Abstract
Simian virus 40 (SV40) large T antigen (LT) can immortalize and transform many cell types. These activities are attributed in large part to the binding and functional inactivation by LT of two major tumor suppressors: p53 and the retinoblastoma protein, pRB. Most effects of LT on pRB have been shown to additionally require an intact J domain, which mediates binding to Hsc70. We show here that the J domain is not required for p53 override in full-length LT. Although LT binds p53, it was shown previously that overcoming a p53-induced cell cycle arrest requires binding to pRB family members (R. S. Quartin et al., J. Virol. 68:1334–1341). We demonstrate that an LT mutant defective for pRB family member binding (K1) can be complemented for efficient override of p53 arrest by a construct encoding the first 135 amino acids of LT with a J domain-inactivating mutation, H42Q. Hence, complementation does not require the J domain, and pRB binding by LT is important for more than dissociating pRB-E2F complexes, which is J dependent. In accordance with this notion, LT alleviates pRB small-pocket-mediated transcriptional repression independently of the J domain. The LT K1 mutant can also be complemented for p53 override by small t antigen (st) in a manner independent of its J domain. Our observations underscore the importance of multiple SV40 functions, two in LT and one in st, that act cooperatively to counteract p53 growth suppression.
Simian virus 40 (SV40) encodes two major early proteins, small tumor antigen (st) and large tumor antigen (LT), that are key players in viral replication as well as regulation of cell growth. LT possesses multifunctional activities, such as site-specific DNA binding, ATP-dependent helicase activity, and multimerization, consistent with its role as a viral initiator of replication (reviewed in reference 23). The virus is often challenged with a quiescent cell environment, and since efficient viral DNA replication depends on S-phase factors, LT has evolved mitogenic properties. These include the ability to immortalize primary cells, transform rodent cells in culture, and induce S phase in quiescent cells (reviewed in reference 41). Fully efficient cellular transformation depends on at least three independent functions of LT: binding to the retinoblastoma protein (pRB) tumor suppressor (14) and other members of this family (19, 22), binding to the p53 tumor suppressor (33, 37), and maintaining the integrity of the first exon (amino acids 1 to 82). This region was first proposed and later demonstrated to be a bona fide DnaJ domain (hereafter referred to as the J domain [7, 11, 31, 66, 68, 86]), mediating interaction with Hsc70 (57). J domain-containing proteins such as LT are thought to function as molecular adapters possessing both a binding site for Hsc70 (the J domain) and a binding site for a target protein such as pRB, which is modified by the action of Hsc70. Evidence also points to the LT binding of p300/CBP and p400 as contributing to deregulation of cell growth (20, 36). The p300 protein binds and modulates p53 transcriptional activity as well as stability (26, 35). Detailed knowledge of cellular signaling pathways has been gained from dissecting the cellular targets of LT. Importantly, related tumor viruses such as adenovirus and human papillomavirus have been shown to target a similar set of proteins as SV40 LT, i.e., p53, pRB, and p300, thus underscoring their critical role in cell growth regulation.
The p53 tumor suppressor has gained an unsurpassed interest given its high frequency of mutation, that is, in more than 50% of human cancers (28). Several types of genotoxic stress activate the p53 protein, which acts as a key regulator of cell cycle progression and apoptosis (reviewed in references 25, 32, and 34). For example, radiation-induced DNA damage causes p53 to arrest the cell cycle in G1 or, depending on cell type and external stimuli, triggers apoptosis (32). The ability of p53 to induce G1 growth arrest is correlated with p53 transcriptional activity, one prominent transcriptional target being the p21CIP1 inhibitor of cyclin-dependent kinases (cdk's) (17, 21). Due to p21CIP1-mediated inhibition of G1 cdk's, pRB phosphorylation is blocked and the cells arrested in G1. This circuit constitutes one of the several known links between the two tumor suppressors p53 and pRB. However, p53 is known to also participate in the regulation of other cell cycle checkpoints such as G2/M (2, 67) and possibly S phase (1), thus underscoring how p53 is central in coordinating external stress signals with intracellular signaling responses. Characterization of p53 function(s) has been aided significantly by a temperature-sensitive allele (Val135) which displays wild-type properties at 32°C (exerting a G1 block) and a mutant configuration at 37°C (43, 45). LT binds, metabolically stabilizes, and functionally inactivates p53, thus rescuing cell growth at 32°C (18, 44, 54). However, p53 binding in this assay is not sufficient to rescue growth at 32°C, since a pRB binding-defective mutant, K1, is totally unable to overcome the p53 arrest (54). Thus, the effects of LT on pRB must play a significant role in overriding p53, perhaps by bypassing the effects of p53 on cdk's. Notably, it was shown that a mutant expressing only the first 121 amino acids of LT, and which binds pRB but not p53, could overcome this p53-dependent cell cycle block although less well than wild-type LT (54).
pRB was initially discovered to be mutant in retinoblastomas but is now known to be mutant or deleted in a broad spectrum of other cancers as well (reviewed in reference 78). Hence, loss of pRB tumor suppressor function is believed to often play a key role in neoplastic transformation. pRB protein functions as a transcriptional repressor containing a core repression motif often referred to as the pocket (59, 79, 80). pRB interacts in a cell cycle- and phosphorylation-dependent manner with several cellular factors, of which the E2F family is considered among the critical targets (78). One intriguing model features pRB being recruited to promoter elements via E2F binding. Promoter repression is accomplished by interaction of pRB with surrounding transcription factors or remodelling of chromatin via histone deacetylase (HDAC) interaction (5, 6, 39, 40). Thus, it is believed that pRB represses key cell cycle-regulatory genes with E2F sites, until a point at mid to late G1 where pRB is released from E2F upon multiple site-specific phosphorylations (78). While the E2F interaction is likely to be critical for growth suppression, there is evidence to indicate that some of the many other identified potential binding proteins may contribute (58, 81, 82).
Notably, binding per se to pRB via an LXCXE motif in LT is necessary but not sufficient for dissociation of pRB-E2F complexes and derepression of many cell cycle genes regulated by E2F. The amino-terminal J domain empowers LT with molecular chaperone capabilities and is strictly required for generation of E2F activity and override of pRB-mediated growth arrest in SAOS-2 cells (60, 68, 86). In addition, as we and others have shown, the J domain plays an important role for viral replication (7) and cell transformation (66, 68, 86). In the case of other pRB family members such as p107 and p130, the J domain is required for modulating their phosphorylation state and targeting them for degradation (68, 69).
st enhances both viral replication and transformation by LT, especially under limiting conditions such as low serum, density arrest, or low levels of LT production (3, 4, 42, 55, 63, 70). Certain cell types such as human diploid fibroblasts require both LT and st for transformation and promotion of cell cycle (52, 53). A unifying theme for st activities is the induction of cell cycle progression in otherwise nondividing cells. The only known cellular targets for st are Hsc70, which binds the J domain, and protein phosphatase 2A (PP2A), which st binds stoichiometrically (49). Experiments in vitro suggest PP2A is inhibited by st (85). Association with PP2A is believed to activate the mitogen-activated protein kinase pathway and induce phosphorylation of the mitogen-responsive Na+/H+ antiporter (29, 65). Another important st function in vivo is the transcriptional activation of some viral (E2A) or cellular (AP-1 [24], cyclin A [52], and cyclin D1 [77]) promoters and repression of others (c-fos [76]). Activation of the E2A and cyclin A promoters does not depend on PP2A binding but rather depends on an intact J domain (integrity of the region from positions 42 to 47) (52). The st J domain function remains poorly understood but is required also for st-dependent transformation activity (52). Recently, st was also reported to downregulate the cdk inhibitor p27KIP1 (53).
This study focuses on the functions that LT and st contribute when challenged with a p53-mediated cell cycle block. It was previously demonstrated that the pRB binding-defective mutant of T (K1) fails to override p53 arrest although this mutant retains p53 binding (54). We find that the LT K1 mutant can be complemented for efficient p53 override either by an amino-terminal mutant of LT expressing residues 1 to 135 (LT1-135) containing a J domain-disruptive H42Q mutation or by st. Neither LT1-135 HQ (LT1-135 with the H42Q mutation) nor st has any activity on its own in this assay. Complementation exhibited by LT1-135 HQ suggests that this construct contributes with J domain-independent, pRB binding-dependent effects; hence LT binding to pRB family members has consequences beyond disruption of pRB family-E2F complexes. Consistent with this view, pRB transcriptional repression as measured with a tetracycline repressor (TETr)-pRB small-pocket (SP) fusion can be relieved by LT in a J-independent manner. Although LT1-135 can bypass the p53 block at reduced efficiency, in this context, override is critically dependent on an intact J domain. Taken together, our results imply that several SV40 functions operate in concert to counteract p53-mediated growth suppression, and this may have important implications for transformation and immortalization by SV40.
