Graphical abstract
Keywords: Cytochrome c oxidase, Goats, Haemonchus contortus, Second internal transcribed spacer, Sheep
Abstract
Haemonchus contortus, a stomach worm, is prevalent in ruminants worldwide. They particularly hamper profitable small ruminant production. Here, we estimate the genetic variation of H. contortus collected from slaughtered goats and sheep from various geographic zones of Bangladesh using multiple genes. To perform this, adult parasites were isolated from the abomasum of slaughtered animals (sheep and goats). Among them, 79 male H. contortus were identified by microscopy. Following the extraction of DNA, ITS-2 and cox1 genes were amplified and subsequently considered for sequencing. After alignment and editing, sequences were analyzed to find out sequence variation, diversity pattern of genes, and population genetics of isolates. Among the sequence data, the analyses identified 19 genotypes of ITS-2 and 77 haplotypes of cox1 genes. The diversity of nucleotides was 0.0103 for ITS-2 and 0.029 for cox1 gene. The dendogram constructed by the genotype and haplotype sequences of H. contortus revealed that two populations were circulating in Bangladesh without any demarcation of host and geographic regions. Analysis of population genetics demonstrated a high flow of genes (89.2 %) within the population of the worm in Bangladesh. The Fst value showed very little amount of genetic difference among the worm populations of Bangladesh but marked genetic variation between different continents. The findings are expected to help explain the risks of anthelmintic resistance and the transmission pattern of the parasite, and also provide a control strategy against H. contortus.
1. Introduction
Sheep and goats are important components of livestock and they play a major role in upgrading the domestic economy of Bangladesh (Alamgir et al., 2023). Globally, the sheep and goat population is 2.1 billion among which about 80 % are present in Asia and Africa (Pitaksakulrat et al., 2021). Currently, 26.2 million goats and 3.5 million sheep are available in Bangladesh. The contribution of livestock to GDP is assessed annually and the GDP growth rate of livestock has gradually progressed from 2.51 % to 3.4 % during fiscal years 2009 and 2018 indicating gradual poverty alleviation and rural development (Hamid, 2019). However, gastrointestinal (GI) parasitic infections are a major constraint to the profitable production of small ruminants worldwide (Claerebout et al., 2018). Among GI parasites, Trichostrongylid nematode, especially Haemonchus Cobbold, 1898 is the major pathogen affecting the abomasum of wild and domestic ruminants reared in various climatic zones of the globe (O’Connor et al., 2006, Charlier et al., 2009). This parasite is prevalent in Bangladesh in various definitive hosts due to favorable climatic conditions for the development and existence of the worm, H. contortus (Dey et al., 2019, Omar et al., 2021). The fecundity of this parasite is very high, generally thousands of eggs in a single day. The blood-feeding nature of Haemonchus results in acute hemorrhagic anemia (0.05 ml blood/ parasite/ day), bottle jaw, and death of animals in severe cases (Taylor et al., 2007). They hamper profitable livestock rearing by causing the decrease of body weight and production as well as through increasing treatment costs. Yearly, treatment costs due to haemonchosis have been estimated in millions in different countries (McLeod, 2004, Waller and Chandrawathani, 2005).
As H. contortus is highly fecund, it shows genetic variability in nuclear and mitochondrial genes (Blouin et al., 1995, Doyle et al., 2020, Troell et al., 2006). Many features are involved to stimulate the genetic structure of Haemonchus such as spatial barrier, large population size, random mobilization of host, and variation of host species. Therefore, appropriate gene selection is vital for genetic analysis. The ribosomal DNA and mitochondrial genes have been extensively used for identification, genetic analysis and evolutionary relationship of the nematode parasite (Jacquiet et al., 1995).
Internal Transcribed Spacer-2 (ITS-2) region is the commonly used molecular marker that reduces the hazard of cross-reaction and distinguishes closely related species due to easy extension, availability of conserved regions, adequate rRNA, fast rate of evolution, and sufficient variation (Gasser et al., 1999). An insignificant amount of intra-specific genetic variation (less than 1 %) can be detected by this marker (Gasser and Newton, 2000). Helminths; especially nematodes, trematodes and cestodes, were specifically identified by amplifying the ITS-2 region (Kralova-Hromadova et al., 2012, Luton et al., 1992, Stevenson et al., 1995). Of the mitochondrial DNA (mtDNA), cox1 gene is a very convenient candidate for studying the genetic diversity pattern of parasites, especially Haemonchus, due to its high maternal inheritance and greater mutation rate (Brasil et al., 2012, Gharamah et al., 2012, Kandil et al., 2018, Yin et al., 2013). The information related to the genetic variation of different populations of H. contortus is useful regarding the associated spreading pattern, the extent of drug resistance alleles, and sustainable control strategy against the parasite (Dey et al., 2019). In our previous communication (Dey et al., 2019), we reported our assessment of population genetics and the pattern of genetic diversity using the nad4 gene in Bangladesh. In this study, we determine the genetic pattern of H. contortus collected from sheep and goats from various geographic regions of Bangladesh using ITS-2 and cox1 genes.
2. Materials and methods
2.1. Parasite collections
The abomasa of goats and sheep were collected from eight different areas of Bangladesh including Mymensingh, Tangail, Sirajgonj, Dinajpur, Barishal, Khulna, Sylhet and Rangamati (Fig. 1) (Supplementary file 1); and the adult H. contortus were recovered following the procedure described by MAFF (MAFF, 1986). In brief, the abomasum was separated, ligated and opened. Contents of the organ were transferred into a beaker and washed with Phosphate Buffered Saline (PBS) until the suspension became transparent. The adult parasites were isolated and repeatedly washed in physiological saline. After that, the microscopic examination was performed to identify male H. contortus conferring the key morphological features (Soulsby, 1982). Microscopically identified male H. contortus parasites were then preserved in 70 % glycerin alcohol.
Fig. 1.
Different geographical regions of Bangladesh from where samples were collected from sheep and goats.
2.2. Genomic DNA (gDNA) extraction
Genomic DNA was isolated from individual parasites by DNA Mini Kit (Qiagen, Germany) through following manufacturer’s guidelines. Concentrations of the extracted gDNA were assessed using a Nanodrop Spectrophotometer (Thermo Fisher Scientific- USA) and kept at −20 °C.
2.3. PCR amplification and visualization of PCR products
A region of ITS-2 (∼320 bp) and mitochondrial cox1 genes of H. contortus were amplified by PCR using appropriate primer sets and thermal conditions (Table 1). Visualization of PCR products (5 μl) of ITS-2 and cox1 genes was performed on 2.0 % agarose gel by adding EZ-Vision® IN-Gel (Amresco, USA).
Table 1.
