Abstract
The phenotype of an organism is the manifestation of its expressed genome. The gcr1 mutant of yeast grows at near wild-type rates on nonfermentable carbon sources but exhibits a severe growth defect when grown in the presence of glucose, even when nonfermentable carbon sources are available. Using DNA microarrays, the genomic expression patterns of wild-type and gcr1 mutant yeast growing on various media, with and without glucose, were compared. A total of 53 open reading frames (ORFs) were identified as GCR1 dependent based on the criterion that their expression was reduced twofold or greater in mutant versus wild-type cultures grown in permissive medium consisting of YP supplemented with glycerol and lactate. The GCR1-dependent genes, so defined, fell into three classes: (i) glycolytic enzyme genes, (ii) ORFs carried by Ty elements, and (iii) genes not previously known to be GCR1 dependent. In wild-type cultures, GCR1-dependent genes accounted for 27% of the total hybridization signal, whereas in mutant cultures, they accounted for 6% of the total. Glucose addition to the growth medium resulted in a reprogramming of gene expression in both wild-type and mutant yeasts. In both strains, glycolytic enzyme gene expression was induced by the addition of glucose, although the expression of these genes was still impaired in the mutant compared to the wild type. By contrast, glucose resulted in a strong induction of Ty-borne genes in the mutant background but did not greatly affect their already high expression in the wild-type background. Both strains responded to glucose by repressing the expression of genes involved in respiration and the metabolism of alternative carbon sources. Thus, the severe growth inhibition observed in gcr1 mutants in the presence of glucose is the result of normal signal transduction pathways and glucose repression mechanisms operating without sufficient glycolytic enzyme gene expression to support growth via glycolysis alone.
In Saccharomyces cerevisiae, glucose is the preferred carbon and energy source. The enzymes of glycolysis, which are required for the utilization of glucose, make up a major fraction of the soluble cellular proteins (22, 25). The genes encoding these enzymes are transcribed at high levels, and the individual transcripts are some of the most abundant in yeast (26, 54). Glycolytic enzyme gene expression is brought about by the concerted action of a number of transcription factors that bind in the upstream activating sequences (UAS) of these genes (3, 4, 9, 10, 38, 48, 55). The centerpiece of glycolytic enzyme gene UAS elements is made up of the closely positioned binding sites for the proteins Gcr1p and Rap1p (3, 18, 29, 39, 55).
The role of Gcr1p in glycolytic enzyme gene expression was first realized when mutations were isolated in the gene encoding it (14). gcr1 mutants exhibited a severe growth defect when grown in the presence of glucose and were shown by enzyme assays to have reduced levels of most glycolytic enzyme activities (14). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis with extracts from wild-type and gcr1 mutant strains indicated that the gcr1 mutation affected the expression of a limited set of several prominent bands that comigrated with purified glycolytic enzymes (13). Since the first gcr1 mutation appeared to affect the expression of a few genes primarily involved in glycolysis, it was named gcr1 to signify its effect on glycolytic enzyme gene expression. At the time, however, it was difficult to state the limits of Gcr1p in global gene expression.
Rap1p is a multifunctional protein that can act in transcription as either an activator or a repressor depending on the sequence context of its binding site (5–7, 49). From the outset, it was recognized that Rap1p is one of a class of general transcription factors whose function is required for expression of many different genes. Rap1p has been implicated in the expression of elongation factors, initiation factors, aminoacyl tRNA synthetases, ribosomal protein genes, tRNA and rRNA genes, nutrient transporters, glycolytic enzyme genes, and expression at HMR and HML (5–7, 49). The role of Rap1p as an activator is best understood for glycolytic enzyme genes. At the UAS elements of these genes, Rap1p achieves its activating function by facilitating the binding of Gcr1p at adjacent binding sites (18). Gcr1p is unable to bind these elements in vivo unless Rap1p is bound to an adjacent site (18, 53). In the absence of functional Gcr1p-binding sites or Gcr1p itself, Rap1p, while able to bind to the UAS elements of glycolytic enzyme genes, is unable to mediate the activation of these genes by itself (18). It has been proposed that Rap1p and Gcr1p function together to mediate ribosomal protein gene expression (44). In the case of ribosomal protein genes, which as a class do not have Gcr1p-binding sites, it has been suggested that Rap1p recruits Gcr1p by complexing with it (57).
The precise mechanism by which Rap1p facilitates the binding of Gcr1p has yet to be elucidated, and the true nature of the relationship between Rap1p and Gcr1p has been the subject of research and debate (18, 34, 44, 48, 50). We have proposed that both Rap1p and Gcr1p may have additional DNA binding partners, with which they interact to mediate their roles in transcription (34). Rap1p is known to make contact with other proteins, namely, Rif1p, Sir3p, and Sir4p (23, 33, 37); however, these proteins are not known to make sequence-specific contact with DNA as is the case for Gcr1p (2, 28, 29, 34). A search of the yeast genome for sequences that match the proposed consensus binding sites for both Rap1p and Gcr1p reveals several hundred sites for each, yet in only a limited number of cases are the two sites found adjacent to one another, most notably at glycolytic enzyme gene UAS elements (34). The large number of potential binding sites for each protein raises the intriguing possibilities that Rap1p may facilitate the DNA binding of other proteins in addition to Gcr1p and that the binding of Gcr1p may be facilitated by proteins other than Rap1p.
The advent of DNA microarray technology (11, 45) affords one the opportunity to understand mutant phenotypes in terms of both the primary and secondary effects of the specific genetic lesion (17, 21, 27, 56). To gain a greater understanding of the extent of the role of Gcr1p in yeast gene expression and to more fully understand the phenotype associated with gcr1 mutants, we used labeled cDNA prepared from RNA isolated from wild-type and gcr1 mutant strains to interrogate high-density DNA microarrays of the yeast genome. These experiments defined a limited set of genes which together make up a significant fraction of the yeast transcriptome and provide an explanation for the glucose inhibition observed with gcr1 mutants.
MATERIALS AND METHODS
Yeast strains and growth conditions.
Isogeneic strains S150-2B (MATa leu2-3,112 his3Δ trp1-289 ura3-52) and HBY4 (MATa gcr1Δ::HIS3 leu2-3,112 his3Δ trp1-289 ura3-52) used in this study have been described previously (47).
