Abstract
Medical data often exhibits long-tail distributions with heavy class imbalance, which naturally leads to difficulty in classifying the minority classes (i.e., boundary regions or rare objects). Recent work has significantly improved semi-supervised medical image segmentation in long-tailed scenarios by equipping them with unsupervised contrastive criteria. However, it remains unclear how well they will perform in the labeled portion of data where class distribution is also highly imbalanced. In this work, we present ACTION++, an improved contrastive learning framework with adaptive anatomical contrast for semi-supervised medical segmentation. Specifically, we propose an adaptive supervised contrastive loss, where we first compute the optimal locations of class centers uniformly distributed on the embedding space (i.e., off-line), and then perform online contrastive matching training by encouraging different class features to adaptively match these distinct and uniformly distributed class centers. Moreover, we argue that blindly adopting a constant temperature in the contrastive loss on long-tailed medical data is not optimal, and propose to use a dynamic via a simple cosine schedule to yield better separation between majority and minority classes. Empirically, we evaluate ACTION++ on ACDC and LA benchmarks and show that it achieves state-of-the-art across two semi-supervised settings. Theoretically, we analyze the performance of adaptive anatomical contrast and confirm its superiority in label efficiency.
Keywords: Semi-Supervised Learning, Contrastive Learning, Imbalanced Learning, Long-tailed Medical Image Segmentation
1. Introduction
With the recent development of semi-supervised learning (SSL) [3], rapid progress has been made in medical image segmentation, which typically learns rich anatomical representations from few labeled data and the vast amount of unlabeled data. Existing SSL approaches can be generally categorized into adversarial training [32,39,16,38], deep co-training [23,43], mean teacher schemes [27,42,14,13,15,7,41,34], multi-task learning [19,11,22,37,35], and contrastive learning [2,29,40,33,24,36].
Contrastive learning (CL) has become a remarkable approach to enhance semi-supervised medical image segmentation performance without significantly increasing the amount of parameters and annotation costs [2,29,36]. In real-world clinical scenarios, since the classes in medical images follow the Zipfian distribution [44], the medical datasets usually show a long-tailed, even heavy-tailed class distribution, i.e., some minority (tail) classes involving significantly fewer pixel-level training instances than other majority (head) classes, as illustrated in Figure 1. Such imbalanced scenarios are usually very challenging for CL methods to address, leading to noticeable performance drop [18].
Fig. 1.
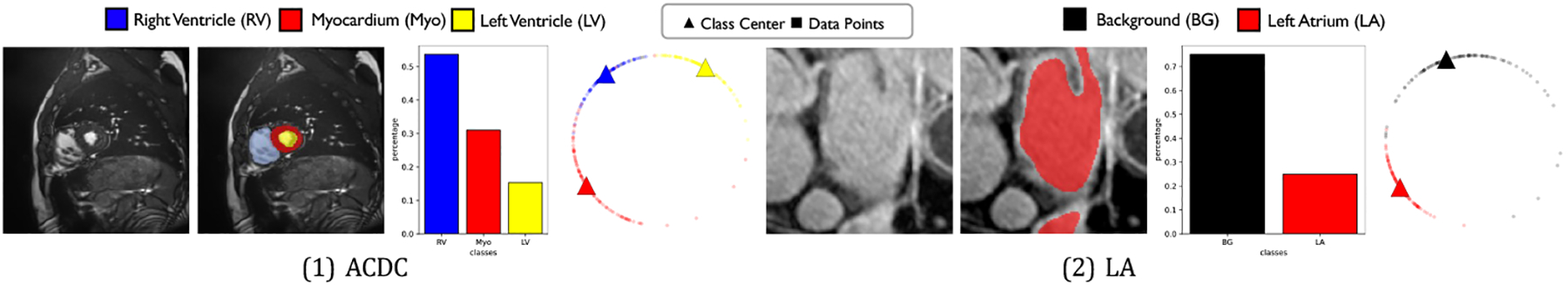
Examples of two benchmarks (i.e., ACDC and LA) with imbalanced class distribution. From left to right: input image, ground-truth segmentation map, class distribution chart, training data feature distribution for multiple classes.