Details about primers and thermal cycler conditions used in this study.
| Target gene | Name of primer | Sequence (5′–3′) | Amplicon size (bp) | PCR protocol | Reference |
|---|---|---|---|---|---|
| ITS-2 | NC1 | ACGTCTGGTTCAGGGTTGTT | ∼320 | 95 °C for 5 min(35 cycles at 95 °C for 30 sec, 55 °C for 30 sec and 72 °C for 1 min) 72 °C for 5 min. | Stevenson et al., 1995 |
| NC2 | TTAGTTTCTTTTCCTCCGCT | ||||
| cox1 | NEMAT-F | CCTACTATAATTGGTGGGTTTGGTAA | ∼720 | 94 °C for 2 min(35 cycles at 94 °C for 30 sec, 50 °C for 30 sec, and 72 °C for 1 min) 72 °C for 10 min | Kandil et al., 2018 |
| NEMAT-R | TAGCCGCAGTAAAAT AAGCACG |
2.4. Sequencing and genetic data analysis
After PCR, amplicons were purified and sequenced in both directions using PCR primers (Wizard PCR-Preps, Promega). The sequences were checked with the BioEdit software, aligned to and edited with MEGA v.10.2.4 software (Tamura et al., 2013) and submitted to GenBank. The nucleotide diversities, haplotype diversities and nucleotide variation were estimated with DnaSP v6 (Rozas, 2009). BioEdit software was used to calculate sequence identities (%) (Hall, 1999). The dendogram was constructed by Neighbor-Joining (NJ), Maximum-Parsimony (MP) and Maximum-Likelihood (ML) methods, respectively with the bootstrap (1000 replicates) (Tamura et al., 2013). The Fst and Nst values were measured with DnaSP v6 (Rozas, 2009) to assess gene flow among different populations. Tajima’s D, Fu and Li’s F, and Fu and Li’s D were calculated to assess the effect of selective forces on H. contortus populations by DnaSP version 6 (Tajima, 1989, Fu, 1997). The network tree was built to study haplotypes' relationship with Network v.10.2.0.0 (Bandelt et al., 1999). A hierarchical analysis of molecular variance (AMOVA) was implemented to calculate the genetic diversity of H. contortus populations using Arlequin (Excoffier et al., 1992). For analysis, 145 sequences from different studies were collected from GenBank.
3. Results
3.1. Species confirmation and genotyping
To confirm the study species as H. contortus, the isolation of adult parasites was followed by amplification and sequencing of the ITS-2 gene. After the alignment and editing of sequences, 231 bp was generated and detected 19 distinct genotypes by analyzing 79 ITS-2 sequences. The overall genotype and nucleotide diversities of H. contortus sequences were 0.905 and 0.0103 respectively from eight geographical locations in Bangladesh (Table 2). Sequence identities (%) among 19 genotypes of ITS-2 sequences varied from 96.9-100 %, when matched with the studied isolates, or with reference sequences retrieved from the GenBank database (Accession no. MT424902.1 and KC415125.1) (Table 3). Eleven (11) single nucleotide polymorphisms (SNPs) at the positions of 18, 20, 21, 22, 55, 59, 63, 93, 196, 199 and 218 were identified after the alignment of 19 studied genotypes with the reference sequence (KC415125.1). Five transversions (one A<–>C, one G<–>C and three A<–>T) and six transitions (one A<–>G and five T<–>C) were recognized at their substitutions (Table 4). The average percentage of GC contents of the ITS-2 sequences was 33.4. The 19 studied genotypes were also aligned to reference sequences of H. placei (LC367242 and LC367244) to detect species variation which found 1.7 % sequences variation only in four nucleotide positions (24th, 123rd, 205th and 219th). Among four nucleotide variations, three (24th, 205th and 219th) were represented by purines (G<–>A) and one by pyrimidine (T<–>C).
Table 2.
Genotypes of ITS-2 and haplotypes of cox1of H. contortus populations from different topographic regions of Bangladesh.
| Geographic regions |
ITS-2 |
CO1 |
||||||
|---|---|---|---|---|---|---|---|---|
| No. of sequences (N) | No. of genotypes (n) | Genotype diversity (Gd) | Nucleotide diversity (Nd) | No. of sequences (N) | No. of genotypes (n) | Genotype diversity (Gd) | Nucleotide diversity (Nd) | |
| Mymensingh | 10 | 7 | 0.933 | 0.0085 | 10 | 9 | 0.978 | 0.032 |
| Sylhet | 10 | 6 | 0.867 | 0.0085 | 10 | 10 | 1 | 0.027 |
| Tangail | 9 | 7 | 0.944 | 0.0118 | 10 | 10 | 1 | 0.029 |
| Sirajgonj | 10 | 9 | 0.978 | 0.0127 | 10 | 10 | 1 | 0.031 |
| Dinajpur | 10 | 6 | 0.844 | 0.0089 | 10 | 10 | 1 | 0.03 |
| Rangamati | 10 | 6 | 0.889 | 0.0097 | 9 | 9 | 1 | 0.031 |
| Khulna | 10 | 7 | 0.933 | 0.0143 | 10 | 10 | 1 | 0.016 |
| Barishal | 10 | 6 | 0.911 | 0.0082 | 10 | 10 | 1 | 0.017 |
| Overall | 79 | 19 | 0.905 | 0.0103 | 79 | 77 | 0.999 | 0.029 |
Table 3.
Pairwise identities (%) among 19 ITS-2 genotypes of H. contortus using reference sequences of H. contortus.