Strains were subcultured and grown at 30°C in medium permissive for growth of the gcr1 mutant. Cultures were grown in YP medium (42) supplemented with 2% lactate and 2% glycerol (YPGL). Cultures in exponential-phase growth at an optical density at 600 nm of approximately 1 were rapidly harvested on ice, and cells were collected by centrifugation at 4°C. In some experiments with strain S150-2B, YPD medium was used to investigate the effect of steady-state growth in medium with glucose.
For glucose induction and repression experiments, cultures were grown in YPGL to an optical density at 600 nm of approximately 1, at which time glucose was added to 2%. Following glucose addition, the cultures were allowed to grow for 4 h before being harvested as described above. Glucose induction and repression growth conditions are denoted throughout the text as YPGL+G.
RNA isolation and cDNA preparation.
Total RNA was isolated using an RNeasy kit from Qiagen (Chatsworth, Calif.) as recommended by the manufacturer.
cDNA was prepared from 1 μg of total RNA using reverse transcriptase following oligo(dT) priming. The cDNA was uniformly labeled using [33P]dCTP during the course of the reverse transcription reaction.
DNA microarrays and hybridization conditions.
DNA microarrays of yeast open reading frames (ORFs) were obtained from Research Genetics (Huntsville, Ala.). In total, four microarray sets, identified by the numbers 11, 77, 83, and 99, were used. The microarrays were prewashed and hybridized as recommended by the manufacturer. Hybridizations were carried out in a Robbins roller drum hybridization chamber at 42°C for a minimum of 16 h. Following hybridization, the filters were washed as specified by the protocol provided by Research Genetics. As recommended by the manufacturer, each array was interrogated, stripped, and reinterrogated to a maximum of five interrogations. Table 1 shows the interrogation schedule for each of the microarray sets used.
TABLE 1.
Microarray interrogation schedulea
| Culture condition (genotype, medium) | Interrogation schedule for microarray setb:
|
|||
|---|---|---|---|---|
| 11 | 83 | 77 | 99 | |
| GCR1, YPGL | i1 | i1 | i2 | i1 |
| gcr1, YPGL | i2 | i2 | i1 | i4 |
| gcr1, YPGL+G | ND | i3 | i3 | i5 |
| GCR1, YPGL+G | ND | i4 | i4 | i2 |
| GCR1, YPD | ND | i5 | i5 | i3 |
The interrogation schedule for each of the microarrays sets used during the course of this study is given.
Each microarray set was interrogated, stripped, and reinterrogated. i1, first interrogation; i2, second interrogation, etc. ND, not done.
Signal detection and analysis.
Hybridization of the radiolabeled cDNA to the immobilized probe DNA on the filters was detected by phosphorimaging using a Molecular Dynamics Storm PhosphorImager scanning at 50 μm. The data obtained from the PhosphorImager were imported into the Pathways 2.01 software package (Research Genetics) for normalization and analysis. For comparison purposes, arrays were normalized between experiments by using the “all data points” method in Pathways 2.01. The normalization method used by Pathways 2.01 in effect adjusts the total hybridization signal between filters such that they are equal and then compares the ratio between adjusted signals at each element on the array.
In total, RNA was isolated and cDNA was prepared from 17 cultures. Each labeled cDNA preparation was used to interrogate one of four DNA microarray sets (sets 11, 83, 77, and 99) used. Comparisons were made between interrogations of a given DNA microarray set. These experiments resulted in five different expression profiles. Two of these highlighted comparisons between mutant and wild-type cultures, i.e., gcr1 versus GCR1, under permissive growth conditions of YPGL (gcr1/GCR1 YPGL) (four data sets) and after a 4-h exposure to glucose (gcr1/GCR1 YPGL+G) (three data sets). The other three profiles examined the effect of exposure to glucose on gene expression: the effect of a 4-h glucose exposure on gene expression in mutant and wild-type cultures was examined in the YPGL+G/YPGL gcr1 and the YPGL+G/YPGL GCR1 comparisons, respectively (three data sets each); and for the wild-type culture, expression profiles were also obtained for comparisons of steady-state growth in YPD versus YPGL (YPD/YPGL GCR1) (three data sets).
Algorithms and databases.
During the course of this work, extensive use was made of the online database resources at the Saccharomyces Genome Database (J. M. Cherry, C. Ball, K. Dolinski, S. Dwight, S. Harris, et al., Saccharomyces Genome Database, 1999 [http:/genome-www.stanford.edu/cgi-bin/SGD/search]) and Proteome (16). Functional groups of genes were classified in accordance with the Martinsried Institute of Protein Sciences classification scheme (36). Gene expression data were subjected to hierarchical clustering analysis and displayed using the algorithms developed by Eisen et al. (20). Common sequence motifs were identified using the algorithm AlignACE, written by Roth et al. (43), and are represented using the logos format of Schneider and Stephens (46).
RESULTS AND DISCUSSION
GCR1-dependent genes.
