Abstract
To determine whether C-C chemokines play an important role in the phenotype switch of human immunodeficiency virus (HIV) from CCR5 to CXCR4 usage during the course of an infection in vivo, macrophage inflammatory protein (MIP)-1α-resistant variants were isolated from CCR5-tropic (R5) HIV-1 in vitro. The selected variants displayed reduced sensitivities to MIP-1α (fourfold) through CCR5-expressing CD4-HeLa/long terminal repeat–β-galactosidase (MAGI/CCR5) cells. The variants were also resistant to other natural ligands for CCR5, namely, MIP-1β (>4-fold) and RANTES (regulated upon activation, normal T-cell expressed and secreted) (6-fold). The env sequence analyses revealed that the variants had amino acid substitutions in V2 (valine 166 to methionine) and V3 (serine 303 to glycine), although the same V3 substitution appeared in virus passaged without MIP-1α. A single-round replication assay using a luciferase reporter HIV-1 strain pseudotyped with mutant envelopes confirmed that mutations in both V2 and V3 were necessary to confer the reduced sensitivity to MIP-1α, MIP-1β, and RANTES. However, the double mutant did not switch its chemokine receptor usage from CCR5 to CXCR4, indicating the altered recognition of CCR5 by this mutant. These results indicated that V2 combined with the V3 region of the CCR5-tropic HIV-1 envelope modulates the sensitivity of HIV-1 to C-C chemokines without altering the ability to use chemokine receptors.
The principal receptor for human immunodeficiency virus (HIV) is the CD4 molecule on T cells and monocytes/macrophages. HIV strains vary greatly in their ability to infect and replicate in CD4-positive T-cell lines or primary T lymphocytes and monocytes/macrophages, defined as T-cell-line-tropic (T-tropic) or macrophage-tropic (M-tropic) strains, respectively. M-tropic strains are often non-syncytium inducing and are commonly recovered from patients in early disease stages, while T-tropic strains are syncytium inducing and are prevalent in patients at advanced disease stages (6, 50), indicating the importance of the cell tropism and phenotypes of HIV strains to the pathogenesis of HIV. Since the discovery that chemokine receptors act as coreceptors for HIV entry, the chemokine receptor usage of each strain has been shown to determine the cell tropism; i.e., CXCR4 serves as the major coreceptor for T-tropic (called X4 or CXCR4-tropic) HIV-1 isolates and CCR5 serves as the major coreceptor for M-tropic (called R5 or CCR5-tropic) HIV-1 isolates (1, 8, 12, 17–19). Natural ligands for CCR5, including macrophage inflammatory protein (MIP)-1α, MIP-1β, and RANTES (regulated upon activation, normal T-cell expressed and secreted), are also shown to block R5 but not X4 HIV-1 infection (9, 17, 18). In some cases, disease progression in HIV-1-infected individuals is associated with a gain of CXCR4 usage (i.e., emergence of R5X4 or X4 virus) (10, 13) and with a loss of sensitivity to these C-C chemokines (26, 44), coinciding with the phenotypic change from M-tropism to T-tropism. These results suggest that the change of the chemokine receptor from CCR5 to CXCR4 may have a key role in the pathogenesis of HIV. However, it remains to be solved how HIV can acquire the ability to use CXCR4 during the course of infection. Although C-C chemokine production levels in HIV-1-infected individuals in different clinical stages vary (3, 36, 56), we hypothesized that the selective pressure by natural ligands for CCR5, including MIP-1α, MIP-1β, and RANTES, may lead to the evolution of HIV-1 variants in vivo. Several studies which have indicated that these C-C chemokines inhibit R5 virus but enhance X4 virus (14, 29) also support this hypothesis. To determine (i) whether X4 or R5X4 viruses emerge from R5 HIV by selective pressure of ligands for CCR5 and (ii) which envelope regions are responsible for the reduced sensitivity to C-C chemokines, MIP-1α-resistant mutants of the R5 HIV strain were selected in vitro. Our resistant variants which had a single amino acid substitution in both the V2 and the V3 region showed a reduced sensitivity to C-C chemokines and no change in coreceptor usage, indicating that HIV-1 variants with a reduced sensitivity to C-C chemokines can emerge without the alteration of chemokine receptor usage.
MATERIALS AND METHODS
Chemokines.
Saccharomyces cerevisiae-derived recombinant MIP-1α (yLD78β [38, 39]), kindly supplied by the Chemo-Sero-Therapeutics Research Institute (Kumamoto, Japan), was used for the selection of MIP-1α-resistant virus. Other chemokines, MIP-1β and RANTES, were purchased from R&D Systems (Minneapolis, Minn.).
Cells and culture conditions.
The CD4-positive T-cell lines MOLT-4#8, CEM, MT-2, and MT-4 were maintained in RPMI 1640 (Gibco BRL, Grand Island, N.Y.) supplemented with 10% heat-inactivated fetal bovine serum (Gibco BRL), 2 mM l-glutamine, and antibiotics. The HeLa-CD4/long terminal repeat (LTR)–β-galactosidase (β-Gal) cell line (28) was kindly provided by M. Emerman through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, and maintained in Dulbecco modified Eagle medium (Gibco BRL) supplemented with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, antibiotics, 0.1 mg of G418 (Gibco BRL) per ml, and 0.05 mg of hygromycin B (Wako, Osaka, Japan) per ml. COS-7 and 293T cells were also maintained in Dulbecco modified Eagle medium supplemented with 10% heat-inactivated fetal bovine serum and antibiotics. A human CD4-expressing glioma cell line, NP-2/CD4 (27), was maintained in Eagle's minimal essential medium (Gibco BRL) supplemented with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, penicillin, and streptomycin.