To address long-tail medical segmentation, our motivations come from the following two perspectives in CL training schemes [2,36]: ➊ Training objective – the main focus of existing approaches is on designing proper unsupervised contrastive loss in learning high-quality representations for long-tail medical segmentation. While extensively explored in the unlabeled portion of long-tail medical data, supervised CL has rarely been studied from empirical and theoretical perspectives, which will be one of the focuses in this work; ➋ Temperature scheduler – the temperature parameter , which controls the strength of attraction and repulsion forces in the contrastive loss [5,4], has been shown to play a crucial role in learning useful representations. It is affirmed that a large emphasizes anatomically meaningful group-wise patterns by group-level discrimination, whereas a small ensures a higher degree of pixel-level (instance) discrimination [28,25]. On the other hand, as shown in [25], group-wise discrimination often results in reduced model’s instance discrimination capabilities, where the model will be biased to “easy” features instead of “hard” features. It is thus unfavorable for long-tailed medical segmentation to blindly treat as a constant hyperparameter, and a dynamic temperature parameter for CL is worth investigating.
In this paper, we introduce ACTION++, which further optimizes anatomically group-level and pixel-level representations for better head and tail class separations, on both labeled and unlabeled medical data. Specifically, we devise two strategies to improve overall segmentation quality by focusing on the two aforementioned perspectives: (1) we propose supervised adaptive anatomical contrastive learning (SAACL) for long-tail medical segmentation. To prevent the feature space from being biased toward the dominant head class, we first pre-compute the optimal locations of class centers uniformly distributed on the embedding space (i.e., off-line), and then perform online contrastive matching training by encouraging different class features to adaptively match these distinct and uniformly distributed class centers; (2) we find that blindly adopting the constant temperature in the contrastive loss can negatively impact the segmentation performance. Inspired by an average distance maximization perspective, we leverage a dynamic via a simple cosine schedule, resulting in significant improvements in the learned representations. Both of these enable the model to learn a balanced feature space that has similar separability for both the majority (head) and minority (tail) classes, leading to better generalization in long-tail medical data. We evaluated our ACTION++ on the public ACDC and LA datasets [1,31]. Extensive experimental results show that our ACTION++ outperforms prior methods by a significant margin and sets the new state-of-the-art across two semi-supervised settings. We also theoretically show the superiority of our method in label efficiency (Appendix A). Code is released at here.
2. Method
2.1. Overview
Problem Statement
Given a medical image dataset , our goal is to train a segmentation model that can provide accurate predictions that assign each pixel to their corresponding -class segmentation labels.
Setup
