Abstract
Background:
Individuals living with human immunodeficiency virus (HIV) who experience virological failure (VF) after combination antiretroviral therapy (cART) initiation may have had low-frequency drug resistance mutations (DRMs) at cART initiation. There are no data on low-frequency DRMs among cART-naïve HIV-positive individuals in Botswana.
Methods:
We evaluated the prevalence of low-frequency DRMs among cART-naïve individuals previously sequenced using Sanger sequencing. The generated pol amplicons were sequenced by next-generation sequencing.
Results:
We observed low-frequency DRMs (detected at <20% in 33/103 (32%) of the successfully sequenced individuals, of whom four also had mutations detected at >20%. K65R was the most common low-frequency DRM detected in 8 individuals. Eighty-two of the 103 individuals had follow-up viral load data while on cART. Twenty-seven of the 82 individuals harbored low-frequency DRMs. Only 12 of 82 individuals experienced VF. The following low-frequency DRMs were observed in four individuals experiencing VF: K65R, K103N, V108I, and Y188C. No statistically significant difference was observed in the prevalence of low-frequency DRMs between individuals experiencing VF (4/12) and those not experiencing VF (23/70) (P = .97). However, individuals with non-nucleoside reverse transcriptase inhibitors-associated low-frequency DRMs were 2.68 times more likely to experience VF (odds ratio, 2.68; 95% confidential interval, 0.4–13.9) compared with those without (P = .22).
Conclusion:
Next-generation sequencing was able to detect low-frequency DRMs in this cohort in Botswana, but these DRMs did not contribute significantly to VF.
Keywords: antiretroviral naïve, HIV drug resistance, next-generation sequencing, low-frequency variants
1. Introduction
Treatment of human immunodeficiency virus (HIV) with combination antiretroviral therapy (cART) is life-long and has high success in suppressing HIV-1 replication.[1,2] However, cART success can be negatively impacted by the emergence of HIV drug resistance mutations (DRMs), leading to virological failure (VF).[3–6] Dolutegravir (DTG)-based regimens have been adopted as the preferred first-line treatment for HIV, replacing non-nucleoside reverse transcriptase (RT) inhibitor (NNRTI)-based regimens.[7,8] However, concerns have emerged about the safety of DTG[9] and its reduced efficacy in patients with DRMs in RT.[10] Furthermore, some countries are opting to continue using efavirenz (EFV)-based first-line cART, especially in patients receiving tuberculosis (TB) treatment due to the risk of increased DTG metabolism, leading to subtherapeutic concentrations.[11,12] With expanded access to cART, there is increased potential for the development and transmission of drug-resistant HIV variants, leading to treatment failure.[13] Population-based Sanger sequencing is widely used in HIV drug resistance testing.[14] However, population-based Sanger sequencing only detects the most dominant viral variants (>20% of the viral quasispecies) and is unable to detect low-frequency DRMs (minority variants).[15,16] Next-generation sequencing (NGS) allows for the effective detection of low-frequency DRMs as low as 1% of the viral population[17–23] and can be cost-effective by using the pooling strategy, which pools index samples into a single library.[24–26]
Previous studies[27–39] have reported that individuals initiating cART with preexisting HIV low-frequency DRMs have a higher likelihood of VF, particularly among those initiating NNRTIs. It has also been reported that individuals with preexisting low-frequency DRMs at baseline have the same DRMs at the time of VF.[40,41] However, other studies have found no association between baseline low-frequency DRMs and VF on cART.[40,42,43]
Conflicting results on the clinical importance of low-frequency DRMs indicate the need for further investigations on the clinical impact of low-frequency DRMs in different settings.
We sought to determine the prevalence of low-frequency DRMs among cART-naïve HIV-positive individuals in Botswana and to assess the impact of baseline pretreatment low-frequency DRMs on VF outcomes once the participants initiated cART.
2. Materials and Methods
2.1. Study design and study population
This was a retrospective longitudinal study aimed at determining the prevalence and impact of low-frequency HIV DRMs in baseline samples of antiretroviral naïve individuals in Botswana. The amplicons used in this study were obtained from a previous Botswana Harvard AIDS Partnership study (BHP063- with protocol title: A novel strategy for HIV drug resistance monitoring in developing countries).[44] Participants were recruited for the study from antenatal clinics and Infectious Disease Care Clinics in 3 different locations in Botswana: Gaborone, Molepolole, and Mochudi. BHP063 enrolled 443 participants between April 2012 and April 2015 and were included in the primary study analysis.[44] An additional 88 participants were enrolled between May 2015 and December 2015, resulting in a total of 531. Before 2016, the standard first-line of ART initiation used included the combination of EFV, tenofovir disoproxil fumarate (TDF), and emtricitabine (FTC) (ATRIPLA) (Gilead Sciences), and there were no patients on DTG-based ART before then. As of June 2016, Botswana adopted DTG-based ART (DTG-TDF-FTC) for all HIV-infected individuals regardless of CD4+ T-cell count or pregnancy status. Most of these participants were probably infected through heterosexuals given their age at the time of enrollment, but mother-to-child transmission cannot be ruled out. Genotyping results for participants in the main cohort who previously developed resistance mutations were communicated to clinicians, and patients were assigned treatment based on baseline mutation status. This would impact their treatment. Of the 531 enrolled in BHP063, 108 (20.3%) participants with available stored HIV-1 RT/PR amplicons and available Sanger sequencing data were included in the present study. Four hundred and twenty-three (423) participants without amplicons were excluded. The characteristics of the included and excluded individuals were compared (Table S1, Supplemental Digital Content, http://links.lww.com/MD/G927). Follow-up clinical, virological, and demographic data for individuals with available amplicons were extracted from the electronic laboratory information system-IPMS (Integrated patient management system). We defined virological suppression as viral load (VL) <400 copies/mL and VF as VL of ≥400 copies/mL at 6 months after initiation of cART as per the Botswana Ministry of Health and Wellness guidelines.
