Therapeutic usage of cytokines in patients is limited by toxicity. The authors report that blocking a cytokine binding protein, IL18BP, to enhance the cytokine’s natural activity yields antitumor activity in preclinical models and shows promise for clinical translation.
Abstract
Recombinant cytokines have limited anticancer efficacy mostly due to a narrow therapeutic window and systemic adverse effects. IL18 is an inflammasome-induced proinflammatory cytokine, which enhances T- and NK-cell activity and stimulates IFNγ production. The activity of IL18 is naturally blocked by a high-affinity endogenous binding protein (IL18BP). IL18BP is induced in the tumor microenvironment (TME) in response to IFNγ upregulation in a negative feedback mechanism. In this study, we found that IL18 is upregulated in the TME compared with the periphery across multiple human tumors and most of it is bound to IL18BP. Bound IL18 levels were largely above the amount required for T-cell activation in vitro, implying that releasing IL18 in the TME could lead to potent T-cell activation. To restore the activity of endogenous IL18, we generated COM503, a high-affinity anti-IL18BP that blocks the IL18BP:IL18 interaction and displaces precomplexed IL18, thereby enhancing T- and NK-cell activation. In vivo, administration of a surrogate anti-IL18BP, either alone or in combination with anti-PD-L1, resulted in significant tumor growth inhibition and increased survival across multiple mouse tumor models. Moreover, the anti-IL18BP induced pronounced TME-localized immune modulation including an increase in polyfunctional nonexhausted T- and NK-cell numbers and activation. In contrast, no increase in inflammatory cytokines and lymphocyte numbers or activation state was observed in serum and spleen. Taken together, blocking IL18BP using an Ab is a promising approach to harness cytokine biology for the treatment of cancer.
Introduction
Immunostimulatory mAbs that block inhibitory checkpoint proteins such as CTLA-4 or PD-(L)1 marked a paradigm shift in the development of anticancer therapies. Nonetheless, most patients do not respond to such treatments and many who respond initially, ultimately develop acquired resistance (1). Thus, there is an unmet need to develop treatments that modulate additional immune pathways to increase response rates and durability. Recombinant cytokines were the first cancer immunotherapy agents that were clinically approved due to their potent immunostimulatory capacity (2, 3). However, cytokines are mostly given systemically and, due to their short half-life, need to be administered at high doses to achieve a concentration sufficient for eliciting potent antitumor responses in the tumor microenvironment (TME; refs. 4, 5). Consequently, recombinant cytokines are endowed with challenging therapeutic windows and their use is limited by systemic toxicities (6). Multiple approaches have been explored to improve the therapeutic window of recombinant cytokines, but to date, preclinical success has not been translated to meaningful improvement in clinical outcome (4).
IL18, a member of the IL1 family of cytokines, is an inflammasome-induced cytokine that is produced by multiple cell types, including myeloid cells and epithelial cells, in response to pathogen or danger signals and activates host defense mechanisms (7–9). Initially identified as IFNγ-inducing factor, IL18 has been shown to stimulate innate lymphoid cells and antigen-experienced T cells through binding to a heterodimeric receptor complex consisting of IL18Rα and IL18Rβ chains (10, 11). Recombinant (r) IL18 administration was evaluated in clinical studies in patients with advanced cancer, where it was well tolerated but showed limited efficacy (12). This is probably not only due to the general limitations of cytokines as therapeutic agents, but also due to the induction of IL18 binding protein (IL18BP) following IL18 administration (12). IL18BP is a soluble high-affinity inhibitor of IL18, which is induced in response to IFNγ upregulation as part of a negative feedback mechanism to prevent excessive IL18 immunostimulatory activity (11, 13). In this study, we computationally identified IL18BP as a potential mechanism by which tumor-associated macrophages (TAM) could mediate resistance to antitumor immunity. Subsequently, we measured high levels of bound IL18 in the TME of samples from patients across multiple cancer indications. To unleash endogenous bound IL18 activity, COM503, a fully human anti-IL18BP blocker was generated and examined in vitro for its ability to enhance T- and NK-cell activity. To assess the effect of IL18BP blockade in vivo, a surrogate anti-mouse IL18BP blocker was generated and tested in multiple mouse tumor models. Our results demonstrate the potential of blocking IL18BP as a promising approach for cancer immunotherapy.
Materials and Methods
Identification of IL18BP as a potential soluble immune checkpoint
Gene arrays and RNA sequencing (RNA-seq) from both human and mouse studies of TAMs, tissue-resident macrophages, and in vitro polarized macrophages were downloaded from the GEO repository and subject to internal QC, normalization, and statistical analysis (Supplementary Table S1). Cancer gene expression profiles were obtained using TCGA primary or metastatic sample data (https://portal.gdc.cancer.gov) through Omicsoft Oncoland platform (http://www.omicsoft.com/oncoland-service/). The data contained >10,000 primary tumor samples and ∼400 metastatic tumor samples across 33 tumor types. Pathologic tumor stage was classified based on The AJCC Cancer Staging System (14). Affymetrix microarrays were processed using MAS5 package, non-Affymetrix microarrays were used as provided by NCBI and next-generation sequencing data were processed from raw data using the Omicsoft platform (http://www.omicsoft.com/). For single cell (sc)RNA-seq data, UMIs were quantified and mapped to the reference transcriptome GRCh38 (GENCODE v32/Ensembl 98) using Cellranger 6.0 (10× Genomics). Subsequent analyses were performed Scanpy (https://github.com/scverse/scanpy/). Standard QC and preprocessing steps were applied to the cells, top variable genes were selected and principal component analysis (PCA) was applied to them. Top principal components were used to construct UMAP embedding and shared nearest neighbor (SNN) graph was calculated with n = 15 followed by Leiden clustering.
Study approval
Between mid-2020 till mid-2023, tumor biopsies, human normal tissue adjacent to the tumor (NAT), and serum samples from healthy donors and patients with cancer were obtained from hospitals in Israel in accordance with the Israel Institutional Review Board (0827–20RMC, 0711–15RMC, 0763–16RMC, and 0333–17-RMB-GN) and purchased from Fidelis Research (Bulgaria, FRT-18101) and Almog diagnostic (Israel; Information about the samples is provided in Supplementary Tables S2, S3, and S4). The tumor-infiltrating lymphocytes (TIL) that were used in the TILs-MEL624 co-culture assay were obtained from Professor Mical Lotem (Center for Melanoma and Cancer Immunotherapy, Department of Oncology, Hadasa Hospital, Jerusalem) in accordance with Israel Helsinki committee (protocol no. 1419). Written informed consent was received from participants prior to inclusion in the study, which was conducted in agreement with the Helsinki Declaration.
Cell lines
B16/Db-hmgp100 cells were kindly provided by Dr. Hanada and colleagues (HHS agency, 2014) and were licensed from NIH. B16/Db-hmgp100 cells were generated by double transduction of B16F10 with H-2Db and a retrovirus that encodes chimeric mouse gp100, which is comprised of the human gp10025–33 and the rest of mouse gp100. Murine colon carcinoma MC38OVAdim cells (clone UC10 4H10) and murine fibrosarcoma MCA100 cells (derived from MCA-treated wild-type mice; ref. 15) were received from the Peter MacCallum cancer center (2015). E0771 cell line was purchased from CH3 BioSystems (Product: #94A001). CT26 (CRL-2638), LLC1 (CRL-1642), Renca (CRL-2947), E.G7-OVA (CRL-2113), and 4T1 (CRL-2539) cell lines were purchased from ATCC. MEL624 cell line was licensed from NIH. All cell lines were routinely checked for Mycoplasma using the MycoAlert Detection Kit (Lonza). E0771, B16/Db-hmgp10, MC38OVAdim, and MEL624 cell lines were reauthenticated using short tandem repeat (STR) analysis as described in ASN-0002–2022 by ATCC in the last year. MCA100, LLC1, 4T1, CT26, Renca, and E.G7-OVA were not reauthenticated in the last year. For E0771, 4T1, CT26, and B16/Db-hmgp100, culture maintenance medium of the cells contained RPMI1640 + 10% FBS + 4 mmol/L glutamine + 1% HEPES + 1% NEAA + 50 μmol/L 2-ME + 1% sodium pyruvate +1% Pen/Strep, for B16/Db-hmgp100, 10 μg/mL blasticidin was added. LLC1 culture medium contained DMEM + 10% FBS+ 4 mmol/L glutamine +1% Pen/Strep. For MCA100 and MC38OVAdim, culture maintenance medium of the cells contained DMEM+10% FBS+ 4 mmol/L glutamine + 1% HEPES + 1% NAAA + 50 μmol/L 2-ME + 1% sodium pyruvate +1% PenStrep. Culture maintenance of Renca, and E.G7-OVA cell lines was according to ATCC guidelines. MEL624 and MEL624 cells overexpressing human PD-L1 culture medium contained DMEM + 10% FBS + 4 mmol/L glutamine +1% Pen/Strep. For MEL624 cells overexpressing human PD-L1 1 μg/mL puromycin was added. MEL624 cells overexpressing human PD-L1 were generated by lentiviral transduction of pcLV-CMV- hPDL-1-IRES-Neo by using 10 MOI in the presence of polybrene (6 μg/mL). Next day, cell medium was replaced and 1 μg/mL puromycin was added to the culture.
In vivo tumor models
All in vivo studies were approved by the Institutional Animal Care and Use Committee at Tel Aviv University (01-22–001). Six-to-8-week-old C57BL/6 and BALB/c female mice (Envigo) were maintained in a specific pathogen-free (SPF) animal facility with food and water provided, ad libitum.
Experiments were conducted with low-passage cultures (<passage 4) of mouse tumor cells. Mice were acclimated for at least 1 week before tumor inoculation. MC38OVAdim (106 or 1.2 × 106), CT26 (2.5 × 105), LLC (2 × 105), MCA100 (5 × 106), or B16/Db-hmgp100 (1 × 105) cells were injected subcutaneously into the right flank of the mouse (50 μL). 4T1 cells (2 × 105) and E0771 cells (5 × 105) were inoculated orthotopically into the left mammary fat pad in a 1:1 mixture with Matrigel (Corning) in a volume of 100 μL. Tumor growth was measured with an electronic caliper every 2 to 3 days and was reported as 0.5 × W2 × L mm3. The experimental endpoint was defined as tumor volume of 1,800 mm3. Mice with body weight loss of above 10% between measurements, or 20% reduction from initial weight were excluded. At a tumor volume of 130 to 260 mm3 for MC38OVAdim, 260 mm3 for E0771, 190 mm3 for MCA100, or on day 4 posttumor inoculation (palpable tumors) for B16/Db-hmgp100, LLC, and 4T1, mice were randomly assigned into treatment groups. For combination studies with anti-PD-L1, treatment initiated at a tumor volume of 330 mm3 for E0771 or on day 4 post-inoculation of CT26. Anti-IL18BP, isotype control or anti-PD-L1 were injected intraperitoneally twice a week for a total of six treatments. For depletion studies, MC38OVAdim engrafted mice were assigned into four groups (Supplementary Table S5) and were administered with anti-CD4 (GK1.5, 300 μg), anti-CD8 (Lyt 3.2, 100 μg), anti-NK1.1 (PK136, 50 μg), or isotype control (50 μg) prior to tumor inoculation (day −1), and on days 6, 13, and 20 for a total of four treatments. At tumor volume of 120 mm3, mice were randomly assigned into treatment groups and treated intraperitoneally with anti-IL18BP or with isotype control (10 mg/kg) twice a week for a total of six treatments. For immune phenotyping studies, C57BL/6 mice were inoculated with MC38OVAdim (1.2 × 106) or with E0771 cells (5 × 105) as described above and treated with anti-IL18BP Ab or isotype control at tumor volume of 120 mm3 for MC38OVAdim and 330 mm3 for E0771 twice a week (15 and 10 mg/kg, respectively). In MC38OVAdim model, tumors, spleens, and sera were collected 24 hours after the fourth treatment. E0771 tumors were collected 24 hours after the third treatment. For cytokine staining, cells were stimulated with 50 ng/mL phorbol myristate acetate (PMA), 1 μg/mL ionomycin and BFA. Then, cells were stained extracellularly for membrane markers and intracellularly for cytokine expression. To examine the capability of anti-IL18BP Ab to induce immunogenic memory, groups of 5 to 10 C57BL/6 tumor-naïve age-matched mice were orthotopically inoculated with E0771. When tumor reached the volume of 250 mm3, mice were treated with either anti-IL18BP Ab or isotype control (15 mg/kg) twice a week for 3 weeks. After 2 months, tumor-free and naïve aged-matched mice were orthotopically re-inoculated with E0771 (5 × 105). Spleens from naïve or rechallenged mice were harvested 12 weeks following primary tumor inoculation, dissociated to single-cell suspension, and stained for lymphoid panel (see Tissue processing).
