Abstract
Coenzyme Q (CoQ) is an isoprenoid quinine that functions as an electron carrier in the mitochondrial respiratory chain in eukaryotes. CoQ having shorter isoprenoid chains, especially CoQ1 and CoQ2, selectively inhibited the in vitro activity of eukaryotic DNA polymerase (pol) γ, which is a mitochondrial pol. These compounds did not influence the activities of nuclear DNA replicative pols such as α, δ and ɛ, and nuclear DNA repair‐related pols such as β, η, ι, κ and λ. CoQ also inhibited DNA topoisomerase II (topo II) activity, although the enzymatic characteristics, including modes of action, amino acid sequences and three‐dimensional structures, were markedly different from those of pol γ. These compounds did not inhibit the activities of procaryotic pols such as Escherichia coli pol I, and other DNA metabolic enzymes such as human immunodeficiency virus reverse transcriptase, T7 RNA polymerase and bovine deoxyribonuclease I. CoQ1, which has the shortest isoprenoid chains, had the strongest inhibitory effect on pol γ and topo II activities among CoQ1–CoQ10, with 50% inhibitory concentration (IC50) values of 12.2 and 15.5 µM, respectively. CoQ1 could prevent the growth of human promyelocytic leukemia cells, HL‐60, and the 50% lethal dose (LD50) value was 14.0 µM. The cells were halted at S phase and G1 phase in the cell cycle, and suppressed mitochondrial proliferation. From these results, the relationship between the inhibition of pol γ/topo II and cancer cell growth by CoQ is discussed. (Cancer Sci 2006; 97: 716–723)
Abbreviations:
- CoQn
coenzyme Qn
- DMSO
dimethyl sulfoxide
- DNAse I
deoxyribonuclease I
- dTTP
2′‐deoxythymidine 5′‐triphosphate
- EDTA
ethylenediamine tetraacetic acid
- HIV‐1
human immunodeficiency virus type‐1
- IC50
50% inhibitory concentration
- Km
Michaelis constant
- LD50
50% lethal dose
- MTT
3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl tetrazolium bromide
- PBS
phosphate‐buffered saline
- pol
DNA‐directed DNA polymerase (EC 2.7.7.7)
- topo
DNA topoisomerase
- Vmax
maximum velocity.
The lipid‐soluble compound CoQ is well known as a biomolecule. It is composed of a quinoid nucleus and a hydrophobic side chain containing a number of monounsaturated trans‐isoprenoid units (–CH2–CH=C[CH3]–CH2–). The isoprenoid chain length of CoQ varies among species. For instance, in primates, CoQ10 (CoQ with 10 isoprenoid units) is the major form, whereas CoQ9 is the major form in mice and rats, and the shorter CoQn is the major form in microorganisms.( 1 ) CoQ analogs having a long hydrophobic tail are likely to become embedded in the mitochondrial membrane and to act as electron carriers in the mitochondrial respiratory chain by reversible oxidation/reduction conversion. The purpose of the present study is to investigate the biochemical action, such as the inhibition of DNA metabolic enzymes, by CoQ in vitro and its use as an antineoplastic agent. In DNA metabolic enzymes, DNA pol catalyzes the addition of deoxyribonucleotides to the 3′‐hydroxyl terminus of primed double‐stranded DNA molecules.( 2 ) The human genome encodes 16 DNA pol that conduct cellular DNA synthesis.( 3 ) Eukaryotic cells reportedly contain three replicative types (α, δ and ɛ), mitochondrial pol γ, and at least 12 repair types (β, δ, ɛ, ζ, η, θ, ι, κ, λ, µ and σ) and REV1.( 4 ) In contrast, DNA topo catalyzes the concerted breaking and rejoining of DNA strands and is involved in producing the necessary topological and conformational changes in DNA.( 2 , 5 ) Therefore, there are no enzymatic similarities between pol and topo, although they are critical to many cellular processes such as DNA replication, repair and recombination, and may act in harmony with each other. The characteristics of both enzymes, including their modes of action, amino acid sequences and three‐dimensional structures, are markedly different from each other. We found that CoQ could inhibit not only the activities of pol γ but also of topo II. Hence, the influence of the compounds on human cancer cell growth, cell cycle arrest and apoptosis induction was investigated. We also observed the effect of differences in the length of the isoprenoid side chains of CoQ.
