Abstract
Hepatitis B virus (HBV) enhancer II (EnII) is a hepatotropic cis element which is responsible for the hepatocyte-specific gene expression of HBV. Multiple transcription factors have been demonstrated to interact with this region. In this study, the region from HBV nucleotides (nt) 1640 to 1663 in EnII was demonstrated to be essential for enhancer activity and to be another target sequence of putative transcription factors. To elucidate the factors which bind to this region, we used a yeast one-hybrid screening system and cloned three transcription factors, HLF, FTF, and E4BP4, from a human adult liver cDNA library. All of these factors had binding affinity to the sequence from nt 1640 to 1663. Investigation of the effects of these factors on transcriptional regulation revealed that HLF and FTF had stimulatory activity on nt 1640 to 1663, whereas E4BP4 had a suppressing effect. FTF coordinately activated both 3.5-kb RNA and 2.4/2.1-kb RNA transcription in a transient transfection assay with an HBV expression vector. HLF, however, activated only 3.5-kb RNA transcription, and in primer extension analysis, HLF strongly stimulated the synthesis of pregenome RNA compared to precore RNA. Thus, FTF stimulated the activity of the second enhancer, while HLF stimulated the activity of the core upstream regulatory sequence, which affects only the core promoter, and had a dominant effect on the pregenome RNA synthesis.
Hepatitis B virus (HBV) causes acute and chronic hepatitis in humans, and chronic infection is closely associated with the development of hepatocellular carcinoma (3). HBV has a partially double-stranded 3.2-kb DNA genome containing four overlapping open reading frames (ORFs) encoding surface antigen (HBsAg), core/e antigen (HBc/eAg), polymerase, and X protein (19, 47). Four promoters (CP, SPI, SPII, and XP) have been identified as cis regulators for transcription of the 3.5-, 2.4-, 2.1-, and 0.8-kb mRNAs, respectively. The 3.5-kb mRNA encodes viral polymerase and HBc/eAg and functions as the pregenome RNA for reverse transcription of the HBV genome as well (8, 45, 54, 56). The 2.4- and 2.1-kb mRNAs are the templates for large and middle/major surface antigens, respectively (7, 36, 41, 43), and the 0.8-kb mRNA is specific for X protein (42, 49). So far, two regions in the HBV genome have been shown to act as transcriptional enhancers. Enhancer I (EnI), which is located upstream of the X gene, displays a preference for hepatocytes (40, 48), while enhancer II (EnII), located just upstream of CP, shows highly restricted hepatocyte-specific activity (53, 57, 60); both enhancers stimulate transcription from the promoters (2, 22, 28, 44, 51, 57, 60). In transfection analysis with cloned HBV DNA, the 3.5-kb mRNAs were detected in well-differentiated human hepatoma cell lines but not in nonhepatic cells (9, 46, 50, 55). This suggests that hepatocyte-specific factors are necessary for the transcription of pregenome RNA and that the hepatotropism of HBV replication might be attributable to the EnII function.
EnII stimulates the transcription from SPI, SPII, and XP in a position- and orientation-independent manner (60). On the other hand, this element regulates the basal CP positively only in a position- and orientation-dependent manner, functioning as a core upstream regulatory sequence (CURS), which coincides with the sequence of EnII (61–63). Yuh et al. have demonstrated that CURS consists of two sequence motifs, a 23-bp box α and a 12-bp box β, that individually regulate transcription activity (61). Hepatocyte-enriched transcription factors, such as hepatocyte nuclear factor 1 (HNF1) (52), HNF3 (29, 31), HNF4 (20, 38), CCAAT/enhancer binding protein (C/EBP) (34, 35, 61), and FTF (fetoprotein transcription factor; hB1F) (32), which are suggested to be responsible for the hepatocyte specificity of EnII activity, and the ubiquitous factor Sp1 (64) have been shown to interact with the EnII sequence and regulate its function (Fig. 1). Several factors were found to regulate HBV EnII, but details of the regulatory mechanism are still unclear, and there remains the possibility that other factors exist.
FIG. 1.
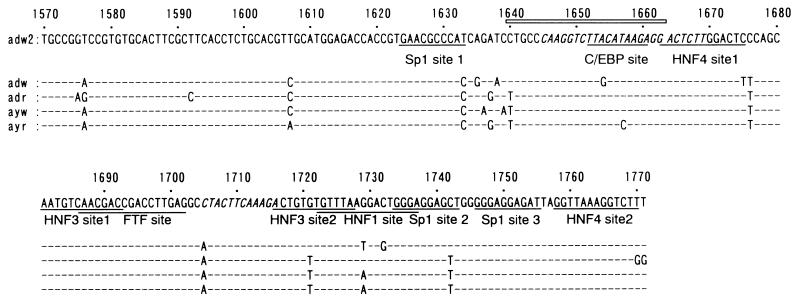
Nucleotide sequences of HBV between nt 1570 to 1771 including the EnII region. The underlined sequences represent Sp1, HNF1, HNF3, HNF4, and FTF (hB1F) recognition sequences, as labeled. Box α (nt 1645 to 1669) and box β (nt 1705 to 1716) in the CURS are shown in italics. The open box indicates the sequence from nt 1640 to 1663 which was used in gel retardation analysis. This region includes the extended C/EBP consensus sequence (T[T/G]NNG[C/T]AA[T/G]).
In this study, transient transfection of HepG2 cells with EnII-minimal TATA-luciferase vector constructs demonstrated that deletion of 20 bp starting with nucleotide (nt) 1640 resulted in a great decrease in the EnII activity. Gel retardation analyses using HepG2 nuclear extract and 32P-labeled double-stranded oligodeoxynucleotides containing HBV nt 1640 to 1663 suggested that multiple factors bound to this region. Next, we used a yeast one-hybrid system to clone transcription factors which interact with HBV nt 1640 to 1663. From a cDNA library of human adult normal liver, we isolated three cDNAs coding HLF (hepatic leukemia factor), FTF, and E4BP4. HLF and E4BP4 are bZIP transcription factors (12, 23, 25), and FTF is characterized as an orphan nuclear receptor (18, 32). Binding motifs for these factors could be found between nt 1640 to 1663. We investigated the interaction of these factors with HBV nt 1640 to 1663 in vitro and their regulatory effect on EnII in vivo. Although all of the factors could bind to nt 1640 to 1663, E4BP4 showed no activating effect and even repressed expression, while HLF and FTF activated reporter gene expression. We further investigated their effects on the transcription of HBV. FTF stimulated the expression of 3.5- and 2.4/2.1-kb RNAs, as determined by Northern blot analysis and HBe/sAg production in the media of cultured cells, but HLF upregulated only the 3.5-kb RNA and HBeAg. In primer extension analysis, HLF strongly activated the transcription of pregenome RNA compared to precore (pre-C) RNA. Thus, we demonstrated that HLF has transcription activity on CURS rather than on EnII and dominantly regulates pregenome RNA transcription, which leads to replication of HBV, and that FTF acts as a trans factor regulating the activity of EnII.
MATERIALS AND METHODS
Plasmids.
The HBV sequence used in this study was of the adw2 subtype (GenBank accession no. X02763). Numbering of the HBV sequence started at the unique EcoRI site. pTATALUC was constructed by removing the chloramphenicol acetyltransferase gene from plasmid pE1bTATACAT (Clontech), followed by insertion of the firefly luciferase reporter gene downstream of the minimal promoter. The luciferase gene was derived from the HindIII-KpnI fragment of pRSV-L (15). Various lengths of fragments of HBV DNA EnII were synthesized or generated by PCR and inserted upstream of the minimal promoter of pTATALUC (Fig. 2 and 4). All of the EnII sequences were inserted in the antisense orientation to evaluate their enhancer activity.
FIG. 2.
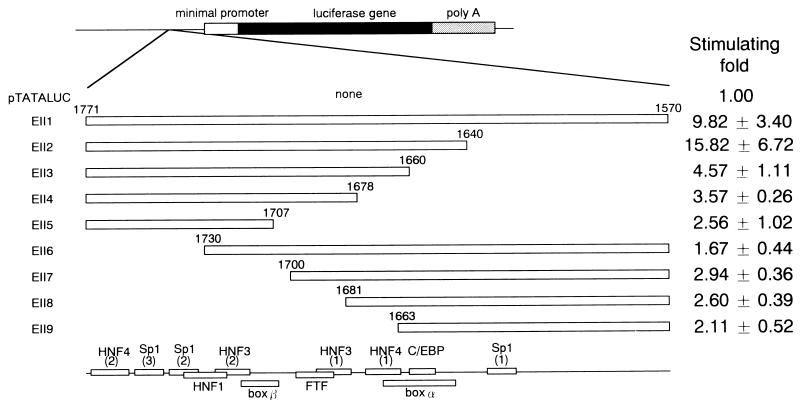
Deletion analysis of HBV EnII. The deletion mutants are shown at the left. Open boxes represent the EnII sequences, and the numerals indicate nucleotides of the HBV genome. The fragment containing the EnII region was cloned into pTATALUC just upstream of the E1b minimal promoter in the antisense orientation. Plasmid DNA was transfected into HepG2 cells, and luciferase activity was determined for the lysate of each sample. Fold stimulation of luciferase activity (mean ± standard deviation) is shown on the right.
