Abstract
Cleptoparasitism, also known as brood parasitism, is a widespread strategy among bee species in which the parasite lays eggs into the nests of the host species. Even though this behavior has significant ecological implications for the dynamics of several species, little is known about the molecular pathways associated with cleptoparasitism. To shed some light on this issue, we used gene expression data to perform a comparative analysis between two solitary neotropical bees: Coelioxoides waltheriae, an obligate parasite, and their specific host Tetrapedia diversipes. We found that ortholog genes involved in signal transduction, sensory perception, learning, and memory formation were differentially expressed between the cleptoparasite and the host. We hypothesize that these genes and their associated molecular pathways are engaged in cleptoparasitism-related processes and, hence, are appealing subjects for further investigation into functional and evolutionary aspects of cleptoparasitism in bees.
Keywords: Comparative transcriptomics, Brood parasitism, Cuckoo bee, Solitary bees
Subject terms: Behavioural genetics, Gene expression, Molecular ecology
Introduction
Parasitism is an interaction between different species, in which one of them, the parasite, benefits at the expense of another, the host1,2. Parasitic species may play an important role in the dynamics of natural populations of host species, for instance, they may affect the susceptibility of their hosts to predation, modify their reproductive patterns, and influence the abundance of endemic and introduced species3,4. Cleptoparasitism, alternatively known as brood parasitism, refers to a parasitic behavior where the parasite lays eggs into the nests of the host species. Then during its larval stage, the parasite offspring thrives by consuming the food resources that have been provided by the adult host, ultimately leading to the demise of the host larva or egg, as it is either killed or eaten by the parasite larva. Finally, an adult parasite emerges from the host nest5.
Cleptoparasitism is widespread in bees (Hymenoptera: Antophila). It is estimated that approximately 13% of the 20,500 bee species in the world6 are obligate cleptoparasites7. Currently, it has been inferred that this behavior has arisen independently 18 times in four out of nine bee families: three distinct times in Apidae8–10; probably five times in Megachilidae11–13; at least once in Colletidae14; and possibly nine in Halictidae5,15. In spite of these multiple and independent origins, most cleptoparasitic bees show important convergent adaptations such as the reduction or complete loss of pollen-collecting (e.g., pollen-manipulating brushes and scopa) and nest-building structures (e.g., basitibial and pygidial plates for ground-nesting species)5. Compared to non-parasitic species, many cleptoparasites also have a heavily sclerotized cuticle in addition to spines, ridges, crests, carinae or lamellae protecting them from the jaws or sting of host females5,7. Moreover, convergent anatomical and physiological changes in the reproductive system of some cleptoparasitic species have been described, such as a greater number of mature oocytes in the ovaries or more ovarioles per ovary16. These adaptations allow parasitic females to lay several eggs in a short period of time5,7.
In addition to their evolutionary relevance, cleptoparasitic species also have an ecological value. Cleptoparasitic bees are considered by far the largest protagonist of solitary bee brood mortality among all natural enemies17. In this context, Sheffield et al.18 suggest that cleptoparasites, especially generalist ones, perform a stabilizing role in bee communities by attacking dominant and abundant host taxa, which may reduce competition among non-parasitic bee species. These authors also argue that cleptoparasites can serve as indicator taxa for assessing the status of the entire bee community, as their diversity and abundance are closely tied to their host species18. However, there is still a deep lack of knowledge regarding the general biology, behavior7, and particularly the molecular mechanisms underlying the evolution and maintenance of cleptoparasitism. This knowledge gap might be driven by the rarity to find cleptoparasitic bees in nature7,19.
The abundance of cleptoparasitic bees often exhibits a positive correlation with the density of available host nests18,20, implying that hosts displaying high levels of gregarious behavior are more susceptible to parasitic attacks. In this context, the species Coelioxoides waltheriae Ducke, 1908 and Tetrapedia diversipes Klug, 1810 comprise a compelling parasite-host species pair to investigate cleptoparasitism. T. diversipes, the host species, has a gregarious behavior, building its nests in naturally pre-existing cavities such as holes in wood21, and trap-nests. Indeed, T. diversipes easy aggregatory behavior in trap-nests allowed the description of the cleptoparasitic behavior of C. waltheriae21, the first recorded for the Coelioxoides genus. Coelioxoides and Tetrapedia are endemic to the Neotropical region22. Both genera have been formerly grouped within the same tribe, Tetrapediini (Apidae), as they share a number of similar morphological traits23. However, recent molecular-based phylogenies have placed Coelioxoides within a large cleptoparasitic clade (Nomadinae: Apidae) not sister to Tetrapedia8–10,24,25. C. waltheriae is considered to be the main natural enemy of T. diversipes26 even though it has been also reported to parasitize nests of other Tetrapedia species27,28.
Recent studies have successfully employed comparative transcriptomic analysis to unveil molecular features related to complex behaviors. For instance, transcriptomics studies in bees have helped to better understand the molecular pathways related to the honeybee (waggle) dance29, Varroa sensitive hygiene behaviour30, olfactory31 and visual32 learning, diapause33,34, and evolution of task division35–37. In this context, herein we use gene expression data to perform a comparative analysis between the correspondent reproductive life stages of C. waltheriae and its host, T. diversipes. We aimed at identifying diverging gene expression patterns between the solitary pollen-collecting host and the cleptoparasitic bee species. We explored the function of enriched diverging pathways and discussed whether these could be related at some level to their ecological interactions. Instead of assuming any of the differences here observed are causative, we focused on relating our findings to molecular processes for which ecological and/or behavioural relevance has been previously described in the literature to shed some light on the molecular mechanisms potentially related to cleptoparasitic behavior.
Results
Transcriptome assembly and annotation
We sampled reproductive females of C. waltheriae and T. diversipes from the same trap-nest location and at correspondent life stages. In total, we used five sample replicates per species, consisting of a pool of whole-body extractions from individuals (see “Materials and methods”).
After transcriptome assemblies and data treatment (see “Materials and methods”) we obtained a set of 18,208 transcripts for C. waltheriae and 11,998 for T. diversipes. The main quality parameters associated with each assembly are summarized in Table 1.
Table 1.
Main quality parameters of C. waltheriae and T. diversipes assemblies.
| Metric | C. waltheriae | T. diversipes |
|---|---|---|
| Total transcripts assembled | 87,358 | 17,866a |
| 40,796b | ||
| Number of clustered SuperTranscriptsc | 18,208 | 11,998 |
| SuperTranscripts ≥ 1000 nt | 10,195 | 8051 |
| p fragments mappedd | 0.93176 | 0.86625 |
| p bases uncoverede | 0.0328 | 0.05047 |
| Transrate optimal scoref | 0.5904 | 0.50711 |
| Complete BUSCOs (%) | 93.4 | 88.5 |
| Missing BUSCOs (%) | 4.1 | 8.2 |
aNumber of transcripts obtained after the reference-based assembly.
bNumber of transcripts obtained after the reference guided de novo assembly.
cAfter removing contaminating sequences.
dProportion of reads pairs that mapped successfully.
eProportion of bases that were not covered by any read.
fThis discrete score represents the accuracy and completeness of the assembly based on the assessment of alignments between contigs and reads. The minimum score is 0, while the maximum is 1.0. For a description of the components used in the calculation of this metric, see Smith-Unna et al.38.
A total of 10,042 (51.2%) and 7447 (61.9%) transcripts in C. waltheriae and T. diversipes, respectively, blasted (e-value < 1e−5) against protein-coding genes in the UniRef90 database. Moreover, 6253 (31.8%) and 2919 (24.2%) transcripts were estimated to be potential non-coding sequences in C. waltheriae and T. diversipes. It is worth mentioning that 1397 (7.1%) transcripts were identified as potential contaminants in C. waltheriae (Supplementary Material 1). Most were attributed to fungi of the family Tubulinosematidae (Microsporidia): Tubulinosema ratisbonensis (1113 transcripts), Anncaliia algerae (15 transcripts), and Tubulinosematidae itself (10 transcripts). In addition, 95 (0.48%) of C. waltheriae transcripts were identified as very similar to plant protein sequences. Overall, the Fabaceae family was the most representative plants among contaminants (52 transcripts), followed by Asteraceae (11 transcripts) and Solanaceae (6 transcripts). Contaminant transcripts of T. diversipes have already been described previously39. Herein, we identified only 31 (0.25%) transcripts as potential contaminants (Supplementary Material 1), far less than that observed in C. waltheriae, which was expected, since the assembly approaches for T. diversipes used a reference genome.
Differential orthologs expression
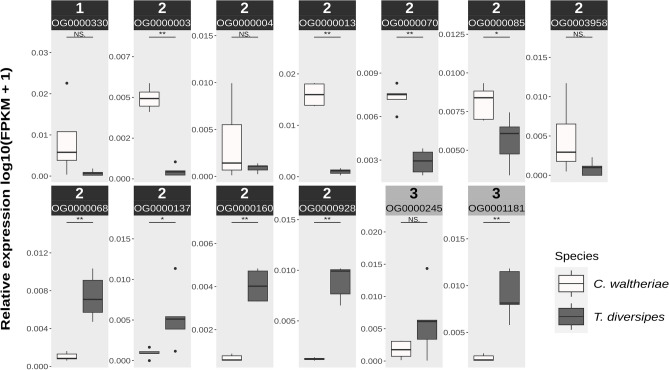
Using Orthofinder40,41 we identified 4859 orthogroups shared between C. waltheriae and T. diversipes, of which 3011 were reported as single-copy orthologues. Using blastp42,43 searches to identify sequences with the highest homology within common orthogroups we additionally selected 1848 one-to-one orthologs, totaling 4154 one-to-one ortholog pairs between the two species. Of these, around half (2096 or 50.45%) were annotated with at least one functional GO term. After correcting for sampling batch effects accounting for species, the number of samples and sampling year (Supplementary Material 2), the significantly differentially expressed orthologs between the species were obtained by overlapping the results from two strategies (1. TMM normalized counts44 and edgeR 3.34.045; 2. Scale Based Normalization [SCBN] method46 and NOISeq47,48). We identified a total of 646 orthologs as differentially expressed (DE) between the two species (Supplementary Material 3), 335 of which were highly expressed in C. waltheriae, and 311 were highly expressed in T. diversipes. Among the annotated DE orthologs, two groups caught our attention prior to GO Enrichment Analysis due to their frequency and function. First, a group of eleven transcripts homologous to sequences of PiggyBac transposable element-derived protein genes (Pgbd4 = 10 transcripts and Pgbd2 = 1 transcript), most of which (7) were highly expressed in C. waltheriae (Fig. 1). Moreover, considering the entire set of transcripts assembled, we found that C. waltheriae has more transcripts (n = 68; 0.34% of the transcriptome) of Pgdb-like genes than T. diversipes (n = 24; 0.19%). Additionally, we found in T. diversipes two sequences identified as homologous to Major Royal Jelly Protein/yellow-like (MRJP/y) that were highly expressed (Fig. 1).
Figure 1.
Relative expression of orthologs differentially expressed between the cleptoparasite Coelioxoides waltheriae and their host Tetrapedia diversipes that were identified as homologs to PiggyBac Transposable Element-derived (1: PGBD2-like; 2: PGBD4-like) and Major Royal Jelly Protein (3) sequences. The respective orthogroups (OGs) are identified according to their IDs in Supplementary Material 3.
