Abstract
Dimorphism in fungi is believed to constitute a mechanism of response to adverse conditions and represents an important attribute for the development of virulence by a number of pathogenic fungal species. We have isolated YlRAC1, a gene encoding a 192-amino-acid protein that is essential for hyphal growth in the dimorphic yeast Yarrowia lipolytica and which represents the first Rac homolog described for fungi. YlRAC1 is not an essential gene, and its deletion does not affect the ability to mate or impair actin polarization in Y. lipolytica. However, strains lacking functional YlRAC1 show alterations in cell morphology, suggesting that the function of YlRAC1 may be related to some aspect of the polarization of cell growth. Northern blot analysis showed that transcription of YlRAC1 increases steadily during the yeast-to-hypha transition, while Southern blot analysis of genomic DNA suggested the presence of several RAC family members in Y. lipolytica. Interestingly, strains lacking functional YlRAC1 are still able to grow as the pseudohyphal form and to invade agar, thus pointing to a function for YlRAC1 downstream of MHY1, a previously isolated gene encoding a C2H2-type zinc finger protein with the ability to bind putative stress response elements and whose activity is essential for both hyphal and pseudohyphal growth in Y. lipolytica.
The members of the Rho family of Ras-related small GTPases (Cdc42, Rac, and Rho proteins) are signaling molecules that, like other Ras proteins, act as molecular switches by transducing signals in the GTP-bound conformation, while being inactive in the GTP-bound state (12, 13). Although Rho family GTPases were originally thought to be involved solely in the organization of the actin cytoskeleton (22, 23, 56, 57, 66, 67, 70), recent evidence has implicated them in an increasing number of vital cellular processes, such as the activation of kinase cascades, regulation of gene expression and membrane trafficking, cell cycle control, and induction of apoptosis, and as critical regulators of oncogenic transformation in mammalian cells (10, 11, 16, 24, 31, 45, 55, 58, 59, 70, 74). Rac proteins have also been shown to regulate the NADPH oxidase system in phagocytes (7) and plants (30) and to induce cell death in rice through a mechanism that shows biochemical and morphological features similar to those of apoptosis in mammalian cells (30).
In yeast, the Rho family has an important function in the budding process and is known to be involved in the control of actin cytoskeleton dynamics in response to extracellular signals (5, 14, 36). This family is represented in Saccharomyces cerevisiae by six proteins (Rho1p, Rho2p, Rho3p, Rho4p, Cdc42p, and Ynl180cp), which are thought to regulate partially overlapping pathways (19). No Rac homolog has yet been described for fungi.
The yeast Yarrowia lipolytica has received increasing attention as a model to study dimorphic transition because of its ability to alternate between a unicellular yeast form and distinct filamentous forms (hyphae and pseudohyphae). This fact, combined with the availability of specific molecular and genetic tools, has provided Y. lipolytica with a number of advantages over S. cerevisiae and Candida albicans for investigation of the molecular mechanisms underlying dimorphic transition (26). The dimorphic switch is induced by environmental signals and is thought to be important for virulence in a number of pathogenic fungi (8, 38, 41, 52, 62).
In this paper, we report the isolation and initial characterization of YlRAC1, a gene encoding the first reported fungal Rac homolog, which is involved in the yeast-to-hypha transition in Y. lipolytica.
MATERIALS AND METHODS
Yeast strains and microbial techniques.
The Y. lipolytica strains used in this study are listed in Table 1. The mutant strain CHY1220 was isolated after chemical mutagenesis of Y. lipolytica E122 cells with 1-methyl-3-nitro-1-nitrosoguanidine, as previously described (51). Media components were as follows: YEPD, 1% yeast extract, 2% peptone, and 2% glucose; YNA, 0.67% yeast nitrogen base without amino acids and 2% sodium acetate; YNBD, 0.67% yeast nitrogen base without amino acids and 2% glucose; YNBGlc, 1.34% yeast nitrogen base without amino acids and 1% glucose; and YNBGlcNAc, 1.34% yeast nitrogen base without amino acids, 1% N-acetylglucosamine, and 50 mM citric acid (pH 6.0). YNA and YNBD were supplemented with uracil, leucine, lysine, and histidine, each at 50 μg/ml, as required. YNBGlc and YNBGlcNAc were supplemented with 2× Complete Supplement Mixture (Bio101, Vista, Calif.) or 2× Complete Supplement Mixture minus leucine, as required. Media, growth conditions, and procedures for mating, sporulation, and transformation of Y. lipolytica have been described previously (9, 51).
TABLE 1.
Y. lipolytica strains used in this study
| Straina | Genotype |
|---|---|
| E122 | MATA ura3-302 leu2-270 lys8-11 |
| 22301-3 | MATB ura3-302 leu2-270 his1 |
| CHY1220 | MATA ura3-302 leu2-270 lys8-11 fi11 |
| CHY1220-A30 | MATA ura3-302 leu2-270 lys8-11 Δrac1::URA3 |
| CHY1220-B36 | MATB ura3-302 leu2-270 his1 Δrac1::URA3 |
| mhy1KO9 | MATA ura3-302 leu2-270 lys8-11 Δmhyl::URA3 |
| E122//22301-3 | MATA/MATB ura3-302/ura3-302 leu2-270/leu2-270 lys8-11/+ his1/+ |
| E122//CHY1220-B36 | MATA/MATB ura3-302/ura3-302 leu2-270/leu2-270 lys8-11/+ his1/+ Δrac1::URA3/+ |
| 22301-3//CHY1220-A30 | MATA/MATB ura3-302/ura3-302 leu2-270/leu2-270 lys8-11/+ his1/+ Δrac1::URA3/+ |
| CHY1220-A30//CHY1220-B36 | MATA/MATB ura3-302/ura3-302 leu2-270/leu2-270 lys8-11/+ his1/+ Δrac1::URA3/Δrac1::URA3 |
Strains E122 and 22301-3 were from C. Gaillardin, Thiverval-Grignon, France. Strain mhy1KO9 was from work described previously (26). All other strains were from this study.
Standard techniques for DNA manipulation and growth of Escherichia coli were used as described previously (6).
Mycelial induction.
Mycelial growth was induced as described previously (20). Cells were grown for 12 h in YNBGlc, harvested by centrifugation at room temperature, washed with sterile distilled water, kept on ice for 15 min in YNB without a carbon source, and inoculated at a final density of 107 cells/ml in prewarmed YNBGlcNAc (for induction of the yeast-to-hypha transition) or YNBGlc (for growth as the yeast form).
Cloning and characterization of the Y. lipolytica RAC1 gene.
The Y. lipolytica RAC1 (YlRAC1) gene was isolated by functional complementation of strain CHY1220 using a Y. lipolytica genomic DNA library contained in the replicative E. coli shuttle vector pINA445 (51). Plasmid DNA was introduced into yeast cells by electroporation, and Leu+ transformants were screened on YNA agar plates for their ability to give rise to filamentous colonies.
Complementing plasmids were recovered by transformation of E. coli, and the smallest fragment capable of restoring hyphal growth was determined. Restriction fragments prepared from the genomic insert of one of these constructs (pRAC1) were subcloned into the vector pGEM-5Zf(+) or pGEM-7Zf(+) (Promega, Madison, Wis.) or pBluescript II SK(+) (Stratagene, La Jolla, Calif.) for dideoxynucleotide sequencing of both strands. The deduced polypeptide sequence, Y. lipolytica Rac1p (YlRac1p), was compared to other known protein sequences using the BLAST Network Service of the National Center for Biotechnology Information (Bethesda, Md.).
