Abstract
Plants exhibit an enormous phenotypic plasticity to adjust to changing environmental conditions. For this purpose, they have evolved mechanisms to detect and measure biotic and abiotic factors in their surroundings. Phytochrome B exhibits a dual function, since it serves as a photoreceptor for red and far-red light as well as a thermosensor. In 1999, it was first reported that phytochromes not only translocate into the nucleus but also form subnuclear foci upon irradiation by red light. It took more than 10 years until these phytochrome speckles received their name; these foci were coined photobodies to describe unique phytochrome-containing subnuclear domains that are regulated by light. Since their initial discovery, there has been much speculation about the significance and function of photobodies. Their presumed roles range from pure experimental artifacts to waste deposits or signaling hubs. In this review, we summarize the newest findings about the meaning of phyB photobodies for light and temperature signaling. Recent studies have established that phyB photobodies are formed by liquid-liquid phase separation via multivalent interactions and that they provide diverse functions as biochemical hotspots to regulate gene expression on multiple levels.
Introduction to phytochrome B
Plant genomes provide the foundation for a striking phenotypic plasticity in response to changes in their environment. While extreme light and temperature conditions can indicate a stressful environment for plants, daily and annual changes of both factors provide important information about the current season and the time of day. Light is one of the most essential environmental factors for plants since it not only serves as an environmental cue but also as their energy source. Therefore, plants have evolved sophisticated systems of photoreceptors to monitor their light environment, which allows them to optimize their growth and development accordingly.
Phytochromes are a family of photoreceptors that can be found in bacteria, fungi, and the entire green lineage from algae to flowering plants. Arabidopsis thaliana's genome encodes for 5 phytochromes that act as red (R) and far-red (FR) light receptors to sense light intensity and quality. Mutant analyses demonstrated that phytochrome B (phyB) is the major phytochrome in Arabidopsis and that it is involved in plant development processes throughout the entire plant life cycle, from germination and seedling de-etiolation to floral induction and seed yield (Franklin and Quail 2010; Ulijasz and Vierstra 2011; Legris et al. 2019).
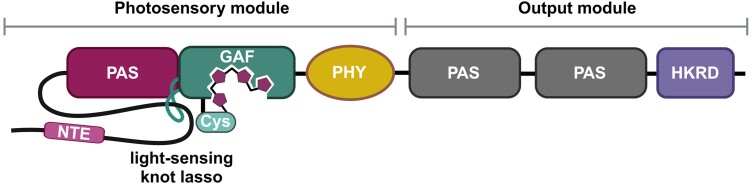
The PHYB apoprotein has a mass of approximately 125 kDa and can be divided into 2 larger domains (or modules) that are linked through a flexible hinge region: the N-terminal photosensory (or signal input) domain and the C-terminal output domain that is necessary for homodimerization or heterodimerization with other phytochromes. These 2 domains can be further divided into several subdomains (Fig. 1). The photosensory domain consists of 4 subdomains: N-terminal extension, PAS (Period/Arnt/Single-Minded) domain, GAF (cGMP phosphodiesterase/adenylyl cyclase/FhlA) domain, and PHY (Phy-specific) domain. The output module is composed of a PAS-related domain and a HKRD (histidine kinase-related domain). The latter subdomain is a feature that points at the bacterial origins of phytochromes as photo-regulated histidine kinases (Rockwell et al. 2006; Nagatani 2010; Burgie and Vierstra 2014).
Figure 1.
Domain structure of Arabidopsis phyB. The photosensory domain consists of a PAS domain, a GAF domain, and a PHY domain. The output module is composed of a PAS-related domain and a HKRD. The C-terminal output module features properties for nuclear localization, dimerization and photobody formation, while the N-terminal photosensory module binds phytochromobilin via a conserved cysteine residue (Cys) and possesses the N-terminal extension (NTE) that is required for photobody formation of the full-length phyB protein. The depicted domain structure is based on (Burgie and Vierstra 2014).
Bacterial phytochromes dimerize in linear and symmetric head-to-head fashion (Li et al. 2010). In contrast, phyB dimers exhibit an asymmetric architecture: in the dimer, the head-to-head association is only retained for the C-terminal histidine-related domains, whereas the N-terminal photosensory modules are arranged head-to-tail (Li et al. 2022b). Interestingly, this likely implies that during evolution, conserved interacting protein surfaces within the photosensory modules must have been abandoned and new ones must have been established to give rise to the PHYB protein clade of phytochromes.
After translation, a chromophore (phytochromobilin) binds covalently to the GAF domain of the photosensory module to form the phyB holoprotein. Absorption of R or FR light triggers the isomerization of the phytochromobilin that transmits into conformational changes of the phyB protein. This allows phyB to reversibly transition between 2 relatively stable conformers: R light induces the biologically active Pfr form, while FR light triggers the transition of Pfr into the Pr conformation, which represents inactive phyB (Klose et al. 2015). The Pfr form is temporarily stable even after the transition to darkness during the onset of night. Here, phyB slowly switches back into its inactive Pr form. This process is called dark reversion or thermal reversion, since it is accelerated by elevated temperatures (Klose et al. 2020). Therefore, phyB exhibits a dual function: not only does it act as R/FR light receptor but also as a thermosensor during the night (Jung et al. 2016) and the day (Legris et al. 2016; Qiu et al. 2019). Additionally, phyB is also able to detect other plants that are potential competitors for sunlight. Photosynthetically active tissues absorb more R than FR light, since the R light is utilized for photosynthesis. In contrast, most of the FR light transmits through or is reflected from leaves of neighboring plants. Therefore, a plant that is surrounded by potential competitors perceives light with a low R to FR light ratio (low R:FR). This reduces phyB activity and hence allows the detection of neighboring vegetation before being exposed to the vegetation's shade. In consequence, shade-avoiding plants like Arabidopsis induce an escape strategy called shade avoidance response that promotes stem and petiole elongation—to outgrow potential competitors—and that induce early flowering (Casal 2012; Ballaré and Pierik 2017). The negative effect on phyB activity is intensified if the plant is exposed to true canopy shade that not only increases the amount of perceived FR light but also decreases the photosynthetically active radiation (PAR), including R light. In consequence, the reduction of the R to FR light ratio, the decrease of phyB activity, and the extent of the shade responses are intensified in true shade conditions (Fiorucci and Fankhauser 2017; Hernando et al. 2021; Martinez-Garcia and Rodriguez-Concepcion 2023).
What are phyB photobodies?
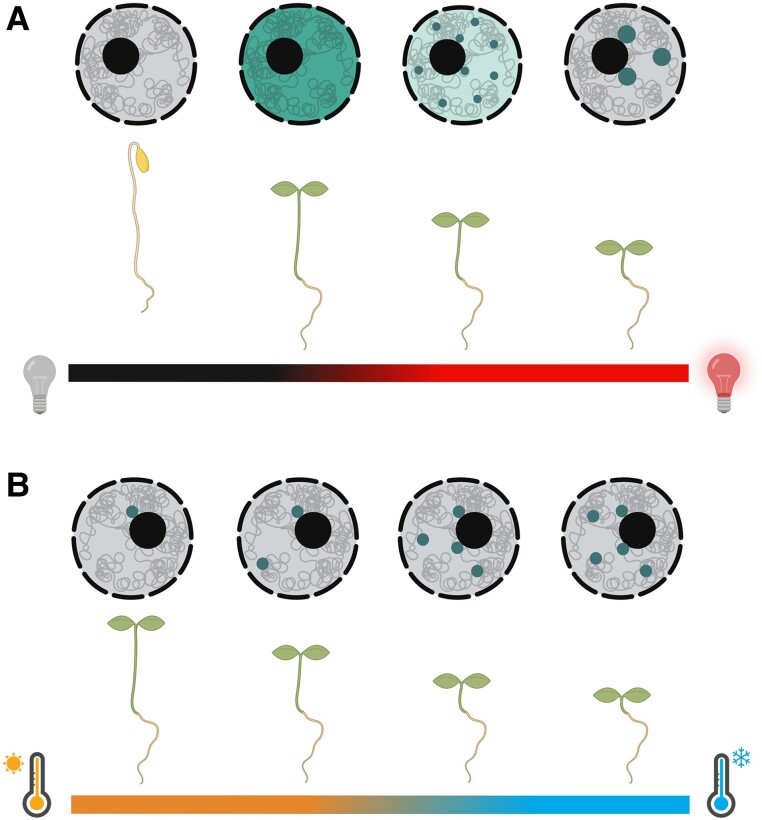
In etiolated seedlings, phyB is inactive and resides within the cytosol. However, the exposure to R light converts phyB into its active Pfr form, which triggers a rise of the cytoplasmic Ca2+ concentration (Zhao et al. 2023). This Ca2+ increase activates 2 calcium-dependent kinases that bind and phosphorylate phyB. Their activity induces the translocation of phyB into the nucleus, where the photoreceptor initiates the physiological responses to R light (Zhao et al. 2023). Under dim R light, phyB is evenly distributed within the nucleoplasm. However, with increasing light intensity, phyB starts to form nuclear speckles or nuclear bodies (Chen et al. 2003). These nuclear speckles formed by phytochromes were first reported in 1999 by Akira Nagatani, Ferenc Nagy, Eberhard Schäfer, and colleagues (Kircher et al. 1999; Yamaguchi et al. 1999). While other light receptors are also able to form nuclear speckles, the term photobodies was originally coined by Joanne Chory in 2011 to specifically describe phytochrome containing nuclear bodies (Chen and Chory 2011). Photobodies formed by phyB are dynamic and increase in size with rising light intensity (Fig. 2). Weaker red light forms many small nuclear speckles, an intermediate R light intensity leads to the production of many small and a few larger photobodies, while strong, saturating R light confines phyB into a handful of large bodies. In contrast, lowering the R light intensity or exposure to low R:FR leads to the opposite behavior. Both promote the redistribution of phyB from large photobodies into smaller speckles or even into the nucleoplasm (Chen et al. 2003; Trupkin et al. 2014; Van Buskirk et al. 2014; Willige et al. 2021). Additionally, photobodies are also temporally regulated. Within minutes, light exposure of etiolated seedlings leads to the formation of early photobodies that are transient and small. In contrast, prolonged light exposure leads to the formation of large and stable photobodies (Bauer et al. 2004). Altogether, smaller photobodies seem to be an intermediate step from phyB's redistribution from the nucleoplasm into large bodies or vice versa. Similarly, the size of photobodies has been reported to decrease in elevated temperatures in diurnal white light conditions (Legris et al. 2016; Murcia et al. 2021). However, in constant red light, inactivation of phyB by elevated temperatures does not seem to mimic the photobody dynamics in response to changes in light intensity or quality. Instead, increasing temperatures lead to the complete disassembly of individual photobodies without the transition to small speckles (Hahm et al. 2020). Here, each photobody within a cell seems to have an unique thermal sensitivity. This demonstrates that photobodies within a cell are not equal, which is supported by the observation that a photobody located in proximity to the nucleolus is most stable (Fig. 2B). Interestingly, when phyB is constitutively active by overexpressing the phyB Y276H mutant (YHB), cotyledon photobodies are shown to be temperature insensitive, indicating that photobody sensitivity is linked to phyB's reversion into its inactive Pr state (Hahm et al. 2020). This observation contrasts with another study that shows temperature sensitivity of photobodies formed by YHB in hypocotyl or HEK293T cells. This would indicate that the temperature sensitivity of photobodies is additionally based on a thermal reversion–independent mechanism (Chen et al. 2022). Future studies will show if these contradicting results are the consequence of different PHYB expression levels or analyzed cell types. However, both studies agree that the successive disassembly of a cell's photobodies could represent a sensor to assess increasing ambient temperatures (Hahm et al. 2020; Chen et al. 2022). Besides light and temperature, photobody stability is also regulated by other external and internal factors such as salt stress or jasmonate signaling (Liu et al. 2023; Peng et al. 2023). This indicates that photobodies have the ability to integrate multiple signaling pathways.
Figure 2.
Schematic illustration of photobody behavior under different red light and temperature conditions. A) Under very dim red light, phyB translocates into the nucleus where it is evenly distributed. An increasing fluence rate leads to small photobodies that fuse and form bigger nuclear bodies with rising light intensities. B) In constant red light, increasing temperatures cause the disassembly of individual photobodies without disintegrating into small nuclear foci. Thermolabile photobodies are only present in lower temperatures, while the most thermostable photobody is located in the proximity of the nucleolus.
How are these dynamics in photobody assembly and disintegration achieved? Two phyB (sub)domains are required for the formation of nuclear bodies. It has been reported that the C-terminal output module alone is sufficient to dimerize, translocate into the nucleus, and form photobodies even in the dark (Nagy et al. 2000; Matsushita et al. 2003). This indicates that the N-terminal photosensory domain prevents the translocation and speckle formation of phyB's Pr form. However, the full-length protein in its Pfr form requires the N-terminal extension to assemble into nuclear foci (Chen et al. 2005). This implies a positive regulatory role of the photosensory domain in speckle formation as well. The N-terminal extension is important to stabilize the Pfr form of phyB by slowing down the dark/thermal reversion, and it harbors an intrinsically disordered region (Burgie et al. 2021). In general, intrinsically disordered regions are often a requirement for biomolecular condensation, since they allow multivalent interactions to form large protein complexes (Emenecker et al. 2020; Borcherds et al. 2021). Similarly, photobody assembly requires the formation of higher order phyB aggregates through interactions of multiple phyB dimers through their N-terminal extensions (Chen et al. 2022).
As stated above, these photobodies are dynamic, since fluorescence recovery after photobleaching experiments indicated that the phyB molecules move from photobody to photobody through the nucleoplasmic phyB pool (Rausenberger et al. 2010). Furthermore, photobodies are spherical in shape and can move and coalesce (Chen et al. 2022). Therefore, it has been suggested that photobodies are formed through the formation of a dense phyB phase (photobodies) within a less dense phyB phase (nucleoplasm) (Fonin et al. 2021; Chen et al. 2022). This process is called phase separation and allows the dynamic assembly of membraneless organelles within the nucleus (Emenecker et al. 2020).
So far, there are 2 estimates of the number of phyB dimers per photobody. Due to the chosen experimental approach or due to photobody dynamics, both estimates provide quite different results: 80 to 150 phyB dimers per 0.4- to 0.8-µm³ photobody and 1,500 dimers per 0.2-µm photobody (Chen et al. 2022; Kim et al. 2023a). Certainly, photobodies consist not only of phyB but of a complex mixture of diverse proteins. A recent mass spectrometry study that used photobodies isolated by fluorescence-activated particle sorting to gain insights into their composition revealed that photobody clients can be classified into 2 groups: proteins that bind phyB directly and proteins that require the presence of the first group to be targeted to photobodies (Kim et al. 2023a). However, the individual photobodies of a single cell are not all the same and do not carry the same protein composition, which is supported by phyB colocalization studies. For example, the blue light receptor cry2 also forms nuclear speckles, but only a subset of these speckles within a nucleus colocalizes with phyB photobodies (Más et al. 2000). This indicates that different photobodies not only have differing thermal sensitivities but are also functional divers.
But what is the significance and function of photobodies? In light conditions where phyB is unable to form photobodies, only very weak R light responses are executed. This suggests that photobodies are not essential for phyB activity, but that they represent a mechanism to potentiate phyB signaling (Chen et al. 2003). In addition, several mutations within PHYB or of structural phyB photobody components demonstrate a causal relationship between photobody formation and phyB activity (Qiu et al. 2017; Huang et al. 2019).
