Abstract
Insertion of an internal DNA fragment into the act1 gene, which encodes one of several ς54-activator proteins in Myxococcus xanthus, produced a mutant defective in fruiting body development. While fruiting-body aggregation appears normal in the mutant, it fails to sporulate (<10−6 the wild-type number of viable spores). The A and C intercellular signals, which are required for sporulation, are produced by the mutant. But, while it produces A-factor at levels as high as that of the wild type, the mutant produces much less C-signal than normal, as measured either by C-factor bioassay or by the total amount of C-factor protein detected with specific antibody. Expression of three C-factor-dependent reporters is altered in the mutant: the level of expression of Ω4414 is about 15% of normal, and Ω4459 and Ω4403 have alterations in their time course. Finally, the methylation of FrzCD protein is below normal in the mutant. It is proposed that Act1 protein responds to C-signal reception by increasing the expression of the csgA gene. This C-signal-dependent increase constitutes a positive feedback in the wild type. The act1 mutant, unable to raise the level of csgA expression, carries out only those developmental steps for which a low level of C-signaling is adequate.
Fruiting body development in Myxococcus xanthus requires the coordination of cell movement and cell differentiation. When starved for nutrients, this gram-negative bacterium initiates a developmental program involving intra- as well as extracellular signal transduction (4, 8). Early in the program, the cells produce and respond to the diffusible A-signal (21, 22). After A-signaling, approximately 105 cells actively move into aggregates that become macroscopic, mound-shaped fruiting bodies. Within a fruiting body, rod-shaped cells differentiate into spherical, environmentally resistant, dormant cells called myxospores. The C-signal, a cell surface-associated protein encoded by the csgA gene (18), is required for aggregation and for sporulation (37). The C-signal response pathway branches, with one segment controlling cell aggregation and the other controlling sporulation (38). The intensity of C-signaling rises during the course of development (18). Moreover, the expression of individual C-signal-dependent genes, the process of aggregation, and the initiation of sporulation have different C-signaling thresholds (17, 24).
Bacteria typically use several different sigma factors, and that multiplicity plays an important role in M. xanthus development. In addition to members of the ς70 family, which appear to initiate transcription of a majority of their genes and thus are vital for growth and development, another sigma factor, ς54, has also been found to be essential for M. xanthus (16). ς54 holo-RNA polymerase transcribes special sets of genes in Escherichia coli, Salmonella spp., and Klebsiella spp., which, for example, adapt them for use of nitrogen sources other than NH4+. A ς54 promoter differs from a ς70 promoter not only in sequence (1) but also in requiring a specific activator protein to work with the sigma factor in transcription initiation (32). Often these activator proteins are connected to a sensory circuit which, for example, is used for adaptation to particular sources of nitrogen in the case of NtrC (NRI) or to oxygen depletion to control NifA (23, 30). The activator, often dependent on phosphorylation, allows the ς54 holoenzyme to form an open promoter complex (32).
Four ς54 promoters have been described for M. xanthus (6, 15, 33, 43). A recent hybridization survey of whole genomes for potential ς54 activator genes (13) yielded 4 different activator clones from Bacillus subtilis, 5 from Rhizobium meliloti, none from Synechococcus sp., and 13 from M. xanthus. Taken to be whole-genome samples, these numbers as well as the unique, vital nature of ς54 may reflect an unusual importance of this sigma factor for M. xanthus. To dissect the role that ς54 promoters play in the transcriptional control of fruiting body development, potential ς54 transcriptional activator proteins derived from the Kaufman and Nixon hybridization survey have been inactivated by insertion of an internal DNA fragment into their genes. One of these insertion mutants that had developmental defects during the period of aggregation and sporulation is the subject of this report.
Cloning and sequencing of the area surrounding the chromosomal insertion has shown that the gene affected has the sequence expected of a ς54 transcriptional activator protein. This protein appears to be involved in the response pathway to C-signal, and to control the level of C-factor produced by means of a positive-feedback circuit.
MATERIALS AND METHODS
Cultures and growth conditions.
The M. xanthus strains used are listed in Table 1. They were grown in the rich Casitone-based medium CTT, as described elsewhere (7), at 32°C. When required, kanamycin was added to a final concentration of 40 μg/ml in agar and 20 μg/ml in liquid. Oxytetracycline was added stepwise, first at 2.5 μg/ml to induce resistance and then at 12.5 μg/ml for selection. To enumerate oxytetracycline-resistant colonies, cells were plated onto media containing 2.5 μg of the drug/ml overnight, then overlaid with soft agar containing enough oxytetracycline to bring the final total-plate concentration to 12.5 μg/ml. To assess development, M. xanthus strains were spotted onto nonnutrient TPM agar (7). Plasmids used are also listed in Table 1. The growth of M. xanthus in liquid medium was monitored by measuring culture turbidity in a Klett-Summerson photoelectric colorimeter equipped with a red filter and was reported in Klett units.
TABLE 1.