Reagents
Commercial ELISA kits were used for detection of human IL18 (MBL), human IL18BP (R&D), and mouse IL18 (MBL) in serum and tumor-derived supernatants (TDS). Human IL18 ELISA Kit was validated to recognize mature IL18 but not pro-IL18. To detect free human IL18 and mouse IL18BP in TDS and serum, ELISA assays were developed (see Free human IL18 ELISA assay establishment and Mouse IL18BP ELISA assay establishment). Cytometric bead array (CBA) human TH1/TH2/TH17 Cytokine Kit (BD Bioscience), CBA human IFNγ Kit (BD Bioscience), CBA Mouse Inflammatory Cytokine Kit (BD Bioscience), CBA mouse Th1, Th2, Th17 Cytokine Kit (BD Bioscience), and LEGENDplex Human Inflammation Panel 1 (BioLegend) were used for cytokine detection. Human IL18 (R&D), human IL18BPa (R&D), human IL12 (R&D), human IL18BPa mIgG1 His monomer (produced by GenScript Biotech), mouse IL18 (R&D), mouse IL18BPd (R&D), and mouse IL12 (R&D) proteins were used for biochemical, affinity measurements, and in vitro assays. A list of Abs to human and mouse lineage markers that were used for immune infiltrate staining in human and murine tumors and in rechallenge experiments is presented in Supplementary Tables S6 to S8.
Free human IL18 ELISA assay establishment
To capture free human IL18, neutralizing antihuman IL18 (clone 12GL, patent US 20140004128A1) was diluted to 1 μg/mL in PBS and coated on ELISA plates overnight at 4°C. Coated plates were washed with PBS (X3) and blocked with 1% BSA/PBS buffer at room temperature (RT) for 2 hours. Blocking buffer was removed, plates were washed, and samples (diluted 1:2 with 1% BSA/PBS) were added for 1 hour incubation at RT. Standard curve was generated by incubating two-fold serial dilutions of recombinant human IL18 (starting at 1 ng/mL; R&D systems) in 1% BSA/ PBS buffer. Following a washing step, biotinylated anti-IL18 (diluted 1/1,000 in 1% BSA/PBS, D045–6; R&D systems) was added for 1 hour incubation at RT. HRP-conjugated streptavidin (50 μL/well, Jackson) was added for 1 hour incubation at RT for detection and ELISA signals were developed by addition of TMB substrate (50 μL; Scytek). Reactions were stopped by addition of HCL solution (50 μL 1N), and absorbance signals were read at 450 nm on a luminescence Reader (EnSpire; Perkin Elmar).
Mouse IL18BP ELISA assay establishment
To capture mouse IL18BP, anti-mouse IL18BPd (R&D systems, AF122) was diluted to 1 μg/mL in PBS and coated on ELISA plates (Maxisorb; Thermo Fisher Scientific) overnight at 4°C. The coated plates were washed with 0.05% Tween PBS buffer (X3) and blocked with 2.5% skim milk/PBS buffer at RT for 2 hours. Blocking buffer was removed, plates were washed, and samples were added for 1 hour incubation at RT. Standard curve was generated by incubating two-fold serial dilutions of recombinant mouse IL18BP-Fc protein (0–100 ng/mL; R&D systems, 122-BP) in 2.5% skim milk/ PBS buffer. Following a washing step, polyclonal anti-IL18BP (diluted 1/1,000 in 1% BSA/PBS, BAF129; R&D systems) was added for 1 hour at RT. HRP-conjugated streptavidin (diluted 1:1,000 in 2.5% skim milk/PBS) was used for detection and ELISA signals were developed by addition of TMB substrate (50 μL). Reactions were stopped by addition of HCL solution (50 μL 1N), and absorbance signals were read at 450 nm on a luminescence Reader.
Tissue processing
Human and mouse serum samples were separated from healthy and cancer patients’ blood (Supplementary Tables S2 and S9) and stored at −80°C. Samples were thawed on ice prior to IL18 and IL18BP testing by ELISA. Fresh human and mouse tumor samples and NAT samples (Supplementary Tables S2, S3, and S9) were dissociated by gentleMACS (Miltenyi Biotec) using Human and Mouse Dissociation Kits (Miltenyi Biotec). NAT samples were collected from the tumor margin and did not contain tumor by histopathologic review. Spleens from C57BL/6 or Balb/C mice were mashed, passed through a 40 μm cell strainer, and centrifuged at 1,200 rpm for 8 minutes. Dissociated cells were stained with Aqua Live Dead (Life Technologies) to distinguish live cells from dead cells and with a cocktail of anti-CD16 (mouse: clone 2.4G2, BD. human3G8; BioLegend), anti-CD32 (mouse: clone 2.4G2, BD. human:FUN2; BioLegend), and anti-CD64 (human:S18012C; BioLegend) to block nonspecific binding to Fcγ receptors. Cells were washed and stained with a target Ab or control isotype cocktail (see Supplementary Tables S10 and S11 for details of the Abs used). All staining was carried out for 30 minutes at 4°C. Samples were acquired on a Fortessa X-20 flow cytometer (BD Biosciences). Analysis was completed using FlowJo (TreeStar LLC) and gated on specific populations. Gating lineages for human and mouse tumor panels are described in Supplementary Tables 6 and 7.
Antibodies
COM503 is a fully human monoclonal anti-IL18BP isolated by panning a yeast display Ab library (Adimab) with human IL18BP FC protein (see Supplementary Materials and Methods). COM503 was optimized for improved affinity via light chain batch shuffle, and then by introducing diversities into the heavy chain and light chain variable regions. Anti-mouse IL18BP mAb was isolated by panning a phage display Ab library (Bio-Rad) with mouse IL18BP FC protein. Pembrolizumab (anti-PD-1 Ab), anti-PD-L1 (mIgG1, Clone YW243.55.S70), and isotype control Abs (mIgG1 D265A and hIgG4 S228P) were produced by GenScript Biotech.
KinExA
The binding affinity (Kd) and kinetics (kon & koff) were measured for human IL18 binding to human IL18BPα-Fc monomer, human IL18BPα-Fc monomer binding to COM503 and mouse IL18 binding to mouse IL18BPd-Fc using KinExA. Human or mouse IL18BP were used as the column binding protein (CBP). Two or three curves with different CBP concentrations were run and analyzed using n-curve analysis to determine the Kd. The low error and well defined 95% confidence intervals give confidence to the accuracy of the Kd measured. The measurements considered the CBP as 100% active and used it as the concentration reference. All measurements were performed using a KinExA 3200 S/N: 5000 and Autosampler S/N: 8064 (Sapidyne Instruments Inc., Boise, Idaho).
Surface plasmon resonance
The binding affinity (Kd) and kinetics (kon and koff) were measured for mouse IL18BP binding to anti-mouse IL18BP using a Biacore S-200 instrument. Each experiment cycle began with an injection (15 seconds at 10 μg/mL) of antimouse IL18BP over the flow cell. Upon capture of anti-mouse IL18BP to the sensor surface, 12 two-fold serial dilutions of mouse I-18BP protein starting from 256 nmol/L were injected over all channels at 30 μL/min for 180 seconds. The dissociation of the IL18BP was monitored for 1,200 seconds. Several blank buffer samples were injected (1,000 seconds at 30 μL/minute) over the flow cell and used for reference surface subtraction. Chip surface was regenerated with 10 μL/min injection of glycine-HCl pH 1.5 for 60 seconds after each cycle. The resulting sensorgrams were processed and double-referenced using Biacore S-200 evaluation software. Where appropriate, the sensorgrams were fit with a simple 1:1 kinetic binding model.
Displacement of IL18 from preformed rIL18:rIL18BP complexes by COM503
COM503 was tested for displacement of human IL18 prebound to IL18BP by ELISA. Human anti-IL18 (clone 12GL) was coated on wells of a high binding plate overnight at 4°C (2.5 μg/mL, Thermo Fisher Scientific). Coated plates were rinsed once with PBS and incubated with blocking buffer (2.5% skim milk/PBS) for 2 hours at RT. Blocking buffer was removed, plates were washed and incubated for 1 hour at 37°C with serial dilutions of COM503 (1:2, 10–1.25 g/mL) mixed with preformed human rIL18:rIL18BP complexes (2 ng/mL human IL18 and 400 ng/mL human IL18BP-Fc incubated 1 hour at 37°C). Plates were washed and incubated with biotinylated anti-human IL18 (1:2,000 in 1% BSA PBS; R&D) for 1 hour at RT. HRP-conjugated streptavidin (Jackson) was added for 1 hour incubation at RT for detection, and ELISA signals were developed by addition of TMB substrate. The HRP reaction was stopped by addition of 1N HCl solution, and absorbance signal was read at 450 nm on a luminescence reader. Percent IL18 rescue was calculated as an addition of free IL18 detected over total IL18:IL18BP complex amount in the presence of an Ab compared with the binding signal in the presence of an isotype control (Human IgG4 with hinge stabilizing mutation S228P).
Co-culture of TIL and MEL624 cells
TIL microcultures were initiated and expanded from tumor specimens taken from resected metastases of patients with melanoma as described elsewhere (16). MEL624 cells and CD8+ TILs were seeded at 0.05 × 106 cells/well each in Iscove's modified Dulbecco medium (IMDM) supplemented with 1% Pen-Strep, 1% MEM-eagle, 1% Glutamax, 1% sodium pyruvate with an effector:target ratio of 1:1. Co-cultured cells were then treated with rhIL18 (30 ng/mL) and rhIL18BP (1 μg/mL) for 30 minutes to allow IL18:IL18BP complex formation before addition of different concentrations of COM503 or relevant isotype control (Human IgG4 S228P). In experiments where COM503 activity was measured against endogenously secreted IL18BP, IL18 was added at 1.2 to 3.7 ng/mL, and COM503 and the isotype control (human IgG4 S228P) were added at 20 mg/mL. Plates were incubated for 24 hours at 37°C, after which supernatants were collected for cytokine secretion evaluation. Remaining cells were analyzed by flow cytometry for CD137 (PE antihuman CD137, clone:4B4–1; BioLegend) expression. Experiments were conducted with low-passage cultures (<passage 5) of human MEL624 cells.
CMV recall assay
CD8+ T cells reactive against human CMV pp65 (495–503) were expanded as described elsewhere (17). Briefly, PBMCs from CMV seropositive donors (purchased from ASTARTE and BIOIVT) were in vitro expanded for 7 to 12 days in the presence of IL2 and IL7 (both from R&D systems), as well as the human CMV pp65 peptide (Anaspec) at 1 μg/mL. Assessment of CMV-specific CD8+ T-cell enrichment was carried out at the end of the culture period by labeling the cells with pp65-loaded MHC-I tetramers (MBL) followed by FACS analysis.
Human MEL624 cells overexpressing PD-L1 were pulsed with 0.01 or 0.03 mg/mL CMV pp65 peptide for 1 hour. Cells were seeded at 0.1 × 106 cells/well in IMDM supplemented with 1% Pen-Strep, 1% MEM-eagle, 1% Glutamax, 1% sodium pyruvate, and incubated for 30 minutes with a combination of IL18 (30 ng/mL) and IL18BP (1 μg/mL) to allow IL18:IL18BP complexes to form. Next, COM503, pembrolizumab or relevant isotype control (10 μg/mL, Human IgG4 S228P) were added to the culture followed by addition of CMV-reactive CD8+ T cells for an effector:target ratio of 1:1. Plates were incubated for 24 hours at 37°C, after which supernatants were collected for cytokine secretion evaluation. Remaining cells were analyzed by flow cytometry for CD137 expression. Experiments were conducted with low-passage cultures (<passage 5) of human MEL624-PD-L1 overexpressing cells.