Nucleic acid manipulation.
Genomic DNA, plasmid DNA, and total RNA were prepared from Y. lipolytica, as described elsewhere (6). Southern and Northern blot analyses were performed with DNA probes prepared with the ECL direct nucleic acid labeling and detection system (Amersham Life Sciences, Oakville, Ontario, Canada). Electrophoresis and transfer to nitrocellulose membranes were carried out as described previously (6). Hybridization, stringency of washes, and signal generation and detection were as recommended by the manufacturer, except that for the analysis of Y. lipolytica genomic DNA, the temperature of hybridization and washes was reduced from 42 to 39°C.
Immunofluorescence microscopy.
F-actin was detected by incubating cells with 1.3 μM Oregon Green 488 phalloidin (Molecular Probes, Eugene, Oreg.), as previously described (2). Images were scanned using SPOT software 1.2.1 (Diagnostic Instruments, Sterling Heights, Mich.), processed in Photoshop 4.0.1 (Adobe Systems, San Jose, Calif.), and printed on a DS8650 PS color printer (Eastman-Kodak, Rochester, N.Y.).
Mutagenesis of CCCCT elements in the YlRAC1 promoter region.
Three-base substitutions were introduced in the putative stress response elements (STREs) found in the promoter region of the YlRAC1 gene (CCCCT to ATTCT in STRE1, CCCCT to AGCTT in STRE2, and CCCCT to GATCT in STRE3) (see Fig. 2) by PCR using the oligonucleotide pairs SE1 and SE2, EH1 and EH2, HB1 and HB2, and BS1 and BS2 (Table 2). The four PCR products (369-bp SpeI-EcoRI, 100-bp EcoRI-HindIII, 197-bp HindIII-BglII, and 122-bp BglII-SalI fragments) were then ligated to the 396-bp SalI-SacII fragment obtained by PCR using the oligonucleotides NT1 and NT2 (Table 2). The resulting SpeI-SacII fragment (∼1.2 kbp) was used to replace its counterpart in the plasmid pRAC1. The integrity of the final construct was confirmed by sequencing.
FIG. 2.
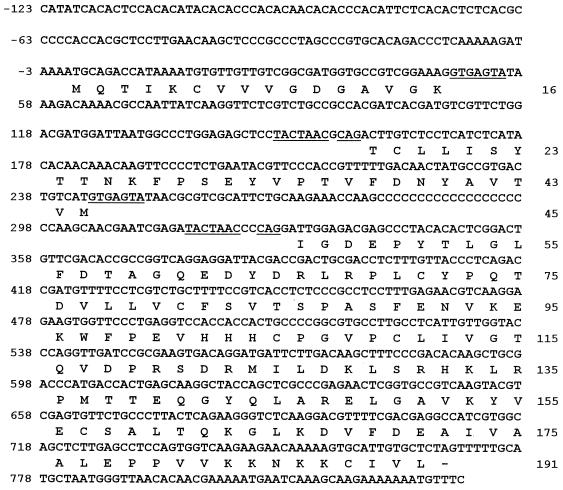
Nucleotide sequence of the YlRAC1 gene and deduced amino acid sequence of YlRac1p. The transcriptional start site of the YlRAC1 gene is indicated by an arrow. Putative STREs are indicated. Consensus sequences for intron splicing are underlined. Putative transcription termination signals are doubly underlined.
TABLE 2.
Oligonucleotide primers used in this study
| Primer | Sequence | Restriction site |
|---|---|---|
| KO1 | 5′-CTC TCC TGA TCT GCA TCT GAT CTG-3′ | |
| KO2 | 5′-TAG CTG AAG ACT CAA TCT GGA GGG-3′ | |
| T3 | 5′-CCA AGC TCG AAA TTA ACC CTC ACT AA-3′ | |
| T7 | 5′-GTA ATA CGA CTC ACT ATA GGG CGA AT-3′ | |
| CDC42U | 5′-CCT AGC CCG TGC ACA GAC CCT CAA-3′ | |
| CDC42M | 5′-TCC GAG TGT GTA GGG CTC GTC TCC-3′ | |
| PR1 | 5′-TTA GGG CCC AAT CTA AGA TAG ACA CAC GCT CAC CAC CCA-3′ | ApaI |
| PR2 | 5′-CTG GTC GAC CAT TTT GGA ACC GGT AGC GAG AGT GGA TGT AGG-3′ | SalI |
| NT1 | 5′-ATG GTC GAC CAG AGT ATA AAA TGT GTC GTC ACT GGC GAC GGG-3′ | SalI |
| NT2 | 5′-GGC CCG CGG TAT CCC AAA GTC CGA GGT TTA TCG GTT TGT-3′ | SacII |
| SE1 | 5′-ACT AGT GAG TCG ATG GGC AAC AAA CCA CAG-3′ | SpeI |
| SE2 | 5′-AGA ATT CAG AGA GCT TAG TGC ACG GCT GGC TTG-3′ | EcoRI |
| EH1 | 5′-TGA ATT CTT GTT GTG CTG AGT TTG TCT TTT TTT CAT CAA-3′ | EcoRI |
| EH2 | 5′-AAA GCT TGT GGT TTG GGT GGT GAG CGT GTG TCT ATC-3′ | HindIII |
| HB1 | 5′-CAA GCT TTC TTT TGC ACA CCA CCC CAC GAC CGA AAC-3′ | HindIII |
| HB2 | 5′-CAG ATC TTG TAG TGA GTG ACG CAA AAA CTG AGA CCG-3′ | BglII |
| BS1 | 5′-AAG ATC TGC ACA AGT CTC AAT CAA GAC ACT CGC AAG-3′ | BglII |
| BS2 | 5′-GGT CGA CCA TTT TGG AAC CGG TAG CGA GAG TGG ATG TA-3′ | SalI |
Nucleotide sequence accession number.
The sequence data reported here for YlRAC1 and Y. lipolytica CDC42 (YlCDC42) are available from EMBL/GenBank/DDBJ under accession numbers AF176831 and AF209750, respectively.
RESULTS
Isolation of the Y. lipolytica mutant strain CHY1220.
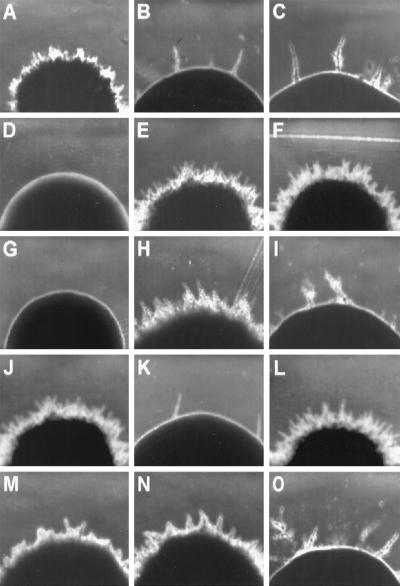
The Y. lipolytica mutant strain CHY1220 (Fig. 1B) was initially isolated by its inability to form wild-type rough-surfaced colonies on YEPD agar plates after 3 days of incubation at 28°C (Fig. 1A and J). Further analysis revealed that, like Δrac1 strains, strain CHY1220 was able to form colonies having a small number of peripheral extensions after prolonged periods of incubation on both rich and minimal media and that these extensions consisted of chains of elongated cells following a pseudohyphal pattern (Fig. 1B, C, K, and O; see Fig. 7).
