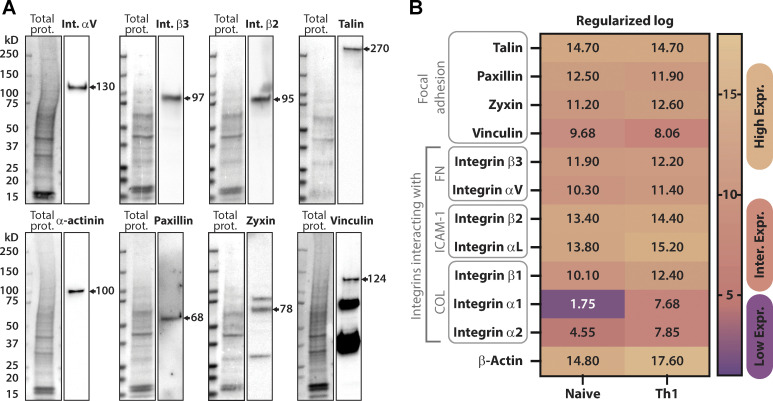
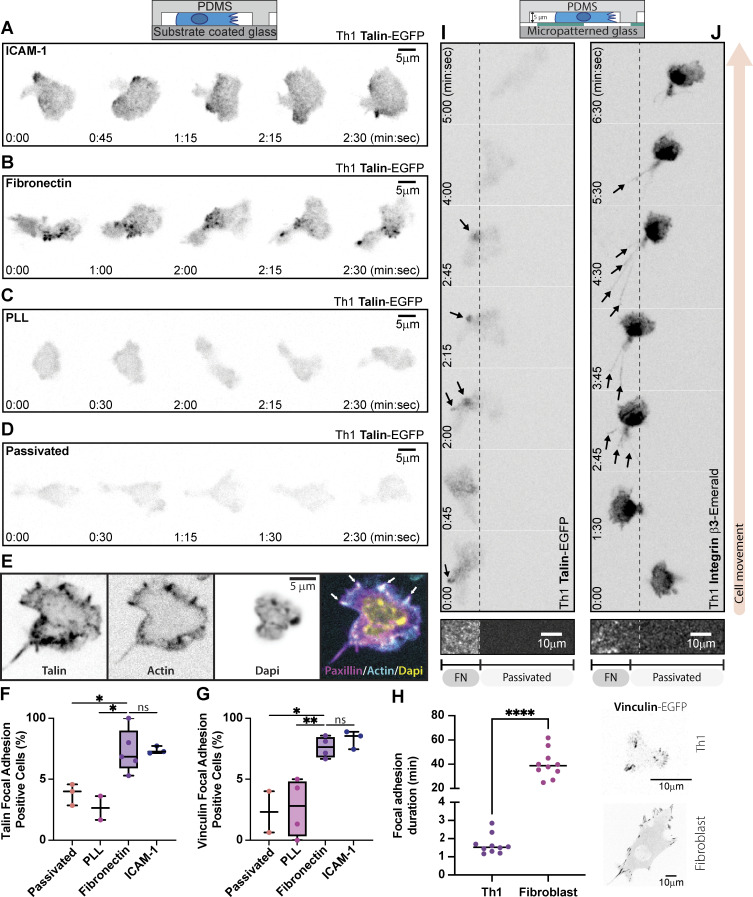
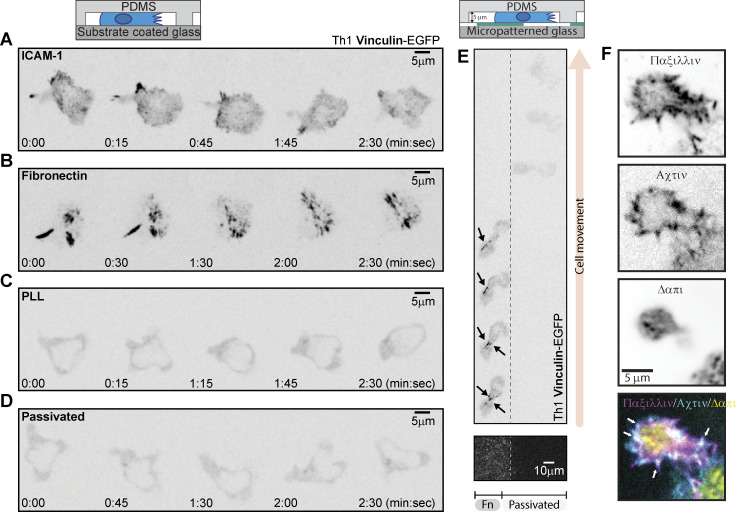
T cells form focal adhesions consisting of integrins, talin, and vinculin when migrating on extracellular matrix proteins. Like mesenchymal cells, these adhesions are sites of contractile force transmission, and T cells use them to facilitate their navigation through complex environments.
Abstract
Immune cells are highly dynamic and able to migrate through environments with diverse biochemical and mechanical compositions. Their migration has classically been defined as amoeboid under the assumption that it is integrin independent. Here, we show that activated primary Th1 T cells require both confinement and extracellular matrix proteins to migrate efficiently. This migration is mediated through small and dynamic focal adhesions that are composed of the same proteins associated with canonical mesenchymal cell focal adhesions, such as integrins, talin, and vinculin. These focal adhesions, furthermore, localize to sites of contractile traction stresses, enabling T cells to pull themselves through confined spaces. Finally, we show that Th1 T cells preferentially follow tracks of other T cells, suggesting that these adhesions modify the extracellular matrix to provide additional environmental guidance cues. These results demonstrate not only that the boundaries between amoeboid and mesenchymal migration modes are ambiguous, but that integrin-mediated focal adhesions play a key role in T cell motility.
Graphical Abstract
Introduction
Migration is a critical component of an efficient immune response. Cells must be ready to respond to diverse signaling cascades and then physically move to various sites throughout the body in response to those signals. While biochemical signals such as chemokines have long been associated with stimulating and facilitating migration, more recent studies have implicated both passive and active biophysical signals as playing equally integral roles (Gaylo et al., 2016; Fowell and Kim, 2021; Tello-Lafoz et al., 2021; Du et al., 2023). Indeed, passive features of the extracellular environment, including its stiffness (Lo et al., 2000; Bangasser et al., 2017; Janmey et al., 2020; Isomursu et al., 2022), composition (Ramos et al., 2016; Hartman et al., 2017), and architecture (Pieuchot et al., 2018; Tien et al., 2020; Nicolas-Boluda et al., 2021) can all influence cell migration (Pathak and Kumar, 2012; Gaylo et al., 2016). Active forces generated internally by the cell and surrounding cells in the tissue can also have an impact (Boyd et al., 1988; Reiss et al., 1998; Zhang et al., 2006; Chen and Zhu, 2013), such as by modulating bond strength and lifetime. Immune cells, which must navigate a vast environmental complexity—from the lymphatic system to the circulatory system and then the surrounding tissues—are particularly well positioned to display adaptive migration mechanisms depending on their given environment.
Much of our foundational knowledge of the molecular mechanisms of migration is derived from studying mesenchymal cells on account of their large, flat morphology, and reasonably slow dynamics (Svitkina, 2018). This combination makes them ideal for high-resolution microscopy. Mesenchymal migration is characterized by a narrow band of actin polymerization at a leading edge, the formation of large integrin-mediated focal adhesion (FA) plaques, and bundled actomyosin stress fibers in the cell body (Gardel et al., 2010). In contrast, immune cells have typically been described as navigating their complex environment using an amoeboid mode of migration (Friedl et al., 1998; Paluch et al., 2016). Amoeboid migration is characterized by a more rounded cell morphology and a dependence on rapid bulk actin polymerization (Paluch et al., 2016). While the term amoeboid was first used to describe migration in amoeba-like Dictyostelium, it has generally evolved to act as a catch-all for migration that is not integrin-mediated (Paluch et al., 2016). The molecular mechanisms and mechanics mediating this form of migration have remained unclear but are generally ascribed to non-specific adhesion and friction forces between the cell and the surrounding environment, enabled by internal fluid flows (Pinner and Sahai, 2009; Lämmermann and Germain, 2014; Bergert et al., 2015; Callan-Jones, 2022; Schick and Raz, 2022).
T cells and other leukocytes express a number of different integrins, including those that bind extracellular matrix (ECM) proteins like fibronectin (FN) and collagen, and cell adhesion proteins like intercellular adhesion molecule 1 (ICAM-1), (Lämmermann et al., 2008; Bertoni et al., 2018; Gaylo-Moynihan et al., 2019). This diverse array of binding proteins should be useful for cells navigating the many complex extracellular environments encountered during an immune response. It was thus surprising when it was shown that dendritic cells could migrate effectively in 3D, but not 2D, geometries when all integrins were knocked out (Lämmermann et al., 2008). Additional works have suggested that integrin-independent migration could arise from non-specific friction generated via the flow of the actin cytoskeleton (Hons et al., 2018; Reversat et al., 2020), which could facilitate immune cell migration through tissue and interstitial spaces after crossing the endothelial layer (Gaertner et al., 2022). In other contexts, however, integrin-mediated interactions have been shown to play crucial roles in immune migration (Hogg et al., 2003; Huse, 2017). Integrin–collagen interactions have previously been shown to be essential for efficient immune response in several mouse models (de Fougerolles et al., 2000; Ray et al., 2004), while LFA-1 plays a crucial role in T cells leaving the lymph node (Reichardt et al., 2013). It was also shown that helper T cells overexpress FN-specific integrins following their activation and that those integrins are required for migration in vivo (Gaylo-Moynihan et al., 2019). More specifically, CD4+ T cell interstitial migration is highly dependent on αv integrin adhesion with the ECM (Overstreet et al., 2013; Fernandes et al., 2020). Integrins have also been shown to be crucial for migration in other immune cells, including mast cells (Kaltenbach et al., 2023) and macrophages (Paterson and Lämmermann, 2022). This array of results demonstrates a need for a deeper molecular and mechanical understanding of how immune cells interact with their extracellular environment during migration (Huse, 2017; Moreau et al., 2018).
In this paper, we demonstrate that primary Th1 T cells form FAs, comprised of integrins and adhesion proteins like talin, and that their formation depends on the extracellular environment composition and geometry. We find that Th1 cells require integrin-mediated adhesion to migrate, and in the case of the FN, cells additionally require confinement to move efficiently. When Th1 cells make FAs, they are consistent in composition to canonical mesenchymal FAs and are sites where cytoskeletal contractile forces are transmitted to the ECM. These findings challenge the traditional framing of T cell migration as both amoeboidal and integrin-independent. Finally, we show that T cells tend to follow in the tracks of other T cells, suggesting additional functional roles for FAs in enabling a rapid and directed multicellular immune response.
Results and discussion
Th1 migration is regulated by environment composition and geometry
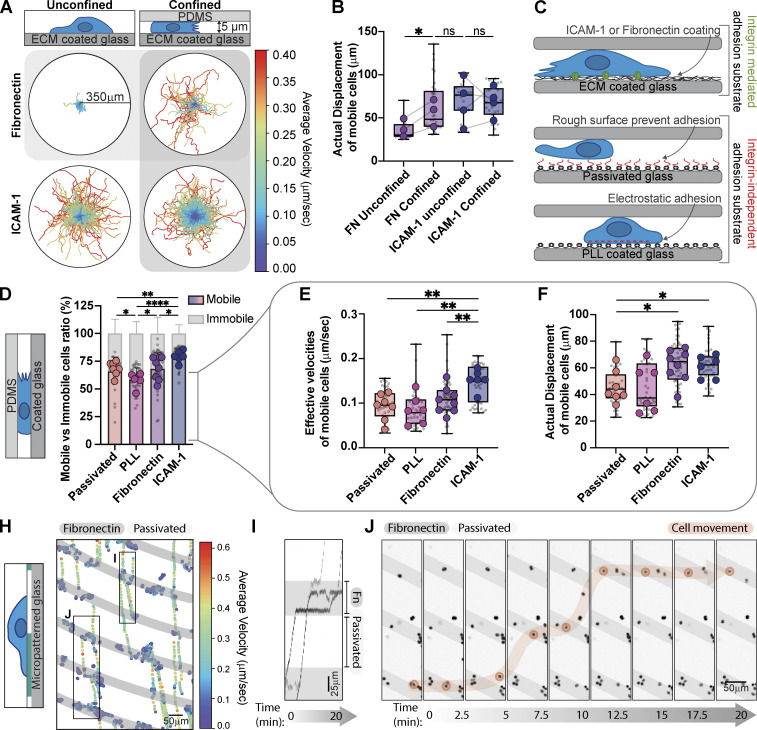
Using activated primary Th1 cells isolated from OTII mice (Fig. S1, A–F), we first tested whether T cells could migrate on glass substrates coated with either FN (likely to be found in the tissue ECM and a ligand for both αvβ3 and α5β1) or ICAM-1 (found on the surface of other cells and a ligand for αLβ2). As expected, we found that Th1 cells migrated fast and efficiently on ICAM-1 (Fig. 1, A and B). On FN, in contrast, cells exhibited Brownian motion and no migration, only transiently interacting with the coverslip surface (Fig. 1, A and B). In vivo, however, cells are restricted by the confinement of the local environment. We, therefore, confined them under a PDMS surface with a gap of 5 µm, which was sufficient to keep them in place without deforming their nucleus. When confined, Th1 cells migrated robustly on both FN and ICAM-1 with speeds and displacements that were indistinguishable from unconfined Th1 cells migrating on ICAM-1 (Fig. 1, A and B).
Figure S1.
