Abstract
The recent isolation of a nonhuman primate hepadnavirus from woolly monkeys prompted an examination of other primates for potentially new hepadnaviruses. A serological analysis of 30 captive gibbons revealed that 47% were positive for at least one marker of ongoing or previous infection with a hepatitis B virus (HBV). The amino acid sequences of the core and surface genes of human and gibbon virus isolates were very similar. Phylogenetic analysis indicated that the gibbon isolates lie within the human HBV family, indicating that these HBV isolates most likely stem from infection of gibbons from a human source.
Hepadnaviruses are classified into two genera representing isolates from birds (Avihepadnavirus) and rodents and humans (Orthohepadnavirus). The avihepadnaviruses include isolates from ducks (8) and heron (11), while rodent viruses have been isolated from the woodchuck (12), ground squirrel (7), and arctic ground squirrel (13). Until recently, the only primate hepadnavirus described was human hepatitis B virus (HBV). Recently, we isolated a hepadnavirus from a woolly monkey experiencing fulminant hepatitis at the Louisville Zoo (WMHBV) (5). A survey of captive woolly monkeys indicated that approximately 50% of the animals at different zoos were positive for at least one marker of current or previous infection with WMHBV. WMHBV is phylogenetically distinct from the human HBV family and thus represents the first nonhuman primate hepadnavirus. The isolation of WMHBV rekindled an interest in the possibility that other primate hepadnaviruses could be identified. Previously, the isolation of a hepadnavirus from a single chimpanzee (14) and a gibbon (10) had been reported. Phylogenetic analysis of the nucleotide sequences of these viruses suggested that they were members of the human HBV family, since they were more closely related to human genotypes A to E than were the human genotype F isolates (5). However, the isolation of a distinctive, nonhuman primate hepadnavirus (WMHBV) suggested that a more in-depth analysis of the gibbon viruses was warranted.
The genus Hylobates (gibbons) contains 11 species and 4 subgenera that occupy forests in distinct geographical regions in Southeast Asia, including eastern India, southern China, Vietnam, Cambodia, Burma, Thailand, Malaysia, Borneo, Java, and Sumatra. Examination of gibbons in the wild was not practical, so we chose to examine captive gibbons housed at the International Center for Gibbon Studies (Santa Clarita, Calif.). A total of 30 animals were examined, which represented six different species and three subgenera of gibbons (Table 1). The gibbons are housed individually or in small monogamous families, thus facilitating the evaluation of common exposures. None of the animals had been involved in any experimental procedures, and some were wild-caught animals. Serum from the animals was examined for the presence of HBV DNA by PCR using primers to the core region that are conserved among all human HBV genotypes (5). Enzyme-linked immunosorbent assays (ELISAs) for HBsAg, anti-HBsAg, and anti-HBcAg were performed with assays purchased from Abbott Laboratories. Two of the animals that were initially negative for all markers were vaccinated with the human HBV vaccine and seroconverted for anti-HBsAg. These animals were considered uninfected with regard to the estimations of the percentage of the animals exposed to HBV. Fourteen of the 30 (46.7%) animals were positive for at least one marker of HBV infection (Table 1), and this included animals in three of the six species of animals examined. Seven of the animals (23.3%) were PCR positive, and all of those tested (n = 6) were positive for HBsAg, indicating that these animals were chronically infected with HBV. All chronic carriers were members of either the Hylobates agilis or Hylobates moloch species. Eight of the animals (26.7%) were positive for antibodies to HBsAg, suggesting viral clearance; however, one of the anti-HBsAg-reactive animals was a chronic carrier (Ling [H. moloch]). Thirteen of the animals (43.3%) were positive for anti-HBcAg, and all anti-HBcAg-positive animals were positive for either HBsAg or anti-HBsAg, demonstrating that the anti-HBcAg positivity was not due to false reactivity. Thus, a high percentage of gibbons at this facility had prior exposure to a hepadnavirus, and 50% of the exposed animals were chronically infected.
TABLE 1.
