Abstract
The existence of novel endogenous retrovirus elements in the chicken genome, designated EAV-HP, with close sequence identity to the env gene of avian leukosis virus (ALV) subgroup J has been reported (L. M. Smith, A. A. Toye, K. Howes, N. Bumstead, L. N. Payne, and K. Venugopal, J. Gen. Virol. 80:261–268, 1999). To resolve the genome structure of these retroviral elements, we have determined the complete sequence of two proviral clones of EAV-HP from a line N chicken genomic DNA yeast artificial chromosome library and from a meat-type chicken line 21 lambda library. The EAV-HP sequences from the two lines were 98% identical and had a typical provirus structure. The two EAV-HP clones showed identical large deletions spanning part of the gag, the entire pol, and part of the env genes. The env region of the EAV-HP clones was 97% identical to the env sequence of HPRS-103, the prototype subgroup J ALV. The 5′ region of EAV-HP comprising the R and U5 regions of the long terminal repeat (LTR), the untranslated leader, and the 5′ end of the putative gag region were 97% identical to the avian retrotransposon sequence, ART-CH. The remaining gag sequence shared less than 60% identity with other ALV sequences. The U3 region of the LTR was distinct from those of other retroviruses but contained some of the conserved motifs required for functioning as a promoter. To examine the ability of this endogenous retroviral LTR to function as a transcriptional promoter, the EAV-HP and HPRS-103 LTR U3 regions were compared in a luciferase reporter gene assay. The low luciferase activity detected with the EAV-HP LTR U3 constructs, at levels close to those observed for a control vector lacking the promoter or enhancer elements, suggested that these elements function as a weak promoter, possibly accounting for their low expression levels in chicken embryos.
Endogenous retrovirus (ERV) sequences inherited as Mendelian genes have been recognized in most of the vertebrate genomes. In the chicken genome, four different families of ERVs have been identified. The CR1 (chicken repeat 1) element, a short interspersed repetitive DNA element belonging to the non-long terminal repeat (LTR) class of retrotransposons, forms one of these families (15). The majority of these elements, existing as approximately 7,000 to 20,000 repeats per haploid genome, have a common 3′ end but show variable 5′ truncations, with a few elements containing open reading frames encoding reverse transcriptase (13). CR1 elements have been identified in several avian and reptilian species, demonstrating that they are ancient sequences that arose before the divergence of birds and reptiles (40).
The second family of chicken retrovirus-like elements, with the best-characterized ev loci, are the avian leukosis virus (ALV) subgroup E (ALV-E) ERVs. There are over 22 ev loci documented in layer-type birds and probably more in meat-type breeds (17), with an average of 5 loci in each bird (33). Although the majority of the ev loci are defective, some of them encode infectious ERVs closely related to the ALVs (reviewed in reference 18). The ev2 locus, for example, encodes Rous-associated virus-0, the prototype of ALV-E (27). The ev loci are considered to represent recent germ line integrations because of their (i) low copy numbers, (ii) segregation in the population, and (iii) distribution restricted in Gallus species to the chicken and the ancestral red jungle fowl (RJF).
ART-CH (avian retrotransposon from the chicken genome), another family of ERVs, is present as approximately 50 genomic copies (22). These elements were identified by PCR amplification of LTRs using primers to conserved ALV sequences. They are composed of functional LTRs and short regions with homology to the avian leukosis and sarcoma virus (ALSV) gag, pol, and env sequences (28). Although ART-CH does not encode functional proteins, the transcribed RNA can be packaged by a helper replication-competent retrovirus to spread in the chicken genome (28). As the data on the distribution of ART-CH among various species of birds within the Gallus genus are not available, the phylogenetic relationship of these elements in relation to other endogenous viruses is not known.
EAV-0, the fourth family of ERVs, is present between 40 and 100 copies per haploid genome. They were first detected in the genome of line 0 chickens by low-stringency hybridization with sequences cloned from an avian sarcoma virus (19, 20). EAV-0 elements had the typical proviral structure of 5′-LTR-gag-pol-env-LTR-3′, although they showed deletions in the env region (12, 19). EAV-0 elements are found in other species of the genus Gallus, indicating that these sequences are more ancient than ev loci and may represent proviral sequences derived from germ line retroviral infections prior to and around the time of Gallus speciation (32).
Several elements related to the EAV-0 elements have been subsequently described. These include the E13, E33, and E51 elements, isolated by low-stringency hybridization of a chicken library with EAV-0 sequences (11). Among these three elements, only E51 appeared to have a complete env region, although this sequence was interrupted by several small deletions and does not encode functional envelope glycoproteins. The E13 element had an unusually long U5 region in its LTR, which was distinct from those of the other EAV elements (11). The variations in the structure between the different elements suggested that the EAV-0 family is a heterogeneous group containing highly diverged retrovirus elements.
Previously, we reported that the chicken genome contained novel sequences closely related to the env gene of HPRS-103, the prototype of ALV subgroup J (ALV-J) (6). As these sequences had a close identity to E51, we considered these elements to be related to EAV-0 and have designated them EAV-HP. Subsequent sequence and phylogenetic analyses of the env region of these elements (39) have also supported their classification as elements related to EAV-0. Recently, the existence of a new family of endogenous viruses, called ev/J, with close homology to ALV-J was reported (8, 36). As the sequences of ev/J were virtually identical to that of EAV-HP (36), we believe that EAV-HP and ev/J are the same endogenous elements.
EAV-HP elements have been suggested to play an important role in the emergence of ALV-J and in the induction of ALV-J-associated disease (41). As the env sequences of these elements were very closely related (over 97% sequence identity) to that of ALV-J (8, 39), it is believed that these elements might have contributed to the origin of ALV-J by recombination. In addition, it was suggested that expression of the EAV-HP env-like sequences in chicken embryonic tissues could result in the induction of tolerance against ALV-J infection (39). As a first step to examine the roles of EAV-HP elements in the emergence of ALV-J and in the development of the disease, we set out to characterize the genomic structure of these novel ERV elements. We wanted to determine how the regions of EAV-HP other than env compared with those of ALV-J, and how the EAV-HP sequences were related to other ERVs, to obtain evidence that justifies their phylogenetic grouping with EAV-0. We also aimed to characterize the transcriptional regulatory elements in the EAV-HP clones so as to examine the nature and functions of their transcriptional elements. We show that the EAV-HP clones analyzed from two different lines of chickens possessed a typical proviral structure with identical deletions of the pol region. We also present data suggesting that part of the 5′ end of EAV-HP clone was identical to that of another endogenous element, ART-CH, and that the unique EAV-HP U3 region contained several transcriptional regulatory elements but had weak promoter activity.
MATERIALS AND METHODS
DNA isolation and genomic library preparation.
Genomic DNA was isolated as previously described (39). Briefly, 10-day-old line 21 chicken embryos (29) were homogenized in DNA extraction buffer (4 M guanidine isothiocyanate, 25 mM sodium citrate, 1% Sarkosyl) in volumes of 5 ml per embryo. The homogenate was phenol-chloroform extracted and precipitated by standard molecular biology techniques (37). DNA was also extracted from cultured chicken embryo fibroblasts (CEFs) prepared from line 0 (4) and brown leghorn chicken embryos and from Sonnerat's (grey) jungle fowl (SJF; Gallus sonneratii) embryos (30). The DNA from RJF (G. gallus) was kindly provided by Jan Salomonson (The Royal Veterinary and Agricultural University, Institute of Veterinary Microbiology, Copenhagen, Denmark).
