Abstract
The genome of lymphocytic choriomeningitis virus (LCMV) consists of two negative-sense single-stranded RNA segments, designated L and S. Both segments contain two viral genes in an ambisense coding strategy, with the genes being separated by an intergenic region (IGR). We have developed a reverse genetic system that allows the investigation of cis-acting signals and trans-acting factors involved in transcription and replication of LCMV. To this end, we constructed an LCMV S minigenome consisting of a negative-sense copy of the chloramphenicol acetyltransferase (CAT) reporter gene flanked upstream by the S 5′ untranslated region (UTR) and IGR and downstream by the S 3′ UTR. CAT expression was detected in LCMV-infected cells transfected with the minigenome RNA. Intracellular coexpression of the LCMV minigenome and LCMV L and NP proteins supplied from cotransfected plasmids driven by the T7 RNA polymerase provided by the recombinant vaccinia virus vTF7-3 resulted in high levels of CAT activity and synthesis of subgenomic CAT mRNA and antiminigenome RNA species. Thus, L and NP represent the minimal viral trans-acting factors required for efficient RNA synthesis mediated by LCMV polymerase.
Lymphocytic choriomeningitis virus (LCMV) is the prototypic member of the family Arenaviridae, which also includes important human pathogens such as Lassa virus and Junin virus. LCMV provides one of the most widely used model systems to study viral persistence and pathogenesis (40, 42). However, the investigation of the molecular mechanisms underlying LCMV persistence and its associated disorders has been hampered by the lack of a genetic system to analyze the structure and function of the LCMV genome and its gene products.
The LCMV genome consists of two negative-sense single-stranded RNA segments, designated L and S, with approximate sizes of 7.2 and 3.4 kb, respectively (48, 52). Each RNA segment has an ambisense coding strategy, encoding two proteins in opposite orientation, separated by an intergenic region (IGR) (3, 4, 56). The S RNA directs synthesis of the three major structural proteins: the nucleoprotein, NP (ca. 63 kDa); and two mature virion glycoproteins, GP-1 (40 to 46 kDa) and GP-2 (35 kDa), that are derived by posttranslational cleavage of a precursor polypeptide, GP-C (75 kDa) (47, 55, 57). Tetramers of GP-1 and GP-2 make up the spikes on the virion envelope. Evidence indicated that GP-1 mediates virus interaction with host cell surface receptor, which has been recently identified as α-dystroglycan (7, 11). The L RNA segment encodes a high-molecular-mass protein (L; ca. 200 kDa) which has the characteristic motifs conserved in all the viral RNA-dependent RNA polymerases and a small polypeptide Z (ca. 11 kDa) which contains a RING finger motif and whose function is unknown (26, 50, 52).
The NP, the most abundant viral protein in virally infected cells, is associated with the viral RNA (vRNA) to form the nucleocapsid (NC) which is the template for the viral RNA polymerase (26). The L protein is thought to be the main viral component of the arenavirus polymerase (25). The NC associated with the viral polymerase constitutes the viral ribonucleoprotein (RNP), which is active in transcription and replication and is the minimum unit of LCMV infectivity. Results from in vitro transcription and immunodepletion studies have implicated Z in both genome replication and mRNA synthesis in Tacaribe virus (29), a member of the Arenaviridae. In addition, biochemical and immunological studies have suggested that Z might be the arenavirus counterpart of the matrix protein found in other negative-strand RNA viruses (50).
All cis-acting signals required for encapsidation and polymerase entry of negative-strand RNA viruses appeared to be located within the 5′- and 3′-terminal untranslated regions (UTRs) (16). LCMV exhibits a short stretch of conserved sequences at the precise 3′ terminus of the L and S genome RNAs (17 of 19 nucleotides [nt] are identical). The inverted complement of this sequence is found at the 5′ termini of the genome RNAs (2, 39, 48). The 3′ terminus of genomic RNAs in highly conserved among other members of the family Arenaviridae, suggesting that these 19 terminal nt may contain cis-acting signals for the replication and transcription. The IGRs in both L and S segments have the potential to form stable stem-loop (hairpin) structures (4, 52, 56). The viral 3′ ends of the mRNAs, which are nonpolyadenylated and heterologous, have been mapped to the base of the hairpin on the distal sides (23, 34), suggesting a possible transcriptional regulatory role of the IGR.
In contrast to positive-strand RNA viruses, manipulation of the genome of negative-strand RNA viruses has been hampered by the nature of their genetic material (16, 17, 27, 28, 49). The minimal infectious unit of a negative-strand RNA virus is the RNP complex which contains the NC template associated with the virus polymerase. Therefore, deproteinized genomic and antigenomic RNAs cannot function as mRNAs and are not infectious. Generation of biologically active synthetic viral genomes for negative-strand RNA viruses need reconstitution of the active viral RNP complexes from synthetic cDNA. However, progress made in this field during the last few years has allowed the rescue of short model genomes for several negative-strand RNA viruses. For this, the required trans-acting viral factors were provided by using either helper viruses or complementation with plasmid-expressed proteins (37, 43, 44). Coexpression of both genome analogs with predetermined termini and viral proteins from transfected plasmids has been achieved using the recombinant vaccinia virus expressing T7 RNA polymerase (vTF7-3) or expression vectors based on RNA polymerase II or I promoters (15, 18–20, 41, 43). This has allowed the analysis of cis- and trans-acting elements required for replication and transcription. Moreover, it has been possible to rescue infectious virus from several families entirely from cloned cDNAs. There are rescued viruses with nonsegmented genomes, such as rabies virus (53), respiratory syncytial virus (RSV) (13), vesicular stomatitis virus (36), and measles virus (46), and also with segmented genomes (bunyavirus [8] and influenza virus [40]).
To establish a reverse genetic system for the analysis in vivo of cis- and trans-acting factors involved in replication and transcription of LCMV, we constructed plasmids expressing genome analogs or a minigenome of LCMV, as well as the viral NP, L, and Z polypeptides under the control of the T7 RNA polymerase promoter. To reconstitute in vivo the replication and transcription of LCMV genome analogs, we infected BHK-21 cells with vTF7-3 and subsequently transfected cells with plasmids encoding the LCMV minigenome, NP, Z, and L (22). We present evidence that the 5′ and 3′ UTRs together with the IGR of the S RNA are sufficient cis-acting signals to allow RNA synthesis mediated by LCMV RNA polymerase, though it remains to be determined which of these elements are strictly required for virus replication and transcription. We also show that NP and L are the minimal trans-acting factors required for replication and transcription of LCMV genome analogs. The system described here will allow a detailed molecular characterization of the cis- and trans-acting elements involved in LCMV replication and transcription. Furthermore, this system provides the basis for investigation of the requirements for LCMV packaging and budding, as well as the rescue of infectious virus entirely from plasmids.
MATERIALS AND METHODS
Cells and virus.
Baby hamster kidney (BHK-21) cells were grown in Dulbecco's minimal essential medium supplemented with 7% fetal calf serum (Life Technologies) in a 5% CO2 atmosphere. Plaque-purified LCMV strain Armstrong (Arm) 53b was used in this study. vTF7-3 was kindly provided by B. Moss (National Institutes of Health, Bethesda, Md.) (24).
Plasmids.
