Abstract
The adenovirus type 5 (Ad5) E4-6/7 protein interacts directly with different members of the E2F family and mediates the cooperative and stable binding of E2F to a unique pair of binding sites in the Ad5 E2a promoter region. This induction of E2F DNA binding activity strongly correlates with increased E2a transcription when analyzed using virus infection and transient expression assays. Here we show that while different adenovirus isolates express an E4-6/7 protein that is capable of induction of E2F dimerization and stable DNA binding to the Ad5 E2a promoter region, not all of these viruses carry the inverted E2F binding site targets in their E2a promoter regions. The Ad12 and Ad40 E2a promoter regions bind E2F via a single binding site. However, these promoters bind adenovirus-induced (dimerized) E2F very weakly. The Ad3 E2a promoter region binds E2F very poorly, even via a single binding site. A possible explanation of these results is that the Ad E4-6/7 protein evolved to induce cellular gene expression. Consistent with this notion, we show that infection with different adenovirus isolates induces the binding of E2F to an inverted configuration of binding sites present in the cellular E2F-1 promoter. Transient expression of the E4-6/7 protein alone in uninfected cells is sufficient to induce transactivation of the E2F-1 promoter linked to chloramphenicol acetyltransferase or green fluorescent protein reporter genes. Further, expression of the E4-6/7 protein in the context of adenovirus infection induces E2F-1 protein accumulation. Thus, the induction of E2F binding to the E2F-1 promoter by the E4-6/7 protein observed in vitro correlates with transactivation of E2F-1 promoter activity in vivo. These results suggest that adenovirus has evolved two distinct mechanisms to induce the expression of the E2F-1 gene. The E1A proteins displace repressors of E2F activity (the Rb family members) and thus relieve E2F-1 promoter repression; the E4-6/7 protein complements this function by stably recruiting active E2F to the E2F-1 promoter to transactivate expression.
The transcription factor E2F plays a pivotal role in the regulation of cellular proliferation in mammalian cells and in Drosophila. Mammalian E2F DNA binding activity is a heterodimer containing one of six known members of the E2F family (E2F-1 through -6) in association with a partner from the DP family (DP-1 or DP-2) (reviewed in reference 11). Different E2F heterodimers are regulated by interactions with members of the retinoblastoma gene family (pRb, p107, and p130) (reviewed in reference 37). E2F-1, -2 and -3–DP complexes bind to pRb, E2F-4–DP heterodimers interact with pRb and p107, and E2F-5–DP is preferentially bound by p130. The association of E2Fs with Rb family members as well as the relative importance of different E2F complexes varies with specific stages of the cell cycle (11, 37). In growth-arrested cells, the predominant E2F activity is an E2F-5–p130 complex. In G1 phase of the cell cycle, E2F-Rb, E2F-p107 and E2F-p130 complexes are evident. The Rb family members are targets of G1-phase cyclin/cyclin-dependent kinases, and their phosphorylation results in their release from E2Fs (11, 37). E2F-1–DP heterodimers are important for cellular proliferation with cells entering the cell cycle from a G0 state, whereas E2F-3–DP complexes are important for cell cycle progression with continuously proliferating cells (29, 34, 56).
E2F binds to the promoter regions of a number of cellular genes involved in DNA synthesis and regulation of the cell cycle. E2F binding sites in the b-myb, cdc2, cdc6, c-myc, cyclin A, cyclin E, dhfr, E2F-1, orc1, and thymidine kinase gene promoters have been shown to be involved in the induction of transcription in resting cells following serum stimulation (2, 3, 9, 18, 25, 28, 33, 40, 44, 45, 50, 55). Ectopic E2F-1 expression is sufficient to induce quiescent cells to enter S phase, although unregulated E2F-1 overexpression may result in the induction of p53-mediated apoptosis (22, 23, 29, 31, 32, 46, 47, 51, 59). Inhibition of cellular proliferation, a hallmark of normal Rb function, is relieved when the Rb-E2F interaction is disrupted by mutation of Rb or dissociation of Rb from E2F by the DNA tumor virus transforming proteins large T antigen, E1A, or E7 (reviewed in references 8 and 37). The Rb family members bind to sequences in E2Fs that overlap the activation domains of these transcription factors (7, 14, 20, 43), and a model whereby Rb family proteins repress E2F activities by physically masking the transactivation domains can be easily envisioned. Further, Rb family members act as dominant repressors of promoter regions to which E2F-Rb complexes are linked by virtue of recruitment of complexes containing histone deacetylases (1, 4, 5, 12, 35, 57). Thus, E2F binding sites serve to repress as well as to activate cellular promoters, depending on the nature of the E2F complexes found in the cell. These observations may explain why E2F-1 may act as either an oncogene or a tumor suppressor, depending on the context in which activity is analyzed (13, 27, 53, 60, 61).
E2F was first described as a nuclear activity that bound to an inverted repeat in the adenovirus type 5 (Ad5) E2a promoter (30). The binding activity of E2F to these sites is stimulated by Ad infection dependent on the activities of two viral proteins. E2F transcriptional activity is positively regulated by the Ad E1A gene products which avidly bind to Rb family members and dissociate them from E2Fs (8). Free E2F is then bound by the Ad E4-6/7 protein, which forms a complex with E2F and induces the cooperative and stable binding of E2F to the inverted binding sites in the Ad5 E2a promoter (Fig. 1A) (16, 17, 26, 36, 48). The induction of E2F binding to the Ad5 E2a promoter in vitro is directly correlated with transcriptional activation of the E2a promoter in vivo (38, 39, 41, 42, 49). The dissociation of Rb family members from E2Fs by the E1A proteins also results in activation of cellular genes containing E2F response elements and the induction of cell cycle progression (8). In this report, we show that while different Ad isolates are capable of induction of E2F dimerization and DNA binding, not all of these viruses carry the inverted binding site targets in their E2a promoter regions. We hypothesize that these viruses may have evolved E4-6/7 products in order to activate other E2F-responsive genes; in confirmation of this idea, we demonstrate that different Ad isolates induce E2F binding to an inverted binding site in the cellular E2F-1 promoter. This induction of E2F binding results in transactivation of the cellular E2F-1 promoter and an increase in E2F-1 protein levels. These results suggest that Ad has evolved two distinct mechanisms to induce expression of the E2F-1 gene. The E1A proteins displace repressors of E2F activity (the Rb family members) and thus relieve E2F-1 promoter repression; the E4-6/7 protein complements this function by stably recruiting active E2F to the E2F-1 promoter to transactivate expression.
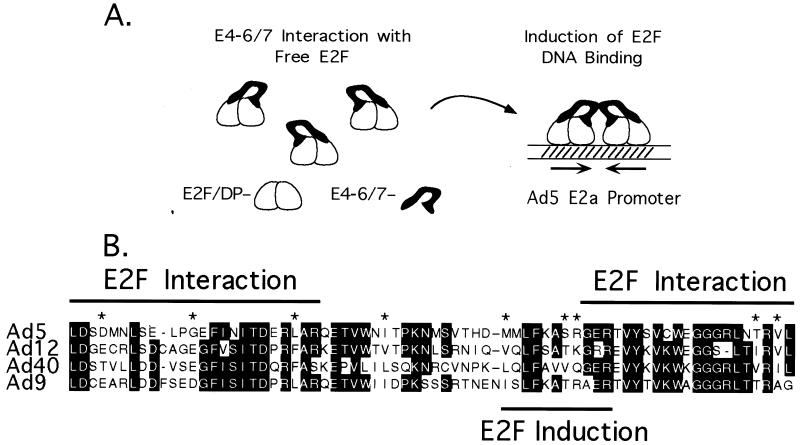
FIG. 1.