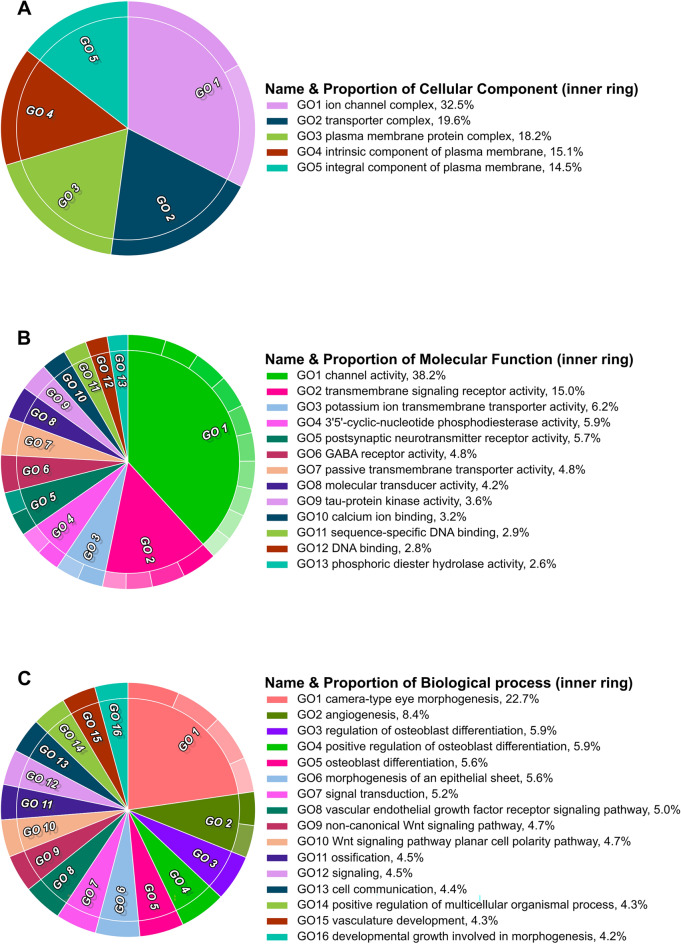
Through the Gene Ontology term enrichment analysis for all categories (BP, CC, and MF), we identified 54 GO terms enriched among the DE orthologs (Supplementary Material 4). The redundant terms of each GO category were summarized in a two-level hierarchical list for visualization (Fig. 2). Top enriched Cellular Component (CC) terms were related to ion channel complex, transporter complex, and plasma membrane protein complex. Consistently, top enriched Molecular Function (MF) terms were related to channel activity, transmembrane signaling receptor activity, and potassium ion transmembrane activity. The orthologs annotated with these CC and MF terms are shown in Table 2.
Figure 2.
Gene Ontology (GO) terms enriched among the Differentially Expressed (DE) Orthologs between Coelioxoides waltheriae and Tetrapedia diversipes. Each pie chart represents one of the three main GO categories: (A) Cellular Component, (B) Molecular Function, and (C) Biological Process. Legends next to each chart indicate the representative color, name, and proportion of DE that received an annotation related to the parental GO term. Child terms from the second hierarchical level are represented in the outermost part of the pie chart using shades of the parental representative term color. Parent–child GO term relationships are listed in Supplementary Material 4.
Table 2.
Differentially expressed (DE) orthologs between Coelioxoides waltheriae and Tetrapedia diversipes grouped according to their association to the top enriched GO terms for Cell Component and Molecular Function categories.
| DEO IDs | Expression patterns | Homologs (Swiss-Prot organism) | GO hierarchical groups |
|---|---|---|---|
| Calcium channels | |||
| OG0000284 |
C. waltheriae ↓ T. diversipes ↑ |
CAC (Apis mellifera) |
Cell component GO1 ion channel complex GO2 transporter complex GO3 plasma membrane protein complex Molecular function GO1 channel activity GO7 passive transmembrane transporter activity GO10 calcium ion binding |
| OG0000168 |
C. waltheriae ↓ T. diversipes ↑ |
Trp (Drosophila melanogaster) |
Cell Component GO1 ion channel complex GO2 transporter complex GO3 plasma membrane protein complex GO4 intrinsic component of plasma membrane GO5 integral component of plasma membrane Molecular Function GO1 channel activity GO7 passive transmembrane transporter activity |
| Sodium channels | |||
| OG0003129 |
C. waltheriae ↑ T. diversipes ↓ |
NaCP60E (Drosophila melanogaster) |
Cell component GO1 ion channel complex GO2 transporter complex GO3 plasma membrane protein complex GO4 intrinsic component of plasma membrane GO5 integral component of plasma membrane Molecular function GO1 channel activity GO7 passive transmembrane transporter activity |
| OG0002153 |
C. waltheriae ↑ T. diversipes ↓ |
unc80 (Drosophila melanogaster) |
Cell component GO1 ion channel complex GO2 transporter complex Molecular Function GO1 channel activity GO7 passive transmembrane transporter activity |
| Potassium channels | |||
| OG0004503 |
C. waltheriae ↓ T. diversipes ↑ |
KCNQ1 (Felis catus, Xenopus laevis) |
Cell component GO1 ion channel complex GO2 transporter complex GO3 plasma membrane protein complex GO4 intrinsic component of plasma membrane GO5 integral component of plasma membrane Molecular function GO1 channel activity GO3 potassium ion transmembrane transporter activity GO7 passive transmembrane transporter activity |
| OG0000260 |
C. waltheriae ↓ T. diversipes ↑ |
Sh (Drosophila melanogaster) |
Cell component GO1 ion channel complex GO2 transporter complex GO3 plasma membrane protein complex GO4 intrinsic component of plasma membrane GO5 integral component of plasma membrane Molecular function GO1 channel activity GO3 potassium ion transmembrane transporter activity GO7 passive transmembrane transporter activity |
| OG0004153 |
C. waltheriae ↑ T. diversipes ↓ |
KCNT1 (Homo sapiens) |
Molecular function GO1 channel activity GO3 potassium ion transmembrane transporter activity GO7 passive transmembrane transporter activity |
| OG0002734 |
C. waltheriae ↑ T. diversipes ↓ |
KCNK9 (Homo sapiens, Rattus norvegicus) |
Molecular function GO1 channel activity GO3 potassium ion transmembrane transporter activity GO7 passive transmembrane transporter activity |
| Chloride channel | |||
| OG0000326 |
C. waltheriae ↓ T. diversipes ↑ |
Grd (Drosophila melanogaster) |
Molecular function GO1 channel activity GO2 transmembrane signaling receptor activity GO6 GABA receptor activity GO7 passive transmembrane transporter activity GO8 molecular transducer activity |
| Ligand-gated ion channels | |||
| OG0000029 |
C. waltheriae ↓ T. diversipes ↑ |
Grik2 (Mus musculus, Xenopus laevis) |
Cell component GO1 ion channel complex GO2 transporter complex GO3 plasma membrane protein complex GO4 intrinsic component of plasma membrane GO5 integral component of plasma membrane Molecular function GO1 channel activity GO2 transmembrane signaling receptor activity GO3 potassium ion transmembrane transporter activity GO5 postsynaptic neurotransmitter receptor activity GO7 passive transmembrane transporter activity GO8 molecular transducer activity |
| OG0000962 |
C. waltheriae ↓ T. diversipes ↑ |
nAChRβ1 (Drosophila melanogaster) |
Cell component GO1 ion channel complex GO2 transporter complex GO3 plasma membrane protein complex GO4 intrinsic component of plasma membrane GO5 integral component of plasma membrane Molecular function GO1 channel activity GO2 transmembrane signaling receptor activity GO5 postsynaptic neurotransmitter receptor activity GO7 passive transmembrane transporter activity GO8 molecular transducer activity |
| G-protein coupled receptors | |||
| OG0002068 |
C. waltheriae ↓ T. diversipes ↑ |
GABBR1 (Homo sapiens, Mus musculus) |
Cell component GO3 plasma membrane protein complex GO4 intrinsic component of plasma membrane GO5 integral component of plasma membrane Molecular function GO2 transmembrane signaling receptor activity GO5 postsynaptic neurotransmitter receptor activity GO6 GABA receptor activity GO8 molecular transducer activity |
| OG0001556 |
C. waltheriae ↑ T. diversipes ↓ |
Gabbr2 (Mus musculus) |
Cell component GO3 plasma membrane protein complex GO4 intrinsic component of plasma membrane GO5 integral component of plasma membrane Molecular function GO2 transmembrane signaling receptor activity GO6 GABA receptor activity GO8 molecular transducer activity |
| OG0000063 |
C. waltheriae ↑ T. diversipes ↓ |
Octβ2R (Drosophila melanogaster) |
Molecular function GO2 transmembrane signaling receptor activity GO8 molecular transducer activity |
| OG0001943 |
C. waltheriae ↑ T. diversipes ↓ |
HTR2C (Mus musculus, Bos taurus) |
Molecular function GO2 transmembrane signaling receptor activity GO8 molecular transducer activity |
| OG0000814 |
C. waltheriae ↓ T. diversipes ↑ |
CG31760 (Drosophila melanogaster) |
Molecular function GO2 transmembrane signaling receptor activity GO8 molecular transducer activity |
| OG0001490 |
C. waltheriae ↓ T. diversipes ↑ |
Gr64f./Gr5a (Drosophila melanogaster) |
Molecular function GO2 transmembrane signaling receptor activity GO8 molecular transducer activity |
| G protein | |||
| OG0004442 |
C. waltheriae ↓ T. diversipes ↑ |
G-sα60A (Anopheles gambiae) |
Cell component GO3 plasma membrane protein complex |
| Translocases | |||
| OG0001155 |
C. waltheriae ↑ T. diversipes ↓ |
Atpα (Drosophila melanogaster) |
Cell component GO2 transporter complex GO3 plasma membrane protein complex GO4 intrinsic component of plasma membrane GO5 integral component of plasma membrane Molecular function GO1 channel activity GO3 potassium ion transmembrane transporter activity |
| OG0001472 |
C. waltheriae ↑ T. diversipes ↓ |
nrv2/nrv1 (Drosophila melanogaster) |
Cell component GO2 transporter complex GO3 plasma membrane protein complex GO4 intrinsic component of plasma membrane GO5 integral component of plasma membrane |
| OG0000864 |
C. waltheriae ↓ T. diversipes ↑ |
UQCRFS1 (Lagothrix lagotricha, Colobus polykomos) |
Molecular function GO1 channel activity |
| DNA-binding protein | |||
| OG0003803 |
C. waltheriae ↑ T. diversipes ↓ |
Hr38 (Drosophila melanogaster) |
Molecular function GO2 transmembrane signaling receptor activity GO8 molecular transducer activity GO11 sequence-specific DNA binding GO12 DNA binding |
| Low-density lipoprotein-receptor-related | |||
| OG0004128 |
C. waltheriae ↑ T. diversipes ↓ |
LRP6 (Homo sapiens, Mus musculus) |
Cell component GO3 plasma membrane protein complex Molecular function GO2 transmembrane signaling receptor activity GO8 molecular transducer activity |
| Cell adhesion molecules | |||
| OG0000727 |
C. waltheriae ↑ T. diversipes ↓ |
NRXN1 (Homo sapiens, Mus musculus) |
Cell component GO4 intrinsic component of plasma membrane GO5 integral component of plasma membrane Molecular function GO2 transmembrane signaling receptor activity GO8 molecular transducer activity GO10 calcium ion binding |
| OG0000165 |
C. waltheriae ↑ T. diversipes ↓ |
Stan (Drosophila melanogaster) |
Molecular function GO2 transmembrane signaling receptor activity GO8 molecular transducer activity GO10 calcium ion binding |
| Protein phosphatase | |||
| OG0000836 |
C. waltheriae ↓ T. diversipes ↑ |
Ptp69D (Drosophila melanogaster) |
Molecular function GO2 transmembrane signaling receptor activity GO8 molecular transducer activity |
| Tyrosine-protein kinase | |||
| OG0002698 |
C. waltheriae ↓ T. diversipes ↑ |
Abl (Drosophila melanogaster) |
Molecular function GO2 transmembrane signaling receptor activity GO8 molecular transducer activity |
The orthologs are represented by the ID of their respective orthogroups. Expression patterns of orthologs comparatively between C. waltheriae and T. diversipes are indicated by arrows: ↑ (upregulated) and ↓ (downregulated). GO hierarchical groups refer to CirGo terms as shown in Fig. 2.
For the Biological Process (BP) category, the most significant terms in the enrichment analysis were related to camera-type eye morphogenesis, angiogenesis, and regulation of osteoblast differentiation. The main DE orthologs annotated within these terms were identified as homologs of Fibrillin-2-like, Low-density lipoprotein receptor-related protein 6, and serine/threonine-protein kinases. Moreover, the highest number of DE orthologs were associated with cell communication, signaling, and signal transduction BP terms (Supplementary Material 5).
Discussion
We employed comparative transcriptomics to investigate molecular distinctions during the reproductive stage of Coelioxoides waltheriae (a cleptoparasite) and its host species, Tetrapedia diversipes. The present study represents an initial effort to obtain some insights about the molecular mechanisms involved in bee cleptoparasitism by comparing orthologs expression data between a cleptoparasitic species and their host, both inhabiting the same location and at equivalent developmental stages. Considering the constraints of cross-species transcriptomic comparisons, our findings shed light on broad molecular variations between the cleptoparasite and its host, suggesting possible ecological implications.
