Abstract
Vibrio cholerae O1, biotype El Tor, accumulates inorganic polyphosphate (poly P) principally as large clusters of granules. Poly P kinase (PPK), the enzyme that synthesizes poly P from ATP, is encoded by the ppk gene, which has been cloned from V. cholerae, overexpressed, and knocked out by insertion-deletion mutagenesis. The predicted amino acid sequence of PPK is 701 residues (81.6 kDa), with 64% identity to that of Escherichia coli, which it resembles biochemically. As in E. coli, ppk is part of an operon with ppx, the gene that encodes exopolyphosphatase (PPX). However, unlike in E. coli, PPX activity was not detected in cell extracts of wild-type V. cholerae. The ppk null mutant of V. cholerae has diminished adaptation to high concentrations of calcium in the medium as well as motility and abiotic surface attachment.
Inorganic polyphosphate poly P is a linear polymer of up to hundreds of orthophosphate (Pi) residues linked by high-energy phosphoanhydride bonds. Among known functions, poly P can serve as a substitute for ATP in kinase reactions, a Pi reservoir, and a chelator of divalent metals (9). Poly P is ubiquitous in nature, having been found in all organisms examined (15), yet little is known about its physiological roles (14).
Several poly P-metabolizing enzymes have been purified, and the genes encoding them have been cloned (14, 28). The enzyme primarily responsible for poly P synthesis in Escherichia coli is poly P kinase (PPK), which catalyzes the polymerization of the γ phosphate of ATP into a poly P chain (1). Poly P can be hydrolyzed to Pi by an exopolyphosphatase (PPX) (3). In E. coli, the encoding genes, ppk and ppx, respectively, form an operon. The inability to accumulate poly P upon deletion of this operon or upon the overproduction of PPX has produced several striking phenotypes in E. coli (6, 20, 25): decreased long-term survival in stationary phase; increased sensitivity to oxidative, osmotic, and thermal stresses; and defects in adaptive growth in minimal medium after a shift from rich medium. These phenotypes are likely due to the decreased expression of the rpoS gene, which encodes the principal stationary-phase sigma factor, ςS, or RpoS (25). These and related results (4) suggest that poly P is an effector signal for responses to acute stringencies and adaptations in the stationary phase.
Recently available genome sequences have revealed that PPK is highly conserved in many bacterial species, including some important pathogens (26). This also implies that PPK and/or poly P has fundamental physiological roles in bacteria. The ppk knockout mutant of Pseudomonas aeruginosa PAO1 shows a dramatic deficiency in motility, both flagellar and pilus mediated, an inability to form biofilms, and a loss of virulence (22, 23, 24). ppk null mutants of several other pathogens and of E. coli also exhibit reduced motility and reduced abiotic surface attachment (22, 24).
Vibrio spp. are among the most common microorganisms in environmental surface waters, such as lakes and rivers. Vibrio cholerae O1 is an enteropathogenic gram-negative bacterium that causes severe diarrheal disease. An rpoS mutant of V. cholerae (29) revealed that RpoS is required for V. cholerae persistence in a medium devised to simulate natural aquatic habitats. A gene highly homologous to E. coli ppk was found in the V. cholerae database of The Institute for Genomic Research (TIGR) (26). The essential role of poly P for the stationary-phase survival of E. coli and the possibility of a similar role in V. cholerae prompted us to study its PPK and to examine the phenotype of a ppk knockout mutant.
V. cholerae O1, biotype El Tor, accumulates much higher levels of poly P than E. coli under normal growth conditions. High accumulations of poly P are stored as granules following a shift from a defined medium lacking Pi to one with an excess (20 mM). The ppk null mutant is defective in motility and abiotic surface attachment and fails to adapt to high concentrations of calcium.
MATERIALS AND METHODS
Strains and plasmids.
E. coli strains MG1655 (λ− F−) and CF5802 (MG1655 Δppk Δppx::Kan [17]) were the wild-type and mutant strains, respectively. Recombinant plasmids based on pBluescript II KS(+) and SK(+) (Stratagene, La Jolla, Calif.) were prepared from DH5α transformants. Suicide plasmids based on pKNG101 (11) were replicated in E. coli strain S17-1(λ pir) (29); pFZY1 (13) was used as a low-copy-number vector. The 92A1552-Rifr wild-type strain of V. cholerae O1 (El Tor, Inaba [29]) and the cosmid library of its genomic DNA were provided by F. Yildiz, Department of Microbiology and Immunology, Stanford University.
Media and growth conditions.
A MOPS (morpholinepropanesulfonic acid)-buffered minimal medium (21) was used to impose Pi-limiting conditions for the growth of V. cholerae. Media were supplemented with ampicillin (100 μg/ml), kanamycin (150 μg/ml), or streptomycin (100 μg/ml) to select for antibiotic-resistant transformants of V. cholerae.
Plasmid construction.
The V. cholerae PPK sequence was obtained by a BLAST search of the TIGR genome sequence database using the E. coli sequence. To obtain the ppk region of V. cholerae, two PCR primers were designed: VCPPKFOR1, CCTTCTAGACAACTCTATGACACTAAAGGCAC, and VC PPKREV1, CCTGTCGACTCTGCCGATGAGATAAAGAC. VCPPKFOR1 contains an XbaI site, and VCPPKREV1 contains a SalI site, each located at the 5′ end (underlined). These primers yielded a 1.95-kb PCR product using genomic DNA prepared from the wild-type V. cholerae strain 92A1552-Rifr as a template. This fragment begins at position −106 relative to the A in the start codon at +1 and ends at position +1829. After digestion with both XbaI and SalI, this PCR product was cloned into XbaI- and SalI-digested pBluescript II KS(+); the resulting plasmid was designated pVCK1.