(A) Model for Ad induction of E2F DNA binding. The left depicts interaction of the E4-6/7 protein with free E2F-DP heterodimers. The right depicts the induction of E2F DNA binding to the Ad5 E2a promoter region by E4-6/7 protein-mediated dimerization. The inverted E2F binding sites in the Ad5 E2a promoter are indicated by inverted arrows. (B) Alignment of E4-6/7 proteins from different Ad serotypes. E4 ORF7 of the Ad5 E4-6/7 protein is shown at the top in alignment with homologous sequences found in Ad12, Ad40, and Ad9. Dark letters indicate amino acid identities; conserved amino acid changes are indicated by asterisks across the top. The two regions in Ad5 E4-6/7 required for stable interaction with a E2F-DP heterodimer are indicated at the top of the sequence (termed E2F interaction), while the region of E4-6/7 that directs dimerization is shown at the bottom of the sequence (termed E2F induction).
MATERIALS AND METHODS
Extract preparation and gel mobility shift assays.
Nuclear and cytoplasmic extracts were prepared according to the method of Dignam and Roeder (10). The cytoplasmic supernatant obtained after isolation of the nuclei was adjusted to 100 mM KCl, spun at 100,000 × g for 1 h, and saved as the cytoplasmic fraction. Cytoplasmic and nuclear fractions were dialyzed against DB (20 mM HEPES [pH 7.5], 100 mM KCl, 10% glycerol, 5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF]), and the dialysate was cleared by centrifugation at 25,000 × g. In vitro DNA binding assays were performed as described previously (43). Briefly, binding reaction mixtures (20 μl) contained 5 to 10 μg of nuclear or cytoplasmic extract, 2 μg of sonicated salmon sperm DNA, and 20,000 cpm (double-site probe) or 40,000 cpm (single-site probe) of 32P-labeled E2F recognition sites (1 to 2 fmol of DNA) in DB supplemented with Nonidet P-40 (final concentration, 0.1%). Binding reactions were incubated for 1 to 2 h at room temperature, followed by electrophoresis on a 4% 30:1 polyacrylamide gel run in 0.5× Tris-borate-EDTA at 4°C. The Ad5 E2a E2F double-site probe contains nucleotides −30 to −73 from the Ad5 E2a promoter plus additional vector sequences; the sequence of one strand is 5′-AATTCGTAGTTTTCGCGCTTAAATTTGAGAAAGGGCGCGAAACTAGTCCCGG-3′. The E2F sites are underlined, and vector sequences are in italics. The E2F single-site probe contains Ad nucleotides 270 to 293 from the E1A enhancer and flanking vector sequences; the sequence of one strand is 5′-AATTCCCCCATTTTCGCGGGAAAACTGAATCCTCGA-3′. The cellular E2F-1 promoter probe corresponds to nucleotides −44 to −12 (25, 28); the sequence of one strand is 5′-AATTCCGGGCTCTTTCGCGGCAAAAAGGATTTGGCGCGTAAAAGG-3′. The sequences of one strand of the probes for the E2a promoter regions of different Ad serotypes are as follows: Ad3, 5′-AATTCGTGATTCCGCCGTTTTCAAAATGAGCGCGGGCAAGGGCTACTC-3′; Ad12, 5′-AATTCGTCACTTTTCCCGCCTGTTGAAAGTCCGCGCGCGGGCTTTTTTACTC-3′; and Ad40, 5′-AATTCGTAATTTGGCGCCTAAAAAAAGCGCGGGTGTTTAGTC-3′. Probe fragments were labeled with [α-32P]dATP and Klenow DNA polymerase; specific activities were 5,000 to 10,000 cpm/fmol.
For bacterial expression of E4-open reading frame 7 (ORF7), a 50-ml overnight culture of Escherichia coli DH5 transformed with pGEX-ORF7 (Ad5 or Ad40) was grown overnight to saturation. A 500-ml culture was then inoculated and allowed to grow for 1 h. Isopropyl-β-d-thiogalactopyranoside was added to 0.1 mM, and incubation was continued for 4 h. Bacteria were harvested, washed with phosphate-buffered saline–(PBS) 1 mM PMSF, and resuspended in 25 ml TNE (10 mM Tris [pH 7.5], 100 mM NaCl, 1 mM EDTA) containing 10 mM dithiothreitol, 1 mM PMSF, and 1% Triton X-100. Aliquots were sonicated, and soluble material was recovered following centrifugation at 25,000 × g for 20 min. The soluble pool was loaded on a column of glutathione-agarose (Sigma), the bound material was washed with DB-100, and glutathione S-transferase (GST)–ORF7 protein was eluted in DB-100–20 mM reduced glutathione.
Virus infections, transfection and microinjection assays, and immunofluorescence assay.
A549 cells were maintained in Dulbecco's modified minimal essential medium (DMEM) containing 10% calf serum. REF52 and ATCC HeLa cells were maintained in DMEM containing 10% fetal bovine serum (FBS). HepG2 cells were maintained in minimal essential medium containing 10% FBS. Different Ad serotypes were purchased from the American Type Culture Collection and propagated on A549 cells. Purified virus particles were purified by CsCl equilibrium gradient centrifugation following standard approaches (54). A549 cells were infected at a multiplicity of 500 particles/cell for 1 h. Nuclear and cytoplasmic extracts were prepared 6 to 8 h after infection.
Plasmid DNA transfections were performed by the calcium phosphate precipitation procedure (58). HepG2 cells were split and plated the day before transfection. The following day, the cells were transfected with 1 μg of the E2a-CAT reporter vector or 2 μg of the E2F-1–CAT reporter plasmid DNA, 18 μg of salmon sperm DNA, and various amounts of effector plasmid per 100-mm-diameter dish, as indicated in the text. The cells were incubated for 4 h with the calcium phosphate precipitate, the medium was removed, and Tris-buffered saline solution containing 20% glycerol was added for 1 min. The cells were then washed three times with Tris-buffered saline, and fresh medium containing 10% serum or 0.5% serum was added, as indicated in the text. Total cell extracts were prepared 24 h later, and chloramphenicol acetyltransferase (CAT) enzymatic activity in cellular extracts was assayed using a fluorescent chloramphenicol substrate (FAST-CAT; Molecular Probes, Eugene, Oreg.). CAT activity was quantified with a phosphorimager. The results presented represent the average of five experiments. The E2a-CAT vector was previously described (42). The E2F-1–CAT vector was constructed from a plasmid provided by Peggy Farnham (University of Wisconsin) and contains positions −176 to +36 of the murine E2F-1 promoter region (25) linked to the CAT gene. The E2F-1–GFP (green fluorescent protein) plasmid contains the same segment of the murine E2F-1 promoter linked to the GFP coding region (62).
For microinjection experiments, REF52 cells plated on gridded glass coverslips were grown to subconfluency and subsequently serum starved for 36 h in DMEM containing 0.5% FBS. DNA was prepared by dilution of microinjection buffer (50 mM HEPES [pH 7.2], 100 mM KCl, 5 mM NaH2PO4). E2F-1–GFP (25 ng/μl, along with empty cytomegalovirus [CMV] vector [25 ng/μl]) was injected into the nucleus followed by maintenance of the cells in DMEM containing 0.5% FBS or with serum stimulation in DMEM containing 20% FBS. Cells coinjected with E2F-1–GFP and CMV–E4-6/7 (42) (each at 25 ng/μl) were maintained in DMEM containing 0.5% FBS. Cells were fixed 16 h after microinjection in 3.7% formaldehyde for 1 h and washed three times in PBS prior to slide mounting. GFP fluorescence was observed using a fluorescein filter on an Axiovert 135 microscope (Zeiss). For immunofluorescence assays, ATCC HeLa cells were seeded on coverslips and infected with 200 to 250 Ad particles/cell for 1 h. Coverslips were harvested at 8 and 16 h after infection, washed with PBS, fixed with 3% formaldehyde, and permeabilized using 0.2% Triton X-100. Blocking as well as primary and secondary antibody application were performed in PBS containing 2% bovine serum albumin. Primary antibodies included E4-6/7 monoclonal antibody M45 (41) and a rabbit polyclonal antibody specific to the C terminus of E2F-1 (sc-193; Santa Cruz Biotechnology). Secondary antibodies were tetramethyl rhodamine isothiocyanate-labeled anti-mouse and fluorescein isothiocyanate-labeled anti-rabbit conjugates. All coverslips were processed with Fluoromount-G (Southern Biotechnologies, Inc.), and immunofluorescence was detected as described above for GFP.
