Abstract
Neurotransmitters and neuromodulators have a wide range of key roles throughout the nervous system. However, their dynamics in both health and disease have been challenging to assess, owing to the lack of in vivo tools to track them with high spatiotemporal resolution. Thus, developing a platform that enables minimally invasive, large-scale and long-term monitoring of neurotransmitters and neuromodulators with high sensitivity, high molecular specificity and high spatiotemporal resolution has been essential. Here, we review the methods available for monitoring the dynamics of neurotransmitters and neuromodulators. Following a brief summary of non-genetically encoded methods, we focus on recent developments in genetically encoded fluorescent indicators, highlighting how these novel indicators have facilitated advances in our understanding of the functional roles of neurotransmitters and neuromodulators in the nervous system. These studies present a promising outlook for the future development and use of tools to monitor neurotransmitters and neuromodulators.
The nervous system is composed of various cell types, primarily neurons and glia. Neurons predominantly communicate with each other via specialized structures called synapses, which include electrical synapses and chemical synapses. In the chemical synapse, neurotransmitters (NTs) and/or neuromodulators (NMs) are released into synaptic cleft from one cell, typically from synaptic vesicles enriched in the presynaptic cell, and bind receptors on the targeted postsynaptic cells1,2. Classical NTs, including glutamate, GABA and acetylcholine (ACh), act through ligand-gated ionotropic receptors and G-protein-coupled receptors (GPCRs) to activate or inhibit the postsynaptic cells. NTs that bind with ionotropic receptors usually initiate rapid, spatially confined, point-to-point synaptic transmission, whereas NMs, including monoamines, nucleotides, neurolipids and neuropeptides, function mainly through GPCRs to initiate molecular signalling cascades, in a relatively slow, long-range and diffused way3,4.
Chemical neurotransmission drives a broad range of physiological processes such as arousal, attention, perception and learning, as well as other complex behaviours5,6. Impaired neurotransmission has been implicated in many brain disorders, including Alzheimer disease, Parkinson disease, schizophrenia, depression and drug addiction7–11. Thus, the ability to monitor the concentration and the dynamics of specific NTs and NMs in the extracellular space with high spatiotemporal precision will provide valuable information regarding the complex processes that underlie intercellular communication in both health and disease.
As the Chinese philosopher Confucius said more than 2,000 years ago, “Good tools are prerequisite to the successful execution of a job.” More recently, the South African biologist and Nobel laureate Sydney Brenner said12 that “Progress in science depends on new techniques, new discoveries and new ideas, probably in that order.” Thanks to the tireless efforts of many innovative researchers, we are entering an era in which NTs and NMs can be tracked at unprecedented spatiotemporal resolution.
Here, we review a range of non-genetically encoded methods (including electrophysiological methods, microdialysis and electrochemical methods) and genetically encoded indicators that have been developed for monitoring neurotransmission. These complementary tools have become indispensable for gaining insights into the dynamic regulation and function of various NTs and NMs in the highly complex nervous system. At the same time, discoveries using these tools are paving the way to explore novel strategies for preventing, diagnosing and treating a wide range of diseases and conditions.
Non-genetically encoded methods
Electrophysiological methods
Electrophysiological methods require the use of discrete electrodes (FIG. 1a), and so information is acquired from a single spatial location (mainly from somas) or, if a multiple-electrode system is used, from a finite number of locations. These recording methods take advantage of fairly standard electrophysiology recording resources and capabilities to measure NT or NM release, which contributes to their wide use13,14. However, electrophysiological recordings are technically demanding, of low throughput and largely limited to in vitro studies. Despite being highly sensitive (for example, detecting changes resulting from quantal presynaptic release of NTs15), electrophysiological readout (current or membrane potential) is not specifically determined by the ligand of interest; for this reason, receptor antagonists, if available, are often used to verify molecular specificity13. Finally, although electrophysiological techniques offer sub-millisecond temporal resolutions, they do not provide a direct measure of release kinetics (FIG. 1a). Nevertheless, electrophysiology can be combined with the direct detection methods to study the release of NTs and NMs and their downstream effects in targeting cells (discussed below).
Fig. 1 |. Non-genetically encoded methods used to measure neurotransmitters and neuromodulators.
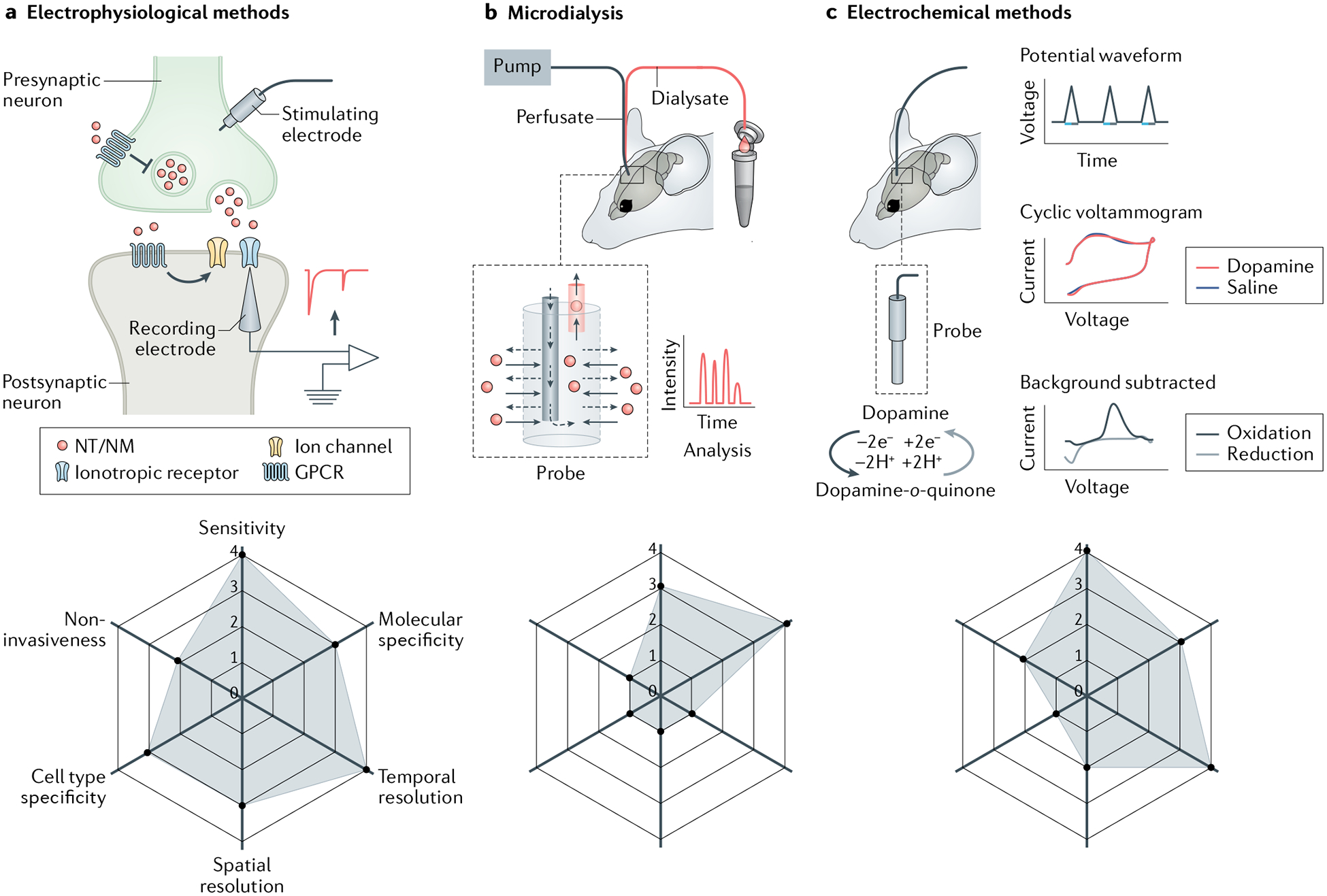
a | Electrophysiological methods. Upon stimulation via the stimulating electrode, neurotransmitters or neuromodulators (NTs/NMs) are released from the presynaptic terminal and activate either ionotropic receptors (which then serve as ion channels) or metabotropic receptors (G protein-coupled receptors (GPCRs)), which in turn can activate other ion channels in postsynaptic membrane. Thus, NTs/NMs induce a current in the postsynaptic cell that can be measured using an electrode. b | In microdialysis, a perfusate is pumped into the brain via a microdialysis probe, and dialysate containing NTs/NMs is collected for analysis. c | Fast-scan cyclic voltammetry (FSCV) as an example of an electrochemical method. FSCV probe shown implanted in the brain, with example of FSCV waveform for dopamine (DA). DA is oxidized to DA-o-quinone (blue arrow), which can be reduced back to DA (grey arrow). Current recorded by the microelectrode is plotted against voltage; by subtracting the large background current, oxidation (blue curve) and reduction (grey curve) peaks of DA at specific voltages can be seen on the cyclic voltammogram. Each panel contains a radar chart summarizing the performance index of methods in arbitrary units ranging from 0 to 4, with larger numbers representing better performance (1, poor; 2, fair; 3, good; 4, excellent). For sensitivity: 3, can measure NTs/NMs at nanomolar level; 4, can measure quantal release of NTs/NMs. For molecular specificity: 3, cannot distinguish structure similar NTs/NMs without manipulations; 4, can distinguish structure similar NTs/NMs. For temporal resolution: 1, can measure NT/NM dynamics in the order of minutes; 3, can measure NT/NM dynamics in the order of milliseconds; 4, can measure NT/NM dynamics in the order of microseconds. For spatial resolution: 1, can only measure NT/NM dynamics at more than 100 μm level; 2, can measure NT/NM dynamics at 10–100 μm level; 3, can measure NT/NM dynamics at cellular level (under 10 μm); 4, can measure NT/NM dynamics at subcellular level (under 1 μm). For cell type specificity: 1, very difficult to measure NTs/NMs from a specific cell type; 4, easy to measure NTs/NMs from a specific cell type. For non-invasiveness: 1, severe damage to tissues or cells; 2, moderate damage to tissues or cells; 4, no detectable effects to cell physiology.
Microdialysis
Since it was developed in the 1960s, in vivo brain microdialysis has become a well-established and widely used tool16. Microdialysis separates the perfusion media from the tissue using a semipermeable membrane with a specific pore size (mimicking the properties of capillary blood vessels) that permits the selective diffusion of soluble chemicals such as NTs and NMs in the dialysate from the extracellular space. The dialysate is then collected for further analysis (FIG. 1b).
Several advantages have led microdialysis to be a popular method for measuring NTs and NMs. First, microdialysis provides continuous sampling. When coupled with sensitive analytical techniques such as liquid chromatography or capillary electrophoresis, together with either an ultraviolet absorption detector or mass spectrometry, microdialysis could achieve nanomolar or even picomolar sensitivity with high molecular specificity17,18. Thus, for several decades, in vivo microdialysis has been extensively used to measure NTs and NMs, including in freely moving animals over several days, across multiple circadian cycles19,20. Second, microdialysis can be used to either simultaneously or sequentially monitor multiple NTs and/or NMs, including electrically inactive transmitters21. When combined with metabolomics, microdialysis can also be used for the discovery of small molecular metabolites with functional importance22. Microdialysis can similarly collect both endogenous molecules and exogenous substances such as drugs in local tissue regions of interest, thus providing clinically relevant information regarding pharmacokinetics and pharmacodynamics23. Moreover, because the movement of molecules through the semipermeable membrane is bidirectional, microdialysis can also be used to deliver compounds locally at defined concentrations and at specific time intervals.
However, despite these advantages, microdialysis has several limitations with respect to NT and NM measurement (FIG. 1b). First, the temporal resolution of microdialysis is relatively low, as minutes are typically needed to collect enough samples to measure specific analytes. In an attempt to increase temporal resolution to 50 s per acquisition, a microfabricated polymer-based probe was used as the droplet collection system24.
Second, similar to electrode-based probes, microdialysis lacks high spatial resolution (for example, at the subcellular level). Third, although microdialysis probes can be extremely small (about 100 μm in diameter), microdialysis is still a relatively invasive procedure and is difficult to apply in small model organisms such as Caenorhabditis elegans and Drosophila. Even in rodents and some larger species, it takes several days for animals to recover from the implantation of the microdialysis probe. Furthermore, placement of the probe for a few days in the brain can induce local changes such as gliosis that can compromise the integrity of the blood–brain barrier25–27.
Last, although in vivo microdialysis has been widely used for measuring small, soluble NTs and NMs, it is not well suited (and thus rarely used) for measuring neuropeptides and neurolipids. Neuropeptides are rapidly diluted in the extracellular space as they diffuse from localized release sites, with their extracellular concentrations often in the picomolar to nanomolar range. Sample volumes for microdialysis are typically in the nanolitre to microlitre range, and the dialysate is often introduced to capillary-scale separations and requires high-sensitivity mass spectrometry (reviewed elsewhere28). Moreover, neurolipids are lipophilic chemicals, and so do not easily diffuse in the aqueous environment of the dialysis probe, and sometimes can even be adsorbed by the probes. These obstacles result in fairly low collection efficiencies and unreliable results29,30. Some efforts have been made to reduce the adsorption of neurolipids on the microdialysis probe, as well as to enhance the solubility and stability of neurolipids by adding ingredients. For example, one group chose probes with polyethersulfone membranes and added β-cyclodextrin (as ingredients) to the perfusion fluid29.
In addition to being combined with microdialysis for monitoring the dynamics of NTs and NMs, mass spectrometry can be used to measure the spatial distribution of these molecules. For example, a recently developed matrix selectively targeting phenolic and primary amine groups31 was coupled with matrix-assisted laser desorption–ionization mass spectrometry imaging to simultaneously measure catecholaminergic and serotonergic signalling molecules — including their precursors and metabolites — in specific brain regions in post-mortem tissue of patients with Parkinson disease31. However, this method can only be used to measure NT or NM levels in a static state in brain sections, not in vivo, and cannot distinguish between extracellular and intracellular signalling molecules.
Electrochemical methods
In the 1970s, Ralph Adams and colleagues demonstrated the use of voltammetry to measure chemicals in tissues by implanting a carbon-paste electrode into the brain of an anaesthetized rat32. Since then, several electrochemical techniques, including amperometry, various potential pulse methods and cyclic voltammetry, have been used to measure fluctuations in NTs and NMs (reviewed elsewhere33). Among these methods, fast-scan cyclic voltammetry (FSCV) using carbon-fibre microelectrodes has become a key method for detecting electrochemically active NTs and NMs — most not ably, dopamine (DA) — in the nanomolar range and on a sub-second timescale34–37. In FSCV, cyclic voltage ramps are applied at a high scan rate (more than 100 V s−1) to rapidly oxidize and reduce the electroactive chemicals at the electrode’s surface; the electrochemical current recorded by the microelectrode is then plotted against the applied voltage. By subtracting the background current, characteristic peaks in oxidation and/or reduction that occur at specific voltages are visible on the cyclic voltammogram, which are then matched to specific chemical species. The current measured at a specific voltage can then be plotted against time to obtain a time course of the chemical’s concentration37 (FIG. 1c).
