Summary
Background
Natural killer (NK) cells are important innate immunity players and have unique abilities to recognize and eliminate cancer cells, particularly in settings of antibody-opsonization and antibody-dependant cellular cytotoxicity (ADCC). However, NK cell-based responses in bladder cancers to therapeutic antibodies are typically immunosuppressed, and these immunosuppressive mechanisms are largely unknown.
Methods
Single cell RNA sequencing (scRNA-seq) and high-dimensional flow cytometry were used to investigate the phenotype of tumour-infiltrating NK cells in patients with bladder cancer. Further, in vitro, and in vivo models of this disease were used to validate these findings.
Findings
NK cells within bladder tumours displayed reduced expression of FcγRIIIa/CD16, the critical Fc receptor involved in ADCC-mediated cytotoxicity, on both transcriptional and protein levels. Transcriptional signatures of transforming growth factor (TGF)-β-signalling, a pleiotropic cytokine known for its immunosuppressive and tissue residency-inducing effects, were upregulated in tumour-infiltrating NK cells. TGF-β mediated CD16 downregulation on NK cells, was further validated in vitro, which was accompanied by a transition into a tissue residency phenotype. This CD16 downregulation was also abrogated by TGF-βR signalling inhibition, which could also restore the ADCC ability of NK cells subject to TGF-β effects. In a humanized mouse model of bladder cancer, mice treated with a TGF-β inhibitor exhibited increased ADCC activity compared to mice treated only with antibodies.
Interpretation
This study highlights how TGF-β-rich bladder cancers inhibit NK cell-mediated ADCC by downregulating CD16. TGF-β inhibition represents new avenues to reverse immunosuppression and enhance the tumoricidal capacity of NK cells in bladder cancer.
Funding
The Guimaraes Laboratory is funded by a US Department of Defense—Breast Cancer Research Program—Breakthrough Award Level 1 (#BC200025), a grant (#2019485) awarded through the Medical Research Future Fund (MRFF, with the support of the Queensland Children's Hospital Foundation, Microba Life Sciences, Richie's Rainbow Foundation, Translational Research Institute (TRI) and UQ), and a grant (#RSS_2023_085) funded by a Metro South Health Research Support Scheme. J.K.M.W. is funded by a UQ Research Training Program PhD Scholarship and N.O. is funded by a NHMRC Postgraduate Scholarship (#2021932).
Keywords: ADCC, NK cells, TGF-β, CD16, Bladder cancer
Research in context.
Evidence before this study
A systematic search was conducted to identify clinical trials in patients with bladder cancer testing IgG1 monoclonal therapeutic antibodies. There were only three identifiable clinical trials NCT03473730, NCT04068896 and NCT00645593. Despite the high expression of tumour targets e.g. HER-2 and EGFR, these antibodies have not shown promising results. NK cells are a subset of innate immune cells that are critical in mounting immune responses to these monoclonal antibodies through a process known as ADCC. The cancer killing capability of these cells is heavily suppressed through a myriad of pathways in cancer. The precise ability of these cells to respond to therapeutic antibodies in bladder cancer has not yet been investigated and their inhibition may contribute to the ineffectiveness of antibody therapies in this disease.
Added value of this study
Our study has revealed a reduction in CD16 on a protein and transcriptional level in NK cells infiltrating bladder tumours, concomitant to expression of tissue residency markers such as CD49a. From a mechanistic standpoint, we have identified TGF-β as a key driver in CD49a upregulation and CD16 downregulation on NK cells, and inhibiting TGF-βR could restore ADCC responses.
Implications of all the available evidence
TGF-β inhibitors could be designed for local administration in combination with monoclonal antibodies to enhance ADCC responses of antibody therapies and immunity against bladder cancer.
Introduction
Natural killer (NK) cells represent an integral arm of the innate immune system. They can inherently recognize and kill tumour cells following activation of germline-encoded receptors.1 This tumoricidal capability has seen NK cells become an emerging target of anti-cancer immunotherapeutic strategies, including antibody-based and NK-cellular therapies.2,3 Multiple studies, however, suggest that NK cells can be suppressed within the solid tumour microenvironment (TME), downregulating critical receptors required for activation.4 In some tumours such as clear cell renal tumours, NK cells were characterized as abundant but displayed poor cytotoxic activity.5 Furthermore, studies have indicated that NK cells can be converted into CD49a-expressing tissue resident ILC1-like cells in response to TGF-β signalling, accompanied by lowered metabolic, proliferative and cytotoxic capacity in both humans and in mice.6, 7, 8, 9
NK cells also possess an ability to perform antibody-dependent cellular cytotoxicity (ADCC) when recognizing antibody opsonized targets.2 This is mediated by their common Fc receptor FcγRIII (CD16), which recognizes the Fc portion of the tumour-bound antibodies.10 Studies have indicated that NK cells lose their cytotoxic capabilities when converted to ILC1-like states in TGF-β rich TMEs,8 however it is unclear whether these cells also lose their ability to efficiently mediate ADCC.
Bladder cancer (urothelial carcinoma) is the ninth most common malignancy in the world.11 Patients who develop muscle invasive bladder cancer (MIBC), an advanced stage where the cancer has invaded the muscle,12 have a significantly lower overall survival rate of 50% compared with 85% for patients with non-MIBC.12 Standard treatments involve radical cystectomy and neoadjuvant chemotherapy, which are overly invasive and ultimately ineffective. New immunotherapeutic options are emerging for the treatment of MIBC, including both preclinical and FDA approved targeted therapies using monoclonal antibodies to induce NK cell ADCC. One such antibody is enfortumab vedotin, which targets Nectin-4.13, 14, 15 Currently, Avelumab is the only FDA approved, fully humanized monoclonal antibody capable of ADCC, and does not show a survival benefit compared to standard chemotherapy, response rates of 17%.16 Many other monoclonal antibodies are undergoing clinical trials but have seen fewer positive results, NCT03473730, NGM120 NCT04068896, cetuximab NCT00645593. As discussed, the effectiveness of these therapeutics may be limited due to immunosuppressive signals within the tumour, inhibiting the cytotoxic function of infiltrating NK cells. Tissue residence, immunosuppression and phenotypic change of tumour infiltrating NK cells may lead to a lowered capacity to perform effector functions like ADCC. Therefore, an in-depth characterization of NK cells within bladder tumours may help to guide the development of NK cell–based immunotherapies for bladder cancers. To address this, we used single cell RNA-sequencing (scRNA-seq) and flow cytometry to characterize NK cells infiltrating human bladder cancers. This analysis identified tumour-infiltrating NK cell subpopulations and highlighted striking differences between tumour-infiltrating and circulating NK cells. Immunosuppressive factors residing within the human bladder TME were further explored, revealing TGF-β as a target for therapeutic strategies such as TGF-β inhibitors to overcome NK cell immunosuppression and facilitate their optimal cytotoxic function.
Methods
Patient samples
Bladder tumour samples were freshly acquired from the surgical oncology unit at the Princess Alexandra Hospital (Woolloongabba, Australia). Samples were cut into small pieces and incubated in RPMI-1640 medium (Gibco, #11875093) containing 10% heat inactivated foetal bovine serum (FBS) (Gibco, #10099141), Normocin (10 μg/ml) (InvivoGen, #ant-nr-1), DNAse I (2 μg/ml) (Roche, #4716728001) and Collagenase IV (1 mg/ml) (Worthington Biochem, #LS004186), for 30 min at 37 °C on a shaker. The cell suspension was then filtered through a 70 μm strainer, and cells were washed with RPMI-1640 media. Cells were then resuspended in 37.5% percoll density gradient media (Cytiva, #17-0891-01) and centrifuged at 690 g for 25 min at room temperature to further purify lymphocytes. Peripheral blood mononuclear cells (PBMCs) were purified using Ficoll–Paque PLUS gradient (Cytiva, #17144003) and LeucoSep tubes according to the instructions of the manufacturer (Greiner Bio-One, #227290). The cells were washed in FACS buffer (PBS containing 2% FBS and EDTA 2 mM) at 4 °C before blocking with human Fc Block (1:100, FACS buffer; Miltenyi Biotec, #130-059-901). They were then incubated with the cocktail of conjugated antibodies (Supplementary Table S1) before acquisition and sorting of the CD45+ cells on a BD FACSAria sorter (Becton Dickinson). RRID tags are provided for all commercial antibodies used. The compensated data were analysed with FlowJo v10.5.2 in a classical supervised manner (exclusion of aggregates/debris/dead cells, gating of the NK cells, analysis of expression and signal intensity for each marker).
