Abstract
The inclusion of retrovirus-derived introns within retrovirus-based expression vectors leads to a fraction of the resulting transcripts being spliced. Such splicing has been shown to markedly improve expression (W. J. Krall et al., Gene Ther. 3:37–48, 1996). One way to improve upon this still further might involve the use of more efficient introns instead of those from the provirus. Currently, however, incorporation of such introns remains self-defeating since they are removed in the nucleus of the producer cell. In the past, elaborate ways to overcome this problem have included the use of alphaviruses to make the vector transcripts within the cytoplasm, thus avoiding the nuclear splicing machinery during vector production (K. J. Li and H. Garoff, Proc. Natl. Acad. Sci. USA 95:3650–3654, 1998). We now present a novel design for the inclusion of introns within a retroviral vector. In essence, this is achieved by exploiting the retroviral replication process to copy not only the U3 promoter but also a synthetic splice donor to the 5′-long-terminal-repeat position during reverse transcription. Once copied, synthesized transcripts then contain a splice donor at their 5′ end capable of interacting with a consensus splice acceptor engineered downstream of the packaging signal. Upon transduction, we demonstrate these vectors to produce enhanced expression from near fully spliced (and thus packaging signal minus) transcripts. The unique design of these high titer and high-expression retroviral vectors may be of use in a number of gene therapy applications.
It is now well established that the inclusion of introns into eukaryotic expression vectors can enhance gene expression by as much as 500 times (2, 5, 7, 12, 26). For retrovirus-based expression vectors, however, the inclusion of such introns remains difficult. This is because such vectors, like the proviruses from which they are derived, undergo an RNA intermediate step during an infective cycle. Consequently, any efficiently spliced introns will always be removed in the nucleus of the first cell prior to packaging and subsequent transduction of the next cell. For this reason the best murine leukemia virus (MLV)-derived expression vectors to date, such as MFG (6, 15, 30) still only contain splice donor (SD) and splice acceptor (SA) sequences of retroviral origin. These sequences are by necessity suboptimal because in the wild-type virus they are used to maintain an equilibrium between full-length, genomic transcripts (required for gag-pol expression and packaging into the virion) and the spliced, subgenomic env-expressing species (see reference 29).
In retroviral vectors such as MFG, where the expressed cDNA effectively replaces env rather than gag-pol, expression of the desired gene is from a spliced transcript and as such is more highly expressed (15). However, because in such constructs only a fraction of the transcripts are ever spliced, there is still a need to optimize expression in the transduced cell. One method by which this is achieved involves the use of internal expression cassettes positioned in a reverse orientation with respect to the 5′ long terminal repeat (LTR) (9, 14, 21). Such vectors therefore make two opposing transcripts: one from the internal cassette with a functional intron, and the second, a genomic transcript, from the LTR. One drawback to this configuration is that the two opposing promoters now effectively make antisense transcripts. A second drawback, and as discussed previously (18, 21), is that reverse intronic and polyadenylation sequences contained within the genomic transcripts are often problematic.
A notable second method for the incorporation of introns into retroviral vectors has involved the use of alphaviruses to express vector transcripts, engineered to contain an intron, within the cytoplasm of the retroviral packaging cell and thus avoid the nuclear splicing machinery (17, 18). This elaborate method, although successful in delivering intron containing vectors with enhanced expression to transduced cells, does introduce a number of extra technical and safety concerns since it involves the use of RNA vectors from another virus (an alphavirus) which is known to shut down translation of cellular proteins (16, 36).
Presented here is a novel third method by which efficient intron inclusion is achieved. For this first prototypic example, we have used an adapted Miller et al. vector as a starting point (22, 32). Into this vector we first added a synthetic SD derived from the simian virus 40 (SV40) small-T antigen gene (st-SD). This was inserted between U3 and R of the 3′ LTR. Next, a synthetic consensus SA (c-SA) was incorporated downstream of the packaging signal. Finally, for some constructs the viral wild-type SD (wt-SD) and a previously unreported cryptic SD (cr-SD; contained within the packaging signal) were disabled (by GU-to-GC alteration). The resulting vector is designed to exploit the reverse transcription (RT) process (see reference 34) to copy not only the 3′ U3 promoter but also the small-T derived SD to the 5′ LTR position. Consequently, upon RT, the resulting LTR-expressed transcripts now contain, at their 5′ termini, the very efficient small-T derived SD (20) capable of interacting with the consensus SA downstream (see Fig. 1 for schematic representation of vector function and design).
