Abstract
Estrogen Receptor 1 (ESR1; also known as ERα, encoded by ESR1 gene) is the main driver and prime drug target in luminal breast cancer. ESR1 chromatin binding is extensively studied in cell lines and a limited number of human tumors, using consensi of peaks shared among samples. However, little is known about inter-tumor heterogeneity of ESR1 chromatin action, along with its biological implications. Here, we use a large set of ESR1 ChIP-seq data from 70 ESR1+ breast cancers to explore inter-patient heterogeneity in ESR1 DNA binding to reveal a striking inter-tumor heterogeneity of ESR1 action. Of note, commonly shared ESR1 sites show the highest estrogen-driven enhancer activity and are most engaged in long-range chromatin interactions. In addition, the most commonly shared ESR1-occupied enhancers are enriched for breast cancer risk SNP loci. We experimentally confirm SNVs to impact chromatin binding potential for ESR1 and its pioneer factor FOXA1. Finally, in the TCGA breast cancer cohort, we can confirm these variations to associate with differences in expression for the target gene. Cumulatively, we reveal a natural hierarchy of ESR1–chromatin interactions in breast cancers within a highly heterogeneous inter-tumor ESR1 landscape, with the most common shared regions being most active and affected by germline functional risk SNPs for breast cancer development.
The Estrogen Receptor 1 (ESR1; also known as ERα, encoded by the ESR1 gene) is the driving force in most breast cancers diagnosed in women—as well as men—worldwide (Waks and Winer 2019). As such, ESR1 is considered the critical drug target in both the adjuvant and metastatic phase of the disease, but resistance to hormonal treatment is common (Early Breast Cancer Trialists’ Collaborative Group [EBCTCG] 2005). ESR1 serves as a hormone-dependent transcription factor (TF), associating to DNA regulatory elements upon ligand-mediated activation to drive activity of responsive genes, ultimately giving rise to tumor growth. DNA binding sites for ESR1 are enriched for the palindromic DNA sequence AGGTCAnnnTGACCT, termed estrogen response elements (EREs), through which the ESR1 homodimer directly interacts with the DNA (Coons et al. 2017). To study ESR1 DNA action in a genome-wide and comprehensive fashion, chromatin immunoprecipitation followed by sequencing (ChIP-seq) was used to profile the receptor's DNA binding pattern in breast cancer cell lines as well as in tumors. From these studies, we now know that the vast majority of ESR1 binding sites are found further away from the genes they control (Carroll et al. 2006), and ∼95% of all ESR1 chromatin binding is found at distal intergenic regions or introns (Carroll et al. 2005; Ross-Innes et al. 2012), positive for the classical enhancer marks H3K27ac and EP300 (Carroll et al. 2006). Notably, the total number of ESR1 sites found at putative enhancers greatly outnumber the genes they control (Fullwood et al. 2009), implying a level of functional redundancy or cooperative action between ESR1 sites that is still poorly understood.
Previous studies have identified ESR1 binding patterns that characterize response to hormonal treatment or prognostication by sex. In cell lines, numerous studies report on plasticity in ESR1 DNA binding in endocrine therapy–sensitive MCF-7 cells, as well as their treatment-resistant derivatives (Hurtado et al. 2011; Ross-Innes et al. 2012; Martin et al. 2017). In patients, Ross-Innes et al. (2012) first reported on distinct ESR1 binding profiles and associated gene expression between patients with good (ESR1+/PGR+) or poor (ESR1+/PGR−) clinical outcome, which associated with FOXA1-mediated ESR1 cistromic reprogramming. Later, our team reported an ESR1 ChIP-seq-based classifier using primary tumor specimens, capable to identify a breast cancer patient's response to aromatase inhibitor (AI) treatment in the metastatic setting (Jansen et al. 2013). Inter-tumor variability in ESR1 DNA binding has also been described to decrease upon neoadjuvant tamoxifen treatment, and again associated binding sites were able to stratify patients on outcome (Severson et al. 2016). ESR1 DNA binding profiles were not found to differ greatly between male or female breast cancer patients, although sites associated with patient outcome were sex specific (Severson et al. 2018). These studies highlight the value of characterizing ESR1 DNA binding between clinically distinguishable groups of patients. However, even within the studied groups, a large variation in ESR1 DNA binding was observed, of which the biological and clinical implications remain unknown.
Because of the observed inter-sample heterogeneity of ESR1 peaks, downstream analyses typically rely on a consensus of peaks. Whether a peak is included in a consensus relies on an (often arbitrarily chosen) threshold for the minimum number of tumors in which the peak is detected. This leaves large amounts of potentially interesting data unused. To the contrary, here we set out to explore inter-patient heterogeneity in ESR1 binding in more detail. We evaluate the biology underlying inter-tumor cistromic heterogeneity of ESR1, in relation to genomic locations and germline variations between breast cancer tumors, to better understand the possible biological implications thereof.
Results
Putative enhancers represent the largest source of inter-patient heterogeneity in ESR1–chromatin interactions
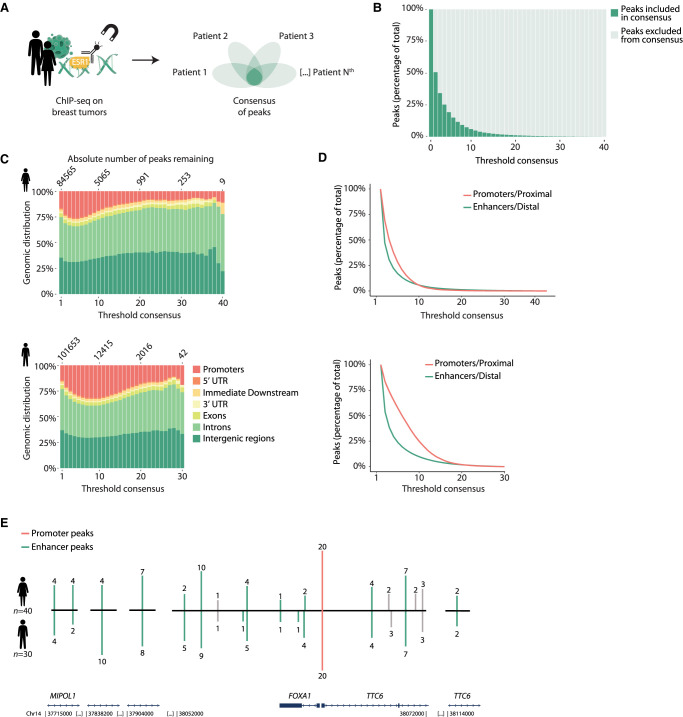
To identify the level of ESR1 chromatin binding heterogeneity in human breast cancer specimens, we used ESR1 ChIP-seq data from 40 female breast cancer patients (Fig. 1A; Supplemental Table S1). Five newly generated ESR1 ChIPs on female tumors were added to this study. The remaining 35 female samples have been described and analyzed, in previous publications, to identify genomic regions that could stratify patients on outcome or sex (Supplemental Table S1; Ross-Innes et al. 2012; Jansen et al. 2013; Severson et al. 2018). Further, in parallel, we used an independent cohort of 30 ESR1+ male breast cancer patients (Supplemental Table S1; Severson et al. 2018) as a validation cohort. All samples were reanalyzed and processed with the same bioinformatics pipeline (for details, see Methods).
Figure 1.
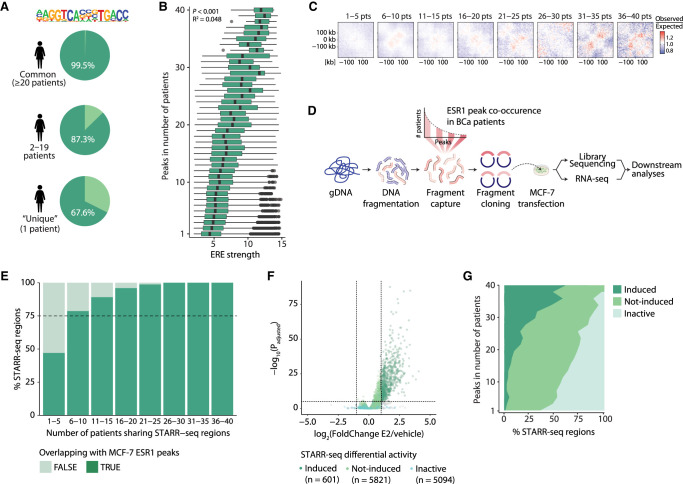
The largest inter-patient heterogeneity in ESR1 chromatin binding is found at putative enhancers. (A) Graphical representation of study design. ESR1 ChIP-seq on tumor samples from 30 male and 40 female breast cancer patients analyzed for the level of overlap and biological features. For sample details, see Supplemental Table S1. (B) Percentage of ESR1 peaks included or excluded in consensus, by varying the threshold of minimal overlap of peaks between female patients. (C) Genomic distribution of ESR1 consensus by varying threshold in females. (D) Percentage of distal and proximal regions retained by varying threshold for consensus in females. (E) ESR1 binding sites in the vicinity of FOXA1, showing the number of patients in which these peaks were called. Green lines represent enhancer regions; red line indicates promoter. Enhancer regions were coupled to FOXA1 on the basis of work by Corces et al. (2018). Gray lines represent peaks that were not coupled to FOXA1 on the basis of work by Corces et al. (2018), but these are shown for completeness as they were located in between peaks that were coupled to FOXA1.
Analyzing these samples, we observed a high level of inter-tumor heterogeneity of ESR1 binding, with the vast majority of ESR1 sites being poorly conserved between tumors from female patients (Fig. 1B). Of note, we were able to identify this high level of heterogeneity despite considering two regions as overlapping with as little as only one base being shared. If we were to reevaluate the aforementioned data by consensus with a lenient threshold of peaks present in at least two patients, 50% of data would be ignored. Previous analyses have been performed with cutoffs as stringent as peaks found in 75% of patients (Ross-Innes et al. 2012). Applying this cutoff to this cohort, merely 0.3% (for female tumors) and 1.1% (for male tumors) of sites would remain. In both female and male breast cancer patients, the level of conservation for ESR1 sites depends on the genomic distribution of the peakset (Fig. 1C), as promoter binding events are more conserved between individual tumors than are those of distal ESR1 binding sites (Fig. 1D).
