Abstract
We provide experimental evidence for RNase III-dependent processing in helix 9 of the 23S rRNA as a general feature of many species in the alpha subclass of Proteobacteria (alpha-Proteobacteria). We investigated 12 Rhodobacter, Rhizobium, Sinorhizobium, Rhodopseudomonas, and Bartonella strains. The processed region is characterized by the presence of intervening sequences (IVSs). The 23S rDNA sequences between positions 109 and 205 (Escherichia coli numbering) were determined, and potential secondary structures are proposed. Comparison of the IVSs indicates very different evolutionary rates in some phylogenetic branches, lateral genetic transfer, and evolution by insertion and/or deletion. We show that the IVS processing in Rhodobacter capsulatus in vivo is RNase III-dependent and that RNase III cleaves additional sites in vitro. While all IVS-containing transcripts tested are processed in vitro by RNase III from R. capsulatus, E. coli RNase III recognizes only some of them as substrates and in these substrates frequently cleaves at different scissile bonds. These results demonstrate the different substrate specificities of the two enzymes. Although RNase III plays an important role in the rRNA, mRNA, and bacteriophage RNA maturation, its substrate specificity is still not well understood. Comparison of the IVSs of helix 9 does not hint at sequence motives involved in recognition but reveals that the “antideterminant” model, which represents the most recent attempt to explain the E. coli RNase III specificity in vitro, cannot be applied to substrates derived from alpha-Proteobacteria.
rRNA and ribosomal DNA (rDNA) sequences are widely used for bacterial classification, phylogenetic studies, and identification purposes. Therefore, it is important to know which sequences are removed from the primary rRNA transcript during rRNA maturation. Ten years ago, it was believed that fragmented 23S rRNAs in bacteria are an exception. Now it is known that this phenomenon is widespread. Fragmented 23S rRNAs are found in representatives of many species of the alpha, gamma, and epsilon subclasses of Proteobacteria (alpha-, gamma-, and epsilon-Proteobacteria, respectively) (4, 9, 12, 13, 16, 19, 21, 23, 28–31, 33, 35, 37) and in Spirochaeta (26). In most cases, the processing sites are characterized by the presence of internal transcribed spacers, or so-called intervening sequences (IVSs), which are removed without splicing. Instead, the resulting fragments are held together by the compact structure of the ribosomes which remain functional.
In all bacteria with fragmented 23S rRNA investigated thus far, with the exception of those belonging to alpha-Proteobacteria, two possible processing sites were found: at positions 540 (helix 25) and 1120 (helix 45) (Escherichia coli numbering). The occurrence of IVSs at these positions is sporadic: only some strains of a bacterial species possess fragmented 23S rRNA, and often even in a given bacterial strain not all rrn operons contain IVSs (4, 12, 21, 23). In alpha-Proteobacteria additional fragmentation sites were found. In Rhodobacter capsulatus and Rhodobacter sphaeroides, the 23S rRNA is fragmented at position 1200 (helix 46; E. coli numbering) due to IVS processing (16, 27). In domain III of 23S rRNA of some Rhizobium and Agrobacterium strains sporadic fragmentation without the involvement of IVSs has been described (28, 30). In all Rhizobium, Agrobacterium, and Bradyrhizobium strains investigated thus far, fragmentation near position 130 (E. coli numbering) of 23S rRNA was found (9, 28–30). In E. coli, nucleotides 130 to 148 of the 23S rRNA form the small helix 9, which is substantially extended due to extra stem-loop structures (IVSs) in rhizobial and agrobacterial strains, both showing fragmentation in this region (28, 30). The occurrence of IVSs in helix 9 of these strains is not sporadic and may be a common feature of a major part of the alpha-Proteobacteria. We investigated the processing of helix 9 of 23S rRNA in 12 Rhodobacter, Rhodopseudomonas, Rhizobium, Sinorhizobium, and Bartonella strains. Recently, fragmentation in helix 9 of 23S rRNA in Rhodopseudomonas palustris was confirmed and such fragmentation in Rhodobacter species was predicted (37). 23S rRNA fragmentation at this position in R. sphaeroides was predicted already 10 years ago (7), but it was not confirmed until now. The processing mechanism of the known helix 9 fragmentation in Rhizobiaceae was also not investigated until now. On the basis of sequence comparisons (EMBL database), we also predicted fragmentation in helix 9 of 23S rRNA in Bartonella bacilliformis (22). To confirm the occurrence of such fragmentation in Bartonella, we investigated the processing of helix 9 of Bartonella henselae ATCC 49882 in vitro.
The IVS processing in helices 25, 45, and 46 of eubacterial 23S rRNA is catalyzed by RNase III (4, 27). This endoribonuclease cleaves rRNA precursors during maturation of rRNA and is also involved in the maturation of some mRNA and bacteriophage RNA species (8, 14). The enzyme is not essential for viability; nevertheless, its primary sequence is highly conserved (27). RNase III recognizes and precisely cleaves double-helical RNA structures with a 20-bp minimal length. These double helices contain some unpaired residues and do not exhibit any consensus sequence (24). The substrate specificity of the enzyme is poorly understood. An attempt to explain the E. coli RNase III substrate specificity was made using the concept of “antideterminants”: a cleavage site is defined by the absence of disfavored sequence motifs in its vicinity (38). The RNases III from E. coli and R. capsulatus (RNase IIIEc and RNase IIIRc, respectively) are very similar in their amino acid sequences, but it was shown that they show significant differences in binding and cleavage of certain substrates in vitro (5). These differences between both enzymes are not well understood. It is also not known which properties make an RNA a “good” substrate for the RNase IIIRc. One way to address these questions is to compare different natural RNase IIIRc specific substrates and their interaction in vitro with both purified enzymes.
The aim of this work was to investigate the occurrence of IVS-dependent fragmentation in helix 9 of 23S rRNA in different phototrophic, symbiotic, and pathogenic alpha-proteobacterial species. Further, we analyzed the mechanism of 23S rRNA processing in this region. The study of the IVSs found in helix 9 of alpha-proteobacterial 23S rRNA and their RNase III-dependent cleavage can help us to better understand the mode of action of this enzyme as well as the evolution of the rRNA genes.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The R. capsulatus strains used in this study were the wild-type strains 37b4 (DSM 938) and B10 (18) and the mutant strain Fm65 (16). R. sphaeroides wild-type strains WS8 (32) and 17023 were used. R. palustris 5D and R. sphaeroides 17023 were obtained from G. Drews, Freiburg, Germany. All Rhodobacter strains as well as the strains R. palustris 5D and Rhodospirillum rubrum DSM 107 were grown in a minimal malate salt medium (6). During growth of the R. capsulatus Fm65 strain, carrying plasmid pRK2fm1 (16), tetracycline was added at a final concentration of 1 μg ml−1.
The rhizobial strains Rhizobium giardinii H152, Rhizobium gallicum R602 (1), and Sinorhizobium fredii MSDJ 1536 were obtained from N. Amarger (INRA-CMSE, Dijon, France). Rhizobium etli strains CFN42 (USDA 9032) and Viking I (USDA 2743) and Rhizobium leguminosarum ATCC 10004 (USDA 2370) were obtained from D. K. Jones (U.S. Department of Agriculture/Agricultural Research Service, Beltsville Rhizobium Germplasm Collection). They were grown on tryptic yeast media (3).
E. coli JM109 (Stratagene) was grown on standard I medium (Difco).
Oligonucleotides (primers).