M. xanthus strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| DK 1622 | Wild type | 10 |
| DK 7837 | pLAG2::act1 | 7 |
| DK 7853 | asgA476; Tn5-132 Ω4560 | 7 |
| DK 5208 | Tn5-132::csgA | 21 |
| DK 7827 | Tn5-132lac Ω4403 | Mx4 transduction→DK 1622 |
| DK 7828 | Tn5-132lac Ω4459 | Mx4 transduction→DK 1622 |
| DK 7160 | Tn5-132lac Ω4000 | Yvonne Cheng, Stanford University |
| DK 5511 | Tn5-132lac Ω4414 (devRS) | 40 |
| DK 7848 | DK 7837; Tn5-132lac Ω4403 | Mx4 transduction→DK 7837 |
| DK 7849 | DK 7837; Tn5-132lac Ω4459 | Mx4 transduction→DK 7837 |
| DK 7852 | DK 7837; Tn5-132lac Ω4400 | φMx4 (DK 7160)→DK 7837 |
| DK 7856 | DK 7837; Tn5-132lac Ω4414 (devRS) | φMx8 (DK 5511)→DK 7837 |
| DK 7873 | Tn5-132lac Ω4491 (fruA) | φP1::Tn5-132→DK 5285 |
| DK 7880 | DK 7837; Tn5-132lac Ω4491 (fruA) | φMx8 (DK 7873)→DK 7837 |
| DK 5285 | Tn5lac Ω4491 (fruA) | 38 |
| Plasmids | ||
| pLAG2 | Kmr (pBGS 18) Mxa259 at EcoRI site | 7 |
| pLAG53 | Clone of the region around the DK 7837 insertion with NotI; Kmr | Fig. 1 |
| pLAG60 | SalI cut and religation of pLAG53 | Fig. 1 |
| pLAG61 | Clone of the region upstream of the DK 7837 insertion with NdeI; Kmr | Fig. 1 |
| pLAG64 | 4.7-kb ClaI fragment from pLAG61 inserted into ClaI site of pBluescript | Fig. 1 |
| pLAG65 | EcoRI cut and religation of pLAG64 | Fig. 1 |
| pLAG66 | 3.7-kb ClaI-EcoRI fragment from pLAG65 blunted and inserted into EcoRV of pBluescript | Fig. 1 |
| pLAG121 | 4.7-kb EcoRI-SalI fragment from pLAG60 inserted into similarly cut pBluescript | Fig. 1 |
| pBGS18 | Kmr | 39 |
| pBluescript | Apr | Stratagene |
Cloning of act1.
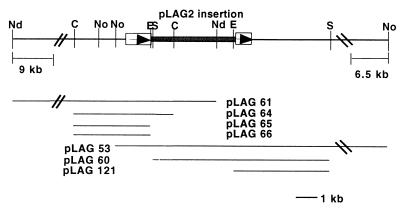
The original Mxa259 mutant had been generated by insertion of pLAG2 into DK 1622 to create strain DK 7837 (act1) (7). To isolate the chromosomal DNA surrounding the act1 insertion, an in situ cloning technique was used (Fig. 1). DNA from DK 7837 was restricted with NotI (for DNA downstream of the insertion) or NdeI (for DNA upstream) and religated. E. coli strain DH10B was transformed with the ligation products by electroporation and then plated onto Luria-Bertani (LB) agar with kanamycin. Plasmid DNA was isolated and digested with the appropriate enzyme to confirm the content of the clones. pLAG53 contains approximately 14 kb of M. xanthus DNA, most of it downstream of the insertion into act1; pLAG61 also contains approximately 14 kb of DNA, but all of it is upstream of the act1 insertion (Fig. 1). Subsequently, both pLAG53 and pLAG61 were subcloned as diagrammed in Fig. 1 and explained in Table 1. pLAG121 carries 4.5 kb of DK 7837 DNA downstream of the plasmid insertion. pLAG66 carries 3 kb of DNA upstream of the original plasmid insertion. Plasmid manipulation and DNA isolation were performed using standard procedures (35).
FIG. 1.
Physical and restriction map of the act1 region in strain DK7837. Open boxes, the act1 coding region. The act1 gene is interrupted by the pLAG2 plasmid insertion (shaded box). Arrows indicate the predicted direction of transcription. The region was cloned in upstream and downstream segments, with the relevant subclones displayed in their pLAG vectors. Plasmids pLAG66 and pLAG121 were used for sequencing. Restriction sites: Nd, NdeI; C, ClaI; No, NotI; E, EcoRI; S, SalI.
Sequencing of act1.
Sequencing was carried out by standard methods using the ABI Prism model 373A at the Stanford University Protein and Nucleic Acid Facility. Combinations of ExoIII deletions and primer walking were used to sequence pLAG66 and pLAG121 to obtain the sequence of act1.
Sporulation assays.
M. xanthus cells (5 10-μl drops at a cell density of Klett 1,000, or 5 × 109 cells/ml) were deposited onto TPM agar, allowed to dry, and then incubated for 3 days at 32°C. The spots were harvested by scraping with a spatula into 1 ml of TPM buffer, sonicated, and heated at 50°C as described elsewhere to disrupt fruiting bodies and kill residual vegetative cells (7). After serial dilution and plating of the spore suspensions, the sporulation efficiencies were calculated as the number of colonies that arose relative to the original number of cells spotted. Efficiencies were compared with those of wild-type controls in each experiment.
To determine the amount of A- and C-signals made by the act1 mutant, the strain in question was developed on TPM agar in coculture with either DK 7853 (asgA) or DK 5208 (csgA). For these assays, cells at a concentration of Klett 1,000 were mixed in a 1:1 ratio, and 5 20-μl drops were placed on TPM agar and then incubated and treated as described above to determine sporulation efficiencies for each strain. For these experiments, the amount of A- or C-factor produced by the tester strain was measured by the sporulation efficiency of the asgA or csgA strain. The amount of A- or C-factor produced was calculated as a percentage of the wild-type (DK 1622) production of those factors.