To construct pLCMVSCAT1, a KpnI-PstI fragment containing the T7 RNA polymerase promoter, 5′ UTR, chloramphenicol acetyltransferase (CAT) open reading frame (ORF) in antisense orientation with respect to T7, and 3′ UTR of plasmid pARM5′IgCAT3′ was inserted between the KpnI and PstI sites of pUC18 (Life Technologies) (Fig. 1A). To generate pARM5′IgCAT3′, a BglII-StuI DNA fragment containing the S IGR (nt 1572 to 1641) was obtained from plasmid pCRIIIg and cloned between the S 5′ and 3′ UTRs of plasmid pARM5′CAT3′ digested with BglII and StuI. To generate pARM5′ICAT3′, the CAT ORF (31) flanked by SalI (upstream) and BglII (downstream) sites was cloned into SalI and BglII unique sites in pARM5′3′, which positioned the CAT ORF in an antisense polarity with respect to the T7 promoter. The S IGR was cloned by reverse transcription-PCR using oligonucleotides that contained BglII and StuI flanking sites and cloned into pCRII to obtain pCRIIIg. Plasmid pARM5′3′ contained the S 5′ UTR (nt 1 to 78) followed by the S 3′ UTR (nt 3316 to 3376). The sequence of the T7 RNA polymerase minimum promoter followed by three G's was placed immediately upstream of the 5′ UTR. Unique BglII and SalI sites were introduced at the junction between the 5′ and 3′ UTR sequences, and an MvnI site was introduced at the 3′ end of the 3′ UTR. Plasmid pUC19 was used as backbone DNA for this construct. Plasmid pLCMVSCAT2 was constructed by inserting an LCMV-specific 3′ hairpin ribozyme (HR) and the T7 RNA polymerase terminator sequences (T7T) into pLCMVSCAT1. The HR sequences were amplified by PCR from plasmid p53MAM (58). After digestion with BamHI and BssHII, the DNA fragment containing the 3′ end of the LCMV S RNA and the 3′ HR was inserted into the BamHI and BssHII sites of pLCMVSCAT1. Finally, a BssHII-HindIII DNA fragment containing the T7T was inserted downstream of the HR to generate pLCMVSCAT2 (Fig. 1B). Plasmids for expression of LCMV NP, L, and Z were obtained as follows. (i) For NP, a cDNA encoding the NP ORF flanked by NcoI and StuI restriction sites was amplified by PCR. The PCR product, digested with NcoI and StuI, was cloned into the NcoI-HincII sites of plasmid pCITE-2a (Novagen). (ii) For Z, a cDNA encoding the Z ORF was cloned into NcoI-StuI sites of plasmid pUCIRES. Plasmid pUCIRES was constructed by inserting the BspEI-ClaI fragment containing the encephalomyocarditis virus internal ribosomal entry site (EMCV IRES) of plasmid pTM1 into the AvaI-AccI sites of pUC19 (Life Technologies) (21). Both pCITE-2a and pUCIRES contain the EMCV IRES downstream of the T7 RNA polymerase promoter, allowing cap-independent translation of mRNA. The cDNA encoding a full-length L protein was cloned by reverse transcription-PCR using RNA from LCMV-infected BHK-21 cells and primers containing KpnI and SalI restriction sites. After digestion of the PCR product with KpnI and SalI, the L cDNA was cloned into the KpnI-SalI sites of pGEM-4Z (Promega). The resulting plasmid was named pGEM-L.
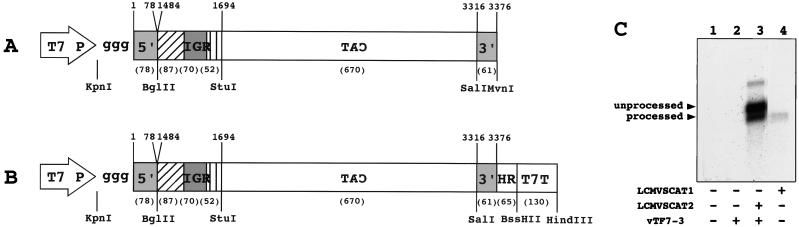
FIG. 1.
Schematic diagrams and characterization of LCMV genomic RNA analogs. (A) Plasmid pLCMVSCAT1 was constructed by combining, in the order indicated in the schematic, the following elements: the minimum T7 RNA polymerase promoter (T7 P) followed by three extra G's, the 5′ UTR of the LCMV S RNA (nt 1 to 78), the IGR of the LCMV S RNA (nt 1484 to 1694), a DNA encoding the full-length CAT ORF in antisense orientation with respect to the T7 promoter, and the 3′ UTR of the LCMV S RNA (nt 3316 to 3376). (B) Plasmid pLCMVSCAT2 was made by inserting an LCMV-specific 3′ HR followed by T7T into pLCMVSCAT1. Viral sequences were derived from LCMV Arm 5. Nucleotide numbers correspond to those of the S RNA. Numbers in parentheses indicate lengths of cDNA fragments. Restriction enzymes used for cloning are indicated at the bottom. Hatched and striped boxes represent the 3′ ends of the GP and NP, respectively. (C) Northern blot analysis of in vitro and intracellularly synthesized LCMV RNA minigenomes. BHK-21 cells were infected with vTF7-3 at an MOI of 3 PFU/cell and subsequently transfected with 2 μg of pLCMVSCAT2. Total cellular RNA was prepared by using TRI reagent at 24 h after infection with vTF7-3. RNA (5 μg) was analyzed by Northern blotting using a CAT sense riboprobe. LCMVSCAT1 RNA was prepared in vitro transcription with T7 RNA polymerase of MvnI-digested template pLCMVSCAT1 DNA. Unprocessed and ribozyme-processed RNA species are indicated by arrowheads at the left.
RNA and DNA transfections and virus infections.
For RNA transfection, 2 × 105 BHK-21 cells were infected with LCMV Arm at a multiplicity of infection (MOI) of either 3 or 0.1 PFU/cell and then transfected with 5 μg of in vitro-transcribed LCMVSCAT1 RNA by using Lipofectin (Life Technologies). For DNA transfection, 4 × 105 BHK-21 cells were seeded into a 35-mm-diameter well to reach ∼50 to 70% confluence at the time of being transfected. Cells were infected with vTF7-3 at an MOI of 3 and subsequently transfected by using Lipofectamine (Life Technologies) with plasmids encoding NP, Z, and L (1.45, 0.005, and 0.1 μg per well, respectively) and 1 μg of the transcribing plasmid pLCMVSCAT2. The total amount of transfected DNA was kept constant by addition of empty pBluescript KS(+) plasmid. All plasmids were prepared according to the protocol for the Qiagen plasmid preparation kit.
CAT assays.
Cells (2 × 105) were washed once with ice-cold phosphate-buffered saline and lysed in 50 μl of 250 mM Tris-HCl (pH 7.5) by three cycles of freezing and thawing; cell lysates were then clarified by centrifugation. For the assay, either 5 μl (DNA transfection) or 12.5 μl (RNA transfection) of cell extract was mixed with 1 μl (0.25 μCi) of [14C]chloramphenicol (53 mCi/mmol; Amersham) and 10 μl of 4 mM acetylcoenzyme A (Boehringer Mannheim Biochemicals). Reaction volumes were adjusted to 0.5 M Tris-HCl (pH 7.5) and incubated at 37°C for either 30 min (DNA transfection) or 3 h (RNA transfection). Products of the CAT reaction were analyzed by ascending thin-layer chromatography (TLC), developed using chloroform-methanol (19:1). The amount of 14C label in each spot of the TLC was quantified using a PhosphorImager (Molecular Dynamics), and percent conversion from [14C]chloramphenicol to its acetylated forms was determined for each sample. When necessary, cell extracts were diluted to obtain values of CAT activity within the linear range (<30% conversion).
Synthesis and analyses of RNA.
