Abstract
Moloney murine leukemia virus-based retroviral vector expression is gradually lost during prolonged in vitro culture of CEMSS T cells. However, when the human beta interferon scaffold attachment region (IFN-SAR) was inserted into the vector immediately upstream of the 3′ long terminal repeat (LTR), expression was maintained for the length of the study (4 months). Clonal analysis of the retrovirus vector-infected CEMSS cells showed that SAR-containing retroviral vector expression levels were positively correlated with the proviral copy numbers (P < 0.0001), while there was no correlation between the proviral copy numbers and expression levels in control vector-infected clones. Thirty-three percent of the CEMSS cell clones infected with the control vector showed evidence of partial or complete methylation in the 5′ LTR region. In sharp contrast, we detected no methylation in the clones infected with the SAR-containing vector. To demonstrate a direct inhibitory effect of methylation on retroviral vector expression, we have transfected 293 cells with in vitro-methylated proviral DNA. In transiently transfected cells, expression of methylated LTR was reduced but not completely inhibited, irrespective of the presence of the IFN-SAR sequence. In stably transfected cells, however, methylation completely abolished expression of the control vector but not of the SAR-containing vector. Furthermore, the expression of the SAR-containing vector was stable over time, indicating the ability of the SAR sequence to alleviate methylation-mediated transcriptional repression of a vector. This study extends our understanding of the mechanisms of retroviral vector inactivation by methylation and provides insight into a functional role for the SAR elements.
Scaffold attachment regions (SARs) or matrix attachment regions are DNA sequences that bind to the isolated nuclear scaffold or matrix with high affinity (9, 14). SAR/matrix attachment region sequences are AT rich (70%) and enriched in DNA topoisomerase II binding sites (9), but no consensus sequence has yet been identified (6). Functional studies have shown that some of the SAR sequences can confer gene expression in a copy number-dependent, position-independent manner when incorporated into a transgene (27, 33). However, other studies have shown the contrary, indicating heterogeneity with respect to function among the SAR elements (41). How SAR elements influence gene expression is not well understood. One hypothesis is that SARs define boundaries of independent chromatin domains and establish local access of transcription factors to the enhancer-promoter sequences within the domain (5, 22). SAR sequences also influence transgene methylation status. Lichtenstein et al. (26) and Kirillov et al. (25) have identified SAR sequences that contribute to B-cell-specific demethylation in the immunoglobulin κ locus.
Retroviral vectors are reliable and efficient vehicles for gene delivery into several cell types, including cells of the hematopoietic system (28). However, one of the major limitations for the use of Moloney murine leukemia virus (MoMuLV)-based vectors in gene therapy is their frequent inability to provide long-term, high-level transgene expression. It has been demonstrated that in murine fibroblasts and in embryonal carcinoma cells the inactivation of the enhancer repeat unit of the viral long terminal repeat (LTR) can be mediated by interactions with negatively acting cellular factors (2, 15, 40, 42). In addition, de novo methylation has also been linked to the silencing of the viral LTR in numerous cell types, including murine embryonic stem cells, fibroblasts, and hematopoietic stem cells (7, 17, 19, 30, 31). In an attempt to improve expression, retroviral vectors derived from the myeloproliferative sarcoma virus, murine stem cell virus, and spleen focus-forming virus have recently been developed (4, 18, 32, 36, 37). In general, these vectors differ from the MoMuLV vectors in the LTR promoter-enhancer region and the primer-binding site. These vectors have been shown to be expressed better than the MoMuLV vectors in mouse hematopoietic cells in vivo (4, 18, 32, 36, 37). In addition, one type of the vector (MND [8]) contains modifications that to some extent reduce de novo methylation of viral LTR (37, 43).
An alternative strategy for improving expression is to incorporate chromatin-controlling DNA elements such as SARs into a vector (1). We have previously reported that the human beta interferon SAR (IFN-SAR) enhances retroviral vector expression in human peripheral blood T cells and in T cells and monocytes generated from retrovirally infected CD34+ hematopoietic progenitor stem cells in vitro and in vivo (1, 3). In the present study, we addressed the mode of action of the IFN-SAR element. Our results indicate that the IFN-SAR confers copy number-dependent expression on the retroviral vector and inhibits de novo methylation of the retroviral 5′-LTR. In addition, when cells were transfected with in vitro-methylated proviral DNA, the SAR element was able to alleviate methylation-mediated transcriptional repression. Thus, the current study extends our understanding of the mechanisms of retroviral vector inactivation by methylation, provides insight into a functional role for the SAR elements, and demonstrates a potent beneficial effect of the IFN-SAR for improving retroviral vector expression.
MATERIALS AND METHODS
Retroviral vector-producing cell lines and infection of CEMSS cells.
