the UCSC Genome Browser Multiz Alignment (Fig. S3E). As expected, both −2.1prox1a and +15.2prox1a are conserved across Actinopterygii (Fig. S4B).
The −87prox1a element, positioned 87 kbp upstream of the TSS and marked as a primed enhancer at 48 hpf (Fig. S1C), drove reporter expression in the facial and trunk lymphatics at 5 dpf (Fig. 3B; Fig. S3F), as well as in the skin and a cell population in the trunk (Fig. S3F). The enhancer contains predicted TF binding sites for vascular regulators such as GATA2, FOX, ETS, NR2F2, TAL1::TCF3 (Cha et al., 2016; Frye et al., 2018; Kazenwadel et al., 2015; Lin et al., 2010; Nicenboim et al., 2015; Niimi et al., 2017; Pham et al., 2007; Scallan et al., 2021; Tang et al., 2006; De Val et al., 2008; Yoshimatsu et al., 2011), as well as actinopterygian-conserved binding sites for RARA (Bowles et al., 2014; Marino et al., 2011) (Fig. 3E; Fig. S4C; Tables S4,S5).
The −71prox1a element is situated 71 kbp upstream of the prox1a TSS and drove reporter expression in the trunk and the facial lymphatics at 5 dpf (Fig. 3C), as well as the PCV, skin and gallbladder (Fig. S3G). It contained predicted binding sites for factors involved in vascular development such as MAFB and ETS factors (Arnold et al., 2022; Koltowska et al., 2015b; Pham et al., 2007; De Val et al., 2008; Yoshimatsu et al., 2011) (Fig. 3E; Fig. S3C; Tables S4,S5). The enhancer also presents a binding site for RARA (Bowles et al., 2014; Marino et al., 2011), FOX (Niimi et al., 2017), SNAI2 (Hultgren et al., 2020) and JUN, which is part of the MAPK activation cascade downstream of VEGF signalling (reviewed by Guo et al., 2020) (Fig. 3E; Fig. S4D; Tables S4,S5).
Marked as a primed enhancer at 48 hpf, −14prox1a is situated 14 kbp upstream of prox1a (Fig. S1C). Within lymphatics, the enhancer activity is restricted to the face (Fig. 3D), but is also expressed in the skin (Fig. S3H). It contains predicted binding sites for vascular relevant factors, such as ETS factors (Pham et al., 2007; De Val et al., 2008; Yoshimatsu et al., 2011), FOX TF (Niimi et al., 2017), TCFs (Cha et al., 2016; Nicenboim et al., 2015) and RARG (Bowles et al., 2014; Marino et al., 2011) (Fig. 3E; Fig. S4E; Tables S4,S5).
In conclusion, we identified three additional lymphatic prox1a enhancers by means of chromatin state. Given that these elements show reduced or absent sequence conservation outside of Actinopterygii, this suggests evolutionary divergence in gene regulation – either through lineage-specific modification in prox1a regulation or through a change at the level of DNA sequence while retaining function.
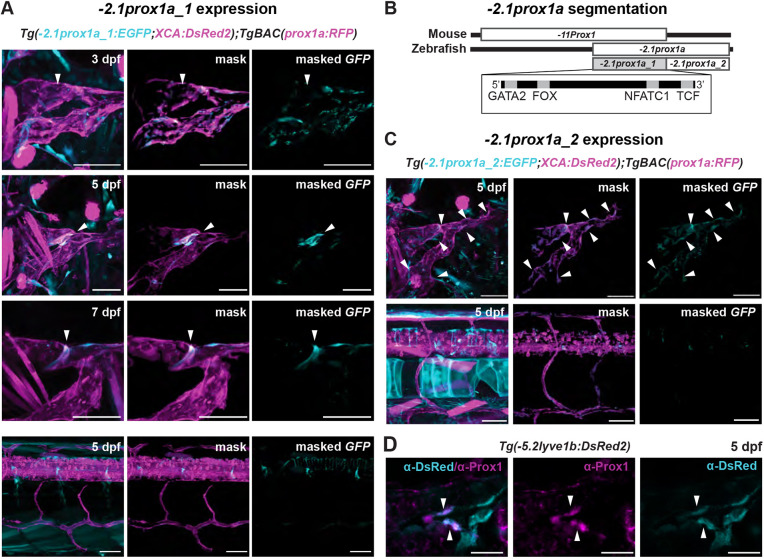
−2.1prox1a_1 drives expression in the lymphatic valve
The −2.1prox1a element partially overlaps with that of a previously described −11Prox1 murine lymphatic valve enhancer (Kazenwadel et al., 2023) in its 5′ portion (Fig. 4B). We hypothesised that this region might be sufficient to induce the valve-specific expression. The 200 bp fragment, called −2.1prox1a_1, was cloned in the ZED vector and tested in vivo. Expression driven by −2.1prox1a_1 appeared at 3 dpf at the future valve position (Fig. 4A; Fig. S5A,B), which mirrors the observations for the full enhancer. The enhancer-driven valve expression was maintained at 5 dpf and 7 dpf (Fig. 4A), aligning with the endogenous Prox1 protein distribution at 5 dpf, which is especially concentrated in the leaflets of the developing valves (Fig. 4D). Similar to −2.1prox1a, −2.1prox1a_1 remained inactive in the trunk lymphatics (Fig. 4A) and all venous tissues (Fig. 4A; Fig. S5D). Furthermore, we explored the potential role for the 3′ portion of −2.1prox1a, here called −2.1prox1a_2, as a complementary contributor to −2.1prox1a_1 activity. Upon in vivo testing, this element showed subtle GFP expression in the facial, but not in the trunk, lymphatics (Fig. 4C; Fig. S5C), and no venous expression (Fig. S5D). This suggests that this segment of −2.1prox1a also plays a role in regulating prox1a expression.
Fig. 4.
−2.1prox1a_1 is the core element driving valve expression. (A) Confocal projections of the facial lymphatics labelled with Tg(−2.1prox1a_1:EGFP; XCA:DsRed2)uu5kk (cyan) and Tg(prox1a:RFP)nim5 (magenta) at 3, 5 and 7 dpf, and lack of expression in the trunk lymphatics (bottom panel) at 5 dpf. Arrowheads show expression in the developing lymphatic valve. (B) Schematic of the −2.1prox1a zebrafish and the −11Prox1 mouse enhancer. The identified sequence overlap between the two enhancers is referred to as −2.1prox1a_1 and the zebrafish unique enhancer part is referred to as −2.1prox1a_2. The TF binding site location in −2.1prox1a_1 is illustrated in the box. (C) Confocal projections of the facial and trunk lymphatics labelled with Tg(−2.1prox1a_2:EGFP;XCA:DsRed2)uu6kk (cyan) and Tg(prox1a:RFP)nim5 (magenta) at 5 dpf. Arrowheads show expression in the facial lymphatics. (D) Confocal projections of the immunostaining against Prox1 (magenta) and Tg(−5.2lyve1b:DsRed2)nz101 (cyan). Prox1 expression is detected in the valve leaflets at 5 dpf (arrowheads). Scale bars: 50 μm (A,C); 20 μm (D).
In summary, −2.1prox1a_1 serves as the core enhancer element of −2.1prox1a, sufficient to drive expression in the developing valve from the early stages to later developmental phases.