Figure 2 illustrates an overview of ACTION++. By default, we build this work upon ACTION pipeline [36], the state-of-the-art CL framework for semi-supervised medical image segmentation. The backbone model adopts the student-teacher framework that shares the same architecture, and the parameters of the teacher are the exponential moving average of the student’s parameters. Hereinafter, we adopt their model as our backbone and briefly summarize its major components: (1) global contrastive distillation pre-training; (2) local contrastive distillation pre-training; and (3) anatomical contrast fine-tuning.
Fig. 2.
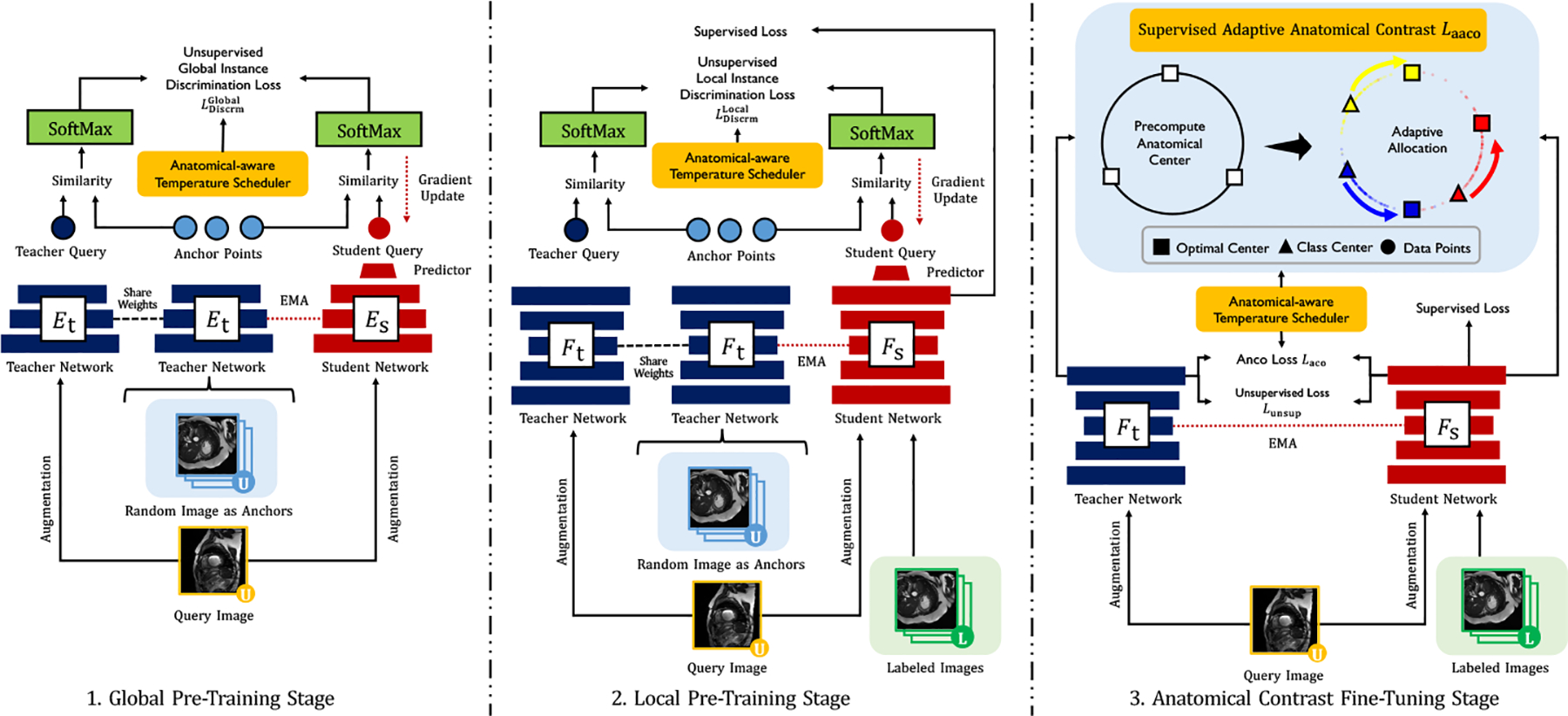
Overview of ACTION++: (1) global and local pre-training with proposed anatomical-aware temperature scheduler, (2) our proposed adaptive anatomical contrast fine-tuning, which first pre-computes the optimal locations of class centers uniformly distributed on the embedding space (i.e., off-line), and then performs online contrastive matching training by encouraging different class features to adaptively match these distinct and uniformly distributed class centers with respect to anatomical features.
Global and Local Pre-training
[36] first creates two types of anatomical views as follows: (1) augmented views - and are augmented from the unlabeled input scan with two separate data augmentation operators; (2) mined views - samples (i.e., ) are randomly sampled from the unlabeled portion with additional augmentation. The pairs are then processed by student-teacher networks that share the same architecture and weight, and similarly, is encoded by . Their global latent features after the encoder (i.e., ) and local output features after decoder (i.e., ) are encoded by the two-layer nonlinear projectors, generating global and local embeddings and from are separately encoded by the non-linear predictor, producing in both global and local manners1. Third, the relational similarities between augmented and mined views are processed by SoftMax function as follows: , where and are two temperature parameters. Finally, we minimize the unsupervised instance discrimination loss (i.e., Kullback-Leibler divergence ) as:
| (1) |
We formally summarize the pretraining objective as the equal combination of the global and local , and supervised segmentation loss (i.e., equal combination of Dice loss and cross-entropy loss).
Anatomical Contrast Fine-tuning
The underlying motivation for the fine-tuning stage is that it reduces the vulnerability of the pre-trained model to long-tailed unlabeled data. To mitigate the problem, [36] proposed to fine-tune the model by anatomical contrast. First, the additional representation head is used to provide dense representations with the same size as the input scans. Then, [36] explore pulling queries to be similar to the positive keys , and push apart the negative keys . The AnCo loss is defined as follows:
| (2) |
where denotes a set of all available classes in the current mini-batch, and is a temperature hyperparameter. For class , we select a query representation set , a negative key representation set whose labels are not in class , and the positive key which is the -class mean representation. Given is a set including all pixel coordinates with the same size as , these queries and keys can be defined as: . We formally summarize the fine-tuning objective as the equal combination of unsupervised , unsupervised cross-entropy loss , and supervised segmentation loss . For more details, we refer the reader to [36].