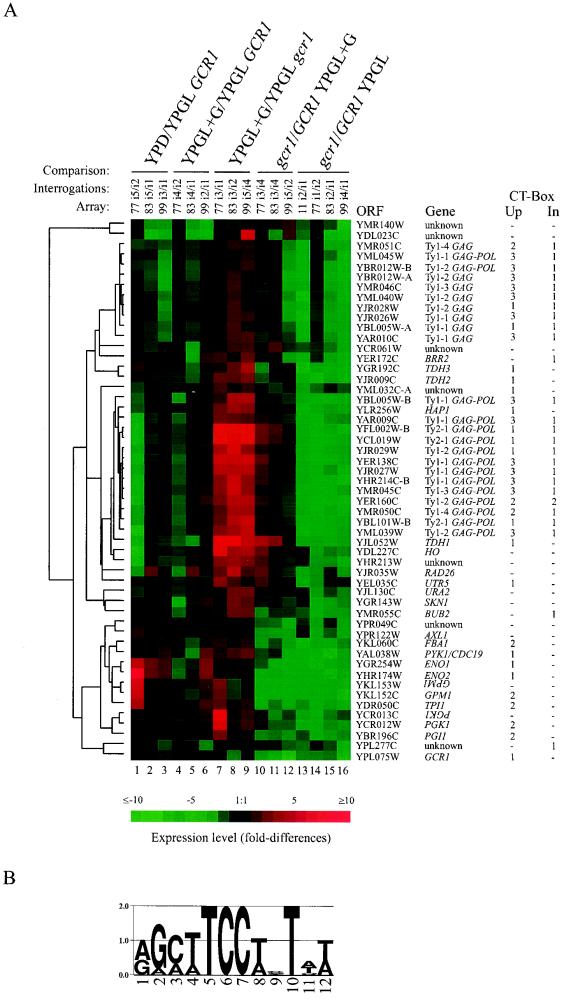
To identify genes dependent on Gcr1p for full expression, we carried out a series of global genomic expression studies with wild-type and gcr1 mutant strains of yeast. In consideration of the severe growth defect that gcr1 mutants exhibit when grown in the presence of glucose and to reduce the difference in gene expression due to differences in the growth rate, we grew yeast cultures in YPGL, which is permissive for the gcr1 mutant (13, 14). In this medium, the doubling times for the wild-type and mutant are ca. 280 and 300 min, respectively. Radiolabeled cDNA, prepared from total RNA isolated from cultures harvested during logarithmic growth, was used to interrogate high-density DNA arrays of yeast ORFs. The relative degree of hybridization to each of the 6,144 ORFs in the array was compared between experiments. From four independent experiments, using a twofold difference cutoff, we identified 53 ORFs that reproducibly displayed lower levels of hybridization when interrogated with cDNA prepared from RNA isolated from the gcr1 mutant than when interrogated with cDNA from the wild type. Figure 1, lanes 13 to 16, show comparisons of the expression profiles of the 53 GCR1-dependent ORFs in YPGL-grown cultures of the gcr1 mutant and the wild type. The complete data set containing more than 104,000 observations and 98,000 comparisons is available on-line at http://cmg.health.ufl.edu/∼bakerlab/genomic.htm.
FIG. 1.
Hierarchical cluster analysis of gene expression patterns of GCR1-dependent genes in wild-type and gcr1 mutant backgrounds in the absence and presence of glucose. (A) Genes whose expression was reduced by twofold or greater in gcr1 mutants growing in YPGL compared to wild-type cells growing in the same medium are represented in the figure by using TreeView (21). The degree of difference between the expression patterns of the various genes is indicated by the length of the branches on the tree. As indicated at the top of the figure, lanes 1 to 9 represent glucose induction-repression experiments and lanes 10 to 16 represent comparisons of gcr1 mutant and wild type. For the glucose induction-repression experiments, the reference condition was growth in YPGL. In comparisons of mutant and wild type, the wild type was set as the reference. Lanes: 1 to 3, YPD/YPGL GCR1 represents gene expression patterns in the wild-type strain growing in YPD medium compared to growth in YPGL medium; 4 to 6, YPGL+G/YPGL GCR1 represents gene expression patterns in the wild-type strain 4 h after the addition of glucose to YPGL (YPGL+G) compared to growth in YPGL medium without the addition of glucose; 7 to 9, YPGL+G/YPGL gcr1 represents gene expression patterns in the gcr1 mutant strain 4 h after the addition of glucose to YPGL compared to growth of the mutant in YPGL without glucose; 10 to 12, gcr1/GCR1 YPGL+G represents a comparison of mutant and wild-type gene expression 4 h after the addition of glucose to each culture growing in YPGL; 13 to 16, gcr1/GCR1 YPGL represents a comparison of mutant and wild-type cultures growing in YPGL medium. The microarray set (Array) and the individual interrogations (Interrogations) from which the ratios were calculated are indicated. The ratios of transcript levels are depicted visually according to the color scale on the figure. Red indicates increased expression relative to the reference, and green indicates decreased expression relative to the reference. On the right of the figure is listed the ORF and gene name for each expression profile. The number of occurrences of the common sequence motif shown in panel B, within 600 nucleotides upstream of (CT-Box Up) or inside (CT-Box In) each ORF is also noted. (B) Logos representation (46) of the common sequence motif found among GCR1-dependent genes by using AlignACE (43). The height of each letter is proportional to its frequency at that position. The overall height of the stack at each position represents the informational content of that position in bits of information ranging from 0 to 2 bits. This sequence motif closely resembles the consensus binding site for Gcr1p proposed by Huie et al. (29), also known as the CT box.
As a group, the ORFs most severely affected by the gcr1 lesion encode glycolytic enzymes. We note that of the enzymes assayed, the phosphoglycerate mutase and enolase activities are the most severely affected in gcr1 mutants (1, 13, 14). These enzymes are encoded by the ORFs most strongly affected in the microarray experiments. Hybridization to YKL152C (GPM1) was reduced 13.8-fold with material from the gcr1 mutant compared to that obtained with material from the wild type. Similar reductions were observed for YHR174W (9.6-fold) and YGR254W (7.9-fold), the ENO2 and ENO1 ORFs, respectively. Two ORFs were identified, YKL153W and YCR013C, which partially overlap ORFs, YKL152C and YCR012W, encoding the glycolytic enzymes phosphoglycerate mutase and phosphoglycerate kinase. The respective ORF pairs are capable of hybridizing to the same cDNAs and thus served as an internal control. The ORF specifying Gcr1p was also identified in the screen and served as a control for our ability to detect differences in genes expressed at low levels; the gcr1 mutant strain used for these experiments harbors a deletion of GCR1 itself. We are therefore unable to comment on whether GCR1 is autoregulated based on this study, although the occurrence of an upstream CT box suggests the possibility. In addition to ORFs encoding glycolytic enzymes, ORFs carried by Ty elements made a second major class of genes identified. All Ty1 and Ty2 ORFs present on the arrays were identified as GCR1 dependent. In Fig. 1, the ORFs encoding Gag and Gag-Pol are listed separately, although the encoded proteins are derived from the same transcript, with the Gag-Pol protein resulting from translational frameshifting. Work from several laboratories previously established the GCR1 dependency of Ty gene expression (19, 51). Fourteen other ORFs, listed in Fig. 1, were also identified as GCR1 dependent using the twofold cutoff criterion with the microarray screen.
The effects of mutations in genes encoding transcription factors on the transcriptional profile of a cell can be classified into three categories. The primary effect would be the loss of expression of the transcription factor itself. The secondary effect would be the loss of expression of genes that are directly dependent on the transcription factor for their expression. The tertiary effect would involve genes whose expression is responding to the altered physiology of the cell resulting from the primary and secondary effects. When classifying genes into sets, two types of errors can be made: type 1 errors exclude genes when they should be included within the set, and type 2 errors include genes when they should be excluded. In the present study, the primary effect is the effect on GCR1 expression itself. The more interesting class of genes contains those which are directly dependent on Gcr1p for their expression; genes of this class would be expected to have common DNA sequence motifs through which Gcr1p acts.
