Abstract
A human immunodeficiency virus (HIV)-negative patient with no risk factor experienced HIV type 1 (HIV-1) primary infection 4 weeks after being hospitalized for surgery. Among the medical staff, only two night shift nurses were identified as HIV-1 seropositive. No exposure to blood was evidenced. To test the hypothesis of a possible nurse-to-patient transmission, phylogenetic analyses were conducted using two HIV-1 genomic regions (pol reverse transcriptase [RT] and env C2C4), each compared with reference strains and large local control sets (57 RT and 41 C2C4 local controls). Extensive analyses using multiple methodologies allowed us to test the robustness of phylogeny inference and to assess transmission hypotheses. Results allow us to unambiguously exclude one HIV-positive nurse and strongly suggest the other HIV-positive nurse as the source of infection of the patient.
Human immunodeficiency virus type 1 (HIV-1) infection risk factors are usually identified directly from a patient's medical history. However, additional evidence is essential when viral transmission occurs during clinical care (8, 16, 25, 37) or is the object of a criminal trial (1, 48). Because nucleotide sequences vary greatly among HIV-1 isolates (28, 41, 49), transmission histories among infected individuals can be investigated through estimated phylogenetic relationships among viral sequences.
Previous studies have shown that phylogenetic methods can lead to accurate reconstruction of known transmission histories (5, 11, 33, 52). Phylogeny inferences have also been used to reconstruct unknown transmission histories (1, 7, 8, 37), although the methodology itself has sometimes been a point of contention. For example, in the Florida dentist case (37), while the original analysis determined HIV transmission, reanalyses of the same data set together with new control sequences did not unequivocally reject the possibility of independent acquisitions of similar local variants (6, 14). However, extensive analyses (using multiple inference methodologies) of an extended data set convincingly confirmed the existence of a dental clade (13, 24). Furthermore, phylogenetic analyses are increasingly used in court (2, 48) and must therefore “rise to a threshold of reliability” (48).
Investigating a possible health care worker-to-patient transmission of HIV-1 without evidence of blood exposure, we performed extensive phylogenetic analyses. The results presented here unambiguously exclude one HIV-seropositive nurse but strongly suggest another HIV-seropositive nurse as the source of transmission of HIV-1 to the patient.
Case report.
At the end of May 1996, a 61-year-old HIV-negative female was admitted in a health care institution in the suburbs of Paris, France, for surgery which did not require blood transfusion. In July 1996, she suffered a severe HIV primary infection associated with hepatitis and massive weight loss. HIV-1 seroconversion occurred at that time, as evidenced by successive Western blot profiles, and was associated with high viral loads (Fig. 1). An epidemiological investigation did not identify any risk factor for HIV infection. No HIV-infected individual was found in the surgical staff, but two night shift nurses (nurse 1 and nurse 2) who had been in contact with the patient during her stay in the clinic (Fig. 1) were found to be HIV-1 seropositive. A virological investigation was requested to determine whether a nurse-to-patient contamination had occurred.
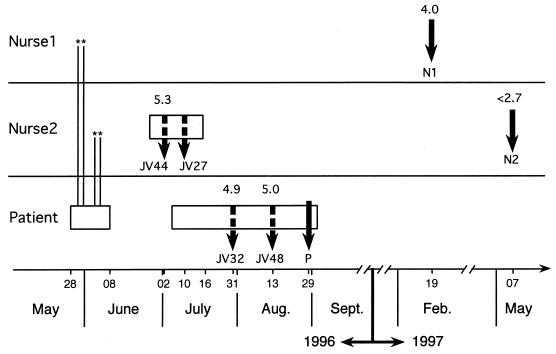
FIG. 1.
Chronology of PBMC and plasma samplings for the two nurses and the patient. Rectangles represent periods of hospitalization; vertical plain and dashed arrows indicate sampling of lymphocytes and plasma, respectively. Nurse 1 and nurse 2 had been in contact, between May 28 and June 8, 1996, each during two nights (indicated by asterisks), with the patient, who had a negative HIV-1 serology and no detectable viral RNA at the time of her hospitalization. Nurse 1 had been aware of his HIV-1 seropositivity for 5 years, and HIV-1 sequences were characterized from lymphocytes (N1) obtained on February 19, 1997, i.e., after the virological investigation was requested. Nurse 2's HIV serological status was unknown until she was diagnosed with HIV-1 and HCV infection during a hospitalization that started on June 20, 1996. The first nurse 2 HIV sequence (N2) used for molecular epidemiology analyses was from lymphocytes obtained on May 7, 1997, 1 month after introduction of antiretroviral treatment. Retrospectively, we also characterized viral sequences from cryopreserved plasma samples (JV44 and JV27) obtained during nurse 2's hospitalization. The second hospitalization (July to August 1996) of the patient was for HIV-1 primary infection. Plasma (JV32 and JV48) and lymphocyte (P) samples were obtained from the patient for viral sequence amplification. HIV-1 viral load, when available, is indicated (logarithm of the number of copies per milliliter) above corresponding sampling pointers.
Nurse 1 was a 30-year-old male, born in former Zaire (now Democratic Republic of Congo). He had been aware of his seropositivity for 5 years. In February 1997, nurse 1 had never received antiretroviral treatment, had a viral load of 104 copies per ml, and had a CD4+ cell count over 500 per mm3.
Nurse 2 was a 51-year-old female unaware of her seropositivity for HIV-1. In June 1996, she was hospitalized for hepatic insufficiency, associated with edema of the lower limbs. A diagnosis of infection with both hepatitis C virus (HCV) and HIV-1 was established. HIV viral load was 1.8 × 105 copies per ml; CD4+ cell count was 94 per mm3.
MATERIALS AND METHODS
Data collection.
