TO THE EDITOR:
Translocations involving the nucleoporin 98 (NUP98) and >30 fusion partner genes are well characterized for their involvement in hematological diseases including myelodysplastic syndromes and acute myeloid leukemia (AML).1 Fusion partners are categorized as homeobox (HOX) class I or class II homeobox genes or nonhomeobox.1 Class I HOX fusions involves ∼10 genes, including HOXA9,2 whereas HOX class II fusion partners are less frequent and includes HHEX.3 Nonhomeobox partner genes, on the contrary, associate with epigenetic regulation and includes NSD1, KDM5A, and KMT2A.4, 5, 6 Expression of fusion transcripts results in oncoproteins with functionality derived from joining of genes. This includes phenylalanine-glycine repeats retained from NUP98 enabling liquid-liquid phase separation and biomolecular condensation of oncoproteins to nuclear puncta combined with functional domains from the partner, including homeodomain, su(var)3-9, enhancer-of-zeste and trithorax, and plant homeodomain fingers.7 The resultant oncoproteins may directly invoke epigenetic dysregulation or recruit additional factors, promoting leukemogenesis, through activation of distal HOX genes and Meis1.8
Although molecular mechanisms behind NUP98 translocations are advancing, there is a gap in the understanding of their clinical significance in adult AML cohorts, with literature focusing on pediatric AML.9,10 This has been reflected with NUP98 translocations only recently incorporated into the World Health Organization and International Consensus Classification diagnostic classifications; enabling diagnosis of AML, based on detection of <20% blasts (>10% if using International Consensus Classification) and clinical presentation.11,12 Consequently, existing estimations regarding adult NUP98 malignancies may underrepresent their true frequency. To address this gap, we performed the most complete screening to date on NUP98 translocations in an adult AML cohort, combining fluorescent in situ hybridization (FISH) and next-generation sequencing (NGS).
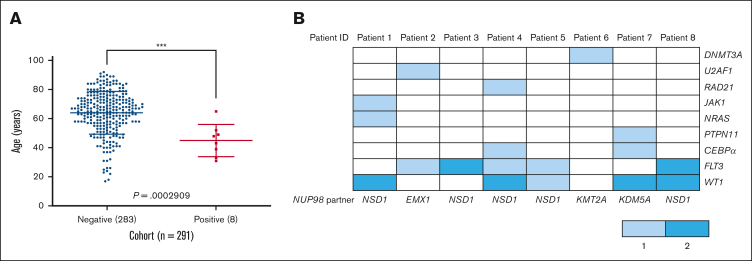
Our sex-matched cohort of 291 adults, with a median age of 63.5 years, included 161 males and 130 females (P = .079, Fischer exact test), consisted of 199 retrospective adult patients with AML diagnosed between 2016 and 2021 and 92 adult patients with AML diagnosed prospectively in 2022. Eight cases of NUP98 translocation (2.8%) were detected (Table 1), significantly affecting younger adults (median, 45 years; range, 31-67), compared with that of patients without NUP98 translocations (P = .0002909, unpaired t test; Figure 1A). A clear male sex bias in patients positive for NUP98 translocation was also observed (7 males; 1 female) (Table 1). Four cases were de novo AML (2 acute myelomonocytic, 1 AML with minimal differentiation, and 1 AML with maturation), with 4 cases of myelodysplasia-related changes AML (Table 1).
Table 1.