RESULTS
Functional conservation of E4-6/7 proteins.
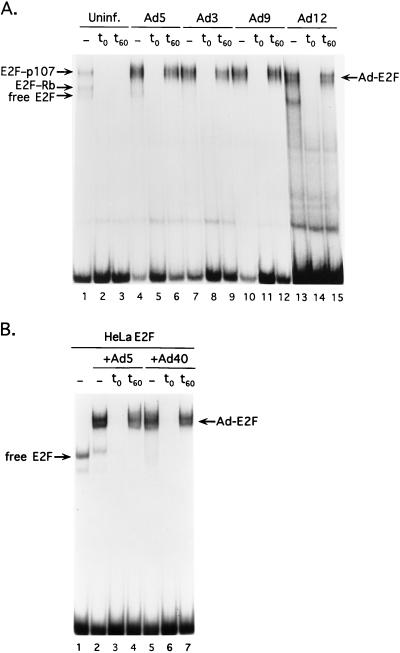
The importance of the function of a viral protein in natural infections can be inferred if that protein is encoded by the genomes of related, but evolutionarily diverse, viruses. The E4 regions of Ad serotypes 2, 5, 9, 12, and 40 have been sequenced. We examined these regions for the capacity to encode proteins homologous to the Ad5 E4 ORF7 coding region, the segment of the Ad5 E4-6/7 protein sufficient to bind to E2F and induce E2F dimerization on an inverted binding site (42). Each of the viruses, representing five of the six Ad subgroups, encoded hypothetical proteins with significant homology to Ad5 ORF7 (Fig. 1B), including the two segments required for stable binding to E2F (E2F interaction) and the region that mediates E2F dimerization (E2F induction) (41). To determine if these viruses exhibited the ability to induce stable E2F binding characteristic of the Ad5 E4-6/7 product, nuclear extracts were prepared from A549 cells infected with Ad3, Ad5, Ad9, and Ad12 and analyzed by mobility shift assay using a probe corresponding to the Ad5 E2a inverted E2F binding sites. A549 cells were used for this experiment since this cell line may be productively infected by all of the Ad serotypes under study. These results are shown in Fig. 2A. Different E2F complexes were observed using nuclear extract from uninfected A549 cells (lane 1) that correspond to free E2F activity as well as E2F in association with pRb and p107. The identification of the different E2F complexes was confirmed using antibodies to E2F-1, E2F-4, and E2F binding partners pRb and p107, which specifically supershift individual complexes evident in lane 1 (data not shown). Infection of A549 cells with the different Ad serotypes induced a new and abundant E2F complex in comparison to nuclear extract from uninfected cells. The presence of the Ad5 E4-6/7 protein in the Ad-E2F complex previously was confirmed using monoclonal antibodies against this viral protein (26, 36, 48). These antibodies recognize the amino-terminal region of the E4-6/7 protein which is variable among Ad serotypes; we found that the anti-E4-6/7 monoclonal antibodies M45 and M41 (41) did not recognize the Ad-E2F complexes formed with Ad serotypes other than 2 or 5 (data not shown). The Ad-induced complexes were efficiently competed when a 500-fold molar excess of a specific E2F binding site was included in the binding reaction when added at the same time as the probe DNA. Each of the Ad-E2F complexes was stable to competitor challenge when the competitor DNA was added after a 1-h preincubation with probe DNA. Thus, each of the Ad serotypes tested induced a stable E2F complex bound to an inverted E2F binding site characteristic of the Ad5-induced E2F/E4-6/7 dimer. We note that the Ad12 E4-6/7 protein is predicted to be 125 amino acids in length, in contrast to the 150-amino-acid Ad2 and Ad5 protein; this is consistent with the faster mobility of the Ad12-induced E2F complex observed in lane 13.
FIG. 2.
Induction of E2F DNA binding to the Ad5 E2a promoter by different Ad serotypes. (A) Nuclear extracts from uninfected A549 cells (Uninf.; lanes 1 to 3) or A549 cells infected with Ad5 (lanes 4 to 6), Ad3 (lanes 7 to 9), Ad9 (lanes 10 to 12), or Ad12 (lanes 13 to 15) were used in gel mobility shift assays with a probe corresponding to the Ad5 E2a inverted E2F binding sites. The first lane of each set (lanes 1, 4, 7, 10, and 13) represents a binding reaction with no specific competitor DNA added. E2F complexes found in uninfected cells are indicated on the left, and the Ad-induced E2F complex is indicated on the right (Ad-E2F). In binding reactions shown in lanes 2, 5, 8, 11, and 14, a 500-fold molar excess of unlabeled DNA corresponding to an E2F single binding site (see Materials and Methods) was added to the binding reaction coincident with the addition of probe DNA. In binding reactions shown in lanes 3, 6, 9, 12, and 15, a 500-fold molar excess of unlabeled E2F single binding site DNA was added after a 60-min preincubation of the binding reaction with probe DNA. The binding reactions were incubated an additional 15 min before loading on the gel. (B) Bacterially expressed E4 ORF7 proteins were used in binding reactions with HeLa cell-free E2F activity. Lane 1 shows a binding reaction with HeLa cell cytoplasmic extract plus the Ad5 E2a probe. Free E2F binding activity is indicated on the left. In lanes 2 to 4, an Ad5 GST-E4-ORF7 fusion protein was added to the HeLa cell extract prior to the addition of probe DNA. In lanes 5 to 7, an Ad40 GST-E4-ORF7 fusion protein was added. Lanes 2 and 5 represent binding reactions without the addition of specific competitor DNA. In lanes 3 and 6, a 500-fold molar excess of unlabeled E2F binding site was added coincident with the probe as described for panel A. In lanes 4 and 7, a 500-fold molar excess of unlabeled E2F binding site was added after a 60-min binding reaction as described in for panel A.
To confirm that the E4-6/7 protein from an Ad other than Ad2 and Ad5 was capable of E2F induction, the region homologous to ORF7 of Ad40 was isolated by PCR and the protein was expressed as a GST fusion product in bacteria. When incubated with uninfected HeLa cell cytoplasmic extract (an abundant source of free E2F activity), bacterially expressed Ad40 ORF7 stabilized E2F binding on the Ad5 E2a inverted binding site in the same manner as the Ad5-encoded protein (Fig. 2B). This experiment confirms that E4 ORF7 from different Ad serotypes is sufficient to direct E2F dimerization. To substantiate the relevance of induction of E2F binding in vitro, we tested promoter transactivation by the E4 ORF7 segments from Ad5 and Ad40 in vivo. A reporter vector containing the Ad5 E2a promoter linked to the CAT gene was transfected into HepG2 cells alone or with the E4 ORF7 expression vectors. HepG2 cells were used for this experiment since the E2a promoter displays very low basal activity in this cell line. Significant transactivation of the E2a promoter (12.8 ± 2.9- and 5.8 ± 0.8-fold above the basal level) was observed with both Ad5 and Ad40 E4 ORF7 products. We conclude that Ad isolates from different subgroups have conserved the ability to induce E2F binding to an inverted E2F binding site and induce expression from a promoter region containing such binding sites.
E2a promoter structure does not correlate with E4-6/7 function.
