Abstract
Apiospora species are widely distributed fungi with diverse lifestyles, primarily functioning as plant pathogens, as well as exhibiting saprophytic and endophytic behaviors. This study reports the discovery of three new species of Apiospora, namely A.gongcheniae, A.paragongcheniae, and A.neogongcheniae, isolated from healthy Poaceae plants in China. These novel species were identified through a multi-gene phylogenetic analysis. The phylogenetic analysis of the combined ITS, LSU, tef1, and tub2 sequence data revealed that the three new species formed a robustly supported clade with A.garethjonesii, A.neogarethjonesii, A.setostroma, A.subrosea, A.mytilomorpha, and A.neobambusae. Detailed descriptions of the newly discovered species are provided and compared with closely related species to enhance our understanding of the genus Apiospora.
Key words: Apiospora , Ascomycota, endophyte, phylogeny, taxonomy
Introduction
Apiospora is an important genus of fungal Sordariomycetes, that produces a basauxic, arthrinium-like conidiogenesis (Hyde et al. 2020). The family Apiosporaceae was established to accommodate the genus Apiospora with the special conidiogenesis (Hyde et al. 1998). Over time, the membership of Apiosporaceae has undergone several revisions. It presently comprises several genera of fungi with similar morphology, including Apiospora, Arthrinium, Nigrospora, and Neoarthrinium (Wang et al. 2017; Pintos and Alvarado 2021; Jiang et al. 2022).
Within the family Apiosporaceae, Apiospora is closely related to Arthrinium and they were once considered as two life stages of a single taxon (Ellis 1965; Crous and Groenewald 2013; Réblová et al. 2016; Jiang et al. 2019). Morphologically, Apiospora and Arthrinium lack clear diagnostic features, although species of Arthrinium often produce conidia of various shapes (Minter and Cannon 2018; Pintos and Alvarado 2021), while most species of Apiospora have rounded lenticular conidia (Li et al. 2023; Liao et al. 2023). Ecologically, most sequenced collections of Arthrinium were found on Cyperaceae or Juncaceae in temperate, cold, or alpine habitats, while those of Apiospora were mainly collected on Poaceae, as well as various other plant host families, in a wide range of habitats, including tropical and subtropical regions (Dai et al. 2016; Jiang et al. 2018; Wang et al. 2018; Feng et al. 2021; Tian et al. 2021; Kwon et al. 2022; Monkai et al. 2022). With the addition of molecular evidence and the expansion of the sample, the latest phylogenetic analysis suggests that Arthrinium s. str. and Apiospora represent independent lineages within Apiosporaceae (Pintos and Alvarado 2021). Consequently, most species of Arthrinium have been reclassified under Apiospora. Furthermore, Pintos and Alvarado defined the exact identity of Apiosporamontagnei (the type species of Apiospora) and delineated the phylogenetic boundaries of Apiospora (Pintos and Alvarado 2022).
Currently, there are 176 records in Apiospora (Index Fungorum; http://www.indexfungorum.org/; accessed on 8 Mar 2024). These fungi primarily act as plant pathogens, causing diseases in a wide range of host plants. For example, A.arundinis is the causal agent for several important plant diseases, such as kernel blight of barley (Martínez-Cano et al. 1992), brown culm streak of Phyllostachyspraecox (Chen et al. 2014), moldy sugarcane (Liao et al. 2022), and leaf spot on Polygonatumcyrtonema (Gong et al. 2023). A.marii causes dieback of olive trees (Gerin et al. 2020), while A.kogelbergense leads to blight of Bambusaintermedi (Yin et al. 2020). Whereas, many Apiospora species are saprophytes, such as A.acutiapica (Senanayake et al. 2020), A.garethjonesii (Dai et al. 2016), A.magnispora (Zhao et al. 2023), A.sasae (Crous et al. 2021), and A.thailandicum (Dai et al. 2017). In addition, certain Apiospora species are reported as endophytes with wide host range, including bamboo (Wang et al. 2018), Camelliasinensis (Wang et al. 2018), Wurfbainiavillosa (Liao et al. 2023), and even hive-stored pollen (Zhao et al. 2018).
Endophytic fungi exhibit rich diversity and play a significant role in the ecosystem. In a previous study, we collected and isolated endophytic fungi from healthy Poaceae plants in China (Liu et al. 2021). In this study, three new endophytic species of Apiospora were identified and described based on morphological characteristics and a multi-gene phylogenetic analysis, utilizing a dataset comprising the combined nuclear ribosomal DNA internal transcribed spacer (ITS), nuclear ribosomal DNA large subunit (LSU), the translation elongation factor 1-alpha (tef1), and β-tubulin (tub2) sequences.
Materials and methods
Fungal isolation
In the present work, Poaceae plant samples were collected from three locations in China: Xilingol Grassland National Nature Reserve in Inner Mongolia, Xishuangbanna, Naban River Watershed National Nature Reserve in Yunnan province, and Baishanzu National Nature Reserve in Zhejiang province (Liu et al. 2021). To isolate endophytic Apiospora strains, healthy tissues of asymptomatic plants were first disinfected for 3 min in 75% ethanol and 10 min in 1% sodium hypochlorite, followed by three washes in sterile distilled water. The disinfected tissues were excised, and then incubated on malt extract agar (MEA) medium at 25 °C. Subsequently, the growing hyphae were transferred to potato dextrose agar (PDA) medium to obtain pure cultures.
All strains of Apiospora were stored in the Ministry of Agriculture Key Laboratory of Molecular Biology of Crop Pathogens and Insects, Institute of Biotechnology, Zhejiang University, Hangzhou, China. In addition, the holotype and ex-type culture were deposited in the Guangdong Microbial Culture Collection Center (GDMCC). Fungal names were registered in the Fungal Names, one of the recognised repositories of fungal taxonomy (https://nmdc.cn/fungalnames/).
Morphological study
Morphological descriptions were recorded on PDA and MEA. The morphological characteristics of the colonies were captured with a digital camera (Canon EOS700D). The fungal structures were observed and photographed using a stereomicroscope (Leica S9D) and a Leica DM2500 microscope equipped with differential interference contrast (DIC). Measurements of conidiogenous cells and conidia were reported as follows: a-b × c-d (mean, n), where “a” and “c” represent the minimum values, “b” and “d” represent the maximum values, and the mean value and number of measurements (n) are shown in parentheses (Wang et al. 2018).
DNA extraction, PCR amplification and sequencing
Fresh fungal mycelia from pure cultures grown on PDA at 25 °C for 5–7 d were used for DNA extraction. Genomic DNA was extracted following the method as described in Chi et al. (2009).
Polymerase chain reaction (PCR) amplification was applied to amplify four gene fragments, including ITS, LSU, tef1, and tub2. The primer pairs were used: ITS1/ITS4 for ITS (White et al. 1990), LR0R/LR5 for LSU (Rehner and Samuels 1995), EF1-728F/EF2 for tef1 (O’Donnell et al, 1998; Carbone and Kohn 1999), and T1/Bt2b for tub2 (Glass and Donaldson 1995; O’Donnell and Cigelnik 1997). PCR program for ITS amplification was conducted with an initial denaturation at 95 °C for 3 min, followed by 35 cycles of 95 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 7 min. The annealing temperatures were adjusted to 56 °C for LSU, tef1, and tub2.
PCR was performed using a Veriti Thermal Cycler (Waltham, MA, USA). Amplification reactions contained 10 μL of 2× Taq Plus Master Mix II (Vazyme, Nanjing, China), 0.8 μL of each primer (10 μM) (Sunya, Hangzhou, China), 0.8 μL of DNA template, and double-distilled water to reach a total volume of 20 μL. Purification and sequencing of PCR products were performed by Sunya Biotechnology Company (Hangzhou, China). All sequences generated in this study were deposited in GenBank (Table 1).
Table 1.
Species of Apiosporaceae used in the phylogenetic analyses. Notes: Strains in this study are marked in bold. “T” indicates a type culture. NA = not available.
