Abstract
Huntington’s disease results from expansion of a polyglutamine tract (polyQ) in mutant huntingtin (mHTT) protein, but mechanisms underlying polyQ expansion-mediated toxic gain-of-mHTT function remain elusive.
Here, deletion and antibody-based experiments revealed that a proline-rich domain (PRD) adjacent to the polyQ tract is necessary for mHTT to inhibit fast axonal transport and promote axonal pathology in cultured mammalian neurons. Further, polypeptides corresponding to subregions of the PRD sufficed to elicit the toxic effect on fast axonal transport, which was mediated by c-Jun N-terminal kinases (JNKs) and involved PRD binding to one or more SH3-domain containing proteins. Collectively, these data suggested a mechanism whereby polyQ tract expansion in mHTT promotes aberrant PRD exposure and interactions of this domain with SH3 domain-containing proteins including some involved in activation of JNKs. In support, biochemical and immunohistochemical experiments linked aberrant PRD exposure to increased JNK activation in striatal tissues of the zQ175 mouse model and from post-mortem Huntington’s disease patients.
Together, these findings support a critical role of PRD on mHTT toxicity, suggesting a novel framework for the potential development of therapies aimed to halt or reduce axonal pathology in Huntington’s disease.
Keywords: Huntington’s disease, huntingtin, JNK3, proline-rich domain, axonal transport, SH3-binding domain
Brady et al. show that axonal toxicity of mutant huntingtin requires a proline rich domain (PRD) adjacent to the polyQ tract which contains SH3-binding motifs and which is normally sequestered in wildtype huntingtin. PolyQ expansion leads to PRD exposure, resulting in aberrant activation of a MAP kinase and axonal degeneration.
Introduction
Huntington’s disease (HD) is a genetic neurodegenerative disorder typically with midlife onset caused by mutations that cause expansion of a CAG repeat in HTT (IT15), the gene encoding the huntingtin (HTT) protein.1 Such mutations result in expansion of a polyglutamine tract (polyQ) near the amino terminus, causing HD when expanded beyond 36 glutamines.2 Remarkably, further increases in the number of glutamines in the polyQ tract affect the age of onset for neurological symptoms, but not the neurological phenotype including motor and cognitive changes. As a result, polyQ expansions exceeding 80 glutamines exhibit a juvenile form of the disease while polyQ expansions of patients in their forties may not present until their fifties or later.2 Although the genetic cause of HD has been defined for decades, the molecular basis underlying mutant HTT (mHTT) mediated neurotoxicity remains unclear. While HTT is expressed ubiquitously in the nervous system, HD pathology preferentially affects a subset of projection neurons in the striatum and cerebral cortex early in course of disease, extending later to neurons in other basal ganglion structures.3,4 HD inheritance is autosomal dominant and mouse models of HD all develop neurodegeneration in adulthood.5 In contrast, homozygous deletion of HTT is embryonic lethal and heterozygotes are largely asymptomatic.6 Thus, polyQ expansion confers upon mHTT a toxic gain of function that manifests clinically later in life. Although mHTT is reported to affect many cellular processes, disease-relevant ones remain a matter of debate.4
In most HD mouse models, including transgenic R6/2 mice, neuronal loss is absent or modest at best and occurs very late in the disease process.7 In contrast, axonal and synaptic pathology are apparent in presymptomatic animals3 and many brain imaging studies have extended these findings to presymptomatic HD patients.8,9 Based on these findings, we examined potential mechanisms underlying mutant HTT-induced axonal pathology. Previous studies identified fast axonal transport as a sensitive readout of mHTT toxicity.10,11 Pathogenic forms of HTT inhibited both anterograde and retrograde fast axonal transport,10 a toxic effect involving activation of axonal JNKs.11 From three JNK isoforms activated, the neuron-specific JNK3 isoform was found to inhibit anterograde fast axonal transport by phosphorylating heavy chains of the major motor protein kinesin-1 (KHC),11 but the question of how mHTT activates JNKs remained unsolved. Canonical activation of JNKs involves phosphorylation by upstream mitogen activated protein kinases (MAPKs),12 but HTT does not have any known enzymatic activity.
To determine how mHTT activates JNKs, we mapped toxic elements in mHTT by perfusing recombinant mHTT fragments into isolated axoplasm from the squid giant axon, a unique ex vivo model for the analysis of fast axonal transport. Consistent with findings from genetic experiments,13 an mHTT fragment encoded by HTT exon 1 sufficed to inhibit fast axonal transport. HTT exon 1 contains three discrete regions, a 17-amino acid (aa) N-terminal sequence, a polymorphic polyQ tract and a proline-rich domain (PRD). To refine the identification of toxic elements in mHTT further, well-characterized antibodies recognizing epitopes within exon 1 were preincubated with mHTT and co-perfused in axoplasm. Significantly, an antibody recognizing polyproline (polyP) tracts in PRD blocked toxic effects of mHTT exon 1 on fast axonal transport, but an antibody recognizing polyQ did not. In addition, several mHTT fragments in which the PRD was deleted lacked toxicity in both squid axoplasm and in primary cultured rodent neurons. Further, both the full PRD and two PRD subregions containing canonical PXXP motifs exhibited toxicity. Prolines are essential components in many protein interaction domains, such as SH3 and WW domains14 and SH3 domains play a role in regulation of many signalling cascades, including the activation of MAPKs upstream of JNKs.15,16 The role of SH3-binding motifs in the PRD was tested by co-perfusing mHTT with a recombinant SH3 domain, which prevented inhibition of fast axonal transport. These studies revealed that the PRD is necessary and sufficient for mHTT to activate a MAPK cascade leading to the activation of JNKs and inhibition of fast axonal transport, an effect dependent on the presence of one or more SH3 binding domains within the PRD.
To determine whether mammalian models of HD pathology also required the presence of a PRD, mHTT proteins lacking the PRD did not exhibit axonal toxicity in cultured neurons. Furthermore, biochemical and immunological studies in a well-established HD mouse model and HD-affected human post-mortem tissues linked aberrant activation of JNKs to increased PRD exposure, which can explain why wild-type HTT fragments do not inhibit fast axonal transport despite containing a PRD. Collectively, results from our work support a mechanism where polyQ expansion in mHTT promotes abnormal PRD exposure, aberrant JNK activation, and toxic effects in axons suggesting new avenues to treat HD.
Materials and methods
Antibodies and reagents
Hybridomas expressing the anti-HTT monoclonal antibodies MW1 (against polyQ) and MW7 (against polyP)17 were obtained from the Iowa State Hybridoma Bank and purified from hybridoma culture supernatants. Supplementary Table 1 lists in detail all antibodies used in this study.
Vesicle motility assays in isolated axoplasm
Giant axons from the squid (Doryteuthis pealeii, Marine Biological Laboratory) were dissected and axoplasm extruded onto a glass coverslip as described previously.18-20 Lack of plasma membrane in this preparation allows the perfusion of proteins and other effectors at precise concentrations.20,21 Recombinant HTT and mHTT fragments, as well as inhibitors, were diluted in X/2 buffer (175 mM potassium aspartate; 65 mM MgCl2, 5 mM EGTA; 1.5 mM CaCl2; 0.5 mM glucose, pH 7.2; supplemented with 2–5 mM ATP) to a final concentration of 100 nM and perfused into axoplasm (20 µl) as done before.10,11,20 Real-time analysis of vesicles moving in anterograde and retrograde directions was analysed using a Zeiss Axiomat microscope equipped with a 100× (1.3 numerical aperture) objective and differential interference contrast optics. Images were acquired using a Model C2400 CCD through a Hamamatsu Argus 20 and processed further with a Hamamatsu Photonics Microscopy C2117 video manipulator for image adjustment as well as generation of calibrated cursors and scale bars. Following perfusion, anterograde and retrograde fast axonal transport rates were measured by matching calibrated cursor movements to the speed of vesicles moving in each direction over 50 min and data plotted as a function of time.18,20 For statistical analysis of independent experiments in isolated axoplasm, individual axonal transport rates recorded during the last 20 min of each individual assay were pooled and analysed.
Recombinant HTT polypeptides for axoplasm studies
Plasmids (pcDNA3.1) encoding FLAG-tagged mHTT (1–969 Q46) with and without deletion of PRD (ΔPRD) were obtained from Dr M. DiFiglia.22 These plasmids were expressed using in vitro transcription/translation (Promega; TnT T7-coupled Reticulocyte Lysate system). To confirm that similar levels of each mHTT construct were expressed, parallel reactions incorporating 35S-labelled methionine (Amersham) were done.10,11
Constructs encoding mHTT exon 1 with a Q49 expansion [mHTT exon 1 (Q49)] and with the PRD deleted [mHTT exon 1 (Q49 ΔPRD)] were a gift from E. Wanker.23 These constructs were subcloned in pGEX-5X1 with an N-terminus GST tag and expressed in Escherichia coli BL21 codon plus (DE3) cells and purified using affinity chromatography (Amersham Glutathione Sepharose 4B). Purified protein was dialysed (10 mM HEPES pH 7.4), and concentration measured via Bradford assay. Proteins were frozen in liquid nitrogen and stored at −80°C until used. Bacterial expression plasmids encoding GST fused to segments P1, P2 and P3 of the HTT PRD were obtained from Dr Hong-Yu Hu and purified as described.24
Primary cortical neuron cultures
Cortical brain regions were dissected from Sprague-Dawley rat embryos at embryonic Day 18 (E18) (Charles River), then treated with 2.5% Trypsin in 1× Hank’s balanced salt solution (HBSS) for 15 min in a 37°C water bath and washed three times with fresh HBSS. Tissue was resuspended in neuronal plating medium [DMEM(++); Dulbecco’s modified Eagle medium (DMEM) medium + 10% bovine calf serum with iron + 1% penicillin/streptomycin] and triturated five times to obtain a single cell suspension. Neurons were counted and plated in either six-channel slides (μ-Slide VI, Ibidi Cat. No. 80601) for morphological assessment in custom microfluidic chambers pre-coated with 0.5 mg/ml poly-L-lysine (Sigma Cat. No. P1399) in borate buffer overnight for axonal outgrowth quantitation. Plating density for Ibidi chambers was 1 × 104 cells per Ibidi well and 5 × 104 cells per somatodendritic compartment (SDC) for microfluidic chambers.
Custom microfluidic chambers
Custom microfluidic chambers were constructed using soft lithography methods with polydimethyl sulphoxide (PDMS), from SU-8 photoresist molds on silicone wafers, and plasma mounted onto nitric acid-treated glass coverslips. Completed chambers were 480 μm in height, 26 mm in length, and 18 mm in width. Microfluidic chambers had three individual SDCs 7 × 7 mm square and one axon terminal compartment (ATC) 20 mm in length. A 1 mm PDMS wall separated the SDC from one another. Microfluidic chambers had an open-top design with a 6 mm hole punched into each SDC and three 4 mm holes in the ATC. There were 106 microchannels connecting each SDC to the ATC. Single microchannels were 500-μm long, 8-μm wide, and 2.5-μm high. Microfluidic chambers were sterilized in ethanol, allowed to air dry in a sterile laminar flow hood, and treated with UV for 5 min, prior to being plasma mounted onto nitric acid-treated, German coverglass (24 × 50 mm).
Live cell imaging
For these experiments, plasmids encoding mCherry-tagged versions of either mHTT exon 1 (Q46) or mHTT exon 1 (Q49 DPRD) on the pcDNA3 vector with insertion of the mCherry sequence following the CMV promoter, and insertion of the N-terminal human HTT, composed of the first 17 amino acids (MATLEKLMKAFESLKSF) followed by a stretch of 46 glutamines either with or without (ΔPRD) the PRD. The sequence deleted is the same as the sequence added to GST for the GST-PRD studies. DNA (0.1 µg) was mixed with 0.3 μl transfection reagent in 10 μl of unsupplemented Neurobasal medium and incubated for 15 min at room temperature (RT), and then mixed with media.
