Abstract
Chronic hepatitis B virus (HBV) infection can lead to liver cirrhosis and hepatocellular carcinoma. Long-term interaction of the immune system with the virus results in the selection of escape mutants and viral persistence. In this work we characterize mutations in the enhancer I region isolated prior to liver transplantation from the HBV genomes of 10 patients with chronic HBV infection. The HBV-genomes were sequenced, and the enhancer I region was cloned into luciferase reporter constructs to determine the transcriptional activity. Functional studies were performed by transfecting HBV replication-competent plasmids into hepatoma cells. Analyses of the replication fitness of the mutant strains were conducted by biochemical analysis. In all HBV genomes the enhancer I region was mutated. Most of these mutations resulted in decreased transcriptional activity. The strongest effects were detectable in strains with mutations in the hepatocyte nuclear factor 3 and 4 (HNF3 and HNF4) binding sites of the enhancer I core domain. Replication-competent HBV constructs containing these mutations demonstrated up to 10-fold-reduced levels of virus replication. Before liver transplantation, when the mutant strains were detected in the patients' sera, low HBV DNA levels were found. After transplantation and reinfection with a wild-type virus, the levels of replication were up to 240-fold higher. Our results show that mutations in the enhancer I region of HBV have a major impact on HBV replication. These mutations may also determine the switch from high to low levels of viral replication which is frequently observed during chronic HBV infection.
Human infection with the hepatitis B virus (HBV) will lead either to an acute, self-limiting disease, to fulminant hepatic failure, or to chronic HBV infection. Despite the fact that only 3 to 10% of all individuals who acquire HBV will become chronic carriers, more than 400 million people worldwide are chronically infected with the virus (2). The risk of developing liver cirrhosis or hepatocellular carcinoma for these persons is 100 to 200 times higher than the risk for the rest of the population (16, 44).
After infection with HBV, a specific T- and B-cell response against viral proteins leads to clearance of the virus from the organism and development of lifelong immunity. However, in the chronic carrier state, the immune system fails to eliminate the virus, leading to a persistent infection with HBV (13, 32). There is growing evidence that escape from elimination by the immune system may be caused by the selection of mutant viruses during the course of HBV infection (7). The high frequency of mutations found in the HBV genome compared to those of other DNA viruses is caused by the inaccuracy of the viral polymerase, which lacks proofreading activity (30, 31, 35).
An example of the selection of a mutant virus has been provided by the replication of HBV in a newborn whose mother was chronically infected. Even though the baby received passive immunization immediately after birth to prevent HBV infection, HBV replication still occurred. Sequence analysis of the HBV S gene revealed a mutation in the “a” determinant, which disrupts antibody binding and thus prevents elimination of the virus by the immune system (8–10).
In addition to mutations which affect the immunogenicity of the virus, changes which influence its replication competence may be of similar importance. During chronic HBV infection, two different stages can be differentiated. The time immediately after the first contact with the virus is characterized by high levels of HBV DNA and a strong inflammatory response in the liver (44). At a later stage, there is a strong decrease in the level of replication, which is frequently associated with seroconversion from hepatitis B virus e antigen (HBeAg) to anti-HBe (28).
The amount of viral gene products, including the pregenomic RNA, is controlled at the level of transcription. Four promoter and two enhancer elements have been characterized in the HBV genome. In these control regions, DNA binding motifs for liver-specific transcription factors have been identified which regulate transcription in a tissue-specific manner and thus directly contribute to the hepatotropism of HBV (1, 18, 23, 30, 34). Transcription of the pregenomic RNA is controlled through the core promoter and the enhancer I region. Besides the pregenomic RNA, the enhancer I region regulates transcription of the core and X genes (1, 19, 21, 26, 46). The activity of enhancer I is regulated by the complex interaction of hepatocyte-specific and ubiquitous transcription factors which can bind in this region (Fig. 1A) (24, 33, 45). The enhancer I region has been described as consisting of three domains (Fig. 1A). The modulator element contains ubiquitous and liver-specific transcription factor binding sites and spans the 5′ end of the enhancer I region. A central region, called the enhancer core domain, is responsible for the strong influence of the enhancer I region on the different promoters in the HBV genome, and the 3′ end overlaps with the X promoter (11, 15, 18, 20, 22, 24, 30, 41). At least four nuclear proteins have been identified which are able to bind to the core domain (3, 18, 22, 30). The external motifs 5′ and 3′ of the inner core domain bind nuclear factor 1 and rheumatoid factor 1, respectively. However, the tissue-specific regulation of enhancer I is mediated by two motifs located in the central core domain which bind proteins from the nuclear receptor family. One motif binds hepatocyte nuclear factor 4 (HNF4) (18, 22); the other motif represents a binding site for HNF3, a transcription factor that is expressed in several isoforms in the liver (24, 30).
FIG. 1.
Enhancer I mutants derived from 10 patients chronically infected with the hepatitis B virus. (A) DNA binding sites in the HBV enhancer I-X promoter between nt 2364 and 2768 of the HBV genome (pre-C ATG = 1) are depicted. The binding motifs of the liver-specific transcription factors HNF3 and HNF4 are marked in boldface letters. Binding sites for other ubiquitous or liver-specific factors are also shown. (B) Sequence analysis of patient-derived enhancer I regions showing deletions and mutations in comparison to the wt HBV enhancer I region. Between nt 2526 and 2558, the enhancer I sequences are highlighted (boldface italics) to show the mutations found in patients 1 and 3.
In this study we tested whether enhancer I mutations have an impact on viral replication. The mutations were derived from patients suffering from chronic HBV infection prior to undergoing liver transplantation for end-stage liver cirrhosis. In addition, we examined the impact of these mutations on the control of viral replication during the early and late stages of HBV infection. Collectively, our results demonstrate that the patient-derived mutations in the binding sites for HNF3 and HNF4 which downregulate enhancer I activity contribute to decreased replication fitness of these mutant viruses. Lower virus replication may result in an advantage for the chronic persistence of HBV.
(Part of this work was contained in the M.D. thesis of Katharinna Köhler.)
MATERIALS AND METHODS
Patients.
The 10 patients included in this study were transplanted for HBV-related end-stage liver disease between September 1985 and July 1992. The patients experienced HBV reinfection, which was defined as the reappearance of hepatitis B virus S antigen (HBsAg). Routinely, 20,000 IU of anti-HBs (hepatitis B virus immune globulin [HBIg]) was administered in the anhepatic phase, and 10,000 IU of HBIg was administered each day for up to 1 week after surgery (7). At later time points, anti-HBs titers were tested, and HBIg was given when the titers fell below 100 IU/liter. Nine out of 10 patients had the subtype adw2, and 1 patient had subtype ayw.
Serological assays.
Serum samples were aliquoted and stored at −20°C until they were defrosted for testing. HBsAg, anti-HBs, HBeAg, and anti-HBe were tested by commercial enzyme immunoassays (EIAs) (Abbott Laboratories, Chicago, Ill.).
Extraction of DNA from sera and amplification of the HBV genome.
