Abstract
To generate synthetic medical data incorporating image-tabular hybrid data by merging an image encoding/decoding model with a table-compatible generative model and assess their utility. We used 1342 cases from the Stony Brook University Covid-19-positive cases, comprising chest X-ray radiographs (CXRs) and tabular clinical data as a private dataset (pDS). We generated a synthetic dataset (sDS) through the following steps: (I) dimensionally reducing CXRs in the pDS using a pretrained encoder of the auto-encoding generative adversarial networks (αGAN) and integrating them with the correspondent tabular clinical data; (II) training the conditional tabular GAN (CTGAN) on this combined data to generate synthetic records, encompassing encoded image features and clinical data; and (III) reconstructing synthetic images from these encoded image features in the sDS using a pretrained decoder of the αGAN. The utility of sDS was assessed by the performance of the prediction models for patient outcomes (deceased or discharged). For the pDS test set, the area under the receiver operating characteristic (AUC) curve was calculated to compare the performance of prediction models trained separately with pDS, sDS, or a combination of both. We created an sDS comprising CXRs with a resolution of 256 × 256 pixels and tabular data containing 13 variables. The AUC for the outcome was 0.83 when the model was trained with the pDS, 0.74 with the sDS, and 0.87 when combining pDS and sDS for training. Our method is effective for generating synthetic records consisting of both images and tabular clinical data.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10278-024-01015-y.
Keywords: Synthetic data generation, Data sharing, Auto-encoding GAN, CTGAN, COVID-19
Introduction
With the advancement of deep-learning technology, its application in the field of radiology is being increasingly recognized [1, 2]. Deep-learning technologies have emerged as essential partners for radiologists, offering significant improvements in both diagnostic accuracy and efficiency [3]. Developing, validating, and maintaining such sophisticated deep-learning models requires a considerable volume of medical data. Therefore, acquiring and managing a large amount of better-quality data is crucial. However, this is not easy owing to the sensitive nature of medical data.
Synthetic medical image generation has received significant attention in response to this challenge. Studies utilizing generative adversarial networks (GANs) have been particularly prevalent for this purpose, and prior research has shown that the use of synthetic images for data augmentation can enhance the performance of algorithms trained on such data [4–7]. Recent reports have described the use of denoising diffusion probabilistic models (DDPM) for data augmentation in medical imaging [8]. In addition, in real-world scenarios, images are often accompanied by descriptive or related information rather than existing in isolation. To address this, technologies for the simultaneous processing and generation of both image and non-image data have been developed [9–11].
Such synthetic data generation technologies are also gaining attention from the perspectives of privacy preservation and data sharing [12]. By effectively mimicking the target data, generative models enable a more secure and private sharing of medical information [13]. This approach is particularly valuable for sharing actual patient data, which is risky because of privacy concerns. However, this requires a large amount of data for successful synthetic data generation [14]. This presents a challenge for small medical researcher groups or single facilities that typically have access to only a few hundred to thousands of data points. Furthermore, in actual medical settings, patient information often comprises a hybrid of data including images, physical findings, blood tests, and other structured data. However, attempts to create comprehensive hybrid patient data are limited. Hybrid data are extremely important for discovering new insights that cannot be obtained with a single modality or for enabling extensive secondary analysis. Therefore, there is a pressing need for innovative ideas to generate more flexible hybrid data with fewer data points, especially in terms of data augmentation and sharing in small studies.
The development of models for generating structured data emerged later than that of image generation models [15–17]. In the medical field, models based on GANs, such as conditional tabular GAN (CTGAN), are widely used [18–20]. As these models deal with inputs of much lower dimensionality than images, they are expected to be successfully trained with far fewer data points [21]. Based on this understanding, we hypothesized that if the dimensionality of images is reduced a priori using a dimensionality encoding/decoding model trained on an external dataset and integrated with tabular data, meaningful synthetic data can be generated using CTGAN, even with a limited number of cases. The purpose of this study is to evaluate the quality and usefulness of a synthetic hybrid dataset generated using the proposed method.
Materials and Methods
Overview of Our Study
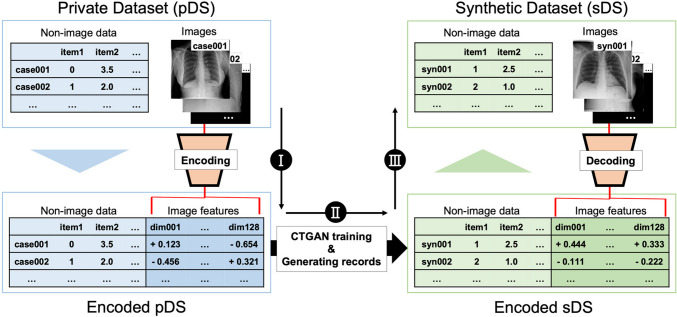
We used a public dataset containing 13 structured variables (tabular clinical data) and a chest X-ray (CXR) with a resolution of 256 × 256 pixels as the private dataset (pDS). Figure 1 provides an overview of the proposed method for generating synthetic hybrid data, which involves (I) dimensional reduction of CXRs; (II) training the CTGAN model, and (III) reconstruct the image features into synthetic CXRs. After generating a synthetic dataset (sDS), the coherence between images and tabular data was evaluated. Additionally, we assessed sDS by examining the area under the receiver operating characteristic curve (AUC) when used to train patient outcome prediction models. Statistical analyses were conducted using R software (version 4.3.1; R Foundation for Statistical Computing, Vienna, Austria). P-values less than 0.05 were considered as statistically significant.
Fig. 1.