The cosmid pVCK20 was obtained by colony hybridization from a V. cholerae genome library using the 1.95-kb PCR fragment as a probe; pVCK20 contained an insert of more than 40 kb in which the ppk homologous region was limited to a 2.5-kb NcoI-BglII fragment (pVCK35 [Fig. 1A]).
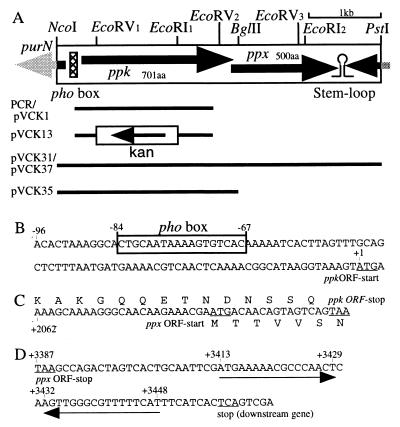
FIG. 1.
The ppk ppx operon in V. cholerae and recombinant plasmids. (A) The large box represents the cloned and sequenced 4-kb NcoI-PstI chromosomal fragment containing the operon. ORFs are indicated by arrows. The lines under the box indicate the portions of the ppk ppx region inserted in the plasmids indicated to the left. The nonunique EcoRI and EcoRV restriction sites in the 4-kb NcoI-PstI fragment are denoted by subscripts. (B) Putative pho box sequence in the ppk promoter region. (C) Sequence overlap between the ppk and ppx ORFs. The center sequence is the nucleotide sequence of the ppk ppx ORF junction, and the top and bottom sequences are the deduced amino acid sequences for PPK and PPX, respectively. (D) Putative transcriptional terminator sequence in the region downstream of ppx. The pair of sequences indicated by the arrows have the potential to form a stem-loop structure. The underlined nucleotides are the indicated start and stop codons.
Plasmids containing the cloned V. cholerae ppk gene were constructed as follows. A 6.5-kb HindIII-PstI fragment containing ppk prepared from pVCK20 was inserted into similarly restricted pBluescript II SK(+). The resulting plasmid, pVCK27, was digested with HindIII and NcoI, and the 7-kb fragment was isolated. After the ends were filled in, this fragment was self-ligated. The resulting plasmid, pVCK31 (Fig. 1A), contains the 4-kb NcoI-PstI fragment in pBluescript II SK(+). A smaller plasmid, pVCK35 (Fig. 1A), was constructed by digestion of pVCK31 with BglII and BamHI, followed by self-ligation of the resulting longer fragment; pVCK35 harbors the 2.5-kb NcoI-BglII fragment in pBluescript II SK(+).
A low-copy-number plasmid containing the V. cholerae ppk ppx loci, pVCK37, was constructed as follows. pVCK31 was digested with BamHI and SalI, and the fragment with the 4-kb NcoI-PstI ppk ppx region was isolated. The BamHI-SalI fragment of about 4 kb was inserted into a similarly restricted low-copy-number vector, pFZY1, resulting in plasmid pVCK37.
Construction of the V. cholerae ppk knockout mutant.
To make the deletion-insertion mutation allele of ppk, a 1.2-kb EcoRV (site 1)-EcoRI (site 1) region of pVCK1 (Fig. 1A) was replaced with a 1.3-kb HincII-EcoRI fragment containing the kanamycin cassette from pVCK4, which was constructed by insertion of a 1.3-kb HincII-HincII fragment with the cassette from pUC4K (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.) into the EcoRV site of pBluescript II KS(+). From the resulting plasmid, pVCK7, the 2.1-kb XbaI-SalI ppk::Kan fragment was transferred into the suicide vector pKNG101, and the resulting plasmid was designated pVCK13 (Fig. 1A).
pVCK13-integrated transformants (single-crossover recombinants) of the V. cholerae wild-type strain 92A1552-Rifr which exhibited both kanamycin and streptomycin resistance phenotypes were obtained. After two successive overnight cultivations of the integrants in Luria broth (LB) supplemented with 5% sucrose, the ppk knockout mutants (double-crossover recombinants) were obtained as kanamycin-resistant but streptomycin-sensitive colonies. The chromosomal mutations were confirmed by Southern blot analysis, and one of the resultant recombinants, KVC3, was selected as the ppk knockout mutant of V. cholerae.
Biochemical assays.
Cell extracts from V. cholerae were prepared as from E. coli (16), modified only by a 2-min sonication after lysozyme treatment. PPK and PPX activities were assayed as described previously (1, 17). Poly P levels were determined by the nonradioactive method (4). The cloned DNA fragment was sequenced by the PAN Facility.
Electron microscopy.
Strains were cultivated as described in the legend to Fig. 3D. Cells were harvested at zero time and 2 h after the addition of Pi and were washed three times with 0.9% sodium chloride (zero time) or phosphate-buffered saline (2 h) solution. For negative staining, the samples were placed on a carbon-coated grid and stained with 1% uranyl acetate. Thin-section samples were prepared by N. Ghori, Department of Microbiology and Immunology, Stanford University.
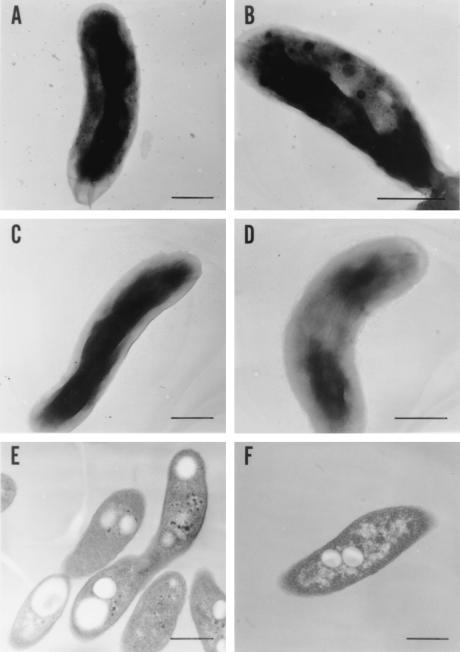
FIG. 3.