Clinical, cytogenetic, and NGS data from patients with AML withNUP98 translocation
| ID | Diagnosis (WHO 2016) | Age | Sex | Year | WBC, ×109/L | HB, g/L | PLT, × 109/L) | Blast % BM | Treatment | CR | Status | Karyotype | FISH | Breakpoint and fusion partner (NUP98::X) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | AML with myelodysplasia-related changes | 43 | M | 2016 | 15.2 | 69 | 50 | 18∗ | CETLAM AML-12 | Yes† | Alive | 46,XY[30] | 1R 1G 1Fus | 12::6 NSD1 |
| 2 | Acute myelomonocityc leukemia | 65 | M | 2017 | 32.4 | 55 | 55 | 35 | Quantumfirst TRIAL | Yes | Died in CR | 46,XY[20] | 1R 1Fus | 12::2 EMX1 |
| 3 | AML with myelodysplasia-related changes | 39 | M | 2020 | 285 | 71 | 29 | 86 | CETLAM AML-12 | No | Died in progression | 45,XY,add(1)(p36),del(1)(p15),? inv(14)(q11q32),der(17)t(17;22)(p11.2;p11.2),?inv(18)(q21.3q23),-22[20] | 1R 1G 1Fus | 12::6 NSD1 |
| 4 | AML with myelodysplasia-related changes | 49 | M | 2020 | 40.75 | 72 | 115 | 55 | CETLAM AML-12 | Yes‡ | Died in progression | 46,XY[20] | 1R 1G 1Fus | 12::6 NSD1 |
| 5 | Acute myelomonocityc leukemia | 52 | F | 2022 | 327 | 43 | 46 | 81 | HOVON 156 TRIAL | No | Died in progression | 46,XX[25] | 1R 2Fus | 12::6 NSD1 |
| 6 | AML with maturation | 31 | M | 2022 | 7.93 | 86 | 160 | 27 | CETLAM AML-12 | No | Died during induction (septicemia) | 46,XY,[inv(11)(p15q23)] | 1R 1G 1Fus | 14::2 KMT2A |
| 7 | AML with minimal differentiation | 33 | M | 2022 | 1.3 | 95 | 285 | 83 | VENAZA | Yes | Alive | 47,XY,inv(11)(q13.5q25),del(12)(p13),+21c[2] / 46,X,-Y,inv(11)(q13.5q25),del(12)(p13),+21c[18] | 1Fus 1R | 12::26 KDM5A |
| 8 | AML with myelodysplasia-related changes | 48 | M | 2022 | 238 | 73 | 47 | 70 | KB-LANRA-1001 Trial | Yes | Alive (relapsed after allo-sct) | 46,XY[30] | 1R 1G 1Fus | 12::6 NSD1 |
add, additional material of unknown origin; CR, complete remission; del, deletion; EMX1, Empty Spiracles Homeobox 1; F, female; Fus, fusion; G, green; HB, hemoglobin; inv, inversion; KDM5A, lysine demethylase 5A; KMT2A, lysine methyltransferase 2A; M, male; NSD1, nuclear receptor binding SET domain protein 1; PLT, platelet count; R, red; VENAZA, venetoclax plus azacitidine protocol; WBC, whole-blood cells.
Peripheral blood 29%.
Relapsed before allogeneic stem cell transplant that was rescued.
Relapsed before allogeneic stem cell transplant that could not be rescued.
Figure 1.
Clinical and genetic characteristics of adult patients with AML with NUP98 translocations. (A) Ages of adult patients with AML without NUP98 rearrangement (n = 283) compared with those with NUP98 rearrangement (n = 8). (B) Comutations detected in adult patients with AML with NUP98 rearrangement through targeted NGS. The scheme shows the number of mutations per patient, with the corresponding NUP98 fusion partner indicated along the lower x-axis, the patient ID along the upper x-axis, and the comutated gene along the y-axis. The number of mutations per gene are determined by color; 1 detected mutation is represented by light blue and 2 mutations in the same gene by dark blue.
Concordant with previous adult studies, most NUP98 translocations were cytogenetically normal (n = 5), with 3 exceptions displaying cytogenetic aberrations to chromosome 11 (Table 1).13, 14, 15, 16, 17 This included an interstitial deletion p15, a pericentric inversion p15-q23, and a paracentric inversion q13.5q25. FISH confirmed NUP98 translocations in all patients (supplemental Figure 1A-G), with a targeted NGS panel detecting fusion breakpoints, comutations, and identification of fusion partners.18 Intronic breakpoints between exons 12 and 13 of NUP98 were the most frequent (87.5%), with 1 breakpoint between exons 14 and 15 (Table 1; supplemental Figure 1H). All patients harbored comutations, in total involving 9 genes (Figure 1B). Comutations in WT1 and FLT3-ITD (internal tandem duplications) were the most frequent, occurring in 5 of 8 patients (62.5%), and were enriched alongside NUP98::NSD1 translocation. Finally, comutations were detected in RAD21, JAK1, and DNMT3A, all previously unreported alongside NUP98 translocations in AML. Overall, 4 fusion partners were detected (Figure 1B), concordantly agreeing with literature that NUP98::NSD1 rearrangements are the most frequent in adult AML (n = 5).14 Additionally, 3 fusions observed in single cases of clinical importance were detected, including an undocumented fusion partner, Empty Spiracles Homeobox 1 (EMX1), named NUP98::EMX1; a rare report of NUP98::KMT2A; and the first adult case of NUP98::KDM5A (Table 1).