FIG. 1.
Colony morphology of various Y. lipolytica strains. (A) Wild-type strain E122; (B) original mutant strain CHY1220; (C) RAC1 disruptant strain CHY1220-A30; (D) MHY1 disruptant strain mhy1KO9; (E) strain CHY1220 transformed with plasmid pRAC1; (F) strain CHY1220-A30 transformed with plasmid pRAC1; (G) strain mhy1KO9 transformed with plasmid pRAC1; (H) strain CHY1220 transformed with plasmid pMHY1; (I) strain CHY1220-A30 transformed with plasmid pMHY1; (J) wild-type strain 22301-3; (K) RAC1 disruptant strain CHY1220-B36; (L) RAC1/RAC1 diploid strain E122//22301-3; (M and N) RAC1/rac1 diploid strains 22301-3//CHY1220-A30 and E122//CHY1220-B36, respectively; (O) rac1/rac1 diploid strain CHY1220-A30//CHY1220-B36. Colonies were photographed after 3 days of incubation at 28°C on YNA agar plates. Magnification, ×100.
FIG. 7.
Disruption of YlRAC1 affects cell morphology and impairs hyphal growth, but not pseudohyphal growth, in Y. lipolytica. Strains were grown in YEPD. Top panels, exponential growth phase (optical density at 600 nm [OD600] = 1). Middle panels, late exponential growth phase (OD600 = 4). Bottom panels and inset, stationary phase (OD600 = 10). WT, wild-type strain E122. Δrac1, strain CHY1220-A30. Bars, 5 μm.
Isolation and characterization of the YlRAC1 gene.
The YlRAC1 gene was isolated from a Y. lipolytica genomic DNA library contained in the replicative E. coli shuttle vector pINA445 (51) by its ability to restore hyphal growth to CHY1220 cells. Of approximately 70,000 transformants screened, 5 showed a restored filamentous phenotype (Fig. 1E). Restriction enzyme analysis demonstrated that all complementing plasmids shared a 2.2-kbp SpeI-ClaI fragment capable of inducing hyphal growth in CHY1220 cells. Sequencing of this fragment revealed an open reading frame (ORF) of 576 bp interrupted by two introns, which are found at codons 12 and 36 (nucleotides +36 to +205 and +278 to +328 relative to the A residue of the potential initiating codon, respectively) (Fig. 2). The putative 5′-splice donor sequences of both introns (GTAAGTPu) diverge at the third and fourth positions from the GTGAGTPu and GTATGT consensus motifs found for Y. lipolytica (39, 65) and S. cerevisiae (69), respectively. As in the Y. lipolytica genes SEC14 (39) and PYK1 (65), a 3′-splice acceptor CAG sequence is found one nucleotide downstream of the consensus TACTAAC box (69) or its abbreviated form CTAAC (Fig. 2).
No obvious TATA box or CT/CA-rich region, which is believed to play a role in transcriptional regulation in Y. lipolytica (50, 71), is seen in the promoter region of YlRAC1. However, analysis of cDNA showed that transcription of the YlRAC1 gene starts preferentially at position −286 relative to the A nucleotide of the first ATG codon and that polyadenylation occurs following the G nucleotide at position +1075. Other features of YlRAC1 include the presence of conserved A nucleotides at positions −1 and −3 relative to the A nucleotide of the initiating ATG and three putative STRE (pentanucleotide CCCCT) (32) in its upstream region (Fig. 2).
The deduced protein product of YlRAC1, YlRac1p, is 192 amino acids in length and has a predicted molecular mass of 21,173 Da (Fig. 2). Comparison of the predicted amino acid sequence of YlRac1p with the sequences of Rac proteins from different sources suggests that its closest homolog is human Rac1 (Fig. 3). In addition, YlRac1p has a relatively high pI (8.47), which is an attribute that distinguishes Rac proteins from Rho and Ras proteins (whose pIs are in the range of 5.0 to 6.5) (17).
FIG. 3.
Amino acid sequence alignment of Rac1p of Y. lipolytica (YlRac1p) and Rac proteins from Homo sapiens (HsRac1, HsRac2, and HsRac3), Mus musculus (MmRac1 and MmRac2), Drosophila melanogaster (DmRac1 and DmRac2), Caenorhabditis elegans (CeRac1 and CeRac2), Canis familiaris (CfRac1), and Xenopus laevis (XlRac). GenBank accession numbers are M29870 (HsRac1), CAB45265 (HsRac2), AAC51667 (HsRac3), CAA40545 (MmRac1), Q05144 (MmRac2), AAA62870 (DmRac1), P48554 (DmRac2), AAA28141 (CeRac1), AAB40386 (CeRac2), P15154 (CfRac1), and AAD50299 (XlRac).
Consensus elements GXXXXGK (GDGAVGK, residues 10 to 16) and DXXG (DTAG, residues 57 to 60) (Fig. 3), which are involved in interactions with the phosphate portion of the GTP molecule, are found in YlRac1p at positions conserved among GTP-binding proteins (15, 18, 28, 46). Conserved motifs are also present at regions implicated in interaction with the GTPase-activating protein (TVFDNY, residues 35 to 40) (61) and in membrane association prior to biological activity (CVIL, residues 189 to 192) (23, 73) (Fig. 3). Notably, YlRac1p also contains the conserved motif TKXD (TKLD, residues 115 to 118), which is responsible for nucleotide specificity in Rac proteins and is involved in the determination of the unusually high intrinsic rate of GTP hydrolysis that distinguishes Rac proteins from other Rho family members (17).
Isolation and characterization of the YlCDC42 gene.
Since no RAC gene had previously been reported for fungi and since the CDC42 gene, which encodes a protein that belongs to the Rac subfamily of Rho GTPases (19), had been shown to be involved in the regulation of filamentous growth in S. cerevisiae and C. albicans (46, 47), we decided to provide further evidence that we had identified a fungal RAC gene by isolating the CDC42 gene of Y. lipolytica (YlCDC42).
We combined the sequence of a partial YlCDC42 clone previously obtained by probing a Y. lipolytica genomic DNA library with an oligonucleotide derived from a highly conserved sequence in the Rab family of proteins (53) with the sequence of a YlCDC42 cDNA obtained by PCR of a Y. lipolytica cDNA library constructed in the ZAP Express vector (Stratagene) with oligonucleotides T3, T7, CDC42U, and CDC42M (Table 2) to obtain the sequence of the YlCDC42 gene. The YlCDC42 gene contains an ORF of 573 bp, which is interrupted by two introns, as is the YlRAC1 gene (Fig. 4). The introns are found between codons 16 and 17 and at codon 45 (nucleotides +49 to +157 and +244 to +327 relative to the A nucleotide of the potential initiating codon, respectively). The putative 5′-splice donor sequences of both introns are identical to the consensus motif GTGAGTPu found in other Y. lipolytica genes (39, 65), and a 3′-splice acceptor CAG sequence is found one or two nucleotides downstream of the consensus TACTAAC box (69) (Fig. 4).