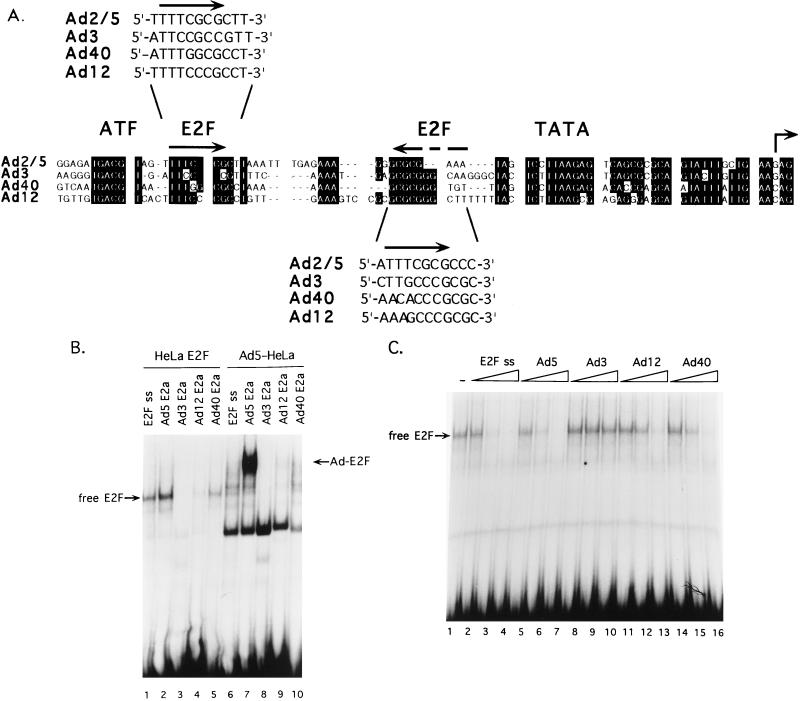
Induction of E2F DNA binding by E4-6/7 in vitro strongly correlates with increased E2a gene expression in vivo with Ad5 (38, 39, 41, 42, 49). Based on the results described above, we anticipated that the E2a promoter regions of different Ad serotypes would retain the inverted configuration of E2F binding sites found in Ad5. Surprisingly, when we compared the E2a promoters of Ad serogroups where the E2a sequence has been determined, this was not found to be the case (Fig. 3A). While the binding site for transcription factor ATF, the TATA box, and the start site regions were highly conserved between Ad serotypes, the interval containing the putative E2F sites was not. Changes in both the upstream and downstream E2F consensus binding sites (5′-TTTCGCGC-3′) as well as alterations in the spacing between these two sites, were evident. It is well established that the correct spacing between the inverted E2F sites in the Ad5 E2a promoter is critical for E4-6/7-mediated induction of E2F DNA binding (17, 48).
FIG. 3.
E2a promoter regions of different Ad serotypes do not contain inverted E2F binding sites. (A) Alignment of the E2a promoter regions from different Ad serotypes. The ATF binding site, TATA box, and potential E2F sites are indicated at the top. Alignment of E2a promoters shows strong conservation of the ATF site, the TATA box, and start site region. The ATF-proximal and TATA-proximal potential E2F sites are shown in a 5′-to-3′ orientation in the insets above and below the sequence. (B) Binding reactions contained HeLa cell E2F and Ad5-induced E2F with probes corresponding to the E2a regions of different Ad serotypes. Lanes 1 to 5 show binding reactions with HeLa cell-free E2F activity and DNA probes corresponding to an E2F single-site (E2F ss) and E2a regions of Ad5, Ad3, Ad12 and Ad40, as indicated above the lanes. Lanes 6 to 10 show binding reactions with Ad5-infected HeLa cell nuclear extract with the same set of probes as described for lanes 1 to 5. The Ad-induced E2F complex is indicated in the right (Ad-E2F). The faster-migrating complex is not specifically competed by an E2F binding site and is an unknown binding activity that serves as an internal control for the integrity of each probe DNA. (C) The binding of HeLa cell free E2F to the E2a regions of different Ad serotypes was analyzed using competition binding assays. Lane 1 shows a binding reaction using HeLa cell-free E2F activity with an E2F single-site probe. Free E2F binding activity is indicated on the left. Lanes 2 to 16 show binding reactions where increasing molar concentrations (5-, 25-, and 125-fold molar excess to the probe DNA) of specific competitor DNAs were added coincident with the probe DNA. The cold competitors corresponded to an E2F single site identical to the probe DNA (lanes 2 to 4) and to E2a DNA of Ad5 (lanes 5 to 7), Ad3 (lanes 8 to 10), Ad12 (lanes 11 to 13), and Ad40 (lanes 14 to 16).
To test if the different Ad E2a promoter regions were capable of binding E2F and Ad-induced E2F activity, two approaches were taken. First, synthetic oligonucleotides corresponding to the putative E2F binding sites of the different E2a promoter regions were used as probes in mobility shift assays with nuclear extract prepared from Ad5-infected Hela cells (Fig. 3B, lanes 6 to 10). The boundaries of the sequences chosen for probes in this analysis overlapped with the 3′ part of the highly conserved ATF binding site and the 5′ part of the highly conserved TATA box. Thus, the entire intervening region was represented in the probe DNA with each Ad serotype. This assay showed that whereas the Ad5 E2a sites bound Ad-E2F in a significant manner, the Ad12 and Ad40 sites only weakly bound Ad-induced E2F activity and no binding was evident with the Ad3 E2a probe. When free E2F activity from uninfected HeLa cells was analyzed with the different probes, the same pattern of binding was found (Fig. 3B, lanes 1 to 5). In a second approach, the oligonucleotides corresponding to the different E2a promoter regions were used as competitor DNAs to test for E2F binding (Fig. 3C). HeLa cell-free E2F was incubated with a probe corresponding to a single E2F binding site with no specific competitor DNA in the binding reaction or increasing amounts of oligonucleotide DNAs corresponding to the different E2a promoters (Fig. 3C). These results demonstrated that Ad5, Ad12, and Ad40 regions were capable of binding E2F comparably when only a single E2F site is required to compete for binding, but that the Ad3 segment did not bind E2F to a significant level. We conclude from these experiments that while the different Ad serotypes evolved E4-6/7 proteins that are capable of dimerizing E2F at the Ad5 E2a inverted E2F binding site, and the Ad40 E4 ORF7 product effectively transactivates the Ad5 E2a promoter (see above), the same viruses did not evolve promoter regions to take advantage of this induction.
Ad induction of E2F binding to the cellular E2F-1 promoter.
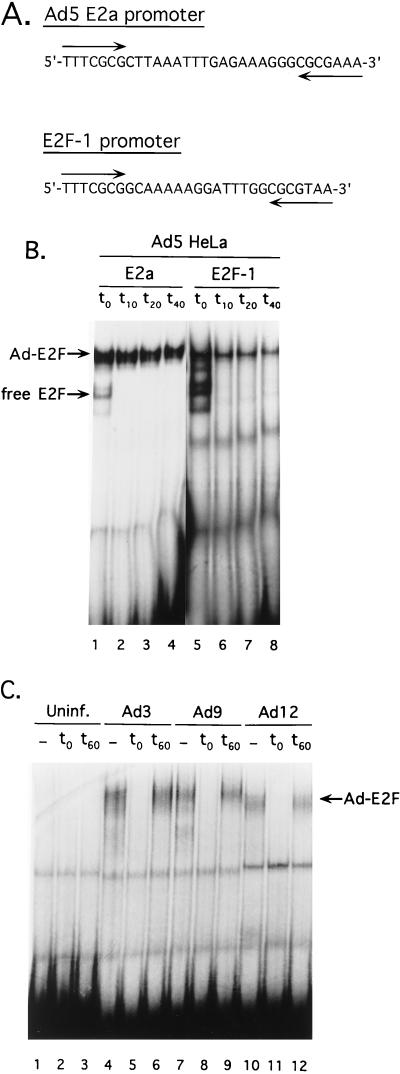
An intriguing explanation of these apparently contradictory observations is that a physiologically relevant target of the E4-6/7 protein is of cellular origin. This possibility stipulates that the putative cellular target gene(s) contains an inverted configuration of E2F binding sites that forms a stable DNA bound E2F complex with E4-6/7. The promoter regions of the human and murine E2F-1 genes contain two pairs of overlapping E2F sites with a configuration similar to that found with the Ad5 E2a promoter region (Fig. 4A) (25, 28). The E2F-1 promoter sequence shown in Fig. 4A is identical in human and mouse cells. The spacing between the E2F binding sites is 3 bp closer than that found with the Ad5 E2a promoter. To determine if the cellular E2F-1 promoter could form a stable E2F complex with E4-6/7, an E2F-1 promoter probe corresponding to this region was used. Ad5-infected HeLa cell extract was incubated with the E2F-1 promoter probe in comparison to an Ad5 E2a probe and analyzed by mobility shift assay (Fig. 4B). A series of complexes were detected with the E2F-1 promoter probe, but only the slowest-migrating complex was stable to competitor challenge in an off-rate analysis (10-, 20-, and 40-min challenges with a 500-fold excess of specific E2F site competitor DNA) in a manner identical to the Ad5 E2a probe fragment. That this slowly migrating complex contained the Ad5 E4-6/7 protein was verified using a specific monoclonal antibody (41) to this viral gene product (data not shown). When extracts from Ad3-, Ad9-, and Ad12-infected A549 cells were analyzed with the E2F-1 promoter probe, a similar complex that was stable to competitor challenge was found (Fig. 4C). We conclude that different Ad serotypes induce stable E2F binding to the cellular E2F-1 promoter.