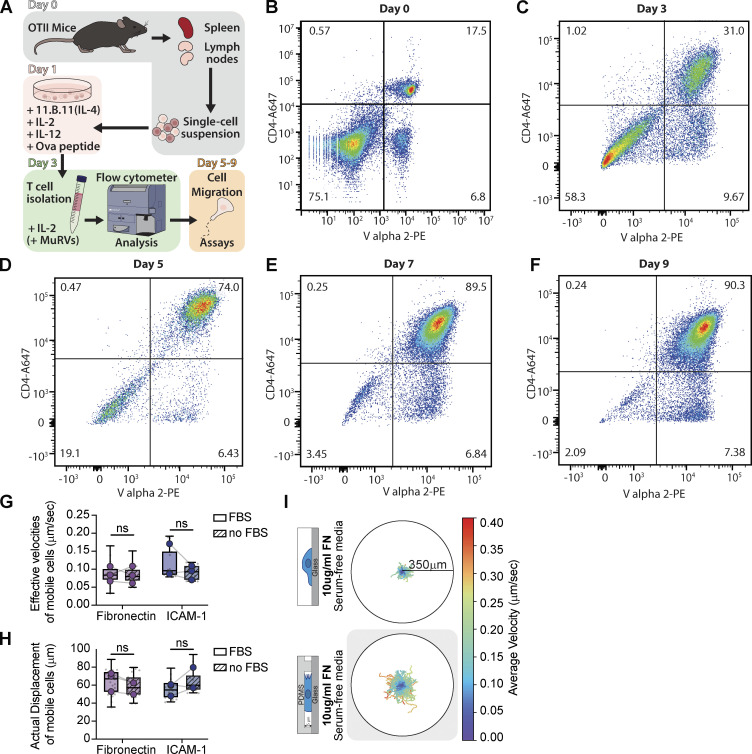
Th1 cell activation protocol and serum-free migration dynamics. (A) Graphical representation of Th1 activation. (B) Flow cytometer analysis of a representative single-cell suspension on the day of extraction from OTII mice using CD4 and Vα2 antibodies as markers. (C) A representative flow cytometer analysis of T cells three days post activation with anti-IL-4 (11B11; 40 µg/ml), IL-2 (20 U/ml), IL-12 (40 ng/ml), OVA peptide (4 µg/ml). T cells were isolated using lymphocyte separation media. (D–F) Representative flow cytometer analyses of Th1 T cells after 5 (D), 7 (E), and 9 (F) days post activation, with continuous treatment of 10 units/ml IL-2 and lymphocyte separation media before each use. (G and H) The actual displacement (G) and effective velocities (H) of mobile cells on fibronectin and ICAM-1 substrate with and without FBS in cell media. (I) Th1 cell tracking in serum-free media on Fibronectin in unconfined versus confined (5 µm) environments. The colormap represents the cell average velocity in µm/sec. Number of replicates for G and H: All conditions n = 3. Samples with and without FBS were performed with the same preparation of cells, on the same day, from the same mice. Each replicate represents the average of 5–10 fields of view (gray dots = average of each field of view), which contain ∼250 cells. Statistical test: one-tail paired parametric t test.
Figure 1.
Th1 migration, velocity, and displacement are regulated by the environment composition and geometry. (A) Th1 cell tracking on ICAM-1 and FN in unconfined versus confined (5 µm) environments. The colormap represents the cell's average velocity in µm/sec. (B) Comparison of the actual displacement, defined as the distance between the starting and end points of a track, of the mobile cells in A. (C) Graphical representation of the different substrates used and their properties. (D) The mobile versus immobile fraction of cells in confinement on ICAM-1, FN, PLL, and Passivated surfaces. Mobile cells are defined by a minimum displacement of 10 µm. (E and F) The effective velocities (E) and actual displacement (F) of mobile cells in D. (H) Cell tracking on a FN micropatterned substrate. Gray = FN, White = Passivated. The colors represent the instantaneous velocities of each cell at a given time point. (I) Kymographs of the regions identified in H show cells stalling on the FN patterned stripes. (J) Snapshots of the movie from H. A single cell is highlighted in orange to illustrate its trajectory over time. Number of biological replicates (colored dots): (B) FN, n = 3; ICAM-1, n = 4. (D-F) FN, n = 8; ICAM-1, n = 6; PLL, n = 6; Passivated, n = 7. Each replicate represents the average of 5–10 fields of view (gray dots = average of each field of view), which each contain ∼250 cells. Statistical tests: (B) one-tail paired parametric t test; (D–F) two-tailed unpaired parametric t test.
To explore this further, we compared the integrin-mediated migration of Th1 cells confined to either FN or ICAM-1, with integrin-independent migration of Th1 cells confined to surfaces coated either with electrostatically charged polymers (PolyL-lysine; PLL) or inert blocking polymers (PLL-PEG or PMOXA; passivated) (Fig. 1 C). We found that over 75% of cells were mobile on ICAM-1, while only ∼50% were mobile on PLL, with cells on FN or passivated surfaces in between (Fig. 1 D and Video 1). If we compared the effective velocity and displacement of the mobile fraction, cells plated on integrin-mediated surfaces moved larger distances than those on integrin-independent surfaces and were fastest on ICAM-1 (Fig. 1, E and F and Video 1).
Video 1.
Th1 cells stained with Hoescht migrate in confined (5 μm) environments coated with ICAM-1, Fibronectin, PLL, or passivated. 10× objective. Frame rate = 15 frames/s. Movie from Fig. 1, D–F.
To confirm that Th1 cells directly interact with FN, we used micropatterning to create stripes of FN in an otherwise passivated surface (Fig. 1 H). Convective flows induced by imaging multiple positions allowed us to track the motion of unconfined cells as they crossed different regions. We found that Th1 cells slowed down and clustered along the FN-coated regions (Fig. 1 H and Video 2), lingering for brief periods of time before detaching and moving on to the next region (Fig. 1, I and J and Video 2). Together, these results indicate that Th1 cells are able to migrate effectively on FN when under confinement and that this migration is mediated through specific interaction with FN.
Video 2.
Left panel: Th1 cells stained with Hoescht migrating on an FN micropatterned substrate (light gray = FN, darker gray = Passivated). Right panel: Cell tracking of the right panel. The colormap represents the cell's average velocity in μm/sec. 10× objective. Frame rate = 15 frames/s. Movie from Fig. 1, H–J.
ECM is necessary for Th1 migration
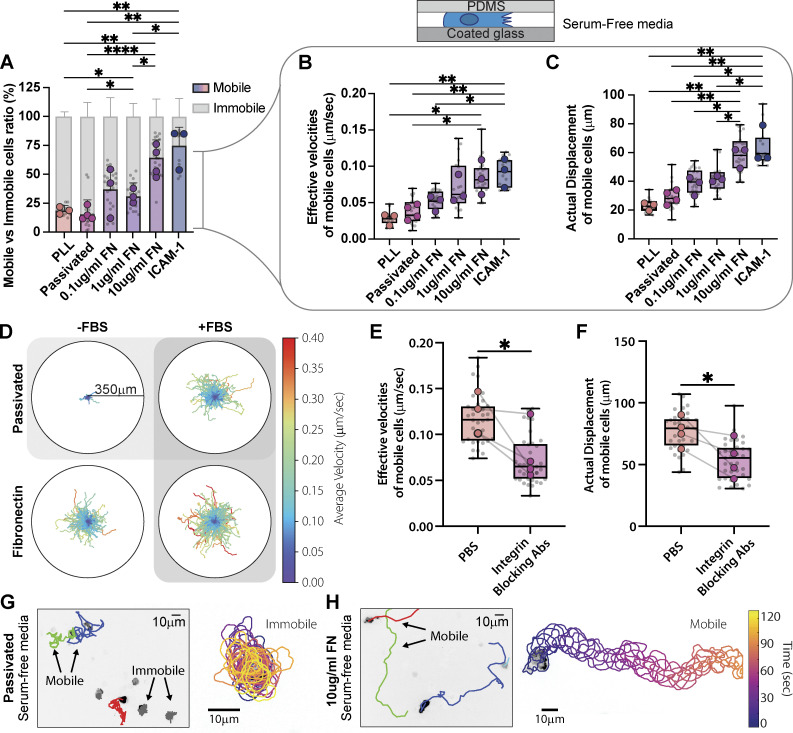
Our initial experiments suggested that Th1 T cells could engage both integrin-mediated and integrin-independent migration modes, although with varying degrees of efficacy when confined on different substrates. Fully passivating a surface is challenging, however, and previous results have shown that these surfaces are still able to adsorb many proteins present in serum, such as ECM (Walkey et al., 2012). To ensure that the ECM present in cell culture media serum (Hayman and Ruoslahti, 1979; Cheng et al., 2020) does not bind to our passivated surfaces and contribute to Th1 migration, we repeated our confined migration experiments, switching the cells to serum-free media immediately prior to confinement (Fig. 2 A). Surprisingly, the mobile fraction dropped precipitously for cells on PLL or passivated substrates (Fig. 2, A and D), and cells exhibited significantly reduced velocities and displacements (Fig. 2, B and C). Th1 cells confined on both 10 µg/ml FN and ICAM-1 in serum-free media, however, exhibited similar mobile fractions (Fig. 2 A and Fig. 1 D), velocities, and displacements as cells plated in full serum media (Fig. 2, B–D; Fig. 1, E and F; Fig. S1, G and H; and Video 3). Moreover, this effect was titratable since the mobile fraction, velocity, and displacement were proportional to the concentration of FN (Fig. 2, A–C). To test whether this migration was integrin-dependent, we incubated the cells with a combination of antibodies specific for αv, β1, β2, and β3 to block integrin binding (Gaylo-Moynihan et al., 2019) and measured the cell velocity and displacement in our migration assay. Cells treated with the integrin-blocking antibodies showed significantly decreased velocities and displacements compared with controls (Fig. 2, E and F). High-resolution imaging of cells migrating in serum-free media on a passivated surface revealed membrane ruffles and protrusions, indicating that cells were remaining active with short-term serum depletion but failing to migrate on the integrin-independent surface (Fig. 2, G and H; and Video 4). Cells migrating on FN-coated surfaces in serum-free media also presented active membrane ruffles and protrusions (Fig. 2, G and H; and Video 4), but the absence of serum had no significant impact on their migration in speed or displacement (Fig. S1, G and H). To confirm that confinement alone impacted Th1 migration on FN, we repeated the experiment in Fig. 1 B, but in serum-free media (Fig. S1 I). Th1 cells in serum-free media still migrated further and faster on FN in confinement than when unconfined. Together these results suggest that integrin-dependent substrates are necessary for robust Th1 migration.
Figure 2.
ECM is essential for Th1 migration. (A) The percentage of mobile versus immobile Th1 cells in confinement on ICAM-1, FN, PLL, and passivated surface in serum-free media. Mobile cells are defined by a minimum displacement of 10 µm. (B and C) Analysis of the (B) effective velocities and the (C) actual displacement of mobile cells in A. (D) Cell tracks on passivated substrates versus on FN in either serum-free media (−FBS) or with serum (+FBS). The colors represent the cell’s average velocity. (E and F) Effective velocities (E) and actual displacement (F) of cells pretreated for 3 h with a mixture of integrin-blocking antibodies compared to control (PBS). (G and H) Tracking of cells and representative trajectories of cells on (G) passivated versus (H) FN-coated substrates in serum-free media. The cell outlines are color-coded for time. Number of biological replicates (colored dots): (A–C) PLL n = 3; Passivated n = 4; 0.1 µg/ml FN n = 3; 1 µg/ml FN n = 3; 10 µg/ml FN n = 4. (E–F) PBS n = 4; Ab-integrin n = 4. Each replicates represent the average of 5–10 fields of view (gray dots = average of each fields of view), which each contain ∼250 cells. Statistical tests: (A–C) Two-tailed unpaired parametric t test; (E and F) One-tail unpaired parametric t test.
Video 3.
Cell tracking of Th1 cells stained with Hoescht migrating in confined (5 μm) environments coated with Fibronectin or Passivated in serum-free media (-FBS) or FBS-supplemented media (+FBS). 10× objective. Frame rate = 15 frames/s. Movie from Fig. 2 D.
Video 4.
Cell tracking of Th1 Lifeact-EGFP cells migrating in confined (5 μm) environments coated with Fibronectin or passivated in serum-free media (−FBS). 63× objective. Frame rate = 10 frames/s. Movie from Fig. 2, G and F.
Th1 cells form focal adhesions
The necessity of integrin-dependent substrates for migration suggests that Th1 T cells form canonical integrin-based adhesions. Western blots of lysates from activated Th1 demonstrate the presence of FA components at the protein level (Fig. S2 A). RNA-seq data from a publicly available database (Th-express.org; Stubbington et al., 2015) corroborate that activated Th1 express RNA for canonical FA proteins (e.g., talin, paxillin, zyxin, vinculin), in addition to the integrins required to bind ECM (Fig. S2 B). To determine whether these proteins coalesce into actual FAs, we imaged Th1 cells transduced with either talin-EGFP (Fig. 3, A–D and Video 5) or vinculin-EGFP (Fig. S3, A–D). On both ICAM-1 (Fig. 3 A and Fig. S3 A) and FN (Fig. 3 B and Fig. S3 B), we observed highly dynamic FA-like structures (Video 5), whereas we were unable to observe any such structures on PLL (Fig. 3 C; Fig. S3 C; and Video 5) or passivated substrates (Fig. 3 D; Fig. S3 D; and Video 5). Endogenous antibody staining of talin (Fig. 3 E) or paxillin (Fig. S3 F) in cells confined on FN revealed similar FA structures colocalizing with actin enrichments around the cell periphery. Quantification of the number of cells containing these types of structures in live movies of cells expressing Talin-EGFP or vinculin-EGFP shows a significant increase in the number of FA-positive cells when on FN and ICAM-1 surfaces (Fig. 3, F and G). As a comparison, we next measured FA lifetime in Th1 cells expressing Vinculin-eGFP compared with the same constructs in a mouse fibroblast cell line. We found that the FAs in Th1 T cells were significantly shorter-lived compared with the fibroblast, with an average lifetime of only 1.7 min compared with 40 min in the fibroblasts (Fig. 3 H).