Runoff transcription of MvnI-linearized plasmid pLCMVSCAT1 was performed with MEGAscript according to the protocols of the supplier (Ambion). Total RNA of BHK-21 cells infected with vTF7-3 and subsequently transfected with various combinations of plasmids pLCMVSCAT2, pCITE-NP, pUCIRES-Z, and pGEM-L was isolated by using TRI reagent (Molecular Biology Center, Inc.) according to supplier's protocol and analyzed by Northern blotting using either CAT sense or antisense strand-specific riboprobes. The RNA was separated on 1.5% agarose gels containing 2.2 M formaldehyde and transferred to nylon membranes. Blots were hybridized with 32P-labeled RNA probes in a solution containing 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 5× Denhardt's solution, 50% formamide, 5% sodium dodecyl sulfate (SDS), yeast tRNA (100 μg/ml) and of single-stranded DNA (100 μg/ml) at 60°C overnight. Filters were washed two times for 15 min each in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–2% SDS at 65°C and subsequently two times for 15 min each in 0.2× SSC–0.2% SDS at 68°C. Bands were visualized by autoradiography. 32P-labeled CAT sense or CAT antisense RNA probes were prepared by in vitro runoff transcription of plasmid pBS-CAT with either T7 or T3 RNA polymerase in the presence of [32P]UTP.
Oligo(dT) chromatography.
RNA (50 μg) dissolved in TE (10 mM Tris-HCl [pH 7.5], 2 mM EDTA) at a concentration of 0.5 μg/μl was heated at 98°C for 10 min, immediately transferred to ice water, and diluted with cold TE to 5 ng/μl. Samples were adjusted to 1× binding buffer (10 mM Tris-HCl [pH 7.5], 2 mM EDTA, 500 mM NaCl, 0.2% Sarkosyl), and oligo(dT) resin (100 mg) was added. After incubation with agitation for 60 min at room temperature, the oligo(dT) RNA-containing pellet was washed three times with 1× binding buffer and twice with low-salt (200 mM NaCl) buffer by resuspension and pelleting. Unbound [poly(A)23] was saved, and bound RNA [poly(A)+] was eluted with hot (75°C) TE using a spin column with filter pad to retain the oligo(dT) resin. Both poly(A)− and poly(A)+ RNAs were precipitated with ethanol, resuspended in 0.5 mM EDTA, and analyzed by Northern blot hybridization as described above. The T7 RNA polymerase expressed by vTF7-3 mediates high levels of intracellular synthesis of the LCMV minigenome, a fraction of which could be polyadenylated by vTF7-3-associated activities. Dilution of RNA following heat denaturation and prior addition of binding buffer was found to be required to avoid artifactual binding through sandwich hybridization of antigenome and subgenomic CAT mRNAs to polyadenylated genomic RNA present in large excess.
Immunoblot analysis.
BHK-21 cells (4 × 105) were infected with vTF7-3 and transfected with plasmids encoding NP, Z, and L of LCMV as described above. Sixteen to eighteen hours postinfection (p.i.), cells were washed twice in phosphate-buffered saline, and extracts were prepared in 1× SDS-gel-loading buffer (50 mM Tris-HCl [pH 6.8], 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, 10% glycerol). Cell lysates were separated by either 10, 16, or 8% SDS-polyacrylamide gel electrophoresis using the Laemmli system (35). Immunoblot analysis was performed as described above. NP was detected using monoclonal antibody 1.1.3 diluted 1:300. Z was detected using a rabbit serum (kindly provided by M. Salvato) at a dilution of 1:100. A rabbit polyclonal antibody against the C-terminal part of L protein (amino acids 1658 to 2210) was raised by immunization with purified recombinant protein expression in bacterial cells. Rabbit anti-L serum was used at dilution 1:50 for the detection of L protein. Secondary antibodies coupled with peroxidase were used at a dilution of 1:20,000. Immunoreactivities were detected using enhanced chemiluminescence (Boehringer Mannheim Biochemicals).
RESULTS
Helper virus-dependent expression of synthetic analog of LCMV genomic S RNA.
We hypothesized that the viral 5′ and 3′ UTR together with the IGR could provide all of the cis-acting signals required for RNA synthesis mediated by LCMV polymerase. To test this hypothesis, we constructed plasmid pLCMVSCAT1 (Fig. 1A), which directs the synthesis, via T7 RNA polymerase, of a synthetic analog of LCMV genomic S RNA. This LCMV minigenome contains a negative-sense copy of the CAT ORF flanked upstream by the S 5′ UTR and IGR and downstream by the S 3′ UTR. To increase the efficiency of T7 transcription, we introduced three G residues between the T7 minimum promoter and the 5′ end of the LCMV minigenome. The precise viral 3′ end was obtained by engineering an MvnI site downstream of the S 3′ UTR in plasmid pLCMVSCAT1. In vitro transcription of MvnI-digested pLCMVSCAT1 DNA generates an LCMV minigenome designated LCMVSCAT1 RNA (Fig. 1C).
We then examined whether LCMVSCAT1 RNA contained all cis-acting sequences required for CAT expression mediated by LCMV polymerase in a helper virus-dependent system. For this purpose, BHK-21 cells infected with LCMV Arm at an MOI of either 0.1 or 3 PFU/cell were transfected with LCMVSCAT1 RNA at 0, 12, and 24 h after LCMV infection. Forty-eight hours after infection with LCMV, we prepared cell extracts for CAT assay. CAT activity would be expected only if the LCMV minigenome RNA was recognized and transcribed by the trans-acting factors provided by LCMV infection. We detected CAT activity only in cells infected at an MOI of 0.1 and transfected with LCMVSCAT1 RNA 24 h after infection with LCMV (Fig. 2A). In four independent experiments, levels of CAT were relatively low but were significant and reproducible. This finding indicated that the 5′ and 3′ UTR together with the IGR contain all of the cis-acting signals required for LCMV RNA synthesis.
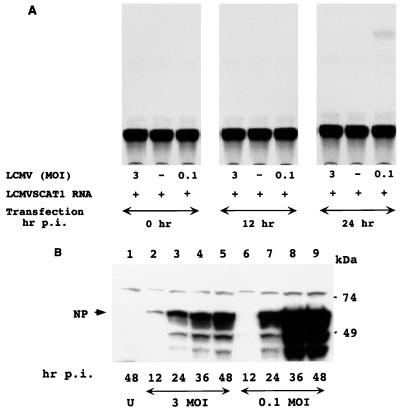
FIG. 2.
Rescue of CAT activity by LCMV helper virus correlates with expression of viral NP. (A) Helper virus-dependent rescue of CAT expression. BHK-21 cells were infected with LCMV at an MOI of either 0.1 or 3 and then transfected with 5 μg of in vitro-transcribed LCMVSCAT1 RNA at 0, 12, and 24 p.i. Cells were harvested at 48 h p.i., and extracts for CAT assay were prepared. CAT assay was performed as described in Materials and Methods. Shown is a representative result of four independent experiments. (B) Kinetics of NP expression in LCMV-infected BHK-21 cells. BHK-21 cells were infected with LCMV at an MOI of either 0.1 or 3. Cells were harvested at 12, 24, 36, and 48 h p.i. Uninfected BHK-21 control cells were harvested at 48 h p.i. Similar amounts of whole cell extracts from each sample were analyzed by Western blotting with a monoclonal antibody to NP. Immunoreactivity was detected by chemiluminescence. U, uninfected cells. Numbers at the bottom correspond to time of harvest. Molecular weight standards are indicated on the right. The position of NP is indicated by an arrow on the left. Consistent with previous reports, additional bands corresponding to specific NP fragments could be seen in extracts from LCMV-infected cells.