In general, research on photobodies collectively indicates that photobodies serve 2 main functions. First, they stabilize phyB's Pfr form and extend the time of phyB activity. Second, they regulate biochemical reactions by concentrating or sequestering enzymes, substrates, and other factors that give these reactions specificity. By this means, phyB inactivates repressors of photomorphogenesis but promotes the activity of positive signaling components. In consequence, phyB regulates different steps of gene expression, including chromatin remodeling, transcription, and pre-mRNA splicing via photobodies. The newest findings about the role of phyB photobodies for light and temperature signaling are summarized below.
PHYTOCHROME-INTERACTING FACTORs are transcription factors regulated by photobodies
Diverse transcription factors and other transcriptional regulators that positively regulate light signaling are found in nuclear bodies—for example, HY5 (ELONGATED HYPOCOTYL 5), HFR1 (LONG HYPOCOTYL IN FAR-RED 1), ELF3 (EARLY FLOWERING 3), and several B-BOX DOMAIN PROTEINs, including BBX4, 21, and 22 (Ang et al. 1998; Jang et al. 2005; Datta et al. 2006, 2007, 2008; Yu et al. 2008). However, the transcription factor family that is best understood in phyB signaling and photobody function is the PIF (PHYTOCHROME-INTERACTING FACTOR) family (Duek and Fankhauser 2005; Leivar and Quail 2011), consisting of a group of 8 basic-helix-loop helix transcription factors. PIFs interact with photoactivated phyB via their Active-PHYB Binding (APB) motif (Khanna et al. 2004). In contrast to PIL1 and PIL2 (PIF3-Like1 and 2, also called PIF2 and 6) (Roig-Villanova et al. 2006; Penfield et al. 2010; Luo et al. 2014), PIF1, 3, 4, 5, 7, and 8 function as antagonists of phytochrome signaling (Huq and Quail 2002; Kim et al. 2003; Bauer et al. 2004; Fujimori et al. 2004; Huq et al. 2004; Oh et al. 2004, 2020; Li et al. 2012). PIFs suppress photomorphogenesis by inhibiting chloroplast biogenesis, stimulating hypocotyl growth, and promoting apical hook formation and maintenance (Leivar et al. 2008b; Shin et al. 2009; Stephenson et al. 2009). Additionally, PIFs also regulate diverse growth and developmental processes throughout a plant's life cycle, especially in response to environmental cues (Leivar and Quail 2011; Balcerowicz 2020; Sharma et al. 2023). PIFs not only mediate the light and temperature information given by light receptors but also integrate these environmental cues and intrinsic hormonal pathways. Connections to most plant hormonal pathways have been identified, including gibberellin (de Lucas et al. 2008; Feng et al. 2008), brassinosteroid (Oh et al. 2012; Bernardo-García et al. 2014), auxin (Franklin et al. 2011; Sun et al. 2012), ethylene (Khanna et al. 2007; Liu et al. 2017b), cytokinin (Richter et al. 2010; Aizezi et al. 2022), abscisic acid (Kim et al. 2016; Qi et al. 2020), and jasmonate (Yang et al. 2012; Fernández-Milmanda et al. 2020). Here, PIFs either regulate the transcription of hormone biosynthesis, transport and signaling genes or physically interact with hormone signaling components. Good examples are the various interactions with the auxin pathway. For instance, PIFs induce the expression of YUCCA genes to promote auxin biosynthesis (Li et al. 2012; Sun et al. 2012) and the transcription of auxin transport activating AGC kinases (Willige et al. 2012; Park et al. 2019). Furthermore, PIFs interact with ARF (AUXIN RESPONSE FACTOR) and AUX/IAA transcriptional regulators (Oh et al. 2014; Xi et al. 2021).
PIFs mainly act in concert with each other, showing at least partial redundancy for the different physiological responses they execute. An example of this redundancy can be seen in hypocotyl growth in response to warm temperatures and low R:FR. These growth responses are mainly stimulated by PIF4, 5, and 7, but PIF4 plays the major role in the context of thermomorphogenesis, while PIF7 is the master regulator in low R:FR and in conditions in which warm temperatures and low R:FR are perceived simultaneously (Burko et al. 2022).
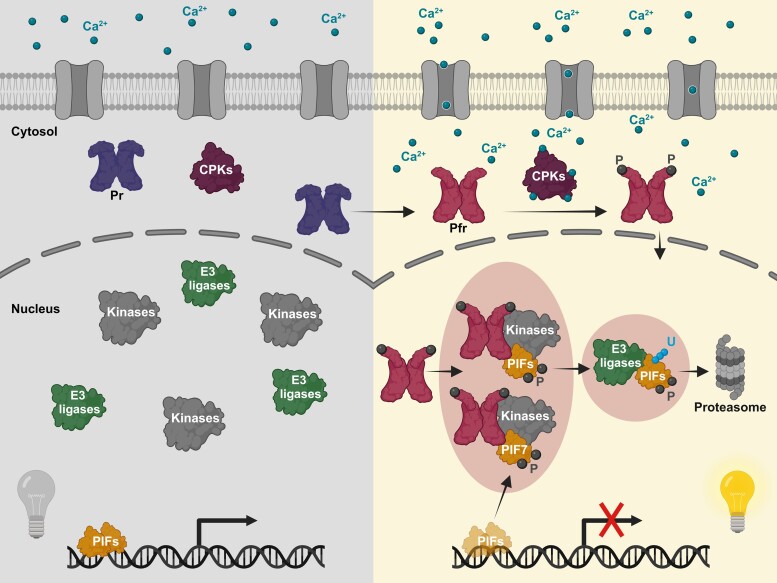
Like the classification of phytochromes as photolabile (phyA-type) and photostable (phyB-type) (Clough and Vierstra 1997), PIFs also fall into 2 separate groups. While PIF1, 3, 4, 5, and 8 are destabilized by R light and phyB activity (Bauer et al. 2004; Park et al. 2004; Shen et al. 2005, 2007; Lorrain et al. 2008; Oh et al. 2020), PIF7 protein levels are stable in response to R light and are not negatively affected by phyB (Leivar et al. 2008a; Willige et al. 2021). Independently of their photostability, photoactivated phyB recruits PIFs to photobodies (Fig. 3). Here, binding by phyB leads to inactivation and phosphorylation of PIFs and subsequently polyubiquitination and, eventually, degradation of red light labile PIFs (Bauer et al. 2004; Al-Sady et al. 2006; Shen et al. 2008; Ni et al. 2013, 2014, 2017). Furthermore, PIFs also seem to play a role in phyB photobody formation. This is supported by the absence of early photobodies in pif3 mutants during de-etiolation in R light (Bauer et al. 2004). In addition, both phyB and PIF7 can independently form nucleolus localized speckles when tethered to nucleoli. Interestingly, when only PIF7 is tethered to nucleoli, it recruits phyB into its speckles, which indicates that PIF7 helps photobody initiation/stabilization (Liu et al. 2014). This is supported by a recent study, where phyB formed faster and larger photobodies in a PIF7 overexpressor line after the transition from low R:FR to white light (Xie et al. 2023).
Figure 3.
In the dark, phyB is located in the cytoplasm in its inactive Pr conformation. Hence, PIFs are abundant and active to regulate their target genes. Upon light activation and conformational switch to its Pfr form, phyB activates calcium channels by an unknown mechanism. Hence, an increased intracellular Ca2+ concentration leads to activation of CPKs (CALCIUM-DEPENDENT PROTEIN KINASEs) that phosphorylate the light receptor and induce its nuclear translocation where it inactivates PIFs. Subsequently, PIFs are phosphorylated and ubiquitinated to determine their degradation via the 26S proteasome. PIF phosphorylation, ubiquitination, and degradation are mediated by the output module, but it is unclear if these processes are all happening within the same photobody. PIF7 is only phosphorylated and inactivated by phyB activity but not targeted for degradation. U denotes ubiquitination, while P indicates phosphorylation events.
The founding member of the PIF family, PIF3, was originally identified in a yeast 2-hybrid screen using the C-terminal output module of phyB as bait protein (Ni et al. 1998). Nevertheless, PIFs do not only interact with phyB's output module but also with the light-sensing knot lasso located in the photosensory domain (Ni et al. 1999; Kikis et al. 2009). It was proposed that only the latter interaction is crucial for phyB signaling and PIF inactivation and degradation; deleting the output domain and forcing the remaining photosensory module to dimerize in the nucleus generates a protein that can trigger various phyB responses without being able to form photobodies (Matsushita et al. 2003; Oka et al. 2008). This suggested that the output domain and photobodies are dispensable for phyB signaling. Nevertheless, subsequent studies demonstrated that PIF interaction with both phyB modules is necessary to execute proper phyB signaling. Using the dimerizing photosensory domain revealed that this N-terminal module sequesters PIF3 and prevents its DNA binding without inducing PIF phosphorylation and degradation (Park et al. 2012). Additionally, this interaction with phyB's N-terminal photosensory module suppresses PIF3's transcriptional activity, which is mediated by a single, evolutionary conserved transactivation domain (Yoo et al. 2021). However, phyB without its C-terminal output module does not properly inhibit hypocotyl elongation in growth conditions with light/dark cycles. Furthermore, a phyB C-terminal mutation that prevents phyB dimerization and photobody formation revealed that it is the output module that is responsible for the induction of PIF phosphorylation and degradation (Qiu et al. 2017; Paik et al. 2019). The N-terminal sequestration function seems to mediate phyB's fast response after photoactivation, while the C-terminal interaction initiates a more extended response that leads to a more profound outcome: the degradation of PIF transcription factors (Park et al. 2018). The sole exception seems to be PIF7 that does not accumulate in phyB mutants and that is not degraded by R light (Leivar et al. 2008a; Willige et al. 2021). Therefore, PIF7 seems to form a stable PIF pool that allows quick responses after phyB inactivation without the necessity of de novo PIF translation. This could explain why PIF7 can bind to its DNA target sites within 5 minutes of low R:FR light exposure (Willige et al. 2021). However, PIF7 is sequestered into photobodies and is phosphorylated by phyB activity, indicating that both phyB's N- and C-terminal modules are also relevant for PIF7 inactivation. Hence, photobodies serve to inactivate all PIFs by posttranslational modifications and for this purpose diverse enzymes are recruited into photobodies.
Recruitment of PIF modifying kinases and phosphatases by photobodies
The current model proposes that PIFs bound by phyB are first phosphorylated and subsequently polyubiquitinated to induce their degradation by the 26S proteasome (Al-Sady et al. 2006; Lorrain et al. 2008; Ni et al. 2013). This raises the possibility that phyB itself phosphorylates the PIF proteins, since bacterial (and fungal) phytochromes bear histidine kinase activity. Interestingly, phyB carries this heritage in its HKRD at its C terminus, which is necessary for PIF phosphorylation (Qiu et al. 2017). However, not all amino acid residues that are crucial for the catalytic activity of histidine kinases are present in phytochrome HKRDs (Boylan and Quail 1996), and too tight packing of phyB's ATP-binding pocket could prevent a potential kinase activity (Li et al. 2022b). In addition to the HKRD, also the hinge region that links both phytochrome modules, the PAS-related domain as well as the photosensory module were proposed to bind ATP to confer serine/threonine kinase activity of plant phytochromes (Wong and Lagarias 1989; Yeh and Lagarias 1998; Shin et al. 2016). Nevertheless, the significance of a potential phyB kinase activity for its signaling processes has been debated for decades.
Several kinases have been reported to phosphorylate PIFs. The MLK/PPK (MUT9P-LIKE-KINASE/PHOTOREGULATORY PROTEIN KINASE) family form a small group of nuclear localized Casein Kinases 1-like proteins that regulate hypocotyl growth and flowering time. The kinases were co-purified as an interactor of PIF3 and phyB in a R light-induced manner. Furthermore, in the dark, MLK4/PPK1 is evenly distributed in the nucleoplasm but concentrates in nuclear speckles after a R light exposure (Ni et al. 2017). However, the recruitment of MLK1/PPK2, MLK2/PPK3, and MLK4/PPK1 into photobodies requires the presence of PIF3. The transcription factor serves as a primary client that is required to relocate MLKs/PPKs into photobodies (Kim et al. 2023a).
Loss of MLK/PPK function attenuates the light-dependent phosphorylation and degradation of PIF3. Additionally, in vitro assays demonstrate that MLK4/PPK1 phosphorylates PIF3 at amino acid residues that were previously identified as light-inducible phospho-sites. Altogether, these results make the MLK/PPK family a good candidate for a phytochrome-activated kinase that induces PIF degradation (Ni et al. 2017). However, surprisingly, higher order mlk/ppk mutants show a reduced hypocotyl growth in contrast to the expected R light hyposensitivity that would go along with PIF stabilization (Huang et al. 2016a; Ni et al. 2017). It was proposed that the underlying cause for the repressed growth are the higher phyB protein levels in mlk/ppk mutants. Alternatively, MLKs/PPKs have pleiotropic functions (Wang et al. 2015, 2021; Liu et al. 2017a; Wirthmueller et al. 2018; Zheng et al. 2018), and the short hypocotyls might not be directly derived from altered phyB signaling. This is also supported by the fact that viable quadruple mlk/ppk knockout mutants could not be generated yet, while the loss of PHYB is certainly not lethal.
SPA1 (SUPPRESSOR OF PHYA-105 1) possesses kinase activity and regulates PIF1 and 4 phosphorylation (Paik et al. 2019; Lee et al. 2020). However, currently, it is unclear if SPA proteins serve as kinases that directly transfer R light-induced phyB activity into PIFs phosphorylation to promote their degradation.
SOS2 (SALT OVERLY SENSITIVE 2) is a serine/threonine kinase that phosphorylates and activates SOS1 under salt stress. Since SOS1 is a plasma membrane–localized Na+/H+ antiporter, its activation leads to a decrease of the cytosolic Na+ concentration (Mahajan et al. 2008). Additionally, it was recently demonstrated that SOS2 is also active within the nucleus, where it phosphorylates PIF1 and 3. Both transcription factors foster salt sensitivity. SOS2 interacts with the 2 PIF proteins and phyB in nuclear bodies, indicating that they altogether colocalize in photobodies. Phytochromes are able to activate SOS2 to induce PIF1 and 3 phosphorylation and reduce their stability to increase salt tolerance (Ma et al. 2023). Notably, there are also reports that describe negative effects of phyB on salt stress tolerance (Kwon et al. 2018; Yang et al. 2018; Liu et al. 2023).
In another study, it was shown that SOS2 also phosphorylates PIF4 and 5 (Han et al. 2023). Surprisingly, this does not lead to increased PIF turnover, as in the case of PIF1 and 3, but to an increased stability of PIF4 and 5. By this means, SOS2 can act as a positive regulator of hypocotyl elongation in low R:FR and counter the reduced growth response during salt stress (Hayes et al. 2019; Han et al. 2023). SOS2 interacts with PIF4 and PIF5 within nuclear bodies, but at this point it is unclear if these are formed by phyB.
The mitogen activated protein kinase MPK6 (MAP KINASE 6) and PIF3 antagonistically regulate cotyledon opening. MPK6 can phosphorylate PIF3 in vitro and is activated by R light through phosphorylation by MKK10 (MAP KINASE KINASE 10). MPK6 and PIF3 are colocalized in nuclear bodies (Xin et al. 2018). Nevertheless, it is not shown yet if these speckles have phyB photobody identity. Additionally, we currently do not understand how MKK10, MPK6, and phyB work together to mediate PIF3 phosphorylation.