Figure S2.
Focal adhesion protein expression in Th1 cells. (A) Representative Western blots showing endogenous expression of various integrin and FA proteins in activated Th1 cells. (B) RNA-sequencing comparison of naive versus activated T cells (Th1) from the publicly available dataset (https://Th-express.org; Stubbington et al., 2015). Genes shown include FA-related proteins, fibronectin-binding integrins, ICAM-1-binding integrins as well as collagen-binding integrins. Their relative levels of expression were calculated from z-log (see Materials and methods) transformed gene expression counts for all genes and presented using a colormap to show high, intermediate to low levels of expression. Source data are available for this figure: SourceData FS2.
Figure 3.
Th1 cells form FAs. (A–D) Representative migrating Th1 cells expressing Talin-EGFP in confinement on (A) ICAM-1, (B) FN, (C) PLL, and (D) PLL-PEG. (E) Representative image of a confined Th1 cell fixed and stained for endogenous talin. (F and G) Quantification of the number of FA-positive cells when expressing (F) Talin-EGFP or (G) vinculin-EGFP compared across surface treatments. (H) A comparison of FA lifetime between Th1 cells and fibroblasts expressing Vinculin eGFP (representative images on right). (I and J) Representative Th1 cells expressing (I) Talin-EGFP or (J) Integrin β3-Emerald in confinement on a micropatterned substrate. The left portion of the field of view is FN coated and the right portion is passivated. Number of biological replicates (colored dots): (F) Passivated, 36 cells from three experiments; PLL, 53 cells from two experiments; FN, 140 cells from five experiments; ICAM-1, 40 cells from three experiments. (G) Passivated, 21 cells from two experiments; PLL, 38 cells from four experiments; FN, 94 cells from four experiments; ICAM-1 169 cells from three experiments. (H) Th1 n = 10; Fibroblast n = 10. Each replicate represents the average turnover of ≥5 FAs per cell. Statistical tests: (F–H) Two-tail unpaired parametric t test was used to compare the replicates.
Video 5.
Th1 Talin-EGFP cells migrating in confined (5 μm) environments coated with ICAM-1, Fibronectin, PLL, or Passivated. 63× objective. Frame rate = 10 frames/s. Movie from Fig. 3, A–D.
Figure S3.
Th1 cells form FAs containing vinculin and paxillin. (A–D) Representative Th1 cells expressing Vinculin-eGFP in confinement on (A) ICAM-1, (B) Fibronectin, (C) PLL, and (D) PLL-PEG. (E) A Th1 expressing Vinculin-eGFP in confinement on a micropatterned substrate. The left portion of the field of view is fibronectin-coated and the right portion is passivated. Arrows indicate vinculin accumulation in FAs. (F) Representative fixed images of Th1 cells in confinement, stained with an anti-paxillin antibody to mark endogenous adhesions, phalloidin to visualize actin, and DAPI to mark the nucleus.
Finally, to confirm that the FAs were specific to the presence of ECM, we imaged cells expressing either talin-EGFP (Fig. 3 I and Video 6), integrin β3-Emerald (Fig. 3 J and Video 6), or vinculin-EGFP (Fig. S3 E) in Th1 cells migrating across boundaries between FN and passivated substrates. In the talin- and vinculin-transduced cells, FA-like structures could be seen forming on the ECM-coated regions and disappearing as the cell moved onto the passivated regions of the substrates (Fig. 3 I; Video 6; and Fig. S3 E). In the integrin β3 transduced cells, it was challenging to identify specific FA structures due to the membrane localization of the probe, but distinct retraction fibers could be seen forming on the ECM-coated portion of the substrate (Fig. 3 J and Video 6), demonstrating clear attachment to the ECM (Baschieri et al., 2023). These results reveal that not only do Th1 cells possess all the necessary components to form FAs, but they only form these structures when ECM or adhesive ligands are present.
Video 6.
Th1 Talin-EGFP (Left panel) and Th1 Integrin β3-Emerald (Right panel) cells migrating in confined (5 μm) environments on a micropatterned substrate. 20× objective. Frame rate = 10 frames/s. Movie from Fig. 3, I and J.
Th1 generate forces on the ECM through focal adhesions
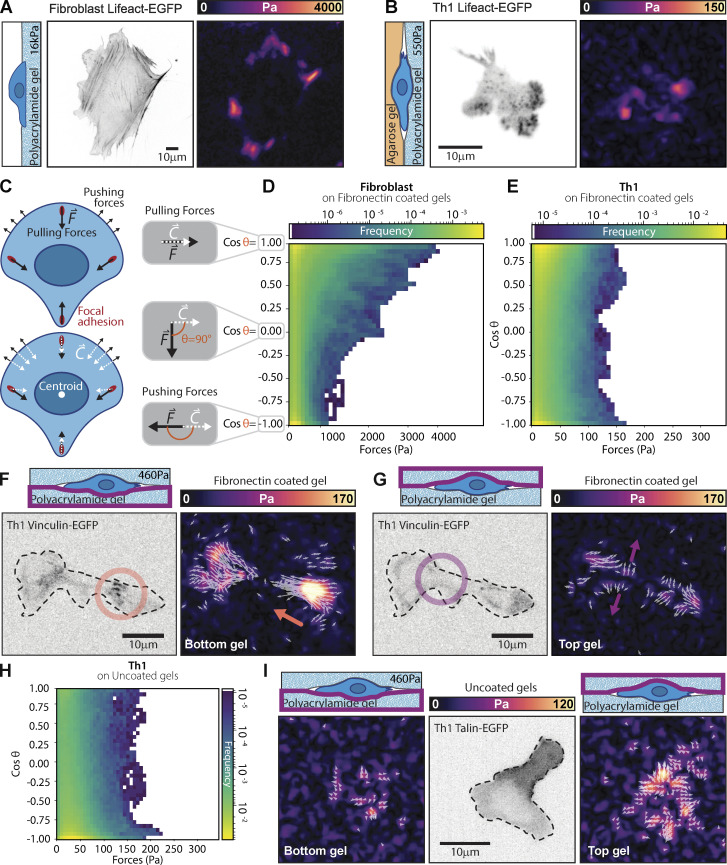
Given the appearance of FA structures, complete with the requisite adhesion proteins (Kanchanawong et al., 2010), we next set out to use traction force microscopy (TFM) (Sabass et al., 2008) to determine whether forces were being exerted at these locations. As Th1 cells require confinement to adhere strongly to FN-coated surfaces, we either layered low-temperature melting agarose on the gel or confined them between two FN-coated acrylamide gels and measured their force production. Most notably, the forces exerted by activated Th1 cells were of a much smaller magnitude than the comparable forces in fibroblasts (Fig. 4, A and B). To compare the different cell types, we measured both the magnitude of traction stresses and the angle between the traction stress direction and the centroid of the cell (Fig. 4 C). While traction stresses in the fibroblasts were in the kPa range (Fig. 4 D), the magnitude of traction stresses produced by Th1 cells was a full order of magnitude lower in the 100 s of Pa range (Fig. 4 E). A similarly strong difference was seen in the distribution of the forces with respect to the cell centroid. In fibroblasts, the traction stresses are almost completely contractile (i.e., skewed heavily toward θ < 90° or directed toward the centroid; Fig. 4 D), consistent with previous work (Dembo and Wang, 1999; Sabass et al., 2008). In the Th1 cells, however, we saw a distribution of traction stresses across all angles with enrichment of traction stresses at both extremities of the distribution. This suggests that the forces were mostly split between pointing toward the cell body (contractile forces, θ < 90°) and away from the cell body (pushing forces, θ > 90°) (Fig. 4 E).
Figure 4.
Th1 cells exert traction stresses at FAs. (A and B) Representative traction maps of a (A) fibroblast or (B) Th1 cell expressing Lifeact-EGFP. The fibroblast is plated on a gel with a shear modulus of 16 kPa, while the Th1 cell is plated under agarose on a gel with a shear modulus of 336 Pa. Both gels are coated with FN. (C) Cartoon illustrating the direction of potential forces (black arrows) and their orientation with respect to the cell centroid (white arrows). (D and E) 2D histograms comparing the magnitude of measured traction stresses and the difference in their direction (θ) from the cell centroid. Pulling (contractile) forces are represented as cosθ = 1, while pushing forces are represented as cosθ = −1. Histograms are shown for (D) fibroblasts and (E) Th1 cells on FN-coated gels. (F) Representative images of a Th1 cell expressing Vinculin-EGFP sandwiched between two acrylamide gels of 460 Pa coated with FN. On the bottom gel, contractile forces (orange arrow) colocalize with vinculin puncta (orange circle). (G) On the top gel, pushing forces (purple arrow) show no colocalization with vinculin (purple circle) and likely arise from the cell pushing the gel out of the way as it is pulled through the gel. (H) 2D histogram comparing the magnitude of measured traction stresses and the difference in their direction from the cell centroid for a cell confined between two passivated gels. (I) Representative images of the pushing forces on both the top and bottom gel for the case when the gels are passivated. Number of replicates: (D) Th1, 12 cells, 45 fields of view from four different experiments; (E) Fibroblast, 12 cells from 12 fields of view. (H) 11 cells, 44 fields of view, from two different experiments. The data for the histograms were pooled from all cells in each condition.
To explore this intriguing finding further, we performed double-sided traction force microscopy where the Th1 cells were sandwiched between two FN-coated polyacrylamide gels to measure how they interacted with their environment. We consistently found that contractile forces coincided with the localization of FA proteins such as vinculin (Fig. 4 F and Video 7), regardless of whether they were on the top or bottom gel. We also observed pushing forces (i.e., θ > 90° or directed outward from the cell centroid) that were generally of smaller magnitude and that did not coincide with any visible enrichment of vinculin (Fig. 4 G and Video 7). In the absence of specific interaction with the substrate, these “pushing” forces likely passively arise in response to the cell body displacing the surrounding gel as the contractile cytoskeletal generated forces exerted at FAs pull the cell forward along its path. When Th1 cells were sandwiched between two uncoated gels (i.e., lacking any ECM protein), we saw pushing forces on both the top and the bottom surfaces (Fig. 4, H and I). The main mechanical interactions driving migration in Th1 cells thus appear very similar to the traditional mesenchymal modes of migration, just at much smaller magnitudes (Fig. 4, A and B) and more rapid timescales (Fig. 3 B).
Video 7.
Top panels: Th1 Vinculin-EGFP cells sandwiched between two acrylamide gels of 460 Pa coated with FN. Bottom panels: Representation of traction stress vector overlay on traction map. Pulling traction stress colocalize with vinculin puncta. Pushing forces show no colocalization with vinculin and seems to outline cell shape. 63× objective. Frame rate = 10 frames/s. Movie from Fig. 4, F and G.
ECM promotes confined migration
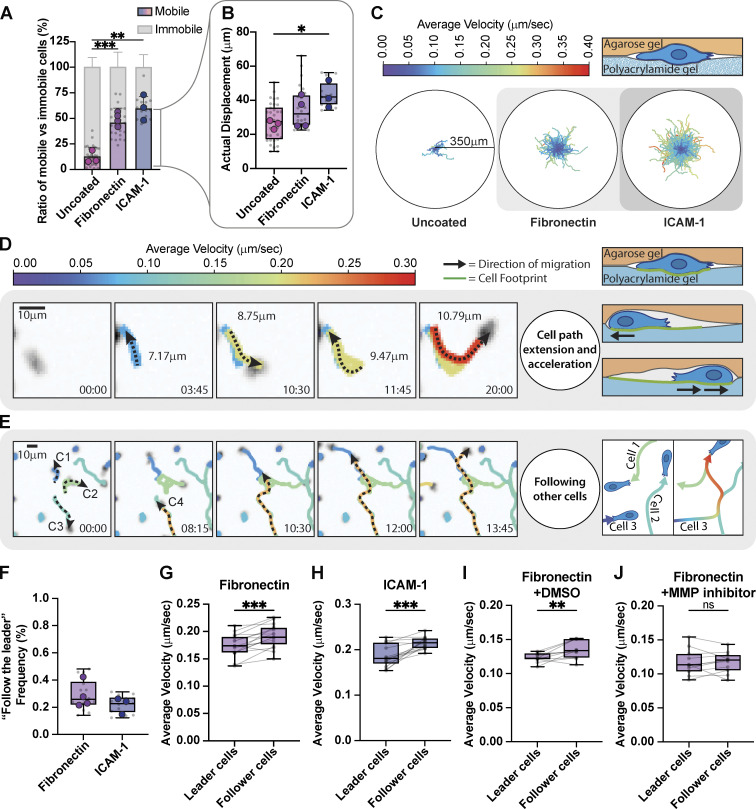
We next measured the migration of activated Th1 T cells confined between a soft acrylamide gel and low-temperature melting agarose. We chose to pour the agar over the cells on the gels so as not to bias our results by only measuring the cells that could squeeze between the gel and agar. Surprisingly, when the polyacrylamide gels were uncoated, only ≈10% of cells were able to migrate (Fig. 5 A), reinforcing our previous results that Th1 cells require adhesive ligands to migrate (Fig. 2). If the acrylamide gel was coated with either FN or ICAM-1, however, around ≈50% of the cells were able to migrate (Fig. 5 A). Comparing only the mobile populations of cells on the different ECM coatings, Th1 cells on ICAM-1 migrated significantly further than those on uncoated acrylamide gels (Fig. 5, B and C). Mobile cells on FN exhibited a similar trend but with a much larger variance (Fig. 5, B and C). These results confirm that ECM is a critical component for the migration of Th1 cells through confined environments.
