Summary
Interactions within the tumor microenvironment (TME) significantly influence tumor progression and treatment responses. While single-cell RNA sequencing (scRNA-seq) and spatial genomics facilitate TME exploration, many clinical cohorts are assessed at the bulk tissue level. Integrating scRNA-seq and bulk tissue RNA-seq data through computational deconvolution is essential for obtaining clinically relevant insights. Our method, ProM, enables the examination of major and minor cell types. Through evaluation against existing methods using paired single-cell and bulk RNA sequencing of human urothelial cancer (UC) samples, ProM demonstrates superiority. Application to UC cohorts treated with immune checkpoint inhibitors reveals pre-treatment cellular features associated with poor outcomes, such as elevated SPP1 expression in macrophage/monocytes (MM). Our deconvolution method and paired single-cell and bulk tissue RNA-seq dataset contribute novel insights into TME heterogeneity and resistance to immune checkpoint blockade.
Subject areas: Microenvironment, Biocomputational method, Cancer systems biology, Cancer, Transcriptomics
Graphical abstract
Highlights
-
•
Introducing ProM: merging marker-based and single-cell RNA-seq deconvolution
-
•
ProM excels in simulations and paired single-cell/bulk RNA sequencing
-
•
Fibroblast subpopulation heterogeneity in tumor subtypes and lymph nodes
-
•
SPP1-high MoMacs correlate with poor urothelial cancer outcomes
Microenvironment; Biocomputational method; Cancer systems biology; Cancer; Transcriptomics
Introduction
Urothelial cancer (UC) is one of the most frequently diagnosed cancer in North America and Europe: a heterogeneous disease that has been molecularly categorized into several subtypes.1 The heterogeneity underlying UC lies not only in the cancer cells, such as basal versus luminal-like cells, but also in the various cellular components comprising the tumor microenvironment (TME). Standard treatment for metastatic UC of the bladder has historically been limited to platinum-based chemotherapy, but recently experienced a major expansion with the introduction of several PD-1/PD-L1 immune checkpoint inhibitors (CPI) into the armamentarium.2 Numerous components of the TME have been linked to intrinsic CPI response and resistance.3,4 A detailed cellular atlas of UC and its TME, may facilitate a refined understanding of molecular UC subtypes and identify cellular states, and cellular interactions, that define treatment response and patient outcomes.
The advent of single cell RNA sequencing (scRNA-seq) technology has dramatically improved the refined characterizing of tumor heterogeneity and microenvironment in multiple cancer types, including UC.5,6,7,8 However, the clinical applicability of existing single cell datasets is limited by relatively small sample sizes and lack of associated treatment and outcome data. On the other hand, bulk tissue RNA sequencing (RNA-seq) datasets of large clinical cohorts with carefully designed and measured clinical endpoints are accumulating, providing unprecedented opportunities to discover biomarkers, dissect resistance mechanisms and identify new therapy strategies.9,10,11 Thus, the integration of single cell and bulk RNA-seq datasets has the potential to markedly accelerate the generation of clinically relevant knowledge and insights.
A key approach to integrate scRNA-seq and bulk tissue RNA-seq data is through computational deconvolution. Several such deconvolution methods have been developed,12,13,14 but their performance and utility in various human tissues and clinical settings remain poorly defined. Considering the heterogeneity and complexity of human tissue and scRNA-seq data, diverse (not “one-size-fits-all”) deconvolution methodologies are needed, and their accuracies need to be evaluated before applying them to particular scenarios.
Here, we report a deconvolution method leveraging both reference profile and cellular markers (ProM) derived from scRNA-seq. To objectively evaluate the performance of ProM and other existing methods, we present a unique dataset of paired single cell and bulk RNA sequencing of human UC samples. The 30 UC samples encompass various molecular subtypes of UC and the paired bulk RNA-seq and scRNA-seq data encompassed in our cohort enables direct linkage of molecular subtypes with single cell profiling.
Finally, applying ProM to three bulk RNA-seq datasets of UC cohorts treated with CPI, we identified pre-treatment cellular features associated with poor outcomes in UC including Macrophage/Monocytes (MM) with higher expression of SPP1.
Results
Overview of the integrated scRNA-seq dataset of UC
We assembled a scRNA-seq dataset of 30 UC tissue samples (20 unique patients) (referred to hereafter as the Sinai scRNA-seq dataset). This cohort includes data from 17 patients included in our prior analyses3,6,15,16 and comprises of a total of 100,667 cells with the number of genes detected per cell ranging from 200 to 6,000 (median of 1,193), and the number of cells detected per sample ranging from 167 to 8,194 (median of 3,548) (See Figure S1A for QC plots). As our approach to sample processing evolved over time, the dataset includes samples processed by fluorescent activated cell sorting: 13 unsorted, 10 CD45+ sorted, 4 CD45− sorted, as well as 3 samples sorted for CD4, CD8 and NK cells, respectively (see Tables S1 and S2 for details). Not surprisingly, we observed “batch effects” between samples generated from different sorting techniques (Figure S1B).
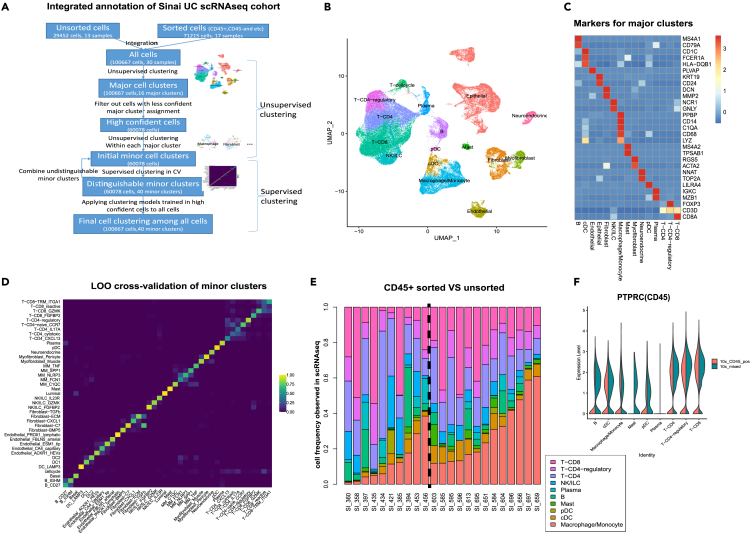
In order to integrate data from these heterogeneous samples, we designed a multistep procedure for cell clustering and annotation, which incorporates both unsupervised and supervised clustering techniques (Figure 1A, see STAR methods). Our cell clustering procedure yielded 14 major cell clusters (Figures 1B and 1C), and 40 minor clusters. Leave-one-out cross-validation confirmed that all but one minor cluster could be found in more than one sample (Figure 1D, see STAR Methods).
Figure 1.
Integrative analysis of UC single cell cohort
(A) Workflow of integration and annotation of scRNA-seq data.
(B) UMAP of all cells in scRNA-seq data.
(C) Markers used to annotate each major cell cluster.
(D) Leave-one-out (LOO) cross-validation of annotation of minor cell cluster using SciBet model with 100 genes per cell cluster. The color represents the percentage of a certain cell subpopulation (x axis) predicted as the other cell subpopulations (y axis) by the model.
(E) Relative cell frequency among immune cells for CD45+ sorted (left of the dash line) and unsorted (right of the dash line) samples. Samples were ordered according to the relative frequency of Macrophage/Monocytes in each group.
(F) Violin plot of expression level of PTPRC (CD45) across different immune cell populations. The expression value was normalized with scale factor of 1000 and then log-transformed.
Cells generated by different sorting techniques can be integrated after reducing “batch effects” (Figure S1C). As expected, compared to unsorted samples, CD45+ sorted samples were enriched with immune cells while CD45− sorted samples were enriched with stromal cells (Figure S1D). However, we also observed differences in immune cell frequency between sorted and unsorted samples. For example, the relative frequency of Monocytes/Macrophages (MoMacs) among immune cells was significantly lower in CD45+ sorted samples than that in unsorted samples (Figure 1E, Wilcoxon rank-sum test p-value = 0.008 and FDR = 0.08). While there are several potential explanations for this observation, including differing expression of CD45 by MoMacs versus other immune cells (Figure 1F), our data suggests while the “batch effect” in cell expression profiles can be minimized by various data integration techniques, systematic shifts in cell frequency caused by technical variation in upstream processing can persist and warrant caution in data integration.
Development of the scRNA-seq-based deconvolution method ProM
Inspired by elements of several commonly employed existing deconvolution methods,13,17 we developed a scRNA-seq-based deconvolution method known as ProM (Figure 2A, see STAR Methods for details). There are three main features of ProM: 1) ProM combines estimation from two distinct deconvolution strategies, i.e., reference profile-based and cell marker-based; 2) In marker-based deconvolution, ProM utilized a semi-supervised learning technique to inform more effective selection of cell marker; 3) In both marker- and profile-based deconvolution, ProM models gene variation in two phases which consider both cross-sample and cross-platform variations (More methodological discussion of ProM can be found in STAR Methods). The performance of ProM was evaluated across several domains as detailed in the following sections.
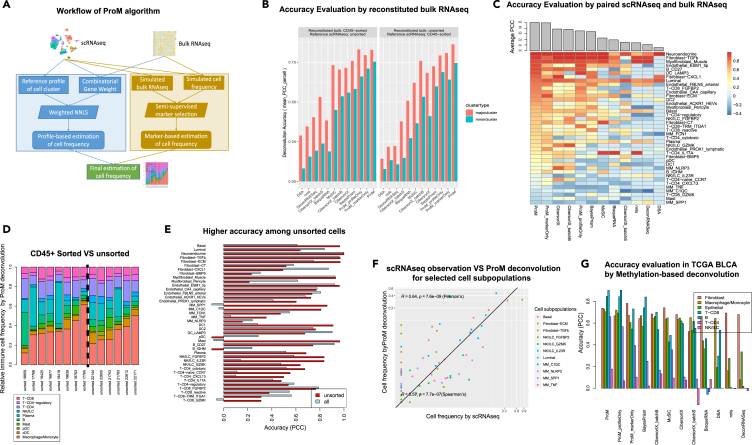
Figure 2.
Evaluation of ProM deconvolution performance
(A) Workflow of ProM algorithm.
(B) Deconvolution accuracy of different algorithms using reconstituted bulk RNA-seq profiles. Y axis indicates PCC (Pearson’s Correlation Coefficient) between deconvoluted and reconstituted cell frequency averaged across all minor or major cell clusters.
