Abstract
Theiler's murine encephalomyelitis virus (TMEV) is a picornavirus of the Cardiovirus genus. Certain strains of TMEV may cause a chronic demyelinating disease, which is very similar to multiple sclerosis in humans, associated with a persistent viral infection in the mouse central nervous system (CNS). Other strains of TMEV only cause an acute infection without persistence in the CNS. It has been shown that sialic acid is a receptor moiety only for the persistent TMEV strains and not for the nonpersistent strains. We report the effect of sialylation on cell surface on entry and the complex structure of DA virus, a persistent TMEV, and the receptor moiety mimic, sialyllactose, refined to a resolution of 3.0 Å. The ligand binds to a pocket on the viral surface, composed mainly of the amino acid residues from capsid protein VP2 puff B, in the vicinity of the VP1 loop and VP3 C terminus. The interaction of the receptor moiety with the persistent DA strain provides new understanding for the demyelinating persistent infection in the mouse CNS by TMEV.
Theiler's murine encephalomyelitis virus (TMEV) belongs to the Cardiovirus genus of the family Picornaviridae based on sequence analysis (34). Based on the characteristics of the disease they cause, TMEV strains are divided into two groups. Viruses of one group, including strain GDVII and FA, are highly neurovirulent, causing a rapid destruction of the neuron and killing their host in a matter of days. These viruses are unable to persist and to induce demyelination in those rare survivors (22). The other group, including strains DA, BeAn, WW, TO4, and Yale, are referred to as the Theiler's original (TO) group (23). Viruses of the TO group are much less virulent than GDVII and FA and cause a biphasic disease with mild central nervous system (CNS) damage. The first phase, or early onset, is acute encephalitis that occurs during the initial few days following intracranial inoculation. At that time, the virus is also found in neurons of the gray matter in the brain and spinal cord. However, the number of infected cells is small and most of the animals survive. Soon after, the neurons are cleared of the virus and the disease enters a second phase, or late onset, during which the virus is found to be persistent in the white matter of the spinal cord. At this stage of the viral persistence, patchy inflammation and demyelination develops in the spinal cord at the sites of infection (7, 23). The symptoms of the demyelination disease of mice caused by TMEV persistence infection in the CNS resemble those of multiple sclerosis (MS), a chronic demyelinating disease of the human CNS. Among the animal models of virus-induced demyelination in the last two decades, TMEV infection has emerged as one of the best models for studying MS.
Although these two groups of TMEV strains have both been shown to replicate well in BHK-21 cells and share high amino acid sequence homology (31, 34–36), they apparently display different pathogenesis characteristics in vivo. Many efforts have been taken to map a determinant for virus persistence, and it has been elucidated that the capsid that carries the host receptor recognition site and the antigenic sites is responsible for viral persistence (4, 11). The capsid of TMEV shares common features of the picornaviruses, a family of small RNA viruses that have three major capsid proteins (VP1, VP2, and VP3) assembled into an icosahedral shell. When the three-dimensional structure of human rhinovirus 14, a picornavirus, was determined, it was hypothesized that the depression surrounding the fivefold vertices, the canyon, was the site for receptor attachment (38). Subsequently, host receptors were identified for several picornaviruses, including rhinovirus (40), poliovirus (28), cardiovirus (14), foot-and-mouth disease virus (FMDV) (2), coxsackievirus B (1), and coxsackievirus A9 (37). The site of receptor recognition for rhinovirus 16 was later shown to be in the surface depression, as demonstrated by cryoelectron microscopy and mutagenesis (32). For FMDV, however, the receptor recognition peptide sequence RGD that interacts with the virus receptor integrin is located on a protruding loop in the virus capsid (24). The binding site for an oligosaccharide receptor of a cell culture adopted FMDV strain was also located on a separate region on the surface of the capsid (10). Despite of the failure to clearly identify the host receptor for TMEV at this time, there is some evidence that the host receptor may be a glycoprotein (20). The sites that influence the capability of TMEV demyelinating persistence in the mouse CNS are located around the depression, a putative receptor recognition site, observed in the crystal structure (12, 25, 26). These results imply that virus entry may be related to viral persistence in the mouse CNS by TMEV.
Efforts to identify the host receptor for Theiler's virus have been reported (9). When membrane proteins of susceptible cells were separated by gel electrophoresis, radiolabeled TMEV was found to bind predominantly to a 34-kDa glycoprotein (20). Wheat germ agglutinin that binds sialic acid specifically could inhibit the infectivity of TMEV or block its attachment to the protein. This suggests that sialic acid may be involved in the virus attachment, which was further approved by an experiment in which removing sialic acid molecules from the cell surface by sialidase treatment could reduce the infectivity of BeAn virus by 90% (9). More evidence was provided by our recent studies on the effects of the sialyl moiety of oligosaccharides on virus attachment and replication with the BeAn virus (44). It was shown that the infection of a demyelinating persistent TMEV strain, BeAn, was reduced by 103-fold when 9 mM sialyllactose was included in the culture medium. In addition, it was demonstrated that the inhibition of the infection of the BeAn virus strain is due to prevention of virus attachment to the cell surface (44). 35S-labeled BeAn virus was used to measure virus attachment to solubilized BHK-21 cells, and sialyllactose was shown to be effective in blocking the virus attachment to BHK-21 cells. There were no observed effects by sialic acids on the infectivity and the receptor attachment of the nonpersistent strain, GDVII, when the experiments of the 3-sialyllactose inhibition and sialidase treatment were performed under the same conditions (9, 44).
Like other picornaviruses, TMEV capsid is composed of 60 copies each of four polypeptides (VP1, VP2, VP3, and VP4), arranged with a pseudo T=3 icosahedral symmetry. Although not obviously related in sequence, VP1, VP2, and VP3 have a similar folded structure and share an antiparallel eight-stranded β-barrel. Among the capsid proteins, only VP1, VP2, and VP3 are exposed on the viral surface and are presumably responsible for cell receptor recognition. Atomic structures of TMEV strains from each group have been determined by X-ray crystallography for strains BeAn, DA, and GDVII (12, 25, 26). Sequence alignment of three major capsid proteins of these two groups of TMEVs shows that the amino acids that are identical in BeAn and DA strains but are different in GDVII are clustered on the outside of the capsid in the loops and corners connecting the β strands. Structure comparisons between GDVII and BeAn or DA revealed that structure differences between these two groups of TMEVs are small and local. Logically, these structural differences may play a functional role in differentiating these phenotypically different viruses by altering the way in which virus binds to its cellular receptor.