MATERIALS AND METHODS
Cells and transfections.
SAOS-2, a human osteosarcoma cell line (pRB−/− p53−/−), was cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS) from JRH Biosciences. The previously described rat embryo fibroblast (REF) cell line (T64-7B [54]) carries a temperature-sensitive p53 allele (Val135 [43, 45]). It was propagated at 37°C under 5% CO2 in DMEM with 10% FCS. The colony formation assay was carried out largely according to the reported protocol (54). Approximately 2 × 105 T64-7B cells were plated on a 10-cm-diameter dish and transfected the following day by the 2-[bis(2-hydroxyethyl)amino]ethanesulfonic acid (BES) calcium phosphate coprecipitation method (12). Briefly, 5 to 10 μg of expression vector was mixed with 450 μl of water and 500 μl of 2× BES-buffered saline (0.05 M BES, 0.28 M NaCl, 0.0015 M Na2HPO4, 2 H2O [pH 6.92]). Subsequently, 50 μl of 2.5 M CaCl2 was added dropwise; the precipitate was allowed to form over 10 min and then added to the cells; 18 to 24 h later, the cells were rinsed once with phosphate-buffered saline (PBS) and once with DMEM and then returned to complete medium. They were also transferred from a 37°C to a 32°C incubator. The medium was changed every 4 to 5 days for approximately 3 weeks, at which point colonies were either picked for cloning and expansion or stained with 0.5% crystal violet.
Pools of puromycin-resistant stable T64-7B cell lines were generated by cotransfection of the relevant expression plasmids with the puromycin resistance vector pEpuro and selection in puromycin (1.5 μg/ml; Calbiochem) for 10 to 14 days at 37°C.
Plasmids and mutagenesis.
All of the full-length LT constructs used for the colony assay (wild type, K1, H42Q, and 83-708) are expressed from the SV40 promoter in pSG5 and have been previously described (68, 86). Cytomegalovirus (CMV) vectors were used for the reporter assays and have also been previously described (7). The pSG5 expression vector for the carboxy-terminally truncated LT (LT1-135) contains the previously reported naturally occurring alternative splice product 17k T encoding the amino-terminal 131 amino acids of LT followed by four unique amino acids in a different reading frame (87). The K1 and H42Q mutations were made into the same background by PCR. All of the LT expression plasmids are cDNA constructs; hence, no st is produced. st was amplified by PCR and also cloned into pSG5. The mutant D44N (abolishing J domain function) was generated by QuikChange mutagenesis according to the protocol of the manufacturer (Stratagene). All mutants were confirmed by automated DNA sequencing.
Fluorescence-activated cell sorting (FACS).
Pools of puromycin-resistant cell lines were shifted to 32°C for 24 h in order to arrest the cell cycle via p53. The cells were washed gently with PBS and then detached with 0.1% EDTA in PBS. Cells were washed with PBS containing 1% FCS and then stained for 30 min at 37°C with 50 μg of propidium iodide per ml in the presence of 10 μg of RNase A per ml. Cell cycle analysis was performed on a Becton Dickinson Flow Cytometer using FACScan software, and all profiles are based on a minimum of 10,000 cells.
Reporter assays.
The pRB promoter repression assay was carried out essentially as previously described (59). Briefly, 2 μg of pGL2ANΔTetO (in which the E2F sites in the E2F promoter have been replaced by tetracycline operator (TETo) sites), 2 μg of pSG5 TetR-RB SP (pRB amino acids 379 to 792), 1 μg of pCMX β-gal as an internal control, and 1 μg of CMV LT, K1, or D44N expression vector were cotransfected into SAOS-2 cells on 6-cm-diameter dishes. The reporter plasmid pGL3 6xE2F (38), containing six consensus E2F sites upstream of a TATA box and a luciferase reporter gene, was used to measure total E2F activity in the presence or absence of pRB. Reporter activity was scored approximately 48 h after transfection, using a luciferase assay kit and buffer from Promega, and then corrected for transfection efficiency based on the β-galactosidase activity in the same samples.
Western blots.
Cells were washed twice in PBS and then extracted with LT extraction buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1.0% Nonidet P-40, 5 μg of leupeptin/ml, 5 μg of pepstatin/ml, 0.5 mM phenylmethylsulfonyl fluoride) for 15 min on ice. Subsequently, extracts were cleared of cell debris by centrifugation at 10,000 × g for 5 min in a microcentrifuge at 4°C. Then the cell extracts were boiled with an equal volume of 2× sodium dodecyl sulfate (SDS) sample buffer (5% SDS, 25% glycerol, 62.5 mM Tris [pH 6.8], 0.0075% bromophenol blue, 0.7 M β-mercaptoethanol) for 3 min. Lysates were resolved on discontinuous 11% SDS-polyacrylamide gels and transferred by Western blotting to nitrocellulose membranes. The immunoblots were probed 2 to 3 h with PAb419 (recognizes an epitope within LT1-82 [27]) tissue culture hybridoma supernatant diluted 1:50. The signal was visualized with enhanced chemiluminescence as specified by the manufacturer (Amersham-Pharmacia).
RESULTS
J domain function is not required for overcoming p53 in full-length SV40 LT but is essential in LT1-135.
Previous experiments have established the absolute requirement of the J domain for several growth-regulatory LT functions, notably those depending on an intact pRB binding site (60, 66, 68, 86). Clearly, binding to p53 and its subsequent functional inactivation correlate with both immortalization and cellular transformation elicited by LT (41, 73, 88). However, it remained unknown if the J domain contributes mechanistically to the targeting of p53 by LT. Hence, we set out to test if the J domain function of LT is required for overcoming p53-mediated growth suppression. To do this, we used a previously described cell line designated T64-7B (54). This REF line expresses a temperature-sensitive p53 allele (Val135) which is functionally wild type at 32°C, causing a G0/G1 arrest, and mutant at 37°C, thus allowing cell growth (43, 45). Previous work has demonstrated the capacity of LT to rescue cell growth at 32°C, resulting in colony formation (18, 44, 54). We tested a commonly used J domain loss of function mutant, H42Q, in this assay and found it to be almost as efficient as wild-type LT in forming colonies at 32°C when introduced by calcium phosphate transfection (Table 1). We also tested an amino-terminal truncation mutant lacking the whole J domain, thus encoding residues 83 to 708, and found it capable of overcoming the p53 cell cycle block, albeit at reduced efficiency (Table 1). The reason for the reduced efficiency by which this mutant overcomes the p53 block may well be related to its relatively poor expression (data not shown).
TABLE 1.
Colony numbers of constructs used in this studya
| Construct | Avg colony no. |
|---|---|
| LT | 1.0 |
| H42Q | 0.937 |
| 83-708 | 0.222 |
| K1 | 0.028 |
| st | 0 |
| K1+st | 0.498 |
| K1+st D44N | 0.419 |
| LT1-135 | 0.07b |
| LT1-135 K1 | 0 |
| LT1-135 H42Q | 0 |
| K1+LT1-135 H42Q | 0.336 |
| K1+LT1-135 | 1.075 |
The constructs indicated were transfected into T64-7B cells, and colonies were scored after approximately 3 weeks at 32°C. For each experiment, the colony number was normalized to that of wild-type LT, and each normalized colony number was averaged from a minimum of three experiments.
This value is based on only one experiment which included LT1-135 and wild-type LT. In several other experiments, comparison of colony number for LT1-135 to that of K1+st or K1+LT1-135 HQ suggests that the average colony number relative to LT is approximately 0.15.
When considering the somewhat surprising result that the J domain plays at most a minor role in full-length LT, we wondered if there were other genetic backgrounds where a J domain dependency could be revealed. It has previously been reported that an amino-terminal construct expressing residues 1 to 121 bypasses a p53 block. Hence, we made a similar amino-terminal construct directing expression of amino acids 1 to 135 of LT (LT1-135) and found it to override p53, albeit at lower efficiency than wild-type LT (Table 1). When the H42Q mutation was placed in the context of LT1-135 (in construct LT1-135 HQ), it abrogated the ability to overcome p53 (Table 1). Similarly, we found that a pRB binding mutant, K1, whether in full-length LT or in LT1-135, was totally defective for bypassing the p53 block (Table 1). Taken together, these data emphasize the importance of the J domain in the context of LT1-135 but not in full-length LT.