HBV status of the gibbons examined in this studya
| Gibbonb | Result byc:
|
|||
|---|---|---|---|---|
| PCR | HBsAg | Anti-HBsAg | Anti-HBcAg | |
| Hylobates agilis | ||||
| Pepino (S) | + | + | − | + |
| Phoebe (D) | + | + | − | + |
| Felix (O) | + | NDd | − | ND |
| Homer | + | + | − | + |
| Sonny (S) | − | − | − | − |
| Elaine (D) | − | − | + | + |
| Betsy (O) | − | − | − | − |
| Mumma (D) | − | − | + | + |
| Albert (O) | − | − | − | − |
| Shorty (O) | − | − | + | + |
| Hylobates moloch | ||||
| Chloe | + | + | − | + |
| Shelby | − | − | + | + |
| Chilibi | + | + | − | + |
| Ling | + | + | + | + |
| Ivan | − | − | + | + |
| Ushko | − | − | + | + |
| Hylobates pileatus | ||||
| Birute (S) | − | − | + | + |
| JR (D) | − | − | − | − |
| Mat (O) | − | − | − | − |
| Cambio | − | − | − | − |
| Koko | − | − | − | − |
| Hylobates syndactylus | ||||
| Kino (S) | − | − | − | − |
| Fatima (D) | − | − | − | − |
| Ella (O) | − | − | − | − |
| Benny (O) | − | − | − | − |
| Hylobates leucogenys | ||||
| Ricky | − | − | − | − |
| Vok | − | − | − | − |
| Hylobates gabriella | ||||
| Kimkhi | − | − | − | − |
The following relationships are not indicated in the table. Sonny is the son of Mumma. Ushko was temporarily caged with both Ling (2 years) and Ivan (5 years). Cambio is the brother of JR. Koko is the son of Birute by a dam other than JR.
S, sire; D, dam; O, offspring.
PCR positivity was determined as described previously (5, 6). DNA was extracted from 10 μl of serum by proteinase K digestion and phenol-chloroform extraction. Primers were from the core region and were conserved among all HBV genotypes (forward primer, 5′-CCTTGGGTGGCTTTGGGGCA-3′; reverse primer, 5′-GGGCATTTGGTGGTCTATA-3′). Amplification was conducted for 35 cycles, and the products were detected by agarose gel electrophoresis and Southern hybridization. HBsAg, anti-HBsAg, and anti-HBcAg were determined by ELISA with assays purchased from Abbott Laboratories.
ND, not determined.
Examination of the family tree of the H. agilis species suggested a pattern of vertical transmission resulting in chronic infection and horizontal transmission resulting in viral clearance. Of the four chronically infected H. agilis animals, Pepino, Phoebe, and Homer shared a common sire that was negative for HBV markers and a dam that was not available for testing, but who was most likely a carrier that resulted in maternal transmission to all three offspring. The other animal with chronic infection among the H. agilis group, Felix, was the offspring of Pepino and Phoebe. Mumma, an anti-HBsAg-, anti-HBcAg-positive dam, mated with Pepino to produce Albert, who possessed no markers of HBV infection. Mumma's offspring by another sire was also negative for all HBV markers. A similar pattern was noted among the H. moloch animals. Two of the chronically infected animals, Chloe and Chilibi, were brother and sister, suggesting transmission from the mother. Shelby, who was first housed with Chloe as an adult, was anti-HBsAg, anti-HBcAg positive. Another chronically infected animal, Ling, had no exposure to Chloe or Chilibi. The antibody-positive status of Ushko could be attributed to being housed first with the sister of Chloe and Chilibi (who was chronically infected) and later with Ling. Thus, chronic HBV infections were found in three independent families. No overt sign of liver disease or mortality due to liver disease was noted in the chronically infected animals; however, since the animals were housed in a sanctuary, liver biopsies were not available for evaluation.