DNA isolated from a yeast artificial chromosome (YAC) clone (YAC62) previously screened with an HPRS-103 env region probe (39) or from line 21 chicken embryos was partially digested with Sau3A1 and size fractionated by gel electrophoresis. DNA fragments ranging from 9 to 20 kbp were electroeluted, phenol-chloroform extracted, and precipitated as described elsewhere (37). The Sau3A1 restriction sites of the size-fractionated DNA were partially filled in and cloned into the LambdaGEM-12 vector (Promega) XhoI half sites according to the manufacturer's instructions. Recombinant phage DNA was packaged into phage by using the Packagene extract (Promega). The chicken line 21 genomic DNA lambda library was amplified as described previously (5).
Screening lambda libraries.
Phage were plated with Escherichia coli KW251, transferred to nitrocellulose filters (Stratagene), and denatured according to manufacturer's instructions. DNA was fixed to filters by UV irradiation for 3 min and prehybridized with 1× hybridization solution with SSC (Sigma) containing 50% formamide for 1 h at 42°C. For preparation of the labeled probe, 25 ng of DNA was incubated with RTS-RadPrime (GIBCO-BRL) and 25 μCi of [α-32P]dCTP at 37°C for 10 min. The probe was purified by using Nick columns (Pharmacia), denatured, and hybridized to filters overnight at 42°C. Filters were washed twice with 2× SSC (30 mM sodium citrate [pH 7.0], 300 mM NaCl)–0.1% (wt/vol) sodium dodecyl sulfate (wt/vol) for 15 min at 42°C and once with 0.2× SSC–0.1% sodium dodecyl sulfate (wt/vol) for 15 min at 65°C before exposure to Kodak BioMax MR film overnight at −70°C with intensifying screens. Positive phage plaques were rescreened as described above. Positive clones were grown according to the plate lysate method (37), and recombinant lambda DNA was isolated by using the Wizard Lambda Preps DNA purification system (Promega) according to the manufacturer's instructions.
Subcloning of lambda DNA inserts.
A positive YAC62 phage lambda library clone was digested with EcoRI, and an approximately 9.5-kb fragment hybridizing to the env probe was gel purified by electroelution (37). The fragment was ligated into EcoRI-digested, dephosphorylated vector pGEM-3Z (Promega) and transformed into JM109 high-efficiency competent cells (Promega). An approximately 9-kb EcoRI fragment hybridizing to the env probe from a positive chicken line 21 lambda library clone was similarly subcloned. Plasmid DNA was isolated by using a Qiagen plasmid miniprep kit.
PCR amplification of env regions.
For preparation of probes for library screening, we used a pGEM-T vector (Promega) which contained an approximately 1.3-kb PCR product (39) derived from genomic DNA amplified with primers H3 (5′-AACAACACCGATTTAGCCAGC-3′) and 37-1 (5′-TCGGAACCTACAGCTGCTCC-3′), corresponding to nucleotides 5659 to 5679 and nucleotides 6983 to 7002, respectively, of the HPRS-103 sequence (6). DNA was amplified by using primers for the T7 and SP6 promoters (Promega) in the vector sequence. Reactions were carried out with 10 pmol of T7 primer and 10 pmol of SP6 primer in PCR buffer (20 mM Tris-HCl [pH 8.4], 50 mM KCl, 0.25 mM deoxynucleoside triphosphates, 2 mM MgCl2) with approximately 1 ng of template DNA. The thermocycling program consisted of 1 cycle of 94°C for 2 min, 25 cycles of 94°C for 30 s, 50°C for 15 s, and 72°C for 3 min, followed by 1 cycle of 72°C for 7 min. The PCR products were purified by agarose gel electrophoresis and eluted from the gel slice with a QIAquick gel extraction kit (Qiagen). Primers H3 and 37-1 were also used to amplify the env regions of EAV-HP elements from RJF and SJF DNA.
DNA sequencing.
DNA was sequenced by using a Thermo Sequenase dye terminator cycle sequencing premix kit (Amersham) and an Applied Biosystems 373 DNA automated sequencing system. The sequence of each clone was determined by using primers designed with the Primer program of the Wisconsin Package version 9.1 (Genetics Computer Group, Madison, Wis.). Database searches and sequence analyses were also carried out with the Wisconsin Package Version 9.1, and potential U3 transcription factor binding sites were determined by using the program Map with the transcription factor sites (tfsites2) data file.
Luciferase reporter gene assays.
The U3 region of the right LTR was amplified from the EAV-HP1 clone by using primers incorporating SstI restriction sites for subsequent cloning. Forward primers EAVFOR (5′-GACGGGAGCTCTCGGCATAGGGAGGGGGAGATGTTG-3′) and ALVFOR (5-GAAATGAGCTCTTGCATAGGGAGGGGGAAATGTAG-3′) included the polypurine tract, while reverse primers EAVREV (5′-TAAGTGAGCTCAAATGGCGTTTATTGCTATAGGCTACG-3′) and ALVREV (5′-GGTGGGAGCTCAAATGGCGTTTATTGTGTCGGGCTAGG-3′) spanned the U3-R border. PCR was carried out as described above, with the following thermocycling conditions: 1 cycle of 94°C for 2 min, 25 cycles of 94°C for 45 s, 60°C for 2 min, and 75°C for 1 min, followed by 1 cycle of 72°C for 10 min. PCR products were digested with SstI, gel purified, ligated into SstI-digested pGL3-Basic (Promega), and transformed into XL1-Blue competent cells (Stratagene). The pGL3-Basic vector lacks eukaryotic promoter and enhancer sequences. Control vector DNAs pGL3-Basic with no inserts, pGL3-Control containing simian virus 40 (SV40) promoter and enhancer sequences, and pGL3-Promoter with SV40 promoter sequences (Promega) were similarly transformed. Positive clones were selected by PCR using the pGL3-specific primers RV2 and GL3 (Promega) and sequenced to rule out PCR-induced errors. DNA was prepared for transfection by using a Qiagen miniprep kit. Plasmid DNA (1 μg) was incubated with 3 μl of FuGENE 6 transfection reagent (Boehringer Mannheim) diluted to 100 μl with serum-free medium and used to transfect secondary CEFs according to the manufacturer's instructions. Cell extracts were prepared after 24 h and assayed for luciferase activity by using the Promega luciferase assay system and an EG & G Berthold Microplate Luminometer LB96V.
Nucleotide sequence accession numbers.
The EMBL accession numbers for the sequences described here are AJ238120 (RJFEAV-HP1), AJ238121 (RJFEAV-HP2), AJ238122 (SJFEAV-HP1), and AJ238123 (SJFEAV-HP2) for EAV-HP env clones from RJF and SJF and AJ238124 (EAV-HP1) and AJ238125 (EAV-HP) for full-length EAV-HP clones from chicken line N and line 21, respectively.
