Abstract
Given the lack of a nucleus in prokaryotic cells, the significance of spatial organization in bacterial chromosome replication is only beginning to be fully appreciated. DnaA protein, the initiator of chromosomal replication in Escherichia coli, is purified as a soluble protein, and in vitro it efficiently initiates replication of minichromosomes in membrane-free DNA synthesis reactions. However, its conversion from a replicatively inactive to an active form in vitro occurs through its association with acidic phospholipids in a lipid bilayer. To determine whether the in situ residence of DnaA protein is cytoplasmic, membrane associated, or both, we examined the cellular location of DnaA using immunogold cryothin-section electron microscopy and immunofluorescence. Both of these methods revealed that DnaA is localized at the cell membrane, further suggesting that initiation of chromosomal replication in E. coli is a membrane-affiliated event.
Initiation of Escherichia coli chromosomal replication occurs at the chromosomal origin, oriC, a unique sequence that contains all necessary cis-acting elements (42, 47). To initiate a round of replication, approximately 20 molecules of DnaA protein bind to the DnaA boxes in oriC (10, 13, 14, 38, 41, 62), promote strand opening of the AT-rich 13-mers (2, 3, 17), and facilitate the loading of DnaB helicase (14, 40). The remaining components of the replisome are subsequently recruited to the sites of the newly formed replication forks (33, 39).
Given the central role of DnaA protein in the initiation process, its regulation is most likely crucial for normal, cell-cycle-controlled chromosomal replication. While dnaA mRNA levels fluctuate throughout the cell cycle, DnaA protein levels remain relatively constant (51). Therefore, other mechanisms must regulate the protein's activity. Prominent among these in vitro is the influence on the replicative action of DnaA protein by the tight binding of ATP and ADP (52, 65). Both the ADP and ATP forms of DnaA protein are capable of binding oriC (52). However, only the binding of ATP-DnaA leads to duplex melting and the ensuing initiation events (52, 53, 65). The ATP bound to DnaA protein is slowly hydrolyzed in a DNA-dependent manner, with the resulting ADP remaining tightly bound, thus producing inactive ADP-DnaA protein. A partially purified factor, termed IdaB, and β-subunit, the processivity factor of DNA polymerase III, stimulate the conversion of ATP-DnaA to ADP-DnaA (28, 29).
Treatment of ADP-DnaA with anionic phospholipids causes the release of the bound nucleotide (8, 54). When exposed to anionic phospholipids in the presence of oriC DNA and excess ATP, replicatively inactive ADP-DnaA is rejuvenated into the active ATP form (9, 54). An examination of DnaA proteolytic fragments demonstrated that a distinct segment of DnaA is necessary for productive interaction with lipid bilayers (15). Amino acid sequence analysis of this region suggests the existence of an amphipathic helix (57), a motif commonly found in peripherally associated membrane proteins. Chemical cross-linking studies with a photoactivatable phospholipid analog demonstrated that the portion of DnaA centered about the putative amphipathic helix efficiently inserts into the hydrophobic interior of acidic phospholipid bilayers (16). A further indication that DnaA may have a membrane residency within cells is that partially purified DnaA protein is found aggregated in acidic phospholipid containing clusters (25, 55), and the protein can be separated and rendered active for replication by treatment with phospholipase A2 (25).
Evidence from in vivo studies also implies a close link between membranes and chromosomal replication. E. coli treated with 3-decynoyl-N-acetylcysteamine (DNAC), a suicide analog, are unable to synthesize unsaturated fatty acids. Their plasma membranes lose fluidity, and the cells are no longer able to initiate new rounds of chromosomal replication (12). Correspondingly, lipids extracted from cells exposed to DNAC are unable to catalyze the release of nucleotide from DnaA protein (66). However, supplementing the growth medium of DNAC-treated cells with oleic acid restores membrane fluidity and DNA replication and imparts nucleotide-releasing properties to the cellular phospholipids (66).
Recent biophysical studies suggest that anionic lipid headgroups are necessary for the reactivation of ADP-DnaA in that they participate in stabilizing electrostatic interactions between DnaA protein and lipid bilayers (30). The physiological relevance of acidic phospholipids being involved in initiation of replication from oriC is strongly supported by the observation that cells unable to synthesize acidic phospholipids are unable to grow once their membrane content of acidic phospholipids drops below a critical level (21, 22). This arrested growth can be suppressed (64) if the cells lack functional RNase H and can therefore bypass normal DnaA-dependent initiations at oriC by carrying out constitutive stable DNA replication (60).
Components involved in cell division and the reproduction and inheritance of the genomes of Bacillus subtilis (34, 35, 59, 61), Caulobacter cresentus (11, 45, 49), and E. coli (1, 6, 18, 23, 46, 48, 50, 63) have been seen to reside at distinct cellular locations, and the spatial organizations of these factors are thought to be key for their proper functioning. Here we demonstrate that the cellular location of the E. coli initiator of chromosomal replication, DnaA protein, is at the cytoplasmic membrane.
MATERIALS AND METHODS
Reagents.
The reagents included glutaraldehyde (25%), paraformaldehyde (16%), osmium tetroxide, and standard hexagonal mesh nickel grids (Electron Microscopy Sciences); Formvar film (Pelco); uranyl acetate (Polysciences); 10-nm gold particles conjugated to goat anti-rabbit immunoglobulin G (IgG), fluorescein isothiocyanate (FITC) conjugated to goat anti-rabbit IgG, tetramethylrhodamine isothiocyanate (TRITC) conjugated to goat anti-rabbit IgG, and 3-aminopropyltriethoxysilane (Sigma Chemical); SlowFade Equilibrium Buffer and SlowFade (Molecular Probes); and Pansorbin (Calbiochem).