We overlapped two statistical approaches to perform a cross-species comparison and to reduce the number of false positives in our analyses. The first of these methods was the SCBN, a recently proposed method for count normalization optimized to deal with cross-species comparisons46. Secondly, we used a traditional differential expression workflow with the edgeR Bioconductor package45. By overlapping the results from these two approaches we consistently retrieved 646 differentially expressed orthologs between the two species, with PiggyBac Transposable Element-derived like (Pgbd-like) genes being the most frequent (Fig. 1). Overall, we found many orthologs annotated as Pgbd-like, and several of them were found to be highly expressed in C. waltheriae. The PiggyBac transposon superfamily is widespread among eukaryotes. The first PiggyBac element was described from a cell culture of Trichoplusia ni (Lepidoptera: Noctuidae), the cabbage looper moth49,50. Since then, several PiggyBac-like sequences have been described in a variety of organisms. Some of these transposable elements lost transposase activity and are called domesticated51. Molecular domestication is a process that occurs due to a transposition inactivating mutation, resulting in the loss of mobility of the transposable element52. These insertions may lead to the emergence of new cellular activities, either by altering the coding and/or regulatory regions in which these elements are inserted in the genome or by the evolution of the former TE genes into new genes52,53. Thus, these findings lead us to hypothesize whether the domestication of PiggyBac-like transcripts and its regulatory consequences could be one of the molecular mechanisms involved in the evolution and adaptation of C. waltheriae, justifying their differential expression between the studied host-parasite species. Indeed, in a recent study, it was observed through comparative genomic analyses that retroviral or transposable elements have undergone a recent or ongoing spread in the genome of a Nomadinae cleptoparasite54. This indicates a possible involvement of TEs in the evolution of cleptoparasitism. Future research should further explore this hypothesis, investigating the evolution of transposable and transposable-derived elements in host-parasites genomes and their expression pattern in other cleptoparasitic species of Apidae.
We also identified two MRJP/Yellow-like sequences as highly expressed in T. diversipes while none were upregulated in C. waltheriae. The MRJP/Yellow gene family encodes multifunctional yellow-like proteins identified in arthropods and in several bacteria55,56. Nonetheless, the MRJP-like genes, as part of this family, seem to be restricted to Hymenoptera56. Even though these genes have been associated with olfactory learning—particularly regarding mrjp1 expression in the mushroom body (Kenyon cells)31,57,58—and functional studies have shown that MRJPs may have immunoregulatory and antibacterial effects56, in bees, MRJPs are mostly associated with larvae feeding, development and with the regulation of phenotypic plasticity and age-polyethism in workers59. In honeybees, MRJPs are known to be essential components of the larvae diet, with MRJP1–3 and 5 being the most abundant proteins of larval food56. These proteins are synthesized by honeybee nurse workers in the specialized hypopharyngeal glands and offered to the immature offspring through a special food jelly (royal jelly)60. In contrast to honeybees, bumblebees do not produce royal jelly. However, their hypopharyngeal glands express one MRJP ortholog61. The production of MRJP is not dependent on brood-feeding activity in bumblebees, and protein signals were identified in the abdominal parts of the digestive tract in queens and workers. This suggests that bumblebee MRJP does not have a nutritional function but is instead involved in food digestion and/or modification, consistent with the putatively ancient function of bee hypopharyngeal glands in food digestion61. In solitary bees the role of MRJP-like genes is currently poorly understood. One hypothesis for the high expression of MRJPs in T. diversipes could be the association of these proteins with larval food. As a solitary bee species, founder females of T. diversipes are also responsible for larvae food provisioning and an increased expression of transcripts related to MRJP-like genes could suggest that these proteins may be component of larval food. Yet, the role of MRJPs-like may not be necessarily nutritional/developmental, as they could alternatively play an antibacterial role and/or aid in the processing of bee products (e.g., formation of pollen-pellets and pollen-bread)62–64.
Several Biological Process (BP) GO terms enriched among DE orthologs were found to be typical or exclusively related to vertebrate species, such as camera-type eye morphogenesis, angiogenesis, and osteoblast differentiation. It is important to note that molecular data banks, including UniRef, predominantly rely on scientific evidence derived from a limited number of model species65. As a result, the nature and extent of GO annotations reflect the aspects investigated in these organisms being only extrapolated to other non-model species under the assumption that putative orthologs are functionally equivalent66. However, when dealing with species that are evolutionarily distant, orthologs may have completely different biological functions67. Therefore, extrapolating the biological significance of these top enriched BP terms to the invertebrate species investigated in this study is challenging. On the other hand, the enriched Molecular Functions (MF) and Cellular Components (CC) GO terms results were more relatable. The top enriched GO terms for the CC category were related to “ion channel complex”, “transporter complex”, and “plasma membrane protein complex”. Consistently, top enriched MF terms were related to “channel activity”, “transmembrane signaling receptor activity”, and “potassium ion transmembrane activity”. Most of the DE orthologs annotated along with these terms (Table 2) express proteins with a variety of functional roles that are putatively relevant for cleptoparasitic adaptations. In summary, in C. waltheriae upregulated orthologs annotated to these terms were related to olfaction, learning, neuronal excitability, synaptic organization, and other neuronal processes. Adaptations in these genetic pathways could be related to the successful exploitation of host resources and the development of specialized behaviors in cleptoparasitic bees. Meanwhile, orthologs highly expressed in T. diversipes were involved in chemosensory and sensory perception, particularly gustatory receptor genes involved in sweet taste response, motor, and mitochondrial activity, as well as neurotransmission, genetic mechanisms that could be associated with foraging and nest provisioning performed by T. diversipes foundresses. In boxes 1 and 2, we detailed the differentially expressed orthologs associated with the top MF and CC GO terms enriched by addressing their putative functions.
Cleptoparasitic bees are thought to rely on olfactory signals to locate host nests68,69. Indeed, morphological differences in antennae sensillae composition have been observed between cleptoparasites and their host species, with cleptoparasites having a greater prevalence of olfactory structures70. Previous studies have also shown that chemosensory-related genes (CRGs) are highly expressed in hymenopteran parasitoids71–74. Chemosensory cues, along with other external stimuli, are processed in sensory systems, triggering the formation of both long-term and short-term memories75. These adaptations may contribute to the localization and memorization of host nest aggregations76. In addition to the sensory-related genes described in box 1, we identified other potential chemosensory-related genes (CRGs) upregulated in the cleptoparasite, including Ionotropic Receptors (IRs), a gene family involved in olfaction, gustation, and other sensory perceptions77,78. Unlike many other chemosensory gene families, IRs exhibit a remarkable degree of conservation across species79. The IR25a homolog was found highly expressed in C. waltheriae. This gene, along with IR8a, functions as a co-receptor (IRco) for the formation of functional IR complexes in conjunction with a ligand-binding receptor protein (IR X)80. Furthermore, we detected an upregulated putative odorant receptor (OR) homologous to the Polistes dominula Or85c-like sequence in C. waltheriae. Although this specific OR's role in odour perception remains unknown, computational analysis in P. dominula suggests its involvement in parasitoid sensory perception. It is important to note that our current approach may have missed lineage-specific CRGs, and further investigation into these lineage-specific CRGs in cleptoparasitic lineages is crucial to gain insights into the underlying processes of parasitism in bees.
C. waltheriae and T. diversipes lineages, both belonging to the Apidae family, diverged over 77 million years ago11. Thus, compared to several cleptoparasites, that prefer hosts from other bee families (e.g., cleptoparasites of the Nomadinae clade)10, the phylogenetic proximity between C. waltheriae and T. diversipes could minimize the “phylogenetic noise” in transcriptomic comparisons. Still, cross-species transcriptomic comparisons present challenges, and certain caveats should be considered in the light of the present results. First, our analytical strategy focused on conserved orthologs, thus neglecting non-orthologous or rapidly evolving genes that may play a crucial role in cleptoparasitism evolution. Additionally, due to the divergent life history trajectories of these two species, the differentially expressed orthologs identified could be associated with non-behavioral or random species-specific adaptations and not directly related to cleptoparasitic adaptations. Thus, we argue that applying the methodology used in this study to other bee host-parasite species pairs should allow the differentiation of species-specific adaptations from shared molecular mechanisms. Lastly, we conducted a broad-scale transcriptome analysis, meaning that the RNA-Seq data used were obtained from whole-body extractions. As an exploratory analysis, this approach allowed some initial broad insights, but it is limited in capturing genes with tissue-specific expression or complex expression patterns across the body81. We also used sample replicates containing a variable number of pooled individuals for C. waltheriae, to account for this sampling strategy we normalized and treated the gene expression counts for batch effects across replicates, and this treatment might have reduced even further our power to detect genes with subtle expression differences. Further research should therefore delve into the obtained results by investigating the expression of these genes in a tissue-specific context, in order to elucidate their functions more accurately. Finally, we analyzed a specific life stage of each species by comparing females performing reproductive activities. We considered that this stage comprises some of the most distinguishing behavioural differences between the host and cleptoparasite species, yet comparative analyses of multiple life stages could provide a more detailed profile of cleptoparasitism across developmental stages.
Box 1: Orthologs upregulated in C. waltheriae and associated to top enriched GO terms
In C. waltheriae, we found upregulated orthologs that may have roles in the olfactory and learning pathways, potentially representing adaptations to cleptoparasitic behavior68,69.
Among the upregulated sequences in C. waltheriae, we discovered homologs to the invertebrate Octβ2R and vertebrate HTR2C, receptors for biogenic amines octopamine (OA) and serotonin (5-HT), respectively. Biogenic amines play important physiological roles in organisms, modulating neuronal, metabolic, and physiological processes82. In hymenopteran species, OA has been linked to locomotor activity, sensory (gustatory, olfactory, and visual) sensitivity, aggressive behavior, and (associative and non-associative) learning76,83,84. Octβ2R has been associated with rewarding reinforcement signaling in Drosophila, indicating its potential role in behavioral responses85. In honeybees, octopaminergic signaling is implicated in appetitive learning86. Overexpression of octopamine receptors in honeybees has also been associated with oxidative stress, neuroinflammation, and olfactory dysfunction87. Similarly, 5-HT acts as a neurotransmitter, neuromodulator, and hormone in insects88, regulating various behaviors such as aggression, mating, feeding, locomotion, and olfaction89–95. Invertebrate 5-HT receptors are found in the central and peripheral nervous systems (CNS and PNS, respectively), mediating both excitatory and inhibitory actions96,97. The signaling of 5-HT receptors in Drosophila is linked to aggression, sleep, circadian behavior, feeding, mating, learning, and memory98–103.
We also observed upregulation of the subunit of a neurotransmitter receptor in C. waltheriae, a homolog of the mammalian Gabbr2 (Gamma-aminobutyric acid type B receptor subunit 2). Meanwhile the Gabbr1 (Gamma-aminobutyric acid type B receptor subunit 1) was upregulated in T. diversipes. Gamma-aminobutyric acid (GABA) is the major neurotransmitter for inhibitory synaptic transmissions in the CNS of both vertebrates and invertebrates104,105. GABA signaling influences insect behaviors such as learning and memory, locomotor activity, and odor processing106–110. GABAB receptors, coupled with ionotropic GABAA receptors, mediate the action of GABA111. These receptors regulate complex behaviors and nervous system functions by inhibiting GABA release and reducing the release of other neurotransmitters112. For instance, in Drosophila and Heliothis virescens, GABAB receptors are involved in the olfactory pathway89,113. In Apis mellifera, GABAB is associated with locomotor behavior106.
In C. waltheriae, we also observed the upregulation of NaCP60E, a voltage-gated sodium channel. This gene has been mainly associated with olfactory function, but it is also expressed in other tissues114,115 and in invertebrates without olfaction116. In Drosophila, decreased expression of NaCP60E has been shown to decrease the olfactory response to benzaldehyde117. Also, a dense concentration of NaCP60E proteins in neuron axons of chemosensory organs suggests that this gene is involved in the olfactory system118. Thus, the upregulation of NaCP60E suggests its involvement in olfaction-related processes in the cleptoparasitic bee.
C. waltheriae also showed increased expression of potassium channel orthologs, specifically homologs of human KCNT1 and KCNK9 genes. KCNT1 encodes a sodium-gated potassium channel that is activated by neuromodulators119. Therefore, it is suggested that it regulates neuronal excitability and may play a role in several behaviors120. KCNK9 encodes a two-pore domain potassium channel that is involved in resting membrane potentials121. It can also be a target for modulatory molecules, such as volatile anesthetics and neurotransmitters122. Additionally, in Drosophila, KCNK9 orthologs (Task6 and Task7) are preferentially expressed in the antennal lobes and may play a role in olfactory memory formation123.