Human NK cell isolation and in vitro culture
Buffy coats were provided by the Australian Red Cross and PBMCs were purified using Ficoll gradient and LeucoSep tubes according to the instructions of the manufacturer. Once PBMCs were isolated, human NK cells were enriched by negative selection using the MojoSort™ Human NK Cell Isolation Kit, as per the manufacturer's instructions (BioLegend, #480053). For in vitro studies, NK cells were seeded into 96-well round bottom plates at 25,000 cells per well in 200 μl of complete NK MACS Medium (Miltenyi Biotec, #130-114-429) supplemented with 5% heat inactivated human AB serum (Red Cross). Cultures included recombinant cytokines including 5 ng/ml human IL-15 (Peprotech, #200-15) and 500 IU/ml of human IL-2 (Peprotech, #200-02), with or without 6.25 ng/ml human TGF-β1 (Peprotech, #100-21) and 25 ng/ml human IL-21 (Peprotech, #200-21), where indicated. Small molecule inhibitor galunisertib (LY2157299, SelleckChem, Houston, TX) was also added at a concentration of 5 μM, respectively, where indicated, as described previously.17 Plates were incubated at 37 °C with 5% CO2 for a total of up to 14 days, where media was refreshed on day 5 and every 3 days after. Cells were processed and analysed by flow cytometry at indicated timepoints.
Quantitative image based cytometry (QIBC)
NK cells, stimulated with culture conditions as indicated on figure legends, were seeded in Poly-d-lysine (Sigma–Aldrich, P6403) coated glass-bottom 96 well plates (CellVis, #P96–1.5H-N) then centrifuged for 5 min at 300 g. Cells were then fixed in PBS containing 4% paraformaldehyde (PFA) (ProSciTech, #C006-100) for 15 min and subsequently permeabilized with Triton X 100 0.5% (Sigma–Aldrich, #T8787) and washed with PBS. Cells were then blocked with PBSG (PBS with 10% goat serum, Sigma–Aldrich, #G9023) for 1 h at RT and stained with mouse anti-human CD16 BUV496 (BD Biosciences, #564653) and incubated for 3.5 h at RT. Finally, cells were stained with Wheat Germ Agglutinin (WGA) (ThermoFisher, #W11261), and DRAQ7 (ThermoFisher, #D15106) in PSBG. Cells were washed and imaged using the High-Content Imaging System IN Cell Analyzer 6500 HS (Cytiva). Four most central fields of view were imaged per well using a Plan Apo 10 × 0.45 NA objective and output as 16-bit TIFF files with 2040 × 2040-pixel resolution and 1.5384 μm pixel size. A 60 10 × 1.0 NA water immersive objective was used for representative images (Supplementary Figure S7A). Exposure time parameters were adjusted on channel basis aiming to maximise use of detector dynamic range. IN Cell Analyzer's default parameters were used for WGA (exc. 488 nm, em. 500–550 nm) and DRAQ7 (exc. 642 nm, em. 650–720 nnm). Custom parameters were used for BUV496 (exc. 405 nm, em. 500–550 nm). Single stained samples were used to confirming no channel leakage between the dyes.
QIBC analysis was based on Besse et al., 202318 with the following alterations. Firstly background subtraction of all channels (rolling ball method, 30-pixel size) was performed in ImageJ (version 1.53t).19 In Cell Profiler 4,20 measurements of pixel intensities of all channels were performed on cell masks generated from WGA channel. Nuclear staining by DRAQ7 was used to further identify cells from staining artifacts of cell debris. Circularity (Form factor) and area measurements were used to filter out missegmented or misidentified nuclei or cells. Data analyses were performed using R scripting (RStudio, Posit) with the following packages: dplyr, ggplot2, readr, plater, gghalves, ggpubr, purr and rstatix. CD16 mean fluorescence intensities (Cell Profiler format) within the WGA mask were plotted and used for CD16 expression analysis and statistics.
Cell lines
The bladder cancer cell line UM-UC-3 was obtained from the American Type Culture Collection (ATCC) and maintained in DMEM (Gibco, #11960044) supplemented with 10% FBS, 1% penicillin-streptomycin (Lonza, #17-745E), 1% Glutamax (Gibco, #35050-061), 1% NEAA (Lonza, #13-114E), and 1% sodium pyruvate (Gibco, #11360-070). Cell lines were grown in a humidified incubator at 37 °C and 5% CO2 and maintained with flask confluence not higher than 80%. All cell lines were tested for Mycoplasma contamination by PCR using standard Mycoplasma testing protocol21 and recently profiled by STR (Supplemental Data–Reagent Validation file). To analyse cell surface expression of GD2, cells were detached using TrypLE Select enzyme (Gibco, #12563029) and washed in FACS buffer (PBS containing 2% FBS and EDTA 2 mM) at 4 °C, before blocking with human Fc Block (1:100 FACS buffer). Cells were subsequently incubated with the anti-GD2 antibody (BioLegend, #357315) for 30 min at 4 °C before acquisition on a BD LSRFortessa flow cytometer (Becton Dickinson). The compensated data were analysed with FlowJo v10.5.2 in a classical supervised manner (exclusion of aggregates/debris/dead cells, gating of the NK cells, analysis of expression and signal intensity for each marker).
NK cell cytotoxicity assays
Target cells (UM-UC-3) were labelled with Calcein AM (15 μM, Invitrogen, #C3099) for 30 min at 37 °C, as previously described.22 Human NK cells, the anti-human-GD2 chimeric mAb ch14.18/dinutuximab (United Therapeutics), and target cells were successively added to each well of a round-bottom 96 well plate and maintained in RPMI without phenol red (Gibco, #11960044) supplemented with 5% heat inactivated human serum, 1% penicillin-streptomycin, 1% Glutamax, 1% NEAA, and 1% sodium pyruvate. After 4 h of co-culture, the supernatant was transferred to an opaque 96-well plate (Costar). CalceinAM released by dead target cells was determined by measuring the number of relative fluorescence units (RFU) with a fluorescence microplate reader (BMG CLARIOstar®; excitation at λ = 495 nm and emission at λ = 516 nm). The percentage specific lysis was calculated with the following formula:
| Specific lysis (%) = (ER (RFU)—SR (RFU))/(MR (RFU)—SR (RFU)) × 100, where ER = experimental release, SR = spontaneous release and MR = maximal release. |
SPLiT-seq scRNA-seq
Immediately after sorting, CD45+ cells from blood or tumour samples were centrifuged at 200 g for 10 min. Supernatant was discarded and pellets were resuspended in 750 μl of Cell Prefixation Buffer (Parse Biosciences, #PBSSB1001). Samples were then fixed using the Parse Biosciences Cell Fixation (v1) kit (Parse Biosciences, #PBSSB1001), as per the manufacturer's instructions and stored at −80°.
Barcoding and library preparation
In advance of cell barcoding and library preparation, fixed samples were removed from −80 °C, thawed in a water bath set to 37 °C, placed on ice, and counted. Samples were barcoded and single cell sequencing libraries were then prepared using the Evercode Whole Transcriptome Mini (v1) kit (Parse Biosciences, #PBSEC-W01010), as per the manufacturer's instructions. Libraries were sequenced using a NovaSeq6000 SP SE100 flow cell.
Raw data processing and quality control
Reads from scRNA-seq experiments were aligned to the GRCh38 (hg38) human reference genome using the Parse biosciences pipeline (split-pipe v0.9.6p) to generate cell by gene counts matrices. Cells with low quality metrics such as high mitochondrial gene content (>7%) and low number of genes detected (<200) were removed (Supplementary Figure S1A–C). RNA counts were log normalized using the standard Seurat workflow.23 To visualize cells based on an unsupervised transcriptomic analysis, we first ran PCA using 2000 variable genes. The integrated sample counts were scaled, and variable features used for principal-component analysis (PCA). With ‘Elbowplot’ function of Seurat the top 30 principal components from this analysis were then used as an input for dimensionality reduction by Uniform Manifold Approximation and Projection (UMAP), as previously described.24 Shared-nearest-neighbour based clustering using the top 30 principal components was used to generate clusters with a resolution = 1.4. After cell clustering, we further filtered out the NK cell cluster using the ‘subset’ function based on cluster expression of NK marker genes NCR1, NKG7 and GNLY. The top highly variable genes were again selected and PCA was used to find principal components. The top 15 PCs were visualized using UMAP with a resolution = 0.6. The signature genes identifying each cluster were found using the ‘FindAllMarkers' function. Single sample gene set enrichment analysis (ssGSEA) was performed using the escape package (v1.8.0). Hallmark gene sets were retrieved from the molecular signature database (MSigDB).
Focussed reanalyses of publicly available single cell sequencing datasets
scRNA-seq data from four samples retrieved from patients with bladder cancer were processed under the accession code HRA000212.25 Raw data processing and quality control was performed as per the original study25 where cells with low quality metrics such as high mitochondrial gene content (>10%) and low number of genes detected (<1000) were removed. To eliminate potential doublets, single cells with over 6000 genes detected were also filtered out (Supplementary Figure S2A–C). RNA counts were log normalized using the standard Seurat workflow.23 To visualize cells based on an unsupervised transcriptomic analysis, we first ran PCA using 2000 variable genes. The integrated sample counts were scaled, and variable features used for principal-component analysis (PCA). With ‘Elbowplot’ function of Seurat the top 20 principal components from this analysis were then used for downstream analysis. After cell clustering, we further filtered out the NK cell cluster using the ‘subset’ function based on cluster expression of NK marker genes NCR1, NKG7 and GNLY. The top highly variable genes were again selected and PCA was used to find principal components. The top 15 PCs were visualized using UMAP with a resolution = 0.5. Tissue residency score was generated using indicated genes with ‘DoHeatmap’ function. Using these cluster defining gene-sets we developed our NK cell gene signatures using the top 20 DEGs, from ‘FindMarkers’ function. |FC| >0.1 and min.pct <0.1 were used as the cut-off criteria.