Because the FSCV probe is relatively small (less than 10 μm in diameter) compared with microdialysis probes, FSCV is less invasive (Fig. 1c) and can be used to measure endogenous NTs and NMs in small animal models such as Drosophila38. Moreover, FSCV-based methods have been used in vivo to measure sub-second fluctuations in NM levels in humans. To minimize damage, FSCV probes were adapted for use in patients receiving deep brain stimulation treatments for Parkinson disease and essential tremor39. By using FSCV data to estimate DA and serotonin (5-HT) concentrations, researchers investigated the role of these NMs in reward-based decision-making40,41 and non-reward-based aspects of cognition and behaviour39 in humans.
Despite their advantages, FSCV-based methods cannot distinguish between structurally similar chemicals that are oxidized or reduced at similar potentials, such as DA and noradrenaline (NA). Thus, new strategies are needed to make more-selective electrodes, and to distinguish signals derived from multiple analytes. In addition, FSCV is not well suited to measuring NTs or NMs that are not readily oxidized, including glutamate, GABA, ACh, adenosine 5′-triphosphate (ATP), neurolipids and most neuropeptides42. Thus, enzyme-modified electrodes have been developed to measure these electrochemically inactive NTs and NMs, particularly glutamate, ATP and ACh42,43. In this approach, enzymes that act specifically on an analyte of interest are immobilized on the electrode’s surface, and the analyte is detected by the formation of an electroactive product (for example, oxidase-generated hydrogen peroxide). For example, to detect extracellular ACh, the enzyme choline oxidase can be used to measure extracellular choline, a product of ACh hydrolysis by acetylcholinesterase. Choline oxidase catalyses the oxidation of choline in the presence of molecular oxygen, generating the electrochemically detectable hydrogen peroxide44. Notably, intermediate products such as choline can also be detected by the enzyme-based electrode. These neurochemical artefacts generate Faradaic currents that can be subtracted using a control biosensor that lacks the first enzyme in the cascade (for example, acetylcholinesterase or adenosine deaminase for measuring ACh44 and adenosine45, respectively) and is therefore not sensitive to the analyte of interest but remains sensitive to downstream products.
Despite recent technical improvements, microdialysis and electrochemical measurements still have relatively low temporal resolution and molecular specificity46. In particular, neither method is suitable for measuring in vivo release of NTs and/or NMs at the single-cell or subcellular level.
Non-genetically encoded optical tools
Compared with the analytical detection methods discussed above, optical imaging methods are more suitable for measuring NTs and NMs, as they typically have higher spatiotemporal resolution and are less invasive. To date, several optical reporters of NTs and NMs have been developed, including chemical dyes (for example, ‘false’ NTs47, radiolabelled metabolites and MRI or functional MRI probes48,49), cell-based sensors such as CNiFERs (cell-based neurotransmitter fluorescent engineered reporters)50 and synthetic nanomaterial-based sensors51,52. For example, a non-genetically encoded fluorescent catecholamine nanosensor called nIRCat was synthesized by non-covalently conjugating single-wall carbon nanotubes to single-strand oligonucleotides, which served as the recognition element and enabled catecholamine-interacting nanotubes to act as near-infrared sensors51. The nIRCat approach could measure DA release in brain slices and was compatible with DA pharmacology. Importantly, the wide availability of sensors in the near-infrared range (for example, 1,000–1,300 nm wavelength sensors) provides the potential for nIRCat to be combined with other spectrally compatible optical sensors to simultaneously measure multiple signals, including calcium signals and the dynamics of other NTs or NMs. Nevertheless, the selectivity of nIRCat needs to be further improved, as it currently responds to both DA and NA with only 3-fold discrimination51.
Although non-genetically encoded tools do not require gene delivery or protein expression, they still need to be delivered to target areas, and cannot be used to measure NTs and NMs in specific cell types.
Genetically encoded methods
Genetically encoded sensors can be expressed in specific cell types for days or even months, enabling researchers to perform long-term imaging of NTs and NMs. In the past few decades, major advances led to the development of various genetically encoded optical tools for imaging neurotransmission and neuronal activity (reviewed elsewhere53). In brief, this versatile toolbox includes indicators of vesicle release such as pHlourin54, pHTomato55, pHmScarlet56 and multiple neuropeptide-release reporters (NPRRs)57–61; genetically encoded Ca2+ indicators (GECIs) such as GCaMPs (which contain green fluorescent protein (GFP), calmodulin and the M13 peptide from myosin light-chain kinase fused together) and jRGECOs (reviewed elsewhere62); the recently developed voltage indicators (reviewed elsewhere63); and NT or NM indicators. The strategies used to develop, optimize and validate genetically encoded indicators — particularly GECIs — have facilitated the design of genetically encoded NT/NM indicators (GENIs). Below, we review the design principles, major properties, applications and limitations of GENIs for directly monitoring NTs and NMs.
Design principles and properties of GENIs
A typical GENI is primarily composed of two components: a ligand-binding module that binds the NT or NM molecule, and a fluorescent module in the form of either a FRET (fluorescence resonance energy transfer) pair of fluorescent proteins or a single fluorescent protein. Upon binding of the appropriate NT or NM, the conformational change of the ligand-binding module will lead to changes in the signal produced by the fluorescent module. Depending on the type of ligand-binding module, GENIs are roughly categorized as bacterial periplasmic-binding protein (PBP)-based sensors or GPCR-based sensors (Fig. 2; Table 1; see Supplementary Table 1).
Fig. 2 |. Genetically encoded indicators used to measure neurotransmitters and neuromodulators.
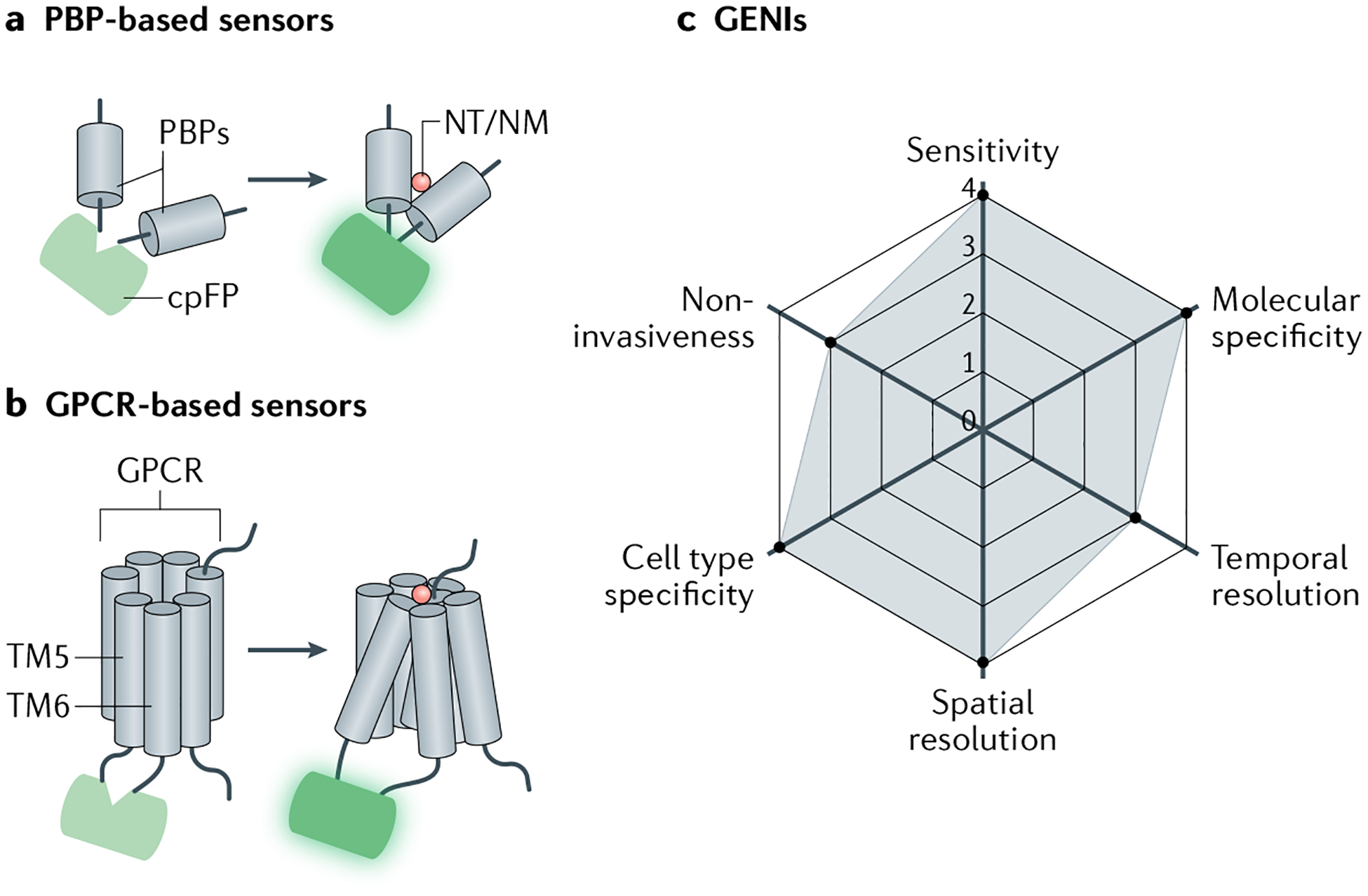
a | Periplasmic-binding protein (PBP)-based genetically encoded neurotransmitter or neuromodulator (NT/NM) indicators (GENIs) contain a circularly permuted fluorescent protein (cpFP) moiety. Following NT or NM binding, conformational changes of PBP will change fluorescence of cpFP. b | G protein-coupled receptor (GPCR)-based GENIs. Similar to PBP-based sensors, cpFP moiety is inserted into the intracellular loop of GPCR between transmembrane domains 5 and 6 (TM5 and TM6). Following NT or NM binding, conformational changes of GPCR induce changes in fluorescence of cpFP. c | Radar chart summarizing performance index of GENIs in arbitrary units, as in Fig. 1.
Table 1 |.
Genetically encoded single-wavelength indicators for neurotransmitters and neuromodulators
| GENI | Ex/Em (nm) | Responses (maximum ΔF/F0) | Affinity (EC50) | In vivo application | Refs |
|---|---|---|---|---|---|
| Glutamate sensors | |||||
| iGluSnFR | 490/510 | 2.0–4.0a 1.03b |
4 μMa 4.9 μMb |
Caenorhabditis elegans, zebrafish, mouse | 65 |
| SF-iGluSnFR. A184S | 490/510 | 3.1a 0.69b |
0.6 μMb | Mouse ferret | 66 |
| R-iGluSnFR | 562/588 | Approximately −0.35a | 18 μMa | – | 67 |
| iGlUu | – | 1.7a | 53 μMa | – | 175 |
| GABA sensors | |||||
| iGABASnFR | 485/510 | ~0.75b | 30 μMb | Mouse | 68 |
| iGABASnFR.F102G | 485/510 | ~1.7b | 42 μMb | Mouse | 68 |
| iGABASnFR.F102Y.Y137L | 485/510 | ~0.45b | 106 μMb | Zebrafish | 68 |
| Acetylcholine sensors | |||||
| GRABACh2.0 (GACh2.0) | 490/510 | 0.76a 0.9b |
2 μMa 0.7 μMb |
Fly, mouse | 76 |
| GRABACh3.0 | – | 2.8a | 2 μMa | Fly mouse | 77 |
| iAChSnFR | 485/510 |
10a 4.5b |
2 μMa 0.4 μMb |
C. elegans, fly zebrafish, mouse | 70 |
| Dopamine sensors | |||||
| GRABDA1m | 490/510 | 0.9a,b | 130 nMa,b | Fly, zebrafish, mouse | 78 |
| GRABDA1h | 490/510 | 0.9a,b | 10 nMa,b | Fly, zebrafish, mouse | 78 |
| GRABDA2m | – | 3.4a,b | 80 nMa,b | Fly mouse | 79 |
| GRABDA2h | 500/520 | 2.8a,b | 7 nMa,b | Fly mouse | 79 |
| rGRABDA1m | 565/595 | 1.5a,b | 100 nMa,b | Fly mouse | 79 |
| rGRABDA1h | 565/595 | 1.0a,b | 4 nMa,b | Fly mouse | 79 |
| dLight1.1 | 490/516 | 2.3a 1.8b |
330 nMa 311 nMb |
Mouse | 92 |
| dLight1.2 | 490/516 | 3.4a 3.0b |
770 nMa 1,157 nMb |
Mouse | 92 |
| dLight1.3b | 490/516 | 9.3a | 1,600 nMa | Mouse, rat | 92 |
| dLight1.4 | 490/516 | 1.7a | 4.1 nMa | Mouse, rat | 92 |
| dLight1.5 | 490/516 | 1.8a | 110 nMa | – | 92 |
| RdLight1 | –/588 | 2.5a 2.3b |
859 nMa 229 nMb |
Mouse | 91 |
| YdLight1 | –/525 | 3.06a | 1,630 nMa | – | 91 |
| Noradrenaline sensors | |||||
| GRABNE1m | 490/510 | 2.3a,b | 930 nMa 1,900 nMb |
Zebrafish, mouse | 80 |
| GRABNE1h | 490/510 | 1.3a,b | 83 nMa 93 nMb |
Zebrafish, mouse | 80 |
| nLight1.3 | 490/516 | 2.1a 1.5b |
764 nMa 919 nMb |
Mouse | 150 |
| Serotonin sensors | |||||
| GRAB5-HT1.0 | – | 2.5a 2.8b |
14 nMa, 22 nMb |
Fly, mouse | 87 |
| iSeroSnFR | 490/512 | 17a | 390 μMa | Mouse | 71 |
| PsychLight | – | 0.8b | 26 nMb | Mouse | 168 |
| Endocannabinoid sensors | |||||
| GRABeCB2.0 | 500/520 | 9.5b (2-AG) 5b (AEA) |
9.0 μMb (2-AG) 0.8 μMb (AEA) |
Mouse | 88 |
| Adenosine sensors | |||||
| GRABAdo1.0 | – | 1.2a ~2.0b |
60 nMb | Mouse | 89,176 |
| GRABAdo1.0m | – | ~3.4b | 3.6 μMb | Mouse | 176 |
| ATP sensors | |||||
| GRABATP1.0 | 500/520 | 5.0a 7.8b |
~6.7 μMa 80 nMb |
Zebrafish, mouse | 90 |
| GRABATP1.0-L | – | ~10b | 32 μMb | Zebrafish | 90 |
| iATPSnFR1.0 | 473/525 | 1.0a 1.5b |
350 μMa 630 μMb |
Mouse | 69 |
| ATPOS | 550/570 | 1.8c | 150 nMc | Mouse | 177 |
| Opioid sensors | |||||
| kLight1.2a | – | 0.6a | – | Mouse | 92,178 |
| GrpLight1.3ER | – | ~8.0b | 198 nMb | Mouse | 179 |
For more details, see Supplementary Table 1. –, not analysed; 2-AG, 2-arachidonoylglycerol; ACh, acetylcholine; Ado, adenosine; AEA, anandamide; ATP, adenosine 5′-triphosphate; DA, dopamine; EC50, half-maximal effective concentration; eCB, endocannabinoid; Ex/Em, peak excitation/emission wavelengths; GENI, genetically encoded NT/NM indicator; Glu, glutamate; GRAB, GPCR activation-based; GRP, gastrin-releasing peptide; h, high affinity; 5-HT, serotonin; iGluSnFR, intensity-based glutamate sensitive fluorescent reporter; k, κ-opioid; I, intensity-based; m, median affinity; NE, norepinephrine (noradrenaline); NM, neuromodulator; NT, neurotransmitter; OS, optical sensor; r, red-shifted; SF, superfolder GFP; SnFR, sensitive fluorescent reporter; u, ultrafast; Y, yellow-shifted.
