Abstract
Introduction
Newborn bloodspot screening (NBS) is a highly successful public health programme that uses biochemical and other assays to screen for severe but treatable childhood-onset conditions. Introducing genomic sequencing into NBS programmes increases the range of detectable conditions but raises practical and ethical issues. Evidence from prospectively ascertained cohorts is required to guide policy and future implementation. This study aims to develop, implement and evaluate a genomic NBS (gNBS) pilot programme.
Methods and analysis
The BabyScreen+ study will pilot gNBS in three phases. In the preimplementation phase, study materials, including education resources, decision support and data collection tools, will be designed. Focus groups and key informant interviews will also be undertaken to inform delivery of the study and future gNBS programmes. During the implementation phase, we will prospectively recruit birth parents in Victoria, Australia, to screen 1000 newborns for over 600 severe, treatable, childhood-onset conditions. Clinically accredited whole genome sequencing will be performed following standard NBS using the same sample. High chance results will be returned by genetic healthcare professionals, with follow-on genetic and other confirmatory testing and referral to specialist services as required. The postimplementation phase will evaluate the feasibility of gNBS as the primary aim, and assess ethical, implementation, psychosocial and health economic factors to inform future service delivery.
Ethics and dissemination
This project received ethics approval from the Royal Children’s Hospital Melbourne Research Ethics Committee: HREC/91500/RCHM-2023, HREC/90929/RCHM-2022 and HREC/91392/RCHM-2022. Findings will be disseminated to policy-makers, and through peer-reviewed journals and conferences.
Keywords: GENETICS, PAEDIATRICS, Health policy
STRENGTHS AND LIMITATIONS OF THIS STUDY.
A prospective feasibility study to offer genomic newborn screening to birth parents during pregnancy.
The study includes a multidisciplinary evaluation approach ascertaining perspectives from the public, health professionals and participating parents.
The study will use online recruitment tools to facilitate education and consent to allow scaling up for a future screening programme. Interpreter-assisted recruitment is available for people who use a language other than English.
Recruitment will encompass birth parents in the private and public healthcare system, however, may not be representative of the wider population as this is a pilot study.
As this is a pilot study, we are unable to fully assess the value of stored genomic data for lifelong healthcare use.
Introduction
Newborn bloodspot screening (NBS) is a well-established and highly successful public health intervention that has enabled the early diagnosis and prompt treatment of many severe childhood conditions.1 Traditionally, NBS programmes select conditions for inclusion using the classic Wilson and Jungner screening principles,2 and employ targeted biochemical and other assays that can be delivered at scale with rapid turnaround times and at a relatively low cost. Wide variability exists between, and even within, countries in the number of conditions included, ranging from 9 in the UK to 80 in California, USA.1 3–5 Currently in Australia, NBS programmes are funded by state and territory governments and screen for around 27 conditions.6
In the last decade, the use of genomic sequencing in the research and clinical setting has transformed rare disease diagnosis.3 At the same time, increased understanding of the underlying molecular mechanisms of disease has accelerated the development of precision treatments.7 The gap between the number of rare diseases now considered ‘treatable’8 and those included in NBS programmes is expanding rapidly. Integrating genomic sequencing into NBS programmes would enable screening for a much broader range of conditions, while also providing the flexibility to quickly add more conditions at low incremental cost.
The concept of performing genomic sequencing in all children at birth, termed genomic newborn screening (gNBS), has been extensively debated. Empirical evidence to guide decision-making is starting to emerge,9–13 addressing important questions of test design, diagnostic performance and clinical utility, as well as parental and healthcare professionals’ (HCP) views and concerns.14–18 However, evidence from prospective cohorts is limited, with the most extensively evaluated cohort, the BabySeq study,19 having recruited 159 infants.
Many concerns have been raised, including reduced sensitivity of genomic testing for some of the metabolic conditions screened via standard NBS (stdNBS),11 and the appropriate timing and means of parental education to ensure informed consent.20 None of the studies to date have fully addressed the key questions of acceptability, feasibility, scalability and cost-effectiveness of gNBS. Most have focused on the clinical, legal, insurance and social contexts in the USA,1 10 11 13 19 20 which have limited applicability to the Australian and other public healthcare systems. Several large-scale newborn genomic screening studies are currently launching worldwide to address these gaps in knowledge.3 21 22
Our prospective cohort study, BabyScreen+, will address many of these questions in an Australian context to inform a future population-scale gNBS programme. We will perform gNBS for a cohort of 1000 Victorian newborns using whole genome sequencing (WGS) that will increase the range of conditions screened in stdNBS to over 600 treatable, childhood-onset conditions. Insights from our research will be used to design laboratory processes that can deliver clinically accredited gNBS in an appropriate time frame and at population scale. In addition, our study will inform the design of pretest and post-test processes, including education, decision support, informed consent and results management, aimed at facilitating understanding and acceptance of gNBS for parents, HCP and the Australian public.