For both sexes, we mapped all ESR1 binding events to genes using promoter–enhancer loops defined by Corces et al. (2018). Interestingly, the gene with the highest number of distal ESR1 binding events identified in our patient cohorts was FOXA1. FOXA1 is the classical pioneer factor, essential for ESR1 to facilitate its chromatin binding (Hurtado et al. 2011). Although the FOXA1 promoter was ESR1-occupied in most samples, ESR1 binding at putative regulatory elements surrounding the FOXA1 locus varied strongly between patients (Fig. 1E).
Common peaks represent 30% of ESR1 binding in a patient ChIP sample
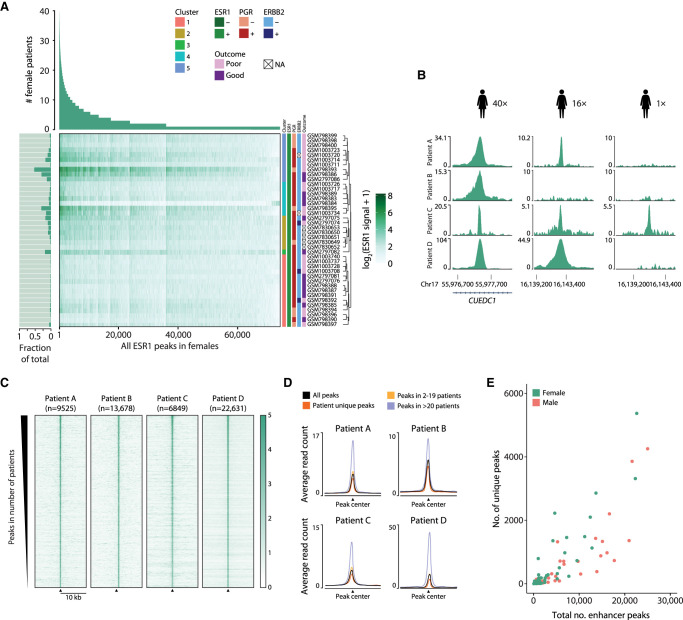
To better appreciate the functional implications of enhancer heterogeneity among patients, we removed all ESR1 binding events at promoters and ranked all distal ESR1 binding events (74,438 in females, 91,712 in males) from common among patients to patient-unique events (Fig. 2A; Supplemental Fig. S1A; Supplemental Tables S2, S3). Fifty-three percent of all ESR1-bound putative enhancers found in female tumors were patient unique, 46% of sites were found in two to 19 female patients, and merely 1.1% were found in more than half of the female cohort (Fig. 2A). Similarly, among males, inter-patient heterogeneity was large, with 46.3% of distal ESR1 binding events being patient unique, 49.5% found in two to 14 patients, and merely 4.3% found in more than half of all male tumors analyzed (Supplemental Fig. S1A).
Figure 2.
Characterization of enhancers ranked from commonly to less frequently bound by ESR1 shows distinct biological features. (A) A ranked overview of 74,438 distal ESR1 peaks showing in how many tumor samples each peak was found in a cohort of 40 female patients. Heatmap showing the average ESR1 ChIP-seq score at a specific peak for each sample. The bar plot (left) indicates the fraction of peaks found in each patient of the total peaks found. Clustering is based on the Pearson correlation at ESR1 peaks for the ESR1 ChIP-seq signal as defined in Supplemental Figure S2A. (B) Examples of ESR1 peaks that were peak-called in tumor samples in all 40 females (left), in 16 females (middle), and in only one female patient (right). (C) Examples of per-patient heatmaps of ESR1 signal, of peaks called in that female patient sample, ranked as in A. (D) For more commonly occurring and unique peaks, examples of the average intensity of ESR1 ChIP-seq signal in four female patients are shown. (E) Correlation plot of the total number of distal peaks in a patient sample (x-axis), versus the percentage of patient-unique peaks in that sample (y-axis).
Differences in peak conservation between patients were not owing to subtle differences in peak calling performance, as partially shared ESR1 sites were genuinely differentially enriched between tumors (Fig. 2B). On patient level, the signal intensity at ESR1 sites is higher at common compared with patient-unique peaks (Fig. 2C,D), with highly common peaks (20 or more patients) showing the strongest signal.
For each patient in our cohort, we looked into the amount of ESR1 ChIP-seq peaks picked up in their respective tissue sample (Supplemental Fig. S1B). When viewing the list of all distal ESR1 ChIP-seq peaks found in a single patient, the minority of that list is made up of peaks that we consider common among individuals in the cohort: 29.3% (in females) and 42% in males (Supplemental Fig. S1C). This implies that the body of ESR1 DNA binding events in a single patient sample occurs at regions considered less common among individuals. Thus, as previous studies have applied stringent consensus-based approaches to identify peaks common among individuals, they do not take into account most of the ESR1 binding in a single individual. Or in other words, a consensus analyzes ESR1 on group level but may inform less on ESR1 behavior in an individual patient.
In an individual patient, peaks that are unique to that patient make up 13.3% (in females) and 8.9% (in males) of all distal ESR1 peaks found in that tissue sample (Supplemental Fig. S1C). Although generally weaker, signal at patient-unique sites is only slightly lower than the average signal for that particular tumor (Fig. 2D). The number of patient-unique peaks in a patient sample increases with the total number of distal peaks picked up in that patient sample (Fig. 2E; Supplemental Fig. S1B,C), implying unique peaks are not a proxy of relatively poor ChIP-seq quality.
Cumulatively, these data confirm a remarkable inter-patient heterogeneity of ESR1 enhancer action that is typically overlooked in consensus-based analyses.
ESR1 cistromic heterogeneity is independent of patient pathological features
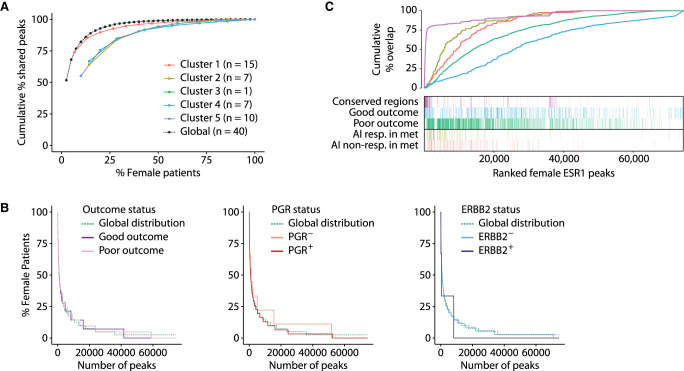
We explored putative biological features that might explain the observed ESR1 peak heterogeneity by clustering the patients by Pearson correlation coefficient of ChIP-seq signal at ESR1 enhancer peaks (Supplemental Fig. S2). In doing so, we defined five clusters based on the unsupervised hierarchical clustering and analyzed the ESR1 binding heterogeneity within each cluster (Fig. 3A; Supplemental Figs. S2, S3A). All the clusters—except for female cluster 1—displayed a lower heterogeneity compared with the global trend, showing that molecularly similar samples share a larger fraction of peaks. Further, we analyzed the clinicopathological and molecular characteristics (Supplemental Table S4) for both the female (Supplemental Figs. S2A, S3B) and male (Supplemental Figs. S2B, S3C) samples. For the female samples, these analyses did not highlight any biological feature that would explain the separation in different clusters (Supplemental Fig. S2A). On the other hand, for the male samples, we identified the male clusters 1, 3, and 4 to be enriched for a specific patient outcome (Supplemental Fig. S3C). However, stratification of the samples by PGR (also known as PR), ERBB2 (also known as HER2), or outcome status was not sufficient to explain the observed ESR1 heterogeneity among the samples (Fig. 3B; Supplemental Fig. S3D).
Figure 3.
ESR1 heterogeneity in female patients is not explained by molecular and clinical features. (A) Cumulative percentage of ESR1 peaks shared among female patients within the five clusters defined in Supplemental Figure S2A. (B) Percentage of female patients sharing ESR1 within groups of patients based on outcome, PGR, and ERBB2 status. The global distribution over all the peaks is depicted by a dashed green line. (C) Cumulative percentages and heatmap of the overlaps between ranked female ESR1 patients and good/poor outcome-associated (Ross-Innes et al. 2012) or aromatase inhibitor (AI) response–associated ESR1 peaks (Jansen et al. 2013).
Our observations that patient outcome in females cannot explain the ESR1 heterogeneity corroborate with the observation that outcome-associated peaks (Ross-Innes et al. 2012) are not specifically enriched with the more common peaks (Fig. 3C). On the other hand, peaks associated to AI response in the metastatic setting (Jansen et al. 2013) were particularly found at the more commonly shared peaks (Fig. 3C).
In summary, we showed that female patients cannot be stratified by their molecular or clinicopathological features, and also less-conserved peaks carry prognostic information on patient outcome.