We were intrigued by the lack of CAT activity when cells were infected at an MOI of 3 or when infected at MOI of 0.1 but transfected at earlier times than 24 h p.i. Levels of NP have been proposed to be implicated in the regulation of LCMV RNA synthesis. Therefore, we examined levels of NP expression under the various experimental conditions used for the rescue of CAT expression. Cell extracts from BHK-21 cells infected with LCMV Arm at an MOI of 0.1 or 3 were prepared at 12, 24, 36, and 48 h p.i., and levels of NP expression were analyzed by Western blotting. At 12 h p.i., low levels of NP expression could be detected in cells infected at an MOI of 3 but not in those infected at an MOI of 0.1 (Fig. 2B, compare lanes 2 and 6). Both in cells infected at an MOI of 3 and those infected at an MOI of 0.1, levels of NP expression increased from 12 to 48 h p.i. (Fig. 2B, lanes 4, 5, 8, and 9). Interestingly and unexpectedly, levels of NP expression at 24, 36, and 48 h p.i. were higher in cells infected at MOI of 0.1 than in those infected at an MOI 3 (Fig. 2B, compare lanes 3 to 5 with lanes 7 to 9). Therefore, it appeared that there was a correlation between intracellular levels of NP and helper virus-dependent expression of CAT activity by the LCMV minigenome. Consistent with previous reports, we observed specific NP breakdown products in LCMV-infected cells. Whether these NP fragments play specific roles is not known.
T7-mediated intracellular production of LCMV RNA genome analog.
Difficulties inherently associated with RNA transfection of cells could have also contributed to low levels of CAT expression. To address this problem, we transfected a plasmid DNA which allowed intracellular synthesis of the LCMV minigenome with predetermined 5′ and 3′ termini through the use of a T7 promoter and ribozyme, respectively. For this purpose, we constructed plasmid pLCMVSCAT2 (Fig. 1B) by inserting the LCMV-specific 3′ HR sequence downstream of the LCMV S RNA 3′ UTR in plasmid pLCMVSCAT1. The HR sequences were followed by T7T sequences.
Intracellular T7-mediated transcription of transfected pLCMVSCAT2 plasmid DNA generates an RNA identical to LCMVSCAT1 but containing the 3′ HR and T7T sequences at its 3′ end. Self-cleavage of the 3′ HR at its 5′ end is predicted to generate a 3′ end in the upstream RNA that corresponds to the precise LCMV S RNA 3′ terminus.
To examine the efficiency of processing of intracellularly produced LCMVSCAT2 RNA, BHK-21 cells infected with vTF7-3 were transfected with plasmid pLCMVSCAT2. At 20 h p.i., total RNA was prepared and analyzed by Northern blot hybridization using a sense CAT riboprobe. Specific hybridization was detected only in RNA samples from cells that had been infected with vTF7-3 and transfected with plasmid pLCMVSCAT2 (Fig. 1C). We detected two main RNA species with approximate sizes of 1,021 and 1,216 nt (Fig. 1C, lane 3). The smaller RNA species had the same electrophoretic mobility as the in vitro-transcribed LCMVSCAT1 RNA (Fig. 1C, compare lanes 3 and 4), which indicated that this RNA species most likely correspond to correctly processed LCMVSCAT2 RNA. The other RNA species had a predicted size compatible with RNA transcripts that were terminated, as expected, at the T7T site but were not processed by the self-cleavage of the 3′ HR. We also detected low levels of larger RNA species (Fig. 1C, lane 3), which likely correspond to read-through transcription of the T7 RNA polymerase across the T7T sequences. Quantitation by densitometry of the unprocessed and processed RNA species indicated that approximately 22% of the intracellularly transcribed LCMVSCAT2 RNA was cleaved by the HR.
Plasmid-mediated expression of LCMV polypeptides.
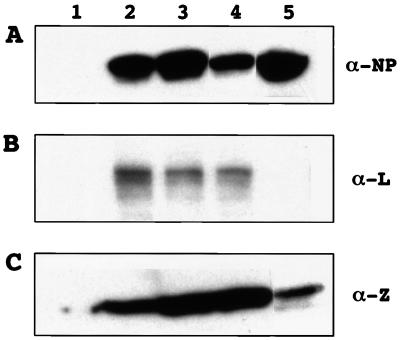
cDNAs encoding NP, Z, and L full-length ORFs of LCMV Arm were cloned in plasmids pCITE-2a, pUCIRES, and pGEM-4Z, respectively, all under the control of T7 RNA polymerase promoter. These plasmids were designated a pCITE-NP, pUCIRES-Z, and pGEM-L, respectively. To analyze plasmid-mediated intracellular expression of LCMV polypeptides, vTF7-3-infected cells were transfected with the corresponding plasmid DNAs, while cell extracts were prepared 20 to 24 h after transfection and analyzed by Western blotting using antibodies specific to NP, L, and Z polypeptides. Cells transfected with both pCITE-NP and pUCIRES-Z expressed proteins which specifically reacted with anti-NP and anti-Z antibodies, respectively (Fig. 3A and C). These proteins had molecular masses similar to those observed for NP and Z in extracts from cells infected with LCMV Arm (Fig. 3A and C, lanes 2 to 5). Cells transfected with pGEM-L expressed a polypeptide with an approximate apparent molecular mass of 200 kDa which was specifically recognized by an anti-L polyclonal antibody (Fig. 3B, lanes 2 to 4). In contrast, under the experimental conditions used here, BHK-21 cells infected with LCMV Arm did not show any specific protein which reacted with the anti-L polyclonal antibody (Fig. 3B, lane 5). This likely affected different levels of expression between transfected and infected cells. Expression levels of each LCMV polypeptide could be modulated by the amount of the corresponding DNA used for transfection. We next examined the amount of each NP and Z plasmid required to coexpress levels of NP and Z similar to those observed in infected cells; 1.2 and 0.4 μg of pCITE-NP and pUCIRES-Z, respectively, provided expression levels of NP and Z similar to those found in LCMV-infected cells at 24 h p.i. A similar comparison was not possible for L because its expression in LCMV-infected cells was below the sensitivity of our Western blot assay.
FIG. 3.
Plasmid-mediated expression of LCMV trans-acting factors. BHK-21 cells (4 × 105) were infected with vTF7-3 at an MOI of 3 and then transfected with pCITE-NP, pUCIRES-Z, and pGEM-L. Transfected cells were harvested at 24 h p.i. BHK-21 cells (2 × 105) were infected with LCMV at an MOI of 0.1 PFU/cell and harvested at 48 h p.i. Whole cell extracts were separated by 10, 16, and 6% SDS-PAGE for NP, Z, and L (top, middle, and bottom, respectively). Monoclonal antibody to NP and polyclonal antibody to Z and L were used for detection of LCMV proteins by Western blotting using chemiluminescence. Lanes: 1, cells infected with vTF7-3, 2, cells infected with vTF7-3 and transfected with 0.4 μg of either pCITE-NP, pUCIRES-Z, or pGEM-L; 3, BHK-21 cells infected with vTF7-3 and transfected with 1.2 μg of either pCITE-NP, pUCIRES-Z, or pGEM-L; 4, BHK-21 cells infected with vTF7-3 and cotransfected with 0.4 μg of each of plasmids pCITE-NP, pUCIRES-Z, and pGEM-L; 5, cells infected with LCMV at an MOI of 0.1. Antibodies used are indicated at the right.