Methods and analysis
Study design
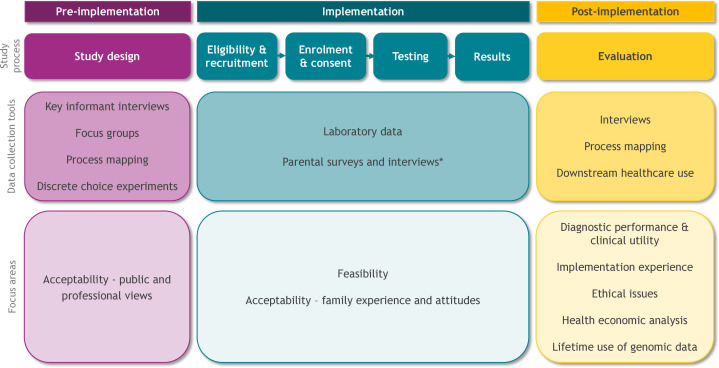
The BabyScreen+study will involve three phases, as outlined in figure 1. Following the design and implementation of education and consent materials, research data collection tools, and laboratory pathways, we will offer and evaluate the feasibility of gNBS. Clinically accredited gNBS will be performed for over 600 treatable, childhood-onset conditions following stdNBS. Evaluation will focus on the health economic, implementation science, psychosocial and ethical aspects of gNBS.
Figure 1.
The three phases of the BabyScreen+study including data collection tools and research focus areas. *Refer to table 1 for survey and interview measures.
Study aims
The overall aim of this study is to design and deliver a gNBS programme to screen 1000 prospectively recruited newborns and assess feasibility.
Secondary aims are to evaluate:
Public preferences and perspectives for the delivery of gNBS.
Ethical issues.
Performance of gNBS alongside stdNBS, comparing diagnostic performance and clinical utility.
Parental screening experience and psychosocial outcomes.
Cost-effectiveness and cost–benefit of gNBS relative to stdNBS.
Implementation of gNBS, including acceptability to HCP, parents and the public.
Value and ethical implications of using genomic data generated at birth as a lifelong healthcare resource.
Study timetable and sites
The preimplementation phase of the study commenced in July 2022 (figure 1). Staged recruitment into the implementation phase of the study began on 20 July 2023 at three sites, including a private obstetrics practice and two public hospitals. The postimplementation phase will commence at the completion of recruitment, and the study will be completed by 31 May 2027.
Preimplementation phase
The preimplementation phase comprises the design, development and testing of study processes and materials. These include HCP education materials; the online enrolment, decision support, education and consent platform; evaluation data collection tools; the list of genes for inclusion in screening and laboratory protocols. Key informant interviews, focus groups and discrete choice experiments (DCEs) will be undertaken to assess public and HCP views on study processes and acceptability of gNBS.
Condition and gene selection
The overarching condition selection principles that are used in stdNBS were maintained when considering which genes and conditions to include in this study.2 Included conditions:
Have a well-established gene–disease association and the majority of known clinically relevant variants are ascertainable by WGS.
Are considered severe (ie, causing mortality or considerable morbidity/disability in the majority of affected children if untreated).
Have early-onset of disease or significantly benefit from early intervention (<5 years of age in the majority).
Have an available treatment (eg, drugs/supplements, enzyme replacement therapy, organ or bone marrow transplant, diet modification, gene therapy) or other intervention that significantly alters the natural progression of the condition. Interventions must be accessible to study participants in Victoria, Australia.
We included all conditions that are currently screened in stdNBS in Australia. The gene selection process and panel content has been informed by prior local and international gNBS projects.21 23 Genes included in the study are publicly available for review and comment on PanelApp Australia (BabyScreen+newborn screening panel: https://panelapp.agha.umccr.org/panels/3931/; online supplemental table 1) and on the study website along with a plain language description of the gene selection process and screening approach.24
bmjopen-2023-081426supp001.pdf (129.3KB, pdf)
Key informant interviews
The implementation phase will be informed by interviews with key stakeholders involved in the current delivery of stdNBS. These include laboratory staff, HCP offering stdNBS, other antenatal HCP and paediatric physicians. Data collection and analysis will be guided by the Action, Actor, Context, Target, Time (AACTT) framework25 to capture views on who should do what and when. Interviews will explore opinions on offering gNBS; education and consent; sample collection and testing; and result management.