DNA was extracted from sera (39) collected before orthotopic liver transplantation (OLT) and at time points after HBV reinfection was evident. Ninety microliters of serum was treated with 10 μl of proteinase K and 300 μl of DNA lysis buffer (0.67% sodium dodecyl sulfate (SDS), 13.33 mM Tris-HCl [pH 8.0], 6.6 mM EDTA [pH 8.0], and 13.3 mg of tRNA/ml). The mixture was incubated at 56°C for 2 h. Four hundred microliters of phenol-chloroform-isoamyl alcohol (25:24:1 [vol/vol]) was added and vortexed. The mixture was centrifuged for 30 min at 13,000 × g and 4°C. The DNA in 300 μl of the upper phase was precipitated by adding 660 μl of 100% ethanol and 30 μl of 3 M sodium acetate (pH 5.2) and centrifuged for 30 min at 13,000 × g and 4°C. After the pellet was briefly washed with 70% ethanol, it was dried and resuspended in 25 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0).
The enhancer I-X promoter region was amplified using the following primers: SF-sense, 5′-AAG CTT GGG TAT ACA TTT AAA CCC T-3′, and SF-antisense, 5′-GAA TTC AGA GTC CTC TTA TGC AAG AC-3′ (spanning the HBV nucleotides [nt] 2229 to 3075). Additionally, the region between nt 3021 and 162 of the HBV genome was amplified by two independent PCRs using the following primers: LF-sense, 5′-GAA TTC GGA AAG AAG TCA GAA GGC AA-3′, and LF-antisense, 5′-AAG CTT AGA CCA CCG TGA ACG CCC A-3′.
In these fragments, the pre-C region, the C promoter, the enhancer II region, and part of the coding region of the X gene are located. Amplification of fragments was performed as described before (40). For each sample, a negative control was treated in parallel starting at the extraction step to lower the probability of cross contamination. Additionally, a positive control was added. In cases where cross contamination was detected, all samples were discarded.
Subcloning and sequencing of PCR products was performed by using the TA cloning kit (Invitrogen, San Diego, Calif.) as described previously (39) and according to the manufacturer's instructions. After EcoRI digestion the sizes of the inserts were determined by agarose gel electrophoresis. Four clones were sequenced in an equimolar mix, and in case of double bands, single sequencing of the clones was performed. Sequencing was done by using an automated sequencer (ALFExpress; Pharmacia) or the Sequenase sequencing kit (United States Biochemicals). SF, LF, or universal primers (Pharmacia) were used for sequencing.
Plasmid construction.
To determine the transcriptional activity of the different enhancer I and enhancer I-X promoter mutants in luciferase assays, the PCR fragments from the serum-derived HBV DNAs of the 10 patients were introduced into the restriction sites KpnI and BglII of the multiple cloning sites of the pGL2 luciferase basic (without any promoter) or promoter reporter (simian virus 40 [SV40] promoter; Stratagene) vector using the following primers: Enh1-sense, 5′-GGG GTA CCC TTC CTG TTA ACA GGC CTA T-3′ (starting at nt 2364), and Enh1-antisense, 5′-GAA GAT CTG CTC CAG ACC GGC TGC GAG C-3′ (terminating at nt 2725). For the enhancer I-X promoter constructs, the following primers were used: Enh1-sense (described above) and Enh1-long: 5′-GAA GAT CTT TTC CGC GAG AGG ACG ACA GA-3′ (terminating at nt 2768). These primers had 5′ extensions with either the KpnI or the BglII restriction enzyme site (underlined sequences) spanning the HBV region from nt 2364 to 2725 or from nt 2364 to 2768, respectively. The enhancer I PCR fragments (nt 2364 to 2725) in all the plasmids were sequenced and checked for the presence of mutations compared to wild-type (wt) HBV (Fig. 1B).
The plasmid pHBV1.2 (subtype adw2) was generated by inserting an NcoI/BspEI HBV fragment from nt 2781 to 518 as a 1.28-mer overlength (4,178 bp) HBV DNA into the EcoRV restriction enzyme site of the multiple cloning site of the pBluescript KS(−) plasmid. The resulting plasmid, pHBV1.2, was checked for HBV replication competence and was used as wt HBV DNA.
To analyze the enhancer I mutations for replication competence, the mutated enhancer I HNF3 and HNF4 sequences were cloned using a two-step procedure with an intermediate cloning vector (HBV 3.2-kb SacII monomer silver vector) to replace the wt sequences with sequences of the enhancer I mutants. PCR fragments were generated from the serum-derived HBV DNA of patients 1 and 3 using the following primers: Enh-repli-sense, 5′-CTTCCTGTTAACAGGCCTATT-3′ (starting at nt 2364 and including the restriction enzyme HpaI site), and Enh-repli-antisense, 5′-AGC AGC CAT GGA AAC GAT GTA TAT TTC CGC GAG AGG ACG ACA GAA-3′ (terminating at nt 2791 and including the restriction enzyme NcoI site). The PCR fragments were digested with HpaI and NcoI. The resulting fragments were introduced into the HpaI and NcoI sites of the silver vector. After amplification of the newly generated vectors, the SacII fragment of the mutant silver vectors replaced the corresponding sequences in the pHBV1.2 plasmid. Each resulting plasmid was digested with several restriction endonucleases and subsequently sequenced to verify the presence of the mutations originally found in patients 1 and 3 (Fig. 1B).
For in vitro translation of HNF3 or HNF4 transcription factors, HNF3α, -β, and -γ, and HNF4 cDNAs, inserted in the EcoRI site of pBluescript KS(+) vector (kindly provided by J. Darnell, Rockefeller University, New York, N.Y.), were used. For protein expression, HNF3 or HNF4 cDNA was excised from the pBluescript vectors by the restriction enzyme EcoRI and transferred into the EcoRI site of the multiple cloning site of pcDNA3 expression vectors (Invitrogen).
Cell culture, transfection experiments, and luciferase assays.
HuH-7 cells (human hepatoma cells [27]), which are negative for HBV markers, were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum under 5% CO2 at 37°C. DNA transfection by the calcium phosphate coprecipitation method was performed as described previously (6). For analysis, the cells were harvested between 24 h and 6 days after transfection. Transfection efficiency was routinely checked by cotransfecting 0.2 μg of the β-galactosidase pCMVβGal vector as an internal standard (29). To verify the results, all transfection experiments were routinely performed in triplicate at a minimum.
For reporter gene experiments, 3 μg of the respective reporter gene construct was transfected alone or with the concentration of the respective expression (HNF4 or HNF3) vector indicated in the figures. The final amount of plasmid DNA was 5 μg of DNA per transfection experiment, containing 0.2 μg of the β-galactosidase pCMVβGal vector as an internal standard. The concentration was kept constant by adding the empty cytomegalovirus expression vector. Forty-eight hours after transfection, the HuH-7 cells were harvested, lysed, and measured exactly as described previously (29).
To monitor HBV replication in HuH-7 cells, 4.8 μg of the HBV replication-competent vector was transfected. The amount of total DNA was kept at 5 μg, containing 0.2 μg of the β-galactosidase pCMVβGal vector as an internal standard.
Intracellular HBeAg and HBsAg expression.
Intracellular HBeAg or HBsAg was evaluated by using whole-cell extracts from lysed transfected cells in buffer (120 mM NaCl, 1 mM EDTA, 10 mM Tris-HCl [pH 8.0], 0.1 mM Pefablock [Boehringer, Mannheim, Germany], 0.1 mM dithiothreitol, 1% Triton X-100). Extracellular HBeAg or HBsAg was collected from the transfected-cell-medium supernatant. Two hundred-microgram aliquots were analyzed by commercial EIAs (Abbott Laboratories).
Preparation of nuclear extracts and in vitro-translated protein.