Measured in cell lines.
Measured in cultured neurons.
Measured using purified proteins.
PBPs belong to a protein superfamily that bind a diverse range of ligands, including NTs and NMs. Ligand binding induces a conformational change in the PBP, providing the basis for GENI design64. PBPs are the ligand-binding module in engineered sensors to measure several NTs, including glutamate65–67, GABA68, ATP69 and ACh70 (Table 1; see Supplementary Table 1). However, naturally occurring PBPs may not be available for all NTs or NMs. Redesigning the binding pocket of an existing PBP-based sensor could be a promising approach to develop a sensor that can selectively bind other NTs or NMs. Computational design combined with machine learning was used to implement this approach recently71 to design a 5-HT sensor called iSeroSnFR, which contains 19 mutations introduced to a previously developed ACh sensor70 called iAChSnFR0.6. The iSeroSnFR sensor has an approximately 15-fold higher response to 5-HT than to ACh or choline.
Most PBP-based sensors have relatively low affinity for NMs, which allows them to respond to NMs with millisecond kinetics. However, their low affinity may also limit their application. For example, the physiological concentration of extracellular ATP is in the tens of nanomoles, and yet the half-maximal effective concentration (EC50) for the PBP-based iATPSnFR sensor is about 350 μM69; therefore, this sensor may not be sensitive enough to measure changes in extracellular ATP in certain conditions. The relatively low specificity of PBP-based GENIs could also limit their application; for example, the iAChSnFR sensor responds to both ACh and its precursor choline70.
GPCRs are the largest family of membrane-spanning proteins and have evolved as natural sensors for detecting extracellular signalling molecules — including NTs and NMs — with remarkably high specificity. Indeed, at least one GPCR has been identified that binds to each small NT, NM and neuropeptide. For example, five subtypes of ACh-binding GPCRs have been identified in humans, each with different affinities and/or pharmacological selectivities. This naturally developed repertoire of diverse GPCRs provides a wide range of choices for engineering GENIs with specific properties and applications. Upon binding of corresponding ligand, the GPCR rapidly (in the order of tens of milliseconds) undergoes a conformational change, with the largest change occurring between the fifth and sixth transmembrane domains72. A series of FRET-based sensors have been engineered73 in which a FRET pair such as CFP and YFP is fused to an intracellular part of the GPCR; upon the conformational change in the GPCR, the distance between the FRET proteins changes, resulting in a change in the FRET signal.
Unlike FRET-based sensors, which use two fluorescent proteins (FPs), sensors that include a single FP rely on detecting a simple increase or decrease in fluorescence intensity. Thus, these single FP-based sensors usually show a relatively high signal-to-noise ratio (SNR) and are more suitable for easy use in vivo. Conformation-sensitive circularly permuted fluorescent proteins (cpFPs)74,75 have been combined with GPCRs to create a series of GENIs called GPCR activation-based (GRAB) sensors for detecting various NMs, including the muscarinic acetylcholine receptor M3 (M3R)-based GRABACh for ACh76,77, the dopamine receptor D2 (D2R)-based GRABDA for DA78,79 and the α2-adrenergic receptor (α2-AR)-based GRABNE for NA80 (Table 1; see Supplementary Table 1). In a GRAB sensor, a cpFP moiety is inserted into the highly conserved third intracellular loop of the GPCR, as this domain undergoes the largest conformational change upon ligand binding81–83.
Here we use the GRABDA sensor78 as an example to summarize how such NT or NM sensors are designed and optimized. The first step is to select a suitable GPCR scaffold for further optimization. There are five subtypes of dopamine receptor (D1R–D5R), so the green cpFP (called cpEGFP) was inserted into each of the five subtypes and the performances of these chimeras evaluated — including their plasma membrane trafficking dynamics, initial dynamic range, affinity and selectivity. The D2R–cpEGFP chimaera was further optimized owing to its superior membrane trafficking and high affinity for DA. To increase the coupling of fluorescence changes to conformational changes induced by ligand binding, the third intracellular loop was systematically truncated to determine the optimal position of cpEGFP insertion. Subsequent site-saturation mutagenesis on the linker region generated the GRABDA1m sensor. The maximal ΔF/F0 (in the presence of a saturating concentration of DA) of GRABDA1m is ~90% and its EC50 is about 130 nM. To generate GRABDA with distinct affinities, to match the concentrations of released DA in different brain regions or conditions, GPCR variants (for example, with alterations in ligand-binding pockets or other sites affecting ligand-binding affinity) were introduced. A high-affinity sensor GRABDA1h (EC50 of about 10 nM) was generated by making a T205M substitution in GRABDA1m. To further improve the dynamic range of the first-generation GRABDA sensors, site-saturation mutagenesis was performed at 32 sites on cpEGFP that were chosen for their potential to improve the folding ability or brightness of the protein, or its structural coupling with the GPCR74,84–86. A screen of about 1,000 site-saturation mutagenesis variants79 yielded GRABDA2h and GRABDA2m, which have a 2-fold to 3-fold greater ΔF/F0 than GRABDA1m.
In fact, the GRAB strategy has been used to develop many other NM sensors, including the serotonin 2C receptor (5-HT2CR)-based GRAB5-HT for 5-HT (ref.87), the cannabinoid receptor type 1 (CB1R)-based GRABeCB for endocannabinoids (eCBs)88, the adenosine 2A receptor (A2AR)-based GRABAdo for adenosine89 and the P2Y1 receptor (P2Y1R)-based GRABATP for ATP90. Furthermore, red-shifted versions of GRAB sensors79, which contain circularly permuted mApple as the fluorescent protein, have been developed for multicolour imaging in vivo together with GFP-based sensors (such as GCaMPs and GFP-based GRAB sensors) and blue light-excited actuators (such as channelrhodopsin 2).
Using the similar strategy above, one group developed the dLight family of DA sensors91,92. A side by side comparison78 of dLight and GRAB sensors expressed in cultured cells revealed that the GRABDA2m sensor has a higher apparent affinity for DA, basal brightness and maximal brightness than do the dLight 1.1, 1.2 and 1.3b sensors; in addition, GRABDA2m has a larger ΔF/F0 than all dLight sensors except dLight1.3b. Furthermore, two-photon imaging in transgenic Drosophila brains showed that GRABDA2m was about 3-fold brighter than dLight1.3b (ref.79). The GRABDA1h sensor is sensitive enough to detect DA release induced by minimal electrical stimulation of a single dopaminergic fibre78. However, dLight family sensors have a faster off-rate than GRABDA2m and are selective to NA79,91. As such, the right sensor to use will depend on the research question93–96.
In native GPCRs, the conformational change induced by ligand binding creates a binding pocket for the Gα subunit of the heterotrimeric G protein. Thus, if a G protein is fused to the carboxyl terminus of the GPCR, it would interact with the activated GPCR upon ligand binding. Accordingly, several GENIs — including neuro peptide sensors — have been generated by inserting cpEGFP between the GPCR and a non-signalling Gα subunit or G protein mimic, including short C-terminal peptides derived from Gα proteins, mini-G proteins and Gα-mimic nanobodies97,98 (TABLE 1; see Supplementary Table 1). The resulting intramolecular engagement of the non-signalling G protein or G protein mimic is believed to compete with endogenous G proteins owing to the former’s relatively high effective local concentration, thus preventing downstream G protein signalling through activated GPCR-based GENIs97. However, these sensors have a relatively limited response and slow kinetics. Importantly, the ability of the GENIs discussed above to detect the dynamics of endogenous NTs or NMs in vivo has not been tested.
Tango GPCR assays99 measure downstream gene expression induced by GPCR activation and usually have good sensitivity. Tango assays detect biased β-arrestin recruitment to GPCRs by ligand binding, enabling comparisons of GPCR activation across whole brain100, as well as the putative detection of NMs if they are released at a relatively constant rate over longer timescales. An improved version called iTango2 (REF.101), as well as SPARK102, can also report NM signalling at a single time point in vivo; however, these methods show relatively slow kinetics (minutes) compared with GRAB sensors (milliseconds to sub-seconds).
In the future, to optimize NT and NM sensors with a wider dynamic range, suitable molecular specificity, desired pharmacological properties and minimal effects on cellular physiology, efforts should be focused on re-engineering domains in existing sensors, including the ligand-binding pocket and linker regions. Moreover, other naturally occurring NT-binding or NM-binding proteins could be used. For example, to develop a specific glycine sensor, a soil phytopathogen-expressed solute-binding protein that displays promiscuous binding activity for glycine, l-serine and GABA was computationally redesigned103. The glycine-selective protein was linked to FRET pairs and underwent linker optimization to generate a sensor called GlyFS. GlyFS has high selectivity for glycine and can detect endogenous glycine release103.
Similar to sensors containing fluorescent proteins, luciferase-based bioluminescent sensors are also genetically encodable and are widely used to measure both intracellular and extracellular molecules, with an extremely low background signal104. For example, Petersen et al. recently developed BLING (bioluminescent indicator of the neurotransmitter glutamate), which was designed with a split luciferase and a glutamate-sensing domain105. The authors showed that BLING reported changes in extracellular glutamate through a change in its luminescence intensity in cultured cells, thus expanding the toolbox of GENIs. However, luciferase-based bioluminescent sensors emit very low numbers of photons, limiting their potential use in imaging NTs or NMs at the cellular scale, particularly in vivo.
Considerations when using GENIs
With the fast expansion of the GENI toolbox, the optical imaging equipment and methods developed for visualizing Ca2+ indicators can be readily used for visualizing NT and NM signals. Numerous GENIs have been developed, each with its own combination of brightness, dynamic range, affinity, selectivity and kinetics (TABLE 1; see Supplementary Table 1). However, as mentioned by other reviews, there is not a ‘one-size-fits-all’ sensor94,96. When designing an experiment using a GENI, it is important to select the appropriate sensor for addressing the specific biological questions of interest (FIG. 3a).
Fig. 3 |. Choosing genetically encoded neurotransmitter or neuromodulator indicators for experiments.
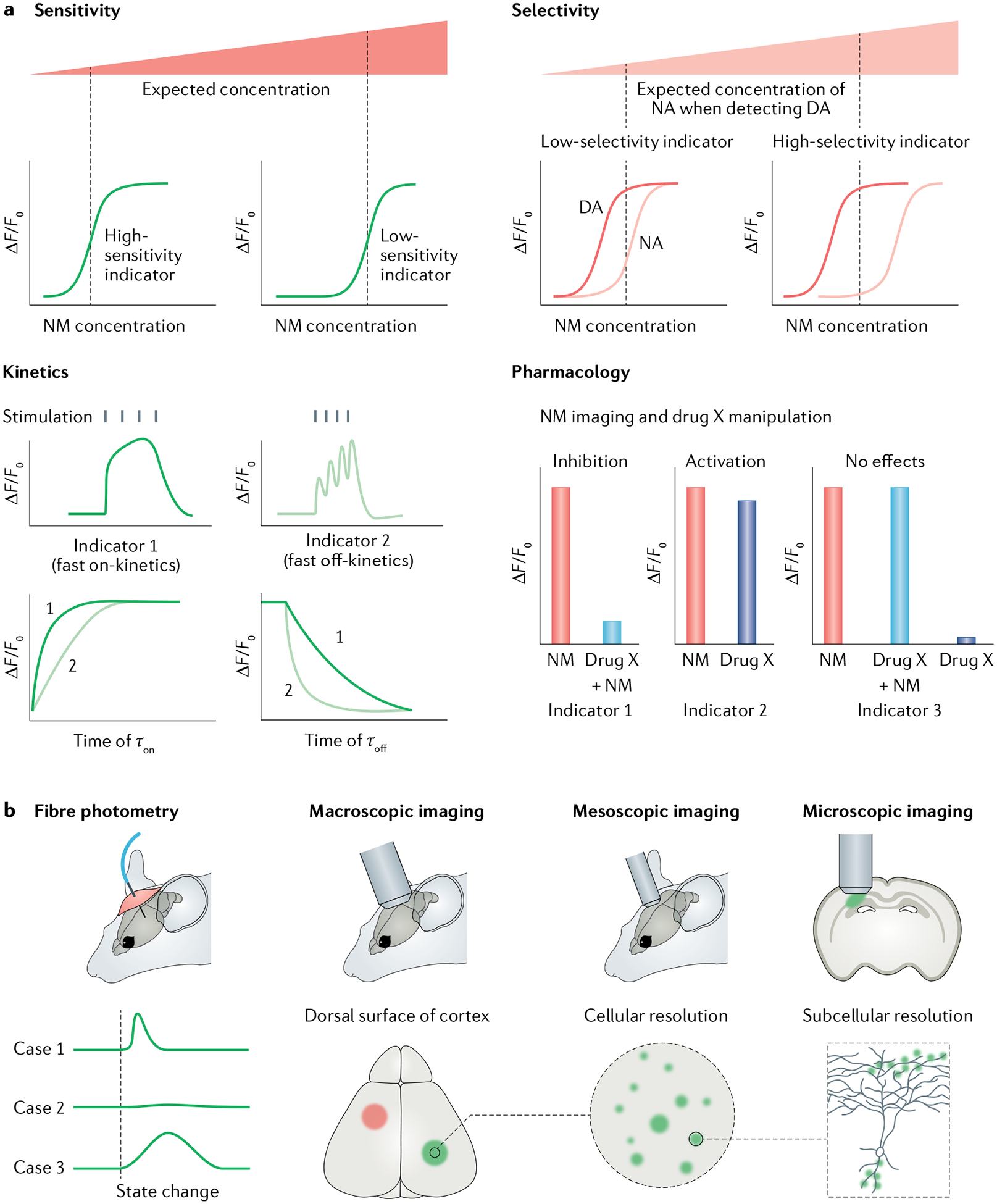
a | Parameters for choosing a genetically encoded neurotransmitter or neuromodulator (NT/NM) indicator (GENI). First, is the GENI sensitive enough for measuring NTs or NMs of interest? Generally, if two sensors showed comparable maximum responses (ΔF/F0), the sensor with higher affinity should be used to detect NT or NM at low concentration. Second, is the sensor selective enough? For example, if users want to detect dopamine (DA) levels, a DA sensor that could better separate DA and noradrenaline (NA) should be chosen. Third, is the sensor fast enough to capture NT or NM dynamics? GENIs with fast on-kinetics (τon) will have relatively large sensor responses to brief release events. GENIs with fast off-kinetics (τoff) are preferred to track the dynamics of NTs and NMs in response to closely related events or rapid changes in behaviour. Fourth, is the GENI compatible with any pharmacological manipulations used? GENIs, especially G protein-coupled receptor (GPCR)-based indicators, are engineered using NT or NM receptors and thus inherit their parental GPCRs’ pharmacological properties. Therefore, GPCR-based indicators are incompatible with certain pharmacological manipulations. b | Imaging modalities and data interpretations. Fibre photometry recording is the method of choice for detecting GENI signals in the brain region of interest, in freely moving animals. When used with GENIs with fast kinetics, fibre photometry recording can report NT or NM dynamics during animal state changes (such as during sleep–wake transitions and learning). Fibre photometry measures bulk fluorescence emitted from a radius of several hundred microns and therefore has relatively poor spatial resolution. One advantage of GENIs is that they can be imaged at multiple spatial scales, ranging from the whole brain to subcellular levels, when combined with different imaging modalities.