| Species | Strain Numbers | Host and Substrates | Locality | GenBank accession numbers | |||
|---|---|---|---|---|---|---|---|
| ITS | LSU | tef1 | tub2 | ||||
| Apiosporaacutiapica | KUMCC 20-0209 | Bambusabambos | China | MT946342 | MT946338 | MT947359 | MT947365 |
| Apiosporaacutiapica | KUMCC 20-0210 T | Bambusabambos | China | MT946343 | MT946339 | MT947360 | MT947366 |
| Apiosporaadinandrae | SAUCC 1282B-1 T | Diseased leaves of Adinandraglischroloma | China | OR739431 | OR739572 | OR753448 | OR757128 |
| Apiosporaadinandrae | SAUCC 1282B-2 | Diseased leaves of Adinandraglischroloma | China | OR739432 | OR739573 | OR753449 | OR757129 |
| Apiosporaagari | KUC21333, SFC20161014-M18 T | Agarumcribrosum | South Korea | MH498520 | MH498440 | MH544663 | MH498478 |
| Apiosporaaquatic | MFLU 18-1628, S-642 T | Submerged wood | China | MK828608 | MK835806 | NA | NA |
| Apiosporaarctoscopi | KUC21331, SFC20200506-M05 T | Eggs of Arctoscopusjaponicus | South Korea | MH498529 | MH498449 | MN868918 | MH498487 |
| Apiosporaarundinis | CBS 124788 | Living leaves of Fagussylvatica | Switzerland | KF144885 | KF144929 | KF145017 | KF144975 |
| Apiosporaarundinis | LC4951 | Dichotomanthestristaniicarpa | China | KY494698 | KY494774 | KY705097 | KY705168 |
| Apiosporaaseptata | KUNCC 23-14169 T | Living roots of Dicranopterispedata | China | OR590341 | OR590335 | OR634949 | OR634943 |
| Apiosporaaurea | CBS 244.83 T | Air | Spain | AB220251 | KF144935 | KF145023 | KF144981 |
| Apiosporabalearica | CBS 145129, AP24118 T | Poaceae plant | Spain | MK014869 | MK014836 | MK017946 | MK017975 |
| Apiosporabambusicola | MFLUCC 20-0144 T | Schizostachyumbrachycladum | Thailand | MW173030 | MW173087 | MW183262 | NA |
| Apiosporabawanglingensis | SAUCC BW0444 T | Leaves of Indocalamuslongiauritus | China | OR739429 | OR739570 | OR753446 | OR757126 |
| Apiosporabiserialis | CGMCC 3.20135 T | Bamboo | China | MW481708 | MW478885 | MW522938 | MW522955 |
| Apiosporacamelliae-sinensis | CGMCC 3.18333, LC5007 T | Camelliasinensis | China | KY494704 | KY494780 | KY705103 | KY705173 |
| Apiosporacamelliae-sinensis | LC8181 | Brassicarapa | China | KY494761 | KY494837 | KY705157 | KY705229 |
| Apiosporacannae | ZHKUCC 22-0139 | Leaves of Canna sp. | China | OR164902 | OR164949 | OR166286 | OR166322 |
| Apiosporacannae | ZHKUCC 22-0127 T | Leaves of Canna sp. | China | OR164901 | OR164948 | OR166285 | OR166321 |
| Apiosporachiangraiense | MFLUCC 21-0053 T | Dead culms of bamboo | Thailand | MZ542520 | MZ542524 | NA | MZ546409 |
| Apiosporachromolaenae | MFLUCC 17-1505 T | Chromolaenaodorata | Thailand | MT214342 | MT214436 | MT235802 | NA |
| Apiosporacordylinae | GUCC 10026 | Cordylinefruticosa | China | MT040105 | NA | MT040126 | MT040147 |
| Apiosporacordylinae | GUCC 10027 T | Cordylinefruticosa | China | MT040106 | NA | MT040127 | MT040148 |
| Apiosporacoryli | CFCC 58978 T | Dead plant culms of Corylusyunnanensis | China | OR125564 | OR133586 | OR139974 | OR139978 |
| Apiosporacoryli | CFCC 58979 T | Dead plant culms of Corylusyunnanensis | China | OR125565 | OR133587 | OR139975 | OR139979 |
| Apiosporacyclobalanopsidis | CGMCC 3.20136 T | Cyclobalanopsidisglauca | China | MW481713 | MW478892 | MW522945 | MW522962 |
| Apiosporacyclobalanopsidis | GZCC 20-0103 | Cyclobalanopsidisglauca | China | MW481714 | MW478893 | MW522946 | MW522963 |
| Apiosporadendrobii | MFLUCC 14-0152 T | Roots of Dendrobiumharveyanum | Thailand | MZ463151 | MZ463192 | NA | NA |
| Apiosporadematiacea | KUNCC 23-14202 T | Living stems of Dicranopterisampla | China | OR590346 | OR590339 | OR634953 | OR634948 |
| Apiosporadescalsii | CBS 145130 T | Ampelodesmosmauritanicus | Spain | MK014870 | MK014837 | MK017947 | MK017976 |
| Apiosporadichotomanthi | CGMCC 3.18332, LC4950 T | Dichotomanthestristaniicarpa | China | KY494697 | KY494773 | KY705096 | KY705167 |
| Apiosporadichotomanthi | LC8175 | Dichotomanthestristaniicarpa | China | KY494755 | KY494831 | KY705151 | KY705223 |
| Apiosporadicranopteridis | KUNCC23-14171 T | Living stems of Dicranopterispedata | China | OR590342 | OR590336 | OR634950 | OR634944 |
| Apiosporadicranopteridis | KUNCC23-14177 | Roots of Dicranopterispedata | China | OR590343 | OR590337 | OR634951 | OR634945 |
| Apiosporadongyingensis | SAUCC 0302 T | Leaves of bamboo | China | OP563375 | OP572424 | OP573264 | OP573270 |
| Apiosporadongyingensis | SAUCC 0303 | Leaves of bamboo | China | OP563374 | OP572423 | OP573263 | OP573269 |
| Apiosporaelliptica | ZHKUCC 22-0131 T | Dead stems of unknown plant | China | OR164905 | OR164952 | OR166284 | OR166323 |
| Apiosporaelliptica | ZHKUCC 22-0140 | Dead stems of unknown plant | China | OR164906 | OR164953 | NA | OR166324 |
| Apiosporaendophytica | ZHKUCC 23-0006 T | Living leaves of Wurfbainiavillosa | China | OQ587996 | OQ587984 | OQ586062 | OQ586075 |
| Apiosporaendophytica | ZHKUCC 23-0007 | Living leaves of Wurfbainiavillosa | China | OQ587997 | OQ587985 | OQ586063 | OQ586076 |
| Apiosporaesporlensis | CBS 145136 T | Phyllostachysaurea | Spain | MK014878 | MK014845 | MK017954 | MK017983 |
| Apiosporaesporlensis | UNIPAMPA010 | Living leaves of the Antarctic Hairgrass Deschampsiaantarctica | Antarctica | MN947641 | genome | genome | genome |
| Apiosporaeuphorbiae | IMI 285638b | Bambusa sp. | Bangladesh | AB220241 | AB220335 | NA | AB220288 |
| Apiosporafermenti | KUC21288, SFC20140423-M86 | Seaweeds | South Korea | MF615230 | NA | MH544668 | MF615235 |
| Apiosporafermenti | KUC21289 T | Seaweeds | South Korea | MF615226 | MF615213 | MH544667 | MF615231 |
| Apiosporagaoyouensis | CFCC 52301T | Phragmitesaustralis | China | MH197124 | NA | MH236793 | MH236789 |
| Apiosporagaoyouensis | CFCC 52302 | Phragmitesaustralis | China | MH197125 | NA | MH236794 | MH236790 |
| Apiosporagarethjonesii | GZCC 20-0115 | Dead culms of bamboo | China | MW481715 | MW478894 | MW522947 | NA |
| Apiosporagarethjonesii | KUMCC 16-0202, JHB004, HKAS 96289 T | Dead culms of bamboo | China | KY356086 | KY356091 | NA | NA |
| Apiosporagarethjonesii | SICAUCC 22-0027 | Bamboo | China | ON228603 | ON228659 | NA | ON237651 |
| Apiosporagarethjonesii | SICAUCC 22-0028 | Bamboo | China | ON228606 | ON228662 | NA | ON237654 |
| Apiosporagelatinosa | GZAAS 20-0107 | Bamboo | China | MW481707 | MW478889 | MW522942 | MW522959 |
| Apiosporagelatinosa | HKAS 11962 T | Bamboo | China | MW481706 | MW478888 | MW522941 | MW522958 |
| Apiosporaglobosa | KUNCC 23-14210 T | Living stems of Dicranopterislinearis | China | OR590347 | OR590340 | OR634954 | NA |
| Apiosporagongcheniae | GDMCC 3.1045, YNE00465 T | Living stems of Oryzameyerianasubsp.granulata | China | PP033259 | PP033102 | PP034683 | PP034691 |
| Apiosporagongcheniae | YNE00565 | Living stems of Oryzameyerianasubsp.granulata | China | PP033260 | PP033103 | PP034684 | PP034692 |
| Apiosporaguangdongensis | ZHKUCC 23-0004 T | Living leaves of Wurfbainiavillosa | China | OQ587994 | OQ587982 | OQ586060 | OQ586073 |
| Apiosporaguangdongensis | ZHKUCC 23-0005 | Living leaves of Wurfbainiavillosa | China | OQ587995 | OQ587983 | OQ586061 | OQ586074 |
| Apiosporaguiyangensis | HKAS 102403 T | Dead culms of Poaceae | China | MW240647 | MW240577 | MW759535 | MW775604 |
| Apiosporaguiyangensis | KUNCC 22-12539 | Poaceae plant | China | OQ029540 | OQ029613 | OQ186444 | OQ186446 |
| Apiosporaguizhouensis | CGMCC 3.18334, LC5322 T | Air in karst cave | China | KY494709 | KY494785 | KY705108 | KY705178 |
| Apiosporaguizhouensis | LC5318 | Air in karst cave | China | KY494708 | KY494784 | KY705107 | KY705177 |
| Apiosporahainanensis | SAUCC 1681 T | Leaves of bamboo | China | OP563373 | OP572422 | OP573262 | OP573268 |
| Apiosporahainanensis | SAUCC 1682 | Leaves of bamboo | China | OP563372 | OP572421 | OP573261 | OP573267 |
| Apiosporahispanica | IMI 326877 T | Beach sands | Spain | AB220242 | AB220336 | NA | AB220289 |
| Apiosporahydei | CBS 114990 T | Culms of Bambusatuldoides | China | KF144890 | KF144936 | KF145024 | KF144982 |
| Apiosporahydei | LC7103 | Leaves of bamboo | China | KY494715 | KY494791 | KY705114 | KY705183 |
| Apiosporahyphopodii | JHB003, HKAS 96288 | Bamboo | China | KY356088 | KY356093 | NA | NA |
| Apiosporahyphopodii | MFLUCC 15-003 T | Bambusatuldoides | Thailand | KR069110 | NA | NA | NA |
| Apiosporahyphopodii | SICAUCC 22-0034 | Bamboo | China | ON228605 | ON228661 | NA | ON237653 |
| Apiosporahysterina | AP12118 | Phyllostachysaurea | Spain | MK014877 | KM014844 | MK017953 | MK017982 |
| Apiosporahysterina | AP29717 | Phyllostachysaurea | Spain | MK014875 | MK014842 | MK017952 | MK017981 |
| Apiosporahysterina | ICPM 6889 T | Bamboo | New Zealand | MK014874 | MK014841 | MK017951 | MK017980 |
| Apiosporaiberica | CBS 145137, AP10118 T | Arundodonax | Portugal | MK014879 | MK014846 | MK017955 | MK017984 |
| Apiosporaintestine | CBS 135835 | Gut of grasshopper | India | KR011352 | MH877577 | KR011351 | KR011350 |
| Apiosporaintestine | MFLUCC 21-0052 T | Dead culms of bamboo | Thailand | MZ542521 | MZ542525 | MZ546406 | MZ546410 |
