Abstract
Asbestos is a potent carcinogen associated with increased risks of malignant mesothelioma and lung cancer in humans. Although the mechanism of carcinogenesis remains elusive, the physicochemical characteristics of asbestos play a role in the progression of asbestos‐induced diseases. Among these characteristics, a high capacity to adsorb and accommodate biomolecules on its abundant surface area has been linked to cellular and genetic toxicity. Several previous studies identified asbestos‐interacting proteins. Here, with the use of matrix‐assisted laser desorption ionization‐time of flight mass spectrometry, we systematically identified proteins from various lysates that adsorbed to the surface of commercially used asbestos and classified them into the following groups: chromatin/nucleotide/RNA‐binding proteins, ribosomal proteins, cytoprotective proteins, cytoskeleton‐associated proteins, histones and hemoglobin. The surfaces of crocidolite and amosite, two iron‐rich types of asbestos, caused more protein scissions and oxidative modifications than that of chrysotile by in situ‐generated 4‐hydroxy‐2‐nonenal. In contrast, we confirmed the intense hemolytic activity of chrysotile and found that hemoglobin attached to chrysotile, but not silica, can work as a catalyst to induce oxidative DNA damage. This process generates 8‐hydroxy‐2′‐deoxyguanosine and thus corroborates the involvement of iron in the carcinogenicity of chrysotile. This evidence demonstrates that all three types of asbestos adsorb DNA and specific proteins, providing a niche for oxidative modification via catalytic iron. Therefore, considering the affinity of asbestos for histones/DNA and the internalization of asbestos into mesothelial cells, our results suggest a novel hypothetical mechanism causing genetic alterations during asbestos‐induced carcinogenesis. (Cancer Sci 2011; 102: 2118–2125)
Asbestos is a natural fibrous mineral that was heavily used in industry during the past century because of its durability, heat resistance and low cost. However, it has become clear that respiratory exposure to asbestos fibers, especially crocidolite and amosite, which have high biopersistence and contain abundant iron, is associated with high risks of developing malignant mesothelioma and lung cancer.( 1 , 2 , 3 ) Many countries anticipate increased numbers of mesothelioma patients in the coming decades because there is an extremely long incubation period (30–40 years) for this fatal disease following asbestos exposure.( 4 )
The molecular mechanism of asbestos‐induced carcinogenesis remains elusive,( 5 ) but both mesothelial cell injury and persistent macrophage activation are thought to be essential, if not sufficient, for mesotheliomagenesis.( 6 ) These two events interact in vivo, leading to genetic mutations, chromosomal aberrations and aneuploidy in mesothelial cells. At least four major hypotheses related to the underlying mechanisms have been proposed.( 6 , 7 )
First, the free radical theory postulates that DNA is injured by reactive oxygen species generated through a foreign body reaction or catalytic action of the asbestos surface.( 8 , 9 , 10 , 11 , 12 ) Asbestos fibers of a large size, especially those that are quite long (>15–20 μm), interrupt macrophage phagocytosis and prohibit them from clearing fibers.( 13 ) In this situation, activated macrophages release cytokines and oxidants, thereby inducing chronic inflammation.( 14 ) Even in the absence of activated phagocytes, asbestos can produce free radicals via the Fenton reaction because some types of amphibole asbestos, for example, crocidolite and amosite, include iron as an integral component of their chemical structure and other types of asbestos contain iron as a surface impurity.( 8 , 15 ) Iron is the most abundant heavy metal in the human body, but excess iron can work as a catalyst for the generation of free radicals, leading to carcinogenesis.( 16 , 17 ) In experiments involving mammalian cells, asbestos fibers produced 8‐hydroxy‐2′‐deoxyguanosine (8‐OHdG),( 18 , 19 , 20 , 21 ) which is a common oxidative modification of DNA involved in mutagenesis, carcinogenesis and aging.( 22 , 23 , 24 ) Asbestos fibers also induce clastogenic events in a free radical‐dependent manner.( 25 )
Second, the mitotic disturbance theory proposes that asbestos fibers physically interact with chromosomes directly and/or via mitotic spindles, thereby inducing chromosomal aberrations.( 26 , 27 , 28 , 29 , 30 , 31 ) This is indeed a specific event caused by fibrous particles and might be involved in the early induction of chromosomal aberrations observed in Syrian hamster cells exposed to asbestos.( 31 )
Third, the molecule adsorption theory suggests that adsorption of various molecules on the surface of asbestos fibers causes the accumulation of intrinsic or extrinsic carcinogenesis‐associated molecules and interferes with intracellular signaling pathways.( 7 ) Certain carcinogenic molecules, such as benzo(a)pyrene in cigarette smoke, are known to have high affinity for asbestos and have a cooperative mutagenic effect.( 32 , 33 , 34 ) Various endogenous molecules, such as vitronectin and tubulin, are also likely to interact with asbestos, playing important roles in fiber internalization( 35 , 36 ) and mitotic disturbances,( 37 ) respectively. The deposition of molecules on asbestos has also attracted attention as a mechanism underlying the formation of asbestos bodies, a process that was extensively investigated in previous studies.( 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 ) These studies suggested the involvement of acid mucopolysaccharides,( 40 ) bilirubin,( 50 ) proteins( 38 ) and iron( 39 , 41 , 43 , 47 , 51 , 52 ) in the development of asbestos bodies. Among these components, the pathobiological contribution of iron should be carefully considered. Iron exists in and near asbestos bodies in various forms: hemosiderin,( 52 ) ferritin( 39 , 51 ) and colloidal iron.( 43 ) Governa et al. ( 42 ) discussed the reactivity of iron inside asbestos bodies and concluded that it can catalyze free radical formation in reductive conditions in vitro. Considering that hemosiderin is capable of inducing hydroxyl radicals under physiological conditions,( 53 , 54 ) iron‐rich asbestos bodies might play a role in oxidative tissue damage.
Finally, the chronic inflammation theory suggests that persistently activated macrophages contribute to the initiation as well as the progression of carcinogenesis.( 55 , 56 ) Particularly in mesotheliomagenesis, chronic inflammation is associated with asbestos‐induced genotoxicity in mesothelial cells. Yang et al. ( 57 ) showed that tumor necrosis factor‐α (TNF‐α), a cytokine persistently released from macrophages during inflammation, inhibits asbestos‐induced mesothelial cell death and might increase the likelihood of transformation.
