Abstract
Notch signaling, renowned for its role in regulating cell fate, organ development, and tissue homeostasis across metazoans, is highly conserved throughout evolution. The Notch receptor and its ligands are transmembrane proteins containing epidermal growth factor-like repeat sequences, typically necessitating receptor-ligand interaction to initiate classical Notch signaling transduction. Accumulating evidence indicates that the Notch signaling pathway serves as both an oncogenic factor and a tumor suppressor in various cancer types. Dysregulation of this pathway promotes epithelial-mesenchymal transition and angiogenesis in malignancies, closely linked to cancer proliferation, invasion, and metastasis. Furthermore, the Notch signaling pathway contributes to maintaining stem-like properties in cancer cells, thereby enhancing cancer invasiveness. The regulatory role of the Notch signaling pathway in cancer metabolic reprogramming and the tumor microenvironment suggests its pivotal involvement in balancing oncogenic and tumor suppressive effects. Moreover, the Notch signaling pathway is implicated in conferring chemoresistance to tumor cells. Therefore, a comprehensive understanding of these biological processes is crucial for developing innovative therapeutic strategies targeting Notch signaling. This review focuses on the research progress of the Notch signaling pathway in cancers, providing in-depth insights into the potential mechanisms of Notch signaling regulation in the occurrence and progression of cancer. Additionally, the review summarizes pharmaceutical clinical trials targeting Notch signaling for cancer therapy, aiming to offer new insights into therapeutic strategies for human malignancies.
Subject terms: Cancer therapy, Tumour biomarkers
Introduction
The Notch locus was initially identified in 1917 through genetic studies involving a mutant strain of Drosophila melanogaster exhibiting notched wings.1 The Drosophila Notch gene was subsequently isolated in 1983.2 It was later revealed that the protein encoded by the Notch gene functions as a transmembrane receptor with multiple epidermal growth factor (EGF)-like repeats, typically activated by transmembrane ligands expressed on adjacent cells.3 To date, Notch receptors and ligands have been discovered in various metazoans, serving as integral components of the Notch signaling cascade and participating in diverse biological processes such as cell fate determination, embryonic development, organ formation, and tissue repair.4,5
Extensive research has been conducted on Notch signaling pathway, investigating its role as an oncogene or tumor suppressor in various cellular contexts. The involvement of the Notch signaling pathway in human malignancies was initially elucidated in T cell acute lymphoblastic leukemia (T-ALL), where the chromosomal translocation t(7;9) (q34;q34.3) results in the truncation of Notch1 transcripts.6 Subsequent cancer genome sequencing has unveiled widespread oncogenic Notch gene mutations in diverse human cancers, such as cutaneous and lung squamous cell carcinoma (LUSC),7 breast cancer,8 anaplastic large cell lymphoma,9 and chronic lymphocytic leukemia (CLL).10 Moreover, accumulating evidence indicates that the dysregulation of the Notch signaling pathway intricately controls the onset and progression of hematologic malignancies and solid tumors in humans.11,12 This occurs through complex mechanisms, including tumor angiogenesis, modulation of the immune microenvironment, epithelial-mesenchymal transition (EMT), cancer energy metabolism, and resistance to chemotherapy. For instance, oncogenic Notch signaling facilitates T-ALL cell proliferation by activating nuclear factor-kappa B (NF-κB) through Asb2 mediation.13 Additionally, activated Notch signaling contributes to the acquisition of stem-like properties in esophageal adenocarcinoma.14 The pivotal role of Notch signaling in cancer biology has garnered significant attention, leading to the exploration of targeted cancer therapies based on Notch signaling. This review offers a systematic exploration of the research advancements in the Notch signaling pathway within the context of tumors. It concentrates on unraveling the molecular mechanisms underlying Notch signaling-mediated tumorigenesis and progression. Furthermore, the review outlines targeted therapeutic strategies for tumors that are rooted in Notch signaling, as evidenced by clinical research endeavors. The systematic insights provided in this review aim to furnish a current and thorough understanding of the Notch signaling pathway in tumors. This knowledge is expected to contribute significantly to the future development of the Notch signaling pathway in both basic research and clinical translation.
Overview of the Notch signaling pathway
The Notch signaling pathway exhibits high conservation throughout evolution, coordinating multiple physiological mechanisms during development and homeostasis in metazoans. Classically, ligand-activated Notch receptors initiate transcription of downstream target genes by interacting with the DNA-bound CSL-co-repressor complex, forming the canonical Notch signaling pathway. Over the years, mounting evidence has demonstrated that Notch can function via non-canonical pathway that is independent of ligands or CSL.15,16 The canonical Notch signaling pathway plays a major physiological function in intercellular interaction and gene transcription regulation, while non-canonical Notch signaling involves the crosstalk between various signaling pathways to execute activation of target genes.17 In this overview, we provide a brief summary of the key components of the Notch signaling pathway and examine the mechanisms underlying both the canonical and non-canonical Notch signaling pathways (Fig. 1).
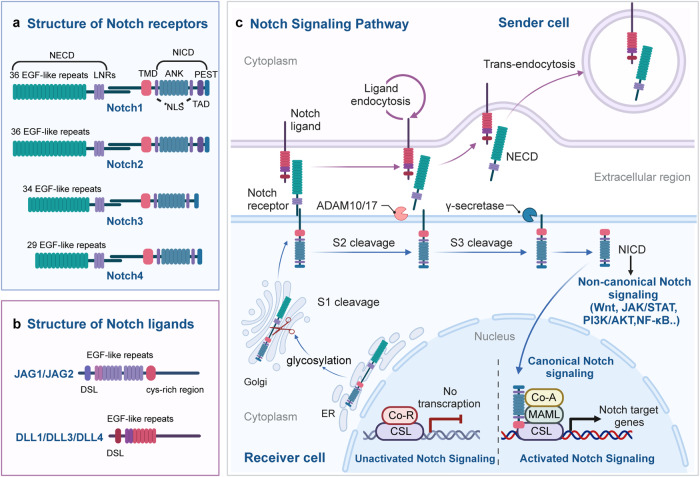
Fig. 1.
Notch signaling overview. a Four Notch receptors (Notch1, Notch2, Notch3, and Notch4) and their respective structures. b Structures of five Notch ligands (JAG1, JAG2, DLL1, DLL3, and DLL4). c Schematic representation of canonical and non-canonical Notch signaling pathways. (Figure created using BioRender.com). NECD Notch extracellular domain, EGF epidermal growth factor, LNRs Lin12-Notch repeats, TMD transmembrane domain, NICD Notch intracellular domain, ANK ankyrin repeat, NLS nuclear localization sequences, TAD transcription activation domain, PEST proline/glutamic acid/serine/threonine, CSL CBF1/suppressor of hairless/Lag1, ADAM a disintegrin and metalloprotease, ER endoplasmic reticulum, Co-R corepressor, CSL CBF1/suppressor of hairless/Lag1, Co-A coactivator, MAML mastermind-like
Components of the Notch signaling pathway
The mammalian Notch signaling pathway comprises three principal components: Notch receptors, ligands that bind to Notch receptors, and downstream effectors of Notch signaling.18 In D. melanogaster, there is a single Notch receptor ortholog, Notch1.19 However, in mammals, three additional Notch receptors exist: Notch2, Notch3, and Notch4. The Notch receptor is a transmembrane protein with three main segments: the Notch extracellular domain (NECD), transmembrane domain (TMD), and Notch intracellular domain (NICD).20 The NECD contains multiple EGF-like repeats and a negative regulatory region (NRR), modified by O-glycans to regulate the Notch receptor’s affinity for different ligands.21 Notch1–4 NECDs have 36, 36, 34, and 29 EGF-like repeats, respectively, crucial for ligand interaction.22 The NRR comprises three cysteine-rich Lin12-Notch repeats, stabilizing NECD and membrane-bound NICD interaction, essential for receptor cleavage.23–25 The TMD includes an extracellular short region and conserved cysteine residues forming heterodimers.26 The NICD consists of an RBPJ [recombination signal binding protein-J] association module (RAM) domain, seven ankyrin repeat (ANK) domains, and two nuclear localization sequences (NLS) on each side of the ANK domain.27 Notch1 and Notch2 have a transcription activation domain (TAD) after the ANK sequence, while Notch3 and Notch4 lack a TAD. The C-terminal of NICD has a “PEST” sequence, rich in proline, glutamic acid, serine, and threonine, crucial for NICD stability.28
Humans and mice possess five ligands binding to extracellular Notch receptor fragments. Based on the presence or absence of the cysteine-rich region, Notch ligands are categorized into Serrate-like ligands Jagged1 (JAG1) and JAG2, and delta-like ligands DLL1, DLL3, and DLL4.29 Notch ligands are cell membrane proteins, sharing structural similarities with the Notch receptor. The extracellular domains of JAG1/2 consist of the DSL [i.e., Delta, Serrate, and LAG-2] domain, EGF-like repeats, and a cysteine-rich region.30 The extracellular domains of DLL1/3/4 are akin to JAG1/2 but lack the cysteine-rich region.
The canonical Notch signaling pathway
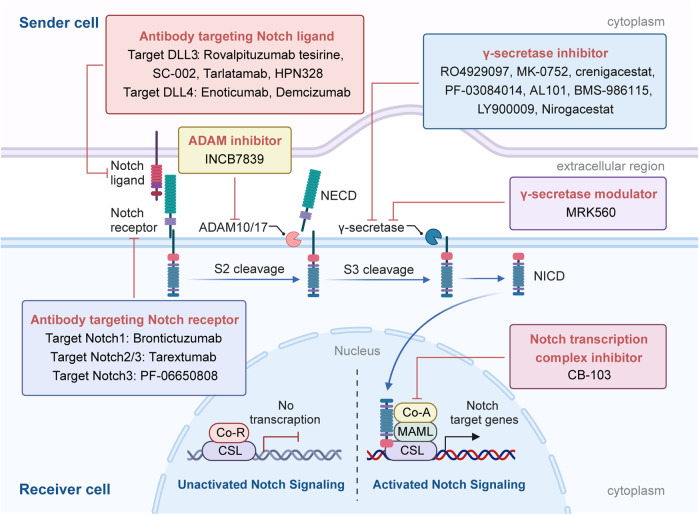
The canonical Notch signaling pathway involves a series of intricate steps in the maturation and activation of Notch proteins. Initially, Notch proteins are transported to the endoplasmic reticulum as single-stranded precursors. Within the endoplasmic reticulum, the EGF-like domain of the Notch receptor undergoes glycosylation.31,32 The glycosylated Notch single-chain precursor is then transported to the Golgi apparatus. In the Golgi apparatus, a furin-like convertase cleaves the S1 site in the extracellular segment of the Notch transmembrane region, resulting in the formation of two distinct fragments: the NECD and the TMD.33,34 These fragments subsequently combine through a Ca2+-dependent non-covalent bond, forming the mature Notch receptor in the shape of a heterodimer. The mature Notch receptor, now a type I transmembrane protein, is then transported to the cell surface. Upon reaching the cell surface, the Notch heterodimeric transmembrane receptor binds to the Notch transmembrane ligand present on adjacent cells. The S2 cleavage site of the Notch receptor is then cleaved by members of the ADAM (a disintegrin and metalloprotease) metalloproteinase family, specifically ADAM10 or ADAM17.35,36 This cleavage releases a partial extracellular fragment, creating a transient intermediate peptide called ‘NeXT’ [Notch extracellular truncation], which consists of the TMD and NICD. The next step involves presenilin-dependent γ-secretase cleaving NeXT at the S3 cleavage site.37 This process leads to the release of the soluble NICD of Notch. Subsequently, NICD translocates to the cellular nucleus, where its RAM domain interacts with the transcription factor CBF1/suppressor of hairless/Lag1 (CSL, also called RBPJ).38 This interaction facilitates the recruitment of co-activator complexes to CSL, including mammalian mastermind-like 1–3 (MAML1–3) proteins. The assembly of these complexes transforms the original “co-repressor complex” into a “co-activator complex,” resulting in the formation of a multi-protein-DNA complex. This complex promotes the transcription of Notch target genes. In the absence of NICD binding, CSL downregulates the expression of target genes by recruiting various co-repressor proteins.39
The non-canonical Notch signaling pathway
In addition to its interaction with CSL, Notch signaling can influence the expression of related genes through non-CSL-dependent regulatory pathways, constituting the non-canonical Notch signaling pathway.40,41 This pathway may be initiated by ligand-independent mechanisms and might not necessitate Notch receptor cleavage. In vertebrates, non-canonical Notch target activation is primarily observed in lineage-restricted progenitors, fate-specific differentiation, and tumorigenesis.42 Notably, studies have revealed that Notch can modulate the Wnt/β-catenin signaling pathway,43 the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway,44 the phosphoinositide 3-kinase/protein kinase B (PI3K/AKT) pathway,45 and the NF-κB pathway at the post-translational level, thereby exerting its non-canonical biological functions. In human breast epithelial cells, Notch-induced expression of Wnt signaling receptor FZD7 requires non-canonical Notch3 Signaling.46 Non-canonical Notch signaling triggered IL-6/JAK/STAT signaling in breast cancer cells and is regulated by IKKα/IKKβ of the NF-κB signaling cascade.47 Additionally, recent studies revealed that the non-canonical Notch signaling cascade, mediated by extracellular vesicles and independent of classical ligand-receptor interactions, may have important implications in the invasive phenotype of breast cancer.48,49 Lee and colleagues discovered that non-canonical Notch signaling interacted with PTEN-induced kinase 1 (PINK1) to impact mitochondrial function and activate mammalian target of rapamycin complex 2 (mTORC2)/AKT signaling, which maintained brain tumor-forming stem cells.50 Perumalsamy et al. identified a novel Notch-mediated non-canonical signaling cascade independent of CBF1/RBPJ, where NICD interacts with the mTOR-Rictor complex, leading to the activation of AKT/PKB to control mammalian cell survival.51 During tumorigenesis and progression, the focus on non-canonical Notch signaling activation is growing due to its significance for tumor cellular function, such as proliferation, neoplastic transformation, and inhibition of apoptosis. For instance, non-canonical activation of Notch1 protein sustained the proliferation of melanoma cells, while non-canonical Notch3 signaling could trigger endothelial cell apoptosis to restrict tumor angiogenesis.52,53 These non-classical mechanisms allow evolutionarily conserved Notch signaling to carry out more specific functions and may uncover new therapeutic targets as additional mechanisms are revealed in cancers.
The Notch signaling pathway and cancer
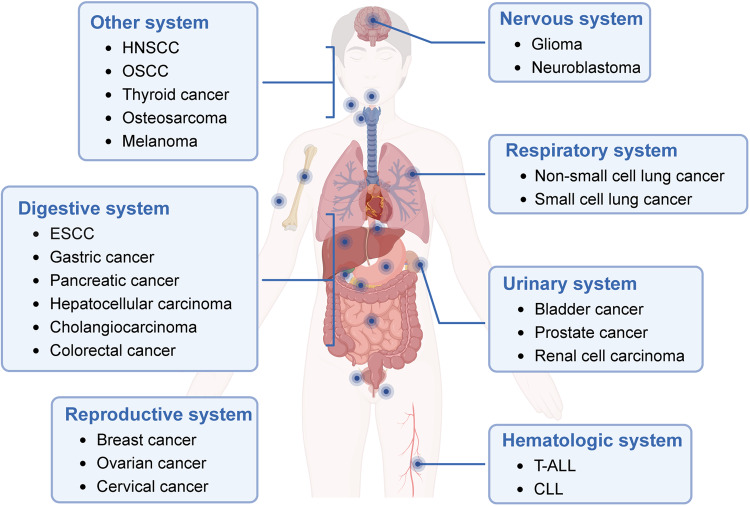
The Notch signaling pathway plays a crucial role in regulating cell fate decisions under physiological conditions, influencing cell proliferation, differentiation, development, and homeostasis. However, dysregulation of the Notch signaling pathway has been increasingly observed in various human malignancies,54,55 spanning digestive system tumors, respiratory system tumors, hematological malignancies, urinary system tumors, reproductive system tumors, nervous system neoplasms, and tumors in other systems (Fig. 2). In this section, we provide a summary of the expression of Notch receptors and ligands in different types of tumors, along with their associations with clinicopathological features and prognosis (Table 1). Furthermore, we review the functions of the dysregulated Notch signaling pathway in different tumors, with the objective of identifying novel diagnostic and prognostic biomarkers based on Notch signaling.
Fig. 2.
Involvement of Notch signaling in the regulation of diverse cancers. Notch signaling plays a role in the regulation of various cancers, encompassing digestive system tumors, respiratory system tumors, hematological malignancies, urinary system tumors, reproductive system tumors, nervous system neoplasms, and tumors in other systems. (Figure created using BioRender.com)
Table 1.
Expression and clinical features of Notch receptor or ligand in cancers
| Category | Cancer type | Notch receptor/ligand | Expression (tumor vs. normal) | Clinical feature | Prognosis | Ref. |
|---|---|---|---|---|---|---|
| Digestive system | CRC | Notch1 | Upregulated | Lymph node metastasis, tumor stage, depth of infiltration, histological differentiation | Poor | 58 |
| CRC | Notch2 | Downregulated | OS | Favorable | 59 | |
| CRC | Notch3 | Upregulated | Tumor differentiation status, tumor recurrence, distant relapse-free survival | Poor | 60 | |
| CRC | Notch4 | Downregulated | Tumor differentiation, invasion, node metastasis, disease-free and OS | Favorable | 61 | |
| CRC | JAG1 | Upregulated | OS, relapse-free survival | Poor | 62 | |
| CRC | JAG2 | Upregulated | Clinical stages | Poor | 63 | |
| CRC | DLL4 | Upregulated | OS, perineural invasion, distant metastasis | Poor | 64 | |
| HCC | Notch1, Notch4 | Upregulated | Disease-specific survival | Poor | 69 | |
| HCC | Notch2 | Upregulated | Clinical stages | Poor | 71 | |
| HCC | Notch3 | Upregulated | Tumor size, TNM stage, OS | Poor | 74 | |
| HCC | JAG1 | Upregulated | Differentiation grade | Poor | 76 | |
| HCC | JAG2 | Upregulated | Intrahepatic metastasis, histological grade, TNM stage | Poor | 78 | |
| ESCC | Notch1 | Upregulated | Tumor grade and stage | Poor | 85 | |
| ESCC | Notch2 | Upregulated | OS and PFS | Poor | 87 | |
| ESCC | Notch3 | N/A | Chemotherapy sensitivity | Favorable | 88 | |
| GC | Notch1 | Upregulated | Lymph node metastasis | Poor | 91 | |
| GC | Notch3 | N/A | Immune tolerance | Favorable | 97 | |
| GC | JAG1 | Downregulated | OS | Favorable | 98 | |
| PC | Notch3 | Upregulated | OS | Poor | 104 | |
| PC | JAG1 | Upregulated | OS | Poor | 106 | |
| PC | DLL4 | N/A | Advanced tumor stage, lymph node metastasis | Poor | 107 | |
| CCA | Notch1 | Upregulated | Tumor size, HBsAg positive | Poor | 123 | |
| CCA | Notch3 | Upregulated | N/A | Poor | 124 | |
| CCA | JAG1 | Upregulated | N/A | Poor | 125 | |
| Respiratory system | NSCLC | Notch1 | Upregulated | Lymph node metastasis, TNM stages | Poor | 129 |
| NSCLC | Notch4, DLL4 | Upregulated | Tumor size, lymph node metastasis, distant metastasis, TNM stage | Favorable | 130 | |
| NSCLC | JAG1, JAG2, DLL1 | Downregulated | N/A | N/A | 131 | |
| SCLC | DLL3 | Upregulated | N/A | N/A | 147 | |
| Postoperative SCLC | DLL3 | Upregulated | PFS, chemoresistance | Poor | 148 | |
| Hematological malignancies | Adult T-ALL | Notch1 | Mutation | OS, event-free survival | Favorable | 168 |
| Pediatric T-ALL | Notch1 | Mutation | OS | Favorable | 169 | |
| CLL | Notch1 | Mutation | PFS | Poor | 179 | |
| CLL | Notch2 | Upregulated | Apoptosis characteristic | Poor | 182 | |
| CLL | JAG1 | Upregulated | N/A | N/A | 184 | |
| Urinary system | Bladder cancer | Notch1–3 | Mutation | OS | Poor | 189 |
| Bladder cancer | Notch2 | Upregulated | Adverse disease parameters | Poor | 191 | |
| Bladder cancer | Notch3 | Upregulated | Cancer-specific mortality | Poor | 193 | |
| Bladder cancer | JAG2 | Upregulated | Tumor size, stage | Poor | 194 | |
| PCa | Notch1 | Upregulated | High-risk, metastasis | Poor | 199 | |
| PCa bone metastasis | Notch3 | Upregulated | Bone metastasis | Poor | 200 | |
| PCa | DLL3 | Upregulated | Survival | Poor | 204 | |
| PCa | JAG1 | Upregulated | Metastasis, recurrence | Poor | 201 | |
| RCC | Notch1 | Downregulated | Tumor stage | N/A | 210 | |
| RCC | Notch3 | Dysregulated | Chromophobe RCC, unbroken capsule, Fuhrman grade, lymph node involvement | N/A | 213 | |
| ccRCC | Notch | Copy number variance | OS | Favorable | 217 | |
| ccRCC | Notch1 | Upregulated | Tumor stage, diameter | Poor | 218 | |
| ccRCC | JAG1 | Upregulated | Tumor size, nuclear grade, TNM stage | Poor | 220 | |
| ccRCC | DLL4 | Upregulated | Tumor grade, tumor stage, survival | Poor | 219 | |
| Reproductive system | Breast cancer | JAG1 and Notch1 | Upregulated | OS, median survival | Poor | 226 |
| Breast cancer | Notch2 | Dysregulated | Subgroups, genotypes | Poor | 231 | |
| Breast cancer | Notch4 | Upregulated | Tumor size, lymph node involvement, metastasis stage | Poor | 227 | |
| Breast cancer | DLL3 | Upregulated | Immune cell infiltration | Poor | 229 | |
| Breast cancer | DLL4 | Upregulated | Nodal and distant metastasis | Poor | 230 | |
| Breast cancer | DLL4, JAG1 | Upregulated | Metastasis, tumor stage | Poor | 228 | |
| OC | Notch1 | Upregulated | Pathology stage, OS | Poor | 176 | |
| OC | Notch2/3, DLL3 | Upregulated | Overall, disease-free survival, stages | Poor | 242 | |
| OC | Notch3 | Upregulated | Progression-free/OS | Poor | 243 | |
| OC | DLL4 | Upregulated | Survival | Poor | 244 | |
| CC | JAG1/Notch1 | Upregulated | Invasion, lymph node metastasis, FIGO system | Poor | 254 | |
| CC | DLL4 | Upregulated | Death and recurrence | Poor | 255 | |
| Nervous system | Oligodendroglioma | Notch1 | Mutation | Survival | Poor | 266 |
| Glioma | Notch1 | Upregulated | OS | Poor | 267 | |
| Glioma | Notch3 | Upregulated | Grade, OS | Poor | 268 | |
| Glioma | DLL3 | Upregulated | Prognosis | Favorable | 269 | |
| Neuroblastoma | Notch1 | Upregulated | Advanced tumor stages, MYCN amplification, undifferentiated histology, low CRT expression level | Poor | 281 | |
| Tumors of other systems | Melanoma | Notch4 | Mutation | OS | Favorable | 293 |
| Osteosarcoma | Notch3 | Upregulated | Survival | Poor | 296 | |
| Osteosarcoma | JAG1 | Upregulated | Metastasis, recurrence | Poor | 297 | |
| Osteosarcoma | DLL4 | Upregulated | Enneking stage and metastasis, tumor differentiation | Poor | 298 | |
| Thyroid cancer | Notch1 | 80.5% positive | Lymph node metastasis | Poor | 307 | |
| Thyroid cancer | DLL4 | 54% positive | Invasion and metastasis | Poor | 308 | |
| Thyroid cancer | Notch3 | Downregulated | Tumor size, distant metastasis, survival | Favorable | 309 | |
| OSCC | Notch1 | Upregulated | T-stage, clinical stage | Poor | 314 | |
| OSCC | Notch3 | 33% positive | Tumor size | Poor | 316 | |
| OSCC | Notch4 | Upregulated | Late stage | Poor | 317 | |
| HNSCC | Notch1/2/3 | Mutation | N/A | N/A | 326 | |
| HNSCC | Notch1 | Upregulated | Early stages, non-recurrent disease, better disease-specific, OS | Favorable | 330 |
CRC colorectal cancer, HCC hepatocellular carcinoma, ESCC esophageal squamous cell carcinoma, GC gastric cancer, PC pancreatic cancer, CCA cholangiocarcinoma, NSCLC non-small cell lung cancer, SCLC small cell lung cancer, T-ALL T cell acute lymphoblastic leukemia, CLL chronic lymphocytic leukemia, PCa prostate cancer, RCC renal cell carcinoma, ccRCC clear cell renal cell carcinoma, OC ovarian cancer, CC cervical cancer, OSCC oral squamous cell carcinoma, HNSCC head and neck squamous cell carcinoma, OS overall survival, PFS progression-free survival, TNM tumor-node-metastasis
Digestive system tumors
Colorectal cancer (CRC)
CRC stands out as one of the most prevalent malignant cancers globally. The expression of members from the Notch family in CRC has been extensively investigated. Numerous studies have demonstrated the high expression of Notch1 in human CRC.56,57 Elevated Notch1 expression has been closely associated with lymph node metastasis, tumor stage, depth of infiltration, and histological differentiation.58 Conversely, the expression of Notch2 in CRC was significantly negatively correlated with Notch1, and reduced Notch2 expression independently predicted a poor prognosis in CRC.59 Additional research has indicated that the overexpression rate of nuclear Notch3 in CRC was 38%, and nuclear Notch3 expression was closely linked to distant relapse-free survival in stage II CRC.60 Furthermore, the co-expression of nuclear Notch3 and Notch1 predicted a worse prognosis than negative subtypes. Regarding Notch4, researchers have verified that Notch4 expression is decreased in CRC, and the Notch4 mRNA level may serve as an independent prognostic factor for disease-free survival and overall survival (OS) in patients with CRC.61 Notch ligands, JAG1,62 JAG2,63 and DLL4,64 have also been reported to be significantly upregulated in CRC, and their high expression can predict a poor prognosis in CRC. Notably, Varga and colleagues revealed that AKT-dependent Notch3 activation led to tumor invasion and metastasis in a Trp53ΔIECAktE17K mice model of mesenchymal CRC subtype, indicating that Notch3 may represent a potential target for patients with this CRC subtype.65 Inhibition of Notch signaling pathway by Aes, an endogenous metastasis suppressor, can block transendothelial migration and intravasation of colon cancer cells.66 Further study is needed to understand the role of Notch signaling in modulating the development of CRC.
Hepatocellular carcinoma (HCC)
The deregulated expression of Notch receptors and their ligands has been noted in HCC.67,68 Ahn and colleagues observed cytoplasmic expression of Notch1, Notch3, and Notch4 in 50.3%, 20.8%, and 59.7% of 288 HCC cases, respectively.69 Notch1 expression and Notch4 overexpression may independently predict poor survival in HCC. Another study revealed that in hepatitis B virus (HBV)-related HCC tissues, the expression of Notch1 or Notch4 was associated with HBV X protein (HBx), suggesting that HBx may play a role in carcinogenesis by regulating the Notch pathway.70 Notch2 is closely linked to liver cancer occurrence. Hayashi et al. found positive Notch2 nuclear staining in 19% of human primary HCC through immunohistochemistry.71 Consistent with this, upregulation of Notch2 was observed in human HCC cell lines.72 Functionally, Michael et al. found that the constitutive Notch2 signaling in the liver accelerated diethylnitrosamine-induced tumorigenesis through promoting proliferation and less differentiated HCC.73 Notch3 is overexpressed in HCC compared to normal liver tissue and is positively correlated with increased invasiveness and shorter survival.74 Another study reported abnormal accumulation of Notch3 and Notch4 in 78% and 68% of HCC tissues, respectively.75 JAG1 is highly expressed in HCC, with expression of JAG1 and DLL4 in HCC cells at 57.2% and 88.9%, respectively.76,77 However, no correlation between DLL4 expression and clinical features has been observed. Upregulated expression of JAG2 was also noted in HCC tissues and was associated with poor clinicopathological features.78 Targeted inhibition of JAG1 and JAG2 is expected to act as a tumor suppressor in HCC.79 In conclusion, these studies suggest that Notch family members may serve as potential prognostic indicators in patients with HCC.
Esophageal squamous cell carcinoma (ESCC)
Mutations of Notch receptors have been reported to dysregulate the Notch pathway in the development of ESCC.80 Li et al. identified an aberrant Notch signaling pathway in 38.3% of ESCC cases, with univariate analysis revealing an association between Notch2 gene mutations and shorter progression-free survival (PFS).81 Jones and colleagues found that Notch1 mutant clones were present in most human normal esophageal epithelium.82,83 A high proportion of biallelic mutations can block Notch1 signaling and hinder carcinogenesis, while wild-type Notch1 is conducive to the development of ESCC.84 Additional studies indicated that Notch1 expression in ESCC was significantly higher than in benign and reactive esophageal epithelium, showing a positive correlation with tumor grade and stage.85 The missense mutation site in the Notch1 gene was found to be located in the region where Notch1 binds to DLL4, enhancing the Notch1-DLL4 interaction.86 This may lead to resistance to neoadjuvant chemotherapy in patients with ESCC by promoting the activation of the Notch1 signaling pathway. Additionally, both mRNA and protein levels of Notch2 were significantly increased in ESCC tissues, serving as an independent predictor of poor OS and PFS.87 In vivo and in vitro experiments demonstrated that chemotherapy resistance in ESCC was associated with the down-regulation of Notch3 and simultaneous activation of EMT process.88 The ectopic expression of Notch3-activated forms inhibited EMT and increased sensitivity to chemotherapy, suggesting that Notch3 could be a potential biomarker for predicting favorable clinical outcomes in ESCC.
Gastric cancer (GC)
The activation of Notch signaling has been identified as a crucial factor in the development of GC. Studies have underscored the significant role of activated Notch signaling in GC development, revealing variations in the level of Notch signaling family molecules.89,90 Huang et al. observed higher levels of Notch1 expression in GC tissues compared to adjacent non-tumor tissues, suggesting a potential carcinogenic role for Notch1 in GC.91 A feedback loop between Notch1 and HGF/c-Met signaling pathways has been proposed, potentially contributing to drug resistance in GC.92 The high nuclear translocation frequency of Notch2 in GC (97.3%) indicates a close association between Notch2 level and GC formation.93 Mechanistically, both activated Notch1 and Notch2 receptors can drive GC progression through cyclooxygenase-2.94 Additionally, studies have reported that Notch signaling regulates the function of LGR5+ gastric stem cells and Cck2r+ antral stem cells, which is associated with gastric tumorigenesis.95,96 In contrast to the roles of Notch1 and Notch2, highly expressed Notch3 is implicated in the immune tolerance of GC, correlating with low infiltration of activated CD8+ T cells and high infiltration of immunosuppressive cells in the tumor microenvironment (TME).97 This suggests Notch3 could serve as a biomarker for a favorable prognosis in GC. The Notch ligand, JAG1, exhibits significantly lower levels in GC tissues than in non-tumor tissues, and its reduced level in both tumors and non-tumors is associated with poor outcomes.98 However, no significant difference was observed between DLL4 expression and clinicopathological features and OS.99 Further research is needed to explore the mechanisms underlying abnormally activated Notch signaling in GC tumorigenesis.
Pancreatic cancer (PC)
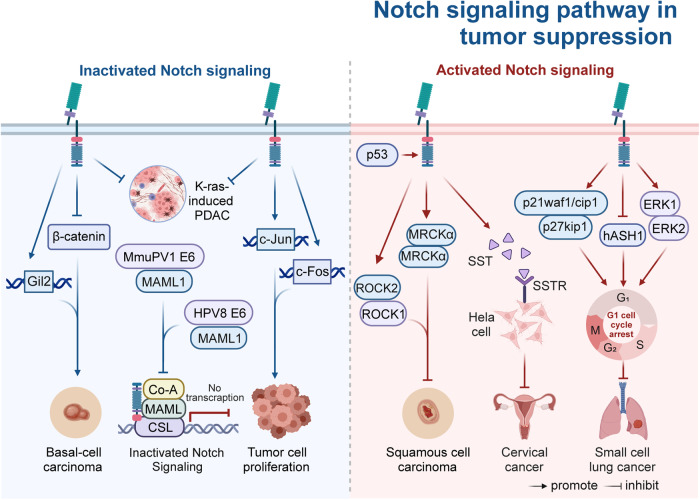
The Notch signaling pathway plays a crucial role in the regulation of pancreatic development and may be implicated in the differentiation, proliferation, and apoptosis of malignant pancreatic cells.100 Initial research indicates that Notch signaling undergoes reactivation during the initiation of pancreatic ductal adenocarcinoma (PDAC), suggesting its involvement in promoting PDAC progression.101 However, during the development of pancreatic intraepithelial neoplasia, Notch receptors demonstrate tumor-suppressive effects. The activated Notch pathway appears to influence the neurovascular development of PC and contributes to maintaining the population of pancreatic cancer stem cells (CSCs).102 In studies involving patients with PDAC, Notch3 is frequently upregulated in the cytoplasm of tumors compared to normal pancreatic ductal tissues.103 In patients with unresectable PC, decreased Notch3 mRNA level is significantly associated with longer OS.104 Inhibiting Notch3 enhances the sensitivity of PC cells to gemcitabine (GEM) chemotherapy by reducing the activity of the PI3K/AKT pathway.105 In both in vivo and in vitro studies, the expression of JAG1 in PC is significantly higher than that in normal pancreatic tissue.106 Combined treatment involving silencing JAG1 and GEM demonstrates a synergistic anti-tumor effect, suggesting that JAG1 may serve as a promising therapeutic target for PC. Furthermore, patients with PC as well as low expression of DLL4 and HES1 exhibit better survival compared to those with high expression.107,108 Low DLL4 abundance in tumor cells can predict the benefits of GEM adjuvant therapy after PDAC resection, and inhibiting DLL4/Notch signaling may represent a novel approach for PC therapy.109,110
Cholangiocarcinoma (CCA)
CCA, an aggressive form of biliary tract cancer with high incidence and mortality rates, can be categorized into intrahepatic (iCCA), perihilar CCA, or distal CCA based on anatomical location.111 Accumulating evidence indicates that the Notch pathway participates in the transformation of mature hepatocytes into malignant cholangiocytes.112,113 Cyclin E gene was identified as a direct transcriptional target of Notch signaling and involved in the formation of CCA caused by over-activated Notch signaling pathway.114 Consequently, the Notch1 pathway has been reported to mediate iCCA cell growth and the transition of the cell cycle from G0/G1 to S-phase.115 Notch2 is recognized as the primary determinant of iCCA formation derived from mouse hepatocytes.116 Mechanistically, Wang et al. uncovered that DLL4-Notch4-Efnb2 signaling mediates the differentiation of hepatic sinusoidal endothelial cells around the portal vein into apical endothelial cells, facilitating the progression of iCCA.117 Additionally, Hu et al. identified a novel Notch-YAP1/TEAD-DNMT1 axis that drives hepatocyte reprogramming into iCCA.118 Another study revealed that elevated fucosylation is a hallmark of human iCCA, promoting cell growth and migration by upregulating Notch and EGFR/NF-κB pathways.119 Simultaneously, the Notch pathway is considered a key indicator of CCA progression and prognosis.120,121 Studies show that Notch1 is upregulated in iCCA, potentially promoting iCCA migration by inducing EMT.122,123 Additionally, Guest et al. identified the differential overactivation of the atypical receptor Notch3 in iCCA in humans, rats, and mice.124 Notch3 activates the PI3K/AKT cascade through a non-classical pathway, maintaining tumor cell survival. Che et al. found that JAG1 is generally up-regulated in human iCCA samples compared with non-neoplastic livers, and inhibiting JAG1 can increase the apoptosis of human iCCA cell lines.125 Importantly, JAG1 is a crucial upstream inducer of Notch signaling in human and mouse iCCA.125 The synergistic overexpression of JAG1 and activated AKT signaling promotes the occurrence of liver cancer.125 In summary, activated Notch signaling is identified as a common carcinogenic event in human CCA. A deeper understanding of the mechanisms triggered by the Notch pathway and its functional crosstalk with other signaling cascades may contribute to the design of new therapies for human CCA.