| Apiosporaitalic | CBS 145138, AP221017 T | Arundodonax | Italy | MK014880 | MK014847 | MK017956 | MK017985 |
| Apiosporaitalic | CBS 145139 | Phragmitesaustralis | Spain | MK014881 | MK014848 | NA | MK017986 |
| Apiosporajatrophae | CBS 134262, MMI00052 T | Living Jatrophapodagrica | India | JQ246355 | NA | NA | NA |
| Apiosporajiangxiensis | CGMCC 3.18381, LC4577 T | Maesa sp. | China | KY494693 | KY494769 | KY705092 | KY705163 |
| Apiosporajiangxiensis | LC4578 | Camelliasinensis | China | KY494694 | KY494770 | KY705093 | KY705164 |
| Apiosporakogelbergensis | CBS 113332 | Cannomoisvirgata | South Africa | KF144891 | KF144937 | KF145025 | KF144983 |
| Apiosporakogelbergensis | CBS 113333 T | Dead culms of Restionaceae | South Africa | KF144892 | KF144938 | KF145026 | KF144984 |
| Apiosporakoreanum | KUC21332, SFC20200506-M06 T | Eggs of Arctoscopusjaponicus | South Korea | MH498524 | MH498444 | MH544664 | MH498482 |
| Apiosporakoreanum | KUC21348 | Eggs of Arctoscopusjaponicus | South Korea | MH498523 | NA | MN868927 | MH498481 |
| Apiosporalageniformis | KUC21686 T | Culms of Phyllostachysnigra | Korea | ON764022 | ON787761 | ON806626 | ON806636 |
| Apiosporalageniformis | KUC21687 | Culms of Phyllostachysnigra | Korea | ON764023 | ON787764 | ON806627 | ON806637 |
| Apiosporalocuta-pollinis | LC11683 T | Brassicacampestris | China | MF939595 | NA | MF939616 | MF939622 |
| Apiosporalongistroma | MFLUCC 11-0479 | Dead culms of bamboo | Thailand | KU940142 | KU863130 | NA | NA |
| Apiosporalongistroma | MFLUCC11-0481 T | Dead culms of bamboo | Thailand | KU940141 | KU863129 | NA | NA |
| Apiosporalophatheri | CFCC 58975 T | Diseased leaves of Lophatherumgracile | China | OR125566 | OR133588 | OR139970 | OR139980 |
| Apiosporalophatheri | CFCC 58976 T | Diseased leaves of Lophatherumgracile | China | OR125567 | OR133589 | OR139971 | OR139981 |
| Apiosporamachili | SAUCC 1175A-4 T | Diseased leaves of Machilusnanmu of Machilusnanmu |
China | OR739433 | OR739574 | OR753450 | OR757130 |
| Apiosporamachili | SAUCC 1175 | Diseased leaves of Machilusnanmu of Machilusnanmu |
China | OQ592560 | OQ615289 | OQ613333 | OQ613307 |
| Apiosporamagnispora | ZHKUCC 22-0001 T | Dead stems of Bambusatextilis | China | OM728647 | OM486971 | OM543543 | OM543544 |
| Apiosporamalaysiana | CBS 102053 T | Macarangahullettii | Malaysia | KF144896 | KF144942 | KF145030 | KF144988 |
| Apiosporamarianiae | AP18219 T | Dead stems of Phleumpratense | Spain | ON692406 | ON692422 | ON677180 | ON677186 |
| Apiosporamarii | CBS 497.90 T | Beach sands | Spain | AB220252 | KF144947 | KF145035 | KF144993 |
| Apiosporamarinum | KUC21328, SFC20140423-M02 T | Seaweeds | South Korea | MH498538 | MH498458 | MH544669 | MH498496 |
| Apiosporamediterranea | IMI 326875 T | Air | Spain | AB220243 | AB220337 | NA | AB220290 |
| Apiosporaminutispora | 1.70E-042 T | Mountain soils | South Korea | LC517882 | NA | LC518889 | LC518888 |
| Apiosporamontagnei | AP19421 | Arundomicrantha | Spain | ON692418 | ON692425 | ON677183 | ON677189 |
| Apiosporamontagnei | AP301120, CBS 148707, PC:0125164 T | Arundomicrantha | Spain | ON692408 | ON692424 | ON677182 | ON677188 |
| Apiosporamori | MFLUCC 20-0181 T | Dead leaves of Morusaustralis | China | MW114313 | MW114393 | NA | NA |
| Apiosporamori | NCYUCC 19-0340 | Dead leaves of Morusaustralis | China | MW114314 | MW114394 | NA | NA |
| Apiosporamukdahanensis | MFLUCC 22-0056 T | Dead leaves of bamboo | Thailand | OP377735 | OP377742 | NA | NA |
| Apiosporamultiloculata | MFLUCC 21-0023 T | Dead culms of Bambusae | Thailand | OL873137 | OL873138 | NA | OL874718 |
| Apiosporamytilomorpha | DAOM 214595 T | Dead blades of Andropogon sp. | India | KY494685 | NA | NA | NA |
| Apiosporaneobambusae | CGMCC 3.18335, LC7106 T | Leaves of bamboo | China | KY494718 | KY494794 | KY806204 | KY705186 |
| Apiosporaneobambusae | LC7107 | Leaves of bamboo | China | KY494719 | KY494795 | KY705117 | KY705187 |
| Apiosporaneobambusae | LC7124 | Leaves of bamboo | China | KY494727 | KY494803 | KY806206 | KY705195 |
| Apiosporaneochinensis | CFCC 53036 T | Fargesiaqinlingensis | China | MK819291 | NA | MK818545 | MK818547 |
| Apiosporaneochinensis | CFCC 53037 | Fargesiaqinlingensis | China | MK819292 | NA | MK818546 | MK818548 |
| Apiosporaneogarethjonesii | KUMCC 18-0192, HKAS 102408 T | Dead culms of Bambusae | China | MK070897 | MK070898 | NA | NA |
| Apiosporaneogongcheniae | GDMCC 3.1047, YNE01248 T | Living stems of Poaceae plant | China | PP033263 | PP033106 | PP034687 | PP034695 |
| Apiosporaneogongcheniae | YNE01260 | Living stems of Poaceae plant | China | PP033264 | PP033107 | PP034688 | PP034696 |
| Apiosporaneosubglobosa | JHB 006 | Bamboo | China | KY356089 | KY356094 | NA | NA |
| Apiosporaneosubglobosa | JHB 007 T | Bamboo | China | KY356090 | KY356095 | NA | NA |
| Apiosporaobovata | CGMCC 3.18331, LC4940 T | Lithocarpus sp. | China | KY494696 | KY494772 | KY705095 | KY705166 |
| Apiosporaobovata | LC8177 | Lithocarpus sp. | China | KY494757 | KY494833 | KY705153 | KY705225 |
| Apiosporaoenotherae | CFCC 58972 | Diseased leaves of Oenotherabiennis | China | OR125568 | OR133590 | OR139972 | OR139982 |
| Apiosporaoenotherae | LS 395 | Diseased leaves of Oenotherabiennis | China | OR125569 | OR133591 | OR139973 | OR139983 |
| Apiosporaovate | CBS 115042 T | Arundinariahindsii | China | KF144903 | KF144950 | KF145037 | KF144995 |
| Apiosporapallidesporae | ZHKUCC 22-0129 T | Dead wood of unknown host | China | OR164903 | OR164950 | NA | NA |
| Apiosporapallidesporae | ZHKUCC 22-0142 | Dead wood of unknown host | China | OR164904 | OR164951 | NA | NA |
| Apiosporaparagongcheniae | GDMCC 3.1046, YNE00992 T | Living stems of Poaceae plant | China | PP033261 | PP033104 | PP034685 | PP034693 |
| Apiosporaparagongcheniae | YNE01259 | Living stems of Poaceae plant | China | PP033262 | PP033105 | PP034686 | PP034694 |
| Apiosporaparaphaeosperma | MFLUCC 13-0644 T | Dead culms of bamboo | Thailand | KX822128 | KX822124 | NA | NA |
| Apiosporaparaphaeosperma | KUC21488 | Culms of bamboo | Korea | ON764024 | ON787763 | ON806628 | ON806638 |
| Apiosporaphragmitis | CPC 18900 T | Phragmitesaustralis | Italy | KF144909 | KF144956 | KF145043 | KF145001 |
| Apiosporaphyllostachydis | MFLUCC 18-1101 T | Phyllostachysheteroclada | China | MK351842 | MH368077 | MK340918 | MK291949 |
| Apiosporapiptatheri | CBS 145149, AP4817A T | Piptatherummiliaceum | Spain | MK014893 | MK014860 | MK017969 | NA |
| Apiosporapiptatheri | SAUCC BW0455 | Diseased leaves of Indocalamuslongiauritus | China | OR739430 | OR739571 | OR753447 | OR757127 |
| Apiosporapseudomarii | GUCC 10228 T | Leaves of Aristolochiadebilis | China | MT040124 | NA | MT040145 | MT040166 |
| Apiosporapseudohyphopodii | KUC21680 T | Culms of Phyllostachyspubescens | Korea | ON764026 | ON787765 | ON806630 | ON806640 |
| Apiosporapseudohyphopodii | KUC21684 | Culms of Phyllostachyspubescens | Korea | ON764027 | ON787766 | ON806631 | ON806641 |
| Apiosporapseudoparenchymatica | CGMCC 3.18336, LC7234 T | Leaves of bamboo | China | KY494743 | KY494819 | KY705139 | KY705211 |
| Apiosporapseudoparenchymatica | LC8173 | Leaves of bamboo | China | KY494753 | KY494829 | KY705149 | KY705221 |
| Apiosporapseudorasikravindrae | KUMCC 20-0208 T | Bambusadolichoclada | China | MT946344 | NA | MT947361 | MT947367 |
| Apiosporapseudosinensis | CPC 21546 T | Leaves of bamboo | Netherlands | KF144910 | KF144957 | KF145044 | MN868936 |
| Apiosporapseudosinensis | SAUCC 0221 | Leaves of bamboo | China | OP563377 | OP572426 | OP573266 | OP573272 |
| Apiosporapseudospegazzinii | CBS 102052 T | Macarangahullettii | Malaysia | KF144911 | KF144958 | KF145045 | KF145002 |
| Apiosporapterosperma | CBS 123185 | Machaerinasinclairii | New Zealand | KF144912 | KF144959 | NA | KF145003 |
| Apiosporapterosperma | CPC 20193, CBS 134000 T | Lepidospermagladiatum | Australia | KF144913 | KF144960 | KF145046 | KF145004 |
| Apiosporapusillispermum | KUC21321 T | Seaweeds | South Korea | MH498533 | MH498453 | MN868930 | MH498491 |
| Apiosporapusillispermum | KUC21357 | Seaweeds | South Korea | MH498532 | NA | MN868931 | MH498490 |
| Apiosporaqinlingensis | CFCC 52303 T | Fargesiaqinlingensis | China | MH197120 | NA | MH236795 | MH236791 |
| Apiosporaqinlingensis | CFCC 52304 | Fargesiaqinlingensis | China | MH197121 | NA | MH236796 | MH236792 |
| Apiosporarasikravindrae | LC8179 | Brassicarapa | China | KY494759 | KY494835 | KY705155 | KY705227 |
| Apiosporarasikravindrae | MFLUCC 21-0051 | Dead culms of bamboo | Thailand | MZ542523 | MZ542527 | MZ546408 | MZ546412 |
| Apiosporasacchari | CBS 372.67 | Air | Not mentioned | KF144918 | KF144964 | KF145049 | KF145007 |
| Apiosporasacchari | CBS 664.74 | Soils under Callunavulgaris | Netherlands | KF144919 | KF144965 | KF145050 | KF145008 |
| Apiosporasaccharicola | CBS 191.73 | Air | Netherlands | KF144920 | KF144966 | KF145051 | KF145009 |
| Apiosporasaccharicola | CBS 831.71 | Not mentioned | Netherlands | KF144922 | KF144969 | KF145054 | KF145012 |
