Abstract
Sex chromosomes are susceptible to the evolution of selfish meiotic drive elements that bias transmission and distort progeny sex ratios. Conflict between such sex-ratio drivers and the rest of the genome can trigger evolutionary arms races resulting in genetically suppressed “cryptic” drive systems. The Winters cryptic sex-ratio drive system of Drosophila simulans comprises a driver, Distorter on the X (Dox), and an autosomal suppressor, Not-much-yang, a retroduplicate of Dox that suppresses via production of endogenous small interfering RNAs (esiRNAs). Here we report that over 22 Dox-like (Dxl) sequences originated, amplified, and diversified over the ~250,000-year history of the three closely related species, D. simulans, D. mauritiana, and D. sechellia. The Dxl sequences encode a rapidly evolving family of protamines. Dxl copy numbers amplified by ectopic exchange among euchromatic islands of satellite DNAs on the X chromosome and separately spawned four esiRNA-producing suppressors on the autosomes. Our results reveal the genomic consequences of evolutionary arms races and highlight complex interactions among different classes of selfish DNAs.
Meiotic drive elements are evolutionarily “selfish” because they can readily invade populations and rise in frequency, even if otherwise deleterious to their bearers and other loci in the genome 1,2. Meiotic drive in the male germline usually involves at least two loci: drive alleles at one locus disrupt formation or function of gametes bearing drive-sensitive alleles at a second target locus 3–5. For such multilocus drive to invade a population, genetic linkage is required to limit the formation of “suicide” haplotypes that combine drive and sensitive target alleles 6,7. For autosomes, multilocus drive can only succeed when recombination is limited by physical proximity, residence in regions of reduced crossing over, and/or chromosomal inversions. For non-recombining sex chromosomes, however, drive alleles anywhere on the X chromosome can target sensitive alleles anywhere on the Y chromosome without risk of suicide genotypes. Sex chromosomes are therefore particularly susceptible to invasion by multilocus drive elements 8–10.
Drive on sex chromosomes can have major downstream effects. First, as sex-linked drivers incapacitate sensitive target-bearing gametes, they reduce fertility and bias progeny sex ratios (a phenotype known as sex-ratio). Second, sex-ratio drivers can increase in frequency and bias the population sex ratio, raising the risk of unisexual population extinction 8,11. Third, as the population frequency of sex-ratio drive increases, so do intensities of selection for resistant target alleles, selection for fertility-restoring suppressors, and Fisherian sex ratio selection for autosomal suppressors 8,12–15. Fourth, drivers are expected to recruit genetically linked enhancers that increase the efficiency and/or strength of drive 16,17. The dynamics of invasion, spread, and suppression can be exceedingly fast 8,18, giving rise to cryptic multilocus systems of interacting sex-ratio drivers, resistant targets, and/or autosomal suppressors that are otherwise difficult to detect. The evolutionary conflict between drivers and enhancers on one side versus targets and suppressors on the other can potentiate molecular arms races characterized by recurrent bouts of innovation and counter-innovation 9,10,15. If sufficiently common, these arms races may have important consequences for the evolution of stable Mendelian transmission, gametogenesis, recombination rates, genome structure, content, and regulation, and speciation 8–10,19–22.
In this paper, we investigate the evolution and genomic consequences of the cryptic Winters sex-ratio drive system first described in Drosophila simulans 23,24. The X-linked Distorter on the X (Dox) gene causes sex-ratio drive by disrupting maturation of Y-bearing spermatids 23, and the autosomal suppressor gene, Not-much-yang (Nmy), silences Dox via RNA interference 24,25. The molecular origins of Dox are obscure, whereas Nmy originated via retroduplication from Dox and now expresses predicted hairpin RNA (hpRNA) precursors in testes that are processed by Dcr-2 and AGO2 into esiRNAs 24,25. Dox and Nmy now segregate at appreciable frequencies in D. simulans and show signatures of selective sweeps, consistent with recent bouts of drive and suppression 26,27. Both genes are also present in the closely related species D. mauritiana 23,24, in which Dox is similarly associated with a very large selective sweep 27–29, but both are absent from D. sechellia 23,24. A second autosomal gene in D. simulans, Too-much-yin (Tmy), suppresses sex-ratio drive 30, but its functional relationship to Dox is unclear— Tmy produces esiRNAs that partially match 25 but fail to suppress Dox 23. This curious observation may hint that Tmy suppresses an as yet unidentified X-linked gene with sequence similarity to Dox. Notably, Dox and Nmy are absent from Sanger and Illumina sequence-based genome assemblies due, in part, to their proximity to difficult-to-map dispersed repetitive DNAs 23,24 (see below). Here we circumvent this problem using new ultrahigh-quality genome assemblies for the three D. simulans clade species based on long single-molecule sequence reads 31. We find three species-specific sex-ratio systems involving different, partially overlapping subsets of at least 18 X-linked Dox-related genes and four autosomal esiRNA-producing suppressor genes.
