Abstract
Background
Circular RNAs (circRNAs) hold potential as diagnostic markers for colorectal cancer (CRC); however, their functional mechanisms remain incompletely elucidated. This work investigates the clinical implications of a unique set comprising six circRNAs derived from serum in CRC. Furthermore, we delve into the role of exosomal circ_0084043, originating from colorectal cancer-associated fibroblasts (CAFs), with a specific focus on its contribution to endothelial cell angiogenesis.
Methods
The study analyzed circRNA levels in serum samples obtained from both CRC and control groups using qRT-PCR. Additionally, exosomes originating from colorectal CAFs and normal fibroblasts (NFs) were purified and confirmed by electron microscopy and Western blotting techniques. The proangiogenic effects of CAF-derived exosomal circ_0084043 were assessed in endothelial cells through proliferation, migration, and in vitro capillary tube formation assays. Gain- and loss-of-function experiments were employed to clarify the role of the circ_0084043/miR-140–3p/HIF-1α axis in endothelial cell angiogenesis, utilizing luciferase reporter assay, Western blotting, and ELISA for mechanism elucidation.
Results
The candidate circRNAs (circ_0060745, circ_001569, circ_007142, circ_0084043, Circ_BANP, and CiRS-7) exhibited notably elevated expression in CRC patient sera compared to the levels observed in healthy individuals. Except for CiRS-7, all circRNAs showed elevated expression in CRC patients with positive lymph node metastasis and advanced tumor stages. Exosomes released by colorectal CAFs augmented endothelial cell proliferation, migration, and angiogenesis by upregulating VEGF expression and secretion. Circ_0084043 was highly detected in endothelial cells treated with CAF-derived exosomes. Silencing circ_0084043 reduced VEGFA expression and diminished CAF exosome-induced endothelial cell processes, indicating its pivotal role in angiogenesis. Circ_0084043 sponges miR-140–3p, regulating HIF-1α, and a reverse relationship was also identified between miR-140–3p and VEGFA in endothelial cells. Inhibiting miR-140–3p mitigated circ_0084043 knockdown effects in CAF exosome-treated endothelial cells. Co-transfection of si-circ_0084043 and a miR-140–3p inhibitor reversed the inhibited migration and angiogenesis caused by circ_0084043 knockdown in CAF exosome-treated endothelial cells. Inhibiting miR-140–3p rescued reduced VEGFA expression due to circ_0084043 knockdown in endothelial cells exposed to CAF-derived exosomes, indicating modulation of the circ_0084043/miR-140–3p/VEGF signaling in CAF-derived exosome-induced angiogenesis.
Conclusions
This study unveiled a distinctive signature of six serum-derived circular RNAs, indicating their potential as promising diagnostic biomarkers for CRC. Importantly, exosomal circ_0084043 originating from colorectal CAFs was identified as playing a crucial role in endothelial cell angiogenesis, exerting its influence through the modulation of the miR-140–3p/HIF-1α/VEGF signaling axis.
Keywords: Colorectal cancer, Cancer-associated fibroblasts, Exosome, Non-coding RNA, Angiogenesis
Highlights
-
•
This study introduces a signature of six serum-derived circRNAs that can distinguish between CRC and healthy individuals.
-
•
Transfer of CAF-derived circ_0084043 triggers endothelial cell angiogenesis through the miR-140–3p/HIF-1α/VEGF signaling axis.
-
•
Th candidate circRNAs can serve as dual biomarkers and therapeutic targets, advancing the understanding of CRC pathogenesis.
1. Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer globally [1], ranking third in frequency among men and second among women [2]. It also claims the second-highest mortality rate among global cancer-associated deaths [3] for both males and females [2]. Despite ongoing efforts in treatments aimed at improving survival chances for CRC patients, the efficacy of treatment strategies often falls short, resulting in a bleak prognosis for individuals with CRC [4]. Approximately one-third of the CRC patients are likely to be identified with metastatic CRC, thus requiring post-surgical intervention [5] to minimize metastasis. Given that information on disease progression is crucial, biomarkers would greatly contribute to monitoring CRC pathology and preventing tumor development.
Among different diagnostic and prognostic biomarkers, non-coding RNAs (ncRNAs), such as microRNA (miRNAs), hold promise as they are tissue-specific [6] and abundant within various body fluids. miRNAs are short, single-stranded nucleotide sequences typically composed of 21–25 nucleotides and serve as gene silencers by inhibiting the translation of mRNAs [7]. These molecules can accurately reflect CRC development at different stages [[8], [9], [10]], underscoring their importance in CRC [11]. Additionally, miRNAs interact with various ncRNAs under different conditions, broadening the repertoire of ncRNAs as biomarkers, including the recently discovered circular RNAs (circRNAs) in CRC [11] which function as miRNA sponges. CircRNAs are resilient and stable RNA molecules characterized by their closed circular RNA structure [12], rendering them more resistant to exonuclease degradation [13,14]. Primarily recognized as miRNA sponges, circRNAs impede miRNAs from executing their inhibitory functions [15]. Due to their widespread presence and exonuclease-resistant structure, circRNAs have emerged as ideal biomarkers for cancer diagnosis, including CRC [11,14]. For instance, circRNA CiRS-7, exhibits abnormally high expression in CRC tissues and is identified as a promising therapeutic target due to its involvement in CRC progression [16], suggesting its potential as a CRC biomarker. However, the precise mechanism by which these molecules alter cell fate, transitioning normal cells into tumor cells, remains elusive.
Exosomes, tools of tumor cells carrying ncRNAs, have gained prominence as pivotal elements of cancer metastasis, influencing normal cell physiology and the microenvironment. These unique extracellular vesicles (50–150 nm in diameter) are released from cellular membranes in the form of multivesicular bodies [17]. Exosomes are pivotal in instigating the establishment of a tumor microenvironment (TME), ultimately supporting the resilience and survival of cancer cells [18,19]. They achieve this by transferring various cargos, including ncRNAs, into recipient cells [[19], [20], [21]]. Cancerous cells, such as CRC cells, are capable of releasing exosomes, which also serve as effective indicators of cellular status [11]. It has been identified that exosomes mediate the conversion of normal fibroblasts (NFs) into cancer-associated fibroblasts (CAFs) [22]. CAFs primarily influence tumorigenesis through the secretion of extracellular vesicles, particularly exosomes, impacting the tumor phenotype and microenvironment [23,24]. These molecules can disrupt the activity of recipient cells, thereby contributing to tumorigenesis by regulating processes such as proliferation, immune escape, drug resistance, and metastasis [24]. However, the precise molecular mechanisms through which CAF-derived exosomes induce endothelial cell angiogenesis remain unclear.
In this work, our first objective was to ascertain the distinctiveness of a specific group of six circRNAs derived from serum (circ_0060745, circ_001569, circ_007142, circ_0084043, Circ-BANP, and CiRS-7) in distinguishing between individuals with CRC and those who are healthy. Our selection of the six-circRNA signature was driven by a blend of bioinformatics analysis and literature review, with a focus on cancer-related pathways. We prioritized circRNAs with potential roles in CRC development and interactions with established miRNAs in CRC. It was concluded that all of these candidate circRNAs can function as biomarkers for distinguishing between tumor and healthy samples. Furthermore, we revealed associations between the candidate circRNAs and clinical features in individuals diagnosed with CRC. Moreover, our study aimed to clarify whether exosomal circ_0084043, released by colorectal CAFs, stimulates endothelial cell migration and angiogenesis. We clarified that this process is primarily facilitated by the miR-140–3p/hypoxia inducible factor 1 (HIF-1α) axis, providing insights into the functional roles of circ_0084043 in CRC tumorigenesis. The findings of this work could offer novel molecular evidence, suggesting that circ_0084043 holds promise as a potential therapeutic target for CRC.
2. Patients, materials and methods
2.1. Subjects and specimens
Forty-five colorectal carcinoma patients at varying stages of the disease progression were selected for this study, along with 45 other healthy individuals as controls. The selected individuals were within the age bracket of 45–70 years and had not been subjected to any pharmacological interventions prior to sample collection. The cohort of healthy control participants was carefully matched for age and sex, and individuals with background of malignancies or chronic illnesses were excluded. CRC diagnoses were verified by histopathological examinations conducted on surgically excised tumor specimens. Tumor staging was assessed using the Tumor Node Metastasis (TNM) criteria, as proposed by pertinent criteria [25]. Supplementary Table S1 outlines the clinicopathological features of CRC patients.
2.2. In silico analyses
To illuminate the regulatory functions of the candidate miRNAs targeted by circRNAs and to highlight prominent molecular pathways from Kyoto Encyclopedia of Genes and Genomes (KEGG) [26], miRTarBase v7.0 algorithm was used [27]. For obtaining a list of candidate miRNAs, the miRTargetLink Human algorithm [28] was applied. This algorithm was useful as well to obtain and visualize the potential interaction networks. Circular RNA Interactome [29] and CircMiMi [30] were used to predict the interactions between miRNAs and circRNAs.
2.3. Cell line and primary human fibroblast culture
Human umbilical vein endothelial cells (HUVECs) were obtained from the National Cell Bank of Iran (NCBI; Code number: C554). The cells were cultivated in DMEM/F12 medium containing 10 % exosome-free FBS and 1 % penicillin/streptomycin. The tumor specimens were surgically collected from patients at Rasoul Akram and Shariati Hospitals. Importantly, the patients involved did not experience chemotherapy or radiotherapy prior to undergoing the surgical resection procedure. Collagenase A was used to isolate the fibroblasts from all samples, according to the previous published protocol [31]. In brief, fresh tissue samples, including both tumor and adjacent non-tumor tissue, were initially rinsed with PBS to remove contaminants. The tissues were cut into small fragments of 1–2 mm and then enzymatically digested using collagenase A (2 mg/mL). Following digestion, the tubes were vigorously shaken for 5 min, and then the tissue suspension was filtered to acquire a single-cell suspension. This suspension was centrifuged to collect the cells, which were then washed with PBS. Lastly, the cells were resuspended in fibroblast-specific culture medium and cultured to promote fibroblast adhesion and proliferation over multiple passages. The assessment of cell morphology was conducted using phase-contrast microscopy. Fibroblasts within the initial six passages were specifically employed. The supernatants were harvested upon fibroblasts reaching over 70 % confluency and were kept at −80 °C.
