Abstract
Background
MRT4 Homolog, Ribosome Maturation Factor (MRTO4) is often upregulated in cancer cells. However, its impact in hepatocellular carcinoma (HCC) is less well understood. Herein, we explored the prognostic and energy metabolism reprogramming role of MRTO4 in HCC.
Material/Methods
Clinical data were obtained from The Cancer Genome Atlas (TCGA), and the expression of MRTO4 in clinical samples was analyzed. The association between different variables and overall survival (OS) was studied, as well as their potential as independent prognostic factors, using Cox regression analysis. We constructed a nomogram including clinical pathological variables and MRTO4 expression to provide a predictive model for prognosis. Heatmaps, Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed the relationship between energy metabolism pathways and MRTO4. We used classic molecular biology research methods, including RT-qPCR, Western blotting, CCK8, TUNEL, Clone formation, Transwell assay, ELISA, and immunohistochemistry, to study the role of MRTO4 in promoting the progression of HCC through glycolysis regulation.
Results
Our study showed that MRTO4 is an independent prognostic risk factor for HCC and that MRTO4 accelerates glycolysis of HCC cells, promotes proliferation and invasion, and suppresses apoptosis of HCC cells. The underlying mechanism involves MRTO4 promoting glycolysis and accelerating HCC by inhibiting ALDOB.
Conclusions
Our study revealed a novel mechanism by which MRTO4 promotes glycolysis and accelerates HCC progression, and suggests that inhibiting MRTO4 could be a potential therapeutic strategy for HCC.
Keywords: Glycolysis, Liver Neoplasms, Cell Proliferation
Introduction
Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, ranking in the top 3 for cancer-related mortality [1]. There were 906 000 new cases of HCC and 830 000 deaths worldwide in 2020, with confirmed cases and mortality rates increasing year by year [1]. The high mortality rate of HCC is largely attributed to delayed diagnosis and limited treatment options [2,3]. Despite the possibility of aggressive treatments such as tumor resection, chemotherapy, and external beam radiation therapy, HCC patients currently face a poor prognosis [4]. Thus, exploring molecular biomarkers and their mechanisms in HCC is of great significance in guiding clinical treatment and improving long-term survival rates of patients.
Research has found that various characteristic metabolic changes, inflammation, immune dysregulation, and fibrosis are closely associated with the progression of HCC [5,6]. Energy metabolism reprogramming is considered one of the major hallmarks of cancer [7]. Adequate energy and biosynthetic intermediates are prerequisites for the progression of cancer cells [8]. Increasing evidence suggests that high levels of glycolytic flux, which consume large amounts of glucose and produce abundant lactate, promote tumor cell proliferation, metastasis, and drug resistance [9,10]. The metabolic intermediates generated during aerobic glycolysis can be used for biosynthesis of tumor biological macromolecules to meet the needs of rapid growth. Moreover, lactate production also provides an acidic environment, suppresses immune cell function, and further promotes the survival of cancer cells, contributing to cancer invasion and metastasis [11,12]. Aldolase has a crucial role in glycolysis; it can convert fructose 1,6-bisphosphate into glyceraldehyde 3-phosphate (G3P) and dihydroxyacetone phosphate (DHAP), thereby controlling fatty acid synthesis and glycolysis to further influence the Warburg effect [13]. ALDOB is one of the major enzymes in the aldolase family, mainly expressed in the liver, kidneys, and small intestine [14]. Human ALDOB mutations have been reported to cause hereditary fructose intolerance (HFI), which can lead to liver and kidney failure, coma, and even death after fructose intake [15]. Moreover, multiple studies have demonstrated that ALDOB is downregulated in HCC tissues [16,17], and stable expression of ALDOB can inhibit the invasiveness and metastatic potential of liver cancer cells [17]. Therefore, ALDOB downregulation causing metabolic reprogramming of cancer cells may be important in accelerating the progression of HCC. However, the upstream signaling events of HCC progression and metabolic reprogramming caused by ALDOB deficiency are still unknown.
MRT4 homolog, MRTO4, is located on human chromosome 17q25.3 and plays a critical role in ribosome biogenesis [7,18]. In recent years, several studies have reported MRTO4 as a biomarker in tumor progression, such as buccal mucosa squamous cell carcinoma and ovarian cancer [19,20]. However, the specific role of MRT4 in tumor progression and the associated molecular mechanisms have not been reported. Therefore, in this study, we investigated the predictive value of MRTO4 in HCC and clarified its relationship with tumor metabolism reprogramming. Additionally, we assessed the role of MRTO4 in HCC glycolysis and metastasis by regulating ALDOB in vitro and in vivo. This study is the first to explore the promoting effect of MRTO4 on HCC by regulating ALDOB, may help develop novel therapies, and provides effective clinical biomarkers for HCC.
Material and Methods
Patient Datasets
We sourced mRNA expression data, encompassing 374 HCC samples (RNAseq, TPM), along with clinical details, from the TCGA database (https://cancergenome.nih.gov). The detailed clinical features were listed in Table 1. To corroborate the expression levels of MRTO4 in HCC, we additionally retrieved mRNA expression datasets GSE121248 (comprising 70 HCC patients and 36 adjacent normal tissues) and GSE45267 (with 48 primary HCC samples and 39 non-cancerous tissues) from the NCBI Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov). Approval of the Ethics Committee of the Second Affiliated Hospital, Shantou University Medical College was obtained for the protocols used in the study (2020–7).
Table 1.
Clinical characteristics of the hepatocellular carcinoma patients.
| Characteristics | Low expression of MRTO4 | High expression of MRTO4 | P value |
|---|---|---|---|
| n | 187 | 187 | |
| Pathologic T stage, n (%) | <0.001 * | ||
| T1 | 112 (30.2%) | 71 (19.1%) | |
| T2 | 35 (9.4%) | 60 (16.2%) | |
| T3 | 32 (8.6%) | 48 (12.9%) | |
| T4 | 6 (1.6%) | 7 (1.9%) | |
| Pathologic N stage, n (%) | 1.000 | ||
| N0 | 125 (48.4%) | 129 (50%) | |
| N1 | 2 (0.8%) | 2 (0.8%) | |
| Pathologic M stage, n (%) | 0.553 | ||
| M0 | 127 (46.7%) | 141 (51.8%) | |
| M1 | 3 (1.1%) | 1 (0.4%) | |
| Pathologic stage, n (%) | <0.001 * | ||
| Stage I | 103 (29.4%) | 70 (20%) | |
| Stage II | 33 (9.4%) | 54 (15.4%) | |
| Stage III | 33 (9.4%) | 52 (14.9%) | |
| Stage IV | 4 (1.1%) | 1 (0.3%) | |
| Tumor status, n (%) | 0.040 * | ||
| Tumor free | 112 (31.5%) | 90 (25.4%) | |
| With tumor | 68 (19.2%) | 85 (23.9%) | |
| Gender, n (%) | 0.740 | ||
| Female | 62 (16.6%) | 59 (15.8%) | |
| Male | 125 (33.4%) | 128 (34.2%) | |
| Age, n (%) | 0.570 | ||
| ≤60 | 86 (23.1%) | 91 (24.4%) | |
| >60 | 101 (27.1%) | 95 (25.5%) | |
| BMI, n (%) | 0.010 * | ||
| ≤25 | 78 (23.1%) | 99 (29.4%) | |
| >25 | 93 (27.6%) | 67 (19.9%) | |
| Histological type, n (%) | 0.117 | ||
| Hepatocellular carcinoma | 182 (48.7%) | 182 (48.7%) | |
| Hepatocholangiocarcinoma (mixed) | 2 (0.5%) | 5 (1.3%) | |
| Fibrolamellar carcinoma | 3 (0.8%) | 0 (0%) | |
| Residual tumor, n (%) | 0.464 | ||
| R0 | 167 (48.4%) | 160 (46.4%) | |
| R1 | 7 (2%) | 10 (2.9%) | |
| R2 | 1 (0.3%) | 0 (0%) | |
| Histologic grade, n (%) | <0.001 * | ||
| G1 | 35 (9.5%) | 20 (5.4%) | |
| G2 | 102 (27.6%) | 76 (20.6%) | |
| G3 | 46 (12.5%) | 78 (21.1%) | |
| G4 | 1 (0.3%) | 11 (3%) | |
| AFP(ng/ml), n (%) | 0.012 * | ||
| ≤400 | 124 (44.3%) | 91 (32.5%) | |
| >400 | 26 (9.3%) | 39 (13.9%) | |
| Albumin(g/dl), n (%) | 0.938 | ||
| <3.5 | 38 (12.7%) | 31 (10.3%) | |
| ≥3.5 | 126 (42%) | 105 (35%) | |
| Prothrombin time, n (%) | 0.124 | ||
| ≤4 | 106 (35.7%) | 102 (34.3%) | |
| >4 | 54 (18.2%) | 35 (11.8%) | |
| Child-Pugh grade, n (%) | 0.809 | ||
| A | 120 (49.8%) | 99 (41.1%) | |
| B | 10 (4.1%) | 11 (4.6%) | |
| C | 1 (0.4%) | 0 (0%) | |
| Fibrosis ishak score, n (%) | 0.006 * | ||
| 0 | 48 (22.3%) | 27 (12.6%) | |
| 1/2 | 14 (6.5%) | 17 (7.9%) | |
| 3/4 | 9 (4.2%) | 19 (8.8%) | |
| 5 | 3 (1.4%) | 6 (2.8%) | |
| 6 | 47 (21.9%) | 25 (11.6%) | |
| Vascular invasion, n (%) | 0.009 * | ||
| No | 119 (37.4%) | 89 (28%) | |
| Yes | 46 (14.5%) | 64 (20.1%) | |
| Adjacent hepatic tissue inflammation, n (%) | 0.254 | ||
| None | 71 (30%) | 47 (19.8%) | |
| Mild | 50 (21.1%) | 51 (21.5%) | |
| Severe | 11 (4.6%) | 7 (3%) | |
| OS event, n (%) | 0.005 * | ||
| Alive | 135 (36.1%) | 109 (29.1%) |
Bold values indicate
P<0.05.