Live cell morphology compared neurons transfected with mCherry-tagged versions of mHTT exon 1 (Q46) or mHTT exon 1 with PRD deletion (Q46 ΔPRD) in Ibidi chambers. At 6 days in vitro (DIV), neurons were transfected using the TransIT-X2 reagent (Mirus Cat. No. MIR6000). Neurons were labelled with a neuron-specific membrane-permeable fluorescent probe (NeuroFluor NeuO, StemCell Technologies Cat. No. 01801) and a Hoechst 33342 cell-permeant nuclear counterstain (NucBlue Live ReadyProbes Reagent, ThermoFisher Scientific Cat. No. R37605). Two hours prior to imaging, the neurons were treated with NeuO at 1:400 dilution in a neuronal maintenance media [NB(+++): neurobasal medium + 2% B27 supplement + 0.5 mM GlutaMAX + 1% penicillin/streptomycin] with 1 drop of NucBlue. Live cell fluorescent microscopy was performed at 13 DIV, using a LSM880 equipped with Airyscan (Zeiss; 20× magnification + 1.8 digital zoom). NeuO treatment of primary cultured neurons had no effect on viability or axonal outgrowth (Supplementary Figs 1 and 2).
Quantitation of axonal outgrowth in living cells
Two hours prior to imaging, neurons were labelled with NeuO as described earlier. At 6 DIV, axons within the microfluidic chamber microchannels were imaged sequentially from the SDC to the ATC just prior to the cellular transfections, and at 8, 11, and 13 DIV after transduction. Live cell fluorescent microscopy was used to capture each 500-μm long microchannel in its entirety; each microchannel contained a single axon (×10 magnification, Zeiss LSM710). A predetermined start and stop point for measuring the axonal outgrowth length exists within the microfluidic chamber design, with the upper boundary being the SDC edge adjacent to microchannels and the lower boundary the ATC edge adjacent to the microchannels. Individual axons were traced from entry into the microchannel from the SDC until they crossed into the ATC using Zen Lite Blue imaging analysis software (Zeiss). Axonal outgrowth was measured throughout the time course to determine whether axons were elongating or undergoing degeneration (decreasing in length over time or showing disconnected, i.e. degenerated, axonal segments within the microchannel). The total number of elongating and degenerating axons were quantified at the end of the time course (GraphPad Prism 7).
Immunocytochemistry
HaloTag® constructs encoding HTT exon 1 (Q18), HTT exon 1 with PRD deletion (Q18 ΔPRD), mHTT exon 1 (Q46), and mHTT exon 1 (Q46 ΔPRD) constructs were developed using the pHTN HaloTag CMV-neo vector (Promega Cat. No. G7721). For transfection, DNA (0.5 μg) was mixed with 1.5 μl X2 in 10 μl unsupplemented neurobasal medium and incubated for 15 min at RT, and then mixed with media in each culture compartment. At 4 DIV, cortical neurons were transfected with HaloTag-expression vectors using the Transit-X2 reagent and labelled with the HaloTag TMR ligand (Promega Cat. No. G8251).
The cell permeant, HaloTag TMR ligand was diluted using neuronal maintenance medium to 200 nM and added to the neuronal samples 24-h incubation prior to fixation. Cortical neurons were fixed 48 h post-transfection with 4% paraformaldehyde, washed 3 × 5 min in PBS, autofluorescence quenched with 50 mM NH4Cl for 15 min, washed 3 × 5 min in PBS, permeabilized with 0.1% Triton X-100 for 10 min, washed 3 × 5 min in PBS, non-specific binding blocked with 5% milk for 1 h, washed twice with PBS, incubated overnight at 4°C with primary antibody (βIII tubulin, 1:1000, Sigma T8660), washed 3 × 5 min in PBS, incubated for 45 min at RT with secondary antibody (AF488, 1:800, Invitrogen Cat. No. A11029), washed 3 × 5 min in PBS, and covered in an aqueous anti-fade mounting medium (Vectashield Cat. No. H-1000) for storage at 4°C until imaged (Zeiss LSM710, 63×). Neuronal morphology was examined in fixed neurons transfected with plasmids encoding HaloTag versions of wild-type mHTT exon 1 (Q18), wild-type-HTT exon 1 with PRD deletion (Q18 ΔPRD), polyQ-expanded mHTT exon 1 (Q46), and mHTT exon 1 with PRD deletion (Q46 ΔPRD) in Ibidi chambers.
Mice
Heterozygous zQ175DN mice on a C57BL/6J background were obtained from the Jackson Laboratory (JAX# 029928, henceforth: zQ175 mice). A colony of these mice was maintained by crossing heterozygous zQ175DN males to C57BL/6J females (JAX# 000664). Heterozygous zQ175DN male mice from this colony were also bred with YFP(J16)+/+ females (C57BL/6J background, JAX# 003709) to generate YFP(J16)+/− control and YFP+/−:zQ175DN littermates (henceforth: ‘YFP mice’ and ‘YFP-zQ175 mice’, respectively), as done before for R6/2 mice.3 Up to five mice per cage were housed in a manner dependent on sex, but genotypes were mixed. Mice had unrestricted access to food and water. The mean and standard deviation for the CAG repeat-sizes for the zQ175 mice that were used for immunohistochemistry and immunoblotting analyses were 199 ± 14 Q repeats. For both biochemical and immunohistochemical analyses, mice were used at 10 months of age. All procedures were approved by the Institutional Animal Care and Use Committee of University of Illinois at Chicago.
Preparation of proteins from mouse striatal tissues
Following euthanasia, mouse brains were dissected and 1 mm coronal sections prepared using a brain matrix (Zivic Instruments, Cat. No. 5325). Striata were collected using a punch, flash-frozen in liquid nitrogen, and stored at −80°C until processing. For preparation of proteins under native conditions, striatal tissues were lysed in buffer X/2 containing 1% Triton X-100 and protease inhibitors (Sigma Cat. No. P8340 1/200). Following clarification, samples were normalized to 1 µg/µl concentration. Aliquots of native protein stocks were also denatured by adding 5% SDS and Laemmli sample buffer. Proteins were separated by SDS-PAGE, transferred to PVDF membranes, and the resulting western blots analysed using quantitative immunoblot analysis, as before.25
Dot blots for mouse lysates
For experiments in Supplementary Fig. 4, 2 µl aliquots of native protein (containing 3.4 µg total protein each) were spotted (in triplicate) onto nitrocellulose membranes (AmershamTM ProtanTM GE Healthcare Life Sciences, Cat. No. 10600002), as done before.26 Samples were left at RT for 30 min before incubation with primary antibodies (see later).
Immunoblotting analysis of mouse striatal tissues
Both western and dot blot membranes (Supplementary Fig. 4B–D) were blocked with Tris-buffered saline (TBS, pH 7.4) containing 1% non-fat dried milk for 1 h at RT. Following this step, membranes were incubated overnight at 4°C with primary antibodies (Supplementary Table 1). After rinsing in 0.1% Tween-20/TBS (TBST), membranes were incubated with secondary antibodies for 1 h at RT (Supplementary Table 1). Following rinsing in TBST, membranes were imaged using a Li-COR Odyssey infrared imaging system and Li-COR ImageStudioLite 5.2 Software was used to quantify immunoreactivity of bands.25
Mouse tissue immunohistochemistry
Mice were transcardially perfused with PBS, followed by PBS containing 4% paraformaldehyde (PFA). Brains were dissected and post-fixed with 4% PFA for 16 h, then rinsed in TBS, cryoprotected through washes in sucrose solutions of increasing concentration (5%, 10%, 20% and 30% sucrose in TBS), and stored at 4°C. Brains were embedded in OCT compound, fast frozen in dry ice plus isopentane and stored at −80°C overnight. Coronal and sagittal brain sections (40 µm) were prepared using a Leica Cryostat 3050 and stored in TBS containing 0.02% sodium azide. Free-floating sections were permeabilized in 0.5% Triton-X (w/v) in TBS, blocked in blocking buffer [1% bovine serum albumin (BSA), 0.1% Triton-X, 1% gelatin, 0.3 M and 0.5% goat serum in TBS], and then incubated with primary antibodies (Supplementary Table 1) overnight at 4°C. Tissue sections were washed three times in TBS, then incubated at RT for 2 h with appropriate secondary antibodies (Supplementary Table 1). After 2 h incubation, sections were washed in TBS, counterstained with DAPI, mounted onto glass slides, and coverslips applied. Negative control sections were prepared where primary antibodies were omitted. Images were collected on a Zeiss LSM 880 Confocal Airyscan.
Human tissue collection, immunohistochemistry, and imaging
Post-mortem human brain tissues were fixed in formalin, embedded in paraffin (see Supplementary Table 3 for demographic information), and stored in the Massachusetts Alzheimer's Disease Research Center at Massachusetts General Hospital, under the strict ethical and Institutional Review Board regulations from the Neuropathology core. Tissue blocks of the striatum (caudate, internal capsule, putamen, and nucleus accumbens) were sectioned at 7-µm thickness and mounted on SuperFrost Plus glass slides. Sections were then processed by Leica Bond Autostainer through steps of bake and dewax, antigen retrieval (with citrate buffer for 30 min, 95°C), permeabilization (0.4% Triton in TBST for 10 min, RT), blocking (10% goat serum and 3% BSA in TBST for 45 min, RT), primary antibody incubation in blocking buffer (see Supplementary Table 1 for a list of antibodies used, for 60 min, RT), washes (TBST), secondary antibody incubation (60 min, RT), washing (TBST), incubation with Tubb3-488 directly conjugated antibody (60 min, RT), washes (TBST), Hoechst staining (1 µg/ml, 5 min, RT), and additional washes (TBS). Slides were then treated with Trueblack (Biotium Cat. No. 23007) for 30 s to eliminate lipofuscin autofluorescence and reduce autofluorescence from other sources, followed by mounting with ProLong™ Glass Antifade Mountant (ThermoFisher P36980).
Epifluorescence imaging of the whole striatum section was performed automatically by Nanozoomer Slide Scanner (Hamamatsu Photonics) to avoid human bias in selecting regions of interest. All images were acquired using exactly the same parameters (e.g. exposure time) for all HD and control samples. Confocal images were acquired on Olympus FV3000 using the same laser power, PMT voltage, gain, and offset for all samples, and fields of view were randomly selected after the striatal region was located based on brightfield imaging. Structured Illumination Microscopy was performed on Zeiss Elyra 7 and processed by SIM2 software (Zeiss) with alignment correction from calibration beads. Images were quantified by QuPath 0.4.3 by the user blinded to the sample genotype.
Dot blot analysis of human samples
Human fresh refrozen tissues were quickly dissected and homogenized in RIPA lysis buffer supplemented with phosphatase and proteases inhibitor cocktail (Cell Signaling Technology Cat. No. 5872). The homogenate was centrifuged, and the supernatant was collected for dot blotting using nitrocellulose membrane and Bio-Dot apparatus (Bio-Rad). Blots were blocked in LI-COR blocking buffer and incubated with primary and secondary antibodies as described in Supplementary Table 1. Images were taken by LI-COR Odyssey Imager and quantified using LI-COR Image Studio software.
Statistical analysis
All experiments were repeated more than three times. Vesicle motility studies in axoplasm were analysed by two sample t-test of µ1-µ2 using DataDesk software or a one sample t and Wilcoxon test with GraphPad Prism software. Mouse quantitative data from immunoblotting and dot blots was analysed by unpaired T-test using GraphPad Prism Software. Human data analysis was performed using either unpaired T-test or Kruskal-Wallis one-way ANOVA with GraphPad Prism as indicated in the figure legends.