(C) Deconvolution accuracy of different deconvolution algorithms estimated using paired single cell and bulk RNA-seq data. The color in the heatmap represents the PCC between deconvoluted and observed frequency for a particular cell subpopulation. The barplot above the heatmap indicates the average PCC across all cell subpopulations.
(D) Relative cell frequency among immune cells according to ProM deconvolution of bulk RNA-seq samples with matched scRNA-seq samples CD45+ sorted (left of the dash line) or unsorted (right of the dash line). Samples were ordered according to the relative frequency of Macrophage/Monocytes in each group.
(E) Deconvolution accuracy of ProM (average PCC) in paired data when all samples or only unsorted samples were considered.
(F) Scatterplot of ProM deconvoluted cell frequencies from bulk RNA-seq samples and the observed cell frequencies via matched scRNA-seq samples for a subset of subpopulations of interest (that are discussed in more details later on).
(G) Deconvolution accuracy of different algorithms in TCGA BLCA dataset by methylation-based deconvolution. Y axis indicates the PCC between deconvolution based on RNA-seq profiles and that based on methylation profiles. Different colors represent the seven common cell types shared by the RNA-seq and methylation-based deconvolution. Algorithms are ordered according to the average PCC across the seven cell types.
Evaluating ProM using reconstituted bulk RNA-seq data
Similar to previous studies,14 we utilized the reconstituted bulk RNA-seq profiles from scRNA-seq profiles to evaluate the performance of ProM in the context of existing deconvolutions methods (see STAR Methods). We employed the “cross-validation” schema, where the 14 CD45+/CD45-sorted scRNA-seq samples were used to reconstitute bulk RNA-seq while the 13 unsorted scRNA-seq samples as reference profiles in deconvolution, and vice versa. Since data generated from different sorting techniques showed obvious “batch effect” in both expression profiles and cell abundance, such a “cross-validation” schema can mimic the cross-platform variation between bulk and reference single cell RNA-seq in real scenario. ProM consistently performed better than nine existing deconvolution methods as measured by both correlation (Figure 2B; Table S3) and Mean Square Error (MSE) (Figure S2A). Of note, methods designed for scRNA-seq based deconvolution, e.g., BayesianPrism,14 CibersortX12 and MuSiC,13 generally outperformed other deconvolution strategies. Furthermore, the contribution of different features of ProM to the overall improved accuracy of this method, i.e., the semi-supervised marker selection and the two-phase variation modeling, were revealed when various versions of the ProM algorithm were compared (Figures S2B and S2C, see Supplementary Methods for details). Lastly, under leave-one-out cross-validation schema,14 a similar comparison of deconvolution approaches was applied to reconstituted bulk samples in nine different cancer types18 (GSE210347, only cancer types with more than 10 samples were analyzed here). As shown in Figure S2D, despite the relative performance of different algorithms varies depending on the cancer type, ProM demonstrated the best overall performance.
Evaluating ProM using paired bulk and scRNA-seq data
When applying deconvolution methods to bulk RNA-seq data, a limitation of most prior studies is the lack of paired scRNA-seq and bulk RNA-seq data from the same samples to serve as ground truth for deconvolution. Our UC cohort, containing matched scRNA-seq and bulk RNA-seq data therefore provided a unique opportunity. Among the 30 Sinai scRNA-seq samples, 18 samples (corresponding to 14 unique patients) had paired bulk RNA-seq data (Table S1). The cell frequency deconvoluted from bulk RNA-seq profiles was compared with observations from paired scRNA-seq samples (see STAR Methods). Though the best performing algorithm varied for different cell subsets, ProM consistently ranked at or near the top (Figures 2C and S2E; Table S3).
A caveat of using scRNA-seq as ground truth is that scRNA-seq profiling is not devoid of potential artifacts. For example, according to our scRNA-seq data, the relative frequency of MoMacs among immune cells was significantly lower in CD45+ sorted samples than in unsorted samples (Figure 1E). However, such differences are not observed in our deconvolution results (Figure 2D, Wilcoxon rank-sum test p-value = 0.47). Thus, the lower proportion of MoMacs in CD45+ sorted samples versus unsorted samples may represent a technical artifact of different sample processing procedures. This highlights the potential underestimation of deconvolution accuracy when integrating datasets employing different processing approaches. Indeed, when sorted samples were excluded from the evaluation, the correlation between scRNA-seq and ProM deconvolution was improved, particularly for some MoMac subsets, such as, MM_C1QC (C1QC high macrophage/monocyte) and MM_SPP1 (SPP1 high macrophage/monocyte) (Figure 2E).
The overall correlation between ProM deconvolution and scRNA-seq observation was 0.79 (Pearson’s, p-value<2.2e-16) and 0.44 (Spearman’s, p-value = 9.8e-13) when all 40 cell subpopulations are included (Figure 2F) and 0.64 (Pearson’s, p-value = 7.6e-9) and 0.57 (Spearman’s, p-value = 7.7e-7) for 11 key cell subpopulations discussed further below (Figure S2F).
Evaluating ProM using orthogonal information
To assess the deconvolution accuracy when applying ProM to external bulk RNA-seq datasets, we relied on orthogonal information since a direct confirmatory measurement of cell proportions was not available in these scenarios. For example, applying ProM and our scRNA-seq profiles to deconvolute the bulk RNA-seq profiles in The Cancer Genome (TCGA) Bladder Cancer (BLCA) dataset, we utilized cell frequencies estimated from methylation profiles19 as an indirect, but orthogonal, evaluation (not the golden standard). Among the common major cell clusters estimated, ProM results correlated with methylation-based results better than other deconvolution methods (Figure 2G; Table S3).
The speed of the ProM algorithm
In addition to improving deconvolution accuracy, our ProM method offers practical advantages in terms of processing requirements and efficiency. For example, ProM deconvolution of the paired bulk RNA-seq data and two external bulk RNA-seq datasets from two CPI clinical trials in UC (IMvigor 210 and CheckMate 275) was completed in 5, 4, and 7 min (CPU speed of 3.1GHz), respectively. In contrast, even with 8-core parallel computing (CPU speed of 3.1GHz), deconvolution of the same datasets with BayesianPrism was completed in 10, 22, and 91 min, respectively.
Cellular deconvolution of molecular subtypes of urothelial cancer
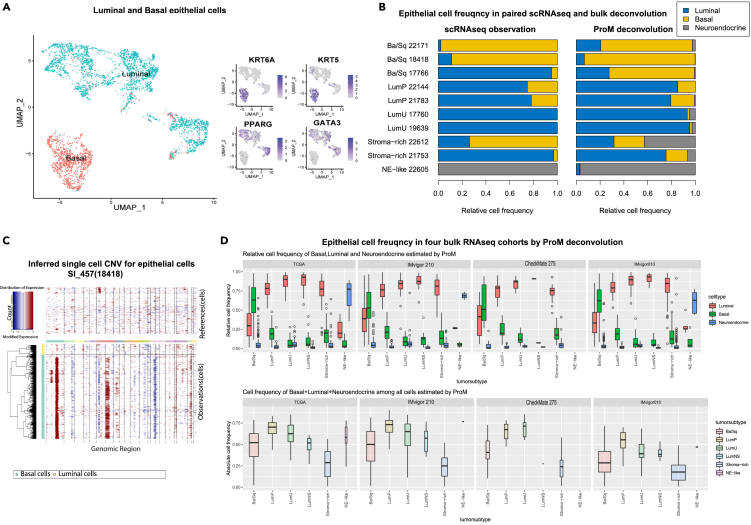
Our paired scRNA-seq and bulk RNA-seq cohort provided an opportunity to investigate the consensus molecular tumor subtypes of UC,1 developed based on bulk RNA-seq data, at the single cell level. Among the ten samples in our cohort with paired bulk RNA-seq and scRNA-seq data, three were classified as basal, four as luminal, two as stromal-enriched and one as neuroendocrine(NE)-like.1 The relative frequency of basal and luminal cells for those ten samples was then derived from their scRNA-seq profiles. Epithelial cells were clustered into two clusters annotated as basal or luminal cells based on markers used to categorize bulk tumor subtypes20 (Figure 3A). The cell frequency of epithelial cells in single cell RNA-seq and ProM deconvolution agreed well with the molecular subtype annotation based on bulk RNA-seq data (Figure 3B). However, according to both our single cell RNA-seq data and ProM deconvolution, there was a considerable fraction of luminal cells in tumors classified by bulk RNA-seq alone as being of basal subtype, and vice versa, indicating the co-existence of basal and luminal cells in most tumors. This co-existence may be due to lineage plasticity of tumor cells21 (Figure S3A) or some non-cancerous luminal epithelial cells scattered in tumor tissue mostly consisting of basal cancer cells. For instance, while the basal epithelial cells of sample SI_457 displayed an obvious pattern of copy number variation (indicating they were cancerous), the luminal epithelial cells do not appear significantly different from the other normal cells (immune and stromal cells) (Figure 3C). Such co-existence of basal and luminal cells across UC subtypes were consistently observed across four external UC cohorts, i.e., TCGA BLCA,20 IMvigor 210,10 CheckMate 2759 and IMvigor 01011 when ProM deconvolution was applied (Figure 3D). Therefore, using paired single cell and bulk RNA-seq data and ProM deconvolution techniques, we uncovered the within-sample heterogeneity of epithelial cells underlying the traditional bulk RNA-seq based UC classification of basal versus luminal tumor subtypes.
Figure 3.
Co-existence of basal and luminal epithelial cells across bulk RNA-seq-derived tumor subtypes
(A) UMAP of epithelial cells annotated into basal and luminal cells based on classical markers.
(B) Relative frequency of luminal, basal and neuroendocrine cells among all epithelial cells based on scRNA-seq observation (left) and ProM deconvolution of the corresponding bulk RNA-seq (right). The bulk RNA-seq-derived tumor subtypes labeled in the left of the bar were predicted using R package consensusMIBC.
(C) The copy number variation (CNV) for each cell inferred from its scRNA-seq profile using R package inferCNV (http://github/broadinstitute/inferCNV) for scRNA-seq sample SI_457 or the paired bulk sample 18418. The upper-panel is the inferred CNV for non-epithelial cells (immune and stromal cells), and the lower-panel is that for epithelial cells split into luminal and basal groups.