Electron micrographs of V. cholerae strains. The bacteria were harvested at 0 and 2 h after the addition of Pi, as for Fig. 2D. Bars, 0.5 μm. Negatively stained samples were prepared from the wild-type culture after 0 (C) and 2 (A and B) h of incubation and from KCV3 (the ppk mutant) after 2 h of incubation (D). Thin-section samples were prepared from the wild type (E) and KVC3 (F) after 2 h of incubation. Magnifications, ×28,000 (A, C, E, and F), ×35,000 (D), and ×45,000 (B).
Surface attachment assay.
Cells (2 μl) cultured overnight in LB were inoculated into 100 μl of LB in 96-well polyvinyl chloride plates (Falcon 3911 Microtester III flexible assay plate; Becton Dickinson, Oxnard, Calif.) and incubated at 30°C without shaking for 24 h. After the medium was removed, each well was rinsed with sterile water, 100 μl of 1% crystal violet solution was added, and the plate was incubated for 15 min at room temperature. The wells were thoroughly washed with water and air dried. The adherent crystal violet was extracted with 200 μl of 95% ethanol, diluted in 95% ethanol, and measured at 595 nm in a microplate spectrophotometer (model 550; Bio-Rad Laboratories, Hercules, Calif.).
Pi uptake assay.
Cells were grown in a MOPS medium with 0.1 mM Pi overnight, collected, washed with a Pi-free MOPS medium, and resuspended in 50 ml of the Pi-free medium to an optical density at 600 nm (OD600) of 0.1. The cultures were shaken for 2 h at 37°C and were readjusted to an OD600 of 0.1 by dilution with the Pi-free MOPS medium. K2HPO4 (0.1 mM) with [32PO4]Pi (1 μCi/ml) was added to the shaking cultures, and samples (0.1 ml) were taken at intervals and immediately filtered through an HA filter (3.5-mm diameter; Millipore). The cells trapped on the filter were washed with 10 ml of Pi-free MOPS medium without glucose, amino acids, or vitamins. The radioactivity on the membrane filter was measured in a liquid scintillation counter; the values of Pi uptake in the cells are indicated as counts per minute per milliliter per OD600 unit.
Purification of V. cholerae PPK.
E. coli strain CF5802 transformed with pVCK31 was used as the starting material (4.2 × 108 U in 610 mg of protein). The purification was performed essentially as described previously (2), modified only by the addition of a phenyl Sepharose column (Amersham Pharmacia Biotech, Inc.) as the final step (yielding 4.6 × 106 U in 0.12 mg of protein).
Nucleotide sequence accession number.
The GenBank accession number for the sequence reported here is AF083928.
RESULTS
Poly P accumulation.
When E. coli was grown in rich medium (LB) under normal conditions (37°C with aeration), poly P levels did not exceed 1 nmol (in Pi residues) per mg of total cell protein at any stage (21). The highest levels of transient poly P accumulation, about 25 nmol/mg of protein, occur when E. coli cells are exposed to specific stresses (4). However, V. cholerae strain 92A1552-Rifr, even when grown in LB, accumulated poly P to levels of more than 50 nmol/mg (Fig. 2A) during logarithmic growth, while in stationary-phase cells the levels were low (∼1 nmol/mg). In MOPS minimal salts medium, V. cholerae accumulates more than 20 nmol of poly P/mg in stationary phase, far exceeding the <1-nmol/mg levels for E. coli (21). Thus, V. cholerae accumulates significantly higher levels of poly P than E. coli at all stages: during growth in rich media and during stationary phase in minimal media.
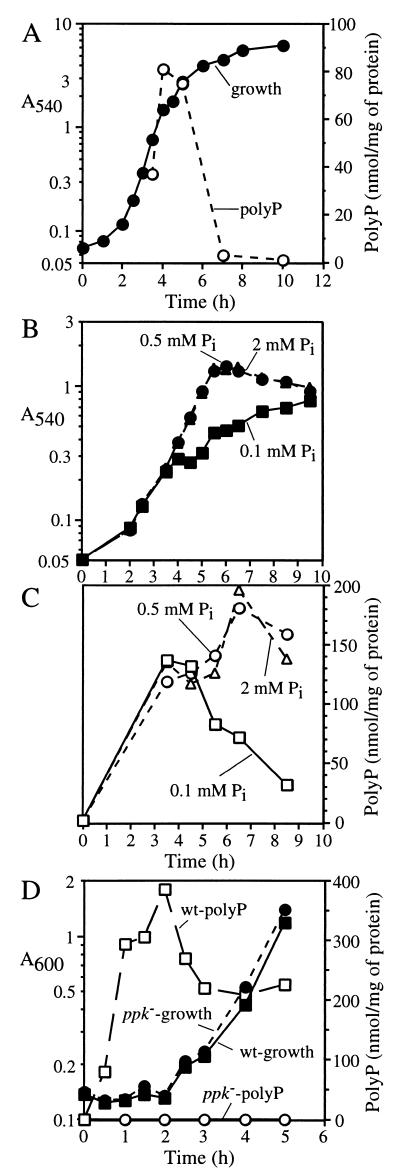
FIG. 2.