Free virus was separated from plasma samples by ultracentrifugation; RNA was extracted and reverse transcribed. DNA was extracted from peripheral blood mononuclear cells (PBMCs) obtained by density gradient centrifugation. Two reverse transcriptase (RT) gene fragments (RT1 [codons 6 to 152] and RT2 [codons 157 to 252]) were amplified from proviral DNA (PBMCs) by nested PCR (32) with external primers A35 and NE-135 as described elsewhere (31). Forward internal primers were A and B (31); reverse internal primers were 5′ GGTGATCCTTTCCATCC 3′ and 5′ TCATTGACAGTCCAGCT 3′ for RT1 and RT2, respectively. One fragment spanning the C2C4 region of the env gene encoding gp120 was amplified by nested PCR as described elsewhere (15) from both proviral DNA and viral RNA (serum). All PCR products were directly sequenced on both strands on a ABI Prism 377 automated sequencer (Applied Biosystems, Foster City, Calif.). Patient and nurse PBMC samples were collected and purified in three different hospitals (one for each individual). All plasma samples were extracted and reverse transcribed in a laboratory where HIV had never been cultured or PCR amplified. Both for the patient and nurse 2, the different cell samples (designated P and N2, respectively) were handled, DNA extracted, and PCR amplified separately. Contamination prevention procedures were strictly observed (29). Ambiguities and discrepancies in DNA-sequencing electropherograms were resolved by two independent readers. To exclude possible contamination with a known HIV strain, each sequence obtained was searched via BLAST (4) against GenBank. We also excluded the possibility of internal contamination by aligning all sequences against routine sequences obtained in the laboratory during the same period.
A chronology of PBMC and plasma samplings for the nurses and the patient is depicted in Fig. 1. Samples from the patient (P, JV32, and JV48) were all obtained during primary infection (Fig. 1). PBMCs from nurse 1 and nurse 2 (N1 and N2) were first obtained, and plasma aliquots from nurse 2 (JV44 and JV27, sampled at dates closer to the patient's primary infection) were retrospectively collected. Local control plasma samples were obtained in 1997 from HIV-infected individuals living in the Paris area. Fifty-nine RT sequences were obtained from 57 patients consulting at the Hôpital Rothschild (RO samples); C2C4 sequences were obtained from 41 patients from Hôpital Jean Verdier (JV samples). Control cohorts included individuals of both sexes (80% males), infected for 1 to 12 years, whose risk factor was homosexual (53%) or heterosexual (20%) activity, intravenous drug use (14%), blood transfusion (3%), mother-to-child transmission (2%), or unknown (8%). None of the sequences resulting from the BLAST searches were more similar to the patient or nurse sequences than were at least one of the local controls. Plasma samples for C2C4 analysis (including patient and nurse 2 samples) were randomly numbered and processed blind for both sequencing and phylogenetic analyses. Reference sequences were chosen from the recommended strains of the Los Alamos National Laboratory (LANL) HIV database (28).
Sequence alignments.
RT sequences were obtained by concatenation of RT1 and RT2 (618 nucleotides). Alignment was trivial (no insertion or deletion detected). Because C2C4 sequences are quite variable, we first aligned the amino acid translated sequences with Clustal W (47) using four different sets of parameters (matrix = Blossum or PAM; gap/extension penalties = 10/0.05 or 20/0.1). Positions at which the four alignments differed were excluded in the subsequent analyses (21). All alignments are available on request from M. C. Milinkovitch.
Phylogenetic analyses. (i) RT.
Using different outgroup and ingroup samplings, all maximum parsimony (MP) analyses were performed with PAUP* (45) with exact branch-and-bound searches when computationally practical (i.e., in all analyses except permutation tail probability [PTP] searches and reanalyses with extended data sets). All characters were first weighted equally and treated as unordered. However, it is now well known that different types of changes can occur at different evolutionary rates, which may, in specific cases, justify differential weighting or encoding (36, 46). We therefore checked for possible saturation of nucleotide substitution types by plotting Tv versus Ti as well as the number of transitions (Ti) and transversions (Tv) versus the uncorrected pairwise distances. We also assessed the outcome of our unweighted analyses by excluding Ti or by using the Goloboff fit criterion (22) (k = 0, 2, 4, and 8). We estimated the reliability of the various inferred clades by bootstrapping (103 to 104 replicates, exact branch-and-bound searches, heuristics for extended data set), although bootstrap values (BV) may be misleading estimates of accuracy under specific conditions (36). For selected branches and using the “constraints” command in PAUP*, we computed Bremer branch support—the number of additional character transformations necessary to collapse an internal branch (10)—as an alternative to BV as an estimate of clade stability. We also performed cladistic PTP and topology-dependent PTP (T-PTP) (17, 18) analyses to statistically compare alternative phylogenetic hypotheses. Using a priori T-PTP for evaluating patient-nurse clade monophyly is valid because these tests are the results of a hypothesis of transmission formulated prior to any phylogenetic analysis.
We used PAUP* heuristic searches to estimate maximum likelihood (ML) trees, with the following settings: nucleotide frequencies computed from the data, invariable sites proportion and Tv/Ti ratio estimated via ML, rates for variable sites assumed to follow a gamma distribution with shape parameter estimated by ML (four rate categories; average rate represented by mean), HKY85 model (23) with rate heterogeneity, molecular clock not enforced, Rogers-Swofford approximation for starting branch lengths, starting tree obtained by stepwise addition (“as-is”), and tree bisection-reconnection (TBR) branch swapping. Alternative phylogenetic hypotheses were compared statistically by means of Kishino-Hasegawa ML ratio tests (27). Using two subsamples of the ingroup sequences, we estimated bootstrap supports for the various nodes on the ML trees.
Neighbor-joining (NJ) analyses of the 59 RT local control sequences were performed using PHYLIP 3.572c (19), with pairwise distances estimated under the HKY ML model (23), base frequencies and Ti/Tv ratio estimated from the data, and sequence input jumbling enabled.
(ii) C2C4.
NJ analysis of P, N1, N2, and 29 reference sequences was performed with PHYLIP as described above. Parsimony analysis and T-PTP monophyly tests were performed with PAUP* using heuristic searches.
We performed heuristic MP analyses of the 41 local control C2C4 sequences with starting tree(s) obtained via stepwise addition. We checked whether “simple”, “closest,” and “random” addition sequences yielded identical trees. Other settings were TBR branch swapping, MULPARS, and zero-length branches collapsed. To avoid local optima, we performed replicates both with random starting trees and with starting trees obtained via stepwise addition with random addition sequence. We estimated the reliability of the various MP-inferred clades by bootstrapping (103 replicates), and we compared alternative hypotheses by calculating Bremer branch support and by performing T-PTP tests (see above).