HuH-7 nuclear extracts were prepared according to the method of Dignam et al. 1983 as described before (14).
For in vitro translation of HNF3 and HNF4, cDNA plasmids were used as described above. In vitro translations of HNF3α, -β, and -γ, and HNF4 proteins were performed as described previously (29) using reticulocyte lysate extracts (Pharmacia) according to the manufacturer's instructions. In vitro-transcribed HNF3 or HNF4 mRNA was used as a template. Newly synthesized proteins were analyzed by SDS-PAGE. Purified proteins were used in gel retardation assays.
Gel retardation assays.
For gel retardation assays, nuclear extracts from HuH-7 cells or in vitro-translated protein was used. Three micrograms of nuclear extracts was used unless otherwise indicated. For binding assays, an oligonucleotide spanning the region of interest was used as a 32P-labeled probe. Approximately 50 fmol, corresponding to 30,000 to 50,000 cpm, was used per binding reaction. The binding reaction was performed for 20 min on ice. Free DNA and DNA-protein complexes were resolved on a 6% polyacrylamide gel as described previously (37). The gels were dried and exposed for autoradiography.
For super-shift experiments, an anti-HNF4 antibody (kindly provided by J. Darnell) was incubated in the binding reaction.
The oligonucleotides used as 32P-labeled probes or as unlabeled oligonucleotides in the competition assays were as follows: The enhancer I HNF3 motif (spanning nt 2528 to 2546) included wt HNF3 sense, 5′-TCT AAG TAA ACA GTA CAT G-3′; wt HNF3 antisense, 5′-CAT GTA CTG TTT ACT TAG A-3′; ΔHNF3 sense, 5′-TCT GTG CAA ACA GTA CAT G-3′; and ΔHNF3 antisense, 5′-CAT GTA CTG TTT GCA CAG A-3′. The enhancer I HNF4 motif (spanning nt 2540 to 2576) included wt HNF4 sense, 5′-GTA CAT GAA CCT TTA CCC CGT TGC TCG GCA ACG GCC T-3′; wt HNF4 antisense, 5′-AGG CCG TTG CCG AGC AAC GGG GTA AAG GTT CAT GTA C-3′; ΔHNF4 sense, 5′-GTA CAT AAA CCT TTA CCC CGT TGC TCG GCA ACG GCC T-3′; and ΔHNF4 antisense, 5′-AGG CCG TTG CCG AGC AAC GGG GTA AAG GTT TAT GTA C-3′.
Northern blot analysis.
Northern blot analysis was performed as described before, according to standard procedures (38). In brief, total RNA was extracted from cells with the RNeasy kit (Qiagen Inc., Hilden, Germany) 2 to 3 days after transfection following the manufacturer's instructions. RNA samples were kept at −80°C until they were used. Twenty micrograms of total RNA was analyzed through a 1% MOPS [3-(N-morpholino)propanesulfonic acid] gel (20 mM MOPS, 5 mM sodium acetate, 1 mM EDTA; pH 6.8), followed by transfer to Hybond N+ nylon membrane (Amersham, Little Chalfont, England). For hybridization, a 32P-labeled monomeric HBV DNA probe was used. Equal loading of RNA samples was verified by reprobing the membrane with a 32P-labeled GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe.
Progeny HBV DNA.
Progeny HBV DNA was isolated from transfected cells as described previously (25). HBV capsids were immunoprecipitated from cell lysates by polyclonal rabbit anti-HBcAg antiserum (DAKO, Hamburg, Germany) as described previously. Residual transfected plasmid HBV DNA was eliminated by Staphylococcus aureus nuclease treatment (100 U per 106 cells; Boehringer). Viral progeny DNA was extracted from the capsids by using 500 μg of proteinase K (Sigma, Heidelberg, Germany)/ml–1% SDS at 56°C for 2 h. The DNA was purified by phenol extraction following separation on a 1% alkaline agarose gel (4). Southern blotting was performed on Hybond N+ nylon membranes.
SDS-PAGE and Western blot analysis.
SDS-PAGE and Western blot analyses were performed with whole-cell nuclear extracts derived from HuH-7 cells transfected with the wt or mutant HBV replication-competent vector. Specific signals were detected by using polyclonal rabbit anti-core antiserum (DAKO) as described previously (5).
Quantification.
Quantification of results was performed with a Fuji imager using the Tina-PCBAS software (Raytest, Straubenhardt, Germany) or by a densitometric evaluation as described before (6).
RESULTS
Distinct mutations in the enhancer I region derived from chronic HBV carriers decrease its activity.
The region of the HBV genome containing most of the crucial regulatory elements for viral replication, the enhancer I and II regions, and the core promoter (nt 2229 to 162) was amplified by PCR and sequenced from sera of 10 patients with chronic HBV infection prior to OLT. Major mutational changes were found in the enhancer I region. Therefore, this region was subcloned and further analyzed to investigate its impact on HBV replication. As shown in Fig. 1B, all of the patients had mutated enhancer I sequences. In four patients (1, 2, 3, and 7), the mutations were localized in the so-called enhancer I core domain.
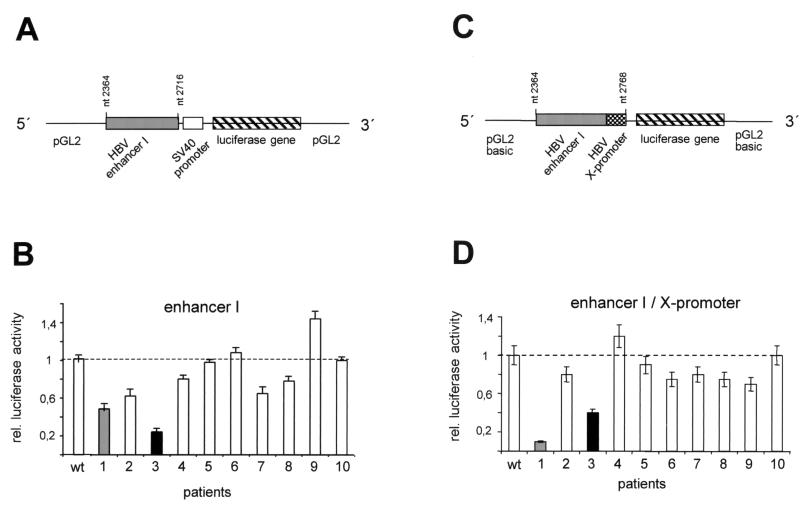
In the HBV genome, the enhancer I region partially overlaps the X promoter, thereby influencing the activity of the promoter. In addition, enhancer I controls the activity of the core promoter, which is located further upstream. In order to investigate both functions separately, we cloned the two regions—the enhancer I and the enhancer I-X-promoter—in front of luciferase reporter gene constructs. To test the enhancer I element (nt 2364 to 2716 [pre-C ATG = 1]) independently of the X promoter, it was cloned into the enhancer plasmid, in which luciferase expression is controlled by the heterologous SV40 promoter (Fig. 2A). In the enhancer I-X promoter constructs, the entire region (nt 2364 to 2768) was cloned in front of the luciferase gene of the basic reporter plasmid (pGL2; Stratagene) (Fig. 2C). To determine the impacts of the different mutations found in the enhancer I region on the transcriptional activities of these constructs, we transfected them into the hepatoma-derived cell line HuH-7 and measured luciferase activity. All experiments were performed a minimum of three times to verify the results.