Respiratory system tumors
Non-small cell lung cancer (NSCLC)
Lung cancer stands as one of the most lethal cancers globally, contributing to ~25% of all tumor-related fatalities.126 Based on histopathological features, lung cancer is broadly categorized into two major types: NSCLC and small cell lung cancer (SCLC), with NSCLC encompassing 80–85% of all lung cancer cases.127 NSCLC further differentiates into two primary subtypes: lung adenocarcinoma (LUAD) and LUSC. Unfortunately, more than half of patients with NSCLC receive a diagnosis at an advanced disease stage, and the efficacy of combination chemotherapy hovers at ~20%. Consequently, comprehending the pathogenesis of NSCLC and overcoming chemotherapy resistance is pivotal to enhancing the prognosis of NSCLC.
Over the decades, the Notch signaling pathway has garnered increasing attention as a promising new target for diagnosing and prognosing NSCLC. Notch1 is detected in 50% of stage I to IV NSCLC cases, predominantly localized to the cell membrane and cytoplasm.128 Meta-analysis reveals that high Notch1 expression correlates positively with lymph node metastasis and higher tumor-node-metastasis (TNM) stage, indicative of poor OS in patients with NSCLC.129 Wang et al. identified significantly higher positive rates of Notch4 and DLL4 in NSCLC compared to normal lung tissues.130 However, contradictory findings emerge from another study, reporting downregulated DLL4 protein levels in NSCLC tissues and lung cancer cell lines. The levels of other Notch ligands, including JAG1, JAG2, and DLL1, in NSCLC were also observed to be lower than that in normal lung tissue.131 Furthermore, DLL4 and Notch1 emerged as independent prognostic factors for NSCLC but exhibited varying effects in LUAD and LUSC.132 The inconsistent results across studies may stem from small sample sizes or variations in sample sources. A comprehensive, large-scale, multi-center study is imperative to thoroughly investigate the expression and function of Notch family members in NSCLC.
Accumulating evidence suggests that the evolutionarily-conserved Notch signaling pathway plays a crucial role in cell specification and fate determination during lung development, and it also mediates the initiation and progression of NSCLC.133 For instance, Xie et al. discovered that Notch1 contributes to the EMT phenotype of NSCLC, promoting acquired resistance in NSCLC.134 Another study demonstrated that the activated Notch1 signal forms a positive feedback loop with the downstream functional transcription target RFC4, conferring metastasis and stemness characteristics to NSCLC cells, as well as resistance to γ-secretase inhibitor (GSI) treatment.135 Furthermore, Baumgart and colleagues established that Notch signaling regulates tumorigenesis in KrasG12D-driven LUAD.136 Surprisingly, Notch1 and Notch2 play distinct roles in NSCLC, where Notch2 mediates differentiation and inhibits tumor formation during lung cancer progression, while Notch1 promotes carcinogenesis. Notably, Zheng et al. identified a rare population of CD24+ITGB4+Notchhi cells from a Kras-driven NSCLC mouse model, which drives tumor progression, and Notch3 has a specific and non-redundant function in mediating the propagation and self-renewal of tumor-propagating cells.137 Importantly, a co-expression analysis revealed that Notch1 exhibits opposite functional effects on angiogenesis and immune pathways in LUAD and LUSC, potentially contributing to the development of Notch1-dependent targeted therapy strategies for specific tumor subgroups within NSCLC.138
Small cell lung cancer
SCLC is a high-grade neuroendocrine cancer that constitutes 13–5% of newly diagnosed lung cancers, with a daunting five-year survival rate of less than 7%.139,140 This aggressive cancer is characterized by high genomic instability, rapid growth, and a substantial potential for metastasis.141 Over 60% of patients with SCLC are diagnosed with extensive-stage SCLC, facing a median survival of less than 10 months.142 Even for those diagnosed with limited-stage SCLC, survival rates are generally poor. While initial responses to standard chemotherapy and radiotherapy are common, rapid relapse due to acquired chemotherapy resistance is a frequent challenge. The uncommon preinvasive histological pattern of SCLC makes traditional early screening strategies ineffective. Therefore, a deeper understanding of SCLC biology, the development of novel predictive biomarkers, and the search for new therapeutic targets are crucial for improving SCLC prognosis.
Dysregulated gene expression patterns and activity of the Notch family have been identified in SCLC. Interestingly, the frequency of gene mutations in the Notch signaling pathway among Chinese patients with SCLC is significantly lower compared to Western populations.143 Almodovar and colleagues reported that 52% of patients with SCLC exhibit inactivating mutations of Notch family genes in their plasma cell-free DNA.144 Another study found that 20–25% of SCLC cases carry loss-of-function Notch mutations.145,146 The cell surface protein DLL3, highly selective for tumors, is expressed in 85% of patients with SCLC.147 Notably, DLL3 expression remains robust across all stages of SCLC and remains stable despite therapeutic interventions. In a study involving postoperative patients with SCLC treated with platinum and etoposide plus anti-programmed cell death ligand 1 antibody, it was observed that SCLC with high DLL3 expression developed resistance to immunochemotherapy due to tumor immunosuppression, despite having a higher load of neoantigens.148 Functionally, DLL3 acts as a regulator of cell-cell interactions in the neuroendocrine state of SCLC.149 Numerous preclinical and clinical studies targeting DLL3 are underway, defining it as a promising treatment strategy for SCLC.150,151
With the exploration of molecular aberrations in SCLC, dysregulation of the Notch pathway has emerged as one of the driving factors in tumorigenesis and intratumoral heterogeneity in SCLC. Activated Notch signaling induces profound G1 cell cycle growth arrest and significantly decreases neoplastic potential. SCLC displays a high degree of heterogeneity, with multiple subtypes coexisting within individual tumors, exhibiting both neuroendocrine cell characteristics and non-neuroendocrine phenotypes in both mouse and human SCLC tumors.152 Ireland et al. demonstrated that MYC mediates the neuroendocrine plasticity of SCLC through the activation of Notch signaling.153 Specifically, endogenous activation of the Notch pathway leads to a fate switch from neuroendocrine to non-neuroendocrine in 10–50% of SCLC cells.154 Notch signaling plays a dual role in SCLC, acting as a tumor suppressor in neuroendocrine cells and as a driver of increased chemoresistance in non-neuroendocrine cells to support SCLC growth.155 Importantly, in preclinical models, the combination of Notch inhibition and chemotherapy effectively suppresses SCLC tumor growth and the generation of non-neuroendocrine cells. The recognition that the Notch pathway initiates tumor heterogeneity and treatment resistance in SCLC has inspired the development of personalized treatment strategies targeting Notch signaling for different SCLC subtypes.
Hematological malignancies
T cell acute lymphoblastic leukemia
T-ALL is an aggressive hematological malignancy, constituting 15% and 25% of ALL cases in children and adults, respectively, with a high recurrence rate and poor prognosis. This malignancy is characterized by acquired chromosomal translocations and genetic alterations, resulting in aberrant expression of transcriptional regulators.156,157 Notch signaling through Notch1 receptors is crucial for T cell lineage development, thymocyte survival, and proliferation of committed T cell progenitors.158,159 A seminal study has identified Notch1-activated point mutations in over 50% of T-ALL cases, underscoring the prominent role of Notch1 signaling cascades in T-ALL pathogenesis.160 For instance, in Ikaros-deficient T-ALL, T cell-specific deletion of floxed Notch1 promoter/exon 1 sequences promotes the activation of oncogenes and accelerates leukemia onset.161 Another study revealed that overexpression of intracellular Notch1 in hematopoietic progenitor cells leads to abnormal lymphatic development, crucial for tumor maintenance.162 Furthermore, abnormal expression of CD44 serves as an early marker of mutant Notch1 signaling and extrathymic T cell development, suggesting that Notch1 signaling may contribute to T-ALL pathogenesis by inducing CD44 expression.163
The presence of Notch1 mutations in patients with T-ALL raises questions regarding the prognostic impact of Notch signaling alterations. F-box and WD40 repeat domain containing-7 (FBXW7), an E3 ubiquitin ligase, has been reported to recognize and bind to the Notch1 PEST domain, leading to degradation of the activated form of Notch1.164,165 FBXW7 mutations stabilize intracellular Notch1 in the nucleus and are thought to work synergistically with the Notch1 PEST mutations.166,167 An early study involving 141 adult T-ALL samples identified 62% with Notch1 mutations and 24% with FBXW7 mutations. The study suggested that activation of the Notch1 pathway due to Notch1/FBXW7 mutations could identify patients with a favorable prognosis.168 Among 162 treated pediatric patients with T-ALL screened in the MRC UKALL2003 trial, those with double mutations of Notch1 and/or FBXW7 exhibited very positive outcomes.169 Overall, these studies indicate that Notch activation may be associated with improved early treatment response in T-ALL, and the impact on prognosis may be influenced by differences in treatment approaches.
Chronic lymphocytic leukemia
CLL is characterized by the expansion of monoclonal CD5+CD23+ B cells in peripheral blood, bone marrow, and secondary lymphoid tissues.170 CLL has a genetic susceptibility, with family members of patients with CLL having a 6–9 times increased risk.171 Recent advancements have unraveled the genetic landscape of CLL, exposing genomic heterogeneity among different patients with CLL.172,173 Approximately 10% of CLL cases carry Notch1 gene mutations at diagnosis.174,175 These mutations, located in the coding region or 3’ untranslated non-coding regions of the Notch1 gene, result in impaired degradation and accumulation of the Notch1 intracellular domain (N1ICD).176 CLL cells with Notch1 mutations display partial chemotherapy resistance in vitro, indicating that Notch1 could be a potential molecular target for CLL. Another study revealed that Notch1 mutations in CLL are associated with relative resistance to low CD20 expression and in vitro anti-CD20 immunotherapy, suggesting epigenetic dysregulation of CD20 expression mediated by histone deacetylases.177 A retrospective analysis of 317 Chinese patients with CLL identified Notch1 mutation as an unfavorable prognostic factor.178 Consistent results were observed in a prospective multicenter COMPLEMENT1 trial, linking Notch1 mutations to poor PFS.179 Further research is needed to explore the molecular mechanisms of Notch1 mutations, their impact on prognosis, and suitable strategies for treating patients with CLL with Notch1 mutations.
Abnormal Notch signaling accelerates the proliferation of CLL cells and contributes to disease progression.180,181 In B-cell CLL cells, the oncogenic gene Notch2 is highly expressed and associated with disease-specific apoptosis failure.182 Notch2 high expression characterizes a subset of patients with CLL, mainly carrying trisomy 12, which is marked by high levels of Mcl-1.183 Silencing Notch2 to reduce Mcl-1 expression can restore the response of CLL cells to venetoclax treatment. Additionally, Filomena et al. provided evidence that JAG1 is constitutively processed in CLL cells, and the activation of Notch1/2 is independent of the up-regulation of JAG1 levels.184 These findings offer new insights into Notch signaling in CLL cells and suggest that targeting the Notch signaling pathway could be developed as a novel therapeutic strategy for CLL.
Urinary system tumors
Bladder cancer
According to the World Health Organization, nearly 600,000 people are diagnosed with bladder cancer each year, with smoking and workplace exposure to suspected carcinogens being the main risk factors for bladder cancer.185,186 Bladder cancer is three to four times more common in men than in women.187 However, women are often diagnosed with advanced disease at the onset and have a poorer prognosis. Currently, there is a lack of ideal treatment methods for bladder cancer. Therefore, exploring the molecular mechanisms of bladder cancer and identifying early diagnosis and treatment targets holds promise for improving the prognosis of patients with bladder cancer. In recent years, the disparate roles of Notch signaling in bladder cancer have been established, with its oncogenic and tumor -suppressive effects depending on tissue type and cellular environment.188 Rampias et al. reported that more than 40% of human bladder cancers carry new inactivation mutations of Notch pathway components.189 Moreover, they found that activated Notch inhibits the proliferation of bladder cancer cells, indicating that the loss of Notch activity is a driver of urothelial carcinoma. Similarly, Maraver et al. revealed that missense mutations in Notch1 and Notch2 in human bladder cancer lead to functional loss of the Notch pathway, favoring the EMT process and promoting the aggressive character of bladder cancer.190
Overall, considerable research supports the function of Notch1 as a tumor suppressor in bladder cancer. In contrast, further work demonstrated that Notch2 functions as an oncogene. Hayashi et al. revealed a high incidence of increased Notch2 copy number in bladder cancer, and Notch2 activation is associated with a poorer prognosis.191 Additionally, the Notch2/HEY1 signaling pathway mediates cancer-associated fibroblasts (CAFs)-derived microfibrillar-associated protein 5 to promote the proliferation and metastasis of bladder cancer.192 In the case of Notch3, a study involving 614 urothelial bladder cancer samples showed that 91.5% of samples expressed Notch3, and the degree of positive Notch3 expression was positively associated with the risk of cancer-specific death.193 Moreover, the gene expression and protein levels of JAG2 were reported to be progressively up-regulated with the increase in primary tumor size and histopathological stage.194 Together, these findings provide evidence that Notch signaling has a dual role in bladder cancer. However, many unsolved problems about the mechanism of Notch signaling in bladder cancer still need further study in the future.
Prostate cancer (PCa)
PCa is the second most common cancer in men, with more than 1.2 million newly diagnosed cases worldwide each year.195 PCa is highly heterogeneous, and its progression can be driven by gene mutations and DNA damage response.196 Although the long-term survival rate of local PCa is satisfactory, metastatic PCa is largely incurable even after intensive comprehensive treatment.197 Over the decades, extensive evidence has suggested that Notch signaling is involved in prostate development and the maintenance of adult prostate homeostasis. Abnormal expression of Notch receptors and ligands leads to Notch signaling dysfunction, which regulates tumor formation and progression in PCa as an oncogene or tumor suppressor gene.198 For example, previous studies have shown that local high-risk PCa and metastatic castration-resistant PCa cells express high levels of Notch1 receptors, and activated Notch1 cooperates with multiple carcinogenic pathways to drive the invasiveness of PCa.199
PCa metastasis primarily occurs in the bone, where it induces a unique osteoblastic response. Studies have found that Notch3 expression is elevated in human PCa bone metastasis.200 Notch3 inhibits osteoclasts and stimulates osteoblastogenesis by inducing MMP-3, thereby promoting osteoblast bone metastasis. A study involving 154 PCa samples indicated that JAG1 expression is higher in metastatic PCa than in localized PCa or benign prostate tissue.201 Additionally, high expression of JAG1 in clinically localized tumors is apparently related to recurrence. Mechanistically, in the phosphatase and tensin homolog (PTEN)-deficient PCa mouse model, overexpression of JAG1 can up-regulate transforming growth factor-beta (TGF-β) signaling in prostate stromal cells and promote the formation of a reactive matrix microenvironment.202 In addition, Tran et al. found that overexpression of JAG1 intracellular domain (JAG1-ICD) enhances the stem-like characteristics and mobility of PCa cells.203 In patients with advanced metastatic PCa, Chou et al. revealed that DLL3 is expressed in de novo and advanced small cell/neuroendocrine carcinoma (SCNC) PCa, and is associated with poor survival rates.204 Immunotherapy targeting DLL3 showed anti-tumor potential in invasive SCNC. Although a large number of studies based on clinical PCa samples, cancer cell lines, and animal models have suggested that Notch pathway elements are dysregulated in PCa, the function of Notch signaling in PCa is still not fully determined.205,206 Based on the current knowledge, more sufficient research is still needed to provide reliable evidence for targeting the Notch signaling pathway in the treatment of PCa.
Renal cell carcinoma (RCC)
RCC is the most common malignancy of the genitourinary system, with a mortality rate of 30–40%.207 Previous studies have shown that several key molecules of the Notch cascade are expressed during nephrogenesis, and dysregulated Notch activity may play a vital role in the development of RCC.208,209 Sun et al. observed that the expression of Notch1 and Notch4 in RCC was either absent or significantly down-regulated compared with adjacent non-tumor tissues.210 Functionally, HES1-mediated down-regulation of microRNA miR-138 maintains the activation of the Notch1 pathway and facilitates the malignant progression of RCC.211 Consistently, selective Notch1 suppression by small interfering RNA could inhibit RCC cell proliferation via the JNK/p38 pathway.212 Another study revealed that Notch3 was positively correlated with chromophobe RCC, unbroken capsule, Fuhrman grade 1, and less lymph node involvement.213 Wang et al. found that DLL4 may participate in the development of RCC by engaging in signal transduction and angiogenesis.214 Blocking DLL4 showed effective antitumor activity in RCC patient-derived xenografts.215 Together, these studies suggest that the Notch pathway may represent previously overlooked treatment opportunities for RCC.
Clear cell RCC (ccRCC) is the most common histological subtype of RCC, accounting for ~75% of all kidney cancers.216 ccRCC is characterized by heterogeneity and potential genetic predisposition. A study involving 415 patients with ccRCC found that 44% of Notch genes had genetic alterations, with copy number variation being the main type of gene variation.217 Additionally, patients with ccRCC with Notch pathway alterations had better OS. Another study found that the expression of Notch1 and JAG1 in ccRCC tissues was higher than in normal adjacent tissues.218 The upregulation of Notch1 signaling promotes the proliferation and migration of tumor cells, increasing the risk of metastasis in T1 stage ccRCC. In addition, the expression of DLL4 and JAG1 in ccRCC were significantly higher than those in normal renal tissues and were positively correlated with poor prognosis.219,220
Reproductive system tumors
Breast cancer
Breast cancer is the most commonly diagnosed malignancy in women, accounting for 31% of all female cancers.221 For nearly half a century, the incidence of breast cancer has continued to rise.222 Although the development of surgery, radiotherapy, chemotherapy, endocrine therapy, and targeted therapy has improved the 10-year survival rate of breast cancer, 30–40% of patients still face significant challenges of metastasis and recurrence. Over the years, Notch has been implicated as a contributor to breast cancer, potentially due to its role in breast cancer stem-like cell (CSLC) characteristics, EMT, resistance to chemotherapy, and other processes.223–225 Studies have revealed that highly expressed Notch1, Notch4, JAG1, DLL3, and DLL4 are observed in breast cancer with poor prognosis, suggesting that Notch signaling is promising as a biomarker for breast cancer prognosis.226–230 The expression of Notch2 in rs11249433 risk genotype (AG/GG) carriers was significantly increased, which may promote the development of estrogen receptor-positive luminal breast cancer.231 Functionally, Notch signaling activates aldehyde dehydrogenase 1A1 (ALDH1A1) by inducing Sirtuin-2, resulting in ALDH1A1 deacetylation and enzymatic catalysis, accelerating breast CSCs.232 Claudin-low breast cancer is thought to originate from breast stem cells, characterized by stemness and an EMT phenotype. Zhang et al. reported that Notch mediates Manic Fringe-induced PIK3CG transcription, promoting the Claudin-low breast cancer phenotype.233 As early as 2006, Myc gene was identified as a direct transcriptional target of Notch1 and a necessary factor for Notch1-induced breast tumorigenesis in mice.234 Besides, Notch1 activation promotes triple-negative breast cancer (TNBC) formation by initiating ATR-CHK1 signaling cascade, restoring S/G2 and G2/M cell cycle checkpoints, and inhibiting mitotic catastrophe.235 Additionally, Notch signaling regulates the EMT process of breast cancer cells through various mechanisms, such as a Slug-dependent manner, the S100A16/Notch1 pathway, the FYN/STAT5 pathway, and the Notch4/STAT3 signaling pathway.236,237 Notch ligands have been proven to play an important role in breast cancer drug resistance. Collectively, Notch signaling plays a carcinogenic function in breast cancer.238,239 Robinson and colleagues identified functionally recurrent rearrangements of Notch gene families in breast cancer, with certain therapeutic implications.240 Given the complexity of the Notch pathway, further exploration is needed to develop successful Notch targeting strategies to prevent and treat breast cancer.
Ovarian cancer (OC)
OC is one of the leading causes of cancer death in women, with a five-year survival rate of ~50% and an even worse prognosis for metastatic disease.241 Early identification of high-risk women for OC is crucial due to the predominance of nonspecific symptoms occurring in the late clinical stages. In recent years, Notch signaling has been increasingly studied in OC, which may be used as a biomarker to predict prognosis. A study of 328 patients with primary OC revealed that high expression of N1ICD in female OC is an independent risk factor for poor prognosis.176 Consistent results were obtained through large data portals, suggesting that upregulation of Notch signaling family proteins in OC is generally related to poorer survival and more advanced cancer stages.242 Another study observed that Notch3 overexpression was related to shorter survival in patients with advanced OC treated with platinum and taxane.243 DLL4 was found to be overexpressed in 72% of OC, which is an independent predictor of poor survival.244
Many studies have linked Notch signaling components to the malignant characteristics of OC. JAG1 promotes the EMT process of OC by crosstalk with the JAK/STAT3 pathway, further enhancing the invasion and migration ability of platinum-resistant OC.245 In OC, tumor-associated neutrophils activate JAG2 to coordinate the intratumoral IL-8-driven immune evasion microenvironment.246 Resistance to standard treatment regimens is one of the main reasons for the poor prognosis of OC. The Notch1 signaling pathway mediates paclitaxel (PTX) resistance in CD44+CD117+ OC cells promoted by chemokine CCL20.247 The activated Notch3 pathway mediates the nuclear receptor NR2F6 to promote epithelial OC (EOC) cells’ resistance to cisplatin.248 Similarly, Notch3 enhances EMT in OC cells and attenuates carboplatin-induced apoptosis.249 Together, these studies elucidate the molecular mechanisms by which the Notch signaling pathway contributes to OC aggressiveness and chemotherapy resistance in vivo and in vitro.
Cervical cancer (CC)
CC is a major public health problem affecting middle-aged women. From 2001 to 2019, 227,062 new cases of CC were reported in the United States.250 There is conclusive evidence that the high-risk subtype of human papillomavirus (HPV) infection is a leading cause of CC.251 Talora et al. reported that the specific down-regulation of Notch1 signaling in CC cells leads to the continuous transcription of HPV-driven E6/E7 viral genes and plays a key role in HPV-induced advanced carcinogenesis.252 In turn, the activated Notch1 signaling inhibits the activity of E47 through the RBP-Jk-dependent mechanism, inducing the growth arrest of HPV-positive CC cells.253 However, Yousif et al. found that Notch1 and JAG1 were overexpressed in CC and were associated with poor OS.254 Another study showed that HPV16 E6 could induce the continuous expression of DLL4 in keratinocytes, and the high expression of DLL4 was closely related to the poor prognosis of CC.255 The role of Notch signaling in CC is complex.256,257 CD66+ cells in primary invasive CC exhibit high Notch signaling and tumorigenicity.258 Activation of Notch signaling can induce cell cycle arrest in human CC cells.259,260 In addition, several research studies have shown that inhibition of Notch signaling can strengthen the sensitivity of CC cells to chemotherapeutic drugs.261 These studies have strengthened the view that dysregulated Notch signaling is associated with the progression of CC and laid the foundation for a detailed exploration of targeted therapy.
Nervous system neoplasms
Glioma
Glioma is a malignant brain tumor derived from glial progenitor cells, accounting for 80.8% of primary central nervous system tumors, resulting in serious morbidity and mortality.262 Previous researches have analyzed the integrated genomic characteristics of gliomas. Notch1 mutations were identified in diffuse lower-grade gliomas, Notch1 and Notch2 mutations were identified in IDH1-mutant gliomas, and grade II and III gliomas carried Notch1–4 mutations.263–265 Furthermore, Halani et al. uncovered that Notch1 mutations were related to disease progression and shorter survival in oligodendroglioma.266 Accumulating studies have highlighted the importance of the Notch signaling pathway in glioma malignancy. Recent research revealed that the expression of Notch1 and Notch3 was significantly increased in glioblastoma and promoted tumor growth activity through the NF-κB pathway.267,268 In addition, DLL3 was found to be up-regulated in IDH1 mutant gliomas and was associated with a better prognosis.269 Overall, the differential expression pattern of Notch1–4 receptors can be used as a marker of glioma differentiation and a possible prognostic factor.270 Functionally, Notch signaling is involved in glioma progression through complex mechanisms. For example, Notch signaling mediates miR-33a-driven self-renewal of glioma-initiating cells.271 Silencing Notch1 can induce autophagy and down-regulate the Notch1/HES1 pathway to inhibit the proliferation of glioma cells.272 Moreover, a large amount of evidence suggests that Notch signaling is involved in maintaining the characteristics of glioma stem cells (GSCs).273 However, Parmigiani et al. found that inhibition of Notch signaling can make proneural glioma cells evade immune monitoring and increase invasiveness.274 In summary, the Notch signaling pathway is heavily involved in the fate determination of glioma cells, which is related to the progression of gliomas. Targeting the Notch pathway may intervene in these processes and potentially bring better therapeutic effects for patients with glioma.
Neuroblastoma
Neuroblastoma is a neuroendocrine tumor originating from the sympathetic nervous system, characterized by genetic, morphological, and clinical heterogeneity.275 Advances in high-throughput technology have contributed to understanding the genetic changes and molecular pathways involved in the pathogenesis of neuroblastoma, including MYCN amplification, PHOX2B mutation, the PI3K/AKT/mTOR pathway, and Notch signaling. Previous studies have shown that Phox2B can control the expression of Delta-Notch pathway genes by regulating HASH1.276 Activation of the Notch signaling pathway leads to growth arrest in neuroblastoma.277 Approximately 20% of neuroblastomas carry MYCN oncogene amplification, which is related to decreased expression of genes encoding gamma-secretase subunits and Notch signaling components.278 In the MYCN transgenic neuroblastoma model, Notch2 signaling mediates Midkine to promote the formation and occurrence of neuroblastoma.279 Axelson’s study demonstrated that the Notch signaling cascade regulates HASH-1/HES-1 to participate in the differentiation of neuroblastoma cells and regulate malignant phenotypes.280 Hooper et al. revealed the presence of N1ICD in the sub-nuclear bodies and primary cortical neurons of SH-SY5Y neuroblastoma. High expression of Notch1 in neuroblastoma indicates a poor prognosis and is expected to be a therapeutic target for patients with neuroblastoma.281 Additionally, the Notch3 feed-forward loop drives the transcriptional reprogramming of neuroblastoma from adrenergic to mesenchymal states.282 Notch3 endows neuroblastoma cells with a highly motile phenotype, and the subpopulation with high expression of Notch3 and its downstream regulatory genes has mesenchymal characteristics, making it prone to metastasis and associated with a worse prognosis.283 These findings reveal the molecular mechanism of Notch signaling in neuroblastoma, which are of strategic significance for improving drug treatments in this cancer type.
Tumors of other systems
Melanoma
Melanoma, the most aggressive form of skin cancer, poses a significant global public health challenge.284 It is estimated that, by 2020, there were a total of 325,000 new cases of melanoma worldwide, resulting in 57,000 deaths.285 If the incidence rate continues at the 2020 level, there is projected to be a ~50% increase in new cases of melanoma and a 68% increase in deaths by 2040. Notch signaling is believed to play a dual role as both oncogenes and tumor suppressor genes in melanoma.286,287 Overexpressed Notch1 signaling promotes melanoma-induced immunosuppression by upregulating TGF-β1.288 Additionally, in vitro studies have revealed that Notch1 signaling in CAFs acts as a molecular switch, reversing the plasticity and stemness of CSCs, thus regulating the heterogeneity and invasiveness of melanoma cells.289 Similarly, miR-146a-5p is transferred to astrocytes via extracellular vesicles, down-regulating NUMB and activating the Notch pathway, thereby promoting melanoma brain metastasis.290 Conversely, in the context of PTEN deficiency, Notch1 and Notch2 exhibit anti-tumor effects in BRAFV600E/PTEN-null-driven melanoma genesis.291 Likewise, Rad et al. reported that Notch4 acts as a tumor suppressor in melanoma.292 In NRAS wild-type melanoma, tumors with Notch4 mutations exhibit a higher tumor mutation burden and tumor neoantigen burden.293 Notch4-mutant tumors enhance anti-tumor immunity, resulting in a better immune therapy response and prognosis. According to the aforementioned studies, the function of Notch signaling in melanoma is highly dependent on the environment, and detailed investigations are still required to elucidate the relevant molecular mechanisms.
Osteosarcoma
Osteosarcoma stands as the most prevalent primary bone malignancy, demonstrating high heterogeneity and primarily affecting children, adolescents, and young adults.294 Despite significant advancements in chemotherapy and surgery, the survival rate of patients with osteosarcoma has shown no improvement in recent decades.295 Studies have revealed that molecules from the Notch signaling family are consistently overexpressed in the majority of clinical osteosarcoma samples, correlating positively with recurrence, metastasis, and poor prognosis.296–298 Both in vivo and in vitro experimental investigations have indicated that Notch signaling plays a crucial role in regulating the cell cycle of osteosarcoma, influencing its recurrence, lung metastasis, and malignant progression.299,300 Furthermore, the up-regulation of JAG1 expression has been linked to promoting the stem-like phenotype and tumor growth of osteosarcoma.301 Conversely, Notch signaling has been found to modulate the sensitivity of osteosarcoma to chemotherapy resistance.302,303 In summary, our current understanding of the intricate function of Notch in osteosarcoma is just scratching the surface, and further comprehensive research holds the potential to facilitate its clinical transformation in tumor therapy.
Thyroid cancer
Thyroid cancer is a prevalent malignancy within the endocrine system, comprising four primary histological subtypes: papillary thyroid cancer (PTC), follicular thyroid cancer (FTC), medullary thyroid cancer (MTC), and anaplastic thyroid cancer.304 The Notch receptor and ligand family have been identified as regulators of tumorigenesis in thyroid cancer.305 Notably, Simonetta et al. observed that the expression pattern of Notch1 varies across different histological types of thyroid cancer.306 Specifically, Notch1 positivity is predominantly limited to papillary carcinoma, rarely detected in follicular carcinoma and medullary carcinoma. Positive expressions of Notch1 and DLL4 were identified in PTC, showing a significant correlation with tumor invasion, metastasis, and poor prognosis.307,308 Conversely, the expression of Notch3 decreased in FTC specimens exhibiting reduced differentiation and increased malignancy, linking to clinicopathological features associated with poor prognosis.309
MTC represents a distinct type of neuroendocrine tumor originating from thyroid C cells. In MTC cells, the role of Notch signaling differs from its function in PTC cells.310 Muthusamy et al. confirmed that overexpression of N1ICD inhibited MTC cell proliferation and altered the neuroendocrine phenotype of MTC cells.311 Similarly, Notch2 also mediated cell apoptosis and inhibited neuroendocrine markers in MTC.312 Despite the limited and at times contradictory nature of research on the Notch pathway in thyroid cancer thus far, in-depth investigations specific to different histological subtypes of thyroid cancer are deemed necessary to elucidate the inconsistent functions of Notch in thyroid cancer comprehensively.
Oral squamous cell carcinoma (OSCC)
OSCC is the most prevalent oral malignant tumor worldwide, with its poor prognosis primarily attributed to metastasis and recurrence.313 Early diagnosis holds the potential to positively impact the survival rates of OSCC. Research indicates that Notch receptors, specifically Notch1,314,315 Notch3,316 and Notch4,317,318 exhibit high levels in human OSCC tissues and are associated with a poor prognosis. This suggests that Notch receptors could serve as biomarkers for the early diagnosis of OSCC. Functionally, the activated Notch-HES1 signaling pathway plays a crucial role in mediating the stem-like phenotype of OSCC and actively contributes to the progression of the disease.319 Furthermore, the up-regulation of Notch signaling demonstrates carcinogenic properties, promoting the proliferation, migration, and invasion of OSCC cells, thereby contributing to the malignant characteristics of OSCC.320,321 Importantly, Notch signaling also interacts with other cell signaling pathways, such as Wnt and Hedgehog, intensifying the aggressiveness of OSCC.322 A comprehensive understanding of the molecular mechanisms underlying Notch signaling in OSCC is imperative for the development of targeted therapeutic strategies aimed at tackling this challenging oral malignancy.
Head and neck squamous cell carcinoma (HNSCC)
HNSCC encompasses a diverse group of tumors originating from the squamous epithelium of the oral cavity, pharynx, and larynx.323 Over the past half-century, there has been a decline in the incidence of smoking-related HNSCC, while cases induced by HPV infection have seen a gradual increase.324 Studies focusing on the long tail genes of HNSCC have revealed that 67% of carcinogenic mutations in human HNSCC cases converge on Notch signaling, establishing Notch inactivation as a marker for HNSCC.325–327 This aligns with the recognized role of Notch1 as a tumor suppressor in HNSCC. Genomic analysis has indicated a significant mutation rate in Notch1, ranking it as the gene with the second-highest mutation frequency after TP53.328,329 In HNSCC tissues, Notch1 is highly expressed compared to normal tissues and is associated with a favorable prognosis.330,331 Targeting hypoxia-inducible factor 1 alpha (HIF1α)/Notch1 signaling has been found to mitigate the stem-like characteristics and chemotherapy resistance of HNSCC CD44+ cells.332 Despite these insights, our understanding of the intricate functions of Notch signaling in HNSCC remains at a preliminary stage.333,334 Elucidating how Notch signaling acts as either an oncogene or a tumor suppressor at different stages of tumorigenesis holds the key to developing new drug targets for HNSCC.