The oligonucleotide used for hybridizations was 5′-GGGTTTCCCCATTCGGAAA (23Sup130, 19 nucleotides [nt], complementary to the highly conserved positions 112 to 130 of the 23S rRNA; E. coli numbering). For PCR amplification of the regions between positions 109 and 205 of rDNA (E. coli numbering), the following primers were used: 5′-GGGGGGAA TTCTAATACGACTCACTATAG(G/A)AT(T/G)TCCGAATGGGGAAACCC- 3′ (23S-IVS-sense primer, 50 nt; the EcoRI site is underlined; the T7 promoter region for transcription initiation is in boldface), corresponding to the rDNA positions 109 to 130, and 5′-GGGGGAAGCTTCTTAG(T/A)(A/T)GTTTC(T/A)GTTCC-3′ (23S-IVS-antisense primer, 30 nt; the HindIII site is underlined), corresponding to the rDNA positions 185 to 205 (E. coli numbering). All oligonucleotides were synthesized on a 380B DNA Synthesizer (Applied Biosystems).
Isolation, amplification, and analysis of nucleic acids.
Total DNA from alpha-Proteobacteria was isolated according to the method of Ausubel et al. (2). Hot phenol RNA isolation, Northern blotting, and hybridization were performed with standard methods (10).
Aliquots of 10 pmol of primer were labeled for 30 to 60 min at 37°C with 20 μCi of [γ-32P]ATP using polynucleotide kinase and subsequently purified with a NucTrap push column (Stratagene). Labeled oligonucleotides were used for hybridization and primer extension analysis.
PCR was carried out in a final volume of 50 μl with 200 ng of total DNA as a template, using 0.8 U of Taq DNA polymerase (Promega) at an annealing temperature of 42°C (45 s), followed by extension at 72°C (30 s). Cycles were repeated 35 times. The resulting PCR products were purified from 3.5% small DNA low-melting-point agarose gels (FMC-Biozym).
The purified PCR products were used directly for in vitro transcription or for direct cycle sequencing using the ABI Prism Dye Terminator Cycle Sequencing kit (Perkin-Elmer). The sequencing reaction was done according to the protocol of the manufacturer. The program used for cycle sequencing was as follows: initial denaturation for 50 s at 96°C, with 25 cycles of 20 s at 96°C, 20 s at 55°C, and 2 min at 60°C. The resulting products were ethanol precipitated and loaded on a 373 DNA Sequencer (Perkin-Elmer).
The agarose gel-purified PCR products encompassing positions 109 to 205 of 23S rDNA (E. coli numbering) were cut with HindIII and EcoRI and cloned into pUC18 vectors. The resulting constructs were purified with Qiagen-tip 100 and used for manual sequencing with the T7Sequencing Kit (Pharmacia Biotech) and [α-35S]dATP.
In vitro transcription of RNAs and the RNase III assays.
In vitro transcriptions using T7 RNA polymerase and purification of the internally labeled transcript on 10% polyacrylamide gel were performed as previously described (5).
Each assay was performed in a 10-μl reaction volume. The cleavage buffer consisted of 30 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 130 or 250 mM KCl, and 5% glycerol. The concentration of RNase III was approximately 3 nM (homodimer), and that of the substrate was 20 or 40 nM. The assays were performed for 3 min (at a 20 nM substrate concentration) or for 5 min (at a 40 nM substrate concentration) at 35°C and stopped by adding 7 μl of formamide-containing dye. Reaction products were denatured at 65°C for 5 min, placed on ice, and analyzed by autoradiography after separation on a 10% polyacrylamide–7 M urea gel. Quantification of the amount of the cleaved transcripts was performed with a Bioimager (Fuji BAS 1000) and TINA software (Raytest).
Mapping of RNA 5′ ends by primer extension.
To determine the exact RNase III cleavage position at the 3′-processing site in helix 9 of 23S rRNA, we used primer extension analysis. The primer for the extension reaction was the 23S-IVS-antisense primer, corresponding to the 23S rDNA positions 185 to 205 (E. coli numbering). Unlabeled in vitro transcripts were incubated with purified R. capsulatus His6-RNase III or E. coli RNase III in cleavage buffer containing 130 mM KCl at 35°C for 15 min. After phenol extraction and ethanol precipitation, the processed RNA substrate was treated as previously described (11). Primer extension reactions were also performed using 2 μg of total RNA to determine the 5′ ends of the large rRNA fragment originating of in vivo processing of helix 9 in the 23S rRNA. Radioactively labeled sequencing reactions of the cloned DNA template were loaded onto the same gel to map the position of the cleavage site for RNase III.
Sequence analysis.
Alignments were performed manually and online using the CLUSTAL W computer program (http://www2.ebi.ac.uk/clustalw/). Putative rRNA secondary structure models were obtained online using the folding program MFOLD (20, 39). For this analysis rDNA sequences obtained in this work, as well as previously published sequences of some other 23S rDNAs taken from the EMBL database, were used.
Accession numbers of nucleotide sequences.
The nucleotide sequences determined in this work were deposited in the EMBL databank under EMBL accession nos. AJ251255 to AJ251267.
RESULTS
Fragmentation in helix 9 of 23S rRNA in some alpha-Proteobacteria.
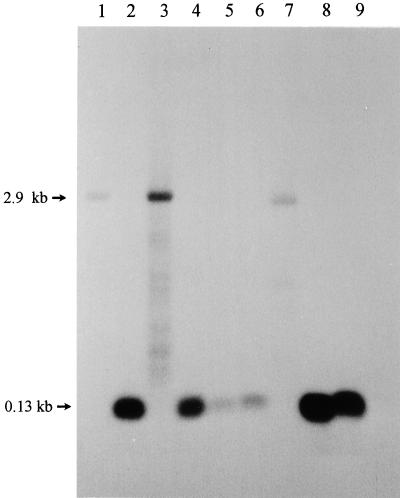
Fragmentation in helix 9 creates a small 5′ segment of 23S rRNA, which corresponds to the first approximately 135 nt of the intact 23S rRNA. This small rRNA migrates in 1.2% agarose formaldehyde gels in one spot, together with the 5S rRNA, and therefore cannot be visualized easily by ethidium bromide staining. Northern hybridization helps to detect this 5′ segment of 23S rRNA (9, 28). Examples of fragmented and intact 23S rRNAs observed in different bacterial strains are shown in Fig. 1. As expected, the radioactive probe complementary to 23S rRNA nt 112 to 130 (E. coli numbering) hybridized to the intact 23S rRNA of E. coli, which is 2.9 kb long (Fig. 1, lane 1). In contrast, the 23S rRNA of R. capsulatus 37b4 is fragmented in helix 9 and the small 5′ segment of the 23S rRNA was detected with this probe (Fig. 1, lane 2). The observed helix 9 fragmentation in R. capsulatus 37b4 was found to be RNase III dependent in vivo. In the RNase III-deficient strain R. capsulatus Fm65, fragmentation is not observed and the probe hybridizes with the unprocessed 2.9-kb 23S rRNA. After complementation of the RNase III-deficient mutant with the plasmid pRK2fm1, which contains the gene encoding RNase IIIRc, the fragmentation of the 23S rRNA was restored to the wild-type R. capsulatus 37b4 pattern (Fig. 1, lanes 2, 3, and 4). The helix 9 fragmentation was observed in all investigated wild-type strains of alpha-Proteobacteria except R. rubrum DSM 107 (Fig. 1, lane 7).
FIG. 1.
Presence of short rRNA corresponding to approximately the first 130 nt of the 23S rRNA in some alpha-Proteobacteria as shown by Northern hybridization of total RNA separated on a 1.2% agarose formaldehyde gel with the radioactively labeled oligonucleotide 23Sup130. Lanes: 1, E. coli JM109; 2, R. capsulatus 37b4; 3, R. capsulatus Fm65; 4, R. capsulatus Fm65 (pRK2fm1); 5, R. sphaeroides 17023; 6, R. palustris 5D; 7, R. rubrum DSM 107; 8, R. gallicum R602; 9, R. giardinii H152.
The lower intensity of the hybridization signals obtained in the lanes containing RNA isolated from E. coli, R. sphaeroides, and R. rubrum (Fig. 1, lanes 1 and 5 to 7) is due to mismatches between the probe and the target sequences.
Primary and proposed secondary structures of helix 9 of 23S rRNA.