2.2. Ribonucleic acid extraction, polymerase chain reaction amplification, and Sanger sequencing
The HIV-1 pol (RT/PR) amplicons were initially generated in a previous study.[44] Briefly, ribonucleic acid (RNA) was extracted from 400 ul plasma samples using an EZ1 Virus Mini Kit v2.0 (Qiagen, Valencia, CA) on an EZ1 Advanced XL (Qiagen) automated instrument. The RT and protease (PR) regions of the HIV-1 pol gene were amplified, and polymerase chain reaction (PCR) products were purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. DNA sequencing of PCR products was performed using BigDye Terminator chemistry on an ABI 3130XL genetic analyzer (Thermo Fisher Scientific, Carlsbad, CA), as previously described.[44] The residual amplicons were stored at −20°C.
2.3. Next-generation sequencing and drug resistance analysis
NGS was conducted at KwaZulu-Natal Research Innovation and Sequencing Platform, Durban, South Africa, and Inqaba Biotechnical Industries, Pretoria, South Africa, using the Illumina MiSeq platform (Illumina, San Diego, CA). Briefly, PCR product concentrations were determined using a Qubit 3.0 fluorometer (Thermo Fisher, Malaysia). Paired-end libraries were generated using the Nextera-XT DNA library preparation kit and Nextera Index kit (Illumina, San Diego, CA), according to the manufacturer’s instructions. Sequencing libraries were purified using Agencourt AMPure XP beads, and quantified, and barcoded libraries were pooled for sequencing on an Illumina MiSeq platform. The generated raw reads (FastQ files) were assembled into contigs using online genome detection tools.[45] NGS sequences were uploaded to the online variant caller polymorphism analysis sequencing (PASeq).[46] HyDRA was also used to confirm the minor variants.[23] Any variants not called by either caller were assumed to have a 0% allele frequency. Low-frequency DRMs were detected at >1% using Geneious software v8.1.9 (Biomatters Ltd, Auckland, New Zealand).[47]
2.4. Statistical analysis
HIV drug resistance was determined based on NGS with detection thresholds of 1%, 5%, 10%, and 20%. Data are presented as medians and interquartile ranges. The demographic characteristics (age and sex) and clinical characteristics (baseline CD4+ T-cell count and baseline VL) of individuals with and without pretreatment low-frequency DRMs were compared using the Wilcoxon rank-sum test and Fisher’s exact test (for continuous and categorical variables). We excluded individuals without follow-up VL data from the analysis to determine the impact of pretreatment low-frequency DRMs on VF. We further excluded pretreatment DRMs detected at ≥20% of mutations. We used a univariate exact logistic regression model[48,49] to assess the association between pretreatment low-frequency DRMs and VF. Odds ratios (OR) were used to describe the association between low-frequency DRMs and VF. We used R version 4.0.3 for statistical analysis. Differences were considered statistically significant at P < .05.
2.5. Ethics
BHP063 study was approved by the Research Ethics Committee of the Ministry of Health and Welfare (HRDC # 00638). In addition, ethical approval was obtained from the Institutional Review Board of the University of Botswana (Reference number: HPDME 13/18/1 Vol 833). Participants provided informed consent for the reuse and storage of their samples for further research.
3. Results
3.1. Participants characteristics
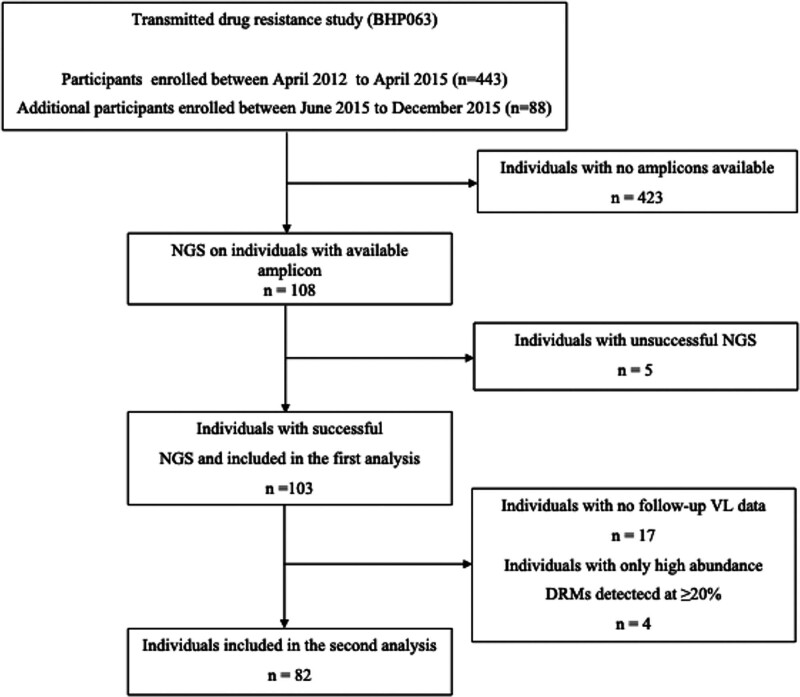
A total of 103 out of 108 samples were successfully sequenced using Illumina MiSeq NGS (Fig. 1) to a mean depth of 20534 reads (min–max: 1849–159407). At baseline, the median VL was 4.1 log10 copies/mL and the median CD4+ T-cell count was 365 cells/mm3. Table 1 summarizes the demographic and clinical characteristics of the participants.
Figure 1.
Flow chart showing individuals included in the analysis. The first analysis focused on determining the prevalence of low-frequency drug resistance mutations. The second analysis only used participants with follow-up viral load data to assess the impact of low-frequency drug resistance mutations on virological outcomes. DRM, drug resistance mutations; NGS, next-generation sequencing.
Table 1.
Baseline characteristics of participants included in the study.
| Characteristics | Participants (n = 108) |
|---|---|
| Age in years, median (IQR) | 27.0 (24–31) |
| Female, n (%) | 107 (99.1%) |
| Male, n (%) | 1 (0.9) |
| VL (log10 copies/mL), median (IQR) | 4.1 (3.5–4.6) |
| CD4+ T-cell count (cells/mm3), median (IQR) | 365 (225–497) |
IQR = interquartile ranges: 25th percentile and 75th percentile; VL, viral load.