All of the aforementioned theories are clearly associated with each other. However, currently there is little information available on asbestos‐interacting proteins. Therefore, in the present study we used matrix‐assisted laser desorption ionization‐time of flight mass spectrometry (MALDI‐TOF/MS) to systematically identify proteins that adsorb to the surface of different asbestos fibers, investigated the modifications of adsorptive molecules and evaluated their possible involvement in mesothelial carcinogenesis.
Materials and Methods
Full materials and methods are provided in Data S1.
Materials. Three types of asbestos (chrysotile, crocidolite and amosite) were acquired from Union for International Cancer Control (UICC; Geneva, Switzerland) and suspended in saline. MeT5A cells were obtained from the American Type Culture Collection (Manassas, VA, USA). Silica (powder, 0.014 μm) was purchased from Sigma Aldrich (St. Louis, MO, USA) and a protease inhibitor cocktail (complete mini) was purchased from Roche diagnostics (Basel, Switzerland).
Collection and analysis of asbestos‐interacting proteins. We modified a previously reported method( 37 ) to collect proteins adsorbed to the surface of asbestos fibers. Briefly, lysates( 58 ) of various rat tissue including lung, liver, kidney, brain and tunica vaginalis or MeT5A human mesothelial cells (200–400 μg) were mixed with each fiber (250 μg), and the total volume was adjusted to 1 mL with radio‐immunoprecipitation assay (RIPA) buffer. After >3 h of incubation at 4 or 37°C, the mixture was centrifuged (20 000 g ) at 4°C for 5 min. The supernatant was discarded and the pellet was washed three times with RIPA buffer. After the final centrifugation, we carefully discarded the supernatant, directly added SDS‐PAGE sample buffer, and heated the sample at 95°C for 10 min. The samples were centrifuged (20 000 g ) at 4°C for 2 min, and the supernatant was analyzed using SDS‐PAGE. We used a silver staining kit to stain the SDS‐PAGE gels but avoided using glutaraldehyde to minimize unnecessary protein modifications.
Results
Adsorption of specific proteins by asbestos. To collect proteins adsorbed by asbestos fibers, we used an assay system similar to immunoprecipitation that we modified from a previously described method.( 37 ) Both silica and asbestos adsorbed a variety of specific proteins (Fig. 1a). The total amount of proteins bound to fibers was highest for chrysotile, followed by crocidolite, amosite and silica. We determined that approximately 1 μg of protein had adsorbed onto 250 μg asbestos by comparison with the original amount of rat lung lysate (2 μg, Fig. 1a). Crocidolite and amosite fibers yielded similar protein profiles on SDS‐PAGE gels, whereas that of chrysotile was distinct, although all three were different from the original profiles of the untreated lysates. Much less protein was adsorbed on silica than asbestos when a protease inhibitor cocktail, which protects proteins from enzymatic degradation, was used for sample preparation (data not shown). Thus, proteins are adsorbed on the surfaces of silica and asbestos via different mechanisms. To test the specificity of the asbestos–protein interaction, we pre‐incubated the fibers with actin or albumin prior to the incubation with tissue lysate. This treatment yielded essentially the same protein profiles, indicating that neither actin nor albumin could inhibit protein adsorption on silica or asbestos (data not shown) and that the protein adsorption was specific. To further confirm the specificity of asbestos–protein interaction, we incubated chrysotile with a various amount of rat lung lysate (10, 100 and 400 μg) and found that there is a difference between proteins in its affinity to asbestos (Fig. 1h). Therefore, there is a specific preference of proteins to asbestos, even though the specificity between proteins and asbestos is much weaker than that of protein–protein interaction because there are many kinds of proteins adsorptive to asbestos simultaneously.
Figure 1.
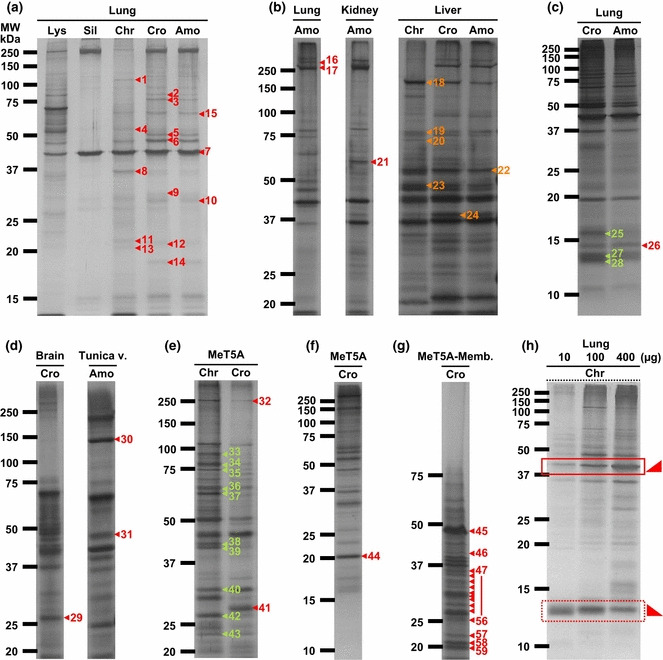
Adsorption of specific proteins by asbestos fibers. Lysates from rat tissues or MeT5A mesothelial cells were incubated with each inorganic material and silver staining was performed after SDS‐PAGE. Asbestos exhibited higher adsorption than silica (Sil), and chrysotile (Chr) was the most adsorptive. (a–h) The panels show the original gels subjected to matrix‐assisted laser desorption ionization‐time of flight mass spectrometry (MALDI‐TOF/MS) analysis. The amount of rat lung lysate (Lys) loaded in panel (a) was 2 μg. Numbers correspond to the identified proteins listed in Table S1. The gels shown in each panel are different in the concentration of polyacrylamide and the combination of inorganic material and cell/tissue lysate. The origin of the rat lysate (e.g. lung, kidney, liver, etc.) used for the incubation is shown at the top and the species of the inorganic material is shown next to the top. (h) Chrysotile was incubated with 10, 100 or 400 μg of rat lung lysate and the proteins adsorbed onto chrysotile were analyzed by the coupling of SDS‐PAGE and silver staining. The square in a continuous line shows an increase in the amount of asbestos‐binding proteins as the total amount of proteins incubated increases. In contrast, the square in a dotted line shows the opposite. Cro, crocidolite; Amo, amosite; Tunica v., tunica vaginalis; MeT5A‐Memb., membrane fraction of MeT5A cells.