Results
Dox is part of a large satellite DNA-associated gene family
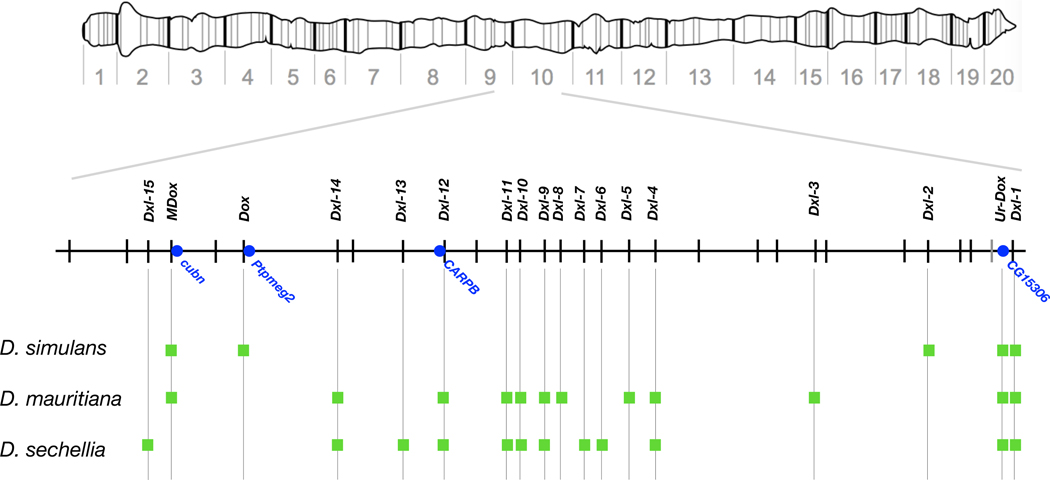
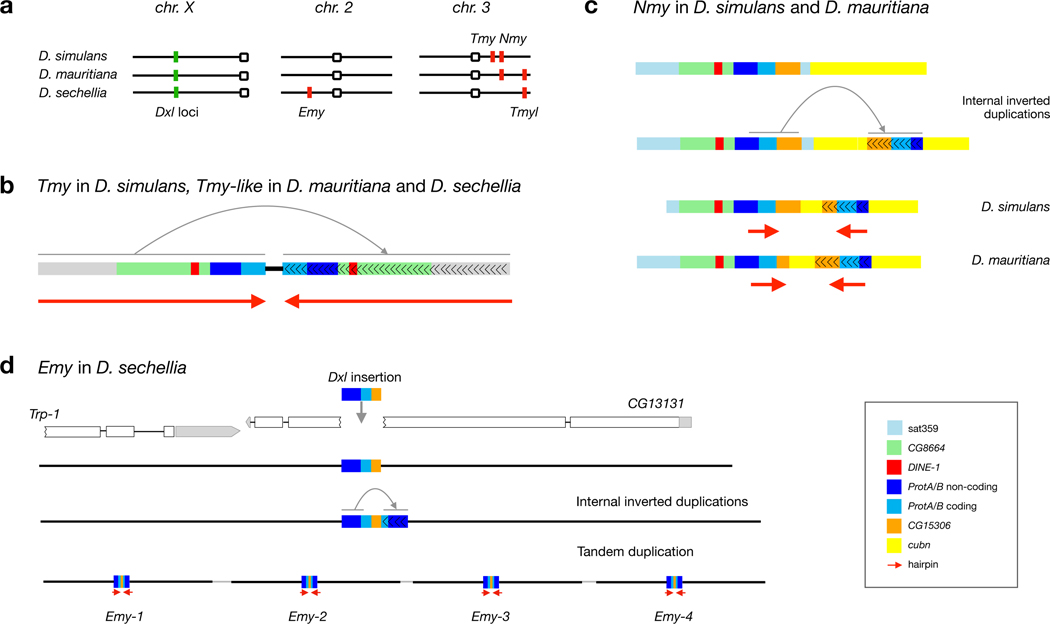
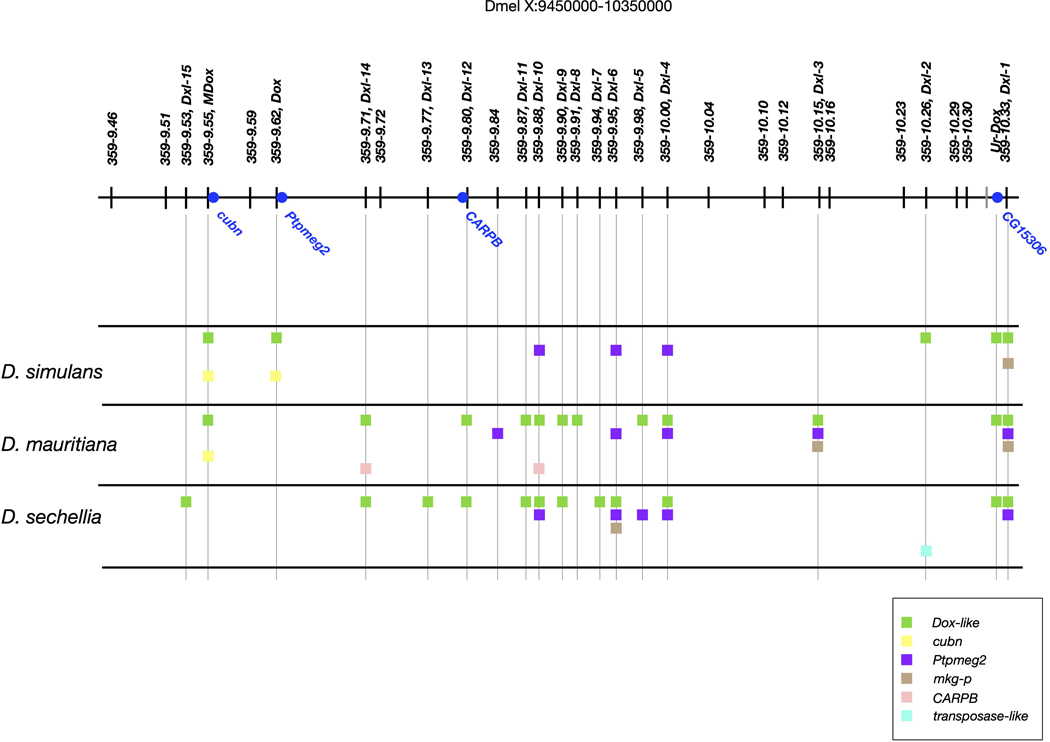
Dox is flanked by multiple copies of the highly abundant 359-bp monomer satellite DNA (hereafter, sat359), making it refractory to genome assembly using short DNA sequence reads. We leveraged new deep-coverage, long read-based genome assemblies that readily span these repetitive sites in all three D. simulans clade species 31. We discovered that a ~1-Mb interval on the X chromosome (~9.4–10.4 Mb using Dmel r6 coordinates) harbors a dispersed family of Dox-like (Dxl) genes at 18 positions, with at least five copies in D. simulans, 12 in D. mauritiana, and 12 in D. sechellia (Fig. 1 and Extended Data Fig. 1; see Methods and Supplementary Text S1). In D. simulans, the five Dxl genes include the previously described Dox and its putative parent gene Mother-of-Dox (MDox) 23 (Fig. 1). The presence of MDox in D. simulans and in D. mauritiana results from recent interspecific introgression 27. All other Dxl genes are previously undescribed, with nine Dxl locations unique to a single species, seven shared among two of the three species, and two shared among all three species (Fig. 1).
Figure 1.
Physical distribution of known Dxl genes in D. simulans, D. mauritiana, and D. sechellia. A schematic of the polytene X chromosome (top) shows location of the Dxl-containing region (Dmel r6 X:9400000–10400000). Tick marks show locations of sat359 islands conserved in all three D. simulans clade species and in the outgroup D. melanogaster; the single gray tick mark distal to Ur-Dox is a sat359 island found in the D. simulans clade species but not in D. melanogaster; the green squares show sat359 islands with a Dxl insertion; and the blue dots show protein-coding genes of interest. While Dxl insertions with the same name occupy orthologous sat359 islands in different species, the Dxl sequences are not necessarily orthologous due to the possibilities of independent, parallel insertion and ectopic exchange.
All but one of the Dxl genes inserted into pre-existing euchromatic islands of sat359 repeats that are conserved among species (Fig. 1). The three D. simulans clade species possess 307–325 islands of sat359s distributed along the euchromatin of the X chromosome, with most (68–89%) comprising 1–4 repeat units 32. These sat359 islands are shared with the outgroup species, D. melanogaster, and mediate X chromosome recognition for sex chromosome dosage compensation in the male soma 33. While homologous sat359 locations are generally conserved, Dxl genes appear to be absent from the more distant outgroup species, D. melanogaster and D. yakuba, indicating that the Dxl gene family originated in the common ancestor of the D. simulans clade and then expanded in each of the three descendant species lineages during the last ~250 Ky 34. In the 9.4–10.4 Mb interval of the X, 17 of 28 syntenic sat359 islands (61%) have been inserted by a Dxl gene in at least one of the D. simulans clade species (Fig. 1). We speculate that the sat359 repeats facilitated Dxl copy number amplification by inserting Dxl genes into paralogous sat359 islands via ectopic gene conversion and/or circular DNA intermediates 32. Assuming Dxl genes have (or once had) the capacity for sex-ratio drive, the expansion of Dxl gene copy number in all three D. simulans clade species suggests that drive may be Dxl gene dose-dependent. The recruitment of additional Dxl gene copies might, for instance, have boosted overall expression and/or contributed to evasion of transcriptional silencing by autosomal suppressors. Under this scenario, additional Dxl gene copies may function as enhancers or co-drivers. The rapid expansion of the Dxl gene family also generates substrate for potential functional diversification among copies (see below).
As the three D. simulans clade species originated via the nearly simultaneous divergence of D. sechellia and D. mauritiana from a D. simulans-like ancestor 34,35, the distribution of the 18 Dxl gene insertions among species is potentially informative about their history. Consistent with origins that predate speciation (>250 Kya), two are present in all three D. simulans clade species. Consistent with origins that postdate speciation (<250 Kya), nine are unique to a single species. Seven Dxl insertions, however, are shared among two of the three species, a configuration consistent with one of three scenarios: Dxl genes once present in all three species were deleted from one; Dxl genes originating in one species were exported to another via gene flow; or, orthologous sat359 islands were inserted by Dxl genes via independent, parallel events in two of the three species. Among the seven Dxl insertions shared by two species, six are shared between the two island endemics species, D. mauritiana and D. sechellia. This observation therefore requires a relative excess of Dxl deletions in D. simulans, an unlikely excess of gene flow between the two island endemic species, or excess parallel insertions in the island endemics.