In this report, we describe our recent findings for the involvement of sialic acid in the entry of Theiler's virus and the crystal structure of a persistent Theiler's virus (strain DA) in complex with sialyllactose. It appears that sialic acid is a receptor moiety required only for the attachment of the persistent Theiler's viruses. This might have implications for the mechanism of in vivo persistence in the mouse CNS by the TO group of the Theiler's virus.
MATERIALS AND METHODS
Virus propagation, purification, and crystallization.
The TMEV DA strain, DAFL3, was kindly provided by R. P. Roos, at the University of Chicago Medical Center. BHK-21 cells were routinely maintained in Dulbecco modified Eagle medium DMEM medium supplemented with 7% fetal bovine serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml in a 37°C incubator with 5% CO2. For production, virus was propagated by infection of confluent BHK-21 cells at a multiplicity of infection (MOI) of approximately 10. The infection was allowed to proceed until extensive cytopathic effect (CPE) was observed (ca. 2 days). The virus titer and CPE assays for sialyllactose effects and sialidase treatment were carried out as described previously (9, 44).
To purify the DA virus, infected cells were scraped off and clarified by centrifugation at 11,300 × g for 10 min at 4°C. The pellet was resuspended in 20 mM Tris buffer with 150 mM NaCl and frozen at −80°C overnight. After thawing and sonication, the virus material was treated with 0.3% sodium dodecyl sulfate at 37°C to dissociate virus from the membranes. The virus was then loaded on the top of a 15 to 30% sucrose step gradient, followed by centrifugation at 72,100 × g for 20 h at 25°C. After centrifugation, a pellet appeared at the bottom of the tube and was treated with DNase and 2% Triton X-100. The viruses were subsequently banded through a 20 to 70% sucrose linear gradient and a Cs2SO4 equilibrium gradient. The purified viruses were dialyzed extensively against 20 mM Tris buffer (pH 8.5) before they were concentrated to a final concentration of ca. 15 mg/ml for crystallization.
The hanging-drop vapor diffusion method was used to crystallize the virus. The precipitant solution in the reservoir contained 1 to 3% polyethylene glycol (PEG)-monomethyl ether (mme) 5,000 and 0.1 M sodium phosphate buffer at pH 6.5. The drop contained 3 μl each of the virus solution and the reservoir solution. The diamond-shaped crystals grew to a maximum size ranging from 0.02 to 0.5 mm in 3 days at 20°C. The suitable crystals were harvested in 7% PEG-mme 5,000 in the same phosphate buffer before they were soaked in the harvest buffer with 30 mM 3′-sialyllactose.
Data collection and structure determination and refinement.
Soaked crystals were transferred to a solution containing 5% PEG-mme 5,000, 35% PEG 400, 30 mM sialyllactose, and 0.1 M sodium phosphate buffer (pH 6.5). Oscillation data were collected at 0.3° per frame at SSRL (Stanford Synchrotron Radiation Laboratory) beam line 7-1. A data set of a total number of 128 images measured from a frozen crystal was obtained. The diffraction data were then processed by using the HKL package (29, 33) data, with a completeness of ca. 72% to a 3.0-Å resolution. The statistics of the data set are listed in Table 1. The crystal symmetry is P21212, the same as the reported DA crystal (12) except that the frozen crystal's unit cells had shrunk ca. 6.5 to 7 Å from each direction.
TABLE 1.
Data collection and refinement statisticsa
| Type of data (parameters)b | Value |
|---|---|
| Data collection (SSRL beamline 7-1, wavelength = 1.08 Å, Mar30 image plate) | |
| Resolution (Å) | 30.0–2.98 |
| High-resolution bin (Å) | 3.09–2.98 |
| Unique reflections | 581,118 |
| Redundancy | 2.1 |
| Completenessc (%) | 71.7 (56.9) |
| <Ι>/<ςΙ>c | 9.8 (3.1) |
| Rsymc | 0.101 (0.328) |
| NCS averaging (6.0–2.98 Å) | |
| Rave | 0.231 (0.306) |
| C.C.ave | 0.880 (0.764) |
| Refinement (6.0–2.98 Å [resolution], NCS strict) | |
| Used reflections (F > 2ςF) | 483,774 |
| R factorc | 0.291 (0.392) |
| R.M.S. deviation from ideal geometry | |
| Bond (Å) | 0.018 |
| Angle (degree) | 2.04 |
| Improper (degree) | 1.88 |
| Dihedral (degree) | 26.9 |
Space group: P21212, a = 354.6 Å, b = 331.8 Å, c = 341.8 Å.
Rsym, residue of symmetry-related reflections; Rave, residue of the averaged map; C.C.ave, correlation coefficient of the averaged map; R.M.S., root mean squares; NCS, noncrystallographic symmetry.
High-resolution shell statistics are given in parentheses.