Strains and plasmids.
The strains were W3110 (F− λ− mcrA mcrB), AQ3519 (F− ilv metB pro rnh::cat dnaA850::Tn10), and MC1061 [hsdR mcrB araD139 Δ(araABC-leu)7679 ΔlacX74 galU galK rpsL thi). Plasmid pZL606 is a pBAD24c-derived overexpression plasmid that contains the dnaA gene under control of the ara promoter (19).
Buffers.
The buffers used included phosphate-buffered saline (PBS), with 10 mM sodium phosphate (pH 7.4) and 137 mM NaCl, and GTE (50 mM glucose, 20 mM Tris-Cl [pH 7.5], 10 mM EDTA). Buffer DB was composed of 25 mM HEPES-KOH (pH 7.5), 0.5 mM magnesium acetate 0.3 mM EDTA, 17% glycerol, 0.005% Triton X-100, and 2 mM dithiothreitol DTT.
Affinity purification of anti-DnaA IgG.
Affinity purified anti-DnaA protein antibody was prepared from New Zealand White rabbits inoculated with purified (55) DnaA protein. Immune and preimmune sera were subjected to ammonium sulfate fractionation and DEAE ion-exchange chromatography (7) to yield an IgG-enriched fraction. The immune IgG preparation was applied to a column of immobilized DnaA protein cross-linked to CNBr-activated Sepharose. IgG bound to DnaA protein was released with phosphoric acid (0.2 M, pH 2.4); the eluted fraction was neutralized and stored in sodium phosphate buffer (0.1 M, pH 6.8) at −80°C.
Immunoprecipitation of DnaA-oriC complexes.
DnaA protein (1.1 pmol) was added to supercoiled [3H]pBSoriC (60 fmol) in buffer DB and incubated at room temperature (10 min). Binding of DnaA to pBSoriC was confirmed with a filter retention assay (67). Samples were returned to ice and incubated (20 min) with anti-DnaA antibody (10 μl), followed by incubation (15 min) with Pansorbin (15 μl). Samples were centrifuged (15,000 × g, 3 min, 4°C), and radiolabeled pBSoriC plasmid remaining in the supernatants was measured by liquid scintillation counting.
Immunogold cryothin-section electron microscopy.
Wild-type cells (W3110) were grown in Luria-Bertani (LB) medium (44) to an optical density at 600 nm of approximately 0.6 to 0.7, and a sample (1 ml) was harvested by centrifugation (15,000 × g, 1 min, 4°C). The cells were resuspended in an equal volume (1 ml) of Karnovsky's fixative (4% glutaraldehyde, 1% paraformaldehyde in 0.1 M cacodylate [pH 7.2]), incubated (10 min) at room temperature, and collected by centrifugation (15,000 × g, 15 min, 4°C), and the resulting pellet was further incubated (50 min) with the supernatant on ice. The supernatant was discarded, the fixed cells were washed twice by resuspension in 0.1 M cacodylate (1 ml, pH 7.2) and centrifugation (15,000 × g, 1 min, 4°C), and the wet cell pellet was stored (for up to 3 days) at 4°C until used.
The cell pellet was transferred to the head of a microtome specimen pin, coated in a solution of sucrose (2.3 M), and rapidly frozen in liquid nitrogen. Thin sections (90 nm) were prepared with a glass blade microtome and immediately transferred to nickel electron microscopy grids that were previously coated in Formvar film (0.45% in dichloroethane), and the grids with thin sections were incubated (15 min, 23°C) in PBS that contained bovine serum albumin (5%). The grids were then incubated with affinity-purified anti-DnaA IgG (10 μl, 1:10 dilution, 1 h), washed in PBS (three times, 3 min per wash), and incubated with goat anti-rabbit antibody conjugated to 10-nm gold particles (10 μl, 1:10 dilution, 1 h). The grids were washed in PBS (three times, 3 min per wash), incubated (3 min) in glutaraldehyde (3%) and paraformaldehyde (1%), washed in PBS (three times, 3 min per wash), incubated (5 min) in osmium tetroxide (1%), washed in distilled H2O (three times, 3 min per wash), and incubated for 5 min in uranyl acetate (2%). The grids were transferred to a solution (10 μl, 0.125% [wt/vol]) of methylcellulose for 10 min, and excess solution was removed by blotting onto filter paper. The grids were incubated for an additional 10 min in an increased concentration of methylcellulose (0.25%), blotted again on filter paper, placed in a covered petri dish, and dried overnight at 37°C. Sections were visualized using a JEOL 1200EX electron microscope.
Immunofluorescence.