Construction of the CCR5 and CXCR4 expression vectors and transfection to the CD4-positive cell line.
The cDNA clones encoding human CCR5 and CXCR4 were obtained by PCR using a human primary lymphocyte cDNA as the template. PCR was performed using LA-Taq (Takara, Tokyo, Japan) (an initial 2 min at 95°C followed by 30 thermal cycles of 98°C for 10 s, 60°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min). The primers used were as follows: 5′-TGCACAGGGTGGAACAAGATGGATTATC-3′ and 5′-TAAGCCATGTGCACAACTCTGACTGGGTCA-3′ for the CCR5 gene and 5′-CCATGGAGGGGATCAGTATAT-3′ and 5′-CTGTGTTAGCTGGAGTGAAAACT-3′ for the CXCR4 gene. The amplified products were ligated into a TA cloning vector, pCR2.1 (Invitrogen, NV Leek, The Netherlands), and then designated pCR2-CCR5 and pCR2-CXCR4, respectively. The complete sequences of the products were verified with an automated DNA sequencer (ABI Prism 377; Applied Biosystems). The CCR5- or CXCR4-encoding fragment was then ligated into a pZeoSV2 expression vector (Invitrogen) using the HindIII and XhoI sites to give pZeoSV-CCR5 or pZeoSV-CXCR4, respectively.
HeLa-CD4/LTR–β-Gal cells were transfected with pZeoSV-CCR5 using the calcium phosphate method (Profection kit; Promega, Madison, Wis.). An R5 HIV-1-sensitive stable transfectant was cloned in the presence of 1.0 mg of G418 per ml, 0.5 mg of hygromycin per ml, and 0.5 mg of zeomycin (Invitrogen) per ml and designated MAGI/CCR5. The CCR5 expression level of MAGI/CCR5 was confirmed with the anti-CCR5 monoclonal antibody 2D7 (Pharmingen, San Diego, Calif.), by flow cytometry, using a FACScan (Becton Dickinson Immunocytometry Systems, San Jose, Calif.). The susceptibility of these cells to R5 HIV was determined by infecting them with the R5 HIV strain JR-FL. CCR5- and CXCR4-expressing NP-2/CD4 cells were also established by the transfection of the expression vectors pZeoSV-CCR5 and pZeoSV-CXCR4, respectively, and stable transfectants were selected in the presence of zeomycin. The expression levels of CCR5 and CXCR4 of the transfectants were confirmed using 2D7 and an anti-CXCR4 monoclonal antibody, 12G5 (R&D Systems), respectively, with a FACScan.
Retrovirus vector construction and transduction of a CD4-positive T-cell line with the CCR5 gene.
The cDNA encoding the CCR5 gene cloned in the pCR2 vector pCR2-CCR5 was ligated into pBluescript II KS(+) (Stratagene, La Jolla, Calif.) using the HindIII and XhoI sites to yield pKS-CCR5, and the CCR5 gene carrying the NotI-EcoRV fragment of pKS-CCR5 was then transferred to the NotI and SnaBI sites of the retrovirus vector pG1TKNeo to produce pG1TKNeo-CCR5. pG1TKNeo-CCR5 was transfected into the murine retrovirus packaging cell line PA317 by the calcium phosphate method, and a retrovirus-producing cell clone was chosen in the presence of G418 (0.8 mg/ml).
The MOLT-4#8 cell line was transduced with the CCR5 gene by coculturing with retrovirus-producing cells. Briefly, MOLT-4#8 cells were cocultured with irradiated retrovirus-producing cells for 2 days and then suspension cells were cultured in the presence of G418 (0.8 mg/ml). The expression levels of CCR5 in transduced cells were confirmed using the anti-CCR5 monoclonal antibody 2D7 with a FACScan. To clone CCR5-expressing cells highly sensitive to R5 HIV infection, limiting dilution was performed, and a clone which was able to induce ballooning after infection of JR-FL was selected for propagation and the selection of a resistant mutant, designated MOLT-4#8/CCR5.
Virus.
The M-tropic virus infectious clone pJR-FL was provided by Y. Koyanagi (unpublished data). The virus was recovered by the transfection of the plasmid into COS-7 cells using Lipofectamine (Gibco BRL) according to the manufacturer's protocol. The cell culture supernatant was further transferred to CCR5-transduced MOLT-4#8 cells (MOLT-4#8/CCR5), and then virus was recovered after 7 to 10 days of culture. Viral titers were determined using MAGI/CCR5 cells.
Isolation of the MIP-1α-resistant mutant from the R5 HIV JR-FL in vitro.
For the selection of MIP-1α-resistant virus, MOLT-4#8/CCR5 cells were treated with various concentrations of MIP-1α and then infected with JR-FL. After the virus was passaged in MOLT-4#8/CCR5 cells, MIP-1α was removed from the virus-infected cells and then the virus was recovered from the cell culture supernatant. The sensitivity of the virus resistant to MIP-1α was determined using MAGI/CCR5 cells as previously described (35). Briefly, MAGI/CCR5 cells (104/well) were seeded in 48-well flat-bottom plates (Iwaki, Chiba, Japan). The following day, the cells were incubated with various concentrations of C-C chemokines for 1 h and then the virus which gave 200 blue cells/well was added. Forty-eight hours after the virus exposure, cells were fixed and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). The blue cells in each well were counted under a microscope. All experiments were done in duplicate. JR-FL was also passaged in MOLT-4#8/CCR5 cells in the absence of MIP-1α for 3 months to exclude the effect of long-term culture.