Identification of asbestos‐interacting proteins with MALDI‐TOF/MS. To identify asbestos‐interacting proteins, we performed in‐gel digestion and subjected the samples to MALDI‐TOF/MS.( 60 ) More than 100 proteins were found to interact with asbestos (Fig. 1a–g; Table 1; Table S1). We classified these proteins into the following eight categories on the basis of their cellular localization and function: chromatin/nucleotide/RNA‐binding proteins; ribosomal proteins; cytoprotective proteins; cytoskeleton‐associated proteins; histones; and hemoglobin.
Table 1.
A summarized list of asbestos‐binding proteins
| Chromatin‐binding proteins |
| ATP‐dependent DNA helicase 2 subunit 2 |
| Coiled‐coil domain‐containing protein 124 |
| DNA replication licensing factor MCM6 |
| DNA replication licensing factor MCM7 |
| DNA‐(apurinic or apyrimidinic site) lyase |
| Flap endonuclease 1 |
| Interleukin enhancer‐binding factor 2 |
| Transcription initiation factor IIE subunit β |
| RNA‐binding proteins |
| Cleavage and polyadenylation specificity factor subunit 5 |
| Eukaryotic translation initiation factor 2 subunit 1 |
| FUS glycine‐rich protein |
| Heterogeneous nuclear ribonucleoprotein A0 |
| Heterogeneous nuclear ribonucleoprotein A1 |
| Heterogeneous nuclear ribonucleoprotein U |
| KH domain‐containing RNA‐binding signal transduction‐associated protein 1 |
| Probable ATP‐dependent RNA helicase DDX5 |
| Putative pre‐mRNA‐splicing factor‐ATP‐dependent RNA helicase DHX15 |
| RNA‐binding protein EWS |
| rRNA 2′‐O‐methyltransferase fibrillarin |
| Splicing factor, proline and glutamine rich |
| THO complex subunit 4 |
| Nucleotide‐binding proteins |
| ATP synthase subunit α |
| ATP synthase subunit O |
| Carbamoyl‐phosphate synthase |
| Developmentally regulated GTP‐binding protein 1 |
| Elongation factor 1‐ α 1 |
| Elongation factor 1‐ α 2 |
| Elongation factor Tu |
| Glutamate dehydrogenase 1 |
| Succinyl‐CoA ligase |
| Cytoprotective proteins |
| 78 kDa glucose‐regulated protein |
| DnaJ homolog subfamily B member 13 |
| Glutathione peroxidase 1 |
| Heat shock 70 kDa protein 1/2 |
| Heat shock cognate 71 kDa protein |
| Peroxiredoxin 1 |
| Peroxiredoxin 2 |
| Superoxide dismutase (Mn) |
| Cytoskeleton‐associated proteins |
| Actin |
| α‐Actinin 1 |
| α‐Actinin 4 |
| Annexin A2 |
| Cytoskeleton‐associated protein 4 |
| Ezrin |
| Filamin‐A |
| Keratin type I cytoskeletal 18 |
| Keratin type II cytoskeletal 8 |
| Moesin |
| Myosin 9 |
| Myosin 10 |
| Myosin 11 |
| Myosin binding protein C |
| Myosin light polypeptide 6 |
| Predicted: similar to Myosin 11 |
| Predicted: similar to septin‐11 |
| Predicted: similar to tubulin polymerization‐promoting protein |
| Radixin |
| Septin‐2 |
| Septin‐7 |
| Spectrin α‐chain |
| Tubulin β‐5 chain |
| Histones |
| Histone H2A type 3 |
| Histone H2B type 1 |
| Histone H3.3 |
| Histone H4 |
| Hemoglobins |
| Hemoglobin subunit α 1/2 |
| Hemoglobin subunit β 1 |
| Hemoglobin subunit β 2 |
| Ribosomal proteins |
| 39S ribosomal protein L28 |
| 39S ribosomal protein L40 |
| 39S ribosomal protein L48 |
| 40S ribosomal protein S2 |
| 40S ribosomal protein S3a |
| 40S ribosomal protein S4, X isoform |
| 40S ribosomal protein S7 |
| 40S ribosomal protein S9 |
| 40S ribosomal protein S16 |
| 60S ribosomal protein L7a |
| 60S ribosomal protein L8 |
| 60S ribosomal protein L9 |
| 60S ribosomal protein L10 |
| 60S ribosomal protein L13 |
| 60S ribosomal protein L17 |
| 60S ribosomal protein L18 |
| 60S ribosomal protein L18a |
| 60S ribosomal protein L22 |
| 60S ribosomal protein L23a |
| 60S ribosomal protein L31 |
Refer to Table S1 for details.
Proteins adsorbed to asbestos are modified by 4‐hydroxy‐2‐nonenal (HNE). We studied the oxidative modifications of proteins adsorbed to asbestos because asbestos can catalyze the Fenton reaction.( 61 ) We analyzed the presence of HNE( 62 ) modifications of the proteins using western blotting.( 63 , 64 ) 4‐Hydroxy‐2‐nonenal is a major lipid peroxidation end‐product associated with a variety of signaling pathways. We found that all three types of asbestos induced HNE modification of adsorbed proteins (Fig. 2a). In particular, crocidolite and amosite, which contain high amounts of iron (approximately 30%), induced higher amounts of HNE modification than chrysotile. Furthermore, actin incubated with amosite was degraded and modified by HNE (Fig. 2b). Interestingly, this phenomenon of actin degradation was only observed when we used amosite, not chrysotile or crocidolite (data not shown), indicating that asbestos–protein interactions can differ even between crocidolite and amosite. Thus, asbestos not only adsorbed proteins on its surface but also modified proteins via the HNE modification and degraded proteins.
Figure 2.