Of the 108 samples with available amplicon, 26 individuals had to be excluded from the second analysis of the study because of either unsuccessful NGS, presence of baseline pretreatment DRMs only, or no follow-up VL data (Fig. 1). Demographic characteristics of individuals included (n = 108) and excluded (n = 423) from the main cohort were summarized, and there was no significant difference in terms of age and VL between the individuals included and those excluded, but a significant difference was observed in sex and CD4 (Table S1, Supplemental Digital Content, http://links.lww.com/MD/G927).
3.2. HIV drug resistance mutations detected in protease and reverse transcriptase
Eight (7.8%) of the 108 individuals with successful NGS sequencing had at least 1 DRM detected at >20% frequency. Seven of the 8 sequences revealed single-class resistance (1 nucleoside RT inhibitor (NRTI), two NNRTI, and four PR inhibitors (PIs), whereas the other harbored both NRTI and NNRTI resistance. The most common mutation was K103N, which was detected in 3/8 sequences. PASeq and HyDRA detected all DRMs (>20% frequency), as determined by Sanger sequencing. The 2 analysis pipelines (PASeq and HyDRA) had a good agreement above 20% and 5% thresholds but gave highly discrepant results around a 1% threshold (Table S2, Supplemental Digital Content, http://links.lww.com/MD/G927). The variants called by PASeq were used to represent the NGS results for subsequent analyses.
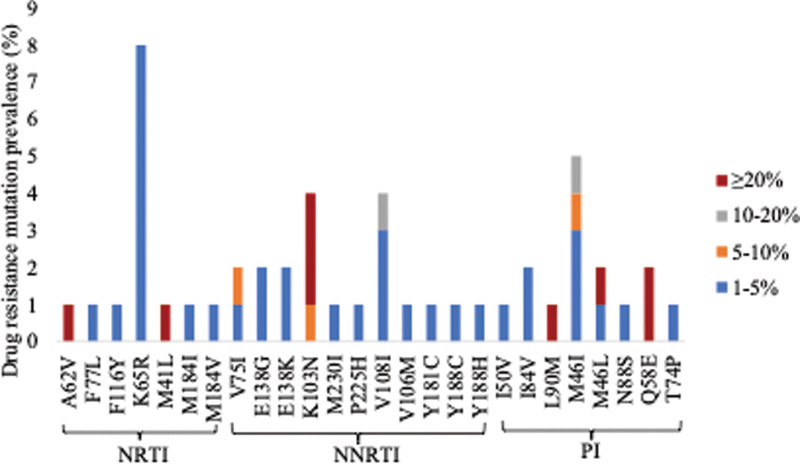
Thirty-three individuals (32.0%) had at least 1 low-frequency DRMs (1%–20% frequency). Four of the 33 participants also had DRMs at a ≥20% threshold (Fig. 2 and Table 2). Among the 33 individuals, NRTI-associated low-frequency DRMs were detected in 12 individuals. In addition, 2 individuals harbored both NRTI-and NNRTI-associated low-frequency DRMs. The most common NRTI-associated low-frequency DRM was K65R, occurring in 8 individuals with frequencies between 1% and 2.96%. Other NRTI-associated low-frequency DRMs identified were V75I, F77L, F116Y, M184I, and M184V at frequencies from 2.43% to 4.06%, respectively. NNRTI-associated low-frequency DRMs were found in 8 individuals. In addition, 2 individuals had both NNRTI-and PI-associated low-frequency DRMs. The most common NNRTI-associated low-frequency DRM was V108I, detected in 3 individuals with frequencies between 1.15% and 11.25%. Other NNRTI-associated low-frequency DRMs identified were K103N, V106M, E138G, E138K, Y181C, Y188C, Y188H, P225H, and M230I at frequencies of 1.01% and 7.7%, respectively. PI-associated low-frequency DRMs were found in 9 individuals, with the most common being M46I found in 5 individuals. Other PI-associated low-frequency DRMs identified were M46L, I50V, T74P, I84V, and N88S at frequencies of 1.13% and 3.38%, respectively.
Figure 2.
Baseline HIV drug resistance mutations detected at different thresholds. Mutations detected at 1 to <20% represent low-frequency mutations detected by next-generation sequencing only. Mutations detected at ≥20% represent mutations detected by both Sanger sequencing and next-generation sequencing. NRTI, Nucleoside reverse-transcriptase inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor; PI, Protease inhibitor.
Table 2.
Drug class mutations observed by NGS at different mutation thresholds.
| Mutations detected | Detection threshold | ||||
|---|---|---|---|---|---|
| 1%–2% | >2%–5% | >5%–10% | >10%–<20% | ≥20% | |
| NRTI-associated | |||||
| M41L | 0 | 0 | 0 | 0 | 1 |
| A62V | 0 | 0 | 0 | 0 | 1 |
| K65R | 6 | 2 | 0 | 0 | 0 |
| V75I | 0 | 1 | 1 | 0 | 0 |
| F77L | 0 | 1 | 0 | 0 | 0 |
| F116Y | 0 | 1 | 0 | 0 | 0 |
| M184I | 0 | 1 | 0 | 0 | 0 |
| M184V | 0 | 1 | 0 | 0 | 0 |
| Total | 6 | 7 | 1 | 0 | 2 |
| NNRTI-associated | |||||
| K103N | 0 | 0 | 1 | 0 | 2 |
| V106M | 1 | 0 | 0 | 0 | 0 |
| V108I | 1 | 1 | 0 | 1 | 0 |
| E138G | 1 | 1 | 0 | 0 | 0 |
| E138K | 1 | 1 | 0 | 0 | 0 |
| Y181C | 1 | 0 | 0 | 0 | 0 |
| Y188C | 0 | 1 | 0 | 0 | 0 |
| Y188H | 1 | 0 | 0 | 0 | 0 |
| P225H | 1 | 0 | 0 | 0 | 0 |
| M230I | 1 | 0 | 0 | 0 | 0 |
| Total | 8 | 4 | 1 | 1 | 2 |
| PI-associated | |||||
| M46I | 2 | 2 | 0 | 1 | 0 |
| M46L | 0 | 1 | 0 | 0 | 0 |
| I50V | 1 | 0 | 0 | 0 | 0 |
| T74P | 1 | 0 | 0 | 0 | 0 |
| I84V | 1 | 1 | 0 | 0 | 0 |
| N88S | 0 | 1 | 0 | 0 | 0 |
| Q58E | 0 | 0 | 0 | 0 | 2 |
| Total | 5 | 5 | 1 | 1 | 2 |
NGS = next-generation sequencing; NRTI = nucleoside reverse-transcriptase inhibitor; NNRTI = non-nucleoside reverse transcriptase inhibitor; PI = Protease inhibitor. Mutations detected at >20% were also detected by Sanger sequencing.