Figure 5.
T cells follow the paths of each other. (A) Comparison of the mobile versus immobile fraction of Th1 cells in confinement between an agarose gel and a polyacrylamide gel coated with FN, ICAM-1, or uncoated. (B) Comparison of actual displacement of the cells from A. Mobile cells are defined by a minimum displacement of 10 µm. (C) Roseplots showing Th1 cell tracks of cells in A and B. The colormap represents the cell's average velocity. (D) Individual cell tracking shows cells extending their path and accelerating after each pass. (E) Individual cell tracking shows three cells extending their paths and a fourth cell (orange) reusing those previously made paths and accelerating while doing so. Colors represent the cell's average velocity. (F) Measurement of the frequency of cells following each other. (G–J) Measurement of the change in velocity of cells as they encounter previous paths of migrating T cells confined between an agarose gel and a polyacrylamide gel coated with (G) FN, (H) ICAM-1, (I) DMSO, or (J) 10 µM Pan MMP Inhibitor. Number of biological replicates (colored dots): (A and B) Uncoated, n = 3; FN, n = 4; ICAM-1, n = 3. (F–H) FN n = 4; ICAM-1 n = 3. (I–J) FN + DMSO n = 2; FN + MMP inhibitor, n = 2. Each replicates represent the average of 5–10 fields of view (gray dots = average of each fields of view). Statistical tests: (A and B) Two-tail unpaired parametric t test; (G–J) One-tail paired parametric t test.
FN provides environmental cues to guide migration
It has been previously shown that many immune cells have developed mechanisms to recruit and guide other cells toward inflammation (Lim et al., 2015). In our in vitro setup, Th1 cells exhibit similar behaviors, following paths apparently demarcated by previous cells. In these cases, we found two interesting observations: First, cells often migrate back and forth along a path, extending it with every pass (Fig. 5 D), similar to the behavior that has been reported previously in vivo (Mrass et al., 2017; Torres et al., 2023). Second, cells that migrate along a previously trodden path, whether made by themselves or another cell, migrate faster (Fig. 5, E, G, and H; and Video 8). In our setup, a cautious minimum estimate (see Materials and methods for details) found that ∼25% of motile cells followed the track of another cell for a distance of at least 10 µm (Fig. 5 F). When we compared the velocities of cells in regions where trajectories overlapped, we found that on both ICAM-1 and FN the cell passing through second moves faster than the first cell that moves through the same area (Fig. 5, G and H). This suggests that cells could potentially modify the confined environment either biochemically (e.g., leaving behind a chemokine or degradation of the ECM) or physically (e.g., mechanical deformation of the ECM or membranous tubular network [Baschieri et al., 2023]). To test this idea further, we repeated the experiment in the presence of a 10-µM pan MMP inhibitor, as previous works have shown that Th1 secrete MMPs to facilitate their migration through gelatinase (Abraham et al., 2005; Oviedo-Orta et al., 2008). Cells treated with the vehicle control (DMSO) exhibited the same increase in speed for the follower cells, while those treated with the MMP inhibitor displayed no difference in speed (Fig. 5, I and J). These results imply that FAs and their associated ECM play crucial roles in facilitating migration of Th1 cells through complex environments, beyond simply anchoring the cells to the substrate.
Video 8.
Cell tracking of Th1 cells stained with Hoechst migrating in confinement between an agarose gel and a polyacrylamide gel coated with fibronectin. The movie shows three cells extending their paths and a fourth cell (orange) reusing those previously made paths and accelerating while doing so. The colormap represents the cell's average velocity in μm/sec. 10× objective. Frame rate = 10 frames/s. Movie from Fig. 5 E.
Redefining the mesenchymal to amoeboid migration spectrum
In this paper, we demonstrated that activated primary Th1 T cells are able to form FAs and use them to transmit contractile forces generated by the cytoskeleton toward the ECM. We used primary murine helper T cells specifically because they have been shown to increase their expression of β3 integrins, which bind to FN, when activated (Overstreet et al., 2013; Gaylo et al., 2016), and they are more likely to encounter ECM proteins as they leave the circulatory system to respond to infection. Because of the heterogeneity found in living tissue (Huse, 2017), we chose to perform these experiments in vitro to maximize our control over the extracellular environment. Traditionally, in vitro assays studying T cell migration have relied on coating the substrate with ICAM-1, a ligand to which T cells bind strongly. We found that T cells can migrate similarly on FN, an important ECM protein, but only when in confinement. Surprisingly, only a small concentration of FN (such as found in the serum added to media) is required for T cells to migrate. T cells plated on integrin-independent substrates failed to migrate but could be rescued simply by coating the substrates with FN or ICAM-1. Similarly, the blocking of integrins via antibody binding reduced migration on FN. This interaction with ECM is facilitated by FAs consisting of the canonical proteins associated with adhesions, including integrins, talin, and vinculin, and coincides with regions of contractile forces. Furthermore, we found that when migrating, Th1 cells will often follow paths formed by other cells or even by themselves, increasing their migration speed in these regions. Previous reports have described T cells turning back on their migration tracks in vivo (Mrass et al., 2017; Torres et al., 2023), while it was also shown that neutrophils can leave behind a chemotractic trail for other immune cells to follow (Lim et al., 2015). These findings, combined with our observations that the use of an MMP inhibitor abrogates this response, suggest that the T cells likely modify the ECM matrix or leave behind biochemical cues for other T cells to interpret. In total, these results demonstrate that specific FA-mediated cell–substrate interactions are vital components for proper T cell migration, and thus an effective immune response.
The literature has predominantly split different forms of migration into two camps: mesenchymal, encompassing integrin-mediated migration, and amoeboid, encompassing everything else (Paluch et al., 2016). Amoeboid migration derives from the study of the seemingly non-specific interactions between the cell membrane protrusions and the substrate of amoeba such as dictyostelium (Fukui and Inoué, 1997; Mijanović and Weber, 2022). Over the years, additional non-specific migration mechanisms, including bleb-based motility (Keller and Bebie, 1996; Poincloux et al., 2011), pressure-driven gradients (Stroka et al., 2014), and even swimming (Aoun et al., 2020) have often been included in the amoeboid category (Petrie and Yamada, 2012). Migrating immune cells, including T cells, display rapid protrusions, morphology changes, and strong actin flows, consistent with descriptions of amoeboid migration, while lacking the big stable adhesion plaques found in mesenchymal cells (De Bruyn, 1946; Wolf et al., 2003; Lämmermann and Sixt, 2009). This narrative has been supported by findings that dendritic cells can migrate in 3D following the knock-out of all integrins (Lämmermann et al., 2008) and that both dendritic cells and T cells can migrate on passivated surfaces when in confinement (Hons et al., 2018; Reversat et al., 2020). In other contexts, however, many immune cells do require integrins (Hogg et al., 2003; Owen et al., 2007; Van Goethem et al., 2011; Gawden-Bone et al., 2014; Paterson and Lämmermann, 2022; Hamoudi et al., 2023; Kaltenbach et al., 2023), and T cells specifically form integrin-mediated adhesions when migrating on the planar surface of blood vessels (Nourshargh and Alon, 2014; Li et al., 2016; Kim and Hammer, 2019) and similar structures at the immunological synapse (Basu and Huse, 2017; Huse, 2017; Wang et al., 2022). These results suggest that immune cells have the ability to use a variety of mechanisms to migrate in response to different environmental conditions.
With their rapid turnover and small size, the FAs in Th1 cells resemble nascent adhesions seen in mesenchymal cells (Nayal et al., 2006; Choi et al., 2008). The lack of bundled actin stress fibers in T cells could facilitate the rapid turnover and short lifetimes of FAs as actin bundling has been previously shown to play a critical role in stabilizing these structures in mesenchymal cells (Choi et al., 2008; Oakes et al., 2012). While T cells can form FAs in 2D on ICAM-1-coated surfaces, confinement is required for T cells to form FAs and migrate on FN-coated substrates. The act of confining cells likely increases the number of integrins interacting with the ECM at any given point, helping to stabilize the entire FA as seen in other systems (Cavalcanti-Adam et al., 2007; Oakes et al., 2018; Bidone et al., 2019). Integrins, including both αvβ3 (Chen et al., 2017) and αLβ2 (Chen et al., 2010), also behave as catch-bonds, with their bond lifetime increasing with load. Confinement may thus increase the interaction of the cell with the substrate, thereby increasing the force of retrograde flow in the cortex and strengthening integrin-ligand bonds. In the tissue, changes in the stiffness of the ECM, which are typically associated with inflammation (Martinez-Vidal et al., 2021), could also increase the load across the bonds, making them last longer.
Strikingly, we find that only a small amount of ECM is required to induce T cell migration. While cells can migrate on passivated surfaces in the presence of serum, migration was significantly reduced when the serum was removed but could be rescued by coating the substrate with FN. Similarly, when T cells were sandwiched between two uncoated acrylamide gels, the vast majority of T cells were unable to migrate. Previous studies have shown that even when passivated, ECM proteins can still adsorb to surfaces (Walkey et al., 2012). Together, these findings suggest that passivated glass and PDMS substrates likely contain a small amount of ECM when there is serum present, and this small amount is sufficient to enable migration. Our data also highlights that the percentage of cells that migrate in a given condition is highly variable and that even in incredibly challenging conditions, some T cells are still able to migrate. This is first and foremost a testament to the robustness of the immune system but also illustrates the large variation in motility across the population and the importance of avoiding bias by only measuring motile cells.
That we see both specific pulling forces at FAs and non-specific pushing forces on the surrounding ECM during migration is consistent with the idea that ECM geometry strongly influences immune cell migration (Adebowale et al., 2023, Preprint; Böhringer et al., 2023, Preprint; Czerwinski et al., 2023, Preprint). Adhesions, as we see here, would be a natural mechanism to sense these different physical properties (Huse, 2017; Moreau et al., 2018) and could be involved in modifying the external environment to allow following cells to move faster. Indeed, such a pathway was recently proposed with modeling results suggesting that secretion or modification of the substrate could strongly bias the paths of migrating cells (Perez Ipiña et al., 2024), consistent with what we see in cells that follow the path of a previous cell (Fig. 5, D–J).
In this study, we focused on activated primary Th1 T cells on account of their known expression of FN ligands and their increased likelihood to encounter ECM protein in tissues as they migrate during an immune response. We believe it likely, however, that other immune cells behave similarly. Importantly, our model does not rule out true integrin-independent migration but rather suggests that if a cell has the components to make an adhesion and ligands are available, it likely will form an FA. Ultimately, the difference between amoeboid and mesenchymal migration is blurry at best as the mechanisms behind true integrin-independent migration remain to be explored. Lastly, it is also likely that these FAs play important additional functional roles, such as leaving behind chemokine trails (Lim et al., 2015) or guiding other cells (Baschieri et al., 2023), and future work will be needed to decipher these important behaviors.
Materials and methods
Primary T cell activation and cell culture
The workflow of primary T cell collection and activation is illustrated in Fig. S1 A. Spleen and lymph node cells were isolated from OTII mice that express a T cell receptor that recognizes ovalbumin. The next day, 60–80 million cells were stimulated with antigen under Th1 conditions: 4 million viable cells per ml in DMEM AB+ media (high glucose DMEM (SH30243.01; HyClone) supplemented with 10% FBS (10437.028; Gibco), 2.5% HEPES solution 1M (SH3023701; Hyclone), 1.3% MEM nonessential amino acids 100x (11-140-050; Thermo Fisher Scientific), 0.1% B-ME 55 mM (2-mercaptoethanol, 21895-023; Gibco), 1.2% L-glutamine (Sh30034.01; HyClone), 1.3% antibiotic–antimycotic (30-004-CL), 40 µg/ml Anti Murine IL-4 (11B11; 05060201; NCI-Frederick), 10 U/ml IL-2 (NCI-Frederick, Teceleukin Recombinant Human Interleukin-2 [rIL-2]—Bulk Ro 23-6019), 2 µg/ml OVA peptide (OVA 323-339 custom order; Biomatik), and 20 ng/ml IL-12 p70 (Cat# 210-12; Peprotech)). 48 h later viable T cells were isolated on a lymphocyte separation media (LSM) gradient (25-072-CV; Corning) and either incubated for cell expansion or transduced with retrovirus (see below). After initial activation, the purity of CD4 T cells expressing the OTII T cell receptor was confirmed by flow cytometry (BD LSRFortessa Cell Analyzer) after staining with Anti-CD4-Alexa647 (557681; BD bioscience) and Anti-Vα2- PE (553289; BD bioscience). Typically, 30–40% of cells were CD4/Vα2 double positive on day 3 (Fig. S1 C) and 70–90% on day 5–9 (Fig. S1, D–F). Fibroblasts were cultivated in high glucose DMEM (SH30243.01; HyClone) supplemented with 10% FBS (10437.028; Gibco), 1.3% antibiotic-antimycotic (30-004-CL).