FIG. 4.
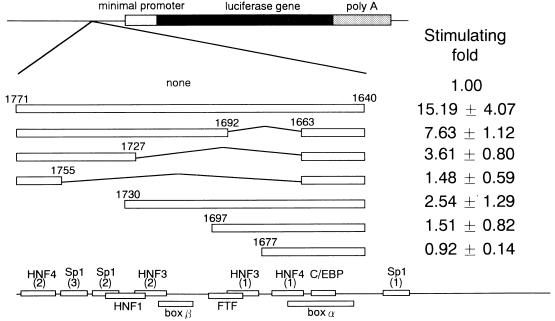
Deletion analysis of HBV nt 1640 to 1771 to determine the correlation between nt 1640 to 1663 and the region from nt 1664 to 1771. The sequences are shown at the left as open boxes. Fold stimulation of luciferase activity (mean ± standard deviation) is shown on the right.
Two oligonucleotides, 5′-GGGCTGCCCAAGGTCTTACATAAGAGGCTGCCCAAGGTCTTACATAAGAGGCTGCCCAAGGTCTTACATAAGAGGCTGCCCAAGGTCTTACATAAGAGGCTGCCCAAGGTCTTACATAAGAGGCCC-3′ and 5′-GGGCCTCTTATGTAAGACCTTGGGCAGCCTCTTATGTAAGACCTTGGGCAGCCTCTTATGTAAGACCTTGGGCAGCCTCTTAT GTAAGACCTTGGGCAGCCTCTTATGTAAGACCTTGGGCAGCCC-3′, containing five tandem 24-bp repeats of HBV nt 1640 to 1663 and some extra nucleotides for cloning to the SmaI site, were synthesized, annealed, and then inserted into the SmaI site of pTATALUC in both sense and antisense orientations to construct pTATALUC-E5 and pTATALUC-E5AS. The m4 sequence, CTGCCCgAGtTCTcACATAgGAGG, was designed to contain four nucleotide mutations (lowercase) in the sequence from nt 1640 to 1663, and plasmids pTATALUC-m4 and pTATALUC-m4AS, containing five tandem repeats of the m4 sequence in the sense and antisense orientations, respectively, were constructed in the same way as pTATALUC-E5 and pTATALUCE5AS (Fig. 3). All fragments were confirmed by sequencing. Plasmids for yeast one-hybrid screening, pHIS-E5 and pLac-E5, were constructed by inserting the double-stranded oligonucleotide upstream of the E1b minimal promoter in pHISi-1 (Clontech) and the CYC1 minimal promoter in pLacZi (Clontech), respectively.
FIG. 3.
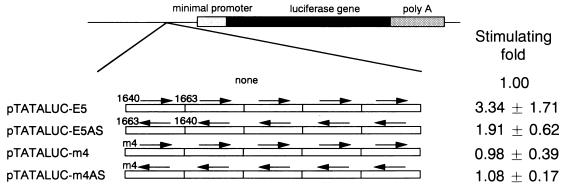
Transcriptional regulation of HBV nt 1640 to 1663 or mutant sequence m4, which has four nucleotide mutations within nt 1640 to 1663. Five tandem repeats of the sequences were inserted upstream of the E1b minimal promoter in the sense or antisense orientation. Fold stimulation of luciferase activity (mean ± standard deviation) is shown on the right.
A HindIII-XbaI fragment containing cDNA of HLF from pSP64-HLF (a gift from Michael L. Cleary) was inserted into the HindIII-XbaI site of pBluescript II SK(+) (Stratagene); then the ClaI-BamHI fragment of this plasmid was subcloned into the ClaI-BamHI site of pFLAG-CMV-2 (Eastman Kodak Company) to produce pCMVHLF. The full-length cDNA of E4BP4 was amplified by PCR using 20 ng of a human liver cDNA library (Clontech), native Pfu DNA polymerase (Stratagene), and primers 5′-GGAAGCTTGATGCAGCTGAGAAAAATGCAG-3′ and 5′-CCGAATTCTTACCCAGAGTCTGAAGCAGAG-3′. The PCR product was gel purified, digested with HindIII and XbaI, and inserted into the HindIII-XbaI site of pFLAG-CMV-2 to produce pCMVE4BP4. To construct the FTF expression vector, the two oligonucleotides 5′-AGCTTATGCAAGTGTCTCAATTTAAAATGGTGAATTACTCCTATGATGAA-3′ and 5′-GATCTTCATCATAGGAGTAATTCACCATTTTAAATTGAGACACTT GCATA-3′, containing the sequence from nt 438 to the unique BglII site (nt 482) of FTF, were synthesized, annealed, and inserted into the HindIII-BglII site of pFLAG-CMV-2. A DNA fragment including the cDNA of FTF from the unique BglII site to the TAA codon (nt 1865) was obtained by digesting pGADFTF, which was isolated by screening with the yeast one-hybrid system, with BglII. This DNA fragment was then inserted into the BglII site of the vector containing the 5′ sequence of the FTF gene mentioned above to produce pCMVFTF. pCMVLUC was constructed by inserting the luciferase gene from pGL3-Basic (Promega) downstream of the cytomegalovirus promoter of pcDNA3 (Invitrogen).
For in vitro transcription-translation, plasmids pGEMHLF, pGEMFTF, and pGEME4BP4 were constructed by inserting HLF, FTF, and E4BP4 cDNAs, respectively, downstream of the SP6 promoter of pGEM-3Zf(+) (Promega).
The nucleotide sequences of HLF, FTF, and E4BP4 referred to in this paper are from GenBank accession no. HUMHLF, HSU93553, and HSE4BP4RN, respectively.
cDNA cloning by the yeast one-hybrid system.
pLac-E5 and pHIS-E5 were linearized by digestion with NcoI and XhoI, respectively, and sequentially integrated into the genome of Saccharomyces cerevisiae YM4271 (MATa ura3-52 his3-200 ade2-101 lys2-801 leu2-3,112 trp1-903 tyr1-501) (Clontech), generating yeast reporter strain YM-E5. Next, the yeast was transformed with the human adult liver MATCHMAKER cDNA library (Clontech), which contains a human liver cDNA library cloned into the EcoRI site of pGAD10 (Clontech), producing a GAL4 activation domain-cDNA fusion protein in yeast cells. The transformants were selected on uracil-, histidine-, and leucine-deficient plates containing 20 mM 3-aminotriazole. Large colonies (His+) were streaked onto another plate with the same amino acid contents and assayed for β-galactosidase activity. The colonies were transferred onto nitrocellulose filters (Hybond C Extra; Amersham Pharmacia Biotech product no. RPN82E). The filters were then submerged into liquid nitrogen for 1 min, placed on filter papers presoaked with a buffer containing 0.8 mM 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside, and incubated at 30°C. Colonies that turned blue (LacZ+) were recovered, and plasmids were isolated from these clones. To confirm that the candidate plasmids were true positives, they were transformed into YM-E5 and retested for His+ phenotype and for β-galactosidase activity. The double-positive plasmids were considered to be true positives, and their insert cDNAs were studied by sequencing.
Cell culture and DNA transfection.
The human hepatocellular carcinoma cell lines HepG2 and HuH7 were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, glucose (1 mg/ml), penicillin G (100 U/ml), streptomycin (100 μg/ml), and amphotericin B (0.25 μg/ml) at 37°C in a 5% CO2 atmosphere.
Approximately 3 × 105 HepG2 cells/well in six-well dishes (Falcon 3046) were transfected with 0.5 μg of luciferase expression vector and 0.5 μg of β-galactosidase expression vector pCMVβ (Clontech), which served as an internal control for transfection efficiency, using Lipofectin (Gibco-BRL). Cells were harvested 3 days after transfection. They were washed three times with phosphate-buffered saline, and the cell lysate was prepared as instructed by the manufacturer of PicaGene (Toyo Ink). Next, 50-μg extracted protein samples were assayed for luciferase and β-galactosidase activities. Transfection efficiency was normalized to the activity of the internal control. Each set of experiments was performed with two different preparations and repeated three to four times for each preparation, and the mean fold stimulating activity relative to that of pTATALUC was calculated.
For cotransfections of luciferase and transcription factor expression vectors, approximately 3 × 105 HuH7 or HepG2 cells/well in six-well dishes were cotransfected with 0.5 μg of luciferase expression vector (pTATALUC, pTATALUC-E5, pTATALUC-EII2, or pTATALUC-EII3), 0.5 μg of pFLAG-CMV-2, pCMVE4BP4, pCMVHLF, or pCMVFTF, and 0.1 μg of pCMVβ, using Lipofectin. Cells were harvested, and cell lysates were prepared and assayed for luciferase activity and for β-galactosidase activity as described above. Each set of experiments was performed with two different preparations and repeated five to six times for each preparation, and the mean fold activity of pTATALUC-E5, pTATALUC-EII2, and pTATALUC-EII3 relative to that of pTATALUC was calculated.