| Sample ID | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. BDMYHC1 (LC716151) | − | ||||||||||||||||||||
| 2. BDMYHC9 (LC716152) | 99.5 | − | |||||||||||||||||||
| 3. BDSYHC12 (LC716153) | 98.2 | 98.7 | − | ||||||||||||||||||
| 4. BDSYHC19 (LC716154) | 99.1 | 98.7 | 98.2 | − | |||||||||||||||||
| 5. BDTAHC22 (LC716155) | 97.8 | 98.2 | 99.5 | 97.8 | − | ||||||||||||||||
| 6. BDTAHC26 (LC716156) | 98.2 | 98.7 | 99.1 | 98.2 | 99.5 | − | |||||||||||||||
| 7. BDDHHC28 (LC716157) | 98.7 | 98.2 | 98.7 | 99.5 | 98.2 | 98.7 | − | ||||||||||||||
| 8. BDSGHC34 (LC716158) | 98.2 | 98.7 | 99.1 | 99.1 | 98.7 | 99.1 | 99.5 | − | |||||||||||||
| 9. BDSGHC36 (LC716159) | 98.2 | 97.8 | 97.4 | 99.1 | 96.9 | 97.4 | 98.7 | 98.2 | − | ||||||||||||
| 10. BDSGHC38 (LC716160) | 99.5 | 99.1 | 97.8 | 98.7 | 97.4 | 97.8 | 98.2 | 97.8 | 97.8 | − | |||||||||||
| 11. BDDPHC41 (LC716161) | 99.1 | 98.7 | 98.2 | 99.1 | 97.8 | 98.2 | 98.7 | 98.2 | 99.1 | 98.7 | − | ||||||||||
| 12. BDDPHC49 (LC716162) | 99.1 | 98.7 | 99.1 | 99.1 | 98.7 | 99.1 | 99.5 | 99.1 | 98.2 | 98.7 | 99.1 | − | |||||||||
| 13. BDRMHC54 (LC716163) | 98.2 | 97.8 | 97.4 | 99.1 | 96.9 | 97.4 | 98.7 | 98.2 | 99.1 | 97.8 | 99.1 | 98.2 | − | ||||||||
| 14. BDRMHC58 (LC716164) | 98.2 | 97.8 | 97.4 | 99.1 | 96.9 | 97.4 | 98.7 | 98.2 | 99.1 | 97.8 | 99.1 | 98.2 | 99.1 | − | |||||||
| 15. BDKHHC66 (LC716165) | 98.2 | 97.8 | 98.2 | 98.2 | 98.7 | 99.1 | 98.7 | 98.2 | 97.4 | 97.8 | 98.2 | 99.1 | 97.4 | 98.2 | − | ||||||
| 16. BDKHHC70 (LC716166) | 99.5 | 99.1 | 97.8 | 99.5 | 97.4 | 97.8 | 99.1 | 98.7 | 98.7 | 99.1 | 98.7 | 98.7 | 98.7 | 98.7 | 97.8 | − | |||||
| 17. BDBSHC73 (LC716167) | 98.7 | 99.1 | 99.5 | 98.7 | 99.1 | 99.5 | 99.1 | 99.5 | 97.8 | 98.2 | 98.7 | 99.5 | 97.8 | 97.8 | 98.7 | 98.2 | − | ||||
| 18. BDBSHC77 (LC716168) | 99.5 | 99.1 | 98.7 | 99.5 | 98.2 | 98.7 | 99.1 | 98.7 | 98.7 | 99.1 | 99.5 | 99.5 | 98.7 | 98.7 | 98.7 | 99.1 | 99.1 | − | |||
| 19. BDBSHC80 (LC716169) | 98.7 | 98.2 | 97.8 | 99.5 | 97.4 | 97.8 | 99.1 | 98.7 | 99.5 | 98.2 | 99.5 | 98.7 | 99.5 | 99.5 | 97.8 | 99.1 | 98.2 | 99.1 | − | ||
| 20. H. contortus (MT424902.1) | 99.1 | 98.7 | 98.2 | 100 | 97.8 | 98.2 | 99.5 | 99.1 | 99.1 | 98.7 | 99.1 | 99.1 | 99.1 | 99.1 | 98.2 | 99.5 | 98.7 | 99.5 | 99.5 | − | |
| 21. H. contortus (KC415125.1) | 99.1 | 98.7 | 99.1 | 99.1 | 98.7 | 99.1 | 99.5 | 99.1 | 98.2 | 98.7 | 99.1 | 100 | 98.2 | 98.2 | 99.1 | 98.7 | 99.5 | 99.5 | 98.7 | 99.1 | − |
Table 4.
Variation of nucleotides at different position of 19 genotypes from 79 H. contortus isolates with the reference sequence of H. contortus of ITS-2 gene.
| Genotypes | Nucleotide position |
No. of isolates | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 18 | 20 | 21 | 22 | 55 | 59 | 63 | 93 | 196 | 199 | 218 | ||
| LC360148.1 | T | G | G | C | C | T | C | A | A | T | T | |
| 1. BDMYHC1(LC716151) | . | . | . | T | . | . | . | C | . | . | . | 1 |
| 2. BDMYHC9(LC716152) | A | . | . | T | . | . | . | C | . | . | . | 2 |
| 3. BDSYHC12(LC716153) | A | . | . | . | . | A | . | . | . | . | . | 2 |
| 4. BDSYHC19(LC716154) | . | . | . | T | . | . | . | . | T | . | . | 4 |
| 5. BDTAHC22(LC716155) | A | . | . | . | . | A | . | . | . | . | . | 4 |
| 6. BDTAHC26(LC716156) | A | . | . | . | . | . | T | . | . | . | . | 3 |
| 7. BDTAHC28(LC716157) | . | . | . | . | . | . | . | . | T | . | . | 2 |
| 8. BDSGHC34(LC716158) | A | . | . | . | . | . | . | . | T | . | . | 1 |
| 9. BDSGHC36(LC716159) | . | A | C | T. | . | . | . | . | T | . | . | 1 |
| 10. BDSGHC38(LC716160) | . | . | . | T | T | . | . | C | . | . | . | 1 |
| 11. BDDPHC41(LC716161) | . | . | T | T | . | . | . | . | . | . | . | 11 |
| 12. BDDPHC49(LC716162) | . | . | . | . | . | . | . | . | . | . | . | 12 |
| 13. BDRMHC54(LC716163) | . | . | C | T. | . | . | . | T | . | . | . | 1 |
| 14. BDRMHC58(LC716164) | . | . | C | T | . | . | . | . | T | . | C | 1 |
| 15. BDKHHC66(LC716165) | . | . | . | . | . | . | T | . | . | . | C | 1 |
| 16. BDKHHC70(LC716166) | . | . | . | T | . | . | C | T | . | . | 1 | |
| 17. BDBSHC73(LC716167) | A | . | . | . | . | . | . | . | . | . | . | 11 |
| 18. BDBSHC77(LC716168) | . | . | . | T | . | . | . | . | . | . | . | 7 |
| 19. BDBSHC80(LC716169) | . | . | C | T | . | . | . | . | T | . | . | 13 |
| Total | 79 | |||||||||||
Dot (.) represents similar position with LC360148.1.