Cells (W3110) were grown in LB medium to an OD (A600) of approximately 0.6 to 0.7 and fixed (1) by the addition (to final concentrations) of paraformaldehyde (2.6%), glutaraldehyde (0.04%), and sodium phosphate (32 mM, pH 7.4) directly to the culture medium. The cells were incubated in the fixative mixture at room temperature (10 min) and then on ice (50 min). Aliquots (1 ml) of the fixed cells were transferred to microcentrifuge tubes and washed three times by centrifugation (15,000 × g, 4°C, 1 min) and suspension in PBS (1 ml). Washed cells were again harvested by centrifugation (15,000 × g, 4°C, 1 min) and permeabilized by resuspension in GTE buffer (50 μl) that contained lysozyme (4 μg/ml). The cells were immediately transferred to silane-treated wells of a multiwell microscope slide (slides were dipped in succession in ethanolic KOH [10%, wt/vol], distilled H2O, methanol, and acetone, then incubated [for 5 min] in 3-aminopropyltriethoxysilane [4%, vol/vol] in acetone, dipped twice in acetone, and finally air dried), and permeabilization was allowed to continue for an additional minute before the liquid was aspirated from the wells. The wells were washed twice with PBS (10 μl), air dried (10 to 15 min), rehydrated in PBS (10 μl, 4 min), and blocked with bovine serum albumin (10 μl, 3% [wt/vol]) in PBS. The blocking solution was removed by aspiration, anti-DnaA IgG (10 μl, 1:10 dilution in PBS) was added and incubated overnight (4°C). The excess antibody was removed, and the wells were washed in PBS (10 times, 1 min per wash) and incubated in the dark (2 h, 23°C) with FITC conjugated to goat anti-rabbit IgG (10 μl, a 1:100 dilution in PBS). The wells were washed (10 times) in PBS, followed by washing in SlowFade Equilibrium Buffer (5 min). SlowFade (1 drop) was added to each well, and the slides stored at −20°C until visualization using a ×100 oil immersion lens on a Zeiss Photoscope II microscope equipped with a 35-mm camera containing Kodak EPH 1600 film. The images were electronically scanned, and figures were produced using Adobe Photoshop 6.0.
Immunoblotting.
Cultures of wild-type (W3100), dnaA null (AQ3519), and DnaA-overproducing (MC1061/pZL606) cells were grown in LB medium (44) to an OD600 of 0.6 to 0.7. Samples of each culture were normalized to a volume of 1.0 ml, and an OD of 0.6. Ice-cold trichloroacetic acid (100 μl, 100%) was added, the mixtures were incubated on ice (15 min), acid-insoluble material was collected by microcentrifugation (15,000 × g, 4°C, 3 min), and each pellet was washed in acetone (70%, 1 ml). Proteins were solubilized, separated by SDS-PAGE, and transferred to nitrocellulose. Membranes were blocked in a solution (5%) of nonfat dried milk, probed with affinity-purified anti-DnaA IgG (1:30 dilution), washed, and probed with goat anti-rabbit IgG antiserum conjugated to horseradish peroxidase. DnaA protein was visualized using enhanced chemiluminescence (Pharmacia).
RESULTS
Affinity-purified anti-DnaA IgG shows high specificity for DnaA.
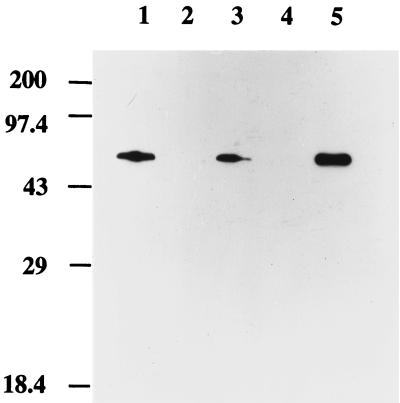
Since the techniques of immunogold electron microscopy and immunofluorescent microscopy employed in these studies rely on antibody detection, the specificity of the affinity-purified anti-DnaA IgG was assessed using Western blot analysis of whole-cell lysates (Fig. 1). Probing of the lysates with immune serum generated numerous bands (data not shown), while the affinity-purified IgG used to probe lysates prepared from wild-type (lane 3) and DnaA-overproducing (lane 5) cells detected a single, discrete band which corresponded to the molecular weight of purified DnaA protein (lane 1). This band was not detected in lysates prepared from dnaA null cells (lane 2). Furthermore, the appearance of this band was dependent on immune IgG; preimmune serum failed to recognize purified DnaA protein or material in the lysates from the wild-type or DnaA-overproducing cells (data not shown).
FIG. 1.
Specificity of affinity-purified anti-DnaA IgG. Purified DnaA protein (1 μg, lane 1), lysates from dnaA null mutant cells (lane 2), wild-type cells (lane 3), and DnaA-overproducing cells (lane 5) are shown. Lane 4 does not contain a sample.
Anti-DnaA antibody recognizes DnaA in DnaA-oriC complexes.
Immunoprecipitation experiments demonstrated that the anti-DnaA antibody is able to recognize the DNA-bound form of DnaA (Table 1). DnaA protein was allowed to bind to radiolabeled (67) supercoiled oriC plasmids. The protein-nucleic acid complexes were incubated with anti-DnaA antibody and then precipitated by the addition of pickled Staphylococcus aureus cells. The radiolabeled plasmid was precipitated only when incubated with DnaA, anti-DnaA antibody, and S. aureus cells (Table 1). Omission of DnaA protein caused the radiolabeled plasmid to remain in solution, as did the omission of anti-DnaA antibody and fixed S. aureus cells (Table 1). Thus, the anti-DnaA antibody is able to recognize, and has access to, DnaA in DnaA-oriC complexes.
TABLE 1.
Immunoprecipitation of oriC-bound DNA
| Immunoprecipitation reaction contained:
|
Amt of oriC plasmid in immunoprecipitation supernatant (fmol)a | |||
|---|---|---|---|---|
| oriC | DnaA | IgG | S. aureus | |
| + | + | + | + | 7 |
| + | − | + | + | 21 |
| + | + | − | − | 40 |
| + | − | − | − | 43 |
Average of two experiments.
Immunogold cryothin-section electron microscopy locates DnaA protein at the cell membrane.