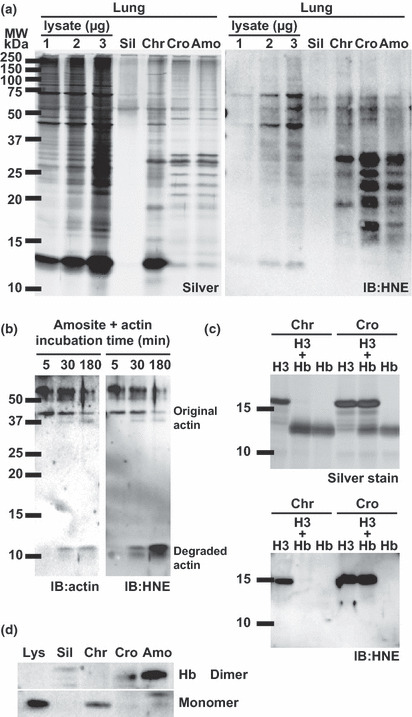
4‐Hydroxy‐2‐nonenal modification of proteins adsorbed on asbestos. (a) After incubation of lung lysates with asbestos at 37°C, notable increases in the 4‐hydroxy‐2‐nonenal (HNE) modification were observed in the samples incubated with crocidolite (Cro), amosite (Amo) and chrysotile (Chr). IB, immunoblot. (b) After incubation with amosite at room temperature for the indicated periods, actin was degraded and modified by HNE in a time‐dependent manner. (c) Competition of histone H3 and hemoglobin for the asbestos surface. When chrysotile (Chr) was used, hemoglobin had a higher affinity than histone H3. Histone H3 was modified by HNE. When crocidolite (Cro) was used, histone H3 and hemoglobin showed similar affinities, although only histone H3 was modified by HNE. (d) All types of asbestos fibers adsorbed hemoglobin, whereas silica showed no adsorptive activity.
Among the various asbestos‐interacting proteins, we focused on histone H3 and hemoglobin to evaluate differences in adsorptive activity and oxidative modification. We therefore incubated asbestos with both histone H3 and hemoglobin and analyzed various parameters (Fig. 2c). Hemoglobin showed a higher binding specificity for chrysotile than histone H3, but the two proteins had similar specificity values for crocidolite. Furthermore, histone H3 was modified by HNE, but hemoglobin was not (Fig. 2c). Hemoglobin specifically bound to asbestos, but not to silica. Finally, hemoglobin dimers were observed after incubation with crocidolite or amosite (Fig. 2d).
Hemoglobin adsorption to asbestos results in high catalytic activity.( 59 ) Asbestos is known to cause hemolysis.( 65 ) We confirmed this property using UICC asbestos and silica (Fig. 3a,b). Silica exhibited the most potent hemolytic activity, followed by chrysotile. Crocidolite and amosite were also hemolytic but with a much lower activity (approximately 200‐fold less). Silica, chrysotile and crocidolite were then evaluated for catalytic activity related to free radical generation in the presence of bleomycin sulfate and DNA. Both asbestos fibers showed significantly higher catalytic activity( 59 ) after incubation with hemoglobin, whereas silica did not (Fig. 3c), consistent with the result shown in Figure 2(d). These results suggest that asbestos induces hemolysis, collects iron‐containing proteins, namely hemoglobin, on its surface and catalyzes free radical generation, whereas silica causes hemolysis but lacks subsequent catalytic activity.
Figure 3.
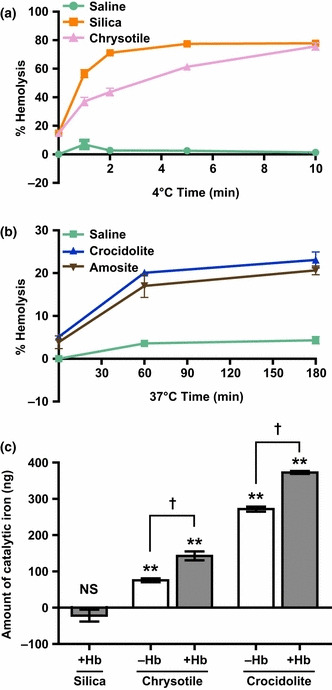
Hemolytic activity of each type of asbestos and increases in catalytic iron resulting from adsorption of hemoglobin on the asbestos surface. (a,b) Silica and chrysotile exhibited prominent hemolysis, whereas crocidolite and amosite showed lower hemolytic activity. Hemolysis was time‐dependent and eventually reached a plateau. Incubations were performed at 4 or 37°C as indicated. (c) Increased catalytic activity of chrysotile and crocidolite after hemoglobin adsorption. Silica did not show catalytic activity even after incubation with hemoglobin. The amount of catalytic iron was calculated based on a standard curve using Fe(NO3)3. **P < 0.01 relative to the negative control (deionized water instead of asbestos or iron). †P < 0.05 between the indicated groups. NS, no significant detection.
Asbestos adsorbs DNA and generates 8‐OHdG on its surface. In addition to proteins, we also studied DNA adsorption to the asbestos surface. We quantified the amount of DNA adsorbed on asbestos fibers (250 or 500 μg). Chrysotile most effectively adsorbed DNA, followed by silica, crocidolite and amosite (Fig. 4a). We then found that the DNA adsorbed by crocidolite was oxidized to generate 8‐OHdG( 66 ) after incubation at 37°C for 3 h (Fig. 4b). Furthermore, we found that adding hemoglobin to the reaction with chrysotile enhanced the oxidation of DNA in the presence of hydrogen peroxide (Fig. 4c). Collectively, every commercial type of asbestos, regardless of its iron content, can utilize iron in various forms to induce oxidative DNA damage.
Figure 4.
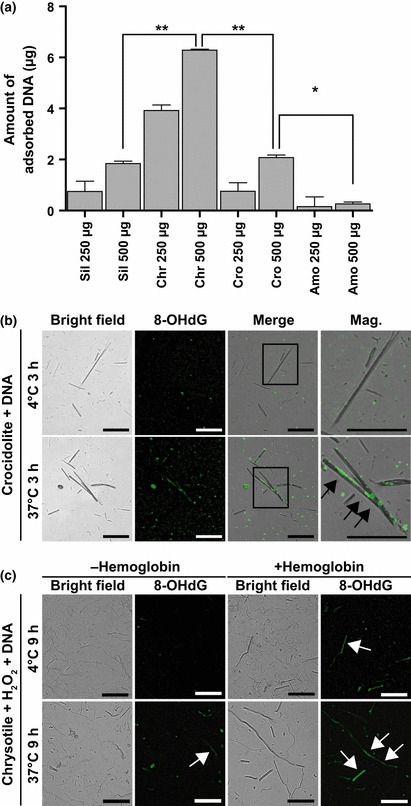
Simultaneous adsorption of DNA and generation of 8‐hydroxy‐2′‐deoxyguanosine (8‐OHdG) on asbestos fibers. (a) Asbestos and silica adsorbed DNA after incubation at 37°C. Chrysotile was the most potent DNA adsorbent, followed by silica, crocidolite and amosite. *P < 0.05. **P < 0.01. (b) Crocidolite was incubated with genomic DNA at 4 or 37°C for 3 h. Fluorescent immunohistochemistry was performed after incubation to show the generation of 8‐OHdG on the asbestos surface (arrows). (c) Formation of 8‐OHdG on the surface of hemoglobin‐treated chrysotile after incubation with DNA in the presence of H2O2 (arrows). Scale bar, 50 μm.