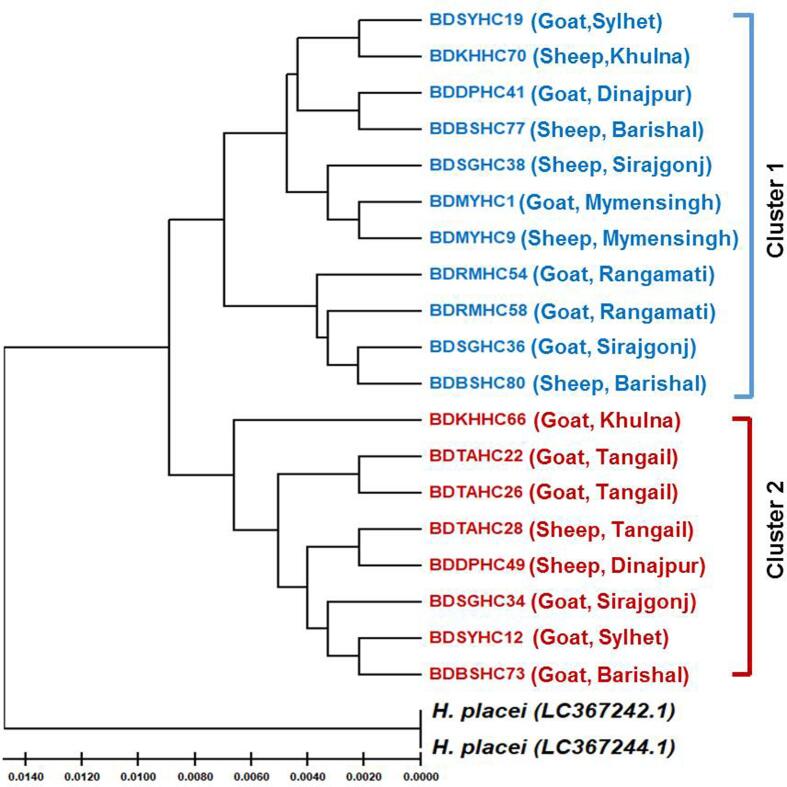
A phylogenetic tree (UPGMA tree) built by 19 ITS-2 genotypes was built using the UPGMA method where two H. placei were used as outgroup. The constructed phylogram formed two distinct clusters without any relation to hosts (sheep and goats) and geographic regions (Fig. 2).
Fig. 2.
Unweighted pair group method with arithmetic mean dendogram showing the relationships among H. contortus genotypes isolated from sheep and goats from different geographical regions of Bangladesh based on ITS-2 and H. placei (GenBank accession nos. LC367242 and LC367244) serve as outgroup.
3.2. Genetic diversity and phylogeny of cox1
A total of 79 cox1 sequences of H. contortus were aligned edited and produced 632 bp length for all cases. Sequences were considered for nucleotide BLAST search and detected high sequence identities (98–99 %) with reference to cox1 gene (Accession no. KJ724366.1) of H. contortus. There were 77 distinct haplotypes identified from 79 amplicons. The number of polymorphic sites was 129 (at different nucleotide positions) where 43 for singleton variable sites and 86 for parsimony informative sites. The haplotype diversity was 0.999 representing a high degree of gene diversity whereas the overall nucleotide diversity was 0.029 (Table 2).
The selective neutrality test of H. contortus populations was also analyzed and found insignificant (P > 0.05) variation from neutrality signifying a lack of selective forces functioning on the studied populations (Table 5).
Table 5.
Selective neutrality test among cox1 sequences from H. contortus populations of different geographic regions of Bangladesh.
| Geographic regions | Tajima’s D | p value | Fu & Li’s D | p value | Fu & Li’s F | p value |
|---|---|---|---|---|---|---|
| Mymensingh | −0.2698 | p ˃ 0.10 | −0.1396 | p ˃ 0.10 | −0.1945 | p ˃ 0.10 |
| Tangail | −0.1397 | p ˃ 0.10 | −0.2495 | p ˃ 0.10 | −0.2508 | p ˃ 0.10 |
| Sirajgonj | −1.02248 | p ˃ 0.10 | −1.0092 | p ˃ 0.10 | −1.1440 | p ˃ 0.10 |
| Dinajpur | −0.4907 | p ˃ 0.10 | −0.8640 | p ˃ 0.10 | −0.8706 | p ˃ 0.10 |
| Sylhet | −0.9949 | p ˃ 0.10 | −0.8640 | p ˃ 0.10 | −1.0130 | p ˃ 0.10 |
| Rangamati | −0.6464 | p ˃ 0.10 | −0.5036 | p ˃ 0.10 | −0.6046 | p ˃ 0.10 |
| Khulna | −1.3220 | p ˃ 0.10 | −1.4700 | p ˃ 0.10 | −1.6180 | p ˃ 0.10 |
| Barishal | −1.1757 | p ˃ 0.10 | −1.1259 | p ˃ 0.10 | −1.2852 | p ˃ 0.10 |
| Overall | −1.3914 | p ˃ 0.10 | −2.0840 | 0.10 ˃ p ˃ 0.05 | −2.1599 | 0.10 ˃ p ˃ 0.05 |
The network tree was built to determine the association between 77 haplotypes data from eight populations of H. contortus (Supplementary file 2). From the network profile, two clusters were formed randomly with the haplotypes of H. contortus excluding a few isolates without any demarcation of host and source of isolates. Interestingly, isolates from Khulna and Barishal were uniquely grouped in a single cluster.
For comparing population genetics, cox1 sequences of H. contortus including studied sequences and reference sequences from Nigeria, Brazil and Pakistan were selected. In the phylogenetic tree constructed by NJ method, three main clades were found with 1000 replicates in bootstrap analysis in which H. placei (KJ724428) served as an outgroup (Supplementary file 3). The isolates from Nigeria and Brazil formed separate clades termed as African and American clades respectively, and the isolates from Pakistan and Bangladesh formed a unique clade termed as Asian clade. The phylogenetic trees built in ML and MP methods also showed a similar clustering pattern.
3.3. Population genetic structure
To determine genetic distinction, pairwise Fst and Nst values were computed among the H. contortus populations of different geographic regions of Bangladesh. An insignificant amount of genetic difference was observed which ranged from −0.03495 to 0.23575 in case of Fst value and −0.03541 to 0.23839 in case of Nst value. Comparatively high genetic variation was detected in H. contortus populations of Khulna (Fst value = 0.05241 to 0.23575, Nst value = 0.0526 to 0.23839) and Barishal (Fst value = 0.05241 to 0.2309, Nst value = 0.0526 to 0.23332) when comparing with other studied populations (Table 6).
Table 6.
Pairwise Fst and Nst value of cox-1 gene between H. contortus populations of different geographic regions of Bangladesh.
| Geographic regions | Mymensingh | Sylhet | Tangail | Sirajgonj | Dinajpur | Rangamati | Khulna | Barishal |
|---|---|---|---|---|---|---|---|---|
| Mymensingh | − | 0.00635 | 0.00076 | 0.00658 | −0.01170 | −0.00910 | 0.20733 | 0.22703 |
| Sylhet | 0.00669 | − | 0.01826 | −0.00396 | 0.04110 | −0.03541 | 0.11431 | 0.13419 |
| Tangail | 0.00109 | 0.01815 | − | −0.00125 | −0.03192 | −0.01248 | 0.21070 | 0.22978 |
| Sirajgonj | 0.00674 | −0.00437 | −0.00129 | − | 0.03006 | −0.02806 | 0.12800 | 0.14411 |
| Dinajpur | −0.01197 | 0.04051 | −0.03122 | 0.02944 | − | 0.00956 | 0.23839 | 0.23332 |
| Rangamati | −0.00869 | −0.03495 | −0.01220 | −0.02805 | 0.00939 | − | 0.13363 | 0.15108 |
| Khulna | 0.20594 | 0.11343 | 0.20820 | 0.12629 | 0.23575 | 0.13194 | − | 0.05260 |
| Barishal | 0.22557 | 0.13322 | 0.22750 | 0.14264 | 0.2309 | 0.14937 | 0.05241 | − |
Fst and Nst are below and above the diagonal, respectively. Negative values signify more nucleotide substitutions within than between populations.