Exponentially growing wild-type E. coli cells were fixed with paraformaldehyde and glutaraldehyde. Drops of the suspension were placed on specimen pins, cryoprotected by treatment with sucrose, rapidly frozen, and cut with a cryomicrotome. The thin sections were transferred to microscope grids and immersed in a solution of bovine serum albumin to prevent nonspecific antibody adsorption. The samples were probed with affinity-purified anti-DnaA IgG, followed by incubation with a secondary antibody conjugated to 10-nm gold particles. The samples were fixed with glutaraldehyde, treated with osmium tetroxide and uranyl acetate to highlight the cell morphology, dried, and visualized. Methods that included embedding the fixed samples in resins so that cellular structures could be best visualized resulted in poor antibody recognition of DnaA protein (data not shown), and thus the less-harsh method of cryothin sectioning was chosen. With cryothin sectioning, 93% of the cell sections examined for DnaA and all of those examined for GroEL (45 of 45) were labeled with gold particles.
Electron micrographs of representative sections show that the gold particles are associated almost exclusively with the cellular membrane, with very few particles found in the interior region of the cells (Fig. 2A). Preimmune IgG prepared from the rabbit used for the production of affinity-purified anti-DnaA protein IgG failed to label the thin section (data not shown). To further demonstrate that the antigen detected in these experiments was DnaA derived, sections of dnaA null cells were also probed with anti-DnaA IgG (Fig. 2B). Few (9%) contained gold particles, and in those that did the particles were found randomly distributed both within and outside the cellular sections.
FIG. 2.
Visualization of DnaA and GroEL proteins with immunogold cryothin-section electron microscopy. Thin sections were prepared from wild-type cells (A and C) or dnaA null cells (B) and probed with primary antibodies to DnaA protein (A and B) and GroEL (C). Representative gold particles are indicated by arrows; the bar equals 100 nm.
To assure that the detection of DnaA protein at the membrane reflects its true cellular location and not an inability to detect cytoplasmic antigens, cryothin sections prepared from wild-type cells were probed with primary antibody of a known cytoplasmic protein, GroEL. GroEL was found distributed throughout the thin sections (Fig. 2C).
The cellular contents of DnaA and GroEL are approximately 1,200 and 23,500 molecules, respectively, based on densitometric scanning of immunoblots of wild-type whole-cell extracts and the purified proteins (reference 55 and data not shown). The efficiency with which immunogold electron microscopy was able to detect these proteins was estimated from the number of gold particles that would be found in cylindrically shaped “cells” (average dimension, 1.1 by 0.4 μm) reconstructed from the 90-nm-thick thin sections. From this calculation, approximately 10% of the cells' DnaA and GroEL were detected by using cryothin-section immunogold electron microscopy (Table 2); these are sensitivities that are commonly found with this technique (7).
TABLE 2.
Cellular location of DnaA and GroEL as detected by cryothin-section immunogold electron microscopy
| Antibody | Cell sections with gold particlesa | Total no. of particles at:
|
Antigen detection (%) | Density (particles/μm2)b
|
||
|---|---|---|---|---|---|---|
| Membrane | Cytosol | Membrane | Cytosol | |||
| Anti-DnaA | 93 | 229 | 93 | 10 | 9,490 | 270 |
| Anti-GroEL | 45 | 129 | 790 | 9 | 2,980 | 1,890 |
Total cell sections examined: DnaA, 100; GroEL, 45.
Approximate areas of membrane and cytosol regions for each cellular cross-section were calculated as follows: cytosol (μm2) = (L − 0.01)(W − 0.01) and membrane region (μm2) = [(L + 0.01)(W + 0.01)] − [(L − 0.01)(W − 0.01)], where L and W equal cell rod length and width, respectively, in microns.
The molecular lengths of the primary IgG and secondary antibody-gold conjugate define an approximately 20-nm tether between a gold particle and a molecule of DnaA protein on the thin sections. Thus, the distribution of gold particles within 10 nm of each side of the membrane versus those found within the cytoplasm was determined (Table 2). A large majority (71%, i.e., 229 of 322) of the gold particles associated with DnaA protein was found at the membrane, in comparison to only 16% (i.e., 129 of 919) of the GroEL-related particles. Furthermore, when we accounted for the substantially smaller area of the membrane region relative to that of the cytosol, the density of DnaA protein detected adjacent to membranes was 35-fold higher than its density in the cytosol. In contrast, the densities of the gold particles attached to GroEL in the cytosol and membrane region were nearly the same (Table 2).
Since the thin sections were prepared from cells growing exponentially in an asynchronous culture, the possibility existed that the location of DnaA would vary from cell to cell in the thin sections. However, such variation was absent, suggesting that rather than transiently associating with the membrane, the majority of DnaA in cells remains in close proximity to the membrane throughout the cell cycle.
Immunofluorescence finds DnaA protein at the membrane.
Although hundreds of sections were examined, a caveat of immunogold electron microscopy is the possibility that during the preparation of sections, foci of interest may be missed or destroyed. Therefore, immunofluorescence was used to independently establish where DnaA protein resides within cells. This technique, which allows for a holistic view of the cell rather than relying on sectioning, has been applied successfully in determining the location of numerous bacterial proteins (1, 6, 20, 59).
