Abstract
Wolbachia pipientis is a maternally transmitted symbiotic bacterium that mainly colonizes arthropods, potentially affecting different aspects of the host's physiology, e.g., reproduction, immunity, and metabolism. It has been shown that Wolbachia modulates glycogen metabolism in mosquito Aedes fluviatilis (Ae. fluviatilis). Glycogen synthesis is controlled by the enzyme GSK3, which is also involved in immune responses in both vertebrate and invertebrate organisms. Here we investigated the mechanisms behind immune changes mediated by glycogen synthase kinase β (GSK3β) in the symbiosis between Ae. fluviatilis and W. pipientis using a GSK3β inhibitor or RNAi-mediated gene silencing. GSK3β inhibition or knockdown increased glycogen content and Wolbachia population, together with a reduction in Relish2 and gambicin transcripts. Furthermore, knockdown of Relish2 or Caspar revealed that the immunodeficiency pathway acts to control Wolbachia numbers in the host. In conclusion, we describe for the first time the involvement of GSK3β in Ae. fluviatilis immune response, acting to control the Wolbachia endosymbiotic population.
Keywords: Wolbachia, symbiosis, immunometabolism, GSK3β, Relish2
In the last 2 decades, a vast array of data revealed that most metazoans are essentially a complex community of microorganisms associated with a metazoan host body. Animals are no longer considered completely biologically autonomous and isolated individuals, given the presence of symbionts that act to complement metabolic pathways and multiple physiological functions (1, 2). Symbionts, specifically bacteria, can play important roles in the metabolism and reproduction of arthropods (3, 4, 5). It is now largely recognized that symbionts exert important influence on the host immune response while maintaining a persistent infection (6, 7).
In insects, the innate immune response is driven by the three most intensively studied signaling pathways, namely Toll, immunodeficiency (Imd), and Jak/Stat pathways (8, 9). In mosquitoes, the Toll and Imd pathways activate two distinct nuclear kappa B-like transcription factors, Relish 1 (REL1) and Relish 2 (REL2), respectively, while the Jak/Stat pathway activates Stat (10, 11, 12, 13). The activation and nuclear translocation of these transcription factors positively regulate the expression of antimicrobial peptides (AMPs) (8, 14, 15). In insect–bacterial symbiosis, AMPs are specific factors produced by the host's immune system to modulate the symbiont population, thus keeping control over the microbiota (16, 17, 18).
Endosymbionts can influence metabolic and immune pathways while coevolving with its host insect. The endosymbiont Wolbachia pipientis is a widespread endosymbiotic alphaproteobacterium that has been associated with several signaling pathways and can impact the metabolism, immunity, and fitness improvement in different hosts, including insects, isopods, spiders, as well as filarial nematodes (19, 20, 21). This diversity of effects triggered by Wolbachia underlies the interest it attracts as a subject for symbiont/host interaction studies.
Previous research by our group has shown that natural infection of the mosquito Aedes fluviatilis by the Wolbachia wFlu strain is implicated in changing the immune response and glycogen metabolism of the host (22, 23, 24). Embryonic cells from Ae. fluviatilis infected with Wolbachia (wAflu1) respond differently to an immunological challenge with heat-killed bacteria when compared with cells without Wolbachia (23). Furthermore, developing embryos and wAflu1 cells have higher glycogen content when compared to Wolbachia-free controls (22, 23); this finding led us to focus on glycogen synthase kinase β (GSK3β), a canonical regulator of glycogen synthesis, acting on glycogen synthase (GS) (25). GSK3β knockdown increased glycogen and positively modulated Wolbachia in Ae. fluviatilis embryos (22). In addition to controlling glycogen synthesis, GSK3β has been associated with immune functions in vertebrates and invertebrates (26, 27, 28, 29). In the present work, we describe an immune-metabolic crosstalk between the GSK3β and immunological signaling, impacting glycogen content, as well as the Imd pathway-mediated immune response controlling Wolbachia infection in its natural host.
Results
GSK3β influences glycogen content and Wolbachia population in wAflu1 cells
To explore the potential impact of Wolbachia infection on GSK3β transcription and glycogen synthesis in Ae. fluviatilis mosquitoes, we conducted a comparison using embryonic cell lines from naturally infected individuals (wAflu1) or devoid of Wolbachia by tetracycline treatment (wAflu1.tet) (Fig. S1A). To establish Wolbachia-free cell culture (wAflu1.tet), wAflu1 cells were treated with 10 μg/ml of tetracycline during three passages, with three days intervals in the absence of the antibiotic to allow for cell recovery. After Wolbachia removal, the cells were maintained antibiotic-free for at least two passages before performing experiments. The presence of Wolbachia was confirmed by PCR amplification of a 650-bp fragment using primers specific to the gene coding for Wolbachia surface protein (WSP) (Fig. S1B). Additionally, RT-qPCR analysis was carried out, revealing the absence of WSP transcripts in tetracycline-treated cells (Fig. S1C). Wolbachia presence was monitored throughout all the experiments.
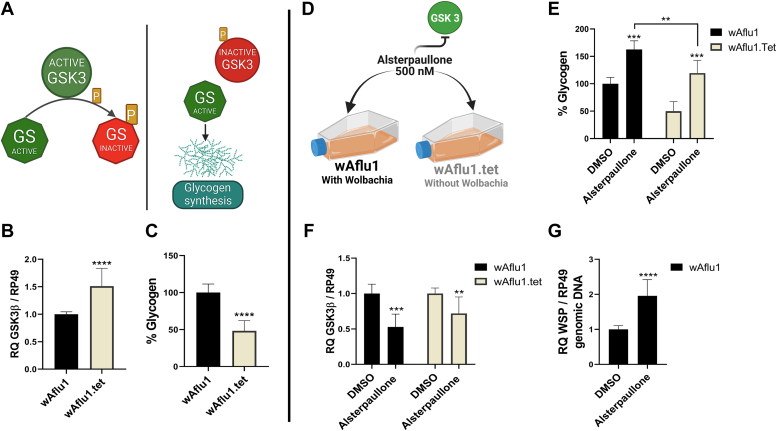
GSK3β downregulates glycogen synthesis by phosphorylating and inactivating GS (Fig. 1A). We investigated whether Wolbachia interfered with transcription of GSK3β gene as detected by RT-qPCR. The relative quantification of GSK3β transcripts revealed approximately 0.5-fold decrease in GSK3β transcripts (Fig. 1B), together with an increase in glycogen content (Fig. 1C) in the Wolbachia-positive wAflu1 cells compared with Wolbachia-free cells.
Figure 1.
GSK3β activity affects glycogen content and increases Wolbachia population in Ae. fluviatilis embryonic cell lines.A, schematic representation (created with BioRender.com) summarizing the role of GSK3β in glycogen synthesis. GSK3β negatively controls glycogen synthesis by phosphorylating glycogen synthase (GS); when GSK3β is phosphorylated, it becomes inactive, favoring glycogen synthesis. B, GSK3β transcript fragment was amplified by RT-PCR using cDNA from cell lines with or without Wolbachia (wAflu1 and wAflu1.tet, respectively). mRNA levels are expressed as mean ± SD (n = 3). Quantitative PCR was analyzed by t test; asterisks indicate significant differences (∗∗∗∗p < 0.0001). C, glycogen quantification was performed on five independent biological samples with three experimental replicates and analyzed by t test (∗∗∗∗p < 0.0001). D, schematic representation of experimental design, and both cell lines were treated with 500 nM of alsterpaullone, a chemical inhibitor of GSK3β, during 24 h. E, glycogen content was measured in three independent biological samples with three experimental replicates each. Statistical analysis was performed by two-way ANOVA followed by the Tukey multiple comparison test. Asterisks indicate significant differences (∗∗∗p < 0.001; ∗∗p < 0.05). F, GSK3β transcript fragment was amplified by RT-PCR using cDNA from each cell line. mRNA levels are expressed as mean ± SD (n = 3) (∗∗∗p < 0.001; ∗∗p < 0.05), and G, WSP gene was quantified by qPCR using DNA from wAflu1 cell line. Relative quantification (WSP/RP49) is expressed as mean ± SD (n = 3). Results were analyzed by t test; asterisks indicate significant differences (∗∗∗∗p < 0.0001). GSK3β, glycogen synthase kinase β; WSP, Wolbachia surface protein.