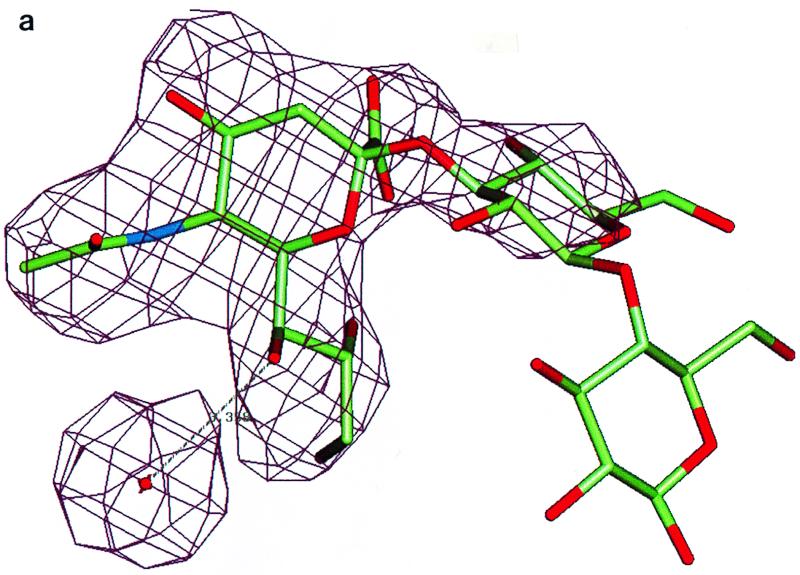
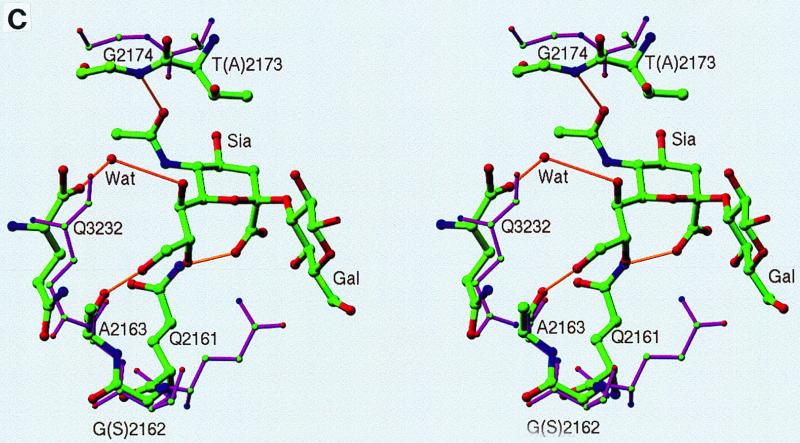
The frozen crystal shares the same packing scheme with the reported DA structure (12). One virus particle sits on the 2-fold crystallographic symmetry axis, which gives rise to the 30-fold noncrystallographic redundancy. Rotation and translation function calculations indicated a particle orientation of 3.97° around the twofold (z) axis relative to a standard icosahedral orientation and the particle center to be close to z = 0.2501. Rigid-body refinement with X-PLOR (3) with the DA native coordinate yielded an R factor of 0.32 for data between resolutions of 20 and 3.0 Å. Phases were improved by 30-fold noncrystallographic symmetry electron density averaging. An averaged difference map (see Fig. 3a) with CCP4 (6) and RAVE (21) between the observed data and DA native structure located unambiguously the position of the sialyllactose. An omit map from the improved phases also showed very clear electron density contributed by the bound 3′-sialyllactose. The galactose has recognizable density, while the glucose is completely disordered. The complex structure was then refined by one round of positional refinement by using X-PLOR to yield an R factor of 29.1% for resolution data from 6.0 to 3.0 Å. No water molecules were included except for the one coordinating with the sialic acid moiety.
FIG. 3.
(a) Averaged (Fo-Fc) electron density map (>5ς) corresponding to the sialic acid moiety in the complex of DA virus and sialyllactose. The galactose moiety has a recognizable density, while the glucose moiety is completely disordered. The complex structure was refined by using X-PLOR to yield an R factor of 29.1% for 6.0- to 3.0-Å resolution data. (b) Stick-and-ball drawing illustrating the detailed interactions of the sialic acid with the DA virus. The hydrogen bonds are indicated by yellow sticks.
RESULTS
Inhibition of DA virus growth by sialyllactose.
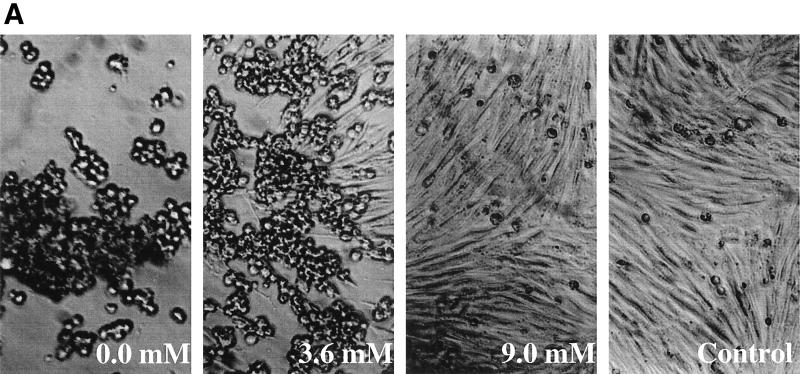
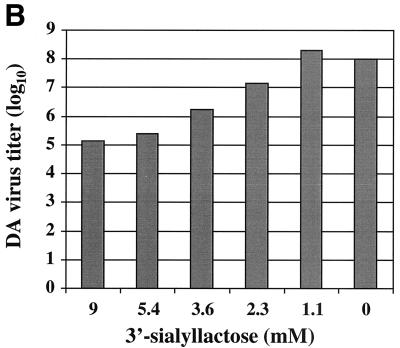
Sialyllactose was used in this experiment to present the sialic acid molecule in the α-configuration that is mostly observed in the natural glycoconjugates. Free sialic acid molecules would mostly assume the β-configuration in aqueous solutions. To verify that a second demyelinating persistent strain, DA, has the same characteristics in receptor attachment as the BeAn virus, the same experiment was performed by including sialyllactose in the growth medium. As clearly demonstrated in Fig. 1, soluble sialyllactose effectively reduced the CPE caused by DA virus infection in BHK-21 cells. The titer of the DA virus was reduced by at least 3 orders of magnitude when 9 mM sialyllactose was included in the growth medium, which was a degree of inhibition similar to that observed with BeAn virus. The results appear to support the notion that sialic acid is a critical part of the host receptor.
FIG. 1.
Effects of the sialyllactose in medium on the infectivity of the DA virus. The virus was purified as described by Zhou et al. (44). (A) At 48 h postinfection, DA virus was able to infect and induce CPE in BHK-21 cells at an MOI of 10 in the presence of 0.0, 1.1, and 2.3 mM sialyllactose, while 5.4 and 9.0 mM sialyllactose prevented CPE. The virus yield titer under the same conditions was then determined by plaque assay in six-well plates containing confluent monolayers of BHK-21 cells. In the presence of 3.6 mM sialyllactose, plaques were observed. (B) The virus yield titer was reduced by roughly 10-fold at each increment of sialyllactose concentration, with 103 reduction at 9.0 mM.