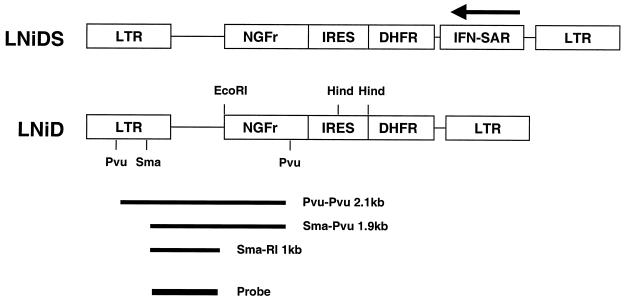
The vectors LNiD and LNiDS (Fig. 1) were described previously (3). Retroviral vector supernatants were prepared using amphotropic ProPak-A packaging cells as described previously (13, 35). All producer cells tested negative for replication-competent retrovirus by S+L− assay on PG4 cells (16). Infection of CEMSS cells was performed by centrifugation at 2,000 × g of 5 × 105 cells with 1 ml of retroviral supernatant supplemented with 8 μg of Polybrene per ml for 3 h at 34°C. At different time intervals after infection, CEMSS cells were stained with immunofluorescent anti-nerve growth factor receptor (NGFr)–fluorescein isothiocyanate antibody and analyzed for expression of NGFr surface marker using a FACScan device (Becton Dickinson). At 3 months after infection, individual CEMSS cell clones were obtained by limited dilution. The clones were screened for the presence of the retroviral transgene by PCR using LTR-specific primers, AGACCCCACCTGTAGGTTTG (upstream primer) and TTGAGCTCGGGGAGCAGAAG (downstream primer).
FIG. 1.
Schematic presentation of the retroviral constructs LNiD and LNiDS. NGFr, truncated human NGFr gene; IRES, internal ribosomal entry site; DHFR, murine dihydrofolate reductase (DHFR) L22Y gene. Restriction sites used for the Southern blot analysis are indicated. Hind, HindIII; Pvu, PvuII; Sma, SmaI. A radioactively labeled SpeI-SphI fragment common to both vectors (“Probe”) was used for all Southern blot hybridizations.
Analysis of proviral DNA methylation status.
Methylation analysis of individual clones was performed as described in reference 8. Twenty micrograms of genomic DNA was digested with restriction enzyme PvuII. One-half of the PvuII-digested sample was then digested with the methylation-sensitive restriction enzyme SmaI. To monitor completeness of the digestion, 1/10 of the SmaI digestion mixture was mixed with 2 μg of control plasmid DNA containing unmethylated SmaI cleavage sites. This mixture was incubated in parallel with main samples at 25°C overnight. Completeness of the SmaI digestions was then determined by resolving bands on an agarose gel. The main PvuII and PvuII-SmaI digests were resolved on a 1% agarose gel and analyzed by Southern blotting using a radioactively labeled SpeI-SphI fragment from the vector LNiD as a probe (Fig. 1). The same method was applied to analyze the DNA methylation status of individual proviral copies, except that, in the first digestion, the EcoRI restriction enzyme was used instead of PvuII.
In vitro methylation of proviral DNA and transfection of 293 cells.
The LNiD and LNiDS plasmid DNAs were methylated in vitro using SssI methylase according to the manufacturer's protocol (Biolabs). Methylated DNA was fully resistant to SmaI digestion. For transient transfections, 10 μg of proviral DNA and 2 μg of green fluorescent protein (GFP)-expressing pGreenLantern-1 plasmid (Gibco BRL) were cotransfected into 293 cells using the CalPhos kit (Clontech). Forty-eight hours posttransfection, cells were harvested, stained with immunofluorescent anti-NGFr–phycoerythrin antibody, and analyzed for GFP and NGFr expression on a FACScan (Becton Dickinson). For stable transfections, 10 μg of proviral DNA and 1 μg of Neo-expressing pCI-neo plasmid (Promega) were used. Transfected cells were selected in 0.8 mg of G418 per ml for 4 weeks and then maintained in a medium without selection for the length of the experiment.
RESULTS
IFN-SAR improves long-term retroviral vector expression in a cultured human T-cell line.
Our initial observation of the ability of the IFN-SAR sequence to enhance retroviral vector expression was in human primary T cells (1). It is not feasible, however, to culture primary T cells over a prolonged period of time, and therefore, to test the long-term effects of the IFN-SAR on retroviral transgene expression we have used an immortalized human T-cell line (CEMSS cells). Retroviral vectors used in this study, LNiD and LNiDS (Fig. 1), were described previously (3). The vectors are MoMuLV based and encode the NGFr surface marker gene and the murine dihydrofolate reductase resistance marker gene. The IFN-SAR element in the LNiDS vector is located in reverse orientation just upstream of the 3′ LTR (Fig. 1). This location was chosen because it gives optimal expression in resting primary T cells (34). Amphotropic producer cell lines were generated using ProPak-A packaging cells (35). Infection efficiencies of the retroviral stocks used in this study were determined by measuring the percent NGFr-positive NIH 3T3 cells 2 days postinoculation with 1:3-diluted viral supernatants (13): LNiD, 71%; and LNiDS, 78%.