Common sequence motif.
We analyzed the upstream DNA region of the 53 ORFs identified for common sequence motifs by using the algorithm AlignACE (43). The algorithm successfully identified a sequence motif that closely resembled the sequence previously proposed as the consensus Gcr1p DNA-binding site (2, 29), known as the CT box (9). The motif shown in Fig. 1B identifies the T at position 10 as having a high informational content. The significance of this position was not appreciated previously. This sequence motif was found at least once within 600 nucleotides of the translational start of 38 of the 53 genes identified above, as indicated in Fig. 1. Genes strongly dependent on GCR1 tended to have multiple copies of the CT-box motif in their regulatory regions. Conversely, the 10 genes in which CT boxes were not found tended to exhibit less dependence on GCR1 than the others did. This latter class of genes without CT boxes is most probably responding to tertiary effects of the gcr1 mutation. Six ORFs having CT boxes in their 5′ noncoding region or within the ORF itself were identified as GCR1 dependent when they had not previously been recognized as such. They are YML032C, YLR256W (HAP1), YEL035C (UTR5), YER172C (BRR2), YMR055C (BUB2), and YPL277C.
CT boxes that score better than the average of the aligned CT boxes identified in front of the GCR1-dependent genes occur elsewhere in the genome. In total, 854 CT boxes were identified, and most of these sites (670 of 854) occur within coding regions. CT boxes are found scattered throughout the yeast genome resident on the long terminal repeats of Ty1 and Ty2 (12). Altogether, 158 CT boxes occur within 600 nucleotides of the initiation codon of ORFs. In some cases, these motifs were found in front of genes that were not identified as GCR1-dependent genes by the above criteria. Two possibilities exist: either these sites are not Gcr1p-binding sites, and the cognate genes are not GCR1-dependent genes, or they are but not under the physiological conditions tested here. We showed previously, in the context of two glycolytic enzyme gene UAS elements, that a CT box alone is not sufficient for Gcr1p binding in vivo (18). In the context of the TPI1 and PYK1 UAS elements, Gcr1p binding in vivo requires Rap1p bound at an appropriately spaced Rap1p-binding site. There are genetic and biochemical indications that regions adjacent to Gcr1p-binding sites in Ty elements are important for GCR1-mediated expression (19, 51). The observations with Ty elements suggest that Gcr1p may have other binding partners in addition to Rap1p. Thus, it is possible to envision that there may be GCR1-dependent genes for which the binding of Gcr1p is regulated by another DNA-binding protein whose expression itself is regulated. If the hypothetical binding partner of Gcr1p was not expressed under the experimental conditions used, the requirement for Gcr1p would be imperceptible.
The sequence-aligning algorithm AlignACE (43) did not identify a sequence motif that closely resembled the consensus Rap1p DNA-binding site among the GCR1-dependent genes, even though Rap1p-binding sites are essential features of several glycolytic enzyme gene UAS elements.
Expression levels of GCR1-dependent genes.
We compared the hybridization intensities obtained with samples from the wild-type strain, S150-2B, growing in YPD medium to the results of serial analysis of gene expression (SAGE), as reported by Velculescu et al., of yeast strain YPH499 growing in the same medium (54). In terms of fraction of total hybridization signal for microarray experiments and occurrences of sequence tags for SAGE experiments, markers for transcripts encoding Ty Gag and Gag-Pol proteins, glycolytic enzymes, and ribosomal proteins were in the top 1% of each list. On the whole, our microarray results are in general agreement with the SAGE results of Velculescu et al. (54), although there are some differences between the relative levels of highly expressed genes, with the most notable being those carried on Ty elements. The microarray result indicates that Ty transcripts make up ca. 18% of polyadenylated RNA in YPD-grown cells, whereas the SAGE results indicate that they make up ca. 0.5% of transcripts (54). Based on Northern analysis, Curcio et al. (16) previously estimated that Ty transcripts may make up as much as 50% of polyadenylated RNA in yeast cells. The reason for the discrepancy in measurements of Ty transcript abundance is not clear and may have to do with peculiarities of the three different assay systems used. However, by whatever measure, Ty transcripts are among the most abundant in yeast.
Apart from ratio comparisons of genes between conditions or strains, hybridization intensities of the individual elements of an array can be used, within limits, as an indication of the relative abundance of the corresponding transcripts within cells. For the wild-type strain growing on YPGL, the 53 ORFs identified above as GCR1-dependent genes accounted for 27% of the total hybridization intensity on the microarray. The same 53 ORFs accounted for only 6% of the total hybridization when cDNA was prepared from the gcr1 mutant grown in the same medium. This result indicates that as a whole, GCR1-dependent genes are expressed at very high levels in wild-type cells, with 0.9% of the genes (53 of 6,144) accounting for 27% of the total hybridization signal. Furthermore, the residual level of expression of the GCR1-dependent genes in gcr1 mutants, comprising 6% of the total hybridization signal, is still quite high compared to the levels of most genes in the genome. Thus, while only a few genes are dependent on Gcr1p for expression, together they make up a sizable proportion of the yeast transcriptome under these growth conditions.
Yeast cells growing on YPGL are dependent on gluconeogenesis, which is required for ribose and cell wall biosynthesis. The GCR1-independent expression of glycolytic enzyme genes in gcr1 mutants suggests that the residual glycolytic enzyme gene expression is sufficient for gluconeogenesis and growth.
Glucose induction and repression.