Overview of the proposal method. Our approach comprises the following three steps: I dimensional reduction of chest X-ray radiographs (CXRs) in a private dataset (pDS) using a pretrained encoder and merging with the remaining tabular clinical data; II training the CTGAN model on these merged tabular data and generating synthetic records that contain encoded image features and tabular clinical data; and III reconstructing the image features to synthetic CXRs using a pretrained decoder
Datasets and Preprocessing
In this study, we used the Stony Brook University Covid-19-positive cases, a publicly available hybrid dataset containing CXRs and non-image clinical data [22, 23]. This hybrid database comprises 1384 COVID–19-positive cases. It features diverse imaging modalities and comprehensive non-image clinical tabular data. Originally, there were 130 variables per case in the non-imaging data. After excluding cases in which the initial CXR was not available, as well as variables with more than 5% missing values and similar items, we obtained data on 1343 cases along with 13 clinical variables (Supplementary file 1). We randomly split the pDS into training, validation, and test datasets at a ratio of 6:2:2. The detailed demographics of each subset of pDS are presented in the “Results” section. For the categorical variables, missing values were replaced with a new category representing the absence of data. For the numerical variables, missing values were imputed using the mean values from the training and validation sets. All images in the pDS were resized to 256 × 256 pixel.
The Radiological Society of North America (RSNA) Pneumonia Detection Challenge dataset [24], hereafter referred to as the RSNA dataset, was used to pretrain the encoding/decoding model. This publicly available dataset contains approximately 30,000 frontal view CXRs, each classified by one to three board-certified radiologists into the following categories: “Normal,” “No Opacity/Not Normal,” or “Opacity.” The “Opacity” category includes images indicative of opacities suggestive of pneumonia, while the “No Opacity/Not Normal” category comprises images showing abnormalities other than pneumonia. The diversity of pathologies included in the RSNA dataset makes it a suitable choice for pretraining, as it is expected to encompass the range of image findings present in the pDS. For our study, 1000 cases from the RSNA dataset were designated as the test set, with the remaining data split in an 8:2 ratio to form the training and validation sets. All the images in the RSNA dataset were resized to 256 × 256 pixels.
Preliminary Study of Dimensional Encoding–Decoding for CXRs
Pretraining of Encoding–Decoding Models on RSNA Dataset
The methods for dimensional encoding of images range from classical machine learning models (such as principal component analysis (PCA) and t-distributed stochastic neighbor embedding (t-SNE)) to modern deep-learning technologies (including convolutional neural networks (CNN), autoencoder (AE), and auto-encoding GAN (αGAN)) [25, 26]. In the proposed method, it is necessary to employ models that are capable of decoding. Therefore, we selected PCA [27] as the baseline, AE [28], and αGAN [29]. Using these models, we validated the 128-dimensional code transformation of a 256 × 256-pixel CXR and its inverse transformation. PCA was trained using a combined training and validation set of the RSNA dataset, while AE and αGAN were trained using the training set, with the validation set used for monitoring the learning process. Descriptions and structures of the models used in our study are provided in Supplementary file 1.
Selection of Encode/Decode Models
To determine the most effective dimensional encoding/decoding model, two radiologists reviewed 10 original images from the test set alongside the reconstructed images produced by each model. A consensus was reached on which model generated the “most authentic-like image set.” Additionally, the peak signal-to-noise ratio (PSNR) [30], structural similarity (SSIM) [31], and mean squared error (MSE) [32] were calculated and tested using paired t-tests. P-values were adjusted using the Holm method.
Synthetic Data Generation and Evaluation
Synthetic Hybrid Data Generation
Figure 1 presents an overview of this study. We adopted CTGAN [18] as the network for synthetic data generation. CTGAN is a GAN-based model for modeling the distribution of tabular data. It performs normalization on each column of complex data distributions with respect to categorical variables and trains the model using a conditional generator and discriminator. We used the stand-alone library, CTGAN (version 0.7.4; https://github.com/sdv–dev/CTGAN). After obtaining the encoded pDS by passing the CXRs to the pretrained encoder of the αGAN model, we trained the CTGAN model using training and validation set of the encoded pDS, and a number of synthetic records corresponding to the training and validation sets of the pDS were generated. These synthetic records encompassed both image features and tabular clinical data (encoded sDS). The CXRs were reconstructed by passing the synthetic image features in encoded sDS to the pretrained generator of the αGAN model. The detailed demographics of the patients with sDS are presented in the “Results” section.
Consistency Evaluation of Images and Tabular Data Via Contrastive Learning
To evaluate the correspondence between image and tabular data for the proposed method, contrastive learning was implemented as follows: after encoding CXRs and tabular clinical data into an 18-dimensional latent code using other encoders (Supplementary file 1), the training was directed such that positive pairs (pairs of image and tabular data from the same patients) moved closer together, whereas negative pairs (pairs of image and tabular data from different patients) diverged. The networks were trained on pDS and sDS independently, and we evaluated the correspondence between the image and tabular data through the following steps using the test set of pDS: (I) calculating the cosine similarity between the encoded results of an image and the corresponding tabular data using the trained encoders, (II) computing the cosine similarity of this image with all the other tabular records, (III) determining the rank of the value from Step I among the values from Step 2, and (IV) repeating this procedure for all images in the test set. The aggregate results were displayed as histograms, and the difference in results when encoders were trained with pDS and when they were trained with sDS was examined using the Wilcoxon signed-rank test.