(D) Boxplots of relative frequency of luminal, basal, and neuroendocrine cells for each tumor cell type by ProM deconvolution in four UC cohorts (upper panel). Boxplots of the sum of frequency of luminal, basal, and neuroendocrine cells among all cells (lower panel).
Heterogeneity of fibroblast subpopulations across tumor subtypes and lymph node
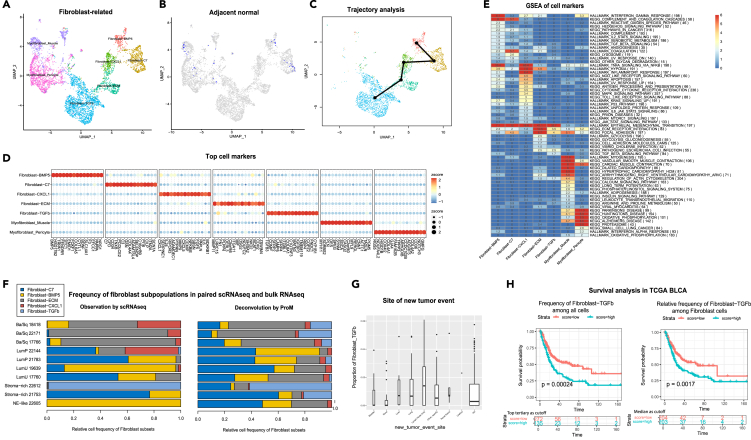
Among the five fibroblast subpopulations and two myofibroblast subpopulations identified in our scRNA-seq cohort (top markers and enriched pathways detailed in Figures 4A, 4D, and 4E), several lines of evidence suggested that Fibroblast-CXCL1, Fibroblast-extracellular_matrix (ECM) and Fibroblast-TGFb were likely to be cancer-specific or “CAF-like”. Mapping to fibroblast annotations in a previous pan-cancer study22 using the SciBet23 model, these three subpopulations largely corresponded to “C10_COMP” (Figure S3B) which was shown to be cancer-specific across multiple cancer types. Additionally, using scRNA-seq data of adjacent normal bladder tissue, these three cellular subpopulations were not detected whereas all the other fibroblast/myofibroblast subpopulations were detected (Figure 4B). Lastly, trajectory analysis24 revealed the gradual progression from the more “normal-like” subpopulation (Fibroblast-BMP5 and Fibroblast-C7), to the three “CAF-like” subpopulations (Figure 4C).
Figure 4.
Heterogeneity of fibroblast subpopulations
(A) UMAP of five fibroblast and two myofibroblast subpopulations.
(B) UMAP with cells from adjacent normal tissue were highlighted in blue color.
(C) Trajectory analysis of five fibroblast subpopulations using R package slingshot.
(D) A set of 15 selected markers for each subpopulation. Marker genes for each subpopulation were obtained using FindAllMarkers() function in R package Seurat.
(E) Enriched pathways for marker genes of each fibroblast subpopulation. The number and the color in each cell of the heatmap is –log10 of p-value for enrichment after multiple test correction (Fisher’s exact test and Holm’s multiple-test correction).
(F) Relative cell frequencies of the five fibroblast subpopulations based on scRNA-seq observation (left) and ProM deconvolution of the corresponding bulk RNA-seq (right). Refer to Figure 3B for more details.
(G) The proportion of Fibroblast-TGFb among all cells by ProM in TCGA BLCA samples grouped by sites of new tumor (NA represents no new tumor information is available).
(H) Kaplan-Meier (KM) curves of overall survival for TCGA BLCA samples grouped by the frequency of Fibroblast-TGFb among all cells (left) or by the relative frequency of Fibroblast-TGFb among all five fibroblast subpopulations (right). Three different cutoffs, i.e., the lower tertiary vs. the median and higher tertiary, were used to split patients into low and high cell frequency groups. The cutoff resulting in the most significant p-value (log-rank test) is shown here. The p-value of likelihood ratio test is 0.002 (left) and 0.0004(right) when cell frequency was treated as a continuous variable in Cox-regression model.
Notably, we discovered an enrichment of the Fibroblast-ECM subpopulation in basal tumor subtypes according to both single cell observation and paired bulk RNA-seq deconvolution (Figure 4F). This phenomenon was consistently observed in four external UC cohorts when ProM deconvolution was applied (Figure S3C).
While the Fibroblast-TGFb subpopulation resembled Fibroblast-ECM in its heavy enrichment of epithelial-mesenchymal transition (EMT) genes, the Fibroblast-TGFb subpopulation had higher expression of the gene TGFB3 and TGFb pathway genes instead of ECM interaction genes (Figure 4E). It likely represented a lymphatic fibroblast subpopulation, as it was only found in the lymph node sample in our cohort. From our ProM deconvolution of the TCGA BLCA cohort, we saw a higher frequency of the Fibroblast-TGFb subpopulation in the cystectomy specimens of patients that had post-cystectomy cancer recurrence in lymph nodes than patients that did not (Figure 4G, Wilcoxon test p-value = 0.027). The absolute and relative frequency of the Fibroblast-TGFb subpopulation was the most significantly associated with worse survival among the five fibroblast subpopulations (Figures 4H, S3D, and S3E).
Deconvoluted MoMac subpopulations are associated with disparate survival outcomes in cohorts treated with PD-1/PD-L1 blockade
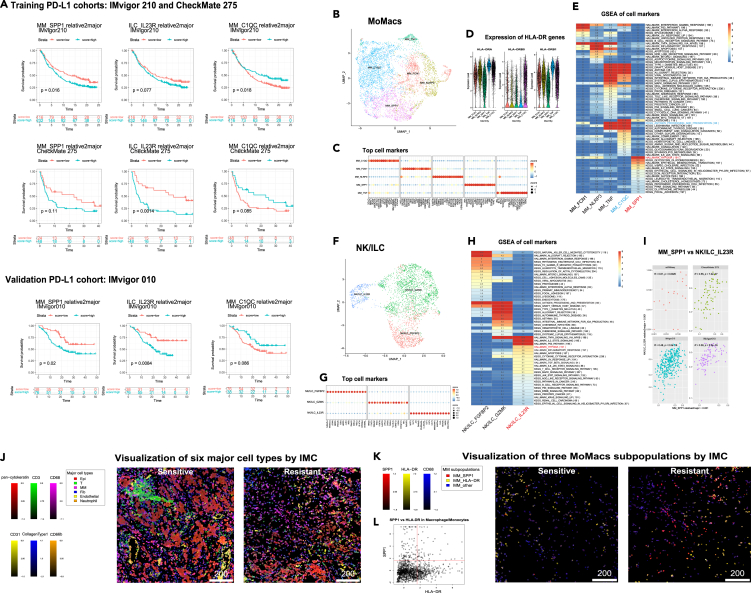
Inspired by the ability of ProM deconvolution to identify cellular subpopulations that comprise previously defined molecular subtypes of bladder cancer, we next conducted a systematic analysis of cell subpopulations that might be associated with outcomes in cohorts of patients with UC treated with PD-1/PD-L1 blockade. Since we are particular interested in dissecting the functional and phenotypic difference of closely related cell subtypes, the following survival association analysis was conducted on the relative cell frequency of each minor cell cluster within the major cell cluster. In the exploratory phase, we assessed the survival association of each cell subpopulation in the two CPI trial cohorts: IMvigor 210 and CheckMate 275. Three (MM_SPP1, NK/ILC_IL23R —“IL23R high NK/ILC cells”, and TCD8_GZMK—“CD8 positive and GZMK high T cells”) and two (MM_C1QC and T-CD8_reactive—” reactive subset of CD8 positive T cells”) cell subpopulations were identified as significantly associated with worse and better survival outcome, respectively (adjusted p-value <0.05 using Fisher’s method to combine both cohorts, see STAR Methods for details). In the validation phase, we assessed their survival association in IMvigor 010 cohort of ctDNA+ patients. The association with worse survival remains significant for two of three subpopulations: MM_SPP1 and NK/ILC_IL23R (Figure 5A, p-value of log-rank test = 0.02 and 0.008, respectively). For the third subpopulation of T-CD8_GZMK, the same trend still holds, but the association is not statistically significant (Figure S4A, p-value of log-rank test = 0.19). The association of MM_C1QC subpopulations with better survival is marginally significant in the validation dataset with consistent trend (Figure 5A, p-value of log-rank test = 0.086), while the association of T-CD8_reactive is not statistically significant and the trend is not clear either (Figure S4A, p-value of log-rank test = 0.54). Since the deconvolution accuracy of T-CD8_GZMK and T-CD8_reactive is relatively low according to ProM evaluation (Figure 2E), they were not investigated further in this study. It is of note that other deconvolution methods failed to detect any reproducible associations except that of MM_C1QC (Figure S4B). Overall, ProM allowed us to deconvolute bulk RNA sequencing data of CPI clinical trial cohorts in UC and revealed distinct cell subpopulations associated with survival and resistance.
Figure 5.
Dichotomy of MoMac and NK/ILC cells in PD-L1 clinical cohorts
(A) Patient overall survival is associated with fractions of cell subpopulations in patients’ tumor tissues. Patients were partitioned according to the ratio of MM_SPP1 vs. MM, NK/ILC_IL23R vs. NK/ILC, MM_C1QC vs. MM in each of cohorts IMvigor210, CheckMate 275, and IMvigor010, respectively. Markers are ordered in columns, and cohorts are separated in rows. The cohorts IMvigor210 and CheckMate275 were used as training data and cohort IMvigor010 as a validation dataset.
(B) UMAP of five Macrophage/Monocyte subpopulations.
(C) A set of 15 selected markers for each MM subpopulation (see legend of Figure 4D for details).
(D) Expression of HLA-DR genes across different MM subpopulations. The expression value was normalized with scale factor of 1000 and then log-transformed.
(E) Enriched pathways for marker genes of each MM subpopulation (see legend of Figure 4E for details). The red color in the labels highlighted worse-survival associated MM subpopulations and some enriched pathways, and the blue color highlighted better-survival associated MM subpopulations and some enriched pathways.
(F) UMAP of three NK/ILC subpopulations.
(G) Similar to (C) except for NK/ILC subpopulations.
(H) Similar to (E) except for NK/ILC subpopulations.
(I) Scatterplots of relative cell frequencies of MM_SPP1 versus that of NK/ILC_IL23R as observed in scRNA-seq cohort or by ProM deconvolution in three PD-1/PD-L1 clinical cohorts.