Effect of sialylation on DA virus infection.
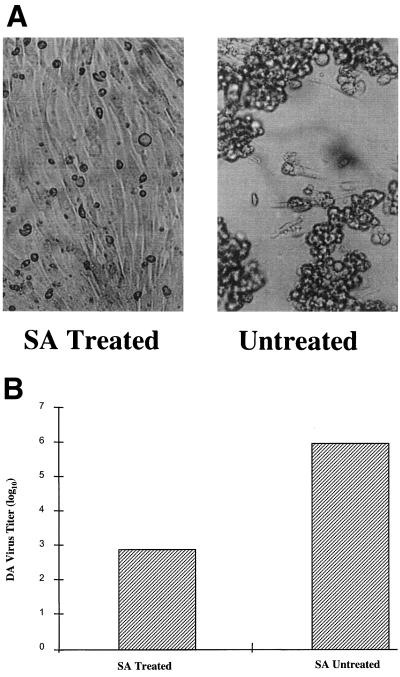
To confirm that the dependence of sialylation for entry is universal for all persistent strains of TMEV, the similar experiment previously performed with BeAn virus was repeated with the DA virus. Monolayers of BHK-21 cells were treated with bacterial sialidase (S. typhimurium, prepared in the lab of M.L. for unrelated crystallization experiments) prior to DA virus infection. Bacterial sialidase effectively removes the terminal sialic acid moieties that are linked to the oligosaccharides by 2,3 or 2,6 linkages. Confluent monolayers of BHK-21 cells were incubated with 1 ml of a sialidase solution diluted in DMEM (1 μg/liter) for 10 min. The sialidase solution was then removed. The bacterial sialidase-treated cells were then infected with an equal amount of the DA virus as with the untreated BHK-21 cells. As shown in Fig. 2, the titer of the DA virus was reduced by 3 orders of magnitudes when the BHK-21 cells were treated with bacterial sialidase. Reduced CPE that has the same appearance as that of DA virus infection in the presence of 9.0 mM sialyllactose in the medium was also observed with the bacterial sialidase treated cells (Fig. 2). Apparently, the DA virus lost its ability to attach efficiently to the host receptor for entry when sialic acids were removed from the receptor.
FIG. 2.
Effects of sialidase (SA) treatment on the infectivity of the DA virus. Confluent monolayers of BHK-21 cells were incubated with 1 ml of a sialidase solution diluted in DMEM (1 μg/liter) for 10 min. The sialidase solution was then removed. Purified DA virus stock has a concentration of 0.1 mg/ml and was diluted 100 times with DMEM medium. One milliliter of the diluted virus was added to each flask of monolayer cells, which were treated with bacterial sialidase and incubated at 37°C in an incubator with 5% CO2 for 2 h. Unattached virus was removed by washing with phosphate-buffered saline. Then, 15 ml of DMEM containing the same amount of antibiotics and bacterial sialidase but only 1% fetal bovine serum was added to the flask and incubated for 48 h at 37°C in an incubator with 5% CO2. Parallel controls were included by using untreated monolayer cells. (A) CPE was easily observed on untreated cells during DA infection, while CPE was significantly reduced when the cells were treated with bacterial sialidase. (B) The virus yield titer was determined for the two infection batches.
Binding site of the sialic acid on DA virus.
Based on the structural analyses with phenotypically different parental as well as chimeric TMEVs, we predicted that, when carried only on the persistent strains, a surface structural component next to the picornavirus putative receptor binding site may influence the ability of TMEV to persist through the differential interaction with the cellular receptor, such as binding to sialic acid (44).
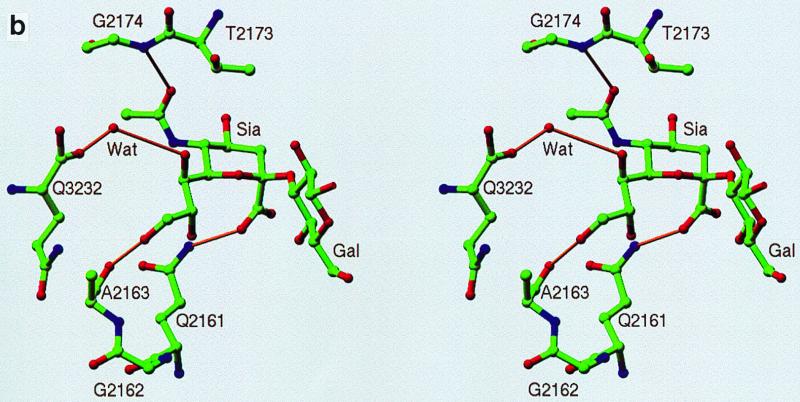
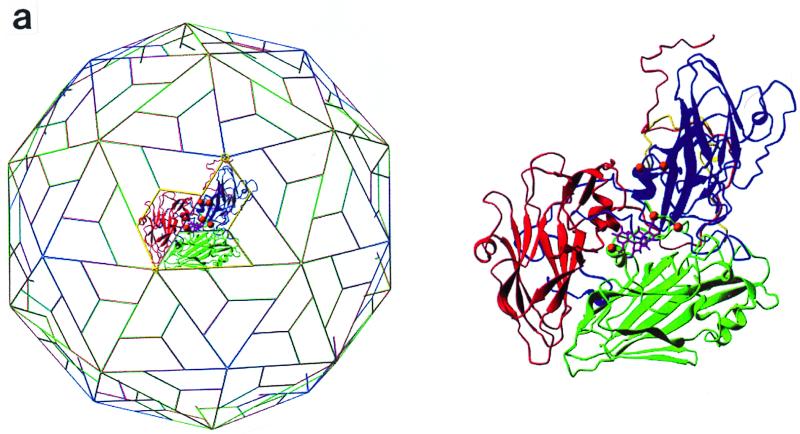
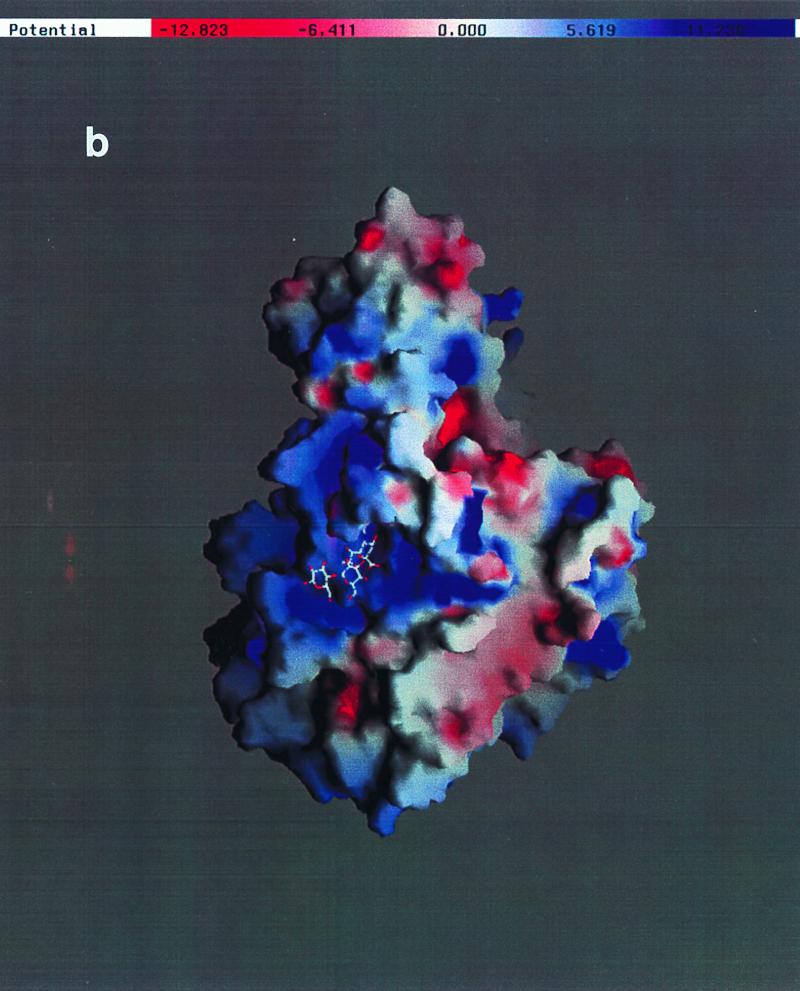
To clearly identify the binding site for sialic acid and to map the three-dimensional interactions between the DA virus and this receptor moiety, the complex structure of DA virus crystals prepared by soaking in a solution containing 30 mM sialyllactose was determined and refined to a resolution of 3 Å. The binding site (Fig. 3a and 4a) for sialic acid is located at the interface between VP1 and VP2, close to the C terminus of VP3. The center of the putative receptor binding site, the “pit,” is about 15 Å from the sialic acid binding site. The sialic acid molecule is accommodated in a positively charged depression formed by a gigantic loop, puff B of VP2 (Fig. 3b and 4b).
FIG. 4.