A pRB binding-defective mutant of LT can be complemented for p53 override by LT1-135 HQ or by st.
Since the results indicate that the pRB binding site is critical for p53 override, we wished to determine what could restore function to the K1 mutant when expressed in trans. Because LT1-135 HQ was, like the K1 mutant of LT, defective, we tested if complementation could occur between LT1-135 HQ and K1. Interestingly, efficient complementation was observed upon coexpression of LT1-135 HQ and K1 (Fig. 1 and Table 1). No complementation was observed between LT1-135 K1 and K1 (data not shown). Hence, complementation is likely to arise from the introduction via LT1-135 HQ of an intact pRB binding site, even in concert with a defective J domain. These observations stress the contribution of activities of LT linked to pRB binding alone and without an accompanying functional J domain. This finding is consistent with the observation that in full-length LT, pRB binding is critical while the J domain is dispensable for overcoming p53-induced arrest (compare K1 to H42Q in Table 1). Also, our data suggest that (at least) two functions in LT cooperate in efficient override of p53 growth inhibition.
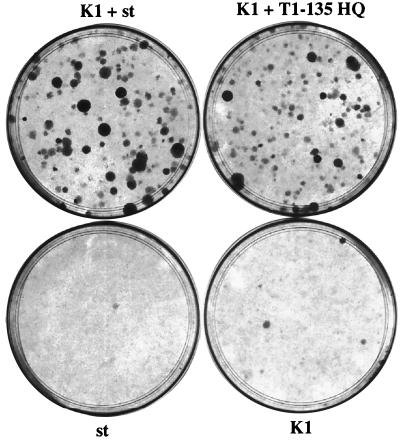
FIG. 1.
Complementation between K1 and LT1-135 HQ or K1 and st for override of p53 arrest. T64-7B cells were transfected by calcium phosphate coprecipitation with K1, st, K1 and st, or K1 and LT1-135 HQ expression vectors. After transfer to 32°C for approximately 3 weeks, colonies were stained with crystal violet for visualization. The LT1-135 HQ dish showed no colonies.
Since st has been associated with cell cycle progression and exerts an LT helper function, we tested whether st can play any role in overcoming p53 growth inhibition. When an st expression vector was transfected into T64-7B cells, it became evident that st independently has no ability to overcome p53 arrest, at least at this expression level. However, we found that st coexpression with K1 rescues cell growth at 32°C, thus displaying complementation (Fig. 1; Table 1). We subsequently used a mutant in st to map the function involved in complementation. The D44N mutant in the conserved HPDK motif is a classical J domain mutant disrupting this function. Analogous st mutants have been shown to be defective in some transcriptional assays (47, 52). We found that the st D44N mutant complemented as well as wild-type st (Table 1). Fig. 2 shows in a schematic drawing four different scenarios for successful override of p53, including the two based on complementation.
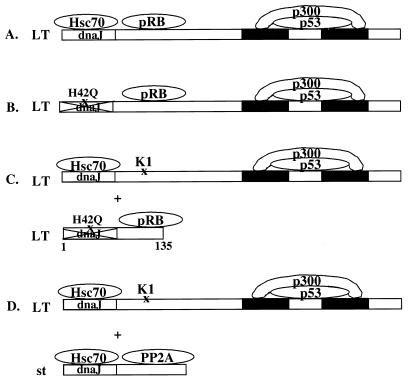
FIG. 2.
Schematic representation of the various complementation scenarios. (A) Wild-type LT with an intact J domain and pRB binding site, which can cooperate with an undefined carboxy-terminal activity, examplified here by p53 or p300, although a novel binding protein cannot be excluded. (B) Diagram demonstrating that even full-length LT with the J domain-inactivating mutation H42Q can override p53, presumably because of cooperation between pRB binding and the carboxy-terminal activity. The complementation is emphasized in panel C, since the pRB binding site when expressed in trans, even without an accompanying functional J domain, can restore p53 override to the K1 mutant that fails to bind pRB. Panel D emphasizes another route to complementation in which st, perhaps due to its PP2A binding capability, can restore p53 override function to K1.
Expression of K1 and small t or K1 and LT1-135 HQ in individual clones.
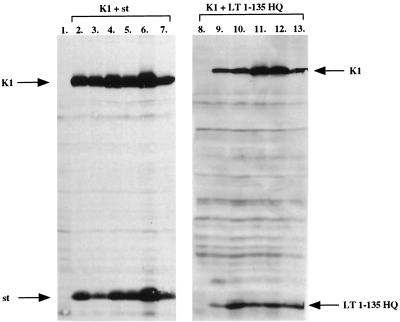
To verify that the complementation is authentic and dependent on continuous expression of each complementation partner, individual clones were picked and expanded into lines maintained at 32°C. As shown in Fig. 3, five of six independent lines from a cotransfection of LT1-135 HQ and K1 expression plasmids expressed both proteins (lanes 8 to 13). Briefly, cell lysates from each line were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and Western blot analysis with antibody PAb419 (27), recognizing an epitope within the first 82 amino acids of LT. The one cell line (lane 8) that expressed neither K1 nor LT1-135 HQ may have arisen due to a mutation, perhaps induced by expression of either complementation partner early on after transfection.
FIG. 3.
Expression in independent clones derived from complementation experiments. Six colonies were cloned and expanded at 32°C from either K1 and LT1-135 HQ or K1 and st cotransfections. Cell lysates were prepared and resolved by SDS-PAGE followed by Western blotting and probing with antibody PAb419, which recognizes an epitope within the first 82 amino acids of LT. Hence, st could be detected on the same blot as K1. Lanes: 1, lysate from untransfected parental T64-7B cells, included as a control; 2 to 7, lysates from K1 and st cotransfections; 8 to 13, lysates from K1 and LT1-135 HQ clones.
All of the six clones from the K1 and st plasmid cotransfection expressed both proteins, albeit at variable levels (Fig. 3, lanes 2 to 7). The same antibody, PAb419, was used for detection, since the first 82 amino acids are in common between st and LT. Overall, the lines derived from K1 and st cotransfection displayed a more robust growth than those derived from K1 and LT1-135 HQ. This observation manifested in a generally greater colony size for K1 and st lines compared to K1 and LT1-135 lines (Fig. 1). Nevertheless, the lines expressing K1 and LT1-135 HQ were continuously growing at 32°C, showing a stable phenotype similar to those lines derived from K1 and st. Taken together, the complementation is very likely real, since all lines except one continue to express both complementation partners. Furthermore, reverse transcription-PCR reveals that no recombination events have taken place in any of the K1+LT1-135 HQ lines (data not shown).
Cell cycle distribution in pools of stable lines after shift to 32°C.