The Dox-like genes have complex chimeric origins
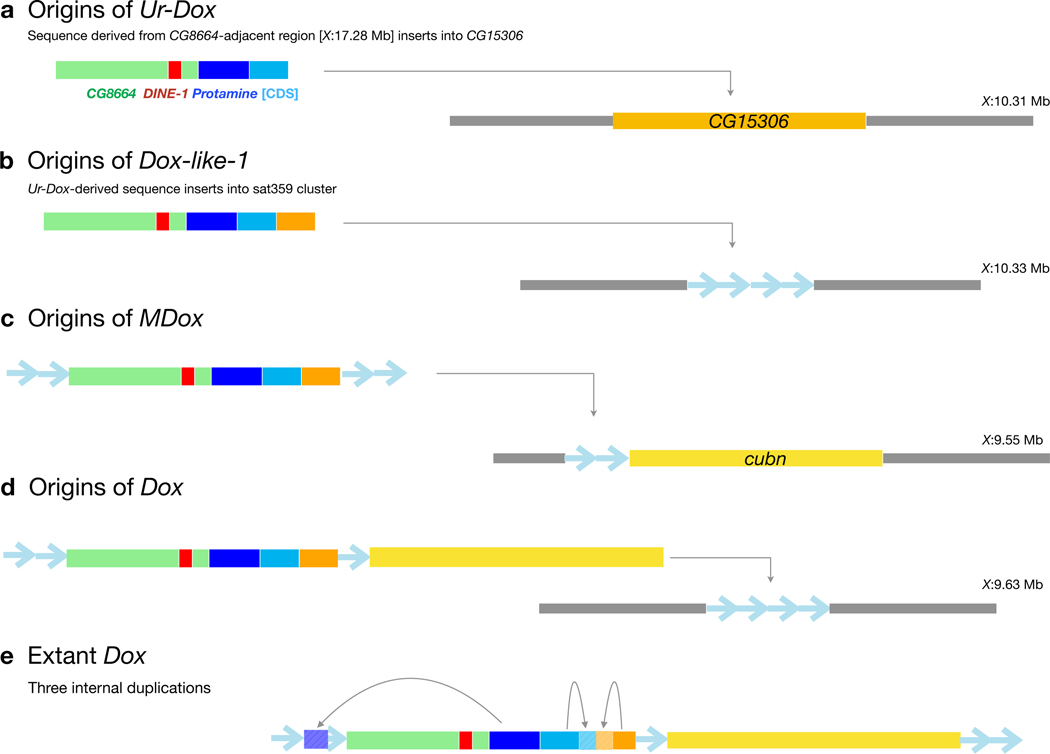
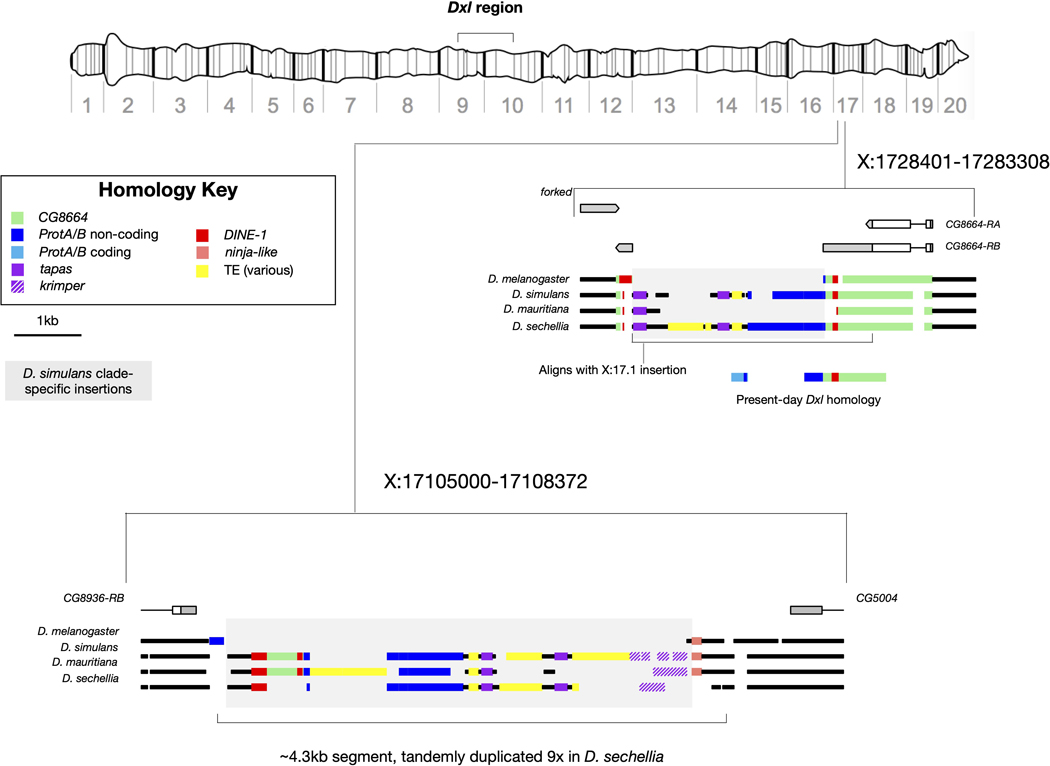
Previous work suggested that Dox has complex chimeric origins and limited coding potential 23. Using sequences from the set of Dxl gene copies reported here, we have inferred the putative stepwise evolutionary origins of Dox and identified a predicted ORF with a clear potential for drive. We speculate that a gene copy we have termed Ur-Dox (X:1033508, Dmel r6) is the founding member of the Dxl gene family, for three reasons. First, Ur-Dox is one of only two Dxl genes present in all three species (Fig. 1), consistent with an early origin in the common ancestor of the D. simulans clade. Second, Ur-Dox is the only Dxl gene that is not in a conserved sat359 island (Fig. 2a and Extended Data Fig. 1). Third, a sequence structure created during the origins of Ur-Dox is present in all other Dxl insertions. Ur-Dox arose via two insertion events: the 3´-UTR of the CG8664 gene was inserted by a ~2-kb sequence with homology to two protein-coding genes, tapas (tap) and Protamine A or Protamine B (ProtA/B) (Extended Data Fig. 2; Supplementary Text S2); subsequently, a ~1.6-kb sequence including the protamine-related sequence, but not tapas, and spanning the proximal insertion breakpoint of CG8664 was inserted into the testis-expressed gene, CG15306, replacing its endogenous single-exon CDS but leaving its 5´- and 3´-UTRs intact (Fig. 2a and Extended Data Fig. 2). The resulting ProtA/B-CG15306 junction (Fig. 2a,b) is preserved in all of the other, putatively descendant, Dxl gene family members. The formation of these other Dxl genes may have been seeded when a ~1.8-kb fragment of Ur-Dox was copied and pasted into the sat359 island at coordinate X:10339982 (Fig. 2b), creating a gene here called, Dxl-1 (Fig. 1, 2b, and Extended Data Fig. 3). Dxl-1 is the only other Dxl gene present in all three species, consistent with early origins in the common ancestor of the D. simulans clade. The Dxl-sat359 junction of Dxl-1 is preserved in all other Dxl insertions. After the three D. simulans clade species diverged from one another, a ~2-kb Dxl sequence became amplified in each of the three lineages (along with other non-native sequence; Extended Data Fig. 1 and Supplementary Text S1), spreading to different local sat359 islands (Fig. 1). One of these Dxl sequences inserted into the sat359 island at coordinate X:9558620, immediately distal to the gene cubulin (cubn), creating MDox (Fig. 2c). Finally, a sequence spanning all of MDox, an internal sat359 sequence, and ~1.8 kb of the 3´-end of cubn inserted into another sat359 island, creating Dox (Fig. 2d). The Dox sequence also contains three internal duplications (Fig. 2e; see also ref. 23). Remarkably, then, Dox evolved via the stepwise recruitment and movement of sequences through at least five insertion events involving flanking sat359 and four different protein-coding genes (ProtA/B, CG8664, CG15306, and cubn; Fig. 2). Other Dxl insertions have similarly complex histories that include the acquisition of gene sequences different from Dox. Overall, among the 18 Dxl insertions, seven have acquired sequence from Protein tyrosine phosphatase Meg2 (Ptpmeg2), three from monkey king protein (mkg-p), two from Carbonic anhydrase-related protein B (CARPB) introns, and two (MDox and Dox) from cubn (Extended Data Fig. 1 and 3). The functional significance of these acquired sequences, if any, is unclear.
Figure 2.
Inferred stepwise historical origins of Dox. Color coding of sequence blocks indicates the putative sequence homology, and light blue arrows represent sat359 repeats.