Eliciting public preferences and perspectives
Public preferences and values will be sought through a multiphase mixed-methods design involving focus groups and DCE surveys with members of the Australian public. Focus groups will explore public preferences and perspectives for (1) implementation of gNBS, (2) risks and benefits of gNBS and (3) use of data generated by gNBS as a lifetime resource and will involve qualitative exploration through facilitated discussion to elicit characteristics of gNBS delivery.26 Outcomes from the focus groups will be used to inform the implementation phase of the study. The DCE surveys will elicit public preferences, values and priorities for gNBS to support the economic evaluation and implementation of gNBS in Australia.
Implementation phase
HCP engagement
We will invite HCP from selected public and private healthcare settings to offer the study to their patients. HCP participation is voluntary and will not be incentivised. HCP who wish to be involved will receive education and support from the study team regarding gNBS and study processes.
Participant eligibility and recruitment
Pregnant individuals will be invited to take part in the third trimester. At one public hospital, recruitment will also occur via advertisement on a digital pregnancy care platform. To be eligible to participate, birth parents must be due to give birth in Victoria, Australia, within the study time frame, and participate in the state-based stdNBS. They must be aged 16 years or over, or have a legal representative capable of providing informed consent on their behalf. Enrolment must be completed by 2 weeks after their baby is born. This time frame allows for exceptional circumstances, for example, premature delivery, where enrolment is not completed before birth. However, in the majority of cases, we expect enrolment to be completed during pregnancy. The study team will follow up incomplete enrolments 2 weeks before the expected due date to ensure that results can be returned in a clinically meaningful time frame.
Enrolment and consent
After receiving a study invitation, birth parents will complete enrolment and consent for the study online. gNBS and all required pretest and post-test support will be offered at no cost to birth parents. Figure 2 provides a detailed outline of the participation process.
Figure 2.
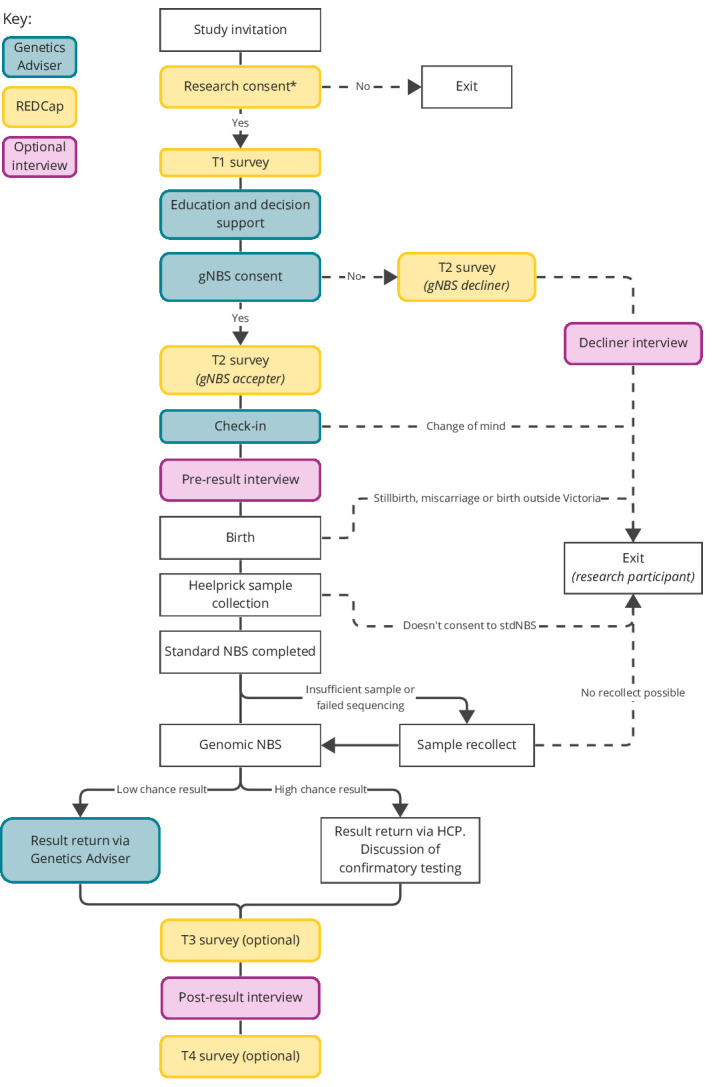
Participation process for birth parents. Solid lines represent the expected end-to-end study pathway (offer to result and final surveys), dashed lines represent anticipated study exit pathways. *Participants not able to complete enrolment in English will be supported by a genetic counsellor and an interpreter. HCP, healthcare professional; gNBS, genomic NBS; NBS, newborn bloodspot screening; REDCap, Research Electronic Data Capture; T1–T4, survey time points/identifiers.