FIG. 4.
Nucleotide sequence of the YlCDC42 gene and deduced amino acid sequence of YlCdc42p. Consensus sequences for intron splicing are underlined.
The deduced protein of YlCDC42, YlCdc42p, is 191 amino acids long and has a predicted molecular mass of 21,336 Da (Fig. 4) and an estimated pI of 6.09, which is characteristic of Cdc42 proteins (17). In addition, YlCdc42p contains all motifs required for the biological function of small GTPases of the Rho family (GDGAVGK, residues 10 to 16; TVFDNY, residues 35 to 40; DTAG, residues 57 to 60; and CIVL, residues 188 to 191) (Fig. 4 and 5). Comparison of YlCdc42p with Cdc42 proteins from a number of different organisms suggests that its closest relative is Schizosaccharomyces pombe Cdc42 (Fig. 5). Importantly, YlCdc42p contains the signature sequence of Cdc42 proteins (27), the motif TQXD (TQVD, residues 115 to 118) in the region responsible for nucleotide specificity.
FIG. 5.
Amino acid sequence alignment of Cdc42p of Y. lipolytica (YlCdc42p) and its homologs in S. cerevisiae (ScCdc42p) (28), C. albicans (CaCdc42p) (46), S. pombe (SpCdc42p) (44), Caenorhabditis elegans (CeCdc42p) (15), Mus musculus (MmCdc42p) (43), and Homo sapiens (HsCdc42p) (48).
Strains with the YlRAC1 gene disrupted are viable and unaffected in mating ability.
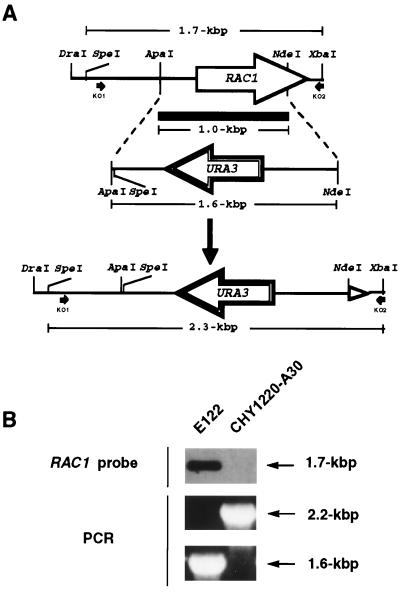
A 1.0-kbp ApaI-NdeI fragment of YlRAC1 was replaced by a 1.6-kbp ApaI-NdeI fragment containing the Y. lipolytica URA3 gene (Fig. 6). This construct was digested with DraI and XbaI to liberate a 2.4-kbp fragment containing the entire URA3 gene flanked by 0.5 and 0.3 kbp of the 5′ and 3′ regions of the YlRAC1 gene, respectively. This linear fragment was used to transform the wild-type Y. lipolytica strain E122 to uracil prototrophy.
FIG. 6.
Integrative disruption of the YlRAC1 gene. (A) Diagram illustrating the replacement of a 1.0-kbp ApaI-NdeI fragment of YlRAC1 by a 1.6-kbp ApaI-NdeI fragment containing the Y. lipolytica URA3 gene. (B) Southern blot analysis of SpeI-HpaI-digested genomic DNA, and PCR analysis of total genomic DNA, from wild-type strain E122 and strain CHY1220-A30, confirming the correct replacement of the YlRAC1 gene with the URA3-containing linear molecule in strain CHY1220-A30. Primers KO1 and KO2 (Table 2) are indicated by black arrows in panel A.
Of 303 Ura+ transformants obtained, 3 showed a smooth phenotype after 3 days on YEPD agar plates. Two of these transformants were confirmed by Southern blot analysis and PCR to have had the YlRAC1 gene correctly replaced by the URA3 gene (Fig. 6), and one of them, CHY1220-A30 (Fig. 1C and Table 1), was selected for further analysis.
Because mating has been found to be intimately connected to dimorphism and, like dimorphism, is a phenomenon that involves dramatic changes in cell morphology in response to environmental conditions (40), we investigated whether disruption of the YlRAC1 gene had any effect on the mating ability of Y. lipolytica. Crossing of the A mating type strain CHY1220-A30 (Δrac1) with the B mating type wild-type strain 22301-3 was readily attained (Fig. 1M), indicating that no mating defect was associated with disruption of YlRAC1 in these strains. To determine whether the lack of effect on mating by disruption of the YlRAC1 gene was confined to A mating type cells, a B mating type strain, CHY1220-B36 (Fig. 1K and Table 1), with its YlRAC1 gene deleted, was obtained by sporulation of the diploid strain 22301-3//CHY1220-A30 (Fig. 1M and Table 1) and selection of haploids for their inability to form hyphal cells. The Δrac1::URA3 genotype of the CHY1220-B36 strain was confirmed by cosegregation of the Ura+ and Fil− phenotypes in random spore analysis (data not shown), and this strain was found to be able to mate to both wild-type E122 and CHY1220-A30 strains (Fig. 1N and O), demonstrating that YlRAC1 is not essential for mating. One copy of the YlRAC1 gene was sufficient to support dimorphic transition in diploid strains of Y. lipolytica, although a slight reduction in the proportion of hyphal cells could be observed in these strains (Fig. 1M and N).
Disruption of the YlRAC1 gene affects cell morphology but does not impair actin polarization or cell invasiveness.
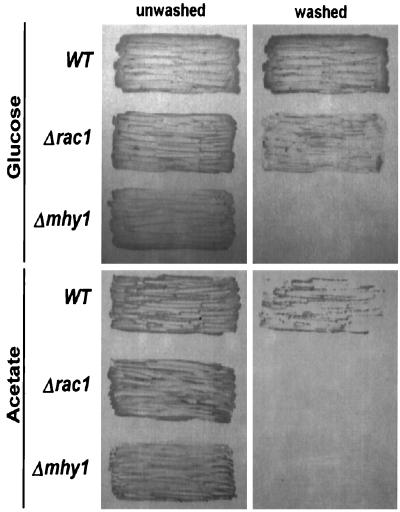
Since the organization of the actin cytoskeleton is directly involved in the determination of cell shape and because Rac proteins play a fundamental role in this process (21, 22, 57, 67), disruption of YlRAC1 was anticipated to result in morphological defects in Y. lipolytica cells. Indeed, exponentially growing Δrac1 mutant cells were found to be round in shape, clearly contrasting with the typically ovoid cells observed for wild-type strains (Fig. 7, top panels). Continued incubation in rich medium yielded wild-type cultures composed of yeast cells, pseudohyphae, and a few germ tubes, while Δrac1 cultures contained only a small proportion of pseudohyphal cells and no germ tubes (Fig. 7, middle panels). In general, pseudohyphal Δrac1 cells were found to be shorter than their wild-type counterparts (Fig. 7, middle panels). As stationary phase was reached, hyphal growth became predominant in wild-type cultures, while only a limited number of chains composed of pseudohyphal cells were seen in the Δrac1 cultures (Fig. 7, bottom panels). Germ tubes arising from pseudohyphal cells were sometimes seen in the wild-type strain (Fig. 7, bottom right panel, inset). Interestingly, invasive pseudohyphal growth was found to be substantially induced in the Δrac1 strain by incubation on minimal medium containing glucose as the sole carbon source (YNBD agar), whereas this effect was not observed in Δrac1 cells grown on minimal medium containing acetate as the sole carbon source (YNA agar) or in mhy1KO9 (Δmhy1) cells incubated under either condition (Fig. 8). No difference in invasiveness was observed between the original mutant strain CHY1220 and the Δrac1 strain CHY1220-A30 when they were subjected to growth under the same conditions (data not shown).