Results
Effects of N-terminal polyQ HTT fragments on fast axonal transport
Although HTT is a large protein (>3100 aa), the polyQ repeat is near the N-terminus, and fragments as small as the one encoded by exon 1 of the HTT gene suffices for toxicity in cell and animal models.11,28,29 The molecular basis of mHTT toxicity is unclear, but HTT contains multiple protein interaction motifs including SH3 binding domains, WW and HEAT motifs.30 As a result, HTT may interact with various proteins.31,32
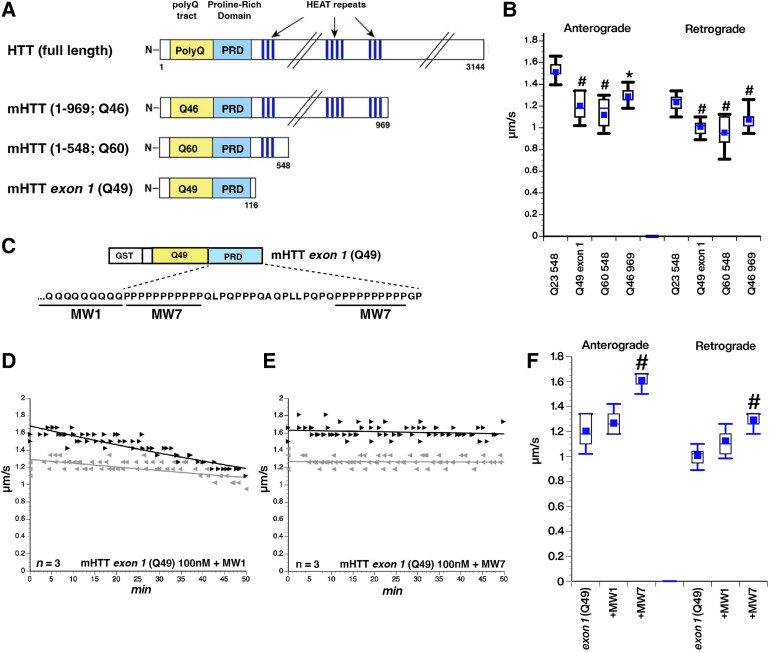
Vesicle motility assays in isolated axoplasm from squid giant axons previously revealed toxic effects of mHTT proteins, but not wild-type HTT proteins, on fast axonal transport,10 a major cellular process sustaining axonal function and health.33 Subsequent studies showed this effect involved abnormal activation of axonal JNKs and phosphorylation-based inhibition of molecular motors by the JNK3 isoform.11 To map discrete regions in mHTT involved in these toxic effects, we compared toxicity of different mHTT fragments.10 Three recombinant mHTT protein fragments containing different numbers of HEAT domains were evaluated in isolated axoplasm for effects on fast axonal transport using vesicle motility assays, as before11,20: mHTTs (1–969, Q46), mHTT (1–548, Q60), and mHTT exon 1 (Q49) (Fig. 1A). Despite differences in size as well as in the number and identity of HEAT domains, all three mHTT fragments had comparable inhibitory effects on both anterograde and retrograde fast axonal transport (Fig. 1B). A control HTT fragment, HTT (1–548, Q23), had no effect on fast axonal transport (Fig. 1B).
Figure 1.
Toxicity of huntingtin fragments on fast axonal transport. (A) Huntingtin (HTT) is a protein of unknown function containing more than 3000 amino acids. The absence of an identified catalytic activity suggests a role for HTT as a scaffold for unknown cellular components. While previous studies have determined that exon 1 is sufficient for neurotoxicity, HTT contains multiple potential protein interaction domains [e.g. polyQ tract, a proline-rich domain (PRD) and HEAT repeats], which could provide additional biological activities. Three different recombinant mHTT fragments ranging from approximately 100 to 1000 amino acids were generated to determine whether different mHTT fragments exhibit different toxicities on fast axonal transport in the isolated axoplasm model. (B) All three mutant HTT (mHTT) constructs perfused at 100 nM have the same inhibitory effect on both anterograde and retrograde fast axonal transport rates. #P < 0.0001; *P < 0.001. (C) Exon 1 of mHTT appears to contain the toxic element causing the inhibitory effect in axons. There are two major motifs in exon 1: the expanded polyQ tract and a PRD. Previous studies generated monoclonal antibodies (Abs) to polyQ (MW1) and to polyP (MW7). (D and E) Plots depicting results from vesicle motility assays in isolated squid axoplasm. mHTT exon 1 (49Q) was perfused into axoplasm (100 nM) and fast axonal transport rates monitored by video microscopy, as in B. Individual rate measurements (µm/s, arrowheads) are plotted as a function of time (min). Both anterograde (black arrowheads and lines) and retrograde (reverse grey arrowheads and lines) fast axonal transport rates are shown. (D) Preincubation of mHTT exon 1 (49Q) with MW1 Ab (polyQ-directed) did not alter the toxic effects of this protein, so both anterograde and retrograde transport were still inhibited. n = number of axoplasms evaluated. (E) In contrast, preincubation with MW7 (polyP directed) completely blocked the effects of HTT exon 1 (Q49) on axonal transport. This suggests that exposure and/or accessibility of the PRD is essential for mHTT toxicity on fast axonal transport. (F) Box plots show that preincubation with M1 Ab did not significantly alter the toxic effects of mHTT exon 1 (Q49) on either anterograde or retrograde fast axonal transport. In contrast, preincubation with MW7 significantly reduced such effects. #P < 0.0001.
Since mHTT exon 1 activated the same JNK MAPK cascade to inhibit fast axonal transport as longer mHTT constructs, it must contain the pathogenic element(s) in mHTT required for activation of the JNK pathway and inhibition of fast axonal transport. Exon 1 contains a short N-terminal sequence followed by the polyQ and an adjacent PRD, which includes two polyP stretches separated by an intervening 17 aa sequence that is 30% prolines (Fig. 1C). The effect of mHTT exon 1 (Q49) on fast axonal transport focused attention on these regions as potential mediators of pathogenicity.
To determine potential contributions of the polyQ and PRD regions to mHTT-induced fast axonal transport inhibition, antibodies (Ab) binding to either the polyQ (MW1 Ab) or polyP (MW7 Ab) domains17 were co-perfused with mHTT exon 1 (Q49), a strategy used before for other pathogenic proteins26,34 (Fig. 1C). If inhibition of fast axonal transport by mHTT involved interactions mediated by the polyQ, MW1 Ab would be protective, while MW7 Ab should be protective if interactions with the PRD are required. Preincubation with MW1 Ab did not prevent inhibition of fast axonal transport by mHTT exon 1 (Q49) (Fig. 1D), but preincubation with MW7 Ab completely abolished the toxic effect of this mHTT fragment (Fig. 1E). Boiling MW7 Ab prior to incubation with mHTT exon 1 (Q49) eliminated protective effects of MW7 (not shown). The protective effect of MW7 Ab was significant from mHTT exon 1 (Q49) alone at P < 0.0001 in a two sample t-test of µ1-µ2, while treatment with MW1 Ab was not (Fig. 1F). These findings suggested that accessibility to the PRD is critical for mHTT-mediated inhibition of fast axonal transport.
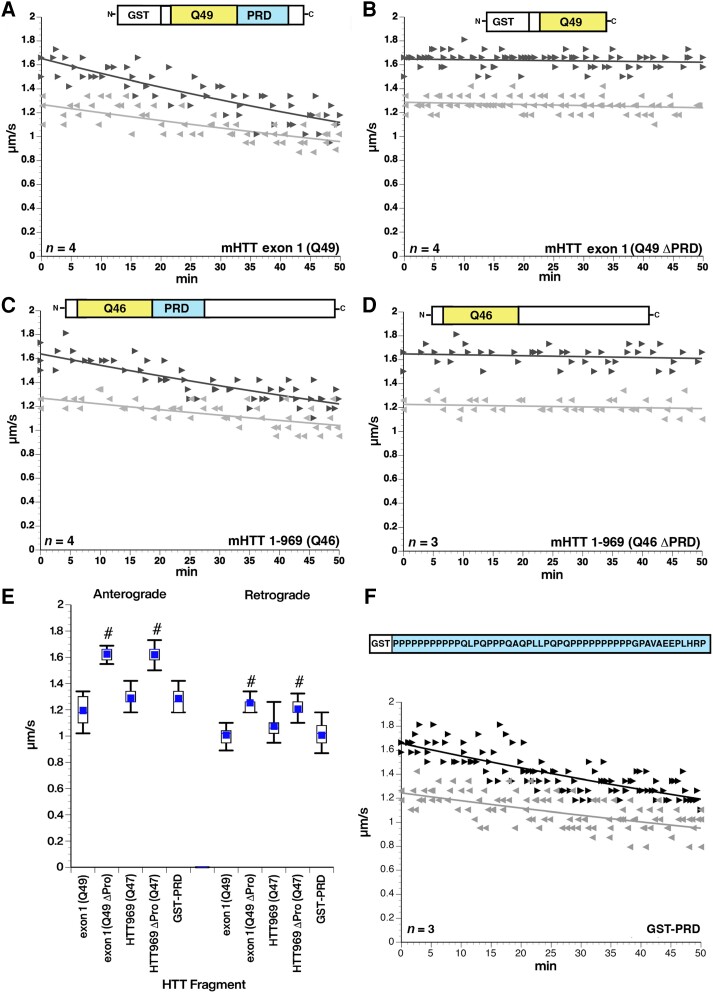
The requirement of PRD for mHTT toxicity on fast axonal transport was confirmed by protein deletion experiments (Fig. 2). Inhibition of fast axonal transport by mHTT exon 1 (Q49) protein (Fig. 2A) was no longer seen with mHTT exon 1 with PRD deletion (Q49 ΔPRD), despite the presence of a Q49 tract (Fig. 2C). Similar results were obtained with a larger mHTT fragment. Specifically, mHTT (1–969, Q46) inhibited fast axonal transport (Fig. 2B), whereas the same mHTT fragment with PRD deleted (1–969, Q46 ΔPRD) did not (Fig. 2D). Inhibitory effects of mHTT exon 1 (Q49) or mHTT 969 (Q46) were significantly different from their counterparts lacking a PRD at P < 0.0001 in a two sample t-test of µ1-µ2 (Fig. 2E), confirming that the PRD is necessary for mHTT to inhibit fast axonal transport (Fig. 2E). Remarkably, perfusion of a GST-tagged protein containing the wild-type PRD sequence (Fig. 2F) inhibited both directions of fast axonal transport, comparable to the effects of mHTT containing the PRD (Fig. 2E), and GST-PRD activated axonal JNK in biochemical experiments (Supplementary Fig. 7). While the pathogenic mutation in mHTT is an expansion of the polyQ tract, these results suggest that expanded polyQ tracts do not affect fast axonal transport in the absence of the PRD and that the PRD alone in the absence of an expanded polyQ tract has toxic effects on fast axonal transport comparable to mHTT.
Figure 2.
Deletion of the PRD eliminate toxic effects of mHTT fragments on fast axonal transport. (A) Perfusion of mHTT exon 1 (Q49) at 100 nM inhibits fast axonal transport in both directions through activation of the MAPK JNK3,35 (B) The same mHTT construct with the proline-rich domain (PRD) deleted [mHTT exon 1 (Q49 ΔPRD)] has no effect on axonal transport in either direction at 100 nM. (C and D) Similar effects are seen with a longer mHTT (1–969 Q46) fragment at 100 nM. Deletion of the PRD [mHTT (1–969 Q46 ΔPRD)] eliminates the toxic effects on fast axonal transport elicited by mHTT (1–969 Q46). (E) Quantitative comparison of rates observed between 30 and 50 min with each construct shows that the PRD is necessary for the toxicity of both mHTT exon 1 (Q49) and mHTT (1–969 Q46). #P < 0.0001. (F) A glutathione-S-transferase (GST) tagged protein in which the PRD from HTT is added to the C-terminus (GST-PRD) has the same effect on both anterograde and retrograde axonal transport without the presence of a polyQ stretch. The inhibitory effect of GST-PRD is not significantly different from the effects of pathogenic forms of HTT with an expanded polyQ. This suggests that the PRD is both necessary and sufficient to produce the effect of pathological mutant HTT on fast axonal transport. n = number of axoplasms.