Materials and Methods
Materials
Coenzyme Q such as CoQ1, CoQ2, CoQ4, CoQ6, and CoQ10 were purchased from Sigma (St Louis, MO, USA). Nucleotides and chemically synthesized DNA template primers such as poly(dA), poly(rA), and oligo(dT)12−18 and [3H]‐dTTP (16TBq/mmol) were purchased from Amersham Biosciences (Buckinghamshire, UK). Supercoiled pUC19 plasmid DNA and pBR322 plasmid DNA were obtained from Takara (Tokyo, Japan). Mito Tracker Green FM was purchased from Molecular Probes (Eugene, OR, USA). All other reagents were of analytical grade and purchased from Nakarai Tesque (Kyoto, Japan). HL‐60, a human promyelocytic leukemia cell line (IFO 050022), was supplied by the Health Science Research Resources Bank (Osaka, Japan). The enzyme Pol α was purified from calf thymus by immunoaffinity column chromatography as described by Tamai et al.( 6 ) Recombinant rat pol β was purified from Escherichia coli JMpβ5 as described by Date et al.( 7 ) The human pol γ catalytic gene was cloned into pFastBac. Histidine‐tagged enzyme was expressed using the BAC‐TO‐BAC HT Baculovirus Expression System according to the supplier's manual (Life Technologies Inc., Gaithersburg, MD, USA) and purified using ProBoundresin (Invitrogen Japan, Tokyo Japan).( 8 ) Human pol δ and ɛ were purified by nuclear fractionation of human peripheral blood cancer cells (Molt‐4) using the second subunit of pol δ and ɛ‐conjugated affinity column chromatography, respectively.( 9 ) Recombinant human pol η and ι tagged with His6 at their C‐terminus were expressed in SF9 insect cells using the baculovirus expression system, and were purified as described previously.( 10 , 11 ) A truncated form of pol κ (hDINB1DC) with 6 × His‐tags attached at the C‐terminus was overproduced using the BAC‐to‐BAC Baculovirus Expression System kit (Gibco BRL, Grand Island, NY, USA) and purified as described previously.( 12 ) Recombinant human His‐pol λ was overexpressed and purified according to a method described previously.( 13 ) Fish pol α and δ were purified from the testis of cherry salmon (Oncorhynchus masou).( 14 ) Fruit fly pol α, δ and ɛ were purified from the early embryos of Drosophila melanogaster as described previously.( 15 , 16 ) Pol I (α‐like) and II (β‐like) from a higher plant, cauliflower inflorescence, were purified according to the methods outlined by Sakaguchi et al.( 17 ) Recombinant rice (Oryza sativa L. cv. Nipponbare) His‐pol λ was overexpressed and purified according to a method described previously.( 18 ) HIV‐1 reverse transcriptase (recombinant) and the Klenow fragment of pol I from E. coli were purchased from Worthington Biochemical Corporation (Freehold, NJ, USA). T4 pol, Taq pol, T7 RNA polymerase and T4 polynucleotide kinase were purchased from Takara (Kyoto, Japan). Calf thymus terminal deoxynucleotidyltransferase and bovine pancreas DNase I were obtained from Stratagene Cloning Systems (La Jolla, CA, USA). Purified human placenta DNA topo I and II were purchased from TopoGen (Columbus, OH, USA).
DNA polymerase assays
The reaction mixtures for pol α, pol β, plant pol and prokaryotic pol were described previously.( 19 , 20 ) Those for pol γ, and pol δ and ɛ were as described by Umeda et al.( 8 ) and Ogawa et al.,( 21 ) respectively. The reaction mixtures for pol η, ι and κ were the same as that for pol α, and the reaction mixture for pol λ was the same as that for pol β. For DNA‐dependent pol, poly(dA)/oligo(dT)12−18 (A/T = 2/1) and dTTP were used as the DNA template‐primer and nucleotide (dNTP) substrate, respectively. For RNA‐dependent pol γ and HIV‐1 reverse transcriptase, poly(rA)/oligo(dT)12−18 (A/T = 2/1) and dTTP were used as the template‐primer and nucleotide substrate, respectively. For terminal deoxynucleotidyl transcriptase, oligo(dT)12−18 (3′‐OH) and dTTP were used as the DNA template‐primer and nucleotide substrate, respectively.
Coenzyme Q were dissolved in distilled DMSO at various concentrations and sonicated for 30 s. Aliquots of 4 µL of sonicated samples were mixed with 16 µL of each enzyme (final amount 0.05 units) in 50 mM Tris‐HCl (pH 7.5) containing 1 mM dithiothreitol, 50% glycerol and 0.1 mM EDTA, and kept at 0°C for 10 min. These inhibitor–enzyme mixtures (8 µL) were added to 16 µL of each of the enzyme standard reaction mixtures, and incubation was carried out at 37°C for 60 min, except for Taq pol, which was incubated at 74°C for 60 min. Activity without the inhibitor was considered 100%, and the remaining activity at each concentration of the inhibitor was determined relative to this value. One unit of pol activity was defined as the amount of enzyme that catalyzed the incorporation of 1 nmol of dNTP (i.e. dTTP) into synthetic DNA template‐primers in 60 min at 37°C under the normal reaction conditions for each enzyme.( 19 , 20 )
DNA topoisomerase assays
The relaxation activity of topo II was determined by detecting the conversion of supercoiled plasmid DNA to its relaxed form.( 22 ) The topo II reaction was carried out in 20 µL of reaction mixture containing 50 mM Tris‐HCl buffer (pH 8.0), 120 mM KCl, 10 mM MgCl2, 0.5 mM ATP, 0.5 mM dithiothreitol, supercoiled pUC19 DNA (0.25 µg), 2 µL of inhibitor solution (10% DMSO) and topo II. The reaction mixtures were incubated at 37°C for 30 min and terminated by adding 2 µL of loading buffer consisting of 5% sarkosyl, 0.0025% bromophenol blue and 25% glycerol. The mixtures were subjected to 1% agarose gel electrophoresis in Tris‐acetate‐EDTA running buffer. The agarose gel was stained with ethidium bromide and DNA was visualized on an ultraviolet transilluminator. The relaxation activity of topo I was analyzed in the same manner as described above except that the reaction mixtures contained 10 mM Tris‐HCl (pH 7.9), pBR322 DNA (0.25 µg), 1 mM EDTA, 150 mM NaCl, 0.1% bovine serum albumin, 0.1 mM spermidine, 5% glycerol and topo I. One unit was defined as the amount of enzyme capable of relaxing 0.25 µg of DNA in 15 min at 37°C.