2.2. Supervised Adaptive Anatomical Contrastive Learning
The general efficacy of anatomical contrast on long-tail unlabeled data has previously been demonstrated by the authors of [36]. However, taking a closer look, we observe that the well-trained shows a downward trend in performance, which often fails to classify tail classes on labeled data, especially when the data shows long-tailed class distributions. This indicates that such well-trained is required to improve the segmentation capabilities in long-tailed labeled data. To this end, inspired by [17] tailored for the image classification tasks, we introduce supervised adaptive anatomical contrastive learning (SAACL), a training framework for generating well-separated and uniformly distributed latent feature representations for both the head and tail classes. It consists of three main steps, which we describe in the following.
Anatomical Center Pre-computation
We first pre-compute the anatomical class centers in latent representation space. The optimal class centers are chosen as positions from the unit sphere in the -dimensional space. To encourage good separability and uniformity, we compute the class centers by minimizing the following uniformity loss :
| (3) |
In our implementation, we use gradient descent to search for the optimal class centers constrained to the unit sphere , which are denoted by . Furthermore, the latent dimension is a hyper-parameter, which we set such that to ensure the solution found by gradient descent indeed maximizes the minimum distance between any two class centers [6]. It is also known that any analytical minimizers of Eqn. 3 form a perfectly regular -vertex inscribed simplex of the sphere [6]. We emphasize that this first step of pre-computation of class centers is completely off-line as it does not require any training data.
Adaptive Allocation
As the second step, we explore adaptively allocating these centers among classes. This is a combinatorial optimization problem and an exhaustive search of all choices would be computationally prohibited. Therefore, we draw intuition from the empirical mean in the K-means algorithm and adopt an adaptive allocation scheme to iteratively search for the optimal allocation during training. Specifically, consider a batch where denotes a set of samples in a batch with class label , for . Define be the empirical mean of class in current batch, where is the feature embedding of sample . We compute assignment by minimizing the distance between pre-computed class centers and the empirical means:
| (4) |
In implementation, the empirical mean is updated using moving average. That is, for iteration , we first compute the empirical mean for batch as described above, and then update by .
Adaptive Anatomical Contrast
Finally, the allocated class centers are well-separated and should maintain the semantic relation between classes. To utilize these optimal class centers, we want to induce the feature representation of samples from each class to cluster around the corresponding pre-computed class center. To this end, we adopt a supervised contrastive loss for the label portion of the data. Specifically, given a batch of pixel-feature-label tuples where is the i-th pixel in the batch, is the feature of the pixel and is its label, we define supervised adaptive anatomical contrastive loss for pixel as:
| (5) |
where is the pre-computed center of class . The first term in Eqn. 5 is supervised contrastive loss, where the summation over refers to the uniformly sampled positive examples from pixels in batch with label equal to . The summation over refers to all features in the batch excluding . The second term is contrastive loss with the positive example being the pre-computed optimal class center.
2.3. Anatomical-aware Temperature Scheduler (ATS)
Training with a varying induces a more isotropic representation space, wherein the model learns both group-wise and instance-specific features [12]. To this end, we are inspired to use an anatomical-aware temperature scheduler in both the supervised and the unsupervised contrastive losses, where the temperature parameter evolves within the range for . Specifically, for iteration with being the total number of iterations, we set as:
| (6) |
3. Experiments
Experimental Setup
We evaluate ACTION++ on two benchmark datasets: the LA dataset [31] and the ACDC dataset [1]. The LA dataset consists of 100 gadolinium-enhanced MRI scans, with the fixed split [29] using 80 and 20 scans for training and validation. The ACDC dataset consists of 200 cardiac cine MRI scans from 100 patients including three segmentation classes, i.e., left ventricle (LV), myocardium (Myo), and right ventricle (RV), with the fixed split2 using 70, 10, and 20 patients’ scans for training, validation, and testing. For all our experiments, we follow the identical setting in [42,19,30,29], and perform evaluations under two label settings (i.e., 5% and 10%) for both datasets.