The Fst value between the studied worm populations reported from Nigeria, Brazil and Pakistan was calculated. The results showed the highest genetic variation between populations of H. contortus of Bangladesh and Brazil (Fst = 0.66752), and the lowest between Bangladesh and Pakistan (Fst = 0.06944) (Table 7).
Table 7.
Pairwise Fst value of cox-1 gene of H. contortus populations between Bangladesh and other three countries.
| Name of countries | No. of sequences | Fst value | Nucleotide diversity |
|---|---|---|---|
| Pakistan | 61 | 0.069 | 0.031 |
| Nigeria | 73 | 0.459 | 0.019 |
| Brazil | 05 | 0.667 | 0.014 |
| Bangladesh | 77 | 0.029 |
A hierarchical AMOVA was estimated which showed 89.2 % of genetic variance circulated within the studied population (Fst = 0.108, p = 0.001) and 7.57 % among groups (Fct = 0.076, p = 0.034) (Table 8).
Table 8.
Analysis of Molecular Variance (AMOVA) and F-statistics of partial CO1 gene for different populations of H. contortus in Bangladesh.
| Source of variation | Percentage of variation | F-statistics | p-value |
|---|---|---|---|
| Among groups | 7.57 | Fct = 0.076 | 0.034* |
| Among populations within groups | 3.23 | Fsc = 0.035 | 0.056 |
| Within populations | 89.2 | Fst = 0.108 | 0.001** |
Eight populations were divided into two groups including northern part (Mymensingh, Tangail, Rajshahi, Rangpur and Sylhet) and southern part (Rangamati, Khulna and Bhola) in Bangladesh.
p < 0.05.
p < 0.01.
4. Discussion
Small ruminants are generally parasitized by numerous endo- and ecto-parasites (Poddar et al., 2017, Shuvo et al., 2021, Dey et al., 2022). Among them, H. contortus is an important blood-feeding stomachworm. The prevalence of this parasite depends on availability of host and host range, zoogeographic location, climatic condition, and habit and habited of the host. The freeliving stages (egg to L3) of H. contortus experience with the fluctuation of climatic conditions that affect the development and spatio-temporal distribution of this parasite (Rose et al., 2016). H. contortus is prevalent all over the world in multiple hosts, however, particularly affects livestock, with a varying infection rate (Jacquiet et al., 1998, Waller and Chandrawathani, 2005, Craig, 2009, Hussain et al., 2014). The climatic conditions such as annual temperature and rainfall are the important abiotic factors impacting on genetic variation of H. contortus (Salle et al., 2019). In our previous communication (Dey et al., 2019), we described the genetic diversity pattern and population genetics of H. contortus using the mitochondrial gene nad4, and Mannan et al (2023) also confirmed the findings using the same genetic markers. Therefore, in this study, the genetic pattern of H. contortus has been validated further using COX1 and we unambiguously proved that two distinct genotypes of H. contortus populations (irrespective to hosts and special distribution) is circulating simultaneously in Bangladesh.
Haemonchus contortus was identified by morphological structures at the genus level (Gareh et al., 2021) and is quite impossible to detect at the species level using microscopic features. Identification at the species level was confirmed by sequencing with the nuclear ribosomal ITS-2 gene. Haemonchus contortus and H. placei have very low intra-specific variation and this insignificant amount of variation can be detected by the analysis of ITS-2 sequences, a suitable gene for the diagnosis of any large range of trichostrongylid parasites (Gasser et al., 1994).
In this study, 1.7 % variation was estimated between H. contortus and H. placei at the 24th, 123rd, 205th and 219th nucleotide positions. According to Stevenson et al.(1995), variation between the closely related species is detected at nucleotide positions 24, 205 and 219. In addition, 2.6 % variation was observed among ITS-2 sequences of H. contortus isolates. Similar results were reported in Kenya, Sweden and Germany (Heise et al., 1999, Troell et al., 2006). Hence the variation was recorded as high (5.2 %) in Australia, France, New Zealand and the UK (Gasser et al., 1998). Among 79 ITS-2 sequences, 19 unique genotypes were determined. However, these numbers were varied in earlier reports including seven among the isolates of Pakistan (Hussain et al., 2014), six of Yemen and Malaysia (Gharamah et al., 2012), and eighteen of China (Yin et al., 2013). The GC nucleotide contents of ITS-2 sequence were 33.4 %. A similar result was reported by previous studies such as 33.0 % from sheep in Switzerland, China and Australia (Stevenson et al., 1995) and 33.4 % from Malaysian and Yemenese goats (Gharamah et al., 2012).
The nucleotide and genotype diversity in our study was 0.01 and 0.905 respectively. The low nucleotide and high genotype diversity were also recorded in H. contortus isolates in Thailand (0.017, 0.832) (Laosutthipong and Eardmusic, 2019) and China (0.006, 0.703) (Yin et al., 2013). Several host-related and parasite-related factors are responsible for the high genotype diversity of the Haemonchus population. Host-related factors include parasites from heterologous host species or cross-infection, mobilization of the host within or between countries, and absence of selection pressure of anthelmintics (Akkari et al., 2013). Parasite-related factors comprise high fecundity with high prevalence, short and direct life cycle, and a large population size with extensive genetic variability (Prichard, 2001). In Bangladesh especially in rural areas, grazing of multiple hosts such as cattle, sheep and goats in a common pasture is very predominant. This grazing pattern enhances the chance of cross-infection of H. contortus infection among the broad range of ruminant hosts (Dey et al., 2021).
MtDNA including nad4 and cox1 genes were more useful genetic markers due to high substitution rates and potential to distinguish closely related species (Anderson et al., 1998, Troell et al., 2006). These genes are also used as basic tools for several studies such as taxonomic studies and population genetics (Blouin, 2002).