| Apiosporasargassi | KUC21228 T | Sargassumfulvellum | South Korea | KT207746 | KT207696 | MH544677 | KT207644 |
| Apiosporasargassi | KUC21232 | Seaweeds | South Korea | KT207750 | NA | MH544676 | KT207648 |
| Apiosporasasae | CPC 38165, CBS 146808 T | Dead culms of Sasaveitchii | Netherlands | MW883402 | MW883797 | MW890104 | MW890120 |
| Apiosporaseptata | CGMCC 3.20134, CS19-8 T | Bamboo | China | MW481711 | MW478890 | MW522943 | MW522960 |
| Apiosporaseptata | GZCC 20-0109 | Bamboo Food | China | MW481712 | MW478891 | MW522944 | MW522961 |
| Apiosporaserenensis | IMI 326869 T | Excipients, atmosphere and home dust | Spain | AB220250 | AB220344 | NA | AB220297 |
| Apiosporasetariae | CFCC 54041 T | Decaying culms of Setariaviridis | China | MT492004 | NA | MW118456 | MT497466 |
| Apiosporasetariae | MT492005 | Setariaviridis | China | MT492005 | NA | MW118457 | MT497467 |
| Apiosporasetostroma | KUMCC 19-0217 | Dead branches of bamboo | China | MN528012 | MN528011 | MN527357 | NA |
| Apiosporasichuanensis | HKAS 107008 T | Dead culms of Poaceae | China | MW240648 | MW240578 | MW759536 | MW775605 |
| Apiosporasorghi | URM 93000, URM 7417 T | Sorghumbicolor | Brazil | MK371706 | NA | NA | MK348526 |
| Apiosporasphaerosperma | CBS 114314 | Leaves of Hordeumvulgare | Iran | KF144904 | KF144951 | KF145038 | KF144996 |
| Apiosporasphaerosperma | CBS 114315 | Leaves of Hordeumvulgare | Iran | KF144905 | KF144952 | KF145039 | KF144997 |
| Apiosporastipae | CPC 38101, CBS 146804 T | Dead culms of Stipagigantea | Spain | MW883403 | MW883798 | MW890082 | MW890121 |
| Apiosporasubglobosa | MFLUCC 11-0397 T | Dead culms of bamboo | Thailand | KR069112 | KR069113 | NA | NA |
| Apiosporasubrosea | CGMCC 3.18337, LC7292 T | Leaves of bamboo | China | KY494752 | KY494828 | KY705148 | KY705220 |
| Apiosporasubrosea | LC7291 | Leaves of bamboo | China | KY494751 | KY494827 | KY705147 | KY705219 |
| Apiosporataeanense | KUC21322T | Seaweeds | South Korea | MH498515 | NA | MH544662 | MH498473 |
| Apiosporataeanense | KUC21359 | Seaweeds | South Korea | MH498513 | NA | MN868935 | MH498471 |
| Apiosporathailandica | MFLUCC 15-0199 | Dead culms of bamboo | Thailand | KU940146 | KU863134 | NA | NA |
| Apiosporathailandica | MFLUCC 15-0202 T | Dead culms of bamboo | Thailand | KU940145 | KU863133 | NA | NA |
| Apiosporatropica | MFLUCC 21-0056 | Dead culms of Bambusoideae | Thailand | OK491657 | OK491653 | NA | OK560922 |
| Apiosporawurfbainiae | ZHKUCC 23-0008 T | Wurfbainiavillosa | China | OQ587998 | OQ587986 | OQ586064 | OQ586077 |
| Apiosporawurfbainiae | ZHKUCC 23-0009 | Wurfbainiavillosa | China | OQ587999 | OQ587987 | OQ586065 | OQ586078 |
| Apiosporavietnamensis | IMI 99670 T | Citrussinensis | Vietnam | KX986096 | KX986111 | NA | KY019466 |
| Apiosporaxenocordella | CBS 478.86 T | Soils from roadway | Zimbabwe | KF144925 | KF144970 | KF145055 | KF145013 |
| Apiosporaxenocordella | CBS 595.66 | Soils | Austria | KF144926 | KF144971 | NA | NA |
| Apiosporaxishuangbannaensis | KUMCC 21-0695 T | Rhinolophuspusillus | China | ON426832 | OP363248 | OR025969 | OR025930 |
| Apiosporaxishuangbannaensis | KUMCC 21-0696 | Rhinolophuspusillus | China | ON426833 | OP363249 | OR025970 | OR025931 |
| Apiosporayunnana | DDQ 00281 | Phyllostachysnigra | China | KU940148 | KU863136 | NA | NA |
| Apiosporayunnana | MFLUCC 15-1002 T | Phyllostachysnigra | China | KU940147 | KU863135 | NA | NA |
| Apiosporayunnanensis | ZHKUCC 23-0014 T | Dead stems of grass | China | OQ588004 | OQ587992 | OQ586070 | OQ586083 |
| Apiosporayunnanensis | ZHKUCC 23-0015 | Dead stems of grass | China | OQ588005 | OQ587993 | OQ586071 | OQ586084 |
| Arthriniumaustriacum | GZU 345004 | Carexpendula | Austria | MW208928 | NA | NA | NA |
| Arthriniumaustriacum | GZU 345006 | Carexpendula | Austria | MW208929 | MW208860 | NA | NA |
| Arthriniumcaricicola | CBS 145127, AP23518 | Carexericetorum | China | MK014871 | MK014838 | MK017948 | MK017977 |
| Arthriniumcaricicola | CBS 145903, CPC33297 T | Dead and attached leaves | Germany | MN313782 | MN317266 | NA | MN313861 |
| Arthriniumcrenatum | AG19066, CBS 146353 T | Carex sp. | France | MW208931 | MW208861 | MW221917 | MW221923 |
| Arthriniumcurvatum | AP25418 | Leaves of Carex sp. | China | MK014872 | MK014839 | MK017949 | NA |
| Arthriniumjaponicum | IFO 30500 | Carexdespalata | Japan | AB220262 | AB220356 | NA | AB220309 |
| Arthriniumjaponicum | IFO 31098 | Leaves of Carexdespalata | Japan | AB220264 | AB220358 | NA | AB220311 |
| Arthriniumluzulae | AP7619-3 | Luzulasylvatica | Spain | MW208937 | MW208863 | MW221919 | MW221925 |
| Arthriniummorthieri | GZU 345043 | Cyperaceaecarex | Austria | MW208938 | MW208864 | MW221920 | MW221926 |
| Arthriniumphaeospermum | AP25619, CBS 146355 | Poaceae plant | Norway | MW208943 | MW208865 | NA | NA |
| Arthriniumpuccinioides | CBS 549.86 | Lepidospermagladiatum | Germany | AB220253 | AB220347 | NA | AB220300 |
| Arthriniumsporophleoides | GZU 345102 | Carex firma | Austria | MW208944 | MW208866 | NA | MW221927 |
| Arthriniumsporophleum | AP21118, CBS 145154 | Dead leaves of Juncus sp. | Spain | MK014898 | MK014865 | MK017973 | MK018001 |
| Nigrosporaguilinensis | CGMCC 3.18124, LC 3481 T | Camelliasinensis | China | KX985983 | KX986113 | KY019292 | KY019459 |
| Nigrosporaguilinensis | LC 7301 | Stems of Nelumbo sp. | China | KX986063 | NA | KY019404 | KY019608 |
| Nigrosporahainanensis | CGMCC 3.18129, LC 7030 T | Leaves of Musaparadisiaca | China | KX986091 | KX986112 | KY019415 | KY019464 |
| Nigrosporahainanensis | LC 6979 | Leaves of Musaparadisiaca | China | KX986079 | NA | KY019416 | KY019586 |
| Nigrosporapyriformis | CGMCC 3.18122, LC 2045 T | Citrussinensis | China | KX985940 | KX986100 | KY019290 | KY019457 |
| Nigrosporapyriformis | LC 2688 | Linderaaggregata | China | KX985941 | NA | KY019297 | KY019468 |
| Nigrosporavesicularis | CGMCC 3.18128, LC 7010 T | Leaves of Musaparadisiaca | China | KX986088 | KX986099 | KY019294 | KY019463 |
| Nigrosporavesicularis | LC 0322 | Unknown host plant | Thailand | KX985939 | NA | KY019296 | KY019467 |
| Neoarthriniumlithocarpicola | CFCC 54456 T | Lithocarpusglaber | China | ON427580 | ON427582 | NA | ON456914 |
| Neoarthriniumlithocarpicola | CFCC 55883 | Lithocarpusglaber | China | ON427581 | ON427583 | NA | ON456915 |
| Neoarthriniumtrachycarpi | CFCC 53038 | Trachycarpusfortune | China | MK301098 | NA | MK303396 | MK303394 |
| Neoarthriniumtrachycarpi | CFCC 53039 | Trachycarpusfortune | China | MK301099 | NA | MK303397 | MK303395 |
| Sporocadustrimorphus | CFCC 55171 | Rose | China | OK655798 | OK560389 | OL814555 | OM401677 |
| Sporocadustrimorphus | ROC 113 | Rose | China | OK655799 | OK560390 | OL814556 | OM401678 |
Phylogenetic analyses
The quality of obtained sequences was assessed using Chromas v.2.6.6 and the sequences were assembled using SeqMan v.7.1.0. The reference sequences were retrieved from GenBank. All sequences, including the reference sequences, were aligned in batches with MAFFT (Katoh and Standley 2013), manually correcting the resulting alignment by MEGA v.11.0.13 where necessary. A single alignment was made using ITS, LSU, tef1 region including partial exon 4 and partial exon 5 (the largest exon), tub2 region including exon 2, exon 3, and partial exon 4. Then phylogenetic analyses were conducted using partial sequences of the above four loci. The sequences were trimmed and concatenated, and subsequent phylogenetic analyses were performed in PhyloSuite platform (Zhang et al. 2020). ModelFinder (Kalyaanamoorthy et al. 2017) was used to select the best-fit partition model (Edge-unlinked) using the BIC criterion. Maximum likelihood (ML) phylogenies were inferred using IQ-TREE (Nguyen et al. 2015) under Edge-linked partition model for 5000 ultrafast (Minh et al. 2013) bootstraps. Bayesian Inference (BI) phylogenies were inferred using MrBayes 3.2.6 (Ronquist et al. 2012) under partition model, in which the initial 27% of sampled data were discarded as burn-in. The resulting phylogenetic tree was visualized in FigTree v1.4.3. (http:/tree.bio.ed.ac.uk/software/figtree/) with maximum likelihood bootstrap proportions (MLBP) greater than 70% and Bayesian inference posterior probabilities (BIPP) greater than 0.90, as shown at the nodes. The phylogram was edited in Adobe Illustrator v.27.5 (Adobe Systems Inc., USA). All GenBank accession numbers of sequences used in this study are provided in Table 1.
Results
Phylogeny
The combined ITS, LSU, tef1, and tub2 dataset encompassed 215 strains, including six newly sequenced strains, with Sporocadustrimorphus CFCC 55171 and ROC 113 serving as the outgroup taxa, and representative species of Arthrinium, Nigrospora, and Neoarthrinium as the sister groups. The multi-locus sequence dataset comprised 2,081 characters, including gaps, with the following character ranges: ITS (1-352), LSU (353-1149), tef1 (1150-1775), and tub2 (1776-2081). The topologies of phylogenetic trees generated by ML and BI analyses were congruent, and the BI tree with MLBP and BIPP is presented in Fig. 1.
Figure 1.