NP and L are the minimal trans-acting factors required for efficient replication and transcription of LCMV RNA genome analogue.
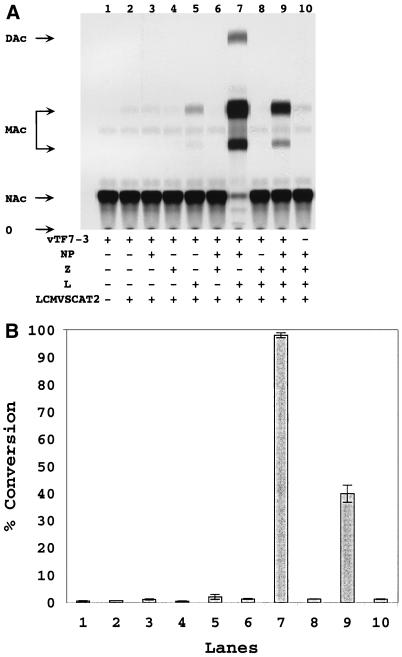
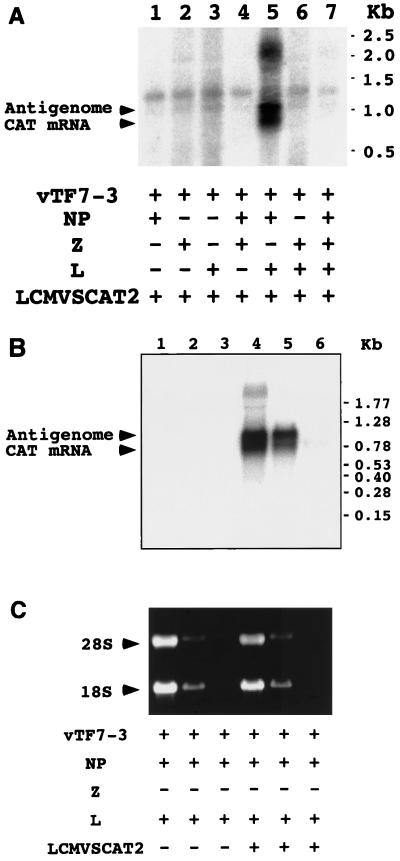
LCMV RNP isolated from infected cells can direct both replication and transcription of LCMV genome RNA in vitro (25). LCMV RNP contain NP and L but not GP-1 and GP-2 polypeptides (25). Whether Z is associated with RNP in RNA synthesis remains to be determined. However, Z has been implicated in both replication and transcription of the genome RNA of the arenavirus Tacaribe virus. Therefore, we reasoned that coexpression of NP, L, and Z would be required to reconstitute intracellular replication and transcription of LCMV minigenome. BHK-21 cells were infected with vTF7-3 and subsequently cotransfected with pLCMVSCAT2 and various combinations of plasmids encoding LCMV NP, Z, and L proteins. Sixteen to twenty hours after vTF7-3 infection, cell extracts were prepared and CAT activity was measured. It would be expected that CAT could be expressed only after formation of a functional LCMV RNP and subsequent transcription of the minigenome by the LCMV polymerase complex. Consistent with this hypothesis, CAT activity was not detected in cells infected with vTF7-3 and transfected only with the transcription plasmid pLCMVSCAT2 (Fig. 4A, lane 2). Plasmid-mediated expression of LCMV NP, L, and Z required vTF7-3 infection to provide T7 RNA polymerase. Accordingly, no CAT activity was detected in cells cotransfected with pLCMVSCAT2 and plasmids expressing NP, L, and Z in the absence of vTF7-3 infection (lane 10). Cells transfected only with NP or Z did not express detectable CAT activity (lanes 3 and 4). In contrast, and unexpectedly, cells transfected only with L showed detectable, although low, levels of CAT activity (lane 5). Percent conversion from [14C]chloramphenicol to its acetylated forms mediated by L alone was about 50-fold lower than that found in cells transfected with L plus NP (compare lanes 5 and 7 in Fig. 4B). No CAT activity was detected in cells cotransfected with NP and Z or L and Z (lanes 6 and 8), but high levels of CAT activities were obtained in cells cotransfected with NP and L (lane 7). These results indicated that NP and L are the minimal trans-acting viral factors required for efficient CAT expression mediated by the LCMV minigenome. Intriguingly, inclusion of Z together with L and NP caused a significant reduction in the level of CAT activity (lane 9).
FIG. 4.
Expression of LCMV minigenome in cells infected with vTF7-3 and transfected with pLCMVSCAT2 and LCMV NP, Z, and L protein expression plasmids. (A) Analysis of CAT activity. BHK-21 cells (4 × 105) were infected with vTF7-3 at an MOI of 3 PFU/cell and then transfected with various combination of plasmids pLCMVSCAT2, pCITE-NP, pUCIRES-Z, and pGEM-L as indicated in Materials and Methods. Cells were harvested at 24 h p.i. and analyzed for CAT activity by TLC as described in Materials and Methods. + and − indicate presence and the absence of plasmid or vTF7-3. NAc, MAc, and DAc indicate the positions of nonacetylated, monoacetylated, and diacetylated forms, respectively, of chloramphenicol in the TLC; O indicates the origin of the TLC. (B) CAT activities were quantified as described in Materials and Methods and presented as percent conversion from nonacetylated to acetylated forms of chloramphenicol. Values correspond to average ± standard deviation of three independent experiments.
We next examined whether detection of CAT activity correlated with synthesis of CAT mRNA derived from the LCMV minigenome and mediated by LCMV NP and L polypeptides. For this, BHK-21 cells infected with vTF7-3 were cotransfected with pLCMVSCAT2 and various combinations of plasmids encoding NP, L, and Z. Total RNA was extracted 16 to 20 h after infection with vTF7-3 and analyzed by Northern blot hybridization using an antisense CAT riboprobe. This riboprobe can hybridize to both the CAT mRNA and LCMVSCAT2 antigenomic predicted RNA species. Samples from cells cotransfected with pLCMVSCAT2 together with NP and L showed two RNA species that specifically hybridized to the antisense CAT riboprobe (Fig. 5A, lane 5). The sizes of these two RNA species, ca. 700 and 1,100 nt, were compatible with being the nonpolyadenylated CAT mRNA and antiminigenome RNA species, respectively, produced from the LCMVSCAT2 minigenome by the LCMV polymerase. Oligo(dT) fractionation confirmed that the two RNA species were nonpolyadenylated (Fig. 5B).
FIG. 5.
Analysis of RNA synthesized intracellularly by LCMV polymerase components. (A) Northern blot analysis of total cellular RNA. BHK-21 cells were infected with vTF7-3 at an MOI of 3 and then transfected with pLCMVSCAT2 and LCMV NP, Z, and L protein expression plasmids as for Fig. 4. Cells were harvested at 24 h p.i., and total RNA was isolated. RNA (5 μg) was analyzed by Northern blotting using an antisense CAT riboprobe. + and − indicate presence and absence of plasmid or vTF7-3. CAT mRNA and antigenome RNA are indicated by arrowheads on the left. (B) Oligo(dT) chromatography analysis. RNA isolated from BHK-21 cells infected with vTF7-3 transfected with the indicated plasmids was fractionated by oligo(dT) chromatography. Total (lanes 1 and 4), poly(A)− (lanes 2 and 5), and poly(A)+ (lanes 3 and 6) RNAs corresponding to equivalent amounts of cell numbers were analyzed by Northern blot hybridization using an antisense CAT riboprobe. (C) Ethidium bromide staining of the formaldehyde agarose gel prior transfer of the RNA samples to the membrane.