Statistical analysis was performed by the Pearson χ2-test. OS – overall survival.
Survival Analysis
Kaplan-Meier curves were constructed using the TCGA database for HCC to investigate the association between MRTO4 expression and patient survival across a range of clinical factors. The ‘survival’ package in R facilitated the analysis of this association. Both the hazard ratio (HR) and the log-rank P value were determined within a 95% confidence interval [21].
Formulation and assessment of nomograms for predicting HCC survival. The ROC diagnostic curve, along with its time-dependent counterpart, was crafted using the ‘pROC’ and ‘time-ROC’ R packages. A nomogram designed to estimate the 1-, 3-, and 5-year overall survival rates for HCC patients was created with the aid of an R package. Key prognostic indicators from the clinical dataset were identified through Cox regression analysis.
Protein–Protein Interaction Network
Data on protein–protein interactions (PPI) were obtained from the STRING database (https://string-db.org) to envision the PPI network for genes co-expressed alongside MRTO4. Interactions that achieved a combined score greater than 0.15 were deemed important. The Cytoscape 3.9.1 tool was employed to create this network [22].
Functional Enrichment Analysis
From the TCGA, we retrieved the top 300 genes linked to MRTO4 in HCC as well as the top 300 genes related to HCC survival. These genes underwent enrichment analysis in the DAVID database, emphasizing Gene Ontology (GO) categories, including biological processes, cellular components, and molecular functions. We also carried out KEGG pathway analyses, and the results were visualized using the ggplot2 package in R. A corrected P value of less than 0.05 was deemed statistically significant [23].
Immune Infiltration Analysis by Single-Sample GSEA (ssGSEA)
We utilized the GSVA package in R (http://www.biocondutor.org/package/release/bioc/html/GSVA.html) to conduct immune infiltration analysis on carcinoma samples, using GSEA to assess 24 distinct immune cell types. To discern the relationships between carcinoma and each immune cell subset, we executed a Spearman’s correlation coefficient analysis.
ROC Curve Methodology
Receiver operating characteristic (ROC) curve analysis was conducted using the ‘pROC’ package in R. This analysis evaluated the sensitivity and specificity of MRTO4 expression levels in distinguishing between HCC and adjacent normal tissues. The area under the curve (AUC) was calculated to assess the diagnostic performance.
LinkedOmics Database Analysis
We gleaned information on the MRTO4 expression pattern from the LinkedOmics database. Using the GSEA function within the Link Interpreter module, we explored the GO and KEGG pathways associated with MRTO4 and genes expressed in conjunction with it [24].
TISIDB Database Analysis
We utilized the TISIDB database (http://cis.hku.hk/TISIDB/) to assess the relationship between MRTO4 mRNA expression and immune checkpoint genes through the “Immunomodulator” module. Additionally, the “chemokine” module facilitated our exploration of the link between MRTO4 mRNA expression and the expression of chemokines and their receptors [25].
Cell Culture
Human hepatoma cell lines HepG2 and MHCC97H were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Two cells were sown in RPMI-1640 media with 10% FBS and 1% penicillin streptomycin, incubated at 37°C in a humidity-saturated incubator with 5% CO2. When the cell fusion rate was 80~90%, the cells were passaged, and the cells with good growth status in generation 3 were taken for subsequent experiments.
In Vivo Tumorigenesis
We obtained 18 BALB/c nude mice, aged from 5 to 8 weeks, weighing 18 to 20 g, from the Experimental Center of Guangzhou Ruige Biological Technology Co. LTD. The mice were placed in a specific pathogen-free room kept at 25°C with 12-h alternating light and dark cycles, fed ad libitum, and assigned to groups after 1 week of adaptive feeding. The mice were randomly divided into the si-NC group and the si-MRTO4 group, with 5 mice in each group. The si-MRTO4 group and si-NC group were injected with 2.0×106 MHCC-97H cells in the left hepatic lobe [26]. The vector group mice were injected with an equal amount of MHCC-97H cells in the same location. The growth of the xenografts was observed every 5 days. After the observation period of 21 days, the mice were euthanized, and the tumor tissues were isolated to measure tumor mass and volume. The tumors were weighed using an electronic laboratory scale. The Experimental Animal Ethics Review Committee of Guangzhou Seyotin Biotechnology Co., LTD granted approval for this animal experiment (SYT2023046).
Immunohistochemistry
Tissue sections were dewaxed in xylene, washed in phosphate-buffered saline (PBS) for 5 min, and then underwent antigen repair and endogenous enzyme activity blocking. Then, they were incubated with 10% fetal bovine serum albumin at 37°C for 20 min. After removing the protein block solution, we added 50 μL at a 1: 200 dilution ratio and MRTO4, ALDOB, and Ki67 (Proteintech Group, Wuhan, China) primary antibodies were added and samples were incubated overnight. The next day, the sections were washed with PBS, and then incubated with secondary antibodies (1: 2000) at 30°C for 30 min. All sections were stained using peroxidase substrate 3, 3′-diaminobenzidine, counterstained with hematoxylin, dehydrated, cleared, and mounted. The staining was observed under a microscope (Olympus, Tokyo, Japan).
TUNEL
The tumor tissues from naked mice were prepared as paraffin sections and stained according to the TUNEL kit (C1090, Beyotime Biotechnology, Shanghai, China) instructions. We added 100 μL TUNEL staining solution, followed by incubation in the dark for 1 h, then added 50 μL DAPI stain, followed by further incubation in the dark for 10 min. Samples were observed and photographed under a microscope (Olympus, Tokyo, Japan).
Cell Viability
The cell viability of HepG2 and MHCC97H cells was measured using the CCK8 assay. After digestion and counting, 1×104 cells per well were seeded in a 96-well plate for each experimental group. Six replicates were set for each concentration, and the cells were cultured for 24, 48, and 72 h. After incubation for 3 h with 10 μL of CCK8 solution (Seyotin, China) added to each well, the absorbance at 450 nm was measured using a multifunctional microplate reader (Thermo Fisher Scientific, USA).