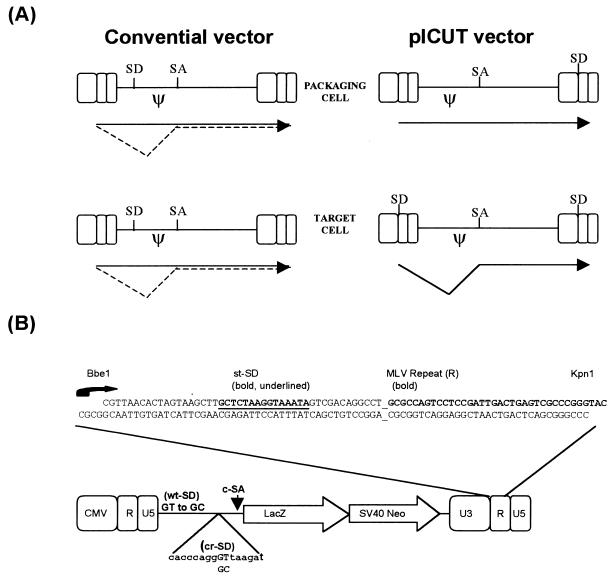
FIG. 1.
Design and function of the pICUT vector. (A) Comparison of splicing events in conventional and pICUT vectors. Conventional vectors such as MFG (6, 15, 30) and pLXSN (22) have suboptimal retroviral splice sites such that only a fraction of transcripts are ever spliced in packaging cells and target cells. For the pICUT vectors, efficient splice sites are instead used and positioned such that splicing occurs only after one round of RT. Consequently, for these vectors the splicing is minimal in transfected packaging cells while maximal in transduced target cells. (B) Construction of vector. A synthetic oligonucleotide containing the splice donor sequence from the SV40 st-SD intron was inserted between the naturally occurring BbeI and KpnI sites of the 3′ LTR of the vector. This oligonucleotide is designed to recreate the start of transcription (represented by a curly arrow and starting with GCGCC) which resides over the BbeI site. Contained within the oligonucleotide is the complete R sequence up to the KpnI site. Also shown is the positioning of the c-SA. This is located downstream of the packaging signal. Because of this overall design, upon one round of RT both the 3′ U3 promoter and the st-SD sequence are copied to the 5′ LTR position (with priming beginning at the 5′ terminus of R). Consequently, transcripts synthesized now contain a functional intron with the 5′ and 3′ splice sites located on either side of the packaging signal (ψ). Certain pICUT vectors also contain further modifications of both wild-type and cryptic SD sites located within the packaging signal (see also Fig. 2).
We demonstrate that the resulting vector, named pICUT (intron created upon transduction), maintains high titers (106 [LFU]/ml) and that upon transduction it produces transcripts of which the majority are spliced. Furthermore, we show this vector to have enhanced expression levels and, because the packaging signal (and marker gene if so desired) is subsequently deleted, we demonstrate it to have significantly improved safety over the parent vectors from which it was derived. Consequently, the pICUT design should have utility in a number of gene therapy applications.
MATERIALS AND METHODS
Cell culture.
All cell lines were maintained in Dulbecco modified Eagle medium containing 10% (vol/vol) fetal calf serum supplemented with penicillin, streptomycin, and l-glutamine.
Transient three-plasmid expression system and viral titration.
Retroviral vector stocks were produced according to the transient-transfection protocol described by Soneoka et al. (32). Briefly, 10 μg each of pHIT456 (4070A envelope), pHIT60 (gag-pol), and appropriate vector were transfected by overnight calcium phosphate treatment onto 293T cells at 70 to 80% confluency in 10-cm-diameter dishes. The following morning the cell medium was replaced with medium containing sodium butyrate (10 mM, final concentration) for 12 h. This medium was then replaced with 5 ml of fresh medium, and the viral supernatants were harvested 12 h later (therefore at 48 h posttransfection).
Plasmid constructs.
The starting plasmid for this construct is pLNCX (22). This was first cut with NheI, and the backbone was religated to make a single LTR plasmid. st-SD was then inserted, as shown in Fig. 1, to make the plasmid 3′LTR-SD. The pHIT111 XbaI-NheI fragment (containing the 5′ CMVLTR-lacZ SV40Neo-3′ppt) was then inserted into the NheI site of 3′LTR-SD. The GU-to-GC mutation of the wt-SD was derived from pBABE (23). The GU-to-GC mutagenesis of the cryptic SD (position 810 in the wild-type MLV provirus) was made by overlapping PCR. The consensus splice acceptor-branchpoint sequence used here was derived from sequence 899 to 992 of pCI (Promega, Madison, Wis.). This in turn is a modified (to consensus) splice acceptor obtained from the heavy-chain immunoglobulin variable region (1). A synthetic double-stranded oligonucleotide was made to this sequence and inserted downstream of the packaging signal (see Fig. 1 and 2). Complete plasmid sequences are available upon request.
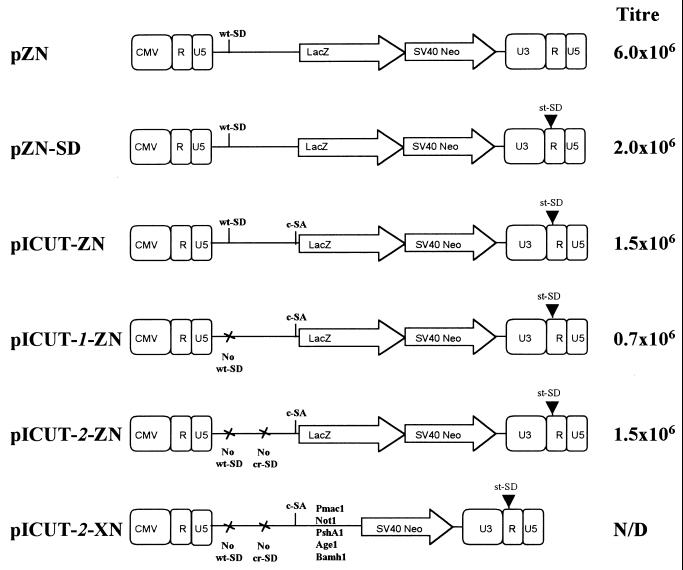
FIG. 2.