(a) Ribbon drawing for the protomer (VP1 in blue, VP2 in green, and VP3 in red) with the bound sialyllactose (magenta) (5). The protomer location on the icosahedral lattice is shown on the left. (b) Electrostatic potential calculated by using the coordinates of a protomer (VP1, VP2, and VP3) and the program GRASP (30). The sialic acid moiety is situated in a largely negatively charged depression near the putative receptor binding site. The areas on the side of the protomer will not be exposed in the intact icosahedral capsid. (c) Superposition of residues of GDVII virus (magenta) with those of DA virus involved in binding sialic acid. Amino acid sequence variations in GDVII virus compared to DA virus are presented in parentheses. VP2 Gly2174 in DA virus forms a hydrogen bond with the sialic acid N-acetyl carbonyl group through its main chain nitrogen. The expanded conformation of this region in GDVII virus does not allow the formation of the same hydrogen bond while maintaining the interactions with other functional groups of sialic acid. The side chain orientation of Gln2161 in GDVII does not favor a similar hydrogen bond formation with the carboxyl group of the sialic acid either. However, the distance from the amide group of Gln2161 in GDVII virus to the carboxyl group of the sialic acid is still within hydrogen bond distance.
Interaction of sialic acid with the capsid of DA virus.
The pyranose ring of the sialic acid assumes the usual chair conformation with all constituent groups at equatorial positions except for the characteristic carboxylate. In several cases, the negatively charged carboxyl group of sialic acid interacts with positively charged side chains when binding to proteins (16, 27, 41). In the DA virus, however, a hydrogen bond between the carboxylate and the side chain of VP2 Gln2161 was found (Fig. 3b). The carbonyl oxygen of the N-acetyl group is stabilized by a hydrogen bond with the main chain amide of VP2 Gly2174. There is no hydrogen bond with the amino nitrogen. The glycerol group is held extended by a hydrogen bond between the 9-hydroxyl group and the carbonyl oxygen of VP2 Ala2163. In addition, the C terminus of VP3 is positioned in the vicinity of the 7-hydroxyl group of the glycerol. The interaction between the 7-hydroxyl group and the VP3 C terminus is mediated by a well-ordered water molecule. No salt bridge is formed with the sialic acid carboxylate. The surface electrostatic potential showed that the sialic acid binding site is within a capsid surface depression with a significant positive potential (Fig. 4b). In addition to the specific hydrogen bonds, the shape complementarity and long-range charge-charge interactions may also play important roles in sialic acid recognition by the DA virus. No contacts have been observed between the capsid proteins and the last two sugar residues in the sialyllactose. No intramolecular hydrogen bond has been observed between sialic acid and either galactose or glucose.
DISCUSSION
Entry is the first interaction of the virus with its host cell. It has been shown that viruses can use proteins or oligosaccharides as host receptors, such as influenza virus that uses sialic acids as the receptor (13, 42) or poliovirus that uses a glycoprotein (28). A virus could use two separate receptors during entry, as shown by human immunodeficiency virus that uses CD4 as the initial receptor and the chemokine receptor, CCR5, as the second receptor (8). In rhinoviruses, two types of receptors are used by different members of the same virus family that are divided into major and minor groups (40). TMEV presents a new scenario in which the same family of viruses binds to the same glycoprotein receptor but in different ways. The binding of the demyelinating persistent group is dependent on the interaction with both a sialyl moiety and the protein surface of the receptor, while the nonpersistent group is only dependent on the interaction with the protein surface of the receptor, even though the two groups bind to the same receptor competitively (20).