Besides the herein described kinases, PIFs were also shown to be phosphorylated by BIN2 (BRASSINOSTEROID-INSENSITIVE 2) (Bernardo-García et al. 2014; Ling et al. 2017) and CK2 (CASEIN KINASE 2) (Bu et al. 2011), but recruitment to photobodies has not been studied yet. Altogether, it is likely that PIFs are phosphorylated by a concert of kinases; some act downstream of phyB in photobodies, while others likely function to integrate PIF activity into connected signaling pathways.
PIF phosphorylation is reversible, but the underlying regulatory mechanisms are very little understood. PP6 phosphatases and TOPP4 (TYPE ONE SERINE/THREONINE PROTEIN PHOSPHATASE 4) were reported to interact with diverse PIFs and regulate their phospho-status and protein stability (Yue et al. 2016; Yu et al. 2019). Currently, it is unknown if PIFs get dephosphorylated within photobodies or after their release from photobodies. Both mechanisms could coexist, as indicated by bimolecular fluorescence complementation; in the dark, TOPP4 binds to PIF3 in the nucleoplasm, while R light relocates their interaction to nuclear speckles (Yue et al. 2016). However, sequestering phosphatases into photobodies could also represent a potential mechanism to inactivate these enzymes and limit PIF activity.
Recruitment of E3 ubiquitin ligases by photobodies
A plethora of E3 ubiquitin ligases initiate proteasomal degradation of proteins by polyubiquitination. Hereby, the substrate recognition by the E3 provides specificity to the degradation process (Mazzucotelli et al. 2006). There are several E3 ligase candidates that could induce PIF degradation within photobodies after phyB activation.
The LRB 1 to 3 (LIGHT-RESPONSE BTB1 to 3) form a small family of BTB (Broad-complex, Tramtrack, and Bric-à-brac) proteins that serve as substrate recognition adaptors of CULLIN3-based E3 complexes. The family was initially identified in a reverse genetic screen and described as negative regulators of phyB protein abundance and hence R light signaling (Christians et al. 2012). However, a subsequent study revealed that LRBs not only promote phyB turnover but also initiate PIF3 degradation after its phosphorylation in response to R light (Ni et al. 2014). In the dark and in R light, LRB1 and 2 are evenly distributed within the nucleoplasm and do not seem to be recruited to photobodies (Christians et al. 2012). This suggests that phyB activity leads to the recruitment of other E3 s into photobodies or that PIF3 is released from photobodies after phosphorylation to initiate its degradation via LRB-mediated polyubiquitination within the nucleoplasm. The second option is supported by the observation that mimicking phosphorylation of PIF3 increases its turnover even in etiolated seedlings (Ni et al. 2013), where photobodies are absent. Nevertheless, both alternatives are certainly not mutually exclusive, especially since in recent years several other E3 ligases were identified that target PIF proteins. For instance, phyB utilizes EBF1 and 2 (EIN3-BINDING F BOX PROTEIN 1 and 2) to induce degradation of PIF3 and of EIN3 (ETHYLENE-INSENSITIVE 3) to promote photomorphogenesis via 2 distinct molecular mechanisms: The phyB-dependent phosphorylation of PIF3 is sufficient to induce the recognition by EBF1/2 without further requirement of phyB activity (Dong et al. 2017). In contrast, phyB seems to directly enhance the binding affinity of EBF1/2 to EIN3 by acting as a scaffold for the transcription factor and the F-box proteins (Shi et al. 2016).
We currently do not know if LRBs and EBFs are recruited to photobodies. However, other E3 ligases that control PIFs were at least shown to be located in some type of nuclear speckles. PIF1 interacts with the F-box protein CTG10 (COLD TEMPERATURE GERMINATING 10) and PIF4 binds to BOP2 (BLADE-ON-PETIOLE 2) within nuclear localized speckles in bimolecular fluorescence complementation assays. CTG10 negatively regulates PIF1 levels to induce germination and promote photomorphogenesis, while BOP1 and 2 reduce PIF4 abundance to lower the response to higher ambient temperatures and promote photomorphogenesis (Zhang et al. 2017; Majee et al. 2018).
The RING finger E3 ubiquitin ligase HOS1 (HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 1) is mainly located at the nuclear envelope, which is in agreement with its function in nuclear-cytoplasmic mRNA export (Tamura et al. 2010; Lazaro et al. 2012; MacGregor et al. 2013). However, interaction with at least 1 of its protein substrates is concentrated in nuclear bodies: HOS1 promotes turnover of the flowering regulator CO (CONSTANS), which seems to be mediated by phyB (Jung et al. 2012; Lazaro et al. 2012, 2015). Interestingly, HOS1 also represses PIF4, however, without inducing its turnover. Instead, HOS1 is phyB dependently recruited to PIF4 DNA binding sites and inhibits PIF4's transactivation activity (Kim et al. 2017).
The best characterized E3 ligases that regulate light responses are COP1-SPA complexes. The Arabidopsis genome encodes for 1 COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1) gene and for 4 SPA paralogues. COP1-SPA complexes form E3 ligases that initiate the degradation of positive regulators of photomorphogenesis. Many COP1-SPA substrates are transcription factors, like HY5, HYH, PIL1, and HFR1 (Osterlund et al. 2000; Holm et al. 2002; Duek et al. 2004; Luo et al. 2014). In addition, COP1-SPA complexes functionally interact with DET1 (DE-ETIOLATED 1) (Nixdorf and Hoecker 2010). Arabidopsis plants with a loss of DET1 function were the very first described mutants that showed a de-etiolated/constitutive photomorphogenic phenotype in darkness (Chory et al. 1989). Like COP1 and SPAs, DET1 forms CULLIN4-based E3 complexes that are necessary to suppress photomorphogenesis in the dark (Lau and Deng 2012). Currently, DET1 is functionally less understood than COP1 and SPA proteins. However, a recent study suggested that DET1 and COP1 jointly work in HY5 degradation, since DET1 is able to interact with both COP1 and HY5 and fosters the interaction between HY5 and COP1 and, hence, HY5 degradation (Cañibano et al. 2021).
In addition to their role in degradation of positive light signaling components, COP1, DET1, and SPAs stabilize PIF transcription factors (Bauer et al. 2004; Leivar et al. 2008b; Dong et al. 2014; Shi et al. 2015; Gangappa and Kumar 2017; Pham et al. 2018). For example, COP1 and SPAs prevent PIF3 phosphorylation by BIN2 kinases that otherwise mark PIF3 for degradation (Ling et al. 2017). Altogether, COP1, DET1, and SPAs act PIF dependently and independently to promote skotomorphogenesis, thermomorphogenesis, and shade avoidance and hence act as negative regulators of phyB activity.
Different light receptors inactivate COP1-SPA complexes by at least 3 mechanisms: (1) nuclear exclusion of COP1, (2) degradation of SPA subunits, and (3) light receptor–SPA protein interaction to prevent COP1-SPA complex formation. COP1 nuclear exclusion by R light was described in the past but is disputed by more recent reports (Osterlund and Deng 1998; Jang et al. 2010; Balcerowicz et al. 2017). Nevertheless, COP1 re-accumulates in the nucleus under canopy shade (Pacín et al. 2013). In addition, phyB can sequester SPA proteins to prevent COP1-SPA binding and initiate the degradation of SPAs (Lu et al. 2015; Sheerin et al. 2015). COP1 and SPA proteins colocalize in nuclear speckles that can have phyB photobody identity (Seo et al. 2003; Zheng et al. 2013; Kim et al. 2023a). This indicates that in analogy to PIF inactivation, photobodies serve to sequester, inactivate, and degrade these negative regulators of photomorphogenesis. In addition, photobodies seem to functionally repurpose COP1 and SPAs by turning them into positive regulators of phyB activity: COP1 is required for the R light-induced degradation of SPA2 and potentially other SPAs (Chen et al. 2015). Additionally, COP1 and SPA proteins are involved in the phosphorylation and degradation of PIFs in response to R light (Zhu et al. 2015; Pham et al. 2018; Paik et al. 2019). However, based on cop1's de-etiolated phenotype, its role in suppressing phyB action is more profound than its role in promoting R light signaling. In agreement, COP1 is also a negative regulator of photobody formation and maintenance by regulating structural components of these subnuclear domains.
Structural photobody components regulate gene expression
As stated above, in the dark, the Pfr form of phyB slowly switches back into its inactive Pr form, a process called dark or thermal reversion. Interestingly, the in vivo dark reversion is much slower than the reversion in vitro (Rausenberger et al. 2010). This indicates that phyB modifications or phyB binding proteins decelerate the dark reversion rate. One of these phyB interactors is PCH1 (PHOTOPERIODIC CONTROL OF HYPOCOTYL 1), a positive regulator of R light-mediated photomorphogenesis (Huang et al. 2016b). PCH1 not only directly interacts with phyB and is part of photobodies but also stimulates the formation of large photobodies and the maintenance of photobodies during the night. Hence, overexpression of PCH1 accelerates the appearance of photobodies after activating phyB and prevents photobody dissociation in the dark. It turned out that PCH1 and its paralogue PCHL (PCH1-LIKE) slow down the dark/thermal reversion of phyB by stabilizing phyB's Pfr form without interfering with its intrinsic R-FR photoconversion (Fig. 4) (Enderle et al. 2017; Huang et al. 2019). Stabilizing the Pfr form and hence photobodies extends phyB activity and therefore R light signaling in the night. Therefore, pch1 mutants show long hypocotyls in short-day conditions and in long days that coincide with elevated temperatures (when the dark/thermal reversion is accelerated). Since the Pfr form of phyB sequesters PIFs and initiates their degradation, PIF4 protein levels in pch1 are increased during the night (Huang et al. 2019). This could also be the consequence of increased PIF4 transcript levels in pch1. Altogether, the increased PIF4 levels go along with higher PIF target gene expression. Furthermore, PCH1 and PCHL seem to regulate PIF activity also by another mechanism, since they not only regulate hypocotyl growth in the night but also various R light responses, including seed germination and chlorophyll biosynthesis (Cheng et al. 2020). Interestingly, PCH1/PCHL negative influence on PIF1 abundance is only partial phyB dependent: PCH1/PCHL directly interact with PIF1 in a phyB-independent way. Additionally, in PCH1/PCHL overexpressors, PIF1 protein levels are lower even in etiolated seedlings before any light inputs and hence prior phytochrome activity. This phyB-independent function has consequences for PIF1 DNA binding and target gene expression: both are reduced in imbibed seeds of PCH1 overexpressors after phyB inactivation by FR light. Nevertheless, phyB binds stronger to PIF1 in the presence of PCH1/PCHL. The latter might be due to a bridging activity by PCH1/PCHL or due to the acceleration of photobody formation by these proteins.
Figure 4.
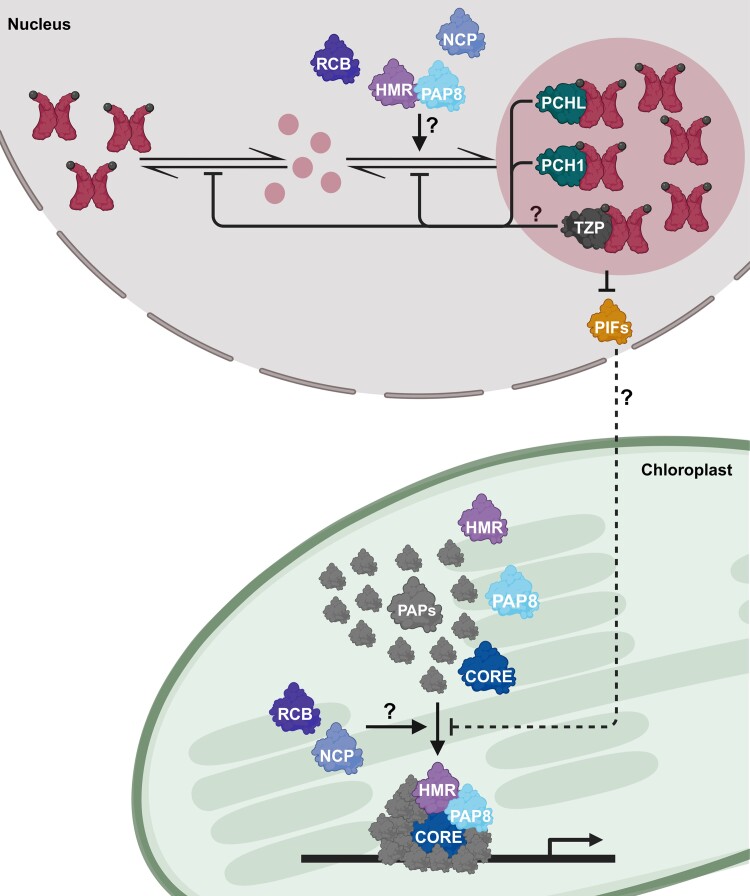
Function of photobody regulators. PCH1 and PCHL stabilize phyB's Pfr form and hence prevent photobody disassembly. TZP also regulates photobody formation and growth, but it is currently unknown if this happens through the stabilization of phyB's Pfr form as well. HMR, PAP8, RCB, and NCP are located within the nucleoplasm and chloroplasts. They exhibit a dual function, since they promote photobody growth and the assembly of plastid-encoded RNA polymerase complexes consisting of core proteins and PAPs. Additionally, the formation of the PEP holoenzyme complex is regulated by PIF-dependent nucleus-to-plastid signaling.
In addition to phyB and PIFs, PCH1 also interacts with members of the TPL/TPR (TOPLESS/TOPLESS-RELATED) family, a group of transcriptional corepressors. These proteins are recruited to target genes by various transcription factors to negatively regulate gene expression (Plant et al. 2021). phyB alone does not bind TPL protein, and hence PCH1 serves as a primary photobody client to pull TPL and related proteins into photobodies. Interestingly, multiple tpl/tpr mutants show reduced hypocotyl growth in R light, indicating that TPL/TPR-dependent gene repression serves to decrease phyB signaling (Kim et al. 2023a). Hence, incorporation of TPL/TPR proteins into photobodies could be a means of preventing their interaction with transcription factors or could serve to isolate transcription factor-TPL/TPR complexes from their DNA binding sites. PCH1/PCHL also interact with COP1, which can lead to COP1-induced PCH1/PCHL ubiquitination and subsequent degradation in the dark (Cheng et al. 2020). In consequence, COP1 could promote the dark/thermal reversion of phyB by lowering PCH1/PCHL levels and thus decreasing Pfr stability.
Through binding of PIFs, COP1-SPA complexes and TPL/TPR proteins, photobodies regulate transcription indirectly. However, photobodies themselves are locations of active gene expression as work on TZP (TANDEM ZINC KNUCKLE PROTEIN) has demonstrated. TZP was originally identified in a study of natural variations for hypocotyl elongation and encodes a protein with 2 types of nucleotide binding domains: 1 PLUS3 and 2 zinc finger domains (Loudet et al. 2008). TZP is recruited by phyB into photobodies which are disrupted by chemicals that are known to block transcription (Kaiserli et al. 2015). This is an indication that photobodies are associated with transcriptional processes. Furthermore, TZP binds single, but not double stranded DNA in vitro, which might indicate that TZP binds to separated DNA strands in active chromatin regions. This is supported by TZP's binding to 2 loci of floral activators (CO and FT). TZP's DNA binding and TZP-dependent gene activation is inhibited in the absence of photobodies (by FR light treatment or loss of phyB function), supporting the hypothesis that TZP-dependent gene regulation is happening in transcriptionally active photobodies (Kaiserli et al. 2015).