FIG. 4.
Ad induction of E2F binding to the cellular E2F-1 promoter. (A) Nucleotide sequence comparison of the Ad5 E2a promoter E2F binding sites and similar sites found in the cellular E2F-1 promoter. Inverted E2F binding sites are indicated by arrows below the sequences. (B) Binding of Ad-induced E2F to the E2a and E2F-1 binding sites. Lanes 1 and 5 show binding reactions using Ad5-infected HeLa cell extract with probes corresponding to Ad5 E2a (lane 1) or the cellular E2F-1 promoter E2F binding sites (lane 5). The positions of free E2F and the Ad-induced E2F (Ad-E2F) complexes are indicated on the left. Following a 60-min binding reaction, a 500-fold molar excess of unlabeled E2F single binding site was added, and aliquots of the binding reaction were removed and loaded on the gel at the times indicated above the lanes (10, 20, and 40 min after the addition of specific competitor DNA). (C) Different Ad serotypes induce E2F binding to the E2F-1 promoter. Nuclear extracts from uninfected (Uninf.) A549 cells or A549 cells infected with Ad3, Ad9, or Ad12 were incubated with the probe corresponding to the E2F-1 promoter. Lanes 1, 4, 7, and 10 show a binding reaction without the addition of specific competitor DNA. In lanes 2, 5, 8, and 11, a 500-fold molar excess of unlabeled E2F single binding site was added coincident with the probe. In lanes 3, 6, 9, and 12, a 500-fold molar excess of unlabeled E2F binding site was added after a 60-min binding reaction. The binding reactions were incubated an additional 15 min before loading on the gel. The position of the Ad-induced E2F complex (Ad-E2F) is indicated on the right.
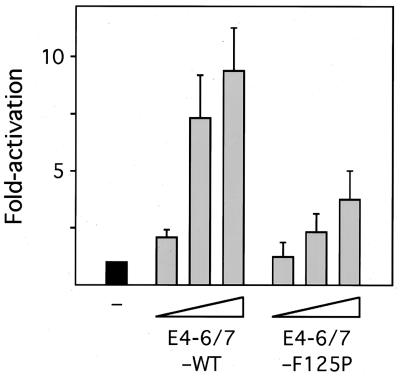
To test the functional consequence of Ad induction of E2F binding to the E2F-1 promoter, three different assays were used. First, a reporter vector containing the cellular E2F-1 promoter linked to the CAT gene was transfected into HepG2 cells alone or with increasing amounts of E4-6/7 expression vectors (Fig. 5). Two E4-6/7 proteins were tested in this analysis: E4-6/7 wild type (E4-6/7-WT) and E4-6/7-F125P, a point mutant that contains a single amino change at position 125 (41). E4-6/7-F125P was previously shown to bind efficiently to E2F, but it is defective for E2F dimerization. This F125P mutant protein was analyzed to distinguish between two possible mechanisms of E2F activation in this assay: relief of Rb family protein repression via displacement of Rb by E4-6/7 interaction with E2F and E2F-1 promoter transactivation via E4-6/7 dimerization of E2F DNA binding activity. The E4-6/7-WT and E4-6/7-F125P mutant proteins were shown to be equally stable when expressed in vivo (41). The level of CAT activity was taken as a measure of promoter induction. These results showed that E4-6/7-WT induced expression from the E2F-1 promoter in HepG2 cells to a significantly higher level that the dimerization mutant E4-6/7-F125P. We attribute transactivation by the mutant protein to relief of Rb repression and the additional increase in activation observed with E4-6/7-WT protein to the induction of E2F dimerization and DNA binding.
FIG. 5.
Transactivation of the E2F-1 promoter by the E4-6/7 protein in HepG2 cells. HepG2 cells were transfected with a reporter vector containing the cellular E2F-1 promoter fused to CAT. The reporter vectors were transfected alone (−) or with increasing concentrations (2, 10, and 50 ng) of a vector expressing Ad2 E4-6/7-WT or E4-6/7-F125P. Following transfection, the cells were maintained in medium containing 0.5% serum for 24 h. The level of CAT activity in cellular extracts was then measured. The level of expression of the reporter vector alone is set at 1, and the results with E4-6/7 coexpression are indicated as fold activation relative to the basal level. The results represent the average of five experiments. Error bars indicate standard deviations.
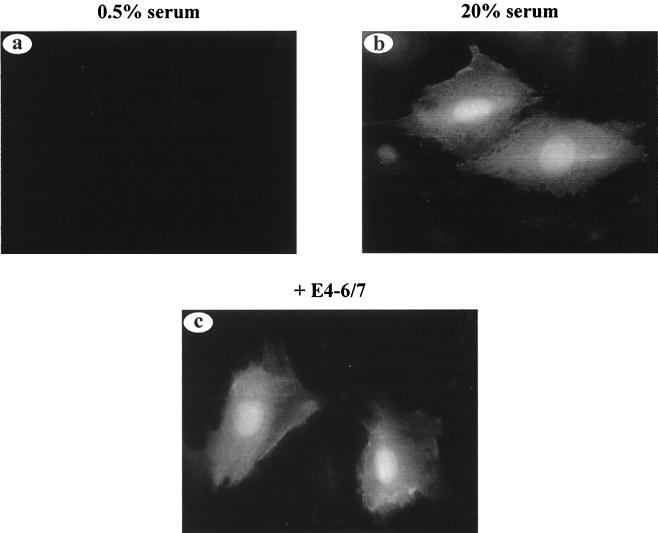
In a second assay, quiescent REF52 cells were microinjected with a plasmid containing the E2F-1 promoter region linked to the GFP coding region with or without coinjection of the E4-6/7-WT expression vector (Fig. 6). REF52 cells were used for this experiment since a majority of the cells establish quiescence under the experimental conditions used and they are readily microinjected. GFP fluorescence was taken as a measure of E2F-1 promoter activity. In quiescent cells, the E2F-1 promoter is repressed by Rb family-E2F bound complexes, and little GFP activity was evident. In contrast, the E4-6/7 protein significantly induced E2F-1 promoter activity to a level similar to that found with serum induction. The dimerization mutant E4-6/7-F125P was capable of transactivation of the E2F-1–GFP reporter vector under these experimental conditions (data not shown). This result is consistent with the ability of E4-6/7 to displace Rb from E2F and thus relieve E2F promoter repression (Fig. 5), particularly under conditions of growth arrest where the majority of E2F is bound by Rb family members. The nonlinear response of a GFP fluorescence signal in relation to GFP protein levels precludes accurate quantitative comparison of the level of induction by E4-6/7-WT versus E4-6/7-F125P, although we note that fewer microinjected cells showed increased GFP fluorescence with E4-6/7-F125P in comparison to E4-6/7-WT. We conclude from these analyses that the E4-6/7 protein can induce stable E2F binding to the cellular E2F-1 promoter via an inverted E2F binding site in vitro and that this induction of E2F binding by E4-6/7 correlates well with transactivation of the E2F-1 promoter by E4-6/7 in vivo.
FIG. 6.