The first fusion observed in a single case involved rearrangement of NUP98 with the antennapedia class homeobox family gene EMX1, detected from a cytogenetically normal patient sample of a 65-year-old male diagnosed with AML (supplemental Figure 2A). FISH detected an atypical split signal, with deletion of a distal 3’ green-end signal (supplemental Figure 2B). NGS revealed the fusion between exon 12 of NUP98 and exon 2 of the Class II HOX gene and comutations in U2AF1 and FLT3. Reverse transcription polymerase chain reaction using complementary DNA produced from patient bone marrow and a forward primer in exon 12 of NUP98 and a reverse primer in exon 2 of EMX1 confirmed expression of the novel fusion vs a NUP98::NSD1 control (supplemental Figure 2C). A full-length NUP98::EMX1 complementary DNA transcript was generated using a forward primer at the start of exon 1 of NUP98 and a reverse primer at the end of exon 3 of EMX1. Subsequent cloning into the pLVX-Puro vector, followed by Sanger sequencing confirmed an in-frame fusion transcript fusion between NUP98 exon 12 with EMX1 exon 2 (supplemental Figure 2D). This included retention of phenylalanine-glycine repeats from NUP98 (amino acids 80-152 and 253-332) and exons 2 and 3 of EMX1, retaining the homeobox domain (amino acids 192-251) and disordered region (amino acids 249-290; supplemental Figure 2E). The second fusion observed in a single case included a 31-year-old male diagnosed with AML with maturation, with pericentric inversion of chromosome 11 and a karyotype of 46,XY,inv(11)(p15q23) (supplemental Figure 3A). NGS revealed NUP98 translocation with KMT2A, with a metaphase FISH confirmation of rearrangement (supplemental Figure 1A). In addition, NGS detected the well-known missense hot spot mutation, c.2645G>A in DNMT3A, representing a very rare genetic event in AML, with DNMT3A comutated alongside NUP98 rearrangement AML. Critically, this rare case, to our knowledge, represents the third reported case in AML of NUP98::KMT2A and, consistent with previous cases, involves the same pericentric inversion of chromosome 11 inv(11)(p15q23).6
Finally, the third fusion observed in a single case includes a 33-year-old male with Down syndrome diagnosed with AML with minimal differentiation and a karyotype of 47,XY,inv(11)(q13.5q25),del(12)(p13),+21c[2]/46,X,-Y,inv(11)(q13.5q25),del(12)(p13),+21c[18] (supplemental Figure 3B). FISH revealed a single joint fusion and deletion of a distal 3’-green probe (supplemental Figure 1F), with NGS detecting translocation with KDM5A and mutations in PTPN11 and CEBPA and 2 mutations in WT1. Despite the characterization of NUP98::KDM5A in pediatric AML, we believe this case represents, to our knowledge, the first reported case of NUP98::KDM5A translocation involvement in adult AML, doubling as the first reported NUP98 translocation AML case with Down syndrome.