Ti/Tv ratio was the only parameter which could be estimated within practical central processing unit time for ML analyses including local controls (48 sequences). Alternative phylogenetic hypotheses were compared statistically by means of Kishino-Hasegawa ML ratio tests (27). Using a subsample of the ingroup sequences, we estimated bootstrap supports for the various nodes on the ML trees. NJ analyses were performed as described above.
Nucleotide sequence accession numbers.
All new sequences are deposited at EMBL under accession no. AF125604 to AF125651 for C2C4 and AF125683 to AF125806 for RT. Accession numbers of sequences used in Fig. 2 are M26727 (OYI), K03455 (HXB2), K02013 (LAI), U63632 (JRFL), M93258 (yu2), U23487 (manc), U21135 (weau), M22639 (Z2Z6), K03454 (eli), M27323 (ndk), M62320 (u455), and L39106 (ibng). Reference sequences used in Fig. 3a are the same as in Fig. 2, with the addition of AF004885, U5190, U51188, U51189, and U54771 (subtype A); AF005494 (subtype F); U88825 and U88826 (subtype G); and AF005496 (subtype H). Reference sequences used in Fig. 3b are U08794, U09127, L22939, L34667, and L22957 (subtype A); U08445, U23112, M93258, U08449, U08797, U27434, and K02013 (subtype B); U08453, U52953, and X65639 (subtype C); M22639 and K03454 (subtype D); U08810, U08456, U08458, and L03700 (subtype E); U37033, U08974, and U27413 (subtype F); U09664, U27426, and U26301 (subtype G); and U09666 and AF005496 (subtype H).
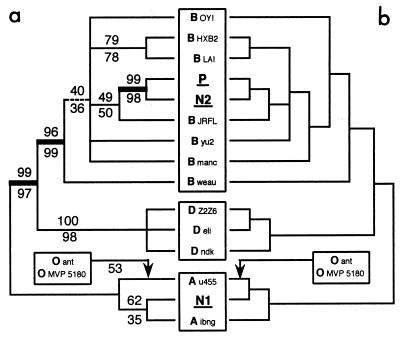
FIG. 2.
MP and ML trees among P, N1, N2, and reference sequences (RT). (a) Strict consensus of the MP trees under no weighting of substitution types. BV under exclusion and inclusion of group O sequences are indicated above and below the branches, respectively. Including the group O sequences collapsed the branch indicated as dashed and rooted the tree (with a BV value of 53) as indicated by the arrow. N1 and P are separated by three branches (thick line) supported by BV > 95. Sequence labels correspond to reference strain names from the LANL website (see text). (b) Strict consensus of the 3 ML trees (−ln L = 2062.54867). Estimation of Ti/Tv ratio, proportion of invariable sites, and gamma shape parameter for variable sites are 4.09, 0.32, and 0.64, respectively. When group O sequences are included, the branching pattern is unchanged but the tree is rooted as indicated by the arrow.
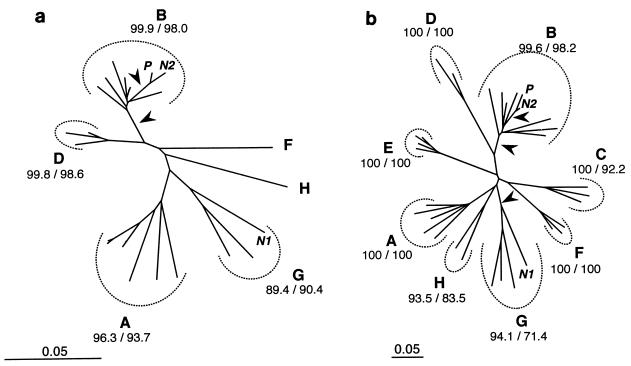
FIG. 3.
NJ trees among P, N1, N2, and reference sequences for RT (a) and C2C4 (b). For each subtype (A to H), BV (in percent) are given for 104 NJ/104 MP replicates. Arrows indicate branches separating P from N1 with BV > 94%. Scale is in percent expected substitution per position. Note the difference of scale between RT and C2C4. Reference sequences used for RT were the same as in Fig. 2, with the addition of nine sequences from subtypes A, F, G, H (see Materials and Methods) and the exclusion of ibng (see text for details). The accession numbers of the 29 reference sequences (subtypes A to H) used for C2C4 are given in the text.
RESULTS
We compared HIV-1 strains infecting the individuals described in the case report over two regions of the viral genome: RT, and env gp120 (C2C4), representing a total of 10.7% of the whole genome. Proviral DNA sequences from lymphocytes are referred as P, N1, and N2. Phylogenies were estimated using both reference sequences and local controls (RO and JV). The stability of our results was assessed using bootstrapping, Bremer support (10), Goloboff weighting (22), Kishino-Hasegawa ML ratio tests (27), and T-PTP analyses (17, 18).
RT: patient, nurse, and reference strains.
Uncorrected pairwise sequence divergence ranges from 0.16% (for comparison of HXB2 with LAI in subtype B) to 11.33% within group M and to 26.37% when group O is included. The unweighted parsimony analysis excluding group O yielded 14 MP trees (tree length = 238) whose strict consensus is shown in Fig. 2a. BV for 104 branch-and-bound replicates are indicated above the branches. P is separated from N1 by three very well supported nodes (BV > 95%). All resolved nodes are stable to Goloboff weighting (22). Constraining not to keep a monophyletic [P + N2] requires a minimum of seven additional substitutions, whereas constraining a monophyletic [P + N1] requires 35 additional evolutionary events. [P + N1] was never observed in 104 bootstrap replicates. A T-PTP test for [P + N2] monophyly yielded a P value of <10−4, indicating that this clade is very unlikely the product of random data. Using the same theoretical framework, the comparison of the best trees containing a [P + N1] clade to the best trees containing a [P + N2] clade yielded very significantly better (P < 10−4) support for the latter. The unweighted MP analysis including group O yielded trees fully compatible with the consensus shown in Fig. 2a and corresponding BV are indicated below the branches. All results under this taxon sampling were very similar to those excluding group O.