FIG. 8.
Invasive filamentous growth by different Y. lipolytica strains. Following 5 days of incubation at 28°C on minimal agar medium containing glucose or acetate as the sole carbon source, plates were washed with running water to remove cells from the agar surface. Pictures were taken before and after washes. WT, wild-type strain E122. Δrac1, strain CHY1220-A30. Δmhy1, strain mhy1KO9.
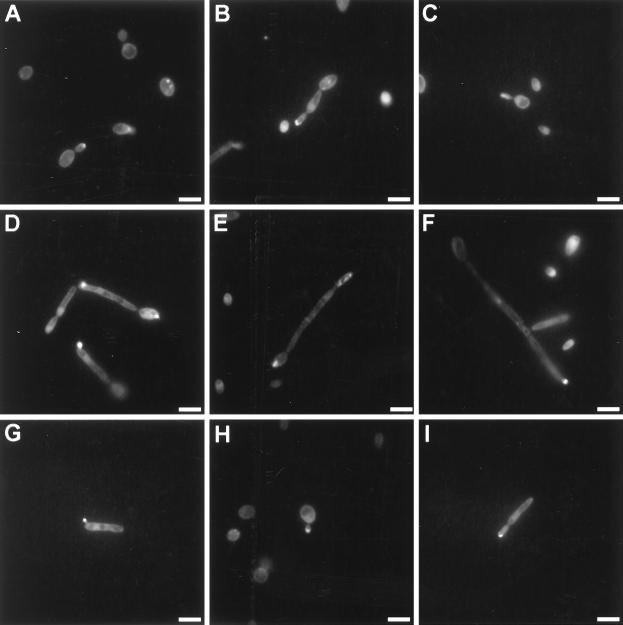
As described for a number of fungi (1, 3, 4, 25, 34, 42, 60, 72), actin-rich zones at the sites of growth (apices of germ tubes, hyphae, pseudohyphae, and yeast cells), combined with a background of diffuse actin staining with punctate actin patches, were observed for wild-type cells of Y. lipolytica (Fig. 9A to G). Interestingly, despite alterations in cell morphology, Δrac1 mutant cells appeared to retain the ability to concentrate actin granules at the apices of pseudohyphal cells and emerging buds (Fig. 9H and I).
FIG. 9.
Actin localization during different stages of development of wild-type and Δrac1 cells. Actin was detected by staining of cells with Oregon Green 488 phalloidin followed by fluorescence microscopy. (A to G) Wild-type strain E122. (H and I) Δrac1 strain CHY1220-A30. (A and H) Yeast-like cells; (B, G, and I) pseudohyphal growth; (C) early germ tube formation; (D) late germ tube formation; (E and F) hyphal growth. Bars, 5 μm.
Transcription of the YlRAC1 gene is increased during the yeast-to-hypha transition.
The dimorphic switch was induced in exponentially growing E122 cells by a 15-min carbon source starvation period at 4°C, followed by transfer to prewarmed (28°C) YNBGlcNAc. Under these conditions, more than 80% of the cell population gave rise to germ tubes after 10 h of incubation, while cells transferred to fresh YNBGlc grew almost exclusively as the budding form, as described previously (20, 26). Northern blot analysis performed with total RNA extracted from cells harvested at 0, 1, 3, and 10 h of incubation in YNBGlcNAc showed that YlRAC1 mRNA levels increased steadily during the yeast-to-hypha transition, but they remained virtually constant during incubation in YNBGlc (Fig. 10).
FIG. 10.
YlRAC1 mRNA levels are increased during the dimorphic transition. Total RNA was isolated from E122 cells incubated at 28°C in YNBGlcNAc (induction of filamentous growth) or YNBGlc (control culture, yeast-like cells) for the times indicated and subjected to Northern blot analysis. Ten micrograms of RNA from each time point was separated on a formaldehyde agarose gel and transferred to nitrocellulose. Blots were hybridized with a probe specific for the YlRAC1 gene (1.0-kbp ApaI-NdeI fragment [see Fig. 6]). Equal loading of RNA was ensured by ethidium bromide staining (data not shown).
Putative STREs in the YlRAC1 gene are not necessary for the induction of hyphal growth.
Genetic interaction between YlRAC1 and MHY1 was initially suggested by the observation that a pINA445-based multicopy plasmid bearing the MHY1 gene (pMHY1) (26) was able to restore hyphal growth upon introduction into CHY1220, but not CHY1220-A30 (Δrac1), cells (Fig. 1H and I). The inability of pRAC1 to induce dimorphic transition in Δmhy1 cells (Fig. 1D and G); the presence of three copies of the pentanucleotide STRE sequence CCCCT in the promoter region of YlRAC1 (Fig. 2), one of which has been shown to specifically bind in vitro-synthesized Mhy1p (26); and the finding that Δrac1 mutant cells can form pseudohyphae, while Δmhy1 mutants are unable to grow as either the hyphal or pseudohyphal form (Fig. 7 and 8) suggested that Mhy1p might act to promote hyphal growth through these regulatory elements via YlRAC1. In order to investigate the role of these putative STRE sequences in the induction of hyphal growth, mutagenesis of these elements in pRAC1 was performed. No defect was observed in the ability to induce the dimorphic transition upon introduction of the plasmid pRAC1-Mut (which contains mutations in all three STREs) into a Δrac1 strain (CHY1220-A30), suggesting that these elements are not necessary for the induction of hyphal growth via YlRAC1 (data not shown).
Genomic DNA analysis of RAC genes in Y. lipolytica.
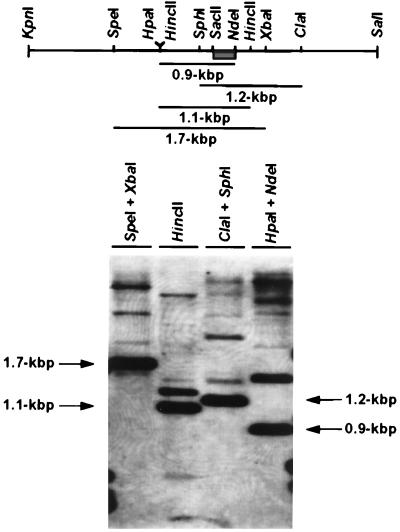
As most organisms have multiple Rac and Rho homologs, we looked for evidence of other RAC genes in Y. lipolytica. Genomic DNA from the E122 strain was digested with various combinations of restriction endonucleases and analyzed by Southern blotting under low-stringency conditions with a labeled 240-bp SacII-NdeI fragment of the YlRAC1 gene. A complex pattern of bands was observed, suggestive of the presence of several RAC-related superfamily members in Y. lipolytica (Fig. 11).