Implementation Details
We use an SGD optimizer for all experiments with a learning rate of 1e-2, a momentum of 0.9, and a weight decay of 0.0001. Following [42,19,30,29] on both datasets, all inputs were normalized as zero mean and unit variance. The data augmentations are rotation and flip operations. Our work is built on ACTION [36], thus we follow the identical model setting except for temperature parameters because they are of direct interest to us. For the sake of completeness, we refer the reader to [36] for more details. We set as 0.2, 128, and regarding all , we use and if not stated otherwise. On ACDC, we use the U-Net model [26] as the backbone with a 2D patch size of 256 × 256 and batch size of 8. For pre-training, the networks are trained for 10K iterations; for fine-tuning, 20K iterations. On LA, we use the V-Net [21] as the backbone. For training, we randomly crop 112 × 112 × 80 patches and the batch size is 2. For pre-training, the networks are trained for 5K iterations. For fine-tuning, the networks are for 15K iterations. For testing, we adopt a sliding window strategy with a fixed stride (18 × 18 × 4). All experiments are conducted in the same environments with fixed random seeds (Hardware: Single NVIDIA GeForce RTX 3090 GPU; Software: PyTorch 1.10.2+cu113, and Python 3.8.11).
Main Results
We compare our ACTION++ with current state-of-the-art SSL methods, including UAMT [42], SASSNet [16], DTC [19], URPC [20], MC-Net [30], SS-Net [29], and ACTION [36], and the supervised counterparts (UNet [26]/VNet [21]) trained with Full/Limited supervisions – using their released code. To evaluate 3D segmentation ability, we use Dice coefficient (DSC) and Average Surface Distance (ASD). Table 2 and Table 1 display the results on the public ACDC and LA datasets for the two labeled settings, respectively. We next discuss our main findings as follows. (1) LA: As shown in Table 1, our method generally presents better performance than the prior SSL methods under all settings. Fig. 4 (Appendix) also shows that our model consistently outperforms all other competitors, especially in the boundary region; (2) ACDC: As Table 2 shows, ACTION++ achieves the best segmentation performance in terms of Dice and ASD, consistently outperforming the previous SSL methods across two labeled settings. In Fig. 3 (Appendix), we can observe that ACTION++ can yield the segmentation boundaries accurately, even for very challenging regions (i.e., RV and Myo). This suggests that ACTION++ is inherently better at long-tailed learning, in addition to being a better segmentation model in general.
Table 2.
Quantitative comparison (DSC[%]/ASD[voxel]) for ACDC under two unlabeled settings (5% or 10%). All experiments are conducted as [42,16,19,20,30,29,36] in the identical setting for fair comparisons. The best results are indicated in bold.
| 3 Labeled (5%) | 7 Labeled (10%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Method | Average | RV | Myo | LV | Average | RV | Myo | LV |
| UNet-F [26] | 91.5/0.996 | 90.5/0.606 | 88.8/0.941 | 94.4/1.44 | 91.5/0.996 | 90.5/0.606 | 88.8/0.941 | 94.4/1.44 |
| UNet-L | 51.7/13.1 | 36.9/30.1 | 54.9/4.27 | 63.4/5.11 | 79.5/2.73 | 65.9/0.892 | 82.9/2.70 | 89.6/4.60 |
| UAMT [42] | 48.3/9.14 | 37.6/18.9 | 50.1/4.27 | 57.3/4.17 | 81.8/4.04 | 79.9/2.73 | 80.1/3.32 | 85.4/6.07 |
| SASSNet [16] | 57.8/6.36 | 47.9/11.7 | 59.7/4.51 | 65.8/2.87 | 84.7/1.83 | 81.8/0.769 | 82.9/1.73 | 89.4/2.99 |
| URPC [20] | 58.9/8.14 | 50.1/12.6 | 60.8/4.10 | 65.8/7.71 | 83.1/1.68 | 77.0/0.742 | 82.2/0.505 | 90.1/3.79 |
| DTC [19] | 56.9/7.59 | 35.1/9.17 | 62.9/6.01 | 72.7/7.59 | 84.3/4.04 | 83.8/3.72 | 83.5/4.63 | 85.6/3.77 |
| MC-Net [30] | 62.8/2.59 | 52.7/5.14 | 62.6/0.807 | 73.1/1.81 | 86.5/1.89 | 85.1/0.745 | 84.0/2.12 | 90.3/2.81 |
| SS-Net [29] | 65.8/2.28 | 57.5/3.91 | 65.7/2.02 | 74.2/0.896 | 86.8/1.40 | 85.4/1.19 | 84.3/1.44 | 90.6/1.57 |
| ACTION [36] | 87.5/1.12 | 85.4/0.915 | 85.8/0.784 | 91.2/1.66 | 89.7/0.736 | 89.8/0.589 | 86.7/0.813 | 92.7/0.804 |
| • ACTION++ (ours) | 88.5/0.723 | 86.9/0.662 | 86.8/0.689 | 91.9/0.818 | 90.4/0.592 | 90.5/0.448 | 87.5/0.628 | 93.1/0.700 |
Table 1.