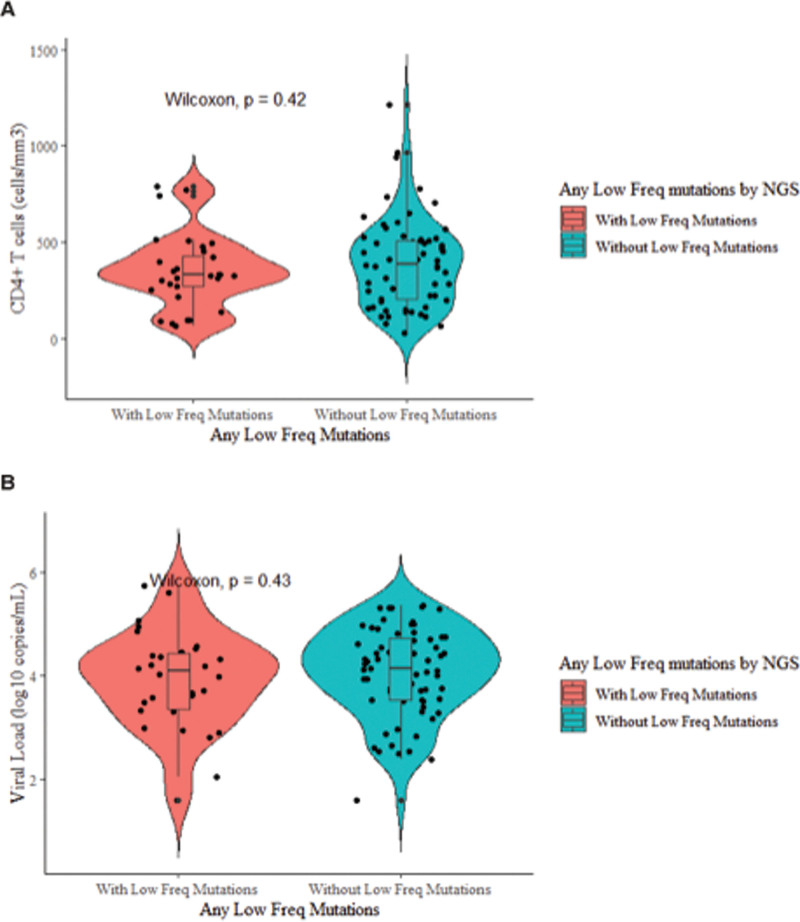
The median CD4+ T cell counts for individuals with (n = 33) and without (n = 70) low-frequency DRMs were 331 cell/mm3 (IQR, 241–429.5) and 365 cell/mm3 (IQR, 227–495), respectively (Fig. 3). In addition, the median VL was 4.2 log10 copies/mL (IQR, 3.6–4.6) and 4.1 log10 copies/mL (IQR, 3.5–4.6), respectively. There was no difference in the VLs (P = .43) and CD4+ T cell counts (P = .42) between individuals with low-frequency DRMs and individuals without low-frequency DRMs.
Figure 3.
The relationship between the CD4+ T-cell count, viral load and low frequency drug resistance mutations. (A) CD4+ T-cell and (B) viral loads in individuals with low-frequency DRMs and individuals without low-frequency DRMs. Low-frequency DRMs are mutations detected at <20%. DRM, drug resistance mutation. NGS, next-generation sequencing.
3.3. Virological outcome
To determine the impact of low-frequency DRMs, we excluded four individuals with only DRMs detected at ≥20% threshold from further analysis. Seventeen individuals without follow-up VL data were excluded from further analysis. A total of 82 individuals had at least 1 follow-up VL data and were used to investigate the association between baseline low-frequency DRMs and VF (Table 3). Sixty-one of the followed-up participants initiated on ATRIPLA (TDF + FTC + EFV) based regimen while 18 initiated on DTG based regimen (Table S3, Supplemental Digital Content, http://links.lww.com/MD/G927). Twenty-seven of the 82 individuals had low-frequency DRMs, whereas 55 individuals did not. Of the 12 individuals experiencing VF, 4 had low-frequency DRMs. Nine of the 12 individuals initiated an Atripla-based regimen and three individuals initiated on DTG based regimen (Table S3, Supplemental Digital Content, http://links.lww.com/MD/G927). Low-frequency DRMs detected in individuals experiencing VF were NRTI-associated, such as K65R and NNRTI-associated K103N, V108I, and Y188C (Table 4).
Table 3.
Virological outcome in individuals with and without low-frequency DRMs.
| n | Individuals with low-frequency DRMs (%) | P-value | |
|---|---|---|---|
| Individuals experiencing VF | 12 | 4 (33.3) | .97 |
| Individuals not experiencing VF | 70 | 23 (32.9) |
DRMs = drug resistance mutations; VF = virological failure.
Table 4.
DRMs observed in individuals experiencing virological failure with low frequency drug resistance mutations.
| Sample identification | Mutation frequency level (%) within the viral | |
|---|---|---|
| NRTI | NNRTI | |
| P26 | K65R (2.9%) | 0 |
| P29 | 0 | V108I (1.15%) |
| P51 | 0 | Y188C (2.2%) |
| P53 | 0 | K103N (7.8%) |
NRTI = nucleoside reverse-transcriptase inhibitor; NNRTI = non-Nucleoside reverse transcriptase inhibitor; PI = Protease inhibitor.