Birth parents will first be asked to provide consent for the research component of the study via Research Electronic Data Capture (REDCap).27 28 This encompasses collection of personal and health-related data and survey responses. Participants who use a language other than English will be assisted to enrol and take part by a research genetic counsellor and an interpreter.
Birth parents who consent to research will receive education and decision support, as well as provide informed consent to clinical gNBS for their baby, via the online Genetics Adviser platform. Genetics Adviser is a patient-centred interactive online tool that provides educational, decisional and counselling support to patients undergoing genetic testing, along with managing consent capture and return of results (https://www.geneticsadviser.com/). Genetics Adviser has been extensively evaluated and improved by iterative codesign and has been shown to improve participant knowledge and decisional confidence.29–32 The study will use a locally deployed version of the platform, adapted in collaboration with the Genetics Adviser team.
A ‘check-in’ module in Genetics Adviser will automatically remind birth parents about the study 4 weeks prior to their estimated due date via email and SMS. Participants will be invited to log back into Genetics Adviser via email and SMS, where they will receive a brief refresher on gNBS, explore how they are currently feeling, and have the option to review their gNBS consent.
Genetic counselling is available at any time, if requested. Consent to research participation can be withdrawn at any time. Consent to clinical gNBS can be withdrawn at any time prior to result reporting.
Sample testing
Sample collection will be performed by the pregnancy service provider as part of stdNBS undertaken by Victorian Clinical Genetics Services (VCGS). StdNBS is expected to take up to 2 weeks. In order to comply with local requirements for access to NBS cards and avoid interference with stdNBS, BabyScreen+ will only have access to the sample once the routine process is complete. Following completion of stdNBS, four 3 mm punches will be taken from a single dried blood spot and processed using clinically accredited protocols and procedures at VCGS. DNA will be extracted using the Omega Biotek Mag-Bind DNA Blood and Tissue kit. Following DNA extraction, PCR-free genome sequencing libraries will be created using the PCR-free DNA prep kit (Illumina) and sequenced using a 2×150 base paired end read configuration to an average depth of 30× on NovaSeq 6000 of X Plus instruments (Illumina). The target time frame for return of results following handover of the punches from the stdNBS laboratory is 2 weeks. Considering the two blocks of 2 weeks, results will be returned within 4 weeks of sample collection. For samples with insufficient quantity or quality of DNA, the study team will contact the participant to arrange sample recollection.
Genome data analysis and interpretation will be performed using validated and clinically accredited, cloud-based analysis pipelines and procedures, using the Dragen and eMedgene (Illumina) analysis tools, with custom in-house filtering configuration. Interpretation will be restricted to genes on the BabyScreen+ gNBS panel, and only variants that are classified as likely pathogenic or pathogenic will be considered for reporting. We will not report carrier status, adult-onset conditions or variants of uncertain significance. All variants that are considered for reporting will be discussed by a team of clinical specialists, genetic pathologists and medical scientists.
Screening results
There will be two categories of result:
A low chance result, where no reportable variants are identified.
A high chance result, where one or more reportable variants are identified.
For low chance results, birth parents will be informed via the online Genetics Adviser platform following notification of result availability via email and SMS. Genetic counselling follow-up will be available on request.
For high chance results, birth parents will receive a phone call from a genetics HCP to discuss the result, arrange an appointment and any required additional testing (including confirmation of reported variants), as well as referral to specialist services. Further genetic counselling support will also be available.
Postimplementation phase
The study will move to the postimplementation phase after recruitment, with a focus on evaluation and data analysis. See figures 1 and 2 for details on the data collection tools, time points and evaluation focus areas.
Evaluation
Feasibility will be evaluated through a set of key performance indicators gathered from laboratory data. This includes data on the percentage of study samples that:
Are successfully identified from stdNBS samples
Require a recollect due to insufficient DNA
Fail sequencing.
Are processed within the expected turn-around time.
Have a concordant high chance result from stdNBS and gNBS.