Developmental β-galactosidase assays.
A series of Tn5lac promoter fusions have been described previously (21) and are included in Table 1. To measure promoter expression in terms of β-galactosidase produced by these and mutant derivatives of these strains, cultures were allowed to develop either on TPM plates or in submerged culture, harvested, and extracted as previously described (12). For development in submerged culture, cells were starved in 24-well polystyrene microtiter plates (7).
Western blotting.
Standardized Western blot hybridization was used to monitor both the methylation state of the FrzCD protein with an anti-FrzCD antibody and the level of CsgA protein with an anti-CsgA antibody. Cells were allowed to develop in submerged culture in A50 buffer as described elsewhere for 0, 6, and 12 h and then were harvested and frozen (37). Cell pellets were resuspended in 50 μl of sodium dodecyl sulfate (SDS) sample buffer, and protein from 5 × 107 input cells was analyzed. Standard SDS-polyacrylamide gel electrophoresis (PAGE) conditions (35) were used to reveal the CsgA protein, and modified (26, 27) conditions were used to resolve the methylated from the nonmethylated FrzCD. The secondary antibody was conjugated to horseradish peroxidase for chemiluminescence.
Introduction of reporter gene fusions and other mutations into the act1 mutant.
Myxophages Mx4 (3) and Mx8 (25) were used to transduce Tn5-132lacZ promoter fusions and other mutant alleles into the act1 strains to create double mutants for epistatic analysis. The structure of all chromosomal insertions was confirmed by Southern blot hybridization.
Nucleotide sequence accession number.
The sequence of act1 has been assigned GenBank accession no. AF230804.
RESULTS
The act1 gene.
A severe developmental defect was produced in strain DK 7837 when plasmid pLAG2 was inserted into the M. xanthus genome (7). This plasmid carried a 475-bp PCR fragment of a sequence potentially encoding a ς54 activator protein and was one of 14 such fragments from M. xanthus (7, 13). Drug resistance on the plasmid facilitated the cloning of those segments of M. xanthus DNA immediately to the left and right of the plasmid insertion point. Sequence similarity searches revealed that pLAG2 had inserted within an open reading frame homologous to the well-studied ς54 transcriptional activator protein NtrC. An alignment of the proposed protein sequences of activator 1 with those of NtrC and PilR, a ς54 transcriptional activator of pilin synthesis in M. xanthus (43), is shown in Fig. 2. Since the new gene has the full sequence of a ς54 transcriptional activator, it will be called act1, replacing the temporary designation Mxa259, which refers to the fragment used to target it (7, 13).
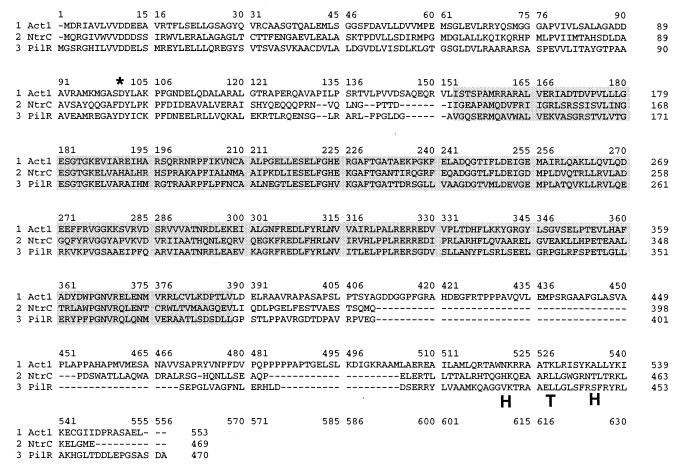
FIG. 2.
Amino acid sequence of Act1 deduced from its DNA sequence shown in an alignment with NtrC from E. coli (28) (GenBank accession no. P06713) and PilR from M. xanthus (42) (GenBank accession no. L39904). The highly conserved central domain in ς54 activator proteins is shaded. The aspartate residue that is phosphorylated in NtrC is marked with an asterisk. The area containing the DNA binding domain of NtrC is indicated as HTH (for helix-turn-helix). A second (potential) ATG start codon is located 27 bases upstream and in frame with the start codon shown in this figure. No data are available to distinguish which translation start site is used in vivo.
Both Act1 and PilR share the domains and subdomains identified in an earlier survey of ς54 transcriptional activator proteins (29). These structures include an N-terminal region where typically these activator proteins are modified by phosphorylation of an aspartate residue, a highly conserved central region of several hundred amino acids containing an ATP-binding motif, and a C-terminal region that contains a helix-turn-helix motif near its end (32). Both Act1 and PilR have an aspartate residue (marked in Fig. 2) in the N-terminal domain that aligns with the aspartate residue in NtrC, which is phosphorylated by NtrB (14).
Despite their similarities, Act1 and PilR are unique proteins. Their protein sequences are predicted to be 47% identical and 58% similar in their central ATP-binding domains. pilR encodes a soluble 51-kDa protein of 470 amino acids; act1 is predicted to encode a soluble 60-kDa protein with 553 amino acids. Guided by the Morett and Segovia comparisons (29), the N-terminal domain of PilR is predicted to be 143 amino acids, its central domain 236 residues, and its C-terminal domain 91 amino acids. On the same basis, Act1 would have an N-terminal domain of 151 amino acids, a central domain of 236 amino acids, and a C-terminal domain of 166 amino acids. Comparison of the three sequences reveals that act1 encodes a stretch of more than 50 amino acids, starting with its residue 411, for which there is no homologous stretch in either NtrC or PilR.