The gcr1 mutant phenotype afforded us the opportunity to investigate alterations in gene expression that result from the addition of glucose in the absence of the rapid growth normally associated with glucose. Therefore, a series of glucose induction-repression experiments were carried out with wild-type and gcr1 mutant strains. Due to the severe growth defect of the gcr1 mutant in the presence of glucose, we chose the 4-h glucose induction protocol that we had previously used to monitor glycolytic enzyme gene expression in gcr1 mutants (1). Figure 1 shows the response of the GCR1-dependent genes to the addition of glucose to the growth medium. In the wild-type strain growing under steady-state conditions in YPD medium, the expression profile of GCR1-dependent genes was similar to the expression profile of the wild-type strain after a 4-h glucose induction (Fig. 1, compare lanes 1 to 3 with lanes 4 to 6). In the gcr1 mutant background, GCR1-dependent genes usually increase their level of expression as a result of exposure to glucose (lanes 7 to 9). However, clear distinctions can be made between the responses of the GCR1-dependent genes, with two classes emerging. The first class is composed primarily of genes carried by Ty elements. In gcr1 mutants, these genes displayed a strong induction ratio when glucose was added to the medium (lanes 7 to 9), whereas in the wild-type background, glucose induction of these genes was not observed (lanes 4 to 6). Accordingly, as can be seen in lanes 10 to 12, the expression levels of the Ty element genes are similar in the gcr1 mutant and wild-type strains upon addition of glucose. The second response class, consisting primarily of glycolytic enzyme genes, displays a modest level of glucose induction in both wild-type and mutant backgrounds. Glucose induction of the residual glycolytic enzyme activities has been noted previously with gcr1 mutant strains (1, 13), Uemura and Fraenkel (52) have recently shown that gcr1 mutants are capable of responding to glucose by increasing their capacity to flux glucose through glycolysis. The residual expression and glucose induction of GCR1-dependent genes in the gcr1 mutant background implies that other regulatory elements are operational in the absence of Gcr1p.
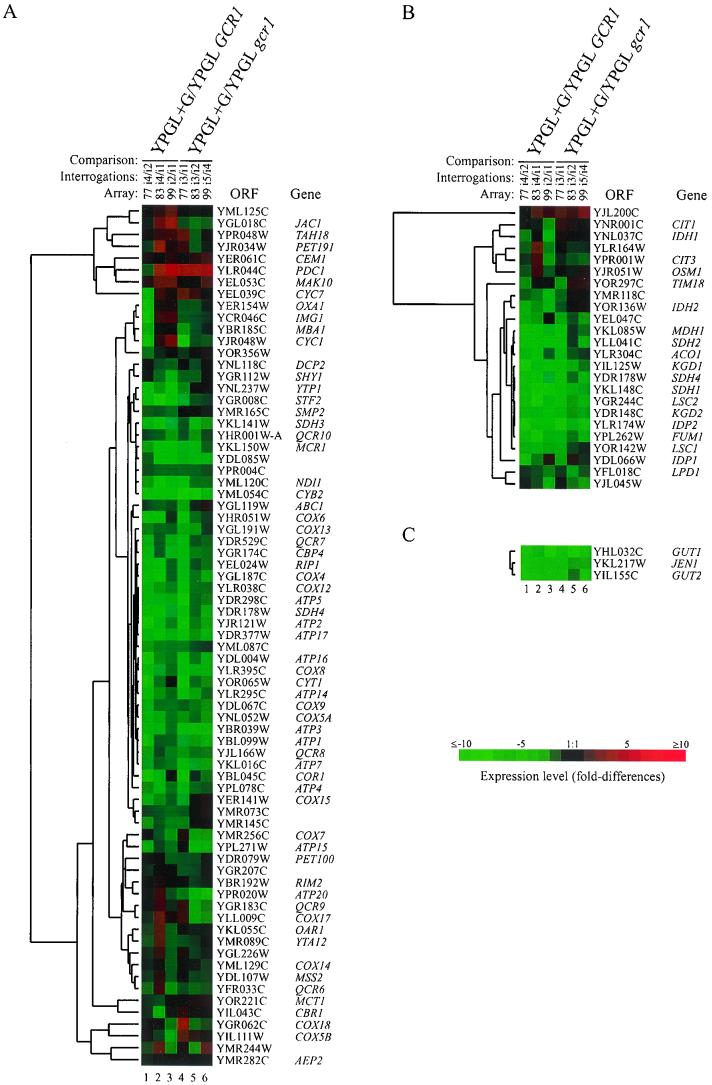
There are similarities and clear differences in the global response of the wild-type and gcr1 mutants to the addition of glucose. In the wild-type background, 444 genes were induced and 711 genes were repressed at the twofold level, whereas in the gcr1 mutant, the response to glucose was more restricted, with 211 genes being induced and 252 genes being repressed. The principal elements of glucose repression and induction remained intact in the gcr1 mutant. Both strains responded to glucose by repressing genes encoding respiratory functions. Of the 73 ORFs specifying respiratory function present on the arrays used, glucose reduced the expression of 31 in the mutant compared to 30 in the wild type by greater than twofold (Fig. 2A, lanes 4 to 6 and lanes 1 to 3, respectively). Likewise, similar patterns of repression were observed in the wild type and mutant for genes encoding tricarboxylic acid pathway functions; 11 out of 24 genes were reduced in expression by greater than twofold in the mutant compared to 13 in the wild type (Fig. 2B, lanes 4 to 6 and lanes 1 to 3, respectively). On the other hand, the expression of glucose transporters appeared to increase after the addition of glucose to the cultures; the hybridization intensities at the elements specifying HXT1 to HXT3 were increased by more than twofold with cDNA prepared from both the wild type and mutant after glucose addition. As a class, genes subject to growth rate-dependent regulation were induced in the wild type and not in the mutant. The most notable examples of growth rate-dependent genes are those encoding ribosomal proteins (24, 30, 31, 35) (see below). The doubling time of the wild-type strain decreased from 280 min in YPGL to 135 min on the addition of glucose. The gcr1 mutant, on the other hand, effectively stopped growing on the addition of glucose. Its doubling time increased from 300 to 780 min after glucose was added to the culture medium.
FIG. 2.
(A) Expression profile of respiratory genes in the wild type and gcr1 mutants. (B) Expression profile of genes encoding tricarboxylic acid pathway function in the wild type and gcr1 mutants. (C) Expression profile of genes encoding glycerol metabolic enzymes and lactate transporter. Total RNA was isolated for expression profiling 4 h after the addition of glucose to wild-type and gcr1 mutant cultures growing in YPGL. Genes were selected for inclusion in panels A and B based on the Martinsried Institute of Protein Sciences classification scheme. The lanes are as described in the legend to Fig. 1. Column headings for panel C are the same as those for panel B. The ratio of transcript levels is depicted visually according to the color scale on the figure. Red indicates increased expression relative to the reference, and green indicates decreased expression relative to the reference.