FIG. 2.
Transcriptional activity of the enhancer I mutants derived from patients 1 to 10. (A and C) The enhancer I sequences between nt 2364 and 2716 and between nt 2364 and 2768 derived from wt HBV and the HBV sequences isolated from patients 1 to 10 were cloned as depicted in combination with the SV40 promoter in the pGL2 vector (A) or in the enhancer I-X promoter constructs (C) with the luciferase reporter gene alone. (B and D) Transfection experiments were performed with the different enhancer I (B) or enhancer I-X promoter constructs (D) in HuH-7 cells. The cells were harvested 48 h after transfection, and luciferase activity was measured. The error bars indicate standard deviations. The relative luciferase activities of the different mutant constructs are shown in comparison to that of the wt constructs, which was set to 1 (dashed lines). The activities for patients 1 and 3 are highlighted.
Six of the 10 patient-derived HBV enhancer I constructs demonstrated significantly reduced reporter gene activity compared to that of wt HBV (Fig. 2B). Only the construct cloned from patient 9 had higher activity (40%) than the wt HBV construct. The sequences derived from patient 1 and patient 3 showed a clear reduction in enhancer I activity. The luciferase activity of patient 1 was only 45% and that of patient 3 was only 22% of the levels expressed when the wt HBV construct was used. Next, we used the enhancer I-X promoter construct (Fig. 2D) to test for the effects of the mutated sequences on this viral promoter. In 7 out of 10 patients, the activity of the enhancer I-X promoter construct was reduced compared to that of the wt (Fig. 2D). In agreement with our previous observations, the HBV sequences isolated from patients 1 and 3 showed the lowest reporter activities. The mutant sequence derived from patient 1 revealed 10% and that from patient 3 revealed 40% of the wt enhancer I-X promoter activity.
Mutations in the enhancer I core domain result in lack of activation through decreased binding of liver-specific transcription factors.
The enhancer I sequences isolated from patient 1 and patient 3 showed the most prominent reduction in enhancer I activity compared to that of wt HBV. We therefore decided to characterize these two HBV variants in more detail. The mutation derived from patient 1 contained a 2-nt deletion between nt 2531 and 2532 and a 2-nt insertion between nt 2535 and 2536 in the enhancer I HNF3 element (ΔHNF3; AAGTA was changed to GTGCA in the HNF3 motif). The mutation derived from patient 3 consisted of a single point mutation at nucleotide 2546 in the enhancer I HNF4 motif (ΔHNF4). Other enhancer I mutations found in the HBV variants of patients 1 and 3 are not located at a known protein-binding site of the enhancer I region (Fig. 1B). Analysis of the other mutants shown in Fig. 1B showed they were not located in known DNA binding sites controlling enhancer I activity.
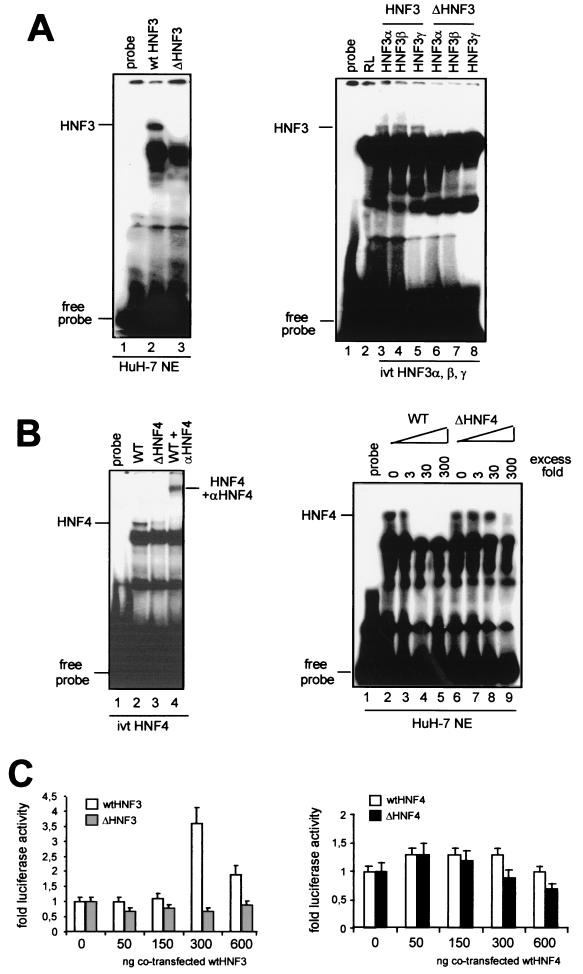
In order to determine whether these mutations in enhancer I alter the binding of HNF3 or HNF4 to their putative binding sites, gel shift experiments were performed using oligonucleotides representing either the wt or the mutated HNF3 and -4 sequences. As shown in Fig. 3A (left), formation of a specific DNA-protein complex was evident when the radiolabeled wt HNF3, but not the mutant (ΔHNF3), oligonucleotide was incubated with HuH-7 cell nuclear extracts. The intensity of this band diminished when the nuclear extracts were preincubated with unlabeled wt HNF3 oligonucleotide but remained unchanged when the mutant sequence (ΔHNF3) was used (data not shown). Three different isoforms of HNF3 (α, β, and γ) expressed in the liver are known to bind to the wt HNF3 binding site. In order to detect differences in their abilities to bind the wt or the mutant HNF3 sequence, we used in vitro-translated proteins of these three isoforms in gel shift experiments. Complex formation was clearly detected when each of the HNF3 proteins was incubated with wt, but not mutant HNF3 (ΔHNF3), oligonucleotides (Fig. 3A; right). These experiments suggest that a 2-nt deletion (nt 2531 to 2532) and a 2-nt insertion between nt 2535 and 2536 in the HBV enhancer I region abolishes binding of all isoforms of HNF3 to this sequence.
FIG. 3.
Mutations in the HBV enhancer I core domain decrease binding of HNF3 and HNF4. (A) 32P-labeled wt (left, lane 2) or mutant (left, lane 3) HNF3 oligonucleotides were used in gel shift experiments. The HNF3 oligonucleotides were incubated with HuH-7 nuclear extracts. The positions of the HNF3 complexes are marked. In vitro-translated (ivt) HNF3α, -β, and -γ proteins were incubated with wt (right, lanes 3 to 5) or mutant (right, lanes 6 to 8) HNF3 oligonucleotides. Incubation of reticulocyte (RL) extracts with wt HNF3 oligonucleotide is shown in lane 2. The positions of HNF3 complexes and free probe are depicted on the left. (B) HNF4 wt (left, lane 2) or mutant (left, lane 3) 32P-labeled oligonucleotides were incubated with in vitro-translated HNF4 protein. HNF4 was supershifted with polyclonal anti-HNF4 antibodies (left, lane 4). Competition experiments were performed with cold wt HNF4 (right, lanes 2 to 5) or mutant HNF4 (right, lanes 6 to 9) in the indicated molar excess in the incubation assay. (C) HuH-7 cells were transfected with wt, ΔHNF3, and ΔHNF4 enhancer I reporter gene constructs. Cotransfection experiments were performed with expression vectors for HNF3 (left) and HNF4 (right). The constructs were cotransfected with increasing amounts (0, 50, 150, 300, or 600 ng) of expression vectors for HNF3 (left) or HNF4 (right). The cells were harvested 48 h after transfection, and the luciferase activity was determined. The error bars indicate standard deviations. The activities of the samples in which only the luciferase reporter construct was transfected were set to 1. Note that dramatic changes were found in the basal activity with ΔHNF3 and -4 compared to that with the wt HBV construct (Fig. 2). The changes are shown as fold activation.