The DNA sequence of the act1 central domain proved to be identical to that of the PCR fragment, Mxa259, used for insertional mutagenesis. Evidently, Mxa259 had inserted into the identical gene, despite the presence of several other genes belonging to the same family of activators in M. xanthus (7, 13). In other words, the DNA sequence of the insertion mutant strain DK 7837 confirms that precise, homologous integration had occurred.
Fruiting body development.
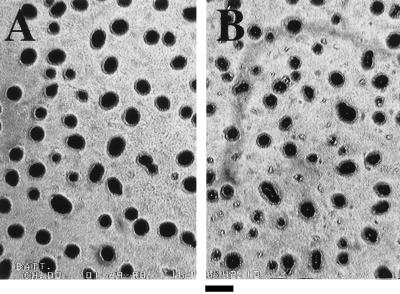
The act1 mutant fails to sporulate: fewer than 10−6 the wild-type number of heat- and sonication-resistant spores are formed, and no colonies were found on the spore assay plates in this experiment, in our previous work (7), or in any of the subsequent repeat experiments. Nevertheless, the mutant forms the same number of mounded aggregates having the same range of sizes as the wild-type fruiting bodies (Fig. 3). It should be emphasized that the same number of mutant and wild-type cells were plated for the experiments reported in Fig. 3. Despite their lack of spores, the aggregates formed by the act1 mutant are darkened like those in the wild type. Frequently, at 24 h, they appear to have a small white dot in the center.
FIG. 3.
Aggregation phenotypes of the wild-type strain DK 1622 (A) and the act1 mutant DK 7837 (B). Photographs of fruiting bodies were taken at 48 h of development on TPM agar. Bar, 0.2 mm.
To be certain that the plasmid insertion in act1 is responsible for all the developmental defects, the original insertion was outcrossed by transduction into a wild-type strain (DK 1622) to create DK 7837 by selection for kanamycin resistance. That drug resistance is carried by the plasmid which inserted within the act1 gene. Resembling the original Mxa259 insertion strain, the backcrossed strain has the same aggregate morphology and the same failure to sporulate. This confirms that act1 is essential for development. Because the insertion mutation splits the act1 gene, strain DK 7837 should be a null mutant.
A- and C-signal production.
Aggregation and sporulation require both extracellular signals A and C (21, 22). The capacity of the act1 mutant to produce the extracellular A- and C-factors was assessed by bioassay. Wild-type cells which produce these factors can rescue, by codevelopment, the sporulation defect of an asgA or a csgA strain, which fails to produce its own A-factor or C-factor, respectively. The results of a bioassay (Table 2) show that the act1 mutant is able to make A-factor, since it induces the A-signal-defective mutant to sporulate, at least to wild-type levels. However, the activator (act1) mutant appears to make no more than a fraction of the normal amount of C-factor. Moreover, the sporulation defect in the act1 mutant cannot be rescued by coculture with wild-type cells, showing that the act1 mutant is unable to respond properly to C-signaling from wild-type cells. If the act1 mutant produces less than the normal level of C-factor, this might also be reflected in the expression levels of C-signal-dependent genes.
TABLE 2.
A- and C-factor production as assessed by rescue
| Strain mixture
|
Sporulation frequency (%)
|
||
|---|---|---|---|
| Test strain | Coculture strain | Test strain | Coculture strain |
| act1 | DK 1622 | <10−4a | 100b |
| DK 7853 (asgA) | DK 1622 | 100c | 100 |
| DK 7853 | act1 | 285 ± 165c | <10−4a |
| DK 5208 (csgA) | DK 1622 | 100c | 100 |
| DK 5208 | act1 | 1.5 ± 1.2c | <10−4a |
The lower limit of the assay is <10−4%. No spores were evident in any of several experiments.
The frequency was normalized to that of the wild-type (DK 1622) control in each experiment.
Taking wild-type rescue of the mutant as 100%, this frequency has been normalized to the amount of rescue the wild-type strain would confer under the same conditions.
Expression of signal-dependent Tn5lac promoter fusions.
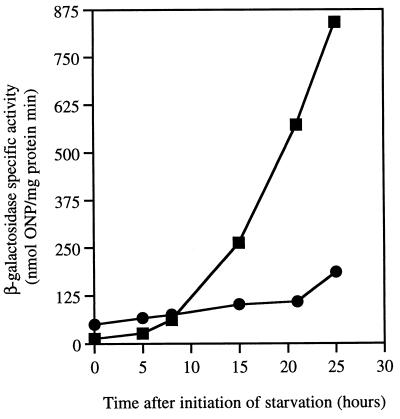
Expression of several different C-signal-dependent genes can be monitored by existing Tn5lac promoter fusions (11, 21). Expression of the dev operon, which is C-signal dependent (21) and can be measured by the Ω4414 reporter (40), is shown in Fig. 4. Expression from the Ω4414 insertion is highly defective in the act1 mutant. In the wild type, dev expression begins to rise at 7 to 8 h. Assuming that the slight rise in the act1 mutant at 25 h is significant, expression is delayed from the normal 7 to 8 h to beyond 21 h, and even then it is less than 15% of that of the wild type.