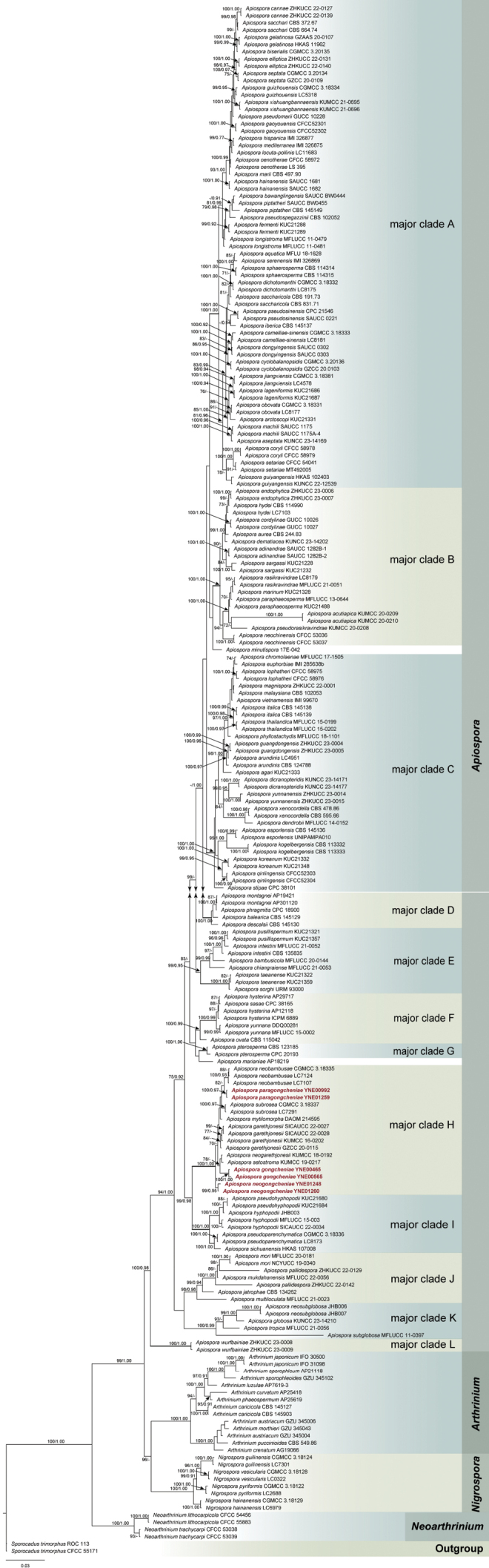
Phylogenetic tree of Apiospora based on the combined ITS, LSU, tef1, and tub2 sequences alignment. Maximum likelihood bootstrap proportions ≥70% (left) and Bayesian inference posterior probability ≥0.90 (right) are indicated at nodes (MLBP/BIPP). Sporocadustrimorphus (CFCC 55171 and ROC 113) are chosen as the outgroup taxa. The novel species from this study are highlighted in red.
The phylogenetic analysis revealed that the species of Apiospora, Arthrinium, Nigrospora, and Neoarthrinium formed four well-supported distinct lineages. Within the genus Apiospora, the 187 strains, encompassing six newly sequenced strains, formed twelve well-supported major clades. The six endophytic strains clustered within one of the major clades H, along with A.garethjonesii, A.neogarethjonesii, A.setostroma, A.subrosea, A.mytilomorpha, and A.neobambusae. Concurrently, the six endophytic strains segregated into three independent clades with robust supported values, indicating the presence of three novel species. These novel taxa are formally described herein and assigned the new names A.gongcheniae, A.paragongcheniae, and A.neogongcheniae.
Taxonomy
. Apiospora gongcheniae
C. L. Zhang sp. nov.
3CB66A0C-DA1A-52F5-B42B-C85EE95BDB90
Fungal Names: FN 571885
Figure 2.
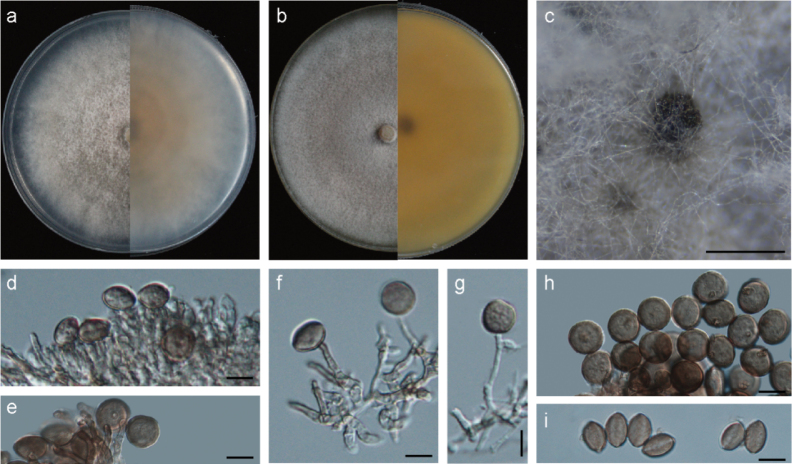
Apiosporagongcheniae (YNE00465, ex-type culture) a colonies after 7 d at 25 °C on PDAb colonies after 7 d at 25 °C on MEAc conidiomata on MEAd-g conidiogenous cells giving rise to conidia h–i conidia with pale germ slit. Scale bars: 500 μm (e); 10 μm (f–k).
Etymology.
Named after Prof. Gongchen Wang in recognition of her significant contribution to the fields of mycology and plant pathology in China.
Type.
China, Yunnan Province: Xishuangbanna, Naban River Watershed National Nature Reserve, 22°04'N, 100°32'E, on the stems of Oryzameyerianasubsp.granulata, Aug 2015, J.J. Chen, YNE00465 (holotype GDMCC 3.1045, stored in a metabolically inactive state); ex-type culture YNE00465.
Description.
Asexual morph: Hyphae hyaline, branched, septate, smooth, 1.1–2.6 μm diameter (mean = 1.7 μm, n = 30). Conidiophores reduced to conidiogenous cells. Conidiogenous cells hyaline to pale brown, erect, verrucose, cylindrical with tiny denticles, clustered in groups, sometimes aggregated in clusters on hyphae or sporodochia, 3.5–9.4 × 1.9–5.2 μm (mean = 5.6 × 3.1 μm, n = 30). Conidia pale brown to dark brown, smooth, granular, globose to subglobose in surface view, lenticular to side view with a pale longitudinal germ slit, with obvious central basal scar, 8.0–17.0 × 6.8–16.1 μm (mean = 13.6 × 11.6 μm, n = 50). Sexual morph: Undetermined.
Culture characteristics.
On PDA, colonies flat, cottony, dense, margin circular, greyish, reverse light orange, covering the 90 mm plate after 7 days at 25 °C. On MEA, colonies dusty pink, dense, covering the 90 mm plate after 7 days at 25 °C. Conidiomata black, globose, abundant, attach to surface of substrate, forming on PDA and MEA after 7–10 days.
Additional specimens examined.
China, Yunnan Province: Xishuangbanna, Naban River Watershed National Nature Reserve, 22°04'N, 100°32'E, on the stems of Oryzameyerianasubsp.granulata, Aug 2015, J.J. Chen, YNE00565.
Note.
Phylogenetic analyses confirmed that A.gongcheniae formed an independent clade, exhibiting a close evolutionary relationship with A.garethjonesii, A.neogarethjonesii and A.subrosea. Based on a BLASTN search of the GenBank database, it was found that A.paragongcheniae shares high similarities with the following strains: A.garethjonesii strain HKAS 96289 (93.76% in ITS, 99.81% in LSU), strain GZCC 20-0115 (93.76% in ITS, 99.24% in LSU, 94.06% in tef1), strain SICAUCC 22-0027 (93.76% in ITS, 99.81% in LSU, 94.51% in tub2), strain SICAUCC 22-0028 (93.76% in ITS, 99.81% in LSU, 93.63% in tub2); A.subrosea strain CGMCC 3.18337 (96.94% in ITS, 99.42% in LSU, 93.47% in tef1, 91.87% in tub2), strain LC7291 (90.09% in ITS, 99.41% in LSU, 93.47% in tef1, 91.87% in tub2); and A.neogarethjonesii strain HKAS 102408 (92.86% in ITS, 99.82% in LSU). The tef1 and tub2 sequence data are currently unavailable for A.neogarethjonesii to compare with A.gongcheniae.
As a synopsis of the morphological characteristics presented in Table 2, A.gongcheniae differs from A.garethjonesii and A.neogarethjonesii in having smaller conidia (8.0–17.0 × 6.8–16.1 μm, mean = 13.6 × 11.6 μm) compared to A.garethjonesii (surface view: 16–19 µm diam, side view: 17–22 µm diam) and A.neogarethjonesii (20–35 × 15–30 µm, mean = 28.5 × 25.6 µm). Additionally, A.gongcheniae exhibits shorter conidiogenous cells (3.5–9.4 × 1.9–5.2 μm, mean = 5.6 × 3.1 μm) in contrast to A.garethjonesii (6–19 × 3–5 µm, mean = 11 × 4 µm) and A.neogarethjonesii (10–48 × 4–5.5 µm, mean = 35.4 × 4.3 µm). While A.gongcheniae shares a similar size range for conidia and conidiogenous cells with A.subrosea, it is distinguished by A.gongcheniae having conidia featuring a central basal scar and cylindrical conidiogenous cells with tiny denticles. Based on molecular and morphological evidence, we propose A.gongcheniae as a new species.
Table 2.