2.4. Exosome isolation and identification
The cellular supernatant underwent a series of centrifugation steps: initially at 500×g for 5 min, followed by 2000×g for 15 min, and finally at 12,000×g for 30 min to eliminate cellular debris. The supernatant was then ultracentrifuged at 120,000×g for 70 min. The resulting pellet was resuspended in ice-cold RNase- and DNase-free PBS. This suspension underwent another round of ultracentrifugation at 120,000×g for 70 min and suspended in PBS. To do imaging, a small portion of purified exosomes was fixed with 2.5 % glutaraldehyde followed by PBS washing. Ethanol was used to dehydrate the fixed exosomes, which were then covered with a thin gold layer on a dry glass surface for microscopic observation. A scanning electron microscope (SEM, KYKY-EM3200, China) was then used for exosome morphology and size observations. The exosomal protein concentration was evaluated using a BCA protein assay. Western blotting was utilized to detect the expression of exosome-specific protein marker CD9.
2.5. Fluorescent labeling of purified exosomes
The purified exosomes underwent labeling using the PKH26 Red Fluorescent labeling reagents (Sigma-Aldrich). Then, HUVECs were treated with these labeled exosomes in a six-well plate for 12 h. Afterward, the cells were fixed with 4 % paraformaldehyde for 4 h at 4 °C. Following fixation, HUVECs were permeabilized with 0.2 % Triton X-100 at room temperature for 10 min. To visualize the cells, the nucleus was stained using 4′-6-diamidino-2-phenylindole (DAPI), and observations were conducted using fluorescent confocal microscopy (ECLIPSE Ti, Nikon, Tokyo, Japan).
2.6. Transfection assays
HUVECs were transfected with the miR-140–3p mimic, miR-140–3p inhibitor, and their corresponding negative controls using Lipofectamine® 2000 (Invitrogen, USA). Additionally, two independent short interference RNAs (siRNAs) targeting circ_0084043 or a nonspecific siRNA negative control (si-NC) (GenePharma) were delivered into the cells using the same material. To do luciferase reporter assays, HUVECs were plated in 96-well plates and cultured for 24 h. The cells later on were transfected with either the miR-140–3p mimic along with the wild-type (WT) sequence of circ_0084043 or the 3′-UTR of HIF-1A. Lipofectamine® 2000 Reagent was employed for this transfection. The relative dual-luciferase activity was assessed 48 h after transfection using the dual-luciferase assay kit (Promega, Wisconsin, WI, USA). The firefly luciferase activity was normalized relative to the Renilla luciferase activity for each sample.
2.7. RNA extraction and qRT-PCR
All RNAs were extracted by TRIzol reagent (Invitrogen) following the manufacturer's recommendations and then were reverse-transcribed into cDNA using the PrimeScript RT reagent kit (TAKARA, Japan). Quantitative real-time reverse transcription polymerase chain reactions (qRT-PCR) were performed on a Step One Detection System (Applied Biosystems, USA) with SYBR Green Master Mix (Ampliqon, Denmark). Meanwhile, U48 small nuclear RNA (snRNA) and GAPDH were utilized as the internal references for miRNA and mRNA, respectively [32,33].
2.8. Western blot analysis
To analyze protein expression in both cellular and exosomal preparations, proteins were isolated using radioimmunoprecipitation assay lysis buffer (RockLand), with the addition of a protease inhibitor cocktail to preserve protein integrity. To facilitate protein detection, the protein samples were then loaded onto polyacrylamide gels and transferred to polyvinylidene difluoride membranes. To reduce non-specific binding, the membrane was blocked using 5 % skimmed milk. Following blocking, the membranes were incubated with specific primary antibodies (1:1000) targeting the proteins of interest. After thorough washing to remove unbound antibodies, the membranes were probed with relevant HRP-conjugated secondary antibodies, enabling visualization of specific protein bands via chemiluminescence detection (ECL, Amersham, Buckinghamshire, UK). To ensure accurate normalization and quantification, β-actin was employed as a loading control throughout the experiment.
2.9. Immunofluorescent staining
Immunochemistry (ICC) was utilized to detect protein markers Alpha Smooth Muscle Actin (α-SMA) and platelet-derived growth factor receptor alpha (PDGFR-α) in isolated CAFs, following the standard protocol. Briefly, after three passages, the cells were fixed using 4 % PFA and permeabilized with Triton X-100. Subsequently, they were incubated overnight at 4 °C with primary antibodies targeting α-SMA and PDGFR-α. Following this, incubation with pertinent secondary antibodies (1:1000) for 1 h allowed to reveal the protein expression in CAFs. Nuclei were counterstained with DAPI, and the stained fibroblasts were imaged using a fluorescence microscope (Olympus, CKX41).
2.10. Enzyme-linked immunosorbent assay (ELISA)
Supernatants harvested from HUVECs treated with CAF- or NF-derived for 48 h, along with a vehicle control (PBS), were utilized for quantitative measurements of secreted VEGF. Following collection, the media underwent centrifugation at 1000×g for 10 min to obtain the supernatant. An ELISA kit designated for human VEGFA (ab100662) was utilized to assess the secretion rate following the manufacturer's instructions. VEGFA secretion was evaluated at a wavelength of 450 nm (and 570 nm as the reference) using a microplate ELISA reader.
2.11. Cell counting and proliferation assessment
HUVECs were cultured in a 96-well plate (1 × 104 cells per well). Subsequently, the cells were exposed to different concentrations of CAF-derived exosomes or transfected with si-Circ_001422, along with their respective controls. We then assessed cell viability using the trypan blue dye at 24- and 48-h post-exposure incubation period. Additionally, cell proliferation in transfected cells was assessed using the cell counting kit-8 (CCK-8) assay. Briefly, endothelial cells were plated at approximately 2 × 103 cells per well in triplicate. After a 2.5-h incubation at 37 °C, 10 μl of CCK-8 solution was added, and the optical density was assessed at a wavelength of 450 nm using the SpectraMax M5 microplate reader.
2.12. Migration assay
Transwell migration chambers (8 μm pore size; BD Biosciences) were initially placed at room temperature, and then 500 μl of serum-free culture medium was added. The chambers were subsequently incubated at 37 °C for 2 h to ensure the complete hydration of the basement membrane. HUVECs were maintained in serum-starved media and then seeded into the transwell chambers. The lower chamber was filled with a medium containing 10 % exosome-free FBS, which acted as a chemoattractant. The cells were then maintained in an environment of 37 °C and 5 % CO2 for a duration of 12 h. Following the incubation period, HUVECs were fixed with 4 % PFA for 30 min. The cells then underwent staining with a 0.1 % methanol-dissolved crystal violet solution (ThermoFisher Scientific). Images were captured using an inverted microscope to facilitate counting.
2.13. Matrigel-based tube formation assay
The endothelial cells were subjected to serum starvation before being seeded onto Matrigel-coated 24-well plates. Subsequently, the cells were grown in high-glucose DMEM containing 10 % exosome-free FBS and incubated with 100 μg/ml of either CAF- or NF-derived exosomes or si-Circ_001422. The assessment of capillary-like structure formation was conducted using an inverted phase-contrast microscope after a 12-h incubation period. To quantify endothelial cell tubulogenesis, branch density and tube length were measured by selecting at least 3 fields per well in a completely random and blinded manner.
2.14. Statistical analyses
The data are expressed as mean values with their respective standard deviations (SD) using GraphPad Prism v9. Statistical analysis involved either two-tailed Student's t-tests or one-way ANOVAs, depending on the type of comparison. To assess the biomarker potential of each circRNA, receiver operating characteristic (ROC) curve analysis was conducted, measuring the area under the curve (AUC). Throughout all analyses, statistical significance was defined as p-values <0.05.
3. Results
3.1. Analysis of expression and diagnostic potential of a panel of six circulating circRNAs in colorectal cancer
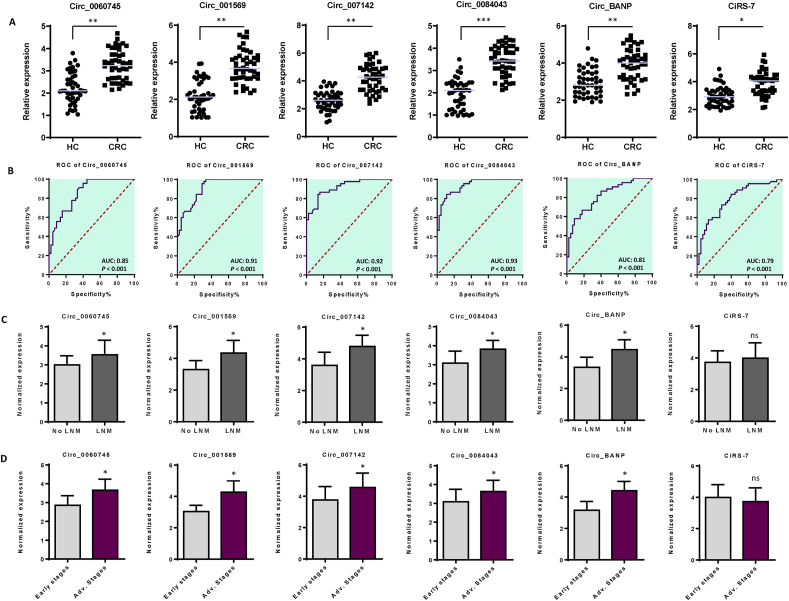
Although circRNAs have become valuable diagnostic and prognostic biomarkers for patients with CRC [34], their specific functions in advanced-stage CRC are still not well understood. Based on prior experimental evidence suggesting their potential oncogenic roles and interactions with established miRNAs in CRC [[35], [36], [37], [38], [39], [40]], we prioritized candidate circRNAs for their potential roles in CRC diagnosis. In this work, we evaluated the expression levels of six circRNAs —circ_0060745, circ_001569, circ_007142, circ_0084043, Circ-BANP, and CiRS-7— in serum specimens obtained from CRC patients and healthy controls. QRT-PCR data indicated elevated expression levels of this panel of circRNAs in CRC patients (Fig. 1A). In order to determine the potential of each circRNA as a diagnostic marker, we used ROC curve analysis, which indicated that all circRNAs could indeed function as biomarkers for CRC diagnosis. Specifically, our results revealed statistically significant AUC values (p-values <0.001) for each circRNA: circ_0060745 at 0.85, circ_001569 at 0.91, circ_007142 at 0.92, circ_0084043 at 0.93, circ-BANP at 0.81, and CiRS-7 at 0.79. These results underscored the effectiveness of these circRNAs in distinguishing between CRC and healthy samples (Fig. 1B and Supplementary Fig. S1).