Saturation plots (data not shown) indicated some transition (Ti) saturation for pairwise comparisons involving group O. Nevertheless, MP analyses with Ti excluded still yield a monophyletic [P + N2] (BV = 97%) separated by several nodes from N1. A [P + N1] clade did not occur among 104 bootstrap replicates and was very significantly less likely than [P + N2] (P < 10−4). Results remain very similar when both Ti substitutions and group O sequences are excluded.
The ML analysis with most parameters estimated from the data (see Materials and Methods) yield three trees (−ln L = 2732.98086 and 2062.54867 when sequences from group O are included and excluded, respectively) whose strict consensus (Fig. 2b) is fully compatible to that obtained by MP (Fig. 2a), aside from one poorly supported branch within subtype A. Constraining not to keep a monophyletic [P + N2] requires a significant decrease in lognormal likelihood by the Kishino-Hasegawa test (27), both under inclusion (Δln L = 28.84419, P = 0.0132) and exclusion (Δln L = 27.58001, P = 0.0140) of group O sequences. This means that any tree not containing a [P + N2] clade is significantly less likely that the ML tree shown in Fig. 2a. Constraining the grouping of N1 with P very significantly decreased the lognormal likelihood (Δln L = 62.03562 and 54.71307 without O; P < 10−4). Because of the high computational intensity of ML estimations, it was practical to perform a bootstrap analysis (1,000 replicates) only on a reduced data set (HXB2, groups O and D excluded), constraining ML parameters values to those obtained in a single ML search with the same taxon sampling. In the bootstrap consensus tree (data not shown), P is separated from N1 by three nodes, two of which are supported by 100 and 99% BV. Violation of the assumptions of stationarity and time reversibility of the substitution probability matrix, which can yield systematic errors in tree estimation (e.g., references in reference 46), is unlikely because none of the sequences included in our analyses differed in nucleotide composition from the frequency distribution of the ML model (PUZZLE 3.1 [44]; PAUP* [45]).
Because the exact placement of N1 in relation to subtype A sequences was unstable (Fig. 2), we reanalyzed, under NJ and MP, the RT data set after inclusion of extra subtype A sequences as well as subtype F, G, and H sequences. Furthermore, ibng was excluded from this analysis, as it may be a recombinant between subtype A and G viruses (reference 28 and the LANL website [http://hiv-web.lanl.gov/ALIGN_98 /ALIGN-INDEX-98.html]). The NJ tree is shown in Fig. 3a, with NJ and MP BV for 104 replicates. N1 strongly clustered with subtype G strains. T-PTP test for monophyly of [N1 + subtype G] indicated significance (P = 6 × 10−4).
Envelope gp120 (C2C4): patient, nurse, and reference strains.
It is recommended that strains in at least two distinct genomic regions be analyzed to determine HIV-1 subtypes (28). Analyses of C2C4 (Fig. 3b) yield results in total agreement with those described above for the RT region. NJ bootstrap, MP bootstrap, and T-PTP analyses strongly support (i) the grouping of P and N2 in a clade (BV = 99.7%, 99.7%, PT-PTP = 5 × 10−3) and (ii) the grouping of N1 with subtype G (BV = 94.1%, 71.4%, PT-PTP = 9 × 10−3).
Conclusion: patient, nurse, and reference strains analyses.
All RT and C2C4 analyses of patient, nurse, and reference sequences strongly support the grouping of N2 and P in a clade (i.e., a monophyletic group), to the exclusion of N1 (Fig. 2 and 3). Given that N1 is separated from P by at least three strongly supported nodes for either region, and clusters with subtype G sequences, these analyses strongly reject N1 as the source of transmission of HIV-1 to the patient. The grouping of N2 and P in a clade is consistent with the hypothesis of HIV-1 transmission from N2 to P. Alternatively, these results are also compatible with the hypothesis that P and N2 independently acquired similar geographic local variants. Hence, both for the RT and the more rapidly evolving C2C4 region, we tested the stability of the [P + N2] clade to the inclusion of local controls. Furthermore, in addition to the proviral DNA sequences obtained from N1, N2, and P lymphocyte samples used in all of the above analyses, we blindly introduced, for C2C4 analyses, viral RNA sequences obtained from the patient (JV32 and JV48) and nurse 2 (JV27 and JV44) plasma samples collected earlier (Fig. 1; Materials and Methods).
RT: patient, nurses, and 57 local controls.
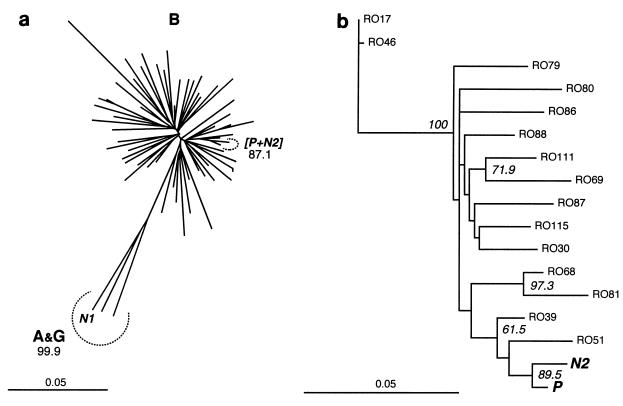
Fifty-five out of the 57 local control (RO) individuals were infected by HIV-1 subtype B. Sequences from the remaining two individuals were subtype A and formed, under NJ and MP analyses, a separate cluster with N1 (Fig. 4a). Local controls did not strictly cluster according to known risk factors. NJ bootstrap and MP T-PTP analyses yielded strong support for a [P + N2] clade excluding all local controls (BV = 87.1%, PT-PTP = 6 × 10−3).
FIG. 4.
Inference of phylogenetic relationships among RT sequences for P, N1, N2, and local controls. All local control sequences were obtained from plasma; P, N1, and N2 were obtained from lymphocytes from the patient, nurse 1, and nurse 2, respectively. Scale is in percent expected substitution per position. (a) NJ tree for the full set of 59 local control sequences (RO). Relevant BV (104 replicates) are indicated. Letters in bold indicate subtypes. (b) ML tree (−ln L = 1913.91065) for the subset including the 15 local controls closest to [P + N2]. BV > 50% (ML, 1,000 replicates) are indicated.