To further explore GSK3β role in glycogen metabolism and Wolbachia population, we used the chemical inhibitor alsterpaullone (Fig. 1D). The exposure time and inhibitor concentration for this experiment were defined in a preliminary viability assay (3-(4,5-dimethylthiazol-2-yl) −2,5-diphenyltetrazolium bromide [MTT]) and glycogen quantification (Fig. S2, A and B). We chose to proceed with the treatment with 500 nM alsterpaullone for 24 h because these conditions resulted in an increase of about 100% in glycogen content with no significant decrease in cell viability (Fig. S2, A and B). GSK3β inhibition increased glycogen content by 50% in wAflu1 cells and 70% in wAflu1.tet cells (Fig. 1E). The wAflu1 cells had more glycogen than wAflu1.tet cells, but glycogen levels increased upon alsterpaullone treatment in both cell lines. (Fig. 1E). In addition to inhibiting GSK3 enzymatic activity, alsterpaullone treatment reduced GSK3β transcript levels in both cell lines (Fig. 1F). The GSK3 inhibitor also increased Wolbachia population in wAflu1 cells, doubling WSP transcript levels (Fig. 1G).
The presence of Wolbachia induces changes in the transcription of immune genes in Ae. fluviatilis
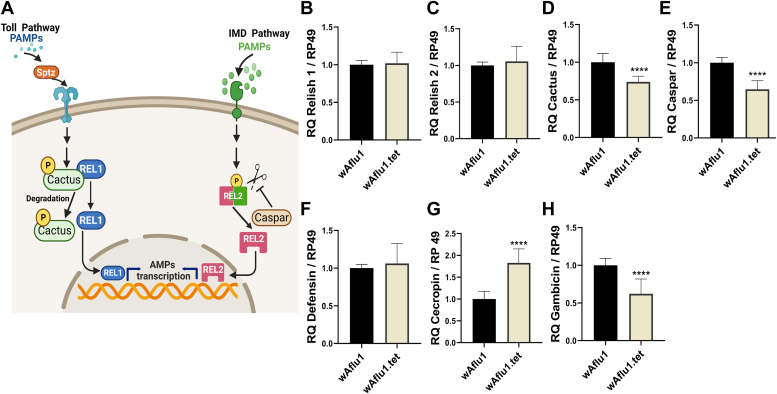
We analyzed two transcription factors (Rel1 and Rel2) activated by Toll and Imd pathways, respectively. In addition, we analyzed their respective negative regulators Cactus and Caspar and downstream effectors genes, such as AMPs (Fig. 2A), namely defensin, cecropin, and gambicin. We observed that in the absence of Wolbachia, there were no significant changes in the relative quantification of REL1 (Fig. 2B) and REL2 (Fig. 2C) transcripts, whereas a reduction in the transcription of Cactus (0.8-fold, Fig. 2D) and Caspar (0.6-fold, Fig. 2E) was observed in tetracycline-treated cells. Interestingly, levels of defensin transcripts were not influenced by Wolbachia presence (Fig. 2F), while cecropin transcripts were upregulated (2-fold, Fig. 2G), and gambicin transcript levels were decreased (0.5-fold, Fig. 2H) in the absence of Wolbachia, suggesting a complex interaction between Wolbachia and the transcription of host immune genes.
Figure 2.
Transcription of immune genes is modulated by Wolbachia in Ae. fluviatilis embryonic cell line.A, schematic representation (created with BioRender.com) summarizing the Toll and Imd immune pathways. When the Toll pathway is activated, it triggers signaling through adapter proteins that result in the phosphorylation and degradation of Cactus, a negative regulator that binds to the transcription factor Relish1 (REL1) in the cytoplasm. Cactus degradation allows REL1 translocation to the nucleus. When the Imd pathway is activated, the transcription factor Relish2 (REL2) is phosphorylated and cleaved by DREDD. DREDD activity is negatively regulated by Caspar. Activated REL2 translocates to the nucleus. Once inside the nucleus, both transcription factors can activate antimicrobial peptides (AMPs) transcription. B–H, REL1 (B), REL2 (C), Cactus (D), Caspar (E), defensin (F), cecropin (G), and gambicin (H) transcript fragments were amplified by RT-qPCR using cDNA from cells with or without Wolbachia (wAflu1 and wAflu1.tet, respectively). mRNA levels are expressed as mean ± SD (n = 3). RT-qPCR results were analyzed by t test; asterisks indicate significant differences (∗∗∗∗p < 0.0001). Imd, immunodeficiency; PAMPs, pathogen-associated molecular patterns.
Chemical inhibition or knockdown of GSK3β induces changes in the transcription of immune genes in Ae. fluviatilis
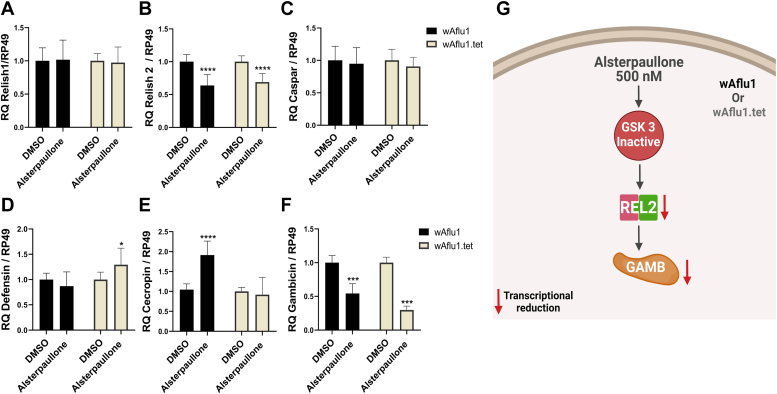
Considering that Wolbachia presence decreased GSK3β expression and GSK3β inhibition increased Wolbachia population, we hypothesized that GSK3β inhibition could attenuate the immune response to favor the presence of Wolbachia. GSK3β inhibition by alsterpaullone treatment did not change REL1 or Caspar transcript levels (Fig. 3, A and C). In contrast, relative levels of REL2 transcripts were significantly decreased by about 0.6-fold in both cell lines treated with alsterpaullone (Fig. 3B). Defensin transcription was subtly increased in alsterpaullone-treated wAflu1.tet cells (Fig. 3D), and a higher cecropin expression (approximately 2-fold) was observed in wAflu1 cells (Fig. 3E). Interestingly, the transcriptional decrease in REL2 was accompanied by a reduction in gambicin transcription by about 0.5-fold in both cell lines (Fig. 3F).
Figure 3.
GSK3β activity reduces levels of REL2 and gambicin transcripts in Ae. fluviatilis embryonic cell line.A–F, REL1 (A), REL2 (B), Caspar (C), defensin (D), cecropin (E), and gambicin (F) transcript fragments were amplified by RT-qPCR using cDNA cells with or without Wolbachia (wAflu1 and wAflu1.tet, respectively). mRNA levels are expressed as mean ± SD (n = 3) and were analyzed by two-way ANOVA followed by Sidak’s multiple comparison test (∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗p < 0.5). G, schematic representation (created with BioRender.com) summarizing the effect on immune gene transcription as a result of GSK3β inhibition. GAMB, gambicin; GSK3β, glycogen synthase kinase β; REL2, Relish 2.
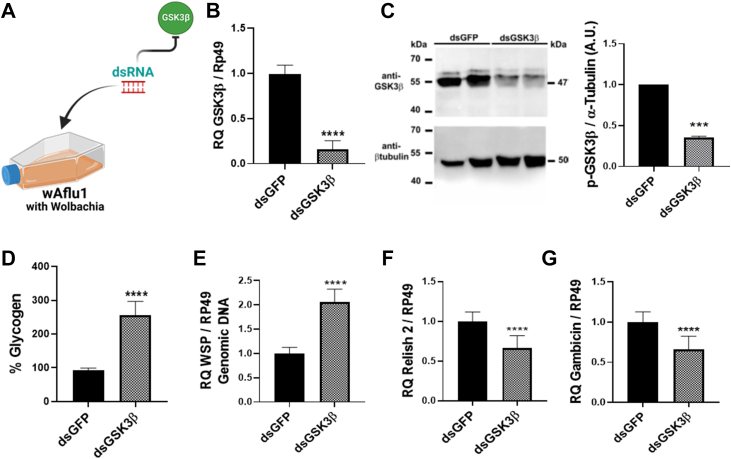
We then used gene silencing by RNAi to further elucidate the role of GSK3β in wAflu1 cells (Fig. 4A). GSK3β knockdown produced an 8-fold decrease in GSK3β transcript levels (Fig. 4B), in line with the reduction of phospho-GSK3β content observed by Western blot (Fig. 4C). This was followed by a 2.5-fold increase in glycogen content (Fig. 4D) and a significantly higher content of Wolbachia (Fig. 4E). Furthermore, we observed a significant reduction of about 0.6-fold in REL2 (Fig. 4F) and gambicin transcription (Fig. 4G). Interestingly, GSK3β gene silencing increased REL1 transcript levels by about 3-fold (Fig. S3A), while not affecting cecropin (Fig. S3B) or defensin transcription (Fig. S3C).
