Abstract
Ensuring a rapid and accurate identification of harmful bacteria is crucial in various fields including environmental monitoring, food safety, and clinical diagnostics. Conventional detection methods often suffer from limitations such as long analysis time, complexity, and the need for qualified personnel. Therefore, a lot of research effort is devoted to developing technologies with the potential to revolutionize the detection of pathogenic bacteria by offering rapid, sensitive, and user-friendly platforms for point-of-care analysis. In this light, biosensors have gained significant commercial attention in recent years due to their simplicity, portability, and rapid analysis capabilities. The purpose of this review is to identify a trend by analyzing which biosensor technologies have become commercially successful in the field of bacteria detection. Moreover, we highlight the characteristics that a biosensor must possess to finally arrive in the market and therefore in the hands of the end-user, and we present critical examples of the market applications of various technologies. The aim is to investigate the reason why certain technologies have achieved commercial success and extrapolate these trends to the future economic viability of a new subfield in the world of biosensing: the development of biomimetic sensor platforms. Therefore, an overview of recent advances in the field of biomimetic bacteria detection will be presented, after which the challenges that need to be addressed in the coming years to improve market penetration will be critically evaluated. We will zoom into the current shortcomings of biomimetic sensors based on imprinting technology and aptamers and try to come up with a recommendation for further development based on the trends observed from previous commercial success stories in biosensing.
Bacterial contamination poses a significant risk to both healthcare and food safety, with a variety of dangerous bacteria capable of causing serious illnesses and diseases, that can eventually lead to death. Exposure to pathogenic bacteria can occur via different sources, including contaminated food and water, contact with animals, and contact with other people. Some of the most common dangerous bacteria that can be found in food or biological samples include Salmonella species, Escherichia coli, Listeria monocytogenes, Campylobacter species, Clostridium botulinum, and Pseudomonas aeruginosa.1 A more comprehensive list of pathogens was published in an open-access database, which also includes the specificities of the microorganisms such as their strains, infectious rates, places where they can be found, and common therapies.2 These bacteria can cause a range of illnesses, including food poisoning, diarrhea, urinary tract infections, and respiratory infections. As estimated by the U.S. Center for Disease Control and Prevention (CDC), more than 48 million people are infected annually by different pathogens, developing dangerous diseases resulting in more than 128,000 hospitalizations in the USA.3 Furthermore, an issue that has become increasingly important in recent years is the increasing resistance that bacteria are developing toward antibiotics, leading to the so-called Antimicrobial Resistance (AMR), thus increasing the urgency of finding novel alternatives to detect the presence of pathogenic bacteria in different types of samples.4
In order to prevent infectious illnesses, both industry and academia put a lot of effort and investments into bacterial detection, with a particular focus devoted to developing innovative ways to detect contaminations using devices that allow for direct assessment. Several benchtop technologies have therefore been developed to detect bacterial contamination, many of which have become standard analytical laboratory techniques. This category is occupied by well-established methods such as gas chromatography (GC), high-pressure liquid chromatography (HPLC), and UV–vis-based technologies for the detection of chemical markers for contamination, but also other platforms based on specifically detecting (part of) the pathogens directly such as polymerase chain reaction (PCR), or loop-mediated isothermal amplification (LAMP).5 These technologies possess the advantage of being highly specific but require expensive equipment that has to be operated in a lab environment by technically trained and qualified personnel. In addition, detailed analysis of samples and regulatory certification still require bacterial culture analysis in a microbiological lab. This process is very time-consuming and expensive, and routine analysis of samples is not possible since the standard procedure relies on taking samples at regular time intervals and sending them off to external laboratories for analysis.6
Technologies that would enable direct point-of-care (PoC) detection of pathogens could increase the number of measurements per unit of time and reduce cost, thereby decreasing the number of infections that go unnoticed leading to a positive impact on the healthcare system and making them highly interesting from a commercial point of view. Therefore, numerous researchers worldwide have developed a wide variety of low-cost, user-friendly, and rapid sensor technologies in the last two decades.7−9 A key driver of these developments is placed on pursuing innovative techniques that offer a valid alternative to conventional testing procedures that are frequently labor- and time-intensive. One of the most promising approaches investigated in the past few years is the development of biosensors. These devices couple a receptor material to a variety of transducers, facilitating the detection of various molecular and biological species.10,11 The recognition element is typically of biological origin, ranging from enzymes or antibodies to small molecules and even whole microorganisms. In this way, these sensors exploit the natural affinity and specificity the receptors have for their target.
This review aims to provide an inventory of lab-based and point-of-care biosensor technologies for the detection of pathogenic bacteria that have made it to the diagnostic market in the past few decades. We will analyze the technical benefit these technologies offer over traditional culture-based approaches and study the commercial aspects of their success in terms of scalability, ease of use, and market penetration. The final goal of this study is to identify trends in these commercial success stories and apply potential trends to a relatively nascent field in bacteria detection: biomimetic sensor platforms. In recent years, multiple reviews have been written that specifically address a subfield of biomimetic pathogen detection but a complete overview of various technologies and a comparison between these technologies and to existing commercial success stories is missing.12−15 Therefore, this research aims to summarize the potential advantages of various biomimetic sensing approaches and frame them within the commercial trends observed in the first part of the study. In this way, this review aims to provide a roadmap to assess the commercial viability of novel biomimetic detection technologies and provide recommendations to guide future research lines on these technologies.
Commercially Available Biosensor Technologies for the Detection of Bacteria
Traditionally, the identification of bacteria in medical, food, and environmental samples is achieved by culture-based techniques in a microbiology laboratory. This approach is very specific and sensitive and is therefore still the gold standard, but it is also time-consuming, requires transport of samples to a lab environment, and the analysis is performed by highly educated staff members.16 To speed up the process and enable point-of-care identification of the presence of bacteria, several technologies have been developed and successfully commercially implemented in standard routine practice in the past few decades (Table 1). To provide meaningful recommendations about the research direction that nascent biomimetic technologies should adopt in the coming years, it is essential to analyze the commercial success of the different technologies summarized in this chapter. Potential trends that facilitate market penetration into the PoC market for bacteria detection are highly relevant to guide future research on emerging technologies and maximize the chance of implementation.17
Table 1. Comparison of Commercial Biosensor Technologies for the Detection of Bacteria.
| Technology | Principle | Detection Time | Sensitivity | Cost | Applications |
|---|---|---|---|---|---|
| Lateral Flow Assays | Antibody–antigen interaction | Minutes | Variable, generally high | Low | Point-of-care diagnostics |
| Enzyme-Linked Immunosorbent Assay (ELISA) | Antibody–antigen interaction | Hours | High | Moderate | Clinical diagnostics |
| ATP Sensors | ATP bioluminescence | Minutes | High | Moderate | Hygiene monitoring |
| Polymerase Chain Reaction (PCR) | Amplification of DNA | Hours | Extremely high | High | Clinical diagnostics, research |
Lateral Flow Assay
In several domains, lateral flow assays (LFAs) have become a popular and efficient biosensing tool for the quick identification of microorganisms, including dangerous bacteria.18 Due to their extensive use and well-established methodology, LFAs are regarded as conventional biosensors. These types of tests depend on the precise interaction between immobilized antibodies and target bacteria to identify bacterial species with high specificity and sensitivity.19 More precisely, in a typical LFA, specific antibodies that are exclusive to the target bacteria of interest are coated on a porous strip. The sample, typically a liquid such as urine, blood, or water, is applied to the strip and flows along the strip by capillary action.20 There are multiple LFA constructs possible but the most widely used is a sandwich-like assembly (Figure 1) in which the liquid sample (potentially after extraction by enzymes) is loaded onto a conjugate pad containing antibodies for a biomarker of interest, conjugated with a label, typically gold nanoparticles. If the target is present in the sample, it will form a complex with the antibody-label conjugate.21 The sample complex moves to the detection line through capillary force. Capture antibodies bind the antigen–antibody-label conjugate, causing a visible color change in the refractive index. Any unbound antibody-label conjugate will be transported to the “control line”, where they will bind to capture antibodies that specifically bind the antibodies in the test solution. This will lead to the appearance of a colored line in this zone. The absence of this line indicates an issue with antibodies or their label conjugate.22
Figure 1.
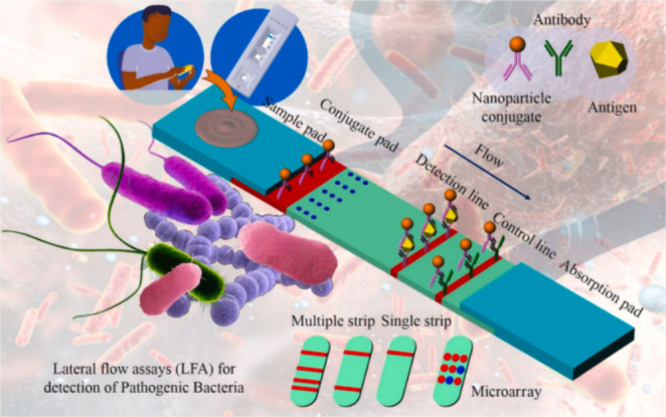
Application of lateral flow assay for the detection of pathogenic bacteria. Reproduced from ref (23). Copyright 2022 American Chemical Society.
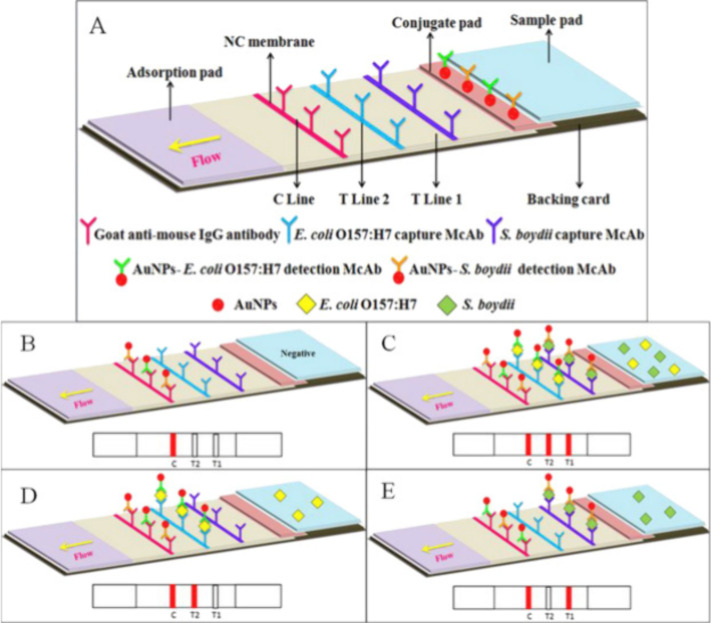
In the last years, several research groups have fabricated lateral-flow assays for the detection of different pathogenic bacteria. In a recently published work, Song et al. developed a lateral flow colloidal gold immunoassay strip for the simultaneous detection of two different pathogenic bacteria, Shigella boydii and Escherichia coli.24 The strip was constructed as a sandwich-like assembly using two types of antibodies, selective for the two different bacteria, respectively (Figure 2). The test strip exhibited excellent sensitivity and selectivity toward the two different bacteria. Moreover, the assay demonstrated its full potential as a PoC tool for the prevention and control of food-borne pathogen diseases by evaluating the strip’s response in bread, milk, and jelly samples with a sensitivity of 4 CFU/mL.
Figure 2.
(A–E) Schematic diagram of the lateral flow colloidal gold immunoassay strip for the detection of two different pathogenic bacteria, Shigella boydii, and Escherichia coli. Reproduced from ref (25). Copyright 2016 American Chemical Society.
Lateral flow assays have become ubiquitous in modern-day diagnostics where they are typically used for diagnosing patients with virus-induced infectious diseases. More recently, several companies such as Hydrosense, Cytiva Life Sciences, and InvivoGen have successfully implemented lateral flow assays for the detection of bacteria and endotoxins in a wide range of applications.26−28 For instance, Hydrosense offers the Legionella Test, designed specifically for detecting Legionella bacteria in water samples. On the other hand, Cytiva Life Sciences provides Lateral Flow Assay Development Tools, allowing researchers to customize lateral flow assays, including those for bacteria detection. Furthermore, many other companies have presented strip-based immunoassays, including Sigma-Aldrich, Thermo Fisher Scientific, Creative Diagnostics, and Rapid Microbiology. These industries offer Lateral Flow Assay Kits designed to detect microbial contaminants in various sample types and with limited costs, thus presenting the best example of PoC devices for the fast detection of pathogens. However, when analyzing the success of these LFAs in pathogen detection by means of (RE)ASSURED criteria it is clear that they mainly excel in providing an affordable, user-friendly, rapid, equipment-free, and deliverable tool for end-users that offers a benefit over fast lab-based technologies such as ELISA and PCR.29 This makes them more suitable for PoC applications, especially in resource-limited settings where a quick, on-site diagnosis is more important than a full microbiological profile of the contamination. Nevertheless, the drawback is that their sensitivity and specificity are limited, and the use of biological receptor elements limits their robustness, as they are vulnerable to changes in temperature and pH and have a limited shelf life. Furthermore, LFAs typically provide the end-user with a 1–0 signal to the presence of a single type of bacterium, which is enough when it comes to diagnosing a patient with infection but for application in environmental screening, hygiene monitoring, and food safety it is often necessary to quantify the amount of bacteria present in a sample.30 It is possible to couple the LFAs to readout with a plate reader but this will rapidly decrease the affordability and user-friendliness of the technology that made it a commercial success in the first place.