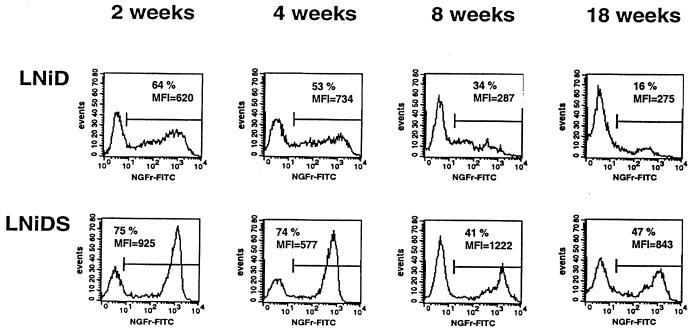
CEMSS cells were infected with the SAR-containing vector LNiDS and the control vector LNiD and maintained in culture for 18 weeks. At no time during the experiment were cells exposed to any selective pressure. Two weeks after inoculation, 75% of LNiDS-infected and 64% of LNiD-infected CEMSS cells expressed the NGFr surface marker (Fig. 2). Mean cell fluorescence intensity (MFI, in arbitrary units) of NGFr staining was used as a measure for the level of transgene expression. At 2 weeks after infection, MFI was 925 U for the LNiDS and 620 U for the LNiD vector (Fig. 2). Over the 18-week observation period, the percentage of NGFr-positive cells decreased for both vectors, but the decrease was much more pronounced in the LNiD- than in the LNiDS-infected cells. In the LNiD-infected CEMSS cells, expression decreased from 64% at 2 weeks to just 16% at 18 weeks (Fig. 2). During this same period, the NGFr MFI also decreased from 620 U at 2 weeks to 275 U at 18 weeks (Fig. 2). The cells infected with the SAR-containing vector LNiDS had a smaller loss of expression during this 18-week period. The expression dropped from 75% at 2 weeks to 47% at 18 weeks. In contrast to the LNiD vector, the NGFr MFI remained stable over the 18-week period in the LNiDS-infected cells (MFI was 925 U at 2 weeks and 843 U at 18 weeks [Fig. 2]).
FIG. 2.
Long-term effects of the SAR element on retroviral vector expression in infected CEMSS cells. CEMSS cells were infected with the SAR-containing LNiDS and the control LNiD retroviral vectors. The expression of the NGFr transgene was measured in pooled infected cells by FACS at different time points as indicated.
In order to determine the level of gene marking, 96 single-cell clones were prepared by limiting dilution at the 12-week point and screened by PCR for the presence of proviral DNA using LTR-specific primers. This analysis showed that 73% of the LNiD-infected and 65% of the LNiDS-infected CEMSS cells carried a vector. Thus, among vector-positive cells 22% of the LNiD-infected and 72% of LNiDS-infected CEMSS cells expressed NGFr at 18 weeks after infection. These data demonstrate the ability of the IFN-SAR sequence to improve long-term retroviral vector expression.
Proviral copy numbers are positively correlated with NGFr expression levels in LNiDS-infected CEMSS cells.
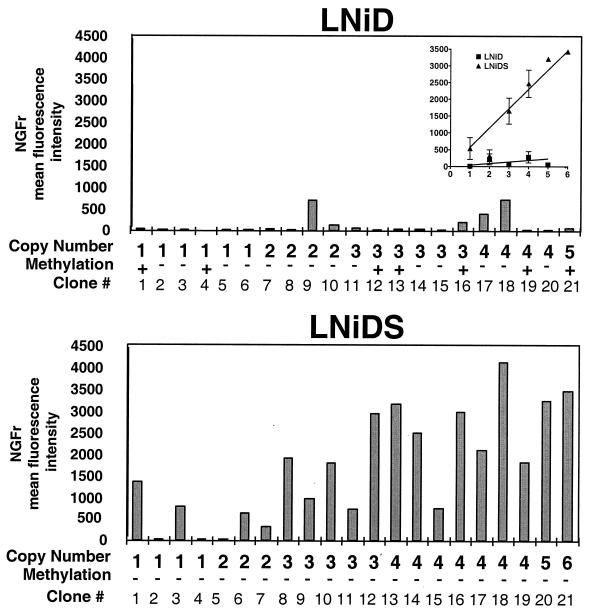
Out of 96 clones that were prepared at the 12-week point, 21 randomly selected provirus-positive clones were analyzed in detail. First, we analyzed provirus integration pattern. Total genomic DNA was extracted from individual clones and digested with the restriction enzymes HindIII and EcoRI, which cleave the central region of the provirus (Fig. 1). Southern blot analysis of the digestion products revealed that each clone had a unique banding pattern, indicating that all the clones represent independent infection events (data not shown). Proviral copy numbers in individual clones varied from one to six copies/cell (Fig. 3), and the average proviral copy number in the 21 clones was 2.5 for LNiD and 2.8 for LNiDS. Thus, the two vectors infected CEMSS cells with similar efficiencies.
FIG. 3.
Expression analysis of the infected CEMSS cell clones. Expression levels in the 21 randomly selected CEMSS clones were determined by FACS analysis and are presented as NGFr MFI. The inset shows linear regression analysis (95% confidence interval) which demonstrates a positive correlation between proviral copy number (x axis) and MFI (y axis) for the LNiDS vector (r2 = 0.59; P < 0.0001) while there is no such correlation for the LNiD vector (r2 = 0.07; P = 0.25). Proviral copy number and the methylation status (+ or −) of the individual clones are also shown. Of 21 LNiD-infected clones, 7 had complete or partial methylation in the LTR region, while there was no methylation in clones infected with the SAR-containing LNiDS vector.
Next, we measured NGFr expression by fluorescence-activated cell sorting (FACS). For each individual clone, 100% of the cells showed uniform staining with the NGFr-specific antibody, indicating that we indeed selected true monoclonal populations. The NGFr MFI, however, varied from clone to clone, indicating differences in expression levels between the clones (Fig. 3). Only a small proportion (5 of 21, or 24%) of LNiD-infected clones had detectable NGFr expression. The NGFr MFI ranged from 62 to 706, with an average of 118 U. In sharp contrast, the majority (18 of 21, or 86%) of the LNiDS-infected clones expressed high NGFr levels. The NGFr MFI in these clones ranged from 321 to 3,428, with an average of 1,680 U. Thus, when we compare the average expression levels, the SAR-containing vector LNiDS gives a 14-fold-higher value than does the control vector LNiD. In addition, the expression levels in LNiDS-infected clones were positively correlated with transgene copy numbers (P < 0.001) while there was no such correlation in the clones infected with the control vector LNiD (P = 0.25) (Fig. 3, inset). These results indicate that the IFN-SAR can reduce the integration site-related effects on retroviral vector expression.