FIG. 11.
Southern blot analysis of E122 genomic DNA. Ten micrograms of DNA per lane was digested with the indicated restriction enzymes, separated by electrophoresis, transferred to nitrocellulose, and probed with a 240-bp SacII-NdeI-labeled fragment from YlRAC1 (boxed), as described in Materials and Methods.
DISCUSSION
Rac proteins have been implicated in the control of a diverse and extensive set of cellular processes (74), including notably the regulation of the assembly and organization of the actin cytoskeleton (21, 22, 49, 56, 57, 66, 67, 70). As a consequence, Rac-mediated changes in actin organization or in gene expression are believed to play a pivotal role in the control of cell shape, cell attachment, cell motility and invasion, cell-cell interaction, and cell proliferation and differentiation (74). In plants, Rac homologs are thought to be involved in the regulation of growth of the pollen tube, a process that shares several characteristics with filamentous growth in fungi. Pollen tube elongation is based on a process known as tip growth, where polarized secretion is restricted to the apex, and cell membrane and cell wall material are delivered exclusively to this location (33, 37, 64, 68). Significantly, plant Rac homologs have been shown to be localized to the plasma membrane of the pollen tube in the region of the tip (37). The localization of YlRac1p in Y. lipolytica has so far remained elusive. Although an epitope-tagged YlRac1p could be detected at the growing tip region of filamentous Y. lipolytica cells (data not shown), this fusion protein was unable to induce hyphal growth in Δrac1 cells, making it impossible to ascertain whether the distribution of the tagged protein was truly representative of that of the natural protein. The availability of antibodies specific for YlRac1p will be extremely useful in addressing this question.
Although no Rac homolog had previously been described for fungi, a newly identified S. cerevisiae ORF, Ynl180c, codes for a protein with a number of features typical of Rac proteins, including the presence of all conserved motifs required for biological function and an estimated pI of 8.76. However, this putative protein, which is only 52.1% identical to YlRac1p, is at 331 amino acids considerably larger than all known Rac proteins, and it is not known at this time whether it is functional.
Disruption of the YlRAC1 gene is not lethal and does not abolish the ability of cells to polarize actin at the site of growth. These findings might be explained by the presence of other genes in Y. lipolytica that are closely related to YlRAC1, as suggested by the complex banding pattern revealed by Southern analysis of Y. lipolytica genomic DNA under conditions of low stringency. Nevertheless, alterations in the cell morphology of Δrac1 mutants and the inability of these strains to grow as the hyphal form strongly suggest that YlRAC1 functions in some aspect of the polarization of cell growth.
We have previously reported the isolation and characterization of the gene MHY1, which codes for a putative transcription factor that binds in vitro to an oligonucleotide containing STRE1 from YlRAC1 (Fig. 2) and is essential for filamentous growth in Y. lipolytica (26). Here we report that while deletion of MHY1 completely abolishes the ability of Y. lipolytica to grow as both the hyphal and pseudohyphal forms on solid minimal medium containing either glucose or acetate as the sole carbon source, strains lacking a functional YlRAC1 gene are still able to form pseudohyphae and invade agar on glucose-based minimal medium, suggesting, as has been suggested for C. albicans (38), that these two morphologies in Y. lipolytica are controlled, at least in part, by two parallel signaling pathways, each with a different and additive input, or that they represent a sequence of events in a single pathway of filamentous growth requiring a quantitatively stronger regulatory input to produce hyphae rather than pseudohyphae. Likewise, the analysis of a Ras homolog in Aspergillus nidulans has suggested a scenario in which several thresholds of Ras concentration exist, each of which allows development to proceed to a certain point, producing the proper cell type while inhibiting further development (63). Our findings that overexpression of MHY1 can restore hyphal growth in CHY1220 but not in CHY1220-A30 (Δrac1) cells, that disruption of YlRAC1 affects only hyphal growth while disruption of MHY1 blocks both hyphal and pseudohyphal growth, and that pseudohyphal cells can give rise to hyphae (Fig. 7, bottom right panel, inset) support such a scenario and suggest that MHY1 acts upstream of YlRAC1 in the filamentous pathway(s).
It is important to point out that regardless of the fact that mutagenesis of all three putative STREs in the promoter of YlRAC1 did not affect its ability to induce hyphal growth in Y. lipolytica (data not shown), a role for these elements in the induction of dimorphism cannot be ruled out. The activity of other unidentified regulatory elements in the YlRAC1 gene or compensation for the loss of transcriptional induction of YlRAC1 by the activation of other related GTPases may explain this negative result. We are currently investigating these possibilities.
Here we also report the isolation and initial characterization of the YlCDC42 gene. In C. albicans, a transient increase in the CDC42 mRNA levels was observed during the switch to hyphal growth (46), but it still remains to be determined whether Cdc42p also plays a role in pseudohyphal formation in this organism. Although no variation in the abundance of Cdc42p has been observed during the cell cycle of S. cerevisiae (73), CDC42 has been shown to be a potent regulator of filamentous growth in this yeast, acting downstream of RAS2 and activating pseudohyphal growth of diploid cells and invasive growth of haploid cells in response to nitrogen starvation via STE20 (35, 47, 54). We are currently investigating whether YlCDC42 also plays a role in the induction of filamentous growth in Y. lipolytica.
In conclusion, we report the isolation and initial characterization of the first fungal Rac homolog and provide evidence that YlRac1p plays an important role in the regulation of hyphal growth in Y. lipolytica. Greater knowledge of the function(s) of YlRAC1 and of its interactions with other Y. lipolytica genes will be important for a better understanding of the mechanisms by which environmental conditions induce changes in the pattern of cell growth, a phenomenon with strong implications for the development of virulence by fungal pathogens (8, 38, 41, 52, 62) and for the elucidation of the molecular mechanisms controlling differentiation in higher eukaryotes. Furthermore, as one of the mechanisms of Ras transformation relies on signaling cascades controlled by Rac and Rho GTPases, the dimorphic yeast Y. lipolytica may represent a more suitable model for the understanding of such complex events in mammalian cells, with important implications for the search for potential drug targets for Ras-mediated malignancies (29, 66, 74).
ACKNOWLEDGMENTS
This work was supported by an International Research Scholarship from the Howard Hughes Medical Institute to R.A.R. R.A.R. is a Medical Research Council of Canada Senior Scientist.