The structure of the sialic acid-DA complex clearly identified the binding site for a receptor moiety that binds at the rim of the pit, which has been postulated as the receptor binding site (25). Mutations in the pit alter the receptor binding of Theiler's virus (S. Hertzler, M. Luo, and H. L. Lipton, unpublished data.) In demyelinating persistent strains of TMEV, the sialic acid moiety is an essential part for sufficient virus attachment to the host receptor that results in virus entry. Without the presence of sialic acid molecules on the cell surface after treatment with sialidase or with the sialic acid binding site of the virus occupied by sialyllactose, the virus can no longer bind to its host receptor tightly enough to induce endocytosis. The location of the sialic acid binding site is consistent with the positions of residues which impact virus replication and its ability to induce a demyelinating persistent infection in the mouse CNS. For instance, the major sequence variation between persistence and nonpersistent virus strains were located at VP2 position 2162 and VP2 positions 2171 to 2173, both of which are on the VP2 puffB. In DA and BeAn strains, VP2 position 2162 is Gly, and VP2 positions 2171 to 2173 are Ser-Arg-Thr. In the nonpersistent GDVII strain, however, the corresponding residues are Ser and Arg-Gln-Ala, respectively. These variable residues are not part of the sialic acid binding site (Fig. 4b and c), but they could induce changes in the residues that directly form hydrogen bonds with sialic acid, as was observed in the GDVII structure (26). The position of the amide nitrogen of Gly2174 was moved away from the sialic acid binding site in GDVII, probably due to the conformation changes induced by residues 2171 to 2173 (Arg-Gln-Ala) (Fig. 4c). The movement of the nitrogen atom of Gly2174 in GDVII is likely to abolish its hydrogen bonding capability to sialic acid. In other examples, mutations in the capsid proteins modified the ability of TMEV's persistence. When VP1 Thr1101 was mutated to Ile or Ala or when VP2 Thr2173 was mutated to Phe, the persistent DA virus either lost or had a reduced ability to persist and demyelinate (39; Y. Wada, M. L. Pierce, and R. S. Fujinami, Abstr. 12th Gen. Meet. Am. Soc. Microbiol. 1993, abstr. A37, 1993). Thr2173 is directly over the bound sialic acid moiety. A mutation to Phe will introduce a steric hindrance to the sialic acid binding site. In another instance, chimeric viruses between DA and GDVII could persist only when residue 2141 in VP2 is Lys in the parental DA virus (17, 18). These surface residues may have changed the interaction of the virus with the host CNS, most likely at the virus entry stage. Their locations suggest a role in sialic acid affinity, even though there is no direct evidence that they reduce the binding of DA virus to sialic acid.
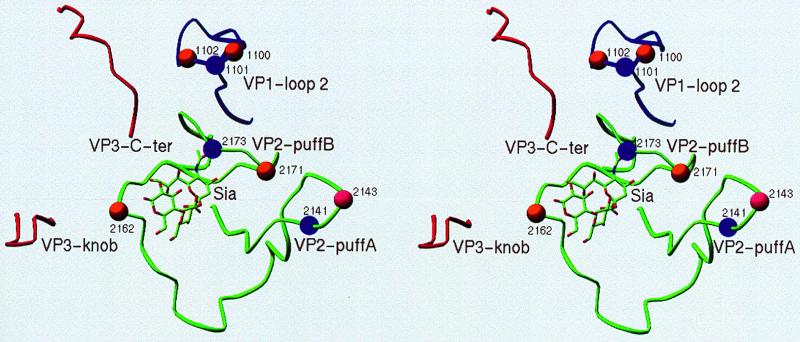
More-direct evidence for the involvement of some of those residues in virus entry is provided by the adaptation of DA viruses from BHK-21 cells to L929 cells. The wild-type DA virus was shown to have a defective entry when infecting L929 cells, which results in a slow replication rate of DA virus in L929 cells. The KJ6 DA variant that had a more rapid CPE in L929 cells was generated by multiple passages in L929 cells (19). This adaptation appears to be related to an enhanced viral entry rather than to an increased genome replication rate. Interestingly, the mutations responsible for the adaptation were found at Gly1100→Asp and Thr1102→Ala in VP1, Gly2162→Ser, Ser2171→Gly, and Thr2173→Ile in VP2. All of these mutational sites are located around the sialic acid binding site (Fig. 5). These residues overlap with the mutations described above that change the virus persistence in the CNS. KJ6 does not show a significant persistence in the mouse CNS. It is possible that these mutations alter the sialic acid binding of the DA virus by causing conformational changes in the VP2 puffB, a finding which is consistent to what we have suggested for the nonpersistent mutants.
FIG. 5.
Loop structures surrounding the sialic acid binding site. Residues that alter the capability of a demyelinating persistent infection by the DA virus are marked by spheres. The purple spheres represent those from the nonpersistent mutants. The orange spheres are those from the adaptation variant KJ6. The pink sphere represents both.
The persistence by the TO group of TMEV in the CNS is strictly an in vivo phenomenon. All strains of TMEV go through a lytic infection cycle in tissue culture. Since the capability of recognizing the sialic acid moiety is the unique feature of the demyelinating persistent group of TMEV, it may be speculated that this special interaction of the virus with a major surface component, the sialyl moiety of the gangliosides and other glycoconjugates, on the glial cells is one of the determinants of virus persistence and demyelination. There is some circumstantial evidence that might support this idea. It has been reported that administration of sialyl gangliosides to mice at the later phase of the TMEV demyelinating persistent infection could suppress the activation of pathogenic Th1 cells and did suppress the TMEV-induced demyelinating disease both clinically and histologically (15). Sialyl gangliosides could either inhibit virus infection in the CNS as that by sialyllacose in tissue culture or else act as decoys to prevent the interactions of TMEV with gangliosides on the glial cells. In the CNS, the virus particles may adhere on the surface of the glial cells by binding to nonspecific sialyl conjugates. This nonproductive attachment to the sialylated surface molecules could reduce the effective infectivity of persistent TMEV (an increased ratio of particle/PFU). It could also prevent the association of several key proteins required for myelination, such as myelin-associated glycoproteins (MAG). MAG mediates the association of myelinating glial cells with neuronal axon. It has been shown that the MAG-mediated cell-cell interactions occur through the recognition and attachment of MAG to sialyl gangliosides, as demonstrated by Yang et al. (43). A recent study on the interaction of the foot-and-mouth disease with heparan sulfate also revealed a receptor binding site for an oligosaccharide moiety on the viral surface (10).