Individuals with NNRTI low-frequency DRMs were 2.68 times more likely to experience VF (OR, 2.68; 95% confidential interval, 0.4–13.9) compared with those without NNRTI low-frequency DRM’s.
4. Discussion
HIV-1 VF due to the presence or development of HIV variants harboring DRMs remains a major challenge for the success of cART. There are conflicting data on the significance of pretreatment low-frequency HIV DRMs. We report the first study from Botswana that evaluated NGS-defined low-frequency DRMs in antiretroviral naïve individuals. NGS identified all DRMs at levels ≥20% that were detected by Sanger sequencing. The prevalence drastically increased when low-frequency DRMs of 1% were included. Low-frequency DRMs were found in 33 individuals with K65R being the most common low-frequency mutation detected in 8 individuals, and these findings are similar to those reported by others.[40,50,51] The K65R mutation is the most important TDF resistance mutation, which makes it a relevant mutation to the current WHO recommended first-line regimen and may compromise its effectiveness.[52] DRMs detected at levels <20% of viral quasi-species were not detected by Sanger sequencing. The K103N mutation was found in 3 individuals above 5% frequency, and this mutation is known to be highly selected by EFV.[52] The presence of NNRTI DRMs has been suggested to be negatively associated with long-term virologic outcomes of both EFV- and DTG-based first-line ART.[10] Therefore, it is important to continue assessing and monitoring pretreatment mutations in antiretroviral naïve patients. It is also vital to continue assessing NNRTI low-frequency DRMs to ensure optimization of treatment regimens for individuals living with HIV/TB for whom an EFV-based ART may be more appropriate. It has been indicated that there is a difference in CD4 and VL between individuals with low-frequency DRMs and those without low-frequency DRMs.[50] Studies have reported successful amplification and characterization of plasma HIV-1 RNA sequences in patients with VLs below 50 copies/mL.[53–55] A similar finding was also reported in a study conducted in Botswana that used samples with undetectable VLs to determine the prevalence of DRMs.[56] In our study, two samples with VLs of <40 copies/mL were successfully sequenced by Sanger sequencing and NGS. Participants were divided into two groups depending on whether they had low-frequency DRMs. The median VLs did not differ significantly between the group with and without low-frequency DRMs (4.2 log10 copies/mL vs 4.1 log10 copies/mL), and these findings are consistent with the results reported by Melanie et al.[57]
Three of the 4 individuals experiencing VF harbored the most common NNRTI mutations (K103N, V108I, and Y188C), which have been shown to occur more frequently in participants experiencing VF.[52] Individuals with low-frequency NNRTIs associated DRMs were 2.68 times more likely to experience VF (OR, 2.68; 95% confidential interval, 0.4–13.9) compared with those without (P = .22) although not statistically significant. This is not unexpected, as it was recently found that the detection of low-frequency DRMs alone was not associated with virologic failure in a South African cohort where HIV-1C also predominates.[40] In a South African study, inclusion of DRMs at >20% to low-frequency DRMs was associated with an increased prediction of VF. In our study, we did not include mutations detected at >20% in our second analysis because genotyping results were communicated in real-time to treat clinicians for appropriate patient management, which might have affected the outcome of our study. It has been previously reported that linked dual-class resistance mutations occurring in a single genome are associated with an increased risk of VF.[58] Since we were not able to analyze viral variants at the single genome level, we could not determine if there was an association of linked dual-class mutations with VF. This study was a retrospective study, and while there were efforts to avoid selection bias, the use of available samples, although it was not intentional. A significant difference was observed in the included versus the excluded in terms of sex and CD4 suggesting that there was some difference between the original cohort and the samples selected for this analysis. Here, we used a threshold of ≥1% frequency, which might have overestimated the prevalence of some mutations, especially K65R; however, the presence of genuine pretreatment DRMs occurring at ≥1% frequency cannot be ruled out.
A limitation of this study was the absence of resistance data at the time of VF. Moreover, we defined VF as a single VL of at least 400 copies/mL, whereas previous studies have used a cutoff of 1000 copies/mL.[51,59] We attempted to amplify samples with low VLs as low as <40 copies/mL, and only 2 samples were successfully amplified. The results should be interpreted with caution, as mutations cannot be reliably detected in such low copies of the VL. Most participants in this study later initiated cART on EFV, a drug that is no longer used in first-line cART therapy in Botswana, so studies investigating the impact of low-frequency DRMS on the current DTG-based first-line regimen are warranted.
In conclusion, the results presented in this study showed that antiretroviral naïve individuals had a high prevalence of low-frequency DRMs that did not have an impact on virologic suppression once they initiated ART. Future studies will need to focus on the role of low-frequency DRMs in DTG-based first-line regimens and to determine the prevalence of low-frequency linked dual-class resistance mutations, which have been found to be more associated with VF.
Author contributions
Conceptualization: Dorcas Maruapula, Christopher F. Rowley, Melvin Leteane, Sikhulile Moyo, and Simani Gaseitsiwe.
Formal analysis: Dorcas Maruapula, Jennifer Giandhari, Kesaobaka Molebatsi, Olorato Morerinyane, Sikhulile Moyo, and Simani Gaseitsiwe.
Funding acquisition: Dorcas Maruapula, Sikhulile Moyo, and Simani Gaseitsiwe.
Investigation: Dorcas Maruapula, Kaelo K. Seatla, and Jennifer Giandhari.
Methodology: Dorcas Maruapula, Christopher F. Rowley, Jennifer Giandhari, Melvin Leteane, Sikhulile Moyo, and Simani Gaseitsiwe.
Project administration: Dorcas Maruapula.
Resources: Dorcas Maruapula, Christopher F. Rowley, Sikhulile Moyo, Simani Gaseitsiwe.
Supervision: Christopher F. Rowley, Melvin Leteane, Sununguko W. Mpoloka, Sikhulile Moyo, and Simani Gaseitsiwe.