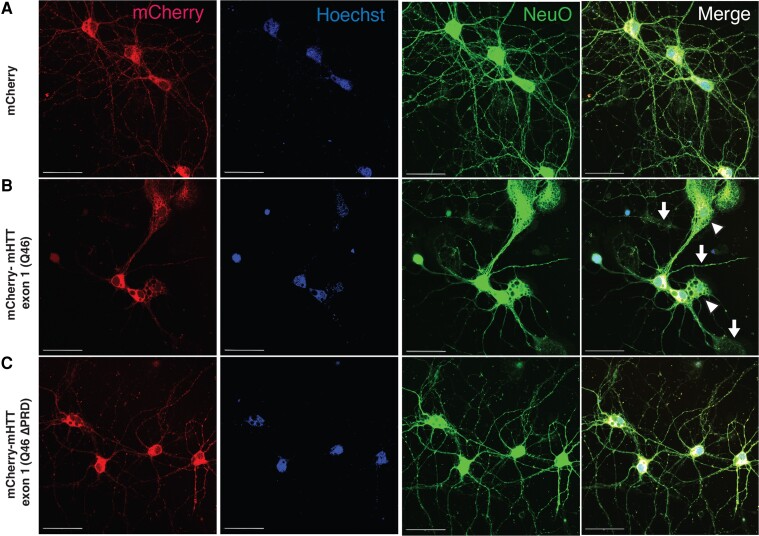
To corroborate axoplasm findings in mammalian neurons, rat primary cortical neurons were plated in chamber slides or custom microfluidic chambers and transfected in situ with mCherry- (Fig. 3) and Halo-tagged (Supplementary Fig. 1) versions of mHTT exon 1 (Q46) and mHTT exon 1 (Q46 ΔPRD). Changes in neuronal morphology, neuronal viability, axonal outgrowth and axonal degeneration in response to mHTT exon 1 (Q46) or mHTT exon 1 (Q46 ΔPRD) expression were examined using live cell imaging (Figs 3 and 4 and Supplementary Fig. 2) or by immunofluorescence using a Halo-ligand tag for mHTT (Supplementary Fig. 1). Live cortical neurons were visualized with NeuO, a fluorescent live cell imaging reagent that specifically labels neurons, Hoechst (for DNA) and mCherry. Neither repeated administration of NeuO (Supplementary Fig. 3) nor transfection with mCherry alone (Fig. 3A) affected neuronal morphologies.
Figure 3.
Toxic effects of mHTT-exon 1 (Q46) on axons of primary cultured cortical neurons depend on the PRD. (A) Cortical neurons, transfected to express mCherry (red), were also visualized with fluorescent membrane-permeant marker NeuO, a live neuron-specific dye (green), and a DNA label (Hoechst, in blue). (A) Cortical neurons were transfected with mCherry empty expression vector (mCherry, control); (B) mCherry fused to mHTT exon 1 (Q46) [mCherry-mHTT exon 1 (Q46)]; or (C) mCherry fused to mHTT exon 1 (Q46) with proline-rich domain (PRD) deletion [mCherry-mHTT exon 1 (Q46 ΔPRD)]. Cortical neurons expressing mCherry-mHTT exon 1 (Q46) displayed membrane/cytoplasmic morphological disturbances in both neurites (B, arrow) and soma (B, arrowheads), whereas neurons expressing mCherry-mHTT exon 1 (Q46 ΔPRD) did not. Scale bars = 50 μm. See also Supplementary Fig. 1.
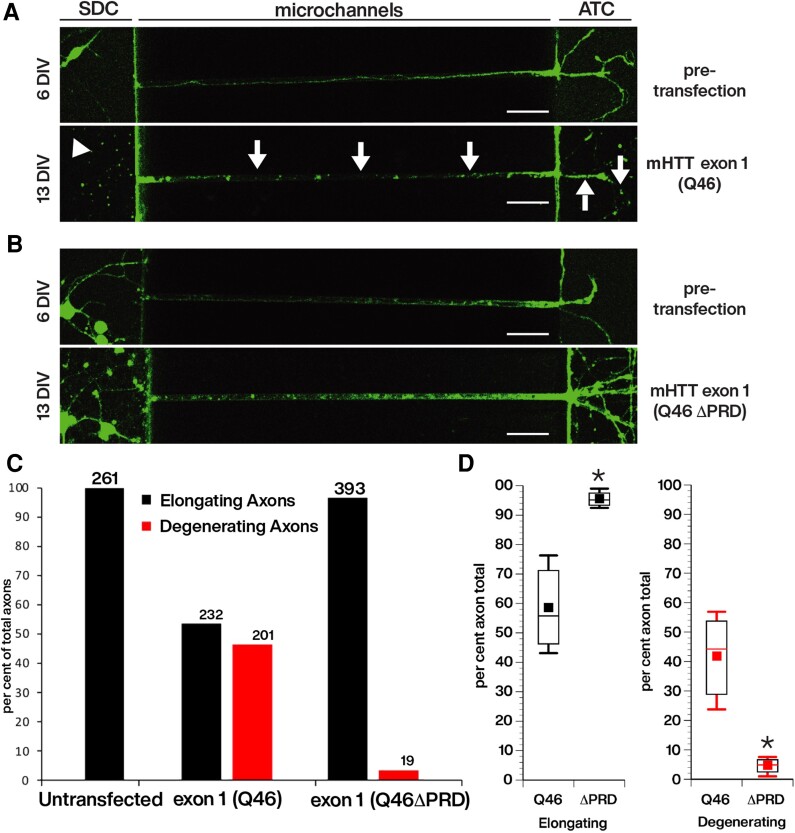
Figure 4.
Reduced outgrowth and degeneration of axons from primary cortical neurons following transfection with cherry-tagged mHTT exon 1, but not with cherry-tagged mHTT exon 1 lacking the proline-rich domain (PRD). Live imaging of axons elongating within the microchannels of our custom microfluidic chambers were visualized with the live neuronal marker, NeuO, beginning just prior to transfection at 6 days in vitro (DIV) after many axons have already entered the microchannels (Supplementary Fig. 2) and continuing until 13 DIV, or 7 days post-transfection. This design evaluates the effect of constructs on the growth and viability of pre-existing axons. Neurons were transfected with (A) mCherry-mHTT exon 1 (Q46) or with (B) mCherry-mHTT exon 1 (Q46 ΔPRD). The length of axons within each microchannel was measured as they entered the microchannel from the somatodendritic compartment (SDC) until it reached the axon terminal compartment (ATC). Cortical neurons expressing mCherry-mHTT exon 1 (Q46) developed axonal discontinuities, observed with the loss of NeuO labelling (A, arrows), illustrating axonal degeneration within the microchannel and in the ATC. There was an apparent reduction of cortical neuron somas in the SDC for cultured neurons transfected with mCherry-mHTT exon 1 (Q46) (A, arrowhead), but degeneration of neuronal soma was not quantitatively evaluated. Scale bar = 50 μm. (C) The total number of axons elongating and degenerating in cortical neurons expressing mHTT exon 1 (Q46) (n = 433) or mHTT exon 1 (Q46 ΔPRD) (n = 412) was quantified over time. Scale bars = 50 μm. Analysis of 261 axons of untransfected neurons had no degenerating axons. See also Supplementary Figs 2 and 3. (D) Comparison of axonal degeneration between neurons expressing mCherry-mHTT exon 1 (Q46) and mCherry-mHTT exon 1 (Q46 ΔPRD) indicates that the fraction of degenerating axons was reduced by >90% (P = 0.0067).
Cortical neurons were transfected after 6 DIV and morphology assessed at 6 DIV to compare effects of mCherrry-mHTT exon 1 (Q46) or mCherry-mHTT exon 1 (Q46 ΔPRD) expression to mCherry (control). Representative photomicrographs illustrate cortical neuron morphology labelled with NeuO (green), mCherry (red) and Hoechst (blue) are shown in Fig. 3. Cortical neurons expressing mCherry alone had normal morphologies (Fig. 3A), but neurons expressing mCherry-mHTT exon 1 (Q46) displayed shorter neurites, neuritic damage (Fig. 3B, arrows), and reduced somal size (Fig. 3B, arrowheads). In contrast, cortical neurons expressing mCherry-mHTT exon 1 (Q46 ΔPRD) (Fig. 3C) exhibited morphologies comparable to mCherry-expressing neurons. Concurrent studies using Halo-tagged versions of mHTT fragments produced the same results (Supplementary Fig. 1). The lack of toxicity for Q46 ΔPRD was consistent with the hypothesis that the PRD is also necessary to cause neurite degeneration in cortical neurons expressing Halo-tagged mHTT exon 1 (Q46) (Supplementary Fig. 1).
To differentiate whether the aberrant neuronal phenotypes associated with mHTT exon 1 (Q46) expression resulted from developmental delay or degeneration, the outgrowth and degeneration of axons within microchannels of custom microfluidic chambers was monitored and quantified over time. Cortical neurons were plated in SDCs, and axons allowed to elongate through the microchannels into the ATC (Fig. 4). Neurons were imaged at 6 DIV, before transfection with mHTT exon 1 (Q46) or exon 1 (Q46 ΔPRD), and changes in axonal outgrowth evaluated at 8, 11, and 13 DIV (Fig. 4A and B). Every microchannel was sequentially imaged to follow growth and degeneration of axons over time. Axons were measured from the time they entered microchannels projecting from the SDC until reaching the ATC (500 μm). Before transfection, neurons displayed normal axonal morphologies with axons elongating from SDC into the ATC (Fig. 4A and B, top). Seven days after transfection (13 DIV), there were dramatic differences in the extent of axonal outgrowth between neurons transfected with mHTT exon 1 (Q46) and mHTT exon 1 (Q46 ΔPRD) (Fig. 4A and B, bottom and Supplementary Fig. 3), as well as differences in the number of axons that converted from growing to degenerating (Fig. 4C and D). Specifically, neurons expressing mHTT exon 1 (Q46) showed loss of many neuronal cell bodies (Fig. 4A, arrowhead, bottom) and axonal discontinuities (arrows), shown by non-uniform NeuO staining within microchannels and ATC. In contrast, cortical neurons expressing mHTT exon 1 (Q46 ΔPRD) exhibited no loss of neuronal cell bodies or signs of axonal degeneration at 13 DIV (Fig. 4B, bottom). Instead, axons continued to elongate normally, as shown by axonal branching in the ATC. Axonal loss was also apparent at the ATC of neurons expressing Halo-tagged mHTT exon 1 (Q46) (Supplementary Fig. 3A and B) compared to extensive axonal outgrowth and branching of neurons expressing Halo-tagged mHTT exon 1 (Q46 ΔPRD) (Supplementary Fig. 3C and D).
Quantification of axonal outgrowth over time was binned to reflect the total number of axons elongating or degenerating (Fig. 4C). Elongating axons were defined as those that either continued to increase in outgrowth length within microchannels from 6 to 13 DIV or maintained their length within microchannels extending from SDC to ATC (Supplementary Fig. 2). Degenerating axons were defined as those that exhibited either a reduction in outgrowth length within the microchannels or axonal discontinuities. Of the 433 total axons (n = 3 experiments) measured over time for neurons expressing mHTT exon 1 (Q46), 232 axons elongated and 201 degenerated at 7 days post-transfection. In contrast, of 412 axons from neurons expressing mHTT exon 1 (Q46 ΔPRD) (n = 4 experiments), 393 axons elongated and only 19 exhibited signs of degeneration (Fig. 4C). The difference between mHTT exon 1 (Q49) and mHTT exon 1 (Q46 ΔPRD) was significant at P = 0.0067 in a pooled t-test of µ1-µ2 (Fig. 4D). No axonal degeneration was observed in control, untransfected neurons, where all 261 axons measured underwent elongation (Supplementary Fig. 2E). Thus, PRD deletion protected cortical neurons from mHTT exon 1 (Q46)-induced axonal degeneration, as seen with vesicle motility assays in axoplasm.