Figure 4.
GSK3β knockdown modulates the transcription of immune genes in Ae. fluviatilis embryonic cell line.A, schematic representation of experimental design: wAflu1 cells were treated with dsGSK3β. B, GSK3β transcript fragment was amplified by qRT-PCR using cDNA from wAflu1 cell lines. mRNA levels are expressed as mean ± SD (n = 3). C, Western blotting and densitometry for anti-phospho-GSK3β and anti-β-tubulin from two independent replicates show the effect of dsGSK3β in phosphor-GSK3β content. Data were normalized by the dsGFP results, and expressed in arbitrary units (A.U.), where each bar shows the mean ± SD. D, glycogen content was analyzed in three independent biological samples with three experimental replicates each, using the t test. Asterisks indicate significant differences (∗∗∗∗p < 0.0001). E, WSP gene was quantified by qPCR using DNA from wAflu1 cell line. Relative quantification (WSP/RP49) is expressed as mean ± SD (n = 3). F, G, REL2 (F) and gambicin (G) transcript fragments were amplified by RT-PCR using cDNA from wAflu1 cell lines. mRNA levels are expressed as mean ± SD (n = 3). Quantitative PCR results were analyzed using the t test; asterisks indicate significant differences (∗∗∗∗p < 0.0001). GSK3β, glycogen synthase kinase β; REL2, Relish 2; WSP, Wolbachia surface protein.
The Imd pathway modulates Wolbachia population and glycogen synthesis in Ae. fluviatilis
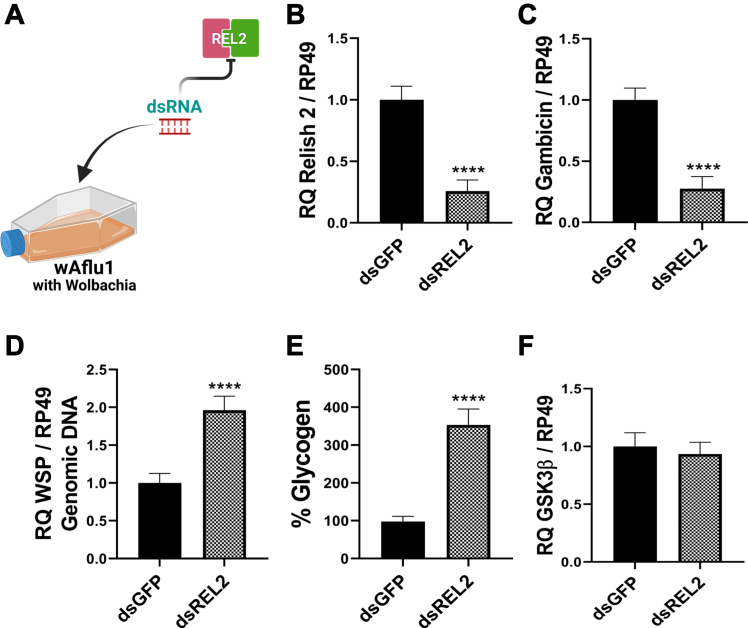
Having determined that GSK3β inhibition or knockdown decreased REL2 and gambicin expression, we next hypothesized that Wolbachia population could be modulated via REL2 and gambicin. To test this hypothesis, we performed REL2 gene knockdown in wAflu1 cells (Fig. 5A). REL2 knockdown showed a decrease of about 8-fold in REL2 transcript abundance (Fig. 5B), resulting in an 8-fold decrease in gambicin transcriptional levels (Fig. 5C) and a 2-fold increase in the Wolbachia marker WSP (Fig. 5D). Furthermore, glycogen content increased 3.5-fold in REL2-silenced cells (Fig. 5E), whereas GSK3β expression remained unaltered (Fig. 5F). In addition, REL2 knockdown induced a 7-fold increase in REL1 transcription together with a 10-fold reduction of cecropin, but no significant change in defensin (Fig. S3, D–F).
Figure 5.
REL2 knockdown induced an increase in Wolbachia population in Ae. fluviatilis embryonic cell line.A, schematic representation of the experimental design; wAflu1 cells were treated with dsREL2. B, C and F, REL2 (B), gambicin (C), and GSK3β (F) transcript fragments were amplified by RT-qPCR using cDNA from wAflu1 cell line. mRNA levels are expressed as mean ± SD (n = 3). D, WSP was quantified by qPCR using DNA from wAflu1 cell line. Relative quantification (WSP/RP49) results are expressed as mean ± SD (n = 3). RT-qPCR data were analyzed using the t test; asterisks indicate significant differences (∗∗∗∗p < 0.0001). E, the glycogen content was analyzed in three independent biological samples with three experimental replicates each. The results were analyzed by t test; asterisks indicate significant differences (∗∗∗∗p < 0.0001). GSK3β, glycogen synthase kinase β; REL2, Relish 2; WSP, Wolbachia surface protein.
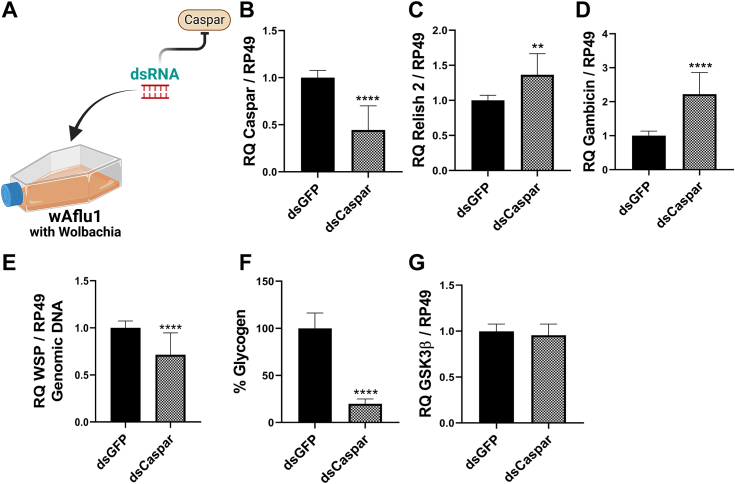
To confirm that Wolbachia population was modulated by the Imd pathway, we activated this pathway by silencing Caspar, its negative regulator (Fig. 6A). Caspar knockdown showed a 0.5-fold decrease in Caspar expression (Fig. 6B). This was accompanied by an increase in REL2 expression (1.5-fold, Fig. 6C) and gambicin (2-fold, Fig. 6D). Imd upregulation resulted in a 0.7-fold decrease in Wolbachia population (Fig. 6E) and a drastic 80% decrease in glycogen content (Fig. 6F). GSK3β expression, in contrast, was not affected (Fig. 6G).
Figure 6.
Caspar knockdown induced Wolbachia population decrease in Ae. fluviatilis embryonic cell line.A, schematic representation of experimental design; wAflu1 cells were treated with dsCaspar. B–D and G, Caspar (B), REL2 (C), gambicin (D), and GSK3β (G) transcript fragments were amplified by RT-qPCR using cDNA from wAflu1 cell line. mRNA levels are expressed as mean ± SD (n = 3). E, WSP was quantified by RT-qPCR using DNA from wAflu1 cell line. Relative quantification (WSP/RP49) results are expressed as mean ± SD (n = 3). Data were analyzed using the t test; asterisks indicate significant differences (∗∗∗∗p < 0.0001; ∗∗p < 0.05). F, the glycogen content was analyzed in three independent biological samples with three experimental replicates each. Results were analyzed by t test; asterisks indicate significant differences (∗∗∗∗p < 0.0001). GSK3β, glycogen synthase kinase β; REL2, Relish 2; WSP, Wolbachia surface protein.