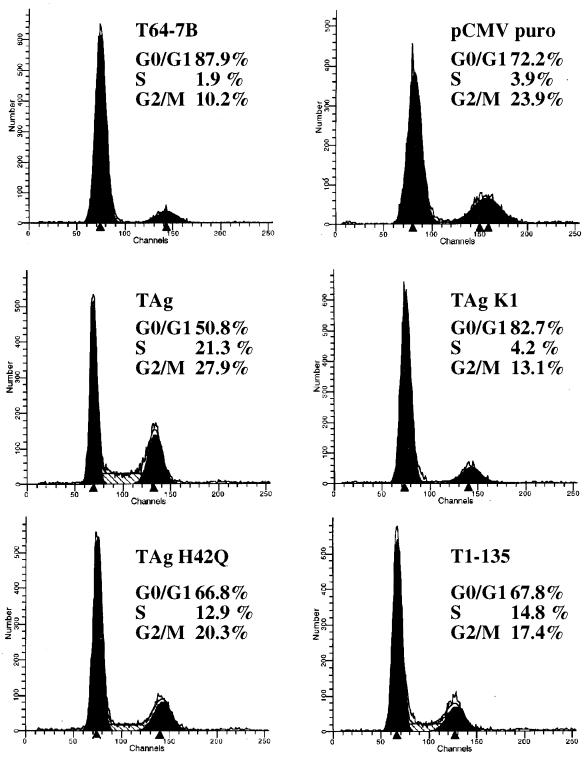
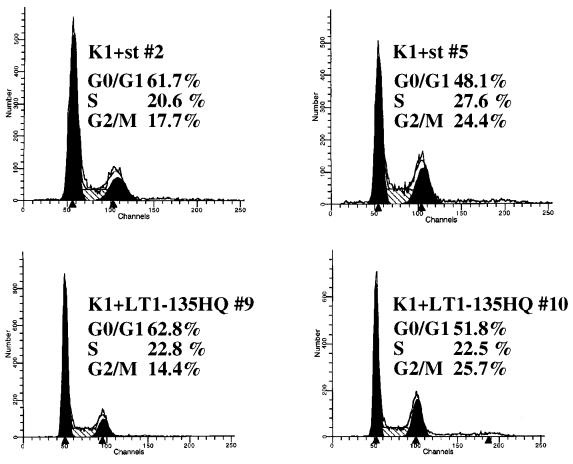
The p53 tumor suppressor protein is known to exert its activity at several independent cell cycle checkpoints, in programmed cell death, and perhaps in yet undiscovered growth suppression pathways. Complementation between K1 and st could reflect cooperation in override of one or more cell cycle checkpoints monitored by p53. To test whether the K1 mutant and st can affect distinct p53 controlled cell cycle checkpoints, FACS analysis was performed on pools of stable lines. The stable cell lines were generated by cotransfection of an empty CMV vector or an LT, K1, H42Q, LT1-135, or st expression plasmid together with a puromycin drug resistance vector. After selection in puromycin for 10 days, the colonies were pooled and shifted to 32°C for 24 h to induce wild-type p53 function. Each pool of stable lines was then stained with propidium iodide and analyzed by FACS. In agreement with published reports, the parental cell line and the vector control were predominantly arrested in the G0/G1 phase of the cell cycle, showing only 2 to 4% of the cells in S phase (Fig. 4). Conversely, the stable line expressing wild-type LT continued to proliferate at 32°C, with 21% of the cells in S phase (Fig. 4). Interestingly, both the K1 and the st stable lines arrested efficiently in G0/G1, with only approximately 4% of the population in S phase (Fig. 4). These results correlate well with the long-term colony assay and indicate that both K1 and st fail to overcome the G0/G1 block elicited by p53. Perhaps the two complementation partners synergize in promoting G0 exit and/or the G1-S cell cycle transition. Both the H42Q allele of LT and LT1-135 displayed an intermediate phenotype, thus overcoming the p53-induced G0/G1 block, with an S-phase content ranging from 13 to 15% (Fig. 4). Again, this degree of sustained cell cycle progression correlates well with their activity in the colony assay. For comparison, two of the K1+st clonal lines (lines 2 and 5) as well as two K1+LT1-135 HQ clonal lines (lines 9 and 10) were included for FACS analysis. Consistent with the colony assay, all of these lines have a significant S-phase population (ranging from 20.6 to 27.6%) at 32°C.
FIG. 4.
Cell cycle distribution in stable lines after shift to 32°C. Pools of stable lines were derived from T64-7B cells by cotransfection of various expression plasmids with a puromycin resistance vector (pCMV puro) and selection in puromycin for about 10 days. Each pool was shifted to 32°C for 24 h, and cell cycle distribution was monitored by propidium iodide staining and FACS analysis. For comparison, two of the K1+st clonal lines (2 and 5) as well as two of the K1+LT1-135 HQ clonal lines (9 and 10), grown at 32°C, were included. Percentages indicate the content of G0/G1, S, and G2/M cells as estimated by using FACScan software. TAg, T antigen.
pRB-mediated repression can be relieved in a partially J domain-independent manner.
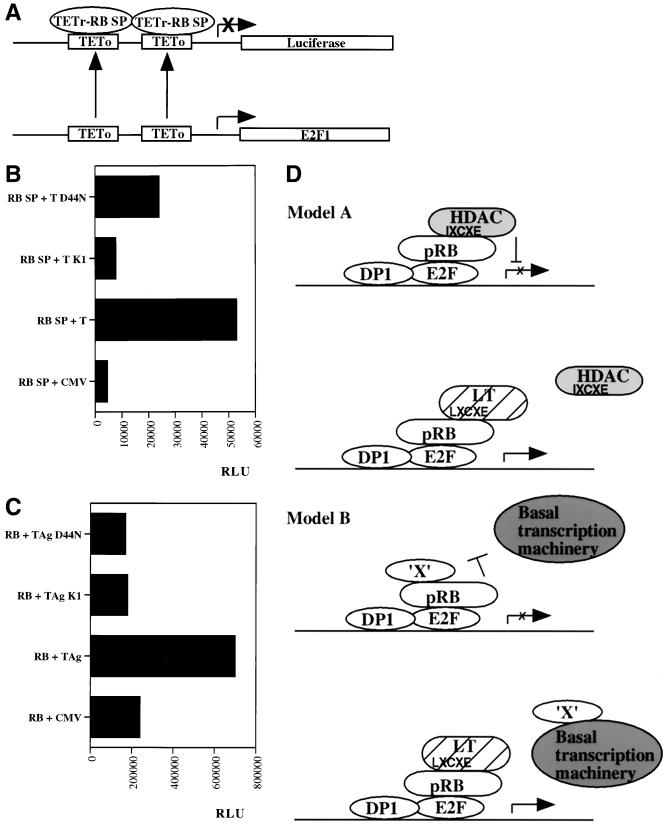
Complementation exhibited by the coexpression of K1 and LT1-135 HQ emphasizes the existence of an activity of LT that is pRB binding site dependent but J domain independent. To be more precise, this complementing activity is partially dependent on the J domain (Table 1). This activity stands in contrast to the effects of LT on modulation of E2F, which are absolutely dependent on the J domain. The pRB protein is a versatile transcriptional modulator, behaving as a repressor of several proliferation-associated genes and an activator of some differentiation-associated genes. Previous reports have documented that derepression of E2F activity and release of free E2F can be accomplished by LT in a manner dependent on binding to pRB and retention of an intact J domain (60, 86). However, pRB can repress via several distinct mechanisms, dependent on promoter context (5). The core repression motif (SP; amino acids 379 to 792) works as a repressor when fused to a heterologous DNA binding protein, even though it does not bind E2F (59). Sellers and coworkers measured pRB repression by using the pRB SP fused to TETr in conjunction with a luciferase reporter construct containing TETo sites in place of the E2F sites in the E2F promoter (Fig. 5A) (59). We used the same system to test whether LT-mediated relief of pRB SP repression is J domain dependent when assayed in pRB-deficient SAOS-2 cells. Luciferase reporter assays were conducted and normalized for transfection efficiency based on β-galactosidase activity from a cotransfected CMV β-galactosidase plasmid. Repression by the pRB SP fusion was relieved efficiently by LT and in a totally pRB binding site-dependent manner, since the K1 mutant is defective (Fig. 5B). Interestingly, the J domain mutant D44N, which is defective in J domain dependent assays, is partially effective at alleviating pRB repression (Fig. 5B).
FIG. 5.
Genetic requirements for LT-mediated relief of pRB repression. (A) Schematic drawing of the reporter construct used in panel B for measurement of pRB SP repression when this minimal repression domain is fused to the heterologous DNA binding domain of the TETr (59). The E2F sites in the E2F1 promoter have been replaced with TETo sites, which function to recruit the TETr-pRB SP repressor complex. (B) The reporter construct pGL2ANΔTETo, depicted in panel A, was cotransfected into SAOS-2 cells with a TETr-pRB SP expression plasmid and a CMV expression vector for wild-type LT, K1, or D44N or an empty CMV vector as a control. Luciferase activity (expressed as relative light units) was assayed to measure reporter activity, and this value was subsequently corrected based on β-galactosidase activity originating from a cotransfected CMV β-galactosidase expression vector. The data shown are from one representative experiment. Three independent reporter assays produced very similar results. (C) The reporter construct pGL3 6xE2F (38) was cotransfected into SAOS-2 cells with a CMV pRB expression plasmid and a CMV vector for LT, K1, or D44N or an empty CMV control. Luciferase activity was measured as described for panel B. The data presented are from one representative experiment out of a series of three with very similar results. TAg, T antigen. (D) The two models outlined may explain how LT can relieve pRB repression in a partially J domain-independent manner. In model A, LT displaces HDAC1 from pRB because of competition from the LXCXE motif present in LT. At least in some promoter contexts it is likely that pRB, although remaining bound to E2F, fails to sustain repression upon release of HDAC1. Previous reports have demonstrated that an LXCXE-containing peptide from SV40 LT suffices to disrupt the pRB-HDAC1 interaction (40). Hence, the J domain is not required. Note that the pRB-HDAC1 interaction is likely to be indirect and bridged by an as yet unidentified protein (6, 40) which is left out of the drawing for simplicity. In model B it is hypothesized that LT may prevent pRB repression arising from its blockade of interactions between certain transcription factors and the basal transcription machinery. For example, it was previously shown that a human papillomavirus E7-derived peptide can disrupt interaction between pRB and upstream binding factor and thereby prevent pRB-mediated repression of RNA polymerase I transcription (9). Similar mechanisms of pRB repression may also exist with regard to some RNA polymerase II-dependent promoters.