First, the sensitivity of the GENI should be considered. GENI sensitivity is mainly determined by their dynamic range, affinity and brightness. In principle, if two GENIs show comparable brightness and maximum responses, the GENIs with higher affinity will be more sensitive for detecting NTs or NMs under physio logical conditions (FIG. 3a). The sensor’s SNR is an important criterion for evaluating the performance of various GENIs, with the goal of achieving the highest SNR possible. Importantly, however, a sensor’s SNR is not determined solely by the sensor’s response; it is also determined by its basal brightness. For example, although the maximum response of dLight1.3b is higher than that of GRABDA2m, GRABDA2m actually has a higher SNR owing to higher basal brightness79.
The second consideration is whether the sensor is selective enough. For example, although current GENIs for DA have better selectivity for DA over NA than FSCV does, GENIs still respond to NA. If researchers want to detect DA dynamics, a DA sensor that could better discriminate DA from NA should be chosen (FIG. 3a).
Third, the kinetics of the GENI should be considered in the context of the research question. Generally, GENIs with fast on-kinetics will have greater responses to brief release events, whereas GENIs with fast off-kinetics are preferable for tracking the dynamics of NTs or NMs associated with closely related events or rapid changes in behaviour (FIG. 3a). In addition, faster off-kinetics may decrease analyte buffering96.
Fourth, GENIs, especially GPCR-based indicators, may also respond to agonists or antagonists that normally bind the corresponding parent GPCR, providing a method for validating the signals recorded using GPCR-based GENIs. For example, a GPCR antagonist should compete and block the signal induced by the corresponding NT or NM. However, this may prevent the use of some GPCR-based indicators in combination with certain pharmacological manipulations (FIG. 3a). This issue can be solved, in part, by using different GPCR subtypes as scaffolds for engineering GENIs. In addition, the availability of the structure of most GPCRs provides important information regarding which residues are crucial for agonist or antagonist binding, thus making it possible to engineer GENIs with different pharmacological selectivity. Alternatively, PBP-based indicators, microdialysis or FSCV can be used, although some PBP-based GENIs also respond to certain pharmacological compounds (for example, the iAChSnFR sensor responds to some acetylcholinesterase inhibitors70).
The biophysical properties of GENIs — including their dynamic range, brightness, kinetics, ligand affinity, chemical specificity, pharmacological properties and photostability — are relatively easy to characterize in vitro. However, given the complexity of in vivo systems, the performance of a candidate GENI should also be validated in vivo using multiple imaging methods — for example, the response of a GENI to optogenetic or chemogenetic stimuli or certain behaviours94.
Another issue when designing an experiment using GENIs is their potential effects on cellular physiology. For example, the GRABACh2.0 sensor, which is based on the M3R, does not completely lack Gq protein coupling, and so expression of this sensor may affect downstream signalling76. Fortunately, optimizing the interface between the M3R and the cpEGFP module led to the next generation of ACh sensor, GRABACh3.0, which has an approximately 3.7-fold larger response than GRABACh2.0, yet negligible coupling with Gq signalling77, thus providing a superior sensor for measuring ACh in vivo with minimal effect on cellular physiology. In fact, most of the GPCR-based GENIs (including GRABs and dLights) show negligible coupling with major GPCR downstream pathways (such as G protein pathways, β-arrestin pathways) that might lead to disrupted cell function, presumably due to the steric hindrance imposed by the bulky cpFP moiety that replaces parts of the third intracellular loop, which is where G proteins and arrestin interact with GPCRs106,107. Importantly, GRAB sensor expression does not seem to perturb animal behaviours88.
Finally, although the GPCR-based indicators sensors are engineered NT or NM receptors, their expression pattern may not mimic that of endogenous receptors, and hence the signal reports NT or NM release but is not suited to infer the spatial organization of NT or NM sensing on target cells108. To more faithfully infer the spatial organization of NT or NM sensing on target cells, GENIs can be selectively expressed in targeted cells with endogenous receptor expression.
Applications for GENIs
Because all GENI sensors convert a neurochemical signal to a change in fluorescence, any techniques that report a change in fluorescence intensity are suitable for measuring the signals generated by GENI sensors. Currently available methods for recording GENI signals include fibre photometry, wide-field epifluorescence imaging, light-sheet fluorescence imaging, multiphoton imaging and the recently developed super-resolution imaging (FIG. 3b). These imaging methods provide different levels of spatial and/or temporal resolution and require instruments that vary widely in both complexity and cost. It is therefore important to find the optimum set of tools to answer the specific research question at hand. Below, we discuss the various imaging methods available for detecting GENIs, and their recent applications.
Detecting GENI signals in freely moving animals.
Fibre photometry has become the method of choice for detecting GENI fluorescent signals in freely moving animals — particularly rodents — owing to its compact size and the relatively simple instruments needed to detect the signal109. Fibre photometry measures bulk fluorescence emitted from a radius of several hundred microns and therefore has relatively poor spatial resolution. The sampling rate of fibre photometry ranges from tens to thousands of hertz and, therefore, is generally not a limiting factor for temporal resolution. However, the level of synchronization among the cells emitting the fluorescent signals does affect temporal resolution. If the changes in fluorescence within a region of interest are highly heterogeneous (for example, in the case of Ca2+ transients of the neurons in the prefrontal cortex), the sum of the fluorescent signal will be difficult to interpret. By contrast, if the changes in fluorescence are relatively well synchronized within the region of interest across the recording area, the bulk change in fluorescence provides an excellent estimate of the underlying unitary responses. Because volume transmission is a major mode of action of many NMs, changes in the amount of NM in a small region are often highly correlated, thus providing a meaningful readout (FIG. 4). Nevertheless, finer details involving spatial and temporal heterogeneity at NM-releasing sites will be inevitably missed by this method108.
Fig. 4 |. Fibre photometry recording of neurotransmitter or neuromodulator dynamics in freely moving mice.
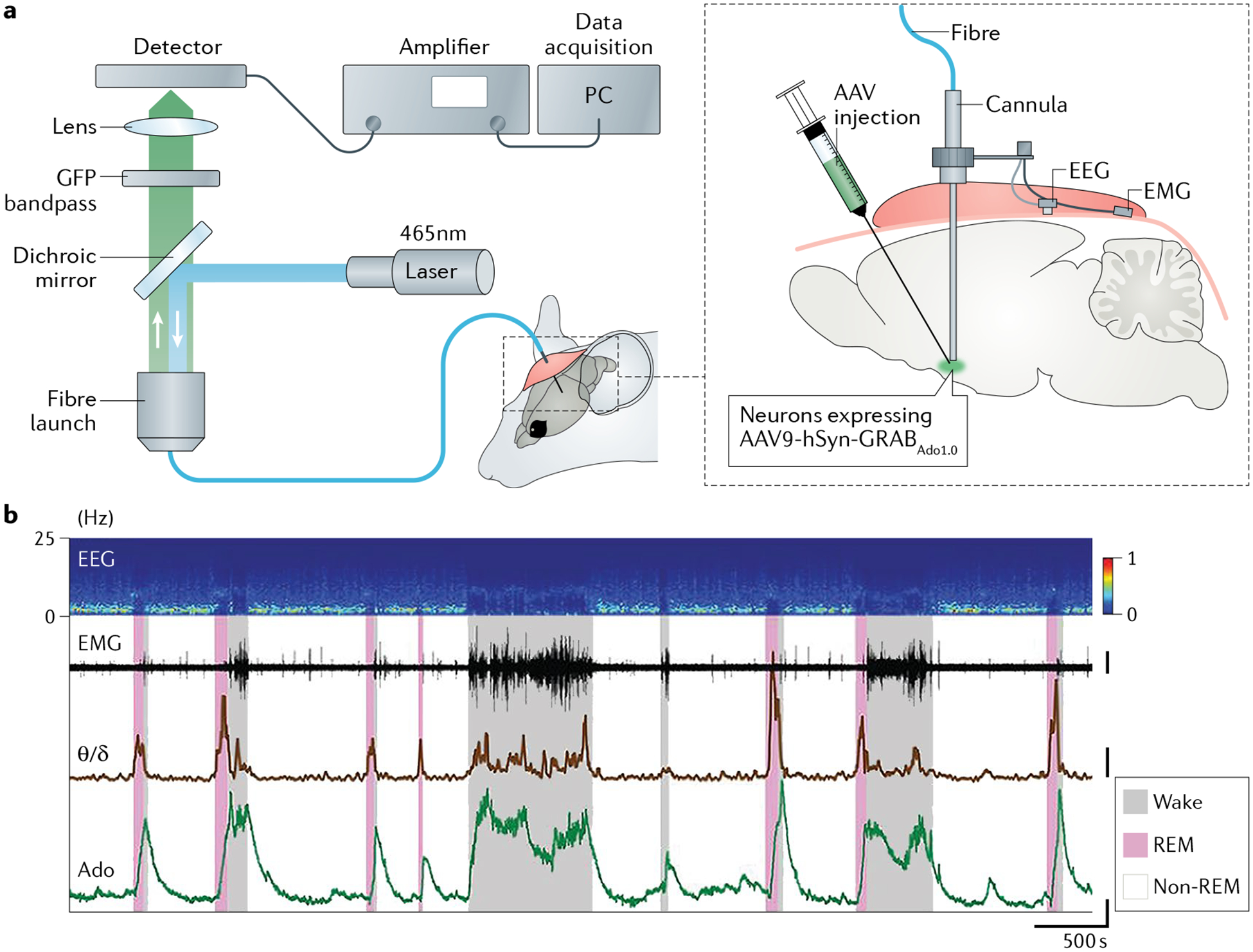
a | G protein-coupled receptor (GPCR) activation-based sensor for adenosine (GRABAdo1.0) expressed in the basal forebrain (under control of the human synapsin (hSyn) promoter encoded by an adeno-associated virus (AAV)) of fibre-attached mice and used to monitor adenosine dynamics. b | Electroencephalogram (EEG), including ratio between EEG theta power (θ) and delta power (δ), and electromyogram (EMG) also recorded to identify specific brain states, namely wake, rapid eye movement (REM) sleep and non-REM sleep. GFP, green fluorescent protein; PC, personal computer. Panel b adapted with permission from REF.89.
To date, fibre photometry has been used to detect the release of a wide range of NMs (including ACh, DA, NA, 5-HT, adenosine and eCB) under various conditions, including during the switch between sleep and wake states77,87,89,110, natural behaviours78,79,92,111–114, aversive and positive learning92,115–123 and stress77,80,88. For example, although adenosine is believed to have a prominent role in sleep homeostasis, how neural activity underlying the sleep–wake cycle controls adenosine release in the brain remains unclear. To address this question, fibre photometry was combined with the GRABAdo sensor in the mouse basal forebrain. This approach revealed that adenosine levels are relatively high during wakefulness and rapid eye movement (REM) sleep, but low during non-REM sleep89 (FIG. 4b). To study the relationship between adenosine dynamics and neuronal activities, the authors performed simultaneous recordings of adenosine levels and Ca2+ signals of different BF neurons, and found that the activity of both cholinergic and glutamatergic neurons correlated highly with adenosine levels. Furthermore, activation of glutamatergic neurons induced an increase in extracellular adenosine, whereas selectively ablating BF glutamatergic neurons decreased extracellular adenosine levels and impaired sleep homeostasis89. More recently, another study used the GRAB5-HT sensor and found that 5-HT levels in the basal forebrain are also high during wakefulness and low during non-REM sleep. Interestingly, in contrast to the increased levels of adenosine during REM sleep89, this study found that 5-HT levels are actually reduced during REM sleep87. Using similar recording strategies, other groups found distinct sleep–wake state-dependent changes in both NA and ACh levels77,110, suggesting that many NMs probably work together to mediate the switch between sleep and wake states.
DA has been implicated in motivation and reward-related behaviours. Soon after the development of the genetically encoded DA sensors, several studies used fibre photometry to monitor fluorescent signals of DA sensors as a real-time readout of DA release under both physiological and pathological conditions. In particular, several studies focused on understanding the dynamics of dopaminergic activity in drug addiction, finding that the administration of morphine, heroin, cocaine, alcohol or nicotine to rodents increased DA levels in the nucleus accumbens124–127. In a separate study, DA release in the nucleus accumbens was found to be greater in mice fed a high-fat diet than in those fed a standard chow diet; moreover, following repeated exposure to the high-fat diet, DA release became diminished when fasted animals consumed the standard diet128. Thus, drugs of addiction and a high-fat diet seem to have similar effects on DA signalling, in that both overstimulate the dopaminergic system and lead to diminished DA responses to typical stimuli.