Given the high computational intensity of ML estimation, it was impractical to perform bootstrapping under ML with such a high number of sequences. We therefore produced two reduced data sets, (i) one including the 15 local controls with sequences most similar to P and N2 and (ii) the other including the 15 sequences defining the closest nodes to the [P + N2] clade in the NJ tree (Fig. 4a). Both ML analyses (most parameters estimated from the data; see Materials and Methods) yielded a [P + N2] clade. The tree obtained with the reduced data set (ii) is shown in Fig. 4b. The ML parameters estimated during that search were constrained in bootstrap analyses (103 replicates) which yielded (i) 87.7% and (ii) 89.5% BV support for the [P + N2] clade.
Envelope gp120 (C2C4): patient, nurses, and 41 local controls.
The high variability of the C2C4 region makes sequence alignment potentially problematic. Amino acid positions sensitive to alignment parameters (6 aligned positions in the C3 segment; 25 in the V4 loop) were excluded from phylogeny inference analyses (see Materials and Methods).
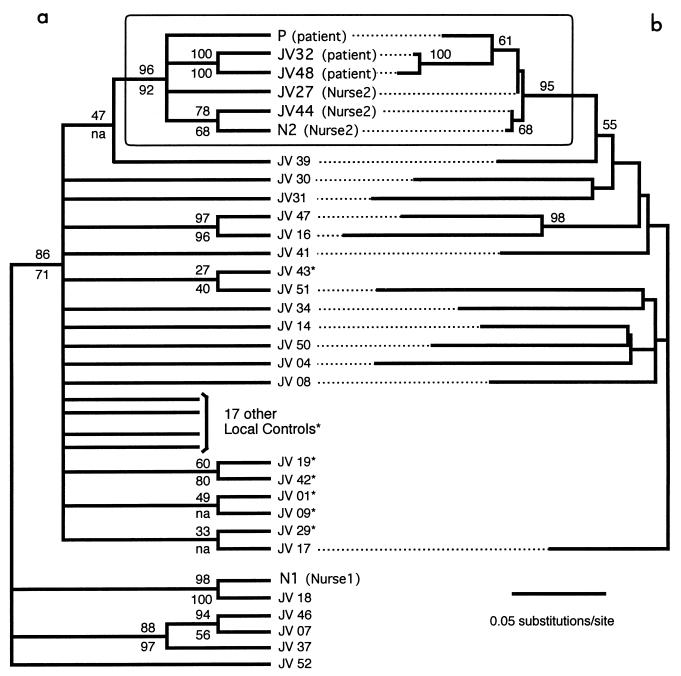
Saturation plots indicate no sign of Ti or Tv saturation within the whole range of sequence divergence. The unweighted parsimony analysis yields 382 MP trees (tree length = 1,191) whose strict consensus is shown in Fig. 5a, with BV (103 replicates) indicated above the branches. The six sequences belonging to the patient and to nurse 2 still form a monophyletic group [patient + nurse 2], despite the fact that numerous local controls were included. This result was obtained whatever methods were used to generate starting trees in heuristic searches. A T-PTP test for [patient + nurse 2] monophyly (103 replicates of 10 random additions each) yielded a P value of 10−3.
FIG. 5.
Phylogenetic relationships among C2C4 sequences from the patient, the two nurses, and multiple local controls. (a) Strict consensus among 382 MP trees (tree length = 1,191). BV obtained under unweighted MP (103 replicates) and under NJ (104 replicates) are indicated above and below the branches, respectively. na, not applicable (the group was not found in the NJ bootstrap consensus tree). (b) ML phylogram (−ln L = 2662.67099) among patient, nurse 2, and a reduced set of local control sequences (i.e., redundant sequences, indicated by asterisks, were excluded from these analyses; see text for details). BV obtained under ML are indicated on the branches. Plasma viral sequences from the patient (JV32 and JV48) and nurse 2 (JV27 and JV44) were processed blind during the analyses.
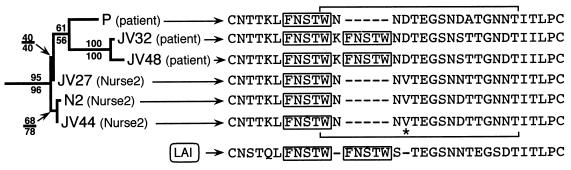
ML analysis yields 3 ML trees which are compatible with the MP consensus tree of Fig. 5a except for the branches supported by BV lower than 50%. To perform ML bootstrap analyses, we reduced the data set by excluding redundant local controls using the following algorithm (36): after excluding patient and nurse 2 sequences and the local control sequence JV39 (i.e., the sequence most closely related to [patient + nurse 2]), we computed the pairwise uncorrected distances between all remaining taxa. We then randomly excluded one of the two sequences defining the lowest distance value. The remaining taxa defined a smaller distance matrix on which a new lowest distance value was identified. The process was repeated iteratively until 12 local controls remained. The ML tree obtained under this taxon sampling (19 sequences, i.e., the 12 remaining controls plus JV39 plus the six sequences from the patient and nurse 2) is shown in Fig. 5b together with associate BV (200 replicates; ML parameters constrained from the initial ML search). The bootstrap consensus tree under unweighted parsimony yielded the same branching pattern within the [patient + nurse 2] clade as that shown in Fig. 5b. The ML branching order and relative branch lengths within the [patient + nurse 2] clade are redrawn in Fig. 6, with both ML and MP BV indicated. Although the branch supporting the paraphyly of nurse 2 sequences is short and supported by a very low bootstrap value (40% [Fig. 6]), this result is consistent with virus transmission from nurse 2 to the patient rather than from the patient to nurse 2. All samples from the patient were obtained during primary infection (Fig. 1), but viral plasma sequences (JV32 and JV48) differed from the lymphocyte proviral DNA (P) by the duplication, in the V4 loop, of a sequence coding for an FNSTW motif (Fig. 6). Among all reference and local control sequences, only the reference sequence LAI presented a similar duplication.