DNA was also extracted from virus-infected cells using Instagene (Bio-Rad) according to the manufacturer's protocol and subjected to PCR using Taq polymerase. The amplified products were cloned into pCR2.1, and then the env regions in both the passaged and selected virus were sequenced using an ABI PRISM 377 automated DNA sequencer (Applied Biosystems).
Construction of mutant envelope expression vectors.
For the construction of mutant envelope expression vectors, we used pCXN2, which has a chicken β-actin promoter. Briefly, the JR-FL env region was cloned by PCR and ligated into pCR2.1, generating pCR2-FLenv wild type. The sequence of the amplified env region of JR-FL was confirmed using an ABI Prism 377 automated DNA sequencer. The EcoRI fragment of pCR2-FLenv containing the entire env region was ligated into pCXN2 to give pCXN-FLenv wild type. For the cloning of the V2 region of env, an NdeI-StuI fragment cloned by PCR with a valine-to-methionine substitution at codon 166 (V166M) was ligated into pCXN-FLenv, generating pCXN-FLenv V166M. For the cloning of the V3 region, a BglII-BglII fragment with a serine-to-glycine substitution at codon 303 (S303G) in the env region cloned by PCR was ligated into pCXN-FLenv, generating pCXN-FLenv S303G. A mutant envelope expression vector which contains both V166M and S303G was also constructed in the same manner, generating pCXN-FLenv V166M/S303G. The NL4-3 env region was also amplified and ligated into pCXN2, generating pCXN-NLenv in the same manner as pCXN-FLenv.
Determination of the chemokine sensitivity and chemokine receptor usage of each mutant clone by single-round replication assay.
Recombinant luciferase reporter virus stocks pseudotyped with various HIV-1 envelopes were generated by cotransfection of 293T cells with 10 μg of HIV-1NLLucΔBgl and 5 μg of pCXN2 plasmids expressing envelope from NL4-3 or JR-FL using the calcium phosphate method. At 3 days posttransfection, the cell culture supernatant was filtered (0.22-μm pore size) and used as pseudotype virus. For the determination of the sensitivities to chemokines, MAGI/CCR5 cells (104/well) in 48-well plates were first incubated with various concentrations of chemokines for 1 h at 37°C and then infected with the above-named luciferase reporter viruses. Three days after the infection, the cells were lysed with 100 μl of luciferase assay buffer (Promega). Luciferase activity was measured by adding 50 μl of the luciferase assay substrate (Promega) to 10 μl of lysate and reading the light activity in a luminometer detector (Lumat LB 9501/16; EG&G Berthold, Bad Wildbad, Germany). The light activity is reported in relative light units. The sensitivity of a chemokine was determined from the 50% inhibitory concentration (IC50) of the virus.
For the determination of chemokine receptor usage, CCR5- or CXCR4-expressing NP-2/CD4 cells were infected with the above-named luciferase reporter viruses pseudotyped with envelope containing the intended mutations. The luciferase activity of cell lysates was also measured 3 days after infection under the conditions mentioned above.
RESULTS
Selection of an MIP-1α-resistant virus from the M-tropic virus JR-FL.
For the isolation of the MIP-1α-resistant mutant from R5 HIV in vitro, CCR5-expressing MOLT-4#8 cells, designated MOLT-4#8/CCR5 cells, were established since this cell line expressed both CXCR4 and CCR5 and was sensitive to both X4 and R5 HIV and accompanied by prominent syncytia (data not shown). This cell line was, therefore, expected to enable a possible shift in coreceptor usage from CCR5 to CXCR4. An R5 HIV strain, JR-FL, which uses CCR5 but not CXCR4 as the coreceptor and is also known to be an M-tropic HIV strain, was used for the selection of MIP-1α-resistant virus. For the determination of the sensitivity, the CCR5-expressing MAGI cell line, designated MAGI/CCR5, was established since this cell line also expresses both CXCR4 and CCR5 and is sensitive to both X4 and R5 HIV (data not shown).
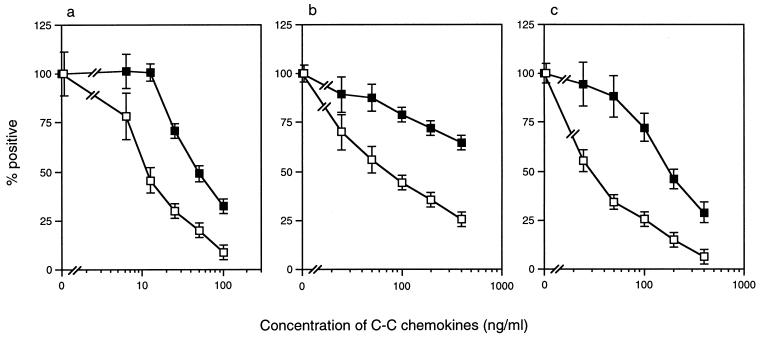
The cell culture supernatant of JR-FL-infected MOLT-4#8/CCR5 cells was first used for the selection of virus. The initial concentration of yeast-derived recombinant MIP-1α (yLD78β) for the selection process was 10 ng/ml, which inhibits JR-FL infection by 50% as determined by the MAGI/CCR5 assay. For the selection of an MIP-1α-resistant mutant, MOLT-4#8/CCR5 cells were first treated with MIP-1α and then infected with JR-FL. After observing the syncytium formation, virus was recovered for the next infection with increasing amounts of MIP-1α. JR-FL was also passaged in MOLT-4#8/CCR5 cells in the absence of MIP-1α to exclude the effect of long-term culture. After 3 months of passage with increasing concentrations of MIP-1α (up to 200 ng/ml), the virus was subjected to an MAGI cell assay using MAGI/CCR5 cells. After removing the residual MIP-1α from the culture by passaging infected cells for 3 days without MIP-1α, MAGI/CCR5 cells were infected with the supernatant from a selected culture after treatment with various concentrations of MIP-1α and then the blue cells were counted 2 days after infection (Fig. 1). The selected virus displayed reduced sensitivity (fourfold) to MIP-1α (Fig. 1a). The IC50 of the selected virus for MIP-1α was 45 ng/ml, while that of wild-type JR-FL was 11 ng/ml. This resistant mutant also displayed reduced sensitivity to MIP-1β (>4-fold) and RANTES (6-fold) (Fig. 1b and c).