Discussion
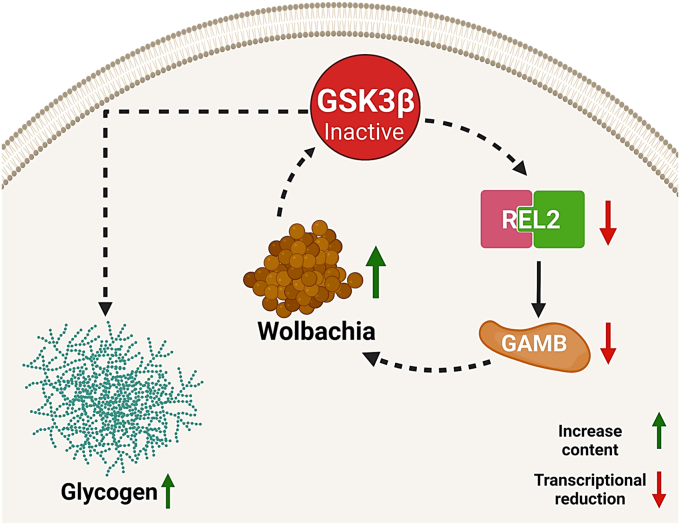
W. pipientis is a widespread alphaproteobacterium endosymbiont that attracts research interest due to its pleiotropic effects on the host physiology, leading to a fitness improvement. Previously, we observed that GSK3β knockdown positively modulated Wolbachia population (22). More recently, we showed an inverse correlation between the increase in Wolbachia population and GSK3β expression during Ae. fluviatilis oogenesis, supporting our hypothesis that GSK3β is involved in controlling this bacterial population (24). Here, we show that GSK3β expression impacts Wolbachia population and that GSK3β acts as a major regulator of an immune metabolic network involving the Imd pathway. Wolbachia downregulates GSK3β transcription, leading to increased glycogen content, and the lowered GSK3β levels result in REL2 and gambicin decrease, thereby allowing Wolbachia proliferation and glycogen synthesis (Fig. 7). This indicates that GSK3β may be an important regulatory hub in the control of the endosymbiotic Wolbachia population.
Figure 7.
The proposed role of GSK3β in modulating Wolbachia population via glycogen metabolism and Imd pathway. Schematic representation (created with BioRender.com) summarizing the relationship between GSK3β and REL2 in the Wolbachia-mediated metabolic immune response. We suggest that Wolbachia can induce metabolic changes leading to GSK3 inhibition. GSK3 inhibition induces glycogen synthesis and impacts REL2 and gambicin transcription, favoring increased Wolbachia population. GSK3β, glycogen synthase kinase β; Imd, immunodeficiency; REL2, Relish 2.
Recently, it was demonstrated that overexpression of GSK3β led to a reduction in the transcription of dorsal (known as REL1 in mosquitoes) and AMPs in arthropod Litopenaeus vannamei infected by the white spot syndrome virus. Conversely, both inhibition and knockdown of GSK3β resulted in an upregulation of dorsal and AMP transcription, indicating its potential immunological role in countering infection (28, 30). Here, we observed that GSK3β inhibition (Fig. 3, B, F and G) or knockdown (Fig. 4, F and G) decreased REL2 and gambicin. This transcriptional response was accompanied by an increase in Wolbachia population (Figs. 1G and 4E), revealing that GSK3β can interfere with the immune response and regulate Wolbachia proliferation. In contrast, GSK3β inhibition by alsterpaullone did not interfere with REL1 and Caspar expression (Fig. 3, A and C) but increased cecropin and defensin transcription in wAflu1 and wAflu1.tet cells, respectively (Fig. 3, E and D). GSK3β knockdown provided distinct results, increasing REL1 transcript levels (Fig. S3A). This may be a compensatory response to the 40% reduction in REL2 transcription (Fig. 4F), considering that REL2 silencing also caused an increase in REL1 (Fig. S4A). However, this response was not observed while using alsterpaullone (Fig. 3A). In addition to GSK3, alsterpaullone also inhibits cyclin-dependent kinases, which suggests that cyclin-dependent kinase inhibition could be preventing REL1 activation in the context of lower GSK3β activity. The GSK3β knockdown did not interfere with cecropin and defensin transcript levels (Fig. S3, B and C). This could be due to the lack of REL1 activation, as the cells were not challenged with appropriate molecular triggers for the Toll pathway. Therefore, we were unable to precisely correlate a transcriptional profile of cecropin and defensin with GSK3β activity and that remains a relevant subject for future research.
GSK3β knockdown or inhibition induced a downregulation on REL2 transcription. REL2 in mosquitoes has been implicated in the responses against infectious agents like Plasmodium falciparum, Plasmodium gallinaceum, and Gram-negative and Gram-positive bacteria (12, 31, 32). In the embryonic cells of Ae. fluviatilis, REL2 was upregulated upon Gram-negative immunological challenge (23). Here we show that REL2 plays a role in controlling Wolbachia population. REL2 knockdown (Fig. 5, A and B) increased Wolbachia population (Fig. 5D), indicating that REL2 can also regulate endosymbiotic bacteria. Additionally, REL2 knockdown induced a decrease in gambicin transcript levels (Fig. 5C) but did not interfere with GSK3β (Fig. 5F), suggesting that gambicin is mainly regulated by REL2 and in accordance with GSK3β being upstream of REL2. Furthermore, the knockdown of REL2 led to an upregulation in REL1 transcription levels (Fig. S4A), suggesting a compensatory response. It also resulted in a reduction in cecropin transcription but had no impact on defensin levels (Fig. S4, B and C). These findings suggest that cecropin, but not defensin, may be subject to regulation by REL2.
The changes observed in AMPs indicate ongoing immune activity. These peptides are usually secreted, and we do not know exactly how they work to control intracellular Wolbachia. The role of AMPs in the immune process is extremely complex. They regulate cell surface receptors such as cytokine receptors, chemokine receptors, and G-protein coupled receptors, including formyl peptide receptors and Toll-like receptors and several intracellular signal pathways such as nuclear factor-κB, extracellular signal-regulated kinase half, p38, JUN N-terminal kinase, mitogen-activated protein kinase, and phosphoinositide 3-kinase (33). While the current literature discusses some aspects of these processes, we recognize the need for further studies to fully understand the intricacies of these interactions.
Previously, several authors have highlighted the significance of the Imd pathway as a pivotal immune signaling pathway in insect antibacterial defense. Moreover, its role in microbiota control has been a subject of discussion and investigation (11, 13, 34). Here, Imd activation by Caspar knockdown (Fig. 6B) led to a decrease in the Wolbachia population (Fig. 6E) and increased REL2 and gambicin transcription (Fig. 6, C and D), indicating that REL2/gambicin mobilization through the Imd pathway can regulate Wolbachia proliferation. Furthermore, there was no change in GSK3β transcription (Fig. 6G), again confirming that GSK3β is upstream of this pathway.
Moreover, many authors provided evidence for a Wolbachia role in the host energy metabolism (35, 36, 37, 38). For example, in the symbiotic relationship with the fly Drosophila melanogaster, it is suggested that Wolbachia acts to increase insulin signaling (39, 40). Typically, the activation of the insulin pathway results in GSK3β inhibition by serine-9 residue phosphorylation, increasing glycogen synthesis by GS enzyme (41, 42). The fact that both inhibition and knockdown of GSK3β increased glycogen content (Figs. 1E and 4C) indicates that this enzyme performs its typical function of controlling glycogen synthesis in the mosquito cells. We observed that Wolbachia presence induced a decrease in the relative amount of GSK3β transcripts (Fig. 1B) and increased glycogen content (Fig. 1C). We postulate that Wolbachia provides metabolites which fuel energy production pathways, including glycogen synthesis, implying that mechanisms sensing metabolite/energy levels might be controlling GSK3B expression. Downstream of PI3 kinase activation, protein kinase A, protein kinase B—also known as Akt—protein kinase C, and p90Rsk contribute to the inactivation of GSK3, which ultimately leads to the dephosphorylation of GSK3 substrates. Thus, the activation of PI3K/AKT signaling pathway usually leads to the inhibition of GSK3β and, consequently, increased glycogen synthesis (43). This mechanism could explain the higher glycogen content observed in cells infected with Wolbachia compared to uninfected counterparts. This indicates that suppression of GSK3β by Wolbachia results in a metabolic remodeling that changes glycogen metabolism.