Transactivation of the E2F-1 promoter by the E4-6/7 protein in REF52 cells. Quiescent REF52 cells were microinjected with the E2F-1–GFP reporter vector. Cells were coinjected with the E2F-1–GFP vector and an empty CMV vector (a and b) or coinjected with the E2F-1–GFP vector and an expression vector for the Ad2 E4-6/7 protein. In panels a and c, the cells were maintained in 0.5% serum; panel b, the cells were treated with medium containing 20% FBS. GFP activity was visualized 16 h after microinjection using a fluorescein filter on an Axiovert 135 microscope (Zeiss).
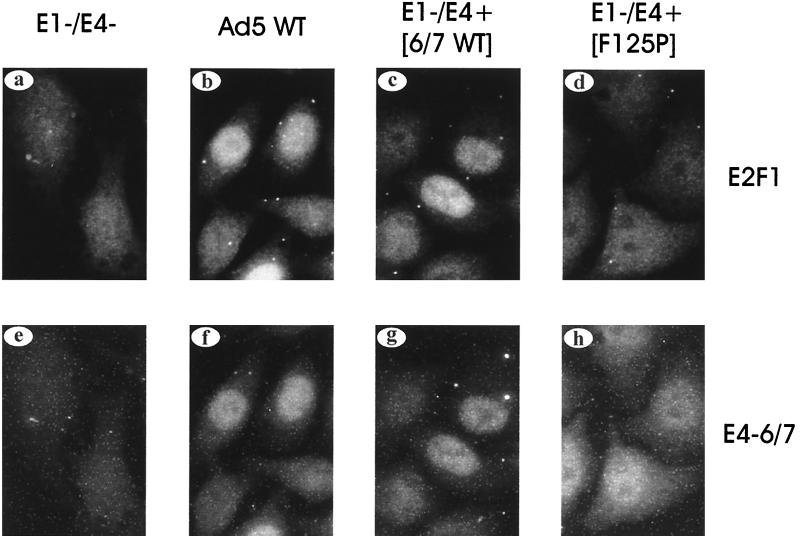
In the third assay, endogenous E2F-1 protein levels were measured in HeLa cells infected with Ad recombinants that express different viral E2F modulators. HeLa cells were used for this experiment since they contain a significant level of free E2F-1 activity, and thus relief of Rb repression of E2F by the E4-6/7 protein should be minimized. HeLa cells were infected with wild-type Ad, an Ad that lacks the E1 region but constitutively expresses E4-6/7-WT, an Ad that lacks the E1 region but constitutively expresses E4-6/7-F125P, or an Ad that lacks E1 and also contains a mutation that disrupts the E4-6/7 protein. E4-6/7 and endogenous E2F-1 protein levels were measured by immunofluorescence using antibodies specific to these products. E4-6/7 expression was observed in cells infected with viruses that express either E4-6/7-WT or E4-6/7-F125P (Fig. 7f to h), whereas no signal was observed in cells infected with a virus that lacks an intact E4-6/7 reading frame (Fig. 7e). Importantly, E4-6/7-WT but not E4-6/7-F125P augmented the level of endogenous E2F-1 activity to a significant extent (compare Fig. 7b and c with Fig. 7d). These results confirm the transfection and microinjection studies (Fig. 5 and 6) and demonstrate that E4-6/7-WT induced E2F-1 expression to a significantly higher level than the dimerization mutant E4-6/7-F125P, consistent with transactivation of the endogenous E2F-1 promoter. In related experiments, the viral E2a gene product DBP showed the same pattern of induction (data not shown), supporting the notion that the regulation of the E2a and E2F-1 promoter regions during infection is similar.
FIG. 7.
Induction of E2F-1 protein levels by the E4-6/7 protein in Ad-infected HeLa cells. HeLa cells were infected with wild-type Adenovirus (expressing E1A and E4-6/7; b and f) or a recombinant that lacks the E1 region and constitutively expresses E4-6/7-WT (c and g), E4-6/7-F125P (d and h), or an E1-lacking virus that also contains a mutation that disrupts the E4-6/7 protein (a and e). Cells were harvested at 8 h after infection, and E2F-1 (a to d) and E4-6/7 (e to h) were visualized by indirect immunofluorescence using antibodies specific to these products.
DISCUSSION
The Ad E4-6/7 protein interacts directly with different members of the E2F family and mediates the cooperative and stable binding of E2F to a unique pair of binding sites in the Ad5 E2a promoter region (16, 17, 26, 36, 48). This induction of E2F DNA binding activity strongly correlates with increased E2a transcription when analyzed in virus infection and transient expression assays (38, 39, 41, 42, 49). E2F heterodimers are typically found associated with members of the Rb family of tumor suppressor proteins that repress E2F activity. In resting cells, where very little free E2F activity is present, these interactions are believed to preclude E4-6/7 binding with E2F due to masking of the marked box region, a segment in E2F-1 required for binding to Rb and the E4-6/7 protein in vivo (7, 14, 19, 20, 43). Ad E1A proteins function to physically displace Rb family proteins from E2Fs, thereby allowing association with E4-6/7 (8). The prevailing model of E4-6/7 protein function proposes that E2F induction is an Ad-specific mechanism for recruiting and sequestering E2F activity at a viral promoter region. The experiments described in this report show that this model may not fully represent the scope of E4-6/7 function during Ad infection.
Previous experiments with an Ad mutant that does not express a functional E4-6/7 protein have shown that the mutant virus grows as well as wild-type Ad5 in HeLa cells (15). Thus, while the E4-6/7 protein augments E2a promoter activity via induction of E2F DNA binding, this function of the E4-6/7 protein is not essential for Ad growth in cultured cells. This appears to be due to the dominant nature of the multifunctional E1A proteins exhibited in cultured cells. The conservation of E2F induction by evolutionarily diverse Ad isolates described in this report argues for a selective advantage for virus growth provided by the E4-6/7 protein. While the precise nature of this advantage is not yet certain, our results suggest that the induction of E2F binding to cellular, rather than viral, promoter regions may represent the basis for the selection of E4-6/7 activity. Whereas all Ad isolates tested were capable of induction of E2F DNA binding activity, not all Ad E2a promoter regions recruit this induced E2F activity. The Ad12 and Ad40 E2a promoter regions bind E2F via a single binding site (Fig. 3C), yet the same promoter intervals very weakly recruit dimerized (Ad-induced) E2F activity (Fig. 3B). The Ad3 E2a promoter region binds E2F very weakly, even via a single binding site. It remains possible that the E4-6/7 proteins from diverse Ad serotypes such as 3, 12, and 40 have evolved stable interactions with E2F and other distinct transcription factors in their promoter regions. While we can not preclude this possibility, experiments to test this idea using matched infected-cell nuclear extracts and E2a promoter regions of different Ad serotypes did not reveal any novel Ad-induced DNA binding complexes (data not shown).
Our data show that adenovirus infection induces the binding of E2F to an inverted configuration of binding sites in the cellular E2F-1 promoter. This induction of E2F binding observed in vitro (Fig. 4B and C) correlates well with the observed transactivation of this promoter by E4-6/7 expression in vivo (Fig. 5 to 7). The results from both transient expression assays (Fig. 5 and 6) support the notion that E4-6/7 can effectively modulate transcription from E2F-responsive promoter elements. However, the reliability of making direct inferences about endogenous promoter activity based solely on heterologous transient expression assays prompted our examination of endogenous E2F-1 activity in Ad-infected cells (Fig. 7). Indeed, we found a striking induction of E2F-1 protein levels following expression of E4-6/7-WT protein but not the dimerization mutant E4-6/7-F125P, supporting the idea that this increase is due to transactivation of the endogenous cellular E2F-1 promoter. The transactivation of the cellular E2F-1 promoter by the E4-6/7 protein appears to complement the relief of repression of this promoter, driven by E1A protein-mediated release of Rb-repressive complexes from endogenous E2F activity. In addition, the ability of E4-6/7-F125P to transactivate E2F-1 promoter expression (Fig. 5) likely reflects the capacity of this protein to displace Rb from E2F-1 via competition for overlapping binding regions, i.e., the marked box region of E2F-1 (19, 21, 43). The inverted configuration of E2F binding sites found in the E2F-1 promoter that serve as a target for E4-6/7-induced E2F DNA binding activity may represent a fortuitous arrangement of these elements. Indeed, while a number of cellular promoter regions carry multiple E2F binding sites, this is the only cellular promoter region that we have identified that contains the inverted configuration of E2F sites and that stably binds Ad-induced E2F activity. It is possible that a subset of cellular E2F-responsive promoter regions carry such inverted E2F binding sites and that these sites may be targets of specific E2F protein complexes. Perhaps the E4-6/7 protein has evolved as an analogue of a cellular gene product which serves the same function. Considering the centrol role that the E2F-1 gene product plays in the regulation of cell cycle progression, a novel regulatory circuit may control its expression.