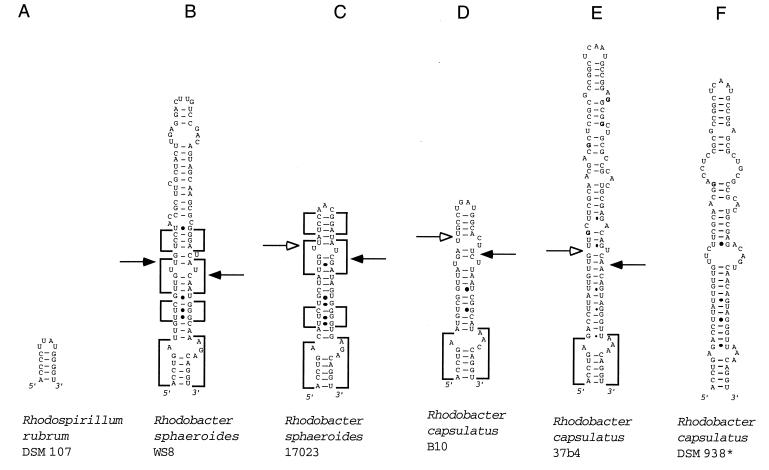
The 23S rDNA region between positions 109 and 205 (E. coli numbering) was amplified and sequenced. IVSs were found in helix 9 of all studied alpha-proteobacterial strains, with the exception of R. rubrum DSM 107. The 23S rRNA sequences were folded with the aid of the MFOLD computer program (20, 39). The proposed secondary structures of the helices 9 are shown in Fig. 2. Previously published helix 9 sequences from some phylogenetically related bacterial strains were also included in the analysis (Fig. 2).
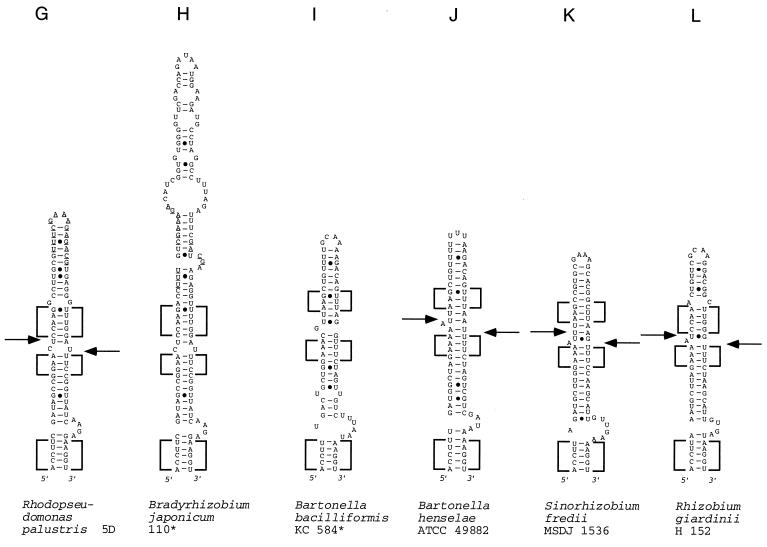
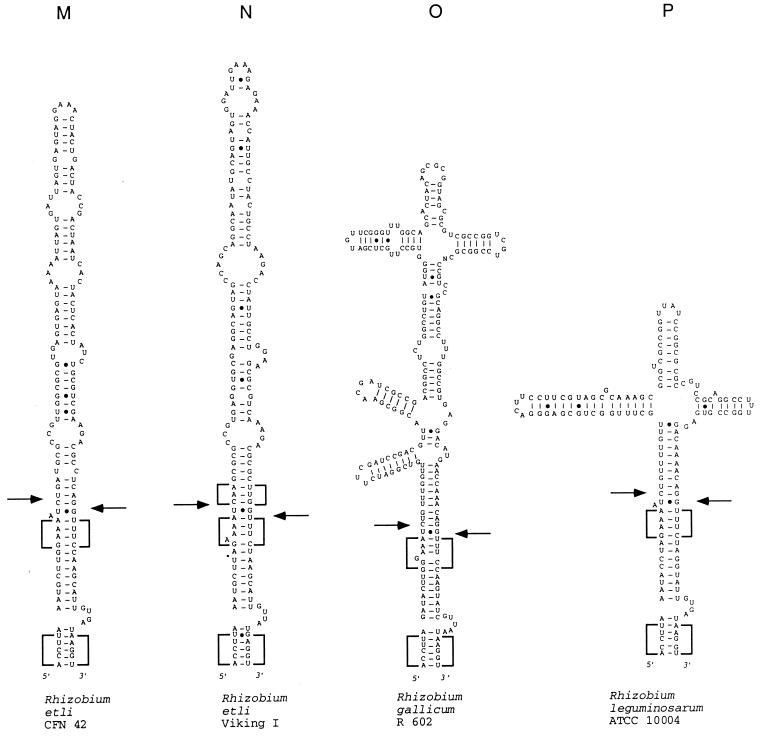
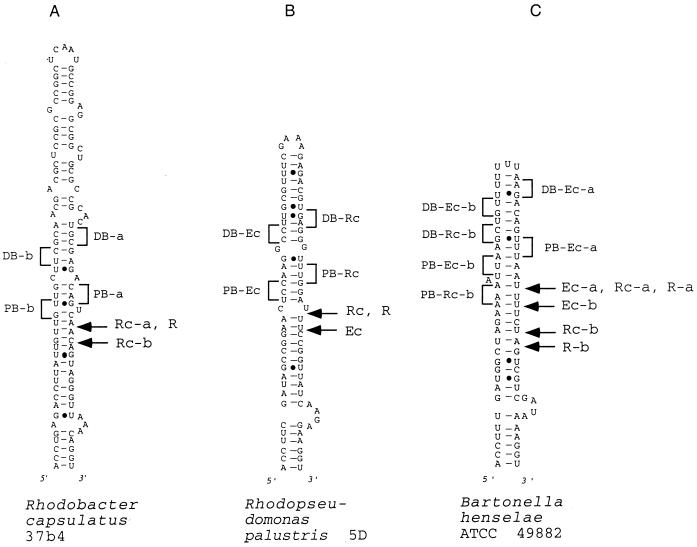
FIG. 2.
Models of potential secondary structure of helix 9 in the 23S rRNA primary transcript of the following strains (asterisks indicate sequences obtained from the data bank, the other sequences were determined in our laboratory, and EMBL accession numbers are in parentheses): A, R. rubrum DSM 107 (AJ251267); B, R. sphaeroides WS8 (AJ251261); C, R. sphaeroides 17023 (AJ251260); D, R. capsulatus B10 (AJ251256); E, R. capsulatus 37b4 (AJ251255); F, R. capsulatus DSM 938* (reference 11); G, R. palustris 5D (AJ251262); H, B. japonicum 110* (reference 17); I, B. bacilliformis KC 584* (reference 22); J, B. henselae ATCC 49882 (AJ251257); K, S. fredii MSDJ 1536 (AJ251258); L, R. giardinii H152 (AJ251263); M, R. etli CFN 42 (AJ251265); R. etli Viking I (AJ251266); O, R. gallicum R602 (AJ251259), R. leguminosarum ATCC 10004 (AJ251264). (B to F) Rhodobacter group of helices. Boxes with highly conservative base pair occupation, specific for this group, are indicated. The differences between the sequences shown in panels E and F are in boldface letters. (G to P) Rhizobium-Bradyrhizobium group of helices. Boxes with highly conservative base pair occupation, specific for this group, are indicated. In panels G and H, sequences of high similarity around the putative deletion and/or insertion site are underlined. Arrows indicate the approximate positions of the RNase III processing sites as determined by RNA fragment length estimation (Table 4). Arrows on the left side of the helices indicate 5′-processing sites; arrows on the right side of the helices indicate 3′-processing sites. Filled arrowheads indicate primary processing sites; empty arrowheads indicate secondary processing sites.