Preparation of nuclear extract and in vitro-translated protein.
The nuclear extract was prepared as previously described (16), with modification. About 108 cells were harvested and washed three times with phosphate-buffered saline. After centrifugation at 1,500 rpm (1.3 × 102 × g) for 5 min, the cell pellet was suspended in 5 volumes of buffer A (10 mM HEPES-KOH [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol [DTT], 0.5 mM phenylmethylsulfonyl fluoride) and incubated on ice for 5 min. After centrifugation at 1,500 rpm for 5 min, the pellet was resuspended in 3 volumes of buffer A; Nonidet P-40 was added to 0.05%, and then homogenization was effected with 20 strokes in a Dounce homogenizer. After pelleting of the nuclei by centrifugation at 1,500 rpm for 5 min, the pellet was resuspended in 1 ml of buffer C (5 mM HEPES-KOH [pH 7.9], 26% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride), and NaCl was added to a final concentration of 300 mM. The mixture was kept on ice for 30 min. After centrifugation at 15,000 rpm (1.3 × 104 × g) for 20 min at 4°C, the supernatant was collected and the nuclear extract was stored in small aliquots at −80°C.
In vitro transcription-translation was performed with a coupled wheat germ extract system (TnT system; Promega) as instructed by the manufacturer. Briefly, cDNAs of E4BP4, HLF, and FTF inserted downstream of the SP6 promoter of pGEM-3Zf(+) were incubated in reaction mixtures with wheat germ extract and SP6 RNA polymerase at 30°C for 120 min. The transcription-translation reactions were stored in small aliquots at −80°C.
Gel retardation analysis.
For gel retardation analysis, the probe was prepared with a double-stranded oligonucleotide, which was cloned into the HindIII site of pBluescript II SK(+) and gel purified after digestion with HindIII (in the following sequence, underlined nucleotides indicate nt 1640 and 1663, respectively, of the HBV sequence):
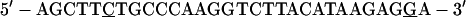 |
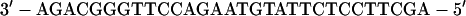 |
The oligonucleotide was dephosphorylated with calf intestine alkaline phosphatase (Toyobo, Osaka, Japan) and end labeled with [γ-32P]ATP (Amersham Co., Ltd., Tokyo, Japan) and T4 polynucleotide kinase (Toyobo). The double-stranded HNF1 oligonucleotide (37)
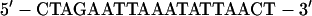 |
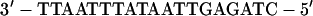 |
was used as the nonspecific competitor or 32P labeled for use as the positive control. The probe was incubated in 20 μl of reaction mixture containing 20 μg of nuclear extract or 0.5 μl of in vitro-translated product, 2 μg of poly(dI-dC) (Pharmacia, Inc.), 20 mM HEPES (pH 7.9), 1 mM MgCl2, 4% Ficoll, and 0.5 mM DTT at room temperature for 30 min. The mixture was resolved on 4% polyacrylamide gel made in 0.25× Tris-borate-EDTA. Electrophoresis was performed at 150 V for 2 h. Gels were dried and autoradiographed. For competition experiments, 10- and 50-fold molar excesses of unlabeled double-stranded oligonucleotides were preincubated with nuclear extracts for 5 min before addition of probe. A double-stranded oligonucleotide containing a mutant sequence of nt 1640 to 1663 (m4)
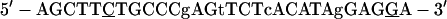 |
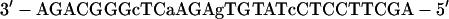 |
(mutated nucleotides are in lowercase letters; nt 1640 and 1663 are underlined) was synthesized for use as a probe or competitor.
Northern blot and primer extension analyses.
Approximately 3 × 106 HuH7 cells in 10-cm-diameter dishes were transfected with DNA (total of 10 μg per plate) consisting of 2.5 μg of pHBV1.5 (6) and 7.5 μg of pFLAG-CMV-2, pCMVHLF, pCMVFTF, or pCMVE4BP4 with Lipofectin (Gibco-BRL). In some experiments, 1 μg of pCMVLUC was cotransfected as an internal control. The media were collected to monitor HBe/sAg expression by radioimmunoassay (RIA), and the cells were harvested 3 days after transfection. Total cellular RNA from the transfected cells was isolated with ISOGEN (Nippon Gene), and then 20 μg of total RNA was analyzed by Northern blotting with the HBV adw2 probe. To compare the precise expression levels of pregenome and pre-C RNAs, primer extension analyses were performed with a primer extension kit (Promega) according to the manufacturer's instructions. Briefly, mRNAs were selected from 100 μg of total RNAs with oligo(dT). Antisense oligonucleotide 5′-GCCCCAAAGCCACCCAAGGCACAGCTTGGA-3′ (nt 1832 to 1861 of HBV adw2) was phosphorylated with [γ-32P]ATP and T4 polynucleotide kinase (Toyobo). Reaction mixtures containing mRNA, the labeled probe, and avian myeloblastosis virus reverse transcriptase were incubated at 42°C for 30 min and then resolved on a 7 M urea–8% acrylamide gel, resulting in detection of 110- and 80-base DNAs corresponding to pre-C and pregenome RNAs, respectively. To determine the level of viral gene transcription, the band intensity of the transcript was measured with an image analyzer (BAS2000; Fuji Film).
RESULTS
Deletion mutant analyses of the EnII element indicates the importance of nt 1640 to 1663 for enhancer activity.
We generated a series of luciferase gene expression vectors containing HBV fragments spanning nt 1570 to 1771 and various deletion mutants, located just upstream of the E1b minimal TATA-luciferase construct. Figure 2 shows a serial 5′ and 3′ deletion analysis of EnII. The contribution of each part of the HBV EnII sequence to enhancer activity was evaluated by transient transfection analyses in the differentiated hepatoma cell line HepG2. The full-length sequence (nt 1570 to 1771) increased the level of gene expression 9.82-fold. When the region from nt 1570 to 1639 was deleted, luciferase expression increased, indicating negative regulation of this region as reported by Lo and Ting (33). Deletion of nt 1640 to 1659 led to a 3.5-fold reduction in luciferase expression. Further deletions of nt 1660 to 1677 and nt 1678 to 1706 resulted in a mild reduction in luciferase expression. These data suggested that the sequence from nt 1640 to 1659 had a significant effect on gene expression. In contrast, none of the deletions from the 3′ end resulted in marked expression of the luciferase gene, indicating that nt 1731 to 1771 had a pivotal role in transcriptional activation. Thus, the sequence from nt 1640 to 1663 was demonstrated to include one of the regions important for enhancer activity.
To investigate the effect of the interaction between the sequence from nt 1640 to 1663 and other regions, we inserted five copies of nt 1640 to 1663 upstream of the minimal promoter (Fig. 3); moderate activities were found to depend to some extent on orientation. The m4 sequence, however, showed no transcriptional activity. These results indicated that the segment from nt 1640 to 1663 was a weak independent regulatory element and that this region could be an independent transcription factor binding site. Next, we constructed luciferase gene expression vectors containing the EnII sequence spanning nt 1640 to 1771 and various deletion mutants thereof (Fig. 4). Removal of nt 1664 to 1691 resulted in reduction of about twofold, and additional removal of nt 1692 to 1726 resulted in a further twofold reduction. In contrast, the sequence lacking nt 1731 to 1771 exhibited a severe decrease in enhancer activity. Thus, HNF4 site 2 and the Sp1 sites seemed to be essential for enhancer activity, and cooperation between HNF3 site 2 to HNF4 site 2 and the sequence from nt 1640 to 1663 seemed to be more important than that between HNF4 site 1 to HNF3 site 1 and the sequence from nt 1640 to 1663 (Fig. 2 and 4).
Detection of specific factors interacting with nt 1640 to 1663 in HepG2 nuclear extract by gel retardation analysis.
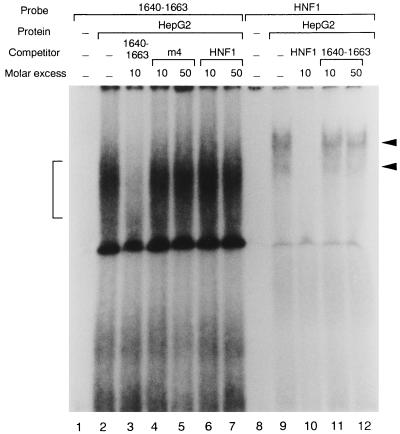
Since deletion mutant analyses showed that the region from nt 1640 to 1663 was a cis-acting element, some transcription factors should be associated with this region. We performed gel retardation analyses using double-stranded oligonucleotides corresponding to the sequence from nt 1640 to 1663 as probes. When the HepG2 nuclear extract was used in the reaction, a shifted band was detected (Fig. 5). It appeared to be specific since there was competition for the signal by the unlabeled sequence from nt 1640 to 1663 but not by the nonspecific competitor, the HNF1 consensus sequence, or the mutated sequence from nt 1640 to 1663. A 32P-labeled double-stranded oligonucleotide of the HNF1 consensus, used as a positive control and incubated with the same nuclear extract, yielded a clear shifted band, confirming that the HepG2 nuclear extract was appropriate for the assay. Compared to the HNF1 band, the shifted band from nt 1640 to 1663 appeared rather broad, probably because of the presence of multiple bands, which suggested that multiple factors which bound to nt 1640 to 1663 existed in the HepG2 nuclear extract and might regulate EnII activity.