In the nematode Brugia malayi, a natural host for Wolbachia wBm strain, studies have demonstrated that treatment with the antibiotic doxycycline reduces Wolbachia fitness. This treatment leads to an increase in glycogen content and reduction of GSK3β transcription, suggesting that Wolbachia may rely on host glycogen content. These findings underscore the potential role of GSK3β transcription in mediating glycogen metabolism remodeling induced by Wolbachia (44). Similarly, in the Ae. fluviatilis mosquito, Wolbachia fitness appears to be closely linked to glycogen metabolism. We show that interfering with GSK3β (Figs. 1E and 4C) or REL2 (Fig. 5E) causes an increase in glycogen content followed by an increase in Wolbachia population (Figs. 1H, 4D, and 5D). Caspar knockdown, in contrast, resulted in reduction of the Wolbachia population (Fig. 6E) and surprisingly was accompanied by a reduction in glycogen content (Fig. 6F). These data strongly suggest a close association between glycogen content and the Wolbachia population. Moreover, it is possible that this bacterium is supplying metabolites that enhance cellular energy efficiency in its natural host.
Apart from the impact on nutritional reserves, Wolbachia also exerts influence on the transcription of immune genes in its transfected hosts. Wolbachia reduces the susceptibility of Ae. aegypti to infection by positive-sense RNA viruses, such as dengue. For example, in Ae. aegypti artificially transfected with Wolbachia strain wMel, there is an increase in the basal immune response within the Toll and Imd pathways. Activation of the Toll pathway induces increased transcription of the AMPs defensins and cecropins, which are involved in the inhibition of DENV virus proliferation (45). On the other hand, suppressing the Imd and Toll pathways reduces the wMel strain population in this mosquito, suggesting that Wolbachia manipulates the host's immune system to compromise virus stability and facilitate persistent infection (46). However, it is known that RNA viruses can react to antiviral selection pressures against which they can adapt to be effectively transmitted by Ae. aegypti that carries Wolbachia. On the other hand, the evolution of the Aedes aegypti genome could attenuate Wolbachia-mediated viral inhibition, adapting to the endosymbiont over time (46). Studies suggest that Wolbachia does not induce the expression of genes that encode AMPs. This has been demonstrated for organisms where Wolbachia establishes natural infection, as in Ae. albopictus, D. melanogaster, and Tetranychus urticae (47, 48, 49). Interestingly, we observed that there was no difference in relative quantification of defensin transcripts (Fig. 2F), REL1, and REL2 (Fig. 2, B and C) between wAflu1 and wAflu1.tet cells. However, in wAflu1.tet cells, there was a decrease in the relative amount of Cactus and Caspar transcripts (Fig. 2, D and E), indicating that Toll and Imd pathways may be more active in cells with Wolbachia. This suggests that the Wolbachia (strain wFlu) differentially manipulates the Ae. fluviatilis immune pathways, and it can be adapted according to the host and bacterium strain.
We also observed that when Wolbachia was removed, there was an increase in the relative quantity of cecropin transcripts (Fig. 2G), and there was no change in defensin transcript levels (Fig. 1F). However, it has already been shown that cecropin and defensin were drastically upregulated by immunological challenge with heat-killed bacteria (23). It is possible that these AMPs are efficient against infecting bacteria and do not act on the endosymbiotic bacteria. Furthermore, there are higher transcription levels of gambicin in wAflu1 cells (Fig. 2H), indicating that gambicin is upregulated in the presence of Wolbachia, regardless of a subtle downregulation of GSK3β transcripts (Fig. 1B). In natural conditions, GSK3β downregulation may not decrease gambicin transcript levels (Fig. 2H) or does not interfere with REL2 relative transcripts (Fig. 2B), thus a stronger interference with GSK3β is necessary to change the transcription of these immune genes and trigger microbiota dysregulation.
Taken together, our studies unveil a novel interaction between GSK3β and Imd pathway. Supporting this hypothesis, the data presented here show that downregulation of GSK3β transcription can reduce Imd pathway/gambicin system, thereby promoting Wolbachia proliferation. Furthermore, our data indicate that Wolbachia may contribute energetically to host glycogen accumulation, though further studies are required to verify this hypothesis.
Experimental procedures
Maintenance of wAflu1 and wAflu1.tet cells
The wAflu1 and wAflu1.tet cell lines were maintained in culture flasks (25 cm2) with 5 ml of supplemented L-15 medium at 28 °C as described previously (23). L-15 medium was supplemented with amino acids, glucose, mineral salts, and vitamins (50). A 5 ml plastic syringe attached to a 22-gauge needle (0.70 × 25 mm) was used to resuspend the cells. Culture density was determined using a Neubauer chamber.
Glycogen quantification
Adhered cells were washed with phosphate buffered saline PBS, pH 7.4 (137 mM NaCl; 10 mM Na2HPO4; 2.7 mM KCl; 1.8 mM KH2PO4), after which they were resuspended in 5 ml of PBS by jetting the liquid onto the cell layer with 5 ml syringe and needle, then centrifuged at 1500g for 3 min. The cell pellet was resuspended in 100 μl of Tris/HCl pH 7.4 buffer containing 0.1% Triton ×100.
Glycogen content was estimated by measuring the glucose generated after glycogen digestion by the enzyme α-amyloglucosidase activity (Sigma A7095; Source Aspergillus niger). Cleared lysed cells (12 μl) were incubated with 1 unit of α-amyloglycosidase in 20 μl of acetate buffer (200 mM, pH 4.8) for 4 h at 40 °C. Then, 100 μl of glucose monoreagent (glucose oxidase-based method Labtest - ref.: 133) was added and incubated for 15 min. Absorbance was determined at 510 nm in a spectrophotometer SpectraMax M3 reader (Molecular devices). The control (without α-amyloglucosidase) was used to determine the free glucose levels of each sample and was subtracted from the absorbances of samples incubated with α-amyloglucosidase. A standard curve was generated to calculate glycogen levels in the samples, and data were normalized by 1 × 105 cells.
DNA extraction
DNA was extracted using a DNeasy blood and tissue kit (Qiagen) according to manufacturer recommendations for cell cultures. Briefly, 5 × 106 cells were pelleted for 5 min at 300g and resuspended in 200 μl PBS buffer. Proteinase K (20 μl) was added, followed by 200 μl of Buffer AL. Samples were mixed thoroughly by vortexing and incubated at 56 °C for 10 min. Ethanol 100% (200 μl) was added and mixed thoroughly by vortexing. The mixture was pipetted onto a DNeasy Mini spin column placed in a 2 ml collection tube and centrifuged at 6000g for 1 min, discarding the flow-through. After washing the column, as recommended by the manufacturer, the DNA was eluted in 200 μl of elution buffer and quantified in spectrophotometer (Thermo Fisher Scientific).
Total RNA extraction and cDNA synthesis
Total RNA was extracted from about 5 × 105 cells and used to investigate differences in target genes transcription between cell lines. Culture medium was discarded, and 500 μl of Trizol reagent (Invitrogen) was added to each well of a 12-well plate. Extraction was performed as recommended by the manufacturer, and RNA concentration and respective purity were determined in a NanoDrop spectrophotometer (Thermo Fisher Scientific) and agarose gel containing ethidium bromide. About 2 μg of RNA was treated with DNAse I (Invitrogen Ambion), as recommended by the manufacturer. After treatment, samples were used as a template for cDNA synthesis using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). The synthesized cDNAs were stored at −20 °C until use.
PCR check for W. pipientis presence
The cDNA obtained from wAflu1 and wAflu1.tet cells was used for WSP gene amplification by PCR using oligonucleotide primers designed from the sequence GQ917108 (NCBI). The WSP fragment (650 bp) was amplified using the following primers: 5′-TGGTCCAATAAGTGATGAAGAAAC-3′ forward and 5′-AAAAATTAAACGCTACTCCA-3′ reverse. The PCR reaction was analyzed on a 1.5% agarose gel containing ethidium bromide.