As described above for PCH1/PCHL, TZP also physically interacts with COP1, which facilitates TZP degradation (Li et al. 2022a). However, this is not the only common feature of PCH1/PCHL and TZP, since tzp pch1 double mutants synergistically suppress R light signaling in the context of hypocotyl growth (Fang et al. 2022). In tzp pch1 double mutants, PIF4 levels do not only accumulate during the dark phase but also during the day, even though PIF4 transcription is barely affected in these mutants. In consequence PIF target gene expression is upregulated in tzp pch1. TZP nuclear body formation is impaired in pch1, which is not surprising, since TZP requires phyB to be incorporated into photobodies and PCH1 promotes phyB photobody formation. However, TZP is a structural component of photobodies as well, since photobody formation and growth are also negatively affected in tzp mutants (Fang et al. 2022), but it is not demonstrated yet if this also happens through the stabilization of phyB's Pfr form (Fig. 4).
The dual function of photobody regulators
After the discovery of phyB photobodies, forward genetic screens led to the identification of regulators of photobody formation. The first protein that was reported to be one of these regulators was HMR (HEMERA), a nucleus and plastid dual-localized protein (Chen et al. 2010). HMR was identified in a confocal-based forward genetic screen in a mutagenized PHYB:GFP background. Loss of HMR does not affect phyB protein levels but affects the size of phyB photobodies, which have a much smaller size than photobodies in the wild-type under continuous red light (Chen et al. 2010). HMR directly interacts with phyB in a light-dependent manner (Galvão et al. 2012). In addition, HMR directly interacts with and regulates the degradation of PIF1 and PIF3 as a transcriptional coactivator (Qiu et al. 2015).
HMR is also known as PAP5/pTAC12 (PEP-ASSOCIATED PROTEIN 5/PLASTID TRANSCRIPTIONALLY ACTIVE 12), since it is one of the associated proteins of the plastid-encoded RNA polymerase (PEP) complex. HMR/PAP5/pTAC12 was co-purified with the plastid transcriptionally active chromosome (pTAC) fraction from chloroplasts (Pfalz et al. 2006; Pfannschmidt et al. 2015). This is why hmr mutants show a combination of tall hypocotyl and albino phenotypes due to defects in phyB signaling in the nucleus and in PEP activity that is essential for chloroplast biogenesis. Interestingly, it has been demonstrated that HMR is first targeted to plastids, where it is processed to its mature form and from there it is relocated to the nucleus (Nevarez et al. 2017). Altogether, these studies propose that once photoactivated phyB is translocated into the nucleus, HMR promotes the formation of large phyB photobodies in the nucleus while HMR is also assembled into the PEP complex in chloroplasts (Fig. 4).
The dual role of HMR in photobody formation in the nucleus and PEP activation in plastids is in line with the nuclear control of chloroplast biogenesis. A recent report showed that photo-activated phytochromes in the nucleus send a nucleus-to-plastid signal to trigger the assembly of PEP complexes and chloroplast biogenesis by promoting PIF degradation (Yoo et al. 2019). Accordingly, forward genetic screens for hmr-like tall-and-albino mutants defective in phytochrome-mediated control of the PEP assembly identified 2 novel phytochrome signaling components: REGULATOR OF CHLOROPLAST BIOGENESIS (RCB) and NUCLEAR CONTROL OF PEP ACTIVITY (NCP) (Yang et al. 2019; Yoo et al. 2019). Like hmr mutants, both rcb and ncp mutants are defective in the formation of large photobodies, leading to the accumulation of PIF1 and PIF3 (Fig. 4) (Yang et al. 2019; Yoo et al. 2019). RCB/NCP-dependent photobody formation and PIF degradation in the nucleus triggers the assembly and activation of the PEP complex in chloroplast biogenesis (Yang et al. 2019; Yoo et al. 2019). Although they are dual-localized proteins, RCB regulates the PEP assembly and complex primarily in the nucleus via nucleus-to-plastid signaling, while NCP located in plastids has an essential role in PEP assembly. Both proteins contain noncatalytic thioredoxin-like domains in their C terminus. The biochemical functions of RCB and NCP in regulating photobody formation and the assembly of the PEP complex are still unknown.
Recently, another subunit of the PEP complex, PAP8/pTAC6, was reported to affect photobody formation presumably by acting together with HMR/PAP5/pTAC12. Mutation of PAP8 suppresses the short hypocotyl phenotype of PHYB:GFP overexpressors under red light and leads to defects in the formation of large photobodies (Liebers et al. 2020). PAP8 is also targeted to both the nucleus and chloroplasts, where it directly interacts with HMR/PAP5. Since the size of nuclear PAP8 is similar to the transit peptide-cleaved form of chloroplast PAP8, this study proposes that PAP8 is translocated from chloroplasts to the nucleus. Here, PAP8 physically interacts with HMR to promote the formation of large photobodies (Fig. 4). The regulation of photobody formation by PAP8 might work through HMR because the direct interaction between PAP8 and phyB has not been demonstrated yet.
It is worth pointing out that all these regulators of photobody stability share similar characteristics. First, HMR, RCB, NCP, and PAP8 are all dual-localized proteins in the nucleus and in chloroplasts. Second, they are required for the formation of large photobodies without affecting phyB protein level or nuclear localization. Third, they are essential for chloroplast biogenesis since all mutants are albinos. Fourth, HMR and PAP8 are essential components of the PEP complex, while RCB and NCP are required for PEP assembly even though they are not thought to be components of the PEP complex. Fifth, none of these factors have been identified as structural components of photobodies. Altogether, these factors suggest that although the regulatory mechanism of photobody stability by these proteins is still unclear, the formation of large photobodies is tightly linked to the formation of the PEP holoenzyme complex via PIF-dependent nucleus-to-plastid signaling (Fig. 4) (Hwang et al. 2022). The nature of this anterograde signal is unknown; however, photobody stability during light exposure must be important to initiate light-triggered chloroplast biogenesis in a seedling's early development. It is also worth mentioning that these regulators are all plant-specific proteins evolved during land adaptation and do not exist in cyanobacteria or green algae (Chen et al. 2010; Nishiyama et al. 2018; Yang et al. 2019). Their regulatory mechanisms on photobody stability might have evolved to optimize photomorphogenesis including chloroplast biogenesis during terrestrial adaptation.
Photobodies recruit splicing factors to regulate gene expression
A cell's precursor mRNAs (pre-mRNAs) need to undergo splicing in order to form mature mRNAs that serve as templates for protein production. Hence, introns must be removed and exons must be fused. These reactions are mediated by the major spliceosome, which consists of over 100 proteins and several small protein–RNA complexes, called small nuclear ribonucleoproteins. These include U1, 2, 4, 5, and 6 that recognize splicing signals within pre-mRNA molecules (Will and Lührmann 2011; Matera et al. 2014; Lee and Rio 2015).
Mandatory retention of exons and removal of introns is called constitutive splicing, while the inclusion of introns, the exclusion of exons, or the use of nonconstitutive splice sites are called alternative splicing. Alternative splicing produces multiple transcripts from a single gene by using different splice sites. Splicing factors such as SR proteins and heterogenous nuclear riboproteins promote or prevent access to these splicing signals. Therefore, they give specificity to each splicing process and determine the produced mature mRNA isoform (Reddy et al. 2013; Lee and Rio 2015). Interestingly, this specificity is regulated by photobodies.
About 7% of all Arabidopsis genes undergo phytochrome A and B-dependent alternative splicing after R light exposure for up to 3 h (Shikata et al. 2014). For example, phytochromes promote alternative splicing of SPA3 transcripts. This leads to a loss of SPA3's C terminus due to the formation of premature termination codons. Unlike its full-length isoform, the truncated SPA3 proteins act as repressors of hypocotyl elongation (Shikata et al. 2014). Another example of phyB-mediated alternative splicing affects PIF3 translation. The 5′UTR of PIF3 contains 2 introns. Within the second intro lies an upstream open reading frame. R light and phyB promote retention of this upstream open reading frame in the spliced PIF3 mRNA, which negatively affects PIF3 translation (Dong et al. 2020).
These 2 examples demonstrate how phytochrome-activity promotes R light signaling through alternative splicing. So far, 6 phyB interacting splicing factors were identified. For 2 heterogenous nuclear riboproteins that regulate alternative splicing in the moss Physcomitrella patens (Shih et al. 2019; Lin et al. 2020), no photobody localization has been assessed yet. However, 4 different splicing factors were identified in Arabidopsis. They colocalize with phyB in nuclear speckles, indicating that photobodies directly influence pre-mRNA splicing.
RRC1 (REDUCED RED-LIGHT RESPONSES IN CRY1CRY2 BACKGROUND 1) was identified in a screen for R light hyposensitive mutants (Shikata et al. 2012). RRC1 encodes for an ortholog of the human splicing factor SR140. RRC1 and SR140 are SR proteins that carry a serine and arginine-rich domain at their C termini (RS domain) and an N-terminal RNA recognition domain (RRM domain). Loss of RRC1's C-terminal RS domain leads to reduced phyB signaling. These mutations or loss of PHYB affect alternative splicing of several SR protein genes in response to R light. However, rrc1 null mutants show pleiotropic developmental defects, which indicates that RRC1's function is not limited to phyB signaling (Shikata et al. 2012).
RRC1 interacts with SFPS (SPLICING FACTOR FOR PHYTOCHROME SIGNALING). Both splicing factors have several features in common (Xin et al. 2017, 2019). Like RRC1, SFPS was also identified in a screen for R light hyposensitive mutants. SFPS is related to the human splicing factor SPF45. The human orthologues of RRC1 and SFPS (SR140 and SPF45) associate with the small nuclear ribonucleoprotein U2 and the same seems to be true for RRC1 and SFPS (Xin et al. 2017, 2019). U2 and its associated factors bind to 3′ splice sites and intronic branch points (typically located up to 50 bp upstream of the 3′ splice site). Hence, pre-mRNA binding by U2 and its associated factors define the actual 3′ splice site of each individual splicing reaction (Wahl et al. 2009; Matera et al. 2014). Interestingly, the interaction of RRC1 and SFPS is conserved, since human SPF45 and SR140 were later found to interact as well (Martín et al. 2021), which supports the notion that these splicing factors act jointly in various signaling pathways.
The above-mentioned similarities between RRC1 and SFPS go even further. Both proteins were shown to interact with phyB and to locate in nuclear speckles that at least partly colocalize with phyB photobodies (Xin et al. 2017, 2019). However, in the case of RRC1 colocalization and interaction with phyB could not be confirmed in all studies (Shikata et al. 2012; Kim et al. 2023b), which could be explained by differing experimental conditions.
In rrc1 and sfps mutants, splicing of genes that are associated with light stimulus, photosynthesis, and the circadian clock is impaired. One example is that ELF3: rrc1 and sfps mutants contain higher levels of not fully spliced ELF3 transcripts that encode for a nonfunctional ELF3 protein (Xin et al. 2017, 2019; Kim et al. 2023b). The alternative splicing of clock gene transcripts in rrc1 leads to misregulation of rhythmic gene expression and in consequence to higher PIF4, PIF5, and PIF7 protein levels and increased hypocotyl growth (Kim et al. 2023b).
Both RRC1 and SFPS bind independently and jointly to the splicing factor SWAP1 (SUPPRESSOR-OF-WHITE-APRICOT/SURP RNA-BINDING DOMAIN-CONTAINING PROTEIN1) (Kathare et al. 2022). As shown for RRC1 and SFPS, SWAP1 also interacts with phyB and colocalizes with phyB and U2 subunits in nuclear speckles. Altogether, phyB might recruit RRC1, SFPS, and SWAP1 to photobodies to concentrate these splicing factors to induce splicing of light signaling and circadian clock genes. The increased concentration of particular splicing factors could enhance the efficiency of specific splicing reactions to promote R light signaling. The association of RRC1, SFPS, and SWAP1 with U2 suggests that they link phyB activity with the selection of 3′ splice sites. It is worth pointing out that an alternative model was proposed in which phyB and RRC1 promote light responses spatially separated. While PHYB expression within the epidermis is sufficient to repress hypocotyl growth, RRC1 seems to act mainly within the endodermis to promote R light responses (Kim et al. 2023b).
While RRC1, SFPS, and SWAP1 act as positive components of phyB signaling, SMP2 (SWELLMAP 2) is a splicing factor that suppresses photomorphogenesis—it antagonizes phyB signaling to promote hypocotyl elongation (Yan et al. 2022). SMP2 was found as a phyB interactor in a yeast two-hybrid screen using the C-terminal output module. It is a homolog of human Slu7 that is also involved in 3′ splice site selection. SMP2 was found to regulate RVE8 (REVEILLE 8) pre-mRNA splicing. RVE8 is a transcription factor that induces the expression of several transcription regulators involved in hypocotyl growth repression (Hsu et al. 2013). Alternative splicing of RVE8 pre-mRNA can lead to nonfunctional mature mRNA isoforms (James et al. 2012). In smp2, more functional RVE8 mRNA isoforms are produced, which might explain the reduced hypocotyl growth in these mutants. SMP2 is localized in the nucleoplasm and in nuclear speckles, and some of these speckles exhibit photobody identity, since they colocalize with phyB (Yan et al. 2022). In analogy to the sequestration of other negative regulators of phyB signaling (like PIFs or SPAs), photobodies might isolate SMP2 from cofactors or pre-mRNA substrates to impair its activity.
Regulation of chromatin dynamics by photobodies
Active phyB also regulates gene expression by modifying gene activity on a macromolecular level. For example, phyB is involved in light-dependent chromatin compaction. Here, phyB activity stimulates the formation of highly condensed heterochromatin domains (chromocenters) that are transcriptionally inactive (Tessadori et al. 2009; van Zanten et al. 2010). In addition to chromatin compaction, phyB modifies the location of light-induced genes within the nucleus as shown for the reposition of the CAB (CHLOROPHYLL A/B BINDING PROTEIN) gene cluster. This gene cluster moves phyB dependently from the nuclear interior to the nuclear periphery. Since this repositioning seems to happen before the gene cluster is fully transcriptionally induced, it might be a mechanism to increase light-dependent gene expression. In contrast, in the dark, PIFs retain these light-inducible loci in the nuclear interior (Feng et al. 2014). This suggests that phyB activation and photobody formation induce PIF degradation to translocate light-inducible loci. It is possible that these loci move into the proximity of nuclear pore complexes to promote transcription, since these structures are not only important for gene activation in yeast and metazoan systems, but also in plants (Tamura 2020). However, it needs to be tested, if phyB-dependent gene reposition colocalizes with photobodies and/or nuclear pore complexes.