When helix 9 sequences are compared, the first 4 to 6 bp are highly conserved. Possibly, they correspond phylogenetically to the canonical helix 9 of bacteria lacking an IVS in this region. A high degree of conservation can also be observed in the next approximately 25 bp of the helix, which are part of the IVS. The first 30 bp of the helices 9 were aligned, and the percentage of identically occupied positions was calculated (Table 1). Values of 50 to 60% indicate difficulties in aligning the sequences; values of <50% indicate that an alignment of the IVS sequences was impossible. The analyzed sequences can be divided into two groups, which reflect well the phylogenetic relationships between the corresponding bacteria.
TABLE 1.
Percentage of sequence identity found by comparison of the first 30 bp of the helix 9 of 23S rRNA of various bacterial strainsa
| Strain | % Sequence identity of strain:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WS8 | 17023 | B10 | 37b4 | 938 | 5D | 110 | 584 | 49882 | 1536 | H152 | CFN42 | Viking | R602 | 10004 | |
| R. sphaeroides WS8 | 100 | 70 | 51 | 57 | 57 | – | – | – | – | – | – | – | – | – | – |
| R. sphaeroides 17023 | 100 | 54 | 60 | 60 | – | – | – | – | – | – | – | – | – | – | |
| R. capsulatus B10 | 100 | – | – | – | – | – | – | – | – | – | – | – | – | ||
| R. capsulatus 37b4 | 100 | 98 | – | – | – | – | – | – | – | – | – | – | |||
| R. capsulatus 938 | 100 | – | – | – | – | – | – | – | – | – | – | ||||
| R. palustris 5D | 100 | 97 | 60 | 60 | 57 | 59 | – | 51 | – | – | |||||
| B. japonicum 110 | 100 | 60 | 59 | 55 | 60 | – | – | – | 52 | ||||||
| B. bacilliformis 584 | 100 | 81 | 70 | 68 | 55 | 55 | 60 | 65 | |||||||
| B. henselae 49882 | 100 | 71 | 68 | 52 | 54 | 55 | 62 | ||||||||
| S. fredii 1536 | 100 | 79 | 82 | 68 | 68 | 67 | |||||||||
| R. giardinii H152 | 100 | 81 | 79 | 65 | 76 | ||||||||||
| R. etli CFN42 | 100 | 81 | 73 | 76 | |||||||||||
| R. etli Viking | 100 | 62 | 67 | ||||||||||||
| R. gallicum R602 | 100 | 81 | |||||||||||||
| R. leguminosarum 10004 | 100 | ||||||||||||||
WS8, R. sphaeroides WS8; 37b4, R. capsulatus 37b4; 938, R. capsulatus DSM 938; 5D, R. palustris 5D; 110, B. japonicum 110; 584, B. bacilliformis KC 584; 49882, B. henselae ATCC 49882; 1536, S. fredii MSDJ 1536; H152, R. giardinii H152; CFN42, R. etli CFN42; Viking, R. etli Viking I; R602, R. gallicum R602; 10004, R. leguminosarum ATCC 10004. The helices 9 of the strains R. sphaeroides 17023 and R. capsulatus B10 are shorter than 30 bp, and their sequences were compared with first 25 bp of the other helices 9. –, Sequence identity values of <50%; alignment was impossible. Boldface letters indicate sequence identity values of >70% found in representatives of different species.
One of them is the Rhodobacter group (Fig. 2B to F, Table 1). The identically occupied positions in the sequences of the R. sphaeroides strains WS8 and 17023 are concentrated in four blocks (Fig. 2B and C). A few differences were found in the sequences of the R. capsulatus strains 37b4 and DSM 938 (Fig. 2E and F). The sequence of R. capsulatus B10 is highly different. High divergence is also observed when R. sphaeroides and R. capsulatus sequences are compared (Table 1). Thus, high variability is observed in the Rhodobacter group of sequences.
The second group includes sequences from representatives of five bacterial genera, i.e., the Rhizobium-Bradyrhizobium group (Fig. 2G to P; Table 1). Frequently, the same high degrees of conservation (≥70%) were observed between sequences belonging to representatives of the same species compared to those between different species and even genera (see boldfaced values in Table 1). In the secondary structure proposals, we found blocks with very conservative base-pair occupation in nearly identical positions (Fig. 2G to P). We conclude that the investigated helix 9 region of the Rhizobium-Bradyrhizobium group is more conservative than that of the Rhodobacter group.
The apical part of helices 9 is highly variable. Obviously, the IVSs of the Bradyrhizobium and Rhodopseudomonas strains evolved from each other by deletion and/or insertion events. The first 30 bp of helices 9 of both strains exhibit 97% sequence identity (Table 1), but the B. japonicum helices 9 are approximately 50 nt longer than those of the R. palustris strains. The additional nucleotide stretch in the B. japonicum helix 9 is inserted in a region with an almost identical sequence in both the R. palustris and the B. japonicum helices (these sequences are underlined in Fig. 2G and H).
We determined the GC contents of the helix 9 sequences and compared them with each other as well as with the GC contents of already-sequenced rrn operons or 23S rRNA sequences of phylogenetically closely related bacterial strains (Table 2). Generally, the GC content of the helices 9 is lower than that of the overall rRNA sequences. Both values are lower than the GC content of genomes of the Rhodobacter or Rhizobium species, which is >65%. Noteworthy is the large difference between the GC content of the Bartonella helix 9 sequences compared to that of the respective 23S rRNA (Table 2).
TABLE 2.
GC content of helix 9 of 23S rRNA in comparison with the overall GC content of complete rrn operons or 23S rRNA
| Strain | % GC contenta of:
|
|||
|---|---|---|---|---|
| rrn operon-23S rRNA | Helix 9 | First 30 bp of helix 9 | Apical part of helix 9 | |
| R. sphaeroides WS8 | 571 | 50 | 50 | 50 |
| R. sphaeroides 17023 | 571 | 45 | 45 | |
| R. capsulatus B10 | 552 | 45 | 45 | |
| R. capsulatus 37b4 | 552 | 64 | 45 | 83 |
| R. palustris 5D | 533 | 50 | 50 | 50 |
| B. japonicum 110 | 543 | 46 | 44 | 48 |
| B. henselae ATCC 49882 | 484 | 27 | 27 | |
| B. bacilliformis KC 584 | 484 | 35 | 35 | |
| S. fredii MSDJ 1536 | 535 | 42 | 42 | |
| R. giardinii H152 | 535 | 40 | 40 | |
| R. etli CFN42 | 535 | 44 | 45 | 43 |
| R. etli Viking I | 535 | 47 | 45 | 50 |
| R. gallicum R602 | 535 | 55 | 40 | 70 |
| R. leguminosarum ATCC 10004 | 535 | 50 | 30 | 70 |
The values marked in bold letters illustrate the mosaic structure of certain helices 9. Superscript numbers: 1, complete R. sphaeroides rrnA operon (7); 2, 23S rRNA of R. capsulatus (11); 3, 23S rRNA of R. palustris and B. japonicum (34); 4, 23S rRNA of B. bacilliformis (22); 5, complete rrn operon of Agrobacterium vitis (25).
The helices 9 of R. capsulatus 37b4 and R. gallicum R602 have the highest overall GC content (Table 2). Separate analysis of the first 30 bp of these helices revealed that these regions have a GC content of 40 to 45%, which is typical for most of the other helices 9 studied here. In contrast, the GC content of the apical helix 9 regions of these two strains is very high (>70%). This remarkable difference between the GC content of the two helix 9 regions was also found in R. leguminosarum ATCC 10004 (Table 2), suggesting that they are of different origin.
In vitro processing of helix 9 of 23S rRNA by RNase III.