Cell Transfection and Grouping
The growth-favorable HepG2 and MHCC97H cells were seeded in a 6-well plate (1×105 cells/well). Liposome (C0535, Beyotime, China) transfection was used to transfect si-NC, si-MRTO4, OE-NC, and OE-MRTO4 into HepG2 or MHCC97H cells, respectively, referred to as the si-NC group, si-MRTO4 group, OE-NC group, and OE-MRTO4 group. Liposome transfection was used to co-transfect si-MRTO4 with si-NC and si-ALDOB into HepG2 or MHCC97H cells, respectively, referred to as the si-MRTO4+si-NC group and si-MRTO4+si-ALDOB group.
Clonogenic Assay
HepG2 and MHCC97H cells from each experimental group were seeded in 6-well plates. After 15–18 days of culture, visible clones were observed. Subsequently, the clones were fixed with paraformaldehyde and stained with crystal violet. Five randomly selected fields were photographed (Nikon, Tokyo, Japan).
Transwell Assay
The upper Transwell chambers were coated with matrix gel. Then, 200 μL of cell suspension was added to the upper chambers, and 500 μL of culture medium containing 10% FBS was added to the lower chambers. After incubation at 37°C for 24 h, the chambers were fixed and stained. Six randomly selected fields were photographed and counted under an optical microscope(Olympus, Tokyo, Japan).
Lactate Assay, Glucose Uptake Assay, and ATP Assay
Lactate, glucose uptake, and ATP levels were measured according to the recommended methods. The lactate assay kit (C0017), glucose uptake assay kit (S0201M), and ATP assay kit (S0026) were purchased from Beyotime Biotechnology (Shanghai, China).
ECAR Assay
Cells from each experimental group were grown in test medium containing 2 mmol/L glutamine at 37°C for 1.5 h and then measured with a ECAR kit (BB-48311, Bestbio, Nanjing, China).
Western Blotting
HepG2 and MHCC97H cell suspensions from each experimental group were added to RIPA lysis buffer, and the proteins were separated by 10% SDS-PAGE. After transfer to PVDF membrane, the membrane was blocked with 50 g/L skim milk for 1 h. Then, primary antibodies including ALDOB (1: 1000,18065-1-AP, Proteintech Group, Wuhan, China), MRTO4 (1: 1000, 201944-AP, Proteintech Group, Wuhan, China), Akt (1: 1000, 80455-1-RR, Proteintech Group, Wuhan, China), p-Akt (66444-1-Ig, Proteintech Group, Wuhan, China), and GAPDH (1: 5000, 60004-1-Ig, Proteintech Group, Wuhan, China) were added and incubated at 4°C for 24 h. After membrane washing, secondary antibodies were added and incubated at room temperature for 2 h. The membranes were then exposed to ECL (Seyotin, China) and visualized in a dark room. Protein relative expression was analyzed using Image J software.
RT-qPCR
Total RNA was extracted from liver cancer cells using the Trizol method. The PrimeScript RT Reagent Kit (Seyotin, China) was used for reverse transcription synthesis of cDNA. PCR was performed using SYBR Green PCR Master Mix (Seyotin, China). GAPDH was used as an internal reference to detect the expression changes of MRTO4 and ALDOB. The relative expression levels of MRTO4 and ALDOB were represented by the 2−ΔΔCt method. The primer sequences are shown in Table 2.
Table 2.
Gene primer sequences.
| Genes | Forward (5′-3′) | Reverse (5′-3′) |
|---|---|---|
| MRTO4 | TGGCCAACATGAGGAACAGC | TTCATCAGATGGGCTCCGAC |
| ALDOB | GCCAGGGAAAGCTGTTCAGA | AGGCCATCAAGCCCTTGAAT |
| GAPDH | AGAAGGCTGGGGCTCATTTG | CAGGGGTGCTAAGCAGTTGG |
Statistical Analysis
For our statistical evaluations, we employed box plots to determine the MRTO4 gene expression levels among HCC patients. The median gene expression served as the demarcation point for MRTO4 expression categorization. To ascertain the association between clinical parameters and MRTO4 expression in HCC, we applied the Wilcoxon signed-rank test and logistic analysis. Univariate Cox analysis was used to identify potential prognostic markers, and multivariate Cox analysis validated the effect of MRTO4 expression on survival, factoring in other clinical indicators. We carried out all statistical computations using R (version 3.5.3) and SPSS (version 24.0) software. The experimental data are expressed as mean±standard deviation (SD). The comparison between 2 groups is done using t tests for comparing means, while the comparison among multiple groups is done using one-way analysis of variance. P<0.05 it indicated that the differences between the 2 groups were statistically significant.
Results
Baseline Characteristics of Patients
Data from a total of 374 HCC with the required clinical features were acquired from the TCGA data portal. The detailed clinical features are listed inTable 1. Among the 374 participants, 253 were male (67.6%), and 121 were female (32.4%). Among them, 196 patients (52.6%) were younger than or equal to 60 years, and 177 patients (47.4%) were older than 60 years. In terms of histologic grade, 55 patients were G1 (14.9%), 178 patients were G2 (48.2%), 124 patients were G3 (33.6%), and 14 patients were G4 (3.3%). In terms of pathologic T stage, 183 patients were T1 (49.3%), 95 patients were T2 (25.6%), 80 patients were T3 (21.5%), and 13 patients were T4 (3.5%). In terms of pathologic N stage, 254 patients were N0 (98.4%) and 4 patients were N1 (1.6%). In terms of pathologic M stage, 268 patients were M0 (98.5%) and 4 patients were M1 (1.5%). In terms of pathologic stage, 173 patients were stage I (49.4%), 87 patients were stage II (25.8%), 85 patients were stage III (24.3%), and 5 patients were T4 (1.4%). In terms of tumor status, 202 patients were tumor free (56.9%) and 153 patients had tumors (43.1%). In those with residual tumors, 327 patients were R0 (94.8%), 17 patients were R1 (4.9%), and 1 patient was R2 (0.3%). In terms of vascular invasion, 208 patients were without vascular invasion (65.4%) and 110 patients had vascular invasion (34.6%). In terms of Child-Pugh grade, 219 patients were Child-Pugh grade A (90.9%), 21 patients were Child-Pugh grade B (8.7%), and 1 patient was Child-Pugh grade C (0.4%). Meanwhile, 199 patients had non-multifocal lesion (83.3%) and 40 had multifocal lesion (16.8%). In terms of tumor necrosis, 71 patients had no necrosis (38.8%), 38 patients had focal necrosis (20.7%), 62 patients had moderate necrosis (33.8%), and 12 patients had no necrosis (6.5%). As to tumor depth, 21 patients had superficial tumors (10.0%) and 188 had deep tumors (90.0%). Furthermore, 120 patients had no metastases (67.1%) and 69 had metastases (33.0%); only 78 received radiation therapy (30.4%) and other 179 patients did not (69.6%). In overall survival, 244 patients (65.2%) were still alive and 130 (34.8%) had died (Table 1).
MRTO4 Is Highly Expressed in HCC
To explore the expression level of MRTO4 in normal and tumor tissues, we downloaded and analyzed the expression levels of MRTO4 mRNA in different tumors and normal tissues from TCGA using the R package. The results showed that MRTO4 expression was significantly higher in tumor tissues such as bladder urothelial carcinoma (BLCA), breast cancer (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrial carcinoma (UCEC) (Figure 1A). The expression level of MRTO4 in HCC tended to be higher than that in normal tissue (Figure 1B–1E). To assess the correlation between MRTO4 expression and clinical characteristics in HCC patients, we analyzed the mRNA expression levels of MRTO4 in different clinical categories in the TCGA database. The results showed that high expression of MRTO4 was significantly related to pathologic T stage (P<0.05), histologic grade (P<0.05), vascular invasion (P<0.05), BMI (P<0.05), weight (P<0.05), tumor status (P< 0.05), AFP (P<0.05), overall survival (OS) event (P<0.05), and disease-specific survival (DSS) event (P<0.05) (Figure 2A–2I). The association identified between MRTO4 expression and clinical features in patients with HCC is summarized in Table 1. These data suggest that MRTO4 is upregulated in HCC.
Figure 1.
The expression level of MRTO4 in different human cancers. (A) Variances in MRTO4 expression in diverse cancer datasets versus normal tissues from the TCGA database. (B) MRTO4 expression in normal versus unmatched HCC samples. (C) MRTO4 expression in normal versus matched HCC samples. (D, E) Expression level of DOCK7 in normal tissues and HCC tissues from GSE121248 and GSE45267. * P<0.05, ** P<0.01, *** P<0.001.