NK-cell assay
Human NK cells, isolated from buffy coats (Magen David Adom National Blood Services) using the RosetteSep Human NK Cell Enrichment Cocktail (Stemcell Technologies), were seeded in RPMI supplemented with 10% FBS+ 4 mmol/L glutamine + 1% HEPES + 1% NAAA + 50 μmol/L 2-ME + 1% sodium pyruvate +1% Pen/Strep at 0.05 × 106 cells/well. Cells were incubated for 30 minutes with a combination of recombinant IL12 (10 ng/mL), IL18 (10 ng/mL) and IL18BP (1 μg/mL), to allow formation of IL18:IL18BP complexes prior to addition of COM503 or relevant isotype control. Plates were incubated for 24 hours at 37°C, after which supernatants were collected and analyzed for IFNγ release.
PBMC assay
Human PBMCs were isolated using density gradient centrifugation from buffy coats that were purchased from the Magen David Adom National Blood Services. The cells were seeded at 200k cells/well and cultured with a combination of IL12 (10 ng/mL), IL18 (2 ng/mL, and COM503 or relevant isotype control. Plates were incubated for 24 hours at 37°C, after which supernatants were collected and analyzed for IFNγ secretion.
Mouse th1 skewing assay
Mouse CD3+ T cells were isolated from mouse splenocytes using the EasySep Mouse T Cell Isolation Kit (from STEMCELL technologies) and seeded in RPMI supplemented with 10% FBS+ 4 mmol/L glutamine + 1% HEPES + 1% NAAA + 50 μmol/L 2-ME + 1% sodium pyruvate +1% Pen/Strep on anti-CD3-coated flasks (10 μg/mL; BD Pharmingen). Cells were additionally supplemented with anti-CD28 (1 μg/mL; BioLegend) at 0.8 × 106 cells/mL for 3 days. Cells were subsequently harvested and cultured in the presence of IL12 (2 ng/mL) for an additional 24 hours. Next day, IL18 (0.5 ng/mL) and IL18BP (2 μg/mL) were allowed to complex for 30 minutes at 37°C, before addition of anti-mouse IL18BP or isotype control (10 μg/mL) for an additional 30 minutes. Cells were harvested, supplemented with IL12 (0.1 ng/mL final) and added to the IL18/IL18BP/anti-IL18BP containing wells (0.04 × 106 cells/well) for 24 hours. Following the 24-hour culture, supernatants were collected for IFNγ secretion analysis.
scRNA-seq
E0771 mouse tumors treated either with anti-mouse IL18BP or isotype control were enzymatically dissociated to generate singe-cell suspensions and enriched for immune cell populations using CD45 MicroBeads (Miltenyi). Individual samples from each mouse were hashed using Total-seq-c antibodies (BioLegend). For each treatment, 6 mice were pooled together, and 40,000 cells were loaded into each channel of chromium single-cell 5′ Chip to create a single cell GEM using the Chromium X machine. cDNA and the subsequent libraries were prepared according to the manufacturer's recommendations. Libraries from two 10× channels were pooled and sequenced together on an Illumina NovaSeq S1 using paired-end sequencing (R1 = 38BP, R2 = 90BP).
Single-cell gene expression analysis preprocessing and QC
The CellRanger single-cell software suite (V7.0) pipeline from 10× genomics was utilized to identify cell hashing and VDJ features, and map reads to the reference transcriptome. Modified gene reference (based on GENCODE) was employed to align the reads, resulting in the generation of unique molecular identifier (UMI) count matrices.
The data underwent downstream analyses using the scanpy package (V1.9.3) including quality control, clustering, cell annotation, and the identification of differentially expressed genes (DEG).
The feature-barcode matrix obtained from 43,615 cells was de-multiplexed using the scanpy implementation of hashsolo. A total of 33,376 cells were recognized as singlets.
Cell quality was further assessed utilizing the total UMI counts per cell, the number of detected genes per cell, and the proportion of mitochondrial gene counts. Cells of low quality, that is, cells with less than 200 genes or more than 10% of expressed genes being mitochondrial genes were filtered out. Genes that were expressed in fewer than three cells were removed. Further analysis excluded cells co-expressing cell type-specific markers (doublets) after which 28,366 cells remained. Some ambient RNA originating from highly abundant myeloid transcripts were noticed during preprocessing and were considered during downstream analysis. Using the cell barcodes, scRNA-seq data could be linked with the scTCR-seq data. Having noticed a high percentage of TCR-negative T cells, additional quality control revealed some low-quality T cells (TCR-negative, low ribosomal gene expression, high mitochondria expression), which were filtered out prior to downstream analysis. For further downstream analysis the remaining 25,713 cells were analyzed.
scRNA-seq clustering and annotation
Feature counts were normalized to the median of total counts for observations (per cell) and natural-log transformation was applied. The scanpy function sc.find_variable_genes() in flavor “Seurat” was employed to identify highly variable genes (HVG), which were used in subsequent dimension reduction steps. The log-normalized data underwent scaling and PCA was then conducted on the basis of the expression matrix of the HVG, with the top 50 principal components (PC) selected for graph-based clustering. Nearest neighbor detection with n = 15 was computed followed by UMAP projections. The data underwent leiden clustering and major cell types were annotated based on conventional marker expression Monocytes & Macrophages (Cd68), dendritic cells (DCs, Zbtb46), T-cells (Cd3e), Natural Killer (NK) cells (Ncr1), Neutrophils (S100a8), Stroma (Col1a1) and B cells (Cd79a). For detailed cell-type analysis, subset of major cell types was isolated and reset to its raw counts. Subsequently, the same preprocessing pipeline as for the initial data were followed to recluster the population.
Statistical analysis
Statistical analysis for IL18BP induction following immune checkpoint blocker treatment and IL18BP expression in inflamed versus noninflamed tumors was performed using Wilcoxon signed-rank test and t test (two-tailed), respectively. Statistical differences for IL18 levels in human healthy donor versus patient with cancer sera were calculated using t test (two-tailed); for IL18 levels in human sera versus TDS using Mann–Whitney test; for IL-8Rα expression on human PBMCs or mouse spleens versus TME using paired t test (two-tailed). Statistical differences for in vitro experiments were calculated using one-way ANOVA followed by paired t test (two-tailed). For Ab dose-titration experiments, EC50 values depicting COM503 functionality were calculated by generating a nonlinear fit (three parameters). Statistical analysis for in vivo studies was performed by two-way ANOVA with repeated measures, followed by two-way ANOVA with repeated measures for selected pairs of groups using JMP (Statistical Discoveries TM) software. Analyses of tumor growth measurements were performed by comparing tumor volumes measured on the last day on which all study animals were alive to the tumor volumes at the randomization day. Statistical differences in percentage of mice tumor free were determined by a log-rank Mantel–Cox test. Experiments were repeated more than two times. For each experiment, the number of animals per group is described in the corresponding figure legend. Statistical differences between naïve and rechallenged mice were calculated using unpaired t test (two tailed). Statistical differences for mouse scRNA-seq and immunophenotyping (IPT) studies were calculated using unpaired t test (two-tailed) for all readout except CD8 effector/mg tumor and CD8/mg tumor counts that were calculated using Mann–Whitney test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Data availability
Primary scRNA-seq data sets have been deposited in the Gene Expression Omnibus under accession code GSE239473. All other data associated with this study are present in the main paper or the Supplementary Materials or Methods or are available from the corresponding author upon reasonable request.
Results
Identification of IL18BP as a potential soluble immune checkpoint in cancer
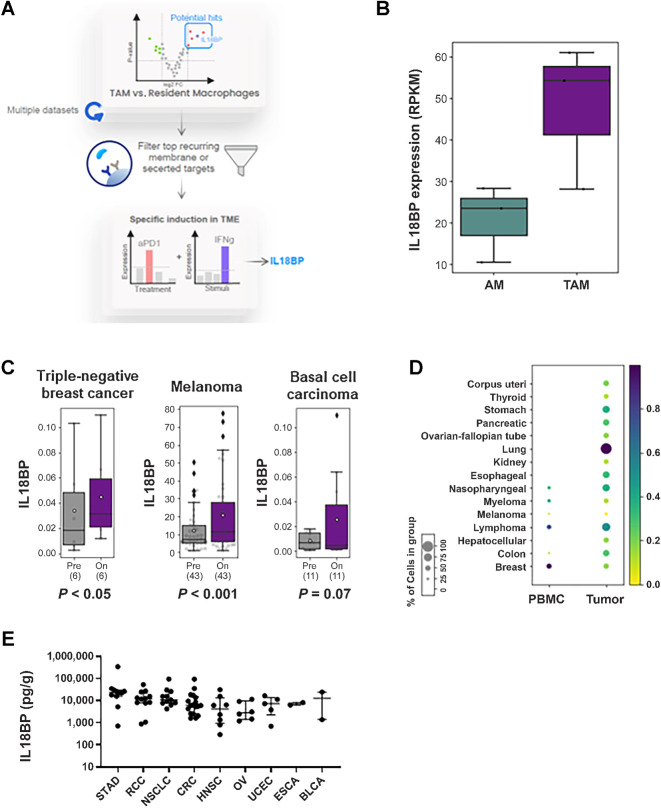
To identify immune checkpoint proteins suitable for Ab targeting that are expressed in myeloid cells in the TME, we screened over 50 gene expression studies and looked for genes that have more than two-fold increased expression in TAMs versus tissue resident macrophage populations (Fig. 1A; Supplementary Table S1). IL18BP was identified to be upregulated in TAMs versus resident macrophages. A representative example of IL18BP upregulated expression in human TAMs from lung adenocarcinoma versus alveolar macrophages from adjacent nontumor tissue is presented in Fig. 1B. We next focused on the top membrane or secreted proteins and queried for those with specific induction in the TME versus peripheral myeloid cells, and those induced following treatment with immune checkpoint inhibitors suggesting a potential immune resistance mechanism. IL18BP met all these criteria and emerged as a promising candidate (Fig. 1C and D). Moreover, IL18BP expression was found to be increased exclusively by IFNγ and not by other treatments used to stimulate macrophages in vitro (59 ligands, Supplementary Fig. S1A). Consistent with this, IL18BP RNA expression was detected to be higher in patients with high levels of IFNγ inflammation signature (18) across indications (Supplementary Fig. S1B). Likewise, IL18BP RNA expression correlated with another dominant IFNγ-induced cancer immune resistance protein, PD-L1 (19), across multiple tumor types (Supplementary Fig. S1C; Supplementary Table S12). Next, we explored IL18BP expression in the TME of patients with cancer. IL18BP RNA levels were consistently observable at all stages of the disease across various indications without significant changes in expression between different stages (Supplementary Table S13). Consistent with this, high levels of IL18BP protein were detected in TDS across indications (Fig. 1E).
Figure 1.
IL18BP is a potential soluble immune checkpoint in cancer. A, Schematic depiction of IL18BP identification process as a potential soluble immune checkpoint. B, Representative example in human tumor associated macrophages (TAM) from lung adenocarcinoma versus alveolar macrophages (AM) from adjacent nontumor tissue (GSE162669) showing IL18BP expression in the two populations in BoxPlot (3 patients, for each three replicates). C, IL18BP expression following immune checkpoint blockade treatment in triple negative breast cancer (anti PD-1, EGAD00001006608_02), melanoma (anti PD-1 + anti CTLA-4, GSE91061), and basal cell carcinoma (anti PD-1, GSE123814). Statistical analysis was preformed using Wilcoxon signed-rank test. Data are shown in BoxPlot, patient sample numbers indicated below. D, scRNA analysis of tumor-infiltrating myeloid cells across indications showing IL18BP expression in myeloid populations in the TME compared with peripheral blood (PBMC). E, IL18BP expression in tumor-derived supernatants from individual patients across indications (n = 76). Tumor biopsies were collected from patients with cancer and dissociated; supernatants were collected and analyzed for IL18BP expression using ELISA assay. Each dot represents an individual sample. The median is depicted by a short black line. In BoxPlots, the bottom of the box indicates the first quartile (Q1), the 25th percentile of the data, and the top of the box indicates the third quartile (Q3), the 75th percentile; the line indicates the median. Whiskers extend to the smallest and largest values within 1.5 times the IQR from the first and third quartiles, respectively.