The purified PCR amplificates were used as templates for in vitro transcription. The transcripts were assayed with RNases III from R. capsulatus and E. coli, purified in our laboratory. Both enzymes differ in their preferences for monovalent cations during catalysis. For RNase IIIEc, the standard assay buffer contains 250 mM KCl; for RNase IIIRc, the optimal KCl concentration is 130 mM. Both enzymes exhibit lower activity at higher KCl concentrations. It is known that RNase IIIEc can cleave suboptimal substrates at monovalent cation concentrations of <250 mM (38). On the other hand, at 250 mM KCl RNase IIIRc does not process in vitro its natural in vivo substrate, an IVS in helix 46 of 23S rRNA (5). We performed activity assays for both enzymes under both KCl concentrations.
In each experiment the amount of radioactivity in distinct product bands was determined. The amount of cleaved substrate was calculated, comparing this value with the decrease in uncleaved substrate. The results are shown in Table 3.
TABLE 3.
Percentage of in vitro-cleaved transcripts containing helix 9 of 23S rRNA using E. coli and R. capsulatus RNases III at 130 mM and 250 mM KCla
| Strain (lane nos. in Fig. 3) | % Transcript cleaved in vitro using:
|
|||
|---|---|---|---|---|
| RNase IIIEc with:
|
RNase IIIRc with:
|
|||
| 130 mM KCl | 250 mM KCl | 130 mM KCl | 250 mM KCl | |
| R. sphaeroides WS8 (21–25) | 0 | 0 | 98 | 43 |
| R. sphaeroides 17023 (56–58) | 0 | 0 | 90 | 10 |
| R. capsulatus B10 (46–50) | 0 | 0 | 97 | 12 |
| R. capsulatus 37b4 (6–10) | 0 | 0 | 94 | 14 |
| R. palustris 5D (1–5) | 96 | 75 | 100 | 99 |
| B. henselae ATCC 49882 (11–15) | 97 | 70 | 99 | 96 |
| S. fredii MSDJ 1536 (31–35) | 2 | 1.5 | 100 | 100 |
| R. giardinii H152 (26–30) | 25 | 0 | 100 | 80 |
| R. etli CFN42 (16–20) | 10 | 0 | 99 | 99 |
| R. etli Viking I (41–45) | 80 | 0 | 98 | 99 |
| R. gallicum R602 (36–40) | 60 | 0 | 100 | 86 |
| R. leguminosarum ATCC 10004 (51–55) | 88 | 2 | 99 | 48 |
The reported values are the average of two experiments; the values did not deviate by more than 15%.
All transcripts were almost completely cleaved by RNase IIIRc at 130 mM KCl. Interestingly, at 250 mM KCl the transcripts from the Rhizobium group are much better substrates for RNase IIIRc than the Rhodobacter transcripts.
The transcripts derived from the Rhodobacter strains are not cleaved by RNase IIIEc at both KCl concentrations. All other transcripts can be cleaved by RNase IIIEc at 130 mM KCl (with the exception of the S. fredii MSDJ 1536 transcript, where only 2% is processed), but most of them cannot be used as efficient substrates for this enzyme under its specific assay conditions of 250 mM KCl. Only two exceptions were found: 70 to 75% of the transcripts from B. henselae ATCC 49882 and R. palustris 5D are processed at a high monovalent ion concentration.
Even transcripts containing helix 9 with very similar primary and secondary structures differ markedly in their interaction with RNase IIIEc. For example, the transcripts derived from the strains S. fredii MSDJ 1536 and R. giardinii H152 have 80% identity in their overall helix 9 sequences. Nevertheless, the first transcript is less reactive with RNase IIIEc than the latter transcript (Table 3). Moreover, the transcripts derived from the R. etli strains CFN 42 and Viking I also show 81% sequence identity in the first 30 bp of their helices 9 (Table 1). This is the region recognized and cleaved by RNase III (see below). Despite their very similar sequences, the reactivities of these substrates with RNase IIIEc are markedly different (Table 3).
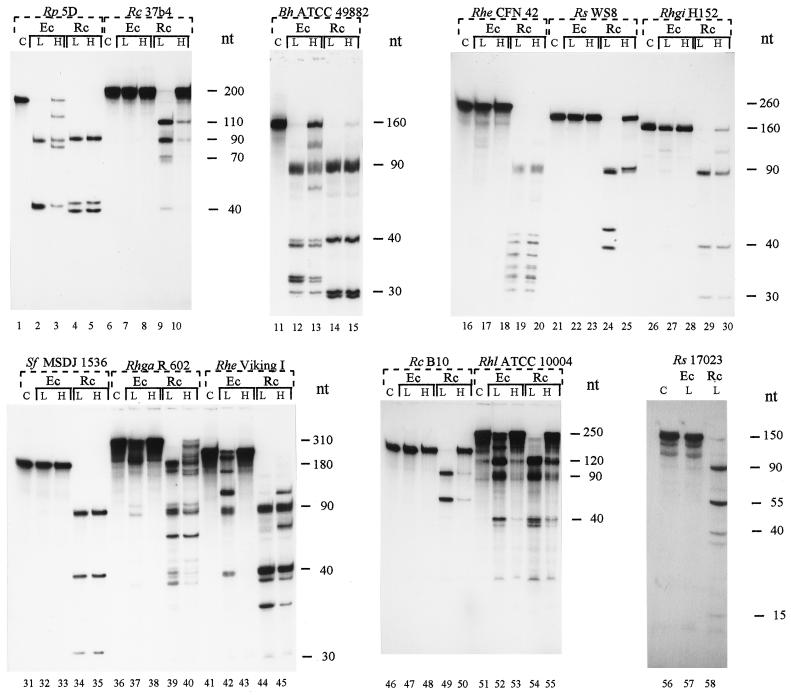
The RNase III cleavage patterns of the different substrates used are shown in Fig. 3. Despite helix 9 variability, they all share bands of similar length because the transcripts also include 22 nt upstream from helix 9 (containing helix 8 sequences) and approximately 70 nt downstream from helix 9 (containing helix 10, helix 11, and downstream primer sequences). Usually, RNase III cleaves in both strands of a duplex, and the 5′- and 3′-processing sites are separated by only 2 bp (24). The scissile bonds in helices 9 studied here are localized after the first 18 bp of the helix (Fig. 2). After complete cleavage at the 5′- and 3′-processing sites, a 5′ fragment of approximately 40 nt, a 3′ fragment of approximately 90 nt, and additional fragments corresponding to internal parts of the processed IVS arise (Fig. 2 and 3). The length of the transcripts and the corresponding fragments are summarized in Table 4.
FIG. 3.
In vitro processing of transcripts containing helix 9 of 23S rRNA by R. capsulatus (Rc) and E. coli (Ec) RNase III at low and high monovalent ion concentrations. L, 130 mM KCl; H, 250 mM KCl; C, uncleaved substrate. Rp, R. palustris; Rc, R. capsulatus; Bh, B. henselae; Rhe, R. etli; Rs, R. sphaeroides; Rhgi, R. giardinii; Sf, S. fredi; Rhga, R. gallicum; Rhl, R. leguminosarum.
TABLE 4.
Summary of the approximate lengths of the helix 9 containing transcripts from the strains studied here and the approximate lengths of the fragments arising after in vitro cleavage of these transcripts by RNases III from R. capsulatus and E. colia
| Transcript source strain (lane nos. in Fig. 3) | Approximate length (no. of nt) of:
|
|||
|---|---|---|---|---|
| Full-length transcript | 5′-End fragment | Internal fragment | 3′-End fragment | |
| R. sphaeroides WS8 (21–25) | 180 | 40 | 50 | 90 |
| R. sphaeroides 17023 (56–58) | 145 | 55 (40) | – (15) | 90 |
| R. capsulatus B10 (46–50) | 145 | 55 (40) | – (15) | 90 |
| R. capsulatus 37b4 (6–10) | 200 | 110 (40) | – (70) | 90 |
| R. palustris 5D (1–5) | 170* | 40 | 40 | 90 |
| B. henselae ATCC 49882 (11–15) | 160* | 40** | 30** | 90** |
| S. fredii MSDJ 1536 (31–35) | 160 | 40 | 30 | 90 |
| R. giardinii H152 (26–30) | 160 | 40 | 30 | 90 |
| R. etli CFN 42 (16–20) | 260* | 40** | 130 (processed in fragments of 30–40 nt) | 90** |
| R. etli Viking I (41–45) | 250* | 40** | 120 (processed in fragments of 30–40 nt) | 90** |
| R. gallicum R602 (36–40) | 310* | 40** | 180 (processed further) | 90** |
| R. leguminosarum ATCC 10004 (51–55) | 250* | 40** | 120** | 90** |
In some of the Rhodobacter strains, the 3′-cleavage site was used as a primary processing site, and only a small amount of the 5′ fragment is further processed at the secondary, 5′-processing site. The length of the resulting 5′ and internal fragments is given in parentheses. ∗, differences in the cleavage sites of both enzymes; ∗∗, multiple fragments which differ in their length by a few nucleotides due to cleavage of different scissile bonds at the 5′- and/or 3′-processing sites.