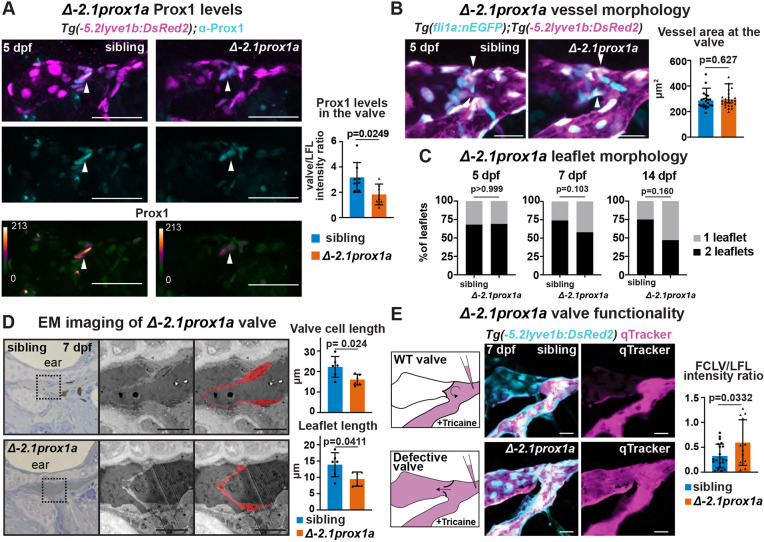
−2.1prox1a_1 is necessary for correct valve morphology and functionality
To test the necessity of these enhancers for the development of lymphatic vasculature, we concentrated on the core valve element −2.1prox1a_1 and used CRISPR/Cas9 to generate the en−2.1prox1auu12kk mutant line, hereafter referred to as Δ−2.1prox1a, carrying a 102 bp deletion covering the predicted NFATC1 binding site in the −2.1prox1a_1 sequence. Homozygous mutants are viable, fertile and show normal body morphology. We focused on the valve area to characterise the phenotype. Immunostainings revealed reduced Prox1a protein levels within the valve of mutant Δ−2.1prox1a compared with siblings (Fig. 5A), suggesting the deletion of the enhancer negatively impacts the regulation of prox1a. We then investigated gross vessel morphology at the valve position, which showed high resemblance between Δ−2.1prox1a homozygous and sibling embryos (Fig. 5B; Fig. S6A-C). Similarly, the volume of the FCLV remained unaffected in Δ−2.1prox1a embryos (Fig. S6D). Using the average phenotype approach (Arnold et al., 2022) at 5 and 7 dpf, we further investigated the vessel morphology. No discernible differences were observed (Fig. S6E), further confirming that the facial lymphatic vessels form accurately in the mutants. Conversely, an analysis of gross valve morphology at 7 and 14 dpf revealed a noticeable trend of leaflet alteration in the mutants compared with the sibling (Fig. 5C). As 7 dpf is the stage in which the valve leaflets are completely formed (Shin et al., 2019), we further investigated the fine morphology of the valve using transmission electron microscopy (TEM). Imaging of three embryos per genotype revealed altered valve morphology in 7 dpf mutant embryos, with a reduction in length of both the cells composing the leaflet and the leaflets themselves (Fig. 5D). Roundness of the valve cell nuclei was unaffected (Fig. S6F).
Fig. 5.
−2.1prox1a is necessary for correct valve development and function. (A) Top and middle: confocal projections of immunostaining against Prox1 (cyan) and Tg(−5.2lyve1b:DsRed2)nz101 (magenta) in sibling and Δ−2.1prox1a mutant embryos at 5 dpf. Bottom: heatmap visualisation of Prox1 protein levels in sibling and Δ−2.1prox1a mutant embryos. Left: quantification of Prox1 protein levels in the valves of 5 dpf embryos. Mean±s.d. Sibling (n=8) versus mutants (n=7). P=0.0249 (unpaired two-tailed Student's t-test). Three technical replicates, biological replicates correspond to the number of data points per condition. (B) Right: confocal projections of Tg(fli1:nEGFP)y7 (cyan) and Tg(−5.2lyve1b:DsRed2)nz101 (magenta) in sibling and Δ−2.1prox1a mutant embryos at 5 dpf. Left: quantification of vessel section at the valve (arrowheads) in Δ−2.1prox1a embryos at 5 dpf. Mean±s.d. Sibling (n=22) versus mutants (n=24). Not significant (ns) (P=0.627; two-tailed Mann–Whitney test). Three technical replicates, biological replicates correspond to the number of data points per condition. (C) Quantification of leaflet morphology in the valves of 5, 7 and 14 dpf sibling and Δ−2.1prox1a embryos. Mean±s.d. 5 dpf siblings (n=19) versus Δ−2.1prox1a (n=26), ns (P>0.999; Fisher's test). 7 dpf siblings (n=27) versus Δ−2.1prox1a (n=29), ns (P=0.103; Fisher's test). 14 dpf siblings (n=12) versus Δ−2.1prox1a (n=21), ns (P=0.160; Fisher's test). Three technical replicates, biological replicates correspond to the number of data points per condition. (D) Left: brightfield and TEM imaging of sibling (n=3) and Δ−2.1prox1a (n=3) valves at 7 dpf. The leaflets in the TEM images are highlighted in red. Right: quantification of cell length: mean±s.d. Siblings (n=6) versus Δ−2.1prox1a (n=6). P=0.024 (unpaired two-tailed Student's t-test). Quantification of leaflet length: mean±s.d. Siblings (n=6) versus Δ−2.1prox1a (n=6). P=0.0411 (unpaired two-tailed Student's t-test). Two technical replicates, biological replicates correspond to the number of data points per condition. (E) Left: schematic and confocal projections of Tg(−5.2lyve1b:DsRed2)nz101 (cyan) and Qtracker (magenta) in sibling and Δ−2.1prox1a mutant embryos at 7 dpf, visualising flow through the vessel. Right: quantification of Qtracker leakage through the valve in sibling and Δ−2.1prox1a embryos at 7 dpf. Mean±s.d. Siblings (n=21) versus mutants (n=12). P=0.0332 (unpaired two-tailed Student's t-test). Two technical replicates, biological replicates correspond to the number of data points per condition. Scale bars: 50 μm (A); 20 μm (B,E); 10 μm (D).
A correctly formed valve should be closed under tricaine anaesthesia and therefore in an anaesthetised WT embryo the Qtracker injected in the lateral facial lymphatic (LFL) should not leak into the FCLV. However, when the valve is not correctly formed, and cannot close properly, the Qtracker can leak into the FCLV (Fig. 5E) (Shin et al., 2019). To assess whether the altered morphology could affect valve function, we performed Qtracker injections in the LFL in anaesthetised 7 dpf larvae and quantified leakage through the valve. Our findings demonstrated increased leakage in Δ−2.1prox1a mutants compared with siblings, confirming a functional impairment of the lymphatic valve in the absence of the enhancer (Fig. 5E; Fig. S6G). Collectively, these data show the necessity for −2.1prox1a_1 for correct valve formation and proper expression of prox1a, highlighting the functional importance of prox1a enhancers for precise lymphatic development.
DISCUSSION
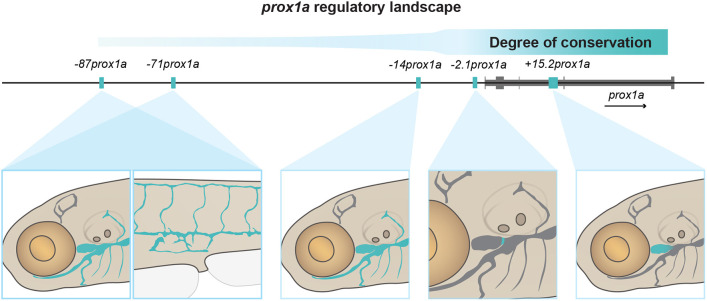
PROX1 is a key factor for LEC development (Koltowska et al., 2015a; Wigle and Oliver, 1999). Despite this, only one cis-regulatory element of Prox1 has been identified so far (Kazenwadel et al., 2023). Here, we used a mixed approach, taking advantage of both evolutionary conservation and tissue-specific chromatin accessibility, to identify prox1a enhancers active in the lymphatic endothelium of zebrafish. Unexpectedly, we could identify five separate enhancers driving reporter expression in the lymphatic vasculature (Fig. 6). Two of the enhancers, −2.1prox1a and +15.2prox1a, show significantly enriched expression in the valve and the FCLV, respectively. These anatomically and functionally distinct subsets of the lymphatics are also regulated by different gene networks than the remaining lymphatics (Hußmann et al., 2023; Meng et al., 2023; Shin et al., 2019). Conversely, the area of activity of the three remaining enhancers is wider and overlaps to a larger extent. Specifically, both −87prox1a and −71prox1a drive expression in trunk and facial lymphatics. Enhancers with high redundancy in their expression patterns are defined as shadow enhancers (Hobert, 2010; Hong et al., 2008). Shadow enhancers are a common feature among developmental genes (Cannavò et al., 2016; Kvon et al., 2021). They are thought to serve as a mechanism aimed at ensuring robustness during the developmental process (Antosova et al., 2016; Kvon et al., 2021; Osterwalder et al., 2018). Various types of interactions – additive, superadditive, subadditive and repressive – among these enhancers have been shown to effectively fine-tune the regulation of target genes into specific patterns (Bothma et al., 2015; El-Sherif and Levine, 2016; Kvon et al., 2021; Lam et al., 2015). Moreover, studies indicate that shadow enhancers can form transcriptional hubs, where multiple enhancers concurrently interact with the gene promoter at the same time (Kvon et al., 2021). Given this intricate interplay, the presence of two enhancers, −87prox1a and −71prox1a, with extensively overlapping expression suggests a potential role for shadow enhancers in prox1a regulation. The complex regulatory logic at the prox1a locus highlights the necessity for tight regulation of prox1a activity to ensure correct lymphatic development.