FIG. 6.
ML phylogram of the [patient + nurse 2] clade. ML BV are indicated above the branches. The branching pattern of the MP bootstrap consensus tree is identical to that shown here, and corresponding MP BV are indicated below the branches. The full V4 loop sequence is indicated for each sample. Duplication of an FNSTW motif (boxed) was observed only in JV32 and JV48 (patient plasma viral sequences) as well as in the unrelated reference sequence LAI. Horizontal brackets indicate amino acid positions sensitive to alignment parameters (see Materials and Methods) when all local control C2C4 sequences are included. These positions have been excluded from phylogeny inference analyses. Note that the position indicated by an asterisk adds support to the grouping of P with JV32 and JV48.
Conclusions: local control analyses.
All of our results strongly support the grouping of viral sequences from the patient and nurse 2 in a clade to the exclusion of nurse 1 and of all local controls (Fig. 4 and 5). Analyses of both RT and C2C4 corroborate the following conclusions: (i) nurse 1 is not the source of transmission of HIV-1 to the patient; (ii) it is very likely that HIV-1 was transmitted from nurse 2 to the patient or from the patient to nurse 2; (iii) the hypothesis that the transmission was from nurse 2 to the patient is strongly suggested by clinical data and consistent with the branching pattern within the [patient + nurse 2] clade.
DISCUSSION
A virological investigation was requested by the Seine Saint-Denis sanitary authorities to evaluate the possibility of a nurse-to-patient transmission of HIV-1 in the absence of documented exposure to blood. We tested the nurse-to-patient transmission hypothesis using extensive phylogenetic analyses of viral sequences.
We could unambiguously exclude nurse 1 as the source of infection because his strain, identified as an African G subtype, was separated from the patient by several strongly supported phylogenetic nodes (Fig. 2 and 3). Our analyses of two distinct genomic regions from free viruses and integrated proviruses combined a large number of geographical and temporal local controls and consistently indicate that HIV-1 sequences from the patient and from nurse 2 share a common ancestor not shared by any other sequence analyzed (Fig. 2 to 6). This result, strongly supported by a wide range of phylogenetic approaches, indicates that HIV-1 was transmitted from nurse 2 to the patient or vice versa. However, given that nurse 2 had a full Western blot profile and a low CD4+ cell count (indicative of advanced infection) at the time of the patient's seroconversion, it is very likely that transmission occurred from nurse 2 to the patient.
Nurse 2 was coinfected with HCV genotype 3a (data not shown). However, no evidence of HCV infection was found for the patient either by RT-PCR or by third-generation serological tests performed 12 months later. Transmission of HIV-1, but not HCV, from coinfected individuals has already been described (9).
HIV-1 variants with an insertion of five amino acids in the V4 loop quickly emerged in the patient plasma, possibly indicating emergence of mutants escaping immunological neutralization. Insertions in the V4 loop were previously described for a case of mother-to-child transmission (11). Selection of variants during primary HIV-1 infection is common (34, 38), and our observation indicates that early samples, preferably from lymphocytes (which usually harbor more ancestral sequences [20]), should be studied when addressing transmission hypotheses.
In the near Paris Seine Saint-Denis district (1.4 million inhabitants), 9,856 AIDS cases had been reported at the end of June 1996. Among those, only 323 (3%) had unrecognized risk factors (30). In two other studies (35, 53), risk factor was not identified in 11.9 to 14.8% of patients over 50 year old. However, Stevenson et al. (43) demonstrated elsewhere that careful reappraisal of clinical data may resolve more than half of these cases. Some of the remaining cases might be associated to yet unidentified transmission routes. Although transmission of blood-borne pathogens is a major concern during health care practice (50, 51), it was suggested (26) that HIV-infected nurses would not be associated with transmission of HIV to a patient because they usually do not perform particularly invasive medical procedures. The case discussed here, indicative of atypical nurse-to-patient HIV-1 transmission, raises the issue of the mandatory screening of nurses. Because such screening would be overwhelmingly cost-ineffective to reduce these rare transmissions (12, 39, 40), suggestions that are acceptable, from both an ethical and a financial point of view, should be proposed. For example, HIV-infected health care workers, when aware of their seropositivity, could place themselves under the guidance of an expert panel which would determine, on a case-by-case basis, whether practice restrictions are appropriate (3).
Finally, utmost care is obviously advised in the divulgence of atypical HIV transmissions in a clinical context because it could lead to unnecessary alarm regarding infected medical staff (42). The case described here may be exceptional, but it indicates nonetheless that safety guidelines established for reducing risks of blood-borne transmission of pathogens between health care workers and patients should be carefully followed.
ACKNOWLEDGMENTS
We thank H. Agut, F. Brun-Vezinet, S. Fegueux, M. Georges, P. Mardulyn, and P. Sonigo for commenting on an earlier version of the manuscript and/or for participating in valuable and informative discussions. We thank N. Consoli and P. Blanche for clinical monitoring of patients; we thank L. Bélec, F. Ferchal, and P. Soussan for providing some of the samples used in this analysis. We are particularly grateful to Dave L. Swofford for giving us the opportunity to use the successive beta versions of Paup*.
Financial support was provided by the Fondation pour la Recherche Médicale (SIDACTION 95). M.C.M. is supported by grants from the National Fund for Scientific Research Belgium, the Free University of Brussels, the Communauté Française de Belgique (ARC 98/03-223), the Defay Fund, and the Van Buuren Fund. C.P.G. is supported by a grant from the Ministère de l'Education, France. C.P.G., V.M.S. and J.-C.N. are supported by EA 2391 (ISARS). J.G. and P.D. are supported by UPRES 1622.