The enhancer I element isolated from patient 3 significantly affected the transcriptional activity of the X promoter (Fig. 2). However, sequence analysis revealed only a single point mutation (nt 2546; G to A) in the putative binding site for HNF4 (Fig. 1). We therefore expected to detect only minor changes in the affinity of HNF4 for this mutated sequence (ΔHNF4). Formation of a specific DNA-protein complex was clearly detectable when in vitro-translated HNF4 protein was incubated with the wt enhancer I element. Furthermore, antibodies directed against HNF4 supershifted this complex, thereby confirming its specificity (Fig. 3B). Unexpectedly, when the mutant HNF4 (ΔHNF4) oligonucleotide was incubated with in vitro-translated HNF4 protein, a dramatic reduction in complex formation was evident (Fig. 3B, left, lane 3). Using nuclear extracts from HuH-7 cells, we were able to confirm the strong differences between complex formation of HNF4 with the wt and with the mutated HNF4 binding sites (data not shown). Additional competition experiments with either an unlabeled wt or mutant HNF4 oligonucleotide showed that the wt element was able to compete against complex formation in low molar excess. However, mutant HNF4 competition is only found with high molar concentrations of unlabeled oligonucleotides (Fig. 3B, right).
In order to determine whether overexpression of HNF3 or HNF4 would result in a significant increase in luciferase activity of the ΔHNF3 and ΔHNF4 reporter constructs, we cotransfected them with expression plasmids of both transcription factors. While the wt enhancer I construct could be stimulated by cotransfecting the expression vector for HNF3, neither of the two mutant constructs (ΔHNF3 or ΔHNF4) could be stimulated by cotransfecting the expression vector for HNF3 or HNF4, respectively (Fig. 3C). Additionally, endogenous wt HNF4 expression in the hepatoma cell lines used seemed to be sufficient to maximally increase wt HNF4 construct activity. Thus, only a minor increase in luciferase activity could be detected if exogenous wt HNF4 was cotransfected.
The ΔHNF3 and ΔHNF4 mutants result in decreased replication efficiency of HBV.
From our initial experiments, we conclude that the described mutations in the enhancer I core domain result in decreased binding of HNF3 and -4 to this regulatory element. The functional unresponsiveness of the enhancer I core domain is responsible for the significantly reduced transcription of the different luciferase constructs tested. In order to determine the impacts of these mutations on HBV replication, we generated replication-competent HBV constructs containing the mutated enhancer I regions of patients 1 and 3 (ΔHNF3 and ΔHNF4) (Fig. 1B).
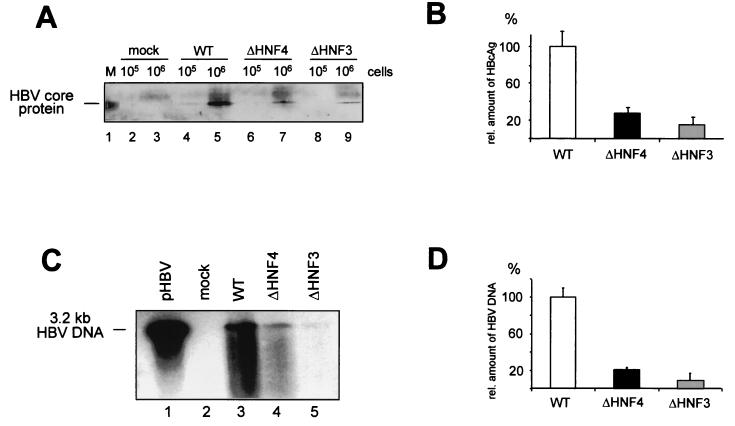
The wt and mutant HNF3 or HNF4 replication-competent plasmids were transfected into HuH-7 cells from which total cellular RNA was isolated 3 days after transfection. Northern blot analysis was performed using radiolabeled full-length HBV DNA to detect HBV-specific RNAs (Fig. 4A). Quantification of the RNA signals showed that both the ΔHNF3 and the ΔHNF4 mutations resulted in decreased X and pregenomic RNA levels compared to the corresponding wt levels (Fig. 4A, right). Pregenomic RNA levels were reduced to approximately 20 (ΔHNF3) or 40% (ΔHNF4) those of the wt construct. The reduction of X mRNA levels was even stronger (25% for ΔHNF3 and 18% for ΔHNF4). A less pronounced effect was evident for the pre-S and S mRNAs (70% for ΔHNF3 and 60% for ΔHNF4) (Fig. 4A). These results demonstrate that both mutations strongly reduce HBV-specific RNA levels.
FIG. 4.
Enhancer I mutants reduce expression of viral gene products. (A) Northern blot analysis was performed with total RNA derived from HuH-7 cells transfected with an empty control vector (mock; pBluescript) (lane 1) or HBV replication-competent vectors for 2 days. The wt (lane 2) and two clones of the enhancer I ΔHNF4 (lanes 3 and 4) and ΔHNF3 (lanes 5 and 6) vectors are shown. Twenty micrograms of RNA was separated on a 1% gel and transferred to nylon membranes. HBV-specific RNA was detected with a 32P-labeled HBV probe. The positions of the HBV-specific RNAs are shown at the right. Differences in the RNA concentrations were visualized by quantification of RNA bands. Pregenomic RNAs, pre-S and S mRNAs, and X mRNA are shown. The intensity of RNA expression found with the wt HBV replication-competent construct was set to 100%, and changes found with the mutant HNF3 and HNF4 constructs were calculated accordingly. The results of three independent experiments were quantified for this analysis. The error bars indicate standard deviations. (B) The amount of intra- and extracellular HBeAg or HBsAg was determined in whole-cell extracts or the supernatant 6 days after transfection by EIA. The expression of the wt HBV construct was set to 100%, and the other samples (mock, ΔHNF3, and ΔHNF4) were calculated accordingly. The error bars indicate standard deviations. The average of three independent experiments is shown. rel. conc., relative concentration.
The amounts of pregenomic RNA and HBV core protein might be rate limiting for HBV replication and thus determine the level of viremia. As the ΔHNF3 and ΔHNF4 HBV mutations reduced the levels of viral pregenomic and C RNAs, we subsequently quantitated the expression of the viral gene products. The levels of HBsAg and HBeAg expression were quantitated intracellularly and extracellularly in whole-cell extracts and supernatants of transfected HuH-7 cells 5 days after transfection (Fig. 4B). These experiments showed that both mutations resulted in a reduction to less than 40% of intracellular and extracellular HBeAg expression compared to that of wt HBV. The decrease in HBsAg was less pronounced, and at least 80% of the wt HBV level was found with both mutant strains (Fig. 4B).
After transfection of HuH-7 cells with wt HBV or the ΔHNF3 or ΔHNF4 replication construct, core protein levels were determined using whole-cell extracts (Fig. 5A). When the ΔHNF3 or ΔHNF4 replication construct was transfected, only 30 or 15%, respectively, of the amount of core protein expressed by wt HBV was detected by Western blot analysis (Fig. 5B).
FIG. 5.