Visualization: Dorcas Maruapula, Sikhulile Moyo, and Simani Gaseitsiwe.
Writing—original draft: Dorcas Maruapula, Christopher F. Rowley, Jennifer Giandhari, Kaelo K. Seatla, Kesaobaka Molebatsi, Olorato Morerinyane, Melvin Leteane, Sununguko W. Mpoloka, Sikhulile Moyo, and Simani Gaseitsiwe.
Writing—review and editing: Dorcas Maruapula, Christopher F. Rowley, Jennifer Giandhari, Kaelo K. Seatla, Kesaobaka Molebatsi, Olorato Morerinyane, Melvin Leteane, Sununguko W. Mpoloka, Rosemary M. Musonda, Tulio de Oliveira, Sikhulile Moyo, and Simani Gaseitsiwe.
Acknowledgments
The authors would like to acknowledge the study participants, principal investigators, and study coordinators from the novel strategy completed the study. We would also like to extend our acknowledgements to the University of Botswana and the Botswana Harvard HIV Reference Laboratory for their support and contribution to the study’s success. NGS sequence analysis was performed with the assistance of the KwaZulu Natal Research Sequencing Platform and Inqaba biotechnology.
Supplementary Material
Abbreviations:
- ANCs =
- antenatal clinics
- BHP =
- Botswana Harvard AIDS Partnership
- cART =
- combination antiretroviral therapy
- DRMs =
- drug resistance mutations
- DTG =
- dolutegravir
- EFV =
- efavirenz
- FTC =
- emtricitabine
- HIV =
- human immunodeficiency virus
- IDCC =
- infectious disease care clinics
- KRISP =
- Kwazulu-Natal Research Innovation and Sequencing Platform
- NGS =
- next generation sequencing
- NNRTI =
- non-nucleoside reverse transcriptase inhibitors
- NRTI =
- nucleoside reverse transcriptase inhitors
- OR =
- odds ratio
- PASeq =
- polymorphism analysis sequencing
- PCR =
- polymerase chain reaction
- PI =
- protease inhibitors
- PR =
- protease
- RNA =
- ribonucleic acid
- RT =
- reverse transcriptase
- TB =
- tuberculosis
- TDF =
- tenofovir disoproxil fumarate
- VF =
- virological failure
- VL =
- viral load
How to cite this article: Maruapula D, Seatla KK, Morerinyane O, Molebatsi K, Giandhari J, de Oliveira T, Musonda RM, Leteane M, Mpoloka SW, Rowley CF, Moyo S, Gaseitsiwe S. Low-frequency HIV-1 drug resistance mutations in antiretroviral naïve individuals in Botswana. Medicine 2022;101:28(e29577).
This work was supported by the Sub-Saharan African Network for TB/HIV Research Excellence (SANTHE), a DELTAS Africa Initiative (grant # DEL-15-006). The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Sciences (AAS)’s Alliance for Accelerating Excellence in Science in Africa (AESA) and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (grant # 107752/Z/15/Z) and the UK government. The views expressed in this publication are those of the author(s) and not necessarily those of AAS, NEPAD Agency, Wellcome Trust, or the UK government. CFR was funded by NIH/NIAIDS (R01 AI089350). This work was also supported by H3ABioNet. H3ABioNet is supported by the National Institutes of Health Common Fund (U41HG006941). H3ABioNet is an initiative of the Human Health and Heredity in Africa Consortium (H3Africa) program of the African Academy of Science (AAS). Dorcas Maruapula was partially supported by the Fogarty International Center (Grant # 5D43TW009610). Kaelo K. Seatla was partially supported by Harvard University Center for AIDS Research (CFAR), an NIH-funded program (P30 AI060354), which is supported by the following NIH Co-Funding and Participating Institutes and Centers: National Institute of Allergy and Infectious Diseases, National Cancer Institute, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, National Institute on Aging, National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of General Medical Sciences, National Institute on Minority Health and Health Disparities, National Institute of Dental and Craniofacial Research, Office of AIDS Research, and Fogarty International Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Kaelo K. Seatla was partially supported by the Fogarty International Center and National Institute of Mental Health, National Institutes of Health under Award Number D43 TW010543. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This study was conducted according to the guidelines of the Declaration of Helsinki.
In the parent study, participants provided written informed consent to reuse and store their samples for further research. Next-generation sequencing was performed on stored leftover amplicons and combined with routine follow-up data.
Data are contained within the article. Sequence data have been deposited in NCBI GenBank under the sequential accession numbers MZ615642-MZ615654 and MZ678268-MZ678357. Raw reads will be available upon request.
Supplemental Digital Content is available for this article.
The authors have no conflicts of interest to disclose.
Contributor Information
Dorcas Maruapula, Email: dmaruapula@gmail.com.
Kaelo K. Seatla, Email: kaeloseatlamd@yahoo.com.
Olorato Morerinyane, Email: oloratomorerinyane@gmail.com.
Kesaobaka Molebatsi, Email: KesaMolebatsi@gmail.com.
Jennifer Giandhari, Email: jennifer.giandhari@gmail.com.
Tulio de Oliveira, Email: tuliodna@gmail.com.
Rosemary M. Musonda, Email: rmusonda@gmail.com.
Melvin Leteane, Email: melvin.leteane@gmail.com.
Sununguko W Mpoloka, Email: mpoloka@ub.ac.bw.
Christopher F. Rowley, Email: crowley1@bidmc.harvard.edu.
Sikhulile Moyo, Email: smoyo@bhp.org.bw.