Bulk sequencing data collection and filtering
The TCGA clinical data for epithelial cancer subsets were obtained from the GDC legacy database using the TCGAbiolinks R package (https://bioconductor.org/packages/release/bioc/html/TCGAbiolinks.html). The legacy mRNA TCGA expression data (Illumina HiSeqV2) was manually downloaded from XenaBrowser (https://xenabrowser.net/datapages/?cohort=TCGA%20Bladder%20Cancer%20(BLCA)&removeHub=https%3A%2F%2Fxena.treehouse.gi.ucsc.edu%3A443). The gene expression data from this database is normalized. Both clinical and expression data was filtered to include “Primary Tumour” sample types only, using the unique descriptive barcode assigned to each patient. Technical assistance was offered by the Queensland Facility for Advanced Bioinformatics (QFAB) to collect and analyse data.
NK score generation and pearson rank correlation analysis
The NKscore gene signature was adapted from.26 Each sample were ranked in the dataset by the increasing abundance of each gene using the ‘rankgenes’ function in the Singscore software package. The scores were determined using ‘simpleScore’ function by providing NKscore marker genes. The optimal threshold for NKscore was determined using median threshold partitioning to establish the association between scores and time to death.
In vivo bladder tumour modelling
Human IL-7 and IL-15 double knock in (KI) nonobese diabetic–severe combined immunodeficient gamma (NSG) mice hIL-7xhIL-15 KI NSG) were used to maintain human NK cell development and function, as described previously.27 1 × 106 UM-UC-3 cells for each mouse were re-suspended in 50:50 PBS and growth factor reduced matrigel (Corning, #354230) (100 μl total injection volume).28 The cell-matrigel mix was injected subcutaneously into hIL-7xhIL-15 KI NSG mice. Tumour volume (measured with digital callipers) and body weight were monitored every 2–3 days. All experiments were performed using age matched cohort of mice (age range, 6–12 weeks). Cohort sizes were described in each figure legends to achieve statistical significance. No biological replicate was excluded based on preestablished criteria.
In vivo treatment
Four days post tumour inoculation, mice bearing tumours were randomly assigned into treatment groups and groups receiving NK cells were injected intravenously with 5 × 106 NK cells. For tumour initial NK cell phenotyping experiments, mice received 1 × 106 NK cells or 1 × 107 human PBMCs. Mice receiving dinutuximab were injected intravenously twice per week with of 500 μg antibody. Mice receiving galunisertib were administered at 75 mg/kg twice a day (BID) by oral gavage. Galunisertib was solubilized in a vehicle solution [1% carboxymethylcellulose sodium salt, 0.5% SDS, 0.085% povidone, and 0.05% antifoam Y-30 emulsion] as previously described by Yingling et al.29 Mice not receiving galunisertib were gavaged BID with the formulated vehicle solution alone.
Endpoint ex vivo bioluminescent imaging
For tumour size control study, NSG mice were euthanized when their primary tumours reached 800 mm3. At endpoint mice received an intraperitoneal injection of luciferin (150 mg/kg, Promega) and were culled 5 min post administration. Lungs were harvested and rinsed in PBS, and covered with 150 μg/ml luciferin in PBS and imaged immediately. Bioluminescence signals of individual organs ex vivo were quantified by drawing regions of interest (ROIs) from which the total flux (photons/second) was obtained.
Tumour processing and flow cytometry
Tumours were harvested, and peripheral blood collected by cardiac puncture in EDTA-containing collection tubes. Tumours were enzymatically and mechanically dissociated using the gentleMACS Dissociator (Miltenyi Biotec) and RPMI, collagenase, DNAse and normicin media, as described earlier. The digested tissue was filtered through a 70-μm cell strainer (Falcon, #352350) to obtain a single-cell suspension. Percoll density gradient media was then used to further purify lymphocytes from tumours. Red blood cells (RBC) were lysed for all tissue compartments by incubation in RBC Lysis Buffer (BioLegend, #420302). The cells from both blood and tumour were washed in FACS buffer at 4 °C before blocking with human Fc Block (1:100 FACS buffer; Miltenyi Biotec, #130-059-901). They were then incubated with the cocktail of conjugated antibodies (Supplementary Table S1) before acquisition on a BD LSRFortessa flow cytometer (Becton Dickinson). The compensated data were analysed with FlowJo v10.5.2 in a classical supervised manner (exclusion of aggregates/debris/dead cells, gating of the NK cells, analysis of expression and signal intensity for each marker).
Statistical analysis
All graphs show mean ± SEM. Statistical analyses were performed in GraphPad Prism 9 and R version 4.3.2. Statistical tests were applied as indicated in figure legends and significant differences annotated on each respective figure. We used Mann–Whitney T test to compare two groups, where data was non-parametric and for parametric data we used Welch's T test. For multiple comparisons tests we used the two-way ANOVA.
Role of funders
The funders have no role in the study design, the data collection, data analysis, interpretation, and the writing of the report. The corresponding author had full access to all the data and the final responsibility to submit for publication.
Ethics
All experiments using human samples were conducted with approval from the Metro South Human Research Ethics Committee (clearances #HREC/2019/QMS/55385 and HREC/2022/QMS/89430), which were ratified by the UQ Human Research Ethics Committee. All patients gave written consent for sample collection. All animal experiments were conducted according to approval by the University of Queensland Molecular Biosciences Animal Ethics Committee (approval number 2021/AE001137) and procedures were carried out in accordance with the regulatory standards of the National Health and Medical Research Council (NHMRC) and the Australian Code for the Responsible Conduct of Research.
Results
Human bladder tumours contain conventional and tissue resident NK cells
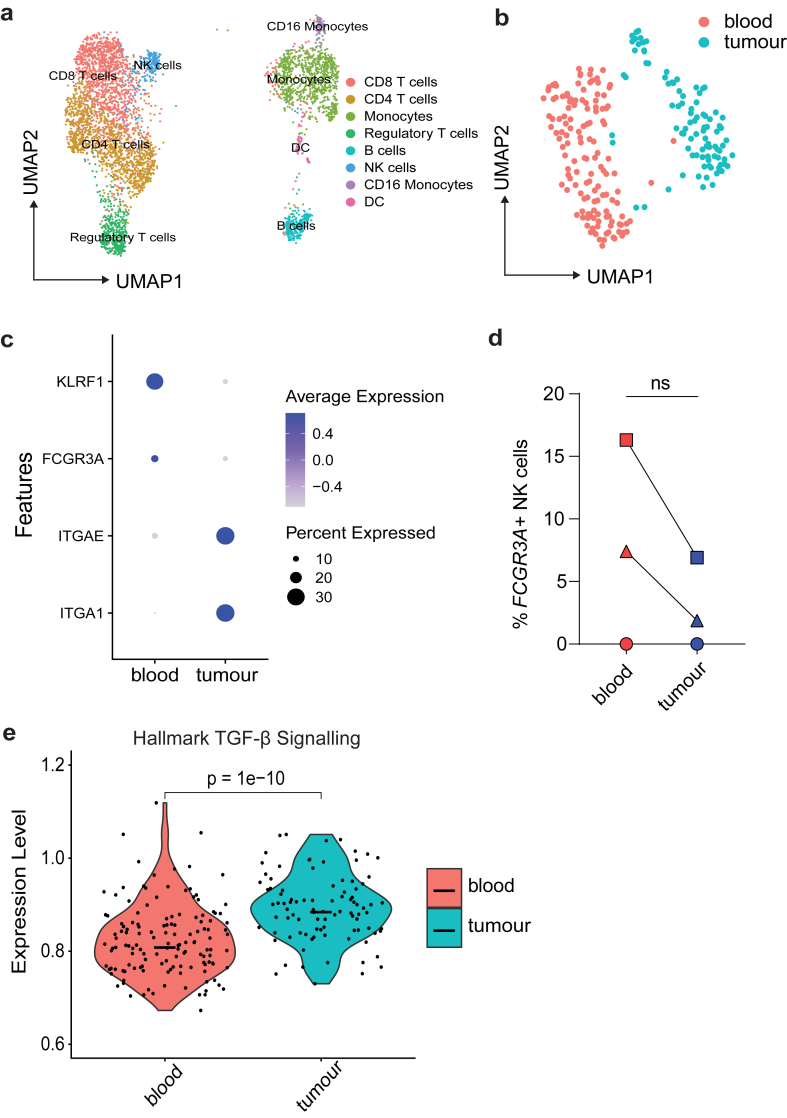
Using scRNA-seq, we took an unbiased approach to evaluate NK cell states within human bladder cancer. We obtained histologically confirmed bladder cancer tumour samples, as well as adjacent bladder tissue and peripheral blood (patient description listed in Supplementary Table S2). Samples for each tissue type from patients with bladder cancer (n = 3) were subject to processing for scRNA-seq. We used the R package Seurat to subset major immune populations into clusters (Fig. 1a), which were annotated based on feature genes (Supplementary Figure S3A and B) as described in previous studies.23 To specifically explore NK cells, the NK cell cluster was subset and re-clustered, which identified three distinct clusters. Notably, clusters 0 and 2 predominately represented NK cells from the blood, whereas cluster 1 predominantly represented NK cells from the tumour (Fig. 1b and Supplementary Figure S3C). When investigating tumour and blood NK cells, blood derived NK cells expressed conventional NK cell genes such as KLRF1, PRF1, GZMB whereas tumour NK cells were defined by an elevated expression of tissue resident NK cell genes such as ITGA1 (encoding CD49a) in agreement with previous reports of tumour infiltrating NK cells30 (Fig. 1c). Direct comparison of NK cells from the blood vs tumour found that NK cells from the blood trended towards a higher expression of FCGR3A, the gene for FcγRIIIa/CD16, however given the limited patient cohort number, this difference was not found significant (Fig. 1d). We subsequently performed single sample gene set enrichment analysis (ssGSEA) of relevant pathways, comparing NK cells from the blood to the tumour in our human bladder cancer scRNA-seq dataset, as previously described.31 We found a significant upregulation in the hallmark pathways for TGF-β signalling in the tumour NK cells (Fig. 1e). As previously described, TGF-β can have many immunosuppressive effects on NK cells,8,32 and we evidence NK cells in bladder cancer upregulate ITGA1, which is known to be a consequence of TGF-β signalling.6,8 Taken together, these results suggest that there is a downregulation of FCGR3A or CD16 in bladder-tumour infiltrating NK cells, which is a potential mechanism impeding NK cell effector function and anti-tumour immunity in this cancer group.