From a market and commercial perspective, LFAs are highly dynamic as they can easily be modified to suit approximately any situation. The ease with which this can occur is highly attractive for rapid development and manufacturing times, meaning that LFAs can quickly be tailored to suit various scenarios. This facilitates great market penetration, primarily due to the fact that the approach is easy to pivot and can be placed accordingly. Another strong factor to consider is the simplistic design, with this being a consistent element across all lateral flow assays. This is highly beneficial when considering both the scaling and the usability of the methodology, with the aspect being one of the main drivers of its success. This main drawback is regarding the accessibility of target-specific antibodies and their relative price. In particular, the approach only works if there is already a receptor available for the detection of a specific pathogenic target because without one the method simply does not function.
Overall, this technology has been proven to be versatile, relatively cheap, and extremely user-friendly, allowing for fast, PoC detection of bacteria which is particularly useful in resource-limited settings or urgent situations where a 1/0 response offers adequate information to the end-user. Limitations surrounding specificity and sensitivity are less important in these circumstances. However, the limited thermal stability of the antibodies and their storage conditions are drawbacks even in these specific, niche applications.
Enzyme-Linked Immunosorbent Assay
The enzyme-linked immunosorbent assay (ELISA) is a widely used technique in immunology and molecular biology that allows the detection and quantification of several targets including proteins, antibodies, and other biological molecules.31 ELISA can also be used for the detection of bacteria in a wide variety of samples by means of antigens or antibodies specifically produced in response to bacterial infections.30 There are multiple approaches to ELISA but all these rely on the same working principle that involves the use of antibodies that present specific regions able to bind to the target antigen and an enzyme that functions as an optical transducer. In the most widely used approach, the sandwich ELISA protocol, a sample containing the target antigen is added to a 96-well plate where the antibody was previously immobilized, allowing for the antigen to bind to the immobilized antibody. A secondary antibody, which is connected to an enzyme, that catalyzes the conversion of a colorless substrate into a colored product, is then added to the well to visually confirm the target presence in the analyzed sample (Figure 3). This assay offers the possibility to detect antigens related to bacteria by using a very small amount of sample, which represents a great advantage of this highly specific technique.32 Moreover, the detection of bacteria via ELISA can be performed in challenging matrices, such as biological or food samples.33
Figure 3.
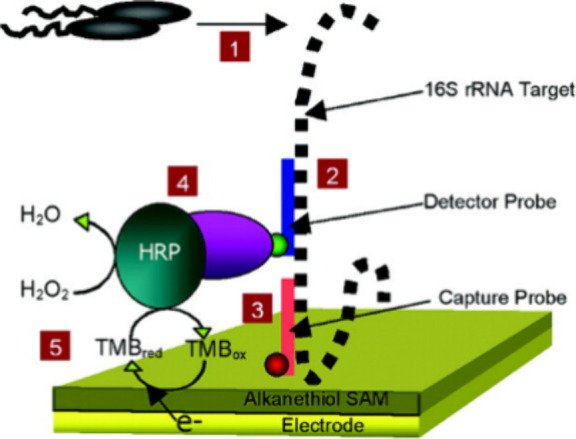
Illustration of the DNA-ELISA method for the detection of bacteria. Reproduced from ref (34). Copyright 2006 American Chemical Society.
The increased versatility and faster throughput time of ELISA have led it to become an increasingly popular technology in microbiology laboratories. It is often used in parallel to traditional culture-based methods to obtain a first, fast answer to a microbiological challenge. The field of ELISA-based bacteria detection is evolving fast, with ready-to-use cartridges coupled to benchtop readout devices such as Bio-Techne’s Ella, enabling automated sample analysis.
The observed trend in this subfield of the market is that most companies try to occupy a specific niche in the market. Abcam for instance, offers monoclonal antibodies that laboratories can implement into their existing ELISA’s. They offer, for instance, antibodies for enterotoxin A and B of S. Aureus. They can develop additional antibodies upon request, tailoring their performance to the application’s needs.43 Other companies such as Biocompare, HycultBiotech, Cygnus Technologies, and Creative Diagnostics tend to provide integrated kits coupled to a readout platform for the fast and ultrasensitive detection of specific bacterial strains.44−52 The main trend in the market, is to gradually miniaturize the technology and try to provide easy-to-use kits and gradually try to work toward PoC versions of the technology.
This is also the main drawback associated with the current state-of-the-art ELISA technologies. The technology is faster but less sensitive and specific than microbiological culturing. It can, therefore, never truly replace it. On the other hand, even the benchtop ELISA devices are still relatively big and expensive. This limits their use in PoC settings and commercial competitiveness with truly low-cost assays such as LFAs. In addition, they suffer from the same stability issues as LFAs and their use in resource-limited settings with e.g. limited temperature control remains challenging. Therefore, the commercial success of ELISA is still mainly confined to analytical and microbiological laboratories where they offer a faster or less expensive alternative to traditional culture or chromatographic approaches.
ATP Sensors
Adenosine triphosphate (ATP) biosensors have emerged as valuable tools for the detection of bacteria, offering a rapid PoC detection method. ATP is a fundamental molecule present in all living cells, including bacteria. ATP-based sensors typically utilize the enzymatic activity of firefly luciferase, which catalyzes the conversion of ATP to produce light.35 Several studies have demonstrated the effectiveness of ATP sensors in detecting dangerous bacteria. For instance, researchers have developed ATP-based assays for the detection of foodborne pathogens, including Salmonella species, Escherichia coli, and Listeria monocytogenes.36 These assays showed high sensitivity and specificity, with detection limits as low as 10–100 colony-forming units per milliliter (CFU/mL).
In addition to food safety applications, ATP sensors have also been employed for the detection of bacterial contaminants in healthcare settings. In one study, an ATP-based rapid hygiene monitoring system was used to detect pathogens, such as methicillin-resistant Staphylococcus aureus (MRSA), in hospital environments.37 The system allowed for a quick assessment of bacterial contamination, facilitating timely intervention and infection control measures. By measuring the emitted luminescence, ATP sensors are able to provide real-time, qualitative, and semiquantitative information about the bacterial load.38 This technology offers several advantages, such as speed, simplicity, and portability, making it particularly suitable for on-site testing, environmental monitoring, and healthcare settings. Furthermore, ATP sensors can be easily integrated with automated systems, facilitating high-throughput screening and analysis. For example, researchers have developed microfluidic platforms that combine ATP sensing with sample processing and analysis, enabling rapid and automated detection of dangerous bacteria.39 The application of ATP sensors in the detection of dangerous bacteria therefore represents a noteworthy tool for improving public health and safety, enabling timely interventions and effective control measures against bacterial infections.
In fact, a wide variety of commercial applications based on ATP-sensing technology are spreading on the market. For instance, Charles River introduced Celsis, a rapid microbial detection system based on ATP monitoring that can detect the presence or absence of microbiological contaminations.40 In addition, the company Hygiena has recently introduced on the market the system MicroSnap, a compact and portable detection and incubation device that allows the detection of microorganisms such as E. coli, Coliforms, or Enterobacteriaceae in less than 8 h.41,42 The EnSURE Touch monitoring platform that can be coupled with the MicroSnap system allows reliable and fast environmental monitoring, thanks to an intuitive luminometer interface with compatibility across several Hygiena test devices (Figure 4– a mock-up of a similar hand-held readout device).
Figure 4.
Artistic representation of a handheld readout device that can currently be used for the rapid sensing of ATP.
The rapid and sensitive nature of ATP-based detection systems allows for quick identification of contaminated samples, reducing the risk of disease outbreaks and transmission. Additionally, the portability and ease of use of ATP sensors make them suitable in resource-limited settings, where access to sophisticated laboratory facilities is limited. To this end, Abcam presented the RubyGlow bacterial assay, a system based on the detection of ATP sensors that can provide an easy and quick method to detect and quantify bacterial cells through the luminescent detection method.43 It is also important to mention that BioThema presented a microbial ATP kit on the market for bacteriological control of liquids, for instance, drinking water, beverages, or beer, but also in pharmaceuticals and cosmetics.44 This system is particularly effective also to determine the antibiotic effects on bacteria since the determination of intracellular ATP in bacterial cells can also prove the efficacy of antibiotic treatments. Moreover, the company Isogen Life Science introduced on the market an ATP assay system that can be used to estimate biomass. In particular, if the amount of ATP per cell is known, consequently the cell number may be estimated.45 The range of ATP detection for this product is 10–12 to 10–6 mol L–1, and it can be used for 200–100 assays depending on the assay volume. The response time is fast (minutes), but the half-life does not exceed 1 h. Numerous other companies offer the possibility of purchasing an ATP kit for the detection of pathogens, including GLbiocontrol, Rapid Microbial Diagnostics, Sigma-Aldrich, Rawlins, Accepta, ThermoFisher Scientific, and many others, allowing this way the detection of contaminations in a wide variety of samples in a limited amount of time.46−48
While the sensitivity and efficacy of the ATP-based sensors are proven by the fact that these devices are the most used biosensor-based devices on the market for bacteria detection, innovations are always investigated due to the drawbacks related to this technology. For instance, when analyzing the PoC use of the technology it is clear that it offers similar advantages as LFAs in terms of being more affordable, user-friendly, rapid, equipment-free, and deliverable than the more expensive lab-based techniques, making them more suitable for PoC application.49 Nonetheless, their specificity and sensitivity are even worse than those of LFAs as they only indicate that there is some organism present in the sample that produces ATP. For most applications, this makes them useless as further analysis using traditional methods would still be necessary to identify the nature of the microbial contamination.50 However, this main drawback is at the same time their main advantage over LFAs. Many hygiene applications require a quick prescreening tool to determine if there is any form of microbial contamination present. The ATP meter is the only technological solution that can provide a PoC answer to this question, which enables it to quickly occupy a nice subfield in the area of microbial sensing.
Polymerase Chain Reaction
The polymerase chain reaction (PCR) is a diffused biological technique based on the amplification of specific DNA sequences of a sample, to allow their analysis and identification. PCR presents a variety of applications, including the detection of bacteria, which has gained more and more importance in fields such as medicine, food safety, and environmental monitoring.51 The working mechanism is based on the amplification of a specific region of DNA, involving three main steps: denaturation, annealing, and extension.52 In the denaturation step, the DNA sample is heated up to separate the two strands of the DNA itself. In the annealing step, DNA primers that are complementary to the chosen DNA strain are added to the sample, giving the possibility to the primer to bind the target sequence, and providing a starting point for the DNA polymerase enzyme to start the amplification. In the extension step, the enzyme adds nucleotides to the primer, thus expanding the DNA strand. The PCR mechanism discussed is then repeated multiple times, and every time the DNA present in the sample doubles. As mentioned, PCR can be used to detect bacteria by targeting unique DNA strains uniquely present in the microorganism of interest.53 Once the DNA has been amplified using the PCR, the presence of the target bacteria can be determined using various methods, for instance with fluorescence probes or electrophoresis
Efforts to accurately detect and identify various pathogens have encouraged innovation in the scientific and diagnostic industries. For instance, ThermoFisher Scientific offers a flexible alternative to accurately detect and identify different types of pathogens.54 In particular, the company introduced a real-time PCR instrument that can expand the testing processes performed in a laboratory thanks to an innovative technology that can also detect slow-growing, difficult-to-cultivate, or uncultivable microorganisms with high sensitivity. Other companies, such as Bio-Rad, Roche, and Qiagen are also pioneers of cutting-edge instrumentations for PCR and real-time PCR applications. Renowned for their commitment to innovation, these industries continually introduce state-of-the-art technologies that not only enhance the precision and sensitivity of PCR assays but also contribute to the growing field of molecular diagnostics. Their comprehensive product portfolios include a wide array of solutions, from high-throughput automated systems to robust multiplexing capabilities, making laboratory-based research faster and more accurate. Moreover, The company Quidel Ortho introduced on the market the Solana Molecular Testing Platform, equipment that combines helicase-dependent amplification (HAD) with fluorescence detection to improve the detection of contaminants in a short time (30 min).55 On the other hand, industries started to develop custom-made approaches for the detection of pathogens through PCR-based alternatives. In particular, the so-called PCR kits have been recently introduced on the market and present an easy-to-use alternative for pathogen detection. An example of this effort is presented by Meizheng, a PerkinElmer company, that officially offers pathogen real-time PCR kits to detect pathogens in food and environmental samples. This kit can detect different bacteria, including Salmonella, Listeria monocytogenes, Bacillus cereus, and E. coli O157.56 However, the samples that can be analyzed do not involve clinical samples. Other companies, including Canvax, Hygiena, Sigma-Aldrich, Invitek Diagnostics, and Biovalley have also introduced in the market PCR kits designed for the detection of pathogens that are compatible with most commercially available PCR detection systems (Figure 5).57−60
Figure 5.