Herein, we present results from analyzing a large series of nearly 300 adult patients with AML using FISH and NGS. We report NUP98 translocations are more frequent in adult AML than the summarized adult studies estimate (supplemental Table 1), reporting an occurrence of 2.8%.13, 14, 15, 16, 17 Our results also challenge previously assumed equal sex distributions of NUP98 translocations in adult AML, reporting a notable male sex bias (7:1).14,16,17 Three clinically interesting cases were also detected, including the novel fusion NUP98::EMX1. Believed to be involved in the determination of cellular identity during corticogenesis, EMX1 and its paralogue EMX2 may also act as tumor suppressors in solid tumors; however, no reports currently link EMX1 to hematological disease.19,20 Finally, we provide, to our knowledge, the first evidence that NUP98::KDM5A is not exclusive to pediatric or non–Down syndrome AML and report a rare case of NUP98::KMT2A in AML.6 Alongside this, extremely rare comutations including DNMT3A, JAK1, and RAD21 were detected, expanding the small mutagenome associated with NUP98 rearrangements. Crucially, through an improved understanding of comutations, this could inform new treatment strategies, including JAK inhibitors or epidrug trials.21 Furthermore, due to the technical success highlighted by this study that NGS and FISH possess in detecting NUP98 translocations, we recommend their use in clinical evaluation, particularly on cytogenetically normal patients with AML. This may prove important in guiding more appropriate treatment strategies, such as myeloablative therapy, which may be more tolerated and benefit the typically younger patients affected by NUP98 translocation AML. To conclude, our findings offer valuable clinical insights into AML, opening potential avenues for therapeutics and research, while providing a clinical reference for NUP98 translocations in adult AML.
All patient samples included in the study were obtained from the unit of collection of samples of Josep Carreras Leukaemia Research Institute after their respective institutional review board and ethical approval (ethics committee at Hospital Universitari Germans Trias I Pujol Ref. PI-20-278; date of approval: 25 September 2020).
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Acknowledgments
Acknowledgments: This project has been supported by funding from the European Union’s Horizon 2020 Program under grant number H2020-MSCA-ITN-2020 (INTERCEPT-MDS Project; number: 953407; M.B. and M.E.) and the Ministerio de Ciencia e Innovación cofunded by European Regional Development Fund (European Regional Development Fund; PID2022-139279OB-I00; M.B.). This work was also supported in part by a grant from the Instituto de Salud Carlos III, Ministerio de Economía y Competitividad, Spain (PI/ 17/0575 and PI 20/00531) (cofunded by European Regional Development Fund, a way to build Europe); Translational Cancer Research (Asociación Española Contra el Cáncer AC 18/000002 and Instituto de Salud Carlos III), 2017 Support Grup de Recerca 288 (Grup Reserca Consolidat) and 2021 Support Grup de Recerca 00560 (Grup Reserca Consolidat) Generalitat de Catalunya; and economical support from CERCA Programme/Generalitat de Catalunya, Fundació Internacional Josep Carreras alongside funding from “la Caixa” Foundation.
Contribution: J.S.H., A.M.L., M.L.P., N.R.-X., I.G., M.C., and L.Z. performed and analyzed the research; M.B., I.G., L.Z., F.S., and M.E. provided critical materials; M.B. designed the study and provided critical scientific advice, along with J.S.H., I.G., L.Z., and F.S.; S.V., R.C., and C.M. managed the patients and also collected clinical data and clinical samples and analyzed clinicopathological data; J.S.H. prepared the tables and figures; J.S.H. and M.B. wrote the manuscript; and all authors have reviewed and agreed to publish the manuscript.
Footnotes
Data are available on reasonable request from the corresponding author, María Berdasco (mberdasco@carrerasresearch.org).
The full-text version of this article contains a data supplement.