FIG. 1.
Sensitivity of a selected mutant to C-C chemokines. MAGI/CCR5 cells were treated with various concentrations of MIP-1α (a), MIP-1β (b), and RANTES (c), followed by an inoculation of wild-type JR-FL (□) and virus selected for 3 months in the presence of increasing concentrations of MIP-1α (■). Blue cells were counted 2 days after infection. The y axis represents the percentages of positive blue cells counted. The x axis represents the concentrations of each C-C chemokine. All experiments were performed in duplicate. The data are expressed as means ± standard deviations.
Sequence analysis of the envelope region of the resistant mutant.
To determine which region is responsible for the reduced sensitivity of this resistant mutant to C-C chemokines, the V1-V2, V3, V4, C3, and C4 regions of the envelope were sequenced after the cloning of the PCR product of each region (Fig. 2) using DNAs from infected cells as templates. Ten to 22 clones from each PCR product were isolated and sequenced. Analyses of the env sequences of the resistant isolate using DNA from selected virus-infected cells revealed that the selected virus had a valine-to-methionine substitution at codon 166 (V166M) in the second variable (V2) region of the envelope (9 out of 10 clones) and also a serine-to-glycine substitution at codon 303 (S303G) in the third variable (V3) region (Fig. 2) (22 out of 22 clones).
FIG. 2.
V2 and V3 amino acid sequences from JR-FL-infected cells passaged and selected in MIP-1α for 3 months. Amplified products from infected MOLT-4#8/CCR5 cells passaged for 2 weeks and 3 months and selected by MIP-1α were cloned, and 8 to 22 clones from each sample were sequenced. The wild-type JR-FL amino acid sequences of V2 and V3 are shown in the top line. Numbers on the right are the numbers of clones with the sequence over the total number of clones tested. In each set of clones, the deduced amino acid sequences of the V2 (a) and V3 (b) regions were aligned by the single-amino-acid code. Dashes denote sequence identity.
Surprisingly, the virus passaged in MOLT-4#8/CCR5 cells for both 2 weeks and 3 months without MIP-1α showed the substitution at codon 303 (S303G) (10 out of 11 clones) but not the V166M substitution (none of 10 clones). This passaged virus without MIP-1α showed almost the same sensitivity to MIP-1α (data not shown), suggesting that the V3 region substitution in selected viruses was not due to a selective pressure of MIP-1α but that it was probably due to an adaptation in MOLT-4#8/CCR5 cells. Other regions, including V1, V4, C3, and C4, had no remarkable changes in the resistant mutant envelope (data not shown).
Determination of the chemokine sensitivity of each mutant clone by single-round replication assay.
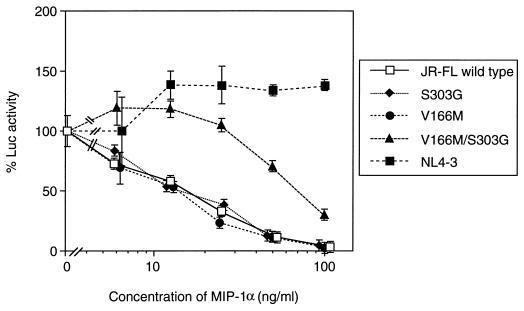
To confirm whether V2 and V3 region mutations in the envelope were responsible for the reduced sensitivity to MIP-1α, an envelope complementation assay was performed. First, the envelope expression vectors with intended mutations were constructed using the pCXN2 vector with the chicken β-actin promoter. Luciferase reporter HIV stocks pseudotyped with HIV envelopes were generated by cotransfecting 293T cells with HIV envelope expression vectors and a luciferase reporter HIV plasmid. Luciferase-reporter viruses were recovered pseudotyped with wild-type JR-FL, singly mutated with V166M in the V2 region, singly mutated with S303G in the V3 region, and doubly mutated with V166M and S303G. A luciferase reporter virus pseudotyped with the NL4-3 envelope, which uses CXCR4 as a coreceptor, was also recovered. As expected, the luciferase reporter HIV strain pseudotyped with the NL4-3 envelope was totally resistant to MIP-1α (Fig. 3). On the other hand, neither the strain with a single mutation at codon 166 (V166M) nor that with a single mutation at codon 303 (S303G) displayed a reduced sensitivity to MIP-1α (Fig. 3). Only the strain with a double mutation of both V166M and S303G (V166M/S303G) displayed a reduced sensitivity to MIP-1α (fourfold) to a level similar to that in the selected virus. This double mutant envelope was also responsible for reduced sensitivities to both MIP-1β and RANTES in the same assay (Table 1). The level of resistance to MIP-1α was similar to that of a selected variant (fivefold).