ACKNOWLEDGMENTS
L.Z. and Y.L. contributed equally to this work.
We are very grateful to Michael Soltis and the staff for their help at SSRL beamline 7-1. We also appreciate the help from Bindong Sha in data collection. We thank Ray P. Roos, University of Chicago Medical Center, for providing the DAFL3 virus stock. We also thank L. Andrew Ball for critical reading of the manuscript.
L. Zhou was supported by a National Research Service Award from the National Institutes of Health.
REFERENCES
- 1.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 2.Berinstein A, Roivainen M, Hovi T, Mason P W, Baxt B. Antibodies to the vitronectin receptor (integrin avβ3) inhibit binding nad infection of foot-and-mouth disease virus to cultured cells. J Virol. 1995;69:2664–2666. doi: 10.1128/jvi.69.4.2664-2666.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunger A T. X-PLOR version 3.1: a system for X-ray crystallography and NMR. New Haven, Conn: Yale University Press; 1992. [Google Scholar]
- 4.Calenoff M A, Faaberg K S, Lipton H L. Genomic regions of neurovirulence and attenuation in Theiler murine encephalomyelitis virus. Proc Natl Acad Sci USA. 1990;87:978–982. doi: 10.1073/pnas.87.3.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carson M. Ribbons 2.0. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- 6.CCP4 (Collaborative Computing Project no. 4) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 7.Chamorro M, Aubert C, Brahic M. Demyelinating lesions due to Theiler's virus are associated with ongoing central nervous system infection. J Virol. 1986;57:992–997. doi: 10.1128/jvi.57.3.992-997.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe H, Martin K A, Farzan M, Sodroski J, Gerard N P, Gerard C. Structural interactions between chemokine receptors, gp120 Env and CD4. Semin Immunol. 1998;10:249–257. doi: 10.1006/smim.1998.0127. [DOI] [PubMed] [Google Scholar]
- 9.Fotiadis C, Kilpatrick D R, Lipton H L. Comparison of the binding characteristics to BHK-21 cells of viruses representing the two Theiler's neurovirulence groups. Virology. 1991;182:365–370. doi: 10.1016/0042-6822(91)90683-3. [DOI] [PubMed] [Google Scholar]
- 10.Fry E E, Lea S M, Jackson T, Newman J W I, Ellard F M, Blakemore W E, Abu-Ghazaleh R, Samuel A, King A M Q, Stuart D I. The structure and function of a foot-and-mouth disease virus-oligosaccharide receptor complex. EMBO J. 1999;18:543–554. doi: 10.1093/emboj/18.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu J, Rodriguez M, Roos R P. Strains from both Theiler's virus subgroups encode a determinant for demyelination. J Virol. 1990;64:6345–6348. doi: 10.1128/jvi.64.12.6345-6348.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grant R A, Filman D J, Fujinami R S, Icenogle J P, Hogle J P. Three-dimensional structure of Theiler's virus. Proc Natl Acad Sci USA. 1992;89:2061–2065. doi: 10.1073/pnas.89.6.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirst G K. The agglutination of red blood cells by allontoic fluid of chick embryos infected with influenza virus. Science. 1941;94:22–23. doi: 10.1126/science.94.2427.22. [DOI] [PubMed] [Google Scholar]
- 14.Huber S A. VCAM-1 is a receptor for encephalomyocarditis virus on murine vascular endothelial cells. J Virol. 1994;68:3453–3458. doi: 10.1128/jvi.68.6.3453-3458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue A, Koh C, Yanagisawa N, Taketomi T, Ishihara Y. Suppression of Theiler's murine encephalomyelitis virus induced demyelinating disease by administration of gangliosides. J Neuroimmunol. 1996;6:45–53. doi: 10.1016/0165-5728(95)00149-2. [DOI] [PubMed] [Google Scholar]
- 16.Janakiraman M N, White C L, Laver W G, Air G M, Luo M. Structure of influenza virus neuraminidase B/Lee/40 complexed with sialic acid and a dehydro analog at 1.8 Å resolution: implications for the catalytic mechanism. Biochemistry. 1994;33:8172–8179. doi: 10.1021/bi00193a002. [DOI] [PubMed] [Google Scholar]
- 17.Jarousse N, Fiette L, Grant R A, Hogle J M, McAllister A, Michiels T, Aubert C, Tangy F, Brahic M, Rossi C P. Chimeric Theiler's virus with altered tropism for the central nervous system. J Virol. 1994;68:2781–2786. doi: 10.1128/jvi.68.5.2781-2786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarousse N, Grant R A, Hogle J M, Zhang L, Senkowski A, Roos R P, Michiels T, Brahic M, McAllister A. A single amino acid change determines persistence of a chimeric Theiler's virus. J Virol. 1994;68:3364–3368. doi: 10.1128/jvi.68.5.3364-3368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jnaoui K, Michiels T. Adaptation of Theiler's virus to L929 cells: mutations in the putative receptor binding site on the capsid map to neutralization sites and modulate viral persistence. Virology. 1998;244:397–404. doi: 10.1006/viro.1998.9134. [DOI] [PubMed] [Google Scholar]
- 20.Kilpatrick D R, Lipton H L. Predominant binding of Theiler's viruses to a 34-kilodalton receptor protein on susceptible cell lines. J Virol. 1991;65:5244–5249. doi: 10.1128/jvi.65.10.5244-5249.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleywegt G J, Jones T A. From first map to final model (CCP4 manual). Warrington, United Kingdom: Daresburg Laboratory; 1994. [Google Scholar]
- 22.Lipton H L. Persistent Theiler's murine encephalomyelitis virus infection in mice depends on plaque size. J Gen Virol. 1980;46:169–177. doi: 10.1099/0022-1317-46-1-169. [DOI] [PubMed] [Google Scholar]