References
- [1].Collaboration HI-C. The effect of combined antiretroviral therapy on the overall mortality of HIV-infected individuals. AIDS. 2010;24:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Weissberg D, Mubiru F, Kambugu A, et al. Ten years of antiretroviral therapy: incidences, patterns and risk factors of opportunistic infections in an urban Ugandan cohort. PLoS One. 2018;13:e0206796e0206796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gupta RK, Jordan MR, Sultan BJ, et al. Global trends in antiretroviral resistance in treatment-naive individuals with HIV after rollout of antiretroviral treatment in resource-limited settings: a global collaborative study and meta-regression analysis. Lancet. 2012;380:1250–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Stadeli KM, Richman DD. Rates of emergence of HIV drug resistance in resource-limited settings: a systematic review. Antivir Ther. 2013;18:115115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ávila-Ríos S, García-Morales C, Matías-Florentino M, et al. Pretreatment HIV-drug resistance in Mexico and its impact on the effectiveness of first-line antiretroviral therapy: a nationally representative 2015 WHO survey. Lancet HIV. 2016;3:e579–91. [DOI] [PubMed] [Google Scholar]
- [6].Naziri H, Baesi K, Moradi A, et al. Antiretroviral drug resistance mutations in naïve and experienced patients in Shiraz, Iran, 2014. Arch Virol. 2016;161:2503–9. [DOI] [PubMed] [Google Scholar]
- [7].WHO, World Health organization. Progress Report 2016: Prevent HIV, Test and Treat All: WHO Support for Country Impact. [access date November 27, 2019]. 2016. World Health Organization.
- [8].Mo HW, Handbook of the Botswana 2016 integrated HIV Clinical Care Guidelines. 2019. [accessed date 27 November 2019].
- [9].Zash R, Jacobson DL, Diseko M, et al., Comparative safety of dolutegravir-based or efavirenz-based antiretroviral treatment started during pregnancy in Botswana: an observational study. Lancet Glob Health. 2018;6: e804–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Siedner MJ, Moorhouse MA, Simmons B, et al. Reduced efficacy of HIV-1 integrase inhibitors in patients with drug resistance mutations in reverse transcriptase. Nat Commun. 2020;11:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].World Health Organization. Transition to New Antiretroviral Drugs in HIV Programmes: Clinical and Programmatic Considerations. World Health Organization, 2017. Available at: http://apps.who.int/iris/bitstream/handle/10665/255888/WHO-HIV-2017.20-eng.pdf;jsessionid=F4224B861D28C35C173E54829631585F?sequence=1. [Google Scholar]
- [12].Maartens G, Boffito M, Flexner CW. Compatibility of next-generation first-line antiretrovirals with rifampicin-based antituberculosis therapy in resource limited settings. Curr Opin HIV AIDS. 2017;12:355–8. [DOI] [PubMed] [Google Scholar]
- [13].Hamers RL, Sigaloff KCE, Kityo C, et al. Emerging HIV-1 drug resistance after roll-out of antiretroviral therapy in sub-Saharan Africa. Curr Opin HIV AIDS. 2013;8:19–26. [DOI] [PubMed] [Google Scholar]
- [14].Paredes R, Clotet B. Clinical management of HIV-1 resistance. Antiviral Res. 2010;85:245–65. [DOI] [PubMed] [Google Scholar]
- [15].Mohamed S, Penaranda G, Gonzalez D, et al. Comparison of ultra-deep versus Sanger sequencing detection of minority mutations on the HIV-1 drug resistance interpretations after virological failure. AIDS. 2014;28:1315–24. [DOI] [PubMed] [Google Scholar]
- [16].Tzou PL, Ariyaratne P, Varghese V, et al. Comparison of an in vitro diagnostic next-generation sequencing assay with sanger sequencing for HIV-1 genotypic resistance testing. J Clin Microbiol. 2018;56:e00105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Samuel R, Paredes R, Parboosing R, et al. Minority HIV-1 drug-resistant mutations and prevention of mother-to-child transmission: perspectives for resource-limited countries. AIDS Rev. 2014;16:187–98. [PubMed] [Google Scholar]
- [18].Beerenwinkel N, Zagordi O. Ultra-deep sequencing for the analysis of viral populations. Curr Opin Virol. 2011;1:413–8. [DOI] [PubMed] [Google Scholar]
- [19].Lapointe HR, Dong W, Lee GQ, et al. HIV drug resistance testing by high-multiplex “wide” sequencing on the MiSeq instrument. Antimicrob Agents Chemother. 2015;59:6824–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tzou PL, Ariyaratne P, Varghese V, et al. Comparison of an in vitro diagnostic next-generation sequencing assay with Sanger sequencing for HIV-1 genotypic resistance testing. J Clin Microbiol. 2018;56:e00105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nicot F, Jeanne N, Raymond D, et al. Performance comparison of deep sequencing platforms for detecting HIV-1 variants in the pol gene. J Med Virol. 2018;90:1486–92. [DOI] [PubMed] [Google Scholar]
- [22].Dudley DM, Bailey AL, Mehta SH, et al. Cross-clade simultaneous HIV drug resistance genotyping for reverse transcriptase, protease, and integrase inhibitor mutations by Illumina MiSeq. Retrovirology. 2014;11:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Taylor T, Lee ER, Nykoluk M, et al. A MiSeq-HyDRA platform for enhanced HIV drug resistance genotyping and surveillance. Sci Rep. 2019;9:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Quiñones-Mateu ME, Avila S, Reyes-Teran G, et al. Deep sequencing: becoming a critical tool in clinical virology. J Clin Virol. 2014;61:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Casadellà M, Paredes R. Deep sequencing for HIV-1 clinical management. Virus Res. 2017;239:69–81. [DOI] [PubMed] [Google Scholar]
- [26].Inzaule SC, Ondoa P, Peter T, et al. Affordable HIV drug-resistance testing for monitoring of antiretroviral therapy in sub-Saharan Africa. Lancet Infect Dis. 2016;16:e267–75. [DOI] [PubMed] [Google Scholar]
- [27].Vandenhende M-A, Bellecave P, Recordon-Pinson P, et al. Prevalence and evolution of low frequency HIV drug resistance mutations detected by ultra deep sequencing in patients experiencing first line antiretroviral therapy failure. PLoS One. 2014;9:e86771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Johnson JA, Li JF, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment–naïve populations and associate with reduced treatment efficacy. PLoS Med. 2008;5:e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li JZ, Paredes R, Ribaudo HJ, et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA. 2011;305:1327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Metzner KJ, Giulieri SG, Knoepfel SA, et al. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and-adherent patients. Clin Infect Dis. 2009;48:239–47. [DOI] [PubMed] [Google Scholar]