Schematic representation of vector constructs and their corresponding titers. The starting control vector (pZN) is a derivative of the pHIT111 (32), which in turn is derived from pLXSN (22). The lacZ reporter marker gene is expressed from the LTR and an internal SV40 promoter promotes Neor gene expression. Both expression cassettes use the same 3′ LTR R/U5 polyadenylation signal. This vector already contains the MLV wt-SD, which suboptimally pairs with a cryptic SA contained elsewhere within the packaging signal (22). The sequential cloning steps to make pICUT-2-ZN from this starting vector are represented by the intermediate vectors shown. First, the st-SD and, next, the c-SA were added (to make the vectors pZN-SD and pICUT-ZN, accordingly); subsequently, the wt-SD only or this as well as the cr-SD were disabled to make the vectors pICUT-1–ZN and pICUT-2–ZN, respectively. Finally, a pICUT-2 vector in which the lacZ marker is replaced with a multiple cloning site is also shown. The corresponding titers are the average LFU per milliliter recorded for each vector from three separate assays (with <5% deviation).
CAT assay.
Chloramphenicol acetyltransferase (CAT) assays were performed by the Quan-T-CAT assay system (Amersham Pharmacia, St. Albans, United Kingdom) according to the manufacturer's instructions, and the resulting 3H-acetylated chloramphenicol was measured in a scintillation counter.
Analysis of pICUT splicing patterns by RT-PCR.
Total RNA was isolated with the RNeasy Kit (Qiagen, Crawley, United Kingdom) and used in RT-PCR reactions according to instructions of the First-Strand cDNA Synthesis Kit (Amersham Pharmacia). PCR amplification was then performed by using the Expand High Fidelity PCR System (Roche Molecular Biochemicals, Lewes, United Kingdom) with 25 cycles of 94°C (1 min), 55°C (1 min), and 72°C (1 min). The forward primer was designed to anneal between the start of transcription and the st-SD of pICUT. The reverse primer was designed to bind 500 bp downstream of the splice acceptor site in the lacZ gene. The resulting RT-PCR products were cloned into pGEM-T Easy (Promega) for subsequent sequence analysis.
FACS analysis.
Fluorescence-activated cell sorting (FACS) analysis of enhanced green fluorescent protein (EGFP) was performed using the FACSCaliber System (Becton Dickinson, Rutherford, N.J.). Samples were prepared from trypsinized cells cultured on 6-cm-diameter dishes and resuspended in 0.5 ml of phosphate-buffered saline (PBS).
EGFP cell photography.
A sterile coverslip was placed in each well of a six-well plate before the addition of target cells and 24 h prior to transduction. The target cells were transduced as described above and, 24 h posttransduction, the cells were washed once in 1× PBS and fixed for 5 min at room temperature with PBS containing 2% (vol/vol) formaldehyde and 0.2% (vol/vol) glutaraldehyde. The coverslips were then removed, mounted on a slide, and sealed with nail-varnish prior to photography.
RESULTS
Vector design.
The first step in designing a retroviral vector containing an intron was to choose appropriate splicing signals. The consensus splice donor sequence is complementary to the U1 snRNA with which it base pairs. Nevertheless, comparative studies have demonstrated that as short donor site insertions into the β-globin pre-mRNA, the SV40 st-SD is actually more effective than a consensus sequence (20). As previously discussed (20), this finding suggests that the intrinsic activity of a SD sequence is not just dependent on its U1 binding affinity but also upon other events. Consequently, for construction of the pICUT vectors, the st-SD was used instead of a consensus equivalent. The positioning of this SD, between the U3 and R is so designed to be downstream of the start of transcription but upstream of the R repeat (Fig. 1B). This design ensures that upon transduction it will be copied with the U3 promoter to the 5′ LTR and thus be located upstream of the consensus SA (c-SA) with which it interacts (Fig. 1A). For certain pICUT vectors (the pICUT-1 series; see Fig. 2), as well as inclusion of the st-SD and c-SA splice sites, the MLV wt-SD was also disabled. This was initially considered important to ensure that the packaging signal of transcripts was not deleted by a splicing reaction between wt-SD and c-SA during vector production in 293T transient “packaging” cells. Subsequent RT-PCR analysis of transcripts produced from these vectors lead to their further modification (see below) to make the pICUT-2 vector.
Transcript splicing in pICUT vectors.