Fig. 1.
A panel of six serum-derived circular RNAs and their potential diagnostic value in CRC. A. Expression levels of circRNAs, including circ_0060745, circ_001569, circ_007142, circ_0084043, Circ_BANP, and CiRS-7, in CRC and healthy control serum samples. B. The area under the ROC curve (AUC) with 95 % of confidence intervals (CIs) was determined to test the diagnostic accuracy of each circular RNA. All circRNAs exhibited satisfactory AUC values, with circ_0060745 at 0.85, circ_001569 at 0.91, circ_007142 at 0.92, circ_0084043 at 0.93, Circ_BANP at 0.81, and CiRS-7 at 0.79. In all cases, p-values were less than 0.001. C. Expression levels of serum-derived circRNAs in CRC patients with positive and negative lymph node metastasis (LNM). All circRNAs (except for CiRS-7) exhibited elevated expression levels in CRC patients with positive LNM. D. Expression levels of serum-derived circRNAs associated with TNM stages. All circRNAs (except for CiRS-7) showed high expression levels in advanced stages of tumorigenesis in CRC patients. The columns represent the means of three independent experiments, with the bars indicating the standard deviation (SD). ns: not significant, *P < 0.05, **P < 0.01. HC: Healthy control, CRC: Colorectal cancer patients, ROC: Receiver operating characteristic, AUC: Area Under the Curve, Early stages: TNM stages I and II, Advanced stages: TNM stage III.
In the subsequent phase, we delved into the potential relatioship between the altered expression levels of each circRNA and lymph node metastasis (LNM) scores in CRC specimens. Notably, all circRNAs, with the exception of CiRS-7, exhibited elevated expression levels in patients with positive LNM. However, a significant correlation was not discerned for CiRS-7 across the conditions of LNM-positive and LNM-negative CRC patients (Fig. 1C). Furthermore, to unravel the significance in a clinical context, we initially investigated the expression levels of the candidate circRNAs with the tumorigenesis-specific score (also known as TNM) in CRC patients. The 'T' (in TNM) signifies tumor malignancy, distinguishing between benign and malignant tumors. 'N' (in TNM) denotes lymph node metastasis, and finally, 'M' represents metastasis, identifying the dissemination of tumor cells from the primary tumor site to other organs [41]. The 'T' classification ranges from I to IV, with a higher number indicating greater malignancy detection [41,42]. Our results demonstrated elevated expression levels of five candidate circRNAs (circ_0060745, circ_007142, 0084043, circ_0060745, and circ-BANP) in patients at advanced stages of tumorigenesis. However, CiRS-7 displayed no correlation with tumorigenesis stages (Fig. 1D).
3.2. Characterization and evaluation of cellular internalization of exosomes obtained from primary fibroblasts
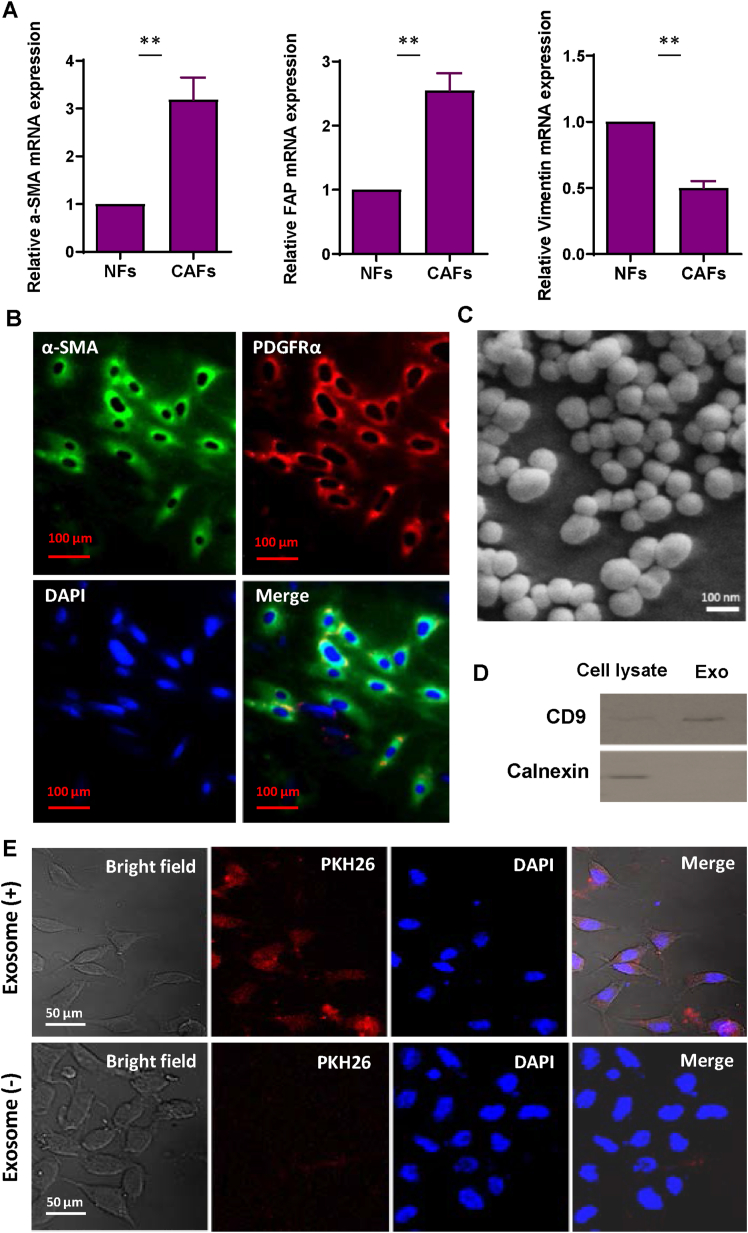
Each specimen from CRC patients underwent detailed morphological examination, with particular attention given to dividing areas containing CAF and NF into separate fragments. The tissues were dissociated, and the cells from the tissues cultured. These cells were differentiated based on their physical characteristics and biomarker profiles. All CAFs displayed a spindle-shaped morphology and a multi-layered growth pattern when reaching a specific cellular density, defining the typical cell morphology for this type cells. Furthermore, fibroblast identity was confirmed through analyzing the expression of fibroblast-specific markers α-SMA, Fibroblast Activation Protein (FAP), and Vimentin via qRT-PCR. As depicted in Fig. 2A, the upregulation of α-SMA and FAP in the isolated CAFs strongly indicated the success of their isolation, while NFs exhibited higher levels of Vimentin expression. Moreover, ICC staining of α-SMA and PDGFR-α confirmed the expression of these positive surface markers in isolated CAFs (Fig. 2B).
Fig. 2.
Characterization of exosomes derived from colorectal cancer-associated fibroblasts isolated from colorectal tumor tissues. A. QRT-PCR analysis of the expression of CAF markers (α-SMA and FAP) and fibroblasts marker (Vimentin) in isolated fibroblasts. Remarkably, CAFs displayed low levels of Vimentin, a marker for NF population. B. Immunocytochemical staining of CAF-related proteins α-SMA and PDGFR-α in primary CAFs isolated from CRC patients' tissues. The scale bar represents 100 μM. C. A scanning electron micrograph of purified exosomes reveals spherical and membrane-encapsulated particles with diameters ranging from 50 to 150 nm. D. Western blot analysis revealed that exosomes derived from CAFs were positive for CD9 but negative for calnexin, confirming the successful purification of fibroblast-derived exosomes. The original bots of Fig. 2D are provided in Supplementary Fig. S5. E. Internalization of purified exosomes into endothelial cells. HUVECs were incubated with CAF-derived exosomes labeled with PKH26 (red) for 12 h. A red fluorescence in the cytoplasm of HUVECs indicates significant uptake of exosomes by the cells. Additionally, HUVECs were exposed to PKH26 alone without exosomes as a negative control to monitor any PKH26 carryover. The columns represent the means of three independent experiments, with the bars indicating the standard deviation (SD). ** p-value <0.01. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The exosomes isolated from CAFs exhibited a morphological profile, characterized by oval or bowl-shaped structures measuring between 50 and 150 nm in diameter. This observation was supported by SEM imaging, as depicted in Fig. 2C. Additionally, Western blot analysis confirmed the presence of CD9, a distinct exosome marker, in the purified exosomes (Fig. 2D). To eliminate the possibility of contamination of isolated exosomes with other extracellular vesicles, the protein levels of Calnexin (a protein that signifies cellular compartments such as endoplasmic reticulum) was assessed in exosome preparations. Our data did not show any expression of Calnexin in isolated exosomes, suggesting that all isolated particles are pure exosomes (Fig. 2D). To assess whether the isolated exosomes are internalized by endothelial cells, the isolated exosomes were labeled by PKH26 and incubated with HUVECs for 12 h. Confocal microscopy imaging revealed that red fluorescently labeled exosomes were situated within the cytoplasm of the endothelial cells. These findings provide evidence that endothelial cells uptake CAF-derived exosomes (Fig. 2E).