DISCUSSION
In this report, we have described a reverse genetic system that allows experimental investigation in vivo of the cis-acting signals and trans-acting factors involved in transcription and replication of LCMV.
In vitro transcription with T7 RNA polymerase of MvnI-linearized plasmid pLCMVSCAT1 generated a synthetic LCMV S RNA analog (LCMVSCAT1 RNA). CAT activity was detected upon transfection of LCMVSCAT1 RNA into LCMV-infected cells. Expression of CAT was strictly dependent on LCMV infection and was not obtained upon complementation with vTF7-3 or vesicular stomatitis virus (data not shown). CAT expression likely resulted from a combination of LCMVSCAT1 RNA primary transcription together with RNA amplication via replication followed by transcription of progeny LCMV minireplicon. This result provides direct experimental evidence to support the hypothesis that conserved sequences at the precise 3′ and 5′ termini of arenavirus genomic RNAs, together with the IGR, contain all of the cis-acting signals required for LCMV RNA synthesis. However, there is evidence that viral genomes with short deletions in their 5′ and 3′ UTRs are generated and accumulated during LCMV natural infection (38). It has been suggested that these truncated RNA genome molecules may play an important role in the outcome of the infection by modulating virus replication and transcription (38).
CAT expression was detected only in LCMV-infected cells transfected with LCMVSCAT1 RNA at the time when intracellular levels of viral NP were relative high (Fig. 2B). The requirement of certain threshold levels of NP or precise NP/Z/L ratios for efficient encapsidation of LCMVSCAT1 RNA might have contributed to this finding. NP expression levels at 24 h p.i. were higher in cells infected at an MOI of 0.1 than of 3 PFU/cell. The reasons for this unexpected finding are unknown. It is possible that the LCMV stock used for these experiments contained defective interfering (DI) particles which contributed to this phenomenon.
Levels of CAT activity in LCMV-infected cells transfected with LCMVSCAT1 RNA were consistently very low. It is worth noting that studies with bunyavirus and influenza virus, two other segmented negative-strand RNA viruses, have shown that naked vRNA analogs cannot be transcribed in virus-infected cells (8, 41), but that the viral trans-acting factors provided as recombinant proteins are required (54). Competition between authentic viral RNP and synthetic vRNA analog for polymerase factors has been proposed as one possible explanation. It is, however, intriguing that vRNA analogs of nonsegmented negative-strand RNA viruses do not appear to be equally affected by this competition.
Continuous plasmid-mediated intracellular synthesis of LCMV RNA analog would help to overcome difficulties intrinsic to RNA transfection and intracellular RNA instability. With this aim, we cloned the ribozyme sequences from the antigenomic strand of the hepatitis delta virus (HDV) downstream of the 3′ end of the pLCMVSCAT1 minireplicon, followed by T7 transcription termination sequence (45). The transcript generated intracellularly by T7 RNA polymerase contained the genomic polarity of HDV ribozyme which provided autolytic cleavage at the 5′ of the HDV ribozyme sequence, generating a 3′ end of the upstream RNA that corresponded to the authentic LCMV S RNA 3′ terminus (58). A similar approach has been used successfully with a variety of negative-strand RNA viruses (8, 19, 32, 41). However, we observed that less than 1% of the synthesized genomic RNA analog was autolytically processed. A possible explanation for this is that the last nucleotide at the 3′ end of the LCMV minigenome is guanosine (G) (52), whereas the efficiency of autolytic cleavage by HDV ribozyme follows the pattern C < U < A < G, with respect to the nucleotide located immediately 5′ to the site of cleavage (45). To solve this problem, we constructed plasmid pLCMVSCAT2, where an LCMV-specific HR was replaced for the HDV ribozyme (58). This ribozyme was designed to carry out self-cleavage, leaving in the upstream RNA the precise 3′ terminus of the LCMV S RNA. In this case we observed that 20 to 40% of the intracellularly transcribed vRNA analog was self-cleaved.
We also sought to establish a system that would facilitate the functional analysis of the trans-acting factors involved in LCMV RNA synthesis. To do so, we investigated whether the LCMVSCAT2 minigenome could serve as template for RNA synthesis mediated by LCMV proteins driven by T7 RNA polymerase produced by the recombinant vTF7-3. We demonstrated that LCMV NP, L, and Z proteins can be coexpressed using this system and that the expression levels of each polypeptide could be modulated by varying the amount of transfected plasmid DNA. This allows determination of the transfection conditions that closely resemble the levels and ratios of viral proteins found in infected cells.
Intracellular coexpression of the LCMVSCAT2 minigenome template and NP, L, and Z proteins resulted in high levels of CAT activity and synthesis of LCMVSCAT2 antiminigenome and subgenomic CAT mRNAs. These RNA species were readily detected by Northern blot using total cellular RNA. Studies with other negative-strand RNA viruses have provided compelling evidence that the template recognized by the viral polymerase is only encapsidated genome and antigenome RNA species. Therefore, it was unexpected to find that L alone was capable of mediating levels of CAT activity that were significantly higher than background levels. This finding suggests that L alone could mediate, though poorly, LCMV RNA synthesis. The biological significance of this observation remains to be determined. Presently, we cannot rule out the possibility that LCMV RNP-like structures are formed in vTF7-3-infected cells, thus providing a template that might be recognized, though very inefficiently, by L.
LCMV mRNAs are nonpolyadenylated (34). Therefore, a faithful recreation of the LCMV transcriptional activity would predict the mRNA of the CAT reporter gene to be also nonpolyadenylated. Vaccinia virus infection can mediate polyadenylation of heterologous mRNAs (30). However, oligo(dT) chromatography and Northern blot analysis showed that the two RNA species synthesized by the LCMV polymerase were nonpolyadenylated and had the sizes predicted for the subgenomic CAT mRNA and antiminigenome RNA. Interestingly, lack of polyadenylation did not prevent an efficient translation of the CAT mRNA. Whether LCMV-specific sequences corresponding to the 3′ UTR of this CAT mRNA contribute to its efficient translation is under investigation.
RNA replication by certain paramyxoviruses is efficient only if the nucleotide length of the genome is a multiple of six (“rule of six”) (10). This likely reflects a requisite association in the nucleocapsid of each NP monomer with exactly six nucleotides. This rule appears to be a feature of viruses which have RNA editing and might prevent the accumulation of genomes and antigenomes containing length changes. Based on the size of the LCMV minireplicon used in these studies, it appears that LCMV does not obey the rule of six, although a detailed analysis remains to be done.
Expression of CAT and synthesis of LCMVSCAT2 antiminigenome and subgenomic CAT RNA species did not require Z. Thus, NP and L constitute the minimal viral trans-acting factors required for RNA synthesis mediated by LCMV polymerase. The 11-kDa Z is a 90-amino-acid protein containing a zinc-binding domain (52). The role of Z in the life cycle of arenaviruses is poorly understood. Based on results from in vitro transcription combined with immunodepletion experiments of the Z protein from Tacaribe virus, it was proposed that Z is required for both genome replication and mRNA synthesis (29). The apparent discrepancies between these findings and our results could be related to possible differences between Tacaribe virus and LCMV with respect to Z functions. It is also possible that the antibody to Tacaribe virus Z protein used in the immunodepletion studies affected cellular factors required for virus RNA synthesis. For several negative-strand RNA viruses, the role of cellular components, especially proteins associated with the cytoskeleton, in virus replication and transcription has been documented (12).