Poly P accumulations in V. cholerae. (A) Following overnight culture in LB, the wild-type strain 92A1552-Rifr was subcultured into fresh LB at a 1:100 dilution and grown at 37°C with aeration. (B and C) Following overnight culture in MOPS medium with 0.1 mM Pi, the wild-type strain was subcultured in MOPS medium with 0.1 (squares), 0.5 (circles), or 2 (triangles) mM Pi. Solid symbols, growth (A540); open symbols, poly P. (D) Following overnight culture in MOPS with 0.1 mM Pi, the wild-type (squares) and KVC3 (ppk mutant [circles]) strains were harvested and washed with MOPS buffer. The cells were transferred to Pi-free MOPS and cultivated for 1 h at 37°C with aeration. Potassium phosphate buffer (pH 7.4) was then added to a final concentration of 20 mM Pi at zero hour. Solid symbols, growth (A600); open symbols, poly P.
To determine whether the high level of poly P accumulation in V. cholerae relative to that in E. coli is due to a higher level of PPK, we measured PPK activity in V. cholerae and found that the membrane fraction activity (Table 1) was quite similar to that of E. coli MG1655. PPK activity levels were virtually the same in logarithmic- and stationary-phase cells (data not shown). Thus, poly P accumulation levels do not reflect the levels of PPK activity, which is also true in E. coli (21). In contrast to E. coli, no detectable activity of PPX was observed in V. cholerae (Table 1). Since a measurable level of PPX activity was detected in an E. coli Δppk Δppx strain bearing a plasmid with the V. cholerae ppk ppx operon (see below), V. cholerae PPX should be expressed. The undetectable level of native PPX activity may explain in part why V. cholerae accumulates large amounts of poly P. The rapid degradation of poly P shown in Fig. 1A, in the absence of significant PPX activity, may be due to the reverse reaction of PPK (27), in which poly P is converted to ATP. A Klebsiella aerogenes ppx mutant strain demonstrated profiles for poly P accumulation and degradation similar to those of the parental wild-type strain (A. Kuroda, personal communication).
TABLE 1.
PPK and PPX activities and poly P levels in V. cholerae and E. colia
| Strain | Genotypeb | PPK activity (U/mg)
|
PPX activity (U/mg) | Poly P level (nmol/mg) | |
|---|---|---|---|---|---|
| Soluble | Membrane | ||||
| V. cholerae | |||||
| 92A1552-Rifr | WT | 1,120 | 17,300 | <30 | 25.3 |
| KVC3 | ppk::Kan | <30 | <300 | <30 | <0.10 |
| E. coli MG1655 | WT | 550 | 14,000 | 1,200 | |
PPK and PPX activities were determined using cell extracts prepared from stationary-phase cultures in LB, and poly P levels were determined using cell extracts prepared from stationary-phase cultures in 2 mM Pi MOPS medium.
WT, wild type.
Poly P overplus.
The influence of Pi concentration on poly P accumulation was determined in cells grown in a defined medium (MOPS) (Fig. 2B and C). Poly P levels were below the detection limit (<0.5 nmol/mg) when cultivated with 0.1 mM Pi overnight (Fig. 2C, 0 h). However, >100 nmol/mg accumulated after 4 h in log phase with 0.1, 0.5, or 2 mM Pi. Cultures with 0.5 or 2 mM Pi maintained poly P levels of >100 nmol/mg for up to 9 h, i.e., after entering stationary phase. However, poly P levels in the 0.1 mM Pi culture decreased gradually along with the decrease in growth rate (Fig. 2B and C, 5 to 9 h). Given the similarity in the growth curves for the 0.5 and 2 mM Pi cultures, the slow growth of the 0.1 mM Pi culture is likely due to Pi depletion. Thus, poly P levels in V. cholerae depend on the Pi concentration in the medium.
When cells were shifted from a MOPS medium with no Pi added to the same medium with 20 mM Pi, dramatic accumulations of poly P occurred immediately after the upshift (Fig. 2D). Within 1 h, poly P accumulated to >300 nmol/mg and to near 400 nmol/mg after 2 h without any concomitant growth. With the resumption of growth, the poly P level decreased to 250 nmol/mg and remained there for at least five more hours. Similarly, large poly P accumulations have been reported in Aerobacter aerogenes as the “poly P overplus” phenomenon (9).
These massive levels of poly P were observed by electron microscopy as numerous bodies of relatively high electron density (Fig. 3). About 1 in 30 cells contained bodies 20 to 40 nm in diameter (Fig. 3B), while the others had smaller bodies, about 5 nm in diameter (Fig. 3A). Such granules were not found in wild-type cells grown in Pi-free medium or in ppk mutant cells (see below) grown in Pi-rich medium for 2 h (Fig. 3C and D). Inasmuch as these bodies were correlated with the dmassive accumulations of poly P, they are presumed to be poly P. Similar granules observed in Myxococcus xanthus (9) and Helicobacter pylori are somewhat localized at the flagellar pole and in association with the inner membrane as well as being dispersed in the cytoplasm (5). As determined by thin-section electron microscopy (Fig. 3E), the granules in V. cholerae are distributed in the cytoplasm but not localized along the inner membrane.
The ppk gene.
The region in the TIGR V. cholerae genome database surrounding a sequence homologous to that of E. coli ppk was cloned from a V. cholerae genome library by colony hybridization using a PCR fragment of the region as a probe (see Materials and Methods). The E. coli Δppk Δppx knockout strain CF5802, transformed with pVCK35 containing the V. cholerae ppk homolog (Fig. 1A), exhibited high levels of PPK activity (38,000 U/mg), demonstrating that this region did in fact contain the V. cholerae ppk gene; this was confirmed subsequently by sequencing. The deduced amino acid sequence of V. cholerae PPK is 701 amino acid residues long, with a calculated molecular mass of 81.6 kDa; it is 64% identical and 83% similar (in conserved residues) to that of E. coli. Sequencing also revealed a ppx homolog downstream of ppk in V. cholerae, as in E. coli, although with a two-cistron overlap. A transformant of the E. coli Δppk Δppx mutant strain CF5802 harboring pVCK31, which contains both the ppk and ppx homologs (Fig. 1A), gave low but significant levels of PPX activity (540 U/mg). Its deduced amino acid sequence is 500 amino acid residues in length, with a calculated molecular mass of 56.4 kDa, and is 51% identical and 70% similar (in conserved residues) to E. coli PPX.