Fig. 6.
Schematic representation of the lymphatic prox1a enhancers identified in this study. The more distal enhancers, not conserved in mammals, drive expression in wide lymphatic domains. The proximal enhancers, conserved in mammals, show instead restricted activity in specific subsets of the lymphatic vasculature.
Our conservation analysis revealed a high density of CNEs surrounding the prox1a locus. As CNEs can be considered putative enhancers, this suggests that cis-regulation plays an important role in controlling prox1a expression in more than just LECs. In fact, among ten tested CNEs, only two functional lymphatic enhancers were identified: −2.1prox1a and +15.2prox1a. In contrast, the three sequences identified by chromatin state, −87prox1a, −71prox1a and −14prox1a, presented varying conservation levels within Actinopterygii. Although distal enhancers have a tendency to be less conserved, the presence of other vertebrate-conserved CNEs spanning the 150 kbp upstream region of prox1a suggests that no major rearrangement has contributed to the loss of these enhancers in tetrapods. In addition, we observed conservation of microsynteny surrounding prox1a in tetrapods and actinopterygians, with the exception of an Otocephala-specific divergence. Such conservation in loci disposition also speaks against major genomic rearrangements in the region. Interestingly, the most conserved enhancers at the sequence level also have the most spatially restricted expression (the lymphatic valve and the FCLV), whereas the less conserved enhancer sequences with tetrapods display broader and overlapping expression patterns. Despite the obvious morphological differences, the molecular programme underlying LEC development in mammals and zebrafish presents strong similarities, such as the role of growth factor signalling and TFs such as PROX1, NFATC1 or GATA2 (Kazenwadel et al., 2015; Koltowska et al., 2015a; Norrmén et al., 2009; Secker and Harvey, 2021; Shin et al., 2019; Wigle and Oliver, 1999). Therefore, it is worth noticing that the majority of the binding sites for these conserved lymphatic regulators are found in the sequence-conserved enhancers. This suggests potential divergence in the molecular code upstream of −87prox1a, −71prox1a and −14prox1a between Actinopterygii and Tetrapoda, possibly involving different TFs. However, it is important to note that evolutionary conservation of functional enhancers does not necessarily require high levels of conservation at a sequence level, as long as key transcription factor motifs endure (Wong et al., 2020). As the nature of the enhancers can be complex, adopting complementary methods for their identification is crucial for a comprehensive understanding of a gene regulatory landscape.
As we just discussed, sequence conservation can serve as an indicator of enhancer presence; however, it does not guarantee complete functional conservation. In the case of −11Prox1/−2.1prox1a, the element drives expression in the lymphatic valve in both mouse and zebrafish. Both in mouse and zebrafish, Prox1/prox1a is a key player in lymphatic valve formation acting in concert with Foxc1, Nfatc1 and Gata2 (Bazigou et al., 2009; Norrmén et al., 2009; Sabine et al., 2012; Shin et al., 2019) and activating key downstream targets for valve development such as integrin alpha 9 (Itga9; Bazigou et al., 2009; Shin et al., 2019) and connexin 37 (Gja4; Sabine et al., 2012). The conserved cluster of binding sites for Foxc, Nfatc1 and Gata2 in the −11Prox1/−2.1prox1a enhancer strongly suggests this enhancer is part of the network. Despite this, the mice and zebrafish phenotypes for −11Prox1/−2.1prox1a mutants are profoundly different. The −2.1prox1a mutants are viable and fertile, and only show developmental defects in the lymphatic valve regarding the morphology of the leaflets. As mentioned above, key factors in the regulation of the leaflet extracellular matrix such as itga9 (Bazigou et al., 2009) have been suggested to be regulated under the Prox1/Foxc/Nfatc1/Gata2 network in zebrafish lymphatic valve formation (Shin et al., 2019), which could explain the observed phenotype. Conversely, mouse −11Prox1 mutants have more severe lymphatic defects and die perinatally (Kazenwadel et al., 2023). The deletion of −11Prox1 is fully phenocopied by the deletion of the GATA2 binding site, which is also responsible for the transition of LECs to HECs in −11Prox1 mutant mice (Kazenwadel et al., 2023). However, the putative GATA2 binding site is not conserved in zebrafish, which could again explain the different severity of the phenotype. The absence of this binding site is also the reason the hematopoietic phenotypes were not further explored in this study. The observed differences between −11Prox1 and −2.1prox1a suggest functional divergence despite their common evolutionary origin, and highlights that functional conservation cannot be solely deduced from enhancer sequence data.
In conclusion, this study has demonstrated that prox1a in the lymphatic vasculature is regulated by a diverse group of distinct enhancers. We uncovered that the distal enhancers drive the expression in large regions of the lymphatic endothelium, whereas the proximal and sequence-conserved enhancers channel the expression to the functionally distinct sub-compartments of the developing lymphatic vascular network. This work represents a first step towards a full characterisation of the cis-regulatory landscape of prox1a and the understanding of the complex mechanisms regulating its lymphatic expression.
MATERIALS AND METHODS
Zebrafish
Zebrafish (Danio rerio) work was carried out with ethical approval from the Swedish Board of Agriculture (5.8.18-10590/2018 and 5.8.18-06282/2023) and in alignment with guidelines set by animal ethics committees at The University of Melbourne, ‘Zebrafish Breeding and Husbandry Ethics (Ethics ID 22235)’. The fish were maintained at the Genome Engineering Zebrafish National Facility (SciLifeLab, Uppsala, Sweden), the CIV (Centre for In Vivo, Uppsala University, Sweden) and the Danio rerio University of Melbourne facility (DrUM, Melbourne, Australia). Adults and embryos were housed according to standard procedures (Aleström et al., 2020). The previously published lines used in this study were Tg(−5.2lyve1b:DsRed2)nz101 (Okuda et al., 2012), TgBAC(prox1a:KalTA4-4xUAS-ADV. E1b:TagRFP)nim5, referred to as Tg(prox1a:RFP)nim5 in this study, (Dunworth et al., 2014; van Impel et al., 2014), Tg(kdr-l:ras-cherry)s916 (Hogan et al., 2009), Tg(fli1a:nEGFP)y7 (Lawson and Weinstein, 2002) and Tg(−2.1prox1a:EGFP;XCA:DsRed2)uu3kk (Kazenwadel et al., 2023).
The Tg(−2.1prox1a_1:EGFP;XCA:DsRed2)uu5kk, Tg(−2.1prox1a_2:EGFP; XCA:DsRed2)uu6kk, Tg(+15.2prox1a:EGFP;XCA:DsRed2)uu7kk, Tg(−2.1prox1a_3:EGFP; XCA:DsRed2)uom119, Tg(−71prox1a:EGFP; XCA:DsRed2)uom121, Tg(−87prox1a:EGFP; XCA:DsRed2)uom122, Tg(−14prox1a:EGFP; XCA:DsRed2)uom120, Tg(−2.1prox1a:basEGFP;ACry:GFP)uu10kk, en.−2.1prox1auu12kk and Tg(gata-i4:GFP)uu11kk, Tg(+15.2prox1a:basEGFP;ACry:GFP)uu13kk, Tg(−87prox1a:basEGFP;ACry:GFP)uu14kk, Tg(−71prox1a:basEGFP;ACry:GFP)uu15kk, Tg(−14prox1a:basEGFP;ACry:GFP)uu16kk lines were generated for this study.
In-silico predictions
To identify sequence-conserved enhancers, the DNA regions between the two loci neighbouring Prox1, prox1a or prox3 were downloaded from ENSEMBL, together with annotations. For prox3, homologs were identified using BLAST of zebrafish prox3 and confirmed by local synteny. The sequences were oriented in the direction of the transcription. The species, assemblies and regions used are listed in Table S1. CNEs were identified using mVISTA non-coding DNA conservation analysis (Dubchak et al., 2000; Frazer et al., 2004; Mayor et al., 2000). The alignment was performed using the LAGAN program (Brudno et al., 2003).