FIG. 5.
Gel retardation analysis of HBV nt 1640 to 1663. The 32P-labeled double-stranded oligonucleotide containing the sequence from nt 1640 to 1663 (lane 1 to 7) or 32P-labeled HNF1 consensus sequence (lanes 8 to 12) was incubated with HepG2 nuclear extract at room temperature for 30 min. The mixture was resolved on a 4% polyacrylamide gel. Lane 1, probe only; lanes 2 to 7, probe and 10 μg of nuclear extract with no competitor (lane 2), with 10-fold molar excess of unlabeled specific oligonucleotide of nt 1640 to 1663 (lane 3), with 10- and 50-fold molar excess of unlabeled nonspecific HNF1 oligonucleotide (lanes 4 and 5), and with 10- and 50-fold molar excess of unlabeled mutant sequence of nt 1640 to 1663, m4 (lanes 6 and 7), respectively; lane 8, probe only; lanes 8 to 12, probe and 10 μg of nuclear extract with no competitor (lane 9), with 10-fold molar excess of unlabeled HNF1 consensus sequence (lane 10), and with 10- and 50-fold molar excess of unlabeled nonspecific oligonucleotide of nt 1640 to 1663 (lanes 11 and 12). The specifically shifted band of nt 1640 to 1663 is indicated by a bracket, and the specifically shifted band of HNF1 is indicated by arrowheads.
Cloning of cDNAs encoding three transcription factors, HLF, FTF, and E4BP4, which bind to HBV nt 1640 to 1663, by the yeast one-hybrid system.
As indicated above, hepatocyte-specific transcription factors should bind to HBV nt 1640 to 1663 and regulate EnII activity. To identify those factors, we used a yeast one-hybrid screening system. Five tandem repeats of HBV nt 1640 to 1663 were used as bait for the interaction with the fusion protein consisting of the putative transcription factor and GAL4 activation domain produced in yeast cells. Approximately 5 × 106 cDNA clones were screened by transforming the yeast reporter strain YM-E5, and 24 double-positive clones were obtained. The plasmids were recovered from the positive yeast clones and retransformed into YM-E5 to confirm these positive tests. Next, three transcription factors, HLF (1 clone), FTF (7 clones), and E4BP4 (16 clones), were identified by sequence analyses as candidates that bind to HBV nt 1640 to 1663.
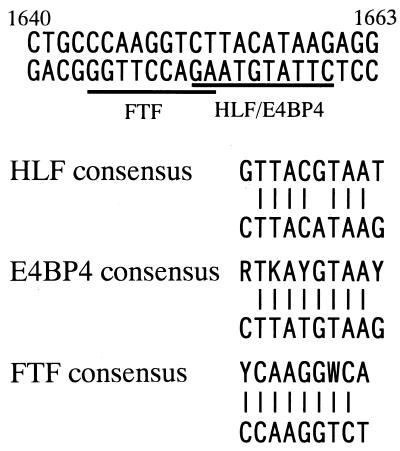
The cloned E4BP4 fragment contained nt 329 to 814 of the ORF in the sense orientation. E4BP4 is a member of bZIP transcription factors originally cloned as a repressor protein for the E4 promoter of adenovirus (12) and as an activator for the interleukin-3 (IL-3) promoter (65). The protein transcribed from the cloned fragment contained the basic region as the DNA binding domain and the leucine zipper domain as the dimerizing domain, suggesting that the cloned protein could bind to the DNA sequence motif. The binding site for E4BP4 was shown to have the consensus sequence (G/A)T(G/T)A(C/T)GTAA(C/T) (12), and the region from nt 1640 to 1663 contained a sequence homologous to the consensus in the half site from the 3′ end (Fig. 6).
FIG. 6.
Nucleotide sequence of HBV nt 1640 to 1663. The underlined sequences represent the regions homologous to FTF and HLF/E4BP4 recognition sequences. Comparison of the HLF, FTF, and E4BP4 binding sequences in HBV EnII and their consensus binding sequences is shown below (R = A or G; K = G or T; Y = C or T; W = A or T). The nucleotide sequence of HBV compared to the E4BP4 consensus is shown in the antisense orientation.
HLF is a hepatocyte-enriched transcription factor with characteristics of the bZIP family. It was initially cloned as a fusion protein, E2A-HLF, produced by genomic translocation in acute lymphoblastic leukemia cells (23, 26). The cloned fragment of HLF contained a downstream sequence from nt 437, a region which included the basic region and the leucine zipper domain. Interestingly, HLF and E4BP4 have been demonstrated to share the same binding motif (Fig. 6) (1, 27). Therefore, both HLF and E4BP4 may regulate viral gene expression through interaction with the same site of HBV EnII.
We obtained two kinds of FTF-encoding plasmids; one had the sequence from nt 444 to 2176 in the antisense orientation, and the other had nt 444 to 1237 in the sense orientation. Li et al. reported the cloning of hB1F, a factor binding to EnII with the yeast one-hybrid system using the EnII B1 region as bait (32). They isolated a plasmid encoding the hB1F ORF in the antisense orientation, suggesting that the gene was transcribed by a cryptic promoter around the ADH1 terminator region of the vector. The fragments of FTF cloned in the present study had both the DNA binding domain and the ligand binding domain. FTF is a homolog of the orphan nuclear receptor fushi tarazu factor I (FTZ-F1) in Drosophila melanogaster and is thought to have the same binding property as FTZ-F1. HBV nt 1640 to 1663 contained a sequence that fits the FTZ-F1 consensus ([T/C]CAAGG[T/A]CA) (Fig. 6) (18), indicating that FTF could bind to that region. Thus, all of the three isolated cDNAs contained the DNA binding domains, and their binding sites lay within the sequence from nt 1640 to 1663. They are likely to be involved in regulation of the transcriptional activity of EnII.
Factors binding specifically to the sequence of HBV nt 1640 to 1663.
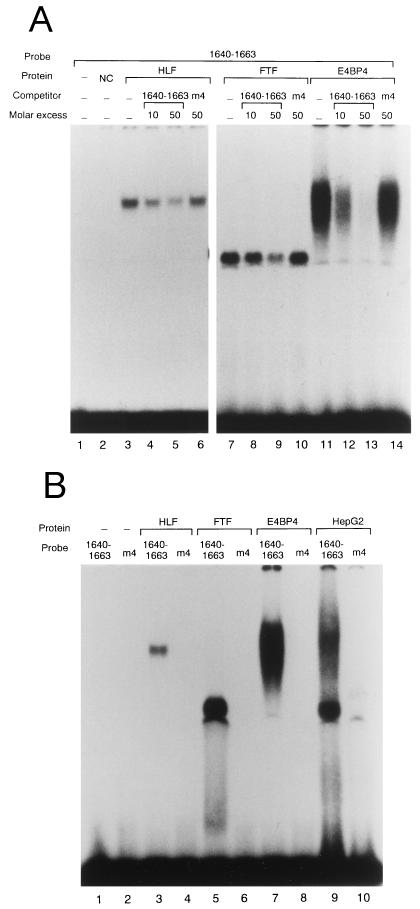
Since homologous sequences of the consensus motifs of the three proteins identified with the yeast one-hybrid system could be found within the region from nt 1640 to 1663, gel retardation analyses were performed to confirm their binding to that sequence. The cDNAs inserted downstream of the SP6 promoter were transcribed with SP6 RNA polymerase and translated with wheat germ extract. As shown in Fig. 7A, all of the in vitro transcription-translation products exhibited complex formation with the probe for nt 1640 to 1663. Complex formation was abolished after preincubation with an unlabeled double-stranded oligonucleotide of nt 1640 to 1663 but not with m4, the mutant sequence of nt 1640 to 1663, which had two nucleotide substitutions in the HLF/E4BP4 binding motif and two more substitutions in the FTF binding motif. Next, to confirm specific binding of the factors to nt 1640 to 1663, gel retardation analyses with a probe containing the m4 sequence were performed (Fig. 7B). None of the proteins showed complex formation with the mutant probe, or the HepG2 nuclear extract contained no factors interacting with the mutant sequence. These results indicated that all three transcription factors bound specifically to the nucleotide sequence from HBV nt 1640 to 1663 in EnII.