Quantitative PCR
Quantification by RT-qPCR analysis using cDNA or by quantitative PCR using bacterial DNA samples was performed in the StepOnePlus Real Time PCR equipment (Applied Biosystems) with the SyGreen Mix HI-ROX kit (PCR Biosystems) according to the following protocol: 95 °C for 2 min, followed by 40 cycles of 95 °C for 7 s and 60 °C for 25 s, plus a step of 95 °C for 15 s, another step of 60 °C for 1 min, and 95 °C for 10 s. cDNA serial dilutions were used to construct a calibration curve. Reaction efficiencies higher than 92% were obtained from calibration curves for each set of primers with final volumes of 15 μl. The oligonucleotide primers for the genes that were amplified by the RT-qPCR reaction are shown in Table 1. The constitutively expressed gene for the 60S ribosomal protein, known as RP49 (MW574127), was used as a reference gene (23). Relative quantification was determined using Ct values from each run in the Software Tool table as described by PFAFFL (51). Statistical analyses were performed on data from three independent experiments in the Prisma GraphPad Prism 7.00 program using Student t test or two-way ANOVA. The graphs show averages along with the respective standard deviation.
Table 1.
List of oligonucleotides used to analyze the immunometabolic crosstalking on Aedes fluviatilis Wolbachia pipientis symbiosis
| Oligonucleotide name | Oligonucleotide | Gene code |
|---|---|---|
| Rp49 qPCR Fw | GATGCAGAACCGTGTCTACT | MW574127 |
| Rp49 qPCR Rv | AGCTTACTCGTTTTCTTGCG | |
| CASPAR qPCR Fw | GCGGATCGAGCAAAGCAAGA | OM830326 |
| CASPAR qPCR Rv | CAGTCGGTGCCCTTATCCTA | |
| CACTUS qPCR Fw | CCGTAATTTTGGGCACTCAT | OM830327 |
| CACTUS qPCR Rv | CATTAACGCAAGGGAAGGAA | |
| WSP Fw qPCR | ATCTTTTATAGCTGGTGGTGGT | GQ917108 |
| WSP Rv qPCR | GGAGTGATAGGCATATCTTCAAT | |
| WSP Fw (Check) | TGGTCCAATAAGTGATGAAGAAAC | GQ917108 |
| WSP Rv (Check) | AAAAATTAAACGCTACTCCA | |
| Cecropin qPCR Fw | CGATTTGACGTTCGAGATGA | MW574131 |
| Cecropin qPCR Rv | CGTTTGCCTACTCCTTCCAA | |
| Defensin qPCR Fw | AACTCTCCTCTCACGCCGTA | MW574130 |
| Defensin qPCR Rv | TACGAGCGAACATCATCAGC | |
| Gambicin qPCR Fw | AGATGCGCTGGTGTTCGTAT | OM830330 |
| Gambicin qPCR Rv | TCACTGCAGGTTCTGATTGC | |
| Rel1 qPCR Fw | CAGCCAATCAGCAACAGAAA | MW574128 |
| Rel1 qPCR Rv | ATTCGTTTGATGGGCGATAG | |
| Rel2 qPCR Fw | ATTGTTTCCGTCTGGATTCG | MW574129 |
| Rel2 qPCR Rv | TCACGCAGAACGTATGAAGC | |
| GSK3 qPCR Fw | TCATCAAAGTCCTCGGAACG | OM830324 |
| GSK3 qPCR Rv | ATCGCATCTGGTGGAGTACG | |
| GFP t7 Fw | TAATACGACTCACTATAGGGACGTAAACGGCCACAAGTTCAGCGTGTC | EF152770.1 |
| GFP t7 Rv | TAATACGACTCACTATAGGGTCACGAACTCCAGCAGGACCATGTCATC | |
| Rel2 t7 Fw | TAATACGACTCACTATAGGGCATTGCCGAGGAACTGAGCA | MW574129 |
| Rel2 t7 Rv | TAATACGACTCACTATAGGGCCCAATCTTGCCATGCTTCTT | |
| GSK3 t7 Fw | TAATACGACTCACTATAGGGCCCGTTATGGGAGGCATGAA | OM830324 |
| GSK3 t7 Rv | TAATACGACTCACTATAGGGTCGGTTCCCCATGCAGAAGT |
Primer sequences used to evaluate immunometabolism-related gene expression and perform dsRNA synthesis to gene knockdown by RNA interference.
MTT viability
Cell viability was measured using MTT assay measuring reduction of tetrazolium dye MTT to its insoluble form formazan (52). After alsterpaullone treatment, 50 μl of MTT (5 mg/ml in PBS) was added to each well. After 2 h incubation at 28 °C, the medium was completely removed, and 1 ml of acid-isopropyl alcohol (0.15% HCl in isopropyl alcohol) was added to dissolve formazan crystals. The mixture was transferred to 1.5-ml tubes and centrifuged at 6000g for 15 min. The supernatant was collected for absorbance measurement at 570 nm in a spectrophotometer (SpectraMax M3 reader, molecular devices).
Alsterpaullone treatment
GSK3 inhibition was performed based on previous work (53, 54, 55). To determine the optimal concentration of GSK3 inhibitor alsterpaullone for the conditions of the present experiment, 5 × 105 cells were seeded in 24-well plates with 500 μl of L-15 medium. After 24 h, the culture medium was changed, and alsterpaullone was added to reach final concentrations of 100, 200, 300, 400, 500, and 600 nM. DMSO (0.6 and 1.2%) was used as a control. Cell viability was tested by MTT after 24 h of alsterpaullone exposure. DMSO treatment was used for data normalization (100% viability). In addition, to test the inhibitor functionality, we performed glycogen quantification at 300 and 500 nM alsterpaullone concentrations.
Double-stranded RNA synthesis
Double-stranded RNA (dsRNA) synthesis used oligonucleotide primers for REL2, GSK3β, and GFP (green fluorescent protein—unrelated gene) containing T7 promoter region in both strand 5′ ends. Oligonucleotide primers (Table 1) generated fragments of 398 bp, 574 bp, and 652 bp for the REL2, GSK3β, and GFP targets, respectively. With the generated fragments, dsRNA synthesis was performed using the T7 RiboMAX Express RNAi System kit (Promega) using 1 μg of template. The resulting dsRNA was analyzed for integrity by electrophoresis on a 1.5% agarose gel and quantified in a spectrophotometer nanodrop (Thermo Scientific). We employed two strategies to assess the specificity of the dsRNA. First, we evaluated the potential off-target effects of GSK-3 RNAi through analysis using Ae. fluviatilis sequences and the Si-Fi21 algorithm (56). Second, we conducted Blast analyses comparing the GSK-3 sequence with Ae. fluviatilis sequences. In both cases, no off-targets were detected.
Western blotting for p-GSK3β
For immunoblotting, 106 cells treated with dsGFP or dsGSK3β were homogenized in 100 μl of lysis buffer containing tris-HCl 10 mM pH 7.4 and 0.1% Triton X-100 in a 1:1 mixture with protease inhibitor cocktail Complete EDTA-free EASYpack (Roche) and phosphatase inhibitor cocktail PhosSTOP EASYpack (Roche). Supernatant was collected by centrifugation at 14,000g for 10 min at 4 °C. The protein concentration in the supernatant was determined by the Lowry method (57) using bovine serum albumin (Sigma) for standard curve. Total protein (25 μg) was separated by 10% SDS-PAGE and electroblotted onto polyvinylidene fluoride membranes (GE Healthcare) with transfer buffer (48 mM Tris and 39 mM glycine). The membrane was then blocked with 5% albumin in Tris buffered saline-Tween (TBS-T) (10 mM Tris pH 8.0, 150 mM sodium chloride and 0.1% Tween-20) for 2 h at room temperature and subsequently incubated with anti-phospho-GSK3β (Santa Cruz) primary antibody overnight at 4 °C. After that, the membrane was washed with TBS-T three times for 5 min and further incubated with anti-mouse secondary antibody linked to horseradish peroxidase (Thermo Fisher Scientific) at a 1:5000 dilution in TBS-T albumin for 2 h at room temperature. After incubation, the membrane was washed with TBS-T, followed by three TBS washes of 5 min each, and the assay was developed by chemiluminescence using the Enhanced Chemiluminescence System (Millipore) and the ChemiDoc MP Imaging System (Bio-Rad) for visualization and imaging. Next, the membrane was stripped twice with 0.1 M of NaOH for 5 min each, followed by TBS washes and TBS-T albumin incubation for 1 h at room temperature. The stripped membrane was incubated with primary anti-β-Tubulin (57) at a 1:5000 dilution for 2 h at room temperature, then washed with TBS-T before further incubation with anti-rabbit secondary antibody (Cell Signaling) at 1:5000 dilution for 1 h at room temperature. Once more the membrane was washed with TBS-T, followed by TBS washes, and developed using the Enhanced Chemiluminescence System. The densitometry of anti-phospho-GSK3β and anti-β-Tubulin Western Blots was carried out using the program ImageJ (58) to determine the ratio between the anti-phospho-GSK3β and anti-β-Tubulin signals, with results from dsGFP-treated cells used for normalization. Antibodies against total unphosphorylated GSK3B failed to recognize the Ae. fluviatilis polypeptide, in contrast to the antibody against Pser-9-GSK3B (Santa Cruz), which was thus used in our experiments. The phosphorylated inactive form of GSK3, Pser-9-GSK3B is the form that leads to activation of glycogen synthesis. Evaluating levels of Pser-9-GSK3B is therefore an effective way to monitor the impact of gene silencing on pathway function.