Upstream of the ppk open reading frame (ORF) (Fig. 1B) lies a putative promoter region which contains a probable pho box sequence with 15 of the 18 consensus base pairs at positions −84 to −67 from the A in the ppk start codon (Fig. 1B). In E. coli, pho boxes are the binding sites of the two-component system regulator protein PhoB. Homologs of phoB and phoR (the cognate sensor) are found in the V. cholerae genome database, suggesting that transcription of the ppk ppx operon may be regulated by this system. Putative pho boxes are also present in the ppk promoter regions of E. coli, K. aerogenes (12), and Acinetobacter sp. strain ADP1 (8). A pair of 17-bp inverted-repeat sequences (from positions 24 to 40 and 43 to 59 from the ppx stop codon) appear to constitute a rho-independent transcriptional terminator site downstream of the ppx ORF (Fig. 1D). These features imply that ppk and ppx form an operon, as in E. coli and other gram-negative bacteria. The E. coli Δppk Δppx transformant with a high copy number of the V. cholerae ppk ppx operon [CF5802(pVCK31)] expressed 690,000 U of PPK and 540 U of PPX activity per mg. We have shown previously that the E. coli wild type transformed with a high copy number of the E. coli ppk ppx operon exhibited 630,000 U of PPK and 50,000 U of PPX activity per mg (2, 3). Thus, V. cholerae PPK was expressed in an E. coli host cell at levels similar to those of E. coli PPK from the E. coli operon, but V. cholerae PPX was expressed about 100-fold less than E. coli PPX. Unlike in E. coli, there is a 20-bp overlap between the ppk and ppx ORFs in the V. cholerae operon (Fig. 1C), which may interfere with translation of the ppx ORF from the ppk-ppx mRNA, accounting for the undetectable level of PPX activity (Table 1).
Null mutant of ppk.
In a ppk knockout mutant in V. cholerae (KVC3) (see Materials and Methods), the levels of PPK activity and poly P accumulation were undetectable (Table 1 and Fig. 2D), in contrast to the parental strain, 92A1552-Rifr, which accumulated more than 300 nmol of poly P per mg when shifted from a Pi-free to a 20 mM Pi defined medium. We tested KVC3 for the reported phenotypes of the E. coli ppk mutant strain CA10 (5, 18, 20). No phenotypes were observed for long-term survival in synthetic medium (for 30 days at 30°C), sensitivity to heat (at 45 and 55°C) and hydrogen peroxide (in 10, 3, and 1 mM), adaptive growth following a shift from rich medium to minimal medium, or long-term (10 days at 30°C) survival in artificial seawater (29).
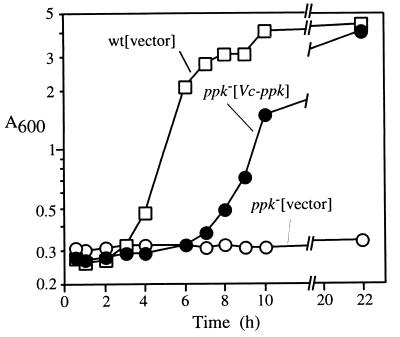
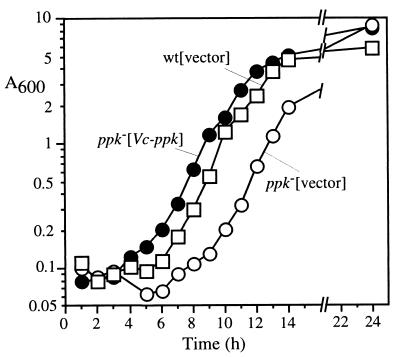
With regard to adaptive growth in a nutrient downshift (Fig. 4), the E. coli wild-type strain MG1655 transformed with the plasmid vector (pFZY1) grew in a minimal medium after a 2-h lag following downshift from a rich medium (LB) whereas the E. coli Δppk Δppx mutant CF5802 harboring pFZY1 was unable to grow even 22 h after the downshift. However, CF5802 bearing the V. cholerae ppk plasmid pVCK37 grew in minimal medium after the downshift with only a 4-h lag, after which the growth rate and final OD were similar to that of the wild type. Thus, the V. cholerae ppk complements the E. coli Δppk mutation in response to this stress condition.
FIG. 4.
V. cholerae ppk complements the adaptive growth defect in an E. coli ppk mutant. E. coli strains MG1655(pFYZ1) (squares), CF5802(pFYZ1) (open circles), and CF5802(pVCK37) (solid circles) were grown in 2×YT medium supplemented with 50 μg of ampicillin/ml to an A600 of 0.5. The cells were harvested and washed twice with MOPS medium devoid of nutrients and resuspended in MOPS medium with 2 mM Pi and 0.4% glucose at zero hour.
A P. aeruginosa PAO1 ppk mutant shows no defects in adaptive responses but is severely impaired in motility and surface attachment (22, 23, 24), and the V. cholerae ppk mutant was found to be deficient in these features as well (23). The swim area of the ppk mutant was 57% that of the wild type on 0.3% agar plates (Table 2). The ppk mutant also exhibited a significantly lower ability for surface attachment (Table 2). Deficiencies in motility and surface attachment have also been observed in ppk mutants of E. coli, K. pneumoniae, and Salmonella spp. (22).
TABLE 2.
Swimming and surface attachment
| Strain | Genotype | Swim area (cm2)a | Surface attachment (A595)b |
|---|---|---|---|
| 92A1552-Rifr | WTc | 1.52 ± 0.07 | 2.14 ± 0.70 |
| KVC3 | ppk::Kan | 0.87 ± 0.07 | 1.46 ± 0.30 |
Data are from Rashid et al. (23).