Furthermore, we identified in C. waltheriae the upregulation of genes associated with neuronal excitability, including circadian rhythm, memory formation, and development. These include Unc80, Hr38, and Lrp6. Unc80 is a regulatory component of the sodium leak channel NALCN complex124, which play a role in modulating resting membrane potential, neuronal excitability, firing rates, and pacemaker activity125,126. In Drosophila, for instance, Unc80 is necessary for circadian rhythmicity124. It is thought that Unc80 may also be involved in bee learning and memory31. The gene Hr38, a nuclear receptor, is involved in the transcriptional control of the dopamine synthesis pathway127, the cuticle gene expression128, and the ecdysteroid signaling pathways129. In addition, it has been suggested that Hr38 may play an important role in high neuronal functions such as memory formation, courtship behavior, and circadian rhythm130–132. Lrp6 belongs to the low-density lipoprotein receptor family of cell surface receptors and is an essential component of the Wnt signaling pathway, which controls several biological processes throughout the development and adult life of metazoans133. Expression of the Drosophila ortholog of Lrp6 (arr) has been associated with CNS morphogenesis and organization134, as well as long-term memory formation135.
We also found upregulated orthologs of cell adhesion molecules (CAMs) genes, including NRXN1 and Fmi/Stan. In Drosophila, NRXN1 gene is required for proper organization and growth at neuromuscular junctions136, and it also regulates sleep and synaptic plasticity137. In honeybees, the expression of NRXN1 is increased under sensory stimulation, suggesting a link between sensory processing and associative learning138. Drosophila Fmi/Stan encodes a cadherin that regulates planar cell polarity139, axon guidance in photoreceptor neurons140, dendrite morphogenesis of sensory neurons141, and neuronal morphogenesis of the mushroom body142. The upregulation of CAMs genes can be involved with their role in synaptic organization and plasticity, which may be relevant to the cleptoparasitic lifestyle of C. waltheriae.
Lastly, we found upregulated orthologs of ATPα and nrv2/nrv1 genes in C. waltheriae, which encode components of the Na + /K + ATPase143,144. Na + /K + ATPase is important for maintaining the balance of sodium and potassium ions across the cell membrane145. Defects in the sodium–potassium pump can lead to neuronal dysfunctions, such as circadian rhythm disturbances, locomotor problems, and auditory mechanosensation146–148.
Box 2: Orthologs upregulated in T. diversipes and associated to top enriched GO terms
T. diversipes exhibited upregulated sequences related to chemosensory perception, particularly in the context of gustatory receptor (GR) genes. One such sequence was identified in T. diversipes as the Gr5a gene while in C. waltheriae, the corresponding sequence was identified as the Gr64f. gene. Despite the difference in annotation, both genes belong to the same conserved subfamily that is involved in the sweet taste response observed in Drosophila149. Nevertheless, they are located on different chromosomes in Drosophila (Gr5a: X chromosome; Gr64f.: third chromosome)150. These receptors are considered the primary basis for sugar reception in Drosophila, and their co-expression in gustatory receptor neurons is necessary for the response to certain sugars149,151. Since bees primarily rely on flower nectar as an energy source, sugar detection is crucial for their survival152. Thus, a higher expression of these receptors in T. diversipes may be attributed to their intense foraging activity while provisioning food in their nests, an activity not performed by the parasite. Another upregulated sequence in T. diversipes was also related to GR, the transcript homologous to the G-sα60A gene of Anopheles gambiae. Studies on female antennae of A. gambiae have suggested that G-sα60A isoforms participate in olfactory signal transduction153. It has also been proposed that Gsα in Drosophila is responsible for Gr5a-mediated sweet taste perception154.
Apart from GR genes, several other upregulated genes in T. diversipes were associated with sensory perception but were notably involved in distinct pathways when compared to those upregulated in C. waltheriae. For example, the Drosophila Sh (Shaker) gene encodes a voltage-gated potassium channel (Kv)155 that is involved in shaping and firing the action potential156. It is also expressed in the retina and different regions of the CNS, including the mushroom body neuropil157,158. Furthermore, evidence suggests that Sh regulation affects olfactory learning and memory159. Another putative Kv upregulated in T. diversipes was a homolog to the vertebrate KCNQ1 gene, which regulates essential physiological processes all over the body160. In Drosophila, KCNQ1 ortholog (dKCNQ) is expressed in the brain cortical neurons, cardia, and in the nurse cells and oocytes in the ovary161,162. Apart from its role in the fly's normal heartbeat162 and early embryonic development161, dKCNQ has been implicated in age-dependent memory impairment and is required in α/β Mushroom Body Neurons for setting short-term memories163.
Another upregulated sequence in T. diversipes, homologous to the Drosophila trp (Transient Receptor Potential) gene, may also be related to sensory perception. TRP proteins have been associated with phototransduction and olfactory response to CO2 in Drosophila164,165.
An upregulated homolog of the Drosophila abl (Abelson) gene, which encodes a non-receptor tyrosine kinase, was also identified upregulated in T. diversipes. The Drosophila abl gene is expressed in the axons of the CNS and plays a very important role in establishing axonal connections during CNS development166. Also, it has been suggested that Abl-mediated phosphorylation is an important mechanism for the fly visual development167.
Motor activity-related genes also exhibited higher expression levels in T. diversipes. One such gene is the cacophony (Cac), which encodes a voltage-gated calcium channel that plays a role in regulating neurotransmitter release at neuromuscular synapses in Drosophila168. Similarly, a homolog of the Drosophila Ptp69D gene, which encodes a receptor of tyrosine phosphatase that is associated with mechanosensory neuron development169, was also upregulated gene in T. diversipes. Ptp69D might influence some elements of motor function in adult T. diversipes females.
Additionally, in T. diversipes, there was a higher expression of a homolog to the UQCRFS1 gene. This gene encodes an iron-sulfur protein (Rieske Fe-S protein—RISP)170 and is involved in electron transfer in bc1 complex171. Furthermore, T. diversipes displayed higher expression of homologs of other genes associated to electron transport activity, including genes with mitochondrial activity, indicating an increased energy demand during the founder life stage. These upregulated genes included, ETFA, MRPS18C, mdh2, Cox6al, and Ndufs2. The ETFA gene encodes an electron transfer flavoprotein subunit alpha, which is involved in mitochondrial fatty acid beta-oxidation and amino acid catabolism172. MRPS18C encodes a mitochondrial ribosomal protein that plays a role in protein synthesis within mitochondria173. The mdh2 gene encodes a malate dehydrogenase, an enzyme involved in the citric acid cycle174. Cox6al encodes a subunit of cytochrome c oxidase175, and Ndufs2 encodes a core subunit of NADH dehydrogenase (Complex I)176, another two of the critical enzymes in the electron transport chain.
Finally, several upregulated genes in T. diversipes were homologous to neurotransmitter receptors, including nAChRβ1, Grik2, and Grd. The nAChRβ1 gene encodes a subunit of the nicotinic acetylcholine receptor (nAChR), which is the primary excitatory neurotransmitter in insects. It is involved in olfactory learning and memory formation, as well as modulation of aggressive behavior177,178. Grik2 encodes the vertebrate glutamate receptor ionotropic kainate 2, which contribute to rapid synaptic transmission179. However, the role of kainate receptors in CNS glutamatergic circuits of insects is not well understood180. The Grd gene encodes a Drosophila chloride-channel homolog of the mammalian GABA receptor delta subunit that may respond to different neurotransmitters181. This gene has been linked to mediating the glycine response181 and forming functional ionotropic GABA receptors182. Additionally, an upregulated homolog of the Drosophila CG31760 gene was found in T. diversipes. CG31760 is a probable G-protein coupled receptor (GPCR), whose expression was highest in the adult fly brain however its exact function is unknown183.
Conclusion
In conclusion, our study provides global insights into the molecular mechanisms of cleptoparasitism in bees. We observed differential gene expression between the cleptoparasite C. waltheriae and its host T. diversipes particularly involving genes related to signal transduction, sensory perception, learning, and memory formation. These findings suggest the importance of sensory adaptations and learning in host-parasite interactions. We also identified a higher abundance of transcripts derived from transposable elements in C. waltheriae transcriptome, indicating these could be involved in gene neofunctionalization for parasite-specific adaptations. Additionally, the host species exhibited highly expressed genes from the Major Royal Jelly Proteins family. In bees, these genes are associated with various functions, including nutritional, immunity, and developmental regulation. We hypothesize that the differential expression of these genes could be related to nest provisioning in T. diversipes, a task not performed by the parasite. Further investigation is required to fully understand the role of all these molecular mechanisms in cleptoparasitism and their potential tissue-specific functions. Moreover, we propose that the methodology employed in the present study should be extended to other bee host-parasite species pairs, allowing for better differentiation of species-specific adaptations and shared molecular mechanisms underlying cleptoparasitism and its convergent traits in bees.
Material and methods
Sample collection and RNA sequencing
T. diversipes samples were collected and sequenced in a previous study33. Briefly, females were collected during their reproductive period (November to December of 2012, here called G1, and March to July of 2013, here called G2), at Universidade de São Paulo (23° 33′ 53.2″ S 46° 43′ 51.7″ W), while provisioning their nests. G1 and G2 represent the foundresses of different reproductive generations. Individuals were always sampled between 10:00 A.M. and 12:30 P.M., immediately frozen in liquid nitrogen and stored at − 80 °C. RNA extractions were performed individually using the whole body. Each sample replicated contained pooled RNA from three individuals. Here we included in the analyses RNA-Seq data from five replicates: three from G1 and two from G2. For more details see33.
Adult females of C. waltheriae were collected at the same location as T. diversipes samples while hovering at the entry of T. diversipes trap nesting spots. This behaviour is typical of C. waltheriae females looking for locations to lay their eggs21. Two individuals were collected in March 2013 (along with T. diversipes samples), and six more were collected later between November 2015 and March 2016. These individuals were also sampled between 10:00 A.M. and 12:30 P.M., immediately frozen in liquid nitrogen, and stored at − 80 °C. Total RNA extractions were performed for each individual separately using the RNeasy® Mini Kit (Qiagen, Austin, Texas, USA), following the manufacturer’s instructions. RNA was extracted from the whole body of the samples to enable a broad-scale transcriptomic analysis. Quantification and quality assessment of RNA was performed initially using NanoDrop™ Lite (Thermo Fisher Scientific, Wilmington, Delaware, USA) and later with the Agilent 2100 Bioanalyzer system (Agilent Technologies, Palo Alto, California, USA) before the library preparations. C. waltheriae samples were divided into five replicates: two containing only one individual in each (samples collected in 2013); and three containing the pooled RNA of two individuals (collected between 2015 and 2016). Library preparation and sequencing of the first two replicates (2013 samples) were performed by Macrogen (Macrogen Inc., Seoul, South Korea), along with T. diversipes samples from33, using the Illumina® HiSeq 2000 platform. The other three replicates of C. waltheriae (sampled from 2015–2016) were sequenced at LaCTAD (Unicamp, Campinas, Brazil), also using an Illumina® HiSeq 2000 sequencer.
C. waltheriae RNA sequencing resulted in 285,806,598 raw reads. After cleaning, read number decreased to 169,961,662. For T. diversipes, 343,059,264 raw reads were used, resulting in 190,339,784 cleaned reads after trimming.
Transcripts assembly
Reads quality assessment was performed using the FastQC 0.11.5184. Trimmomatic 0.38185 was used to quality trimming the raw reads (options: SLIDINGWINDOW:4:30 TRAILING:3 MINLEN:80 AVGQUAL:30 HEADCROP:14). Cleaned reads were then digitally normalized (20 × coverage) using a script (insilico read normalization) implemented within the Trinity toolbox186,187. Transcriptome assembly was performed differently for each species. C. waltheriae transcriptome was assembled following Trinity 2.10.0186,187 de novo assembly protocol with default parameters. For T. diversipes, a draft genome was available (Santos et al., unpublished), so two different approaches were used: (1) a genome-guided transcriptome assembly, using STAR 2.7.3188 and Cufflinks 2.2.1189; and (2) a reference guided de novo assembly with Trinity, using the bam file generated by the STAR software. The transcripts resulting from these two assemblies were clustered into SuperTranscripts190 by using CD-Hit191, Corset192, and Lace190. This clustering approach was also applied to the de novo assembly of C. waltheriae, to make the datasets more comparable between the two species and to reduce transcript redundancy in C. waltheriae. Downstream differential expression analyses were performed using the SuperTranscripts non-redundant datasets. Quality parameters of the transcriptomes were analyzed with Transrate 1.0.338 and BUSCO 5.2.2193 (using hymenoptera_odb10 database).