ESR1 cistrome converges to common gene regulation programs
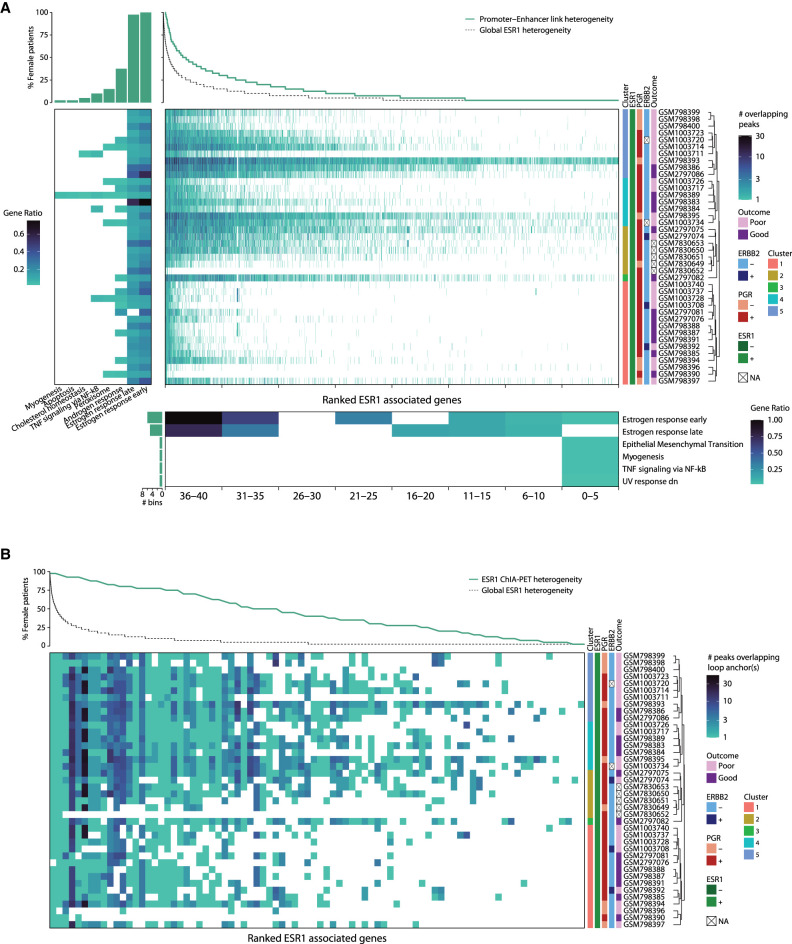
Because clinicopathological features of the patients do not explain ESR1 binding heterogeneity, we hypothesized that ESR1 occupancy, despite being highly variable between patients, might occur at regions that regulate similar cellular programs. Of note, only eight peaks were found being conserved among all female patients (Fig. 2A) and 34 in all male patients (Supplemental Fig. S1A). However, analyzing the genes associated to each ESR1 peak in each sample, using the promoter–enhancer linkage data as defined by Corces et al. (2018), we found that the ESR1 profiles for all patients were enriched for gene signatures associated to estrogen-receptor response (Fig. 4A; Supplemental Fig. S4A). Further, we considered the genes linked with ESR1 peaks depending on the degree with which these peaks are shared among patients, and we identified the estrogen-receptor response to be among the top enriched signatures also in this case (Fig. 4A; Supplemental Fig. S4A). A few well-known estrogen-related genes are proximal to top common peaks, including PGR, RARA, IGFBP4, CUEDC1, and GREB1 (Supplemental Table S3). Common peaks relate to canonical early and late estrogen response genes more frequently (Fig. 4A; Supplemental Fig. S4A), although less common and unique peaks were also associated to classical estrogen-related signaling genes such as CCND1 and TFF1 (Supplemental Table S3). Interestingly, the percentage of putative ESR1-regulated genes shared among patients is higher than the percentage of shared ESR1 peaks (Fig. 3C; Supplemental Fig. S3C). To confirm these observations, we performed similar analyses using physical enhancer–promoter chromatin loops detected by ESR1–chromatin interaction analysis with paired-end tag (ChIA-PET) in MCF-7 cells (Fig. 4B; Supplemental Fig. S4B; Fullwood et al. 2009; The ENCODE Project Consortium 2012). These data highlighted a drastically lower variability in the ESR1-linked genes in each patient compared with the ESR1 binding heterogeneity (Fig. 4B; Supplemental Fig. S4B).
Figure 4.
ESR1 female peaks converge to redundant enhancers regulating estrogen response genes. (A) Heatmap shows the number of ESR1 peaks that are overlapping with a region associated to a gene (x-axis) (Corces et al. 2018) per each female patient (y-axis). Each gene is ranked by decreasing number of patients carrying ESR1 peaks associated to that specific gene. The number of patients sharing a gene is shown by the line above the heatmap. The global distribution of ESR1 peak conservation among samples is depicted by a black dashed line. Ranked genes are grouped in seven bins depending on the degree of coregulation among patients. For each bin, the statistically significantly enriched cancer hallmark gene sets are shown (bottom heatmap), and the bar plot on the bottom left shows the number of bins sharing a given hallmark. The left heatmap depicts the cancer hallmarks enriched in each patient; above this heatmap, a bar plot indicates the percentage of patients showing the enrichment of each hallmark. (B) Same heatmap as in A, but in this case, the gene associated is based on chromatin loops identified by ESR1 ChIA-PET in MCF-7 (Fullwood et al. 2009; The ENCODE Project Consortium 2012).
Overall, ESR1 peak–gene association analyses showed that, despite the high variability of ESR1 chromatin occupancy, the transcriptional program converges in the estrogen response owing to a redundancy of distal regulatory element binding by ESR1.
ERE strength weakly associates with the level of inter-tumor conservation of ESR1 sites
From cell lines, ESR1 DNA binding is known to be enriched at EREs, although indirect chromatin associations by tethering through other TFs may also occur (Coons et al. 2017). In our ranked list of peaks in female tumors, an ERE could be found in 99.5% of common peaks (20 or more patients), in 87.3% of peaks found in two to 19 patients, and in 67.6% of patient-unique peaks (Fig. 5A). We used HOMER (Heinz et al. 2010) to determine the strength of the EREs; that is, sequences most similar to the consensus ERE receive a higher score versus those deviating from the consensus ERE. When present, the average strength of the ERE was found to correlate with how often a binding site was bound by ESR1 in our cohort of female patients (P < 0.001) (Fig. 5B). A similar observation was made performing TF binding enrichment analyses (GIGGLE) (Layer et al. 2018), displaying that ESR1 is the top enriched TF and that its enrichment is stronger at highly conserved peaks (Supplemental Fig. S5). Nonetheless, the variance in ERE strength per patient number is large, and strong EREs were still observed at less common sites and vice versa (Fig. 5B). In a regression model in which strength of the ERE in a peak was evaluated as predictor for the number of patients in which an ESR1 peak was found, the R2 (goodness of fit) was only 0.048, suggesting that although ERE strength is statistically significantly associated with ESR1 site conservation, it is not a powerful predictor of heterogeneity in ESR1 binding.
Figure 5.
Common ESR1 peaks are associated with stronger ERE motif, increased chromatin interactions, and higher enhancer activity. (A) The percentage of common, less common, and patient-unique ESR1 peaks in females that contain an estrogen response element (ERE). (B) The strength of those EREs as determined by HOMER, ranked from those in common to those in more patient-unique ESR1 peaks. Black dots represent outliers. (C) Aggregate region analyses (ARAs) showing the average Hi-C contacts (observed over expected scores) at ESR1 binding sites shared by an increasing number of patients from left to right. The matrices include a window of ±250 kb from the ESR1 peak centers. (D) Schematic overview of STARR-seq methodology. (E) Stacked bar plot showing the overlap between STARR-seq regions and MCF-7 ESR1 peaks (Ross-Innes et al. 2012) in bins of female patient STARR-seq shared regions. (F) Volcano plot of STARR-seq results in the cell line MCF-7 upon 6 h of 10 nM estradiol (E2) stimulation. (G) Distribution of enhancer activity as determined by STARR-seq upon 6 h of 10 nM estradiol stimulation, from common to more patient-unique peaks. Details on cutoffs for categories induced, not-induced, and inactive are described in the Methods section.
ESR1 does not act independently but requires activity of other proteins for its function. One essential ESR1 interactor is the forkhead protein FOXA1, which serves as a pioneer factor rendering the chromatin accessible for ESR1 to bind (Hurtado et al. 2011). The consensus FOXA1 motif TGTTTAC is generally found close to ERE sequences, yet does not overlap (Serandour et al. 2013). In our female cohort, 87.1% of common ESR1 peaks, 79.6% of peaks in two to 19 patients, and 62.1% of patient-unique peaks carried a forkhead motif (P < 0.001) (Supplemental Fig. S6A), but no relationship between strength of the forkhead motif and heterogeneity in ESR1 DNA binding was observed (Supplemental Fig. S6B).
Estrogen-induced enhancer activity is highest for commonly shared ESR1 binding sites
We next questioned if the ESR1 binding profiles found in the most commonly used ESR1+ breast cancer cell lines MCF-7, T-47D, and ZR-75-1 are representative of the ESR1 DNA binding identified in primary human tumors. Comparing ESR1 peaks from these cell lines in full medium conditions to peaks found in patients, we found that these cell lines capture 99.7% of peaks present in 19 or more patients and 65.3% of peaks present in at least two to 19 patients (Supplemental Fig. S6C,D). The 29% of the “patient-unique” peaks was also called in at least one cell line, further solidifying the confidence in the ESR1 signal at these locations. ESR1 ChIP-seq from MCF-7, T-47D, and ZR-75-1 appears to recapitulate a significant number of ESR1 peaks found among 40 patients (Supplemental Fig. S6D), and we therefore deemed these cell lines adequate models to investigate the functional consequences of ESR1 enhancer heterogeneity.
Using one of these cell line models, MCF-7, we further investigate the biological features at commonly and less frequently bound ESR1 sites. For this, we first evaluated chromatin conformation behavior of common and more patient-unique sites by means of high-throughput chromatin conformation analyses (Hi-C), which illustrated a direct positive association of long-range chromatin interaction frequency with the level of ESR1 site conservation among patients (Fig. 5C). Although classical ESR1 target genes were represented among the most commonly shared regions (Supplemental Table S2), no enrichment for essentiality was found for genes proximal to ESR1 sites (Dempster et al. 2021; https://doi.org/10.6084/m9.figshare.22765112.v2), relative to the level of inter-tumor heterogeneity (Supplemental Fig. S6E).
We then surveyed enhancer activity across the ranked peaks using self-transcribing active regulatory region sequencing (STARR-seq) (Fig. 5D; Arnold et al. 2013). This method allows for the massive parallel testing of intrinsic enhancer activity of DNA fragments by cloning these sequences downstream from a core promoter and then quantifying the enhancer activity based on the self-transcription in mRNA transcripts. We generated a library of 11,147 regions, which included 7922 peaks that were called in at least seven or more patients and a random sampling of less common peaks. All subsets of regions used for STARR-seq analyses—except for the least shared peaks (bin 1–5)—were showing a high fraction (>75%) of overlap with MCF-7 ESR1 ChIP-seq peaks (Fig. 5E), excluding therefore any cell type–specific bias in the further analyses. The library was transfected into MCF-7 cells, and reporter read-out was generated under beta-estradiol (E2) stimulation or vehicle control. Out of all 11,147 ESR1 sites cloned in the library, 597 (5.2%) were found to be E2-induced, 5777 (51.8%) were not-induced, and 5053 (44.1%) were inactive (for details on these categories, see Methods) (Fig. 5F). Of note, these distributions of observed activities were comparable to that of androgen receptor (AR) sites studied in LNCaP prostate cancer cells (Huang et al. 2021; Kneppers et al. 2022) or glucocorticoid receptor (GR) sites studied in lung cancer A549 cells (Vockley et al. 2016). Overall, this suggests that only a small fraction of nuclear receptor sites is actively engaged in transcriptional regulation. By ranking the peaks from common to less frequently bound by ESR1 in patients, we observed that these binding sites have distinct intrinsic properties. The more common ESR1 peaks are among patients, the higher the percentage of E2-induced enhancer activity (Fig. 5G). Enhancer activity at patient-unique sites is slightly more often constitutively active, acting in an ESR1-independent manner, although the receptor does bind.