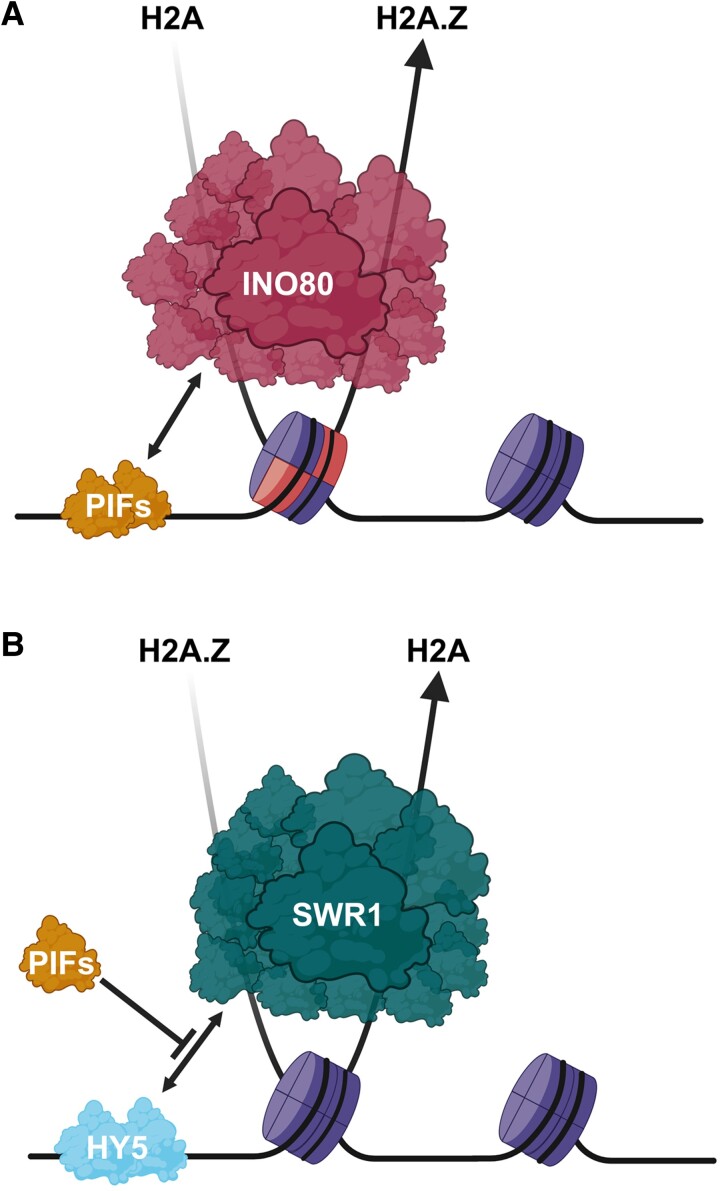
Another antagonistic function of phyB and PIFs is the control of nucleosome occupancy by the histone variant H2A.Z. Nucleosomes consist of a histone octamer and approximately 146 base pairs of DNA that are wrapped around it (Luger et al. 1997). The octamer is formed by 4 different histones, including H2A. H2A can be exchanged by H2A.Z within gene bodies, and by this means, gene expression and gene responsiveness is regulated. In Arabidopsis, higher H2A.Z levels within gene bodies are found to correlate with reduced gene activity (Coleman-Derr and Zilberman 2012; Yelagandula et al. 2014; Sura et al. 2017; Carter et al. 2018). Two evolutionary conserved chromatin remodeling complexes can exchange H2A for H2A.Z and vice versa. The SWR1 complex incorporates H2A.Z into nucleosomes, while the INO80 complex evicts H2A.Z and deposits H2A (Mizuguchi et al. 2004; Papamichos-Chronakis et al. 2011). These H2A-H2A.Z dynamics enable transcriptional responses to various environmental stimuli in plants, including light. Mutations of INO80 cause R light hypersensitivity, in part due to reduced H2A.Z occupancy at the HY5 gene body, which leads to increased expression of HY5 (Yang et al. 2020). However, SWR1 and INO80 complexes need to be recruited to light-responsive genes. Several recent studies have identified antagonistic phyB and PIF-dependent mechanisms to regulate H2A.Z occupancy. phyB was found to be able to bind 2 subunits of the SWR1 complex (SWC6 and ARP6) in a R light-dependent fashion (Wei et al. 2021). Using in vitro assays, the photosensory module and the output module interacted independently with both subunits. However, in planta interactions were found only in the presence of R light, indicating that the photobody forming Pfr form is necessary for these interactions. Like loss of PHYB function, mutations of both SWR1 subunits show light-hyposensitive growth and increased expression of auxin biosynthesis and response genes, suggesting that the SWR1 complex represses growth by lowering auxin biosynthesis. This is supported by the R light and SWR1-dependent deposition of H2A.Z at the YUCCA9 locus. Interestingly, this incorporation of H2A.Z is not only phyB but also HY5 dependent (Wei et al. 2021). The cause for the HY5 dependency is likely HY5's ability to bind to SWC6 and ARP6 as well. This interaction allows the recruitment of SWR1 complexes to HY5 target loci (Fig. 5) (Mao et al. 2021). Altogether, this indicates that light-dependent H2A-H2A.Z dynamics happen upstream (Yang et al. 2020) and downstream (Mao et al. 2021; Wei et al. 2021) of HY5.
Figure 5.
Negative regulation of H2A.Z occupancy by PHYTOCHROME-INTERACTING FACTORs. A) PIF4 and 7 recruit the INO80 complex to promote H2A.Z eviction and gene activity of PIF target genes in response to low R:FR or elevated temperatures. PIF4, 5, and 7 can interact with the small INO80 complex subunit EEN (EIN6 ENHANCER), a homolog of Ies6 (Ino eighty subunit 6) found in yeast. PIF4 was also shown to interact with the N-terminal region of INO80 itself. B) HY5 interacts with 2 SWR1 complex subunits, ARP6 and SWC6, to recruit the chromatin remodeling complex. This promotes H2A.Z incorporation into nucleosomes. Several PIFs physically interact with SWC6 as well, which seems to compete with the HY5's SWR1 recruitment.
PIF7 is the major PIF for the induction of hypocotyl growth in response to low R:FR (Li et al. 2012; Willige et al. 2021). After exposure to low R:FR, PIF7 gets dephosphorylated and changes its subnuclear localization. While PIF7 is concentrated in large photobodies in white light, it is released from these domains after low R:FR exposure (Willige et al. 2021; Xie et al. 2023). These responses likely precede PIF7 DNA binding to its target genes. These target genes are highly enriched in transcriptional regulators, suggesting that PIF7 acts upstream of a transcription factor cascade. Interestingly, 5 minutes of low R:FR exposure is sufficient to induce PIF7 DNA binding, while longer exposure times intensify this binding and lead to more binding events (Willige et al. 2021). This rapid DNA binding is accompanied by H2A.Z removal from gene bodies of PIF7 target genes. Hereby, PIF7 DNA binding and H2A.Z seem to precede the low R:FR-induced transcriptional activation of these genes. Importantly, this H2A.Z eviction in response to low R:FR is strongly compromised in pif457 mutants, demonstrating its PIF dependency.
EEN (EIN6 ENHANCER), the ortholog of the yeast and human Ies6 (INO80 Subunit 6) protein, is a small but essential subunit of the INO80 complex (Zander et al. 2019). Based on available cryo-EM structures of the INO80 complex, Ies6 is directly involved in nucleosome binding by the INO80 complex (Watanabe et al. 2015; Ayala et al. 2018; Eustermann et al. 2018). At least PIF4, 5, and 7 can interact with EEN, and een mutants show hyposensitivity to low R:FR. Additionally, een exhibits defects in low R:FR induced H2A.Z eviction (Willige et al. 2021). This indicates that PIFs recruit the INO80 complex via interaction with EEN to regulate H2A.Z removal and hence gene expression (Fig. 5). However, PIFs seem not only bind to EEN, since at least PIF4 also interacts with the N-terminal region of INO80, the largest subunit of the INO80 complex. In response to warm temperatures, these interactions mediate H2A.Z eviction at PIF4 target genes to facilitate gene activity and hence thermomorphogenesis (Xue et al. 2021).
Promoting H2A.Z eviction is not the only way PIFs regulate H2A.Z occupancy. It was shown that several PIFs are able to physically interact with SWC6, the aforementioned subunit of the SWR1 complex. This interaction happens within nuclear speckles, but at this point it is not clear if these foci are photobodies. Interestingly, PIF-SWC6 binding seems to compete with the HY5-SWC6 interaction that serves to recruit the SWR1 complex to HY5 target loci (Chen et al. 2023). Hence, besides facilitating H2A.Z eviction, PIFs also seem to prevent H2A.Z deposition that is promoted by HY5 (Fig. 5).
Overall, PIFs play an opposing role to phyB and HY5 in regulating H2A.Z occupancy. The promoters of photomorphogenesis interact with the SWR1 complex to deposit H2A.Z to reduce gene expression. In contrast, environmental signals such as low R:FR and warm temperatures reduce photobody formation/maintenance. In consequence, HY5 levels are reduced, but PIF activity is promoted. This leads to reduced H2A.Z occupancy and hence increased expression of PIF target genes.
Conclusions and perspectives
As summarized above, the current state of knowledge indicates that phyB photobodies are biochemical hotspots that concentrate factors to accelerate enzymatic reactions and to give these reactions specificity. Conversely, this does not necessarily mean that these biochemical processes are inactive in the absence of photobodies since muted phyB responses can be observed even without photobody formation. Nevertheless, these nuclear foci are required to adjust the growth and development of plants to environmental conditions. In this context it is noteworthy that photobodies respond not only to light and temperature changes that directly influence the Pr-Pfr conversion. For example, photobodies are also influenced by salinity and jasmonate. Relatively little is understood about how these other abiotic and potentially biotic factors influence photobodies, if they modify the stability of phyB's Pfr form, or if they utilize other mechanisms to influence photobodies. In addition, it is unclear how all these signals are integrated to regulate the outcome of phyB signaling.
One key factor to unravel photobody signal integration is the understanding of the functional and compositional complexity of differing photobodies. One open question is if multiple speckles within a single nucleus are functionally similar and have overlapping protein compositions. These foci might interact with different genomic sites and fusion of photobodies could lead to intrachromosomal but also interchromosomal interactions. By this means, regulatory DNA elements could be brought into proximity of distant protein coding regions to allow flexible gene regulation. In contrast, multiple photobodies within a nucleus could be functionally specialized and might focus on certain functions of phyB signaling.
The finding that multiple speckles within a nucleus seem to have differing thermal stabilities indicates that they are compositionally diverse. The recently published mass spectrometry analysis of photobodies is a huge step in our understanding photobody composition. However, there are still many unknowns regarding not only photobody protein composition but also DNA and RNA sequences that might be structural components and determine photobody stability and size. Furthermore, we currently do not know if tissue- and cell type–specific photobodies exist that might provide another level of phyB signaling complexity.
Acknowledgments
The authors express their gratitude to Dr. Joanne Chory (Salk Institute for Biological Studies, Howard Hughes Medical Institute) and Dr. Meng Chen (University of California, Riverside) for their previous mentorship and to the anonymous reviewers whose comments and suggestions helped improve this manuscript. All figures were created with BioRender.com.
Contributor Information
Björn Christopher Willige, Department of Soil and Crop Sciences, College of Agricultural Sciences, Colorado State University, Fort Collins, CO 80521, USA.
Chan Yul Yoo, School of Biological Sciences, University of Utah, UT 84112, USA.
Jessica Paola Saldierna Guzmán, Department of Soil and Crop Sciences, College of Agricultural Sciences, Colorado State University, Fort Collins, CO 80521, USA.
Author contributions
All authors contributed to writing and figure preparation.