RESULTS
EAV-HP proviruses show identical deletions of the pol region.
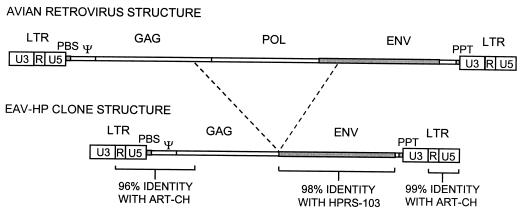
To characterize the genomic structure of EAV-HP elements, we have isolated several proviral clones from a line N chicken genomic YAC library and line 21 chicken genomic lambda library, using a probe derived from the env region of EAV-HP. One positive clone from each of these libraries was subcloned and sequenced completely. The provirus sequences were 4,202 bp from line N (clone EAV-HP1) and 4,216 bp from line 21 (clone EAV-HP2). The complete DNA sequence of EAV-HP1 is shown in Fig. 1 with the deduced amino acid sequence. The two EAV-HP sequences were 98% identical to each other, with a deleted pol region. The deletion junctions were identical between the two proviruses. However, as the flanking sequences of these two clones were different, they should represent integrations at distinct loci in the genome. A schematic diagram of the EAV-HP structure in comparison to a typical avian retrovirus such as ALV is shown in Fig. 2. The difference in length of the two EAV-HP proviral clones was due to the presence of four codons encoding an additional TGAQ repeat unit and an insertion of two residues in a poly(G) tract in the gag region of EAV-HP2. The EAV-HP1 clone showed a long ORF capable of encoding a continuous Gag-Env fusion protein (Fig. 1), while the EAV-HP2 clone encoded a truncated Gag protein due to a frameshift resulting from the 2-bp insertion.
FIG. 1.
Complete DNA sequence and deduced amino acid sequence of the EAV-HP1 provirus. Two identical LTRs flank the 4.2-kbp EAV-HP provirus. U3, R, and U5 region boundaries of the LTRs are indicated above the sequence. The 5-bp inverted repeat sequences of the LTR termini are shown in bold, and the 5-bp direct repeat sequences in flanking host DNA are underlined. The conserved tRNATrp primer-binding site (PBS) and polypurine tract (PPT) are also underlined. Asterisks above sequence (nucleotides 842 to 865) mark the boundaries of a repeat region for which one additional TGAQ repeat was found in the EAV-HP2 provirus sequence, and an arrow points to the poly(G) tract where two extra G residues were found in EAV-HP2.
FIG. 2.
Schematic diagram of the EAV-HP provirus structure compared to a typical ALV provirus. The overall structure of the full-length EAV-HP sequences is homologous to the avian retrovirus structure with a large deletion including the pol region. Dashed lines align the deletion junction of the EAV-HP provirus with the typical ALV provirus structure (not to scale). Abbreviations are as for Fig. 1.
Two additional line 21 EAV-HP clones (clones EAV-HP3 and EAV-HP4) have been isolated and characterized by partial sequencing of PCR products of different regions including a region spanning the gag-env junction (data not shown). These two clones also had the same deletion as the two completely sequenced clones. The two clones likely represent different loci since the sequenced regions were only 97% identical to EAV-HP1 and EAV-HP2. This conclusion was further supported by the distinct flanking DNA sequence of the EAV-HP3 clone, confirming that it was located in a different integration site. A 5-bp direct repeat was located in the flanking DNA in both the EAV-HP1 and EAV-HP2 integration sites. In the case of ALV, similar proviral integration creates a 6-bp direct repeat (23); ART-CH, on the other hand, is flanked by 3-bp direct repeats (28).
The EAV-HP LTR has a distinct U3 region.
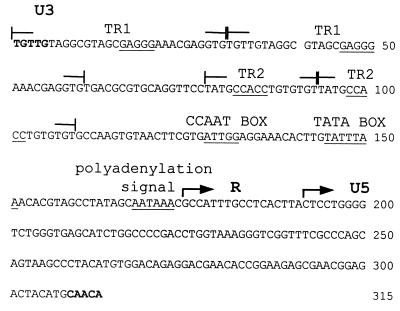
EAV-HP clones from both lines of chickens showed the typical provirus structure with LTR-like sequences at either ends. These sequences, comprised of 315 bp, were slightly shorter than the 325-bp HPRS-103 LTR (6) but longer than the 243-bp EAV-0 LTR (12). On the basis of the homology to other retrovirus LTR sequences, the EAV-HP LTR also could be divided into U3, R, and U5 regions (Fig. 3). The 174-bp U3 region in the EAV-HP LTR was shorter than that of HPRS-103 (224 bp) but longer than that of EAV-0 (144 bp). The R region in the two EAV-HP clones consisted of 17 bp (nucleotides 175 to 191), and the LTR U5 region contained 124 bp (nucleotides 192 to 315). The corresponding regions in the HPRS-103 LTR were 21 and 80 bp, respectively (6).
FIG. 3.
Sequence of the unique EAV-HP LTR U3 region. The U3 region has a TATA box and polyadenylation signal sequence, which are conserved in position and sequence compared to ALV LTRs. A CCAAT box motif is located upstream of the TATA box. A 30-bp tandem repeat is located at the 5′ end of the EAV-HP LTR (TR1), and a 16-bp tandem repeat is located in the center of the U3 region (TR2). The inverted repeat sequences at the termini of the LTR are indicated in bold. Putative elements for DNA binding proteins within the tandem repeat regions, a CCCTC motif and a pentanucleotide repeat element (GGTGG), are underlined.
While several elements found in a typical retrovirus LTR were conserved in the U3 region of the EAV-HP LTR, the sequence of the EAV-HP U3 region was distinct from those of other avian endogenous and exogenous retrovirus sequences. The conserved elements within the U3 region included a TATA box and polyadenylation signal, which were located in conserved positions at the 3′ end of the U3 region, and a CCAAT box motif 13 bp upstream of the TATA box. The U3 region also had several potential transcriptional regulatory elements that differed from those present in previously characterized retrovirus LTRs. The 5′ end of the U3 region, beginning with the first nucleotide of the provirus, was comprised of a 30-bp sequence repeated in tandem (TR1 [Fig. 3]). A putative transcription regulatory element (5′-GAGGG-3′) within this region was identical to the core sequence of elements recognized by the CCCTC-binding factor (CTCF) in the c-myc transcriptional regulatory element. CTCF has been shown to function in the regulation of tissue-specific expression of c-myc (24). A second tandem repeat of 16 bp (TR2, nucleotides 78 to 93 and 94 to 109 [Fig. 3]) containing a putative pentanucleotide response element (PRE) element (5′-GGTGG-3′) was located 36 bp upstream of the TATA box. Two PRE motifs which function as transcriptional enhancers during coinfection with Marek's disease virus are present in ALSV LTRs (7, 31), although the neighboring sequences were unrelated to those of the EAV-HP PRE motifs. A purine-rich sequence (5′-GAGGAA-3′) overlapping the putative CCAAT box (nucleotides 131 to 136) was identical to the core sequence of the avian PU.1 box, which is involved in the macrophage- and B-cell-specific expression of the chicken lysozyme gene (2).