Cumulatively, these results imply direct biological consequences of the observed inter-patient heterogeneity of ESR1, with enhancers showing hormone-induced activity being mostly conserved among patients.
Breast cancer risk SNP loci are enriched at ESR1 sites commonly shared by tumors
Although we observed a relationship between average ERE strength and the commonness of ESR1 binding to enhancers among patients, this was not sufficient to explain the large observed variation. We therefore hypothesized that genetic variation at enhancer elements contributes to this inter-patient heterogeneity of ESR1 enhancer action. To test this hypothesis, we turned to a known source in variation of breast cancer risk and analyzed our data for a possible overlap with breast cancer risk single-nucleotide polymorphisms (rSNPs) and small indels. Importantly, rSNPs for different cancer types have been found previously to be enriched in enhancer regions, with prostate cancer risk SNPs found enriched at AR sites (Morova et al. 2020), but also breast cancer rSNPs have been found enriched at ESR1-bound regulatory elements (Cowper-Sal lari et al. 2012; Li et al. 2013; Fachal et al. 2020). Any association of rSNPs or small indels with ESR1 site heterogeneity between tumors remains unexplored.
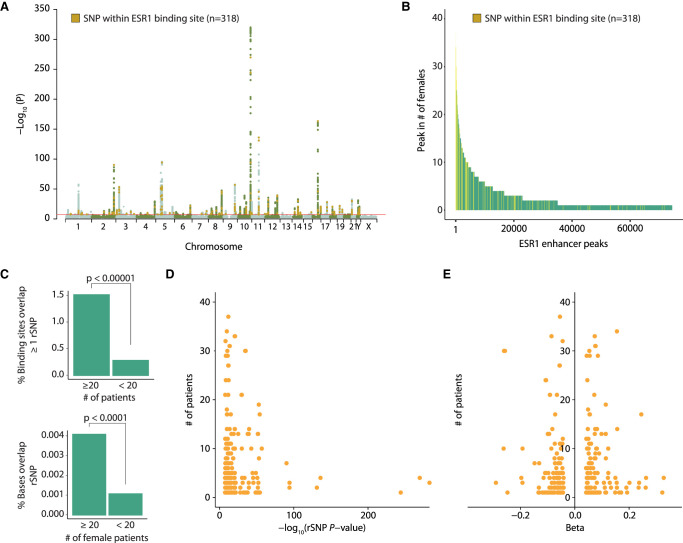
We hypothesized that alterations in DNA sequence by rSNPs or small indels may affect ESR1 binding and thereby facilitate inter-patient heterogeneity. rSNPs and indels with a significant (P < 10−6) correlation with ESR1+ breast cancer risk as published by Michailidou et al. (2017) were tested for overlap with ESR1 binding sites in our cohort, yielding a combined list of 318 rSNPs and a small number of indels that overlap with ESR1 sites identified in our patient samples (Fig. 6A). Surprisingly, the rSNPs and indels showed relative enrichment at the top of the ranked peaks, converging more often at ESR1 binding sites common between patients (P < 0.0001) (Fig. 6B,C). The relative enrichment of rSNPs and some indels at common ESR1 binding sites was also seen in the male breast cancer samples (P < 0.001) (Supplemental Fig. S7A,B). When normalized to the number of bases in peaks (to exclude enrichment being an artifact of peak width, as genomic location of common sites may be slightly broader owing to merging of more info) (Fig. 6C; Supplemental Fig. S7B) or when leaving out patient-unique peaks (Supplemental Fig. S7C), the statistically significant association holds. Of note, in both sexes, the more commonly bound an ESR1 site is, with which an rSNP/indel coordinate overlaps, the stronger the P-value for the association with breast cancer for that rSNP/indel (Fig. 6D; Supplemental Fig. S7D), although the protective (negative beta) or risk effects (positive beta) of the rSNPs/indels at common sites are modest (Fig. 6E).
Figure 6.
ESR1+ breast cancer rSNPs are enriched at regions with low inter-patient heterogeneity in ESR1. (A) Manhattan plot of ESR1+ breast cancer risk SNPs (rSNPs) with genome-wide significance originating from Michailidou et al. (2017). Highlighted in orange are 318 rSNPs, for which the coordinates intersect with one of the 74,438 ESR1 peaks found among 40 female breast cancer patients. (B) The position of these 318 rSNPs in the ranked peaks introduced in Figure 2A. (C, top) Comparison (Fisher's exact test) of the percentage of ESR1 peaks with which coordinates overlap with at least one rSNP coordinate, for common and less common ESR1 peaks. (Bottom) Comparison (Fisher's exact test) of the percentage of bases, present in common or less common ESR1 peaks, that overlap with at least one rSNP coordinate. (D) Correlation between the P-value of rSNP (x-axis) and its position in the ranking of ESR1 peaks introduced in Figure 2A (y-axis). If multiple rSNPs overlapped the same ESR1 peak, the strongest P-value was used for analysis. (E) Overview of beta values corresponding to rSNPs with which a coordinate intersected an ERE. Negative beta values correspond with rSNPs that confer less risk to breast cancer, whereas positive beta values correspond to increased risk of ESR1+ breast cancer.
In accordance with literature showing that rSNPs often lie in regions with TF motifs (Cowper-Sal lari et al. 2012; Li et al. 2013; Fachal et al. 2020), ESR1 peaks that overlap with rSNP/indel coordinates do hold an ERE more often than those that do not overlap with rSNP/indel coordinates (Supplemental Fig. S7E). These EREs also tend to be slightly stronger than EREs without rSNPs/indels (P = 0.06) (Supplemental Fig. S7F). Although ERE strength was a statistically significant but not a powerful predictor of observed heterogeneity in ESR1 binding, we nonetheless checked if the enrichment of rSNPs/indels at common peaks was confounded by ERE strength, but we found no evidence to this (Supplemental Fig. S7G).
Breast cancer risk SNPs affect ESR1 inter-patient heterogeneity through TF motif perturbation, with biological implications on gene expression
Of the 318 rSNP/indel coordinates overlapping with ESR1 peaks in our cohort, 25 of those rSNP/indel coordinates fall within an ERE palindromic sequence (Supplemental Table S5). Of those, we focused our attention on rSNPs/indels in sites that showed enhancer activity in the STARR-seq data (E2-induced, constitutively active, or not-induced) (Supplemental Fig. S7H). To determine which remaining rSNPs/indels had the potential to significantly and directly affect the strength of the ERE and thereby ESR1 binding, we used the in silico prediction tool SNP2TFBS (Kumar et al. 2017), which compares the position weight matrix of a TF binding motif in reference format and when including the rSNP/indel variant. This resulted in a short list of three rSNPs, rs9952980, rs11695384, and rs11665924, which SNP2TFBS predicted to affect a hormone response element (Supplemental Table S5).
To validate the effects of these rSNPs on ESR1 binding experimentally, we designed two 50-bp oligonucleotides containing the WT and rSNP-affected ERE, which were both biotin-labeled and pulled through an MCF-7 lysate to detect interacting proteins via mass spectrometry in an unbiased fashion (Supplemental Table S5; Vermeulen 2012). For rs11695384 and rs11665924, we were unable to confirm differential binding of ESR1, leaving rs9952980 for further study.
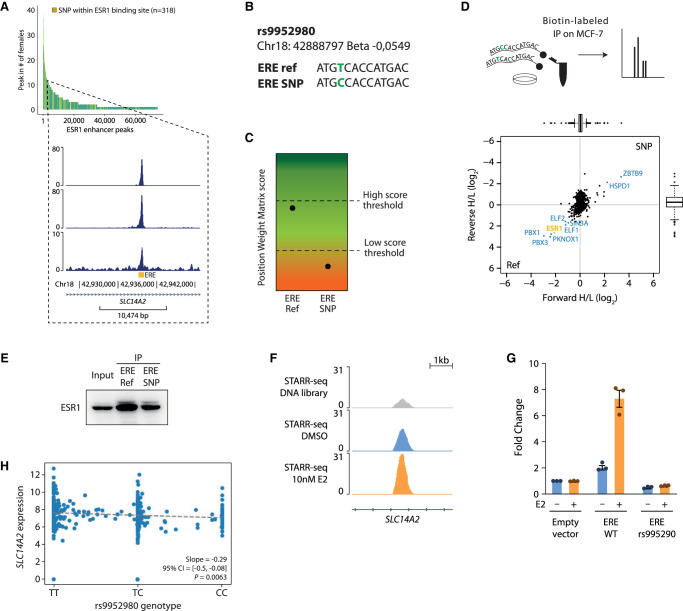
rs9952980 is an rSNP located in an intron of the gene SLC14A2, located on Chromosome 18. This region was bound by ESR1 in 11 female (exemplified in Fig. 7A) and six male tumor samples. STARR-seq data confirmed the region to be active and induced upon E2 treatment (Supplemental Fig. S7H). Considering the reference genome, the region holds a relatively strong ERE at a log odds motif score of 12.3. rs9952980 affects the fifth nucleotide of the ERE (Fig. 7B), which is a position of high importance for strong ESR1 affinity (Fig. 5A), as it facilitates direct DNA–protein interaction with ESR1 (Coons et al. 2017). Accordingly, in silico analysis using SNP2TFBS (Kumar et al. 2017) predicted rs9952980 to significantly decrease ESR1's binding affinity (Fig. 7C; Supplemental Table S5). Mass spectrometry (Fig. 7D) and western blot (Fig. 7E) of DNA-oligo pulled-down proteins confirmed diminished ESR1 binding in the rSNP condition. Moreover, we investigated the effect of rs9952980 on enhancer activity of the region carrying this risk SNP by using luciferase reporter assays in MCF-7 cells. We observed that although the wild-type sequence displays an increased enhancer activity in the presence of active ESR1 (E2-inducible) (Fig. 7F,G), rs9952980 was sufficient to completely abolish the enhancer activity of this genomic locus (Fig. 7G).