(J) Visualization of six cell types within regions of interest (ROIs) from one patient sensitive (left) or resistant (right) to checkpoint inhibitor using Imaging Mass Cytometry. The color of the cell boundary indicates the six cell types. The color within each cell corresponds to the scaled intensity of the six markers summarized for each cell.
(K) Visualization of Macrophage/Monocytes in the same two ROIs as shown in (J). The color of cell boundary indicates the three MM subpopulations. The color within the cell corresponds to the scaled intensity of the three markers summarized for each cell.
(L) Scatterplot of cell-level intensity of SPP1 and HLA-DR for each MM cell in the two ROIs. The red line indicates the cutoff used to annotate MM_SPP1 and MM_HLA-DR.
Intriguingly, among the five macrophage/monocyte (MM) subpopulations identified in our scRNA-seq cohorts, MM_SPP1 was associated with worse survival while MM_C1QC was associated with better survival. Investigation of their top markers and enriched gene sets (Figures 5B–5E) revealed that MM_C1QC was enriched in antigen-presenting pathway genes such as human leukocyte antigen (HLA) genes, while MM_SPP1 was enriched in Hallmark_Hypoxia signaling pathway and epithelial mesenchymal transformation pathway genes. Our observation is concordant with the dichotomy of C1QC+ tumor associated macrophages (TAM) and SPP1+ TAM identified in colon cancer.25 Interestingly, the association of SPP1-exressing MoMacs with worse patient outcome has been observed across multiple cancer types.26,27,28
Similarly, amongst our three NK/ILC subpopulations, only the NK/ILC_IL23R subset (IL23R and KIT) resembling an ILC3 subset29 was associated with worse survival (Figures 5F and 5G). Interestingly, this NK/ILC subpopulation was also enriched in hypoxia pathway, bearing striking similarity to the MM_SPP1 subpopulation. (Figure 5H). In fact, we observed a significant positive correlation between the MM_SPP1 and NK/ILC_IL23R cell frequency in all three larger cohorts by ProM estimation (Figure 5I). Taken together, our study uncovered a coordinated immune network in the TME encompassing myeloid and lymphoid subpopulations and illustrated that an enrichment in hypoxia genes across these cell types is associated with resistance to PD-1/PD-L1 blockade.
Exploratory analysis of MM-SPP1 subpopulation using imaging mass cytometry
To begin to understand the function of the MM_SPP1 subpopulation, we investigated their spatial organization in the TME and its relationship with other cells. We used imaging mass cytometry (IMC) to quantify the expression of 16 proteins with subcellular spatial resolution (see STAR Methods). In this exploratory study, we examined two exemplary regions of interest (ROIs) from two UC patients, one who was sensitive and the other resistant to the PD-1 CPI, pembrolizumab.
Cells were annotated into six major types according to the expression of cell marker expression. The cell-level expression of selected markers can be visualized in Figure 5J. (The pixel level data can be visualized in Figure S5). There were obvious differences in cell type distribution between the two patients. For instance, a dense cluster of T-cells was observed in the CPI-sensitive patient, while none were found in the CPI-resistant patient. In addition, the proportion of MM cells is noticeably higher in resistant ROI than the sensitive one. We then further split MM cells into SPP1 high (MM_SPP1), HLA-DR high (MM_HLA-DR) and others (MM_other) (Figures 5K and 5L). Based on our prior data, we hypothesized that MM_HLA-DR MMs represented better-survival associated MM subpopulations with higher expression of HLA genes and antigen-presentation pathway genes (Figure 5F). Supporting our earlier ProM findings, we found a larger proportion of MM_SPP1 cells in the resistant ROI (20% of all MM cells) than in the sensitive ROI (<1%) (Figures 5K and S5), but more MM_HLA-DR cells in the sensitive ROI (27% of all MM cells) than in the resistant ROI (18%), consistent with results in Figures 5A and S5. Furthermore, MM_HLA-DR was enriched in the neighborhood of T cells while MM_SPP1 was depleted from T cell neighborhood (p < 0.05 in permutation test). Thus, the IMC data revealed the separation of MM-SPP1 subpopulation and T cells in an immunotherapy resistant tumor. These data supported our earlier ProM findings of an MM subpopulation dichotomy corresponding to CPI response and resistance.
Discussion
In this study, we present one of the largest scRNA-seq cohort in UC to date with paired bulk RNA-seq profiles. Leveraging our paired single cell and bulk RNA-seq cohort, we uncovered the heterogeneity of epithelial and fibroblast subpopulations, challenging canonical tumor subtypes defined based on bulk RNA-seq profiles. Although previous studies also suggested such heterogeneity,7,30 our paired datasets gave the first direct evidence of the true diversity of bulk RNA-seq based molecular subtypes of UC at a single cell level. In addition, we uncovered a lymphatic fibroblast subpopulation with higher TGFB3 expression and enriched in TGFb pathway genes associated with worse survival that also correlated with tumor recurrence in lymph nodes in TCGA BLCA dataset. Comparing the immune cell frequencies between sorted and unsorted scRNA-seq samples, we also highlighted potential technical bias caused by cell sorting procedures. Although more comparisons of similar datasets are needed to confirm such a bias, it supports the importance of consistency in upstream processing techniques in scRNA-seq experimental design. Finally, we found that paired datasets provide a much-needed resource to evaluate the performance of existing and future computational deconvolution algorithms in complex TMEs.
We developed our novel bulk RNA-seq deconvolution algorithm, named ProM, by combining two classical deconvolution strategies, i.e., marker-based and profile-based. Leveraging the complementary nature of the two strategies together with a few other technical improvements, ProM showed attractive advantages over multiple existing deconvolution methods by being more accurate and faster. However, there is no “one-size-fits-all” approach in the field of computational deconvolution. Different methods have been found to excel in different scenarios.31 We hope to continue to assess the utility of ProM in other applications in future studies.
Applying ProM and our scRNA-seq cohort to three CPI-treated cohorts, we systematically identified and validated cell subpopulations associated with response and resistance. Our analysis uncovered a clinical dichotomy in MM subpopulations in UC. While the CPI response associated MM_C1QC subpopulation was enriched in antigen-presentation pathway genes, the CPI resistant associated MM_SPP1 and NK/ILC_IL23R subpopulations were enriched in hypoxia pathway. Tumor hypoxia has been associated with resistance to immunotherapy in multiple cancer types.32,33,34,35 In addition, the relative abundance of MM_SPP1 and NK/ILC_IL23R were significantly correlated among patients.
The shared activation of similar pathways among different cell types and subtypes suggested a coordinated cellular network in TME where cells of different lineages are under the same external and inter-cellular signals. Some of these signals, such as hypoxia and tumor-promoting inflammation, are tightly linked to the abundance of certain immune cell subpopulations, e.g., MM_SPP1 and NK/ILC_IL23R, which can be used to predict poor patient outcome under immunotherapy. Because of such cellular interactions, spatial studies of the TME are critical. Our initial exploratory spatial analysis of MM subpopulations by IMC indicates MM_SPP1 were located away from cell subpopulations generally associated with better survival, such as MM with high HLA-DR or T cell nest. To build off our bioinformatic findings and validate the importance of these cell subsets to CPI response, we are continuing to expand our spatial datasets as well as assess the function of these MM and NK/ILC cells through in vitro model systems.
Limitations of the study
Although ProM was shown to rank at the top of existing deconvolution methods evaluated in this study, the performances of ProM and other deconvolution methods should be interpreted in context. First, many of the previous deconvolution methods were only evaluated by reconstituted or simulated data, which can cause significant over-estimation of the accuracy. We propose that more paired datasets in different tissues and disease conditions are necessary to improve their accuracy. Second, even real cell count data used for validation, commonly gathered by flow cytometry with broad cell lineage panels, are often very coarse cell counts of known pre-existing major cell populations.14 This unfortunately may not capture the cellular complexity in tissues like tumor and may limit the uncovering of novel cellular subsets. Finally, because of the limited and varied deconvolution accuracy depending on the cell types and tissues, the results should be interpreted cautiously. For instance, we only focused here a few subpopulations, such as MM_SPP1, which, according to our evaluation, had relatively optimal deconvolution accuracy. For the same reasoning, we might miss some important cell-subpopulations genuinely associated with clinical outcome in PD-L1 cohorts because ProM deconvolution for these cell types were poor. Additional paired scRNA-seq and bulk RNA-seq datasets in diverse tissues and disease states are critical to further access accuracy of different deconvolution methods and identify critical associations of cell subpopulations in disease progression or drug response.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD45 | Abcam | ab10559 |
| SMA | Fluidigm | 3141017D |
| CD14 | Abcam | ab226121 |
| Cytokeratin 5 | Biolegend | 905501 |
| CD16 | Abcam | ab256582 |
| Pan-Cytokeratin | Fluidigm | 3148020D |
| CD31 | Fluidigm | 3151025D |
| Collagen 3 | Proteintech | 22734-1-AP |
| SPP1 | Abcam | ab69498 |
| CD8a | Fluidigm | 3162034D |
| CD66b | Biolegend | 396902 |
| Collagen Type 1 | Fluidigm | 3169023D |
| CD3 | Fluidigm | 3170019D |
| GATA3 | BD Biosciences | BDB558686 |
| CD68 | Abcam | ab233172 |
| HLA-DR | Fluidigm | 3174025D |
| DNA 1 | Fluidigm | 201192A |
| DNA 2 | Fluidigm | 201192A |
| ICSK1 Cell segmentation | Fluidigm | TIS-00001 |
| ICSK2 Cell segmentation | Fluidigm | TIS-00001 |
| ICSK3 Cell segmentation | Fluidigm | TIS-00001 |
| Deposited data | ||
| Paried bulk RNAseq data of Urothelial cancer | This paper | GSE223971 |
| scRNAseq data of Urothelial cancer | Salomé et al.6 | Mendeley at https://doi.org/10.17632/7yb7s9769c.1. |
| scRNAseq data of Urothelial cancer | Yu et al.16 | GSE211388 |
| scRNAseq data of Urothelial cancer | Wang et al.15 | GSE130001 |
| The assembled and reanalyzed scRNAseq dataset with meta information including the cell annotation | This paper | https://zenodo.org/records/10009351 |
| Software and algorithms | ||
| ProM | This paper | https://github.com/integrativenetworkbiology/ProM |
| BayesianPrism (v 1.4) | Chu et al.14 | https://github.com/Danko-Lab/TED |
| MuSiC (v 0.2.0) | Wang et al.13 | https://github.com//xuranw/MuSiC |
| CibersortX | Newman et al.12 | https://cibersortx.stanford.edu/ |
| BisqueRNA (v 1.0.5) | Jew et al.36 | https://cran.r-project.org/web/packages/BisqueRNA/index.html |
| DeconRNAseq (v 1.32.0) | Gong et al.37 | https://www.bioconductor.org/packages/release/bioc/html/DeconRNASeq.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jun Zhu (junzhu_99@yahoo.com).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Paired bulk RNAseq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper assembles and reanalyzes existing publicly available scRNAseq data. These accession numbers for the datasets are listed in the key resources table. The reanalyzed scRNAseq dataset with meta information including the cell annotation generated in this study have been deposited at Zenodo and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
R package of ProM algorithm together with script to reproduce the cross-validation comparison of different deconvolution algorithms in pan cancer datasets has been deposited at github and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Primary urothelial bladder cancer tumor tissue was obtained after obtaining informed consent in the context of an Institutional Review Board (IRB)-approved genitourinary cancer clinical database and specimen collection protocol (IRB #10-1180) at the Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai (New York, NY). The assembled scRNAseq cohort consists of 13 male patients and 7 female patients. The participant age ranges from 46 to 87 with the median age of 70. Details on participant race, age and gender can be found in Table S2.