The replication competence of enhancer I mutants is impaired. HuH-7 cells were transfected with the empty vector control (pBS; mock), the wt, and the enhancer I ΔHNF3 and ΔHNF4 HBV replication-competent constructs. (A and B) Quantification of intracellular core protein expression. (A) Core protein was precipitated by monoclonal mouse anti-HBcAg-specific antibodies. The precipitated protein was run on SDS-PAGE and transferred to nylon membranes. Filters were probed with polyclonal rabbit anti-HBcAg antiserum. Either 105 or 106 cells were used in the experiments as indicated. In lane 1, recombinant core protein was used. (B) The relative amounts of HBcAg were determined by Fuji phosphoimager or densitometric evaluation. The amount found with the wt HBV construct was set to 100%. The error bars indicate standard deviations. (C and D) The amount of newly synthesized progeny DNA was determined from cytoplasmic HBV core particles by using anti-HBc antibodies for immunoprecipitation from 106 cells. (C) HBV DNA was separated on a 1% alkaline agarose gel and visualized by Southern blot analysis as described previously (4). A 3.2-kb linear HBV DNA was used as a reference (lane 1). (D) The concentration of progeny HBV DNA was quantified; wt values were set to 100%, and the amounts of the HNF3 and HNF4 mutants were calculated. The error bars indicate standard deviations.
As HBV core protein is essential for nucleocapsid assembly and thus for HBV replication, we next examined intracellular HBV progeny DNA levels. As shown in Fig. 5C, the amount of progeny DNA was significantly reduced for both mutant constructs. Quantification of the DNA signals revealed a reduction of 25% for the ΔHNF4 and 8% for the ΔHNF3 mutant (Fig. 5D).
The occurrence of the ΔHNF4 and ΔHNF3 enhancer I mutants correlates with reduced HBV replication.
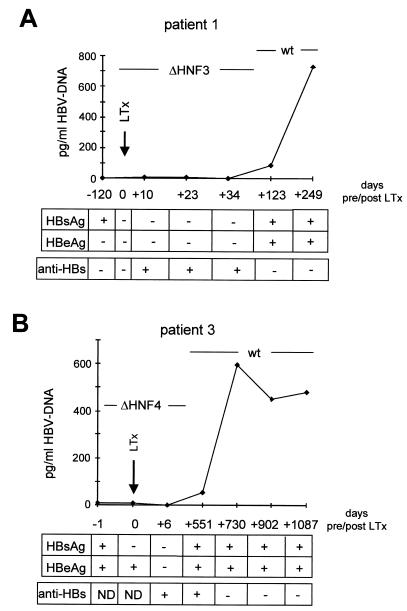
The ΔHNF4 and ΔHNF3 enhancer I core domain mutants were isolated from sera derived immediately before liver transplantation (OLT). Here, we studied HBV replication in both patients at time points before and after liver transplantation (Fig. 6).
FIG. 6.
Clinical course of patients 1 (ΔHNF3) and 3 (ΔHNF4). Virological parameters (HBV DNA [pg/ml], HBsAg, HBeAg, and anti-HBs) of patient 1 (A) and patient 3 (B) were determined at different time points before and after liver transplantation. The time points are indicated based on the day of OLT, which was set to 0. Sequencing of the HBV DNAs at different time points revealed the dominant DNA strains as indicated. +, present; −, absent; LTx, liver transplantation.
The HBV DNA level of patient 1 before OLT, when ΔHNF3 DNA was isolated, was 3 pg/ml (Fig. 6A). One hundred twenty-three days after OLT, HBV reinfection occurred. Sequencing of the enhancer I region revealed a wt HNF3 motif. At this time point, HBV replication increased to 730 pg/ml. At both time points, before and after OLT, the sera were HBsAg positive. However, before OLT no HBeAg was detected, but it was positive at the time of HBV reinfection (Fig. 6A).
In the sera of the second patient carrying the ΔHNF4 motif, 15 pg of HBV DNA/ml was detectable before OLT (Fig. 6B). HBV reinfection occurred 730 days after liver transplantation, and HBV DNA levels varied between 450 and 580 pg/ml as determined at time points between 730 and 1,087 days after OLT. Analysis of the HBV enhancer I region after OLT revealed a wt HNF4 motif. In this patient at both time points, before and after OLT, HBsAg and HBeAg were positive (Fig. 6B).
DISCUSSION
Chronic HBV infection reflects the permanent interaction of the immune system with viral epitopes (12, 32). Escape mutants, which are selected in the viral genome during the course of infection, contribute to the persistence of the virus and may also result in a more progressive form of liver disease. During chronic HBV infection, two stages of viremia can frequently be distinguished. Directly after HBV infection, high HBV DNA levels are found associated with a strong inflammatory response in the liver. Later, during infection, HBV replication decreases and may be associated with seroconversion from HBeAg to anti-HBe (28). However, seroconversion from HBeAg to anti-HBe does not necessarily indicate that HBV DNA levels have decreased. For example, selection of a stop mutation in the pre-C region results in a lack of HBeAg expression, leading to an anti-HBe status and increased viral replication (20, 25).
The switch between a high- and a low-replicative period during chronic HBV infection has not been addressed so far. This clinical observation led us to hypothesize that mutations resulting in decreased HBV replication may also be able to escape the host's immune response. Ideally, an escape mutation would lead to both decreased expression of viral epitopes and diminished replication of the virus. Such a mutation would most likely affect the regulation of several viral genes rather than affecting the function of a single protein. A candidate region for this type of mutation is the enhancer I sequence within the HBV genome. Originally identified as a regulatory motif able to influence the expression of different viral genes (34), the enhancer I region may be a potential target for mutations leading to escape from the immune system.
We therefore sequenced the enhancer I regions of 10 patients with chronic HBV infection prior to OLT. In the majority of these viral sequences, the enhancer I region was mutated, resulting in decreased transcriptional activity of viral and heterologous promoters. The lowest activity was detected when enhancer I sequences with mutations in the previously described HNF3 and -4 transcription factor binding sites were tested (11, 18, 30).
These sites are located in the enhancer I core domain, which has the strongest impact on transcriptional activation of HBV genes. To characterize the molecular mechanism responsible for this loss of activity in more detail, we examined the abilities of HNF3 and -4 to bind to these mutated sequences.
Surprisingly, the minor sequence changes we detected in the enhancer I regions of these two patients almost entirely abolished the binding of these transcription factors. In agreement with the loss of transcriptional activity, we detected dramatically decreased levels of pregenomic RNA (40% for ΔHNF3 and 20% for ΔHNF4) and an even stronger reduction in X mRNA expression (25% for ΔHNF3 and 18% for ΔHNF4). A less pronounced effect was evident for the pre-S and S mRNAs (70% for ΔHNF3 and 60% for ΔHNF4), supporting the observation that an element outside of the enhancer I region is essential for the control of the two S promoters (47). Not surprisingly, these remarkable reductions in transcriptional activity resulted in significant changes in the expression of core protein levels (ΔHNF3, 30%, and ΔHNF4, 15%) and nucleocapsid assembly (25% progeny DNA for ΔHNF4 and 8% for the ΔHNF3 mutant) compared with a wt HBV genome.