Finally, why have only certain Ad serotypes evolved inverted E2F binding sites that are capable of recruiting Ad-induced E2F activity? Ad2 and Ad5 were originally isolated from long-term cultures of adenoid tissue. A large proportion of the cell mass of adenoids consists of lymphoid cells. Group C viruses (Ad1, Ad2, Ad5, and Ad6) establish persistent infections of both lymphoid and monocyte cultures in vitro (6, 52). In addition, DNA sequences of group C Ad have been isolated from peripheral blood lymphocytes from healthy individuals (24). While not yet proven, these results have suggested that group C Ad isolates establish persistent infections of circulating lymphocytes. The stable binding of E2F–E4-6/7 complexes to the viral E2a promoter region may help to mediate viral DNA replication in these cells via activated expression of the replication genes driven by the E2a promoter. When free E2F activity is available in late G1 and early S phase, the resident Ad in a latent infection may need an active mechanism to recruit E2F to the viral E2a promoter. In this situation, limiting viral genomes are in competition with numerous cellular E2F binding sites for E2F activity. This may contrast with lytic infection where amplified viral DNA may readily compete for limiting concentrations of cellular transcription factors to allow for efficient E2a transcription. This very speculative possibility awaits further investigation.
ACKNOWLEDGMENTS
The first two authors contributed equally to this work.
We thank Mike Hayman, Todd Miller, Nancy Reich, Peter Tegtmeyer, and our laboratory colleagues for many helpful discussions, and we thank Gia Feeney and Tina Philipsberg for excellent technical help. We thank Bayar Thimmappaya for the E2a-CAT vector, Peggy Farnham for the E2F-1 promoter clone, and Sergei Zolotukhin for the GFP clone.
This research was supported by Public Health Service grant CA28146 from the National Cancer Institute to D.B. and P.H. R.J.O., J.S., and L.J.T. were supported by NIH training grant CA09176.
REFERENCES
- 1.Adnane J, Shao Z, Robbins P D. The retinoblastoma susceptibility gene product represses transcription when directly bound to the promoter. J Biol Chem. 1995;270:8837–8843. doi: 10.1074/jbc.270.15.8837. [DOI] [PubMed] [Google Scholar]
- 2.Blake M C, Azizkhan J C. Transcription factor E2F is required for efficient expression of the hamster dihydrofolate reductase gene in vitro and in vivo. Mol Cell Biol. 1989;9:4994–5002. doi: 10.1128/mcb.9.11.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botz J, Zerfass-Thome K, Spitkovsky D, Delius H, Vogt B, Eilers M, Hatzigeorgiou A, Jansen-Durr P. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol Cell Biol. 1996;16:3401–3409. doi: 10.1128/mcb.16.7.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brehm A, Miska E A, McCance D J, Reid J L, Bannister A J, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 5.Bremner R, Cohen B L, Sopta M, Hamel P A, Ingles C J, Gallie B L, Phillips R A. Direct transcriptional repression by pRB and its reversal by specific cyclins. Mol Cell Biol. 1995;15:3256–3265. doi: 10.1128/mcb.15.6.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chu Y, Sperber K, Mayer L, Hsu M T. Persistent infection of human adenovirus type 5 in human monocyte cell lines. Virology. 1992;188:793–800. doi: 10.1016/0042-6822(92)90534-v. [DOI] [PubMed] [Google Scholar]
- 7.Cress W D, Johnson D G, Nevins J R. A genetic analysis of the E2F1 gene distinguishes regulation by Rb, p107, and adenovirus E4. Mol Cell Biol. 1993;13:6314–6325. doi: 10.1128/mcb.13.10.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cress W D, Nevins J R. Use of the E2F transcription factor by DNA tumor virus regulatory proteins. Curr Top Microbiol Immunol. 1996;208:63–78. doi: 10.1007/978-3-642-79910-5_3. [DOI] [PubMed] [Google Scholar]
- 9.Dalton S. Cell cycle regulation of the human cdc2 gene. EMBO J. 1992;11:1797–1804. doi: 10.1002/j.1460-2075.1992.tb05231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 12.Ferreira R, Magnaghi-Jaulin L, Robin P, Harel-Bellan A, Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc Natl Acad Sci USA. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Field S J, Tsai F Y, Kuo F, Zubiaga A M, Kaelin W G, Jr, Livingston D M, Orkin S H, Greenberg M E. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- 14.Hagemeier C, Cook A, Kouzarides T. The retinoblastoma protein binds E2F residues required for activation in vivo and TBP binding in vitro. Nucleic Acids Res. 1993;21:4998–5004. doi: 10.1093/nar/21.22.4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halbert D N, Cutt J R, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy S, Engel D A, Shenk T. An adenovirus early region 4 gene product is required for induction of the infection-specific form of cellular E2F activity. Genes Dev. 1989;3:1062–1074. doi: 10.1101/gad.3.7.1062. [DOI] [PubMed] [Google Scholar]
- 17.Hardy S, Shenk T. E2F from adenovirus-infected cells binds cooperatively to DNA containing two properly oriented and spaced recognition sites. Mol Cell Biol. 1989;9:4495–4506. doi: 10.1128/mcb.9.10.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hateboer G, Wobst A, Petersen B O, Le Cam L, Vigo E, Sardet C, Helin K. Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol Cell Biol. 1998;18:6679–6697. doi: 10.1128/mcb.18.11.6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helin K, Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 is required for binding to the adenovirus E4 (ORF6/7) protein. J Virol. 1994;68:5027–5035. doi: 10.1128/jvi.68.8.5027-5035.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helin K, Harlow E, Fattaey A. Inhibition of E2F1 transactivation by direct binding of the retinoblastoma protein. Mol Cell Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helin K, Wu C L, Fattaey A R, Lees J A, Dynlacht B D, Ngwu C, Harlow E. Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 1993;7:1850–1861. doi: 10.1101/gad.7.10.1850. [DOI] [PubMed] [Google Scholar]
- 22.Hiebert S W, Packham G, Strom D K, Haffner R, Oren M, Zambetti G, Cleveland J L. E2F-1:DP-1 induces p53 and overrides survival factors to trigger apoptosis. Mol Cell Biol. 1995;15:6864–6874. doi: 10.1128/mcb.15.12.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holmberg C, Helin K, Sehested M, Karlstrom O. E2F-1-induced p53-independent apoptosis in transgenic mice. Oncogene. 1998;17:143–155. doi: 10.1038/sj.onc.1201915. [DOI] [PubMed] [Google Scholar]
- 24.Horvath J, Palkonyay L, Weber J. Group C adenovirus DNA sequences in human lymphoid cells. J Virol. 1986;59:189–192. doi: 10.1128/jvi.59.1.189-192.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsiao K M, McMahon S L, Farnham P J. Multiple DNA elements are required for the growth regulation of the mouse E2F1 promoter. Genes Dev. 1994;8:1526–1537. doi: 10.1101/gad.8.13.1526. [DOI] [PubMed] [Google Scholar]
- 26.Huang M-M, Hearing P. The adenovirus early region 4 open reading frame 6/7 protein regulates the DNA binding activity of the cellular transcription factor, E2F, through a direct complex. Genes Dev. 1989;3:1699–1710. doi: 10.1101/gad.3.11.1699. [DOI] [PubMed] [Google Scholar]
- 27.Johnson D G, Cress W D, Jakoi L, Nevins J R. Oncogenic capacity of the E2F1 gene. Proc Natl Acad Sci USA. 1994;91:12823–12827. doi: 10.1073/pnas.91.26.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson D G, Ohtani K, Nevins J R. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle expression. Genes Dev. 1994;8:1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 29.Johnson D G, Schwartz J K, Cress W D, Nevins J R. Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature. 1993;365:349–352. doi: 10.1038/365349a0. [DOI] [PubMed] [Google Scholar]