Asbestos interacts with red blood cells after instillation. Finally, we investigated how asbestos fibers interact with red blood cells in vivo. We instilled chrysotile or crocidolite suspension through the airways of mice or rats and 3 h later their lungs were then collected and histopathological specimens were prepared. We found that both types of asbestos fibers were surrounded by inflammatory cells and red blood cells (Fig. 5, top panels; hematoxylin and eosin‐stained sections). Immunohistochemistry using an antibody against hemoglobin revealed that chrysotile was directly interacting with red blood cells and that hemoglobin was colocalized with crocidolite, suggesting that the surface of asbestos fibers can accommodate erythrocytes as well as hemoglobin in vivo.
Figure 5.
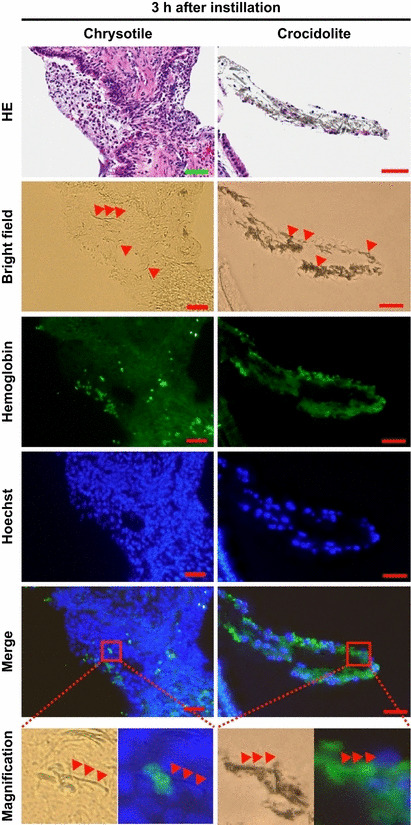
Direct interaction of red blood cells/hemoglobin with asbestos fibers in vivo. Three hours after instillation of asbestos fiber suspension to the airways of mice or rats, asbestos fibers were surrounded by inflammatory cells as well as red blood cells (in hematoxylin and eosin [HE]‐stained sections). Immunohistochemistry to detect hemoglobin revealed that chrysotile fibers directly interacted with erythrocytes and that hemoglobin was colocalized with crocidolite with serial sections. Arrowheads indicate asbestos fibers. Hoechst, hoechst 33342; scale bar, 50 μm.
Discussion
We identified asbestos‐interacting proteins using a method similar to immunoprecipitation. This method allowed us to measure the selective adsorption of proteins on the asbestos surface using cell or tissue lysates. All of the proteins identified in the present study are listed in Table S1. We classified 99 out of 128 asbestos‐interacting proteins into the following eight categories: nine chromatin‐binding proteins; 10 nucleotide‐binding proteins; 14 RNA‐binding proteins; 24 ribosomal proteins; nine cytoprotective proteins; 26 cytoskeleton‐associated proteins; four histones; and three hemoglobin subunits (Table S1). These results indicate that not only DNA (Fig. 4a) but also many types of DNA‐interacting proteins have an affinity for asbestos. Indeed, physical interactions between the mitotic spindle and asbestos might cause mitotic disturbances and chromosomal aberrations.( 7 , 37 ) Moreover, our identification of RNA‐binding and ribosomal proteins as asbestos‐interacting proteins indicates that asbestos might interfere with chromosomal replication, transcription and translation.
The different affinities of each asbestos type for DNA can be partially explained by their surface charges. The surface of chrysotile is positively charged, whereas crocidolite and amosite are negatively charged.( 3 ) Thus, chrysotile provides more suitable surface area for a negatively charged biomolecule such as DNA.
Among the cytoskeleton‐associated proteins, tubulin, actin, vimentin and cytoskeleton‐associated protein 4 were previously reported to interact with asbestos.( 7 , 37 ) In addition, we identified many other cytoskeleton‐associated proteins, including α actinin 1 and 4, filamin‐A, keratin 8 and 18, myosin 9, 10 and 11, septin 2 and 7, spectrin, ezrin, radixin and moesin. Adsorption of these proteins onto the surface of asbestos might affect cytoskeletal regulation.
We identified several cytoprotective proteins that metabolize reactive oxygen species (ROS), including manganese superoxide dismutase, glutathione peroxidase 1, and peroxiredoxin 1 and 2,( 67 ) suggesting that asbestos might disturb the redox state of cells not only by generating ROS but also by adsorbing proteins that metabolize and reduce ROS.
Hemoglobin was identified as an asbestos‐interacting protein. This interaction was specific to asbestos at body temperature (37°C) in vitro and did not occur on silica. Hemoglobin is a major oxygen‐transporting protein and is released from red blood cells during hemolysis. Adsorption of hemoglobin on the asbestos surface has been mentioned previously,( 65 ) but the present study is the first to demonstrate that this event augments asbestos‐induced free radical generation (3, 4). We also investigated the direct interaction of red blood cells/hemoglobin and asbestos fibers by immunohistochemistry (Fig. 5), although to what extent this direct interaction contributes to oxidative damage in surrounding tissue still remains elusive.
In addition to enhancement of oxidative damage, this direct interaction might be the first step in the formation of asbestos bodies. Governa et al. ( 50 ) suggested that bilirubin was present in the innermost layer of the asbestos body. Because bilirubin is a metabolite of heme, our results support this idea. Thus, we propose a model in which asbestos utilizes heme iron from erythrocytes to enhance free radical generation, thereby inducing DNA damage in vivo, at least temporarily. Hemoglobin metabolism is mediated by heme oxygenase‐1, which is upregulated when animals or cells are treated with asbestos.( 68 , 69 ) Therefore, this enzyme might play a role in the conversion of hemoglobin into bilirubin on asbestos.