The 5′ ends of EAV-HP and ART-CH genomes are identical.
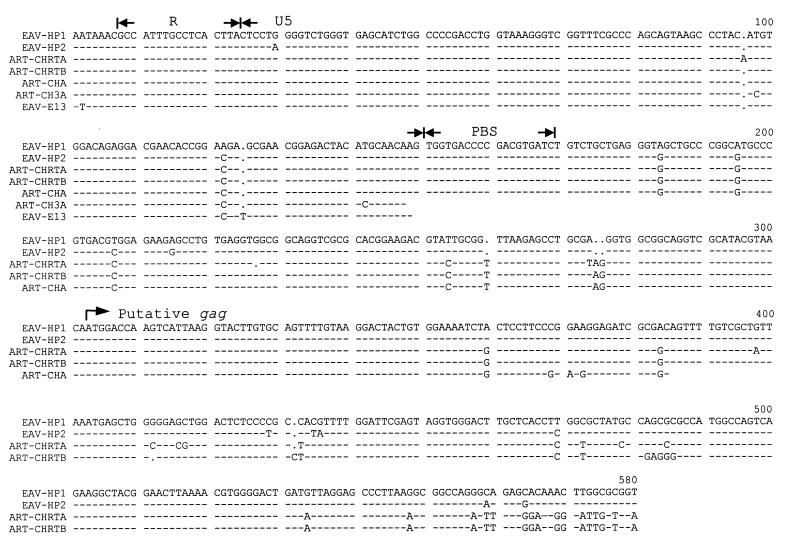
The R and U5 regions of EAV-HP were 99% identical to those of the ART-CH retrotransposon LTRs (clones A, RTA, and RTB) and 98% identical to those of ART-CH clone 3A and the EAV-E13 LTR (Fig. 4). The EAV-0 LTR U5, on the other hand, showed only 70% sequence similarity to that of EAV-HP. The 3′-R boundary of EAV-HP transcripts present in total RNA extracted from the chicken embryo, as determined by rapid amplification of 3′ cDNA ends, was found to be identical to that of ART-CH (22) (data not shown). The 5′-R boundary is also likely to be the same as for ART-CH since the sequences downstream of the polyadenylation signal were identical and the TATA boxes were at the same distance from the transcription initiation site. A 5-bp inverted repeat located at the LTR termini of EAV-HP clones was also conserved among the ART-CH and EAV family LTRs, suggesting the importance of these sequences for integration.
FIG. 4.
Nucleotide sequence alignment of EAV-HP and homologous regions from ART-CH and EAV-E13. Regions of high sequence identity include the polyadenylation signal (AATAAA), the R and U5 regions of the LTR, primer-binding site (PBS), untranslated leader, and 5′ end of the putative ART-CH gag gene. Dashes indicate sequence identity with EAV-HP1, and dots indicate nucleotides that are absent from the sequence in the alignment.
The high degree of sequence identity between EAV-HP and ART-CH at the 5′ ends of their genomes was observed to extend beyond the LTR when a pairwise comparison was performed (Fig. 4). The EAV-HP sequences were 96 to 97% identical to the ART-CH sequences over a length of 555 bp (nucleotides 218 to 768 [Fig. 1]), with sequence similarity in the polyadenylation signal, R region, U5, untranslated leader sequence, and the region encoding the putative ART-CH gag sequence (28). The untranslated leader also contained the tRNATrp primer-binding site, which is conserved among ALSVs. A packaging signal (Ψ) involved in the transfer of ART-CH sequences to mammalian cells by Rous sarcoma virus (RSV) (28) was also conserved in EAV-HP. The published sequence for EAV-0 including the untranslated leader and gag region had only 59% identity with the EAV-HP sequence. This region had low sequence identity with HPRS-103 in the leader but 60% identity in the gag gene sequence.
The EAV-HP Gag contains several highly conserved motifs.
The EAV-HP1 provirus contained a nearly full-length gag gene with a short region (nucleotides 515 to 768) showing close identity to the ART-CH putative gag region. The remainder of EAV-HP1 gag region (nucleotides 769 to 2490) had a maximum of 58% identity with gag sequences from myeloblastosis-associated virus and 57% identity with HPRS-103. The ART-CH is only 53% identical in this region of the EAV-HP clone.
The Gag proteins encoded by EAV-HP1 and HPRS-103 had short stretches with high degrees of sequence similarity, although the proteins were only 55% similar overall. Several elements of the gag sequence that are highly conserved among retroviruses were identified in the EAV-HP gag. The PPPPY motif of p2, which has been shown to be essential for retrovirus budding (44), is encoded by the EAV-HP sequences (nucleotides 989 to 1003). The capsid major homology region, the only well-conserved region of capsid protein sequences from diverse retroviruses (16), contained 14 of 20 amino acids identical to that of HPRS-103. Amino acids that were different from that of HPRS-103, however, were identical to the residues found in major homology regions of other retroviruses. Two nucleocapsid zinc finger motifs (CX2CX4HX4C) which are required for ALSV particle assembly (26) are encoded by nucleotides 1928 to 1969 and 1997 to 2038. The sequence encoding the three amino acids (DSG) of the protease active site (3) were also conserved within a stretch of 19 amino acids with 100% sequence similarity to the ALV sequence (nucleotides 2234 to 2290). By comparison with the ALV Gag sequences, the last eight amino acids of the protease encoded by gag of the EAV-HP clones were found to be missing due to the deletion between gag and env.
The EAV-HP env region is highly conserved in Gallus species.
The EAV-HP env sequences (nucleotides 2491 to 3889 of EAV-HP [Fig. 1]) determined from the two clones were 97% identical to the sequence in HPRS-103. Figure 5 shows a comparison of the amino acid sequence translated from the env sequences. The EAV-HP env sequences are truncated at the 5′ end, resulting in a deletion of 117 bp from the gp85 sequence, in addition to the 174-bp region encoding the N-terminal endoplasmic reticulum signal peptide. At the 3′ end, the identity between EAV-HP and HPRS-103 ended abruptly 25 bp downstream of the env stop codon, within the 3′ untranslated region (3′-UT). The 3′-UT is only 45 bp long in the two EAV-HP clones, compared to the approximately 200-bp sequences found in EAV-0, and is missing the highly conserved direct repeat 1 (DR1) region found in all ALSVs, EAV-0, and ART-CH. The E (or XSR) element sequence present in RSV and HPRS-103 (6) is also missing from the EAV-HP 3′-UT. The 11-bp polypurine tract (5′-AGGGAGGGGGA-3′), another conserved region in ALSVs, was identical to that of HPRS-103 and ART-CH.
FIG. 5.
Amino acid sequence comparison of EAV-HP and HPRS-103 env regions. The deduced amino acid sequences of the env region of the EAV-HP1 and EAV-HP2 full-length clones and homologous region in HPRS-103 env are shown aligned with the amino acid sequences deduced from two different cloned 1.3-kbp PCR products from RJF (RJFEAV-HP1 and RJFEAV-HP2) and SJF (SJFEAV-HP1 and SJFEAV-HP2). Amino acids encoded by PCR primers were not included. Dashes indicate sequence identity to EAV-HP1, and an asterisk marks the env stop codon.