To determine the zebrafish-specific binding sites in the identified prox1a enhancers, we conducted a motif discovery analysis of zebrafish enhancer sequences using FIMO in the MEME Suite (Grant et al., 2011). Our motif prediction was constrained to a search for motifs of size 7, with a P-value cut-off of 1e-04. A comprehensive list of predicted motifs is shown in Table S6.
To identify evolutionarily conserved motifs and binding sites within the specified enhancers, we retrieved the identified conservation peaks and subjected them to analysis using the MEME Suite (Bailey and Elkan, 1994) and TOMTOM (Gupta et al., 2007). A list of the conserved motifs identified can be found in Table S4. To validate the predicted conserved binding sites, we compared the results of this analysis with the outcomes of the FIMO motif discovery analysis, using a P-value cut-off of 1e-02. The decision to use a higher P-value stems from the conservation of these sequences and aims to capture an informative overview of the potential binding sites. Binding sites for known endothelial factors that appear in both analyses have been reported, and a list can be found in Table S5.
snATAC-seq data processing and analysis
For snATAC-seq data analysis, we included only the 4 dpf WT cells from the publicly available dataset (Grimm et al., 2023). The analysis was performed and data were processed as previously described (Grimm et al., 2023). Briefly, the LEC cluster was defined by combined high accessibility at the prox1a, cdh6 and lyve1b loci. The venous endothelial cells cluster by combined high accessibility at the cdh5, kdrl and stab2 or lyve1b loci, and arterial endothelial cells by combined high accessibility at the cdh5, kdrl, flt1 and dll4 loci but low accessibility at lybe1b. Mural lymphatic endothelial cells (MuLECs) were identified based on the high accessibility at osr2 in addition to the standard LEC loci but low accessibility at cdh6, and the endocardium by the accessibility at the cdh5 and hand2 loci.
All GO analyses were performed using Panther.db (Thomas et al., 2006) (Biological Process Complete). The complete list of the predicted GO terms is shown in Table S7.
Cloning and transgenesis
The conserved elements of interest (Tables S2 and S3) were cloned into the ZED vector as previously described (Bessa et al., 2009) (Addgene plasmids 218205, 218206, 218207 and 218208). The Tg(−2.1prox1a:basEGFP;ACry:GFP)uu10kk line (Addgene plasmid 218209) was generated by cloning the −2.1prox1a sequence in a p5E-MCS vector (Quillien et al., 2017) (Addgene vector 26029) using In-Fusion cloning (Takara Bio, primers are listed in Table S3) and then inserted into a pDestTol2ACryGFP backbone (Berger and Currie, 2013) (Addgene vector 64022), with a pENTRbasEGFP (Villefranc et al., 2007) (Addgene plasmid 22453) and a p3E-polyA (Tol2kit v1.2 #302) vector using the Gateway cloning method (Invitrogen). ATAC-identified enhancers were inserted into the ZED vector by In-Fusion cloning (#638910, In-Fusion HD Cloning Plus Kits, Takara Bio) using BspEI and BmgBI cutting sites for linearisation. Primers used are listed in Table S3. Vectors used to generate the Tg(+15.2prox1a:basEGFP;ACry:GFP)uu13kk (Addgene plasmid 218210), Tg(−87prox1a:basEGFP;ACry:GFP)uu14kk (Addgene plasmid 218213), Tg(−71prox1a:basEGFP;ACry:GFP)uu15kk (Addgene plasmid 218212), Tg(−14prox1a:basEGFP;ACry:GFP)uu16kk (Addgene plasmid 218211) stable lines, as well as the transient −2.1prox1a:basEGFP;ACry:GFP with WT, NFATC1-mutated (Addgene plasmid 218215) and scrambled-sequence (Addgene plasmid 218216) transient transgenic embryos (Fig. 2E) were ordered from GenScript using the same backbone used for −2.1prox1a:basEGFP;ACry:GFP (Addgene plasmid 218214). To generate transgenic lines, 1 μl of construct at 20 ng/μl and tol2 transposase mRNA at 100 ng/μl were injected into the one-cell stage WT zebrafish embryos. F0 embryos were screened for reporter expression and F1 embryos were screened using confocal microscopy for GFP expression in the lymphatic structures. The numbers of F0 founders screened for each tested CRE are listed in Table S2. Bleed-through in the GFP channel was excluded by imaging the embryos that were negative for TgBAC(prox1a: RFP)nim5 (Fig. S5E). F1 fish with lymphatic GFP expression were used to establish the stable lines. The Tg(gata2a-i4-1.1kb:GFP)uu11kk was created by injecting the construct (Dobrzycki et al, 2020) as previously described. For embryos imaged at F0, injections were performed as described for stable transgenic lines. The plasmids produced in this study have been deposited in Addgene.
Imaging and image processing
For the conserved enhancer reporter and mutant lines, transgenic embryos were anaesthetised with tricaine, mounted in 1% low-melting agarose, and face or trunk was imaged using a Leica TCS SP8 DLS microscope with a Fluotar VISR 25× water objective (objective number: 11506375). For enhancers identified by snATAC-seq, transgenic embryos were anaesthetised with tricaine, mounted in 0.5% low-melting agarose and imaging was conducted at the Centre for Advanced Histology and Microscopy (Peter MacCallum Cancer Centre, Melbourne, Australia). Live samples were imaged using a Zeiss LSM 780 FCS confocal microscope. Images were processed using ImageJ software (v. 2.9.0). Immunostained embryos were mounted in clearing solution Omnipaque (350 mg/l concentration per 1 ml iohexol, GE Healthcare) and imaged using the Leica TCS SP8 DLS microscope as described above. All representative images are maximum intensity projections of the z-stack generated using ImageJ (v. 2.9.0). The skin signal in the GFP channel was manually removed to allow the visualisation of the vascular tissues underneath.
Mutant line generation
Mutants were generated using CRISPR/Cas9 as described previously (Carrington et al., 2015; Varshney et al., 2016). The guides were designed to flank the target enhancer sequences. Zebrafish embryos were injected at the one-cell stage with 70-140 ng/μl of each gRNA and 200 ng/μl Cas9 mRNA. These guides are listed in Table S8. Fish were genotyped by PCR, followed by gel electrophoresis, and the deletion was confirmed by Sanger sequencing (Eurofins). F1 embryos with identical mutations were used to establish the stable mutant line.
Genotyping
The Δ−2.1prox1a embryos were genotyped by PCR, followed by gel electrophoresis. PCR was performed as previously described for FLA genotyping (Carrington et al., 2015) using the primers listed in Table S3. Fluorescent primers were omitted from the reaction. mafba and mafbb mutant embryos were genotyped as previously described (Arnold et al., 2022).
Immunostaining
Immunostaining was performed as previously described (Le Guen et al., 2014; Shin et al., 2016), with the addition of a 45 min at room temperature digestion step in proteinase K (PK) as described previously (Koltowska et al., 2015b). The primary antibodies used were anti-Prox1 rabbit (AngioBio, #11-002P, 1:100, Lot: GR3247830-10) (Koltowska et al., 2015a) and anti-mCherry chicken (AvesLabs, #MCHERRY-0020, 1:100, Lot: MC87977980). For the anti-mCherry primary antibody, we verified that the observed signal recapitulated the endogenous transgenic line expression. The secondary antibodies used were anti-rabbit IgG HRP (Cell Signaling Technology, #7076, 1:1000, Lot: 28) and anti-chicken 488 (Jackson ImmunoResearch, #703-545-155, 1:200, Lot: 158347). Signal amplification was performed using the TSA™ Plus Cyanine 3 System (Perkin Elmer, #NEL744001KT), with a development time of 3 h. Imaging was performed as described above.
Electron microscopy imaging
For TEM, Δ−2.1prox1a embryos were fixed in 2.5% Glutaraldehyde (Ted Pella)+1% Paraformaldehyde (Merck) in 0.1 M Phosphate buffer (PB; pH 7.4), then embedded in 8% agar and a 300 μm sagittal section was cut on a Microm M 650V vibratome (Thermo Fisher Scientific) and stored at 4°C until further processing. The tails were then used for genotyping. Samples were rinsed with 0.1 M PB for 10 min before 1 h incubation in 1% osmium tetroxide (TAAB) in 0.1 M PB. After rinsing in 0.1 M PB, samples were dehydrated using increasing concentrations of ethanol (50%, 70%, 95% and 99.9%) for 10 min each step, followed by 5 min incubation in propylene oxide (TAAB). The samples were then placed in a mixture of Epon resin (Ted Pella) and propylene oxide (1:1) for 1 h, followed by 100% resin and left overnight. Subsequently, samples were embedded in capsules in newly prepared Epon resin and left for 1-2 h, and then polymerized at 60°C for 48 h.