The mechanism of Notch signaling patyway-mediated tumorigenesis and progression
Over the past two decades, extensive investigations have revealed that the Notch signaling pathway is intricately involved in various facets of cancer biology.335–337 This includes its role in EMT, angiogenesis, the acquisition of CSLC properties, metabolic reprogramming, regulation of the TME, and mediation of chemotherapy resistance. Dysregulation of Notch signaling can function either as an oncogene or a tumor suppressor, exerting influence over the progression of tumors. In the following section, we have compiled and emphasized the molecular mechanisms underlying Notch signaling-mediated tumorigenesis and progression (Table 2). Our aim is to offer new insights into potential targeted therapies for various types of cancers.
Table 2.
Mechanisms of Notch signaling pathway-mediated tumorigenesis and progression
| Cancer | Notch signaling component | Role | Involved biological process | Mechanism | Related molecule | Cell line | Ref. |
|---|---|---|---|---|---|---|---|
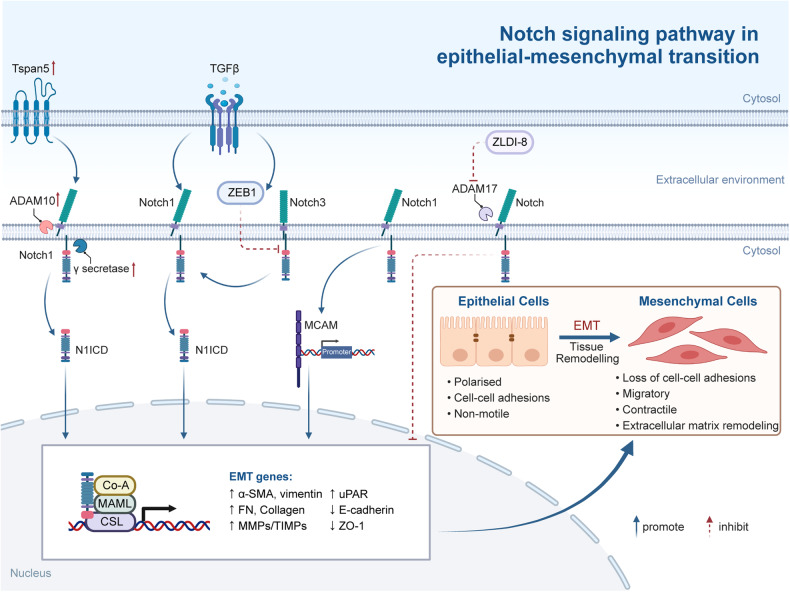
| ESCC | Notch1, Notch3 | Oncogene | EMT | TGF-β→active Zeb1→represses Notch3→limiting terminal differentiation; TGF-β→active Notch1→drive EMT→promote SCC tumor initiation | TGF-β, ZEB1 | TE11, EN60, EPC2T | 346 |
| HCC | Notch1 | Oncogene | EMT | Tspan5 ↑ →Activation of Notch signaling→enhance EMT and actin skeleton rearrangement→promote tumor metastasis | Tspan5 | HL7702, BEL7402, Hep3B, HUH7, MHCC97H, MHCC97L, PLC, QGY7701, SK-Hep1 | 345 |
| SCLC | Notch2, HES1 | Oncogene | EMT | ZLDI-8→inhibit the Notch signaling→inhibit migration, invasion and EMT phenotype of drug-resistant lung cancer | ZLDI-8 | A549 cells, resistance A549/Taxol cells | 354 |
| Glioblastoma | RBPJ | Oncogene | EMT, chemoresistance | RBPJ ↓ →blocks EMT activators→reduce cellular invasion and resistance | ZEB1 | GBM1, BTSC407/407p, JHH520/JHH | 353 |
| HNSCC | Notch4, HEY1 | Oncogene | EMT | Notch4-HEY1 pathway ↑ →promote EMT→induce proliferation and cisplatin resistance | E-cadherin, Vimentin, Fibronectin, TWIST1, and SOX2 | SKN3, Cal27, SCC61, and SCC090 | 347 |
| ACC | HEY1, Notch1 | Oncogene | EMT | Notch1-HEY1 pathway ↑ →drive self-renewal and EMT→increase proliferation, invasion and metastasis | MMPs | The SACC-LM cell line | 348 |
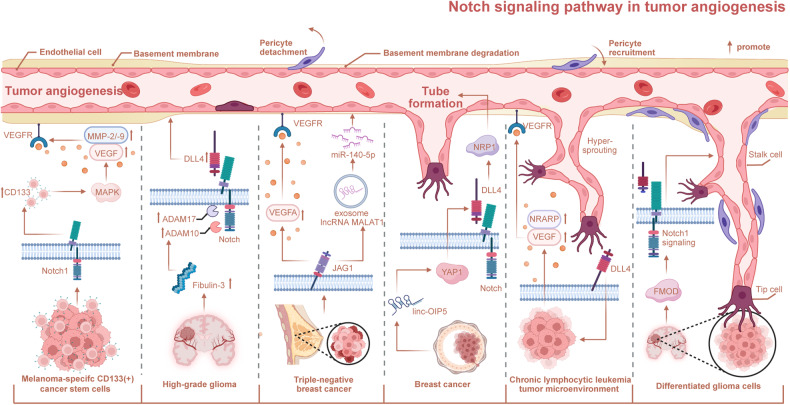
| NSCLC | Notch1, DLL4 | Oncogene | Angiogenesis | ZLDI-8→suppress Notch1-HIF1α-VEGF signaling pathway→inhibit angiogenesis and vasculogenic mimicry | ZLDI-8, HIF1α, VEGF | The HUVECs, resistance A549/Taxol cells | 377 |
| CLL | DLL4, Notch1 | Oncogene | Angiogenesis | DLL4→active Notch signaling→cell migration and angiogenesis↑ | CXCR4 | OP9-DLL1 and -DLL4 cells, Mouse OP9 BM stromal cells | 363 |
| Breast cancer | JAG1, DLL4, Notch1 | Oncogene | Angiogenesis | linc-OIP5 ↓ → and JAG1 ↓ →disrupted DLL4/Notch/NRP1 signaling→suppress proliferation, migration, and tube formation | Linc-OIP5, YAP1, NRP1 | MCF-10 cells, MDA-MB-231 cells, MCF-7 cells, HUVECs | 371 |
| Breast cancer | JAG1 | Oncogene | Angiogenesis | JAG1 ↑ → MALAT1-miR-1405p-JAG1/VEGFA pathway ↑ →microenvironment angiogenesis↑ | MALAT1, miR-140-5p, VEGFA | MCF-10A, T47D, MCF-7, MDA-MB-231 (231), MDA-MB-231 Bone (231B), HUVEC | 370 |
| Glioma | Notch1 | Oncogene | Angiogenesis | DGC-secreted FMOD→activate integrin-dependent Notch1 signaling→promote angiogenesis | FMOD | GSCs MGG4, MGG6, and MGG8, DBT-Luc cells, ST1 endothelial cells, B.End3 cell line, The U251/U87/LN229 cell line | 376 |
| Glioma | DLL4, Notch1, NICD | Oncogene | Angiogenesis | Fibulin-3→active Notch signaling→DLL4 ↑ →promotes glioma angiogenesis | Fibulin-3 | CNS1, GSCs GBM8 and GBM34, HBMECs | 364 |
| Melanoma | Notch1, N1ICD | Oncogene | Angiogenesis | Notch1 ↑ →CD133 ↑ →mitogen-activated protein kinase activation ↑ →melanoma growth, angiogenesis, and lung metastasis↑ | CD133, p-p38 | CD133 + , CD133- or unsorted B16F10-Luc cells | 375 |
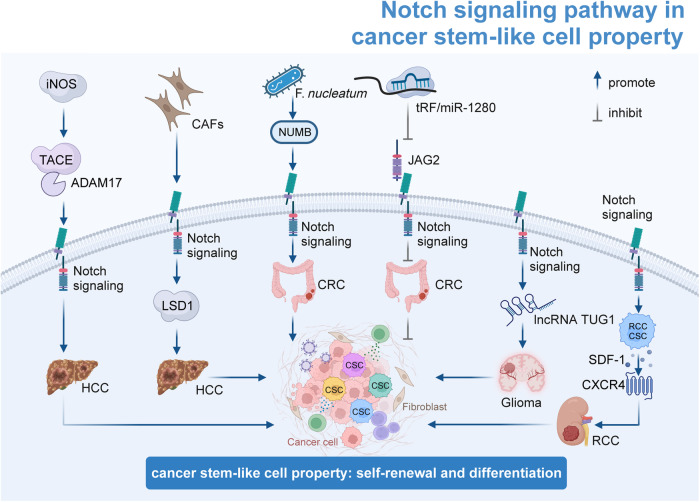
| CRC | NICD | Oncogene | Stem-like properties | Lipids→recruit Numb→MDM2 degrade Numb→activate Notch signaling→promote stem-like cell features | Lipids, Numb, MDM2 | HCT116, HT29 | 387 |
| CRC | JAG2, Notch1, Notch2 | Oncogene | Stem-like properties | tRF/miR-1280 ↑ →inhibit Notch signaling→suppress CSC phenotypes→suppress CRC growth and metastasis | tRF, miR-1280 | HCT116, HCT15, HT29, Panc-1, 293 T cells | 388 |
| Liver cancer | Notch1 | Oncogene | Stem-like properties | iNOS/NO→activate TACE/ADAM17→activate Notch1 signaling→aggressive cancer phenotype | iNOS/NO, TACE/ADAM17 | HLE, MHCC-97H, PLC/PRF/5, HepG2 | 393 |
| LUAD | Notch | Oncogene | Stem-like properties | HIF1ɑ-regulated miR-1275→ activate Wnt/beta-catenin and Notch signaling→enhance the stemness of LUAD→promote tumorigenicity, recurrence, and metastasis | HIF1ɑ, miR-1275 | EBAS-2B, L78, H460, A549, GLC-82, SPC-A1, PC9, H1299, H1975, H2228, A549-luc, H1299-luc | 397 |
| Breast cancer | Notch1, Notch3, Notch4, Hey1 | Oncogene | Stem-like properties | Syndecan-1→regulate IL-6/STAT3, Notch and EGFR signaling pathways→modulate CSC phenotype | Syndecan-1 | SUM-149, SKBR3 | 399 |
| Glioma | Notch1 | Oncogene | Glioma stem cells stemness | Notch1 ↑ →lncRNA TUG1 ↑ →sponging miR-145 and recruiting polycomb→maintain stemness features | lncRNA TUG1, miR-145 | 1228-GSC, 222-GSC | 389 |
| Breast cancer | NICD, Hey2 | Oncogene | Stem-like properties, chemoresistance | HIF2α ↑ →activate Wnt and Notch pathways→ promote stem phenotype conversion and induce resistance | HIF2α | MCF7, MDA-MB-231 | 398 |
| PDAC | Notch3, Notch4 | Oncogene | Stem-like properties | KLF10 ↓ →activate Notch signaling→promote stem cell phenotype and tumorigenesis | KLF10 | BCRC 60284, BCRC 60139 | 403 |
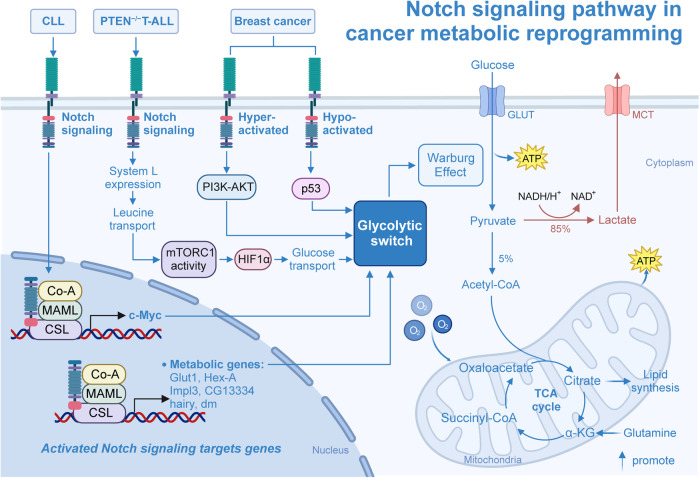
| GC | Notch1 | Oncogene | Glycolysis, chemoresistance | Pyrimidine biosynthesis→augmente Notch signaling→critical glycolytic enzymes ↑ →enhance aerobic glycolysis→confer chemoresistance | c-Myc and PKM2 | AGS, HGC-27 | 434 |
| Lung cancer | Notch1 | Oncogene | Glycolysis | Notch1/TAZ axis→inhibit cytotoxic T cell activity, promote aerobic glycolysis and immune escape | TAZ | A549, PC9 | 416 |
| T-ALL | Notch1 | Oncogene | Metabolic reprogramming | Notch1 signaling ↓ →inhibition of glutaminolysis→induce a metabolic shutdown | PTEN | (HEK) 293 T, HPB-ALL, DND41 | 428 |
| OC | NICD | Tumor suppressor | Glycolysis | miR-101 and miR-26a ↑ → imposed glucose restriction on T cells→EZH2 ↓ →inhibit the Notch pathway→inhibit T cell survival→poor antitumor immunity | miR-101 and miR-26a, EZH2 | HEK293T | 419 |
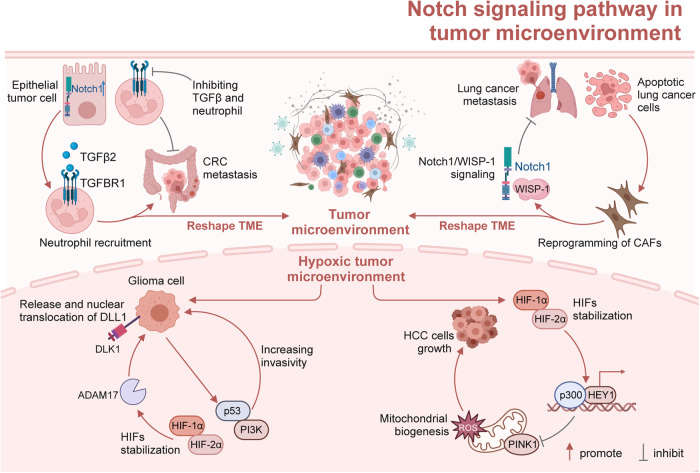
| CRC | Notch1 | Oncogene | Tumor microenvironment | Notch1 ↑ →promote TGF-β-mediated neutrophil infiltration recruitment→drive metastasis | TGF-β | villinCreER; KrasG12D/+; Trp53fl/fl Rosa26N1icd/+ organoids; villinCreER; Apcfl/fl; KrasG12D/+; Trp53fl/fl TgfbrIfl/fl organoids | 442 |
| HCC | HEY1 | Oncogene | Hypoxia | Hypoxia→HIF-1 ↑ → HEY1 ↑ → PINK1 ↓ →reduced oxidative stress, increased HCC growth | HIF-1, PINK1 | HeLa, PLC/PRF/5, HepG2, Hep3B | 461 |
| Lung cancer | Notch1 | Tumor suppressor | Tumor microenvironment | Apoptotic lung cancer cells→trigger Notch1 signaling→WISP-1 production ↑ →preventing the migration and invasion | WISP-1 | 344SQ, A549, HCT116 | 444 |
| Glioma | Notch1 | Oncogene | Tumor immune microenvironment | NKAP ↑ →Notch1 ↑ →active Notch1-dependent immune-suppressive tumor microenvironment→promote glioma growth and invasion | NKAP, SDF-1, M-CSF | U251, U87, Gl261 | 447 |
| Glioma | DLL1 | Oncogene | Hypoxia | hypoxia→Notch signaling ↓ →HES1 and Hey1 ↓ →promote neuroendocrine differentiation | NSE, β3-tubulin | LNCaP, PC-3, Du145 | 458 |
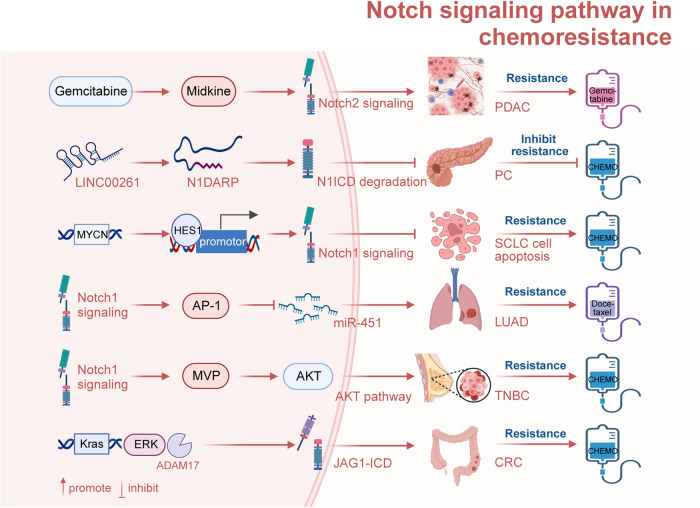
| CRC | Notch2, RBPJ | Oncogene | Chemoresistance | miR195-5p→Notch2 and RBPJ ↓ →reduced CRC cell sternness and chemoresistance | miR195-5p | SW480, SW620, HT-29, HCT-160 | 486 |
| PC | Notch2 | Oncogene | Chemoresistance | Midkine–Notch2 interaction→activated Notch signaling→increase chemoresistance | Midkine | PaCa 5061, 5072, and 5156; L3.6pl (19), BxPC-3, PANC-1 cells | 472 |
| PC | N1ICD | Oncogene | Chemoresistance | LINC00261 → N1DARP→inhibit USP10-Notch1 oncogenic signaling →inhibit chemoresistance | LINC00261, N1DARP, USP10 | Aspc1, Bxpc3, Capan1, MiaPaCa2, Panc1, Sw1990, and Patu8988; HPNE; MDA-MB-231 and MCF-10A; A549 and BEAS-2B; HCT116 and NCM460 | 476 |
| PC | Notch1 | Oncogene | Chemoresistance | GAS41 → H2A.Z.2→Notch1 ↑ →GEM resistance | GAS41, H2A.Z.2 | PANC-1, BXPC3, Capan-1, MiaPaCa-2, SW1990 | 475 |
| LUAD | Notch1 | Oncogene | Chemoresistance | Notch1/AP-1/miR-451/MDR-1 signaling axis→influence chemoresistance of LUAD | AP-1, miR-451, MDR-1 | SPC-A1 and H1299 | 480 |
| Breast cancer | Notch1 | Oncogene | Chemoresistance | Exosomal miR-378a3p and miR-378d → EZH2/STAT3 signaling→activate WNT and Notch stemness pathways→induce drug resistance | miR-378a3p, miR-378d, EZH2, STAT3 | CAL51, MDA-MB231, MCF-7 | 483 |
| TNBC | Notch1 | Oncogene | Chemoresistance | Notch1→MVP→activate AKT pathway→promote EMT→promote chemoresistance | MVP | MDA-MB-231, BT549, MCF-7, T47D, SKBR-3, HCC1937, ZR751 | 484 |
| Skin tumor | Notch1 | Tumor suppressor | N/A | Notch1 ↓ →Gli2 ↑ →basal-cell carcinoma-like tumors ↑ ; Notch1 ↓ →beta-catenin ↑ →skin carcinogenesis | Gli2, beta-catenin | Primary keratinocytes | 496 |
| Skin tumor | Notch1 | Tumor suppressor | Tumor microenvironment | Notch1 ↓ →epidermal differentiation/barrier formation defects→skin carcinogenesis | N/A | N/A | 498 |
| Multifocal epithelial tumor | CSL/RBP-Jκ | Tumor suppressor | N/A | CSL/RBP-Jκ ↓ →c-Jun and c-Fos ↑ →promote tumor cell proliferation | c-Jun, c-Fos | Dermal fibroblasts | 499 |
| SCC | Notch1 | Tumor suppressor | N/A | Notch1 ↑ →ROCK1/2 and MRCK α kinases ↓ →suppress SCC | p53, ROCK1/2, MRCK α kinases | SCC011, SCC012, SCC022, SCC028 | 500 |
| SCLC | Notch | Tumor suppressor | N/A | Notch ↑ →hASH1 ↓ →cell cycle arrest | hASH1, p21, p27kip1, ERK1, ERK2, raf/MEK/MAPK | DMS53, NCI-H209, Low passage 293, NCI-H209/ΔRaf-1:ER cells | 502 |
| Forebrain tumor subtypes | Notch1, Notch2, RBPJ | Tumor suppressor | N/A | Notch1/Notch2/RBPJ ↓ →accelerate glioma | p53, Trp53 | Platinum-E cells | 503 |
| Cervical cancer | Notch1 | Tumor suppressor | N/A | Notch1 ↑ →SST, SSTR1 and SSTR2 ↑ →inhibit cervical cancer | SST, SSTR1 and SSTR2 | HeLa, HeLa-GFP, HeLa-ICN1 | 257 |
| K-ras–induced PDAC | Notch1 | Tumor suppressor | N/A | Notch1 ↓ →increase tumor incidence and progression | K-ras | Primary pancreatic ductal cells | 504 |
| B-ALL | HES1 | Tumor suppressor | N/A | HES1 ↑ → PARP1 ↑ →cell apoptosis | PARP1, bHLH | JM1, Nalm6, 697, SupT1, Molt4, HEK-293 | 505 |
ESCC esophageal squamous cell carcinoma, HCC hepatocellular carcinoma, SCLC small cell lung cancer, HNSCC head and neck squamous cell carcinoma, ACC adenoid cystic carcinoma, NSCLC non-small cell lung cancer, CLL chronic lymphocytic leukemia, CRC colorectal cancer, LUAD lung adenocarcinoma, PDAC pancreatic ductal adenocarcinoma, GC gastric cancer, T-ALL T cell acute lymphoblastic leukemia, B-ALL B cell acute lymphoblastic leukemia, OC ovarian cancer, PC pancreatic cancer, TNBC triple-negative breast cancer, EMT epithelial-mesenchymal transition, TGF-β transforming growth factor beta, Zeb1 zinc finger E-box-binding homeobox 1, SCC squamous cell carcinoma, Tspan5 tetraspanin-5, MCAM melanoma cell adhesion molecule, RBPJ recombining binding protein suppressor of hairless, HEY1 hairy and enhancer of split 1, HIF1α hypoxia-inducible factor 1-alpha, VEGF vascular endothelial growth factor, DLL4 delta-like ligand 4, JAG1 Jagged-1, linc-OIP5 long intergenic non-protein coding RNA OIP5-AS1, MALAT1 metastasis-associated lung adenocarcinoma transcript 1, miR microRNA, FMOD fibromodulin, DGC discoidin domain-containing receptor 1, CD133 cluster of differentiation 133, MDM2 mouse double minute 2, tRF transfer RNA-derived fragment, CSC cancer stem cell, iNOS inducible nitric oxide synthase, NO nitric oxide, TACE tumor necrosis factor-alpha converting enzyme, ADAM17 a disintegrin and metalloprotease 17, IL-6 interleukin 6, STAT3 signal transducer and activator of transcription 3, EGFR epidermal growth factor receptor, lncRNA TUG1 long non-coding RNA taurine upregulated gene 1, KLF10 Krüppel-like factor 10, USP24 ubiquitin-specific protease 24, PLK1 polo-like kinase 1, GSI gamma-secretase inhibitor, AKT protein kinase B, mTOR mammalian target of rapamycin, EZH2 enhancer of Zeste homolog 2, PINK1 PTEN-induced putative kinase 1, WISP-1 Wnt-induced signaling protein 1, NKAP NF-kappa-B-activating protein, HES1 hairy and enhancer of split 1, Hey1 Hes-related family bHLH transcription factor with YRPW motif 1, RBPJ recombining binding protein suppressor of hairless, N1ICD Notch1 intracellular domain, GAS41 growth arrest-specific 41, H2A.Z.2 histone H2A.Z.2, GEM gemcitabine, MVP major vault protein, SST somatostatin, SSTR somatostatin receptor, PARP1 poly ADP-ribose polymerase1, bHLH basic-helix-loop-helix
Notch signaling pathway in EMT
EMT, originally described by Elizabeth Hays in the 1980s, denotes the intricate process wherein epithelial cells undergo a transformation, losing their characteristic features and adopting mesenchymal phenotypes.338 EMT is a fundamental occurrence in events such as embryogenesis and tissue repair.339,340 Over the years, it has been observed that EMT is reactivated during tumor progression, emerging as a pivotal mechanism for cancer cells to acquire malignant properties.341,342 Various signaling pathways participate in the regulation of EMT, including the Notch signaling pathway (Fig. 3).
Fig. 3.
Notch signaling pathway in epithelial-mesenchymal transition (EMT). EMT is a complex process wherein epithelial cells undergo a transition, losing their inherent characteristics and adopting a mesenchymal phenotype. Notch signaling plays a crucial role in regulating EMT, representing a significant mechanism for tumor cells to acquire malignant properties. (Figure created using BioRender.com). ADAM a disintegrin and metalloprotease, N1ICD Notch1 intracellular domain, ZEB1 zinc finger E-box-binding homeobox 1, MCAM melanoma cell adhesion molecule, Co-A coactivator, MAML mastermind-like, CSL CBF1/suppressor of hairless/Lag1
Numerous studies have indicated that the activation of Notch1 promotes EMT in HCC, contributing to the acquisition of stem-like characteristics, as well as facilitating migration, invasion, and chemoresistance.343,344 Mechanistically, Xie and colleagues have noted that tetraspanin5 activates Notch signaling by enhancing the γ-secretase-catalyzed cleavage of the Notch1 receptor.345 This activation further promotes EMT and rearrangement of the actin cytoskeleton, ultimately fostering the metastasis of HCC. In squamous cell carcinoma (SCC), emerging evidence suggests that Notch1 functions as an EMT-promoting factor driven by TGF-β, while Notch3-mediated signaling restricts terminal differentiation.346 Another study has demonstrated that the Notch4-HEY1 pathway is specifically up-regulated in HNSCC, inducing proliferation, cisplatin resistance, and promoting EMT.347 Similarly, Xie et al. have shown that the Notch1-HEY1 pathway is specifically up-regulated in salivary adenoid cystic carcinoma (ACC), driving cell self-renewal and EMT.348 These findings hold significant potential to broaden our comprehension of the role of the Notch pathway in tumor EMT and may guide the development of new strategies to reverse EMT by targeting the Notch signaling pathway.
Primary drug resistance is commonly observed in cancer cells exhibiting mesenchymal differentiation.349,350 EMT is recognized as a contributor to chemotherapy resistance in various tumors, including NSCLC, breast cancer, and glioma.351,352 Consequently, effective inhibition of the Notch signaling pathway emerges as a promising strategy to overcome chemoresistance. CBF1, also known as RBPJ, has been identified as a participant in the EMT-like phenotype of glioma cells.353 Maciaczyk et al. demonstrated that inhibiting CBF1 can impede EMT activators, such as zinc finger E-box-binding homeobox 1, resulting in decreased cell invasiveness and chemoresistance in EMT-like glioblastoma cells. ZLDI-8, a novel inhibitor of ADAM17, has been reported to inhibit the Notch pathway and reverse the EMT process, thereby inhibiting migration and invasion in chemotherapy-resistant NSCLC.354 Notch1 induces EMT and chemoresistance in TNBC cells by directly activating the MCAM promoter. Down-regulation of Notch1 significantly inhibits MCAM expression, thereby reversing EMT and cisplatin chemotherapy resistance in TNBC cells. These studies collectively provide molecular evidence highlighting the impact sof Notch signaling-mediated EMT on tumor chemoresistance. Consequently, Notch inhibitors may prove to be effective anti-EMT therapies, offering a potential avenue to prevent chemoresistance in tumor cells.
Notch signaling pathway in tumor angiogenesis
The Notch signaling pathway in tumor angiogenesis is a significant aspect of the multi-stage process involved in the formation of new blood vessels from the original ones.355 This process is crucial for embryonic development, normal tissue growth, bone formation, and wound healing. Abnormal angiogenesis, a distinctive feature of the TME, provides essential nutrients for tumor growth and creates an opportunity for malignant cells to enter the circulation, forming distant metastases.356,357 Vascular endothelial growth factor (VEGF) is considered the central signaling mediator for angiogenesis.358,359 Moreover, the Notch signaling cascade has been demonstrated to play a crucial role in regulating tumor angiogenesis (Fig. 4).360 DLL4, a Notch ligand, modulates angiogenesis by controlling endothelial cell activation, vascular development, and maturation.361,362 Recent research by Mónica et al. indicates that DLL4 expressed in the TME can induce Notch signaling activation in Notch1-mutated CLL cells.363 Additionally, DLL4 triggers the expression of Notch-regulated ankyrin repeat protein and VEGF, leading to increased angiogenesis. Nandhu et al. discovered that Fibulin-3, a protein secreted by glioma cells, acts as a paracrine activator of Notch signaling, motivating angiogenesis in high-grade glioma.364 Mechanistically, Fibulin-3 enhances the expression of DLL4 in an ADAM10/17-dependent manner, thereby activating DLL4-Notch signaling.
Fig. 4.
Notch signaling pathway in tumor angiogenesis. Abnormal angiogenesis, a distinctive feature of the tumor microenvironment, provides essential nutrients for tumor growth and facilitates the entry of malignant cells into circulation, leading to distant metastases. The Notch signaling cascade plays a crucial role in mediating tumor angiogenesis. (Figure created using BioRender.com). VEGFR vascular endothelial growth factor receptor, VEGF vascular endothelial growth factor, MAPK mitogen-activated protein kinase, DLL4 delta-like ligand 4, ADAM a disintegrin and metalloprotease, VEGFA vascular endothelial growth factor A, JAG1 Jagged1, NRP1 neuropilin 1, NRARP Notch-regulated ankyrin repeat protein, FMOD fibromodulin
JAG1, as a classic ligand of the Notch pathway, has been identified as playing a role in angiogenesis. However, accumulating evidence suggests that JAG1 and DLL4 influence different downstream signaling pathways, resulting in distinct vascular phenotypes.365,366 Generally, DLL4 inhibits endothelial cell sprouting by activating Notch signaling, leading to a sparse network of large-caliber vessels.367 In contrast, JAG1 mediates signal transduction in both tumor cells and endothelial cells, promoting vascular sprouting and higher vascular density.368,369 Liu et al. have preliminary evidence confirming JAG1’s pro-angiogenic effect in TNBC, possibly participating in angiogenesis through the enhancement of the MALAT1-miR-1405p-JAG1/VEGFA pathway.370 This suggests a potential synergistic effect between JAG1 and VEGFA in promoting angiogenesis. Another study has reported that JAG1 may mediate the long intergenic non-coding RNA linc-OIP5 to regulate the DLL4/Notch/NRP1 signaling pathway in human umbilical vein endothelial cells, affecting angiogenesis in the breast cancer microenvironment.371 However, further experiments are needed to explore how JAG1 interacts with Notch ligands such as DLL4 to regulate tumor angiogenesis.
Activated Notch1 signaling is frequently observed in endothelial cells of various human cancers, and this is positively correlated with worsened prognosis.372 Continuous activation of Notch1 alters the morphology and function of endothelial cells, promoting the migration of tumor cells across the vascular wall. Additionally, Notch1 signaling participates in the angiogenesis phenotype.373,374 Kumar et al. reported that Notch1 drives the expression of CD133, activates MAPK, and regulates the expression of MMP-2/-9 and VEGF in melanoma-specific CD133+ CSCs, leading to melanoma angiogenesis.375 Sengupta et al. revealed that differentiated glioma cells secrete the proteoglycan fibromodulin to promote glioma angiogenesis by activating Notch1 signaling.376 In drug-resistant NSCLC, the inactivation of the Notch1-HIF1α-VEGF pathway by ZLDI-8 suppresses angiogenesis and vasculogenic mimicry.377 Collectively, these findings contribute to a comprehensive understanding of the mechanism of the Notch pathway in mediating tumor angiogenesis and may enrich the therapeutic targets for tumors.
Notch signaling pathway in CSLC properties
CSCs, a subgroup of tumor cells with notable self-renewal potential and multidirectional differentiation ability, are increasingly recognized in various solid tumors.378–380 Their presence is considered a driver of malignancy initiation, metastasis, and chemotherapy resistance. Recent evidence suggests that non-CSCs can acquire stem-like properties in certain processes, such as EMT, abnormal activation pathways, expression of specific stem cell biomarkers, and immune escape.381–383 Abnormal activation of key signaling pathways controlling stem cell self-renewal, including the Notch signaling pathway, is deemed a crucial factor in regulating CSLC properties (Fig. 5).384,385 For instance, Xiao et al. reported that in RCC, activated Notch signaling can maintain the stemness of CSCs and promote their chemotaxis through the SDF-1/CXCR4 axis.386 This study provides new insights into how RCC CSCs maintain stemness through the Notch pathway. Liu et al. found that Fusobacterium nucleatum infection promotes the degradation of Numb mediated by lipid droplets, resulting in activated Notch signaling and the acquisition of stem-like properties in CRC cells.387 Targeting the Notch ligand JAG2, tRF/miR-1280 inactivates Notch signaling, suppressing the CRC stem-like phenotype and inhibiting tumor formation and metastasis.388 Katsushima et al. revealed the role of Notch signaling in maintaining the stemness of GSCs.389 Specifically, activated Notch1 in GSCs induces the expression of the long non-coding RNA TUG1, influencing the stemness of GSCs. In HCC, CSCs are implicated in treatment resistance and poor survival outcomes.390,391 Liu et al. demonstrated that Notch3 is essential for liver CSC self-renewal and tumor proliferation.392 CAFs maintain the stability of lysine-specific histone demethylase 1 A (LSD1) by inducing LSD1 deacetylation through Notch3 activation, accelerating the self-renewal of liver CSCs. Additionally, highly expressed inducible nitric oxide synthase activates Notch1 through the TACE/ADAM17 pathway, promoting the CSC phenotype and enhancing HCC aggressiveness.393 These groundbreaking findings illuminate the role of the Notch signaling pathway in coordinating the self-renewal of liver CSCs, with potential implications for improving treatment strategies and limiting recurrence.
Fig. 5.