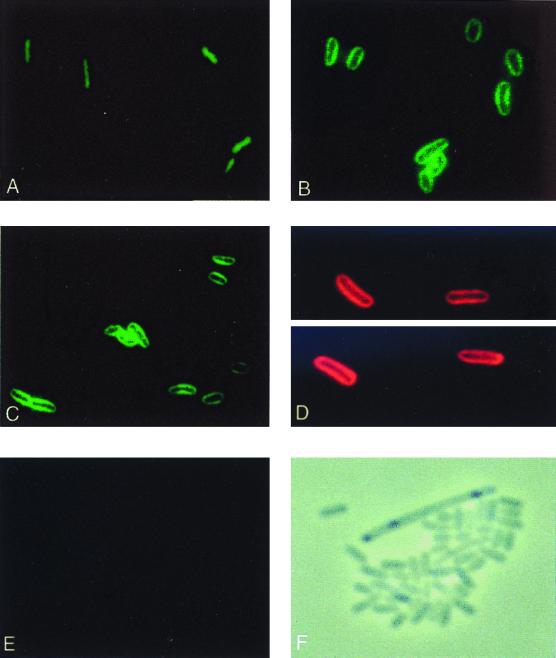
Similar to the examination with immunogold electron microscopy (Fig. 2 and Table 2), the fluorescent patterns of a cytosolic versus membrane protein were established. Probing fixed wild-type cells with anti-GroEL as the primary antibody generated a complete and uniform labeling (Fig. 3A). In contrast, if the cells were probed with an antibody to leader peptidase, a transmembrane protein of the inner membrane, a very distinct “halo” pattern of fluorescence was observed (Fig. 3B). Probing for DnaA protein produced an identical fluorescent halo. Moreover, this pattern was not affected by the choice of fluorescent label conjugated to the secondary antibody (Fig. 3C and D). The same pattern of fluorescence was seen in a number of different dnaA wild-type E. coli strains (data not shown), whereas dnaA null cells (Fig. 3F) probed with anti-DnaA as the primary antibody failed to produce a fluorescent signal (Fig. 3E).
FIG. 3.
Immunofluorescent visualization of DnaA, GroEL, and leader peptidase proteins. Wild-type cells (A to D) or dnaA null cells (E and F) were probed with anti-GroEL (A), anti-leader peptidase (B), or anti-DnaA (C to E) as primary antibodies. Goat anti-rabbit IgG conjugated to FITC (panels A to C and panel E) or to TRITC (D) were used as secondary antibodies. Panel F shows phase-contrast microscopy of the field of dnaA null cells (E).
Also, as was seen with the immunogold electron microscopy, DnaA protein appears to be associated with the cellular membrane in all of the cells in an exponentially growing asynchronous culture. Thus, throughout the cell cycle the majority of DnaA seems to be evenly distributed at the cell membrane.
DISCUSSION
Immunogold electron microscopy of cryothin sections and immunofluorescence of whole cells revealed that in exponentially growing E. coli, DnaA protein is not cytosolic, as might have been expected, but instead is a membrane-associated protein. Inasmuch as the examined cells were from an asynchronous culture and DnaA was found near the perimeter of virtually all of the hundreds of cells examined, it appears that DnaA remains at the membrane as cells progress through their growth cycle.
It is important to realize, however, that with these techniques the location of the bulk of an examined protein is what is readily apparent, while potential different locations of small subpopulations of the protein may be overlooked. For DnaA protein, one such small population may be molecules involved in initiating a round of replication at the chromosomal origin. In an E. coli cell there are approximately 1,000 DnaA protein molecules, of which only 20 to 80 (depending on the growth rate) are thought to participate in initiations at oriC during a specific time in the cell cycle. If chromosomal replication occurs in the cytosol independent of the cell membrane, a small fraction of DnaA protein would be expected to be cytosolic and may have gone undetected. Some insight into this possibility may come from examining the localization of DnaA in cells containing multiple copies of oriC plasmids, plasmids that replicate in synchrony with the host chromosome. In such cells, a redistribution of DnaA protein at the time of initiation, if it occurs, may become apparent.
However, accumulating evidence suggests that chromosomal replication in bacteria is a membrane-related event. In E. coli, recently replicated origins are prevented from being reinitiated by a process termed sequestration. Newly replicated origins are transiently hemimethylated and while in that state are sequestered at the membrane by SeqA protein (4, 5, 37, 58). In addition, origins have been observed at the cell poles in E. coli, B. subtilis, and C. crescentus (18, 27, 61). The recruitment of origins to the poles may be critical not only for proper chromosomal partitioning but also to facilitate interaction between the origins and membrane-bound initiator proteins.
In support of an initiator protein acting at the membrane, specific mutations in the site of DnaA protein shown to be essential for lipid binding in vitro (15, 16) are able to suppress the arrested growth of cells lacking sufficient levels of acidic phospholipids (W. Zheng, Z. Li, and E. Crooke, unpublished data).
Along with initiation, DNA replication itself may occur at the cell membrane. The catalytic subunit of B. subtilis DNA polymerase was found at a fixed site near the bacterium mid-cell, and it was proposed that the polymerase is recruited to this site by initiating proteins (34). Anchorage to a specific site, rather than being randomly dispersed in the cell, indicates that the polymerase is tethered, through some means, to the membrane, while chromosomal DNA is processed through the replisome.
Many essential factors involved in accurate inheritance of replicated DNA and cellular division are also seen to be spatially organized in a membrane-mediated fashion. Included in these are (i) the E. coli proteins SopAB and MukBEF involved in the segregation of plasmid and chromosomal DNA (24, 36, 46, 56); (ii) the SeqA protein, the sequester of hemimethylated oriC, which forms discrete foci that rapidly migrate in opposite directions immediately following duplication (23, 48); and (iii) the cell division proteins FtsZ, FtsA, FtsI (PBP3), FtsN, FtsQ, ZipA, FtsW (1, 6, 20, 63), and MinCDE (50).
By being localized to the membrane, DnaA protein itself may play a role in directing proper chromosomal inheritance. There are numerous DnaA boxes, binding sites for DnaA protein, throughout the E. coli chromosome. Association of DnaA boxes with membrane-bound DnaA protein may serve to help organize and position the chromosome for accurate partitioning and segregation. The possibility that DnaA, as an initiator protein, has an additional function in chromosomal inheritance needs to be tested.