REFERENCES
- 1.Albert J, Wahlberg J, Leitner T, Escanilla D, Uhlen M. Analysis of a rape case by direct sequencing of the human immunodeficiency virus type 1 pol and gag genes. J Virol. 1994;68:5918–5924. doi: 10.1128/jvi.68.9.5918-5924.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert J, Wahlberg J, Uhlen M. Forensic evidence by DNA sequencing. Nature. 1993;361:595–596. doi: 10.1038/361595b0. [DOI] [PubMed] [Google Scholar]
- 3.Allerberger F, Luthe R. HIV-testing of health care workers: unethical request or moral obligation? Wien Klin Wochenschr. 1995;107:91–94. [PubMed] [Google Scholar]
- 4.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnold C, Balfe P, Clewley J P. Sequence distances between env genes of HIV-1 from individuals infected from the same source: implications for the investigation of possible transmission events. Virology. 1995;211:198–203. doi: 10.1006/viro.1995.1391. [DOI] [PubMed] [Google Scholar]
- 6.Barr S. The 1990 Florida Dental Investigation: is the case really closed? Ann Intern Med. 1996;124:250–254. doi: 10.7326/0003-4819-124-2-199601150-00009. [DOI] [PubMed] [Google Scholar]
- 7.Belec L, Mohamed A S, Muller-Trutwin M C, Gilquin J, Gutmann L, Safar M, Barre-Sinoussi F, Kazatchkine M D. Genetically related human immunodeficiency virus type 1 in three adults of a family with no identified risk factor for intrafamilial transmission. J Virol. 1998;72:5831–5839. doi: 10.1128/jvi.72.7.5831-5839.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchard A, Ferris S, Chamaret S, Guetard D, Montagnier L. Molecular evidence for nosocomial transmission of human immunodeficiency virus from a surgeon to one of his patients. J Virol. 1998;72:4537–4540. doi: 10.1128/jvi.72.5.4537-4540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brambilla A, Pristera R, Salvatori F, Poli G, Vicenzi E. Transmission of HIV-1 and HBV [corrected] by head-butting. Lancet. 1997;350:1370. doi: 10.1016/s0140-6736(05)65143-4. [DOI] [PubMed] [Google Scholar]
- 10.Bremer K. Branch support and tree stability. Cladistics. 1994;10:295–304. [Google Scholar]
- 11.Burger H, Weiser B, Flaherty K, Gulla J, Nguyen P N, Gibbs R A. Evolution of human immunodeficiency virus type 1 nucleotide sequence diversity among close contacts. Proc Natl Acad Sci USA. 1991;88:11236–11240. doi: 10.1073/pnas.88.24.11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chavey W E, Cantor S B, Clover R D, Reinarz J A, Spann S J. Cost-effectiveness analysis of screening health care workers for HIV. J Fam Pract. 1994;38:249–257. [PubMed] [Google Scholar]
- 13.Crandall K A. Intraspecific phylogenetics: support for dental transmission of human immunodeficiency virus. J Virol. 1995;69:2351–2356. doi: 10.1128/jvi.69.4.2351-2356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeBry R W, Abele L G, Weiss S H, Hill M D, Bouzas M, Lorenzo E, Graebnitz F, Resnick L. Dental HIV transmission? Nature. 1993;361:691. doi: 10.1038/361691a0. [DOI] [PubMed] [Google Scholar]
- 15.Delwart E L, Shpaer E G, McCutchan F E, Louwagie J, Grez M, Rübsamen-Waigmann H, Mullins J I. Genetic relationships determined by a heteroduplex mobility assay: analysis of HIV env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 16.Dickinson G M, Morhart R E, Klimas N G, Bandea C I, Laracuente J M, Bisno A L. Absence of HIV transmission from an infected dentist to his patients. An epidemiologic and DNA sequence analysis. JAMA. 1993;269:1802–1806. [PubMed] [Google Scholar]
- 17.Faith D. Cladistic permutation tests for monophyly and nonmonophyly. Syst Zool. 1991;40:366–375. [Google Scholar]
- 18.Faith D P, Trueman J W H. When the topology-dependent permutation test (T-PTP) for monophyly returns significant support for monophyly, should that be equated with (a) rejecting a null hypothesis of nonmonophyly, (b) rejecting a null hypothesis of “no structure”, (c) failing to falsify a hypothesis of monophyly, or (d) none of the above? Syst Biol. 1996;45:580–586. [Google Scholar]
- 19.Felsenstein J. PHYLIP—Phylogeny Inference Package. Cladistics. 1989;5:164–166. [Google Scholar]
- 20.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 21.Gatesy J, DeSalle R, Wheeler W. Alignment-ambiguous nucleotide sites and the exclusion of systematic data. Mol Phylogenet Evol. 1993;2:152–157. doi: 10.1006/mpev.1993.1015. [DOI] [PubMed] [Google Scholar]
- 22.Goloboff P A. Estimating character weights during tree search. Cladistics. 1993;9:83–91. doi: 10.1111/j.1096-0031.1993.tb00209.x. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 24.Hillis D M, Huelsenbeck J P, Swofford D L. Hobgoblin of phylogenetics? Nature. 1994;369:363–364. doi: 10.1038/369363a0. [DOI] [PubMed] [Google Scholar]
- 25.Holmes E C, Zhang L Q, Simmonds P, Rogers A S, Brown A J. Molecular investigation of human immunodeficiency virus (HIV) infection in a patient of an HIV-infected surgeon. J Infect Dis. 1993;167:1411–1414. doi: 10.1093/infdis/167.6.1411. [DOI] [PubMed] [Google Scholar]
- 26.Jackson M M, Russell B S. What if the health-care worker is the one infected with HIV or HBV? Todays OR Nurse. 1991;13:21–24. [PubMed] [Google Scholar]
- 27.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 28.Korber B, Hahn B, Foley B, Mellors J W, Leitner T, Myers G, McCutchan F, Kuiken C, Bradac J. HIV sequence compendium 1997. Theoretical Biology and Biophysics Group T10. Los Alamos, N.Mex: Los Alamos National Laboratory; 1997. [Google Scholar]
- 29.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. . (Erratum, 339:490.) [DOI] [PubMed] [Google Scholar]