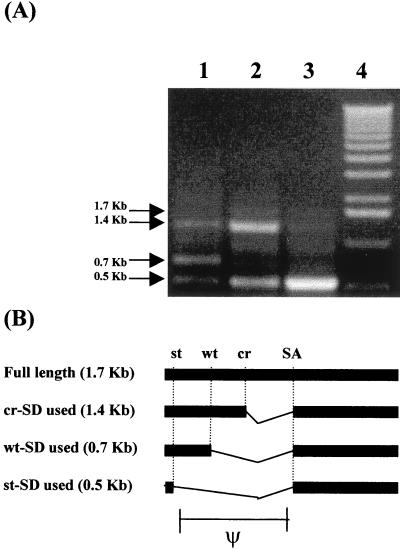
Initially, RT-PCR analysis was undertaken to investigate pICUT and pICUT-1 vector transcript splicing in transduced cells. The results of this analysis are presented in Fig. 3 and reveal that while three prominent products from the original pICUT vector are visible, only two are observed from pICUT-1. Subsequent cloning and sequencing of these products was performed to investigate them further. The results of this revealed first that in both instances the smallest (0.5-kb) product was indeed that of a splicing reaction between the 5′ st-SD and the c-SA. Second, we found that the mid-sized (0.7-kb) band present in only the pICUT vector was due to a splicing event between the wt-SD and c-SA (pICUT-1 vector has a disabled wt-SD and no such product; compare lanes 1 and 2 of Fig. 3A); and third, that the largest of three products (1.4 kb) is not of full length but instead due to an interaction between a previously unreported cryptic-SD (cr-SD) and the c-SA. A faint band above the 1.4-kb product (see lane 2 of Fig. 3A) was identified as the expected full-length 1.7-kb product. As a result of this analysis it was decided to make new vectors with both wt-SD and the newly identified cr-SD disabled. These were named the pICUT-2 vector series (see Fig. 2). Subsequent RT-PCR and sequence analysis of transcripts produced in transduced cells from these vectors was then performed. This revealed that the major product now identified was from the desired splicing reaction between that of the 5′ st-SD and the c-SA (see Fig. 3A, lane 3). Interestingly, mutation of cr-SD by GU-to-GC mutagenesis did not lead to its complete inactivation (unpublished observations and also see lane 3, where the 1.4-kb band is still present).
FIG. 3.
The splicing pattern of pICUT vectors in transduced cells. (A) RT-PCR analysis. The forward oligonucleotide was designed to prime upstream of the st-SD but downstream of the start of transcription (see also Fig. 1), while the reverse oligonucleotide was designed to prime within lacZ. Subsequent RT-PCR analysis was performed on cells transduced with vectors pICUT-ZN (lane 1), pICUT-1–ZN (lane 2), and pICUT-2–ZN (lane 3). Also shown (lane 4) is the 1-kb DNA marker (Gibco-BRL, Paisley, United Kingdom) which, in ascending order, contains the markers of sizes 0.5, 1, 1.6, 2, 3, and 4 kb, etc. Blank negative controls (untransduced cells or no RT step) are not shown. For the pICUT-ZN vector (lane 1), three major RT-PCR products predominate. Mutation of the wt-SD in such vectors to make pICUT-1–ZN leads to the loss of the 0.7-kb product (lane 2), while subsequent mutation of an identified cr-SD leads to the loss of also the 1.4-kb product (lane 3). (B) Schematic representation of the corresponding splicing events identified in the pICUT vectors. The identities of all spliced products were confirmed by cloning and/or sequencing. A faint 1.7-kb product (see lane 2 of panel A) was also observed. This was identified as the unspliced full-length transcript.
Titer comparison in pICUT vectors.
Titers were compared between the starting vector pZN with that of test vectors pZN-SD, pICUT-ZN, pICUT-1-ZN, and pICUT-2-ZN. The results of this analysis are presented in Fig. 2 and illustrate that both st-SD inclusion and wt-SD removal are the most notable modifications affecting the titer: insertion of the st-SD within the 3′ LTR might lower the titer by reducing vector transcript polyadenylation, for it has previously been demonstrated that an unpaired 5′ splice site (SD) at the 3′ end of a transcript can have this effect (35). Alternatively, it may hinder other functions of retroviral replication. Mutation of the wt-SD might disrupt the packaging signal secondary structure or instead increase cr-SD–c-SA splicing because subsequent mutation of cr-SD restores the titer to that observed prior to wt-SD modification.
Collectively, these data illustrate that it is possible to retain high titer (>106 LFU/ml) in a retroviral construct containing both of the SD and SA splice site modifications described here.
Splicing-mediated packaging signal deletion: potential for improved safety.
With the advent of retrovirus-mediated in vivo gene delivery has come the requirement for safer, self-inactivating retroviral expression vectors. Such vectors are designed to undergo only one round of replication in order to reduce the chances of mobilization in transduced cells by helper virus. To date, examples of such vectors include both those with disabled 3′ U3 promoters (37) and those with direct repeats flanking the packaging signal (13). For the former, after one round of replication the disabled U3 promoter is copied to the 5′ end of the provirus, and thus subsequent full-length genomic RNA is never transcribed. Such vectors contain internal promoters for cDNA transcription. For the latter, after entry into the transduced cell and during the RT event, the sequence (the packaging signal) between the direct repeats is skipped. Consequently, transcripts from these integrated proviruses contain no packaging signal.