A striking example of the importance of spatial organization acting in concert with temporal control in regulating chromosomal replication is seen in C. crescentus. CtrA, a protein responsible for repressing the initiation of DNA replication, is under the control of the histidine kinase, CckA. The cellular location of CckA is dynamic. It is located at the poles in a membrane-dependent manner in early predivisional cells, at which point it can act upon CtrA to prevent premature reinitiations of recently replicated DNA (26, 45).
Surprisingly, in the studies presented here both immunogold electron microscopy and immunofluorescence did not detect enhanced and distinct DnaA foci that were expected to illuminate the DAT locus, a site on the chromosome that has been proposed to be a titration sink for DnaA protein (31, 32).
Also of interest, cardiolipin-enriched domains, found mostly at cell poles and septal regions, were recently visualized in living cells with the fluorescent dye 10-N-nonyl acridine orange (43). Given the affinity DnaA protein has for anionic phospholipids (30), there is the possibility that DnaA protein may reside at these lipid-enriched domains. However, here DnaA protein was found to be evenly distributed about the cell membrane. If DnaA is preferentially localized to the cell poles and septa, the failure to detect an enrichment of DnaA protein at these sites may arise from the treatment of the cells in preparation for immunofluorescence: lateral heterogeneity of membrane lipids may well be disrupted in semi-intact cells. An independent and direct detection of the location of DnaA through the use of a chromosomally encoded green fluorescent protein-DnaA fusion in living cells will help answer whether DnaA is found preferentially at lipid-enriched domains and help to define the role that the spatial arrangement of this initiator protein plays in the control of chromosomal replication.
ACKNOWLEDGMENTS
We thank Susette Mueller and Maozheng Dai for guidance in the techniques of immunogold cryothin-section electron microscopy and immunofluorescence, William Wickner for the kind gift of anti-leader peptidase antibody, E. Mileykovskaya and W. Dowhan for sharing of data prior to publication, and Hiroshi Nakai for critical reading of the manuscript.
The studies, in part, utilized the Microscopy and Imaging shared resource of the Lombardi Cancer Center (P30CA51008). This work was funded in part by grants from the National Institutes of Health (GM49700 to E.C.) and the National Science Foundation (MCB9408830 to E.C.).
REFERENCES
- 1.Addinall S, Erfei B, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker T A, Sekimizu K, Funnell B E, Kornberg A. Extensive unwinding of the plasmid template during staged enzymatic initiation of DNA replication from the origin of the Escherichia coli chromosome. Cell. 1986;45:53–64. doi: 10.1016/0092-8674(86)90537-4. [DOI] [PubMed] [Google Scholar]
- 3.Bramhill D, Kornberg A. Duplex opening by DnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988;52:743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- 4.Brendler T, Abeles A, Austin S. A protein that binds to the P1 origin core and the oriC 13mer region in a methylation-specific fashion is the product of the host seqA gene. EMBO J. 1995;14:4083–4089. doi: 10.1002/j.1460-2075.1995.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brendler T, Austin S. Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. EMBO J. 1999;18:2304–2310. doi: 10.1093/emboj/18.8.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buddelmeijer N, Aarsman M E, Kolk A H, Vicente M, Nanninga N. Localization of cell division protein FtsQ by immunofluorescence microscopy in dividing and nondividing cells of Escherichia coli. J Bacteriol. 1998;180:6107–6116. doi: 10.1128/jb.180.23.6107-6116.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell D H, Garvey J S, Cremer N E, Sussdorf D H. Methods in immunology. London, England: Benjamin; 1970. [Google Scholar]
- 8.Castuma C E, Crooke E, Kornberg A. Fluid membranes with acidic domains activate DnaA, the initiator protein of replication in Escherichia coli. J Biol Chem. 1993;268:24665–24668. [PubMed] [Google Scholar]
- 9.Crooke E, Castuma C E, Kornberg A. The chromosome origin of E. coli stabilizes DnaA protein during rejuvenation by phospholipids. J Biol Chem. 1992;267:16779–16782. [PubMed] [Google Scholar]
- 10.Crooke E, Thresher R, Hwang D S, Griffith J, Kornberg A. Replicatively active complexes of DnaA protein and the Escherichia coli chromosomal origin observed in the electron microscope. J Mol Biol. 1993;233:16–24. doi: 10.1006/jmbi.1993.1481. [DOI] [PubMed] [Google Scholar]
- 11.Din, N., E. M. Quardokus, M. J. Sackett, and Y. V. Brun. Dominant C-terminal deletions of FtsZ that affect its ability to localize in Caulobacter and its interaction with FtsA. Mol. Microbiol. 27:1051–1063. [DOI] [PubMed]
- 12.Endo K, Helmkamp G M, Bloch K. Mode of inhibition of β-hydroxydecanoyl thioester dehydrase by 3-decynoyl-N-acetylcysteamine. J Biol Chem. 1970;245:4293–4296. [PubMed] [Google Scholar]
- 13.Fuller R S, Funnell B E, Kornberg A. The DnaA protein complex with the E. coli chromosomal origin (oriC) and other DNA sites. Cell. 1984;38:889–900. doi: 10.1016/0092-8674(84)90284-8. [DOI] [PubMed] [Google Scholar]