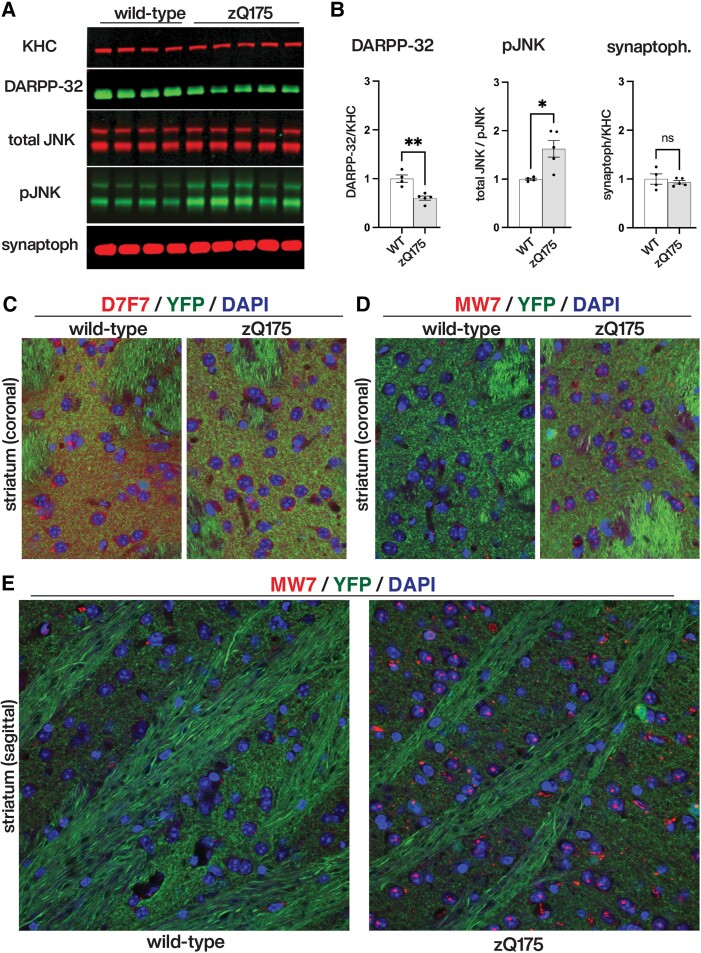
The fact that non-pathogenic (wild-type) HTT proteins did not elicit toxic effects on fast axonal transport or axonal outgrowth despite containing the PRD was intriguing, raising the possibility that polyQ tract expansion in mHTT might promote abnormal exposure of the toxic PRD.35 To evaluate this possibility, we performed biochemical and immunohistochemical experiments in striatal tissues of zQ175, a well-established mouse model of HD36 (Fig. 5). Striatal protein lysates prepared from wild type and zQ175 mice were separated by SDS-PAGE and western blots analysed by quantitative immunoblotting (Fig. 5A). Quantitative data from these experiments in Fig. 5B show reduced levels of the striatum-enriched protein DARPP32.37 As previously reported before for the HdhQ111 knock in mouse model of HD,11 these experiments also showed strong activation of JNKs in striatal tissues of zQ175mice, compared to wild-type mouse littermates, whereas total levels of kinesin heavy chains (KHC) and synaptophysin remained unchanged.
Figure 5.
Aberrant activation of JNKs and increased PRD exposure in the zQ175 mouse model of Huntington’s disease. (A) Immunoblots of striatal protein lysates prepared from wild type (n = 4) and heterozygous zQ175 (n = 5) mice were analysed with antibodies that recognize kinesin heavy chains (KHC), the striatum-enriched protein DARPP-32, total (total JNK) and active (pJNK) forms of JNKs, and synaptophysin (synaptoph). (B) Li-COR quantitation of immunoreactive bands in A revealed that, compared to wild-type mice, zQ175 mice striata has lower levels of striatal DARPP32 (DARPP32/KHC ratios), increased activation of JNKs (total JNK/pJNK ratios), and similar levels of synaptophysin (synaptoph/KHC ratios). (C–E) Immunohistochemical analyses of striatal tissues from YFP (control) and YFP-zQ175 mice littermates (n = 4 mice/genotype). Nuclei were stained using DAPI (in blue) in all panels. (C) An antibody that recognizes an epitope close to the P1218 residue in HTT (D7F7, in red) displayed increased immunoreactivity in YFP mice, compared to YFP-zQ175 mice littermates. This finding was confirmed by quantitative dot blot experiments in Supplementary Fig. 4A, suggesting reduced levels of total HTT expression in zQ175 mice, compared to wild-type (WT) mice. (D and E) Conversely, and despite this reduction, MW7 antibody immunoreactivity was markedly increased in striata of YFP-zQ175 mice, compared to YFP mice littermates. Results from quantitative dot blot experiments in Supplementary Fig. 4C supported and extended this finding. Collectively, data in C–E suggested increased exposure of the PRD in mHTT, compared to wild-type HTT.
To evaluate whether activation of JNKs was associated with increased exposure of PRD domain in mHTT, we performed immunohistochemistry experiments (Fig. 5). As done before for the R6/2 mouse model of HD,3 we first generated YFP (control) and YFP-zQ175 ‘reporter’ littermates, a genetic strategy that facilitates visualization of axonal fibres that pass through the striatum and neuropil that arrive from the cortex and other brain structures. Consistent with immunoblotting data in Fig. 5A, striatal sections prepared from YFP-zQ175 mice displayed reduced D7F7 antibody immunoreactivity, compared to sections prepared from YFP littermates (Fig. 5C). This observation was further confirmed quantitatively using dot blot analysis of striatal tissue lysates (Supplementary Fig. 4B). Despite lower levels of D7F7-immunoreactive HTT in YFP-zQ175 mice, there was a marked increase in MW7 immunoreactivity in these mice, compared to YFP littermates (Fig. 5D and E). These findings were confirmed by quantitative dot blot analysis of striatal lysates using antibodies MW7 (Supplementary Fig. 4C) and 4C9, another antibody whose epitope maps to the PRD (Supplementary Fig. 4C and D). Taken together, results from comparative analysis of wild-type and zQ175 mouse striata strongly supported increased exposure of the PRD in mHTT, compared to wild-type-HTT. Further, this event correlated with abnormal activation of JNKs.
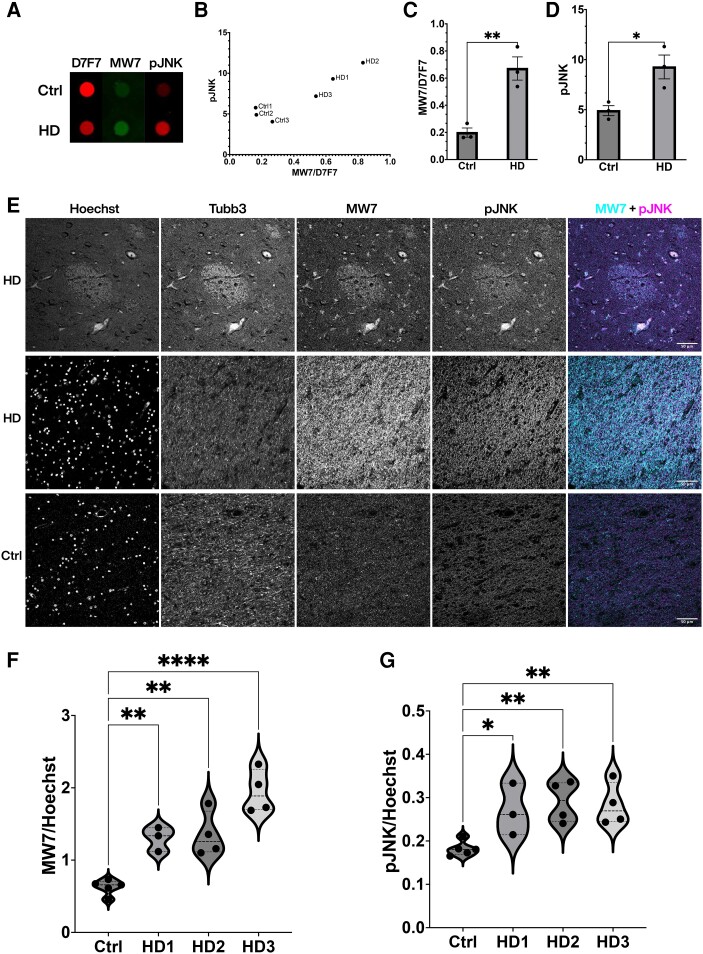
To evaluate further the disease relevance of findings from squid axoplasm (Figs 1 and 3), cultured mammalian neurons (Figs 3 and 4) and the zQ175 mouse model (Fig. 5 and Supplementary Fig. 4), PRD exposure and JNK activation were both examined in HD-affected human samples (Fig. 6 and Supplementary Fig. 5). Human post-mortem caudate tissues from HD patients (n = 3) and non-HD controls (n = 3, see Supplementary Table 2 for demographic information) were prepared and quantified using dot blot (Fig. 6A). While there was a moderate reduction in the total HTT protein level in HD cases (as indicated by D7F7 Ab immunoreactivity), PRD exposure and JNK activity increased as indicated by the MW7 and pJNK immunoreactivity ratios, respectively. When individual HD cases were examined (Fig. 6B), there was a correlation between JNK activity and PRD exposure, both of which were higher than non-HD controls (Fig. 6C and D). Consistent with the biochemistry results, the immunohistochemistry on patient FFPE sections showed higher MW7 and pJNK levels in HD striatum (n = 11) than in non-HD controls (n = 5) (Fig. 6E–G, see Supplementary Table 3 for demographic information). Increased PRD exposure and active JNK levels were detected in striosomes, axonal tracts, and nuclei, as demonstrated in confocal images (Fig. 6E) and enhanced resolution SIM images (Supplementary Fig. 5). These results are consistent with studies in the squid axoplasm, primary neurons, and HD mouse models, suggesting aberrant activation of JNK and increased exposure of PRD in human HD striatum.
Figure 6.
Aberrant activation of JNKs and increased PRD exposure in striatal tissue from HD patients. (A) A representative dot blot of human postmortem caudate tissue (see Supplementary Table 2 for patient demographic information) showed increased immunoreactivity of MW7 and pJNK in Huntington’s disease (HD) patients, despite a moderate reduction in the total level of HTT protein as detected by D7F7 antibody. (B) Quantification of dot blots was plotted for each case (n = 3 for control and n = 3 for HD) to demonstrate a correlation between MW7/D7F7 ratio and JNK activity in HD cases, with controls clustered at the lower quadruple. (C and D) Relative immunoreactivity of MW7 (shown as ratio of MW7/D7F7) and pJNK from the same blots was quantified and plotted to show increases in HD as compared with the control samples (**P = 0.0068, *P = 0.0276, unpaired two-sample T-test). (E) Representative confocal images of the striatum indicated associated and increased immunostaining of MW7 and pJNK in HD patients compared with their controls. Tissues were counterstained with Hoechst as a nuclear marker and Tubb3 as a neuronal marker. (F and G) Quantification was performed on whole striatum images stitched from single images captured by wide-field epifluorescence imaging and showed higher MW7 and pJNK immunoreactivities in HD patients (n = 11) than in controls (n = 5) and the differences were statistically significant (****P < 0.0001, **P < 0.01, *P < 0.05 Kruskal-Wallis one-way ANOVA). HD cases were graded according to neuropathology27 (see Supplementary Table 3 for patient demographic information of cases examined in E–G).