FIG. 4.
Expression of the C-signal-dependent dev operon, measured by the level of β-galactosidase from the Tn5lac fusion Ω4414. For the 0-h sample, cells were taken immediately after transfer from growth medium into starvation buffer and before incubation to start development. Squares, expression in a wild-type background; circles, fusion Ω4414 in an act1 mutant background.
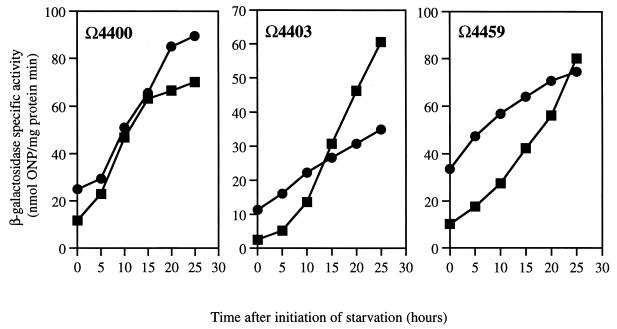
An act1 defect is also evident in the expression pattern of the C-signal-dependent fusions Ω4403 and Ω4459. Their expression approaches, but differs significantly from, that of the wild type (Fig. 5). In the act1 mutant, Ω4459 expression is significantly higher than that in the wild type at all times prior to 25 h. Differences between the mutant and the wild type may not be significant for Ω4400 and may indicate that the basal level of C-factor expression in the act1 mutant is sufficient for full expression of Ω4400. The facts that the act1 mutant fails to sporulate and that three different C-signal-dependent reporters are altered either in the ultimate level (Ω4414) or in the time course (Ω4403 and Ω4459) implies that the act1 mutant gives an aberrant C-signal response. This view is further supported by the methylation of FrzCD protein.
FIG. 5.
Expression of the C-signal-dependent Tn5lac promoter fusions Ω4400, Ω4403, and Ω4459. Squares, activity in a wild-type background; circles, activity in an act1 mutant background.
FrzCD methylation.
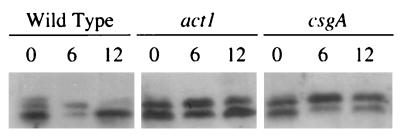
Mutants defective in a Frz protein aggregate in an unusual way (2). The seven proteins encoded by the frz locus form a phosphorelay, whose activity regulates cell movement behavior, including the average frequency of reversal (2, 41). During development, this phosphorelay modulates movement in response to C-signaling (37). The FrzCD protein, which resembles the cytoplasmic domain of the methyl-accepting chemotaxis proteins of E. coli and Salmonella, becomes methylated and demethylated as fruiting body aggregates form (27). These changes in the state of the protein are starvation and C-signal dependent. The effect of act1 on the methylation of FrzCD is shown in Fig. 6, where each lane shows two bands, often of unequal intensities. The methylated and nonmethylated forms have different electrophoretic mobilities: the lower of the two FrzCD bands is methylated, and the upper band is nonmethylated.
FIG. 6.
FrzCD methylation during development depends on act1 and csgA. Cells were developed and lysed, and their proteins were separated by electrophoresis. The Western blots were probed with anti-FrzCD antiserum. Each lane contains protein from 5 × 107 cells. Separated this way, the upper band is unmethylated FrzCD, and the lower band is methylated FrzCD.
A large fraction of the FrzCD of the wild-type cells (at 0 h [Fig. 6]), which had been growing in rich medium before transfer to starvation buffer, is methylated, as previously observed (27, 37). Within the first 6 h of starvation-induced development, FrzCD loses methyl groups, until the majority becomes demethylated. Then, by 12 h, FrzCD is almost fully remethylated. It has been shown that the early (<6-h) demethylation is induced by starvation, while the later remethylation (12 h) depends on C-signaling (37). While the wild-type strain shows almost all of its FrzCD methylated by 12 h into development, the act1 mutant remethylates only part of the FrzCD, leaving almost half in the nonmethylated form. In its failure to remethylate, the act1 mutant resembles the csgA mutant, which produces no C-factor (Fig. 6). The failure to remethylate FrzCD fully can be rescued by adding purified C-factor to the csgA mutant cells, demonstrating that remethylation depends on C-signaling (37). The failure of the act1 mutant to fully remethylate its FrzCD protein suggests that it suffers from a deficiency of C-factor. The act1 mutant remethylates a greater proportion of its FrzCD protein by 12 h than the csgA mutant, consistent with the presence of some C-factor in the act1 strain. Complicating the picture, the act1 mutant has more total FrzCD protein (methylated plus nonmethylated) at both 6 and 12 h of development than the wild type, to judge from the intensities of the bands. As a consequence of the larger amount of FrzCD protein in the act1 mutant, the absolute amounts of methylated FrzCD protein in the act1 mutant and the wild type do not seem to differ. However, the ratios of methylated to nonmethylated protein at 12 h do differ: the ratio is very much greater than 1 in the wild type, close to 1 in the act1 strain, and less than 1 in the csgA strain (Fig. 6). In terms of the ratio, the act1 mutant falls between the wild type and the csgA mutant, as if the act1 defect had decreased the level of C-signaling.
Expression of csgA.