Figure 7.
rs9952980 affects SLC14A2 expression via reduced ESR1 binding by impacting ERE. (A) Snapshots of ESR1 peak intersecting the coordinate of rs9952980. The peak, positioned in an intron of SLC14A2, was found in 11 female patients. (B) ERE at this peak, in reference allele and rSNP format. (C) Predicted score of position weight matrix for WT and rSNP ERE, by SNP2TFBS (Kumar et al. 2017). (D) Using MCF-7 lysate, an immunoprecipitation (IP) was performed with 50-bp biotin-labeled oligos containing the WT or the rs9952980 variant of the ERE, followed by mass spectrometry. (E) ESR1 western blot (WB) of IP by 50-bp biotin-labeled oligos containing the reference allele or the rs9952980 variant of the ERE. (F) Snapshot of STARR-seq normalized signal at the SLC14A2 locus. (G) Luciferase reporter assay in MCF-7 cells stimulated or not by estradiol (E2) for the SLC14A2 locus enhancer activity containing or not the rs995290 variant. Bar plot represents the fold change of luciferase expression over the untreated empty vector condition. (H) TCGA gene expression of SLC14A, which rs9952980 is predicted to affect, by homozygous or heterozygous genotype.
Clinically, carriers of the alternative allele (T) have less risk (beta: −0.0549) of developing breast cancer than do homozygous carriers of the reference allele (C). rs9952980 was previously predicted to regulate expression of its target gene SLC14A2 (Fachal et al. 2020), and in TCGA data, we indeed find rs9952980 significantly associates (P = 0.0063) with a reduced expression of SLC14A2 (Fig. 7H), likely mediated via the rSNPs direct impact on ESR1–DNA binding.
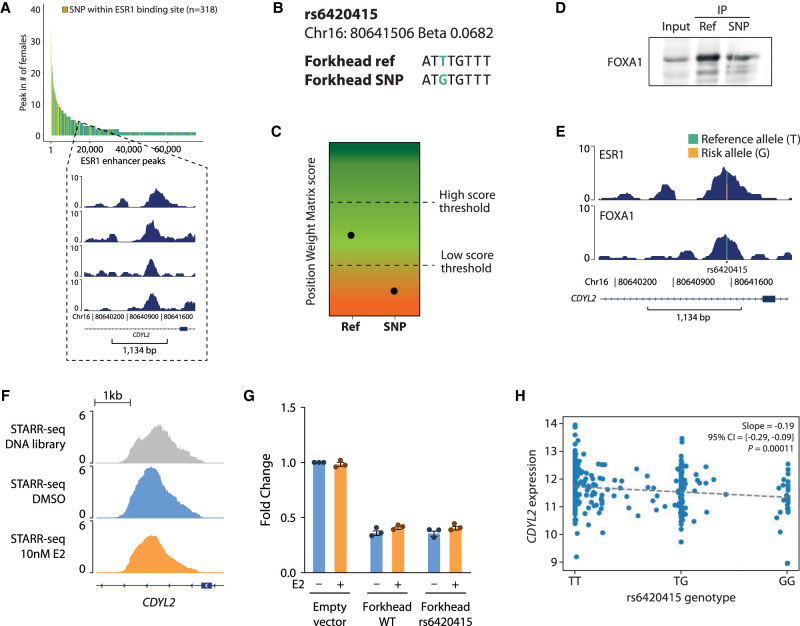
Few rSNPs/indels that intersected with our ranked peaks directly overlapped an ERE, although they were often located in close proximity. Such rSNPs/indels may affect ESR1 binding indirectly by affecting affinity of ESR1's partners, such as FOXA1. An example for this is found for the rSNP rs6420415, located in an intron of CDYL2 and of which the region was occupied by ESR1 in only four females (Fig. 8A) and four males. The rs6420415 is predicted to perturb the forkhead motif such that FOXA1 binding is negatively affected (Fig. 8B,C). Indeed, western blot of oligo-mediated immunoprecipitation of the local forkhead motif in a reference allele and rSNP format confirmed diminished FOXA1 binding (Fig. 8D). Coinciding with these in vitro data, in a female tumor sample for which both ESR1 and FOXA1 ChIP-seq data were available, we noted about half of the reads from the reference allele (T) and 50% from the SNP allele (G) in the ESR1 ChIP, whereas reads from the FOXA1 ChIP-seq were dominated by the reference allele T (Fig. 8E).
Figure 8.
rs6420415 affects CDYL2 expression via reduced FOXA1 binding by impacting the forkhead motif. (A) Snapshots of ESR1 peak intersecting the coordinate of rs6420415. The peak, positioned in an intron of CDYL2, was found in four female patients. (B) Forkhead motif at this peak, in reference allele and rSNP format. (C) Predicted score of position weight matrix for reference allele and rSNP forkhead motif by SNP2TFBS (Kumar et al. 2017). (D) FOXA1 western blot of pulldown by 50-bp biotin-labeled oligos containing the WT or rs6420415 variant of the forkhead motif. (E) Distribution of reads in the ESR1 and FOXA1 ChIP-seq peak performed on tumor tissue from the same breast cancer patient, at the locus surrounding rs6420415. (F) Snapshot of STARR-seq normalized signal at the CDYL2 locus. (G) Luciferase reporter assay in MCF-7 cells stimulated or not by estradiol (E2) for the CDYL2 locus enhancer activity containing or not the rs6420415 variant. Bar plot represents the fold change of luciferase expression over the untreated empty vector condition. (H) TCGA gene expression of CDYL2, which rs6420415 is predicted to affect, by homozygous or heterozygous genotype for rs6420415.
We investigated the effect of rs6420415 on the enhancer activity by using luciferase reporter assays in MCF-7 cells. In this case, we did not observe differences in the enhancer activity in the presence of the rs6420415 variant (Fig. 8F,G). The different behavior of the two rSNPs might be explained by the fact that, as reported previously (Hurtado et al. 2011), FOXA1 expression status does not impact ESR1–DNA binding on nonchromatinized templates.
Carriers of rs6420415's G allele are thought to have an elevated risk of developing breast cancer (beta: 0.0682) (Michailidou et al. 2017), possibly mediated via reduced expression of CDYL2 (Fig. 8H). CDYL2 has been described to exert both tumor-suppressing and oncogenic effects, depending on its isoform (Siouda et al. 2020; Yang et al. 2020), although this distinction was not made in GWAS studies (Michailidou et al. 2017).
Nonetheless, our data cumulatively illustrate that breast cancer risk SNPs/indels are enriched at commonly shared active enhancer elements and can perturb binding of ESR1 or its pioneer factor FOXA1, for instance, via allele preferential binding, which is associated with affected expression of the genes they control.
Discussion
Breast cancer is a heterogenous disease, with clearly distinct inter-tumor differences on subtype, aggressiveness, and, ultimately, patient prognostication. Here, we show that on epigenetic scale, substantial inter-patient heterogeneity of ESR1 chromatin binding capacity is found. Therefore, consensus-based analysis of ESR1 ChIP-seq on patient samples, despite being a strategy often applied in the field to reduce data complexity and limit noise, eliminates potentially interesting and biologically meaningful data. Here, we queried the full spectrum of inter-patient ESR1 enhancer heterogeneity by ranking all peaks in patients from commonly shared to patient unique. Many peaks could only be found in a handful of patients, and around half of all the peaks identified were patient unique. This level of heterogeneity was substantially higher for putative enhancer elements as opposed to promoters. After investigating the genes associated to these heterogenous enhancers, our data suggested functional redundancy between enhancers in regulating the same gene. These results are in line with recently shown data of combinatorial CRISPR screening analyses (Carleton et al. 2017).
Although extensive quality control (QC) analyses were performed on the ChIP-seq data sets, we cannot exclude that a fraction of the ESR1 heterogeneity between the patients in our cohort may be of technical origin. The female samples in this cohort were produced in different laboratories, but a single antibody was used. Also, a similar degree of inter-patient heterogeneity was seen in the male cohort, for which ChIP samples were exclusively produced in our laboratory with the same (batch of) antibody. Further, although contamination of the signal derived from stromal cells cannot be excluded, evidence that the tumor-surrounding microenvironment does not significantly contribute to the overall ESR1 ChIP-seq signal is provided by analyses on ESR1-negative breast tumors as performed by Ross-Innes et al. (2012), in which ESR1 ChIP-seq analyses on these tumors did not detect any peaks. Furthermore, as current technologies do not allow for precise single-cell TF profiling in tumor tissues, it remains elusive to what degree intra-tumor heterogeneity of ESR1–chromatin interaction profiles impacts inter-tumor heterogeneity, as we reported in this study.
Although inter-patient heterogeneity of ESR1 signal was high, our analyses on ERE strength, enhancer activity (STARR-seq data), and rSNP/indel analyses suggest that the most functional activity, hormone-induced action, and clinically relevant information are found at the commonly shared ESR1 sites. In particular, we identified that the most patient-conserved ESR1 peaks, carrying stronger ERE motifs, are engaged in more chromatin–chromatin interactions in the surrounding regions and display a higher enhancer activity potential. However, most enhancers analyzed by STARR-seq were found to be not-induced by E2 stimulation or to be completely inactive. These results are in line with previous studies that reported a similar behavior for other steroid hormone receptors (SHRs), such as AR in the prostate (Huang et al. 2021; Kneppers et al. 2022) and GR in lung cancer (Vockley et al. 2016). These findings indicate that chromatin binding of SHRs cannot be directly translated to genuine enhancer activity but rather that a relatively small fraction of SHR-chromatin bound sites is actively induced in activity following stimulation, as determined by massive parallel reporter assays. These findings, in combination with the observed enhancer redundancy, may suggest a model in which the strongest EREs are more commonly bound and drive activation of the estrogen response, whereas “weaker” enhancers cooperate to maintain this transcriptional program, in support of a previously reported phase-separation model of ligand-activated enhancers (Nair et al. 2019).
On the other hand, less commonly shared peaks found in female patients revealed enrichment of other pathways that are reported to be associated with therapy resistance, such as epithelial-mesenchymal transition (EMT) and TNF/NF-kB (Kastrati et al. 2020). As inhibitors of NF-kB show potential in targeting endocrine therapy resistance (Kastrati et al. 2020), it is relevant to consider that the intrinsic intra-tumor heterogeneity of ESR1 action and downstream transcriptional programs may also impact which particular patients may respond to these inhibitors. Thus, based on these observations, we conclude that the observed enhancer heterogeneity represents a biological hierarchy of ESR1 action, with biological and clinical consequences. This also underlines the importance of considering the whole ESR1 binding spectrum, because these less common sites, often discarded when analyzing consensi of peaks, do harbor biologically meaningful information.