Performance of gNBS will be further evaluated against stdNBS where a genetic diagnosis is confirmed. We will collect data on subsequent clinical management, including the timing of commencement of therapies and downstream healthcare utilisation. Where possible, we will compare this against historical controls from VCGS and the Royal Children’s Hospital (RCH) (patients diagnosed with the same disorder in the last 10 years). Cost-effectiveness and cost–benefit analyses will be conducted to evaluate whether the additional cost of gNBS relative to stdNBS is outweighed by longer-term cost savings and improvements in diagnostic, clinical and personal outcomes for children and families. Clinical and laboratory data from RCH and VCGS will be accessed and examined to establish if and for what purpose genomic data generated at birth has been accessed and/or reanalysed within 5 years postresult.
Views on acceptability and participant experiences will be collected during the first two phases of the study. Participant, public and HCP views will be captured through key informant interviews, focus groups, surveys and DCEs (see figures 1 and 2 and table 1). Participants will complete compulsory surveys at enrolment (T1) and after consent (T2), followed by optional surveys at 3 months postresult (T3) and ~12 months postresult (T4). Participants will also be invited to take part in interviews before and after receiving their gNBS results. Participants who decline gNBS will be asked to complete a modified T2 survey and invited to take part in an interview. Refer to table 1 for survey and interview measures.
Table 1.
Survey and interview measures used to ascertain parental experiences and preferences throughout the study
| Measure | Description | T1 | T2 | Preresult int | T3 | Postresult int | T4 | Decliner int |
| Details of pregnancy and demographics | Estimated due date, intended birth hospital, sex of baby (if known), twin/triplet pregnancy details (if applicable), education, income, language, marital status, number of children, prior experience with genomic testing, family history, ancestry. | X | ||||||
| Acceptability E-scale (gNBS accepter) |
Seven-item scale to determine the acceptability and usability of computerised health-related programmes.37 | X | ||||||
| Information in Genetics Adviser (gNBS accepter) |
Three study-specific questions to assess the perceived bias of Genetics Adviser content and review/influence of the study gene list. | X | ||||||
| Knowledge of gNBS (gNBS accepter) |
Eight study-specific true/false questions to determine participants’ understanding of gNBS. | X | X | X | ||||
| Difficulty and deliberation of decision (gNBS accepter) |
Three study-specific questions to assess the level of difficulty making a decision, length of deliberation and sources of information used when consenting to gNBS. | X | ||||||
| Reasons for accepting or declining (gNBS accepter) |
One study‐specific question addressing reasons for consenting to gNBS. Participants are asked to rate a selection of reasons on a 5-point Likert scale and asked to comment if there are other reasons not listed. | X | ||||||
| Willingness to pay (gNBS accepter) |
Dynamic triple-bounded dichotomous choice contingent valuation, also known as a ‘bidding game’, to assess the value participants place on gNBS. | X | X | X | ||||
| State trait anxiety index (STAI-AD) (gNBS accepter) |
26-item scale measuring state and trait anxiety. Copyright 1968, 1977 by Charles D. Spielberger. All rights reserved in all media. Published by Mind Garden,Inc. www.mindgarden.com Note: Only the six questions that form the STAI-6 short form38 are repeated at each time point. |
X | X | X | ||||
| Reasons for declining (gNBS decliner) |
One study-specific question exploring the reasons for declining. Participants are presented with 11 reasons and asked to rate how much each one influenced their decision on a scale of 1 (did not influence) to 5 (strongly influenced). | X | ||||||
| Result recall | One study-specific question to determine if participants recall their baby’s result correctly. | X | X | |||||
| Decision regret scale | Five-item scale to measure regret after a healthcare decision.39 | X | X | |||||
| Health system utilisation | One study-specific question to determine health system utilisation postresult. | X | X | |||||
| Result dissemination (high chance result only) |
One study-specific question for people to determine who participants have shared their result with and why. | X | ||||||
| Genomics Outcome Scale (high chance result only) |
Six-item scale based on the Genetic Counselling Outcome Scale-24 adapted to assess outcomes of genetic counselling and patient empowerment.40 | X | ||||||
| Service delivery preferences | 10 study-specific questions assessing opinions on if, how, and when gNBS should be offered, and what conditions should be screened. | X | ||||||
| Experience of gNBS | Questions on prior knowledge of genetic or genomic testing and experiences taking part in the study. | X | X | X | ||||
| Expectations of results | Questions on expected result and how they plan to use the result. | X | ||||||
| Receiving results, understanding and making meaning | Questions on result including knowledge of outcomes and how they plan to use the result. | X | ||||||
| Attitudes and perceptions | Questions on if, how and when gNBS should be offered, and what conditions should be screened. | X | X | X | ||||
| Decision making | Questions on reasons for declining gNBS. | X | ||||||
| Data reuse | Study-specific questions around preferences for additional analyses of gNBS data. | X | ||||||
| Future gNBS decision | One study-specific question asking preference for gNBS for future children | X |
gNBS, genomic newborn screening; STAI-6, Six-item State-Trait Anxiety Inventory; STAI-AD, STAI for Adults.