Figure 2.
The expression level of MRTO of HCC patients with different clinical characteristics in TCGA. (A) pathologic T stage, (B) histologic grade, (C) vascular invasion, (D) BMI, (E) weight, (F) tumor status, (G) AFP, (H) OS event, (I) DSS event. * P<0.05, ** P<0.01, *** P<0.001.
High Expression of MRTO4 Is an Independent Prognostic Factor for Overall Survival of HCC Patients
To identify whether MRTO4 expression affected patient survival, we classified HCC patients in the TCGA database into a high MRTO4 expression group (the top 50% of samples with the highest expression) and a low MRTO4 expression group (the remaining 50% of the samples) to perform survival analysis according to the mean expression value of MRTO4. Kaplan-Meier survival analysis showed that high expression of MRTO4 was related to the poor prognosis in overall survival (P<0.001), disease-specific survival (P<0.001), and progression-free interval (P<0.001) of HCC patients (Figure 3A–3C). Subgroup analysis showed that high MRTO4 expression was significantly correlated with poor prognosis in HCC in the following cases: pathologic T1 stage (P=0.019), pathologic T2 stage (P<0.001), pathologic T3 stage (P=0.002), pathologic stage I (P=0.043), pathologic Stage II (P<0.001), pathologic Stage III (P<0.001), histologic G2 grade (P=0.032), histologic G3 grade (P=0.001), male (P<0.001), Female (P=0.037), age ≤60 years old (P≤0.001), age >60 years old (P=0.002), AFP >400 ng/ml (P=0.041), patients without vascular invasion (P=0.025), patients with vascular invasion (P=0.011), albumin <3.5 g/dl (P=0.024), and prothrombin time >4 (P=0.001) (Figure 3D–3G, Table 3). Univariate and multivariate Cox regression analyses also showed that MRTO4 was independently associated with overall survival of HCC patients (Table 4). These data suggest that high expression of MRTO4 is an independent prognostic factor for the overall survival (OS) of HCC patients.
Figure 3.
Kaplan-Meier survival curve analysis of the prognostic significance of MRTO4 with different expression in HCC. (A) Kaplan-Meier estimates of the overall survival probability of TCGA patients in HCC patients. (B) Kaplan-Meier estimates of the disease-specific survival. (C) Kaplan-Meier estimates of the progression-free interval. (D–T) Subgroup analysis for pathologic T1 stage (D), pathologic T2 stage (E), pathologic T3 stage (F), pathologic Stage I (G), pathologic Stage II (H), pathologic Stage III (I), histologic G2 grade (J), histologic G3 grade (K), male (L), Female (M), age ≤60 years old (N), age >60 years old (O), AFP >400 ng/ml (P), patients without vascular invasion (Q), patients with vascular invasion (R), Albumin <3.5 g/dl (S), prothrombin time >4 (T). * P<0.05, ** P<0.01, *** P<0.001.
Table 3.
Logistic analysis of the association between MRTO4 expression and clinical characteristics.
| Characteristics | Total (N) | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| Hazard ratio (95% CI) | P value | Hazard ratio (95% CI) | P value | ||
| Pathologic T stage | 370 | ||||
| T1 | 183 | Reference | Reference | ||
| T2 | 94 | 1.431 (0.902–2.268) | 0.128 | 1.234 (0.759–2.004) | 0.396 |
| T3 | 80 | 2.674 (1.761–4.060) | <0.001 * | 2.030 (1.295–3.180) | 0.002 * |
| T4 | 13 | 5.386 (2.690–10.784) | <0.001 * | 4.191 (2.041–8.606) | <0.001 * |
| Tumor status | 354 | ||||
| Tumor free | 202 | Reference | Reference | ||
| With tumor | 152 | 2.317 (1.590–3.376) | <0.001 * | 1.780 (1.203–2.632) | 0.004 * |
| Gender | 373 | ||||
| Female | 121 | Reference | |||
| Male | 252 | 0.793 (0.557–1.130) | 0.200 | ||
| Age | 373 | ||||
| ≤60 | 177 | Reference | |||
| >60 | 196 | 1.205 (0.850–1.708) | 0.295 | ||
| BMI | 336 | ||||
| ≤25 | 177 | Reference | |||
| >25 | 159 | 0.798 (0.550–1.158) | 0.235 | ||
| Residual tumor | 344 | ||||
| R0 | 326 | Reference | |||
| R1 & R2 | 18 | 1.604 (0.812–3.169) | 0.174 | ||
| Histologic grade | 368 | ||||
| G1 | 55 | Reference | |||
| G2 | 178 | 1.162 (0.686–1.969) | 0.576 | ||
| G3 | 123 | 1.185 (0.683–2.057) | 0.545 | ||
| G4 | 12 | 1.681 (0.621–4.549) | 0.307 | ||
| AFP(ng/ml) | 279 | ||||
| ≤400 | 215 | Reference | |||
| >400 | 64 | 1.075 (0.658–1.759) | 0.772 | ||
| Albumin(g/dl) | 299 | ||||
| <3.5 | 69 | Reference | |||
| ≥3.5 | 230 | 0.897 (0.549–1.464) | 0.662 | ||
| Prothrombin time | 296 | ||||
| ≤4 | 207 | Reference | |||
| >4 | 89 | 1.335 (0.881–2.023) | 0.174 | ||
| Child-Pugh grade | 240 | ||||
| A | 218 | Reference | |||
| B & C | 22 | 1.643 (0.811–3.330) | 0.168 | ||
| Vascular invasion | 317 | ||||
| No | 208 | Reference | |||
| Yes | 109 | 1.344 (0.887–2.035) | 0.163 | ||
| Adjacent hepatic tissue inflammation | 236 | ||||
| None | 118 | Reference | |||
| Mild & severe | 118 | 1.194 (0.734–1.942) | 0.475 | ||
| MRTO4 | 373 | ||||
| Low | 187 | Reference | Reference | ||
| High | 186 | 2.258 (1.582–3.223) | <0.001 * | 1.974 (1.355–2.877) | <0.001 * |
Bold values indicate
P<0.05.
OR – odds ratio; CI – confidence interval
Table 4.
Univariate and multivariate Cox regression analyses of clinical characteristics associated with overall survival.
| Characteristics | Total (N) | OR (95% CI) | P value |
|---|---|---|---|
| Pathologic T stage (T2 & T3 & T4 vs T1) | 371 | 2.485 (1.637–3.773) | <0.001 * |
| Pathologic N stage (N1 vs N0) | 258 | 0.969 (0.134–6.986) | 0.975 |
| Pathologic M stage (M1 vs M0) | 272 | 0.300 (0.031–2.923) | 0.300 |
| Pathologic stage (Stage III & Stage IV vs Stage I & Stage II) | 350 | 1.571 (0.967–2.552) | 0.068 |
| Tumor status (with tumor vs tumor free) | 355 | 1.556 (1.019–2.374) | 0.040* |
| Gender (Male vs Female) | 374 | 1.076 (0.698–1.660) | 0.740 |
| Age (>60 vs ≤60) | 373 | 0.889 (0.592–1.335) | 0.570 |
| Race (Black or African American & White vs Asian) | 362 | 0.591 (0.389–0.898) | 0.014 * |
| Histological type (fibrolamellar carcinoma & hepatocholangiocarcinoma (mixed) vs hepatocellular carcinoma) | 374 | 1.000 (0.285–3.513) | 1.000 |
| BMI (>25 vs ≤25) | 337 | 0.568 (0.368–0.874) | 0.010 * |
| Residual tumor (R1 & R2 vs R0) | 345 | 1.305 (0.502–3.389) | 0.585 |
| Histologic grade (G3 & G4 vs G1 & G2) | 369 | 2.702 (1.742–4.193) | <0.001 * |
| AFP(ng/ml) (>400 vs ≤400) | 280 | 2.044 (1.161–3.597) | 0.013 * |
| Albumin(g/dl) (≥3.5 vs <3.5) | 300 | 1.022 (0.595–1.754) | 0.938 |
| Prothrombin time (>4 vs ≤4) | 297 | 0.674 (0.407–1.116) | 0.125 |
| Child-Pugh grade (B & C vs A) | 241 | 1.212 (0.504–2.914) | 0.667 |
| Vascular invasion (Yes vs No) | 318 | 1.860 (1.165–2.970) | 0.009 * |
| Adjacent hepatic tissue inflammation (Mild & Severe vs None) | 237 | 1.436 (0.858–2.403) | 0.168 |
| Weight (>70 vs ≤70) | 346 | 0.427 (0.277–0.657) | <0.001 * |
| Height (≥170 vs <170) | 341 | 0.748 (0.485–1.153) | 0.189 |
Bold values indicate
P<0.05.