Average of 16 measurements; two-tail P value by t test with a 0.05 threshold is 0.0012.
WT, wild type.
In view of the capacity of poly P to function as a chelator of divalent metals (9), the calcium sensitivity of the V. cholerae ppk mutant was tested (Fig. 5). The growth lag time of the wild-type strain (92A1552-Rifr) in LB containing 200 mM CaCl2 was 5 h, more than 4 h longer than in the absence of CaCl2 (Fig. 2A). The lag time of the ppk mutant KVC3 was significantly longer, at 7 h. When complemented with the high-copy-number plasmid pVCK31 harboring the ppk gene, the lag time was shortened to 3 h. As both the wild type and the ppk mutant of V. cholerae had the same lag time when grown in LB in the presence of 400 mM NaCl (data not shown), these results imply that the ppk gene is involved in an adaptation to excess levels of calcium but not chloride or osmolality.
FIG. 5.
Growth adaptation to an excess amount of calcium. V. cholerae strains 92A1552-Rifr(pBluescript II) (squares), KVC3(pBluescript II) (open circles), and KVC3(pVCK31) (solid circles) were grown in LB medium supplemented with 10 mM KH2PO4 and 50 μg of ampicillin/ml overnight. The cells were inoculated in LB supplemented with 200 mM CaCl2 and shaken at 37°C.
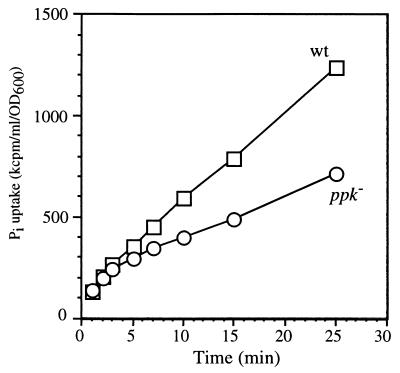
Another phenotype of the ppk mutant was observed with regard to Pi uptake (Fig. 6). After growth in a Pi-free medium for 2 h, wild-type cells displayed a linear (nonsaturable) Pi uptake for up to 25 min when incubated in 0.1 mM Pi (the Pi-limited condition) (Fig. 2B and C). The Δppk mutant had unique saturable profiles for Pi uptake. It showed a rate of uptake similar to that of the wild type from 0 to 3 min, but the uptake after 5 min was minimal and the rate was decreased significantly. These data suggest that ppk is required for the continual high-rate uptake of Pi under low-Pi conditions.
FIG. 6.
Pi uptake of V. cholerae. Pi uptake activities of 92A1552-Rifr (squares) and KVC3 (circles) were tested as described in Materials and Methods.
Characterization of PPK.
PPK was purified from a transformant of the E. coli CF5802 strain (Δppk Δppx::Kan) bearing the V. cholerae ppk ppx operon on a high-copy-number vector, pVCK31. Homogeneity of the purified protein was verified as a single band on a Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel (data not shown) with a molecular mass estimated at 87 kDa, compared to the calculated mass of 81.6 kDa. Comparison of the PPK retention time to those of reference proteins in a high-performance liquid chromatography gel filtration column showed a molecular mass of 310 kDa (data not shown), indicating that V. cholerae PPK is a homotetramer like E. coli PPK (1). The final fraction has a specific activity of 40 × 106 U/mg, slightly higher than that of E. coli PPK (29 × 106 U/mg) (1).
The optimal reaction conditions for V. cholerae PPK are almost the same as those for E. coli PPK, e.g., a pH optimum of 7.2 in HEPES buffer and 4 mM Mg2+, and 40 mM ammonium sulfate increases activity twofold. Autophosphorylated PPK was observed in a reaction with [γ-32P]ATP. Like the E. coli enzyme (16), the V. cholerae enzyme can catalyze the synthesis of ATP from ADP and poly P and can catalyze the synthesis of GTP and ppppG (linear guanosine 5′-tetraphosphate) from GDP and poly P. Significant differences in the kinetic parameters for the PPKs of V. cholerae and E. coli are shown by the 40-fold decrease in the V. cholerae Km value for ATP for the forward (poly P synthesis) reaction (Table 3). The kcat/Km ratio for the ATP synthesis reaction of V. cholerae PPK is more than 10 times higher than that for E. coli. On the other hand, the kcat/Kms ratios for the GTP and ppppG synthesis reactions of V. cholerae PPK are 5 and 20 times lower, respectively, than those for E. coli. These data suggest that the V. cholerae PPK is more specific for the generation of ATP than GTP or ppppG.
TABLE 3.
Kinetic parameters of PPK
| Reaction and source | Km (mM) | Vmax (pmol mg−1 min−1) | kcat (s−1) | kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| Poly P synthesis | ||||
| V. cholerae | 0.05 | 1.6 × 106 | 8.2 | 164 |
| E. colia | 2 | 51 × 106 | 59 | 29.5 |
| ATP synthesis | ||||
| V. cholerae | 0.12b | 95 × 106 | 65.3 | 544 |
| E. colic | 0.25 | 3.7 × 106 | 10.5 | 42 |
| GTP synthesis | ||||
| V. cholerae | 2.5b | 1.5 × 104 | 0.076 | 0.03 |
| E. colic | 0.63 | 5.7 × 104 | 0.088 | 0.14 |
| ppppG synthesis | ||||
| V. cholerae | 2.3b | 1.1 × 103 | 0.006 | 0.002 |
| E. coli | 0.46 | 9.0 × 103 | 0.027 | 0.04 |
DISCUSSION
Accumulations of poly P in V. cholerae are remarkable for being so great and sustained compared to those in E. coli. The levels in excess of 50 nmol/mg of protein during exponential growth in a rich medium (Fig. 2A) and 150 nmol/mg in stationary phase in a defined medium (Fig. 2C) are roughly 100 times those in E. coli. Yet the PPK activities in extracts are nearly the same (Table 1). Some of the reason may lie in the undetectable levels of PPX activity in extracts of V. cholerae (Table 1). In that organism, as in E. coli (3), an operon contains the ppk as well as the ppx gene (Fig. 1). Unlike in E. coli, where the ppx gene is separated by 7 bp from the upstream ppk gene, the amino terminus-encoding region of ppx in V. cholerae overlaps the carboxy terminus-encoding region of ppk by 20 bp (Fig. 1C). The E. coli transformant with the V. cholerae ppx gene on a high-copy-number plasmid did express PPX activity. How these genes and possibly others are regulated pre- and posttranscriptionally remains to be determined.