3.3. Exosomes released by colorectal cancer-associated fibroblasts enhance endothelial cell angiogenesis and upregulate VEGF in HUVECs
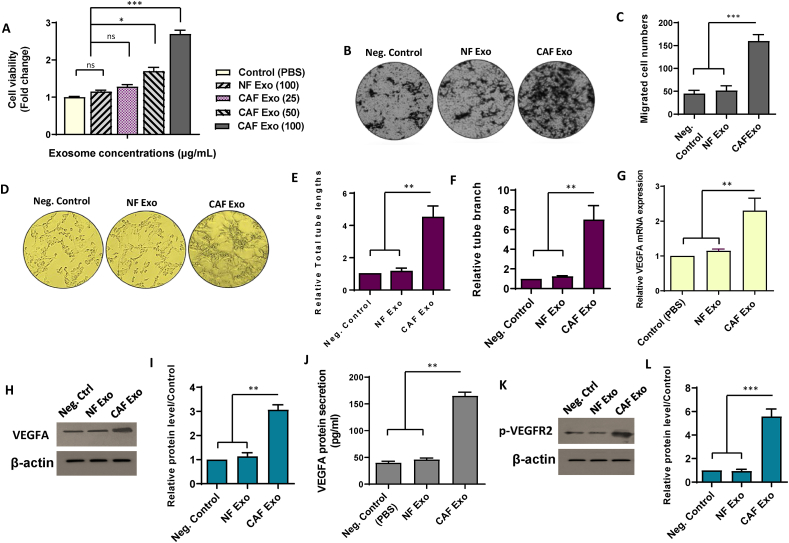
To evaluate how colorectal CAF-derived exosomes influence endothelial cell characteristics, we conducted an initial investigation, considering the possibility of CAF-induced alterations in endothelial cell proliferation and migration. To this end, we utilized varying concentrations (25, 50, and 100 μg/ml) of exosomes obtained from colorectal CAFs to examine how CAFs affect HUVEC proliferation through a cell counting kit-8 (CCK-8) assay. The findings demonstrated that 100 μg/ml of CAF Exo significantly enhanced the viability of HUVECs compared to lower concentrations of CAF Exo or 100 μg/ml of NF Exo after a 24-h incubation period (Fig. 3A). Consequently, 100 μg/ml CAF Exo was selected for use in all subsequent experiments. Moreover, HUVECs incubated with CAF-derived exosomes (100 μg/ml) exhibited significantly enhanced cell migration compared to those exposed to the same concentration of NF-derived exosomes (p-value <0.001) (Fig. 3B and C). The findings indicated the potential of CAF-derived exosomes to significantly promote both endothelial cell proliferation and migration. Conversely, administration of NF-derived exosomes did not lead to observable changes in endothelial cell characteristics compared to the negative controls treated with PBS.
Fig. 3.
Exosomes released by colorectal cancer-associated fibroblasts enhance endothelial cell angiogenesis and up regulate VEGF in HUVECs. A. CAF-derived exosomes enhanced the proliferation rate of HUVECs in a dose-dependent manner. B–C. Treatment of HUVECs with 100 μg/ml of CAF-derived exosomes resulted in increased cell migration in comparison with cells incubated with PBS or an equivalent of NF-derived exosomes. D. Representative photomicrographs illustrate that HUVECs treated with Exo CAF (100 μg/mL) form more and longer tube-like structures on Matrigel compared to control conditions. E, F. Quantitative assessment of the tube length and branch. Treatment of the CAF-derived exosomes (100 μg/mL) resulted in the increased total tube length (E) and the number of tube branches (F) in HUVECs. G-I. Assessment of VEGFA levels at both mRNA (G) and protein (H) levels in HUVECs treated with 100 μg/mL exosomes derived from either CAF or NFs. The original bots of Fig. 3H are provided in Supplementary Fig. S6. For quantitative analysis, the density of bands was assessed using ImageJ software and represented as relative intensity (I). J. Quantitative measurements of secreted VEGF using ELISA revealed elevated secretion of VEGFA in HUVECs treated with 100 μg/mL CAF-derived exosomes compared to control conditions and those treated with 100 μg/mL NF-derived exosomes. K, L. Western blot analysis showed the increased phosphorylated level of VEGFR2 in HUVECs incubated with 100 μg/mL CAF-derived exosomes. There was no significant difference in VEGFR2 phosphorylation levels between HUVECs incubated with NF-derived exosomes and those treated with PBS. The original bots of Fig. 3K are provided in Supplementary Fig. S7. For quantitative analysis, the density of bands was assessed using ImageJ software and represented as relative intensity (L). The columns represent the means of three independent experiments, with the bars indicating the standard deviation (SD). ns: not significant, * p-value <0.05, ** p-value <0.01, *** p-value <0.001.
Considering the undeniable significance of angiogenesis in tumor progression, our investigation aimed to evaluate the impact of CAF-derived exosomes on endothelial cell angiogenesis. The results of the matrigel-based tube formation assay showed that exposure to CAF Exo (100 μg/ml) significantly increased in vitro tubulogenesis of HUVECs (p-value <0.01). However, treatment of HUVECs with NF-derived exosomes (100 μg/ml) or PBS did not result in any noticeable changes in the capacity of endothelial cells to form capillary structures (Fig. 3D–F). These findings indicated the potential of CAF-derived exosomes to enhance both the tube length and number of branches in recipient endothelial cells.
Building upon the suggestion that stromal cells enhance endothelial cell angiogenesis through the secretion of proteins, such as VEGF [43], our investigation proceeded to assess whether a similar pattern could manifest in HUVECs treated with CAF-derived exosomes. QRT-PCR results revealed an upregulation of VEGFA mRNA levels in HUVECs treated with CAF-derived exosomes (Fig. 3G). To corroborate these findings at the protein level, we conducted Western blot analysis to assess VEGFA expression in the three experimental groups. The results demonstrated an increase in VEGFA protein levels in endothelial cells incubated with CAF Exo compared to both the control and NF-derived exosome-treated cells (Fig. 3H and I). The noted increase in VEGFA levels, observed both at mRNA and protein levels, sparked additional exploration into the potential impact on VEGFA secretion in HUVECs following treatment with CAF-derived exosomes. To investigate this further, the secretion of VEGFA was quantified using ELISA. The findings revealed a heightened secretion of VEGFA in endothelial cells incubated with CAF-derived exosomes compared to both the control group and those incubated with NF-derived exosomes (p-value <0.01) (Fig. 3J).
The significance of VEGFR-2 as a pivotal endothelial cell-specific receptor implicated in VEGF-triggered angiogenesis was also emphasized [44]. To identify the potential paracrine effects of exosomes derived from CAFs on endothelial cell angiogenesis, HUVECs were exposed to 100 μg/ml of CAF-derived exosomes, and the activation of VEGFR2 in HUVECs was evaluated. Western blot analysis revealed increased VEGFR2 phosphorylation in HUVECs exposed to CAF-derived exosomes compared to controls (Fig. 3K and L). on the other hand, there was no notable disparity observed in the phosphorylated levels of VEGFR2 between HUVECs exposed to NF-derived exosomes and those exposed to PBS alone (Fig. 3K and L). Altogether, the collective findings substantiate the hypothesis that CAF-derived exosomes may potentially contribute to enhancing endothelial cell angiogenesis by activating the VEGF signaling pathway.
3.4. Circ_0084043 participates in the pro-angiogenic effects of CAF-derived exosomes in endothelial cells
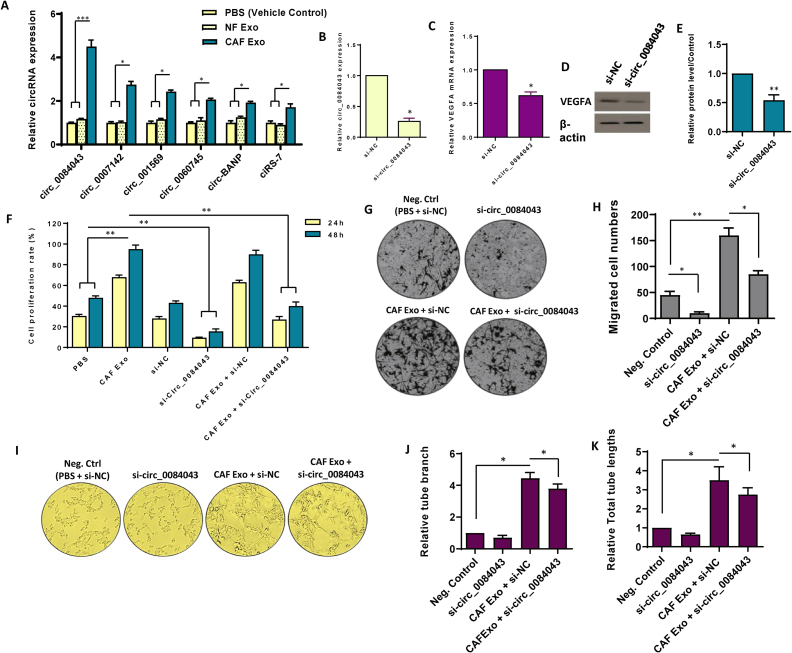
Growing evidence proposes that circRNAs take crucial parts in regulating cellular functions and are implicated in the tumorigenesis of several cancer types, including CRC [45]. Endothelial cells are crucial in tumor angiogenesis, and circRNAs affecting these cells can significantly influence tumor development [46]. Here, we began by assessing the expression of specific circRNAs —circ_0060745, circ_0007142, circ_001569, circ_0084043, ciRS-7, and circ-BANP— in HUVECs after exposure to 100 μg/ml CAF Exo. Among these circRNAs, circ_0084043 was found to have highest expression level (Fig. 4A). Furthermore, to verify whether circ_0084043 is transferred from CAFs to endothelial cells instead of being transcriptionally induced, we exposed HUVECs to either CAF-derived exosomes or PBS, along with α-amanitin, a component that represses transcriptional activation. As depicted in Supplementary Fig. S2, we observed a gradual increase in the level of circ_0084043 in HUVECs treated with 100 μg/mL CAF Exo, while its level remained relatively unchanged in the control endothelial cells. These findings confirm the transfer of circ_0084043 from CAFs to endothelial cells via exosomes.
Fig. 4.