Artistic representation of a PCR Kit for the detection of bacteria with the corresponding legend for results interpretation.
PCR-based bacterial recognition systems are becoming increasingly popular, and industries and academia are investing in these alternatives thanks to their high reliability. Despite the great advantage of its accurate sensitivity and the possibility of obtaining a strain identification for bacteria, PCR can present weaknesses such as the need for qualified personnel to execute the analysis, or the time and costs needed to perform the studies. To date, only a few PoC devices have been developed and are currently monitored before being officially introduced as detection equipment, but the main drawback always remains the cost for the implementation of such devices in multiple locations.61 Therefore, PCR-based technologies mainly attribute their commercial success in this field, as they can provide a faster, almost equally reliable alternative to traditional culture-based methods. However, this limits their use in PoC settings.
In summary, PCR, ELISA, LFAs, and ATP meters have all significantly changed and improved the diagnostic market when it comes to pathogenic detection. They offer faster, sometimes almost equally reliable results and have therefore turned into lab-based analysis tools that are utilized in addition to the traditional culture-based methods. On top of that, they are very versatile and can be applied to multiple analytes. Other technologies have adopted a place in the market due to their ease of use and affordability rather than their performance. LFAs are often used when a fast answer is needed, while ATP meters are used to assess general hygiene levels. Although these technologies serve their purpose, they also have their limitations. Emerging technologies that want to take up their niche in the PoC market should offer an alternative that is equally as good in terms of usability, deliverability, and affordability but at the same time overcomes the drawbacks associated with existing technologies. The limited stability of biological receptors, for instance, limits the use of these devices in resource-limited settings. Besides, niche single-use devices do not tend to be as commercially successful as they rely on very specific needs and uses, and unless they have a huge market request (e.g., glucose sensors) it is unlikely that they meet the market needs.
Emerging Biomimetic Sensor Technologies for Bacteria Detection
In addition to research focusing on the continuous development of already existing biosensor technologies for pathogen detection, new technologies are continuously being developed in academic research. Most of these technologies are well established and many of them are unlikely to reach the market due to limiting factors, often related to scalability. Often biosensor applications also suffer from stability issues relating to the nature of the biological receptors that are inherently unstable when used under nonphysiological conditions, which is a problem for PoC applications, especially in resource-deprived settings.62
Therefore, several interesting emerging technologies are now focusing on developing so-called biomimetic sensor platforms based on synthetic, bioinspired receptors such as aptamers or imprinted polymers. Implementing these receptors into sensor applications will further decrease the cost price of these devices and increase their robustness, making them interesting for PoC applications. In this chapter, we will therefore summarize the most recent advances in this field but also analyze the current market. As these technologies are less established, smaller companies and spin-offs typically occupy the market and champion them. The goal is to identify niche applications to enable market penetration. It is important to identify these trends to come up with recommendations for future research and business development.
Aptamers
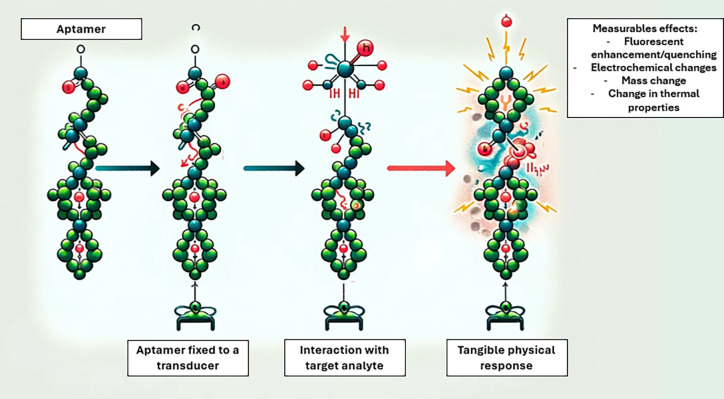
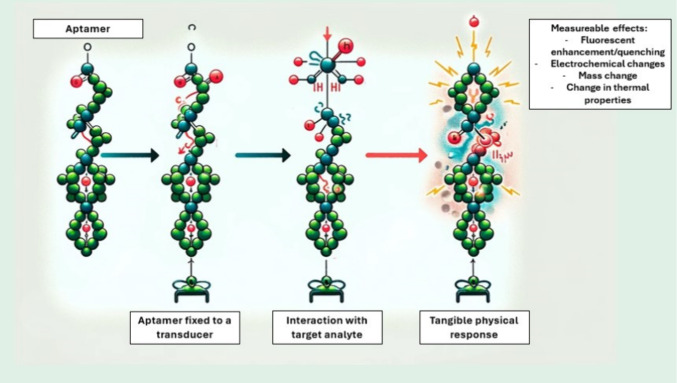
Aptamers are synthetic receptors that have been developed for the detection of bacteria in biomimetic sensor platforms.63 Briefly, aptamers are short single-stranded oligonucleotides capable of binding to a target molecule of interest with high affinity (Figure 6).
Figure 6.
Schematic representation of aptamer–target interaction.
The synthesis of aptamers is based on an iterative selection process called SELEX (systematic evolution of ligands by exponential enrichment). This synthetic process involves the incubation of the target molecule with a nucleic acid library consisting of 1014 - 1015 oligonucleotides. Subsequently, the target-bound oligonucleotide DNA or RNA strands are separated from the unbound strands and then replicated via PCR to start a new pool of nucleic acids. This whole process is repeated for up to 15 cycles thereby increasing the separation accuracy, ensuring that the obtained nucleic acid has the highest possible affinity with the chosen target molecule.64,65 In order to sense Gram-negative and Gram-positive bacteria, these synthetic receptors can be tuned to specifically bind to bacterial proteins, nucleic acids, and other relevant molecules present in the bacterial cell.66 These synthetic recognition elements exhibit several advantages over traditional antibodies for bacteria detection, namely they are smaller in size, have higher specificity and affinity, and can be generated quickly and cost-effectively. Aptamers also have the potential to be used in multiplexed assays, which can allow for the simultaneous detection of multiple bacterial species.67 More critically, aptamers can be produced to detect bacterial antigens that are not accessible to antibodies, opening the possibility for the detection of pathogenic bacterial species that have proteins or other molecular targets that are not accessible on the surface of the bacteria.68
One of the most effective ways to use aptamers for bacterial identification is to implement them as recognition elements in biomimetic sensor platforms analogous to the biosensor platforms previously discussed. Aptasensors can be used with different readout methods, which in general depend on the transducing element chosen, and on the analyte. Such methods include electrochemical,69,70 fluorometric,71 or optical systems.62,72 It is therefore understandable that the use of aptamers has been extensively studied in both laboratory and clinical settings, proving their applicability for fast and reliable bacteria detection. Recent academic studies further explain and demonstrate that aptamers can be used to detect bacterial species such as Escherichia coli,73Staphylococcus aureus,74Pseudomonas aeruginosa,75,76 and Salmonella spp.77
Evidently, aptamers have emerged as promising molecular recognition tools for various applications, including bacteria sensing. An example of the huge application potential of aptasensors is portrayed in a study presenting an aptamer-based colorimetric assay for S. aureus detection.78 In this research, Yu et al. designed an S. aureus-specific aptamer that exhibited high affinity and selectivity toward the target bacteria. The assay utilized a biotin-labeled captured probe anchored on a 96-well plate through a streptavidin–biotin system. The S. aureus-aptamer complex was therein detected through colorimetric readout, and the color released is proportional to the added bacteria concentration. This study exhibited a wide linear range, high specificity, and remarkable sensitivity, with a LoD of 81 CFU mL–1 in PBS buffer solutions. Moreover, the sensor’s potential for the detection of S. aureus in real-life samples was demonstrated by analyzing the colorimetric response in milk samples. The approach is straightforward and can be coupled to a plate reader to provide a high throughput alternative for ELISA, omitting the need for using less stable natural receptors. However, the concept offers a high degree of sensitivity and can therefore also be combined with hand-held readout technologies, such as miniaturized spectrophotometers, for PoC application.79 Similar aptasensor platforms were developed for the detection of other bacterial strains based on electrochemical, gravimetrical, and optical transducer technologies, an overview of which can be found in Table 2.
Table 2. Further Examples of Aptamer-Based Sensors for the Detection of Bacteria.
| Target | Application | Readout | LoD | Linear Range | Reference |
|---|---|---|---|---|---|
| Escherichia coli | Buffer solution | Gravimetric (QCM) | 1.46 × 103 CFU/mL | 102 to 107 CFU/mL | (80) |
| Salmonella enteritidis | Environmental samples | Colorimetric | N/A | 101 to 107 cells | (81) |
| Staphylococcus aureus | Milk, urine, wastewater, tap water, and lake water samples | Colorimetric | 10 CFU/mL | 10 to 106 CFU/mL | (82) |
| Pseudomonas aeruginosa | Serum samples | Electrochemical (EIS) | 33 CFU/mL | 102 to 107 CFU/mL | (83) |
| Legionella pneumophila | Aqueous medium | Optical (SPR) | 104 cells/mL | 104 to 108 CFU/mL | (84) |
The studies summarized in Table 2 illustrate the potential use of aptamers in diagnostic applications, where they can serve as sensitive and specific probes for the detection of various bacteria and other pathogens.85 These results show that aptasensors have the technical potential to become a commercially successful product as several platforms reach low detection limits and can perform in challenging matrices. As mentioned earlier, aptamers offer several advantages over antibodies that might lead them to replace their unstable natural counterparts that require biological hosts for their production.
When considering the commercial viability of the technology, currently, the main limitation for market penetration is the affordability and scalability of their synthesis process, which in most cases has been an academic exercise. However, numerous companies are attempting to upscale the synthesis process and develop large batches of aptamers toward the commercial sensing of bacterial pathogens. Base Pair Biotechnologies is one such company, specializing in the development of aptamer-based solutions for bacterial detection and diagnostics.86 Their platform utilizes aptamers to specifically capture and identify bacteria from complex samples. Likewise, Apto-gen is a company founded in 2019 that is also utilizing aptamers for bacterial detection, and they offer an array of molecular tools to perform this task.87 This vision is also shared by many other companies such as Optimer, BasePair, Bioaptus, Novaptech, and Achiko AG, offering clear examples of the effective implementation of aptamer technology for food production and diagnostics applications.86,88−91
In conclusion, the aptasensor market is currently mainly dominated by companies that are trying to focus on creating industrial processes that allow for the mass production of aptamers. Once that has been achieved, aptamers can start to compete with antibodies in terms of commercial implementation, and technology companies can start implementing them into biosensor production lines. However, several other pitfalls still have to be addressed to achieve market penetration. The stability of aptamers, especially in challenging circumstances would make them theoretically advantageous over their natural counterparts but in practice, they suffer from degradation by nucleases. An issue that needs to be overcome before successful implementation can occur. Even more important is market need, several biosensor technologies have been demonstrated in academic literature but never reached the market because they do not address any pressing market need. Companies that design aptamers should focus first on trying to replace antibodies in LFAs but should also look for applications in which they can occupy a niche in the field similar to ATP meters. These sensors offer the benefit of being able to quantify the amount of target present in a sample (which typical LFAs struggle with) while offering superior selectivity to ATP meters.
