Abstract
Deletion of the entire leader polypeptide of the GDVII strain of Theiler's murine encephalomyelitis virus (TMEV) results in the production of an attenuated virus that grows in baby hamster kidney (BHK) cells but cannot grow at all in mouse L-929 cells. This study examined the reasons for the failure of dl-L, the GDVII variant that lacks the leader polypeptide, to grow in mouse cells. At low multiplicities of infection, it was difficult to detect any viral proteins in mouse cells. However, levels of positive- and negative-strand RNA molecules were only moderately reduced in these infections. Viral RNA showed no major defect in translatability, as the mutant viral RNA was nearly as efficient as that of the wild-type (WT) virus in directing protein synthesis in vitro in assays using extracts prepared from mouse L cells. Viral protein synthesis was detected in dl-L-infected mouse cells as multiplicities of infection were increased and approached the levels observed in WT infections. Despite this, there was a total lack of virus production in high-multiplicity infections, and this was found to correlate with the failure of viral proteins and early virion precursors to assemble into virions in mouse cells. Thus, the inability of dl-L to grow in mouse cells reflects complex effects on various stages of the virus infection but is primarily a defect in virus assembly.
Translation of picornavirus genomes occurs through utilization of long 5′ noncoding regions that contain internal ribosome entry sequences (10, 11). A primary initiating AUG is used during translation, which results in the production of a single polyprotein translation product that is cleaved by viral proteases as translation proceeds along the full length of the viral RNA. All picornaviruses share a general genetic organization in which the first region translated consists of precursors to virion capsid proteins, while the second and third regions encode proteases, such as 2A and 3C, as well as known enzymes of RNA synthesis, such as 3CD/3Dpol (33).
Some picornavirus subgroups express a leader protein that precedes the capsid coding region. In foot-and-mouth-disease virus (FMDV), two forms of the leader protein exist, and these are proteases which are cleaved autocatalytically from the polyprotein (12, 29). Leader proteins are also expressed by cardioviruses such as encephalomyocarditis virus (EMCV) and Theiler's murine encephalomyelitis virus (TMEV). These leader proteins have no sequence homology to those of FMDV, and their function is largely unknown. The EMCV leader is known to be released rapidly from the viral polyprotein by the principal viral protease, 3C (27, 28), and rapid processing has been observed for TMEV as well (17, 31). Neither the EMCV leader (28) nor the TMEV leader (31) has detectable autocatalytic activity. An unusual feature of cardiovirus leaders is that they are very acidic proteins. For example, the GDVII strain of TMEV has a leader protein that is only 76 amino acids in size but has an overall negative charge of −16 (4). There is also genetic evidence that binding of the leader protein to zinc ions may be important in its function (6).
Various strains of TMEV cause either an acute lethal encephalitis or a more slowly progressing demyelinating disease (16, 36). The demyelinating strains like DA, but not the encephalitis strain GDVII, synthesize an alternate form of leader protein, L*, from an internal out-of-frame AUG within the leader coding sequence (14). This protein enhances growth and demyelination by the DA strain of TMEV and appears to be membrane associated in DA-infected cells (7). The ability to express L* may also correlate with the induction of apoptosis in virus infections (9).
To avoid difficulties in interpretation imposed by the presence of L* in some TMEV strains, we have approached the function of the leader protein by using the virulent GDVII strain, which does not encode the alternate AUG. In a previous study, we precisely deleted the leader sequence of GDVII, positioning the initiating AUG and the authentic 5′ noncoding region immediately upstream of the P1 capsid region sequences in a construct named dl-L. The complete deletion of the GDVII leader only slightly affected the growth of GDVII in baby hamster kidney (BHK) cells but strongly attenuated the neurovirulence of this virus in mice (3). In complete contrast to the growth of dl-L in BHK cells, we were unable to detect any virus replication in intracerebrally inoculated mice. Given this contrast, we were intrigued by the report that mutations in leader sequences of the DA strain of TMEV affected host range and that growth of the mutants in mouse L cells was far more dependent on the leader than was growth in BHK cells (13).
In this report we show that the same host range restriction exists for GDVII dl-L. At low multiplicities of infection (MOIs), the absence of the leader protein correlated with reduced viral protein synthesis. Slight reductions in viral RNA synthesis were also observed. At high MOIs, defects in viral protein synthesis were less pronounced and did not seem sufficiently severe to account for the complete block to virus production found in L cells. Rather, a far greater defect was found in the assembly of higher-order virion structures.
MATERIALS AND METHODS
Cell culture and virus assays.
BHK-21 cells and L-929 cells (murine fibroblasts) were grown in 60-mm-diameter plates in Dulbecco's modified Eagle medium and minimal essential medium, respectively, both supplemented with 2 mM l-glutamine, and 7.5% fetal bovine serum. Plaque assays were performed at 33°C for 4 days as described previously (4, 15).
Viral protein synthesis and analysis.
Intracellular viral protein synthesis was analyzed by incubating infected cells with 50 μCi of [35S]methionine (Tran35S-label; ICN) in methionine-free medium for 30 min. Viral proteins were extracted in buffer containing 20 mM Tris-HCl [pH 7.4], 100 mM sodium chloride, and 0.5% NP-40; then soluble proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a sulfate-borate system (34).
Immunoprecipitation of viral proteins was performed in radioimmunoprecipitation assay buffer (buffer containing 1% NP-40, 1% deoxycholate, and 0.1% SDS), using a monoclonal antibody (MAb) against GDVII capsid protein VP2 (GDVII MAb4, kindly provided by Raymond Roos, University of Chicago) (23). This antibody is a conformationally sensitive antibody that cannot recognize viral proteins following denaturation (23). Immunoprecipitates were collected by using formalin-treated, heat-inactivated Staphylococcus aureus Cowan strain. Proteins were released from bacteria by boiling in buffer containing 2% SDS and then analyzed by SDS-PAGE.
Viral RNA extraction from infected cells.