Semi-thin sections were cut, stained with Toluidine Blue and examined using light-microscopy to identify the area of interest. Ultrathin sections (60-70 nm) were cut using an EM UC7 Ultramicrotome (Leica) and placed on a grid. The sections were subsequently contrasted with 5% uranyl acetate and Reynold's lead citrate and visualised with Tecnai™ G2 Spirit BioTwin transmission electron microscope (Thermo Fisher Scientific/FEI) at 80 kV with an ORIUS SC200 CCD camera and Gatan Digital Micrograph software (both from Gatan).
Qtracker injections
Microangiography was performed following a previously published protocol (Arnold et al., 2022; Shin et al., 2019). Embryos were anaesthetised with Tricane and injected with 1 nl of Qtracker™ 655 Vascular labels (Thermo Fisher Scientific) at 7 dpf in the LFL for valve leakage experiments and imaged on a Leica TCS SP8 DLS microscope at ∼5 min post-injection.
Image quantification
Intensity of +15.2prox1 activity in WT and mafba/b mutant embryos was calculated in Imaris v9.3.0. Intensity of signal in the FCLV was calculated by manually segmenting the FCLV and extracting the intensity measurement. Intensity was normalised using the intensity signal in the prox1a-postive ganglia over the ear.
The mean intensity of GFP signal in the valve of embryos injected with WT and binding site-mutated −2.1prox1a embryos was calculated in Imaris v9.3.0. The GFP+ valve cells were isolated using a surface detail of 0.454 and a fixed thresholding value of 14. As no GFP+ cells were observed in the scrambled controls, the intensity could not be calculated in these embryos.
The relative levels of Prox1 protein in the valve were calculated as the ratio between the representative valve and LFL nucleus. Briefly, the chosen nuclei were masked in Imaris v9.3.0 as described above. The average intensity was calculated and used to compile the ratio. Nuclei with a maximum intensity of 255 were excluded from the calculations.
Vessel sections were calculated in ImageJ (v. 2.9.0) using a custom-made script, available at https://github.com/virpa81/Vessel-section-calculation. The z-stack was rotated along its main axis to ensure that the vessel section was perpendicular to the cutting planes. The stack was then re-sliced along the yz-plane, and the valve position was selected. Thresholding was used to mask off the vessel area, which was subsequently measured.
The FCLV volume was calculated in Imaris v9.3.0, by manually creating a surface spanning the 400 pixels of FCLV past the valve position and calculating its volume. To generate average phenotypes, images were acquired and processed as previously described (Arnold et al., 2022).
Valve leaflet morphology was scored in Imaris v9.3.0, looking at the projections of the stack in x, y and z, and scoring whether one or two leaflets were visible in any of them. To calculate the valve cell length in the TEM images, the cells composing the leaflets were traced manually and used to create a mask in ImageJ. The mask was then skeletonised and secondary branches were removed to obtain the length of the cell. Nuclei roundness was calculated in ImageJ using the ‘Round’ image descriptor. Leaflet length was calculated by tracing the internal edge of the leaflet in ImageJ and measuring.
Valve leakage in the injected embryos was quantified in ImageJ (v2.9.0). Intensity of the Qtracker signal was calculated in a representative slice by tracing a region of interest encompassing the FCLV or the LFL. Leakage was quantified as the ratio between these two measures.
Statistical analysis
For all analysis, data were collected across at least three technical replicates, except for the TEM imaged embryos, the 3 dpf Δ−2.1prox1a embryos and the Qtracker injections, which were collected from two technical replicates. The number of biological replicates for each experiment is indicated in the figure legends.
Embryos presenting clear sign of altered general development were excluded from the analysis. n size was set as 10 per group, unless technical limitations prevented it. For injections of WT, mutated and scrambled vectors in Fig. 2E, injections were randomised by injecting different constructs into the same clutch. Quantifications of mutant embryos were performed before the assignment of the genotype, to ensure that experimenters were unaware of the conditions. The normality of all numerical datasets was tested with a Shapiro-Wilk test. For pair-wise comparison, an unpaired two-tailed Student's t-test was run on normally distributed data, while a Mann–Whitney test was run if normality was not confirmed. For multiple comparison, an Anova or Kruskal–Wallis test was used, depending on normality of the distribution. For frequency data, Fisher's exact test was used.
Supplementary Material
Acknowledgements
We thank the DANIO-CODE consortium (https://danio-code.zfin.org) for providing zebrafish epigenomics data. The DanioReadout, Department of Immunology, Genetics and Pathology, Uppsala University, provided the en-2.1 prox1a_1 mutant line. Electron microscopy was performed at BioVis at Uppsala University. We thank the BioImage Informatics Facility at SciLifeLab for image analysis support. DanioReadout and the BioImage Informatics Facility are supported by SciLifeLab, Sweden. Biovis is supported by the Disciplinary Domain of Medicine and Pharmacy, Uppsala University, Sweden.
Footnotes
Author contributions
Conceptualization: V.P., J.K., N.L.H., T.H., K.K.; Methodology: V.P., D.P., K.S., M.H., B.F.-G., R.S., A.A., T.H.; Formal analysis: V.P., H.Y., D.P., A.A., K.K.; Resources: B.M.H., K.K.; Data curation: V.P., H.Y., K.K.; Writing - original draft: V.P., H.Y., B.M.H., K.K.; Writing - review & editing: V.P., K.K.; Supervision: E.M., B.M.H., K.K.; Project administration: K.K.; Funding acquisition: K.K.
Funding
This work was supported by Knut och Alice Wallenbergs Stiftelse (2017.0144), Ragnar Söderbergs stiftelse (M13/17), Vetenskapsrådet (VR-MH-2016-01437) and Kjell och Märta Beijers Stiftelse. N.L.H. and J.K. were supported by the National Health and Medical Research Council (1146706). Open Access funding provided by the Knut och Alice Wallenbergs Stiftelse. Deposited in PMC for immediate release.
Data availability
Custom script has been deposited in GitHub (https://github.com/virpa81/Vessel-section-calculation).