Quantitative comparison (DSC[%]/ASD[voxel]) for LA under two unlabeled settings (5% or 10%). All experiments are conducted as [42,16,19,20,30,29,36] in the identical setting for fair comparisons. The best results are indicated in bold. VNet-F (fully-supervided) and VNet-L (semi-supervided) are considered as the upper bound and the lower bound for the performance comparison.
| 4 Labeled (5%) | 8 Labeled (10%) | |||
|---|---|---|---|---|
| Method | DSC[%]↑ | ASD[voxel]↓ | DSC[%]↑ | ASD[voxel]↓ |
| VNet-F [21] | 91.5 | 1.51 | 91.5 | 1.51 |
| VNet-L | 52.6 | 9.87 | 82.7 | 3.26 |
| UAMT [42] | 82.3 | 3.82 | 87.8 | 2.12 |
| SASSNet [16] | 81.6 | 3.58 | 87.5 | 2.59 |
| DTC [19] | 81.3 | 2.70 | 87.5 | 2.36 |
| URPC [20] | 82.5 | 3.65 | 86.9 | 2.28 |
| MC-Net [30] | 83.6 | 2.70 | 87.6 | 1.82 |
| SS-Net [29] | 86.3 | 2.31 | 88.6 | 1.90 |
| ACTION [36] | 86.6 | 2.24 | 88.7 | 2.10 |
| • ACTION++ (ours) | 87.8 | 2.09 | 89.9 | 1.74 |
Ablation Study
We first perform ablation studies on LA with 10% label ratio to evaluate the importance of different components. Table 3 shows the effectiveness of supervised adaptive anatomical contrastive learning (SAACL). Table 4 (Appendix) indicates that using anatomical-aware temperature scheduler (ATS) and SAACL yield better performance in both pre-training and fine-tuning stages. We then theoretically show the superiority of our method in Appendix A.
Table 3.
Ablation studies of Supervised Adaptive Anatomical Contrast (SAACL).
Finally, we conduct experiments to study the effects of cosine boundaries, cosine period, different methods of varying , and in Table 5, Table 6 (Appendix), respectively. Empirically, we find that using our settings (i.e., , cosine scheduler, ) attains optimal performance.
4. Conclusion
In this paper, we proposed ACTION++, an improved contrastive learning framework with adaptive anatomical contrast for semi-supervised medical segmentation. Our work is inspired by two intriguing observations that, besides the unlabeled data, the class imbalance issue exists in the labeled portion of medical data and the effectiveness of temperature schedules for contrastive learning on long-tailed medical data. Extensive experiments and ablations demonstrated that our model consistently achieved superior performance compared to the prior semi-supervised medical image segmentation methods under different label ratios. Our theoretical analysis also revealed the robustness of our method in label efficiency. In future, we will validate CT/MRI datasets with more foreground labels and try t-SNE.
Supplementary Material
Table 4.
Effect of cosine boundaries in with the largest difference between and .
| ╲ | 0.2 | 0.3 | 0.4 | 0.5 | 1.0 |
|---|---|---|---|---|---|
| 0.07 | 84.1 | 85.0 | 86.9 | 87.9 | 89.7 |
| 0.1 | 84.5 | 85.9 | 87.1 | 88.3 | 89.9 |
| 0.2 | 84.2 | 84.4 | 85.8 | 87.1 | 87.6 |
Footnotes
For simplicity, we omit details of local instance discrimination in the following.