RNAi silencing
The wAflu1 cells (3 × 106 cells) were maintained for 5 days in 75 cm2 bottles with L-15 medium supplemented with 10% fetal bovine serum at 28 °C. Cells were transfected using the cell line Nucleofector kit V (VCA-1003) according to the manufacturer's instructions (Amaxa Biosystems). About 5 × 106 cells were centrifuged and carefully resuspended in 100 μl of transfection reagent (82 μl of cell line plus and 18 μl supplement) containing 1 μg of dsRNA (dsGFP, dsGSK3, or dsREL2). Suspended cells were transferred to a Lonza certified cuvette and transfected in the Nucleofector I Device (Lonza) with the G-030 transfection preset program. After transfection, the cell suspension was added to 3 ml of supplemented L-15 medium and was split: 1.5 ml was seeded into three wells (500 μl per well) of a 6-well plate for RNA extraction; and 1.5 ml was seeded in a 25 cm2 bottle for glycogen quantification. After seeding, cells were incubated for 24 h at 28 °C. Silencing was verified by RT-qPCR of the target genes.
Statistical analysis
The experiments were performed in biological triplicate with technical triplicate. Averages are presented along with standard deviations. GraphPad Prism 8.3 was used to perform unpaired t test or two-way ANOVA followed by multiple comparisons, when applicable.
Data availability
All data are described within the manuscript.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
J. N. d. S., C. C. C., and C. L. conceptualization; J. N. d. S., C. C. C., and C. L. methodology; J. N. d. S., C. C. C., G. C. R. d. B., A. A., C. R. d. O. D. F., and A. B. W. N. investigation; J. N. d. S., C. C. C., O. A. C. T., I. d. S. V., C. L. formal analysis; J. N. d. S., A. A., I. d. S. V., and O. A. C. T. resources; J. N. d. S., C. C. C., I. d. S. V., and C. L. writing-original draft; P. L. d. O., L. A. M., I. d. S. V., C. L. supervision; P. L. d. O., L. A. M., I. d. S. V., C. L. writing-review and editing.
Funding and additional information
This work was supported by Brazilian grants from CNPq- Conselho Nacional de Desenvolvimento Científico e Tecnológico (3085672020), INCT- Entomologia Molecular (465678/2014-9), and FAPERJ (E-26/200.334/2023).
Reviewed by members of the JBC Editorial Board. Edited by Qi-Qun Tang
Supporting information
References
- 1.Moran N.A., Ochman H., Hammer T.J. Evolutionary and ecological consequences of gut microbial communities. Annu. Rev. Ecol. Evol. Syst. 2019;50:451. doi: 10.1146/annurev-ecolsys-110617-062453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilbert S.F., Sapp J., Tauber A.I. A symbiotic view of life: we have never been individuals. Q. Rev. Biol. 2012;87:325–341. doi: 10.1086/668166. [DOI] [PubMed] [Google Scholar]
- 3.Jiménez-Cortés J.G., García-Contreras R., Bucio-Torres M.I., Cabrera-Bravo M., Córdoba-Aguilar A., Benelli G., et al. Bacterial symbionts in human blood-feeding arthropods: patterns, general mechanisms and effects of global ecological changes. Acta Trop. 2018;186:69–101. doi: 10.1016/j.actatropica.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Jiménez N.E., Gerdtzen Z.P., Olivera-Nappa Á., Salgado J.C., Conca C. A systems biology approach for studying Wolbachia metabolism reveals points of interaction with its host in the context of arboviral infection. PLoS Negl. Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Douglas A.E. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 6.Gerardo N.M., Hoang K.L., Stoy K.S. Evolution of animal immunity in the light of beneficial symbioses. Philos. Trans. R. Soc. B Biol. Sci. 2020;375 doi: 10.1098/rstb.2019.0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoang K.L., King K.C. Symbiont-mediated immune priming in animals through an evolutionary lens. Microbiology (Reading) 2022;168 doi: 10.1099/mic.0.001181. [DOI] [PubMed] [Google Scholar]
- 8.Zhang W., Tettamanti G., Bassal T., Heryanto C., Eleftherianos I., Mohamed A. Regulators and signalling in insect antimicrobial innate immunity: functional molecules and cellular pathways. Cell. Signal. 2021;83 doi: 10.1016/j.cellsig.2021.110003. [DOI] [PubMed] [Google Scholar]
- 9.Tsakas S., Marmaras V.J. Insect immunity and its signalling: an overview. Invertebr. Surviv. J. 2010;7:228–238. [Google Scholar]
- 10.Bian G., Shin S.W., Cheon H.M., Kokoza V., Raikhel A.S. Transgenic alteration of Toll immune pathway in the female mosquito Aedes Aegypti. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13568–13573. doi: 10.1073/pnas.0502815102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barletta A.B.F., Nascimento-Silva M.C.L., Talyuli O.A.C., Oliveira J.H.M., Pereira L.O.R., Oliveira P.L., et al. Microbiota activates IMD pathway and limits Sindbis infection in Aedes aegypti. Parasit. Vectors. 2017;10:103. doi: 10.1186/s13071-017-2040-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zakovic S., Levashina E.A. NF-κB-Like signaling pathway REL2 in immune defenses of the Malaria vector Anopheles gambiae. Front. Cell. Infect. Microbiol. 2017;7:258. doi: 10.3389/fcimb.2017.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sim S., Jupatanakul N., Dimopoulos G. Mosquito immunity against Arboviruses. Viruses. 2014;6:4479. doi: 10.3390/v6114479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mylonakis E., Podsiadlowski L., Muhammed M., Vilcinskas A. Diversity, evolution and medical applications of insect antimicrobial peptides. Philos. Trans. R. Soc. B Biol. Sci. 2016;371 doi: 10.1098/rstb.2015.0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Viljakainen L. Evolutionary genetics of insect innate immunity. Brief. Funct. Genomics. 2015;14:407. doi: 10.1093/bfgp/elv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuesta A., Chaves-Pozo E., St S., Aczek ˛, Cytry'nska M., Cytry'nska C., et al. Unraveling the role of antimicrobial peptides in insects. Int. J. Mol. Sci. 2023;24:5753. doi: 10.3390/ijms24065753. 5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanson M.A., Lemaitre B. New insights on Drosophila antimicrobial peptide function in host defense and beyond. Curr. Opin. Immunol. 2020;62:22–30. doi: 10.1016/j.coi.2019.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Mergaert P. Role of antimicrobial peptides in controlling symbiotic bacterial populations. Nat. Prod. Rep. 2018;35:336–356. doi: 10.1039/c7np00056a. [DOI] [PubMed] [Google Scholar]
- 19.Bi J., Wang Y.F. The effect of the endosymbiont Wolbachia on the behavior of insect hosts. Insect Sci. 2020;27:846. doi: 10.1111/1744-7917.12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaur R., Shropshire J.D., Cross K.L., Leigh B., Mansueto A.J., Stewart V., et al. Living in the endosymbiotic world of Wolbachia: a centennial review. Cell Host Microbe. 2021;29:879–893. doi: 10.1016/j.chom.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werren J.H. Biology of Wolbachia. Annu. Rev. Entomol. 1997;42:587–609. doi: 10.1146/annurev.ento.42.1.587. [DOI] [PubMed] [Google Scholar]
- 22.da Rocha Fernandes M., Martins R., Pessoa Costa E., Casagrande Pacidônio E., Araujo de Abreu L., da Silva Vaz I., et al. The modulation of the symbiont/host interaction between Wolbachia pipientis and Aedes fluviatilis embryos by glycogen metabolism. PLoS One. 2014;9 doi: 10.1371/journal.pone.0098966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conceição C.C., Da Silva J.N., Arcanjo A., Nogueira C.L., de Abreu L.A., de Oliveira P.L., et al. Aedes fluviatilis cell lines as new tools to study metabolic and immune interactions in mosquito-Wolbachia symbiosis. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-98738-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nascimento da Silva J., Calixto Conceição C., Cristina Ramos de Brito G., Costa Santos D., Martins da Silva R., Arcanjo A., et al. Wolbachia pipientis modulates metabolism and immunity during Aedes fluviatilis oogenesis. Insect Biochem. Mol. Biol. 2022;146 doi: 10.1016/j.ibmb.2022.103776. [DOI] [PubMed] [Google Scholar]