Method details
Single cell RNAseq cohort
Most of scRNAseq data were described in previous studies3,6,15,16 (listed in the key resources Table S1).
Specimen preparation
In brief, all scRNAseq libraries were constructed from fresh tumor tissues. Patients undergoing transurethral resection of bladder tumor had a portion of their tumor placed immediately into RPMI medium in the operating room. The specimen was then transferred to the laboratory for further processing. Patients undergoing radical cystectomy and lymph node dissection had their bladder and lymph node tissues sent directly to the pathology suite upon completion of the lymph node dissection. The bladder was bivalved and a portion of visible tumor was then placed in media as above. Adjacent normal tissue was identified in a subset of specimens on the basis of visual inspection. The specimen was then transferred to the laboratory for further processing.
Unsorted scRNAseq sample preparation
Tissue specimens were processed immediately upon receipt and dissociated into single-cell suspensions using the GentleMACS Octodissociator with kit matched to the tissue type (Miltenyi Biotec) following the manufacturer's instructions. scRNA-seq libraries were prepared on these samples using the Chromium Platform (10x Genomics) with the 3′ gene expression V3 kit, using an input of approximately 10,000 cells.
CD45 sorted scRNAseq sample preparation
The fresh bladder tumor samples were minced to 1–2-mm chunks via mechanical dissociation. The samples were subsequently undergone mechanical/chemical dissociation using the Mintenyi Biotec Gentlemacs 130-096-427 Octo Dissociator at the h_2 dissociation program. Upon completion of the dissociation, the dissociated tumor and buffer was filtered to single-cell suspension, using the 70-μm Gentelmacs filter. The suspension was then spun down and the dissociation buffer was removed, followed by the addition of 1% ACK lysis buffer for RBC lysis for 1 min. The RBC lysis buffer was then diluted with DBPS, and the bladder cancer cells were then spun down at 200–300 G. After a final wash, the supernatant was discarded, and the pellet was resuspended in cold FACS buffer (DPBS containing 2% BSA and 2 mM EDTA). Bladder cancer single cells were analyzed by fluorescence-activated cell sorting (FACS) to determine phenotype. In brief, cells were stained with fixable viability dye blue to ascertain viability (ThermoFisher) and BV510-labeled anti-CD45 (BioLegend). For the sort, cells were gated on live singlets followed by the CD45 makers.
scRNA sequencing
Prior to the preparation of single-cell sequencing libraries, the cell density was determined using a hemocytometer using a mixture of trypan blue and cell suspension was examined under the microscope at low magnification using the Evos Cell Imaging System-digital inverted microscope. This assessment of cell viability ensured successful preparation. The single-cell Chromium chip loading, gel emulsion (GEM) generation and barcoding, post GEM_RT and cDNA amplification, and library construction were performed according to the Chromium™ Single Cell 3′ Protocol - Chemistry version 2 (10X Genomics). Libraries were quantified using Bioanalyzer (Agilent Technologies) and QuBit (Thermo Fisher Scientific) analysis. Libraries were then sequenced in paired-end mode on a NovaSeq Instrument (Illumina) targeting a depth of 5 × 104–1 × 105 reads per cell.
Paired single cell and bulk RNA sequencing cohort
We assembled a scRNAseq dataset of 100,667 cells from 30 UC tissue samples (20 unique patients). Among 20 patients, data from 17 patients were included in our prior studies.3,6,15,16 Bulk RNA sequencing was performed on a subset of patients in the single-cell RNA sequencing cohort due to tissue availability. Total RNA from UC tissue stored in RNAlater or embedded in OCT was isolated with the Qiagen miRNeasy Mini Kit (217084, Qiagen, Germantown, MD, USA), eluted in water and treated with DNAse, at The Biorepository and Pathology Core Facility at the Icahn School of Medicine at Mount Sinai. Total RNA was sequenced at the Genomics Core Facility at Icahn School of Medicine at Mount Sinai. Agilent RNA Direct SureSelect XT V6 kit was used for library prep and NovaSeq 6000 S2 Reagent Kit v1.5 (300 cycles) was used for sequencing.
MSulti-step integration and annotation of scRNAseq dataset
Unsupervised clustering of major cell clusters
29,452 unsorted cells from 13 UC samples and 71,215 sorted cells from 17 UC samples were assembled from published and unpublished scRNAseq datasets at Sinai (Table S1). To remove “batch effects” between sorted and unsorted samples, we followed the Seurat data integration strategy38 where 4,000 most variable genes and 20 reduced dimensions were used in the “anchor”-based integration procedure. Following the Seurat integration procedure, enhanced mixing of sorted and unsorted cells was observed, as evidenced by two metrics39: Firstly, the mean acceptance rate, assessed using the k-nearest neighbor batch-effect test (kBET),40 increased from 0.21 to 0.56, with a 10% sample size considered and the analysis repeated ten times. Secondly, the batch's 1 - adjusted Rand index (ARI)41 improved from 0.94 to 0.98, where batch labels were compared to K-means clustering labels, also with ten repetitions. Graph-based unsupervised clustering method implemented in Seurat was used to cluster cells into 16 major cell clusters after the effect of read count and mitochondrial gene percentage per cell were removed (resolution parameter set to 0.3).
Unsupervised clustering of minor cell clusters among high confident cells
Minor cell clusters within a major cluster are usually more difficult to derive. It requires better resolution of gene expression profiles to dissect the more subtle difference among cell subsets. In addition, the technical artificial effects could be more prominent and harder to discern from the genuine biological signals. Under these considerations, we constructed a high confident cell set by filtering out the following three types of cells: 1) cells of lower resolution (<800 genes detected per cells); 2) “unexpected” cells in the sorted samples, e.g., stromal and epithelial cells in CD45+ sorted samples, immune cells in CD45−sorted samples; 3) doublets that express markers of more than one major clusters. After filtering, 60,078 cells of high confidence were used for the unsupervised clustering of minor cell clusters. In particular, graph-based unsupervised clustering with 2,000 most variable genes and 20 reduced dimensions was performed within each major cluster separately.
Distinguishable minor cell clusters evaluated by cross-validation
To determine the optimal number of minor cell clusters for each major cluster, we employed the strategy of cross-validation (CV) similar to a previous study.42 We started by generating more minor cell clusters than ideal, and gradually combined similar minor clusters together based on CV results. To be specific, we employed leave-one-sample-out CV where in each iteration, the cells of one sample were considered as a testing set and the rest were considered as a training set. The automatic cell annotation algorithm SciBet23 was applied to the training set to learn the predictive models and then evaluated in the testing dataset. Under such a leave-one-sample-out CV schema, minor cell clusters detected predominantly only in one sample are unlikely to give a good CV performance when that sample was held out to be the testing set. In other words, we are prioritizing cell clusters that exist in more than one sample, which arguably is less likely due to sample-specific artifact or being false positives. However, a caveat is that we might miss some genuine cell subsets appearing in only one sample and increase false negatives. That is the consideration by which we kept the Fibroblast-TGFb subset despite its poor CV performance. Since our dataset contains only one lymph node sample, the requirement of cell subset existing in more than one sample will exclude us from detecting any lymph node-specific subset. We relied on the paired bulk RNAseq data to confirm the Fibroblast-TGFb subset in this case. In summary, a total of 40 distinguishable minor cell clusters were generated from the high confident cell set.
Annotating all cells via supervised cell annotation
SciBet annotation models for the 40 minor cell clusters were trained using the high confident set above, then applied to automatically annotate all the 100,667 cells in Sinai scRNAseq dataset. The major cell cluster annotation for each cell was derived according to its minor cell annotation. These annotations were the final ones used throughout our paper.
ProM method
ProM leverages the advantages of two different deconvolution strategies, i.e, marker-based and profile-based, which will be detailed in following paragraphs. The final ProM result is the average of the cell frequencies estimated by the two methods. The rationale behind the combination of two strategies is that: while the reference profiles which usually contain hundreds and thousands of genes can capture a more complete picture of a cell type compared with marker-based method, profile-based algorithms mostly relies on some sort of regression models on the reference profiles of all cell type/subtypes together and could face the issue of collinearity especially when the profiles of cell subtypes are very similar to each other. On the other hand, marker-based methods can focus on a few marker genes that distinguish cell subtypes from its close neighboring subtypes, and the cell abundance is estimated based on the collective expression of those markers without the collinearity problem. Thus, the two strategies are complementary to each other in that profile-based methods are more powerful in “globally” estimating cell components and marker-based methods can further refine the estimation by “locally” distinguishing the cell subtypes of high similarity.
Marker-based deconvolution
In marker-based deconvolution, the frequency of the cell type in the bulk sample can be estimated by the mean expression of its marker genes through a simple linear expression (formula 1) where is a coefficient to proportionally transform gene expression to cell frequency, and is the intercept. We average the marker expression at the log-scale of the transcripts per million (TPM) and then transform back to the original scale to avoid dominance of markers with higher expression.
| (Equation 1) |
Allowing the intercept in the formula 1 will result in some negative estimation of cell abundance, while fixing the intercept =0 will result in positive estimation of cell abundance for almost every bulk sample. To balance these two effects, we set =0 for cell subtypes that exist in more than half of the bulk samples and allowing estimation of for the rest subtypes. We then set all the negative estimation of to be zero. To estimate and in formula 1, we simulate bulk RNAseq with known cell proportion. The two-phase simulation procedure are detailed in the following paragraphs.