Whole-cell RNA was harvested by the guanidinium isothiocyanate method (35). Briefly, phosphate-buffered saline (PBS)-washed cells were scraped with a rubber policeman into 3 ml of lysis buffer (4 M guanidium isothiocyanate, 25 mM sodium citrate [pH 7.0], 0.1 M β-mercaptoethanol). The lysate was passed several times through a syringe with an 18-gauge needle to reduce viscosity and then layered on a cushion of 2.2 ml of 5.7 M cesium chloride–25 mM sodium acetate (pH 5.5). RNA was pelleted by centrifugation at 150,000 × g (35,000 rpm in an SW50.1 rotor) for 18 to 22 h at 18°C, resuspended in diethyl pyrocarbonate-treated water to which sodium acetate (pH 5.2) was added to a final concentration of 0.3 M, and ethanol precipitated.
RNase protection assays.
For viral positive-strand RNA detection, a standard RNase protection assay was used (1). Probes were generated from GDVII constructs in plasmid pGEM4 (Promega). Transcription of these constructs with SP6 polymerase in the presence of [α-32P]CTP produced probes with negative polarity. Cytoplasmic RNA was hybridized to the labeled probe (5 × 106 cpm per sample) overnight at 60°C in 30 μl of hybridization buffer {40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); pH 6.4], 400 mM sodium chloride, 1 mM EDTA, 80% formamide}. The unhybridized RNA was removed by RNase digestion at 10°C for 60 min in RNase mixture (300 mM sodium chloride, 10 mM Tris-HCl [pH 7.5], 5 mM EDTA, 350 U of RNase T1 per ml). The samples were then treated with proteinase K, phenol extracted, ethanol precipitated with 10 μg of tRNA as carrier, and then loaded onto 6% polyacrylamide–7 M urea gels.
A two-stage RNase protection assay (25) was used for negative-strand detection. In this method, excess viral positive strands were removed before probing. Briefly, cytoplasmic RNA was first hybridized overnight in the absence of probe so that all negative strands were driven into double-stranded hybrids. Excess unhybridized positive strands were then removed by RNase digestion. Double-stranded RNA was recovered by ethanol precipitation, denatured by heating to 85°C for 10 min, hybridized overnight to a radiolabeled positive-sense probe, and then treated with RNase and analyzed as described above. Radiolabeled probes were generated by T7 transcription of various pGEM4-GDVII plasmids. The samples were treated with proteinase K, phenol extracted, ethanol precipitated with 10 μg of tRNA, and loaded onto 6% polyacrylamide–7 M urea gels.
In both negative- and positive-strand analyses, the signals obtained were proportional to the amount of starting RNA. This was confirmed by analyzing three dilutions of each RNA sample, and signals were further quantified by densitometric analysis.
Virus purification and viral RNA extraction.
For in vitro translation, RNA was extracted from purified virus. Virus was purified by using 20 to 70% continuous sucrose gradients centrifuged at 210,000 × g (35,000 rpm in an SW41 rotor) for 3 h at 25°C. Virus-containing fractions from the sucrose gradient were further purified by layering them over a 2.2-ml cushion of cesium sulfate (1 g/ml) and sedimenting at 180,000 × g (40,000 rpm in an SW50.1 rotor) for 20 h at 4°C. Virus was recovered by dilution and centrifugation at 210,000 × g (35,000 rpm in an SW41 rotor) for 3 h at 4°C and was immediately processed for RNA extraction. After a 45-min incubation with 1% SDS and proteinase K (100 μg/ml), RNA was purified by passage through silica gel membrane columns (RNeasy Total RNA kit; Qiagen, Inc.) as specified by the manufacturer.
Cell-free translation using L-cell extracts.
Extracts of L-929 cells were prepared by an adaptation of the lysolecithin method (2, 21) developed by Caroll and Lucas-Lenard (5). L-929 cells grown to confluence in a 850-cm2 roller bottle were trypsinized and plated in 20 100-mm-diameter polystyrene plates, to which cells do not readily attach. The following day, cells were harvested into 50-ml conical tubes and pelleted by centrifugation at 5,000 × g for 5 min. Cell pellets were washed with 12 ml of ice-cold PBS, collected by centrifugation, and resuspended in 1 ml of PBS. They were then transferred to microcentrifuge tubes, sedimented for 10 s, and then resuspended at a concentration of 8 × 107 cells/ml in buffer (20 mM HEPES-KOH [pH 7.4], 100 mM potassium acetate, 2.2 mM magnesium acetate, 2 mM dithiothreitol [DTT]) containing 0.1 mg of lysolecithin (l-α-lysophosphatidylcholine, palmitoyl; Sigma Chemicals) per ml. The cells were held on ice for 50 s and then centrifuged for 10 s. The pellet was resuspended at a concentration of 8 × 107 cell/ml in a mixture containing 25 mM HEPES-KOH (pH 7.4), 125 mM potassium acetate, 2.8 mM magnesium acetate, 2.5 mM DTT, 1.25 mM ATP, 0.25 mM GTP, 6.25 U of creatine phosphokinase per ml, 37.5 mM creatine phosphate, and 0.125 mM amino acid mixture minus methionine. The cells were lysed by pipetting them 20 to 25 times with a micropipette. The lysed cells were left on ice for 5 min and then centrifuged for 20 s. The postmitochondrial supernatant was collected and then stored in small aliquots at −80°C before use as the translation extract.
For translation in vitro, the L-929 cell extracts were incubated in a mixture adjusted to final concentrations of 20 mM HEPES-KOH (pH 7.4), 100 mM potassium acetate, 2.2 mM magnesium acetate, 2 mM DTT, 1 mM ATP, 0.2 mM GTP, 5 U of creatine phosphokinase per ml, 30 mM creatine phosphate, 0.1 mM amino acids mixture minus methionine, and 0.4 mCi of [35S]methionine (SJ 1515; Amersham) per ml. The concentration of magnesium ions was critical for translation in L-929 cell extracts and was optimized each time the lysate was made. Optimal magnesium concentrations were usually between 2.0 and 2.5 mM. Viral RNA was added in amounts indicated for the individual experiments, and the reaction mixtures (25-μl volumes) were incubated for various times as long as 18 h at 30°C before analysis by SDS-PAGE.