Molecularly Imprinted Polymers
An alternative approach to creating synthetic receptors that overcome the drawbacks typically associated with biological affinity reagents lies in the development of so-called Molecularly Imprinted Polymers (MIPs). These polymeric materials are essentially molds formed around a target containing highly specific cavities tailored toward the recognition of an analyte of interest, including molecules related to the metabolism or the quorum sensing of bacteria.92 In addition to unique binding properties, the highly cross-linked polymeric nature of these receptors yields highly desirable physical characteristics (e.g., resistance to high temperature/pH) enabling them to effectively function in challenging PoC environments. MIPs have been effectively coupled with different cost-effective readout technologies, and it is possible to produce them on a large scale in a low-cost manner. Therefore, they are excellent candidates to replace biological recognition elements in PoC detection platforms.93,94
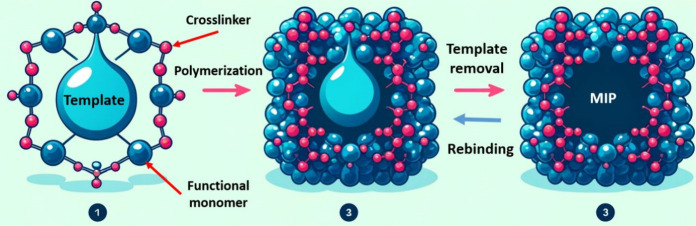
The working principle of these materials relies on the formation of a polymeric network around a template, forming specific cavities that mimic the analyte shape and provide complementary interactions to the chosen target molecule (Figure 7). Generally, this is facilitated by the use of one or more functional monomers and a functional cross-linker dispersed in a porogenic solvent, which is subsequently polymerized by means of a photo/thermal initiator that yields the network. As MIPs intrinsically focus on molecular targets, the most effective technologies developed for applications in this field confirm the presence of bacteria inside a sample by the indirect detection of a specific toxin, metabolite, or autoinducer molecule associated with a specific pathogen of interest.95
Figure 7.
Schematic representation of the concept behind molecularly imprinted polymers.
Once the MIP has been obtained, it can be coupled with a variety of readout methods, including electrochemical,96,97 thermal,98,99 colorimetric,100 chromatographic,101 and piezoelectric.102 The research in the field of bacterial detection has yielded both positive and negative outcomes, showcasing novel technologies with potential while also facing certain limitations. For instance, some studies have focused on the detection of Gram-negative bacteria by targeting quorum-sensing molecules produced by the bacteria, particularly N-acyl-homoserine-lactones (AHLs).103 This approach shows promise in achieving highly specific and sensitive detection, providing a valuable tool for the early identification of bacterial infections. Similarly, another noteworthy advancement involves the indirect detection of Pseudomonas aeruginosa, a notorious pathogen, through its main toxin, pyocyanin.104 The sensor is capable of detecting pyocyanin in biological samples at medically relevant concentrations at a fraction of the cost price of current lab analysis tools that make use of expensive chromatographic techniques to detect this biomarker for P. aeruginosa infection in cystic fibrosis (CF) patients. This sensor could therefore be very interesting as a prescreening tool in CF patients, leading to faster results and a more adequate treatment, prior to meticulous lab analysis. Several other MIP-based sensors have been developed for detecting important toxins secreted by various species of Aspergillus, Byssochlamys, and Penicillium as summarized in Table 3.
Table 3. Further Publications That Utilize MIPs for Bacterial Detection.
| Target | Application | Readout | LoD | Linear Range | Reference |
|---|---|---|---|---|---|
| Listeria monocytogenes | Detection in buffer solutions | Electrochemical (PAD) | 70 CFU/mL | 300 to 6700 CFU/mL | (105) |
| Klebsiella pneumonia | Detection in human urine | Electrochemical (DPV) | 1.352 CFU/mL | 1 to 105 CFU/mL | (70) |
| Staphylococcus aureus | Detection in lettuce and shrimp | Electrochemical (EIS) | 4 CFU/mL | 10 to 107 CFU/mL | (106) |
| Salmonella spp. | Detection in aqueous media | Electrochemical (EIS) | 23 CFU/mL | 103 to 107 CFU/mL | (107) |
| Pseudomonas aeruginosa | Biofilm inhibition | Culture-based | - | - | (108) |
| Pseudomonas aeruginosa | Detection in saliva | Thermal (HTM) | 0.569 ± 0.063 μM (PYO) | 0.05–2 mM (PYO) | (109) |
The results summarized in the table above show that MIPs, very much like aptamers, are able to drive the molecular recognition in relatively simple electrochemical and thermal sensor applications that achieve competitive LoDs in medical, food, and environmental samples. They achieve this by detecting bacterial toxins rather than the entire microbiological entity, which allows them to compete with commercial endotoxin sensors when it comes to performance.
It is for these reasons commercially MIPs are an extremely interesting prospect when it comes to PoC endotoxin (and hence micro-organism) detection. Their low-cost nature is especially interesting in resource-limited settings, where their stability makes them additionally advantageous over natural receptors. However, the main challenge lays in the batch-to-batch variation observed in the traditional bulk polymerization methods. Alternative approaches give producers more control over the production process, but these are more expensive and therefore less desirable. Therefore, the current market in MIP-based PoC applications is currently focusing on creating spin-off companies that aim to create scalable, affordable approaches for creating large batches of homogeneous MIP particles.110 MIP Diagnostics Ltd. was a such spin-off company of Leicester University that is currently focusing on industrial-grade MIP synthesis.111 They have developed a method for the automated production of MIPs in solid-phase extraction (SPE) columns. The method separates high- and low-affinity MIPs from each other, thereby diminishing the batch-to-batch variation. The company has developed MIP-based products for the detection of bacterial pathogens such as Campylobacter spp. and Salmonella spp. in food samples. In 2019, MIP Diagnostics signed a licensing agreement with AgPlus Diagnostics to develop a range of MIP-based lateral flow assays (LFAs) for the detection of bacterial infections.112 The company solely focuses on MIP production and has strategic partnerships with universities all over the world that benchmark the reliability of their MIP products and demonstrate their integration into sensor platforms not only for diagnostics but also for food safety and environmental safety. This has inspired them to rebrand the company in 2022 to MIP Discovery Ltd. In addition to academic spin-offs, other companies such as Biotage and Sigma-Merck are also showing great interest in MIPs with their MIP Technologies subsidiary and Supelco Analytical Products branches, respectively.113
However, this focus on MIP production requires other companies to utilize these synthetic reagents and integrate them into their biosensor platforms instead of antibodies, increasing market penetration and enabling the MIP technology to get a foothold in the market as it becomes more prevalent. The license agreement with AgPlus Diagnostics is a first step in the right direction, but convincing key players in the diagnostic market to do so will require time and a lot of scientific evidence that confirms the commercial benefit of MIPs over their natural counterparts. Therefore, it is of the uttermost importance that academic research efforts continue to explore and understand the working principles of MIPs and demonstrate their potential application in various industrial fields and for different applications. Private-public collaboration will continue to play a vital role in maximizing the chance of penetration into the diagnostic market in the coming decades. Additionally, MIP discovery is still relatively young, and the technology is still in the scaling phase. Current procedures generate MIPs on the mg scale, whereas for true market acceptance, these volumes need to be much larger. This sentiment is also reflected in the price of these receptors, with current pricing proving much more costly than their biological counterparts.
It is essential to address these issues in the coming few years for MIPs to fully achieve their potential and replace natural receptors in biosensing solutions creating sensors that are more affordable and robust than the current PoC biosensor solutions. Their stability will allow them to be used in certain niche applications, particularly in challenging conditions where natural receptors, and probably aptamers, fall short. In addition, MIPs can be tailored to be very selective or, similar to an ATP meter, less selective in the targets they detect. Therefore, they can offer PoC alternatives to both high-end lab devices such as HPLC or ELISA as well as current commercial PoC products that are less specific, qualitative, and display limited robustness.
It is therefore clear that the reason why MIPs are still an emerging technology is because of the inherent inconsistencies with the scaling of the technology. When considering market penetration, usability, and cost, MIP technology is highly competitive and could compete with some of the technologies currently accepted onto the market.
Surface Imprinted Polymers
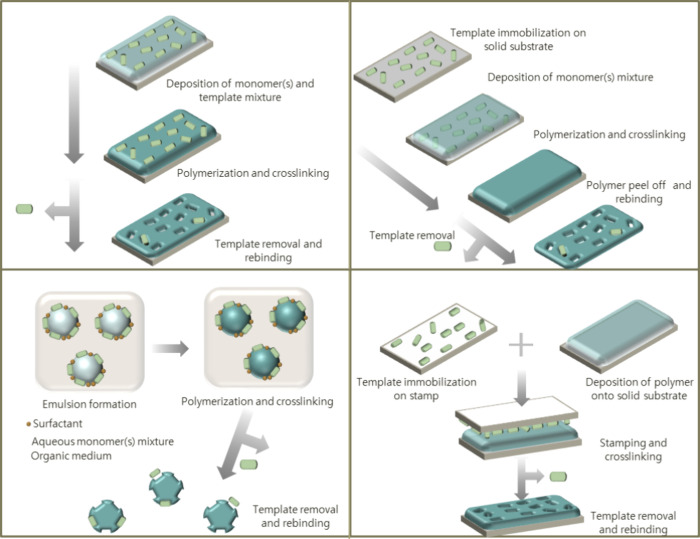
Akin to MIPs, Surface Imprinted Polymers (SIPs) are polymeric-based materials developed and used for a wide range of applications, including the detection of proteins and antibodies, but they are particularly interesting for the detection of cells and microorganisms.114 The working principle of SIPs for bacterial detection relies on the possibility to form whole bacterium cell imprint on the surface of a polymer, in order to allow the bacterium’s shape and functionalities to be captured. The most common way of achieving this to date is microcontact imprinting (stamping), where the cells are introduced to the softened polymer and physically forced into the surface. Nonetheless, alternative approaches have been developed in recent years, for instance, the interfacial imprinting method.115 This approach entails the deposition of a drop of buffer solution containing the target bacteria on a polymeric matrix, allowing this way the bacteria to deposit in the polymeric layer, thus imprinting their functionalities (Figure 8). This novel approach allows to achieve of a SIP without the need for a template-immobilized substrate, thus yielding a straightforward and more reproducible SIP platform. The fabricated biomimetic platform showed excellent sensitivity (LoD = 80 ± 10 CFU/mL) and specificity toward E. coli. Moreover, the potential of this biomimetic platform in detecting E. coli contaminations in food samples was demonstrated by analyzing the response in fruit juice samples.
Figure 8.
Schematic representation of different methods for the preparation of SIPs. Reproduced from ref (116). Copyright 2021 American Chemical Society.
Once the bacteria are imprinted on the surface, they can be washed away in order to leave microcavities able to selectively detect the same bacteria inside a sample.117 This washing step is usually performed using a medium that is able to denature the template bacteria, thereby breaking the bonds between them and the polymer. For this purpose, various solvents have been used ranging from hot water to ethanol, methanol, or a sodium dodecyl sulfate (SDS) solution. Once the SIPs are synthesized, they can be coupled with a variance of read-out methods, including electrochemical, optical, or thermal methods for the creation of sensors that are able to directly detect a wide range of pathogenic bacteria (Table 4).118,119 All these sensors allow for the direct identification of bacterial strains and quantification of the bacterial load in liquid samples. As these devices are typically very easy to use, deliver fast results, and are very cheap, they have a lot of potential for PoC bacteria detection when compared to the gold-standard culture-based methods.
Table 4. Further Publications That Utilize SIPs and Their Associated Readout Technologies for Bacterial Detection.
| Target | Application | Readout | LoD | Linear Range | Imprinting method | Reference |
|---|---|---|---|---|---|---|
| Klebsiella pneumoniae | Detection in hospitals | Electrochemical (CV) | 0.012 CFU/mL | 101–105 CFU/mL | Self-assembly | (120) |
| Escherichia coli | Detection in milk | Thermal and Electrochemical (HTM and EIS) | 1070 CFU/mL (HTM) and 120 CFU/mL (EIS) | 102–105 CFU/mL | Microcontact imprinting | (121) |
| Listeria monocytogenes | Detection in milk and pork | Optical | 103 CFU/mL | 103–105 CFU/mL | Pickering emulsion | (122) |
| Staphylococcus aureus | Detection in milk and rice | Optical | 103 CFU/mL | 102–106 CFU/mL | Imprinting on magnetic particles | (123) |
| Escherichia coli | Detection in water | Plate culturing | 103 CFU/mL | 103–104 CFU/mL | Bulk polymerization | (124) |
Considering the potential of this technology from a commercial perspective, the drawbacks of SIPs are very much parallel to that of MIPs. Likewise, it is therefore understandable why there are companies actively pursuing the development of industrial processes to mass produce homogeneous batches of SIPs. In contrast to the MIP-based LFAs being developed by MIP discovery, companies such as the Dutch SME Sensip Dx B.V. are seeking to detect bacteria directly utilizing SIPs. This spin-off from Maastricht University aims to couple SIP technology with their proprietary thermal sensing technology (coined the “Heat-Transfer Method” (HTM)) for the rapid analysis of bacterial contaminations of biological and food samples.125 A lot like MIP Discovery, Sensip Dx is currently focusing on the scalability of their receptors, mainly by investigating stamp-free production methods in the short term themselves, while focusing on the promise of template-free methods in the longer run by partnering up with various universities.