- 14.Funnell B E, Baker T, Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J Biol Chem. 1987;262:10327–10334. [PubMed] [Google Scholar]
- 15.Garner J, Crooke E. Membrane regulation of the chromosomal replication activity of E. coli DnaA requires a discrete site on the protein. EMBO J. 1996;15:3477–3485. [PMC free article] [PubMed] [Google Scholar]
- 16.Garner J, Durrer P, Kitchen J, Brunner J, Crooke E. Membrane-mediated release of nucleotide from an initiator of chromosomal replication, Escherichia coli DnaA, occurs with insertion of a distinct region of the protein into the lipid bilayer. J Biol Chem. 1998;273:5167–5173. doi: 10.1074/jbc.273.9.5167. [DOI] [PubMed] [Google Scholar]
- 17.Gille H, Messer W. Localized DNA melting and structural perturbations in the origin of replication, oriC, of Escherichia coli in vitro and in vivo. EMBO J. 1991;10:1579–1584. doi: 10.1002/j.1460-2075.1991.tb07678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon G S, Sitnikov D, Webb C, Teleman A, Straight A, Losick R, Murray A, Wright A. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 19.Guzman L, Belin D, Carson M, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hale C A, de Boer P A. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol. 1999;181:167–176. doi: 10.1128/jb.181.1.167-176.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heacock P N, Dowhan W. Alteration of the phospholipid composition of Escherichia coli through genetic manipulation. J Biol Chem. 1989;264:14972–14977. [PubMed] [Google Scholar]
- 22.Heacock P N, Dowhan W. Construction of a lethal mutation in the synthesis of the major acidic phospholipids of Escherichia coli. J Biol Chem. 1987;262:13044–13049. [PubMed] [Google Scholar]
- 23.Hiraga S, Ichinose C, Niki H, Yamazoe M. Cell cycle-dependent duplication and bidirectional migration of SeqA-associated DNA-protein complexes in E. coli. Mol Cell. 1998;1:381–387. doi: 10.1016/s1097-2765(00)80038-6. [DOI] [PubMed] [Google Scholar]
- 24.Hirano M, Mori H, Onogi T, Yamazoe M, Niki H, Ogura T, Hiraga S. Autoregulation of the partition genes of the mini-F plasmid and the intracellular localization of their products in Escherichia coli. Mol Gen Genet. 1998;257:392–403. doi: 10.1007/s004380050663. [DOI] [PubMed] [Google Scholar]
- 25.Hwang D S, Crooke E, Kornberg A. Aggregated DnaA protein is dissociated and activated for DNA replication by phospholipase or DnaK protein. J Biol Chem. 1990;265:19244–19248. [PubMed] [Google Scholar]
- 26.Jacobs C, Domian I, Maddock J, Shapiro L. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell. 1999;97:111–120. doi: 10.1016/s0092-8674(00)80719-9. [DOI] [PubMed] [Google Scholar]
- 27.Jensen R B, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci USA. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katayama T, Crooke E. DnaA protein is sensitive to a soluble factor and is specifically inactivated for initiation of in vitro replication of the Escherichia coli minichromosome. J Biol Chem. 1995;270:9265–9271. doi: 10.1074/jbc.270.16.9265. [DOI] [PubMed] [Google Scholar]
- 29.Katayama T, Kubota T, Kurokawa K, Crooke E, Sekimizu K. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell. 1998;94:61–71. doi: 10.1016/s0092-8674(00)81222-2. [DOI] [PubMed] [Google Scholar]
- 30.Kitchen J, Li Z, Crooke E. Electrostatic interactions during acidic phospholipid reactivation of DnaA protein, the Escherichia coli initiator of chromosomal replication. Biochemistry. 1999;38:6213–6221. doi: 10.1021/bi982733q. [DOI] [PubMed] [Google Scholar]
- 31.Kitagawa R, Ozaki T, Moriya S, Ogawa T. Negative control of replication initiation by a novel chromosomal locus exhibiting exceptional affinity for Escherichia coli DnaA protein. Genes Dev. 1998;12:3032–3043. doi: 10.1101/gad.12.19.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitagawa R, Mitsuki H, Okazaki T, Ogawa T. A novel DnaA protein-binding site at 94.7 min on the Escherichia coli chromosome. Mol Microbiol. 1996;19:1137–1147. doi: 10.1046/j.1365-2958.1996.453983.x. [DOI] [PubMed] [Google Scholar]
- 33.Kornberg A, Baker T A. DNA replication. W. H. New York, N.Y: Freeman and Co.; 1992. [Google Scholar]
- 34.Lemon K, Grossman A. Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science. 1998;282:1516–1519. doi: 10.1126/science.282.5393.1516. [DOI] [PubMed] [Google Scholar]
- 35.Levin P A, Shim J J, Grossman A. Effect of minCD on FtsZ ring position and polar septation in Bacillus subtilis. J Bacteriol. 1998;180:6048–6051. doi: 10.1128/jb.180.22.6048-6051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lockhart A, Kendrick-Jones J. Nucleotide-dependent interaction of the N-terminal domain of MukB with microtubules. J Struct Biol. 1998;124:303–310. doi: 10.1006/jsbi.1998.4056. [DOI] [PubMed] [Google Scholar]
- 37.Lu M, Campbell J L, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 38.Margulies C, Kaguni J M. Ordered and sequential binding of DnaA protein to oriC, the chromosomal origin of Escherichia coli. J Biol Chem. 1996;271:17035–17040. doi: 10.1074/jbc.271.29.17035. [DOI] [PubMed] [Google Scholar]
- 39.Marians K. Prokaryotic DNA replication. Annu Rev Biochem. 1992;61:673–719. doi: 10.1146/annurev.bi.61.070192.003325. [DOI] [PubMed] [Google Scholar]
- 40.Marszalek J, Kaguni J M. DnaA protein directs the binding of DnaB protein in initiation of DNA replication in Escherichia coli. J Biol Chem. 1994;269:4883–4890. [PubMed] [Google Scholar]