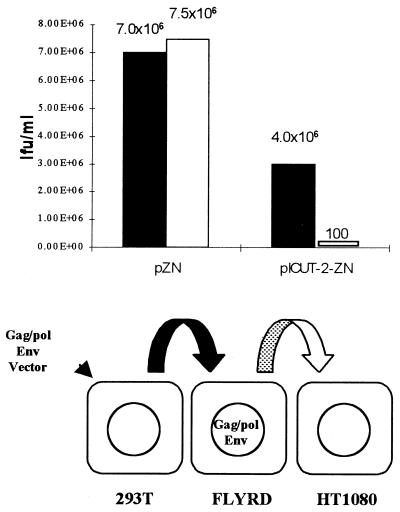
We predicted that after one round of RT and due to a splicing-mediated event, pICUT vector transcripts would be integrated as proviral genomes into target cells that are subsequently unable to generate RNA capable of being packaged. To test this hypothesis, an RD114-enveloped packaging line (FLYRD18; see reference 8) was used as a target cell line capable of vector mobilization. This was transduced with 4070A pseudotyped vector, the resulting supernatant was harvested, and its viral titer was established. The results of one such experiment are presented in Fig. 4 and demonstrate that after one round of replication the majority of pICUT vector transcripts in transduced cells are no longer capable of mobilization, with titers dropping 10,000-fold relative to the control. This therefore demonstrates that the pICUT vectors are safer than the equivalent LTR-based vectors from which they are derived.
FIG. 4.
Mobilization of pICUT vectors after one round of transduction into a packaging cell line. The 4070A envelope pseudotyped virus, made by transient transfection of 293T cells, was initially used to transduce the FLYRD18 packaging cell line at a multiplicity of infection of 1. At 48 h posttransduction, the supernatant from these cells was then harvested, and the titers were determined on HT1080 cells. The corresponding primary and secondary titers of the pICUT-2–ZN vector and the control pZN vector were established on HT1080 cells as LFU per milliliter. The primary titers were established from the harvested supernatant taken from transfected 293T cells (shown as black bars) and secondary titers from supernatant harvested from the transduced FLYRD18 cells (shown as white bars). This figure illustrates that while secondary titers for the pZN control remain high, such titers drop 10,000-fold in the pICUT vectors after one round of RT.
The residual vector transcripts still “mobilized” from the packaging cells transduced with pICUT-2–ZN (Fig. 4; 100 LFU/ml) remain uncharacterized but are nevertheless of interest. One possible explanation for their persistence is that prior to integration into the packaging line, occasional reverse transcriptase copying errors might lead to splice site mutations and thus a splicing inactivation event in a small fraction of vectors which would subsequently give rise to packageable transcripts.
Use of pICUT vectors to inhibit expression of marker genes in transduced cells.
To further demonstrate splicing in pICUT vectors, it was decided to construct a vector containing the EGFP gene upstream of the c-SA such that upon transduction it, along with the packaging signal, will be contained within a functional intron and therefore not translated. By such a design it should thus be possible to limit EGFP gene expression to the initial, transfected cell. This may be of use since often certain genes are only required for the selection of high-titer packaging lines and, consequently, any subsequent expression in transduced cells is undesirable.
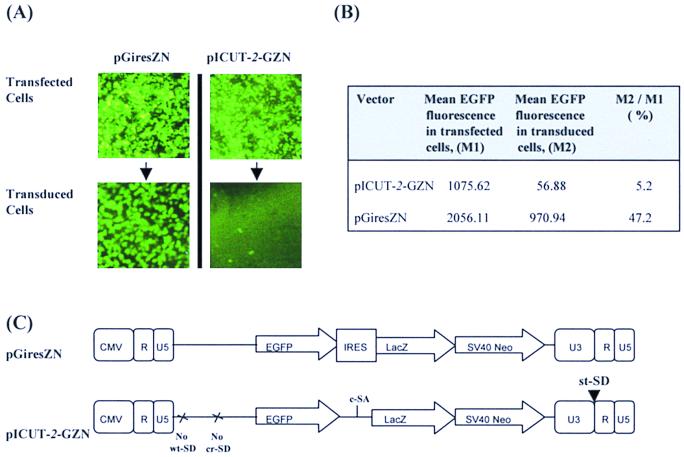
To investigate the effectiveness of the pICUT vectors to inhibit EGFP expression, the EGFP gene was cloned upstream of the c-SA in pICUT-2–ZN to make the construct pICUT-2–GZN. The EGFP expression levels from this vector were monitored in both transfected and transduced cells and then compared to that of a control vector pGiresZN. The results of this analysis are presented in Fig. 5 and reveal that compared to the control, EGFP expression from pICUT-2–GZN is significantly decreased in transduced cells, thus demonstrating EGFP gene removal from transcripts in such cells. However, one concern regarding this result is that if splicing in pICUT-2–GZN is nearing 100% efficiency, as expected from previous analysis performed on the parental pICUT-2–ZN (Fig. 3 and 4), almost no EGFP expression would be expected. Therefore, the fact that residual EGFP expression does exist suggests that inclusion of an extra sequence (the EGFP cDNA) upstream of the c-SA might affect splicing efficiency between 5′ and 3′ splice sites of the intron in which EGFP is found. In support of this it has been previously reported that intron size can affect splicing efficiency (33). Figure 5 also shows that, relative to control, EGFP expression from pICUT-2–GZN is also lower upon transfection and thus prior to intron creation. Preliminary RT-PCR analysis (data not shown) indicates that this may be due to as-yet-unidentified cryptic SDs contained within EGFP activated by the downstream consensus SA. To conclude, although EGFP marker gene expression is reduced in the pICUT-2–GZN upon transduction, further investigation is required to resolve why such residual EGFP expression persists.