Peer review history
The peer review history is available online at https://journals.biologists.com/dev/lookup/doi/10.1242/dev.202525.reviewer-comments.pdf
References
- Aday, A. W., Zhu, L. J., Lakshmanan, A., Wang, J. and Lawson, N. D. (2011). Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites. Dev. Biol. 357, 450-462. 10.1016/j.ydbio.2011.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleström, P., D'Angelo, L., Midtlyng, P. J., Schorderet, D. F., Schulte-Merker, S., Sohm, F. and Warner, S. (2020). Zebrafish: housing and husbandry recommendations. Lab. Anim. 54, 213-224. 10.1177/0023677219869037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antosova, B., Smolikova, J., Klimova, L., Lachova, J., Bendova, M., Kozmikova, I., Machon, O. and Kozmik, Z. (2016). The gene regulatory network of lens induction is wired through meis-dependent shadow enhancers of Pax6. PLoS Genet. 12, e1006441. 10.1371/journal.pgen.1006441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, H., Panara, V., Hußmann, M., Filipek-Gorniok, B., Skoczylas, R., Ranefall, P., Gloger, M., Allalou, A., Hogan, B. M., Schulte-Merker, S.et al. (2022). mafba and mafbb differentially regulate lymphatic endothelial cell migration in topographically distinct manners. Cell Rep. 39, 110982. 10.1016/j.celrep.2022.110982 [DOI] [PubMed] [Google Scholar]
- Bailey, T. L. and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc. Second Int. Conf. Intell. Syst. Mol. Biol. 2, 28-36. [PubMed] [Google Scholar]
- Bazigou, E., Xie, S., Chen, C., Weston, A., Miura, N., Sorokin, L., Adams, R., Muro, A. F., Sheppard, D. and Makinen, T. (2009). Integrin-α9 is required for fibronectin matrix assembly during lymphatic valve morphogenesis. Dev. Cell 17, 175-186. 10.1016/j.devcel.2009.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, J. and Currie, P. D. (2013). 503unc, a small and muscle-specific zebrafish promoter. Genesis 51, 443-447. 10.1002/dvg.22385 [DOI] [PubMed] [Google Scholar]
- Bessa, J., Tena, J. J., de la Calle-Mustienes, E., Fernández-Miñán, A., Naranjo, S., Fernández, A., Montoliu, L., Akalin, A., Lenhard, B., Casares, F.et al. (2009). Zebrafish Enhancer Detection (ZED) vector: a new tool to facilitate transgenesis and the functional analysis of cis-regulatory regions in zebrafish. Dev. Dyn. 238, 2409-2417. 10.1002/dvdy.22051 [DOI] [PubMed] [Google Scholar]
- Bogdanović, O., Fernandez-Miñán, A., Tena, J. J., de la Calle-Mustienes, E., Hidalgo, C., van Kruysbergen, I., van Heeringen, S. J., Veenstra, G. J. C. and Gómez-Skarmeta, J. L. (2012). Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 22, 2043-2053. 10.1101/gr.134833.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonn, S., Zinzen, R. P., Girardot, C., Gustafson, E. H., Perez-Gonzalez, A., Delhomme, N., Ghavi-Helm, Y., Wilczyński, B., Riddell, A. and Furlong, E. E. M. (2012). Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat. Genet. 44, 148-156. 10.1038/ng.1064 [DOI] [PubMed] [Google Scholar]
- Bothma, J. P., Garcia, H. G., Ng, S., Perry, M. W., Gregor, T. and Levine, M. (2015). Enhancer additivity and non-additivity are determined by enhancer strength in the Drosophila embryo. eLife 4, e07956. 10.7554/eLife.07956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles, J., Secker, G., Nguyen, C., Kazenwadel, J., Truong, V., Frampton, E., Curtis, C., Skoczylas, R., Davidson, T.-L., Miura, N.et al. (2014). Control of retinoid levels by CYP26B1 is important for lymphatic vascular development in the mouse embryo. Dev. Biol. 386, 25-33. 10.1016/j.ydbio.2013.12.008 [DOI] [PubMed] [Google Scholar]
- Brudno, M., Do, C. B., Cooper, G. M., Kim, M. F., Davydov, E., NISC Comparative Sequencing Program, Green, E. D., Sidow, A. and Batzoglou, S. (2003). LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 13, 721-731. 10.1101/gr.926603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann, J., Bos, F. L., Urasaki, A., Kawakami, K., Duckers, H. J. and Schulte-Merker, S. (2010). Arteries provide essential guidance cues for lymphatic endothelial cells in the zebrafish trunk. Development 137, 2653-2657. 10.1242/dev.048207 [DOI] [PubMed] [Google Scholar]
- Cannavò, E., Khoueiry, P., Garfield, D. A., Geeleher, P., Zichner, T., Gustafson, E. H., Ciglar, L., Korbel, J. O. and Furlong, E. E. M. (2016). Shadow enhancers are pervasive features of developmental regulatory networks. Curr. Biol. 26, 38-51. 10.1016/j.cub.2015.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington, B., Varshney, G. K., Burgess, S. M. and Sood, R. (2015). CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic Acids Res. 43, e157. 10.1093/nar/gkv802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha, B., Geng, X., Mahamud, M. R., Fu, J., Mukherjee, A., Kim, Y., Jho, E.-H., Kim, T. H., Kahn, M. L., Xia, L.et al. (2016). Mechanotransduction activates canonical Wnt/β-catenin signaling to promote lymphatic vascular patterning and the development of lymphatic and lymphovenous valves. Genes Dev. 30, 1454-1469. 10.1101/gad.282400.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang, I. K.-N., Fritzsche, M., Pichol-Thievend, C., Neal, A., Holmes, K., Lagendijk, A., Overman, J., D'Angelo, D., Omini, A., Hermkens, D.et al. (2017). SoxF factors induce Notch1 expression via direct transcriptional regulation during early arterial development. Development 144, 3847-3848. 10.1242/dev.159715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton, M. P., Cheng, A. W., Welstead, G. G., Kooistra, T., Carey, B. W., Steine, E. J., Hanna, J., Lodato, M. A., Frampton, G. M., Sharp, P. A.et al. (2010). Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 107, 21931-21936. 10.1073/pnas.1016071107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val, S., Chi, N. C., Meadows, S. M., Minovitsky, S., Anderson, J. P., Harris, I. S., Ehlers, M. L., Agarwal, P., Visel, A., Xu, S.-M.et al. (2008). Combinatorial regulation of endothelial gene expression by Ets and forkhead transcription factors. Cell 135, 1053-1064. 10.1016/j.cell.2008.10.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich, L. C., Klein, S., Mathelier, A., Sliwa-Primorac, A., Ma, Q., Hong, Y.-K., Shin, J. W., Hamada, M., Lizio, M., Itoh, M.et al. (2015). DeepCAGE transcriptomics reveal an important role of the transcription factor MAFB in the lymphatic endothelium. Cell Rep. 13, 1493-1504. 10.1016/j.celrep.2015.10.002 [DOI] [PubMed] [Google Scholar]
- Dobrzycki, T., Mahony, C. B., Krecsmarik, M., Koyunlar, C., Rispoli, R., Peulen-Zink, J., Gussinklo, K., Fedlaoui, B., de Pater, E., Patient, R. and Monteiro, R. (2020). Deletion of a conserved Gata2 enhancer impairs haemogenic endothelium programming and adult zebrafish haematopoiesis. Commun. Biol. 3, 71. 10.1038/s42003-020-0798-3. PMID: 32054973; PMCID: PMC7018942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubchak, I., Brudno, M., Loots, G. G., Pachter, L., Mayor, C., Rubin, E. M. and Frazer, K. A. (2000). Active conservation of noncoding sequences revealed by three-way species comparisons. Genome Res. 10, 1304-1306. 10.1101/gr.142200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunworth, W. P., Cardona-Costa, J., Bozkulak, E. C., Kim, J.-D., Meadows, S., Fischer, J. C., Wang, Y., Cleaver, O., Qyang, Y., Ober, E. A.et al. (2014). Bone morphogenetic protein 2 signaling negatively modulates lymphatic development in vertebrate embryos. Circ. Res. 114, 56-66. 10.1161/CIRCRESAHA.114.302452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sherif, E. and Levine, M. (2016). Shadow enhancers mediate dynamic shifts of gap gene expression in the Drosophila embryo. Curr. Biol. 26, 1164-1169. 10.1016/j.cub.2016.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng, T. C. Y., Chen, W., Okuda, K. S., Misa, J. P., Padberg, Y., Crosier, K. E., Crosier, P. S., Hall, C. J., Schulte-Merker, S., Hogan, B. M.et al. (2019). Zebrafish facial lymphatics develop through sequential addition of venous and non-venous progenitors. EMBO Rep. 20, e47079. 10.15252/embr.201847079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- François, M., Caprini, A., Hosking, B., Orsenigo, F., Wilhelm, D., Browne, C., Paavonen, K., Karnezis, T., Shayan, R., Downes, M.et al. (2008). Sox18 induces development of the lymphatic vasculature in mice. Nature 456, 643-647. 10.1038/nature07391 [DOI] [PubMed] [Google Scholar]