Fig. 1.
scRNA-seq of human bladder cancer identifies immunosuppressed tumour infiltrating NK cells. Bladder tumours harvested from patients and matched blood were analysed by scRNA-seq. (a) UMAP plot of CD45+ cells coloured clusters defined by canonical immune markers, where each dot represents a single cell. (b) UMAP plots of NK cell clusters where clusters separated by tissue compartment. (c) Dot plots showing expression of indicated genes across tumour or blood derived NK cell clusters. (d) Individual patient raw gene counts for indicated gene FCGR3A plotted as a percentage of total NK cells (n = 3). Data from one experiment (n = 3 biological samples) where each symbol represents an individual patient, graphs show mean value ± SEM. Groups were compared using Mann–Whitney t test, where P > 0.05 was deemed not significant. (e) Violin plots showing relative enrichment of indicated Hallmark gene sets, calculated by ssGSEA (n = 3). ssGSEA groups were compared using Welch's t test, where P > 0.05 was deemed not significant.
FCRG3A/CD16 surface expression is reduced on bladder tumour-infiltrating NK cells
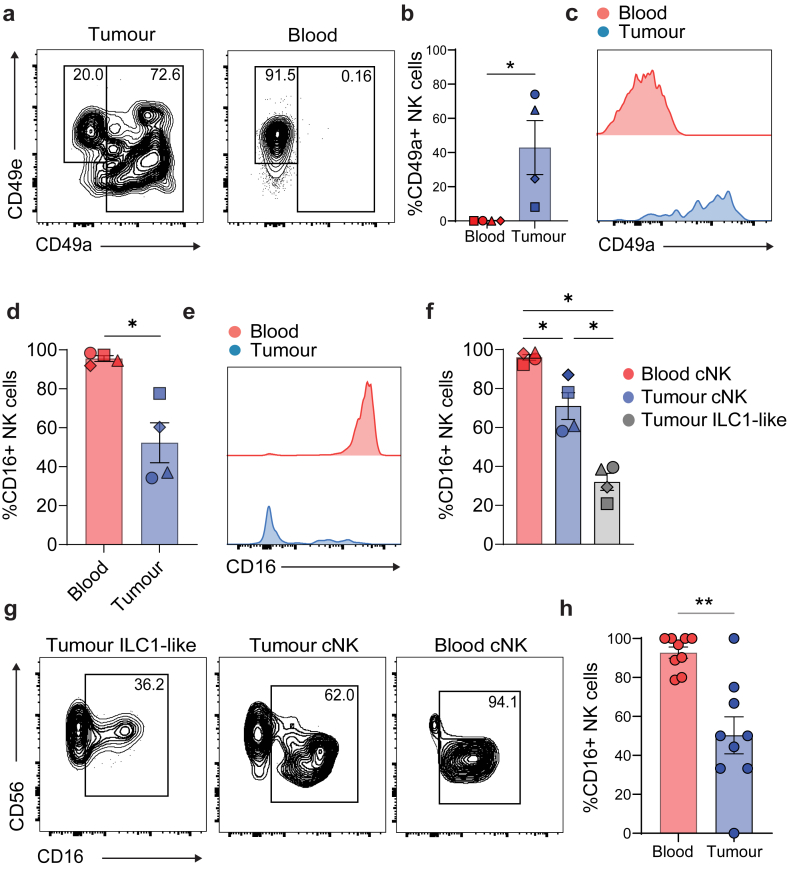
To validate downregulation of FCGR3A gene expression in tumour NK cells corresponded with downregulation of CD16 surface protein, we performed flow cytometry on the matched patient samples from the scRNA-seq experiments (Fig. 1). NK cell activating markers were analysed during flow cytometry sorting (Supplementary Figure S4A). We observed a significant upregulation of the tissue residency marker and collagen binding protein CD49a in NK cells infiltrating the tumour, which conversely was not seen in circulating NK cells in the blood (Fig. 2a–c). This validated the elevated ITGA1 gene expression observed in the tumour from the scRNA-seq analysis. Upregulation of CD49a on NK cells has previously been linked to a phenotypic switch from conventional NK (cNK) to tissue resident ILC1-like cells.6,8,9,33 In association with the upregulation of CD49a within the tumour NK cells, we also observed a drastically decreased expression of CD16 compared to blood circulating NK cells (Fig. 2d–e). Interestingly, a lower proportion of tumour infiltrating NK cells expressed CD16 and this was significantly lower in ILC1-like CD49a+ NK cells compared to conventional CD49a−CD49e+ NK cells (Fig. 2f–g). Previous reports indicate that CD49a is a classical biomarker of immunosuppressive TGF-β signalling in NK cells,6,8 and we show that this pathway can also be associated with other features such as the observed CD16 loss in NK cells infiltrating bladder tumours. To assess if downregulation of CD16 on NK cells could be replicated in vivo, human bladder cancer cells were inoculated in hIL-7/hIL-15 KI NSG mice reconstituted with human PBMCs. At the indicated endpoint, we then analysed the NK cell expression of CD16 from the blood and tumour homogenates 3 days post PBMCs infusion. We observed a downregulation of CD16 expression in intratumoural NK cells compared to circulating blood NK cells (Fig. 2h) experimentally supporting the hypothesis that the TME can induce CD16-downregulation in tumour-infiltrating NK cells.
Fig. 2.
Validation of CD16 downregulation on NK cells within bladder tumours. CD16 marker expression on NK cells from human bladder tumours and matched blood was analysed by multiparametric flow cytometry. (a) Representative flow cytometry plots showing CD49a (ILC1-like marker) and CD49e (conventional NK cell marker) expression from one patient resected tumour and blood. (b) CD49a expression as a bar plot and (c) representative histogram plot for both tumour and blood NK (CD45+lin−CD56+NKp46+) cells. (d) CD16 expression as a bar plot and (e) representative histogram plot for both tumour and blood NK (CD45+lin−CD56+NKp46+) cell. (f) CD16 expression as a percentage of each gated cell type; conventional blood NK cell (CD45+lin−CD56+NKp46+CD49e+CD49a−) (cNK), tumour conventional NK cell (CD45+lin−CD56+NKp46+CD49e+CD49a−) and tumour ILC1-like NK cell (CD45+lin−CD56+NKp46+CD49e−CD49a+) displayed as bar plots and (g) representative contour plots. Data from 4 independent experiments (n = 4) and each symbol represents an individual patient bar graphs show mean value ± SEM Statistical P values determined by Mann–Whitney t test, where ∗P < 0.05. (H) Tumour bearing NSG IL-7/IL-15 KI mice were injected with isolated human PBMC and after 3 days blood and tumours were taken to analyse NK cell populations. Expression of CD16 on NK cells across each compartment. Data are from two separate pooled experiments, denoted by symbols (n = 9). Each symbol represents an individual mouse, bar graphs show mean value ± SEM. Statistical P values determined by Mann–Whitney t test, where ∗P < 0.05, and ∗∗P < 0.01.