Identification of virions and assembly intermediates.
Virions and assembly intermediates were detected as described previously (30). Briefly, cells were infected for 5.5 h and then labeled for 3 h with 100 μCi of [35S]methionine in methionine-free medium. Lysates were then prepared in radioimmunoassay buffer. For detection of capsid and virion peaks, the cell lysates were layered over 15 to 30% (wt/vol) sucrose gradients and centrifuged at 60,000 × g (18,000 rpm in an SW41 rotor) for 10 h at 15°C. For detection of the virion assembly intermediates, protomers and pentamers, cell lysates were fractionated through 5 to 20% (wt/vol) sucrose gradients centrifuged at 150,000 × g (35,000 rpm in an SW50.1 rotor) for 16 h at 15°C. Fractions (0.5 ml) were collected by pumping from the bottom of the gradient, and aliquots of each fraction were counted in a scintillation counter. For detection of protomer and pentamer peaks, each fraction was immunoprecipitated with 5 μl of polyclonal rabbit anti-BeAn serum, kindly provided by Howard L. Lipton, Northwestern University. Final pellets were resuspended in 50 μl of sample buffer, boiled, and counted.
RESULTS
Growth of dl-L in L-929 mouse cells.
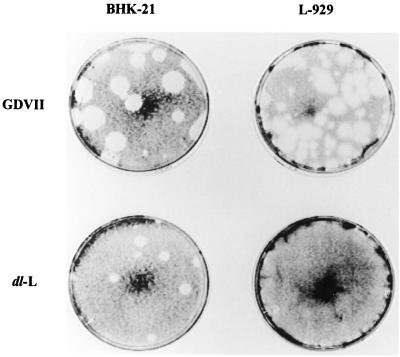
To approach the function of the leader protein of the cardioviruses, we decided to study this protein in the neurovirulent GDVII strain of TMEV because interpretations of the behavior of mutant viruses would not be complicated by the alternate leader protein which is encoded in an alternate reading frame in the more attenuated TMEV strains (14). Accordingly, we deleted the entire leader polypeptide of GDVII to produce a virus, dl-L. Although dl-L virus was readily grown in BHK cells, viral constructs in which the entire leader coding region was deleted failed to replicate in mice and were, consequently, avirulent (3). A subsequent study with the DA strain of TMEV found that deletions of the leader sequences led to a species-specific block in tissue culture growth. Accordingly, we decided to examine the ability of the GDVII-derived mutant dl-L to plaque on L-929 monolayers. As shown in Fig. 1, dl-L does not form plaques on L-929 monolayers although plaques are readily observed when virus is plated on BHK cells.
FIG. 1.
Plaque formation on BHK-21 and L-929 cells. WT GDVII and the dl-L mutant were diluted and used to infect either BHK-21 cells or mouse L-929 cells. Results shown were obtained with a 10−7 dilution of a GDVII lysate and a 10−6 dilution of a dl-L lysate.
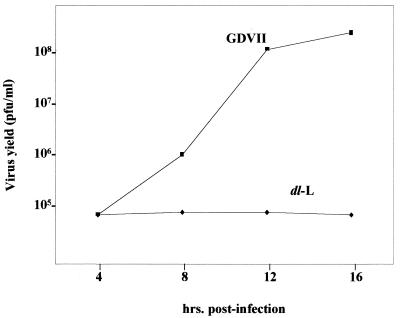
To extend these findings, GDVII and dl-L viruses grown in BHK cells were used to develop growth curves from single-cycle infections. In BHK-21 cells, GDVII virus reaches a final titer of over 109 PFU/ml whereas dl-L grows to around 108 PFU/ml, approximately a 1-log decrease (3). The growth of wild-type (WT) GDVII and dl-L viruses in L-929 cells was studied in the experiment shown in Fig. 2. GDVII grows less well in L-929 cells than was reported originally for BHK-21 cells but reaches a final titer of about 108 PFU/ml. In contract, dl-L shows no growth at all, and the titers of virus recovered were actually lower than the starting amounts of input virus (5 × 106 PFU/ml).
FIG. 2.
Growth of WT GDVII and dl-L mutant in L-929 cells. L-929 cells were infected with virus at an MOI of 5 and incubated at 37°C. At various times p.i., lysates were prepared and virus titers were determined by plaque assays on BHK-21 monolayers.
The defect in dl-L growth in L-929 cells could not be overcome by increasing the MOI. In the experiment shown in Table 1, cells were infected with 5, 20, or 80 PFU/cell; then final virus yields were measured and expressed as virus produced on a per-cell basis. Although there was a slight increase in dl-L yields with increasing MOI, the recovery of dl-L was always less than the amount of the input virus. For example, following infection of cells with 80 PFU/cell, only 31 PFU/cell was found 24 h later. This suggests that essentially no replication of dl-L occurs in L-929 cells. In contrast, cells infected with only 5 PFU of WT GDVII per cell produced over 1,200 PFU/cell in the same 24-h period. Although not shown here, further incubation of infected cells for 24 to 48 h did not result in greater virus production.
TABLE 1.
Effect of increasing MOI on virus yields from L-929 cells
| Variant | MOI | Virus yielda (PFU/cell) |
|---|---|---|
| GDVII | 5 | 1,261 |
| 20 | 1,190 | |
| 80 | 2,459 | |
| dl-L | 5 | 5 |
| 20 | 18 | |
| 80 | 31 |
L-929 cells were infected with TMEV variants at various MOIs and incubated at 37°C. At 24 h p.i., virus yields were determined by plaquing on BHK-21 monolayers.
Viral protein synthesis.