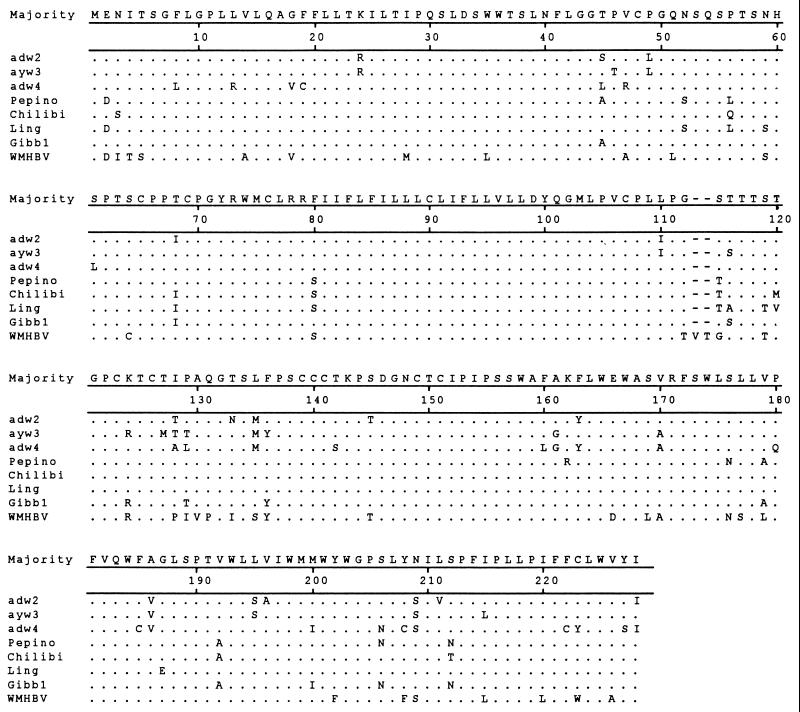
To examine the relationship of the viruses from gibbons to human HBV isolates, the core and surface genes were PCR amplified from serum-derived virion DNA and sequenced directly from the PCR products. The amino acid sequences from the gibbons were aligned with three human isolates of different genotypes: ayw3/France/genotype D, adw2/USA/genotype A, and adw4/Colombia/genotype F. Also included in the alignments were the sequences from the WMHBV isolate and the previously characterized gibbon sequence (Gibb1) (10). The sequences of all isolates are shown in reference to the consensus sequence. The sequences from Pepino, Phoebe, and Homer were identical for both genes, as were the sequences from Chloe and Chilibi, so only the sequences from Pepino and Chilibi are shown in the alignment. The core antigen amino acid sequences (Fig. 1) were similar for all isolates. The most notable difference was the 2-amino-acid insertion in the adw2 isolate at the same position as a size polymorphism in the WMHBV sequence (3-amino-acid deletion with respect to adw2, 1-amino-acid deletion with respect to all other isolates). Genotype F is the most divergent of the human genotypes (1), and the representative isolate of this genotype had unique amino acids in three positions not found in other isolates. In contrast, the WMHBV core gene contained unique amino acids in 14 positions. The core sequences of the gibbon isolates were very similar to those of the human isolates. Pepino contained unique residues at two positions and shared with Chilibi and Ling a single residue (asparagine 91) not found in the human isolates examined; however, this could not be considered a signature sequence for gibbon isolates, since it was lacking in the Gibb1 sequence. Pepino differed from Chilibi and Ling at two residues (amino acids 67 and 174), while Chilibi and Ling differed from each other by a single residue (amino acid 49).
FIG. 1.
Alignment of core gene amino acid sequences from primate hepadnaviruses. The core gene sequences from three human and four gibbon hepadnaviruses and one woolly monkey hepadnavirus were aligned by using the MEGALIGN program of LASERGENE (DNAstar, Madison, Wis.). All sequences are compared to the consensus sequence, with dots indicating identity and dashes indicating deletions. GenBank accession numbers are given in the legend to Fig. 3.
The surface gene amino acid sequences (Fig. 2) were more divergent than the core genes; however, the gibbon isolates were still very similar to the human isolates. The WMHBV sequence has a 2-amino-acid insertion not seen in any of the other isolates. The genotype F isolate contained unique amino acids at 16 positions, while the WMHBV isolate differed from all other isolates at 28 positions. In contrast, the sequences of isolates from Pepino, Chilibi, and Ling were unique at one, four, and three positions, respectively. No signature changes were present in the sequences of the different gibbon isolates. Pepino differed from Chilibi at 12 positions, while Chilibi differed from Ling at 8 positions.
FIG. 2.
Alignment of surface gene amino acid sequences from primate hepadnaviruses. The surface gene sequences from three human and four gibbon hepadnaviruses and one woolly monkey hepadnavirus were aligned as described in the legend to Fig. 1. All sequences are compared to the consensus sequence, with dots indicating identity and dashes indicating deletions. GenBank accession numbers are given in the legend to Fig. 3.