- Frazer, K. A., Pachter, L., Poliakov, A., Rubin, E. M. and Dubchak, I. (2004). VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 32, W273-W279. 10.1093/nar/gkh458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye, M., Taddei, A., Dierkes, C., Martinez-Corral, I., Fielden, M., Ortsäter, H., Kazenwadel, J., Calado, D. P., Ostergaard, P., Salminen, M.et al. (2018). Matrix stiffness controls lymphatic vessel formation through regulation of a GATA2-dependent transcriptional program. Nat. Commun. 9, 1511. 10.1038/s41467-018-03959-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng, X., Cha, B., Mahamud, M. R., Lim, K.-C., Silasi-Mansat, R., Uddin, M. K. M., Miura, N., Xia, L., Simon, A. M., Engel, J. D.et al. (2016). Multiple mouse models of primary lymphedema exhibit distinct defects in lymphovenous valve development. Dev. Biol. 409, 218-233. 10.1016/j.ydbio.2015.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacco, L. D., Pistocchi, A. and Ghilardi, A. (2010). prox1b activity is essential in zebrafish lymphangiogenesis. PLoS ONE 5, e13170. 10.1371/journal.pone.0013170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow, E. and Tomarev, S. I. (1998). Restricted expression of the homeobox gene prox1 in developing zebrafish. Mech. Dev. 76, 175-178. 10.1016/S0925-4773(98)00121-X [DOI] [PubMed] [Google Scholar]
- Grant, C. E., Bailey, T. L. and Noble, W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017-1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, L., Mason, E., Yu, H., Dudczig, S., Panara, V., Chen, T., Bower, N. I., Paterson, S., Rondon Galeano, M., Kobayashi, S.et al. (2023). Single–cell analysis of lymphatic endothelial cell fate specification and differentiation during zebrafish development. EMBO J. 42, e112590. 10.15252/rc.2022603105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Y. J., Pan, W. W., Liu, S. B., Shen, Z. F., Xu, Y. and Hu, L. L. (2020). ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 19, 1997-2007. 10.3892/etm.2020.8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., Stamatoyannopoulos, J. A., Bailey, T. L. and Noble, W. S. (2007). Quantifying similarity between motifs. Genome Biol. 8, R24. 10.1186/gb-2007-8-2-r24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman, N. D., Stuart, R. K., Hon, G., Fu, Y., Ching, C. W., Hawkins, R. D., Barrera, L. O., Van Calcar, S., Qu, C., Ching, K. A.et al. (2007). Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311-318. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- Hobert, O. (2010). Gene regulation: enhancers stepping out of the shadow. Curr. Biol. 20, R697-R699. 10.1016/j.cub.2010.07.035 [DOI] [PubMed] [Google Scholar]
- Hogan, B. M., Bos, F. L., Bussmann, J., Witte, M., Chi, N. C., Duckers, H. J. and Schulte-Merker, S. (2009). Ccbe1 is required for embryonic lymphangiogenesis and venous sprouting. Nat. Genet. 41, 396-398. 10.1038/ng.321 [DOI] [PubMed] [Google Scholar]
- Hong, J.-W., Hendrix, D. A. and Levine, M. S. (2008). Shadow enhancers as a source of evolutionary novelty. Science (80-.) 321, 1314-1314. 10.1126/science.1160631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultgren, N. W., Fang, J. S., Ziegler, M. E., Ramirez, R. N., Phan, D. T. T., Hatch, M. M. S., Welch-Reardon, K. M., Paniagua, A. E., Kim, L. S., Shon, N. N.et al. (2020). Slug regulates the Dll4-Notch-VEGFR2 axis to control endothelial cell activation and angiogenesis. Nat. Commun. 11, 5400. 10.1038/s41467-020-18633-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hußmann, M., Schulte, D., Weischer, S., Carlantoni, C., Nakajima, H., Mochizuki, N., Stainier, D. Y. R., Zobel, T., Koch, M. and Schulte-Merker, S. (2023). Svep1 is a binding ligand of Tie1 and affects specific aspects of facial lymphatic development in a Vegfc-independent manner. eLife 12, e82969. 10.7554/eLife.82969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazenwadel, J., Betterman, K. L., Chong, C.-E., Stokes, P. H., Lee, Y. K., Secker, G. A., Agalarov, Y., Demir, C. S., Lawrence, D. M., Sutton, D. L.et al. (2015). GATA2 is required for lymphatic vessel valve development and maintenance. J. Clin. Invest. 125, 2879-2994. 10.1172/JCI78888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazenwadel, J., Venugopal, P., Oszmiana, A., Toubia, J., Arriola-Martinez, L., Panara, V., Piltz, S. G., Brown, C., Ma, W., Schreiber, A. W.et al. (2023). A Prox1 enhancer represses haematopoiesis in the lymphatic vasculature. Nature 614, 343-348. 10.1038/s41586-022-05650-9 [DOI] [PubMed] [Google Scholar]
- Koltowska, K., Lagendijk, A. K., Pichol-Thievend, C., Fischer, J. C., Francois, M., Ober, E. A., Yap, A. S. and Hogan, B. M. (2015a). Vegfc regulates bipotential precursor division and Prox1 expression to promote lymphatic identity in zebrafish. Cell Rep. 13, 1828-1841. 10.1016/j.celrep.2015.10.055 [DOI] [PubMed] [Google Scholar]
- Koltowska, K., Paterson, S., Bower, N. I., Baillie, G. J., Lagendijk, A. K., Astin, J. W., Chen, H., Francois, M., Crosier, P. S., Taft, R. J.et al. (2015b). Mafba is a downstream transcriptional effector of Vegfc signaling essential for embryonic lymphangiogenesis in zebrafish. Genes Dev. 29, 1618-1630. 10.1101/gad.263210.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küchler, A. M., Gjini, E., Peterson-Maduro, J., Cancilla, B., Wolburg, H. and Schulte-Merker, S. (2006). Development of the Zebrafish lymphatic system requires Vegfc signaling. Curr. Biol. 16, 1244-1248. 10.1016/j.cub.2006.05.026 [DOI] [PubMed] [Google Scholar]
- Kvon, E. Z., Waymack, R., Gad, M. and Wunderlich, Z. (2021). Enhancer redundancy in development and disease. Nat. Rev. Genet. 22, 324-336. 10.1038/s41576-020-00311-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, D. D., de Souza, F. S. J., Nasif, S., Yamashita, M., López-Leal, R., Otero-Corchon, V., Meece, K., Sampath, H., Mercer, A. J., Wardlaw, S. L.et al. (2015). Partially redundant enhancers cooperatively maintain mammalian Pomc expression above a critical functional threshold. PLoS Genet. 11, e1004935. 10.1371/journal.pgen.1004935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, N. D. and Weinstein, B. M. (2002). In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307-318. 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- Le Guen, L., Karpanen, T., Schulte, D., Harris, N. C., Koltowska, K., Roukens, G., Bower, N. I., van Impel, A., Stacker, S. A., Achen, M. G.et al. (2014). Ccbe1 regulates Vegfc-mediated induction of Vegfr3 signaling during embryonic lymphangiogenesis. Development 141, 1239-1249. 10.1242/dev.100495 [DOI] [PubMed] [Google Scholar]
- Lin, F.-J., Chen, X., Qin, J., Hong, Y.-K., Tsai, M.-J. and Tsai, S. Y. (2010). Direct transcriptional regulation of neuropilin-2 by COUP-TFII modulates multiple steps in murine lymphatic vessel development. J. Clin. Invest. 120, 1694-1707. 10.1172/JCI40101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long, H. K., Prescott, S. L. and Wysocka, J. (2016). Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167, 1170-1187. 10.1016/j.cell.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino, D., Dabouras, V., Brändli, A. W. and Detmar, M. (2011). A role for all-trans-retinoic acid in the early steps of lymphatic vasculature development. J. Vasc. Res. 48, 236-251. 10.1159/000320620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor, C., Brudno, M., Schwartz, J. R., Poliakov, A., Rubin, E. M., Frazer, K. A., Pachter, L. S. and Dubchak, I. (2000). Vista: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 16, 1046-1047. 10.1093/bioinformatics/16.11.1046 [DOI] [PubMed] [Google Scholar]