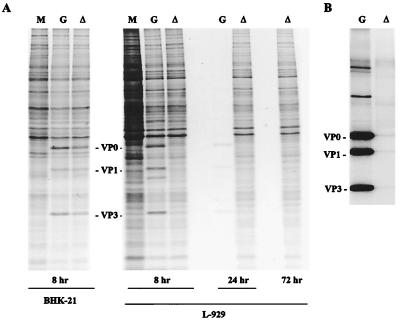
We began studies of the dl-L defect by examining viral protein synthesis in L-929 cells. BHK-21 or L-929 cells were infected with dl-L at an MOI of 5 and labeled with [35S]methionine for 1 h at various times postinfection (p.i.). Viral proteins in cell lysates were then examined by SDS-PAGE. In BHK-21 cells, both GDVII and dl-L proteins were readily detectable at 8 h p.i. (Fig. 3A). In contrast, viral proteins were not apparent in L-929 cells infected with dl-L virus at 8 h p.i. This finding did not simply reflect a slower time course for dl-L viral protein synthesis, because viral proteins were not found at 24 or 72 h p.i. Furthermore, dl-L-infected cells showed no cytopathic effect even by 72 h p.i., while cytopathic effect was clearly present by 8 h p.i. with GDVII (data not shown); this accounts for the extremely poor labeling of GDVII infected cells at 24 h p.i. It is also interesting that despite the absence of viral proteins in dl-L infected cells, the background of host proteins synthesized was lower than in uninfected cells.
FIG. 3.
Viral protein synthesis in BHK-21 and L-929 cells. BHK-21 and L-929 cells were infected with WT GDVII (G) or dl-L (▵) at an MOI of 5 and incubated at 37°C. (A) At 8, 24, or 72 h p.i., the cells were labeled with [35S]methionine for 1 h; then cell lysates were prepared and analyzed by SDS-PAGE. Lane M shows the patterns of proteins synthesized in mock-infected cells. (B) Lysates from L-929 cells radiolabeled 8 h p.i. were used for immunoprecipitation as described in Materials and Methods.
To increase the sensitivity of detecting viral proteins, immunoprecipitations were also used (Fig. 3B). A MAb to VP2 was used in these experiments, and as expected, the immunoprecipitates show the presence of VP0, a precursor that is not cleaved to produce VP2 plus VP4 until late in the virion assembly process (33). Other high-molecular-weight products processed from the polyprotein were also observed. The proteins VP1 and VP3 were present in immunoprecipitates because VP0, VP1, and VP3 remain associated in viral protomers once they are cleaved from the P1 precursor. Thus, precipitation of VP0 with antibody to VP2 results in the coimmunoprecipitation of VP1 and VP3. Immunoprecipitates of extracts of cells infected with WT GDVII show the presence of large amounts of VP0, VP1, and VP3. In contrast, little evidence of viral proteins was found following immunoprecipitation of extracts of L-929 cells infected with dl-L.
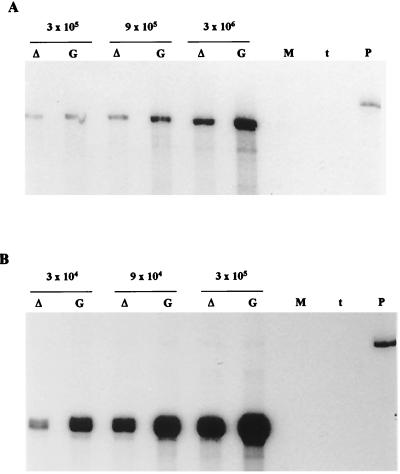
To determine whether a defect in viral RNA synthesis might account for the reduced levels of viral proteins found in dl-L-infected cells, L-929 cells were infected with GDVII or dl-L at an MOI of 5 and incubated at 37°C. At 8 h p.i., cytoplasmic RNA from infected L-929 cells was harvested and subjected to two-cycle RNase protection using a positive-sense probe to detect negative-strand RNA (Fig. 4A). Reduced levels of negative strands were detected in dl-L-infected cells, but these levels were only about threefold lower than WT levels, an estimate confirmed by densitometric analysis. Serial dilutions of the RNA samples confirmed that the RNase protection signal was proportional to the amount of viral RNA in the sample. The synthesis of positive strands of viral RNA was quantitated in parallel, using different aliquots of the same cytoplasmic RNA samples, by standard RNase protection of a negative-sense radiolabeled probe (Fig. 4B). As was the case for negative strands, the levels of positive strands were also around threefold lower in dl-L infections than in WT infections.
FIG. 4.
Viral RNA synthesis in L-929 cells. L-929 cells were infected with WT GDVII (G) or dl-L (▵) at an MOI of 5 and incubated at 37°C. At 8 h p.i., cytoplasmic RNA was extracted and purified as described in Materials and Methods. Amounts of RNA used in each analysis were derived from set numbers of cells, as indicated above the pairs of lanes. RNA from mock-infected cells (lane M) was used as a control. (A) Extracted RNA was subjected to a two-cycle RNase protection assay for the detection of negative strands as described in Materials and Methods. Lane P, starting probe; lane t, tRNA. (B) Lower amounts of the same RNA preparations were used to probe for positive strands of viral RNA by using a negative-strand probe as described in Materials and Methods.
Translation of dl-L viral RNA in L-929 cell extracts.