We also provided evidence suggesting that asbestos can modify proteins with HNE. In particular, incubation with asbestos promoted the accumulation of HNE modifications on both histone H3 and actin, leading to subsequent degradation of the latter. We believe that membrane lipids in the lysate and contaminant lipids in commercial proteins are the source of HNE in this setting. Here, we propose that asbestos not only adsorbs proteins but also provides a niche for oxidative reactions. This situation is also true in the case of DNA. DNA is adsorbed on the surface of asbestos (Fig. 4a) and oxidatively modified to 8‐OHdG (Fig. 4b,c). This reaction might occur in the nuclei of dividing cells, leading to prominent genomic alterations.
In conclusion, we have identified and classified a variety of asbestos‐interacting proteins. Among these proteins, we believe that hemoglobin and chromatin constituents such as DNA and histones are especially important; hemoglobin is an iron source and histones lie in close proximity to genomic DNA. DNA also showed a high affinity for asbestos, especially chrysotile. Taken together, we propose that asbestos provides a niche for oxidative reactions by specifically adsorbing various important proteins and DNA and subsequently generating local iron overload (Fig. 6). Among the three types of asbestos, chrysotile induced mesothelioma most rapidly when injected intraperitoneally to rats (Li Jiang, Hirotaka Nagai and Shinya Toyokuni, unpublished data, 2011). With these results, further studies are necessary to re‐evaluate the risk posed by chrysotile exposure in the development of lung cancer and mesothelioma.
Figure 6.
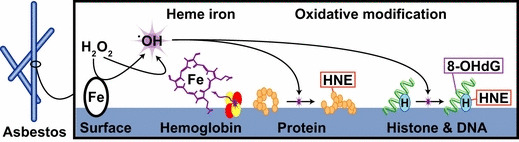
Hypothetical scheme of asbestos providing a niche for oxidative modification. The asbestos surface offers a niche for oxidative chemical reactions by concentrating substrates and iron. In the case of crocidolite and amosite, iron as an asbestos component plays a major role as a catalyst. However, in the case of chrysotile, hemolysis and subsequent hemoglobin attachment are important because the adsorbed hemoglobin possesses catalytic activity. Numerous important biomolecules, including DNA and nuclear proteins, are oxidatively modified by these mechanisms, a process closely associated with carcinogenesis. 8‐OHdG, 8‐hydroxy‐2′‐deoxyguanosine; HNE, 4‐hydroxy‐2‐nonenal.
Disclosure Statement
The authors have no conflict of interest.
Supporting information
Table S1. A list of asbestos‐binding proteins.
Data S1. Materials and methods.
Supporting info item
Supporting info item
Acknowledgments
We thank Nobuaki Misawa (Toyokuni laboratory) and Yi Zhong (Graduate School of Medicine, Kyoto University) for technical assistance and Seishiro Hirano (National Institute for Environmental Studies), Yasushi Shinohara (National Institute of Occupational Health and Safety) and Norihiko Kohyama (National Science Laboratory, Faculty of Economics, Toyo University) for the kind gift of asbestos. We acknowledge the support of the Division for Medical Research Engineering, Nagoya University Graduate School of Medicine, for fluorescent microscopy. This study was supported by a MEXT grant (Special Coordination Funds for Promoting Science and Technology), a research grant from the Princess Takamatsu Cancer Research Fund (10‐24213), a grant‐in‐aid for cancer research from the Ministry of Health, Labour and Welfare of Japan, a grant‐in‐aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan, a grant from the Takeda Science Foundation and a grant‐in‐aid from the Japan Society for the Promotion of Science Fellows (H. N.).
References
- 1. Berry G. Models for mesothelioma incidence following exposure to fibers in terms of timing and duration of exposure and the biopersistence of the fibers. Inhal Toxicol 1999; 11: 111–30. [DOI] [PubMed] [Google Scholar]
- 2. Pass HI, Vogelzang NJ, Carbone M, eds. Malignant Mesothelioma: Advances in Pathogenesis, Diagnosis, and Translational Therapies. New York, NY: Springer Science + Business Media Inc., 2005. [Google Scholar]
- 3. Roggli VL, Oury TD, Sporn TA, eds. Pathology of Asbestos‐Associated Diseases. New York: Springer Verlag, 2004. [Google Scholar]
- 4. Robinson BW, Lake RA. Advances in malignant mesothelioma. N Engl J Med 2005; 353: 1591–603. [DOI] [PubMed] [Google Scholar]
- 5. Sekido Y. Genomic abnormalities and signal transduction dysregulation in malignant mesothelioma cells. Cancer Sci 2010; 101: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nagai H, Toyokuni S. Biopersistent fiber‐induced inflammation and carcinogenesis: lessons learned from asbestos toward safety of fibrous nanomaterials. Arch Biochem Biophys 2010; 502: 1–7. [DOI] [PubMed] [Google Scholar]
- 7. Toyokuni S. Mechanisms of asbestos‐induced carcinogenesis. Nagoya J Med Sci 2009; 71: 1–10. [PMC free article] [PubMed] [Google Scholar]
- 8. Kamp DW, Graceffa P, Pryor WA, Weitzman SA. The role of free radicals in asbestos‐induced diseases. Free Radic Biol Med 1992; 12: 293–315. [DOI] [PubMed] [Google Scholar]
- 9. Maples KR, Johnson NF. Fiber‐induced hydroxyl radical formation: correlation with mesothelioma induction in rats and humans. Carcinogenesis 1992; 13: 2035–9. [DOI] [PubMed] [Google Scholar]
- 10. Mossman BT, Marsh JP, Sesko A et al. Inhibition of lung injury, inflammation, and interstitial pulmonary fibrosis by polyethylene glycol‐conjugated catalase in a rapid inhalation model of asbestosis. Am Rev Respir Dis 1990; 141: 1266–71. [DOI] [PubMed] [Google Scholar]
- 11. Vallyathan V, Shi X. The role of oxygen free radicals in occupational and environmental lung diseases. Environ Health Perspect 1997; 105(Suppl. 1): 165–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weitzman SA, Graceffa P. Asbestos catalyzes hydroxyl and superoxide radical generation from hydrogen peroxide. Arch Biochem Biophys 1984; 228: 373–6. [DOI] [PubMed] [Google Scholar]