The key to marker-based deconvolution is to select a good set of marker genes for each cell cluster based on scRNAseq profiles. We start with a strategy to select the most over-expressed genes in one particular cell cluster as compared to the remaining cell clusters, referred to as “differentially expressed genes” (DEGs). In particular, for each minor cell cluster, we use the function of FindAllMarkers() in Seurat package to identify the marker genes by comparing with the other minor cell clusters within the same major cluster since we focus on distinguishing similar cell subtypes in this marker-based strategy. For the major cluster with no minor clusters, we obtain the DEGs by comparing it with all the other major clusters. When there are more than 200 markers, only the top 200 markers (by p-value of DEG analysis) are kept in further analyses. We then employ a semi-supervised strategy to rank those DEGs by their correlations with the targeted cell abundance in the simulated “pseudo-bulk” RNAseq.
Thus, we utilize simulated bulk RNAseq in both selecting effective cell markers and estimating and in formula 1. Particularly, the pseudo-bulk simulation procedure is split into two phases which differ in how the gene variance is modeled.
Phase 1) Simulating the training dataset of pseudo-bulk samples with gene variation estimated from scRNAseq only. 1,000 pseudo-bulk samples are simulated according to the (Equation 2), (Equation 3), (Equation 4) where cell type-specific gene variations between samples are estimated from scRNAseq dataset.
| (Equation 2) |
| (Equation 3) |
| (Equation 4) |
where represents the simulated pseudo-bulk expression of gene for simulated sample , represents the simulated expression of gene in cell type for sample , represents the simulated frequency of cell type for sample . Specifically, is simulated according to normal distribution (formula 3) with mean and variance estimated based on scRNAseq observation. denotes the between-sample mean and variance of expression of gene in cell type as estimated in the multi-sample scRNAseq dataset, respectively (calculated the same as in MuSiC13). is simulated according to a zero-inflated Beta distribution as shown in formula 4, where denotes the probability of bulk samples without cell type . Both and the two shape parameters in the Beta distribution can be estimated from the input scRNAseq data directly or through an independent cell frequency data provided by user. Note that any negative value in simulated will be set to zero before the next step.
Using the above simulated pseudo-bulk dataset, the initial gene markers (DEGs by Seurat) for each cell type are ranked based on their expression correlation with the cell frequency in the pseudo-bulk data. A heurist cutoff is chosen to select the top-ranked markers to estimate mean marker expression in formula 1 for each pseudo-bulk sample. (More discussion of cutoff can be found later). Coefficients and in formula 1 are then estimated by fitting the simulated dataset through least square error.
We then predict cell frequency in the input bulk sample using formula 1 where the cell markers and coefficients are determined above using simulated bulk datasets in phase 1.
Phase 2) Besides the between-sample gene variation simulated above, there is also cross-platform gene variation caused by difference between scRNAseq and bulk RNAseq platforms, annotated as . Given the initial estimation of cell frequency of input bulk RNAseq data (Phase 1), can be calculated by formula 5.
| (Equation 5) |
The pseudo-bulk sample is then simulated according to (Equation 6), (Equation 7).
| (Equation 6) |
| (Equation 7) |
where represents the added cross-platform variation, and are simulated the same way as in phase 1: formula 3 and 4.
Using the above simulated pseudo-bulk data, cell markers are re-selected, and coefficients in formula 1 are re-estimated in the same way as phase 1. Finally, cell frequency of the input bulk dataset is updated according to formula 1
Profile-based deconvolution
In profile-based deconvolution, the cell frequency is estimated by minimizing the weighted NNLS of formula 8. Here, the reference profile is computed as in marker-based deconvolution. Note that only gene markers of each minor and major cell cluster identified by Seurat FindAllMarkers() function are considered.
| (Equation 8) |
The key here is to design the effective weight for each gene which correctly reflects the gene variation in deconvolution for weighted NNLS estimation. As discussed in the marker-based deconvolution, the gene variation in deconvolution consists of two components: between-sample variation and cross-platform variation . Similarly, the profile-based deconvolution is also split into two phases.
Phase 1) Gene weight is computed with only between-sample variation considered (formula 9). The cell frequency and weight is updated in an iterative fashion using formula 8 and 9 until convergence or a maximum of 1000 iterations (similar to what is implemented in MuSiC,13 the initial weight is set to 1).
| (Equation 9) |
Phase 2) The output of Phase 1) is used to estimate cross-platform variation according to formula 5. Then a combinatory gene weight was calculated by considering both the between-sample and cross-platform variations (formula 10). and is then updated in an iterative fashion using formula 8 and 10, similar to phase 1).
| (Equation 10) |
The final estimation of cell frequency is the average of marker-based and profile-based deconvolution results, and then normalized to be sum of one for each bulk sample. Note that the estimation of requires multiple samples in the scRNAseq reference profiles. In addition, because (formula 5) is estimated cross all the input bulk RNAseq samples, ProM deconvolution results may vary slightly with inclusion or exclusion a set of bulk samples.
Comparison of marker selection strategies
To illustrate the advantage of semi-supervised marker selection strategy, we compared it with the DEGs-based selection strategy in their performance of deconvoluting reconstituted bulk RNAseq data. In marker-based ProM deconvolution, cell markers (output from FindAllMarkers() function in Seurat) were ordered according to their correlation with simulated pseudo-bulk RNAseq data, and the top-ranking markers were chosen. As a comparison, cell markers were also ordered according to p-values output from FindAllMarkers(), and the same number of top-ranking markers were chosen as in ProM. As shown in Figure S2B, semi-supervised marker selection strategy generally performed better than DEG-based marker selection.
As to the cutoff for marker selection, ProM allows users to input the number of top-ranking markers for each cell cluster. As an alternative, users can also specify a cutoff of correlation coefficient (CC) so that all markers whose correlation with the targeted cell frequency in pseudo-bulk RNAseq larger than the cutoff will be selected in ProM simulation steps. In order to avoid too few markers selected, a minimum of 20 markers per cell cluster are selected (or all markers when the number of markers are less than 20). This alternative cutoff allows varied number of markers for different cell types. In this study, we used a heuristic cutoff CC>0.3 for deconvolution of the real bulk RNAseq datasets, which corresponds to a median marker number of 34 per cell cluster with median absolute deviation of 11. We used a cutoff of top 20 markers for all the reconstituted bulk RNAseq datasets, for controlling the marker numbers facilitates the methodology comparison in different scenarios. In practice, both cutoff selection strategies work well. A marker number less than 50 per cell cluster or a correlation cutoff larger than 0.3 is recommended and generally give reasonably good results. Users can reconstitute bulk RNAseq data using their own scRNAseq data, and then investigate the effect of marker cutoff selection as shown in Figure S2B if the best cutoff is desired.
The advantage of modeling cross-platform variation
One difference between ProM and other existing deconvolution methods, such as MuSiC, is to incorporate the cross-platform gene variations explicitly in phase2 of both marker-based and profile-based deconvolution. To illustrate the benefits of modeling cross-platform variation, noise at different levels was then added to the reconstituted bulk profiles to represent cross-platform variations, and the performance of ProM with and without phase 2 was evaluated. To simulate the cross-platform variation that mimics empirical data, we took advantage of our paired bulk and single cell RNAseq data. In particular, the difference between the reconstituted bulk profiles (summed from scRNAseq) and the corresponding paired empirical bulk profiles was calculated. The mean and standard deviation of the difference were estimated for each gene, which was then used to simulate the noise added to the reconstituted bulk profiles. Finally, the simulated noise was multiplied by a factor to represent the different noise levels. At each noise level, five runs of simulation corresponding to five sets of reconstituted bulk samples were conducted. The final deconvolution accuracy was measured by the average of the five runs of simulation. As shown in Figure S2C, ProM with phase 2 performed consistently better than with phase 1 only. In addition, the improvement by phase 2 increased as the added cross-platform noise level increased.
Evaluating deconvolution accuracy using paired single cell and bulk RNAseq
The cell frequency output from ProM and that observed from scRNAseq dataset were properly transformed to ensure comparability. To be specific, for the CD45+ sorted scRNAseq samples, all the non-immune cells were removed before the relative cell frequency of each immune subset among all immune cells were calculated. To be comparable, the non-immune cells from the corresponding deconvolution result were also removed. Similarly, for CD45- sorted scRNAseq samples and their paired bulk RNAseq deconvolution results, all immune cells were removed before the relative cell frequency among non-immune cells were calculated. No transformation was applied for unsorted scRNAseq samples and the paired deconvolution results.
Other deconvolution methods
The following deconvolution methods were evaluated in our study. R packages for BayesianPrism14 (v 1.4) and MuSiC13 (v 0.2.0) were downloaded from github (Danko-Lab/TED and xuranw/MuSiC). CibersortX12 and its two batch-corrected versions were run through the docker downloaded from (https://cibersortx.stanford.edu/). The R packages for BisqueRNA36 (v 1.0.5) and DeconRNAseq37 (v 1.32.0) was obtained from CRAN and Bioconductor, respectively. The methods of non-negative least square(nnls)31 and DSA17 were implemented by ourselves based on R package of nnls and NMF according to the description of the original study. To have a fair comparison with ProM, gene markers used in ProM (identified by FindAllMarkers() function in Seurat package) were input into the above algorithms when markers were allowed.
Applying ProM to external bulk RNAseq datasets
Independent urothelial cancer cohorts were used in this study, i.e., TCGA BLCA cohort20 and three PD-L1 blockade cohorts (IMvigor 210,10 CheckMate 2759 and IMvigor 01011) consisting of 408, 348, 72, and 114 samples, respectively. The detailed description of the preprocessing of TCGA, IMvigor 210 and CheckMate 275 datasets can be found in our previous study.3 The IMvigor 010 dataset was downloaded from European Genome-Phenome Archive under accession number EGAS00001004997. In total, 114 samples that are positive for ctDNA and under atezolizumab treatment were included in our association study since ctDNA negative samples did not show benefit from the treatment.11 For all datasets, TPM measurements of the bulk RNAseq were used as input for ProM deconvolution.