We also examined LT-mediated relief of repression using another reporter. The reporter construct pGL3 6xE2F contains six E2F sites upstream of a basic TATA box and a luciferase gene (38). This reporter is more likely to measure pRB repression due to sequestration and inactivation of the E2F activation domain. As expected, LT relieves repression by pRB, but this time in a strictly pRB binding and J domain-dependent manner (Fig. 5C). This observation is concordant with published data, since this reporter is more likely to measure free E2F activity. Taken together, our data indicate that LT targets all of the various pRB repression mechanisms, but some of these require only binding to pRB whereas others, such as release of E2F, additionally require an intact J domain.
DISCUSSION
Our data suggest that two functions, one located in the amino terminus and the second in the carboxy terminus of LT, cooperate in efficient override of a p53-mediated cell cycle block. Three lines of evidence support this hypothesis. First, a construct encoding amino acids 1 to 135 of LT (LT1-135) with the J domain-inactivating mutation H42Q can efficiently complement the K1 mutant in full-length LT (Fig. 1; Table 1). Both of these constructs are inactive by themselves. This complementation in trans strongly suggests there are indeed two independent functions. Second, while a construct encoding LT1-135 itself has some activity in the assay, it is dramatically complemented by a K1 mutant. Again, this suggests the existence of two functions, with the second function residing to the carboxyl side of residue 135. Finally, we have unpublished data that the mutant dl434–444 is defective in this assay although it has wild-type function in its J domain and pRB binding site. Our interpretation of experimental data stands in some contrast to that of Quartin et al., who attributed LT activity in this assay to its amino terminus and pRB binding site (54).
In this study, we have concentrated on the amino-terminal function defined by the K1 mutant in LT. This function appears to be pRB family binding dependent but does not strictly depend on the J domain. Notably, pRB family binding is restored in trans when cotransfecting LT1-135 HQ. Interestingly, it has been reported, and our own findings are in agreement, that J domain function is strictly required for disrupting pRB-E2F complexes and promoting E2F-dependent transcription (86) (Fig. 5C). In addition, LT-mediated degradation and modulation of phosphorylation state of the other pRB family members p107 and p130 absolutely require an intact J domain (68, 69). Since LT1-135 HQ can complement a K1 mutant in p53 override, our data support the notion that LT binding to pRB has important consequences beyond dissociation of pRB-E2F complexes and release of free E2F.
What function of pRB might underlie the effects of LT in this assay? pRB is implicated in a plethora of cellular processes, including cell cycle regulation, differentiation, and apoptosis (78), presumably via its roles in transcriptional control. pRB is a versatile transcriptional regulator, repressing the majority of target genes but activating a subset. Mechanistically, pRB repression is believed to be accomplished in several different ways dependent on the surrounding promoter elements. In one common model, it is assumed that E2F tethers pRB to the promoter region, where pRB repression may occur in either of two ways: (i) Recruitment of an HDAC modifying the surrounding chromatin to promote repression (model A in Fig. 5D) or (ii) disruption of contacts between other bound transcription factors and the basal transcription machinery (model B in Fig. 5D).
One plausible hypothesis to account for biological activity due to J domain-independent pRB binding is the derepression of some E2F-regulated promoters due to disruption of a repressive pRB-HDAC complex (Fig. 5D). It is in fact known that the pRB-HDAC complex relies on interactions between a HDAC LXCXE-like motif and the small pocket of pRB (6, 40). Importantly, SV40 LT disrupts this interaction, presumably by competition via its own LXCXE, since a peptide containing the LXCXE motif of LT is sufficient to abrogate the pRB-HDAC1 association (Fig. 4D) (40). We have found that repression mediated by the SP (amino acids 379 to 792) of pRB when fused to the heterologous DNA binding domain of TETr can be relieved by LT in a manner entirely dependent on pRB binding but not strictly dependent on a functional J domain. In this particular promoter context (the E2F promoter where E2F sites are replaced by TETo sites [59]), pRB SP repression may accur via several different mechanisms that are all targeted by LT, but only a subset requires an intact J domain.
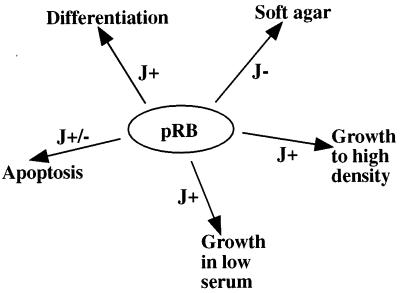
An alternative interpretation, that we cannot exclude at this point, is that another pRB growth suppression pathway, independent of E2F binding, is targeted by LT in a partially J domain-independent manner. In the literature there are reports suggesting the existence of growth-inhibitory functions of pRB in addition to E2F binding (58, 81). Furthermore, pRB can repress the polymerase I and polymerase III promoters (82), both of which constitute potential targets for LT in dismantling pRB-inhibitory function(s). Consistent with our findings, others have made similar discoveries. First, LT-induced soft agar growth of mouse embryo fibroblasts (MEFs) depends on pRB binding but is J domain independent (68). Second, LT confers a growth advantage to p107−/− p130−/− MEFs, and this is pRB binding but not J dependent (68). Finally, Tevethia and coworkers have reported that MEFs expressing an LT construct lacking the J domain and encoding residues 128 to 708 can be restored for growth to high saturation density, but not growth in low serum and without anchorage, by addition at the carboxy terminus of an pRB binding site (72). Interestingly, LT targets multiple pRB activities involved in transformation, differentiation, and apoptosis; the LT J domain is required for some of these pRB effects but not for others (Fig. 6). The varying requirements for a functional J domain in addition to the LXCXE motif is likely to reflect LT-mediated disruption of pRB interaction with distinct binding partners, each specific for a particular biological response.
FIG. 6.
LT abrogates multiple pRB-dependent biological functions, and only some require a functional J domain. pRB has been implicated in control of diverse biological processes such as proliferation, differentiation, and apoptosis. LT interferes with all of these, but the mechanism seems to differ with regard to requirements for a functional J domain. For example, LT induces growth in soft agar depending on its pRB binding site but not on its J domain (indicated as J− [68]). In contrast, growth to high saturation density and in low serum requires both pRB binding and a functional J domain of LT (indicated as J+ [68]). The polyomavirus LT-mediated block in myoblast differentiation requires both pRB binding and an intact J domain (B. Schaffhausen and Q. Sheng, personal communication). However, polyomavirus LT induction of apoptosis in C2C12 cells is pRB binding site dependent but only partially J domain dependent (Schaffhausen and Sheng, personal communication). As shown in this study, override of p53 growth suppression requires pRB binding but not the J domain. Taken together, the universal requirement for pRB binding, but variable dependency on the J domain, suggests interconnections of distinct biological processes with particular pRB interaction partners.
We find that in full-length LT the J domain mutation H42Q has only a modest effect on override of p53 (Table 1). A construct encoding LT 83-708 and thus lacking the whole J domain also points to the same conclusion, that J function is not required in full-length LT for overcoming p53 growth inhibition. While the J domain is dispensable in the context of full-length LT for overriding p53 arrest, it is required in the LT1-135 context (Table 1). Concordant with our results, Srinivasan et al. also found a requirement for the J domain in LT1-136 but not in full-length LT when analyzing focus formation (66). In our scenario, we interpret this to mean that in full-length LT pRB binding, with or without J function, can cooperate with a carboxy-terminal function. We do not believe that there are additional carboxy-terminal targets for the J domain involved in p53 override, since a K1/DN double mutant in full-length LT performs in complementation assays (with either st or LT1-135HQ) as well as the K1 single mutant (unpublished data). The carboxy-terminal cooperating function(s) remains to be identified, but the most prominent candidates are likely to be p53 and/or p300/p400 binding. In fact, adenovirus E1A-mediated override of p53 depends on p300 binding (35, 64). Interestingly, Tevethia et al. found that two activities of LT, one mapping to the N terminus and the other in the C terminus, individually extend the life span of C57BL/6 MEFs and together cooperate to immortalize (71). It remains possible that this complementation is reminiscent of the one we observe, since functional inactivation of p53 by LT has been correlated well with its capacity to immortalize (73, 88). In fact, immortalization may be associated at some level with overcoming cell cycle-inhibitory function(s) of p53. Also, our observed complementation for override of a p53 cell cycle block may be linked to oncogenic transformation. Polyomavirus middle T antigen fails to override a p53 growth arrest (18) and also fails to transform REF52 cells unless p53 is functionally inactivated by polyomavirus LT or a dominant negative p53 (46).