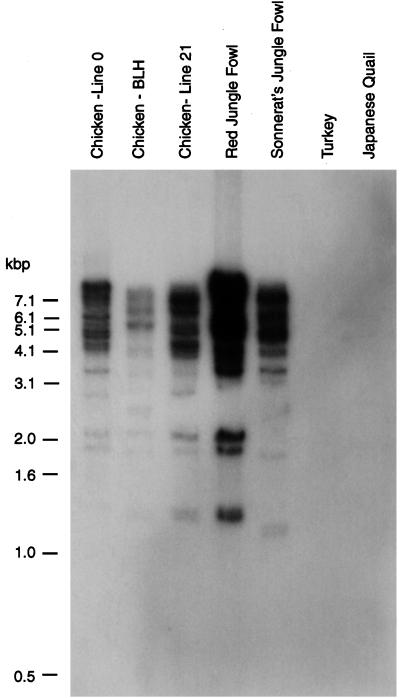
Apart from chickens, EAV-HP sequences were also detected in red and Sonnerat's jungle fowl DNA by Southern blot hybridization with env probes (39). These sequences were also detected by Southern blot analysis using a PCR product amplified with primers EAVFOR and EAVREV as a probe including the polypurine tract, U3 region, and first half of the R region of the EAV-HP LTR (Fig. 6). This confirmed that EAV-HP sequences were present in jungle fowl DNA but not in the phylogenetically more distant turkey and quail genomes, and that previously detected bands were not due to cross-hybridization with similar env sequences, such as E51-related elements. To examine the relationships of the EAV-HP env sequences in these birds, we determined the sequence of the 1.3-kb env region amplified by PCR using primers H3 and 37-1. The EAV-HP env sequences from the two jungle fowl species were 98 to 99% identical to each other and to those of the chicken and encoded a continuous ORF in the region analyzed (Fig. 5). Alignment of the deduced amino acid sequences of the EAV-HP env region from chicken lines and jungle fowl showed that the variations were mainly clustered between amino acids 78 and 111 and amino acids 147 and 171 (Fig. 5), corresponding to the regions of the gp85 which were most variable among ALV-J variant strains (42). These regions, however, differ from the variable and hypervariable regions defined for the more distantly related env sequences of ALV-A to ALV-E (10, 42).
FIG. 6.
Detection of EAV-HP LTR sequences in genomic DNA from several avian species. Genomic DNA from line 21, line 0, and brown leghorn (BLH) chickens, RJF SJF, turkeys, and Japanese quail was digested with EcoRI, separated by agarose gel electrophoresis, and blotted onto a Duralon-UV membrane (Stratagene) according to standard molecular biology techniques (37). DNA was probed by using the PCR product derived by amplification of EAV-HP1 cloned DNA with primers EAVFOR and EAVREV. The PCR product includes the polypurine tract, U3 region, and part of the R region of the 3′ LTR. Stringency washes were performed as described in the text. Sizes of the DNA ladder (GIBCO-BRL) are indicated on the left.
The EAV-HP U3 is a weak promoter of transcription.
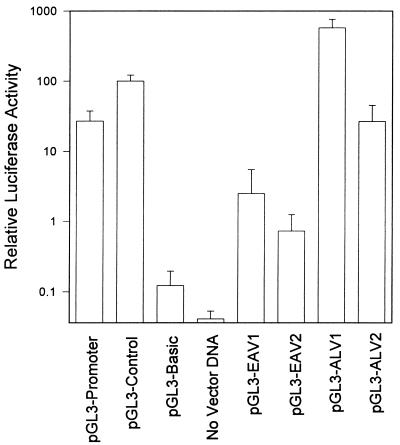
EAV-HP transcripts are expressed in chicken embryonic tissues or CEFs at low levels not detectable by Northern blotting (reference 8 and our unpublished data). Analysis of the LTR sequence revealed several motifs that may function as promoter elements, including a TATA box identical to that of ALVs. Since other endogenous retroviruses, including several ev loci, have been demonstrated to have little expression due to positional effects and silencing by methylation (21), the EAV-HP LTRs were examined further to determine whether the low levels of EAV-HP transcripts may be due to the defectiveness of the promoter or due to DNA positional effects. The EAV-HP and HPRS-103 LTR U3 regions were cloned into the pGL3-Basic vector, which lacks any promoter or enhancer elements, upstream of the luciferase gene. To examine the promoter functions of these constructs, clones in both orientations were selected and used in a transient transfection assay alongside pGL3-Basic and pGL3 vectors with the SV40 promoter (pGL3-Promoter) or the SV40 promoter and enhancer (pGL3-Control). Luciferase activity was determined relative to that of the pGL3-Control cell extracts (Fig. 7). The ALV and EAV-HP LTR U3 regions were seen to act as orientation-dependent promoters, as observed by the higher luciferase activity in cell extracts transfected with plasmid DNA containing the U3 region in the forward orientation. Interestingly, both ALV and EAV LTR U3 constructs in the reverse orientation showed levels of luciferase activity approximately 10% of the levels from the forward constructs, suggesting that this region might contain regulatory elements that could function in the reverse direction. Cell extracts from CEFs transfected with the EAV-HP U3 sequences in either orientation had comparatively very low levels of luciferase activity. Extracts from cells transfected with pGL3-EAV1, with the EAV-HP U3 in the forward orientation, had luciferase activity levels approximately 10-fold lower than those in extracts of CEFs transfected with pGL3-Promoter and approximately 230-fold lower than those in extracts of CEFs transfected with pGL3-ALV1, with the HPRS-103 U3 in the same orientation. These data suggested that the U3 region of the EAV-HP LTR functioned only as a very weak promoter supporting the low transcriptional activity of these elements in CEFs.
FIG. 7.
Assay of promoter activity of the EAV-HP LTR U3 region. Secondary CEFs were transiently transfected with the pGL3 vectors indicated or with pGL3-Basic constructs with the EAV-HP U3 in forward (pGL3-EAV1) or reverse (pGL3-EAV2) orientation. The same region of HPRS-103 cloned into pGL3-Basic in forward and reverse orientations (pGL3-ALV1 and pGL3-ALV2, respectively) was included for comparison of the endogenous provirus LTR strength with the promoter activity of an exogenous ALV LTR. Luciferase activity for each plasmid indicated represents the mean ± standard deviation of six independent transfections with two different plasmid preparations relative to pGL3-Control.
DISCUSSION
In this paper, we report the molecular characterization and sequence of the novel endogenous avian retrovirus element EAV-HP. These endogenous elements are important, as they were shown to have intact env-like sequences with very high sequence identity to the ALV-J env (6, 39), and are potentially transcribed in chicken embryonic tissues (8). Because of this close sequence homology, it is thought that these elements have contributed to the origin of the new J subgroup ALV by recombination (41). The 4.2-kbp EAV-HP clones from two chicken lines had a typical proviral DNA structure with LTR sequences and regions showing similarity to retroviral genes (Fig. 2).