Health economic analyses will be performed from the perspective of the Australian healthcare system, using a lifetime horizon, and based on the outcomes of cost per additional diagnosis, cost per quality-adjusted life-year gained and net benefit. A budget impact analysis will be conducted to provide policy-relevant insights into the affordability and sustainability of implementing gNBS.
Potential barriers and enablers to implementation of gNBS will be identified to generate strategies to support successful delivery and inform future studies. These will be ascertained through theory-informed interviews. This will include capturing the experiences of professionals including HCP involved in the delivery of gNBS, and those impacted by the implementation of gNBS, including perinatal HCP and paediatric physicians. In addition, process mapping will be undertaken with laboratory staff and study team members at intervals throughout the study to understand adaptations required to deliver gNBS at scale and within clinically meaningful time frames.
Data analysis
Personal identifying information, clinical and evaluation data will be collected using recording devices, and electronic data collection forms in the online Genetics Adviser platform32 and REDCap,27 28 both hosted at the Murdoch Children’s Research Institute, Melbourne, Australia.
Findings from interviews and surveys will be used to provide qualitative and statistical information on the associations between a particular behaviour and opinions or attitudes about the programme. Qualitative data will be either deductively analysed where a theoretical framework has been employed, for example, AACTT25 or analysed using inductive content analysis33 or thematic analysis to generate rich descriptions responding to the research aims, for example, acceptability of testing. Economic evaluation and DCEs will be conducted according to the best practice recommendations.34–36 Quotes from participants will be used to illustrate qualitative findings in scientific presentations and other publications. Statistical analysis will use appropriate methods and tests depending on the nature of the data and its normality. Probability values of <0.05 will be considered statistically significant, and CIs will be calculated where possible.
Patient and public involvement
The preimplementation phase of the study involved extensive consultation with the public via focus groups26 and DCE surveys. These have informed the design of the implementation phase. Condition selection has been done in conjunction with patient support groups.24 The study also uses Genetics Adviser, a decision support platform with extensive public input into design and development.29–32
Ethics and dissemination
This study is governed and administered by the Murdoch Children’s Research Institute (MCRI), Melbourne, Australia. All genetic testing is performed by VCGS, Melbourne, Australia, a wholly owned not-for-profit subsidiary of MCRI. VCGS is clinically accredited (NATA/RCPA) to ISO15189;2012 to carry out genetic and genomic testing. The project has received ethics approval from the Royal Children’s Hospital Melbourne Human Research Ethics Committee (main BabyScreen+ protocol: HREC/91500/RCHM-2023; key informant interviews: HREC/90929/RCHM-2022; and focus groups and DCE: HREC/91392/RCHM-2022). Findings from this study will be published in peer-reviewed scientific journal articles and presented at appropriate clinical and research conferences. The findings will also be disseminated to policy-makers including the standing committee on screening. All participants have provided informed consent to be involved in this study.
Supplementary Material
Acknowledgments
The research conducted at the Murdoch Children’s Research Institute was supported by the Victorian Government's Operational Infrastructure Support Program. The Chair in Genomic Medicine awarded to JChristodoulou is generously supported by The Royal Children’s Hospital Foundation. YB holds the Canada Research Chair in Genomics Health Services and Policy, funded by the Canada Research Chairs Program.
Footnotes
Contributors: All authors contributed to the design of the protocol, including its concept and the critical review and final approval of the manuscript. SL, SEB and ZS drafted the manuscript. SS, SE, SH, CL, MW, RFG, PDF and SL contributed to the design of the laboratory processes. DJA, LCD, AA, JChristodoulou, AY, ZS and SL contributed to the design of the gene list. YB, MC, JCaruana, AA, LCD, CG, SEB, NL, AK-P and ZS contributed to the design of participant and HCP education materials. DFV, FL and CG contributed to the design of the ethical evaluation. ET, AK-P, AA and SB contributed to the design of the psychosocial and implementation evaluation. RP and IG contributed to the design of the health economic evaluation.
Funding: The BabyScreen+ study is funded by the Australian Government’s Medical Research Future Fund as part of the Genomics Health Futures Mission, grant MRF2015937 titled ‘Genomic newborn screening for personalised lifelong healthcare in Australian babies’.
Competing interests: YB and MC are cofounders of the Genetics Adviser. The other authors declare no relevant disclosures.