Biochemical and immunological studies have shown that Z is a structural component of the virion, where it is apparently closely associated with NP (51). Therefore, immunodepletion of Z might have cause depletion of NP too. However, treatment of LCMV with nonionic detergents showed that Z partitions into the hydrophobic phase rather than remaining associated with the viral nucleocapsid. These findings have led to the proposal that Z might be the arenavirus counterpart of the matrix proteins found in other negative-strand RNA viruses. Moreover, Z has been shown to interact with the promyelocytic leukemia (PML) protein, leading to the relocation of PML nuclear bodies to the cytoplasm, which has been proposed to be responsible for the noncytolytic nature of LCMV (5, 6). Together, these observations raise intriguing questions about a spectrum of potential functions played by the Z protein in the biology of arenaviruses. The investigation of some of these questions will benefit from the use of the system described here.
Finally, our results also suggest that Z might have an inhibitory effect on replication and transcription of the LCMV minigenome. Detailed studies defining this inhibition are in progress. Precendents for proteins with roles as negative regulatory factors can be found in other negative-strand viruses. The RSV NS1 protein has been shown to be a potent inhibitor of transcription and replication of the RSV minigenome (1). In addition, the RSV M2 mRNA encodes a negative regulatory factor from its downstream ORF (14). The Sendai virus V protein was shown to inhibit the replication of a DI genome; whereas the Sendai virus C protein strongly inhibited the amplification of an internal deletion type of DI genome as well as a complete infectious genome (9, 33). It should be also considered that Z may play roles in vRNA synthesis that are not recreated in the minigenome system.
The capability to manipulate cis-acting RNA sequences and trans-acting proteins should facilitate studies of the molecular mechanisms of LCMV transcription and replication. Moreover, the system here described provides the foundation for the rescue of infectious LCMV entirely from plasmid DNA. The ability to generate predetermined specific mutations within the LCMV genome and analyze their phenotypic expression will significantly contribute to the elucidation of the molecular mechanisms underlying arenavirus-host interactions, including the bases of persistence and associated disease.
ACKNOWLEDGMENTS
I. S. Novella and M. N. Teng contributed equally to this work.
This work was supported by grants AI 09484 and AG 04342. I. S. Novella was supported by a fellowship from the Stanley Foundation.
We thank M. Salvato for the antibody to LCMV Z protein and P. Collins for helpful suggestions.
Footnotes
Publication 12708-NP from The Scripps Research Institute.
REFERENCES
- 1.Atreya P L, Peeples M E, Collins P L. The NS1 protein of human respiratory syncytial virus is a potent inhibitor of minigenome transcription and RNA replication. J Virol. 1998;72:1452–1461. doi: 10.1128/jvi.72.2.1452-1461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auperin D D, Compans R W, Bishop D H. Nucleotide sequence conservation at the 3′ termini of the virion RNA species of New World and Old World arenaviruses. Virology. 1982;121:200–203. doi: 10.1016/0042-6822(82)90130-1. [DOI] [PubMed] [Google Scholar]
- 3.Auperin D D, Romanowski V, Galinski M, Bishop D H. Sequencing studies of pichinde arenavirus S RNA indicate a novel coding strategy, an ambisense viral S RNA. J Virol. 1984;52:897–904. doi: 10.1128/jvi.52.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auperin D D, Sasso D R, McCormick J B. Nucleotide sequence of the glycoprotein gene and intergenic region of the Lassa virus S genome RNA. Virology. 1986;154:155–167. doi: 10.1016/0042-6822(86)90438-1. [DOI] [PubMed] [Google Scholar]
- 5.Borden K L, Campbell Dwyer E J, Salvato M S. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J Virol. 1998;72:758–766. doi: 10.1128/jvi.72.1.758-766.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borden K L, Campbell Dwyer E J, Carlile G W, Djavani M, Salvato M S. Two RING finger proteins, the oncoprotein PML and the arenavirus Z protein, colocalize with the nuclear fraction of the ribosomal P proteins. J Virol. 1998;72:3819–3826. doi: 10.1128/jvi.72.5.3819-3826.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrow P, Oldstone M B. Mechanism of lymphocytic choriomeningitis virus entry into cells. Virology. 1994;198:1–9. doi: 10.1006/viro.1994.1001. [DOI] [PubMed] [Google Scholar]
- 8.Bridgen A, Elliott R M. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc Natl Acad Sci USA. 1996;93:15400–15404. doi: 10.1073/pnas.93.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, Curran J, Kolakofsky D. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70:5067–5074. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao W, Henry M D, Borrow P, Yamada H, Elder J H, Ravkov E V, Nichol S T, Compans R W, Campbell K P, Oldstone M B. Identification of alpha-dystroglycan as a receptor for lymphocytic choriomeningitis virus and Lassa fever virus. Science. 1998;282:2079–2081. doi: 10.1126/science.282.5396.2079. [DOI] [PubMed] [Google Scholar]
- 12.Ciampor F. The role of cytoskeleton and nuclear matrix in virus replication. Acta Virol. 1988;32:168–189. [PubMed] [Google Scholar]
- 13.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins P L, Hill M G, Cristina J, Grosfeld H. Transcription elongation factor of respiratory syncytial virus, a nonsegmented negative-strand RNA virus. Proc Natl Acad Sci USA. 1996;93:81–85. doi: 10.1073/pnas.93.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collins P L, Mink M A, Stec D S. Rescue of synthetic analogs of respiratory syncytial virus genomic RNA and effect of truncations and mutations on the expression of a foreign reporter gene. Proc Natl Acad Sci USA. 1991;88:9663–9667. doi: 10.1073/pnas.88.21.9663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conzelmann K K. Genetic manipulation of non-segmented negative-strand RNA viruses. J Gen Virol. 1996;77:381–389. doi: 10.1099/0022-1317-77-3-381. [DOI] [PubMed] [Google Scholar]
- 17.Conzelmann K K. Nonsegmented negative-strand RNA viruses: genetics and manipulation of viral genomes. Annu Rev Genet. 1998;32:123–162. doi: 10.1146/annurev.genet.32.1.123. [DOI] [PubMed] [Google Scholar]
- 18.Conzelmann K K, Schnell M. Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J Virol. 1994;68:713–719. doi: 10.1128/jvi.68.2.713-719.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De B P, Banerjee A K. Rescue of synthetic analogs of genome RNA of human parainfluenza virus type 3. Virology. 1993;196:344–348. doi: 10.1006/viro.1993.1486. [DOI] [PubMed] [Google Scholar]