Several approaches have been described to find out the genetic pattern of Haemonchus populations in different countries of the world (Brasil et al., 2012, Gharamah et al., 2012, Yin et al., 2013, Pitaksakulrat et al., 2021). Our previous research on H. contortus populations has been performed by the nad4 gene to find out the pattern of genetic diversity (Dey et al., 2019). Along with other authors (Brasil et al., 2012, Hussain et al., 2014), the cox1 gene was used to detect the level of genetic diversity of Haemonchus populations in Bangladesh. In our study, the nucleotide diversity was 0.029. The value is in line with the previous study in Thailand (0.043) and Pakistan (0.036) (Hussain et al., 2014, Pitaksakulrat et al., 2021). Our 79 cox1 sequences from Bangladesh represented 77 haplotypes with high haplotype diversity (0.999). The high haplotype diversity has also been found in H. contortus isolates originating from Thailand and Pakistan. For example, 73 haplotypes among 74 Pakistani isolates (Hussain et al., 2014) and 122 haplotypes among 130 Thailand isolates (Pitaksakulrat et al., 2021) were detected. Haemonchus follows a high level of intra-population diversity, the key feature of trichostrongylid parasites (Blouin et al., 1998, Archie and Ezenwa, 2011).
In the present study, Tajima’s D and Fu and Li’s D showed statistically significant negative values for H. contortus populations of eight different geographical regions. The negative value indicates a high number of low-frequency polymorphisms attributing a recent selective sweep or population expansion (Belanger and Perkins, 2010).
The phylogram built by 77 sequences of the cox1 gene revealed two populations of H. contortus circulating in Bangladesh. Particularly, the clustering pattern was not unique according to the geographical origin of isolates or the host species. A similar clustering pattern was also found previously relating to nad4 gene (Dey et al., 2019, Mannan et al., 2023). Possibly, two populations of H. contortus existed during the onset of infection in Bangladesh (Cerutti et al., 2010). Within the country, there is no strong geographical barrier. Moreover, extensive management systems and practice of common grazing fields for multiple host species namely sheep, goats, cattle and buffaloes are responsible for the fast transmission of infection and breakdown of the host barrier; signifying the circulation of two H. contortus populations.
Globally, clustering patterns were observed based on continental demarcation such as Asian clade including Pakistani and Bangladeshi H. contortus isolates, African clade including Nigerian isolates and American isolates including Brazilian isolates. Also, noticeable genetic differentiation was observed between continents. Lack of opportunity for regular movement of hosts restricts the spreading of parasites; thus, contributing to the distinct continental clustering pattern (Troell et al., 2006).
A low genetic differentiation was noted between the populations of the same continent (Bangladeshi and Pakistani populations) (Blouin et al., 1995). This might be due to the poor control strategy of nematodes and the frequent entrance of hosts within the continent for trading purposes.
In population genetics, host is an essential component for detecting the gene flow and genetic variability (McCoy et al., 2003). The genetic analysis of the cox1 gene revealed that most of the genetic variation (89.2 %) was dispersed within the H. contortus populations, signifying high gene flow without strong terrestrial obstacles among the populations. The high gene flow favors the transmission of resistant parasites within the populations. High variation within populations, followed by variation among groups and population within the group is in line with the earlier report in Thailand (Pitaksakulrat et al., 2021), Pakistan (Hussain et al., 2014) and Brazil (Brasil et al., 2012). These findings are related to the rapid evolutionary rate of mtDNA in nematodes (Blouin et al., 1995).
In conclusion, the present study confirms that two H. contortus populations are circulating in Bangladesh without any host and spatial discrimination. The insignificant amount of genetic differentiation was found within the studied H. contortus populations. The analysis of COX1 gene showed high gene flow within the populations of H. contortus irrespective to geographical barriers. The generated knowledge of the study will help assess the risk of anthelmintic resistance and eventually formulate an effective and sustainable control strategy against H. contortus.
CRediT authorship contribution statement
Shanaz Parvin: Writing – original draft, Methodology, Funding acquisition, Formal analysis, Data curation. Anita Rani Dey: Writing – review & editing, Validation, Formal analysis. Nusrat Nowrin Shohana: Writing – review & editing, Formal analysis. Anisuzzaman: Writing – review & editing, Formal analysis. Md. Hasanuzzaman Talukder: Writing – review & editing, Formal analysis. Mohammad Zahangir Alam: Validation, Supervision, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors greatly acknowledge financial support from the project funded by Bangladesh Agricultural University (2020/957/BAU). The work was also supported by the grants of the Ministry of Education (LS2018710) and Ministry of Science and Technology (SRG-221109).
Footnotes
Supplementary material to this article can be found online at https://doi.org/10.1016/j.sjbs.2024.104030.
Contributor Information
Shanaz Parvin, Email: shanaz.fvas@gonouniversity.edu.bd.
Anita Rani Dey, Email: anitadey@bau.edu.bd.
Nusrat Nowrin Shohana, Email: shohana.vpar@bau.edu.bd.
Anisuzzaman, Email: zaman.a@bau.edu.bd.
Md. Hasanuzzaman Talukder, Email: talukdermhasan@bau.edu.bd.
Mohammad Zahangir Alam, Email: mzalam@bau.edu.bd.
Appendix A. Supplementary material
The following are the Supplementary material to this article:
Supplementary file 1.

Supplementary file 2.