Retroviral production and infection
The empty backbone MIGR1 (a gift from Warren Pear (University of Pennsylvania, Philadelphia, PA, USA), #27490; Addgene) was used to make retroviral constructs to express Lifeact-EGFP, Integrin β3-Emerald, Vinculin-EGFP, and Talin-EGFP (mEGFP-Lifeact-7 [#54610; Addgene] and mEmerald-Beta3-N-18 [#54130; Addgene] were gifts from Michael Davidson (Florida State University, Tallahassee, FL, USA); pEGFP Vinculin [#50513; Addgene] was a gift from Kenneth Yamada (National Institute of Health, Bethesda, MD, USA); and GFP-Talin1 [#26724; Addgene] was a gift from Anna Huttenlocher (University of Wisconsin, Madison, WI, USA)). Retroviral DNA constructs were transiently transfected into the ecotropic retroviral packaging line, Phoenix. In some cases, Phoenix cells stably producing ecotropic retrovirus were generated by first producing VSV-G pseudotyped virus and using that to transduce ecotropic Phoenix cells. Viral supernatants were collected and concentrated 20x using Retro-X concentrator (PT5063-2; Takara Bio) and stored at 80°C. For T cell transduction, on day 3 of activation (Fig. S1 A), 250 μl of the concentrated virus was preincubated with 8 µg/ml polybrene (TR-1003-G; EMD Millipore) for 30 min on ice, mixed with 1–4 million density purified T cells (see above), and spun for 60 min at 2,000 RPM at 4°C in a 24-well plate.
In vitro live cell imaging
Experiments with Th1 cells were done in DMEM AB+ media while fibroblasts were plated in DMEM (see Cell culture section). All imaging was performed at 37°C with 5% CO2 on an Axio Observer 7 inverted microscope (Zeiss) attached to a W1 Confocal Spinning Disk (Yokogawa) with Mesa field flattening (Intelligent Imaging Innovations), a motorized X,Y stage (ASI), and a Prime 95B sCMOS (Photometrics) camera. Illumination was provided by a TTL-triggered multifiber laser launch (Intelligent Imaging Innovations) consisting of 405, 488, 561, and 637 nm lasers using either a 63X, 1.4 NA Plan Apochromat or 10X 0.45NA Plan Apochromat objective (Zeiss). Temperature and humidity were maintained using a Bold Line full enclosure incubator (Oko Labs). The microscope was controlled using Slidebook 6 Software (Intelligent Imaging Innovations). All in vitro imaging was performed as single confocal slices. For the traction and force microscopy experiment, FA proteins and the gel were imaged at the same focal plane. Cells were imaged for 20 min at 15-s intervals. When cells were imaged in confinement, confinement was applied to the cells immediately before mounting the cell chamber on the microscope in a humidified and tempered chamber, then allowed to stabilize for 5–10 min prior to commencing imaging. All imaging was performed in DMEM High glucose with L-glutamine without sodium pyruvate (SH30022.01; Cytiva) supplemented with 10% FBS (unless specified otherwise, 10437-028; Gibco), 0.02 M Hepes (SH30237.01; Cytiva), 1x MEM Nonessential Amino Acids (SH30238.01; HyClone), 0.05 mM 2-mercaptoethanol (21985023; Gibco), and 1x antibiotic–antimycotic solution (ABL02; Caisson).
Cell confinement experiments
Cells in Figs. 1, 2, and 3 were confined under a PDMS surface with a gap of 5 µm as previously described (Le Berre et al., 2014; Liu et al., 2015), also called static confiner. Briefly, we used SU8 as a mold (courtesy of Matthieu Piel, Insitut Curie, Paris, France) to make a confinement slide (CS) that was covered with 5 µm height pillars. Cells were plated on a glass coverslip in a cell chamber. A big PDMS pillar attached to a magnetic lid was used to push down the CS onto the cells. The bottom coverslip in the cell chamber was coated prior to an experiment (see Surface coatings section). In Figs. 4 and 5, we confined cells by using a soft acrylamide gel on the bottom and either 1% low melting agarose (16520-100; Invitrogen) or a second soft acrylamide gel on the top. The protocol for soft polyacrylamide gel preparation is described in the section Traction Forces Microscopy Experiments. For confinement between two soft acrylamide gel, we used the same methodology as for the static confiner but replaced the glass coverslips with gels. For confinement between one soft acrylamide gel and 1% low meltin agarose, 1 million cells in 1 ml of DMEM AB+ with 1:10,000 Hoechst 33342 (H3570; Invitrogen) were plated on a soft polyacrylamide gel in a coverslip cell chamber and incubated for 10 min at 37°C or until the cells settled down. The media was removed and 1 ml of soft agarose (1% d [16520-100; Invitrogen] in DMEM AB+), previously tempered at 37°C, was poured on top of the cells. The cell chamber was kept at 37°C for 10–20 min to let the cells settle down again while the agarose gel was still unpolymerized. Right before imaging, the cell chamber was placed on the ventilation of the tissue culture hood to help reduce the temperature faster and allow the agarose to polymerize for 5 min. Finally, 1 ml of DMEM AB+ was added on top of the agarose gel. For Fig. 5, I and J, cells were pretreated with a pan MMP inhibitor (GM6001; Sigma-Aldrich) at 10 μM for 10 min before confinement under an agarose pad. We added fresh media with the same concentration of MMP inhibitor and then repeated our migration measurements. Our control cells were treated identically with the same volume of DMSO.
Antibody blocking
Cells were pretreated with a combination of antibodies (integrin αv [cat:153202; Biolegend], integrin β1 [cat:102202; Biolegend], integrin β2 [cat: 101401; Biolegend], and integrin β3 [cat:104302; Biolegend]). The antibodies (or PBS for control) were added at a final concentration of 1/50 in the cell culture media and placed at 37°C for 3 h. Once the pretreatment was done, cells were detached by pipetting up and down and kept in their antibody-supplemented (or PBS-supplemented) media. Cells were plated in a cell chamber on a glass coverslip previously coated with FN (see Surface coatings section). The cell chamber was placed back at 37°C for 10 min to allow cells to settle and then cells were confined following the static confiner method (see Cell confinement experiments).
Surface coatings
For FN, glass coverslips were coated with 10 µg/ml human plasma FN (FC010; Millipore) overnight at 37°C. For ICAM-1, glass coverslips were first coated with 10 µg/ml recombinant protein A (10-110-0; Novex) overnight at 37°C, then blocked with 2% BSA (Bovine Serum Albumin, 9048-46-8; Research Products International) for 30 min at RT, then coated with 5 µg/ml ICAM-1 (50440-M03H; Sino-Biological) for 2 h at RT, and finally blocked with 2% BSA (Bovine Serum Albumin, 9048-46-8; Research Products International) for 30 min at RT. For PLL-coated substrates, glass was coated with 0.01% PLL (Poly-L-Lysine, P4707; Sigma-Aldrich) for 20–45 min at RT. For passivated substrates, glass was coated with either 1 mg/ml PLL-PEG (PLL20K-G35-PEG2K; JenKem Technology) or 1 mg/ml PMOXA (SuSoS; PAcrAm-g-[PMOXA, amine, silane]) for 20–45 min at RT. For each coating, the coverslips were rinsed and kept in PBS at 4°C until used (same day for PLL and passivated coverslips, same or next day for FN and ICAM-1). For polyacrylamide gels, after polymerization, gels were rehydrated overnight in ddH2O, treated with crosslinker Sulfo-Sanpah (22589; Pierce Scientific), and photoactivated for 5 min, and then washed in ddH2O. Polyacrylamide gels were then immediately coupled to either human plasma FN (1 mg/ml overnight at 37°C; FC010; Millipore) or ICAM-1 (0.1 mg/ml Recombinant Protein A [10-110-0; Novex] overnight at 37°C, 2% BSA [9048-46-8; Research Products International] for 30 min at RT, 37.5 µg/ml ICAM-1 [50440-M03H; Sino-Biological] for 2 h at RT, and 2% BSA [9048-46-8; Research Products International] 30 min at RT).
Micropatterning experiments
Desired micropatterns were designed in AutoCAD and sent to the Nanotechnology Core Facility of the University of Illinois Chicago (https://ncf.uic.edu) to make a quartz chrome mask. Before usage, the mask was thoroughly washed with water, then isopropanol, and then air dried with an air gun. Glass coverslips were treated with plasma (Plasma Cleaner, Harrick Plasma, PDC32G) for 2 min, coated with PMOXA (SuSoS; PAcrAm-g-[PMOXA, amine, silane]) at 1 mg/ml for 45 min at room temperature, and then rinsed in ddH2O. The coverslip was placed on the mask, the PMOXA-coated side facing the chrome side of the mask. A piece of glass, with similar dimensions to the mask, is then placed on top of the coverslips to hold it in place. The mask-coverslip-glass sandwich is then held together in a custom magnetic frame. The sandwich was then placed in a preheated UVO cleaner oven (Model 342; Jelight), quartz side up, and exposed to deep UV for 4 min. The coverslips were then rinsed in water and coated with 20 µg/ml FN 1/10 Rhodamine FN (FNR01; Cytoskeleton), 9/10 FN (FC010; Millipore) for 30 min at RT. Micropatterned coverslips were placed in the coverslip’s cell chamber with 250–500 k cells. Cells were then allowed to settle for 10 min before imaging or before confinement.
Focal adhesion turnover analysis
FA turnover was measured in a minimum of 12 individual cells for each cell type (Th1 and Fibroblast). For each individual cell, the turnover of five focal adhesions was analyzed per cell. To analyze turnover, a region of interest (ROI) was drawn around an FA at its largest size. We manually tracked the FA over time starting one frame before it appeared until its complete disappearance and recorded that length of time as the FA lifetime.
Th1 fixed and stained under confinement
A four-well glass bottom 35-mm dish (C4-1.5H-N; Celvis) was coated with 10 µg/ml human plasma FN (FC010; Millipore) overnight at 37°C. Th1 cells were plated for 1 h, and then the media was slowly removed and 100 μl of soft agarose (1% Low Melting Agarose [16520-100; Invitrogen] in DMEM AB+), previously tempered at 37°C, was poured on top of the cells. The agarose was quickly polymerized under continuous airflow for 3 min at room temperature. 1 ml of DMEM AB+ was added on top of the gel before the dish was placed back at 37°C to allow cells to start migrating normally for 1 h. With the gel still in place, the cell culture media was gently removed and rinsed once with cytoskeleton buffer (CB) (19.52 g MES [0.1 M], 6.10 g MgCl2 [0.03 M], 102.88 g KCl [1.38 M], 7.6 g EGTA [0.02 M]) for 30 s. CB was gently removed and 250 µl of fixing solution (0.15 g BSA, 2.5 ml 16% paraformaldehyde solution, 50 μl Triton, and 7.5 ml CB) was added on top of the gel and kept at room temperature for 15 min. We slowly removed the fixing solution and rinsed it three times for 30 s with 1x PBS. Primary antibody (Talin [ab157808; Abcam] and Paxillin [05-417; Millipore]) were diluted 1:200 in 200 μl of antibody solution (0.15 g BSA, 50 μl Triton, and 10 ml CB) and incubated for 1 h at room temperature. We slowly removed the primary antibody and rinsed it three times for 30 s with 1x PBS. Rabbit fluorescent secondary antibody (111-545-144; Jackson Immunoresearch) was diluted 1:1,000 in antibody solution with 1:2,000 Phalloidin (65906-10NMOL; Sigma-Aldrich) and 1:5,000 Hoechst 33342 (H3570; Invitrogen). PBS was removed and secondary antibody was added and kept for 1 h at room temperature in the dark. A final PBS wash was performed (three times for 30 s) before immediately imaging the fixed cells.
Western blot
Protein samples were prepared using 4 million Th1 cells, spun down, rinsed with 1X PBS (21-040-CV; Corning), and extracted in 500 μl 1X Laemmli Sample Buffer (1610737; Bio-Rad). The samples were then boiled at 90°C for 5 min. The protein quantity was measured using RC DC protein assay Kit II (5000121EDU; Bio-Rad). For each protein of interest, 10 µg of the sample was loaded in a 4–15% Tris-Glycine gel (4568083; Bio-Rad). The gel was transferred onto the PVDF membrane (L00726; GenScript) using Genscript eBlot (L00686; GenScript) for 16 min. The membrane was air dried and then blocked using 5% nonfat dry milk (1706404; Bio-Rad) in 1X PBS (21-040-CV; Corning) with 0.1% Tween 20 (BP337-500; Thermo Fisher Scientific) (PBS-T) for 30 min and then rinsed 3 × 5 min with PBS-T. The PVDF membranes were incubated overnight at 4°C in 2% BSA (9048-46-8; Research Products International) with primary antibody: 1:1,000 Anti-Talin 1 (ab157808; Abcam), 1:1,000 Anti-Zyxin (ABC1463; Millipore Sigma), 1:500 Anti-Paxillin, (clone 5H11, 05-417; Millipore Sigma), 1:1,000 Integrin β2 (47598; Cell Signaling), 1:1,000 Integrin β3 (13166; Cell Signaling), 1:1,000 Integrin αv (A19071; Abclonal), 1:1,000 α-actinin (6487T; Cell Signaling), and vinculin (V4139; Sigma-Aldrich). They were then rinsed 3 × 5 min with PBS-T before incubation for 30 min in 5% non-fat dry milk in PBS-T with secondary antibody: 1:5,000 anti-Mouse HRP antibody (5178-2504; Bio-rad) and 1:5,000 anti-Rabbit HRP antibody (5196-2504; Bio-rad), and then washed 3 × 5 min with PBS-T. Finally, the membranes were revealed with Clarity ECL (1705061; Bio-Rad) for Talin, Zyxin, Paxillin, Integrin β3, Integrin αv, α-actinin, vinculin, and Clarity-Max ECL (1705062; Bio-Rad) for Integrin β2 using a ChemiDoc (Bio-Rad) imaging system.