- 41.Matsui M, Oka A, Takanami M, Yasuda S, Hirota Y. Sites of DnaA protein-binding in the replication origin of the E. coli K-12 chromosome. J Mol Biol. 1985;184:529–533. doi: 10.1016/0022-2836(85)90299-2. [DOI] [PubMed] [Google Scholar]
- 42.Messer W, Meijer M, Bergmans H E, Hansen F G, von Meyenburg K, Beck E, Schaller H. Origin of replication, oriC, of the Escherichia coli K-12 chromosome: nucleotide sequence. Cold Spring Harbor Symp Quant Biol. 1979;43:139–145. doi: 10.1101/sqb.1979.043.01.020. [DOI] [PubMed] [Google Scholar]
- 43.Mileykovskaya E, Dowhan W. Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol. 2000;182:1172–1175. doi: 10.1128/jb.182.4.1172-1175.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller J. Experiments in molecular genetics. New York, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 45.Mohl D, Gober J. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 46.Niki H, Hiraga S. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell. 1997;90:951–957. doi: 10.1016/s0092-8674(00)80359-1. [DOI] [PubMed] [Google Scholar]
- 47.Oka A, Sugimoto K, Takanami M, Hirota Y. Replication origin of the Escherichia coli K-12 chromosome: the size and structure of the minimum DNA segment carrying the information for autonomous replication. Mol Gen Genet. 1980;178:9–20. doi: 10.1007/BF00267207. [DOI] [PubMed] [Google Scholar]
- 48.Onogi T, Niki H, Yamazoe M, Hiraga S. The assembly and migration of SeqA-Gfp fusion in living cells of Escherichia coli. Mol Microbiol. 1999;31:1775–1782. doi: 10.1046/j.1365-2958.1999.01313.x. [DOI] [PubMed] [Google Scholar]
- 49.Quardokus E, Din N, Brun Y V. Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc Natl Acad Sci USA. 1996;93:6314–6319. doi: 10.1073/pnas.93.13.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raskin D, de Boer P. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakakibara Y, Yuasa S. Continuous synthesis of the dnaA gene product of Escherichia coli in the cell cycle. Mol Gen Genet. 1982;186:87–94. doi: 10.1007/BF00422917. [DOI] [PubMed] [Google Scholar]
- 52.Sekimizu K, Bramhill D, Kornberg A. ATP activates DnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987;50:259–265. doi: 10.1016/0092-8674(87)90221-2. [DOI] [PubMed] [Google Scholar]
- 53.Sekimizu K, Bramhill D, Kornberg A. Sequential early stages in the in vitro initiation of replication at the origin of the Escherichia coli chromosome. J Biol Chem. 1988;263:7124–7130. [PubMed] [Google Scholar]
- 54.Sekimizu K, Kornberg A. Cardiolipin activation of DnaA protein, the initiation protein of replication in Escherichia coli. J Biol Chem. 1988;263:7131–7135. [PubMed] [Google Scholar]
- 55.Sekimizu K, Yung B Y M, Kornberg A. The DnaA protein of Escherichia coli: abundance, improved purification, and membrane binding. J Biol Chem. 1988;263:7136–7140. [PubMed] [Google Scholar]
- 56.Sharpe M E, Errington J. Upheaval in the bacterial nucleoid. An active chromosome segregation mechanism. Trends Genet. 1999;15:70–74. doi: 10.1016/s0168-9525(98)01660-6. [DOI] [PubMed] [Google Scholar]
- 57.Skarstad K, Boye E. The initiator protein DnaA: evolution, properties and function. Biochim Biophys Acta. 1994;1217:111–120. doi: 10.1016/0167-4781(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 58.Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in replication initiation and origin sequestration. Cell. 1995;82:927–936. doi: 10.1016/0092-8674(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 59.Teleman A A, Graumann P L, Lin D C H, Grossman A D, Losick R. Chromosome arrangement within a bacterium. Curr Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- 60.von Meyenburg K, Boye E, Skarstad K, Koppes L, Kogoma T. Mode of initiation of constitutive stable DNA replication in RNase H-defective mutants of Escherichia coli K12. J Bacteriol. 1987;169:2650–2658. doi: 10.1128/jb.169.6.2650-2658.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Webb C D, Teleman A, Gordon S, Straight A, Belmont A, Lin D C, Grossman A D, Wright A, Losick R. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 62.Weigel C, Schmidt A, Ruckert B, Lurz R, Messer W. DnaA protein binding to individual DnaA boxes in the Escherichia coli replication origin, oriC. EMBO J. 1997;16:6574–6583. doi: 10.1093/emboj/16.21.6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiss D, Chen J, Ghigo J, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xia W, Dowhan W. In vivo evidence for the involvement of anionic phospholipids in initiation of DNA replication in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:783–787. doi: 10.1073/pnas.92.3.783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yung B Y M, Crooke E, Kornberg A. Fate of the DnaA initiator protein in replication at the origin of the Escherichia coli chromosome in vitro. J Biol Chem. 1990;265:1282–1285. [PubMed] [Google Scholar]
- 66.Yung B Y M, Kornberg A. Membrane attachment activates DnaA protein, the initiation protein of chromosome replication in Escherichia coli. Proc Natl Acad Sci USA. 1988;85:7202–7205. doi: 10.1073/pnas.85.19.7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yung B Y M, Kornberg A. The DnaA initiator protein binds separate domains in the replication origin of Escherichia coli. J Biol Chem. 1989;264:6146–6150. [PubMed] [Google Scholar]