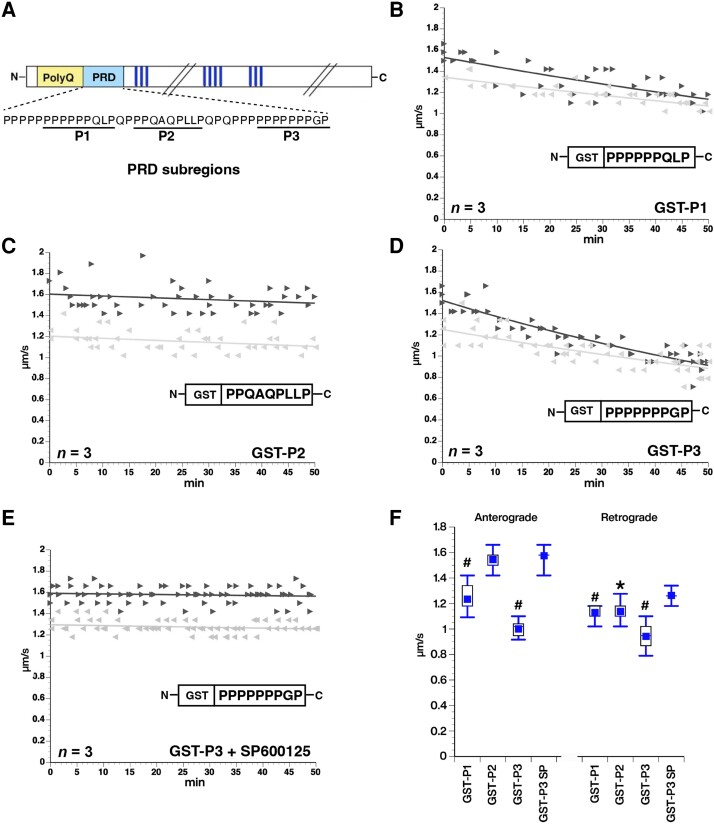
Given that the PRD alone was sufficient to inhibit fast axonal transport (Fig. 2F), and activate JNKs (Supplementary Fig. 7), we sought to refine the mapping of toxic elements within the PRD. GST-tagged proteins containing sequences corresponding to N-terminal (GST-P1), middle (GST-P2), and C-terminal (GST-P3) subregions of the PRD (Fig. 7A)24 were purified, perfused into axoplasm and their effects on fast axonal transport evaluated. Both GST-P1 and GST-P3 inhibited fast axonal transport (Fig. 7B and D), whereas GST-P2 had no effect (Fig. 7C). These effects did not involve the GST tag, as synthetic P1 or P3 peptides without GST elicited the same effects (see Supplementary Fig. 6A and B for results with P1). Notably, P1 and P3 include polyP repeats (Fig. 7A), which is the epitope recognized by the MW7 antibody that blocked the inhibition of fast axonal transport by mHTT exon1 (Q49) (Fig. 1E). To test whether P1 and P3 also inhibited fast axonal transport by activation of JNKs, as GST-PRD did, they were co-perfused with SP600125, a specific inhibitor of JNK family kinases. As previously reported for larger mHTT fragments,11,38 SP600125 prevented inhibition of fast axonal transport by both GST-P3 (Fig. 7E) and P1 (Supplementary Fig. 6C and D). Fast axonal transport rates in both directions were significantly lower after either GST-P1 or GST-P3 perfusion than those measured after non-toxic mHTT (1–548) Q23 perfusion at P < 0.0001 in a two sample t-test of µ1-µ2, whereas GST-P3 co-perfused with SP600125 was not (Fig. 7F). The effect of GST-P2 on anterograde fast axonal transport was not significantly different from that of the non-pathogenic mHTT (1–548) Q23 construct. Although GST-P2 retrograde axonal transport in the 30–50-min window was different from non-pathogenic mHTT (1–548) Q23 retrograde axonal transport (P = 0.001), there was no change over the course of 50 min (Fig. 7C), suggesting that GST-P2 did not inhibit retrograde axonal transport although basal retrograde axonal transport rates in these axoplasms were slower than typical. Collectively, data in Fig. 7 and Supplementary Fig. 6 suggest that P1 and P3 regions of mHTT contain toxic elements that suffice to inhibit both anterograde and retrograde fast axonal transport by activating the JNK pathway in axons.
Figure 7.
Subdomains P1 and P3 of the PRD suffice to inhibit fast axonal transport via JNKs. (A) The proline-rich domain (PRD) contains three subregions previously shown to interact with SH3 domain- and WW motif-containing proteins: a variable length polyP (P1), a proline rich sequence that also contains Q, L and A residues (P2) and a second polyP stretch (P3). GST-tagged versions of these subregions were perfused in axoplasm and their effect on fast axonal transport evaluated using vesicle motility assays as in Fig. 1. (B) Perfusion of a GST-P1 (100 nM) construct inhibited both anterograde and retrograde fast axonal transport rates. (C) In contrast, a GST-P2 (100 nM) construct has no effect on either direction of fast axonal transport. (D) Effects of the GST-P3 (100 nM) construct on fast axonal transport are comparable to P1, the full-length PRD alone (Fig. 1F) and HTT exon 1 Q49 with an intact PRD (Fig. 1). (E) SP600125 (500 nM), a highly specific pharmacological inhibitor of JNKs, blocked the effect of GST-P3 (100 nM) on transport, just as it does for mHTT exon 1 (Q49) or other pathogenic mHTT fragments. (F) Quantitative comparisons for effects of PRD subdomains on axonal transport. Effects of each peptide on transport were compared to effects of the control wild-type HTT 1–548 (Q23) polypeptide. GST-P1 and GST-P3 significantly inhibited both anterograde and retrograde fast axonal transport as shown in the box plots (P < 0.0001) and the slopes in B and D reflecting a decline in transport. In contrast, GST-P2 did not differ significantly from mHTT (548) Q23 in anterograde transport (P = 0.2599). Although retrograde fast axonal transport was lower than with HTT 1–548 (Q23) (P = 0.0014) (see Figure 1B), there was no decline in retrograde transport rate and GST-P2 started out less that HTT 1–548 (Q23) retrograde, so effects of GST-P2 on axonal transport are likely negligible. Inhibition of JNK MAPK activity by SP600125 prevented the effect so GST-P3 on fast axonal transport. #P < 0.0001, *P < 0.001.
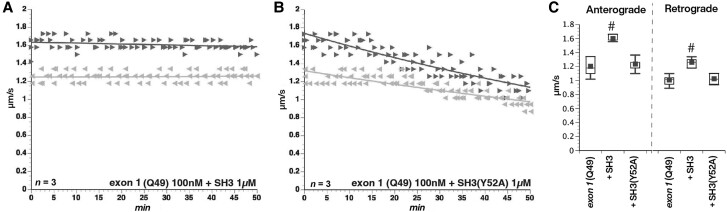
Proline-rich motifs are involved in a variety of protein-protein interactions, including many involved in signalling pathways.14 These include polyP helices, WW domains and SH3-binding domains, all present in the PRD of HTT. Both P1 and P3 have polyP motifs and MW7 Ab that recognizes the polyP motif in the PRD blocks inhibition of fast axonal transport by mHTT exon 1 (Q49) (Fig. 1E). P1 and P3 peptides also contain multiple SH3-binding domains, based on the consensus PXXP motif. To test the role of SH3-binding motifs in the toxicity of mHTT, we co-perfused axoplasm with mHTT exon 1 (Q49) (100 nM) plus 1 µM of a GST-tagged, recombinant SH3 domain (YANPVWTALFDYEPSGQDELALRKGDRVEVLSRDAAISGDEGWWAGQVGGQVGIFPSNYVSRGGG).39 In the presence of this SH3 domain, mHTT exon 1 (Q49) no longer inhibited fast axonal transport. In contrast, co-perfusion of mHTT exon 1 (Q49) with a mutant SH3 domain (Y52A) that fails to interact with SH3-binding domains40 did not prevent inhibition of fast axonal transport (Fig. 8B and C). These results suggested that binding of an SH3-containing protein to P1 and/or P3 subdomains is required for the PRD to activate the JNK pathway and inhibit fast axonal transport.
Figure 8.
Toxic effects of mHTT on fast axonal transport involve binding to SH3 domain-containing proteins. Proline-rich domains are implicated in a variety of protein–protein interaction domains. (A) SH3-binding domains are a common motif with a characteristic PXXP conserved sequence. The PRD contains multiple PXXP motifs, so we co-perfused mHTT exon 1 (Q49) (100 nM) with a peptide containing an SH3 motif at 1 µM. Excess SH3 peptide prevents mHTT exon 1 (Q49) effects on fast axonal transport. (B) In contrast, a mutant SH3 domain that no longer binds PXXP motifs (SH3-Y52A) at 1 µM fails to protect fast axonal transport. (C) Preincubation with a recombinant peptide encoding an SH3 motif blocked effects of mHTT exon 1 (Q49) on both anterograde and retrograde transport rates (P < 0.0001), whereas a mutant SH3 peptide unable to bind SH3-binding motifs (Y52A) did not (P = 0.41 for anterograde rates; P = 0.1964 for retrograde rates). #P < 0.0001.
Discussion
Homozygous deletion of the HTT gene is embryonic lethal41 suggesting that wild-type HTT is an essential gene for development,42 but physiological functions of HTT protein have not yet been established.43 For example, mHTT associates with vesicles44 and is reported to interact with a wide range of proteins including several SH3 domain-containing proteins involved in endocytosis and trafficking,45 but the role of these interactions in HD or normal functions of HTT is not known.32 HTT has been proposed to play roles in many cellular processes including gene transcription, BDNF trafficking46 and regulation of metabolism.47 While specific biological roles of HTT are poorly defined, the role of mHTT in HD is well established.1 Although a loss of HTT function may contribute to HD pathology, animal models have clearly established a toxic gain of function for mHTT in HD pathogenesis.48 Activation of a MAPK pathway leading to activation of JNKs, including JNK3, represents one toxic gain of function for mHTT.11,49-51 Dysregulation of JNKs could affect a wide range of cellular processes, including gene transcription52 and inhibition of fast axonal transport,10,11 but the question of how mHTT activates the JNK pathway remained unresolved.
Isolated axoplasm from squid giant axons has been used for decades to study fast axonal transport.20,53 Such studies illuminated the complexity of fast axonal transport regulation mediated by protein kinases and demonstrated a remarkable conservation in molecular mechanisms for regulation of fast axonal transport from squid to mammalian neurons.33,54 Isolated axoplasm provides a unique preparation20 for evaluating effects of pathogenic proteins and pharmacological reagents on fast axonal transport and axonal signalling as well as characterizing axon-specific candidate mechanisms.33,55
Given the large size of HTT and presence of multiple protein interaction motifs including HEAT repeats, WW domains and SH3 binding domains, the first question addressed was whether mHTT fragments of different sizes similarly affect JNK signalling and fast axonal transport. Three different fragments of mHTT, ranging in size from 118 to 969 aa, had comparable effects on fast axonal transport (Fig. 1A and B), and these effects were blocked by inhibiting JNK kinase activity.12 This was consistent with the finding that mHTT exon 1 was sufficient to produce neurodegeneration in cellular and animal models13,29 and focused attention on elements within mHTT exon 1. Exon 1 of the HTT gene encodes three well-defined domains: a 17-aa N-terminal sequence, the polyQ sequence and a PRD. The HEAT repeats present in full-length HTT29 are not in exon 1, indicating that they are not required for mHTT to inhibit fast axonal transport.