Circ_0084043 orchestrates pro-angiogenic effects in endothelial cells induced by exosomes originating from colorectal cancer-associated fibroblasts. A. The differential expression of six candidate circRNAs was assessed in endothelial cells following treatment with 100 μg/mL exosomes derived from cancer-associated fibroblasts (CAF), exosomes derived from normal fibroblasts (NF), and PBS as a vehicle control. Notably, circ_0084043 exhibited the most substantial differences in expression levels among the tested conditions. B. The relative expression of circ_0084043 was examined after treating the cells with siRNA targeting circ_0084043 (si-circ_0084043) for 24 h. C-E. Following the inhibition of circ_0084043 using siRNA, both mRNA (C) and protein (D) levels of VEGFA were evaluated, indicating that circ_0084043 induced VEGFA expression in endothelial cells. The original bots of Fig. 4D are provided in Supplementary Fig. S8. For quantitative analysis, the density of bands was assessed using ImageJ software and represented as relative intensity (E). F. The relationship between the expression levels of CAF-derived exosomal circ_0084043 and cell proliferation in endothelial cells was observed over 24 and 48 h. Upon the inhibition of circ_0084043, cell proliferation decreased. G, H. Microscopic images, along with quantitative analysis, revealed a heightened rate of cell migration in endothelial cells treated with 100 μg/mL CAF-derived exosomes. However, upon inhibition of circ_0084043, cell migration decreased. I. The tube formation assay demonstrated that treating endothelial cells with CAF-derived exosomes (100 μg/mL) increased the formation of tubes, the number of branches, and the overall length. Conversely, inhibiting circ_0084043 significantly decreased the angiogenesis rate in endothelial cells. J, K. Quantitative assessment of the tube length and branch points calculated in three randomly selected fields. The columns represent the means of three independent experiments, with the bars indicating the standard deviation (SD). * p-value <0.05, ** p-value <0.01, *** p-value <0.001.
To delineate the possible function of circ_0084043 in endothelial cells, circ_0084043 was specifically targeted using siRNA in HUVECs. As shown in Fig. 4B, siRNAs directed against circ_0084043 (si-circ_0084043) exhibited significant suppression in the expression of circ_0084043, underscoring the efficacy of si-circ_0084043 in targeting its intended molecular target. Initially, our focus was to determine whether circ_0084043 positively regulated VEGFA. Results from both qRT-PCR and western blotting indicated that silencing circ_0084043 significantly reduced VEGFA expression in CAF Exo-treated HUVECs (Fig. 4C–E).
To further corroborate circ_0084043's involvement in endothelial cell proliferation, HUVECs were exposed to CAF-derived exosomes (100 μg/ml), and cell proliferation was assessed at 24- and 48-h post-treatment. The findings demonstrated a notable enhancement in cell proliferation at both time points following treatment with CAF-derived exosomes in comparison with the negative control group treated with PBS (p-value <0.01) (Fig. 4F). To ascertain whether the observed proliferative effect is primarily attributed to circ_0084043, HUVECs were transfected with si-circ_0084043 to specifically inhibit circ_0084043. The data illustrated a decrease in cell proliferation in the si-circ_0084043-treated group at both time points compared to the PBS-treated group (p-value <0.01), emphasizing the crucial role of circ_0084043 in endothelial cell proliferation (Fig. 4F). To confirm the specific involvement of CAF-secreted exosomal circ_0084043 in this phenomenon, si-circ_0084043 was introduced alongside CAF-derived exosomes and compared with the combination of si-NC and CAF-derived exosome treatment. The findings indicated that inhibiting circ_0084043 significantly diminished the proliferative effects of CAF-derived exosomes in endothelial cells (p-value <0.01) (Fig. 4F). As endothelial cell migration is a crucial aspect of angiogenesis, we investigated the role of circ_0084043 in this process by initially exanimating the correlation between cell migration and circ_0084043. Our findings demonstrated that incubation of HUVECs with 100 μg/mL CAF-derived exosomes significantly increased endothelial cell migration (Fig. 4G and H). To confirm the specific involvement of CAF-secreted exosomal circ_0084043 in this migratory phenomenon, we employed siRNA to inhibit circ_0084043. The data showed that inhibiting circ_0084043 resulted in a significant decrease in cell migration, highlighting the influential role of circ_0084043 in endothelial cell migration. Crucially, transfection of HUVECs with si-circ_0084043 resulted in a partial attenuation of the stimulatory effect of CAF-derived exosomes on endothelial cell migration. (Fig. 4G and H).
To delve deeper into the complex role of circ_0084043 in angiogenesis, we performed a matrigel-based tube formation assay. Our results unveiled a notable increase in tube formation when cells were exposed to 100 μg/ml CAF-derived exosomes along with si-NC, as illustrated in Fig. 4I. To establish the connection between this angiogenic phenomenon and circ_0084043, we employed siRNA to suppress the expression of this circRNA. Intriguingly, a significant decrease in tube formation was observed upon circ_0084043 inhibition, underscoring the indispensable role of this circRNA in orchestrating angiogenesis. More importantly, we found that transfection of HUVECs with si-circ_0084043 led to a partial reduction of the enhancing effect exerted by CAF-derived exosomes on tube formation in endothelial cells (p-value <0.05) (Fig. 4I–K). Altogether, these data underscore the crucial involvement of CAF-secreted circ_0084043 in regulating fundamental aspects of angiogenesis in endothelial cells.
3.5. Circular RNA circ_0084043 functions as a mir-140–3p sponge, thereby inhibiting VEGF signaling
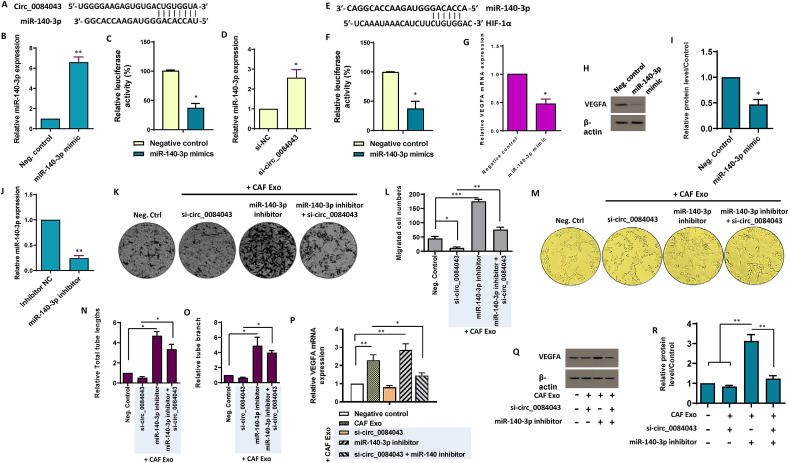
CircRNAs play a pivotal role in modulating cellular function, primarily by serving as targeted sponges for miRNAs, operating as competing endogenous RNA molecules [47]. Notably, circ_0084043 has demonstrated proficiency as an efficient miRNA sponge. Utilizing the https://circinteractome.nia.nih.gov/, we uncovered potential targets for the candidate circRNAs. Among a myriad of candidates, our focus honed in on miR-140–3p, given its established roles in hematopoiesis and angiogenesis [[48], [49], [50]]. Significantly, miR-140–3p demonstrated a strong association with circ_0084043 (Supplementary Fig. S3A). To enhance reliability, we also utilized CancerMIRNome to analyze the gene expression profiles of the candidate miRNA across CRC patients. The results showed a consistent decrease in the expression patterns of miR-140–3p in CRC patients (Supplementary Fig. S3B). To explore a potential association between circ_0084043 and miR-140–3p, as predicted by TargetScan and miRanda databases (Fig. 5A), we conducted a luciferase assay in vitro. Our findings revealed that when miR-140–3p was overexpressed (Fig. 5B), the luciferase intensity of the circ_0084043 reporter construct significantly decreased in HUVECs. This reduction was notably observed compared to cells transfected with the circ_0084043 reporter vector along with a negative control scramble (Fig. 5C). To further substantiate the targeting relationship between circ_0084043 and miR-140–3p, we performed an inhibition experiment in endothelial cells. As shown in Fig. 5D, the obtained data revealed that the knockdown of circ_0084043 led to an increased level of miR-140–3p. These findings suggest a regulatory interplay between circ_0084043 and miR-140–3p, supporting the idea that circ_0084043 can target and modulate the expression of miR-140–3p in endothelial cells.
Fig. 5.
Exosomes derived from CAFs promote endothelial cell angiogenesis by modulating the circ_0084043/miR-140-3p/VEGF axis. A-D. Circular RNA circ-0084043 acts as the sponge of miR-140–3p. A.In silico predictions indicated the complementary binding site between circ_0084043 and miR-140–3p. B. The relative expression of miR-140–3p increased in transfected endothelial cells with the miR-140–3p mimic, confirming the functionality of the miR-140–3p mimic. C. A significant reduction in luciferase activity was observed when the miR-140–3p mimic was introduced, suggesting the binding affinity between miR-140–3p and circ_0084043. The results suggest that circ_0084043 effectively functions as a sponge for miR-140–3p in endothelial cells. D. SiRNA-mediated knockdown of circ_0084043 resulted in an increase in the expression levels of miR-140–3p in HUVECs, as indicated by qRT-PCR. E-I. miR-140–3p directly targets HIF-1α to suppress VEGFA. E.In silico predictions indicated the complementary binding site between miR-140–3p and the 3′-UTR of HIF-1α transcript. F. By introducing the miR-140–3p, luciferase activity decreased, indicating the binding affinity between miR-140–3p and the 3′-UTR of HIF-1α. The results confirmed the targeting of HIF-1α by miR-140–3p in endothelial cells. G. The transcript expression of VEGFA decreased when HUVECs were treated with the miR-140–3p mimic, suggesting a suppressive effect of this miRNA on VEGFA expression. H, I. Western blot analysis showed that protein levels of VEGFA decreased in endothelial cells transfected with the miR-140–3p mimic. The original bots of Fig. 5H are provided in Supplementary Fig. S9. For quantitative analysis, the density of bands was assessed using ImageJ software and represented as relative intensity (I). J. The expression of miR-140–3p was measured in endothelial cells treated with a miR-140–3p inhibitor, confirming the functionality of the inhibitor. K, L. When miR-140–3p was inhibited in HUVECs exposed to CAF Exo (100 μg/mL), there was an increase in endothelial cell migration. However, co-inhibition of miR-140–3p and circ_0084043 diminished the promoting effect of miR-140–3p suppression on endothelial cell migration. M-O. Exosomes derived from CAFs enhanced tubularization of endothelial cells through shuttling of circ_001422. M. Representative micrographs showing that when si-circ_001422 was transfected, the ability of CAF Exo-treated endothelial cells to form capillary-like tubular structures was hindered. Importantly, the suppression of angiogenesis in CAF Exo-treated HUVECs, resulting from circ_0084043 knockdown, was rescued by co-transfection of si-circ_0084043 and the miR-153–3p inhibitor. Quantitative analyses of the tube length (N) and branch (O) of endothelial cells were calculated in three randomly selected fields. P–R. Inhibiting miR-140–3p in endothelial cells treated with 100 μg/mL CAF-derived exosomes increased the expression of VEGFA at both mRNA (P) and protein (Q) levels. Results indicated that the reduction of VEGF expression in CAF Exo-treated HUVECs, resulting from circ_0084043 knockdown, was rescued by co-transfection of si-circ_0084043 and the miR-153–3p inhibitor to some extent. The original bots of Fig. 5Q are provided in Supplementary Fig. S10. For quantitative analysis, the density of bands was assessed using ImageJ software and represented as relative intensity (R). The columns represent the means of three independent experiments, with the bars indicating the standard deviation (SD). * p-value <0.05, ** p-value <0.01, *** p-value <0.001.