- 20.Dimock K, Collins P L. Rescue of synthetic analogs of genomic RNA and replicative-intermediate RNA of human parainfluenza virus type 3. J Virol. 1993;67:2772–2778. doi: 10.1128/jvi.67.5.2772-2778.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elroy-Stein O, Fuerst T R, Moss B. Cap-independent translation of mRNA conferred by encephalomyocarditis virus 5′ sequence improves the performance of the vaccinia virus/bacteriophage T7 hybrid expression system. Proc Natl Acad Sci USA. 1989;86:6126–6130. doi: 10.1073/pnas.86.16.6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elroy-Stein O, Moss B. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc Natl Acad Sci USA. 1990;87:6743–6747. doi: 10.1073/pnas.87.17.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francis S J, Southern P J. Molecular analysis of viral RNAs in mice persistently infected with lymphocytic choriomeningitis virus. J Virol. 1988;62:1251–1257. doi: 10.1128/jvi.62.4.1251-1257.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fuller-Pace F V, Southern P J. Detection of virus-specific RNA-dependent RNA polymerase activity in extracts from cells infected with lymphocytic choriomeningitis virus: in vitro synthesis of full-length viral RNA species. J Virol. 1989;63:1938–1944. doi: 10.1128/jvi.63.5.1938-1944.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuller-Pace F V, Southern P J. Temporal analysis of transcription and replication during acute infection with lymphocytic choriomeningitis virus. Virology. 1988;162:260–263. doi: 10.1016/0042-6822(88)90419-9. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Sastre A. Negative-strand RNA viruses: applications to biotechnology. Trends Biotechnol. 1998;16:230–235. doi: 10.1016/s0167-7799(98)01192-5. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Sastre A, Palese P. Genetic manipulation of negative-strand RNA virus genomes. Annu Rev Microbiol. 1993;47:765–790. doi: 10.1146/annurev.mi.47.100193.004001. [DOI] [PubMed] [Google Scholar]
- 29.Garcin D, Rochat S, Kolakofsky D. The Tacaribe arenavirus small zinc finger protein is required for both mRNA synthesis and genome replication. J Virol. 1993;67:807–812. doi: 10.1128/jvi.67.2.807-812.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gershon P D, Ahn B Y, Garfield M, Moss B. Poly(A) polymerase and a dissociable polyadenylation stimulatory factor encoded by vaccinia virus. Cell. 1991;66:1269–1278. doi: 10.1016/0092-8674(91)90048-4. [DOI] [PubMed] [Google Scholar]
- 31.Gorman C M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He B, Paterson R G, Ward C D, Lamb R A. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- 33.Horikami S M, Smallwood S, Moyer S A. The Sendai virus V protein interacts with the NP protein to regulate viral genome RNA replication. Virology. 1996;222:383–390. doi: 10.1006/viro.1996.0435. [DOI] [PubMed] [Google Scholar]
- 34.Iapalucci S, Lopez N, Franze-Fernandez M T. The 3′ end termini of the Tacaribe arenavirus subgenomic RNAs. Virology. 1991;182:269–278. doi: 10.1016/0042-6822(91)90670-7. [DOI] [PubMed] [Google Scholar]
- 35.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Lawson N D, Stillman E A, Whitt M A, Rose J K. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. . (Erratum, 92:9009.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mena I, Vivo A, Perez E, Portela A. Rescue of a synthetic chloramphenicol acetyltransferase RNA into influenza virus-like particles obtained from recombinant plasmids. J Virol. 1996;70:5016–5024. doi: 10.1128/jvi.70.8.5016-5024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meyer B J, Southern P J. A novel type of defective viral genome suggests a unique strategy to establish and maintain persistent lymphocytic choriomeningitis virus infections. J Virol. 1997;71:6757–6764. doi: 10.1128/jvi.71.9.6757-6764.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyer B J, Southern P J. Sequence heterogeneity in the termini of lymphocytic choriomeningitis virus genomic and antigenomic RNAs. J Virol. 1994;68:7659–7664. doi: 10.1128/jvi.68.11.7659-7664.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nathaanson N. Viral pathogenesis. Philadelphia, Pa: Lippincott-Raven; 1997. [Google Scholar]
- 41.Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez D R, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci USA. 1999;96:9345–9350. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oldstone M B, Ahmed R, Byrne J, Buchmeier M J, Riviere Y, Southern P. Virus and immune responses: lymphocytic choriomeningitis virus as a prototype model of viral pathogenesis. Br Med Bull. 1985;41:70–74. doi: 10.1093/oxfordjournals.bmb.a072029. [DOI] [PubMed] [Google Scholar]
- 43.Palese P. Genetic engineering of infectious negative-strand RNA viruses. Trends Microbiol. 1995;3:123–125. doi: 10.1016/s0966-842x(00)88897-6. [DOI] [PubMed] [Google Scholar]
- 44.Pekosz A, He B, Lamb R A. Reverse genetics of negative-strand RNA viruses: closing the circle. Proc Natl Acad Sci USA. 1999;96:8804–8806. doi: 10.1073/pnas.96.16.8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perrotta A T, Been M D. The self-cleaving domain from the genomic RNA of hepatitis delta virus: sequence requirements and the effects of denaturant. Nucleic Acids Res. 1990;18:6821–6821. doi: 10.1093/nar/18.23.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, Christiansen G, Billeter M A. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riviere Y, Ahmed R, Southern P J, Buchmeier M J, Dutko F J, Oldstone M B. The S RNA segment of lymphocytic choriomeningitis virus codes for the nucleoprotein and glycoproteins 1 and 2. J Virol. 1985;53:966–968. doi: 10.1128/jvi.53.3.966-968.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Romanowski V, Bishop D H. Conserved sequences and coding of two strains of lymphocytic choriomeningitis virus (WE and ARM) and Pichinde arenavirus. Virus Res. 1985;2:35–51. doi: 10.1016/0168-1702(85)90058-9. [DOI] [PubMed] [Google Scholar]
- 49.Rose J K. Positive strands to the rescue again: a segmented negative-strand RNA virus derived from cloned cDNAs. Proc Natl Acad Sci USA. 1996;93:14998–5000. doi: 10.1073/pnas.93.26.14998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salvato M, Shimomaye E, Oldstone M B. The primary structure of the lymphocytic choriomeningitis virus L gene encodes a putative RNA polymerase. Virology. 1989;169:377–384. doi: 10.1016/0042-6822(89)90163-3. [DOI] [PubMed] [Google Scholar]
- 51.Salvato M S, Schweighofer K J, Burns J, Shimomaye E M. Biochemical and immunological evidence that the 11 kDa zinc-binding protein of lymphocytic choriomeningitis virus is a structural component of the virus. Virus Res. 1992;22:185–198. doi: 10.1016/0168-1702(92)90050-j. [DOI] [PubMed] [Google Scholar]
- 52.Salvato M S, Shimomaye E M. The completed sequence of lymphocytic choriomeningitis virus reveals a unique RNA structure and a gene for a zinc finger protein. Virology. 1989;173:1–10. doi: 10.1016/0042-6822(89)90216-x. [DOI] [PubMed] [Google Scholar]
- 53.Schnell M J, Mebatsion T, Conzelmann K K. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seong B L, Brownlee G G. A new method for reconstituting influenza polymerase and RNA in vitro: a study of the promoter elements for cRNA and vRNA synthesis in vitro and viral rescue in vivo. Virology. 1992;186:247–260. doi: 10.1016/0042-6822(92)90079-5. [DOI] [PubMed] [Google Scholar]
- 55.Southern P J, Singh M K, Riviere Y, Jacoby D R, Buchmeier M J, Oldstone M B. Molecular characterization of the genomic S RNA segment from lymphocytic choriomeningitis virus. Virology. 1987;157:145–155. doi: 10.1016/0042-6822(87)90323-0. [DOI] [PubMed] [Google Scholar]
- 56.Wilson S M, Clegg J C. Sequence analysis of the S RNA of the African arenavirus Mopeia: an unusual secondary structure feature in the intergenic region. Virology. 1991;180:543–552. doi: 10.1016/0042-6822(91)90068-m. [DOI] [PubMed] [Google Scholar]
- 57.Wright K E, Spiro R C, Burns J W, Buchmeier M J. Post-translational processing of the glycoproteins of lymphocytic choriomeningitis virus. Virology. 1990;177:175–183. doi: 10.1016/0042-6822(90)90471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xing Z, Whitton J L. Ribozymes which cleave arenavirus RNAs: identification of susceptible target sites and inhibition by target site secondary structure. J Virol. 1992;66:1361–1369. doi: 10.1128/jvi.66.3.1361-1369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]