REFERENCES
- 1.Adams A E M, Pringle J R. Relationship of actin and tubulin distribution to bud growth in wild-type and morphogenetic-mutant Saccharomyces cerevisiae. J Cell Biol. 1984;98:934–945. doi: 10.1083/jcb.98.3.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams A E M, Pringle J R. Staining of actin with fluorochrome-conjugated phalloidin. Methods Enzymol. 1991;194:729–731. doi: 10.1016/0076-6879(91)94054-g. [DOI] [PubMed] [Google Scholar]
- 3.Akashi T, Kanbe T, Tanaka K. The role of the cytoskeleton in the polarized growth of the germ tube in in Candida albicans. Microbiology. 1994;140:271–280. doi: 10.1099/13500872-140-2-271. [DOI] [PubMed] [Google Scholar]
- 4.Alfa C E, Hyams J S. Distribution of tubulin and actin through the cell division cycle of the fission yeast Schizosaccharomyces japonicus var. versitalis: comparison with Schizosaccharomyces pombe. J Cell Sci. 1990;96:71–77. doi: 10.1242/jcs.96.1.71. [DOI] [PubMed] [Google Scholar]
- 5.Arellano M, Coll P M, Perez P. RHO GTPases in the control of cell morphology, cell polarity, and actin localization in fission yeast. Microsc Res Tech. 1999;47:51–60. doi: 10.1002/(SICI)1097-0029(19991001)47:1<51::AID-JEMT5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel F J, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates; 1989. [Google Scholar]
- 7.Baggiolini B, Wymann M P. Turning on the respiratory burst. Trends Biochem Sci. 1990;15:69–72. doi: 10.1016/0968-0004(90)90179-f. [DOI] [PubMed] [Google Scholar]
- 8.Banuett F. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu Rev Genet. 1995;29:179–208. doi: 10.1146/annurev.ge.29.120195.001143. [DOI] [PubMed] [Google Scholar]
- 9.Barth G, Weber H. Improvement of sporulation in the yeast Yarrowia lipolytica. Antonie Leewenhoek J Microbiol Serol. 1986;51:167–177. doi: 10.1007/BF02310010. [DOI] [PubMed] [Google Scholar]
- 10.Bazenet C E, Mota M A, Rubin L L. The small GTP-binding protein Cdc42 is required for nerve growth factor withdrawal-induced neuronal death. Proc Natl Acad Sci USA. 1998;95:3984–3989. doi: 10.1073/pnas.95.7.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Block C, Wittinghofer A. Switching to Rac and Rho. Structure. 1995;3:1281–1284. doi: 10.1016/s0969-2126(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 12.Boguski M S, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 13.Bourne H R, Sanders D A, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 14.Bussey H. Cell shape determination: a pivotal role for Rho. Science. 1996;272:224–225. doi: 10.1126/science.272.5259.224. [DOI] [PubMed] [Google Scholar]
- 15.Chen W, Lim H H, Lim L. The CDC42 homologue from Caenorhabditis elegans. J Biol Chem. 1993;268:13280–13285. [PubMed] [Google Scholar]
- 16.Chuang T-H, Hahn K M, Lee J-D, Danley D E, Bokoch G M. The small GTPase Cdc42 initiates an apoptotic signaling pathway in Jurkat T lymphocytes. Mol Biol Cell. 1997;8:1687–1698. doi: 10.1091/mbc.8.9.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delmer D P, Pear J P, Andrawis A, Stalker D M. Genes encoding small GTP-binding proteins analogous to mammalian rac are preferentially expressed in developing cotton fibers. Mol Gen Genet. 1995;248:43–51. doi: 10.1007/BF02456612. [DOI] [PubMed] [Google Scholar]
- 18.Dever T E, Glynias M J, Merrick W C. GTP-binding domain: three consensus sequence elements with distinct spacing. Proc Natl Acad Sci USA. 1987;84:1814–1818. doi: 10.1073/pnas.84.7.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Ranea J A, Valencia A. Distribution and functional diversification of the ras superfamily in Saccharomyces cerevisiae. FEBS Lett. 1998;434:219–225. doi: 10.1016/s0014-5793(98)00967-3. [DOI] [PubMed] [Google Scholar]
- 20.Guevara-Olvera L, Calvo-Mendez C, Ruiz-Herrera J. The role of polyamine metabolism in dimorphism of Yarrowia lipolytica. J Gen Microbiol. 1993;193:485–493. doi: 10.1099/00221287-139-3-485. [DOI] [PubMed] [Google Scholar]
- 21.Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- 22.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 23.Hancock J F, Magee A I, Childs J E, Marshall C J. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 24.Harii Y, Beeler J E, Sakaguchi K, Tachibana M, Miki T. A novel oncogene, ost, encodes a guanine nucleotide exchange factor that potentially links Rho and Rac signalling pathways. EMBO J. 1994;13:4776–4786. doi: 10.1002/j.1460-2075.1994.tb06803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heath I B. Preservation of a labile cortical array of actin microfilaments in growing hyphal tips of the fungus Saprolegnia ferax. Eur J Cell Biol. 1987;44:10–16. [Google Scholar]
- 26.Hurtado C A R, Rachubinski R A. MHY1 encodes a C2H2-type zinc finger protein that promotes dimorphic transition in the yeast Yarrowia lipolytica. J Bacteriol. 1999;181:3051–3057. doi: 10.1128/jb.181.10.3051-3057.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson D I. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson D I, Pringle J R. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joneson T, Bar-Sagi D. Suppression of Ras-induced apoptosis by the Rac GTPase. Mol Cell Biol. 1999;19:5892–5901. doi: 10.1128/mcb.19.9.5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K. The small GTP-binding protein Rac is a regulator of cell death in plants. Proc Natl Acad Sci USA. 1999;96:10922–10926. doi: 10.1073/pnas.96.19.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khosravi-Far R, Chrzanowska-Wodnicka M, Solski P A, Alessandra E, Burridge K, Der C J. Dbl and Vav mediate transformation via mitogen-activated protein kinase pathways that are distinct from those activated by oncogenic Ras. Mol Cell Biol. 1994;14:6848–6857. doi: 10.1128/mcb.14.10.6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi N, McEntee K. Identification of cis and trans components of a novel heat shock stress regulatory pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:248–256. doi: 10.1128/mcb.13.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kost B, Lemichez E, Spielhofer P, Hong Y, Tolias K, Carpenter C, Chua N-H. Rac homologues and compartmentalized phosphatidylinositol 4,5-biphosphate act in a common pathway to regulate polar pollen tube growth. J Cell Biol. 1999;145:317–330. doi: 10.1083/jcb.145.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon Y H, Hoch H C, Staples R C. Cytoskeletal organization in Uromyces urediospore germling apices during appressorium formation. Protoplasma. 1991;165:37–50. [Google Scholar]