Similar to many of the other endogenous viruses, EAV-HP proviral clones from both lines were defective for replication, mainly due to the deletion of an identical region of the pol region in the EAV-HP clones. The EAV-HP sequences of the two full-length clones, as well as the two partially sequenced clones, were very closely related to each other, with a divergence of only less than 3% among them. The identical sequences of the 5′ and 3′ LTRs of the EAV-HP1 clone also indicated that they are not very ancient endogenous elements. Demonstration of EAV-HP sequences in the genomes of chickens and ancestral jungle fowls, but not in closely related avian species such as turkeys and quails, suggested that they were acquired around the time of separation of Gallus species.
The EAV-HP clones have an env region related to EAV-E51, while part of the LTR and the gag region are identical to those of ART-CH and EAV-E13. EAV-E51 and related clones were previously designated as a subfamily of the EAV-0 family (11). The relationship among the EAV-0 elements and the related E51, E13, and E33 clones, as well as EAV-HP clones reported here, suggests that these elements would best be regarded as members of one family of ERVs, designated EAV. There were two recent reports suggesting the existence of new endogenous elements in the chicken genome with close sequence identity to the ALV-J env (8, 36). Although the env sequences of these elements initially published are identical to that of EAV-HP, the authors have designated these elements as a new family of endogenous viruses called ev/J. To avoid the confusion of assigning different names for the same elements, there is a need to systematically look at the characteristics of the different endogenous elements to devise a universal system of nomenclature for these elements.
The ART-CH sequence was reported to be a chimera of sequences derived from multiple recombinations between exogenous ALSV sequences (28). Our data showing that part of the EAV-HP was identical to that of ART-CH and the close sequence identity of ART-CH with EAV elements such as E13 and E51 support the view that ART-CH has a chimeric genome. A comparison of the limited sequence available for EAV-E13 and its env gene deletion (11) with the full-length ART-CH sequence (28) suggests that these clones may represent nearly identical proviruses integrated in different loci. Characterization of full-length provirus sequences of the E51-related elements will be important to determine whether the ART-CH elements are a separate family of endogenous retroelements or another member of the diverse EAV family of ERVs in the chicken genome. Examination of the ART-CH sequences present in other Gallus species may help to define their origin and relationship to the EAV family of ERVs. We note that the number of ART-CH sequences in the chicken genome originally reported was based on the results of Southern hybridization with a 570-bp 3′-LTR probe which included 119 bp of the region shared with EAV-HP. Therefore, it is likely that there are fewer ART-CH copies in the genome than the 25 to 50 copies originally estimated from this experiment (22).
The LTRs from both full-length EAV-HP clones had a unique U3 region, distinct from those of previously isolated endogenous and exogenous retrovirus sequences. However, the R and U5 regions of EAV-HP were virtually identical to those of the ART-CH and EAV-E13 LTRs but differed greatly from those of other members of the EAV family (Fig. 8). The U3 regions of ART-CH and EAV-E13 are closely related (more than 92% identical) to EAV-E51. This distinct combination of EAV LTRs may have resulted from recombination between different avian retroviruses, resulting in LTR domain shuffling (11). The existence of EAV-0 and EAV-HP sequences in all Gallus species together with recombinant EAV LTR suggest these retroviruses were active at a similar time and may have coinfected an ancestral species to give rise to recombinant retrovirus genomes, such as the EAV-E13.
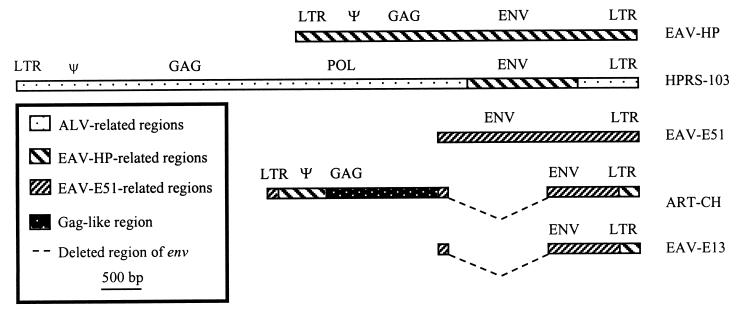
FIG. 8.
Schematic diagram of the avian retrovirus elements showing related regions. Regions of retrovirus sequences which are related to EAV-HP include the env gene of ALV-J, the U5, R, and 5′ end of ART-CH, and the U5 and R of EAV-E13. The deletion breakpoint indicated for the env of EAV-E13 is shown as described by Boyce-Jacino et al. (11). The env deletion in ART-CH was determined by comparison with the E51 sequence. The gag-like region of ART-CH had little similarity with any of the sequenced retrovirus elements.
Although we have not determined the structure of all EAV-HP loci in the genome, the structures of the two fully sequenced and two partially sequenced clones show that all of these EAV-HP proviral clones share the same large pol deletion. The genome structure of ev/J proviruses (36) also showed the internal deletion of this region. This suggests that this deletion may represent the most common form of defectiveness in the EAV-HP genome. Since the EAV-HP clones reported here are defective, the spread and multiple germ line integrations are likely to have occurred with infectious virus acting in trans as a helper virus. Such a mechanism has been suggested for the spread of other defective endogenous retroelements, including VL30 sequence in mice and the ART-CH retrotransposons in chickens (1, 28). A study of the ability of the EAV-HP genomic RNA to be packaged into virus particles may provide support for this hypothesis.
The ALV-J subgroup is thought to have arisen by a nonhomologous recombination event between EAV-HP and an exogenous ALV (6). Such a recombination would also require the expression and packaging of EAV-HP RNA. Expression of EAV-HP env-specific transcripts has been demonstrated by reverse transcriptase (RT)-mediated PCR (RT-PCR) on total RNA extracted from chicken embryos (reference 8 and our unpublished results). As the EAV-HP clones described here showed a packaging signal (Ψ) similar to those of ART-CH, it is possible that these transcripts can be copackaged with ALSV RNA into virions, facilitating recombination with heterologous RNAs. While the EAV-HP sequences include the env gene unique to the J subgroup of exogenous ALVs, the clones analyzed thus far have a deletion at the 5′ end of the env gene resulting in the loss of 97 amino acids of the amino-terminal portion of the envelope precursor protein. These clones also have a short, possibly deleted 3′-UT. A comparison of the nucleotide sequence of the 3′-UT sequence adjacent to the env region showed no stretch of high sequence identity with available ALSV sequences other than HPRS-103. Isolation of an EAV-HP provirus with an intact env region may be useful to delineate sequences at both the 5′ and 3′ ends of env that may have been involved in the recombination event(s) from which ALV-J may have been derived.