- [31].Paredes R, Lalama CM, Ribaudo HJ, et al. Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis. 2010;201:662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Alidjinou EK, Deldalle J, Hallaert C, et al. RNA and DNA Sanger sequencing versus next-generation sequencing for HIV-1 drug resistance testing in treatment-naive patients. J Antimicrob Chemother. 2017;72:2823–30. [DOI] [PubMed] [Google Scholar]
- [33].Cozzi-Lepri A, Noguera-Julian M, Di Giallonardo F, et al. Low-frequency drug-resistant HIV-1 and risk of virological failure to first-line NNRTI-based ART: a multicohort European case–control study using centralized ultrasensitive 454 pyrosequencing. J Antimicrob Chemother. 2015;70:930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kyeyune F, Gibson RM, Nankya I, et al. Low-frequency drug resistance in HIV-infected Ugandans on antiretroviral treatment is associated with regimen failure. Antimicrob Agents Chemother. 2016;60:3380–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mohamed S, Ravet S, Camus C, et al. Clinical and analytical relevance of NNRTIs minority mutations on viral failure in HIV-1 infected patients. J Med Virol. 2014;86:394–403. [DOI] [PubMed] [Google Scholar]
- [36].Li JZ, Kuritzkes DR. Clinical implications of HIV-1 minority variants. Clin Infect Dis. 2013;56:1667–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rowley CF, Boutwell CL, Lee EJ, et al. Ultrasensitive detection of minor drug-resistant variants for HIV after nevirapine exposure using allele-specific PCR: clinical significance. AIDS Res Hum Retroviruses. 2010;26:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Boltz VF, Zheng Y, Lockman S, et al. Role of low-frequency HIV-1 variants in failure of nevirapine-containing antiviral therapy in women previously exposed to single-dose nevirapine. Proc Natl Acad Sci USA. 2011;108:9202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jakobsen MR, Tolstrup M, Søgaard OS, et al. Transmission of HIV-1 drug-resistant variants: prevalence and effect on treatment outcome. Clin Infect Dis. 2010;50:566–73. [DOI] [PubMed] [Google Scholar]
- [40].Li JZ, Stella N, Choudhary MC, et al. Impact of pre-existing drug resistance on risk of virological failure in South Africa. J Antimicrob Chemother. 2021;76:1558–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li JZ, Paredes R, Ribaudo HJ, et al. Relationship between minority nonnucleoside reverse transcriptase inhibitor resistance mutations, adherence, and the risk of virologic failure. AIDS. 2012;26:185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Boltz VF, Bao Y, Lockman S, et al. Low-frequency nevirapine (NVP)–resistant HIV-1 variants are not associated with failure of antiretroviral therapy in women without prior exposure to single-dose NVP. J Infect Dis. 2014;209:703–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zoufaly A, Jochum J, Hammerl R, et al. Virological failure after 1 year of first-line ART is not associated with HIV minority drug resistance in rural Cameroon. J Antimicrob Chemother. 2015;70:922–5. [DOI] [PubMed] [Google Scholar]
- [44].Rowley C.F., MacLeod IJ, Maruapula D, et al., Sharp increase in rates of HIV transmitted drug resistance at antenatal clinics in Botswana demonstrates the need for routine surveillance. Antimicrob Chemother. 2016;71(5)(1460-2091 (Electronic)): 1361–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Vilsker M, Moosa Y, Nooij S, et al. Genome detective: an automated system for virus identification from high-throughput sequencing data. Bioinformatics. 2019;35:871–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].PASEQ. Available at: https://paseq.org/ [access date March 25, 2021].
- [47].Kearse M, Moir R, Wilson A, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Mehta CR, Patel NR. Exact logistic regression: theory and examples. Stat Med. 1995;14:2143–60. [DOI] [PubMed] [Google Scholar]
- [49].Hsieh FY. Sample size tables for logistic regression. Stat Med. 1989;8:795–802. [DOI] [PubMed] [Google Scholar]
- [50].Li M, Liang S, Zhou C, et al. HIV drug resistance mutations detection by next-generation sequencing during antiretroviral therapy interruption in China. Pathogens. 2021;10:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Chimukangara B, Giandhari J, Lessells R, et al. Impact of pretreatment low-abundance HIV-1 drug-resistant variants on virological failure among HIV-1/TB-co-infected individuals. J Antimicrob Chemother. 2020;75:3319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006;42:1608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Lopez CA, Vazquez M, Hill MD, et al. Characterization of HIV-1 RNA forms in the plasma of patients undergoing successful HAART. Arch Virol. 2010;155:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gupta S, Taylor T, Patterson A, et al. A robust PCR protocol for HIV drug resistance testing on low-level viremia samples. Biomed Res Int. 2017;2017:4979252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kantor R, DeLong A, Schreier L, et al. HIV second-line failure and drug resistance at high-and low-level viremia in Western Kenya. AIDS. 2018;32:2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Moyo S., Gaseitsiwe S, Zahralban-Steele M, et al. Low rates of nucleoside reverse transcriptase inhibitor and nonnucleoside reverse transcriptase inhibitor drug resistance in Botswana. AIDS (London, England). 2019;33:1073–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Balduin M, Oette M, Däumer MP, et al. Prevalence of minor variants of HIV strains at reverse transcriptase position 103 in therapy-naive patients and their impact on the virological failure. J Clin Virol. 2009;45:34–8. [DOI] [PubMed] [Google Scholar]
- [58].Boltz VF, Shao W, Bale MJ, et al. Linked dual-class HIV resistance mutations are associated with treatment failure. JCI Insight. 2019;4:e130118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Hassan AS, Bibby DF, Mwaringa SM, et al. Presence, persistence and effects of pre-treatment HIV-1 drug resistance variants detected using next generation sequencing: A Retrospective longitudinal study from rural coastal Kenya. PLoS One. 2019;14:e0210559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.