NK cell tissue residency correlates with TGF-β signalling and FCGR3A downregulation
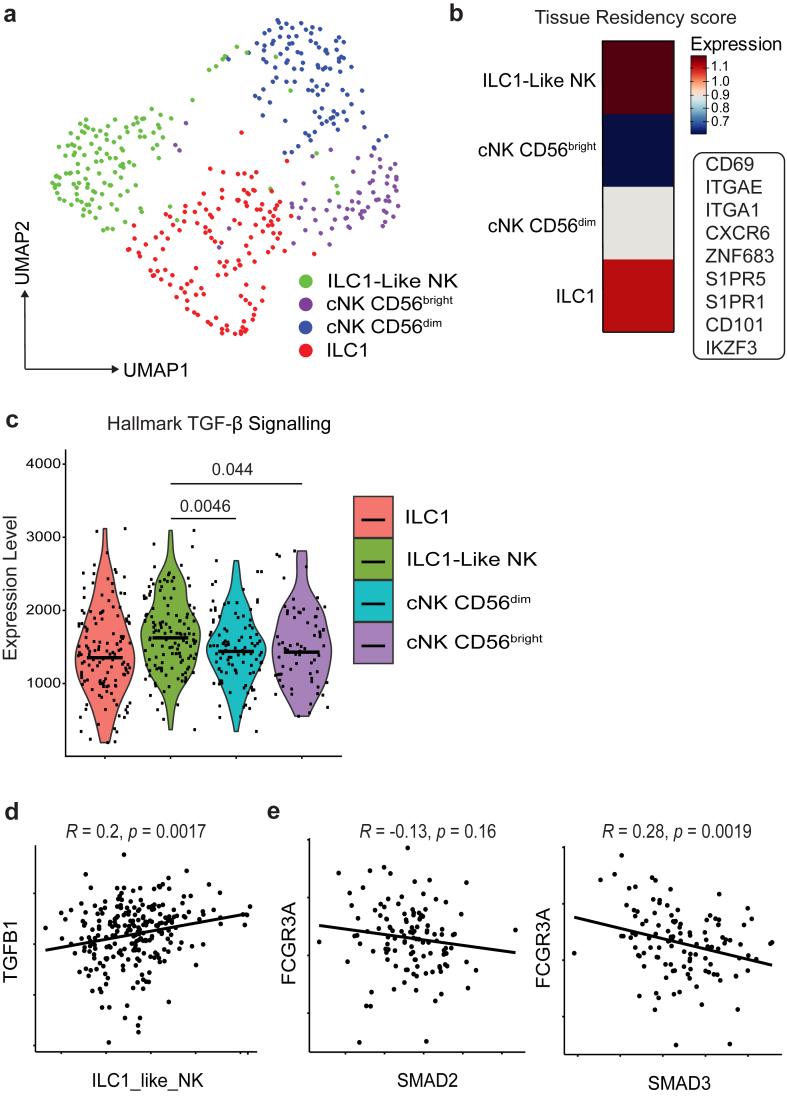
To investigate potential mechanisms behind FCGR3A downregulation and upregulation of ITGA1, we analysed a publicly available scRNA-seq bladder cancer dataset with higher sequencing depth by Chen et al., 202025 to identify cell states more stringently. Similar processing was performed as our own dataset, to identify NK cells by their major identifying markers in humans NCR1, NKG7 and GNLY (Supplementary Figure S5A and B). We were able to identify four ILC/NK cell subsets, most notably cNK CD56dim and ILC1-like cell subsets (Fig. 3a). These subsets were defined by marker genes as defined in Supplementary Figure S6A. The specific tissue residency status of each cluster was identified using a set of NK cell tissue residency genes defined by Herman et al., 202334 (Fig. 3b). The ILC1-like cell cluster also presented the highest expression of tissue residency genes. Using ssGSEA, as described earlier, we evidence that the ILC1-like NK cluster did have a significant upregulation of TGF-β signalling genes when compared to the cNK CD56dim cluster (Fig. 3c). We developed two gene signatures (Supplementary Table S3) derived from the ILC1-like cell subset and cNK CD56dim cell subset. To validate these NK signatures, we applied the gene-sets to a pan-cancer human NK cell atlas developed by Tang et al., 202335 (Supplementary Figure S6B). Our ILC1-like NK cell signature highly corroborated with a tumour specific NK cluster C5 CD56bright CD16lo CREM (Supplementary Figure S6C) and the cNK CD56dim signature matching with a blood specific NK cell cluster C1 CD56bright CD16lo GZMH (Supplementary Figure S6D). We applied our now validated gene signatures to a bulk RNA sequencing dataset obtained from the TCGA, which was normalized RNA sequencing from patients with bladder cancer (TCGA BLD). TGFB1 expression positively correlated with the ILC1-like NK cell signature (Fig. 3d) and after thresholding high NKscore patients, FCGR3A expression negatively correlated with TGF-β signalling genes SMAD2/3 (Fig. 3e). We identified a subset of FCGR3Aneg ILC1-like NK cells within bladder cancer that we suggest are inducible by TGF-β mediated signalling.
Fig. 3.
scRNA-seq and bulk sequencing datasets in bladder cancer further identifies TGF-β as a potential cause for CD16 loss. (a) UMAP plot showing NK cell clusters, integrated from 5scRNA-seq datasets from samples retrieved from patients with bladder cancer (n = 5). (b) Heatmap showing tissue residency score for each cluster as defined by indicated genes within the panel. (c) Violin plots showing relative enrichment of indicated Hallmark gene sets, calculated by ssGSEA. ssGSEA groups were compared using Welch's t test. (d) Correlation between ILC1-like NK score and TGFB1 subgroups in TCGA BLD cohort. (e) Correlation between FCRG3A and SMAD2/3 subgroups in TCGA BLD partitioned by high expression of NKscore. Coefficient was calculated with Pearson correlation analysis. Significant differences are indicated where P > 0.05 was deemed not significant.
TGF-β signalling downregulates CD16 expressing NK cells and limits ADCC capability in vitro
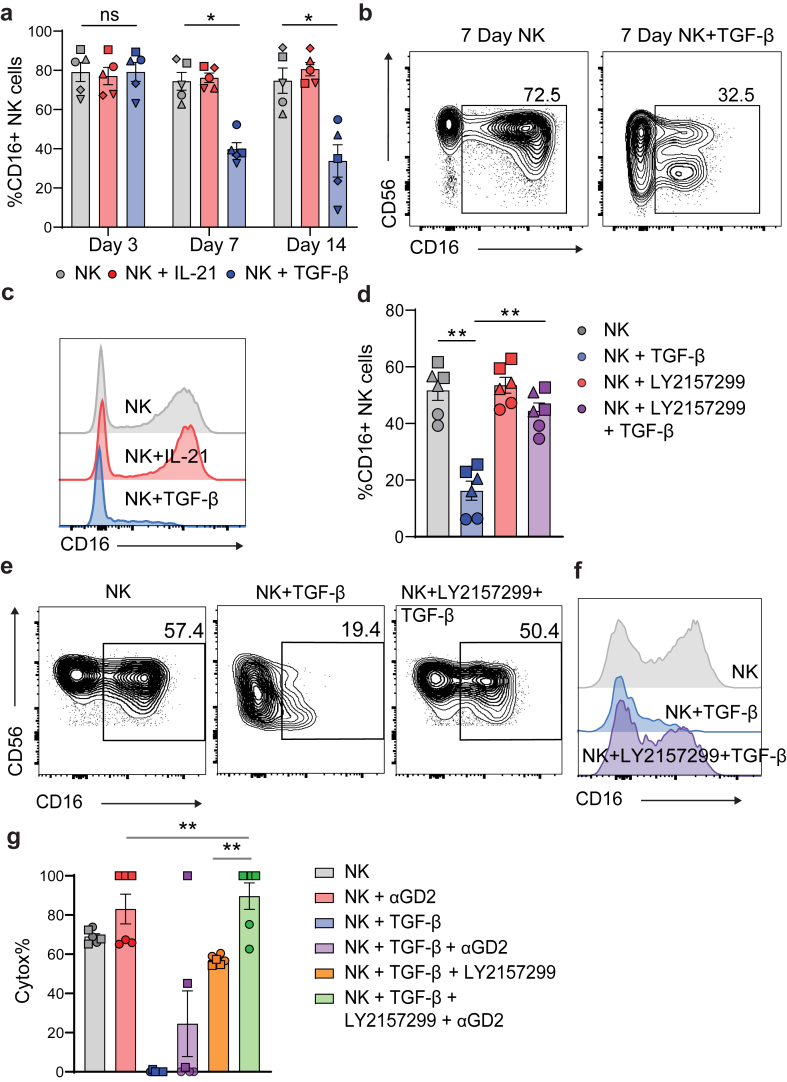
To confirm TGF-β induced downregulation of CD16, we cultured human PBMC-derived NK cells with or without recombinant (r) TGF-β, using IL-21 as a benchmark condition of sustained CD16 expression.36 Adding TGF-β was able to reduce the proportion of CD16+ NK cells by day 7, when compared to NK cells without TGF-β (Fig. 4a–c). CD16 downregulation was also verified using quantitative image based cytometry (QIBC), which has been rigorously applied in this context elsewhere (Supplementary Figure S7A–C).18 As expected, CD49a was also upregulated on NK cells after treatment with TGF-β in vitro (Supplementary Figure S4B). CD16 expression was dramatically reduced on CD49a+ cells, compared to CD49aneg, confirming that TGF-β induced mechanisms downregulate CD16 (Supplementary Figure S4C and D). Galunisertib (LY2157299 monohydrate) is a well-studied, small-molecule inhibitor of TGF-βR1.29 Galunisertib (LY2157299) binds antagonistically to TGFβ-R1, preventing the intracellular phosphorylation of SMAD2 and SMAD3. The addition of this inhibitor to the culture was able to block CD16 downregulation (Fig. 4d–f). Having observed that LY2157299 significantly inhibits effects of TGF-β1 on NK cells, we examined whether it also prevents inhibition of their cytotoxicity by TGF-β1. We evidence that the commonly used bladder cancer cell line UM-UC-3 highly expressed Disialoganglioside (GD2), a clinically relevant target for the ADCC antibody dinutuximab (αGD2) (Supplementary Figure S8A). Importantly, NK cells treated with αGD2showed an ADCC effect against UM-UC-3 cells, compared to untreated NK cells. TGF-β treatment significantly decreased the cytotoxic capability of NK cells, however treatment with αGD2 was able to restore both the cytotoxic and ADCC ability of the TGF-β treated cells (Fig. 4g). These findings demonstrate that LY2157299 inhibits the suppressive effects of TGF-β signalling on NK cell-mediated direct cytotoxicity and ADCC against bladder cancer cells.