Synopsis of morphological characteristics of related Apiospora species. Notes: ND = Not determined.
| Strains | Apiosporagarethjonesii (D.Q. Dai & H.B. Jiang) Pintos & P. Alvarado (2021) | A.neogarethjonesii (D.Q. Dai & K.D. Hyde) Pintos & P. Alvarado (2021) | A.subrosea (M. Wang & L. Cai) Pintos & P. Alvarado (2021) | A.neobambusae Pintos & P. Alvarado (2021) (=Arthriniumbambusae M. Wang & L. Cai (2018)) | A.gongcheniae | A.paragongcheniae | A.neogongcheniae |
|---|---|---|---|---|---|---|---|
| Host / Substrate | Dead culms of bamboo | Dead culms of bamboo | Leaves of bamboo | Leaves of bamboo | Stems of Oryzameyerianasubsp.granulata | Stems of unidentified Poaceae plant | Stems of unidentified Poaceae plant |
| Known lifestyle | Saprobe | Saprobe | Endophyte | Endophyte | Endophyte | Endophyte | Endophyte |
| Asci | 125–154 × 35–42 μm (x– = 139 × 38 μm, n = 20), 8-spored | 95–125 × 20–25 μm (x– = 97.6 × 21.3 μm, n = 20), 8-spored | ND | ND | ND | ND | ND |
| Ascospores | 30–42 × 11–16 μm (x– = 39 × 13 μm, n = 20), 2-seriate, 1-septate, ellipsoidal | 25–30 × 9.5–11 μm (x– = 29.1 × 10.3 μm, n = 20), 2-seriate, overlapping, 1-septate, ellipsoidal, 3–10 µm wide | ND | ND | ND | ND | ND |
| Conidiomata | Black, with hair-like setae | Black, ellipsoid to irregular, coriaceous | Black, irregular | Black, irregular | Black, globose, abundant, attach to the surface of the substrate | Black, globose to irregular shape, sparse, semi-immersed in the substrate | ND |
| Conidiophores | Reduced to conidiogenous cells | 4.5–6 × 3.5–4.5 µm (x– = 5.4 × 4.3 µm, n = 20), cylindrical, aseptate | Hyaline to pale brown, smooth, erect or ascending, simple, flexuous, subcylindrical, clustered in groups, aggregated in brown sporodochia, up to 20 µm long, 2–4.5 µm width | Reduced to conidiogenous cells | Reduced to conidiogenous cells | Hyaline, erect, basauxic, doliiform, subspherical to barrel-shaped, aggregated in clusters on pale brown sporodochia, sometimes reduced to conidiogenous cells, 12.2–35.1 × 2.1–8.8 μm (x– = 24.5 × 4.3 μm, n = 30) | ND |
| Conidiogenous cells | Hyaline to pale brown, smooth, ampulliform, aggregated in black sporodochia, (5−) 6–19 (−20) µm × (2−) 3–5 (−7) µm (x– = 11 µm × 4 µm, n = 20) | Basauxic, cylindrical, discrete, smooth-walled, 10–48 × 4–5.5 µm (x– = 35.4 × 4.3 µm, n = 20) | Pale brown, smooth, doliiform to subcylindrical, 3.0–6.5 × 2.0–5.0 µm (x– = 4.7 ± 1.2 × 3.7 ± 0.9, n = 30) | Hyaline to pale brown, erect, aggregated in clusters on hyphae, smooth, doliiform to ampulliform, or lageni-form, 4.0–12.0 × 3.0–7.0 µm (x– = 6.6 ± 1.8 × 4.8 ± 0.9, n = 30) | Hyaline to pale brown, erect, verrucose, cylindrical with tiny denticles, clustered in groups, sometimes aggregated in clusters on hyphae or sporodochia, 3.5–9.4 × 1.9–5.2 μm (x– = 5.6 × 3.1 μm, n = 30) | Hyaline, ampulliform, doliiform to clavate, verrucose, 5.0–13.1 × 2.1–6.0 μm (x– = 8.2 × 3.9 μm, n = 30) | ND |
| Conidia | (14–)16–19 (–20) µm diam, brown, smooth, granular, globose to subglobose in surface view, and (16−) 17–22 (−23) µm diam, with pale equatorial slit in side view | Dark brown, globose to subglobose, smooth-walled, with a truncate basal scar, 20–35 × 15–30 µm (x– = 28.5 × 25.6 µm, n = 20) | Pale brown to dark brown, smooth, globose to subglobose or ellipsoidal, 12.0–17.5 × 9.0–16.0 µm (x– = 14.9 ± 1.4 × 11.8 ± 1.8, n = 50) | Olivaceous to brown, smooth to finely roughened, subglobose to ellipsoid, 11.5–15.5 × 7.0–14.0 µm (x– = 13.2 ± 0.8 × 11.4 ± 1.2, n = 50) | Pale brown to dark brown, smooth, granular, globose to subglobose in surface view, lenticular to side view with a pale longitudinal germ slit, with obvious central basal scar, 8.0–17.0 × 6.8–16.1 μm (x– = 13.6 × 11.6 μm, n = 50) | Pale brown to dark brown, smooth to granular, subglobose to oval, occasionally swollen into pyriform to reniform, with a pale longitudinal germ slit in side view, 8.2–18.7 × 6.4–13.4 μm (x– = 12.4 × 10.0 μm, n = 50) | ND |
| References | (Dai et al. 2016; Feng et al. 2021) | (Hyde et al. 2020) | (Wang et al. 2018) | (Wang et al. 2018) | This study | This study | This study |
. Apiospora paragongcheniae
C. L. Zhang sp. nov.
4B35590E-B30E-5BC6-8608-CEE8CD1E6423
Fungal Names: FN 571886
Figure 3.
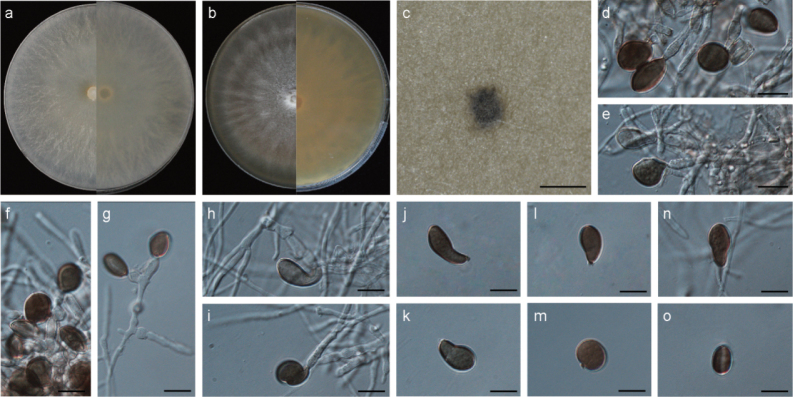
Apiosporaparagongcheniae (YNE00992, ex-type culture) a colonies after 7 d at 25 °C on PDAb colonies after 6 d at 25 °C on MEAc conidioma on MEAd–i conidiogenous cells giving rise to conidia j–o conidia. Scale bars: 500 μm (c); 10 μm (d–o).
Etymology.
Named after its phylogenetic close related to A.gongcheniae.
Type.
China, Yunnan Province: Xishuangbanna, Naban River Watershed National Nature Reserve, 22°04'N, 100°32'E, on the stems of unidentified Poaceae plant, Sep 2016, J.J. Chen, YNE00992 (Holotype GDMCC 3.1046, stored in a metabolically inactive state); ex-type culture YNE00992.
Description.
Asexual morph: Hyphae hyaline, branched, septate, smooth, 1.1–2.2 μm diameter (mean = 1.6 μm, n = 30). Conidiophores hyaline, erect, basauxic, doliiform, subspherical to barrel-shaped, aggregated in clusters on pale brown sporodochia, sometimes reduced to conidiogenous cells, 12.2–35.1 × 2.1–8.8 μm (mean = 24.5 × 4.3 μm, n = 30). Conidiogenous cells hyaline, ampulliform, doliiform to clavate, verrucose, 5.0–13.1 × 2.1–6.0 μm (mean = 8.2 × 3.9 μm, n = 30). Conidia pale brown to dark brown, smooth to granular, subglobose to oval, occasionally swollen into pyriform to reniform, with a pale longitudinal germ slit in side view, 8.2–18.7 × 6.4–13.4 μm (mean = 12.4 × 10.0 μm, n = 50). Sexual morph: Undetermined.
Culture characteristics.
On PDA, colonies flat, rounded, initially white, becoming yellowish-white, with sparse aerial mycelia, mycelium partly immersed in the medium, covering the 90 mm plate after 6 days at 25 °C. On MEA, colonies white, more abundant aerial mycelia, covering the 90 mm plate after 6 days at 25 °C. Conidiomata black, globose to irregular shape, sparse, solitary, semi-immersed in the substrate, observed on MEA after 21–30 days.
Additional specimens examined.
China, Yunnan Province: Xishuangbanna, Naban River Watershed National Nature Reserve, 21°10'N, 99°55'E, on the stems of unidentified Poaceae plant, Oct 2018, X.X. Feng, YNE001259.
Note.
Phylogenetic analyses confirmed that A.paragongcheniae formed an independent clade, exhibiting a close evolutionary relationship with A.subrosea, A.neobambusae and A.neogarethjonesii. Based on a BLASTN search of the GenBank database, it was found that A.paragongcheniae shares high similarities to the following strains: A.subrosea strain CGMCC 3.18337 (98.05% in ITS, 99.23% in LSU, 95.93% in tef1, 93.63% in tub2), strain LC7291 (98.05% in ITS, 99.22% in LSU, 95.93% in tef1, 93.63% in tub2); A.neobambusae strain CGMCC 3.18335 (98.05% in ITS, 100% in LSU, 97.13% in tef1, 93.48% in tub2), strain LC7107 (98.03% in ITS, 100% in LSU, 94.44% in tef1, 93.48% in tub2), strain LC7124 (98.05% in ITS, 100% in LSU, 96.82% in tef1, 93.47% in tub2); and A.neogarethjonesii strain HKAS 102408 (95.43% in ITS, 99.63% in LSU). The tef1 and tub2 sequence data are currently unavailable for A.neogarethjonesii to compare with A.paragongcheniae.
As a synopsis of morphological characteristics presented in Table 2, A.paragongcheniae distinguishes itself from A.neobambusae, A.neogarethjonesii, and A.subrosea in the shapes and sizes of its conidia. The conidia of A.paragongcheniae range from subglobose to oval, occasionally swollen into pyriform to reniform shapes, measuring 8.2–18.7 × 6.4–13.4 μm. This contrasts with A.neobambusae (subglobose to ellipsoid, 11.5–15.5 × 7.0–14.0 µm), A.neogarethjonesii (globose to subglobose, 20–35 × 15–30 µm), and A.subrosea (globose to subglobose or ellipsoidal, 12.0–17.5 × 9.0–16.0 µm). Furthermore, A.paragongcheniae exhibits elongated conidiogenous cells (5.0–13.1 × 2.1–6.0 μm, mean = 8.2 × 3.9 μm) compared to A.neobambusae (4.0–12.0 × 3.0–7.0 µm, mean = 6.6 × 4.8 μm) and A.subrosea (3.0–6.5 × 2.0–5.0 µm, mean = 4.7 × 3.7 μm). Additionally, A.paragongcheniae exhibits shorter conidiogenous cells (5.0–13.1 × 2.1–6.0 μm) compared to A.neogarethjonesii (10–48 × 4–5.5 µm). Moreover, these species differ in the morphology of their conidiophores. A.paragongcheniae displays hyaline, basauxic, doliiform, subspherical to barrel-shaped conidiophores, whereas A.neogarethjonesii has shorter conidiophores, and A.subrosea has hyaline to pale brown, simple, subcylindrical conidiophores. Notably, the conidiophores of A.neobambusae have reduced to conidiogenous cells.
. Apiospora neogongcheniae
C. L. Zhang sp. nov.
658C79C8-D19C-517C-BED6-D851C9B7EDF9
Fungal Names: FN 571887
Figure 4.