Notch signaling pathway in cancer stem-like cell (CSLC) properties. CSLC, a subset of tumor cells with notable self-renewal potential and multidirectional differentiation ability, are regulated by the abnormal activation of the Notch signaling pathway. (Figure created using BioRender.com). iNOS inducible nitric oxide synthase, TACE TNF-alpha converting enzyme, ADAM a disintegrin and metalloprotease, HCC hepatocellular carcinoma, CAFs cancer-associated fibroblasts, LSD1 lysine-specific histone demethylase 1A, CRC colorectal cancer, JAG2 Jagged2, SDF-1 stromal cell-derived factor-1, CXCR4 CXC chemokine receptor 4, RCC renal cell carcinoma, CSC cancer stem cell
CSLC properties rely on a complex interplay of multiple signaling pathways that form an interacting network.394,395 Studies have demonstrated that Notch signaling can synergistically interact with other biological processes, such as the WNT and EGFR pathways, to regulate CSLC phenotypes.396 For instance, Jiang et al. reported that HIF1α mediates the overexpression of miR-1275, activating both Wnt/β-catenin and Notch signaling pathways, thereby enhancing the stemness of LUAD cells.397 In a hypoxic TME, Yan et al. found that overexpressed HIF2α induces stem-like phenotypic transformation through the activation of Wnt and Notch pathways, increasing the resistance of breast cancer cells to PTX.398 Syndecan-1, identified as a novel molecular marker in inflammatory breast cancer, was shown to modulate CSLCphenotypes through the IL-6/STAT3, Notch, and EGFR signaling pathways.399 In ACC, a population of CD133+ cells with neural stem cell properties was identified, and Notch1 and SOX10 were found to drive the proliferation and radiation-resistance of CD133+ CSCs.400 Lin et al. observed significant upregulation of Notch4 protein in melanoma CSLCs (MCSLCs), where Notch4+ MCSLCs promoted metastasis and invasion by initiating the EMT process.401 Krüppel-like factor 10 (KLF10), a zinc finger-containing transcription factor, was revealed to inhibit Notch3 and Notch4 transcription by binding to the promoter of E74-like ETS transcription factor 3.402 KLF10 deficiency led to the development of a PDAC stem-like phenotype and tumorigenesis by promoting the Notch signaling pathway.403 Up-regulation of KLF10 or inhibition of Notch signaling at the gene level or pharmacologically reduced the stem-like phenotype and tumor growth in PDAC. To fully exploit the therapeutic potential of targeted Notch signaling in malignant tumors, further research is required to explore the intricate crosstalk between Notch signaling and core components of other pathways. This exploration aims to identify potential balances for regulating stem-like phenotypes in cancer cells.
Notch signaling pathway in cancer metabolic reprogramming
Metabolic reprogramming is a prominent feature of cancer, encompassing alterations in glucose, lipid, and amino acid metabolism. The Warburg effect, first described by Warburg in the 1920s, highlighted that tumor cells preferentially undergo glycolysis even in the presence of oxygen.404–406 Although less efficient in ATP production, aerobic glycolysis supports the rapid proliferation and survival of malignant cells.407,408 The Notch signaling pathway plays a pivotal role in the metabolic reprogramming of cancer cells (Fig. 6). Previous studies have demonstrated that Notch signaling becomes active during the glycolytic switch in cells.409 Several genes involved in controlling glycolysis and the tricarboxylic acid cycle, such as Glut1, Hex-A, Ecdysone-inducible gene L3, Impl3, and hairy, are direct transcriptional targets of the Notch pathway, mediating the transition of cellular metabolism toward the Warburg effect. Jitschin et al. found that stromal cells facilitate glycolytic switching in CLL cells through the Notch-c-Myc signaling pathway, leading to an increase in aerobic glycolysis.410,411 Another study showed that hyper-activated and hypo-activated Notch signaling induces glycolytic switching in breast tumor cells through different mechanisms.412 Specifically, hyper-activated Notch signaling promotes glycolysis in a PI3K/AKT-dependent manner, while hypo-activated Notch signaling weakens mitochondrial respiratory chain activity and enhances glycolysis via the p53-dependent pathway. In T-ALL cells, activated Notch signaling drives high System L amino acid transporter activity, promoting leucine transport and uptake, which, in turn, enhances glucose transport via the mTORC1/HIF1α pathway.413 Additionally, Notch coordinates c-Myc and mTORC1-controlled metabolic reprogramming, increasing glutamine transport. Sellers et al.414 found that the two main subtypes of NSCLC, SCCs and adenocarcinomas, exhibit distinct metabolic reprogramming. Upregulated Notch activity is associated with altered metabolic phenotypes in lung SCC. Mouse lung tumors driven by Notch and MYC reproduce the SCC-specific metabolic reprogramming characteristics. These studies collectively suggest that Notch signaling is a crucial regulator of energy metabolism in malignant cells and is required to maintain metabolic flexibility.
Fig. 6.
Notch signaling pathway in cancer metabolic reprogramming. Notch signaling plays a crucial role in the metabolic reprogramming of cancer cells, particularly during the glycolytic switch. The Notch pathway is active during this transition in cancer cells, with several genes directly regulated as transcriptional targets. This regulation mediates the shift in cellular metabolism towards the Warburg effect. (Figure created using BioRender.com). CLL chronic lymphocytic leukemia, PTEN phosphatase and tensin homolog, T-ALL T cell acute lymphoblastic leukemia, PI3K/AKT phosphoinositide 3-kinase/protein kinase B, HIF1α hypoxia-inducible factor 1α, Co-A coactivator, MAML mastermind-like, CSL CBF1/suppressor of hairless/Lag1, GLUT glucose transporters, MCT monocarboxylate transporter, ATP adenosine triphosphate, NADH nicotinamide adenine dinucleotide hydride, NAD nicotinamide adenine dinucleotide, TCA tricarboxylic acid, α-KG alpha-ketoglutarate
Lactic acid, a byproduct of aerobic glycolysis, contributes to the increased acidity of the TME. In the presence of a low pH environment, anti-tumor effector cells, such as T cells, are prone to functional loss and apoptosis. Growing evidence suggests that the acidity of the TME plays a crucial role in regulating tumor immunity, orchestrating local and systemic immunosuppression.415 Consequently, oncogene-induced metabolic reprogramming may be linked to immune escape. For instance, Xie and colleagues uncovered that Notch1 promotes the expression of glycolytic genes through interaction with histone acetyltransferases p300 and pCAF.416 Moreover, Notch1 signaling and PDZ-binding motif form a positive feedback loop that elevates extracellular lactate levels, inhibiting the activity of cytotoxic T cells and ultimately contributing to the malignant behavior of lung cancer. The shift from oxidative phosphorylation to glycolysis is known to be crucial for the activation and effector function of memory T cells.417 Given the limited nutrient availability in tumors, it is reasonable to hypothesize that T cell glycolytic metabolism undergoes changes in the TME.418 Zhao et al. found that Notch signaling is implicated in effector T cell dysfunction mediated by the methyltransferase EZH2 in OC.419 Mechanistically, OC cells restrict the aerobic glycolysis of T cells by maintaining high expression of miR-101 and miR-26a. These specific miRNAs inhibit EZH2 expression, target Notch repressors, and promote Notch activation, thereby attenuating T cell-mediated anti-tumor immunity. Collectively, these studies unveil the connection between Notch signaling, cancer metabolic reprogramming, and immune escape. Targeting Notch signaling holds the potential to enhance the efficacy of immunotherapy by inhibiting aerobic glycolysis.
The heightened Warburg effect not only fosters the proliferation and metastasis of tumor cells but also bestows various tumor characteristics that contribute to drug resistance.420,421 These characteristics include increased drug efflux, mutations in drug targets, inactivation of drug metabolism, and enhanced DNA damage repair, among others. Recent studies have shed light on specific mechanisms linking the Warburg effect to drug resistance in different cancers. For example, a study revealed that the ACYP1/HSP90/MYC/LDHA axis promotes the Warburg effect, driving sorafenib resistance in HCC.422 Another investigation found that Aldo-keto reductase family 1 B10 enhances the Warburg effect, marked by excessive lactic acid production, leading to acquired chemotherapy resistance in lung cancer brain metastasis to pemetrexed.423 Additionally, inhibition of HIF1α-mediated aerobic glycolysis and mitochondrial dysfunction can restore the sensitivity of tamoxifen in the treatment of breast cancer.424 Notch signaling has also been implicated in mediating chemotherapy resistance through metabolic reprogramming.425 Notch1-induced glutaminolysis is identified as a key carbon source for T-ALL and a determinant of anti-Notch1 therapeutic response in vivo.426,427 Inhibition of Notch1 in T-ALL results in metabolic impairment, significant inhibition of glutaminolysis, and induction of autophagy to provide essential metabolites to leukemia cells.428 The mutational loss of the tumor suppressor PTEN, a negative regulator of the PI3K/AKT signaling pathway, can promote glycolysis and induce drug resistance against anti-Notch1 treatment in T-ALL.428,429 This emphasizes the critical role of Notch1 signaling in controlling leukemia cell metabolism and glutaminolysis as a therapeutic target in Notch1-induced T-ALL. Furthermore, cancer cells have a high demand for pyrimidine nucleotides to support accelerated DNA and RNA synthesis.430,431 Thus, they heavily rely on the de novo synthesis approach of pyrimidine.432,433 He et al. found that de novo pyrimidine synthesis enhances the expression of key glycolytic enzymes and promotes aerobic glycolysis by activating Notch signaling and c-Myc gene transcription, conferring GC cells with chemotherapy resistance.434 This underscores the pivotal role of pyrimidine de novo synthesis in aerobic glycolysis and identifies it as a metabolic vulnerability that can be targeted to overcome chemotherapy resistance in GC.
Notch signaling pathway in TME
The malignant characteristics of cancer rely on the bidirectional interaction between cancer cells and their environment, giving rise to a well-organized complex ecosystem known as the TME.435 The TME consists of tumor cells, blood vessels, immune cells, stromal cells, and extracellular matrix, forming a dynamic and intricate network.436 The crosstalk between cancer cells and their environment involves various signaling pathways, including NF-κB,437 TGF-β,438 cGAS-STING signaling,439,440 and the Notch pathway.441 Notably, the Notch signaling pathway plays a crucial role in shaping the components of the TME, regulating it through paracrine or autocrine signals (Fig. 7). In a recent study by Jackstadt et al., the activation of Notch1 signaling in mouse intestinal epithelium was found to reshape the TME of CRC, showing a close association with poor prognosis.442 Moreover, activated Notch1 signaling was shown to promote the metastasis of KrasG12D-driven serrated cancer through TGF-β-dependent neutrophil recruitment. Tumor growth and metastasis driven by locally activated neutrophils was also observed in the lung microenvironment, which is governed by enhanced Notch signaling.443 Significantly, targeting Notch-driven neutrophil recruitment might be an effective strategy in preventing cancer metastasis. CAFs represent crucial stromal cells in the TME, capable of reshaping the extracellular matrix environment and promoting tumor progression and metastasis through paracrine communication. Kim et al.444 demonstrated that apoptotic lung cancer cells, induced by ultraviolet irradiation, can reprogram CAFs, enhance the secretion of Wnt-induced signaling protein 1 by activating Notch1 signaling, and subsequently inhibit the migration and invasion of both cancer cells and CAFs. This study underscores the context-dependent role of activated Notch signaling in either promoting or inhibiting carcinogenesis within the TME.
Fig. 7.
Notch signaling pathway in tumor microenvironment (TME). The Notch signaling pathway actively participates in the components of the TME, regulating TME through both paracrine and autocrine signals. (Figure created using BioRender.com). TGF-β2 transforming growth factor-beta2, TGF- βR1 transforming growth factor-beta receptor 1, CRC colorectal cancer, DLL1 delta-like ligand 1, ADAM a disintegrin and metalloprotease, HIF hypoxia-inducible factor, PI3K phosphoinositide 3-kinase, WISP Wnt-induced signaling protein 1, CAFs cancer-associated fibroblasts, HCC hepatocellular carcinoma, PINK1 PTEN-induced putative kinase 1, ROS reactive oxygen species
The immune components within tumors collectively form the tumor immune microenvironment (TIME), which includes innate immune cells, adaptive immune cells, and cytokines.445,446 The TIME has been shown to play a crucial role in the initiation and progression of tumors. Gu et al. reported that the overexpression of NF-kappa B activating protein directly binds to the Notch1 promoter and transactivates it, contributing to glioma growth by promoting the Notch1-dependent immunosuppressive TME.447 Advancements in high-throughput and high-dimensional technologies, such as spatial transcriptome and proteome analyses, have allowed researchers to describe the spatial architecture of the TIME and explore its functions in tumor biology.448 Single-cell RNA sequencing has revealed the remodeling of myeloid cells and lymphocytes in the TIME during tumor dormancy.449 Specifically, the JAG1/Notch signaling pathway was found to regulate immune homeostasis during dormant minimal residual disease.449 In another study, the single-cell landscapes of the human liver, from development to disease, were examined. The research found that VEGF/Notch signaling pathways mediate an immunosuppressive onco-fetal TME in HCC.450 Further investigation revealed a common immunosuppressive microenvironment between fetal liver and HCC, particularly involving VEGF/Notch signaling in the re-emergence of fetal-associated endothelial cells (i.e., PLVAP/VEGFR2) and fetal-like (i.e., FOLR2) tumor-associated macrophages.451 This concept of an immunosuppressive onco-fetal TME mediated by VEGF/Notch signaling provides a potential new target for immunotherapy of HCC.
The hypoxic microenvironment is a prominent and common feature in many solid tumors, including PC,452 HCC,453 breast cancer,454 and melanoma.455 Hypoxia plays a role in mediating the malignant biological behavior of cancer cells and can impact the therapeutic outcomes of tumors through complex mechanisms.456,457 In glioma cells, Grassi et al.458 revealed that hypoxia induces the release of intracellular fragments of DLL1, a Notch ligand, which is dependent on ADAM17 and HIF1α/HIF2α. Interestingly, hypoxic glioma cells exhibit unexpected nuclear translocation of DLL1, leading to altered activation of the p53 and PI3K pathways and increased aggressiveness of gliomas. Hypoxia often leads to inefficient electron transfer in the mitochondrial electron transport chain, resulting in the accumulation of reactive oxygen species (ROS). The downstream signaling triggered by HIF activation is a key molecular mechanism for cells to adapt to hypoxia.459,460 Chiu et al. found that the HIF1 and Notch signaling pathways interact to control mitochondrial biogenesis in cancer cells and maintain redox balance.461 Specifically, HIF1 directly binds to the hypoxia response element of HEY1 in the Notch signaling pathway, activating the transcription of HEY1 in HCC. HEY1, in turn, inhibits the expression of PTEN-induced putative kinase 1 (PINK1), reducing the production of mitochondrial ROS and promoting the growth of HCC. Therefore, the HIF1/HEY1/PINK1 pathway confers a survival advantage on HCC in the hypoxic microenvironment.
Notch signaling pathway in chemoresistance
Chemotherapy is the traditional treatment for all types of cancer.462 Despite the development of numerous novel chemotherapy strategies, response rates to treatment for many advanced tumors remain low due to the emergence of intrinsic or acquired chemoresistance.463,464 Various mechanisms can confer chemoresistance in cancer,465,466 including decreased drug activity, elevated efflux of anticancer agents, alterations in drug targets, changes in DNA repair mechanisms, and evasion of drug-induced apoptosis. Emerging evidence suggests that acquired chemoresistance may also involve complex mechanisms, such as the development of EMT-like phenotypes in cancer cells, metabolic reprogramming, stem cell characterization, and alterations in molecular pathways.467,468 Identifying specific signaling pathways that are abnormally activated in chemoresistance is crucial for adjusting therapeutic regimens. Therefore, exploring the molecular processes behind chemoresistance is essential for improving tumor treatment outcomes. Aberrant expression of components in the Notch pathway is known to play a crucial role in contributing to chemoresistance (Fig. 8).469,470
Fig. 8.
Notch signaling pathway in chemoresistance. The aberrant expression and overactivation of Notch pathway components play crucial roles in contributing to chemoresistance. (Figure created with BioRender.com). PDAC pancreatic ductal adenocarcinoma, N1DARP Notch1 degradation-associated regulatory polypeptide, N1ICD Notch1 intracellular domain, CHEMO chemotherapy, SCLC small cell lung cancer, AP-1 activator protein-1, LUAD lung adenocarcinoma, MVP major vault protein, AKT protein kinase B, TNBC triple-negative breast cancer, ADAM a disintegrin and metalloprotease, JAG1-ICD Jagged1 intracellular domain, CRC colorectal cancer
Previous research has shown that excessive activation of the DLL4/Notch pathway in PDAC causes defective angiogenesis within tumors, resulting in low efficiency of chemotherapeutic drug delivery in vivo and enhanced multi-drug chemoresistance.471 Another study revealed that GEM, a first-line chemotherapy agent for PDAC, induces Midkine expression in a dose-dependent manner.472 Furthermore, Midkine interacts with Notch2 to activate Notch signaling, driving PDAC resistance to GEM.472 Similar to PDAC, chemotherapy resistance is a major challenge for PC. Cumulative data suggest that activation of the Notch signaling pathway contributes to PC resistance to GEM. Inhibition of Notch signaling is reported to enhance the chemosensitivity of PC to GEM by activating the intrinsic apoptotic pathway.473 Glioma amplified sequence 41 (GAS41) is reported to be a novel regulator of Notch signaling by controlling H2A.Z deposition.474 Han et al. found that GAS41 binds to H2A.Z.2 to activate Notch1 signaling and its downstream mediators, driving PC stemness and GEM resistance.475 Given the function of widely activated Notch signaling in chemotherapy resistance of PC cells, Notch signaling is expected to become a potential therapeutic target for PC. Zhai et al. identified a new microprotein, Notch1 degradation-associated regulatory polypeptide (N1DARP), encoded by LINC00261.476 N1DARP promotes N1ICD degradation by destroying USP10-N1ICD interactions, thereby suppressing chemoresistance in Notch1-overactivated PC. These findings provide a promising alternative strategy for PC and may have widespread application in a variety of malignancies.
Chemoresistance in lung cancer is a multifactorial process involving the dysfunction of oncogenes and tumor suppressors in various signaling pathways, such as Notch.477,478 A recent study revealed that MYCN binds to the HES1 promoter, activating the Notch pathway.479 This activation inhibits drug-induced apoptosis and enhances chemotherapy resistance in SCLC. Similarly, Li et al. reported that chemo-resistant NSCLC cells acquire a more invasive phenotype through EMT and dysregulated Notch pathways. Furthermore, molecular evidence demonstrated that terfenadine reverses epirubicin sensitization via EMT and the Notch pathway. Huang and colleagues uncovered that Notch1 negatively regulates miR-451 through activator protein-1, influencing the proliferation and apoptosis of LUAD and conferring chemoresistance to docetaxel (DTX).480 Inhibition of Notch1 sensitized LUAD to DTX, suggesting that combining DTX with a GSI could be a novel strategy for treating DTX-resistant LUAD. Notably, beyond PC and lung cancer, Notch signaling also mediates the sensitivity of breast cancer and CRC cells to chemotherapy agents.481,482 Yang et al. demonstrated that cytotoxic drugs used in neoadjuvant therapy for breast cancer can stimulate the secretion of exosomes by cancer cells, promoting chemotherapy resistance through the activation of WNT/β-catenin and Notch stem cell pathways in vivo.483 Additionally, Xiao et al. revealed that Notch1 positively regulates the transcription of major vault protein, activating the AKT pathway, promoting the EMT process, and participating in chemotherapy resistance in TNBC cells.484 In CRC with a Kras mutation, JAG1 has been shown to trigger intrinsic reverse signal transduction through its nuclear-targeted JAG1-ICD, maintaining the progression and chemoresistance of CRC.485 Another study demonstrated that miR-195-5p inhibits CRC cell stemness and 5-fluorouracil resistance by inhibiting Notch2 and RBPJ.486 In summary, inhibiting Notch pathways holds promise for restoring chemotherapy efficacy in CRC.487
Notch signaling pathway in tumor suppression
Research on the role of Notch signaling pathway during tumorigenesis primarily centers on its function as an oncogene. Increasing evidence suggests that Notch signaling also acts as a tumor suppressor in various malignancies, including SCC, hematological malignancies, cervical cancer, and forebrain glioma (Fig. 9).488–491 The oncogenic or tumor-suppressive role of the Notch signaling pathway is believed to be greatly dependent on the environment.492,493 As a crucial form of intercellular communication, Notch signaling regulates the differentiation of keratinocytes and maintains skin homeostasis.494,495 Nicolas and colleagues found that Notch1 functions as a tumor suppressor gene in mammalian skin.496 Deficiency of Notch1 in mouse skin and primary keratinocytes results in elevated Gli2 expression and improper activation of beta-catenin signaling, ultimately leading to skin carcinogenesis. Another study found that E6 proteins of the cancer associated human papillomavirus (HPV) 8 and Mus musculus papillomavirus 1(MmuPV1) can bind to the Notch co-activator MAML to inhibit Notch signaling, which is associated with delayed differentiation and sustained keratinocyte proliferation.497 Additionally, Demehri et al. demonstrated that the tumor-promoting effect of Notch1 deletion in epidermal keratinocytes involves the impaired skin-barrier integrity and wound-like skin microenvironment.498 In human skin samples, suppressed Notch/CSL signaling was observed in stromal fields surrounding multifocal premalignant actinic keratosis lesions, while gene expression of c-Jun and c-Fos was upregulated.499 Moreover, Notch1 is a p53 target gene and participates in the inhibition of human aggressive SCC by negatively regulating ROCK1/2 and MRCKα kinases.500 Wu et al. found that PTC124 (Ataluren) could help HNSCC cells re-express functional Notch1 to substitute the nonsense mutation Notch1, thus preventing the proliferation of HNSCC cells.501 Taken together, these findings provide evidence that Notch functions as tumor suppressor in relation to SCC, and SCC may be a cancer subtype that could benefit from specific activation of the Notch receptor.
Fig. 9.
Notch signaling pathway in tumor suppression. Earlier studies provided evidence supporting Notch-mediated tumor suppression in various malignancies, including SCC, cervical cancer, and SCLC. (Figure created using BioRender.com). PDAC pancreatic ductal adenocarcinoma, MmuPV1 Mus musculus papillomavirus 1, HPV8 human papillomavirus, Co-A coactivator, MAML mastermind-like, CSL CBF1/suppressor of hairless/Lag1, hASH1 human achaete-scute homolog-1, SST somatostatin, SSTR somatostatin receptor, SCC squamous cell carcinoma, SCLC small cell lung cancer
In addition to SCC, earlier studies have offered evidence supporting Notch-mediated tumor suppression in several solid malignancies. SCLC exhibits typical neuroendocrine characteristics, dependent on the involvement of the basic-helix-loop-helix transcription factor known as human achaete-scute homolog-1 (hASH1). Previous research has indicated that the activated Notch signaling pathway suppresses the expression of hASH1 in SCLC cells, leading to cell cycle arrest and growth inhibition linked to the p21waf/cip1 and ras signaling pathway.502 Giachino et al. discovered that Notch signaling acts as a tumor suppressor in forebrain tumor subtypes.503 Their findings indicate that Notch signaling can collaborate with p53 to inhibit cell proliferation and tumor growth in grades II-III astrocytoma, proneural glioblastoma, and supratentorial primitive neuroectodermal tumor. In a stable Notch1-activated cervical cancer HeLa cell line established by Laura and colleagues, activation of Notch1 led to apoptosis, cell cycle arrest, and tumor suppression.257 Mechanistically, Notch1-mediated tumor suppression in cervical cancer may be partly achieved by up-regulating somatostatin (SST) signaling. A prior study also revealed an unforeseen tumor suppressor role for Notch1 in a K-ras-induced PDAC murine model, where K-ras is activated and Notch1 is deleted.504 In this model, the absence of Notch1 results in increased tumor occurrence and advancement, suggesting that Notch1 may act as a tumor suppressor in K-ras-induced PDAC. With respect to B-ALL, Notch target HES1 triggers the activation of poly ADP-ribose polymerase1 (PARP1), leading to B-ALL cell apoptosis in a cell type-specific manner.505 These findings suggest that Notch signaling might regulate the fate of tumor cells in a context-dependent way through various intricate mechanisms. Importantly, the functions of Notch signaling as a tumor suppressor could contribute to the advancement of Notch agonist-based cancer treatments.
Ongoing therapeutic strategies targeting Notch signaling in human malignancies
Efforts to develop therapeutic strategies targeting Notch signaling continues unabated, with numerous drug studies currently progressing through preclinical or clinical trials for various human malignancies. Researchers have devised a range of Notch-targeted therapies for each stage of the Notch signaling cascade, as illustrated in Fig. 10. In this context, we provide a summary of specific inhibitors and blocking antibodies currently undergoing clinical trials for Notch signaling, encompassing GSIs, ADAM inhibitors, antibodies targeting Notch receptors or ligands, Notch transcription complex inhibitors, and γ-secretase modulators (GSMs) (Table 3).
Fig. 10.
Therapeutic strategies targeting Notch signaling in human malignancies. Various pharmacological agents aimed at the Notch pathway have been developed, including γ-secretase inhibitors (GSIs), ADAM inhibitors, antibodies against Notch receptors or ligands, inhibitors targeting the Notch transcription complex, and γ-secretase modulators (GSMs). (Figure created with BioRender.com). DLL3 delta-like ligand 3, DLL4 delta-like ligand 4, ADAM a disintegrin and metalloprotease, NECD Notch extracellular domain, NICD Notch intracellular domain, Co-A coactivator, MAML mastermind-like, CSL CBF1/suppressor of hairless/Lag1
Table 3.
Therapeutic strategies targeting Notch signaling in clinical trials against human malignancies
| Type | Agents | Cancer | Identifier | Enrollment (N) | Phase | Status | Country | First posted | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| γ-Secretase inhibitor | RO4929097 | Metastatic CRC | NCT01116687 | 37 | Phase II | Completed | United States | 2010 | 506 |
| RO4929097 | Previously treated metastatic PDAC | NCT01232829 | 37 | Phase II | Completed | United States | 2010 | 507 | |
| RO4929097 | Metastatic melanoma | NCT01120275 | 36 | Phase II | Terminated | United States | 2010 | 508 | |
| RO4929097 | Recurrent platinum-resistant EOC | NCT01175343 | 45 | Phase II | Completed | United States, Canada | 2010 | 509 | |
| RO4929097 | Recurrent/progressive Glioblastoma | NCT00980343 | 47 | Phase II | Completed | United States | 2010 | 510 | |
| RO4929097 | Advanced solid tumors | NCT01198184 | 18 | Phase Ib | Completed | Canada | 2010 | 511 | |
| RO4929097 | Glioma | NCT01119599 | 21 | Phase 0/I | Completed | United States | 2010 | 512 | |
| RO4929097 | Recurrent malignant glioma | NCT01189240 | 13 | Phase I | Terminated | United States | 2010 | 513 | |
| RO4929097 | Metastatic breast cancer | NCT01149356 | 15 | Phase Ib | Terminated | United States | 2010 | N/A | |
| RO4929097 | Advanced sarcoma | NCT01154452 | 78 | Phase Ib/II | Completed | United States | 2010 | 514 | |
| MK-0752 | PDAC | NCT01098344 | 44 | Phase I | Completed | United Kingdom | 2010 | 518 | |
| MK-0752 | Advanced solid tumors | NCT01243762 | 47 | Phase I | Terminated | USA, Canada, Israel | 2010 | 520 | |
| MK-0752 | Advanced solid tumors | NCT01295632 | 28 | Phase I | Completed | United States; France; etc. | 2011 | 519 | |
| Crenigacestat (LY3039478) | Advanced or metastatic ACC | NCT01695005 | 22 | Phase I | Completed | United States, Denmark, etc. | 2012 | 521 | |
| Crenigacestat | Advanced or metastatic cancer | NCT01695005 | 28 | Phase I | Completed | United States, Denmark, etc. | 2012 | 522 | |
| Crenigacestat | Advanced or metastatic solid tumors | NCT02784795 | 63 | Phase Ib | Completed | United States, Denmark, etc. | 2016 | 523 | |
| Crenigacestat | Advanced solid tumors | NCT02836600 | 16 | Phase I | Completed | Japan | 2016 | 524 | |
| Crenigacestat | Advanced or metastatic solid tumors | NCT02784795 | 31 | Phase Ib | Completed | United States, Denmark, etc. | 2016 | 525 | |
| Crenigacestat | T-ALL and T-LLy | NCT02518113 | 36 | Phase I | Completed | United States, France, etc. | 2015 | 527 | |
| PF-03084014 | T-ALL and T-LLy | NCT00878189 | 8 | Phase I | Completed | United States, Italy | 2009 | 528 | |
| PF-03084014 | Advanced solid malignancies | NCT00878189 | 64 | Phase I | Completed | United States, Italy | 2009 | 526 | |
| AL101 | TNBC | NCT04461600 | 67 | Phase II | Active, not recruiting | United States, Belgium, etc. | 2020 | N/A | |
| AL101 | ACC | NCT04973683 | 14 | Phase Ib | Recruiting | Texas | 2021 | N/A | |
| Selective γ-secretase inhibitor | BMS-986115 | Advanced solid tumors | NCT01986218 | 36 | Phase I | Terminated | United States, Australia, etc. | 2013 | 529 |
| LY900009 | Advanced cancer | NCT01158404 | 35 | Phase I | Completed | United States | 2010 | 530 | |
| Nirogacestat (PF-03084014) | Desmoid tumor/aggressive fibromatosis | NCT03785964 | 142 | Phase 3 | Active, not recruiting | United States, Belgium, etc. | 2019 | 531 | |
| ADAM inhibitor | INCB7839 | Metastatic HER2+ breast cancer | NCT01254136 | 20 | Phase I/II | Terminated | United States | 2010 | N/A |
| INCB7839 | HER2-positive metastatic breast cancer | NCT00864175 | 68 | Phase I/II | Terminated | India | 2007 | N/A | |
| INCB7839 | Recurrent/ progressive high-grade gliomas | NCT04295759 | 13 | Phase I | Active, not recruiting | United States | 2020 | N/A | |
| mAb targeting Notch1 | Brontictuzumab (OMP-52M51) | Solid tumors | NCT01778439 | 48 | Phase I | Completed | United States | 2013 | 535 |
| Cross-reactive antibody targeting Notch2/3 | Tarextumab (OMP-59R5) | Solid tumors | NCT01277146 | 42 | Phase I | Completed | United States | 2010 | 536 |
| Tarextumab | Untreated metastatic PC | NCT01647828 | 177 | Phase II | Completed | United States | 2012 | 537 | |
| ADC targeting Notch3 | PF-06650808 | Breast cancer and other advanced solid tumors | NCT02129205 | 40 | Phase I | Terminated | United States | 2014 | 538 |
| ADC targeting DLL3 | Rovalpituzumab tesirine (Rova-T) | Recurrent SCLC | NCT01901653 | 82 | Phase I | Completed | United States | 2013 | 539 |
| Rovalpituzumab tesirine | Advanced, recurrent SCLC | NCT03086239 | 29 | Phase I | Completed | Japan | 2017 | 540 | |
| Rovalpituzumab tesirine | Extensive-stage SCLC | NCT02819999 | 28 | Phase I | Terminated | United States | 2016 | 541 | |
| Rovalpituzumab tesirine | DLL3-expressing, relapsed/refractory SCLC | NCT02674568 | 342 | Phase II | Completed | United States, France, etc. | 2016 | 542 | |
| Rovalpituzumab tesirine | Previously-treated extensive-stage SCLC | NCT03026166 | 42 | Phase I-II | Terminated | United States, France, etc. | 2017 | 543 | |
| Rovalpituzumab tesirine | Extensive-stage SCLC | NCT03033511 | 748 | Phase III | Terminated | United States, Australia, etc. | 2017 | 544 | |
| Rovalpituzumab tesirine | DLL3-high SCLC | NCT03061812 | 444 | Phase III | Completed | United States, Australia, etc. | 2017 | 545 | |
| Rovalpituzumab tesirine | DLL3-expressing advanced solid tumors | NCT02709889 | 200 | Phase I/II | Terminated | United States | 2016 | 546 | |
| SC-002 | Relapsed or refractory SCLC and LCNEC | NCT02500914 | 35 | Phase I | Terminated | United States | 2015 | 547 | |
| BiTE targeting DLL3 | Tarlatamab (AM757) | SCLC | NCT03319940 | 392 | Phase I | Recruiting | United States, Australia, etc. | 2017 | N/A |
| TriTAC targeting DLL3 | HPN328 | Advanced cancers | NCT04471727 | 162 | Phase I/II | Recruiting | United States | 2020 | N/A |
| mAb targeting DLL4 | Enoticumab (REGN421) | Advanced solid tumors | NCT00871559 | 83 | Phase I | Completed | United States | 2009 | 548 |
| Demcizumab (OMP-21M18) | Previously-treated solid tumors | NCT00744562 | 42 | Phase I | Completed | United States | 2008 | 549 | |
| Demcizumab (OMP-21M18) | Metastatic non-squamous NSCLC | NCT01189968 | 46 | Phase Ib | Completed | Australia, New Zealand, etc. | 2010 | 550 | |
| Demcizumab (OMP-21M18) | Platinum-resistant EOC | NCT01952249 | 20 | Phase Ib | Terminated | United States | 2013 | 551 | |
| Notch transcription complex inhibitor | CB-103 | Advanced or metastatic solid tumors and hematological malignancies | NCT03422679 | 79 | Phase I/II | Terminated | United States, France, etc. | 2017 | 553 |
CRC colorectal cancer, PDAC pancreatic ductal adenocarcinoma, EOC epithelial ovarian cancer, ACC adenoid cystic carcinoma, T-ALL T cell acute lymphoblastic leukemia, T-LLy T cell lymphoblastic lymphoma, TNBC triple-negative breast cancer, PC pancreatic cancer, SCLC small cell lung cancer, LCNEC large cell neuroendocrine carcinoma, ADAM a disintegrin and metalloprotease, mAb monoclonal antibody, ADC antibody-drug conjugate, BiTE bispecific T cell engager, TriTAC tri-specific T cell activating construct, DLL3 delta-like ligand 3
γ-Secretase inhibitors
Given their therapeutic potential in inhibiting Notch signaling in specific cancers, GSIs are actively being explored as cancer therapeutic drugs. Over the past decade, the antitumor activity of at least eight GSIs has undergone investigation in early-stage clinical trials across various tumor types. RO4929097, a Notch signaling GSI, has been a focus of clinical studies since 2010, assessing its efficacy in patients with advanced tumors. Phase II trials revealed limited clinical activity for RO4929097 as a standalone treatment in advanced tumors, including metastatic CRC,506 previously treated metastatic PDAC,507 metastatic melanoma,508 recurrent platinum-resistant EOC,509 and recurrent/progressive glioblastoma.510 As a result, RO4929097 was deemed insufficient for further single-drug study, with common mild toxicity including fatigue, nausea, and anemia. However, when combined with immunosuppressants or monoclonal antibodies, RO4929097 demonstrated good tolerance in the treatment of advanced solid tumors.511–513 Notably, its combination with endocrine therapy for endocrine-resistant Erα-positive breast cancer warrants further investigation. Unfortunately, a Phase Ib/II trial investigating the effects of RO4929097 and the hedgehog inhibitor vismodegib in advanced sarcoma did not observe any objective responses in patients.514 MK0752, used as a single drug in Phase I clinical trials for advanced solid tumors and children with refractory CNS malignancies, exhibited good tolerance.515–517 The most common drug-related adverse events included diarrhea, nausea, vomiting, and fatigue. In PDAC, the combination of MK-0752 with GEM showed a satisfactory evaluation of tumor response in 44 patients receiving the recommended Phase II dose.518 However, combining MK-0752 with the mTOR inhibitor ridaforolimus or insulin growth factor 1 receptor pathway inhibitors demonstrated clinical activity accompanied by drug-related adverse events such as diarrhea and rash.519,520 Crenigacestat (LY3039478) underwent Phase I clinical trials, alone or in combination with different anticancer drugs, in advanced or metastatic solid tumors and T-ALL and T cell lymphoblastic lymphoma (T-Lly).521–527 However, the clinical efficacy was disappointing, and crenigacestat treatment frequently resulted in dose-limiting toxicities such as fatigue, diarrhea, nausea, and vomiting. Oral GSI PF-03084014 demonstrated antitumor activity in advanced solid malignancies and T-ALL/T-Lly, supporting further evaluation of its clinical application.528 The primary drug-related toxicities of PF-03084014 include diarrhea, nausea, fatigue, and hypophosphatemia, typically ranging from mild to moderate in severity. Ongoing clinical trials are assessing the efficacy and safety of AL101 monotherapy in patients with Notch-activated recurrent or metastatic TNBC, as well as the potential benefits of AL101 before surgery for treating Notch-activated ACC. Results from these trials are awaited. Selective GSIs BMS-986115 and LY900009, explored in Phase I clinical trials, have shown safety and good tolerance for advanced tumors, exhibiting sustained targeting and biological activity in inhibiting Notch signaling.529,530 Furthermore, a Phase III clinical trial of the oral selective GSI nirogacestat (PF-03084014) demonstrated significant benefits in PFS and objective reflection of progressing desmoid tumors.531
ADAM inhibitor
The metalloproteinases ADAM10 and ADAM17 play a crucial role in cleaving Notch receptors, initiating downstream signaling that contributes to maintaining the invasive characteristics of tumors.532,533 Consequently, targeted inhibition of ADAM10 or ADAM17 represents a crucial approach to halting the progression of malignant tumors. INCB7839, an inhibitor of ADAM10 and ADAM17 proteases, has undergone assessment in Phase I–II clinical trials for previously treated solid tumors and breast cancer. However, Phase I clinical trials revealed that INCB7839 monotherapy displayed restrictive toxicity, including deep vein thrombosis, along with adverse events such as fatigue and nausea. An ongoing multi-center Phase I clinical trial is investigating INCB7839 targeting microenvironmental neuroligin-3 in the treatment of recurrent or progressive high-grade gliomas, with results yet to be reported. More recently, Nayanendu et al. developed a human anti-ADAM10 monoclonal antibody (mAb) named 1H5.534 Preclinical studies have shown that 1H5, when combined with the chemotherapeutic drug Irinotecan, effectively inhibits tumor growth in colon cancer mice without causing obvious toxic effects. Consequently, researchers hypothesize that mAb-mediated ADAM10 inhibition is a promising method to specifically prevent drug resistance and metastasis in CRC.