References
- 1.Bernard O, Lalande A, Zotti C, Cervenansky F, Yang X, Heng PA, Cetin I, Lekadir K, Camara O, Ballester MAG, et al. : Deep learning techniques for automatic mri cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Transactions on Medical Imaging (2018) [DOI] [PubMed] [Google Scholar]
- 2.Chaitanya K, Erdil E, Karani N, Konukoglu E: Contrastive learning of global and local features for medical image segmentation with limited annotations. In: NeurIPS; (2020) [Google Scholar]
- 3.Chapelle O, Scholkopf B, Zien A: Semi-supervised learning (chapelle o. et al. , eds.; 2006)[book reviews]. IEEE Transactions on Neural Networks (2009) [Google Scholar]
- 4.Chen T, Kornblith S, Norouzi M, Hinton G: A simple framework for contrastive learning of visual representations. In: ICML. pp. 1597–1607. PMLR (2020) [Google Scholar]
- 5.Chen X, Fan H, Girshick R, He K: Improved baselines with momentum contrastive learning. arXiv preprint arXiv:2003.04297 (2020) [Google Scholar]
- 6.Graf F, Hofer C, Niethammer M, Kwitt R: Dissecting supervised contrastive learning. In: ICML. PMLR (2021) [Google Scholar]
- 7.He Y, Lin F, Tzeng NF, et al. : Interpretable minority synthesis for imbalanced classification. In: IJCAI; (2021) [Google Scholar]
- 8.Huang W, Yi M, Zhao X: Towards the generalization of contrastive self-supervised learning. arXiv preprint arXiv:2111.00743 (2021) [Google Scholar]
- 9.Kang B, Li Y, Xie S, Yuan Z, Feng J: Exploring balanced feature spaces for representation learning. In: ICLR; (2021) [Google Scholar]
- 10.Kang B, Xie S, Rohrbach M, Yan Z, Gordo A, Feng J, Kalantidis Y: Decoupling representation and classifier for long-tailed recognition. arXiv preprint arXiv:1910.09217 (2019) [Google Scholar]
- 11.Kervadec H, Dolz J, Granger É, Ben Ayed I: Curriculum semi-supervised segmentation. In: MICCAI. Springer; (2019) [Google Scholar]
- 12.Kukleva A, Böhle M, Schiele B, Kuehne H, Rupprecht C: Temperature schedules for self-supervised contrastive methods on long-tail data. In: ICLR; (2023) [Google Scholar]
- 13.Lai Z, Wang C, Cheung S.c., Chuah CN: Sar: Self-adaptive refinement on pseudo labels for multiclass-imbalanced semi-supervised learning. In: CVPR. pp. 4091–4100 (2022) [Google Scholar]
- 14.Lai Z, Wang C, Gunawan H, Cheung SCS, Chuah CN: Smoothed adaptive weighting for imbalanced semi-supervised learning: Improve reliability against unknown distribution data. In: ICML. pp. 11828–11843 (2022) [Google Scholar]
- 15.Lai Z, Wang C, Oliveira LC, Dugger BN, Cheung SC, Chuah CN: Joint semi-supervised and active learning for segmentation of gigapixel pathology images with cost-effective labeling. In: ICCV. pp. 591–600 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, Zhang C, He X: Shape-aware semi-supervised 3d semantic segmentation for medical images. In: MICCAI. pp. 552–561. Springer; (2020) [Google Scholar]
- 17.Li T, Cao P, Yuan Y, Fan L, Yang Y, Feris RS, Indyk P, Katabi D: Targeted supervised contrastive learning for long-tailed recognition. In: CVPR; (2022) [Google Scholar]
- 18.Li Z, Kamnitsas K, Glocker B: Analyzing overfitting under class imbalance in neural networks for image segmentation. IEEE Transactions on Medical Imaging (2020) [DOI] [PubMed] [Google Scholar]
- 19.Luo X, Chen J, Song T, Wang G: Semi-supervised medical image segmentation through dual-task consistency. In: AAAI; (2020) [Google Scholar]
- 20.Luo X, Liao W, Chen J, Song T, Chen Y, Zhang S, Chen N, Wang G, Zhang S: Efficient semi-supervised gross target volume of nasopharyngeal carcinoma segmentation via uncertainty rectified pyramid consistency. In: MICCAI Springer; (2021) [Google Scholar]
- 21.Milletari F, Navab N, Ahmadi SA: V-net: Fully convolutional neural networks for volumetric medical image segmentation. In: 3DV. pp. 565–571. IEEE; (2016) [Google Scholar]
- 22.Oliveira LC, Lai Z, Siefkes HM, Chuah CN: Generalizable semi-supervised learning strategies for multiple learning tasks using 1-d biomedical signals. In: NeurIPS Workshop on Learning from Time Series for Health (2022) [Google Scholar]
- 23.Qiao S, Shen W, Zhang Z, Wang B, Yuille A: Deep co-training for semi-supervised image recognition. In: ECCV; (2018) [Google Scholar]