Annotation and expression analysis of orthologs
Non-redundant transcripts were annotated with Annocript 2.0.1194 pipeline using the UniProt Reference Clusters (UniRef90)195 and UniProtKB/Swiss-Prot196 databases from February 2021. Transcripts with significant blast hits (e-value < 1e−5) against possible contaminants (i.e., Acari, Alveolata, Archaea, Bacteria, Fungi, Rhizaria, Rhodophyta, Viridiplantae, and Viruses) in the UniRef90 database were removed from the final data set using custom scripts (https://github.com/PauloCseri/Annotation.git).
To identify orthologs between the two species the amino acid sequences of the resulting ORFs predicted in the Annocript pipeline were used as input to Orthofinder 2.1.140,41. Then, we selected the best matching transcripts per species in each orthogroup to get one-to-one ortholog data. This selection was performed by filtering the transcripts of the highest bitscore value (better alignment) between sequences of the two species in the same orthogroup. These alignments were performed using the blastp algorithm42,43 with default parameters. Differential expression (DE) analyses were then performed using only the orthologs identified in both species.
To estimate gene expression levels, we used Bowtie 2.3.5.1197 to align the cleaned reads to their respective ortholog transcripts set, i.e., C. waltheriae sample reads were aligned to C. waltheriae ortholog transcripts and T. diversipes reads to its corresponding set of ortholog transcripts. Then, we used RSEM 1.3.3198 for read counting. The significant DE orthologs between the two species were identified by combining the following approaches: (1) using the edgeR 3.34.045 with TMM normalized counts44; and (2) using NOISeq 2.36.047,48 with the normalization factor calculated through the SCBN method to normalize read counts46. This normalization strategy is optimized for cross-species DE analyses46 and is implemented in the Bioconductor package SCBN 1.10.0. Only significant DE orthologs (|Log2FC|≥ 2, adjusted p-value < 0.05) commonly identified as so in these two analyses were selected for the resulting set of DEs. It is worth mentioning that as the samples were not sequenced in the same batch (different years), we additionally adjusted these counts for batch effects using the ComBat-seq method199 available on the Bioconductor package sva 3.38.0200. In detail, for the batch parameter, we used the different sequencing times as factors (0: initial sequencing of C. waltheriae and T. diversipes samples; 1: later sequencing of C. waltheriae samples only) and for the group parameter (biological condition), we used the respective species (0: C. waltheriae; 1: T. diversipes).
GO functional analysis
Assuming that sequences from the same orthogroup descend from a single gene in the last common ancestor40,41 and hence they likely have similar functions, the ensemble of functional GO annotations from all SuperTranscripts belonging to the same orthogroup were used for functional analyses. The Bioconductor package PloGO2 1.4.0201 was used to plot and visualize the GO annotations of DE orthologs. To test whether any GO term was enriched among the DE orthologs in comparison to all other orthologs identified we used the Bioconductor package TopGO 2.20.0202. Redundant GO enriched terms were summarized in a two-level hierarchical GO set using the REVIGO web server203 for simplification, and these hierarchical sets were represented in charts generated by the CirGO 2.0 software204.
Supplementary Information
Acknowledgements
The authors acknowledge Susy Coelho Oliveira and Thiago Geronimo Pires Alegria for technical assistance, Isabel Alves dos-Santos and Guaraci Duran Cordeiro for support in specimen sampling, and Priscila Karla Ferreira dos Santos for initial assistance in data analysis.
Author contributions
P.C.R.: Methodology, investigation, data analyses and interpretation. N.S.A.: Conceptualization, methodology, sampling, assistance in data analyses and interpretation. M.C.A.: Conceptualization, supervision, fund acquisition. All authors contributed to the preparation of the manuscript.
Funding
This study was financed in part by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—CAPES, Brasil (scholarship to PCR, Proc. 1783961 and 88882.377416/2019-01, Finance Code 001), CNPq—Conselho Nacional de Desenvolvimento Científico e Tecnológico (research sponsorship to MCA—306932/2016-4) and Fundação de Amparo à Pesquisa do Estado de São Paulo—FAPESP (Proc. 2013/12530-4, 2016/24669-5 and 2019/23186-9). NSA is supported by the Fonds de la Recherche Scientifique—FNRS (Grant ID 40005980).
Data availability
The raw sequence reads have been deposited in the NCBI Sequence Read Archive (SRA) under the following accession numbers: SRR24187037, SRR24187038, SRR24187039, SRR24187040, SRR24187041, SRR24187042, SRR24187043, SRR24187044, SRR24187045, SRR24187046. The corresponding BioProject accession number is PRJNA955762. The transcriptome assemblies, as well as the sets of SuperTranscripts, are available on FigShare (10.6084/m9.figshare.23264771.v1).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-56261-5.
References
- 1.Schmidt GD, Roberts LS. Foundations of Parasitology. McGraw-Hill; 2009. [Google Scholar]
- 2.Phillips RS. Parasitism: The Variety of Parasites. Wiley; 2012. [Google Scholar]
- 3.Lefèvre T, et al. The ecological significance of manipulative parasites. Trends Ecol. Evol. 2009;24:41–48. doi: 10.1016/j.tree.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Dobson AP, Hudson PJ. Parasites, disease and the structure of ecological communities. Trends Ecol. Evol. 1986;1:11–15. doi: 10.1016/0169-5347(86)90060-1. [DOI] [PubMed] [Google Scholar]
- 5.Michener CD. The Bees of the World. The Johns Hopkins University Press; 2007. [Google Scholar]
- 6.Ascher, J. S. & Pickering, J. Discover Life Bee Species Guide and World Checklist (Hymenoptera: Apoidea: Anthophila). http://www.discoverlife.org/mp/20q?guide=Apoidea_species (2020).
- 7.Danforth BN, Minckley RL, Neff JL. The Solitary Bees. Princeton University Press; 2019. [Google Scholar]
- 8.Cardinal S, Straka J, Danforth BN. Comprehensive phylogeny of apid bees reveals the evolutionary origins and antiquity of cleptoparasitism. Proc. Natl. Acad. Sci. USA. 2010;107:16207–16211. doi: 10.1073/pnas.1006299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bossert S, et al. Combining transcriptomes and ultraconserved elements to illuminate the phylogeny of Apidae. Mol. Phylogenet. Evol. 2019;130:121–131. doi: 10.1016/j.ympev.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Sless TJL, et al. Phylogenetic relationships and the evolution of host preferences in the largest clade of brood parasitic bees (Apidae: Nomadinae) Mol. Phylogenet. Evol. 2022;166:107326. doi: 10.1016/j.ympev.2021.107326. [DOI] [PubMed] [Google Scholar]
- 11.Litman JR, Praz CJ, Danforth BN, Griswold TL, Cardinal S. Origins, evolution, and diversfication of cleptoparasitic lineages in long-tongued bees. Evolution. 2013;67:2982–2998. doi: 10.1111/evo.12161. [DOI] [PubMed] [Google Scholar]
- 12.Trunz V, Packer L, Vieu J, Arrigo N, Praz CJ. Comprehensive phylogeny, biogeography and new classification of the diverse bee tribe Megachilini: Can we use DNA barcodes in phylogenies of large genera? Mol. Phylogenet. Evol. 2016;103:245–259. doi: 10.1016/j.ympev.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez VH, Gustafson GT, Engel MS. Morphological phylogeny of Megachilini and the evolution of leaf-cutter behavior in bees (Hymenoptera: Megachilidae) J. Melittol. 2019;85:1–123. doi: 10.17161/jom.v0i85.11541. [DOI] [Google Scholar]
- 14.Magnacca KN, Danforth BN. Evolution and biogeography of native Hawaiian Hylaeus bees (Hymenoptera: Colletidae) Cladistics. 2006;22:393–411. doi: 10.1111/j.1096-0031.2006.00119.x. [DOI] [Google Scholar]
- 15.Danforth BN, et al. Phylogeny of Halictidae with an emphasis on endemic African Halictinae. Apidologie. 2008;39:86–101. doi: 10.1051/apido:2008002. [DOI] [Google Scholar]
- 16.Rozen JG. Eggs, ovariole numbers, and modes of parasitism of cleptoparasitic bees, with emphasis on neotropical species (Hymenoptera: Apoidea) Am. Mus. Novit. 2003;3413:1–36. doi: 10.1206/0003-0082(2003)413<0001:EONAMO>2.0.CO;2. [DOI] [Google Scholar]
- 17.Minckley RL, Danforth BN. Sources and frequency of brood loss in solitary bees. Apidologie. 2019;50:515–525. doi: 10.1007/s13592-019-00663-2. [DOI] [Google Scholar]
- 18.Sheffield CS, Pindar A, Packer L, Kevan PG. The potential of cleptoparasitic bees as indicator taxa for assessing bee communities. Apidologie. 2013;44:501–510. doi: 10.1007/s13592-013-0200-2. [DOI] [Google Scholar]
- 19.Oertli S, Müller A, Dorn S. Ecological and seasonal patterns in the diversity of a species-rich bee assemblage (Hymenoptera: Apoidea: Apiformes) Eur. J. Entomol. 2005;102:53–63. doi: 10.14411/eje.2005.008. [DOI] [Google Scholar]
- 20.Polidori C, Borruso L, Boesi R, Andrietti F. Segregation of temporal and spatial distribution between kleptoparasites and parasitoids of the eusocial sweat bee, Lasioglossum malachurum (Hymenoptera: Halictidae, Mutillidae) Entomol. Sci. 2009;12:116–129. doi: 10.1111/j.1479-8298.2009.00311.x. [DOI] [Google Scholar]
- 21.Alves-dos-Santos I, Melo GAR, Rozen JG. Biology and immature stages of the Bee Tribe Tetrapediini (Hymenoptera: Apidae) Am. Mus. Novit. 2002;3377:1–45. doi: 10.1206/0003-0082(2002)377<0001:BAISOT>2.0.CO;2. [DOI] [Google Scholar]
- 22.Moure, J. S. Tetrapediini Michener & Moure, 1957. In Catalogue of Bees (Hymenoptera, Apoidea) in the Neotropical Region—online version (2012).