When cells are switched from a Pi starvation medium to one with adequate Pi, there is a massive accumulation of poly P (Fig. 2D), as observed in A. aerogenes (9) and designated the poly P overplus phenomenon. The accumulation of poly P is evident as electron-dense granules up to 40 nm in diameter with an ordered matrix structure as in crystal complexes (Fig. 3B). The granules appear to be largely cytoplasmic (Fig. 3E), unlike some of the granules in H. pylori, which have polar and membrane-oriented locations (5). The dynamic accumulation and removal of poly P and its mobilization at a molecular level, as well as the nature of the granules and their cellular locations, need to be clarified.
V. cholerae PPK resembles that of E. coli in size and in its multiple activities: processive poly P synthesis from ATP, nucleoside diphosphate kinase action on ADP and GDP by donor poly P, pyrophosphoral transfer to GDP to form ppppG, and autophosphorylation by ATP. The most notable differences are in the kinetic parameters (Table 3), e.g., a Km for ATP of 0.2 mM for V. cholerae PPK compared to 2.0 mM for E. coli PPK. Also, kinetic parameters for ATP, GTP, and ppppG synthesis reactions indicate that V. cholerae PPK is more specific for the generation of ATP than for GTP or ppppG compared to E. coli PPK. These enzyme characteristics are similar to those of H. pylori PPK purified as a recombinant protein (27; C.-M. Tzeng and A. Kornberg, unpublished data).
The strong PPK sequence homologies of 20 or more bacterial species include a number of the major pathogens (26), V. cholerae among them. In view of the striking dependence of E. coli on PPK for a variety of adaptive responses in the stationary phase and the expression of virulence factors in the stationary phases of some pathogens (7), the phenotypes of null mutants of ppk in these pathogens have been sought. Furthermore, there is the attractive possibility that PPK might prove a novel target for an antimicrobial drug with a broad spectrum and minimal side effects, inasmuch as PPK has not been found in animal species.
Among the pathogenic features of the PPK null mutants, a decrease in motility (commonly associated with a loss of virulence [19]) and weakened attachment to abiotic surfaces (a frequent correlate of poor biofilm formation) have been observed in several pathogens. Particularly striking are mutants of P. aeruginosa (22, 23, 24) which are defective in quorum sensing and have also lost their virulence in mouse models. The V. cholerae ppk mutant also had diminished attachment to an abiotic surface (Table 2), with the activity decreased by 68% compared to that of the wild type. Although this was not a dramatic reduction as with P. aeruginosa (22, 24), it could be emphasized by using the rugose colony variant of V. cholerae O1 (30), which prefers to form biofilm, compared to the smooth colony variant which was used in this study.
The failure to adapt to stress and the lack of survival in stationary phase observed in the E. coli ppk mutant (6) have not been apparent in the V. cholerae mutant. In addition to decreased motility and decreased attachment to abiotic surfaces (Table 2), other defects have been observed. One is a delayed adaptation to high calcium levels in the medium (Fig. 5); complementation of the mutant with the ppk gene more than corrects for the extended lag in growth. It is plausible that a large amount of poly P can trap excess calcium in the cell and maintain the calcium level at a low enough level to permit growth. Another defect of the ppk mutant is an abnormal rate of uptake of Pi from the medium (Fig. 6). Whereas the initial rate resembled that of the wild type, the subsequent rate was considerably reduced. Not enough is known about Pi uptake in V. cholerae to identify which system might be affected by the lack of PPK function. Interestingly, budding-yeast mutants deficient in poly P accumulation also showed a similar saturable curve for Pi uptake (18a).
The phenotypic tests of the V. cholerae ppk mutant were patterned on those performed on bacterial species which differ sharply from V. cholerae in their physiologic features, commensal interactions, and host invasiveness (10). In view of the unique aspects of the aquatic ecology of V. cholerae, much needs to be studied to evaluate what effect the lack of PPK and poly P might have on its survival and pathogenesis.
ACKNOWLEDGMENTS
We thank F. H. Yildiz and G. Schoolnik in the Department of Microbiology and Immunology for providing strains, plasmids, the gene library, and technical advice, N. Ghori in the same department for electron microscopy, S. Handy in the Department of Chemistry for high-performance liquid chromatography analyses, N. Rao in our laboratory for performing surface attachment assays, and L. Bertsch for help with the manuscript.
This work was supported by a grant from the National Institute of General Medical Sciences.