- 35.Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall J E, Thomas D Y. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R, Zheng Y, Drubin D G. Regulation of cortical actin cytoskeleton assembly during polarized cell growth in budding yeast. J Cell Biol. 1995;128:599–615. doi: 10.1083/jcb.128.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin Y, Wang Y, Zhu J-K, Yang Z. Localization of a Rho GTPase implies a role in tip growth and movement of the generative cell in pollen tubes. Plant Cell. 1996;8:293–303. doi: 10.1105/tpc.8.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lo H-S, Köhler J R, DiDomenico B, Loebenberg D, Cacciapuoti A, Fink G R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 39.Lopez M C, Nicaud J-M, Skinner H B, Vergnolle C, Kader J C, Bankaitis V A, Gaillardin C. A phosphatidylinositol/phosphatidylcholine transfer protein is required for differentiation of the dimorphic yeast Yarrowia lipolytica from the yeast to the mycelial form. J Cell Biol. 1994;124:113–117. doi: 10.1083/jcb.125.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madhani H D, Fink G R. The control of filamentous differentiation and virulence in fungi. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- 41.Maresca B, Kobayashi G S. Dimorphism in Histoplasma capsulatum: a model for the study of cell differentiation in pathogenic fungi. Microbiol Rev. 1989;53:186–209. doi: 10.1128/mr.53.2.186-209.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marks J, Hyams J S. Localization of F-actin through the cell division cycle of Schizosaccharomyces pombe. Eur J Cell Biol. 1985;39:27–32. [Google Scholar]
- 43.Marks P W, Kwiatkowski D J. Genomic organization and chromosomal location of murine Cdc42. Genomics. 1996;38:13–18. doi: 10.1006/geno.1996.0586. [DOI] [PubMed] [Google Scholar]
- 44.Miller P J, Johnson D I. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol Cell Biol. 1994;14:1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Minden A, Lin A, Claret F-X, Abo A, Karin M. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 46.Mirbod F, Nakashima S, Kitajima Y, Cannon R D, Nozawa Y. Molecular cloning of a Rho family, CDC42Ca gene from Candida albicans and its mRNA expression changes during morphogenesis. J Med Vet Mycol. 1997;135:173–179. [PubMed] [Google Scholar]
- 47.Mösch H-U, Roberts R L, Fink G R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Munemitsu S, Innis M A, Clark R, McCormick F, Ullrich A, Polakis P. Molecular cloning and expression of a G25K cDNA, the human homolog of the yeast cell cycle gene CDC42. Mol Cell Biol. 1990;10:5977–5982. doi: 10.1128/mcb.10.11.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nobes C D, Hall A. Rho, rac and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 50.Nuttley W M, Brade A M, Eitzen G A, Veenhuis M, Aitchison J D, Szilard R K, Glover J R, Rachubinski R A. PAY4, a gene required for peroxisome assembly in the yeast Yarrowia lipolytica, encodes a novel member of a family of putative ATPases. J Biol Chem. 1994;269:556–566. [PubMed] [Google Scholar]
- 51.Nuttley W M, Brade A M, Gaillardin C, Eitzen G A, Glover J R, Aitchison J D, Rachubinski R A. Rapid identification and characterization of peroxisomal assembly mutants in Yarrowia lipolytica. Yeast. 1993;9:507–517. [Google Scholar]
- 52.Odds F C. Candida and candidosis. A review and bibliography. London, United Kingdom: Bailliere Tindal; 1988. pp. 42–59. [Google Scholar]
- 53.Pertuiset B, Beckerich J-M, Gaillardin C. Molecular cloning of Rab-related genes in the yeast Yarrowia lipolytica. Analysis of RYL1, an essential gene encoding a SEC4 homologue. Curr Genet. 1995;27:123–130. doi: 10.1007/BF00313425. [DOI] [PubMed] [Google Scholar]
- 54.Peter M, Neiman A M, Park H O, van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- 55.Qiu R-G, Chen J, Kirn D, McCormick F, Symons M. An essential role for Rac in Ras transformation. Nature. 1995;374:457–459. doi: 10.1038/374457a0. [DOI] [PubMed] [Google Scholar]
- 56.Ridley A J. Membrane ruffling and signal transduction. Bioessays. 1994;16:321–327. doi: 10.1002/bies.950160506. [DOI] [PubMed] [Google Scholar]
- 57.Ridley A J. Rho-related proteins: actin cytoskeleton and cell cycle. Curr Opin Genet Dev. 1995;5:24–30. doi: 10.1016/s0959-437x(95)90049-7. [DOI] [PubMed] [Google Scholar]
- 58.Ridley A J, Comoglio P M, Hall A. Regulation of scatter factor/hepatocyte growth factor responses by Ras, Rac, and Rho in MDCK cells. Mol Cell Biol. 1995;15:1110–1122. doi: 10.1128/mcb.15.2.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ridley A J, Paterson H F, Johnston C, Diekmann D, Hall A. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 60.Roberson R W. The actin cytoskeleton in hyphal cells of Sclerotium rolfsii. Mycologia. 1992;84:41–51. [Google Scholar]
- 61.Sekine A, Fujiwara M, Narumiya S. Asparagine residue in the rho gene product is the modification site for botulinum ADP-ribosyltransferase. J Biol Chem. 1989;264:8602–8605. [PubMed] [Google Scholar]
- 62.Shepherd M G. Morphogenetic transformation in fungi. Curr Top Med Mycol. 1988;2:278–304. doi: 10.1007/978-1-4612-3730-3_8. [DOI] [PubMed] [Google Scholar]
- 63.Som T, Kolaparthi V S. Developmental decisions in Aspergillus nidulans are modulated by Ras activity. Mol Cell Biol. 1994;14:5333–5348. doi: 10.1128/mcb.14.8.5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Steer M W, Steer J M. Pollen tube tip growth. New Phytol. 1989;111:323–358. doi: 10.1111/j.1469-8137.1989.tb00697.x. [DOI] [PubMed] [Google Scholar]
- 65.Strick C A, James L C, O'Donnell M M, Gollaher M G, Franke A E. The isolation and characterization of the pyruvate kinase-encoding gene from the yeast Yarrowia lipolytica. Gene. 1992;118:65–72. doi: 10.1016/0378-1119(92)90249-o. [DOI] [PubMed] [Google Scholar]
- 66.Symons M. The Rac and Rho pathways as a source of drug targets for Ras-mediated malignancies. Curr Opin Biotechnol. 1995;6:668–674. doi: 10.1016/0958-1669(95)80110-3. [DOI] [PubMed] [Google Scholar]
- 67.Tapon N, Hall A. Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton. Curr Opin Cell Biol. 1997;9:86–92. doi: 10.1016/s0955-0674(97)80156-1. [DOI] [PubMed] [Google Scholar]
- 68.Taylor L P, Hepler P K. Pollen germination and tube growth. Annu Rev Plant Physiol. 1997;48:461–491. doi: 10.1146/annurev.arplant.48.1.461. [DOI] [PubMed] [Google Scholar]
- 69.Teem J L, Abovich N, Kaufer N F, Schwindinger W F, Warner J R, Levy A, Woolford J, Leer R J, van Raamsdonk-Duin M M C, Mager W H, Planta R J, Schultz L, Friesen J D, Fried H, Rosbach M. A comparison of yeast ribosomal protein gene DNA sequences. Nucleic Acids Res. 1984;12:8295–8312. doi: 10.1093/nar/12.22.8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Aelst L, D'Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–2322. doi: 10.1101/gad.11.18.2295. [DOI] [PubMed] [Google Scholar]
- 71.Xuan J-W, Fournier P, de Clerck N, Chasles M, Gaillardin C. Overlapping reading frames at the LYS5 locus in the yeast Yarrowia lipolytica. Mol Cell Biol. 1990;10:4795–4806. doi: 10.1128/mcb.10.9.4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yokoyama K, Kaji H, Nishimura K, Miyaji M. The role of microfilaments and microtubules in apical growth and dimorphism of Candida albicans. J Gen Microbiol. 1990;136:1067–1075. doi: 10.1099/00221287-136-6-1067. [DOI] [PubMed] [Google Scholar]
- 73.Ziman M, Preuss D, Mulholland J, O'Brien J M, Botstein D, Johnson D I. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol Biol Cell. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zohn I M, Campbell S L, Khosravi-Far R, Rossman K L, Der C J. Rho family proteins and Ras transformation: the RHOad less traveled gets congested. Oncogene. 1998;17:1415–1438. doi: 10.1038/sj.onc.1202181. [DOI] [PubMed] [Google Scholar]