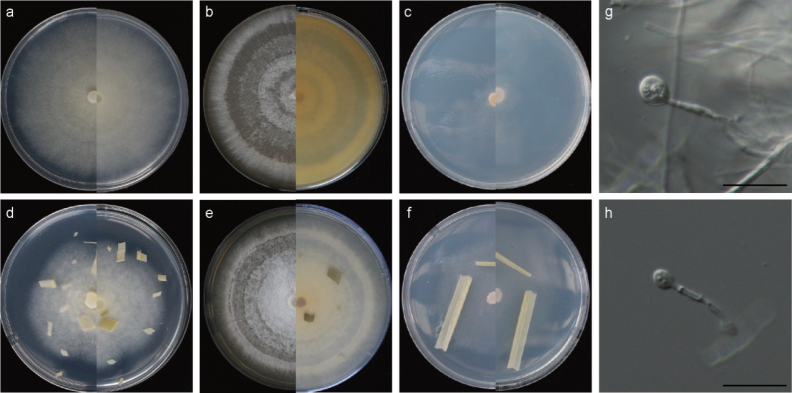
Apiosporaneogongcheniae (YNE01248, ex-type culture) a colonies after 7 d at 25 °C on PDAb colonies after 7 d at 25 °C on MEAc colonies after 7 d at 25 °C on SNA d colonies after 7 d at 25 °C on PDA with rice leaves e colonies after 7 d at 25 °C on MEA with rice leaves f colonies after 7 d at 25 °C on SNA with rice leaves g–h chlamydospores. Scale bars: 20 μm.
Etymology.
Named after its phylogenetic close related to A.gongcheniae.
Type.
China, Yunnan Province: Xishuangbanna, Naban River Watershed National Nature Reserve, 21°10'N, 99°55'E, on the stems of unidentified Poaceae plant, Oct 2018, X.X. Feng, YNE01248 (holotype GDMCC 3.1047, stored in a metabolically inactive state); ex-type culture YNE01248.
Description.
Asexual morph: Hyphae hyaline, branched, septate, smooth, 1.0–2.5 μm diameter (mean = 1.5 μm, n = 30). Conidia not observed. Chlamydospores single, terminal, globose, rare. Sexual morph: Undetermined.
Culture characteristics.
On PDA, colonies flat, rounded, initially white, becoming yellowish-white, cottony, with moderate aerial mycelia, covering the 90 mm plate after 7 days at 25 °C. On MEA, colonies white, dense aerial mycelia, forming multiple circles around the center, covering the 90 mm plate after 7 days at 25 °C. Conidiomata were not observed.
Additional specimens examined.
China, Yunnan Province: Xishuangbanna, Naban River Watershed National Nature Reserve, 21°10'N, 99°55'E, on the stems of unidentified Poaceae plant, Oct 2018, X.X. Feng, YNE001260.
Note.
Phylogenetic analyses confirmed that A.neogongcheniae formed an independent clade, exhibiting a close evolutionary relationship with A.garethjonesii, A.neogarethjonesii and A.subrosea. Based on a BLASTN search of the GenBank database, it was found that A.neogongcheniae shares high similarities with the following strains: A.garethjonesii strain HKAS 96289 (94.88% in ITS, 100% in LSU), strain GZCC 20-0115 (94.88% in ITS, 99.41% in LSU, 96.67% in tef1), strain SICAUCC 22-0027 (94.88% in ITS, 100% in LSU, 96.69% in tub2), strain SICAUCC 22-0028 (94.88% in ITS, 100% in LSU; 96.79% in tub2); A.subrosea strain CGMCC 3.18337 (98.35% in ITS, 99.80% in LSU, 94.61% in tef1, 94.99% in tub2), strain LC7291 (91.41% in ITS, 99.80% in LSU, 94.38% in tef1, 94.99% in tub2); and A.neogarethjonesii strain HKAS 102408 (93.97% in ITS, 100% in LSU). The tef1 and tub2 sequence data are currently unavailable for A.neogarethjonesii to compare with A.neogongcheniae.
Due to the absence of sexual and asexual sporulation characters in A.neogongcheniae, a comparison of its culture characteristics with those of A.garethjonesii, A.neogarethjonesii and A.subrosea was conducted. On PDA, A.neogongcheniae exhibits a yellowish-white surface and reverse color, whereas A.garethjonesii displays a white surface with a reddish reverse, A.neogarethjonesii shows a white to black surface coloration, and A.subrosea presents a light pink surface with a peach-puff reverse. Phylogenetically, A.neogongcheniae strains YNE01248 and YNE01260 form a distinct branch with 99% MLBP and 0.95 BIPP. Therefore, we propose A.neogongcheniae as a novel species.
Discussion
In the present study, three new species of endophytic Apiospora were examined: A.gongcheniae, A.paragongcheniae, and A.neogongcheniae, all of them isolated from the stems of Poaceae plants in Yunnan province of China. According to morphological and molecular identification, the taxonomic position of the three new species was verified.
The generic circumscription of Apiospora was primarily defined through phylogenetic analysis, given the limited morphological characteristics of Apiospora and Arthrinium. The results of a multi-locus phylogenetic analysis in this study, utilizing a combined dataset of ITS, LSU, tef1, and tub2 sequences, supported the previous classification that Apiospora and Arthrinium are distinct lineages rather than synonyms (Pintos and Alvarado 2021). Unlike the six major clades identified in a previous study (Pintos and Alvarado 2022), the current study revealed twelve major clades with robust support through the phylogenetic analysis of 114 Apiospora species, including all known species with available sequences. Apiosporaminutispora (Das et al. 2020) and Apiosporamarianiae AP18219 (Pintos and Alvarado 2022) were not classified within these twelve major clades due to their representation by a single record. The delineation of most Apiospora species into major clades remained consistent across both studies. Notably, A.garethjonesii, A.neogarethjonesii, A.neobambusae, A.mytilomorpha, A.subrosea, and A.setostroma clustered together in a strongly supported major clade H, aligning with findings from previous studies (Crous et al. 2021; Monkai et al. 2022; Pintos and Alvarado 2022; Liao et al. 2023; Liu et al. 2024). Within this major clade, three distinct clades representing three new species were identified (Fig. 1). A.gongcheniae is distinguished from A.garethjonesii by 34/545 nucleotides in the ITS sequences, from A.neogarethjonesii by 39/546, and from A.subrosea by 13/425. A.paragongcheniae is distinguished from A.subrosea by 10/512, from A.neobambusae by 10/512, and from A.neogarethjonesii by 24/525 nucleotides in the ITS sequences. A.neogongcheniae is distinguished from A.garethjonesii by 28/547, from A.neogarethjonesii by 34/547, and from A.subrosea by 7/425 nucleotides in the ITS sequences.
Apiospora exhibits ecological diversity, as evidenced by its wide host ranges. Most reported Apiospora species show a host preference within the Poaceae family, as noted by Monkai et al. (Monkai et al. 2022). Our new species were also found growing on plant hosts of the Poaceae family. Specifically, A.gongcheniae was discovered on the stems of Oryzameyerianasubsp.granulata, a member of the plant family Poaceae. The other two new species, A.paragongcheniae and A.neogongcheniae, were found on the stems of unidentified Poaceae plants. Their close relatives, A.garethjonesii, A.neogarethjonesii, A.neobambusae, and A.subrosea, were found on bamboo plants. Most Apiospora species exhibit saprobic and endophytic lifestyles, which are likely associated with the prevalence of Apiospora (Liao et al. 2023). Our new species occurred as endophytic fungi. Further investigation into endophytic Apiospora species will significantly enhance the diversity within the Apiospora genus.
Morphological characteristics, including asexual and sexual structures, serve as a fundamental basis for fungal systematics and phylogenetic studies, playing a vital role in the comprehensive examination of fungi. However, many endophytes do not form distinct asexual and sexual structures, as observed in A.neogongcheniae in this study, posing challenges in determining their taxonomic status based on morphological features. Recent advances in fungal taxonomy and phylogeny have provided new insights into many species with limited morphological features. Future taxonomic efforts necessitate the integration of morphological traits with molecular evidence to elucidate the natural and stable phylogenetic relationships among Apiospora species and their related Arthrinium species.
Supplementary Material
Citation
Yan X-N, Zhang C-L (2024) Three new endophytic Apiospora species (Apiosporaceae, Amphisphaeriales) from China. MycoKeys 105: 295–316. https://doi.org/10.3897/mycokeys.105.122583
Additional information
Conflict of interest
The authors have declared that no competing interests exist.
Ethical statement
No ethical statement was reported.
Funding
This work was financed by the National Natural Science Foundation of China (Grant No. 31870010).
Author contributions
Xiao-Ni Yan: Investigation, data curation, formal analysis and writing-original draft. Chu-Long Zhang: Conceptualization, methodology, validaiton, formal analysis, supervision, writing-review & editing, funding acquistition.
Author ORCIDs
Xiao-Ni Yan https://orcid.org/0009-0009-9984-3617
Chu-Long Zhang https://orcid.org/0000-0001-5180-0348
Data availability
All of the data that support the findings of this study are available in the main text.