Sample preparation for IMC
Two surgical UC luminal subtype baseline specimens prior to PD-1 checkpoint blockade treatment were identified for IMC. Tumor tissues were formalin-fixed, paraffin-embedded, and 5μm tissue sections were mounted on positively charged glass slides. A section of tissue was stained by Hematoxylin and eosin (H&E) to evaluate structures by two pathologists (R.B. and C.C.C.) and identify representative areas within tumors for ROI selection (1mm2). Samples were also evaluated by pan-cytokeratin immunostaining for tumor nest identification and quality control of the FFPE sections prior to IMC staining. Multiple ROIs per specimen were manually selected based on features of the tissue (e.g. immune infiltrates, tumor nests) that were representative of the entire specimen for data acquisition.
IMC antibody conjugation
The design of our custom antibody panel was hypothesis driven. BSA-free antibodies were used and conjugated to pure rare earth metal isotopes from the lanthanide series using the MaxPar X8 Multimetal Labeling Kits (Fluidigm, Ontario, CAN) according to the manufacturer’s instructions. After conjugation, antibody yield was measured with a Nanodrop (Thermo Scientific, Waltham, MA, USA) and diluted in Candor Antibody Stabilizer (CANDOR Bioscience Gmbh, Wangen im Allgäu, Germany) for long-term storage at 4°C. IMC Antibody dilution and specificity were assessed using control FFPE tissues, including UC and normal tonsil, before proceeding with study samples. Samples were stained following the IMC staining protocol for FFPE sections from the manufacturer (Fluidigm, South San Francisco, CA, USA).
IMC data acquisition
Data acquisition was performed by laser ablation of the selected ROIs at a resolution of 1mm2 using the Hyperion Imaging System at Fluidigm Corporation (South San Francisco, CA, USA).
IMC data processing
The raw IMC data was processed using the python package steinbock (https://github.com/BodenmillerGroup/steinbock). The deep-learning based cell segmentation was conducted based on DNA and cell membrane markers.43 The expression readout of the 16 proteins summarized at the cell level was arcsine transformed and scaled into Z-score before any downstream analysis. The Z-score of 14 (out of 16) cell marker proteins were used as input for PhenoGraph44 to cluster and annotate cells into six major types: Epithelial cells ("pan-cytokeratin-148Nd","GATA3-171Yb","Cytokeratin5-144Nd"), T cells ("CD45-89Y","CD3-170Er","CD8a-162Dy"), Macrophage/Monocytes ("CD45-89Y","CD68-173Yb","CD16-147Sm","CD14-143Nd"), Neutrophil ("CD45-89Y","CD66b-164Dy"), Fibroblast ("CollagenType1-169Tm","Collagen3-158Gd","SMA-141Pr") and Endothelial cells ("CD31-151Eu"). Two additional markers “HLA-DR-174Yb" and "SPP1-161Dy" were used to refined categorization of Macrophage/Monocytes into MM_SPP1 (Z-score of SPP1>1.64 corresponding to p-value of 0.05), MM_HLA-DR (Z-score of HLA-DR>1.64) and MM_others.
Quantification and statistical analysis
Two PD-1/PD-L1 cohorts, i.e., IMvigor 210 and CheckMate 275, were used as exploratory cohorts to identify subpopulations significantly associated with survival outcome. The third PD-1/PD-L1 cohort, i.e., IMvigor 010, was used as an independent cohort to validate the discoveries made in exploratory cohorts.
For each of the minor scRNAseq clusters, the relative cell frequency was calculated by the ratio of its abundance versus all cells that belong to the same major cluster (relative2major) For example, the relative frequency of MM_SPP1 was calculated by the abundance of MM_SPP1 versus that of all MM cells.
To assess the survival associations of a particular cell subpopulation in each exploratory cohort, p-value of the likelihood ratio test by the Cox-regression model was computed where the cell frequency was treated as a continuous variable (We treated cell frequency as continuous variable since the optimal cutoff used to discretize samples were unknown) . We then used Fisher’s combined probability test45 to combine p-values from the two exploratory cohorts into one p-value for each cell population. Finally, we adjusted p-value for multiple testing correction using FDR method.46 We used the adjusted p-value < 0.05 to select significant cell subpopulations. To avoid the Fisher’s combined test to be dominated by one cohort, we also require the nominal p-value before combination (likelihood ratio test) in each cohort to be at least marginally significant (p-value<0.1). Under such criteria, we obtained three cell subpopulations associated with worse survival: ILC_IL23R (nominal p-value =0.092 and 0.0062 in IMvigor 210 and CheckMate 275 dataset, respectively, and combined p-value after adjustment =0.034), MM_SPP1 (nominal p-value = 0.071 and 0.038, combined and adjusted p-value=0.044), T-CD8_GZMK (nominal p-value=0.037 and 0.028, combined and adjusted p-value= 0.034). Similarly, we obtained two cell subpopulations associated with better survival: MM_C1QC (nominal p-value=0.029 and 0.0021, combined and adjusted p-value =0.0066), T-CD8_reactive (nominal p-value =0.044 and 0.0025, combined and adjusted p-value =0.0066).
To illustrate the survival association of a particular cell subpopulation using KM curves, samples were split into high and low groups according to their relative cell frequency. Three different cutoffs, i.e., the lower tertiary vs. the median and higher tertiary, were assessed, and the cutoff resulted in the most significant log rank test p-value (combined both exploratory cohorts) was chosen to plot KM curves (Figure 5A). To be consistent, the same cutoff was applied when plotting KM curves in the validation cohort.
Acknowledgments
This research was supported by the National Cancer Institute (NCI) of the National Institutes of Health (NIH) under award R01 CA249175-01. S.I. is supported by the Training Program in Cancer Biology, NCI, NIH (T32-CA078207) and the Loan Repayment Program, The National Center for Advancing Translational Sciences, NIH. M.T. is supported by the NCI, NIH under award CA275269-01.
Author contributions
Conceptualization, L.W., J.Z., and M.D.G.; Computational methodology and data analysis, L.W. and J.Z.; Sample acquisition and data generation: S.I., J.P.S., M.T., K.G.B., R.B., C.C.-C., A.H., and R.S.; Writing – Original Draft, L.W.; Writing – Review and Editing, L.W., J.Z., M.D.G., S.I., J.P.S., M.T., K.G.B., R.B., C.C.-C., A.H., R.S., W.K.O., and N.B.; Supervision, J.Z. and M.D.G; Funding acquisition: N.B., M.D.G., and J.Z.
Declaration of interests
R.S. is a paid consultant and shareholder of GeneDx, Stamford, CT. A.H. receives research funds from Zumutor Biologics and is on the advisory boards of HTG Molecular Diagnostics, Immunorizon, UroGen, and Takeda. N.B. is an extramural member of the Parker Institute for Cancer Immunotherapy; receives research funds from Regeneron, Harbor Biomedical, DC Prime, and Dragonfly Therapeutics; and is on the advisory boards of Neon Therapeutics, Novartis, Avidea, Boehringer Ingelheim, Rome Therapeutics, Rubius Therapeutics, Roswell Park Comprehensive Cancer Center, BreakBio, Carisma Therapeutics, CureVac, Genotwin, BioNTech, Gilead Therapeutics, Tempest Therapeutics, and the Cancer Research Institute. M.D.G. has received grants or contracts from Bristol Myers Squibb, Novartis, Dendreon, AstraZeneca, and Merck; and has received consulting fees from Bristol Myers Squibb, Merck, Genentech, Inc., AstraZeneca, Pfizer, EMD Serono, Seagen, Janssen, Numab, Dragonfly, GlaxoSmithKline, Basilea, UroGen, RapptaTherapeutics, Alligator, Silverback, Fujifilm, and Curis.
Published: May 7, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.109928.
Contributor Information
Matthew D. Galsky, Email: matthew.galsky@mssm.edu.
Jun Zhu, Email: junzhu_99@yahoo.com.