- 24.Quan Q, Yao Q, Li J, Zhou S.k.: Information-guided pixel augmentation for pixel-wise contrastive learning. arXiv preprint arXiv:2211.07118 (2022) [Google Scholar]
- 25.Robinson J, Sun L, Yu K, Batmanghelich K, Jegelka S, Sra S: Can contrastive learning avoid shortcut solutions? In: NeurIPS; (2021) [PMC free article] [PubMed] [Google Scholar]
- 26.Ronneberger O, Fischer P, Brox T: U-net: Convolutional networks for biomedical image segmentation. In: MICCAI. Springer; (2015) [Google Scholar]
- 27.Tarvainen A, Valpola H: Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results. In: NeurIPS. pp. 1195–1204 (2017) [Google Scholar]
- 28.Wang F, Liu H: Understanding the behaviour of contrastive loss. In: CVPR; (2021) [Google Scholar]
- 29.Wu Y, Wu Z, Wu Q, Ge Z, Cai J: Exploring smoothness and class-separation for semi-supervised medical image segmentation. In: MICCAI; (2022) [Google Scholar]
- 30.Wu Y, Xu M, Ge Z, Cai J, Zhang L: Semi-supervised left atrium segmentation with mutual consistency training. In: MICCAI; (2021) [Google Scholar]
- 31.Xiong Z, Xia Q, Hu Z, Huang N, Bian C, Zheng Y, Vesal S, Ravikumar N, Maier A, Yang X, et al. : A global benchmark of algorithms for segmenting the left atrium from late gadolinium-enhanced cardiac magnetic resonance imaging. Medical Image Analysis (2021) [DOI] [PubMed] [Google Scholar]
- 32.Xue Y, Xu T, Zhang H, Long LR, Huang X: Segan: Adversarial network with multi-scale l 1 loss for medical image segmentation. Neuroinformatics (2018) [DOI] [PubMed] [Google Scholar]
- 33.You C, Dai W, Liu F, Su H, Zhang X, Staib L, Duncan JS: Mine your own anatomy: Revisiting medical image segmentation with extremely limited labels. arXiv preprint arXiv:2209.13476 (2022) [Google Scholar]
- 34.You C, Dai W, Min Y, Liu F, Zhang X, Clifton DA, Zhou SK, Staib LH, Duncan JS: Rethinking semi-supervised medical image segmentation: A variance-reduction perspective. arXiv preprint arXiv:2302.01735 (2023) [PMC free article] [PubMed] [Google Scholar]
- 35.You C, Dai W, Min Y, Staib L, Duncan JS: Implicit anatomical rendering for medical image segmentation with stochastic experts. arXiv preprint arXiv:2304.03209 (2023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.You C, Dai W, Staib L, Duncan JS: Bootstrapping semi-supervised medical image segmentation with anatomical-aware contrastive distillation. In: IPMI; (2023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.You C, Xiang J, Su K, Zhang X, Dong S, Onofrey J, Staib L, Duncan JS: Incremental learning meets transfer learning: Application to multi-site prostate mri segmentation. In: International Workshop on Distributed, Collaborative, and Federated Learning (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.You C, Yang J, Chapiro J, Duncan JS: Unsupervised wasserstein distance guided domain adaptation for 3d multi-domain liver segmentation. In: Interpretable and Annotation-Efficient Learning for Medical Image Computing. pp. 155–163. Springer International Publishing; (2020) [Google Scholar]
- 39.You C, Zhao R, Liu F, Dong S, Chinchali S, Topcu U, Staib L, Duncan J: Class-aware adversarial transformers for medical image segmentation. In: NeurIPS; (2022) [PMC free article] [PubMed] [Google Scholar]
- 40.You C, Zhao R, Staib LH, Duncan JS: Momentum contrastive voxel-wise representation learning for semi-supervised volumetric medical image segmentation. In: MICCAI; (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.You C, Zhou Y, Zhao R, Staib L, Duncan JS: Simcvd: Simple contrastive voxel-wise representation distillation for semi-supervised medical image segmentation. IEEE Transactions on Medical Imaging (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu L, Wang S, Li X, Fu CW, Heng PA: Uncertainty-aware self-ensembling model for semi-supervised 3d left atrium segmentation. In: MICCAI; (2019) [Google Scholar]
- 43.Zhou Y, Wang Y, Tang P, Bai S, Shen W, Fishman E, Yuille A: Semi-supervised 3d abdominal multi-organ segmentation via deep multi-planar co-training. In: WACV. IEEE; (2019) [Google Scholar]
- 44.Zipf GK: The psycho-biology of language: An introduction to dynamic philology. Routledge; (2013) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