Evaluating the Utility of sDS Through Predictive Modeling
To evaluate the utility and quality of sDS for pDS, we trained and tested a predictive model with Last Status as an independent variable. The original purpose of pDS is described as “building AI systems for diagnostic and prognostic modeling” (https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=89096912), and the Last Status, indicating that patients are deceased/discharged, was considered the most critical variable. We constructed a network using both image and tabular data as inputs, with the output being the prediction of the Last Status (Supplementary file 1). The model was trained multiple times using different data combinations. For each training session, we systematically replaced the records in pDS with those in sDS. This replacement was performed in increments of 25%, progressively shifting the composition from 100% pDS (1:0) to 100% sDS (0:1). The records that were replaced in each training session were chosen randomly. In addition, we trained the model using all the training and validation sets of pDS and sDS. We then compared the AUC results from different training scenarios. We used the DeLong test and adjusted P-values using the Holm method. The results of the classification and regression for items other than the Last Status using the same model are presented in Supplementary file 1.
Results
Table 1 summarizes the preprocessed pDS used in this study. The training, validation, and test sets comprised 800, 272, and 271 patients, respectively.
Table 1.
Summary of the contents of the dataset used as private dataset
| Non-image items |
Training set n = 800 |
Validation set n = 272 |
Test set n = 271 |
|---|---|---|---|
| Categorical variables (number of patients) | |||
| Last status | |||
| Discharged | 692 | 237 | 235 |
| Deceased | 108 | 35 | 36 |
| Age splits | |||
| [18,59] | 428 | 168 | 149 |
| (59, 74] | 218 | 53 | 72 |
| (74, 90] | 154 | 51 | 50 |
| Gender concept name | |||
| Male | 439 | 152 | 153 |
| Female | 343 | 110 | 114 |
| NA | 18 | 10 | 4 |
| Visit concept name | |||
| Inpatient visit | 588 | 204 | 203 |
| Outpatient visit | 2 | 0 | 0 |
| Emergency room visit | 210 | 68 | 68 |
| Is ICU | |||
| True | 144 | 57 | 57 |
| False | 656 | 215 | 214 |
| Was ventilated | |||
| Yes | 120 | 47 | 46 |
| No | 680 | 225 | 225 |
| Acute kidney injury | |||
| Yes | 147 | 51 | 50 |
| No | 653 | 221 | 221 |
| Numeric variables (mean ± standard deviation) | |||
| Length of stay (day) | 10.0 ± 12.7 | 8.9 ± 9.6 | 10.1 ± 13.2 |
| Oral temperature (°C) | 37.5 ± 0.9 | 37.5 ± 0.8 | 37.6 ± 0.9 |
| Oxygen saturation (%) | 93.8 ± 5.5 | 93.5 ± 6.5 | 93.8 ± 5.6 |
| Respiratory rate (/min) | 21.7 ± 7.5 | 22.2 ± 7.6 | 21.1 ± 6.7 |
| Heart rate (/min) | 97.9 ± 19.4 | 100.6 ± 19.4 | 98.8 ± 22.0 |
| Systolic blood pressure (mmHg) | 129.4 ± 23.1 | 128.7 ± 22.1 | 129.0 ± 22.4 |
*This dataset is a subset of the Stony Brook University Covid-19 Positive Cases. The description for each variable is provided in Supplementary file 1. Please also refer to the information provided in the original dataset: https://wiki.cancerimagingarchive.net/pages/viewpage.action?pageId=89096912
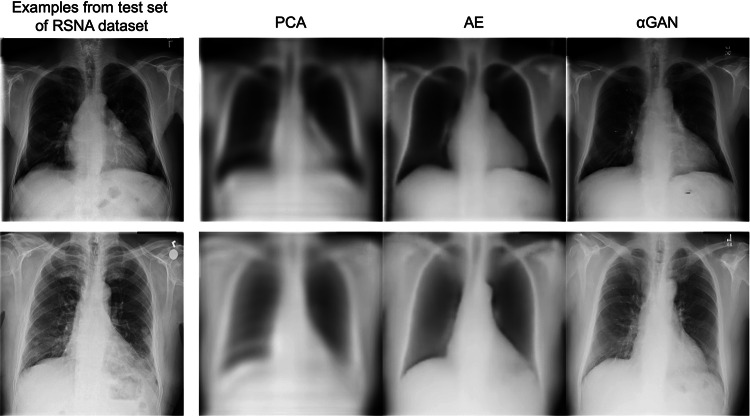
Figure 2 presents the samples of the encode–decode process using the actual test set of the RSNA dataset. During the stage of selecting the encoder/decoder model to be used, a consensus was reached by two radiologists that the reconstructed images from the αGAN were closest to the actual images. Consequently, the αGAN was employed in subsequent implementations. The SSIM, PSNR, and MSE values of each model tested on the test set of the RSNA datasets are summarized in Table 2. Although quantitative metrics indicated that the AE delivered the best results, visual evaluation by radiologists was prioritized during model selection.
Fig. 2.
Samples of encode/decode results using three models. From left to right, the original image, reconstructed image using PCA, the AE, and the αGAN
Table 2.