Traction force microscopy experiments
Traction force microscopy was performed as described previously (Sabass et al., 2008; Sala and Oakes, 2021). Coverslips were prepared by incubating with a 2% solution of (3-aminopropyl)trimethyoxysilane (313255000; Acros Organics) diluted in isopropanol. Coverslips were washed with DI water 5X for 10 min and cured overnight at 37°C. Coverslips were incubated with 1% glutaraldehyde (16360; Electron Microscopy Sciences) in ddH20 for 30 min at room temperature and washed 3X for 10 min in distilled water, air dried, and stored at RT. Polyacrylamide gels with a shear modulus of 336 or 460 Pa (for Th1 cells) and 16 kPa (for fibroblast) were embedded with 0.04-µm fluorescent microspheres (F8789; Invitrogen) and polymerized on activated glass coverslips for 30 min–1 h at room temperature. After polymerization, gels were rehydrated overnight in ddH2O, treated with crosslinker Sulfo-Sanpah (22589; Pierce Scientific), photoactivated for 5 min, and then washed in ddH2O. Polyacrylamide gels were then immediately coupled to matrix proteins, human plasma FN (1 mg/ml overnight at 37°C; FC010; Millipore), or adhesion protein, ICAM-1 (0.1 mg/ml Recombinant Protein A [10-110-0; Novex] overnight at 37°C, 2% BSA [9048-46-8; Research Products International] 30 min at RT, 37.5 µg/ml ICAM-1 [50440-M03H; Sino-Biological] for 2 h at RT, and 2% BSA [9048-46-8; Research Products International] for 30 min at RT). Following matrix protein crosslinking, gels were rinsed in PBS and kept at 4°C in PBS until used (either the same day or the next day). For fibroblast cells, they were plated on the gels, allowed to adhere overnight, and imaged the following day. For Th1 cells, they were plated on the gels 10–20 min before confinement by low-melting agarose or a second polyacrylamide gel. A reference image of unstrained gel was obtained by waiting for the cell to migrate out of the field of view.
For 336 Pa gels, a standard solution was prepared with 1.25 ml of 40% Acrylamide (1610140; Biorad), 583 μl 2% bisAcrylamide (1610142; Biorad), and 3.16 ml ddH2O. To make the gel, 150 μl of the standard solution was mixed with 341.75 μl water, 5 μl 0.04-µm fluorescent microspheres (F8789; Invitrogen), 0.75 μl TEMED (110-18-9; Fisher Bioregents), and 2.5 μl of 10% ammonium persulfate (APS; BP179-25; Fisher Bioregents). 7 μl of the mixture was used for a 22 × 30 mm coverslip.
For 460 Pa gels, a standard solution was prepared with 1.25 ml of 40% Acrylamide (1610140; Biorad), 666 μl 2% bisAcrylamide (1610142; Biorad), and 3.08 ml ddH2O. To make the gel, 150 μl of the standard solution was mixed with 341.75 μl water, 5 μl 0.04-µm fluorescent microspheres (F8789; Invitrogen), 0.75 μl TEMED (110-18-9; Fisher Bioregents), and 2.5 μl of 10% APS (BP179-25; Fisher Bioregents). 7 μl of the mixture was used for a 22 × 30 mm coverslip.
For 16 kPa gels, a standard solution was prepared with 2.5 ml of 40% Acrylamide (1610140; Biorad), 604 μl 2% bisAcrylamide (1610142; Biorad), and 1.896 ml ddH2O. To make the gel, 300 μl of standard solution was mixed with 191.75 μl water, 5 μl 0.04-µm fluorescent microspheres (F8789; Invitrogen), 0.75 μl TEMED (110-18-9; Fisher Bioregents) and 2.5 μl 10% APS (BP179-25; Fisher Bioregents). 7 μl of the mixture was used for a 22 × 30 mm coverslip.
Analysis of traction forces was performed using code written in Python according to previously described approaches (Sabass et al., 2008; Hanke et al., 2018; Sala and Oakes, 2021). The code is available at https://github.com/OakesLab/TFM. Prior to processing, images were flat-field corrected and aligned to the reference bead image with the cell detached. Other acquired channels were shifted using the same alignment measurements from the bead channel. Displacements in the beads were calculated using an optical flow algorithm in OpenCV (Open Source Computer Vision Library, https://github.com/itseez/opencv) with a window size of eight pixels. Traction stresses were calculated using the Fourier Transform Traction Cytometry (FTTC) approach (Sabass et al., 2008; Huang et al., 2019) as previously described, with a regularization parameter of 8.87 × 10−5 for the soft (336 and 460 Pa) gels and a regularization parameter of 3.74 × 10−7 for the 16 kPa gels.
For the 2D histograms (Fig. 4, D and E), we calculated the centroid of a binary cell mask, and then for each pixel in the cell mask, we calculated the vector that pointed from that pixel toward the centroid. If represents the traction stress vector at a pixel and represents the vector toward the centroid, we used the dot product of two vectors to calculate . The magnitude of the traction stress was calculated as . The 2D histogram was then constructed from these two values on a per-pixel basis using all pixels contained in the cell masks across all the cells measured in the dataset.
Gel stiffness measurements
Gels were fabricated as described above, with the only difference being that a spacer was used during polymerization to create a thicker gel of ≈300–350 µm in height. Gel stiffness was measured by measuring the deformation caused by a stainless steel ball-bearing 1.5 mm in diameter, as previously described (Lee et al., 2015; Schmitt et al., 2024). Briefly, the gel height was measured by taking the difference between the bottom and top of the gel. A confocal z-stack with a step size of 1.25 µm was then taken through the top of the gel and the deformation was determined by finding the center of the indentation and fitting a circle with a radius equivalent to the bearing. This depth measurement was repeated in two orthogonal directions and averaged. The gel Young’s modulus was then determined using a modified Hertz model (Dimitriadis et al., 2002; Lee et al., 2015) to account for the gel being thin gels bonded to a surface. At least two measurements were taken per gel, with at least two gels per replicate, and experiments were repeated in triplicate. The gel stiffness value represents the shear modulus.
Cell migration tracking
All cell tracking was performed using trackpy (Allan et al., 2023). Tracks were only included if they lasted more than five frames and tracked cells were not allowed to skip frames. Once the track positions were calculated, track parameters were calculated as follows: instantaneous velocity was the difference between positions between frames; path duration was the total number of frames multiplied by the frame interval; path length was the sum of the displacements between each point along the track; the average velocity was the path length divided by the path duration; the actual displacement was the geometric distance between the first and last points of the path; and the effective velocity was the actual displacement divided by the path duration. Cells were classified as immobile if the actual displacement was <10 µm (Fig. 2 F).
To determine overlapping tracks, we only considered instances of two separate cells traversing the same area, leaving out those cells that crossed back upon their own trajectory. We first identified all tracks with a path length>10 µm. All possible pair permutations of these tracks were compared with each other by calculating the Euclidean distance between each point in the track. Overlaps were identified as regions where points were <1 µm apart. Following identification, the list of overlaps was filtered to only consider those overlaps that contained at least three time points and that were >10 µm. This was done to ensure that random crossings and coincidental overlaps were not considered. From this list, the leader cell was identified as the cell that appeared first in the overlap region, and the average velocity in the overlap region was calculated. The second cell that passed through an overlap region was labeled as the follower and the velocity was calculated as the average in the overlap region. To calculate the frequency of overlaps, we counted the number of unique cells that followed another cell’s track and divided this by the total number of tracks in a movie. Given the stringent constraints imposed, this value should be taken as a cautious minimum estimate.
Statistical analysis
For all analyses, statistically different results are defined as follows: P value < 0.05 = *; P value < 0.005 = **; P value < 0.0005 = ***; P value < 0.0001 = ****. Non-statistically different results (P value > 0.05) are either identified with “ns” or without a star. For all analyses, cells that moved <10 µm were considered immobile. All comparisons were made using two-tail unpaired parametric t tests, except for Fig. 1 B; Fig. 2, E and F; Fig. 5, G–J; and Fig. S1, G and H, which used one-tail paired parametric t tests. For each replicate, unless otherwise specified, between 1,000 and 2,500 cells were analyzed spread across ≈5 fields of view per experiment, and the data point represents the mean of the value plotted. All graphical representations (except for Fig. 1 D; Fig. 2 A; and Fig. 5 A) used box and whiskers to represent the spread of the average of each field of view (gray dots). The whiskers represent the minimum and maximum value, the box represents the median as well as the 25th and 75th percentile. Overlays (colored dots) are the average of each biological replicate. For Fig. 1 D; Fig. 2 A; and Fig. 5 A, the ratio of mobile versus immobile cells is represented by a stacked bar graph. The whiskers represent the standard deviation. Gray dots = average of each field of view. Colored dots = average of each replicate. The number of biological replicates for the data presented in the figures is detailed below.
-
•
Fig. 1 B: FN, n = 3; ICAM-1, n = 4.
•Fig. 1, D–F: FN, n = 8; ICAM-1, n = 6; PLL, n = 6; Passivated, n = 7.
•Fig. 2, A–C: PLL, n = 3; Passivated, n = 4; 0.1 µg/ml FN, n = 3; 1 µg/ml FN, n = 3; 10 µg/ml FN, n = 4.
•Fig. 2, E and F: PBS n = 4; Ab-integrin n = 4.
•Fig. 3 F: Passivated, 36 cells from three experiments; PLL, 53 cells from two experiments; FN, 140 cells from five experiments; and ICAM-1, 40 cells from three experiments.
•Fig. 3 G: Passivated, 21 cells from two experiments; PLL, 38 cells from four experiments; FN, 94 cells from four experiments; ICAM-1 169 cells from 3 experiments.
•Fig. 3 H: Th1 n = 10; Fibroblast n = 10. Each replicate compared the turnover of at least five FAs per cell.
•Fig. 4, D and E: Th1, 12 cells, 45 fields of view from four different experiments; fibroblast, 12 cells from 12 fields of view. The data for the histogram was pooled from all cells resulting in ≈6 × 105 vectors for the Th1 cells, and 2 × 106 vectors for the fibroblasts. Both cell types were on FN-coated gels.
•Fig. 4 H: Th1 on uncoated gels, 11 cells, 44 fields of view from two different experiments. The data for the histogram was pooled from all cells resulting in ≈4 × 105 vectors.
•Fig. 5, A and B: Uncoated, n = 3; FN, n = 4; ICAM-1, n = 3.
•Fig. 5 F: FN, n = 4; ICAM-1, n = 3.
•Fig. 5 G: FN, n = 4.
•Fig. 5 H: ICAM-1, n = 3.
•Fig. 5 I: FN + DMSO, n = 2.
•Fig. 5 J: FN + MMP inhibitor, n = 2.
•Fig. S1, G and H. All conditions n = 3. Samples with and without FBS were performed with the same preparation of cells, on the same day, from the same mice.
RNAseq data analysis
We used RNA-seq data from the publicly available database (https://Th-express.org [Stubbington et al., 2015]) to compare a selection of different proteins involved in focal adhesion and integrins. The gene name of each of the selected proteins was searched on https://Th-express.org database. The regularized-log (rlog) measure was collected for each gene. The regularized-log (Rlog) represents transformed gene expression counts for all genes to ensure that they are homoskedastic (Love et al., 2014). The results were presented using a colormap to show high expression, intermediate expression, and low expression levels based on the description of gene frequency in the database.
Code
All codes used to analyze data is available at https://github.com/OakesLab.
Online supplemental material
The supplement contains three figures. Fig. S1 contains the Th1 activation protocol and flow cytometry data as well as data related to Fig. 1. Fig. S2 contains Western blots and RNAseq data to represent the endogenous focal adhesion proteins present in Th1. Fig. S3 shows additional focal adhesion protein localization to FAs similar to Fig. 3. Finally, the supplement also contains movies representing data in the following figures: Video 1 represents data shown in Fig. 1, D–F; Video 2 represents data shown in Fig. 1, H–J; Video 3 represents data shown in Fig. 2, A–D; Video 4 represents data shown in Fig. 2, G and H; Video 5 represents data shown in Fig. 3, A–D; Video 6 represents data shown in Fig. 3, I and J; Video 7 represents data shown in Fig. 4, F and G; and Video 8 represents data shown in Fig. 5 E.
Supplementary Material
is the source file for Fig. S2.
Acknowledgments
We want to thank all the members of the Beach and Oakes laboratories at Loyola, the Topham and Kim labs at the University of Rochester, and the Fowell lab at Cornell for their helpful feedback. We also want to thank the Piel lab from Institut Curie for their help in the confinement techniques and Dr. Stefano Sala for assistance with the gel stiffness measurements. A. Caillier acknowledges support from the Company of Biologist for financing a training visit to the Piel lab. We thank the Frederick National Laboratory BRB Preclinical Repository for supplying IL-2 and IIB.II (Anti-Murine IL-4).
D.J. Fowell, J. Miller, and P.W. Oakes acknowledge support from the National Institutes of Health National Institute of Allergy and Infectious Disease award P01-AI102851.