De novo methylation of retroviral LTR is inhibited by the IFN-SAR sequence.
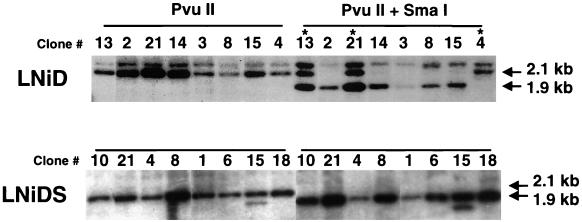
The absence of expression in cells infected with MoMuLV-based vectors is thought to be the result of inactivation of the MoMuLV LTR (2). Studies with embryonal carcinoma cells (40) and in mouse bone marrow transplant experiments (7) have linked methylation of the MoMuLV LTR to its inactivation. We investigated the possibility that de novo methylation may be the cause for the lack of expression in the NGFr-negative CEMSS clones. Total genomic DNA was extracted from the two sets of 21 clones described above. DNA was first digested with the restriction enzyme PvuII and then with the methylation-sensitive enzyme SmaI (Fig. 1), and the digestion products were analyzed by Southern blotting. Digestion with PvuII alone produces a 2.1-kb fragment, while digestion with PvuII and SmaI should produce a 1.9-kb fragment. The presence of the 2.1-kb fragment in the PvuII-SmaI digest is indicative of the methylation at the SmaI site. Representative Southern blots are shown in Fig. 4, and the entire data set is summarized in Fig. 3. Seven out of 21 (33%) LNiD-infected clones showed complete (e.g., clone 4 [Fig. 4]) or partial (e.g., clones 13 and 21 [Fig. 4]) methylation at the SmaI site. All of the methylated clones had no detectable expression, but there were also clones that had no expression and no evidence of methylation at the SmaI site (Fig. 3). In sharp contrast, we found no evidence of methylation in the LNiDS-infected clones. Thus, the IFN-SAR element can inhibit de novo methylation of the retroviral LTR in CEMSS cells.
FIG. 4.
De novo methylation of retroviral LTR is observed in LNiD-infected clones but not in LNiDS-infected clones. Chromosomal DNA was digested with PvuII or PvuII-SmaI restriction enzymes (Fig. 1) and analyzed by Southern blotting. Results with eight LNiD- and LNiDS-infected clones are shown. Clone numbers are the same as in Fig. 3. Methylated proviral DNA is resistant to SmaI digestion and is detected as the 2.1-kb-length PvuII product in the PvuII-SmaI digestion. Asterisks indicate methylated clones.
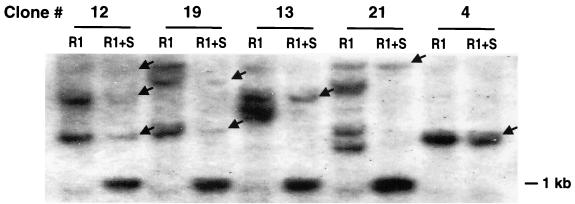
Some of the LNiD clones with multiple proviral copies were only partially methylated (e.g., clones 13 and 21 [Fig. 4]). We therefore investigated the methylation status of individual integrants in five methylated clones (clones 4, 12, 13, 19, and 21). The number of proviral copies in these clones ranged from one to five. Total genomic DNA was digested first with the restriction enzyme EcoRI, which cleaves the central region of the provirus, and then with the methylation-sensitive enzyme SmaI (Fig. 1). The EcoRI digest produces a banding pattern, where each band represents individual proviral integration. Upon SmaI digestion, all of the bands are reduced to a 1-kb fragment (Fig. 1). If the provirus is methylated, the band produced by EcoRI digest will remain intact after SmaI digestion. As shown in Fig. 5, different proviral integrants within a single clone are methylated to different extents. The only exception is clone 4, which carries one fully methylated proviral copy. This observation indicates that de novo methylation of CpG islands in the retroviral LTR occurs randomly.
FIG. 5.
De novo methylation of individual LNiD proviral copies. DNA samples from five partially or completely methylated clones infected with the LNiD vector were digested with EcoRI or EcoRI-SmaI restriction enzymes (Fig. 1) and analyzed by Southern blotting. Methylated proviral DNA is resistant to SmaI digestion and is detected as the full-length EcoRI product after SmaI digestion (indicated by arrows). Clone numbers are the same as in Fig. 3, R1, EcoRI; S, SmaI.
Methylation inhibits retroviral LTR expression in transiently transfected 293 cells irrespective of the presence of the IFN-SAR sequence.