Aggregated results of PSNR, SSIM, and MSE for the three models
| PCA | AE | αGAN | P-value | |
|---|---|---|---|---|
| PSNR | 26.51 ± 1.83 | 29.16 ± 1.40 | 25.36 ± 1.68 |
PCA–AE: p < 0.001 AE–αGAN: p < 0.001 PCA–αGAN: p < 0.001 |
| SSIM | 0.73 ± 0.05 | 0.80 ± 0.04 | 0.74 ± 0.04 |
PCA–AE: p < 0.001 AE–αGAN: p < 0.001 PCA–αGAN: p = 0.052 |
| MSE | 158.44 ± 69.60 | 83.05 ± 28.13 | 204.27 ± 86.12 |
PCA–AE: p < 0.001 AE–αGAN: p < 0.001 PCA–αGAN: p < 0.001 |
PCA principal component analysis, AE autoencoder, αGAN auto-encoding generative adversarial networks, PSNR peak signal-to-noise ratio, SSIM structural similarity, MSE mean squared error
P-values were adjusted using the Holm method
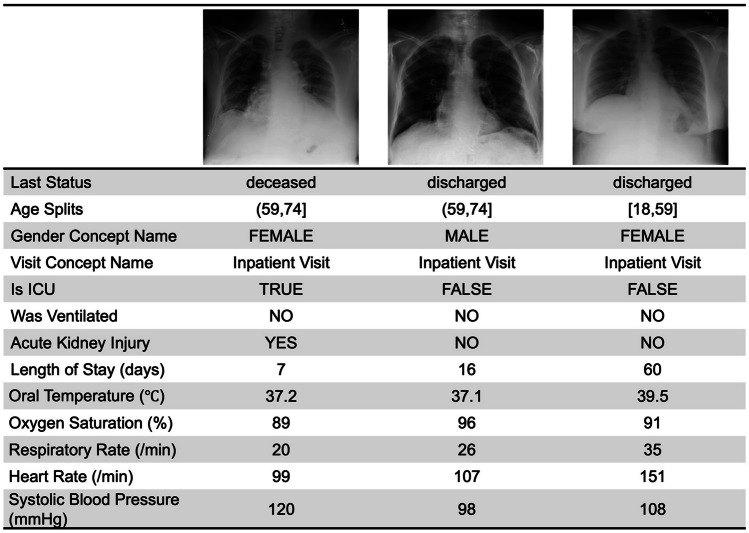
Table 3 presents the demographic information of the synthetic data generated during implementation. The number of synthesized records was matched to that of the training and validation sets of the pDS. Figure 3 displays three examples of synthetic hybrid records.
Table 3.
Summary of the contents of the synthetic dataset
| Non-image items |
Synthetic training set n = 800 |
Synthetic validation set n = 272 |
|---|---|---|
| Categorical variables (number of patients) | ||
| Last status | ||
| Discharged | 663 | 224 |
| Deceased | 137 | 48 |
| Age splits | ||
| [18,59] | 414 | 135 |
| (59, 74] | 236 | 84 |
| (74, 90] | 150 | 53 |
| Gender concept name | ||
| Male | 441 | 147 |
| Female | 323 | 105 |
| NA | 36 | 20 |
| Visit concept name | ||
| Inpatient visit | 595 | 210 |
| Outpatient visit | 6 | 3 |
| Emergency room visit | 199 | 59 |
| Is ICU | ||
| True | 207 | 75 |
| False | 593 | 197 |
| Was ventilated | ||
| Yes | 134 | 46 |
| No | 666 | 566 |
| Acute kidney injury | ||
| Yes | 201 | 94 |
| No | 599 | 178 |
| Numeric variables (mean ± standard deviation) | ||
| Length of stay (day) | 15.3 ± 18.7 | 16.1 ± 18.8 |
| Oral temperature (°C) | 37.7 ± 0.9 | 37.7 ± 0.9 |
| Oxygen saturation (%) | 92.3 ± 8.1 | 92.8 ± 7.1 |
| Respiratory rate (/min) | 21.6 ± 8.0 | 22.0 ± 8.1 |
| Heart rate (/min) | 106.8 ± 25.87 | 105.9 ± 23.6 |
| Systolic blood pressure (mmHg) | 121.6 ± 27.0 | 121.0 ± 24.9 |
The description for each variable is provided in Supplementary file 1
Fig. 3.
Three samples of synthetic hybrid records
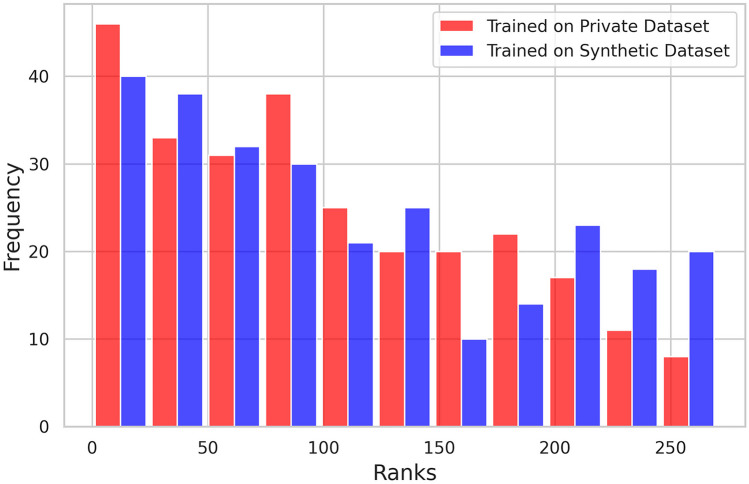
Figure 4 illustrates the results of contrastive learning conducted independently on the pDS and sDS and evaluated using the pDS test set, showing histograms of similarity rankings for positive pairs. The histograms display higher plots on the left side for training on both pDS and sDS, suggesting that contrastive learning was successful and that the consistency between the images and tabular data was preserved, even in the sDS. However, the slope of the histogram resulting from training on the sDS was less steep than that on the pDS. The Wilcoxon signed-rank test revealed a statistically significant difference in the median ranks, which were higher (indicating poorer performance) when learning on the sDS (p = 0.041), as shown in Table 4.
Fig. 4.
Evaluation of metric learning using the pDS test set. Red represents the training and validation performed on the pDS, whereas blue represents the training performed on the sDS and validation conducted on the pDS
Table 4.