FIG. 3.
Sensitivities of luciferase reporter HIV strains pseudotyped with mutant envelope to MIP-1α. MAGI/CCR5 cells were infected with luciferase reporter HIVs pseudotyped with the JR-FL envelope wild type (□), an S303G mutant (⧫), a V166M mutant (●), an S303G/V166M mutant (▴), and the NL4-3 envelope (■). Luciferase (Luc) activity was measured 3 days after infection. The y axis represents the relative luciferase activity of each virus. All experiments were performed in duplicate. The data are expressed as means ± standard deviations.
TABLE 1.
Sensitivities of luciferase reporter HIV strains pseudotyped with mutant envelope to C-C chemokines
| Envelope | IC50 (ng/ml)a of:
|
||
|---|---|---|---|
| MIP-1α | MIP-1β | RANTES | |
| JR-FL | |||
| Wild type | 16 ± 0.5 | 260 ± 7.1 | 130 ± 9.2 |
| S303G | 14 ± 1.3 | 290 ± 42 | 160 ± 6.4 |
| V166M | 13 ± 1.2 | 220 ± 7.1 | 110 ± 0.7 |
| V166M/S303G | 69 ± 1.4 | >400 | 320 ± 35 |
| NL4-3 | >100 | ND | ND |
All assays were performed in duplicate, and the mean IC50s (± standard deviations) are shown. ND, not done.
Determination of the chemokine receptor usage of each mutant clone and cellular tropism.
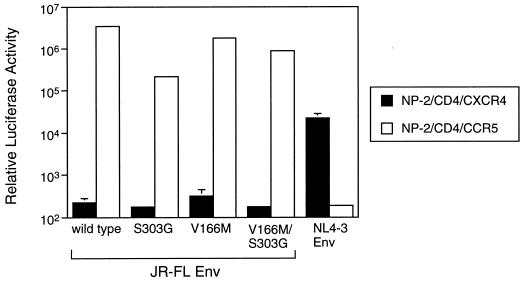
We used MOLT-4#8/CCR5 and MAGI/CCR5 cells for determination of sensitivity to C-C chemokines and the selection of an MIP-1α-resistant mutant, respectively. If the resistant mutant has acquired the ability to use another major coreceptor, CXCR4, which was expressed on both cell lines, the selected variant would display resistance to C-C chemokines because MIP-1α, MIP-1β, and RANTES are not able to interact with CXCR4. To determine whether these resistant mutants acquire the ability to use CXCR4, we used a CD4-expressing glioma cell line, NP-2/CD4, since this cell line does not allow replication of any HIV strain as described previously (27). First, we established CCR5- or CXCR4-expressing NP-2/CD4 cells, designated NP-2/CD4/CCR5 or NP-2/CD4/CXCR4 cells, respectively. The expression of each chemokine receptor in these cells was verified using anti-CCR5 and anti-CXCR4 monoclonal antibodies. NP-2/CD4/CCR5 cells expressed only CCR5 on their surfaces, while NP-2/CD4/CXCR4 expressed only CXCR4 (data not shown). These cells were infected with luciferase reporter viruses pseudotyped with the wild-type or mutant JR-FL envelope or NL4-3 envelope. Luciferase reporter HIV pseudotyped with the wild-type JR-FL envelope was able to infect only NP-2/CD4/CCR5 cells, while HIV pseudotyped with the NL4-3 envelope was able to infect only NP-2/CD4/CXCR4 cells, verifying the functional expression of the chemokine receptors of each cell for HIV infection (Fig. 4). Luciferase activity after the infection of pseudotyped viruses with mutant JR-FL envelope clones, including S303G, V166M, and S303G/V166M mutants, showed that none of the mutants changed their chemokine receptor phenotype (Fig. 4). We also attempted to infect MOLT-4#8, CEM, MT-2, and MT-4 cells, which have only CXCR4 and not CCR5 for the coreceptor of HIV entry. The infection was determined from both p24 antigen production in the cell culture supernatant and an indirect immunofluorescence assay using an anti-p24 antigen monoclonal antibody. The cell lines used were not infected with the S303G/V166M mutant (data not shown), confirming that this resistant mutant does not display acquisition of other coreceptor usage.
FIG. 4.
Chemokine receptor usage of luciferase reporter HIVs pseudotyped with mutant envelope. CXCR4 (filled bar)- or CCR5 (open bar)-expressing NP-2/CD4 cells were infected with luciferase reporter HIVs pseudotyped with the JR-FL envelope with the indicated mutations or the NL4-3 envelope. Luciferase activity was measured 3 days after infection. The data are geometric means ± standard deviations of duplicate determinations.
DISCUSSION
Our in vitro resistant mutant which had reduced sensitivity to MIP-1α showed amino acid substitutions in both V2 (V166M) and V3 (S303G). Importantly, the amino acid change in V3 (S303G) also occurred in long-term culture without MIP-1α, indicating that this substitution might be necessary for the adaptation in MOLT-4#8/CCR5 cells and not for resistance. However, we confirmed that both mutations in V2 and V3 are crucial for the reduced sensitivity to MIP-1α using luciferase reporter HIV pseudotyped with molecularly cloned mutant envelopes. Neither a single mutation in V2 nor a single mutation in V3 displayed reduced sensitivity to C-C chemokines, suggesting that a single amino acid change in the envelope region is not sufficient to obtain mutants resistant to C-C chemokines.