- 23.Roig-Alsina A. Coelioxoides Cresson, a parasitic genus of Tetrapediini (Hymenoptera: Apoidea) J. Kans. Entomol. Soc. 1990;63:279–287. [Google Scholar]
- 24.Danforth BN, Cardinal S, Praz C, Almeida EAB, Michez D. The impact of molecular data on our understanding of bee phylogeny and evolution. Annu. Rev. Entomol. 2013;58:57–78. doi: 10.1146/annurev-ento-120811-153633. [DOI] [PubMed] [Google Scholar]
- 25.Martins AC, Luz DR, Melo GAR. Palaeocene origin of the Neotropical lineage of cleptoparasitic bees Ericrocidini-Rhathymini (Hymenoptera, Apidae) Syst. Entomol. 2018;43:510–521. doi: 10.1111/syen.12286. [DOI] [Google Scholar]
- 26.Rocha-Filho LC, Garófalo CA. Natural history of Tetrapedia diversipes (Hymenoptera: Apidae) in an Atlantic semideciduous forest remnant surrounded by coffee crops, Coffea arabica (Rubiaceae) Ann. Entomol. Soc. Am. 2016;109:183–197. doi: 10.1093/aesa/sav153. [DOI] [Google Scholar]
- 27.Araújo PCS, Lourenço AP, Raw A. Trap-nesting bees in montane grassland (Campo Rupestre) and Cerrado in Brazil: Collecting generalist or specialist nesters. Neotrop. Entomol. 2016;45:482–489. doi: 10.1007/s13744-016-0395-9. [DOI] [PubMed] [Google Scholar]
- 28.Lima R, Oliveira DM, Garófalo CA. Interaction network and niche analysis of natural enemy communities and their host bees (Hymenoptera: Apoidea) in fragments of Cerrado and Atlantic forest. Sociobiology. 2018;65:591. doi: 10.13102/sociobiology.v65i4.3386. [DOI] [Google Scholar]
- 29.Feng W, et al. Understanding of waggle dance in the honey bee (Apis mellifera) from the perspective of long non-coding RNA. Insects. 2022;13:111. doi: 10.3390/insects13020111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mondet F, et al. Antennae hold a key to Varroa-sensitive hygiene behaviour in honey bees. Sci. Rep. 2015;5:10454. doi: 10.1038/srep10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fahad Raza M, et al. Differential gene expression analysis following olfactory learning in honeybee (Apis mellifera L.) PLoS ONE. 2022;17:e0262441. doi: 10.1371/journal.pone.0262441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L, et al. Large-scale transcriptome changes in the process of long-term visual memory formation in the bumblebee, Bombus terrestris. Sci. Rep. 2018;8:534. doi: 10.1038/s41598-017-18836-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Araujo NS, Santos PKF, Arias MC. RNA-Seq reveals that mitochondrial genes and long non-coding RNAs may play important roles in the bivoltine generations of the non-social Neotropical bee Tetrapedia diversipes. Apidologie. 2018;49:3–12. doi: 10.1007/s13592-017-0542-2. [DOI] [Google Scholar]
- 34.Santos PKF, de Souza Araujo N, Françoso E, Zuntini AR, Arias MC. Diapause in a tropical oil-collecting bee: Molecular basis unveiled by RNA-Seq. BMC Genom. 2018;19:305. doi: 10.1186/s12864-018-4694-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Araujo NS, Arias MC. Gene expression and epigenetics reveal species-specific mechanisms acting upon common molecular pathways in the evolution of task division in bees. Sci. Rep. 2021;11:3654. doi: 10.1038/s41598-020-75432-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berens AJ, Hunt JH, Toth AL. Comparative transcriptomics of convergent evolution: Different genes but conserved pathways underlie caste phenotypes across lineages of eusocial insects. Mol. Biol. Evol. 2015;32:690–703. doi: 10.1093/molbev/msu330. [DOI] [PubMed] [Google Scholar]
- 37.Saleh NW, Ramírez SR. Sociality emerges from solitary behaviours and reproductive plasticity in the orchid bee Euglossa dilemma. Proc. R. Soc. B. 2019;286:20190588. doi: 10.1098/rspb.2019.0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith-Unna R, Boursnell C, Patro R, Hibberd JM, Kelly S. TransRate: Reference-free quality assessment of de novo transcriptome assemblies. Genome Res. 2016;26:1134–1144. doi: 10.1101/gr.196469.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Araujo NS, Zuntini AR, Arias MC. Getting useful information from RNA-Seq contaminants: A case of study in the oil-collecting bee Tetrapedia diversipes transcriptome. OMICS. 2016;20:491–492. doi: 10.1089/omi.2016.0054. [DOI] [PubMed] [Google Scholar]
- 40.Emms DM, Kelly S. OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emms DM, Kelly S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul S. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Y, et al. A statistical normalization method and differential expression analysis for RNA-seq data between different species. BMC Bioinform. 2019;20:163. doi: 10.1186/s12859-019-2745-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarazona S, et al. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015;43:gkv711. doi: 10.1093/nar/gkv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tarazona S, García-Alcalde F, Dopazo J, Ferrer A, Conesa A. Differential expression in RNA-seq: A matter of depth. Genome Res. 2011;21:2213–2223. doi: 10.1101/gr.124321.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fraser MJ, Smith GE, Summers MD. Acquisition of host cell DNA sequences by baculoviruses: Relationship between Host DNA insertions and FP mutants of Autographa californica and Galleria mellonella nuclear polyhedrosis viruses. J. Virol. 1983;47:287–300. doi: 10.1128/jvi.47.2.287-300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cary LC, et al. Transposon mutagenesis of baculoviruses: Analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–169. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- 51.Bouallègue M, Rouault JD, Hua-Van A, Makni M, Capy P. Molecular evolution of piggyBac superfamily: From selfishness to domestication. Genome Biol. Evol. 2017;9:323–339. doi: 10.1093/gbe/evw292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Volff J-N. Turning junk into gold: Domestication of transposable elements and the creation of new genes in eukaryotes. BioEssays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- 53.Jangam D, Feschotte C, Betrán E. Transposable element domestication as an adaptation to evolutionary conflicts. Trends Genet. 2017;33:817–831. doi: 10.1016/j.tig.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sless TJL, Searle JB, Danforth BN. Genome of the bee Holcopasites calliopsidis: A species showing the common apid trait of brood parasitism. G3. 2022;12:160. doi: 10.1093/g3journal/jkac160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ferguson LC, Green J, Surridge A, Jiggins CD. Evolution of the insect Yellow gene family. Mol. Biol. Evol. 2011;28:257–272. doi: 10.1093/molbev/msq192. [DOI] [PubMed] [Google Scholar]
- 56.Buttstedt A, Moritz RFA, Erler S. Origin and function of the major royal jelly proteins of the honeybee (Apis mellifera) as members of the yellow gene family. Biol. Rev. 2014;89:255–269. doi: 10.1111/brv.12052. [DOI] [PubMed] [Google Scholar]
- 57.Hojo M, Kagami T, Sasaki T, Nakamura J, Sasaki M. Reduced expression of major royal jelly protein 1 gene in the mushroom bodies of worker honeybees with reduced learning ability. Apidologie. 2010;41:194–202. doi: 10.1051/apido/2009075. [DOI] [Google Scholar]
- 58.Kucharski R, Maleszka R, Hayward DC, Ball EE. A royal jelly protein is expressed in a subset of kenyon cells in the mushroom bodies of the honey bee brain. Naturwissenschaften. 1998;85:343–346. doi: 10.1007/s001140050512. [DOI] [PubMed] [Google Scholar]
- 59.Dobritzsch D, Aumer D, Fuszard M, Erler S, Buttstedt A. The rise and fall of major royal jelly proteins during a honeybee (Apis mellifera) workers’ life. Ecol. Evol. 2019;9:8771–8782. doi: 10.1002/ece3.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winkler P, Sieg F, Buttstedt A. Transcriptional control of honey bee (Apis mellifera) major royal jelly proteins by 20-hydroxyecdysone. Insects. 2018;9:122. doi: 10.3390/insects9030122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kupke J, Spaethe J, Mueller MJ, Rössler W, Albert Š. Molecular and biochemical characterization of the major royal jelly protein in bumblebees suggest a non-nutritive function. Insect Biochem. Mol. Biol. 2012;42:647–654. doi: 10.1016/j.ibmb.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 62.Fratini F, Cilia G, Mancini S, Felicioli A. Royal Jelly: An ancient remedy with remarkable antibacterial properties. Microbiol. Res. 2016;192:130–141. doi: 10.1016/j.micres.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 63.Park HG, et al. Antibacterial activity of major royal jelly proteins of the honeybee (Apis mellifera) royal jelly. J. Asia Pac. Entomol. 2019;22:737–741. doi: 10.1016/j.aspen.2019.06.005. [DOI] [Google Scholar]
- 64.Šimúth J. Some properties of the main protein of honeybee (Apis mellifera) royal jelly. Apidologie. 2001;32:69–80. doi: 10.1051/apido:2001112. [DOI] [Google Scholar]
- 65.Hassani-Pak K, Rawlings C. Knowledge discovery in biological databases for revealing candidate genes linked to complex phenotypes. J. Integr. Bioinform. 2017;14:20160002. doi: 10.1515/jib-2016-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaudet P, Dessimoz C. Gene ontology: Pitfalls, biases, and remedies. Methods Mol. Biol. 2017;1446:189–205. doi: 10.1007/978-1-4939-3743-1_14. [DOI] [PubMed] [Google Scholar]
- 67.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 68.Cane JH. Olfactory evaluation of Andrena host nest suitability by kleptoparasitic Nomada bees (Hymenoptera: Apoidea) Anim. Behav. 1983;31:138–144. doi: 10.1016/S0003-3472(83)80181-X. [DOI] [Google Scholar]
- 69.Dotterl S. Antennal responses of an oligolectic bee and its cleptoparasite to plant volatiles. Plant Signal. Behav. 2008;3:296–297. doi: 10.4161/psb.3.5.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Galvani GL, González-Vaquero RA, Guerra-Navarro C, Settembrini BP. Antennal sensilla of cleptoparasitic and non-parasitic bees in two subfamilies of Apidae. Apidologie. 2017;48:437–449. doi: 10.1007/s13592-016-0486-y. [DOI] [Google Scholar]
- 71.Qi Y, et al. Transcriptome analysis of an endoparasitoid wasp Cotesia chilonis (Hymenoptera: Braconidae) reveals genes involved in successful parasitism. Arch Insect Biochem. Physiol. 2015;88:203–221. doi: 10.1002/arch.21214. [DOI] [PubMed] [Google Scholar]
- 72.Zhou C-X, Min S-F, Yan-Long T, Wang M-Q. Analysis of antennal transcriptome and odorant binding protein expression profiles of the recently identified parasitoid wasp, Sclerodermus sp. Comp. Biochem. Physiol. Part D. 2015;16:10–19. doi: 10.1016/j.cbd.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 73.Zhao Y, et al. Transcriptome and expression patterns of chemosensory genes in antennae of the parasitoid wasp Chouioia cunea. PLoS ONE. 2016;11:e0148159. doi: 10.1371/journal.pone.0148159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nie XP, et al. Antennal transcriptome and odorant binding protein expression profiles of an invasive mealybug and its parasitoid. J. Appl. Entomol. 2018;142:149–161. doi: 10.1111/jen.12417. [DOI] [Google Scholar]
- 75.Leavell BC, Bernal XE. The cognitive ecology of stimulus ambiguity: A predator-prey perspective. Trends Ecol. Evol. 2019;34:1048–1060. doi: 10.1016/j.tree.2019.07.004. [DOI] [PubMed] [Google Scholar]
- 76.Haverkamp A, Smid HM. A neuronal arms race: The role of learning in parasitoid–host interactions. Curr. Opin. Insect Sci. 2020;42:47–54. doi: 10.1016/j.cois.2020.09.003. [DOI] [PubMed] [Google Scholar]
- 77.Rimal S, Lee Y. The multidimensional ionotropic receptors of Drosophila melanogaster. Insect. Mol. Biol. 2018;27:1–7. doi: 10.1111/imb.12347. [DOI] [PubMed] [Google Scholar]
- 78.van Giesen L, Garrity PA. More than meets the IR: The expanding roles of variant Ionotropic Glutamate Receptors in sensing odor, taste, temperature and moisture. F1000Res. 2017;6:1753. doi: 10.12688/f1000research.12013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eyun S, et al. Evolutionary history of chemosensory-related gene families across the arthropoda. Mol. Biol. Evol. 2017;34:1838–1862. doi: 10.1093/molbev/msx147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Breer H, Fleischer J, Pregitzer P, Krieger J. Olfactory Concepts of Insect Control: Alternative to insecticides. Springer International Publishing; 2019. Molecular mechanism of insect olfaction: Olfactory receptors; pp. 93–114. [Google Scholar]
- 81.Johnson BR, Atallah J, Plachetzki DC. The importance of tissue specificity for RNA-seq: Highlighting the errors of composite structure extractions. BMC Genom. 2013;14:586. doi: 10.1186/1471-2164-14-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sinakevitch IT, Wolff GH, Pflüger H-J, Smith BH. Editorial: Biogenic amines and neuromodulation of animal behavior. Front. Syst. Neurosci. 2018;12:31. doi: 10.3389/fnsys.2018.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blenau W, Baumann A. Trace Amines and Neurological Disorders. Elsevier; 2016. Octopaminergic and tyraminergic signaling in the honeybee (Apis mellifera) Brain; pp. 203–219. [Google Scholar]