IL18 is upregulated in sera from patients with cancer and the pathway is elevated in the TME
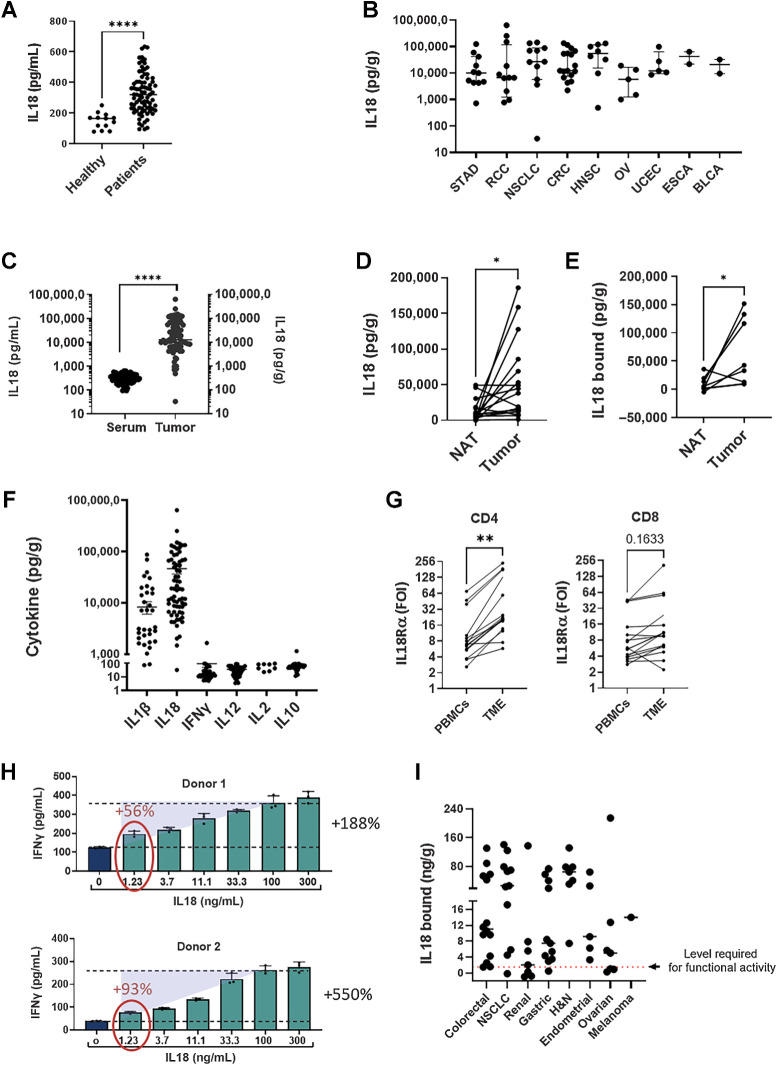
Because IL18BP is a natural inhibitor of the effector cytokine IL18 (11), we speculated that its predicted activity as a myeloid-derived immune resistance mechanism in cancer is through inhibition of IL18 activity. For this mechanism to take place, both IL18 and IL18BP should be naturally expressed at high levels in the TME. Having shown that IL18BP is maintained at high levels in the TME (Fig. 1E), we explored the expression of IL18 protein in patients with cancer. Because both mature and pro-IL18 could be found in TDS and sera samples, we first validated that the human IL18 ELISA Kit recognized mature IL18 but not pro-IL18 (Supplementary Fig. S2A). IL18 levels were measured in serum samples from a number cancer patients and compared to levels in sera from healthy donors. As shown in Fig. 2A, IL18 levels were detectable in cancer patients’ sera and were significantly elevated compared with levels in sera from healthy donors. In line with this, IL18 was found to be highly expressed in TDS of patients across multiple cancer indications (Fig. 2B). As for IL18BP, expression of IL18 RNA was detected in all stages of the disease across various indications, with no notable variations in expression observed between different stages (Supplementary Table S13). In addition, IL18 serum levels were relatively low even in patients with cancer and were significantly upregulated in the TME compared with sera (Fig. 2C) and to NATs (Fig. 2D). Furthermore, we demonstrated that IL18BP-bound IL18 (i.e., inactive) levels in the TME were higher compared with matched NAT samples (Fig. 2E). We next examined the expression of IL18 in the TME across different tumor types compared with other inflammatory cytokines including IL2, IL10, IFNγ, IL12, and IL1β. Although the inflammasome-induced cytokines IL1β and IL18 were abundant in the TME, expression of other cytokines was below the detection limit in most evaluated tumor samples (Fig. 2F).
Figure 2.
IL18 is upregulated in patients with cancer and in the TME and mostly inactive. Tumor biopsies, normal tissues adjacent to the tumors (NAT) and serum samples from patients with cancer and healthy donors were analyzed. Tumor biopsies and NATs were dissociated, and supernatant was collected. IL18 and cytokine expression were analyzed using ELISA, CBA, and LEGENDplex kits. A, IL18 levels in sera from patients with cancer (n = 81) and healthy donors(n = 13). B, IL18 expression in tumor-derived supernatants (TDS) from individual patients across indications (n = 75). C, IL18 expression in sera and TDS. D, IL18 expression in matched NAT and tumor-derived supernatant samples (n = 17). E, Levels of IL18BP-bound IL18 in matched NAT and tumor-derived supernatant samples (n = 7). IL18BP-bound IL18 levels (n = 65) were calculated by deducting IL18 free from total IL18 measured for each sample by two separate ELISA assays. F, Cytokine expression in TDS (n = 56) were analyzed using CBA TH1/TH2/TH17 and LEGENDplex Human Inflammation Panel 1 kits. G, T cells were purified from tumor biopsies and matched PBMC (n = 16) and stained for IL18Ra expression by flow cytometry. H,Ex vivo stimulated human melanoma CD8+ TILs (n = 2) were co-cultured with melanoma antigen-expressing MEL624 cells in the presence of rIL18 (0–300 ng/mL) for 24 hours, after which supernatants were analyzed for IFNγ secretion. I, Levels of bound IL18 in tumor derived supernatants across indications. Tumor biopsies were dissociated, and supernatants were collected and analyzed using free and total IL18 ELISA assays. IL18BP-bound IL18 levels were calculated by deducting IL18 free from total IL18 measured for each sample by two separate ELISA assays. Dashed red line represents the level required for functional activity (1.2 ng/g). Each dot represents 1 patient. The median is depicted by a short black line. *, P < 0.05; **, P < 0.01; ***, P < 0.0001 by two-tailed t test or Mann–Whitney test.
Binding of IL18 to its receptor triggers a signaling cascade that activates an inflammatory response in T and NK cells. To confirm that T cells in the TME can respond to IL18 signaling, we evaluated IL18Rα expression on T cells in the TME and in peripheral blood of patients with cancer. Compared with PBMCs, tumor-infiltrating T cells showed considerably higher levels of IL18Rα, with the most prominent induction in CD4+ T cell, but also in CD8+ T cells (Fig. 2G). IL18 and IL18Rα upregulation in the tumor may potentially induce an enhanced response to IL18 in the TME compared with the periphery.
Levels of IL18BP-bound IL18 in TME are mostly above the level required for T-cell activation in vitro
Next, to explore whether IL18BP-bound IL18 (i.e., inactive) levels in the TME are sufficient for inducing CD8+ TIL responses, we designed a co-culture assay using ex vivo–expanded CD8+ TILs and tumor cells. Addition of rhIL18 to the co-culture induced elevated secretion of IFNγ from CD8+ TILs in a dose-dependent manner, starting from a concentration of 1.2 ng/mL (Fig. 2H). To correlate the levels of IL18 required for T-cell responses in vitro with those detected in the TME, bound IL18 levels were calculated by deducting free IL18 levels from total IL18 levels measured for each tumor sample. The calculated bound IL18 levels should reflect the potential release of IL18 in the TME upon displacing it from the IL18:IL18BP complex. Accordingly, levels of calculated bound IL18 in TDS were mostly above the level required for in vitro T-cell activation (1.2 ng/mL) across multiple cancer types (Fig. 2I). Thus, our results show that IL18 is abundant in the TME but is mostly inactivated by binding to IL18BP. Therefore, IL18BP blockade could trigger robust immune stimulation by releasing IL18 in the TME resulting in potent antitumor responses.
COM503 displaces IL18 from IL18:IL18BP complexes to stimulate CD8+ TILs and NK cells
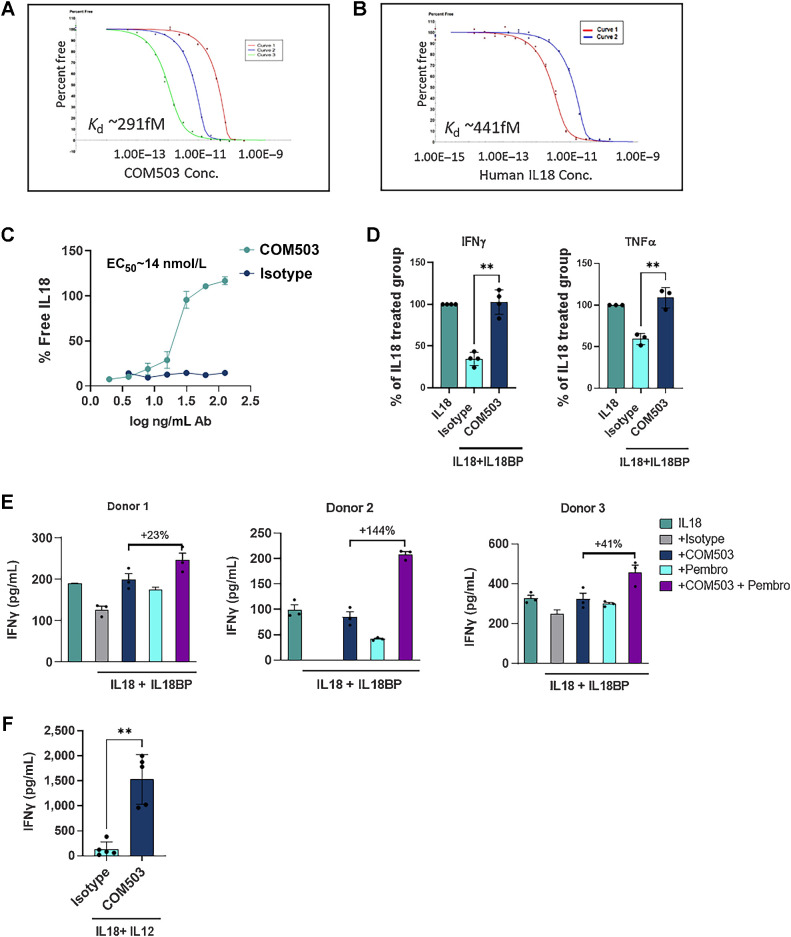
To unleash endogenous IL18 activity, COM503, a fully human anti-IL18BP Ab was generated. COM503 was optimized for improved affinity to human IL18BP, as measured by a kinetic exclusion assay (KinExA, Kd was improved from 1.8 nmol/L to 0.291 pmol/L; Fig. 3A), to outcompete the affinity between the naturally interacting human IL18 and IL18BP (Kd = 0.441 pmol/L; Fig. 3B). COM503 specificity was examined using a human plasma membrane protein cell array (Retrogenix). COM503 showed a specific interaction with IL18BP on both fixed and live cell microarrays, which was further confirmed in a follow-on flow cytometry study (Supplementary Fig. S2B). No other significant interactions were detected. The capacity of COM503 to block the IL18BP:IL18 interaction was assessed biochemically in a free IL18 ELISA assay. COM503 displaced IL18 from preformed IL18BP:IL18 complexes in a dose-dependent manner (EC50 = 14 nmol/L; Fig. 3C).
Figure 3.