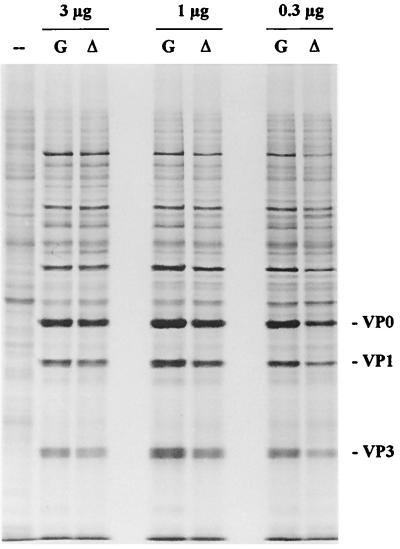
It seemed unlikely that the 3-fold reduction in RNA levels could account for the at least 200-fold reduction in dl-L yields from L-929 cells. A second possibility was that the deletion of leader sequences affected the translatability of viral RNA. To determine whether the primary defect resulted from reduced translatability of dl-L viral RNA, viral RNA was translated in the presence of [35S]methionine in extracts prepared from L-929 cells. Translation mixtures were incubated for 18 h at 30°C to ensure complete processing of the newly synthesized viral polyprotein, and then aliquots of the reactions were analyzed by SDS-PAGE. The pattern of viral proteins synthesized in this in vitro translation system is shown in Fig. 5. To our surprise, there was no significant difference in the levels of viral proteins generated in reactions programmed with either WT GDVII or dl-L RNA over the range of RNA concentrations tested.
FIG. 5.
Translation of viral RNA in L-929 cell extracts. The indicated amounts of GDVII (G) or dl-L (▵) viral RNA prepared from purified virus were translated for 18 h at 30°C, using extracts prepared from L-929 cells as described in Materials and Methods. The pattern in lane –– is the result of translation without exogenous RNA.
Reevaluation of viral protein synthesis in vivo.
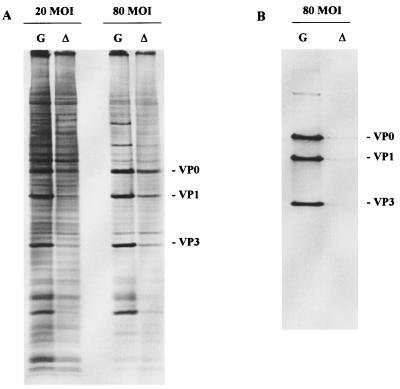
Our initial results had shown that dl-L viral protein synthesis could not be detected in L-929 cells infected at an MOI of 5 (Fig. 3). This observation was difficult to reconcile with the relatively normal levels of viral RNAs and translatability of the RNA in mouse cell extracts and led us to reevaluate the status of viral protein synthesis in infected L-929 cells. To visualize viral proteins more readily, infections were initiated with a much higher MOI. As shown in Fig. 6, protein synthesis directed by dl-L could be detected when cells are infected at a high MOI. Levels of dl-L proteins were lower than those of GDVII at all MOIs, but the degree of difference was less pronounced at MOIs of 20 and 80 than we had estimated in assays using an MOI of 5. Densitometric analysis of the viral protein bands revealed that dl-L protein levels relative to GDVII were reduced three- to fourfold at the higher MOI. As was the case for viral RNA synthesis, it seemed unlikely that these differences could account for the more than 200-fold reduction in viral yields from L-929 cells, a reduction that was found regardless of input multiplicity (Table 1).
FIG. 6.
Viral protein expression at increased MOIs. L-929 cells were infected with GDVII (G) or dl-L (▵) at the indicated MOI and then labeled with [35S]methionine from 8 to 9 h p.i. (A) Cell lysates were prepared and analyzed directly by SDS-PAGE. (B) Extracts of cells in panel A that were infected at an MOI of 80 were used for immunoprecipitation as described in Materials and Methods.
To visualize dl-L proteins more clearly, the infected cell lysates were immunoprecipitated before analysis by SDS-PAGE. Surprisingly, dl-L proteins were barely detectable following immunoprecipitation (Fig. 6B) even when cells were infected at an MOI of 80, a level at which dl-L proteins were readily detectable without immunoprecipitation (Fig. 6A). The antibody used for the immunoprecipitations, MAb4, is a MAb against GDVII capsid protein VP2 which was kindly provided by Raymond P. Roos (University of Chicago). While MAb4 has GDVII-neutralizing activity, it is very sensitive to the conformation of VP2 and its precursor VP0. MAb4 is unable to recognize isolated VP2 or denatured viral proteins on Western blots. These findings suggest that the antibody requires an intact virion conformation for epitope recognition (23). The failure of dl-L proteins to react with this antibody raised the possibility that dl-L proteins fail to assemble into higher-order structures.
Virion assembly.
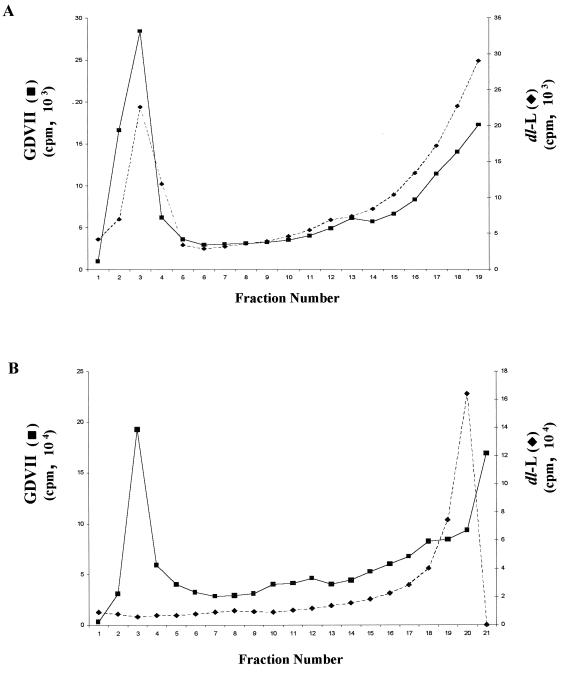
To examine virion assembly, BHK or L-929 cells were infected with GDVII or dl-L at an MOI of 5 or 80, respectively, and viral proteins were labeled with [35S]methionine between 5.5 and 8.5 h p.i. Cytoplasmic extracts were prepared and then fractionated on 15 to 30% sucrose gradients. Virions were readily detected in infected BHK cells (Fig. 7A) or in L-929 cells infected with GDVII (Fig. 7B). In contrast, no dl-L virions or empty capsids were detected in L-929 cells (Fig. 7B). Similar results were obtained with extracts from dl-L infected cells prepared 24 h p.i. (data not shown).