We expected that this variant would have the ability to use CXCR4 as a coreceptor for HIV-1 entry. However, it is unlikely that this resistant mutant uses CXCR4 and other chemokine receptors, including CCR2b, CCR3, CCR8, GPR15 (BOB), STRL33 (Bonzo), V28 (CX3CR1), and Apj (reviewed in reference 23), since the mutant was not able to replicate in parental MOLT-4#8 cells. Our chemokine receptor usage experiment with pseudotype HIV confirmed that the double mutant clone (V166M/S303G), which displayed reduced sensitivity to MIP-1α, did not acquire the ability to use CXCR4.
Recent studies using chimeric chemokine receptors showed that multiple sites of the extracellular domains were involved in the interaction of HIV-1 but that the chemokine binding site was limited to only the N-terminal domain of the receptor (2, 4, 16, 34, 40, 43, 53). Thus, it is possible that the double mutant interacts with another portion(s) of CCR5. An SDF-1α-resistant virus isolated from X4 virus by another group did not switch coreceptors (45), also supporting our speculation. Alternatively, the affinity of the resistant mutant envelope for CCR5 after CD4 binding may compete the binding of MIP-1α to CCR5 even if it uses the same portion of CCR5.
Previous studies have shown that the V3 configuration is crucial to the cellular tropism of HIV-1 (7, 11, 15, 22, 25, 47, 48, 51, 52). Other studies have suggested, however, that the V1/V2 configuration was also involved in cellular tropism. Several amino acid changes in the V2 region were able to alter cellular tropism (5, 21, 30, 31). We also observed that the V1/V2 configuration was important to cellular tropism and soluble CD4 sensitivity in combination with the V3 configuration (37). Since the discovery of chemokine receptors for HIV-1 entry, several studies have shown the importance of the V3 region in the determination of coreceptor usage. More recently, V1/V2 and other variable portions have been shown to influence or alter the usage (24, 42). However, the mechanism(s) by which the coreceptor usage changes is still poorly understood. The present study showed that the V2 region modulated sensitivity to C-C chemokines in combination with the V3 region without affecting chemokine receptor usage.
Cocchi et al. (9) reported that the blockade of R5 HIV-1 by C-C chemokines was determined by the V3 region. Jansson et al. (26) further showed that a serine-to-glycine substitution in the V3 region, which was also found in our mutant (S303G in our case), was associated with a loss of sensitivity to C-C chemokines together with an additional amino acid substitution (glutamic acid to arginine) in the V3 region of some HIV-1-infected individuals during disease progression. Previous studies showed that positively charged amino acid substitutions in the V3 region were correlated with the syncytium-inducing phenotype of HIV-1 isolates (11, 20). Our resistant mutant, however, did not acquire positively charged amino acid changes in V3 during the selection, suggesting that it displays an intermediate preference for the chemokine receptors CCR5 and CXCR4. This serine-to-glycine substitution might increase the replicative ability of HIV since it occurred in passaged virus in MOLT-4#8/CCR5 cells. Further, several amino acid changes, especially of positively charged amino acids in the V3 region, might be necessary to change the coreceptor usage. On the other hand, our chimeric envelope experiments revealed that the sensitivity of HIV to C-C chemokines was dependent on the cooperative interaction of CCR5 with both V2 and V3. Previous studies have shown that the V2 configuration is associated with disease progression in combination with the V3 configuration (21, 46, 49, 55), suggesting that this amino acid position in V2 has a role in the evolution of HIV. It is also of note that some primary isolates in the Los Alamos database (32) have methionine at position 166 in the V2 region although the sensitivities to C-C chemokines of those isolates are not known. Thus, both the V2 and V3 region may be associated with a loss of sensitivity to C-C chemokines and a phenotype switch from CCR5 to CXCR4 usage during the evolution of HIV and disease progression in vivo.
Recently, the structure of the HIV gp120 cores crystallized in a ternary complex with a two-domain fragment of CD4 and the 17b Fab, which recognizes the gp120 epitope after soluble CD4 binding, was solved (33, 54) and a conserved structure in gp120 for CCR5 binding was determined (41). The gp120 core is composed of an inner domain, an outer domain, and a “bridging sheet,” including a V1/V2 stem. The V1/V2 stem and fourth conserved region make up the CD4-induced epitope. CD4 binding to gp120 distorts the V1/V2 loop, and then repositioning of the bridging sheet allows for CCR5 binding. The mutations in the V1/V2 stem combined with V3 mutations might affect the formation of the bridging sheet, allowing this mutant to interact with CCR5 differently after CD4 binding. Alternatively, some residues in the V2 stem may also be directly involved in chemokine receptor interaction, as previously suggested (41, 54). It is also conceivable that this V2 mutation combined with the V3 mutation alters the binding affinity of gp120 for CD4, resulting in conformational changes in the bridging sheet.
Our resistant mutant did not alter cellular tropism (data not shown) probably because of its low level of resistance. It is also possible that another factor is necessary for the phenotype switch from CCR5 to CXCR4 usage. Further selection of mutants resistant to C-C chemokines might elucidate the role of C-C chemokines in the ability to switch chemokine receptors and in cellular tropism during the course of HIV infection in vivo.