Following this hierarchy, we found rSNP/indel coordinates to be enriched at the most commonly shared ESR1 peaks, both in a cohort of 40 female samples and in 30 male samples. Yet, peculiarly, GWAS P-values corresponding to these rSNPs/indels are stronger at more common ESR1 sites, whereas corresponding beta values were relatively modest. One could be tempted to interpret these rSNPs/indels as statistically significant but biologically unimportant, but this does not reconcile with their enrichment at common ESR1 sites. The rSNPs with strong P-values and small effect sizes have been hypothesized to result from heterogeneity in the studied cohort (Hodge and Greenberg 2016). In this case, breast cancer risk may indeed be caused by a large number of variants and associated genes (i.e., polygenic risk model), but rather than each locus contributing a small amount to breast cancer risk, loci commonly bound by ESR1 contribute relatively more to breast cancer risk. Indeed, the patients in our cohorts have been described to have different prognoses, despite all having ESR1-positive breast cancer. When bound by ESR1 in almost all tumors, this also allows common sites to contribute differently to different subtypes of ESR1 breast cancer and thereby cause an attenuation in effect size in GWAS studies. Neither in work by Michailidou et al. (2017) nor in this work was a distinction in subtypes of ESR1-positive breast cancer made. However, we can speculate that other non-risk-associated variants, or somatic mutations at regulatory elements, further contribute to the observed inter-patient heterogeneity of ESR1–chromatin interactions. Previously we reported, in a concise CRISPR-screen studying 99 ESR1 binding sites, that only a rather small fraction of ESR1 sites individually impact breast cancer cell proliferation capacity (Korkmaz et al. 2016). Therefore, other variants may further contribute to the observed ESR1 heterogeneity, for which the biological consequences are yet to be understood.
As proof of concept, here we reported the rSNP rs9952980, occurring at the SLC14A2 locus, to be sufficient to abrogate the enhancer activity owing to disruption of an ERE motif. On the other end, the rSNP rs6420415, occurring at the CYL2 locus, disrupts a forkhead motif.
Overall, we did not observe any striking differences between both sexes on ESR1 chromatin action, risk SNP enrichment, or any other genomic features reported in this paper. Differences in ESR1 regulation and features of the chromatin context bound by ESR1 between the two sexes were extensively studied in our previous publication (Severson et al. 2018), but also here, very limited differences were observed.
Cumulatively, these findings contribute to our basic understanding how sequence variants at specific regulatory elements contribute to ESR1+ breast cancer development. Recently, we reported a comparable observation in prostate cancer when analyzing inter-tumor heterogeneity of AR action between primary tumors, revealing not only somatic mutations but also rSNPs being enriched at more commonly shared regions, which were more active on transcriptional level (Kneppers et al. 2022). In that setting, we also observed that less commonly shared regions were associated with disease progression and may become engaged later on, at the metastatic disease stage. Future studies should address whether this phenomenon would also occur in breast cancer and whether the selectivity and plasticity of enhancer action are more general features in hormone-driven cancers.
In conclusion, our analyses suggest a hierarchy of ESR1–chromatin interactions in breast cancers, resulting in a high degree of inter-patient heterogeneity in ESR1 enhancer action. We find that the most commonly shared ESR1 regions are the most-hormone inducible enhancers and serve as hotspots for germline functional risk SNPs/indels for ESR1+ breast cancer development, highlighting a new perspective in better understanding the biological basis of risk variants in breast cancer.
Methods
Patient cohorts
ESR1 ChIP-seq on 30 male breast cancer samples was previously published by our group (Severson et al. 2018). The female cohort was compiled of previously published ESR1 ChIPs performed in our laboratory (Severson et al. 2018), five newly generated samples, and ESR1 ChIPs published by others (Ross-Innes et al. 2012; Jansen et al. 2013). Only primary tumors were included. Sample details are described in Supplemental Table S1.
Cell culture and chemicals
MCF-7 human breast cancer cell lines have been cultured in Dulbecco's Modified Eagle Medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (Capricorn Scientific FBS-12A) and penicillin/streptomycin (100 μg/mL, Gibco). Cell lines were subjected to regular Mycoplasma testing and underwent authentication by short tandem repeat profiling (Eurofins Genomics).
For hormone stimulation, cells were precultured for 3 d in phenol red-free DMEM (Gibco) supplied with 5% dextran-coated charcoal (DCC) stripped FBS, 2 mM L-glutamine (Gibco), and penicillin/streptomycin (100 μg/mL, Gibco) and then stimulated with 10 nM DMSO-solubilized 17beta-estradiol (MedChemExpress HY-B0141) for 6 h.
Publicly available cell line data analysis
Called peaks of ESR1 ChIP-seq for MCF-7 (GSM798423, GSM631484, GSM1967545), T-47D (GSE68359, GSE32222) (Ross-Innes et al. 2012; Mohammed et al. 2015), and ZR-75-1 (GSE25710) were downloaded from the Cistrome Data Browser (Mei et al. 2017; Zheng et al. 2019) and subsequently lifted over from hg38 to hg19 by UCSC liftOver (Haeussler et al. 2019). Sample details are described in Supplemental Table S1. To assess how well these cell lines represented the heterogeneous ESR1 cistrome found in patients, a union of cell line peaks was generated in DiffBind v.2.9.0 (Ross-Innes et al. 2012) and subsequently intersected with the list of patient peaks by BEDTools (Quinlan 2014). The intersected peakset was subsequently used to create heatmaps shown in Supplemental Figure S4D, using EaSeq (Lerdrup et al. 2016).
ChIP-seq library preparation
Five newly generated ESR1 samples were generated on fresh-frozen female breast cancer tissues as previously described (Zwart et al. 2013; Singh et al. 2019). Technical details are available in the Supplemental Material, and sample information is described in Supplemental Table S1.
Publicly available data access
ESR1 ChIP-seq on 30 male breast cancer samples are available from the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) (Barrett et al. 2013) under accession number GSE104399 (Severson et al. 2018). The female cohort raw data can be found at GEO under accession numbers GSE104399 (Severson et al. 2018), GSE32222 (Ross-Innes et al. 2012), and GSE40867 (Jansen et al. 2013).
ChIP-seq data analysis
Data of all patient samples were aligned to hg19/GRCh37 using Burrows–Wheeler Aligner (BWA, v0.7.5a) (Li and Durbin 2009), with mapping quality above 20, and were peak-called with MACS2 (v1.4) (Zhang et al. 2008) using the pipeline available at GitHub (https://github.com/csijcs/snakepipes) (Bhardwaj et al. 2019). For the peak calling, the corresponding input sample was used as background, a filter on the Q-value was applied (q < 0.01), and reads were extended using the fragment size identified by phantompeakqualtools (v1.2.2) (Kharchenko et al. 2008; Landt et al. 2012). ChIP-seq signal was normalized to 1× coverage and expressed as reads per genomic content (RPGC; bamCoverage from deepTools) (Ramírez et al. 2016). Heatmaps and peak snapshots were generated with EaSeq (Lerdrup et al. 2016) and all other plots using ggplot2 (Wickham 2016) under R v4.0.1. The percentage of peaks included or left out at varying thresholds (Fig. 1B,D) was analyzed in DiffBind (Ross-Innes et al. 2012). Genomic distribution (Fig. 1C) was assessed with package ChIPpeakAnno (v3.15.1) (Zhu et al. 2010).
To determine in how many patients a putative enhancer peak was called, per-patient BED files were separated into promoter (defined as peak between 1 and 1000 bp upstream of TSS, from RefSeq hg19) and putative enhancer files. A union of all patient putative enhancer peaks was generated by DiffBind. The union was next intersected with the per-patient enhancer files again, using the option -C of BEDTools (v2.29.2) intersect (Quinlan 2014), producing a list discriminating between patients with and without signal for each peak in the union. Subsetting this to patients with signal and then using BEDTools intersect -C option again produced the final list of peaks and patient counts used in this manuscript.
Heterogeneity plots have been generated using Rseb’s (v0.3.3) (Gregoricchio et al. 2022) function evaluate.heterogeneity. Pearson correlations of ESR1 ChIP-seq signal at enhancer ESR1 peaks were computed using deepTools (Ramírez et al. 2016). The 1 − correlation values were used as distance to perform a hierarchical clustering (method “complete”) of the samples. Clustering dendrograms were plotted using ggh4x (v0.2.4, https://cran.r-project.org/web/packages/ggh4x/index.html).
Motif analysis
Presence and strength of EREs (HOMER's MC00355) or forkhead motifs (also HOMER's) was defined by HOMER using a minimal log odds threshold of two (Heinz et al. 2010). In case of multiple EREs or forkhead motifs in a peak, the strongest was used for analyses. To assess the relationship between motif strength and heterogeneity in ESR1 binding, linear regression (with dummy variables) was performed in SPSS.
TF binding enrichment analyses
GIGGLE (Layer et al. 2018) analyses have been performed using the toolkit available at the CistromeDB website (http://dbtoolkit.cistrome.org/). Top 1000 peaks according to peak enrichment have been used for each publicly available data set.
Gene coupling, patterns, and dependency
Breast cancer–specific ATAC-seq-based enhancer–promoter loops published by Corces et al. (2018) were used to associate distal ESR1 binding sites to genes. Enrichment of gene patterns was assessed with GSEA 4.3.2 (Mootha et al. 2003; Subramanian et al. 2005) For the purpose of enrichment analysis, if multiple enhancers looped to the same gene, only the binding site with the highest patient binding score was included. DepMap's Chronos 23Q2 (Broad Institute; https://depmap.org/) data set was used to assess the relationship between commonness of ESR1 binding and the dependency of breast cancer cells to the associated gene.
Replicates of ESR1 ChIA-PET (GSM970212, ENCSR000BZZ) (Fullwood et al. 2009; The ENCODE Project Consortium 2012) have been merged and overlapped to ESR1 breast ranked peaks using Rseb’s (v0.3.3) (Gregoricchio et al. 2022) function intersect.regions.
Hi-C library preparation and data processing
Hi-C single-index library preparation of MCF-7 cells was performed as previously described using MboI (New England Biolabs) restriction enzyme (Donaldson-Collier et al. 2019).