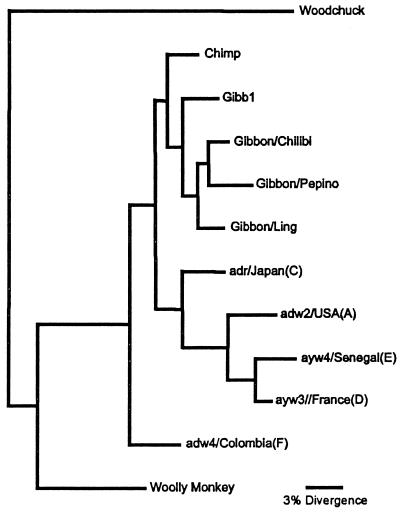
Phylogenetic analysis was performed to evaluate the evolutionary relationship of the various primate isolates of HBV. Analyses were performed with the PAUP* program, to compare the nucleotide sequences (approximately 549 to 555 bp) of the core gene, as implemented previously (5). Although the surface gene is larger and generally more variable than the core gene, the core gene sequences generated a more robust phylogenetic tree (only one tree versus four different versions when surface gene sequences were used) and contained a greater number of parsimony-informative characters (118 versus 100). An exhaustive search for the most parsimonious trees for the core gene revealed a single tree (Fig. 3). The nucleotide sequences of the core genes of Pepino, Chilibi, and Ling were compared along with those of the previously reported isolates from a chimpanzee and a gibbon. Five human virus isolates representative of five of the six human genotypes (9) were included, as was the WMHBV sequence. A woodchuck virus isolate was used as the outgroup to root the trees. The analyses indicated that the gibbon and chimpanzee isolates were members of the human HBV family, since their sequences were more closely related to the genotypes A, C, D, and E, than was the sequence of the genotype F isolate. A similar phylogenetic tree was obtained from the analysis of the surface antigen gene nucleotide sequence (data not shown). Genotype F is the most divergent of the human genotypes. Genotype F isolates are typically found in South and Central America and have been postulated to be the original New World strains (1). Although the nonhuman primate viruses were within the human HBV family, they were more closely related to each other than to other human genotypes. As previously reported, the WMHBV sequence is not within the human HBV family.
FIG. 3.
Phylogenetic analyses of the nucleotide sequence of core genes from primate hepadnaviruses. Phylogenetic analyses were performed with the test version, 4.0b2, of the PAUP∗ program written by David Swofford (Sinauer Associates, Sunderland, Mass.). Exhaustive searches made the use of statistical support for trees (e.g., BOOTSTRAP) unnecessary and gave the single tree shown here. The nucleotide sequences of the core antigen genes from gibbons Pepino, Chilibi, and Ling were compared with the previously reported isolates from a chimpanzee (Chimp) (14) and a gibbon (Gibb1) (10). Five human isolates representative of five of the six human genotypes were included with the genotype given in parentheses. The sequence of WMHBV (5) was included as an example of a primate HBV sequence outside of the human HBV family. A woodchuck isolate was used as the outgroup to root the trees. The distance along the horizontal axis among isolates is proportional to genetic divergence. The GenBank accession numbers for the sequences of the isolates shown here are as follows: adw2/USA (A), X02763; ayw4/Senegal(E), X75664; ayw3/France(D), V01460; adr/Japan(C), L08805; adw4/Colombia(F), X75663; Gibb1, U46935; Chimp, D00220; Woolly Monkey, AF046996; Woodchuck, J02442; Gibbon/Chilibi/Core, AF213008; Gibbon/Chilibi/Surface, AF213006; Gibbon/Pepino/Core, AF213010; Gibbon/Pepino/Surface, AF213007; Gibbon/Ling/Core, AF213009; Gibbon/Ling/Surface, AF213005.
Several possibilities exist to explain the phylogenetic relationship between the gibbon and human viruses. The genomic sequences of greater apes are very closely related and difficult to distinguish based on limited sequence information. It is possible that the gibbon and human hepadnaviruses are derived from a common ancestral virus and that the sequences have not diverged sufficiently to distinguish the human and gibbon viruses. This is unlikely considering the divergence of the genotype F viruses from the remainder of the human family. Infection of gibbons with human HBV could have occurred some time in the distant or not so distant past, and the virus has become established in gibbons living in the wild. Finally, the transmission of human HBV to gibbons could be a current and ongoing problem due to close contact between these primates and humans in an area in which a high percentage of the population is chronically infected with HBV. Based on the serological information, this appears to be the most likely scenario. Although seven animals were chronic carriers, the relationships of the animals suggest this could be accounted for by vertical transmission from three chronically infected dams. The other seven HBV-exposed animals had cleared their infections and thus were more likely to have been exposed as adults.