ACKNOWLEDGMENTS
We thank Y. Eda for supplying yeast-derived recombinant MIP-1α (yLD78β). We also thank I. S. Y. Chen, Y. Koyanagi, M. Shimada, J. Miyazaki, H. Hoshino, and M. Emerman for kindly providing HIV NL-Luc plasmid, JR-FL plasmid, pG1TKNeo plasmid, pCXN2 plasmid, NP-2/CD4 cells, and CD4-HeLa/LTR–β-Gal cells, respectively. We thank K. Morizono, A. Koito, K. Obaru, K. Yusa, and H. Maeda for helpful discussion and K. Nanke for technical assistance.
This work was supported by grants from the Ministry of Education, Science, Sports and Culture and the Ministry of Health and Welfare, Tokyo, Japan.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 3.Aukrust P, Muller F, Froland S S. Circulating levels of RANTES in human immunodeficiency virus type 1 infection: effect of potent antiretroviral therapy. J Infect Dis. 1998;177:1091–1096. doi: 10.1086/517402. [DOI] [PubMed] [Google Scholar]
- 4.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S, Caron M G, Cullen B R. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 co-receptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R A. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng-Mayer C, Seto D, Tateno M, Levy J A. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 7.Chesebro B, Wehrly K, Nishio J, Perryman S. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J Virol. 1992;66:6547–6554. doi: 10.1128/jvi.66.11.6547-6554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 9.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1α, and MIP-1β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 10.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Jong J J, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 13.Dittmar M T, McKnight A, Simmons G, Clapham P R, Weiss R A, Simmonds P. HIV-1 tropism and co-receptor use. Nature. 1997;385:495–496. doi: 10.1038/385495a0. [DOI] [PubMed] [Google Scholar]
- 14.Dolei A, Biolchini A, Serra C, Curreli S, Gomes E, Dianzani F. Increased replication of T-cell-tropic HIV strains and CXC-chemokine receptor-4 induction in T cells treated with macrophage inflammatory protein (MIP)-1α, MIP-1β and RANTES β-chemokines. AIDS. 1998;12:183–190. doi: 10.1097/00002030-199802000-00008. [DOI] [PubMed] [Google Scholar]
- 15.Donaldson Y K, Bell J E, Holmes E C, Hughes E S, Brown H K, Simmonds P. In vivo distribution and cytopathology of variants of human immunodeficiency virus type 1 showing restricted sequence variability in the V3 loop. J Virol. 1994;68:5991–6005. doi: 10.1128/jvi.68.9.5991-6005.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doranz B J, Lu Z H, Rucker J, Zhang T Y, Sharron M, Cen Y H, Wang Z X, Guo H H, Du J G, Accavitti M A, Doms R W, Peiper S C. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 18.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 19.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 20.Fouchier R A, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groenink M, Fouchier R A, Broersen S, Baker C H, Koot M, van't Wout A B, Huisman H G, Miedema F, Tersmette M, Schuitemaker H. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- 22.Henkel T, Westervelt P, Ratner L. HIV-1 V3 envelope sequences required for macrophage infection. AIDS. 1995;9:399–401. [PubMed] [Google Scholar]
- 23.Hoffman T L, Doms R W. Chemokines and coreceptors in HIV/SIV-host interactions. AIDS. 1998;12(Suppl. A):S17–S26. [PubMed] [Google Scholar]
- 24.Hoffman T L, Stephens E B, Narayan O, Doms R W. HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors. Proc Natl Acad Sci USA. 1998;95:11360–11365. doi: 10.1073/pnas.95.19.11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 26.Jansson M, Popovic M, Karlsson A, Cocchi F, Rossi P, Albert J, Wigzell H. Sensitivity to inhibition by b-chemokines correlates with biological phenotypes of primary HIV-1 isolates. Proc Natl Acad Sci USA. 1996;93:15382–15387. doi: 10.1073/pnas.93.26.15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jinno A, Shimizu N, Soda Y, Haraguchi Y, Kitamura T, Hoshino H. Identification of the chemokine receptor TER1/CCR8 expressed in brain-derived cells and T cells as a new coreceptor for HIV-1 infection. Biochem Biophys Res Commun. 1998;243:497–502. doi: 10.1006/bbrc.1998.8130. [DOI] [PubMed] [Google Scholar]
- 28.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinter A, Catanzaro A, Monaco J, Ruiz M, Justement J, Moir S, Arthos J, Oliva A, Ehler L, Mizell S, Jackson R, Ostrowski M, Hoxie J, Offord R, Fauci A S. CC-chemokines enhance the replication of T-tropic strains of HIV-1 in CD4(+) T cells: role of signal transduction. Proc Natl Acad Sci USA. 1998;95:11880–11885. doi: 10.1073/pnas.95.20.11880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koito A, Harrowe G, Levy J A, Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994;68:2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koito A, Stamatatos L, Cheng-Mayer C. Small amino acid sequence changes within the V2 domain can affect the function of a T-cell line-tropic human immunodeficiency virus type 1 envelope gp120. Virology. 1995;206:878–884. doi: 10.1006/viro.1995.1010. [DOI] [PubMed] [Google Scholar]
- 32.Korber B, Hahn B, Foley B, Mellors J W, Leitner T, Myers G, McCutchan F, Kuiken C L. Human retroviruses and AIDS 1997: a compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N. Mex: Los Alamos National Laboratory; 1997. [Google Scholar]