The Dox-like genes encode a rapidly evolving sperm-specific histone
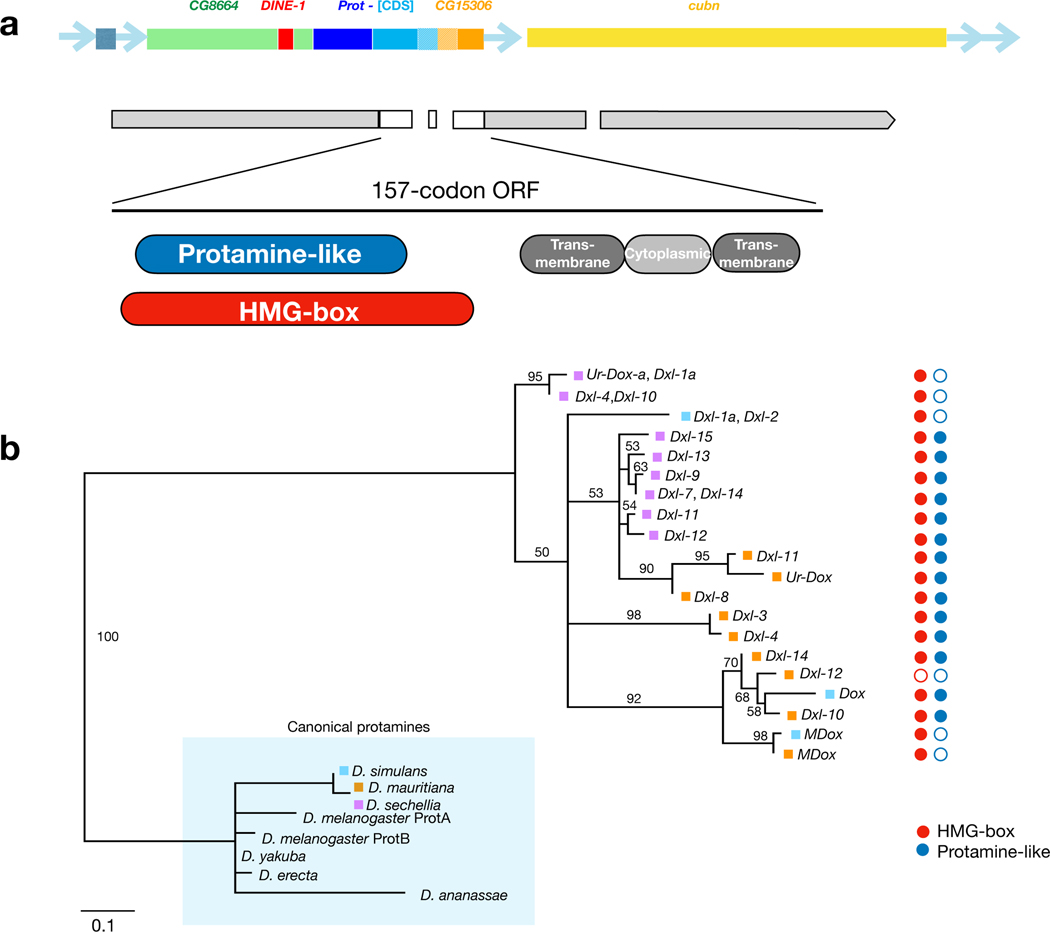
Although four protein-coding genes contributed sequence to Dox, most of the original open reading frames (ORFs) are disrupted or absent entirely. Among seven Dox ORFs with potential to encode polypeptides of ≥ 100 amino acids 23, one is 157 amino acids long, with N-terminal positions 2–79 corresponding to a predicted protamine-like domain that comprises a high mobility group (HMG) box and C-terminal positions 90–150 corresponding to two transmembrane domains and a cytoplasmic domain (Fig. 3a; Methods). The Dox protamine-encoding sequence is a highly diverged, recent derivative of Protamine A/B (ProtA/B). In Drosophila, HMG-box containing protamines are small, arginine-rich sperm-specific proteins that replace canonical histones during the remodeling and condensation of chromatin in late spermatogenesis 36. Interestingly, perturbation of HMG-box protamine function, including ProtA and ProtB, causes autosomal drive in the Segregation Distorter (SD) system of D. melanogaster 37. The Dox-encoded HMG-box protamine domains thus represent strong candidates for Dox-mediated sex-ratio drive that could for instance differentially affect chromatin remodeling of X- versus Y-bearing spermatid nuclei.
Figure 3.
Dxl genes encode a rapidly evolving protamine. a. Dox gene structure, validated splice structure for functional transcript, and the inferred protamine-like, HMG-box, cytoplasmic, and transmembrane domains of the predicted 157-aa protein. b. Phylogeny of amino acid sequence positions 1–66 predicted for 24 Dxl genes aligned with known protamines, annotated with presence (filled circles) or absence (open circles) of “protamine-like” (blue) and “HMG-box” (red) domains. Dxl genes lacking intact ORFs are not included in the phylogeny.
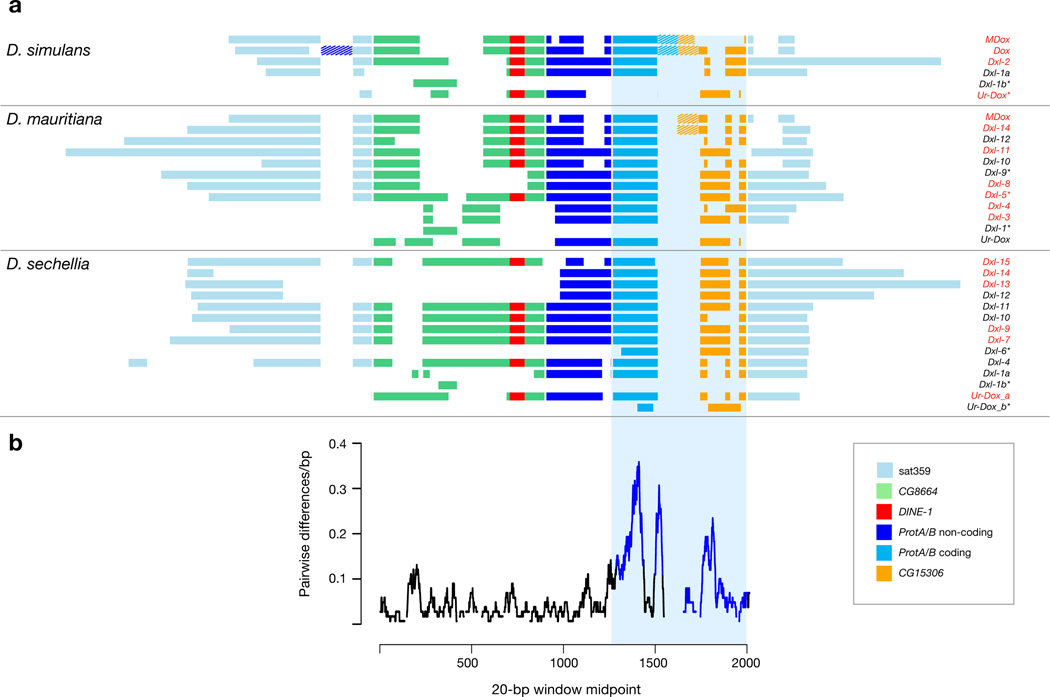
Like Dox, the other Dxl genes have complex histories of structural evolution (Fig. 4a). However, despite considerable structural diversity, 24 extant Dxl gene copies have retained an ORF in this region (assuming conservation of local splice sites relative to Dox; Fig. 4a). At the nucleotide level, the Dxl HMG-box protamine domain has evolved faster than immediately surrounding non-coding sequence (Fig. 4b). We used the translated coding sequence (CDS) of the protamine-like domain region to infer phylogenetic distances among Dxl gene copies (Fig. 3b and Supplementary Text S3). This analysis underlines the observation that the Dxl protamine domain has an extraordinary rate of sequence evolution, even compared to the rapidly evolving ProtA and ProtB genes included in the phylogeny. Despite this rapid protein evolution, 23 of the 24 putative Dxl proteins included in the phylogeny retain identifiable HMG-box homology, with 15 identifiable as protamines (Fig. 3b). Detailed evolutionary inferences about genealogical history or gene-specific rates of substitution are complicated by paralogous gene conversion among Dxl genes (see also ref. 26). Ectopic exchange among paralogous copies within species overwrites and obscures the phylogenetic history of orthologous Dxl gene copies. Indeed, most Dxl gene copies in D. mauritiana and D. sechellia cluster together by species, as expected if most copies originated within each lineage and/or experienced concerted evolution. In contrast, Dox and MDox in D. simulans cluster with D. mauritiana Dxl genes, as expected given their recent interspecific introgression 27.
Figure 4.