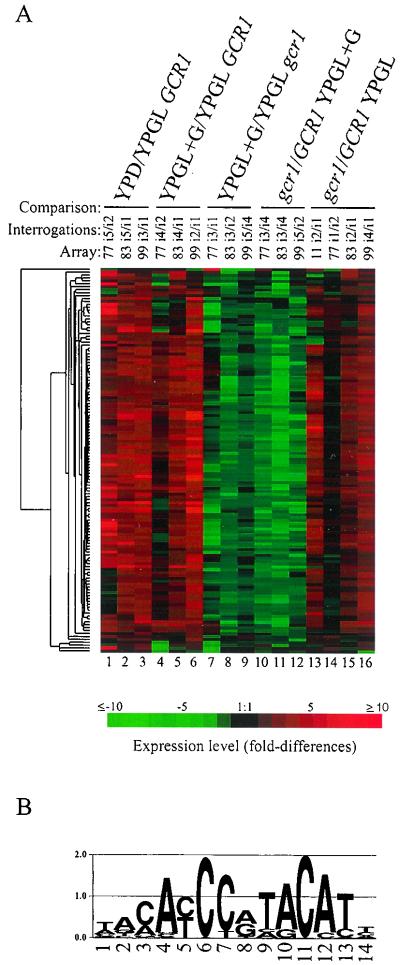
Ribosomal protein gene expression in the wild type and gcr1 mutants.
Santangelo and Tornow (44) previously reported that ribosomal protein genes are dependent on Gcr1p acting through Rap1p-binding sites which are known as UAS(RPG) sites for ribosomal protein genes. These workers went on to propose that Gcr1p is recruited to UAS(RPG) via contacts with Rap1p (57). Figure 3 shows the expression pattern of each of the 132 ribosomal protein genes represented on the arrays that we used. Figure 3, lanes 13 to 16, show that as a class, ribosomal protein gene expression appears to be slightly enhanced in the gcr1 mutant growing in YPGL compared to that in wild-type strains growing in the same medium. Elements on the array specifying cytoplasmic ribosomal protein genes accounted for 14% of the total hybridization signal in the mutant compared to 9% for the wild type in the permissive medium YPGL. Rap1p-binding sites, as noted in Fig. 3B, are the most prominent feature in the regulatory regions of ribosomal protein genes (32). The expression profile of ribosomal protein genes in YPGL indicates that Gcr1p is not required for the expression of ribosomal protein genes per se. The apparent increase in expression of ribosomal protein genes under this condition in the gcr1 mutant may be due to increased availability of the transcriptional apparatus that otherwise would be engaged in transcribing GCR1-dependent genes.
FIG. 3.
(A) Expression profile of 132 cytoplasmic ribosomal protein genes in wild-type and gcr1 mutant strains growing in the absence and presence of glucose. The lanes are as described in the legend to Fig. 1. The ratio of transcript levels is depicted visually according to the color scale on the figure. Red indicates increased expression relative to the reference, and green indicates decreased expression relative to the reference. (B) Logos representation (46) of the common sequence motif found 166 times among the 132 cytoplasmic ribosomal protein genes by using AlignACE (43). The height of each position represents the informational content of that position in bits of information. This sequence motif closely resembles the consensus binding site for Rap1p as recently refined by Lascaris et al. (32).
The expression of cytoplasmic ribosomal protein genes in wild-type and gcr1 mutant strains differed markedly on the addition of glucose to the growth medium (Fig. 3, compare lanes 4 to 6 with lanes 7 to 9). With wild-type cells, exposure to glucose resulted in an increase in the expression level of ribosomal protein genes to 21% of the total hybridization signal, which is in agreement with the SAGE results of Velculescu et al. (54) as noted by Lascaris et al. (32). By contrast, addition of glucose to cultures of gcr1 mutants resulted in a reduction in the level of ribosomal protein gene expression from 14 to 11% of the total hybridization signal. It is important to recall, as noted above, that addition of glucose to the growth medium of wild-type cells results in an increased growth rate whereas a similar addition to mutant cultures results in drastic reductions in the growth rate. Both growth rate differences and differences in ribosomal protein gene expression patterns were magnified in comparisons between wild-type and mutant strains after glucose induction (Fig. 3, lanes 10 to 12). The differences in ribosomal protein gene expression between the wild type and the gcr1 mutant can best be explained by differences in growth rates between the two strains. Ribosomal protein gene expression is known to be subject to growth rate-dependent regulation (24, 30, 31, 35). The expression profiles of ribosomal protein genes presented here argue for an important but indirect role of Gcr1p in ribosomal protein gene expression. The expression profile of ribosomal protein genes in gcr1 mutants growing in the presence of glucose is a manifestation of a tertiary effect of the gcr1 mutation.
Gene expression and growth phenotype.
One of the most striking features of gcr1 mutants is the severe growth defect that they exhibit when grown on media containing glucose, whereas they grow relatively normally under gluconeogenic conditions (13). The gene expression pattern observed with the gcr1 mutant provides an explanation for the growth phenotypes of the mutant. Figure 2 shows that upon addition of glucose to a medium otherwise permissive for growth, such as YP supplemented with glycerol plus lactate, the mutant, like the wild-type strain, responded by repressing the expression of genes encoding key respiratory enzymes, genes involved in trichloroacetic acid cycle function, and genes involved in the metabolism of alternative carbon sources. Repression of GUT1, GUT2, and JEN1 is relevant to the discussion here (Fig. 2C). These genes encode functions that are required for the utilization of glycerol and lactate. GUT1 and GUT2 encode the activities required for the conversion of glycerol to dihydroxyacetone-phosphate (40, 41), and JEN1 encodes a lactate-proton symporter required to transport lactate into the cell (8). In the wild-type and gcr1 mutant strains, each of the aforementioned genes was repressed by fourfold or greater on addition of glucose. Thus, the gcr1 mutant responds to glucose, as do wild-type cells, by reprogramming its gene expression profile to take advantage of the available glucose; however, the gcr1 mutant does not have sufficient glycolytic enzyme gene expression to permit normal rates of growth when utilizing glucose alone. The plight of the mutant is further aggravated by glucose repression, which suppresses the expression of genes involved in metabolism of other energy sources, thereby robbing the cell of any potential of utilizing the available carbon sources, notably lactate and minor constituents of YP.