Analyzing the mutated enhancer I sequence within the background of a wt HBV genome helped us to specifically determine the roles of mutations in this region. The coding sequence of the viral polymerase, however, overlaps the enhancer I sequence (17). The mutations we described result in amino acid changes in the corresponding polymerase sequences for ΔHNF3 (Leu to Pro and Ser to Cys) and for the single ΔHNF4 point mutation (Met to Ile). These mutations were located in the reverse-transcription domain of the viral polymerase. These polymerase mutations may lead to a reduction in HBV polymerase activity. However, our results as shown in Fig. 5 indicate that in the mutated viral strains the amounts of viral RNA, viral capsids, and HBV progeny DNA are significantly reduced, leading to a comparable decrease. While the expression of viral RNA and HBV core protein is independent of the HBV polymerase activity, our results also indicate that the capsid-to-progeny DNA ratio is not changed between the wt and the mutant strains. Therefore, these results revealed that the encapsidated wt and mutant HBV polymerase seem to have comparable activities, and thus the change in the polymerase has no major influence on the replication level of the mutant viruses.
The in vitro studies presented here illustrate the effects ΔHNF3 and ΔHNF4 mutations have on the expression of viral proteins and the assembly of virus particles. They are important for understanding the molecular mechanisms which may be responsible for the decreased replication and immunogenicity of HBV in chronic carriers. The clinical significance of the enhancer mutations, however, is very difficult to assess. When we first detected the ΔHNF3 and ΔHNF4 mutations, the amount of HBV DNA was measured as 3 pg/ml in patient 1 and 15 pg/ml in patient 3, possibly reflecting the effects of these mutations on virus replication in vivo. After liver transplantation, however, both patients were reinfected with HBV. Patient 1 showed a 240-fold increase and patient 3 showed a 40-fold increase in HBV DNA levels. Sequencing of the enhancer I regions in both patients revealed the wt HBV sequence. In general, in patients undergoing liver transplantation HBV replication increases compared to the pre-OLT level (36). These patients receive lifelong immunosuppressive therapy, and it has been demonstrated that the glucocorticoids especially increase HBV replication through a glucocorticoid-responsive element in the HBV genome (42, 43). At the time of reinfection, however, both patients received only very low doses (2.5 and 5 mg, respectively) of this immunosuppressive drug. Thus, the increase in HBV replication after OLT cannot simply be explained by glucocorticoid responsiveness.
All 10 patients with chronic active HBV infection investigated in this study had mutations in the enhancer I region. We believe that these mutations may influence replication and immunogenicity of the virus to different degrees. While mutations like ΔHNF3 and ΔHNF4 abolish the binding of important transcription factors, other mutations might only exert an effect in association with mutations in the enhancer I region or elsewhere in the HBV genome. Mutating regulatory sequences in the HBV genome allows the virus to affect different functions, such as protein expression, virus assembly, and expression of epitopes, simultaneously, which under the selective pressure of the host's immune system might provide an efficient escape mechanism.
In summary, our experiments describe a new form of escape mechanism which controls the level of HBV replication. Therefore, enhancer I mutants seem to be involved in explaining the clinical observation that two stages of viremia during chronic HBV infection can be differentiated: a high- and a low-replicative period.
ACKNOWLEDGMENTS
We thank H. A. Sundberg for critical reading of the manuscript and R. Raupach and P. Magerstedt for excellent technical assistance.
This work was supported by SFB 265/C5.
Footnotes
This work is dedicated to K. H. Meyer zum Büschenfelde on the occasion of his 70th birthday.
REFERENCES
- 1.Anttonucci T K, Rutter W J. Hepatitis B virus (HBV) promoters are regulated by the HBV enhancer in a tissue-specific manner. J Virol. 1989;63:579–583. doi: 10.1128/jvi.63.2.579-583.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beasley R P, Lin C-C, Hwang L Y, Chen C-S. Hepatocellular carcinoma and hepatitis B virus: a prospective study of 22,707 men in Taiwan. Lancet. 1981;iii:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 3.Blake M, Niklinski J, Zayjac-Kaye M. Interactions of the transcription factors MIBP1 and RFX1 with the EP element of the hepatitis B virus enhancer. J Virol. 1996;70:7060–7066. doi: 10.1128/jvi.70.9.6060-6066.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock C-T, Schranz P, Schroeder C-H, Zentgraf H. The hepatitis B virus genome is organized into nucleosomes in the nucleus of the infected cell. Virus Genes. 1994;8:215–229. doi: 10.1007/BF01703079. [DOI] [PubMed] [Google Scholar]
- 5.Bock C-T, Schwinn S, Schroeder C-H, Velhagen I, Zentgraf H. The hepatitis B virus core protein mediates the transport of the viral genome into the nucleus of the infected cell. Virus Genes. 1996;12:53–63. doi: 10.1007/BF00370001. [DOI] [PubMed] [Google Scholar]
- 6.Bock C T, Tillmann H L, Maschek H J, Manns M P, Trautwein C. A preS mutation isolated from a patient with chronic hepatitis B infection leads to virus retention and misassembly. Gastroenterology. 1997;113:1976–1982. doi: 10.1016/s0016-5085(97)70018-0. [DOI] [PubMed] [Google Scholar]
- 7.Buendia M A. Hepatitis B viruses and hepatocellular carcinoma. Adv Cancer Res. 1992;59:167–226. doi: 10.1016/s0065-230x(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 8.Carman W F, Korula J, Wallace L, MacPhee R, Mimms L, Decker R. Fulminant reactivation of hepatitis B due to envelope protein mutant that escaped detection by monoclonal HBsAg ELISA. Lancet. 1995;345:1406–1407. doi: 10.1016/s0140-6736(95)92599-6. [DOI] [PubMed] [Google Scholar]
- 9.Carman W F, Thomas H, Esteban D. Viral genetic variation: hepatitis B as a clinical example. Lancet. 1993;341:349–352. doi: 10.1016/0140-6736(93)90146-8. [DOI] [PubMed] [Google Scholar]
- 10.Carman W F, Zanetti A R, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman A J, Thomas H C. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325–329. doi: 10.1016/0140-6736(90)91874-a. [DOI] [PubMed] [Google Scholar]
- 11.Chen M, Hieng S, Qian X, Costa R, Ou J-H. Regulation of hepatitis B virus ENI enhancer activity by hepatocyte-enriched transcription factor HNF3. Virology. 1994;205:127–132. doi: 10.1006/viro.1994.1627. [DOI] [PubMed] [Google Scholar]
- 12.Chisari F V, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 13.Chisari F V, Klopchin K, Moriyama T, Pasquinelli C, Dunsford H A, Sell S, Pinkert C A, Brinster R L, Palmiter R D. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989;59:1145–1156. doi: 10.1016/0092-8674(89)90770-8. [DOI] [PubMed] [Google Scholar]