Structural and sequence evolution among Dxl gene copies. a. Schematic alignment of all known Dxl copies, organized by species, and color-coded by putative sequence homology. Internally duplicated segments are indicated by dashed fill. Dxl gene copies with RNA-seq evidence for expression are indicated by red font and those lacking an intact ORF are indicated by *. b. The average number of pairwise differences per bp among all Dxl copies is shown calculated in 20-bp windows across the region of Dxl homology. The blue box shows the alignment of sequences represented in panels a and b.
The rapid structural and sequence evolution of the Dxl gene family raises questions about maintenance of gene function, especially given that drivers that are fixed in the population and/or silenced by suppressors are expected to degenerate. Using diagnostic SNPs in testes RNA-seq data, we find evidence of expression for 4 of 5 Dxl genes in D. simulans, 7 of 12 in D. mauritiana, and 8 of 12 in D. sechellia (Fig. 3b; Supplementary Table 1), whereas overall Dxl expression is absent or negligible in females and in early-stage embryos (Supplementary Table 2). These numbers are conservative for two reasons. First, it is possible that more Dxl genes are expressed but fail to produce reads with diagnostic sequence features. Second, our RNA-seq data come from wildtype flies in which Dxl gene expression is silenced by autosomal suppressors (see below). It will be of considerable interest to characterize Dxl expression in males in which autosomal suppressors are disrupted.
The Dxl genes generated four autosomal esiRNA-producing suppressors
The three D. simulans clade species possess different autosomal suppressor genotypes (Fig. 5a). In D. simulans, Nmy and Tmy encode predicted hairpin RNAs (hpRNAs) that are processed into ~21-nt esiRNAs 25. Nmy originated via retroduplication of Dox, as evidenced by its lack of introns, its cubn-derived sequences (among Dxl genes, only Dox and MDox have cubn sequence), and subsequently experienced internal rearrangements that created the inverted repeats required for hpRNA formation (Fig. 5b; see ref. 24). Tmy originated via a DNA-based duplication event, as evidenced by the presence of Dxl introns and subsequently acquired a complete inverted duplication of both the Dxl sequence and additional, apparently unrelated, sequence (Fig. 5c and Supplementary Text S4; see ref. 25). However, while both Nmy and Tmy-derived esiRNAs show sequence matching with Dox 25 and both suppress sex-ratio drive in D. simulans 24,30, Tmy does not suppress Dox-mediated sex-ratio drive 24. This difference in the ability to suppress Dox, in particular, is consistent with differences in the extent of Dox sequence-matching by Nmy- and Tmy-derived esiRNAs: Nmy esiRNAs are on average 87% identical to Dox, whereas Tmy esiRNAs are only 54% identical to Dox (see Methods). Based on our discovery of a multicopy Dxl gene family, we speculated that Tmy suppresses one or more of the other Dxl genes. Indeed, Tmy esiRNAs show 94% and 92% identity to Dxl-1 and Dxl-2, respectively, whereas Nmy shows only 75% matching to Dxl-1 and Dxl-2. These findings strongly implicate Dxl-1 and/or Dxl-2 as the previously unidentified candidate genes for the sex-ratio phenotype suppressed by Tmy 30 (Supplementary Table 3).
Figure 5.
Autosomal hpRNA suppressor loci in the D. simulans clade species. a. The three species have different, partially overlapping systems of Dxl genes and esiRNA-producing autosomal suppressors. D. simulans has Tmy and Nmy; D. mauritiana has Nmy and Tmyl; and D. sechellia has Tmyl and Emy. Structural features of the esiRNA-producing putative autosomal suppressors, Tmy and Tmyl (b), Nmy (c), and Emy (d). Each putative suppressor originated via the insertion of Dxl material into an autosomal location followed by internal duplication and inversion of sequence (gray arrows), allowing formation of hpRNA precursor molecules (red arrows). In D. sechellia, a ~7-kb region that includes Emy has been tandemly amplified four times (d).
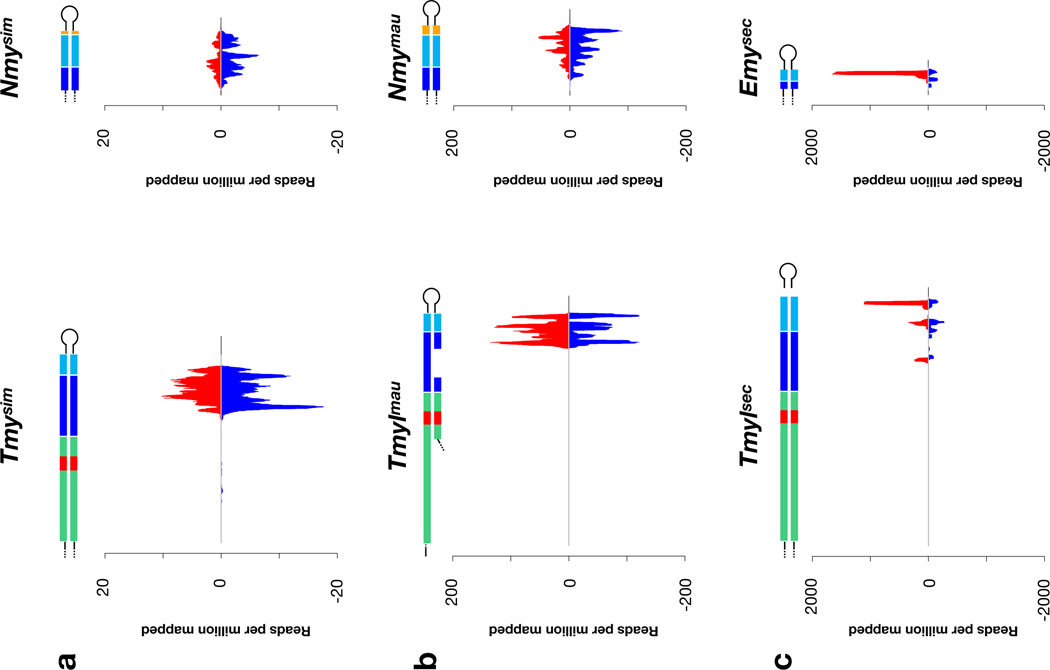
D. mauritiana has Nmy but lacks Tmy, whereas D. sechellia lacks both, suggesting that other suppressor loci exist in D. sechellia. We discovered two. First, another DNA-based duplication, here termed Tmy-like (Tmyl), exists in D. sechellia and in D. mauritiana but not in D. simulans (Fig. 5b and Supplementary Text S4). The Tmylmau and Tmylsec loci reside in offset positions on 3R, show hallmarks of target-site duplication (in D. mauritiana), and have an internal inverted duplication that gives rise to predicted hpRNAs. We confirmed using diagnostic SNPs that esiRNAs are produced in testes by Nmy and Tmy in D. simulans (see also ref. 25), by Nmy and Tmyl in D. mauritiana, and by Tmyl in D. sechellia testes (Fig. 6). In D. mauritiana, the Tmyl and Nmy esiRNAs show sequence similarity to different subsets of Dxl gene copies (Supplementary Table 3). Second, another novel hpRNA gene was revealed when we discovered that the most abundant Dxl-matching esiRNAs in D. sechellia do not originate from Tmyl. Instead, they are produced by a locus on 2L, here termed Even more yin (Emy). The structural origin of Emy differs from the other suppressors: a small ~200-bp Dxl-derived fragment inserted into exon 2 of the testes-expressed gene, CG13131 (2L:10014186), disrupting the downstream endogenous ORF; the ~200-bp fragment was partially duplicated and inverted to produce a 421-bp predicted hairpin structure; and, finally, the new Dxl-derived sequence plus ~7 kb of flanking sequence including the promoter region of CG13131 was amplified, creating four tandem copies of Emy spanning some ~28 kb (Fig. 5d). As in the other two species, Tmyl and Emy esiRNAs in D. sechellia show highest sequence matching to different subsets of Dxl gene copies (Supplementary Table 3). Overall, our findings reveal that at least four autosomal suppressor loci— Nmy, Tmy, Tmyl, and Emy— have evolved among the three D. simulans clade species. Each of the three species now possesses divergent, multilocus systems of Dxl (Fig. 1) and autosomal esiRNA-suppressor genotypes (Fig. 5a, 6), with the different esiRNA-suppressors appearing to target different subsets of Dxl genes for silencing.