Other enzyme assays
The DNA primase activity of pol α, the activities of T7 RNA polymerase, T4 polynucleotide kinase and bovine DNase I were measured in standard assays according to the manufacturer's specifications as described by Tamiya‐Koizumi et al.( 23 ) Nakayama and Saneyoshi,( 24 ) Soltis and Uhlenbeck,( 25 ) and Lu and Sakaguchi,( 26 ) respectively.
Investigation of cytotoxicity on cultured cells
For investigation of the effects of CoQ in cultured cells, we used the human cancer cell line HL‐60, derived from a cancer patient. The cells were cultured routinely in RPMI‐1640 medium supplemented with 10% fetal bovine serum, 100 µg/mL streptomycin, 100 units/mL penicillin, and 1.6 µg/mL NaHCO3. The cells were cultured at 37°C in standard medium in a humidified atmosphere of 5% CO2−95% air. The cytotoxicity of the compound was investigated as follows: High concentrations (10 mM) of the compounds were dissolved in DMSO and stored. Approximately 1 × 104 cells per well were inoculated in 96‐well microplates, then the compound stock solution was diluted to various concentrations and applied to each well. After incubation for 24 h, the survival rate was determined by MTT assay.( 27 )
Cell cycle analysis
Cellular DNA content for cell cycle analysis was determined as follows. Aliquots of 3 × 105 HL‐60 cells were harvested into a 35 mm dish, and incubated with medium containing CoQ1 for various times. The cells were then washed with ice‐cold PBS three times with centrifugation. The cells were fixed with 70% (v/v) ethanol and stored at −20°C. DNA was stained with DAPI staining solution for at least 10 min at room temperature in the dark. Fluorescence intensity was measured by flow cytometry (Partec cell counter analyzer, CCA; Partec GmbH, Munster, Germany). Cell cycle distribution was analyzed using the Multicycle software program (version 2.5; Phoenix Flow Systems, San Diego, CA, USA).
Quantitative analysis of mitochondria in cancer cells
HL‐60 cell density was adjusted to 1 × 105 cells/mL, and cells were treated with a 50% inhibitory concentration of CoQ1 (i.e. 14.0 µM) for various times. The cells were centrifuged to obtain a cell pellet and the supernatant was aspirated. The cells were suspended in prewarmed (37°C) medium containing Mito Tracker Green FM, and then incubated 15 min under growth conditions. The cells were washed twice with ice‐cold PBS with centrifugation. The fluorescence intensity of the stained cells was analyzed using a FACSCanto flow cytometer in combination with FACSDiVa software (Becton Dickinson, Rutherford, NY, USA).
Analysis of DNA fragmentation
Apoptosis was determined using a DNA fragmentation assay by means of agarose electrophoresis. Total DNA was extracted from 6 × 105 HL‐60 cells following the method of Sambrook et al.( 28 ) and 5‐µg aliquots were separated by 1.5% (w/v) agarose gel electrophoresis in 40 mM Tris−5 mM sodium acetate−1 mM EDTA (pH 7.8) and stained with ethidium bromide. DNA bands were visualized under ultraviolet light.
Results and Discussion
Effects of CoQ on pol activities
First, inhibition of the activities of mammalian pol by CoQ was investigated (Table 1). Of the five CoQ tested, CoQ1, CoQ2 and CoQ4 indicated pol γ inhibitory activity, but CoQ6 and CoQ10 did not influence the activity of pol γ. CoQ1 had the strongest inhibitory effect among the CoQ, and in order of their effect, the CoQ were ranked as follows: CoQ1 > CoQ2 > CoQ4 > CoQ6 > CoQ10. Fig. 1 shows the dose–response curves of CoQ1 for mammalian pol α–ɛ. CoQ1 selectively inhibited pol γ activity, and did not suppress the activities of other mammalian pol at all. Pol γ is the sole pol in animal mitochondria. Biochemical and genetic evidence document a key role for pol γ in mitochondrial DNA replication, whereas DNA repair and recombination were thought to be limited or absent in animal mitochondria.( 29 ) As pol γ has not only DNA‐dependent pol activity but also RNA‐dependent pol activity,( 29 ) the influence of both activities by CoQ1 was investigated. The inhibitory activity of CoQ1 on RNA‐dependent pol γ was as strong as that on DNA‐dependent pol γ, with 50% inhibition observed at doses of 14.5 and 12.2 µM, respectively (Fig. 1). When activated DNA (i.e. DNA digested by bovine DNase I) was used as the template‐primer, the mode of inhibition by these compounds did not change (data not shown).
Table 1.