ACKNOWLEDGMENTS
We thank Frank Rosenzweig, Hiroshi Uemura, and Phil Farabaugh for advice and comments. We thank Eisen et al. (20) and Roth et al. (43) for making their algorithms freely available.
This work was supported in part by a grant from the National Science Foundation (MCB 9816990).
REFERENCES
- 1.Baker H V. Glycolytic gene expression in Saccharomyces cerevisiae: nucleotide sequence of GCR1, null mutants, and evidence for expression. Mol Cell Biol. 1986;6:3774–3784. doi: 10.1128/mcb.6.11.3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker H V. GCR1 of Saccharomyces cerevisiae encodes a DNA binding protein whose binding is abolished by mutations in the CTTCC sequence motif. Proc Natl Acad Sci USA. 1991;88:9443–9447. doi: 10.1073/pnas.88.21.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bitter G A, Chang K K, Egan K M. A multi-component upstream activation sequence of the Saccharomyces cerevisiae glyceraldehyde-3-phosphate dehydrogenase gene promoter. Mol Gen Genet. 1991;231:22–32. doi: 10.1007/BF00293817. [DOI] [PubMed] [Google Scholar]
- 4.Brindle P K, Holland J P, Willett C E, Innis M A, Holland M J. Multiple factors bind the upstream activation sites of the yeast enolase genes ENO1 and ENO2: ABFI protein, like repressor activator protein RAP1, binds cis-acting sequences which modulate repression or activation of transcription. Mol Cell Biol. 1990;10:4872–4885. doi: 10.1128/mcb.10.9.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchman A R, Kimmerly W J, Rine J, Kornberg R D. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchman A R, Lue N F, Kornberg R D. Connections between transcriptional activators, silencers, and telomeres as revealed by functional analysis of a yeast DNA-binding protein. Mol Cell Biol. 1988;8:5086–5099. doi: 10.1128/mcb.8.12.5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Capieaux E, Vignais M L, Sentenac A, Goffeau A. The yeast H+-ATPase gene is controlled by the promoter binding factor TUF. J Biol Chem. 1989;264:7437–7446. [PubMed] [Google Scholar]
- 8.Casal M, Paiva S, Andrade R P, Gancedo C, Leao C. The lactate-proton symport of Saccharomyces cerevisiae is encoded by JEN1. J Bacteriol. 1999;181:2620–2623. doi: 10.1128/jb.181.8.2620-2623.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chambers A, Stanway C, Kingsman A J, Kingsman S M. The UAS of the yeast PGK gene is composed of multiple functional elements. Nucleic Acids Res. 1988;16:8245–8260. doi: 10.1093/nar/16.17.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambers A, Stanway C, Tsang J S, Henry Y, Kingsman A J, Kingsman S M. ARS binding factor 1 binds adjacent to RAP1 at the UASs of the yeast glycolytic genes PGK and PYK1. Nucleic Acids Res. 1990;18:5393–5399. doi: 10.1093/nar/18.18.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chee M, Yang R, Hubbell E, Berno A, Huang X C, Stern D, Winkler J, Lockhart D J, Morris M S, Fodor S P. Accessing genetic information with high-density DNA arrays. Science. 1996;274:610–614. doi: 10.1126/science.274.5287.610. [DOI] [PubMed] [Google Scholar]
- 12.Cherry J M, Adler C, Ball C, Chervitz S A, Dwight S S, Hester E T, Jia Y, Juvik G, Roe T, Schroeder M, Weng S, Botstein D. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clifton D, Fraenkel D G. The gcr (glycolysis regulation) mutation of Saccharomyces cerevisiae. J Biol Chem. 1981;256:13074–13078. [PubMed] [Google Scholar]
- 14.Clifton D, Weinstock S B, Fraenkel D G. Glycolysis mutants in Saccharomyces cerevisiae. Genetics. 1978;88:1–11. doi: 10.1093/genetics/88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costanzo M C, Hogan J D, Cusick M E, Davis B P, Fancher A M, Hodges P E, Kondu P, Lengieza C, Lew-Smith J E, Lingner C, Roberg-Perez K J, Tillberg M, Brooks J E, Garrels J I. The Yeast Proteome Database (YPD) and Caenorhabditis elegans Proteome Database (WormPD): comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curcio M J, Hedge A M, Boeke J D, Garfinkel D J. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Mol Gen Genet. 1990;220:213–221. doi: 10.1007/BF00260484. [DOI] [PubMed] [Google Scholar]
- 17.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 18.Drazinic C M, Smerage J B, Lopez M C, Baker H V. Activation mechanism of the multifunctional transcription factor repressor-activator protein 1 (Rap1p) Mol Cell Biol. 1996;16:3187–3196. doi: 10.1128/mcb.16.6.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudley A M, Gansheroff L J, Winston F. Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4-912delta promoter in Saccharomyces cerevisiae. Genetics. 1999;151:1365–1378. doi: 10.1093/genetics/151.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen M B, Spellman P T, Brown P O, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferea T L, Botstein D, Brown P O, Rosenzweig R F. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Natl Acad Sci USA. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fraenkel D G. Carbohydrate metabolism. In: Strathern J N, Jones E W, Broach J R, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 1–37. [Google Scholar]
- 23.Hardy C F, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 24.Herruer M H, Mager W H, Woudt L P, Nieuwint R T, Wassenaar G M, Groeneveld P, Planta R J. Transcriptional control of yeast ribosomal protein synthesis during carbon-source upshift. Nucleic Acids Res. 1987;15:10133–10144. doi: 10.1093/nar/15.24.10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hess B, Boiteux A, Kruger J. Cooperation of glycolytic enzymes. Adv Enzyme Regul. 1969;7:149–169. doi: 10.1016/0065-2571(69)90016-8. [DOI] [PubMed] [Google Scholar]
- 26.Holland M J, Holland J P. Isolation and identification of yeast messenger ribonucleic acids coding for enolase, glyceraldehyde-3-phosphate dehydrogenase, and phosphoglycerate kinase. Biochemistry. 1978;17:4900–4907. doi: 10.1021/bi00616a007. [DOI] [PubMed] [Google Scholar]