In R. capsulatus 37b4 (Fig. 3, lanes 6 to 10) and B10 (Fig. 3, lanes 46 to 50) transcripts and in the R. sphaeroides 17023 (Fig. 3, lanes 56 to 58) transcript the 3′-processing site is the primary cleavage site used by RNase IIIRc, and only a small amount of these substrates is also cleaved at the 5′-processing site (indicated also in Fig. 2 and Table 4). In the R. sphaeroides WS8 (Fig. 3, lanes 21 to 25) transcript, as well as in all other transcripts used here, both sites are processed by RNase IIIRc at 130 mM KCl (Fig. 2 and 3; Table 4).
Both RNases III create different cleavage patterns when they process some of the transcripts (marked by asterisks in Table 4; compare with Fig. 3). This may be due to the processing of different scissile bonds. Apparently, multiple scissile bonds are cleaved at the processing sites in most of these transcripts. In their cleavage patterns multiple 5′-end, 3′-end, and internal fragments occur which differ in their lengths by a few nucleotides (marked by two asterisks in Table 4; compare with Fig. 3). A typical example for these phenomena is the processing pattern of the B. henselae ATCC 49882 transcript (Fig. 3, lanes 11 to 15). We performed primer extension to analyze the 3′-processing sites of this transcript in vitro. In addition, we determined the exact in vitro and in vivo RNase III 3′-processing sites in helix 9 of the strains R. capsulatus 37b4 and R. palustris 5D.
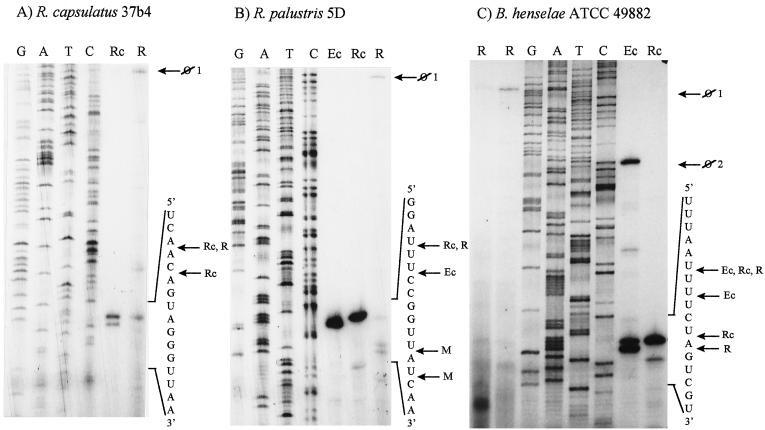
Primer extension determines the exact RNase III 3′ cleavage sites.
The results of primer extension analyses are shown in Fig. 4 and are schematically summarized in Fig. 5.
FIG. 4.
Primer extension analysis determines the rRNA 5′ ends obtained during in vitro cleavage of the transcripts from R. capsulatus 37b4 (A), R. palustris 5D (B), and B. henselae ATCC 49882 (C) with R. capsulatus (lanes Rc) and E. coli (lanes Ec) RNases III at the 3′-processing site in helix 9. The corresponding 5′ ends in vivo were detected by primer extension analysis using total RNA (lanes R) isolated from these strains. Lanes G, A, T, and C each refer to the corresponding nucleotide of the DNA template (cloned 23S rDNA region), as determined by sequencing. Parts of the in vitro transcripts and of the pre-rRNA sequences are indicated on the right side of each panel. The detected 5′ ends are marked by labeled arrows as follows: ⊘1, 5′ end of the 23S rRNA unprocessed in helix 9; ⊘2, 5′ end of the unprocessed in vitro transcript; Ec, 5′ end after RNase IIIEc cleavage in vitro; Rc, 5′ end after RNase IIIRc cleavage in vitro; R, 5′ end corresponding to the RNase III processing site in helix 9, detected in vivo; M, 5′ end arising from further maturation of rRNA in vivo.
FIG. 5.
Schematic representation of the RNase III 3′-processing sites in vitro and the in vivo 5′ ends found in helix 9 of 23S rRNA with primer extension analysis shown in Fig. 4. (A) R. capsulatus 37b. (B) R. palustris 5D. (C) B. henselae ATCC 49882. Labeled arrows: Ec, 5′ end after RNase IIIEc cleavage in vitro; Rc, 5′ end after RNase IIIRc cleavage in vitro; R, 5′ end corresponding to the RNase III processing site in helix 9, detected in vivo; PB and DB, proximal and distal boxes, respectively, found in the vicinity of the corresponding scissile bonds (compare with references 24 and 38).
At the 3′-processing site in helix 9 of the R. capsulatus 37b4 transcript two scissile bonds were cleaved by RNase IIIRc in vitro, but only the upstream one was detected in vivo (Fig. 4A and Fig. 5A). The primer extension analysis also confirmed the existence of multiple scissile bonds in the B. henselae ATCC 49882 transcript.
In the R. palustris 5D and B. henselae ATCC transcripts, both RNases III can cleave different scissile bonds in vitro. In vivo, cleavage sites identical to the RNase IIIRc processing sites were detected, probably because the endogenous RNases III of these bacteria are more similar to the RNase IIIRc than to RNase IIIEc (Fig. 4B and C and Fig. 5B and C).
The intensity of the signals which represent 5′ ends created by RNase III cleavage in vivo is similar to the intensity of the signals corresponding to the unprocessed 23S rRNA (Fig. 4). Additional 5′ ends downstream of the in vivo processed helix 9 of R. palustris 5D were detected (Fig. 4B), probably arising due to further maturation of the 5′ end of the large rRNA fragment. Much stronger signals, different from those corresponding to the RNase III cleavage sites, were obtained by primer extension analysis of total RNA of all three strains. They correspond to structural obstacles for cDNA synthesis at the GC-rich stem of the helix 11, as well as to 5′ ends at the stem of helix 10 (data not shown; see also reference 37). They were not detectable in primer extension reactions when in vitro assays were used.
DISCUSSION
We found that in Rhodobacter, Rhodopseudomonas, Rhizobium, Sinorhizobium, and Bartonella strains 23S rRNA is fragmented near position 130 (E. coli numbering, helix 9) due to RNase III-dependent processing of IVSs. These IVSs are present in all operons and in all strains of a species (this work and references 28 and 29). They do not occur sporadically only, in contrast to all other IVSs found in other rRNA regions until now (4, 9, 12, 23). This suggests that these IVSs are of ancient origin and already occur in the last common ancestor of modern alpha-Proteobacteria. The ubiquitous distribution of IVSs in the helices 9 of 23S rRNA of certain alpha-proteobacterial species probably reflects an evolutionary pressure for fragmentation in this region. It was recently reported that the 5′ end at the base of the helix 10 stem in R. palustris represents the real 5′ end of the mature large rRNA segment. The excision of the IVS from helix 9 is followed by complete removal of the helix 9 and 10 sequences from the 23S pre-rRNA (37). The authors of that study also discuss the consequences of such extensive processing for the ribosome structure (37). We were able to detect the in vivo 3′-processing sites of RNase III in helices 9 of three different bacterial species by primer extension analysis (Fig. 4). Comparison of the intensity of the obtained signals leads to the conclusion that the amount of the large rRNA segments with a 5′ end corresponding to the RNase III cleavage site is comparable to the amount of the unprocessed 23S pre-rRNA (Fig. 4). In contrast, much stronger signals were obtained which correspond to downstream 5′ ends at the stem of helix 10 (not shown). These results are in accordance with observations of others (37) and suggest that the ubiquitous distribution of IVS in helix 9 of certain bacterial species may reflect the need for an initial processing signal leading to further downstream 23S rRNA processing necessary to create functionally optimal ribosomes.