50% inhibitory concentration (IC50) values of coenzyme Q (CoQ) for the activities of various DNA polymerases and other DNA metabolic enzymes
| Enzyme | IC50 values of coenzyme Q (µM) | ||||
|---|---|---|---|---|---|
| CoQ1 | CoQ2 | CoQ4 | CoQ6 | CoQ10 | |
| Mammalian DNA polymerases | |||||
| Calf DNA polymerase α | >200 | >200 | >200 | >200 | >200 |
| Rat DNA polymerase β | >200 | >200 | >200 | >200 | >200 |
| Human DNA polymerase γ | |||||
| *DNA polymerase activity | 12.2 | 45.6 | 145 | >200 | >200 |
| **Reverse transcriptase activity | 14.5 | 49.0 | 151 | >200 | >200 |
| Human DNA polymerase δ | >200 | >200 | >200 | >200 | >200 |
| Human DNA polymerase ɛ | >200 | >200 | >200 | >200 | >200 |
| Human DNA polymerase η | >200 | >200 | >200 | >200 | >200 |
| Human DNA polymerase ι | >200 | >200 | >200 | >200 | >200 |
| Human DNA polymerase κ | >200 | >200 | >200 | >200 | >200 |
| Human DNA polymerase λ | >200 | >200 | >200 | >200 | >200 |
| Fish DNA polymerases | |||||
| Cherry salmon DNA polymerase α | >200 | >200 | >200 | >200 | >200 |
| Cherry salmon DNA polymerase δ | >200 | >200 | >200 | >200 | >200 |
| Insect DNA polymerases | |||||
| Fruit fly DNA polymerase α | >200 | >200 | >200 | >200 | >200 |
| Fruit fly DNA polymerase δ | >200 | >200 | >200 | >200 | >200 |
| Fruit fly DNA polymerase ɛ | >200 | >200 | >200 | >200 | >200 |
| Plant DNA polymerases | |||||
| Cauliflower DNA polymerase I (α‐like) | >200 | >200 | >200 | >200 | >200 |
| Cauliflower DNA polymerase II (β‐like) | >200 | >200 | >200 | >200 | >200 |
| Rice DNA polymerase λ | >200 | >200 | >200 | >200 | >200 |
| Procaryotic DNA polymerases | |||||
| E. coli DNA polymerase I (Klenow fragment) | >200 | >200 | >200 | >200 | >200 |
| Taq DNA polymerase | >200 | >200 | >200 | >200 | >200 |
| T4 DNA polymerase | >200 | >200 | >200 | >200 | >200 |
| Other DNA metabolic enzymes | |||||
| Calf primase of DNA polymerase α | >200 | >200 | >200 | >200 | >200 |
| Calf terminal deoxynucleotidyltransferase | >200 | >200 | >200 | >200 | >200 |
| HIV‐1 reverse transcriptase | >200 | >200 | >200 | >200 | >200 |
| T7 RNA polymerase | >200 | >200 | >200 | >200 | >200 |
| Human DNA topoisomerase I | >200 | >200 | >200 | >200 | >200 |
| Human DNA topoisomerase II | 12.5 | >55.0 | >200 | >200 | >200 |
| T4 polynucleotide kinase | >200 | >200 | >200 | >200 | >200 |
| Bovine deoxyribonuclease I | ≥200 | ≥200 | ≥200 | ≥200 | ≥200 |
These compounds were incubated with each enzyme (0.05 units). The enzymatic activity was measured as described in Materials and Methods. The enzymatic activity in the absence of the compounds was taken as 100%. *The template‐primer is poly(dA)/oligo(dT)12−18, A/T = 2/1. **The template‐primer is poly(rA)/oligo(dT)12−18, A/T = 2/1. HIV‐1, human immunodeficiency virus type‐1.
Figure 1.
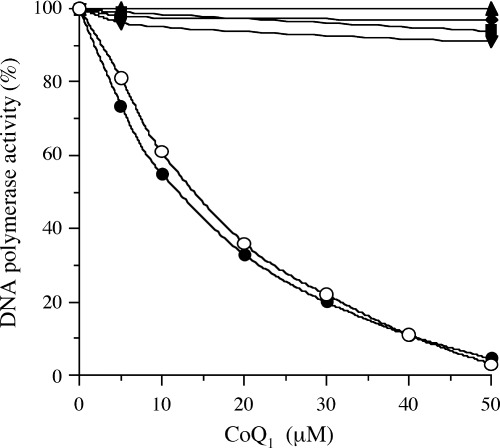
Dose–response curves of coenzyme Q1 (CoQ; 0–50 µM) for mammalian DNA polymerases (pol). The enzymes used (0.05 units each) were calf pol α (▪), rat pol β (▴), DNA‐dependent pol activity of human pol γ (•), RNA‐dependent pol activity of human pol γ (○), human pol δ (◆), and human pol ɛ (▾). The pol activities were measured as described in the text. Enzymatic activity in the absence of CoQ1 was taken as 100%.
Coenzyme Q had no inhibitory effect on fish pol α and δ, insect pol α, δ and ɛ, plant pol I (α‐like), pol II (β‐like) and pol λ, or procaryotic pols such as the Klenow fragment of E. coli pol I, Taq pol and T4 pol (Table 1).
Mode of pol γ inhibition by CoQ1
Next, to elucidate the mechanism of inhibition, the extent of inhibition as a function of the DNA template‐primer or dNTP substrate concentrations was studied (Table 2). In kinetic analysis, poly(dA)/oligo(dT)12−18 (A/T = 2/1) and dTTP were used as the DNA template‐primer and dNTP substrate, respectively. Double reciprocal plots of the results showed that the CoQ1‐induced inhibition of human pol γ activity was competitive with both the DNA template‐primer and dNTP substrate. In the case of the DNA template‐primer, the apparent Km was unchanged at 6.06 µM, whereas 68.8%, 81.6% and 87.0% decreases in the Vmax were observed in the presence of 5.0, 10 and 15 µM CoQ1, respectively. The Km for the dNTP substrate was 1.54 µM, and the Vmax for the dNTP substrate decreased from 47.6 to 9.26 pmol/h in the presence of 15 µM CoQ1. The inhibition constant value, obtained from Dixon plots, was found to be 5.0 µM and 3.3 µM for the DNA template‐primer and dNTP substrate, respectively. When activated DNA and four deoxynucleoside triphosphates were used as the DNA template‐primer and dNTP substrate, respectively, the inhibition of human pol γ by CoQ1 was competitive with both the DNA template‐primer and dNTP substrate. These results suggest that CoQ1 binds directly to the DNA template‐primer binding site and dNTP binding site of pol γ.