The increasing variety of GENIs for detecting neurochemicals should rapidly advance our understanding of the neural mechanisms that underlie various diseases, particularly mental illness. Indeed, nearly all drugs developed to treat psychiatric disorders target a neuromodulatory system129. In a recent study, hallucination-like perceptions in mice were assessed using a behavioural task, and a DA sensor revealed a correlation between excessive levels of DA in the tail of the striatum before the presentation of sensory cues and hallucination-like perception130. Moreover, this study suggests that excessive DA in the tail of the striatum encodes perceptual expectation, and its increase leads to ‘overconfidence’ of a false perception (that is, the hallucination-like percept)130.
Developments in fibre photometry, such as multifibre photometry with high-density arrays of optical fibres131, wireless fibre photometry132 and depth-resolved fibre photometry with tapered optical fibres133, will provide further flexibility for experimental design and expand our understanding of how NTs or NMs contribute to different behavioural states or dysfunction in animal models, with high spatial selectivity.
Another popular imaging tool used in freely moving animals is the miniaturized epifluorescence microscope (miniscope)134, which can — at least in principle — provide cellular resolution if the sensors are expressed primarily in the cell body. However, as GPCRs are expressed in the cell membrane, it is difficult to discern individual cells expressing current NM sensors owing to the high level of fluorescence signals from axons and dendrites. Nevertheless, inspired by the design of soma-targeted and axon-targeted Ca2+ sensors135–137, as well as soma-targeted voltage sensors138–141, the future development of compartment-specific NM sensors may present opportunities for using miniscopes to monitor NM signals with subcellular resolution in freely moving animals. On the hardware side, miniaturized two-photon microscopes can provide high spatial resolution and reduce the out-of-plane background signal, and thus have the potential to discern GENI signals emitted from individual somas and processes in freely moving animals77,142,143.
Detecting GENI signals in head-fixed animals.
More sophisticated microscopy tools and methods can be used when the imaged sample is stationary or fixed than when imaging in freely moving animals. Thus, current methods focus on either detecting GENIs over a large surface area or with high spatial resolution. For example, mesoscopic imaging (an imaging technique that provides details regarding biological systems in the context of an organ, body part or entire organism) collects the fluorescence signal from the brain surface via a microscope-coupled camera (FIG. 5a). Using this approach, a typical field of view can cover 100 mm2 with about 512×512 pixels, yielding spatial resolution in the order of a few tens of microns per pixel. Moreover, the images are acquired at 10–50 frames per second, which is sufficient for detecting changes in fluorescence based on the kinetics of most GENIs. Although this method cannot be used to resolve single-cell activity, it can be used to monitor activity across a large area, such as the entire dorsal surface of the cerebral cortex144. For example, intensity-based glutamate sensitive fluorescent reporter (iGluSnFR) has been used to produce low-resolution maps of extracellular glutamate concentration across the surface of the entire mouse cortex145 (FIG. 5b). In another study, Ca2+ signalling and ACh levels were measured across the dorsal surface of the cortex in mice expressing jRCaMP1b and GRABACh3.0, respectively. This approach revealed that both the ACh and Ca2+ signals exhibit behavioural state-dependent and spatially heterogeneous fluctuations across the surface of the cortex146.
Fig. 5 |. Wide-field mesoscopic imaging of neurotransmitter or neuromodulator dynamics across the neocortex in awake mice.
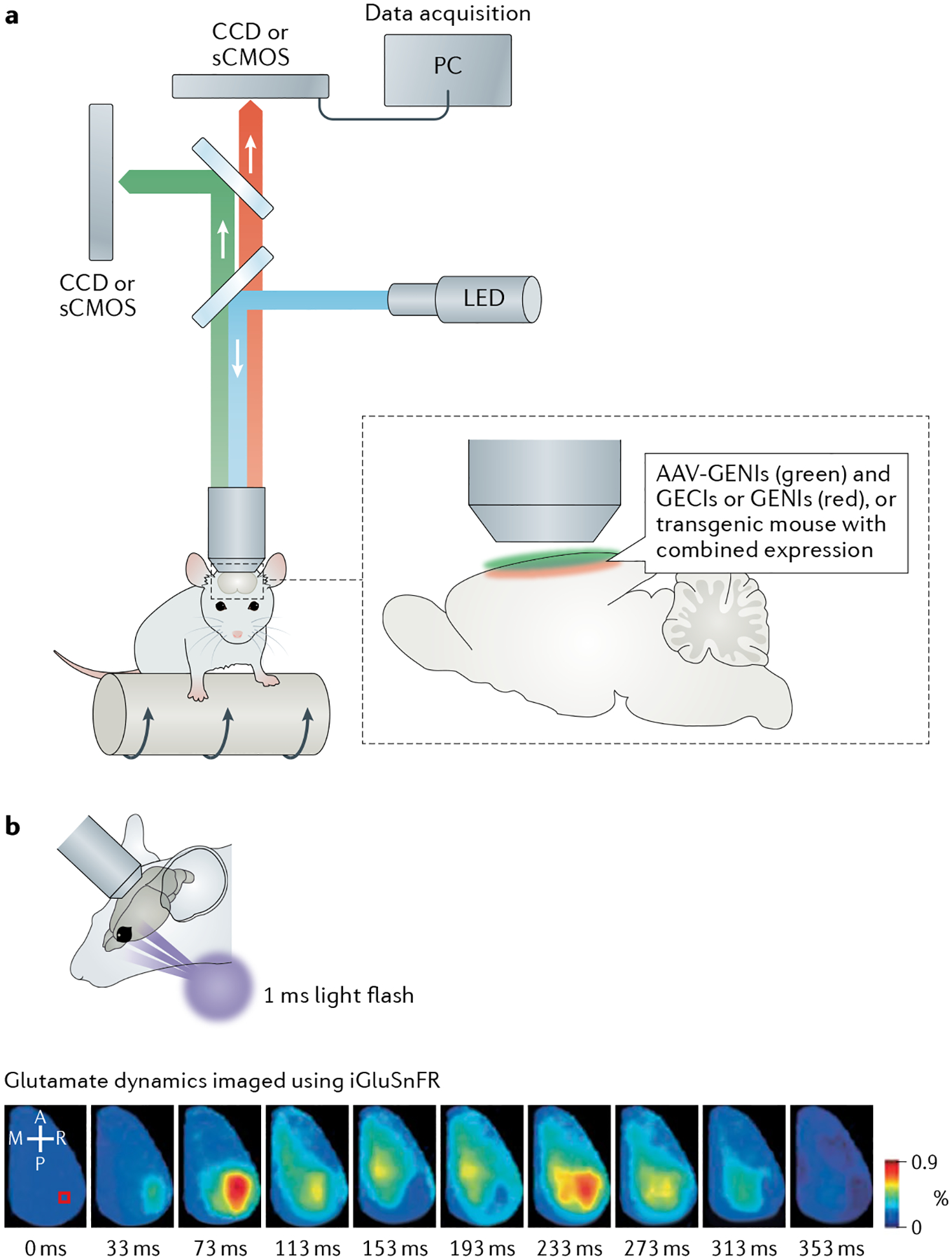
a | Dual-wavelength, wide-field imaging set-up, with mouse on a moving platform. Genetically encoded neurotransmitter or neuromodulator (NT/NM) indicators (GENIs) and genetically encoded Ca2+ indicators (GECIs) with separated spectra can be used to monitor NT or NM and Ca2+ dynamics, respectively; GENIs with separated spectra could be used to monitor dynamics of different NTs or NMs simultaneously. b | Imaging glutamate dynamics with intensity-based glutamate sensitive fluorescent reporter (iGluSnFR) across cortical surface. Top: experimental set-up of visual stimulation in awake mouse. Bottom: representative montage of cortical iGluSnFR responses after visual stimulation with 1 ms flash of light; this induces an initial rapid response, followed by a clearly separated, delayed secondary response in awake mouse. Scale bar, 2 mm. Panel b adapted with permission from Xie et al.145. A, anterior; AAV, adeno-associated virus; CCD, charge-coupled device camera; sCMOS, scientific complementary metal–oxide–semiconductor camera; LED, light-emitting diode; M, medial; P, posterior; PC, personal computer; R, right.
Multiphoton imaging — in this context, primarily two-photon imaging — excites the GENI using focused, high-intensity light at infrared wavelengths, resulting in the absorption of two or more photons and the emission of photons at a visible wavelength. Because this method requires point by point scanning, it has a relatively limited field of view (generally less than 1 mm2) and temporal resolution (with sampling typically in the range of 1–30 Hz); however, it provides excellent spatial resolution (under 1 μm). Thus, two-photon microscopy has been used to measure the spatial patterns of NM release in high detail68,77,78,147–152. For example, it was recently used to track the fluorescence of GRABACh3.0 in direction-selective retinal ganglion cells, which receive input from cholinergic starburst cells147. Increases of extracellular ACh at ganglion cell dendrites were observed to be tuned to light direction at the local level (within about 1 μm) but heterogeneous across the dendritic tree, with no relationship to the ganglion cell’s preferred direction (FIG. 6A), suggesting that cholinergic input has a minor role in determining the ganglion cell’s direction selectivity147. As another example, extracellular glutamate and Ca2+ signals of mitral cells in the mouse main olfactory bulb were simultaneously recorded using iGluSnFR and GCaMP6f, respectively. This approach revealed that the temporal dynamics of extracellular glutamate in the olfactory bulb is highly diverse and can account — to a large extent — for the heterogeneity of mitral cell responses, suggesting a faithful and linear input–output transformation of mitral cell activity152.
Fig. 6 |. Two-photon imaging of neurotransmitter or neuromodulator dynamics in different preparations and model organisms.
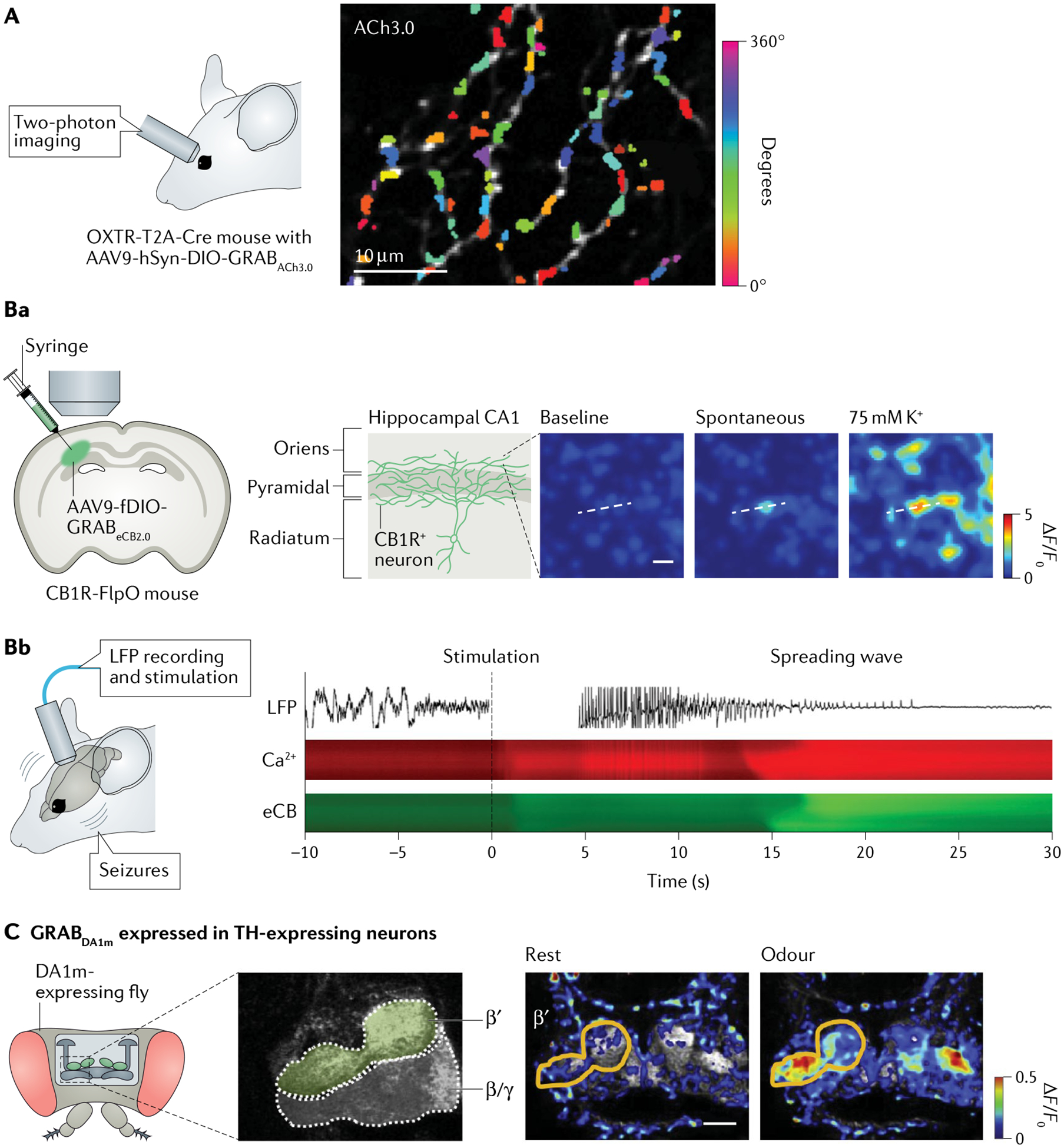
A | Two-photon imaging of acetylcholine (ACh) dynamics at subcellular level. Direction-selective ganglion cells of mouse retina were selectively labelled with G protein-coupled receptor (GPCR)-activation-based sensor for acetylcholine (GRABACh3.0; ACh3.0), using intravitreal injection of adeno-associated virus 9 (AAV9) expressing the sensor under control of the human synapsin promoter (hSyn) into Oxtr-T2A-Cre transgenic mice. Colours indicate preferred direction for each site on dendrites, quantified from responses to spots of light moving in eight directions. B | Imaging of endocannabinoid (eCB) dynamics with good spatiotemporal resolution. Fluorescence images of local eCB dynamics at single axonal boutons by expressing GRABeCB2.0 (eCB2.0) in cannabinoid receptor type 1-expressing (CB1R+) neurons in CA1 region of hippocampus. Scale bar, 5 μm (part Ba). Example local field potential (LFP) trace (top), Ca2+ dynamics (assessed using genetically encoded calcium indicator jRGECO1a; middle) and eCB dynamics imaged using eCB2.0 (bottom) of imaged mediolateral projections during stimulus-induced non-convulsive seizures and subsequent spreading wave. Dashed vertical line at time 0 indicates stimulus onset (part Bb). C | Imaging of odorant-evoked dopamine (DA) release in olfactory mushroom body in a live fly measured using two-photon microscopy. To express GRABDA1m (DA1m) in dopaminergic neurons, transgenic UAS-DA1m flies were crossed with tyrosine hydroxylase (TH)-GAL4 flies. The odorant elicited fluorescence increases prominently in the β′ lobes, not the β/γ lobes. Scale bar, 20 μm. Panel A adapted with permission from REF.147. Panel B adapted with permission from REF.88. Panel C adapted with permission from REF.78.