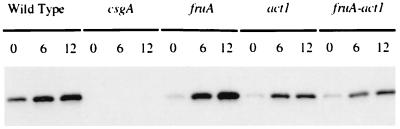
To evaluate more directly the total level of csgA protein, extracts of developing cells were fractionated by gel electrophoresis and proteins in the gel were reacted with an anti-CsgA antibody (Fig. 7). The specificity of the antibody used is confirmed by the absence of any CsgA band reaction in the csgA null mutant extract and the presence of a band in all csgA+ strains. In both the single act1 mutant and the double act1 fruA mutant, the intensity of the CsgA band is lower than in the wild type at each time point, and it remains at its 6-h level at 12 h, indicating that C-factor protein is produced but that its level is significantly lower at 12 h as a consequence of the insertion mutation in act1 (DK 7837). Ellehauge et al. have shown that FruA lies in the C-signal response circuit after signal reception and before the branch that separates aggregation from sporulation (5). Since the fruA mutant produces the normal, high levels of CsgA, it is apparent that act1 but not fruA controls the level of C-factor and that the effect of act1 is epistatic to that of fruA.
FIG. 7.
Amount of CsgA protein during development of the wild type, three single mutants, and one double mutant. After development, the cells were extracted, and their proteins were separated by electrophoresis, before blotting. The Western blot was probed with anti-CsgA antiserum. Each lane contains protein from 5 × 107 cells.
DISCUSSION
The act1 mutant produces some C-factor protein, which is evident in the Western blot of Fig. 7. Moreover, this protein is biologically active; it increases the sporulation frequency of the csgA mutant at least 10,000-fold (Table 2). A csgA mutant is unable to sporulate because C-signaling is essential for sporulation (21, 24, 40). Addition of partially purified C-factor to the csgA mutant rescues its capacity to sporulate (18).
It is also clear from several observations that the act1 mutant produces substantially less C-factor than wild-type cells. First, the direct assay of CsgA protein with specific antibody shows a reduction in amount. While the wild type increases the CsgA protein level from 6 to 12 h, act1 continues the 6-h level through 12 h (Fig. 7). Second, the act1 mutant only partially rescues the sporulation defect of a csgA mutant when the two strains are mixed 1:1 and allowed to develop together. The sporulation rescued by the act1 mutant is 70-fold less than the sporulation rescued by admixed wild-type cells under the same conditions (Table 2). Third, expression of the C-signal-dependent reporter Ω4414 is greatly lowered in an act1 mutant.
The consistently higher expression of Ω4403 and Ω4459 in the act1 mutant than in the wild type at early times, including vegetative cells (time zero), was unexpected. It might be due to an inhibitory action of the Act1 protein on these genes in growing cells.
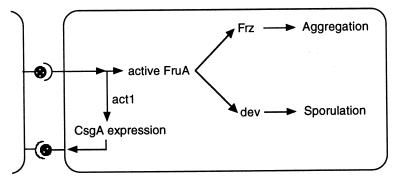
The C-signal response pathway is branched, with one branch leading to aggregation and the other to sporulation (38). The FruA response regulator (31), which receives C-signal input, occupies the branch point (38). On the one hand, activated FruA sends a signal through the Frz phosphorelay that changes cell movement behavior and that causes the cells to aggregate (9). frz gene null mutants are aggregation defective but are still able to sporulate efficiently (37, 44). On the other hand, activated FruA initiates expression of the dev operon. Dev mutants can aggregate but are sporulation defective (40). Dev operon expression depends on both C-signal and FruA, and that expression in turn is believed to initiate sporulation. The third response of the C-signaling pathway is augmentation of csgA expression. Since a defect in act1 decreases the expression of dev (Fig. 4) and diminishes the signal through FrzCD (Fig. 6), the defect must precede the separation between aggregation and sporulation. Because the act1 fruA double mutant makes the same low level of CsgA protein as an act1 single mutant, act1 must be upstream of the activation of FruA in the C-signal response pathway. Finally, since a fruA mutant has a severe aggregation defect compared to the near-normal aggregation in the act1 mutant, act1 is apparently not on the line leading to fruA. These observations and arguments are embodied in the C-signaling response circuit shown in Fig. 8.
FIG. 8.
C-signal response pathway, including the proposed act1-dependent step, which increases the expression of csgA. Evidence for this and the other steps is detailed in the text. CsgA protein on the surfaces of the donor cells is represented as filled lollipops projecting from the ends of both cells. The sensors, not yet identified, that transduce C-signal to the responding cell are represented as cups that engage CsgA and carry the signal to FruA and Act1.
Starting from a low level at 6 h, near the beginning of development, expression of the csgA gene is found to rise during the aggregation phase of development (17). This rise can be explained by the process of C-signaling itself, as indicated in Fig. 8. First, either partially purified C-factor or wild-type cells presenting C-factor have been shown to induce a rise in csgA expression monitored with a lacZ transcriptional fusion (17). Second, C-factor is not released to the medium but is located on the cell surface (20, 36). It has been experimentally verified that C-signaling requires contact between cells (19). Aggregation by increasing the local cell density would be expected to increase the frequency of contacts between cells that continue to move within the nascent aggregate (34). Together, these conditions bring about positive feedback, which might explain the developmental rise in csgA expression in wild-type cells that is evident in Fig. 7. We propose that activator 1, encoded by the act1 gene, is an essential component of the positive-feedback loop.