References
- Akkari H., Jebali J., Gharbi M., Mhadhbi M., Awadi S., Darghouth M.A. Epidemiological study of sympatric Haemonchus species and genetic characterization of Haemonchus contortus in domestic ruminants in Tunisia. Vet. Parasitol. 2013;193(1–3):118–125. doi: 10.1016/j.vetpar.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Alamgir M., Uddin M.N., Hasan M.M., Wang X., Shiru M.S., Shahid S. Prioritization of sectoral adaptation strategies and practices: A case study for Bangladesh. Environ. Dev. 2023;45:45. [Google Scholar]
- Anderson T.J., Blouin M.S., Beech R.N. Population biology of parasitic nematodes: applications of genetic markers. Adv. Parasitol. 1998;41:219–283. doi: 10.1016/s0065-308x(08)60425-x. [DOI] [PubMed] [Google Scholar]
- Archie E.A., Ezenwa V.O. Population genetic structure and history of a generalist parasite infecting multiple sympatric host species. Int. J. Parasitol. 2011;41(1):89–98. doi: 10.1016/j.ijpara.2010.07.014. [DOI] [PubMed] [Google Scholar]
- Bandelt H.J., Forster P., Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16(1):37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Belanger D.H., Perkins S.L. Wolbachia infection and mitochondrial diversity in the canine heartworm (Dirofilaria immitis) Mitochondrial DNA. 2010;21(6):227–233. doi: 10.3109/19401736.2010.533765. [DOI] [PubMed] [Google Scholar]
- Blouin M.S. Molecular prospecting for cryptic species of nematodes: mitochondrial DNA versus internal transcribed spacer. Int. J. Parasitol. 2002;32(5):527–531. doi: 10.1016/s0020-7519(01)00357-5. [DOI] [PubMed] [Google Scholar]
- Blouin M.S., Yowell C.A., Courtney C.H., Dame J.B. Host movement and the genetic structure of populations of parasitic nematodes. Genetics. 1995;141(3):1007–1014. doi: 10.1093/genetics/141.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouin M.S., Yowell C.A., Courtney C.H., Dame J.B. Substitution bias, rapid saturation, and the use of mtDNA for nematode systematics. Mol. Biol. Evol. 1998;15(12):1719–1727. doi: 10.1093/oxfordjournals.molbev.a025898. [DOI] [PubMed] [Google Scholar]
- Brasil B.S., Nunes R.L., Bastianetto E., Drummond M.G., Carvalho D.C., Leite R.C., Oliveira D.A. Genetic diversity patterns of Haemonchus placei and Haemonchus contortus populations isolated from domestic ruminants in Brazil. Int. J. Parasitol. 2012;42(5):469–479. doi: 10.1016/j.ijpara.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Cerutti M., Citterio C., Bazzocchi C., Epis S., D'amelio S., Ferrari N., Lanfranchi P. Genetic variability of Haemonchus contortus (Nematoda: Trichostrongyloidea) in alpine ruminant host species. J. Helminthol. 2010;84(3):276–283. doi: 10.1017/S0022149X09990587. [DOI] [PubMed] [Google Scholar]
- Charlier J., Hoglund J., von Samson-Himmelstjerna G., Dorny P., Vercruysse J. Gastrointestinal nematode infections in adult dairy cattle: impact on production, diagnosis and control. Vet. Parasitol. 2009;164(1):70–79. doi: 10.1016/j.vetpar.2009.04.012. [DOI] [PubMed] [Google Scholar]
- Claerebout E., Charlier J., Vande Velde F. Farmer behavior and gastrointestinal nematodes in ruminant livestock–uptake of sustainable control approaches. Front. Vet. Sci. 2018;5:255. doi: 10.3389/fvets.2018.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig T.M. Food Animal Practice. Elsevier; 2009. Helminth parasites of the ruminant gastrointestinal tract; pp. 78–91. [Google Scholar]
- Dey A.R., Zhang Z., Begum N., Alim M.A., Hu M., Alam M.Z. Genetic diversity patterns of Haemonchus contortus isolated from sheep and goats in Bangladesh. Infect. Genet. Evol. 2019;68:177–184. doi: 10.1016/j.meegid.2018.12.021. [DOI] [PubMed] [Google Scholar]
- Dey A.R., Begum N., Biswas H., Alam M.Z. Prevalence and factors influencing gastrointestinal parasitic infections in sheep in Bangladesh. Ann. Parasitol. 2021;67(2):187–194. doi: 10.17420/ap6702.328. [DOI] [PubMed] [Google Scholar]
- Dey A.R., Begum N., Islam M.T., Alam M.Z. A large-scale epidemiological investigation on trematode infections in small ruminants in Bangladesh. Vet. Med. Sci. 2022;8:1219–1228. doi: 10.1002/vms3.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S.R., Tracey A., Laing R., Holroyd N., Bartley D., Bazant W., Brooks K. Genomic and transcriptomic variation defines the chromosome-scale assembly of Haemonchus contortus, a model gastrointestinal worm. Commun. Biol. 2020;3(1):1–16. doi: 10.1038/s42003-020-01377-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Smouse P.E., Quattro J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131(2):479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147(2):915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gareh A., Elhawary N.M., Tahoun A., Ramez A.M., El-Shewehy D., Elbaz E., Dyab A.K. Epidemiological, morphological and morphometric study on Haemonchus spp. recovered from goats in Egypt. Front. Vet. Sci. 2021;8 doi: 10.3389/fvets.2021.705619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser R.B., Chilton N.B., Hoste H., Stevenson L.A. Species identification of trichostrongyle nematodes by PCR-linked RFLP. Int. J. Parasitol. 1994;24(2):291–293. doi: 10.1016/0020-7519(94)90041-8. [DOI] [PubMed] [Google Scholar]
- Gasser R.B., Newton S.E. Genomic and genetic research on bursate nematodes: significance, implications and prospects. Int. J. Parasitol. 2000;30(4):509–534. doi: 10.1016/s0020-7519(00)00021-7. [DOI] [PubMed] [Google Scholar]
- Gasser R.B., Zhu X., Chilton N.B., Newton L.A., Nedergaard T., Guldberg P. Analysis of sequence homogenisation in rDNA arrays of Haemonchus contortus by denaturing gradient gel electrophoresis. Electrophoresis. 1998;19(14):2391–2395. doi: 10.1002/elps.1150191405. [DOI] [PubMed] [Google Scholar]
- Gasser R.B., Zhu X., McManus D.P. NADH dehydrogenase subunit 1 and cytochrome c oxidase subunit I sequences compared for members of the genus Taenia (Cestoda) Int. J. Parasitol. 1999;29(12):1965–1970. doi: 10.1016/s0020-7519(99)00153-8. [DOI] [PubMed] [Google Scholar]
- Gharamah A., Azizah M.S., Rahman W. Genetic variation of Haemonchus contortus (Trichostrongylidae) in sheep and goats from Malaysia and Yemen. Vet. Parasitol. 2012;188:268–276. doi: 10.1016/j.vetpar.2012.04.003. [DOI] [PubMed] [Google Scholar]
- Hall T.A. Vol. 41. 1999. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT; pp. 95–98. (Nucleic Acids Symposium Series). [Google Scholar]
- Hamid M. Biological diversity of farm animals in Bangladesh: A Review. SAARC J. Agric. 2019;17(2):15–29. [Google Scholar]