Table 2.
Kinetic analysis of the inhibitory effects of coenzyme Q1 (CoQ1) on the activities of DNA polymerase γ, as a function of the DNA template‐primer dose and the dNTP substrate concentration
| Substrate | CoQ1 (µM) | Km (µM) | Vmax (pmol/h) | Ki (µM) | Inhibitory mode |
|---|---|---|---|---|---|
| DNA template‐primer † | 0 | 41.7 | |||
| 5.0 | 6.06 | 13.0 | 5.0 | Competitive | |
| 10 | 7.69 | ||||
| 15 | 5.43 | ||||
| dNTP substrate ‡ | 0 | 47.6 | |||
| 5.0 | 1.54 | 20.4 | 3.3 | Competitive | |
| 10 | 12.8 | ||||
| 15 | 9.26 |
Poly(dA)/oligo(dT)12−18.
dTTP. The amount of calf polymerase γ in the assay mixture was 0.05 units. Ki, inhibition constant; Km, Michaelis constant; Vmax, maximum velocity.
Inhibitory effect of CoQ on the activities of topo and other DNA metabolic enzymes
Coenzyme Q did not inhibit the activities of other DNA‐metabolic enzymes such as calf primase of pol α, HIV‐1 reverse transcriptase, T7 RNA polymerase, T4 polynucleotide kinase, bovine deoxyribonuclease I, or human topo I, except for human topo II (Table 1). As shown in Fig. 2, 100 µM of CoQ1 and CoQ2 completely inhibited the relaxation activity of topo II, with IC50 values of 15.5 and 55.0 µM, respectively, although CoQ4, CoQ6 and CoQ10 had no influence. The inhibitory effect of CoQ1 was the strongest among the CoQs tested, and the IC50 value for pol γ was almost the same as that for topo II. These results suggest that longer isoprenoid chains of the benzquinone ring in CoQ weaken the inhibitory effect on pol γ and topo II.
Figure 2.
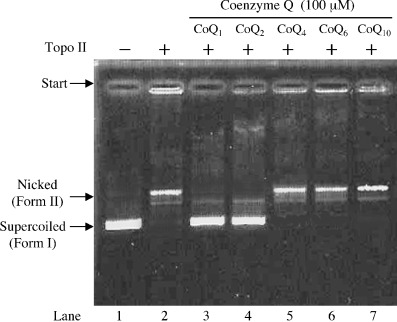
Inhibitory effects of coenzyme Q (CoQ) on human DNA topoisomerase (topo) II. pUC19 supercoiled plasmid DNA was mixed with the enzyme and inhibitor. Lanes 3–7, 100 µM of CoQ1, CoQ2, CoQ4, CoQ6 and CoQ10, respectively; lanes 2–7, 0.05 units of topo II; lane 1, no enzyme. Plasmid DNA (0.25 µg) was added in each of the lanes. A photograph of the ethidium bromide‐stained gel is shown.
From the thermal transition of double‐stranded DNA with or without CoQ, CoQ did not intercalate with DNA, and the compound might bind directly to pol γ and topo II and then inhibit the activity (data not shown). Three‐dimensional structural analysis of the binding shorter chain CoQ and pol γ/topo II should be examined in future studies.
Effects of CoQ on cultured human cancer cells
Polymerases and topo have emerged recently as important cellular targets for chemical intervention in the development of anticancer agents. CoQ‐related compounds could therefore be useful in chemotherapy. We tested the cytotoxic effects of the five compounds against the human promyelocytic leukemia cell line HL‐60.
As shown in Table 3, CoQ1 to CoQ4 had potent growth inhibitory effects on this cancer cell line, whereas CoQ6 and CoQ10 did not prevent cell growth. The inhibitory effect of each CoQ varied markedly in the following order: CoQ1 > CoQ2 > CoQ4 > CoQ6 > CoQ10, and the suppression of cell growth had the same tendency as the inhibition of pol γ and topo II among the compounds. CoQ1 was the strongest inhibitor of human cancer cell growth, and the concentration required for LD50 was 14.0 µM. As the IC50 values of CoQ1 were 12.2 µM for pol γ and 15.5 µM for topo II (Table 1), the LD50 value was clearly the same as the IC50 values in vitro for these enzymes. Therefore, we concentrated on the properties of CoQ1 in the latter part of this study.
Table 3.
Inhibitory effect of coenzyme Q on the proliferation of human cancer cells
| Compound | LD50 values (µM) |
|---|---|
| Coenzyme Q1 | 14.0 1.2 |
| Coenzyme Q2 | 56.5 1.8 |
| Coenzyme Q4 | 168 2.5 |
| Coenzyme Q6 | >200 |
| Coenzyme Q10 | ≥200 |
Human promyelocytic leukemia cells, HL‐60, were incubated for 24 h and then assayed with 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl tetrazolium bromide.( 27 ) Data are expressed as the mean SD (n = 5). LD50, 50% lethal dose.