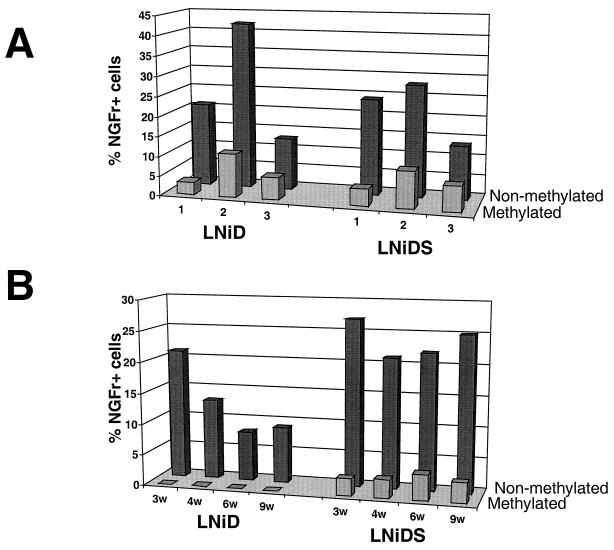
To demonstrate a direct inhibitory effect of methylation on retroviral vector expression, we have methylated proviral DNAs in vitro using SssI methylase. Ten micrograms of methylated or nonmethylated DNA, together with 2 μg of GFP-expressing pGreenLantern-1 plasmid, was then cotransfected into 293 cells. GFP expression was used to normalize transfection efficiency between samples. NGFr expression in a GFP-positive fraction of transfected cells was measured 48 h posttransfection by FACS analysis. The results from three independent experiments are shown in Fig. 6A. Transfection with nonmethylated plasmids yielded 13 to 42% NGFr-expressing cells, and there was no significant difference between the LNiD and LNiDS vectors. Transfection with methylated plasmids yielded two- to sevenfold-less expression (3 to 11% NGFr-expressing cells), and again there was no significant difference between the LNiD and the LNiDS vectors. Thus, methylation directly inhibits retroviral vector expression, irrespective of the presence of the IFN-SAR sequence.
FIG. 6.
Effect of methylation on retroviral vector expression in transiently and stably transfected 293 cells. (A) LNiD and LNiDS plasmid DNA was methylated in vitro using SssI methylase. Ten micrograms of methylated or nonmethylated plasmid was transfected into 293 cells together with 2 μg of GFP-expressing pGreenLantern-1 plasmid. Percent NGFr-expressing cells in a GFP-positive fraction of transfected cells measured 48 h posttransfection is shown. Numerals 1, 2, and 3 indicate three independent experiments. (B) Methylated or nonmethylated LNiD and LNiDS plasmid DNAs (10 μg) were cotransfected into 293 cells together with 2 μg of a Neo selectable marker-expressing pCI-neo plasmid. Transfected cells were selected in 0.8 mg of G418 per ml for 3 weeks. Percent NGFr-expressing cells in pools of G418-resistant cells was determined at 3, 4, 6, and 9 weeks posttransfection by FACS analysis.
IFN-SAR can partially relieve methylation-mediated repression of retroviral vector expression in stably transfected cells.
Next we analyzed the effect of methylation on LNiD and LNiDS vector expression in stably transfected 293 cells. Ten micrograms of methylated or nonmethylated LNiD and LNiDS plasmids together with 1 μg of a Neo selectable marker-expressing pCI-neo plasmid was cotransfected into 293 cells. Transfected cells were selected in 0.8 mg of G418 per ml for 4 weeks. NGFr expression in pools of G418-resistant clones was measured at 3, 4, 6, and 9 weeks posttransfection by FACS analysis. Cells transfected with nonmethylated LNiD and LNiDS DNA expressed comparable levels (21 and 26%, respectively) of NGFr surface marker at 3 weeks posttransfection (Fig. 6B). The expression of the LNiD vector decreased over time, from 21% at 3 weeks to 8 to 9% at 6 to 9 weeks. In contrast, the LNiDS vector expression remained quite stable; it was 26% at 3 weeks and 22 to 25% at 6 to 9 weeks.
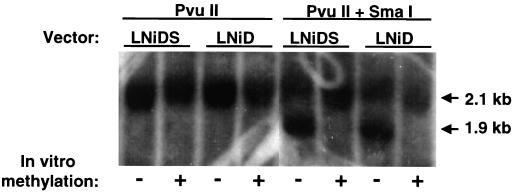
Most notably, methylation completely abolished expression of the LNiD vector, such that at no time during the experiment did we detect any NGFr-expressing cells (Fig. 6B). In sharp contrast, we did detect NGFr-positive cells (3 to 4%) in cultures transfected with the methylated LNiDS vector. Although the level of expression was six- to eightfold lower than that of nonmethylated DNA, it remained stable over the length of the experiment (9 weeks [Fig. 6B]). Southern blot analysis showed that, at 5 weeks posttransfection, integrated LNiD and LNiDS proviral DNAs were still fully methylated (Fig. 7). These data demonstrate a strong inhibitory effect of methylation on retroviral expression in stably transfected cells and also indicate the ability of the IFN-SAR element to alleviate to some extent methylation-mediated transcriptional repression of a retroviral vector.
FIG. 7.
Methylation status of proviral LTR in stably transfected 293 cells. LNiD and LNiDS plasmid DNA was methylated in vitro using SssI methylase (indicated by “+”). Methylated and nonmethylated DNAs were then transfected into 293 cells together with 2 μg of a Neo selectable marker-expressing pCI-neo plasmid. Transfected cells were selected in 0.8 mg of G418 per ml for 3 weeks. Genomic DNA was isolated 5 weeks after transfection, digested with PvuII or PvuII-SmaI restriction enzymes (Fig. 1), and analyzed by Southern blotting. Proviral DNAs which were methylated in vitro prior to transfection are fully resistant to SmaI digestion, as indicated by the absence of the 1.9-kb fragment in the PvuII-SmaI digestion.