Figure 6.
Species-specific small RNAs (≤22nt) map to Dxl-matching hairpin regions of each species’ autosomal suppressors. (a) Tmy and Nmy in D. simulans; (b) Tmyl and Nmy in D. mauritiana; and (c) Tmyl and Emy in D. sechellia. Reads per million reads mapped are shown (red = plus strand, blue = minus strand), with schematics of predicted hairpin arms shown for reference. Predicted hairpin stems are drawn to scale, hairpin loops are not.
Discussion
We infer that the progenitors of the Winters sex-ratio driver, Dox, originated de novo in the common ancestor of the three D. simulans clade species (Fig. 1). The Dxl gene family was founded via the chimeric origins of Ur-Dox, and its subsequent amplification was seeded when Dxl-1 inserted into a euchromatic island of sat359 (Fig. 2 and Extended Data Fig. 2). Ur-Dox and Dxl-1 are the only two Dxl genes present in all three D. simulans clade species, consistent with their early origins. The spread of all other Dxl genes to uninserted sat359 islands, and the evidence for paralogous gene exchange among Dxl genes (see also ref. 26), is consistent with similar dynamics characterizing the spread and exchange of repeat sequences among satDNA islands 32. We therefore infer that copy number amplification of Dxl genes occurred by insertion into uninserted sat359 islands and selection for novel insertions owing to Dxl dose-dependent sex-ratio drive. Additional Dxl copies may have increased the strength of drive and/or facilitated escape from esiRNA-mediated silencing by autosomal suppressors. Once established, the multicopy Dxl genes can provide substrate for functional diversification through rapid sequence and structural evolution (Fig. 3b, 4). Any Dxl sequence diversification must however be constrained by selection to maintain drive capacity against targets on the Y chromosome. As each species possesses two autosomal suppressors that match differing subsets of Dxl genes, we tentatively infer that the Dxl genes have diversified into two functional classes per species. Most Dxl genes are expressed in testes and, of these, most encode a highly divergent HMG-box protamine, which we posit as a strong candidate for the basis of the drive phenotype. In the SD system of D. melanogaster 38, drive occurs by disruption of protamine function in sperm nuclei possessing large blocks of the Responder satellite DNA 37,39. It seems likely that the Dxl-encoded HMG-box protamines disrupt sperm nuclei with an as yet unidentified Y chromosome-enriched satellite DNA. Last, during their brief history, the Dxl genes have spawned at least four autosomal suppressor genes that now produce Dxl-matching esiRNAs. Thus, while the recruitment of additional Dxl gene copies on the X chromosome might facilitate drive and/or escape from suppression, it also incidentally increases the number of potential parent copies for— and hence the mutation rate to— new autosomal suppressors.
The findings reported here highlight complex interactions among different classes of selfish DNAs. SatDNA islands that have been coopted for male somatic sex chromosome dosage compensation enabled the proliferation of, and ectopic exchange between, Dxl genes. The origins of the Dxl genes involved the initial chance acquisition of transposon-associated sequences, although their role (if any) in drive remains obscure (Fig. 2, Extended Data Fig. 2). The duplication of Nmy, Tmy, and Tmyl all appear to involve either transposon sequences, products, or processes including, e.g., evidence for target-site duplications (Tmyl), retroduplication via reverse transcriptase (Nmy), or the processing of circular DNAs. The Dxl-10 and Dxl-14 gene copies in D. mauritiana, for instance, show clear hallmarks of sequence transfer via circular DNA intermediates (Extended Data Fig. 4) that could implicate the activity of the abundant DINE-1 transposon machinery 40. And, last, all four esiRNA-producing autosomal suppressors have co-opted the existing transposon surveillance machinery to silence Dxl drivers.
Despite its idiosyncrasies, several features of the cryptic Winters sex-ratio system nevertheless reinforce parallels emerging among different drive systems from fungi, insects, and mammals. First, drive systems are characterized by gene copy number amplification, consistent with a gene dose-dependent basis for drive intensity and/or efficacy 41–43. Second, there are multiple routes to copy number amplification. In some drive systems, multicopy drive genes have amplified via tandem duplication, giving rise to massive Mb-scale runs of ampliconic, testes-expressed genes, like the Slxl1 and Sly co-amplified genes on mouse X and Y chromosomes, respectively 42,43. In other drive systems, multicopy drive genes are dispersed through mechanisms involving repetitive or transposable sequences, like the sat359 of Dxl reported here, the long terminal repeats that flank the multicopy wtf spore killers in Schizosaccharomyces fission yeast 44,45, and the Spok spore killers in Podospora filamentous fungi 46. Third, meiotic drive gene sequences evolve rapidly and often bear population genetic signatures of selection, including large-scale selective sweeps 26–28,47–49, indistinguishable from those resulting from organismal adaptation. Fourth, suppressors can exploit small RNA-mediated silencing of drive genes (Fig. 5, 6), so that drivers and suppressors share extensive sequence similarity 41,50. The sharing of sequences among multicopy drivers and their suppressors may accelerate cycles of conflict, as ectopic conversion can spread sequence innovations among different drive genes as well as to suppressors 45,51. Fifth, the chromatin remodeling of sperm pronuclei appears to be vulnerable to manipulation by drive elements, at least in Drosophila. All but one Dxl gene copy encodes a divergent protamine, and both SD in D. melanogaster 37 and the Wolbachia cytoplasmic incompatibility enzyme cidB perturb the histone-to-protamine exchange 52. It will be of interest to determine how many other male drive systems similarly exploit protamine function. Finally, the sex chromosomes of mammals and fruitflies— and doubtlessly many other taxa— are enriched for repetitive DNAs, multicopy genes, signatures of selection, and other genetic evidence consistent with histories of recurrent drive. Drive may therefore be a pervasive evolutionary force 1 with wide-ranging effects on sex chromosome evolution, regulation, and consequences for speciation 8,21.
Methods
Initial alignment and identification of 359-inserted sequences
The full region corresponding to D. melanogaster X:9400000–10400000 (D. simulans r2.02, Scf_X:8829511–9754201) was roughly aligned using Mauve (build 2015–02-13). Locations of sat359 repeats were identified iteratively by BLAST (v2.7.1), using species-specific sat359 unit sequences identified in ref. [54] as an initial bait sequence. The four-species alignment was then re-aligned by hand using Geneious (v6.18), and all sequences inserted into sat359 islands were extracted and aligned by hand. Components of inserts were further annotated using BLAST, informed by a similar alignment and annotation of two other regions on the X described below.
Identification of two X:17Mb regions and potential autosomal repressors
Using the consensus Dxl sequence from the insertion alignments as bait, we identified several regions sharing substantial homology with Dxl, including the CG15306-location insert, now annotated as Ur-Dox, two regions near D. melanogaster X:17Mb, and four potential autosomal suppressors, which include the previously identified Nmy, Tmy, and the new candidate suppressors Tmyl (in D. mauritiana and D. sechellia) and Emy (in D. sechellia).
RNA-seq and small RNA-seq
Data collection and filtering:
We used new and publicly available RNA-seq data to assess Dxl expression in testes. For the new data, flies were reared at room temperature on standard cornmeal agar medium, and testes from 20 males, aged 3–5 days, of D. simulans wXD1 and the D. sechellia reference strain (Rob3c / Tucson 14021–0248.25) were dissected into cold Ringer’s solution, transferred to 350 μl of cold lysis buffer, and then stored at −80oC. RNA was extracted using the Nucleospin RNA XS isolation kit (Macherey-Nagel). Sequencing libraries were prepared using the TruSeq Stranded Total RNA kit with 150-bp inserts. Paired-end reads were sequenced using the Illumina NextSeq 550 at the University of Rochester Genomics Research Center. Raw reads are deposited in (SRR14777834–14777837). For D. mauritiana w12, we used three publicly-available cDNA RNA-seq D. mauritiana datasets (SRR9025052-SRR9025054). To assess Dxl expression in females and early embryos, we used publicly available datasets from D. simulans, D. sechellia, and D. mauritiana (Extended Data Table 2). To assess Dxl-matching esiRNA expression in testis, we used four publicly-available small RNA fastq datasets from D. simulans (SRR410589 and SRR410590), D. mauritiana (SRR7961897) and D. sechellia (SRR6667444). All fastq files were cleaned using Trim Galore (v0.2.6), discarding bases with Q<33. To enrich for esiRNAs, the small RNA datasets were further filtered to exclude reads >22 nt.