Fig. 4.
TGF-β causes downregulation of CD16 on NK cells and limits ADCC capability in vitro. NK cells from human blood were isolated and cultured for 3, 7 or 14 days in the presence of 5 ng/ml IL-15 and 500 IU/ml IL-2 plus the indicated cytokines. (a) CD16 expression on NK cells (CD45+lin−CD56+NKp46+) across indicated timepoints. (b) representative flow cytometry plots and (c) histogram plots at day 7. Data are from a 5 biological replicates (n = 5). Error bars indicate mean ± SEM. Groups were compared using Mann–Whitney test, where P > 0.05 was deemed not significant, where ∗P < 0.05. (D) CD16 expression on NK cells (CD45+CD3−NKp46+) treated with TGF-βR inhibitor (LY2157299) cultured for 7 days. (e) representative flow cytometry plots and (f) histogram plots at Day 7. Data are from 3 biological replicates, where symbols denote different patients, with two technical replicates each (n = 3). Error bars indicate mean ± SEM. Groups were compared using Mann–Whitney test, where P > 0.05 was deemed not significant, where ∗P < 0.05, and ∗∗P < 0.01. (g) cytotoxicity of NK cells against bladder cell line UM-UC-3 in the presence of indicated conditions. Data are from 2 biological replicates, where symbols denote different patients, with 3 technical replicates each (n = 2). Error bars indicate mean ± SEM. Groups were compared using Mann–Whitney t test, where P > 0.05 was deemed not significant, where ∗P < 0.05, and ∗∗P < 0.01.
TGF-β1 inhibition can enhance in vivo NK cell ADCC
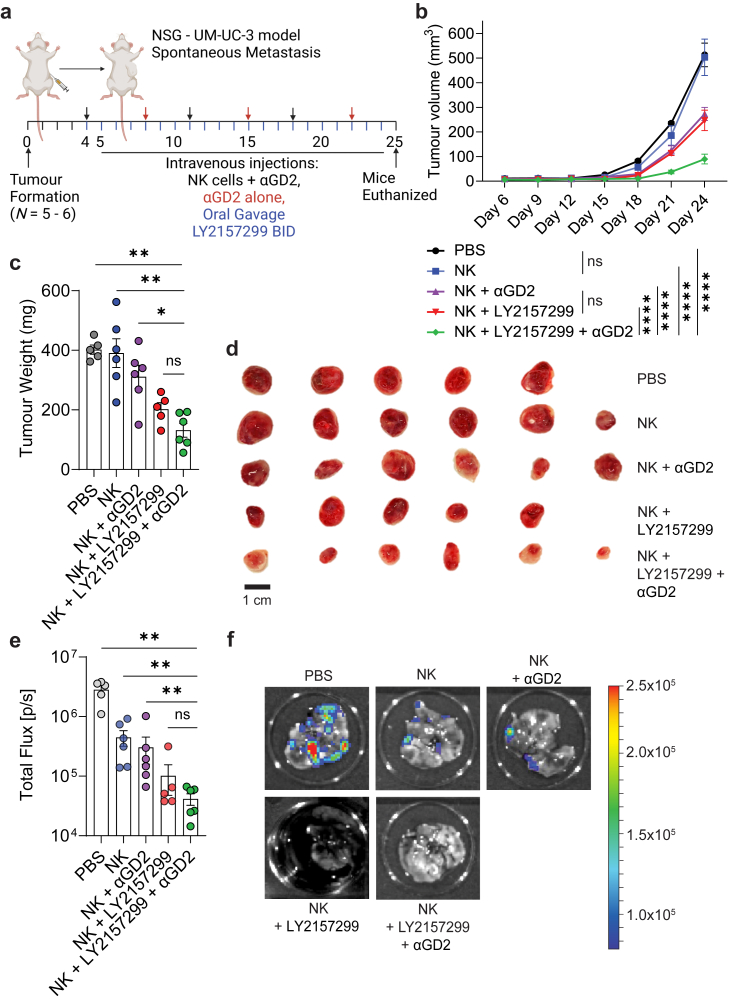
The ability of galunisertib to inhibit TGF-β mediated suppressive effects on NK cells in vivo was tested using a xenogeneic mouse model of bladder cancer, formed via subcutaneous injection of the bladder cancer cell line UM-UC-3. Mice bearing UM-UC-3 tumours were subjected to LY2157299 + αGD2 treatment, single-drug controls, or vehicle (Fig. 5a). Both single-drug controls slowed down tumour growth, where the combination of LY2157299 and αGD2 nearly arrested tumour growth (Fig. 5b). The superior combinatorial efficacy of LY2157299 and αGD2 was also evident in the final tumour weights (Fig. 5c) and tumour sizes (Fig. 5d). In concert, single-drug controls also reduced metastatic burden to the endpoint from circulating tumour cells that migrated from the primary tumours lungs, where the combination of LY2157299 and αGD2 proved superior in metastatic reduction than most groups (Fig. 5e–f). These findings demonstrate that suppression of NK cell-mediated ADCC by TGF-β signalling in vivo, can be restored by inhibiting this suppressive pathway.
Fig. 5.
Concurrent in vivo anti-GD2 mAb treatment with TGF-β inhibition arrests tumour growth and abrogates metastasis. (a) hIL7/IL15 KI NSG mice bearing subcutaneous UM-UC-3 tumours were humanized with 5 × 106 PBMC derived NK cells and reinjected once per week. Mice were injected with dinutuximab (αGD2, 500 μg) twice per week and gavaged with a TGF-β inhibitor galunisertib BID (LY2157299, 75 mg/kg). (b) The combination of αGD2 and LY2157299 was effective in reducing tumour growth, with the growth of combination-treated tumours being essentially arrested, groups (n = 5–6) were compared using two-way ANOVA, where ∗∗∗∗P < 0.0001. NS, not significant. (c and d) This was reflected in final tumour weights and sizes. (e) Lung metastatic lesions were quantified using bioluminescent imaging and the total flux was measured. (f) Representative bioluminescent images of the lungs are shown for each group. Error bars indicate mean ± SEM. (c) and (e) Groups were compared using Mann–Whitney t test, where P > 0.05 was deemed not significant, where ∗P < 0.05, and ∗∗P < 0.01. All data in this figure were generated with UM-UC-3 cells in NSG mice. (a) was created with BioRender.
Discussion
Natural killer cell activation is tightly governed by activating and inhibitory signals, and the TME is known to be heralded with suppressive factors such as TGF-β which can drive NK cell inhibition.1,7,37,38 These suppressive factors and their effects on NK cells in the context of human bladder cancer have not been thoroughly explored. Particularly, in the context of NK cell responses to monoclonal antibody therapies, and whether TME-mediated immunosuppression also inhibits ADCC. In this study, we showed that intratumoral NK cells in patients with bladder cancer upregulate genes and surface proteins (ITGA1/CD49a) resembling an ILC1-like phenotype, simultaneous to downregulation of critical activating receptors such as FCGR3A/CD16. We suggest a correlative link between TGF-β signalling and this NK cell phenotype switch, which has previously been observed in other cancer types,35 but not in bladder cancers. We also observed that loss of CD16 expression after treatment with TGF-β, could be restored as a result of TGF-βR1 specific inhibition, and this could also restore the ADCC capacity of NK cells in vitro. Finally, we observed that CD16 reduction of tumour infiltrating NK cells could be experimentally replicated in vivo and TGF-β inhibition combined with anti-GD2 treatment could dramatically reduce tumour growth and metastatic burden.