- Heise M., Epe C., Schnieder T. Differences in the second internal transcribed spacer (ITS-2) of eight species of gastrointestinal nematodes of ruminants. J. Parasitol. 1999;85:431–435. [PubMed] [Google Scholar]
- Hussain T., Periasamy K., Nadeem A., Babar M.E., Pichler R., Diallo A. Sympatric species distribution, genetic diversity and population structure of Haemonchus isolates from domestic ruminants in Pakistan. Vet. Parasitol. 2014;206(3–4):188–199. doi: 10.1016/j.vetpar.2014.10.026. [DOI] [PubMed] [Google Scholar]
- Jacquiet P., Humbert J., Comes A., Cabaret J., Thiam A., Cheikh D. Ecological, morphological and genetic characterization of sympatric Haemonchus spp. parasites of domestic ruminants in Mauritania. Parasitology. 1995;110(4):483–492. doi: 10.1017/s0031182000064829. [DOI] [PubMed] [Google Scholar]
- Jacquiet P., Cabaret J., Thiam E., Cheikh D. Host range and the maintenance of Haemonchus spp. in an adverse arid climate. Int. J. Parasitol. 1998;28(2):253–261. doi: 10.1016/s0020-7519(97)00185-9. [DOI] [PubMed] [Google Scholar]
- Kandil O.M., Abdelrahman K.A., Eid N.A., Elakabawy L.M., Helal M.A. Epidemiological study of genetic diversity and patterns of gene flow in Haemonchus species affecting domestic ruminants in Egypt. Bull. Natl. Res. Cent. 2018;42(1):1–6. [Google Scholar]
- Kralova-Hromadova I., Bazsalovicsova E., Oros M., Scholz T. Sequence structure and intragenomic variability of ribosomal ITS2 in monozoic tapeworms of the genus Khawia (Cestoda: Caryophyllidea), parasites of cyprinid fish. Parasitol. Res. 2012;111(4):1621–1627. doi: 10.1007/s00436-012-3001-z. [DOI] [PubMed] [Google Scholar]
- Laosutthipong C., Eardmusic S. Genetic characterization of Haemonchus contortus from slaughtered goats in Cha-am District, Phetchaburi Province, Thailand. Songklanakarin J. Sci. Technol. 2019;41:81–88. [Google Scholar]
- Luton K., Walker D., Blair D. Comparisons of ribosomal internal transcribed spacers from two congeneric species of flukes (Platyhelminthes: Trematoda: Digenea) Mol. Biochem. Parasitol. 1992;56(2):323–327. doi: 10.1016/0166-6851(92)90181-i. [DOI] [PubMed] [Google Scholar]
- MAFF . vol. 418. HM Stationery Office; Great Britain: 1986. (Manual of Veterinary Parasitological Laboratory Techniques). [Google Scholar]
- Mannan M.A., Chowdhury S., Hossain M.A., Kabir M.H.B. Genetic variability of Haemonchus contortus isolates in small ruminants from slaughterhouses in Bangladesh. Parasitol. Res. 2023;122(12):3101–3107. doi: 10.1007/s00436-023-08000-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy K.D., Boulinier T., Tirard C., Michalakis Y. Host-dependent genetic structure of parasite populations: differential dispersal of seabird tick host races. Evolution. 2003;57(2):288–296. doi: 10.1111/j.0014-3820.2003.tb00263.x. [DOI] [PubMed] [Google Scholar]
- McLeod R. Economic impact of worm infections in small ruminants in South East Asia, India and Australia. Worm Control Small Ruminants Tropical Asia. 2004;23 [Google Scholar]
- O’Connor L.J., Walkden-Brown S.W., Kahn L.P. Ecology of the free-living stages of major trichostrongylid parasites of sheep. Vet. Parasitol. 2006;142(1–2):1–15. doi: 10.1016/j.vetpar.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Omar A.I., Dey A.R., Alam M.B.B., Mondal M.M.H., Khan M.Y.A., Faruque M.O. Prevalence of common gastrointestinal parasite infection under natural grazing condition in black Bengal Goat of Bangladesh. IJACR. 2021;1(2):63–72. [Google Scholar]
- Pitaksakulrat O., Chaiyasaeng M., Artchayasawat A., Eamudomkarn C., Boonmars T., Kopolrat K.Y., Sithithaworn P. Genetic diversity and population structure of Haemonchus contortus in goats from Thailand. Infect. Genet. Evol. 2021;95 doi: 10.1016/j.meegid.2021.105021. [DOI] [PubMed] [Google Scholar]
- Poddar P.R., Begum N., Alim M.A., Dey A.R., Hossain M.S., Labony S.S. Prevalence of gastrointestinal helminths of sheep in Sherpur, Bangladesh. J. Adv. Vet. Anim. Res. 2017;4(3):274–280. [Google Scholar]
- Prichard R. Genetic variability following selection of Haemonchus contortus with anthelmintics. Trends Parasitol. 2001;17:445–453. doi: 10.1016/s1471-4922(01)01983-3. [DOI] [PubMed] [Google Scholar]
- Rose H., Caminade C., Bolajoko M.B., Phelan P., van Dijk J., Baylis M., Williams D., Morgan E.R. Climate-driven changes to the spatio-temporal distribution of the parasitic nematode, Haemonchus contortus, in sheep in Europe. Glob. Change Biol. 2016;22(3):1271–1285. doi: 10.1111/gcb.13132. [DOI] [PubMed] [Google Scholar]
- Rozas J. DNA sequence polymorphism analysis using DnaSP. Bioinf. DNA Seq. Anal. 2009;537:337–350. doi: 10.1007/978-1-59745-251-9_17. [DOI] [PubMed] [Google Scholar]
- Salle G., Doyle S.R., Cortet J., Cabaret J., Berriman M., Holroyd N., Cotton J.A. The global diversity of Haemonchus contortus is shaped by human intervention and climate. Nat. Commun. 2019;10(1):4811. doi: 10.1038/s41467-019-12695-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuvo S.M., Siddiqui T.R., Hoque M.R., Begum N., Paul D.R., Alim M.A., Dey A.R. The prevalence and potential factors associated with ecto-parasitic infestations in Black Bengal Goats in Mymensingh, Bangladesh. Bangladesh J. Vet. Med. 2021;19:93–105. [Google Scholar]
- Soulsby E.J.L. 7th ed. Bailliere Tindal and Cassell Ltd.; London: 1982. Helminths, Arthropod and Protozoa of Domesticated Animals. [Google Scholar]
- Stevenson L.A., Chilton N.B., Gasser R.B. Differentiation of Haemonchus placei from H. contortus (Nematoda: Trichostrongylidae) by the ribosomal DNA second internal transcribed spacer. Int. J. Parasitol. 1995;25:483–488. doi: 10.1016/0020-7519(94)00156-i. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M., Coop R., Wall R. 3rd ed. Blackwell Publishing; Oxford, UK: 2007. Veterinary Parasitology. [Google Scholar]
- Troell K., Engstrom A., Morrison D.A., Mattsson J.G., Hoglund J. Global patterns reveal strong population structure in Haemonchus contortus, a nematode parasite of domesticated ruminants. Int. J. Parasitol. 2006;36:1305–1316. doi: 10.1016/j.ijpara.2006.06.015. [DOI] [PubMed] [Google Scholar]
- Waller P., Chandrawathani P. Haemonchus contortus: parasite problem No. 1 from tropics-Polar Circle. Problems and prospects for control based on epidemiology. Trop. Biomed. 2005;22:131–137. [PubMed] [Google Scholar]
- Yin F., Gasser R.B., Li F., Bao M., Huang W., Zou F., Zhou Y. Genetic variability within and among Haemonchus contortus isolates from goats and sheep in China. Parasit. Vect. 2013;6:279. doi: 10.1186/1756-3305-6-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.