COM503 binds IL18BP with high affinity and restores IL18 activity in biochemical and functional assays. A and B, Affinity of COM503 (A) and IL18 (B) to human IL18BP measured by KinExA. Three or two curves with different human IL18BP concentrations were run and analyzed using n-curve analysis to determine the Kd. Y-axis represents the free IL18BP. Free fraction of human IL18BP is measured pre-equilibrium, and the signal is a function of time and concentration of the tittered COM503/ IL-18. C, % IL18 release by COM503 in a free IL18 ELISA assay. % IL18 rescue was calculated as the addition of free IL18 detected over total IL18:IL18BP complex amount in the presence of the Ab compared with the binding signal in the presence of an isotype control. D,Ex vivo stimulated human melanoma CD8+ TILs were co-cultured with melanoma antigen-expressing MEL624 cells and incubated with rIL18BP (1 μg/mL) and rIL18 (30 ng/mL) to allow the formation of an IL18:IL18BP complex before addition of COM503 (10 μg/mL) for 24 hours. Supernatants were collected and analyzed for IFNγ (two independent experiments, four donors) and TNFα (two independent experiments, three donors) secretion. Levels of cytokines in treatment groups were normalized to those detected in the IL18-treated group. E, CD8+ CMV-specific T cells (four independent experiments, three donors) were co-cultured with PD-L1-overexpressing MEL624 cells that were pre-pulsed with CMV pp65 peptide and incubated with rIL18BP (1 μg/mL) and rIL18 (30 ng/mL) to allow the formation of an IL18:IL18BP complex before addition of COM503 (10 μg/mL), pembrolizumab (aPD-1 Ab, 10 μg/mL), or both agents for 24 hours. Supernatants were collected and analyzed for cytokine secretion. F, Human PBMCs (six independent experiments, five donors) were stimulated with rIL18 (2 ng/mL) and rIL12 (10 ng/mL) and incubated with COM503 (6 μg/mL) for 24 hours, after which supernatants were collected and analyzed for IFNγ secretion. Bar graphs show the mean ± SEM; **, P < 0.01 by two-tailed t test or by one-way ANOVA followed by two-tailed t test.
To evaluate whether COM503 could restore IL18 function and enhance CD8+ TIL activity, melanoma CD8+ TILs were co-cultured with melanoma antigen–expressing MEL624 cells and incubated with IL18 and IL18BP to allow the formation of IL18:IL18BP complexes before addition of COM503. COM503 addition resulted in significantly increased IFNγ and TNFα secretion (Fig. 3D; Supplementary Fig. S2C) as well as significant upregulation of CD137 on CD8+ TILs (Supplementary Fig. S2D). Furthermore, COM503 significantly restored IL18 stimulatory activity in NK cells pre-incubated with IL12 and preformed IL18:IL18BP complexes, as evident by increased IFNγ secretion (Supplementary Fig. S2E). To assess COM503 in combination with PD-1 blockade, CMV-reactive CD8+ T cells were co-cultured with PD-L1-overexpressing MEL624 cells that were pulsed with CMV pp65 peptide, in the presence or absence of COM503, pembrolizumab, or the combination of both. COM503 was able to block IL18BP and to fully restore IL18 function, as reflected by induction of IFNγ and TNFα secretion in all tested donors (Fig. 3E; Supplementary Fig. S2F). Combination of COM503 with pembrolizumab resulted in further activation, as evident by increased cytokine secretion on top of single-agent treatment.
COM503 restores IL18 activity in the presence of endogenously secreted IL18BP
To evaluate the capacity of COM503 to restore IL18 activity in more physiologic settings, PBMCs, which endogenously secrete IL18BP following stimulation (Supplementary Fig. S2G), were stimulated with IL18 and IL12 in the presence or absence of COM503. COM503 significantly increased IFNγ secretion with an average EC50 value of 1.6 nmol/L in all tested donors (Fig. 3F; Supplementary Fig. S2H). COM503 also enhanced CD8+ TIL function by blockade of endogenously secreted IL18BP in a modified version of the CD8+ TIL-melanoma co-culture assay. Blockade of IL18BP by COM503 resulted in increased IFNγ secretion in the presence of IL18 at concentrations of 1.2 and 3.7 ng/mL (donors 1 and 2, respectively, Supplementary Fig. S2I). These concentrations of IL18 were within the range expected to be found in the TME (Fig. 2), suggesting a beneficial effect of COM503 on CD8+ TIL activity at physiologically relevant IL18 concentrations. Collectively, these data show that releasing IL18 by blocking IL18BP:IL18 interactions with COM503 enhances lymphocyte cytokine secretion and activation. Moreover, this effect can be magnified by combing COM503 with blockade of other checkpoints, such as PD-1.
Anti-IL18BP induces tumor growth inhibition in mouse models
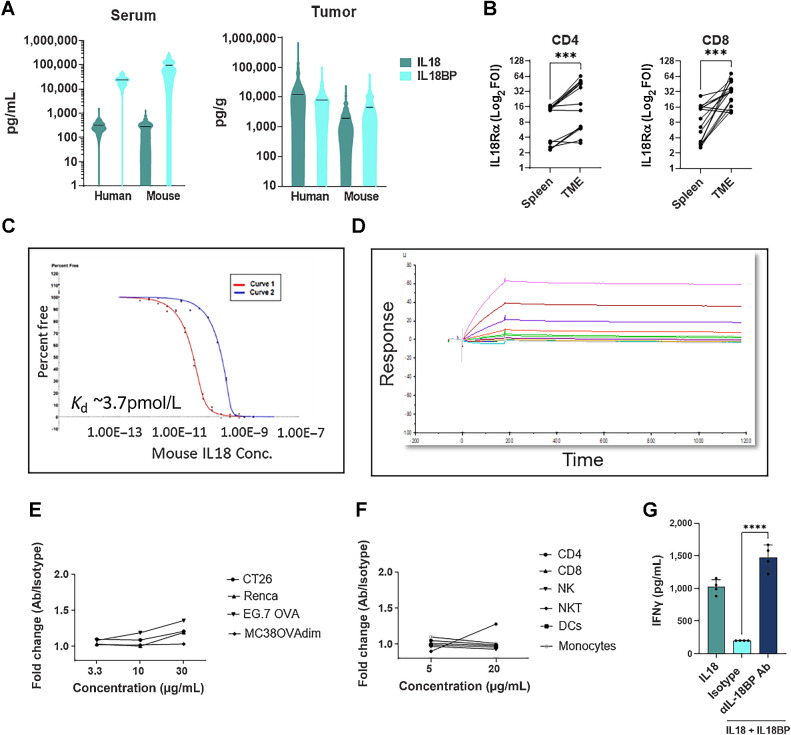
To evaluate whether mouse tumors are relevant models for human cancer, we first analyzed IL18 and IL18BP expression across tumor models (Supplementary Table S9 indicates the number of mice per model analyzed). In line with our human data, IL18 levels in murine TDS were higher when compared with sera, with comparable levels with those measured in human TDS (Fig. 4A). Furthermore, IL18BP levels were comparable in TDS from human and mouse, whereas IL18BP levels detected in sera of tumor bearing mice were overall slightly higher (Fig. 4A). In addition, IL18Rα was shown to be expressed in the TME on tumor-infiltrating T cells across various syngeneic tumor models (Fig. 4B). A significant induction of IL18Rα was detected on CD4+ and CD8+ T cells in the TME compared with the spleen. This observation is consistent with human IL18Rα induction in the TME compared with matched PBMCs (Fig. 2G). Finally, like human IL18BP, we found that mouse IL18BP binds mouse IL18 with high affinity (Kd = 3.7 pmol/L, Fig. 4C). Taken together, we show that most attributes of the IL18 pathway in human cancer biology are preserved in mouse tumors.
Figure 4.
Mouse and human IL18 pathway share similar biological properties. A, IL18 and IL18BP levels were measured in sera and TDS taken from patients with cancer and tumor-bearing mice using ELISA kits (mouse sample information is indicated in Supplementary Table S9). B, Mouse T cells were purified from mouse tumors and matched spleens (n = 16) and stained for IL18Rα expression by flow cytometry. Each dot represents one mouse. C, Affinity of mouse IL18 to mouse IL18BP measured by KinExA. Two curves with different mouse IL18BP concentrations were run and analyzed using n-curve analysis to determine the Kd. Y-axis represents the free IL18BP. Free fraction of mouse IL18BP is measured pre-equilibrium, and the signal is a function of time and concentration of the tittered IL18. D, Affinity of anti-mouse IL18BP Ab to mouse IL18BP measured by surface plasmon resonance (SPR). Different colors represent the different concentrations of mouse IL18BP (0.125–256 nmol/L). E and F, Mouse cell lines (E) and mouse splenocytes, purified from mouse spleens (F), were stained with anti-mouse IL18BP Ab. Staining was analyzed by flow cytometry. G, Mouse CD3+ T cells were isolated from mouse splenocytes, activated with anti-CD3 and anti-CD28 and incubated with IL12 (2 ng/mL), and IL18:IL18BP (0.5 ng/mL:2 μg/mL) preformed complexes before addition of anti-mouse IL18BP Ab (10 μg/mL). Following the 24-hour culture, supernatant was collected for IFNγ secretion analysis. The median is depicted by a short black line in the violin plots. Bar graph shows the mean ± SEM; ***, P < 0.001 and ****, P < 0.0001 by two-tailed t test or by one-way ANOVA followed by two-tailed t test.
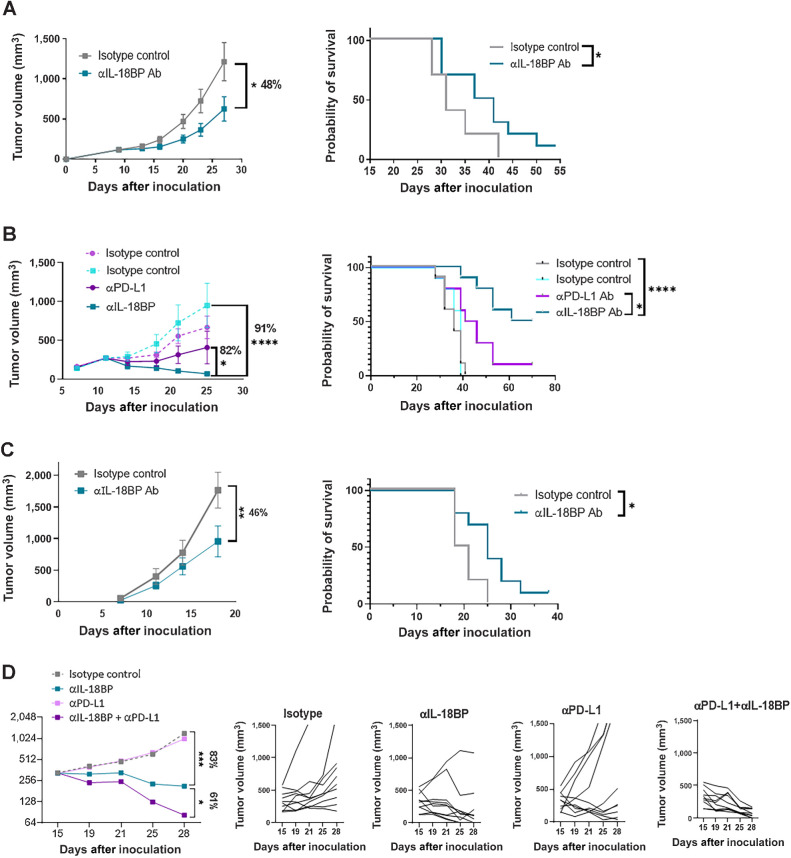
To study the effects of IL18BP blockade in mice, we developed a surrogate high affinity blocking Ab (Kd < 1 pmol/L, Fig. 4D). Anti-mouse IL18BP showed no significant binding to mouse cell lines and mouse splenocytes, confirming the Ab specificity (Fig. 4E and F). The ability of the anti-mouse IL18BP to displace IL18 from preformed IL18:IL18BP complexes was examined in a Th1 mouse skewing assay, and we found that it was able to fully restore IL18 activity, as reflected by increased IFNγ secretion, similarly to COM503 activity in human functional assays (Figs. 3 and 4G). To evaluate its in vivo antitumor activity, the anti-mouse IL18BP or isotype control were administered to MC38OVAdim tumor-bearing mice after tumors were established. Significant tumor growth inhibition and a statistically significant improved survival were demonstrated with IL18BP blockade as a single agent (Fig. 5A). To further evaluate the antitumor activity of anti-mouse IL18BP in a more physiologic tumor model, mice bearing established orthotopic E0771 breast tumors were administered anti-mouse IL18BP or isotype control. IL18BP blockade induced significant and robust antitumor activity (Fig. 5B). Moreover, anti-mouse IL18BP monotherapy inhibited the growth of breast tumors more effectively than anti-PD-L1 monotherapy. These potent antitumor responses translated into statistically significant improved survival (Fig. 5B). We further explored the antitumor activity of anti-mouse IL18BP in the less immunogenic mouse melanoma tumor model B16/Db-hmgp100. In this model as well, IL18BP blockade resulted in significant antitumor responses, which translated to increased survival (Fig. 5C). The antitumor activity of anti-mouse IL18BP was explored in additional tumor models. Although monotherapy with anti-mouse IL18BP induced antitumor activity in the MCA100 tumor model (Supplementary Fig. S3A), we found that the 4T1 and LLC tumor models, which are considered poorly infiltrated by the immune system (20), and thus have relatively low numbers of tumor-infiltrating T cells and NK cells (Supplementary Fig. S3B), were resistant to the treatment (Supplementary Figs. S3C and S3D).