Supplemental information
References
- 1.Kamoun A., de Reyniès A., Allory Y., Sjödahl G., Robertson A.G., Seiler R., Hoadley K.A., Groeneveld C.S., Al-Ahmadie H., Choi W., et al. A consensus molecular classification of muscle-invasive bladder cancer. Eur. Urol. 2020;77:420–433. doi: 10.1016/j.eururo.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niglio S.A., Jia R., Ji J., Ruder S., Patel V.G., Martini A., Sfakianos J.P., Marqueen K.E., Waingankar N., Mehrazin R., et al. Programmed death-1 or programmed death ligand-1 blockade in patients with platinum-resistant metastatic urothelial cancer: a systematic review and meta-analysis. Eur. Urol. 2019;76:782–789. doi: 10.1016/j.eururo.2019.05.037. [DOI] [PubMed] [Google Scholar]
- 3.Wang L., Sfakianos J.P., Beaumont K.G., Akturk G., Horowitz A., Sebra R.P., Farkas A.M., Gnjatic S., Hake A., Izadmehr S., et al. Myeloid Cell–associated Resistance to PD-1/PD-L1 Blockade in Urothelial Cancer Revealed Through Bulk and Single-cell RNA SequencingMyeloid Cell-Associated Resistance to PD-1/PD-L1 Blockade. Clin. Cancer Res. 2021;27:4287–4300. doi: 10.1158/1078-0432.CCR-20-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L., Saci A., Szabo P.M., Chasalow S.D., Castillo-Martin M., Domingo-Domenech J., Siefker-Radtke A., Sharma P., Sfakianos J.P., Gong Y., et al. EMT-and stroma-related gene expression and resistance to PD-1 blockade in urothelial cancer. Nat. Commun. 2018;9:3503. doi: 10.1038/s41467-018-05992-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tran M.A., Farkas A., Beaumont K., O’Donnell T., Mehrazin R., Wiklund P., Horowitz A., Galsky M., Sfakianos J., Bhardwaj N. Characterization of urine-derived immune cells from bladder cancer patients and comparison to tumor and peripheral blood. J. Immunol. 2022;208 [Google Scholar]
- 6.Salomé B., Sfakianos J.P., Ranti D., Daza J., Bieber C., Charap A., Hammer C., Banchereau R., Farkas A.M., Ruan D.F., et al. NKG2A and HLA-E define an alternative immune checkpoint axis in bladder cancer. Cancer Cell. 2022;40:1027–1043.e1029. doi: 10.1016/j.ccell.2022.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z., Zhou L., Liu L., Hou Y., Xiong M., Yang Y., Hu J., Chen K. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat. Commun. 2020;11:5077. doi: 10.1038/s41467-020-18916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sacher A.G., St Paul M., Paige C.J., Ohashi P.S. Cytotoxic CD4+ T Cells in Bladder Cancer—A New License to Kill. Cancer Cell. 2020;38:28–30. doi: 10.1016/j.ccell.2020.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Sharma P., Retz M., Siefker-Radtke A., Baron A., Necchi A., Bedke J., Plimack E.R., Vaena D., Grimm M.-O., Bracarda S., et al. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol. 2017;18:312–322. doi: 10.1016/S1470-2045(17)30065-7. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman-Censits J.H., Grivas P., Van Der Heijden M.S., Dreicer R., Loriot Y., Retz M., Vogelzang N.J., Perez-Gracia J.L., Rezazadeh A., Bracarda S. IMvigor 210, a phase II trial of atezolizumab (MPDL3280A) in platinum-treated locally advanced or metastatic urothelial carcinoma (mUC) American Society of Clinical Oncology. 2016;34 doi: 10.1200/jco.2016.34.2_suppl.355. [DOI] [Google Scholar]
- 11.Powles T., Assaf Z.J., Davarpanah N., Banchereau R., Szabados B.E., Yuen K.C., Grivas P., Hussain M., Oudard S., Gschwend J.E., et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature Cancer. 2021;595:432–437. doi: 10.1038/s41586-021-03642-9. [DOI] [PubMed] [Google Scholar]
- 12.Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X., Park J., Susztak K., Zhang N.R., Li M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun. 2019;10:380. doi: 10.1038/s41467-018-08023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu T., Wang Z., Pe’er D., Danko C.G. Cell type and gene expression deconvolution with BayesPrism enables Bayesian integrative analysis across bulk and single-cell RNA sequencing in oncology. Nat. Cancer. 2022;3:505–517. doi: 10.1038/s43018-022-00356-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L., Sebra R.P., Sfakianos J.P., Allette K., Wang W., Yoo S., Bhardwaj N., Schadt E.E., Yao X., Galsky M.D., Zhu J. A reference profile-free deconvolution method to infer cancer cell-intrinsic subtypes and tumor-type-specific stromal profiles. Genome Med. 2020;12:24. doi: 10.1186/s13073-020-0720-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H., Sfakianos J.P., Wang L., Hu Y., Daza J., Galsky M.D., Sandhu H.S., Elemento O., Faltas B.M., Farkas A.M., et al. Tumor-Infiltrating Myeloid Cells Confer De Novo Resistance to PD-L1 Blockade through EMT-Stromal and Tgfβ-Dependent Mechanisms. Mol. Cancer Ther. 2022;21:1729–1741. doi: 10.1158/1535-7163.Mct-22-0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong Y., Wan Y.-W., Pang K., Chow L.M.L., Liu Z. Digital sorting of complex tissues for cell type-specific gene expression profiles. BMC Bioinf. 2013;14:89. doi: 10.1186/1471-2105-14-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo H., Xia X., Huang L.B., An H., Cao M., Kim G.D., Chen H.N., Zhang W.H., Shu Y., Kong X., et al. Pan-cancer single-cell analysis reveals the heterogeneity and plasticity of cancer-associated fibroblasts in the tumor microenvironment. Nat. Commun. 2022;13:6619. doi: 10.1038/s41467-022-34395-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakravarthy A., Furness A., Joshi K., Ghorani E., Ford K., Ward M.J., King E.V., Lechner M., Marafioti T., Quezada S.A., et al. Pan-cancer deconvolution of tumour composition using DNA methylation. Nat. Commun. 2018;9:3220. doi: 10.1038/s41467-018-05570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson A.G., Kim J., Al-Ahmadie H., Bellmunt J., Guo G., Cherniack A.D., Hinoue T., Laird P.W., Hoadley K.A., Akbani R., et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171:540–556.e25. doi: 10.1016/j.cell.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sfakianos J.P., Daza J., Hu Y., Anastos H., Bryant G., Bareja R., Badani K.K., Galsky M.D., Elemento O., Faltas B.M., Mulholland D.J. Epithelial plasticity can generate multi-lineage phenotypes in human and murine bladder cancers. Nat. Commun. 2020;11:2540. doi: 10.1038/s41467-020-16162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qian J., Olbrecht S., Boeckx B., Vos H., Laoui D., Etlioglu E., Wauters E., Pomella V., Verbandt S., Busschaert P., et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020;30:745–762. doi: 10.1038/s41422-020-0355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C., Liu B., Kang B., Liu Z., Liu Y., Chen C., Ren X., Zhang Z. SciBet as a portable and fast single cell type identifier. Nat. Commun. 2020;11:1818. doi: 10.1038/s41467-020-15523-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Street K., Risso D., Fletcher R.B., Das D., Ngai J., Yosef N., Purdom E., Dudoit S. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L., Li Z., Skrzypczynska K.M., Fang Q., Zhang W., O'Brien S.A., He Y., Wang L., Zhang Q., Kim A., et al. Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell. 2020;181:442–459.e29. doi: 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 26.Cheng S., Li Z., Gao R., Xing B., Gao Y., Yang Y., Qin S., Zhang L., Ouyang H., Du P., et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell. 2021;184:792–809.e23. doi: 10.1016/j.cell.2021.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Ojalvo L.S., King W., Cox D., Pollard J.W. High-density gene expression analysis of tumor-associated macrophages from mouse mammary tumors. Am. J. Pathol. 2009;174:1048–1064. doi: 10.2353/ajpath.2009.080676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen P., Zhao D., Li J., Liang X., Li J., Chang A., Henry V.K., Lan Z., Spring D.J., Rao G., et al. Symbiotic macrophage-glioma cell interactions reveal synthetic lethality in PTEN-null glioma. Cancer Cell. 2019;35:868–884.e6. doi: 10.1016/j.ccell.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazzurana L., Czarnewski P., Jonsson V., Wigge L., Ringnér M., Williams T.C., Ravindran A., Björklund Å.K., Säfholm J., Nilsson G., et al. Tissue-specific transcriptional imprinting and heterogeneity in human innate lymphoid cells revealed by full-length single-cell RNA-sequencing. Cell Res. 2021;31:554–568. doi: 10.1038/s41422-020-00445-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai H., Cheng X., Liu Q., Luo W., Liu M., Zhang M., Miao J., Ji Z., Lin G.N., Song W., et al. Single-cell RNA sequencing reveals the epithelial cell heterogeneity and invasive subpopulation in human bladder cancer. Int. J. Cancer. 2021;149:2099–2115. doi: 10.1002/ijc.33794. [DOI] [PubMed] [Google Scholar]
- 31.Avila Cobos F., Alquicira-Hernandez J., Powell J.E., Mestdagh P., De Preter K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat. Commun. 2020;11:5650. doi: 10.1038/s41467-020-19015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scharping N.E., Menk A.V., Whetstone R.D., Zeng X., Delgoffe G.M. Efficacy of PD-1 Blockade Is Potentiated by Metformin-Induced Reduction of Tumor HypoxiaMetformin Improves PD-1 Blockade Immunotherapy. Cancer Immunol. Res. 2017;5:9–16. doi: 10.1158/2326-6066.CIR-16-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Najjar Y.G., Menk A.V., Sander C., Rao U., Karunamurthy A., Bhatia R., Zhai S., Kirkwood J.M., Delgoffe G.M. Tumor cell oxidative metabolism as a barrier to PD-1 blockade immunotherapy in melanoma. JCI insight. 2019;4 doi: 10.1172/jci.insight.124989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daniel S.K., Sullivan K.M., Labadie K.P., Pillarisetty V.G. Hypoxia as a barrier to immunotherapy in pancreatic adenocarcinoma. Clin. Transl. Med. 2019;8:10–17. doi: 10.1186/s40169-019-0226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noman M.Z., Desantis G., Janji B., Hasmim M., Karray S., Dessen P., Bronte V., Chouaib S. PD-L1 is a novel direct target of HIF-1α, and its blockade under hypoxia enhanced MDSC-mediated T cell activation. J. Exp. Med. 2014;211:781–790. doi: 10.1084/jem.20131916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jew B., Alvarez M., Rahmani E., Miao Z., Ko A., Garske K.M., Sul J.H., Pietiläinen K.H., Pajukanta P., Halperin E. Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nat. Commun. 2020;11:1971. doi: 10.1038/s41467-020-15816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong T., Szustakowski J.D. DeconRNASeq: a statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-Seq data. Bioinformatics. 2013;29:1083–1085. doi: 10.1093/bioinformatics/btt090. [DOI] [PubMed] [Google Scholar]
- 38.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran H.T.N., Ang K.S., Chevrier M., Zhang X., Lee N.Y.S., Goh M., Chen J. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 2020;21:12. doi: 10.1186/s13059-019-1850-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Büttner M., Miao Z., Wolf F.A., Teichmann S.A., Theis F.J. A test metric for assessing single-cell RNA-seq batch correction. Nat. Methods. 2019;16:43–49. doi: 10.1038/s41592-018-0254-1. [DOI] [PubMed] [Google Scholar]
- 41.Hubert L., Arabie P. Comparing partitions. J. Classif. 1985;2:193–218. doi: 10.1007/BF01908075. [DOI] [Google Scholar]
- 42.Miao Z., Moreno P., Huang N., Papatheodorou I., Brazma A., Teichmann S.A. Putative cell type discovery from single-cell gene expression data. Nat. Methods. 2020;17:621–628. doi: 10.1038/s41592-020-0825-9. [DOI] [PubMed] [Google Scholar]
- 43.Greenwald N.F., Miller G., Moen E., Kong A., Kagel A., Dougherty T., Fullaway C.C., McIntosh B.J., Leow K.X., Schwartz M.S., et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 2022;40:555–565. doi: 10.1038/s41587-021-01094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine J.H., Simonds E.F., Bendall S.C., Davis K.L., Amir E.a.D., Tadmor M.D., Litvin O., Fienberg H.G., Jager A., Zunder E.R., et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015;162:184–197. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fisher R.A. Springer; 1992. Statistical Methods for Research Workers. [Google Scholar]
- 46.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Paired bulk RNAseq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. This paper assembles and reanalyzes existing publicly available scRNAseq data. These accession numbers for the datasets are listed in the key resources table. The reanalyzed scRNAseq dataset with meta information including the cell annotation generated in this study have been deposited at Zenodo and are publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
R package of ProM algorithm together with script to reproduce the cross-validation comparison of different deconvolution algorithms in pan cancer datasets has been deposited at github and is publicly available as of the date of publication. DOIs are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.