- 13. Donaldson K, Murphy FA, Duffin R, Poland CA. Asbestos, carbon nanotubes and the pleural mesothelium: a review of the hypothesis regarding the role of long fibre retention in the parietal pleura, inflammation and mesothelioma. Part Fibre Toxicol 2010; 7: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Donaldson K, Brown GM, Brown DM, Bolton RE, Davis JM. Inflammation generating potential of long and short fibre amosite asbestos samples. Br J Ind Med 1989; 46: 271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harington JS. Chemical studies of asbestos. Ann N Y Acad Sci 1965; 132: 31–47. [DOI] [PubMed] [Google Scholar]
- 16. Toyokuni S. Iron‐induced carcinogenesis: the role of redox regulation. Free Radic Biol Med 1996; 20: 553–66. [DOI] [PubMed] [Google Scholar]
- 17. Toyokuni S. Role of iron in carcinogenesis: cancer as a ferrotoxic disease. Cancer Sci 2009; 100: 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chao CC, Park SH, Aust AE. Participation of nitric oxide and iron in the oxidation of DNA in asbestos‐treated human lung epithelial cells. Arch Biochem Biophys 1996; 326: 152–7. [DOI] [PubMed] [Google Scholar]
- 19. Kasai H. Analysis of a form of oxidative DNA damage, 8‐hydroxy‐2′‐deoxyguanosine, as a marker of cellular oxidative stress during carcinogenesis. Mutat Res 1997; 387: 147–63. [DOI] [PubMed] [Google Scholar]
- 20. Takeuchi T, Morimoto K. Crocidolite asbestos increased 8‐hydroxydeoxyguanosine levels in cellular DNA of a human promyelocytic leukemia cell line, HL60. Carcinogenesis 1994; 15: 635–9. [DOI] [PubMed] [Google Scholar]
- 21. Xu A, Wu LJ, Santella RM, Hei TK. Role of oxyradicals in mutagenicity and DNA damage induced by crocidolite asbestos in mammalian cells. Cancer Res 1999; 59: 5922–6. [PubMed] [Google Scholar]
- 22. Harman D. The aging process. Proc Natl Acad Sci USA 1981; 78: 7124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuchino Y, Mori F, Kasai H et al. Misreading of DNA templates containing 8‐hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature 1987; 327: 77–9. [DOI] [PubMed] [Google Scholar]
- 24. Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation‐damaged base 8‐oxodG. Nature 1991; 349: 431–4. [DOI] [PubMed] [Google Scholar]
- 25. Poser I, Rahman Q, Lohani M et al. Modulation of genotoxic effects in asbestos‐exposed primary human mesothelial cells by radical scavengers, metal chelators and a glutathione precursor. Mutat Res 2004; 559: 19–27. [DOI] [PubMed] [Google Scholar]
- 26. Ault JG, Cole RW, Jensen CG, Jensen LC, Bachert LA, Rieder CL. Behavior of crocidolite asbestos during mitosis in living vertebrate lung epithelial cells. Cancer Res 1995; 55: 792–8. [PubMed] [Google Scholar]
- 27. Barrett JC, Lamb PW, Wiseman RW. Multiple mechanisms for the carcinogenic effects of asbestos and other mineral fibers. Environ Health Perspect 1989; 81: 81–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dopp E, Saedler J, Stopper H, Weiss DG, Schiffmann D. Mitotic disturbances and micronucleus induction in Syrian hamster embryo fibroblast cells caused by asbestos fibers. Environ Health Perspect 1995; 103: 268–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dopp E, Schiffmann D. Analysis of chromosomal alterations induced by asbestos and ceramic fibers. Toxicol Lett 1998; 97: 155–62. [DOI] [PubMed] [Google Scholar]
- 30. Hesterberg TW, Barrett JC. Induction by asbestos fibers of anaphase abnormalities: mechanism for aneuploidy induction and possibly carcinogenesis. Carcinogenesis 1985; 6: 473–5. [DOI] [PubMed] [Google Scholar]
- 31. Oshimura M, Hesterberg TW, Barrett JC. An early, nonrandom karyotypic change in immortal Syrian hamster cell lines transformed by asbestos: trisomy of chromosome 11. Cancer Genet Cytogenet 1986; 22: 225–37. [DOI] [PubMed] [Google Scholar]
- 32. DiPaolo JA, DeMarinis AJ, Doniger J. Asbestos and benzo(a)pyrene synergism in the transformation of Syrian hamster embryo cells. Pharmacology 1983; 27: 65–73. [DOI] [PubMed] [Google Scholar]
- 33. Lakowicz JR, Hylden JL. Asbestos‐mediated membrane uptake of benzo[a]pyrene observed by fluorescence spectroscopy. Nature 1978; 275: 446–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mossman BT, Eastman A, Bresnick E. Asbestos and benzo[a]pyrene act synergistically to induce squamous metaplasia and incorporation of [3H]thymidine in hamster tracheal epithelium. Carcinogenesis 1984; 5: 1401–4. [DOI] [PubMed] [Google Scholar]
- 35. Boylan AM, Sanan DA, Sheppard D, Broaddus VC. Vitronectin enhances internalization of crocidolite asbestos by rabbit pleural mesothelial cells via the integrin alpha v beta 5. J Clin Invest 1995; 96: 1987–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu W, Ernst JD, Broaddus VC. Phagocytosis of crocidolite asbestos induces oxidative stress, DNA damage, and apoptosis in mesothelial cells. Am J Respir Cell Mol Biol 2000; 23: 371–8. [DOI] [PubMed] [Google Scholar]
- 37. MacCorkle RA, Slattery SD, Nash DR, Brinkley BR. Intracellular protein binding to asbestos induces aneuploidy in human lung fibroblasts. Cell Motil Cytoskeleton 2006; 63: 646–57. [DOI] [PubMed] [Google Scholar]
- 38. Blount M, Holt PF, Leach AA. The protein coating of asbestos bodies. Biochem J 1966; 101: 204–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Davis JM. Further observations on the ultrastructure and chemistry of the formation of asbestos bodies. Exp Mol Pathol 1970; 13: 346–58. [DOI] [PubMed] [Google Scholar]
- 40. Davis JM. Asbestos dust as a nucleation center in the calcification of old fibrous tissue lesions, and the possible association of this process to the formation of asbestos bodies. Exp Mol Pathol 1970; 12: 133–47. [DOI] [PubMed] [Google Scholar]