An NK cell phenotypic switch to a tissue resident ILC1-like phenotype has been extensively described in many other human tumours.7,35 In our study, we validate the presence of an ILC1-like phenotype in human bladder tumours. It has been suggested that the immunosuppressive cytokine TGF-β can drive an NK cell switch to a less cytotoxic, ILC1-like phenotype, expressing markers such as CD49a.6,8 We used correlative studies of an ILC1-like NK cell score and conventional NK cells showing an upregulation in TGF-β signalling pathway genes in tumour NK cells, and correlation of this score with ITGA1 upregulation. Similarly, FCGR3A expression correlated negatively with TGF-β signalling genes SMAD2/3. The diminished cytotoxic profile of ILC1-like human NK cells has been validated in in vitro systems,8 however the exact ability of these ILC1-like human NK cells to respond to antibody therapies and perform ADCC is still unclear. We replicated the transition of NK cells into ILC1-like NK cells using TGF-β in an in vitro system. Interestingly, we also observed a downregulation of CD16 in this transition, which we could reverse using a TGF-βR inhibitor, allowing enhanced ADCC responses. In other cancers such as neuroblastoma, it has also been reported that TGF-β inhibition can enhance NK cell ADCC responses,17 however the specific mechanisms underpinning this are unknown.
A previous study by Trotta et al. highlighted the suppressive effects of TGF-β on NK cell ADCC capacity,38 however did not observe a downregulation of CD16 expression, over a short TGF-β stimulation. They evidence a SMAD3 dependent, TGF-β mediated inhibition of IFN-γ production in an ADCC response. In our study we investigated the responses to TGF-β, and conversely to Trotta et al. we observed both in human patient samples and in a humanised mouse model of bladder cancer a downregulation of CD16, which is more prominent in CD49a+ NK cells, and could be reversed using a TGFBR1 specific small molecule inhibitor.
Other studies have elegantly shown the impacts of SMAD3 dependant, TGF-β signalling on NK cells, whereby deletion of SMAD3, led to improved tumour control in mice.39 Another study used genetic engineering to generate a stable SMAD3-silencing human NK cell line and reported enhanced tumoricidal capabilities of NK cells under TGF-β conditions.40 Our study was limited in a clear mechanistic identification of the signalling pathway involved in TGF-β mediated CD16 downregulation on NK cells. Due to the complexity of SMAD2/3/4 TGF-β signalling, as described,41 galunisertib alone cannot reveal the specific signalling pathway involved. Nonetheless we have uncovered that TGF-β can downregulate CD16 and opened the field for further exploration into the specific signalling involved.
Using a xenogeneic mouse model of human bladder cancer, we also validated downregulation of CD16 on tumour infiltrating NK cells. Using galunisertib and the clinical anti-GD2 antibody dinutuximab, we showed that dual treatment could demonstrate dramatic reduction in tumour volume and metastatic burden by unleashing the ADCC capacity of NK cells in vivo. Our work highlights the importance of clinical trials such as NCT02423343,42 which has already shown promising patient outcomes for therapeutic antibodies and galunisertib dual therapy in solid tumours including lung cancer, and serve as rational for future considerations of this approach for malignant bladder cancers.
In conclusion, we unveil distinct NK cell states within human bladder cancer through scRNA-seq, revealing blood-derived and tissue-resident NK cell clusters. CD49a elevation in tumour NK cells correlates with reduced CD16 expression, suggestive of a phenotypic shift. ssGSEA highlights heightened TGF-β signalling in tumour NK cells, aligning with reduced FCGR3A expression and elevated ITGA1. Collectively, our findings underline the intricate interplay of TGF-β signalling, CD16 modulation, and NK cell dysfunction in bladder cancer, offering insights for potential therapeutic interventions to enhance immune responses against solid tumours.
Code and data availability
scRNA-seq data are available through the Dryad. The dryad data sharing link is as follows https://doi.org/10.5061/dryad.vt4b8gtz9.
Contributors
J.W., G.R.R, and F.S.F.G designed research and wrote the paper. J.W., G.R.R., T.R.M., L.A., A.B.M., and F.S.F.G., performed research and J.W., analysed data. A.N., E.C., F.S. H.R., and N.O. provided critical patient samples or reagents, clinical or research advice. A.M and Z.K.T provided critical support in bioinformatics analysis. F.S.F.G. supervised work. J.K.M.W, G.R.R. and F.S.F.G have directly accessed and verified the underlying data. All authors have read and approved the final version of the manuscript.
Data sharing statement
All relevant data can be found within the figures and supplementary materials of this publication. Should there be a need for any further information to facilitate reanalysis of the data presented in this paper, it can be obtained from the lead author upon request.
Declaration of interests
A/Prof. F. Simpson declares support from NHMRC Investigator grant (#2026628) and holds patents unrelated to this manuscript. She serves on advisory boards (Elsevier, UQ Academic Board, Victorian Cancer Council and NSW Cancer Council); and none of these roles' present conflicts with this study. A/Prof. Guimaraes' laboratory is funded by multiple grants, including the US Department of Defense and Medical Research Future Fund. He consults for Microba Life Sciences and Prescient Therapeutics, with all payments made to his institution and unrelated to this study. Both authors confirm that these disclosures do not influence the findings of this manuscript. The remaining authors declared that there is no conflict of interest related to this work.
Acknowledgements
We thank all members of the Guimaraes laboratory, A/Prof. John Hooper and Prof. Kiarash Khosrotehrani for their critical comments and advice. We also acknowledge Prof. Fumihiko Ishikawa for providing the NSG hIL-7/IL-15 KI mice, and the Decode Science team and CARF genomics team for their support in sequencing experiments and analysis. We also acknowledge Dr. Janelle Munns, Dr. John Preston and Dr. Malcolm Lawson for assistance in bladder sample collection and Dr. Kyla Driscoll for helpful technical advice on preparing the vehicle formulation for galunisertib. We also thank the TRI flow cytometry core and BRF facilities for their support. This research was carried out at the Translational Research Institute, Woolloongabba, QLD 4102, Australia. The Translational Research Institute is supported by grants from the Australian and Queensland Governments.
The Guimaraes Laboratory is funded by a US Department of Defense—Breast Cancer Research Program—Breakthrough Award Level 1 (#BC200025), a grant (#2019485) awarded through the Medical Research Future Fund (MRFF, with the support of the Queensland Children's Hospital Foundation, Microba Life Sciences, Richie's Rainbow Foundation, Translational Research Institute (TRI) and UQ), and a grant (#RSS_2023_085) funded by a Metro South Health Research Support Scheme. J.K.M.W. is funded by a UQ Research Training Program PhD Scholarship and N.O. is funded by a NHMRC Postgraduate Scholarship (#2021932).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105176.