Historically, the polyQ repeat has been the primary focus of studies of HD pathology, given that polyQ expansion beyond Q36 causes HD. In addition, polyP repeats in the PRD have attracted some interest.56 To determine whether polyQ, polyP, or both are required for toxicity, we preincubated mHTT exon 1 (Q49) with antibodies whose epitopes mapped to either the polyQ or the polyP (Fig. 1D and E). Only the antibody against polyP blocked effects on fast axonal transport (Fig. 1E), indicating that access to the PRD was necessary for toxicity. This is consistent with findings that polyQ expansion increased exposure of the PRD in living cells.57 Deleting the PRD in mHTT fragments eliminated their effects on fast axonal transport (Fig. 2) which confirmed that the PRD was necessary for activation of JNKs and inhibition of fast axonal transport.
To validate this observation in mammalian models, we transfected rat primary cortical neurons with mHTT exon 1 (Q46) either with or without the PRD (Figs 3 and 4 and Supplementary Figs 1 and 3). Neurons expressing mHTT exon 1 (Q46) with an intact PRD exhibited axonal degeneration (Fig. 6 and Supplementary Fig. 3) and neurotoxicity (Supplementary Fig. 2), while neurons expressing mHTT exon 1 (Q46 ΔPRD) could not be distinguished from neurons transfected with an empty vector (Fig. 3) or with non-pathogenic wild-type HTT exon 1 (Q18) (Supplementary Fig. 1A). Thus, the PRD was necessary for neurotoxicity of mHTT exon 1 in both isolated axoplasm and mammalian cortical neurons. Furthermore, increased PRD exposure and aberrant JNK activation were confirmed in an HD mouse model (Fig. 5 and Supplementary Fig. 4) and in human post-mortem HD cases (Fig. 6 and Supplementary Fig. 5), by both biochemical and immunohistochemical methods. We also found a noticeable heterogeneity in human HD cases at different neuropathological stages, partially due to neuronal loss and structural changes in the axons and nuclei, as shown by the neuronal marker Tubb3 staining (Fig. 6E and Supplementary Fig. 5). However, the detailed characterization of individual cases at different stages is beyond the scope of the current study and is currently under active investigation.
Prolines are unique amino acids, with side chains cyclized onto the backbone nitrogen, restricting available conformations.14 PolyP domains tend to adopt a polyproline II (PPII) helix, an extended structure with 3 aa per turn.58 In a PPII helix, prolines form a hydrophobic strip on the helix surface and backbone carbonyls are conformationally restricted. As a result, PPII helices have easily accessible polar and hydrophobic surfaces for potential interacting partners (SH3, WW domains, etc.). PPII helices are commonly found on protein surfaces, where side chains are available to interacting partners, forming a stiff ‘sticky arm’ for binding other proteins.58,59
Evidence that the HTT PRD contains a PPII helix comes from a crystal structure of HTT exon 1 showing the first polyP repeat in HTT exon 1 as a PPII helix.60 Only the first polyP region (roughly corresponding to P1) was visible in the structure (Supplementary Fig. 8) and the PPII helix interacted with the polyQ tract through a region of polyQ in random coil conformation.60 These observations suggested that the PRD is normally sequestered in the HTT interior by normal length polyQ domains. Expansion of the polyQ domain might destabilize this interaction, increasing exposure of the PRD or PRD subregions. This would explain both why wild-type HTT with a normal length polyQ does not affect fast axonal transport despite the presence of a PRD; and the ability of anti-PolyP MW7, but not anti-PolyQ MW1 antibody, to prevent inhibition of fast axonal transport (Fig. 1D and E). This would also explain why mHTT, but not HTT, preferentially interacts with SH3 domain-containing proteins in affinity purification studies.24 Studies in 293 cells expressing mHTT transfected with single-chain variable regions of MW1 Ab and MW7 Ab found that intracellular expression of MW1 increased mHTT-induced cell death, whereas expression of MW7 reduced cell death.61 Further, MW7 and other intrabodies against epitopes in the PRD reduced neuronal toxicity induced by mHTT in a rat corticostriatal slice model of HD61 and in mouse models of HD.62 These authors suggested that MW7 effects were due to increased turnover of mHTT, but there is no evidence of mHTT turnover in axoplasm and inhibition of fast axonal transport was observed at sub-nanomolar concentrations of mHTT.11 In light of observations presented here, including data in Fig. 8, a more likely explanation is that MW7 reduced mHTT toxicity by preventing protein–protein interactions with the PRD.
SH3 and WW motifs are both involved in many proline-mediated interactions, particularly in cell signalling cascades.63 For example, Src is inactivated by intramolecular interaction of its SH3 domain with a proline-rich region.64 The preferred binding partner for SH3 domains is a domain with a PXXP core, but there is considerable variability in surrounding amino acids that confers specificity to specific SH3 domains.14 Multiple potential SH3-binding consensus sites are present in the PRD that may interact with various proteins containing SH3 domains, including some involved in activation of the JNK MAPK pathway.
NMR studies mapped specific interactions of HTT with the SH3 domain of SH3GL3 (endophilin) and a WW domain of the HYPA protein24 showing that HTT can interact directly with proteins containing SH3 or WW domains but did not determine whether these are physiological partners in vivo. Several additional proteins containing SH3 domains and WW motifs are reported to interact preferentially with mHTT over wild-type HTT, again supporting a scenario where PRD becomes more accessible with polyQ expansion.35
In an effort to map toxic components of the PRD more precisely, two subregions (P1 and P3) were found to inhibit fast axonal transport (Fig. 7), and their effects were blocked by a JNK inhibitor (Fig. 7E and Supplementary Fig. 7C and D), focusing attention on specific motifs within these peptides. Both P1 and P3 contain a polyP sequence and multiple PXXP motifs known to interact with SH3 binding domains. A recombinant protein encoding an SH3 domain blocked effects of mHTT exon 1 (Q49) on fast axonal transport (Fig. 8A), but a mutant version of this domain that cannot interact with SH3-binding motifs did not (Fig. 8B). Thus, the SH3 domain acted as a competitive inhibitor by preventing interactions of the PRD with one or more SH3 domain-containing protein(s). This suggests that the toxicity of polyQ-expanded mHTT involves aberrant exposure of one or more SH3-binding motifs in the PRD. Such exposure would allow mHTT interaction with SH3 domain-containing proteins that can activate a MAPK pathway leading to JNK3 activation.12 This could represent a long-sought mechanism by which expansion of the polyQ domain confers upon mHTT a hypermorphic, toxic gain of function contributing to HD pathogenesis.
One challenge presented by adult-onset neurodegenerative diseases like HD is the lack of specific molecular mechanisms to explain neurotoxicity. Although these diseases are typically associated with disease-specific misfolded proteins, there have been few indications of how such misfolded proteins produce a given disease.32 The evidence that misfolding of mHTT causes exposure of a biologically active SH3-binding motif in the PRD, which interacts with SH3 domains involved in activation of a JNK MAPK pathway, provides a mechanistic basis linking protein misfolding to a disease-specific toxic gain of function that helps explain several puzzling aspects of HD pathology and suggests novel avenues for therapeutic intervention.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Eric Wanker and Mariam DiFiglia for providing cDNA constructs encoding GST- and FLAG-tagged HTT proteins, respectively, as well as Dr Gosia Kokoszka for production of GST-PRD proteins and Bin Wang, who provided technical support for imaging and cell culture studies.
Contributor Information
Scott T Brady, Department of Anatomy and Cell Biology, University of Illinois at Chicago, Chicago, IL 60612, USA; Marine Biological Laboratory, Woods Hole, MA 02543, USA.
Nichole A Mesnard-Hoaglin, Department of Anatomy and Cell Biology, University of Illinois at Chicago, Chicago, IL 60612, USA.
Sarah Mays, Department of Anatomy and Cell Biology, University of Illinois at Chicago, Chicago, IL 60612, USA; Marine Biological Laboratory, Woods Hole, MA 02543, USA.
Mercedes Priego, Department of Anatomy and Cell Biology, University of Illinois at Chicago, Chicago, IL 60612, USA.
Joanna Dziechciowska, Department of Anatomy and Cell Biology, University of Illinois at Chicago, Chicago, IL 60612, USA.
Sarah Morris, Department of Anatomy and Cell Biology, University of Illinois at Chicago, Chicago, IL 60612, USA; Marine Biological Laboratory, Woods Hole, MA 02543, USA.
Minsu Kang, Department of Anatomy and Cell Biology, University of Illinois at Chicago, Chicago, IL 60612, USA; Marine Biological Laboratory, Woods Hole, MA 02543, USA.
Ming Ying Tsai, Department of Anatomy and Cell Biology, University of Illinois at Chicago, Chicago, IL 60612, USA.
Jennifer L Purks, Marine Biological Laboratory, Woods Hole, MA 02543, USA.
Alison Klein, Marine Biological Laboratory, Woods Hole, MA 02543, USA.
Angelica Gaona, Department of Neurology, Massachusetts General Hospital, Charlestown, MA 02129, USA.
Alexandra Melloni, Department of Neurology, Massachusetts General Hospital, Charlestown, MA 02129, USA.
Theresa Connors, Department of Neurology, Massachusetts General Hospital, Charlestown, MA 02129, USA.
Bradley Hyman, Department of Neurology, Massachusetts General Hospital, Charlestown, MA 02129, USA; Department of Neurology, Harvard Medical School, Boston, MA 02129, USA.
Yuyu Song, Marine Biological Laboratory, Woods Hole, MA 02543, USA; Department of Neurology, Massachusetts General Hospital, Charlestown, MA 02129, USA; Department of Neurology, Harvard Medical School, Boston, MA 02129, USA.
Gerardo A Morfini, Department of Anatomy and Cell Biology, University of Illinois at Chicago, Chicago, IL 60612, USA; Marine Biological Laboratory, Woods Hole, MA 02543, USA.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.
Funding
The authors would like to acknowledge funding from CHDI (grants #020311 and #A-11872), the Brain Research Foundation, 2007/2008 MBL fellowships a 2019 Whitman Fellowship from the Marine Biological Laboratory; National Institutes of Health grants R01NS023868, R01NS041170 and a Zenith Award from the Alzheimer’s Association to S.T.B., as well as a Jack Satter Foundation grant, Pape Adams Award, an AARG grant from the Alzheimer’s Association, NIH R21AG072516 and P30AG062421 Development Project Award to Y.S., and P30AG062421 support of the ADRC brain bank.
Competing interests
The authors report no competing interests
Supplementary material
Supplementary material is available at Brain online.