Figure 5.
Anti-mouse IL18BP Ab demonstrates monotherapy and combo activity with anti PD-L1 Ab across murine syngeneic tumor models. A–C, MC38OVAdim (representative experiment out of 6; A), E0771 (representative experiment out of 4; B), and B16F10-hmgp100 (representative experiment out of 3; C) mouse tumor cells were inoculated in C57Bl/6 mice. At tumor volume of 120 mm3 (MC38OVAdim), 250 mm3 (E0771), and on day 4 after inoculation (B16F10-hmgp100), mice were randomized (n = 10 per group) and treated either with anti-IL18BP Ab or with isotype control (15 mg/kg) twice a week for a total of six treatments. Tumor volumes are represented as the mean volume ± SEM. Kaplan–Meier survival curves for each group are shown. D, E0771 tumor cells were inoculated in C57Bl/6 mice as described in material and methods. At tumor volume of 330 mm3, mice were randomized (n = 10 per group) and treated either with anti-IL18BP Ab, isotype control (15 mg/kg), anti-PD-L1 Ab (5 mg/kg), or with both agents twice a week for a total of six treatments. Representative experiment out of 3 is shown. Tumor volumes are represented as the mean volume ± SEM. Individual tumors measurements for each mouse are depicted. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by two-way ANOVA with repeated measures and log-rank Mantel–Cox test for survival analysis.
Next, to determine the contribution of specific immune cell populations to the antitumor effects induced by anti-mouse IL18BP, we performed Ab-mediated depletion studies in the MC38OVAdim model (Supplementary Fig. S3E). The effects of IL18BP blockade were completely abrogated by the depletion of CD8+ T cells, whereas the depletion of CD4+ T cells only partially abrogated these antitumor effects (Supplementary Figs. S3F–S3H). The antitumor effects of IL18BP blockade were abolished also by NK-cell depletion (Supplementary Fig. S3I). Finally, we correlated the percent of CD8+ T-cell infiltration into the tumor mouse models used in this study, to the degree of antitumor activity induced by IL18BP blockade. This analysis revealed that the response to anti-mouse IL18BP treatment was correlated with the infiltration of CD8+ T cells in most responsive models (Supplementary Fig. S3J). The antitumor activity observed in the B16/Db-hmgp100 tumor model might be attributed to the relatively high percentage of NK cells (Supplementary Fig. S3B). Collectively, the activity of IL18BP blockade in vivo was found to depend on CD8+ T and on NK cells and partially on CD4+ T cells, which are the main cells expressing the IL18Rα (Supplementary Fig. S3K). Next, we aimed to assess whether combination with PD-L1 blockade would further increase the antitumor activity induced by anti-mouse IL18BP. Anti-PD-L1 monotherapy had minor effects on the growth of large established E0771 tumors. In contrast, anti-mouse IL18BP monotherapy induced a significant inhibition in tumor growth compared with isotype control or with anti-PD-L1 monotherapy. Moreover, combining both agents resulted in potentiated antitumor activity compared with monotherapies (Fig. 5D). Subsequently, the combination was tested in an additional mouse model, the CT26 colon tumor model. In this model neither anti-PD-L1 alone nor anti-mouse IL18BP alone affected tumor growth. However, the combined blockade of IL18BP and PD-L1 induced a significant inhibition in tumor growth compared with monotherapies and led to a significant increase in mouse survival (Supplementary Fig. S3L). Collectively, these results demonstrate the potential of IL18BP blockade in combination with PD-L1 blockade to effectively inhibit tumor growth in vivo.
Because monotherapy with anti-mouse IL18BP resulted in complete rejection of E0771 tumors in mice, we examined whether the treatment induced generation of immunologic memory by rechallenging mice with E0771 cells. Two months after primary tumor inoculation, mice with no evident residual tumors and tumor-naïve age-matched mice were inoculated with E0771 tumor cells (Supplementary Fig. S4A). Eight out of nine rechallenged mice rejected the tumors, in contrast to 1 of 9 tumor-naïve mice (Supplementary Fig. S4B). Moreover, we encountered a significant increase in percentage of CD19+ cells and CD44+CD62L−CD8+ effector T cells in rechallenged mice compared with tumor-naïve mice (Supplementary Figs. S4C and S4D). Taken together, these results show that systemic memory was induced by anti-mouse IL18BP treatment.
IL18BP blockade modulates the TME without affecting the periphery
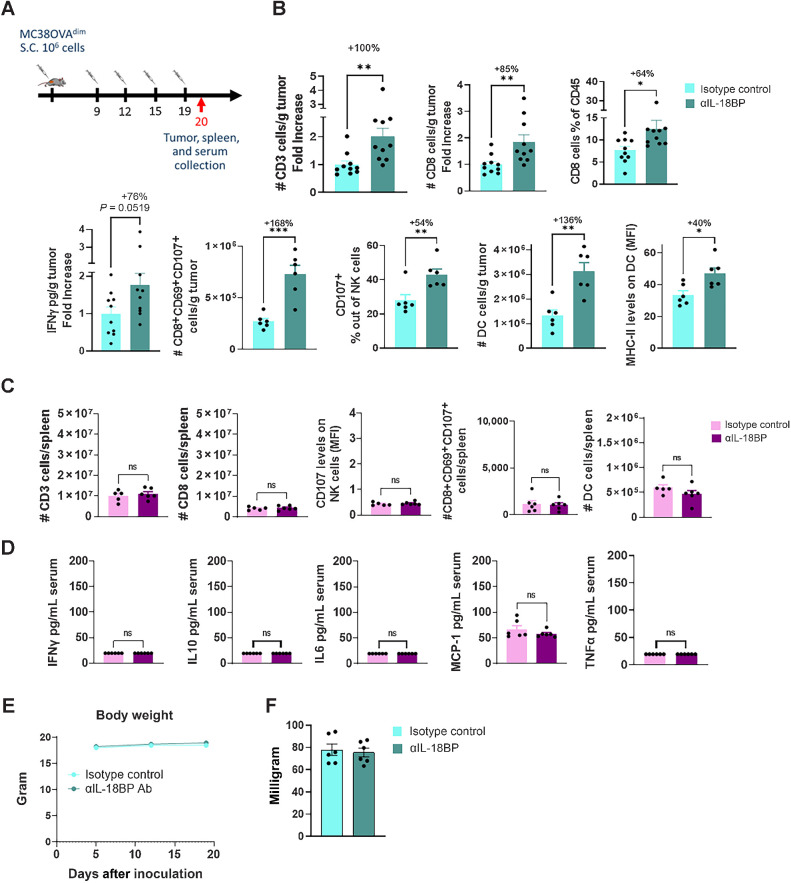
To elucidate the mechanism by which IL18BP blockade modulates the TME and to evaluate its potential therapeutic window, we performed immunophenotyping studies of TME versus blood and spleens in mice bearing MC38OVAdim tumors, following anti-mouse IL18BP treatment (Fig. 6A). Anti-mouse IL18BP monotherapy resulted in a significant increase in CD3+ and CD8+ T-cell numbers, as well as in IFNγ levels in the TME (Fig. 6B). Furthermore, significant increases in activated CD8+CD69+CD107+ T cells and CD107+ NK cells were observed (Fig. 6B). Following treatment there was a significant increase in the number DCs as well as in their MHC-II expression levels, suggesting increased capacity to stimulate T cells (Fig. 6B). No immune activation was apparent in mouse blood and spleens following the treatment, as evident by the similar number and activation state of immune cells in the spleen (Fig. 6C), and by no induction of inflammatory cytokines in the blood (Fig. 6D). Consistent with this, IL18BP blockade had no impact on either body weights or spleen weights (Fig. 6E and F). Thus, blocking IL18BP is expected to induce TME-localized immune modulation without affecting the periphery, resulting in a favorable therapeutic window in patients with cancer.
Figure 6.
Anti-mouse IL18BP Ab alters the immune infiltrate composition of MC38OVAdim tumors without affecting the periphery. A, MC38OVAdim tumors were inoculated in C57Bl/6 mice. At tumor volume of 120 mm3, mice were randomized (n = 6 per group) and treated either with anti-IL18BP Ab or with isotype control (15 mg/kg) twice a week for a total of four treatments. Tumors and spleen were harvested 24 hours after the fourth treatment and immune composition was examined. Tumor supernatants and blood sera were collected at the same timepoint and analyzed for cytokine concentrations. B–D, Immune composition and cytokine secretion in the TME (B), spleens (C), and in the sera (D) of treated mice. E, Effect of treatment on body weight. F, Effect of treatment on spleen weight. Pooled data from two experiments is shown. Bar graphs show the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by two-tailed t test.