Antibodies targeting Notch receptors or ligands
While GSIs have exhibited robust therapeutic potential in clinical trials, their significant limitation lies in the inhibition of all Notch receptors. Consequently, highly specific mAbs targeting individual Notch receptors or ligands have been developed to address this challenge. Brontictuzumab (OMP-52M51) is a mAb that specifically targets Notch1, inhibiting the activation of the Notch pathway. In a Phase I clinical trial involving 48 subjects with solid tumors, the investigation focused on determining the maximum tolerated dose (MTD) and preliminary efficacy of brontictuzumab.535 The results indicated that brontictuzumab was well tolerated at the MTD, with diarrhea identified as the main adverse reaction, attributed to the targeted effect of Notch1 inhibition. Tarextumab (OMP-59R5) is a novel cross-reactive antibody that binds to and selectively inhibits Notch2 and Notch3 signaling pathways. In the treatment of solid tumors, tarextumab demonstrated general tolerability with dose-limited diarrhea.536 However, when combined with nab-PTX and GEM, tarextumab did not improve the survival of untreated metastatic PC.537 Additionally, PFS in patients treated with tarextumab was statistically worse. PF-06650808 is a novel anti-Notch3 antibody-drug conjugate (ADC). In a Phase I clinical trial involving 40 patients with advanced breast cancer and other advanced solid tumors, PF-06650808 displayed early signs of manageable safety and anti-tumor activity.538 The most common adverse reactions in patients treated with PF-06650808 were gastrointestinal symptoms such as decreased appetite, nausea, and abdominal pain, as well as fatigue, alopecia, and pruritus. Rovalpituzumab tesirine (Rova-T) is an ADC targeting DLL3, expressed in over 80% of SCLC. While the Phase I clinical trial of Rova-T monotherapy for recurrent SCLC demonstrated encouraging anti-tumor activity and manageable safety, subsequent Phase II and Phase III trials indicated a lack of survival benefits in extensive-stage SCLC.539–545 Additionally, Rova-T was associated with toxicities such as serosal effusion, photosensitivity, and peripheral edema. In another Phase I/II clinical trial involving 200 patients with DLL3-expressing advanced solid tumors, Rova-T exhibited controllable toxicity at the recommended Phase II dose.546 Anti-tumor activity was observed in patients with neuroendocrine carcinomas/neuroendocrine tumors, melanoma, MTC, and glioblastoma. SC-002, another DLL3-directed ADC, showed systemic toxicity and limited efficacy in Phase I clinical trials for the treatment of advanced SCLC and large cell neuroendocrine carcinoma.547 Tarlatamab (AM757) is a half-life extended bispecific T cell engager (BiTE®) targeting DLL3. A Phase I study is currently evaluating the safety, tolerability, and pharmacokinetics of tarlatamab in patients with SCLC. HPN328, a tri-specific T cell activating construct (TriTAC®) targeting DLL3, is undergoing a Phase I/II trial to assess safety, tolerability, and pharmacokinetics, both as monotherapy and in combination with atezolizumab, in patients with advanced cancer associated with DLL3 expression. Enoticumab (REGN421) is a fully human IgG (1) mAb that binds to human DLL4, disrupting Notch-mediated signal transduction. In a Phase I trial, enoticumab was well tolerated in the treatment of advanced solid tumors, with observed treatment responses.548 Demcizumab (OMP-21M18) is an IgG2 humanized mAb targeting DLL4. Phase I clinical trials suggested that demcizumab is generally well tolerated and exhibits anti-tumor activity in previously treated solid tumors.549 Subsequent Phase IB clinical trials revealed that 50% of patients with metastatic non-squamous NSCLC had an objective tumor response after treatment with the truncated demcizumab regimen.550 In platinum-resistant EOC, demcizumab combined with PTX demonstrated controllable toxicity and activity in patients with severely pretreated platinum-resistant patients with OC.551
Notch transcription complex inhibitors
Notch signaling initiates downstream cascades by guiding the formation of core transcriptional activation complexes. In addition to targeting the upstream components of the Notch signaling cascade through GSIs or antibodies that disrupt the interaction between Notch receptors and ligands, inhibiting transcriptional activation complexes offer an attractive approach to prevent Notch signal transduction.552 CB-103, the first small molecule drug developed to effectively inhibit the Notch transcription complex, underwent a Phase I/II clinical trial involving 79 adult patients with advanced or metastatic solid tumors and hematological malignancies.553 The trial demonstrated that CB-103 was well-tolerated, with 19% of patients experiencing grade 3–4 treatment-related adverse events, including elevated liver function, anemia, and visual changes, 6% of patients discontinued treatment due to toxicity. Recent studies have revealed that CB-103 has in vitro antitumor activity in a small subset of lymphoma cell lines from various lymphoma subtypes, with activity surpassing that achieved by GSIs.554 In preclinical models of endocrine-resistant and TNBC, CB-103, when combined with fulvestrant or PTX, exhibits synergistic effects, effectively inhibiting the formation of breast spheroids.555 Moellering et al. introduced a hydrocarbon-stapled peptide named SAHM1, which can prevent the assembly of active transcription complexes.556 In vivo and in vitro experiments have demonstrated that SAHM1 can treat T-ALL cells by globally suppressing Notch-activated genes. Another study identified a small molecule inhibitor, mastermind recruitment-1 (IMR-1), which targets the inhibition of the Notch transcription-activating complex.557 IMR-1 represents a potential new paradigm for Notch-based anticancer therapy.
γ-Secretase modulators
GSMs have emerged as preferable drug candidates in response to the observed toxicity linked to non-selective GSIs in clinical trials. Unlike GSIs, GSMs do not inhibit gamma-secretase itself; instead, they are designed to regulate the catalytic activity of gamma-secretase, thereby influencing the function of Notch signaling.558,559 The initial objective in the quest for GSMs was to diminish the production of the 42-amino acid amyloid β peptide variant in the brains of patients with Alzheimer’s disease, without impeding the hydrolysis of Notch protein or causing the accumulation of carboxy-terminal fragments of the amyloid precursor protein.560,561 A preclinical study demonstrated that MRK560, a GSM targeting PSEN1, effectively reduced the processing of Notch1 mutants and induced cell cycle arrest without causing intestinal toxicity in T-ALL animal models.562 In certain scenarios, GSMs may present a potential alternative to GSIs, but further preclinical trials and clinical studies are still required to validate their efficacy and safety.
Conclusions and perspectives
Since the initial discovery of the Notch protein family, our understanding of the Notch signaling pathway has deepened significantly. Despite the simple structure of the Notch signaling cascade, which involves only a few steps from ligand binding to initiating downstream target gene transcription, the biological functions of the Notch signaling pathway are complex and diverse in different systems. Overall, the effects of the Notch signaling heavily rely on the cellular environment and involve intricate crosstalk with other signaling pathways. The studies summarized in this review provide compelling evidence that Notch signaling pathway plays a considerable role in human malignancies. Given the complex oncogenic or tumor suppressive functions of the Notch signaling pathway in different malignancies, it is of great significance to focus on understanding the mechanisms through which the Notch signaling pathway regulates tumorigenesis and development.
Present research areas include the regulation of the Notch signaling pathway in tumors via biological processes like EMT, angiogenesis, and cancer metabolic reprogramming. However, it is vital to acknowledge that this investigation field is complicated, and related molecular mechanisms have not been extensively studied. In particular, the Notch signaling cascade is a crucial tumor suppressor in multiple cellular contexts and cancer types. Further studies are needed to delve into the precise mechanism of Notch-mediated tumor suppression, which will be beneficial for developing novel therapeutic strategies.
Currently, various inhibitors targeting γ-secretase, ADAM, and the Notch transcription complex, as well as antibodies targeting Notch receptors and ligands, have been proposed to control tumor progression. While early clinical trials have shown that therapies targeting the Notch signaling pathway exhibit some antitumor activity, the development of safe, effective, and tumor-specific Notch-targeted drugs for clinical use remains a significant challenge. It is important to note that tumors with Notch inactivating mutations, like HNSCC, are not appropriate for “anti-Notch” treatment approaches. Conversely, Notch receptor-specific antibody agonizts could be clinically valuable in tumors where Notch signaling acts as a tumor suppressor. Moreover, solely targeting the Notch signaling pathway may prove insufficient for effective cancer treatment. The combination of Notch-targeted drugs with immune checkpoint inhibitors, anti-angiogenic agents, or chemotherapy holds the promise of enhancing synergistic therapeutic effects. Continued research in this area is essential for unlocking the full potential of Notch-targeted therapies in cancer treatment.
To optimize drug development efforts based on Notch signaling, several strategies can be considered. First, combining Notch-targeted therapy with carrier-based nanomaterials may enhance drug delivery efficiency.563 Second, a deeper exploration of the role of Notch signaling in regulating the TME can inform the design of immune therapies centered on Notch signaling. For instance, packaging Notch-targeted drugs into oncolytic viruses and releasing them into the TME can inhibit the recruitment and activation of immune-suppressive cells.564 Moreover, comprehensive research on the complex interaction networks between Notch signaling and pathways such as Hedgehog and Wnt can provide more compelling evidence for rational combination therapies.565
It is noteworthy that the advancement of high-throughput sequencing technology and artificial intelligence holds the potential to elucidate the molecular mechanisms of the Notch signaling pathway in specific tumors. This could offer new perspectives on the pathogenesis and therapeutic targets of cancer. The exponential growth of our understanding of the Notch signaling pathway in tumor biology over the past two decades underscores the need to translate this fundamental science into clinical practice. The time is ripe for harnessing this knowledge to advance more effective and targeted cancer therapies.
Acknowledgements
This work was supported by the Fundamental Research Funds for the Central Universities (grant no. 2022ZFJH003); the Opening Foundation of the State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, Zhejiang University School of Medicine (grant no. SKLID2024KF01).
Author contributions
L.J.L. and D.H.Z. conceived the central concept of this review and delineated its overall structure. Q.M.S., C.X., and Y.F.Z. contributed to the literature search and original draft preparation. X.Y., Q.F.C., and S.W.J. participated in figure visualization and table organizing. J.Z.W. and Y.Q.Z. helped with the investigation and outline development. L.J.L. and D.H.Z. provided guidance and supervision during the writing process, and critically reviewed and edited the manuscript. All authors have read and approved the article.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Qingmiao Shi, Chen Xue, Yifan Zeng
Contributor Information
Danhua Zhu, Email: zhudanhua@zju.edu.cn.
Lanjuan Li, Email: ljli@zju.edu.cn.
References
- 1.Metz CW, Bridges CB. Incompatibility of mutant races in Drosophila. Proc. Natl Acad. Sci. USA. 1917;3:673–678. doi: 10.1073/pnas.3.12.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Muskavitch MA, Yedvobnick B. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 1983;80:1977–1981. doi: 10.1073/pnas.80.7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wharton KA, Johansen KM, Xu T, Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- 4.Perron M, Harris WA. Determination of vertebrate retinal progenitor cell fate by the Notch pathway and basic helix-loop-helix transcription factors. Cell Mol. Life Sci. 2000;57:215–223. doi: 10.1007/PL00000685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao J, et al. Cell-fate transition and determination analysis of mouse male germ cells throughout development. Nat. Commun. 2021;12:6839. doi: 10.1038/s41467-021-27172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellisen LW, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-B. [DOI] [PubMed] [Google Scholar]
- 7.Wang NJ, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc. Natl Acad. Sci. USA. 2011;108:17761–17766. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang K, et al. PEST domain mutations in Notch receptors comprise an oncogenic driver segment in triple-negative breast cancer sensitive to a γ-secretase inhibitor. Clin. Cancer Res. 2015;21:1487–1496. doi: 10.1158/1078-0432.CCR-14-1348. [DOI] [PubMed] [Google Scholar]
- 9.Larose H, et al. Whole Exome Sequencing reveals NOTCH1 mutations in anaplastic large cell lymphoma and points to Notch both as a key pathway and a potential therapeutic target. Haematologica. 2021;106:1693–1704. doi: 10.3324/haematol.2019.238766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Ianni M, et al. A new genetic lesion in B-CLL: a NOTCH1 PEST domain mutation. Br. J. Haematol. 2009;146:689–691. doi: 10.1111/j.1365-2141.2009.07816.x. [DOI] [PubMed] [Google Scholar]
- 11.Lujambio A, Maina F. Turning up our understanding of liver cancer by a notch. J. Hepatol. 2021;74:502–504. doi: 10.1016/j.jhep.2020.10.027. [DOI] [PubMed] [Google Scholar]
- 12.Mangolini M, et al. Viral transduction of primary human lymphoma B cells reveals mechanisms of NOTCH-mediated immune escape. Nat. Commun. 2022;13:6220. doi: 10.1038/s41467-022-33739-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu W, Nie L, Zhang L, Li Y. The notch pathway promotes NF-κB activation through Asb2 in T cell acute lymphoblastic leukemia cells. Cell Mol. Biol. Lett. 2018;23:37. doi: 10.1186/s11658-018-0102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, et al. Activation of NOTCH signaling via DLL1 is mediated by APE1-redox-dependent NF-κB activation in oesophageal adenocarcinoma. Gut. 2023;72:421–432. doi: 10.1136/gutjnl-2022-327076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shawber C, et al. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 16.Shin HM, et al. Notch1 augments NF-kappaB activity by facilitating its nuclear retention. EMBO J. 2006;25:129–138. doi: 10.1038/sj.emboj.7600902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siebel C, Lendahl U. Notch signaling in development, tissue homeostasis, and disease. Physiol. Rev. 2017;97:1235–1294. doi: 10.1152/physrev.00005.2017. [DOI] [PubMed] [Google Scholar]
- 18.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kidd S, Lockett TJ, Young MW. The Notch locus of Drosophila melanogaster. Cell. 1983;34:421–433. doi: 10.1016/0092-8674(83)90376-8. [DOI] [PubMed] [Google Scholar]
- 20.Chillakuri CR, Sheppard D, Lea SM, Handford PA. Notch receptor-ligand binding and activation: insights from molecular studies. Semin. Cell Dev. Biol. 2012;23:421–428. doi: 10.1016/j.semcdb.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rana NA, Haltiwanger RS. Fringe benefits: functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr. Opin. Struct. Biol. 2011;21:583–589. doi: 10.1016/j.sbi.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovall RA, Gebelein B, Sprinzak D, Kopan R. The canonical notch signaling pathway: structural and biochemical insights into shape, sugar, and force. Dev. Cell. 2017;41:228–241. doi: 10.1016/j.devcel.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon WR, et al. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009;113:4381–4390. doi: 10.1182/blood-2008-08-174748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephenson NL, Avis JM. Direct observation of proteolytic cleavage at the S2 site upon forced unfolding of the Notch negative regulatory region. Proc. Natl Acad. Sci. USA. 2012;109:E2757–E2765. doi: 10.1073/pnas.1205788109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li K, et al. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J. Biol. Chem. 2008;283:8046–8054. doi: 10.1074/jbc.M800170200. [DOI] [PubMed] [Google Scholar]
- 26.Deatherage CL, Lu Z, Kim JH, Sanders CR. Notch transmembrane domain: secondary structure and topology. Biochemistry. 2015;54:3565–3568. doi: 10.1021/acs.biochem.5b00456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Majumder S, et al. Targeting notch in oncology: the path forward. Nat. Rev. Drug Discov. 2021;20:125–144. doi: 10.1038/s41573-020-00091-3. [DOI] [PubMed] [Google Scholar]
- 28.Bhanushali AA, et al. Mutations in the HD and PEST domain of Notch-1 receptor in T-cell acute lymphoblastic leukemia: report of novel mutations from Indian population. Oncol. Res. 2010;19:99–104. doi: 10.3727/096504010X12864748215007. [DOI] [PubMed] [Google Scholar]
- 29.Sprinzak D, Blacklow SC. Biophysics of Notch signaling. Annu Rev. Biophys. 2021;50:157–189. doi: 10.1146/annurev-biophys-101920-082204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishna BM, et al. Notch signaling in breast cancer: from pathway analysis to therapy. Cancer Lett. 2019;461:123–131. doi: 10.1016/j.canlet.2019.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varshney S, Stanley P. Multiple roles for O-glycans in Notch signalling. FEBS Lett. 2018;592:3819–3834. doi: 10.1002/1873-3468.13251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsumoto K, Luther KB, Haltiwanger RS. Diseases related to Notch glycosylation. Mol. Asp. Med. 2021;79:100938. doi: 10.1016/j.mam.2020.100938. [DOI] [PubMed] [Google Scholar]
- 33.Logeat F, et al. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl Acad. Sci. USA. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lake RJ, Grimm LM, Veraksa A, Banos A, Artavanis-Tsakonas S. In vivo analysis of the Notch receptor S1 cleavage. PLoS One. 2009;4:e6728. doi: 10.1371/journal.pone.0006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lieber T, Kidd S, Young MW. Kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 2002;16:209–221. doi: 10.1101/gad.942302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zolkiewska A. ADAM proteases: ligand processing and modulation of the Notch pathway. Cell Mol. Life Sci. 2008;65:2056–2068. doi: 10.1007/s00018-008-7586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Strooper B, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 38.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 39.Kim GS, Park HS, Lee YC. OPTHiS identifies the molecular basis of the direct interaction between CSL and SMRT corepressor. Mol. Cells. 2018;41:842–852. doi: 10.14348/molcells.2018.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanders PG, et al. Ligand-independent traffic of Notch buffers activated Armadillo in Drosophila. PLoS Biol. 2009;7:e1000169. doi: 10.1371/journal.pbio.1000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon C, et al. Notch post-translationally regulates β-catenin protein in stem and progenitor cells. Nat. Cell Biol. 2011;13:1244–1251. doi: 10.1038/ncb2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanalkumar R, Dhanesh SB, James J. Non-canonical activation of Notch signaling/target genes in vertebrates. Cell Mol. Life Sci. 2010;67:2957–2968. doi: 10.1007/s00018-010-0391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andersen P, Uosaki H, Shenje LT, Kwon C. Non-canonical Notch signaling: emerging role and mechanism. Trends Cell Biol. 2012;22:257–265. doi: 10.1016/j.tcb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hurlbut GD, Kankel MW, Lake RJ, Artavanis-Tsakonas S. Crossing paths with Notch in the hyper-network. Curr. Opin. Cell Biol. 2007;19:166–175. doi: 10.1016/j.ceb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 45.Liu L, et al. Non-canonical notch signaling regulates actin remodeling in cell migration by activating PI3K/AKT/Cdc42 pathway. Front Pharm. 2019;10:370. doi: 10.3389/fphar.2019.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhat V, Sun YJ, Weger S, Raouf A. Notch-induced expression of FZD7 requires noncanonical NOTCH3 signaling in human breast epithelial cells. Stem Cells Dev. 2016;25:522–529. doi: 10.1089/scd.2015.0315. [DOI] [PubMed] [Google Scholar]
- 47.Jin S, et al. Non-canonical Notch signaling activates IL-6/JAK/STAT signaling in breast tumor cells and is controlled by p53 and IKKα/IKKβ. Oncogene. 2013;32:4892–4902. doi: 10.1038/onc.2012.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Q, Lu Q. Plasma membrane-derived extracellular microvesicles mediate non-canonical intercellular NOTCH signaling. Nat. Commun. 2017;8:709. doi: 10.1038/s41467-017-00767-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.González-King H, et al. Non-classical Notch signaling by MDA-MB-231 breast cancer cell-derived small extracellular vesicles promotes malignancy in poorly invasive MCF-7 cells. Cancer Gene Ther. 2022;29:1056–1069. doi: 10.1038/s41417-021-00411-8. [DOI] [PubMed] [Google Scholar]
- 50.Lee KS, et al. Roles of PINK1, mTORC2, and mitochondria in preserving brain tumor-forming stem cells in a noncanonical Notch signaling pathway. Genes Dev. 2013;27:2642–2647. doi: 10.1101/gad.225169.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perumalsamy LR, Nagala M, Banerjee P, Sarin A. A hierarchical cascade activated by non-canonical Notch signaling and the mTOR-Rictor complex regulates neglect-induced death in mammalian cells. Cell Death Differ. 2009;16:879–889. doi: 10.1038/cdd.2009.20. [DOI] [PubMed] [Google Scholar]
- 52.Ma J, et al. Noncanonical activation of Notch1 protein by membrane type 1 matrix metalloproteinase (MT1-MMP) controls melanoma cell proliferation. J. Biol. Chem. 2014;289:8442–8449. doi: 10.1074/jbc.M113.516039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin S, et al. Non-canonical NOTCH3 signalling limits tumour angiogenesis. Nat. Commun. 2017;8:16074. doi: 10.1038/ncomms16074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aster JC, Pear WS, Blacklow SC. The varied roles of notch in cancer. Annu. Rev. Pathol. 2017;12:245–275. doi: 10.1146/annurev-pathol-052016-100127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hashemi M, et al. Non-coding RNAs targeting notch signaling pathway in cancer: from proliferation to cancer therapy resistance. Int. J. Biol. Macromol. 2022;222:1151–1167. doi: 10.1016/j.ijbiomac.2022.09.203. [DOI] [PubMed] [Google Scholar]
- 56.Fender AW, Nutter JM, Fitzgerald TL, Bertrand FE, Sigounas G. Notch-1 promotes stemness and epithelial to mesenchymal transition in colorectal cancer. J. Cell Biochem. 2015;116:2517–2527. doi: 10.1002/jcb.25196. [DOI] [PubMed] [Google Scholar]
- 57.Xiong Y, et al. Correlation of over-expressions of miR-21 and Notch-1 in human colorectal cancer with clinical stages. Life Sci. 2014;106:19–24. doi: 10.1016/j.lfs.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 58.Li G, et al. The expression profile and clinicopathological significance of Notch1 in patients with colorectal cancer: a meta-analysis. Future Oncol. 2017;13:2103–2118. doi: 10.2217/fon-2017-0178. [DOI] [PubMed] [Google Scholar]
- 59.Chu D, et al. Notch1 and Notch2 have opposite prognostic effects on patients with colorectal cancer. Ann. Oncol. 2011;22:2440–2447. doi: 10.1093/annonc/mdq776. [DOI] [PubMed] [Google Scholar]
- 60.Ozawa T, et al. Nuclear Notch3 expression is associated with tumor recurrence in patients with stage II and III colorectal cancer. Ann. Surg. Oncol. 2014;21:2650–2658. doi: 10.1245/s10434-014-3659-9. [DOI] [PubMed] [Google Scholar]
- 61.Zhang Z, et al. NOTCH4 regulates colorectal cancer proliferation, invasiveness, and determines clinical outcome of patients. J. Cell Physiol. 2018;233:6975–6985. doi: 10.1002/jcp.26619. [DOI] [PubMed] [Google Scholar]
- 62.Sugiyama M, et al. High expression of the Notch ligand Jagged-1 is associated with poor prognosis after surgery for colorectal cancer. Cancer Sci. 2016;107:1705–1716. doi: 10.1111/cas.13075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.He W, et al. Mutual regulation of JAG2 and PRAF2 promotes migration and invasion of colorectal cancer cells uncoupled from epithelial-mesenchymal transition. Cancer Cell Int. 2019;19:160. doi: 10.1186/s12935-019-0871-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim G, Jung J, Kim JW, Kim JY. Low HES-1 and positive DLL4 expression predicts poor prognosis of colorectal cancers. Pathology. 2023;55:52–57. doi: 10.1016/j.pathol.2022.07.008. [DOI] [PubMed] [Google Scholar]
- 65.Varga, J. et al. AKT-dependent NOTCH3 activation drives tumor progression in a model of mesenchymal colorectal cancer. J. Exp. Med. 217, e20191515 (2020). [DOI] [PMC free article] [PubMed]
- 66.Sonoshita M, et al. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell. 2011;19:125–137. doi: 10.1016/j.ccr.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 67.Kawaguchi K, et al. Jagged1 DNA copy number variation is associated with poor outcome in liver cancer. Am. J. Pathol. 2016;186:2055–2067. doi: 10.1016/j.ajpath.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 68.Liu J, et al. Loss of function of Notch1 identifies a poor prognosis group of early-stage hepatocellular carcinoma following hepatectomy. Oncol. Rep. 2015;34:3174–3186. doi: 10.3892/or.2015.4300. [DOI] [PubMed] [Google Scholar]
- 69.Ahn S, Hyeon J, Park CK. Notch1 and Notch4 are markers for poor prognosis of hepatocellular carcinoma. Hepatobiliary Pancreat. Dis. Int. 2013;12:286–294. doi: 10.1016/S1499-3872(13)60046-6. [DOI] [PubMed] [Google Scholar]
- 70.Gao J, et al. Hepatitis B virus X protein activates Notch signaling by its effects on Notch1 and Notch4 in human hepatocellular carcinoma. Int. J. Oncol. 2016;48:329–337. doi: 10.3892/ijo.2015.3221. [DOI] [PubMed] [Google Scholar]
- 71.Hayashi Y, Osanai M, Lee GH. NOTCH2 signaling confers immature morphology and aggressiveness in human hepatocellular carcinoma cells. Oncol. Rep. 2015;34:1650–1658. doi: 10.3892/or.2015.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu WR, et al. Notch2 is a crucial regulator of self-renewal and tumorigenicity in human hepatocellular carcinoma cells. Oncol. Rep. 2016;36:181–188. doi: 10.3892/or.2016.4831. [DOI] [PubMed] [Google Scholar]
- 73.Dill MT, et al. Constitutive Notch2 signaling induces hepatic tumors in mice. Hepatology. 2013;57:1607–1619. doi: 10.1002/hep.26165. [DOI] [PubMed] [Google Scholar]
- 74.Hu L, Xue F, Shao M, Deng A, Wei G. Aberrant expression of Notch3 predicts poor survival for hepatocellular carcinomas. Biosci. Trends. 2013;7:152–156. [PubMed] [Google Scholar]
- 75.Gramantieri L, et al. Aberrant Notch3 and Notch4 expression in human hepatocellular carcinoma. Liver Int. 2007;27:997–1007. doi: 10.1111/j.1478-3231.2007.01544.x. [DOI] [PubMed] [Google Scholar]
- 76.Gao J, et al. Expression of Jagged1 and its association with hepatitis B virus X protein in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2007;356:341–347. doi: 10.1016/j.bbrc.2007.02.130. [DOI] [PubMed] [Google Scholar]
- 77.Kunanopparat A, Hirankarn N, Issara-Amphorn J, Tangkijvanich P, Sanpavat A. The expression profile of Jagged1 and Delta-like 4 in hepatocellular carcinoma. Asian Pac. J. Allergy Immunol. 2021;39:44–52. doi: 10.12932/AP-040818-0388. [DOI] [PubMed] [Google Scholar]
- 78.Zhang J, et al. Evaluation of Jagged2 and Gli1 expression and their correlation with prognosis in human hepatocellular carcinoma. Mol. Med. Rep. 2014;10:749–754. doi: 10.3892/mmr.2014.2246. [DOI] [PubMed] [Google Scholar]
- 79.Ren K, et al. miR-199a-3p inhibits cell proliferation and induces apoptosis by targeting YAP1, suppressing Jagged1-Notch signaling in human hepatocellular carcinoma. J. Biomed. Sci. 2016;23:79. doi: 10.1186/s12929-016-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao YB, et al. Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 2014;46:1097–1102. doi: 10.1038/ng.3076. [DOI] [PubMed] [Google Scholar]
- 81.Li L, et al. Clinical outcome-related cancer pathways and mutational signatures in patients with unresectable esophageal squamous cell carcinoma treated with chemoradiotherapy. Int J. Radiat. Oncol. Biol. Phys. 2023;115:382–394. doi: 10.1016/j.ijrobp.2022.07.1835. [DOI] [PubMed] [Google Scholar]
- 82.Colom B, et al. Spatial competition shapes the dynamic mutational landscape of normal esophageal epithelium. Nat. Genet. 2020;52:604–614. doi: 10.1038/s41588-020-0624-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Colom B, et al. Mutant clones in normal epithelium outcompete and eliminate emerging tumours. Nature. 2021;598:510–514. doi: 10.1038/s41586-021-03965-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Abby E, et al. Notch1 mutations drive clonal expansion in normal esophageal epithelium but impair tumor growth. Nat. Genet. 2023;55:232–245. doi: 10.1038/s41588-022-01280-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lubin DJ, Mick R, Shroff SG, Stashek K, Furth EE. The notch pathway is activated in neoplastic progression in esophageal squamous cell carcinoma. Hum. Pathol. 2018;72:66–70. doi: 10.1016/j.humpath.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 86.Gao K, et al. The Notch1 gene may control cell chemoresistance in esophageal squamous cell cancer. Transl. Cancer Res. 2021;10:3278–3285. doi: 10.21037/tcr-21-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang C, et al. Notch2 as a promising prognostic biomarker for oesophageal squamous cell carcinoma. Sci. Rep. 2016;6:25722. doi: 10.1038/srep25722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matsuura N, et al. NOTCH3 limits the epithelial-mesenchymal transition and predicts a favorable clinical outcome in esophageal cancer. Cancer Med. 2021;10:3986–3996. doi: 10.1002/cam4.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ye Y, et al. COX-2 regulates Snail expression in gastric cancer via the Notch1 signaling pathway. Int. J. Mol. Med. 2017;40:512–522. doi: 10.3892/ijmm.2017.3011. [DOI] [PubMed] [Google Scholar]
- 90.Hsu KW, et al. Activation of the Notch1/STAT3/Twist signaling axis promotes gastric cancer progression. Carcinogenesis. 2012;33:1459–1467. doi: 10.1093/carcin/bgs165. [DOI] [PubMed] [Google Scholar]
- 91.Huang B, et al. Elevated expression of NOTCH1 associates with lymph node metastasis of gastric cancer and knock-down of NOTCH1 attenuates tumor cell progression. Med. Sci. Monit. 2019;25:9939–9948. doi: 10.12659/MSM.918703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang KH, et al. Correlation between HGF/c-Met and Notch1 signaling pathways in human gastric cancer cells. Oncol. Rep. 2018;40:294–302. doi: 10.3892/or.2018.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun Y, et al. Differential Notch1 and Notch2 expression and frequent activation of Notch signaling in gastric cancers. Arch. Pathol. Lab Med. 2011;135:451–458. doi: 10.5858/2009-0665-OA.1. [DOI] [PubMed] [Google Scholar]
- 94.Yeh TS, et al. The activated Notch1 signal pathway is associated with gastric cancer progression through cyclooxygenase-2. Cancer Res. 2009;69:5039–5048. doi: 10.1158/0008-5472.CAN-08-4021. [DOI] [PubMed] [Google Scholar]
- 95.Demitrack ES, et al. Notch signaling regulates gastric antral LGR5 stem cell function. EMBO J. 2015;34:2522–2536. doi: 10.15252/embj.201490583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang W, et al. Hormonal suppression of stem cells inhibits symmetric cell division and gastric tumorigenesis. Cell Stem Cell. 2020;26:739–754.e738. doi: 10.1016/j.stem.2020.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cui Y, et al. NOTCH3 is a prognostic factor and is correlated with immune tolerance in gastric cancer. Front. Oncol. 2020;10:574937. doi: 10.3389/fonc.2020.574937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu H, et al. Expression of Jagged1 predicts postoperative clinical outcome of patients with gastric cancer. Int. J. Clin. Exp. Med. 2015;8:14782–14792. [PMC free article] [PubMed] [Google Scholar]
- 99.Segami K, et al. Clinical significance of TAP1 and DLL4 expression in patients with locally advanced gastric cancer. In Vivo. 2021;35:2771–2777. doi: 10.21873/invivo.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 101.Mullendore ME, et al. Ligand-dependent Notch signaling is involved in tumor initiation and tumor maintenance in pancreatic cancer. Clin. Cancer Res. 2009;15:2291–2301. doi: 10.1158/1078-0432.CCR-08-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Büchler P, et al. The Notch signaling pathway is related to neurovascular progression of pancreatic cancer. Ann. Surg. 2005;242:791–800. doi: 10.1097/01.sla.0000189115.94847.f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Doucas H, et al. Expression of nuclear Notch3 in pancreatic adenocarcinomas is associated with adverse clinical features, and correlates with the expression of STAT3 and phosphorylated Akt. J. Surg. Oncol. 2008;97:63–68. doi: 10.1002/jso.20894. [DOI] [PubMed] [Google Scholar]