Our results showing only modest effects of J domain mutations on p53 override (Table 1) stand in apparent contrast to the reported serious defect of mutants dl1135 (deletion of amino acids 17 to 27) and 2831 (deletion of amino acids 5 to 35 + insertion of five amino acids) in the same assay (54). We believe that these mutants not only affect the J domain but sustain multiple structural defects which may affect carboxy-terminal activities. This claim is supported by several past and current observations. First, dl1135 affects ATPase activity although this activity maps to the carboxy terminus (13). Second, Lill et al. drew a similar conclusion with regard to dl1135, since this mutant fails to complement an E1A p300 binding mutant (84), yet the p300 binding site is located largely within the carboxy terminus (36). Third, Srinivasan et al. were unsuccessful in obtaining complementation between dl1135 and a construct encoding the first 136 amino acids of LT (66). Fourth, in REF52 transformation assays the dl1135 mutation has a more severe phenotype than the structurally less distorting J point mutations H42Q and D44N (50, 51, 66). Fifth, Cavender et al. suggested that this class of amino-terminal mutants (for example, dl1135) may distort a portion of the region from positions 251 to 708, since they fail to cooperate with oncogenic Ras for transformation of REFs (10). Finally, structural determination of the J domain of LT suggests that the regions deleted in dl1135 and 2831 are critical for folding of the entire J domain (J. A. DeCaprio and G. Wagner, unpublished data). Further support for our conclusion that J function is not required in full-length LT for overcoming p53 comes from the construct entirely lacking the J domain (LT 83-708), which overrides moderately well in spite of low expression (data not shown). Quartin et al. reported complementation between the mutants K1 and 2831 (54). We believe, based on our current observations, that this complementation is analogous to that found for K1+LT1-135 HQ. The mutant 2831 belongs to the same class as dl1135 and is likely to retain pRB binding, but its J domain is severely disrupted. Supporting this notion, we find that K1 can also be complemented by LT1-135 dl1135 (data not shown), and one earlier report found that dl1135 indeed complements an E1A pRB binding mutant for transformation (84).
It may appear seemingly paradoxical that pRB family member binding is absolutely critical for bypassing a cell cycle block whereas p53 binding is not. These results are consistent with previous findings, however, and underscore the importance of interconnections between the p53 and pRB growth-suppressive pathways. Past reports have indicated that pRB binding is critical for SV40 LT, polyomavirus LT, adenovirus E1A, or human papillomavirus E7 override of a p53 cell cycle block (16, 18, 44, 54, 62, 74). One level of interconnection is likely to be p21CIP1, which is transcriptionally induced by p53 and blocks G1 cell cycle progression by inhibiting cdk phosphorylation of pRB (21). Another interface is likely to involve p19ARF, which is induced by oncogene-mediated release of E2F from pRB and in turn triggers p53 activation, resulting in either G1 arrest or apoptosis (61). The cellular oncoprotein MDM2 is a negative regulator of p53 which suffices to bypass both pRB and p53 growth arrest (83), suggesting that it may in fact link the two pathways somehow. Additional examples of cross talk between pRB and p53 are likely to arise. Interestingly, blocking p53-dependent transcription may not be sufficient for overcoming a p53 cell cycle block, since the K1 mutant has been reported to abrogate p53 transcription as efficiently as wild-type LT (30) yet fails to override (reference 54 and our data). Future experiments will be aimed at dissecting the mechanistic details of cooperation. Traditionally, the ability of p53 to arrest the cell cycle in G1 has been correlated with its transcriptional ability (17, 21), but several other cell cycle and growth-suppressive properties appear independent of p53 transcriptional control (15, 48, 56, 75). Indeed, in one study, some mutants of p53 resulted in an extended life span of mammary epithelial cells and others conferred full immortalization, but all mutants abrogated p53-mediated transactivation to similar extents (8). Hence, full biological activity of p53 may require more than its transcriptional function(s).
Another important finding from this study is that st also can complement the K1 mutant in overcoming p53 (Fig. 1; Table 1). Several earlier reports have noted that st can provide a helper function to LT in both replication and transformation assays, especially in particular cell types or under demanding growth conditions for the virus (3, 4, 42, 53, 55, 63, 70). It is possible that the ability of st and LT to cooperate in p53 override is a contributing factor in some of these cases. Since st D44N complements as well as wild-type st, the st J domain appears to play no role in complementation. We have attempted to address any role that PP2A binding may play in complementation, but our results with the st PP2A binding mutants C97S and C103S have, due to low expression under the particular assay conditions, failed to provide unequivocal conclusions as to the role of PP2A binding (data not shown). While it remains a formal possibility that st complementation is partially dependent on the LT/st common region, we favor the notion that the unique st region is required, especially given previous results implicating this st function, and not the J domain, in induction of cell cycle progression (29). Furthermore, our unpublished data indicate that a chimeric LT (HSJ1-T [86]), where the LT J domain is replaced with that of HSJ1, is as effective as wild-type LT in overcoming p53 growth arrest. This finding may suggest that for this particular assay, the only relevant function residing within the first exon is that of the J domain. Experiments are in progress to address the mechanistic details of st complementation.
ACKNOWLEDGMENTS
This work was supported by NIH grants to T.M.R. (PO1-CA50661 and CA30002) and J.A.D. (PO-CA50661 and RO1CA 63113).
We thank Arnold Levine for providing T64-7B cells and William Sellers for contributing several of the plasmids used in reporter assays. We are grateful to Brian Schaffhausen for critical reading of the manuscript.
T.M.R. has consulting relationships with Upstate Biotechnology and Novartis Pharmaceuticals, Inc.
REFERENCES
- 1.Agarwal M L, Agarwal A, Taylor W R, Chernova O, Sharma Y, Stark G R. A p53-dependent S-phase checkpoint helps to protect cells from DNA damage in response to starvation for pyrimidine nucleotides. Proc Natl Acad Sci USA. 1998;95:14775–14780. doi: 10.1073/pnas.95.25.14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agarwal M L, Agarwal A, Taylor W R, Stark G R. p53 controls both the G2/M and the G1 cell cycle checkpoints and mediates reversible growth arrest in human fibroblasts. Proc Natl Acad Sci USA. 1995;92:8493–8497. doi: 10.1073/pnas.92.18.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bikel I, Mamon H, Brown E L, Boltax J, Agha M, Livingston D M. The t-unique coding domain is important to the transformation maintenance function of the simian virus 40 small t antigen. Mol Cell Biol. 1986;6:1172–1178. doi: 10.1128/mcb.6.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bikel I, Montano X, Agha M E, Brown M, McCormack M, Boltax J, Livingston D M. SV40 small t antigen enhances the transformation activity of limiting concentrations of SV40 large T antigen. Cell. 1987;48:321–330. doi: 10.1016/0092-8674(87)90435-1. [DOI] [PubMed] [Google Scholar]
- 5.Brehm A, Kouzarides T. Retinoblastoma protein meets chromatin. Trends Biochem Sci. 1999;24:142–145. doi: 10.1016/s0968-0004(99)01368-7. [DOI] [PubMed] [Google Scholar]
- 6.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 7.Campbell K S, Mullane K P, Aksoy I A, Stubdal H, Zalvide J, Pipas J M, Silver P A, Roberts T M, Schaffhausen B S, DeCaprio J A. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 1997;11:1098–1110. doi: 10.1101/gad.11.9.1098. [DOI] [PubMed] [Google Scholar]
- 8.Cao Y, Gao Q, Wazer D E, Band V. Abrogation of wild-type p53-mediated transactivation is insufficient for mutant p53-induced immortalization of normal human mammary epithelial cells. Cancer Res. 1997;57:5584–5589. [PubMed] [Google Scholar]