Compared with classical water-soluble NTs and NMs, the understanding of signalling through neurolipids (such as eCBs) in vivo is limited by the inability to measure their changes at timescales commensurate with the high lability of lipid signals. A genetically encoded GRAB sensor was recently developed for directly detecting eCB dynamics. Researchers selectively expressed the GRABeCB sensor in CB1R+ neurons and observed eCB transients from single axonal boutons in acute brain slices, suggesting constrained and localized eCB signalling88 (FIG. 6Ba). GRABeCB could also resolve the molecular identity and spatiotemporal dynamics of hippocampal eCB signalling in living mice at the timescale of physiological and pathological neural activity153 (FIG. 6Bb).
Although two-photon calcium imaging has become a standard tool for functional studying of neuronal populations in awake behaving animals, most two-photon imaging protocols measure calcium and NT or NM dynamics in single areas, limiting the ability to link the function of local circuits to global cortical dynamics. However, a relatively new method for performing both two-photon imaging and mesoscopic wide-field imaging simultaneously provides a powerful strategy for linking the function of a local circuit to the global cortical state, defined by the dynamics of various neurochemicals154.
Two-photon microscopy is also well suited for imaging animals with a relatively small brains, such as Drosophila and zebrafish. For example, transgenic flies expressing the DA sensor GRABDA1m selectively in dopaminergic cells were used to record the odorant-evoked DA release in the olfactory mushroom body using two-photon imaging78 (FIG. 6C). Further studies using this transgenic Drosophila line revealed that, on odorant-induced activation, dopaminergic cells suppress a pair of GABAergic cells that normally negatively regulate aversive olfactory learning, thereby allowing efficient learning155. A separate study investigated the DA-mediated plasticity that underlies the temporal sensitivity of associative learning using a DA sensor151. Together, these studies provide mechanistic and circuit insights through the use of GENI sensors.
Light-sheet fluorescence microscopy (LSFM) illuminates the plane of interest with a thin sheet of light while the camera is focused onto the plane156. A 3D volumetric image can be formed by moving the co-aligned excitation and detection planes. This approach allows researchers to rapidly image a large volume at cellular resolution. Furthermore, owing to the focused illumination at the plane of interest, LSFM causes minimal light-induced toxicity and makes it possible to measure fluorescence for long periods of time. Recently, Liang et al. measured DA signalling in ring neurons in the Drosophila ellipsoid body over 24 h using LSFM and the GRABDA2m sensor; they observed a daily oscillation in the DA signal and found that the peak in dopaminergic activity coincided with the daily increase in locomotor activity, suggesting that dopaminergic neurons have a role in controlling daily rhythmic activity157.
Because LSFM uses single-photon illumination, the light does not penetrate far into non-transparent tissue (but see two-photon LSFM158,159). Thus, LSFM has been used primarily to record GENIs in relatively trans parent organisms such as Drosophila larvae, zebrafish larvae and C. elegans160. Several groups have used ‘closed-loop’ behavioural paradigms in which behavioural responses such as tail movement are used to update sensory inputs, thus providing a realistic experience in head-fixed animals161,162. These innovative behavioural paradigms have enabled researchers to monitor whole-brain neural responses in head-fixed zebrafish larvae during ‘naturalistic’ behaviours. Additionally, two groups developed LSFM methods to image freely swimming zebrafish larvae while performing volumetric imaging at near-cellular resolution163,164. To date, whole-brain LSFM has been used primarily to measure Ca2+ signals; however, the generation of transgenic zebrafish lines that express various GENIs has opened the door to examining the dynamics of various neurochemicals65,78,80.
Additional applications for GENIs.
GENIs have also been used to address fundamental questions regarding NM release. For example, two studies used high-resolution imaging to measure the release and diffusion of ACh and monoamines in mouse brain slices. High-resolution imaging achieves sub-diffraction-limit resolution (below 200 nm) on the basis of cooperative capture of many photons165. By combining such imaging with ACh, NA, 5-HT and DA sensors, ACh and monoamines were found to diffuse from their respective releasing sites over a distance of approximately 0.75 μm; however, when release sites are packed closely and activate simultaneously, the NMs can diffuse over a larger space, giving rise to volume transmission, suggesting that nonvolume transmission may be an important mode of action for NMs166. Similar results were reported by a separate group70.
Besides understanding physiological roles of NTs and NMs, GENIs also present potential for facilitating drug discovery. For example, one group developed iNicSnFRs, a series of genetically encoded fluorescent sensors for detecting nicotine, and used these sensors to examine the pharmacokinetics of nicotinic drugs in the endoplasmic reticulum of multiple mammalian cell lines derived from human or mouse167. Combined with pharmacokinetic and pharmacodynamic simulations of human smoking, these experiments suggest that, in typical smokers, nicotinic ACh receptors in the endoplasmic reticulum are probably continuously activated at a level of more than 75%, leading to an upregulation of these receptors in the plasma membrane167. Another example is PsychLight, a recently developed fluorescent sensor based on the serotonin 2A receptor, a major target for classic hallucinogens, atypical antipsychotics and psychoplastogens (a relatively new class of compounds that rapidly promote structural and functional neural plasticity)168. The researchers predicted the hallucinogenic effects of different compounds, on the basis of their activation of PsychLight, and identified a non-hallucinogenic psychedelic analogue with both short-term and long-term antidepressant effects168. These examples illustrate the promise of GENI application in advancing our understanding of drug actions at the subcellular level, thereby facilitating drug discovery.
Lastly, it is worth noting that GENIs can also be used to study neuromodulation in non-traditional model organisms or non-human primates, as they can be delivered into the brain of virtually any species using viral infection. Indeed, GENIs have been used to study DA release in the high vocal centre (the song generation centre) of zebra finches during song learning148. Also, the GRABDA sensor was used to examine dopaminergic activity in the nucleus accumbens during motivated behaviours in monogamous prairie voles169. In addition, glutamate sensors have been used in macaque monkeys to examine the organization of the excitatory synaptic inputs that project different visual features onto the dendrites of neurons in the primary visual cortex170. Given their wide range of applications, GENIs provide a highly versatile toolkit that can be used to study both microscopic and macroscopic neuromodulation during various behaviours under physiological and pathological conditions, in various species.
Finishing remarks
Although the field of biosensors for measuring NTs and NMs is rapidly developing, there is still room to improve sensors’ sensitivities, selectivities, kinetics and pharmacological properties, as well as the techniques used to image the sensors and process the resulting data. To reach these goals, further efforts are needed from tool developers and end users.
Limitations and further improvement of GENIs
Further innovations by tool developers are needed to improve current methods for measuring NTs and NMs. First, although current GENIs provide high spatiotemporal resolution, overexpression of GENIs may cause buffering effects and interfere with endogenous signals. To minimize this effect, the expression level of GENIs should be controlled, although not so much that the SNR falls to suboptimal levels. Existing methods must be optimized to achieve even higher sensitivity. Second, relative to the vast number of extracellular signalling molecules identified to date (for example, the mammalian brain expresses hundreds of different neuropeptides), the number of existing biosensors is not nearly sufficient. Therefore, the repertoire of biosensors for measuring various chemicals (including neurolipids and neuropeptides) or other neuronal signals (such as mechanical tension) must be expanded.
Third, although microdialysis-based methods and electrochemical methods can measure small changes in NT or NM concentrations — even under basal conditions — most imaging probes rely on intensity and can therefore only qualitatively monitor relative changes in NT or NM levels. Therefore, a new class of sensors (such as ratiometric sensors) are needed to quantitatively measure NTs and NMs. Fourth, similar to most non-genetically encoded methods, GENIs only measure the presence of the NTs or NMs in the extracellular space, but not their downstream effects. Therefore, GENIs could be combined with electrophysiological recordings or Ca2+ imaging to study NT or NM release as well as their downstream signals.
Fifth, simultaneous measurements of multiple NTs and/or NMs will provide comprehensive information regarding communication in the brain. Indeed, NMs and NTs probably form parallel communication channels, and their temporal relationships could carry important information and only be revealed with simultaneous recordings; as such, sensors with high molecular specificity and non-overlapping spectra could be combined for use under the same conditions. Currently, the only red-shifted GENIs that have been developed for potential combination with green GENIs for dual-colour imaging are for glutamate67 and DA79,91. We anticipate that the colour spectrum of GENIs can be expanded even further, particularly into the near-infrared region171–173, for measuring multiple NTs and NMs simultaneously. Finally, given the nearly unlimited potential for sensor development, structure-guided high-throughput screening will probably increase the success rate. For example, obtaining an atomic-resolution structure of GENIs — particularly GPCR-b ased sensors — will help accelerate sensor development.
Future directions using GENIs
Having these new GENIs in hand will allow researchers to study the dynamic properties of NTs and NMs in unprecedented detail in cultured cells, tissues and living organisms. These experiments will provide important new insights, including different spatial and temporal information regarding NTs and NMs in health and disease; the mechanisms that underlie the release and modulation of non-classical NTs and NMs such as lipids, nucleotides and neuropeptides; and the communication between neurons, as well as communication between neurons and non-neuronal cell types such as glial cells, immune cells, endothelial cells and muscle cells. Furthermore, given that GPCRs are the most extensively studied family of drug targets, GRAB sensors may provide tools for screening drugs and for providing in vivo validation.
Building on the highly fruitful collaborations between sensor developers and end users will be essential for driving the development and optimization of new sensors. Nevertheless, it is also important to note that GENIs are just one of many groundbreaking tools and techniques developed in recent years. Other notable techniques, reviewed recently elsewhere174, include other optical sensors (such as voltage sensors, pH sensors, Ca2+ sensors, cAMP sensors and protein kinase A activation sensors), imaging set-ups, optogenetic and chemogenetic tools, CRISPR–Cas9-mediated gene editing, genetic models and pharmacological methods. These robust tools can be combined to interpret the data obtained using GENIs and to dissect chemical signalling in complex systems in health and disease.
Supplementary Material
Acknowledgements
The authors thank all scientists whose studies were reviewed in this paper, and apologize to those whose work was not cited owing to space limitations. The authors thank the Li laboratory members for fruitful discussions. This research was supported by the Beijing Municipal Science & Technology Commission (Z181100001318002 and Z181100001518004), the National Natural Science Foundation of China (81821092), the National Key Research and Development Program of China (2020YFE0204000), the Feng Foundation of Biomedical Research, the Peking-Tsinghua Center for Life Sciences and the State Key Laboratory of Membrane Biology at Peking University School of Life Sciences (Y.L.); the US National Institutes of Health (NIH) Brain Research Through Advancing Innovative Neurotechnologies (BRAIN) Initiative (NS103558; Y.L. and D.L.); the NIH (R01MH101377, 1R01HD092596 and U19NS107616; D.L.); and the Boehringer Ingelheim-Peking University Postdoctoral Program (Z.W.).
Glossary
- Gliosis
The hypertrophy of glial cells
- Voltammetry
An electrochemical method used to measure the concentration of neurochemicals by detecting the oxidation and reduction processes; signals are calculated in terms of applied potential
- Amperometry
An electrochemical method used to measure the concentration of neurochemicals by detecting the oxidation and reduction processes; signals are determined at a fixed voltage
- Cyclic voltammetry
A voltammetric method in which the current is measured while a linearly cycled potential is swept over the range of interest
- Faradaic currents
Currents generated by the reduction or oxidation of a chemical substance at an electrode
- Half-maximal effective concentration
(EC50). The concentration of a chemical (for example, dopamine (DA)) which induces a response halfway between the baseline and the maximum
- Dynamic range
The ratio between the largest signal and the lowest one induced by neurochemicals
- Site-saturation mutagenesis
A powerful mutagenesis strategy for protein engineering and directed evolution, which allows the substitution of predetermined protein sites against all 20 possible amino acids at once
- ΔF/F0
A commonly used equation to quantify the fluorescent intensity changes of fluorescent indicators, in which F is the signal trace from each detector and F0 is the fluorescence baseline
- Ring neurons
Named for their circumferential ring-like axonal arborisation patterns that form several circular laminae in the anterior shell of the ellipsoid body in Drosophila brain
Footnotes
Competing interests
Y.L. is listed as an inventor on a pending patent application filed by Peking University (international patent no. PCT/CN2018/107533), the value of which might be affected by this publication. The remaining authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at https://doi.org/10.1038/s41583-022-00577-6.