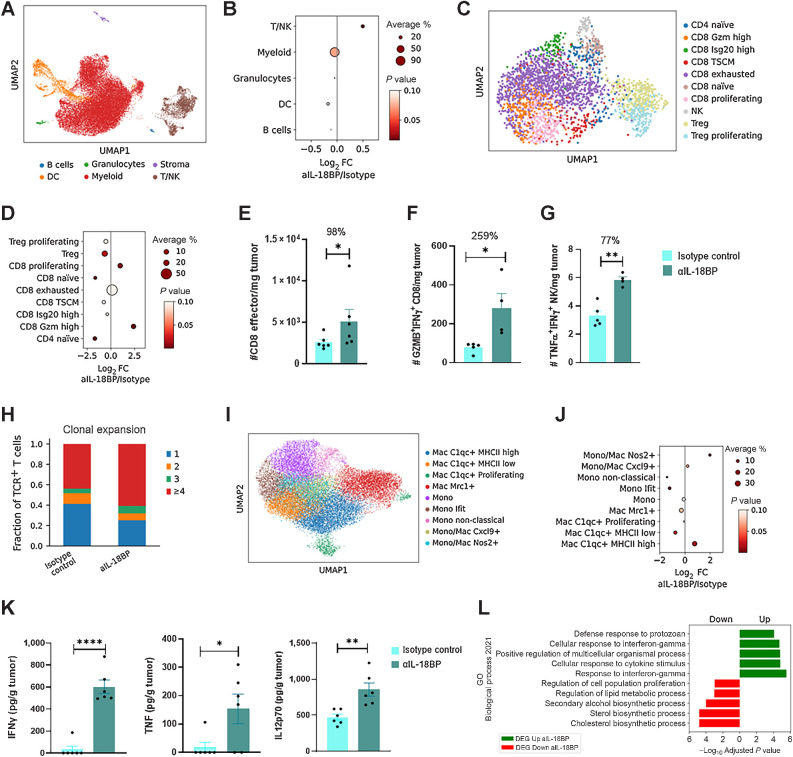
To further dissect, in high-resolution, IL18BP blockade effects on the TME, we performed scRNA-seq of E0771 tumors following treatment with anti-mouse IL18BP. Major cell types were annotated on the basis of conventional lineage markers (Fig. 7A). Anti-mouse IL18BP treatment increased the fraction of T cells, with no major effects on other cell types (Fig. 7B). In line with the scRNA-seq results, a significant increase in CD3+ CD4+ and CD8+ T-cell numbers was observed by flow cytometry analysis following IL18BP blockade (Supplementary Fig. S5A). Further examination of T-cell subsets revealed reshaping of TME T-cell populations following treatment (Fig. 7C and D; Supplementary Figs. S5B and S5C). A significant increase in an effector polyfunctional CD8+ T-cell population (expressing perforin, multiple granzymes, and IFNγ) as well as proliferating CD8+ T cells were evident following treatment, whereas the frequencies of naïve CD8+ and CD4+ T cells were reduced (Fig. 7D). Moreover, exhausted T cells were not increased by the treatment (Fig. 7D). Likewise, flow cytometry analysis confirmed a significant induction of effector CD44+CD62L–CD8+ T cells, polyfunctional GZMB+IFNγ+CD8+ T cells, IL2+ and TNFα+ CD4+ T cells, and TNFα+IFNγ+ polyfunctional NK cells following treatment (Fig. 7E–G; Supplementary Figs. S5D–S5F). A significant reduction in CD4+Foxp3+ Treg cells was also observed by scRNA-seq (Fig. 7D). Furthermore, TCR-seq analysis revealed that the increase in effector T cells following IL18BP blockade was also associated with an increase in T-cell clonal expansion, suggesting induction of an antigen-specific, potentially tumor-specific, immune response (Fig. 7H). In addition, although exhausted T cells were clonally expanded in the TME as expected (21, 22), GZMB-high and proliferating CD8+ T cells, which were expanded by treatment, exhibited the most pronounced clonal expansion among the various T-cell subtypes suggesting an expansion of polyfunctional tumor-specific clones by the treatment (Fig. 7D; Supplementary Fig. S5G).
Figure 7.
IL18BP blockade alters the immune infiltrate composition of E0771 tumors. E0771 tumor cells were inoculated in C57Bl/6 mice and treated either with anti-IL18BP Ab or isotype control (n = 6 per group, 15 mg/kg) twice a week for a total of three treatments. TME modulation was assessed by flow cytometry, scRNA-seq, and cytokines profiling. Cytokine intracellular staining was done following ex vivo stimulation with phorbol myristate acetate and ionomycin. A, UMAP projection showing major cell types. B, Enrichment of major immune population frequencies in anti-IL18BP Ab treatment compared with the control group. Depicted is the log2 fold change of the mean frequency. The size of the dots indicates the average fraction of the cell population between treatments, whereas the color of the dots represents the P values. C, UMAP projection showing T cells present in E0771 tumors treated mice. D, As in B for T-cell populations. E–G, Numbers of polyfunctional effector CD8+ T cells (CD44+CD62L−; E) as well as GZMB+IFNγ+CD8+ T cells (F) and TNFα+IFNγ+-secreting NK cells (G) were analyzed using flow cytometry. H, Quantification of clonal expansion frequencies in anti-IL18BP Ab treatment compared to the control group in TCR+ T cells. I, UMAP projection showing tumor-associated monocyte and macrophage subpopulations present in E0771 tumors treated mice. J, As in B for tumor-associated monocytes and macrophages. K, IFNγ, TNFα, and IL12p70, levels in tumor derived supernatant were analyzed using CBA Inflammation Kit. L, GSEA on DEGs of myeloid cells (DCs, monocytes, Macs) from control versus treatment group querying GO Biological Process 2021 gene sets. Bar graphs show the mean ± SEM. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 by two-tailed t test or Mann–Whitney test.
Next, we examined the myeloid compartment and identified nine myeloid-cell subpopulations and six DC populations (Fig. 7I; Supplementary Figs. S5H–S5J). IL18BP blockade induced significant increases in inflammatory MHCIIhighC1qa+ and Nos2+ macrophages and in activated DCs (Fig. 7J; Supplementary Fig. S5K). Accordingly, a significant decrease in MHCIIlowC1qa+ macrophages, suppressive Mrc1+ macrophages, Ifit+ MonoMacs, and low-activated DCs was evident in treated mice (Fig. 7J; Supplementary Fig. S5K). Consistent with the increase in inflammatory myeloid cells, significantly enhanced levels of inflammatory cytokines including IFNγ, TNFα, IL12p70, CXCL9, MIP1a, and a decrease in IL1β in tumor supernatants were detected following IL18BP blockade (Fig. 7K; Supplementary Fig. S5L). Gene set enrichment analysis (GSEA) of upregulated DEGs in myeloid-cell populations following anti-mouse IL18BP treatment showed a significant enrichment of genes in GO Biological process gene sets such as “Response to interferon gamma” and “Cellular response to cytokine stimulus.” In contrast, DEGs downregulated following the anti-mouse IL18BP treatment group were significantly associated with gene sets like “cholesterol biosynthesis process” or “sterol biosynthesis process” (Fig. 7L), which were recently reported to be associated with an immunosuppressive and tolerogenic state of these cell types (23). In summary, monotherapy with anti-mouse IL18BP induced pronounced TME-localized modulation and anticancer immunity spanning multiple adaptive and innate immune cells.
Discussion
Cytokines have an important role in regulating innate and adaptive immune responses (24). Because of their potent immunostimulatory activity, there is high interest in harnessing cytokines for treatment of cancer (4). Although in preclinical studies recombinant cytokine administration has demonstrated potent antitumor activity (25–27), therapeutic usage in patients is limited by cytokines’ fast clearance, pleiotropic activity, and cytotoxicity (27, 28). The resulting challenging therapeutic window is reflected by systemic toxicities that stem from the high doses of cytokine that are required to be administered for achieving sufficient TME concentration for efficient antitumor immune responses (4–6).
IL18 is a potent proinflammatory cytokine that is tightly regulated by a natural inhibitor, IL18BP (11). Here we demonstrated that unlike some other proinflammatory cytokines, IL18 as well as IL1β are abundant in the TME. Therefore, although most other cytokines must be exogenously administered in recombinant form to tentatively reach active concentrations, cytokines with high endogenous expression in the TME, such as IL1β and IL18, could be therapeutically manipulated in situ. Unlike IL1β, whose activity is largely tumor promoting (and indeed IL1 pathway antagonists are being pursued clinically; refs. 29, 30), IL18 is a potent stimulator of T and NK cells and could promote tumor regression (31). Both IL18 and IL1β are produced as inert pro-cytokines and undergo proteolytic maturation and extracellular secretion by caspase-1 upon inflammasome activation (8). Inflammasomes are activated in the TME, which is highly enriched with danger signals due to the inflamed environment (32–34). Therefore, IL1β and IL18 are most probably abundantly expressed in the TME due to their regulated secretion following inflammasome activation. Although our data show high levels of total mature IL18 in the TME across multiple cancer indications, most of it is bound by IL18BP and therefore inactive. Of note, the levels of IL18BP-bound IL18 in the TME are shown here to be above those required for TIL activation in vitro, implying that IL18BP blockade has the potential to exploit endogenous IL18 in the tumor, thereby enhancing anticancer immunity.
To this end, we generated COM503, a fully human high-affinity anti-IL18BP blocker and demonstrated its capacity to displace IL18 from preformed IL18:IL18BP complexes and restore IL18 stimulatory activity on T and NK cells. Furthermore, the therapeutic potential of this approach is reflected in our mouse studies in which an anti-mouse IL18BP surrogate blocker showed robust antitumor activity across multiple models, an effect that was ameliorated by addition of anti-PD-L1. The therapeutic potential of combining IL18BP and PD-(L)1 blockers was also demonstrated in vitro, where combination of both agents resulted in greater activation of CMV-reactive CD8+ T cells. Moreover, we showed that IL18BP is induced following PD-(L)1 blockade in patients with cancer and a recent study has shown that serum levels of IL18BP are negatively correlated with clinical response to pembrolizumab and lenvatinib in patients with renal cancer, suggesting an induced resistance mechanism (35). Although mouse models are not fully reflective of human tumor biology, we showed that the IL18 pathway is preserved in mouse with similar expression patterns to human. Therefore, our data support the potential of COM503 as monotherapy or in combination with PD-(L)1 blockade to overcome a tumor immune resistance mechanism and enable effective immunotherapy in patients not responding to currently approved checkpoint inhibitors.
Unlike other T-/NK-cell–stimulating cytokines (e.g., IL2, IL15, IL12), IL18 signals through the MYD88 signaling pathway (11). MYD88 signaling has been shown to maintain CD8+ T cells in an early effector stage, enabling them to better expand and retain their cytolytic activity after chronic antigen stimulation (36, 37). Accordingly, our data demonstrated prominent induction of polyfunctional nonexhausted T and NK cells following IL18 release by IL18BP blockade in mouse models. Thus, following treatment, naive T-cell populations were reduced while proliferating and polyfunctional effector CD8+ T cells were increased, without induction of T-cell exhaustion. These results suggest a treatment-induced T-cell differentiation that maintains T cells in an early functional effector state, as shown by other studies mentioned above (36, 37). Moreover, TCR-seq analysis revealed clonal expansion of these polyfunctional effector T-cell populations, implying an antigen-specific effective immune response that persisted to mediate systemic memory upon tumor rechallenge. This effect on effector T and NK cells was directly associated to the antitumor outcome, as depletion of these populations abrogated anti-mouse IL18BP activity, and no activity was evident in models with poor tumor infiltration by T and NK cells. Moreover, the TME modulation effect was not restricted to T and NK cells, which are the main cell populations bearing the IL18R, but also extended to myeloid-cell populations with increased levels of inflammatory macrophages and activated DCs. This is probably a secondary effect to the potent TME inflammatory induction following treatment. Likewise, IL18BP blockade induced multiple proinflammatory cytokines in the TME. Interestingly, examining downregulated gene signatures in myeloid cells following anti-IL18BP treatment showed enrichment in gene sets such as “Cholesterol Biosynthesis Pathway” and “Mevalonate arm of cholesterol biosynthesis pathway.” Cholesterol biosynthesis and mevalonate pathways in macrophages, monocytes. and DCs were recently reported to be associated with an immunosuppressive and tolerogenic state of these cell types (23, 38). Altogether these results further demonstrate mechanistically the potent TME immune modulation that underlies the antitumor growth effects of IL18BP blockade.
Recently, an alternative approach to overcome IL18BP inhibition was reported. In this study, an engineered IL18 that retains binding to IL18 receptor but does not bind IL18BP showed efficacy in preclinical tumor models (31). Although this approach might overcome some of the challenges encountered by the GSK rIL18 clinical trials (12) described above, engineered IL18 clinical activity might still be limited by the challenges faced by other cytokines moving from preclinical to clinical studies. Namely, fast clearance, peripheral toxicity and the resultant narrow therapeutic window might impose challenges in clinical translation of engineered IL18 administered systemically to patients. In contrast, our study demonstrated the potential of blocking IL18BP with an Ab to unleash endogenous IL18 activity in mediating potent TME-localized antitumor immunity, with no increase in inflammatory cytokines and lymphocyte numbers or activation state in serum and spleen. This observation is in line with the biology of the IL18 pathway, with IL18 being induced by inflammasome signals in TME, IL18BP induced by negative feedback loop following IFNγ increase and IL18R induced on activated T cells (11, 13, 39, 40). Accordingly, our expression studies showed low levels of IL18 in patients’ blood, induction of IL18BP in tumor myeloid cells and upregulation of both IL18 and IL18Rα in the TME compared with the periphery. Moreover, both IL18 and IL18BP-bound IL18 levels were significantly higher in the TME compared with matched NAT samples, further supporting the idea that IL18BP blockade will induce TME-localized immune modulation.