The added benefit is that they are working in parallel on their direct integration into sensing platforms, which makes them independent of major diagnostic companies and allows them to create industrial prototypes to attract market attention.126 As they are one of the few companies working on the scaling and synthesis of SIPs, alongside working on their integration, this puts them in a unique position where they can be genuinely unique and lead the advancements in the field. The downside to this approach is that all sensor and SIP development is in-house, leading to long development times and a limited team working on the SIPs progression. This said, if successful, this SIP-based sensing technology has the potential to generate a huge impact and change the face of bacterial detection in numerous fields. The main bottleneck, however, like with MIPs, remains that SIP materials also show high heterogeneity, and thus newer optimized imprinting techniques need to be established to address some of the currently associated drawbacks. Currently, there is limited control over imprint distribution, depth, and reproducibility. Tackling all these issues is tedious, but Sensip Dx states that they will have a SIP-based product on the market within the coming few years due to their unique position in the market, the adaptability of their sensor strategy, and their continuous focus on improving SIP production and integration in sensing technologies.
SIP-based sensing is the youngest alternative of all the proposed biomimetic alternative receptor technologies and is therefore clearly the furthest away from the market. However, the field can benefit from the same biosensor market lessons; they have a unique selling point in that they target the whole bacterium. This has drawbacks when comparing it to the ATP sensor, but the selectivity can also be lowered to create a more generic bacterial detection platform. On the other hand, SIP-based sensors can also be aimed at very selectively targeting specific pathogenic strains rather than providing a false positive signal when a harmless species is encountered. This illustrates that these sensors could have their own set of niche applications in the very broad field of bio(mimetic) bacteria detection. It is important to work on improving the mechanical part of the technology but at the same time evaluate where this technology can be optimally positioned in the market.
Synthetic receptors represent a novel frontier in biosensing, offering distinct advantages over natural counterparts, particularly in robustness and scalability in challenging environments, such as extremely warm or cold countries or zones where it is hard to properly store pathogen detection kits. Furthermore, their tunable properties allow for precise customization, which is still a challenge for natural receptors that can be stable only in specific conditions of temperature, pressure, or pH. The durability of synthetic receptors ensures extended operational lifetimes, crucial for maintaining consistency in industrial processes. Finally, reduced environmental impact thanks to straightforward production processes and scalability are an additional benefit. The optimization of production processes contributes to a more sustainable approach compared to the extraction or production of natural receptors which need to be performed in controlled and qualified environments, thus aligning with the growing emphasis on environmental-friendly practices.
Recent Innovations in Other Types of Biomimetic Sensors for Bacterial Detection
Due to the increasing need for the identification of infected samples in a wide variety of environments, novel technologies have been introduced to present preliminary proofs of application for the detection of pathogens. In this section, it is possible to identify interesting research based on Metal–Organic Frameworks (MOFs).127 These structures present a novel sensing approach, offering tailored receptivity toward specific targets, akin to Molecularly Imprinted Polymers (MIPs). The technology of MOFs lies on a specific synthetic pathway: a template-driven polymeric network formation, able to form cavities that mirror the target analyte’s geometry and promote specific interactions.128 It was also shown that it is possible to couple MOFs and MIPs technologies for the efficient detection of bacteria.129 On the other hand, MOFs can also be coupled with aptamers to enhance their sensitivity to detect bacteria like S. aureus or Acinetobacter baumannii reaching a very low limit of detection in the range of 5–7 CFU/mL.130,131 By encapsulating target molecules within their framework, MOFs have highly precise cavities, finely tuned for recognizing key bacterial elements. Moreover, their robust polymeric composition confers good stability to harsh environmental conditions, rendering them adept for Point-of-Care (PoC) applications. MOFs seamlessly integrate with diverse, cost-effective readout technologies, promising scalability at an economical cost.132 Another innovation for the rapid and sensitive detection of bacteria is presented by Bacterial Rapid Detection using Optical Scattering Technology (BARDOT), namely a method that couples the culture-based detection methods with imaging performed through a laser, thus able to discriminate the presence of bacteria, and also understanding specific information about the colonies present within the sample.133 Applications of this technology have shown to be effective for the detection of bacteria like Salmonella species or Pseudomonas aeruginosa.134,135 Moreover, the high interest in finding novel portable alternatives for the detection of microorganisms lead to the miniaturization of BARDOT-based technologies, leading this way to portable alternatives such as adaptations in Lab-on-A-Chip format and other point-of-care systems.136,137
Overall, all the emerging biomimetic sensors including the technologies based on aptamers, molecularly imprinted polymers, surface imprinted polymers, metal–organic-frameworks, and others, besides being very well-studied alternatives, still did not fully reach the market. In particular, the spin-off companies mentioned in this chapter and the academic groups focused on the engineering of biomimetic sensors are still studying better alternatives to overcome the most urgent problems related to these technologies, for instance, the fact that they suffer from mass production and heterogeneity manufacturing issues. However, compared to immunosensors and enzymatic biosensors, this category of technologies is still considered relatively new, and it could not take advantage of decades of research on natural receptors and continuous improvements as the better-established technologies had since their market introduction 50 years ago. Synthetic receptors have been used for about 30 years in the sensing field and the prior knowledge they build on was less developed, besides, the market competition generated by the well-established biosensors is a factor that can slow down the development of new technologies as the latter cannot take advantage of the urgent demand of the product.
Future Outlook and Conclusions
The literature study performed in this review elucidates that application potential does not automatically result in market penetration. New technologies have to address an existing demand for which no adequate solution exists, or they should at least offer significant benefits over already existing technologies used in the field. In a field as big as bacterial detection, that finds its applications in medical care, food safety, or environmental screening, this often results in technologies occupying niche subfields. Alternatively, different technologies having their limitations and advantages are often used in parallel so they can complement each other.
For applications where a full bacterial identification profile and quantification are necessary, culture-based methods are still considered the gold standard. It is possible to collect virtually any sample, solid or liquid, and plate them in a lab to identify strains, bacterial loads, and even antibiotic resistance. However, this process does not allow for PoC diagnosis or on-site screening as it is slow, time-consuming, and expensive. Therefore, faster lab-based tests have been employed in addition to culture-based techniques, for instance, complementary ELISA tests, to treat a patient with a potential infection or to throw away food products that are potentially contaminated. Likewise, LFAs have been used to determine whether a specific bacterial strain might be present in a sample and ATP sensors are used to detect general contamination of products that need to be sterile. However, when a product or a sample always contains a background microbial load, biosensors that can quickly quantify specific bacterial strains could occupy a niche market. Biosensors based on natural receptors that allow for quantification have shown to be able to offer a solution, but they lack the stability needed to perform under challenging conditions, such as temperature in low-income countries, in-line measurements in environmental or industrial applications, and others.
The study in this paper demonstrates that sensors based on MIPs, SIPs, and aptamers have been shown to bring highly interesting alternatives in academic studies in recent years. These receptors are a lot more stable and can be constructed synthetically for nearly every target. In addition, they can be coupled to the same transducer/readout methods as their natural counterparts. However, to date, there are just a few biomimetic sensing platforms on the market based on these receptors. The reason is that these technologies are relatively new when compared to natural receptors, especially when it comes to valorization and validation studies. Their production process needs to be optimized to be able to create large batches of receptors that do not suffer from batch-to-batch variations. Both academia and industry are in the process of addressing the issue by focusing on the production of these receptors to bridge the gap with antibodies and enzymes. These novel alternatives can bridge the gap and even outperform their natural counterparts as they are purely synthetic and therefore do not always require living organisms for their production and have outstanding robustness. At the same time, these new MIPs need to be implemented into sensor platforms to demonstrate the superiority of these receptors.138 Therefore, the strategy to collaborate with both academic and industrial partners specialized in sensor engineering seems to be logical. The field is at a crucial crossroads and advancements in the coming decade or two will determine if biomimetic sensing will claim its commercial niche in pathogen sensing or remain just an academic exercise.
Acknowledgments
This work was supported by the European Regional Development Fund through the AgrEU food project, funded by the Interreg VA Flanders-The Netherlands program, CCI grant no. 2014TC16RFCB046.
Glossary
Vocabulary Section
- Point of care (PoC)
Device that refers to immediate diagnostic equipment that can be directly used on site by the patient, thus allowing rapid diagnosis and a fast decision-making process.
- Transducer
A component that can convert the binding event of a specific target into a measurable signal.
- Polymeric network
A structure formed by interconnected polymer chains that create three-dimensional networks. These networks influence the mechanical properties of the material, for instance, its strength, elasticity, or resistance to harsh conditions.
- Receptor
A dedicated structure that senses and responds to specific signals. It plays a key role in transmitting information and initiating the detection process.
- Pathogen
A microorganism, like a virus or a bacterium, that causes diseases by invading and reproducing within a host organism, leading to adverse health effects.
- Detection
The process of identification or discovery of an entity, such as a molecule or a microorganism, often involving the recognition of signals generated by the contact between the entity and the receptor.
The authors declare no competing financial interest.
References
- Ramírez-Arcos S.; Goldman M. Bacterial Contamination. Pract. Transfus. Med. Sixth Ed. 2022, 221–228. 10.1002/9781119665885.ch18. [DOI] [Google Scholar]
- Pathogenic Bacteria - Database - GlobalRPH. https://globalrph.com/bacteria/ (accessed 2024-01-10).
- Foodborne Germs and Illnesses | CDC. https://www.cdc.gov/foodsafety/foodborne-germs.html (accessed 2023-07-20).