The quality and quantification of the Hi-C libraries were assessed using the 2100 Bioanalyzer (Agilent, DNA 7500 kit). Four biological replicates have been pooled in an equimolar manner and subjected to sequencing using the Illumina NextSeq 550 system in a 75-bp paired-end setup. Demultiplexed FASTQ data were analyzed at 10-kb resolution using the snHiC pipeline (v0.2.0) (Gregoricchio and Zwart 2023), applying default parameters and the hg19/GRCh37 genome assembly. Aggregate analyses at ESR1 binding sites have been performed using GENOVA (v1.0.1) (van der Weide et al. 2021).
ESR1-focused STARR-seq capture library design
A custom oligonucleotide probe pool (Agilent) was designed to capture ESR1 binding regions from clinical ChIP-seq. We selected 11,463 regions, which included all peaks that were called in at least seven or more patients (n = 7922), all regions for which coordinates intersected with rSNP coordinates (n = 217), and a random sampling of less common peaks.
Pooled human genomic DNA (Coriell Institute for Medical Research NA13421) was randomly sheared into 500- to 800-bp fragments and ligated with Illumina compatible IDT xGen CS stubby adaptors that contain 3-bp unique molecular identifiers (UMIs). After the hybridization of the adaptor-ligated gDNA fragments to the biotinylated probe pool, the target regions were captured with Dynabeads M-270 streptavidin beads (Invitrogen). The postcapture was PCR-amplified with STARR_in-fusion_Fw and STARR_in-fusion_Rv primers (5′-TAGAGCATGCACCGGACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ and 5′-GGCCGAATTCGTCGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′), and cloned into AgeI-HF (New England Biolabs [NEB]) and SalI-HF (NEB) digested hSTARR-ORI plasmid (Addgene plasmid 99296) using the NEBuilder HiFi DNA assembly master mix (NEB). The ESR1-focused STARR-seq capture library was transformed into MegaX DH10B T1R electrocompetent cells (Thermo Fisher Scientific), and the plasmid DNA was extracted using the Qiagen plasmid maxi kit.
ESR1 STARR-seq library preparation and analyses
MCF-7 cells (more than 2 × 108 cells/replica for three biological replica) were grown for 48 h under hormone-deprivation conditions. The ESR1-focused STARR-seq capture library was transfected in the cells, and after 24 h, cells were stimulated with 10 nM E2. The poly(A) mRNA was isolated using the oligo (dT)25 Dynabeads (Thermo Fisher Scientific), converted to cDNA, and used for library preparation. The detailed protocol is available in the Supplemental Material.
Bioinformatics analyses for the ESR1-dependent STARR-seq status of tested regions are described in detail in the Supplemental Material.
Luciferase reporter assay
For luciferase assays, the regions of interest (WT) were PCR-amplified from pooled male human genomic DNA (Promega). The amplified regions were cloned by Gibson assembly into a STARR luciferase vector ORI empty plasmid (Addgene 99298) (Muerdter et al. 2018). Variants were either introduced by site-directed mutagenesis PCR or found endogenously in the genomic DNA pool. Reporter plasmids were transfected in MCF-7 cells stimulated or not for 24 h with 10 nM E2. Cells were lysed and luciferase activity quantified using the dual-luciferase reporter assay kit (Promega) according to the manufacturer's instructions. Firefly luciferase values were normalized to the Renilla luciferase. The detailed protocol is described in the in the Supplemental Material.
rSNP analysis
Accompanying breast cancer risk SNPs (Michailidou et al. 2017) were downloaded from the Breast Cancer Association Consortium (BCAC) website (https://bcac.ccge.medschl.cam.ac.uk/). rSNP information was used from the combined Oncoarray, iCOGS GWAS meta-analysis results for ESR1-positive disease, and only considering rSNPs with a P-value < 10−6, excluding rSNPs that also had a significant association with ESR1-negative disease.
DNA affinity purification and LC-MS analysis
MCF-7 cells were harvested and washed twice with ice-cold PBS, and nuclear extracts were prepared as described previously (Vermeulen 2012); ∼50-bp oligonucleotide probes, encompassing the ERE roughly at the center with either WT or SNP sequence, were ordered with the forward strand containing a 5′-biotin moiety (Integrated DNA Technologies) (Supplemental Table S5). DNA affinity purifications, on-bead trypsin digestion, and dimethyl labeling were performed as previously described (Makowski et al. 2016). Matching light and medium labeled samples were then combined and analyzed using a gradient from 7% to 30% buffer B in buffer A over 44 min, followed by a further increase to 95% in the next 16 min at a flow rate of 250 nL/min using an easy-nLC 1000 (Thermo Fisher Scientific) coupled online to an Orbitrap Exploris 480 (Thermo Fisher Scientific). MS1 spectra were acquired at 120,000 resolution with a scan range from 350 to 1300 m/z, normalized AGC target of 300%, and maximum injection time of 20 msec. The top 20 most intense ions with a charge state of two to six from each MS1 scan were selected for fragmentation by HCD. MS2 resolution was set at 15,000 with a normalized AGC target of 75%. Raw MS spectra were analyzed using MaxQuant software (v1.6.0.1) with standard settings, with multiplicity set to two, dimethyl Lys 0 and N-term 0 as light labels, and dimethyl Lys 4 and N as medium labels, and requantify enabled (Cox and Mann 2008; Makowski et al. 2016). Data were searched against the human UniProt database (FASTA file downloaded the 2017.06) using the integrated search engine.
Statistics and computational analyses
Statistical and computational analyses have been performed using R (v4.0.3) (R Core Team 2021).
Data access
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE244845. The patient-related raw data generated in this study have been submitted to the European Genome-phenome Archive (EGA; https://ega-archive.org) under accession number EGAS50000000008. The STARR-seq raw data generated in this study have been submitted to EGA under accession number EGAS50000000009, and processed data are included in Supplemental Table S6. The mass spectrometry proteomics data generated in this study have been submitted to the ProteomeXchange Consortium (https://www.proteomexchange.org) (Deutsch et al. 2017) via the PRIDE (Perez-Riverol et al. 2022) partner repository under the data set identifier PXD045526.
Supplementary Material
Acknowledgments
We thank the patients who donated tumor material for scientific research and all the researchers involved in the generation and availability of the public data sets used in this manuscript. This project was funded by Alpe d'HuZes/Dutch Cancer Society (NKI-2014-7140). The Vermeulen and Zwart laboratories are part of the Oncode Institute, which is partly funded by the Dutch Cancer Society. Per BCAC terms of use, we emphasize their analyses were supported by the Government of Canada through Genome Canada and the Canadian Institutes of Health Research, the “Ministère de l’Économie, de la Science et de l'Innovation du Québec” through Genome Québec and grant PSR-SIIRI-701, The National Institutes of Health (U19 CA148065, X01HG007492), Cancer Research UK (C1287/A10118, C1287/A16563, C1287/A10710), and The European Union (HEALTH-F2-2009-223175 and H2020 633784 and 634935).
Author contributions: S.E.P.J. and W.Z. designed the study. S.E.P.J., S.G., S.S., E.Y., C.-C.F.H., K.Y., and M.D.C. performed the experiments. S.E.P.J., S.G., T.M., U.B.A., Y.K., and S.C. analyzed the data. C.B.M. and P.J.v.D. analyzed the clinical data. S.E.P.J., S.G., and W.Z. wrote the manuscript. P.J.v.D., G.K., N.A.L., M.V., S.C.L., and W.Z. supervised the project. S.E.P.J., S.G., G.K., S.S., M.D.C., G.K., N.A.L., M.V., S.C.L., and W.Z. critically read and edited the manuscript. All authors reviewed and approved the manuscript before submission.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.278680.123.
Competing interest statement
The authors declare no competing interests.