Read mapping and data analysis:
The base sequence set (Supplementary Text S5) used for all read mapping consisted of curated D. simulans transcripts (Flybase r2.02) and transposable elements, from which any sequences with Dxl homology had been identified via BLAST and removed. This base was then augmented by sequences of interest for each analysis (Supplementary Information). Overall Dxl expression was quantified using kallisto (v0.43.1) and a reference genome consisting of the base set plus species-appropriate Dxl consensus sequence. For small RNA-seq analysis in each species, we added consensus hairpin sequence for that species’ putative autosomal suppressors to the base set, while for the cDNA-selected and total RNA-seq (expression analysis) datasets we added the species-appropriate consensus Dxl sequence as a mapping target. Reads were mapped using the “local” options in Bowtie2 (v2.3.5.1) for the small RNA datasets and “very-fast-local” options for expression analysis. Read pileups were assessed using BEDtools (v2.26.0), IGV (v2.3.88), and R (v3.6.0). In the expression analysis, SNPs unique to individual Dxl genes were identified using species-specific Dxl alignments (Supplemental Information). BEDtools and IGV were then used to visually inspect alignments, count reads matching those unique SNPs, and thus assess the evidence for expression of particular Dxl genes (Supplementary Text S5 and Supplementary Table 1).
Extended Data
Extended Data Fig. 1 |. Schematic alignment of inserted sat359 islands in X:9.4–10.4 Mb, color-coded by putative sequence homology.
Segments of the same color aligned vertically are high-confidence nucleotide alignments, whereas segments of different color do not share sequence homology regardless of vertical alignment in the figure. One insertion into a sat359 island, the transposase-like sequence inserted into the D. sechellia Dxl-2 location, does not share sequence homology with any of the other loci, and has been omitted from this figure.
Extended Data Fig. 2 |. Alignment of the putative source material for Dxl genes found at approximate coordinates X:17.1 and X:17.2 Mb.
Segments of the same color aligned vertically are high-confidence nucleotide alignments. At X:17.2, the three D. simulans clade species all have remnants of an insertion (total span = ~3 kb) that interrupts the CG8664 gene. This sequence, along with some of CG8664 and additional material, is alignable with insertion at the second position, X:17.1 Mb. Putative homology of the inserted sequence at both locations, along with CG8664, are color-coded and the span of present-day Dxl homology is indicated.
Extended Data Fig. 3. Chromosomal distribution of sat359 islands in X:9.4–10.4 Mb region with all identified inserted sequence, including Dxl genes, in D. simulans, D. mauritiana, and D. sechellia.
Tick marks indicate locations of conserved sat359 islands, blue dots indicate protein-coding genes of interest in the region, and the different colored squares represent the homologies of sequences inserted into sat359 islands (green=Dxl; purple=Ptpmeg2 fragment; brown=mkg-p retrotransposition; red=transposase-like sequence; yellow=cubn fragment; pink=CARPB intron fragments).
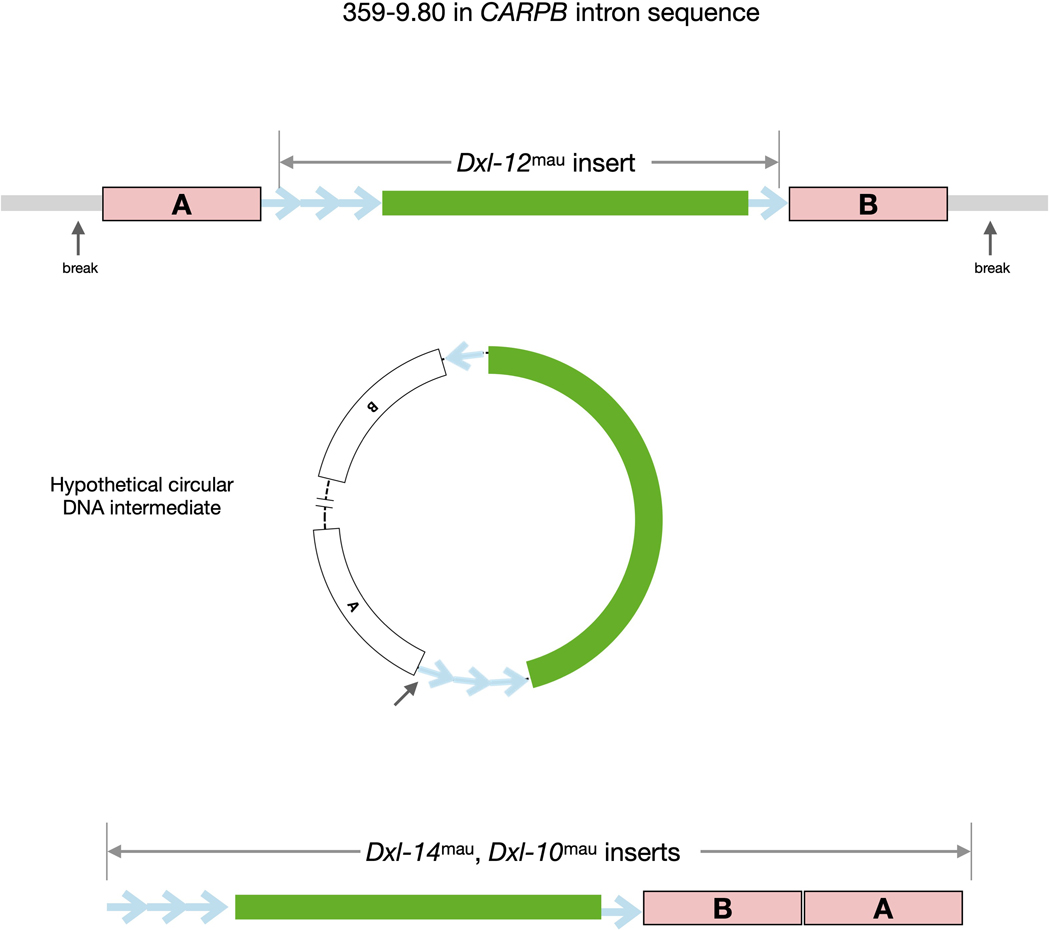
Extended Data Fig. 4.
Evidence for transfer of Dxl material via a circular DNA. Intermediate molecule. Dxl-12mau (green) is flanked by sat359 repeats 835 (blue arrows) and by CARPB intronic sequence (boxes labelled A and B). These flanking sequence match sequences present in Dxl-10mau and Dxl-14mau, but 836 the order of the two CARPB segments A and B differs from Dxl-12mau. The re-ordering of homologous sequences, as well as their intervening sequence, is 837 consistent with a transfer of material via circular DNA intermediate.
Supplementary Material
Acknowledgements
This work was supported by funds from NIH grant no. R01 GM123194 and the University of Rochester to D.C.P. We thank J.J. Emerson, Amanda Larracuente, Colin Meiklejohn, Kristi Montooth, and Jeffrey Vedanayagam for early access to the PacBio-based genome assemblies. And we thank Beatriz Navarro Dominguez, Colin Meiklejohn, and Aaron Vogan for valuable feedback on the manuscript.
Footnotes
Competing interests
The authors declare no competing interests.
Data availability
All data used in our analyses are publicly available via the Sequence Read Archive. Data accessions are listed in Supplementary Tables 2 and 4.