The obviously different primary structures of many of the studied IVSs, the fact that RNase IIIRc processes the Rhodobacter transcripts less efficiently than those derived from the Rhizobium-Bradyrhizobium group (Table 3), and the differences between the GC content of the IVSs and overall 23S rRNA sequences (Table 2) can be explained by lateral genetic transfer events. On the other hand, AU-rich duplexes are preferred substrates for RNase III (24).
It is interesting that the proximal 30 bp of helix 9 in representatives of five different genera are very similar (Table 1). We showed that the RNase III cleavage sites are positioned approximately in the middle of this region (Fig. 2 and 5). The RNase III binding sites are located in the vicinity of the cleavage sites (24). Obviously, these facts contribute to the relative sequence conservation of this part of helix 9. The slower divergence of these sequences may also reflect stronger constraints in the recognition of the cleavage sites by the Rhizobium-type RNases III in comparison to the Rhodobacter-type RNases III.
The mosaic structure of the IVS found in the helices 9 of the strains R. capsulatus 37b4, R. gallicum R602, and R. leguminosarum ATCC 10004 (Table 2), together with the above-described evolution of the B. japonicum and R. palustris helix 9 sequences from each other and the presence of blocks with very conservative base pair occupation, show that IVSs often evolve via insertion and/or deletion events.
The helix 9 secondary structures shown in Fig. 2 are based only on computer-assisted folding (MFOLD [20, 39]). When only the sequences including the 23S rRNA helices 8, 9, and 10 were folded, we always obtained correct secondary structures for helices 8 and 10 compared with the universal secondary structure models for the 23S rRNAs of E. coli, R. capsulatus, R. sphaeroides, and R. palustris (23S rRNA Comparative Structure Database, http://www.rna.icmb.utexas.edu/). Alternative secondary structures for the relatively short helices 9 shown in Fig. 2A to L were not obtained. The differences in the alternative structures obtained for the very long rhizobial helices 9 (Fig. 2M to P) were localized apical to their first 20 to 30 bp, these first 20 to 30 bp remaining invariable. For the overall length of the transcripts, we obtained many different secondary structures, but the differences were localized mainly outside of helix 9. Only the first 6 to 8 bp of the short helices 9 presented in Fig. 2B to L were involved in alternative structures. The 20-bp region, which harbors the RNase III cleavage sites, remained invariable. We therefore suggest that our secondary structure models can be used for analysis of enzyme-substrate recognition.
Based on the secondary-structure proposals, we analyzed whether our RNase III substrates, shown in Fig. 5, fit into the pattern proposed by the “antideterminant” model (24, 38). This model explains recognition and cleavage of perfect double-helical substrates by RNase IIIEc in vitro. According to this model the positions −4 to −6 relative to the scissile bond must not have the sequence G(G,C)G (proximal box) and the positions −11 and −12 must not have the sequence UC (distal box). The presence of only one antideterminant base at positions −4 or −5 in the proximal box reduces the cleavage of the substrate by the RNase IIIEc to 7 to 15%; the presence of only two antideterminant bases at both positions reduces it to 3%. The use of lower salt concentrations in the reaction mix (<160 mM KCl) can promote cleavage of less-reactive substrates (38).
In the case of the R. capsulatus 37b4 transcript, the putative proximal boxes (determined in relation to both RNase IIIRc cleavage sites) are involved in, or are positioned immediately next to, predicted helix distortions (Fig. 5A). The irregular double-helical structure of this transcript could explain why it is not processed by RNase IIIEc even at 130 mM KCl. It is more difficult to understand the ability of the RNase IIIEc to cleave the R. palustris 5D transcript at 250 mM KCl, despite the presence of two strong antideterminant bases (GG at positions −5 and −6 relative to the scissile bond) in the proximal box (Fig. 5B). Using the antideterminant model, it is also not possible to explain why RNase IIIEc cannot use the RNase IIIRc unique processing site in the B. henselae ATCC 49882 transcript (Fig. 5C).
The antideterminant model may be limited to a subset of all possible RNase IIIEc-specific substrates. In addition to the presence or absence of certain nucleotides near the cleavage site, the overall secondary and tertiary structure of this substrate may be responsible for recognition by the enzyme. It is possible that transcripts with alternative structures were present in our in vitro assays, which differ in their cleavage sites and efficiency of the cleavage. This could explain the detection of multiple scissile bonds at the RNase III processing sites in some of the studied transcripts (Table 3). We also cannot exclude the possibility of sequence differences between the various rrn operons of a bacterial strain. The observation that RNase III cleaves an additional site in vitro in R. capsulatus 37b4 transcript (Fig. 4A) could be explained by the existence of transcripts with alternative secondary structures in the assay, as well as by differences in RNase III specificities in vitro and in vivo.
We describe here important differences in the substrate specificities and cleavage sites of the RNases III from R. capsulatus and E. coli. Nothing is known about the way RNase IIIRc recognizes its substrates. Until now, only one R. capsulatus RNase III-specific substrate was available (5). Our set of natural substrates provides new perspectives for studying this enzyme. The exact determination of RNase IIIRc cleavage sites in many different substrates should at least allow us to find the position-exclusion of base pairs near the processing site for this enzyme. This should make possible the construction of an uncleavable IVS which can be used to replace wild-type IVSs in a model alpha-proteobacterial strain. The availability of such mutant strains with intact 23S rRNA and otherwise-unchanged genetic backgrounds will for the first time allow study of the consequences of the 23S rRNA fragmentation in bacteria.
ACKNOWLEDGMENTS
We thank C. Conrad for providing purified RNases III and many buffers. We are grateful to N. Amarger (INRA-CMSE, Dijon, France), and D. K. Jones (USDA/ARS Beltsville Rhizobium Germplasm Collection), for sending us bacterial strains. We thank O. Fuhrmann (Charite, Berlin) for providing DNA from B. henselae ATCC 49882 and R. Rauhut for reading the manuscript and for help with the computing programs.