- 84.Manfredini F, Brown MJF, Toth AL. Candidate genes for cooperation and aggression in the social wasp Polistes dominula. J. Comp. Physiol. A. 2018;204:449–463. doi: 10.1007/s00359-018-1252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Burke CJ, et al. Layered reward signalling through octopamine and dopamine in Drosophila. Nature. 2012;492:433–437. doi: 10.1038/nature11614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Agarwal M, et al. Dopamine and octopamine influence avoidance learning of honey bees in a place preference assay. PLoS ONE. 2011;6:e25371. doi: 10.1371/journal.pone.0025371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Farooqui T. A potential link among biogenic amines-based pesticides, learning and memory, and colony collapse disorder: A unique hypothesis. Neurochem. Int. 2013;62:122–136. doi: 10.1016/j.neuint.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 88.Monastirioti M. Biogenic amine systems in the fruit fly Drosophila melanogaster. Microsc. Res. Tech. 1999;45:106–121. doi: 10.1002/(SICI)1097-0029(19990415)45:2<106::AID-JEMT5>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 89.Dacks AM, Green DS, Root CM, Nighorn AJ, Wang JW. Serotonin modulates olfactory processing in the antennal lobe of Drosophila. J. Neurogenet. 2009;23:366–377. doi: 10.3109/01677060903085722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang X, Gaudry Q. Functional integration of a serotonergic neuron in the Drosophila antennal lobe. Elife. 2016;5:16836. doi: 10.7554/eLife.16836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ro J, et al. Serotonin signaling mediates protein valuation and aging. Elife. 2016;5:16843. doi: 10.7554/eLife.16843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Neckameyer WS, Coleman CM, Eadie S, Goodwin SF. Compartmentalization of neuronal and peripheral serotonin synthesis in Drosophila melanogaster. Genes Brain Behav. 2007;6:756–769. doi: 10.1111/j.1601-183X.2007.00307.x. [DOI] [PubMed] [Google Scholar]
- 93.Kamyshev NG, Smirnova GP, Savvateeva EV, Medvedeva AV, Ponomarenko VV. The influence of serotonin and p-chlorophenylalanine on locomotor activity of Drosophila melanogaster. Pharmacol. Biochem. Behav. 1983;18:677–681. doi: 10.1016/0091-3057(83)90005-9. [DOI] [PubMed] [Google Scholar]
- 94.Alekseyenko OV, et al. Single serotonergic neurons that modulate aggression in Drosophila. Curr. Biol. 2014;24:2700–2707. doi: 10.1016/j.cub.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pooryasin A, Fiala A. Identified serotonin-releasing neurons induce behavioral quiescence and suppress mating in Drosophila. J. Neurosci. 2015;35:12792–12812. doi: 10.1523/JNEUROSCI.1638-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tierney AJ. Structure and function of invertebrate 5-HT receptors: A review. Comp. Biochem. Physiol. A. 2001;128:791–804. doi: 10.1016/S1095-6433(00)00320-2. [DOI] [PubMed] [Google Scholar]
- 97.Barnes NM, Sharp T. A review of central 5-HT receptors and their function. Neuropharmacology. 1999;38:1083–1152. doi: 10.1016/S0028-3908(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 98.Nichols CD. 5-HT2 receptors in Drosophila are expressed in the brain and modulate aspects of circadian behaviors. Dev. Neurobiol. 2007;67:752–763. doi: 10.1002/dneu.20370. [DOI] [PubMed] [Google Scholar]
- 99.Yuan Q, Joiner WJ, Sehgal A. A sleep-promoting role for the Drosophila serotonin receptor 1A. Curr. Biol. 2006;16:1051–1062. doi: 10.1016/j.cub.2006.04.032. [DOI] [PubMed] [Google Scholar]
- 100.Johnson O, Becnel J, Nichols CD. Serotonin 5-HT2 and 5-HT1A-like receptors differentially modulate aggressive behaviors in Drosophila melanogaster. Neuroscience. 2009;158:1292–1300. doi: 10.1016/j.neuroscience.2008.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lyu Y, et al. Drosophila serotonin 2A receptor signaling coordinates central metabolic processes to modulate aging in response to nutrient choice. Elife. 2021;10:1–67. doi: 10.7554/eLife.59399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Johnson O, Becnel J, Nichols CD. Serotonin receptor activity is necessary for olfactory learning and memory in Drosophila melanogaster. Neuroscience. 2011;192:372–381. doi: 10.1016/j.neuroscience.2011.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Becnel J, Johnson O, Luo J, Nässel DR, Nichols CD. The serotonin 5-HT7Dro receptor is expressed in the brain of Drosophila, and is essential for normal courtship and mating. PLoS ONE. 2011;6:e20800. doi: 10.1371/journal.pone.0020800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Goetz T, Arslan A, Wisden W, Wulff P. GABAA receptors: Structure and function in the basal ganglia. Prog. Brain Res. 2007;160:21–41. doi: 10.1016/S0079-6123(06)60003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lummis SCR. GABA receptors in insects. Comp. Biochem. Physiol. C. 1990;95:1–8. doi: 10.1016/0742-8413(90)90073-I. [DOI] [PubMed] [Google Scholar]
- 106.Mustard JA, Jones L, Wright GA. GABA signaling affects motor function in the honey bee. J. Insect Physiol. 2020;120:103989. doi: 10.1016/j.jinsphys.2019.103989. [DOI] [PubMed] [Google Scholar]
- 107.Leal SM, Neckameyer WS. Pharmacological evidence for GABAergic regulation of specific behaviors in Drosophila melanogaster. J. Neurobiol. 2002;50:245–261. doi: 10.1002/neu.10030. [DOI] [PubMed] [Google Scholar]
- 108.Raccuglia D, Mueller U. Temporal integration of cholinergic and GABAergic inputs in isolated insect mushroom body neurons exposes pairing-specific signal processing. J. Neurosci. 2014;34:16086–16092. doi: 10.1523/JNEUROSCI.0714-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Choudhary AF, Laycock I, Wright GA. γ-Aminobutyric acid receptor A-mediated inhibition in the honeybee’s antennal lobe is necessary for the formation of configural olfactory percepts. Eur. J. Neurosci. 2012;35:1718–1724. doi: 10.1111/j.1460-9568.2012.08090.x. [DOI] [PubMed] [Google Scholar]
- 110.Dupuis JP, et al. Homomeric RDL and heteromeric RDL/LCCH3 GABA receptors in the honeybee antennal lobes: Two candidates for inhibitory transmission in olfactory processing. J. Neurophysiol. 2010;103:458–468. doi: 10.1152/jn.00798.2009. [DOI] [PubMed] [Google Scholar]
- 111.Bettler B, Kaupmann K, Mosbacher J, Gassmann M. Molecular structure and physiological functions of GABAB receptors. Physiol. Rev. 2004;84:835–867. doi: 10.1152/physrev.00036.2003. [DOI] [PubMed] [Google Scholar]
- 112.Pinard A, Seddik R, Bettler B. GABAB receptors: Physiological functions and mechanisms of diversity. Adv. Pharmacol. 2010;58:231–255. doi: 10.1016/S1054-3589(10)58010-4. [DOI] [PubMed] [Google Scholar]
- 113.Pregitzer P, Schultze A, Raming K, Breer H, Krieger J. Expression of a GABAB: Receptor in olfactory sensory neurons of sensilla trichodea on the male antenna of the Moth Heliothis virescens. Int. J. Biol. Sci. 2013;9:707–715. doi: 10.7150/ijbs.6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gosselin-Badaroudine P, et al. Biophysical characterization of the honeybee DSC1 orthologue reveals a novel voltage-dependent Ca2+ channel subfamily: Cav4. J. Gen. Physiol. 2016;148:133–145. doi: 10.1085/jgp.201611614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hong C, Ganetzky B. Spatial and temporal expression patterns of two sodium channel genes in Drosophila. J. Neurosci. 1994;14:5160–5169. doi: 10.1523/JNEUROSCI.14-09-05160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liebeskind BJ, Hillis DM, Zakon HH. Evolution of sodium channels predates the origin of nervous systems in animals. Proc. Natl. Acad. Sci. USA. 2011;108:9154–9159. doi: 10.1073/pnas.1106363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kulkarni NH, Yamamoto AH, Robinson KO, Mackay TFC, Anholt RRH. The DSC1 channel, encoded by the smi60E locus, contributes to odor-guided behavior in Drosophila melanogaster. Genetics. 2002;161:1507–1516. doi: 10.1093/genetics/161.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Castella C, Amichot M, Bergé J-B, Pauron D. DSC1 channels are expressed in both the central and the peripheral nervous system of adult Drosophila melanogaster. Invertebr. Neurosci. 2001;4:85–94. doi: 10.1007/s101580100010. [DOI] [PubMed] [Google Scholar]
- 119.Santi CM. Opposite regulation of slick and slack K+ channels by neuromodulators. J. Neurosci. 2006;26:5059–5068. doi: 10.1523/JNEUROSCI.3372-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Budelli G, et al. SLO2 channels are inhibited by all divalent cations that activate SLO1 K+ channels. J. Biol. Chem. 2016;291:7347–7356. doi: 10.1074/jbc.M115.709436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Goldstein SAN, Bockenhauer D, O’Kelly I, Zilberberg N. Potassium leak channels and the KCNK family of two-p-domain subunits. Nat. Rev. Neurosci. 2001;2:175–184. doi: 10.1038/35058574. [DOI] [PubMed] [Google Scholar]
- 122.Talley EM, Bayliss DA. Modulation of TASK-1 (Kcnk3) and TASK-3 (Kcnk9) potassium channels. J. Biol. Chem. 2002;277:17733–17742. doi: 10.1074/jbc.M200502200. [DOI] [PubMed] [Google Scholar]
- 123.Döring F, Scholz H, Kühnlein RP, Karschin A, Wischmeyer E. Novel Drosophila two-pore domain K+ channels: Rescue of channel function by heteromeric assembly. Eur. J. Neurosci. 2006;24:2264–2274. doi: 10.1111/j.1460-9568.2006.05102.x. [DOI] [PubMed] [Google Scholar]
- 124.Lear BC, et al. UNC79 and UNC80, putative auxiliary subunits of the NARROW ABDOMEN ion channel, are indispensable for robust circadian locomotor rhythms in Drosophila. PLoS ONE. 2013;8:e78147. doi: 10.1371/journal.pone.0078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Flourakis M, et al. A conserved bicycle model for circadian clock control of membrane excitability. Cell. 2015;162:836–848. doi: 10.1016/j.cell.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shi Y, et al. Nalcn is a ‘leak’ sodium channel that regulates excitability of brainstem chemosensory neurons and breathing. J. Neurosci. 2016;36:8174–8187. doi: 10.1523/JNEUROSCI.1096-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sekine Y, et al. p38 MAPKs regulate the expression of genes in the dopamine synthesis pathway through phosphorylation of NR4A nuclear receptors. J. Cell Sci. 2011;124:3006–3016. doi: 10.1242/jcs.085902. [DOI] [PubMed] [Google Scholar]
- 128.Kozlova T, Lam G, Thummel CS. Drosophila DHR38 nuclear receptor is required for adult cuticle integrity at eclosion. Dev. Dyn. 2009;238:701–707. doi: 10.1002/dvdy.21860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Baker KD, et al. The Drosophila orphan nuclear receptor DHR38 mediates an atypical ecdysteroid signaling pathway. Cell. 2003;113:731–742. doi: 10.1016/S0092-8674(03)00420-3. [DOI] [PubMed] [Google Scholar]
- 130.Fujita N, et al. Visualization of neural activity in insect brains using a conserved immediate early gene, Hr38. Curr. Biol. 2013;23:2063–2070. doi: 10.1016/j.cub.2013.08.051. [DOI] [PubMed] [Google Scholar]
- 131.Takayanagi-Kiya S, Kiya T. Activity-dependent visualization and control of neural circuits for courtship behavior in the fly Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2019;116:5715–5720. doi: 10.1073/pnas.1814628116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu W, et al. Dibutyl phthalate disrupts conserved circadian rhythm in Drosophila and human cells. Sci. Total Environ. 2021;783:147038. doi: 10.1016/j.scitotenv.2021.147038. [DOI] [PubMed] [Google Scholar]
- 133.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 134.Koizumi K, et al. RNA interference screen to identify genes required for Drosophila embryonic nervous system development. Proc. Natl. Acad. Sci. USA. 2007;104:5626–5631. doi: 10.1073/pnas.0611687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tan Y, Yu D, Busto GU, Wilson C, Davis RL. Wnt signaling is required for long-term memory formation. Cell Rep. 2013;4:1082–1089. doi: 10.1016/j.celrep.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Banerjee S, Venkatesan A, Bhat MA. Neurexin, Neuroligin and Wishful Thinking coordinate synaptic cytoarchitecture and growth at neuromuscular junctions. Mol. Cell. Neurosci. 2017;78:9–24. doi: 10.1016/j.mcn.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Larkin A, et al. Neurexin-1 regulates sleep and synaptic plasticity in Drosophila melanogaster. Eur. J. Neurosci. 2015;42:2455–2466. doi: 10.1111/ejn.13023. [DOI] [PubMed] [Google Scholar]
- 138.Biswas S, et al. Sensory regulation of Neuroligins and Neurexin I in the honeybee brain. PLoS ONE. 2010;5:e9133. doi: 10.1371/journal.pone.0009133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Usui T, et al. Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of frizzled. Cell. 1999;98:585–595. doi: 10.1016/S0092-8674(00)80046-X. [DOI] [PubMed] [Google Scholar]