- Dadgostar P.Antimicrobial Resistance: Implications and Costs. Infection and Drug Resistance; Dove Medical Press Ltd., 2019; pp 3903–3910. 10.2147/IDR.S234610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y.; Ye C.; Zhang C.; Zhang G.; Hu H.; Zhang Z.; Fang H.; Zheng J.; Liu H. Simultaneous Detection of Multiple Foodborne Bacteria by Loop-Mediated Isothermal Amplification on a Microfluidic Chip through Colorimetric and Fluorescent Assay. Food Control 2022, 134, 108694 10.1016/j.foodcont.2021.108694. [DOI] [Google Scholar]
- Ali Q.; Zheng H.; Rao M. J.; Ali M.; Hussain A.; Saleem M. H.; Nehela Y.; Sohail M. A.; Ahmed A. M.; Kubar K. A.; Ali S.; Usman K.; Manghwar H.; Zhou L. Advances, Limitations, and Prospects of Biosensing Technology for Detecting Phytopathogenic Bacteria. Chemosphere 2022, 296, 133773 10.1016/j.chemosphere.2022.133773. [DOI] [PubMed] [Google Scholar]
- Petrovszki D.; Valkai S.; Gora E.; Tanner M.; Bányai A.; Fürjes P.; Dér A. An Integrated Electro-Optical Biosensor System for Rapid, Low-Cost Detection of Bacteria. Microelectron. Eng. 2021, 239–240, 111523. 10.1016/j.mee.2021.111523. [DOI] [Google Scholar]
- Mazur F.; Tjandra A. D.; Zhou Y.; Gao Y.; Chandrawati R. Paper-Based Sensors for Bacteria Detection. Nat. Rev. Bioeng. 2023, 1 (3), 180–192. 10.1038/s44222-023-00024-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.; Sharma R.; Bhawna; Gupta A.; Singh P.; Kalia S.; Thakur P.; Kumar V. Prospects of Biosensors Based on Functionalized and Nanostructured Solitary Materials: Detection of Viral Infections and Other Risks. ACS Omega 2022, 7 (26), 22073–22088. 10.1021/acsomega.2c01033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClements J.; Bar L.; Singla P.; Canfarotta F.; Thomson A.; Czulak J.; Johnson R. E.; Crapnell R. D.; Banks C. E.; Payne B.; Seyedin S.; Losada-Pérez P.; Peeters M. Molecularly Imprinted Polymer Nanoparticles Enable Rapid, Reliable, and Robust Point-of-Care Thermal Detection of SARS-CoV-2. ACS Sensors 2022, 7 (4), 1122–1131. 10.1021/acssensors.2c00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çimen D.; Bereli N.; Denizli A. Advanced Plasmonic Nanosensors for Monitoring of Environmental Pollutants. Curr. Anal. Chem. 2023, 19 (1), 2–17. 10.2174/1573411018666220618155324. [DOI] [Google Scholar]
- Pilvenyte G.; Ratautaite V.; Boguzaite R.; Ramanavicius S.; Chen C. F.; Viter R.; Ramanavicius A. Molecularly Imprinted Polymer-Based Electrochemical Sensors for the Diagnosis of Infectious Diseases. Biosens. 2023, Vol. 13, Page 620 2023, 13 (6), 620. 10.3390/bios13060620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. F.; Zhao X.; Yang Z. Aptasensors for the Detection of Infectious Pathogens: Design Strategies and Point-of-Care Testing. Mikrochim. Acta 2022, 189 (12), 3. 10.1007/s00604-022-05533-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.; Zhang W.; Yin Y.; Fang W.; Xue H. Metal-Organic Framework-Based Sensors for the Detection of Toxins and Foodborne Pathogens. Food Control 2022, 133, 108684 10.1016/j.foodcont.2021.108684. [DOI] [Google Scholar]
- Bhunia A. K.; Singh A. K.; Parker K.; Applegate B. M. Petri-Plate, Bacteria, and Laser Optical Scattering Sensor. Front. Cell. Infect. Microbiol. 2022, 12, 1087074 10.3389/fcimb.2022.1087074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. P.; Erb-Downward J. R.; Prescott H. C.; Martinez F. J.; Curtis J. L.; Lama V. N.; Huffnagle G. B. Analysis of Culture-Dependent versus Culture-Independent Techniques for Identification of Bacteria in Clinically Obtained Bronchoalveolar Lavage Fluid. J. Clin. Microbiol. 2014, 52 (10), 3605–3613. 10.1128/JCM.01028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicle Y.; Karamese M. Biosensors for the Detection of Pathogenic Bacteria: Current Status and Future Perspectives 2024, 19, 281–291. 10.2217/fmb-2023-0182. [DOI] [PubMed] [Google Scholar]
- Tominaga T. Rapid Detection of Coliform Bacteria Using a Lateral Flow Test Strip Assay. J. Microbiol. Methods 2019, 160, 29–35. 10.1016/j.mimet.2019.03.013. [DOI] [PubMed] [Google Scholar]
- Sohrabi H.; Majidi M. R.; Khaki P.; Jahanban-Esfahlan A.; de la Guardia M.; Mokhtarzadeh A. State of the Art: Lateral Flow Assays toward the Point-of-Care Foodborne Pathogenic Bacteria Detection in Food Samples. Compr. Rev. Food Sci. Food Saf. 2022, 21 (2), 1868–1912. 10.1111/1541-4337.12913. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Xu T.; Wang W.; Wen Y.; Li K.; Qian L.; Zhang X.; Liu G. Lateral Flow Biosensors Based on the Use of Micro- and Nanomaterials: A Review on Recent Developments. Microchim. Acta 2020, 10.1007/s00604-019-3822-x. [DOI] [PubMed] [Google Scholar]
- Batra A. R.; Dike C. C.; Mantri N.; Ball A. S. Recombinase Polymerase Amplification-Lateral Flow Assay (RPA-LFA) as a Rapid and Sensitive Test for Escherichia Coli O157:H7 Detection in Food and Beverage: A Comparative Study. Food Control 2024, 155, 110076 10.1016/j.foodcont.2023.110076. [DOI] [Google Scholar]
- Srisrattakarn A.; Charoensri N.; Prompipak J.; Ouancharee W.; Saiboonjan B.; Tippayawat P.; Chanawong A.; Wonglakorn L.; Kanwattanee E.; Piyapatthanakul S.; Masmalai T.; Ariyapim A.; Kendal R. P.; Lulitanond A. Rapid Detection of Staphylococcus Aureus in Blood Culture Samples Using Human IgG-Based Lateral Flow Assay. Microbiol. Spectr. 2024, 10.1128/spectrum.03046-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabi H.; Majidi M. R.; Fakhraei M.; Jahanban-Esfahlan A.; Hejazi M.; Oroojalian F.; Baradaran B.; Tohidast M.; Guardia M. de la; Mokhtarzadeh A. Lateral Flow Assays (LFA) for Detection of Pathogenic Bacteria: A Small Point-of-Care Platform for Diagnosis of Human Infectious Diseases. Talanta 2022, 243, 123330 10.1016/j.talanta.2022.123330. [DOI] [PubMed] [Google Scholar]
- Song C.; Liu C.; Wu S.; Li H.; Guo H.; Yang B.; Qiu S.; Li J.; Liu L.; Zeng H.; Zhai X.; Liu Q. Development of a Lateral Flow Colloidal Gold Immunoassay Strip for the Simultaneous Detection of Shigella Boydii and Escherichia Coli O157: H7 in Bread, Milk and Jelly Samples. Food Control 2016, 59, 345–351. 10.1016/j.foodcont.2015.06.012. [DOI] [Google Scholar]
- Song C.; Liu C.; Wu S.; Li H.; Guo H.; Yang B.; Qiu S.; Li J.; Liu L.; Zeng H.; Zhai X.; Liu Q. Development of a Lateral Flow Colloidal Gold Immunoassay Strip for the Simultaneous Detection of Shigella Boydii and Escherichia Coli O157:H7 in Bread, Milk and Jelly Samples. Food Control 2016, 59, 345–351. 10.1016/j.foodcont.2015.06.012. [DOI] [Google Scholar]
- Hydrosense. Hydrosense Testing System.
- InvivoGen: Reagents and Tools for Cell Biology Research. https://www.invivogen.com/ (accessed 2023-10-09).
- Cytiva. https://www.cytivalifesciences.com/en/nl (accessed 2023-10-09).
- Land K. J.; Boeras D. I.; Chen X. S.; Ramsay A. R.; Peeling R. W. REASSURED Diagnostics to Inform Disease Control Strategies, Strengthen Health Systems and Improve Patient Outcomes. Nat. Microbiol. 2019, 4 (1), 46–54. 10.1038/s41564-018-0295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodashenas S.; Khalili S.; Forouzandeh Moghadam M. A Cell ELISA Based Method for Exosome Detection in Diagnostic and Therapeutic Applications. Biotechnol. Lett. 2019, 41 (4–5), 523–531. 10.1007/s10529-019-02667-5. [DOI] [PubMed] [Google Scholar]
- Iha K.; Inada M.; Kawada N.; Nakaishi K.; Watabe S.; Tan Y. H.; Shen C.; Ke L. Y.; Yoshimura T.; Ito E.. Ultrasensitive ELISA Developed for Diagnosis. Diagnostics; MDPI AG, September 1, 2019. 10.3390/diagnostics9030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W.; Zhu L.; Xu H.; Tang Q.; Ma Y.; Chou S. H.; He J. Bio-Hybrid Nanoarchitectonics of Nanoflower-Based ELISA Method for the Detection of Staphylococcus Aureus. Sensors Actuators B Chem. 2022, 366, 132005 10.1016/j.snb.2022.132005. [DOI] [Google Scholar]
- Wu L.; Li G.; Xu X.; Zhu L.; Huang R.; Chen X. Application of Nano-ELISA in Food Analysis: Recent Advances and Challenges. TrAC Trends Anal. Chem. 2019, 113, 140–156. 10.1016/j.trac.2019.02.002. [DOI] [Google Scholar]
- Liao J. C.; Mastali M.; Gau V.; Suchard M. A.; Møller A. K.; Bruckner D. A.; Babbitt J. T.; Li Y.; Gornbein J.; Landaw E. M.; McCabe E. R. B.; Churchill B. M.; Haake D. A. Use of Electrochemical DNA Biosensors for Rapid Molecular Identification of Uropathogens in Clinical Urine Specimens. J. Clin. Microbiol. 2006, 44 (2), 561–570. 10.1128/JCM.44.2.561-570.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Pérez-Cejuela H.; Calabretta M. M.; Bocci V.; D’Elia M.; Michelini E. Super-Stable Metal–Organic Framework (MOF)/Luciferase Paper-Sensing Platform for Rapid ATP Detection. Biosensors 2023, 13 (4), 451. 10.3390/bios13040451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Cao C.; Bi W.; Lin J.; Tan L.; Gan N. On-Site Enrichment and Detection of Live Salmonella Typhimurium Using a Bioluminescence Sensor Coupled with a Hyperbranched Aptamer Probe-Labelled Stir-Bars Array. Sensors Actuators B Chem. 2022, 364, 131862 10.1016/j.snb.2022.131862. [DOI] [Google Scholar]
- van Arkel A.; Willemsen I.; Kluytmans J. The Correlation between ATP Measurement and Microbial Contamination of Inanimate Surfaces. Antimicrob. Resist. Infect. Control 2021, 10 (1), 1–5. 10.1186/s13756-021-00981-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A.; Jiao T.; Ali S.; Xu Y.; Ouyang Q.; Chen Q. SERS Sensors Based on Aptamer-Gated Mesoporous Silica Nanoparticles for Quantitative Detection of Staphylococcus Aureus with Signal Molecular Release. Anal. Chem. 2021, 93 (28), 9788–9796. 10.1021/acs.analchem.1c01280. [DOI] [PubMed] [Google Scholar]
- Chen C.; Hong W. Recent Development of Rapid Antimicrobial Susceptibility Testing Methods through Metabolic Profiling of Bacteria. Antibiotics 2021, 10 (3), 311. 10.3390/antibiotics10030311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapid Microbial Detection | Charles River. https://www.criver.com/products-services/qc-microbial-solutions/microbial-detection (accessed 2023-12-05).
- MicroSnap by Hygiena - Indicator Organism Test. https://campaigns.hygiena.com/acton/media/38777/microsnap-indicator-organism-test?matchtype=b&device=c&keyword=bacteriatestkit&network (accessed 2023-12-05).
- EnSURE Touch | Hygiena. https://www.hygiena.com/hygiene-monitoring/atp-cleaning-verification/ensure-touch (accessed 2024-01-10).
- RubyGlowTM Bacterial assay (Luminescent) (ab189820) | Abcam. https://www.abcam.com/en-nl/products/chip-kits/rubyglow-bacterial-assay-luminescent-ab189820 (accessed 2023-12-05).
- Microbial ATP Kit HS - BioThema. https://biothema.com/shop/kits/microbial-atp-kit-hs/ (accessed 2023-12-06).
- ATP Kits - Isogen Lifescience. https://isogen-lifescience.com/products/reagents-and-kits/atp-kits (accessed 2023-12-06).
- ATP Determination Kit, 200–1,000 assays. https://www.thermofisher.com/order/catalog/product/A22066 (accessed 2023-12-06).
- ATP test kit for microbial analysis of surfaces - GL Biocontrol. https://www.gl-biocontrol.com/en/produits/atp-test-kit-for-microbial-analysis-of-surfaces/ (accessed 2023-12-06).
- High-Accuracy ATP Testing Kits | Quick and Reliable Results - Rawlins. https://www.rawlins.co.uk/cleaning-equipment/atp-testing-kit.html (accessed 2023-12-06).
- Calabretta M. M.; Álvarez-Diduk R.; Michelini E.; Roda A.; Merkoçi A. Nano-Lantern on Paper for Smartphone-Based ATP Detection. Biosens. Bioelectron. 2020, 150, 111902. 10.1016/j.bios.2019.111902. [DOI] [PubMed] [Google Scholar]
- Eed H. R.; Abdel-Kader N. S.; El Tahan M. H.; Dai T.; Amin R. Bioluminescence-Sensing Assay for Microbial Growth Recognition. J. Sensors 2016, 2016, 1. 10.1155/2016/1492467. [DOI] [Google Scholar]
- Zhu H.; Zhang H.; Xu Y.; Laššáková S.; Korabečná M.; Neužil P.. PCR Past, Present and Future. BioTechniques; Future Science Ltd., October 1, 2020; pp 317–325. 10.2144/BTN-2020-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralik P.; Ricchi M.. A Basic Guide to Real Time PCR in Microbial Diagnostics: Definitions, Parameters, and Everything. Frontiers in Microbiology; Frontiers Research Foundation, February 2, 2017. 10.3389/fmicb.2017.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toze S. PCR and the Detection of Microbial Pathogens in Water and Wastewater. Water Res. 1999, 33 (17), 3545–3556. 10.1016/S0043-1354(99)00071-8. [DOI] [Google Scholar]
- Infectious Disease Research Solutions | Thermo Fisher Scientific - NL. https://www.thermofisher.com/nl/en/home/clinical/clinical-genomics/pathogen-detection-solutions/infectious-disease-research-solutions (accessed 2023-12-11).
- Solana Molecular Testing Platform | QuidelOrtho. https://www.quidelortho.com/global/en/products/solana-molecular-testing-platform (accessed 2024-04-17).
- pathogen Real-Time PCR Kits. https://mzfoodtest.com/product-category/analytes/microorganism/real-time-pcr/pathogen-real-time-pcr/ (accessed 2023-12-11).
- PCR Detection Kits | Canvax. https://www.canvaxbiotech.com/product-category/pcr-detection-kits/ (accessed 2023-12-11).
- Pathogen Detection | Hygiena. https://www.hygiena.com/food-safety/pathogen-detection (accessed 2023-12-11).
- Foodborne Pathogen Detection | Invitek Diagnostics. https://www.invitek.com/en/foodborne-pathogen-detection (accessed 2023-12-11).
- Cell Culture Contamination/Bacteria Detection/Onar Bacteria. https://minervabiolabs.us/bacteria-detection-kits/1219-onar-bacteria.html (accessed 2024-01-10).