FIG. 5.
Efficiency of EGFP marker loss in pICUT vectors. The two vectors schematically represented in panel C were constructed and used to make retroviral stocks by transient transfection. This virus was then used to transduce HT1080 cells. (A) Photographic record of EGFP expression in both transfected and transduced cells. (B) Results of EGFP analysis by FACS. For each cell type (transfected or transduced with either pGiresZN or pICUT-GZN vectors) the mean count for cell fluorescence was established and used to assess EGFP expression levels. For both vectors analyzed, the EGFP genes are expressed from a cytomegalovirus-based LTR promoter upon transfection and from a U3-based LTR upon transduction. To ensure normalization of transfection and transduction efficiencies between vectors, FACS analysis was performed on only the “gated” fluorescent-cell population. Titers of pGiresZN and pICUT-GZN for this experiment were 9 × 105 and 1 × 105 LFU/ml, respectively. This figure illustrates that upon transduction and relative to control, the EGFP expression from the pICUT vector is decreased.
Enhanced CAT expression in cells infected with pICUT vectors.
In transgenic experiments, the presence of a flanking intron is required to promote high-level expression of virtually all cDNAs, even those that are apparently intron independent when assayed in transfected cell lines (2, 5, 7, 12, 26). Moreover, introns positioned upstream to the cDNA are more effective at this than those located downstream (10, 19). In retroviruses it has previously been suggested that the secondary structure of the 5′ packaging signal can cause ribosome “stutter,” thus reducing the expression of downstream genes (15, 31). These two observations suggest that an ideal retroviral expression vector would be one that produced transcripts containing both a 5′ intron and a deleted packaging signal. The vectors described here produce transcripts with both such characteristics.
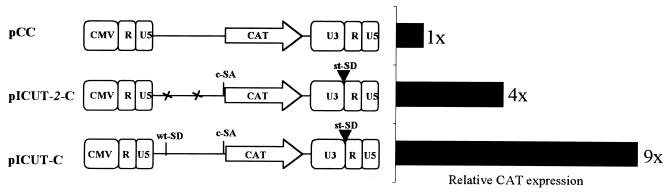
In the tissue culture environment, certain marker genes are more intron dependent than others (4). Because the CAT gene is one such marker, it was chosen to examine any intron augmented expression effects by the pICUT vectors. Initially, this was undertaken by replacing lacZ with CAT in the pICUT-2 vectors. However, no CAT expression above background levels could be observed. Subsequent RT-PCR and sequence analysis revealed that this was most likely because of a splicing reaction between CAT and a splice acceptor in the downstream SV40 promoter sequence which apparently becomes activated only upon transduction (data not shown). This suggests that in the pICUT vectors, upon creation of the upstream intron, downstream splicing events within CAT may be activated. This idea is in agreement with previous observations demonstrating CAT to possess cryptic splice sites (10) and multiple introns to augment overall splicing through cooperation (24). Consequently, for the pICUT-CAT vectors the SV40 Neo cassette (between the BamHI site upstream of the SV40 promoter and the RsrII site in Neo) was deleted to make the vectors pICUT-C and pICUT-2–C, as well as the control pC. For expression analysis, CAT activity was measured in equivalent transduced cells. The results are presented in Fig. 6 and demonstrate that the pICUT vectors have enhanced expression levels relative to the starting control vector pC. Of interest is the observation that the presence the wild-type MLV SD further enhances the augmented expression of pICUT-2. We are currently investigating this further by more extensive mutational analysis of the wild-type SD sequence in these vectors.
FIG. 6.
CAT expression analysis. Viral preparation of the pCC, pICUT-C, and pICUT-2–C vectors schematically described in the figure were produced by transient transfection. This was then used to transduce 293T cells (with a multiplicity of infection of 1; calculated from established titers of equivalent vectors with a marker gene in place of CAT), and the CAT activity from cell lysates was subsequently assayed. The average fold increase in CAT activity observed by the two pICUT vectors over that of the pCC control vector is shown.