- 27.Holstege F C, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 28.Huie M A, Baker H V. DNA-binding properties of the yeast transcriptional activator, Gcr1p. Yeast. 1996;12:307–317. doi: 10.1002/(sici)1097-0061(19960330)12:4<307::aid-yea912>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 29.Huie M A, Scott E W, Drazinic C M, Lopez M C, Hornstra I K, Yang T P, Baker H V. Characterization of the DNA-binding activity of GCR1: in vivo evidence for two GCR1-binding sites in the upstream activating sequence of TPI of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2690–2700. doi: 10.1128/mcb.12.6.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kief D R, Warner J R. Coordinate control of syntheses of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraakman L S, Griffioen G, Zerp S, Groeneveld P, Thevelein J M, Mager W H, Planta R J. Growth-related expression of ribosomal protein genes in Saccharomyces cerevisiae. Mol Gen Genet. 1993;239:196–204. doi: 10.1007/BF00281618. [DOI] [PubMed] [Google Scholar]
- 32.Lascaris R F, Mager W H, Planta R J. DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics. 1999;15:267–277. doi: 10.1093/bioinformatics/15.4.267. [DOI] [PubMed] [Google Scholar]
- 33.Liu C, Mao X, Lustig A J. Mutational analysis defines a C-terminal tail domain of RAP1 essential for telomeric silencing in Saccharomyces cerevisiae. Genetics. 1994;138:1025–1040. doi: 10.1093/genetics/138.4.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopez M C, Smerage J B, Baker H V. Multiple domains of repressor activator protein 1 contribute to facilitated binding of glycolysis regulatory protein 1. Proc Natl Acad Sci USA. 1998;95:14112–14117. doi: 10.1073/pnas.95.24.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mager W H, Planta R J. Coordinate expression of ribosomal protein genes in yeast as a function of cellular growth rate. Mol Cell Biochem. 1991;104:181–187. doi: 10.1007/BF00229818. [DOI] [PubMed] [Google Scholar]
- 36.Mewes H W, Albermann K, Bahr M, Frishman D, Gleissner A, Hani J, Heumann K, Kleine K, Maierl A, Oliver S G, Pfeiffer F, Zollner A. Overview of the yeast genome. Nature. 1997;387:7–65. doi: 10.1038/42755. [DOI] [PubMed] [Google Scholar]
- 37.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 38.Nishizawa M, Araki R, Teranishi Y. Identification of an upstream activating sequence and an upstream repressible sequence of the pyruvate kinase gene of the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:442–451. doi: 10.1128/mcb.9.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ogden J E, Stanway C, Kim S, Mellor J, Kingsman A J, Kingsman S M. Efficient expression of the Saccharomyces cerevisiae PGK gene depends on an upstream activation sequence but does not require TATA sequences. Mol Cell Biol. 1986;6:4335–4343. doi: 10.1128/mcb.6.12.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pavlik P, Simon M, Schuster T, Ruis H. The glycerol kinase (GUT1) gene of Saccharomyces cerevisiae: cloning and characterization. Curr Genet. 1993;24:21–25. doi: 10.1007/BF00324660. [DOI] [PubMed] [Google Scholar]
- 41.Ronnow B, Kielland-Brandt M C. GUT2, a gene for mitochondrial glycerol 3-phosphate dehydrogenase of Saccharomyces cerevisiae. Yeast. 1993;9:1121–1130. doi: 10.1002/yea.320091013. [DOI] [PubMed] [Google Scholar]
- 42.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1990. [Google Scholar]
- 43.Roth F P, Hughes J D, Estep P W, Church G M. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation. Nat Biotechnol. 1998;16:939–945. doi: 10.1038/nbt1098-939. [DOI] [PubMed] [Google Scholar]
- 44.Santangelo G M, Tornow J. Efficient transcription of the glycolytic gene ADH1 and three translational component genes requires the GCR1 product, which can act through TUF/GRF/RAP binding sites. Mol Cell Biol. 1990;10:859–862. doi: 10.1128/mcb.10.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schena M, Shalon D, Davis R W, Brown P O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 46.Schneider T D, Stephens R M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott E W, Allison H E, Baker H V. Characterization of TPI gene expression in isogenic wild-type and gcr1-deletion mutant strains of Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:7099–7107. doi: 10.1093/nar/18.23.7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott E W, Baker H V. Concerted action of the transcriptional activators REB1, RAP1, and GCR1 in the high-level expression of the glycolytic gene TPI. Mol Cell Biol. 1993;13:543–550. doi: 10.1128/mcb.13.1.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- 50.Tornow J, Zeng X, Gao W, Santangelo G M. GCR1, a transcriptional activator in Saccharomyces cerevisiae, complexes with RAP1 and can function without its DNA binding domain. EMBO J. 1993;12:2431–2437. doi: 10.1002/j.1460-2075.1993.tb05897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Turkel S, Liao X B, Farabaugh P J. GCR1-dependent transcriptional activation of yeast retrotransposon Ty2-917. Yeast. 1997;13:917–930. doi: 10.1002/(SICI)1097-0061(199708)13:10<917::AID-YEA148>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 52.Uemura H, Fraenkel D G. Glucose metabolism in gcr mutants of Saccharomyces cerevisiae. J Bacteriol. 1999;181:4719–4723. doi: 10.1128/jb.181.15.4719-4723.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uemura H, Koshio M, Inoue Y, Lopez M C, Baker H V. The role of Gcr1p in the transcriptional activation of glycolytic genes in yeast Saccharomyces cerevisiae. Genetics. 1997;147:521–532. doi: 10.1093/genetics/147.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Velculescu V E, Zhang L, Zhou W, Vogelstein J, Basrai M A, Bassett D E, Jr, Hieter P, Vogelstein B, Kinzler K W. Characterization of the yeast transcriptome. Cell. 1997;88:243–251. doi: 10.1016/s0092-8674(00)81845-0. [DOI] [PubMed] [Google Scholar]
- 55.Willett C E, Gelfman C M, Holland M J. A complex regulatory element from the yeast gene ENO2 modulates GCR1-dependent transcriptional activation. Mol Cell Biol. 1993;13:2623–2633. doi: 10.1128/mcb.13.4.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyrick J J, Holstege F C, Jennings E G, Causton H C, Shore D, Grunstein M, Lander E S, Young R A. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- 57.Zeng X, Deminoff S J, Santangelo G M. Specialized Rap1p/Gcr1p transcriptional activation through Gcr1p DNA contacts requires Gcr2p, as does hyperphosphorylation of Gcr1p. Genetics. 1997;147:493–505. doi: 10.1093/genetics/147.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]