Effects of CoQ1 on the cell cycle of human cancer cells
Coenzyme Q1 was suggested to inhibit the activities of pol γ and topo II in intact cells. These observations suggested that cell growth inhibition occurs in a manner dependent on enzyme inhibition, and that enzyme inhibition influences cell growth in vivo. To confirm this suggestion in detail, we examined the effect of this compound on the cell cycle of HL‐60 cells by flow cytometry. As shown in Fig. 3a,c, the cells were arrested in S phase (increase of 9.7%) and decreased the rate of G1 phase and G2/M phases (decreases of 6.0 and 3.7%) by 14.0 µM CoQ1 (i.e. the LD50 value) with incubation for 12 h. After 24 h and 48 h treatment with CoQ1, the S block was partially released and G1 phase was increased (Fig. 3d,e). Etoposide (VP‐16), which is a DNA topo II inhibitor, blocked the cell cycle at G2/M phase.( 30 ) It was suggested that CoQ1 inhibited the activity of pol γ in the cell, and then could arrest the cell cycle in S phase for 12 h, although the enzymatic inhibitory activity of pol γ by CoQ1 was as strong as that of topo II (Table 1). Furthermore, CoQ1 may influence the second step of the cell cycle at G1 phase by pol γ after 24 h and 48 h exposure to CoQ1.
Figure 3.
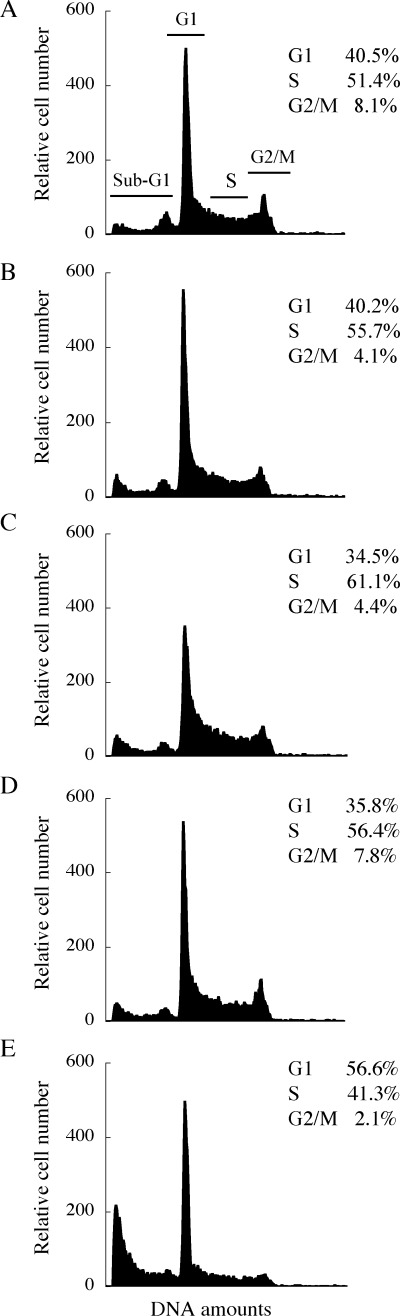
Flow cytometric analysis of cell cycle perturbation by coenzyme Q1 (CoQ1). (A–E) HL‐60 human cancer cells were incubated with 14.0 µM CoQ1 for 0, 6, 12, 24 and 48 h, respectively. DNA was stained with 4′,6‐diamino‐2‐phenyl‐indole (DAPI) solution. Fluorescence intensity was measured by flow cytometry.
Effect of CoQ1 on mitochondrial amplification of human cancer cells
Next, the effect of CoQ1 on the amount of mitochondria in the human cancer cells was investigated (Fig. 4). After HL‐60 cells were exposed to 14.0 µM of CoQ1 for various times (0, 6, 12, 24 and 48 h), the cells were stained with Mito Tracker Green FM, which selectively stains the plasma membrane in mitochondria.( 31 ) Flow cytometric analysis showed that the amount of mitochondria in the cells was significantly reduced after 12 h incubation (Fig. 4a), and the decreased value was approximately 60% (Fig. 4b). These results suggest that CoQ1 must inhibit cell growth mainly by blocking the primary step of mitochondrial DNA replication, proliferation of mitochondria, and then cell division, by acting as pol γ. As CoQ are present at high concentrations in the mitochondrial matrix,( 32 ) CoQ might regulate pol γ activity and mitochondrial proliferation.
Figure 4.
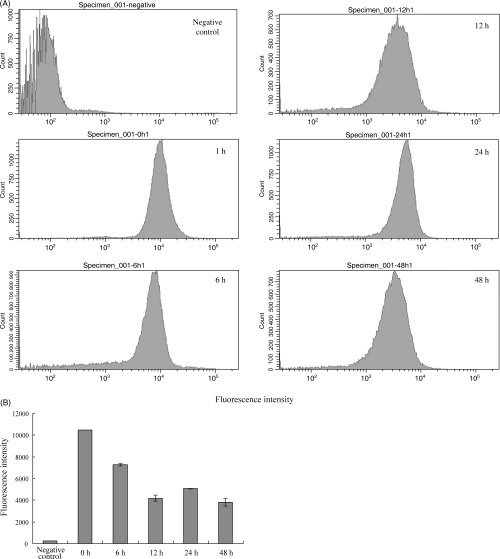
The effect of coenzyme Q1 (CoQ1) on the amount of mitochondria in human cancer cells. HL‐60 human cancer cells were incubated with 14.0 µM CoQ1 for 0, 6, 12, 24 and 48 h. There were no stained cells at 0 h (negative control). (A) Fluorescence intensity of MitoTracker Green FM per stained cells was measured by flow cytometry. (B) The mean values of the stained cells are indicated. Data are shown as the mean ± SEM for five independent experiments.