HR – hazard ratio; CI – confidence interval.
Diagnostic Value of MRTO4 Expression in HCC
To analyze the diagnostic value of MRTO4 expression in HCC, we performed ROC curve and nomogram analyses on the MRTO4 gene expression data from the TCGA database to evaluate the diagnostic value of the gene. The area under the ROC curve (AUC) was 0.914, suggesting a higher diagnostic value, as shown in Figure 4A. The time-dependent AUC curve shows that the predicted AUC values of patients in the next 5 years were all above 0.6. A time-dependent survival ROC curve of MRTO4 was created to predict the 1-, 3-, and 5-year survival rates. All these AUC values were above 0.6, which is considered suitable for prediction (Figure 4B). The time-dependent AUC curve showed that the predicted AUC values of patients in the next 5 years were all above 0.6 (Figure 4C). Then, we combined the expression level of MRTO4 with the clinical variables to construct a nomogram to predict the survival probability of patients at 1, 3, and 5 years. The nomogram indicated that the prognostic prediction of the expression level of MRTO4 was better than that of the traditional clinical features of pathologic T stage, tumor status, sex, age, BMI, residual tumor, histologic grade, AFP, albumin, prothrombin time, Child-Pugh grade, vascular invasion, and adjacent hepatic tissue inflammation (Figure 4E, Table 4). Calibration curves were plotted to compare the predicted probabilities of survival from the nomogram against the observed outcomes. The goodness-of-fit was assessed using the Hosmer-Lemeshow test in R, ensuring the model’s reliability in clinical predictions. The result of the calibration curve demonstrated that the predictive probability of 1-, 3-, and 5-year survival was close to the actual 1-year survival in clinical practice (Figure 4D).
Figure 4.
Diagnostic value of MRTO4 expression in hepatocellular carcinoma. (A) Receiver operating characteristic (ROC) curve analysis for MRTO4 expression in HCC and adjacent tissue. (B) Time-dependent survival ROC analysis for MRTO4 expression in HCC and adjacent tissue to predict 1-, 3-, and 5-year survival rates. (C) Time-dependent AUC curve analysis for MRTO4 expression in HCC and adjacent tissue. (D) Calibration plots of the nomogram for predicting the probability of OS at 1, 3, and 5 years. (E) Nomogram survival prediction chart for predicting the 1-, 3-, and 5-year overall survival rates.
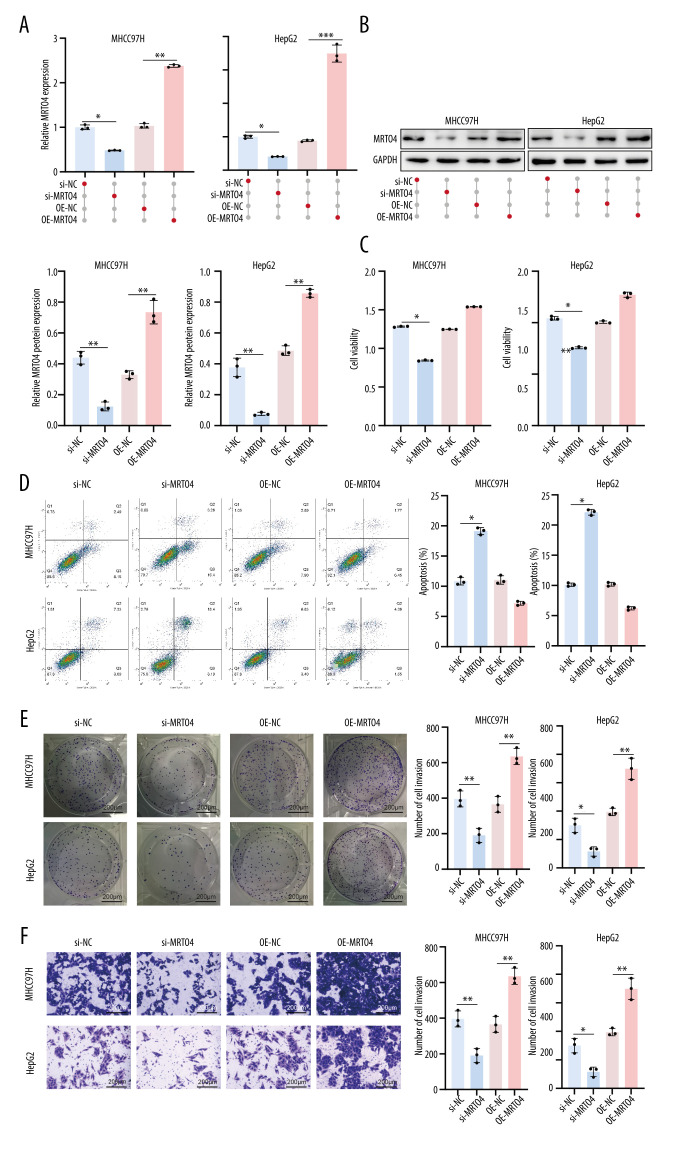
Knockdown of MRTO4 Inhibited Proliferation and Invasion of HCC Cells
Next, we investigated the impact of MRTO4 on the function of HCC cells lines. First, we transfected si-NC, si-MRTO4, OE-NC, and OE-MRTO4 into HepG2 and MHCC97H cells and assessed transfection efficiency by RT-qPCR and protein analysis. The results in Figure 5A, 5B show that si-MRTO4 significantly decreased the expression of MRTO4 in HepG2 and MHCC97H cells (P<0.05), while OE-MRTO4 significantly enhanced the expression of MRTO4 in HepG2 and MHCC97H cells (P<0.01). Subsequently, we investigated the impact of MRTO4 on the function of HCC cells. Our experimental results demonstrated that si-MRTO4 weakened the viability of HepG2 and MHCC97H cells (P<0.05) and promoted apoptosis in these cells (P<0.05). Conversely, OE-MRTO4 reversed this trend, enhancing the viability of HepG2 and MHCC97H cells and suppressing apoptosis (Figure 5C, 5D). Furthermore, si-MRTO4 significantly inhibited aggregation (P<0.05) and invasion (P<0.05) of HepG2 and MHCC97H cells. Consistently, this trend was reversed in the OE-MRTO4 group, as OE-MRTO4 obviously increased aggregation (P<0.01) and invasion (P<0.01) capabilities in HepG2 and MHCC97H cells (Figure 5E, 5F). These results indicated that knockdown of MRTO4 inhibits the proliferation and invasion of HCC cells.
Figure 5.
Knockdown of MRTO4 inhibited proliferation and invasion of HCC cells. (A) RT-qPCR analysis of MRTO4 expression. (B) Protein blotting analysis of MRTO4 expression. (C) CCK 8 assessed the viability of HepG2 and MHCC97H. (D) Apoptosis was measured by flow cytometry. (E) Cloning experiments assessing cell integration ability. (F) Transwell assay was used for cell invasion ability. All results are presented as mean±SD (n=3).* P<0.05, ** P<0.01, *** P<0.001.