- 104.Eto K, et al. Human equilibrative nucleoside transporter 1 and Notch3 can predict gemcitabine effects in patients with unresectable pancreatic cancer. Br. J. Cancer. 2013;108:1488–1494. doi: 10.1038/bjc.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yao J, Qian C. Inhibition of Notch3 enhances sensitivity to gemcitabine in pancreatic cancer through an inactivation of PI3K/Akt-dependent pathway. Med. Oncol. 2010;27:1017–1022. doi: 10.1007/s12032-009-9326-5. [DOI] [PubMed] [Google Scholar]
- 106.Lee J, Lee J, Kim JH. Association of Jagged1 expression with malignancy and prognosis in human pancreatic cancer. Cell Oncol. 2020;43:821–834. doi: 10.1007/s13402-020-00527-3. [DOI] [PubMed] [Google Scholar]
- 107.Chen HT, et al. High expression of delta-like ligand 4 predicts poor prognosis after curative resection for pancreatic cancer. Ann. Surg. Oncol. 2012;19:S464–S474. doi: 10.1245/s10434-011-1968-9. [DOI] [PubMed] [Google Scholar]
- 108.Cao F, et al. HES 1 is essential for chemoresistance induced by stellate cells and is associated with poor prognosis in pancreatic cancer. Oncol. Rep. 2015;33:1883–1889. doi: 10.3892/or.2015.3789. [DOI] [PubMed] [Google Scholar]
- 109.Drouillard A, et al. DLL4 expression is a prognostic marker and may predict gemcitabine benefit in resected pancreatic cancer. Br. J. Cancer. 2016;115:1245–1252. doi: 10.1038/bjc.2016.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yen WC, et al. Anti-DLL4 has broad spectrum activity in pancreatic cancer dependent on targeting DLL4-Notch signaling in both tumor and vasculature cells. Clin. Cancer Res. 2012;18:5374–5386. doi: 10.1158/1078-0432.CCR-12-0736. [DOI] [PubMed] [Google Scholar]
- 111.Rizvi S, Khan SA, Hallemeier CL, Kelley RK, Gores GJ. Cholangiocarcinoma—evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 2018;15:95–111. doi: 10.1038/nrclinonc.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sekiya S, Suzuki A. Intrahepatic cholangiocarcinoma can arise from Notch-mediated conversion of hepatocytes. J. Clin. Investig. 2012;122:3914–3918. doi: 10.1172/JCI63065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cigliano A, Wang J, Chen X, Calvisi DF. Role of the Notch signaling in cholangiocarcinoma. Expert Opin. Ther. Targets. 2017;21:471–483. doi: 10.1080/14728222.2017.1310842. [DOI] [PubMed] [Google Scholar]
- 114.Zender S, et al. A critical role for notch signaling in the formation of cholangiocellular carcinomas. Cancer Cell. 2013;23:784–795. doi: 10.1016/j.ccr.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 115.Li JH, et al. MFAP5 facilitates the aggressiveness of intrahepatic Cholangiocarcinoma by activating the Notch1 signaling pathway. J. Exp. Clin. Cancer Res. 2019;38:476. doi: 10.1186/s13046-019-1477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang J, et al. Notch2 controls hepatocyte-derived cholangiocarcinoma formation in mice. Oncogene. 2018;37:3229–3242. doi: 10.1038/s41388-018-0188-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang T, et al. Cellular heterogeneity and transcriptomic profiles during intrahepatic cholangiocarcinoma initiation and progression. Hepatology. 2022;76:1302–1317. doi: 10.1002/hep.32483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hu S, et al. NOTCH-YAP1/TEAD-DNMT1 axis drives hepatocyte reprogramming into intrahepatic cholangiocarcinoma. Gastroenterology. 2022;163:449–465. doi: 10.1053/j.gastro.2022.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ament CE, et al. Aberrant fucosylation sustains the NOTCH and EGFR/NF-κB pathways and has a prognostic value in human intrahepatic cholangiocarcinoma. Hepatology. 2023;78:1742–1754. doi: 10.1097/HEP.0000000000000322. [DOI] [PubMed] [Google Scholar]
- 120.Wu WR, et al. Clinicopathological significance of aberrant Notch receptors in intrahepatic cholangiocarcinoma. Int. J. Clin. Exp. Pathol. 2014;7:3272–3279. [PMC free article] [PubMed] [Google Scholar]
- 121.Yoon HA, et al. Clinicopathological significance of altered Notch signaling in extrahepatic cholangiocarcinoma and gallbladder carcinoma. World J. Gastroenterol. 2011;17:4023–4030. doi: 10.3748/wjg.v17.i35.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhou Q, Wang Y, Peng B, Liang L, Li J. The roles of Notch1 expression in the migration of intrahepatic cholangiocarcinoma. BMC Cancer. 2013;13:244. doi: 10.1186/1471-2407-13-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wu WR, et al. Notch1 is overexpressed in human intrahepatic cholangiocarcinoma and is associated with its proliferation, invasiveness and sensitivity to 5-fluorouracil in vitro. Oncol. Rep. 2014;31:2515–2524. doi: 10.3892/or.2014.3123. [DOI] [PubMed] [Google Scholar]
- 124.Guest RV, et al. Notch3 drives development and progression of cholangiocarcinoma. Proc. Natl Acad. Sci. USA. 2016;113:12250–12255. doi: 10.1073/pnas.1600067113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Che L, et al. Jagged 1 is a major Notch ligand along cholangiocarcinoma development in mice and humans. Oncogenesis. 2016;5:e274. doi: 10.1038/oncsis.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Leiter A, Veluswamy RR, Wisnivesky JP. The global burden of lung cancer: current status and future trends. Nat. Rev. Clin. Oncol. 2023;20:624–639. doi: 10.1038/s41571-023-00798-3. [DOI] [PubMed] [Google Scholar]
- 127.Howlader N, et al. The effect of advances in lung-cancer treatment on population mortality. N. Engl. J. Med. 2020;383:640–649. doi: 10.1056/NEJMoa1916623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nguyen D, et al. Notch1 phenotype and clinical stage progression in non-small cell lung cancer. J. Hematol. Oncol. 2015;8:9. doi: 10.1186/s13045-014-0104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yuan X, et al. Meta-analysis reveals the correlation of Notch signaling with non-small cell lung cancer progression and prognosis. Sci. Rep. 2015;5:10338. doi: 10.1038/srep10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang Y, et al. Evaluation of the correlation of vasculogenic mimicry, Notch4, DLL4, and KAI1/CD82 in the prediction of metastasis and prognosis in non-small cell lung cancer. Medicines. 2018;97:e13817. doi: 10.1097/MD.0000000000013817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Pancewicz-Wojtkiewicz J, et al. Prognostic significance of Notch ligands in patients with non-small cell lung cancer. Oncol. Lett. 2017;13:506–510. doi: 10.3892/ol.2016.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Donnem T, et al. Prognostic impact of Notch ligands and receptors in nonsmall cell lung cancer: coexpression of Notch-1 and vascular endothelial growth factor—a predicts poor survival. Cancer. 2010;116:5676–5685. doi: 10.1002/cncr.25551. [DOI] [PubMed] [Google Scholar]
- 133.Sharif A, Shaji A, Chammaa M, Pawlik E, Fernandez-Valdivia R. Notch transduction in non-small cell lung cancer. Int. J. Mol. Sci. 2020;21:5691. doi: 10.3390/ijms21165691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xie M, He CS, Wei SH, Zhang L. Notch-1 contributes to epidermal growth factor receptor tyrosine kinase inhibitor acquired resistance in non-small cell lung cancer in vitro and in vivo. Eur. J. Cancer. 2013;49:3559–3572. doi: 10.1016/j.ejca.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 135.Liu L, et al. An RFC4/Notch1 signaling feedback loop promotes NSCLC metastasis and stemness. Nat. Commun. 2021;12:2693. doi: 10.1038/s41467-021-22971-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Baumgart A, et al. Opposing role of Notch1 and Notch2 in a Kras(G12D)-driven murine non-small cell lung cancer model. Oncogene. 2015;34:578–588. doi: 10.1038/onc.2013.592. [DOI] [PubMed] [Google Scholar]
- 137.Zheng Y, et al. A rare population of CD24(+)ITGB4(+)Notch(hi) cells drives tumor propagation in NSCLC and requires Notch3 for self-renewal. Cancer Cell. 2013;24:59–74. doi: 10.1016/j.ccr.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sinicropi-Yao SL, et al. Co-expression analysis reveals mechanisms underlying the varied roles of NOTCH1 in NSCLC. J. Thorac. Oncol. 2019;14:223–236. doi: 10.1016/j.jtho.2018.10.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Oser MG, Niederst MJ, Sequist LV, Engelman JA. Transformation from non-small-cell lung cancer to small-cell lung cancer: molecular drivers and cells of origin. Lancet Oncol. 2015;16:e165–e172. doi: 10.1016/S1470-2045(14)71180-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 2015;29:1447–1462. doi: 10.1101/gad.263145.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Thai AA, Solomon BJ, Sequist LV, Gainor JF, Heist RS. Lung cancer. Lancet. 2021;398:535–554. doi: 10.1016/S0140-6736(21)00312-3. [DOI] [PubMed] [Google Scholar]
- 142.Yu Y, Chen K, Fan Y. Extensive-stage small-cell lung cancer: current management and future directions. Int. J. Cancer. 2023;152:2243–2256. doi: 10.1002/ijc.34346. [DOI] [PubMed] [Google Scholar]
- 143.Hu J, et al. Comprehensive genomic profiling of small cell lung cancer in Chinese patients and the implications for therapeutic potential. Cancer Med. 2019;8:4338–4347. doi: 10.1002/cam4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Almodovar K, et al. Longitudinal cell-free DNA analysis in patients with small-cell lung cancer reveals dynamic insights into treatment efficacy and disease relapse. J. Thorac. Oncol. 2018;13:112–123. doi: 10.1016/j.jtho.2017.09.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hong D, et al. Plasticity in the absence of NOTCH uncovers a RUNX2-dependent pathway in small cell lung cancer. Cancer Res. 2022;82:248–263. doi: 10.1158/0008-5472.CAN-21-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.George J, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524:47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rojo F, et al. International real-world study of DLL3 expression in patients with small cell lung cancer. Lung Cancer. 2020;147:237–243. doi: 10.1016/j.lungcan.2020.07.026. [DOI] [PubMed] [Google Scholar]
- 148.Shirasawa M, et al. Tumor microenvironment-mediated immune profiles and efficacy of anti-PD-L1 antibody plus chemotherapy stratified by DLL3 expression in small-cell lung cancer. Br. J. Cancer. 2023;129:2003–2013. doi: 10.1038/s41416-023-02427-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim JW, Ko JH, Sage J. DLL3 regulates Notch signaling in small cell lung cancer. iScience. 2022;25:105603. doi: 10.1016/j.isci.2022.105603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rudin CM, et al. Emerging therapies targeting the delta-like ligand 3 (DLL3) in small cell lung cancer. J. Hematol. Oncol. 2023;16:66. doi: 10.1186/s13045-023-01464-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Owen DH, et al. DLL3: an emerging target in small cell lung cancer. J. Hematol. Oncol. 2019;12:61. doi: 10.1186/s13045-019-0745-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Rudin CM, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat. Rev. Cancer. 2019;19:289–297. doi: 10.1038/s41568-019-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ireland AS, et al. MYC drives temporal evolution of small cell lung cancer subtypes by reprogramming neuroendocrine fate. Cancer Cell. 2020;38:60–78.e12. doi: 10.1016/j.ccell.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lim JS, et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature. 2017;545:360–364. doi: 10.1038/nature22323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hassan KA. Small cell lung cancer heterogeneity: elevated a Notch above the Rest! J. Thorac. Dis. 2018;10:554–556. doi: 10.21037/jtd.2018.01.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Reynolds TC, Smith SD, Sklar J. Analysis of DNA surrounding the breakpoints of chromosomal translocations involving the beta T cell receptor gene in human lymphoblastic neoplasms. Cell. 1987;50:107–117. doi: 10.1016/0092-8674(87)90667-2. [DOI] [PubMed] [Google Scholar]
- 157.Ferrando AA, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/S1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 158.Bray SJ. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 159.Van Vlierberghe P, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J. Clin. Investig. 2012;122:3398–3406. doi: 10.1172/JCI61269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 161.Jeannet R, et al. Oncogenic activation of the Notch1 gene by deletion of its promoter in Ikaros-deficient T-ALL. Blood. 2010;116:5443–5454. doi: 10.1182/blood-2010-05-286658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Li X, Gounari F, Protopopov A, Khazaie K, von Boehmer H. Oncogenesis of T-ALL and nonmalignant consequences of overexpressing intracellular NOTCH1. J. Exp. Med. 2008;205:2851–2861. doi: 10.1084/jem.20081561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.García-Peydró M, et al. The NOTCH1/CD44 axis drives pathogenesis in a T cell acute lymphoblastic leukemia model. J. Clin. Investig. 2018;128:2802–2818. doi: 10.1172/JCI92981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gupta-Rossi N, et al. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- 165.Thompson BJ, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.O’Neil J, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J. Exp. Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Maser RS, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Asnafi V, et al. NOTCH1/FBXW7 mutation identifies a large subgroup with favorable outcome in adult T-cell acute lymphoblastic leukemia (T-ALL): a Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL) study. Blood. 2009;113:3918–3924. doi: 10.1182/blood-2008-10-184069. [DOI] [PubMed] [Google Scholar]
- 169.Jenkinson S, et al. Impact of NOTCH1/FBXW7 mutations on outcome in pediatric T-cell acute lymphoblastic leukemia patients treated on the MRC UKALL 2003 trial. Leukemia. 2013;27:41–47. doi: 10.1038/leu.2012.176. [DOI] [PubMed] [Google Scholar]
- 170.Hallek M, Shanafelt TD, Eichhorst B. Chronic lymphocytic leukaemia. Lancet. 2018;391:1524–1537. doi: 10.1016/S0140-6736(18)30422-7. [DOI] [PubMed] [Google Scholar]
- 171.Houlston RS, Catovsky D, Yuille MR. Genetic susceptibility to chronic lymphocytic leukemia. Leukemia. 2002;16:1008–1014. doi: 10.1038/sj.leu.2402538. [DOI] [PubMed] [Google Scholar]
- 172.Fabbri G, Dalla-Favera R. The molecular pathogenesis of chronic lymphocytic leukaemia. Nat. Rev. Cancer. 2016;16:145–162. doi: 10.1038/nrc.2016.8. [DOI] [PubMed] [Google Scholar]
- 173.Edelmann J, et al. Genomic alterations in high-risk chronic lymphocytic leukemia frequently affect cell cycle key regulators and NOTCH1-regulated transcription. Haematologica. 2020;105:1379–1390. doi: 10.3324/haematol.2019.217307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Fabbri G, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J. Exp. Med. 2011;208:1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Alniaimi AN, et al. Increased Notch1 expression is associated with poor overall survival in patients with ovarian cancer. Int. J. Gynecol. Cancer. 2015;25:208–213. doi: 10.1097/IGC.0000000000000359. [DOI] [PubMed] [Google Scholar]
- 177.Pozzo F, et al. NOTCH1 mutations associate with low CD20 level in chronic lymphocytic leukemia: evidence for a NOTCH1 mutation-driven epigenetic dysregulation. Leukemia. 2016;30:182–189. doi: 10.1038/leu.2015.182. [DOI] [PubMed] [Google Scholar]
- 178.Zou Y, et al. NOTCH1 mutation and its prognostic significance in Chinese chronic lymphocytic leukemia: a retrospective study of 317 cases. Cancer Med. 2018;7:1689–1696. doi: 10.1002/cam4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tausch E, et al. Prognostic and predictive role of gene mutations in chronic lymphocytic leukemia: results from the pivotal phase III study COMPLEMENT1. Haematologica. 2020;105:2440–2447. doi: 10.3324/haematol.2019.229161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tardivon D, et al. Notch signaling promotes disease initiation and progression in murine chronic lymphocytic leukemia. Blood. 2021;137:3079–3092. doi: 10.1182/blood.2020006701. [DOI] [PubMed] [Google Scholar]
- 181.Biran A, et al. Activation of notch and Myc signaling via B-cell-restricted depletion of Dnmt3a generates a consistent murine model of chronic lymphocytic leukemia. Cancer Res. 2021;81:6117–6130. doi: 10.1158/0008-5472.CAN-21-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hubmann R, et al. Notch2 is involved in the overexpression of CD23 in B-cell chronic lymphocytic leukemia. Blood. 2002;99:3742–3747. doi: 10.1182/blood.V99.10.3742. [DOI] [PubMed] [Google Scholar]
- 183.Fiorcari S, et al. Notch2 increases the resistance to venetoclax-induced apoptosis in chronic lymphocytic leukemia B cells by inducing Mcl-1. Front. Oncol. 2021;11:777587. doi: 10.3389/fonc.2021.777587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.De Falco F, et al. IL-4-dependent Jagged1 expression/processing is associated with survival of chronic lymphocytic leukemia cells but not with Notch activation. Cell Death Dis. 2018;9:1160. doi: 10.1038/s41419-018-1185-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bladder cancer. Nat. Rev. Dis. Primers. 9, 59 (2023). [DOI] [PMC free article] [PubMed]
- 186.Grayson M. Bladder cancer. Nature. 2017;551:S33. doi: 10.1038/551S33a. [DOI] [PubMed] [Google Scholar]
- 187.Dobruch J, et al. Gender and bladder cancer: a collaborative review of etiology, biology, and outcomes. Eur. Urol. 2016;69:300–310. doi: 10.1016/j.eururo.2015.08.037. [DOI] [PubMed] [Google Scholar]
- 188.Greife A, Hoffmann MJ, Schulz WA. Consequences of disrupted notch signaling in bladder cancer. Eur. Urol. 2015;68:3–4. doi: 10.1016/j.eururo.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 189.Rampias T, et al. A new tumor suppressor role for the Notch pathway in bladder cancer. Nat. Med. 2014;20:1199–1205. doi: 10.1038/nm.3678. [DOI] [PubMed] [Google Scholar]
- 190.Maraver A, et al. NOTCH pathway inactivation promotes bladder cancer progression. J. Clin. Investig. 2015;125:824–830. doi: 10.1172/JCI78185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Hayashi T, et al. Not all NOTCH is created equal: the oncogenic role of NOTCH2 in bladder cancer and its implications for targeted therapy. Clin. Cancer Res. 2016;22:2981–2992. doi: 10.1158/1078-0432.CCR-15-2360. [DOI] [PubMed] [Google Scholar]
- 192.Zhou Z, et al. CAFs-derived MFAP5 promotes bladder cancer malignant behavior through NOTCH2/HEY1 signaling. FASEB j. 2020;34:7970–7988. doi: 10.1096/fj.201902659R. [DOI] [PubMed] [Google Scholar]
- 193.Ristic Petrovic A, et al. The association between NOTCH3 expression and the clinical outcome in the urothelial bladder cancer patients. Bosn. J. Basic Med Sci. 2022;22:523–530. doi: 10.17305/bjbms.2021.6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Chen YT, et al. Jagged2 progressively increased expression from Stage I to III of bladder cancer and melatonin-mediated downregulation of Notch/Jagged2 suppresses the bladder tumorigenesis via inhibiting PI3K/AKT/mTOR/MMPs signaling. Int. J. Biol. Sci. 2020;16:2648–2662. doi: 10.7150/ijbs.48358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Prostate cancer. Nat. Rev. Dis. Primers. 7, 8 (2021). [DOI] [PubMed]
- 196.Haffner MC, et al. Genomic and phenotypic heterogeneity in prostate cancer. Nat. Rev. Urol. 2021;18:79–92. doi: 10.1038/s41585-020-00400-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wang G, Zhao D, Spring DJ, DePinho RA. Genetics and biology of prostate cancer. Genes Dev. 2018;32:1105–1140. doi: 10.1101/gad.315739.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Deng G, et al. Notch signaling in the prostate: critical roles during development and in the hallmarks of prostate cancer biology. J. Cancer Res. Clin. Oncol. 2016;142:531–547. doi: 10.1007/s00432-015-1946-x. [DOI] [PubMed] [Google Scholar]
- 199.Stoyanova T, et al. Activation of Notch1 synergizes with multiple pathways in promoting castration-resistant prostate cancer. Proc. Natl Acad. Sci. USA. 2016;113:E6457–e6466. doi: 10.1073/pnas.1614529113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ganguly SS, et al. Notch3 promotes prostate cancer-induced bone lesion development via MMP-3. Oncogene. 2020;39:204–218. doi: 10.1038/s41388-019-0977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Santagata S, et al. JAGGED1 expression is associated with prostate cancer metastasis and recurrence. Cancer Res. 2004;64:6854–6857. doi: 10.1158/0008-5472.CAN-04-2500. [DOI] [PubMed] [Google Scholar]
- 202.Su Q, et al. Jagged1 upregulation in prostate epithelial cells promotes formation of reactive stroma in the Pten null mouse model for prostate cancer. Oncogene. 2017;36:618–627. doi: 10.1038/onc.2016.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tran TT, Lee K. JAG1 Intracellular domain enhances AR expression and signaling and promotes stem-like properties in prostate cancer cells. Cancers. 2022;14:5714. doi: 10.3390/cancers14225714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chou J, et al. Immunotherapeutic targeting and PET imaging of DLL3 in small-cell neuroendocrine prostate cancer. Cancer Res. 2023;83:301–315. doi: 10.1158/0008-5472.CAN-22-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Cheng JW, et al. Bone marrow mesenchymal stem cells promote prostate cancer cell stemness via cell-cell contact to activate the Jagged1/Notch1 pathway. Cell Biosci. 2021;11:87. doi: 10.1186/s13578-021-00599-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Revandkar A, et al. Inhibition of Notch pathway arrests PTEN-deficient advanced prostate cancer by triggering p27-driven cellular senescence. Nat. Commun. 2016;7:13719. doi: 10.1038/ncomms13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 208.Chen L, Al-Awqati Q. Segmental expression of Notch and Hairy genes in nephrogenesis. Am. J. Physiol. Ren. Physiol. 2005;288:F939–F952. doi: 10.1152/ajprenal.00369.2004. [DOI] [PubMed] [Google Scholar]
- 209.Xiao W, Gao Z, Duan Y, Yuan W, Ke Y. Downregulation of miR-19a exhibits inhibitory effects on metastatic renal cell carcinoma by targeting PIK3CA and inactivating Notch signaling in vitro. Oncol. Rep. 2015;34:739–746. doi: 10.3892/or.2015.4041. [DOI] [PubMed] [Google Scholar]
- 210.Sun S, et al. Expression and clinical significance of Notch receptors in human renal cell carcinoma. Pathology. 2009;41:335–341. doi: 10.1080/00313020902885003. [DOI] [PubMed] [Google Scholar]
- 211.Liu S, et al. HES1-mediated down-regulation of miR-138 sustains NOTCH1 activation and promotes proliferation and invasion in renal cell carcinoma. J. Exp. Clin. Cancer Res. 2023;42:72. doi: 10.1186/s13046-023-02625-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wu K, Hu L, Hou J. Selective suppression of Notch1 inhibits proliferation of renal cell carcinoma cells through JNK/p38 pathway. Oncol. Rep. 2016;35:2795–2800. doi: 10.3892/or.2016.4687. [DOI] [PubMed] [Google Scholar]
- 213.Aparicio LM, et al. Expression of Notch1 to -4 and their ligands in renal cell carcinoma: a tissue microarray study. Cancer Genom. Proteom. 2011;8:93–101. [PubMed] [Google Scholar]
- 214.Wang X, et al. Potential biomarkers and the molecular mechanism associated with DLL4 during renal cell carcinoma progression. Am. J. Med. Sci. 2022;364:220–228. doi: 10.1016/j.amjms.2022.03.001. [DOI] [PubMed] [Google Scholar]
- 215.Miles KM, et al. Dll4 blockade potentiates the anti-tumor effects of VEGF inhibition in renal cell carcinoma patient-derived xenografts. PLoS One. 2014;9:e112371. doi: 10.1371/journal.pone.0112371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Jonasch E, Walker CL, Rathmell WK. Clear cell renal cell carcinoma ontogeny and mechanisms of lethality. Nat. Rev. Nephrol. 2021;17:245–261. doi: 10.1038/s41581-020-00359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Feng C, et al. Genetic alteration in Notch pathway is associated with better prognosis in renal cell carcinoma. Biofactors. 2016;42:41–48. doi: 10.1002/biof.1250. [DOI] [PubMed] [Google Scholar]
- 218.Ai Q, et al. High-level expression of Notch1 increased the risk of metastasis in T1 stage clear cell renal cell carcinoma. PLoS One. 2012;7:e35022. doi: 10.1371/journal.pone.0035022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wang W, et al. Delta-like ligand 4: a predictor of poor prognosis in clear cell renal cell carcinoma. Oncol. Lett. 2014;8:2627–2633. doi: 10.3892/ol.2014.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wu K, Xu L, Zhang L, Lin Z, Hou J. High Jagged1 expression predicts poor outcome in clear cell renal cell carcinoma. Jpn J. Clin. Oncol. 2011;41:411–416. doi: 10.1093/jjco/hyq205. [DOI] [PubMed] [Google Scholar]
- 221.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J. Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 222.Giaquinto AN, et al. Breast cancer statistics, 2022. CA Cancer J. Clin. 2022;72:524–541. doi: 10.3322/caac.21754. [DOI] [PubMed] [Google Scholar]
- 223.Stylianou S, Clarke RB, Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66:1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 224.Guo S, Liu M, Gonzalez-Perez RR. Role of Notch and its oncogenic signaling crosstalk in breast cancer. Biochim. Biophys. Acta. 2011;1815:197–213. doi: 10.1016/j.bbcan.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jhappan C, et al. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 1992;6:345–355. doi: 10.1101/gad.6.3.345. [DOI] [PubMed] [Google Scholar]
- 226.Reedijk M, et al. High-level coexpression of JAG1 and NOTCH1 is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 227.Wang JW, et al. The association between Notch4 expression, and clinicopathological characteristics and clinical outcomes in patients with breast cancer. Oncol. Lett. 2018;15:8749–8755. doi: 10.3892/ol.2018.8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zohny SF, Zamzami MA, Al-Malki AL, Trabulsi NH. Highly expressed DLL4 and JAG1: their role in incidence of breast cancer metastasis. Arch. Med. Res. 2020;51:145–152. doi: 10.1016/j.arcmed.2019.12.019. [DOI] [PubMed] [Google Scholar]
- 229.Yuan C, Chang K, Xu C, Li Q, Du Z. High expression of DLL3 is associated with a poor prognosis and immune infiltration in invasive breast cancer patients. Transl. Oncol. 2021;14:101080. doi: 10.1016/j.tranon.2021.101080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kontomanolis E, et al. Delta-like ligand 4 (DLL4) in the plasma and neoplastic tissues from breast cancer patients: correlation with metastasis. Med. Oncol. 2014;31:945. doi: 10.1007/s12032-014-0945-0. [DOI] [PubMed] [Google Scholar]
- 231.Fu YP, et al. NOTCH2 in breast cancer: association of SNP rs11249433 with gene expression in ER-positive breast tumors without TP53 mutations. Mol. Cancer. 2010;9:113. doi: 10.1186/1476-4598-9-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zhao D, et al. NOTCH-induced aldehyde dehydrogenase 1A1 deacetylation promotes breast cancer stem cells. J. Clin. Investig. 2014;124:5453–5465. doi: 10.1172/JCI76611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zhang S, et al. Manic fringe promotes a claudin-low breast cancer phenotype through notch-mediated PIK3CG induction. Cancer Res. 2015;75:1936–1943. doi: 10.1158/0008-5472.CAN-14-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Klinakis A, et al. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc. Natl Acad. Sci. USA. 2006;103:9262–9267. doi: 10.1073/pnas.0603371103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Miao K, et al. NOTCH1 activation compensates BRCA1 deficiency and promotes triple-negative breast cancer formation. Nat. Commun. 2020;11:3256. doi: 10.1038/s41467-020-16936-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Shao S, et al. Notch1 signaling regulates the epithelial-mesenchymal transition and invasion of breast cancer in a Slug-dependent manner. Mol. Cancer. 2015;14:28. doi: 10.1186/s12943-015-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Zhou W, et al. Up-regulation of S100A16 expression promotes epithelial-mesenchymal transition via Notch1 pathway in breast cancer. J. Biomed. Sci. 2014;21:97. doi: 10.1186/s12929-014-0097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Kumar S, et al. Estrogen-dependent DLL1-mediated Notch signaling promotes luminal breast cancer. Oncogene. 2019;38:2092–2107. doi: 10.1038/s41388-018-0562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19:192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Robinson DR, et al. Functionally recurrent rearrangements of the MAST kinase and Notch gene families in breast cancer. Nat. Med. 2011;17:1646–1651. doi: 10.1038/nm.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Horackova K, Janatova M, Kleiblova P, Kleibl Z, Soukupova J. Early-onset ovarian cancer <30 years: what do we know about its genetic predisposition? Int. J. Mol. Sci. 2023;24:17020. doi: 10.3390/ijms242317020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Jia D, Underwood J, Xu Q, Xie Q. NOTCH2/NOTCH3/DLL3/MAML1/ADAM17 signaling network is associated with ovarian cancer. Oncol. Lett. 2019;17:4914–4920. doi: 10.3892/ol.2019.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Rahman MT, et al. Notch3 overexpression as potential therapeutic target in advanced stage chemoresistant ovarian cancer. Am. J. Clin. Pathol. 2012;138:535–544. doi: 10.1309/AJCPKDLRQ8F3EWNS. [DOI] [PubMed] [Google Scholar]
- 244.Hu W, et al. Biological roles of the Delta family Notch ligand Dll4 in tumor and endothelial cells in ovarian cancer. Cancer Res. 2011;71:6030–6039. doi: 10.1158/0008-5472.CAN-10-2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yang J, et al. Role of Jagged1/STAT3 signalling in platinum-resistant ovarian cancer. J. Cell Mol. Med. 2019;23:4005–4018. doi: 10.1111/jcmm.14286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yang M, et al. Tumour-associated neutrophils orchestrate intratumoural IL-8-driven immune evasion through Jagged2 activation in ovarian cancer. Br. J. Cancer. 2020;123:1404–1416. doi: 10.1038/s41416-020-1026-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Chen M, et al. Chemokine CCL20 promotes the paclitaxel resistance of CD44(+)CD117(+) cells via the Notch1 signaling pathway in ovarian cancer. Mol. Med. Rep. 2021;24:635. doi: 10.3892/mmr.2021.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Li H, et al. Nuclear orphan receptor NR2F6 confers cisplatin resistance in epithelial ovarian cancer cells by activating the Notch3 signaling pathway. Int J. Cancer. 2019;145:1921–1934. doi: 10.1002/ijc.32293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Gupta N, Xu Z, El-Sehemy A, Steed H, Fu Y. Notch3 induces epithelial-mesenchymal transition and attenuates carboplatin-induced apoptosis in ovarian cancer cells. Gynecol. Oncol. 2013;130:200–206. doi: 10.1016/j.ygyno.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 250.Shahmoradi Z, et al. Cervical cancer incidence among US women, 2001-2019. JAMA. 2022;328:2267–2269. doi: 10.1001/jama.2022.17806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393:169–182. doi: 10.1016/S0140-6736(18)32470-X. [DOI] [PubMed] [Google Scholar]
- 252.Talora C, Sgroi DC, Crum CP, Dotto GP. Specific down-modulation of Notch1 signaling in cervical cancer cells is required for sustained HPV-E6/E7 expression and late steps of malignant transformation. Genes Dev. 2002;16:2252–2263. doi: 10.1101/gad.988902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Talora C, et al. Constitutively active Notch1 induces growth arrest of HPV-positive cervical cancer cells via separate signaling pathways. Exp. Cell Res. 2005;305:343–354. doi: 10.1016/j.yexcr.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 254.Yousif NG, et al. Notch1 ligand signaling pathway activated in cervical cancer: poor prognosis with high-level JAG1/Notch1. Arch. Gynecol. Obstet. 2015;292:899–904. doi: 10.1007/s00404-015-3694-1. [DOI] [PubMed] [Google Scholar]
- 255.Khelil M, et al. Delta-like ligand-Notch1 signaling is selectively modulated by HPV16 E6 to promote squamous cell proliferation and correlates with cervical cancer prognosis. Cancer Res. 2021;81:1909–1921. doi: 10.1158/0008-5472.CAN-20-1996. [DOI] [PubMed] [Google Scholar]
- 256.Chen Y, Wu Q, Lin J, Wei J. DARS-AS1 accelerates the proliferation of cervical cancer cells via miR-628-5p/JAG1 axis to activate Notch pathway. Cancer Cell Int. 2020;20:535. doi: 10.1186/s12935-020-01592-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Franko-Tobin LG, et al. Notch1-mediated tumor suppression in cervical cancer with the involvement of SST signaling and its application in enhanced SSTR-targeted therapeutics. Oncologist. 2012;17:220–232. doi: 10.1634/theoncologist.2011-0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Bajaj J, et al. Notch signaling in CD66+ cells drives the progression of human cervical cancers. Cancer Res. 2011;71:4888–4897. doi: 10.1158/0008-5472.CAN-11-0543. [DOI] [PubMed] [Google Scholar]
- 259.Yao J, Duan L, Fan M, Yuan J, Wu X. Notch1 induces cell cycle arrest and apoptosis in human cervical cancer cells: involvement of nuclear factor kappa B inhibition. Int. J. Gynecol. Cancer. 2007;17:502–510. doi: 10.1111/j.1525-1438.2007.00872.x. [DOI] [PubMed] [Google Scholar]