The feedback circuit would operate as follows. After an initial developmental phase of starvation evaluation, the cells proceed to aggregate. Aggregation requires a higher level of C-signaling than the prior phase (17). Sporulation requires a still-higher level (17, 24). For these reasons, the mounting levels of C-signaling are responsible for a natural progression from nutrient evaluation to aggregation, and finally to sporulation. The process of aggregation in act1 mutants is normal, judging by the number and gross morphology of the fruiting body aggregates that it forms (Fig. 3). The failure of an act1 mutant to form spores implies that its low level of C-signaling, while sufficient for aggregation, is not sufficient to initiate expression of dev (Ω4414 [Fig. 4]), which is needed to induce sporulation.
It is not obvious how act1 might control the expression of the csgA gene, since Li et al. (24) have suggested that the promoter upstream of the csgA gene is of the ς70 type. Nevertheless, those authors also reported that as many as 930 bases upstream of the csgA transcriptional start site are needed for development and maximal csgA transcription, suggesting that there are additional regulatory factors. The act1 transcriptional activator may have a direct or indirect action on that upstream region to augment expression of the csgA gene during development. Although act1 mutant cells show less than 10−4% sporulation when mixed with wild-type cells (Table 2), csgA mutant cells show 1.5% sporulation when mixed with act1 mutant cells. The much-lower efficiency of sporulation in act1 mutant cells may indicate that activator 1 is needed not only for regulating the intensity of C-signaling but for another sporulation function as well.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant GM23441 from the National Institute of General Medical Sciences to D.K. and postdoctoral fellowship GM16344 to L.G. from the National Institute of General Medical Sciences.
We are grateful to Lotte Sogaard-Andersen for antibody to CsgA protein and to David Zusman for antibody to FrzCD protein.
REFERENCES
- 1.Barrios H, Valderrama B, Morett E. Compilation and analysis of ς54-dependent promoter sequences. Nucleic Acids Res. 1999;27:4305–4313. doi: 10.1093/nar/27.22.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackhart B D, Zusman D. The frizzy genes of Myxococcus xanthus control directional movement of gliding motility. Proc Natl Acad Sci USA. 1985;82:8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campos J, Geisselsoder J, Zusman D. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978;119:167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- 4.Dworkin M. Recent advances in the social and developmental biology of the Myxobacteria. Microbiol Rev. 1996;60:70–102. doi: 10.1128/mr.60.1.70-102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ellehauge E, Norregaard-Madsen M, Sogaard-Andersen L. The FruA signal transduction protein provides a checkpoint for the temporal coordination of intercellular signals in M. xanthus development. Mol Microbiol. 1998;30:807–813. doi: 10.1046/j.1365-2958.1998.01113.x. [DOI] [PubMed] [Google Scholar]
- 6.Garza A, Pollack J S, Harris B Z, Lee A, Keseler I M, Licking E F, Singer M. SdeK is required for early fruiting body development in Myxococcus xanthus. J Bacteriol. 1998;180:4628–4637. doi: 10.1128/jb.180.17.4628-4637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorski L, Kaiser D. Targeted mutagenesis of ς54 activator proteins in Myxococcus xanthus. J Bacteriol. 1998;180:5896–5905. doi: 10.1128/jb.180.22.5896-5905.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris B Z, Kaiser D, Singer M. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 1998;12:1022–1035. doi: 10.1101/gad.12.7.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jelsbak L, Sogaard-Andersen L. The cell surface-associated C-signal induces behavioral changes in individual M. xanthus cells during fruiting body morphogenesis. Proc Natl Acad Sci USA. 1999;96:5031–5036. doi: 10.1073/pnas.96.9.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaiser D, Kroos L. Intercellular signaling. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C.: American Society for Microbiology; 1993. pp. 257–283. [Google Scholar]
- 12.Kaplan H B, Kuspa A, Kaiser D. Suppressors that permit A-signal-independent developmental gene expression in Myxococcus xanthus. J Bacteriol. 1991;173:1460–1470. doi: 10.1128/jb.173.4.1460-1470.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaufman R I, Nixon B T. Use of PCR to isolate genes encoding ς54-dependent activators from diverse bacteria. J Bacteriol. 1996;178:3967–3970. doi: 10.1128/jb.178.13.3967-3970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keener J, Kustu S. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc Natl Acad Sci USA. 1988;85:4976–4980. doi: 10.1073/pnas.85.14.4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keseler I M, Kaiser D. An early A-signal-dependent gene in Myxococcus xanthus has a ς54-like promoter. J Bacteriol. 1995;177:4638–4644. doi: 10.1128/jb.177.16.4638-4644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keseler I M, Kaiser D. Sigma-54, a vital protein for Myxococcus xanthus. Proc Natl Acad Sci USA. 1997;94:1979–1984. doi: 10.1073/pnas.94.5.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S K, Kaiser D. C-factor has distinct aggregation and sporulation thresholds during Myxococcus development. J Bacteriol. 1991;173:1722–1728. doi: 10.1128/jb.173.5.1722-1728.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S K, Kaiser D. C-factor: a cell-cell signalling protein required for fruiting body morphogenesis of M. xanthus. Cell. 1990;61:19–26. doi: 10.1016/0092-8674(90)90211-v. [DOI] [PubMed] [Google Scholar]