FIG. 7.
Gel retardation analyses detecting complex formation of in vitro-translated protein or HepG2 nuclear extract and the probe containing the sequence from nt 1640 to 1663 or m4, the mutant sequence of nt 1640 to 1663. (A) Lane 1, probe only; lane 2, negative control [NC; probe and 0.5 μl of in vitro translation reaction of pGEM-3Zf(+)]; lanes 3 to 6, probe and 0.5 μl of in vitro translation reaction of HLF without competitor (lane 3), with 10- and 50-fold molar excess of unlabeled specific competitor of nt 1640 to 1663 (lanes 4 and 5), and with 50-fold molar excess of m4 (lane 6); lanes 7 to 10, probe and 0.5 μl of in vitro translation reaction of FTF without competitor (lane 7), with 10- and 50-fold molar excess of unlabeled specific competitor of nt 1640 to 1663 (lanes 8 and 9), and with 50-fold molar excess of m4 (lane 10); lanes 11 to 14, probe and 0.5 μl of in vitro translation reaction of E4BP4 without competitor (lane 11), with 10- and 50-fold molar excess of unlabeled specific competitor of nt 1640 to 1663 (lanes 12 and 13), and with 50-fold molar excess of m4 (lane 14). (B) Probes used in the assays were the sequence from nt 1640 to 1663 (lane 1, 3, 5, 7, and 9) and m4 (lane 2, 4, 6, 8, and 10). Lanes 1 and 2, probe only; lanes 3 and 4, probe and 0.5 μl of in vitro translation reaction of HLF; lanes 5 and 6, probe and 0.5 μl of in vitro translation reaction of FTF; lanes 7 and 8, probe and 0.5 μl of in vitro translation reaction of E4BP4; lanes 9 and 10, probe and 10 μg of HepG2 nuclear extract.
Activation of HBV EnII by HLF and FTF and suppression by E4BP4.
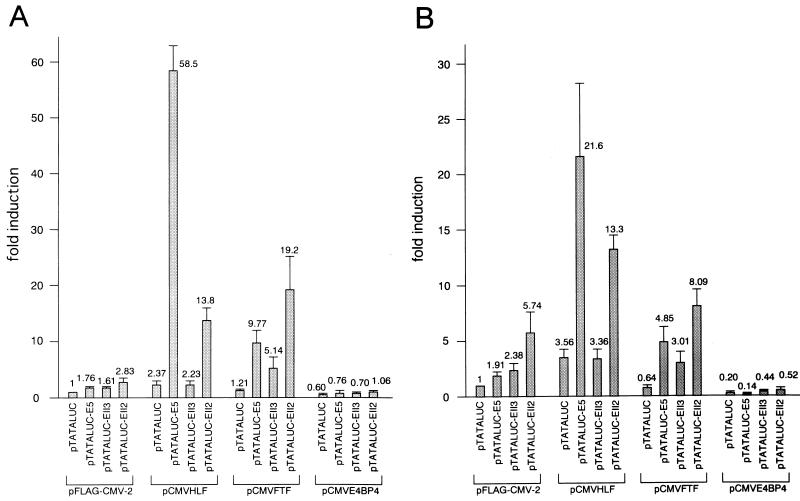
As all three transcription factors were shown to be able to bind to EnII and to be expressed in human liver tissue (25, 30, 32), we measured luciferase activities in cells transfected with pTATALUC, pTATALUC-E5, pTATALUC-EII2, and pTATALUC-EII3 in the presence of overexpression of each factor. As shown in Fig. 8, luciferase gene expression by pTATALUC increased after transfection with pCMVHLF and decreased after transfection with pCMVE4BP4. This alteration of baseline luciferase expression occurred probably because the original vector sequence of pTATALUC contained several motifs homologous to the HLF/E4BP4 consensus motif (data not shown). With respect to the luciferase activity of pTATALUC-E5 compared to that of pTATALUC, the pCMVHLF transfection in HuH7 cells showed that the presence of five iterations of the sequence between nt 1640 to 1663 increased luciferase activity about 25-fold. pCMVFTF demonstrated a more than eightfold increase in luciferase activity. As luciferase activity was induced 1.76-fold in the mock transfection experiment, HLF and FTF were shown to activate the minimal promoter via interaction with nt 1640 to 1663. In contrast, E4BP4 had a suppressive effect compared to the mock transfection experiment.
FIG. 8.
Transcriptional regulation of nt 1640 to 1663 by HLF, FTF, and E4BP4 in HuH7 cells (A) and HepG2 cells (B). pTATALUC, pTATALUC-E5, which contains five iterations of nt 1640 to 1663 in tandem, pTATALUC-EII2, which contains HBV nt 1640 to 1771, or pTATALUC-EII3, which contains HBV nt 1660 to 1771, was cotransfected with pFLAG-CMV-2, pCMVHLF, pCMVFTF, or pCMVE4BP4, and the cell lysate was assayed for luciferase activity. The fold activity of each transfectant relative to the cells transfected with pTATALUC and pFLAG-CMV-2 was calculated.
We investigated the effects of these factors by cotransfection of a vector containing the EnII configuration, pTATALUC-EII2 or pTATALUC-EII3. Alteration of the luciferase activity of pTATALUC-EII3, which did not include the HLF/E4BP4 binding motif in the EnII region, was observed again in the experiments using pCMVHLF and pCMVE4BP4. As pTATALUC-EII3 included another FTF binding site, its luciferase expression was upregulated by pCMVFTF cotransfection. Comparing the ratio of the luciferase activity of pTATALUC-EII2 to that of pTATALUC-EII3, we found that both HLF and FTF stimulated luciferase expression in HuH7 cells about 6- and 4-fold, respectively, while the value for mock transfectants was 1.76-fold. E4BP4 showed a slight suppressive effect. Though the extents of induction in the HLF and FTF experiments were lower than in the experiments comparing pTATALUC and pTATALUC-E5, the results seemed reasonable since pTATALUC-EII2 contained only one iteration of nt 1640 to 1663. In experiments using HepG2 cells, luciferase activities were similar to those for HuH7 cells, though somewhat lower. Taken together, these effects reflected the enhancer activity of the EnII region, suggesting that the three factors are involved in the regulation of HBV gene expression in hepatocytes.
Effects of HLF, FTF, and E4BP4 on HBV transcription.
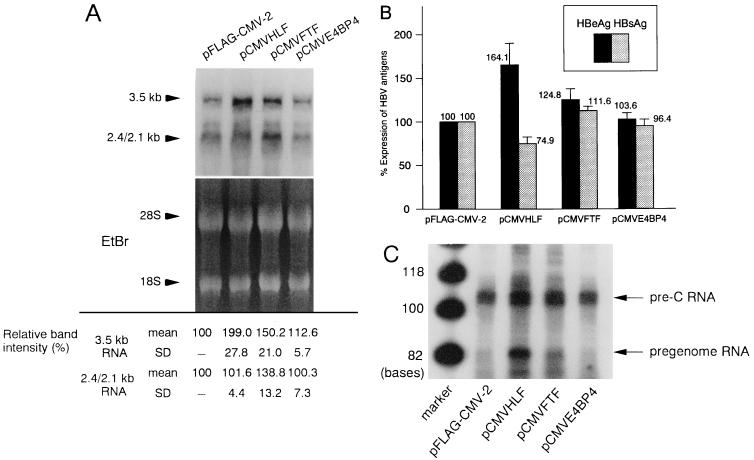
To assess the effects of HLF, FTF, and E4BP4 on the transcription of HBV, we performed transient cotransfection experiments with pHBV1.5, an HBV expression vector, and pFLAG-CMV-2, pCMVHLF, pCMVFTF, or pCMVE4BP4. With cotransfection of pCMVLUC as an internal control, no significant difference was observed between the samples. As shown in Fig. 9A, FTF stimulated both 3.5- and 2.4/2.1-kb RNAs while E4BP4 had no significant effect on transcription. Interestingly, HLF stimulated only 3.5-kb RNA transcription. The levels of HBeAg and HBsAg, which were translated from 3.5- and 2.4/2.1-kb RNAs, respectively, were the same as the levels of their own transcripts including the HLF transfectant (Fig. 9B). As HLF stimulated only transcription from CP, we evaluated the two classes of transcripts specific to the 3.5-kb RNA, the pregenome and pre-C RNAs, by the primer extension method (Fig. 9C). The levels of pre-C RNA were upregulated in the lanes representing pCMVHLF (about 1.5-fold) and pCMVFTF (about 1.3-fold), consistent with the results for HBeAg. Among the pregenome RNAs, however, the band for the HLF transfectant was specifically amplified (about 5.9-fold). Thus, the transcriptional activity of HLF may be specific to CP; it upregulated pregenome RNA more strongly than pre-C RNA.
FIG. 9.