- 30.Laporte A, Gouezel P, Lot F, Pillonel J, Bouraoui L, Pinget R, Cazein F. Surveillance du SIDA, situation au 30 juin 1996, Région, Ile de France. Saint-Maurice Cedex, France: Réseau National de Santé Publique; 1996. [Google Scholar]
- 31.Larder B A, Boucher C A B. PCR detection of human immunodeficiency virus drug resistance mutations. In: Persing D H, editor. Diagnostic molecular microbiology: principles and applications. Vol. 1. Washington, D.C.: American Society for Microbiology; 1993. pp. 527–533. [Google Scholar]
- 32.Larder B A, Kellam P, Kemp S D. Zidovudine resistance predicted by direct detection of mutations in DNA from HIV-infected lymphocytes. AIDS. 1991;5:137–144. doi: 10.1097/00002030-199102000-00002. [DOI] [PubMed] [Google Scholar]
- 33.Leitner T, Escanilla D, Franzen C, Uhlen M, Albert J. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proc Natl Acad Sci USA. 1996;93:10864–10869. doi: 10.1073/pnas.93.20.10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markham R B, Schwartz D H, Templeton A, Margolick J B, Farzadegan H, Vlahov D, Yu X F. Selective transmission of human immunodeficiency virus type 1 variants to SCID mice reconstituted with human peripheral blood monoclonal cells. J Virol. 1996;70:6947–6954. doi: 10.1128/jvi.70.10.6947-6954.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez Hernandez P L, Valencia Ortega M E, Pena Sanchez de Rivera J M, Soriano Vazquez V, Gonzalez-Lahoz J, Arnalich Fernandez F, Lopez Gay D, Poveda Gomez F, Camacho Siles J, Magallon Martinez M, Gonzalez Garcia J, Vazquez Rodriguez J J. HIV infection in old age: an epidemiological and clinical study in 42 patients in the Community of Madrid. Rev Clin Esp. 1997;197:684–689. . (In Spanish.) [PubMed] [Google Scholar]
- 36.Milinkovitch M C, LeDuc R G, Adachi J, Farnir F, Georges M, Hasegawa M. Effects of character weighting and species sampling on phylogeny reconstruction: a case study based on DNA sequence data in cetaceans. Genetics. 1996;144:1817–1833. doi: 10.1093/genetics/144.4.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ou C Y, Ciesielski C A, Myers G, Bandea C I, Luo C C, Korber B T, Mullins J I, Schochetman G, Berkelman R L, Economou A N, et al. Molecular epidemiology of HIV transmission in a dental practice. Science. 1992;256:1165–1171. doi: 10.1126/science.256.5060.1165. [DOI] [PubMed] [Google Scholar]
- 38.Price D A, Goulder P J, Klenerman P, Sewell A K, Easterbrook P J, Troop M, Bangham C R, Phillips R E. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci USA. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhodes R S, Telford G L, Hierholzer W, Jr, Barnes M. Bloodborne pathogen transmission from healthcare worker to patients. Legal issues and provider perspectives. Surg Clin North Am. 1995;75:1205–1217. doi: 10.1016/s0039-6109(16)46792-7. [DOI] [PubMed] [Google Scholar]
- 40.Robert L M, Chamberland M E, Cleveland J L, Marcus R, Gooch B F, Srivastava P U, Culver D H, Jaffe H W, Marianos D W, Panlilio A L, et al. Investigations of patients of health care workers infected with HIV. The Centers for Disease Control and Prevention database. Ann Intern Med. 1995;122:653–657. doi: 10.7326/0003-4819-122-9-199505010-00002. [DOI] [PubMed] [Google Scholar]
- 41.Saag M S, Hahn B H, Gibbons J, Li Y, Parks E S, Parks W P, Shaw G M. Extensive variation of human immunodeficiency virus type-1 in vivo. Nature. 1988;334:440–444. doi: 10.1038/334440a0. [DOI] [PubMed] [Google Scholar]
- 42.Salluzzo R F, Bartfield J M, Freed H, Graber M, Peters T. Attitude of emergency department patients toward HIV-infected health care workers. Am J Emerg Med. 1997;15:141–144. doi: 10.1016/s0735-6757(97)90085-1. [DOI] [PubMed] [Google Scholar]
- 43.Stevenson E M, Thompson S C, Crofts N. Ascertaining exposure categories of HIV-infected individuals with previously unrecorded risk data. Med J Aust. 1995;163:66–69. doi: 10.5694/j.1326-5377.1995.tb126116.x. [DOI] [PubMed] [Google Scholar]
- 44.Strimmer K, von Haeseler A. Quartet puzzling: a quartet maximum likelihood method for reconstructing tree topologies. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 45.Swofford D. PAUP*: Phylogenetic Analysis Using Parsimony (and other methods), version 4.0d64 (in progress). Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 46.Swofford D, Olsen G, Waddell P, Hillis D. Phylogenetic inference. In: Hillis D, Moritz C, Mable B, editors. Molecular systematics. 2nd ed. Sunderland, Mass: Sinauer Associates; 1996. pp. 407–514. [Google Scholar]
- 47.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogel G. Phylogenetic analysis: getting its day in court. Science. 1997;275:1559–1560. doi: 10.1126/science.275.5306.1559. [DOI] [PubMed] [Google Scholar]
- 49.Wain-Hobson S. HIV genome variability in vivo. AIDS. 1989;3(Suppl. 1):S13–S18. doi: 10.1097/00002030-198901001-00003. [DOI] [PubMed] [Google Scholar]
- 50.Weiss S H. Risks and issues for the health care worker in the human immunodeficiency virus era. Med Clin North Am. 1997;81:555–575. doi: 10.1016/s0025-7125(05)70531-9. [DOI] [PubMed] [Google Scholar]
- 51.Wicher C P. AIDS & HIV: the dilemma of the health care worker. J Neurosci Nurs. 1993;25:118–124. doi: 10.1097/01376517-199304000-00009. [DOI] [PubMed] [Google Scholar]
- 52.Wolfs T F, Zwart G, Bakker M, Goudsmit J. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology. 1992;189:103–110. doi: 10.1016/0042-6822(92)90685-i. [DOI] [PubMed] [Google Scholar]
- 53.Wutoh A K, Hidalgo J, Rhee W, Bareta J. A characterization of older AIDS patients in Maryland. J Natl Med Assoc. 1998;90:369–373. [PMC free article] [PubMed] [Google Scholar]