In summary, we demonstrated that IL18 is upregulated in the TME and is mostly bound by IL18BP. COM503, a potential first-in-class, high-affinity anti-IL18BP blocker, induces human T- and NK-cell responses in vitro. A surrogate anti-mouse IL18BP induces potent antitumor responses and pronounced TME-localized immune modulation, which contrasts with systemically administered therapeutic cytokines, which generate a nonlocalized inflammatory response. Taken together, blocking IL18BP is a promising approach to harness cytokine biology for the treatment of cancer.
Supplementary Material
Supplemental Figure S1. IL-18BP is a potential soluble immune checkpoint in cancer.
Supplemental Figure S2. COM503 releases IL-18 activity in biochemical and functional assays.
Supplemental Figure S3. Antitumor activity of anti-mouse-IL-18BP Ab in mice studies.
Supplemental Figure S4. Anti-IL-18BP Ab induced immunogenic memory response in E0771 tumor model.
Supplemental Figure S5. IL-18BP blockade alters the immune infiltrate composition of E0771 tumors.
Supplementary Methods
Footnotes
Note: Supplementary data for this article are available at Cancer Immunology Research Online (http://cancerimmunolres.aacrjournals.org/).
Authors' Disclosures
A. Menachem reports employment with Compugen and ownership of Compugen stock. Z. Alteber reports employment with Compugen and ownership of Compugen stock. T. Fridman-Kfir reports employment with Compugen and ownership of Compugen stock. D. Blat reports ownership of Compugen stock and employment with Compugen. M. Galperin reports employment with Compugen and ownership of Compugen stock. L. Sever reports employment with Compugen and ownership of Compugen stock. N. Cohen reports employment with Compugen and ownership of Compugen stock. K. Cohen reports employment with Compugen and ownership of Compugen stock. R.Z. Granit reports employment with Compugen Ltd. S. Vols reports employment with Compugen and ownership of Compugen stock. M. Frenkel reports employment with Compugen and ownership of Compugen stock. L. Soffer reports employment with Compugen and ownership of Compugen stock. K. Menachem reports employment with Compugen and ownership of Compugen stock. H. Galon-Tilleman reports employment with Compugen with ownership of Compugen stock. I. Borukhov reports employment with Compugen and ownership of Compugen stock. P. Ferre reports personal fees from Compugen during the conduct of the study. E. Ophir reports employment with Compugen Ltd. and ownership of Compugen stock. No disclosures were reported by the other authors.
Authors' Contributions
A. Menachem: Conceptualization, visualization, methodology, writing–original draft, project administration. Z. Alteber: Conceptualization, supervision, visualization, methodology, writing–review and editing. G. Cojocaru: Conceptualization, formal analysis, visualization. T. Fridman-Kfir: Conceptualization, supervision, writing–review and editing. D. Blat: Supervision, methodology, writing–review and editing. O. Leiderman: Validation, investigation, visualization, methodology. M. Galperin: Investigation, visualization, methodology. L. Sever: Investigation, visualization. N. Cohen: Investigation, visualization. K. Cohen: Investigation, visualization. R.Z. Granit: Formal analysis, supervision, visualization, writing-review and editing. S. Vols: Formal analysis, visualization. M. Frenkel: Investigation, visualization. L. Soffer: Validation, investigation, visualization. K. Meyer: Investigation, visualization. K. Menachem: Validation, investigation, visualization. H. Galon Tilleman: Investigation, visualization. D. Morein: Investigation, visualization. I. Borukhov: Visualization. A. Toporik: Visualization. M. Perpinial Shahor: Investigation, visualization. E. Tatirovsky: Investigation, visualization. A. Mizrachi: Resources. A. Levy-Barda: Resources. E. Sadot: Resources. Y. Strenov: Resources. R. Eitan: Resources. A. Jakobson-Setton: Resources. N. Yanichkin: Resources. P. Ferre: Writing–review and editing. E. Ophir: Conceptualization, supervision, methodology, writing–review and editing.
References
- 1. Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jiang T, Zhou C, Ren S. Role of IL-2 in cancer immunotherapy. Oncoimmunology 2016;5:e1163462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aricò E, Castiello L, Capone I, Gabriele L, Belardelli F. Type I interferons and cancer: an evolving story demanding novel clinical applications. Cancers (Basel) 2019;11:1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Propper DJ, Balkwill FR. Harnessing cytokines and chemokines for cancer therapy. Nat Rev Clin Oncol 2022;19:237–53. [DOI] [PubMed] [Google Scholar]
- 5. Schwartz RN, Stover L, Dutcher JP. Managing toxicities of high-dose interleukin-2. Oncology (Williston Park) 2002;16:11–20. [PubMed] [Google Scholar]
- 6. Leonard JP, Sherman ML, Fisher GL, Buchanan LJ, Larsen G, Atkins MB, et al. Effects of single-dose interleukin-12 exposure on interleukin-12–associated toxicity and interferon-γ production. Blood 1997;90:2541–8. [PubMed] [Google Scholar]
- 7. Swain SL. Interleukin 18: tipping the balance towards a T helper cell 1 response. J Exp Med 2001;194:F11–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mantovani A, Dinarello CA, Molgora M, Garlanda C. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity 2019;50:778–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fabbi M, Carbotti G, Ferrini S. Context-dependent role of IL-18 in cancer biology and counter-regulation by IL-18BP. J Leukoc Biol 2015;97:665–75. [DOI] [PubMed] [Google Scholar]
- 10. Guo L, Junttila IS, Paul WE. Cytokine-induced cytokine production by conventional and innate lymphoid cells. Trends Immunol 2012;33:598–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dinarello CA, Novick D, Kim S, Kaplanski G. Interleukin-18 and IL-18 binding protein. Front Immunol 2013;4:289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Robertson MJ, Kirkwood JM, Logan TF, Koch KM, Kathman S, Kirby LC, et al. A dose-escalation study of recombinant human interleukin-18 using two different schedules of administration in patients with cancer. Clin Cancer Res 2008;14:3462–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Paulukat J, Bosmann M, Nold M, Garkisch S, Kämpfer H, Frank S, et al. Expression and release of IL-18 binding protein in response to IFN-gamma. J Immunol 2001;167:7038–43. [DOI] [PubMed] [Google Scholar]
- 14. Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 2018;173:400–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andrews DM, Sullivan LC, Baschuk N, Chan CJ, Berry R, Cotterell CL, et al. Recognition of H2-M3 by Ly49A regulates natural killer cell licensing and activation. Nat Immunol 2012;13:1171–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Uzana R, Eisenberg G, Sagi Y, Frankenburg S, Merims S, Amariglio N, et al. Trogocytosis is a gateway to characterize functional diversity in melanoma-specific CD8+ T cell clones. J Immunol 2012;188:632–40. [DOI] [PubMed] [Google Scholar]
- 17. Hansen K, Kumar S, Logronio K, Whelan S, Qurashi S, Cheng H-Y, et al. COM902, a novel therapeutic antibody targeting TIGIT augments anti-tumor T cell function in combination with PVRIG or PD-1 pathway blockade. Cancer Immunol Immunother 2021;70:3525–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 2017;127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, et al. Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep 2017;19:1189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mosely SIS, Prime JE, Sainson RCA, Koopmann J-O, Wang DYQ, Greenawalt DM, et al. Rational selection of syngeneic preclinical tumor models for immunotherapeutic drug discovery. Cancer Immunol Res 2017;5:29–41. [DOI] [PubMed] [Google Scholar]
- 21. Aoki H, Shichino S, Matsushima K, Ueha S. Revealing clonal responses of tumor-reactive T-cells through T cell receptor repertoire analysis. Front Immunol 2022;13:807696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell 2019;176:775–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Plebanek MP, Xue Y, Nguyen Y-V, DeVito NC, Wang X, Holtzhausen A, et al. A SREBF2-dependent gene program drives an immunotolerant dendritic cell population during cancer progression. Biorxiv 2023;2023.04.26.538456. [Google Scholar]
- 24. Liu C, Chu D, Kalantar-Zadeh K, George J, Young HA, Liu G. Cytokines: from clinical significance to quantification. Adv Sci 2021;8:2004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rhode PR, Egan JO, Xu W, Hong H, Webb GM, Chen X, et al. Comparison of the superagonist complex, ALT-803, to IL15 as cancer immunotherapeutics in animal models. Cancer Immunol Res 2016;4:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Connor J, Bannerji R, Saito S, Heston W, Fair W, Gilboa E, et al. Regression of bladder tumors in mice treated with interleukin 2 gene-modified tumor cells. J Exp Med 1993;177:1127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tugues S, Burkhard SH, Ohs I, Vrohlings M, Nussbaum K, Vom Berg J, et al. New insights into IL-12-mediated tumor suppression. Cell Death Differ 2015;22:237–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Berraondo P, Sanmamed MF, Ochoa MC, Etxeberria I, Aznar MA, Pérez-Gracia JL, et al. Cytokines in clinical cancer immunotherapy. Br J Cancer 2019;120:6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oberstein PE, Rahma OE, Beri N, Stoll-D'Astice AC, Duliege A-M, Nazeer S, et al. A phase 1b study evaluating IL-1β and PD-1 targeting with chemotherapy in metastatic pancreatic cancer (PanCAN-SR1). JCO 2022;40:557. [Google Scholar]
- 30. Lewis AM, Varghese S, Xu H, Alexander HR. Interleukin-1 and cancer progression: the emerging role of interleukin-1 receptor antagonist as a novel therapeutic agent in cancer treatment. J Transl Med 2006;4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou T, Damsky W, Weizman O-E, McGeary MK, Hartmann KP, Rosen CE, et al. IL-18BP is a secreted immune checkpoint and barrier to IL-18 immunotherapy. Nature 2020;583:609–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lillo S, Saleh M. Inflammasomes in cancer progression and anti-tumor immunity. Front Cell Dev Biol 2022;10:839041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Z, Li X, Wang Y, Wei Y, Wei X. Involvement of inflammasomes in tumor microenvironment and tumor therapies. J Hematol Oncol 2023;16:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krysko O, Løve Aaes T, Bachert C, Vandenabeele P, Krysko DV. Many faces of DAMPs in cancer therapy. Cell Death Dis 2013;4:e631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee C-H, Motzer RJ, Glen H, Michaelson MD, Larkin J, Minoshima Y, et al. Correlative serum biomarker analyses in the phase 2 trial of lenvatinib-plus-everolimus in patients with metastatic renal cell carcinoma. Br J Cancer 2021;124:237–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Prinzing B, Schreiner P, Bell M, Fan Y, Krenciute G, Gottschalk S. MyD88/CD40 signaling retains CAR T cells in a less differentiated state. JCI Insight 2020;5:e136093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lange S, Sand LGL, Bell M, Patil SL, Langfitt D, Gottschalk S. A chimeric GM-CSF/IL18 receptor to sustain CAR T-cell function. Cancer Discov 2021;11:1661–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huang B, Song B, Xu C. Cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat Metab 2020;2:132–41. [DOI] [PubMed] [Google Scholar]
- 39. Mutala LB, Deleine C, Karakachoff M, Dansette D, Ducoin K, Oger R, et al. The caspase-1/IL-18 axis of the inflammasome in tumor cells: a modulator of the Th1/Tc1 response of tumor-infiltrating T lymphocytes in colorectal cancer. Cancers (Basel) 2021;13:189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yoshimoto T, Takeda K, Tanaka T, Ohkusu K, Kashiwamura S, Okamura H, et al. IL-12 up-regulates IL-18 receptor expression on T cells, Th1 cells, and B cells: synergism with IL-18 for IFN-γ production1. J Immunol 1998;161:3400–7. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. IL-18BP is a potential soluble immune checkpoint in cancer.
Supplemental Figure S2. COM503 releases IL-18 activity in biochemical and functional assays.
Supplemental Figure S3. Antitumor activity of anti-mouse-IL-18BP Ab in mice studies.
Supplemental Figure S4. Anti-IL-18BP Ab induced immunogenic memory response in E0771 tumor model.
Supplemental Figure S5. IL-18BP blockade alters the immune infiltrate composition of E0771 tumors.
Supplementary Methods
Data Availability Statement
Primary scRNA-seq data sets have been deposited in the Gene Expression Omnibus under accession code GSE239473. All other data associated with this study are present in the main paper or the Supplementary Materials or Methods or are available from the corresponding author upon reasonable request.