Patient and public involvement: Patients and/or the public were involved in the design, or conduct, or reporting, or dissemination plans of this research. Refer to the Methods section for further details.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Ethics statements
Patient consent for publication
Not applicable.
References
- 1. Therrell BL, Padilla CD, Loeber JG, et al. Current status of newborn screening worldwide: 2015. Semin Perinatol 2015;39:171–87. 10.1053/j.semperi.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 2. Wilson JMG, Jungner G, World Health O. Principles and Practice of Screening for Disease. Geneva: World Health Organization, 1968. [Google Scholar]
- 3. Stark Z, Scott RH. Genomic newborn screening for rare diseases. Nat Rev Genet 2023;24:755–66. 10.1038/s41576-023-00621-w [DOI] [PubMed] [Google Scholar]
- 4. Koracin V, Mlinaric M, Baric I, et al. Current status of newborn screening in southeastern Europe. Front Pediatr 2021;9:648939. 10.3389/fped.2021.648939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sikonja J, Groselj U, Scarpa M, et al. Towards achieving equity and innovation in newborn screening across Europe. Int J Neonatal Screen 2022;8:31. 10.3390/ijns8020031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Department of Health and Aged Care . What is screened in the program 2023. 2023. Available: https://www.health.gov.au/our-work/newborn-bloodspot-screening/what-is-screened
- 7. Tambuyzer E, Vandendriessche B, Austin CP, et al. Therapies for rare diseases: therapeutic modalities, progress and challenges ahead. Nat Rev Drug Discov 2020;19:93–111. 10.1038/s41573-019-0049-9 [DOI] [PubMed] [Google Scholar]
- 8. Bick D, Bick SL, Dimmock DP, et al. An online compendium of treatable genetic disorders. Am J Med Genet C Semin Med Genet 2021;187:48–54. 10.1002/ajmg.c.31874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berg JS, Agrawal PB, Bailey DB, et al. Newborn sequencing in genomic medicine and public health. Pediatrics 2017;139:e20162252. 10.1542/peds.2016-2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Roman TS, Crowley SB, Roche MI, et al. Genomic sequencing for newborn screening: results of the NC NEXUS project. Am J Hum Genet 2020;107:596–611. 10.1016/j.ajhg.2020.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Adhikari AN, Gallagher RC, Wang Y, et al. The role of exome sequencing in newborn screening for inborn errors of metabolism. Nat Med 2020;26:1392–7. 10.1038/s41591-020-0966-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ceyhan-Birsoy O, Machini K, Lebo MS, et al. A curated gene list for reporting results of newborn genomic sequencing. Genet Med 2017;19:809–18. 10.1038/gim.2016.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pereira S, Robinson JO, Gutierrez AM, et al. Perceived benefits, risks, and utility of newborn genomic sequencing in the babyseq project. Pediatrics 2019;143:S6–13. 10.1542/peds.2018-1099C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Downie L, Halliday J, Lewis S, et al. Principles of genomic newborn screening programs: a systematic review. JAMA Netw Open 2021;4:e2114336. 10.1001/jamanetworkopen.2021.14336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cao M, Notini L, Ayres S, et al. Australian healthcare professionals' perspectives on the ethical and practical issues associated with genomic newborn screening. J Genet Couns 2023;32:376–86. 10.1002/jgc4.1645 [DOI] [PubMed] [Google Scholar]
- 16. Iskrov G, Angelova V, Bochev B, et al. Prospects for expansion of universal newborn screening in Bulgaria: a survey among medical professionals. Int J Neonatal Screen 2023;9:57. 10.3390/ijns9040057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goldenberg AJ, Dodson DS, Davis MM, et al. Parents' interest in whole-genome sequencing of newborns. Genet Med 2014;16:78–84. 10.1038/gim.2013.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Waisbren SE, Bäck DK, Liu C, et al. Parents are interested in newborn genomic testing during the early postpartum period. Genet Med 2015;17:501–4. 10.1038/gim.2014.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ceyhan-Birsoy O, Murry JB, Machini K, et al. Interpretation of genomic sequencing results in healthy and ill newborns: results from the babyseq project. Am J Hum Genet 2019;104:76–93. 10.1016/j.ajhg.2018.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Timmins GT, Wynn J, Saami AM, et al. Diverse parental perspectives of the social and educational needs for expanding newborn screening through genomic sequencing. Public Health Genomics 2022;2022:1–8. 10.1159/000526382 [DOI] [PubMed] [Google Scholar]