Appendix A. Supplementary data
References
- 1.Souza-Fonseca-Guimaraes F., Cursons J., Huntington N.D. The emergence of natural killer cells as a major target in cancer immunotherapy. Trends Immunol. 2019;40(2):142–158. doi: 10.1016/j.it.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Wong J.K.M., Dolcetti R., Rhee H., Simpson F., Souza-Fonseca-Guimaraes F. Weaponizing natural killer cells for solid cancer immunotherapy. Trends in Cancer. 2023;9(2):111–121. doi: 10.1016/j.trecan.2022.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Vivier E., Rebuffet L., Narni-Mancinelli E., Cornen S., Igarashi R.Y., Fantin V.R. Natural killer cell therapies. Nature. 2024;626(8000):727–736. doi: 10.1038/s41586-023-06945-1. [DOI] [PubMed] [Google Scholar]
- 4.Cózar B., Greppi M., Carpentier S., Narni-Mancinelli E., Chiossone L., Vivier E. Tumor-infiltrating natural killer cells. Cancer Discov. 2021;11(1):34–44. doi: 10.1158/2159-8290.CD-20-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckl J., Buchner A., Prinz P.U., et al. Transcript signature predicts tissue NK cell content and defines renal cell carcinoma subgroups independent of TNM staging. J Mol Med. 2012;90(1):55–66. doi: 10.1007/s00109-011-0806-7. [DOI] [PubMed] [Google Scholar]
- 6.Gao Y., Souza-Fonseca-Guimaraes F., Bald T., et al. Tumor immunoevasion by the conversion of effector NK cells into type 1 innate lymphoid cells. Nat Immunol. 2017;18(9):1004–1015. doi: 10.1038/ni.3800. [DOI] [PubMed] [Google Scholar]
- 7.Hawke L.G., Mitchell B.Z., Ormiston M.L. TGF-β and IL-15 synergize through MAPK pathways to drive the conversion of human NK cells to an innate lymphoid cell 1-like phenotype. J Immunol. 2020;204(12):3171–3181. doi: 10.4049/jimmunol.1900866. [DOI] [PubMed] [Google Scholar]
- 8.Rautela J., Dagley L.F., de Oliveira C.C., et al. Therapeutic blockade of activin-A improves NK cell function and antitumor immunity. Sci Signal. 2019;12(596) doi: 10.1126/scisignal.aat7527. [DOI] [PubMed] [Google Scholar]
- 9.Souza-Fonseca-Guimaraes F., Rossi G.R., Dagley L.F., et al. TGFβ and CIS inhibition overcomes NK-cell suppression to restore antitumor immunity. Cancer Immunol Res. 2022;10(9):1047–1054. doi: 10.1158/2326-6066.CIR-21-1052. [DOI] [PubMed] [Google Scholar]
- 10.Wolf N.K., Kissiov D.U., Raulet D.H. Roles of natural killer cells in immunity to cancer, and applications to immunotherapy. Nat Rev Immunol. 2022;23(2):90–105. doi: 10.1038/s41577-022-00732-1. [DOI] [PubMed] [Google Scholar]
- 11.Sung H., Ferlay J., Siegel R.L., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 12.Tran L., Xiao J.-F., Agarwal N., Duex J.E., Theodorescu D. Advances in bladder cancer biology and therapy. Nat Rev Cancer. 2021;21(2):104–121. doi: 10.1038/s41568-020-00313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ranti D., Bieber C., Wang Y.-S., Sfakianos J.P., Horowitz A. Natural killer cells: unlocking new treatments for bladder cancer. Trends Cancer. 2022;8(8):698–710. doi: 10.1016/j.trecan.2022.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Mollica V., Rizzo A., Montironi R., et al. Current strategies and novel therapeutic approaches for metastatic urothelial carcinoma. Cancers. 2020;12(6):1449. doi: 10.3390/cancers12061449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Powles T., Rosenberg J.E., Sonpavde G.P., et al. Enfortumab vedotin in previously treated advanced urothelial carcinoma. N Engl J Med. 2021;384(12):1125–1135. doi: 10.1056/NEJMoa2035807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witjes J.A., Bruins H.M., Cathomas R., et al. European association of urology guidelines on muscle-invasive and metastatic bladder cancer: summary of the 2020 guidelines. Eur Urol. 2021;79(1):82–104. doi: 10.1016/j.eururo.2020.03.055. [DOI] [PubMed] [Google Scholar]
- 17.Tran H.C., Wan Z., Sheard M.A., et al. TGFβR1 blockade with galunisertib (LY2157299) enhances anti-neuroblastoma activity of the anti-GD2 antibody dinutuximab (ch14.18) with natural killer cells. Clin Cancer Res. 2017;23(3):804–813. doi: 10.1158/1078-0432.CCR-16-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Besse L., Rumiac T., Reynaud-Angelin A., et al. Protocol for automated multivariate quantitative-image-based cytometry analysis by fluorescence microscopy of asynchronous adherent cells. STAR Protoc. 2023;4(3) doi: 10.1016/j.xpro.2023.102446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stirling D.R., Swain-Bowden M.J., Lucas A.M., Carpenter A.E., Cimini B.A., Goodman A. CellProfiler 4: improvements in speed, utility and usability. BMC Bioinformatics. 2021;22(1):433. doi: 10.1186/s12859-021-04344-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young L., Sung J., Stacey G., Masters J.R. Detection of Mycoplasma in cell cultures. Nat Protoc. 2010;5(5):929–934. doi: 10.1038/nprot.2010.43. [DOI] [PubMed] [Google Scholar]
- 22.Neri S., Mariani E., Meneghetti A., Cattini L., Facchini A. Calcein-acetyoxymethyl cytotoxicity assay: standardization of a method allowing additional analyses on recovered effector cells and supernatants. Clin Diagn Lab Immunol. 2001;8(6):1131–1135. doi: 10.1128/CDLI.8.6.1131-1135.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao Y., Hao S., Andersen-Nissen E., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z., Zhou L., Liu L., et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat Commun. 2020;11(1):5077. doi: 10.1038/s41467-020-18916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cursons J., Souza-Fonseca-Guimaraes F., Foroutan M., et al. A gene signature predicting natural killer cell infiltration and improved survival in melanoma patients. Cancer Immunol Res. 2019;7(7):1162–1174. doi: 10.1158/2326-6066.CIR-18-0500. [DOI] [PubMed] [Google Scholar]
- 27.Matsuda M., Ono R., Iyoda T., et al. Human NK cell development in hIL-7 and hIL-15 knockin NOD/SCID/IL2rgKO mice. Life Sci Alliance. 2019;2(2) doi: 10.26508/lsa.201800195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramakrishnan S., Huss W., Foster B., et al. Transcriptional changes associated with in vivo growth of muscle-invasive bladder cancer cell lines in nude mice. Am J Clin Exp Urol. 2018;6(3):138–148. [PMC free article] [PubMed] [Google Scholar]
- 29.Yingling J.M., McMillen W.T., Yan L., et al. Preclinical assessment of galunisertib (LY2157299 monohydrate), a first-in-class transforming growth factor-β receptor type I inhibitor. Oncotarget. 2018;9(6):6659–6677. doi: 10.18632/oncotarget.23795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun H., Liu L., Huang Q., et al. Accumulation of tumor-infiltrating CD49a+ NK cells correlates with poor prognosis for human hepatocellular carcinoma. Cancer Immunol Res. 2019;7(9):1535–1546. doi: 10.1158/2326-6066.CIR-18-0757. [DOI] [PubMed] [Google Scholar]
- 31.Borcherding N., Vishwakarma A., Voigt A.P., et al. Mapping the immune environment in clear cell renal carcinoma by single-cell genomics. Commun Biol. 2021;4(1):122. doi: 10.1038/s42003-020-01625-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zaiatz-Bittencourt V., Finlay D.K., Gardiner C.M. Canonical TGF-β signaling pathway represses human NK cell metabolism. J Immunol. 2018;200(12):3934–3941. doi: 10.4049/jimmunol.1701461. [DOI] [PubMed] [Google Scholar]
- 33.Marquardt N., Kekäläinen E., Chen P., et al. Unique transcriptional and protein-expression signature in human lung tissue-resident NK cells. Nat Commun. 2019;10(1):3841. doi: 10.1038/s41467-019-11632-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herman N., Aline P., Jodie P.G., et al. Pan-cancer profiling of tumor-infiltrating natural killer cells through transcriptional reference mapping. bioRxiv. 2023 doi: 10.1101/2023.10.26.564050. [DOI] [Google Scholar]
- 35.Tang F., Li J., Qi L., et al. A pan-cancer single-cell panorama of human natural killer cells. Cell. 2023;186(19):4235–4251.e20. doi: 10.1016/j.cell.2023.07.034. [DOI] [PubMed] [Google Scholar]
- 36.Wagner J., Pfannenstiel V., Waldmann A., et al. A two-phase expansion protocol combining interleukin (IL)-15 and IL-21 improves natural killer cell proliferation and cytotoxicity against rhabdomyosarcoma. Front Immunol. 2017;8:676. doi: 10.3389/fimmu.2017.00676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viel S., Marçais A., Guimaraes F.S., et al. TGF-β inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci Signal. 2016;9(415) doi: 10.1126/scisignal.aad1884. [DOI] [PubMed] [Google Scholar]
- 38.Trotta R., Col J.D., Yu J., et al. TGF-β utilizes SMAD3 to inhibit CD16-mediated IFN-γ production and antibody-dependent cellular cytotoxicity in human NK Cells1. J Immunol. 2008;181(6):3784–3792. doi: 10.4049/jimmunol.181.6.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang P.M., Zhou S., Meng X.M., et al. Smad3 promotes cancer progression by inhibiting E4BP4-mediated NK cell development. Nat Commun. 2017;8 doi: 10.1038/ncomms14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Q.M., Tang P.M., Lian G.Y., et al. Enhanced cancer immunotherapy with smad3-silenced NK-92 cells. Cancer Immunol Res. 2018;6(8):965–977. doi: 10.1158/2326-6066.CIR-17-0491. [DOI] [PubMed] [Google Scholar]
- 41.Lucarelli P., Schilling M., Kreutz C., et al. Resolving the combinatorial complexity of smad protein complex formation and its link to gene expression. Cell Syst. 2018;6(1):75–89.e11. doi: 10.1016/j.cels.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 42.Nadal E., Saleh M., Aix S.P., et al. A phase Ib/II study of galunisertib in combination with nivolumab in solid tumors and non-small cell lung cancer. BMC Cancer. 2023;23(1):708. doi: 10.1186/s12885-023-11153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq data are available through the Dryad. The dryad data sharing link is as follows https://doi.org/10.5061/dryad.vt4b8gtz9.