Regulation of HBV transcripts and of HBeAg and HBsAg by HLF, FTF, E4BP4. HuH7 cells (3 × 106 per 10-cm-diameter dish) were transfected with 2.5 μg of pHBV1.5 and 7.5 μg of pFLAG-CMV-2, pCMVHLF, pCMVFTF, or pCMVE4BP4. (A) Northern blot analysis of HBV transcripts. Total RNA (20 μg) was hybridized with an HBV adw2 probe. Arrows indicate the 3.5- and 2.4/2.1-kb transcripts. The band intensity of the HBV transcripts was measured with an image analyzer. The band intensity of the reaction using pFLAG-CMV-2 was considered 100%, and the band intensity relative to this value was calculated. (B) Quantitation of the degree of expression of HBeAg and HBsAg measured by RIA. The expression level of the reaction using pFLAG-CMV-2 was considered 100%, and percent expression relative to this value was calculated. (C) Synthesis of pre-C and pregenome RNAs. mRNAs selected from 100 μg of total RNAs with oligo(dT) were used for primer extension. The products corresponding to pre-C and pregenome RNAs are indicated by arrows.
DISCUSSION
HBV EnII is a cis-acting element essential for the expression of HBV genes and viral replication. It is located just upstream of the pregenome RNA start site; the sequence is highly (more than 94%) conserved among HBV subtypes (Fig. 1) and thought to be involved mainly in synthesis of this RNA. This activity displays differentiated liver cell specificity; it is strong in such differentiated hepatoma cell lines as HepG2, HuH7, and HuH6 but not in such nonliver cell lines as HeLa and CV-1 (53, 57, 60), indicating the involvement of hepatocyte-specific factors. As previous studies have demonstrated, hepatocyte-enriched transcription factors HNF1, HNF3, HNF4, FTF (hB1F), and C/EBP, and other ubiquitous transcription factors such as Sp1, have been thought to regulate gene expression via interaction with EnII (20, 29, 31, 32, 34, 35, 38, 52, 64).
In this study, we demonstrated that the region from nt 1640 to 1663 of HBV is an independent sequence which is sufficient for the enhancer activity of EnII; deletion of this sequence resulted in a great decrease in luciferase gene expression under the control of the minimal promoter, and five tandem repeats of nt 1640 to 1663 increased luciferase gene expression, which indicated that this sequence itself possessed a modest cis-activating effect. The orientation had some effect on this activity, implying that it does not conform to the original definition of the enhancer. Gel retardation analyses suggested that multiple putative transcription factors in the HepG2 nuclear extract bound to nt 1640 to 1663. When we performed gel retardation analyses with the nonhepatic cell line HeLa nuclear extract, the signal intensity of the band was much weaker than that of the HepG2 nuclear extract, suggesting that the factors are hepatocyte enriched (data not shown). Though the activity is modest, the sequence from nt 1640 to 1663 is considered important because (i) none of the transcription factor binding sites within EnII can be a definite cis element, and EnII activity is exhibited by the cooperative action of multiple transcription factors; (ii) considering that lack of this region resulted in great decrease in EnII activity, factors binding to this element may play a key role in transcription, complementing or cooperating with other factors such as HNF4; and (iii) EnII and CURS activities do not necessarily coincide, and it is possible that EnII largely influences the function of CP.
In the study reported by Yuh and Ting, two putative transcription factors in HepG2 cells were found to bind to box α, which partially overlaps with nt 1640 to 1663 (61). Since this region appeared to have weak homology to the extended consensus for a C/EBP binding site (T[T/G]NNG[C/T]AA[T/G]) (39), C/EBP may be one of the hepatocyte-enriched transcription factors which bind to this sequence. However, at a high concentration, C/EBPα, which is expressed abundantly in hepatocytes, repressed the expression from CP (34). Moreover, although the sequence of box α appeared to have low homology to the extended consensus for a C/EBP binding site, it showed transcription activity in HepG2 cells, in which C/EBP α is expressed at a much lower level than in differentiated hepatocytes (17). These data indicated that some transcription factors other than C/EBPα might bind to box α. Alternatively, there may be another factor binding to the region, not overlapping with box α, between nt 1640 and 1663. To identify the trans factors regulating HBV gene expression, we used a yeast one-hybrid system to clone these factors from a human adult liver cDNA library. In that screening, five tandem repeats of nt 1640 to 1663 were used as bait, and three cDNAs identical to transcription factors HLF, FTF, and E4BP4 were obtained; again C/EBPα was not picked up in our study.
HLF and E4BP4 belong to the family of bZIP transcription factors, which are characterized by a region rich in basic amino acids followed by a leucine zipper domain, through which they form a homo- or heterodimer and interact with the target DNA motif. HLF was initially cloned as a chimeric bZIP transcription factor, E2A-HLF, created by the t(17;19)(q22;p13) chromosomal translocation in some cases of acute leukemia involving pro-B lymphocytes (23–25). Expression of the native form of HLF was shown to be restricted in liver and kidney tissue but not in normal or transformed lymphoid cells. There is a report that HLF may be involved in the expression of factor IX in the liver (5), but the details of its function remain unclear. As shown in Fig. 8, HLF displayed a strong trans-activating effect on EnII activity.
E4BP4 binds to the E4 promoter of adenovirus, the IL-1β promoter, the gamma interferon promoter, and the A site of the IL-3 promoter (11, 12, 65). Interestingly, E4BP4 has been shown to have two antagonistic transcriptional properties, serving as a repressor of the E4 promoter of adenovirus and an activator of the IL-3 promoter. These different characteristics are thought to depend on the cellular context. Recently, Lai and Ting demonstrated that E4BP4 repressed the stimulating activity of box α in CURS and EnII (30). Our results from the cotransfection experiment, which showed that overexpressed E4BP4 suppressed transcription from a minimal promoter followed by five tandem repeats of nt 1640 to 1663 or EnII sequence, agreed with their data. Though luciferase activity decreased in the presence of E4BP4 overexpression, the effect was relatively small when the promoter region included only one copy of the E4BP4 binding motif. Upon cotransfection with the HBV expression vector pHBV1.5, there was no significant change in viral gene expression. As previously reported, repression of E4BP4 is caused by interaction of the E4BP4 repression domain with the TATA-binding protein-binding repressor protein Dr1 (13, 14), rendering E4BP4 an active transcriptional repressor. On the other hand, Lai and Ting suggested that E4BP4 repressed transcription by binding site occlusion rather than by acting as an active repressor (30). They mentioned the possibility that other factors binding to overlapping or identical sites influence the regulation of E4BP4. Considering that HLF and E4BP4 bind to the same DNA motif, and HLF is substantially expressed in liver and HepG2 cells, it is likely that E4BP4 represses transcription by competing with HLF for their shared binding motif.
FTF is categorized as a homolog of the orphan nuclear receptor FTZ-F1. In previous studies, the rat FTZ-F1 homolog rFTF was found to activate the α-fetoprotein gene and to be expressed in the liver and pancreas (4, 18, 32), but there has been no report of its effect on transcription in human hepatocytes. Recently, Li et al. reported that they had cloned a transcription factor binding to the B1 region within EnII (they named it hB1F, for human B1-binding factor) that was identical to FTF (32). The B1 region is located downstream of nt 1640 to 1663, which means that EnII contains two FTF binding sites. It is not surprising that more than two binding motifs of some transcription factor exist nearby, as both HNF3 and HNF4 have two binding sites and Sp1 has three binding sites in EnII. The gel retardation assay showed that FTF could bind to nt 1640 to 1663 (Fig. 7); in cotransfection analyses, it stimulated transcription from the minimal promoter, resulting in an increase in luciferase activity (Fig. 8). In gel retardation analyses using HepG2 nuclear extract with mutant double-stranded oligonucleotide m4 as a competitor (Fig. 5, lanes 6 and 7), no change was observed in the shifted band; when m4 was used as a probe, no complex formation was observed (Fig. 7B, lanes 9 and 10). As demonstrated in Fig. 7B, the mutations introduced within nt 1640 to 1663 abolished binding affinity to HLF, FTF, and E4BP4, which are expressed in HepG2 cells (23, 32), nor did it show complex formation with HepG2 nuclear extract. In addition, m4 revealed no transcriptional activity in HepG2 cells. These findings suggest that the band may be produced by HLF, FTF, and E4BP4 in addition to C/EBP, and the transcriptional activity of nt 1640 to 1663 is likely to be exhibited by such factors.
Since these factors were demonstrated to have transcriptional regulatory activity, we further investigated their effects on HBV RNA transcription. After transfection with HBV expression vector pHBV1.5, FTF coordinately increased the expression levels of both 3.5- and 2.4/2.1-kb RNAs, and the HBeAg and HBsAg levels in the media monitored by RIA were consistent with the HBV transcripts. The regulation by HLF, however, was different from the pattern for FTF: it strongly stimulated transcription of the 3.5-kb RNA but had no significant effect on the 2.4/2.1-kb RNA. The EnII sequence is believed to have two roles when acting as a cis element, as an enhancer of all promoters of HBV and as a CURS to CP. The CURS region coincides with the sequence of EnII but is applicable only to CP, functioning in a position- and orientation-dependent manner (62). Taken together, the data indicate that FTF upregulated the global enhancer activity of EnII and HLF upregulated the CURS activity to specifically affect CP.