- 9.Cavanaugh A H, Hempel W M, Taylor L J, Rogalsky V, Todorov G, Rothblum L I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 10.Cavender J F, Conn A, Epler M, Lacko H, Tevethia M J. Simian virus 40 large T antigen contains two independent activities that cooperate with a ras oncogene to transform rat embryo fibroblasts. J Virol. 1995;69:923–934. doi: 10.1128/jvi.69.2.923-934.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheetham M E, Brion J P, Anderton B H. Human homologues of the bacterial heat-shock protein DnaJ are preferentially expressed in neurons. Biochem J. 1992;284:469–476. doi: 10.1042/bj2840469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark R, Peden K, Pipas J M, Nathans D, Tjian R. Biochemical activities of T-antigen proteins encoded by simian virus 40 A gene deletion mutants. Mol Cell Biol. 1983;3:220–228. doi: 10.1128/mcb.3.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeCaprio J A, Ludlow J W, Figge J, Shew J Y, Huang C M, Lee W H, Marsilio E, Paucha E, Livingston D M. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 15.Del Sal G, Ruaro E M, Utrera R, Cole C N, Levine A J, Schneider C. Gas1-induced growth suppression requires a transactivation-independent p53 function. Mol Cell Biol. 1995;15:7152–7160. doi: 10.1128/mcb.15.12.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demers G W, Foster S A, Halbert C L, Galloway D A. Growth arrest by induction of p53 in DNA damaged keratinocytes is bypassed by human papillomavirus 16 E7. Proc Natl Acad Sci USA. 1994;91:4382–4386. doi: 10.1073/pnas.91.10.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Leonardo A, Linke S P, Clarkin K, Wahl G M. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 18.Doherty J, Freund R. Polyomavirus large T antigen overcomes p53 dependent growth arrest. Oncogene. 1997;14:1923–1931. doi: 10.1038/sj.onc.1201025. [DOI] [PubMed] [Google Scholar]
- 19.Dyson N, Buchkovich K, Whyte P, Harlow E. The cellular 107K protein that binds to adenovirus E1A also associates with the large T antigens of SV40 and JC virus. Cell. 1989;58:249–255. doi: 10.1016/0092-8674(89)90839-8. [DOI] [PubMed] [Google Scholar]
- 20.Eckner R, Ludlow J W, Lill N L, Oldread E, Arany Z, Modjtahedi N, DeCaprio J A, Livingston D M, Morgan J A. Association of p300 and CBP with simian virus 40 large T antigen. Mol Cell Biol. 1996;16:3454–3464. doi: 10.1128/mcb.16.7.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.el-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 22.Ewen M E, Ludlow J W, Marsilio E, DeCaprio J A, Millikan R C, Cheng S H, Paucha E, Livingston D M. An N-terminal transformation-governing sequence of SV40 large T antigen contributes to the binding of both p110Rb and a second cellular protein, p120. Cell. 1989;58:257–267. doi: 10.1016/0092-8674(89)90840-4. [DOI] [PubMed] [Google Scholar]
- 23.Fanning E, Knippers R. Structure and function of simian virus 40 large tumor antigen. Annu Rev Biochem. 1992;61:55–85. doi: 10.1146/annurev.bi.61.070192.000415. [DOI] [PubMed] [Google Scholar]
- 24.Frost J A, Alberts A S, Sontag E, Guan K, Mumby M C, Feramisco J R. Simian virus 40 small t antigen cooperates with mitogen-activated kinases to stimulate AP-1 activity. Mol Cell Biol. 1994;14:6244–6252. doi: 10.1128/mcb.14.9.6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giaccia A J, Kastan M B. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 26.Grossman S R, Perez M, Kung A L, Joseph M, Mansur C, Xiao Z X, Kumar S, Howley P M, Livingston D M. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- 27.Harlow E, Crawford L V, Pim D C, Williamson N M. Monoclonal antibodies specific for simian virus 40 tumor antigens. J Virol. 1981;39:861–869. doi: 10.1128/jvi.39.3.861-869.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollstein M, Rice K, Greenblatt M S, Soussi T, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montesano R, Harris C C. Database of p53 gene somatic mutations in human tumors and cell lines. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 29.Howe A K, Gaillard S, Bennett J S, Rundell K. Cell cycle progression in monkey cells expressing simian virus 40 small t antigen from adenovirus vectors. J Virol. 1998;72:9637–9644. doi: 10.1128/jvi.72.12.9637-9644.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang D, Srinivasan A, Lozano G, Robbins P D. SV40 T antigen abrogates p53-mediated transcriptional activity. Oncogene. 1993;8:2805–2812. [PubMed] [Google Scholar]
- 31.Kelley W L, Landry S J. Chaperone power in a virus? Trends Biochem Sci. 1994;19:277–278. doi: 10.1016/0968-0004(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 32.Ko L J, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 33.Lane D P, Crawford L V. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 34.Levine A J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 35.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 36.Lill N L, Tevethia M J, Eckner R, Livingston D M, Modjtahedi N. p300 family members associate with the carboxyl terminus of simian virus 40 large tumor antigen. J Virol. 1997;71:129–137. doi: 10.1128/jvi.71.1.129-137.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linzer D I, Levine A J. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell. 1979;17:43–52. doi: 10.1016/0092-8674(79)90293-9. [DOI] [PubMed] [Google Scholar]
- 38.Lukas J, Herzinger T, Hansen K, Moroni M C, Resnitzky D, Helin K, Reed S I, Bartek J. Cyclin E-induced S phase without activation of the pRb/E2F pathway. Genes Dev. 1997;11:1479–1492. doi: 10.1101/gad.11.11.1479. [DOI] [PubMed] [Google Scholar]
- 39.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 40.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain J P, Troalen F, Trouche D, Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- 41.Manfredi J J, Prives C. The transforming activity of simian virus 40 large tumor antigen. Biochim Biophys Acta. 1994;1198:65–83. doi: 10.1016/0304-419x(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 42.Martin R G, Chou J Y. Simian virus 40 functions required for the establishment and maintenance of malignant transformation. J Virol. 1975;15:599–612. doi: 10.1128/jvi.15.3.599-612.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martinez J, Georgoff I, Levine A J. Cellular localization and cell cycle regulation by a temperature-sensitive p53 protein. Genes Dev. 1991;5:151–159. doi: 10.1101/gad.5.2.151. [DOI] [PubMed] [Google Scholar]
- 44.Michael-Michalovitz D, Yehiely F, Gottlieb E, Oren M. Simian virus 40 can overcome the antiproliferative effect of wild-type p53 in the absence of stable large T antigen-p53 binding. J Virol. 1991;65:4160–4168. doi: 10.1128/jvi.65.8.4160-4168.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michalovitz D, Halevy O, Oren M. Conditional inhibition of transformation and of cell proliferation by a temperature-sensitive mutant of p53. Cell. 1990;62:671–680. doi: 10.1016/0092-8674(90)90113-s. [DOI] [PubMed] [Google Scholar]
- 46.Mor O, Read M, Fried M. p53 in polyoma virus transformed REF52 cells. Oncogene. 1997;15:3113–3119. doi: 10.1038/sj.onc.1201549. [DOI] [PubMed] [Google Scholar]
- 47.Mungre S, Enderle K, Turk B, Porras A, Wu Y Q, Mumby M C, Rundell K. Mutations which affect the inhibition of protein phosphatase 2A by simian virus 40 small-t antigen in vitro decrease viral transformation. J Virol. 1994;68:1675–1681. doi: 10.1128/jvi.68.3.1675-1681.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Notterman D, Young S, Wainger B, Levine A J. Prevention of mammalian DNA reduplication, following the release from the mitotic spindle checkpoint, requires p53 protein, but not p53- mediated transcriptional activity. Oncogene. 1998;17:2743–2751. doi: 10.1038/sj.onc.1202210. [DOI] [PubMed] [Google Scholar]
- 49.Pallas D C, Shahrik L K, Martin B L, Jaspers S, Miller T B, Brautigan D L, Roberts T M. Polyoma small and middle T antigens and SV40 small t antigen form stable complexes with protein phosphatase 2A. Cell. 1990;60:167–176. doi: 10.1016/0092-8674(90)90726-u. [DOI] [PubMed] [Google Scholar]
- 50.Peden K W, Pipas J M. Simian virus 40 mutants with amino-acid substitutions near the amino terminus of large T antigen. Virus Genes. 1992;6:107–118. doi: 10.1007/BF01703060. [DOI] [PubMed] [Google Scholar]