- 14.Dignam J, Lebowitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian cells. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dikstein R, Faktor O, Shaul Y. Hierarchic and cooperative binding of the rat liver nuclear protein C/EBP at the hepatitis B virus enhancer. Mol Cell Biol. 1990;10:4427–4440. doi: 10.1128/mcb.10.8.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganem D. Oncogenic viruses. Of marmots and men. Nature. 1990;347:230–232. doi: 10.1038/347230b0. [DOI] [PubMed] [Google Scholar]
- 17.Ganem D. Hepadnaviridae and their replication. New York, N.Y: Raven Press; 1996. pp. 2703–2737. [Google Scholar]
- 18.Garcia A D, Ostapchuck P, Hearing P. Functional interaction of nuclear factors EF-C, HNF-4, and RXRα with hepatitis B virus enhancer I. J Virol. 1993;67:3940–3950. doi: 10.1128/jvi.67.7.3940-3950.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo W, Bell K D, Ou J H. Characterization of the hepatitis B virus EnhI enhancer and X promoter complex. J Virol. 1991;65:6686–6692. doi: 10.1128/jvi.65.12.6686-6692.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasegawa K, Huang J, Rogers S A, Blum H E, Liang T J. Enhanced replication of a hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. J Virol. 1994;68:1651–1659. doi: 10.1128/jvi.68.3.1651-1659.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu K-Q, Siddiqui A. Regulation of the hepatitis B virus gene expression by the enhancer element I. Virology. 1991;181:721–726. doi: 10.1016/0042-6822(91)90906-r. [DOI] [PubMed] [Google Scholar]
- 22.Huan B, Kosovsky M J, Siddiqui A. Retinoid X receptor a transactivates the hepatitis B virus enhancer I element by forming a heterodimer complex with the peroxisome proliferator-activated receptor. J Virol. 1995;69:547–551. doi: 10.1128/jvi.69.1.547-551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jameel S, Siddiqui A. The human hepatitis B virus enhancer requires trans-acting cellular factor(s) for activity. Mol Cell Biol. 1986;6:710–715. doi: 10.1128/mcb.6.2.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kosovsky M J, Huan B, Siddiqui A. Purification and properties of rat liver nuclear proteins that interact with the hepatitis B virus enhancer I. J Biol Chem. 1996;71:21859–21869. doi: 10.1074/jbc.271.36.21859. [DOI] [PubMed] [Google Scholar]
- 25.Lamberts C, Nassal M, Velhagen I, Zentgraf H, Schroder C H. Precore-mediated inhibition of hepatitis B virus progeny DNA synthesis. J Virol. 1993;67:3756–3762. doi: 10.1128/jvi.67.7.3756-3762.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Cabrera M, Letovsky J, Hu K-Q, Siddiqui A. Transcriptional factor C/EBP binds to and transactivates the enhancer II element of the hepatitis B virus. Virology. 1999;183:825–829. doi: 10.1016/0042-6822(91)91019-d. [DOI] [PubMed] [Google Scholar]
- 27.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 28.Niederau C, Heintges T, Lange S, Goldmann G, Niederau C M, Mohr L, Haussinger D. Long-term follow-up of HBeAg-positive patients treated with interferon alfa for chronic hepatitis B. N Engl J Med. 1996;334:1422–1427. doi: 10.1056/NEJM199605303342202. [DOI] [PubMed] [Google Scholar]
- 29.Niehof M, Manns M P, Trautwein C. CREP controls LAP/C/EBP transcription. Mol Cell Biol. 1997;17:3600–3613. doi: 10.1128/mcb.17.7.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ori A, Shaul Y. Hepatitis B virus enhancer binds and is activated by the hepatocyte nuclear factor 3. Virology. 1995;207:98–106. doi: 10.1006/viro.1995.1055. [DOI] [PubMed] [Google Scholar]
- 31.Ori A, Zauberman A, Doitsh G, Paran N, Oren M, Shaul Y. p53 binds and represses the HBV enhancer: an adjacent enhancer element can reverse the transcription effect of p53. EMBO J. 1998;17:544–553. doi: 10.1093/emboj/17.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rehermann B, Chang K M, McHutchinson J, Kokka R, Houghton M, Rice C M, Chisari F V. Differential cytotoxic T-lymphocyte responsiveness to the hepatitis B and C viruses in chronically infected patients. J Virol. 1996;70:7092–7102. doi: 10.1128/jvi.70.10.7092-7102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaller H, Fischer M. Transcriptional control of hepadnavirus gene expression. Curr Top Microbiol Immunol. 1991;168:21–39. doi: 10.1007/978-3-642-76015-0_2. [DOI] [PubMed] [Google Scholar]
- 34.Shaul Y, Rutter W J, Laub O. A human hepatitis B viral enhancer element. EMBO J. 1985;4:427–430. doi: 10.1002/j.1460-2075.1985.tb03646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Summers J, Mason W. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell. 1982;29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 36.Todo S, Demetris A J, Van Thiel D, Teperman L, Fung J J, Starzl T E. Orthotopic liver transplantation for patients with hepatitis B virus-related liver disease. Hepatology. 1991;13:619–626. [PMC free article] [PubMed] [Google Scholar]
- 37.Trautwein C, Caelles C, van der Geer P, Hunter T, Karin M, Chojkier M. Transactivation by LAP/NF-IL6 is enhanced by phosphorylation of its activation domain. Nature (London) 1993;364:544–547. doi: 10.1038/364544a0. [DOI] [PubMed] [Google Scholar]
- 38.Trautwein C, Rakemann T, Malek N P, Plumpe J, Tiegs G, Manns M P. Concanavalin A-induced liver injury triggers hepatocyte proliferation. J Clin Investig. 1998;101:1960–1969. doi: 10.1172/JCI504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trautwein C, Rakemann T, Pietrangelo A, Pluempe J, Montosi G, Manns M P. C/EBP-beta/LAP controls downregulation of albumin gene expression during liver regeneration. J Biol Chem. 1996;271:22262–22270. doi: 10.1074/jbc.271.36.22262. [DOI] [PubMed] [Google Scholar]
- 40.Trautwein C, Schrem H, Tillmann H L, Kubicka S, Walker D, Boker K H, Maschek H J, Pichlmayr R, Manns M P. Hepatitis B virus mutations in the pre-S genome before and after liver transplantation. Hepatology. 1996;24:482–488. doi: 10.1002/hep.510240303. [DOI] [PubMed] [Google Scholar]
- 41.Trujillo M A, Letovsky J, Maguire H F, Lopez-Cabrera M, Siddiqui A. Functional analysis of a liver-specific enhancer of the hepatitis B virus. Proc Natl Acad Sci USA. 1991;88:3797–3801. doi: 10.1073/pnas.88.9.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tur-Kaspa R, Burk R D, Shaul Y, Shafritz D A. Hepatitis B virus DNA contains a glucocorticoid-responsive element. Proc Natl Acad Sci USA. 1986;83:1627–1631. doi: 10.1073/pnas.83.6.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tur-Kaspa R, Shaul Y, Moore D D, Burk R D, Okret S, Poellinger L, Shafritz D A. The glucocorticoid receptor recognizes a specific nucleotide sequence in hepatitis B virus DNA causing increased activity of the HBV enhancer. Virology. 1988;167:630–633. [PubMed] [Google Scholar]
- 44.Wright T L, Lau J Y N. Clinical aspects of hepatitis B virus infection. Lancet. 1993;342:1340–1344. doi: 10.1016/0140-6736(93)92250-w. [DOI] [PubMed] [Google Scholar]
- 45.Yen T S B. Regulation of hepatitis B virus gene expression. Semin Virol. 1993;4:33–42. [Google Scholar]
- 46.Zhang P, Raney A K, McLachlan A. Characterization of the hepatitis B virus X- and nucleocapsid gene transcriptional regulatory elements. Virology. 1992;191:31–41. doi: 10.1016/0042-6822(92)90163-j. [DOI] [PubMed] [Google Scholar]
- 47.Zhou D-X, Yen T S B. Differential regulation of the hepatitis B surface gene promoters by a second viral enhancer. J Biol Chem. 1990;265:20731–20734. [PubMed] [Google Scholar]