Knockdown of MRTO4 Negatively Regulated ALDOB to Inhibit Glycolysis
To understand the biological function of MRTO4 in HCC, we used the LinkFinder module of the LinkedOmics website to detect the co-expression pattern of MRTO4 in HCC in the TCGA database. The red dot indicates the top 20 genes that were positively correlated with MRTO4, and the blue dot represents the bottom 15 genes that were negatively correlated with MRTO4 (Figure 6A). Interestingly, ALDOB, a key enzyme in energy metabolism (the glycolytic process), is found in these negatively regulated genes. Previous studies have confirmed that downregulation of ALDOB promotes the invasiveness and metastatic potential of HCC [27,28]. We used DAVID Functional Annotation Bioinformatics Microarray Analysis to identify the enriched GO functional enrichment and KEGG pathways among the MRTO4-related 600 genes (top 300 correlation value) and found that pentose phosphate pathway, fructose and mannose metabolism, and cysteine and methionine metabolism were enriched among these genes (Figure 6B, Table 5). These pathways are closely related to energy metabolism and glycolysis, revealing the important role of MRTO4 in mediating metabolism and glycolysis. Given the important role of ALDOB in the glycolysis process [29], we next investigated whether MRTO4 could mediate the expression of ALDOB in HCC. Our results clearly indicated that si-MRTO4 upregulates the expression of ALDOB in HepG2 and MHCC97H (P<0.05), while OE-MRTO4 inhibits the expression of ALDOB in HepG2 and MHCC97H (P<0.05) (Figure 6C–6E). Subsequently, we further studied the impact of MRTO4 on glycolysis. We evaluated the extracellular acidification rate (ECAR) of HepG2 and MHCC97H using a Seahorse extracellular flux analyzer, as ECAR indirectly reflects the glycolytic capacity of cells. The results showed that si-MRTO4 inhibited the ECAR of HepG2 and MHCC97H, while OE-MRTO4 increased the ECAR levels of HepG2 and MHCC97H (Figure 6F). Specifically, glucose consumption, lactate production, and ATP are important indicators of the glycolysis process [30]. In addition, we found that si-MRTO4 reduced the glucose consumption (P<0.05), lactate production rate (P<0.05), and ATP levels (P<0.05) in HepG2 and MHCC97H cells, while OE-MRTO4 reversed the effects of si-MRTO4, promoting glucose consumption (P<0.05), increasing lactate levels (P<0.05) and ATP levels (P<0.05) in HepG2 and MHCC97H cells (Figure 6G–6I). In all, these results demonstrated that knockdown of MRTO4 inhibits glycolysis in liver cancer cells by upregulating ALDOB.
Figure 6.
Knockdown of MRTO4 negatively regulated ALDOB to inhibit glycolysis. (A) Heatmap showing the top 35 genes in liver cancer that were positively and negatively related to MRTO4. Red represents positively related genes and blue represents negatively related genes. (B) Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of MRTO4-related genes in HCC. (C) RT-qPCR assayed the expression of ALDOB. (D) Immunofluorescence analyzed the expression levels of ALDOB in the cells. (E) Protein imprint measured ALDOB expression in HCC cells. (F) Seahorse extracellular flux analyzer assessed the ECAR of HepG2 and MHCC97H. (G) ELISA assessed the cells for glucose consumption. (H) ELISA assessed the cellular lactate production capacity. (I) ELISA assessed the level of ATP released from the cells. All results are presented as mean±SD (n=3).* P<0.05, ** P<0.01, *** P<0.001.
Table 5.
Gene sets enriched in the high MRTO4 expression phenotype by GO & KEGG analysis.
| Ontology | ID | Description | GeneRatio | BgRatio | P value | P.adjust |
|---|---|---|---|---|---|---|
| BP | GO: 0030388 | Fructose 1,6-bisphosphate metabolic process | 2/33 | 10/18800 | 0.0001 * | 0.0916 |
| BP | GO: 0006000 | Fructose metabolic process | 2/33 | 13/18800 | 0.0002 * | 0.0916 |
| MF | GO: 0005319 | Lipid transporter activity | 4/34 | 159/18410 | 0.0002 * | 0.0298 * |
| KEGG | hsa00030 | Pentose phosphate pathway | 2/15 | 30/8164 | 0.0013 * | 0.0274 * |
| KEGG | hsa00051 | Fructose and mannose metabolism | 2/15 | 33/8164 | 0.0016 * | 0.0274 * |
| KEGG | hsa00270 | Cysteine and methionine metabolism | 2/15 | 51/8164 | 0.0038 * | 0.0432 * |
| KEGG | hsa00010 | Glycolysis/gluconeogenesis | 2/15 | 67/8164 | 0.0065 * | 0.0550 |
| KEGG | hsa03320 | PPAR signaling pathway | 2/15 | 75/8164 | 0.0081 * | 0.0550 |
Bold values indicate
P<0.05.
BP – biological process; CC – cellular component; MF – molecular function.
Knockdown of ALDOB Activated Akt and Reversed the Low Expression of MRTO4 on HCC Cell Function
Due to the negative regulation of MRTO4 on ALDOB, ALDOB is a key enzyme in glycolysis. Therefore, we began to investigate whether MRTO4 exerts its function in HCC through the regulation of ALDOB. We established stable HepG2 and MHCC97H cell lines co-transfected with si-MRTO4 and si-ALDOB. The results showed that si-ALDOB clearly inhibited the expression of ALDOB in HepG2 and MHCC97H (P<0.05) (Figure 7A). In addition, knocking down ALDOB almost completely rescued the inhibition of lactate production (P<0.01) and ATP release (P<0.05) in HepG2 and MHCC97H cells with si-MRTO4 (Figure 7B, 7C). At the same time, compared with the si-MRTO4 group, knocking down ALDOB obviously restored the vitality of HepG2 and MHCC97H cells (Figure 7D) and reversed the colony formation (P<0.01) and cell invasion ability (P<0.001) of HepG2 and MHCC97H cells (Figure 7E, 7F). The promoting effect of activated Akt in glycolysis has been extensively studied [31,32]. Recent research has shown that the loss of ALDOB activates Akt and promotes the occurrence of HCC [33]. We speculated that MRTO4 activates Akt in HCC by inducing the loss of ALDOB. As expected, the protein imprinting results showed that, compared with the si-MRTO4 group, the ratio of p-Akt/Akt in HepG2 and MHCC97H cells in the si-ALDOB group was significantly increased (P<0.001) (Figure 7G). Overall, these results revealed that knocking down ALDOB activates Akt and reverses the low expression of MRTO4 on HCC cell function.
Figure 7.
Knockdown of ALDOB activated Akt and reverses the low expression of MRTO4 on HCC cell function. (A) Protein blot analysis of ALDOB expression in HCC cells. (B) ELISA evaluation of cell lactate production capacity. (C) ELISA evaluation of cell ATP release levels. (D) CCK8 evaluation of HepG2 and MHCC97H vitality. (E) Clone formation experiment evaluating cell integration ability. (F) Transwell assay was used to assay HCC cell invasion ability. (G) Protein imprint analysis of p-Akt and Akt expression in HCC cells. All results are presented as mean±SD (n=3).* P<0.05, ** P<0.01, *** P<0.001.
Knockdown of MRTO4 Suppressed HCC Progression In Vivo
We further clarified the effect of MRTO4 on HCC by transplanting MHCC97H cells transfected with si-MRTO4 into nude mice. First, we measured the transplanted tumors and consistently discovered si-MRTO4 significantly inhibited tumor growth (P<0.05) (Figure 8A, 8B). Furthermore, TUNEL staining showed that si-MRTO4 significantly promoted apoptosis of HCC cells (P<0.001) (Figure 8C, 8D). More Importantly, we found that si-MRTO4 significantly inhibited ki67 expression and promoted ALDOB positive expression (P<0.05) (Figure 8E, 8F). In conclusion, these results indicated that knockdown of MRTO4 suppresses HCC progression by upregulating ALDOB in vivo (Figure 8G).
Figure 8.
Knockdown of MRTO4 suppressed HCC progression in vivo. (A) Tumor photograph. (B) The tumor was weighed. (C, D) TUNEL analysis of the number of positive cells in HCC tissue. (E, F) Immunohistochemical evaluation of Ki67 and ALDOB expression in HCC tissue. (G) Mechanism diagram.* P<0.05, ** P<0.01, *** P<0.001.