Data availability
There are no new data associated with this article.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Aizezi Y, Shu H, Zhang L, Zhao H, Peng Y, Lan H, Xie Y, Li J, Wang Y, Guo H, et al. Cytokinin regulates apical hook development via the coordinated actions of EIN3/EIL1 and PIF transcription factors in Arabidopsis. J Exp Bot. 2022:73(1):213–227. 10.1093/jxb/erab403 [DOI] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006:23(3):439–446. 10.1016/j.molcel.2006.06.011 [DOI] [PubMed] [Google Scholar]
- Ang L-H, Chattopadhyay S, Wei N, Oyama T, Okada K, Batschauer A, Deng X-W. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998:1(2):213–222. 10.1016/S1097-2765(00)80022-2 [DOI] [PubMed] [Google Scholar]
- Ayala R, Willhoft O, Aramayo RJ, Wilkinson M, McCormack EA, Ocloo L, Wigley DB, Zhang X. Structure and regulation of the human INO80-nucleosome complex. Nature. 2018:556(7701):391–395. 10.1038/s41586-018-0021-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcerowicz M. PHYTOCHROME-INTERACTING FACTORS at the interface of light and temperature signalling. Physiol Plant. 2020:169(3):347–356. 10.1111/ppl.13092 [DOI] [PubMed] [Google Scholar]
- Balcerowicz M, Kerner K, Schenkel C, Hoecker U. SPA proteins affect the subcellular localization of COP1 in the COP1/SPA ubiquitin ligase complex during photomorphogenesis. Plant Physiol. 2017:174(3):1314–1321. 10.1104/pp.17.00488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL, Pierik R. The shade-avoidance syndrome: multiple signals and ecological consequences. Plant Cell Environ. 2017:40(11):2530–2543. 10.1111/pce.12914 [DOI] [PubMed] [Google Scholar]
- Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KCS, Adám E, Fejes E, Schäfer E, et al. Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell. 2004:16(6):1433–1445. 10.1105/tpc.021568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo-García S, de Lucas M, Martínez C, Espinosa-Ruiz A, Davière J-M, Prat S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev. 2014:28(15):1681–1694. 10.1101/gad.243675.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borcherds W, Bremer A, Borgia MB, Mittag T. How do intrinsically disordered protein regions encode a driving force for liquid-liquid phase separation? Curr Opin Struct Biol. 2021:67:41–50. 10.1016/j.sbi.2020.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan MT, Quail PH. Are the phytochromes protein kinases? Protoplasma. 1996:195(1–4):12–17. 10.1007/BF01279182 [DOI] [Google Scholar]
- Bu Q, Zhu L, Dennis MD, Yu L, Lu SX, Person MD, Tobin EM, Browning KS, Huq E. Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis. J Biolo Chem. 2011:286(14):12066–12074. 10.1074/jbc.M110.186882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie ES, Gannam ZTK, McLoughlin KE, Sherman CD, Holehouse AS, Stankey RJ, Vierstra RD. Differing biophysical properties underpin the unique signaling potentials within the plant phytochrome photoreceptor families. Proc Natl Acad Sci U S A. 2021:118(22):e2105649118. 10.1073/pnas.2105649118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgie ES, Vierstra RD. Phytochromes: an atomic perspective on photoactivation and signaling. Plant Cell. 2014:26(12):4568–4583. 10.1105/tpc.114.131623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burko Y, Willige BC, Seluzicki A, Novák O, Ljung K, Chory J. PIF7 is a master regulator of thermomorphogenesis in shade. Nat Commun. 2022:13(1):4942. 10.1038/s41467-022-32585-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañibano E, Bourbousse C, García-León M, Garnelo Gómez B, Wolff L, García-Baudino C, Lozano-Durán R, Barneche F, Rubio V, Fonseca S. DET1-mediated COP1 regulation avoids HY5 activity over second-site gene targets to tune plant photomorphogenesis. Mol Plant. 2021:14(6):963–982. 10.1016/j.molp.2021.03.009 [DOI] [PubMed] [Google Scholar]
- Carter B, Bishop B, Ho KK, Huang R, Jia W, Zhang H, Pascuzzi PE, Deal RB, Ogas J. The chromatin remodelers PKL and PIE1 act in an epigenetic pathway that determines H3K27me3 homeostasis in Arabidopsis. Plant Cell. 2018:30(6):1337–1352. 10.1105/tpc.17.00867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. Shade avoidance. Arabidopsis Book. 2012:10:e0157. 10.1199/tab.0157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Lyu M, Kou X, Li J, Yang Z, Gao L, Li Y, Fan L-M, Shi H, Zhong S. Integration of light and temperature sensing by liquid-liquid phase separation of phytochrome B. Mol Cell. 2022:82(16):3015–3029.e6. 10.1016/j.molcel.2022.05.026 [DOI] [PubMed] [Google Scholar]
- Chen H, Wang W, Chen X, Niu Y, Qi Y, Yu Z, Xiong M, Xu P, Wang W, Guo T, et al. Phytochrome-interacting factors interact with SWI2/SNF2-related 1 complex subunit 6 to regulate H2A.Z deposition and photomorphogenesis in Arabidopsis. J Genet Genomics. 2023:50(12):983–992. 10.1016/j.jgg.2023.04.008 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 2011:21(11):664–671. 10.1016/j.tcb.2011.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, Chory J. Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell. 2010:141(7):1230–1240. 10.1016/j.cell.2010.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Schwab R, Chory J. Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc Natl Acad Sci U S A. 2003:100(24):14493–14498. 10.1073/pnas.1935989100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Tao Y, Lim J, Shaw A, Chory J. Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr Biol. 2005:15(7):637–642. 10.1016/j.cub.2005.02.028 [DOI] [PubMed] [Google Scholar]
- Chen S, Lory N, Stauber J, Hoecker U. Photoreceptor specificity in the light-induced and COP1-mediated rapid degradation of the repressor of photomorphogenesis SPA2 in Arabidopsis. PLoS Genet. 2015:11(9):e1005516. 10.1371/journal.pgen.1005516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M-C, Enderle B, Kathare PK, Islam R, Hiltbrunner A, Huq E. PCH1 and PCHL directly interact with PIF1, promote its degradation, and inhibit its transcriptional function during photomorphogenesis. Mol Plant. 2020:13(3):499–514. 10.1016/j.molp.2020.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Peto C, Feinbaum R, Pratt L, Ausubel F. Arabidopsis thaliana mutant that develops as a light-grown plant in the absence of light. Cell. 1989:58(5):991–999. 10.1016/0092-8674(89)90950-1 [DOI] [PubMed] [Google Scholar]
- Christians MJ, Gingerich DJ, Hua Z, Lauer TD, Vierstra RD. The light-response BTB1 and BTB2 proteins assemble nuclear ubiquitin ligases that modify phytochrome B and D signaling in Arabidopsis. Plant Physiol. 2012:160(1):118–134. 10.1104/pp.112.199109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough RC, Vierstra RD. Phytochrome degradation. Plant Cell Environ. 1997:20(6):713–721. 10.1046/j.1365-3040.1997.d01-107.x [DOI] [Google Scholar]
- Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 2012:8(10):e1002988. 10.1371/journal.pgen.1002988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi C, Johansson H, Holm M. SALT TOLERANCE HOMOLOG2, a B-box protein in Arabidopsis that activates transcription and positively regulates light-mediated development. Plant Cell. 2007:19(10):3242–3255. 10.1105/tpc.107.054791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi GHCM, Deng X-W, Holm M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell. 2006:18(1):70–84. 10.1105/tpc.105.038182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Johansson H, Hettiarachchi C, Irigoyen ML, Desai M, Rubio V, Holm M. LZF1/SALT TOLERANCE HOMOLOG3, an Arabidopsis B-box protein involved in light-dependent development and gene expression, undergoes COP1-mediated ubiquitination. Plant Cell. 2008:20(9):2324–2338. 10.1105/tpc.108.061747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière J-M, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008:451(7177):480–484. 10.1038/nature06520 [DOI] [PubMed] [Google Scholar]
- Dong J, Chen H, Deng XW, Irish VF, Wei N. Phytochrome B induces intron retention and translational inhibition of PHYTOCHROME-INTERACTING FACTOR3. Plant Physiol. 2020:182(1):159–166. 10.1104/pp.19.00835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Ni W, Yu R, Deng XW, Chen H, Wei N. Light-dependent degradation of PIF3 by SCF promotes a photomorphogenic response in Arabidopsis. Curr Biol. 2017:27(16):2420–2430.e6. 10.1016/j.cub.2017.06.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Tang D, Gao Z, Yu R, Li K, He H, Terzaghi W, Deng XW, Chen H. Arabidopsis DE-ETIOLATED1 represses photomorphogenesis by positively regulating phytochrome-interacting factors in the dark. Plant Cell. 2014:26(9):3630–3645. 10.1105/tpc.114.130666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Elmer MV, van Oosten VR, Fankhauser C. The degradation of HFR1, a putative bHLH class transcription factor involved in light signaling, is regulated by phosphorylation and requires COP1. Curr Biol. 2004:14(24):2296–2301. 10.1016/j.cub.2004.12.026 [DOI] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C. bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 2005:10(2):51–54. 10.1016/j.tplants.2004.12.005 [DOI] [PubMed] [Google Scholar]
- Emenecker RJ, Holehouse AS, Strader LC. Emerging roles for phase separation in plants. Dev Cell. 2020:55(1):69–83. 10.1016/j.devcel.2020.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderle B, Sheerin DJ, Paik I, Kathare PK, Schwenk P, Klose C, Ulbrich MH, Huq E, Hiltbrunner A. PCH1 and PCHL promote photomorphogenesis in plants by controlling phytochrome B dark reversion. Nat Commun. 2017:8(1):2221. 10.1038/s41467-017-02311-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S, Schall K, Kostrewa D, Lakomek K, Strauss M, Moldt M, Hopfner K-P. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature. 2018:556(7701):386–390. 10.1038/s41586-018-0029-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Vellutini E, Perrella G, Kaiserli E. TANDEM ZINC-FINGER/PLUS3 regulates phytochrome B abundance and signaling to fine-tune hypocotyl growth. Plant Cell. 2022:34(11):4213–4231. 10.1093/plcell/koac236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C-M, Qiu Y, Van Buskirk EK, Yang EJ, Chen M. Light-regulated gene repositioning in Arabidopsis. Nat Commun. 2014:5(1):3027. 10.1038/ncomms4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008:451(7177):475–479. 10.1038/nature06448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Milmanda GL, Crocco CD, Reichelt M, Mazza CA, Köllner TG, Zhang T, Cargnel MD, Lichy MZ, Fiorucci A-S, Fankhauser C, et al. A light-dependent molecular link between competition cues and defence responses in plants. Nat Plants. 2020:6(3):223–230. 10.1038/s41477-020-0604-8 [DOI] [PubMed] [Google Scholar]
- Fiorucci A-S, Fankhauser C. Plant strategies for enhancing access to sunlight. Curr Biol. 2017:27(17):R931–R940. 10.1016/j.cub.2017.05.085 [DOI] [PubMed] [Google Scholar]
- Fonin AV, Antifeeva IA, Shpironok OG, Stepanenko OV, Silonov SA, Stepanenko OV, Antifeev IE, Romanovich AE, Kuznetsova IM, Kim J-I, et al. Photo-dependent membrane-less organelles formed from plant phyB and PIF6 proteins in mammalian cells. Int J Biol Macromol. 2021:176:325–331. 10.1016/j.ijbiomac.2021.02.075 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci U S A. 2011:108(50):20231–20235. 10.1073/pnas.1110682108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010:61(1):11–24. 10.1093/jxb/erp304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimori T, Yamashino T, Kato T, Mizuno T. Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 2004:45(8):1078–1086. 10.1093/pcp/pch124 [DOI] [PubMed] [Google Scholar]
- Galvão RM, Li M, Kothadia SM, Haskel JD, Decker PV, Van Buskirk EK, Chen M. Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes Dev. 2012:26(16):1851–1863. 10.1101/gad.193219.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Kumar SV. DET1 and HY5 control PIF4-mediated thermosensory elongation growth through distinct mechanisms. Cell Rep. 2017:18(2):344–351. 10.1016/j.celrep.2016.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm J, Kim K, Qiu Y, Chen M. Increasing ambient temperature progressively disassembles Arabidopsis phytochrome B from individual photobodies with distinct thermostabilities. Nat Commun. 2020:11(1):1660. 10.1038/s41467-020-15526-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R, Ma L, Lv Y, Qi L, Peng J, Li H, Zhou Y, Song P, Duan J, Li J, et al. SALT OVERLY SENSITIVE2 stabilizes phytochrome-interacting factors PIF4 and PIF5 to promote Arabidopsis shade avoidance. Plant Cell. 2023:35(8):2972–2996. 10.1093/plcell/koad119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes S, Pantazopoulou CK, van Gelderen K, Reinen E, Tween AL, Sharma A, de Vries M, Prat S, Schuurink RC, Testerink C, et al. Soil salinity limits plant shade avoidance. Curr Biol. 2019:29(10):1669–1676.e4. 10.1016/j.cub.2019.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando CE, Murcia MG, Pereyra ME, Sellaro R, Casal JJ. Phytochrome B links the environment to transcription. J Exp Bot. 2021:72(11):4068–4084. 10.1093/jxb/erab037 [DOI] [PubMed] [Google Scholar]
- Holm M, Ma L-G, Qu L-J, Deng X-W. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 2002:16(10):1247–1259. 10.1101/gad.969702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Devisetty UK, Harmer SL. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife. 2013:2:e00473. 10.7554/eLife.00473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Alvarez S, Bindbeutel R, Shen Z, Naldrett MJ, Evans BS, Briggs SP, Hicks LM, Kay SA, Nusinow DA. Identification of evening Complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol Cell Proteomics. 2016a:15(1):201–217. 10.1074/mcp.M115.054064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, McLoughlin KE, Sorkin ML, Burgie ES, Bindbeutel RK, Vierstra RD, Nusinow DA. PCH1 regulates light, temperature, and circadian signaling as a structural component of phytochrome B-photobodies in. Proc Natl Acad Sci U S A. 2019:116(17):8603–8608. 10.1073/pnas.1818217116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Yoo CY, Bindbeutel R, Goldsworthy J, Tielking A, Alvarez S, Naldrett MJ, Evans BS, Chen M, Nusinow DA. PCH1 integrates circadian and light-signaling pathways to control photoperiod-responsive growth in Arabidopsis. eLife. 2016b:5:e13292. 10.7554/eLife.13292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson M, Kim C, Apel K, Quail PH. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004:305(5692):1937–1941. 10.1126/science.1099728 [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2002:21(10):2441–2450. 10.1093/emboj/21.10.2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y, Han S, Yoo CY, Hong L, You C, Le BH, Shi H, Zhong S, Hoecker U, Chen X, et al. Anterograde signaling controls plastid transcription via sigma factors separately from nuclear photosynthesis genes. Nat Commun. 2022:13(1):7440. 10.1038/s41467-022-35080-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AB, Syed NH, Brown JWS, Nimmo HG. Thermoplasticity in the plant circadian clock: how plants tell the time-perature. Plant Signal Behav. 2012:7(10):1219–1223. 10.4161/psb.21491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I-C, Henriques R, Seo HS, Nagatani A, Chua N-H. Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell. 2010:22(7):2370–2383. 10.1105/tpc.109.072520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang I-C, Yang J-Y, Seo HS, Chua N-H. HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev. 2005:19(5):593–602. 10.1101/gad.1247205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J-H, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. Phytochromes function as thermosensors in Arabidopsis. Science. 2016:354(6314):886–889. 10.1126/science.aaf6005 [DOI] [PubMed] [Google Scholar]
- Jung J-H, Seo PJ, Park C-M. The E3 ubiquitin ligase HOS1 regulates Arabidopsis flowering by mediating CONSTANS degradation under cold stress. J Biol Chem. 2012:287(52):43277–43287. 10.1074/jbc.M112.394338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiserli E, Páldi K, O’Donnell L, Batalov O, Pedmale UV, Nusinow DA, Kay SA, Chory J. Integration of light and photoperiodic signaling in transcriptional nuclear foci. Dev Cell. 2015:35(3):311–321. 10.1016/j.devcel.2015.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathare PK, Xin R, Ganesan AS, June VM, Reddy ASN, Huq E. SWAP1-SFPS-RRC1 splicing factor complex modulates pre-mRNA splicing to promote photomorphogenesis in Arabidopsis. Proc Natl Acad Sci U S A. 2022:119(44):e2214565119. 10.1073/pnas.2214565119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004:16(11):3033–3044. 10.1105/tpc.104.025643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schäfer E, Quail PH. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell. 2007:19(12):3915–3929. 10.1105/tpc.107.051508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikis EA, Oka Y, Hudson ME, Nagatani A, Quail PH. Residues clustered in the light-sensing knot of phytochrome B are necessary for conformer-specific binding to signaling partner PIF3. PLoS Genet. 2009:5(1):e1000352. 10.1371/journal.pgen.1000352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Kwon Y, Jeong J, Kang M, Lee GS, Moon JH, Lee H-J, Park Y-I, Choi G. Phytochrome B photobodies are comprised of phytochrome B and its primary and secondary interacting proteins. Nat Commun. 2023a:14(1):1708. 10.1038/s41467-023-37421-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kim J, Choi G. Epidermal phyB requires RRC1 to promote light responses by activating the circadian rhythm. New Phytol. 2023b:238(2):705–723. 10.1111/nph.18746 [DOI] [PubMed] [Google Scholar]