REFERENCES
- 1.Ahn K, Kornberg A. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J Biol Chem. 1990;265:11734–11739. [PubMed] [Google Scholar]
- 2.Akiyama M, Crooke E, Kornberg A. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. J Biol Chem. 1992;267:22556–22561. [PubMed] [Google Scholar]
- 3.Akiyama M, Crooke E, Kornberg A. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J Biol Chem. 1993;268:633–639. [PubMed] [Google Scholar]
- 4.Ault-Riché D, Fraley C D, Tzeng C-M, Kornberg A. Novel assay reveals multiple pathways regulating stress-induced accumulation of inorganic polyphosphate in Escherichia coli. J Bacteriol. 1998;180:1841–1847. doi: 10.1128/jb.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bode G, Mauch F, Ditschuneit H, Malfertheiner P. Identification of structures containing polyphosphate in Helicobacter pylori. J Gen Microbiol. 1993;139:3029–3033. doi: 10.1099/00221287-139-12-3029. [DOI] [PubMed] [Google Scholar]
- 6.Crooke E, Akiyama M, Rao N N, Kornberg A. Genetically altered levels of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1994;269:6290–6295. [PubMed] [Google Scholar]
- 7.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geissdörfer W, Ratajczak A, Hillen W. Transcription of ppk from Acinetobacter sp. strain ADP1, encoding a putative polyphosphate kinase, is induced by phosphate starvation. Appl Environ Microbiol. 1998;64:896–901. doi: 10.1128/aem.64.3.896-901.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harold F M. Inorganic polyphosphates in biology: structure, metabolism, and function. Bacteriol Rev. 1966;30:772–794. doi: 10.1128/br.30.4.772-794.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Islam M S, Drasar B S, Sack R B. The aquatic flora and fauna as reservoirs of Vibrio cholerae: a review. J Diarrhoeal Dis Res. 1994;12:87–96. [PubMed] [Google Scholar]
- 11.Kaniga K, Delor I, Cornelis G R. A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria: inactivation of the blaA gene of Yersinia enterocolitica. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]
- 12.Kato J, Yamamoto T, Yamada K, Ohtake H. Cloning, sequence and characterization of the polyphosphate kinase-encoding gene (ppk) of Klebsiella aerogenes. Gene. 1993;137:237–242. doi: 10.1016/0378-1119(93)90013-s. [DOI] [PubMed] [Google Scholar]
- 13.Koop A H, Hartley M E, Bourgeois S. A low-copy-number vector utilizing beta-galactosidase for the analysis of gene control elements. Gene. 1987;52:245–256. doi: 10.1016/0378-1119(87)90051-5. [DOI] [PubMed] [Google Scholar]
- 14.Kornberg A. Inorganic polyphosphate: a molecule of many functions. Prog Mol Subcell Biol. 1999;23:1–18. doi: 10.1007/978-3-642-58444-2_1. [DOI] [PubMed] [Google Scholar]
- 15.Kulaev I S, editor. The biochemistry of inorganic polyphosphates. New York, N.Y: John Wiley & Sons, Inc.; 1979. [Google Scholar]
- 16.Kuroda A, Kornberg A. Polyphosphate kinase as a nucleoside diphosphate kinase in Escherichia coli and Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1997;94:439–442. doi: 10.1073/pnas.94.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 18.Kuroda A, Tanaka S, Ikeda T, Kato J, Takiguchi N, Ohtake H. Inorganic polyphosphate kinase is required to stimulate protein degradation and for adaptation to amino acid starvation in Escherichia coli. Proc Natl Acad Sci USA. 1999;96:14264–14269. doi: 10.1073/pnas.96.25.14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Ogawa, N., J. DeRisi, and P. O. Brown. New components of a system for phosphate acquisition and polyphosphate metabolism of Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell, in press. [DOI] [PMC free article] [PubMed]
- 19.Ottemann K M, Miller J F. Roles for motility in bacterial-host interactions. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 20.Rao N N, Kornberg A. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J Bacteriol. 1996;178:1394–1400. doi: 10.1128/jb.178.5.1394-1400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao N N, Liu S, Kornberg A. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J Bacteriol. 1998;180:2186–2193. doi: 10.1128/jb.180.8.2186-2193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rashid M H, Kornberg A. Inorganic polyphosphate is needed for swimming, swarming and twitching motilities of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2000;97:4885–4890. doi: 10.1073/pnas.060030097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rashid M H, Rao N N, Kornberg A. Inorganic polyphosphate is required for motility of bacterial pathogens. J Bacteriol. 2000;182:225–227. doi: 10.1128/jb.182.1.225-227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rashid M H, Rumbaugh K, Passador L, Davies D G, Hamood A N, Iglewski B H, Kornberg A. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2000;97:9636–9641. doi: 10.1073/pnas.170283397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shiba T, Tsutsumi K, Yano H, Ihara Y, Kameda A, Tanaka K, Takahashi H, Munekata M, Rao N N, Kornberg A. Inorganic polyphosphate and the induction of rpoS expression. Proc Natl Acad Sci USA. 1997;94:11210–11215. doi: 10.1073/pnas.94.21.11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tzeng C-M, Kornberg A. Polyphosphate kinase is highly conserved in many bacterial pathogens. Mol Microbiol. 1998;29:381–382. doi: 10.1046/j.1365-2958.1998.00887.x. [DOI] [PubMed] [Google Scholar]
- 27.Tzeng C-M, Kornberg A. The multiple activities of polyphosphate kinase of Escherichia coli and their subunit structure determined by radiation target analysis. J Biol Chem. 2000;275:3977–3983. doi: 10.1074/jbc.275.6.3977. [DOI] [PubMed] [Google Scholar]
- 28.Wood H G, Clark J E. Biological aspects of inorganic polyphosphates. Annu Rev Biochem. 1988;57:235–260. doi: 10.1146/annurev.bi.57.070188.001315. [DOI] [PubMed] [Google Scholar]
- 29.Yildiz F H, Schoolnik G K. Role of rpoS in stress survival and virulence of Vibrio cholerae. J Bacteriol. 1998;180:773–784. doi: 10.1128/jb.180.4.773-784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yildiz F H, Schoolnik G K. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine, and biofilm formation. Proc Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]