Discussion
MRTO4 is involved in various oncogenic processes, such as tumor cell proliferation, metastatic progression, tumor immune evasion, and modulation of the tumor microenvironment [19,34]. MRTO4 is upregulated in IGC tissues and exerts pro-proliferative and oncogenic functions [35]. MRTO4 is an important ribosome biogenesis protein, which regulate the sensitivity of tumors to HDACis inhibitors in solid tumors [36]. We downloaded and analyzed the expression levels of MRTO4 mRNA in different tumors and normal tissues from the TCGA using the R package. Our findings elucidate the significant role of MRTO4 in the pathogenesis of various cancers, particularly HCC. Across multiple tumor types, MRTO4 overexpression was consistently observed, with HCC demonstrating one of the most pronounced elevations. Delving deeper into HCC-specific findings, an upregulated MRTO4 was demonstrably associated with numerous clinical indices. Our data firmly establish that augmented MRTO4 levels had strong associations with parameters like advanced pathologic T stage, indicative of a more aggressive tumor phenotype. Similarly, an elevated histologic grade, a marker of tumor differentiation and invasiveness, was also correlated with increased MRTO4 expression. Notably, systemic factors such as a high body mass index (BMI) and high body weight were also linked to elevated MRTO4 levels, suggesting broader physiological implications. High MRTO4 expression was also associated with overall and disease-specific survival events, implying its prognostic potential. The consistent patterns identified across diverse datasets bolster the hypothesis that MRTO4 is a potentially valuable biomarker in diagnosis and prognosis of HCC. Importantly, this hypothesis has been adequately validated in our in vitro and in vivo experiments. We found that knockdown of MRTO4 significantly suppressed liver cancer cell proliferation, invasion, and tumor growth, whereas overexpression of MRTO4 reversed this trend. Given the profound clinical implications of these observations, it becomes paramount to further explore the precise mechanistic role of MRTO4 in tumor biology.
It is known that energy metabolism reprogramming is a hallmark of cancer. Even under sufficient oxygen conditions, tumor cells’ metabolism can reprogram from oxidative phosphorylation (OXPHOS) to aerobic glycolysis, which is known as the “Warburg effect” [37]. Studies have shown that tumor cells primarily obtain energy through aerobic glycolysis, meeting the requirements for proliferation and survival in adverse environments, ultimately leading to cancer progression [38]. Interestingly, in our study, we identified genes closely related to MRTO4 expression by using co-expression patterns obtained from the TCGA database. The 15 genes negatively associated with MRTO 4 were BTNL 9, TFR 2, TAT, SLC10A1, ALDOB, F9, FBP 1, CD300LG, SLC27A2, G6PC, CFHR 4, RBP 4, CLEC3B, APOC3, and HPX. Among them, ALDOB, TAT, SLC10A1, FBP 1, G6PC, and RBP 4 have been reported to be closely related with glycolysis. We found that these genes were notably involved in vital metabolic pathways, such as pentose phosphate pathway, fructose and mannose metabolism, and cysteine and methionine metabolism. These pathways are intimately related to glycolysis [39,40]. Aerobic glycolysis was first reported in hepatocellular carcinoma (HCC) in the 1920s, and it enhances the role of aerobic glycolysis in HCC. The proliferation, growth, and immune evasion of HCC are influenced by aerobic glycolysis [41,42]. There are 3 rate-limiting enzymes involved in aerobic glycolysis – hexokinase (HK), phosphofructokinase (PFK), and pyruvate kinase (PK). The expression of these enzymes can greatly impact the progression of HCC [5]. In this study, we investigated the possible regulatory effect of MRTO4 on glycolysis, considering the importance of Warburg effects as a marker of tumor metabolism in tumorigenesis and progression. We knocked down MRTO4 expression in hepatoma cells and found that low expression of MRTO4 significantly inhibited glycolysis in HCC cells, manifested by decreased ECAR, glucose consumption, lactate production and ATP levels, while the overexpressed MRTO4 largely reversed this the trend. These results reveal MRTO4 influences the progression of HCC by modulating glycolysis.
Moreover, the aberrant expression and regulation of related enzymes in the glycolytic pathway plays a crucial role in the development and recurrence of HCC [43]. For example, glucose transporter type 1 (GLUT1) expression was significantly upregulated in HCC tissues and was positively correlated with tumor size [44]. ZEB1 can stimulate glycolysis in hepatoma cells by upregulating PFKM (the rate-limiting enzyme in glycolysis), and then promotes the proliferation and metastasis of HCC [26]. ALDOB is an important member of the aldolase family that plays an important role in glycolysis. Loss of ALDOB has been reported to promote HCC through the release of inhibition of glucose-6-phosphate dehydrogenase (G6PD) [28], and to induce chemoresistance by remodeling glucose metabolism in glucose-PDAC [45]. In this study, we revealed that MRTO4 promotes glycolysis in HCC cells by downregulating ALDOB expression, and subsequently induces the proliferative and invasive phenotype of HCC cells.
Aberrant activation of Akt signaling promotes multiple cancer progression by regulating cell proliferation, metabolism, and survival, including HCC [46], and activated Akt enhances the expression and activity of several glycolytic enzymes such as glucose transporters, hexokinase, and phosphofructokinase, thus upregulating glucose uptake and glycolytic process [47,48]. Loss of ALDOB has been reported to activate Akt and promote hepatocellular carcinogenesis by destabilizing the ALDOB/Akt/PP2A protein complex [33]. We investigated whether MRTO4 affects Akt activation by modulating ALDOB. Consistently, here, we found that MRTO4 induces glycolysis in HCC cells by activating the Akt pathway through downregulation of ALDOB. We did not determine whether MRTO4 directly promotes Akt activation, and we did not assess specific molecular interactions between MRTO4, ALDOB, and Akt, which will be the focus of our subsequent work to improve the role of MRTO 4 in HCC progression by regulating glycolysis and related mechanisms.
Conclusions
Our study found that MRTO4 is an independent prognostic factor for overall survival in HCC. MRTO4 is highly expressed in HCC tissue samples and is closely associated with glycolytic pathways through bioinformatics analysis. Moreover, our results showed that MRTO4 can promote glycolysis to accelerate HCC progression by activating the Akt pathway through ALDOB downregulation. These results suggest that MRTO4 is a potential molecular therapeutic target and biomarker in HCC.
Abbreviations
- MRT4
Homolog, Ribosome Maturation Factor (MRTO4)
- HCC
hepatocellular carcinoma
- TCGA
The Cancer Genome Atlas
- AFP
α-fetoprotein
- GEO
Gene Expression Omnibus
- HR
hazard ratio
- PPI
protein–protein interactions
- DAVID
The Database for Annotation, Visualization and Integrated Discovery
- GO
gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OS
overall survival
- DEGs
differentially expressed genes
- ssGSEA
single-sample gene set enrichment analysis
- MF
molecular function
- BP
biological pathways
Footnotes
Conflict of interest: None declared
Declaration of Figures’ Authenticity: All figures submitted have been created by the authors who confirm that the images are original with no duplication and have not been previously published in whole or in part.