DISCUSSION
The principal findings in this report are (i) the SAR has the ability to inhibit de novo methylation of retroviral LTR, (ii) it can alleviate methylation-mediated repression of retroviral vector transcription, and (iii) it confers long-term, copy number-dependent expression on the retroviral vector. Expression of MoMuLV-based retroviral vectors is unpredictable and varies with the chromosomal integration site (11, 12). Indeed, out of 21 CEMSS clones infected with the LNiD vector, only 5 showed expression at 3 months after infection (Fig. 3). Furthermore, there was no correlation between the expression levels and the proviral copy numbers, and even five proviral copies were not sufficient to ensure LNiD vector expression (e.g., LNiD clone 21 [Fig. 3]). In sharp contrast, there was a significant (P < 0.0001) positive correlation between the expression levels and proviral copy numbers in CEMSS cell clones infected with the SAR-containing LNiDS vector (Fig. 3, inset). With the exception of a few clones that had no expression, all of the LNiDS clones showed high NGFr expression levels that increased with the increase in the proviral copy number per cell (Fig. 3). Thus, the IFN-SAR can overcome influences of neighboring chromatin and confer position-independent, copy number-dependent expression on a retroviral vector in CEMSS cells. We have previously analyzed expression and marking in T cells derived from retrovirus-infected hematopoietic stem cells in the SCIDhu Thy/Liv model and did not observe position-independent expression with a SAR vector (1). Thompson et al. (41) have also reported that SARs stimulate transgene expression in preimplantation embryos but not in differentiated tissues. It appears, therefore, that the IFN-SAR element can protect the retroviral vector from influences of neighboring chromatin in defined cell types but cannot induce formation of a transcriptionally active chromatin, a feature that would be required to protect the vector from major changes in chromatin which occur during cell differentiation.
Methylation plays an important role in controlling gene expression in plants and animals (reviewed in reference 24), and it has been implicated as one of the mechanisms responsible for inactivation of retroviral genomes (7, 17, 19, 30). We have also observed correlation between the lack of retroviral vector expression and the presence of methylation. In LNiD-infected CEMSS cells, methylation was observed only in expression-negative clones, and in stably transfected 293 cells, methylation completely abolished LNiD vector expression. However, in CEMSS cells the level of methylation was not very high. Only 7 out of 16 (or 44%) expression-negative LNiD CEMSS clones showed evidence of de novo methylation at the SmaI site in the LTR. Furthermore, in clones with multiple proviral integrations only some integrants were methylated, and the level of methylation of most of these integrants was less than 100% (Fig. 5). Nevertheless, there was no expression. Also in 293 cells stably transfected with LNiD DNA expression decreased over time (Fig. 6), but we did not detect de novo methylation (Fig. 7). One explanation for these findings is that a very low level of methylation is sufficient to inactivate MoMuLV LTR. In the case of the Rous sarcoma virus LTR, as little as 7% methylation can significantly inhibit expression (20). Alternatively, LTR expression could have been suppressed by some other mechanism and de novo methylation could have occurred subsequently. There are a total of 19 CpG sites in the LTR sequence. Time course analysis correlating vector expression and the level of de novo methylation on all 19 CpG sites (43) will be required to answer whether methylation is the only cause for LTR inactivation in CEMSS cells.
Most notably, we detected no methylation and high-level expression in CEMSS clones infected with the SAR-containing LNiDS vector. If we assume that de novo methylation of different proviral integrants is a random process, we would predict that the extent of methylation in LNiDS-infected clones must have been less than 0.25% (this is the maximum value for which there is 95% probability of observing no methylation at the SmaI site in 21 clones). Thus, the IFN-SAR was able to significantly delay or completely inhibit de novo methylation of proviral DNA in CEMSS cells. It remains to be determined whether the IFN-SAR can directly inhibit methylation machinery or whether it indirectly prevents methylation by keeping the LTR transcriptionally active.
In 293 cells, the IFN-SAR was able to relieve some of the methylation-mediated repression of transcription. In stably transfected 293 cells, methylated LNiDS proviral DNA was expressed (although at six- to eightfold-lower levels than nonmethylated DNA), while the control LNiD vector was completely silenced by methylation. Two mechanisms through which methylation could inhibit expression have been described in the literature (reviewed in reference 39). First, methylation could directly reduce LTR promoter activity by inhibiting transcription factor binding (10, 21) or by recruiting inhibitory factors that bind methylated DNA and prevent transcription (38). Second, methylated proviral DNA can be bound by proteins such as MeCP2, which recruit histone deacetylase complex, leading to modification of chromatin structure to a transcriptionally inactive state (23, 29). Our data indicate that both mechanisms may be involved in silencing expression of methylated proviral DNA in 293 cells. Methylation reduced proviral expression in transiently transfected 293 cells two- to sevenfold, presumably by preventing binding of transcription factors to the LTR enhancer-promoter region. Once proviral DNA was integrated, methylation induced formation of transcriptionally inactive chromatin, which completely silenced LTR expression (Fig. 6). It is tempting to speculate that the IFN-SAR was able to prevent formation of transcriptionally inactive chromatin and in this way to rescue vector expression in 293 cells. But because the LTR region was still methylated (Fig. 7), and the methylation does directly affect transcription factor binding, the overall expression levels were lower than those for nonmethylated DNA.