Supplementary Material
References
- 1.Michmerhuizen NL, Klco JM, Mullighan CG. Mechanistic insights and potential therapeutic approaches for NUP98-rearranged hematologic malignancies. Blood. 2020;136(20):2275–2289. doi: 10.1182/blood.2020007093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamura T, Largaespada DA, Lee MP, et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12(2):154–158. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 3.Jankovic D, Gorello P, Liu T, et al. Leukemogenic mechanisms and targets of a NUP98/HHEX fusion in acute myeloid leukemia. Blood. 2008;111(12):5672–5682. doi: 10.1182/blood-2007-09-108175. [DOI] [PubMed] [Google Scholar]
- 4.Jaju RJ, Fidler C, Haas OA, et al. A novel gene, NSD1, is fused to NUP98 in the t(5;11)(q35;p15.5) in de novo childhood acute myeloid leukemia. Blood. 2001;98(4):1264–1267. doi: 10.1182/blood.v98.4.1264. [DOI] [PubMed] [Google Scholar]
- 5.van Zutven LJCM, Onen E, Velthuizen SCJM, et al. Identification of NUP98 abnormalities in acute leukemia: JARID1A (12p13) as a new partner gene. Genes Chromosomes Cancer. 2006;45(5):437–446. doi: 10.1002/gcc.20308. [DOI] [PubMed] [Google Scholar]
- 6.Kaltenbach S, Soler G, Barin C, et al. NUP98-MLL fusion in human acute myeloblastic leukemia. Blood. 2010;116(13):2332–2335. doi: 10.1182/blood-2010-04-277806. [DOI] [PubMed] [Google Scholar]
- 7.Terlecki-Zaniewicz S, Humer T, Eder T, et al. Biomolecular condensation of NUP98 fusion proteins drives leukemogenic gene expression. Nat Struct Mol Biol. 2021;28(2):190–201. doi: 10.1038/s41594-020-00550-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang GG, Song J, Wang Z, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459(7248):847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertrums EJM, Smith JL, Harmon L, et al. Comprehensive molecular and clinical characterization of NUP98 fusions in pediatric acute myeloid leukemia. Haematologica. 2023;108(8):2044–2058. doi: 10.3324/haematol.2022.281653. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cerveira N, Correia C, Dória S, et al. Frequency of NUP98-NSD1 fusion transcript in childhood acute myeloid leukaemia. Leukemia. 2003;17(11):2244–2247. doi: 10.1038/sj.leu.2403104. [DOI] [PubMed] [Google Scholar]
- 11.Arber DA, Orazi A, Hasserjian RP, et al. International consensus classification of myeloid neoplasms and acute leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140(11):1200–1228. doi: 10.1182/blood.2022015850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khoury JD, Solary E, Abla O, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36(7):1703–1719. doi: 10.1038/s41375-022-01613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollink IHIM, van den Heuvel-Eibrink MM, Arentsen-Peters STCJM, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118(13):3645–3656. doi: 10.1182/blood-2011-04-346643. [DOI] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research Network. Ley TJ, Miller C, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie W, Raess PW, Dunlap J, et al. Adult acute myeloid leukemia patients with NUP98 rearrangement have frequent cryptic translocations and unfavorable outcome. Leuk Lymphoma. 2022;63(8):1907–1916. doi: 10.1080/10428194.2022.2047672. [DOI] [PubMed] [Google Scholar]
- 16.Thol F, Kölking B, Hollink IHI, et al. Analysis of NUP98/NSD1 translocations in adult AML and MDS patients. Leukemia. 2013;27(3):750–754. doi: 10.1038/leu.2012.249. [DOI] [PubMed] [Google Scholar]
- 17.Fasan A, Haferlach C, Alpermann T, Kern W, Haferlach T, Schnittger S. A rare but specific subset of adult AML patients can be defined by the cytogenetically cryptic NUP98-NSD1 fusion gene. Leukemia. 2013;27(1):245–248. doi: 10.1038/leu.2012.230. [DOI] [PubMed] [Google Scholar]
- 18.Gargallo P, Molero M, Bilbao C, et al. Next-generation DNA sequencing-based gene panel for diagnosis and genetic risk stratification in onco-hematology. Cancers (Basel) 2022;14(8) doi: 10.3390/cancers14081986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida M, Suda Y, Matsuo I, et al. Emx1 and Emx2 functions in development of dorsal telencephalon. Development. 1997;124(1):101–111. doi: 10.1242/dev.124.1.101. [DOI] [PubMed] [Google Scholar]
- 20.Jimenez-García MP, Lucena-Cacace A, Otero-Albiol D, Carnero A. Regulation of sarcomagenesis by the empty spiracles homeobox genes EMX1 and EMX2. Cell Death Dis. 2021;12(6):515. doi: 10.1038/s41419-021-03801-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berdasco M, Esteller M. Clinical epigenetics: seizing opportunities for translation. Nat Rev Genet. 2019;20(2):109–127. doi: 10.1038/s41576-018-0074-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.