References
- 1.Luo L Principles of Neurobiology (Garland Science, 2020). [Google Scholar]
- 2.Sudhof TC Neurotransmitter release. Handb. Exp. Pharmacol 10.1007/978-3-540-74805-2_1 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Nadim F & Bucher D Neuromodulation of neurons and synapses. Curr. Opin. Neurobiol 29, 48–56 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marder E Neuromodulation of neuronal circuits: back to the future. Neuron 76, 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lovinger DM Neurotransmitter roles in synaptic modulation, plasticity and learning in the dorsal striatum. Neuropharmacology 58, 951–961 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma S, Hangya B, Leonard CS, Wisden W & Gundlach AL Dual-transmitter systems regulating arousal, attention, learning and memory. Neurosci. Biobehav. Rev 85, 21–33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarter M, Bruno JP & Parikh V Abnormal neurotransmitter release underlying behavioural and cognitive disorders: toward concepts of dynamic and function-specific dysregulation. Neuropsychopharmacology 32, 1452–1461 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Lotharius J & Brundin P Pathogenesis of Parkinson’s disease: dopamine, vesicles and α-synuclein. Nat. Rev. Neurosci 3, 932–942 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Higley MJ & Picciotto MR Neuromodulation by acetylcholine: examples from schizophrenia and depression. Curr. Opin. Neurobiol 29, 88–95 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Chiara G et al. Dopamine and drug addiction: the nucleus accumbens shell connection. Neuropharmacology 47 (Suppl 1), 227–241 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Ferreira-Vieira TH, Guimaraes IM, Silva FR & Ribeiro FM Alzheimer’s disease: targeting the cholinergic system. Curr. Neuropharmacol 14, 101–115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robertson M Biology in the 1980s, plus or minus a decade. Nature 285, 358–359 (1980). [DOI] [PubMed] [Google Scholar]
- 13.Ting JT & Phillips PE Neurotransmitter release. Wiley Ency. Chem. Biol 10.1002/9780470048672.wecb385 (2007). [DOI] [Google Scholar]
- 14.Sakmann B Single-Channel Recording (Springer Science & Business Media, 2013). [Google Scholar]
- 15.Del Castillo J & Katz B Quantal components of the end-plate potential. J. Physiol 124, 560–573 (1954). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bito L, Davson H, Levin E, Murray M & Snider N The concentrations of free amino acids and other electrolytes in cerebrospinal fluid, in vivo dialysate of brain, and blood plasma of the dog. J. Neurochem 13, 1057–1067 (1966). [DOI] [PubMed] [Google Scholar]
- 17.Buck K, Voehringer P & Ferger B Rapid analysis of GABA and glutamate in microdialysis samples using high performance liquid chromatography and tandem mass spectrometry. J. Neurosci. Methods 182, 78–84 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Hogan BL, Lunte SM, Stobaugh JF & Lunte CE On-line coupling of in vivo microdialysis sampling with capillary electrophoresis. Anal. Chem 66, 596–602 (1994). [DOI] [PubMed] [Google Scholar]
- 19.Sun X, Deng J, Liu T & Borjigin J Circadian 5-HT production regulated by adrenergic signaling. Proc. Natl Acad. Sci. USA 99, 4686–4691 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borjigin J & Liu T Application of long-term microdialysis in circadian rhythm research. Pharmacol. Biochem. Behav 90, 148–155 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Li C, Yan ZY, Yang J & Chen H Simultaneous monitoring multiple neurotransmitters and neuromodulators during cerebral ischemia/reperfusion in rats by microdialysis and capillary electrophoresis. J. Neurosci. Methods 189, 162–168 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Kao CY, Anderzhanova E, Asara JM, Wotjak CT & Turck CW NextGen brain microdialysis: applying modern metabolomics technology to the analysis of extracellular fluid in the central nervous system. Mol. Neuropsychiatry 1, 60–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westerhout J, Ploeger B, Smeets J, Danhof M & de Lange EC Physiologically based pharmacokinetic modeling to investigate regional brain distribution kinetics in rats. AAPS J. 14, 543–553 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petit-Pierre G et al. In vivo neurochemical measurements in cerebral tissues using a droplet-based monitoring system. Nat. Commun 8, 1239 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson CJ, Venton BJ & Kennedy RT In vivo measurements of neurotransmitters by microdialysis sampling. Anal. Chem 78, 1391–1399 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Benveniste H & Diemer NH Cellular reactions to implantation of a microdialysis tube in the rat hippocampus. Acta Neuropathol. 74, 234–238 (1987). [DOI] [PubMed] [Google Scholar]
- 27.Hascup ER et al. Histological studies of the effects of chronic implantation of ceramic-based microelectrode arrays and microdialysis probes in rat prefrontal cortex. Brain Res. 1291, 12–20 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L & Sweedler JV Peptides in the brain: mass spectrometry-based measurement approaches and challenges. Annu. Rev. Anal. Chem 1, 451–483 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Bequet F et al. CB1 receptor-mediated control of the release of endocannabinoids (as assessed by microdialysis coupled with LC/MS) in the rat hypothalamus. Eur. J. Neurosci 26, 3458–3464 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Buczynski MW & Parsons LH Quantification of brain endocannabinoid levels: methods, interpretations and pitfalls. Br. J. Pharmacol 160, 423–442 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shariatgorji M et al. Comprehensive mapping of neurotransmitter networks by MALDI-MS imaging. Nat. Methods 16, 1021–1028 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Adams RN Probing brain chemistry with electroanalytical techniques. Anal. Chem 48, 1126A–1138A (1976). [DOI] [PubMed] [Google Scholar]
- 33.Bucher ES & Wightman RM Electrochemical analysis of neurotransmitters. Annu. Rev. Anal. Chem 8, 239–261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wightman RM Detection technologies. Probing cellular chemistry in biological systems with microelectrodes. Science 311, 1570–1574 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Ganesana M, Lee ST, Wang Y & Venton BJ Analytical techniques in neuroscience: recent advances in imaging, separation, and electrochemical methods. Anal. Chem 89, 314–341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puthongkham P & Venton BJ Recent advances in fast-scan cyclic voltammetry. Analyst 145, 1087–1102 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venton BJ & Cao Q Fundamentals of fast-scan cyclic voltammetry for dopamine detection. Analyst 145, 1158–1168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vickrey TL, Condron B & Venton BJ Detection of endogenous dopamine changes in Drosophila melanogaster using fast-scan cyclic voltammetry. Anal. Chem 81, 9306–9313 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bang D et al. Sub-second dopamine and serotonin signaling in human striatum during perceptual decision-making. Neuron 108, 999–1010.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates the use of FSCV-based methods to measure dopamine and 5-HT dynamics in human participants during a visual motion-discrimination task.
- 40.Kishida KT et al. Subsecond dopamine fluctuations in human striatum encode superposed error signals about actual and counterfactual reward. Proc. Natl Acad. Sci. USA 113, 200–205 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moran RJ et al. The protective action encoding of serotonin transients in the human brain. Neuropsychopharmacology 43, 1425–1435 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ou Y, Buchanan AM, Witt CE & Hashemi P Frontiers in electrochemical sensors for neurotransmitter detection: towards measuring neurotransmitters as chemical diagnostics for brain disorders. Anal. Methods 11, 2738–2755 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Llaudet E, Hatz S, Droniou M & Dale N Microelectrode biosensor for real-time measurement of ATP in biological tissue. Anal. Chem 77, 3267–3273 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Mitchell KM Acetylcholine and choline amperometric enzyme sensors characterized in vitro and in vivo. Anal. Chem 76, 1098–1106 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Frenguelli BG, Llaudet E & Dale N High-resolution real-time recording with microelectrode biosensors reveals novel aspects of adenosine release during hypoxia in rat hippocampal slices. J. Neurochem 86, 1506–1515 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Disney AA & Higley MJ Diverse spatiotemporal scales of cholinergic signaling in the neocortex. J. Neurosci 40, 720–725 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gubernator NG et al. Fluorescent false neurotransmitters visualize dopamine release from individual presynaptic terminals. Science 324, 1441–1444 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuominen L, Nummenmaa L, Keltikangas-Jarvinen L, Raitakari O & Hietala J Mapping neurotransmitter networks with PET: an example on serotonin and opioid systems. Hum. Brain Mapp 35, 1875–1884 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ghosh S, Harvey P, Simon JC & Jasanoff A Probing the brain with molecular fMRI. Curr. Opin. Neurobiol 50, 201–210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen QT et al. An in vivo biosensor for neurotransmitter release and in situ receptor activity. Nat. Neurosci 13, 127–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reports the design of CNiFERs through activation of M1 muscarinic receptors for detecting ACh release in living rodents.
- 51.Beyene AG et al. Imaging striatal dopamine release using a nongenetically encoded near infrared fluorescent catecholamine nanosensor. Sci. Adv 5, eaaw3108 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reports the design of a synthetic catecholamine nanosensor with fluorescent emission in the near-infrared range (1,000–1,300 nm), named nIRCat, which was compatible with DA pharmacology and could be used to measure DA release in brain tissue.
- 52.Jeong S et al. High-throughput evolution of near-infrared serotonin nanosensors. Sci. Adv 5, eaay3771 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin MZ & Schnitzer MJ Genetically encoded indicators of neuronal activity. Nat. Neurosci 19, 1142–1153 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miesenbock G, De Angelis DA & Rothman JE Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394, 192–195 (1998). [DOI] [PubMed] [Google Scholar]
- 55.Li Y & Tsien RW pHTomato, a red, genetically encoded indicator that enables multiplex interrogation of synaptic activity. Nat. Neurosci 15, 1047–1053 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu A et al. pHmScarlet is a pH-sensitive red fluorescent protein to monitor exocytosis docking and fusion steps. Nat. Commun 12, 1413 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ding K et al. Imaging neuropeptide release at synapses with a genetically engineered reporter. eLife 8, e46421 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rao S, Lang C, Levitan ES & Deitcher DL Visualization of neuropeptide expression, transport, and exocytosis in Drosophila melanogaster. J. Neurobiol 49, 159–172 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Xia X, Lessmann V & Martin TF Imaging of evoked dense-core-vesicle exocytosis in hippocampal neurons reveals long latencies and kiss-and-run fusion events. J. Cell Sci 122, 75–82 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wong MY, Cavolo SL & Levitan ES Synaptic neuropeptide release by dynamin-dependent partial release from circulating vesicles. Mol. Biol. Cell 26, 2466–2474 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dominguez N, van Weering JRT, Borges R, Toonen RFG & Verhage M Dense-core vesicle biogenesis and exocytosis in neurons lacking chromogranins A and B. J. Neurochem 144, 241–254 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen Y, Nasu Y, Shkolnikov I, Kim A & Campbell RE Engineering genetically encoded fluorescent indicators for imaging of neuronal activity: progress and prospects. Neurosci. Res 152, 3–14 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Knopfel T & Song C Optical voltage imaging in neurons: moving from technology development to practical tool. Nat. Rev. Neurosci 20, 719–727 (2019). [DOI] [PubMed] [Google Scholar]
- 64.Okumoto S et al. Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors. Proc. Natl Acad. Sci. USA 102, 8740–8745 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marvin JS et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 10, 162–170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reports the design of a PBP-based genetically encoded glutamate sensor, iGluSnFR, and the validation of its utility for visualizing glutamate release in vitro and in vivo.
- 66.Marvin JS et al. Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nat. Methods 15, 936–939 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu J et al. Genetically encoded glutamate indicators with altered color and topology. ACS Chem. Biol 13, 1832–1837 (2018). [DOI] [PubMed] [Google Scholar]
- 68.Marvin JS et al. A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nat. Methods 16, 763–770 (2019). [DOI] [PubMed] [Google Scholar]
- 69.Lobas MA et al. A genetically encoded single-wavelength sensor for imaging cytosolic and cell surface ATP. Nat. Commun 10, 711 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Borden PM et al. A fast genetically encoded fluorescent sensor for faithful in vivo acetylcholine detection in mice, fish, worms and flies. Preprint at bioRxiv 10.1101/2020.02.07.939504 (2020). [DOI] [Google Scholar]
- 71.Unger EK et al. Directed evolution of a selective and sensitive serotonin sensor via machine learning. Cell 183, 1986–2002.e26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study describes the development and application of a binding-pocket redesign strategy, guided by machine learning, to create a fluorescent 5-HT sensor, iSeroSnFR, which enabled optical detection of 5-HT release in freely behaving animals.
- 72.Vilardaga JP, Bunemann M, Krasel C, Castro M & Lohse MJ Measurement of the millisecond activation switch of G protein-coupled receptors in living cells. Nat. Biotechnol 21, 807–812 (2003). [DOI] [PubMed] [Google Scholar]
- 73.Hoffmann C et al. A FlAsH-based FRET approach to determine G protein-coupled receptor activation in living cells. Nat. Methods 2, 171–176 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Baird GS, Zacharias DA & Tsien RY Circular permutation and receptor insertion within green fluorescent proteins. Proc. Natl Acad. Sci. USA 96, 11241–11246 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kostyuk AI, Demidovich AD, Kotova DA, Belousov VV & Bilan DS Circularly permuted fluorescent protein-based indicators: history, principles, and classification. Int. J. Mol. Sci 20, 4200 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jing M et al. A genetically encoded fluorescent acetylcholine indicator for in vitro and in vivo studies. Nat. Biotechnol 36, 726–737 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study describes the design of a genetically encoded GPCR activation-based ACh fluorescent sensor, GRABACh, and the validation of its utility for visualizing ACh release in vitro and in vivo.
- 77.Jing M et al. An optimized acetylcholine sensor for monitoring in vivo cholinergic activity. Nat. Methods 17, 1139–1146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun F et al. A genetically encoded fluorescent sensor enables rapid and specific detection of dopamine in flies, fish, and mice. Cell 174, 481–496.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reports the development of a first-generation 2R-based dopamine sensor, GRABDA, which enabled fast, sensitive DA detection with molecular and cellular specificity in multiple organisms and during complex behaviours.
- 79.Sun F et al. Next-generation GRAB sensors for monitoring dopaminergic activity in vivo. Nat. Methods 17, 1156–1166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reports on developed red-fluorescent GPCR activation-based dopamine (rGRABDA) sensors and optimized versions of greenfluorescent GRABDA sensors. The new sensors reveal compartmental DA release in flies and report mesoaccumbens dopaminergic activity during sexual behaviour in freely behaving mice.
- 80.Feng J et al. A genetically encoded fluorescent sensor for rapid and specific in vivo detection of norepinephrine. Neuron 102, 745–761.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kruse AC et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature 504, 101–106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hilger D, Masureel M & Kobilka BK Structure and dynamics of GPCR signaling complexes. Nat. Struct. Mol. Biol 25, 4–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weis WI & Kobilka BK The molecular basis of G protein-coupled receptor activation. Annu. Rev. Biochem 87, 897–919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC & Waldo GS Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol 24, 79–88 (2006). [DOI] [PubMed] [Google Scholar]
- 85.St-Pierre F et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat. Neurosci 17, 884–889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bajar BT et al. Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci. Rep 6, 20889 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wan J et al. A genetically encoded sensor for measuring serotonin dynamics. Nat. Neurosci 24,746–752 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dong A et al. A fluorescent sensor for spatiotemporally resolved imaging of endocannabinoid dynamics in vivo. Nat. Biotechnol 10.1038/s41587-021-01074-4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Peng W et al. Regulation of sleep homeostasis mediator adenosine by basal forebrain glutamatergic neurons. Science 369, eabb05566 (2020). [DOI] [PubMed] [Google Scholar]; This study describes the development and application of a new genetically encoded adenosine sensor (GRABAdo) to monitor adenosine dynamics with fibre photometry recordings during sleep–wake cycles in mice.
- 90.Wu Z et al. A sensitive GRAB sensor for detecting extracellular ATP in vitro and in vivo. Neuron 110, 770–782.e5 (2022). [DOI] [PubMed] [Google Scholar]
- 91.Patriarchi T et al. An expanded palette of dopamine sensors for multiplex imaging in vivo. Nat. Methods 17, 1147–1155 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Patriarchi T et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360, eaat4422 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reports the development of D1R-based and other dopamine receptor-based DA sensors, dLight1 sensors, to visualize spatial and temporal release of DA in rodents.