In summary, it appears that the ability of the IFN-SAR to prevent de novo methylation and also to alleviate methylation-mediated transcriptional repression of a vector is one reason for the long-term stability of the SAR vector expression in cell lines. An inverse correlation between CpG methylation in the LTR and vector expression has been well documented in a mouse bone marrow transplantation model (7, 37). Additional studies using this model will determine if the IFN-SAR element also can inhibit methylation in primary hematopoietic cells.
ACKNOWLEDGMENTS
We thank Gabor Veres for critical reading of the manuscript. CEMSS cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
REFERENCES
- 1.Agarwal M, Austin T W, Morel F, Chen J, Böhnlein E, Plavec I. Scaffold attachment region-mediated enhancement of retroviral vector expression in primary T cells. J Virol. 1998;72:3720–3728. doi: 10.1128/jvi.72.5.3720-3728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akgun E, Ziegler M, Grez M. Determinants of retrovirus gene expression in embryonal carcinoma cells. J Virol. 1991;65:382–388. doi: 10.1128/jvi.65.1.382-388.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auten J, Agarwal M, Chen J, Sutton R, Plavec I. Effect of scaffold attachment region on transgene expression in retrovirus vector transduced primary T cells and macrophages. Hum Gene Ther. 1999;10:1389–1399. doi: 10.1089/10430349950018058. [DOI] [PubMed] [Google Scholar]
- 4.Baum C, Hegewisch-Becker S, Eckert H G, Stocking C, Ostertag W. Novel retroviral vectors for efficient expression of the multidrug resistance (mdr-1) gene in early hematopoietic cells. J Virol. 1995;69:7541–7547. doi: 10.1128/jvi.69.12.7541-7547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bode J, Schlake T, Ríos-Ramírez M, Mielke C, Stengert M, Kay V, Klehr-Wirth D. Scaffold/matrix-attachment regions (S/MAR): structural properties creating transcriptionally active loci. Orlando, Fla: Academic Press, Inc.; 1995. [DOI] [PubMed] [Google Scholar]
- 6.Boulikas T. Nature of DNA sequences at the attachment regions of genes to the nuclear matrix. J Cell Biochem. 1993;52:14–22. doi: 10.1002/jcb.240520104. [DOI] [PubMed] [Google Scholar]
- 7.Challita P M, Kohn D B. Lack of expression from a retroviral vector after transduction of murine hematopoietic stem cells is associated with methylation in vivo. Proc Natl Acad Sci USA. 1994;91:2567–2571. doi: 10.1073/pnas.91.7.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Challita P M, Skelton D, El-Khoueiry A, Yu X J, Weinberg K, Kohn D B. Multiple modifications in cis elements of the long terminal repeat of retroviral vectors lead to increased expression and decreased DNA methylation in embryonic carcinoma cells. J Virol. 1995;69:748–755. doi: 10.1128/jvi.69.2.748-755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cockerill P N, Garrard W T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 10.Comb M, Goodman H M. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–3982. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duch M, Paludan K, Jørgensen P, Pedersen F S. Lack of correlation between basal expression levels and susceptibility to transcriptional shutdown among single-gene murine leukemia virus vector proviruses. J Virol. 1994;68:5596–5601. doi: 10.1128/jvi.68.9.5596-5601.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duch M, Paludan K, Lovmand J, Pedersen L, Jøorgensen P, Pedersen F S. A correlation between dexamethasone inducibility and basal expression levels of retroviral vector proviruses. Nucleic Acids Res. 1993;21:4777–4782. doi: 10.1093/nar/21.20.4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forestell S P, Böhnlein E, Rigg R J. Retroviral end-point titer is not predictive of gene transfer efficiency: implications for vector production. Gene Ther. 1995;2:723–730. [PubMed] [Google Scholar]
- 14.Gasser S M, Laemmli U K. Cohabitation of scaffold binding regions with upstream enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986;46:521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- 15.Gorman C M, Rigby P W. Negative regulation of viral enhancers in undifferentiated embryonic stem cells. Cell. 1985;42:519–526. doi: 10.1016/0092-8674(85)90109-6. [DOI] [PubMed] [Google Scholar]
- 16.Haapala D K, Robay S D, Oroszlan S D, Tsai W P. Isolation from cats of an endogenous type C virus with a novel glycoprotein. J Virol. 1985;53:827–833. doi: 10.1128/jvi.53.3.827-833.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harbers K, Schnieke A, Stuhlman H, Jahner D, Jaenisch R. DNA methylation and gene expression: endogenous retroviral genome becomes infectious after molecular cloning. Proc Natl Acad Sci USA. 1981;78:7609–7613. doi: 10.1073/pnas.78.12.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawley R G, Lieu F H L, Fong A Z C, Hawley T S. Versatile retroviral vectors for potential use in gene therapy. Gene Ther. 1994;1:136–138. [PubMed] [Google Scholar]