- Meng, Y., Lv, T., Zhang, J., Shen, W., Li, L., Li, Y., Liu, X., Lei, X., Lin, X., Xu, H.et al. (2023). Temporospatial inhibition of Erk signaling is required for lymphatic valve formation. Signal Transduct. Target. Ther. 8, 342. 10.1038/s41392-023-01571-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicenboim, J., Malkinson, G., Lupo, T., Asaf, L., Sela, Y., Mayseless, O., Gibbs-Bar, L., Senderovich, N., Hashimshony, T., Shin, M.et al. (2015). Lymphatic vessels arise from specialized angioblasts within a venous niche. Nature 522, 56-61. 10.1038/nature14425 [DOI] [PubMed] [Google Scholar]
- Niimi, K., Ueda, M., Fukumoto, M., Kohara, M., Sawano, T., Tsuchihashi, R., Shibata, S., Inagaki, S. and Furuyama, T. (2017). Transcription factor FOXO1 promotes cell migration toward exogenous ATP via controlling P2Y1 receptor expression in lymphatic endothelial cells. Biochem. Biophys. Res. Commun. 489, 413-419. 10.1016/j.bbrc.2017.05.156 [DOI] [PubMed] [Google Scholar]
- Niimi, K., Kohara, M., Sedoh, E., Fukumoto, M., Shibata, S., Sawano, T., Tashiro, F., Miyazaki, S., Kubota, Y., Miyazaki, J.-I.et al. (2019). FOXO1 regulates developmental lymphangiogenesis by upregulating CXCR4 in the mouse-tail dermis. Development 147, dev181545. 10.1242/dev.181545 [DOI] [PubMed] [Google Scholar]
- Norrmén, C., Ivanov, K. I., Cheng, J., Zangger, N., Delorenzi, M., Jaquet, M., Miura, N., Puolakkainen, P., Horsley, V., Hu, J.et al. (2009). FOXC2 controls formation and maturation of lymphatic collecting vessels through cooperation with NFATc1. J. Cell Biol. 185, 439-457. 10.1083/jcb.200901104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda, K. S., Astin, J. W., Misa, J. P., Flores, M. V., Crosier, K. E. and Crosier, P. S. (2012). Lyve1 expression reveals novel lymphatic vessels and new mechanisms for lymphatic vessel development in zebrafish. Development 139, 2381-2391. 10.1242/dev.077701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterwalder, M., Barozzi, I., Tissières, V., Fukuda-Yuzawa, Y., Mannion, B. J., Afzal, S. Y., Lee, E. A., Zhu, Y., Plajzer-Frick, I., Pickle, C. S.et al. (2018). Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239-243. 10.1038/nature25461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, D.-Y., Lee, J., Park, I., Choi, D., Lee, S., Song, S., Hwang, Y., Hong, K. Y., Nakaoka, Y., Makinen, T.et al. (2014). Lymphatic regulator PROX1 determines Schlemm's canal integrity and identity. J. Clin. Invest. 124, 3960-3974. 10.1172/JCI75392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham, V. N., Lawson, N. D., Mugford, J. W., Dye, L., Castranova, D., Lo, B. and Weinstein, B. M. (2007). Combinatorial function of ETS transcription factors in the developing vasculature. Dev. Biol. 303, 772-783. 10.1016/j.ydbio.2006.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistocchi, A., Gaudenzi, G., Carra, S., Bresciani, E., Del Giacco, L. and Cotelli, F. (2008). Crucial role of zebrafish prox1 in hypothalamic catecholaminergic neurons development. BMC Dev. Biol. 8, 27. 10.1186/1471-213X-8-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillien, A., Abdalla, M., Yu, J., Ou, J., Zhu, L. J. and Lawson, N. D. (2017). Robust identification of developmentally active endothelial enhancers in zebrafish using FANS-assisted ATAC-seq. Cell Rep. 20, P709-P720. 10.1016/j.celrep.2017.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabine, A., Agalarov, Y., Maby-El Hajjami, H., Jaquet, M., Hägerling, R., Pollmann, C., Bebber, D., Pfenniger, A., Miura, N., Dormond, O.et al. (2012). Mechanotransduction, PROX1, and FOXC2 cooperate to control Connexin37 and calcineurin during lymphatic-valve formation. Dev. Cell 22, 430-445. 10.1016/j.devcel.2011.12.020 [DOI] [PubMed] [Google Scholar]
- Scallan, J. P., Knauer, L. A., Hou, H., Castorena-Gonzalez, J. A., Davis, M. J. and Yang, Y. (2021). Foxo1 deletion promotes the growth of new lymphatic valves. J. Clin. Invest. 131, e142341. 10.1172/JCI142341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secker, G. A. and Harvey, N. L. (2021). Regulation of VEGFR signalling in lymphatic vascular development and disease: an update. Int. J. Mol. Sci. 22, 7760. 10.3390/ijms22147760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, M., Male, I., Beane, T. J., Villefranc, J. A., Kok, F. O., Zhu, L. J. and Lawson, N. D. (2016). Vegfc acts through ERK to induce sprouting and differentiation of trunk lymphatic progenitors. Development 143, 3785-3795. 10.1242/dev.137901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, M., Nozaki, T., Idrizi, F., Isogai, S., Ogasawara, K., Ishida, K., Yuge, S., Roscoe, B., Wolfe, S. A., Fukuhara, S.et al. (2019). Valves are a conserved feature of the zebrafish lymphatic system. Dev. Cell 51, 374-386.e5. 10.1016/j.devcel.2019.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitz, F. and Furlong, E. E. M. (2012). Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 13, 613-626. 10.1038/nrg3207 [DOI] [PubMed] [Google Scholar]
- Srinivasan, R. S., Geng, X., Yang, Y., Wang, Y., Mukatira, S., Studer, M., Porto, M. P. R., Lagutin, O. and Oliver, G. (2010). The nuclear hormone receptor Coup-TFII is required for the initiation and early maintenance of Prox1 expression in lymphatic endothelial cells. Genes Dev. 24, 696-707. 10.1101/gad.1859310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai-Nagara, I., Hasumi, Y., Kusumoto, D., Hasumi, H., Okabe, K., Ando, T., Matsuzaki, F., Itoh, F., Saya, H., Liu, C.et al. (2020). Blood and lymphatic systems are segregated by the FLCN tumor suppressor. Nat. Commun. 11, 6314. 10.1038/s41467-020-20156-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, T., Shi, Y., Opalenik, S. R., Brantley-Sieders, D. M., Chen, J., Davidson, J. M. and Brandt, S. J. (2006). Expression of the TAL1/SCL transcription factor in physiological and pathological vascular processes. J. Pathol. 210, 121-129. 10.1002/path.2028 [DOI] [PubMed] [Google Scholar]
- Thomas, P. D., Kejariwal, A., Guo, N., Mi, H., Campbell, M. J., Muruganujan, A. and Lazareva-Ulitsky, B. (2006). Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 34, W645-W650. 10.1093/nar/gkl229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Impel, A., Zhao, Z., Hermkens, D. M. A., Roukens, M. G., Fischer, J. C., Peterson-Maduro, J., Duckers, H., Ober, E. A., Ingham, P. W. and Schulte-Merker, S. (2014). Divergence of zebrafish and mouse lymphatic cell fate specification pathways. Development 141, 1228-1238. 10.1242/dev.105031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney, G. K., Carrington, B., Pei, W., Bishop, K., Chen, Z., Fan, C., Xu, L., Jones, M., LaFave, M. C., Ledin, J.et al. (2016). A high-throughput functional genomics workflow based on CRISPR/Cas9-mediated targeted mutagenesis in zebrafish. Nat. Protoc. 11, 2357-2375. 10.1038/nprot.2016.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman, M. B. and Lin, S. (2012). Etsrp/Etv2 is directly regulated by Foxc1a/b in the Zebrafish Angioblast. Circ. Res. 110, 220-229. 10.1161/CIRCRESAHA.111.251298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villefranc, J. A., Amigo, J. and Lawson, N. D. (2007). Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 236, 3077-3087. 10.1002/dvdy.21354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigle, J. T. and Oliver, G. (1999). Prox1 function is required for the development of the murine lymphatic system. Cell 98, P769-P778. 10.1016/S0092-8674(00)81511-1 [DOI] [PubMed] [Google Scholar]
- Wong, E. S., Zheng, D., Tan, S. Z., Bower, N. I., Garside, V., Vanwalleghem, G., Gaiti, F., Scott, E., Hogan, B. M., Kikuchi, K.et al. (2020). Deep conservation of the enhancer regulatory code in animals. Science (80-.). 370, eaax8137. 10.1126/science.aax8137 [DOI] [PubMed] [Google Scholar]
- Yaniv, K., Isogai, S., Castranova, D., Dye, L., Hitomi, J. and Weinstein, B. M. (2006). Live imaging of lymphatic development in the zebrafish. Nat. Med. 12, 711-716. 10.1038/nm1427 [DOI] [PubMed] [Google Scholar]
- Yoshimatsu, Y., Yamazaki, T., Mihira, H., Itoh, T., Suehiro, J., Yuki, K., Harada, K., Morikawa, M., Iwata, C., Minami, T.et al. (2011). Ets family members induce lymphangiogenesis through physical and functional interaction with Prox1. J. Cell Sci. 124, 2753-2762. 10.1242/jcs.083998 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Custom script has been deposited in GitHub (https://github.com/virpa81/Vessel-section-calculation).