The env sequence of the EAV-HP clones from chickens and those determined by PCR from the two jungle fowl DNAs shared 98 to 99% sequence identity, demonstrating a high degree of sequence conservation among these closely related avian species. The retrovirus envelope, including that of ALV-J, is comprised of two functional regions, designated the surface (SU) and transmembrane (TM) domains. Alignment of the EAV-HP envelope sequences from the chicken and jungle fowl shows a clustering of sequence changes within regions of the SU domain (Fig. 5). Antigenic variants of ALV-J showing sequence changes within these regions have been described (42). It is thought that these variants arose by mutations induced by polymerase errors and selection from immune pressure. However, the possibility does exist that some of these variant viruses were generated by recombination with expressed env regions of EAV-HP, by mechanisms similar to those described for feline leukemia viruses (34). The presence of identical amino acid residues in the variable regions of the EAV-HP sequences and in some of the antigenic variants (42) supports this notion. However, extensive sequence analysis and molecular studies are required to prove this hypothesis.
Only low levels of EAV-HP transcripts have been detected by RT-PCR (8). The U3 region of the EAV-HP LTR is much smaller than the exogenous ALSV U3 and does not have the enhancer region defined for the exogenous retrovirus LTR (35). The absence of a DR1 element in the EAV-HP also may contribute to the low expression level, as these elements have been shown to be important in the accumulation of unspliced RNA of RSV (38). Together with the weak promoter activity of the EAV-HP U3 observed in luciferase assays, these data may indicate that the EAV-HP LTR is defective and incapable of promoting higher levels of transcription. On the other hand, the EAV-HP U3 sequence may contain elements that could function in cell type-specific transcriptional modulation. For example, the EAV-HP U3 contains two putative DNA elements for binding by CTCF (24), which has been shown to function as a negative regulator of transcription in chicken lysozyme gene expression in nonmyeloid tissues (14). The CTCF DNA elements in the EAV-HP U3 may also play a role in the downregulation of transcription in CEFs, as part of a myeloid cell-specific regulatory unit. A detailed analysis of the EAV-HP LTR is required to determine if these motifs do function as negative regulatory elements in CEFs and promote increased levels of transcription in a tissue-specific manner before concluding that the EAV-HP LTR is a defective promoter.
Compared to the layer-type brown leghorn birds, meat-type chickens are immunologically more tolerant to ALV-J, which may result from expression of endogenous envelope proteins in the embryonic tissues (39). It is not known whether this is due to different EAV-HP loci being present in different breeds or due to differences in expression of the same EAV-HP loci. Retrovirus envelope glycoproteins are translated from spliced subgenomic mRNAs (25). RT-PCR methods used to demonstrate the env-specific cDNA of EAV-HP do not distinguish between the genomic and subgenomic transcripts. The EAV-HP clones described here have deletions at the 5′ end of the env gene and hence are unlikely to be able to generate spliced subgenomic env transcripts. It will be important in future to determine whether there is an intact, functional env gene that is present and expressed only in meat-type birds, or whether differences in expression from the retroviral U3 exist between meat-type and layer-type chickens.
Retrovirus-like particles containing EAV-0 RNA and RT activity have been identified recently in CEF supernatants and live attenuated vaccines produced in chicken cells (9, 43). These particles remain uncharacterized because they are present at only very low levels. The characterization of an EAV-HP sequence with an intact gag gene containing conserved elements identified as important for retrovirus particle assembly and formation may provide a starting point for further dissection of these RT-associated particles.
ACKNOWLEDGMENTS
This work was partly funded by the Ministry of Agriculture, Fisheries and Food, United Kingdom and the National Institute for Biological Standards and Control.
We thank Jim Kaufman and Jim Payne for critical reading of the manuscript. We are grateful to Jim Robertson (National Institute for Biological Standards and Control) and Peter Russell (Royal Veterinary College, University of London) for helpful discussion and support.
REFERENCES
- 1.Adams S E, Rathjen P D, Stanway C A, Fulton S M, Malim M H, Wilson W, Ogden J, King L, Kingsman S M, Kingsman A J. Complete nucleotide sequence of a mouse VL30 retro-element. Mol Cell Biol. 1988;8:2989–2998. doi: 10.1128/mcb.8.8.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahne B, Stratling W H. Characterization of a myeloid-specific enhancer of the chicken lysozyme gene. Major role for an Ets transcription factor-binding site. J Biol Chem. 1994;269:17794–17801. [PubMed] [Google Scholar]
- 3.Arad G, Chorev M, Shtorch A, Goldblum A, Kotler M. Point mutation in avian sarcoma leukaemia virus protease which increases its activity but impairs infectious virus production. J Gen Virol. 1995;76:1917–1925. doi: 10.1099/0022-1317-76-8-1917. [DOI] [PubMed] [Google Scholar]
- 4.Astrin S M, Buss E G, Hayward W S. Endogenous viral genes are non-essential in chicken. Nature. 1979;282:339–341. doi: 10.1038/282339a0. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 6.Bai J, Payne L N, Skinner M A. HPRS-103 (exogenous avian leukosis virus, subgroup J) has an env gene related to those of endogenous elements EAV-0 and E51 and an E element found previously only in sarcoma viruses. J Virol. 1995;69:779–784. doi: 10.1128/jvi.69.2.779-784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banders U T, Coussens P M. Interactions between Marek's disease virus encoded or induced factors and the Rous sarcoma virus long terminal repeat promoter. Virology. 1994;199:1–10. doi: 10.1006/viro.1994.1092. [DOI] [PubMed] [Google Scholar]
- 8.Benson S J, Ruis B L, Fadly A M, Conklin K F. The unique envelope gene of the subgroup J avian leukosis virus derives from ev/J proviruses, a novel family of avian endogenous viruses. J Virol. 1998;72:10157–10164. doi: 10.1128/jvi.72.12.10157-10164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boni J, Stalder J, Reigel F, Schupbach J. Detection of reverse transcriptase activity in live attenuated virus vaccines. Clin Diagn Virol. 1996;5:43–53. doi: 10.1016/0928-0197(95)00159-x. [DOI] [PubMed] [Google Scholar]
- 10.Bova C A, Manfredi J P, Swanstrom R. env genes of avian retroviruses: nucleotide sequence and molecular recombinants define host range determinants. Virology. 1986;152:343–354. doi: 10.1016/0042-6822(86)90137-6. [DOI] [PubMed] [Google Scholar]
- 11.Boyce-Jacino M T, O'Donoghue K, Faras A J. Multiple complex families of endogenous retroviruses are highly conserved in the genus Gallus. J Virol. 1992;66:4919–4929. doi: 10.1128/jvi.66.8.4919-4929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyce-Jacino M T, Resnick R, Faras A J. Structural and functional characterization of the unusually short long terminal repeats and their adjacent regions of a novel endogenous avian retrovirus. Virology. 1989;173:157–166. doi: 10.1016/0042-6822(89)90231-6. [DOI] [PubMed] [Google Scholar]