Author contributions: A. Caillier: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing—original draft, Writing—review and editing, D. Oleksyn: Resources, D.J. Fowell: Conceptualization, Funding acquisition, Resources, Writing—review and editing, J. Miller: Conceptualization, Methodology, Resources, Writing—review and editing, P.W. Oakes: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing—original draft, Writing—review and editing.
Data availability
The data are available from the corresponding author upon reasonable request.
References
- Abraham, M., Shapiro S., Karni A., Weiner H.L., and Miller A.. 2005. Gelatinases (MMP-2 and MMP-9) are preferentially expressed by Th1 vs. Th2 cells. J. Neuroimmunol. 163:157–164. 10.1016/j.jneuroim.2005.02.001 [DOI] [PubMed] [Google Scholar]
- Adebowale, K., Ha B., Saraswathibhatla A., Indana D., Popescu M.C., Demirdjian S., Yang J., Bassik M.C., Franck C., Bollyky P.L., and Chaudhuri O.. 2023. Monocytes use protrusive forces to generate migration paths in viscoelastic collagen-based extracellular matrices. bioRxiv. 10.1101/2023.06.09.544394 (Preprint posted June 11, 2023). [DOI] [Google Scholar]
- Allan, D.B., Caswell T., Keim N.C., van der Wel C.M., and Verweij R.W.. 2023. Soft-matter/trackpy: v0.6.1. Zenodo. 10.5281/zenodo.7670439 [DOI] [Google Scholar]
- Aoun, L., Farutin A., Garcia-Seyda N., Nègre P., Rizvi M.S., Tlili S., Song S., Luo X., Biarnes-Pelicot M., Galland R., et al. 2020. Amoeboid swimming is propelled by molecular paddling in lymphocytes. Biophys. J. 119:1157–1177. 10.1016/j.bpj.2020.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangasser, B.L., Shamsan G.A., Chan C.E., Opoku K.N., Tüzel E., Schlichtmann B.W., Kasim J.A., Fuller B.J., McCullough B.R., Rosenfeld S.S., and Odde D.J.. 2017. Shifting the optimal stiffness for cell migration. Nat. Commun. 8:15313. 10.1038/ncomms15313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baschieri, F., Illand A., Barbazan J., Zajac O., Henon C., Loew D., Dingli F., Vignjevic D.M., Lévêque-Fort S., and Montagnac G.. 2023. Fibroblasts generate topographical cues that steer cancer cell migration. Sci. Adv. 9:eade2120. 10.1126/sciadv.ade2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu, R., and Huse M.. 2017. Mechanical communication at the immunological synapse. Trends Cell Biol. 27:241–254. 10.1016/j.tcb.2016.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergert, M., Erzberger A., Desai R.A., Aspalter I.M., Oates A.C., Charras G., Salbreux G., and Paluch E.K.. 2015. Force transmission during adhesion-independent migration. Nat. Cell Biol. 17:524–529. 10.1038/ncb3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertoni, A., Alabiso O., Galetto A.S., and Baldanzi G.. 2018. Integrins in t cell physiology. Int. J. Mol. Sci. 19:485. 10.3390/ijms19020485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidone, T.C., Skeeters A.V., Oakes P.W., and Voth G.A.. 2019. Multiscale model of integrin adhesion assembly. PLOS Comput. Biol. 15:e1007077. 10.1371/journal.pcbi.1007077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhringer, D., Cóndor M., Bischof L., Czerwinski T., Gampl N., Ngo P.A., Bauer A., Voskens C., López-Posadas R., Franze K., et al. 2023. Dynamic traction force measurements of migrating immune cells in 3D matrices. bioRxiv. 10.1101/2022.11.16.516758 (Preprint posted August 24, 2023). [DOI] [Google Scholar]
- Boyd, A.W., Wawryk S.O., Burns G.F., and Fecondo J.V.. 1988. Intercellular adhesion molecule 1 (ICAM-1) has a central role in cell-cell contact-mediated immune mechanisms. Proc. Natl. Acad. Sci. USA. 85:3095–3099. 10.1073/pnas.85.9.3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan-Jones, A. 2022. Self-organization in amoeboid motility. Front. Cell Dev. Biol. 10:1000071. 10.3389/fcell.2022.1000071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalcanti-Adam, E.A., Volberg T., Micoulet A., Kessler H., Geiger B., and Spatz J.P.. 2007. Cell spreading and focal adhesion dynamics are regulated by spacing of integrin ligands. Biophys. J. 92:2964–2974. 10.1529/biophysj.106.089730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., and Zhu C.. 2013. Mechanical regulation of T-cell functions. Immunol. Rev. 256:160–176. 10.1111/imr.12122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W., Lou J., and Zhu C.. 2010. Forcing switch from short- to intermediate- and long-lived states of the alphaA domain generates LFA-1/ICAM-1 catch bonds. J. Biol. Chem. 285:35967–35978. 10.1074/jbc.M110.155770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Lee H., Tong H., Schwartz M., and Zhu C.. 2017. Force regulated conformational change of integrin αVβ3. Matrix Biol. 60-61:70–85. 10.1016/j.matbio.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, N.-C., Tu Y.-K., Lee N.-H., and Young T.-H.. 2020. Influence of human platelet lysate on extracellular matrix deposition and cellular characteristics in adipose-derived stem cell sheets. Front. Cell Dev. Biol. 8:558354. 10.3389/fcell.2020.558354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, C.K., Vicente-Manzanares M., Zareno J., Whitmore L.A., Mogilner A., and Horwitz A.R.. 2008. Actin and alpha-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10:1039–1050. 10.1038/ncb1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerwinski, T., Bischof L., Böhringer D., Kara S., Wittmann E., Winterl A., Gerum R., Nusser G., Wiesinger M., Budday S., et al. 2023. Immune cells employ intermittent integrin-mediated traction forces for 3D migration. bioRxiv. 10.1101/2023.04.20.537658 (Preprint posted August 3, 2023). [DOI] [Google Scholar]
- De Bruyn, P.P.H. 1946. The amoeboid movement of the mammalian leukocyte in tissue culture. Anat. Rec. 95:177–191. 10.1002/ar.1090950209 [DOI] [PubMed] [Google Scholar]
- de Fougerolles, A.R., Sprague A.G., Nickerson-Nutter C.L., Chi-Rosso G., Rennert P.D., Gardner H., Gotwals P.J., Lobb R.R., and Koteliansky V.E.. 2000. Regulation of inflammation by collagen-binding integrins α1β1 and α2β1 in models of hypersensitivity and arthritis. J. Clin. Invest. 105:721–729. 10.1172/JCI7911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dembo, M., and Wang Y.-L.. 1999. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys. J. 76:2307–2316. 10.1016/S0006-3495(99)77386-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitriadis, E.K., Horkay F., Maresca J., Kachar B., and Chadwick R.S.. 2002. Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophys. J. 82:2798–2810. 10.1016/S0006-3495(02)75620-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, H., Bartleson J.M., Butenko S., Alonso V., Liu W.F., Winer D.A., and Butte M.J.. 2023. Tuning immunity through tissue mechanotransduction. Nat. Rev. Immunol. 23:174–188. 10.1038/s41577-022-00761-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes, N.R.J., Reilly N.S., Schrock D.C., Hocking D.C., Oakes P.W., and Fowell D.J.. 2020. Cd4+ t cell interstitial migration controlled by fibronectin in the inflamed skin. Front. Immunol. 11:1501. 10.3389/fimmu.2020.01501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowell, D.J., and Kim M.. 2021. The spatio-temporal control of effector T cell migration. Nat. Rev. Immunol. 21:582–596. 10.1038/s41577-021-00507-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl, P., Entschladen F., Conrad C., Niggemann B., and Zänker K.S.. 1998. CD4+ T lymphocytes migrating in three-dimensional collagen lattices lack focal adhesions and utilize beta1 integrin-independent strategies for polarization, interaction with collagen fibers and locomotion. Eur. J. Immunol. 28:2331–2343. [DOI] [PubMed] [Google Scholar]
- Fukui, Y., and Inoué S.. 1997. Amoeboid movement anchored by eupodia, new actin-rich knobby feet in Dictyostelium. Cell Motil. Cytoskeleton. 36:339–354. [DOI] [PubMed] [Google Scholar]
- Gaertner, F., Reis-Rodrigues P., de Vries I., Hons M., Aguilera J., Riedl M., Leithner A., Tasciyan S., Kopf A., Merrin J., et al. 2022. WASp triggers mechanosensitive actin patches to facilitate immune cell migration in dense tissues. Dev. Cell. 57:47–62.e9. 10.1016/j.devcel.2021.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel, M.L., Schneider I.C., Aratyn-Schaus Y., and Waterman C.M.. 2010. Mechanical integration of actin and adhesion dynamics in cell migration. Annu. Rev. Cell Dev. Biol. 26:315–333. 10.1146/annurev.cellbio.011209.122036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawden-Bone, C., West M.A., Morrison V.L., Edgar A.J., McMillan S.J., Dill B.D., Trost M., Prescott A., Fagerholm S.C., and Watts C.. 2014. A crucial role for β2 integrins in podosome formation, dynamics and Toll-like-receptor-signaled disassembly in dendritic cells. J. Cell Sci. 127:4213–4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaylo, A., Schrock D.C., Fernandes N.R., and Fowell D.J.. 2016. T cell interstitial migration: Motility cues from the inflamed tissue for micro-and macro-positioning. Front. Immunol. 7:428. 10.3389/fimmu.2016.00428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaylo-Moynihan, A., Prizant H., Popović M., Fernandes N.R.J., Anderson C.S., Chiou K.K., Bell H., Schrock D.C., Schumacher J., Capece T., et al. 2019. Programming of distinct chemokine-dependent and-independent search strategies for th1 and th2 cells optimizes function at inflamed sites. Immunity. 51:298–309.e6. 10.1016/j.immuni.2019.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoudi, C., Muheidli A., and Aoudjit F.. 2023. β1 Integrin induces adhesion and migration of human Th17 cells via Pyk2-dependent activation of P2X4 receptor. Immunology. 168:83–95. 10.1111/imm.13563 [DOI] [PubMed] [Google Scholar]
- Hanke, J., Probst D., Zemel A., Schwarz U.S., and Köster S.. 2018. Dynamics of force generation by spreading platelets. Soft Matter. 14:6571–6581. 10.1039/C8SM00895G [DOI] [PubMed] [Google Scholar]
- Hartman, C.D., Isenberg B.C., Chua S.G., and Wong J.Y.. 2017. Extracellular matrix type modulates cell migration on mechanical gradients. Exp. Cell Res. 359:361–366. 10.1016/j.yexcr.2017.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman, E.G., and Ruoslahti E.. 1979. Distribution of fetal bovine serum fibronectin and endogenous rat cell fibronectin in extracellular matrix. J. Cell Biol. 83:255–259. 10.1083/jcb.83.1.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg, N., Laschinger M., Giles K., and McDowall A.. 2003. T-Cell integrins: More than just sticking points. J. Cell Sci. 116:4695–4705. 10.1242/jcs.00876 [DOI] [PubMed] [Google Scholar]
- Hons, M., Kopf A., Hauschild R., Leithner A., Gaertner F., Abe J., Renkawitz J., Stein J.V., and Sixt M.. 2018. Chemokines and integrins independently tune actin flow and substrate friction during intranodal migration of T cells. Nat. Immunol. 19:606–616. 10.1038/s41590-018-0109-z [DOI] [PubMed] [Google Scholar]
- Huang, Y., Schell C., Huber T.B., Şimşek A.N., Hersch N., Merkel R., Gompper G., and Sabass B.. 2019. Traction force microscopy with optimized regularization and automated Bayesian parameter selection for comparing cells. Sci. Rep. 9:539. 10.1038/s41598-018-36896-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse, M. 2017. Mechanical forces in the immune system. Nat. Rev. Immunol. 17:679–690. 10.1038/nri.2017.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isomursu, A., Park K.-Y., Hou J., Cheng B., Mathieu M., Shamsan G.A., Fuller B., Kasim J., Mahmoodi M.M., Lu T.J., et al. 2022. Directed cell migration towards softer environments. Nat. Mater. 21:1081–1090. 10.1038/s41563-022-01294-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey, P.A., Fletcher D.A., and Reinhart-King C.A.. 2020. Stiffness sensing by cells. Physiol. Rev. 100:695–724. 10.1152/physrev.00013.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbach, L., Martzloff P., Bambach S.K., Aizarani N., Mihlan M., Gavrilov A., Glaser K.M., Stecher M., Thünauer R., Thiriot A., et al. 2023. Slow integrin-dependent migration organizes networks of tissue-resident mast cells. Nat. Immunol. 24:915–924. 10.1038/s41590-023-01493-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong, P., Shtengel G., Pasapera A.M., Ramko E.B., Davidson M.W., Hess H.F., and Waterman C.M.. 2010. Nanoscale architecture of integrin-based cell adhesions. Nature. 468:580–584. 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller, H.U., and Bebie H.. 1996. Protrusive activity quantitatively determines the rate and direction of cell locomotion. Cell Motil. Cytoskeleton. 33:241–251. [DOI] [PubMed] [Google Scholar]