FIG. 7.
Virion assembly in BHK and L-929 cells. (A) BHK cells were infected with GDVII or dl-L virus stocks at an MOI of 5; then proteins were labeled with [35S]methionine from 5.5 to 8.5 h p.i. Lysates of infected cells were then fractionated through separate 15 to 30% sucrose gradients. (B) L-929 cells were infected with GDVII or dl-L virus stocks at an MOI of 80 and labeled from 5.5 to 8.5 h p.i.; then lysates were fractionated on 15 to 30% gradients. Fractions were immunoprecipitated, and then aliquots of each precipitate were counted in a scintillation counter. Intact virions sediment to the bottom of these gradients (fractions 1 to 5), and empty capsids migrate midway through the gradient (fractions 9 to 13).
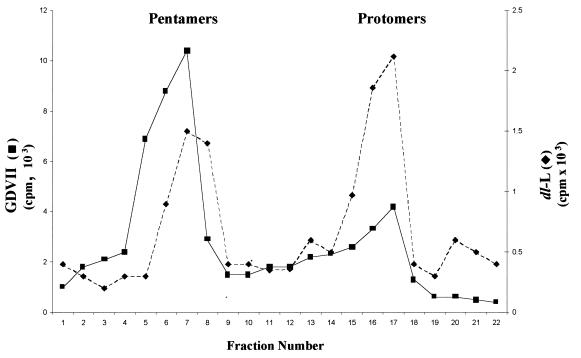
As neither virions nor empty capsids were seen in dl-L infection of L-929 cells, we wished to determine the stage in viral assembly at which the block occurred. As the capsid precursor is translated during polyprotein synthesis, it must fold into a form which allows cleavage to produce VP0, VP1, and VP3. These three products of the P1 precursor remain associated in an assembly intermediate called the protomer. Five protomers then assemble to produce a larger intermediate, termed the pentamer, the precursor of empty capsids (8, 33). To detect protomer and pentamer assembly intermediates, cell lysates were fractionated through 5 to 20% gradients. Both protomer and pentamer peaks were detected in dl-L-infected L-929 cell lysates (Fig. 8). These peaks were two- and fourfold lower than those seen in the GDVII gradient profiles, consistent with the reductions in dl-L protein and RNA synthesis. Thus, dl-L exhibits a severe defect in virus assembly in L-929 cells, a defect that is far more severe than defects in viral RNA and protein synthesis, and this may be the major contributing factor to the failure of the virus to reproduce in mouse cells.
FIG. 8.
Capsid precursors synthesized during GDVII and dl-L infection of L-929 cells. L-929 cells were infected with GDVII or dl-L virus stocks at an MOI of 80 and labeled from 5.5 to 8.5 h p.i.; then lysates were fractionated on 5 to 20% gradients. Fractions were immunoprecipitated with rabbit polyclonal antiviral serum; then aliquots of each precipitate were counted in a scintillation counter. Data are plotted so that the bottom of the gradient is at the left. Note that the y-axis scales are different for GDVII and dl-L gradients.
DISCUSSION
Although previous studies had shown that the GDVII strain of TMEV did not require a leader polypeptide for productive growth in BHK cells (3), effects of the leader on virus growth in mouse L cells were reexamined because deletions in this region of an attenuated strain of TMEV, DA, affected virus growth in mouse cells (13). As was the case for DA virus, the absence of the leader sequences completely blocked the growth of GDVII in mouse L cells. Thus, the growth properties of the leader variant in L cells more closely approximates the behavior of this virus in animals, where it is completely avirulent and where no virus replication in the mouse brain or spinal cord could be detected at all (3).
Studies described here explore the basis for the failure of the dl-L to grow in mouse cells. We conclude from these studies that deletion of the leader sequences reduces viral translation, but that an even more striking defect is at the level of virus assembly. It has not been possible to determine the nature of the defect in viral protein synthesis because we were unable to reproduce this defect in cell extracts prepared from mouse L-929 cells. Rather, the modest two- to threefold decreases in in vitro translatability of dl-L RNA seem unlikely to explain the complete absence of virus production observed in mouse cells.
Similarly, reduced levels of viral protein synthesis did not result in dramatic alterations in viral RNA synthesis. Both negative- and positive-strand RNAs were found in dl-L-infected cells at levels within three- to fourfold those found in GDVII infections. Again, it seemed unlikely that the modest reduction in RNA synthesis would be sufficient to explain the complete block to virus production observed in L cells. In addition, it was possible to greatly increase expression of dl-L viral proteins at increased MOIs. Despite the appearance of viral proteins in such infections, no infectious virus was found and an apparent block to virus assembly became apparent. Indeed, given that the processes of translation of viral proteins, viral RNA replication, and capsid assembly are closely coupled (24, 26, 32), the observed reductions in protein and RNA levels may well be secondary effects of defective viral assembly.
Sucrose gradient fractionation of dl-L-infected L-929 cell lysates revealed the presence of early viral assembly intermediates like protomers and pentamers which were unable to assemble into higher-order structures like empty capsids or mature virions. Thus, in L-929 cells the leader appears to be required for viral assembly at the postpentameric stages. Deletion of the leader could adversely affect viral assembly by perturbing the folding of the nascent P1 capsid region precursor as it emerges from the ribosome.
A modification that might influence polypeptide folding and subsequent assembly is the myristoylation of VP4 which occurs just as the leader is cleaved from the emerging P1 precursor. Members of all four subgroups of picornaviruses have a myristate moiety covalently linked to the N terminus of VP4 and its precursors VP0 and P1 (8). Myristoylation occurs cotranslationally (39) and plays an important role in several stages of viral assembly (22). Poliovirus mutants lacking myristoylation are nonviable and fail to make viral capsids or mature virions (19, 20). However, in cells transfected with such mutant RNAs, protomers and pentamers have been identified. This pattern of assembly defect is similar to our findings in dl-L infection of L-929 cells.