The 3.5-kb RNA is subdivided into two classes, pregenome RNA and pre-C RNA. The 5′ end of pregenome RNA is located within 30 bases downstream from the beginning of the pre-C RNA. As previously demonstrated, the syntheses of these two RNAs are regulated by two separate promoters, and the expressions of these promoters are differentially regulated by several factors (10, 58, 59). Our results of primer extension analyses indicated that HLF upregulated both pre-C and, to an even greater extent, pregenome RNA. Synthesis of the pregenome RNA is a pivotal step in the replicative cycle of HBV, as it encodes proteins C and P, which are essential for the formation of nucleocapsids, and serves as the template for viral DNA synthesis. These findings imply that HLF plays an important role in the viral replication in hepatocytes, in which HLF is highly expressed. Differential regulation of the RNAs transcribed from CP was demonstrated previously by Yu and Mertz (59). HNF4, chicken ovalbumin upstream promoter transcription factor 1 (COUP-TF1), human testicular receptor 2 (TR2), and peroxisome proliferator-activated receptors (PPARs) as heterodimers with retinoid X receptors (RXRs) can regulate transcription via interaction with hormone response element DR1. HNF4 and TR2 repressed synthesis of the pre-C RNA, PPARγ-RXRα activated synthesis of the pregenome RNA, and COUP-TF1 coordinately repressed synthesis of both the pre-C and pregenome RNAs. A recent study by Lai and Ting (30) suggested that the C/EBP family and E4BP4 can bind to box α and may be involved in the regulation of viral gene expression. Therefore, HLF is another factor regulating RNA transcription from CP differently via interaction with sites other than the HNF4 site. In addition, HLF is one of the bZIP transcription factors regulating EnII activity which may interact with each other (forming heterodimers, competing for the binding site), leading to a subtle change in transcriptional regulation depending on the cellular context (21).
Finally, EnII activity is a result of cooperative action of various transcription factors such as HNF1, HNF3, HNF4, Sp1, C/EBP, HLF, FTF, and E4BP4. The factors which stimulate transcription of HBV gene must be expressed in hepatocytes because the absence of even one of them results in a great decrease in transcription. Since HLF and FTF were demonstrated to be involved in HBV gene expression, though their function in human hepatocytes is not yet established, and HBV replication can be determined by the combined action of hepatocyte-enriched factors leading to hepatotropism of HBV, we suggest that controlling the functions of these factors may contribute to the suppression of HBV replication and offer a therapeutic method for treating HBV infection.
REFERENCES
- 1.Altura R A, Inukai T, Ashmun R A, Zambetti G P, Roussel M F, Look A T. The chimeric E2A-HLF transcription factor abrogates p53-induced apoptosis in myeloid leukemia cells. Blood. 1998;92:1397–1405. [PubMed] [Google Scholar]
- 2.Antonucci T K, Rutter W J. Hepatitis B virus (HBV) promoters are regulated by the HBV enhancer in a tissue-specific manner. J Virol. 1989;63:579–583. doi: 10.1128/jvi.63.2.579-583.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beasley R P, Hwang L-Y, Lin C-C, Chien C-S. Hepatocellular carcinoma and hepatitis B virus: a prospective study of 22,707 men in Taiwan. Lancet. 1981;ii:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 4.Bernier D, Thomassin H, Allard D, Guertin M, Hamel D, Blaquière M, Beauchemin M, LaRue H, Estable-Puig M, Bélanger L. Functional analysis of developmentally regulated chromatin-hypersensitive domains carrying the α1-fetoprotein gene promoter and the albumin/α 1-fetoprotein intergenic enhancer. Mol Cell Biol. 1993;13:1619–1633. doi: 10.1128/mcb.13.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boccia L M, Lillicrap D, Newcombe K, Mueller C R. Binding of the Ets factor GA-binding protein to an upstream site in the factor IX promoter is critical event in transactivation. Mol Cell Biol. 1996;16:1929–1935. doi: 10.1128/mcb.16.5.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruss V, Ganem D. The role of envelope proteins in hepatitis B virus assembly. Proc Natl Acad Sci USA. 1991;88:1059–1063. doi: 10.1073/pnas.88.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cattaneo R, Will H, Hernandez N, Schaller H. Signals regulating hepatitis B surface antigen transcription. Nature. 1983;305:336–338. doi: 10.1038/305336a0. [DOI] [PubMed] [Google Scholar]
- 8.Cattaneo R, Will H, Schaller H. Hepatitis B virus transcription in the infected liver. EMBO J. 1984;3:2191–2196. doi: 10.1002/j.1460-2075.1984.tb02113.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang C, Jeng K-S, Hu C-P, Lo S J, Su T-S, Ting L-P, Chou C-K, Han S-H, Pfaff E, Salfeld J, Schaller H. Production of hepatitis B virus in vitro by transient expression of cloned HBV DNA in a hepatoma cell line. EMBO J. 1987;6:675–680. doi: 10.1002/j.1460-2075.1987.tb04807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen I-H, Huang C-J, Ting L-P. Overlapping initiator and TATA box functions in the basal core promoter of hepatitis B virus. J Virol. 1995;69:3647–3657. doi: 10.1128/jvi.69.6.3647-3657.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen W-J, Lewis K S, Chandra G, Cogswell J P, Stinnett S W, Kadwell S H, Gray J G. Characterization of human E4BP4, a phosphorylated bZIP factor. Biochim Biophys Acta. 1995;1264:388–396. doi: 10.1016/0167-4781(95)00182-4. [DOI] [PubMed] [Google Scholar]
- 12.Cowell I G, Skinner A, Hurst H C. Transcriptional repression by a novel member of the bZIP family of transcription factors. Mol Cell Biol. 1992;12:3070–3077. doi: 10.1128/mcb.12.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowell I G, Hurst H C. Transcriptional repression by the human bZIP factor E4BP4: definition of a minimal repression domain. Nucleic Acids Res. 1994;22:59–65. doi: 10.1093/nar/22.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowell I G, Hurst H C. Protein-protein interaction between the transcriptional repressor E4BP4 and the TBP-binding protein Dr1. Nucleic Acids Res. 1996;18:3607–3613. doi: 10.1093/nar/24.18.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Wet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman A D, Landschulz W H, McKnight S L. CCAAT/enhancer binding protein activates the promoter of the serum albumin gene in cultured hepatoma cells. Genes Dev. 1989;3:1314–1322. doi: 10.1101/gad.3.9.1314. [DOI] [PubMed] [Google Scholar]
- 18.Galarneau L, Paré J-F, Allard D, Hamel D, Lévesque L, Tugwood J D, Green S, Bélanger L. The α1-fetoprotein locus is activated by a nuclear receptor of the Drosophila FTZ-F1 family. Mol Cell Biol. 1996;16:3853–3865. doi: 10.1128/mcb.16.7.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganem D, Varmus H E. The molecular biology of the hepatitis B virus. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 20.Guo W, Chen M, Yen T S B, Ou J-H. Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol Cell Biol. 1993;13:443–448. doi: 10.1128/mcb.13.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas N B, Cantwell C A, Johnson P F, Burch J B E. DNA-binding specificity of the PAR basic leucine zipper protein VBP partially overlaps those of the C/EBP and CREB/ATF families and is influenced by domains that flank the core basic region. Mol Cell Biol. 1995;15:1923–1932. doi: 10.1128/mcb.15.4.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honigwachs J, Faktor O, Dikstein R, Shaul Y, Laub O. Liver-specific expression of hepatitis B virus is determined by the combined action of the core gene promoter and the enhancer. J Virol. 1989;63:919–924. doi: 10.1128/jvi.63.2.919-924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunger S P, Ohyashiki K, Toyama K, Cleary M L. Hlf, a novel hepatic bZIP protein, shows altered DNA-binding properties following fusion to E2A in t(17;19) acute lymphoblastic leukemia. Genes Dev. 1992;6:1608–1620. doi: 10.1101/gad.6.9.1608. [DOI] [PubMed] [Google Scholar]
- 24.Hunger S P, Brown R, Cleary M L. DNA-binding and transcriptional regulatory properties of hepatic leukemia factor (HLF) and the t(17;19) acute lymphoblastic leukemia chimera E2A-HLF. Mol Cell Biol. 1994;14:5986–5996. doi: 10.1128/mcb.14.9.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inaba T, Roberts W M, Shapiro L H, Jolly K W, Raimondi S C, Smith S D, Look A T. Fusion of the leucine zipper gene HLF to the E2A gene in human acute B-lineage leukemia. Science. 1992;257:531–534. doi: 10.1126/science.1386162. [DOI] [PubMed] [Google Scholar]