References
- Arnold CD, Gerlach D, Stelzer C, Boryń LM, Rath M, Stark A. 2013. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science 339: 1074–1077. 10.1126/science.1232542 [DOI] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. 2013. NCBI GEO: archive for functional genomics data sets: update. Nucleic Acids Res 41: D991–D995. 10.1093/nar/gks1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj V, Heyne S, Sikora K, Rabbani L, Rauer M, Kilpert F, Richter AS, Ryan DP, Manke T. 2019. snakePipes: facilitating flexible, scalable and integrative epigenomic analysis. Bioinformatics 35: 4757–4759. 10.1093/bioinformatics/btz436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carleton JB, Berrett KC, Gertz J. 2017. Multiplex enhancer interference reveals collaborative control of gene regulation by estrogen receptor α-bound enhancers. Cell Syst 5: 333–344.e5. 10.1016/j.cels.2017.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43. 10.1016/j.cell.2005.05.008 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. 2006. Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38: 1289–1297. 10.1038/ng1901 [DOI] [PubMed] [Google Scholar]
- Coons LA, Hewitt SC, Burkholder AB, McDonnell DP, Korach KS. 2017. DNA sequence constraints define functionally active steroid nuclear receptor binding sites in chromatin. Endocrinology 158: 3212–3234. 10.1210/en.2017-00468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, Silva TC, Groeneveld C, Wong CK, Cho SW, et al. 2018. The chromatin accessibility landscape of primary human cancers. Science 362: eaav1898. 10.1126/science.aav1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowper-Sal lari R, Zhang X, Wright JB, Bailey SD, Cole MD, Eeckhoute J, Moore JH, Lupien M. 2012. Breast cancer risk-associated SNPs modulate the affinity of chromatin for FOXA1 and alter gene expression. Nat Genet 44: 1191–1198. 10.1038/ng.2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Dempster JM, Boyle I, Vazquez F, Root DE, Boehm JS, Hahn WC, Tsherniak A, McFarland JM. 2021. Chronos: a cell population dynamics model of CRISPR experiments that improves inference of gene fitness effects. Genome Biol 22: 343. 10.1186/s13059-021-02540-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Csordas A, Sun Z, Jarnuczak A, Perez-Riverol Y, Ternent T, Campbell DS, Bernal-Llinares M, Okuda S, Kawano S, et al. 2017. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res 45: D1100–D1106. 10.1093/nar/gkw936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson-Collier MC, Sungalee S, Zufferey M, Tavernari D, Katanayeva N, Battistello E, Mina M, Douglass KM, Rey T, Raynaud F, et al. 2019. EZH2 oncogenic mutations drive epigenetic, transcriptional, and structural changes within chromatin domains. Nat Genet 51: 517–528. 10.1038/s41588-018-0338-y [DOI] [PubMed] [Google Scholar]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG). 2005. Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365: 1687–1717. 10.1016/S0140-6736(05)66544-0 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachal L, Aschard H, Beesley J, Barnes DR, Allen J, Kar S, Pooley KA, Dennis J, Michailidou K, Turman C, et al. 2020. Fine-mapping of 150 breast cancer risk regions identifies 191 likely target genes. Nat Genet 52: 56–73. 10.1038/s41588-019-0537-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, et al. 2009. An oestrogen-receptor-α-bound human chromatin interactome. Nature 462: 58–64. 10.1038/nature08497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoricchio S, Zwart W. 2023. snHiC: a complete and simplified snakemake pipeline for grouped Hi-C data analysis. Bioinform Adv 3: vbad080. 10.1093/bioadv/vbad080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregoricchio S, Polit L, Esposito M, Berthelet J, Delestré L, Evanno E, Diop MB, Gallais I, Aleth H, Poplineau M, et al. 2022. HDAC1 and PRC2 mediate combinatorial control in SPI1/PU.1-dependent gene repression in murine erythroleukaemia. Nucleic Acids Res 50: 7938–7958. 10.1093/nar/gkac613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Zweig AS, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Hinrichs AS, Gonzalez JN, et al. 2019. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res 47: D853–D858. 10.1093/nar/gky1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge SE, Greenberg DA. 2016. How can we explain very low odds ratios in GWAS? I: polygenic models. Hum Hered 81: 173–180. 10.1159/000454804 [DOI] [PubMed] [Google Scholar]
- Huang CF, Lingadahalli S, Morova T, Ozturan D, Hu E, Yu IPL, Linder S, Hoogstraat M, Stelloo S, Sar F, et al. 2021. Functional mapping of androgen receptor enhancer activity. Genome Biol 22: 149. 10.1186/s13059-021-02339-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. 2011. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet 43: 27–33. 10.1038/ng.730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen MP, Knijnenburg T, Reijm EA, Simon I, Kerkhoven R, Droog M, Velds A, van Laere S, Dirix L, Alexi X, et al. 2013. Hallmarks of aromatase inhibitor drug resistance revealed by epigenetic profiling in breast cancer. Cancer Res 73: 6632–6641. 10.1158/0008-5472.CAN-13-0704 [DOI] [PubMed] [Google Scholar]
- Kastrati I, Joosten SEP, Semina SE, Alejo LH, Brovkovych SD, Stender JD, Horlings HM, Kok M, Alarid ET, Greene GL, et al. 2020. The NF-κB pathway promotes tamoxifen tolerance and disease recurrence in estrogen receptor–positive breast cancers. Mol Cancer Res 18: 1018–1027. 10.1158/1541-7786.MCR-19-1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Tolstorukov MY, Park PJ. 2008. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 26: 1351–1359. 10.1038/nbt.1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneppers J, Severson TM, Siefert JC, Schol P, Joosten SEP, Yu IPL, Huang CF, Morova T, Altintaş UB, Giambartolomei C, et al. 2022. Extensive androgen receptor enhancer heterogeneity in primary prostate cancers underlies transcriptional diversity and metastatic potential. Nat Commun 13: 7367. 10.1038/s41467-022-35135-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz G, Lopes R, Ugalde AP, Nevedomskaya E, Han R, Myacheva K, Zwart W, Elkon R, Agami R. 2016. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat Biotechnol 34: 192–198. 10.1038/nbt.3450 [DOI] [PubMed] [Google Scholar]
- Kumar S, Ambrosini G, Bucher P. 2017. SNP2TFBS: a database of regulatory SNPs affecting predicted transcription factor binding site affinity. Nucleic Acids Res 45: D139–D144. 10.1093/nar/gkw1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. 2012. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 22: 1813–1831. 10.1101/gr.136184.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer RM, Pedersen BS, DiSera T, Marth GT, Gertz J, Quinlan AR. 2018. GIGGLE: a search engine for large-scale integrated genome analysis. Nat Methods 15: 123–126. 10.1038/nmeth.4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerdrup M, Johansen JV, Agrawal-Singh S, Hansen K. 2016. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat Struct Mol Biol 23: 349–357. 10.1038/nsmb.3180 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Seo JH, Stranger B, McKenna A, Pe'er I, Laframboise T, Brown M, Tyekucheva S, Freedman ML. 2013. Integrative eQTL-based analyses reveal the biology of breast cancer risk loci. Cell 152: 633–641. 10.1016/j.cell.2012.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski MM, Willems E, Fang J, Choi J, Zhang T, Jansen PW, Brown KM, Vermeulen M. 2016. An interaction proteomics survey of transcription factor binding at recurrent TERT promoter mutations. Proteomics 16: 417–426. 10.1002/pmic.201500327 [DOI] [PubMed] [Google Scholar]
- Martin LA, Ribas R, Simigdala N, Schuster E, Pancholi S, Tenev T, Gellert P, Buluwela L, Harrod A, Thornhill A, et al. 2017. Discovery of naturally occurring ESR1 mutations in breast cancer cell lines modelling endocrine resistance. Nat Commun 8: 1865. 10.1038/s41467-017-01864-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei S, Qin Q, Wu Q, Sun H, Zheng R, Zang C, Zhu M, Wu J, Shi X, Taing L, et al. 2017. Cistrome data browser: a data portal for ChIP-seq and chromatin accessibility data in human and mouse. Nucleic Acids Res 45: D658–D662. 10.1093/nar/gkw983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michailidou K, Lindström S, Dennis J, Beesley J, Hui S, Kar S, Lemaçon A, Soucy P, Glubb D, Rostamianfar A, et al. 2017. Association analysis identifies 65 new breast cancer risk loci. Nature 551: 92–94. 10.1038/nature24284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed H, Russell IA, Stark R, Rueda OM, Hickey TE, Tarulli GA, Serandour AA, Birrell SN, Bruna A, Saadi A, et al. 2015. Progesterone receptor modulates ERα action in breast cancer. Nature 523: 313–317. 10.1038/nature14583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Morova T, McNeill DR, Lallous N, Gönen M, Dalal K, Wilson DM III, Gürsoy A, Keskin O, Lack NA. 2020. Androgen receptor-binding sites are highly mutated in prostate cancer. Nat Commun 11: 832. 10.1038/s41467-020-14644-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerdter F, Boryń Ł M, Woodfin AR, Neumayr C, Rath M, Zabidi MA, Pagani M, Haberle V, Kazmar T, Catarino RR, et al. 2018. Resolving systematic errors in widely used enhancer activity assays in human cells. Nat Methods 15: 141–149. 10.1038/nmeth.4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair SJ, Yang L, Meluzzi D, Oh S, Yang F, Friedman MJ, Wang S, Suter T, Alshareedah I, Gamliel A, et al. 2019. Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nat Struct Mol Biol 26: 193–203. 10.1038/s41594-019-0190-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M, et al. 2022. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 50: D543–D552. 10.1093/nar/gkab1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR. 2014. BEDTools: the Swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics 47: 11.12.1–11.12.34. 10.1002/0471250953.bi1112s47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/. [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481: 389–393. 10.1038/nature10730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serandour AA, Brown GD, Cohen JD, Carroll JS. 2013. Development of an Illumina-based ChIP-exonuclease method provides insight into FoxA1-DNA binding properties. Genome Biol 14: R147. 10.1186/gb-2013-14-12-r147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson TM, Nevedomskaya E, Peeters J, Kuilman T, Krijgsman O, van Rossum A, Droog M, Kim Y, Koornstra R, Beumer I, et al. 2016. Neoadjuvant tamoxifen synchronizes ERα binding and gene expression profiles related to outcome and proliferation. Oncotarget 7: 33901–33918. 10.18632/oncotarget.8983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson TM, Kim Y, Joosten SEP, Schuurman K, van der Groep P, Moelans CB, Ter Hoeve ND, Manson QF, Martens JW, van Deurzen CHM, et al. 2018. Characterizing steroid hormone receptor chromatin binding landscapes in male and female breast cancer. Nat Commun 9: 482. 10.1038/s41467-018-02856-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AA, Schuurman K, Nevedomskaya E, Stelloo S, Linder S, Droog M, Kim Y, Sanders J, van der Poel H, Bergman AM, et al. 2019. Optimized ChIP-seq method facilitates transcription factor profiling in human tumors. Life Sci Alliance 2: e201800115. 10.26508/lsa.201800115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siouda M, Dujardin AD, Barbollat-Boutrand L, Mendoza-Parra MA, Gibert B, Ouzounova M, Bouaoud J, Tonon L, Robert M, Foy JP, et al. 2020. CDYL2 epigenetically regulates MIR124 to control NF-κB/STAT3-dependent breast cancer cell plasticity. iScience 23: 101141. 10.1016/j.isci.2020.101141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weide RH, van den Brand T, Haarhuis JHI, Teunissen H, Rowland Benjamin D, de Wit E. 2021. Hi-C analyses with GENOVA: a case study with cohesin variants. NAR Genom Bioinform 3: lqab040. 10.1093/nargab/lqab040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M. 2012. Identifying chromatin readers using a SILAC-based histone peptide pull-down approach. Methods Enzymol 512: 137–160. 10.1016/B978-0-12-391940-3.00007-X [DOI] [PubMed] [Google Scholar]
- Vockley CM, D'Ippolito AM, McDowell IC, Majoros WH, Safi A, Song L, Crawford GE, Reddy TE. 2016. Direct GR binding sites potentiate clusters of TF binding across the human genome. Cell 166: 1269–1281.e19. 10.1016/j.cell.2016.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waks AG, Winer EP. 2019. Breast cancer treatment: a review. JAMA 321: 288–300. 10.1001/jama.2018.19323 [DOI] [PubMed] [Google Scholar]
- Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York. [Google Scholar]
- Yang LF, Yang F, Zhang FL, Xie YF, Hu ZX, Huang SL, Shao ZM, Li DQ. 2020. Discrete functional and mechanistic roles of chromodomain Y-like 2 (CDYL2) transcript variants in breast cancer growth and metastasis. Theranostics 10: 5242–5258. 10.7150/thno.43744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, Wan C, Mei S, Qin Q, Wu Q, Sun H, Chen CH, Brown M, Zhang X, Meyer CA, et al. 2019. Cistrome data browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res 47: D729–D735. 10.1093/nar/gky1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR. 2010. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11: 237. 10.1186/1471-2105-11-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart W, Koornstra R, Wesseling J, Rutgers E, Linn S, Carroll JS. 2013. A carrier-assisted ChIP-seq method for estrogen receptor-chromatin interactions from breast cancer core needle biopsy samples. BMC Genomics 14: 232. 10.1186/1471-2164-14-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.