The hypoxia-responsive transcriptional activator HIF-1 (hypoxia-inducible factor-1) is vital in orchestrating cellular responses to low oxygen levels [51]. It is well-established that the transcriptional activation of VEGF is finely controlled by the transcription factor HIF-1α [52]. As several studies have indicated a connection between hypoxia and miR-140–3p, with HIF-1α being recognized as an authentic target of miR-140–3p [[53], [54], [55]], we sought to identify a potential miR-140–3p binding site within HIF-1α (Fig. 5E). Our investigation revealed that co-transfection of the HIF-1α reporter vector with the miR-140–3p mimic, but not the scrambled oligonucleotide sequence, significantly reduced luciferase activity (Fig. 5F). This finding was consistent with qRT-PCR, which showed a marked decrease in HIF-1α transcript levels in HUVECs transfected with the miR-140–3p mimic (Supplementary Fig. S4). Considering HIF-1α′s function as a transcriptional activator of VEGFA expression, we proceeded to assess the mRNA and protein levels of VEGF after transfection with the miR-140–3p mimic. As demonstrated in Fig. 5G–I, the results indicated that the miR-140–3p mimic substantially reduced VEGFA expression. Overall, these findings suggest a regulatory mechanism in which circ_0084043 modulates VEGF signaling by serving as a sponge for miR-140–3p, thereby influencing HIF-1α expression in endothelial cells.
3.6. The pro-angiogenic effects of CAF exosomes could potentially be mediated by the modulation of the circ_0084043/miR-140–3p/VEGF axis in endothelial cells
To evaluate whether inhibiting miR-140–3p could mitigate the effects induced by suppressing circ_0084043 in CAF Exo-treated HUVECs, a set of functional assays was conducted. The efficacy of miR-140–3p inhibitor transfection was confirmed by qRT-PCR (Fig. 5J). Transwell migration assays indicated that co-transfection of si-circ_0084043 and the miR-140–3p inhibitor significantly reversed the inhibited migration caused by circ_0084043 knockdown in CAF Exo-treated HUVECs (Fig. 5K and L). Furthermore, the matrigel-based tube formation assay illustrated that the suppression of angiogenesis in CAF Exo-treated HUVECs resulting from circ_0084043 knockdown was rescued by co-transfecting si-circ_0084043 and the miR-153–3p inhibitor (Fig. 5M − O). Exploring the interplay among circ_0084043, miR-140–3p, and VEGFA in CAF Exo-treated HUVECs, we examined whether the reduced expression of VEGFA resulting from circ_0084043 knockdown could be restored by inhibiting miR-140–3p in HUVECs incubated with 100 μg/ml CAF Exo. Results from qRT-PCR and western blotting suggested that the reduction in VEGFA caused by circ_0084043 knockdown could be inhibited by co-transfecting si-circ_0084043 and the miR-140–3p inhibitor in CAF Exo-treated HUVECs (Fig. 5P-R). These data collectively propose that the pro-angiogenic effects of exosomes originating from CAFs may occur through the regulation of the circ_0084043/miR-140–3p/VEGF signaling axis in endothelial cells.
4. Discussion
Detecting CRC early is essential for minimizing morbidity and mortality [56]. Unfortunately, most CRC cases are diagnosed at advanced stages, often being asymptomatic, thereby restricting opportunities for timely and effective early treatment [57]. Hence, it is imperative to pinpoint novel therapeutic targets and biomarkers that can aid in early detection, personalized treatment, and precise monitoring of CRC, ultimately leading to an improved prognosis [58]. Recently, there has been an increasing fascination with the realm of exosome biology and its clinical relevance in CRC, spurred by changes in genetic and epigenetic factors [11]. Within the diverse landscape of ncRNAs, circular RNAs, a recently uncovered class, have emerged as noteworthy entities implicated in both the initiation and progression of tumors, while also holding potential as biomarkers [14]. Aligning with this, the primary objective of our study was to identify reliable diagnostic biomarkers for CRC and unravel the mechanism(s) through which the most prominent biomarker influences CRC development.
We noted higher expression levels of serum-derived circ_0060745, circ_001569, circ_007142, circ_0084043, circ-BANP, and CiRS-7 in CRC patients than healthy controls. This observation implies that all candidate circRNAs may function as oncogenes in the context of CRC. Subsequently, our aim was to clarify how these candidates function as potential biomarkers in CRC patients. In total, all of these circRNAs effectively distinguish between CRC patients and healthy controls, as confirmed by ROC curve analysis. Notably, circ_0084043 exhibited the most promising results, boasting an AUC 0.93 among the various candidates. It has been identified that circ_001569 not only exhibits increased expression in CRC patients but also effectively predicts a poor prognosis for CRC patients [59]. Furthermore, circ_007142 [37], circ_0060745 [35], circ-BANP [60], and CiRS-7 [40] have been acknowledged as potential biomarkers owing to their heightened expression in CRC patients. In the case of circ_0084043, there remains a significant gap in understanding its functions in CRC development. Interestingly, our investigations unveiled clinicopathological associations between circRNAs and stages of tumorigenesis; we demonstrated a positive correlation between all candidate circRNAs (excluding CiRS-7) and the advanced stages of tumor development. These collective data bolster the probability of these circRNAs as promising biomarkers for diagnosing and prognosticating CRC. We were also intrigued by the potential associations between these circRNAs and LNM conditions in CRC patients. Our data showed that all circRNAs (except CiRS-7) could distinguish between LNM-positive and LNM-negative CRC patients, further supporting their role as effective biomarkers for CRC.
CAFs emerge as the predominant components of the stromal and play essential roles in shaping the TME [61]. However, there still exists a gap in our understanding of the mechanism through which CAFs communicate with other component of the TME, resulting in a change in their fate [62]. The secretion of exosomes represents a crucial mechanism through which CAFs influence the behavior of the TME components [24]. Exosomes released by CAFs can initiate metastasis and confer chemotherapy resistance in CRC [63,64]. While CAFs have been shown to induce angiogenesis [65]; the particular involvement of CAF-derived exosomes in tumor angiogenesis has largely remained unexplored.
To elucidate the role of exosomal circ_0084043 originating from colorectal CAFs in endothelial cell angiogenesis, we initially isolated CAFs and their normal counterparts. CAFs and NFs displayed noticeable differences in their morphology and gene expression pattern. Classified as α-SMA+ myofibroblasts, CAFs also show high levels of FAP and platelet-derived growth factor receptor α (PDGFRα). Our findings depicted that isolated CAFs exhibited elevated levels of both α-SMA and FAP expression, whereas NFs predominantly expressed Vimentin, a specific fibroblast marker [66]. In the subsequent step, exosomes were isolated from each cell population, CAFs and NFs, to assess the gene expression pattern of each candidate circRNA. Given that CAF-derived exosomes have been shown to enhance the cell viability and alter the microenvironment of normal cells [65], our results demonstrated that treating endothelial cells with CAF-derived exosomes resulted in increased endothelial cell viability. CAFs actively contribute to the degradation of the extracellular matrix and the disruption of the endothelial barrier, thereby taking a part during metastatic growth and cancer progression [67]. When endothelial cells were exposed to CAF-derived exosomes, we observed an increase in their migration. As suggested by Lamalice et al., endothelial cell migration is crucial for vascularization [68]. Therefore, in the next step, we aimed to identify changes in endothelial cell angiogenesis driven by CAF-derived exosomes. Our data revealed that CAF-derived exosomes can indeed promote angiogenesis in endothelial cells. It was identified that this phenomenon is primarily mediated by enhanced VEGFA. However, it remained unclear which exosomal circRNA originating from CAFs could specifically promote angiogenesis in recipient endothelial cells. In addressing this question, we meticulously assessed the expression levels of circRNAs in isolated exosomes. While all candidate circRNAs exhibited increased expression, circ_0084043 emerged with the most promising expression pattern, particularly in CAF-derived exosomes. Notably, circ_0084043 has been identified as an oncogene in melanoma [69,70]. Furthermore, our experimental validation confirmed that inhibiting circ_0084043 leads to a decrease in endothelial cell proliferation, and the predominant source of circ_0084043 is exogenous, specifically CAF-derived exosomal circ_0084043. Building upon these findings, our subsequent investigations delved into the significance of circ_0084043 in endothelial cell migration. Inhibition of circ_0084043 resulted in a noticeable decrease in endothelial cell migration. Chen et al. previously demonstrated that the suppressing circ_0084043 in human melanoma models inhibits melanoma cell proliferation and migration, while enhancing cell apoptosis [69]. However; knowledge regarding the roles of this circRNA in CRC development is limited or nonexistent. Our study represents a pioneering effort, being the first to illuminate the crucial role of circ_0084043 in induced endothelial cell proliferation and migration triggered by CAF-derived exosomes in CRC. Subsequently, we explored the involvement of circ_0084043 in induced angiogenesis driven by CAF-derived exosomes, revealing that exosomes derived from CAFs can indeed promote angiogenesis in endothelial cells, primarily through the actions of circ_0084043.