- 21. Kingsmore SF, Smith LD, Kunard CM, et al. A genome sequencing system for universal newborn screening, diagnosis, and precision medicine for severe genetic diseases. Am J Hum Genet 2022;109:1605–19. 10.1016/j.ajhg.2022.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spiekerkoetter U, Bick D, Scott R, et al. Genomic newborn screening: are we entering a new era of screening J Inherit Metab Dis 2023;46:778–95. 10.1002/jimd.12650 [DOI] [PubMed] [Google Scholar]
- 23. Downie L, Halliday JL, Burt RA, et al. A protocol for whole-exome sequencing in newborns with congenital deafness: a prospective population-based cohort. BMJ Paediatr Open 2017;1:e000119. 10.1136/bmjpo-2017-000119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Downie L, Bouffler SE, Amor DJ, et al. Gene selection for Genomic newborn screening: moving toward consensus? Genetics in Medicine 2024;26:101077:S1098-3600(24)00010-8. 10.1016/j.gim.2024.101077 [DOI] [PubMed] [Google Scholar]
- 25. Presseau J, McCleary N, Lorencatto F, et al. Action, actor, context, target, time (AACTT): a framework for specifying behaviour. Implement Sci 2019;14:102. 10.1186/s13012-019-0951-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lynch F, Best S, Gaff C, et al. Australian public perspectives on Genomic newborn screening: risks, benefits, and preferences for implementation. International Journal of Neonatal Screening 2024;10:6. 10.3390/ijns10010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harris PA, Taylor R, Minor BL, et al. The redcap consortium: building an international community of software platform partners. J Biomed Inform 2019;95:103208. 10.1016/j.jbi.2019.103208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (Redcap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377–81. 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bombard Y, Clausen M, Shickh S, et al. Effectiveness of the genomics adviser decision aid for the selection of secondary findings from Genomic sequencing: a randomized clinical trial. Genet Med 2020;22:727–35. 10.1038/s41436-019-0702-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shickh S, Rafferty SA, Clausen M, et al. The role of digital tools in the delivery of genomic medicine: enhancing patient-centered care. Genet Med 2021;23:1086–94. 10.1038/s41436-021-01112-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bombard Y, Clausen M, Mighton C, et al. The genomics adviser: development and usability testing of a decision aid for the selection of incidental sequencing results. Eur J Hum Genet 2018;26:984–95. 10.1038/s41431-018-0144-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shickh S, Hirjikaka D, Clausen M, et al. Genetics adviser: a protocol for a mixed-methods randomised controlled trial evaluating a digital platform for genetics service delivery. BMJ Open 2022;12:e060899. 10.1136/bmjopen-2022-060899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vears DF, Gillam L. Inductive content analysis: a guide for beginning qualitative researchers. FoHPE 2022;23:111–27. 10.11157/fohpe.v23i1.544 [DOI] [Google Scholar]
- 34. Briggs A, Claxton K, Sculpher M. Decision Modelling for Health Economic Evaluation. Oxford: Oxford University Press, Available: https://academic.oup.com/book/52453 [Google Scholar]
- 35. Hauber AB, González JM, Groothuis-Oudshoorn CGM, et al. Statistical methods for the analysis of discrete choice experiments: a report of the ISPOR conjoint analysis good research practices task force. Value Health 2016;19:300–15. 10.1016/j.jval.2016.04.004 [DOI] [PubMed] [Google Scholar]
- 36. Reed Johnson F, Lancsar E, Marshall D, et al. Constructing experimental designs for discrete-choice experiments: report of the ISPOR conjoint analysis experimental design good research practices task force. Value Health 2013;16:3–13. 10.1016/j.jval.2012.08.2223 [DOI] [PubMed] [Google Scholar]
- 37. Tariman JD, Berry DL, Halpenny B, et al. Validation and testing of the acceptability E-scale for web-based patient-reported outcomes in cancer care. Appl Nurs Res 2011;24:53–8. 10.1016/j.apnr.2009.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marteau TM, Bekker H. The development of a six-item short-form of the state scale of the spielberger state-trait anxiety inventory (STAI). Br J Clin Psychol 1992;31:301–6. 10.1111/j.2044-8260.1992.tb00997.x [DOI] [PubMed] [Google Scholar]
- 39. Brehaut JC, O’Connor AM, Wood TJ, et al. Validation of a decision regret scale. Med Decis Making 2003;23:281–92. 10.1177/0272989X03256005 [DOI] [PubMed] [Google Scholar]
- 40. Grant PE, Pampaka M, Payne K, et al. Developing a short-form of the genetic counselling outcome scale: the genomics outcome scale. Eur J Med Genet 2019;62:324–34. 10.1016/j.ejmg.2018.11.015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2023-081426supp001.pdf (129.3KB, pdf)