Aggregated results of similarity rank for positive pairs post-metric learning
| Trained on pDS | Trained on sDS | P-value | |
|---|---|---|---|
| Rank | 92 [38 − 163] | 96 [43 − 186] | 0.041 |
Values represent the median [interquartile range]
Figure 5 and Table 5 show the receiver operating characteristic curves and AUCs for predicting Last Status with varying ratios of pDS and sDS in the training and validation sets. An increase in the proportion of the sDS corresponded with a decrease in the AUC (from 0.83 to 0.74). However, when comparing combinations using the DeLong test with pDS:sDS = 1:0 as reference, no significant differences were observed. Additionally, the AUC for the model trained with both pDS and sDS was 0.87, which was higher than that for the model trained with only the pDS (AUC = 0.83), although this difference was not statistically significant (p = 0.346). Supplementary file 1 presents the classification and regression results for items other than Last Status.
Fig. 5.
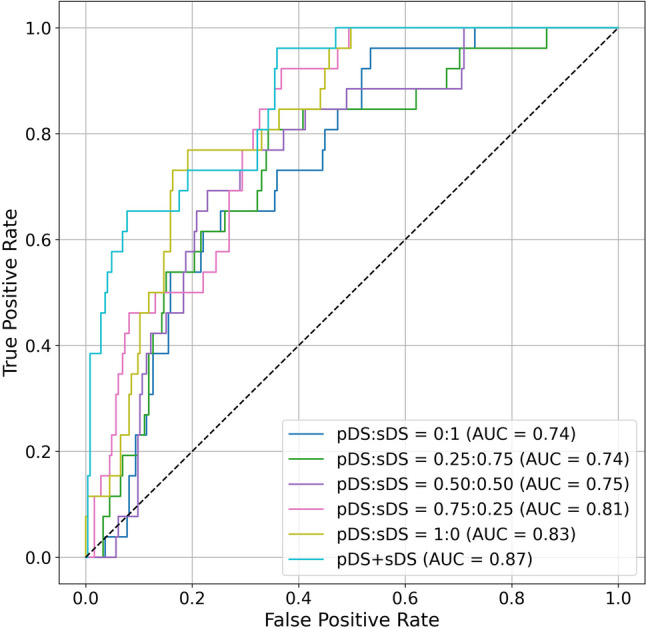
Receiver operating characteristic curves and AUCs for predicting Last Status. The models were evaluated on the test set or pDS for six different pDS and sDS ratios
Table 5.
AUC values and results of DeLong’s test
| Dataset (pDS:sDS) | AUC | P-value (compared with pDS:sDS = 1:0) |
|---|---|---|
| 0:1 | 0.74 | 0.143 |
| 0.25:0.75 | 0.74 | 0.092 |
| 0.5:0.5 | 0.75 | 0.149 |
| 0.75:0.75 | 0.81 | 0.576 |
| 1:0 | 0.83 | Reference |
| pDS + sDS | 0.87 | 0.346 |
P-values were adjusted using the Holm method
Discussion
This study illustrated the practicality of generating hybrid image-tabular records by merging encoded image features from the pretrained encoder of αGAN with tabular clinical data, which were then input into the CTGAN. Pretraining the encoding/decoding model for dimensionality compression using external large-scale datasets can circumvent the requirement for a large pDS. As a result, we successfully generated records comprising CXRs with a resolution of 256 × 256 pixels and tabular data with 13 variables derived from 1072 cases in the training and validation sets. In the downstream prediction task, the decrease resulting from replacing the pDS with the sDS was slight, further suggesting the possibility of using the sDS for data augmentation.
In addition to the αGAN used herein, various advanced deep-learning models have been developed in the field of image encoding and decoding. This includes a range of evolutions and integrations of architectures such as the AE, the GAN, and transformer-based architectures [33–35]. In this study, we adopted the αGAN, based on implementations from previous research that successfully reconstructed images with a similar dataset and image modality using a relatively small number of latent codes [36]. For comparative analysis, we conducted preliminary investigations with models of simpler structures and mechanisms, specifically, simple AE and PCA. When the encoded dimensionality was set to 128, the AE exhibited superior performance in quantitative metrics, including SSIM, PSNR, and MSE. However, the αGAN was clearly superior in reconstructing images that were vivid and closer to the actual CXRs, leading to its adoption in subsequent processes. Consequently, it was confirmed that the synthetic CXRs in the sDS were also well reconstructed, as presented in Fig. 3. Table 3 summarizes the geometric properties of the tabular clinical data from the sDS (800 and 272 records from the training and validation sets, respectively). The distribution order of categorical variables in the sDS was unchanged from that in the pDS, and continuous variables did not show unrealistic mean values. An evaluation of the sDS by contrastive learning suggested that the sDS may be a slightly degraded dataset compared to the pDS in terms of consistency between the image and tabular data.
In medicine, deep generative models are primarily used for data augmentation and data sharing purposes [14, 37–39]. However, the simultaneous generation of both image and non-image data in small-scale databases, as demonstrated herein, remains limited. We believe that it is important to address such scenarios considering the actual clinical situation wherein imaging and non-imaging information always coexist. Furthermore, there has been an increase in medical research that incorporates simultaneous analysis of images and tabular data to obtain new clinical insights [40–42]. Consequently, hybrid datasets have garnered attention as valuable research sources. Our technique for generating hybrid records can enhance integrated analyses in terms of data augmentation and sharing. In our experiments, the AUC for the Last Status task was 0.83 when employing only the pDS and 0.74 when using only the sDS. When the pDS and sDS were combined, the AUC was 0.87. In addition to the variations in their ratios, the changes in the AUCs were not statistically significant according to DeLong’s test. However, considering the results presented in Supplementary file 1, when the sDS completely replaced the pDS, the classification and regression performance deteriorated in 7 out of 13 items, indicating that a perfect sDS had not been achieved at this point. However, replacing up to 25% of the pDS with the sDS did not degrade performance in any of the tasks. Furthermore, in 2 out of 14 tasks, data augmentation with the sDS was found to be significantly successful.