- 23.Lipton H L. Theiler's virus infection in mice: an unusual biphasic disease process leading to demyelination. Infect Immun. 1975;11:1147–1155. doi: 10.1128/iai.11.5.1147-1155.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Logan D, Abu-Ghazaleh R, Blakemore W, Curry S, Jackson T, King A, Lea S, Lewis R, Newman J, Parry N, Rowlands D, Stuart D, Fry E. Structure of a major immunogenic site on foot-and-mouth disease virus. Nature. 1993;362:566–568. doi: 10.1038/362566a0. [DOI] [PubMed] [Google Scholar]
- 25.Luo M, He C, Toth K S, Zhang C X, Lipton H L. Three-dimensional structure of Theiler's murine encephalomyelitis virus (BeAn strain) Proc Natl Acad Sci USA. 1992;89:2409–2413. doi: 10.1073/pnas.89.6.2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo M, Toth K S, Zhou L, Pritchard A, Lipton H L. The structure of a highly virulent Theiler's murine encephalomyelitis virus (GDVII strain) and implications for determinants of viral persistence. Virology. 1996;220:246–250. doi: 10.1006/viro.1996.0309. [DOI] [PubMed] [Google Scholar]
- 27.May A P, Robinson R C, Vinson M, Crocker P R, Jones E Y. Crystal structure of the N-terminal domain of sialoadhesin in complex with 3′ siallylactose at 1.85 Å resolution. Mol Cell. 1998;1:719–728. doi: 10.1016/s1097-2765(00)80071-4. [DOI] [PubMed] [Google Scholar]
- 28.Mendelsohn C L, Wimmer E, Racaniello V R. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell. 1989;13:881–887. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 29.Minor W. XdisplyF Program. West Lafayette, Ind: Purdue University; 1993. [Google Scholar]
- 30.Nicholls A. GRASP: graphical representation and analysis of surface properties. New York, N.Y: Columbia University; 1992. [Google Scholar]
- 31.Ohara Y, Stein S, Fu J, Stillman L, Klaman L, Roos R P. Molecular cloning and sequence determination of DA strain in the native murine encephalomyelitis virus. Virology. 1988;164:245–255. doi: 10.1016/0042-6822(88)90642-3. [DOI] [PubMed] [Google Scholar]
- 32.Olson N H, Kolatkar P R, Oliveira M A, Cheng R H, Greve J M, McClelland A, Baker T S, Rossmann M G. Structure of a human rhinovirus complexed with its receptor molecule. Proc Natl Acad Sci USA. 1993;90:507–511. doi: 10.1073/pnas.90.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otwinowski Z. Proceedings of the CCP4 Study Weekend. Warrington, United Kingdom: SERC Daresbury Laboratory; 1993. p. 56. [Google Scholar]
- 34.Ozden S, Tangy F, Chamorro M, Brahic M. Theiler's virus genome is closely related to that of encephalomyocarditis virus, the prototype cardiovirus. J Virol. 1986;60:1163–1165. doi: 10.1128/jvi.60.3.1163-1165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pevear D C, Borkowski J, Calenoff M, Oh C K, Ostrowski B, Lipton H L. Insights into Theiler's virus neurovirulence based on a genomic comparison of the beurovirulent GDVII and less virulent BeAn strains. Virology. 1988;164:1–12. doi: 10.1016/0042-6822(88)90652-6. [DOI] [PubMed] [Google Scholar]
- 36.Pevear D C, Calenoff M, Rozhon E, Lipton H L. Analysis of the complete nucleotide sequence of the picornavirus Theiler's murine encephalomyelitis virus indicates that it is closely related to cardioviruses. J Virol. 1987;61:1507–1516. doi: 10.1128/jvi.61.5.1507-1516.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roivainen M, Piirainen L, Hovi T, Virtanen I, Riikonen T, Heino T, Hyypia T. Entry of coxsackievirus A9 into host cells: specific interactions with αvβ3 integrin, the vitronectin receptor. Virology. 1994;203:357–365. doi: 10.1006/viro.1994.1494. [DOI] [PubMed] [Google Scholar]
- 38.Rossmann M G, Arnold E, Erockson J W, Frankenberger E A, Griffith J P, Hecht H J, Johnson J E, Kamer G, Luo M, Mosser A G, Rueckert R R, Sherry B, Vriend G. Structure of a human common cold virus and functional relationships to other picornaviruses. Nature. 1985;317:145–153. doi: 10.1038/317145a0. [DOI] [PubMed] [Google Scholar]
- 39.Sato S, Zhang L, Kim J, Jakob J, Grabt R A, Wollmann R, Roos R P. A neutralization site of DA strain of Theiler's murine encephalomyelitis virus important for disease phenotype. Virology. 1996;226:327–337. doi: 10.1006/viro.1996.0660. [DOI] [PubMed] [Google Scholar]
- 40.Staunton D E, Merluzzi V J, Rothlein R, Barton R, Marlin S D, Springer T A. A cell adhesion molecule, ICAM-1, is the major surface receptor for rhinoviruses. Cell. 1989;61:849–853. doi: 10.1016/0092-8674(89)90689-2. [DOI] [PubMed] [Google Scholar]
- 41.Stehle T, Harrison S C. Crystal structures of murine polyomavirus in complex with straight-chain and branched-chain sialyloligosaccharide receptor fragments. Structure. 1996;4:183–194. doi: 10.1016/s0969-2126(96)00021-4. [DOI] [PubMed] [Google Scholar]
- 42.Weis W, Brown J H, Cusack S, Paulson J C, Skehel J J, Wiley D C. Structure of the influenza virus haemaglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 43.Yang L J, Zeller C B, Shaper N J, Kiso M, Hasegawa A, Shapiro R E, Schnaar R L. Gangliosides are neuronal ligands for myelin-associated glycoprotein. Proc Natl Acad Sci USA. 1996;93:814–818. doi: 10.1073/pnas.93.2.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou L, Lin X, Green T J, Lipton H L, Luo M. Role of sialyloligosaccharide binding in Theiler's virus persistence. J Virol. 1997;71:9701–9712. doi: 10.1128/jvi.71.12.9701-9712.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]