The lingering question pertained to identifying the mechanism of circRNA action, specifically targeting miRNAs that play a role in tumor angiogenesis. To uncover these targets, we employed in silico analysis and found miR-140–3p to be the most significant miRNA interacting with circ_0084043. Liu et al. revealed that miR-140–3p suppresses CRC progression and acts as a tumor suppressor gene [71], as further corroborated by another investigation [72]. While there is limited information on how miR-140–3p specifically influences angiogenesis in CRC, Wan et al. identified that upregulation of miR-140–3p can suppress angiogenesis in lung adenocarcinoma [50]. Our luciferase assay confirmed the sponging interaction between circ_0084043 and miR-140–3p. Therefore, we hypothesized that circ_0084043 might exhibit a reverse effect on cell proliferation, migration, and angiogenesis compared to miR-140–3p. Our data indeed demonstrated that circ_0084043 not only enhances endothelial cell proliferation and migration, but also its inhibition can inhibit angiogenesis though the upregulation of miR-140–3p in endothelial cells. Our findings validated an upregulation in VEGFA expression following the inhibition of miR-140–3p. According to Lee et al. VEGFA binds to and activates both VEGFR-1 and VEGFR-2, stimulating vascular permeability and endothelial cell angiogenesis [73]. We propose that circ_0084043 sponges miR-140–3p, which is a negative regulator of endothelial cell angiogenesis. Indeed, we identified that miR-140–3p binds to and suppresses the HIF-1A expression. HIF-1α is an activator of VEGFA and promotes the transcription of VEGFA. Thus, if circ_0084043 is upregulated in CRC, it can subsequently decrease miR-140–3p level, relieving the inhibitory effects of this miRNA on angiogenesis (i.e., enhancing the expression levels of HIF-1α), endothelial cell proliferation and migration, and in turn, promoting angiogenesis in endothelial cells (Fig. 6).
Fig. 6.
Proposed mechanism of exosome-mediated regulation of endothelial cell angiogenesis in CRC. Exosomes originating from cancer-associated fibroblasts (CAFs) encapsulate circ_0084043, which functions as a sponge for miR-140–3p, leading to a reduction in the functional levels of miR-140–3p. Consequently, this reduction increases HIF-1α levels due to the relieved inhibitory effects of miR-140–3p. This proposed model suggests that exosome-transmitted circ_0084043 promotes endothelial cell migration and angiogenesis by regulating the miR-140–3p/HIF-1α/VEGF signaling axis.
The role of circ_0084043 seems to extend beyond endothelial cell angiogenesis and may also involve the metastasis of colorectal tumor cells. The pathology of CRC development highlights enhanced metastasis in the advanced stages of CRC [74,75]. With circ_0084043 identified to promote endothelial cell migration and angiogenesis, and CAF derived exosomes described for their disruptive properties of the endothelial barrier [67], it could suggest that CAF derived exosomal circ_0084043 could potentially contribute to the promotion of metastasis in CRC cells. The notably increased expression of circ_0084043 in late stages of CRC, along with its significantly higher expression in conditions with LNM compared to those without, implies a potential involvement of circ_0084043 in late-stage CRC, potentially promoting metastasis. However, further information is needed to draw a definitive conclusion. Hence, angiogenesis propelled by circ_0084043 through the miR-140–3p/HIF-1α/VEGF signaling axis lays down the groundwork for CRC cells by initially developing vessel pathways from the primary tumor for metastasis while also acquiring nutrients for sustenance and survival. As the disease progresses, the tumor becomes neoplastic and can metastasize, potentially involving circ_0084043 in facilitating the metastasis of CRC cells once more. Nevertheless, However, further studies are needed to validate this conjecture.
5. Conclusions
In summary, the present study unveiled a distinctive profile of six serum-derived circular RNAs, comprising circ_0060745, circ_001569, circ_007142, circ_0084043, Circ-BANP, and CiRS-7. This distinctive circRNA signature holds promise as a valuable biomarker for diagnostic purposes. Notably, our investigation delved into the intricate molecular mechanisms, highlighting the pivotal role of circ_0084043 in CAF-derived exosomes. We demonstrated its function as a miR-140–3p sponge, unleashing endothelial cell migration and angiogenesis by modulating the miR-140–3p/HIF-1α/VEGF axis. While further exploration is necessary to clarify the precise roles of the circ_0084043/miR-140–3p/VEGF signaling pathway, our study has established a foundation for understanding CRC pathogenesis and emphasized the potential of circRNAs as diagnostic biomarkers and therapeutic targets in cancer biology.
Funding
Funding for this study was provided by the Islamic Azad University, Tehran Medical Sciences, Tehran, Iran.
Ethic declarations
The study was approved by the ethics committee of Islamic Azad University, Iran (Number: IR.IAU.PS.REC.1400.245). The study was conducted according to established ethical guidelines and all participants provided written consent before starting the experiment.
Data availability
Data included in this work have not been deposited into a publicly available repository. The data supporting the findings of this study are available from the corresponding author upon reasonable request.
CRediT authorship contribution statement
Nafiseh Payervand: Methodology, Investigation. Katayoon Pakravan: Writing – review & editing, Methodology, Investigation. Ehsan Razmara: Writing – original draft, Software, Methodology. Kailash Kumar Vinu: Writing – review & editing, Writing – original draft, Methodology. Sara Ghodsi: Resources, Investigation. Masoumeh Heshmati: Supervision, Methodology. Sadegh Babashah: Writing – review & editing, Supervision, Resources, Project administration, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors wish to sincerely express gratitude to the research participants for their valuable contributions to this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e31584.
Contributor Information
Masoumeh Heshmati, Email: heshmati.m@iaups.ac.ir.
Sadegh Babashah, Email: babashah@modares.ac.ir.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Calu V., Ionescu A., Stanca L., Geicu O.I., Iordache F., Pisoschi A.M., Serban A.I., Bilteanu L. Sci. Rep. 2021;11:7940. doi: 10.1038/s41598-021-86941-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Delattre J.-F., Selcen Oguz Erdogan A., Cohen R., Shi Q., Emile J.-F., Taieb J., Tabernero J., André T., Meyerhardt J.A., Nagtegaal I.D., Svrcek M. Cancer Treat Rev. 2022;103 doi: 10.1016/j.ctrv.2021.102325. [DOI] [PubMed] [Google Scholar]
- 4.Curtin J.C. Expet Opin. Drug Discov. 2013;8:1153–1164. doi: 10.1517/17460441.2013.807249. [DOI] [PubMed] [Google Scholar]
- 5.Gilard-Pioc S., Abrahamowicz M., Mahboubi A., Bouvier A.-M., Dejardin O., Huszti E., Binquet C., Quantin C. Cancer epidemiology. 2015;39:447–455. doi: 10.1016/j.canep.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Ogata-Kawata H., Izumiya M., Kurioka D., Honma Y., Yamada Y., Furuta K., Gunji T., Ohta H., Okamoto H., Sonoda H., Watanabe M., Nakagama H., Yokota J., Kohno T., Tsuchiya N. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C.M. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raut J.R., Schöttker B., Holleczek B., Guo F., Bhardwaj M., Miah K., Schrotz-King P., Brenner H. Nat. Commun. 2021;12:4811. doi: 10.1038/s41467-021-25067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo X., Burwinkel B., Tao S., Brenner H. Cancer Epidemiol. Biomarkers Prev. 2011;20:1272–1286. doi: 10.1158/1055-9965.Epi-11-0035. [DOI] [PubMed] [Google Scholar]
- 10.Rafiee R., Razmara E., Motavaf M., Mossahebi-Mohammadi M., Khajehsharifi S., Rouhollah F., Babashah S. J. Cancer Res. Therapeut. 2022;18:S383–S390. doi: 10.4103/jcrt.JCRT_752_20. [DOI] [PubMed] [Google Scholar]
- 11.Ghafouri I., Pakravan K., Razmara E., Montazeri M., Rouhollah F., Babashah S. J. Cancer Res. Clin. Oncol. 2023;149:12227–12240. doi: 10.1007/s00432-023-05095-1. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y., Luo J., Yang W., Ye W.-C. Cell Death Dis. 2023;14:353. doi: 10.1038/s41419-023-05881-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.-L., Yang L., Chen L.-L. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 14.Almouh M., Razmara E., Bitaraf A., Ghazimoradi M.H., Hassan Z.M., Babashah S. Life Sci. 2022 doi: 10.1016/j.lfs.2022.120975. [DOI] [PubMed] [Google Scholar]
- 15.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 16.Weng W., Wei Q., Toden S., Yoshida K., Nagasaka T., Fujiwara T., Cai S., Qin H., Ma Y., Goel A. Clin. Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.Ccr-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahgoub E.O., Razmara E., Bitaraf A., Norouzi F.-S., Montazeri M., Behzadi-Andouhjerdi R., Falahati M., Cheng K., Haik Y., Hasan A. Mol. Biol. Rep. 2020;47:7229–7251. doi: 10.1007/s11033-020-05715-w. [DOI] [PubMed] [Google Scholar]
- 18.Shu S.L., Yang Y., Allen C.L., Maguire O., Minderman H., Sen A., Ciesielski M.J., Collins K.A., Bush P.J., Singh P. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-31323-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobb R.J., Lima L.G., Möller A. Seminars in Cell & Developmental Biology. Elsevier; 2017. pp. 3–10. [DOI] [PubMed] [Google Scholar]