- 51.Pipas J M, Peden K W, Nathans D. Mutational analysis of simian virus 40 T antigen: isolation and characterization of mutants with deletions in the T-antigen gene. Mol Cell Biol. 1983;3:203–213. doi: 10.1128/mcb.3.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porras A, Bennett J, Howe A, Tokos K, Bouck N, Henglein B, Sathyamangalam S, Thimmapaya B, Rundell K. A novel simian virus 40 early-region domain mediates transactivation of the cycle A promoter by small-t antigen and is required for transformation in small-t antigen-dependent assays. J Virol. 1996;70:6902–6908. doi: 10.1128/jvi.70.10.6902-6908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Porras A, Gaillard S, Rundell K. The simian virus 40 small-t and large-T antigens jointly regulate cell cycle reentry in human fibroblasts. J Virol. 1999;73:3102–3107. doi: 10.1128/jvi.73.4.3102-3107.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Quartin R S, Cole C N, Pipas J M, Levine A J. The amino-terminal functions of the simian virus 40 large T antigen are required to overcome wild-type p53-mediated growth arrest of cells. J Virol. 1994;68:1334–1341. doi: 10.1128/jvi.68.3.1334-1341.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubin H, Figge J, Bladon M T, Chen L B, Ellman M, Bikel I, Farrell M, Livingston D M. Role of small t antigen in the acute transforming activity of SV40. Cell. 1982;30:469–480. doi: 10.1016/0092-8674(82)90244-6. [DOI] [PubMed] [Google Scholar]
- 56.Sabbatini P, Lin J, Levine A J, White E. Essential role for p53-mediated transcription in E1A-induced apoptosis. Genes Dev. 1995;9:2184–2192. doi: 10.1101/gad.9.17.2184. [DOI] [PubMed] [Google Scholar]
- 57.Sawai E T, Butel J S. Association of a cellular heat shock protein with simian virus 40 large T antigen in transformed cells. J Virol. 1989;63:3961–3973. doi: 10.1128/jvi.63.9.3961-3973.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G., Jr Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sellers W R, Rodgers J W, Kaelin W G., Jr A potent transrepression domain in the retinoblastoma protein induces a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci USA. 1995;92:11544–11548. doi: 10.1073/pnas.92.25.11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheng Q, Denis D, Ratnofsky M, Roberts T M, DeCaprio J A, Schaffhausen B. The DnaJ domain of polyomavirus large T antigen is required to regulate Rb family tumor suppressor function. J Virol. 1997;71:9410–9416. doi: 10.1128/jvi.71.12.9410-9416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sherr C J. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 62.Slebos R J, Lee M H, Plunkett B S, Kessis T D, Williams B O, Jacks T, Hedrick L, Kastan M B, Cho K R. p53-dependent G1 arrest involves pRB-related proteins and is disrupted by the human papillomavirus 16 E7 oncoprotein. Proc Natl Acad Sci USA. 1994;91:5320–5324. doi: 10.1073/pnas.91.12.5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sleigh M J, Topp W C, Hanich R, Sambrook J F. Mutants of SV40 with an altered small t protein are reduced in their ability to transform cells. Cell. 1978;14:79–88. doi: 10.1016/0092-8674(78)90303-3. [DOI] [PubMed] [Google Scholar]
- 64.Somasundaram K, El-Deiry W S. Inhibition of p53-mediated transactivation and cell cycle arrest by E1A through its p300/CBP-interacting region. Oncogene. 1997;14:1047–1057. doi: 10.1038/sj.onc.1201002. [DOI] [PubMed] [Google Scholar]
- 65.Sontag E, Fedorov S, Kamibayashi C, Robbins D, Cobb M, Mumby M. The interaction of SV40 small tumor antigen with protein phosphatase 2A stimulates the MAP kinase pathway and induces cell proliferation. Cell. 1993;75:887–897. doi: 10.1016/0092-8674(93)90533-v. [DOI] [PubMed] [Google Scholar]
- 66.Srinivasan A, McClellan A J, Vartikar J, Marks I, Cantalupo P, Li Y, Whyte P, Rundell K, Brodsky J L, Pipas J M. The amino-terminal transforming region of simian virus 40 large T and small t antigens functions as a J domain. Mol Cell Biol. 1997;17:4761–4773. doi: 10.1128/mcb.17.8.4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stewart N, Hicks G G, Paraskevas F, Mowat M. Evidence for a second cell cycle block at G2/M by p53. Oncogene. 1995;10:109–115. [PubMed] [Google Scholar]
- 68.Stubdal H, Zalvide J, Campbell K S, Schweitzer C, Roberts T M, DeCaprio J A. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol Cell Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stubdal H, Zalvide J, DeCaprio J A. Simian virus 40 large T antigen alters the phosphorylation state of the RB-related proteins p130 and p107. J Virol. 1996;70:2781–2788. doi: 10.1128/jvi.70.5.2781-2788.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sugano S, Yamaguchi N, Shimojo H. Small t protein of simian virus 40 is required for dense focus formation in a rat cell line. J Virol. 1982;41:1073–1075. doi: 10.1128/jvi.41.3.1073-1075.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tevethia M J, Lacko H A, Conn A. Two regions of simian virus 40 large T-antigen independently extend the life span of primary C57BL/6 mouse embryo fibroblasts and cooperate in immortalization. Virology. 1998;243:303–312. doi: 10.1006/viro.1998.9056. [DOI] [PubMed] [Google Scholar]
- 72.Tevethia M J, Lacko H A, Kierstead T D, Thompson D L. Adding an Rb-binding site to an N-terminally truncated simian virus 40 T antigen restores growth to high cell density, and the T common region in trans provides anchorage-independent growth and rapid growth in low serum concentrations. J Virol. 1997;71:1888–1896. doi: 10.1128/jvi.71.3.1888-1896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tevethia M J, Pipas J M, Kierstead T, Cole C. Requirements for immortalization of primary mouse embryo fibroblasts probed with mutants bearing deletions in the 3′ end of SV40 gene A. Virology. 1988;162:76–89. doi: 10.1016/0042-6822(88)90396-0. [DOI] [PubMed] [Google Scholar]
- 74.Vousden K H, Vojtesek B, Fisher C, Lane D. HPV-16 E7 or adenovirus E1A can overcome the growth arrest of cells immortalized with a temperature-sensitive p53. Oncogene. 1993;8:1697–1702. [PubMed] [Google Scholar]
- 75.Walker K K, Levine A J. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci USA. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang W B, Bikel I, Marsilio E, Newsome D, Livingston D M. Transrepression of RNA polymerase II promoters by the simian virus 40 small t antigen. J Virol. 1994;68:6180–6187. doi: 10.1128/jvi.68.10.6180-6187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Watanabe G, Howe A, Lee R J, Albanese C, Shu I W, Karnezis A N, Zon L, Kyriakis J, Rundell K, Pestell R G. Induction of cyclin D1 by simian virus 40 small tumor antigen. Proc Natl Acad Sci USA. 1996;93:12861–12866. doi: 10.1073/pnas.93.23.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weinberg R A. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 79.Weintraub S J, Chow K N, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 80.Weintraub S J, Prater C A, Dean D C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature. 1992;358:259–261. doi: 10.1038/358259a0. [DOI] [PubMed] [Google Scholar]
- 81.Whitaker L L, Su H, Baskaran R, Knudsen E S, Wang J Y. Growth suppression by an E2F-binding-defective retinoblastoma protein (RB): contribution from the RB C pocket. Mol Cell Biol. 1998;18:4032–4042. doi: 10.1128/mcb.18.7.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.White R J. Regulation of RNA polymerases I and III by the retinoblastoma protein: a mechanism for growth control? Trends Biochem Sci. 1997;22:77–80. doi: 10.1016/s0968-0004(96)10067-0. [DOI] [PubMed] [Google Scholar]
- 83.Xiao Z X, Chen J, Levine A J, Modjtahedi N, Xing J, Sellers W R, Livingston D M. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature. 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 84.Yaciuk P, Carter M C, Pipas J M, Moran E. Simian virus 40 large-T antigen expresses a biological activity complementary to the p300-associated transforming function of the adenovirus E1A gene products. Mol Cell Biol. 1991;11:2116–2124. doi: 10.1128/mcb.11.4.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang S I, Lickteig R L, Estes R, Rundell K, Walter G, Mumby M C. Control of protein phosphatase 2A by simian virus 40 small-t antigen. Mol Cell Biol. 1991;11:1988–1995. doi: 10.1128/mcb.11.4.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zalvide J, Stubdal H, DeCaprio J A. The J domain of simian virus 40 large T antigen is required to functionally inactivate RB family proteins. Mol Cell Biol. 1998;18:1408–1415. doi: 10.1128/mcb.18.3.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zerrahn J, Knippschild U, Winkler T, Deppert W. Independent expression of the transforming amino-terminal domain of SV40 large I antigen from an alternatively spliced third SV40 early mRNA. EMBO J. 1993;12:4739–4746. doi: 10.1002/j.1460-2075.1993.tb06162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhu J Y, Abate M, Rice P W, Cole C N. The ability of simian virus 40 large T antigen to immortalize primary mouse embryo fibroblasts cosegregates with its ability to bind to p53. J Virol. 1991;65:6872–6880. doi: 10.1128/jvi.65.12.6872-6880.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]