Although the animals housed at the International Center for Gibbon Studies have not participated in medical experimentation, the wild-born animals were often captive pets of people in the area before being acquired by a refuge. Close contact with infected humans could easily account for the exposures, with gibbons being exposed to contaminated blood through biting and/or scratching of infected humans, perhaps at the time of capture. A serological analysis of a large population of gibbons living in the wild was not feasible, leaving open the question of whether hepadnavirus infections persist in gibbons in the absence of current contact with humans. The transmission of viruses between nonhuman primates and humans can have dire consequences, as is expected to be the case for human immunodeficiency virus (3) and Ebola virus (2, 4). The transmission of human HBV to gibbons does not appear to have resulted in more severe infections in gibbons than those observed in humans. As the habitat of nonhuman primates continues to diminish, contact between humans and our closest relatives will increase, as will the cross-species transmission of viruses unique to each species. The outcomes cannot be predicted and may be benign or catastrophic. A continued surveillance of both humans and nonhuman primates for such occurrences is warranted.
Acknowledgments
This work was supported by grants CA53246 (R.E.L.), AI01124 (R.R.-H.), and P51 RR13986 from the National Institutes of Health.
REFERENCES
- 1.Arauz-Ruiz P, Norder H, Visona K A, Magnius L O. Molecular epidemiology of hepatitis B virus in Central America reflected in the genetic variability of the small S Gene. J Infect Dis. 1997;176:851–858. doi: 10.1086/516507. [DOI] [PubMed] [Google Scholar]
- 2.Formenty P, Boesch C, Wyers M, Steiner C, Donati F, Dind F, Walker F, Le Guenno B. Ebola virus outbreak among wild chimpanzees living in a rain forest of Cote d'Ivoire. J Infect Dis. 1999;179:S120–S126. doi: 10.1086/514296. [DOI] [PubMed] [Google Scholar]
- 3.Gao F, Bailes B, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, Sharp P M, Hahn B H. Origin of HIV-1 in the chimpanzee Pan Troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 4.Georges-Courbot M-C, Sanchez A, Lu C-Y, Baize S, Leroy E, Lansout-Soukate J, Tevi-Benissan C, Georges A J, Trappier S G, Zaki S R, Swanepoel R, Leman P A, Rollin P E, Peters C J, Nichol S T, Ksiazek T G. Isolation and phylogenetic characterization of Ebola viruses causing different outbreaks in Gabon. Emerg Infect Dis. 1997;3:59–62. doi: 10.3201/eid0301.970107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lanford R E, Chavez D, Brasky K M, Burns III R B, Rico-Hesse R. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc Natl Acad Sci USA. 1998;95:5757–5761. doi: 10.1073/pnas.95.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanford R E, Michaels M G, Chavez D, Brasky K, Fung J, Starzl T E. Persistence of extrahepatic hepatitis B virus DNA in the absence of hepatic replication in patients with baboon liver transplants. J Med Virol. 1995;46:207–212. doi: 10.1002/jmv.1890460307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marion P L, Oshiro L S, Regnery D C, Scullard G H, Robinson W S. A virus in Beechey ground squirrels that is related to hepatitis B virus of humans. Proc Natl Acad Sci USA. 1980;77:2941–2945. doi: 10.1073/pnas.77.5.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mason W S, Seal G, Summers J. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J Virol. 1980;36:829–836. doi: 10.1128/jvi.36.3.829-836.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Norder H, Couroucé A-M, Magnius L O. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 10.Norder H, Ebert J W, Fields H A, Mushahwar I K, Magnius L O. Complete sequencing of a Gibbon hepatitis B virus genome reveals a unique genotype distantly related to the chimpanzee hepatitis B virus. Virology. 1996;218:214–223. doi: 10.1006/viro.1996.0181. [DOI] [PubMed] [Google Scholar]
- 11.Sprengel R, Kaleta E F, Will H. Isolation and characterization of a hepatitis B virus endemic in herons. J Virol. 1988;62:3832–3839. doi: 10.1128/jvi.62.10.3832-3839.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Summers J, Smolec J M, Snyder R. A virus similar to human hepatitis B virus associated with hepatitis and hepatoma in woodchucks. Proc Natl Acad Sci USA. 1978;75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Testut P, Renard C-A, Terradillos O, Vitvitski-Trepo L, Tekaia F, Degott C, Blake J, Boyer B, Buendia M A. A new hepadnavirus endemic in arctic ground squirrels in Alaska. J Virol. 1996;70:4210–4219. doi: 10.1128/jvi.70.7.4210-4219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vaudin M, Wolstenholme A J, Tsiquaye K N, Zuckerman A J, Harrison T J. The complete nucleotide sequence of the genome of a hepatitis B virus isolated from a naturally infected chimpanzee. J Gen Virol. 1988;69:1383–1389. doi: 10.1099/0022-1317-69-6-1383. [DOI] [PubMed] [Google Scholar]