- Kim, S.H.J., and Hammer D.A.. 2019. Integrin crosstalk allows CD4+ T lymphocytes to continue migrating in the upstream direction after flow. Integr. Biol.. 11:384–393. 10.1093/intbio/zyz034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämmermann, T., and Germain R.N.. 2014. The multiple faces of leukocyte interstitial migration. Semin. Immunopathol. 36:227–251. 10.1007/s00281-014-0418-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lämmermann, T., and Sixt M.. 2009. Mechanical modes of “amoeboid” cell migration. Curr. Opin. Cell Biol. 21:636–644. 10.1016/j.ceb.2009.05.003 [DOI] [PubMed] [Google Scholar]
- Lämmermann, T., Bader B.L., Monkley S.J., Worbs T., Wedlich-Söldner R., Hirsch K., Keller M., Förster R., Critchley D.R., Fässler R., and Sixt M.. 2008. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature. 453:51–55. 10.1038/nature06887 [DOI] [PubMed] [Google Scholar]
- Le Berre, M., Zlotek-Zlotkiewicz E., Bonazzi D., Lautenschlaeger F., and Piel M.. 2014. Methods for two-dimensional cell confinement. Methods Cell Biol. 121:213–229. 10.1016/B978-0-12-800281-0.00014-2 [DOI] [PubMed] [Google Scholar]
- Lee, D., Rahman M.M., Zhou Y., and Ryu S.. 2015. Three-Dimensional confocal microscopy indentation method for hydrogel elasticity measurement. Langmuir. 31:9684–9693. 10.1021/acs.langmuir.5b01267 [DOI] [PubMed] [Google Scholar]
- Li, Q., Huth S., Adam D., and Selhuber-Unkel C.. 2016. Reinforcement of integrin-mediated T-Lymphocyte adhesion by TNF-induced Inside-out Signaling. Sci. Rep. 6:30452. 10.1038/srep30452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, K., Hyun Y.-M., Lambert-Emo K., Capece T., Bae S., Miller R., Topham D.J., and Kim M.. 2015. Neutrophil trails guide influenza-specific CD8+ T cells in the airways. Science. 349:aaa4352. 10.1126/science.aaa4352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.-J., Le Berre M., Lautenschlaeger F., Maiuri P., Callan-Jones A., Heuzé M., Takaki T., Voituriez R., and Piel M.. 2015. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 160:659–672. 10.1016/j.cell.2015.01.007 [DOI] [PubMed] [Google Scholar]
- Lo, C.-M., Wang H.-B., Dembo M., and Wang Y.L.. 2000. Cell movement is guided by the rigidity of the substrate. Biophys. J. 79:144–152. 10.1016/S0006-3495(00)76279-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Vidal, L., Murdica V., Venegoni C., Pederzoli F., Bandini M., Necchi A., Salonia A., and Alfano M.. 2021. Causal contributors to tissue stiffness and clinical relevance in urology. Commun. Biol. 4:1011. 10.1038/s42003-021-02539-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mijanović, L., and Weber I.. 2022. Adhesion of Dictyostelium amoebae to surfaces: A brief history of attachments. Front. Cell Dev. Biol. 10:910736. 10.3389/fcell.2022.910736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau, H.D., Piel M., Voituriez R., and Lennon-Duménil A.-M.. 2018. Integrating physical and molecular insights on immune cell migration. Trends Immunol. 39:632–643. 10.1016/j.it.2018.04.007 [DOI] [PubMed] [Google Scholar]
- Mrass, P., Oruganti S.R., Fricke G.M., Tafoya J., Byrum J.R., Yang L., Hamilton S.L., Miller M.J., Moses M.E., and Cannon J.L.. 2017. ROCK regulates the intermittent mode of interstitial T cell migration in inflamed lungs. Nat. Commun. 8:1010. 10.1038/s41467-017-01032-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayal, A., Webb D.J., Brown C.M., Schaefer E.M., Vicente-Manzanares M., and Horwitz A.R.. 2006. Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J. Cell Biol. 173:587–589. 10.1083/jcb.200509075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas-Boluda, A., Vaquero J., Vimeux L., Guilbert T., Barrin S., Kantari-Mimoun C., Ponzo M., Renault G., Deptula P., Pogoda K., et al. 2021. Tumor stiffening reversion through collagen crosslinking inhibition improves T cell migration and anti-PD-1 treatment. Elife. 10:e58688. 10.7554/eLife.58688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourshargh, S., and Alon R.. 2014. Leukocyte migration into inflamed tissues. Immunity. 41:694–707. 10.1016/j.immuni.2014.10.008 [DOI] [PubMed] [Google Scholar]
- Oakes, P.W., Beckham Y., Stricker J., and Gardel M.L.. 2012. Tension is required but not sufficient for focal adhesion maturation without a stress fiber template. J. Cell Biol. 196:363–374. 10.1083/jcb.201107042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes, P.W., Bidone T.C., Beckham Y., Skeeters A.V., Ramirez-San Juan G.R., Winter S.P., Voth G.A., and Gardel M.L.. 2018. Lamellipodium is a myosin-independent mechanosensor. Proc. Natl. Acad. Sci. USA. 115:2646–2651. 10.1073/pnas.1715869115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overstreet, M.G., Gaylo A., Angermann B., Hughson A., Hyun Y., Lambert K., Acharya M., Billroth-Maclurg A.C., Rosenberg A.F., Topham D.J., et al. 2013. Inflammation-induced effector cd4+ t cell interstitial migration is alpha-v integrin dependent. Nat. Immunol. 14:949. 10.1038/ni.2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oviedo-Orta, E., Bermudez-Fajardo A., Karanam S., Benbow U., and Newby A.C.. 2008. Comparison of MMP-2 and MMP-9 secretion from T helper 0, 1 and 2 lymphocytes alone and in coculture with macrophages. Immunology. 124:42–50. 10.1111/j.1365-2567.2007.02728.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen, K.A., Pixley F.J., Thomas K.S., Vicente-Manzanares M., Ray B.J., Horwitz A.F., Parsons J.T., Beggs H.E., Stanley E.R., and Bouton A.H.. 2007. Regulation of lamellipodial persistence, adhesion turnover, and motility in macrophages by focal adhesion kinase. J. Cell Biol. 179:1275–1287. 10.1083/jcb.200708093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paluch, E.K., Aspalter I.M., and Sixt M.. 2016. Focal adhesion-independent cell migration. Annu. Rev. Cell Dev. Biol. 32:469–490. 10.1146/annurev-cellbio-111315-125341 [DOI] [PubMed] [Google Scholar]
- Paterson, N., and Lämmermann T.. 2022. Macrophage network dynamics depend on haptokinesis for optimal local surveillance. Elife. 11:e75354. 10.7554/eLife.75354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak, A., and Kumar S.. 2012. Independent regulation of tumor cell migration by matrix stiffness and confinement. Proc. Natl. Acad. Sci. USA. 109:10334–10339. 10.1073/pnas.1118073109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez Ipiña, E., d’Alessandro J., Ladoux B., and Camley B.A.. 2024. Deposited footprints let cells switch between confined, oscillatory, and exploratory migration. Proc. Natl. Acad. Sci. USA. 121:e2318248121. 10.1073/pnas.2318248121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie, R.J., and Yamada K.M.. 2012. At the leading edge of three-dimensional cell migration. J. Cell Sci. 125:5917–5926. 10.1242/jcs.093732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieuchot, L., Marteau J., Guignandon A., Dos Santos T., Brigaud I., Chauvy P.-F., Cloatre T., Ponche A., Petithory T., Rougerie P., et al. 2018. Curvotaxis directs cell migration through cell-scale curvature landscapes. Nat. Commun. 9:3995. 10.1038/s41467-018-06494-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinner, S.E., and Sahai E.. 2009. Integrin-independent movement of immune cells. F1000 Biol. Rep. 1:67. 10.3410/B1-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poincloux, R., Collin O., Lizárraga F., Romao M., Debray M., Piel M., and Chavrier P.. 2011. Contractility of the cell rear drives invasion of breast tumor cells in 3D Matrigel. Proc. Natl. Acad. Sci. USA. 108:1943–1948. 10.1073/pnas.1010396108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, G.O., Bernardi L., Lauxen I., Sant’Ana Filho M., Horwitz A.R., and Lamers M.L.. 2016. Fibronectin modulates cell adhesion and signaling to promote single cell migration of highly invasive oral squamous cell carcinoma. PLoS One. 11:e0151338. 10.1371/journal.pone.0151338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray, S.J., Franki S.N., Pierce R.H., Dimitrova S., Koteliansky V., Sprague A.G., Doherty P.C., de Fougerolles A.R., and Topham D.J.. 2004. The collagen binding alpha1beta1 integrin VLA-1 regulates CD8 T cell-mediated immune protection against heterologous influenza infection. Immunity. 20:167–179. 10.1016/S1074-7613(04)00021-4 [DOI] [PubMed] [Google Scholar]
- Reichardt, P., Patzak I., Jones K., Etemire E., Gunzer M., and Hogg N.. 2013. A role for LFA-1 in delaying T-lymphocyte egress from lymph nodes. EMBO J. 32:829–843. 10.1038/emboj.2013.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss, Y., Hoch G., Deutsch U., and Engelhardt B.. 1998. T cell interaction with ICAM-1-deficient endothelium in vitro: Essential role for ICAM-1 and ICAM-2 in transendothelial migration of T cells. Eur. J. Immunol. 28:3086–3099. [DOI] [PubMed] [Google Scholar]
- Reversat, A., Gaertner F., Merrin J., Stopp J., Tasciyan S., Aguilera J., de Vries I., Hauschild R., Hons M., Piel M., et al. 2020. Cellular locomotion using environmental topography. Nature. 582:582–585. 10.1038/s41586-020-2283-z [DOI] [PubMed] [Google Scholar]
- Sabass, B., Gardel M.L., Waterman C.M., and Schwarz U.S.. 2008. High resolution traction force microscopy based on experimental and computational advances. Biophys. J. 94:207–220. 10.1529/biophysj.107.113670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala, S., and Oakes P.W.. 2021. Stress fiber strain recognition by the LIM protein testin is cryptic and mediated by RhoA. Mol. Biol. Cell. 32:1758–1771. 10.1091/mbc.E21-03-0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick, J., and Raz E.. 2022. Blebs—formation, regulation, positioning, and role in amoeboid cell migration. Front. Cell Dev. Biol. 10:926394. 10.3389/fcell.2022.926394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, M.S., Colen J., Sala S., Devany J., Seetharaman S., Caillier A., Gardel M.L., Oakes P.W., and Vitelli V.. 2024. Machine learning interpretable models of cell mechanics from protein images. Cell. 187:481–494.e24. 10.1016/j.cell.2023.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroka, K.M., Jiang H., Chen S.-H., Tong Z., Wirtz D., Sun S.X., and Konstantopoulos K.. 2014. Water permeation drives tumor cell migration in confined microenvironments. Cell. 157:611–623. 10.1016/j.cell.2014.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbington, M.J., Mahata B., Svensson V., Deonarine A., Nissen J.K., Betz A.G., and Teichmann S.A.. 2015. An atlas of mouse CD4(+) T cell transcriptomes. Biol. Direct. 10:14. 10.1186/s13062-015-0045-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina, T. 2018. The actin cytoskeleton and actin-based motility. Cold Spring Harb. Perspect. Biol. 10:a018267. 10.1101/cshperspect.a018267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tello-Lafoz, M., Srpan K., Sanchez E.E., Hu J., Remsik J., Romin Y., Calò A., Hoen D., Bhanot U., Morris L., et al. 2021. Cytotoxic lymphocytes target characteristic biophysical vulnerabilities in cancer. Immunity. 54:1037–1054.e7. 10.1016/j.immuni.2021.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien, J., Ghani U., Dance Y.W., Seibel A.J., Karakan M.Ç., Ekinci K.L., and Nelson C.M.. 2020. Matrix pore size governs escape of human breast cancer cells from a microtumor to an empty cavity. iScience. 23:101673. 10.1016/j.isci.2020.101673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres, D.J., Mrass P., Byrum J., Gonzales A., Martinez D.N., Juarez E., Thompson E., Vezys V., Moses M.E., and Cannon J.L.. 2023. Quantitative analyses of T cell motion in tissue reveals factors driving T cell search in tissues. Elife. 12:e84916. 10.7554/eLife.84916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Goethem, E., Guiet R., Balor S., Charrière G.M., Poincloux R., Labrousse A., Maridonneau-Parini I., and Le Cabec V.. 2011. Macrophage podosomes go 3D. Eur. J. Cell Biol. 90:224–236. 10.1016/j.ejcb.2010.07.011 [DOI] [PubMed] [Google Scholar]
- Walkey, C.D., Olsen J.B., Guo H., Emili A., and Chan W.C.. 2012. Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake. J. Am. Chem. Soc. 134:2139–2147. 10.1021/ja2084338 [DOI] [PubMed] [Google Scholar]
- Wang, M.S., Hu Y., Sanchez E.E., Xie X., Roy N.H., de Jesus M., Winer B.Y., Zale E.A., Jin W., Sachar C., et al. 2022. Mechanically active integrins target lytic secretion at the immune synapse to facilitate cellular cytotoxicity. Nat. Commun. 13:3222. 10.1038/s41467-022-30809-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, K., Müller R., Borgmann S., Bröcker E.-B., and Friedl P.. 2003. Amoeboid shape change and contact guidance: T-Lymphocyte crawling through fibrillar collagen is independent of matrix remodeling by MMPs and other proteases. Blood. 102:3262–3269. 10.1182/blood-2002-12-3791 [DOI] [PubMed] [Google Scholar]
- Zhang, X., Wojcikiewicz E.P., and Moy V.T.. 2006. Dynamic adhesion of T lymphocytes to endothelial cells revealed by atomic force microscopy. Exp. Biol. Med. 231:1306–1312. 10.1177/153537020623100804 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is the source file for Fig. S2.
Data Availability Statement
The data are available from the corresponding author upon reasonable request.