References
- 1. The Huntington's Disease Collaborative Research Group . A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993; 72:971–983. [DOI] [PubMed] [Google Scholar]
- 2. Gusella JF, MacDonald ME. Molecular genetics: Unmasking polyglutamine triggers in neurodegenerative disease. Nat Rev Neurosci. 2000;1:109–115. [DOI] [PubMed] [Google Scholar]
- 3. Gatto RG, Chu Y, Ye AQ, et al. . Analysis of YFP(J16)-R6/2 reporter mice and postmortem brains reveals early pathology and increased vulnerability of callosal axons in Huntington’s disease. Hum Mol Genet. 2015;2:5285–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han I, You Y, Kordower JH, Brady ST, Morfini GA. Differential vulnerability of neurons in Huntington’s disease: The role of cell type-specific features. J Neurochem. 2010;113:1073–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pouladi MA, Morton AJ, Hayden MR. Choosing an animal model for the study of Huntington’s disease. Nat Rev Neurosci. 2013;14:708–721. [DOI] [PubMed] [Google Scholar]
- 6. White JK, Auerbach W, Duyao MP, et al. . Huntingtin is required for neurogenesis and is not impaired by the Huntington’s disease CAG expansion. Nature Genet. 1997;17:404–410. [DOI] [PubMed] [Google Scholar]
- 7. Rattray I, Smith E, Gale R, Matsumoto K, Bates GP, Modo M. Correlations of behavioral deficits with brain pathology assessed through longitudinal MRI and histopathology in the R6/2 mouse model of HD. PLoS One. 2013;8:e60012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li JY, Plomann M, Brundin P. Huntington’s disease: A synaptopathy? Trends Mol Med. 2003;9:414–420. [DOI] [PubMed] [Google Scholar]
- 9. Rosas HD, Lee SY, Bender AC, et al. . Altered white matter microstructure in the corpus callosum in Huntington’s disease: Implications for cortical “disconnection”. research support, N.I.H., extramural. NeuroImage. 2010;49:2995–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Szebenyi G, Morfini GA, Babcock A, et al. . Neuropathogenic forms of huntingtin and androgen receptor inhibit fast axonal transport. Neuron. 2003;40:41–52. [DOI] [PubMed] [Google Scholar]
- 11. Morfini GA, You YM, Pollema SL, et al. . Pathogenic huntingtin inhibits fast axonal transport by activating JNK3 and phosphorylating kinesin. Nat Neurosci. 2009;12:864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gallo KA, Johnson GL. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat Rev Mol Cell Biol. 2002;3:663–672. [DOI] [PubMed] [Google Scholar]
- 13. Yang H, Yang S, Jing L, et al. . Truncation of mutant huntingtin in knock-in mice demonstrates exon1 huntingtin is a key pathogenic form. Nat Commun. 2020;11:2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kay BK, Williamson MP, Sudol M. The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. Faseb J. 2000;14:231–241. [PubMed] [Google Scholar]
- 15. Zhang H, Gallo KA. Autoinhibition of mixed lineage kinase 3 through its src homology 3 domain. J Biol Chem. 2001;276:45598–45603. [DOI] [PubMed] [Google Scholar]
- 16. Kay BK. SH3 domains come of age. FEBS Lett. 2012;586:2606–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ko J, Ou S, Patterson PH. New anti-huntingtin monoclonal antibodies: Implications for huntingtin conformation and its binding proteins. Brain Res Bull. 2001;56(3–4):319–329. [DOI] [PubMed] [Google Scholar]
- 18. Brady ST, Lasek RJ, Allen RD. Video microscopy of fast axonal transport in isolated axoplasm: A new model for study of molecular mechanisms. Cell Motil. 1985;5:81–101. [DOI] [PubMed] [Google Scholar]
- 19. Leopold PL, Lin J-W, Sugimori M, Llinas R, Brady ST. The nervous system of Loligo pealei provides multiple models for analysis of organelle motility. In: Abbott NJ, Williamson R, Maddock L, eds. Cephalopod neurobiology: Neuroscience studies in squid, octopus and cuttlefish. Oxford University Press; 1994:15–34. [Google Scholar]
- 20. Song Y, Kang M, Morfini G, Brady ST. Fast axonal transport in isolated axoplasm from the squid giant axon. Methods Cell Biol. 2016;131:331–348. [DOI] [PubMed] [Google Scholar]
- 21. Kang M, Baker L, Song Y, Brady ST, Morfini G. Biochemical analysis of axon-specific phosphorylation events using isolated squid axoplasms. Methods Cell Biol. 2016;131:199–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qin ZH, Wang Y, Sapp E, et al. . Huntingtin bodies sequester vesicle-associated proteins by a polyproline-dependent interaction. J Neurosci. 2004;24:269–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hollenbach B, Scherzinger E, Schweiger K, Lurz R, Lehrach H, Wanker EE. Aggregation of truncated GST-HD exon 1 fusion proteins containing normal range and expanded glutamine repeats. Philos Trans R Soc Lond B Biol Sci. 1999;354:991–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gao YG, Yan XZ, Song AX, et al. . Structural insights into the specific binding of huntingtin proline-rich region with the SH3 and WW domains. Structure. 2006;14:1755–1765. [DOI] [PubMed] [Google Scholar]
- 25. Combs B, Christensen KR, Richards C, et al. . FTLD mutant tau impairs axonal transport through a protein phosphatase 1gamma-dependent mechanism. J Neurosci. 2021;41:1914–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanaan NM, Morfini GA, Lapointe NE, et al. . Pathogenic forms of tau inhibit kinesin-dependent axonal transport through a mechanism involving activation of axonal phosphotransferases. J Neurosci. 2011;31:9858–9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP Jr. Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44:559–577. [DOI] [PubMed] [Google Scholar]
- 28. Huang B, Schiefer J, Sass C, Kosinski CM, Kochanek S. Inducing huntingtin inclusion formation in primary neuronal cell culture and in vivo by high-capacity adenoviral vectors expressing truncated and full-length huntingtin with polyglutamine expansion. J Gene Med. 2008;10:269–279. [DOI] [PubMed] [Google Scholar]
- 29. Mangiarini L, Sathasivam K, Seller M, et al. . Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87:493–506. [DOI] [PubMed] [Google Scholar]
- 30. Takano H, Gusella JF. The predominantly HEAT-like motif structure of huntingtin and its association and coincident nuclear entry with dorsal, an NF-kB/rel/dorsal family transcription factor. BMC Neurosci. 2002;3:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hosp F, Vossfeldt H, Heinig M, et al. . Quantitative interaction proteomics of neurodegenerative disease proteins. Cell Rep. 2015;11:1134–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tourette C, Li B, Bell R, et al. . A large scale huntingtin protein interaction network implicates rho GTPase signaling pathways in Huntington disease. J Biol Chem. 2014;289:6709–6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brady ST, Morfini GA. Regulation of motor proteins, axonal transport deficits and adult-onset neurodegenerative diseases. Neurobiol Dis. 2017;105:273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bosco DA, Morfini G, Karabacak NM, et al. . Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat Neurosci. 2010;13:1396–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Caron NS, Desmond CR, Xia J, Truant R. Polyglutamine domain flexibility mediates the proximity between flanking sequences in huntingtin. Proc Natl Acad Sci U S A. 2013;110:14610–14615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Southwell AL, Smith-Dijak A, Kay C, et al. . An enhanced Q175 knock-in mouse model of Huntington disease with higher mutant huntingtin levels and accelerated disease phenotypes. Hum Mol Genet. 2016;25:3654–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deng Y, Wang H, Joni M, Sekhri R, Reiner A. Progression of basal ganglia pathology in heterozygous Q175 knock-in Huntington’s disease mice. J Comp Neurol. 2021;529:1327–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morfini G, Pigino G, Szebenyi G, You Y, Pollema S, Brady ST. JNK mediates pathogenic effects of polyglutamine-expanded androgen receptor on fast axonal transport. Nat Neurosci. 2006;9:907–916. [DOI] [PubMed] [Google Scholar]
- 39. Kokoszka ME, Kall SL, Khosla S, McGinnis JE, Lavie A, Kay BK. Identification of two distinct peptide-binding pockets in the SH3 domain of human mixed-lineage kinase 3. J Biol Chem. 2018;293:13553–13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Weng Z, Rickles RJ, Feng S, et al. . Structure-function analysis of SH3 domains: SH3 binding specificity altered by single amino acid substitutions. Mol Cell Biol. 1995;15:5627–5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nasir J, Floresco SB, O’Kusky JR, et al. . Targeted disruption of the Huntington’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. [DOI] [PubMed] [Google Scholar]
- 42. Liu JP, Zeitlin SO. Is huntingtin dispensable in the adult brain? J Huntingtons Dis. 2017;6:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bates GP, Dorsey R, Gusella JF, et al. . Huntington disease. Nat Rev Dis Primers. 2015;1:15005. [DOI] [PubMed] [Google Scholar]
- 44. DiFiglia M, Sapp E, Chase K, et al. . Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. [DOI] [PubMed] [Google Scholar]
- 45. Truant R, Atwal R, Burtnik A. Hypothesis: Huntingtin may function in membrane association and vesicular trafficking. Biochem Cell Biol. 2006;84:912–917. [DOI] [PubMed] [Google Scholar]
- 46. Zuccato C, Cattaneo E. Huntington’s disease. Handb Exp Pharmacol. 2014;220:357–409. [DOI] [PubMed] [Google Scholar]
- 47. Duan W, Jiang M, Jin J. Metabolism in HD: Still a relevant mechanism? Mov Disord. 2014;29:1366–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rubinsztein DC. Lessons from animal models of Huntington’s disease. Trends Genet. 2002;18:202–209. [DOI] [PubMed] [Google Scholar]
- 49. Perrin V, Dufour N, Raoul C, et al. . Implication of the JNK pathway in a rat model of Huntington’s disease. Research support, non-U.S. Gov’t. Exp Neurol. 2009;215:191–200. [DOI] [PubMed] [Google Scholar]
- 50. Taylor DM, Moser R, Regulier E, et al. . MAP kinase phosphatase 1 (MKP-1/DUSP1) is neuroprotective in Huntington’s disease via additive effects of JNK and p38 inhibition. J Neurosci. 2013;33:2313–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Garcia M, Charvin D, Caboche J. Expanded huntingtin activates the c-jun terminal kinase/c-jun pathway prior to aggregate formation in striatal neurons in culture. Neuroscience. 2004;127:859–870. [DOI] [PubMed] [Google Scholar]
- 52. Mielke K, Herdegen T. JNK and p38 stresskinases–degenerative effectors of signal-transduction-cascades in the nervous system. Prog Neurobiol. 2000;61:45–60. [DOI] [PubMed] [Google Scholar]
- 53. Brady ST, Lasek RJ, Allen RD. Fast axonal transport in extruded axoplasm from squid giant axon. Science. 1982;218:1129–1131. [DOI] [PubMed] [Google Scholar]
- 54. Gibbs KL, Greensmith L, Schiavo G. Regulation of axonal transport by protein kinases. Trends Biochem Sci. 2015;40:597–610. [DOI] [PubMed] [Google Scholar]
- 55. Morfini GA, Burns M, Binder LI, et al. . Axonal transport defects in neurodegenerative diseases. J Neurosci. 2009;29:12776–12786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Khoshnan A, Ko J, Patterson PH. Effects of intracellular expression of anti-huntingtin antibodies of various specificities on mutant huntingtin aggregation and toxicity. Proc Natl Acad Sci U S A. 2002;99:1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Caron NS, Hung CL, Atwal RS, Truant R. Live cell imaging and biophotonic methods reveal two types of mutant huntingtin inclusions. Hum Mol Genet. 2014;23:2324–2338. [DOI] [PubMed] [Google Scholar]
- 58. Rath A, Davidson AR, Deber CM. The structure of “unstructured” regions in peptides and proteins: Role of the polyproline II helix in protein folding and recognition. Biopolymers. 2005;80(2–3):179–185. [DOI] [PubMed] [Google Scholar]
- 59. Adzhubei AA, Sternberg MJ, Makarov AA. Polyproline-II helix in proteins: Structure and function. J Mol Biol. 2013;425:2100–2132. [DOI] [PubMed] [Google Scholar]
- 60. Kim MW, Chelliah Y, Kim SW, Otwinowski Z, Bezprozvanny I. Secondary structure of huntingtin amino-terminal region. Structure. 2009;17:1205–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Southwell AL, Khoshnan A, Dunn DE, Bugg CW, Lo DC, Patterson PH. Intrabodies binding the proline-rich domains of mutant huntingtin increase its turnover and reduce neurotoxicity. J Neurosci. 2008;28:9013–9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Southwell AL, Ko J, Patterson PH. Intrabody gene therapy ameliorates motor, cognitive, and neuropathological symptoms in multiple mouse models of Huntington’s disease. J Neurosci. 2009;29:13589–13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Li SS. Specificity and versatility of SH3 and other proline-recognition domains: Structural basis and implications for cellular signal transduction. Biochem J. 2005;390(Pt 3):641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Roskoski R Jr. Src protein-tyrosine kinase structure and regulation. Biochem Biophys Res Commun. 2004;324:1155–1164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material.