- Kim J-H, Lee H-J, Jung J-H, Lee S, Park C-M. HOS1 facilitates the phytochrome B-mediated inhibition of PIF4 function during hypocotyl growth in Arabidopsis. Mol Plant. 2017:10(2):274–284. 10.1016/j.molp.2016.11.009 [DOI] [PubMed] [Google Scholar]
- Kim J, Kang H, Park J, Kim W, Yoo J, Lee N, Kim J, Yoon T-Y, Choi G. PIF1-Interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of PIF1. Plant Cell. 2016:28(6):1388–1405. 10.1105/tpc.16.00125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song P-S, Choi G. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell. 2003:15(10):2399–2407. 10.1105/tpc.014498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Schafer E, Nagy F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell. 1999:11(8):1445–1456. 10.1105/tpc.11.8.1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C, Nagy F, Schäfer E. Thermal reversion of plant phytochromes. Mol Plant. 2020:13(3):386–397. 10.1016/j.molp.2019.12.004 [DOI] [PubMed] [Google Scholar]
- Klose C, Venezia F, Hussong A, Kircher S, Schäfer E, Fleck C. Systematic analysis of how phytochrome B dimerization determines its specificity. Nat Plants. 2015:1(7):15090. 10.1038/nplants.2015.90 [DOI] [PubMed] [Google Scholar]
- Kwon C-T, Song G, Kim S-H, Han J, Yoo S-C, An G, Kang K, Paek N-C. Functional deficiency of phytochrome B improves salt tolerance in rice. Environ Exp Bot. 2018:148:100–108. 10.1016/j.envexpbot.2017.12.020 [DOI] [Google Scholar]
- Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012:17(10):584–593. 10.1016/j.tplants.2012.05.004 [DOI] [PubMed] [Google Scholar]
- Lazaro A, Mouriz A, Piñeiro M, Jarillo JA. Red light-mediated degradation of CONSTANS by the E3 ubiquitin ligase HOS1 regulates photoperiodic flowering in Arabidopsis. Plant Cell. 2015:27(9):2437–2454. 10.1105/tpc.15.00529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaro A, Valverde F, Piñeiro M, Jarillo JA. The Arabidopsis E3 ubiquitin ligase HOS1 negatively regulates CONSTANS abundance in the photoperiodic control of flowering. Plant Cell. 2012:24(3):982–999. 10.1105/tpc.110.081885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Paik I, Huq E. SPAs promote thermomorphogenesis by regulating the phyB-PIF4 module in. Development. 2020:147(19):dev189233. 10.1242/dev.189233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rio DC. Mechanisms and regulation of alternative Pre-mRNA splicing. Annu Rev Biochem. 2015:84(1):291–323. 10.1146/annurev-biochem-060614-034316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Ince YÇ, Fankhauser C. Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants. Nat Commun. 2019:10(1):5219. 10.1038/s41467-019-13045-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Rojas CCR, Neme M, Hiltbrunner A, Wigge PA, Schäfer E, Vierstra RD, Casal JJ. Phytochrome B integrates light and temperature signals in Arabidopsis. Science. 2016:354(6314):897–900. 10.1126/science.aaf5656 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell. 2008a:20(2):337–352. 10.1105/tpc.107.052142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008b:18(23):1815–1823. 10.1016/j.cub.2008.10.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011:16(1):19–28. 10.1016/j.tplants.2010.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qi L, Zhang S, Dong X, Jing Y, Cheng J, Feng Z, Peng J, Li H, Zhou Y, et al. Mutual upregulation of HY5 and TZP in mediating phytochrome A signaling. Plant Cell. 2022a:34(1):633–654. 10.1093/plcell/koab254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebers M, Gillet F-X, Israel A, Pounot K, Chambon L, Chieb M, Chevalier F, Ruedas R, Favier A, Gans P, et al. Nucleo-plastidic PAP8/pTAC6 couples chloroplast formation with photomorphogenesis. EMBO J. 2020:39(22):e104941. 10.15252/embj.2020104941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Burgie ES, Gannam ZTK, Li H, Vierstra RD. Plant phytochrome B is an asymmetric dimer with unique signalling potential. Nature. 2022b:604(7904):127–133. 10.1038/s41586-022-04529-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang J, Vierstra RD, Li H. Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy. Proc Natl Acad Sci U S A. 2010:107(24):10872–10877. 10.1073/pnas.1001908107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung H-S, et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012:26(8):785–790. 10.1101/gad.187849.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B-Y, Shih C-J, Hsieh H-Y, Chen H-C, Tu S-L. Phytochrome coordinates with a hnRNP to regulate alternative splicing via an exonic splicing silencer. Plant Physiol. 2020:182(1):243–254. 10.1104/pp.19.00289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling J-J, Li J, Zhu D, Deng XW. Noncanonical role of COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc Natl Acad Sci U S A. 2017:114(13):3539–3544. 10.1073/pnas.1700850114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wang Q, Deng W, Wang X, Piao M, Cai D, Li Y, Barshop WD, Yu X, Zhou T, et al. Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2. Nat Commun. 2017a:8(1):15234. 10.1038/ncomms15234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang W, Li Y, Nie H, Cui L, Li R, Tan L, Peng L, Li C, Luo J, et al. FERONIA coordinates plant growth and salt tolerance via the phosphorylation of phyB. Nat Plants. 2023:9(4):645–660. 10.1038/s41477-023-01390-4 [DOI] [PubMed] [Google Scholar]
- Liu X, Liu R, Li Y, Shen X, Zhong S, Shi H. EIN3 and PIF3 form an interdependent module that represses chloroplast development in buried seedlings. Plant Cell. 2017b:29(12):3051–3067. 10.1105/tpc.17.00508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Liu Q, Yan Q, Shi L, Fang Y. Nucleolus-tethering system (NoTS) reveals that assembly of photobodies follows a self-organization model. Mol Biol Cell. 2014:25(8):1366–1373. 10.1091/mbc.e13-09-0527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 2008:53(2):312–323. 10.1111/j.1365-313X.2007.03341.x [DOI] [PubMed] [Google Scholar]
- Loudet O, Michael TP, Burger BT, Le Metté C, Mockler TC, Weigel D, Chory J. A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008:105(44):17193–17198. 10.1073/pnas.0807264105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997:389(6648):251–260. 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Luo Q, Lian H-L, He S-B, Li L, Jia K-P, Yang H-Q. COP1 and phyB physically interact with PIL1 to regulate its stability and photomorphogenic development in Arabidopsis. Plant Cell. 2014:26(6):2441–2456. 10.1105/tpc.113.121657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X-D, Zhou C-M, Xu P-B, Luo Q, Lian H-L, Yang H-Q. Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis. Mol Plant. 2015:8(3):467–478. 10.1016/j.molp.2014.11.025 [DOI] [PubMed] [Google Scholar]
- MacGregor DR, Gould P, Foreman J, Griffiths J, Bird S, Page R, Stewart K, Steel G, Young J, Paszkiewicz K, et al. HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 is required for circadian periodicity through the promotion of nucleo-cytoplasmic mRNA export in Arabidopsis. Plant Cell. 2013:25(11):4391–4404. 10.1105/tpc.113.114959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Pandey GK, Tuteja N. Calcium- and salt-stress signaling in plants: shedding light on SOS pathway. Arch Biochem Biophys. 2008:471(2):146–158. 10.1016/j.abb.2008.01.010 [DOI] [PubMed] [Google Scholar]
- Majee M, Kumar S, Kathare PK, Wu S, Gingerich D, Nayak NR, Salaita L, Dinkins R, Martin K, Goodin M, et al. KELCH F-BOX protein positively influences Arabidopsis seed germination by targeting PHYTOCHROME-INTERACTING FACTOR1. Proc Natl Acad Sci U S A. 2018:115(17):E4120–E4129. 10.1073/pnas.1711919115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Han R, Yang Y, Liu X, Li H, Zhao X, Li J, Fu H, Huo Y, Sun L, et al. Phytochromes enhance SOS2-mediated PIF1 and PIF3 phosphorylation and degradation to promote Arabidopsis salt tolerance. Plant Cell. 2023:35(8):2997–3020. 10.1093/plcell/koad117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Wei X, Li L, Xu P, Zhang J, Wang W, Guo T, Kou S, Wang W, Miao L, et al. Arabidopsis cryptochrome 1 controls photomorphogenesis through regulation of H2A.Z deposition. Plant Cell. 2021:33(6):1961–1979. 10.1093/plcell/koab091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín E, Vivori C, Rogalska M, Herrero-Vicente J, Valcárcel J. Alternative splicing regulation of cell-cycle genes by SPF45/SR140/CHERP complex controls cell proliferation. RNA. 2021:27(12):1557–1576. 10.1261/rna.078935.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Rodriguez-Concepcion M. Molecular mechanisms of shade tolerance in plants. New Phytol. 2023:239(4):1190–1202. 10.1111/nph.19047 [DOI] [PubMed] [Google Scholar]
- Más P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000:408(6809):207–211. 10.1038/35041583 [DOI] [PubMed] [Google Scholar]
- Matera AG, Gregory Matera A, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014:15(2):108–121. 10.1038/nrm3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita T, Mochizuki N, Nagatani A. Dimers of the N-terminal domain of phytochrome B are functional in the nucleus. Nature. 2003:424(6948):571–574. 10.1038/nature01837 [DOI] [PubMed] [Google Scholar]
- Mazzucotelli E, Belloni S, Marone D, De Leonardis A, Guerra D, Di Fonzo N, Cattivelli L, Mastrangelo A. The e3 ubiquitin ligase gene family in plants: regulation by degradation. Curr Genom. 2006:7(8):509–522. 10.2174/138920206779315728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu W-H, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004:303(5656):343–348. 10.1126/science.1090701 [DOI] [PubMed] [Google Scholar]
- Murcia G, Enderle B, Hiltbrunner A, Casal JJ. Phytochrome B and PCH1 protein dynamics store night temperature information. Plant J. 2021:105(1):22–33. 10.1111/tpj.15034 [DOI] [PubMed] [Google Scholar]
- Nagatani A. Phytochrome: structural basis for its functions. Curr Opin Plant Biol. 2010:13(5):565–570. 10.1016/j.pbi.2010.07.002 [DOI] [PubMed] [Google Scholar]
- Nagy F, Kircher S, Schäfer E. Nucleo-cytoplasmic partitioning of the plant photoreceptors phytochromes. Semin Cell Dev Biol. 2000:11(6):505–510. 10.1006/scdb.2000.0202 [DOI] [PubMed] [Google Scholar]
- Nevarez PA, Qiu Y, Inoue H, Yoo CY, Benfey PN, Schnell DJ, Chen M. Mechanism of dual targeting of the phytochrome signaling component HEMERA/pTAC12 to plastids and the nucleus. Plant Physiol. 2017:173(4):1953–1966. 10.1104/pp.16.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998:95(5):657–667. 10.1016/S0092-8674(00)81636-0 [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999:400(6746):781–784. 10.1038/23500 [DOI] [PubMed] [Google Scholar]
- Ni W, Xu S-L, Chalkley RJ, Pham TND, Guan S, Maltby DA, Burlingame AL, Wang Z-Y, Quail PH. Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell. 2013:25(7):2679–2698. 10.1105/tpc.113.112342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu S-L, González-Grandío E, Chalkley RJ, Huhmer AFR, Burlingame AL, Wang Z-Y, Quail PH. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat Commun. 2017:8:15236. 10.1038/ncomms15236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni W, Xu S-L, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang Z-Y, Quail PH. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science. 2014:344(6188):1160–1164. 10.1126/science.1250778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Sakayama H, de Vries J, Buschmann H, Saint-Marcoux D, Ullrich KK, Haas FB, Vanderstraeten L, Becker D, Lang D, et al. The chara genome: secondary complexity and implications for plant terrestrialization. Cell. 2018:174(2):448–464.e24. 10.1016/j.cell.2018.06.033 [DOI] [PubMed] [Google Scholar]
- Nixdorf M, Hoecker U. SPA1 and DET1 act together to control photomorphogenesis throughout plant development. Planta. 2010:231(4):825–833. 10.1007/s00425-009-1088-y [DOI] [PubMed] [Google Scholar]
- Oh E, Kim J, Park E, Kim J-I, Kang C, Choi G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell. 2004:16(11):3045–3058. 10.1105/tpc.104.025163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu J-Y, Bai M-Y, Arenhart RA, Sun Y, Wang Z-Y. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife. 2014:3:e03031. 10.7554/eLife.03031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu J-Y, Wang Z-Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012:14(8):802–809. 10.1038/ncb2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Park E, Song K, Bae G, Choi G. PHYTOCHROME INTERACTING FACTOR8 inhibits phytochrome A-mediated far-red light responses in Arabidopsis. Plant Cell. 2020:32(1):186–205. 10.1105/tpc.19.00515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oka Y, Matsushita T, Mochizuki N, Quail PH, Nagatani A. Mutant screen distinguishes between residues necessary for light-signal perception and signal transfer by phytochrome B. PLoS Genet. 2008:4(8):e1000158. 10.1371/journal.pgen.1000158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Deng X-W. Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 1998:16(2):201–208. 10.1046/j.1365-313x.1998.00290.x [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000:405(6785):462–466. 10.1038/35013076 [DOI] [PubMed] [Google Scholar]
- Pacín M, Legris M, Casal JJ. COP1 re-accumulates in the nucleus under shade. Plant J. 2013:75(4):631–641. 10.1111/tpj.12226 [DOI] [PubMed] [Google Scholar]
- Paik I, Chen F, Ngoc Pham V, Zhu L, Kim J-I, Huq E. A phyB-PIF1-SPA1 kinase regulatory complex promotes photomorphogenesis in Arabidopsis. Nat Commun. 2019:10(1):4216. 10.1038/s41467-019-12110-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011:144(2):200–213. 10.1016/j.cell.2010.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Kim J, Lee Y, Shin J, Oh E, Chung W-I, Liu JR, Choi G. Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol. 2004:45(8):968–975. 10.1093/pcp/pch125 [DOI] [PubMed] [Google Scholar]
- Park E, Kim Y, Choi G. Phytochrome B requires PIF degradation and sequestration to induce light responses across a wide range of light conditions. Plant Cell. 2018:30(6):1277–1292. 10.1105/tpc.17.00913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Park J, Kim J, Nagatani A, Lagarias JC, Choi G. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 2012:72(4):537–546. 10.1111/j.1365-313X.2012.05114.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y-J, Lee H-J, Gil K-E, Kim JY, Lee J-H, Lee H, Cho H-T, Vu LD, De Smet I, Park C-M. Developmental programming of thermonastic leaf movement. Plant Physiol. 2019:180(2):1185–1197. 10.1104/pp.19.00139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Josse E-M, Halliday KJ. A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol Biol. 2010:73(1-2):89–95. 10.1007/s11103-009-9571-1 [DOI] [PubMed] [Google Scholar]
- Peng K-C, Siao W, Hsieh H-L. FAR-RED INSENSITIVE 219 and phytochrome B corepress shade avoidance via modulating nuclear speckle formation. Plant Physiol. 2023:192(2):1449–1465. 10.1093/plphys/kiad103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz K-J, Oelmüller R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell. 2006:18(1):176–197. 10.1105/tpc.105.036392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T, Blanvillain R, Merendino L, Courtois F, Chevalier F, Liebers M, Grübler B, Hommel E, Lerbs-Mache S. Plastid RNA polymerases: orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J Exp Bot. 2015:66(22):6957–6973. 10.1093/jxb/erv415 [DOI] [PubMed] [Google Scholar]
- Pham VN, Kathare PK, Huq E. Dynamic regulation of PIF5 by COP1-SPA complex to optimize photomorphogenesis in Arabidopsis. Plant J. 2018:96(2):260–273. 10.1111/tpj.14074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant AR, Larrieu A, Causier B. Repressor for hire! the vital roles of TOPLESS-mediated transcriptional repression in plants. New Phytol. 2021:231(3):963–973. 10.1111/nph.17428 [DOI] [PubMed] [Google Scholar]