The potential for generalization of our method likely hinges on the availability of large external image datasets (not hybrid datasets) with the same imaging modalities and imaged regions for training encoder/decoder models. In our scenario, the availability of the RSNA dataset, which is large and considered to encompass the radiographic findings present in the pDS dataset, contributed to the good results. Recently, the availability of large-scale medical image datasets has increased. As the number of these datasets increases, the potential to generalize the proposed methods may also increase. Furthermore, the scale of the datasets used herein (~ 30,000 images in the external dataset and ~ 1343 records in the pDS), image resolution (256 × 256 pixels), and number of data items (seven categorical and six continuous variables) could provide a baseline of information for future research.
A significant potential competitor to our research is the language learning model (LLM). Recent advances in LLMs have enabled them to handle multimodal inputs [43, 44]. To a certain extent, the capacity to manage and manipulate abstract information within latent space overlaps with the objectives of our current methodology. However, our approach distinctly focuses on the generation of synthesized data that closely align with the characteristics of the original data. This outcome distinguishes the proposed method. Although both LLMs and our technique share a common underpinning in the form of advanced learning mechanisms, their roles remain divergent because of their different goals. As this field continues to evolve, we anticipate that the functionalities of these two methodologies will retain their individuality and serve complementary roles in various applications.
In future studies, we will consider extending to three-dimensional medical images. Modalities such as CT and MRI provide rich three-dimensional information and have a more significant impact on diagnosis [45]. If this method can be adapted to three-dimensional images, such as CT scans, it can provide even greater utility. Another potential application of our method is the introduction of a more rigorous notion of privacy. Although generative models are often used to protect privacy, they are not entirely free from the risk of privacy leakage [13]. Recently, the concept of differential privacy (DP) has been applied to deep learning to guarantee strict privacy protection [46, 47]. There is research in which a CTGAN was implemented as a DP-CTGAN [48], and the fusion of this technology with ours is an important future theme.
There are several limitations in this study. First, we used αGAN and CTGAN models in our main implementation, which can be replaced with other networks. Our study utilized the αGAN and CTGAN based on their proven capabilities in specific medical contexts [20, 36]. However, recent advancements in transformer-based methods, diffusion models, and other GAN alternatives may enhance performance. We acknowledge that exploring these newer models could further optimize our approach. This consideration is particularly important because our current synthetic dataset did not achieve the best possible results or quality, suggesting room for improvement in future iterations. Secondly, the limits regarding the pDS, such as the minimum number of records or the upper limit of column numbers that enable meaningful synthetic data generation, is unknown. Our experimental results serve as indicators for future research. Third, in this study, one of the primary limitations is the lack of well-established metrics for evaluating consistency between tabular data and corresponding images. While we have utilized existing methods such as contrastive learning approaches to assess this consistency, these metrics are still in nascent stages, particularly in the context of hybrid datasets that combine radiographic images and clinical records. The development of more refined and specialized metrics that can accurately measure the alignment and representational accuracy between different data modalities remains an area for future research.
Conclusion
Using αGAN and CTGAN models, we generated synthetic hybrid records consisting of CXRs with a size of 256 × 256 pixels and tabular data with 13 variables. When another large image dataset was available, the proposed method enabled the creation of synthetic hybrid data from approximately 1000 records.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
TK and SH conceived the study. TN and YN significantly contributed to preparing and managing data sets. TT, HM, and TY contributed to the interpretation of the results. All authors reviewed the manuscript draft and critically revised the intellectual content. All authors approved the final version of the manuscript to be published.
Funding
This study was partially supported by the JST CREST (Grant Number JPMJCR21M2).
Data Availability
The datasets utilized in our paper are publicly available and can be accessed through the references cited in the text. While the models developed and the data generated for this study will not be publicly released, they can be obtained from the corresponding author upon reasonable request.
Declarations
Ethical Statement
The authors declare that the work described herein did not involve experimentation with humans or animals.