- 19.Hoeben R C, Migchielsen A A, van der Jagt R C, van Ormondt H, van der Eb A J. Inactivation of the Moloney murine leukemia virus long terminal repeat in murine fibroblast cell lines is associated with methylation and dependent on its chromosomal position. J Virol. 1991;65:904–912. doi: 10.1128/jvi.65.2.904-912.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsieh C L. Dependence of transcriptional repression on CpG methylation density. Mol Cell Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iguchi-Ariga S M, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 22.Jenuwein T, Forrester W C, Fernández-Herrero L A, Laibe G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- 23.Jones P L, Veenstra G J, Wade P A, Vermaak D, Kass S U, Landsberger N, Strouboulis J, Wolffe A P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 24.Kass S U, Pruss D, Wolffe A P. How does DNA methylation repress transcription? Trends Genet. 1997;13:444–449. doi: 10.1016/s0168-9525(97)01268-7. [DOI] [PubMed] [Google Scholar]
- 25.Kirillov A, Kistler B, Mostoslavsky R, Ceder H, Wirth T, Bergman Y. A role for nuclear NF-kappaB in B-cell-specific demethylation of the Ig kappa locus. Nat Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- 26.Lichtenstein M, Keini G, Cedar H, Bergman Y. B cell-specific demethylation: a novel role for the intronic kappa chain enhancer sequence. Cell. 1994;76:913–923. doi: 10.1016/0092-8674(94)90365-4. [DOI] [PubMed] [Google Scholar]
- 27.McKnight R A, Shamay A, Sankaran L, Wall R J, Hennighausen L. Matrix-attachment regions can impart position-independent regulation of a tissue-specific gene in transgenic mice. Proc Natl Acad Sci USA. 1992;89:6943–6947. doi: 10.1073/pnas.89.15.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulligan R C. The basic science of gene therapy. Science. 1993;260:926–932. doi: 10.1126/science.8493530. [DOI] [PubMed] [Google Scholar]
- 29.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 30.Niwa O, Yokota Y, Ishida H, Sugahara T. Independent mechanisms involved in suppression of the Moloney murine leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983;32:1105–1113. doi: 10.1016/0092-8674(83)90294-5. [DOI] [PubMed] [Google Scholar]
- 31.Palmer T D, Rosman G J, Osborne W R, Miller A D. Genetically modified skin fibroblasts persist long after transplantation but gradually inactivate introduced genes. Proc Natl Acad Sci USA. 1991;88:1330–1334. doi: 10.1073/pnas.88.4.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pawliuk R, Eaves C J, Humphries R K. Sustained high-level reconstitution of the hematopoietic system by preselected hematopoietic cells expressing a transduced cell-surface antigen. Hum Gene Ther. 1997;8:1595–1604. doi: 10.1089/hum.1997.8.13-1595. [DOI] [PubMed] [Google Scholar]
- 33.Phi-Van L, von Kries J P, Ostertag W, Stratling W H. The chicken lysozyme 5′ matrix attachment region increases transcription from a heterologous promoter in heterologous cells and dampens position effects on the expression of transfected genes. Mol Cell Biol. 1990;10:2302–2307. doi: 10.1128/mcb.10.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plavec, I., and M. Agarwal. Utility of scaffold attachment regions for enhanced retroviral vector expression in human hematopoietic cells. In A. C.-A. A. Garcia (ed.), Viral vectors: basic science and gene therapy, in press. Eaton Publishing, Natick, Mass.
- 35.Rigg J R, Chen J, Dando J S, Forestell S P, Plavec I, Böhnlein E. A novel human amphotropic packaging cell line: high titer, complement resistance, and improved safety. Virology. 1996;218:290–295. doi: 10.1006/viro.1996.0194. [DOI] [PubMed] [Google Scholar]
- 36.Rivière I, Brose K, Mulligan R C. Effect of retroviral vector design on expression of human adenosine deaminase in murine bone marrow transplant recipients engrafted with genetically modified cells. Proc Natl Acad Sci USA. 1995;92:6733–6737. doi: 10.1073/pnas.92.15.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robbins P B, Skelton D C, Yu X J, Halene S, Leonard E H, Kohn D B. Consistent, persistent expression from modified retroviral vectors in murine hematopoietic stem cells. Proc Natl Acad Sci USA. 1998;95:10182–10187. doi: 10.1073/pnas.95.17.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singal R, Ferris R, Little J A, Wang S Z, Ginder G D. Methylation of the minimal promoter of an embryonic globin gene silences transcription in primary erythroid cells. Proc Natl Acad Sci USA. 1997;94:13724–13729. doi: 10.1073/pnas.94.25.13724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singal R, Ginder G D. DNA methylation. Blood. 1999;93:4059–4070. [PubMed] [Google Scholar]
- 40.Stewart C L, Stuhlman H, Jahner D, Jaenisch R. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc Natl Acad Sci USA. 1982;79:4098–4102. doi: 10.1073/pnas.79.13.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson E M, Christians E, Stinnakre M-G, Renard J-P. Scaffold attachment regions stimulate HSP70.1 expression in mouse preimplantation embryos but not in differentiated tissues. Mol Cell Biol. 1994;14:4694–4703. doi: 10.1128/mcb.14.7.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsukiyama T, Ueda H, Hirose S, Niwa O. Embryonal long terminal repeat-binding protein is a murine homolog of FTZ-F1, a member of the steroid receptor superfamily. Mol Cell Biol. 1992;12:1286–1291. doi: 10.1128/mcb.12.3.1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Robbins P B, Carbonaro D A, Kohn D B. High-resolution analysis of cytosine methylation in the 5′ long terminal repeat of retroviral vectors. Hum Gene Ther. 1998;9:2321–2330. doi: 10.1089/hum.1998.9.16-2321. [DOI] [PubMed] [Google Scholar]