- 25.Wang L., Li J., Di L.J. Glycogen synthesis and beyond, a comprehensive review of GSK3 as a key regulator of metabolic pathways and a therapeutic target for treating metabolic diseases. Med. Res. Rev. 2022;42:946–982. doi: 10.1002/med.21867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beurel E., Michalek S.M., Jope R.S. Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3) Trends Immunol. 2010;31:24–31. doi: 10.1016/j.it.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Souder D.C., Anderson R.M. An expanding GSK3 network: implications for aging research. Geroscience. 2019;41:369–382. doi: 10.1007/s11357-019-00085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang S., Zhu L., Hou C., Yuan H., Yang S., Dehwah M.A.S., et al. GSK3β plays a negative role during white spot syndrome virus (WSSV) infection by regulating NF-κB activity in Shrimp Litopenaeus vannamei. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.607543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piazzi M., Bavelloni A., Cenni V., Faenza I., Blalock W.L. Revisiting the role of GSK3, A modulator of innate immunity, in Idiopathic Inclusion Body Myositis. Cells. 2021;10:3255. doi: 10.3390/cells10113255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruan L., Liu H., Shi H. Characterization and function of GSK3β from Litopenaeus vannamei in WSSV infection. Fish Shellfish Immunol. 2018;82:220–228. doi: 10.1016/j.fsi.2018.08.032. [DOI] [PubMed] [Google Scholar]
- 31.Khan M.B., Liew J.W.K., Leong C.S., Lau Y.L. Role of NF-kβ factor Rel2 during Plasmodium falciparum and bacterial infection in Anopheles dirus. Parasit. Vectors. 2016;9:1–7. doi: 10.1186/s13071-016-1810-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pike A., Vadlamani A., Sandiford S.L., Gacita A., Dimopoulos G. Characterization of the Rel2-regulated transcriptome and proteome of Anopheles stephensi identifies new anti-Plasmodium factors. Insect Biochem. Mol. Biol. 2014;52:82–93. doi: 10.1016/j.ibmb.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q.Y., Yan Z.B., Meng Y.M., Hong X.Y., Shao G., Ma J.J., et al. Antimicrobial peptides: mechanism of action, activity and clinical potential. Mil. Med. Res. 2021;81:1–25. doi: 10.1186/s40779-021-00343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Georgel P., Naitza S., Kappler C., Ferrandon D., Zachary D., Swimmer C., et al. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev. Cell. 2001;1:503–514. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- 35.Dutra H.L.C., Deehan M.A., Frydman H. Wolbachia and Sirtuin-4 interaction is associated with alterations in host glucose metabolism and bacterial titer. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caragata E.P., Rancès E., O’Neill S.L., McGraw E.A. Competition for amino acids between Wolbachia and the mosquito host, Aedes aegypti. Microb. Ecol. 2014;67:205–218. doi: 10.1007/s00248-013-0339-4. [DOI] [PubMed] [Google Scholar]
- 37.Ponton F., Wilson K., Holmes A., Raubenheimer D., Robinson K.L., Simpson S.J. Macronutrients mediate the functional relationship between Drosophila and Wolbachia. Proc. Biol. Sci. 2015;282 doi: 10.1098/rspb.2014.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saucereau Y., Valiente Moro C., Dieryckx C., Dupuy J.W., Tran F.H., Girard V., et al. (2017) Comprehensive proteome profiling in Aedes albopictus to decipher Wolbachia-arbovirus interference phenomenon. BMC Genomics. 2017;181:1–14. doi: 10.1186/s12864-017-3985-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Currin-Ross D., Husdell L., Pierens G.K., Mok N.E., O’Neill S.L., Schirra H.J., et al. The metabolic response to infection with Wolbachia Implicates the insulin/insulin-like-Growth factor and Hypoxia signaling pathways in Drosophila melanogaster. Front. Ecol. Evol. 2021;9 [Google Scholar]
- 40.Ikeya T., Broughton S., Alic N., Grandison R., Partridge L. The endosymbiont Wolbachia increases insulin/IGF-like signalling in Drosophila. Proc. Biol. Sci. 2009;276:3799–3807. doi: 10.1098/rspb.2009.0778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doble B.W., Woodgett J.R. GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen P., Alessi D.R., Cross D.A.E. PDK1, one of the missing links in insulin signal transduction? FEBS Lett. 1997;410:3–10. doi: 10.1016/s0014-5793(97)00490-0. [DOI] [PubMed] [Google Scholar]
- 43.He R., Du S., Lei T., Xie X., Wang Y. Glycogen synthase kinase 3β in tumorigenesis and oncotherapy. Oncol. Rep. 2020;44:2373. doi: 10.3892/or.2020.7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voronin D., Bachu S., Shlossman M., Unnasch T.R., Ghedin E., Lustigman S. Glucose and glycogen metabolism in Brugia malayi is associated with Wolbachia symbiont fitness. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan X., Zhou G., Wu J., Bian G., Lu P., Raikhel A.S., et al. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E23–E31. doi: 10.1073/pnas.1116932108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pan X., Pike A., Joshi D., Bian G., McFadden M.J., Lu P., et al. The bacterium Wolbachia exploits host innate immunity to establish a symbiotic relationship with the dengue vector mosquito Aedes aegypti. ISME J. 2018;12:277. doi: 10.1038/ismej.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wong Z.S., Hedges L.M., Brownlie J.C., Johnson K.N. Wolbachia-mediated antibacterial protection and immune gene regulation in Drosophila. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rancès E., Ye Y.H., Woolfit M., McGraw E.A., O’Neill S.L. The relative importance of innate immune priming in Wolbachia-mediated dengue interference. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bourtzis K., Pettigrew M.M., O’Neill S.L. Wolbachia neither induces nor suppresses transcripts encoding antimicrobial peptides. Insect Mol. Biol. 2000;9:635–639. doi: 10.1046/j.1365-2583.2000.00224.x. [DOI] [PubMed] [Google Scholar]
- 50.Munderloh U.G., Kurtti T.J. Formulation of medium for tick cell culture. Exp. Appl. Acarol. 1989;7:219–229. doi: 10.1007/BF01194061. [DOI] [PubMed] [Google Scholar]
- 51.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Meerloo J., Kaspers G.J.L., Cloos J. Cell sensitivity assays: the MTT assay. Methods Mol. Biol. 2011;731:237–245. doi: 10.1007/978-1-61779-080-5_20. [DOI] [PubMed] [Google Scholar]
- 53.Fabres A., Pinto De Andrade C., Guizzo M., Henrique M., Sorgine F., De G., et al. Effect of GSK-3 activity, enzymatic inhibition and gene silencing by RNAi on tick oviposition and egg hatching. Parasitology. 2010;137:1537–1546. doi: 10.1017/S0031182010000284. [DOI] [PubMed] [Google Scholar]
- 54.Mury F.B., Lugon M.D., Nunes Fonseca R.D.A., Silva J.R., Berni M., Araujo H.M., et al. Glycogen Synthase Kinase-3 is involved in glycogen metabolism control and embryogenesis of Rhodnius prolixus. Parasitology. 2016;143:1569–1579. doi: 10.1017/S0031182016001487. [DOI] [PubMed] [Google Scholar]
- 55.Meijer L., Flajolet M., Greengard P. Pharmacological inhibitors of glycogen synthase kinase 3. Trends Pharmacol. Sci. 2004;25:471–480. doi: 10.1016/j.tips.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 56.Lück S., Kreszies T., Strickert M., Schweizer P., Kuhlmann M., Douchkov D. siRNA-finder (si-Fi) Software for RNAi-target design and off-target Prediction. Front. Plant Sci. 2019;10:1023. doi: 10.3389/fpls.2019.01023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 58.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;97:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are described within the manuscript.