Competing Interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, Kadoury S, Tang A. Deep learning: a primer for radiologists. Radiographics. 2017;37:2113–2131. doi: 10.1148/rg.2017170077. [DOI] [PubMed] [Google Scholar]
- 2.Cheng PM, Montagnon E, Yamashita R, Pan I, Cadrin-Chênevert A, Perdigón Romero F, Chartrand G, Kadoury S, Tang A. Deep learning: an update for radiologists. Radiographics. 2021;41:1427–1445. doi: 10.1148/rg.2021200210. [DOI] [PubMed] [Google Scholar]
- 3.van Leeuwen KG, Schalekamp S, Rutten MJCM, van Ginneken B, de Rooij M. Artificial intelligence in radiology: 100 commercially available products and their scientific evidence. Eur Radiol. 2021;31:3797–3804. doi: 10.1007/s00330-021-07892-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frid-Adar M, Diamant I, Klang E, Amitai M, Goldberger J, Greenspan H. GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification. Neurocomputing. 2018;321:321–331. doi: 10.1016/j.neucom.2018.09.013. [DOI] [Google Scholar]
- 5.Onishi Y, Teramoto A, Tsujimoto M, Tsukamoto T, Saito K, Toyama H, Imaizumi K, Fujita H. Automated pulmonary nodule classification in computed tomography images using a deep convolutional neural network trained by generative adversarial networks. Biomed Res Int. 2019;2019:6051939. doi: 10.1155/2019/6051939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gadermayr M, Li K, Müller M, Truhn D, Krämer N, Merhof D, Gess B. Domain-specific data augmentation for segmenting MR images of fatty infiltrated human thighs with neural networks. J Magn Reson Imaging. 2019;49:1676–1683. doi: 10.1002/jmri.26544. [DOI] [PubMed] [Google Scholar]
- 7.Russ T, Goerttler S, Schnurr A-K, Bauer DF, Hatamikia S, Schad LR, Zöllner FG, Chung K. Synthesis of CT images from digital body phantoms using CycleGAN. Int J Comput Assist Radiol Surg. 2019;14:1741–1750. doi: 10.1007/s11548-019-02042-9. [DOI] [PubMed] [Google Scholar]
- 8.Müller-Franzes G, Niehues JM, Khader F, Arasteh ST, Haarburger C, Kuhl C, Wang T, Han T, Nolte T, Nebelung S, Kather JN. A multimodal comparison of latent denoising diffusion probabilistic models and generative adversarial networks for medical image synthesis. Sci Rep. 2023;13:12098. doi: 10.1038/s41598-023-39278-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee H, Park S, Lee J, Choi E: Unconditional image-text pair generation with multimodal cross quantizer. arXiv preprint, 10.48550/arXiv.2204.07537 (October 14, 2022)
- 10.Hu M, Zheng C, Zheng H, Cham T-J, Wang C, Yang Z, Tao D, Suganthan PN: Unified discrete diffusion for simultaneous vision-language generation. arXiv prepirnt, 10.48550/arXiv.2211.14842 (November 27, 2022)
- 11.Chambon P, Bluethgen C, Delbrouck J-B, Van der Sluijs R, Połacin M, Chaves JMZ, Abraham TM, Purohit S, Langlotz CP, Chaudhari A: RoentGen: vision-language foundation model for chest X-ray generation. arXiv preprint, 10.48550/arXiv.2211.12737 (November 23, 2022)
- 12.Giuffrè M, Shung DL. Harnessing the power of synthetic data in healthcare: innovation, application, and privacy. Npj Digital Medicine. 2023;6:1–8. doi: 10.1038/s41746-023-00927-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen RJ, Lu MY, Chen TY, Williamson DFK, Mahmood F. Synthetic data in machine learning for medicine and healthcare. Nat Biomed Eng. 2021;5:493–497. doi: 10.1038/s41551-021-00751-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goncalves A, Ray P, Soper B, Stevens J, Coyle L, Sales AP. Generation and evaluation of synthetic patient data. BMC Med Res Methodol. 2020;20:108. doi: 10.1186/s12874-020-00977-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodriguez-Almeida AJ, Fabelo H, Ortega S, Deniz A, Balea-Fernandez FJ, Quevedo E, Soguero-Ruiz C, Wagner AM, Callico GM: Synthetic patient data generation and evaluation in disease prediction using small and imbalanced datasets. IEEE J Biomed Health Inform. 10.1109/JBHI.2022.3196697, 2022 [DOI] [PubMed]
- 16.Wang J, Yan X, Liu L, Li L, Yu Y: CTTGAN: traffic data synthesizing scheme based on conditional GAN. Sensors. 10.3390/s22145243, 2022 [DOI] [PMC free article] [PubMed]
- 17.Kotelnikov A, Baranchuk D, Rubachev I, Babenko A: TabDDPM: modelling tabular data with diffusion models. arXiv preprint, 10.48550/arXiv.2209.15421 (September 30, 2022)
- 18.Xu L, Skoularidou M, Cuesta-Infante A, Veeramachaneni K: Modeling tabular data using conditional GAN. arXiv preprint, 10.48550/arXiv.1907.00503 (October 28, 2019)
- 19.Bourou S, El Saer A, Velivassaki T-H, Voulkidis A, Zahariadis T. A review of tabular data synthesis using GANs on an IDS dataset. Information. 2021;12:375. doi: 10.3390/info12090375. [DOI] [Google Scholar]
- 20.Hameed MAB, Alamgir Z. Improving mortality prediction in acute pancreatitis by machine learning and data augmentation. Comput Biol Med. 2022;150:106077. doi: 10.1016/j.compbiomed.2022.106077. [DOI] [PubMed] [Google Scholar]
- 21.Fonseca J, Bacao F. Tabular and latent space synthetic data generation: a literature review. J Big Data. 2023;10:115. doi: 10.1186/s40537-023-00792-7. [DOI] [Google Scholar]
- 22.The cancer imaging archive. Available at https://doi.org/10.7937/TCIA.BBAG-2923. Accessed February 8, 2024
- 23.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, Tarbox L. The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26:1045–1057. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shih G, Wu CC, Halabi SS, Kohli MD, Prevedello LM, Cook TS, Sharma A, Amorosa JK, Arteaga V, Galperin-Aizenberg M, Gill RR, Godoy MC, Hobbs S, Jeudy J, Laroia A, Shah PN, Vummidi D, Yaddanapudi K, Stein A: Augmenting the national institutes of health chest radiograph dataset with expert annotations of possible pneumonia. Radiol Artif Intell 1:e180041, 2019 [DOI] [PMC free article] [PubMed]
- 25.Marlapalli K, Bandlamudi RSBP, Busi R, Pranav V, Madhavrao B: A review on image compression techniques, Singapore: Springer Singapore, 2021
- 26.Mishra D, Singh SK, Singh RK. Deep architectures for image compression: A critical review. Signal Processing. 2022;191:108346. doi: 10.1016/j.sigpro.2021.108346. [DOI] [Google Scholar]
- 27.Ng SC. Principal component analysis to reduce dimension on digital image. Procedia Comput Sci. 2017;111:113–119. doi: 10.1016/j.procs.2017.06.017. [DOI] [Google Scholar]
- 28.Bank D, Koenigstein N, Giryes R. Autoencoders. In: Rokach L, Maimon O, Shmueli E, editors. Machine Learning for Data Science Handbook: Data Mining and Knowledge Discovery Handbook. Cham: Springer International Publishing; 2023. pp. 353–374. [Google Scholar]
- 29.Rosca M, Lakshminarayanan B, Warde-Farley D, Mohamed S: Variational approaches for auto-encoding generative adversarial networks. arXiv preprint, 10.48550/arXiv.1706.04987 (October 21, 2017)
- 30.Deshpande RG, Ragha LL, Sharma SK. Video quality assessment through PSNR estimation for different compression standards. Indones J Electr Eng Comput Sci. 2018;11:918–924. [Google Scholar]
- 31.Wang Z, Simoncelli EP, Bovik AC: Multiscale structural similarity for image quality assessment. Proc. 37th IEEE Asilomar Conference on Signals, Systems and Computers, 2003.