- 33.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu Z, Berson J F, Chen Y, Turner J D, Zhang T, Sharron M, Jenks M H, Wang Z, Kim J, Rucker J, Hoxie J A, Peiper S C, Doms R W. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeda Y, Venzon D J, Mitsuya H. Altered drug sensitivity, fitness, and evolution of human immunodeficiency virus type 1 with pol gene mutations conferring multi-dideoxynucleoside resistance. J Infect Dis. 1998;177:1207–1213. doi: 10.1086/515282. [DOI] [PubMed] [Google Scholar]
- 36.McKenzie S W, Dallalio G, North M, Frame P, Means R T., Jr Serum chemokine levels in patients with non-progressing HIV infection. AIDS. 1996;10:F29–F33. doi: 10.1097/00002030-199610090-00001. [DOI] [PubMed] [Google Scholar]
- 37.Morikita T, Maeda Y, Fujii S, Matsushita S, Obaru K, Takatsuki K. The V1/V2 region of human immunodeficiency virus type 1 modulates the sensitivity to neutralization by soluble CD4 and cellular tropism. AIDS Res Human Retrovir. 1997;13:1291–1299. doi: 10.1089/aid.1997.13.1291. [DOI] [PubMed] [Google Scholar]
- 38.Obaru K, Fukuda M, Maeda S, Shimada K. A cDNA clone used to study mRNA inducible in human tonsillar lymphocytes by a tumor promoter. J Biochem (Tokyo) 1986;99:885–894. doi: 10.1093/oxfordjournals.jbchem.a135549. [DOI] [PubMed] [Google Scholar]
- 39.Obaru K, Hattori T, Yamamura Y, Takatsuki K, Nomiyama H, Maeda S, Shimada K. A cDNA clone inducible in human tonsillar lymphocytes by a tumor promoter codes for a novel protein of the b-thromboglobulin superfamily. Mol Immunol. 1989;26:423–426. doi: 10.1016/0161-5890(89)90131-4. [DOI] [PubMed] [Google Scholar]
- 40.Picard L, Simmons G, Power C A, Meyer A, Weiss R A, Clapham P R. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J Virol. 1997;71:5003–5011. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 42.Ross T M, Cullen B R. The ability of HIV type 1 to use CCR-3 as a coreceptor is controlled by envelope V1/V2 sequences acting in conjunction with a CCR-5 tropic V3 loop. Proc Natl Acad Sci USA. 1998;95:7682–7686. doi: 10.1073/pnas.95.13.7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, Doms R W, Parmentier M. Regions in β-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 44.Scarlatti G, Tresoldi E, Bjorndal A, Fredriksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyo E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 45.Schols D, Este J A, Cabrera C, De Clercq E. T-cell-line-tropic human immunodeficiency virus type 1 that is made resistant to stromal cell-derived factor 1α contains mutations in the envelope gp120 but does not show a switch in coreceptor use. J Virol. 1998;72:4032–4037. doi: 10.1128/jvi.72.5.4032-4037.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuitemaker H, Fouchier R A, Broersen S, Groenink M, Koot M, van't Wout A B, Huisman H G, Tersmette M, Miedema F. Envelope V2 configuration and HIV-1 phenotype: clarification. Science. 1995;268:115. doi: 10.1126/science.7755774. [DOI] [PubMed] [Google Scholar]
- 47.Shioda T, Levy J A, Cheng-Mayer C. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature. 1991;349:167–169. doi: 10.1038/349167a0. [DOI] [PubMed] [Google Scholar]
- 48.Shioda T, Levy J A, Cheng-Mayer C. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1992;89:9434–9438. doi: 10.1073/pnas.89.20.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shioda T, Oka S, Xin X, Liu H, Harukuni R, Kurotani A, Fukushima M, Hasan M K, Shiino T, Takebe Y, Iwamoto A, Nagai Y. In vivo sequence variability of human immunodeficiency virus type 1 envelope gp120: association of V2 extension with slow disease progression. J Virol. 1997;71:4871–4881. doi: 10.1128/jvi.71.7.4871-4881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tersmette M, Lange J M, de Goede R E, de Wolf F, Eeftink-Schattenkerk J K, Schellekens P T, Coutinho R A, Huisman J G, Goudsmit J, Miedema F. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet. 1989;1:983–985. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- 51.Westervelt P, Trowbridge D B, Epstein L G, Blumberg B M, Li Y, Hahn B H, Shaw G M, Price R W, Ratner L. Macrophage tropism determinants of human immunodeficiency virus type 1 in vivo. J Virol. 1992;66:2577–2582. doi: 10.1128/jvi.66.4.2577-2582.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willey R L, Theodore T S, Martin M A. Amino acid substitutions in the human immunodeficiency virus type 1 gp120 V3 loop that change viral tropism also alter physical and functional properties of the virion envelope. J Virol. 1994;68:4409–4419. doi: 10.1128/jvi.68.7.4409-4419.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu L, LaRosa G, Kassam N, Gordon C J, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, Moore J P, Mackay C R. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 55.Yoshimura K, Matsushita S, Hayashi A, Takatsuki K. Relationship of HIV-1 envelope V2 and V3 sequences of the primary isolates to the viral phenotype. Microbiol Immunol. 1996;40:277–287. doi: 10.1111/j.1348-0421.1996.tb03347.x. [DOI] [PubMed] [Google Scholar]
- 56.Zanussi S, D'Andrea M, Simonelli C, Tirelli U, De Paoli P. Serum levels of RANTES and MIP-1α in HIV-positive long-term survivors and progressor patients. AIDS. 1996;10:1431–1432. doi: 10.1097/00002030-199610000-00018. [DOI] [PubMed] [Google Scholar]