- 41. Davis JM, Gross P, De Treville RT. “Ferruginous bodies” in guinea pigs. Fine structure produced experimentally from minerals other than asbestos. Arch Pathol 1970; 89: 364–73. [PubMed] [Google Scholar]
- 42. Governa M, Amati M, Fontana S et al. Role of iron in asbestos‐body‐induced oxidant radical generation. J Toxicol Environ Health A 1999; 58: 279–87. [DOI] [PubMed] [Google Scholar]
- 43. Governa M, Rosanda C. A histochemical study of the asbestos body coating. Br J Ind Med 1972; 29: 154–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Governa M, Rosanda Vadala C. Histochemical study of asbestos fibre coating in experimental carrageenin granulomas. Br J Ind Med 1973; 30: 248–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koerten HK, de Bruijn JD, Daems WT. The formation of asbestos bodies by mouse peritoneal macrophages. An in vitro study. Am J Pathol 1990; 137: 121–34. [PMC free article] [PubMed] [Google Scholar]
- 46. Koerten HK, Hazekamp J, Kroon M, Daems WT. Asbestos body formation and iron accumulation in mouse peritoneal granulomas after the introduction of crocidolite asbestos fibers. Am J Pathol 1990; 136: 141–57. [PMC free article] [PubMed] [Google Scholar]
- 47. Langer AM, Rubin IB, Selikoff IJ. Chemical characterization of asbestos body cores by electron microprobe analysis. J Histochem Cytochem 1972; 20: 723–34. [DOI] [PubMed] [Google Scholar]
- 48. Suzuki Y, Churg J. Structure and development of the asbestos body. Am J Pathol 1969; 55: 79–107. [PMC free article] [PubMed] [Google Scholar]
- 49. Suzuki Y, Churg J. Formation of the asbestos body. A comparative study with three types of asbestos. Environ Res 1970; 3: 107–18. [DOI] [PubMed] [Google Scholar]
- 50. Governa M, Vadala CR. Histochemical demonstration of hematoidin in the innermost layers of human asbestos body coating. Int Arch Arbeitsmed 1972; 30: 273–82. [DOI] [PubMed] [Google Scholar]
- 51. Fubini B, Barcelo F, Otero Arean C. Ferritin adsorption on amosite fibers: possible implications in the formation and toxicity of asbestos bodies. J Toxicol Environ Health 1997; 52: 343–52. [DOI] [PubMed] [Google Scholar]
- 52. Pooley FD. Asbestos bodies, their formation, composition and character. Environ Res 1972; 5: 363–79. [DOI] [PubMed] [Google Scholar]
- 53. O’Connell M, Halliwell B, Moorhouse CP, Aruoma OI, Baum H, Peters TJ. Formation of hydroxyl radicals in the presence of ferritin and haemosiderin. Is haemosiderin formation a biological protective mechanism? Biochem J 1986; 234: 727–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. O’Connell MJ, Ward RJ, Baum H, Peters TJ. Iron release from haemosiderin and ferritin by therapeutic and physiological chelators. Biochem J 1989; 260: 903–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Coussens LM, Werb Z. Inflammation and cancer. Nature 2002; 420: 860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010; 140: 883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang H, Bocchetta M, Kroczynska B et al. TNF‐alpha inhibits asbestos‐induced cytotoxicity via a NF‐kappaB‐dependent pathway, a possible mechanism for asbestos‐induced oncogenesis. Proc Natl Acad Sci USA 2006; 103: 10397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dutta KK, Nishinaka Y, Masutani H et al. Two distinct mechanisms for loss of thioredoxin‐binding protein‐2 in oxidative stress‐induced renal carcinogenesis. Lab Invest 2005; 85: 798–807. [DOI] [PubMed] [Google Scholar]
- 59. Gutteridge JM, Rowley DA, Halliwell B. Superoxide‐dependent formation of hydroxyl radicals in the presence of iron salts. Detection of ‘free’ iron in biological systems by using bleomycin‐dependent degradation of DNA. Biochem J 1981; 199: 263–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jensen ON, Podtelejnikov A, Mann M. Delayed extraction improves specificity in database searches by matrix‐assisted laser desorption/ionization peptide maps. Rapid Commun Mass Spectrom 1996; 10: 1371–8. [DOI] [PubMed] [Google Scholar]
- 61. Jiang L, Nagai H, Ohara H et al. Characteristics and modifying factors of asbestos‐induced oxidative DNA damage. Cancer Sci 2008; 99: 2142–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Uchida K. 4‐Hydroxy‐2‐nonenal: a product and mediator of oxidative stress. Prog Lipid Res 2003; 42: 318–43. [DOI] [PubMed] [Google Scholar]
- 63. Toyokuni S, Kawaguchi W, Akatsuka S, Hiroyasu M, Hiai H. Intermittent microwave irradiation facilitates antigen‐antibody reaction in Western blot analysis. Pathol Int 2003; 53: 259–61. [DOI] [PubMed] [Google Scholar]
- 64. Toyokuni S, Miyake N, Hiai H et al. The monoclonal antibody specific for the 4‐hydroxy‐2‐nonenal histidine adduct. FEBS Lett 1995; 359: 189–91. [DOI] [PubMed] [Google Scholar]
- 65. Macnab G, Harington JS. Haemolytic activity of asbestos and other mineral dusts. Nature 1967; 214: 522–3. [DOI] [PubMed] [Google Scholar]
- 66. Toyokuni S, Tanaka T, Hattori Y et al. Quantitative immunohistochemical determination of 8‐hydroxy‐2′‐deoxyguanosine by a monoclonal antibody N45.1: its application to ferric nitrilotriacetate‐induced renal carcinogenesis model. Lab Invest 1997; 76: 365–74. [PubMed] [Google Scholar]
- 67. Woo HA, Yim SH, Shin DH, Kang D, Yu DY, Rhee SG. Inactivation of peroxiredoxin I by phosphorylation allows localized H2O2 accumulation for cell signaling. Cell 2010; 140: 517–28. [DOI] [PubMed] [Google Scholar]
- 68. Nagatomo H, Morimoto Y, Ogami A et al. Change of heme oxygenase‐1 expression in lung injury induced by chrysotile asbestos in vivo and in vitro . Inhal Toxicol 2007; 19: 317–23. [DOI] [PubMed] [Google Scholar]
- 69. Nagatomo H, Morimoto Y, Oyabu T et al. Expression of heme oxygenase‐1 in the lungs of rats exposed to crocidolite asbestos. Inhal Toxicol 2005; 17: 293–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. A list of asbestos‐binding proteins.
Data S1. Materials and methods.
Supporting info item
Supporting info item