References
- 1.Sandler L. & Novitski E. Meiotic drive as an evolutionary force. American Naturalist 91, 105–110 (1957). [Google Scholar]
- 2.Lindholm AK et al. The Ecology and Evolutionary Dynamics of Meiotic Drive. Trends Ecol Evol 31, 315–326, doi: 10.1016/j.tree.2016.02.001 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Lyttle TW Segregation Distorters. Annual Review of Genetics 25, 511–557 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Lyttle TW Cheaters sometimes prosper: distortion of mendelian segregation by meiotic drive. Trends in Genetics 9, 205–210 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Presgraves DC in Sperm Biology: An Evolutionary Perspective (eds Birkhead TR, Hosken DJ, & Pitnick S) (Elsevier Press, 2008). [Google Scholar]
- 6.Hartl DL Genetic dissection of segregation distortion. I. Suicide combinations of SD genes. Genetics 76, 477–486 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Charlesworth B. & Hartl DL Population dynamics of the segregation distorter polymorphism of Drosophila melanogaster. Genetics 89, 171–192 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamilton WD Extraordinary sex ratios. Science 156, 477–488 (1967). [DOI] [PubMed] [Google Scholar]
- 9.Hurst LD & Pomiankowski A. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane’s rule and related phenomena. Genetics 128, 841–858 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank SH Divergence of meiotic drive-suppressors as an explanation for sex-biased hybrid sterility and inviability. Evolution 45, 262–267 (1991). [DOI] [PubMed] [Google Scholar]
- 11.Gershenson S. A new sex ratio abnormality in Drosophila obscura. Genetics 13, 488–507 (1928). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher RA The Genetical Theory of Natural Selection. (Oxford University Press, 1930). [Google Scholar]
- 13.Jaenike J. Sex chromosome meiotic drive. Annual Review of Ecology and Systematics 32, 25–49 (2001). [Google Scholar]
- 14.Vaz SC & Carvalho AB Evolution of autosomal suppression of the sex-ratio trait in Drosophila. Genetics 166, 265–277 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall DW Meiotic drive and sex chromosome cycling. Evolution 58, 925–931 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Hartl DL Modifier theory and meiotic drive. Theoretical Population Biology 7, 168–174 (1975). [DOI] [PubMed] [Google Scholar]
- 17.Thomson GJ & Feldman MW Population genetics of modifiers of meiotic drive. II. Linkage modification in the Segregation Distorter system. Theoretical Population Biology 5, 155–162 (1974). [DOI] [PubMed] [Google Scholar]
- 18.Bastide H, Gerard PR, Ogereau D, Cazemajor M. & Montchamp-Moreau C. Local dynamics of a fast-evolving sex-ratio system in Drosophila simulans. Molecular Ecology 22, 5352–5367 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Haig D. & Grafen A. Genetic scrambling as a defence against meiotic drive. Journal of Theoretical Biology 153, 531–558 (1991). [DOI] [PubMed] [Google Scholar]
- 20.Burt A. & Trivers RA Genes in conflict. (Harvard University Press, 2006). [Google Scholar]
- 21.Meiklejohn CD & Tao Y. Genetic conflict and sex chromosome evolution. Trends in Ecology and Evolution 25, 215–223 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachtrog D. The Y Chromosome as a Battleground for Intragenomic Conflict. Trends Genet 36, 510–522, doi: 10.1016/j.tig.2020.04.008 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao Y. et al. A sex-ratio meiotic drive system in Drosophila simulans. II: An X-linked distorter. Public Library of Science Biology 5, e293 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao Y, Masly JP, Araripe L, Ke Y. & Hartl DL A sex-ratio meiotic drive system in Drosophila simulans. I: An autosomal suppressor. Public Library of Science Biology 5, e292 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin CJ et al. The hpRNA/RNAi Pathway Is Essential to Resolve Intragenomic Conflict in the Drosophila Male Germline. Dev Cell 46, 316–326 e315, doi: 10.1016/j.devcel.2018.07.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kingan SB, Garrigan D. & Hartl DL Recurrent selection on the Winters sex-ratio genes in Drosophila simulans. Genetics 184, 253–265, doi: 10.1534/genetics.109.109587 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meiklejohn CD et al. Gene flow mediates the role of sex chromosome meiotic drive during complex speciation. eLife 7, e35468 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nolte V, Pandey RV, Kofler R. & Schlotterer C. Genome-wide patterns of natural variation reveal strong selective sweeps and ongoing genomic conflict in Drosophila mauritiana. Genome Res 23, 99–110, doi: 10.1101/gr.139873.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garrigan D, Kingan SB, Geneva AJ, Vedanayagam JP & Presgraves DC Genome diversity and divergence in Drosophila mauritiana: multiple signatures of faster X evolution. Genome Biology and Evolution 6, 2444–2458, doi: 10.1093/gbe/evu198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao Y, Hartl DL & Laurie CC Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proceedings of the National Acadamy of Sciences 98, 13183–13188 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakraborty M. et al. Evolution of genome structure in the Drosophila simulans species complex. Genome Research 31, 380–396, doi: 10.1101/2020.02.27.968743 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sproul JS et al. Dynamic Evolution of Euchromatic Satellites on the X Chromosome in Drosophila melanogaster and the simulans Clade. Molecular Biology and Evolution 37, 2241–2256 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joshi SS & Meller VH Satellite Repeats Identify X Chromatin for Dosage Compensation in Drosophila melanogaster Males. Current Biology 27, 1393–1402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrigan D. et al. Genome sequencing reveals complex speciation in the Drosophila simulans clade. Genome Research 22, 1499–1511 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kliman RM et al. The population genetics of the origin and divergence of the Drosophila simulans complex species. Genetics 156, 1913–1931 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller D, Brinkworth M. & Iles D. Paternal DNA packaging in spermatozoa: more than the sum of its parts? DNA, histones, protamines and epigenetics. Reproduction 139, 287–301 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Gingell LF & McLean JR A Protamine Knockdown Mimics the Function of Sd in Drosophila melanogaster. G3 (Bethesda) 10, 2111–2115, doi: 10.1534/g3.120.401307 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Larracuente AM & Presgraves DC The selfish Segregation Distorter complex of Drosophila melanogaster. Genetics 192, 33–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu C-I, Lyttle TW, Wu M-L & Lin GF Association between DNA satellite sequences and the responder of Segregation Distorter in D. melanogaster. Cell 54, 179–189 (1988). [DOI] [PubMed] [Google Scholar]
- 40.Thomas J, Phillips CD, Baker RJ & Pritham EJ Rolling-circle transposons catalyze genomic innovation in a mammalian lineage. Genome Biol Evol 6, 2595–2610, doi: 10.1093/gbe/evu204 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hurst LD Is Stellate a relict meiotic driver? Genetics 130, 229–230 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cocquet J. et al. The Multicopy Gene Sly Represses the Sex Chromosomes in the Male Mouse Germline after Meiosis. PLoS Genetics 7, e1000244 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kruger AN et al. A Neofunctionalized X-Linked Ampliconic Gene Family Is Essential for Male Fertility and Equal Sex Ratio in Mice. Curr Biol 29, 3699–3706 e3695, doi: 10.1016/j.cub.2019.08.057 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu W. et al. A large gene family in fission yeast encodes spore killers that subvert Mendel’s law. eLife 6, doi: 10.7554/eLife.26057 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eickbush MT, Young JM & Zanders SE Killer Meiotic Drive and Dynamic Evolution of the wtf Gene Family. Mol Biol Evol 36, 1201–1214, doi: 10.1093/molbev/msz052 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogan AA et al. Combinations of Spok genes create multiple meiotic drivers in Podospora. eLife 8, doi: 10.7554/eLife.46454 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derome N, Metayer K, Montchamp-Moreau C. & Veuille M. Signature of selective sweep associated with the evolution of sex-ratio drive in Drosophila simulans. Genetics 1166, 1357–1366 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Presgraves DC, Gerard PR, Cherukuri A. & Lyttle TW Large-Scale Selective Sweep among Segregation Distorter Chromosomes in African Populations Drosophila melanogaster. PLoS Genetics 5, e1000463, Doi 10.1371/Journal.Pgen.1000463 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nam K. et al. Extreme selective sweeps independently targeted the X chromosomes of the great apes. Proceedings of the National Acadamy of Sciences 112, 6413–6418 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aravin AA et al. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11, 1017–1027, doi: 10.1016/s0960-9822(01)00299-8 (2001). [DOI] [PubMed] [Google Scholar]
- 51.Daugherty MD & Zanders SE Gene conversion generates evolutionary novelty that fuels genetic conflicts. Curr Opin Genet Dev 58–59, 49–54, doi: 10.1016/j.gde.2019.07.011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beckmann JF, Sharma GD, Mendez L, Chen H. & Hochstrasser M. The Wolbachia cytoplasmic incompatibility enzyme CidB targets nuclear import and protamine-histone exchange factors. eLife 8, doi: 10.7554/eLife.50026 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in our analyses are publicly available via the Sequence Read Archive. Data accessions are listed in Supplementary Tables 2 and 4.