The myristate-modified N termini of VP4 are located along the fivefold axis of each pentamer within the virion (8) and are believed to stabilize the interactions between the five protomers that make up the pentamer. Myristoylation-deficient mutants can still generate pentamers from unmodified protomers (20), but there is a kinetic preference for myristoylated proteins during the formation of pentamers (22). In this context, the 1:1 ratio of pentamers to protomer peaks generated in dl-L infection of L-929 cells in contrast to the 5:1 ratio in GDVII infection may be interesting. The hypothesis, therefore, would be that the N-myristoyltransferase (NMT) of mouse cells is less able to recognize the conformationally altered P1 that lacks the leader sequences. NMT modifies a consensus sequence Gly-X-X-X-Ser/Thr (8). This sequence is found in all picornaviruses, and mutations in this consensus sequence have demonstrated differences in substrate specificity of NMT derived from different species (22, 37, 38). Although this is a intriguing possibility, we were unable to show that dl-L assembly intermediates were devoid of [3H]myristate (data not shown). It remains possible, however, that rates of myristoylation could be reduced sufficiently to prevent virion production even though some residual myristoylation could be detected. This will require a more detailed, quantitative comparison of molar ratios of virion proteins and associated myristate residues.
Alternatively, although the leader is removed fairly rapidly from the nascent P1 capsid precursor (17, 31), the absence of the highly charged leader at the amino terminus of P1 may cause misfolding of partially synthesized polyprotein. The sucrose gradient patterns suggest that protomer structures (VP0-VP1-VP3) can be formed in dl-L-infected mouse cells, and so the absence of the leader does not alter the initial proteolytic cleavages of P1 or the formation of these earliest assembly intermediates. Similarly, pentamers were observed, although the amounts appear to be reduced relative to protomers. The complete failure to detect empty capsid or mature virions suggests that protomers and pentamers that form in dl-L infections may be conformationally altered, blocking their further assembly into higher-order structures. In this case, the basis for the host range difference could be in the cellular chaperonins available to the nascent precursor polypeptide. Along these lines, it is interesting that heat shock protein 70 is found to be associated with the capsid precursor P1 in HeLa cells infected with two other picornaviruses, poliovirus and coxsackievirus B (18). Thus, a detailed comparison of the molar amounts and the nature of the cellular chaperonins found with newly translated P1 in BHK and L-929 cells might be informative.
Although the molecular basis for the failure of dl-L virions to assemble in mouse L-929 cells remains unknown, the effect of the leader sequence in promoting assembly is quite striking, and the use of altered leader sequences may contribute to future studies of structural and conformational aspects of the cardiovirus assembly process.
ACKNOWLEDGMENTS
Early stages of this investigation were supported by National Multiple Sclerosis Society pilot project award PP 3028 to M.A.C. Salary for M.A.C. was provided by NIH grant P01 NS23349.
We thank Howard L. Lipton and his laboratory for helpful discussions throughout the course of this work. We also thank Howard L. Lipton and Raymond Roos for monoclonal and polyclonal antibodies.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K S, editors. Current protocols in molecular biology. 4th ed. Vol. 1. New York, N.Y: John Wiley & Sons; 1990. [Google Scholar]
- 2.Brown G D, Peluso R W, Moyer S A, Moyer R W. A simple method for the preparation of extracts from animal cells which catalyze efficient in vitro protein synthesis. J Biol Chem. 1983;258:14309–14314. [PubMed] [Google Scholar]
- 3.Calenoff M A, Badshah C S, Dal Canto M C, Lipton H L, Rundell M K. The leader polypeptide of Theiler's virus is essential for neurovirulence but not for virus growth in BHK cells. J Virol. 1995;69:5544–5549. doi: 10.1128/jvi.69.9.5544-5549.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Calenoff M A, Faaberg K S, Lipton H L. Genomic regions of neurovirulence and attenuation in Theiler's murine encephalomyelitis virus. Proc Natl Acad Sci USA. 1990;87:978–982. doi: 10.1073/pnas.87.3.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caroll R, Lucas-Lenard J. Preparation of a cell-free translation system with minimal loss of initiation factor eIF-2/eIF-2B activity. Anal Biochem. 1993;212:17–23. doi: 10.1006/abio.1993.1284. [DOI] [PubMed] [Google Scholar]
- 6.Chen H H, Kong W P, Roos R P. The leader peptide of Theiler's murine encephalomyelitis virus is a zinc-binding protein. J Virol. 1995;69:8076–8078. doi: 10.1128/jvi.69.12.8076-8078.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen H H, Kong W P, Zhang L, Ward P L, Roos R P. A picornaviral protein synthesized out of frame with the polyprotein plays a key role in a virus-induced immune-mediated demyelinating disease. Nat Med. 1995;1:927–931. doi: 10.1038/nm0995-927. [DOI] [PubMed] [Google Scholar]
- 8.Chow M, Newman J F E, Filman D, Hogle J M, Rowlands D J, Brown F. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature. 1987;327:482–486. doi: 10.1038/327482a0. [DOI] [PubMed] [Google Scholar]