- 30.Kovesdi I, Reichel R, Nevins J R. Identification of a cellular transcription factor involved in E1A trans-activation. Cell. 1986b;45:219–228. doi: 10.1016/0092-8674(86)90386-7. [DOI] [PubMed] [Google Scholar]
- 31.Kowalik T F, DeGregori J, Leone G, Jakoi L, Nevins J R. E2F1-specific induction of apoptosis and p53 accumulation, which is blocked by Mdm2. Cell Growth Differ. 1998;9:113–118. [PubMed] [Google Scholar]
- 32.Kowalik T F, DeGregori J, Schwarz J K, Nevins J R. E2F1 overexpression in quiescent fibroblasts leads to induction of cellular DNA synthesis and apoptosis. J Virol. 1995;69:2491–2500. doi: 10.1128/jvi.69.4.2491-2500.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam E W-F, Watson R J. An E2F binding site mediates cell cycle regulated repression of mouse B-myb transcription. EMBO J. 1993;12:2705–2713. doi: 10.1002/j.1460-2075.1993.tb05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leone G, DeGregori J, Yan Z, Jakoi L, Ishida S, Williams R S, Nevins J R. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 1998;12:2120–2130. doi: 10.1101/gad.12.14.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 36.Marton M J, Baim S B, Ornelles D A, Shenk T. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulation E1A-independent accumulation of E2 mRNA. J Virol. 1990;64:2345–2359. doi: 10.1128/jvi.64.5.2345-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulligan G, Jacks T. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- 38.Neill S D, Hemstrom C, Virtanen A, Nevins J R. An adenovirus E4 gene product trans-activates E2 transcription and stimulates stable E2F binding through a direct association with E2F. Proc Natl Acad Sci USA. 1990;87:2008–2012. doi: 10.1073/pnas.87.5.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neill S D, Nevins J R. Genetic analysis of the adenovirus E4 6/7 trans activator: interaction with E2F and induction of a stable DNA-protein complex are critical for activity. J Virol. 1991;65:5364–5373. doi: 10.1128/jvi.65.10.5364-5373.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neuman E, Flemington E K, Sellers W R, Kaelin W G., Jr Transcription of the E2F-1 gene is rendered cell cycle dependent by E2F DNA-binding sites within its promoter. Mol Cell Biol. 1994;14:6607–6615. doi: 10.1128/mcb.14.10.6607. . (Erratum, 15:4660, 1995.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obert S, O'Connor R J, Schmid S, Hearing P. The adenovirus E4-6/7 protein transactivates the E2 promoter by inducing dimerization of a heteromeric E2F complex. Mol Cell Biol. 1994;14:1333–1346. doi: 10.1128/mcb.14.2.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O'Connor R J, Hearing P. The C-terminal 70 amino acids of the adenovirus E4-ORF6/7 protein are essential and sufficient for E2F complex formation. Nucleic Acids Res. 1991;19:6579–6586. doi: 10.1093/nar/19.23.6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Connor R J, Hearing P. Mutually exclusive interaction of the adenovirus E4-6/7 protein and the retinoblastoma gene product with internal domains of E2F-1 and DP-1. J Virol. 1994;68:6848–6862. doi: 10.1128/jvi.68.11.6848-6862.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohtani K, DeGregori J, Leone G, Herendeen D R, Kelly T J, Nevins J R. Expression of the HsOrc1 gene, a human ORC1 homolog, is regulated by cell proliferation via the E2F transcription factor. Mol Cell Biol. 1996;16:6977–6984. doi: 10.1128/mcb.16.12.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohtani K, DeGregori J, Nevins J R. Regulation of the cyclin E gene by transcription factor E2F1. Proc Natl Acad Sci USA. 1995;92:12146–12150. doi: 10.1073/pnas.92.26.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phillips A C, Bates S, Ryan K M, Helin K, Vousden K H. Induction of DNA synthesis and apoptosis are separable functions of E2F-1. Genes Dev. 1997;11:1853–1863. doi: 10.1101/gad.11.14.1853. [DOI] [PubMed] [Google Scholar]
- 47.Qin X Q, Livingston D M, Kaelin W G, Jr, Adams P D. Deregulated transcription factor E2F-1 expression leads to S-phase entry and p53-mediated apoptosis. Proc Natl Acad Sci USA. 1994;91:10918–10922. doi: 10.1073/pnas.91.23.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raychaudhuri P, Bagchi S D, Neill S, Nevins J R. Activation of the E2F transcription factor in adenovirus-infected cells involves E1A-dependent stimulation of DNA-binding activity and induction of cooperative binding mediated by an E4 gene product. J Virol. 1990;64:2702–2710. doi: 10.1128/jvi.64.6.2702-2710.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reichel R, Neill S D, Kovesdi I, Simon M C, Raychaudhuri P, Nevins J R. The adenovirus E4 gene, in addition to the E1A gene, is important for trans-activation of E2 transcription and for E2F activation. J Virol. 1989;63:3643–3650. doi: 10.1128/jvi.63.9.3643-3650.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schulze A, Zerfass K, Spitkovsky D, Middendorp S, Berges J, Helin K, Jansen-Durr P, Henglein B. Cell cycle regulation of the cyclin A gene promoter is mediated by a variant E2F site. Proc Natl Acad Sci USA. 1995;92:11264–11268. doi: 10.1073/pnas.92.24.11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shan B, Lee W H. Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol Cell Biol. 1994;14:8166–8173. doi: 10.1128/mcb.14.12.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silver L, Anderson C W. Interaction of human adenovirus serotype 2 with human lymphoid cells. Virology. 1988;165:377–387. doi: 10.1016/0042-6822(88)90582-x. [DOI] [PubMed] [Google Scholar]
- 53.Singh P, Wong S H, Hong W. Overexpression of E2F-1 in rat embryo fibroblasts leads to neoplastic transformation. EMBO J. 1994;13:3329–3338. doi: 10.1002/j.1460-2075.1994.tb06635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tollefson A E, Herminston T W, Wold W S M. Preparation and titration of CsCl-banded adenovirus stock. In: Wold W S M, editor. Methods in molecular medicine. 1999. pp. 1–10. Adenovirus methods and protocols. Edited by Humana Press, Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 55.Tommasi S, Pfeifer G P. In vivo structure of the human cdc2 promoter: release of a p130–E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol Cell Biol. 1995;15:6901–6913. doi: 10.1128/mcb.15.12.6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z M, Yang H, Livingston D M. Endogenous E2F-1 promotes timely G0 exit of resting mouse embryo fibroblasts. Proc Natl Acad Sci USA. 1998;95:15583–15586. doi: 10.1073/pnas.95.26.15583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weintraub S J, Chow K N, Luo R X, Zhang S H, He S, Dean D C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature. 1995;375:812–815. doi: 10.1038/375812a0. [DOI] [PubMed] [Google Scholar]
- 58.Wigler M, Silverstein S, Lee L S, Pellicer A, Cheng Y C, Axel R. Transfer of purified herpes simplex virus thymidine kinase gene to cultured mouse cells. Cell. 1977;11:223–232. doi: 10.1016/0092-8674(77)90333-6. [DOI] [PubMed] [Google Scholar]
- 59.Wu X, Levine A J. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci USA. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu G, Livingston D M, Krek W. Multiple members of the E2F transcription factor family are the products of oncogenes. Proc Natl Acad Sci USA. 1995;92:1357–1361. doi: 10.1073/pnas.92.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson N J. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 62.Zolotukhin S, Potter M, Hauswirth W W, Guy J, Muzyczka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–4654. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]