DISCUSSION
The precise mechanism by which introns enhance cDNA gene expression remains to be defined. Nevertheless, the cumulative evidence is compelling, particularly when the in vivo environment is considered. To date, it is likely that introns and splicing help in identifying mRNA for polyadenylation, nuclear export, and translation. In support of this idea, our laboratory has previously demonstrated that splicing can facilitate the recruitment of mRNAs for translation (3), while other researchers have implicated splicing factors as substrates for nuclear-cytoplasmic transport of mRNA (28). Furthermore, it has also been demonstrated that the presence of an upstream intron can enhance mRNA polyadenylation and that the two processes might possibly be coupled (12, 25, 27, 35). Consequently, it is not surprising that cDNAs, made from reverse-transcribed intron-free mRNA, often require intron addition for effective and sustained levels of expression.
The suboptimal splicing reaction observed in retroviral genomic transcripts is essential since it removes both the upstream packaging and gag-pol sequence to generate an env-expressing mRNA. Such a splicing event has been shown to augment gene expression (15) and might explain why retroviruses generally prefer suboptimal splicing, rather than internal ribosome entry sites, to express their envelope proteins. By inclusion of these virus-derived intronic sequences, the best retroviral expression vectors make use of this splicing reaction to express heterologous cDNAs from a spliced message. The main drawback to such vectors is that this splicing reaction must still remain suboptimal (see above). Consequently, upon transduction there remains an abundance of unspliced messages in transduced cells. This is problematic for a number of reasons. First, expression is not optimal. Second, any one of the numerous upstream translation initiation signals (AUGs) might lead to aberrant and possibly immunogenic polyprotein synthesis. Finally, such unspliced RNAs can be packaged by helper virus and “mobilized.”
The vectors described here are the first to exploit the RT cycle to overcome these problems. Upon transduction, the packaging signal is now found within an efficient intron and consequently removed. These vectors can therefore produce nearly fully spliced, packaging-signal-free transcripts and thus, for the first time, exploit the same intron-mediated expression enhancement utilized by other expression systems.
The full utility of these vectors is at present restricted to intron sequences that lack cryptic splice sites. In most applications this is completely resolved by identifying and altering the sites, as we have done for the CAT gene constructs. In conclusion, the enhanced level of expression in target cells that can be obtained with the pICUT vectors and the enhanced safety may be particularly useful for a number of applications of retroviral vectors ranging from gene therapy to functional genomics.
ACKNOWLEDGMENTS
S.I.I. was supported by The Karim Rida Said Foundation and has also been awarded an Overseas Research Studentship.
We gratefully acknowledge Melvin Yap, Kyriacos Mitrophanous, and Ekaterini Kotsopoulou for excellent technical help and advice.
REFERENCES
- 1.Bothwell A L, Paskind M, Reth M, Imanishi-Kari T, Rajewsky K, Baltimore D. Heavy chain variable region contribution to the NPb family of antibodies: somatic mutation evident in a gamma 2a variable region. Cell. 1981;24:625–637. doi: 10.1016/0092-8674(81)90089-1. [DOI] [PubMed] [Google Scholar]
- 2.Brinster R L, Allen J M, Behringer R R, Gelinas R E, Palmiter R D. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci USA. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braddock M, Muckenthaler M, White M R, Thorburn A M, Sommerville J, Kingsman A J, Kingsman S M. Intron-less RNA injected into the nucleus of Xenopus oocytes accesses a regulated translation control pathway. Nucleic Acids Res. 1994;22:5255–5264. doi: 10.1093/nar/22.24.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brondyk B. PCI and pSI mammalian expression vectors. Promega Notes. 1994;49:7. [Google Scholar]
- 5.Buchman A R, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byun J, Kim S H, Kim J M, Yu S S, Robbins P D, Yim J, Kim S. Analysis of the relative level of gene expression from different retroviral vectors used for gene therapy. Gene Ther. 1996;3:780–788. [PubMed] [Google Scholar]
- 7.Choi T, Huang M, Gorman C, Jaenisch R A. A generic intron increases gene expression in transgenic mice. Mol Cell Biol. 1991;11:3070–3074. doi: 10.1128/mcb.11.6.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosset F L, Takeuchi Y, Battini J L, Weiss R A, Collins M K. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dzierzak E A, Papayannopoulou T, Mulligan R C. Lineage-specific expression of a human beta-globin gene in murine bone marrow transplant recipients reconstituted with retrovirus-transduced stem cells. Nature. 1988;331:35–41. doi: 10.1038/331035a0. [DOI] [PubMed] [Google Scholar]
- 10.Evans M J, Scarpulla R C. Introns in the 3′-untranslated region can inhibit chimeric CAT and beta-galactosidase gene expression. Gene. 1989;84:135–142. doi: 10.1016/0378-1119(89)90147-9. [DOI] [PubMed] [Google Scholar]