- 260.Sun L, et al. Notch signaling activation in cervical cancer cells induces cell growth arrest with the involvement of the nuclear receptor NR4A2. J. Cancer. 2016;7:1388–1395. doi: 10.7150/jca.15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Li Y, Wang J, Gao C, Hu Q, Mao X. Integral membrane protein 2A enhances sensitivity to chemotherapy via notch signaling pathway in cervical cancer. Bioengineered. 2021;12:10183–10193. doi: 10.1080/21655979.2021.2001218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wang GM, et al. Importance of the intersection of age and sex to understand variation in incidence and survival for primary malignant gliomas. Neuro Oncol. 2022;24:302–310. doi: 10.1093/neuonc/noab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Brat DJ, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N. Engl. J. Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Bai H, et al. Integrated genomic characterization of IDH1-mutant glioma malignant progression. Nat. Genet. 2016;48:59–66. doi: 10.1038/ng.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Suzuki H, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat. Genet. 2015;47:458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- 266.Halani SH, et al. Multi-faceted computational assessment of risk and progression in oligodendroglioma implicates NOTCH and PI3K pathways. NPJ Precis. Oncol. 2018;2:24. doi: 10.1038/s41698-018-0067-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hai L, et al. Notch1 is a prognostic factor that is distinctly activated in the classical and proneural subtype of glioblastoma and that promotes glioma cell survival via the NF-κB(p65) pathway. Cell Death Dis. 2018;9:158. doi: 10.1038/s41419-017-0119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Su LP, et al. The expression of ASAP3 and NOTCH3 and the clinicopathological characteristics of adult glioma patients. Open Med. 2022;17:1724–1741. doi: 10.1515/med-2022-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Noor H, Whittaker S, McDonald KL. DLL3 expression and methylation are associated with lower-grade glioma immune microenvironment and prognosis. Genomics. 2022;114:110289. doi: 10.1016/j.ygeno.2022.110289. [DOI] [PubMed] [Google Scholar]
- 270.Dell’albani P, et al. Differential patterns of NOTCH1-4 receptor expression are markers of glioma cell differentiation. Neuro Oncol. 2014;16:204–216. doi: 10.1093/neuonc/not168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Wang H, et al. miR-33a promotes glioma-initiating cell self-renewal via PKA and NOTCH pathways. J. Clin. Investig. 2014;124:4489–4502. doi: 10.1172/JCI75284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Yao J, Zheng K, Li C, Liu H, Shan X. Interference of Notch1 inhibits the growth of glioma cancer cells by inducing cell autophagy and down-regulation of Notch1-Hes-1 signaling pathway. Med. Oncol. 2015;32:610. doi: 10.1007/s12032-015-0610-2. [DOI] [PubMed] [Google Scholar]
- 273.Man J, et al. Hypoxic induction of vasorin regulates notch1 turnover to maintain glioma stem-like cells. Cell Stem Cell. 2018;22:104–118.e106. doi: 10.1016/j.stem.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Parmigiani E, et al. Interferon-γ resistance and immune evasion in glioma develop via Notch-regulated co-evolution of malignant and immune cells. Dev. Cell. 2022;57:1847–1865.e1849. doi: 10.1016/j.devcel.2022.06.006. [DOI] [PubMed] [Google Scholar]
- 275.Qiu B, Matthay KK. Advancing therapy for neuroblastoma. Nat. Rev. Clin. Oncol. 2022;19:515–533. doi: 10.1038/s41571-022-00643-z. [DOI] [PubMed] [Google Scholar]
- 276.van Limpt V, Chan A, Schramm A, Eggert A, Versteeg R. Phox2B mutations and the Delta-Notch pathway in neuroblastoma. Cancer Lett. 2005;228:59–63. doi: 10.1016/j.canlet.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 277.Zage PE, et al. Notch pathway activation induces neuroblastoma tumor cell growth arrest. Pediatr. Blood Cancer. 2012;58:682–689. doi: 10.1002/pbc.23202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Agarwal, P. et al. MYCN amplification is associated with reduced expression of genes encoding γ-secretase complex and NOTCH signaling components in neuroblastoma. Int. J. Mol. Sci. 24, 8141 (2023). [DOI] [PMC free article] [PubMed]
- 279.Kishida S, et al. Midkine promotes neuroblastoma through Notch2 signaling. Cancer Res. 2013;73:1318–1327. doi: 10.1158/0008-5472.CAN-12-3070. [DOI] [PubMed] [Google Scholar]
- 280.Axelson H. The Notch signaling cascade in neuroblastoma: role of the basic helix-loop-helix proteins HASH-1 and HES-1. Cancer Lett. 2004;204:171–178. doi: 10.1016/S0304-3835(03)00453-1. [DOI] [PubMed] [Google Scholar]
- 281.Chang HH, et al. Notch1 expression predicts an unfavorable prognosis and serves as a therapeutic target of patients with neuroblastoma. Clin. Cancer Res. 2010;16:4411–4420. doi: 10.1158/1078-0432.CCR-09-3360. [DOI] [PubMed] [Google Scholar]
- 282.van Groningen T, et al. A NOTCH feed-forward loop drives reprogramming from adrenergic to mesenchymal state in neuroblastoma. Nat. Commun. 2019;10:1530. doi: 10.1038/s41467-019-09470-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.van Nes J, et al. A NOTCH3 transcriptional module induces cell motility in neuroblastoma. Clin. Cancer Res. 2013;19:3485–3494. doi: 10.1158/1078-0432.CCR-12-3021. [DOI] [PubMed] [Google Scholar]
- 284.Owens B. Melanoma. Nature. 2014;515:S109. doi: 10.1038/515S109a. [DOI] [PubMed] [Google Scholar]
- 285.Arnold M, et al. Global burden of cutaneous melanoma in 2020 and projections to 2040. JAMA Dermatol. 2022;158:495–503. doi: 10.1001/jamadermatol.2022.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Zhang JP, et al. Overexpression of Notch ligand Dll1 in B16 melanoma cells leads to reduced tumor growth due to attenuated vascularization. Cancer Lett. 2011;309:220–227. doi: 10.1016/j.canlet.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 287.Pekkonen P, et al. Lymphatic endothelium stimulates melanoma metastasis and invasion via MMP14-dependent Notch3 and β1-integrin activation. Elife. 2018;7:e32490. doi: 10.7554/eLife.32490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Yang Z, et al. Notch1 signaling in melanoma cells promoted tumor-induced immunosuppression via upregulation of TGF-β1. J. Exp. Clin. Cancer Res. 2018;37:1. doi: 10.1186/s13046-017-0664-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Du Y, et al. Intracellular notch1 signaling in cancer-associated fibroblasts dictates the plasticity and stemness of melanoma stem/initiating cells. Stem Cells. 2019;37:865–875. doi: 10.1002/stem.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Rigg E, et al. Inhibition of extracellular vesicle-derived miR-146a-5p decreases progression of melanoma brain metastasis via Notch pathway dysregulation in astrocytes. J. Extracell. Vesicles. 2023;12:e12363. doi: 10.1002/jev2.12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Mikheil D, et al. Notch signaling suppresses melanoma tumor development in BRAF/Pten mice. Cancers. 2023;15:519. doi: 10.3390/cancers15020519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Bonyadi Rad E, et al. Notch4 signaling induces a mesenchymal-epithelial-like transition in melanoma cells to suppress malignant behaviors. Cancer Res. 2016;76:1690–1697. doi: 10.1158/0008-5472.CAN-15-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Li H, et al. NOTCH4 mutation as predictive biomarker for immunotherapy benefits in NRAS wildtype melanoma. Front. Immunol. 2022;13:894110. doi: 10.3389/fimmu.2022.894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Rojas GA, Hubbard AK, Diessner BJ, Ribeiro KB, Spector LG. International trends in incidence of osteosarcoma (1988–2012) Int. J. Cancer. 2021;149:1044–1053. doi: 10.1002/ijc.33673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Meltzer PS, Helman LJ. New horizons in the treatment of osteosarcoma. N. Engl. J. Med. 2021;385:2066–2076. doi: 10.1056/NEJMra2103423. [DOI] [PubMed] [Google Scholar]
- 296.Tang XF, et al. Overexpression of Notch3 is associated with metastasis and poor prognosis in osteosarcoma patients. Cancer Manag. Res. 2019;11:547–559. doi: 10.2147/CMAR.S185495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Zhang J, et al. The role of Notch ligand Jagged1 in osteosarcoma proliferation, metastasis, and recurrence. J. Orthop. Surg. Res. 2021;16:226. doi: 10.1186/s13018-021-02372-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Xie H, et al. High expression of Dll4 and CD44V6 is associated with clinicopathological characteristics and poor prognosis in osteosarcoma patients. Transl. Cancer Res. 2021;10:1065–1072. doi: 10.21037/tcr-21-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Engin F, et al. Notch signaling contributes to the pathogenesis of human osteosarcomas. Hum. Mol. Genet. 2009;18:1464–1470. doi: 10.1093/hmg/ddp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Yu L, et al. The notch pathway promotes osteosarcoma progression through activation of ephrin reverse signaling. Mol. Cancer Res. 2019;17:2383–2394. doi: 10.1158/1541-7786.MCR-19-0493. [DOI] [PubMed] [Google Scholar]
- 301.Lu J, et al. MiR-26a inhibits stem cell-like phenotype and tumor growth of osteosarcoma by targeting Jagged1. Oncogene. 2017;36:231–241. doi: 10.1038/onc.2016.194. [DOI] [PubMed] [Google Scholar]
- 302.Wang L, et al. Targeting Notch1 signaling pathway positively affects the sensitivity of osteosarcoma to cisplatin by regulating the expression and/or activity of Caspase family. Mol. Cancer. 2014;13:139. doi: 10.1186/1476-4598-13-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Li C, et al. Notch1 is associated with the multidrug resistance of hypoxic osteosarcoma by regulating MRP1 gene expression. Neoplasma. 2016;63:734–742. doi: 10.4149/neo_2016_510. [DOI] [PubMed] [Google Scholar]
- 304.Chen DW, Lang BHH, McLeod DSA, Newbold K, Haymart MR. Thyroid cancer. Lancet. 2023;401:1531–1544. doi: 10.1016/S0140-6736(23)00020-X. [DOI] [PubMed] [Google Scholar]
- 305.Traversi F, Stooss A, Dettmer MS, Charles RP. BRAF(V600E) overrides NOTCH signaling in thyroid cancer. Thyroid. 2021;31:787–799. doi: 10.1089/thy.2019.0595. [DOI] [PubMed] [Google Scholar]
- 306.Piana S, et al. Expression of NOTCH1 in thyroid cancer is mostly restricted to papillary carcinoma. Endocr. Connect. 2019;8:1089–1096. doi: 10.1530/EC-19-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Park HS, et al. Notch1 receptor as a marker of lymph node metastases in papillary thyroid cancer. Cancer Sci. 2012;103:305–309. doi: 10.1111/j.1349-7006.2011.02161.x. [DOI] [PubMed] [Google Scholar]
- 308.Zhang YZ, et al. Prognostic significance of DLL4 expression in papillary thyroid cancer. Eur. Rev. Med. Pharm. Sci. 2015;19:2901–2905. [PubMed] [Google Scholar]
- 309.Somnay YR, et al. Notch3 expression correlates with thyroid cancer differentiation, induces apoptosis, and predicts disease prognosis. Cancer. 2017;123:769–782. doi: 10.1002/cncr.30403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Cook M, Yu XM, Chen H. Notch in the development of thyroid C-cells and the treatment of medullary thyroid cancer. Am. J. Transl. Res. 2010;2:119–125. [PMC free article] [PubMed] [Google Scholar]
- 311.Kunnimalaiyaan M, Vaccaro AM, Ndiaye MA, Chen H. Overexpression of the NOTCH1 intracellular domain inhibits cell proliferation and alters the neuroendocrine phenotype of medullary thyroid cancer cells. J. Biol. Chem. 2006;281:39819–39830. doi: 10.1074/jbc.M603578200. [DOI] [PubMed] [Google Scholar]
- 312.Truong M, Cook MR, Pinchot SN, Kunnimalaiyaan M, Chen H. Resveratrol induces Notch2-mediated apoptosis and suppression of neuroendocrine markers in medullary thyroid cancer. Ann. Surg. Oncol. 2011;18:1506–1511. doi: 10.1245/s10434-010-1488-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Chamoli A, et al. Overview of oral cavity squamous cell carcinoma: risk factors, mechanisms, and diagnostics. Oral. Oncol. 2021;121:105451. doi: 10.1016/j.oraloncology.2021.105451. [DOI] [PubMed] [Google Scholar]
- 314.Yoshida R, et al. The pathological significance of Notch1 in oral squamous cell carcinoma. Lab Investig. 2013;93:1068–1081. doi: 10.1038/labinvest.2013.95. [DOI] [PubMed] [Google Scholar]
- 315.Cierpikowski P, Lis-Nawara A, Bar J. Prognostic value of WNT1, NOTCH1, PDGFRβ, and CXCR4 in oral squamous cell carcinoma. Anticancer Res. 2023;43:591–602. doi: 10.21873/anticanres.16195. [DOI] [PubMed] [Google Scholar]
- 316.Kayamori K, et al. NOTCH3 is induced in cancer-associated fibroblasts and promotes angiogenesis in oral squamous cell carcinoma. PLoS One. 2016;11:e0154112. doi: 10.1371/journal.pone.0154112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Mk H, Prince S, Mohan AM, Krishnan KV, Devi A. Association of Notch4 with metastasis in human oral squamous cell carcinoma. Life Sci. 2016;156:38–46. doi: 10.1016/j.lfs.2016.05.019. [DOI] [PubMed] [Google Scholar]
- 318.Harishankar MK, Mohan AM, Krishnan AV, Devi A. Downregulation of Notch4 - a prognostic marker in distinguishing oral verrucous carcinoma from oral squamous cell carcinoma. Braz. J. Otorhinolaryngol. 2019;85:11–16. doi: 10.1016/j.bjorl.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Lee SH, et al. TNFα enhances cancer stem cell-like phenotype via Notch-Hes1 activation in oral squamous cell carcinoma cells. Biochem. Biophys. Res. Commun. 2012;424:58–64. doi: 10.1016/j.bbrc.2012.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Lv L, Wang Q, Yang Y, Ji H. MicroRNA‑495 targets Notch1 to prohibit cell proliferation and invasion in oral squamous cell carcinoma. Mol. Med. Rep. 2019;19:693–702. doi: 10.3892/mmr.2018.9616. [DOI] [PubMed] [Google Scholar]
- 321.Zheng Y, et al. Membrane-tethered Notch1 exhibits oncogenic property via activation of EGFR-PI3K-AKT pathway in oral squamous cell carcinoma. J. Cell Physiol. 2019;234:5940–5952. doi: 10.1002/jcp.27022. [DOI] [PubMed] [Google Scholar]
- 322.Patni AP, et al. Comprehending the crosstalk between Notch, Wnt and Hedgehog signaling pathways in oral squamous cell carcinoma - clinical implications. Cell Oncol. 2021;44:473–494. doi: 10.1007/s13402-021-00591-3. [DOI] [PubMed] [Google Scholar]
- 323.Johnson DE, et al. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Prim. 2020;6:92. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.McDermott JD, Bowles DW. Epidemiology of head and neck squamous cell carcinomas: impact on staging and prevention strategies. Curr. Treat. Options Oncol. 2019;20:43. doi: 10.1007/s11864-019-0650-5. [DOI] [PubMed] [Google Scholar]
- 325.Loganathan SK, et al. Rare driver mutations in head and neck squamous cell carcinomas converge on NOTCH signaling. Science. 2020;367:1264–1269. doi: 10.1126/science.aax0902. [DOI] [PubMed] [Google Scholar]
- 326.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature517, 576–582 (2015). [DOI] [PMC free article] [PubMed]
- 328.Fukusumi T, Califano JA. The NOTCH Pathway in Head and Neck Squamous Cell Carcinoma. J. Dent. Res. 2018;97:645–653. doi: 10.1177/0022034518760297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Grilli G, et al. Impact of notch signaling on the prognosis of patients with head and neck squamous cell carcinoma. Oral. Oncol. 2020;110:105003. doi: 10.1016/j.oraloncology.2020.105003. [DOI] [PubMed] [Google Scholar]
- 331.Wirth M, Jira D, Ott A, Piontek G, Pickhard A. High NOTCH1 mRNA expression is associated with better survival in HNSCC. Int. J. Mol. Sci. 2018;19:830. doi: 10.3390/ijms19030830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Byun JY, et al. Targeting HIF-1α/NOTCH1 pathway eliminates CD44(+) cancer stem-like cell phenotypes, malignancy, and resistance to therapy in head and neck squamous cell carcinoma. Oncogene. 2022;41:1352–1363. doi: 10.1038/s41388-021-02166-w. [DOI] [PubMed] [Google Scholar]
- 333.Shah PA, et al. NOTCH1 signaling in head and neck squamous cell carcinoma. Cells. 2020;9:2677. doi: 10.3390/cells9122677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Porcheri C, Meisel CT, Mitsiadis T. Multifactorial contribution of notch signaling in head and neck squamous cell carcinoma. Int. J. Mol. Sci. 2019;20:1520. doi: 10.3390/ijms20061520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.D’Assoro AB, Leon-Ferre R, Braune EB, Lendahl U. Roles of notch signaling in the tumor microenvironment. Int. J. Mol. Sci. 2022;23:6241. doi: 10.3390/ijms23116241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Bi P, Kuang S. Notch signaling as a novel regulator of metabolism. Trends Endocrinol. Metab. 2015;26:248–255. doi: 10.1016/j.tem.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Takebe N, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: clinical update. Nat. Rev. Clin. Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Greenburg G, Hay ED. Cytoskeleton and thyroglobulin expression change during transformation of thyroid epithelium to mesenchyme-like cells. Development. 1988;102:605–622. doi: 10.1242/dev.102.3.605. [DOI] [PubMed] [Google Scholar]
- 339.Pei D, Shu X, Gassama-Diagne A, Thiery JP. Mesenchymal-epithelial transition in development and reprogramming. Nat. Cell Biol. 2019;21:44–53. doi: 10.1038/s41556-018-0195-z. [DOI] [PubMed] [Google Scholar]
- 340.Marconi GD, et al. Epithelial-mesenchymal transition (EMT): the type-2 EMT in wound healing, tissue regeneration and organ fibrosis. Cells. 2021;10:1587. doi: 10.3390/cells10071587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 342.Ang HL, et al. Mechanism of epithelial-mesenchymal transition in cancer and its regulation by natural compounds. Med. Res. Rev. 2023;43:1141–1200. doi: 10.1002/med.21948. [DOI] [PubMed] [Google Scholar]
- 343.Jing L, et al. Epithelial-mesenchymal transition induced cancer-stem-cell-like characteristics in hepatocellular carcinoma. J. Cell Physiol. 2019;234:18448–18458. doi: 10.1002/jcp.28480. [DOI] [PubMed] [Google Scholar]
- 344.Jin M, et al. MCUR1 facilitates epithelial-mesenchymal transition and metastasis via the mitochondrial calcium dependent ROS/Nrf2/Notch pathway in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019;38:136. doi: 10.1186/s13046-019-1135-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Xie Q, et al. Tspan5 promotes epithelial-mesenchymal transition and tumour metastasis of hepatocellular carcinoma by activating Notch signalling. Mol. Oncol. 2021;15:3184–3202. doi: 10.1002/1878-0261.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Natsuizaka M, et al. Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma. Nat. Commun. 2017;8:1758. doi: 10.1038/s41467-017-01500-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Fukusumi T, et al. The NOTCH4-HEY1 pathway induces epithelial-mesenchymal transition in head and neck squamous cell carcinoma. Clin. Cancer Res. 2018;24:619–633. doi: 10.1158/1078-0432.CCR-17-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Xie J, et al. The NOTCH1-HEY1 pathway regulates self-renewal and epithelial-mesenchymal transition of salivary adenoid cystic carcinoma cells. Int. J. Biol. Sci. 2020;16:598–610. doi: 10.7150/ijbs.36407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Debaugnies M, et al. RHOJ controls EMT-associated resistance to chemotherapy. Nature. 2023;616:168–175. doi: 10.1038/s41586-023-05838-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017;14:611–629. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Raoof S, et al. Targeting FGFR overcomes EMT-mediated resistance in EGFR mutant non-small cell lung cancer. Oncogene. 2019;38:6399–6413. doi: 10.1038/s41388-019-0887-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Wu DP, et al. Cx43 deficiency confers EMT-mediated tamoxifen resistance to breast cancer via c-Src/PI3K/Akt pathway. Int. J. Biol. Sci. 2021;17:2380–2398. doi: 10.7150/ijbs.55453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Maciaczyk D, et al. CBF1 is clinically prognostic and serves as a target to block cellular invasion and chemoresistance of EMT-like glioblastoma cells. Br. J. Cancer. 2017;117:102–112. doi: 10.1038/bjc.2017.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Lu HY, et al. Novel ADAM-17 inhibitor ZLDI-8 inhibits the proliferation and metastasis of chemo-resistant non-small-cell lung cancer by reversing Notch and epithelial mesenchymal transition in vitro and in vivo. Pharm. Res. 2019;148:104406. doi: 10.1016/j.phrs.2019.104406. [DOI] [PubMed] [Google Scholar]
- 355.Eelen G, Treps L, Li X, Carmeliet P. Basic and therapeutic aspects of angiogenesis updated. Circ. Res. 2020;127:310–329. doi: 10.1161/CIRCRESAHA.120.316851. [DOI] [PubMed] [Google Scholar]
- 356.Dudley AC, Griffioen AW. Pathological angiogenesis: mechanisms and therapeutic strategies. Angiogenesis. 2023;26:313–347. doi: 10.1007/s10456-023-09876-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Khan JA, Maki RG, Ravi V. Pathologic angiogenesis of malignant vascular sarcomas: implications for treatment. J. Clin. Oncol. 2018;36:194–201. doi: 10.1200/JCO.2017.74.9812. [DOI] [PubMed] [Google Scholar]
- 358.Huang C, et al. BICC1 drives pancreatic cancer progression by inducing VEGF-independent angiogenesis. Signal Transduct. Target Ther. 2023;8:271. doi: 10.1038/s41392-023-01478-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Patel SA, et al. Molecular mechanisms and future implications of VEGF/VEGFR in cancer therapy. Clin. Cancer Res. 2023;29:30–39. doi: 10.1158/1078-0432.CCR-22-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Wang Z, et al. Using apelin-based synthetic Notch receptors to detect angiogenesis and treat solid tumors. Nat. Commun. 2020;11:2163. doi: 10.1038/s41467-020-15729-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Lu P, et al. Perinatal angiogenesis from pre-existing coronary vessels via DLL4-NOTCH1 signalling. Nat. Cell Biol. 2021;23:967–977. doi: 10.1038/s41556-021-00747-1. [DOI] [PubMed] [Google Scholar]
- 362.Hultgren NW, et al. Slug regulates the Dll4-Notch-VEGFR2 axis to control endothelial cell activation and angiogenesis. Nat. Commun. 2020;11:5400. doi: 10.1038/s41467-020-18633-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.López-Guerra M, et al. Specific NOTCH1 antibody targets DLL4-induced proliferation, migration, and angiogenesis in NOTCH1-mutated CLL cells. Oncogene. 2020;39:1185–1197. doi: 10.1038/s41388-019-1053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Nandhu MS, et al. Novel paracrine modulation of Notch-DLL4 signaling by fibulin-3 promotes angiogenesis in high-grade gliomas. Cancer Res. 2014;74:5435–5448. doi: 10.1158/0008-5472.CAN-14-0685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Tiemeijer LA, et al. Engineered patterns of Notch ligands Jag1 and Dll4 elicit differential spatial control of endothelial sprouting. iScience. 2022;25:104306. doi: 10.1016/j.isci.2022.104306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Pedrosa AR, et al. Endothelial Jagged1 antagonizes Dll4 regulation of endothelial branching and promotes vascular maturation downstream of Dll4/Notch1. Arterioscler. Thromb. Vasc. Biol. 2015;35:1134–1146. doi: 10.1161/ATVBAHA.114.304741. [DOI] [PubMed] [Google Scholar]
- 367.Ubezio B, et al. Synchronization of endothelial Dll4-Notch dynamics switch blood vessels from branching to expansion. Elife. 2016;5:e12167. doi: 10.7554/eLife.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Zhou ZY, et al. Microglial Galectin3 enhances endothelial metabolism and promotes pathological angiogenesis via Notch inhibition by competitively binding to Jag1. Cell Death Dis. 2023;14:380. doi: 10.1038/s41419-023-05897-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Suchting S, Eichmann A. Jagged gives endothelial tip cells an edge. Cell. 2009;137:988–990. doi: 10.1016/j.cell.2009.05.024. [DOI] [PubMed] [Google Scholar]
- 370.Liu J, et al. JAG1 enhances angiogenesis in triple-negative breast cancer through promoting the secretion of exosomal lncRNA MALAT1. Genes Dis. 2023;10:2167–2178. doi: 10.1016/j.gendis.2022.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Zhu Q, et al. Linc-OIP5 in the breast cancer cells regulates angiogenesis of human umbilical vein endothelial cells through YAP1/Notch/NRP1 signaling circuit at a tumor microenvironment. Biol. Res. 2020;53:5. doi: 10.1186/s40659-020-0273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Wieland E, et al. Endothelial notch1 activity facilitates metastasis. Cancer Cell. 2017;31:355–367. doi: 10.1016/j.ccell.2017.01.007. [DOI] [PubMed] [Google Scholar]
- 373.Lu H, et al. Angiotensin-converting enzyme inhibitor promotes angiogenesis through Sp1/Sp3-mediated inhibition of notch signaling in male mice. Nat. Commun. 2023;14:731. doi: 10.1038/s41467-023-36409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Kim HS, et al. Cell-membrane-derived nanoparticles with notch-1 suppressor delivery promote hypoxic cell-cell packing and inhibit angiogenesis acting as a two-edged sword. Adv. Mater. 2021;33:e2101558. doi: 10.1002/adma.202101558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Kumar D, et al. Notch1-MAPK signaling axis regulates CD133(+) cancer stem cell-mediated melanoma growth and angiogenesis. J. Investig. Dermatol. 2016;136:2462–2474. doi: 10.1016/j.jid.2016.07.024. [DOI] [PubMed] [Google Scholar]
- 376.Sengupta S, et al. Differentiated glioma cell-derived fibromodulin activates integrin-dependent Notch signaling in endothelial cells to promote tumor angiogenesis and growth. Elife. 2022;11:e78972. doi: 10.7554/eLife.78972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Lu H, Wu C, Jiang XW, Zhao Q. ZLDI-8 suppresses angiogenesis and vasculogenic mimicry in drug-resistant NSCLC in vitro and in vivo. Lung Cancer. 2023;182:107279. doi: 10.1016/j.lungcan.2023.107279. [DOI] [PubMed] [Google Scholar]
- 378.Babaei G, Aziz SG, Jaghi NZZ. EMT, cancer stem cells and autophagy; the three main axes of metastasis. Biomed. Pharmacother. 2021;133:110909. doi: 10.1016/j.biopha.2020.110909. [DOI] [PubMed] [Google Scholar]
- 379.Huang T, et al. Stem cell programs in cancer initiation, progression, and therapy resistance. Theranostics. 2020;10:8721–8743. doi: 10.7150/thno.41648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Marquardt S, Solanki M, Spitschak A, Vera J, Pützer BM. Emerging functional markers for cancer stem cell-based therapies: understanding signaling networks for targeting metastasis. Semin. Cancer Biol. 2018;53:90–109. doi: 10.1016/j.semcancer.2018.06.006. [DOI] [PubMed] [Google Scholar]
- 381.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Wu K, Wu M, Yang H, Diao R, Zeng H. Hypoxia promotes conversion to a stem cell phenotype in prostate cancer cells by activating HIF-1α/Notch1 signaling pathway. Clin. Transl. Oncol. 2023;25:2138–2152. doi: 10.1007/s12094-023-03093-w. [DOI] [PubMed] [Google Scholar]
- 383.Wong ALA, Bellot GL, Hirpara JL, Pervaiz S. Understanding the cancer stem cell phenotype: a step forward in the therapeutic management of cancer. Biochem. Pharm. 2019;162:79–88. doi: 10.1016/j.bcp.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 384.Aoki S, et al. Aberrant activation of Notch signaling in extrahepatic cholangiocarcinoma: clinicopathological features and therapeutic potential for cancer stem cell-like properties. BMC Cancer. 2016;16:854. doi: 10.1186/s12885-016-2919-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Wang X, et al. miR-181b/Notch2 overcome chemoresistance by regulating cancer stem cell-like properties in NSCLC. Stem Cell Res. Ther. 2018;9:327. doi: 10.1186/s13287-018-1072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Xiao W, Gao Z, Duan Y, Yuan W, Ke Y. Notch signaling plays a crucial role in cancer stem-like cells maintaining stemness and mediating chemotaxis in renal cell carcinoma. J. Exp. Clin. Cancer Res. 2017;36:41. doi: 10.1186/s13046-017-0507-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Liu H, et al. Fusobacterium nucleatum promotes colorectal cancer cell to acquire stem cell-like features by manipulating lipid droplet-mediated numb degradation. Adv. Sci. 2022;9:e2105222. doi: 10.1002/advs.202105222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Huang B, et al. tRF/miR-1280 suppresses stem cell-like cells and metastasis in colorectal cancer. Cancer Res. 2017;77:3194–3206. doi: 10.1158/0008-5472.CAN-16-3146. [DOI] [PubMed] [Google Scholar]
- 389.Katsushima K, et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat. Commun. 2016;7:13616. doi: 10.1038/ncomms13616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Wang J, et al. N6-Methyladenosine-mediated up-regulation of FZD10 regulates liver cancer stem cells’ properties and lenvatinib resistance through WNT/β-catenin and hippo signaling pathways. Gastroenterology. 2023;164:990–1005. doi: 10.1053/j.gastro.2023.01.041. [DOI] [PubMed] [Google Scholar]
- 391.Leung HW, et al. EPHB2 activates β-catenin to enhance cancer stem cell properties and drive sorafenib resistance in hepatocellular carcinoma. Cancer Res. 2021;81:3229–3240. doi: 10.1158/0008-5472.CAN-21-0184. [DOI] [PubMed] [Google Scholar]
- 392.Liu C, et al. LSD1 stimulates cancer-associated fibroblasts to drive Notch3-dependent self-renewal of liver cancer stem-like cells. Cancer Res. 2018;78:938–949. doi: 10.1158/0008-5472.CAN-17-1236. [DOI] [PubMed] [Google Scholar]
- 393.Wang R, et al. iNOS promotes CD24(+)CD133(+) liver cancer stem cell phenotype through a TACE/ADAM17-dependent Notch signaling pathway. Proc. Natl Acad. Sci. USA. 2018;115:E10127–E10136. doi: 10.1073/pnas.1722100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Clara JA, Monge C, Yang Y, Takebe N. Targeting signalling pathways and the immune microenvironment of cancer stem cells—a clinical update. Nat. Rev. Clin. Oncol. 2020;17:204–232. doi: 10.1038/s41571-019-0293-2. [DOI] [PubMed] [Google Scholar]
- 395.Xie C, et al. Sulforaphane inhibits the acquisition of tobacco smoke-induced lung cancer stem cell-like properties via the IL-6/ΔNp63α/Notch axis. Theranostics. 2019;9:4827–4840. doi: 10.7150/thno.33812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Wang Y, et al. Targeting EGFR enriches stem cell-like properties in salivary adenoid cystic carcinoma by activating the Notch1 pathway. Cancer Manag. Res. 2020;12:6655–6663. doi: 10.2147/CMAR.S253500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Jiang N, et al. HIF-1ɑ-regulated miR-1275 maintains stem cell-like phenotypes and promotes the progression of LUAD by simultaneously activating Wnt/β-catenin and Notch signaling. Theranostics. 2020;10:2553–2570. doi: 10.7150/thno.41120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Yan Y, et al. HIF-2α promotes conversion to a stem cell phenotype and induces chemoresistance in breast cancer cells by activating Wnt and Notch pathways. J. Exp. Clin. Cancer Res. 2018;37:256. doi: 10.1186/s13046-018-0925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Ibrahim SA, et al. Syndecan-1 is a novel molecular marker for triple negative inflammatory breast cancer and modulates the cancer stem cell phenotype via the IL-6/STAT3, Notch and EGFR signaling pathways. Mol. Cancer. 2017;16:57. doi: 10.1186/s12943-017-0621-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Panaccione A, et al. NOTCH1 and SOX10 are essential for proliferation and radiation resistance of cancer stem-like cells in adenoid cystic carcinoma. Clin. Cancer Res. 2016;22:2083–2095. doi: 10.1158/1078-0432.CCR-15-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Lin X, et al. Notch4+ cancer stem-like cells promote the metastatic and invasive ability of melanoma. Cancer Sci. 2016;107:1079–1091. doi: 10.1111/cas.12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Mishra VK, et al. Krüppel-like transcription factor KLF10 suppresses TGFβ-induced epithelial-to-mesenchymal transition via a negative feedback mechanism. Cancer Res. 2017;77:2387–2400. doi: 10.1158/0008-5472.CAN-16-2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Tsai YC, et al. Krüppel-like factor 10 modulates stem cell phenotypes of pancreatic adenocarcinoma by transcriptionally regulating notch receptors. J. Biomed. Sci. 2023;30:39. doi: 10.1186/s12929-023-00937-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Warburg O. The metabolism of carcinoma cells1. J. Cancer Res. 1925;9:148–163. doi: 10.1158/jcr.1925.148. [DOI] [Google Scholar]
- 405.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J. Gen. Physiol. 1927;8:519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Cori CF, Cori GT. The carbohydrate metabolism of tumors. I. The free sugar, lactic acid, and glycogen content of malignant tumors. J. Biol. Chem. 1925;64:11–22. doi: 10.1016/S0021-9258(18)84944-4. [DOI] [Google Scholar]
- 407.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Slaninova V, et al. Notch stimulates growth by direct regulation of genes involved in the control of glycolysis and the tricarboxylic acid cycle. Open Biol. 2016;6:150155. doi: 10.1098/rsob.150155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Jitschin R, et al. Stromal cell-mediated glycolytic switch in CLL cells involves Notch-c-Myc signaling. Blood. 2015;125:3432–3436. doi: 10.1182/blood-2014-10-607036. [DOI] [PubMed] [Google Scholar]
- 411.Moreno C. Chronic lymphocytic leukemia and the Warburg effect. Blood. 2015;125:3368–3369. doi: 10.1182/blood-2015-04-636332. [DOI] [PubMed] [Google Scholar]
- 412.Landor SK, et al. Hypo- and hyperactivated Notch signaling induce a glycolytic switch through distinct mechanisms. Proc. Natl Acad. Sci. USA. 2011;108:18814–18819. doi: 10.1073/pnas.1104943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Grzes KM, et al. Control of amino acid transport coordinates metabolic reprogramming in T-cell malignancy. Leukemia. 2017;31:2771–2779. doi: 10.1038/leu.2017.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Sellers K, et al. Metabolic reprogramming and Notch activity distinguish between non-small cell lung cancer subtypes. Br. J. Cancer. 2019;121:51–64. doi: 10.1038/s41416-019-0464-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Huber V, et al. Cancer acidity: an ultimate frontier of tumor immune escape and a novel target of immunomodulation. Semin Cancer Biol. 2017;43:74–89. doi: 10.1016/j.semcancer.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 416.Xie M, Fu XG, Jiang K. Notch1/TAZ axis promotes aerobic glycolysis and immune escape in lung cancer. Cell Death Dis. 2021;12:832. doi: 10.1038/s41419-021-04124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Gubser PM, et al. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat. Immunol. 2013;14:1064–1072. doi: 10.1038/ni.2687. [DOI] [PubMed] [Google Scholar]