- 140.Hakeda-Suzuki S, et al. Golden Goal collaborates with Flamingo in conferring synaptic-layer specificity in the visual system. Nat. Neurosci. 2011;14:314–323. doi: 10.1038/nn.2756. [DOI] [PubMed] [Google Scholar]
- 141.Kimura H, Usui T, Tsubouchi A, Uemura T. Potential dual molecular interaction of the Drosophila 7-pass transmembrane cadherin Flamingo in dendritic morphogenesis. J. Cell Sci. 2006;119:1118–1129. doi: 10.1242/jcs.02832. [DOI] [PubMed] [Google Scholar]
- 142.Reuter JE, et al. A mosaic genetic screen for genes necessary for Drosophila mushroom body neuronal morphogenesis. Development. 2003;130:1203–1213. doi: 10.1242/dev.00319. [DOI] [PubMed] [Google Scholar]
- 143.Lebovitz RM, Takeyasu K, Fambrough DM. Molecular characterization and expression of the (Na+ + K+)-ATPase alpha-subunit in Drosophila melanogaster. EMBO J. 1989;8:193–202. doi: 10.1002/j.1460-2075.1989.tb03364.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sun B, Salvaterra PM. Two Drosophila nervous system antigens, Nervana 1 and 2, are homologous to the beta subunit of Na+, K+-ATPase. Proc. Natl. Acad. Sci. USA. 1995;92:5396–5400. doi: 10.1073/pnas.92.12.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Palladino MJ, Bower JE, Kreber R, Ganetzky B. Neural dysfunction and neurodegeneration in Drosophila Na+ /K+ ATPase alpha subunit mutants. J. Neurosci. 2003;23:1276–1286. doi: 10.1523/JNEUROSCI.23-04-01276.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Roy M, Sivan-Loukianova E, Eberl DF. Cell-type–specific roles of Na+ /K+ ATPase subunits in Drosophila auditory mechanosensation. Proc. Natl. Acad. Sci. USA. 2013;110:181–186. doi: 10.1073/pnas.1208866110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Damulewicz M, Rosato E, Pyza E. Circadian regulation of the Na+/K+-Atpase alpha subunit in the visual system is mediated by the pacemaker and by retina photoreceptors in Drosophila melanogaster. PLoS ONE. 2013;8:e73690. doi: 10.1371/journal.pone.0073690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Trotta N, Rodesch CK, Fergestad T, Broadie K. Cellular bases of activity-dependent paralysis in Drosophila stress-sensitive mutants. J. Neurobiol. 2004;60:328–347. doi: 10.1002/neu.20017. [DOI] [PubMed] [Google Scholar]
- 149.Dahanukar A, Lei Y-T, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56:503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ueno K, et al. Trehalose sensitivity in Drosophila correlates with mutations in and expression of the gustatory receptor gene Gr5a. Curr. Biol. 2001;11:1451–1455. doi: 10.1016/S0960-9822(01)00450-X. [DOI] [PubMed] [Google Scholar]
- 151.Jiao Y, Moon SJ, Wang X, Ren Q, Montell C. Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr. Biol. 2008;18:1797–1801. doi: 10.1016/j.cub.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jung JW, Park KW, Ahn Y-J, Kwon HW. Functional characterization of sugar receptors in the western honeybee, Apis mellifera. J. Asia Pac. Entomol. 2015;18:19–26. doi: 10.1016/j.aspen.2014.10.011. [DOI] [Google Scholar]
- 153.Rützler M, Lu T, Zwiebel LJ. Gα encoding gene family of the malaria vector mosquito Anopheles gambiae: Expression analysis and immunolocalization of AGαq and AGαo in female antennae. J. Comp. Neurol. 2006;499:533–545. doi: 10.1002/cne.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Ueno K, et al. Gsα is involved in sugar perception in Drosophila melanogaster. J. Neurosci. 2006;26:6143–6152. doi: 10.1523/JNEUROSCI.0857-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Papazian DM, Schwarz TL, Tempel BL, Jan YN, Jan LY. Cloning of genomic and complementary DNA from Shaker, a putative potassium channel gene from Drosophila. Science. 1987;1979(237):749–753. doi: 10.1126/science.2441470. [DOI] [PubMed] [Google Scholar]
- 156.Wicher D, Walther C, Wicher C. Non-synaptic ion channels in insects: Basic properties of currents and their modulation in neurons and skeletal muscles. Prog. Neurobiol. 2001;64:431–525. doi: 10.1016/S0301-0082(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 157.Mottes JR, Iverson LE. Tissue-specific alternative splicing of hybrid Shaker/lacZ genes correlates with kinetic differences in Shaker K+ currents in vivo. Neuron. 1995;14:613–623. doi: 10.1016/0896-6273(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 158.Rogero O, Hämmerle B, Tejedor FJ. Diverse expression and distribution of Shaker potassium channels during the development of the Drosophila nervous system. J. Neurosci. 1997;17:5108–5118. doi: 10.1523/JNEUROSCI.17-13-05108.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cowan TM, Siegel RW. Drosophila mutations that alter ionic conduction disrupt acquisition and retention of a conditioned odor avoidance response. J. Neurogenet. 1986;3:187–201. doi: 10.3109/01677068609106849. [DOI] [PubMed] [Google Scholar]
- 160.Jespersen T, Grunnet M, Olesen S-P. The KCNQ1 potassium channel: From gene to physiological function. Physiology. 2005;20:408–416. doi: 10.1152/physiol.00031.2005. [DOI] [PubMed] [Google Scholar]
- 161.Wen H, et al. A Drosophila KCNQ channel essential for early embryonic development. J. Neurosci. 2005;25:10147–10156. doi: 10.1523/JNEUROSCI.3086-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ocorr K, et al. KCNQ potassium channel mutations cause cardiac arrhythmias in Drosophila that mimic the effects of aging. Proc. Natl. Acad. Sci. USA. 2007;104:3943–3948. doi: 10.1073/pnas.0609278104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cavaliere S, Malik BR, Hodge JJL. KCNQ channels regulate age-related memory impairment. PLoS ONE. 2013;8:e62445. doi: 10.1371/journal.pone.0062445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Agam K, et al. Metabolic stress reversibly activates the Drosophila light-sensitive channels TRP and TRPL in vivo. J. Neurosci. 2000;20:5748–5755. doi: 10.1523/JNEUROSCI.20-15-05748.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Badsha F, et al. Mutants in Drosophila TRPC channels reduce olfactory sensitivity to carbon dioxide. PLoS ONE. 2012;7:e49848. doi: 10.1371/journal.pone.0049848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gertler FB, Bennett RL, Clark MJ, Hoffmann FM. Drosophila abl tyrosine kinase in embryonic CNS axons: A role in axonogenesis is revealed through dosage-sensitive interactions with disabled. Cell. 1989;58:103–113. doi: 10.1016/0092-8674(89)90407-8. [DOI] [PubMed] [Google Scholar]
- 167.Xiong W, Dabbouseh NM, Rebay I. Interactions with the abelson tyrosine kinase reveal compartmentalization of eyes absent function between nucleus and cytoplasm. Dev. Cell. 2009;16:271–279. doi: 10.1016/j.devcel.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kawasaki F. Active zone localization of presynaptic calcium channels encoded by the cacophony locus of Drosophila. J. Neurosci. 2004;24:282–285. doi: 10.1523/JNEUROSCI.3553-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sun Q, Schindelholz B, Knirr M, Schmid A, Zinn K. Complex genetic interactions among four receptor tyrosine phosphatases regulate axon guidance in Drosophila. Mol. Cell. Neurosci. 2001;17:274–291. doi: 10.1006/mcne.2000.0939. [DOI] [PubMed] [Google Scholar]
- 170.Pennacchio LA, et al. Structure, sequence and location of the UQCRFS1 gene for the human Rieske Fe-S protein. Gene. 1995;155:207–211. doi: 10.1016/0378-1119(94)00683-J. [DOI] [PubMed] [Google Scholar]
- 171.Saraste M. Oxidative phosphorylation at the fin de siècle. Science. 1999;1979(283):1488–1493. doi: 10.1126/science.283.5407.1488. [DOI] [PubMed] [Google Scholar]
- 172.Henriques BJ, Katrine Jentoft Olsen R, Gomes CM, Bross P. Electron transfer flavoprotein and its role in mitochondrial energy metabolism in health and disease. Gene. 2021;776:145407. doi: 10.1016/j.gene.2021.145407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Marygold SJ, et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007;8:R216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang L, Lam G, Thummel CS. Med24 and Mdh2 are required for Drosophila larval salivary gland cell death. Dev. Dyn. 2010;239:954–964. doi: 10.1002/dvdy.22213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Grossman LI, Rosenthal NH, Akamatsu M, Erickson RP. Cloning, sequence analysis, and expression of a mouse cDNA encoding cytochrome c oxidase subunit VIa liver isoform. Biochim. Biophys. Acta. 1995;1260:361–364. doi: 10.1016/0167-4781(94)00232-R. [DOI] [PubMed] [Google Scholar]
- 176.Dunham-Snary KJ, et al. Ndufs2, a core subunit of mitochondrial complex I, is essential for acute oxygen-sensing and hypoxic pulmonary vasoconstriction. Circ Res. 2019;124:1727–1746. doi: 10.1161/CIRCRESAHA.118.314284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gauthier M, Grünewald B. Honeybee Neurobiology and Behavior. Springer; 2012. Neurotransmitter systems in the honey bee brain: Functions in learning and memory; pp. 155–169. [Google Scholar]
- 178.Alekseyenko OV, et al. Serotonergic modulation of aggression in Drosophila involves GABAergic and cholinergic opposing pathways. Curr. Biol. 2019;29:2145–2156.e5. doi: 10.1016/j.cub.2019.05.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tikhonov DB, Magazanik LG. Origin and molecular evolution of ionotropic glutamate receptors. Neurosci. Behav. Physiol. 2009;39:763–773. doi: 10.1007/s11055-009-9195-6. [DOI] [PubMed] [Google Scholar]
- 180.Li Y, et al. Novel functional properties of Drosophila CNS glutamate receptors. Neuron. 2016;92:1036–1048. doi: 10.1016/j.neuron.2016.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Frenkel L, et al. Organization of circadian behavior relies on glycinergic transmission. Cell Rep. 2017;19:72–85. doi: 10.1016/j.celrep.2017.03.034. [DOI] [PubMed] [Google Scholar]
- 182.Henry C, et al. Heterogeneous expression of GABA receptor-like subunits LCCH3 and GRD reveals functional diversity of GABA receptors in the honeybee Apis mellifera. Br. J. Pharmacol. 2020;177:3924–3940. doi: 10.1111/bph.15135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bastian FB, et al. The Bgee suite: Integrated curated expression atlas and comparative transcriptomics in animals. Nucleic Acids Res. 2021;49:D831–D847. doi: 10.1093/nar/gkaa793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2018).
- 185.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Grabherr MG, et al. Trinity: Reconstructing a full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2013;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Davidson NM, Hawkins ADK, Oshlack A. SuperTranscripts: A data driven reference for analysis and visualisation of transcriptomes. Genome Biol. 2017;18:148. doi: 10.1186/s13059-017-1284-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Huang Y, Niu B, Gao Y, Fu L, Li W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics. 2010;26:680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Davidson NM, Oshlack A. Corset: Enabling differential gene expression analysis for de novoassembled transcriptomes. Genome Biol. 2014;15:410. doi: 10.1186/s13059-014-0410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 194.Musacchia F, Basu S, Petrosino G, Salvemini M, Sanges R. Annocript: A flexible pipeline for the annotation of transcriptomes able to identify putative long noncoding RNAs. Bioinformatics. 2015;31:2199–2201. doi: 10.1093/bioinformatics/btv106. [DOI] [PubMed] [Google Scholar]
- 195.Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH. UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Bairoch A. The SWISS-PROT protein sequence data bank and its new supplement TREMBL. Nucleic Acids Res. 1996;24:21–25. doi: 10.1093/nar/24.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323–333. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Zhang Y, Parmigiani G, Johnson WE. ComBat-seq: Batch effect adjustment for RNA-seq count data. NAR Genom. Bioinform. 2020;2:1–10. doi: 10.1093/nargab/lqaa078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Pascovici, D. & Wu, J. PloGO2: Plot Gene Ontology and KEGG Pathway Annotation and Abundance. R Package Version 1.4.0 (2021).
- 202.Alexa, A. & Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology. R Package Version 2.20.0 (2020).
- 203.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kuznetsova I, Lugmayr A, Siira SJ, Rackham O, Filipovska A. CirGO: An alternative circular way of visualising gene ontology terms. BMC Bioinform. 2019;20:84. doi: 10.1186/s12859-019-2671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence reads have been deposited in the NCBI Sequence Read Archive (SRA) under the following accession numbers: SRR24187037, SRR24187038, SRR24187039, SRR24187040, SRR24187041, SRR24187042, SRR24187043, SRR24187044, SRR24187045, SRR24187046. The corresponding BioProject accession number is PRJNA955762. The transcriptome assemblies, as well as the sets of SuperTranscripts, are available on FigShare (10.6084/m9.figshare.23264771.v1).