- Jiang Y.; Li B.; Wu W. Application of Automatic Feedback Photographing by Portable Smartphone in PCR. Sensors Actuators, B Chem. 2019, 298, 126782 10.1016/j.snb.2019.126782. [DOI] [Google Scholar]
- Chen Q.; Gao R.; Jia L. Enhancement of the Peroxidase-like Activity of Aptamers Modified Gold Nanoclusters by Bacteria for Colorimetric Detection of Salmonella Typhimurium. Talanta 2021, 221, 121476. 10.1016/j.talanta.2020.121476. [DOI] [PubMed] [Google Scholar]
- Li D.; Liu L.; Huang Q.; Tong T.; Zhou Y.; Li Z.; Bai Q.; Liang H.; Chen L. Recent Advances on Aptamer-Based Biosensors for Detection of Pathogenic Bacteria. World J. Microbiol. Biotechnol. 2021, 10.1007/s11274-021-03002-9. [DOI] [PubMed] [Google Scholar]
- Stefan G.; Hosu O.; De Wael K.; Lobo-Castañón M. J.; Cristea C. Aptamers in Biomedicine: Selection Strategies and Recent Advances. Electrochim. Acta 2021, 376, 137994 10.1016/j.electacta.2021.137994. [DOI] [Google Scholar]
- Kong H. Y.; Byun J. Nucleic Acid Aptamers: New Methods for Selection, Stabilization, and Application in Biomedical Science. Biomol. Ther. 2013, 21 (6), 423–434. 10.4062/biomolther.2013.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C.; Gao X.; Chen Y.; Ren J.; Liu C. Aptamer-Based Lateral Flow Test Strip for the Simultaneous Detection of Salmonella Typhimurium, Escherichia Coli O157:H7 and Staphylococcus Aureus. Anal. Lett. 2020, 53, 646–659. 10.1080/00032719.2019.1663528. [DOI] [Google Scholar]
- Trunzo N. E.; Hong K. L. Recent Progress in the Identification of Aptamers against Bacterial Origins and Their Diagnostic Applications. Int. J. Mol. Sci. 2020, 21 (14), 5074. 10.3390/ijms21145074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. W.; Wang H. X.; Jia G. C.; Li Z. Application of Aptamer-Based Biosensor for Rapid Detection of Pathogenic Escherichia Coli. Sensors (Switzerland) 2018, 18, 2518. 10.3390/s18082518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Mohamed M. A.; Vinu Mohan A. M.; Zhu Z.; Sharma V.; Mishra G. K.; Mishra R. K. Application of Electrochemical Aptasensors toward Clinical Diagnostics, Food, and Environmental Monitoring: Review. Sensors (Switzerland) 2019, 19, 5435. 10.3390/s19245435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R.; Lakshmi G. B. V. S.; Kumar A.; Solanki P. Polypyrrole Based Molecularly Imprinted Polymer Platform for Klebsiella Pneumonia Detection. ECS Sensors Plus 2022, 1 (1), 010603 10.1149/2754-2726/ac612c. [DOI] [Google Scholar]
- Chinnappan R.; AlAmer S.; Eissa S.; Rahamn A. A.; Abu Salah K. M.; Zourob M. Fluorometric Graphene Oxide-Based Detection of Salmonella Enteritis Using a Truncated DNA Aptamer. Microchim. Acta 2018, 10.1007/s00604-017-2601-9. [DOI] [PubMed] [Google Scholar]
- Kim S. H.; Lee J.; Lee B. H.; Song C. S.; Gu M. B. Specific Detection of Avian Influenza H5N2 Whole Virus Particles on Lateral Flow Strips Using a Pair of Sandwich-Type Aptamers. Biosens. Bioelectron. 2019, 134, 123–129. 10.1016/j.bios.2019.03.061. [DOI] [PubMed] [Google Scholar]
- Gupta R.; Kumar A.; Kumar S.; Pinnaka A. K.; Singhal N. K. Naked Eye Colorimetric Detection of Escherichia Coli Using Aptamer Conjugated Graphene Oxide Enclosed Gold Nanoparticles. Sensors Actuators, B Chem. 2021, 329, 129100. 10.1016/j.snb.2020.129100. [DOI] [Google Scholar]
- Huang Z.; Yu X.; Yang Q.; Zhao Y.; Wu W. Aptasensors for Staphylococcus Aureus Risk Assessment in Food. Frontiers in Microbiology 2021, 10.3389/fmicb.2021.714265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X.; Zhang J.; He F. A New Aptamer/Polyadenylated DNA Interdigitated Gold Electrode Piezoelectric Sensor for Rapid Detection of Pseudomonas Aeruginosa. Biosens. Bioelectron. 2019, 132, 224–229. 10.1016/j.bios.2019.02.053. [DOI] [PubMed] [Google Scholar]
- Zheng X.; Gao S.; Wu J.; Hu X. Recent Advances in Aptamer-Based Biosensors for Detection of Pseudomonas Aeruginosa. Frontiers in Microbiology 2020, 10.3389/fmicb.2020.605229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai G.; Li Z.; Luo F.; Ai S.; Chen B.; Wang Q. Electrochemical Determination of Salmonella Typhimurium by Using Aptamer-Loaded Gold Nanoparticles and a Composite Prepared from a Metal-Organic Framework (Type UiO-67) and Graphene. Microchim. Acta 2019, 10.1007/s00604-019-3724-y. [DOI] [PubMed] [Google Scholar]
- Yu T.; Xu H.; Zhao Y.; Han Y.; Zhang Y.; Zhang J.; Xu C.; Wang W.; Guo Q.; Ge J. Aptamer Based High Throughput Colorimetric Biosensor for Detection of Staphylococcus Aureus. Sci. Rep. 2020, 10 (1), 1–6. 10.1038/s41598-020-66105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh R. O.; almajidi Y. Q.; Mansouri S.; Hammoud A.; Rodrigues P.; Mezan S. O.; maabreh H. G.; Deorari M.; Shakir M. N.; alasheqi M. qasim Dual-Mode Colorimetric and Fluorescence Biosensors for the Detection of Foodborne Bacteria. Clin. Chim. Acta 2024, 553, 117741 10.1016/j.cca.2023.117741. [DOI] [PubMed] [Google Scholar]
- Yu X.; Chen F.; Wang R.; Li Y. Whole-Bacterium SELEX of DNA Aptamers for Rapid Detection of E.Coli O157:H7 Using a QCM Sensor. J. Biotechnol. 2018, 266, 39–49. 10.1016/j.jbiotec.2017.12.011. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Liu D.; Bai Y.; Cui Y.; Wang D.; Shi X. Identification and Characterization of Two High Affinity Aptamers Specific for Salmonella Enteritidis. Food Control 2019, 106 (March), 106719. 10.1016/j.foodcont.2019.106719. [DOI] [Google Scholar]
- Zhang H.; Yao S.; Song X.; Xu K.; Wang J.; Li J.; Zhao C.; Jin M. One-Step Colorimetric Detection of Staphylococcus Aureus Based on Target-Induced Shielding against the Peroxidase Mimicking Activity of Aptamer-Functionalized Gold-Coated Iron Oxide Nanocomposites. Talanta 2021, 232, 122448 10.1016/j.talanta.2021.122448. [DOI] [PubMed] [Google Scholar]
- Roushani M.; Sarabaegi M.; Pourahmad F. Impedimetric Aptasensor for Pseudomonas Aeruginosa by Using a Glassy Carbon Electrode Modified with Silver Nanoparticles. Microchim. Acta 2019, 10.1007/s00604-019-3858-y. [DOI] [PubMed] [Google Scholar]
- Saad M.; Castiello F. R.; Faucher S. P.; Tabrizian M. Introducing an SPRi-Based Titration Assay Using Aptamers for the Detection of Legionella Pneumophila. Sensors Actuators B Chem. 2022, 351, 130933 10.1016/j.snb.2021.130933. [DOI] [Google Scholar]
- Das R.; Dhiman A.; Kapil A.; Bansal V.; Sharma T. K. Aptamer-Mediated Colorimetric and Electrochemical Detection of Pseudomonas Aeruginosa Utilizing Peroxidase-Mimic Activity of Gold NanoZyme. Anal. Bioanal. Chem. 2019, 411 (6), 1229–1238. 10.1007/s00216-018-1555-z. [DOI] [PubMed] [Google Scholar]
- About Base Pair Biotechnologies – Base Pair Biotechnologies. https://www.basepairbio.com/about-base-pair/ (accessed 2023-07-20).
- Apto-Gen - Home. https://apto-gen.com/ (accessed 2023-07-20).
- About - Aptamer Group. https://aptamergroup.com/about/ (accessed 2023-07-20).
- BIOAPTUS - A Biotecnologia dos Anticorpos Sintéticos - ANFITECH. http://www.bioaptus.com.br/ (accessed 2023-07-20).
- Achiko AG. https://www.achiko.com/ (accessed 2023-07-20).
- Aptamer services & Aptamer-based assays | Novaptech. https://novaptech.com/ (accessed 2023-09-01).
- Zhang J.; Wang Y.; Lu X. Molecular Imprinting Technology for Sensing Foodborne Pathogenic Bacteria. Anal. Bioanal. Chem. 2021, 413 (18), 4581–4598. 10.1007/s00216-020-03138-x. [DOI] [PubMed] [Google Scholar]
- Elfadil D.; Lamaoui A.; Della Pelle F.; Amine A.; Compagnone D. Molecularly Imprinted Polymers Combined with Electrochemical Sensors for Food Contaminants Analysis. Molecules 2021, 26 (15), 4607. 10.3390/molecules26154607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldara M.; Kulpa J.; Lowdon J. W.; Cleij T. J.; Dilien H.; Eersels K.; Grinsven B. v. Recent Advances in Molecularly Imprinted Polymers for Glucose Monitoring: From Fundamental Research to Commercial Application. Chemosensors 2023, 11 (1), 32. 10.3390/chemosensors11010032. [DOI] [Google Scholar]
- Frigoli M.; Lowdon J. W.; Caldara M.; Arreguin-Campos R.; Sewall J.; Cleij T. J.; Diliën H.; Eersels K.; van Grinsven B. Thermal Pyocyanin Sensor Based on Molecularly Imprinted Polymers for the Indirect Detection of Pseudomonas Aeruginosa. ACS Sensors 2023, 8, 353. 10.1021/acssensors.2c02345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowdon J. W.; Diliën H.; Singla P.; Peeters M.; Cleij T. J.; van Grinsven B.; Eersels K. MIPs for Commercial Application in Low-Cost Sensors and Assays – An Overview of the Current Status Quo. Sensors Actuators, B Chem. 2020, 325, 128973. 10.1016/j.snb.2020.128973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.; Jiang D.; Shao J.; Sun X. Magnetic Molecularly Imprinted Polymer Nanoparticles Based Electrochemical Sensor for the Measurement of Gram-Negative Bacterial Quorum Signaling Molecules (N-Acyl-Homoserine-Lactones). Biosens. Bioelectron. 2016, 75, 411–419. 10.1016/j.bios.2015.07.045. [DOI] [PubMed] [Google Scholar]
- Caldara M.; Lowdon J. W.; Royakkers J.; Peeters M.; Cleij T. J.; Diliën H.; Eersels K.; van Grinsven B. A Molecularly Imprinted Polymer-Based Thermal Sensor for the Selective Detection of Melamine in Milk Samples. Foods 2022, 11 (18), 2906. 10.3390/foods11182906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerets B.; Peeters M.; van Grinsven B.; Bers K.; de Ceuninck W.; Wagner P. Optimizing the Thermal Read-out Technique for MIP-Based Biomimetic Sensors: Towards Nanomolar Detection Limits. Sensors (Basel). 2013, 13 (7), 9148–9159. 10.3390/s130709148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowdon J. W.; Eersels K.; Arreguin-Campos R.; Caldara M.; Heidt B.; Rogosic R.; Jimenez-Monroy K. L.; Cleij T. J.; Diliën H.; van Grinsven B. A Molecularly Imprinted Polymer-Based Dye Displacement Assay for the Rapid Visual Detection of Amphetamine in Urine. Molecules 2020, 25 (22), 5222. 10.3390/molecules25225222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse Sum Bui B.; Auroy T.; Haupt K. Fighting Antibiotic-Resistant Bacteria: Promising Strategies Orchestrated by Molecularly Imprinted Polymers. Angewandte Chemie - International ed. 2022, 10.1002/anie.202106493. [DOI] [PubMed] [Google Scholar]
- Haghdoust S.; Arshad U.; Mujahid A.; Schranzhofer L.; Lieberzeit P. A. Development of a MIP-Based QCM Sensor for Selective Detection of Penicillins in Aqueous Media. Chemosensors 2021, 9 (12), 362. 10.3390/chemosensors9120362. [DOI] [Google Scholar]
- Piletska E. V.; Stavroulakis G.; Larcombe L. D.; Whitcombe M. J.; Sharma A.; Primrose S.; Robinson G. K.; Piletsky S. A. Passive Control of Quorum Sensing: Prevention of Pseudomonas Aeruginosa Biofilm Formation by Imprinted Polymers. Biomacromolecules 2011, 12 (4), 1067–1071. 10.1021/bm101410q. [DOI] [PubMed] [Google Scholar]
- Rajpal S.; Bhakta S.; Mishra P. Biomarker Imprinted Magnetic Core-Shell Nanoparticles for Rapid, Culture Free Detection of Pathogenic Bacteria. J. Mater. Chem. B 2021, 9 (10), 2436–2446. 10.1039/D0TB02842H. [DOI] [PubMed] [Google Scholar]
- Liustrovaite V.; Pogorielov M.; Boguzaite R.; Ratautaite V.; Ramanaviciene A.; Pilvenyte G.; Holubnycha V.; Korniienko V.; Diedkova K.; Viter R.; Ramanavicius A. Towards Electrochemical Sensor Based on Molecularly Imprinted Polypyrrole for the Detection of Bacteria—Listeria Monocytogenes. Polymers (Basel) 2023, 15 (7), 1597. 10.3390/polym15071597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Lin X.; Liu T.; Zhang Z.; Kong J.; Yu H.; Yan J.; Luan D.; Zhao Y.; Bian X. Reusable and Universal Impedimetric Sensing Platform for the Rapid and Sensitive Detection of Pathogenic Bacteria Based on Bacteria-Imprinted Polythiophene Film. Analyst 2022, 147 (20), 4433–4441. 10.1039/D2AN01122K. [DOI] [PubMed] [Google Scholar]
- Wang O.; Jia X.; Liu J.; Sun M.; Wu J. Rapid and Simple Preparation of an MXene/Polypyrrole-Based Bacteria Imprinted Sensor for Ultrasensitive Salmonella Detection. J. Electroanal. Chem. 2022, 918, 116513 10.1016/j.jelechem.2022.116513. [DOI] [Google Scholar]
- Tajani A. S.; Soheili V.; Moosavi F.; Ghodsi R.; Alizadeh T.; Fazly Bazzaz B. S. Ultra Selective and High-Capacity Dummy Template Molecular Imprinted Polymer to Control Quorum Sensing and Biofilm Formation of Pseudomonas Aeruginosa. Anal. Chim. Acta 2022, 1199, 339574 10.1016/j.aca.2022.339574. [DOI] [PubMed] [Google Scholar]
- Frigoli M.; Lowdon J. W.; Caldara M.; Arreguin-Campos R.; Sewall J.; Cleij T. J.; Diliën H.; Eersels K.; Van Grinsven B. Thermal Pyocyanin Sensor Based on Molecularly Imprinted Polymers for the Indirect Detection of Pseudomonas Aeruginosa. ACS Sensors 2023, 8 (1), 353–362. 10.1021/acssensors.2c02345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BelBruno J. J. Molecularly Imprinted Polymers. Chem. Rev. 2019, 119 (1), 94–119. 10.1021/acs.chemrev.8b00171. [DOI] [PubMed] [Google Scholar]
- We design recognition. - MIP Discovery. https://mipdiscovery.com/ (accessed 2023-07-20).