- 9.Ghadge G D, Ma L, Sato S, Kim J, Roos R P. A protein critical for a Theiler's virus-induced immune system-mediated demyelinating disease has a cell type-specific antiapoptotic effect and a key role in virus persistence. J Virol. 1998;72:8605–8612. doi: 10.1128/jvi.72.11.8605-8612.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang S K, Wimmer E. Cap-independent translation of encephalomyocarditis virus: structural element of the internal ribosome entry site and involvement of a cellular 57-kD RNA binding protein. Genes Dev. 1990;4:1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- 12.Kleina L G, Grubman M J. Antiviral effects of a thiol protease inhibitor on foot-and-mouth disease virus. J Virol. 1992;66:7168–7175. doi: 10.1128/jvi.66.12.7168-7175.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong W P, Ghadge G D, Roos R P. Involvement of cardiovirus leader in host cell-restricted virus expression. Proc Natl Acad Sci USA. 1994;91:1796–1800. doi: 10.1073/pnas.91.5.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong W P, Roos R P. Alternative translation initiation site in the DA strain of Theiler's murine encephalomyelitis virus. J Virol. 1991;65:3395–3399. doi: 10.1128/jvi.65.6.3395-3399.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lipton H L. Persistent Theiler's murine encephalomyelitis virus infection in mice depends on plaque size. J Gen Virol. 1980;46:169–177. doi: 10.1099/0022-1317-46-1-169. [DOI] [PubMed] [Google Scholar]
- 16.Lipton H L. Theiler's virus infection in mice: an unusual biphasic disease process leading to demyelination. Infect Immun. 1975;11:1147–1155. doi: 10.1128/iai.11.5.1147-1155.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipton H L, Rozhon E J, Black D. Theiler's virus-specified polypeptides made in BHK-21 cells. J Gen Virol. 1984;65:1095–1100. doi: 10.1099/0022-1317-65-6-1095. [DOI] [PubMed] [Google Scholar]
- 18.Macejak D, Sarnow P. Association of heat shock protein with enterovirus capsid precursor P1 in infected human cells. J Virol. 1992;66:1520–1527. doi: 10.1128/jvi.66.3.1520-1527.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marc D, Drugeon G, Haenni A L, Girard M, van der Werf S. Role of myristoylation of poliovirus capsid protein VP4 as determined by site-directed mutagenesis of its N-terminal sequence. EMBO J. 1989;8:2661–2668. doi: 10.1002/j.1460-2075.1989.tb08406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marc D, Masson G, Girard M, van der Werf S. Lack of myristoylation of poliovirus capsid polypeptide VP0 prevents the formation of virions or results in the assembly of noninfectious virus particles. J Virol. 1990;64:4099–4107. doi: 10.1128/jvi.64.9.4099-4107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller M R, Castellot J J, Pardee A B. A permeable animal cell preparation for studying macromolecular synthesis, DNA synthesis and the role of deoxyribonucleotides in S phase initiation. Biochem. 1978;17:1073–1080. doi: 10.1021/bi00599a021. [DOI] [PubMed] [Google Scholar]
- 22.Moscufo N, Simons J, Chow M. Myristoylation is important in multiple stages in poliovirus assembly. J Virol. 1991;65:2372–2380. doi: 10.1128/jvi.65.5.2372-2380.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nitayaphan S, Toth M A, Roos R P. Neutralizing monoclonal antibodies to Theiler's murine encephalomyelitis virus. J Virol. 1985;53:651–657. doi: 10.1128/jvi.53.2.651-657.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novak J E, Kirkegaard K. Coupling between genome translation and replication in an RNA virus. Genes Dev. 1994;8:1726–1737. doi: 10.1101/gad.8.14.1726. [DOI] [PubMed] [Google Scholar]
- 25.Novak J E, Kirkegaard K. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J Virol. 1991;65:3384–3387. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nugent C I, Johnson K L, Sarnow P, Kirkegaard K. Functional coupling between replication and packaging of poliovirus replicon RNA. J Virol. 1999;73:427–435. doi: 10.1128/jvi.73.1.427-435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmenberg A. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 28.Parks G D, Duke G M, Palmenberg A C. Encephalomyelitis virus 3C protease: efficient cell-free expression from clones which link 5′ noncoding sequences to the P3 region. J Virol. 1986;60:376–384. doi: 10.1128/jvi.60.2.376-384.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piccone M E, Zellner M, Kumosinski T F, Mason P W, Grubman M J. Identification of the active-site residues of the L proteinase of foot-and-mouth disease virus. J Virol. 1995;69:4950–4956. doi: 10.1128/jvi.69.8.4950-4956.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pritchard A E, Jensen K, Lipton H L. Assembly of Theiler's virus recombinants used in mapping determinants of neurovirulence. J Virol. 1993;67:3901–3907. doi: 10.1128/jvi.67.7.3901-3907.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roos R P, Kong W, Semler B L. Polyprotein processing of Theiler's murine encephalomyelitis virus. J Virol. 1989;63:5344–5353. doi: 10.1128/jvi.63.12.5344-5353.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rueckert R R. Picornaviridae and their replication. In: Fields B N, Knipe D M, et al., editors. Fundamental virology. New York, N.Y: Raven Press, Ltd.; 1991. pp. 409–450. [Google Scholar]
- 33.Rueckert R R, Wimmer E. Systematic nomenclature of picornaviral proteins. J Virol. 1984;50:957–959. doi: 10.1128/jvi.50.3.957-959.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rundell K, Collins K, Tegtmeyer P, Ozer H L, Lai C J, Nathans D. Identification of simian virus 40 protein A. J Virol. 1977;21:636–646. doi: 10.1128/jvi.21.2.636-646.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Theiler M. Spontaneous encephalomyelitis of mice, a new virus disease. J Exp Med. 1937;55:705–719. doi: 10.1084/jem.65.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Towler D A, Adams S P, Eubanks S R, Towery D S, Jackson-Machelski E, Glaser L, Gordon J L. Myristoyl CoA:protein N-myristoyltransferase activities from rat liver and yeast possess overlapping yet distinct peptide substrate specificities. J Biol Chem. 1988;263:1784–1790. [PubMed] [Google Scholar]
- 38.Towler D A, Eubanks S R, Towery D S, Adams S P, Glaser L. Amino-terminal processing of proteins by N-myristoylation. Substrate specificity of N-myristoyl transferase. J Biol Chem. 1987;263:1030–1036. [PubMed] [Google Scholar]
- 39.Wilcox C, Hu J S, Olson E N. Acylation of proteins with myristic acid occurs cotranslationally. Science. 1987;238:1275–1278. doi: 10.1126/science.3685978. [DOI] [PubMed] [Google Scholar]