- 93.Labouesse MA, Cola RB & Patriarchi T GPCR-based dopamine sensors — a detailed guide to inform sensor choice for in vivo imaging. Int. J. Mol. Sci 21, 8048 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Labouesse MA & Patriarchi T A versatile GPCR toolkit to track in vivo neuromodulation: not a one-size-fits-all sensor. Neuropsychopharmacology 46, 2043–2047 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leopold AV, Shcherbakova DM & Verkhusha VV Fluorescent biosensors for neurotransmission and neuromodulation: engineering and applications. Front. Cell Neurosci 13, 474 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sabatini BL & Tian L Imaging neurotransmitter and neuromodulator dynamics in vivo with genetically encoded indicators. Neuron 108, 17–32 (2020). [DOI] [PubMed] [Google Scholar]; This review discusses the development, optimization and applications of optical approaches to monitor NT and NM dynamics in the brain using GENIs.
- 97.Wu Z, Feng J, Jing M & Li Y G protein-assisted optimization of GPCR-activation based (GRAB) sensors. Proc. Spie 10.1117/12.2514631 (2019). [DOI] [Google Scholar]
- 98.Kroning K & Wang W Designing a single proteinchain reporter for opioid detection at a cellular resolution. Angew. Chem. Int. Ed. Engl 133, 13470–13477 (2021). [DOI] [PubMed] [Google Scholar]
- 99.Barnea G et al. The genetic design of signalling cascades to record receptor activation. Proc. Natl Acad. Sci. USA 105, 64–69 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Inagaki HK et al. Visualizing neuromodulation in vivo: TANGO-mapping of dopamine signalling reveals appetite control of sugar sensing. Cell 148, 583–595 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee D et al. Temporally precise labeling and control of neuromodulatory circuits in the mammalian brain. Nat. Methods 14, 495–503 (2017). [DOI] [PubMed] [Google Scholar]
- 102.Kim MW et al. Time-gated detection of protein–protein interactions with transcriptional readout. eLife 6, e30233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang WH et al. Monitoring hippocampal glycine with the computationally designed optical sensor GlyFS. Nat. Chem. Biol 14, 861–869 (2018). [DOI] [PubMed] [Google Scholar]
- 104.Zhou X, Mehta S & Zhang J Genetically encodable fluorescent and bioluminescent biosensors light up signaling networks. Trends Biochem. Sci 45, 889–905 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Petersen ED et al. Bioluminescent genetically encoded glutamate indicator for molecular imaging of neuronal activity. Preprint at bioRxiv 10.1101/2021.06.16.448690 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Neves SR, Ram PT & Iyengar R G protein pathways. Science 296, 1636–1639 (2002). [DOI] [PubMed] [Google Scholar]
- 107.Luttrell LM & Lefkowitz RJ The role of β-arrestins in the termination and transduction of G-protein-coupled receptor signals. J. Cell Sci 115, 455–465 (2002). [DOI] [PubMed] [Google Scholar]
- 108.Liu C, Goel P & Kaeser PS Spatial and temporal scales of dopamine transmission. Nat. Rev. Neurosci 22, 345–358 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Y, DeMarco EM, Witzel LS & Keighron JD A selected review of recent advances in the study of neuronal circuits using fiber photometry. Pharmacol. Biochem. Behav 201, 173113 (2021). [DOI] [PubMed] [Google Scholar]
- 110.Kjaerby C et al. Dynamic fluctuations of the locus coeruleus–norepinephrine system underlie sleep state transitions. Preprint at bioRxiv 10.1101/2020.09.01.274977 (2020). [DOI] [Google Scholar]
- 111.Mayer FP et al. There’s no place like home? Return to the home cage triggers dopamine release in the mouse nucleus accumbens. Neurochem. Int 142, 104894 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang M et al. The SC–SNc pathway boosts appetitive locomotion in predatory hunting. Preprint at bioRxiv 10.1101/2020.11.23.395004 (2020). [DOI] [Google Scholar]
- 113.Augustine V et al. Temporally and spatially distinct thirst satiation signals. Neuron 103, 242–249.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dai B, Sun F, Kuang A, Li Y & Lin D Dopamine release in nucleus accumbens core during social behaviors in mice. Preprint at bioRxiv 10.1101/2021.06.22.449478 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mohebi A et al. Dissociable dopamine dynamics for learning and motivation. Nature 570, 65–70 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.de Jong JW et al. A neural circuit mechanism for encoding aversive stimuli in the mesolimbic dopamine system. Neuron 101, 133–151.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lutas A et al. State-specific gating of salient cues by midbrain dopaminergic input to basal amygdala. Nat. Neurosci 22, 1820–1833 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim HR et al. A unified framework for dopamine signals across timescales. Cell 183, 1600–1616.e25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yuan L, Dou YN & Sun YG Topography of reward and aversion encoding in the mesolimbic dopaminergic system. J. Neurosci 39, 6472–6481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lin R et al. The raphe dopamine system controls the expression of incentive memory. Neuron 106, 498–514 e498 (2020). [DOI] [PubMed] [Google Scholar]
- 121.Sturgill JF et al. Basal forebrain-derived acetylcholine encodes valence-free reinforcement prediction error. Preprint at bioRxiv 10.1101/2020.02.17.953141 (2020). [DOI] [Google Scholar]
- 122.Gallo EF et al. Dopamine D2 receptors modulate the cholinergic pause and inhibitory learning. Mol. Psychiatry 10.1038/s41380-021-01364-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee SJ et al. Cell-type-specific asynchronous modulation of PKA by dopamine in learning. Nature 590, 451–456 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Corre J et al. Dopamine neurons projecting to medial shell of the nucleus accumbens drive heroin reinforcement. eLife 7, e39945 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu Y et al. The mesolimbic dopamine activity signatures of relapse to alcohol-seeking. J. Neurosci 40, 6409–6427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Alhadeff AL et al. Natural and drug rewards engage distinct pathways that converge on coordinated hypothalamic and reward circuits. Neuron 103, 891–908.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lefevre EM et al. Interruption of continuous opioid exposure exacerbates drug-evoked adaptations in the mesolimbic dopamine system. Neuropsychopharmacology 45, 1781–1792 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mazzone CM et al. High-fat food biases hypothalamic and mesolimbic expression of consummatory drives. Nat. Neurosci 23, 1253–1266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Reid WH, Balis GU & Sutton BJ The Treatment of Psychiatric Disorders (Routledge, 2013). [Google Scholar]
- 130.Schmack K, Bosc M, Ott T, Sturgill JF & Kepecs A Striatal dopamine mediates hallucination-like perception in mice. Science 372, eabf4740 (2021). [DOI] [PubMed] [Google Scholar]
- 131.Sych Y, Chernysheva M, Sumanovski LT & Helmchen F High-density multi-fiber photometry for studying large-scale brain circuit dynamics. Nat. Methods 16, 553–560 (2019). [DOI] [PubMed] [Google Scholar]
- 132.Burton A et al. Wireless, battery-free subdermally implantable photometry systems for chronic recording of neural dynamics. Proc. Natl Acad. Sci. USA 117, 2835–2845 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pisano F et al. Depth-resolved fiber photometry with a single tapered optical fiber implant. Nat. Methods 16, 1185–1192 (2019). [DOI] [PubMed] [Google Scholar]
- 134.Ghosh KK et al. Miniaturized integration of a fluorescence microscope. Nat. Methods 8, 871–878 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shemesh OA et al. Precision calcium imaging of dense neural populations via a cell-body-targeted calcium indicator. Neuron 107, 470–486.e11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen Y et al. Soma-targeted imaging of neural circuits by ribosome tethering. Neuron 107, 454–469.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Broussard GJ et al. In vivo measurement of afferent activity with axon-specific calcium imaging. Nat. Neurosci 21, 1272–1280 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Adam Y et al. Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature 569, 413–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Piatkevich KD et al. Population imaging of neural activity in awake behaving mice. Nature 574, 413–417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Villette V et al. Ultrafast two-photon imaging of a high-gain voltage indicator in awake behaving mice. Cell 179, 1590–1608.e23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Abdelfattah AS et al. Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science 365, 699–704 (2019). [DOI] [PubMed] [Google Scholar]
- 142.Zong W et al. Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice. Nat. Methods 14, 713–719 (2017). [DOI] [PubMed] [Google Scholar]
- 143.Zong W et al. Miniature two-photon microscopy for enlarged field-of-view, multi-plane and long-term brain imaging. Nat. Methods 18, 46–49 (2021). [DOI] [PubMed] [Google Scholar]
- 144.Cardin JA, Crair MC & Higley MJ Mesoscopic imaging: shining a wide light on large-scale neural dynamics. Neuron 108, 33–43 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xie Y et al. Resolution of high-frequency mesoscale intracortical maps using the genetically encoded glutamate sensor iGluSnFR. J. Neurosci 36, 1261–1272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lohani S et al. Dual color mesoscopic imaging reveals spatiotemporally heterogeneous coordination of cholinergic and neocortical activity. Preprint at bioRxiv 10.1101/2020.12.09.418632 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sethuramanujam S et al. Rapid multi-directed cholinergic transmission in the central nervous system. Nat. Commun 12, 1374 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tanaka M, Sun F, Li Y & Mooney R A mesocortical dopamine circuit enables the cultural transmission of vocal behaviour. Nature 563, 117–120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Parker PD et al. Non-canonical glutamate signalling in a genetic model of migraine with aura. Neuron 109, 611–628.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Oe Y et al. Distinct temporal integration of noradrenaline signaling by astrocytic second messengers during vigilance. Nat. Commun 11, 471 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Handler A et al. Distinct dopamine receptor pathways underlie the temporal sensitivity of associative learning. Cell 178, 60–75.e19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Moran AK, Eiting TP & Wachowiak M Dynamics of glutamatergic drive underlie diverse responses of olfactory bulb outputs in vivo. eNeuro 8, ENEURO.0110–21.2021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Farrell JS et al. In vivo endocannabinoid dynamics at the timescale of physiological and pathological neural activity. Neuron 109, 2398–2403.e4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Barson D et al. Simultaneous mesoscopic and two-photon imaging of neuronal activity in cortical circuits. Nat. Methods 17, 107–113 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhou M et al. Suppression of GABAergic neurons through D2-like receptor secures efficient conditioning in Drosophila aversive olfactory learning. Proc. Natl Acad. Sci. USA 116, 5118–5125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Santi PA Light sheet fluorescence microscopy: a review. J. Histochem. Cytochem 59, 129–138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liang X et al. Morning and evening circadian pacemakers independently drive premotor centers via a specific dopamine relay. Neuron 102, 843–857.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Truong TV, Supatto W, Koos DS, Choi JM & Fraser SE Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat. Methods 8, 757–760 (2011). [DOI] [PubMed] [Google Scholar]
- 159.Mahou P, Vermot J, Beaurepaire E & Supatto W Multicolor two-photon light-sheet microscopy. Nat. Methods 11, 600–601 (2014). [DOI] [PubMed] [Google Scholar]
- 160.Hillman EMC, Voleti V, Li W & Yu H Light-sheet microscopy in neuroscience. Annu. Rev. Neurosci 42, 295–313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kawashima T, Zwart MF, Yang CT, Mensh BD & Ahrens MB The serotonergic system tracks the outcomes of actions to mediate short-term motor learning. Cell 167, 933–946.e20 (2016). [DOI] [PubMed] [Google Scholar]
- 162.Bianco IH & Engert F Visuomotor transformations underlying hunting behavior in zebrafish. Curr. Biol 25, 831–846 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Cong L et al. Rapid whole brain imaging of neural activity in freely behaving larval zebrafish (Danio rerio). eLife 6, e28158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kim DH et al. Pan-neuronal calcium imaging with cellular resolution in freely swimming zebrafish. Nat. Methods 14, 1107–1114 (2017). [DOI] [PubMed] [Google Scholar]
- 165.Lin L, Gupta S, Zheng WS, Si K & Zhu JJ Genetically encoded sensors enable micro-and nano-scopic decoding of transmission in healthy and diseased brains. Mol. Psychiatry 26, 443–455 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zhu PK et al. Nanoscopic visualization of restricted nonvolume cholinergic and monoaminergic transmission with genetically encoded sensors. Nano Lett. 20, 4073–4083 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Shivange AV et al. Determining the pharmacokinetics of nicotinic drugs in the endoplasmic reticulum using biosensors. J. Gen. Physiol 151, 738–757 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Dong C et al. Psychedelic-inspired drug discovery using an engineered biosensor. Cell 184, 2779–2792 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Burt E Developing Novel Methods to Investigate Real-time In Vivo Dopamine Dynamics in the Monogamous Prairie Vole. Thesis, Univ. of Colorado Boulder Libraries (2019). [Google Scholar]
- 170.Ju N et al. Spatiotemporal functional organization of excitatory synaptic inputs onto macaque V1 neurons. Nat. Commun 11, 697 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates the use of GENIs in non-human primates. The authors used two-photon imaging to map the excitatory synaptic inputs on dendrites of individual V1 superficial layer neurons in awake monkeys.
- 171.Kim B, Kim H, Kim S & Hwang YR A brief review of non-invasive brain imaging technologies and the near-infrared optical bioimaging. Appl. Microsc 51, 9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Qian Y et al. A genetically encoded near-infrared fluorescent calcium ion indicator. Nat. Methods 16, 171–174 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Deo C et al. The HaloTag as a general scaffold for far-red tunable chemigenetic indicators. Nat. Chem. Biol 17, 718–723 (2021). [DOI] [PubMed] [Google Scholar]
- 174.Vázquez-Guardado A, Yang Y, Bandodkar AJ & Rogers JA Recent advances in neurotechnologies with broad potential for neuroscience research. Nat. Neurosci 23, 1522–1536 (2020). [DOI] [PubMed] [Google Scholar]
- 175.Helassa N et al. Ultrafast glutamate sensors resolve high-frequency release at Schaffer collateral synapses. Proc. Natl Acad. Sci. USA 115, 5594–5599 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wu Z et al. A GRAB sensor reveals activity-dependent non-vesicular somatodendritic adenosine release. Preprint at bioRxiv 10.1101/2020.05.04.075564 (2020). [DOI] [Google Scholar]
- 177.Kitajima N et al. Real-time in vivo imaging of extracellular ATP in the brain with a hybrid-type fluorescent sensor. eLife 9, e57544 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Abraham AD et al. Release of endogenous dynorphin opioids in the prefrontal cortex disrupts cognition. Neuropsychopharmacology 46, 2330–2339 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Melzer S et al. Bombesin-like peptide recruits disinhibitory cortical circuits and enhances fear memories. Cell 184, 5622–5634.e25 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