- 11.Hamer D H, Leder P. Splicing and the formation of stable RNA. Cell. 1979;18:1299–1302. doi: 10.1016/0092-8674(79)90240-x. [DOI] [PubMed] [Google Scholar]
- 12.Huang M T, Gorman C M. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Julias J G, Hash D, Pathak V K. E-vectors: development of novel self-inactivating and self-activating retroviral vectors for safer gene therapy. J Virol. 1995;69:6839–6846. doi: 10.1128/jvi.69.11.6839-6846.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsson S, Papayannopoulou T, Schweiger S G, Stamatoyannopoulos G, Nienhuis A W. Retroviral-mediated transfer of genomic globin genes leads to regulated production of RNA and protein. Proc Natl Acad Sci USA. 1987;84:2411–2415. doi: 10.1073/pnas.84.8.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krall W J, Skelton D C, Yu X J, Riviere I, Lehn P, Mulligan R C, Kohn D B. Increased levels of spliced RNA account for augmented expression from the MFG retroviral vector in hematopoietic cells. Gene Ther. 1996;3:37–48. [PubMed] [Google Scholar]
- 16.Lachmi B, Kaariainen L. Control of protein synthesis in Semliki Forest virus-infected cells. J Virol. 1977;22:142–149. doi: 10.1128/jvi.22.1.142-149.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li K J, Garoff H. Production of infectious recombinant Moloney murine leukemia virus particles in BHK cells using Semliki Forest virus-derived RNA expression vector. Proc Natl Acad Sci USA. 1996;93:11658–11663. doi: 10.1073/pnas.93.21.11658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li K J, Garoff H. Packaging of intron-containing genes into retrovirus vectors by alphavirus vectors. Proc Natl Acad Sci USA. 1998;95:3650–3654. doi: 10.1073/pnas.95.7.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto K, Wassarman K M, Wolffe A P. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998;17:2107–2121. doi: 10.1093/emboj/17.7.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayeda A, Ohshima Y. Short donor site sequences inserted within the intron of beta-globin pre-mRNA serve for splicing in vitro. Mol Cell Biol. 1988;8:4484–4491. doi: 10.1128/mcb.8.10.4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller A D, Bender M A, Harris E A, Kaleko M, Gelinas R E. Design of retrovirus vectors for transfer and expression of the human beta-globin gene. J Virol. 1988;62:4337–4345. doi: 10.1128/jvi.62.11.4337-4345.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–982. , 984–986, 989–990. [PMC free article] [PubMed] [Google Scholar]
- 23.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neel H, Weil D, Giansante C, Dautry F. In vivo cooperation between introns during pre-mRNA processing. Genes Dev. 1993;7:2194–2205. doi: 10.1101/gad.7.11.2194. [DOI] [PubMed] [Google Scholar]
- 25.Niwa M, Rose S D, Berget S M. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;9:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 26.Palmiter R D, Sandgren E P, Avarbock M R, Allen D D, Brinster B L. Heterologous introns can enhance expression of transgenes in mice. Proc Natl Acad Sci USA. 1991;88:478–482. doi: 10.1073/pnas.88.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey N B, Chodchoy N, Liu T J, Marzluff W F. Introns in histone genes alter the distribution of 3′ ends. Nucleic Acids Res. 1990;18:3161–3170. doi: 10.1093/nar/18.11.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 29.Rabson A B, Graves B J. Synthesis and processing of viral RNA. In: Coffin J M, Hughes S, Varmus H, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 205–261. [PubMed] [Google Scholar]
- 30.Riviere I, Brose K, Mulligan R C. Effects of retroviral vector design on expression of human adenosine deaminase in murine bone marrow transplant recipients engrafted with genetically modified cells. Proc Natl Acad Sci USA. 1995;92:6733–6737. doi: 10.1073/pnas.92.15.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sagliocco F A, Vega Laso M R, Zhu D, Tuite M F, McCarthy J E, Brown A J. The influence of 5′-secondary structures upon ribosome binding to mRNA during translation in yeast. J Biol Chem. 1993;268:26522–26530. [PubMed] [Google Scholar]
- 32.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sterner D A, Carlo T, Berget S M. Architectural limits on split genes. Proc Natl Acad Sci USA. 1996;93:15081–15085. doi: 10.1073/pnas.93.26.15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Telesnitsky A, Goff S P. Reverse transcriptase and the generation of retroviral DNA. In: Coffin J M, Hughes S, Varmus H, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 35.Wassarman K M, Steitz J A. Association with terminal exons in pre-mRNAs: a new role for the U1 snRNP? Genes Dev. 1993;7:647–659. doi: 10.1101/gad.7.4.647. [DOI] [PubMed] [Google Scholar]
- 36.Wengler G. Protein synthesis in BHK-21 cells infected with Semliki Forest virus. J Virol. 1975;17:10–19. doi: 10.1128/jvi.17.1.10-19.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu S F, von Ruden T, Kantoff P W, Garber C, Seiberg M, Ruther U, Anderson W F, Wagner E F, Gilbon E. Self-inactivating retroviral vectors designed for transfer of whole genes into mammalian cells. Proc Natl Acad Sci USA. 1986;83:3194–3198. doi: 10.1073/pnas.83.10.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]