References
- Carbone I, Kohn LM. (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91(3): 553–556. 10.1080/00275514.1999.12061051 [DOI] [Google Scholar]
- Chen K, Wu XQ, Huang MX, Han YY. (2014) First report of brown culm streak of Phyllostachyspraecox caused by Arthriniumarundinis in Nanjing, China. Plant Disease 98(9): 1274. 10.1094/PDIS-02-14-0165-PDN [DOI] [PubMed] [Google Scholar]
- Chi M, Park S, Lee Y. (2009) A quick and safe method for fungal DNA extraction. The Plant Pathology Journal 25(1): 108–111. 10.5423/PPJ.2009.25.1.108 [DOI] [Google Scholar]
- Crous PW, Groenewald JZ. (2013) A phylogenetic re-evaluation of Arthrinium. IMA Fungus 4(1): 133–154. 10.5598/imafungus.2013.04.01.13 [DOI] [PMC free article] [PubMed]
- Crous PW, Hernández-Restrepo M, Schumacher RK, Cowan DA, Maggs-Kölling G, Marais E, Wingfield MJ, Yilmaz N, Adan OCG, Akulov A, Duarte EÁ, Berraf-Tebbal A, Bulgakov TS, Carnegie AJ, de Beer ZW, Decock C, Dijksterhuis J, Duong TA, Eichmeier A, Hien LT, Houbraken JAMP, Khanh TN, Liem NV, Lombard L, Lutzoni FM, Miadlikowska JM, Nel WJ, Pascoe IG, Roets F, Roux J, Samson RA, Shen M, Spetik M, Thangavel R, Thanh HM, Thao LD, van Nieuwenhuijzen EJ, Zhang JQ, Zhang Y, Zhao LL, Groenewald JZ. (2021) New and Interesting Fungi. 4. Fungal Systematics and Evolution 7(1): 255–343. 10.3114/fuse.2021.07.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DQ, Jiang HB, Tang LZ, Bhat DJ. (2016) Two new species of Arthrinium (Apiosporaceae, Xylariales) associated with bamboo from Yunnan, China. Mycosphere : Journal of Fungal Biology 7(9): 1332–1345. 10.5943/mycosphere/7/9/7 [DOI] [Google Scholar]
- Dai DQ, Phookamsak R, Wijayawardene NN, Li WJ, Bhat DJ, Xu JC, Taylor JE, Hyde KD, Chukeatirote E. (2017) Bambusicolous fungi. Fungal Diversity 82(1): 1–105. 10.1007/s13225-016-0367-8 [DOI] [Google Scholar]
- Das K, Lee SY, Choi HW, Eom AH, Cho YJ, Jung HY. (2020) Taxonomy of Arthriniumminutisporum sp. nov., Peziculaneosporulosa, and Acrocalymmapterocarpi: New Records from Soil in Korea. Mycobiology 48(6): 450–463. 10.1080/12298093.2020.1830741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MB. (1965) Dematiaceous Hyphomycetes. VI. Mycological Papers 103(29): 1–46. [Google Scholar]
- Feng Y, Liu JK, Lin CG, Chen YY, Xiang MM, Liu ZY. (2021) Additions to the genus Arthrinium (Apiosporaceae) from bamboos in China. Frontiers in Microbiology 12: 661281. 10.3389/fmicb.2021.661281 [DOI] [PMC free article] [PubMed]
- Gerin D, Nigro F, Faretra F, Pollastro S. (2020) Identification of Arthriniummarii as causal agent of olive tree dieback in Apulia (Southern Italy). Plant Disease 104(3): 694–701. 10.1094/PDIS-03-19-0569-RE [DOI] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61(4): 1323–1330. 10.1128/aem.61.4.1323-1330.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Yang Y, Zhang L, Luo Q, Luo J, Zhang Y. (2023) First report of Apiosporaarundinis causing leaf spot on Polygonatumcyrtonema in China. Plant Disease 108(2): 524. 10.1094/PDIS-08-23-1580-PDN [DOI] [Google Scholar]
- Hyde KD, Fröhlich J, Taylor JE. (1998) Fungi from palms. XXXVI. Reflections on unitunicate ascomycetes with apiospores. Sydowia 50(1): 21–80. [Google Scholar]
- Hyde KD, Norphanphoun C, Maharachchikumbura S, Bhat DJ, Jones E, Bundhun D, Chen YJ, Bao DF, Boonmee S, Calabon MS, Chaiwan N, Chethana K, Dai DQ, Dayarathne MC, Devadatha B, Dissanayake AJ, Dissanayake LS, Doilom M, Dong W, Fan XL, Goonasekara ID, Hongsanan S, Huang SK, Jayawardena RS, Jeewon R, Karunarathna A, Konta S, Kumar, Lin CG, Liu JK, Liu NG, Luangsaard J, Lumyong S, Luo ZL, Marasinghe DS, McKenzie E, Niego A, Niranjan M, Perera RH, Phukhamsakda C, Rathnayaka AR, Samarakoon MC, Samarakoon S, Sarma VV, Senanayake IC, Shang QJ, Stadler M, Tibpromma S, Wanasinghe DN, Wei DP, Wijayawardene NN, Xiao YP, Yang J, Zeng XY, Zhang SN, Xiang MM. (2020) Refined families of Sordariomycetes. Mycosphere 11(1): 305–1059. 10.5943/mycosphere/11/1/7 [DOI] [Google Scholar]
- Jiang N, Li J, Tian CM. (2018) Arthrinium species associated with bamboo and reed plants in China. Fungal Systematics and Evolution 2: 1–9. 10.3114/fuse.2018.02.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HB, Hyde KD, Doilom M, Karunarathna SC, Xu JC, Phookamsak R. (2019) Arthriniumsetostromum (Apiosporaceae, Xylariales), a novel species associated with dead bamboo from Yunnan, China. Asian Journal of Mycology 2(1): 254–268. 10.5943/ajom/2/1/16 [DOI] [Google Scholar]
- Jiang N, Voglmayr H, Ma CY, Xue H, Piao CG, Li Y. (2022) A new Arthrinium-like genus of Amphisphaeriales in China. MycoKeys 92: 27–43. 10.3897/mycokeys.92.86521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong T, von Haeseler A, Jermiin LS. (2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nature Methods 14(6): 587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Molecular Biology and Evolution 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SL, Cho M, Lee YM, Lee H, Kim C, Kim GH, Kim JJ. (2022) Diversity of the bambusicolous fungus Apiospora in Korea: Discovery of new Apiospora species. Mycobiology 50(5): 302–316. 10.1080/12298093.2022.2133808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Peng C, Yuan R, Tian C. (2023) Morphological and phylogenetic analyses reveal three new species of Apiospora in China. MycoKeys 99: 297–317. 10.3897/mycokeys.99.108384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Jiang W, Wu X, He J, Li H, Wang T, Cheng L, Chen W, Mo L. (2022) First report of Apiospora Mold on sugarcane in China caused by Apiosporaarundinis (Arthriniumarundinis). Plant Disease 106(3): 1058. 10.1094/PDIS-02-21-0386-PDN [DOI] [PubMed] [Google Scholar]
- Liao C, Senanayake IC, Dong W, Thilini Chethana KW, Tangtrakulwanich K, Zhang Y, Doilom M. (2023) Taxonomic and phylogenetic updates on Apiospora: Introducing four new species from Wurfbainiavillosa and grasses in China. Journal of Fungi (Basel, Switzerland) 9(11): 1087. 10.3390/jof9111087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WT, Chen JJ, Feng JW, Xia CY, Shao YX, Zhu YX, Liu F, Cai HM, Yang KB, Zhang CL. (2021) Diversity of endophytic fungi associated with plants of Poaceae from Yunnan, Zhejiang and Inner Mongolia. Mycosystema 40(3): 502–513. 10.13346/j.mycosystema.200240 [DOI] [Google Scholar]
- Liu X, Zhang Z, Wang S, Zhang X. (2024) Three new species of Apiospora (Amphisphaeriales, Apiosporaceae) on Indocalamuslongiauritus, Adinandraglischroloma and Machilusnanmu from Hainan and Fujian, China. Journal of Fungi (Basel, Switzerland) 10(1): 74. 10.3390/jof10010074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Cano C, Grey WE, Sands DC. (1992) First report of Arthriniumarundinis causing kernel blight on barley. Plant Disease 76(10): 1077B. 10.1094/PD-76-1077B [DOI]
- Minh BQ, Nguyen MA, von Haeseler A. (2013) Ultrafast approximation for phylogenetic bootstrap. Molecular Biology and Evolution 30(5): 1188–1195. 10.1093/molbev/mst024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minter DW, Cannon PF. (2018) Arthrinium minus. Descriptions of Fungi and Bacteria 2155. 10.1079/DFB/20183347366 [Descriptions of Fungi and Bacteria] [DOI]
- Monkai J, Phookamsak R, Tennakoon DS, Bhat DJ, Xu S, Li Q, Xu J, Mortimer PE, Kumla J, Lumyong S. (2022) Insight into the taxonomic Resolution of Apiospora: Introducing novel species and records from bamboo in China and Thailand. Diversity 14(11): 918. 10.3390/d14110918 [DOI] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Molecular Biology and Evolution 32(1): 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Cigelnik E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7(1): 103–116. 10.1006/mpev.1996.0376 [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC. (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proceedings of the National Academy of Sciences of the United States of America 95(5): 2044–2049. 10.1073/pnas.95.5.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintos Á, Alvarado P. (2021) Phylogenetic delimitation of Apiospora and Arthrinium. Fungal Systematics and Evolution 7(1): 197–221. 10.3114/fuse.2021.07.10 [DOI] [PMC free article] [PubMed]
- Pintos Á, Alvarado P. (2022) New studies on Apiospora (Amphisphaeriales, Apiosporaceae): Epitypification of Sphaeriaapiospora, proposal of Ap.marianiae sp. nov. and description of the asexual morph of Ap.sichuanensis. MycoKeys 92: 63–78. 10.3897/mycokeys.92.87593 [DOI] [PMC free article] [PubMed]
- Réblová M, Miller AN, Rossman AY, Seifert KA, Crous PW, Hawksworth DL, Abdel-Wahab MA, Cannon PF, Daranagama DA, De Beer ZW, Huang S-K, Hyde KD, Jayawardena R, Jaklitsch W, Jones EBG, Ju Y-M, Judith C, Maharachchikumbura SSN, Pang K-L, Petrini LE, Raja HA, Romero AI, Shearer C, Senanayake IC, Voglmayr H, Weir BS, Wijayawarden NN. (2016) Recommendations for competing sexual-asexually typified generic names in Sordariomycetes (except Diaporthales, Hypocreales, and Magnaporthales). IMA Fungus 7(1): 131–153. 10.5598/imafungus.2016.07.01.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehner SA, Samuels GJ. (1995) Molecular systematics of the Hypocreales: A teleomorph gene phylogeny and the status of their anamorphs. Canadian Journal of Botany 73(S1): 816–823. 10.1139/b95-327 [DOI]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61(3): 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senanayake IC, Bhat JD, Cheewangkoon R, Xie N. (2020) Bambusicolous Arthrinium species in Guangdong Province, China. Frontiers in Microbiology 11: 602773. 10.3389/fmicb.2020.602773 [DOI] [PMC free article] [PubMed]
- Tian X, Karunarathna SC, Mapook A, Promputtha I, Xu J, Bao D, Tibpromma S. (2021) One new species and two new host records of Apiospora from bamboo and maize in northern Thailand with thirteen new combinations. Life 11(10): 1071. 10.3390/life11101071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Liu F, Crous PW, Cai L. (2017) Phylogenetic reassessment of Nigrospora: Ubiquitous endophytes, plant and human pathogens. Persoonia 39(1): 118–142. 10.3767/persoonia.2017.39.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tan XM, Liu F, Cai L. (2018) Eight new Arthrinium species from China. MycoKeys 34: 1–24. 10.3897/mycokeys.34.24221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T, Bruns T, Lee S, Taylor J, Innis M, Gelfand D, Sninsky J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A guide to Methods and Applications 18: 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI] [Google Scholar]
- Yin C, Luo F, Zhang H, Fang X, Zhu T, Li S. (2020) First report of Arthriniumkogelbergense causing blight disease of bambusa intermedia in Sichuan province, China. Plant Disease 105(1): 214. 10.1094/PDIS-06-20-1159-PDN [DOI] [PubMed] [Google Scholar]
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. (2020) PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources 20(1): 348–355. 10.1111/1755-0998.13096 [DOI] [PubMed] [Google Scholar]
- Zhao YZ, Zhang ZF, Cai L, Peng WJ, Liu F. (2018) Four new filamentous fungal species from newly-collected and hivestored bee pollen. Mycosphere : Journal of Fungal Biology 9(6): 1089–1116. 10.5943/mycosphere/9/6/3 [DOI] [Google Scholar]
- Zhao HJ, Dong W, Shu YX, Mapook A, Manawasinghe IS, Doilom M, Luo M. (2023) Bambusicolous fungi in Guangdong, China: Establishing Apiosporamagnispora sp. nov. (Apiosporaceae, Amphisphaeriales) based on morphological and molecular evidence. Current Research in Environmental & Applied Mycology 13(1): 1–15. 10.5943/cream/13/1/1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the data that support the findings of this study are available in the main text.