- 32.Søgaard J, Krasula L, Shahid M, Temel D, Brunnström K, Razaak M. Applicability of existing objective metrics of perceptual quality for adaptive video streaming. IS&T Int Symp Electron Imaging. 2016;28:1–7. [Google Scholar]
- 33.van den Oord A, Vinyals O, Kavukcuoglu K: Neural discrete representation learning. arXiv preprint, 10.48550/arXiv.1711.00937 (May 30, 2018)
- 34.He K, Chen X, Xie S, Li Y, Dollár P, Girshick R: Masked autoencoders are scalable vision learners. arXiv preprint, 10.48550/arXiv.2111.06377 (December 19, 2021)
- 35.Esser P, Rombach R, Ommer B: Taming transformers for high-resolution image synthesis. Proc. 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 2021
- 36.Nakao T, Hanaoka S, Nomura Y, Murata M, Takenaga T, Miki S, Watadani T, Yoshikawa T, Hayashi N, Abe O. Unsupervised deep anomaly detection in chest radiographs. J Digit Imaging. 2021;34:418–427. doi: 10.1007/s10278-020-00413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhagat V, Bhaumik S: Data augmentation using generative adversarial networks for pneumonia classification in chest Xrays. Proc. Fifth International Conference on Image Information Processing (ICIIP), 2019
- 38.Osuala R, Kushibar K, Garrucho L, Linardos A, Szafranowska Z, Klein S, Glocker B, Diaz O, Lekadir K: Data synthesis and adversarial networks: A review and meta-analysis in cancer imaging. Med Image Anal 84:102704, 2022 [DOI] [PubMed]
- 39.Dayarathna S, Islam KT, Uribe S, Yang G, Hayat M, Chen Z. Deep learning based synthesis of MRI, CT and PET: Review and analysis. Med Image Anal. 2023;92:103046. doi: 10.1016/j.media.2023.103046. [DOI] [PubMed] [Google Scholar]
- 40.Wiemken TL, Kelley RR. Machine learning in epidemiology and health outcomes research. Annu Rev Public Health. 2020;41:21–36. doi: 10.1146/annurev-publhealth-040119-094437. [DOI] [PubMed] [Google Scholar]
- 41.Yin Q, Chen W, Zhang C, Wei Z. A convolutional neural network model for survival prediction based on prognosis-related cascaded Wx feature selection. Lab Invest. 2022;102:1064–1074. doi: 10.1038/s41374-022-00801-y. [DOI] [PubMed] [Google Scholar]
- 42.Kikuchi T, Hanaoka S, Nakao T, Nomura Y, Yoshikawa T, Alam MA, Mori H, Hayashi N. Relationship between thyroid CT density, volume, and future TSH elevation: A 5-year follow-up study. Life. 2023;13:2303. doi: 10.3390/life13122303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Acosta JN, Falcone GJ, Rajpurkar P, Topol EJ. Multimodal biomedical AI. Nat Med. 2022;28:1773–1784. doi: 10.1038/s41591-022-01981-2. [DOI] [PubMed] [Google Scholar]
- 44.Koh JY, Fried D, Salakhutdinov R: Generating images with multimodal language models. arXiv preprint, 10.48550/arXiv.2305.17216 (October 13, 2023)
- 45.Hussain S, Mubeen I, Ullah N, Shah SSUD, Khan BA, Zahoor M, Ullah R, Khan FA, Sultan MA. Modern diagnostic imaging technique applications and risk factors in the medical field: a review. Biomed Res Int. 2022;2022:5164970. doi: 10.1155/2022/5164970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dwork C. Differential privacy: A survey of results. In International conference on theory and applications of models of computation. Berlin, Heidelberg: Springer Berlin Heidelberg, 2008
- 47.Ziller A, Usynin D, Braren R, Makowski M, Rueckert D, Kaissis G. Medical imaging deep learning with differential privacy. Sci Rep. 2021;11:13524. doi: 10.1038/s41598-021-93030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang ML, Dhami DS, Kersting K: DP-CTGAN: Differentially private medical data generation using CTGANs. Proc. 20th International Conference on Artificial Intelligence in Medicine (AIME 2022), 2022
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets utilized in our paper are publicly available and can be accessed through the references cited in the text. While the models developed and the data generated for this study will not be publicly released, they can be obtained from the corresponding author upon reasonable request.