Effect of CoQ1 on apoptotic cell death
To examine whether the detection of sub‐G1 phase in the cells treated with CoQ1 (Fig. 3) was due to apoptosis, DNA fragmentation was analyzed by electrophoresis. Time‐dependent DNA ladder formation was observed in HL‐60 cells treated with 14.0 µM CoQ1 (Fig. 5). Such ladders were not evident for the initial 6 h but were apparent at 12 h and thereafter. CoQ1 could be a strong inducer of apoptosis. The effect of CoQ1 must occur in combination with growth arrest, mitochondrial proliferation and cell death. Mitochondria are central to activating apoptosis;( 32 ) pol γ inhibition by CoQ may therefore lead to cell death.
Figure 5.
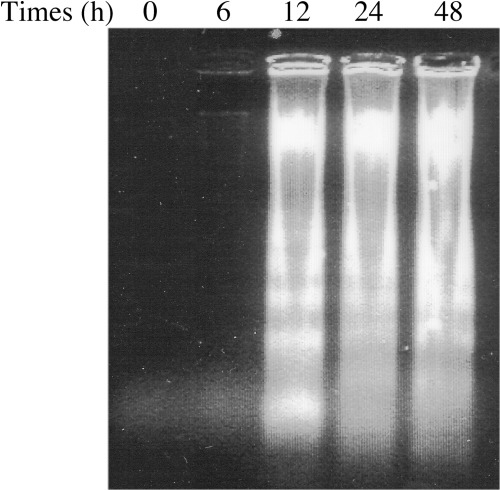
DNA fragmentation of coenzyme Q1 (CoQ1)‐treated cells by agarose gel electrophoresis. HL‐60 human promyelocytic leukemia cells were incubated with 14.0 µM CoQ1 for the times indicated. Total DNA was then extracted and analyzed by 1.5% agarose gel electrophoresis. A photograph of the ethidium bromide‐stained gel is shown.
These results indicate that inhibition of pol γ and/or topo II leads to cell growth inhibition in vivo. CoQ‐related compounds such as CoQ1 could be novel cancer chemotherapy agents.
Acknowledgments
We are grateful for the donations of rat pol β, human pols δ and ɛ, human pols η and ι, human pol κ, and human pol λ by Dr A. Matsukage of Japan Women's University (Tokyo, Japan), Dr K. Sakaguchi of Tokyo University of Science (Chiba, Japan), Dr C. Masutani and Dr F. Hanaoka of Osaka University (Osaka, Japan), Dr E. Ohashi and Dr H. Ohmori of Kyoto University (Kyoto, Japan), and Dr N. Shimazaki and Dr O. Koiwai of Tokyo University of Science (Chiba, Japan), respectively. This work was supported in part by a Grant‐in‐Aid for Kobe‐Gakuin University Joint Research (A) (Y. M. and H. Y.). Y. M. acknowledges Grants‐in‐Aid from the Takeda Science Foundation (Japan), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (Japan), the Japan Food Chemical Research Foundation (Japan), and Grant‐in‐Aid 16710161 for Scientific Research, MEXT (Ministry of Education, Culture, Sports, Science and Technology, Japan).
References
- 1. Turunen M, Olsson J, Dallner G. Metabolism and function of coenzyme Q. Biochim Biophys Acta 2004; 1660: 171–99. [DOI] [PubMed] [Google Scholar]
- 2. Kornberg A, Baker TA. DNA Replication, 2nd edn. New York: W.D. Freeman and Co., 1992. [Google Scholar]
- 3. Bebenek K, Kunkel TA. Eukaryotic DNA polymerase. In: W Yang, ed. DNA Repair and Replication, Advances in Protein Chemistry. San Diego: Elsevier, 2004; 137–65. [DOI] [PubMed] [Google Scholar]
- 4. Friedberg EC, Feaver WJ, Gerlach VL. The many faces of DNA polymerases: strategies for mutagenesis and for mutational avoidance. Proc Natl Acad Sci USA 2000; 97: 5681–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang JC. DNA topoisomerases. Annu Rev Biochem 1996; 65: 635–91. [DOI] [PubMed] [Google Scholar]
- 6. Tamai K, Kojima K, Hanaichi T et al. Structural study of immunoaffinity‐purified DNA polymerase α‐DNA primase complex from calf thymus. Biochim Biophys Acta 1988; 950: 263–73. [DOI] [PubMed] [Google Scholar]
- 7. Date T, Yamaguchi M, Hirose F, Nishimoto Y, Tanihara K, Matsukage A. Expression of active rat DNA polymerase β in Escherichia coli . Biochemistry 1988; 27: 2983–90. [DOI] [PubMed] [Google Scholar]
- 8. Umeda S, Muta T, Ohsato T, Takamatsu C, Hamasaki N, Kang D. The D‐loop structure of human mtDNA is destabilized directly by 1‐methyl‐4‐phenylpyridinium ion (MPP+), a parkinsonism‐causing toxin. Eur J Biochem 2000; 267: 200–6. [DOI] [PubMed] [Google Scholar]
- 9. Oshige M, Takeuchi R, Ruike R, Kuroda K, Sakaguchi K. Subunit protein‐affinity isolation of Drosophila DNA polymerase catalytic subunit. Protein Expr Purif 2004; 35: 248–56. [DOI] [PubMed] [Google Scholar]
- 10. Masutani C, Kusumoto R, Iwai S, Hanaoka F. Mechanisms of accurate translesion synthesis by human DNA polymerase η. EMBO J 2000; 19: 3100–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tissier A, Frank EG, McDonald JP, Iwai S, Hanaoka F, Woodgate R. Misinsertion and bypass of thymine–thymine dimers by human DNA polymerase ι. EMBO J 2000; 19: 5259–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohashi E, Ogi T, Kusumoto R et al. Error‐prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes Dev 2000; 14: 1589–94. [PMC free article] [PubMed] [Google Scholar]