- 20.Ghaffari-Makhmalbaf P., Sayyad M., Pakravan K., Razmara E., Bitaraf A., Bakhshinejad B., Goudarzi P., Yousefi H., Pournaghshband M., Nemati F. Life Sci. 2021;264 doi: 10.1016/j.lfs.2020.118719. [DOI] [PubMed] [Google Scholar]
- 21.Razmara E., Bitaraf A., Karimi B., Babashah S. Ann. N. Y. Acad. Sci. 2021;1503:5–22. doi: 10.1111/nyas.14668. [DOI] [PubMed] [Google Scholar]
- 22.Peng L., Wang D., Han Y., Huang T., He X., Wang J., Ou C. Front. Immunol. 2022;12 [Google Scholar]
- 23.Bhatta B., Cooks T. Carcinogenesis. 2020;41:1461–1470. doi: 10.1093/carcin/bgaa107. [DOI] [PubMed] [Google Scholar]
- 24.Li C., Teixeira A.F., Zhu H.-J., Ten Dijke P. Mol. Cancer. 2021;20:1–19. doi: 10.1186/s12943-021-01463-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang S.H., Xu W., Waldron J., Siu L., Shen X., Tong L., Ringash J., Bayley A., Kim J., Hope A. J. Clin. Oncol. 2015;33:836–845. doi: 10.1200/JCO.2014.58.6412. [DOI] [PubMed] [Google Scholar]
- 26.Kanehisa M., Goto S. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang H.-Y., Lin Y.-C.-D., Li J., Huang K.-Y., Shrestha S., Hong H.-C., Tang Y., Chen Y.-G., Jin C.-N., Yu Y. Nucleic Acids Res. 2020;48:D148–D154. doi: 10.1093/nar/gkz896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamberg M., Backes C., Fehlmann T., Hart M., Meder B., Meese E., Keller A. Int. J. Mol. Sci. 2016;17:564. doi: 10.3390/ijms17040564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiang T.-W., Mai T.-L., Chuang T.-J. BMC Bioinf. 2022;23:1–14. doi: 10.1186/s12859-022-04692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pakravan K., Mossahebi-Mohammadi M., Ghazimoradi M.H., Cho W.C., Sadeghizadeh M., Babashah S. J. Transl. Med. 2022;20:1–21. doi: 10.1186/s12967-022-03780-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bitaraf A., Babashah S., Garshasbi M. J. Clin. Lab. Anal. 2020;34 doi: 10.1002/jcla.23063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nosaeid M.H., Mahdian R., Jamali S., Maryami F., Babashah S., Maryami F., Karimipoor M., Zeinali S. Clin. Biochem. 2009;42:1291–1299. doi: 10.1016/j.clinbiochem.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 34.Wang M., Xie F., Lin J., Zhao Y., Zhang Q., Liao Z., Wei P. Front. Med. 2021;8 doi: 10.3389/fmed.2021.649383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., Ren Y., Ma S., Wang S. OncoTargets Ther. 2020;1941–1951 doi: 10.2147/OTT.S240642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xie H., Ren X., Xin S., Lan X., Lu G., Lin Y., Yang S., Zeng Z., Liao W., Ding Y.-Q. Oncotarget. 2016;7 doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin W., Xu J., Li C., Dai X., Wu T., Wen J. vols. 3689–3701. 2020. (OncoTargets and Therapy). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai G., Zou R., Yang H., Xie J., Chen X., Zheng C., Luo S., Wei N., Liu S., Chen R. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.891476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ni Y., Lu C., Wang W., Gao W., Yu C. Molecular Therapy-Oncolytics. 2021;21:119–133. doi: 10.1016/j.omto.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weng W., Wei Q., Toden S., Yoshida K., Nagasaka T., Fujiwara T., Cai S., Qin H., Ma Y., Goel A. Clin. Cancer Res. 2017;23:3918–3928. doi: 10.1158/1078-0432.CCR-16-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bignold L.P. In: Principles of Tumors. second ed. Bignold L.P., editor. Academic Press; 2020. pp. 279–315. [Google Scholar]
- 42.Hoorens A. In: The Lymphatic System in Colorectal Cancer. Ceelen W.P., editor. Academic Press; 2022. pp. 115–130. [Google Scholar]
- 43.Hughes C.C. Curr. Opin. Hematol. 2008;15:204. doi: 10.1097/MOH.0b013e3282f97dbc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abhinand C.S., Raju R., Soumya S.J., Arya P.S., Sudhakaran P.R. Journal of cell communication and signaling. 2016;10:347–354. doi: 10.1007/s12079-016-0352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ghafouri-Fard S., Taheri M., Hussen B.M., Vafaeimanesh J., Abak A., Vafaee R. Biomed. Pharmacother. 2021;140 doi: 10.1016/j.biopha.2021.111721. [DOI] [PubMed] [Google Scholar]
- 46.Ma Z., Shuai Y., Gao X., Wen X., Ji J. Mol. Cancer. 2020;19:1–22. doi: 10.1186/s12943-019-1113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang S., Fu R., Shi J., Wu H., Mai J., Hua X., Chen H., Liu J., Lu M., Li N. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.609397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolfson B., Eades G., Zhou Q. World J. Stem Cell. 2014;6:591. doi: 10.4252/wjsc.v6.i5.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang N., Liu X., Tang Z., Wei X., Dong H., Liu Y., Wu H., Wu Z., Li X., Ma X. J. Nanobiotechnol. 2022;20:97. doi: 10.1186/s12951-022-01267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wan S., Liu Z., Chen Y., Mai Z., Jiang M., Di Q., Sun B. Bioengineered. 2021;12:11959–11977. doi: 10.1080/21655979.2021.2009422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yfantis A., Mylonis I., Chachami G., Nikolaidis M., Amoutzias G.D., Paraskeva E., Simos G. Cells. 2023;12:798. doi: 10.3390/cells12050798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramakrishnan S., Anand V., Roy S. J. Neuroimmune Pharmacol. 2014;9:142–160. doi: 10.1007/s11481-014-9531-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang S., Ren K., Li B., Li F., Liang Z., Hu J., Xu B., Zhang A. Mol. Cell. Biochem. 2020;465:1–11. doi: 10.1007/s11010-019-03662-0. [DOI] [PubMed] [Google Scholar]
- 54.Zheng H., Wang N., Li L., Ge L., Jia H., Fan Z. Int. J. Oral Sci. 2021;13:41. doi: 10.1038/s41368-021-00148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen F., Chu L., Li J., Shi Y., Xu B., Gu J., Yao X., Tian M., Yang X., Sun X. Thoracic cancer. 2020;11:570–580. doi: 10.1111/1759-7714.13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siegel R., Naishadham D., Jemal A. CA: a cancer journal for clinicians. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 57.De Divitiis C., Nasti G., Montano M., Fisichella R., Iaffaioli R.V., Berretta M. World J. Gastroenterol.: WJG. 2014;20 doi: 10.3748/wjg.v20.i41.15049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maminezhad H., Ghanadian S., Pakravan K., Razmara E., Rouhollah F., Mossahebi-Mohammadi M., Babashah S. Life Sci. 2020;258 doi: 10.1016/j.lfs.2020.118226. [DOI] [PubMed] [Google Scholar]
- 59.Mai S., Zhang Z., Mi W. Ann. Clin. Lab. Sci. 2021;51:55–60. [PubMed] [Google Scholar]
- 60.Zhu M., Xu Y., Chen Y., Yan F. Biomed. Pharmacother. 2017;88:138–144. doi: 10.1016/j.biopha.2016.12.097. [DOI] [PubMed] [Google Scholar]
- 61.Niland S., Riscanevo A.X., Eble J.A. Int. J. Mol. Sci. 2021;23:146. doi: 10.3390/ijms23010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu F., Yang J., Liu J., Wang Y., Mu J., Zeng Q., Deng S., Zhou H. Signal Transduct. Targeted Ther. 2021;6:218. doi: 10.1038/s41392-021-00641-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Masoudi-Khoram N., Soheilifar M.H., Ghorbanifar S., Nobari S., Hakimi M., Hassani M. Crit. Rev. Oncol. Hematol. 2023 doi: 10.1016/j.critrevonc.2023.103967. [DOI] [PubMed] [Google Scholar]
- 64.Ren J., Ding L., Zhang D., Shi G., Xu Q., Shen S., Wang Y., Wang T., Hou Y. Theranostics. 2018;8:3932. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang H., Yue X., Chen Z., Liu C., Wu W., Zhang N., Liu Z., Yang L., Jiang Q., Cheng Q. Mol. Cancer. 2023;22:159. doi: 10.1186/s12943-023-01860-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Giannoni E., Bianchini F., Masieri L., Serni S., Torre E., Calorini L., Chiarugi P. Cancer Res. 2010;70:6945–6956. doi: 10.1158/0008-5472.CAN-10-0785. [DOI] [PubMed] [Google Scholar]
- 67.Kim K., Sohn Y.J., Lee R., Yoo H.J., Kang J.Y., Choi N., Na D., Yeon J.H. Int. J. Mol. Sci. 2020;21:8153. doi: 10.3390/ijms21218153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lamalice L., Le Boeuf F., Huot J. Circ. Res. 2007;100:782–794. doi: 10.1161/01.RES.0000259593.07661.1e. [DOI] [PubMed] [Google Scholar]
- 69.Chen Z., Chen J., Wa Q., He M., Wang X., Zhou J., Cen Y. Life Sci. 2020;243 doi: 10.1016/j.lfs.2020.117323. [DOI] [PubMed] [Google Scholar]
- 70.Cai G., Zou R., Xie J., Chen X., Zheng C., Luo S., Wei N., Liu S., Chen R. Front. Oncol. 2022;12 doi: 10.3389/fonc.2022.891476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu D., Chen C., Cui M., Zhang H. Cancer Med. 2021;10:3358–3372. doi: 10.1002/cam4.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jiang W., Li T., Wang J., Jiao R., Shi X., Huang X., Ji G. OncoTargets Ther. 2019;12 doi: 10.2147/OTT.S226465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shibuya M. Genes & cancer. 2011;2:1097–1105. doi: 10.1177/1947601911423031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Väyrynen V., Wirta E.V., Seppälä T., Sihvo E., Mecklin J.P., Vasala K., Kellokumpu I. BJS Open. 2020;4:685–692. doi: 10.1002/bjs5.50299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hernandez Dominguez O., Yilmaz S., Steele S.R. J. Clin. Med. 2023;12 doi: 10.3390/jcm12052072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in this work have not been deposited into a publicly available repository. The data supporting the findings of this study are available from the corresponding author upon reasonable request.