- 13.Burch J B, Davis D L, Haas N B. Chicken repeat 1 elements contain a pol-like open reading frame and belong to the non-long terminal repeat class of retrotransposons. Proc Natl Acad Sci USA. 1993;90:8199–8203. doi: 10.1073/pnas.90.17.8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, Lottspeich F, Filippova G N, Lobanenkov V V, Renkawitz R. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol Cell Biol. 1997;17:1281–1288. doi: 10.1128/mcb.17.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Z-Q, Ritzel R G, Lin C C, Hodgetts R B. Sequence conservation in avian CR1: an interspersed repetitive DNA family evolving under functional constraints. Proc Natl Acad Sci USA. 1991;88:5814–5818. doi: 10.1073/pnas.88.13.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Craven R C, Leure-duPree A E, Weldon R A, Jr, Wills J W. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crittenden L B, Salter D A. Spontaneous mobility of subgroup A avian leukosis virus proviruses in the genome of the chicken. Avian Pathol. 1998;27:S26–S35. [Google Scholar]
- 18.Crittenden L B. Retroviral elements in the genome of the chicken: implications for poultry genetics and breeding. Crit Rev Poult Biol. 1991;3:73–91. [Google Scholar]
- 19.Dunwiddie C T, Resnick R, Boyce-Jacino M, Alegre J N, Faras A J. Molecular cloning and characterization of gag-, pol-, and env-related gene sequences in the ev− chicken. J Virol. 1986;59:669–675. doi: 10.1128/jvi.59.3.669-675.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunwiddie C, Faras A J. Presence of retrovirus reverse transcriptase-related gene sequences in avian cells lacking endogenous avian leukosis viruses. Proc Natl Acad Sci USA. 1985;82:5097–5101. doi: 10.1073/pnas.82.15.5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groudine M, Eisenman R, Weintraub H. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature. 1981;292:311–317. doi: 10.1038/292311a0. [DOI] [PubMed] [Google Scholar]
- 22.Gudkov A V, Komarova E A, Nikiforov M A, Zaitsevskaya T E. ART-CH, a new chicken retroviruslike element. J Virol. 1992;66:1726–1736. doi: 10.1128/jvi.66.3.1726-1736.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hishinuma F, DeBona P J, Astrin S, Skalka A M. Nucleotide sequence of acceptor site and termini of integrated avian endogenous provirus ev1: integration creates a 6 bp repeat of host DNA. Cell. 1981;23:155–164. doi: 10.1016/0092-8674(81)90280-4. [DOI] [PubMed] [Google Scholar]
- 24.Klenova E M, Nicolas R H, Paterson H F, Carne A F, Heath C M, Goodwin G H, Neiman P E, Lobanenkov V V. CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-Zn-finger protein differentially expressed in multiple forms. Mol Cell Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luciw P A, Leung N J. Mechanisms of retrovirus replication. In: Levy J A, editor. The Retroviridae. Vol. 1. New York, N.Y: Plenum Press; 1992. pp. 159–298. [Google Scholar]
- 26.Meric C, Gouilloud E, Spahr P F. Mutations in Rous sarcoma virus nucleocapsid protein p12 (NC): deletions of Cys-His boxes. J Virol. 1988;62:3328–3333. doi: 10.1128/jvi.62.9.3328-3333.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motta J V, Crittenden L B, Purchase H G, Stone H A, Witter R L. Low oncogenic potential of avian endogenous RNA tumor virus infection or expression. J Natl Cancer Inst. 1975;55:685–689. doi: 10.1093/jnci/55.3.685. [DOI] [PubMed] [Google Scholar]
- 28.Nikiforov M A, Gudkov A V. ART-CH: a VL30 in chickens? J Virol. 1994;68:846–853. doi: 10.1128/jvi.68.2.846-853.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Payne L N, Gillespie A M, Howes K. Myeloid leukaemogenicity and transmissibility of the HPRS-103 strain of avian leukosis virus. Leukemia. 1992;6:1167–1176. [PubMed] [Google Scholar]
- 30.Payne L N, Gillespie A M, Howes K, Smith L M. Host range of Rous sarcoma virus pseudotype RSV(HPRS-103) in 12 avian species: support for a new avian retrovirus envelope subgroup, designated J. J Gen Virol. 1992;73:2995–2997. doi: 10.1099/0022-1317-73-11-2995. [DOI] [PubMed] [Google Scholar]
- 31.Pulaski J T, Tieber V L, Coussens P M. Marek's disease virus-mediated enhancement of avian leukosis virus gene expression and virus production. Virology. 1992;186:113–121. doi: 10.1016/0042-6822(92)90065-w. [DOI] [PubMed] [Google Scholar]
- 32.Resnick R M, Boyce-Jacino M T, Fu Q, Faras A J. Phylogenetic distribution of the novel avian endogenous provirus family EAV-0. J Virol. 1990;64:4640–4653. doi: 10.1128/jvi.64.10.4640-4653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rovigatti V G, Astrin S M. Avian endogenous viral genes. Curr Top Microbiol Immunol. 1983;103:1–21. doi: 10.1007/978-3-642-68943-7_1. [DOI] [PubMed] [Google Scholar]
- 34.Roy-Burman P. Endogenous env elements: partners in generation of pathogenic feline leukemia viruses. Virus Genes. 1995;11:147–161. doi: 10.1007/BF01728655. [DOI] [PubMed] [Google Scholar]
- 35.Ruddell A. Transcription regulatory elements of the avian long terminal repeat. Virology. 1995;10:1–7. doi: 10.1016/s0042-6822(95)80013-1. [DOI] [PubMed] [Google Scholar]
- 36.Ruis B L, Benson S J, Conklin K F. Genome structure and expression of the ev/J family of avian endogenous viruses. J Virol. 1999;73:5345–5355. doi: 10.1128/jvi.73.7.5345-5355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Simpson S B, Zhang L, Craven R C, Stoltzfus C M. Rous sarcoma virus direct repeat cis elements exert effects at several points in the virus life cycle. J Virol. 1997;71:9150–9156. doi: 10.1128/jvi.71.12.9150-9156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith L M, Toye A A, Howes K, Bumstead N, Payne L N, Venugopal K. Novel endogenous retroviral sequences in the chicken genome closely related to HPRS-103 (subgroup J) avian leukosis virus. J Gen Virol. 1999;80:261–268. doi: 10.1099/0022-1317-80-1-261. [DOI] [PubMed] [Google Scholar]
- 40.Vandergon T L, Reitman M. Evolution of chicken repeat 1 (CR1) elements: evidence for ancient subfamilies and multiple progenitors. Mol Biol Evol. 1994;11:886–898. doi: 10.1093/oxfordjournals.molbev.a040171. [DOI] [PubMed] [Google Scholar]
- 41.Venugopal K. Avian leukosis virus subgroup J: a rapidly evolving group of oncogenic retroviruses. Res Vet Sci. 1999;67:113–119. doi: 10.1053/rvsc.1998.0283. [DOI] [PubMed] [Google Scholar]
- 42.Venugopal K, Smith L M, Howes K, Payne L N. Antigenic variants of J subgroup avian leukosis virus: sequence analysis reveals multiple changes in the env gene. J Gen Virol. 1998;79:757–766. doi: 10.1099/0022-1317-79-4-757. [DOI] [PubMed] [Google Scholar]
- 43.Weissmahr R N, Schupbach J, Boni J. Reverse transcriptase activity in chicken embryo fibroblast culture supernatants is associated with particles containing endogenous avian retrovirus EAV-0 RNA. J Virol. 1997;71:3005–3012. doi: 10.1128/jvi.71.4.3005-3012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang Y, Cameron C E, Wills J W, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]