- 19.Kim S K, Kaiser D. Cell alignment required in differentiation of Myxococcus xanthus. Science. 1990;249:926–928. doi: 10.1126/science.2118274. [DOI] [PubMed] [Google Scholar]
- 20.Kim S K, Kaiser D. Purification and properties of Myxococcus xanthus C-factor, an intercellular signaling protein. Proc Natl Acad Sci USA. 1990;87:3635–3639. doi: 10.1073/pnas.87.10.3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kroos L, Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987;1:840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- 22.Kuspa A, Kroos L, Kaiser D. Intercellular signaling is required for developmental gene expression in Myxococcus xanthus. Dev Biol. 1986;117:267–276. doi: 10.1016/0012-1606(86)90369-6. [DOI] [PubMed] [Google Scholar]
- 23.Kustu S, Santero E, Keener J, Popham D, Weiss D. Expression of ς54 (ntrA)-dependent genes is probably united by a common mechanism. Microbiol Rev. 1989;53:367–376. doi: 10.1128/mr.53.3.367-376.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Lee B U, Shimkets L. csgA expression entrains Myxococcus xanthus development. Genes Dev. 1992;6:401–410. doi: 10.1101/gad.6.3.401. [DOI] [PubMed] [Google Scholar]
- 25.Martin S, Sodergren E, Masuda T, Kaiser D. Systematic isolation of transducing phages for Myxococcus xanthus. Virology. 1978;88:44–53. doi: 10.1016/0042-6822(78)90108-3. [DOI] [PubMed] [Google Scholar]
- 26.McBride M, Kohler T, Zusman D. Methylation of FrzCD, a methyl-accepting taxis protein of Myxococcus xanthus, is correlated with factors affecting cell behavior. J Bacteriol. 1992;174:4246–4257. doi: 10.1128/jb.174.13.4246-4257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCleary W R, McBride M J, Zusman D R. Developmental sensory transduction in Myxococcus xanthus involves methylation and demethylation of FrzCD. J Bacteriol. 1990;172:4877–4887. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miranda-Rios J, Sanchez-Pescador R, Urdea M, Covarrubias A A. The complete sequence of the glnALG operon of Escherichia coli K-12. Nucleic Acids Res. 1987;15:2757–2770. doi: 10.1093/nar/15.6.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morett E, Segovia L. The ς54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ninfa A J, Atkinson M R, Kamberov E S, Feng J, Ninfa E G. Control of nitrogen assimilation by the NRI-NRII two-component system of enteric bacteria. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. pp. 67–88. [Google Scholar]
- 31.Ogawa M, Fujitani S, Mao X, Inouye S, Komano T. FruA, a putative transcription factor essential for the development of Myxococcus xanthus. Mol Microbiol. 1996;22:757–767. doi: 10.1046/j.1365-2958.1996.d01-1725.x. [DOI] [PubMed] [Google Scholar]
- 32.Rombel I, North A, Hwang I, Wyman C, Kustu S. The bacterial enhancer-binding protein NtrC as a molecular machine. Cold Spring Harbor Symp Quant Biol. 1998;63:157–166. doi: 10.1101/sqb.1998.63.157. [DOI] [PubMed] [Google Scholar]
- 33.Romeo J M, Zusman D R. Transcription of the myxobacterial hemagglutinin gene is mediated by a ς54-like promoter and a cis-acting upstream regulatory region of DNA. J Bacteriol. 1991;173:2969–2976. doi: 10.1128/jb.173.9.2969-2976.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sager B, Kaiser D. Two cell-density domains within the Myxococcus xanthus fruiting body. Proc Natl Acad Sci USA. 1993;90:3690–3694. doi: 10.1073/pnas.90.8.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Shimkets L J, Rafiee H. CsgA, an extracellular protein essential for Myxococcus xanthus development. J Bacteriol. 1990;172:5299–5306. doi: 10.1128/jb.172.9.5299-5306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Søgaard-Andersen L, Kaiser D. C-factor, a cell-surface-associated intercellular signaling protein, stimulates the cytoplasmic Frz signal transduction system in Myxococcus xanthus. Proc Natl Acad Sci USA. 1996;93:2675–2679. doi: 10.1073/pnas.93.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Søgaard-Andersen L, Slack F, Kimsey H, Kaiser D. Intercellular C-signaling in Myxococcus xanthus involves a branched signal transduction pathway. Genes Dev. 1996;10:740–754. doi: 10.1101/gad.10.6.740. [DOI] [PubMed] [Google Scholar]
- 39.Spratt B G, Hedge P J, te Heesen S, Edelman A, Broome-Smith J K. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8, and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 40.Thony-Meyer L, Kaiser D. devRS, an autoregulated and essential genetic locus for fruiting body development in Myxococcus xanthus. J Bacteriol. 1993;175:7450–7462. doi: 10.1128/jb.175.22.7450-7462.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward M J, Zusman D R. Regulation of directed motility in Myxococcus xanthus. Mol Microbiol. 1997;24:885–893. doi: 10.1046/j.1365-2958.1997.4261783.x. [DOI] [PubMed] [Google Scholar]
- 42.Wu S S, Kaiser D. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 43.Wu S S, Kaiser D. Regulation of expression of the pilA gene in Myxococcus xanthus. J Bacteriol. 1997;179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zusman D. Frizzy mutants: a new class of aggregation-defective developmental mutants of Myxococcus xanthus. J Bacteriol. 1982;150:1430–1437. doi: 10.1128/jb.150.3.1430-1437.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]