- 26.Inaba T, Shapiro L H, Funabiki T, Sinclair A E, Jones B G, Ashmun R A, Look A T. DNA-binding specificity and trans-activating potential of the leukemia-associated E2A-hepatic leukemia factor fusion protein. Mol Cell Biol. 1994;14:3403–3413. doi: 10.1128/mcb.14.5.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikura S, Inukai T, Inaba T, Nimer S D, Cleveland J L, Look A T. Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc Natl Acad Sci USA. 1997;94:2609–2614. doi: 10.1073/pnas.94.6.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jammel S, Siddiqui A. The human hepatitis B virus enhancer requires trans-acting cellular factor(s) for activity. Mol Cell Biol. 1986;6:710–715. doi: 10.1128/mcb.6.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson J L, Raney A K, McLachlan A. Characterization of a functional hepatocyte nuclear factor 3 binding site in the hepatitis B virus nucleocapsid promoter. Virology. 1995;208:147–158. doi: 10.1006/viro.1995.1138. [DOI] [PubMed] [Google Scholar]
- 30.Lai C-K, Ting L-P. Transcriptional repression of human hepatitis B virus genes by a bZIP family member, E4BP4. J Virol. 1999;73:3197–3209. doi: 10.1128/jvi.73.4.3197-3209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li M, Xie Y, Wu X, Kong Y, Wang Y. HNF3 binds and activates the second enhancer, ENII, of hepatitis B virus. Virology. 1995;214:371–378. doi: 10.1006/viro.1995.0046. [DOI] [PubMed] [Google Scholar]
- 32.Li M, Xie Y-H, Kong Y-Y, Wu X, Zhu L, Wang Y. Cloning and characterization of a novel human hepatocyte transcription factor, hB1F, which binds and activates enhancer II of hepatitis B virus. J Biol Chem. 1998;273:29022–29031. doi: 10.1074/jbc.273.44.29022. [DOI] [PubMed] [Google Scholar]
- 33.Lo W-Y, Ting L-P. Repression of enhancer II activity by a negative regulatory element in the hepatitis B virus genome. J Virol. 1994;68:1758–1764. doi: 10.1128/jvi.68.3.1758-1764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.López-Cabrera M, Letovsky J, Hu K-Q, Siddiqui A. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1990;87:5069–5073. doi: 10.1073/pnas.87.13.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.López-Cabrera M, Letovsky J, Hu K-Q, Siddiqui A. Transcriptional factor C/EBP binds to and transactivates the enhancer element II of the hepatitis B virus. Virology. 1991;183:825–829. doi: 10.1016/0042-6822(91)91019-d. [DOI] [PubMed] [Google Scholar]
- 36.Malpièce Y, Michel M L, Carloni G, Revel M, Tiollais P, Weissenbach J. The gene S promoter of hepatitis B virus confers constitutive gene expression. Nucleic Acids Res. 1983;11:4645–4654. doi: 10.1093/nar/11.13.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mendel D B, Crabtree G R. HNF-1, a member of a novel class of dimerizing homeodomain proteins. J Biol Chem. 1991;266:677–680. [PubMed] [Google Scholar]
- 38.Raney A K, Johnson J L, Palmer C N A, McLachlan A. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J Virol. 1997;71:1058–1071. doi: 10.1128/jvi.71.2.1058-1071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ryden T A, Beemon K. Avian retroviral long terminal repeats bind CCAAT/enhancer-binding protein. Mol Cell Biol. 1989;9:1155–1164. doi: 10.1128/mcb.9.3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaul Y, Rutter W J, Laub O. A human hepatitis B viral enhancer element. EMBO J. 1985;4:427–430. doi: 10.1002/j.1460-2075.1985.tb03646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siddiqui A, Jameel S, Mapoles J. Transcriptional control elements of hepatitis B surface antigen gene. Proc Natl Acad Sci USA. 1986;83:566–570. doi: 10.1073/pnas.83.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siddiqui A, Jameel S, Mapoles J. Expression of the hepatitis B virus X gene in mammalian cells. Proc Natl Acad Sci USA. 1987;84:2513–2517. doi: 10.1073/pnas.84.8.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Standring D N, Rutter W J, Varmus H E, Ganem D. Transcription of the hepatitis B surface antigen gene in cultured murine cells initiates within the presurface region. J Virol. 1984;50:563–571. doi: 10.1128/jvi.50.2.563-571.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su H, Yee J-K. Regulation of hepatitis B virus gene expression by its two enhancers. Proc Natl Acad Sci USA. 1992;89:2708–2712. doi: 10.1073/pnas.89.7.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Summers J, Mason W S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 46.Sureau C, Romet-Lemonne J L, Mullins J I, Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell. 1986;47:37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- 47.Tiollais P, Pourcel C, Dejean A. The hepatitis B virus. Nature. 1985;317:489–495. doi: 10.1038/317489a0. [DOI] [PubMed] [Google Scholar]
- 48.Tognoni A, Cattaneo R, Serfling E, Schaffner W. A novel expression selection approach allows precise mapping of the hepatitis B virus enhancer. Nucleic Acids Res. 1985;13:7457–7472. doi: 10.1093/nar/13.20.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Treinin M, Laub O. Identification of a promoter element located upstream from the hepatitis B virus X gene. Mol Cell Biol. 1987;7:545–548. doi: 10.1128/mcb.7.1.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsurimoto T, Fujiyama A, Matsubara K. Stable expression and replication of hepatitis B virus genome in an integrated state in a human hepatoma cell line transfected with the cloned viral DNA. Proc Natl Acad Sci USA. 1987;84:444–448. doi: 10.1073/pnas.84.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vannice J L, Levinson A D. Properties of the human hepatitis B virus enhancer: position effects and cell-type nonspecificity. J Virol. 1988;62:1305–1313. doi: 10.1128/jvi.62.4.1305-1313.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang W-X, Li M, Wu X, Wang Y, Li Z-P. HNF1 is critical for the liver-specific function of HBV enhancer II. Res Virol. 1998;149:99–108. doi: 10.1016/s0923-2516(98)80085-x. [DOI] [PubMed] [Google Scholar]
- 53.Wang Y, Chen P, Wu X, Sun A-L, Wang H, Zhu Y-A, Li Z-P. A new enhancer element, ENII, identified in the X gene of hepatitis B virus. J Virol. 1990;64:3977–3981. doi: 10.1128/jvi.64.8.3977-3981.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Will H, Reiser W, Weimer T, Pfaff E, Büscher M, Sprengel R, Cattaneo R, Schaller H. Replication strategy of human hepatitis B virus. J Virol. 1987;61:904–911. doi: 10.1128/jvi.61.3.904-911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yaginuma K, Koike K. Identification of a promoter region for 3.6-kilobase mRNA of hepatitis B virus and specific cellular binding protein. J Virol. 1989;63:2914–2920. doi: 10.1128/jvi.63.7.2914-2920.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yee J-K. A liver-specific enhancer in the core promoter region of human hepatitis B virus. Science. 1989;246:658–661. doi: 10.1126/science.2554495. [DOI] [PubMed] [Google Scholar]
- 58.Yu X, Mertz J E. Promoters for synthesis of the pre-C and pregenomic mRNAs of human hepatitis B virus are genetically distinct and differentially regulated. J Virol. 1996;70:8719–8726. doi: 10.1128/jvi.70.12.8719-8726.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu X, Mertz J E. Differential regulation of the pre-C and pregenomic promoters of human hepatitis B virus by members of the nuclear receptor superfamily. J Virol. 1997;71:9366–9374. doi: 10.1128/jvi.71.12.9366-9374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuh C-H, Ting L-P. The genome of hepatitis B virus contains a second enhancer: cooperation of two elements within this enhancer is required for its function. J Virol. 1990;64:4281–4287. doi: 10.1128/jvi.64.9.4281-4287.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuh C-H, Ting L-P. C/EBP-like proteins binding to the functional box-α and box-β of the second enhancer of hepatitis B virus. Mol Cell Biol. 1991;11:5044–5052. doi: 10.1128/mcb.11.10.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuh C-H, Chang Y-L, Ting L-P. Transcriptional regulation of precore and pregenomic RNAs of hepatitis B virus. J Virol. 1992;66:4073–4084. doi: 10.1128/jvi.66.7.4073-4084.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yuh C-H, Ting L-P. Differentiated liver cell specificity of the second enhancer of hepatitis B virus. J Virol. 1993;67:142–149. doi: 10.1128/jvi.67.1.142-149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang P, Raney A K, McLachlan A. Characterization of functional Sp1 transcription factor binding sites in the hepatitis B virus nucleocapsid promoter. J Virol. 1993;67:1472–1481. doi: 10.1128/jvi.67.3.1472-1481.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang W, Zhang J, Kornuc M, Kwan K, Frank R, Nimer S D. Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Mol Cell Biol. 1995;15:6055–6063. doi: 10.1128/mcb.15.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]