This work was supported by Deutsche Forschungsgemeinschaft (Kl 563/11-1) and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Amarger N, Macheret V, Laguerre G. Rhizobium gallicum sp. nov. and Rhizobium giardinii sp. nov., from Phaseolus vulgaris nodules. Int J Syst Bacteriol. 1997;47:996–1006. doi: 10.1099/00207713-47-4-996. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. I. New York, N.Y: Greene Publishing Associates and Wiley-Interscience; 1989. [Google Scholar]
- 3.Beringer J E. R-factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 4.Burgin A B, Parodos K, Lane D J, Pace N R. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell. 1990;60:405–414. doi: 10.1016/0092-8674(90)90592-3. [DOI] [PubMed] [Google Scholar]
- 5.Conrad C, Rauhut R, Klug G. Different cleavage specificities of RNases III from Rhodobacter capsulatus and Escherichia coli. Nucleic Acids Res. 1998;26:4446–4453. doi: 10.1093/nar/26.19.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drews G. Mikrobiologisches Praktikum. 3rd ed. Berlin, Germany: Springer-Verlag; 1976. [Google Scholar]
- 7.Dryden S C, Kaplan S. Localization and structural analysis of the ribosomal RNA operons of Rhodobacter sphaeroides. Nucleic Acids Res. 1990;18:7267–7277. doi: 10.1093/nar/18.24.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn J J. Ribonuclease III. In: Boyer P D, editor. The enzymes, vol. XV. Nucleic acids, part B. New York, N.Y: Academic Press, Inc.; 1982. pp. 485–499. [Google Scholar]
- 9.Evguenieva-Hackenberg E, Selenska-Pobell S. Variability of the 5′-end of the large subunit rDNA and the presence of a new short class of rRNA in Rhizobiaceae. Lett Appl Microbiol. 1995;21:402–405. doi: 10.1111/j.1472-765x.1995.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 10.Heck C, Rothfuchs R, Jäger A, Rauhut R, Klug G. Effect of the pufQ-pufB intercistronic region on puf mRNA stability in Rhodobacter capsulatus. Mol Microbiol. 1996;20:1165–1178. doi: 10.1111/j.1365-2958.1996.tb02637.x. [DOI] [PubMed] [Google Scholar]
- 11.Höpfl P, Ludwig W, Schleifer K H. Complete nucleotide sequence of a 23S ribosomal RNA gene from Rhodobacter capsulatus. Nucleic Acids Res. 1988;16:2343. doi: 10.1093/nar/16.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu D, Zee Y Z, Ingreham J, Shih L-M. Diversity of cleavage patterns of Salmonella 23S rRNA. J Gen Microbiol. 1992;138:199–203. doi: 10.1099/00221287-138-1-199. [DOI] [PubMed] [Google Scholar]
- 13.Hurtado A, Clewley J P, Linton D, Owen R J, Stanley J. Sequence similarities between large subunit ribosomal RNA gene intervening sequences from different Helicobacter species. Gene. 1997;194:69–75. doi: 10.1016/s0378-1119(97)00158-3. [DOI] [PubMed] [Google Scholar]
- 14.Jemiolo D K. Processing of procaryotic ribosomal RNA. In: Zimmermann R, Dahlberg A, editors. Ribosomal RNA. Boca Raton, Fla: CRC Press; 1996. pp. 453–468. [Google Scholar]
- 15.Klug G, Drews G. Construction of a gene bank of Rhodopseudomonas capsulata using a broad host range cloning system. Arch Microbiol. 1984;139:319–325. doi: 10.1007/BF00408373. [DOI] [PubMed] [Google Scholar]
- 16.Kordes E, Jock S, Fritsch J, Bosch F, Klug G. Cloning of a gene involved in rRNA precursor processing and 23S rRNA cleavage in Rhodobacter capsulatus. J Bacteriol. 1994;176:1121–1127. doi: 10.1128/jb.176.4.1121-1127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kundig C, Beck C, Hennecke H, Gottfert M. A single rRNA gene region in Bradyrhizobium japonicum. J Bacteriol. 1995;177:5151–5154. doi: 10.1128/jb.177.17.5151-5154.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marrs B. Mobilization of the genes for photosynthesis from Rhodopseudomonas capsulata by a promiscuous plasmid. J Bacteriol. 1981;146:1003–1012. doi: 10.1128/jb.146.3.1003-1012.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marrs B L, Kaplan S. 23S precursor ribosomal RNA of Rhodopseudomonas sphaeroides. J Mol Biol. 1970;49:297–317. doi: 10.1016/0022-2836(70)90247-0. [DOI] [PubMed] [Google Scholar]
- 20.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 21.Mattatall N R, Sanderson K E. Salmonella typhimurium LT2 possesses three distinct 23S rRNA intervening sequences. J Bacteriol. 1996;178:2272–2278. doi: 10.1128/jb.178.8.2272-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minnick M F, Mitchell S J, McAllister S J, Battisti J M. Nucleotide sequence analysis of the 23S ribosomal RNA-encoding gene of Bartonella bacilliformis. Gene. 1995;162:75–79. doi: 10.1016/0378-1119(95)00362-a. [DOI] [PubMed] [Google Scholar]
- 23.Miller W L, Pabbaraju K, Sanderson K. Fragmentation of 23S rRNA in strains of Proteus and Providencia results from intervening sequences in the rrn (rRNA) genes. J Bacteriol. 2000;182:1109–1117. doi: 10.1128/jb.182.4.1109-1117.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolson A W. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol Rev. 1999;23:371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 25.Otten L, De Ruffray P, de Lajudie P, Michot B. Sequence and characterization of a ribosomal RNA operon from Agrobacterium vitis. Mol Gen Genet. 1996;251:99–107. doi: 10.1007/BF02174350. [DOI] [PubMed] [Google Scholar]
- 26.Ralph D, McClelland M. Intervening sequence with conserved open reading frame in eubacterial 23S rRNA genes. Proc Natl Acad Sci USA. 1993;90:6864–6868. doi: 10.1073/pnas.90.14.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rauhut R, Jäger A, Conrad C, Klug G. Identification and analysis of the rnc gene for RNase III in Rhodobacter capsulatus. Nucleic Acids Res. 1996;24:1246–1251. doi: 10.1093/nar/24.7.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selenska-Pobell S, Evguenieva-Hackenberg E. Fragmentations of the large-subunit rRNA in the family Rhizobiaceae. J Bacteriol. 1995;177:6993–6998. doi: 10.1128/jb.177.23.6993-6998.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selenska-Pobell S, Döring H, Evguenieva-Hackenberg E. Unusual organization of the 23S rRNA genes in Rhizobiaceae. Soil Biol Biochem. 1996;29:905–909. [Google Scholar]
- 30.Selenska-Pobell S, Döring H. Sequences around the fragmentation sites of the large subunit ribosomal RRNA in the family Rhizobiaceae. Antonie Leeuwenhoek. 1998;73:55–67. doi: 10.1023/a:1000540023194. [DOI] [PubMed] [Google Scholar]
- 31.Skurnik M, Toivanen P. Intervening sequences (IVSs) in the 23S ribosomal RNA genes of pathogenic Yersinia enterocolitica strains. The IVSs in Y. enterocolitica and Salmonella typhimurium have a common origin. Mol Microbiol. 1991;5:585–593. doi: 10.1111/j.1365-2958.1991.tb00729.x. [DOI] [PubMed] [Google Scholar]
- 32.Sistrom W R. Transfer of chromosomal genes mediated by plasmid R68. J Bacteriol. 1977;131:526–532. doi: 10.1128/jb.131.2.526-532.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song X M, Forsgren A, Janson H. Fragmentation heterogeneity of 23S ribosomal RNA in Haemophilus species. Gene. 1999;230:287–293. doi: 10.1016/s0378-1119(99)00063-3. [DOI] [PubMed] [Google Scholar]
- 34.Springer N, Ludwig W, Hardarson G. A 23S rRNA targeted specific hybridization probe for Bradyrhizobium japonicum. Syst Appl Microbiol. 1993;16:468–470. [Google Scholar]
- 35.Trust T J, Logan S M, Gustafson C E, Romaniuk P J, Kim N W, Chan V L, Ragan M A, Guerry P, Gutell R R. Phylogenetic and molecular characterization of a 23S rRNA gene positions the genus Campylobacter in the epsilon subdivision of the Proteobacteria and shows that the presence of transcribed spacers is common in Campylobacter spp. J Bacteriol. 1994;176:4597–4609. doi: 10.1128/jb.176.15.4597-4609.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Gabain A, Belasco J G, Schottel J L, Chang A C Y, Cohen S N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci USA. 1983;80:653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zahn K, Inui M, Yukawa H. Characterization of a separate small domain derived from the 5′ end of 23S rRNA of an α-proteobacterium. Nucleic Acids Res. 1999;27:4241–4250. doi: 10.1093/nar/27.21.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang K, Nicolson A. Regulation of ribonuclease III processing by double-helical sequence antideterminants. Proc Natl Acad Sci USA. 1997;94:13437–13441. doi: 10.1073/pnas.94.25.13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In: Barciszewski J, Clark B F C, editors. RNA biochemistry and biotechnology. Dordrecht, The Netherlands: NATO ASI series. Kluwer Academic Publishers; 1999. pp. 11–43. [Google Scholar]