- 13. Shimazaki N, Yoshida K, Kobayashi T, Toji S, Tamai T, Koiwai O. Over‐expression of human DNA polymerase 1 in E. coli and characterization of the recombinant enzyme. Genes Cells 2000; 7: 639–51. [DOI] [PubMed] [Google Scholar]
- 14. Tomikawa A, Seno M, Sato‐Kiyotaki K et al. Synthetic nucleosides and nucleotides. 40. Selective inhibition of eukaryotic DNA polymerase α by 9‐(β‐d‐arabinofuranosyl)‐2‐(p‐n‐butylanilino) adenine 5′‐triphosphate (BuAaraATP) and its 2′‐up azido analog: synthesis and enzymatic evaluations. Nucleosides Nucleotides 1998; 17: 487–501. [DOI] [PubMed] [Google Scholar]
- 15. Aoyagi N, Matsuoka S, Furunobu A, Matsukage A, Sakaguchi K. Drosophila DNA polymerase δ: Purification and characterization. J Biol Chem 1994; 269: 6045–50. [PubMed] [Google Scholar]
- 16. Aoyagi N, Oshige M, Hirose F, Kuroda K, Matsukage A, Sakaguchi K. DNA polymerase ɛ from Drosophila melanogaster . Biochem Biophys Res Commun 1997; 230: 297–301. [DOI] [PubMed] [Google Scholar]
- 17. Sakaguchi K, Hotta Y, Stern H. Chromatin‐associated DNA polymerase activity in meiotic cells of lily and mouse. Cell Struct Funct 1980; 5: 323–34. [Google Scholar]
- 18. Uchiyama Y, Kimura S, Yamamoto T, Ishibashi T, Sakaguchi K. Plant DNA polymerase 1, a DNA repair enzyme that functions in plant meristematic and meiotic tissues. Eur J Biochem 2004; 271: 2799–807. [DOI] [PubMed] [Google Scholar]
- 19. Mizushina Y, Tanaka N, Yagi H et al. Fatty acids selectively inhibit eukaryotic DNA polymerase activities in vitro . Biochim Biophys Acta 1996; 1308: 256–62. [DOI] [PubMed] [Google Scholar]
- 20. Mizushina Y, Yoshida S, Matsukage A, Sakaguchi K. The inhibitory action of fatty acids on DNA polymerase β. Biochim Biophys Acta 1997; 1336: 509–21. [DOI] [PubMed] [Google Scholar]
- 21. Ogawa A, Murate T, Suzuki M, Nimura Y, Yoshida S. Lithocholic acid, a putative tumor promoter, inhibits mammalian DNA polymerase β. Jpn J Cancer Res 1998; 89: 1154–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spitzner JR, Chung IK, Muller MT. Eukaryotic topoisomerase II preferentially cleaves alternating purine–pyrimidine repeats. Nucl Acids Res 1990; 18: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tamiya‐Koizumi K, Murate T, Suzuki M et al. Inhibition of DNA primase by sphingosine and its analogues parallels with their growth suppression of cultured human leukemic cells. Biochem Mol Biol Int 1997; 41: 1179–89. [DOI] [PubMed] [Google Scholar]
- 24. Nakayama C, Saneyoshi M. Inhibitory effects of 9‐β‐d‐xylofuranosyladenine 5′‐triphosphate on DNA‐dependent RNA polymerase I and II from cherry salmon (Oncorhynchus masou). J Biochem (Tokyo) 1985; 97: 1385–9. [DOI] [PubMed] [Google Scholar]
- 25. Soltis DA, Uhlenbeck OC. Isolation and characterization of two mutant forms of T4 polynucleotide kinase. J Biol Chem 1982; 257: 11332–9. [PubMed] [Google Scholar]
- 26. Lu BC, Sakaguchi K. An endo‐exonuclease from meiotic tissues of the basidiomycete Coprinus cinereus. Its purification and characterization. J Biol Chem 1991; 266: 21060–6. [PubMed] [Google Scholar]
- 27. Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Meth 1983; 65: 55–63. [DOI] [PubMed] [Google Scholar]
- 28. Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 1989. [Google Scholar]
- 29. Kaguni LS. DNA polymerase γ, the mitochondrial replicase. Annu Rev Biochem 2004; 73: 293–320. [DOI] [PubMed] [Google Scholar]
- 30. Nishida K, Seto M, Ishida R. Different susceptibilities of postmitotic checkpoint‐proficient and ‐deficient Balb/3T3 cells to ICRF‐193, a catalytic inhibitor of DNA topoisomerase II. Jpn J Cancer Res 2001; 92: 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bkaily G, Pothier P, D’Orleans‐Juste P et al. The use of confocal microscopy in the investigation of cell structure and function in the heart, vascular endothelium and smooth muscle cells. Mol Cell Biochem 1997; 172: 171–94. [PubMed] [Google Scholar]
- 32. Chan TS, Wilson JX, O’Brien PJ. Coenzyme Q cytoprotective mechanisms. Meth Enzymol 2004; 382: 89–104. [DOI] [PubMed] [Google Scholar]