- 418.Chang CH, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Zhao E, et al. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat. Immunol. 2016;17:95–103. doi: 10.1038/ni.3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Icard P, et al. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist. Updat. 2018;38:1–11. doi: 10.1016/j.drup.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 421.Bhattacharya B, Mohd Omar MF, Soong R. The Warburg effect and drug resistance. Br. J. Pharm. 2016;173:970–979. doi: 10.1111/bph.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Wang S, et al. Targeting ACYP1-mediated glycolysis reverses lenvatinib resistance and restricts hepatocellular carcinoma progression. Drug Resist. Updat. 2023;69:100976. doi: 10.1016/j.drup.2023.100976. [DOI] [PubMed] [Google Scholar]
- 423.Duan W, et al. Warburg effect enhanced by AKR1B10 promotes acquired resistance to pemetrexed in lung cancer-derived brain metastasis. J. Transl. Med. 2023;21:547. doi: 10.1186/s12967-023-04403-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Chen Y, et al. Baicalein resensitizes tamoxifen-resistant breast cancer cells by reducing aerobic glycolysis and reversing mitochondrial dysfunction via inhibition of hypoxia-inducible factor-1α. Clin. Transl. Med. 2021;11:e577. doi: 10.1002/ctm2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Pi M, et al. Targeting metabolism to overcome cancer drug resistance: a promising therapeutic strategy for diffuse large B cell lymphoma. Drug Resist. Updat. 2022;61:100822. doi: 10.1016/j.drup.2022.100822. [DOI] [PubMed] [Google Scholar]
- 426.Nguyen TL, et al. Downregulation of glutamine synthetase, not glutaminolysis, is responsible for glutamine addiction in Notch1-driven acute lymphoblastic leukemia. Mol. Oncol. 2021;15:1412–1431. doi: 10.1002/1878-0261.12877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Baran N, et al. Inhibition of mitochondrial complex I reverses NOTCH1-driven metabolic reprogramming in T-cell acute lymphoblastic leukemia. Nat. Commun. 2022;13:2801. doi: 10.1038/s41467-022-30396-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Herranz D, et al. Metabolic reprogramming induces resistance to anti-NOTCH1 therapies in T cell acute lymphoblastic leukemia. Nat. Med. 2015;21:1182–1189. doi: 10.1038/nm.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Mollick T, Laín S. Modulating pyrimidine ribonucleotide levels for the treatment of cancer. Cancer Metab. 2020;8:12. doi: 10.1186/s40170-020-00218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Wang W, Cui J, Ma H, Lu W, Huang J. Targeting pyrimidine metabolism in the era of precision cancer medicine. Front. Oncol. 2021;11:684961. doi: 10.3389/fonc.2021.684961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Pal S, et al. A druggable addiction to de novo pyrimidine biosynthesis in diffuse midline glioma. Cancer Cell. 2022;40:957–972.e910. doi: 10.1016/j.ccell.2022.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Liu F, et al. Oncogenic β-catenin stimulation of AKT2-CAD-mediated pyrimidine synthesis is targetable vulnerability in liver cancer. Proc. Natl Acad. Sci. USA. 2022;119:e2202157119. doi: 10.1073/pnas.2202157119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.He D, et al. De novo pyrimidine synthesis fuels glycolysis and confers chemoresistance in gastric cancer. Cancer Lett. 2022;549:215837. doi: 10.1016/j.canlet.2022.215837. [DOI] [PubMed] [Google Scholar]
- 435.Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79:4557–4566. doi: 10.1158/0008-5472.CAN-18-3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Elhanani O, Ben-Uri R, Keren L. Spatial profiling technologies illuminate the tumor microenvironment. Cancer Cell. 2023;41:404–420. doi: 10.1016/j.ccell.2023.01.010. [DOI] [PubMed] [Google Scholar]
- 437.He R, et al. Revisiting of TAMs in tumor immune microenvironment: Insight from NF-κB signaling pathway. Biomed. Pharmacother. 2023;165:115090. doi: 10.1016/j.biopha.2023.115090. [DOI] [PubMed] [Google Scholar]
- 438.Zhao H, Wei J, Sun J. Roles of TGF-β signaling pathway in tumor microenvirionment and cancer therapy. Int Immunopharmacol. 2020;89:107101. doi: 10.1016/j.intimp.2020.107101. [DOI] [PubMed] [Google Scholar]
- 439.Liu Z, et al. cGAS-STING signaling in the tumor microenvironment. Cancer Lett. 2023;577:216409. doi: 10.1016/j.canlet.2023.216409. [DOI] [PubMed] [Google Scholar]
- 440.Li J, Bakhoum SF. The pleiotropic roles of cGAS-STING signaling in the tumor microenvironment. J. Mol. Cell Biol. 2022;14:mjac019. doi: 10.1093/jmcb/mjac019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Meurette O, Mehlen P. Notch signaling in the tumor microenvironment. Cancer Cell. 2018;34:536–548. doi: 10.1016/j.ccell.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 442.Jackstadt R, et al. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell. 2019;36:319–336.e317. doi: 10.1016/j.ccell.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Nolan E, et al. Radiation exposure elicits a neutrophil-driven response in healthy lung tissue that enhances metastatic colonization. Nat. Cancer. 2022;3:173–187. doi: 10.1038/s43018-022-00336-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Kim HJ, et al. Reprogramming of cancer-associated fibroblasts by apoptotic cancer cells inhibits lung metastasis via Notch1-WISP-1 signaling. Cell Mol. Immunol. 2022;19:1373–1391. doi: 10.1038/s41423-022-00930-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Liu Z, et al. Immunosuppression in tumor immune microenvironment and its optimization from CAR-T cell therapy. Theranostics. 2022;12:6273–6290. doi: 10.7150/thno.76854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Lei X, et al. Immune cells within the tumor microenvironment: biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–133. doi: 10.1016/j.canlet.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 447.Gu G, et al. NKAP alters tumor immune microenvironment and promotes glioma growth via Notch1 signaling. J. Exp. Clin. Cancer Res. 2019;38:291. doi: 10.1186/s13046-019-1281-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Fu T, et al. Spatial architecture of the immune microenvironment orchestrates tumor immunity and therapeutic response. J. Hematol. Oncol. 2021;14:98. doi: 10.1186/s13045-021-01103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Janghorban M, et al. Single-cell analysis unveils the role of the tumor immune microenvironment and notch signaling in dormant minimal residual disease. Cancer Res. 2022;82:885–899. doi: 10.1158/0008-5472.CAN-21-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Sharma A, et al. Onco-fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell. 2020;183:377–394.e321. doi: 10.1016/j.cell.2020.08.040. [DOI] [PubMed] [Google Scholar]
- 451.Chew SC, Choo SY, Chow PK. A new perspective on the immune escape mechanism in HCC: onco-foetal reprogramming. Br. J. Cancer. 2021;124:1897–1899. doi: 10.1038/s41416-021-01286-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Tao J, et al. Targeting hypoxic tumor microenvironment in pancreatic cancer. J. Hematol. Oncol. 2021;14:14. doi: 10.1186/s13045-020-01030-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Niu Y, et al. Loss-of-function genetic screening identifies aldolase a as an essential driver for liver cancer cell growth under hypoxia. Hepatology. 2021;74:1461–1479. doi: 10.1002/hep.31846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Tang K, et al. Hypoxia promotes breast cancer cell growth by activating a glycogen metabolic program. Cancer Res. 2021;81:4949–4963. doi: 10.1158/0008-5472.CAN-21-0753. [DOI] [PubMed] [Google Scholar]
- 455.D’Aguanno S, Mallone F, Marenco M, Del Bufalo D, Moramarco A. Hypoxia-dependent drivers of melanoma progression. J. Exp. Clin. Cancer Res. 2021;40:159. doi: 10.1186/s13046-021-01926-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Zandberg DP, et al. Tumor hypoxia is associated with resistance to PD-1 blockade in squamous cell carcinoma of the head and neck. J. Immunother. Cancer. 2021;9:e002088. doi: 10.1136/jitc-2020-002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Jayaprakash P, et al. Targeted hypoxia reduction restores T cell infiltration and sensitizes prostate cancer to immunotherapy. J. Clin. Investig. 2018;128:5137–5149. doi: 10.1172/JCI96268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Grassi ES, Pantazopoulou V, Pietras A. Hypoxia-induced release, nuclear translocation, and signaling activity of a DLK1 intracellular fragment in glioma. Oncogene. 2020;39:4028–4044. doi: 10.1038/s41388-020-1273-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Wu Q, et al. Hypoxia-inducible factors: master regulators of hypoxic tumor immune escape. J. Hematol. Oncol. 2022;15:77. doi: 10.1186/s13045-022-01292-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Missiaen R, Lesner NP, Simon MC. HIF: a master regulator of nutrient availability and metabolic cross-talk in the tumor microenvironment. EMBO J. 2023;42:e112067. doi: 10.15252/embj.2022112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Kung-Chun Chiu D, et al. Hypoxia regulates the mitochondrial activity of hepatocellular carcinoma cells through HIF/HEY1/PINK1 pathway. Cell Death Dis. 2019;10:934. doi: 10.1038/s41419-019-2155-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Pomeroy AE, Schmidt EV, Sorger PK, Palmer AC. Drug independence and the curability of cancer by combination chemotherapy. Trends Cancer. 2022;8:915–929. doi: 10.1016/j.trecan.2022.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Liu W, et al. Glutathione peroxidase 4-dependent glutathione high-consumption drives acquired platinum chemoresistance in lung cancer-derived brain metastasis. Clin. Transl. Med. 2021;11:e517. doi: 10.1002/ctm2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Cao T, et al. A CGA/EGFR/GATA2 positive feedback circuit confers chemoresistance in gastric cancer. J. Clin. Investig. 2022;132:e154074. doi: 10.1172/JCI154074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Kannampuzha S, Gopalakrishnan AV. Cancer chemoresistance and its mechanisms: associated molecular factors and its regulatory role. Med Oncol. 2023;40:264. doi: 10.1007/s12032-023-02138-y. [DOI] [PubMed] [Google Scholar]
- 466.Sun Y, et al. YTHDF1 promotes breast cancer cell growth, DNA damage repair and chemoresistance. Cell Death Dis. 2022;13:230. doi: 10.1038/s41419-022-04672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Meng X, et al. CircPTK2/PABPC1/SETDB1 axis promotes EMT-mediated tumor metastasis and gemcitabine resistance in bladder cancer. Cancer Lett. 2023;554:216023. doi: 10.1016/j.canlet.2022.216023. [DOI] [PubMed] [Google Scholar]
- 468.Dong S, et al. ROS/PI3K/Akt and Wnt/β-catenin signalings activate HIF-1α-induced metabolic reprogramming to impart 5-fluorouracil resistance in colorectal cancer. J. Exp. Clin. Cancer Res. 2022;41:15. doi: 10.1186/s13046-021-02229-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Wu Q, et al. YAP drives fate conversion and chemoresistance of small cell lung cancer. Sci. Adv. 2021;7:eabg1850. doi: 10.1126/sciadv.abg1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Martins-Neves SR, Sampaio-Ribeiro G, Gomes CMF. Self-renewal and pluripotency in osteosarcoma stem cells' chemoresistance: notch, hedgehog, and Wnt/β-catenin interplay with embryonic markers. Int. J. Mol. Sci. 2023;24:8401. doi: 10.3390/ijms24098401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Kang M, et al. Delta like ligand 4 induces impaired chemo-drug delivery and enhanced chemoresistance in pancreatic cancer. Cancer Lett. 2013;330:11–21. doi: 10.1016/j.canlet.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 472.Güngör C, et al. Notch signaling activated by replication stress-induced expression of midkine drives epithelial-mesenchymal transition and chemoresistance in pancreatic cancer. Cancer Res. 2011;71:5009–5019. doi: 10.1158/0008-5472.CAN-11-0036. [DOI] [PubMed] [Google Scholar]
- 473.Du X, et al. Alteration of the intrinsic apoptosis pathway is involved in Notch-induced chemoresistance to gemcitabine in pancreatic cancer. Arch. Med. Res. 2014;45:15–20. doi: 10.1016/j.arcmed.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 474.Giaimo BD, et al. Histone variant H2A.Z deposition and acetylation directs the canonical Notch signaling response. Nucleic Acids Res. 2018;46:8197–8215. doi: 10.1093/nar/gky551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Han S, et al. GAS41 mediates proliferation and GEM chemoresistance via H2A.Z.2 and Notch1 in pancreatic cancer. Cell Oncol. 2022;45:429–446. doi: 10.1007/s13402-022-00675-8. [DOI] [PubMed] [Google Scholar]
- 476.Zhai S, et al. A microprotein N1DARP encoded by LINC00261 promotes Notch1 intracellular domain (N1ICD) degradation via disrupting USP10-N1ICD interaction to inhibit chemoresistance in Notch1-hyperactivated pancreatic cancer. Cell Discov. 2023;9:95. doi: 10.1038/s41421-023-00592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Augert A, et al. Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition. Sci. Signal. 2019;12:eaau2922. doi: 10.1126/scisignal.aau2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Shi L, et al. Cancer-associated fibroblast-derived exosomal microRNA-20a suppresses the PTEN/PI3K-AKT pathway to promote the progression and chemoresistance of non-small cell lung cancer. Clin. Transl. Med. 2022;12:e989. doi: 10.1002/ctm2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Tong Q, Ouyang S, Chen R, Huang J, Guo L. MYCN-mediated regulation of the HES1 promoter enhances the chemoresistance of small-cell lung cancer by modulating apoptosis. Am. J. Cancer Res. 2019;9:1938–1956. [PMC free article] [PubMed] [Google Scholar]
- 480.Huang J, et al. Notch-1 confers chemoresistance in lung adenocarcinoma to taxanes through AP-1/microRNA-451 mediated regulation of MDR-1. Mol. Ther. Nucleic Acids. 2016;5:e375. doi: 10.1038/mtna.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Yun J, et al. Crosstalk between PKCα and Notch-4 in endocrine-resistant breast cancer cells. Oncogenesis. 2013;2:e60. doi: 10.1038/oncsis.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Omar M, et al. Notch-based gene signature for predicting the response to neoadjuvant chemotherapy in triple-negative breast cancer. J. Transl. Med. 2023;21:811. doi: 10.1186/s12967-023-04713-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Yang Q, et al. Chemotherapy-elicited exosomal miR-378a-3p and miR-378d promote breast cancer stemness and chemoresistance via the activation of EZH2/STAT3 signaling. J. Exp. Clin. Cancer Res. 2021;40:120. doi: 10.1186/s13046-021-01901-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Xiao YS, et al. Major vault protein is a direct target of Notch1 signaling and contributes to chemoresistance in triple-negative breast cancer cells. Cancer Lett. 2019;440-441:156–167. doi: 10.1016/j.canlet.2018.09.031. [DOI] [PubMed] [Google Scholar]
- 485.Pelullo M, et al. Kras/ADAM17-dependent Jag1-ICD reverse signaling sustains colorectal cancer progression and chemoresistance. Cancer Res. 2019;79:5575–5586. doi: 10.1158/0008-5472.CAN-19-0145. [DOI] [PubMed] [Google Scholar]
- 486.Jin Y, et al. Overcoming stemness and chemoresistance in colorectal cancer through miR-195-5p-modulated inhibition of notch signaling. Int. J. Biol. Macromol. 2018;117:445–453. doi: 10.1016/j.ijbiomac.2018.05.151. [DOI] [PubMed] [Google Scholar]
- 487.Citarella A, et al. Hedgehog-GLI and notch pathways sustain chemoresistance and invasiveness in colorectal cancer and their inhibition restores chemotherapy efficacy. Cancers. 2023;15:1471. doi: 10.3390/cancers15051471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell. 2014;25:318–334. doi: 10.1016/j.ccr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Nowell CS, Radtke F. Notch as a tumour suppressor. Nat. Rev. Cancer. 2017;17:145–159. doi: 10.1038/nrc.2016.145. [DOI] [PubMed] [Google Scholar]
- 490.Lobry C, Oh P, Aifantis I. Oncogenic and tumor suppressor functions of Notch in cancer: it’s NOTCH what you think. J. Exp. Med. 2011;208:1931–1935. doi: 10.1084/jem.20111855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Zhang M, et al. Does Notch play a tumor suppressor role across diverse squamous cell carcinomas? Cancer Med. 2016;5:2048–2060. doi: 10.1002/cam4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Lobry C, Oh P, Mansour MR, Look AT, Aifantis I. Notch signaling: switching an oncogene to a tumor suppressor. Blood. 2014;123:2451–2459. doi: 10.1182/blood-2013-08-355818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Huang T, et al. NOTCH receptors in gastric and other gastrointestinal cancers: oncogenes or tumor suppressors? Mol. Cancer. 2016;15:80. doi: 10.1186/s12943-016-0566-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Lefort K, Dotto GP. Notch signaling in the integrated control of keratinocyte growth/differentiation and tumor suppression. Semin. Cancer Biol. 2004;14:374–386. doi: 10.1016/j.semcancer.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 495.Dotto GP. Notch tumor suppressor function. Oncogene. 2008;27:5115–5123. doi: 10.1038/onc.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Nicolas M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 497.Meyers JM, Uberoi A, Grace M, Lambert PF, Munger K. Cutaneous HPV8 and MmuPV1 E6 proteins target the NOTCH and TGF-β tumor suppressors to inhibit differentiation and sustain keratinocyte proliferation. PLoS Pathog. 2017;13:e1006171. doi: 10.1371/journal.ppat.1006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Demehri S, Turkoz A, Kopan R. Epidermal Notch1 loss promotes skin tumorigenesis by impacting the stromal microenvironment. Cancer Cell. 2009;16:55–66. doi: 10.1016/j.ccr.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Hu B, et al. Multifocal epithelial tumors and field cancerization from loss of mesenchymal CSL signaling. Cell. 2012;149:1207–1220. doi: 10.1016/j.cell.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Lefort K, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Wu MH, et al. PTC124 rescues nonsense mutation of two tumor suppressor genes NOTCH1 and FAT1 to repress HNSCC cell proliferation. Biomedicines. 2022;10:2948. doi: 10.3390/biomedicines10112948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Sriuranpong V, et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 2001;61:3200–3205. [PubMed] [Google Scholar]
- 503.Giachino C, et al. A tumor suppressor function for notch signaling in forebrain tumor subtypes. Cancer Cell. 2015;28:730–742. doi: 10.1016/j.ccell.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 504.Hanlon L, et al. Notch1 functions as a tumor suppressor in a model of K-ras-induced pancreatic ductal adenocarcinoma. Cancer Res. 2010;70:4280–4286. doi: 10.1158/0008-5472.CAN-09-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Kannan S, et al. Notch/HES1-mediated PARP1 activation: a cell type-specific mechanism for tumor suppression. Blood. 2011;117:2891–2900. doi: 10.1182/blood-2009-12-253419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Strosberg JR, et al. A phase II study of RO4929097 in metastatic colorectal cancer. Eur. J. Cancer. 2012;48:997–1003. doi: 10.1016/j.ejca.2012.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.De Jesus-Acosta A, et al. A phase II study of the gamma-secretase inhibitor RO4929097 in patients with previously treated metastatic pancreatic adenocarcinoma. Investig. N. Drugs. 2014;32:739–745. doi: 10.1007/s10637-014-0083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Lee SM, et al. Phase 2 study of RO4929097, a gamma-secretase inhibitor, in metastatic melanoma: SWOG 0933. Cancer. 2015;121:432–440. doi: 10.1002/cncr.29055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Diaz-Padilla I, et al. A phase II study of single-agent RO4929097, a gamma-secretase inhibitor of Notch signaling, in patients with recurrent platinum-resistant epithelial ovarian cancer: a study of the Princess Margaret, Chicago and California phase II consortia. Gynecol. Oncol. 2015;137:216–222. doi: 10.1016/j.ygyno.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 510.Peereboom DM, et al. A phase II and pharmacodynamic trial of RO4929097 for patients with recurrent/progressive glioblastoma. Neurosurgery. 2021;88:246–251. doi: 10.1093/neuros/nyaa412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Diaz-Padilla I, et al. A phase Ib combination study of RO4929097, a gamma-secretase inhibitor, and temsirolimus in patients with advanced solid tumors. Investig. N. Drugs. 2013;31:1182–1191. doi: 10.1007/s10637-013-0001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Xu R, et al. Molecular and clinical effects of notch inhibition in glioma patients: a phase O/I trial. Clin. Cancer Res. 2016;22:4786–4796. doi: 10.1158/1078-0432.CCR-16-0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Pan E, et al. Phase I study of RO4929097 with bevacizumab in patients with recurrent malignant glioma. J. Neurooncol. 2016;130:571–579. doi: 10.1007/s11060-016-2263-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Gounder MM, et al. A Phase Ib/II randomized study of RO4929097, a gamma-secretase or notch inhibitor with or without Vismodegib, a hedgehog inhibitor, in advanced sarcoma. Clin. Cancer Res. 2022;28:1586–1594. doi: 10.1158/1078-0432.CCR-21-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Krop I, et al. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J. Clin. Oncol. 2012;30:2307–2313. doi: 10.1200/JCO.2011.39.1540. [DOI] [PubMed] [Google Scholar]
- 516.Hoffman LM, et al. Phase I trial of weekly MK-0752 in children with refractory central nervous system malignancies: a pediatric brain tumor consortium study. Childs Nerv. Syst. 2015;31:1283–1289. doi: 10.1007/s00381-015-2725-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Fouladi M, et al. Phase I trial of MK-0752 in children with refractory CNS malignancies: a pediatric brain tumor consortium study. J. Clin. Oncol. 2011;29:3529–3534. doi: 10.1200/JCO.2011.35.7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Cook N, et al. A phase I trial of the γ-secretase inhibitor MK-0752 in combination with gemcitabine in patients with pancreatic ductal adenocarcinoma. Br. J. Cancer. 2018;118:793–801. doi: 10.1038/bjc.2017.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Piha-Paul SA, et al. Results of a phase 1 trial combining ridaforolimus and MK-0752 in patients with advanced solid tumours. Eur. J. Cancer. 2015;51:1865–1873. doi: 10.1016/j.ejca.2015.06.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Brana I, et al. A parallel-arm phase I trial of the humanised anti-IGF-1R antibody dalotuzumab in combination with the AKT inhibitor MK-2206, the mTOR inhibitor ridaforolimus, or the NOTCH inhibitor MK-0752, in patients with advanced solid tumours. Br. J. Cancer. 2014;111:1932–1944. doi: 10.1038/bjc.2014.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Even C, et al. Safety and clinical activity of the Notch inhibitor, crenigacestat (LY3039478), in an open-label phase I trial expansion cohort of advanced or metastatic adenoid cystic carcinoma. Investig. N. Drugs. 2020;38:402–409. doi: 10.1007/s10637-019-00739-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Azaro A, et al. Phase 1 study of 2 high dose intensity schedules of the pan-Notch inhibitor crenigacestat (LY3039478) in combination with prednisone in patients with advanced or metastatic cancer. Investig. N. Drugs. 2021;39:193–201. doi: 10.1007/s10637-020-00944-z. [DOI] [PubMed] [Google Scholar]
- 523.Azaro A, et al. A phase 1b study of the Notch inhibitor crenigacestat (LY3039478) in combination with other anticancer target agents (taladegib, LY3023414, or abemaciclib) in patients with advanced or metastatic solid tumors. Investig. N. Drugs. 2021;39:1089–1098. doi: 10.1007/s10637-021-01094-6. [DOI] [PubMed] [Google Scholar]
- 524.Doi T, et al. A phase 1 study of crenigacestat (LY3039478), the Notch inhibitor, in Japanese patients with advanced solid tumors. Investig. N. Drugs. 2021;39:469–476. doi: 10.1007/s10637-020-01001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Massard C, et al. A phase 1b study of crenigacestat (LY3039478) in combination with gemcitabine and cisplatin or gemcitabine and carboplatin in patients with advanced or metastatic solid tumors. Cancer Chemother. Pharm. 2022;90:335–344. doi: 10.1007/s00280-022-04461-z. [DOI] [PubMed] [Google Scholar]
- 526.Messersmith WA, et al. A Phase I, dose-finding study in patients with advanced solid malignancies of the oral γ-secretase inhibitor PF-03084014. Clin. Cancer Res. 2015;21:60–67. doi: 10.1158/1078-0432.CCR-14-0607. [DOI] [PubMed] [Google Scholar]
- 527.Borthakur G, et al. Phase 1 study to evaluate Crenigacestat (LY3039478) in combination with dexamethasone in patients with T-cell acute lymphoblastic leukemia and lymphoma. Cancer. 2021;127:372–380. doi: 10.1002/cncr.33188. [DOI] [PubMed] [Google Scholar]
- 528.Papayannidis C, et al. A Phase 1 study of the novel gamma-secretase inhibitor PF-03084014 in patients with T-cell acute lymphoblastic leukemia and T-cell lymphoblastic lymphoma. Blood Cancer J. 2015;5:e350. doi: 10.1038/bcj.2015.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Aung KL, et al. A multi-arm phase I dose-escalating study of an oral NOTCH inhibitor BMS-986115 in patients with advanced solid tumours. Investig. N. Drugs. 2018;36:1026–1036. doi: 10.1007/s10637-018-0597-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Pant S, et al. A first-in-human phase I study of the oral Notch inhibitor, LY900009, in patients with advanced cancer. Eur. J. Cancer. 2016;56:1–9. doi: 10.1016/j.ejca.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 531.Gounder M, et al. Nirogacestat, a γ-secretase inhibitor for desmoid tumors. N. Engl. J. Med. 2023;388:898–912. doi: 10.1056/NEJMoa2210140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 532.Mueller AC, et al. Induction of ADAM10 by radiation therapy drives fibrosis, resistance, and epithelial-to-mesenchyal transition in pancreatic cancer. Cancer Res. 2021;81:3255–3269. doi: 10.1158/0008-5472.CAN-20-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Chi M, et al. Novel structured ADAM17 small-molecule inhibitor represses ADAM17/Notch pathway activation and the NSCLC cells’ resistance to anti-tumour drugs. Front. Pharm. 2023;14:1189245. doi: 10.3389/fphar.2023.1189245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Saha N, et al. Fully human monoclonal antibody targeting activated ADAM10 on colorectal cancer cells. Biomed. Pharmacother. 2023;161:114494. doi: 10.1016/j.biopha.2023.114494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Ferrarotto R, et al. A phase I dose-escalation and dose-expansion study of brontictuzumab in subjects with selected solid tumors. Ann. Oncol. 2018;29:1561–1568. doi: 10.1093/annonc/mdy171. [DOI] [PubMed] [Google Scholar]
- 536.Smith DC, et al. A phase 1 dose escalation and expansion study of Tarextumab (OMP-59R5) in patients with solid tumors. Investig. N. Drugs. 2019;37:722–730. doi: 10.1007/s10637-018-0714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Hu ZI, et al. A randomized phase II trial of nab-paclitaxel and gemcitabine with tarextumab or placebo in patients with untreated metastatic pancreatic cancer. Cancer Med. 2019;8:5148–5157. doi: 10.1002/cam4.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Rosen LS, et al. A phase I, dose-escalation study of PF-06650808, an anti-Notch3 antibody-drug conjugate, in patients with breast cancer and other advanced solid tumors. Investig. N. Drugs. 2020;38:120–130. doi: 10.1007/s10637-019-00754-y. [DOI] [PubMed] [Google Scholar]
- 539.Rudin CM, et al. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol. 2017;18:42–51. doi: 10.1016/S1470-2045(16)30565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Udagawa H, et al. Phase I safety and pharmacokinetics study of rovalpituzumab tesirine in Japanese patients with advanced, recurrent small cell lung cancer. Lung Cancer. 2019;135:145–150. doi: 10.1016/j.lungcan.2019.07.025. [DOI] [PubMed] [Google Scholar]
- 541.Hann CL, et al. A phase 1 study evaluating rovalpituzumab tesirine in frontline treatment of patients with extensive-stage SCLC. J. Thorac. Oncol. 2021;16:1582–1588. doi: 10.1016/j.jtho.2021.06.022. [DOI] [PubMed] [Google Scholar]
- 542.Morgensztern D, et al. Efficacy and safety of rovalpituzumab tesirine in third-line and beyond patients with DLL3-expressing, relapsed/refractory small-cell lung cancer: results from the phase II TRINITY study. Clin. Cancer Res. 2019;25:6958–6966. doi: 10.1158/1078-0432.CCR-19-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Malhotra J, et al. A phase 1-2 study of rovalpituzumab tesirine in combination with nivolumab plus or minus ipilimumab in patients with previously treated extensive-stage SCLC. J. Thorac. Oncol. 2021;16:1559–1569. doi: 10.1016/j.jtho.2021.02.022. [DOI] [PubMed] [Google Scholar]
- 544.Johnson ML, et al. Rovalpituzumab tesirine as a maintenance therapy after first-line platinum-based chemotherapy in patients with extensive-stage-SCLC: results from the phase 3 MERU study. J. Thorac. Oncol. 2021;16:1570–1581. doi: 10.1016/j.jtho.2021.03.012. [DOI] [PubMed] [Google Scholar]
- 545.Blackhall F, et al. Efficacy and safety of rovalpituzumab tesirine compared with topotecan as second-line therapy in DLL3-High SCLC: results from the phase 3 TAHOE study. J. Thorac. Oncol. 2021;16:1547–1558. doi: 10.1016/j.jtho.2021.02.009. [DOI] [PubMed] [Google Scholar]
- 546.Mansfield AS, et al. A phase I/II study of rovalpituzumab tesirine in delta-like 3-expressing advanced solid tumors. NPJ Precis. Oncol. 2021;5:74. doi: 10.1038/s41698-021-00214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Morgensztern D, et al. SC-002 in patients with relapsed or refractory small cell lung cancer and large cell neuroendocrine carcinoma: phase 1 study. Lung Cancer. 2020;145:126–131. doi: 10.1016/j.lungcan.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Chiorean EG, et al. A phase I first-in-human study of enoticumab (REGN421), a fully human delta-like ligand 4 (Dll4) Monoclonal antibody in patients with advanced solid tumors. Clin. Cancer Res. 2015;21:2695–2703. doi: 10.1158/1078-0432.CCR-14-2797. [DOI] [PubMed] [Google Scholar]
- 549.Smith DC, et al. A phase I dose escalation and expansion study of the anticancer stem cell agent demcizumab (anti-DLL4) in patients with previously treated solid tumors. Clin. Cancer Res. 2014;20:6295–6303. doi: 10.1158/1078-0432.CCR-14-1373. [DOI] [PubMed] [Google Scholar]
- 550.McKeage MJ, et al. Phase IB trial of the anti-cancer stem cell DLL4-binding agent demcizumab with pemetrexed and carboplatin as first-line treatment of metastatic non-squamous NSCLC. Target Oncol. 2018;13:89–98. doi: 10.1007/s11523-017-0543-0. [DOI] [PubMed] [Google Scholar]
- 551.Coleman RL, et al. Demcizumab combined with paclitaxel for platinum-resistant ovarian, primary peritoneal, and fallopian tube cancer: the SIERRA open-label phase Ib trial. Gynecol. Oncol. 2020;157:386–391. doi: 10.1016/j.ygyno.2020.01.042. [DOI] [PubMed] [Google Scholar]
- 552.Pan B, et al. Targeted inhibition of RBPJ transcription complex alleviates the exhaustion of CD8(+) T cells in hepatocellular carcinoma. Commun. Biol. 2023;6:123. doi: 10.1038/s42003-023-04521-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Hanna GJ, et al. A phase I study of the pan-notch inhibitor CB-103 for patients with advanced adenoid cystic carcinoma and other tumors. Cancer Res. Commun. 2023;3:1853–1861. doi: 10.1158/2767-9764.CRC-23-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Spriano F, et al. In vitro anti-lymphoma activity of the first-in-class pan-NOTCH transcription inhibitor CB-103. Br. J. Haematol. 2023;200:669–672. doi: 10.1111/bjh.18576. [DOI] [PubMed] [Google Scholar]
- 555.Vigolo M, et al. The efficacy of CB-103, a first-in-class transcriptional notch inhibitor, in preclinical models of breast cancer. Cancers. 2023;15:3957. doi: 10.3390/cancers15153957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Moellering RE, et al. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Astudillo L, et al. The small molecule IMR-1 inhibits the notch transcriptional activation complex to suppress tumorigenesis. Cancer Res. 2016;76:3593–3603. doi: 10.1158/0008-5472.CAN-16-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Wong E, Frost GR, Li YM. γ-Secretase modulatory proteins: the guiding hand behind the running scissors. Front. Aging Neurosci. 2020;12:614690. doi: 10.3389/fnagi.2020.614690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Rivkin A, et al. Piperazinyl pyrimidine derivatives as potent gamma-secretase modulators. Bioorg. Med. Chem. Lett. 2010;20:1269–1271. doi: 10.1016/j.bmcl.2009.11.101. [DOI] [PubMed] [Google Scholar]
- 560.Kukar TL, et al. Substrate-targeting gamma-secretase modulators. Nature. 2008;453:925–929. doi: 10.1038/nature07055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Hur JY. γ-Secretase in Alzheimer’s disease. Exp. Mol. Med. 2022;54:433–446. doi: 10.1038/s12276-022-00754-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Habets RA, et al. Safe targeting of T cell acute lymphoblastic leukemia by pathology-specific NOTCH inhibition. Sci. Transl. Med. 2019;11:eaau6246. doi: 10.1126/scitranslmed.aau6246. [DOI] [PubMed] [Google Scholar]
- 563.Medina E, Perez DH, Antfolk D, Luca VC. New tricks for an old pathway: emerging Notch-based biotechnologies and therapeutics. Trends Pharm. Sci. 2023;44:934–948. doi: 10.1016/j.tips.2023.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 564.Li X, et al. The Notch signaling pathway: a potential target for cancer immunotherapy. J. Hematol. Oncol. 2023;16:45. doi: 10.1186/s13045-023-01439-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Chatterjee S, Sil PC. Targeting the crosstalks of Wnt pathway with Hedgehog and Notch for cancer therapy. Pharm. Res. 2019;142:251–261. doi: 10.1016/j.phrs.2019.02.027. [DOI] [PubMed] [Google Scholar]