- Qi L, Liu S, Li C, Fu J, Jing Y, Cheng J, Li H, Zhang D, Wang X, Dong X, et al. PHYTOCHROME-INTERACTING FACTORS interact with the ABA receptors PYL8 and PYL9 to orchestrate ABA signaling in darkness. Mol Plant. 2020:13(3):414–430. 10.1016/j.molp.2020.02.001 [DOI] [PubMed] [Google Scholar]
- Qiu Y, Li M, Kim RJ-A, Moore CM, Chen M. Daytime temperature is sensed by phytochrome B in Arabidopsis through a transcriptional activator HEMERA. Nat Commun. 2019:10(1):140. 10.1038/s41467-018-08059-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Li M, Pasoreck EK, Long L, Shi Y, Galvão RM, Chou CL, Wang H, Sun AY, Zhang YC, et al. HEMERA couples the proteolysis and transcriptional activity of PHYTOCHROME INTERACTING FACTORs in Arabidopsis photomorphogenesis. Plant Cell. 2015:27(5):1409–1427. 10.1105/tpc.114.136093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Pasoreck EK, Reddy AK, Nagatani A, Ma W, Chory J, Chen M. Mechanism of early light signaling by the carboxy-terminal output module of Arabidopsis phytochrome B. Nat Commun. 2017:8(1):1905. 10.1038/s41467-017-02062-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausenberger J, Hussong A, Kircher S, Kirchenbauer D, Timmer J, Nagy F, Schäfer E, Fleck C. An integrative model for phytochrome B mediated photomorphogenesis: from protein dynamics to physiology. PLoS One. 2010:5(5):e10721. 10.1371/journal.pone.0010721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy ASN, Marquez Y, Kalyna M, Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell. 2013:25(10):3657–3683. 10.1105/tpc.113.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C. The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 2010:24(18):2093–2104. 10.1101/gad.594910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockwell NC, Su Y-S, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006:57(1):837–858. 10.1146/annurev.arplant.56.032604.144208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou J, Sorin C, Devlin PF, Martínez-García JF. Identification of primary target genes of phytochrome signaling. Early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol. 2006:141(1):85–96. 10.1104/pp.105.076331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Yang J-Y, Ishikawa M, Bolle C, Ballesteros ML, Chua N-H. LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature. 2003:423(6943):995–999. 10.1038/nature01696 [DOI] [PubMed] [Google Scholar]
- Sharma A, Samtani H, Sahu K, Sharma AK, Khurana JP, Khurana P. Functions of Phytochrome-Interacting Factors (PIFs) in the regulation of plant growth and development: a comprehensive review. Int J Biol Macromol. 2023:244:125234. 10.1016/j.ijbiomac.2023.125234 [DOI] [PubMed] [Google Scholar]
- Sheerin DJ, Menon C, zur Oven-Krockhaus S, Enderle B, Zhu L, Johnen P, Schleifenbaum F, Stierhof Y-D, Huq E, Hiltbrunner A. Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex. Plant Cell. 2015:27(1):189–201. 10.1105/tpc.114.134775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 2005:44(6):1023–1035. 10.1111/j.1365-313X.2005.02606.x [DOI] [PubMed] [Google Scholar]
- Shen H, Zhu L, Castillon A, Majee M, Downie B, Huq E. Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell. 2008:20(6):1586–1602. 10.1105/tpc.108.060020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007:145(3):1043–1051. 10.1104/pp.107.105601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih C-J, Chen H-W, Hsieh H-Y, Lai Y-H, Chiu F-Y, Chen Y-R, Tu S-L. Heterogeneous nuclear ribonucleoprotein H1 coordinates with phytochrome and the U1 snRNP Complex to regulate alternative splicing in Physcomitrella patens. Plant Cell. 2019:31(10):2510–2524. 10.1105/tpc.19.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Shen X, Liu R, Xue C, Wei N, Deng XW, Zhong S. The red light receptor phytochrome B directly enhances substrate-E3 ligase interactions to attenuate ethylene responses. Dev Cell. 2016:39(5):597–610. 10.1016/j.devcel.2016.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Mo X, Tang C, Zhong S, Deng XW. Arabidopsis DET1 degrades HFR1 but stabilizes PIF1 to precisely regulate seed germination. Proc Natl Acad Sci U S A. 2015:112(12):3817–3822. 10.1073/pnas.1502405112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H, Hanada K, Ushijima T, Nakashima M, Suzuki Y, Matsushita T. Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc Natl Acad Sci U S A. 2014:111(52):18781–18786. 10.1073/pnas.1407147112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata H, Shibata M, Ushijima T, Nakashima M, Kong S-G, Matsuoka K, Lin C, Matsushita T. The RS domain of Arabidopsis splicing factor RRC1 is required for phytochrome B signal transduction. Plant J. 2012:70(5):727–738. 10.1111/j.1365-313X.2012.04937.x [DOI] [PubMed] [Google Scholar]
- Shin A-Y, Han Y-J, Baek A, Ahn T, Kim SY, Nguyen TS, Son M, Lee KW, Shen Y, Song P-S, et al. Evidence that phytochrome functions as a protein kinase in plant light signalling. Nat Commun. 2016:7(1):11545. 10.1038/ncomms11545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee C-H, Lee D, Choi G. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci U S A. 2009:106(18):7660–7665. 10.1073/pnas.0812219106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson PG, Fankhauser C, Terry MJ. PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci U S A. 2009:106(18):7654–7659. 10.1073/pnas.0811684106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating arabidopsis hypocotyl growth. PLoS Genet. 2012:8(3):e1002594. 10.1371/journal.pgen.1002594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sura W, Kabza M, Karlowski WM, Bieluszewski T, Kus-Slowinska M, Pawełoszek Ł, Sadowski J, Ziolkowski PA. Dual role of the histone variant H2A.Z in transcriptional regulation of stress-response genes. Plant Cell. 2017:29(4):791–807. 10.1105/tpc.16.00573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. Nuclear pore complex-mediated gene expression in Arabidopsis thaliana. J Plant Res. 2020:133(4):449–455. 10.1007/s10265-020-01177-0 [DOI] [PubMed] [Google Scholar]
- Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell. 2010:22(12):4084–4097. 10.1105/tpc.110.079947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessadori F, van Zanten M, Pavlova P, Clifton R, Pontvianne F, Snoek LB, Millenaar FF, Schulkes RK, van Driel R, Voesenek LACJ, et al. Phytochrome B and histone deacetylase 6 control light-induced chromatin compaction in Arabidopsis thaliana. PLoS Genet. 2009:5(9):e1000638. 10.1371/journal.pgen.1000638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trupkin SA, Legris M, Buchovsky AS, Tolava Rivero MB, Casal JJ. Phytochrome B nuclear bodies respond to the low red to far-red ratio and to the reduced irradiance of canopy shade in Arabidopsis. Plant Physiol. 2014:165(4):1698–1708. 10.1104/pp.114.242438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulijasz AT, Vierstra RD. Phytochrome structure and photochemistry: recent advances toward a complete molecular picture. Curr Opin Plant Biol. 2011:14(5):498–506. 10.1016/j.pbi.2011.06.002 [DOI] [PubMed] [Google Scholar]
- Van Buskirk EK, Reddy AK, Nagatani A, Chen M. Photobody localization of phytochrome B is tightly correlated with prolonged and light-dependent inhibition of hypocotyl elongation in the dark. Plant Physiol. 2014:165(2):595–607. 10.1104/pp.114.236661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zanten M, Tessadori F, McLoughlin F, Smith R, Millenaar FF, van Driel R, Voesenek LACJ, Peeters AJM, Fransz P. Photoreceptors CRYTOCHROME2 and phytochrome B control chromatin compaction in Arabidopsis. Plant Physiol. 2010:154(4):1686–1696. 10.1104/pp.110.164616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009:136(4):701–718. 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Wang Z, Casas-Mollano JA, Xu J, Riethoven J-JM, Zhang C, Cerutti H. Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2015:112(27):8487–8492. 10.1073/pnas.1423325112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kang J, Armando Casas-Mollano J, Dou Y, Jia S, Yang Q, Zhang C, Cerutti H. MLK4-mediated phosphorylation of histone H3T3 promotes flowering by transcriptional silencing of FLC/MAF in Arabidopsis thaliana. Plant J. 2021:105(5):1400–1412. 10.1111/tpj.15122 [DOI] [PubMed] [Google Scholar]
- Watanabe S, Tan D, Lakshminarasimhan M, Washburn MP, Hong E-JE, Walz T, Peterson CL. Structural analyses of the chromatin remodelling enzymes INO80-C and SWR-C. Nat Commun. 2015:6(1):7108. 10.1038/ncomms8108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Wang W, Xu P, Wang W, Guo T, Kou S, Liu M, Niu Y, Yang H-Q, Mao Z. Phytochrome B interacts with SWC6 and ARP6 to regulate H2A.Z deposition and photomorphogensis in Arabidopsis. J Integr Plant Biol. 2021:63(6):1133–1146. 10.1111/jipb.13111 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. Spliceosome structure and function. Cold Spring Harbor Perspect Biol. 2011:3(7):a003707. 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige BC, Ogiso-Tanaka E, Zourelidou M, Schwechheimer C. WAG2 represses apical hook opening downstream from gibberellin and PHYTOCHROME INTERACTING FACTOR 5. Development. 2012:139(21):4020–4028. 10.1242/dev.081240 [DOI] [PubMed] [Google Scholar]
- Willige BC, Zander M, Yoo CY, Phan A, Garza RM, Trigg SA, He Y, Nery JR, Chen H, Chen M, et al. PHYTOCHROME-INTERACTING FACTORs trigger environmentally responsive chromatin dynamics in plants. Nat Genet. 2021:53(7):955–961. 10.1038/s41588-021-00882-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirthmueller L, Asai S, Rallapalli G, Sklenar J, Fabro G, Kim DS, Lintermann R, Jaspers P, Wrzaczek M, Kangasjärvi J, et al. Arabidopsis downy mildew effector HaRxL106 suppresses plant immunity by binding to RADICAL-INDUCED CELL DEATH1. New Phytol. 2018:220(1):232–248. 10.1111/nph.15277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong Y-S, Lagarias JC. Affinity labeling of Avena phytochrome with ATP analogs. Proc Natl Acad Sci U S A. 1989:86(10):3469–3473. 10.1073/pnas.86.10.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Liu W, Liang W, Ling X, Ma J, Yang C, Li L. Phytochrome B inhibits the activity of phytochrome-interacting factor 7 involving phase separation. Cell Rep. 2023:42(12):113562. 10.1016/j.celrep.2023.113562 [DOI] [PubMed] [Google Scholar]
- Xin R, Kathare PK, Huq E. Coordinated regulation of Pre-mRNA splicing by the SFPS-RRC1 complex to promote photomorphogenesis. Plant Cell. 2019:31(9):2052–2069. 10.1105/tpc.18.00786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin R, Zhu L, Salomé PA, Mancini E, Marshall CM, Harmon FG, Yanovsky MJ, Weigel D, Huq E. SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis. Proc Natl Acad Sci U S A. 2017:114(33):E7018–E7027. 10.1073/pnas.1706379114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin X, Chen W, Wang B, Zhu F, Li Y, Yang H, Li J, Ren D. Arabidopsis MKK10-MPK6 mediates red-light-regulated opening of seedling cotyledons through phosphorylation of PIF3. J Exp Bot. 2018:69(3):423–439. 10.1093/jxb/erx418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Yang Y, Yang J, Zhang X, Pan Y, Guo H. IAA3-mediated repression of PIF proteins coordinates light and auxin signaling in Arabidopsis. PLoS Genet. 2021:17(2):e1009384. 10.1371/journal.pgen.1009384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Zhang H, Zhao F, Zhao T, Li H, Jiang D. The INO80 chromatin remodeling complex promotes thermomorphogenesis by connecting H2A.Z eviction and active transcription in Arabidopsis. Mol Plant. 2021:14(11):1799–1813. 10.1016/j.molp.2021.07.001 [DOI] [PubMed] [Google Scholar]
- Yamaguchi R, Nakamura M, Mochizuki N, Kay SA, Nagatani A. Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J Cell Biol. 1999:145(3):437–445. 10.1083/jcb.145.3.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Yin L, Xie F, Ma M, Huang S, Zeng Y, Shen W-H, Dong A, Li L. AtINO80 represses photomorphogenesis by modulating nucleosome density and H2A.Z incorporation in light-related genes. Proc Natl Acad Sci U S A. 2020:117(52):33679–33688. 10.1073/pnas.2001976117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D-L, Yao J, Mei C-S, Tong X-H, Zeng L-J, Li Q, Xiao L-T, Sun T-P, Li J, Deng X-W, et al. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci U S A. 2012:109(19):E1192–E1200. 10.1073/pnas.1201616109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang EJ, Yoo CY, Liu J, Wang H, Cao J, Li F-W, Pryer KM, Sun T-P, Weigel D, Zhou P, et al. NCP activates chloroplast transcription by controlling phytochrome-dependent dual nuclear and plastidial switches. Nat Commun. 2019:10(1):2630. 10.1038/s41467-019-10517-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Lv R, Li J, Lin H, Xi D. Phytochrome A and B negatively regulate salt stress tolerance of Nicotiana tobacum via ABA-jasmonic acid synergistic cross-talk. Plant Cell Physiol. 2018:59(11):2381–2393. 10.1093/pcp/pcy164 [DOI] [PubMed] [Google Scholar]
- Yan T, Heng Y, Wang W, Li J, Deng XW. SWELLMAP 2, a phyB-interacting splicing factor, negatively regulates seedling photomorphogenesis in. Front Plant Sci. 2022:13:836519. 10.3389/fpls.2022.836519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh K-C, Lagarias JC. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci U S A. 1998:95(23):13976–13981. 10.1073/pnas.95.23.13976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelagandula R, Stroud H, Holec S, Zhou K, Feng S, Zhong X, Muthurajan UM, Nie X, Kawashima T, Groth M, et al. The histone variant H2A.W defines heterochromatin and promotes chromatin condensation in Arabidopsis. Cell. 2014:158(1):98–109. 10.1016/j.cell.2014.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CY, He J, Sang Q, Qiu Y, Long L, Kim RJ-A, Chong EG, Hahm J, Morffy N, Zhou P, et al. Direct photoresponsive inhibition of a p53-like transcription activation domain in PIF3 by Arabidopsis phytochrome B. Nat Commun. 2021:12(1):5614. 10.1038/s41467-021-25909-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CY, Pasoreck EK, Wang H, Cao J, Blaha GM, Weigel D, Chen M. Phytochrome activates the plastid-encoded RNA polymerase for chloroplast biogenesis via nucleus-to-plastid signaling. Nat Commun. 2019:10(1):2629. 10.1038/s41467-019-10518-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Qin Q, Meng S, Jing H, Gou X, Li J, Hou S. TOPP4 regulates the stability of PHYTOCHROME INTERACTING FACTOR5 during photomorphogenesis in Arabidopsis. Plant Physiol. 2016:170(3):1381–1397. 10.1104/pp.15.01729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J-W, Rubio V, Lee N-Y, Bai S, Lee S-Y, Kim S-S, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, et al. COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell. 2008:32(5):617–630. 10.1016/j.molcel.2008.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Dong J, Deng Z, Jiang Y, Wu C, Qin X, Terzaghi W, Chen H, Dai M, Deng XW. PP6 phosphatases dephosphorylate PIF proteins to repress photomorphogenesis. Proc Natl Acad Sci U S A. 2019:116(40):20218–20225. 10.1073/pnas.1907540116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander M, Willige BC, He Y, Nguyen TA, Langford AE, Nehring R, Howell E, McGrath R, Bartlett A, Castanon R, et al. Epigenetic silencing of a multifunctional plant stress regulator. eLife. 2019:8:e47835. 10.7554/eLife.47835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Holmlund M, Lorrain S, Norberg M, Bakó L, Fankhauser C, Nilsson O. BLADE-ON-PETIOLE proteins act in an E3 ubiquitin ligase complex to regulate PHYTOCHROME INTERACTING FACTOR 4 abundance. eLife. 2017:6:e26759. 10.7554/eLife.26759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Shi H, Pan Y, Lyu M, Yang Z, Kou X, Deng XW, Zhong S. Sensory circuitry controls cytosolic calcium-mediated phytochrome B phototransduction. Cell. 2023:186(6):1230–1243.e14. 10.1016/j.cell.2023.02.011 [DOI] [PubMed] [Google Scholar]
- Zheng H, Zhang F, Wang S, Su Y, Ji X, Jiang P, Chen R, Hou S, Ding Y. MLK1 and MLK2 coordinate RGA and CCA1 activity to regulate hypocotyl elongation in Arabidopsis thaliana. Plant Cell. 2018:30(1):67–82. 10.1105/tpc.17.00830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Wu S, Zhai H, Zhou P, Song M, Su L, Xi Y, Li Z, Cai Y, Meng F, et al. Arabidopsis phytochrome B promotes SPA1 nuclear accumulation to repress photomorphogenesis under far-red light. Plant Cell. 2013:25(1):115–133. 10.1105/tpc.112.107086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Bu Q, Xu X, Paik I, Huang X, Hoecker U, Deng XW, Huq E. CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nat Commun. 2015:6(1):7245. 10.1038/ncomms8245 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no new data associated with this article.