Financial support: This work was funded by the Special Fund Project for Science and Technology of Guangdong Province (No. 2020035)
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2021;71(3):209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Altekruse SF, Henley SJ, Cucinelli JE, McGlynn KA. Changing hepatocellular carcinoma incidence and liver cancer mortality rates in the United States. Am J Gastroenterol. 2014;109(4):542–53. doi: 10.1038/ajg.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. Cancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34:153–59. doi: 10.1053/j.semdp.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Feng J, Li J, Wu L, et al. Emerging roles and the regulation of aerobic glycolysis in hepatocellular carcinoma. J Exp Clin Cancer Res. 2020;39(1):126. doi: 10.1186/s13046-020-01629-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apostolo D, Ferreira LL, Vincenzi F, et al. From MASH to HCC: The role of Gas6/TAM receptors. Front Immunol. 2024;15:1332818. doi: 10.3389/fimmu.2024.1332818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7(1):11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharya D, Scimè A. Metabolic regulation of epithelial to mesenchymal transition: Implications for endocrine cancer. Front Endocrinol (Lausanne) 2019;10:773. doi: 10.3389/fendo.2019.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chelakkot C, Chelakkot VS, Shin Y, Song K. Modulating glycolysis to improve cancer therapy. Int J Mol Sci. 2023;24(3):2606. doi: 10.3390/ijms24032606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaupel P, Schmidberger H, Mayer A. The Warburg effect: Essential part of metabolic reprogramming and central contributor to cancer progression. Int J Radiat Biol. 2019;95(7):912–19. doi: 10.1080/09553002.2019.1589653. [DOI] [PubMed] [Google Scholar]
- 12.Gatenby RA, Gawlinski ET. The glycolytic phenotype in carcinogenesis and tumor invasion: Insights through mathematical models. Cancer Res. 2003;63(14):3847–54. [PubMed] [Google Scholar]
- 13.Chang YC, Tsai HF, Huang SP, et al. Enrichment of aldolase c correlates with low non-mutated IDH1 expression and predicts a favorable prognosis in glioblastomas. Cancers (Basel) 2019;11(9):1238. doi: 10.3390/cancers11091238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salvatore F, Izzo P, Paolella G. Aldolase gene and protein families: Structure, expression and pathophysiology. Horiz Biochem Biophys. 1986;8:611–65. [PubMed] [Google Scholar]
- 15.Oppelt SA, Sennott EM, Tolan DR. Aldolase-B knockout in mice phenocopies hereditary fructose intolerance in humans. Mol Genet Metab. 2015;114(3):445–50. doi: 10.1016/j.ymgme.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Kinoshita M, Miyata M. Underexpression of mRNA in human hepatocellular carcinoma focusing on eight loci. Hepatology. 2002;36(2):433–38. doi: 10.1053/jhep.2002.34851. [DOI] [PubMed] [Google Scholar]
- 17.Tao QF, Yuan SX, Yang F, et al. Aldolase B inhibits metastasis through Ten-Eleven Translocation 1 and serves as a prognostic biomarker in hepatocellular carcinoma. Mol Cancer. 2015;14:170. doi: 10.1186/s12943-015-0437-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. doi: 10.1016/s0378-1119(03)00629-2. [DOI] [PubMed] [Google Scholar]
- 19.Patel KD, Vora HH, Trivedi TI, et al. Transcriptome profiling and pathway analysis in squamous cell carcinoma of buccal mucosa. Exp Mol Pathol. 2020;113:104378. doi: 10.1016/j.yexmp.2020.104378. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Dong L, Cui H, et al. Up-regulation of mitochondrial antioxidation signals in ovarian cancer cells with aggressive biologic behavior. J Zhejiang Univ Sci B. 2011;12(5):346–56. doi: 10.1631/jzus.B1000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomczak K, Czerwińska P, Wiznerowicz M. The Cancer Genome Atlas (TCGA): An immeasurable source of knowledge. Contemp Oncol (Pozn) 2015;19(1A):A68–77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D12. doi: 10.1093/nar/gkaa1074. [Erratum in: Nucleic Acids Res. 2021;49(18):10800] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherman BT, Hao M, Qiu J, et al. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50(W1):W216–W21. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–D63. doi: 10.1093/nar/gkx1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–2. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y, Lin F, Wan T, et al. ZEB1 enhances Warburg effect to facilitate tumorigenesis and metastasis of HCC by transcriptionally activating PFKM. Theranostics. 2021;11(12):5926–38. doi: 10.7150/thno.56490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia W, Wu Q, Yu X, et al. Prognostic values of ALDOB expression and 18F-FDG PET/CT in hepatocellular carcinoma. Front Oncol. 2022;12:1044902. doi: 10.3389/fonc.2022.1044902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu G, Wang N, Zhang C, et al. Fructose-1,6-bisphosphate aldolase B depletion promotes hepatocellular carcinogenesis through activating insulin receptor signaling and lipogenesis. Hepatology. 2021;74(6):3037–55. doi: 10.1002/hep.32064. [DOI] [PubMed] [Google Scholar]
- 29.Chang YC, Yang YC, Tien CP, et al. Roles of aldolase family genes in human cancers and diseases. Trends Endocrinol Metab. 2018;29(8):549–59. doi: 10.1016/j.tem.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Li Z, Peng Y, Li J, et al. N6-methyladenosine regulates glycolysis of cancer cells through PDK4. Nat Commun. 2020;11(1):2578. doi: 10.1038/s41467-020-16306-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu X, Xu Q, Wan H, et al. PI3K-Akt-mTOR/PFKFB3 pathway mediated lung fibroblast aerobic glycolysis and collagen synthesis in lipopolysaccharide-induced pulmonary fibrosis. Lab Invest. 2020;100(6):801–11. doi: 10.1038/s41374-020-0404-9. [DOI] [PubMed] [Google Scholar]
- 32.Weng ML, Chen WK, Chen XY, et al. Fasting inhibits aerobic glycolysis and proliferation in colorectal cancer via the Fdft1-mediated AKT/mTOR/HIF1α pathway suppression. Nat Commun. 2020;11(1):1869. doi: 10.1038/s41467-020-15795-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.He X, Li M, Yu H, et al. Loss of hepatic aldolase B activates Akt and promotes hepatocellular carcinogenesis by destabilizing the Aldob/Akt/PP2A protein complex. PLoS Biol. 2020;18(12):e3000803. doi: 10.1371/journal.pbio.3000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michalec-Wawiorka B, Wawiorka L, Derylo K, et al. Molecular behavior of human Mrt4 protein, MRTO4, in stress conditions is regulated by its C-terminal region. Int J Biochem Cell Biol. 2015;69:233–40. doi: 10.1016/j.biocel.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 35.Zhang LH, Zhuo HQ, Hou JJ, et al. Proteomic signatures of infiltrative gastric cancer by proteomic and bioinformatic analysis. World J Gastrointest Oncol. 2022;14(11):2097–107. doi: 10.4251/wjgo.v14.i11.2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hao BB, Ma K, Xu JY, et al. Proteomics analysis of histone deacetylase inhibitor-resistant solid tumors reveals resistant signatures and potential drug combinations. Acta Pharmacol Sin. 2024 doi: 10.1038/s41401-024-01236-5. [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baik SH, Kang S, Lee W, et al. A breakdown in metabolic reprogramming causes microglia dysfunction in Alzheime’s disease. Cell Metab. 2019;30(3):493–507e6. doi: 10.1016/j.cmet.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Wu L, Jin Y, Zhao X, et al. Tumor aerobic glycolysis confers immune evasion through modulating sensitivity to T cell-mediated bystander killing via TNF-α. Cell Metab. 2023;35(9):1580–96e9. doi: 10.1016/j.cmet.2023.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Patel JH, Ong DJ, Williams CR, et al. Elevated pentose phosphate pathway flux supports appendage regeneration. Cell Rep. 2022;41(4):111552. doi: 10.1016/j.celrep.2022.111552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathew R, Iacobas S, Huang J, Iacobas DA. Metabolic deregulation in pulmonary hypertension. Curr Issues Mol Biol. 2023;45(6):4850–74. doi: 10.3390/cimb45060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–37. doi: 10.1038/nrc3038. [Erratum in: Nat Rev Cancer. 2011;11(8):618] [DOI] [PubMed] [Google Scholar]
- 42.Xu S, Herschman HR. A Tumor agnostic therapeutic strategy for hexokinase 1-Null/Hexokinase 2-positive cancers. Cancer Res. 2019;79(23):5907–14. doi: 10.1158/0008-5472.CAN-19-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan YT, Lin JF, Li T, et al. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond) 2021;41(2):109–20. doi: 10.1002/cac2.12108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun HW, Yu XJ, Wu WC, et al. GLUT1 and ASCT2 as predictors for prognosis of hepatocellular carcinoma. PLoS One. 2016;11(12):e0168907. doi: 10.1371/journal.pone.0168907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y, Tang S, Shi X, et al. Metabolic classification suggests the GLUT1/ALDOB/G6PD axis as a therapeutic target in chemotherapy-resistant pancreatic cancer. Cell Rep Med. 2023;4(9):101162. doi: 10.1016/j.xcrm.2023.101162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Manning BD, Toker A. AKT/PKB Signaling: Navigating the network. Cell. 2017;169(3):381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elstrom RL, Bauer DE, Buzzai M, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64(11):3892–99. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 48.Robey RB, Hay N. Is Akt the “Warburg kinase”?-Akt-energy metabolism interactions and oncogenesis. Semin Cancer Biol. 2009;19(1):25–31. doi: 10.1016/j.semcancer.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]