- AgPlus diagnostics . Differentiation of Bacterial versus Viral Infection in Acute Respiratory Conditions Using a Point of Care Assay to Detect Human Neutrophil Lipocalin; 2018. https://agplusdiagnostics.com/wp-content/uploads/dobv-eral-0248-nwrc-5234.pdf (accessed 2023-05-01).
- Supelco Analytical Products. https://www.sigmaaldrich.com/NL/en/life-science/supelco (accessed 2023-07-20).
- Cui F.; Zhou Z.; Zhou H. S. Molecularly Imprinted Polymers and Surface Imprinted Polymers Based Electrochemical Biosensor for Infectious Diseases. Sensors (Switzerland) 2020, 20, 996. 10.3390/s20040996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongabi D.; Khorshid M.; Losada-Pérez P.; Eersels K.; Deschaume O.; D’Haen J.; Bartic C.; Hooyberghs J.; Thoelen R.; Wübbenhorst M.; Wagner P. Cell Detection by Surface Imprinted Polymers SIPs: A Study to Unravel the Recognition Mechanisms. Sensors Actuators, B Chem. 2018, 255, 907–917. 10.1016/j.snb.2017.08.122. [DOI] [Google Scholar]
- Arreguin-Campos R.; Jiménez-Monroy K. L.; Diliën H.; Cleij T. J.; van Grinsven B.; Eersels K. Imprinted Polymers as Synthetic Receptors in Sensors for Food Safety. Biosensors 2021, 11 (2), 46. 10.3390/bios11020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arreguin-Campos R.; Eersels K.; Rogosic R.; Cleij T. J.; Diliën H.; Van Grinsven B. Imprinted Polydimethylsiloxane-Graphene Oxide Composite Receptor for the Biomimetic Thermal Sensing of Escherichia Coli. ACS Sensors 2022, 7 (5), 1467–1475. 10.1021/acssensors.2c00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eersels K.; Lieberzeit P.; Wagner P. A Review on Synthetic Receptors for Bioparticle Detection Created by Surface-Imprinting Techniques - From Principles to Applications. ACS Sensors 2016, 1 (10), 1171–1187. 10.1021/acssensors.6b00572. [DOI] [Google Scholar]
- Jenik M.; Seifner A.; Krassnig S.; Seidler K.; Lieberzeit P. A.; Dickert F. L.; Jungbauer C. Sensors for Bioanalytes by Imprinting-Polymers Mimicking Both Biological Receptors and the Corresponding Bioparticles. Biosens. Bioelectron. 2009, 25 (1), 9–14. 10.1016/j.bios.2009.01.019. [DOI] [PubMed] [Google Scholar]
- Pintavirooj C.; Vongmanee N.; Sukjee W.; Sangma C.; Visitsattapongse S. Biosensors for Klebsiella Pneumoniae with Molecularly Imprinted Polymer (MIP) Technique. Sensors 2022, 22 (12), 4638. 10.3390/s22124638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arreguin-Campos R.; Eersels K.; Lowdon J. W.; Rogosic R.; Heidt B.; Caldara M.; Jiménez-Monroy K. L.; Diliën H.; Cleij T. J.; van Grinsven B. Biomimetic Sensing of Escherichia Coli at the Solid-Liquid Interface: From Surface-Imprinted Polymer Synthesis toward Real Sample Sensing in Food Safety. Microchem. J. 2021, 169, 106554 10.1016/j.microc.2021.106554. [DOI] [Google Scholar]
- Zhao X.; Cui Y.; Wang J.; Wang J. Preparation of Fluorescent Molecularly Imprinted Polymers via Pickering Emulsion Interfaces and the Application for Visual Sensing Analysis of Listeria Monocytogenes. Polymers (Basel) 2019, 11 (6), 984. 10.3390/polym11060984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezdekova J.; Zemankova K.; Hutarova J.; Kociova S.; Smerkova K.; Adam V.; Vaculovicova M. Magnetic Molecularly Imprinted Polymers Used for Selective Isolation and Detection of Staphylococcus Aureus. Food Chem. 2020, 321, 126673 10.1016/j.foodchem.2020.126673. [DOI] [PubMed] [Google Scholar]
- Akhtarian S.; Doostmohammadi A.; Youssef K.; Kraft G.; Kaur Brar S.; Rezai P. Metal Microwires Functionalized with Cell-Imprinted Polymer for Capturing Bacteria in Water. ACS Appl. Polym. Mater. 2023, 5 (5), 3235–3246. 10.1021/acsapm.2c01886. [DOI] [Google Scholar]
- van Grinsven B.; Eersels K.; Peeters M.; Losada-Pérez P.; Vandenryt T.; Cleij T. J.; Wagner P. The Heat-Transfer Method: A Versatile Low-Cost, Label-Free, Fast, and User-Friendly Readout Platform for Biosensor Applications. ACS Appl. Mater. Interfaces 2014, 6 (16), 13309–13318. 10.1021/am503667s. [DOI] [PubMed] [Google Scholar]
- Sensip-dx | Food quality. https://www.sensipdx.com/ (accessed 2023-07-20).
- An Y.; Fang X.; Cheng J.; Yang S.; Chen Z.; Tong Y. Research Progress of Metal–Organic Framework Nanozymes in Bacterial Sensing, Detection, and Treatment. RSC Med. Chem. 2024, 15 (2), 380–398. 10.1039/D3MD00581J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik B.; Sahoo R.; Das M. C. PH-Stable MOFs: Design Principles and Applications. Coord. Chem. Rev. 2023, 493, 215301 10.1016/j.ccr.2023.215301. [DOI] [Google Scholar]
- Liu Y.; Meng X. Z.; Luo X.; Gu H. W.; Yin X. L.; Han W. L.; Yi H. C.; Chen Y. Molecularly Imprinted Polymer Combined with MOF-Assisted Redox Recycling Amplification: A Powerful Electrochemical Sensing Strategy for Pathogenic Bacteria. Sensors Actuators B Chem. 2024, 410, 135682 10.1016/j.snb.2024.135682. [DOI] [Google Scholar]
- Li R.; Yan J.; Feng B.; Sun M.; Ding C.; Shen H.; Zhu J.; Yu S. Ultrasensitive Detection of Multidrug-Resistant Bacteria Based on Boric Acid-Functionalized Fluorescent MOF@COF. ACS Appl. Mater. Interfaces 2023, 15 (15), 18663–18671. 10.1021/acsami.3c00632. [DOI] [PubMed] [Google Scholar]
- Yan J.; Chen L.; Teng M.; Hao M.; Feng B.; Yang F.; Shen H.; Yu S.; Wang L. Dual Recognition Strategy for the Rapid and Precise Detection of Bacillus Cereus Using Post-Modified Nano-MOF and Aptamer. Sensors Actuators B Chem. 2023, 386, 133745 10.1016/j.snb.2023.133745. [DOI] [Google Scholar]
- Shang Y.; Xing G.; Lin H.; Chen S.; Xie T.; Lin J. M. Portable Biosensor with Bimetallic Metal-Organic Frameworks for Visual Detection and Elimination of Bacteria. Anal. Chem. 2023, 95 (35), 13368–13375. 10.1021/acs.analchem.3c02841. [DOI] [PubMed] [Google Scholar]
- Tago H.; Maeda Y.; Tanaka Y.; Kohketsu H.; Lim T. K.; Harada M.; Yoshino T.; Matsunaga T.; Tanaka T. Line Image Sensor-Based Colony Fingerprinting System for Rapid Pathogenic Bacteria Identification. Biosens. Bioelectron. 2024, 249, 116006 10.1016/j.bios.2024.116006. [DOI] [PubMed] [Google Scholar]
- Kirti N.; Krishna S. S.; Shukla D.; Kirti N.; Krishna S. S.; Shukla D. Salmonella Infections: An Update, Detection and Control Strategies. Salmonella - Curr. Trends Perspect. Detect. Control [Working Title] 2024, 10.5772/intechopen.1004835. [DOI] [Google Scholar]
- Rattray J. B.; Lowhorn R. J.; Walden R.; Márquez-Zacarías P.; Molotkova E.; Perron G.; Solis-Lemus C.; Pimentel Alarcon D.; Brown S. P. Machine Learning Identification of Pseudomonas Aeruginosa Strains from Colony Image Data. PLOS Comput. Biol. 2023, 19 (12), e1011699 10.1371/journal.pcbi.1011699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W.; Venkat E.; Ceylan Koydemir H. Optical Point of Care Devices for Diagnosis of Urinary Tract Infections. Curr. Opin. Biomed. Eng. 2023, 28, 100513 10.1016/j.cobme.2023.100513. [DOI] [Google Scholar]
- Zimina T. M.; Pinchuk O. A.; Kaplun D. I.; Kraeva L. A.; Sitkov N. O. Study of Laser Light Scattering Methods in Rapid Viability Assessment of Microorganisms under Antibiotics Exposure for Adaptation in Lab-on-A-Chip Format. Diagnostics 2023, Vol. 13, Page 1130 2023, 13 (6), 1130. 10.3390/diagnostics13061130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldara M.; van Wissen G.; Cleij T. J.; Diliën H.; van Grinsven B.; Eersels K.; Lowdon J. W. Deposition Methods for the Integration of Molecularly Imprinted Polymers (MIPs) in Sensor Applications. Adv. Sens. Res. 2023, 2 (7), 1–18. 10.1002/adsr.202200059. [DOI] [Google Scholar]